Abstract
RNA aptamers that bind the reverse transcriptase (RT) of human immunodeficiency virus (HIV) compete with nucleic acid primer/template for access to RT, inhibit RT enzymatic activity in vitro, and suppress viral replication when expressed in human cells. Numerous pseudoknot aptamers have been identified by sequence analysis, but relatively few have been confirmed experimentally. In this work, a screen of nearly 100 full-length and >60 truncated aptamer transcripts established the predictive value of the F1Pk and F2Pk pseudoknot signature motifs. The screen also identified a new, nonpseudoknot motif with a conserved unpaired UCAA element. High-throughput sequence (HTS) analysis identified 181 clusters capable of forming this novel element. Comparative sequence analysis, enzymatic probing and RT inhibition by aptamer variants established the essential requirements of the motif, which include two conserved base pairs (AC/GU) on the 5′ side of the unpaired UCAA. Aptamers in this family inhibit RT in primer extension assays with IC50 values in the low nmol/l range, and they suppress viral replication with a potency that is comparable with that of previously studied aptamers. All three known anti-RT aptamer families (pseudoknots, the UCAA element, and the recently described “(6/5)AL” motif) are therefore suitable for developing aptamer-based antiviral gene therapies.
Keywords: RNA aptamer, antiviral gene therapy, high-throughput sequencing, HIV, reverse transcriptase
Introduction
The reverse transcriptase (RT) from type 1 human immunodeficiency virus (HIV-1) plays an essential role in viral replication by copying the RNA genome into double-stranded DNA (dsDNA) before insertion of the DNA into the host genome. RT inhibition is a proven therapeutic strategy for clinical treatment of HIV infection, although limitations in drug tolerance and the selection of drug-resistant viral strains continue to motivate the search for new therapeutic approaches. Nucleic acid aptamers composed of RNA or single-stranded DNA (ssDNA) that bind RT have been identified through the SELEX process (Systematic Evolution of Ligands by Exponential enrichment).1,2,3,4 Many of these aptamers compete with the natural primer/template for access to RT, inhibiting the DNA polymerization and RNaseH activities of RT at low nanomolar concentrations in enzymatic assays.3,4,5,6,7,8,9,10,11,12,13 In addition, several RNA aptamers strongly suppress viral replication when expressed in cells14,15,16,17 and could potentially be adapted for hematopoietic stem cell gene therapy. These findings have prompted substantial interest in developing RNA aptamer inhibitors of HIV-1.
Structural diversity among nucleic acid aptamers that bind RT correlates with significant functional diversity. For example, pseudoknot (Pk) RNA aptamers have been loosely categorized as being either “family 1” (F1Pk)—defined primarily by the presence of UCCG/CGGG in stem 1 and first identified by Tuerk3—or “family 2” (F2Pk)—originally a catch-all classification for any potential pseudoknot not matching the F1Pk definition.4 In one study,12 two F1Pk and two F2Pk aptamers strongly inhibited RT from HIV strains in which position 277 was Arg, but only the F2Pk aptamers continued to inhibit when position 277 was Lys, which is a common polymorphism among circulating strains of HIV-1. In two other studies,9,11 the ssDNA aptamers RT1t49, R1T and variants of these two strongly inhibited all the members of a phylogenetically diverse panel of purified recombinant RT, whereas another ssDNA aptamer, RT8, was highly specific for RT from a subtype B strain of HIV-1. We recently used mass spectrometry footprinting to establish that the broad-spectrum ssDNA aptamers protect essentially the same surfaces of RT as those protected by dsDNA, whereas a pseudoknot RNA aptamer (F1Pk) protects a substantially smaller surface area.18 Aptamer diversity also has implications for diagnostic applications. Li et al. identified two nonpseudoknot RNA aptamers (M302 and 12.01) that discriminated between wild-type RT and a particular octamutated, drug-resistant RT. Both aptamers M302 and 12.01 bind RT with low nmol/l affinity, but neither one inhibits RT enzymatic activity or competes with pseudoknot aptamers or primer/template for binding to RT.1 Functional variations among aptamers of different structural classes could also lead to differences in susceptibility to the emergence of de novo resistance mutations and to the differences in potential off-target effects. It is therefore important to understand the diversity of structural motifs that can bind RT.
RNA pseudoknots, especially the F1Pk, have long been recognized as high affinity ligands for HIV-1 RT, and they dominated the first three populations of RT-binding RNA aptamers that were described.3,4,13 Interestingly, no such convergence on pseudoknots occurred among RNA and ssDNA aptamers from other selections, such as RNA aptamers selected to bind RT from Moloney murine leukemia virus, feline immunodeficiency virus or avian myeloblastosis virus,19,20 ssDNA aptamers selected to bind HIV-1 RT, and RNA aptamers selected to differentiate between drug-resistant and wild-type HIV-1 RT.1 These observations together suggest that there are likely additional, nonpseudoknot structural motifs present in these populations. Indeed, by applying high-throughput sequencing (HTS) and a newly developed bioinformatics pipeline to the 70HRT14 population of HIV-1 RT aptamers,4 we recently identified another structural element termed “(6/5)AL,” in which an asymmetric internal loop with six nucleotides in one strand and five in the other is flanked by generic stems with different length requirements.13
The 32N population from the first RT-aptamer selection3 and the subsequent 70HRT14 and 80HRT14 populations4 were all originally selected to bind RT from HIV-1 strain BH10. Low-throughput sequence (LTS) analysis of these three populations identified 18, 46, and 44 nonidentical published sequences (108 total), respectively, from among 194 total reads (95, 54, and 45, respectively, for the three populations). Potential pseudoknot-forming elements were identified within most of the sequences from all three selections. More than half (61 of 108) contained the F1Pk signature sequence (11, 31, and 19 aptamer sequences, respectively, for the three populations). Alternative F2Pk lacking this signature sequence were proposed4 for another 36 sequences (11 and 25 from populations 70HRT14 and 80HRT14, respectively). A small handful of F1Pk and relatively compact F2Pk have been confirmed experimentally,3,4,5,6,7,12,21 but several of the manually assigned F2Pk have very large loops or very short stems that may be incompatible with pseudoknot formation, leaving open the possibility that portions of those transcripts other than the putative pseudoknots may be responsible for RT-binding affinity. In addition, nearly all of the sequences in the 70HRT14 and 80HRT14 populations were sampled only once, indicating that significant untapped sequence diversity remains within both populations.
These observations raised two immediate questions: (i) whether the 30–50 nucleotide F1Pk and F2Pk identified by sequence gazing represent the core RT-binding segments within the original 118–134 nucleotide transcripts, and (ii) whether additional RT-binding structures might be present within these populations. By screening nearly 100 full-length aptamers and >60 truncated variants, we established that the original F1Pk definition is highly reliable in defining the RT-binding module within aptamers that contain this sequence, and that most but not all of the original F2Pk account for RT binding by those RNAs. Importantly, this work also identified several nonpseudoknot RNAs, including two aptamers that form similar secondary structures with a conserved UCAA internal bulge and that inhibit RT with IC50 values below 10 nmol/l. HTS analysis identified >150 independent examples of this structural element and, in conjunction with enzymatic digestion and mutational analysis, defined the sequence requirements for forming the RT-binding module. The UCAA aptamers reduce infectivity of virus produced in the presence of aptamer and display a potency that is at least comparable to RNA aptamers with other structural motifs. This new UCAA family of aptamers represents one of the few published examples of nonpseudoknot RNA structures that inhibit RT, and illustrates the ability of structurally unrelated RNA aptamers to bind and inhibit the same protein target.
