Yeast kinetochore protein Slk19 is required for kinetochore clustering, and nocodazole exposure to slk19 mutant cells causes impaired kinetochore capture and delayed chromosome bipolar attachment after nocodazole washout.
Abstract
In all eukaryotic cells, DNA is packaged into multiple chromosomes that are linked to microtubules through a large protein complex called a kinetochore. Previous data show that the kinetochores are clustered together during most of the cell cycle, but the mechanism and the biological significance of kinetochore clustering are unknown. As a kinetochore protein in budding yeast, the role of Slk19 in the stability of the anaphase spindle has been well studied, but its function in chromosome segregation has remained elusive. Here we show that Slk19 is required for kinetochore clustering when yeast cells are treated with the microtubule-depolymerizing agent nocodazole. We further find that slk19Δ mutant cells exhibit delayed kinetochore capture and chromosome bipolar attachment after the disruption of the kinetochore–microtubule interaction by nocodazole, which is likely attributed to defective kinetochore clustering. In addition, we show that Slk19 interacts with itself, suggesting that the dimerization of Slk19 may mediate the interaction between kinetochores for clustering. Therefore Slk19 likely acts as kinetochore glue that clusters kinetochores to facilitate efficient and faithful chromosome segregation.
INTRODUCTION
An eukaryotic cell contains multiple chromosomes, but the centromeric regions from these chromosomes have been shown to be grouped together in yeast and human cells (Funabiki et al., 1993; Goh and Kilmartin, 1993; Goshima and Yanagida, 2000; Solovei et al., 2004; Duan et al., 2009). In budding yeast, chromosomes are connected to microtubules and form clusters during most of the cell cycle (Goshima and Yanagida, 2000; Tanaka et al., 2007; Liu et al., 2008). Previous work shows that the centromere clustering in yeast cells is reduced in kinetochore mutants (Jin et al., 2000; Janke et al., 2001; Anderson et al., 2009). One explanation is that kinetochore clustering is maintained by the association of kinetochores with microtubules. Recent evidence suggests that kinesin-5 sliding motor proteins Cin8 and Kip1 promote kinetochore clustering during mitosis by controlling the dynamics of kinetochore microtubules (Gardner et al., 2008). Another motor protein, Kip3, is also involved in kinetochore clustering at metaphase (Wargacki et al., 2010). However, the molecular mechanism for kinetochore clustering during other cell cycle stages and the biological function of this clustering remain unclear.
The budding yeast SLK19 gene was isolated because the loss of its function causes lethality in cells lacking a motor protein Kar3. Given the fact that Kar3 promotes chromosome biorientation (Liu et al., 2011; Jin et al., 2012), the genetic interaction suggests a possible kinetochore function for Slk19. Indeed, Slk19 has been shown to associate with centromeric DNA (Zeng et al., 1999; Zhang et al., 2006). As a kinetochore protein, Slk19 is not essential for growth, and slk19Δ mutant cells show almost normal chromosome segregation kinetics compared with wild-type (WT) cells (Sullivan and Uhlmann, 2003; Jin et al., 2008). Nevertheless, slk19Δ mutant is lethal in the absence of the spindle assembly checkpoint that monitors flawed kinetochore–microtubule (KT-MT) interactions, indicating the role of Slk19 in faithful chromosome segregation (Tong et al., 2004; Ye et al., 2005; Collins et al., 2007). However, the kinetochore function of Slk19 has remained as a mystery at the molecular level.
Slk19 is also a component of a mitotic exit pathway FEAR (Cdc14 early-anaphase release), which promotes the release of the phosphatase Cdc14 from the nucleolus during early anaphase. In addition to Slk19, the FEAR pathway includes separase Esp1, protein phosphatase 2A, Spo12, Fob1, and the polo-like kinase Cdc5 (Stegmeier et al., 2002; Queralt et al., 2006; Wang and Ng, 2006; Yellman and Burke, 2006). The FEAR-induced Cdc14 release reverses the protein phosphorylation imposed by S-phase CDK (cyclin-dependent kinase), which facilitates spindle elongation (Jin et al., 2008). Moreover, Slk19 localizes to the spindle midzone during anaphase, where it stabilizes the spindle (Pereira and Schiebel, 2003; Khmelinskii et al., 2007). Similar to the cohesin subunit Scc1/Mcd1, Slk19 is cleaved by separase Esp1 upon anaphase onset, and this cleavage is essential for Slk19’s spindle-midzone localization (Sullivan et al., 2001). A series of slk19 in-frame deletion mutants was identified, and they are defective for either FEAR signaling or kinetochore function. Deletion of a fragment at the central region of SLK19 leads to the synthetic lethality with kar3 mutant without disrupting the FEAR function, suggesting the kinetochore-specific role of this region (Havens et al., 2010).
Here we show that the absence of a yeast kinetochore protein Slk19 leads to kinetochore declustering after treatment with nocodazole, a microtubule-depolymerizing agent, even though untreated slk19Δ mutant cells exhibit normal kinetochore clustering. slk19Δ mutants exhibit normal chromosome segregation during an undisturbed cell cycle, but we observed a dramatic anaphase entry delay after nocodazole treatment. Data with live-cell imaging indicate that slk19Δ mutant cells exhibit defects in both kinetochore capture and chromosome biorientation. Our observation that Slk19 directly interacts with itself supports a possibility that Slk19 dimerization mediates kinetochore–kinetochore interaction and clustering, which likely facilitates kinetochore capture and chromosome bipolar attachment, especially after the disruption of kinetochore–microtubule interaction.
RESULTS
Kinetochore clustering is resistant to nocodazole treatment
Budding yeast kinetochores have been shown to form clusters (Goh and Kilmartin, 1993; Goshima and Yanagida, 2000). We confirmed the cluster formation in live cells with green fluorescent protein (GFP)–tagged Mtw1, a protein located in the center portion of the kinetochore (De Wulf et al., 2003). Kinetochores form a single cluster before metaphase and appear as two clusters, presumably after the establishment of chromosome bipolar attachment (Supplemental Figure S1). To examine whether this clustering depends on KT-MT interaction, we examined the kinetochore distribution in cells that were first synchronized in preanaphase and then treated with nocodazole, a microtubule-depolymerizing drug. cdc13-1 mutant cells arrest in preanaphase when incubated at nonpermissive temperatures due to the activation of the DNA-damage checkpoint by unprotected telomeres (Liang and Wang, 2007). Cells in cdc13-1 background were first incubated at 34°C to achieve preanaphase arrest. After nocodazole treatment, cdc13-1 MTW1-GFP SPC42-mApple cells showed two adjacent spindle poles (Spc42-mApple). However, kinetochores marked by Mtw1-GFP formed a ring-like structure with two Spc42-mApple dots residing at each side of the ring (Figure 1A). Because the two spindle poles are still separated, and the Mtw1-GFP foci are in very close proximity to the spindle poles, we speculate that the nocodazole treatment of cells arrested in preanaphase cannot disrupt the KT-MT interaction completely. One possibility is that nocodazole-mediated microtubule depolymerization fails to disrupt some KT-MT interactions once they are established. If that is the case, we need to treat cells with nocodazole before the establishment of KT-MT interaction in order to disconnect kinetochores from microtubules more efficiently.
FIGURE 1:
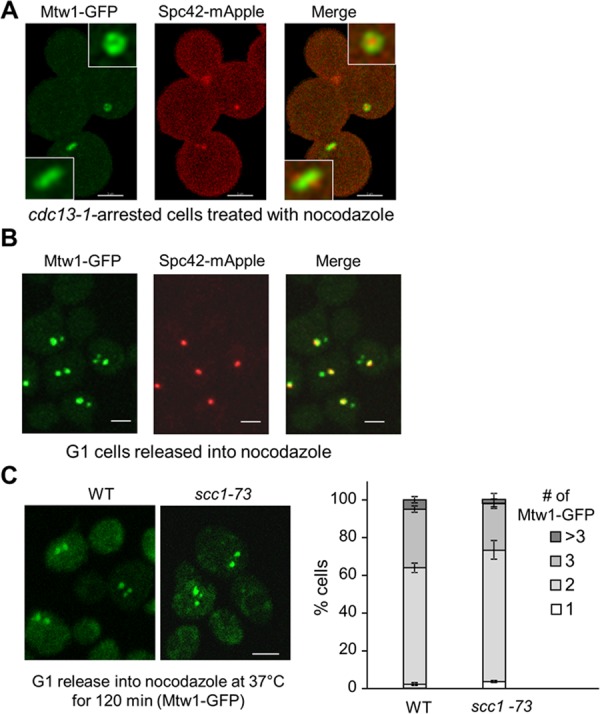
Kinetochores can form clusters in the absence of KT-MT interaction and sister-chromatid cohesion. (A) Most of the kinetochores remain connected to spindle poles after nocodazole treatment in cells first arrested in preanaphase. G1-arrested cdc13-1 MTW1-GFP SPC42-mApple cells were released into YPD medium at 34°C for 120 min to achieve preanaphase arrest. Nocodazole was then added into the medium to 20 μg/ml, and the cells were harvested and fixed after 45 min of incubation at 34°C. Confocal microscopy was used to project a maximum-intensity image. Bar, 3 μm. (B) Some kinetochore clusters are disconnected from the spindle poles after G1-arrested cells are released into nocodazole. G1-arrested MTW1-GFP SPC42-mApple cells were released into medium containing 20 μg/ml nocodazole. After 120 min of incubation at 25°C, cells were fixed and subjected to confocal microscopy. Scale bar, 3 μm. (C) Kinetochore clustering after nocodazole treatment is normal in cohesin mutants. WT and scc1-73 cells with MTW1-GFP were grown to log phase at permissive temperatures. After G1 arrest, the cells were released into 37°C medium containing 20 μg/ml nocodazole for 120 min. More than 100 cells were counted for the number of Mtw1-GFP foci per cell for each sample, and the average from triplicates is shown (right). Left, representative images acquired by confocal microscopy. Scale bar, 3 μm.
