Abstract
Atg6 (beclin 1 in mammals) is a core component of the Vps34 complex that is required for autophagy. Beclin 1 (Becn1) functions as a tumor suppressor, and Becn1+/- tumors in mice possess elevated cell stress and p62 levels, altered NF-κB signaling and genome instability. The tumor suppressor function of Becn1 has been attributed to its role in autophagy, and the potential functions of Atg6/Becn1 in other vesicle trafficking pathways for tumor development have not been considered. Here, we generate Atg6 mutant Drosophila and demonstrate that Atg6 is essential for autophagy, endocytosis and protein secretion. By contrast, the core autophagy gene Atg1 is required for autophagy and protein secretion, but it is not required for endocytosis. Unlike null mutants of other core autophagy genes, all Atg6 mutant animals possess blood cell masses. Atg6 mutants have enlarged lymph glands (the hematopoietic organ in Drosophila), possess elevated blood cell numbers, and the formation of melanotic blood cell masses in these mutants is not suppressed by mutations in either p62 or NFκB genes. Thus, like mammals, altered Atg6 function in flies causes hematopoietic abnormalities and lethality, and our data indicate that this is due to defects in multiple membrane trafficking processes.
Keywords: Autophagy, Endocytosis, Protein secretion, Drosophila
INTRODUCTION
Macroautophagy (autophagy) is an evolutionarily conserved catabolic process that is induced in response to cell stress, such as nutrient restriction, organelle damage and protein aggregation. During autophagy, double-membrane vesicles, known as autophagosomes, sequester cytoplasmic components, such as proteins and organelles, and deliver them to the lysosome for degradation (Mizushima and Komatsu, 2011). Protein turnover by the lysosome enables recycling of amino acids to be utilized for protein synthesis, and breakdown of damaged organelles prevents accumulation of toxic reactive oxygen species in the cell (Yang et al., 2006; Zhang et al., 2007). Autophagy has been implicated in many processes, including the mitigation of cell stress and genome instability, tissue remodeling during development, and clearance of intracellular pathogens (Berry and Baehrecke, 2007; Deretic, 2011; Karantza-Wadsworth et al., 2007).
Genetic screens in the yeast Saccharomyces cerevisiae identified autophagy-related (Atg) genes (Harding et al., 1995; Thumm et al., 1994; Tsukada and Ohsumi, 1993). These genes are required for autophagy and many are conserved in higher animals, including humans. A class III phosphoinositide 3-kinase (PI3K) complex, which includes the class III PI3K vacuolar protein sorting 34 (Vps34; also known as Pik3c3), the serine-threonine kinase Vps15 (p150 in mammals; also known as Pik3r4), and Atg6/Becn1 (also known as Vps30 in yeast), regulates autophagosome formation in yeast and mammals (Funderburk et al., 2010; Kihara et al., 2001b). The substrate of Vps34, phosphatidylinositol (PtdIns or PI), is converted to PI 3-phosphate [PI3P], and this membrane-associated lipid is bound by proteins containing either FYVE or PX domains (Ellson et al., 2002; Stenmark et al., 2002). PI3P-containing membranes include autophagosome isolation membranes, which serve as precursors to double-membrane autophagosomes prior to membrane expansion (Kirisako et al., 1999; Simonsen et al., 2004). In addition to their functions in autophagy, Vps34 and PI3P also regulate sorting of hydrolases to the yeast vacuole and mammalian lysosome, endocytic trafficking, and potentially multiple other vesicle trafficking processes (Juhász et al., 2008; Thoresen et al., 2010).
Two distinct Atg6/Becn1 protein complexes have been described in yeast and mammals (Itakura et al., 2008; Kihara et al., 2001b; Liang et al., 2008). Atg6, Vps34 and Vps15 form a core complex, which recruits other proteins to modulate the specific biological function of this complex. The Atg6, Vps34 and Vps15 complex interacts with Atg14L to promote autophagosome formation, whereas it interacts with Uvrag/Vps38 to regulate vacuolar protein sorting. Recent work also indicates that the complex containing Uvrag is involved in ligand-receptor degradation and cytokinesis (Thoresen et al., 2010). S. cerevisiae and Caenorhabditis elegans Atg6 are also required for retrograde transport from endosomes to the Golgi complex (Ruck et al., 2011; Seaman et al., 1997). In addition, Rubicon, Ambra1 and Bif1 (Zbtb24) function as regulators of these complexes (Fimia et al., 2007; Matsunaga et al., 2009; Takahashi et al., 2007; Zhong et al., 2009).
The function of beclin 1 as a tumor suppressor has influenced our understanding of the role of autophagy in cancer. BECN1 is monoallelically deleted in sporadic breast, ovarian and prostate cancers (Aita et al., 1999), and allelic loss of Becn1 in mice leads to lymphomas and carcinomas (Qu et al., 2003; Yue et al., 2003). At the cellular level, Becn1+/- tumors have decreased autophagy, elevated cell stress and genome instability (Mathew et al., 2007). Moreover, decreased beclin 1 function in oncogene-expressing tissues is associated with the accumulation of the autophagy cargo binding protein p62 (SQSTM1), altered NFκB signaling, and inflammation (Mathew et al., 2009). Combined, these results indicate that the tumor suppressor function of beclin 1 is related to its role in autophagy, but do not consider the potential functions of Atg6/Becn1 in other vesicle trafficking pathways for tumor initiation and progression.
Drosophila has a single beclin 1 ortholog, Atg6, which shares 71% amino acid identity with the evolutionarily conserved domain of mammalian beclin 1, and 50% overall identity. Atg6 protein interacts with Vps34 (Pi3K59F - FlyBase) in vivo, and co-expression of Atg6 with either Vps34 or Vps15 (ird1 - FlyBase) is sufficient to induce autophagy (Juhász et al., 2008). Both Vps34 and Vps15 are required for starvation-induced autophagy and adult viability in Drosophila (Juhász et al., 2008; Lindmo et al., 2008; Wu et al., 2007). However, the lack of a null Atg6 mutant has precluded full functional analysis of the Vps34 complex in flies.
Here, we use gene targeting to generate a Drosophila Atg6 null mutant, and show that Atg6 mutant fat body cells exhibit defects in autophagy and endocytosis. In addition, we show that Atg6, Vps34 and Atg1 function in protein secretion. Consistent with the role of beclin 1 as a tumor suppressor, loss of Atg6 caused over-production of blood cells, a failure in proper blood cell differentiation and the formation of melanotic blood cell masses.
MATERIALS AND METHODS
Fly stocks and culture
Flies were reared at 25°C on standard cornmeal/molasses/agar media. The following Drosophila melanogaster stocks were used: P{PZ} Atg600096, y w hs-FLP;FRT82B ubi-GFP, w;SgsΔ3-GFP, y w hs-FLP; FRT42DGFPnls, y w hs-FLP;FRT42D mRFPnls, y w hs-FLP; FRT80B mRFPnls, y w hs-FLP; FRT80B ubi-GFP, y w eye-FLP1, w; SgsΔ3-GFP, y w hs-FLP; actin >cd2 >GAL4::UAS-dsRed, hmlGAL4::UAS-GFP, CgGAL4 (all from Bloomington Drosophila Stock Center); y w hs-FLP;CgGAL4 UASmCherryatg8a; FRT82B UAS-GFPnls, UAS-GFP-2x-FYVE, UAS-Rab5-GFP, y w hs-hflp; r4GAL4FRT82B UAS-mCherry, y w hs-FLP; FRT80B Atg1Δ3D (all from T. Neufeld, University of Minnesota, MN, USA; y w hs-FLP; FRT42DVps25n55 y+/CyO actin GFP (from A. Bergmann, University of Massachusetts Medical School, MA, USA); Atg6 IR (TID 22123) (from Vienna Drosophila RNAi Center); Ref(2)P OD2, Ref(2)P OD3 (both from I. Nezis, University of Warwick, UK); w; rele20, w;Dif1 cn bw/CyO, w;J4/CyO, dlI5 (all from T. Ip); w;Atg7 d77/Cyo actin-GFP, w; Atg7 d14/Cyo actin-GFP, y1 Atg8a KG07569/FM7c-actin-GFP, Atg13=Atg13D74 (all from T. Neufeld); and FRT42D Vps32G5/Cyo twist-Gal4 UAS-GFP (from D. Bilder, University of California, Berkeley, CA, USA).
Larval staging
For lymph gland analyses, larvae were obtained from collections of 4-hour-old eggs and aged at 25°C. Classification of third instar larvae was based on the number of teeth on the mandibular hooks (Bodenstein, 1965) and developmental age. For eye imaginal disc experiments, larvae were raised on food supplemented with 0.05% Bromophenol Blue (Maroni and Stamey, 1983). Stationary larvae with clear guts were used for dissection of eye imaginal discs from Atg6 and Vps mutants.
