Background: The sensitivity of non-S-phase cells to Top1-targeting drugs has been under-explored.
Results: Quiescent Top2β-deficient cells are specifically hypersensitive to Top1-targeting drugs.
Conclusion: Top2β deficiency promotes camptothecin-induced apoptosis as a result of RNA polymerase II depletion and p53 accumulation.
Significance: This study has identified Top2β as a novel determinant for camptothecin sensitivity.
Keywords: Apoptosis, DNA Topoisomerase, p53, RNA Polymerase II, Transcription, RNA Polymerase Large Subunit, Apoptosis, Cytotoxicity, Topoisomerase I-targeting Drug, Topoisomerase IIβ
Abstract
Camptothecin (CPT), a topoisomerase (Top) I-targeting drug that stabilizes Top1-DNA covalent adducts, can induce S-phase-specific cytotoxicity due to the arrest of progressing replication forks. However, CPT-induced non-S-phase cytotoxicity is less well characterized. In this study, we have identified topoisomerase IIβ (Top2β) as a specific determinant for CPT sensitivity, but not for many other cytotoxic agents, in non-S-phase cells. First, quiescent mouse embryonic fibroblasts (MEFs) lacking Top2β were shown to be hypersensitive to CPT with prominent induction of apoptosis. Second, ICRF-187, a Top2 catalytic inhibitor known to deplete Top2β, specifically sensitized MEFs to CPT. To explore the molecular basis for CPT hypersensitivity in Top2β-deficient cells, we found that upon CPT exposure, the RNA polymerase II large subunit (RNAP LS) became progressively depleted, followed by recovery to nearly the original level in wild-type MEFs, whereas RNAP LS remained depleted without recovery in Top2β-deficient cells. Concomitant with the reduction of the RNAP LS level, the p53 protein level was greatly induced. Interestingly, RNAP LS depletion has been well documented to lead to p53-dependent apoptosis. Altogether, our findings support a model in which Top2β deficiency promotes CPT-induced apoptosis in quiescent non-S-phase cells, possibly due to RNAP LS depletion and p53 accumulation.
Introduction
Eukaryotic DNA topoisomerase (Top)2 I catalyzes relaxation of supercoiled DNA by performing a cleavage/religation reaction that transiently nicks one strand of the DNA double helix with Top1 covalently attached to the 3′-phosphate of the cleaved strand (1, 2). The subsequent religation reaction reseals the nicked strand and reverses the covalent Top1-DNA interaction to a noncovalent interaction. The unique catalytic activity of Top1 enables Top1 to suppress unconstrained DNA supercoiling, known to be generated during various DNA tracking activities (3), which may affect normal DNA metabolism and induce genetic instability (4–6). Specifically, the role of Top1 in removing unconstrained DNA supercoiling during transcription has been demonstrated in both yeast and mammalian cells (7–9). Besides Top1, Top2 also carries DNA relaxation activity by transferring one DNA duplex through a Top2-linked double strand break in an ATP-dependent manner (10, 11). In mammals, there are two highly homologous Top2 isozymes, Top2α and Top2β (12). Top2α is a well known proliferation marker with peak expression found at the late S and G2/M phase of the cell cycle, and serum deprivation can drive cells entering the G0 state in which the Top2α level is greatly reduced (13, 14). By contrast, Top2β is present at a constant level throughout the cell cycle (14) and is expressed in all cell types, including terminally differentiated cells such as neurons and cardiomyocytes (15, 16). Top2β has been proposed to participate in transcription regulation with binding sites located at both the promoter and transcribed regions (17, 18). Recent studies have also indicated a potential overlapping function of both Top1 and Top2 in transcription (6, 19) and thus suggest that Top2β may regulate transcription in collaboration with Top1 in nonproliferating cells.
Camptothecin (CPT), a Top1-targeting drug, can interfere with the catalytic cycle of Top1 by inhibiting the religation reaction and therefore increases the half-life of the transient Top1-DNA covalent adducts (also known as Top1 cleavage complexes) (1, 20). The Top1 cleavage complex itself is highly reversible, and the single strand nick within the complex is concealed by the covalently linked Top1 protein and thus remains undetected in the absence of cellular processing. The two major intracellular helix-tracking processes, DNA replication and transcription, have been implicated in eliciting DNA damage response upon CPT treatment (21–23). CPT is known to selectively kill S-phase cells (20, 24). It has been demonstrated that the collision of Top1 cleavage complexes with DNA replication machineries and the subsequently generated DNA double strand breaks are responsible for the S-phase-specific cytotoxicity of CPT (22). Slowly growing cells are more resistant to CPT treatment, possibly due to the reduced replication-mediated Top1 cleavage complex processing (25). This S-phase-selective cytotoxicity enables Top1-based chemotherapy to be efficacious in treating several types of cancers, including ovarian, lung, and colon cancers (20, 24, 26). Currently, two water-soluble CPT analogs, topotecan (TPT) and irinotecan (CPT-11), are being used in the clinic (20, 24).
Recent studies have also elucidated the interaction of the transcription machinery and Top1 cleavage complexes in activating a DNA repair pathway that requires the proteasome-dependent degradation of Top1 (21, 23, 27). It has been shown that in the absence of DNA replication, DNA single strand breaks are generated upon proteasomal degradation of Top1 that is covalently trapped on DNA in the presence of CPT (23). In addition to proteasome, repair of Top1 cleavage complexes also involves tyrosyl-DNA phosphodiesterase (Tdp1) that catalyzes the hydrolysis of the tyrosyl-3′-phosphate linkage between a protein (peptide) and DNA (28–31). It has been shown that either the inhibition of proteasome or a mutation in Tdp1 can enhance CPT sensitivity (27, 32, 33). The predominant single strand breaks generated upon CPT treatment in non-S-phase cells can largely explain the fact that non-S-phase or postmitotic cells are generally much more resistant to CPT. Other than the involvement of various repair pathways, it remains unclear how CPT sensitivity is determined in the absence of DNA replication.
In this study, we observed that Top2β deficiency could increase CPT cytotoxicity in quiescent Top2β-deficient (Top2β−/−) as well as ICRF-187-treated wild-type (Top2β+/+) mouse embryonic fibroblasts (MEFs). The increased sensitivity is associated with the depletion of RNA polymerase II large subunit (RNAP LS), p53 accumulation, general transcription inhibition, and apoptosis induction.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), VP-16 (etoposide), CPT (camptothecin), TPT (topotecan), ICRF-187, bleomycin, staurosporine, and cisplatin were purchased from Sigma. MG132 was purchased from Selleck Chemicals LLC. The anti-α-tubulin antibodies were purchased from the Developmental Studies Hybridoma Bank, University of Iowa. The mouse anti-γ-H2AX (clone JBW301) antibodies and the mouse anti-phospho-ATM (at Ser-1981) (clone 10H11.E12) antibodies were purchased from Millipore. The anti-Top2β (H-286), anti-polymerase II (N-20, recognizes RNAP LS), and anti-p53 (FL-393) antibodies were purchased from Santa Cruz Biotechnology. Anti-Top1 antibodies were obtained from the sera of scleroderma 70 patients as described previously (34). The anti-PARP-1, anti-caspase 3, and anti-β-actin antibodies were purchased from Cell Signaling Technology. The annexin V-FITC apoptosis detection kit was purchased from BD Biosciences. The tissue culture media, DMEM, was purchased from Invitrogen. Penicillin, streptomycin, and trypsin (0.25% trypsin, 2.21 mm EDTA) were purchased from Cellgro. 5-Ethynyl-2′-deoxyuridine (EdU), 5-ethynyl uridine (EU), and the Click-iT® EdU Alexa Fluor® 488 flow cytometry assay kit were purchased from Invitrogen. Tritium-labeled UTP was purchased from PerkinElmer Life Sciences.
Cell Culture and Cytotoxicity Assay
Top2β+/+ and Top2β−/− primary MEFs were isolated from E13.5 mouse embryos as described previously (35). MEFs were cultured in DMEM containing 10% serum (FetalPlex animal serum complex, Gemini Bio-Products), l-glutamine (2 mm), penicillin (100 units/ml), and streptomycin (0.1 mg/ml) in a humidified incubator with 5% CO2 at 37 °C. For all experiments, 24 h after plating in 10% serum-containing DMEM, MEFs were cultured for another 24 h in 0.2% serum-containing DMEM before various drug treatments. For MTT cytotoxicity assays, primary MEFs were plated in 96-well plates (1.0 × 104 cells/well) for 24 h followed by culturing in DMEM supplemented with 0.2% serum for another 24 h. Cells were then treated continuously with various doses of different drugs for 4 days. At the end of drug treatments, 30 μl of MTT (1 μg/ml) was added, and cells were further incubated for 4 h in the humidified CO2 incubator at 37 °C. Upon removal of media, 150 μl of dimethyl sulfoxide (DMSO) was added to each well, and the absorbance at 570 nm was determined for each well using a microplate reader. All assays were performed at least twice in triplicate.