When centromeric DNA is being duplicated by the replication machinery, kinetochore proteins are displaced from the centromere, which disrupts the KT-MT interaction (Kitamura et al., 2007). Cells can replicate their DNA in the presence of nocodazole, but the reassembled kinetochores may fail to reestablish the connection to microtubules due to repressed microtubule polymerization by nocodazole. Therefore we released G1-arrested yeast cells with Mtw1-GFP and Spc42-mApple into nocodazole (20 μ/ml) for 120 min, a condition that allows DNA synthesis but prevents microtubule polymerization. We examined the kinetochore distribution in these cells and found that most cells exhibited two or three GFP foci that mark kinetochores. Of interest, only one of the GFP dots colocalized with the collapsed spindle poles (Spc42-mApple) and others appeared to float away from the spindle poles, indicating that some kinetochores are not connected to microtubules (Figure 1B). In most of the cases, mApple-marked spindle poles colocalize with the bigger GFP dot, indicating that some KT-MT interactions are resistant to nocodazole treatment or some kinetochores reestablish the interaction with microtubules after disconnection even in the presence of nocodazole.
To further verify that all GFP dots observed are clusters of functional kinetochores, we examined the localization of GFP-marked centromere on chromosome IV (CEN4-GFP) with mCherry-labeled Nuf2, another kinetochore protein. After G1 cells were released into nocodazole for 120 min, we found that the CEN4-GFP dot colocalized with either the bigger or the smaller mCherry dot (Supplemental Figure S2), indicating that even the smaller fluorescent dots are real kinetochores. This result is in agreement with a previous observation that some kinetochores detach from the spindle microtubules after nocodazole treatment (Gillett et al., 2004).
A smaller Mtw1-GFP focus in nocodazole-treated yeast cells could be a single kinetochore or a cluster. If the smaller kinetochore focus away from the spindle poles is from a single chromosome, we expect the fluorescence intensity of the larger foci to be 15 times larger than that of the smaller ones, as a haploid yeast cell contains 16 chromosomes. Therefore we analyzed the fluorescence intensity in cells that contain two GFP dots after nocodazole treatment and found that the average ratio is 2.25 (n = 100). Obviously, this ratio is much less than 15. Thus it is very likely that more than one kinetochore consists of the smaller kinetochore foci that are not linked to microtubules. In other words, kinetochores are clustered in the absence of KT-MT interaction.
Previous work showed that pericentric chromatin in budding yeast is organized into an intramolecular loop, and the loops from the 16 chromosomes form a cylindrical array. Although cohesin is not required for loop formation, it was proposed that cohesin may contribute to the stability or proximity of the intramolecular loops (Yeh et al., 2008). To test whether cohesin facilitates kinetochore clustering, we examined kinetochore distribution in cohesin mutant cells (scc1-73/mcd1) that were released into 37°C nocodazole medium for 120 after G1 arrest. We did not detect an obvious difference in the number of Mtw1-GFP foci per cell in WT and scc1-73 mutant cells, suggesting that cohesin may not be directly involved in kinetochore clustering upon nocodazole treatment (Figure 1C). In addition, we failed to detect a dramatic kinetochore clustering defect in nocodazole-treated GAL-SCC1 cells growing in glucose medium, which represses the expression of cohesin Scc1/Mcd1 (Supplemental Figure S3A). Furthermore, ctf8Δ, ctf18Δ, and dcc1Δ mutant cells were shown to have a cohesion defect (Mayer et al., 2001), but they did not exhibit an obvious kinetochore clustering defect after nocodazole treatment (Supplemental Figure S3B).
Slk19 is required for kinetochore clustering after nocodazole treatment
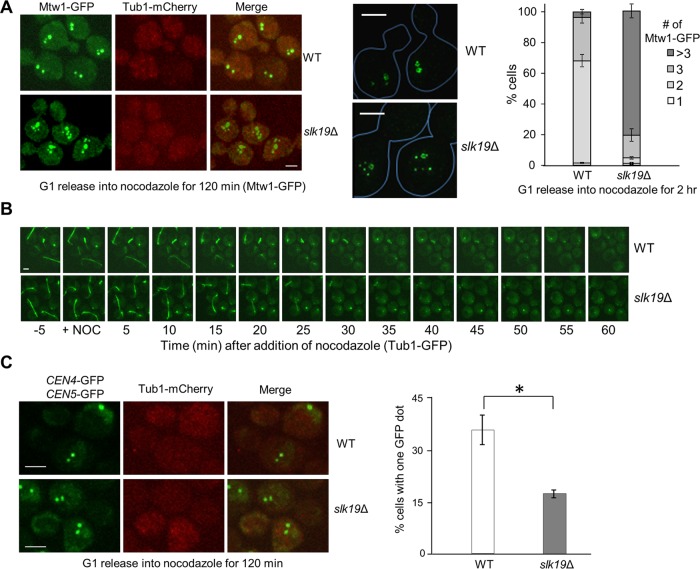
Because kinetochores form clusters when they are away from spindle poles, kinetochore protein(s) is likely responsible for this clustering. Thus we analyzed kinetochore clustering in some yeast mutant cells lacking a nonessential kinetochore protein. For this purpose, we first constructed WT and slk19Δ kinetochore mutant strains in MTW1-GFP TUB1-mCherry background in order to visualize the kinetochore localization and the spindle structure. Like WT cells, slk19Δ mutant cells showed normal kinetochore clustering in an undisturbed cell cycle, with one GFP dot in G1 and S phase and two after metaphase (Supplemental Figure S4). After G1-arrested cells were released into nocodazole for 120 min, most WT cells showed two or three Mtw1-GFP foci, whereas slk19Δ mutant cells exhibited an obvious clustering defect, as evidenced by the increased number of Mtw1-GFP foci per cell. We observed that 79.5% of slk19Δ cells showed more than three Mtw1-GFP foci, compared with only 6.0% for WT cells (Figure 2A). No spindle structure (Tub1-mCherry) was observed in both WT and slk19Δ mutant cells, indicating microtubule depolymerization under this condition. We further used superresolution microscopy to examine the kinetochore distribution in nocodazole-treated cells, and it was obvious that slk19Δ cells exhibited an increased number of Mtw1-GFP foci (Figure 2A, middle). We noticed the presence of several low-intensity GFP foci in some slk19Δ mutant cells. Further experiments are needed to clarify whether the smaller foci represent single pairs of sister kinetochores. We also examined the kinetochore distribution in other nonessential kinetochore mutant cells. In contrast to slk19Δ, no obvious kinetochore clustering defect was observed in ctf3Δ and ydr532cΔ (kre28Δ) mutant cells (Supplemental Figure S5).
FIGURE 2:
Slk19 is required for kinetochore clustering after nocodazole treatment. (A) slk19Δ mutants show a kinetochore clustering defect. G1-arrested WT and slk19Δ cells in a MTW1-GFP TUB1-mCherry background were released into 20 μg/ml nocodazole for 120 min. After fixation, >100 cells were counted for the number of Mtw1-GFP foci per cell. Right, the average of three separate experiments. Left, representative images for Mtw1-GFP and Tub1-mCherry signals from confocal microscopy. Middle, representative images for Mtw1-GFP signals from superresolution microscopy. Bar, 3 μm. (B) slk19Δ mutant cells show similar microtubule-depolymerizing kinetics as WT cells. WT and slk19Δ cells with Tub1-GFP were loaded into a chamber where YPD medium containing 20 μg/ml nocodazole started to flow over the cells at time 0. Maximum-projection images were then created to show the spindle morphology over time. (C) Fewer slk19Δ cells show colocalized centromeres from chromosomes IV and V after nocodazole treatment. WT and slk19Δ cells in a TUB1-mCherry background with GFP-marked centromeres of chromosome IV and V (CEN4-GFP CEN5-GFP) were grown to log phase at 25°C. After G1 arrest, the cells were released into 20 μg/ml nocodazole for 120 min and then harvested and fixed for fluorescence microscopy. The number of GFP dots per cell was counted in >100 cells, and the average from triplicates is shown. Confocal microscopy was used to project representative maximum-intensity images for CEN4-GFP, CEN5-GFP, and Tub1-mCherry. Bar, 3 μm.