Results
Sequence and structural diversity within 70HRT14 and 80HRT14 aptamer populations
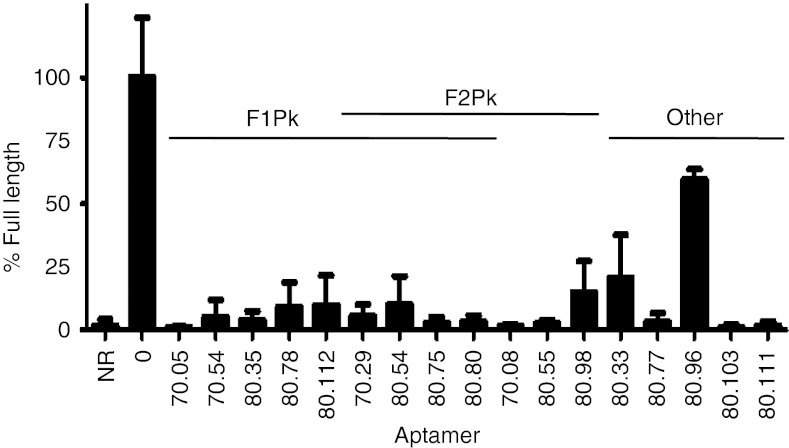
Sixty additional aptamer plasmid sequences were obtained to augment the published LTS data set and gain new insights into 70HRT14 and 80HRT14 aptamer population diversity.4 Thirty-five of these represent sequences that had not previously been sampled, bringing the total published LTS data set to 143 independent sequences from 254 reads (Supplementary Figure S1 and refs. [3,4]). Inhibition of primer extension by RT from HIV-1 subtype B strain HXB2 was evaluated for full-length aptamer transcripts from 98 plasmid isolates from the 70HRT14 and 80HRT14 populations (approximately 118 and 134 nt, respectively), and these aptamers were grouped according to their relative potency (Figure 1, Supplementary Figure S2 and data not shown). When small transcripts (26–48 nt) corresponding to putative pseudoknot cores were similarly tested, all of the F1Pk cores (12 of 12) inhibited RT as potently as their corresponding full-length versions, confirming that the F1Pk signature sequence accurately identifies the RT-binding elements within the full-length 116–134 nt transcripts. In contrast, only 60% of the F2Pk cores (9 of 15) identified manually at the time of the original selection inhibited RT as well as their corresponding full-length transcripts (Supplementary Figure S3). In some cases, manual screens of truncated transcripts for the other 40% identified new inhibitory F2Pk that had not been recognized previously (Supplementary Figure S2), but even after this analysis there remained 19 inhibitory aptamers for which no pseudoknots were evident (Supplementary Figure S2). Two of the most inhibitory nonpseudoknot aptamers were 80.103 and 80.111, which suppressed full-length product formation by RT to below the detection limit of this assay (Figure 1). These two aptamers were therefore chosen for further study.
Figure 1.
Inhibition of DNA polymerization by HIV-1 reverse transcriptase by RNA aptamers. Quantification of primer extension assays showing fraction of primer converted into full-length product in the absence of RT (“NR”), control reactions without aptamer (“0”), and various full-length aptamers, as indicated below the graph. Plotted values and vertical error bars represent the mean and SD of the fraction extended to full length (normalized to the fraction extended to full length in the no-aptamer control, set to 100%) for three independent experiments. “F1Pk” and “F2Pk” indicate aptamers that carry sequences that match the consensus motif definitions of these two families of pseudoknots.13 Several aptamers meet motif definitions for both pseudoknot families. No pseudoknots were evident in the sequences of the five samples on the right. NR, no reaction.
Deletion analysis was used to define the segment within aptamer 80.103 that is required for binding RT. Several RNA transcripts that began at nucleotide 21 or 25 inhibited RT to essentially the same degree as did the full-length aptamer, whereas all transcripts that began at nucleotide 31 failed to inhibit (Figure 2a). Similarly, transcripts of aptamer 80.103 that ended at position 84 or 94 inhibited RT, whereas all transcripts that ended at position 74 failed to inhibit (Figure 2a). Thus, the functional core of aptamer 80.103 is fully contained between nucleotides 25 and 84. A similar analysis for aptamer 80.111 established that its functional core is fully contained between nucleotides 9 and 68 (Figure 2a). Potential secondary structures consistent with these functional boundaries are shown in Figure 2b.
Figure 2.
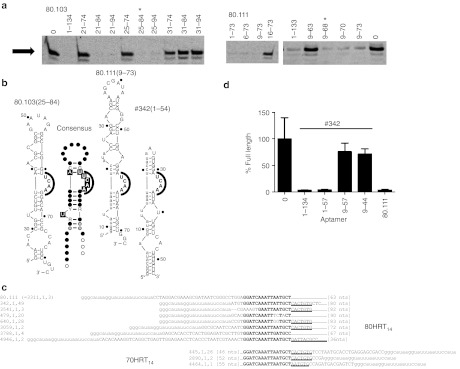
Functional cores of UCAA family aptamers. (a) Primer extension assays were carried out in the presence of truncated aptamer 80.103 (left) and aptamer 80.111 (right), bearing the nucleotide segments indicated above the lanes. Gel images show only the region near the full-length product (arrow). “0”, reaction product formed in the absence of aptamer. *The indicated functional core lies between nucleotides 25 and 84 for aptamer 80.103 and between nucleotides 9 and 68 for aptamer 80.111. (b) Predicted secondary structures of aptamers 80.103(25–84), 80.111(9–73) and #342(1–54). Numbers after the aptamer names indicate the 5′ and 3′ nucleotides in the functional cores. Lower case nucleotides indicate segments derived from the constant primer-binding segments of the library. Numbering on the secondary structures in this and subsequent figures indicates nucleotide positions in the aptamers from which they are derived to facilitate comparison among truncated variants. Equivalent inhibition results were obtained from 80.103(25–84) when two additional G's were appended to the 5′ end to increase transcription efficiency (not shown). Structures of aptamers 80.111 and #342 represent the two major pairing pattern observed for sequences that paired with the constant region. Also shown in this panel is the conservation patterns among the most abundant sequences within each of the 42 clusters for the UCAA family isolates that are fully contained within the 80N random region of the library. Black boxes, >95% conservation within cluster and black dots; Black letters and gray dots, 90–95%, conservation; Grey letters and open circles, 85–90% conservation; R or Y >85% purine or pyrimidine, respectively. (c) Alignments of 80.111 with other sequences in the 70HRT14 and 80HRT14 populations revealed a shared 16 nt sequence that can base pair with part of the constant primer-binding region (bold). Underlined segment extends the shared sequence element to approximately 23 nt. The three numbers in the sequence identifiers indicate (rank number of the cluster within the population), (rank number of this specific sequence within the cluster), (number of repeat samplings of that sequence). Thus, sequence “342.1.49” is the most abundant sequence within cluster #342, and this exact sequence was sampled 49 times. (d) Four truncations of #342 were in vitro transcribed to test base pairing hypothesis. These truncations were used in primer extension assays, and the percent of primer converted to full-length product were plotted relative to the no-aptamer control as in Figure 1. Full-length aptamer 80.111 was included for comparison.
High-throughput comparative sequence analysis reveals a novel conserved UCAA bulge motif
During the course of the analysis above, we completed a separate study of the 70HRT14 aptamer populations by HTS analysis, including development of a bioinformatics pipeline specifically adapted for SELEX data sets.13 HTS reads from the 80HRT14 population were parsed into the “clusters” of closely related sequences with edit distance <7 to capture essentially all mutational variants that share common ancestry with the surviving sequences.13 The 5,000 most abundant clusters collectively comprised 613,313 quality-filtered sequence reads, and all clusters contained at least two such reads. As with the 70HRT14 population,13 the HTS data set for the 80HRT14 population is dominated by pseudoknots (data not shown). The cluster containing aptamer 80.103 is the 27th most abundant cluster in the population, with 768 unique sequences (2,666 total sequence reads) representing 0.43% of the population. These sequences were aligned using the software Mafft, and an initial secondary structure of the functional core was predicted from the observed patterns of conservation and covariation using RNalifold. The stems and a single-stranded UCAA bulge are highly conserved within the cluster, whereas the sequences in the terminal loop, single nucleotide bulges, and sequences near the 5′ and 3′ predicted boundaries are less conserved.
The cluster containing aptamer 80.111 is much more rare (3,311th most abundant cluster), with only six unique sequences and eight total sequence reads, representing 0.001% of the population. The limited sample number and diversity precluded meaningful evaluation of intracluster conservation and covariation.
