Abstract
Cloning by somatic cell nuclear transfer is an important technology, but remains limited due to poor rates of success. Identifying genes supporting clone development would enhance our understanding of basic embryology, improve applications of the technology, support greater understanding of establishing pluripotent stem cells, and provide new insight into clinically important determinants of oocyte quality. For the first time, a systems genetics approach was taken to discover genes contributing to the ability of an oocyte to support early cloned embryo development. This identified a primary locus on mouse chromosome 17 and potential loci on chromosomes 1 and 4. A combination of oocyte transcriptome profiling data, expression correlation analysis, and functional and network analyses yielded a short list of likely candidate genes in two categories. The major category—including two genes with the strongest genetic associations with the traits (Epb4.1l3 and Dlgap1)—encodes proteins associated with the subcortical cytoskeleton and other cytoskeletal elements such as the spindle. The second category encodes chromatin and transcription regulators (Runx1t1, Smchd1, and Chd7). Smchd1 promotes X chromosome inactivation, whereas Chd7 regulates expression of pluripotency genes. Runx1t1 has not been associated with these processes, but acts as a transcriptional repressor. The finding that cytoskeleton-associated proteins may be key determinants of early clone development highlights potential roles for cytoplasmic components of the oocyte in supporting nuclear reprogramming. The transcriptional regulators identified may contribute to the overall process as downstream effectors.
Keywords: reprogramming, oocyte, somatic cell nuclear transfer, oocyte activation, gene expression, pluripotency, quantitative trait loci, oocyte cytoskeleton
THE oocyte is a remarkable cell. It harbors essential stored mRNAs, proteins, and other macromolecules to sustain and direct normal development until embryonic gene expression commences and until external nutrient supplies become available. The oocyte also has the unique ability to reprogram somatic cell nuclei to a totipotent state, a capacity that reflects its unique role during normal embryogenesis of uniting the two gamete genomes and converting them into an embryonic genome. A variety of approaches since the late 1800s investigated the nuclear reprogramming capacity of oocytes by manipulating the ooplasm–nucleus dialog, using blastomere ligature experiments, embryo splitting, embryonic cell nuclear transfer, and ultimately somatic cell nuclear transfer (SCNT). These methodologies have also been useful in determining the timing and mechanisms underlying other processes such as cell-fate restriction and lineage determination in the early embryo. Cloning studies can thus provide unique insight into the early formative processes that are essential to creating each new individual.
Ooplasmic components must mediate a myriad of key events to make cloned embryogenesis possible, and observing the execution of these events in cloned embryos can reveal previously unappreciated aspects of normal development. The first step of the cloning process entails chemical disruption of the cytoskeleton to enable removal of the spindle-chromosome complex (SCC). Next, the SCC is removed and discarded along with associated proteins and other macromolecules. The cytoskeletal architecture, which is key to supporting correct protein trafficking, intracellular signaling, and other essential processes, must eventually be repaired. The SCC-associated factors either are replenished or remain deficient (Miyara et al. 2006). Upon introduction of the donor cell nucleus by either fusion or microinjection, the oolemma must be repaired. The oocyte must then disassemble the nuclear envelope of the donor nucleus, condense the chromosomes, and form a new pseudo-meiotic SCC (pmSCC) by reestablishing a spindle architecture and gathering the chromosomes onto the metaphase plate. This process recapitulates many key aspects of oocyte maturation, but chromosome homologs are not paired, chromosome congression is slow or incomplete, and the pmSCC is defective in many respects (Miyara et al. 2006; Han et al. 2010b). Clone development is initiated by the artificial activation of the oocyte, either using electrical pulses or chemical mediators, and the mode of activation can alter later gene expression relative to embryos activated by fertilization (Ozil et al. 2006). Polar body extrusion is prevented during the activation process through a second round of cytoskeletal disruption to maintain a diploid chromosome complement. After activation, the cloned embryo must undergo DNA replication and correct mitotic divisions during cleavage stages. Gene transcription must initiate before supportive ooplasmic macromolecules become depleted. The donor genome must be reprogrammed, a process that is believed to initiate with chromosome condensation in the oocyte, but likely continues well into cleavage, given the observed persistent differences between cloned and normal embryonic gene expression (Latham 2005; Vassena et al. 2007a,b). Essential epigenetic information must be retained during the reprogramming process, but some epigenetic information may be lost during somatic development and will be absent in cloned embryos. The leisurely pace of nuclear reprogramming relative to the onset of embryonic gene transcription in clones results in many somatic cell-like features being manifested throughout cleavage development (Chung et al. 2002; Gao et al. 2003). Because normal embryos differ markedly from somatic cells with respect to physiology and in vitro culture requirements, the persistence of these somatic characteristics means that cloned embryos likely must adapt to a less than optimum environment in vitro, and even in vivo following embryo transfer (Gao and Latham 2004; Latham 2004, 2005; Latham et al. 2007).
Cloning methodologies have substantial practical value, enabling the propagation of valuable livestock and endangered species, and potentially the production of stem cells for therapeutic application. Since the birth of Dolly in 1996 (Campbell et al. 1996), much effort has been invested in attempting to enhance the production of cloned animals by SCNT. Given the complex series of events that must occur for cloning to succeed, it is not surprising that many barriers to cloning success have been identified, including incomplete nuclear reprogramming, failure to reactivate X chromosomes and aberrant X chromosome inactivation, deficiencies in spindle formation and function, aneuploidy, loss of genomic imprints, aberrant regulation of DNA methyltransferases, and somatic cell-like features leading to altered culture requirements and metabolism (Eggan et al. 2000; Ohgane et al. 2001; Chung et al. 2002, 2003; Humpherys et al. 2002; Gao et al. 2003, 2004; Mann et al. 2003; Gao and Latham 2004; Latham 2005; Nolen et al. 2005; Miyara et al. 2006; Vassena et al. 2007a,b; Jiang et al. 2008; Han et al. 2010b, 2008; Inoue et al. 2010; Matoba et al. 2011; Mizutani et al. 2012). Deficiencies in spindle formation and embryonic aneuploidy have been addressed at least partially by augmenting the ooplasm supply of spindle-associated proteins (Han et al. 2010b). Some genes are imprinted in early embryos but lose their imprints in somatic cells (Ogura et al. 2002; Wagschal and Feil 2006; Guillomot et al. 2010; Chavatte-Palmer et al. 2012). Some imprints are normally retained only in the placenta. The loss of placenta-specific genomic imprinting in somatic donor nuclei could thus affect the placenta in clones. Loss of other imprints cell type specifically could affect cloning outcome in a donor cell type-specific manner. No clear remedy to imprinting defects or other defects has emerged. Limitations in nuclear reprogramming have remained a significant barrier in cloning by SCNT and have consequently received a great deal of attention by those seeking to enhance outcomes. Chemical agents that modify chromatin structure and alterations in the methods for removing the recipient oocyte spindle and chromatin have enhanced success in some situations (Akagi et al. 2011; Bui et al. 2011; Jafari et al. 2011; Kim et al. 2011; Wang et al. 2011; Whitworth et al. 2011; Terashita et al. 2012). However, while exogenous chemicals or genetic manipulations of donor cell gene expression may enhance the practical application of cloning technologies, these innovations fall short of unveiling the endogenous molecules and mechanisms that an oocyte normally employs to create an embryonic genome. Moreover, enhancing other cellular processes in the cloned embryo could also enhance cloning success.
Identifying the endogenous oocyte factors that promote early cloned embryo development thus remains a valuable objective, for devising ways to improve cloning and for understanding normal development. Obviously, understanding the process of nuclear reprogramming is of widespread interest. Recent advances in identifying exogenous nuclear factors that can promote the establishment of pluripotency in cultured cells have provided new insight into potential reprogramming mechanisms. However, some of the proteins designated as pluripotency or stemness genes are not expressed in the early embryo (Mtango et al. 2011), illustrating the importance of identifying the endogenous ooplasmic factors that regulate nuclear programming. It is equally vital to identify the ooplasmic factors that drive other crucial processes, such as energy production, macromolecular processing, cell division, cellular signaling, and maintenance of cell structure in the early embryos. Ooplasmic components that repair the cytoskeleton and cellular architecture, repair the plasma membrane, promote proper pmSCC formation, enable correct mitoses, initiate correct cell cycle transit, and suppress apoptosis may be key for cloned embryogenesis, with parallel roles in the normal embryo. Because the oxidative state of the cell can be compromised by in vitro manipulation and this can lead to long-term phenotypic change (Banrezes et al. 2011), ooplasmic factors that regulate the oxidative state in clones may also be key. With so many processes potentially contributing to cloned embryogenesis, and the fundamental interest in understanding these processes in normal embryos, there is clear value in pursuing a systematic approach to identify genes that enable successful cloned embryo development.
We reasoned that identifying endogenous factors in oocytes that drive early development in SCNT embryos would be achievable using an unbiased genome scanning approach that can capture genes contributing quantitatively to SCNT outcome. We combined a mouse recombinant inbred (RI) mapping strategy with MII and GV stage oocyte gene expression data, gene network data and molecular pathway data, and subsequent quantitative analysis of mRNA expression. Using this composite systems genetics approach, we identified three chromosomal intervals that exert genetic control over SCNT embryo preimplantation development and a fourth potential interacting interval. Within these three quantitative trait loci (QTL) we identified a number of candidate genes for which differential expression is significantly associated with SCNT embryonic progression. The strongest genetic candidates correspond to cytoskeletal scaffolding proteins. Additional genes identified in the study encode known pluripotency regulators, transcription factors, and signaling proteins. Collectively, these results provide new insight into the cellular pathways that contribute to the viability of early stage embryos.
Materials and Methods
Mice, embryo culture, and SCNT
Females of 29 BXD recombinant inbred strains were obtained from the Jackson Laboratory (Bar Harbor, Maine) at the age of 8–10 weeks. Females of the C57BL/6, DBA/2J (abbreviated as B6 and D2) and B6D2 F1 hybrid genotypes were also used. B6 stock was obtained from Harlan Sprague-Dawley. D2 stock was obtained from the Jackson Laboratory. B6D2 F1 hybrids were obtained from the National Cancer Institute. MII stage oocytes were obtained by superovulation using 5 IU equine chorionic gonadotropin (eCG) followed 48 hr later by 5 IU human chorionic gonadotropin (hCG). Cloned constructs and parthenogenetically activated embryos were created as described (Chung et al. 2006; Han et al. 2010a). Clones were prepared using B6D2 F1 cumulus cells as donors for all recipients. SCNT constructs and parthenotes were cultured in MEMα medium supplemented with sodium pyruvate and BSA in a humidified 37° incubator with 5% CO2 in air as described (Han et al. 2010a). This culture medium supports efficiently the development of SCNT embryos (superior to embryo culture formulations) and also supports B6D2 F1 parthenote development (Gao et al. 2004).
All studies were approved by the Temple University Institutional Animal Care and Use Committee, consistent with National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals, and with AAALAC accreditation.
BXD phenotype analysis
Twelve traits were analyzed using the suite of tools available at http://GeneNetwork.org (Andreux et al. 2012). We used three of the mapping functions:
Interval mapping with the permutation test option was used to identify chromosome intervals and associated genes and markers linked to BXD trait values, and which haplotypes positively and negatively affected the traits. This tool uses a rapid regression method that compares trait values to the known genotype at a marker or the probability of a genotype at given location.
Marker regression analysis was used to compute the correlation between genetic variants at loci and phenotypes, and the fraction of strain variance accounted for by each locus. The analysis uses a Pearson product momentum correlation. Data were reported as likelihood ratio statistic (LRS), which provides a measurement of the association between a trait and genetic markers; a significance threshold of P < 0.05 is used. The threshold for suggestive LRS values, indicative of potential correlations worthy of further examination, is set at P < 0.63. Analyses were performed with and without parental strain data.
Composite interval mapping was used to search for possible secondary loci that might be masked by primary loci with genome-wide significant LRS scores. This utility factors out the effect of a primary locus so that any relevant secondary loci can detected. The GeneNetwork utility uses mouse genome build 37.2.
qRT–PCR and statistical analysis
Quantitative real-time RT–PCR was performed using the Applied Biosystems (ABI, Life Technologies, Carlsbad, CA) Step One Plus system. Primers employed are described in Supporting Information, Table S1. A minimum of three pools of 15–25 oocytes were obtained for each strain. The zonae pellucidae of MII stage oocytes were removed using acidified Tyrode’s buffer (pH 2.5). Total RNA was isolated using the Picopure RNA isolation kit (Life Technologies) according to the manufacturer’s recommendation and reverse transcribed using the Superscript III kit (Life Technologies). To provide maximum reproducibility and highest quality data, the PCR analysis was performed without further library amplification. Quantitative real-time PCR was performed using TaqMan gene expression assays according to the ABI protocol. The abundance of each target gene mRNA was normalized to the endogenous Laptm4b mRNA for sample-to-sample comparisons. The relative expression ratios among different groups were obtained by the comparative CT method (Livak and Schmittgen 2001) using the Step One software v. 2.1. Pearson correlation coefficients were calculated to evaluate the relationships among the expression levels for the various candidate genes. Gene expression levels were also analyzed for a relationship to the 12 phenotypic traits and to each other.
