Abstract
Francisella tularensis is a facultative intracellular bacterial pathogen and the causative agent of tularemia. After infection of macrophages, the organism escapes from its phagosome and replicates to high density in the cytosol, but the bacterial factors required for these aspects of virulence are incompletely defined. Here, we describe the isolation and characterization of Francisella tularensis subsp. tularensis strain Schu S4 mutants that lack functional iglI, iglJ, or pdpC, three genes of the Francisella pathogenicity island. Our data demonstrate that these mutants were defective for replication in primary human monocyte-derived macrophages and murine J774 cells yet exhibited two distinct phenotypes. The iglI and iglJ mutants were similar to one another, exhibited profound defects in phagosome escape and intracellular growth, and appeared to be trapped in cathepsin D-positive phagolysosomes. Conversely, the pdpC mutant avoided trafficking to lysosomes, phagosome escape was diminished but not ablated, and these organisms replicated in a small subset of infected macrophages. The phenotype of each mutant strain was reversed by trans complementation. In vivo virulence was assessed by intranasal infection of BALB/c mice. The mutants appeared avirulent, as all mice survived infection with 108 CFU iglJ- or pdpC-deficient bacteria. Nevertheless, the pdpC mutant disseminated to the liver and spleen before being eliminated, whereas the iglJ mutant did not. Taken together, our data demonstrate that the pathogenicity island genes tested are essential for F. tularensis Schu S4 virulence and further suggest that pdpC may play a unique role in this process, as indicated by its distinct intermediate phenotype.
INTRODUCTION
Tularemia is a potentially fatal illness caused by the facultative intracellular Gram-negative bacterium Francisella tularensis (1). Infection can occur following inhalation of as few as 10 bacteria, after ingestion of contaminated food or water, or, most commonly, after introduction of bacteria into the skin via the bite of infected arthropod vectors (2). The two subspecies of the organism that account for nearly all cases of human tularemia differ in geographic distribution and virulence, with highly virulent Francisella tularensis subsp. tularensis (type A strains) restricted to North America and less virulent Francisella tularensis subsp. holarctica (type B strains) distributed throughout the Northern Hemisphere. An attenuated live vaccine strain (LVS) of F. tularensis subsp. holarctica was developed several decades ago in the Soviet Union but is not currently licensed for use in the United States because the genetic and molecular explanations of attenuation are unknown (1). A related organism, F. novicida, causes a tularemia-like disease in mice but is avirulent in healthy humans (1, 3).
Macrophages are phagocytes that play a central role in immunity and host defense and that can also be used by certain pathogens, including Francisella, as vehicles for bacterial amplification and dissemination (1, 4). For this reason, many studies have focused on elucidating the Francisella life cycle in this cell type. Receptors that mediate phagocytosis of opsonized and unopsonized bacteria have been described and include complement receptor 3 (CR3) (CD11b/CD18), complement receptor 4 (CR4) (CD11c/CD18), scavenger receptor A, mannose receptor, and nucleolin (5–10). Shortly after uptake, both F. tularensis and F. novicida disrupt phagosome maturation so that phagosome-lysosome fusion is impaired and bacteria escape from a late-endosome-like compartment to replicate in the macrophage cytosol (11–14). In this regard, it is noteworthy that LVS and all strains of virulent F. tularensis and F. novicida examined to date can replicate in primary human and murine macrophages and macrophage-like cell lines in vitro (4) despite the differences in host range and in vivo virulence noted above, and what accounts for this is unknown.
Central to Francisella virulence is a 30-kb segment of the genome called the Francisella pathogenicity island (FPI) that contains 16 to 19 predicted open reading frames organized in two large operons (15). The FPI was described first by Broms et al. and Nano et al. (15, 16) and is present in a single copy in F. novicida, yet it is duplicated in the genomes of all type A and type B F. tularensis strains, including LVS. Since F. novicida can be used in biosafety level 2 (BSL-2) laboratories, contains only a single copy of the FPI, and is more amenable to genetic manipulation than F. tularensis but still causes lethal disease in mice, the vast majority of studies to date have used the organism to define the roles of FPI genes in pathogenesis. Thus, it is established that the FPI regulatory factors MglA and FevR and at least some genes in the FPI, such as those within the intracellular growth locus (iglABCD) operon, are essential for Francisella virulence in mice and are required for phagosome escape, intracellular growth, and blockade of phagosome maturation in macrophages (13, 16–19). These studies have been extended to LVS and human-pathogenic F. tularensis strains with similar results (20–22).
Other genes within the FPI are less well characterized. However, it has been proposed that pdpB, vgrG, dotU, iglI, and iglJ may encode a type VI-like secretion system, with IglI as a candidate secreted effector (17, 23). Nevertheless, the phenotypes of iglI mutants generated in F. novicida and LVS are distinct, and explanations for these differences are lacking. Thus, whereas F. novicida iglI mutants are avirulent in mice and Drosophila melanogaster (17, 24, 25) and cannot escape the phagosome or replicate in J774A.1 macrophage-like cells in vitro (17, 23, 25, 26), this is not true for LVS iglI mutants, as these organisms grow normally in J774 cells, although their replication was constrained to some extent in peritoneal exudate macrophages (26). The gene encoding IglJ is adjacent to and downstream of iglI in the FPI. The phenotypes of F. novicida iglJ and iglI mutants are similar to one another, as strains lacking functional IglJ are also defective for phagosome escape and growth in both J774 cells and primary murine bone marrow-derived macrophages and do not cause disease in mice or flies (18, 23–25). We recently demonstrated that iglI and iglJ are not required for blockade of neutrophil NADPH oxidase activity by Schu S4 (27), but the effects of these genes on other type A F. tularensis virulence mechanisms have not yet been examined.
The FPI pathogenicity determinant protein C gene (pdpC) (15) is also of interest, as data in the literature are conflicting and argue both for and against a role for pdpC in regulation of F. novicida in vivo virulence, as well as bacterial growth in macrophages and mosquito cells in vitro (23–25, 28, 29). What accounts for these divergent findings is unclear. To date, the role of pdpC in type A F. tularensis virulence and intramacrophage interactions remains unexamined and awaits work to clarify these contradictory findings.
Previously, we reported the use of a transposon mutagenesis and screening strategy to identify genes in the virulent type A F. tularensis strain Schu S4 that are required for bacterial growth in human monocyte-derived macrophages (MDMs) (30). Although we did not expect to identify FPI genes in our mutagenesis strategy because of the FPI duplication, our results suggested that mutants with one functional and one disrupted copy of certain FPI genes may have subtle phenotypes. For this reason, and because the roles of iglI, iglJ, and pdpC in type A F. tularensis virulence were unknown, we sought to determine the fate of Schu S4 iglI, iglJ, and pdpC double-FPI-copy mutants in human MDMs and in mice after intranasal infection. Our results demonstrate that these FPI genes differentially contribute to Schu S4 virulence in vivo and in vitro. In particular, we report that the pdpC mutant has a distinct phenotype that differs from those of strains lacking functional iglI and iglJ and from most other FPI gene mutants characterized to date, as indicated by its ability to replicate in a subset of macrophages in vitro and its ability to disseminate in vivo before being eliminated by the host immune response.
MATERIALS AND METHODS
Macrophage isolation and culture.
Heparinized venous blood was obtained from healthy volunteers following a protocol approved by the Institutional Review Board for human subjects at the University of Iowa. Peripheral blood mononuclear cells were isolated by density gradient separation with Ficoll-Hypaque; washed twice in HEPES-buffered, endotoxin-free RPMI 1640 medium (here called RPMI) supplemented with l-glutamine (both from Lonza, Walkersville, MD); and incubated in RPMI plus 20% autologous human serum at a concentration of 2 × 106 cells/ml in Teflon jars for 5 to 7 days at 37°C, 5% CO2 for differentiation of monocytes into macrophages (31). MDMs were collected, washed with RPMI, counted on a hemocytometer, and plated according to experimental procedures described below. Replicate experiments were performed using cells from different donors.
Bacterial growth and opsonization.
Frozen stocks of wild-type F. tularensis subsp. tularensis strain Schu S4 were used to streak cultures on cysteine heart agar plates (Difco, Sparks, MD) supplemented with 9% defibrinated sheep blood (Remel, Lenexa, KS), and cultures were grown for 2 days at 37°C, 5% CO2. Trans-complemented mutant strains were grown on medium containing 25 μg/ml spectinomycin. Bacteria collected from agar plates were suspended and washed twice in Hanks' balanced salt solution (HBSS) with divalent cations (HBSS2+) (Mediatech, Inc., Manassas, VA). Opsonization was performed by incubation of bacteria in 50% human serum (pooled from a minimum of 14 donors with no history of tularemia) for 30 min at 37°C, followed by washing three times in HBSS. All work with F. tularensis was performed in the University of Iowa Carver College of Medicine Biosafety Level 3 Facility in accordance with all Centers for Disease Control and Prevention guidelines. Experimental protocols were approved by the Carver College of Medicine Biosafety Level 3 Oversight Committee, and recombinant DNA work was approved by the Institutional Biosafety Committee.
Construction of a pdpC mutant.