Apoptosis Assay (Annexin V Staining)
Primary MEFs cultured in 6-well plates were treated with DMSO (0.1%), CPT, or VP-16 for various times. At the end of the treatment, cells were harvested from tissue culture plates using 0.25% trypsin, 2.21 mm EDTA. After washing once with 1× phosphate-buffered saline (PBS), cells were subjected to staining by annexin V using the annexin V-FITC apoptosis detection kit I (BD Biosciences) following the manufacturer's protocol. Briefly, cells were resuspended in 100 μl of 1× annexin V binding buffer, followed by a 15-min staining in the dark with FITC-conjugated annexin V for the detection of phosphatidylserine flipping and propidium iodine for the determination of membrane integrity. Then, an additional 400 μl of 1× annexin V binding buffer was added to each reaction immediately before the flow cytometry analysis.
Band Depletion Assay
2.5 × 105 primary Top2β+/+ and Top2β−/− MEFs were treated with 0, 1, or 10 μm CPT for 20 min. Cells were lysed immediately following CPT treatment upon aspirating CPT-containing DMEM tissue culture media using the 3× SDS sample buffer (175 mm Tris-HCl, pH 6.8, 15% glycerol, 5% SDS, 300 mm dithiothreitol, 0.006% bromphenol blue). Cell lysates were then analyzed by Western blotting using anti-Top1 and anti-α-tubulin antibodies.
DNA Synthesis Using the EdU Labeling Assay
To determine the level of DNA synthesis in Top2β+/+ and Top2β−/− primary MEFs, 6 × 105 cells were plated in 60-mm tissue culture plates, and 24 h later, cells were pulse-labeled with EdU (10 μm) for 1 h. For measuring EdU incorporation in quiescent MEFs, 24 h after plating, MEFs were cultured in DMEM supplemented with 0.2% serum for an additional 24 h before pulse-labeling with EdU. Cells were harvested from tissue culture plates by incubating in 0.25% trypsin, 2.21 mm EDTA and subsequently washed once with ice-cold 1× PBS. Cells were then pelleted and resuspended in 0.1 ml of cold 1× PBS, 0.1% glucose solution and immediately fixed by adding 1 ml of ice-cold 70% ethanol, and the fixation reaction was incubated in 4 °C for 1 h. Fixed cells were then subjected to the Click-iT reaction to reveal the incorporated EdU in newly synthesized DNA as described in the manufacturer's manual (Click-iT® EdU Alexa Fluor® 488 flow cytometry assay kit, Invitrogen). Cells were then treated with RNase A followed by DNA staining with 7-aminoactinomycin D (7AAD) for 1 h at 37 °C. Both DNA content and EdU incorporation were scored using the flow cytometry analysis.
RNA Synthesis Using the Uridine Incorporation Assay
At the end of each treatment, primary MEFs were subjected to pulse labeling for 6 h with 5 μCi of [5,6-3H]uridine (47 Ci/mmol; ICN)/ml. Following uridine labeling, cells were lysed with a solution containing 4 m isothioguanidine, 0.5% Sarkosyl, 2 mm sodium citrate, and 0.1 m β-mercaptoethanol. Cell lysates were then directly spotted onto separate DE81 DEAE-cellulose filter papers. Filters were washed three times (30 min each) with 0.3 m ammonium formate followed by a single ethanol wash (10 min). Filters were then dried, and the amount of uridine present on individual filters was determined using a Beckman liquid scintillation counter following standard procedures.
RNA Synthesis Using the EU Labeling Assay
At the end of each treatment, primary MEFs were subjected to pulse abeling with 0.5 mm EU for 6 h. Following the EU labeling, cells were trypsinized with 0.25% trypsin, 2.21 mm EDTA and processed for Click-iT reaction as described in the EdU labeling assay, except that RNase A and 7-aminoactinomycin D were not used. EU incorporation was then determined by flow cytometry.
RESULTS
Top2β Deficiency Sensitizes Quiescent MEFs to Camptothecin-induced Cell Death
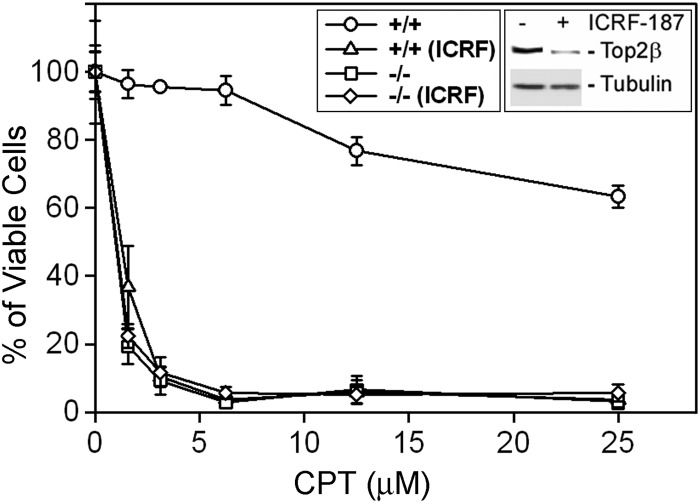
Top1-targeting drugs (e.g. CPT) are known to induce S-phase-specific cytotoxicity; this is largely attributed to the arrest of the advancing replication forks by the drug-induced Top1 cleavage complexes (22). However, the mechanism of non-S-phase cytotoxicity of Top1-targeting drugs is less well characterized. Using serum-starved primary MEFs (quiescent cells that are not in S-phase) as a model, we have previously shown that Top2β knock-out MEFs (Top2β−/−) were highly resistant to Top2-targeting drugs (e.g. VP-16) as compared with wild-type MEFs (Top2β+/+) (36, 37). In addition, we have also examined the cytotoxicity of Top1-targeting drugs in Top2β−/− MEFs. Surprisingly, sensitivity to CPT was much higher in Top2β−/− MEFs than in wild-type MEFs. As shown in Fig. 1, the viability of MEFs upon CPT treatment (4-day continuous treatment) showed a clear dependence on the presence of Top2β. Top2β+/+ MEFs are relatively resistant to CPT, with an IC50 greater than 25 μm (Fig. 1 and Table 1). However, Top2β−/− MEFs were much more sensitive to CPT, with an IC50 less than 2 μm (Fig. 1 and Table 1). To determine whether this hypersensitivity phenomenon was specific to Top1-targeting drugs, a variety of agents was examined using the 4-day MTT assay. As shown in Table 1, Top2β−/− MEFs were hypersensitive only to Top1-targeting drugs (i.e. CPT and TPT) but not other cytotoxic agents such as staurosporine, hydrogen peroxide (H2O2), and bleomycin. The reduced sensitivity to VP-16 (etoposide, a Top2-targeting drug) is expected because Top2β−/− MEFs lack the drug target. These results suggest a specific link between Top2β deficiency and enhanced sensitivity toward Top1-targeting drugs.
FIGURE 1.
Top2β-deficient MEFs are hypersensitive to CPT. Wild-type (Top2β+/+) and Top2β−/− MEFs, cultured in DMEM supplemented with 0.2% serum, were treated with increasing doses of CPT (0, 1.5625, 3.125, 6.25, 12.5 or 25 μm) in the presence or absence of 200 μm ICRF-187 for 4 days. Cellular viability was determined by the MTT assay as described under “Experimental Procedures.” Inset, Western blotting results showing the decreased Top2β protein level in wild-type cells treated with ICRF-187 (200 μm).
TABLE 1.
Top2β-deficient MEFs are hypersensitive to Top1-targeting drugs
Quiescent primary Top2β+/+ and Top2β−/− MEFs were treated with various compounds. MTT assays were performed at the end of the 4th day as described under “Experimental Procedures.” Each experiment was performed in triplicate and repeated at least twice. The IC50 values of each treatment were calculated by regression curve fitting and are listed in the table.
| CPT | TPT | VP-16 | Staurosporine | H2O2 | Bleomycin | |
|---|---|---|---|---|---|---|
| μm | μm | μm | μm | μm | μm | |
| Top2β+/+ | >20 | >25 | 22 ± 1.5 | 0.4 ± 0.1 | 22 ± 3 | 92 ± 5 |
| Top2β−/− | 1.7 ± 0.5 | 6.5 ± 0.3 | >200 | 0.3 ± 0.2 | 20 ± 2 | 99 ± 4 |
In addition to Top2β−/− knock-out MEFs, we have also manipulated the protein level of Top2β by treating wild-type MEFs with ICRF-187 to investigate the relationship between sensitivity to Top1-targeting drugs and the Top2β protein level. ICRF-187 is a catalytic inhibitor of Top2 and is able to induce Top2β degradation in various cells (37, 38). As shown in Fig. 1 (inset), a 24-h treatment of ICRF-187 was able to significantly down-regulate Top2β in primary MEFs. Under this condition, sensitivity to CPT and TPT, but not staurosporine, was greatly increased (>5-fold) in ICRF-187-treated Top2β+/+ MEFs (Table 2). By contrast, VP-16-induced cytotoxicity was reduced at least 7-fold by ICRF-187 treatment (Fig. 1 and Table 2). Our results indicate that Top2β is involved in determining the cytotoxicity of Top1-targeting drugs in quiescent MEFs.