Generation of Atg6 targeting construct
The ‘ends-out’ gene disruption approach (Rong and Golic, 2000) was used to target the open reading frame (ORF) of Atg6 in the isogenic w1118 parental line. The resultant strain contained a w+ mini-gene in place of the Atg6 ORF.
RT-PCR
RNA was collected from third instar larvae (n=10) using Trizol Reagent (Invitrogen) and was treated with DNase. cDNA was generated from 1 μg of RNA, using Superscript II Reverse Transcriptase (Invitrogen), following standard protocols. cDNA was used as PCR template, using the following primers to amplify Atg6 and flanking gene sequences: Atg6: 5′-CGAGCAGCTGGAGAAGATTAG-3′ and 5′-GCGTTGATCTCTGACCAGTC-3′; CG5991-RA: 5′-CATTGCCTAATTGTGTCCGC-3′ and 5′-GGAGAATTGGCGCAAGTGAC-3′; CG5991-RB: 5′-GCACAGCGATACGGAAGCAA-3′ and 5′-GGAGAATTGGCGCAAGTGAC-3′; CG5991-RC: 5′-GCCTCTTCGCATTTGACGAC-3′ and 5′-GGAGAATTGGCGCAAGTGAC-3′; CG5986: 5′-GGCGATAACGCTTGCATCAC-3′ and 5′-CGTTGATATCCCGCAAACGG-3′. Quantitative real-time PCR was performed as described (Denton et al., 2009).
Induction of mutant clones of cells
Standard methods were used for the induction of mutant clones of cells (Xu and Rubin, 1993). To induce loss-of-function mutant cell clones, we used y w hsFlp; FRT42D Ubi-nlsGFP, y w hsFlp; FRT42D mRFP-nls, y w hs-FLP; +; FRT80B Ubi-nlsGFP, y w hs-FLP; +; FRT80B Ubi-mRFP, y w hs-FLP; +; FRT82B Ubi-nlsGFP, y w hsFlp; CgGAL4; FRT82B UAS-mCherry and y w hsFlp; FRT42D Ubi-nlsGFP. Four-hour egg lays were maintained at 37°C for 1 hour to induce clones in the larval salivary glands, and 10-hour egg lays were maintained at 37°C for 1 hour to induce clones in the larval fat body.
Nutrient restriction
Second and third instar larvae were fed 20% sucrose in PBS, pH 7.4, and maintained at 25°C for 4 hours.
Texas Red-avidin assay
To visualize endocytosis, the fat body was dissected from third instar larvae and incubated ex vivo with Texas Red-avidin (Invitrogen) diluted in Schneider’s media to a concentration of 80 μg/ml for 20 minutes, then chased with 0.5% BSA in cold PBS for 10 minutes prior to overnight fixation in 4% formaldehyde. The tissue was washed three times (10 minutes per wash) with 0.1% Tween-20 in PBS and mounted in Vectashield (Vector Laboratories). Images were collected on a Zeiss AxioImager Z1 equipped with an Apotome. Images were acquired with Axiocam and processed using Zeiss Axiovision Suite 4.8 and Photoshop CS4 and Adobe Illustrator CS4 14.0.0.
Immunostaining
For fat body immunofluorescence experiments, tissues were dissected in PBS and fixed overnight in 4% formaldehyde in PBS, pH 7.4, at 4°C. Following fixation, tissues were washed with 0.1% Triton X-100 in PBS (PBST) for 2 hours, then blocked in 0.5% BSA in PBST (PBSBT) for 2 hours at room temperature. Primary antibody incubations were performed overnight at 4°C in PBSBT followed by washing in PBSBT for 2 hours at room temperature. Secondary antibody was added at a dilution of 1:200, and tissues were incubated for 4 hours at room temperature. Following a series of short PBSBT washes, tissues were mounted in Vectashield with DAPI (Vector Laboratories). The following primary antibodies were used: rabbit anti-Ref (2)P (1:1000; I. Nezis) (Nezis et al., 2008), mouse anti-P1/NimC1 (1:10; I. Ando) (Asha et al., 2003) and mouse anti-L1 (1:10; I. Ando) (Asha et al., 2003). Secondary antibodies from Invitrogen were used at 1:200: goat anti-rabbit Oregon Green 488, goat anti-rabbit Alexa Fluor 546, goat anti-mouse Alexa Fluor 546 and donkey anti-rat FITC.
For lymph gland analyses, larval fillets were prepared as previously described (Budnik et al., 2006), and lymph glands were dissected and stained as previously described (Lebestky et al., 2000).
For immunohistochemistry of paraffin sections, third instar larvae were fixed and dehydrated for histology according to published methods (Muro et al., 2006). Histological sections were de-waxed with a series of xylene washes, and rehydrated through a series of decreasing percentage ethanol washes. Following rehydration, antigen retrieval was performed by heating slides in 10 mM sodium citrate, pH 6.0. Specimens were blocked in 5% non-fat dry milk containing 1% BSA and horse serum (Vector Laboratories). Rabbit anti-GFP (Novus) antibody was used at 1:500. The Vectastain Elite ABC Kit (Vector Laboratories) was used for immunohistochemical detection, and the signal was visualized by diaminobenzadine staining. Tissue was counterstained with Weigert’s Hematoxylin and Permount mounting media was applied. Images were collected on a Zeiss Axiophot microscope. Images were minimally processed using Adobe Photoshop CS4.
Hemocyte quantification
Individual third instar larvae of similar age (± 3 hours) were bled into 20 μl of PBS. From this, 10 μl was loaded onto a standard hemocytometer and the average number of cells per milliliter was calculated for 20 animals per genotype. A one-tailed Student’s t-test was used to determine statistical significance.
Quantification and statistical analyses
Zeiss Image Measurement Software was used for the quantification of Atg8a, Ref(2)P and Rab5 puncta, as well as the eye-antennal disc area and the lymph gland area. Statistical analyses were performed using GraphPad prism software. Student’s t-test for two samples assuming unequal variances was used to determine the statistical significance of the data.
Transmission electron microscopy
Tissues were dissected and fixed overnight at 4°C in 4.0% paraformaldehyde, 2.0% glutaraldehyde, 1% sucrose and 0.028% CaCl2 in 0.1 M sodium cacodylate, pH 7.4, thoroughly washed in cacodylate buffer, post-fixed in 2.0% osmium tetroxide for 1 hour, and embedded in SPI-pon/Araldite resin (Polysciences) according to manufacturer’s recommendations. Ultrathin sections (80 nm) were stained with uranyl acetate and lead citrate before examination in a Philips CM10 transmission electron microscope.
Protein secretion assay
Salivary glands were dissected from control and mutant animals at 4 hours after puparium, fixed for 30 minutes in 4% paraformaldehye in PBS, washed three times (5 minutes per wash) in PBS, and mounted in Vectashield with DAPI.
RESULTS
Atg6 is required for autophagy in Drosophila
A transposable P-element, P{PZ}Atg600096, is located in the 5′ untranslated regions of Atg6 and the neighboring gene CG5991 (Fig. 1A). Starvation-induced autophagy was not consistently altered in the fat body of homozygous P{PZ}Atg600096 larvae (Scott et al., 2004). Based on this result and the fact that P{PZ}Atg600096 does not disrupt the Atg6 ORF, we generated an Atg6 mutant using a gene-targeting approach (Rong and Golic, 2000).
Fig. 1.