We previously reported generation of isogenic Schu S4 fevR, iglI, and iglJ mutants and trans-complemented strains (27). For this study, we again used group II intron retargeting (30, 32, 33) to disrupt pdpC in Schu S4. To this end, the Schu S4 pdpC sequence was analyzed for insertion sites using the Web-based TargeTron algorithm (Sigma-Aldrich, St. Louis, MO), and a site was selected for mutagenesis at nucleotide (nt) 608 of pdpC. A DNA fragment encoding a retargeted group II intron was generated by PCR and cloned into the XhoI and BsrGI sites of pKEK1140, and the plasmid was introduced into F. tularensis Schu S4 by cryotransformation (30). Transformants were initially grown on modified Mueller-Hinton (MMH) agar plates with 25 μg kanamycin at 30°C and then passaged twice on nonselective MMH agar at 40°C to cure the temperature-sensitive plasmid. After confirmation of plasmid curing, clones were analyzed by PCR across the insertion site using primers CTATGGGGATTAGTTGATTTCGTAATAG and GCTTTTGAGAAAAAAATATCATTTGCG. To create a complementation plasmid, the wild-type pdpC coding sequence was amplified by PCR from the Schu S4 chromosome so that EcoRI and SalI sites flanked the gene, and this amplicon was cloned into pBB103. The plasmid generated was confirmed by restriction digestion and designated pSL150. This plasmid was introduced into pdpC608 by cryotransformation to generate the trans-complemented pdpC608c strain (referred to as pdpC+).
Reverse transcription (RT)-PCR.
RNA was isolated from bacteria grown for 2 days on cysteine heart agar plates supplemented with 9% defibrinated sheep blood using TRIzol Reagent (Invitrogen, Life Technologies, CA) following the manufacturer's protocol. The RNA was treated with Ambion (Austin, TX) DNA-free rDNase I, and cDNA was made following the standard protocol for the Applied Biosystems (Foster City, CA) high-capacity cDNA reverse transcription kit. cDNA was used for subsequent PCR with primers designed to detect a partial fragment (nucleotides 81 to 599) (GCATGATATTAAAGAACTCAGAGTAGATGC and GCTCTTACCCAATATGGATCGGGA) or a full-length transcript (nucleotides 1503 to 1749) (TCTTCCTGGCTTCTTGAGCTCTGT and AGCAGTTGTTGAGGCAAATGCACC) of pdpC. Primers CCGTGCCAATTACCAGCAAA and GGCCCAGCCATCAGCTATAC were used for pdpE.
Serum sensitivity.
Sensitivity to lysis by serum complement was tested by incubating bacteria (1 × 107/ml) in either 50% human serum or phosphate-buffered saline (PBS) (mock treated) for 90 min at 37°C with shaking. After serial dilution, the bacteria were plated for CFU enumeration.
Intramacrophage growth.
MDMs were plated at 100,000 cells/well in 24-well tissue culture dishes in triplicate. The cells were allowed to adhere overnight in RPMI plus 10% human serum and then rinsed with warm PBS to remove nonadherent cells. Opsonized bacteria were added to MDMs at a multiplicity of infection (MOI) of 20:1 in RPMI plus 10% human serum, and samples were incubated for 1 h at 37°C to allow bacterial uptake. After rinsing three times with PBS, the cells were incubated for 45 min at 37°C in fresh RPMI plus 2.5% human serum and 100 μg/ml gentamicin to inhibit the growth of any remaining extracellular bacteria. Subsequently, the wells were washed three times with PBS to remove the antibiotic, and RPMI plus 2.5% human serum was added to each well. Some cells were lysed at this time point (2 h postinfection [p.i.]) by addition of 1% saponin, and serial dilutions were spotted onto agar plates for CFU enumeration. Other samples were processed similarly at 20 to 26 h p.i. Infection of MDMs with unopsonized bacteria was performed in a similar manner, except that heat-inactivated human serum was used instead of fresh serum, and the MOI was increased to 100:1.
Bacterial growth in J774A.1 macrophage-like cells was also quantified. J774 cells were plated at 200,000 cells/well in antibiotic-free Dulbecco's modified Eagle's medium (DMEM) (Lonza) plus 10% heat-inactivated fetal bovine serum (FBS) (HyClone, Logan, UT) and then infected with unopsonized bacteria at an MOI of 200:1. The initial infection efficiency and intracellular growth were quantified as described for MDMs.
For reinfection experiments, MDMs were infected at an MOI of 20:1 in RPMI containing 10% human serum. After 1 h, the cells were washed to remove free bacteria and returned to 37°C in medium supplemented with 2.5% human serum. Cell lysates prepared at 1 and 20 h p.i. were inoculated onto cysteine heart agar plates supplemented with 9% defibrinated sheep blood for CFU enumeration. Colonies from the 20-h samples were used to infect new MDMs, with quantitation of live intracellular bacteria at 1 and 20 h p.i.
Immunofluorescence microscopy.
MDMs were plated at 40,000 cells/well in Nunc Lab-Tek glass chamber slides (Thermo Fisher Scientific, Rochester, NY) and then infected with opsonized bacteria at an MOI of 20:1 as described above. At 7.5 and 20 h p.i., cells were processed for microscopy using our established methods (21, 34). In brief, samples were fixed for 25 min at room temperature using 10% formalin (Sigma-Aldrich) and then permeabilized in a 1:1 mixture of −20°C methanol-acetone for 15 min. The fixed and permeabilized cells were blocked at 4°C in PBS supplemented with 0.5 mg/ml NaN3, 5 mg/ml bovine serum albumin (BSA), and 10% horse serum. Samples were then double stained to detect bacteria (using rabbit anti-F. tularensis antiserum [BD Diagnostics] and lysosome-associated membrane protein 1 [LAMP-1] [human anti-mouse MAb H4A3 from the Developmental Studies Hybridoma Bank at the University of Iowa, Iowa City, IA] or cathepsin D [MAb G-19; Santa Cruz Biotechnology, Santa Cruz, CA]). Dylight 488-conjugated AffiniPure F(ab′)2 donkey anti-mouse and rhodamine-conjugated AffiniPure F(ab′)2 donkey anti-rabbit secondary antibodies were from Jackson Immuno Research Laboratories (West Grove, PA). All samples were analyzed using a Zeiss LSM-510 laser scanning confocal microscope (Carl Zeiss, Inc., Thornwood, NY). The number of infected cells and the number of bacteria per infected cell were determined after counting at least 100 infected cells within random fields of view. Colocalization of wild-type and mutant bacterial strains with LAMP-1 and cathepsin D was determined by analysis of at least 50 infected cells in triplicate samples at each time point. The data shown are the means and standard deviations (SD) from at least three independent experiments.
Phagosome integrity assay.
Phagosome disruption and bacterial access to the cytosol were quantified using the method of Checroun et al. with minor modifications (35). In brief, MDMs were plated in chamber slides and infected asynchronously with green fluorescent protein (GFP)-expressing wild-type or mutant bacterial strains as described above. At 1.5, 7.5, or 20 h p.i., 50 μg/ml digitonin in warm buffer (110 mM potassium acetate, 20 mM HEPES, 2 mM MgCl2, pH 7.3) was added for 1 min to selectively permeabilize the plasma membrane. The wells were then rinsed to remove residual digitonin, and anti-F. tularensis antiserum was added for 12 min at 37°C to label bacteria with access to the cytosol. Cells were fixed and stained with rhodamine-conjugated F(ab′)2 secondary antibodies. By this assay, all bacteria are GFP positive, whereas bacteria in disrupted phagosomes or free in the cytosol are both rhodamine and GFP positive. At least 100 infected cells for each bacterial strain and time point were analyzed, and the data shown are the means and SD from three independent experiments.
In vivo virulence and bacterial dissemination.
Animal studies were performed using 6- to 8-week-old female BALB/c mice obtained from the National Cancer Institute (Frederick, MD). In preparation to inoculate the mice intranasally, each animal was anesthetized using intraperitoneal avertin. Inocula were delivered in a total of 50 μl of PBS to each nare of an anesthetized mouse. Each strain was also diluted and plated onto agar plates to determine the actual number of CFU that were administered for each group of mice. Lethality studies utilized at least 5 mice per group. The mice were carefully monitored daily, and all animals showing signs of fatal disease were sacrificed. Dissemination was quantified by harvesting organs (lungs, liver, and spleen) from 3 mice per group on days 3, 5, 7, and 14 after infection. The organs were homogenized in PBS containing 3% saponin, and dilutions of each homogenate were spotted onto agar plates to quantitate the bacterial load in each organ by enumeration of CFU.
Statistical analysis.
The statistical significance of data sets (P < 0.05) was determined using analysis of variance (ANOVA) with Dunnett's or Tukey's posttest for multiple comparisons, Student's t test for paired samples, and log rank analysis using the Mantel-Cox test for analysis of Kaplan-Meier survival curves. All analyses were performed using GraphPad Prism 4 software.
RESULTS
F. tularensis Schu S4 iglI, iglJ, and pdpC mutants are defective for growth in primary human macrophages and murine J774A.1 cells.