TABLE 2.
ICRF-187 sensitizes wild-type MEFs to Top1-targeting drugs
Quiescent primary Top2β+/+ were pretreated with ICRF-187 (200 μm) for 24 h followed by co-treatments with various compounds. MTT assays were performed at the end of the 4th day as described under “Experimental Procedures.” Each experiment was performed in triplicate and repeated at least twice. The IC50 values of each treatment were calculated by regression curve fitting and are listed in the table.
| CPT | TPT | VP-16 | Staurosporine | |
|---|---|---|---|---|
| μm | μm | μm | μm | |
| DMSO | 18 ± 1 | >25 | 29 ± 3 | 0.5 ± 0.3 |
| ICRF-187 | 2.7 ± 0.2 | 7.2 ± 0.3 | >200 | 0.7 ± 0.5 |
CPT Selectively Induced Apoptosis in Top2β-deficient Cells
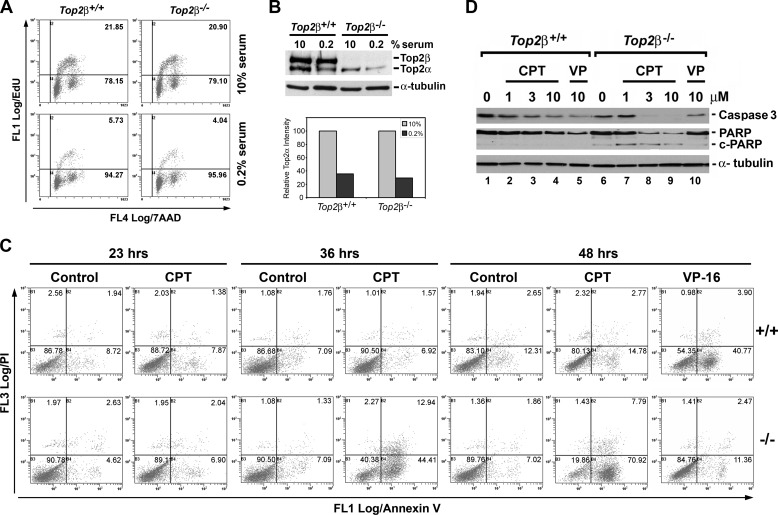
In the absence of high level DNA replication, there was much less collision between the replication fork and CPT-trapped Top1 cleavage complexes, and quiescent or postmitotic cells are in general more resistant to CPT. It is possible that the increased CPT sensitivity in Top2β−/− MEFs could be due to the increased DNA synthesis, and thus replication collision with Top1 cleavage complexes in these cells. We therefore pulse-labeled primary MEFs with the thymidine analog EdU for determining the population of cells that were in S-phase and undergoing active DNA synthesis. As shown in the upper panel of Fig. 2A, the population of cells that were in S-phase (EdU-positive) was similar in wild-type (Top2β+/+, 21.85%) and Top2β-deficient (Top2β−/−, 20.90%) MEFs when they were cultured in media containing 10% serum. When primary MEFs were cultured in media containing 0.2% serum, the S-phase population in both Top2β+/+ (5.73%) and Top2β−/− (4.04%) MEFs was similarly reduced (Fig. 2A, lower panel), indicating that the increased sensitivity of Top2β−/− MEFs to CPT and TPT was not due to the increased replication in these cells. In addition, serum starvation also led to a reduction in the Top2α protein level (Fig. 2B), in agreement with the notion that Top2α is a proliferation marker (13, 14).
FIGURE 2.
CPT preferentially induces apoptosis in Top2β-deficient MEFs. A, wild-type (Top2β+/+) and Top2β−/− MEFs, cultured in DMEM supplemented with either 10% (top panel) or 0.2% (bottom panel) serum, were pulse-labeled with EdU for 1 h followed by flow cytometry analysis as described under “Experimental Procedures.” Percentage of EdU-positive cells was indicated in the dotted plot (upper right-hand corner). B, cell lysates prepared from Top2β+/+ and Top2β−/− MEFs cultured in DMEM supplemented with either 10% serum or 0.2% serum were analyzed by immunoblotting using anti-Top2α/Top2β and anti-α-tubulin (loading control, served as the normalization standard) antibodies. The relative amount of Top2α in MEFs cultured in 0.2% serum as compared with that cultured in 10% serum (arbitrarily set to 1) was determined, plotted, and shown in the bottom panel. C, annexin V/propidium iodine (PI) staining of MEFs treated with CPT (3 μm) for 23, 36, or 48 h or VP-16 (100 μm) for 48 h were analyzed by flow cytometry (upper panel, Top2β+/+ MEFs; lower panel, Top2β−/− MEFs). Individual fluorescence intensity of ∼10,000 cells for each treatment was plotted in a dot plot. The percentage of cell population in each quadrant is indicated by a number located at the upper right-hand corner of each quadrant. Annexin V-positive cells that are in early stages of apoptosis are clustered in the lower right-hand quadrant. D, wild-type and Top2β−/− MEFs cultured in DMEM containing 0.2% serum were treated with CPT (1, 3, or 10 μm) or VP-16 (10 μm) for 36 h. Immunoblotting was performed using anti-caspase 3, anti-PARP-1, and anti-α-tubulin antibodies.
It has been shown that CPT (10–25 μm) can induce apoptosis in nonproliferating cells such as neurons (39–43). We therefore investigated whether the increased CPT sensitivity in Top2β−/− MEFs was due to increased apoptosis. One of the early events taking place during apoptosis is the flipping of phosphatidylserines from the cytoplasmic side of the plasma membrane to the cell surface. Therefore, apoptotic cells that contain surface phosphatidylserine can be specifically labeled by annexin V. Both Top2β+/+ and Top2β−/− MEFs were treated with 3 μm CPT for 23, 36, or 48 h, and then the annexin V-positive cell population was determined by flow cytometry. The results showed that CPT did not induce noticeable changes in the number of annexin V-positive cells in both Top2β+/+ and Top2β−/− MEFs when cells were treated for 23 h. However, lengthening CPT treatment to 36 and 48 h led to an obvious increase of annexin V-positive cells only in Top2β−/− MEFs (Fig. 2C, from 7.09% in control cells to 44.41% in cells treated with CPT for 36 h, and from 7.02% in control cells to 70.92% in cells treated with CPT for 48 h) but not in Top2β+/+ MEFs (Fig. 2C, from 7.09% in control cells to 6.92% in cells treated CPT for 36 h, and from 12.31% in control cells to 14.78% in cells treated with CPT for 48 h). These results indicate that Top2β−/− MEFs were highly sensitive to CPT-induced apoptosis. By contrast, VP-16 (100 μm), a Top2-targeting drug, only induced apoptosis in Top2β+/+ MEFs (from 12.31% in control cells to 40.77% in VP-16-treated cells), whereas Top2β−/− MEFs (from 7.02% in control cells to 11.36% in VP-16-treated cells) were highly resistant (Fig. 2C, 48 hrs). We have also monitored other apoptotic end points such as PARP-1 and caspase-3 cleavage in CPT-treated (36 h) Top2β+/+ and Top2β−/− MEFs. As shown in Fig. 2D, the full-length caspase-3 and PARP-1 were significantly decreased in Top2β−/− MEFs as compared with that observed in Top2β+/+ MEFs, indicating the selective induction of PARP-1 and caspase-3 cleavage in Top2β−/− MEFs upon CPT treatment. These results suggest that CPT selectively kills Top2β−/− MEFs by inducing apoptosis.