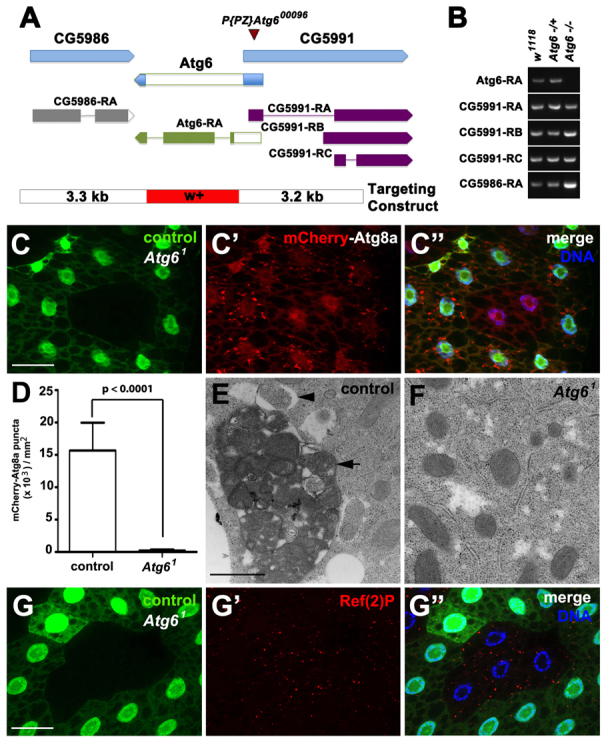
Atg6 is required for autophagy. (A) Atg6 genomic locus and donor targeting construct (not to scale). The donor construct consisted of a w+ mini-gene flanked by 3.5 kb of genomic sequence (blue) from each side of Atg6. Flp recombination target (FRT) sites and I-SceI endonuclease recognition sites were present on each side of flanking sequence, to facilitate double-stranded break and homologous recombination at the target site. The resultant flies contained the w+ minigene (red) in place of the Atg6 ORF (white). (B) RT-PCR of Atg6 and flanking gene transcripts indicates knockdown of Atg6 transcript expression in homozygous Atg61 third instar larvae, and maintenance of flanking gene RNA levels. (C-C″) mCherry-Atg8a puncta reflect starvation-induced autophagosome formation in control (GFP-positive) larval fat body cells, whereas cytoplasmic mCherry-Atg8a localization reflects a defect in autophagy in Atg61 (GFP-negative) fat body cells (n=10). (D) Quantification of mCherry-Atg8a puncta in control and Atg61 mutant fat body cells following starvation. A two-tailed t-test was used for statistical analysis and the P-value relative to control was 8.98×10-8. Error bars represent s.d. (E,F) TEM images reveal abundant autophagosomes (arrowhead) and autolysomes (arrow) in control (E) but not in (F) Atg61 mutant fat body cells following 4 hours of starvation. (G-G″) Ref(2)P accumulated in Atg61 (GFP negative) fat body cells, indicative of defective autophagy, compared with control cells (GFP positive) (n=13). Scale bars: 50 μm in C-C″,G-G″; 500 nm in E,F.
Homologous recombination was used to replace the Atg6 ORF with a w+ mini-gene (Fig. 1A). RT-PCR confirmed the absence of Atg6 RNA in homozygous mutant third instar larvae, whereas the RNA levels of neighboring genes were not altered in the homozygous mutant Atg6 (hereafter termed as Atg61) mutant larvae (Fig. 1B). In addition, PCR was used to confirm the presence of a w+ mini-gene in the Atg6 genomic locus (data not shown). Both homozygous Atg61 and animals transheterozygous for Atg61 and the Df(3R)Exel 6197 deficiency for this region died during the late third larval instar and early pupal stages of development (supplementary material Fig. S1A; data not shown). Significantly, expression of a UAS-GFP-Atg6 transgene under the control of a ubiquitously expressed actin-GAL4 driver rescued the lethality of Atg61 homozygous mutants (supplementary material Fig. S1A). In addition, Atg61 complements P{PZ}Atg600096 even though this is a lethal P-element insertion, indicating that P{PZ}Atg600096 is not an Atg6 mutant. These data indicate that we have isolated a loss-of-function Atg6 mutant.
Atg6, Vps15 and Vps34 are core components of all known Vps34 complexes (Funderburk et al., 2010), and loss of either Vps34 or Vps15 inhibits starvation-induced autophagy in the larval fat body of Drosophila (Juhász et al., 2008). To determine whether Atg6 is required for starvation-induced autophagy, we monitored the localization of the autophagosome marker mCherry-Atg8a in larval fat body. Atg8a, the Drosophila ortholog of mammalian LC3 (microtubule-associated protein 1 light chain 3), displays diffuse cytoplasmic localization in the fat body of feeding Drosophila larvae, but becomes incorporated into autophagosome membranes during starvation and is visualized as punctate spots in the cytoplasm (Scott et al., 2004). We utilized FLP recombinase-mediated recombination at FLP recombination target (FRT) sites to generate Atg61 mitotic mutant cell clones in the fat body, resulting in tissue composed of control (either wild type or heterozygous Atg61/wild type) and homozygous Atg61/Atg61 mutant cells. Following 4 hours of starvation, control fat body cells, which were marked by the presence of green fluorescent protein (GFP) contained several mCherry-Atg8a puncta, whereas homozygous Atg61 mutant cells (lacking GFP) displayed diffuse localization of mCherry-Atg8a (Fig. 1C-C″,D).
Two additional approaches were used to determine the influence of Atg6 function on autophagy. We used transmission electron microscopy (TEM) to investigate Atg61 mutants at the ultrastructural level. Following 4 hours of starvation, many autophagosomes (Fig. 1E, arrowhead) and autophagolysosomes (Fig. 1E, arrow) were observed in control fat body cells, whereas homozygous Atg61 mutant fat body cells lacked these autophagic structures (Fig. 1F). Ref(2)P is the Drosophila ortholog of p62 (SQSTM1) and is known to bind ubiquitylated substrates and aid in their recruitment into autophagosomes to be targeted for degradation (Nezis et al., 2008). Homozygous Atg61 mutant cells lacking GFP accumulated Ref(2)P compared with neighboring control fat body cells (Fig. 1G-G″; supplementary material Fig. S1B). Importantly, the accumulation of Ref(2)P aggregates could be rescued by expressing an Atg6 transgene in the Atg61 mutant cells (supplementary material Fig. S1C-C″). These results indicate that Atg6 is required for autophagy in vivo.
Atg6 functions in multiple vesicle trafficking processes
Vps34 is required for the formation of PI3P in Drosophila and other species (Juhász et al., 2008; Lindmo and Stenmark, 2006). To examine the influence of Atg6 function on PI3P, we used a transgenic reporter consisting of GFP fused to the FYVE domain of hepatocyte growth factor regulated tyrosine kinase substrate (Hrs) (Gillooly et al., 2000). GFP-FYVE localized to punctate structures in the cytoplasm of control (red) cells of the larval fat body (Fig. 2A-A″). By contrast, homozygous Atg61/Atg61 mutant cells (lacking red) had no detectable GFP-FYVE puncta (Fig. 2A-A″).
Fig. 2.
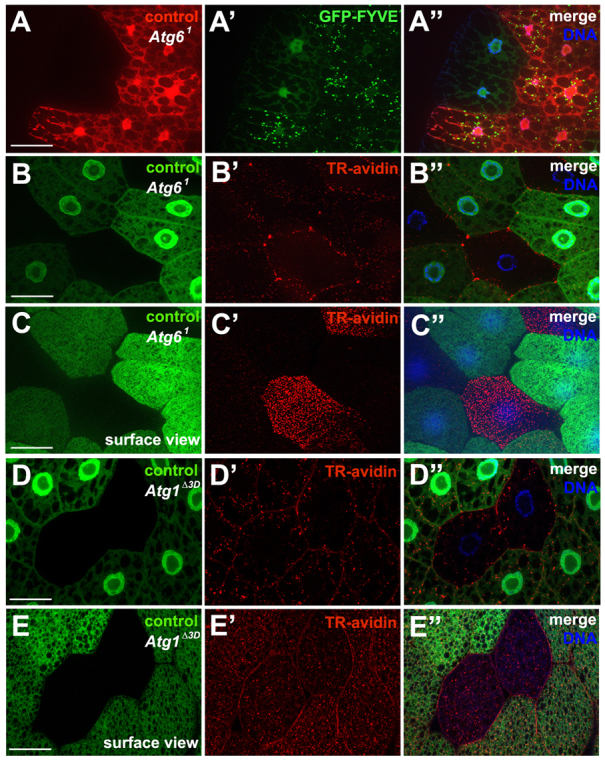
Loss of Atg6 leads to defects in PI3P formation and endocytosis. (A-A″) GFP-FYVE localizes to perinuclear puncta, reflecting Vps34 activity and PI(3)P formation in control (mCherry-positive) fat body cells, but remains cytoplasmic in Atg61 mutant (mCherry-negative) fat body cells (n=24). (B-B″) Texas Red-avidin is endocytosed by control (GFP-positive) fat body cells, but is largely excluded from Atg61 mutant cells (GFP negative). (C-C″) Surface view of the fat body depicted in B showing accumulation of Texas Red-avidin at the surface of the Atg61 cells but not in control cells (n=13). (D-D″) Texas Red-avidin is endocytosed in control (GFP-positive) fat body cells, as well as in Atg1Δ3D mutant cells (GFP negative). (E-E″) Surface view of the fat body depicted in D showing Texas Red-avidin in both control and Atg1Δ3D mutant cells (n=10). Scale bars: 50 μm.
Vps34 is required for endocytosis in Drosophila (Juhász et al., 2008). To test whether Atg6 functions in endocytosis, uptake of Texas Red (TR)-avidin was used to monitor fluid-phase endocytosis in larval fat body. Control (GFP-positive) cells contained TR-avidin-positive puncta throughout the cytosol, whereas homozygous Atg61 mutant (GFP-negative) cells possessed little to no endocytic tracer (Fig. 2B-B″) and instead TR-avidin was often more abundant on the surface of these mutant cells (Fig. 2C-C″).