We previously reported the construction of a transposon mutant library that was used to identify genes in F. tularensis strain Schu S4 that are required for bacterial growth and survival in human MDMs (30). Schu S4 contains two identical copies of the FPI, and in our screen, we identified mutants that contained one intact and one disrupted copy of iglI or iglJ, although mutating only one of the two copies of these genes only marginally reduced Schu S4 growth in MDMs (reference 30 and data not shown). To better define the possible roles of these FPI genes in virulence of type A F. tularensis, we constructed Schu S4 iglI-153 and iglJ-189 mutants (referred to here as iglI and iglJ), as well as trans-complemented strains (referred to here as iglI+ and iglJ+) (27). In addition, we also constructed a Schu S4 pdpC-608 mutant (referred to here as pdpC) using a type II integron mutagenesis strategy (see Materials and Methods) (Fig. 1). Disruption of pdpC was confirmed by PCR (Fig. 1B). A complemented strain that expresses wild-type pdpC in trans (referred to as pdpC+) was also prepared. As disruption of pdpC at position 608 may have caused polar effects on the downstream gene pdpE or resulted in expression of a truncated (and perhaps partially functional) product, we also isolated bacterial RNA and used RT-PCR to assess gene expression. Primers were designed to amplify the region between nucleotides 81 and 599, which should be present in both truncated and full-length transcripts, as well as the region between nucleotides 1503 and 1749 that is specific for full-length pdpC, and a third set of primers was designed to detect expression of pdpE. The data in Fig. 1C show that Schu S4 expressed both pdpC and pdpE, as amplicons were generated using all three primer sets. In contrast, disruption of pdpC appeared to ablate expression of the gene, as we found no evidence of full-length or truncated transcripts by RT-PCR. At the same time, expression of pdpE in the pdpC mutant strain appeared slightly diminished but was not ablated (Fig. 1C). These results demonstrate successful mutagenesis of pdpC in a manner that did not appear to result in synthesis of a truncated product and had at most a slight polar effect on the expression of pdpE.
Fig 1.
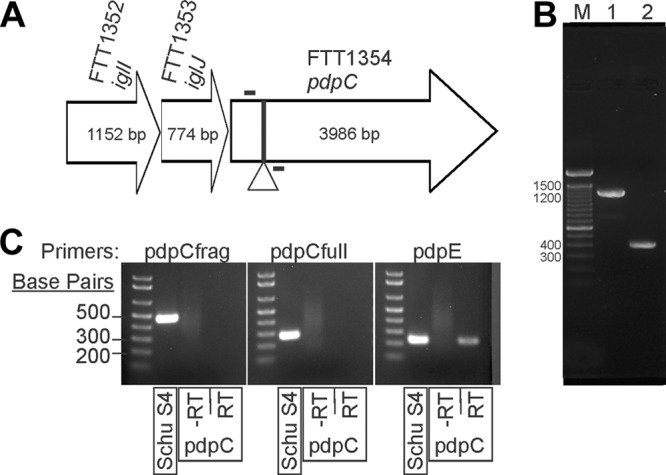
Three FPI genes with unknown contributions to F. tularensis Schu S4 virulence. (A) Schematic of iglI, iglJ, and pdpC genes within the FPI. The site of insertion is marked with a triangle (nt 608). (B) PCR amplification across the target region (the primers are marked by bars above and below the arrow in panel A) shows group II intron-mediated insertions (915 bp) into both copies of pdpC (lane 1) compared to the DNA control without insertions (lane 2). Lane M, molecular weight marker. (C) Primers were designed to amplify nucleotides 81 to 599 (pdpCfrag) or 1503 to 1749 (pdpCfull) of pdpC or a region of pdpE. Each primer set was used for RT-PCR of the wild type or a pdpC mutant. −RT, no-reverse transcriptase controls.
To characterize our mutants, we first determined if disruption of iglI, iglJ, or pdpC rendered the mutants sensitive to complement-mediated killing. To this end, bacteria were left in PBS or incubated in 50% pooled human serum and then plated for enumeration of CFU. By this assay, all three mutant strains were indistinguishable from wild-type Schu S4 and remained serum resistant (data not shown).
Next, we infected MDMs with serum-opsonized bacteria and quantified intracellular growth by enumeration of CFU at 2 and 20 h p.i., using wild-type Schu S4 and a fevR mutant as positive and negative controls, respectively. As shown in Fig. 2A, similar numbers of Schu S4 and the fevR, iglI, iglI+, iglJ, iglJ+, pdpC, and pdpC+ strains were recovered from MDMs at 2 h p.i., indicating similar initial infection efficiencies. By 20 h p.i., Schu S4 had undergone more than 2 log10 growth, whereas the viability of the fevR mutant declined, confirming a defect in intramacrophage replication and survival (36). On the other hand, the numbers of viable iglI and iglJ mutant organisms recovered at 2 and 20 h p.i. were similar, indicating a significant defect in intracellular growth relative to Schu S4. Nevertheless, these strains were not eliminated, suggesting greater resistance to intracellular killing than the fevR mutant. In marked contrast to the other strains examined, the pdpC mutant appeared to undergo limited replication in MDMs, as the number of viable bacteria recovered increased 1.93- ± 1.68-fold between 2 and 20 h p.i. As recent data suggest that the mechanism of uptake can affect intracellular growth (37), we also infected MDMs with unopsonized bacteria and obtained similar results (Fig. 2B). Evidence that the mutations in the genes of interest were responsible for the phenotypes observed was obtained by trans complementation, although intra-MDM growth of the iglI+, iglJ+, and pdpC+ strains did not reach wild-type levels (Fig. 2A and B). What accounts for this is unclear, but it is noteworthy that the complementation plasmid utilizes GroES rather than the native promoters, and the correct stoichiometry of FPI proteins is likely not achieved under these conditions.
Fig 2.
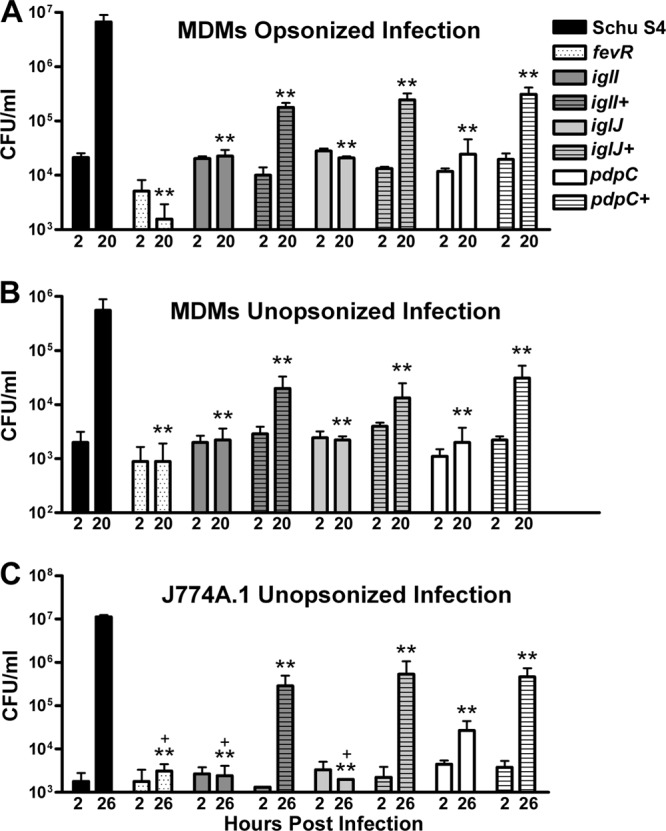
All three mutants are defective for growth in macrophages. (A) Human MDMs were infected with opsonized wild-type or mutant bacteria at an MOI of 20:1, and viable intracellular bacteria were quantified at 2 and 20 h postinfection by enumeration of CFU. The data shown are the means and SD of triplicate samples from one experiment representative of three. **, P < 0.01 versus Schu S4. (B) Similar to panel A, except MDMs were infected at an MOI of 100:1 in medium supplemented with heat-inactivated human serum. **, P < 0.01 versus Schu S4. (C) J774A.1 cells were infected with unopsonized bacteria at an MOI of 200:1, and viable intracellular bacteria were quantified at 2 and 26 h postinfection. The data are the means and SD from one experiment representative of three. **, P < 0.01 versus Schu S4; +, P < 0.05 versus the pdpC mutant.
As we have shown that a subset of Francisella mutants that are defective for growth in primary macrophages can still replicate in macrophage-like cell lines (38), we also quantified the replication of our unopsonized mutant strains in J774A.1 cells. These cells were infected to similar extents by all the strains examined; the pdpC mutant replicated to a limited extent (6.17- ± 3.75-fold) by 26 h p.i., whereas the other mutants did not; and intracellular growth was rescued by trans complementation of the iglI, iglJ, and pdpC mutants (Fig. 2C). Taken together, these data demonstrate that the FPI genes iglI, iglJ, and pdpC play important but perhaps distinct roles in intramacrophage growth of F. tularensis Schu S4.
The pdpC mutant replicates in a subset of infected macrophages.