The difference in kinetochore clustering between WT and slk19Δ cells might be attributed to their differential microtubule-depolymerizing dynamics after nocodazole treatment, as previous data indicate the role of Slk19 in spindle stability during anaphase (Zeng et al., 1999; Pereira and Schiebel, 2003). Therefore we trapped TUB1-GFP and slk19Δ TUB1-GFP cells in a chamber with flowed yeast extract/peptone/dextrose (YPD) medium. We used live-cell imaging to follow the Tub1-GFP signal after addition of nocodazole to the flowed medium (20 μg/ml). WT and slk19Δ mutant cells exhibited similar kinetics for the disappearance of spindle structure after nocodazole addition (Figure 2B), indicating that the kinetochore declustering in nocodazole-treated slk19Δ mutant cells is unlikely due to faster microtubule-depolymerizing kinetics. Unlike the TUB1-mCherry strains used in Figure 2A, we found that some yeast cells with Tub1-GFP showed a GFP dot after nocodazole treatment, which likely represents the short microtubules associated with the spindle pole body. We speculate that these residual microtubules may contribute to the clustering of kinetochores that colocalize with the spindle pole after nocodazole treatment, but the clustering of kinetochores away from the spindle pole in nocodazole-treated cells is likely independent of the microtubules that connect kinetochores to the spindle pole. However, we cannot exclude the possibility that Slk19 promotes kinetochore clustering through the interaction with kinetochore-derived microtubules (Ortiz et al., 2009; Kitamura et al., 2010).
As a kinetochore protein, Slk19 is also a component of FEAR, one of the mitotic exit pathways. To test the possibility that functional FEAR contributes to kinetochore clustering, we examined Mtw1-GFP distribution in cells lacking Spo12, which also functions in the FEAR pathway (Stegmeier et al., 2002). In contrast to slk19Δ, most of spo12Δ mutant cells showed two or three kinetochore clusters after nocodazole treatment, like WT cells (Supplemental Figure S6). Therefore the kinetochore clustering defect in slk19Δ mutant is likely independent of its role in mitotic exit.
To further confirm the kinetochore clustering defect in slk19Δ mutant cells, we used a strain with two GFP-marked centromeres, CEN4-GFP and CEN5-GFP. Our rationale is that if Slk19 participates in kinetochore clustering, then we should observe a lower percentage of cells with colocalized CEN4-GFP and CEN5-GFP in slk19Δ mutant cells after nocodazole treatment compared with WT cells. After G1-arrested cells were released into media containing nocodazole for 120 min, 35.6% of WT cells showed colocalization of CEN4-GFP and CEN5-GFP, whereas only 17.3% of slk19Δ cells did (Figure 2C). Thus the chance of colocalization of kinetochores from chromosome IV and V in slk19Δ cells is significantly lower than that in WT cells, confirming that Slk19 is required to cluster kinetochores from separated chromosomes in the presence of nocodazole.
slk19Δ mutant cells exhibit a delay in the reestablishment of the KT-MT interaction
Our data suggest that Slk19 functions as a glue to mediate the interaction of kinetochores from separate chromosomes. We next asked how Slk19’s kinetochore function facilitates chromosome segregation. Previous data indicate that slk19Δ mutant cells exhibit normal chromosome segregation kinetics (Sullivan et al., 2001; Jin et al., 2008), but the synthetic lethality between slk19Δ and spindle assembly checkpoint mutants suggests a role of Slk19 in faithful chromosome segregation. Therefore we first assessed the sensitivity of slk19Δ mutants to the microtubule poison benomyl. Although slk19Δ cells grew normally on YPD plates, a rich medium for yeast cells, the mutant cells showed a clear slow-growth phenotype on plates containing 15 μg/ml benomyl (Figure 3A). In contrast, spo12Δ cells did not show any noticeable benomyl sensitivity, suggesting that the slow-growth phenotype of slk19Δ mutants is not due to the defect in mitotic exit. Therefore the kinetochore function of Slk19 might become essential when cells are challenged with microtubule-depolymerizing agents.
FIGURE 3:
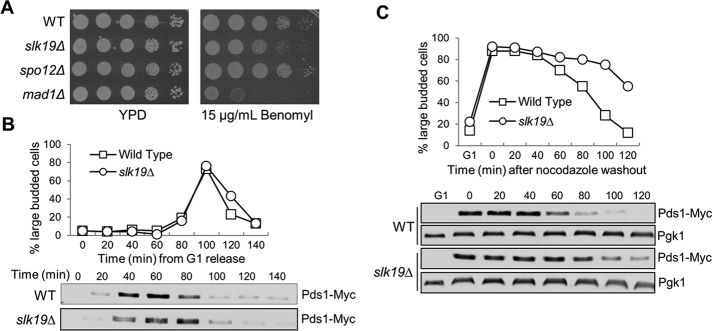
slk19 mutant cells show delayed anaphase entry after nocodazole treatment. (A) slk19Δ mutants show sensitivity to benomyl. WT, slk19Δ, spo12Δ, and mad1Δ mutant cells were grown to saturation, 10-fold diluted, and spotted onto YPD with and without 15 μg/ml benomyl. The plates were scanned after incubation at 30°C for 3 d. (B) slk19Δ mutants do not show an obvious cell cycle defect in an unperturbed cell cycle. WT and slk19Δ cells with PDS1-18myc were released from G1 into YPD at 25°C. Cells were harvested every 20 min and examined for their budding index and Pds1 protein levels. (C) slk19Δ mutants show an anaphase entry delay after exposure to nocodazole. G1-arrested WT and slk19Δ cells with PDS1-18myc were released into YPD containing 20 μg/ml nocodazole at 25°C for 120 min. Nocodazole was then washed off, and the cells were released into YPD medium at 25°C. Cells were harvested every 20 min for budding index and Pds1 protein levels. Pgk1 protein level is shown as a loading control.
Defects in KT-MT interaction trigger the spindle assembly checkpoint to delay the cell cycle through the stabilization of the anaphase inhibitor Pds1 (Cohen-Fix et al., 1996). In the absence of nocodazole treatment, slk19Δ cells exhibited only a slight delay in Pds1 degradation (Figure 4B), which is in agreement with previous data (Sullivan et al., 2001). Next we released the G1-arrested WT and slk19Δ mutant cells into 20 μg/ml nocodazole for 120 min and then washed off the nocodazole to monitor the cell cycle response to nocodazole treatment. In the presence of nocodazole, slk19Δ budded normally, similar to WT cells. After nocodazole washout, however, slk19Δ cells showed an obvious delay in the transition from large-budded to unbudded cells. Consistent with the budding index, slk19Δ cells also showed stabilized Pds1 protein after nocodazole exposure (Figure 3C). Because the disappearance of Pds1 protein marks anaphase entry (Cohen-Fix et al., 1996), this result suggests that slk19Δ cells may have difficulty in establishing correct KT-MT interaction after nocodazole treatment. Previous work indicates that slk19Δ mutant cells lose viability quickly after treatment with nocodazole (Pfiz et al., 2002), but we did not observe significant viability loss with our slk19Δ strains, which could be attributed to different genetic backgrounds.
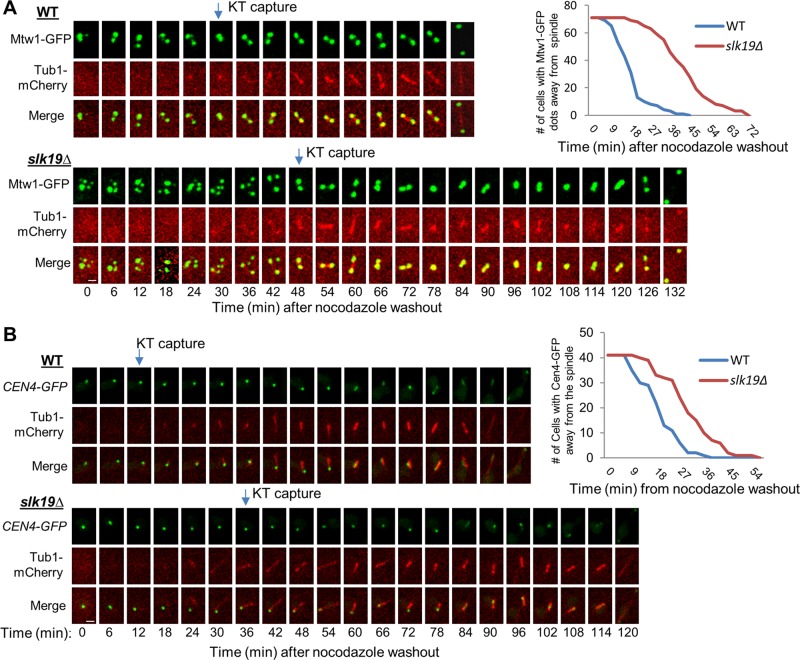
FIGURE 4:
slk19Δ mutant cells show kinetochore capture defect after nocodazole treatment. (A) slk19Δ mutant cells show a dramatic delay in kinetochore capture after the disruption of KT-MT interaction by nocodazole. G1-arrested WT and slk19Δ cells with MTW1-GFP TUB1-mCherry were released into YPD containing 20 μg/ml nocodazole for 100 min. After nocodazole was washed off, the cells were transferred onto an agarose pad on a microscope slide to perform live-cell imaging at 25°C. Although it took 8 min to prepare the slide and start the live-cell imaging after nocodazole washout, we set the movie starting point as time 0 for convenience. The localization of Mtw1-GFP and the spindle structure over the time course in a representative cell of WT and slk19Δ are shown. The timing of kinetochore capture was assessed based on the movies made from the live-cell imaging, and we define the point of kinetochore capture as when all Mtw1-GFP signals colocalize with the spindle (Tub1-mCherry). Right, kinetochore-capture kinetics in the WT and slk19Δ cells (n = 70). Scale bar, 3 μm. (B) slk19Δ mutant cells exhibit delayed capture of a GFP marked chromosome IV (CEN4-GFP) by spindle microtubule. WT and slk19Δ cells with CEN4-GFP TUB1-mCherry were treated the same as in A. The point when the CEN4-GFP colocalizes with the spindle was defined as kinetochore capture. Left, localization of CEN4-GFP and spindle morphology in representative WT and slk19Δ cells over time. Right, kinetics of kinetochore capture for chromosome IV in WT and slk19Δ mutant cells (n = 41). Scale bar, 3 μm.
