Abstract
In tissues with complex architectures such as bone, it is often difficult to purify and characterize specific cell types via molecular profiling. Single cell gene expression profiling is an emerging technology useful for characterizing transcriptional profiles of individual cells isolated from heterogeneous populations. In this study we describe a novel procedure for the isolation and characterization of gene expression profiles of single osteoblast lineage cells derived from cortical bone. Mixed populations of different cell types were isolated from adult long bones of C57BL/6J mice by enzymatic digestion, and subsequently subjected to FACS to purify and characterize osteoblast lineage cells via a selection strategy using antibodies against CD31, CD45, and Alkaline Phosphatase (AP), specific for mature osteoblasts. The purified individual osteoblast lineage cells were then profiled at the single cell level via nanofluidic PCR. This method permits robust gene expression profiling on single osteoblast lineage cells derived from mature bone, potentially from anatomically distinct sites. In conjunction with this technique, we have also shown that it is possible to carry out single cell profiling on cells purified from fixed and frozen bone samples without compromising the gene expression signal. The latter finding means the technique can be extended to biopsies of bone from diseased individuals. Our approach for single cell expression profiling provides a new dimension to the transcriptional profile of the primary osteoblast lineage population in vivo, and has the capacity to greatly expand our understanding of how these cells may function in vivo under normal and diseased states.
1. Introduction
1.1 Background
The skeleton is a complex organ system containing a number of tissues comprised of unique cell populations involved in maintaining structure and function. Within the long bones of the appendicular skeleton, calcified bone can be divided into the trabecular and cortical tissues. Recently, there has been tremendous progress in understanding and treating age-related disorders to prevent the loss of cortical bone tissue [1]. However, there have been few studies that examine primary bone cell populations derived from in vivo sources. One potential reason for this is the inherent difficulty in studying the cell types involved in maintaining bone, as these cells are typically encased within an ossified matrix. Consequently, characterization of these cell types has usually been performed upon differentiated osteoblast culture models [2–4], often after multiple passages in vitro. Thus, there is always the uncertainty of how faithfully the cells cultivated in vitro mirror the behavior of osteoblasts functioning in vivo. It is also not well understood how osteoblasts transcriptionally vary from one type of bone to another [5], nor even within the same bone. For example, it has been demonstrated that the gene expression profiles from calvarial bone are quite different then those derived from long bones [6]. There are also questions about potential differences in osteoblast function in vivo as a consequence of specific location or physiological state; periosteal versus endosteal for the former, and degree of mechanical loading or physiological age for the latter.
1.2 The need for single cell methods to study cells involved in bone formation
There have been a number of recent advances in nucleic acid manipulation and amplification technology that allow for the quantitative assessment of multiple genes using high throughput PCR platforms [7]. Similarly, there have been recent advances in the analysis of single cell data in specific cell populations to elucidate subtle expression differences between cell types in developmental or pathological processes such as tumor progression [8,9]. These approaches have not yet been widely used in studying age-related changes, and are particularly underutilized in the context of bone tissue [10,11]. This may partially be due to a lack of robust procedures to obtain specific cell types from bone for gene expression profiling. To begin to address these issues, we sought to develop methods to isolate and purify cells from the cortical bone matrix, using mouse long bones (femurs), with the eventual goal of developing methods to transcriptionally profile dozens of genes of interest, or potentially carry out whole genome profiling at the single cell level, similar to procedures we recently reported for the cardiomyocyte [12]. Briefly, the method isolates cells from the cortical bone, sorts them based upon canonical markers for osteoblast lineage, pre-amplifies the message using a targeted amplification, and finally analyzes the expression profiles of scores of cells simultaneously using nanofluidic qPCR. Subsequent analysis of single cell data provides data about the variance and co-expression of transcripts that are not observable in bulk tissue preparations.
2. Materials and Methods
2.1 Animals
Five-month-old female C57BL/6J mice were used in this study. The mice were sacrificed via CO2 overdose and cervical dislocation. Femurs were then immediately isolated. The bones were stripped of muscle and immediately placed in ice cold PBS (pH 7.4). The tissue was then prepared for immediate tissue digestion or prepared for long term preservation as described in 2.2. All animal procedures were carried out under approved IACUC protocols of the Buck Institute for Research on Aging.
2.2 Cortical Bone Isolation and Preservation
The collected bone samples were maintained in PBS on ice after their removal from the animal. The samples were submerged in a petri dish of PBS where any remaining soft tissue was stripped from the bone. The ends of the femur were then cut from the shaft of the bone using small scissors or a scalpel. The shaft of the femur was then thoroughly flushed using a 21-gauge needle and syringe of PBS to remove as much marrow material as possible. Note the difference in the appearance before and after (Fig.S1, top) cleaning the bone sample (Fig.S1, bottom). Once the bone samples are prepared it is possible to proceed directly to the digestion phase of the cortical bone cell isolation or preservation procedure for later analysis.
To preserve the bone for later analysis a method was modified from a procedure by Oh et al. [13] where the bone was quickly prepared for cryopreservation. Briefly, after the femurs were flushed with PBS to remove marrow they were placed into 2 ml cryovials containing a solution of M199 media containing 10% DMSO and 10% FBS. These samples were then placed into a 4°C refrigerator to equilibrate for 30 min. The sample tubes were then transferred to a −80°C freezer for storage for a minimum of 48 hours. Upon removal from the freezer the tubes were immediately thawed in a 37°C water bath. The sample preserving solution was replaced with stepwise 5 min washes of M199 media containing 10% FBS supplemented with sucrose (0.5M) to wash out any remaining DMSO from the tissue sample. This was followed by two more washes in media with 0.25M sucrose and finally M199 media alone. From this point, the tissue sample was immediately processed using the digestion procedure described in section 2.3.