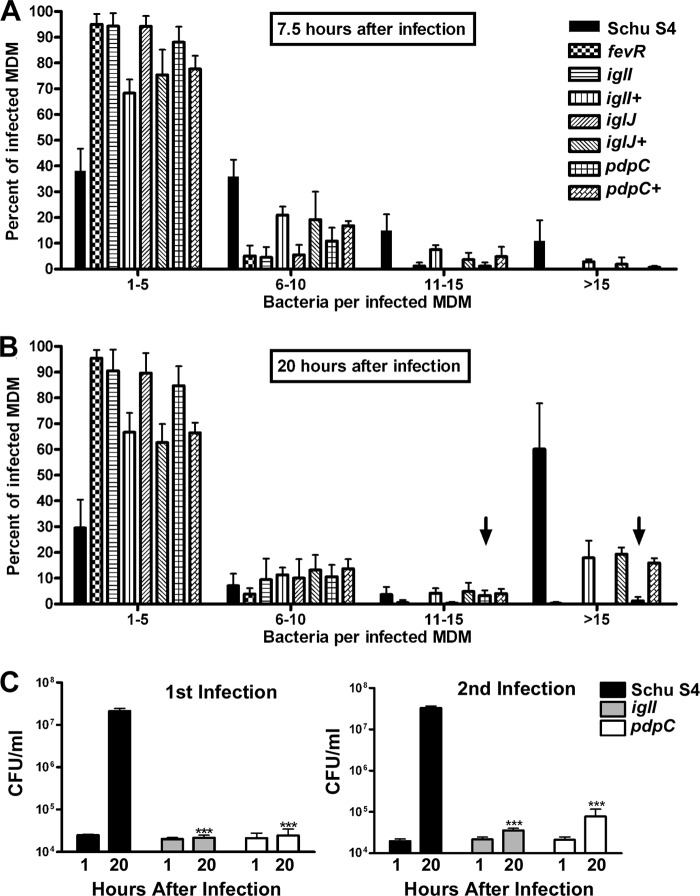
We used immunofluorescence microscopy to further characterize the intracellular phenotype and fate of mutant organisms by analysis of individual infected cells. To this end, MDMs were processed for confocal analysis at 7.5 and 20 h p.i. Pooled data from multiple experiments are shown in Fig. 3, and representative images of MDMs at 20 h p.i. are shown in Fig. 4 and 5.
Fig 3.
Quantitation of bacterial growth in macrophages using microscopy to analyze individual infected cells. (A and B) MDMs were infected for 7.5 h (A) or 20 h (B) with the indicated strains of bacteria and then analyzed by immunofluorescence microscopy. The number of bacteria per infected MDM was scored, and the data were grouped into four categories: 1 to 5, 6 to 10, 11 to 15, and >15 bacteria per cell. At least 100 infected MDMs were counted at each time point per experiment (n ≥ 3). Arrows highlight data that suggest low- level replication of the pdpC mutant strain at 20 h. The percentages of MDMs infected were similar for all strains (data not shown). (C) MDMs were infected for 1 h at an MOI of 20:1 in medium containing 10% human serum and washed, and CFU were enumerated at 1 and 20 h p.i. Viable recovered bacteria were then used to infect a second set of MDMs. ***, P < 0.001 versus Schu S4. The error bars indicate SD.
Fig 4.
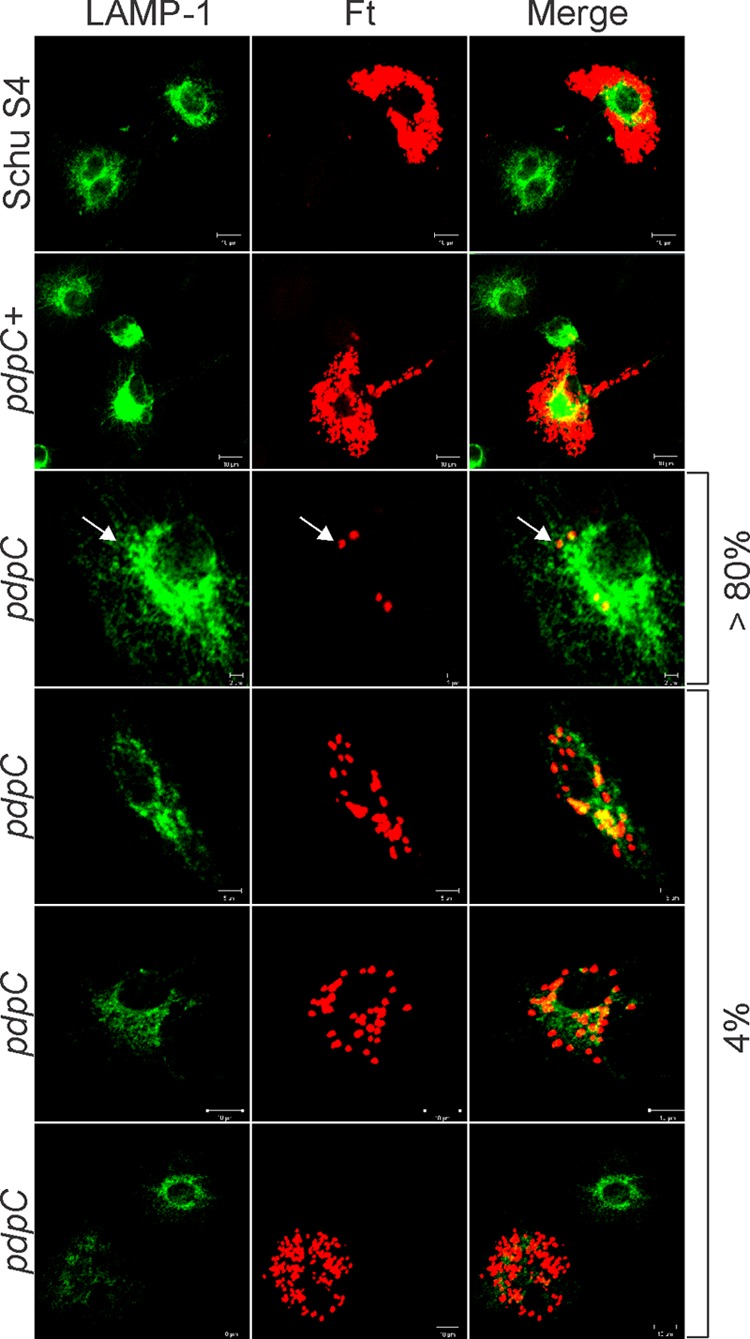
Two distinct intracellular phenotypes of the pdpC mutant. Shown are confocal images of MDMs infected for 20 h with Schu S4, the pdpC mutant, or the complemented pdpC+ strain. Bacteria are shown in red, and LAMP-1 is shown in green as an indicator of overall macrophage morphology. Schu S4 and the pdpC+ strain grew well in MDMs, whereas the pdpC mutant was defective for growth in 80% of infected cells and grew to a limited extent in ∼4% of infected MDMs, as indicated. The arrows indicate nonreplicating pdpC mutants that do not colocalize with LAMP-1. The data shown are representative of results from more than three independent experiments.
Fig 5.
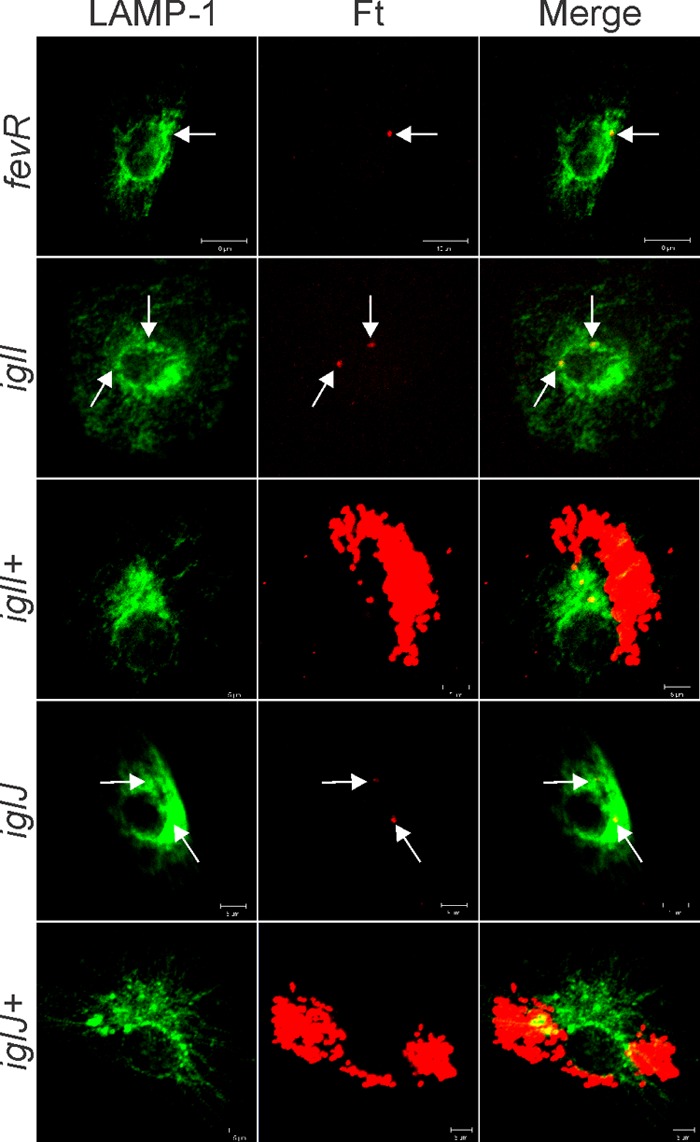
The iglI and iglJ mutants do not replicate in MDMs and resemble bacteria lacking functional fevR. Shown are confocal images of MDMs 20 h after infection with the indicated strain of bacteria. LAMP-1 is shown in green, and bacteria are shown in red. Note that the iglI+ and iglJ+ strains replicate in the MDM cytosol, whereas the iglI, iglJ, and fevR mutants (arrows) do not. The data shown are representative of at least three independent experiments.