Top2β Deficiency Did Not Affect CPT-induced Top1 Cleavage Complex Formation, Top1 Degradation, and Activation of DNA Damage Response
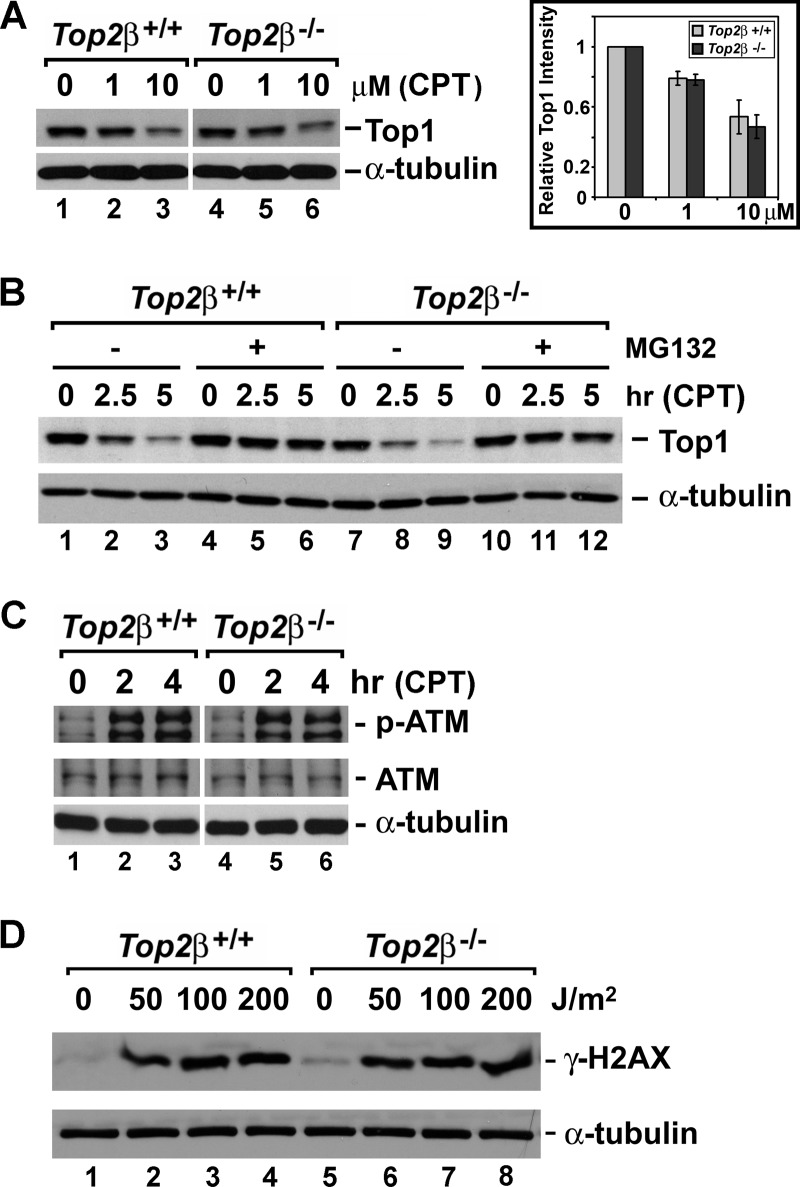
The key determinant of CPT cytotoxicity is the amount of Top1 cleavage complexes, and it is known that elevated Top1 expression can lead to increased sensitivity toward Top1-targeting drugs (44–46). It is possible that the significant increase of CPT sensitivity in Top2β-deficient cells is due to the elevated Top1 protein level and much increased Top1 cleavage complex formation. However, as shown in Fig. 3A (lanes 1 and 4), Top2β+/+ and Top2β−/− MEFs contained similar levels of Top1. In addition, we performed the band depletion assay to estimate the amount of CPT-induced Top1 cleavage complexes in Top2β+/+ and Top2β−/− MEFs. Because of their covalent linkage to genomic DNA, Top1 cleavage complexes are too bulky to enter the SDS-polyacrylamide gel during electrophoresis. Therefore the amount of free Top1 (not linked to DNA) to be detected by immunoblotting will decrease upon CPT treatment. As shown in Fig. 3A (left panel, immunoblotting; right panel, quantification), the immunoreactive band representing the free Top1 protein showed similar patterns of decrease in a dose-dependent manner upon CPT treatment in both Top2β+/+ and Top2β−/− MEFs. Thus, there seemed to be no significant impact of Top2β deficiency on CPT-induced Top1 cleavage complex formations.
FIGURE 3.
Top2β deficiency does not affect the formation or cellular processing of CPT-induced Top1 cleavage complexes, as well as DNA damage response. A, amount of Top1 cleavage complexes was estimated by the band depletion assay as described under “Experimental Procedures.” The decrease in the intensity of the Top1 immunoreactive band indicates the increased covalent trapping of Top1 on DNA due to the formation of Top1 cleavage complexes. The intensity of the Top1 immunoreactive band representing the free Top1 (not covalently trapped on DNA) was first normalized to the intensity of the respective α-tubulin immunoreactive band (loading control) under the same treatment. Then the relative intensity of the free Top1 in CPT-treated MEFs to that in DMSO-treated MEFs was determined from the ratio of the normalized free Top1 intensity in CPT-treated MEFs to the normalized free Top1 intensity in DMSO-treated MEFs (arbitrarily set to 1). The mean relative intensity calculated from two independent experiments was plotted in the bar graph (right panel). The error bar represents the standard deviation. B, processing of Top1 cleavage complexes is similar in both Top2β+/+ and Top2β−/− MEFs. Primary Top2β+/+ and Top2β−/− MEFs were treated with CPT (10 μm) for 0, 2.5, or 5 h in the presence or absence of MG132 (2 μm). For all MG132 treatments, MG132 was added 30 min prior to the addition of CPT. At the end of the drug treatment, cells were washed three times with fresh media and further incubated in fresh media for an additional 30 min in a humidified 37 °C incubator with 5% CO2. Cell lysates were then prepared for the subsequent immunoblotting analysis using anti-Top1 or anti-α-tubulin antibodies following standard protocols. C, similar induction of ATM autophosphorylation in Top2β+/+ and Top2β−/− MEFs. Top2β+/+ and Top2β−/− MEFs were treated with CPT (10 μm) for 2 or 4 h. Cell lysate were then analyzed by immunoblotting using anti-phospho-ATM (Ser-1987, labeled p-ATM), anti-ATM, and anti-α-tubulin antibodies. D, similar UV irradiation-induced DNA damage signal in Top2β+/+ and Top2β−/− MEFs. Top2β+/+ and Top2β−/− MEFs were UV-irradiated at doses of 0, 50, 100, or 200 J/m2, followed by an additional 1-h recovery in a humidified 37 °C incubator with 5% CO2. Cell lysates were prepared and immunoblotted with anti-γ-H2AX and anti-α-tubulin antibodies.
Our previous findings have shown that Top1 cleavage complexes are processed and repaired through proteasome-dependent mechanisms (21, 23). It was proposed that Top1 cleavage complexes can arrest transcription elongation and induce a 26 S proteasome-mediated degradation of Top1, leading to the activation of ATM-dependent DNA damage response and DNA repair (23). Cells defective in repair of Top1 cleavage complexes are hypersensitive to CPT (32). Thus, we next compared CPT-induced degradation of Top1 cleavage complexes in Top2β+/+ and Top2β−/− MEFs. As shown in Fig. 3B, CPT-induced Top1 degradation showed similar kinetics in Top2β+/+ (lanes 2 and 3) and Top2β−/− (lanes 8 and 9) MEFs. In addition, MG132, a proteasome inhibitor, could largely abolish Top1 degradation (Fig. 3B, lanes 5 and 6 and lanes 11 and 12) in both Top2β+/+ and Top2β−/− MEFs. Our data suggest that proteasome-mediated degradation of Top1 covalently linked to DNA, and thus the initial repair of CPT-induced Top1 cleavage complexes was not altered in the absence of Top2β.
Top1 cleavage complexes can elicit DNA damage response, and cellular sensitivity to DNA- damaging agents can be largely affected by the DNA damage repair pathways. ATM is a major DNA damage sensor, and it becomes activated through the autophosphorylation at Ser-1981 (Ser-1987 in mouse) in response to DNA damage. Autophosphorylated ATM subsequently activates a cascade of proteins that are involved in DNA repair. It has been shown previously that ATM autophosphorylation can be induced by a variety of DNA-damaging agents, including CPT in both proliferating and nonproliferating cells (23, 47). We therefore monitored ATM autophosphorylation in both Top2β+/+ and Top2β−/− MEFs treated with CPT. As shown in Fig. 3C, ATM autophosphorylation was efficiently induced in both Top2β+/+ and Top2β−/− MEFs. In addition, both primary MEFs exposed to UV irradiation showed a similar induction of the DNA damage marker γ-H2AX (Fig. 3D). These results, together with the finding that the sensitivity (IC50) of Top2β+/+ and Top2β−/− MEFs to a variety of agents (e.g. H2O2, bleomycin, and staurosporine) was almost identical, suggest that the general DNA damage response was most likely not affected in Top2β−/− MEFs.