Our data suggest that Slk19 protein keeps kinetochores together after their disconnection from the spindle poles. To reestablish the KT-MT interaction, the disconnected kinetochores are first captured by a microtubule and then moved to the vicinity of one spindle pole through Cik1/Kar3-mediated transport (Tanaka et al., 2005, 2007), which may facilitate chromosome bipolar attachment (Liu et al., 2011). Clustered kinetochores might be transported to a spindle pole as a group, thereby facilitating the reestablishment of KT-MT interaction. To test this idea, we compared the kinetochore capture process in WT and slk19Δ cells after nocodazole treatment. The G1-arrested cells were released into nocodazole medium for 100 min to disrupt KT-MT interaction. After nocodazole was washed off, the cells were subjected to live-cell microscopy, and the colocalization of kinetochore (Mtw1-GFP) and spindle (Tub1-mCherry) was examined. Note that spindle reformation varied slightly from cell to cell, but the average time was similar between WT and slk19Δ cells. As described earlier, most WT cells showed two or three Mtw1-GFP foci just after the release from nocodazole, but slk19Δ mutant cells exhibited more GFP foci. The kinetochore capture process in representative WT and slk19Δ cells is shown in Figure 4A. In the WT cell, the mCherry cluster appeared 24 min after the movie was started, and all Mtw1-GFP signals colocalized with Tub1-mCherry at 30 min. Thus this cell finished the kinetochore capture within 6 min. In contrast, the visible Tub1-mCherry cluster appeared at 30 min in the slk19Δ cell, but the complete colocalization of all the GFP-marked kinetochores and mCherry-labeled spindle occurred at 48 min, showing that this cell needs 18 min to achieve kinetochore capture. On average, it took WT cells 16 min for all the GFP foci to colocalize with the spindle after nocodazole washout. It is striking that slk19Δ mutant cells took an average of 40 min to achieve the colocalization (n = 70). We noticed that four slk19Δ cells failed to achieve complete colocalization during the entire course of the experiment (150 min), and these cells were excluded for the calculation of the average capture time. Furthermore, we found that many of the cells formed a single GFP dot at first, and then two separated foci appeared before spindle elongation, presumably indicating the time window between chromosome capture and the establishment of chromosome bipolar attachment.
Using the same approach, we also compared the capture process for a single kinetochore in WT and slk19Δ cells with CEN4-GFP and TUB1-mCherry. Consistently, slk19Δ cells showed an obvious delay in the colocalization of CEN4-GFP with the spindle compared with WT cells (Figure 4B), but the delay was not as dramatic as that from the strains with Mtw1-GFP. We speculate that this difference is due to the fact that CEN4-GFP represents one of the 16 pairs of sister kinetochores. Taken together, these results suggest that Slk19-dependent kinetochore clustering facilitates chromosome capture after the disruption of the KT-MT interaction.
slk19Δ mutant cells exhibit delayed chromosome bipolar attachment after exposure to nocodazole
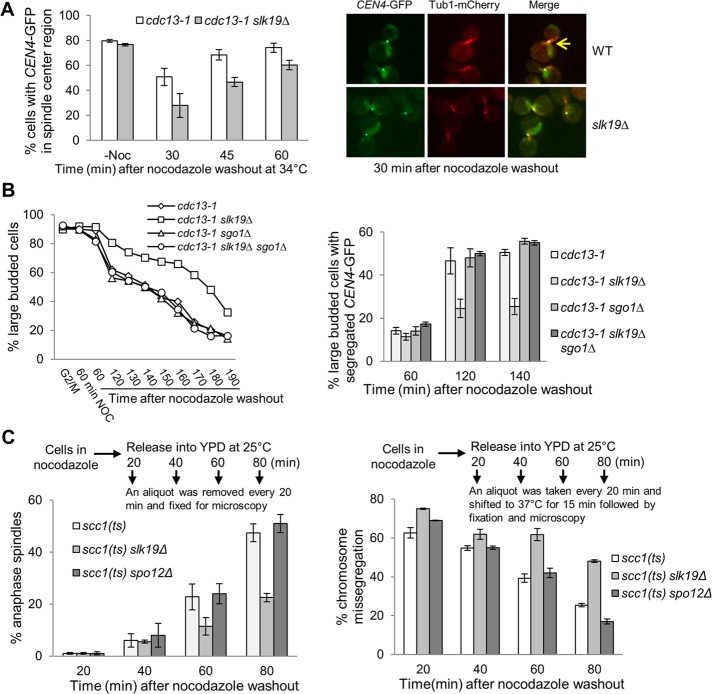
The live-cell microscopy data indicate that slk19Δ cells show a delay in kinetochore capture after nocodazole treatment, but the delay in anaphase onset in slk19Δ cells as indicated by spindle elongation is more dramatic (Figure 4A). It is possible that Slk19 also plays a role in chromosome bipolar attachment in addition to the kinetochore capturing. To test this idea, we first grew cdc13-1 and cdc13-1 slk19Δ cells at 34°C to achieve preanaphase arrest, as loss of Cdc13 function creates unprotected telomeres that activate the DNA-damage checkpoint (Lin and Zakian, 1996). We then treated the cells with nocodazole. The nocodazole treatment of cdc13-1–arrested cells did not cause kinetochore declustering, and that was also true for cdc13-1 slk19Δ cells. After nocodazole was washed off, we examined the localization of CEN4-GFP relative to the spindle over time while keeping the cells at 34°C to retain the preanaphase arrest. The localization of a single or separated GFP dots at the center region of the spindle indicates bipolar attachment on chromosome IV. We counted cells containing a CEN4-GFP dot within the center region of a spindle (one-third) in this category (Figure 5A, arrow), whereas the localization of a CEN4-GFP dot at the end of a spindle suggests failed bipolar attachment. Just after nocodazole release, no spindle structure appeared in both cdc13-1 and cdc13-1 slk19Δ cells. After release from nocodazole treatment for 30 min, 56.8% of cdc13-1 cells exhibited CEN4-GFP localization at the center region of the spindle, whereas only 27.9% of cdc13-1 slk19Δ cells showed this phenotype. cdc13-1 slk19Δ cells also showed a lower percentage of cells with the CEN4-GFP dot at the spindle center after nocodazole release for 45 and 60 min, and thus slk19Δ mutant cells appeared to take longer time to achieve bipolar attachment (Figure 5A). These results suggest that slk19Δ mutants may display a chromosome bipolar attachment defect after nocodazole treatment. Because treatment of cdc13-1 slk19Δ cells growing at 34°C did not cause defective kinetochore clustering, the delayed bipolar attachment in slk19Δ cells is likely independent of the kinetochore capture defect.
FIGURE 5:
slk19Δ mutant cells show capture-independent defect in chromosome bipolar attachment. (A) slk19Δ mutants show a lower percentage of cells with the CEN4-GFP in the middle of the spindle after nocodazole treatment of cells arrested in preanaphase. cdc13-1 and cdc13-1 slk19Δ cells with CEN4-GFP TUB1-mCherry were first incubated in 34°C medium to achieve preanaphase arrest. Then we added 20 μg/ml nocodazole into the medium for 60 min to disrupt the spindle structure. After nocodazole was washed off, the cells were released into 34°C YPD medium, and the relative localization of CEN4-GFP to the spindle was assessed over time. We counted the percentage of cells with the CEN4-GFP dot within the one-third of the center region of the spindle, and at least 100 cells were counted for each sample. The average from three independent experiments is shown. Right, representative images. (B) slk19Δ mutants show tension checkpoint–dependent anaphase entry delay after nocodazole exposure. G1-arrested WT, slk19Δ, sgo1Δ, and slk19Δ sgo1Δ in a cdc13-1 CEN4-GFP background were released to 34°C YPD medium for 120 min to achieve preanaphase arrest. The cells were then shifted to a medium containing 20 μg/ml nocodazole and incubated at 25°C for 60 min. After nocodazole was washed off, the cells were resuspended in fresh YPD media at 25°C and harvested over time to examine budding index and CEN4-GFP segregation. More than 100 cells were counted for each sample. Left, percentage of large-budded cells. Right, percentage of cells with segregated CEN4-GFP dots (one GFP dot in each mother and daughter cell) at the indicated time points. (C) slk19Δ mutant cells exhibit more chromosome missegregation after nocodazole exposure in a cohesin mutant. G1-arrested scc1-73, scc1-73 slk19Δ, and scc1-73 spo12Δ cells with CEN5-GFP TUB1-mCherry were released into YPD containing 20 μg/ml nocodazole for 120 min at 25°C. After nocodazole washout, cells were released into 25°C YPD medium and removed every 20 min to examine the spindle structure. In parallel, an aliquot of the cells was taken every 20 min and shifted to 37°C for 15 min to inactivate cohesin. After fixation, we examined the localization of CEN5-GFP only in cells with an anaphase spindle, and the presence of one or two GFP dots within a single cell body indicates CEN5-GFP missegregation. Left, average percentage of cells with anaphase spindle. Right, average percentage of cells with cosegregated CEN5-GFP after the shift to 37°C. More than 100 cells were counted for each sample, and the average is from three separate experiments.