At 7.5 h p.i. (Fig. 3A) nearly 40% of the Schu S4-infected MDMs contained 1 to 5 bacteria each. A similar fraction of infected cells contained 6 to 10 bacteria each, and the remainder of the cells contained 11 to 15 or >15 bacteria each, consistent with the onset of Schu S4 replication in MDMs by this time point (14, 30). In contrast, nearly all MDMs infected with mutant organisms contained only 1 to 5 bacteria. Results obtained for the iglI+, iglJ+, and pdpC+ strains were intermediate between those for wild-type and mutant organisms, as ∼70 to 80% of infected cells contained 1 to 5 bacteria, 16 to 20% contained 6 to 10 bacteria, and 3 to 8% contained 11 to 15 bacteria. Consistent with the robust growth of wild-type F. tularensis in MDMs shown in Fig. 2A, a majority of Schu S4-infected MDMs contained more than 15 bacteria each by 20 h p.i. (Fig. 3B and 4), whereas results for the fevR, iglI, and iglJ mutants were essentially unchanged (Fig. 3B and 5).
In contrast, we observed two distinct phenotypes within the population of MDMs infected with the pdpC mutant (Fig. 4). Approximately 85% of these cells contained few bacteria (1 to 5 each) without evidence of intracellular growth and in this way resembled MDMs infected with the other mutants examined in this study. Nevertheless, we also consistently observed a small subpopulation of infected MDMs (∼4.4% of the total) that contained large numbers of mutant bacteria that appeared to be replicating in the cytosol. Specifically, we found that 3.3% and 1.1% of MDMs infected with the pdpC mutant contained 11 to 15 bacteria or >15 bacteria, respectively, at 20 h p.i. (Fig. 3B, arrows). These results confirm and extend the data in Fig. 2, which indicate that some pdpC mutant organisms undergo limited replication in macrophages whereas the other mutant strains do not possess this ability. Although the data in Fig. 1C argue against expression of truncated, partially functional PdpC by our mutant, we also sought to determine whether our infection strategy may select for revertants or suppressor mutations that could also account for the atypical phenotype of the strain. To this end, MDMs were infected with the pdpC mutant; viable bacteria were recovered at 20 h p.i., quantified, and then used to reinfect new cells; and wild-type bacteria and the iglI mutant were used as controls. The data obtained for primary infection and reinfection were nearly identical for all strains tested (Fig. 3C) and are within the range of variability that we observe between different experiments. It is therefore highly unlikely that revertant or suppressor mutations account for the pdpC mutant phenotype.
Finally, our microscopy data also confirm the ability of trans complementation to restore rapid intracellular growth, as shown for pdpC+, iglI+, and iglJ+ strains in Fig. 3 to 5. Taken together, our CFU and microscopy data demonstrate that FPI genes iglI, iglJ, and pdpC are required for growth of type A F. tularensis in MDMs and that disruption of pdpC results in a phenotype that differs from those of mutant organisms lacking functional iglI, iglJ, or fevR.
Distinct roles of PdpC, IglI, and IglJ in phagosome escape.
To determine if the intramacrophage growth defects we observed could be attributed to defects in phagosome escape, we quantified phagosome integrity using an assay based on the method of Checroun et al. (35). By this assay, 50% of live, wild-type Schu S4 bacteria had access to the cytosol by 90 min postinfection (Fig. 6A). In contrast, only low-level background staining was detected when cells were infected with killed Schu S4 bacteria or when digitonin was omitted (∼13% cytosolic bacteria) (Fig. 6). Under the same conditions, the ability of the iglI, iglJ, and fevR mutants to breach the phagosome membrane was markedly impaired (P < 0.01 versus live Schu S4), and access of these organisms to the cytosol was increased by only 5 to 7% over the background obtained for the negative controls (Fig. 6), which was not statistically significant. Of note, our results for killed Schu S4 and the fevR mutant are in agreement with published data (14, 39). Moreover, the extent of phagosome escape we report for strains lacking functional iglI and iglJ is nearly identical to that for a Schu S4 ΔiglC mutant (14). In marked contrast, the ability of the pdpC mutant to breach the phagosome membrane and reach the cytosol was diminished but not ablated. Thus, at 90 min postinfection, ∼35% of the pdpC mutants were cytosolic, which represents a statistically significant increase (P < 0.01) relative to the killed Schu S4 control. We therefore conclude that PdpC contributes to but is not essential for phagosome escape, in contrast to IglI, IglJ, and FevR.
Fig 6.
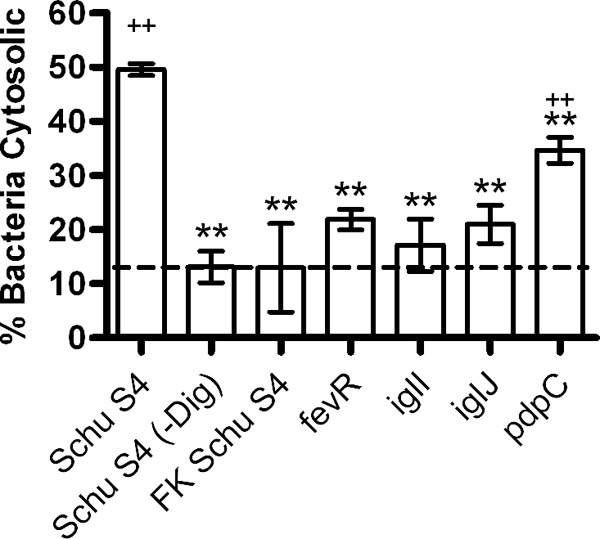
Different roles of PdpC, IglI, and IglJ in phagosome escape. Phagosome integrity was quantified 90 min after infection of human MDMs with GFP-expressing bacteria using digitonin to selectively permeabilize the plasma membrane and introduce anti-F. tularensis antibodies into the MDM cytosol. Background staining was determined using formalin-killed (FK) Schu S4 and by omission of digitonin. The data are the means ± SD from three independent experiments. **, P < 0.01 versus Schu S4; ++, P < 0.01 versus FK Schu S4.
Enhanced colocalization of mutant organisms with LAMP-1.
Next, we quantified the extent of bacterial colocalization with LAMP-1, a glycoprotein that is abundant in the membranes of late endosomes and lysosomes (11, 19, 21). At 7.5 h p.i., only ∼15% of wild-type bacteria colocalized with LAMP-1, in keeping with published findings (11, 14, 19) and the data shown in Fig. 3A and 6. In contrast, we and others have previously reported that fevR mutants in F. tularensis LVS and Schu S4 are defective for phagosome escape and are trapped in LAMP-1-positive compartments (21, 36), and we confirm these data here (Fig. 7A and B). In addition, we extended these results to show that colocalization of the iglI, iglJ, and pdpC mutants, but not the complemented strains, with LAMP-1 was also significantly enhanced relative to the wild type (Fig. 7A and B).
Fig 7.
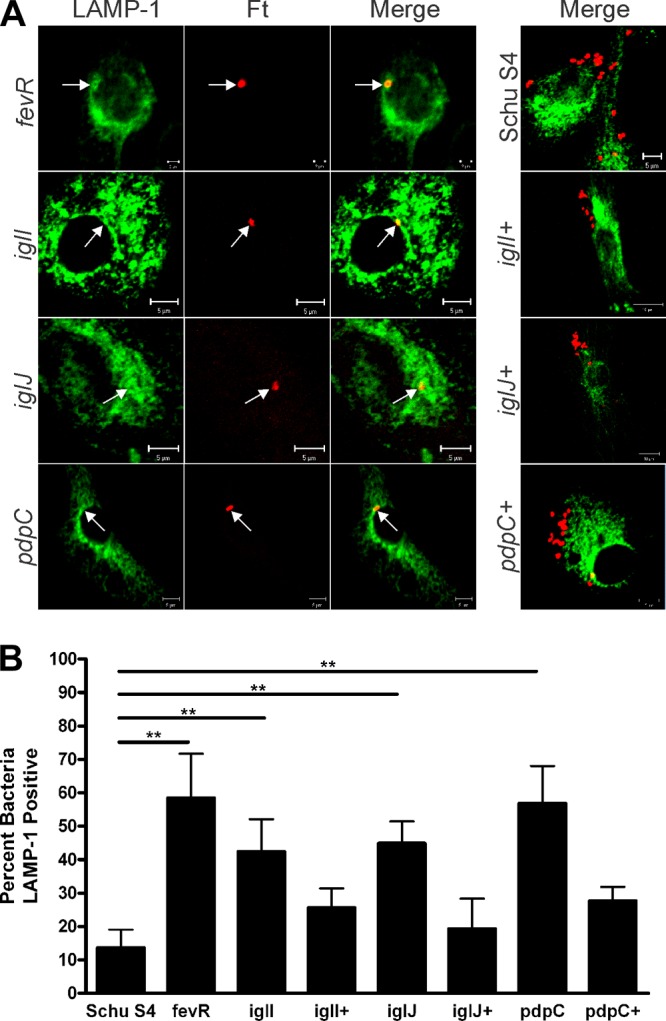
Colocalization of all three mutants with LAMP-1. MDMs were infected in parallel with Schu S4, fevR, iglI, iglI+, iglJ, iglJ+, pdpC, or pdpC+ organisms. At 7.5 h postinfection, samples were processed for confocal microscopy and double stained to detect bacteria (red) and LAMP-1 (green). (A) Representative images. The arrows indicate bacteria that colocalize with LAMP-1. (B) Pooled data are the means and SD from multiple independent experiments (n ≥ 4). **, P < 0.01 versus Schu S4.