2.3 Enzymatic removal of bone matrix
After preparing the bone sample as described above, the tissue is digested to yield a cell suspension. Two femur shafts are placed into one 1.6 ml conical tube that is then filled with collagenase digestion solution consisting of 0.2% collagenase in PBS (Worthington Biochemical, Type 2). This tube is then placed into a 37° C shaking incubator for 30 minutes. After this incubation, the supernatant is removed and discarded. The bone is then pulverized using small scissors or an appropriate sized pestle. The bone was then further broken into smaller pieces leaving no large fragments. The tube is then filled with collagenase digestion solution and incubated for another hour at 37°C with gentle shaking. After one hour of digestion, the tubes are removed from the incubator and the solution is gently triturated with a pipette. The solution should then appear opaque with digested material. The supernatant is carefully removed and transferred to a clean sterile 1.6 ml conical tube. The supernatant containing the cell population is then spun at 750 rpm for 8 minutes to pellet the cells. The supernatant is then removed taking care not to disturb the cell pellet. The cells were then resuspended with M199 media supplemented with 10% FBS, pH 7.4 to neutralize the collagenase. From this point, the bone-derived cells were prepared for either FACS analysis or immunocytochemistry staining.
2.4 Immunohistochemistry and Immunocytochemistry
To identify bone specific markers within the bone cells, we prepared cell smears of the total population isolated from bone, or tissue sections of cortical bone. First, a concentrated cell suspension was placed onto a glass superfrost® plus charged slide and allowed to dry at room temperature. Once completely dry, the slide was rinsed with PBS and then incubated with 4% paraformaldehyde solution overnight. The next day the slides were processed for the presence of the osteoblast marker, alkaline phosphatase (AP) according to an immunohistochemistry protocol (see supplement for detailed protocol). The optimal concentration for the primary AP antibody was a 1:100 dilution (Sigma Anti-mouse IgG (whole molecule), Cat #A4312). The biotinylated secondary antibody was used at a concentration of 1:200, and visualized with VectaStain Elite DAB kit. Once stained, cells were mounted with Permount® and allowed to dry overnight. The following day the cells were imaged on a bright field microscope at 20× and 60× magnification.
For fluorescence immunocytochemistry of cortical bone sections we obtained paraffin embedded sections of a three month old C57Bl6 mouse from Zyagen laboratories. These sections were stained using the same directly conjugated Alkaline phosphatase antibody used in the FACS experiments (1:100; R & D systems, Cat #FAB1448P) and an antibody against Osteocalcin (1:100; Thermo Scientific, Cat #PA1-85754) with an Alexa Fluor 647 nm secondary antibody (1:1000; Life Technologies, Cat #A21447) using standard staining procedures. After staining, the sections were mounted with Prolong Gold Antifade reagent (Invitrogen), and dried for 48 hours prior to imaging. These sections were subsequently imaged using a Zeiss Confocal LSM 780 under 20× objective.
2.5 Cell Staining and FACS analysis
The flow cytometry procedure was developed to determine the optimal treatment of cells to capture the osteoblast lineage cell population while maintaining the RNA quality so that it was suitable for nanofluidic PCR. The cells isolated from long bones were first incubated on ice for 20 minutes in purified rat anti-mouse CD16/CD32 Fc Block (BD Pharmingen, Cat #553142) diluted 1:100 in media. After incubation the cells were spun down at 250 × g for 7 minutes. The supernatant was removed and the cells were resuspended with PBS. All wash steps were carried out in a similar manner. We chose CD31 and CD45 as cell surface markers to select against cells derived from the hematopoietic lineage, to enrich for osteoblasts in the resultant population. For the cell surface marker labeling of CD31 (BD Pharmingen PE-Cy7 rat anti-mouse CD31, Cat #561410) and CD45 (BD Pharmingen FITC rat anti-mouse CD45, Cat #553080), cells were incubated on ice with directly conjugated antibody for 30 minutes at a 1:100 dilution followed by washing. Labeling with the third antibody for alkaline phosphatase to define the osteoblast lineage cells requires a gentle fixation and permeabilization treatment to achieve sufficient labeling of this intracellular marker and to preserve the mRNA quality. This was performed using the BD Cytofix/Cytoperm Kit (Cat #554714) after staining for CD31 and CD45. Cells were incubated in the Fix/Perm solution for 20 minutes on ice, spun down, and stained with anti-human/mouse/rat Alkaline Phosphatatse-Phycoerythrin (R & D systems, Cat #FAB1448P) diluted 1:100 in PBS. NB: This step was omitted in some experiments to examine the qPCR signal from unfixed cells. After incubation, the cells were washed once more and resuspended prior to sorting on the FACS machine. All the direct conjugated antibodies were also tested with isotype controls to ensure the specificity of the signal. The following isotype controls were used for each respective fluorescence channel: PE-Cy7 rat IgG2a k (BD Pharmingen, Cat #552784), FITC mouse IgG2a k (BD Pharmingen, Cat #556652), and mouse IgG1 Isotype Control-PE (R & D systems, Cat #IC002P). All isotype control signals showed negligible non-specific staining, and each individual antibody stained positive cells that were detectable above background signal.
The flow cytometry strategy to isolate pure populations of osteoblast lineage cells was to identify CD31/CD45 negative cells via a P2 gate and select cells positive for Alkaline Phosphatase via a P3 gate in the PE channel. The cells were sorted on a BDFACSAria II Special Order System with 488 nm and 633 nm lasers for excitation, using an 80 micron nozzle for sorting. Control and initial analysis was performed using BD FACSDiva Software v6.1.1. Sort statistics and images were created offline using FloJo analysis software (version 7.6.5).
2.6 Single Cell Gene Expression
Individual cells were assayed with a single cell qPCR procedure using a method similar to that of Flynn et al [14]. Briefly, individual cells were isolated using a micromanipulator and placed into PCR tubes in a small volume of 1–2 µl. These tubes were then immediately frozen on dry ice and stored at −80°C. These cells were thawed on ice and then pre-amplified using specific target amplification containing all assay primer sets for 18 cycles using the Cell-Direct RT-qPCR kit (Invitrogen). The amplified material was then cleaned up using Affymetrix ExoSAP-IT enzymatic solution. The samples were then diluted 1:4 with DNA suspension buffer and assayed using Fluidigm’s 96.96 nanofluidic qPCR arrays on a Biomark HD system. Biotium’s EvaGreen DNA binding dye was used to detect amplified product according to Fluidigm’s advanced development protocol #30 [15]. For a detailed description of the qPCR procedure refer to Supplemental Methods.