Rab5, a small GTPase of the Ras superfamily, is associated with endosomes and functions as a key regulator of vesicle trafficking (Wucherpfennig et al., 2003). To monitor Rab5 localization in fat body cells, we used a GFP-Rab5 transgenic reporter. In control fat body cells, GFP-Rab5 localizes to the plasma membrane and has characteristic puncta with perinuclear localization. In cells with RNAi knockdown of Atg6 (Atg6IR), GFP-Rab5 localized to the plasma membrane in most fat body cells; however, the perinuclear localization was significantly reduced (supplementary material Fig. S2A-D). These data suggest that in larval fat body cells, Atg6 is required for either recruitment of or stable association of Rab5 with the perinuclear endosomal compartment.
To determine whether the endocytosis phenotypes observed in Atg6 and Vps34 mutant cells are due to defects in autophagy, we performed the TR-avidin uptake assay in Atg1 mutant fat cells. Atg1 is a kinase and a core component of the autophagy pathway that is both necessary and sufficient for inducing autophagy (Scott et al., 2007). Both control (GFP-positive) cells and homozygous Atg1Δ3D (GFP-negative) mutant cells contained TR-avidin-positive puncta throughout the cytosol (Fig. 2D-D″). Furthermore, TR-avidin did not accumulate at the surface of these cells (Fig. 2E-E″), indicating that Atg1 function is not required for fluid-phase endocytosis. These results indicate that Atg6, but not Atg1, is required for fluid-phase endocytosis in vivo.
Endosomal sorting complex required for transport (ESCRT) proteins are required for recruitment of ubiquitylated cargo proteins to the endosome, sorting to multivesicular bodies (MVBs), and subsequent degradation by the lysosome (Henne et al., 2011). Like Atg6 mutant cells, mutations in either Vps25 (ESCRT-II) or Vps32 (ESCRT-III; shrb - FlyBase) (GFP-negative cells) suppress TR-avidin fluid-phase endocytic tracer uptake compared with control cells (GFP-positive) (supplementary material Fig. S3A-D″). Loss of ESCRT components in the developing Drosophila eye impairs MVB formation and leads to defects in receptor degradation that cause tissue overgrowth (Fig. 3A,D) (Herz et al., 2006; Herz et al., 2009; Thompson et al., 2005; Vaccari and Bilder, 2005; Vaccari et al., 2009). By contrast, eye imaginal disc tissue isolated from homozygous Atg61 animals at the same third larval instar stage is normal in size (Fig. 3B). Although Atg6 and ESCRT component double mutant eye tissue is disorganized, loss of Atg6 significantly suppressed the overgrowth of either Vps25 or Vps32 mutant eye tissue (Fig. 3C,E,F). Combined, our data indicate that Atg6 is required for fluid-phase endocytosis, and suggest the possibility that an important relationship might exist between Atg6 and the ESCRT pathway.
Fig. 3.
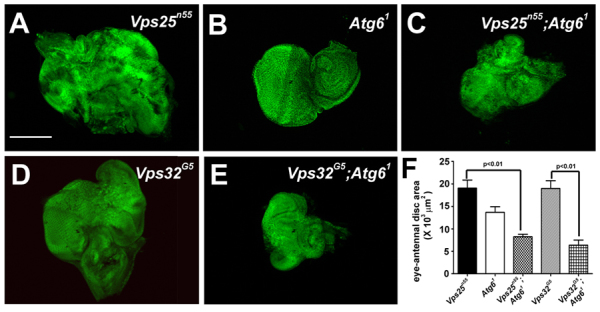
Atg6 suppresses the Vps25 and Vps32 mutant eye phenotypes. (A) Vps25n55 mutant clone cells (GFP-negative) in late third instar eye imaginal discs cause non-autonomous over-proliferation of neighboring (GFP-positive) cells leading to overgrowth of the developing eye tissue (n=11). (B) Homozygous Atg61 mutant late third larval instar eye imaginal discs appear normal in size and morphology (DAPI is shown in green, n=10). (C) Homozygous loss of Atg6 suppressed overgrowth of the eye caused by Vps25 mutant cells (GFP negative) (n=6). (D) Vps32G5 mutant clones (GFP-negative) in late third instar eye imaginal disc cause non-autonomous overproliferation of neighboring (GFP-positive) cells leading to overgrowth of the developing eye tissue (n=11). (E) Vps32G5 mutant clones (GFP negative) in Atg61 mutant eye disc leads to suppression of overgrowth of the eye discs (n=10). (F) Quantification of eye-antennal disc area in each of the genotypes depicted in A-E. A two-tailed t-test was used for statistical analyses. The P-value between Vps25n55 and Vps25n55; Atg61 was 6.6×10-6 and the P-value between Vps32G5 and Vps32G5; Atg61was 1.97×10-5. Error bars represent s.d. Scale bar: 50 μm.
Recent work has implicated autophagy in protein secretion (Deretic et al., 2012), but direct genetic analyses of the role of key autophagy regulatory factors, including Atg6, in protein secretion within animals are lacking. The Drosophila larval salivary gland secretes large quantities of glue proteins in response to steroid at the end of larval development. A transgenic fusion of the secreted glue protein Sgs3 and GFP proteins provides an excellent means to follow protein secretion in this tissue in vivo (Biyasheva et al., 2001). Both control (Fig. 4A,A′) and homozygous Atg61 mutant animal (Fig. 4C,C′) salivary glands are able to synthesize glue protein in the salivary glands based on the presence of GFP. By 4 hours after puparium formation, control animals had secreted most of the glue protein, based on the absence of GFP (Fig. 4B,B′), whereas homozygous Atg61 mutant animals retained GFP, suggesting that they have a protein secretion defect (Fig. 4D,D′). It is possible that this Atg61 mutant animal defect is caused by a failure to arrest protein synthesis that is associated a general delay in development. To address this possibility, homozygous Atg61 mutant clones of cells (mCherry negative) were produced in salivary glands. These Atg6 mutant cells retained Sgs3-GFP, whereas neighboring control cells (mCherry positive) were devoid of GFP reporter (Fig. 4E-E″). Furthermore, Sgs3-GFP was also retained in salivary gland cells homozygous for Vps34dm22 (a null allele of Vps34, mRFP negative) as well as in cells with RNAi knockdown of Vps34, whereas neighboring control cells (mRFP positive) lacked Sgs3-GFP (Fig. 3F-F″; data not shown). Interestingly, Atg1Δ3D mutant cells also retained Sgs3-GFP, whereas neighboring control cells (mRFP positive) secreted Sgs3-GFP (Fig. 3G-G″). These data suggest that Atg6, Vps34 and Atg1 are each required for salivary gland protein secretion.
Fig. 4.
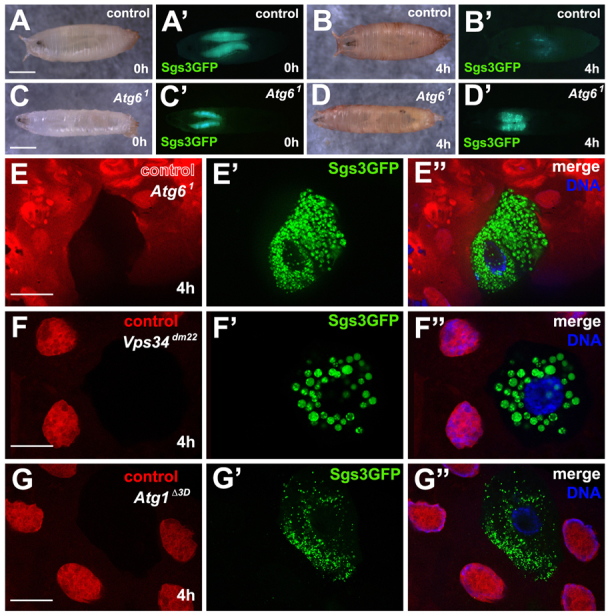
Loss of Atg6 leads to defects in protein secretion. (A,A′) Control pupa (0 hour) showing expression of SgsΔ3-GFP in salivary glands (n=10). (B,B′) Four hours later the same pupa as in A lacked SgsΔ3-GFP. (C-D′) Homozygous Atg61 mutants possess SgsΔ3-GFP protein at 0 hour (C-C′) (n=12), but are unable to secrete it 4 hours after puparium formation (D-D′). (E-E″) Control cells (mCherry positive) are able to secrete Sgs3Δ3-GFP whereas homozygous Atg61 mutant cells (mCherry negative) retain SgsΔ3-GFP in the cytoplasm (n=12). (F-F″) Control cells (mCherry positive) secrete SgsΔ3-GFP whereas Vps34dm22 mutant cells retain SgsΔ3-GFP granules in the cytoplasm (n=11). (G-G″) Control cells (mCherry positive) secrete SgsΔ3-GFP whereas Atg1Δ3D expressing cells retain SgsΔ3-GFP granules in the cytoplasm (n=10). Scale bars: 1 mm in A,C; 50 μm in E-G.