The delay of chromosome bipolar attachment in slk19Δ cells could increase the chance of syntelic attachments, in which both sister kinetochores are connected to microtubules emanating from a single spindle pole. Sgo1 is a component of the tension checkpoint that senses syntelic attachment and delays anaphase onset (Indjeian et al., 2005; Jin et al., 2012). If syntelic attachments occur in slk19Δ cells after nocodazole treatment, these cells will show a Sgo1-dependent cell cycle delay. Because sgo1Δ cells are sensitive to nocodazole treatment in G1 phase but are insensitive when first arrested at G2/M phase (Indjeian and Murray, 2007), we generated slk19Δ, sgo1Δ, and slk19Δ sgo1Δ strains in cdc13-1 CEN4-GFP background. After achieving G2/M arrest by growing the cells at 34°C, we shifted the cells to 25°C medium containing 20 μg/ml nocodazole. After incubation for 60 min, we washed off nocodazole and released the cells into YPD medium at 25°C to follow the cell cycle progression. Compared to cdc13-1 single mutant, more cdc13-1 slk19Δ cells remained as large budded. Of interest, the delayed M-to-G1 transition in cdc13-1 slk19Δ mutants was completely suppressed by sgo1Δ. Moreover, cdc13-1 slk19Δ cells exhibited a significant delay in segregation CEN4-GFP, and this delay was also suppressed by sgo1Δ (Figure 5B). Consistently, cdc13-1 slk19Δ sgo1Δ double mutants exhibited increased viability loss after nocodazole treatment (unpublished data). Given the established checkpoint function of Sgo1 in sensing syntelic attachments, we speculate that the loss of function of Slk19 delays the establishment of chromosome bipolar attachment after nocodazole treatment and increases the frequency of syntelic attachment.
Faulty KT-MT attachment activates the spindle checkpoint to prevent spindle elongation. In cells lacking sister chromatid cohesion, however, the spindle will elongate even in the presence of these faulty attachments, which will allow us to visualize chromosome missegregation. Thus we created scc1-73 (mcd1), scc1-73 slk19Δ, and scc1-73 spo12Δ strains in CEN5-GFP TUB1-mCherry background, which will lose cohesion when incubated at 37°C (Michaelis et al., 1997). Cells arrested in G1 phase were released into nocodazole for 120 min at 25°C to disrupt the spindle structure. After nocodazole washout, the cells were released into 25°C YPD to examine the spindle morphology over time. At 80 min after nocodazole washout, 47.4% of WT cells, but only 22.5% of slk19Δ cells, showed an elongated spindle, indicating an anaphase entry delay in slk19Δ mutants. In contrast, spo12Δ mutant cells did not show any delay in spindle elongation compared with WT cells (Figure 5C, left). Thus slk19Δ mutant cells showed delayed metaphase–anaphase transition after nocodazole exposure.
In parallel, a portion of the cells was removed every 20 min after nocodazole washout and shifted to 37°C for 15 min to inactivate cohesin (see the diagram in Figure 5C for an illustration of the experiment). After fixation, CEN5-GFP segregation was only scored in cells with an elongated spindle. The rationale for this experiment is that the abolition of cohesion will allow spindle elongation regardless of proper chromosome attachment. If slk19Δ cells take longer to form bipolar attachment after nocodazole washout, then these cells will be more prone to chromosome missegregation after cohesin inactivation. After nocodazole washout for 20 and 40 min, the temperature shift caused a high frequency of CEN5-GFP cosegregation in all three strains, indicating the failure of chromosome attachment. After nocodazole release for 80 min, however, the difference in the frequency of CEN5-GFP missegregation between WT, spo12Δ, and slk19Δ mutant cells became very significant (25.4% for WT vs. 48.1% for slk19Δ; Figure 5C, right), indicating that Slk19 is required for the efficient establishment of chromosome bipolar attachment after nocodazole exposure. Given that our data indicate that most of WT and slk19Δ mutant cells can finish chromosome capture 80 min after nocodazole washout (Figure 4A), we speculate that Slk19 also promotes bipolar attachment in addition to its role in chromosome capture. Because spo12Δ mutant cells did not show any defect in chromosome bipolar attachment after nocodazole exposure, we conclude that the kinetochore function of Slk19 is independent of its role in mitotic exit.
Dimerization of Slk19 might be required for its kinetochore function
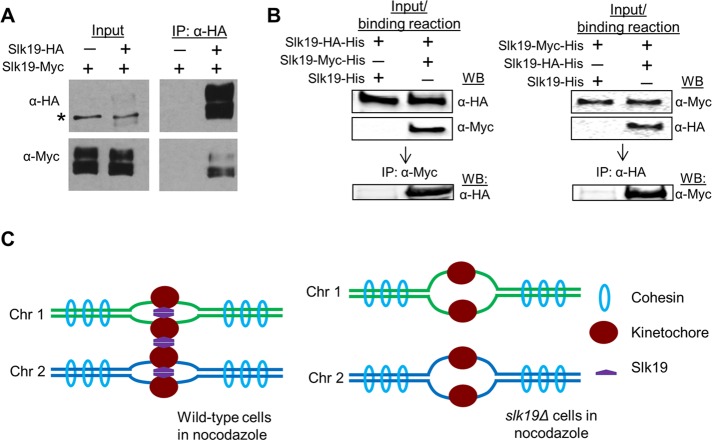
Slk19 is a protein with seven coiled-coil motifs that localize in its central region and C-terminus (Zeng et al., 1999; Zhang et al., 2006). The coiled-coil motifs are made up of α-helices that usually mediate protein dimerization (Lupas and Gruber, 2005). Indeed, genome-wide studies indicate that Slk19 can interact with itself (Newman et al., 2000; De Wulf et al., 2003; Wong et al., 2007; Tarassov et al., 2008). To confirm the Slk19–Slk19 interaction, we generated a SLK19-HA/SLK19-Myc diploid strain. Anti-hemagglutinin (HA) antibody was used to immunoprecipitate the cell lysate from cycling cells, and then proteins were probed with anti-Myc and anti-HA antibodies after separation. In the immunoprecipitate, we detected both full-length and fragments of Slk19-Myc (Figure 6A). The short forms of Slk19 are likely cleavage products, based on previous studies (Sullivan et al., 2001). The coimmunoprecipitation result indicates the Slk19–Slk19 interaction and Slk19 cleavage do not regulate this interaction.
FIGURE 6:
Slk19 protein forms dimers. (A) Both full-length and cleaved fragment of Slk19 physically interact with themselves. Diploid yeast strains containing either SLK19/SLK19-13Myc or SLK19-6HA/SLK19-13Myc were grown to log phase. The cells were harvested to prepare the whole cell extracts with a bead beater. The extracts were first incubated with anti-HA antibody, and then protein A/G-coated agarose beads were added. Western blotting was performed to detect Slk19-6HA and Slk19-13Myc in the whole-cell extracts and the immunoprecipitates. Asterisk indicates a nonspecific band. (B) Slk19 interacts with itself in vitro. Purified Slk19-HA-His was mixed with Slk19-His or Slk19-Myc-His, and then the mixtures were pulled down with anti-Myc antibody. Slk19 proteins with different tags in the mixture and immunoprecipitates were detected using Western blotting with anti-Myc and anti-HA antibodies. A similar pull-down assay was performed after the mixture of Slk19-Myc-His with Slk19-His or Slk19-HA-His. (C) The working model for Slk19-dependent kinetochore clustering and sister kinetochore cohesion.
To further test whether Slk19 directly interacts with itself in vitro, we purified Slk19-histidine (His), Slk19-HA-His, and Slk19-Myc-His from bacterial lysates (Supplemental Figure S7). We first mixed purified Slk19-HA-His proteins with either Slk19-His or Slk19-Myc-His. We then used anti-Myc to immunoprecipitate the mixtures and performed Western blotting by probing with anti-Myc and anti-HA antibodies. We detected a strong Slk19-HA-His band from both mixtures; however, only the mixture containing Slk19-Myc-His and Slk19-HA-His showed an anti-HA band after immunoprecipitation with anti-Myc antibody (Figure 6B, left). Similarly, anti-HA antibody could also pull down Slk19-Myc-His in the mixture containing Slk19-HA-His and Slk19-Myc-His (Figure 6B, right). Together these in vivo and in vitro results demonstrate that Slk19 directly interacts with itself. We speculate that Slk19 dimerization leads to the formation of bridges between kinetochores (Figure 6C).
DISCUSSION
The kinetochore clustering in budding yeast has been documented for many years, but the biological significance of this clustering has remained a mystery. Moreover, it is unclear whether this clustering is totally dependent upon the KT-MT interaction. Here we report that the kinetochore protein Slk19 clusters kinetochores, which is likely independent of KT-MT interaction. We further show that, in the absence of Slk19, kinetochore capture and the establishment of chromosome bipolar attachment are delayed after the disruption of the KT-MT interaction by nocodazole. We also show in vivo and in vitro evidence indicating that Slk19 protein interacts with itself. Therefore we propose a model in which Slk19–Slk19 interaction leads to kinetochore clustering (Figure 6C), a mechanism that becomes critical for chromosome capture and bipolar attachment after the disruption of KT-MT interaction.