2.7 qPCR analysis of Single Cells
Data analysis was performed using Fluidigm’s Real-Time PCR analysis software (v. 3.0.2). Raw image files were processed similarly to previously published studies [14,16], with visual inspection to ensure proper alignment in the reaction chambers. The data was then transformed using linear baseline correction and auto (detectors) method of Ct (Cycle threshold) normalization. The resulting data was then examined for assay specificity using melt curve analysis on each reaction chamber to ensure that all PCR products had a consistent peak melt temperature indicative of the desired PCR product. Assays that did not display a consistent melt peak were discarded from the dataset. The data was subsequently analyzed using the R statistical computing environment (v. 2.15.0) and the Fluidigm Single-Cell Data analysis package available through Fluidigm <www.fluidigm.com>. Heatmap data was expressed as Limit of Detection (LoD) Ct values set at the recommended limit of 26. The analyzed dataset of all cells was subsequently used to create the violin plots within the single cell R package.
3. Results
3.1 Overview of procedure
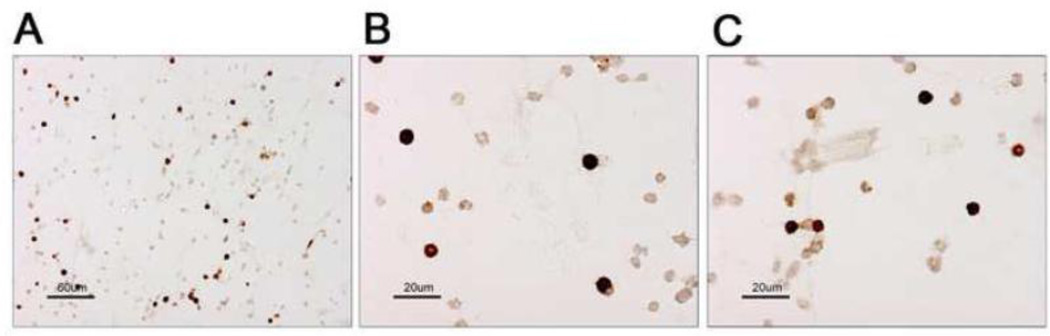
We developed a series of procedures that maintain the fidelity of gene expression in a targeted sub-population of osteoblast like cells derived from mature adult long bones of mice. Initially, the femurs were removed from mice and the surrounding tissue was removed by careful dissection. The ends of the bones were sheared away leaving the shaft of the bone (Supplemental Figure 1). The marrow cavity was then thoroughly flushed to remove any non-bone cells, and the diaphyseal bone placed in collagenase digestion buffer. After the 1st digestion step, the bone was crushed and then further digested, resulting in a suspension of live and viable bone cells. To confirm that this cell population included cells expressing a canonical bone marker, we performed IHC for alkaline phosphatase (AP) on these cells. We showed that thousands of cells staining for AP could be isolated from a relatively small amount of starting material (Figure 1).
Figure 1. Immunocytochemistry of isolated cells.
Alkaline Phosphatase staining of isolated bone cells. (A) 20× image of the stained cells showing a mixture of positive and negative cells. AP positive cells appear as dark brown cells that are also typically larger then the negatively staining cells. (B, C) Two high power (60×) images of the isolated cells again showing the cells that stain positive for the presence of Alkaline Phosphatase enzyme. The cells appear as rounded in shape, sometimes projecting small processes outward.
3.2 Purification of a specific sub-population of cells from cortical bone
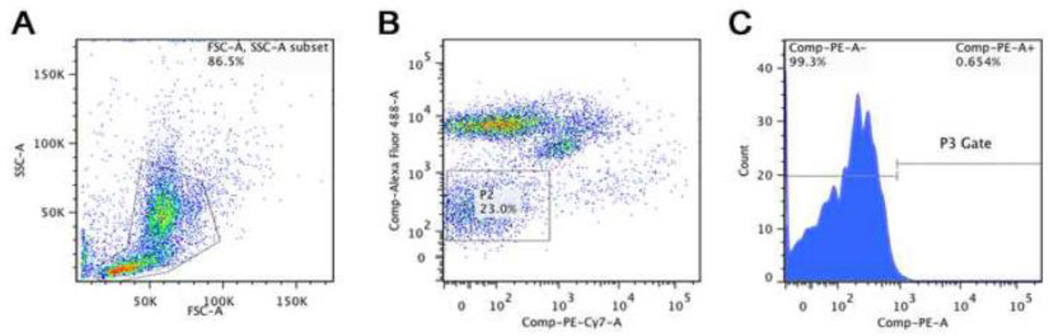
After obtaining viable cells it was critical to develop methodology to separate and purify single cells from the larger heterogeneous population of cells isolated from mature bone. We therefore employed FACS in conjunction with antibody staining to identify cells negative for hematopoietic lineage (CD31− and CD45−) while at the same time positive for an osteoblastic cell marker (AP+). Bone cells were selected through a general P1 gate to remove doublet cells or cellular debris (Figure 2A), and then further purified through sequential gates for non-hematopoietic origin (Figure 2B) and positivity for the osteoblast lineage marker AP (Figure 2C). This resulted in a small subset of cells highly enriched for osteoblasts, approximately 0.6% of all cells enumerated by FACS. The specificity of the AP antibody was confirmed by dual label immune-fluorescence of cortical bone sections identifying cells that express both Osteocalcin and AP within the bone matrix (Supplemental Figure 2). All FACS antibodies were also tested with the proper isotype controls to ensure the specificity of the signal (Supplemental Figure 3). All antibodies produced a signal that was above the background fluorescent signal.
Figure 2. Osteoblast lineage cell isolation and enrichment procedure for single cell qPCR.