CPT-induced Down-regulation of RNAP LS and p53 Accumulation in Top2β-deficient Cells
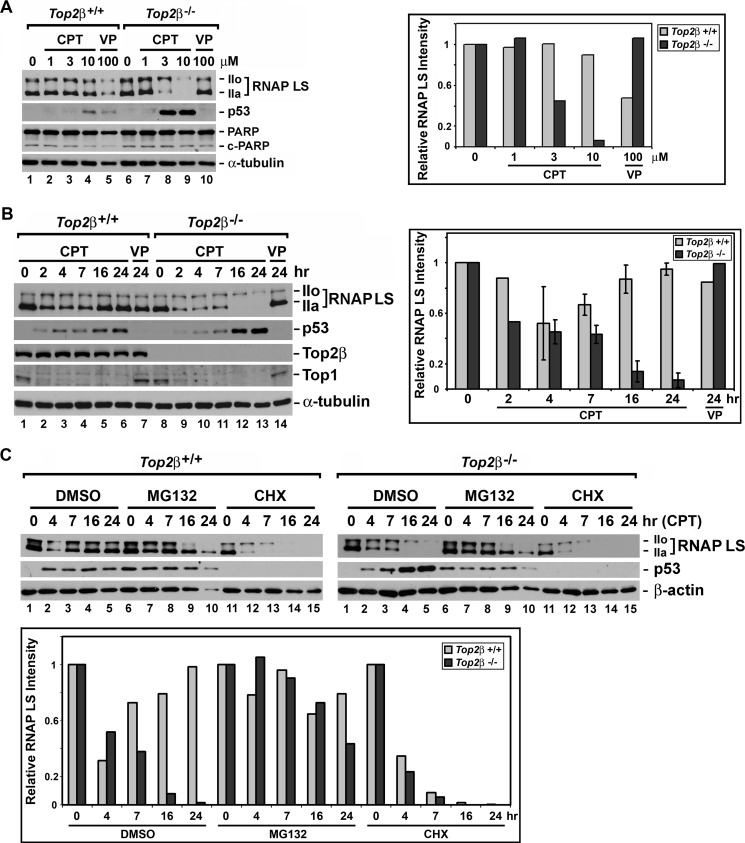
CPT-induced Top1-DNA covalent adducts are known to block transcription. In addition, CPT has been shown to induce proteasome-mediated degradation of the RNAP LS (also known as POLR2A and RPB1) (21, 48). RNAP LS degradation in response to CPT treatment is reminiscent of that observed in cells exposed to UV irradiation, hydrogen peroxide, and α-amanitin (49–52). It appears that the blockade of the RNA polymerase II elongation complex can trigger polyubiquitination and degradation of RNAP LS. We next investigated the effect of CPT treatment on RNAP LS stability in both Top2β+/+ and Top2β−/− MEFs. As shown in Fig. 4A (left panel), RNAP LS in Top2β+/+ MEFs treated with 1, 3, and 10 μm CPT for 24 h retained a level similar to that observed in DMSO-treated cells. However, the level of RNAP LS apparently decreased in Top2β−/− MEFs treated with 3 and 10 μm CPT for 24 h. Especially in Top2β−/− MEFs treated with 10 μm CPT, the hypophosphorylated form (IIa) of RNAP LS was almost undetectable, and the hyperphosphorylated form (IIo) of RNAP LS was present at a much decreased level. The decrease in RNAP LS levels in CPT-treated Top2β−/− MEFs can be better visualized in the bar graph that plots the intensities (normalized against the respective α-tubulin intensity) of immunoreactive bands corresponding to RNAP LS (both IIo and IIa forms) for each treatment relative to that in control MEFs (Fig. 4A, right panel). Moreover, CPT did not induce significant increase in PARP-1 cleavage (Fig. 4A), which was consistent with the annexin V staining pattern showing no induction of apoptosis when Top2β−/− MEFs were treated with CPT for 23 h (Fig. 2C, 23 h).
FIGURE 4.
RNAP LS degradation and p53 accumulation in CPT-treated Top2β-deficient MEFs. A, Top2β+/+ and Top2β−/− MEFs were treated with 0, 1, 3, or 10 μm CPT or 100 μm VP-16 (labeled VP) for 24 h. Cell lysates were analyzed by SDS-PAGE and then immunoblotted using anti-polymerase II (for detecting RNAP LS), anti-p53, and anti-α-tubulin (loading control, served as the normalization standard) antibodies. The amount of RNAP LS in CPT- or VP-16-treated MEFs relative to that in DMSO-treated MEFs (the control, has an arbitrarily value of 1) was calculated and plotted in a bar graph shown in the right panel. B, Top2β+/+ and Top2β−/− MEFs were treated with 10 μm CPT for 2, 4, 7, 16, or 24 h or 10 μm VP-16 (labeled VP) for 24 h. Cell lysates were analyzed using anti-Top2β, anti-Top1, anti-polymerase II, anti-p53, anti-PARP-1 (labeled PARP) and anti-α-tubulin (loading control) antibodies. The amount of RNAP LS in drug-treated MEFs relative to that in DMSO-treated MEFs (the control, has an arbitrarily value of 1) was quantified and plotted (right panel). The error bar-containing histograms represent the average determined from two to three independent experiments, and the error bars represent the standard deviations. C, Top2β+/+ (upper left panel) and Top2β−/− (upper right panel) MEFs were treated with CPT (10 μm) for 4, 7, 16, and 24 h in the presence of either DMSO (control), MG132 (2 μm), or cycloheximide (labeled CHX, 20 μg/ml). Cell lysates were analyzed using anti-polymerase II, anti-p53, and anti-β-actin (loading control) antibodies. The relative amount of RNAP LS in CPT-treated MEFs in the presence of DMSO, MG132, or CHX as compared with that in their respective control MEFs (i.e. DMSO-treated, MG132-treated and CHX-treated MEFs, respectively) was calculated and plotted (lower panel).
The difference in the level of RNAP LS in Top2β+/+ and Top2β−/− MEFs upon CPT treatment was surprising. We therefore further performed experiments to monitor the level of RNAP LS in Top2β+/+ and Top2β−/− MEFs treated with 10 μm CPT for various times. Consistent with previous findings, the level of RNAP LS was decreased in early time points (2 and 4 h) during CPT treatment in both Top2β+/+ (Fig. 4B, left panel, lanes 2 and 3) and Top2β−/− (Fig. 4B, left panel, lanes 9 and 10) MEFs. However, the level of RNAP LS rebounded in Top2β+/+ MEFs (Fig. 4B, left panel, lanes 5 and 6) at later time points of CPT treatment (e.g. 16 and 24 h), whereas a further decrease of RNAP LS levels was observed in Top2β−/− MEFs (Fig. 4B, left panel, lanes 12 and 13). These results indicate that RNAP LS underwent degradation upon CPT treatment initially, and although the level of RNAP LS recovered in Top2β+/+ MEFs, the level of RNAP LS in Top2β−/− MEFs continued to decrease when the treatment time was increased to 16 or 24 h. This dynamic change in RNAP LS levels in MEFs treated with CPT for various times is further illustrated in a bar graph (Fig. 4B, right panel). To investigate whether the disappearance of the RNAP LS was due to proteasome-mediated degradation, various CPT treatments were performed in the presence of the proteasome inhibitor MG132. As shown in Fig. 4C, the level of disappearance of the RNAP LS was much reduced in the presence of MG132 in both Top2β+/+ (compare lane 7 with 2, upper left panel) and Top2β−/− (compare lane 7 with 2, upper right panel) MEFs at an early time point of CPT treatment (4 h). MG132 also attenuated degradation of RNAP LS in Top2β−/− MEFs treated with CPT for 7 h (Fig. 4C, compare lane 8 with 3, upper right panel). At later time points (16 and 24 h), although significant cell loss was observed (as indicated by the decreased β-actin levels in Top2β−/− MEFs) (Fig. 4C, upper right panel, β-actin blot), it is clear that some protection of RNAP LS from degradation indeed took place in the presence of MG132 (Fig. 4C, upper right panel, compare lane 9 with 4 and lane 10 with 5). These results indicate that in response to CPT treatment, RNAP LS underwent proteasome-mediated degradation initially, and its level eventually recovered in wild-type MEFs; however, Top2β deficiency prevented this recovery. To determine whether the recovery of RNAP LS in Top2β+/+ MEFs was due to new protein synthesis, we have treated MEFs with CPT in the presence of the protein synthesis inhibitor cycloheximide (CHX). As shown in Fig. 4C (see upper panel for immunoblotting and lower panel for quantification), continuous CPT treatment in the presence of CHX caused a progressive decrease of RNAP LS levels in both Top2β+/+ (Fig. 4C, lanes 11–15, upper left panel) and Top2β−/− (lanes 11–15, upper right panel) MEFs. It suggests that the observed recovery of RNAP LS in Top2β+/+ MEFs was due to new protein synthesis, and the re-synthesis of RNAP LS was blocked in Top2β-deficient MEFs.
It has been previously documented that transcription stress imposed by stalled RNA polymerase or decreased RNAP LS level can lead to p53 accumulation and p53-dependent apoptosis (53, 54). Because CPT treatment led to a decrease in RNAP LS levels in Top2β−/− MEFs, we thus asked the question whether CPT treatment could also lead to p53 accumulation. As shown in Fig. 4A (left panel), in both untreated Top2β+/+ (lane 1) and Top2β−/− (lane 6) MEFs, the p53 protein level was very low and could not be detected by immunoblotting. Top2β−/− MEFs treated with 3 and 10 μm CPT for 24 h had greatly elevated p53 protein levels (Fig. 4A, lanes 8 and 9), whereas much limited p53 elevation was observed for Top2β+/+ MEFs treated with 10 μm CPT (lane 4). Next, the time course of p53 induction in response to CPT treatment was determined. As shown in Fig. 4B (left panel), 10 μm CPT treatment for 16 and 24 h showed much higher p53 induction in Top2β−/− MEFs (lanes 12 and 13) than in Top2β+/+ MEFs (lanes 5 and 6). However, when RNAP LS degradation was blocked by MG132, p53 accumulation was also largely abolished (Fig. 4C, upper right panel, compare lane 8 with 3, lane 9 with 4, and lane 10 with 5). These findings suggest that prolonged CPT treatment in Top2β−/− MEFs could induce p53 accumulation as a result of RNAP LS degradation.