Results
Study design
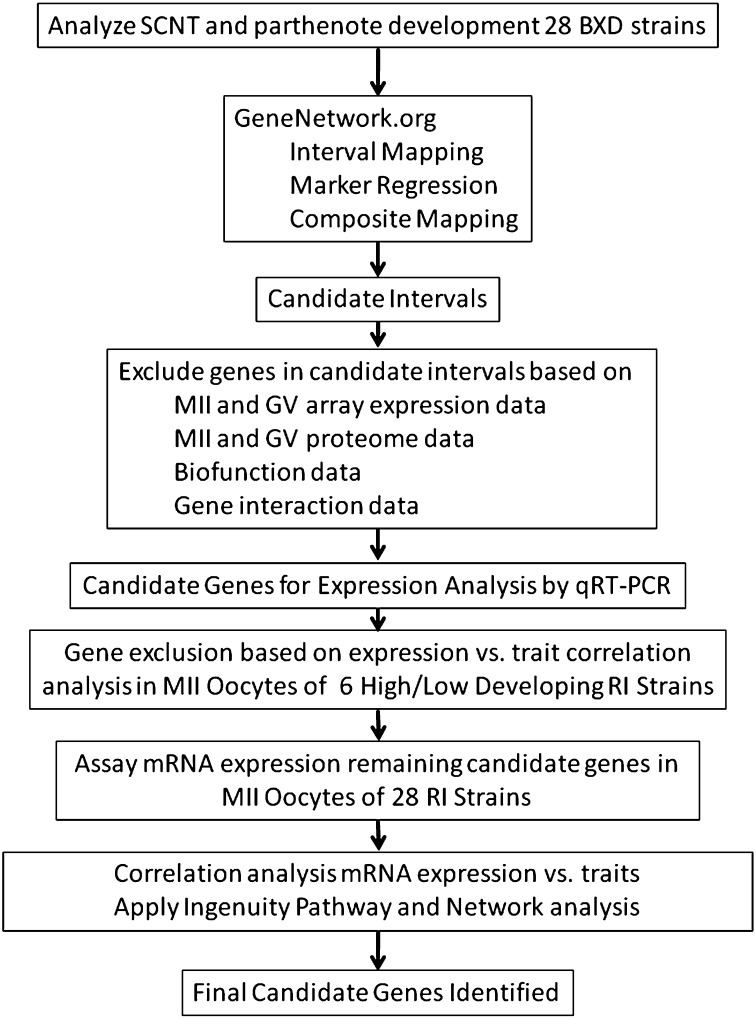
Our goal was to identify genes expressed in the oocyte that underlie successful early development in SCNT embryos. Achieving this would advance cloning biology, our understanding of oocyte biology and determinants of oocyte quality, and our understanding of early formative processes in the embryo. The unique capacity of oocytes to reprogram differentiated somatic genomes to an embryonic state implies that unique nuclear reprogramming factors may be expressed in the oocyte. Genes that meet the specific energy needs of nuclear reprogramming could be key facilitators of the process. Genes that code for cytoplasmic components that facilitate protein localization and intracellular signaling could be vital factors to successful cloned embryogenesis. Additionally, genes that support embryo health and viability, inhibit apoptosis, regulate meiotic and cell-cycle progression, and promote homeostasis could participate in determining the success of early cloned embryogenesis and could be sensitive to the dialog between donor genome and ooplasm. In view of the extensive and unique array of ooplasmic factors that could contribute to nuclear reprogramming and early cloned embryo development, we reasoned that an unbiased genome-wide approach was the only plausible means of identifying the major ooplasmic factors that control early SCNT outcome. We therefore adopted a genome scanning approach to identify QTL that contribute to successful development of SCNT constructs to the blastocyst stage. A schematic summary of our overall strategy is provided in Figure 1.
Figure 1.
Schematic summary of approach for identifying candidate genes for oocyte factors regulating early development of SCNT embryos. The pathway from phenotype analysis of 28 BXD strains, interval mapping, and then use of diverse sources of published data (Table S5) and qRT–PCR data generated in this study (Tables S6 and S7) to narrow progressively the number of candidate genes from 195 to 26 (Table 3) is shown.
The success of any genome scanning approach is determined in part by the genetic resolution offered by the genetic reference population and by the number of genotypes assayed. Mouse RI strains have been used for >25 years for identifying loci that control many phenotypic traits, and one of the most extensive families of RI mice available is the BXD family. The BXD family of RI strains encompasses ∼150 lines (∼80 available from the Jackson Laboratory and 70 available from the University of Tennessee Health Science Center) and segregate for ∼4.8 million SNPs and 500,000 insertions/deletions, copy number variants, and inversions (Wang et al. 2010b) providing a high degree of precision in mapping. Our previous studies revealed a genetic difference between B6, D2, and B6D2 F1 oocytes to support development of SCNT embryos made with B6D2 F1 cumulus cells (Gao et al. 2004). Oocytes from B6D2 F1females support superior preimplantation development and successful term development. Oocytes from B6 females also support preimplantation development but at a lower rate compared to B6D2 F1 oocytes. Oocytes from D2 females do not support efficient clone development beyond the two-cell stage. Thus, the BXD RI panel of mice provided the appropriate genetic resource for quantifying, mapping, and ultimately identifying genes that determine the ability of an oocyte to support early development in SCNT embryos.
Development was scored for cloned constructs as follows: percentage progression from cloned construct to the two-cell stage, percentage progression from two-cell to four-cell stage, percent progression from four-cell to blastocyst stage, and percentage progression from two-cell to blastocyst stage. The first three measures examine progression through particular windows of development, whereas the progression from two-cell to blastocyst stage examines the overall conversion rate from the time of embryonic genome activation to blastocyst. The transition to the two-cell stage indicates that the embryo had the capacity to form pronuclei (pseudopronuclei in SCNT embryos), replicate DNA and transit the cell cycle, and complete mitotic division successfully. Passage through the first cell cycle is controlled by maternal stores of mRNA, proteins, and other macromolecules, as gene transcription is not required (Mtango et al. 2008). The first S phase of mouse embryogenesis is required to establish the ability to transcrib the genome (Latham and Schultz 2001). The two-cell stage is notable for the occurrence of the major transcriptional activation event, degradation of much of the remaining maternal mRNA, and the transition to reliance on embryonically encoded mRNAs for development. The second S phase in the mouse embryo is required to establish the ability to regulate gene expression, as transcriptional enhancers become necessary (Latham and Schultz 2001). The successful transition from two-cell to four-cell stage indicates successful activation of the embryonic genome. A second major transition in gene expression accompanies progression through the eight-cell stage (Latham et al. 1992; Hamatani et al. 2006), and as development progresses from four-cell to morula and blastocyst stages, cells in the embryo acquire developmental bias in fate, with innermost cells being more likely to contribute to the inner cell mass and the definitive embryonic lineages, while the outer cells are more likely to contribute to the extraembryonic trophoblast lineage (Chen et al. 2010). Conversion from four-cell to blastocyst stage reflects the successful establishment within SCNT embryos of the ability to accomplish such key early developmental benchmarks. By assessing developmental progression through these different developmental windows, insight can be gained concerning genes that may contribute to these key events and which transitions particular genes may be promoting. It is noted that the effects of maternally inherited mRNAs and proteins deposited in the oocyte need not be limited to the first cell cycle. Such factors can execute early actions that have long-term secondary effects (e.g., modulating embryonic gene expression) or can persist during cleavage and exert effects at later stages.
The SCNT developmental values reveal quantitative differences in the overall success of SCNT preimplantation development. The same developmental parameters were calculated for parthenogenetic embryos. The parthenogenetic values provide insight into genetic variations affecting oocyte quality and activation and subsequent embryo cleavage. Parthenotes were chosen because they are subjected to the same chemical activation procedure and are derived from the same pools of oocytes used to generate the cloned embryos. The inclusion of parthenotes in this study also provided an opportunity to observe genetic effects on these processes aside from addressing the genetic control of SCNT outcome. Additionally, the ratios of SCNT:parthenote values were calculated for each of the four parameters. The SCNT:parthenote ratio provides potential insight into effects on processes or nucleus–ooplasmic interactions in SCNT embryos independent of those that affect parthenote development. Essentially, the parthenote data reveal a baseline effect of each genotype on early development, and the SCNT:parthenote ratio provides an indication of genetic variation in cloning outcome that is distinct from this baseline. Collectively, these calculations yielded 12 traits that were subjected to genetic analysis.
Due to cost limitations and labor inherent in determining oocyte strain effects on SCNT outcome, the number of BXD strains that could be tested was limited. We obtained females from 29 of the original BXD strains; one of these (BXD20) was later reported by the vendor to be genetically contaminated and was excluded from the study. Oocytes from the remaining 28 strains were tested for their ability to support SCNT embryo development and their ability to support parthenogenetic embryo development. We monitored development for cloned constructs and diploid parthenotes prepared with oocytes from these 28 BXD strains, from B6 and D2 strains, and from B6D2 F1 hybrids.
QTL mapping of oocyte traits related to SCNT and parthenogenetic embryo development
To identify QTL associated with cloning outcome, we tested the abilities of oocytes for each BXD strain to support developmental progression in clones in three to five separate replicates. The universal somatic cell donor nucleus was a B6D2 F1 cumulus cell. Between 138 and 307 cloned constructs were prepared for each strain, with a total of 5730 cloned constructs being assayed. An additional 2793 constructs were assayed for the B6, D2, and B6D2 F1 genotypes, for a total of >8500 cloned constructs assayed. A total of 2612 parthenotes were tested for the BXD strains and 472 for the B6, D2, and B6D2 F1 genotypes.
For SCNT embryos, most BXD strains supported development to the two-cell stage, some even exceeding the value for B6D2 F1 oocytes, but some strains supported two-cell development very poorly (BXD12, BXD33, BXD42) (Table 1). Progression from two-cell to four-cell stages was >50% for eight strains, between 20 and 50% for nine strains, and < 20% for 11 strains. Progression from the two-cell stage to blastocyst stage was >20% for only two strains (BXD11 and BXD27), which is comparable to rates of 20.9 and 27.4% for the B6 and B6D2 F1 genotypes. This suggests that multiple loci are likely to control this overall transition from first cleavage to blastocyst formation. For progression from the four-cell stage to blastocyst stage, seven strains supported development at >30%, comparable to 37.9 and 36.3% for B6 and B6D2 F1 oocytes, indicating that a small number of loci may control the four-cell to blastocyst transition.
Table 1. Percentage developmental progression of cloned and parthenogenetic embryos prepared with eggs from different strains.
|
BXD Strain | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SCNT | 1 | 2 | 5 | 6 | 8 | 9 | 11 | 12 | 13 | 14 | 15 | 16 | 18 | 19 | 21 | 22 |
| Two cell/construct | 59.6 | 75.4 | 77.4 | 38.2 | 80.7 | 97.7 | 79.8 | 11.6 | 57.1 | 71.2 | 80.6 | 82 | 66 | 84.4 | 85.2 | 77.4 |
| Four cell/two cell | 0 | 20.5 | 52.3 | 1.2 | 28.1 | 15 | 64.2 | 15.3 | 53.9 | 17.4 | 4 | 15.2 | 42.1 | 49.7 | 14.7 | 54.5 |
| Blastocyst/two cell | 0 | 7.6 | 11.9 | 1.2 | 6.8 | 4.5 | 27.5 | 0 | 16.5 | 5.7 | 0 | 0 | 10.4 | 4.6 | 4.5 | 11 |
| Blastocyst/four cell | 0 | 29.5 | 17.9 | 25 | 16.8 | 31.9 | 43 | 0 | 34.5 | 27.3 | 0 | 0 | 25.4 | 9.1 | 31.3 | 19.3 |
| 27 | 28 | 29 | 31 | 32 | 33 | 34 | 36 | 38 | 39 | 40 | 42 | B6 | D2 | F1 | ||
| Two cell/construct | 51 | 82.1 | 78.5 | 43.8 | 70.1 | 15.6 | 84.8 | 86.3 | 56.9 | 72.4 | 93.5 | 23.1 | 68.3 | 64.7 | 87.3 | |
| Four cell/two cell | 59 | 29.8 | 10.7 | 7.3 | 50.6 | 0 | 48.9 | 37.2 | 45.8 | 3 | 66 | 39.2 | 53.6 | 43 | 66 | |
| Blastocyst/two cell | 23.7 | 8.7 | 0.9 | 5.2 | 8 | 0 | 13.9 | 17.5 | 4.3 | 1.2 | 16.1 | 7 | 20.9 | 0 | 27.4 | |
| Blastocyst/four cell | 37.9 | 32.9 | 5.8 | 55.6 | 21.5 | 0 | 26.6 | 54.4 | 6.9 | 27.8 | 24.2 | 10.7 | 37.9 | 0 | 36.3 | |
| Parthenote | 1 | 2 | 5 | 6 | 8 | 9 | 11 | 12 | 13 | 14 | 15 | 16 | 18 | 19 | 21 | 22 |
| Two cell/activated | 85.4 | 97.4 | 91.9 | 98.2 | 91.7 | 97.7 | 95.7 | 22.7 | 97.4 | 78.4 | 97.5 | 99.2 | 80.2 | 100 | 98.6 | 95.7 |
| Four cell/two cell | 5.3 | 100 | 59.3 | 83.1 | 52.9 | 86.4 | 95 | 79.5 | 77.8 | 55 | 49.2 | 9.4 | 73.6 | 81.4 | 79.3 | 78 |
| Blastocyst/two cell | 0 | 91.5 | 5.6 | 0 | 2.5 | 38.8 | 83.6 | 7.3 | 38.7 | 11.7 | 33.7 | 0 | 40.9 | 18.8 | 57.5 | 12.6 |
| Blastocyst/four cell | 0 | 91.5 | 7.9 | 6.1 | 3.6 | 44.6 | 88.8 | 7.5 | 46.9 | 16.7 | 62.8 | 0 | 54.8 | 23.4 | 72 | 16.4 |
| 27 | 28 | 29 | 31 | 32 | 33 | 34 | 36 | 38 | 39 | 40 | 42 | B6 | D2 | F1 | ||
| Two cell/construct | 66.7 | 97.5 | 91.3 | 95.1 | 94.3 | 19.4 | 100 | 90.1 | 66.3 | 84.4 | 95.7 | 39.6 | 84 | 72.6 | 100 | |
| Four cell/two cell | 100 | 97.1 | 31.7 | 85.2 | 61.3 | 46.7 | 89.8 | 60.1 | 13.5 | 84.6 | 100 | 80 | 100 | 59 | 99.6 | |
| Blastocyst/two cell | 77.1 | 64.7 | 3.3 | 39.9 | 22.5 | 12.5 | 48.6 | 14.7 | 3.8 | 61.5 | 74.5 | 8.3 | 36.8 | 2.1 | 72.2 | |
| Blastocyst/four cell | 77.1 | 66.4 | 6.7 | 46.7 | 39.5 | 17 | 54.7 | 26.3 | 35 | 73.8 | 74.5 | 8.3 | 36.8 | 2.5 | 72.6 | |
| SCNT:parthenote | 1 | 2 | 5 | 6 | 8 | 9 | 11 | 12 | 13 | 14 | 15 | 16 | 18 | 19 | 21 | 22 |
| Two cell/construct | 69.8 | 77.5 | 84.2 | 38.9 | 88 | 100 | 83.4 | 50.9 | 58.7 | 90.8 | 82.6 | 82.7 | 82.3 | 84.4 | 86.4 | 80.9 |
| Four cell/two cell | 0 | 20.5 | 88.2 | 1.4 | 53.2 | 17.4 | 67.5 | 19.2 | 69.3 | 31.7 | 8 | 163 | 57.2 | 61.1 | 18.6 | 69.9 |
| Blastocyst/two cell | 0 | 8.3 | 215 | 0 | 273 | 11.5 | 32.9 | 0 | 42.7 | 48.9 | 0 | 0 | 25.4 | 24.7 | 7.9 | 87.7 |
| Blastocyst/four cell | 0 | 32.3 | 227 | 408 | 469 | 71.4 | 48.4 | 0 | 73.5 | 164 | 0 | 0 | 46.2 | 39.1 | 43.4 | 118 |
| 27 | 28 | 29 | 31 | 32 | 33 | 34 | 36 | 38 | 39 | 40 | 42 | B6 | D2 | F1 | ||
| Two cell/construct | 76.5 | 84.2 | 86.1 | 46.1 | 74.3 | 80.3 | 84.8 | 95.7 | 85.8 | 85.9 | 97.7 | 58.4 | 81.3 | 89 | 87.3 | |
| Four cell/two cell | 59 | 30.7 | 33.7 | 8.6 | 82.5 | 0 | 54.4 | 61.9 | 339 | 3.6 | 66 | 49.1 | 53.6 | 72.9 | 66.3 | |
| Blastocyst/two cell | 30.7 | 13.4 | 28.9 | 13 | 35.7 | 0 | 28.6 | 119 | 115 | 2 | 21.6 | 84.5 | 56.8 | 0 | 37.9 | |
| Blastocyst/four cell | 49.2 | 49.6 | 87.7 | 119 | 54.5 | 0 | 48.7 | 207 | 19.8 | 37.7 | 32.5 | 129 | 103 | 0 | 49.9 | |
Values show the percentage progression from SCNT construct to two-cell stage, from two-cell to four-cell stage, from two-cell stage to blastocyst stage, and from four-cell stage to blastocyst. Development was scored for SCNT embryos and for parthenotes made with oocytes from the 28 strains. The ratio of SCNT:parthenote development was also calculated for each strain.