(A) Scatter plot for unstained cells showing forward and side scatter for each object. The P1 gate is outlined to eliminate unwanted sample. (B) Scatter plot showing the signal for the CD31 and CD45 signal. The P2 gate was positioned over the CD31/CD45 double negative population. (C) Histogram of Alkaline Phosphatase signal showing the position of the P3 gate, which isolates approximately 0.6% of cells from the entire parent population. These cells represent the purified osteoblast lineage cells that are selected for single cell analysis. The total gene expression quality from isolated cells was examined using histograms of Ct values. These histograms display the distribution of Ct values from all assays from 24 individual cells using various preparation methods.
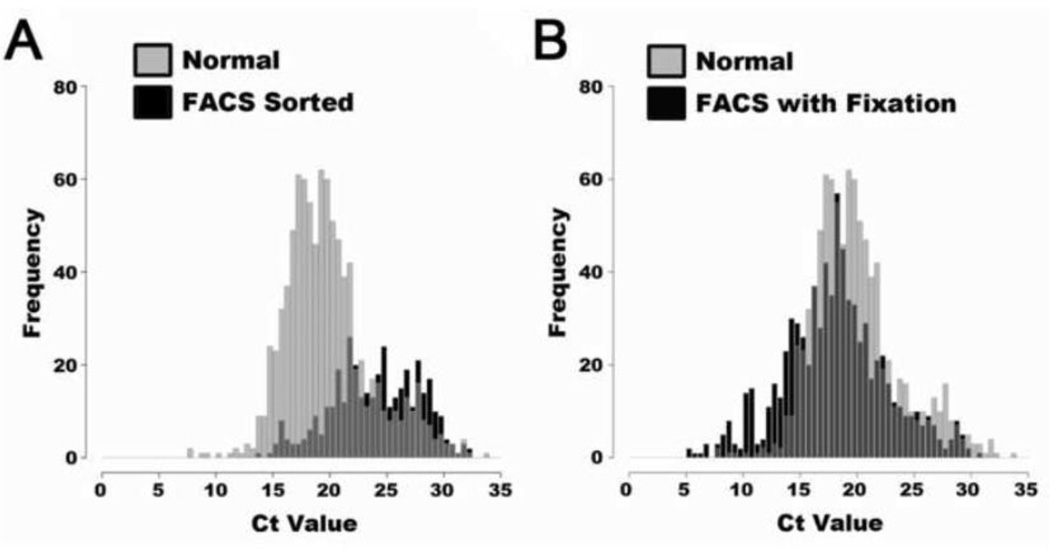
In previous studies it had been reported that the isolation of live cells via FACS may affect the quality of the mRNA for downstream analysis [17,18]. Given that it is possible to carry out gene expression profiling on fixed cell samples, (often demonstrated in paraffin embedded tissues), we reasoned that fixing the cells prior to FACS might be a necessary step to obtain more robust gene expression in the purified cell type. To test this possibility, we isolated cells under three conditions: live unsorted cells, live sorted cells, and fixed sorted cells. We determined that processing live cells via FACS can negatively affect the quality and quantity of mRNA signal which can be captured via single cell qPCR when compared to cells which were not sorted prior to qPCR (Figure 3A). In contrast however, if the cells are treated with gentle fixation prior to flow sort, a substantial increase in gene expression signal was observed, similar to the intensity of expression signal seen in live unsorted single cells (Figure 3B). An expression heatmap of this data demonstrates how the fixation procedure retains a similar expression profile across a population of individual cells (Supplemental Figure 4). The specificity of the qPCR assays were also validated using melt curve analysis after amplification. This ensures that the primer sets used were unambiguous for gene targets under all conditions (Supplemental Figure 5). This quality control procedure allows for the inclusion of only primers sets that met this pass/fail specificity criterion, which are listed for all genes assayed in the cells isolated from bone (Supplemental Table 1).
Figure 3. Distribution of Ct values from cells with and without fixation prior to FACS sorting.
The qPCR assay signal after FACS sorting was tested against unsorted and unfixed cells (A) Distribution of unfixed cells that were either selected randomly prior to sorting, or FACS sorted then selected. The FACS sorted cells are compromised in the signal quality resulting in a blunted peak shifted to the right. (B) Display of Ct distribution with fixation procedure prior to FACS sorting which preserves the signal retrieved from the individual cells, and prevents much of the mRNA degradation caused by FACS sorting.
3.3 Assessment of procedure on frozen samples
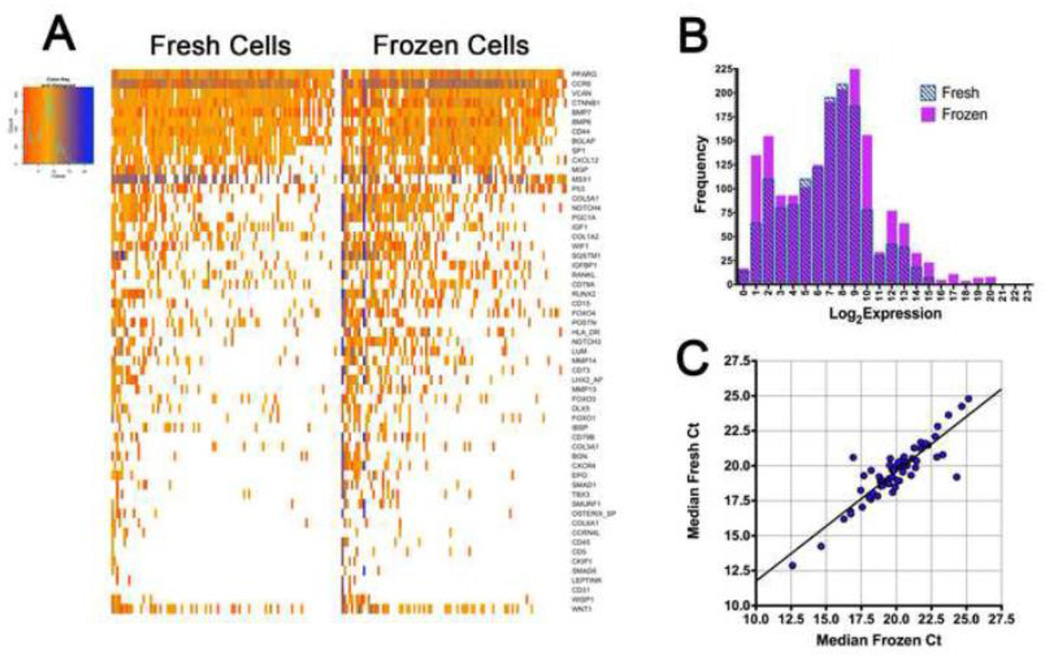
We sought to determine whether our protocol would also work with frozen bone samples; an extension of the method that would add flexibility to the single cell procedure and could potentially allow for analysis of osteoblasts or other bone cell types derived from clinical biopsy samples from patients. A modified method of Oh et al.