However, a search for matching character strings within the experimentally determined functional boundaries and within the original 80N random region identified a stretch of 16 nucleotides (GGATCAAATTAATGCT) within the functional core of 80.111 that is highly conserved among 16 other independent clusters (perfectly conserved in nine), and that extends to 23 nucleotides of conservation in 12 (Figure 2c and Supplementary Figure 4a). Potential pairing of this element with the 5′ primer-binding segment (used for amplifying the library during the selection) produces a UCAA bulge that is analogous to the conserved feature within the 80.103 cluster (Figure 2b). The most abundant clusters to contain this 16 nt conserved element was aptamer #342, so called because it is the 342nd most abundant within the 80HRT14 population (123 unique sequences among 199 reads). Its putative secondary structure includes the full 23 nucleotide version of the conserved element and is fully contained within the first 54 nucleotides. Transcripts comprising nucleotides 1–134 (full length) or nucleotides 1–54 strongly inhibited primer extension by RT, whereas truncations comprising nucleotides 9–54 or 9–44 did not inhibit (Figure 2d), consistent with pairing between the 5′ constant region and the full 23 nucleotide version of the conserved element (Figure 2b).
UCAA aptamer family
The apparent convergence of aptamers 80.103, 80.111, and #342 on a previously unrecognized RT-binding structural motif led us to apply a more rigorous informatics analysis in four stages. The first stage involved two rounds of curated search and refinement. We first built a covariation model (CM) based on the sequences of these three aptamers, aligned in the CM to preserve the predicted UCAA bulge. The CM search identified 28 new sequences that conformed to the CM, including two sequences that formed the two stems with completely different sequences than those observed in the three seed sequences. These two sequences were added to the alignment of the three original seed sequences to generate a new, refined CM that was again searched against the 80HRT14 population. This second search identified a total of 57 sequences that conformed to the refined CM, including four sequences that formed the UCAA structure entirely within the 80 nt random region, and were thus not subject to the constraints of the constant primer-binding sequences.
The second phase of the analysis was a semiautomated CM search of only the 80N random region of the 80HRT14 population, after first removing the 5′ and 3′ constant regions from the data set. The four sequences that formed the UCAA structure without the involvement of the constant regions were then used to seed multiple rounds of semiautomated CM searching and refinement. This process identified 42 clusters that formed the UCAA element entirely within the random region, including the cluster that contains aptamer 80.103. The consensus structure based on these 42 clusters, which is free from sequence constraints imposed by the primer-binding regions, is a simplified stem-loop structure in which the UCAA bulge is flanked on one side by two highly conserved base pairs (AC/GU) and on the other by a largely generic helix that is interrupted by a single unpaired U residue (Figure 2b).
In the third phase of the analysis, the 5′ and 3′ constant regions were re-appended to the sequences in the full 80HRT14 population, and the CM built from sequences that form the UCAA structural element entirely within the random region was then used to search this data set. This search identified an additional 109 clusters that form the UCAA bulge motif secondary structures by pairing with elements in the 5′ primer-binding segment, most frequently in the pairing combination AAU^UCC/GGAUCAAAUU, in which “^” represents the site across from the UCAA element in bold (Figure 3b). The frequent utilization of a specific pairing register with the constant region suggests that the constant region contributes modestly to satisfying the required sequence information content of the motif. Among the aptamers that carried the 16 nt conserved element that pairs with the 5′ constant regions in aptamers 80.111 and #342, only one of these (#947) was also present in this final set of sequences from the CM search based on the conserved sequence features that were not constrained by the 5′ or 3′ constant regions. These three sets of hits therefore yield a total of 167 clusters within the UCAA family from the 80HRT14 population (Supplementary Figure S4a,b).
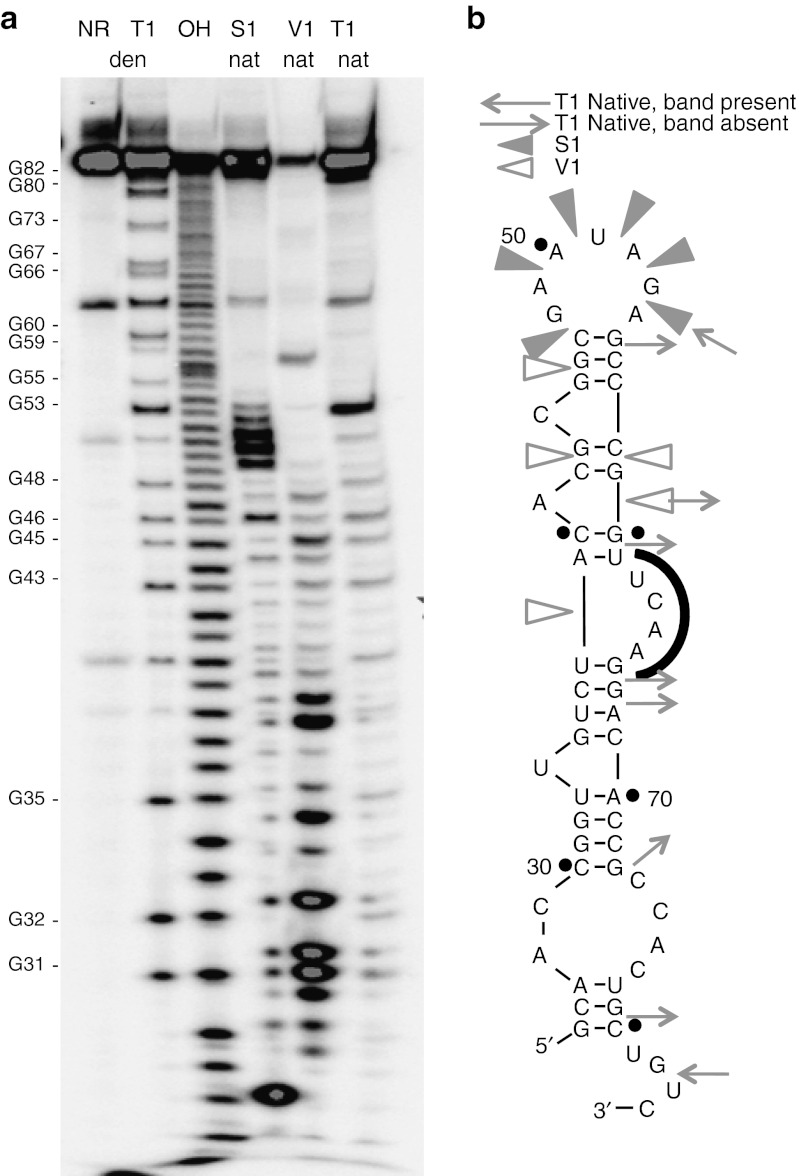
Figure 3.
Enzymatic probing of aptamer 80.103(25–84). (a) Electrophoretic analysis of the products of digestion reactions utilizing nucleases T1, V1 and S1. NR, no reaction; den, digestion carried out under denaturing conditions; nat, digestion carried out under native conditions; OH, alkaline digestion at elevated pH. Note that alkaline and T1 digestions leave 2′,3′-cyclic phosphate products, whereas S1 and V1 digestions leave 3′-OH products;31 hence, S1 and V1 digestion products are shifted approximately 1 nucleotide upward relative to alkaline and T1 digestion products. G residues marked on the left are numbered according to their positions in the full-length aptamer transcripts. (b) Digestion results were mapped onto secondary structure of aptamer 80.103(25–84). Open wedges, cleavage sites for nuclease V1; filled wedges, cleavage sites for nuclease S1; arrows pointing inward, cleavage sites for nuclease T1; arrows pointing outward, G residues that are not cleaved in the native structure. Similar analysis for aptamer 80.111 is presented in Supplementary Figure S5.
Finally, in the fourth phase of the analysis, similar searches were applied to several 70HRT populations described previously.13 This process identified 14 additional UCAA structures, 11 of which were identified using the consensus structural model derived from the 80N random region and three of which contained the 16 nt conserved sequence noted above (Supplementary Figure S4c). Although the 70HRT14 aptamers are from an independent selection, their 3′ primer-binding segment is identical to the 5′ primer-binding segment of the 80HRT14 population4 and can pair with the conserved 16 nucleotide sequence. The 70HRT240s population experienced more stringent selection conditions than the 70HRT14 population. Under the more stringent conditions, the relative abundance of sequences that carried the (6/5)AL and (6/5)ALcp motifs significantly increased with selection stringency, whereas the relative abundances of sequences with F1Pk and F2Pk motifs significantly decreased, and these trends are readily explained by their respective RT-binding affinities.13 Applying a similar analysis to the UCAA-motif aptamers revealed that their relative abundance increases slightly in the 70HRT240s population, suggesting that the UCAA aptamers are intermediate between pseudoknots and (6/5)AL aptamers.