Our data indicate that the Slk19-mediated kinetochore clustering is likely independent of the microtubules that connect kinetochores to the spindle poles. First, the scattered kinetochores in nocodazole-treated slk19Δ mutant cells are away from the spindle pole, suggesting the loss of connection with the spindle poles. Moreover, the slk19Δ mutant cells show similar microtubule depolymerizing kinetics as WT cells in response to nocodazole treatment (Figure 2B). In addition to spindle poles, recent data indicate that kinetochores also generate microtubules (Ortiz et al., 2009; Kitamura et al., 2010). Thus it is possible that Slk19 promotes kinetochore clustering by associating and/or stabilizing these microtubules. Another possibility is that Slk19 binds to other kinetochore proteins directly, and the Slk19–Slk19 interaction leads to kinetochore clustering.
Previous work showed abnormal nuclear morphology in slk19Δ mutant cells (Zhang et al., 2006). One interesting question is whether the kinetochore clustering defect is secondary to the abnormal chromatin structure. Because nocodazole treatment abolishes the abnormal nuclear morphology seen in slk19Δ cells (Zhang et al., 2006), the kinetochore declustering in nocodazole-treated slk19Δ cells is unlikely a consequence of the abnormal nuclear morphology. Instead, the enriched localization of Slk19 at kinetochores suggests that the kinetochore function of Slk19 likely contributes to kinetochore clustering. In addition, the data from the Dawson lab show the colocalization of Slk19 with another kinetochore protein, Mtw1 (Havens et al., 2010). We also performed live-cell imaging in cells with SLK19-GFP NUF2-mCherry and found the colocalized Slk19 and kinetochore protein Nuf2 until anaphase, when the spindle midzone localization of Slk19 was noticed (unpublished data). Because we showed direct Slk19–Slk19 interaction, one possibility is that the interaction between Slk19 proteins from different kinetochores mediates kinetochore clustering (Figure 6C), but more experiments are needed to confirm this model.
In nocodazole-treated slk19Δ mutant cells, in addition to a major kinetochore cluster that associated with the spindle poles, more small Mtw1-GFP foci were detected. One interesting question is whether the smaller Mtw1-GFP dot represents a single pair of sister kinetochores. In some slk19Δ cells, we observed as many as nine Mtw1-GFP foci. Given that yeast cells have only 16 pairs of sister chromatids in G2/M phase, some of the smaller foci have to be a single pair of sister kinetochores. In nocodazole-treated slk19Δ cells, some kinetochores may stay together even when they are disconnected from the spindle pole. One possibility is that these kinetochores just colocalize by chance but without any linkage. Alternatively, an unknown mechanism clusters these kinetochores in the absence of Slk19 and KT-MT interaction. Therefore further analysis is necessary to determine whether kinetochore clustering is abolished completely in slk19Δ mutant cells once they become disconnected from the spindle pole.
Kinetochore declustering was also observed in other kinetochore mutants, such as nuf2-60 and ndc80-1, even without treatment with nocodazole (Janke et al., 2001; Anderson et al., 2009). Nuf2 and Ndc80 are components of the Ndc80 kinetochore complex, which is required for the KT-MT interaction. Recent data show that the association of Slk19 with kinetochore is abolished in some temperature-sensitive kinetochore mutant strains when incubated at 37°C, including spc105ts, ndc80-1, and mtw1-1 (Pagliuca et al., 2009). Therefore it is possible that the abolished KT-MT interaction alone causes kinetochore clustering. Alternatively, both the loss of the KT-MT interaction and the absence of Slk19 at kinetochores are necessary for kinetochore declustering.
What is the biological significance of this Slk19-dependent kinetochore clustering? For the kinetochores that lose their connection to the spindle pole, the highly dynamic spindle microtubules are responsible for their capture. Then the minus end–directed motor complex Cik1/Kar3 transports the captured kinetochores toward the vicinity of the spindle pole (Tanaka et al., 2005). The increased distance between a kinetochore and the spindle pole will significantly reduce the chance of kinetochore capture. A whole kinetochore cluster will be moved to the spindle pole once only one kinetochore in this cluster is captured. This mechanism is expected to increase the efficiency for the transport of detached kinetochores toward the vicinity of a spindle pole, where they have a much higher chance to be captured by spindle microtubules. In support of this speculation, we observed a dramatic kinetochore capturing delay in slk19Δ mutant cells after the disruption of KT-MT interaction. Therefore kinetochore clustering could be an unidentified mechanism that facilitates kinetochore capture.
In budding yeast, an interesting observation is that sister kinetochores separate before anaphase entry (Goshima and Yanagida, 2000; He et al., 2000). Because separase is inactive before anaphase onset, cohesin cleavage is unlikely the cause for this sister centromere separation. Work from the Bloom lab showed a cruciform structure of the pericentric chromatin (Yeh et al., 2008). This observation indicates that the enriched cohesin at the pericentric region unlikely links sister centromeres, and instead it might stabilize the cruciform structure by introducing cohesion within a single chromatin at pericentric regions. Thus a unique mechanism may contribute to sister chromatid cohesion at centromeric regions. Our evidence suggests that Slk19 mediates interaction between kinetochores from different chromosomes. It is possible that Slk19 also mediates interaction between sister kinetochores, which contributes to sister-chromatid cohesion at centromeric regions. To test this possibility, we looped out a pair of GFP-marked centromeres of chromosome IV in nocodazole-treated cells and found that slk19Δ cells showed higher frequency of separated sister centromeres (Supplemental Figure S8). However, the result could not support a solid conclusion because of the lower frequency of the excision of the centromere from chromosome IV, as indicated by only 40% viability loss after the induction of recombination. If the Slk19–Slk19 interaction contributes to sister kinetochore cohesion, the tension resulting from chromosome bipolar attachment can separate sister kinetochores and centromeres by breaking up the Slk19-Slk19 interaction in the absence of separase-dependent cohesin cleavage. Moreover, this Slk19-mediated sister centromere cohesion should be reversible. Indeed, separated sister centromeres in metaphase cells reunite after nocodazole treatment, and this reunion depends on Slk19 (Zhang et al., 2006). Therefore both kinetochore clustering and sister chromatid cohesion at centromere regions could be attributed to Slk19-mediated interkinetochore interaction (Figure 6C).
Our data also indicate the role of Slk19 in the establishment of chromosome bipolar attachment. The exposure of cdc13-1 slk19Δ cells to nocodazole delayed the chromosome bipolar attachment significantly after nocodazole was washed off, and this delay depends on a functional tension checkpoint, indicating the presence of syntelic attachments. Because we did not observe obvious kinetochore declustering in these cells, it is likely that Slk19 also facilitates chromosome bipolar attachment in a kinetochore-clustering–independent manner. One explanation is that the Slk19–Slk19 interaction between sister kinetochores ensures their opposite orientation, which facilitates the bipolar attachment. In the absence of Slk19, however, the presence of misorientated sister kinetochores increases the frequency of syntelic attachment, especially after the disruption of KT-MT interaction.
Kinetochore clustering is also observed in higher eukaryotes (Solovei et al., 2004), indicating that this mechanism could be conserved. Although a Slk19 homologue is lacking in higher organisms based upon protein sequence homology, several functional orthologues have been suggested due to its diverse functions (Sato et al., 2003; Vos et al., 2006; Ohkuni et al., 2008). One of the candidates is the mammalian protein CENP-F/mitosin, which localizes to the kinetochore from late G2 to early anaphase but moves to spindle midzone in late anaphase, a pattern similar to that of Slk19 (Rattner et al., 1993; Liao et al., 1995; Zhu et al., 1995; Yang et al., 2003). Of interest, knockdown of CENP-F leads to weakened centromeric-specific cohesion (Holt et al., 2005). Moreover, more-scattered chromosome distribution in CENP-F knockdown cells is consistent with the function of CENP-F in kinetochore clustering, although further experiments are needed to confirm this notion (Holt et al., 2005). Similar to Slk19, CENP-F has also been shown to dimerize (Zhu et al., 1995). These similarities suggest that CENP-F could be the functional orthologue of yeast Slk19 in mammalian cells.
MATERIALS AND METHODS
Yeast strains and growth
The yeast strains used in this study are listed in Supplemental Table S1. All strains are isogenic to Y300, a W303 derivative. Yeast cells were grown in YPD or indicated synthetic medium. To arrest cells in G1 phase, 5 μg/ml α-factor was added into cell cultures. After 120 min of incubation, the G1-arrested cells were washed twice with water and then released into fresh medium to start cell cycle. Nocodazole was used at 20 μg/ml in 1% dimethyl sulfoxide. To induce the expression of SCC1 (MCD1) from a galactose-inducible promoter control, we first grew cells in raffinose medium and then added galactose to the medium to a final concentration of 2%.