[13] was used to preserve bone tissue while freezing femurs from individual mice (n=4 per group) for a minimum period of 48 hours at −80°C. These samples were then thawed, and osteoblast lineage cells isolated in parallel to those from freshly harvested mouse femurs at ambient temperature. Single cell qPCR was performed on 95 single cells from either the ambient temperature or from previously frozen bone samples. Both data sets are remarkably similar in their expression profiles and quality of signal (Figure 4A and 4B), suggesting that the fixation/freezing procedure was not deleteriously affecting the subsequent expression profiling. A comparison of the frozen cells’ log2 expression distribution across all gene assays significantly overlaps that of the non-frozen cells. Similarly, the median Ct value for each qPCR assay was compared between the frozen cells and the non-frozen cells (Figure 4C). The expression levels between the fresh and frozen tissue samples are highly correlative for each gene assayed (R2=0.780). Additionally, the expression data from each cell was examined with cluster analysis to attempt to identify any grouping based upon preparation method or animal replicates (Supplemental Figure 6). The analysis showed no significant grouping of the mice based upon prior freezing or a variation in the animals within these groups. This indicates that the isolation procedure maintains the relevant expression profile within individual cells. Overall, these experimental results indicate that the cryo-preservation method of cortical bone we describe here does not significantly degrade mRNA quality or qPCR sensitivity.
Figure 4. Quality control of gene expression from frozen bone samples.
Using a nanofluidic high-throughput qPCR approach, we performed gene expression on bone tissue which had been frozen for 48 hours at −80°C in a preservative solution. Each column represents a single cell, and each row a gene. (A) Heatmap of single cells from frozen tissue shows a comparable expression to that of the fresh material. This indicates that with proper preparation gene expression analysis is possible on single cells isolated from frozen bone tissue. (B) Histogram of Log2 expression values from fresh and frozen cells generated from the R single cell analysis package. The two populations are approximately equal in signal intensity. (C) Median Ct for each qPCR assay from the fresh cells and the frozen cells. The observed Ct values for each assay are highly correlated (R2=0.780).
3.4 Expression analysis of 190 individual osteoblast lineage cells
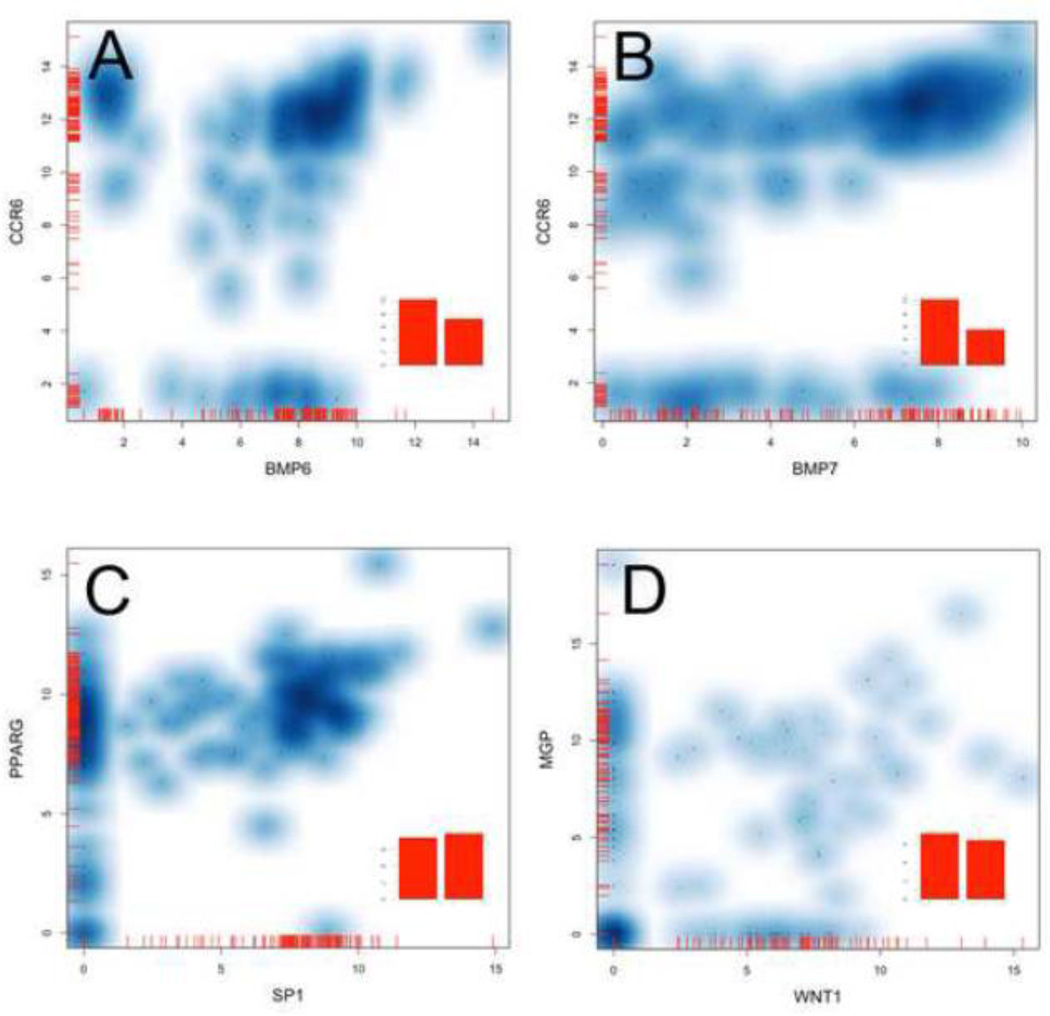
Finally, using the log2 expression distributions, we visualized the gene expression across all osteoblast lineage cells that were isolated (190 cells in total). This data is displayed as violin plots that show a probability density displayed against the expression level for each gene (Supplemental Figure 7). From this data we can observe different dynamics of expression in particular genes across a population of cells. Cells can be grouped based upon their expression of a particular gene of interested. This is demonstrated with scatterplots of Ccr6 and Bmp6 which partitions into cell subsets expressing either gene alone or more commonly they co-express these genes at high levels (Figure 5A). Ccr6’s relationship to the expression of Bmp7 is far less correlated showing a wide range of expression values rather than a discrete on or off pattern (Figure 5B). This approach may also have utility in identifying interacting expression patterns. Sp1 is highly expressed in a subset of cells. For almost every cell expressing this gene, it is also expressing Pparg (Figure 5C). Conversely, the majority of cells expressing Mgp are not expressing Wnt1, shown as a clusters along the x and y axis. As a means of simulating the traditional bulk preparation of mRNA from a tissue the mean gene expression from the 190 single cells is shown as a bar graph inset to each plot. This highlights the depth of data loss that occurs when hundreds or thousands of cells are combined in an analysis. Single cell analysis can provide new insights into the complex transcriptional events and relationships occurring in normal and diseased states within specific cell populations.