Experimental confirmation of secondary structures
Aptamer 80.103(25–84) was analyzed using enzymatic probing to determine which portions of the molecule show susceptibility to cleavage by endonucleases S1, V1, or T1 (Figure 3a), and the results were mapped onto the proposed secondary structure (Figure 3b). Nucleotides 40, 52, and 64 are partially cleaved in the absence of endonuclease (Figure 3a, lane 1), suggesting that these nucleotides are unstructured under native conditions and that they are susceptible to in-line attack by the 2′-OH. These positions map to a single nucleotide bulge, to the terminal loop, and to the UCAA bulge, respectively. Nucleotides 47 and 49–53 in the terminal loop are susceptible to cleavage by nuclease S1 (Figure 3a, lane 4), which preferentially cleaves single-stranded regions within folded RNA. Likewise, nuclease T1 cleaves after each G residue when the RNA is fully denatured (Figure 3a, lane 2) but only after the limited subset of G's that remain unstructured under native conditions (Figure 3a, lane 6). Comparing these two lanes identifies G residues that are predominately single or double stranded in the native structure. Nucleotides 38, 45, and 58–59 are susceptible to cleavage by nuclease V1 (Figure 3a, lane 5), which preferentially cleaves double-stranded RNA. These cleavages are again consistent with the predicted secondary structure. Aptamer 80.111 was subjected to similar enzymatic nuclease treatments and the cleavage patterns were also consistent with the predicted UCAA bulge structure (Supplementary Figure S5).
Mutational analysis reveals simplified sequence requirements for the members of the UCAA aptamer family
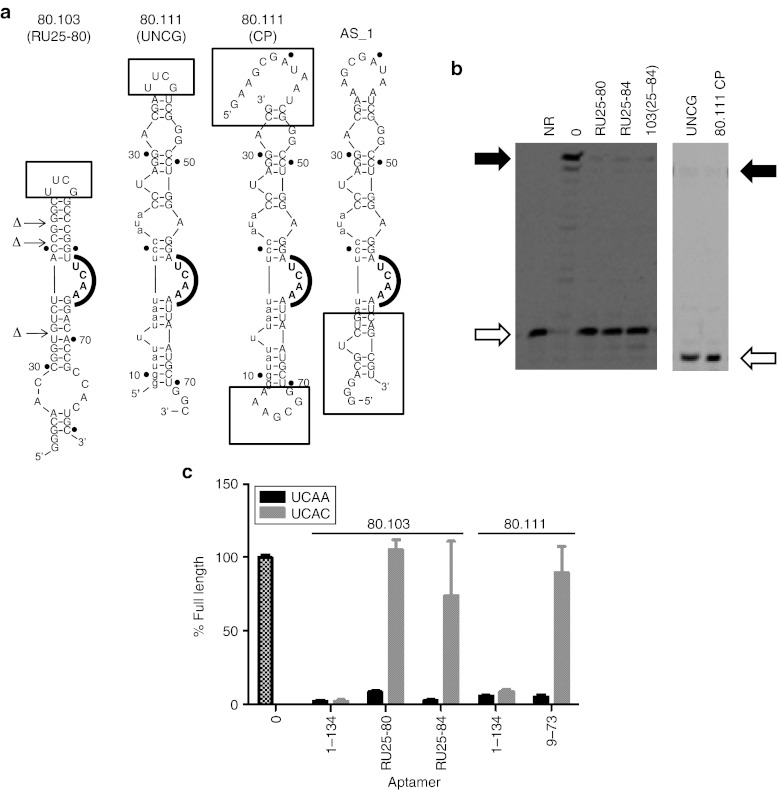
Results from the comparative sequence analysis and enzymatic probing were used to guide the construction of terminal and internal mutations that were introduced to refine the nucleotide requirements of the UCAA aptamer family. Disrupting two of the helical elements of the 60-nt truncated aptamer 80.103(25–84) (mutants “A Stem” and “C Stem,” respectively) abolished RT inhibition, whereas restoring base pairing potential (“B Stem” and “D Stem,” respectively) partially rescued RT inhibition (Supplementary Figure S6a). Additional variants strongly inhibited primer extension by RT when the terminal heptanucleotide loop (GAAUAGA) was simplified to a stable UUCG tetraloop (80.103UNCG), when the unpaired nucleotides on the 5′ side of the stem were removed (80.103RB1), and when the 3′ end was trimmed to nucleotide 80 (data not shown). Combining these three separate mutations yielded the simplified, 50 nucleotide-long RU25-80 variant (Figure 4), which retains strong RT inhibition in primer extension assays.
Figure 4.
Utilization of UCAA bulge and peripheral flanking sequences. (a) Sequences and projected secondary structures of some of the aptamer variants analyzed here. Boxed portions indicate segments addressed by individual mutants. (b) Two gels showing results of simplifying aptamers 80.103 (left) and 80.111 (right). The “RU” designation indicates that three internal unpaired nucleotides were removed from the corresponding truncations of aptamer 80.103. 80.111 mutants “UNCG” and “CP” are shown in panel A. Open and filled arrows indicate positions of unextended primer and full-length product bands, respectively. Results from additional internal mutations are given in Supplementary Figure S6. (c) Partial disruption of UCAA element. Primer extension by RT was measured in the presence of several variants of aptamers 80.103 and 80.111. Black bars indicate parental sequences. Gray bars indicate mutated sequences in which UCAA bulges were mutated to UCAC. Error bars indicate the SDs among three independent measurements. NR, no reaction.
A similar analysis was carried out for aptamer 80.111 (Figure 4 and Supplementary Figure S6b). RT was strongly inhibited by a mutant in which the 9 nt apical loop sequence was changed to a stable UUCG tetraloop (“80.111UNCG”), and by a circularly permutated mutant in which the original 5′ and 3′ ends were joined through an additional GAAA segment and transcription was initiated from an internal position (Figure 4b, right). These observations establish that there is no requirement for specific sequence or structural features at the helical termini. Disrupting a helical element just below the UCAA motif (“16–17 Disrupt”) abolished RT inhibition, whereas restoring base pairing potential (“16–17 Restore”) rescued inhibition (Supplementary Figure S6b). Two mutants that lacked the single unpaired U in the lower stem (“BPU” and “Delta U”) both inhibited RT slightly less than the original 80.111.
To understand the contribution of the UCAA bulge, this element was mutated to UCAC for the full-length and truncated forms of aptamers 80.103 and 80.111. In the context of the truncated aptamers, these mutations abolished RT inhibition (Figure 4c), consistent with the postulated functional importance of the UCAA element. In contrast, the full-length molecules were not sensitive to mutating the UCAA to UCAC, potentially indicating that peripheral elements in the full-length molecules form additional stabilizing contacts that compensate for partial disruption of the UCAA element.
A final series of transcripts interchanged upper and lower helical elements from different aptamers. For this analysis, two categories of UCAA aptamers were considered: those that form the lower stem by pairing with the primer-binding segment using the 16 nucleotide element identified above, and those that form their secondary structures fully within the random region. Strong inhibition was observed for chimeric aptamer AS1, in which the lower portion represents a consensus among aptamers within the first category and the upper portion is derived from aptamer 80.111UNCG (Figure 4a and data not shown). In contrast, no appreciable inhibition was observed when the lower portion was derived from 80.111 and the upper portion was derived from aptamers from the second category, or when any further truncations or modifications were introduced in the distal helical element that is closed by the tetraloop (AS2 through AS23, Supplementary Figure S7). The 60 to 63 nucleotide aptamers 80.111UNCG and AS1 therefore represent minimal versions of aptamer 80.111.