Parthenogenetic development proceeded for most strains, but some strains displayed severely reduced rates of activation and progression to the two-cell stage (BXD12, BXD33, BXD42) and/or reduced development beyond the two-cell or four-cell stages (Table 1). The D2 strain displayed efficient parthenogenetic development to the two-cell stage but restricted development thereafter. These data thus reveal significant genetic effects on oocyte activation and cleavage that are likely independent of the SCNT procedure, but that nevertheless contribute to the overall outcome of the procedure.
The data from Table \1 were subjected to interval mapping and marker regression analysis using the tools at GeneNetwork.org. Analysis of the results for the four SCNT embryo development traits yielded a major controlling locus on chromosome (chr) 17, and additional loci on chr 1 and 4 (Table 2). The region on chr 17 (map position 69.1492–71.10156 Mb) was returned by trait analysis with a significant LRS value for both the two-cell to four-cell conversion and the two-cell to blastocyst conversion, when the BXD data were analyzed without including parental strain data, and displayed a suggestive LRS value with parental strain data included. A second region with a significant LRS value was seen on chr 4 (map position 13.03102–13.76499 Mb) for the two-cell to four-cell conversion trait without parental strain data and displayed a suggestive LRS value when parental strain data were included. For both the chr 4 and chr 17 regions, a D2 haplotype was positively associated with SCNT development. A third region showing a significant LRS value, seen with the transition from the two-cell to blastocyst stage, was observed for chr 1 (map position 40.99094–42.21652 Mb) when parental strain data were included and was just below the significance threshold (but retained a suggestive LRS value) without parental strain data included. For the chr 1 region, a B6 haplotype was positively associated with SCNT embryo development. Each of these regions was flanked by additional markers that displayed suggestive LRS values.
Table 2. Marker regression analysis and interval mapping of cloned embryo development.
| With parentsFour cell/two cell |
Without parentsFour cell/two cell |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Chr | Mb | Mb | Haplotype incr. trait | LRS | Chr | Mb | Mb | Haplotype incr. trait | LRS |
| 1 | 40.990937 | 42.216521 | B6 | 10.723 | 4 | 6.820702 | 11.507152 | D2 | 12.98 |
| 4 | 6.820702 | 11.507152 | D2 | 11.606 | 4 | 13.031015 | 13.764991 | D2 | 19.826* |
| 4 | 13.031015 | 13.764991 | D2 | 17.548 | 17 | 66.785539 | 68.482569 | D2 | 13.914 |
| 17 | 66.785539 | 68.482569 | D2 | 11.486 | 17 | 69.149197 | 71.101564 | D2 | 20.652* |
| 17 | 69.149197 | 71.101564 | D2 | 17.121 | 17 | 71.350424 | 72.001676 | D2 | 13.914 |
| 17 | 71.350424 | 72.001676 | D2 | 11.486 | 17 | 76.561294 | D2 | 11.731 | |
| Suggestive LRS | 10.35 | Suggestive LRS | 11.03 | ||||||
| Significant | 18.29 | Significant | 19.49 | ||||||
| Highly significant | 22.66 | Highly significant | 24.69 | ||||||
| Blastocyst/two cell | Blastocyst/two cell | ||||||||
| Chr | Mb | Mb | Haplotype incr. trait | LRS | Chr | Mb | Mb | Haplotype incr. trait | LRS |
| 1 | 29.231425 | 30.978257 | B6 | 13.63 | 1 | 29.231425 | 30.978257 | B6 | 11.149 |
| 1 | 38.079087 | 39.05313 | B6 | 12.315 | 1 | 39.578771 | B6 | 10.027 | |
| 1 | 39.578771 | B6 | 12.583 | 1 | 40.990937 | 42.216521 | B6 | 15.158 | |
| 1 | 40.990937 | 42.216521 | B6 | 17.992* | 4 | 13.031015 | 13.764991 | D2 | 13.282 |
| 9 | 59.835172 | 59.934576 | B6 | 10.33 | 17 | 66.785539 | 68.482569 | D2 | 13.946 |
| 9 | 72.030418 | 72.952358 | B6 | 11.781 | 17 | 69.149197 | 71.101564 | D2 | 18.806* |
| 9 | 73.368885 | 76.983761 | B6 | 10.331 | 17 | 71.350424 | 72.001676 | D2 | 13.946 |
| 9 | 77.217283 | 79.902981 | B6 | 11.725 | |||||
| 9 | 79.991491 | 81.159484 | B6 | 10.061 | |||||
| 17 | 69.149197 | 71.101564 | D2 | 9.997 | |||||
| Suggestive LRS | 9.87 | Suggestive LRS | 9.94 | ||||||
| Significant | 16.2 | Significant | 16.02 | ||||||
| Highly significant | 19.65 | Highly significant | 19.51 | ||||||
| Blastocyst/two cell | Blastocyst/two cell | ||||||||
| Chr | Mb | Mb | Haplotype Incr. Trait | LRS | Chr | Mb | Mb | Haplotype Incr. Trait | LRS |
| 1 | 29.231425 | 30.978257 | B6 | 11.227 | 1 | 40.990937 | 42.216521 | B6 | 13.428 |
| 1 | 40.990937 | 42.216521 | B6 | 16.274 | 9 | 69.810185 | 70.880253 | B6 | 11.217 |
| 9 | 68.191194 | 69.455899 | B6 | 12.427 | 9 | 71.331037 | 71.798886 | B6 | 10.603 |
| 9 | 69.810185 | 70.880253 | B6 | 13.589 | 9 | 72.030418 | 72.952358 | B6 | 10.433 |
| 9 | 71.331037 | 71.798886 | B6 | 12.973 | 9 | 73.368885 | 76.983761 | B6 | 11.109 |
| 9 | 72.030418 | 72.952358 | B6 | 12.739 | 9 | 79.991491 | 81.159484 | B6 | 11.851 |
| 9 | 73.368885 | 76.983761 | B6 | 13.526 | |||||
| 9 | 77.217283 | 79.902981 | B6 | 12.255 | |||||
| 9 | 79.991491 | 81.159484 | B6 | 14.437 | |||||
| Suggestive LRS | 10.23 | Suggestive LRS | 10.42 | ||||||
| Significant | 16.66 | Significant | 16.96 | ||||||
| Highly Significant | 20.13 | Highly Significant | 19.67 | ||||||
, significant correlation of region with trait.
The above three regions on chr 1, 4, and 17 were the only regions to achieve statistical significance in any of the 12 traits assayed. An additional region on chr 9 displayed suggestive LRS values for three of the SCNT traits and also displayed suggestive LRS values for parthenogenetic development from two- or four-cell stage to blastocyst, accompanied by a region on distal chr 13 (Table S2). Additional suggestive LRS values were obtained for other regions on chr 6, 8, 13, and 16 when analyzing the SCNT:parthenote ratios (Table S3). To test whether any of these regions displayed significant interactions with the regions on chr 1, 4, and 17, we applied composite interval mapping to the SCNT trait data with or without parental strain data included (Table S4). Without parental strain data included, we identified two small regions on chr 6 that yielded suggestive LRS values for interactions with markers in the chr 17 region when examining the two-cell to blastocyst conversion trait without parental strain data. With parental strain data included, the same trait yielded markers on chr 7 and 11 with suggestive LRS values for interactions with markers in the chr 1 region.
Overall, these data indicate that there is a primary QTL on mouse chr 17 that is correlated with multiple trait measures of preimplantation SCNT developmental progression, that additional intervals on chr 1 and 4 display significant LRS values for associations with SCNT development, and that other interacting loci (particularly the ones on chr 6) may also exist. Additional genotypes would need to be tested to provide a robust test of the relevance of these other loci to SCNT embryo development; hence these loci were not pursued further in this study.
It is noted that the BXD strains and the B6 and B6D2 F1 mice employed in these analyses encompass different B6 substrains; BXD mice are based on B6J substrain while Harlan B6 and NCI B6D2 F1 mice are based on the B6N substrain. However, the LRS intervals on chr 4 and 17 were identified as significant using the BXD strain data without the parental data and were scored as suggestive with the parental data added, while the chr 1 locus was significant with parental data and scored as suggestive without the parental data included. Hence, the identification of these three candidate intervals was unlikely to be affected by minor genetic variation between the B6N and B6J substrains. It is also noted that, while genetic polymorphisms exist between these substrains and can affect phenotype, the two substrains are quite close genetically. One study reported just 11 of 1446 SNPs variant between B6J and B6N (Mekada et al. 2009), and another reported just 12 of 1449 SNPs as variant (Zurita et al. 2011). Another study reported the two substrains differ in 12 of 342 microsatellite markers surveyed (Bothe et al. 2004). Genome sequencing revealed just 150 SNPs between the two substrains (Bryant 2011) and commercial vendors advertise 95–128 diagnostic SNP panels for discriminating B6 substrains. These studies also reveal that the different B6 substrains are closely related to each other. By contrast, the B6 and D2 strains are genetically distant from one another (Taylor 1972). We observed little difference in mRNA expression ratios for three mRNAs selected to compare results for oocytes from females obtained from the Jackson Laboratory and Harlan Sprague-Dawley (data not shown). The dramatic difference between B6 and D2 inbred genotypes in cloning outcome would make an effect of substrain genetic background on the QTL mapping result unlikely, accounting for the emergence of the candidate loci with and without parental strain data included in the analysis.
Initial evaluation of candidate genes
The combined three significant LRS regions and the flanking suggestive regions collectively encompassed 195 genes. Our next goal was to identify a subset of these genes to be examined in detail for association with oocyte cloning phenotype. Using a systems genetics approach, QTL mapping can be combined with other data (gene expression, polymorphisms, gene ontology) to achieve an identification of specific genes or combinations of genes that may contribute to a specific phenotype, in this case the ability of the oocyte to support early SCNT embryo development. We incorporated available transcriptome profiling data for B6, D2, and B6D2 F1 hybrid MII and germinal vesicle stage oocytes, as well as knowledge of biological function and cellular compartment localization and oocyte proteome data (Wang et al. 2010a), to narrow the lists of candidate genes for further study. Candidate genes were further scrutinized on the basis of sequential qRT–PCR expression analyses. The combination of these additional data for individual gene characteristics narrowed markedly the array of candidate genes. The presence and numbers of SNPs upstream of the 5′-UTR and within protein-coding regions were also noted, as polymorphisms within the coding regions could affect a trait independent of gene expression level. From the three candidate intervals defined by the significant LRS values and flanking suggestive LRS value intervals, we identified 36 genes for detailed study (Table 3).