Loss of Atg6 leads to melanotic blood cell mass formation
Whereas the parental control w1118 and heterozygous Atg61/wild-type animals exhibited no obvious phenotypes, all homozygous Atg61 mutant larvae displayed a striking melanotic blood cell mass phenotype (visible as black masses) (Fig. 5A-C). Although single melanotic blood cell masses are often present in Atg61 mutant larvae (Fig. 5C), many homozygous mutant animals possess multiple melanotic blood cell masses (Fig. 6B). Significantly, this homozygous Atg6 mutant melanotic blood cell tumor phenotype was completely rescued by ubiquitous expression of Atg6 (Fig. 5D).
Fig. 5.
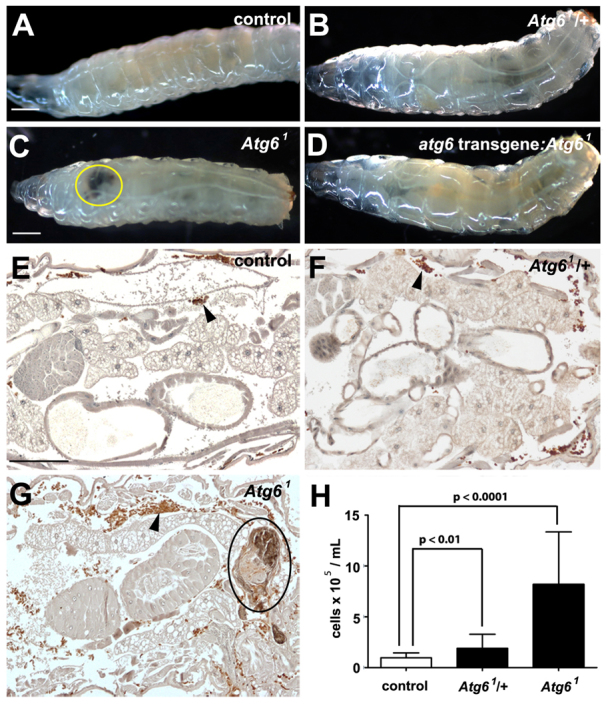
Loss of Atg6 leads to melanotic blood cell mass formation. (A-D) White-light images of wandering third instar larvae. Parental control w1118 (A) and heterozygous Atg61 (B) larvae are phenotypically similar. Homozygous Atg61 mutant larvae (C) contain melanotic blood cell masses (yellow ring), and this phenotype is rescued by ubiquitous expression of a GFP-Atg6 transgene (D). (E-G) Hemocytes were visualized by immunohistochemistry in third instar larvae expressing GFP, driven by hmlΔ-GAL4 (arrowheads). Histological sections of w1118 (E) and Atg61 heterozygous (F) larvae contain few hemocytes (brown) and no visible melanotic masses, whereas sections of homozygous Atg6 mutant larvae (G) reveal many hemocytes, which surround melanotic masses (black ring). (H) Quantification of hemocytes from w1118, Atg61/+ and Atg61/Atg61 larvae revealed a significant increase in hemocyte number in both heterozygous and homozygous Atg6 mutant larvae compared with control w1118 larvae. For each genotype, n=20 animals. Error bars represent s.e.m. A one-tailed t-test was used for statistical analysis and P-values relative to w1118 are: Atg61/+, P=0.004; Atg61/Atg61, P=2.6×10-6. Scale bars: 500 μm in A,C; 100 μm in E.
Fig. 6.
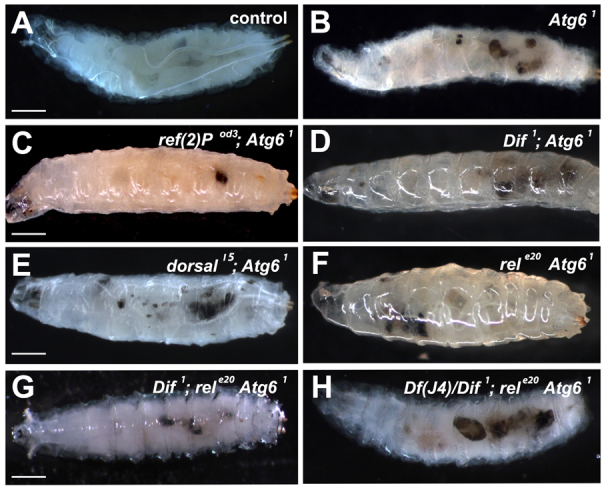
Ref(2)P and NFκB are not required for melanotic mass formation in Atg61 mutant animals. (A) Control late third instar larvae lack melanotic masses. (B) All homozygous Atg61 mutant late third instar larvae contain melanotic masses. (C-F) Double mutant combinations of homozygous Atg61 with ref(2)p (C), dif (D), dorsal (E) and Relish (F) do not suppress melanotic masses. (G) Triple mutant combination of Dif, rel and Atg61 showing presence of melanotic masses. (H) dif, dorsal-/+, Rel and Atg61 mutants exhibit melanotic masses. n≥10 for each genotype. Scale bars: 250 μm.
We investigated whether mutations in other core autophagy genes results in melanotic blood cell mass formation. All homozygous Atg7 and Atg13 mutant animals lack melanotic blood cell masses (supplementary material Fig. S4A; data not shown). Although 18% of Atg8aKG07569 mutant pupae possess melanotic masses (supplementary material Fig. S4A-C), such blood cell masses were not observed in Atg8 mutant larvae. Combined, these data indicate that Atg6 mutants are different from other autophagy mutants in their predisposition to the formation of melanotic blood cell masses.
We investigated whether blood cells are the source of melanotic masses in Atg6 mutant larvae. Blood cell-specific hemolectin (hmlΔ)-GAL4 was used to drive GFP expression in parental w1118 control, heterozygous Atg61/wild-type and homozygous Atg61 mutant larvae. Immunohistochemical analyses of paraffin sections with a GFP antibody revealed that these masses were indeed composed of blood cells in homozygous Atg61 mutants, whereas significantly fewer blood cells were observed in either w1118 or Atg61/wild-type control animals (Fig. 5E-G). Quantification of blood cells revealed that Atg61/wild-type animals contained approximately twice as many blood cells as parental w1118 larvae, and homozygous Atg61/Atg61 mutants contained nearly ten times as many blood cells as control w1118 animals (Fig. 5H). These data indicate that loss of Atg6 results in an increase in blood cell numbers, and like beclin 1 mutant mice, loss of Atg6 causes an increase in the number of circulating blood cells.
Two approaches were taken to determine whether the melanotic mass phenotype is blood cell autonomous. First, we expressed UAS-Atg6 using two different blood cell-specific drivers, either hmlΔ-Gal4 or croquemort-Gal4, in Atg61 mutant animals. Expression of the Atg6 transgene using these approaches failed to rescue the phenotype, suggesting that ectopic blood cell masses are either due to a blood cell-independent effect of Atg6 or that these Gal4 drivers are expressed too late during blood cell development to rescue the mutant phenotype (data not shown). Second, we induced Atg6 loss-of-function mutant cell clones in lymph glands using hs-FLP. Although these animals possess melanotic masses (data not shown), they also have Atg6 mutant cell clones in other tissues. Thus, we cannot conclude that the Atg6 melanotic mass phenotype is blood cell lineage specific.
The current model for beclin 1 function during tumor progression suggests that decreased autophagy leads to elevated p62 signaling and activation of the NFκB pathway (Mathew et al., 2009). In addition, the NFκB pathway has been implicated in the formation of melanotic blood cell masses in Drosophila (Minakhina and Steward, 2006). Therefore, we investigated whether mutations in either ref(2)P (Drosophila p62) or different combinations of NFκB genes suppress melanotic blood cell mass formation in homozygous Atg61 mutant larvae. All homozygous Atg61 mutant larvae possessed melanotic blood cell masses (black masses), whereas control larvae did not contain such structures (Fig. 6A,B). Significantly, homozygous ref(2)P mutants failed to suppress the Atg6 mutant melanotic mass phenotype (Fig. 6C). Drosophila has three NFκB proteins named Dif, Dorsal and Relish. Double mutant analyses of homozygous Atg61 with either homozygous dif, dorsal or Relish mutants indicated that mutations in each individual NFκB failed to suppress the Atg6 mutant melanotic mass phenotype (Fig. 6D-F). Therefore, we constructed a triple mutant containing homozygous dif, Relish and Atg6, and mutation of these two NFκB genes failed to suppress the formation of melanotic masses associated with Atg61 (Fig. 6G). Finally, we constructed a strain containing homozygous dif, Relish and Atg6 and also lacking one allele of dorsal (loss of all four genes was lethal at an earlier developmental stage), and this combination of NFκB mutations failed to suppress the Atg61 melanotic blood cell mass phenotype (Fig. 6H). These results suggest that neither Ref(2)P nor NFκB proteins play a role in melanotic blood cell mass formation in Atg6 mutants.