IglI and IglJ are required to prevent phagosome-lysosome fusion.
Previous studies have shown that wild-type F. tularensis escapes from a LAMP-1-positive and cathepsin D-negative compartment to replicate in the macrophage cytosol (11), and some regulator or FPI gene mutants that are defective for phagosome escape are also unable to prevent phagosome-lysosome fusion (13, 21, 40). Therefore, we also quantified phagosome maturation by analyzing the extent of mutant colocalization with cathepsin D. As expected, only about 10% of Schu S4 colocalized with cathepsin D at 7.5 h p.i. (Fig. 8A and B). In contrast, colocalization with this lysosomal enzyme was moderately but significantly increased by disruption of fevR and more profoundly affected by disruption of iglI or iglJ (Fig. 8A and B). Colocalization of the pdpC mutant with cathepsin D was also enhanced but did not reach statistical significance (P > 0.05) when all samples were analyzed in parallel using ANOVA (Fig. 8A and B). A similar pattern was obtained when each mutant was compared to the wild type by Student's t test, though in this case the results for the pdpC strain did reach statistical significance: P = 0.0001 for iglJ, P = 0.0016 for iglI, P = 0.0022 for fevR, and P = 0.0185 for pdpC. In contrast, the complemented strains resembled wild-type Schu S4. These data strongly suggest that only a subset of FPI genes are essential to prevent F. tularensis trafficking to lysosomes and that IglI and IglJ play critical roles in this process, whereas PdpC may not.
Fig 8.

iglI, iglJ, and pdpC differentially affect phagosome maturation. MDMs were infected in parallel with Schu S4, fevR, iglI, iglI+, iglJ, iglJ+, pdpC, or pdpC+ organisms. At 7.5 h postinfection, samples were processed for confocal microscopy and double stained to detect bacteria (red) and cathepsin D (green). (A) Representative images. The arrows indicate bacteria that colocalize with cathepsin D. (B) Pooled data are the means and SD from multiple independent experiments (n ≥ 4). *, P < 0.05, and **, P < 0.01 versus Schu S4; ns, not significant [P > 0.05]).
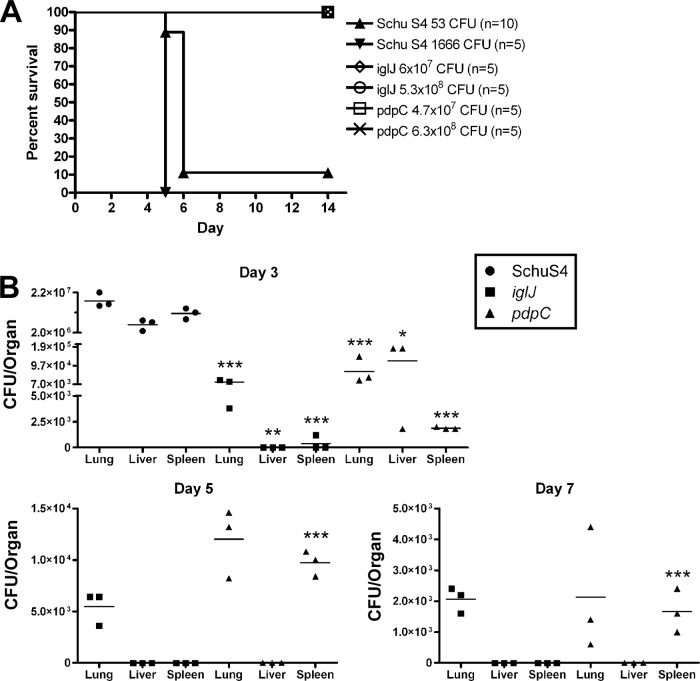
pdpC and iglJ mutants are attenuated for virulence in vivo, despite dissemination of bacteria lacking PdpC.
Finally, we performed intranasal infections of BALB/c mice to evaluate the contributions of IglJ and PdpC to in vivo virulence. In our hands, mice succumbed to infection with wild-type Schu S4 within 6 days after inoculation with 53 or 1,666 CFU (Fig. 9A). The virulence of the iglJ and pdpC mutants was examined by challenging mice in parallel with approximately 107 to 108 CFU. Despite the high inocula, all mice remained healthy, did not show signs of disease, and were alive at day 14 after infection. These data extend our in vitro findings to demonstrate that IglJ and PdpC are essential for virulence of Schu S4 in a mouse intranasal-infection model.
Fig 9.
In vivo virulence and dissemination assays confirm the distinct phenotype of the pdpC mutant. (A) Groups of 5 or 10 BALB/c mice were infected intranasally with the indicated dose of wild-type or mutant bacteria. The data shown are from a representative experiment and demonstrate that both the iglJ and pdpC strains are highly attenuated at ∼108 CFU while wild-type Schu S4 is lethal at 53 CFU, and mice succumb to infection by day 5 or 6. (B) Mice in groups of three were intranasally infected with 5.67 × 106 CFU iglJ, 3.0 × 106 CFU pdpÇ or 2.0 × 103 CFU Schu S4 strain. Dissemination from the lungs to the liver and spleen was determined by plating organ homogenates for enumeration of CFU on days 3, 5, 7, and 14 postinfection. Schu S4 data are shown only for day 3, as all mice succumbed by day 5. By day 14, iglJ and pdpC bacteria had been cleared and were not detected (data not shown). Horizontal lines indicate the mean results for each group of mice at each time point. *, P < 0.05; **, P < 0.01; ***, P < 0.001 versus Schu S4 (day 3) or iglJ (day 5 and 7) samples.
Although both mutants were avirulent at a dose of 108 CFU, we wanted to further examine the hypothesis that the pdpC mutant may be able to undergo limited growth or dissemination in mice, as suggested by the in vitro phenotypic differences we observed with the strain. To this end, mice were infected with ∼106 CFU of the iglJ and pdpC mutants or 103 CFU of Schu S4, and lungs, livers, and spleens were harvested on days 3, 5, 7, and 14 after infection. By day 3, Schu S4 had grown to very high levels in the lung relative to the initial inoculum and had also disseminated to the liver and spleen (Fig. 9B), confirming published data (20). At the same time point, both the iglJ and pdpC mutants were recovered from the lung, albeit at levels lower than the initial inocula. The iglJ mutant was defective for dissemination, as it was not detected in the liver and was just above the threshold of detection in the spleen of one mouse. In contrast, the pdpC mutant was readily detected in both the liver and the spleen.
By day 5, all Schu S4-infected mice had succumbed to infection or were euthanized due to moribund status. In contrast, none of the mice infected with either the iglJ or pdpC mutant displayed signs of disease through day 14. Despite this, the iglJ mutant was detected in the lung, but not the liver or spleen, on days 5 and 7 postinfection. On the other hand, the pdpC mutant was recovered in even greater numbers from the spleen on day 5 than on day 3 and persisted in the lung yet was cleared from the liver. The pdpC mutant was also recovered from mouse lungs and spleens on day 7, though bacterial burdens in these tissues had begun to decline, and by day 14 both mutants had been cleared from all three tissues (Fig. 9B and data not shown). The ability of the pdpC mutant to disseminate, particularly to the spleen, contrasts with the fate of the iglJ strain and is consistent with the phenotypic differences that we observed in vitro. A role for PdpC in F. tularensis virulence in vivo was further assessed by infecting mice with 500 CFU of the complemented pdpC+ strain, and all five animals succumbed by day 5 or 6 (data not shown). Considered together, the results of this study indicate that although both IglJ and PdpC are required for virulence, the ability of the pdpC mutant to replicate to a limited extent in murine organs in vivo, and in murine and human macrophages in vitro, indicates that disruption of pdpC in fully virulent type A F. tularensis results in a previously unappreciated and distinct phenotype compared with other FPI mutants examined to date.
DISCUSSION
Significant research efforts over the past decade have focused on understanding the molecular and genetic features responsible for the pathogenesis of tularemia. These studies have utilized isolates of several different Francisella species and subspecies, including F. novicida strain U112, F. tularensis subsp. holarctica LVS, and F. tularensis subsp. tularensis strain Schu S4, and have examined the fates of these organisms in insect and mammalian cells and in animal models. Each of these experimental systems has both desirable and undesirable aspects. Several seminal discoveries in the field were made using F. novicida, and this was facilitated by the relative ease of genetic manipulation of the organism, its ability to cause a tularemia-like disease in mice, and the ability to perform experiments without the precautions and containment required for studies of select agent strains of F. tularensis (23, 41–43). LVS has some of the same advantages, but its mechanism of attenuation is not defined, and neither F. novicida nor LVS causes disease in healthy humans, despite retaining the ability to replicate in human macrophages in vitro (3). A major challenge remains in the field to identify and characterize genes and other traits that account for the broad host range of the organism in general and the pronounced virulence of type A F. tularensis strains in humans. It is possible that individual Francisella genes may affect virulence in one host or cell type but not another. Mouse models are attractive due to the powerful genetic tools available; however, it should be noted that murine and human macrophages are not identical in their cellular physiology, especially in response to pathogenic microbes (44, 45). The results of this study demonstrate that FPI genes, iglI, iglJ, and pdpC, differentially contribute to virulence of the human-pathogenic type A F. tularensis strain Schu S4.