Table 3. Number of genes evaluated and included in further detailed study by chromosome region.
| Chr | Region and LRS significance | No. genes | No. tested on 6 BXD strains | No. tested on 28 BXD strains |
|---|---|---|---|---|
| 1 | Significant | 1 | 0 | 0 |
| Suggestive | 47 | 11 | 9 | |
| 4 | Significant | 3 | 1 | 1 |
| Suggestive | 57 | 10 | 7 | |
| 17 | Significant | 13 | 3 | 3 |
| Suggestive | 74 | 11 | 6 | |
| Total | 195 | 36 | 26 |
A detailed summary of the gene characteristics and expression data supporting our focus on this set of 36 genes is given in Table S5. Because the http://GeneNetwork.org application uses genome build 37.2, Table S5 incorporates map locations for this build as well as build 38; assignment to intervals with significant LRS values was based on build 37.2 positions, for consistency with the http://GeneNetwork.org application. We first characterized genes on the basis of detection and maximum raw intensity values observed with Affymetrix MOE430v2 arrays corresponding to MII stage oocytes from B6, D2, and B6D2 F1 strains (note that these arrays employed mice from the Jackson laboratory). The array data set (unpublished) incorporated four replicate arrays for each strain for the MII and germinal vesicle stages. Array data displayed robust quality control parameters consistent with previously reported arrays for oocyte samples from mouse and other species (Pan et al. 2005; Lee et al. 2008). Genes with no detected expression on the MII array were excluded from further consideration. The remaining genes were evaluated as candidates for further study on the basis of maximum array average intensity values, array fold changes between strains, qRT–PCR expression ratios between B6, D2, and B6D2 F1 oocytes, biological functions, and cellular compartmentalization. Genes meeting a combination of at least 500 maximum raw intensity value on array, at least twofold change on array, and at least 10% change in qRT–PCR among oocyte samples for B6, D2, and B6D2F1 genotypes were included for further study. For genes showing weak or no expression in MII stage oocytes, germinal vesicle stage arrays were consulted to account for possible protein contributions to MII oocyte phenotypes, where the mRNAs might be degraded during oocyte maturation. Pseudogenes were excluded. Absence of functional annotation was also a criterion for excluding genes lying outside of the intervals with significant LRS values. The miRNA genes were excluded due to the limited role of miRNAs in oocytes and embryos (Svoboda 2010). Genes encoding noncoding RNAs outside of the significant LRS intervals and with no probe coverage on the array were not examined. Genes encoding secreted proteins, components of the extracellular matrix, mitochondrial proteins, cell adhesion proteins, and DNA repair proteins were deemed of lower priority. Germinal vesicle and MII oocyte proteome data were also examined to identify genes with expression at the protein level. Most genes excluded from detailed study were excluded on the basis of a combination of criteria.
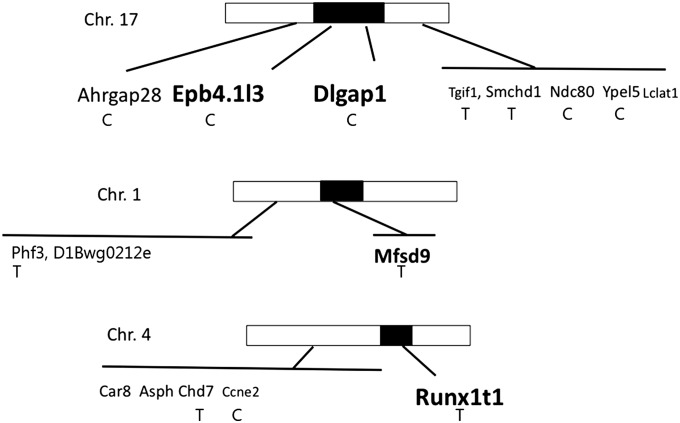
Highest priority for further study was assigned to genes in the regions with significant LRS values. The chr 1 region encompassed one pseudogene. This region was flanked by another pseudogene and one noncoding RNA gene for which expression was not detected in oocytes by qRT–PCR. Additional chr 1 genes centromeric to this region were included in the analysis, as described below. The chr 4 region encompassed two pseudogenes and a single protein-coding gene, Runx1t1. The chr 17 region encompassed 13 genes. Four of these were pseudogenes, one was a noncoding RNA poorly expressed on the array, and three were uncharacterized sequences or characterized as predicted protein coding gene. Two genes were excluded on the basis of a weak signal and small expression differences on arrays. The remaining chr 17 genes included in the study were Epb4.1l3, Zfp161, and Dlgap1 (Table 3 and Table S5).
Other genes within the flanking suggestive intervals on chr 1, 4, and 17 were evaluated for further study. For the chr 1 region, an immediately flanking noncoding RNA (4930448I06Rik) with no reported expression in oocytes or embryos was examined by qRT–PCR with no detected expression and was therefore excluded. Eleven other genes centromeric to the significance interval were examined, including genes immediately adjacent to the noncoding RNA gene. For the chr 4 interval, an adjacent long noncoding RNA gene (Gm11818) was examined, but excluded due to little expression difference between B6, D2, and B6D2 F1 oocytes by qRT–PCR. Nine additional candidates were selected from this flanking suggestive interval. For the chr 17 flanking suggestive intervals, 11 genes were selected for detailed study. Two genes (Myl12a and Myl12b) related to myosin contractility and cellular shape change were not included in the initial analysis due to uncertain functional relevance and limited difference in expression on arrays; one of these (Myl12a) was subsequently tested on the B6, D2, and B6D2 F1 genotypes by qRT–PCR (Table S5) but was not subjected to full analysis. Some genes located in the suggestive chr 17 intervals were excluded from further consideration because of reported cell type-specific expression and functions. One additional gene (Memo1) on chr 17 related to cell motility was excluded on the basis of its reported function. Taking into account the available array data, and the other data summarized in Table S5, including data obtained by qRT–PCR for expression B6, D2, and B6D2 F1 oocytes, we incorporated an additional 32 genes from these chr 1, 4, and 17 suggestive intervals, for a total of 36 genes studied in detail (Table 3 and Table S5, green highlight in column 1).
These 36 genes were first tested for differences in expression between three BXD strains that displayed maximum developmental outcomes (BXD11, BXD22, and BXD40) and three that displayed minimum developmental outcomes (BXD1, BXD16, and BXD29) for cloned embryo preimplantation development. Limiting our initial analysis to genotypes at the two extremes of phenotype distribution minimized the costs associated with collecting sufficient numbers of MII stage oocytes needed for qRT–PCR without benefit of mRNA amplification. Of these 36 genes tested (Table S6), 10 were excluded from further study because they showed either no or limited correlations with the four SCNT traits monitored. Four of the remaining 26 genes failed to display a strong correlation with the phenotypes of these six BXD strains, but were retained for further study because they encode biological functions deemed of high potential relevance or because they lie within or very near the intervals displaying significant LRS values. This yielded a final list of 26 genes for analysis on the entire set of 28 BXD strains used in our analysis (Table 3).
Single gene expression correlations with SCNT developmental traits for Chr 1, 4, 17 regions
To identify those genes most likely to contribute to cloning outcome, our next goal was to identify from among this list of 26 candidates, the genes that manifested significant correlations between expression and phenotype. We evaluated correlations between gene expression and the SCNT embryo and SCNT:parthenote developmental phenotypes for the 28 BXD lines (Table S7). We initially excluded B6, D2, and B6D2 F1 genotype expression data, and then examined the effects of including these genotypes. We also compared associations with SCNT traits alone and associations seen in the SCNT:parthenote ratios. This provided a means of detecting relationships that were specific to SCNT development. Enhancement or acquisition of a significant association revealed in the SCNT:parthenote values relative to SCNT trait values indicates a partial component of the SCNT phenotype that is specific to SCNT biology. Disappearance of an association indicates a predominant effect that is not specific to SCNT biology. An association seen in both SCNT and SCNT:parthenote traits indicates an association with a large SCNT-specific component. Because such associations are quantitative and reflect additive components, effects related specifically to SCNT development could be seen even without significant associations in parthenotes.
The chr 1 interval displaying a significant LRS value encompasses one pseudogene and is adjacent to another pseudogene and a noncoding RNA gene (4930448I06Rik, no expression detected). These genes were excluded from further consideration. Immediately centromeric to this region, however, are two genes, Mfsd9 and Tmem182. The Tmem182 gene, which lies immediately adjacent to the significance interval, displayed highest expression in B6 oocytes (Table 4). The expression of Tmem182 was strongly and negatively correlated with SCNT embryo development for three of the SCNT traits scored (two-cell conversion to four-cell and blastocyst, and four-cell conversion to blastocyst) (Table 4 and Table S7), which is at odds with the positive effect of the B6 haplotype in this region with outcome. With B6, D2, and B6D2 F1 genotypes included, the Tmem182 association retained significance only for the conversion from four cell to blastocyst. No associations were seen for SCNT:parthenote developmental ratios, indicating that the associations seen with SCNT embryos were largely not specific for SCNT development. The Mfsd9 mRNA was also expressed more highly in B6 than D2 oocytes, but to a lesser degree. Mfsd9 expression displayed a single significant negative correlation with SCNT embryo progression from the four-cell stage to blastocyst. With B6, D2, and B6D2 F1 genotypes included this association fell below the level of significance. With the SCNT:parthenote ratio, however, Mfsd9 gene expression displayed a strong positive association with development from the two-cell to four-cell stage and then strong negative associations with development from the two-cell and four-cell stages to blastocyst, both with and without B6, D2, and B6D2 F1 genotypes included. Thus, of these two genes, only Mfsd9 displayed a positive association between expression and any developmental trait measured, consistent with the positive effect of the B6 haplotype, and this was for early progression to the four-cell stage and specific to cloned embryo development. This effect seems to overcome a negative effect seen for parthenotes. The later negative SCNT-specific association, however, indicates stage specificity to the SCNT-specific component.
Table 4. Summary of correlations of gene expression and traits assayed using the panel of 28 BXD strains.
| SCNT |
SCNT:Parthenote |
Parthenotes |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Chr | 2C/ 1C | 4C/ 2C | BL/ 2C | BL/ 4C | 2C/ 1C | 4C/ 2C | BL/ 2C | BL/ 4C | 2C/ 1C | 4C/ 2C | BL/ 2C | BL/ 4C | MII Expr. RatioB6:D2:F1 |
| Genes in or near significant LRS intervals | ||||||||||||||
| Mfsd9 | 1 | (\) | X | (X) | (X) | (\) | 1:0.71:0.97 | |||||||
| Tmem182 | 1 | (\) | (\) | (X) | (X) | (X) | (X) | 1:0.32:0.49 | ||||||
| Runx1t1 | 4 | (\)* | / | (X) | 1:1.09:1.37 | |||||||||
| Arhgap28 | 17 | (X) | (X) | (X) | (X) | (\) | 1:0.07:0.77 | |||||||
| Epb4.1l3 | 17 | X | X | (X) | (X) | 1:2.6:1.98 | ||||||||
| Dlgap1 | 17 | (\) | /,(\) | (\) | / | 1:0.15:0.61 | ||||||||
| Tgif1 | 17 | (X) | \ | X | (X) | (X) | (X) | 1:1.28:0.92 | ||||||
| Other genes in suggestive LRS intervals | ||||||||||||||
| Phf3 | 1 | (/) | (X) | X | X | 1:0.95:0.99 | ||||||||
| Txndc9 | 1 | (X) | (X) | (X) | X | (X) | (X) | (X) | 1:5.23:3.12 | |||||
| Eif5b | 1 | (X) | (X) | (X) | (\) | (X) | (X) | (X) | 1:1.23:1.25 | |||||
| Aff3 | 1 | (X) | (X) | (\) | 1:1.7:1.7 | |||||||||
| Pdcl3 | 1 | \ | \ | (\) | X | \ | 1:0.69:1.01 | |||||||
| Tbc1d8 | 1 | (X) | 1:1.11:1.08 | |||||||||||
| D1Bwg0212e | 1 | (\) | (X) | X | (X) | (X) | (X) | 1:0.82:0.89 | ||||||
| Car8 | 4 | (X) | (X) | (X) | X | (\) | 1:0.14:0.61 | |||||||
| Rab2a | 4 | / | / | 1:0.87:1.01 | ||||||||||
| Chd7 | 4 | / | / | (X) | (X) | 1:1.09:1.39 | ||||||||
| Asph | 4 | X | X | X | X | (X) | (X) | (\) | 1:1.26:1.35 | |||||
| Ccne2 | 4 | (X) | (X) | X | X | (X) | (X) | (X) | (/) | 1:2.36:1.37 | ||||
| Dpy19l4 | 4 | / | 1:1.15:1.02 | |||||||||||
| Esrp1 | 4 | / | / | / | X | \ | 1:0.86:0.89 | |||||||
| Smchd1 | 17 | / | (X) | (X) | (X) | (\) | (\) | 1:0.91:1.54 | ||||||
| Ndc80 | 17 | X | (X) | (X) | (X) | (X) | X | (X) | 1:0.23:0.57 | |||||
| Ypel5 | 17 | X | X | X | X | 1:1.39:1.26 | ||||||||
| Lclat1 | 17 | (/) | (X) | X | X | (X) | (X) | (X) | (X) | 1:1.51:1.15 | ||||
| Spast | 17 | (X) | (X) | (\) | / | 1:0.69:1.21 | ||||||||
/, significant association with trait with B6, D2 and B6D2F1 data included; \, significant association with trait without B6, D2 and B6D2F1 data included; X, significant association with trait with and without B6, D2, and B6D2F1 data included; ( ), negative correlation of expression with developmental trait; *, marginal significance with parental data included.
The chr 4 interval having a significant LRS value contains a single gene, Runx1t1. There was not a strong difference in expression between B6 and D2 oocytes, and the mRNA was only modestly elevated in F1 oocytes (Table 4). Runx1t1 expression displayed a significant negative association with progression from the four-cell to blastocyst stage for SCNT and SCNT:parthenote traits without B6, D2, and B6D2 F1 genotypes included (Table 4 and Table S7). With B6, D2, and B6D2 F1 genotypes included, Runx1t1 expression displayed a marginally significant negative association with SCNT embryo development from the four-cell to blastocyst stage (P = 0.0639), a significant positive association with the SCNT:parthenote two-cell to four-cell development and a negative association with SCNT:parthenote four-cell to blastocyst development. Thus, there was an enhancement of associations in the SCNT:parthenote traits, indicating a significant negative association specific to cloning.