UCAA aptamer family inhibits RT in the low nanomolar range
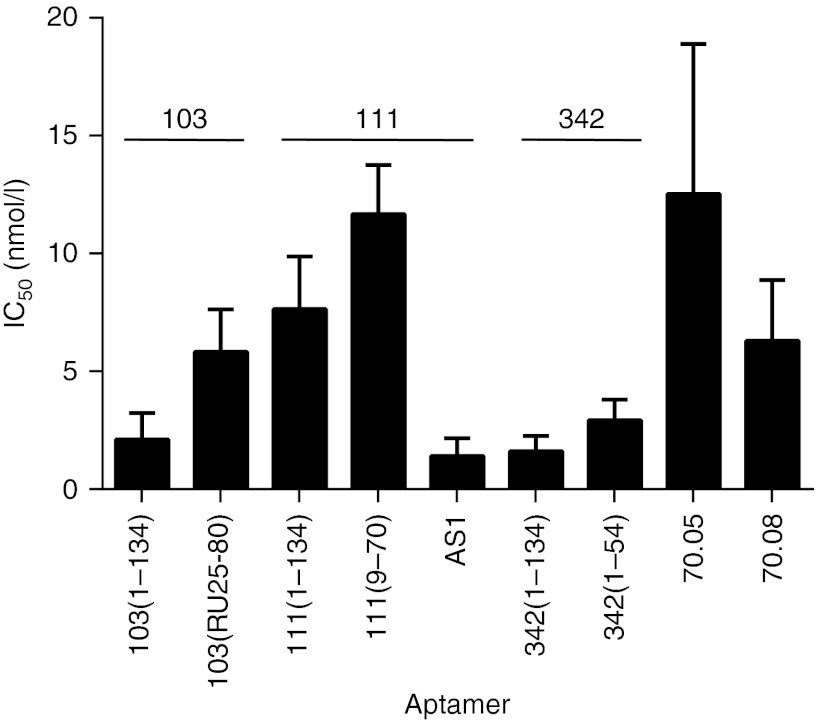
To compare quantitatively the functional cores of each aptamer with the corresponding full-length transcripts, the concentration dependence of the inhibition was determined for both the full-length and minimal cores of aptamers 80.103, 80.111, and #342 (Supplementary Figure S8 and Figure 5). The aptamer concentration required to inhibit 50% of RT activity (IC50) was slightly lower for the full-length aptamers than for the cores. Aptamer #342 had the lowest IC50 value of 1.6 ± 0.3 nmol/l for the full-length and 2.9 ± 0.4 nmol/l for aptamer #342(1–54). Inhibition by aptamer 80.103 was similar, with an IC50 value of 2.1 ± 0.5 nmol/l for the full length and 5.8 ± 0.8 nmol/l for the 50 nt functional core RU25-80. Aptamer 80.111 was slightly weaker, with an IC50 value of 7.6 ± 1.0 nmol/l for the full length and 11.7 ± 1.0 nmol/l for 80.111(9–70), but hybrid aptamer AS1 was as potent as aptamer #342, with an IC50 value of 1.4 ± 0.3 nmol/l. Under these same assay conditions, the full-length transcripts for F1Pk aptamer 70.05 and for F2Pk aptamer 70.08 had IC50 values of 13 ± 3 nmol/l and 6.3 ± 1.1 nmol/l, respectively (Figure 5). The aptamers in the UCAA structural family are therefore comparable to, if not slightly better inhibitors than, these two pseudoknots.
Figure 5.
Concentration dependence of UCAA aptamer inhibition. IC50 values and SDs were calculated from triplicate assays are shown for each aptamer.
UCAA aptamers inhibit HIV-1 replication
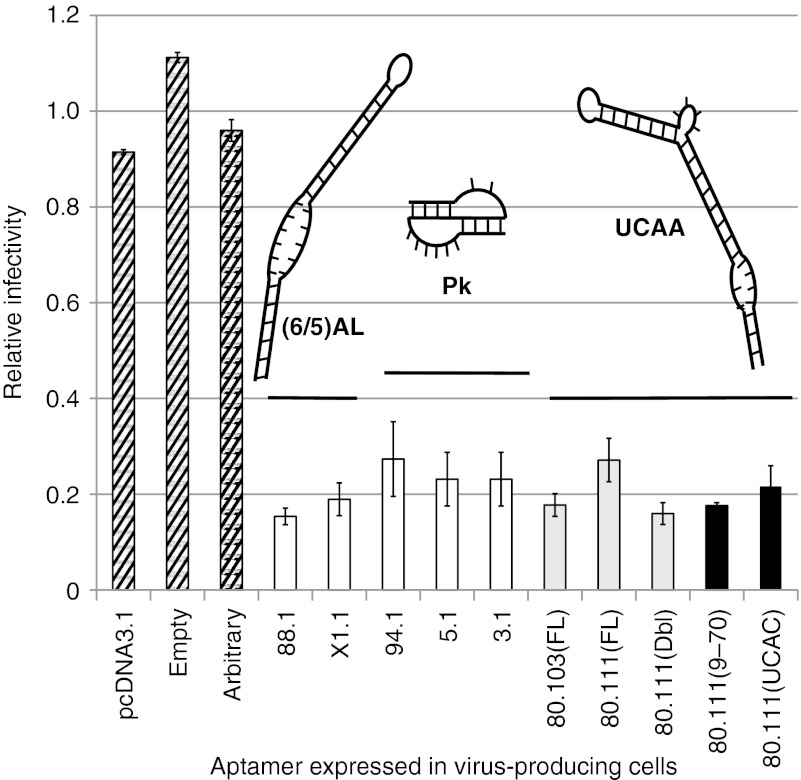
Inhibition of HIV-1 replication in single-cycle infectivity assays was measured to evaluate antiviral bioactivity of these aptamers. We recently demonstrated that aptamers can be expressed to high levels from a cassette in which stable RNA structures flank the expressed aptamer on both the 5′ and 3′ sides. The aptamers become packaged within the nascent virus and inhibit viral replication in the subsequent cycle of replication17 in a manner that is strongly correlated with RT inhibition in vitro.13 Various full-length and truncated aptamer-encoding DNAs and controls were therefore cloned into the aptamer expression cassette, and HIV-1 infectivity was determined for pseudotyped virus produced from cells expressing these aptamers. No viral inhibition was evident for controls representing the parental plasmid vector (“pcDNA3.1”), a modified expression platform containing all nonaptamer components (“Empty”), and a plasmid carrying an arbitrary 70 nt fragment of luciferase mRNA in place of the aptamer (“Arbitrary”). In contrast, infectivity was significantly reduced (P < 0.001) for all aptamers tested in comparison with controls. The UCAA aptamers inhibited as well as, or slightly better than, the pseudoknot aptamers (Figure 6, compare Pk versus UCAA), and inhibition by aptamer 80.103 was greater than that of aptamer 80.111. For aptamer 80.111, inhibition increased when two copies of the aptamer were expressed within the same transcript (Figure 6, 80.111(FL) versus 80.111(Dbl)), raising the possibility that polyaptamer constructs may provide a general strategy of increasing local aptamer concentration and augmenting viral suppression. The UCAA functional core of aptamer 80.111 was slightly more inhibitory than the full-length aptamer (Figure 6, compare 80.111(FL) with 80.111(9–70)). Expressing the UCAC mutant of the functional core provided as much viral inhibition as expressing the UCAA functional core, in contrast with the strong difference in enzyme inhibition between these two constructs (Figure 5c). We speculate that the flanking RNA structures within the expression cassette may provide additional contacts that compensate for partial disruption of the UCAA element.
Figure 6.
UCAA aptamers inhibit HIV in single-cycle viral replication assays. The effects of intracellular expression of aptamers on HIV-1 replication were determined by a recently described single-cycle infectivity assay.17 The y-axis represents the relative fraction of target cells that became infected with VSV-pseudotyped virus, with normalization to the average of the three noninhibitory controls (diagonal stripes). “pcDNA,” parent vector in which the expression cassette was built; “Empty,” plasmid carries an empty expression cassette with no aptamer; “Arbitrary,” plasmid directs expression of a 70-nucleotide fragment of the luciferase gene flanked by the same primer-binding sequences as those used for the 70HRT14 library. Previously studied reference constructs (white bars)17 were included for comparison with the indicated full-length (gray bars) and minimal core or mutant aptamers (black bars). The 80.111(Dbl) construct cotranscribes two expression cassettes for aptamer 80.111. Aptamer 80.111(UCAC) is a mutant in which the UCAA of aptamer 80.111(9–70) was replaced with UCAC. Structural families of each group of aptamers are indicated schematically above the bars: left to right, (6/5)AL, pseudoknots, UCAA. Plotted values represent the mean ± SDs for triplicate transfections. Pk, pseudoknots.