To gain insight into the kinetics of melanotic blood cell mass formation, we followed the lymph gland (the larval hematopoietic organ) and melanotic mass development. We used hmlΔ-GAL4 to drive GFP in blood cells in control (n=107) and homozygous Atg61 mutant (n=157) larvae. Compared with synchronized control animals, homozygous Atg61 mutant larvae displayed enlarged lymph glands (yellow boxes) between 80 hours and 104 hours after egg lay (Fig. 7A,B,E). During the same developmental interval, homozygous Atg61 mutant larvae progressively accumulated GFP-positive blood cell masses, whereas control animals lacked these structures. By 126 hours, all of the Atg61 mutant larvae contained numerous ectopic blood cell masses that were absent in controls (Fig. 7C,D,E). These Atg61 mutant animal blood cell masses initiated melanization 104 hours after egg lay (following the formation of GFP-positive blood cell aggregates) and continued to do so until 160 hours after egg lay (Fig. 7E). Whereas control animals formed prepupae by 120 hours after egg lay, most Atg61 mutants did not pupariate.
Fig. 7.
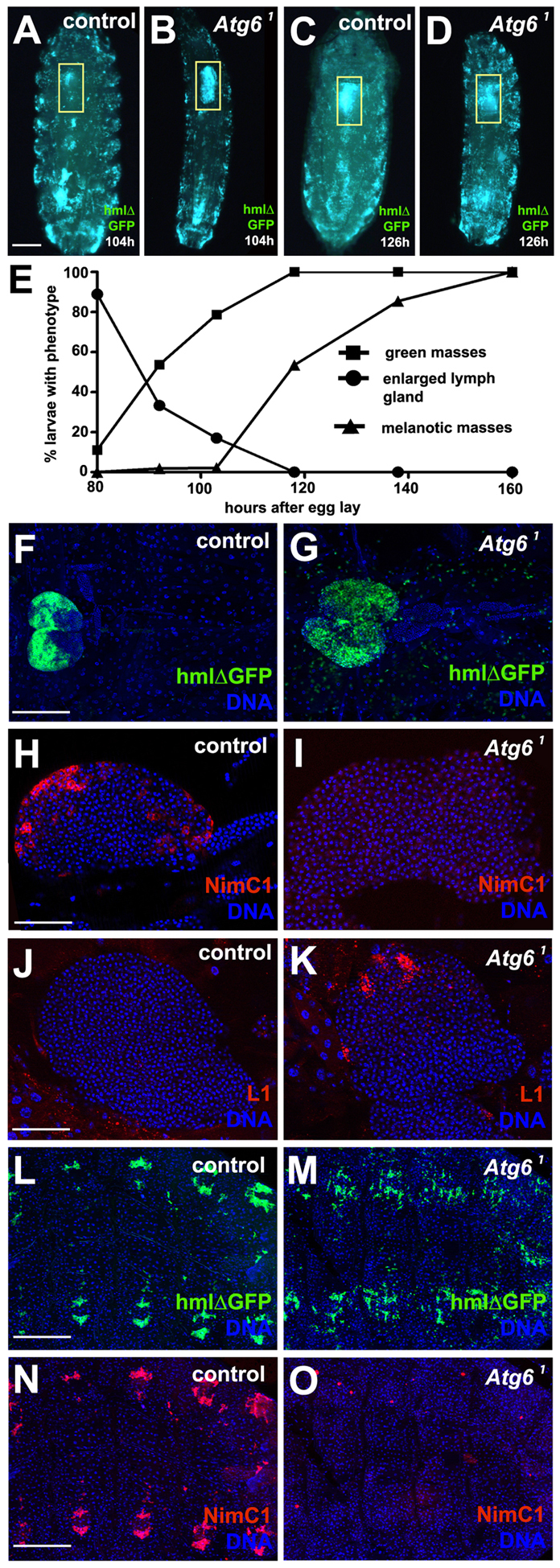
Loss of Atg6 leads to enlargement of lymph gland and altered blood cell development. (A) Control larva at 104 hours after egg lay expressing GFP in blood cells. (B) Homozygous Atg61 mutant larva at 104 hours after egg lay expressing GFP in blood cells. Note the enlarged lymph gland (yellow box). (C) Same control animal as in A at 126 hours after egg lay has formed a prepupa. (D) Same Atg61 mutant animal as in B at 126 hours after egg lay did not pupariate, and appeared to have an increased number of circulating blood cells. (E) Graph showing progression of different phenotypes exhibited by Atg6 mutants during larval development. The larval stage-specific numbers of teeth on mouth hooks of control and Atg6 mutant larvae were used to normalize development of these genotypes. As larvae progress through development there is an increase in formation of blood cell aggregates followed by melanotic masses (n=157). (F) Control hmlΔ-GAL4 UAS-GFP lymph gland at third instar larval stage (n=10). (G) Atg61 mutant hmlΔ-GAL4 UAS-GFP lymph gland at third instar stage (n=10). (H) Control third instar lymph gland stained for NimrodC1 (NimC1) showing expression in the cortical zone (n=7). (I) Atg61 mutant third instar lymph gland stained for NimC1 showing a complete lack of expression in the cortical zone (n=15). (J) Control third instar larval lymph gland stained for lamellocyte specific antigen L1 showing no expression in the cortical zone (n=7). (K) Atg61 mutant third instar animal lymph gland with increased expression of L1 in the cortical zone (n=7). (L) Control hmlΔ-GAL4 UAS-GFP animal showing sessile blood cells that are located in a reiterated pattern along abdominal segments (n=7). (M) hmlΔ-GAL4 UAS-GFP-expressing Atg61 mutant animals possess less patterned sessile blood cells along the abdominal segments than controls (n=7). (N) NimC1 staining of animal shown in L. (O) NimC1 staining of animal shown in M indicates that this blood cell antigen is missing in sessile blood cells. Yellow boxes in A-D delineate lymph glands. Scale bars: 250 μm in A; 100 μm in F; 50 μm in H,J; 200 μm in L,N.
We analyzed the lymph glands of age-matched control and homozygous Atg61 mutant third instar larval animals to obtain a better understanding of how hematopoiesis is altered. Homozygous Atg61 mutant lymph glands were larger than those of control animals (Fig. 7F,G; supplementary material Fig. S5). As expected, the plasmatocyte-specific blood cell differentiation marker NimrodC1 (NimC1) could be detected in the cortical zone in control lymph gland of control animals (Fig. 7H), but this marker could not be detected in the lymph glands of homozygous Atg61 mutant animals (Fig. 7I). L1 is a lamellocyte-specific blood cell differentiation marker that we did not detect in the cortical zone of control animal lymph glands (Fig. 7J). By contrast, homozygous Atg61 mutant lymph glands displayed increased L1 staining (Fig. 7K). In addition, blood cells that are present in clusters in abdominal segments expressed NimC1 in control animals, whereas those in Atg61 mutant animals appeared to lack the NimC1 antigen (Fig. 7L-O). Taken together, these data indicate that Atg6 plays an important role in blood cell development, and that an altered differentiation program probably contributes to melanotic blood cell masses.
DISCUSSION
Here we describe the genetic characterization of Atg6 in Drosophila. Atg6 is an essential gene, and most homozygous Atg6 null mutant animals die at the end of larval development. Drosophila lacking Atg6 function possess melanotic blood cell masses, as well as defects in several vesicle trafficking pathways.
Atg6 is a core component of the Vps34 complex. Studies in yeast and mammalian systems have identified Vps34 as an essential protein regulating a wide variety of vesicular trafficking events, including autophagy, endocytosis, and anterograde and retrograde transport between Golgi and the lysosome (Lindmo and Stenmark, 2006). Therefore, it is logical that Atg6 mutant cells not only have a defect in starvation-induced autophagy, but also fail to produce PI3P and have defects in endocytosis and protein secretion. Our data are consistent with reports from other animal systems in which beclin 1 mutants exhibit endocytosis defects (Ruck et al., 2011; Thoresen et al., 2010).
The accumulation of an endocytic tracer at the periphery of homozygous Atg6 mutant cells suggests that Atg6 functions at an early step of endocytosis. This conclusion is supported by the similarity between Atg6 and ESCRT II and III endocytic tracer phenotypes in the fat body. Therefore, it is possible that loss of Atg6 is similar to ESCRT mutants in flies, and that the Vps34 complex regulates receptor downregulation because of similar defects in endocytosis (Herz et al., 2006; Herz et al., 2009; Thompson et al., 2005; Vaccari and Bilder, 2005; Vaccari et al., 2009). However, the lack of an obvious Atg6 mutant eye over-growth phenotype suggests that Atg6 mutants are different from ESCRT pathway mutants. In addition, unlike ESCRT mutants, Atg6 mutant cell clones neither accumulated Notch intracellular domain antigen nor possessed alter the expression of the m2.61lacZ Notch reporter (data not shown). An alternative explanation for the difference between Atg6 and ESCRT mutants is that maternally contributed Atg6 mRNA may enable normal Atg6 mutant eye imaginal disc development. Although loss of Atg6 suppresses ESCRT mutant developing eye tissue size, the pattern of these structures remains disrupted, suggesting that some aspects of the ESCRT phenotype cannot be suppressed. It is tempting to speculate that Atg6 functions at an earlier stage than these ESCRT genes in endocytosis, but additional studies are needed to understand the relationship of these factors during endocytosis.