The chr 17 interval with a significant LRS value encompasses two genes analyzed on all 28 strains (Epb4.1l3 and Dlgap1); a third gene (Zfp161) failed to show significant associations with the first 6 BXD lines and so was excluded from further study (Table S6). This region is flanked by an immediately distal gene (Tgif1) and a more centromeric gene (Arhgap28), both of which were analyzed. The Epb4.1l3 gene displayed a much higher level of expression in D2 oocytes than either B6 or B6D2 F1 oocytes, and B6D2 F1 oocytes expressed this mRNA nearly twice as highly as B6 oocytes. The Dlgap1 and Arhgap28 mRNAs were greatly reduced in D2 oocytes and more modestly reduced in B6D2 F1 oocytes. The Epb4.1l3 gene displayed significant positive associations of expression with SCNT development from two-cell stage to four-cell and blastocyst stages with and without B6, D2, and B6D2 F1 genotypes included (Table 4 and Table S7), consistent with the elevated expression in D2 oocytes and the positive effect of the D2 haplotype on SCNT traits (Table 2). Despite the positive association with SCNT traits, a negative association was seen between Epb4.1l3 and the SCNT:parthenote four-cell to blastocyst development, with or without B6, D2, and B6D2 F1 genotypes included. This was not seen in the SCNT traits, and thus was a specific component related to cloned embryo biology. The Dlgap1 gene displayed several negative expression associations with SCNT two-cell to four-cell and blastocyst development. These were not apparent for the SCNT:parthenote traits, but a significant negative association was seen for SCNT:parthenote two-cell to blastocyst conversion without B6, D2, and B6D2 F1 genotypes included. These associations were consistent with lower expression in D2 oocytes and the positive effect of D2 haplotype on development. Dlgap1 displayed a positive association between expression and SCNT two-cell to blastocyst conversion with B6, D2, and B6D2 F1 genotypes included, but this was also absent with the SCNT:parthenote traits. Arhgap28 displayed strong, significant negative associations of expression with the SCNT two-cell conversion to four-cell and blastocyst and four-cell to blastocyst conversion both with and without B6, D2, and B6D2 F1 genotypes included. This was also evident in the two-cell to four-cell SCNT:parthenote developmental values with and without B6, D2, and B6D2 F1 genotypes, indicating that this association may be largely specific to cloned embryos. This association is consistent with the low expression in D2 oocytes and the positive effect of the D2 haplotype for this region (Table 4 and Table S7). Tgif1 expression displayed a significant negative association of expression with SCNT four-cell to blastocyst conversion and a positive association with SCNT:parthenote two-cell to four-cell conversion rate, both with and without B6, D2, and B6D2 F1 genotypes included, and a positive association with the SCNT:parthenote zygote to two-cell conversion rate indicating a SCNT-specific relationship early during cleavage (Table 4 and Table S7). Thus, of the genes within or near the chr 17 significance interval, Epb4.1l3, Dlgap1, and Arhgap28 display the most extensive relationships between expression and developmental outcomes.
In addition to genes within the intervals with significant LRS values (and those immediately flanking the significance region on chr 1), some genes within the flanking suggestive intervals displayed significant associations of expression with one or more of the SCNT and SCNT:parthenote traits (Table 4 and Table S7). Most prominent among these were chr 17 genes Ndc80 and Smchd1, and the transcriptionally coupled chr 1 genes Txndc9 and Eif5b, each negatively correlated with three to four SCNT traits, with and without B6, D2, and B6D2 F1 genotypes included.
The negative associations seen for Txndc9 and Eif5b are consistent with higher expression in D2 oocytes but positive effects of the B6 haplotype at this region. Txndc9 and Eif5b were generally coregulated as expected for genes that share regulatory elements, having strong associations of expression with three of the SCNT traits, but for the SCNT:parthenote traits these associations were not apparent, except for Txndc9 for the four-cell to blastocyst conversion and Eif5b for the two-cell to four-cell conversion. The associations between expression and development for these two genes were thus not specific to cloned embryogenesis.
The negative association seen for chr 17 gene Ndc80 expression is consistent with the lower expression level in D2 oocytes and the positive effect of the D2 haplotype at this region. Conservation of this relationship in the SCNT:parthenote two-cell to blastocyst conversion trait indicates a specific component of cloned embryo development. A similar agreement is not as apparent for Smchd1, for which expression was only slightly different between strains. A significant negative association of Smchd1 expression was seen for the SCNT:parthenote two-cell to four-cell conversion trait without B6, D2, and B6D2 F1 genotypes included. Although parthenote development from two-cell to four-cell stage also displayed this association, its persistence at the level of SCNT:parthenote ratio indicates a specific negative contribution to clone development.
The chr 1 gene D1Bwg0212e (also known as C2orf29) displayed a significant negative association of expression with SCNT two-cell to blastocyst conversion without B6, D2, and B6D2 F1 genotypes included and with SCNT four-cell to blastocyst conversion, both with and without B6, D2, and B6D2 F1 genotypes included. A positive association of expression was acquired, however, for SCNT:parthenote two-cell to blastocyst conversion with B6, D2, and B6D2 F1 genotypes included, indicating a potential beneficial effect specific to cloned embryos. D1Bwg0212e expression was slightly elevated in B6 compared to D2 oocytes, potentially contributing to a positive effect of the B6 haplotype. The chr 1 gene Phf3 displayed no significant difference in expression between B6, D2, and B6D2 F1oocytes, a single negative association for the two-cell to four-cell SCNT conversion, and conflicting associations among the SCNT:parthenote traits indicating possible stage specific effects in cloned embryos.
The chr 4 gene Chd7 displayed elevated expression in B6D2 F1 oocytes, plus a positive association of expression with SCNT two-cell to four-cell conversion with B6, D2, and B6D2 F1 genotypes included. For the SCNT:parthenote traits, Chd7 mRNA expression was positively associated with formation of two-cell embryos. Thereafter, it was negatively associated with SCNT:parthenote traits of two-cell and four-cell conversions to blastocyst stage with and without B6, D2, and B6D2 F1 genotypes included, indicating a significant SCNT-specific function in later cleavage. The chr 4 gene Ccne2 displayed significant negative associations of expression with the two SCNT traits of zygote to two-cell and four-cell to blastocyst conversion, at odds with the higher expression value for D2 oocytes. However, Ccne2 displayed positive associations with the SCNT:parthenote traits for two-cell conversion to four-cell and blastocyst, with and without B6, D2, and B6D2 F1 genotypes included, indicating an SCNT-specific component of its actions. Another chr 4 gene, Car8, displayed lower expression in D2 oocytes plus significant negative associations of expression with the two SCNT traits of two-cell to four-cell and two-cell to blastocyst conversion with and without B6, D2, and B6D2 F1 genotypes included. The negative association was reiterated for SCNT:parthenote ratio of two-cell to four-cell conversion with and without B6, D2, and B6D2 F1 genotypes included. A positive association for Car8 expression was seen for SCNT:parthenote four-cell to blastocyst conversion, indicating a second SCNT-specific component to its effect. Asph displayed elevated expression in D2 and F1 oocytes, positive associations with SCNT:parthenote ratios for development, and negative associations with parthenote development.
Another chr 17 gene, Ypel5, displayed a slightly increased expression value for D2 compared to B6 oocytes, along with positive associations of expression with two-cell to four-cell and two-cell to blastocyst conversions for both SCNT and SCNT:parthenote traits, both with and without B6, D2, and B6D2 F1 genotypes included. The effects of Ypel5 thus appeared to have a strong SCNT-specific component independent of more general effects occurring in parthenotes (none observed). The Spast gene displayed modestly reduced expression in D2 compared to B6 oocytes, along with significant negative associations with two-cell to four-cell and two-cell to blastocyst SCNT conversions, with and without B6, D2, and B6D2 F1 genotypes, an association with four-cell to blastocyst conversion without B6, D2, and B6D2 F1 genotypes included, but no associations with SCNT:parthenote traits, indicating that its effects are relatively nonspecific for SCNT development.
Single gene correlations between expression and parthenote development
Processes shared between SCNT embryos and parthenotes may affect SCNT developmental characteristics. The data for parthenote development provide an opportunity to delineate further gene expression effects that are attributable specifically to nuclear reprogramming and SCNT development and an opportunity to dissect effects on oocyte activation and development in the absence of paternally derived chromosomes. With respect to genes associated with the intervals having significant LRS values, gene expression associations for Runx1t1, Epb4.1l3, Mfsd9, and Arhgap28 were generally not consistently seen in the parthenote traits, whereas associations observed for Tmem182 and Tgif1 were seen for parthenotes, with and without B6, D2, and B6D2 F1 genotypes included (Table 4 and Table S8). There was a negative association of Epb4.1l3 expression with parthenote conversion to the two-cell stage. Many of the effects seen for Smchd1, Ypel5, Dlgap1, Spast, and two of the Ndc80 associations were also not seen extensively for parthenotes, with or without B6, D2, and B6D2 F1 genotypes included. Effects seen for chr1 genes Eif5b, Txndc9, and D1Bwg0212e were also present for parthenotes, and thus were not specific to SCNT embryos. With respect to unique associations of gene expression with parthenote development, the most prominent effect was seen for Lclat1, having significant negative associations with all four parthenote traits, but also for two SCNT:parthenote traits.
Testing of correlations in expression between genes
The genetic data indicate cooperative interactions between multiple loci. Additionally, the gene expression data above indicate that more than one of the genes tested likely contribute to processes that specifically affect cloning outcome. We therefore tested for cooperativity between genes at the level of mRNA expression that could contribute to phenotype. This analysis focused on genes displaying individual expression correlations with phenotype. The primary significant LRS interval on chr 17 encompasses two genes (Epb4.1l3 and Dlgap1) and nearby Arhgap28, all of which fulfill expectations of differential expression between B6 and D2 oocytes, haplotype effect, and expected associations between expression and developmental outcomes among BXD strains. Five other chr 17 genes (Smchd1, Tgif1, Ndc80, Lclat1, Ypel5) exerted lesser effects or effects only partially specific to SCNT embryos. For the chr 4 interval, the Runx1t1 gene remains as the only candidate within the defined significance interval, and although it does not show a strong fit to the expected pattern, it displays an association with SCNT four-cell to blastocyst conversion independent of any parthenote effects, indicating that it likely contributes to overall outcome for cloned embryos. Four other chr 4 genes (Car8, Asph, Chd7, Ccne2) may also contribute to SCNT embryo development. For the chr 1 interval, the Mfsd9 gene from near the significance interval appears most likely to contribute to outcome. Two other genes from chr 1 (Phf3 and D1Bwg0212e) may contribute to clone development, whereas Txndc9 and Eif5b may exert nonspecific effects. Thus, of the 26 genes tested on all 28 strains, 5 stand out as major candidates, and another 11 display an elevated likelihood of providing supportive contributions (Figure 2). To gain further insight, we tested for cooperativity among genes in terms of expression correlation with developmental outcomes (Table S8).
Figure 2.
Summary of identified candidate genes. Genes having the most significant genetic and expression associations with phenotype are denoted by boldface type. Progressively smaller type sizes denote genes with progressively lesser associations but apparent SCNT-specific phenotype effects. C denotes cytoskeletal or spindle associated proteins. T denotes transcription and chromatin regulators.
The Epb4.1l3 mRNA displayed significant positive expression correlations with Dlgap1, Mfsd9, Ypel5, Runx1t1, Chd7, and Esrp1 and significant negative correlations with Arhgap28, Ndc80, Spast, Txndc9, Smchd1, Phf3, and Eif5b. The Dlgap1 mRNA displayed significant positive correlations in expression with Epb4.1l3, Arhgap28, Mfsd9, Ndc80, Chd7, Esrp1, Car8, and Rab2a and negative correlations for Tmem182, Txndc9, Ccne2, Pdcl3, Tgif1, and Eif5b. The Arhgap28 gene showed significant positive expression correlations with Dlgap1, Ndc80, Ypel5, Spast, Smchd1, Phf3, and Car8 and negative correlations with Epb4.1l3, Ccne2, Pdcl3, and Lclat1. Thus there are significant correlations between the expression patterns of all three of the primary chr 17 genes with each other and with other genes. Correlations in expression between Epb4.13, Runx1t1, and Mfsd9 are consistent with their effects on conversion from four-cell to blastocyst stage in the SNCT:parthenote data (Table 4) and provide a potential avenue for a cooperative interaction among the three significance intervals on chr 1, 4, and 17.
Ingenuity pathway analysis
To test further for cooperativity between the genes examined in this study, we undertook a biofunction and pathway analysis using the Ingenuity Pathway Analysis (IPA) program. One utility in the IPA program is a search of the IPA database for overrepresentation of members of a gene list among known biological functions and pathways. This analysis reveals known associations with two or more genes in a common biological function and indicates whether such functions are associated with a given experimental condition. Several potential gene associations were identified using the list of 26 genes displaying significant associations between expression and cloning traits (Table S9). One was the combination of Dlgap1, Phf3, Rab2a, and Spast, encompassing genes from chr 1, 4, and 17 regions. Another combination was Aff3, Asph, and Chd7 from chromosomes 1 and 4. These combinations were retrieved for functions related to cell signaling and morphogenesis. There were many other combinations of just two genes. The Epb4.1l3 gene occupied combinations with Dlgap1, Asph, Spast, and Car8 and was connected indirectly to other genes via the other biofunction combinations.