CPT Inhibited General Transcription in Top2β-deficient MEFs
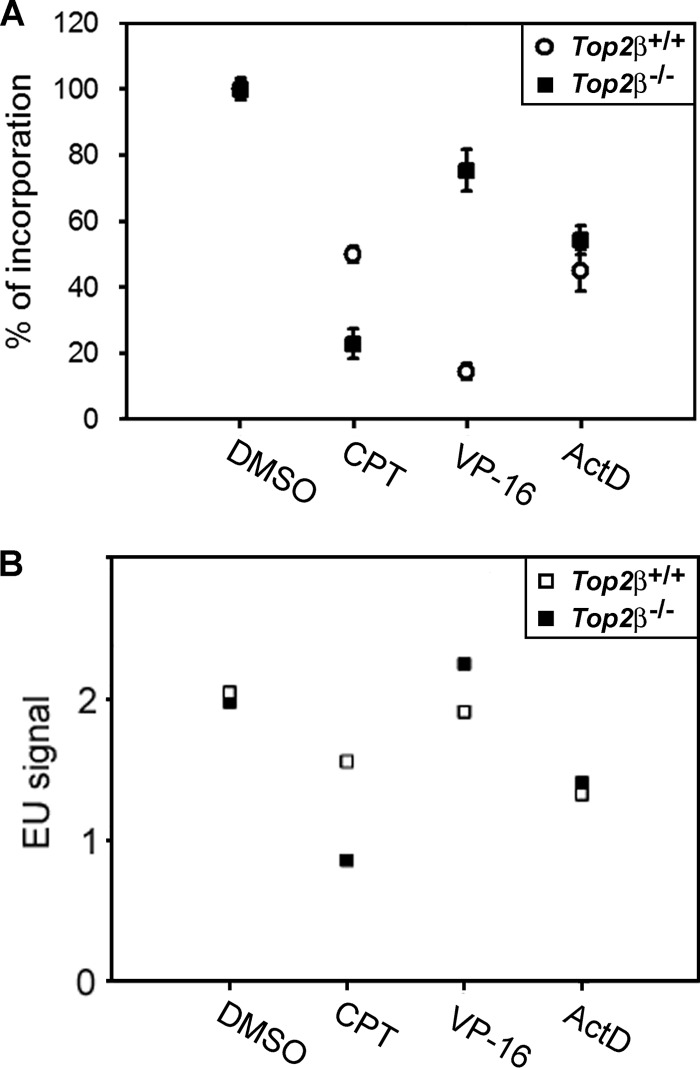
As road blocks, CPT-induced Top1-DNA covalent adducts can impede DNA helix tracking activities such as DNA replication and transcription (20, 55). Under our experimental conditions, the majority of cells were in a quiescent state with minimal DNA replication activity (Fig. 2A, lower panel), and therefore, we compared the effect of CPT on total RNA synthesis in Top2β+/+ and Top2β−/− MEFs upon CPT treatment for 24 h using the uridine incorporation assay. As shown in Fig. 5A, it appeared that the general transcription in the absence of CPT is similar in Top2β+/+ and Top2β−/− MEFs, as indicated by the amount of [3H]uridine incorporation. It suggests that Top2β deficiency did not affect general transcription. However, CPT selectively caused a greater decrease in uridine incorporation in Top2β−/− MEFs when compared with Top2β+/+ MEFs. Conversely, VP-16 preferentially inhibited uridine incorporation in Top2β+/+ MEFs, whereas actinomycin D (ActD, well known for its ability to inhibit transcription) similarly inhibited transcription in both Top2β+/+ and Top2β−/− MEFs. Furthermore, we have also compared the level of general transcription in CPT-treated Top2β+/+ and Top2β−/− MEFs by monitoring EU incorporation into newly synthesized total RNAs. As shown in Fig. 5B, CPT selectively blocked EU incorporation in Top2β−/− MEFs as compared with Top2β+/+ MEFs. By contrast, VP-16 did not significantly block EU incorporation in Top2β−/− MEFs, whereas actinomycin D induced comparable inhibition of EU incorporation in both Top2β+/+ and Top2β−/− MEFs. Together, these results suggest that the general transcription was blocked in Top2β-deficient MEFs after prolonged CPT treatment.
FIGURE 5.
CPT inhibits total RNA synthesis in Top2β-deficient MEFs. Quiescent primary MEFs were treated with CPT (2.5 μm), VP-16 (100 μm), or actinomycin D (100 ng/ml) for 24 h followed by a 6-h pulse-labeling with [3H]UTP (A) and EU (B). The amount of the uridine analog being incorporated was measured as described under “Experimental Procedures.” Percentage of incorporation (A) and mean EU labeling (B) was plotted.
DISCUSSION
Top1-targeting drugs (e.g. CPT, TPT, CPT-11, and ARC-111) are known to induce S-phase-specific cytotoxicity due to double strand breaks generated upon collision of Top1 cleavage complexes and advancing replication machineries (20, 24, 55). Nonreplicating cells are generally more resistant to CPT due to lack of replication collision and therefore minimal induction of double strand breaks (23). There are several factors that can determine CPT cytotoxicity. An elevation in Top1 protein level is known to enhance CPT cytotoxicity due to increased Top1 cleavage complex formation. Mutations in DNA repair proteins such as Mre11 and Tdp1, as well as protein factors that modulate the ubiquitin-proteasome pathway, are also known to sensitize cells to CPT, most likely due to the failed repair of lesions generated from Top1 cleavage complexes (28, 32, 56–59). A recent genome-wide CPT sensitivity screen has identified replication stress regulators, the chromatin remodeling complex FACT and MCM proteins, as major components in suppressing CPT sensitivity (60). These protein complexes are involved in DNA repair and provide resistance to replication stress. In this study, we have identified Top2β as a novel modulator that controls CPT cytotoxicity in quiescent cells with low DNA replication activity. In the absence of Top2β, cellular sensitivity to CPT increased 4–10-fold and was associated with apoptosis induction. Our results further show that this increase in CPT sensitivity in Top2β-deficient cells is not due to the increased formation of CPT-induced Top1 cleavage complexes or the lack of proteasomal processing of Top1 cleavage complexes.
Consistent with previous findings, we have shown that CPT can induce proteasome-mediated degradation of RNAP LS in both wild-type and Top2β-deficient MEFs. However, the level of RNAP LS recovers after prolonged CPT exposure in wild-type MEFs, probably as a result of proteasome-mediated removal of Top1 cleavage complexes and the re-synthesis of RNAP LS. Indeed, when protein synthesis was inhibited by cycloheximide, RNAP LS was degraded and could no longer recover in wild-type MEFs (Fig. 4C). However, levels of RNAP LS continued to decrease to almost undetectable levels after a 24-h treatment with CPT in Top2β-deficient MEFs. It appeared that RNAP LS underwent degradation initially in response to Top1 cleavage complexes, but somehow failed to be re-synthesized in the absence of Top2β. As a consequence of this failed re-synthesis, RNAP LS becomes depleted and Top2β-deficient MEFs could subsequently undergo p53-dependent apoptosis.
The lack of re-synthesis of RNAP LS in Top2β-deficient cells could be related to the observed decrease in general transcription upon prolonged CPT exposure. CPT is a prominent transcription inhibitor, most likely by inducing Top1 cleavage complex formation in the promoter and transcribed regions of active genes (61, 62). CPT can quickly attenuate uridine incorporation into nascent RNAs within minutes upon CPT exposure. As Top1 cleavage complexes are cleared from the chromosome by the proteasome, ∼50% of original transcription activity eventually resumes even in the presence of continuous exposure to CPT (21). We have monitored total RNA synthesis in cells treated with CPT for 24 h, and we observed much decreased uridine/EU incorporation in Top2β-deficient cells as compared with that in Top2β-proficient wild-type cells. It is possible that although Top2β-deficient cells were proficient in removing DNA-linked Top1 cleavage complexes initially (Fig. 3B), the completion of the repair process could somehow be defective. The incompletely repaired DNA lesions could continue to impede all forms of transcription, including mRNA synthesis. Because RNAP LS is initially degraded upon CPT exposure, blockage in mRNA synthesis could lead to the inhibition of RNAP LS re-synthesis and its subsequent depletion in Top2β-deficient cells; RNAP LS depletion can in turn further contribute to the decreased mRNA synthesis and thus starts a vicious cycle in Top2β-deficient cells. As a result of mRNA synthesis inhibition, levels of proteins with relatively short half-lives (e.g. RNAP LS) could decrease rapidly. In addition, because rRNA synthesis is the major component of active transcription, a decrease in total RNA synthesis can reflect in large part the decrease in rRNA production. This decrease may have a direct impact on translation. Thus, decreased re-synthesis of RNAP-LS could be also due to translation inhibition as a result of reduced rRNA levels. Further experiments will be necessary to determine the precise mechanism responsible for the RNAP LS re-synthesis blockage in Top2β-deficient cells upon prolonged CPT treatment.