Recent work indicates that autophagy regulates protein secretion (Deretic et al., 2012). To our knowledge, this is the first report of Atg6 regulating protein secretion. In addition, we show that loss of either Vps34 or Atg1 in salivary gland cells also leads to disruption of protein secretion. Therefore, our data indicate that protein secretion might be an autophagy-dependent process. Interestingly, we noted differences in the size of Sgs3-GFP vesicles in Atg6, Vps34 and Atg1 mutant cells (Fig. 4E-G″). Although these genes might function at distinct steps in the maturation of secretory vesicles, it is also possible that differences in maternal contribution of mRNAs in these mutants are responsible for the differences in these mutant phenotypes. Beclin 1 and PI3P localize to the trans Golgi network (Gillooly et al., 2000; Kihara et al., 2001a). Thus, it is also possible that Vps34 and Atg6 are part of a third Vps34 complex that can regulate protein secretion, although it is also possible that the Vps34 complex that regulates autophagy participates in this process.
Atg6 mutant larvae possess excess hemocytes, the Drosophila equivalent of macrophages, and contain melanotic blood cell masses prior to their death. Melanotic masses are thought to be caused by at least two possible mechanisms: (1) tissue damage that recruits blood cells to encapsulate the unhealthy tissue and potentially protect the organism, and (2) over-proliferation of the blood cell lineage due to a defect in the hematopoietic stem cell niche (Minakhina and Steward, 2006). In support of the latter possibility, a recent study showed that hemocytes with decreased autophagy have decreased recruitment to epidermal wounds because of impaired cortical remodeling in the blood cells (Kadandale et al., 2010). Although blood cells clearly surround the melanotic masses in Atg6 mutant larvae, it is unclear whether the masses themselves are composed strictly of hemocytes and whether the masses result from hemocyte over-proliferation, or if hemocytes are induced to proliferate by the presence of melanotic masses. Our data indicate that Atg6 mutants have enlarged hematopoietic organs, more blood cells and altered blood cell differentiation and that blood cell aggregations precede the formation of melanotic masses. However, we cannot exclude the possibility that cells that are not of hematopoietic origin are involved in the initiation of melanotic masses.
It is interesting that, like beclin 1 mutant mice, loss of Atg6 in Drosophila results in expansion of the hematopoietic lineage. NF-κBs are known to regulate hematopoiesis in both Drosophila and mammals. In flies, Toll and cactus are key regulators of NF-κB signaling, and either Toll gain-of-function or cactus loss-of-function mutants lead to over-proliferation of hemocytes, in particular lamellocytes, resulting in the formation of melanotic masses (Qiu et al., 1998). Given the connection between beclin 1, p62 and NF-κB (Mathew et al., 2009), we speculated that the melanotic mass phenotype in Atg6 mutants could be due to misregulation of p62 and NF-κB. We systematically removed either ref(2)p (fly p62) or the three Drosophila NF-κBs dorsal, dif and Rel in combination with loss of Atg6. Surprisingly, mutations in these genes failed to suppress the Atg6 melanotic mass phenotype.
Numerous reports indicate that beclin 1 plays an important role in cancer, and most studies attribute this function to a defect in autophagy (White and DiPaola, 2009). Although autophagy is likely to contribute to tumor progression, it is also possible that the influence of beclin 1 on other vesicle trafficking pathways may promote tumor development. Consistent with this possibility, loss of Atg5 and autophagy leads to benign adenomas in livers that fail to cause cancer, but this phenotype is not observed in other tissues (Takamura et al., 2011). Here, we show that Atg6 influences multiple trafficking pathways in flies, and that Atg6 mutant animals possess hematopoietic defects and melanotic blood cell masses. Future studies of Drosophila Atg6 mutants should resolve a number of questions that are relevant to both the fundamental cellular function of this protein, as well as potentially advance our understanding of how beclin 1 functions as a tumor suppressor.
Supplementary Material
Acknowledgments
We thank Y. Rong for advice about gene targeting; C. Evans and U. Banerjee for advice about lymph gland analyses; I. Ando, A. Bergmann, D. Bilder, M. Freeman, T. Ip, T. Neufeld, I. Nezis, N. Silverman, H. Stenmark, the Bloomington Stock Center and the Vienna Drosophila RNAi Center for flies and antibodies; T. Fortier for technical support; and the Baehrecke lab for constructive comments.
Footnotes
Funding
This work was supported by National Institutes of Health grants [CA159314 to E.H.B. and S10RR027897 to the UMass EM Core]. E.H.B. is an Ellison Medical Foundation Scholar and a member of the UMass DERC [DK32520]. Deposited in PMC for release after 12 months.
Author contributions
B.V.S., J.H.H. and E.H.B. designed experiments; B.V.S., J.H.H. and C.M.P. performed experiments; and all authors wrote and discussed the manuscript.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.089490/-/DC1
References
- Aita V. M., Liang X. H., Murty V. V., Pincus D. L., Yu W., Cayanis E., Kalachikov S., Gilliam T. C., Levine B. (1999). Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics 59, 59–65 [DOI] [PubMed] [Google Scholar]
- Asha H., Nagy I., Kovacs G., Stetson D., Ando I., Dearolf C. R. (2003). Analysis of Ras-induced overproliferation in Drosophila hemocytes. Genetics 163, 203–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry D. L., Baehrecke E. H. (2007). Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell 131, 1137–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biyasheva A., Do T. V., Lu Y., Vaskova M., Andres A. J. (2001). Glue secretion in the Drosophila salivary gland: a model for steroid-regulated exocytosis. Dev. Biol. 231, 234–251 [DOI] [PubMed] [Google Scholar]
- Bodenstein D. (1965). The postembryonic development of Drosophila. In Biology of Drosophila (ed. Demerec M.), pp. 275–367 New York, NY: Hafner Publishing; [Google Scholar]
- Budnik V., Gorczyca M., Prokop A. (2006). Selected methods for the anatomical study of Drosophila embryonic and larval neuromuscular junctions. Int. Rev. Neurobiol. 75, 323–365 [DOI] [PubMed] [Google Scholar]
- Denton D., Shravage B., Simin R., Mills K., Berry D. L., Baehrecke E. H., Kumar S. (2009). Autophagy, not apoptosis, is essential for midgut cell death in Drosophila. Curr. Biol. 19, 1741–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V. (2011). Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol. Rev. 240, 92–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V., Jiang S., Dupont N. (2012). Autophagy intersections with conventional and unconventional secretion in tissue development, remodeling and inflammation. Trends Cell Biol. 22, 397–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellson C. D., Andrews S., Stephens L. R., Hawkins P. T. (2002). The PX domain: a new phosphoinositide-binding module. J. Cell Sci. 115, 1099–1105 [DOI] [PubMed] [Google Scholar]
- Fimia G. M., Stoykova A., Romagnoli A., Giunta L., Di Bartolomeo S., Nardacci R., Corazzari M., Fuoco C., Ucar A., Schwartz P., et al. (2007). Ambra1 regulates autophagy and development of the nervous system. Nature 447, 1121–1125 [DOI] [PubMed] [Google Scholar]
- Funderburk S. F., Wang Q. J., Yue Z. (2010). The Beclin 1-VPS34 complex - at the crossroads of autophagy and beyond. Trends Cell Biol. 20, 355–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly D. J., Morrow I. C., Lindsay M., Gould R., Bryant N. J., Gaullier J. M., Parton R. G., Stenmark H. (2000). Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 19, 4577–4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding T. M., Morano K. A., Scott S. V., Klionsky D. J. (1995). Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 131, 591–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne W. M., Buchkovich N. J., Emr S. D. (2011). The ESCRT pathway. Dev. Cell 21, 77–91 [DOI] [PubMed] [Google Scholar]