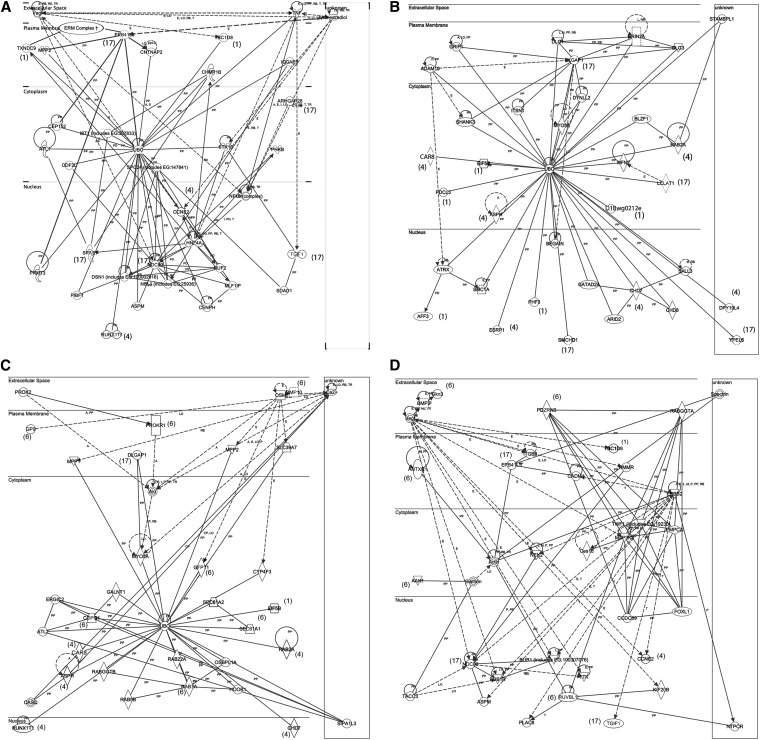
A second utility offered by the IPA program is the ability to observe networks and pathways of interactions, both among members of the query gene list and with genes not on the list. Two networks were retrieved for the 26-gene query. One network contained 9 members of the list and another contained 15 members, indicating strong functional association among the 26 genes. Both networks have at their centers ubiquitin C (UBC), indicating that many of the candidate genes tested are regulated directly or indirectly by protein ubiquitination. Additionally, Network 1 (Figure 3A) indicated significant connectivity for EPB4.1L3 and NDC80, as well as HNF4A and NFKB. Network 1 was modified by the inclusion of the ezrin–radixin–moesin complex, known to associate with EPB4.1L3 and with other actin-associated molecules such as TXNDC9 and TBC1D8, particularly in association with the plasma membrane where microfilaments and microtubules can provide crucial structure to facilitate signaling interactions. Network 2 (Figure 3B) included DLGAP1 as another key regulator of signaling at the plasma membrane. Note that the two networks can be connected via UBC. Each of the two networks contains one of the top chr 17 candidates from the region showing significant LRS values (Epb4.1l3 and Dlgap1) connected to UBC and downstream to many potential mediators, including a range of nuclear factors that include Runx1t1 from the chr 4 interval with a significant LRS value and Tgif1 from near the chr 17 significance interval. Neither network incorporated Mfsd9 or Tmem182, the two genes located adjacent to the chr 1 significance region, but other chr 1 genes (Phf3, Txndc9, Aff3, Pdcl3, and Tbc1d8) were present.
Figure 3.
Ingenuity pathway analysis networks summarizing gene interactions among the 26 genes assayed for expression on all 28 strains (A and B) and these same 26 genes combined with genes from the potential interacting locus on chr 6 (C and D). Numbers in parentheses indicate chr assignments. PP, protein–protein interaction; PD, protein–DNA interaction; E, expression; A, activation; T, transcription; RB, regulation of binding; I, inhibition; LO, localization; CP, chemical–protein interaction; M, biochemical modification; solid line, binding only; line with arrow, downstream effect; broken line, indirect interaction; vertical diamond, enzyme; horizontal diamond, peptidase; horizontal oval, transcription factor; downward triangle, kinase; upward triangle, phosphatase; circle, other.
Because the genetic analysis indicated a possible interaction of the chr 17 interval with a region on chr 6, we also combined the 26 genes here with genes from the potential interacting region on chr 6 and submitted this combined list for IPA analysis. This yielded four networks, one of which included Dlgap1 and one that included Epb4.1l3 (Figure 3, C and D). In general, the chr 6 gene associations were via UBC and HNF4A as above (Figure 3C, Figure S1, and Figure S2). None of the four networks indicated direct dependent interactions between the chr 6 genes and any of the 26 genes tested. Array data indicated that the Aak1, Antxr1, Gfpt1, Copg, Rab7, Sec61a1, and Pdzrn3 mRNAs are expressed in MII oocytes, with Antxr1 (lower in D2) and Pdzrn3 (higher in D2) displaying >25% differences in expression between B6 and D2, both of which reside in the network shown in Figure 2, which incorporates actin and clathrin as key components.
Gene sequence variations between haplotypes for candidate genes
Because sequence variation could affect protein activity and hence gene function apart from differences in mRNA expression abundance, we compared the B6 and D2 sequences for polymorphisms (Tables 5 and Table S10). Seventeen of the 26 genes studied in detail display polymorphisms in the 3′-UTR. Five of the genes studied (Phf3, Pdcl3, Arhgap28, Ndc80, and Spast) display polymorphisms affecting protein-coding region. Notable among these is the Arhgap28 gene, which displays numerous polymorphisms in the 3′-UTR and two genetic variants in the coding region. This is the only gene among the 26 candidates and lying in or near the intervals with significant LRS values that displays variants in the coding region. The Phf3, Pdcl3, Ndc80, and Spast genes also display genetic variants in the coding regions. Polymorphisms in the 3′-UTRs for some mRNAs may also contribute to the observed differences in oocyte expression.
Table 5. Summary of polymorphisms between B6 and D2 haplotypes for selected genes.
| Gene | Gene ID | Chr | No. variants | NS SNPs coding region | No. insertions or deletions coding region | No. SNPs 3′-UTR in variants from column 4 | No. insertions or deletions 3′-UTR in variants from column 4 |
|---|---|---|---|---|---|---|---|
| Genes in or near significance intervals | |||||||
| Mfsd9 | 211798 | 1 | 1 | ||||
| Tmem182 | 381339 | 1 | 1 | ||||
| Runx1t1 | 12395 | 4 | 3 | 2, 2, 2 | |||
| Arhgap28 | 268970 | 17 | 1 | 1 | 1 | 28 | 3 |
| Epb4.1l3 | 13823 | 17 | 1 | ||||
| Dlgap1 | 224997 | 17 | 4 | 0, 1, 1, 1 | 4, 1, 4, 1 | ||
| Tgif1 | 21815 | 17 | 5 | 2, 2, 2, 2, 2 | |||
| Other genes in suggestive intervals | |||||||
| Phf3 | 213109 | 1 | 1 | 3 | 1 | 2 | |
| Txndc9 | 98258 | 1 | 1 | 45 | 9 | ||
| Eif5b | 226982 | 1 | 1 | ||||
| Aff3 | 16764 | 1 | 1 | ||||
| Pdcl3 | 68833 | 1 | 1 | 1 | 1 | ||
| Tbc1d8 | 54610 | 1 | 1 | ||||
| D1Bwg0212e | 52846 | 1 | 1 | ||||
| Car8 | 12319 | 4 | 1 | 65 | 9 | ||
| Rab2a | 59021 | 4 | 1 | 1 | |||
| Chd7 | 320790 | 4 | 1 | 1 | 1 | ||
| Asph | 65973 | 4 | 10 | 0, 4, 4, 5, 5, 5, 5, 5, 5, 8 | 2, 0, 2, 1, 1, 1, 1, 0, 1, 1 | ||
| Ccne2 | 12448 | 4 | 2 | 1, 1 | |||
| Dpy19l4 | 381510 | 4 | 1 | ||||
| Esrp1 | 207920 | 4 | 1 | ||||
| Smchd1 | 74355 | 17 | 1 | 1 | 1 | ||
| Ndc80 | 67052 | 17 | 1 | 1 | 2 | ||
| Ypel5 | 383295 | 17 | 1 | 6 | 2 | ||
| Lclat1 | 225010 | 17 | 3 | 3, 3, 3 | |||
| Spast | 50850 | 17 | 2 | 1, 1 | 9, 9 | ||
| Zfp161 | 22666 | 17 | 1 | ||||
Discussion
For the first time, a systems genetics approach has been applied to the analysis of variation in oocyte composition associated with the ability of different genotypes of oocytes to support early development of cloned embryos made by somatic cell nuclear transfer. The results provide new insight into clone biology, oocyte quality, and key early processes in mammalian embryogenesis. Overall, the analysis yielded polymorphic candidate genes related to two functional categories: (1) microtubule- and microfilament-associated proteins associated with the subcortical cytoskeletal network and spindle, and (2) a small number of proteins related to transcriptional control and chromatin remodeling. The analysis revealed a major locus controlling cloning outcome on chr 17, with additional regions on chr 1 and 4 (Figure 3).
The strongest genetic and expression-phenotype associations were derived for two genes on chr 17 that encode cytoskeletal scaffolding proteins, Epb4.1l3 and Dlgap1, yielding the novel discovery that cytoskeletal architecture plays a key and specific role in meeting specific developmental requirements of cloned embryos. Both Dlgap1 and Epb4.1l3 are components of the cortical cytoskeleton network and likely play roles in supporting cellular signaling. The Epb4.1 family comprises scaffolding proteins that interact with the ezrin–radixin–moesin complex and play a role in tethering the cortical actin–spectrin complex to the plasma membrane, regulating cell shape, intercellular junctions, ion balance, signaling, and control of cellular proliferation (Kuns et al. 2005; Terada et al. 2005; Cifuentes-Diaz et al. 2011). Dlgap1 encodes a scaffold protein also known as GKAP/SAPAP1, which controls receptor functioning as well as microtubule dynamics and organization near the cell cortex and promotes centrosome positioning (Manneville et al. 2010). Hence, these two proteins are positioned to exert profound effects on diverse cellular processes and are the strongest candidates for playing a role in cloned embryo development. Arhgap28 encodes a signal transduction protein that may interact with the cortical cytoskeletal complex. The Epb4.1l3 gene was expressed more highly in D2 oocytes whereas the Dlgap1 gene was expressed less in the D2 oocytes than in the B6 and B6D2 F1 oocytes. The Epb4.1l3 gene expression was positively associated with SCNT embryo from two cell to four cell and blastocyst but its SCNT-specific component appears to be negative. The Dlgap1 gene was expressed at a greatly reduced level in D2 oocytes and displayed a negative SCNT-specific effect and generally negative effects that were not limited to clones. These data are most consistent with a positive effect of the D2 haplotype in this region on cloned embryo development mediated by lower Dlgap1 expression and non-SCNT-specific positive effects of Epb4.1l3 at early cleavage stages.
The expression Arhgap28 was also associated strongly with cloning phenotype, displaying a specific negative association with cloning outcome. Other candidate genes were related to cytoskeletal function as well. TXNDC9 (aka PHLP3), a phosducin, is required for proper actin and tubulin function (Stirling et al. 2006), and hence could interact directly or indirectly with Epb4.1l3 and Dlgap1 at the cortex. Interestingly, another gene studied on chr 1, Pdcl3, encodes another phosducin that also promotes cytoskeletal remodeling (Hayes et al. 2011) and could likewise play a role in controlling the cortical cytoskeleton. EIF5B could positively affect protein translation (Lebaron et al. 2012). Spast encodes a microtubule-severing protein (Fassier et al. 2012) and thereby contributes to cytoskeleton regulation. Three other expressed chr 17 genes from a suggestive interval but not included in our analysis due to uncertain functional relevance (Memo1 and Myl12a, Myl12b) encode regulators of myosin contractility and cellular motility. The qRT–PCR results for one of these genes on B6, D2, and B6D2 F1 oocytes revealed only modest differences in mRNA expression.
This novel putative role for cytoskeletal proteins to meet the specific developmental needs of cloned embryos could reflect a specific need to restore cytoskeletal architecture after SCNT. This architecture may provide essential scaffolding functions that enable autocrine and paracrine signals to be transmitted inwardly to the nucleus, as well as enabling crucial homeostatic processes at the surface. Asph encodes a protein that post-translationally modifies epidermal growth factor domains in many proteins, deficiency for which is associated with later developmental defects (Dinchuk et al. 2002). The cytoskeletal architecture may also play crucial roles in controlling spindle formation and positioning or the positioning of organelles such as endoplasmic reticulum and mitochondria. ER positioning affects protein trafficking as well as calcium signaling (Soboloff et al. 2012). Mitochondrial positioning in close proximity to the genome could be important for providing ATP for reprogramming.
Another interesting possibility is that the cytoskeletal architecture plays a key role in establishing cellular potency. Other studies have pointed to essential roles played by a so-called subcortical maternal complex and cytoplasmic lattices, with potential roles in translational control and ultimately regulation of subsequent embryonic processes in mouse two-cell embryos (Li et al. 2008; Ohsugi et al. 2008; Yurttas et al. 2008; Kim et al. 2010; Kan et al. 2011). Even earlier studies reported interesting associations between microtubule acetylation, localization to the cortical region at the basal side of blastomeres, and subsequent partitioning to the inner cells, so that the microtubules might contribute to determining cell fate (Houliston and Maro 1989). It should prove interesting to discover whether the candidate gene products identified here, particularly EPB4.1L3 and DLGAP1, interact with subcortical maternal complex or other cytoskeletal elements to control early embryo development and whether this cytoskeletal architecture plays a regulative role in determining developmental potency and early cellular specializations.
It is also intriguing that some of the significant associations revealed here relate to spindle formation and function. There are significant protein composition deficits in the spindles that form in cloned constructs before activation and in mitotic spindles of at least the first couple of cell cycles (Miyara et al. 2006). Chromosome congression defects, mitotic errors, and aneuploidies in individual blastomeres of cloned embryos may be one consequence of this (Nolen et al. 2005; Miyara et al. 2006; Mizutani et al. 2012), reducing the number of viable cells and overall embryo viability. One major defect in these spindles is a deficiency in clathrin heavy chain, which bundles together microtubules and promotes spindle formation. Enhancement of clathrin heavy-chain expression improves chromosome congression in cloned embryos (Han et al. 2010b). The data here suggest that variations in expression of other proteins may compound these spindle defects, so that the rate of aneuploidies observed could vary with recipient oocyte genotype. One gene examined here, Ndc80, regulates oocyte spindle formation, chromosome alignment, and cell-cycle progression (Sun et al. 2011) and is associated with the kinetochore where it regulates microtubule dynamics (Umbreit et al. 2012). Ccne2 is associated with Ndc80 in the IPA analysis and could be associated with mediating checkpoint control. YPEL5 associates with the centrosome and spindle and promotes cell proliferation (Hosono et al. 2010). Proteins such as NDC80 and SPAST along with DLGAP1 may work together to regulate spindle or centrosome formation and function in cloned constructs. Further studies of how genetic variation in the expression of these proteins interacts with spindle deficiencies arising from the SCNT method should provide new insight into how the overall process of spindle formation is regulated. Moreover, understanding the roles played by these genes in oocyte spindle biology could provide new understanding related to the exponential increase in oocytes having defective spindles and chromosomal aneuploidies with age and onset of female reproductive senescence (Hunt and Hassold 2008) and may enable better management of this decline based on patient genotype information.