Discussion
This work identifies the UCAA element as a novel, potent, nonpseudoknot RNA module that inhibits DNA polymerization by HIV-1 RT and that suppresses HIV-1 replication in human cells. In the 20 years since RNA aptamer inhibitors of HIV-1 RT were first described,3 only the pseudoknots have received serious attention, in part because they strongly dominated LTS data sets from the early in vitro selections.3,4,13 The structural simplicity of the F1Pk and F2Pk motifs gives them a numerical advantage over other structural motifs in random sequence libraries that results in an over-abundance of pseudoknots in the final selected populations, even though more complex structural motifs with equivalent or improved binding abilities are clearly also present. Aptamers 80.103 and 80.111 were first identified through manual screening of transcripts from approximately 100 aptamer isolates, and the core regions responsible for RT inhibition were identified through enzymatic probing and additional screening of RT inhibition by truncated variants.
HTS analysis of aptamer population has made it possible to describe SELEX populations more completely, as demonstrated in several recent studies.13,22,23,24,25,26 Comparative approaches have proven especially powerful for extracting structural inferences based on the divergent cloud of mutations within individual lineages, identifying convergence on specific structural motifs and evaluating the relative fitness among and within these structural motifs.13 In the present work, comparative sequence analysis of HTS data proved instrumental in generalizing the findings beyond the two exemplars identified from manual screening and in defining the motif as a whole, including identification of UCAA family aptamers in 167 clusters from the 80HRT14 population and 14 from among 70HRT populations. The most abundant cluster, which includes aptamer 80.103, represents only 0.43% of the total 80HRT14 HTS data set and could easily have been missed in the manual screen. It was even more fortuitous that the exceedingly rare aptamer 80.111 was sampled in the manual screen, given that its cluster was sampled in only 8 reads. As with the recently described (6/5)AL motif, which was similarly rare in the 70HRT14 data set (approximately 3%),13 the large data sets associated with HTS analysis greatly accelerated identification of the UCAA element as a cohesive motif and provided a high-resolution view of sequence requirements and subclasses. The (6/5)AL motif13 and the UCAA motif expand the known major classes of RT-inhibiting RNAs from one (pseudoknots only) to three, each of which include identifiable subclasses.
The core of the UCAA-motif element is an eight-nucleotide signature that includes the unpaired UCAA portion and highly conserved CG and AU pairs on the 5′ side of this segment (Figure 2b). Modeling with the Rosetta web server (http://rosie.rosettacommons.org) indicates a sharply bent structure (Figure 7) in which the first three nucleotides of the UCAA sequence interact with the each other and with the two conserved flanking base pairs to establish the stable bend, and the fourth nucleotide stacks on the downstream helix. In the crystal structure of RT in complex with dsDNA,27 approximately 18 bp fill the cleft between the polymerase and RNaseH active sites. DNA near the polymerase active site forms a short A-form helix that abruptly bends near the base of the “thumb” domain, followed by a longer B-form helix that extends to the RNaseH active site. It is intriguing to speculate that a UCAA-induced bend may be similarly located near the RT thumb domain.
Figure 7.
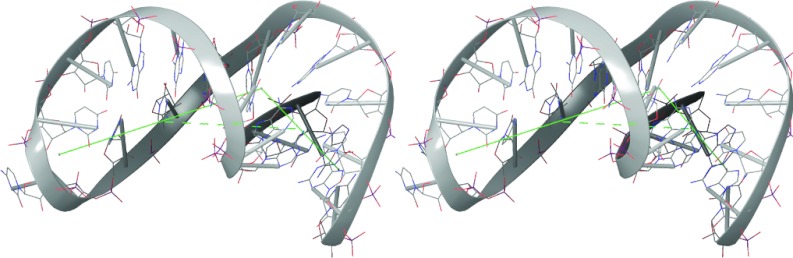
Stereo view of UCAA core as modeled by Rosetta. A 32 nt segment of 80.103 core RU25-80 (5′-UGUCU ACCGGGC UUCG GCCCGGU UCAA GGACA-3′) was modeled by the Rosetta web server. Straight lines indicate approximate trajectory of helical axis.
The subfamilies of UCAA aptamers differ in their helical requirements. About 75% of the UCAA clusters from the 80HRT14 population (125 of 167) utilize the 5′ constant region to form the overall secondary structure. The remaining 25%, including aptamer 80.103, form the conserved secondary structure utilizing only nucleotides derived from the 80N random region. Among the UCAA aptamers and sequence variants, all inhibitory constructs contain one long stem and one short one, with discontinuities built into the long stem. For aptamers 80.103 and #342, the long stem lies “below” the UCAA element in the structural depictions used here, and both aptamers include an asymmetric A/C-rich internal loop. For aptamer 80.111, the long stem lies “above” the UCAA element, and there is an asymmetric A/GG internal loop near the end of the stem. The fact that the long arm is found on both sides of the UCAA sequence suggests that the UCAA sequence does not dictate orientation of the aptamer with respect to RT, and instead serves primarily to establish the bend between the two stems. This is in contrast with the helical length requirements of aptamers in the (6/5)AL family, for which stems 1 and 2, which are also defined relative to the internal loop, have different length requirements. The long arms of UCAA family aptamers carry irregularities that also appear to be important for RT recognition. The long arms of aptamers 80.103 and #342 include an asymmetric A/C-rich internal loop, whereas the long arm of aptamer 80.111 includes an asymmetric A/GG internal loop near the end of the stem. RT inhibition was disrupted when the A/GG internal loop of aptamer 80.111 was converted into two base pairs (AS14; Supplementary Figure S7), or when the A/C-rich internal loop in the long arm of aptamer #342 inhibition was converted into three base pairs (data not shown). Irregularity near the end of the “long” helix may position the end of that helix for specific contacts, perhaps near the RNaseH domain. HTS analysis did not resolve length requirements and details of these distortions because of the high variability in their composition and placement relative to the overall structure.
The strong correlation between RT in vitro and antiviral bioactivity in cell culture supports the use of anti-RT aptamers as tools for exploring viral pathogenesis and as potential therapeutic agents. These results extend and strengthen previously observed correlations for pseudoknots16 and for the (6/5)AL element,13 and is consistent with a model in which intracellular and intraviral RT-aptamer interactions are responsible for the observed antiviral effects. It is likely that there are many additional RNA structural families that bind HIV-1 RT with high affinity and that can be accessed through a combination of specialized selection methods. Comparative sequence analysis of HTS data sets has proven especially adept at accelerating the identification and optimization of such new aptamers, especially against the high backdrop of the dominant pseudoknot F1Pk and F2Pk motifs. Each new structural family enhances the possibilities for developing aptamer antagonists of HIV-1.
Materials and Methods
Materials. Synthetic DNA was purchased from Integrated DNA Technologies (Coralville, IA). Radiolabeled nucleotides for 5′ labeling ([γ-32P]ATP) were purchased from Perkin-Elmer (Waltham, MA). The p51 and p66 subunits of RT from HIV-1 subtype B (HXB2 strain GenBank accession number K03455) were cloned into the protein expression construct pRT-Dual, kindly provided by Dr Stefanos G. Sarafianos, expressed in Escherichia coli strain BL21(DE3) and purified essentially as described.28 Aptamer RNA was transcribed in vitro from synthetic DNA oligonucleotides or from polymerase chain reaction products amplified from plasmids4 using phage T7 RNA polymerase. Transcripts were gel-purified as described7 and resuspended in deionized water.