- Herz H. M., Chen Z., Scherr H., Lackey M., Bolduc C., Bergmann A. (2006). vps25 mosaics display non-autonomous cell survival and overgrowth, and autonomous apoptosis. Development 133, 1871–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz H. M., Woodfield S. E., Chen Z., Bolduc C., Bergmann A. (2009). Common and distinct genetic properties of ESCRT-II components in Drosophila. PLoS ONE 4, e4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., Kishi C., Inoue K., Mizushima N. (2008). Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell 19, 5360–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhász G., Hill J. H., Yan Y., Sass M., Baehrecke E. H., Backer J. M., Neufeld T. P. (2008). The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila. J. Cell Biol. 181, 655–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadandale P., Stender J. D., Glass C. K., Kiger A. A. (2010). Conserved role for autophagy in Rho1-mediated cortical remodeling and blood cell recruitment. Proc. Natl. Acad. Sci. USA 107, 10502–10507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karantza-Wadsworth V., Patel S., Kravchuk O., Chen G., Mathew R., Jin S., White E. (2007). Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 21, 1621–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A., Kabeya Y., Ohsumi Y., Yoshimori T. (2001a). Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2, 330–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A., Noda T., Ishihara N., Ohsumi Y. (2001b). Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 152, 519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T., Baba M., Ishihara N., Miyazawa K., Ohsumi M., Yoshimori T., Noda T., Ohsumi Y. (1999). Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147, 435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebestky T., Chang T., Hartenstein V., Banerjee U. (2000). Specification of Drosophila hematopoietic lineage by conserved transcription factors. Science 288, 146–149 [DOI] [PubMed] [Google Scholar]
- Liang C., Lee J. S., Inn K. S., Gack M. U., Li Q., Roberts E. A., Vergne I., Deretic V., Feng P., Akazawa C., et al. (2008). Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat. Cell Biol. 10, 776–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindmo K., Stenmark H. (2006). Regulation of membrane traffic by phosphoinositide 3-kinases. J. Cell Sci. 119, 605–614 [DOI] [PubMed] [Google Scholar]
- Lindmo K., Brech A., Finley K. D., Gaumer S., Contamine D., Rusten T. E., Stenmark H. (2008). The PI 3-kinase regulator Vps15 is required for autophagic clearance of protein aggregates. Autophagy 4, 500–506 [DOI] [PubMed] [Google Scholar]
- Maroni G., Stamey S. C. (1983). Use of blue food to select synchronous, late third instar larvae. Dros. Inf. Serv. 59, 142–143 [Google Scholar]
- Mathew R., Kongara S., Beaudoin B., Karp C. M., Bray K., Degenhardt K., Chen G., Jin S., White E. (2007). Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 21, 1367–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R., Karp C. M., Beaudoin B., Vuong N., Chen G., Chen H. Y., Bray K., Reddy A., Bhanot G., Gelinas C., et al. (2009). Autophagy suppresses tumorigenesis through elimination of p62. Cell 137, 1062–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga K., Saitoh T., Tabata K., Omori H., Satoh T., Kurotori N., Maejima I., Shirahama-Noda K., Ichimura T., Isobe T., et al. (2009). Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 11, 385–396 [DOI] [PubMed] [Google Scholar]
- Minakhina S., Steward R. (2006). Melanotic mutants in Drosophila: pathways and phenotypes. Genetics 174, 253–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Komatsu M. (2011). Autophagy: renovation of cells and tissues. Cell 147, 728–741 [DOI] [PubMed] [Google Scholar]
- Muro I., Berry D. L., Huh J. R., Chen C. H., Huang H., Yoo S. J., Guo M., Baehrecke E. H., Hay B. A. (2006). The Drosophila caspase Ice is important for many apoptotic cell deaths and for spermatid individualization, a nonapoptotic process. Development 133, 3305–3315 [DOI] [PubMed] [Google Scholar]
- Nezis I. P., Simonsen A., Sagona A. P., Finley K., Gaumer S., Contamine D., Rusten T. E., Stenmark H., Brech A. (2008). Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J. Cell Biol. 180, 1065–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu P., Pan P. C., Govind S. (1998). A role for the Drosophila Toll/Cactus pathway in larval hematopoiesis. Development 125, 1909–1920 [DOI] [PubMed] [Google Scholar]
- Qu X., Yu J., Bhagat G., Furuya N., Hibshoosh H., Troxel A., Rosen J., Eskelinen E. L., Mizushima N., Ohsumi Y., et al. (2003). Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 112, 1809–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Y. S., Golic K. G. (2000). Gene targeting by homologous recombination in Drosophila. Science 288, 2013–2018 [DOI] [PubMed] [Google Scholar]
- Ruck A., Attonito J., Garces K. T., Núnez L., Palmisano N. J., Rubel Z., Bai Z., Nguyen K. C., Sun L., Grant B. D., et al. (2011). The Atg6/Vps30/Beclin 1 ortholog BEC-1 mediates endocytic retrograde transport in addition to autophagy in C. elegans. Autophagy 7, 386–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R. C., Schuldiner O., Neufeld T. P. (2004). Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev. Cell 7, 167–178 [DOI] [PubMed] [Google Scholar]
- Scott R. C., Juhász G., Neufeld T. P. (2007). Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr. Biol. 17, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M. N., Marcusson E. G., Cereghino J. L., Emr S. D. (1997). Endosome to Golgi retrieval of the vacuolar protein sorting receptor, Vps10p, requires the function of the VPS29, VPS30, and VPS35 gene products. J. Cell Biol. 137, 79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A., Birkeland H. C., Gillooly D. J., Mizushima N., Kuma A., Yoshimori T., Slagsvold T., Brech A., Stenmark H. (2004). Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. J. Cell Sci. 117, 4239–4251 [DOI] [PubMed] [Google Scholar]
- Stenmark H., Aasland R., Driscoll P. C. (2002). The phosphatidylinositol 3-phosphate-binding FYVE finger. FEBS Lett. 513, 77–84 [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Coppola D., Matsushita N., Cualing H. D., Sun M., Sato Y., Liang C., Jung J. U., Cheng J. Q., Mulé J. J., et al. (2007). Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat. Cell Biol. 9, 1142–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamura A., Komatsu M., Hara T., Sakamoto A., Kishi C., Waguri S., Eishi Y., Hino O., Tanaka K., Mizushima N. (2011). Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 25, 795–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson B. J., Mathieu J., Sung H. H., Loeser E., Rørth P., Cohen S. M. (2005). Tumor suppressor properties of the ESCRT-II complex component Vps25 in Drosophila. Dev. Cell 9, 711–720 [DOI] [PubMed] [Google Scholar]
- Thoresen S. B., Pedersen N. M., Liestøl K., Stenmark H. (2010). A phosphatidylinositol 3-kinase class III sub-complex containing VPS15, VPS34, Beclin 1, UVRAG and BIF-1 regulates cytokinesis and degradative endocytic traffic. Exp. Cell Res. 316, 3368–3378 [DOI] [PubMed] [Google Scholar]
- Thumm M., Egner R., Koch B., Schlumpberger M., Straub M., Veenhuis M., Wolf D. H. (1994). Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 349, 275–280 [DOI] [PubMed] [Google Scholar]
- Tsukada M., Ohsumi Y. (1993). Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333, 169–174 [DOI] [PubMed] [Google Scholar]
- Vaccari T., Bilder D. (2005). The Drosophila tumor suppressor vps25 prevents nonautonomous overproliferation by regulating notch trafficking. Dev. Cell 9, 687–698 [DOI] [PubMed] [Google Scholar]
- Vaccari T., Rusten T. E., Menut L., Nezis I. P., Brech A., Stenmark H., Bilder D. (2009). Comparative analysis of ESCRT-I, ESCRT-II and ESCRT-III function in Drosophila by efficient isolation of ESCRT mutants. J. Cell Sci. 122, 2413–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E., DiPaola R. S. (2009). The double-edged sword of autophagy modulation in cancer. Clin. Cancer Res. 15, 5308–5316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Randle K. E., Wu L. P. (2007). ird1 is a Vps15 homologue important for antibacterial immune responses in Drosophila. Cell. Microbiol. 9, 1073–1085 [DOI] [PubMed] [Google Scholar]
- Wucherpfennig T., Wilsch-Bräuninger M., González-Gaitán M. (2003). Role of Drosophila Rab5 during endosomal trafficking at the synapse and evoked neurotransmitter release. J. Cell Biol. 161, 609–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Rubin G. M. (1993). Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 117, 1223–1237 [DOI] [PubMed] [Google Scholar]
- Yang Z., Huang J., Geng J., Nair U., Klionsky D. J. (2006). Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol. Biol. Cell 17, 5094–5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z., Jin S., Yang C., Levine A. J., Heintz N. (2003). Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. USA 100, 15077–15082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Qi H., Taylor R., Xu W., Liu L. F., Jin S. (2007). The role of autophagy in mitochondria maintenance: characterization of mitochondrial functions in autophagy-deficient S. cerevisiae strains. Autophagy 3, 337–346 [DOI] [PubMed] [Google Scholar]
- Zhong Y., Wang Q. J., Li X., Yan Y., Backer J. M., Chait B. T., Heintz N., Yue Z. (2009). Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 11, 468–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