Although the major result from this analysis is a focus on cytoskeleton-associated proteins, this is not to discount the potential importance of transcription regulators such as Runx1t1, Smchd1, and Chd7. A significant role for these proteins is indicated by the identification of the intervals that contain these genes as having significant (Runx1t1) or suggestive (Chd7, Smchd1) LRS values. Stronger associations of Chd7 and Smchd1 might emerge with analysis of additional genotypes. Even lacking significant LRS values for these suggestive regions, the correlations between their expression and phenotypic traits among the BXD strains and the known biofunctions of these genes indicate that further study of these genes for roles in normal and cloned development is warranted.
Runx1t1 displayed a significant negative expression association with the SCNT and SCNT:parthenote four-cell to blastocyst transition traits. This indicates that the Runx1t1 gene exerts a specific negative effect on overall cloned embryo development. Runx1t1 appears in a gene network along with Epb4.1l3 and Arhgap28 (Figure 2A). RUNX1T1 (aka ETO) is a transcription factor that is joined to AML1 by chromosomal translocation to generate a leukemia gene, binds to DNA, and recruits histone deacetylase to repress gene expression (Erickson et al. 1994; Gelmetti et al. 1998). Increased RUNX1T1 activity could thus inhibit development.
The specific negative relationships seen between expression and clone development for Chd7 and Smchd1 may reflect effects on nuclear reprogramming or X chromosome regulation. CHD7 is a chromodomain helicase that either negatively or positively modulates gene expression, including ES-cell-specific genes, although negative regulation is described as the more direct effect (Schnetz et al. 2010). CHD7 colocalizes in ES cells with POU5F1 (OCT4), SOX2, and NANOG and negatively modulates many genes selectively expressed in ES cells. SMCHD1 plays a key role in X-chromosome inactivation (Blewitt et al. 2008; Gendrel et al. 2012), which is also aberrant in cloned embryos (Nolen et al. 2005; Jiang et al. 2008; Inoue et al. 2010). It will be very interesting to determine the roles of these two factors in reprogramming during cloning and whether they might aid in vitro reprogramming by exogenous factors to make induced pluripotent stem cells.
Another transcription factor gene, Zfp161, lies within the chr 17 significance interval but was excluded on the basis of a lack of expression correlation with developmental phenotypes of the initial 6 BXD lines studied. This gene represses Myc (Sobek-Klocke et al. 1997). Myc promotes cell proliferation in stem cells and at later stages (Yamanaka 2008), but overexpression has little effect on early cleavage (Pan and Schultz 2011). While MYC may contribute to cell-cycle regulation and possible stress responses, particularly in advanced stage preimplantation embryos (Xie et al. 2007), the lack of a correlation between Zfp161 expression and clone development argues against a specific role here. Additionally, the absence of genetic polymorphism in the coding region (Table 4 and Table S9) argues against a genetic effect at the level of protein structure.
We note the recent discovery that genome scanning approaches correlating genetic variants with phenotypic differences, such as that employed here, have the potential for yielding novel noncoding regulatory elements, such as enhancers and other chromatin regulatory elements (Dunham et al. 2012). The interactions of these elements can extend over large distances along the chromosome. We note, however, that the sizes of the combined significant and suggestive candidate intervals included in our study are large (12.7-cM chr 1, 7-cM chr 4, and 9.5-cM chr 17) and so provided the opportunity to detect significant variations in affected target genes. By evaluating the levels of expression and comparative expression levels between B6, D2, and B6D2 F1 genotypes for genes throughout these regions, we have provided a rigorous coverage of known gene products most likely to be affected by regulatory elements that could exist in the intervals with significant LRS values. Moreover, our identification of several genes that display strong and significant correlations between expression and phenotype, both individually and in combination, while also demonstrating known functional relationships in gene networks, provides high confidence in the newly ascribed relationships of these genes to clone biology.
Other genes examined in this study display relationships between expression and development and thus are likely to contribute to the overall success of cloned embryo development. Four genes displayed notable associations between expression and development that were not specific to cloned embryos (Txndc9, Eif5b, Tmem182, Spast). The Txndc9 and Eif5b genes are a bidirectionally coregulated gene pair (Garcia and Nagai 2011). These two genes displayed negative expression associations with development of both cloned and parthenogenetic embryos, so the associations were not specific to cloned embryos. Some of the associations seen were not in agreement for the two genes, raising some question about their relevance. However, it is noted that the zygotic REDOX state can affect postnatal phenotype (Banrezes et al. 2011). Participation of Txndc9 in such regulation could contribute to SCNT development. Other genes displayed a combination of specific and nonspecific associations between expression and development at different stages. Early, specific associations were seen for genes on chr 4 (Ccne2) and chr 17 (Tgif1, Smchd1, Car8, Ypel5, Esrp1, Lclat1). Later stage-specific associations were seen for genes on chr 1 (D1Bwg0212e), chr 4 (Chd7, Car8, Asph), and chr 17 (Ndc80).
Still other genes not selected for detailed study but lying within the candidate intervals could be proposed to play a role in cloned embryo development. DNA repair genes, for example, could be important for repairing DNA damage in the somatic donor nuclei, or damage that might arise during SCNT, and could also play roles in DNA replication. DNA base excision repair can contribute to epigenetic transitions (Sarkies et al. 2010; Seisenberger et al. 2013). Of the two DNA repair genes residing within the candidate intervals, Rev1 displayed little difference in expression between B6, D2, and B6D2 F1 genotypes and was excluded on that basis, while Rad54b mRNA was expressed at a low level in MII oocytes and displayed a modest difference in expression between B6, D2, and B6D2 F1 oocytes on arrays. Noncoding RNAs present in the ooplasm could be proposed to play important roles. We examined noncoding RNA genes in or near the intervals with significant LRS values without positive results. As described above, these genes were typically excluded as candidates on the basis of two or more criteria including available expression data, making them far less likely candidates than those selected for analysis.
One final point to consider is what these results tell us about the determinants of oocyte quality and how this relates to clinical reproductive medicine outcomes. The differences in expression between MII oocytes of the different genotypes highlight the importance of not assuming that all oocytes are created equal. Indeed, there is substantial genetic variation in oocyte phenotype revealed among these BXD strains, which would be analogous to variation among family members; the amount of variation among a broader spectrum of individuals should be at least this great. This suggests that microsurgical approaches that assume uniformity among oocytes from different patients may reap unexpected outcomes. This also suggests that genetic data for patients could be useful in evaluating relative risks or likely success of clinical procedures and may be useful to modify clinical procedures to optimize them for specific individuals.
Acknowledgments
The authors are grateful to Robert W. Williams and Lu Lu, University of Tennessee Health Science Center, and Fernando Pardo-Manuel de Vilena for their guidance and encouragement in analyzing the data. We also thank Robert W. Williams and Susannah Varmuza for comments on the manuscript. This work was supported by a grant from the National Institutes of Health (NIH), National Institute of Child Health and Human Development, funded under the American Recovery and Reinvestment Act (ARRA), RC1HD063371, by the grant HD43092, and by the NIH Office of the Director, Comparative Medicine Branch, Office of Research Infrastructure Programs (ORIP) R24OD012221-12 to K.E.L.
Footnotes
Communicating editor: J. C. Schimenti
Literature Cited
- Akagi S., Matsukawa K., Mizutani E., Fukunari K., Kaneda M., et al. , 2011. Treatment with a histone deacetylase inhibitor after nuclear transfer improves the preimplantation development of cloned bovine embryos. J. Reprod. Dev. 57: 120–126. [DOI] [PubMed] [Google Scholar]
- Andreux P. A., Williams E. G., Koutnikova H., Houtkooper R. H., Champy M. F., et al. , 2012. Systems genetics of metabolism: the use of the bxd murine reference panel for multiscalar integration of traits. Cell. 150: 1287–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banrezes B., Sainte-Beuve T., Canon E., Schultz R. M., Cancela J., et al. , 2011. Adult body weight is programmed by a redox-regulated and energy-dependent process during the pronuclear stage in mouse. PLoS ONE 6: e29388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blewitt M. E., Gendrel A. V., Pang Z., Sparrow D. B., Whitelaw N., et al. , 2008. SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat. Genet. 40: 663–669. [DOI] [PubMed] [Google Scholar]
- Bothe G. W., Bolivar V. J., Vedder M. J., Geistfeld J. G., 2004. Genetic and behavioral differences among five inbred mouse strains commonly used in the production of transgenic and knockout mice. Genes Brain Behav. 3: 149–157. [DOI] [PubMed] [Google Scholar]
- Bryant C. D., 2011. The blessings and curses of C57BL/6 substrains in mouse genetic studies. Ann. N. Y. Acad. Sci. 1245: 31–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui H. T., Seo H. J., Park M. R., Park J. Y., Thuan N. V., et al. , 2011. Histone deacetylase inhibition improves activation of ribosomal RNA genes and embryonic nucleolar reprogramming in cloned mouse embryos. Biol. Reprod. 85: 1048–1056. [DOI] [PubMed] [Google Scholar]
- Campbell K. H., McWhir J., Ritchie W. A., Wilmut I., 1996. Sheep cloned by nuclear transfer from a cultured cell line. Nature 380: 64–66. [DOI] [PubMed] [Google Scholar]
- Chavatte-Palmer P., Camous S., Jammes H., Le Cleac’h N., Guillomot M., et al. , 2012. Review: placental perturbations induce the developmental abnormalities often observed in bovine somatic cell nuclear transfer. Placenta 33(Suppl): S99–S104. [DOI] [PubMed] [Google Scholar]
- Chen L., Wang D., Wu Z., Ma L., Daley G. Q., 2010. Molecular basis of the first cell fate determination in mouse embryogenesis. Cell Res. 20: 982–993. [DOI] [PubMed] [Google Scholar]
- Chung Y. G., Mann M. R., Bartolomei M. S., Latham K. E., 2002. Nuclear-cytoplasmic “tug of war” during cloning: effects of somatic cell nuclei on culture medium preferences of preimplantation cloned mouse embryos. Biol. Reprod. 66: 1178–1184. [DOI] [PubMed] [Google Scholar]
- Chung Y. G., Ratnam S., Chaillet J. R., Latham K. E., 2003. Abnormal regulation of DNA methyltransferase expression in cloned mouse embryos. Biol. Reprod. 69: 146–153. [DOI] [PubMed] [Google Scholar]
- Chung Y. G., Gao S., Latham K. E., 2006. Optimization of procedures for cloning by somatic cell nuclear transfer in mice. Methods Mol. Biol. 348: 111–124. [DOI] [PubMed] [Google Scholar]
- Cifuentes-Diaz C., Chareyre F., Garcia M., Devaux J., Carnaud M., et al. , 2011. Protein 4.1B contributes to the organization of peripheral myelinated axons. PLoS ONE 6: e25043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinchuk J. E., Focht R. J., Kelley J. A., Henderson N. L., Zolotarjova N. I., et al. , 2002. Absence of post-translational aspartyl beta-hydroxylation of epidermal growth factor domains in mice leads to developmental defects and an increased incidence of intestinal neoplasia. J. Biol. Chem. 277: 12970–12977. [DOI] [PubMed] [Google Scholar]
- Dunham I., Kundaje A., Aldred S. F., Collins P. J., Davis C. A., et al. , 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggan K., Akutsu H., Hochedlinger K., Rideout W., 3rd, Yanagimachi R., et al. , 2000. X–chromosome inactivation in cloned mouse embryos. Science 290: 1578–1581. [DOI] [PubMed] [Google Scholar]
- Erickson P. F., Robinson M., Owens G., Drabkin H. A., 1994. The ETO portion of acute myeloid leukemia t(8;21) fusion transcript encodes a highly evolutionarily conserved, putative transcription factor. Cancer Res. 54: 1782–1786. [PubMed] [Google Scholar]
- Fassier C., Tarrade A., Peris L., Courageot S., Mailly P., et al. , 2012. Microtubule-targeting drugs rescue axonal swellings in cortical neurons from spastin knock-out mice, Dis. Model Mech; 6: 72–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S., Latham K. E., 2004. Maternal and environmental factors in early cloned embryo development. Cytogenet. Genome Res. 105: 279–284. [DOI] [PubMed] [Google Scholar]
- Gao S., Chung Y. G., Williams J. W., Riley J., Moley K., et al. , 2003. Somatic cell-like features of cloned mouse embryos prepared with cultured myoblast nuclei. Biol. Reprod. 69: 48–56. [DOI] [PubMed] [Google Scholar]
- Gao S., Czirr E., Chung Y. G., Han Z., Latham K. E., 2004. Genetic variation in oocyte phenotype revealed through parthenogenesis and cloning: correlation with differences in pronuclear epigenetic modification. Biol. Reprod. 70: 1162–1170. [DOI] [PubMed] [Google Scholar]
- Garcia S. A., Nagai M. A., 2011. Transcriptional regulation of bidirectional gene pairs by 17-beta-estradiol in MCF-7 breast cancer cells. Braz. J. Med. Biol. Res. 44: 112–122. [DOI] [PubMed] [Google Scholar]