In our hands, disruption of either iglI or iglJ in Schu S4 had no effect on bacterial uptake by MDMs or J774 macrophage-like cells in the presence or absence of opsonins yet ablated intracellular growth. At the cellular level, both mutants had profound defects in phagosome escape, as judged by differential staining, and were unable to prevent phagosome maturation, as indicated by their enhanced colocalization with cathepsin D, as well as LAMP-1. These data are consistent with studies by two groups that reported defective replication of F. novicida ΔiglI mutants in murine bone marrow-derived macrophages (17) and J774 cells (23) and are also in keeping with a model in which IglI is translocated into the macrophage cytosol by a type VI-like secretion system encoded within the FPI (17). However, a different phenotype has been reported for ΔiglI mutants generated in the LVS background (26). This ΔiglI mutant is defective for growth in murine peritoneal exudate macrophages but not in J774 cells by CFU analysis, although, interestingly, it exhibited increased colocalization with LAMP-1 and impaired phagosome escape, as judged by transmission electron microscopy (26). Also, in contrast to our findings and to parallel studies of iglI in this organism, deletion of iglJ in F. novicida has little or no effect on bacterial growth in J774 cells, after correcting for diminished uptake relative to the wild type (23). What accounts for these diverse phenotypes is unclear, as they cannot be attributed to the FPI copy number, the macrophage species, the diminished microbicidal capacity of cell lines, or uptake in the presence or absence of opsonins, and we extended our in vitro studies to show that iglJ is also required for Schu S4 virulence in a mouse intranasal-infection model. We therefore conclude that iglI and iglJ are essential for Schu S4 virulence. In future studies, it will be important to elucidate the specific functions of IglI and IglJ with a focus on bacterial fate in macrophages and to determine whether their concerted actions are particularly important for the extreme virulence of type A F. tularensis.
The literature regarding the role of pdpC in Francisella virulence is also complex and somewhat contradictory. A report from Weiss et al. used an F. novicida transposon mutagenesis library to identify genes that were negatively selected in a mouse intraperitoneal-infection model and identified, in addition to pdpC, all FPI genes, including iglI and iglJ, as required for in vivo virulence (24). de Bruin et al. also found that pdpC is required for in vivo virulence of F. novicida yet at the same time reported that the gene is dispensable for bacterial replication in J774 cells (23). In contrast, Barker and Klose reported that pdpC is important for F. novicida growth in the same cell type, though the data were not shown in the publication (42). Additional studies in insects also obtained divergent results. Thus, one screen identified pdpC as being required for virulence of F. novicida in D. melanogaster (29), whereas another, similar screen identified all FPI genes as important except pdpC, pdpE, pdpD, and anmK (25). pdpC is also dispensable for F. novicida infection of Sua1B cells, which are derived from the Anopheles gambiae mosquito (28). Altogether, these data suggest that, at least for F. novicida, pdpC may play a role in virulence under certain conditions and/or during infection of cell types other than macrophages.
FSC043 is a spontaneous Schu S4 variant that is attenuated in mice (20). Subsequent analysis revealed that the FSC043 strain contains four insertions/deletions not present in Schu S4 in FTT0615 and FTT0918/FTT0919, as well as single-base-pair deletions in each copy of pdpC—FTT1354 (pdpC1) and FTT1709 (pdpC2) (46). Disruption of pdpC and the other affected genes in this manner likely introduced frameshift mutations, and growth of the FSC043 strain in peritoneal exudate macrophages is markedly impaired compared to Schu S4, but the relative contribution of each mutation to this phenotype is unknown (20).
For this study, we generated a pdpC mutant in F. tularensis Schu S4. Our mutagenesis strategy appeared to ablate pdpC, expression of the downstream gene pdpE was only slightly diminished, and we report subtle yet significant in vitro phenotypic deficits in the strain that are consistent with the results of our in vivo studies. Specifically, our data indicate that loss of functional pdpC diminished but did not ablate phagosome escape and that the mutant organisms replicated in a small subset of infected macrophages. As ∼35% of these organisms reached the cytosol yet growth occurred in only ∼4% of infected cells, our results indicate that phagosome escape is necessary but not sufficient for replication of the mutant in MDMs. At the same time, our data also suggest that pdpC may not be essential for the blockade of phagosome-lysosome fusion, as mutants that did not reach the cytosol resided in LAMP-1-positive compartments but did not exhibit markedly enhanced colocalization with cathepsin D, unlike strains lacking functional iglI or iglJ. These data are consistent with the idea that dissolution of the phagosome membrane and inhibition of phagosome maturation are distinct aspects of the Francisella life cycle in host cells and as such may be controlled by different subsets of virulence factors. Moreover, the distinct pdpC mutant phenotype we report here could easily have been overlooked if bacterium-macrophage interactions had been analyzed only by measurement of CFU or if our in vivo studies had not included quantitation of bacterial dissemination to the liver and spleen. It is also noteworthy that in vivo virulence was restored by complementation with a wild-type copy of pdpC expressed in trans. These data are strong evidence that the phenotypes we report are predominantly, if not exclusively, attributable to disruption of pdpC rather than diminished expression of pdpE. As at least some capacity for intracellular and in vivo growth appears to be essential for generation of protective immunity to the organism (20, 47), it will be of interest in future studies to determine whether pdpC-deficient strains of type A F. tularensis may be effective vaccines.
The function of PdpC is unknown. Kinetic analyses of F. tularensis gene expression in murine macrophages suggest that pdpC is downregulated at all time points examined between 1 and 24 h after infection, whereas expression of all other FPI genes is enhanced, albeit to different extents (36). As judged by immunoblotting, PdpC is relatively stable, whereas IglC increased sharply during the first 4 h of infection (36). These data suggest that pdpC expression is regulated differently than that of other FPI genes, and our findings suggest that PdpC may play a distinct role in virulence. These data raise several questions. Specifically, what are the events that allow replication in such a small subset of pdpC mutant-organism-infected cells? Is there a unique subset of macrophages that support this outcome? To what extent is PdpC required for F. tularensis growth in other cell types, including dendritic cells, epithelial cells, and neutrophils? Perhaps more interesting will be to further characterize the fate of the pdpC mutant in vivo by examining what cell populations it is able to infect and how the organism disseminates.
In summary, the results of this study demonstrate that iglI, iglJ, and pdpC are all required for intramacrophage growth of the type A F. tularensis strain Schu S4. IglI and IglJ are essential for phagosome escape and blockade of bacterial trafficking to lysosomes in MDMs. In contrast, PdpC contributes to, but is not essential for, disruption of the phagosome membrane, and pdpC mutants replicate in a subset of infected macrophages in vitro. Moreover, the pdpC mutant disseminates from the lung to the liver and spleen in mice before being eliminated in vivo, whereas bacteria lacking functional iglJ do not. Our findings are noteworthy, as the phenotype we report for pdpC is atypical relative to other FPI genes examined to date and because we define potentially important differences between human-pathogenic type A F. tularensis strains, F. novicida, and LVS.
ACKNOWLEDGMENTS
This study utilized the Carver College of Medicine BSL-3 Laboratory Core Facility at the University of Iowa, which is supported by user fees and the generous support of the Roy J. and Lucille A. Carver College of Medicine. The study was supported in part by funds from the NIH/NIAID (P01 AI044642) to L.-A.H.A. and by grant U54 AI057160 awarded to L.-A.H.A. and B.D.J. by the Midwest Regional Center for Excellence for Biodefense and Emerging Infectious Diseases Research (MRCE).
We thank Jason Barker (University of Iowa) for helpful discussions regarding experimental procedures and results.