Both Top1 and Top2 have been implicated in transcription regulation. The increased CPT sensitivity in Top2β-deficient cells may also be related to the function of both Top1 and Top2 in transcription. Chromatin immunoprecipitation (ChIP) analysis has revealed the binding of both Top1 and Top2 to chromosomal regions that maintain active transcription in yeasts and mammalian cells (6, 17, 18, 63). The inactivation of both Top1 and Top2 in yeast reduces RNA polymerase I density, an indication of slowed transcription elongation, on rDNA (19). Recent studies have suggested a possible overlapping function of Top1 and Top2 in transcription. It has been shown that both Top1 and Top2 can enhance RNA polymerase II recruitment to nucleosome-free DNA regions in yeast (6). It is thus possible that Top2 can functionally substitute Top1 during transcription and vice versa. In mammalian cells, the essential biological functions of Top2 were carried out by the two highly homologous Top2 isozymes, Top2α and Top2β. Although the expression of Top2α oscillates during the cell cycle (with the peak expression occurring in the late S- and G2/M-phase), Top2β level remains constant through all the cell cycle. When cells enter terminal differentiation or quiescence state, Top2α is rapidly degraded to reach a level that is beyond detection; Top2β, however, remains to be expressed in nondividing cells (13, 16, 64, 65). The main biological function of Top2α has been linked to cell cycle events such as DNA replication, chromosome condensation/decondensation, and sister chromatid segregation, whereas that of Top2β has been mostly implicated in transcription regulation (2, 11). It is plausible that the overlapping function shared by Top1 and Top2 during transcription in yeast is principally orchestrated by Top1 and Top2β in mammalian cells. Therefore, although CPT-induced degradation of Top1 cleavage complexes can lead to the depletion of Top1 and result in transcription impairment, the loss of the essential function of Top1 in transcription can be mostly compensated by Top2β. In Top2β-deficient cells, however, this transcription rescue is jeopardized due to the absence of a functional substitute of Top1 during transcription; as a result, uridine incorporation is decreased, and RNAP LS becomes depleted upon prolonged CPT treatment. This interpretation is especially well suited to explain our finding that in quiescent MEFs, due to the absence (or reduced level) of Top2α, the transcription function that requires Top2 is largely dependent on Top2β, and Top2β-deficient cells become hypersensitive to CPT as a consequence of RNAP LS depletion. In proliferating cells, an increased non-S-phase CPT sensitivity may require the loss of both Top2α and Top2β, if Top2α, in addition to Top2β, also functions during transcription. Clearly, answers to this question will rely on further experiments.
Top1-based chemotherapy has been successfully used in the clinic to treat a variety of cancers (20). However, slowly growing cells are known to be resistant to CPT treatment due to the lack of replication-mediated Top1 cleavage complex processing (25). In fact, certain cancer cells are intrinsically more resistant to chemotherapeutic agents that target S-phase, due to their residence in a quiescent state (66). In this study, we showed that Top2β deficiency could significantly sensitize quiescent MEFs to CPT-induced apoptosis. In addition, a Top2 catalytic inhibitor that is known to induce Top2β degradation can also enhance CPT sensitivity in quiescent MEFs. Therefore, Top2 catalytic inhibitors, such as ICRF-187 and ICRF-193, may provide an additional edge to Top1-based chemotherapy when used in conjunction with Top1-targeting drugs. This treatment combination may be able to target quiescent, chemo-, and radio-resistant cancer stem cells.
Acknowledgments
We thank Theresa Hyejeong Choi and Joan Dubois for assistance with the flow cytometry analysis.
This work was supported, in whole or in part, by an National Institutes of Health Grant R01 CA039662 (to L. F. L.) and a Department of Defense Idea Award W81XWH-07-1-0407 (to Y. L. L.).
- Top
- topoisomerase
- CPT
- camptothecin
- TPT
- topotecan
- CPT-11
- irinotecan
- VP
- VP-16 (etoposide)
- RNAP LS
- RNA polymerase II large subunit
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- MEF
- mouse embryonic fibroblast
- EdU
- 5-ethynyl-2′-deoxyuridine
- EU
- 5-ethynyl uridine
- CHX
- cycloheximide
- PARP-1
- poly(ADP-ribose) polymerase-1
- ATM
- ataxia telangiectasia mutated.
REFERENCES
- 1. Champoux J. J. (2001) DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70, 369–413 [DOI] [PubMed] [Google Scholar]
- 2. Wang J. C. (2002) Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 3, 430–440 [DOI] [PubMed] [Google Scholar]
- 3. Liu L. F., Wang J. C. (1987) Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. U.S.A. 84, 7024–7027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lippert M. J., Kim N., Cho J. E., Larson R. P., Schoenly N. E., O'Shea S. H., Jinks-Robertson S. (2011) Role for topoisomerase 1 in transcription-associated mutagenesis in yeast. Proc. Natl. Acad. Sci. U.S.A. 108, 698–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Takahashi T., Burguiere-Slezak G., Van der Kemp P. A., Boiteux S. (2011) Topoisomerase 1 provokes the formation of short deletions in repeated sequences upon high transcription in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 108, 692–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sperling A. S., Jeong K. S., Kitada T., Grunstein M. (2011) Topoisomerase II binds nucleosome-free DNA and acts redundantly with topoisomerase I to enhance recruitment of RNA pol II in budding yeast. Proc. Natl. Acad. Sci. U.S.A. 108, 12693–12698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brill S. J., DiNardo S., Voelkel-Meiman K., Sternglanz R. (1987) Need for DNA topoisomerase activity as a swivel for DNA replication for transcription of ribosomal RNA. Nature 326, 414–416 [DOI] [PubMed] [Google Scholar]
- 8. Brill S. J., Sternglanz R. (1988) Transcription-dependent DNA supercoiling in yeast DNA topoisomerase mutants. Cell 54, 403–411 [DOI] [PubMed] [Google Scholar]
- 9. Shaiu W. L., Hsieh T. S. (1998) Targeting to transcriptionally active loci by the hydrophilic N-terminal domain of Drosophila DNA topoisomerase I. Mol. Cell. Biol. 18, 4358–4367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang J. C. (1998) Moving one DNA double helix through another by a type II DNA topoisomerase: the story of a simple molecular machine. Q. Rev. Biophys. 31, 107–144 [DOI] [PubMed] [Google Scholar]
- 11. Nitiss J. L. (2009) DNA topoisomerase II and its growing repertoire of biological functions. Nat. Rev. Cancer 9, 327–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Austin C. A., Marsh K. L. (1998) Eukaryotic DNA topoisomerase IIβ. Bioessays 20, 215–226 [DOI] [PubMed] [Google Scholar]
- 13. Hsiang Y. H., Wu H. Y., Liu L. F. (1988) Proliferation-dependent regulation of DNA topoisomerase II in cultured human cells. Cancer Res. 48, 3230–3235 [PubMed] [Google Scholar]
- 14. Woessner R. D., Mattern M. R., Mirabelli C. K., Johnson R. K., Drake F. H. (1991) Proliferation- and cell cycle-dependent differences in expression of the 170 and 180-kDa forms of topoisomerase II in NIH-3T3 cells. Cell Growth Differ. 2, 209–214 [PubMed] [Google Scholar]
- 15. Capranico G., Tinelli S., Austin C. A., Fisher M. L., Zunino F. (1992) Different patterns of gene expression of topoisomerase II isoforms in differentiated tissues during murine development. Biochim. Biophys. Acta 1132, 43–48 [DOI] [PubMed] [Google Scholar]
- 16. Watanabe M., Tsutsui K., Tsutsui K., Inoue Y. (1994) Differential expressions of the topoisomerase IIα and IIβ mRNAs in developing rat brain. Neurosci. Res. 19, 51–57 [DOI] [PubMed] [Google Scholar]
- 17. Ju B. G., Lunyak V. V., Perissi V., Garcia-Bassets I., Rose D. W., Glass C. K., Rosenfeld M. G. (2006) A topoisomerase IIβ-mediated dsDNA break required for regulated transcription. Science 312, 1798–1802 [DOI] [PubMed] [Google Scholar]