Figure 5. Smoothed scatter plots of single cell expression data.
To examine the distribution of expression profiles that could be observed in the osteoblast lineage cell population, we plotted log2 expression values from 190 cells for specific genes. These plots display the expression of two genes within single cells as a cloud where the darker the blue color the more dense the grouping of single cell expression values. The mean expression across all cells are plotted as bar graphs (inset) which simulates the expression patterning from a bulk mRNA preparation. This data visualization displays the complex expression patterns can be seen. Ccr6 and Bmp6 are expressed in three distinct groupings where cells co-express these genes or express only on of these genes alone (A). This patterning is not observed with Bmp7 which has a more scattered expression profile rather than a discrete grouping (B). The single cell data can be used to identify genes which may have biologically relevant interaction such as PPAR-gamma and transcription factor Sp1 (C), or have very little co-expression like Mgp and Wnt1(D). This visualization has utility for defining subsets of single cells based upon the expression patterns of particular genes of interest.
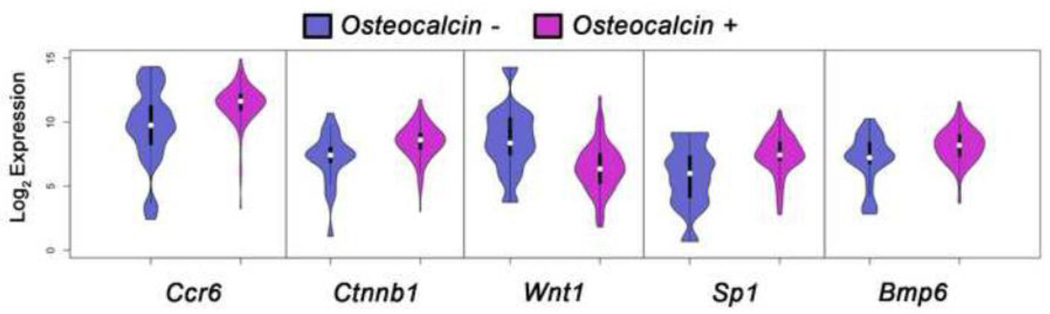
To gain a better understanding of how these relationships occur within cells of an osteoblast lineage we sought to further define the cell population. The total cell population was divided between cells that detectably express Osteocalcin and cells that do not. We then asked among the detectable expression values what genes are differentially expressed between the Osteocalcin+ and Osteocalcin− cells. Through a non-parametric t-test (p<0.05, FDR of 5%) of the expression in each group, we found 5 genes that are differentially expressed based upon their Osteocalcin status: Ccr6, Ctnnb1, Wnt1, Sp1, and Bmp6. This Log2 expression data is shown as violin plots of each group for these genes (Figure 6). Through the selection of specific transcriptional markers it is possible to gain insight into genes that may act as co-expression markers for a specific cell sub-population.
Figure 6. Differentially expressed genes in Osteocalcin expressing cells.
To further dissect subsets of osteoblastic lineage cells; cells which had detectable levels of Ostocalcin (Osteocalcin+) were divided from the non Osteocalcin expressing cells (Osteocalcin−). The differential gene expression was then determined between these two groups by non-parametric t-test resulting in 5 significantly differentially expressed genes (p<0.05, FDR 5%). The normalized expression data above background between these two groups was plotted as violin plots of Log2 Expression for each gene showing the detectable expression profiles between the cell populations.
4. Discussion
4.1 The utility of the single cell approach
Single cell analysis is an expanding area in transcriptomic research with the potential to uncover new insights into the proteomic, transcriptional and metabolic functions of individual cells [19]. Traditionally gene expression has been performed on tissue lysates of thousands if not hundreds of thousands to millions of cells and contains multiple cell types. This approach provides an “average” expression level for a particular gene within a tissue, and can miss rare gene expression when other cells dilute this signal, or be confounded by small numbers of cells expressing high levels of message, while in fact the majority of the population may be quiescent for specific gene expression. There is an increasing interest in examining gene expression within individual cells to gain a deeper understanding of stochastic gene expression variance, which is not observed when studying bulk cell populations, either in cell culture, or from primary tissue preparations. We have developed a method applicable to cortical bone to characterize single cell osteoblast lineage cell expression profiles (or other cell types for which there are immunogenic markers) with an ability to translate the methodology to clinically obtained bone samples.
4.2 Use of mild fixation to maintain gene expression in flow sorted cells
We were initially able to isolate cells from the bone matrix using a serial collagenase digestion steps followed by FACS in which we could purify cells expressing a canonical osteoblast marker. We found that subjecting cells to FACS can have a profound effect on the quality of the mRNA, preventing accurate evaluation of the downstream expression. This technical limitation was overcome with the use of a mild fixation prior to sorting the cells. Gentle fixation maintains the quality of the target message, and did not alter the expression profiles from individual cells. This finding reinforces the need to carefully handle the experimental material when examining gene expression from single cells.