Footnotes
Published ahead of print 28 December 2012
REFERENCES
- 1.McLendon MK, Apicella MA, Allen L-AH. 2006. Francisella tularensis: taxonomy, genetics, and immunopathogenesis of a potential agent of biowarfare. Annu. Rev. Microbiol. 60:167–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saslaw S, Eigelsbach HT, Prior JA, Wilson HE, Carhart S. 1961. Tularemia vaccine study. II. Respiratory challenge. Arch. Intern. Med. 107:702–714 [DOI] [PubMed] [Google Scholar]
- 3.Ellis J, Oyston PC, Green M, Titball RW. 2002. Tularemia. Clin. Microbiol. Rev. 15:631–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sjostedt A. 2007. Tularemia: history, epidemiology, pathogen physiology, and clinical manifestations. Ann. N. Y. Acad. Sci. 1105:1–29 [DOI] [PubMed] [Google Scholar]
- 5.Schulert GS, Allen L-AH. 2006. Differential infection of mononuclear phagocytes by Francisella tularensis: role of the macrophage mannose receptor. J. Leukoc. Biol. 80:563–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clemens DL, Lee BY, Horwitz MA. 2005. Francisella tularensis enters macrophages via a novel process involving pseudopod loops. Infect. Immun. 73:5892–5902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pierini LM. 2006. Uptake of serum-opsonized Francisella tularensis by macrophages can be mediated by class A scavenger receptors. Cell. Microbiol. 8:1361–1370 [DOI] [PubMed] [Google Scholar]
- 8.Balagopal AA, . MacFarlane S, Mohapatra N, Soni S, Gunn JS, Schlesinger LS. 2006. Characterization of the receptor-ligand pathways important for entry and survival of Francisella tularensis in human macrophages. Infect. Immun. 74:5114–5125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barel M, Hovanessian AG, Meibom K, Briand JP, Dupuis M, Charbit A. 2008. A novel receptor-ligand pathway for entry of Francisella tularensis in monocyte-like THP-1 cells: interaction between surface nucleolin and bacterial elongation factor Tu. BMC MIcrobiol. 8:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwartz JT, Barker JH, Long ME, Kaufman J, McCracken J, Allen L-AH. 2012. Natural IgM mediates complement-dependent uptake of Francisella tularensis by human neutrophils via complement receptors 1 and 3 in nonimmune serum. J. Immunol. 189:3064–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clemens DL, Lee BY, Horwitz MA. 2004. Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect. Immun. 72:3204–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Golovliov I, Baranov V, Krocova Z, Kovarova H, Sjostedt A. 2003. An attenuated strain of the facultative intracellular bacterium Francisella tularensis can escape the phagosome of monocytic cells. Infect. Immun. 71:5940–5950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santic M, Molmeret M, Klose KE, Jones S, Abu Kwaik Y. 2005. The Francisella tularensis pathogenicity island protein IglC and its regulator MglA are essential for modulating phagosome biogenesis and subsequent bacterial escape into the cytoplasm. Cell. Microbiol. 7:969–979 [DOI] [PubMed] [Google Scholar]
- 14.Chong A, Wehrly TD, Nair V, Fischer ER, Barker JR, Klose KE, Celli J. 2008. The early phagosomal stage of Francisella tularensis determines optimal phagosomal escape and Francisella pathogenicity island protein expression. Infect. Immun. 76:5488–5499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Broms JE, Sjostedt A, Lavander M. 2010. The role of the Francisella tularensis pathogenicity island in type VI secretion, intracellular survival, and modulation of host cell signaling. Front. Microbiol. 1:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nano FE, Zhang N, Cowley SC, Klose KE, Cheung KK, Roberts MJ, Ludu JS, Letendre GW, Meierovics AI, Stephens G, Elkins KL. 2004. A Francisella tularensis pathogenicity island required for intramacrophage growth. J. Bacteriol. 186:6430–6436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barker JR, Chong A, Wehrly TD, Yu JJ, Rodriguez SA, Liu J, Celli J, Arulanandam BP, Klose KE. 2009. The Francisella tularensis pathogenicity island encodes a secretion system that is required for phagosome escape and virulence. Mol. Microbiol. 74:1459–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brotcke A, Weiss DS, Kim CC, Chain P, Malfatti S, Garcia E, Monack DM. 2006. Identification of MglA-regulated genes reveals novel virulence factors in Francisella tularensis. Infect. Immun. 74:6642–6655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmerk CL, Duplantis BN, Howard PL, Nano FE. 2009. A Francisella novicida pdpA mutant exhibits limited intracellular replication and remains associated with the lysosomal marker LAMP-1. Microbiology 155:1498–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Twine S, Bystrom M, Chen W, Forsman M, Golovliov I, Johansson A, Kelly J, Lindgren H, Svensson K, Zingmark C, Conlan W, Sjostedt A. 2005. A mutant of Francisella tularensis strain SCHU S4 lacking the ability to express a 58-kilodalton protein is attenuated for virulence and is an effective live vaccine. Infect. Immun. 73:8345–8352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buchan BW, McCaffrey RL, Lindemann SR, Allen L-AH, Jones BD. 2009. Identification of migR, a regulatory element of the Francisella tularensis live vaccine strain iglABCD virulence operon required for normal replication and trafficking in macrophages. Infect. Immun. 77:2517–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kadzhaev K, Zingmark C, Golovliov I, Bolanowski M, Shen H, Conlan W, Sjostedt A. 2009. Identification of genes contributing to the virulence of Francisella tularensis SCHU S4 in a mouse intradermal infection model. PLoS One 4:e5463 doi:10.1371/journal.pone.0005463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Bruin OM, Duplantis BN, Ludu JS, Hare RF, Nix EB, Schmerk CL, Robb CS, Boraston AB, Hueffer K, Nano FE. 2011. The biochemical properties of the Francisella pathogenicity island (FPI)-encoded proteins, IglA, IglB, IglC, PdpB and DotU, suggest roles in type VI secretion. Microbiology 157:3483–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weiss DS, Brotcke A, Henry T, Margolis JJ, Chan K, Monack DM. 2007. In vivo negative selection screen identifies genes required for Francisella virulence. Proc. Natl. Acad. Sci. U. S. A. 104:6037–6042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahlund MK, Ryden P, Sjostedt A, Stoven S. 2010. Directed screen of Francisella novicida virulence determinants using Drosophila melanogaster. Infect. Immun. 78:3118–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Broms JE, Lavander M, Meyer L, Sjostedt A. 2011. IglG and IglI of the Francisella pathogenicity island are important virulence determinants of Francisella tularensis LVS. Infect. Immun. 79:3683–3696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCaffrey RL, Schwartz JT, Lindemann SR, Moreland JG, Buchan BW, Jones BD, Allen L-AH. 2010. Multiple mechanisms of NADPH oxidase inhibition by type A and type B Francisella tularensis. J. Leukoc. Biol. 88:791–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Read A, Vogl SJ, Hueffer K, Gallagher LA, Happ GM. 2008. Francisella genes required for replication in mosquito cells. J. Med. Entomol. 45:1108–1116 [DOI] [PubMed] [Google Scholar]
- 29.Moule MG, Monack DM, Schneider DS. 2010. Reciprocal analysis of Francisella novicida infections of a Drosophila melanogaster model reveal host-pathogen conflicts mediated by reactive oxygen and imd-regulated innate immune response. PLoS Pathog. 6:e1001065 doi:10.1371/journal.ppat.1001065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindemann SR, Peng K, Long ME, Hunt JR, Apicella MA, Monack DM, Allen L-AH, Jones BD. 2011. Francisella tularensis Schu S4 O-antigen and capsule biosynthesis gene mutants induce early cell death in human macrophages. Infect. Immun. 79:581–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horwitz MA, Silverstein SC. 1980. Legionnaires' disease bacterium (Legionella pneumophila) multiplies intracellularly in human monocytes. J. Clin. Invest. 66:441–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez SA, Davis G, Klose KE. 2009. Targeted gene disruption in Francisella tularensis by group II introns. Methods 49:270–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodriguez SA, Yu JJ, Davis G, Arulanandam BP, Klose KE. 2008. Targeted inactivation of Francisella tularensis genes by group II introns. Appl. Environ. Microbiol. 74:2619–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwartz JT, Allen L-AH. 2006. Role of urease in megasome formation and Helicobacter pylori survival in macrophages. J. Leukoc. Biol. 79:1214–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Checroun C, Wehrly TD, Fischer ER, Hayes SF, Celli J. 2006. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc. Natl. Acad. Sci. U. S. A. 103:14578–14583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wehrly TD, Chong A, Virtaneva K, Sturdevant DE, Child R, Edwards JA, Brouwer D, Nair V, Fischer ER, Wicke L, Curda AJ, Kupko JJ, III, Martens C, Crane DD, Bosio CM, Porcella SF, Celli J. 2009. Intracellular biology and virulence determinants of Francisella tularensis revealed by transcriptional profiling inside macrophages. Cell Microbiol. 11:1128–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geier H, Celli J. 2011. Phagocytic receptors dictate phagosomal escape and intracellular proliferation of Francisella tularensis. Infect. Immun. 79:2204–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schulert GS, McCaffrey RL, Buchan BW, Lindemann SR, Hollenback C, Jones BD, Allen L-AH. 2009. Francisella tularensis genes required for inhibition of the neutrophil respiratory burst and intramacrophage growth identified by random transposon mutagenesis of strain LVS. Infect. Immun. 77:1324–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Child R, Wehrly TD, Rockx-Brouwer D, Dorward DW, Celli J. 2010. Acid phosphatases do not contribute to the pathogenesis of type A Francisella tularensis. Infect. Immun. 78:59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bonquist L, Lindgren H, Golovliov I, Guina T, Sjostedt A. 2008. MglA and Igl proteins contribute to the modulation of Francisella tularensis live vaccine strain-containing phagosomes in murine macrophages. Infect. Immun. 76:3502–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nano FE, Schmerk C. 2007. The Francisella pathogenicity island. Ann. N. Y. Acad. Sci. 1105:122–137 [DOI] [PubMed] [Google Scholar]
- 42.Barker JR, Klose KE. 2007. Molecular and genetic basis of pathogenesis in Francisella tularensis. Ann. N. Y. Acad. Sci. 1105:138–159 [DOI] [PubMed] [Google Scholar]
- 43.Lyons CR, Wu TH. 2007. Animal models of Francisella tularensis infection. Ann. N. Y. Acad. Sci. 1105:238–265 [DOI] [PubMed] [Google Scholar]
- 44.Mosser DM, Edwards JP. 2008. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 8:958–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gavrilin MA, Wewers MD. 2011. Francisella recognition by inflammasomes: differences between mice and men. Front. Microbiol. 2:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sjodin A, Svensson K, Lindgren M, Forsman M, Larsson P. 2010. Whole-genome sequencing reveals distinct mutational patterns in closely related laboratory and naturally propagated Francisella tularensis strains. PLoS One 5:e11556 doi:10.1371/journal.pone.0011556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barry EM, Cole LE, Santiago AE. 2009. Vaccines against tularemia. Hum. Vaccin. 5:832–838 [DOI] [PMC free article] [PubMed] [Google Scholar]