RT enzymatic inhibition assays. Primer extension was carried out essentially as described.7 Briefly, a Cy3-labeled, 18-mer DNA oligonucleotide corresponding to the 3′ end of tRNALys, 3 was mixed with a 31-mer template in a 1:3 ratio to ensure that all primer was pre-bound to template. This mixture was heated to 90 °C in a heat block for 2 minutes and then annealed by cooling to room temperature. A reaction mster mix was assembled to contain (final concentrations) 30 µmol/l dNTPs, 0.5 mmol/l ethylenediaminetetraacetic acid (EDTA), 50 mmol/l Tris–HCl pH 7.8, 50 mmol/l NaCl2, 10 mmol/l dithiothreitol and 20 nmol/l RT, with the RT and dithiothreitol added last. The reaction master mix of 14 µl was aliquoted to each tube, along with either 2 µl of aptamer solution (final concentration 100 nmol/l unless otherwise noted) or water. Reactions were initiated by adding 4 µl of a solution containing annealed primer/template and MgCl2 ((final concentration 20 nmol/l and 6 mmol/l, respectively). After incubating at 37 °C for 10 min, reactions were stopped by adding 20 µl of 90% formamide, 50 mmol/l EDTA, and a trace amount of bromophenol blue. Samples were heated to 90 °C for 2 min immediately before loading onto a 15% polyacrylamide, 8 mol/l urea denaturing gel. Gels were scanned for Cy3 fluorescence with a FLA9000 phosphorimager (Fujifilm, Valhalla, NY). The fraction of primer converted to full-length product was determined by quantifying band intensities using ImageQuant software (Pharmacia, Piscataway, NJ) and normalized by setting the fraction converted to full-length product in the absence of aptamer to 100%. Aptamer concentrations required for half-maximal inhibition (IC50 values) were calculated as described7 by fitting the data with GraphPad Prism 6 software to a standard two-state sigmoidal dose response curve: Y = 1/(1 + 10^[x − log(IC50)]), where Y is the normalized fraction full-length product at a given aptamer concentration (x). Enzyme inhibition assays were performed in triplicate for all reactions from which IC50 values were calculated.
HTS and analysis pipeline. HTS data for the 80HRT14 population were obtained and analyzed as described previously for 70HRT14 and other populations.13 Flanking sequences required for Illumina sequencing and indexing were appended during two sequential polymerase chain reaction amplification steps using plaque-forming unit DNA polymerase. Final amplified products from 80HRT14 were pooled with other populations, loaded onto a single lane, bridge amplified and then run through 100 sequencing cycles. Illumina's analysis software was used to generate fastq files with sequence calls and associated quality scores for >3 million raw sequence reads per population. 613,313 quality-filtered reads for the 80HRT14 populations were aligned, clustered, and used to identify converged structures.
Enzymatic probing. Secondary structures in solution were assessed by enzymatic digestion as described.29 For each reaction, 50,000–200,000 cpm of 5′ radiolabeled RNA was digested under native conditions at 37 °C with ribonuclease T1 (0.005 U/µl for 2 min; Ambion; Life Technologies, Grand Island, NY), or S1 nuclease (4.75 U/µl for 10 min; New England Biolabs, Ipswich, MA), or ribonuclease V1 (5 × 10–5 U/µl for 8 min; Ambion). All reactions were quenched with equal volumes of colorless gel loading buffer (10 mol/l urea, 15 mmol/l EDTA) and quickly cooled in a dry ice/ethanol bath. Products of digestions were separated on 8 mol/l urea denaturing 15% polyacrylamide gels and analyzed as above.
Cell lines, plasmids and viral assays. Plasmids for directing aptamer expression were constructed as previously described17 and utilize a human cytomegalovirus (CMV) immediate early promoter. Proviral plasmid (pNL4-3-Δenv-CMV-EGFP) was kindly provided by Vineet KewalRamani (National Cancer Institute [NCI], Fredrick, MD) and carries the genome of HIV-1 strain NL4-3, in which the genes encoding vif, vpr, vpu, nef, and env have been deleted, and a CMV-driven enhanced green fluorescent protein (EGFP) reporter gene replaces nef. Cell culture, virus production and evaluation of single-cycle viral infectivity were carried out as described.17,30 The human cell line, 293FT (Invitrogen, Carlsbad, CA), was transfected with polyethyleneimine in 6-well cell culture dishes. Aptamer-expressing plasmids were transfected first (1 µg), followed four hours later by transfection with a mixture of pNL4-3-Δenv-CMV-EGFP (250 ng) and pMD-G (125 ng; Invitrogen) to produce pseudotyped HIV-1 in the presence of aptamer. Medium was changed between transfections and again four hours after the second transfection. Virus was harvested 48 hours posttransfection by filtering the medium through 0.45 µm filters, and was quantified by p24 enzyme-linked immunosorbent assay. Fresh 293FT cells were infected with 25 µl of filtrate so that 5–10% of the target cells would become infected in the no-aptamer control; this level of infection provides sensitive readout of aptamer-mediated viral suppression.17 Cells were collected 48 hours post-infection, fixed with 4% paraformaldehyde, and analyzed for EGFP fluorescence on an Accuri Flow Cytometer (BD Biosciences, San Jose, CA). The percentage of infected (EGFP positive) cells was normalized to p24 levels in each sample, and the average of the control samples was set to 1. One-way analysis of variance and Student's t test were used to determine statistical significance between samples.
SUPPLEMENTARY MATERIAL Figure S1. Complete sequences of isolates from aptamer populations 70HRT14 (a) and 80HRT14 (b), as determined by low-throughput sequencing (LTS) of plasmids that were shotgun cloned from the library. Figure S2. Initial screen and prioritization of 99 aptamers for inhibition of RT from HIV-1 strain HXB2. Figure S3. Results from screen of proposed F1Pk and F2Pk cores. Figure S4. Aligned sequences of the UCAA family members. Figure S5. Enzymatic probing of aptamer 80.111(9–73). Figure S6. Effects of internal mutations of 80.103 and 80.111 on RT inhibition. Figure S7. Effects of helical swaps among subfamilies of UCAA aptamers. Figure S8. Aptamer concentration dependence of RT inhibition used in calculating IC50 values.
Acknowledgments
This work was supported by the National Institute of Allergy and Infectious Disease under award numbers R21AI62513 (D.H.B.), R01AI074389 (D.H.B.), and F32AI085627 (M.J.L.), and by a grant from Trail to a Cure. The authors thank Kamlendra Singh for assistance in evaluating helical offset and generating Figure 7, and Shi Jie Chen and Song Cao for providing helpful insights into potential alternative pseudoknot aptamer structures in the early phases of the work. The authors also thank Daniel M Held, Dayal Saran, Ferrill Rose, Robin Cutler, Xiangmei Xi, Debojit Bose, Ying Wan, Adeyemi Adedeji, Jeremy Harris, and Joshua Willix for help in the initial screens of the panel of aptamer transcripts. The authors declare no competing financial interest.
Supplementary Material
Complete sequences of isolates from aptamer populations 70HRT14 (a) and 80HRT14 (b), as determined by low-throughput sequencing (LTS) of plasmids that were shotgun cloned from the library.
Initial screen and prioritization of 99 aptamers for inhibition of RT from HIV-1 strain HXB2.
Results from screen of proposed F1Pk and F2Pk cores.
Aligned sequences of the UCAA family members.
Enzymatic probing of aptamer 80.111(9–73).
Effects of internal mutations of 80.103 and 80.111 on RT inhibition.
Effects of helical swaps among subfamilies of UCAA aptamers.
Aptamer concentration dependence of RT inhibition used in calculating IC50 values.