Cytological techniques
Cells with GFP-, mApple-, or mCherry-tagged proteins were fixed with 3.7% formaldehyde for 5 min at room temperature, and then washed twice with 1× phosphate-buffered saline (PBS) buffer and resuspended in PBS buffer for fluorescence microscopy (Carl Zeiss MicroImaging, Jena, Germany). Live-cell microscopy was carried out with an Andor Revolution SD imaging system that uses a Nikon Eclipse Ti microscope (Andor Technology, South Windsor, CT). Glass depression slides were used to prepare an agarose pad filled with synthetic or complete medium. All live-cell images were acquired at 25°C with a 100× objective lens. Twelve Z-sections were collected at each time point, and each optical section was set at 0.5 μm thickness. The time-lapse interval was set at 3 or 5 min as indicated. Maximum projections from applicable time points were created using Andor IQ2 software. To visualize microtubule-depolymerizing dynamics, filtered YPD medium containing 20 μg/ml nocodazole was flowed over cells with Tub1-GFP trapped in a chamber (CellASIC, Hayward, CA). Superresolution images were taken from the DeltaVision OMX system from Applied Precision (Issaquah, WA). The error bars in all the figures represent the SEM.
Protein techniques
Cell pellets from 1.5 ml of cell culture were resuspended in 200 μl of 0.1 N NaOH and incubated at room temperature for 5 min. After centrifugation, the cells were resuspended in equal volume (30 μl) of double-distilled H2O and SDS protein-loading buffer. The samples were then boiled for 5 min and resolved with 10% SDS-polyacrylamide gel. Proteins were detected with enhanced chemiluminescence (PerkinElmer LAS, Waltham, MA) after probing with anti-Myc antibody (Covance Research Products, Berkeley, CA) and horseradish peroxidase–conjugated secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA).
Construction of slk19 bacterial-expression plasmids
To construct bacterial expression plasmids, we cloned the Slk19 open reading frame from yeast genomic DNA using PCR. The primers contain cut sites for the NdeI and NotI restriction enzymes. After PCR, the amplified product was digested with NdeI and NotI enzymes (New England BioLabs, Ipswich, MA) and ligated into the NdeI/NotI cloning site of the bacterial expression plasmid pET-20b(+) (Invitrogen, Carlsbad, CA) in frame with a C-terminal His tag. The cloned Slk19 was fully sequenced to make sure that no mutations were introduced during the cloning procedure. The resulting plasmid is referred to as pET-Slk19-His. To construct pET-Slk19-HA-His and pET-Slk19-Myc-His expression plasmids, we subcloned a double-stranded DNA oligonucleotide containing the coding sequence for HA or Myc tag (with NotI overhangs) into the NotI restriction site of the pET-Slk19-His plasmid. This results in an HA or Myc tag at the C-terminus of Slk19, in frame with the His tag. The correct frame and sequence of the tags were confirmed by sequencing.
Bacterial expression and purification of slk19 proteins
Slk19 plasmids were transformed into Rosetta (DE3) cells and grown in LB Miller broth medium (EMD Chemicals, San Diego, CA) with ampicillin (100 μg/ml final concentration) overnight at 37ºC. The overnight bacterial culture was diluted 1:6 in LB–ampicillin and grown for 1 h at 37ºC, then for 6 h at room temperature to allow for optimal expression of the Slk19 proteins. Bacterial cells were then pelleted by centrifugation and resuspended in ice-cold PBS, pH 7.5, supplemented with a cocktail of protease inhibitors (Sigma-Aldrich, St. Louis, MO). The suspension was sonicated on ice (three bursts, 15 s each, with 2-min intervals between sonication bursts to allow cooling). Bacterial lysates were cleared by high-speed centrifugation.
To purify the His-tagged proteins, the cleared bacterial lysates were incubated with nickel beads (Qiagen, Valencia, CA), prewashed, and equilibrated in PBS, with rotation for 2 h at 4°C. The resulting slurry of beads and lysates was centrifuged at 500 × g for 2 min at 4°C. The beads were washed four times with cold PBS, with 5-min rocking for every wash. The beads were further washed twice with PBS containing 20 mM imidazole and twice with PBS containing 50 mM imidazole. Slk19-His, Slk19-HA-His, or Slk19-Myc-His was then eluted at 4°C from the beads in PBS containing 100 mM Imidazole. The eluted proteins were then dialyzed in PBS at 4°C for 2 h. The purified proteins were stored at −80°C until used in the binding/dimerization assay. Samples of cleared bacterial lysates and purified proteins were analyzed on an SDS–PAGE gel and visualized with Coomassie blue staining. In addition, the expression of the HA and Myc tags was confirmed by Western blot analysis of the purified proteins with anti-HA and anti-Myc antibodies.
Slk19 binding/dimerization assays
For the binding assays, purified Slk19-HA-His was mixed with purified Slk19-Myc-His or Slk19-His (negative control) and incubated for 2 h at 4°C. The mixtures were then diluted with PBS up to 1 ml and incubated with anti-Myc antibody (Santa Cruz Biotechnology, Santa Cruz, CA) overnight at 4°C. Protein A/G beads were then added for 1 h. The protein-bead complexes were then pelleted by centrifugation at (1000 × g for 2 min at 4°C), washed four times with cold PBS, and boiled in 2× SDS–PAGE loading buffer before loading on the gel and subsequent Western blotting, as indicated in Figure 6B. This experiment was alternatively performed by incubating purified Slk19-Myc-His with purified Slk19-HA-His or Slk19-His (negative control), followed by immunoprecipitation with anti-HA antibody (Sigma-Aldrich).
Supplementary Material
Acknowledgments
We thank Angelika Amon, Jeff Bachant, Frank Uhlmann, and Paul Megee for yeast strains and plasmids. We thank Xianying Tang, who constructed some of the yeast strains. We are grateful to Ruth Dider, who helped with live-cell imaging and superresolution microscopy. We also thank the members of the yeast community at Florida State University for reagents and suggestions. This work was supported by Research Scholar Grant RSG-08-104-010CCG from the American Cancer Society, Grant R15GM097326-01 from the National Institutes of Health/National Institute of General Medical Sciences, and research grants from the College of Medicine at Florida State University to Y.W.
Abbreviations used:
- KT-MT
kinetochore-microtubule
- WT
wild type
- YPD
yeast peptone dextrose
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-07-0552) on January 2, 2013.
REFERENCES
- Anderson M, Haase J, Yeh E, Bloom K. Function and assembly of DNA looping, clustering, and microtubule attachment complexes within a eukaryotic kinetochore. Mol Biol Cell. 2009;20:4131–4139. doi: 10.1091/mbc.E09-05-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Fix O, Peters JM, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- Collins SR, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- De Wulf P, McAinsh AD, Sorger PK. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 2003;17:2902–2921. doi: 10.1101/gad.1144403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, Lee C, Shendure J, Fields S, Blau CA, Noble WS. A three-dimensional model of the yeast genome. Nature. 2009;465:363–367. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Hagan I, Uzawa S, Yanagida M. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J Cell Biol. 1993;121:961–976. doi: 10.1083/jcb.121.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MK, et al. The microtubule-based motor Kar3 and plus end-binding protein Bim1 provide structural support for the anaphase spindle. J Cell Biol. 2008;180:91–100. doi: 10.1083/jcb.200710164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillett ES, Espelin CW, Sorger PK. Spindle checkpoint proteins and chromosome-microtubule attachment in budding yeast. J Cell Biol. 2004;164:535–546. doi: 10.1083/jcb.200308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh PY, Kilmartin JV. NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J Cell Biol. 1993;121:503–512. doi: 10.1083/jcb.121.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Yanagida M. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell. 2000;100:619–633. doi: 10.1016/s0092-8674(00)80699-6. [DOI] [PubMed] [Google Scholar]
- Havens KA, Gardner MK, Kamieniecki RJ, Dresser ME, Dawson DS. Slk19p of Saccharomyces cerevisiae regulates anaphase spindle dynamics through two independent mechanisms. Genetics. 2010;186:1247–1260. doi: 10.1534/genetics.110.123257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Asthana S, Sorger PK. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell. 2000;101:763–775. doi: 10.1016/s0092-8674(00)80888-0. [DOI] [PubMed] [Google Scholar]
- Holt SV, Vergnolle MA, Hussein D, Wozniak MJ, Allan VJ, Taylor SS. Silencing Cenp-F weakens centromeric cohesion, prevents chromosome alignment and activates the spindle checkpoint. J Cell Sci. 2005;118:4889–4900. doi: 10.1242/jcs.02614. [DOI] [PubMed] [Google Scholar]
- Indjeian VB, Murray AW. Budding yeast mitotic chromosomes have an intrinsic bias to biorient on the spindle. Curr Biol. 2007;17:1837–1846. doi: 10.1016/j.cub.2007.09.056. [DOI] [PubMed] [Google Scholar]
- Indjeian VB, Stern BM, Murray AW. The centromeric protein Sgo1 is required to sense lack of tension on mitotic chromosomes. Science. 2005;307:130–133. doi: 10.1126/science.1101366. [DOI] [PubMed] [Google Scholar]