- Gelmetti V., Zhang J., Fanelli M., Minucci S., Pelicci P. G., et al. , 1998. Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol. Cell. Biol. 18: 7185–7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel A. V., Apedaile A., Coker H., Termanis A., Zvetkova I., et al. , 2012. Smchd1-dependent and -independent pathways determine developmental dynamics of CpG island methylation on the inactive x chromosome. Dev. Cell 23: 265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillomot M., Taghouti G., Constant F., Degrelle S., Hue I., et al. , 2010. Abnormal expression of the imprinted gene Phlda2 in cloned bovine placenta. Placenta 31: 482–490. [DOI] [PubMed] [Google Scholar]
- Hamatani T., Ko M., Yamada M., Kuji N., Mizusawa Y., et al. , 2006. Global gene expression profiling of preimplantation embryos. Hum. Cell 19: 98–117. [DOI] [PubMed] [Google Scholar]
- Han Z., Vassena R., Chi M. M., Potireddy S., Sutovsky M., et al. , 2008. Role of glucose in cloned mouse embryo development. Am. J. Physiol. Endocrinol. Metab. 295: E798–E809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z., Cheng Y., Liang C. G., Latham K. E., 2010a Nuclear transfer in mouse oocytes and embryos. Methods Enzymol. 476: 171–184. [DOI] [PubMed] [Google Scholar]
- Han Z., Liang C. G., Cheng Y., Duan X., Zhong Z., et al. , 2010b Oocyte spindle proteomics analysis leading to rescue of chromosome congression defects in cloned embryos. J. Proteome Res. 9: 6025–6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes N. V., Josse L., Smales C. M., Carden M. J., 2011. Modulation of phosducin-like protein 3 (PhLP3) levels promotes cytoskeletal remodelling in a MAPK and RhoA-dependent manner. PLoS ONE 6: e28271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosono K., Noda S., Shimizu A., Nakanishi N., Ohtsubo M., et al. , 2010. YPEL5 protein of the YPEL gene family is involved in the cell cycle progression by interacting with two distinct proteins RanBPM and RanBP10. Genomics 96: 102–111. [DOI] [PubMed] [Google Scholar]
- Houliston E., Maro B., 1989. Posttranslational modification of distinct microtubule subpopulations during cell polarization and differentiation in the mouse preimplantation embryo. J. Cell Biol. 108: 543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humpherys D., Eggan K., Akutsu H., Friedman A., Hochedlinger K., et al. , 2002. Abnormal gene expression in cloned mice derived from embryonic stem cell and cumulus cell nuclei. Proc. Natl. Acad. Sci. USA 99: 12889–12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt P. A., Hassold T. J., 2008. Human female meiosis: What makes a good egg go bad? Trends Genet. 24: 86–93. [DOI] [PubMed] [Google Scholar]
- Inoue K., Kohda T., Sugimoto M., Sado T., Ogonuki N., et al. , 2010. Impeding Xist expression from the active X chromosome improves mouse somatic cell nuclear transfer. Science 330: 496–499. [DOI] [PubMed] [Google Scholar]
- Jafari S., Hosseini M. S., Hajian M., Forouzanfar M., Jafarpour F., et al. , 2011. Improved in vitro development of cloned bovine embryos using S-adenosylhomocysteine, a non-toxic epigenetic modifying reagent. Mol. Reprod. Dev. 78: 576–584. [DOI] [PubMed] [Google Scholar]
- Jiang L., Lai L., Samuel M., Prather R. S., Yang X., et al. , 2008. Expression of X-linked genes in deceased neonates and surviving cloned female piglets. Mol. Reprod. Dev. 75: 265–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan R., Yurttas P., Kim B., Jin M., Wo L., et al. , 2011. Regulation of mouse oocyte microtubule and organelle dynamics by PADI6 and the cytoplasmic lattices. Dev. Biol. 350: 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B., Kan R., Anguish L., Nelson L. M., Coonrod S. A., 2010. Potential role for MATER in cytoplasmic lattice formation in murine oocytes. PLoS ONE 5: e12587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. J., Ahn K. S., Kim M., Shim H., 2011. Comparison of potency between histone deacetylase inhibitors trichostatin A and valproic acid on enhancing in vitro development of porcine somatic cell nuclear transfer embryos. In Vitro Cell. Dev. Biol. Anim. 47: 283–289. [DOI] [PubMed] [Google Scholar]
- Kuns R., Kissil J. L., Newsham I. F., Jacks T., Gutmann D. H., et al. , 2005. Protein 4.1B expression is induced in mammary epithelial cells during pregnancy and regulates their proliferation. Oncogene 24: 6502–6515. [DOI] [PubMed] [Google Scholar]
- Latham K. E., 2004. Cloning: questions answered and unsolved. Differentiation 72: 11–22. [DOI] [PubMed] [Google Scholar]
- Latham K. E., 2005. Early and delayed aspects of nuclear reprogramming during cloning. Biol. Cell 97: 119–132. [DOI] [PubMed] [Google Scholar]
- Latham K. E., Schultz R. M., 2001. Embryonic genome activation. Front. Biosci. 6: D748–D759. [DOI] [PubMed] [Google Scholar]
- Latham K. E., Garrels J. I., Chang C., Solter D., 1992. Analysis of embryonic mouse development: construction of a high-resolution, two-dimensional gel protein database. Appl. Theor. Electrophor. 2: 163–170. [PubMed] [Google Scholar]
- Latham K. E., Gao S., Han Z., 2007. Somatic cell nuclei in cloning: strangers traveling in a foreign land. Adv. Exp. Med. Biol. 591: 14–29. [DOI] [PubMed] [Google Scholar]
- Lebaron S., Schneider C., van Nues R. W., Swiatkowska A., Walsh D., et al. , 2012. Proofreading of pre-40S ribosome maturation by a translation initiation factor and 60S subunits. Nat. Struct. Mol. Biol. 19: 744–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. S., Latham K. E., Vandevoort C. A., 2008. Effects of in vitro maturation on gene expression in rhesus monkey oocytes. Physiol. Genomics 35: 145–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Baibakov B., Dean J., 2008. A subcortical maternal complex essential for preimplantation mouse embryogenesis. Dev. Cell 15: 416–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D., 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)). Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Mann M. R., Chung Y. G., Nolen L. D., Verona R. I., Latham K. E., et al. , 2003. Disruption of imprinted gene methylation and expression in cloned preimplantation stage mouse embryos. Biol. Reprod. 69: 902–914. [DOI] [PubMed] [Google Scholar]
- Manneville J. B., Jehanno M., Etienne-Manneville S., 2010. Dlg1 binds GKAP to control dynein association with microtubules, centrosome positioning, and cell polarity. J. Cell Biol. 191: 585–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba S., Inoue K., Kohda T., Sugimoto M., Mizutani E., et al. , 2011. RNAi-mediated knockdown of Xist can rescue the impaired postimplantation development of cloned mouse embryos. Proc. Natl. Acad. Sci. USA 108: 20621–20626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekada K., Abe K., Murakami A., Nakamura S., Nakata H., et al. , 2009. Genetic differences among C57BL/6 substrains. Exp. Anim. 58: 141–149. [DOI] [PubMed] [Google Scholar]
- Miyara F., Han Z., Gao S., Vassena R., Latham K. E., 2006. Non-equivalence of embryonic and somatic cell nuclei affecting spindle composition in clones. Dev. Biol. 289: 206–217. [DOI] [PubMed] [Google Scholar]
- Mizutani E., Yamagata K., Ono T., Akagi S., Geshi M., et al. , 2012. Abnormal chromosome segregation at early cleavage is a major cause of the full-term developmental failure of mouse clones. Dev. Biol. 364: 56–65. [DOI] [PubMed] [Google Scholar]
- Mtango N. R., Potireddy S., Latham K. E., 2008. Oocyte quality and maternal control of development. Int. Rev. Cell. Mol. Biol. 268: 223–290. [DOI] [PubMed] [Google Scholar]
- Mtango N. R., VandeVoort C. A., Latham K. E., 2011. Ontological aspects of pluripotency and stemness gene expression pattern in the rhesus monkey. Gene Expr. Patterns 11: 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen L. D., Gao S., Han Z., Mann M. R., Gie Chung Y., et al. , 2005. X chromosome reactivation and regulation in cloned embryos. Dev. Biol. 279: 525–540. [DOI] [PubMed] [Google Scholar]
- Ogura A., Inoue K., Ogonuki N., Lee J., Kohda T., et al. , 2002. Phenotypic effects of somatic cell cloning in the mouse. Cloning Stem Cells 4: 397–405. [DOI] [PubMed] [Google Scholar]
- Ohgane J., Wakayama T., Kogo Y., Senda S., Hattori N., et al. , 2001. DNA methylation variation in cloned mice. Genesis 30: 45–50. [DOI] [PubMed] [Google Scholar]
- Ohsugi M., Zheng P., Baibakov B., Li L., Dean J., 2008. Maternally derived FILIA-MATER complex localizes asymmetrically in cleavage-stage mouse embryos. Development 135: 259–269. [DOI] [PubMed] [Google Scholar]
- Ozil J. P., Banrezes B., Toth S., Pan H., Schultz R. M., 2006. Ca2+ oscillatory pattern in fertilized mouse eggs affects gene expression and development to term. Dev. Biol. 300: 534–544. [DOI] [PubMed] [Google Scholar]
- Pan H., Schultz R. M., 2011. Sox2 modulates reprogramming of gene expression in two-cell mouse embryos. Biol. Reprod. 85: 409–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H., O’Brien M. J., Wigglesworth K., Eppig J. J., Schultz R. M., 2005. Transcript profiling during mouse oocyte development and the effect of gonadotropin priming and development in vitro. Dev. Biol. 286: 493–506. [DOI] [PubMed] [Google Scholar]
- Sarkies P., Reams C., Simpson L. J., Sale J. E., 2010. Epigenetic instability due to defective replication of structured DNA. Mol. Cell 40: 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnetz M. P., Handoko L., Akhtar-Zaidi B., Bartels C. F., Pereira C. F., et al. , 2010. CHD7 targets active gene enhancer elements to modulate ES cell-specific gene expression. PLoS Genet. 6: e1001023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seisenberger S., Peat J. R., Hore T. A., Santos F., Dean W., et al. , 2013. Reprogramming DNA methylation in the mammalian life cycle: building and breaking epigenetic barriers. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368: 20110330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobek-Klocke I., Disque-Kochem C., Ronsiek M., Klocke R., Jockusch H., et al. , 1997. The human gene ZFP161 on 18p11.21-pter encodes a putative c-myc repressor and is homologous to murine Zfp161 (Chr 17) and Zfp161-rs1 (X Chr). Genomics 43: 156–164. [DOI] [PubMed] [Google Scholar]
- Soboloff J., Rothberg B. S., Madesh M., Gill D. L., 2012. STIM proteins: dynamic calcium signal transducers. Nat. Rev. Mol. Cell Biol. 13: 549–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling P. C., Cuellar J., Alfaro G. A., El Khadali F., Beh C. T., et al. , 2006. PhLP3 modulates CCT-mediated actin and tubulin folding via ternary complexes with substrates. J. Biol. Chem. 281: 7012–7021. [DOI] [PubMed] [Google Scholar]
- Sun S. C., Zhang D. X., Lee S. E., Xu Y. N., Kim N. H., 2011. Ndc80 regulates meiotic spindle organization, chromosome alignment, and cell cycle progression in mouse oocytes. Microsc. Microanal. 17: 431–439. [DOI] [PubMed] [Google Scholar]
- Svoboda P., 2010. Why mouse oocytes and early embryos ignore miRNAs? RNA Biol. 7: 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor B. A., 1972. Genetic relationships between inbred strains of mice. J. Hered. 63: 83–86. [DOI] [PubMed] [Google Scholar]
- Terada N., Ohno N., Yamakawa H., Ohara O., Ohno S., 2005. Topographical significance of membrane skeletal component protein 4.1 B in mammalian organs. Anat. Sci. Int. 80: 61–70. [DOI] [PubMed] [Google Scholar]
- Terashita Y., Wakayama S., Yamagata K., Li C., Sato E., et al. , 2012. Latrunculin a can improve the birth rate of cloned mice and simplify the nuclear transfer protocol by gently inhibiting actin polymerization. Biol. Reprod. 86: 180. [DOI] [PubMed] [Google Scholar]
- Umbreit N. T., Gestaut D. R., Tien J. F., Vollmar B. S., Gonen T., et al. , 2012. The Ndc80 kinetochore complex directly modulates microtubule dynamics. Proc. Natl. Acad. Sci. USA 109: 16113–16118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassena R., Han Z., Gao S., Baldwin D. A., Schultz R. M., et al. , 2007a Tough beginnings: alterations in the transcriptome of cloned embryos during the first two cell cycles. Dev. Biol. 304: 75–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassena R., Han Z., Gao S., Latham K. E., 2007b Deficiency in recapitulation of stage-specific embryonic gene transcription in two-cell stage cloned mouse embryos. Mol. Reprod. Dev. 74: 1548–1556. [DOI] [PubMed] [Google Scholar]
- Wagschal A., Feil R., 2006. Genomic imprinting in the placenta. Cytogenet. Genome Res. 113: 90–98. [DOI] [PubMed] [Google Scholar]
- Wang L. J., Zhang H., Wang Y. S., Xu W. B., Xiong X. R., et al. , 2011. Scriptaid improves in vitro development and nuclear reprogramming of somatic cell nuclear transfer bovine embryos. Cell Reprogram 13: 431–439. [DOI] [PubMed] [Google Scholar]
- Wang S., Kou Z., Jing Z., Zhang Y., Guo X., et al. , 2010a Proteome of mouse oocytes at different developmental stages. Proc. Natl. Acad. Sci. USA 107: 17639–17644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Agarwala R., Capra J. A., Chen Z., Church D. M., et al. , 2010b High-throughput sequencing of the DBA/2J mouse genome. BMC Bioinformatics 11(Suppl. 4): 1–2.20043860 [Google Scholar]
- Whitworth K. M., Zhao J., Spate L. D., Li R., Prather R. S., 2011. Scriptaid corrects gene expression of a few aberrantly reprogrammed transcripts in nuclear transfer pig blastocyst stage embryos. Cell Reprogram 13: 191–204. [DOI] [PubMed] [Google Scholar]
- Xie Y., Zhong W., Wang Y., Trostinskaia A., Wang F., et al. , 2007. Using hyperosmolar stress to measure biologic and stress-activated protein kinase responses in preimplantation embryos. Mol. Hum. Reprod. 13: 473–481. [DOI] [PubMed] [Google Scholar]
- Yamanaka S., 2008. Induction of pluripotent stem cells from mouse fibroblasts by four transcription factors. Cell Prolif. 41(Suppl. 1): 51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurttas P., Vitale A. M., Fitzhenry R. J., Cohen-Gould L., Wu W., et al. , 2008. Role for PADI6 and the cytoplasmic lattices in ribosomal storage in oocytes and translational control in the early mouse embryo. Development 135: 2627–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurita E., Chagoyen M., Cantero M., Alonso R., Gonzalez-Neira A., et al. , 2011. Genetic polymorphisms among C57BL/6 mouse inbred strains. Transgenic Res. 20: 481–489. [DOI] [PubMed] [Google Scholar]