4.3 Preservation of bone samples for subsequent purification of cell types of interest
After developing a robust amplification procedure it was important to refine it, so that the sample material could be preserved and potentially shipped from a clinical setting, facilitating evaluation of samples derived from a wide variety of clinical contexts. To accomplish this we used a previously established cryopreservation procedure [13] and stored the samples in a freezer for a minimum of 48 hours prior to isolating cells, thereby mimicking a procedure in which samples would be obtained by biopsy, then frozen and shipped for gene expression evaluation. We then compared the resultant expression data to cells that were extracted from bone that remained at ambient temperatures prior to isolation. We found that expression profiles are largely unaffected by the cryopreservation. The median gene expression values from ambient and frozen osteoblast lineage cells maintain a strong correlation despite being derived from completely independent biological replicates. This indicates that cryopreserved material provides an accurate representation of the transcriptional profile to that of bone directly excised from an animal. Furthermore, this cryopreservation could be useful in collection of bone biopsies from animal models of bone disease at differing stages for later simultaneous extraction and qPCR of individual cells.
4.4 Analysis of gene expression data in cortical osteoblast lineage cells
Previous studies have employed data normalization on gene expression at the bulk population level to a single gene [20]. However this method requires that the normalizer gene be invariant in every cell. This normalization approach can actually increase the variation within a dataset due to the inherent stochastic nature of gene expression within single cells. A more non-biased approach uses the mean expression of all genes in a dataset [21]. In this study, we have employed this approach to normalize the data across the entire 190-cell dataset using a single cell analysis package in the R statistical software environment. This approach also allowed the normalized expression to be displayed in violin plots, showing the probability density of expression values for each gene. We were able to visualize the range of expression within a group of animals by pooling single cells together from these biological replicates. From this data visualization it is possible to see groups of genes behaving in a similar manner at the single cell level. This analysis revealed for the first time in osteoblast lineage cells derived from mature bone, classes of genes that are largely always transcriptionally active, as well as genes that maintain a low expression level except for transcriptional bursting in a small subset of cells. Additionally, it is possible to correlate the expression of gene pairs to identify specific subgroups of cells. These types of analysis may provide unique insights into specific gene regulatory mechanisms that control various aspects of osteoblast function in vivo [22]. As an example of this, we showed that the gross majority of cells expressing the transcription factor Sp1 also highly expressed PPAR-γ (Pparg). This expression is biologically relevant as Sp1 maintains the expression of this gene by binding to its promoter site [23] and as a feedback regulatory mechanism PPAR-γ can reduce the activity of SP1 [24].
We also sought to identify genes that are differentially expressed relative to Osteocalcin’s expression status in osteoblastic lineage cells. To do this the cell population was divided into cells that express Osteocalcin, versus those which do not. We then examined which genes are significantly differentially expressed between these two groups of osteoblast lineage cells, and found 5 differentially expressed genes. Interestingly 2 of the 5 genes modulate the Wnt signaling pathway, Ctnnb1 and Wnt1. β-catenin is upregulated in Osteocalcin+ cells while Wnt1 is downregulated. This data is similar to results from a transgenic mouse model which showed that the removal of β-catenin (Ctnnb1) shifts cells from a osteoblastic cell fate to an adipocyte cell lineage within bone [25]. The dissection of transcriptional differences in specific subsets of cells based upon expression may prove useful in uncovering the regulation of osteoblastic fate cells without the need for external genetic manipulations. Future studies using this single cell approach in normal and diseased states of bone should provide information about transcriptional derangements which are acutely affecting small groups of cells in a pathological situation that are not detectable by studying only the mean expression level of bulk populations.
4.5 Overall summary
In summary, we have developed a new method for the isolation of specific subpopulations of cells from mature bone that can be used to determine expression profile differences as a function of location within bone. It also permits the assessment of variation relating to different physiological and pathophysiological states, or address inherent stochastic variability between cells. The methodology represents a robust approach for profiling in a high throughput format with flexibility regarding cell type, and sample handling amendable to various animal model studies of bone disease. It also potentially permits the evaluation of transcriptional changes over time within targeted cell types from bones of individual patients before and after therapeutic treatments.
Supplementary Material
Highlights.
Method of isolating osteoblastic lineage cells from cortical bone for gene expression via FACS.
Gene expression analysis of single cells from fresh or frozen bone samples.
Data normalization and visualization for single cell expression analysis.
Identify expression pattern and variance of 60+ genes in single cortical osteoblast lineage cells
Acknowledgments
The authors would like to express their gratitude to Marc Unger, Ken Livak and Tze-Howe Charn who provided assistance with Fluidigm’s single cell analysis package for R. This work was supported in part by the NIH (RL1AG032117), a Glenn Foundation fellowship to JF, and an award to SM from Merck (Investigator Initiated Studies Program, # 39326).
Author Roles:
SM conceived the project, performed data analysis, and prepared the manuscript; JF generated methodological procedures, isolated bone cells, performed gene expression and data analysis, and prepared the manuscript. CJR provided critical input on various aspects of experimental design. SS conceived cell-labeling method and performed FACS analysis. Approving final version of manuscript: JM, SS, CJR, and SM.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Discloser: All authors state that they have no conflicts of interest or competing financial interests.