- Janke C, Ortiz J, Lechner J, Shevchenko A, Magiera MM, Schramm C, Schiebel E. The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J. 2001;20:777–791. doi: 10.1093/emboj/20.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F, Liu H, Li P, Yu HG, Wang Y. Loss of function of the cik1/kar3 motor complex results in chromosomes with syntelic attachment that are sensed by the tension checkpoint. PLoS Genet. 2012;8:e1002492. doi: 10.1371/journal.pgen.1002492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F, Liu H, Liang F, Rizkallah R, Hurt MM, Wang Y. Temporal control of the dephosphorylation of Cdk substrates by mitotic exit pathways in budding yeast. Proc Natl Acad Sci USA. 2008;105:16177–16182. doi: 10.1073/pnas.0808719105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin QW, Fuchs J, Loidl J. Centromere clustering is a major determinant of yeast interphase nuclear organization. J Cell Sci. 2000;113:1903–1912. doi: 10.1242/jcs.113.11.1903. [DOI] [PubMed] [Google Scholar]
- Khmelinskii A, Lawrence C, Roostalu J, Schiebel E. Cdc14-regulated midzone assembly controls anaphase B. J Cell Biol. 2007;177:981–993. doi: 10.1083/jcb.200702145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura E, Tanaka K, Kitamura Y, Tanaka TU. Kinetochore microtubule interaction during S phase in Saccharomyces cerevisiae. Genes Dev. 2007;21:3319–3330. doi: 10.1101/gad.449407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura E, Tanaka K, Komoto S, Kitamura Y, Antony C, Tanaka TU. Kinetochores generate microtubules with distal plus ends: their roles and limited lifetime in mitosis. Dev Cell. 2010;18:248–259. doi: 10.1016/j.devcel.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F, Wang Y. DNA damage checkpoints inhibit mitotic exit by two different mechanisms. Mol Cell Biol. 2007;27:5067–5078. doi: 10.1128/MCB.00095-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H, Winkfein RJ, Mack G, Rattner JB, Yen TJ. CENP-F is a protein of the nuclear matrix that assembles onto kinetochores at late G2 and is rapidly degraded after mitosis. J Cell Biol. 1995;130:507–518. doi: 10.1083/jcb.130.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JJ, Zakian VA. The Saccharomyces CDC13 protein is a single-strand TG1–3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc Natl Acad Sci USA. 1996;93:13760–13765. doi: 10.1073/pnas.93.24.13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Jin F, Liang F, Tian X, Wang Y. The Cik1/Kar3 motor complex is required for the proper kinetochore-microtubule interaction after stressful DNA replication. Genetics. 2011;187:397–407. doi: 10.1534/genetics.110.125468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Liang F, Jin F, Wang Y. The coordination of centromere replication, spindle formation, and kinetochore-microtubule interaction in budding yeast. PLoS Genet. 2008;4:e1000262. doi: 10.1371/journal.pgen.1000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas AN, Gruber M. The structure of alpha-helical coiled coils. Adv Protein Chem. 2005;70:37–78. doi: 10.1016/S0065-3233(05)70003-6. [DOI] [PubMed] [Google Scholar]
- Mayer ML, Gygi SP, Aebersold R, Hieter P. Identification of RFC(Ctf18p, Ctf8p, Dcc1p): an alternative RFC complex required for sister chromatid cohesion in S. cerevisiae. Mol Cell. 2001;7:959–970. doi: 10.1016/s1097-2765(01)00254-4. [DOI] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Newman JR, Wolf E, Kim PS. A computationally directed screen identifying interacting coiled coils from Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2000;97:13203–13208. doi: 10.1073/pnas.97.24.13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkuni K, Abdulle R, Tong AH, Boone C, Kitagawa K. Ybp2 associates with the central kinetochore of Saccharomyces cerevisiae and mediates proper mitotic progression. PLoS One. 2008;3:e1617. doi: 10.1371/journal.pone.0001617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz J, Funk C, Schafer A, Lechner J. Stu1 inversely regulates kinetochore capture and spindle stability. Genes Dev. 2009;23:2778–2791. doi: 10.1101/gad.541309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliuca C, Draviam VM, Marco E, Sorger PK, De Wulf P. Roles for the conserved spc105p/kre28p complex in kinetochore-microtubule binding and the spindle assembly checkpoint. PLoS One. 2009;4:e7640. doi: 10.1371/journal.pone.0007640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G, Schiebel E. Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science. 2003;302:2120–2124. doi: 10.1126/science.1091936. [DOI] [PubMed] [Google Scholar]
- Pfiz S, Zimmermann J, Hilt W. The yeast kinetochore protein Slk19 is required to prevent aberrant chromosome segregation in meiosis and mitosis. Genes Cells. 2002;7:1033–1042. doi: 10.1046/j.1365-2443.2002.00583.x. [DOI] [PubMed] [Google Scholar]
- Queralt E, Lehane C, Novak B, Uhlmann F. Downregulation of PP2A(Cdc55) phosphatase by separase initiates mitotic exit in budding yeast. Cell. 2006;125:719–732. doi: 10.1016/j.cell.2006.03.038. [DOI] [PubMed] [Google Scholar]
- Rattner JB, Rao A, Fritzler MJ, Valencia DW, Yen TJ. CENP-F is a .ca 400 kDa kinetochore protein that exhibits a cell-cycle dependent localization. Cell Motil Cytoskeleton. 1993;26:214–226. doi: 10.1002/cm.970260305. [DOI] [PubMed] [Google Scholar]
- Sato M, Koonrugsa N, Toda T, Vardy L, Tournier S, Millar JB. Deletion of Mia1/Alp7 activates Mad2-dependent spindle assembly checkpoint in fission yeast. Nat Cell Biol. 2003;5:764–766. doi: 10.1038/ncb0903-764. [DOI] [PubMed] [Google Scholar]
- Solovei I, Schermelleh L, During K, Engelhardt A, Stein S, Cremer C, Cremer T. Differences in centromere positioning of cycling and postmitotic human cell types. Chromosoma. 2004;112:410–423. doi: 10.1007/s00412-004-0287-3. [DOI] [PubMed] [Google Scholar]
- Stegmeier F, Visintin R, Amon A. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 2002;108:207–220. doi: 10.1016/s0092-8674(02)00618-9. [DOI] [PubMed] [Google Scholar]
- Sullivan M, Lehane C, Uhlmann F. Orchestrating anaphase and mitotic exit: separase cleavage and localization of Slk19. Nat Cell Biol. 2001;3:771–777. doi: 10.1038/ncb0901-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M, Uhlmann F. A non-proteolytic function of separase links the onset of anaphase to mitotic exit. Nat Cell Biol. 2003;5:249–254. doi: 10.1038/ncb940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Kitamura E, Kitamura Y, Tanaka TU. Molecular mechanisms of microtubule-dependent kinetochore transport toward spindle poles. J Cell Biol. 2007;178:269–281. doi: 10.1083/jcb.200702141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Mukae N, Dewar H, van Breugel M, James EK, Prescott AR, Antony C, Tanaka TU. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature. 2005;434:987–994. doi: 10.1038/nature03483. [DOI] [PubMed] [Google Scholar]
- Tarassov K, et al. An in vivo map of the yeast protein interactome. Science. 2008;320:1465–1470. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- Tong AH, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Vos LJ, Famulski JK, Chan GK. How to build a centromere: from centromeric and pericentromeric chromatin to kinetochore assembly. Biochem Cell Biol. 2006;84:619–639. doi: 10.1139/o06-078. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ng TY. Phosphatase 2A negatively regulates mitotic exit in Saccharomyces cerevisiae. Mol Biol Cell. 2006;17:80–89. doi: 10.1091/mbc.E04-12-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wargacki MM, Tay JC, Muller EG, Asbury CL, Davis TN. Kip3, the yeast kinesin-8, is required for clustering of kinetochores at metaphase. Cell Cycle. 2010;9:2581–2588. doi: 10.4161/cc.9.13.12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J, et al. A protein interaction map of the mitotic spindle. Mol Biol Cell. 2007;18:3800–3809. doi: 10.1091/mbc.E07-06-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZY, Guo J, Li N, Qian M, Wang SN, Zhu XL. Mitosin/CENP-F is a conserved kinetochore protein subjected to cytoplasmic dynein-mediated poleward transport. Cell Res. 2003;13:275–283. doi: 10.1038/sj.cr.7290172. [DOI] [PubMed] [Google Scholar]
- Ye P, Peyser BD, Pan X, Boeke JD, Spencer FA, Bader JS. Gene function prediction from congruent synthetic lethal interactions in yeast. Mol Syst Biol. 2005;1:2005.0026. doi: 10.1038/msb4100034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Haase J, Paliulis LV, Joglekar A, Bond L, Bouck D, Salmon ED, Bloom KS. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr Biol. 2008;18:81–90. doi: 10.1016/j.cub.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yellman CM, Burke DJ. The role of Cdc55 in the spindle checkpoint is through regulation of mitotic exit in Saccharomyces cerevisiae. Mol Biol Cell. 2006;17:658–666. doi: 10.1091/mbc.E05-04-0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Kahana JA, Silver PA, Morphew MK, McIntosh JR, Fitch IT, Carbon J, Saunders WS. Slk19p is a centromere protein that functions to stabilize mitotic spindles. J Cell Biol. 1999;146:415–425. doi: 10.1083/jcb.146.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Lim HH, Cheng CS, Surana U. Deficiency of centromere-associated protein Slk19 causes premature nuclear migration and loss of centromeric elasticity. J Cell Sci. 2006;119:519–531. doi: 10.1242/jcs.02757. [DOI] [PubMed] [Google Scholar]
- Zhu X, Chang KH, He D, Mancini MA, Brinkley WR, Lee WH. The C terminus of mitosin is essential for its nuclear localization, centromere/kinetochore targeting, and dimerization. J Biol Chem. 1995;270:19545–19550. doi: 10.1074/jbc.270.33.19545. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.