References
- Li N, Wang Y, Pothukuchy A, Syrett A, Husain N, Gopalakrisha S.et al. (2008Aptamers that recognize drug-resistant HIV-1 reverse transcriptase Nucleic Acids Res 366739–6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannoush RN, Carriero S, Min KL., and, Damha MJ. Selective inhibition of HIV-1 reverse transcriptase (HIV-1 RT) RNase H by small RNA hairpins and dumbbells. Chembiochem. 2004;5:527–533. doi: 10.1002/cbic.200300831. [DOI] [PubMed] [Google Scholar]
- Tuerk C, MacDougal S., and, Gold L. RNA pseudoknots that inhibit human immunodeficiency virus type 1 reverse transcriptase. Proc Natl Acad Sci USA. 1992;89:6988–6992. doi: 10.1073/pnas.89.15.6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke DH, Scates L, Andrews K., and, Gold L. Bent pseudoknots and novel RNA inhibitors of type 1 human immunodeficiency virus (HIV-1) reverse transcriptase. J Mol Biol. 1996;264:650–666. doi: 10.1006/jmbi.1996.0667. [DOI] [PubMed] [Google Scholar]
- Green L, Waugh S, Binkley JP, Hostomska Z, Hostomsky Z., and, Tuerk C. Comprehensive chemical modification interference and nucleotide substitution analysis of an RNA pseudoknot inhibitor to HIV-1 reverse transcriptase. J Mol Biol. 1995;247:60–68. doi: 10.1006/jmbi.1994.0122. [DOI] [PubMed] [Google Scholar]
- Kensch O, Connolly BA, Steinhoff HJ, McGregor A, Goody RS., and, Restle T. HIV-1 reverse transcriptase-pseudoknot RNA aptamer interaction has a binding affinity in the low picomolar range coupled with high specificity. J Biol Chem. 2000;275:18271–18278. doi: 10.1074/jbc.M001309200. [DOI] [PubMed] [Google Scholar]
- Held DM, Kissel JD, Saran D, Michalowski D., and, Burke DH. Differential susceptibility of HIV-1 reverse transcriptase to inhibition by RNA aptamers in enzymatic reactions monitoring specific steps during genome replication. J Biol Chem. 2006;281:25712–25722. doi: 10.1074/jbc.M604460200. [DOI] [PubMed] [Google Scholar]
- Schneider DJ, Feigon J, Hostomsky Z., and, Gold L. High-affinity ssDNA inhibitors of the reverse transcriptase of type 1 human immunodeficiency virus. Biochemistry. 1995;34:9599–9610. doi: 10.1021/bi00029a037. [DOI] [PubMed] [Google Scholar]
- Kissel JD, Held DM, Hardy RW., and, Burke DH. Single-stranded DNA aptamer RT1t49 inhibits RT polymerase and RNase H functions of HIV type 1, HIV type 2, and SIVCPZ RTs. AIDS Res Hum Retroviruses. 2007;23:699–708. doi: 10.1089/aid.2006.0262. [DOI] [PubMed] [Google Scholar]
- Kissel JD, Held DM, Hardy RW., and, Burke DH. Active site binding and sequence requirements for inhibition of HIV-1 reverse transcriptase by the RT1 family of single-stranded DNA aptamers. Nucleic Acids Res. 2007;35:5039–5050. doi: 10.1093/nar/gkm420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalowski D, Chitima-Matsiga R, Held DM., and, Burke DH. Novel bimodular DNA aptamers with guanosine quadruplexes inhibit phylogenetically diverse HIV-1 reverse transcriptases. Nucleic Acids Res. 2008;36:7124–7135. doi: 10.1093/nar/gkn891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held DM, Kissel JD, Thacker SJ, Michalowski D, Saran D, Ji J.et al. (2007Cross-clade inhibition of recombinant human immunodeficiency virus type 1 (HIV-1), HIV-2, and simian immunodeficiency virus SIVcpz reverse transcriptases by RNA pseudoknot aptamers J Virol 815375–5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditzler MA, Lange MJ, Bose D, Bottoms CA, Virkler KF, Sawyer AW.et al. (2012High-throughput sequence analysis reveals structural diversity and improved potency among RNA inhibitors of HIV reverse transcriptase Nucleic Acids Res (epub ahead of print) [DOI] [PMC free article] [PubMed]
- Chaloin L, Lehmann MJ, Sczakiel G., and, Restle T. Endogenous expression of a high-affinity pseudoknot RNA aptamer suppresses replication of HIV-1. Nucleic Acids Res. 2002;30:4001–4008. doi: 10.1093/nar/gkf522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi PJ, North TW., and, Prasad VR. Aptamers directed to HIV-1 reverse transcriptase display greater efficacy over small hairpin RNAs targeted to viral RNA in blocking HIV-1 replication. Mol Ther. 2005;11:677–686. doi: 10.1016/j.ymthe.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Joshi P., and, Prasad VR. Potent inhibition of human immunodeficiency virus type 1 replication by template analog reverse transcriptase inhibitors derived by SELEX (systematic evolution of ligands by exponential enrichment) J Virol. 2002;76:6545–6557. doi: 10.1128/JVI.76.13.6545-6557.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange MJ, Sharma TK, Whatley AS, Landon LA, Tempesta MA, Johnson MC.et al. (2012Robust Suppression of HIV Replication by Intracellularly Expressed Reverse Transcriptase Aptamers Is Independent of Ribozyme Processing Mol Ther 202304–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditzler MA, Bose D, Shkriabai N, Marchand B, Sarafianos SG, Kvaratskhelia M.et al. (2011Broad-spectrum aptamer inhibitors of HIV reverse transcriptase closely mimic natural substrates Nucleic Acids Res 398237–8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., and, Gold L. Selection of high-affinity RNA ligands to reverse transcriptase: inhibition of cDNA synthesis and RNase H activity. Biochemistry. 1994;33:8746–8756. doi: 10.1021/bi00195a016. [DOI] [PubMed] [Google Scholar]
- Chen H, McBroom DG, Zhu YQ, Gold L., and, North TW. Inhibitory RNA ligand to reverse transcriptase from feline immunodeficiency virus. Biochemistry. 1996;35:6923–6930. doi: 10.1021/bi9600106. [DOI] [PubMed] [Google Scholar]
- Held DM, Kissel JD, Patterson JT, Nickens DG., and, Burke DH. HIV-1 inactivation by nucleic acid aptamers. Front Biosci. 2006;11:89–112. doi: 10.2741/1782. [DOI] [PubMed] [Google Scholar]
- Cho M, Xiao Y, Nie J, Stewart R, Csordas AT, Oh SS.et al. (2010Quantitative selection of DNA aptamers through microfluidic selection and high-throughput sequencing Proc Natl Acad Sci USA 10715373–15378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoon S, Zhou B, Janda KD, Brenner S., and, Scolnick J. Aptamer selection by high-throughput sequencing and informatic analysis. BioTechniques. 2011;51:413–416. doi: 10.2144/000113786. [DOI] [PubMed] [Google Scholar]
- Kupakuwana GV, Crill JE., 2nd, , McPike MP., and, Borer PN. Acyclic identification of aptamers for human alpha-thrombin using over-represented libraries and deep sequencing. PLoS ONE. 2011;6:e19395. doi: 10.1371/journal.pone.0019395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayrac AT, Sefah K, Parekh P, Bayrac C, Gulbakan B, Oktem HA.et al. (2011In vitro Selection of DNA Aptamers to Glioblastoma Multiforme ACS Chem Neurosci 2175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel WH, Bair T, Peek AS, Liu X, Dassie J, Stockdale KR.et al. (2012Rapid identification of cell-specific, internalizing RNA aptamers with bioinformatics analyses of a cell-based aptamer selection PLoS ONE 7e43836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobo-Molina A, Ding J, Nanni RG, Clark AD, Jr, Lu X, Tantillo C.et al. (1993Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA Proc Natl Acad Sci USA 906320–6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckmann MM, Marchand B, Hachiya A, Kodama EN, Kirby KA, Singh K.et al. (2010The N348I mutation at the connection subdomain of HIV-1 reverse transcriptase decreases binding to nevirapine J Biol Chem 28538700–38709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biondi E, Nickens DG, Warren S, Saran D., and, Burke DH. Convergent donor and acceptor substrate utilization among kinase ribozymes. Nucleic Acids Res. 2010;38:6785–6795. doi: 10.1093/nar/gkq433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange M., and, Burke D.2013Screening inhibitory potential of anti-HIV RT RNA aptamers Methods Mol Biolin the press). [DOI] [PubMed]
- Knapp G. Enzymatic approaches to probing of RNA secondary and tertiary structure. Meth Enzymol. 1989;180:192–212. doi: 10.1016/0076-6879(89)80102-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complete sequences of isolates from aptamer populations 70HRT14 (a) and 80HRT14 (b), as determined by low-throughput sequencing (LTS) of plasmids that were shotgun cloned from the library.
Initial screen and prioritization of 99 aptamers for inhibition of RT from HIV-1 strain HXB2.
Results from screen of proposed F1Pk and F2Pk cores.
Aligned sequences of the UCAA family members.
Enzymatic probing of aptamer 80.111(9–73).
Effects of internal mutations of 80.103 and 80.111 on RT inhibition.
Effects of helical swaps among subfamilies of UCAA aptamers.
Aptamer concentration dependence of RT inhibition used in calculating IC50 values.