References
- 1.Rosen CJ. Exploiting new targets for old bones. J Bone Miner Res. 2010;25:934–936. doi: 10.1002/jbmr.91. [DOI] [PubMed] [Google Scholar]
- 2.Kulterer B, Friedl G, Jandrositz A, Sanchez-Cabo F, Prokesch A, Paar C, Scheideler M, Windhager R, Preisegger KH, Trajanoski Z. Gene expression profiling of human mesenchymal stem cells derived from bone marrow during expansion and osteoblast differentiation. BMC Genomics. 2007;8:70. doi: 10.1186/1471-2164-8-70. [pii] 10.1186/1471-2164-8-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foster LJ, Zeemann PA, Li C, Mann M, Jensen ON, Kassem M. Differential expression profiling of membrane proteins by quantitative proteomics in a human mesenchymal stem cell line undergoing osteoblast differentiation. Stem Cells. 2005;23:1367–1377. doi: 10.1634/stemcells.2004-0372. [pii] 10.1634/stemcells.2004-0372. [DOI] [PubMed] [Google Scholar]
- 4.Luo W, Friedman MS, Hankenson KD, Woolf PJ. Time series gene expression profiling and temporal regulatory pathway analysis of BMP6 induced osteoblast differentiation and mineralization. BMC Syst Biol. 2011;5:82. doi: 10.1186/1752-0509-5-82. [pii] 10.1186/1752-0509-5-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Varanasi SS, Olstad OK, Swan DC, Sanderson P, Gautvik VT, Reppe S, Francis RM, Gautvik KM, Datta HK. Skeletal site-related variation in human trabecular bone transcriptome and signaling. PLoS One. 2010;5:e10692. doi: 10.1371/journal.pone.0010692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rawlinson SC, McKay IJ, Ghuman M, Wellmann C, Ryan P, Prajaneh S, Zaman G, Hughes FJ, Kingsmill VJ. Adult rat bones maintain distinct regionalized expression of markers associated with their development. PLoS One. 2009;4:e8358. doi: 10.1371/journal.pone.0008358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ståhlberg A, Andersson D, Aurelius J, Faiz M, Pekna M, Kubista M, Pekny M. Defining cell populations with single-cell gene expression profiling: correlations and identification of astrocyte subpopulations. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalerba P, Kalisky T, Sahoo D, Rajendran PS, Rothenberg ME, Leyrat AA, Sim S, Okamoto J, Johnston DM, Qian D, Zabala M, Bueno J, Neff NF, Wang J, Shelton AA, Visser B, Hisamori S, Shimono Y, van de Wetering M, Clevers H, Clarke MF, Quake SR. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotechnol. 2011;29:1120–1127. doi: 10.1038/nbt.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pina C, Fugazza C, Tipping AJ, Brown J, Soneji S, Teles J, Peterson C, Enver T. Inferring rules of lineage commitment in haematopoiesis. Nat Cell Biol. 2012;14:287–294. doi: 10.1038/ncb2442. [DOI] [PubMed] [Google Scholar]
- 10.Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bahar R, Hartmann CH, Rodriguez KA, Denny AD, Busuttil RA, Dollé ME, Calder RB, Chisholm GB, Pollock BH, Klein CA, Vijg J. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature. 2006;441:1011–1014. doi: 10.1038/nature04844. [DOI] [PubMed] [Google Scholar]
- 12.Flynn JM, Santana LF, Melov S. Single cell transcriptional profiling of adult mouse cardiomyocytes. J Vis Exp. 2011:e3302. doi: 10.3791/3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oh JH, Zoller JE, Kubler A. A new bone banking technique to maintain osteoblast viability in frozen human iliac cancellous bone. Cryobiology. 2002;44:279–287. doi: 10.1016/s0011-2240(02)00034-2. [pii] [DOI] [PubMed] [Google Scholar]
- 14.Flynn JM, Santana LF, Melov S. Single cell transcriptional profiling of adult mouse cardiomyocytes. J Vis Exp. 2011:e3302. doi: 10.3791/3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mao F, Leung WY, Xin X. Characterization of EvaGreen and the implication of its physicochemical properties for qPCR applications. BMC Biotechnol. 2007;7:76. doi: 10.1186/1472-6750-7-76. [pii] 10.1186/1472-6750-7-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Golden TR, Hubbard A, Dando C, Herren MA, Melov S. Age-related behaviors have distinct transcriptional profiles in Caenorhabditis elegans. Aging Cell. 2008;7:850–865. doi: 10.1111/j.1474-9726.2008.00433.x. [pii] 10.1111/j.1474-9726.2008.00433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Esser C, Göttlinger C, Kremer J, Hundeiker C, Radbruch A. Isolation of full-size mRNA from ethanol-fixed cells after cellular immunofluorescence staining and fluorescence-activated cell sorting (FACS) Cytometry. 1995;21:382–386. doi: 10.1002/cyto.990210411. [DOI] [PubMed] [Google Scholar]
- 18.Barrett MT, Glogovac J, Prevo LJ, Reid BJ, Porter P, Rabinovitch PS. High-quality RNA and DNA from flow cytometrically sorted human epithelial cells and tissues. Biotechniques. 2002;32:888–890. doi: 10.2144/02324rr06. 892, 894, 896. [DOI] [PubMed] [Google Scholar]
- 19.Kim SH, Fourmy D, Fujii T. Expanding the horizons for single-cell applications on lab-on-a-chip devices. Methods Mol Biol. 2012;853:199–210. doi: 10.1007/978-1-61779-567-1_15. [DOI] [PubMed] [Google Scholar]
- 20.Guo G, Huss M, Tong GQ, Wang C, Li Sun L, Clarke ND, Robson P. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell. 2010;18:675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 21.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munsky B, Neuert G, van Oudenaarden A. Using gene expression noise to understand gene regulation. Science. 2012;336:183–187. doi: 10.1126/science.1216379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nebbaki SS, El Mansouri FE, Afif H, Kapoor M, Benderdour M, Duval N, Pelletier JP, Martel-Pelletier J, Fahmi H. Egr-1 contributes to IL-1-mediated down-regulation of peroxisome proliferator-activated receptor γ expression in human osteoarthritic chondrocytes. Arthritis Research and Therapy. 2012;14:R69. doi: 10.1186/ar3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang Q, Chen A. Curcumin inhibits srebp-2 expression in activated hepatic stellate cells in vitro by reducing the activity of specificity protein-1. Endocrinology. 2009;150:5384–5394. doi: 10.1210/en.2009-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song L, Liu M, Ono N, Bringhurst FR, Kronenberg HM, Guo J. Loss of Wnt/b-Catenin signaling causes cell fate shift of preosteoblasts from osteoblasts to adipocytes. J Bone Miner Res. 2012;27:2344–2358. doi: 10.1002/jbmr.1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.