- 18. Lyu Y. L., Lin C. P., Azarova A. M., Cai L., Wang J. C., Liu L. F. (2006) Role of topoisomerase IIβ in the expression of developmentally regulated genes. Mol. Cell. Biol. 26, 7929–7941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. French S. L., Sikes M. L., Hontz R. D., Osheim Y. N., Lambert T. E., El Hage A., Smith M. M., Tollervey D., Smith J. S., Beyer A. L. (2011) Distinguishing the roles of topoisomerases I and II in relief of transcription-induced torsional stress in yeast rRNA genes. Mol. Cell. Biol. 31, 482–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pommier Y. (2006) Topoisomerase I inhibitors: camptothecins and beyond. Nat. Rev. Cancer 6, 789–802 [DOI] [PubMed] [Google Scholar]
- 21. Desai S. D., Zhang H., Rodriguez-Bauman A., Yang J. M., Wu X., Gounder M. K., Rubin E. H., Liu L. F. (2003) Transcription-dependent degradation of topoisomerase I-DNA covalent complexes. Mol. Cell. Biol. 23, 2341–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hsiang Y. H., Lihou M. G., Liu L. F. (1989) Arrest of replication forks by drug-stabilized topoisomerase I-DNA cleavable complexes as a mechanism of cell killing by camptothecin. Cancer Res. 49, 5077–5082 [PubMed] [Google Scholar]
- 23. Lin C. P., Ban Y., Lyu Y. L., Desai S. D., Liu L. F. (2008) A ubiquitin-proteasome pathway for the repair of topoisomerase I-DNA covalent complexes. J. Biol. Chem. 283, 21074–21083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li T. K., Liu L. F. (2001) Tumor cell death induced by topoisomerase-targeting drugs. Annu. Rev. Pharmacol. Toxicol. 41, 53–77 [DOI] [PubMed] [Google Scholar]
- 25. D'Arpa P., Beardmore C., Liu L. F. (1990) Involvement of nucleic acid synthesis in cell killing mechanisms of topoisomerase poisons. Cancer Res. 50, 6919–6924 [PubMed] [Google Scholar]
- 26. Rasheed Z. A., Rubin E. H. (2003) Mechanisms of resistance to topoisomerase I-targeting drugs. Oncogene 22, 7296–7304 [DOI] [PubMed] [Google Scholar]
- 27. Desai S. D., Li T. K., Rodriguez-Bauman A., Rubin E. H., Liu L. F. (2001) Ubiquitin/26S proteasome-mediated degradation of topoisomerase I as a resistance mechanism to camptothecin in tumor cells. Cancer Res. 61, 5926–5932 [PubMed] [Google Scholar]
- 28. El-Khamisy S. F., Saifi G. M., Weinfeld M., Johansson F., Helleday T., Lupski J. R., Caldecott K. W. (2005) Defective DNA single strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature 434, 108–113 [DOI] [PubMed] [Google Scholar]
- 29. Liu C., Pouliot J. J., Nash H. A. (2002) Repair of topoisomerase I covalent complexes in the absence of the tyrosyl-DNA phosphodiesterase Tdp1. Proc. Natl. Acad. Sci. U.S.A. 99, 14970–14975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Interthal H., Champoux J. J. (2011) Effects of DNA and protein size on substrate cleavage by human tyrosyl-DNA phosphodiesterase 1. Biochem. J. 436, 559–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Katyal S., el-Khamisy S. F., Russell H. R., Li Y., Ju L., Caldecott K. W., McKinnon P. J. (2007) TDP1 facilitates chromosomal single strand break repair in neurons and is neuroprotective in vivo. EMBO J. 26, 4720–4731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Desai S. D., Wood L. M., Tsai Y. C., Hsieh T. S., Marks J. R., Scott G. L., Giovanella B. C., Liu L. F. (2008) ISG15 as a novel tumor biomarker for drug sensitivity. Mol. Cancer Ther. 7, 1430–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Interthal H., Chen H. J., Kehl-Fie T. E., Zotzmann J., Leppard J. B., Champoux J. J. (2005) SCAN1 mutant Tdp1 accumulates the enzyme–DNA intermediate and causes camptothecin hypersensitivity. EMBO J. 24, 2224–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Desai S. D., Liu L. F., Vazquez-Abad D., D'Arpa P. (1997) Ubiquitin-dependent destruction of topoisomerase I is stimulated by the antitumor drug camptothecin. J. Biol. Chem. 272, 24159–24164 [DOI] [PubMed] [Google Scholar]
- 35. Zhang A., Lyu Y. L., Lin C. P., Zhou N., Azarova A. M., Wood L. M., Liu L. F. (2006) A protease pathway for the repair of topoisomerase II-DNA covalent complexes. J. Biol. Chem. 281, 35997–36003 [DOI] [PubMed] [Google Scholar]
- 36. Azarova A. M., Lyu Y. L., Lin C. P., Tsai Y. C., Lau J. Y., Wang J. C., Liu L. F. (2007) From the cover: Roles of DNA topoisomerase II isozymes in chemotherapy and secondary malignancies. Proc. Natl. Acad. Sci. U.S.A. 104, 11014–11019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lyu Y. L., Kerrigan J. E., Lin C. P., Azarova A. M., Tsai Y. C., Ban Y., Liu L. F. (2007) Topoisomerase IIβ-mediated DNA double strand breaks: implications in doxorubicin cardiotoxicity and prevention by dexrazoxane. Cancer Res. 67, 8839–8846 [DOI] [PubMed] [Google Scholar]
- 38. Xiao H., Mao Y., Desai S. D., Zhou N., Ting C. Y., Hwang J., Liu L. F. (2003) The topoisomerase IIβ circular clamp arrests transcription and signals a 26S proteasome pathway. Proc. Natl. Acad. Sci. U.S.A. 100, 3239–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Enokido Y., Araki T., Tanaka K., Aizawa S., Hatanaka H. (1996) Involvement of p53 in DNA strand break-induced apoptosis in postmitotic CNS neurons. Eur. J. Neurosci. 8, 1812–1821 [DOI] [PubMed] [Google Scholar]
- 40. Keramaris E., Stefanis L., MacLaurin J., Harada N., Takaku K., Ishikawa T., Taketo M. M., Robertson G. S., Nicholson D. W., Slack R. S., Park D. S. (2000) Involvement of caspase 3 in apoptotic death of cortical neurons evoked by DNA damage. Mol. Cell. Neurosci. 15, 368–379 [DOI] [PubMed] [Google Scholar]
- 41. Morris E. J., Geller H. M. (1996) Induction of neuronal apoptosis by camptothecin, an inhibitor of DNA topoisomerase-I: evidence for cell cycle-independent toxicity. J. Cell Biol. 134, 757–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Park D. S., Morris E. J., Greene L. A., Geller H. M. (1997) G1/S cell cycle blockers and inhibitors of cyclin-dependent kinases suppress camptothecin-induced neuronal apoptosis. J. Neurosci. 17, 1256–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang Y., Qu D., Morris E. J., O'Hare M. J., Callaghan S. M., Slack R. S., Geller H. M., Park D. S. (2006) The Chk1/Cdc25A pathway as activators of the cell cycle in neuronal death induced by camptothecin. J. Neurosci. 26, 8819–8828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sorensen M., Sehested M., Jensen P. B. (1995) Characterisation of a human small-cell lung cancer cell line resistant to the DNA topoisomerase I-directed drug topotecan. Br. J. Cancer 72, 399–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kapoor R., Slade D. L., Fujimori A., Pommier Y., Harker W. G. (1995) Altered topoisomerase I expression in two subclones of human CEM leukemia selected for resistance to camptothecin. Oncol. Res. 7, 83–95 [PubMed] [Google Scholar]
- 46. Pommier Y., Pourquier P., Urasaki Y., Wu J., Laco G. S. (1999) Topoisomerase I inhibitors: selectivity and cellular resistance. Drug Resist. Updat. 2, 307–318 [DOI] [PubMed] [Google Scholar]
- 47. Das B. B., Antony S., Gupta S., Dexheimer T. S., Redon C. E., Garfield S., Shiloh Y., Pommier Y. (2009) Optimal function of the DNA repair enzyme TDP1 requires its phosphorylation by ATM and/or DNA-PK. EMBO J. 28, 3667–3680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sordet O., Larochelle S., Nicolas E., Stevens E. V., Zhang C., Shokat K. M., Fisher R. P., Pommier Y. (2008) Hyperphosphorylation of RNA polymerase II in response to topoisomerase I cleavage complexes and its association with transcription- and BRCA1-dependent degradation of topoisomerase I. J. Mol. Biol. 381, 540–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bregman D. B., Halaban R., van Gool A. J., Henning K. A., Friedberg E. C., Warren S. L. (1996) UV-induced ubiquitination of RNA polymerase II: a novel modification deficient in Cockayne syndrome cells. Proc. Natl. Acad. Sci. U.S.A. 93, 11586–11590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Luo Z., Zheng J., Lu Y., Bregman D. B. (2001) Ultraviolet radiation alters the phosphorylation of RNA polymerase II large subunit and accelerates its proteasome-dependent degradation. Mutat. Res. 486, 259–274 [DOI] [PubMed] [Google Scholar]
- 51. Lee K. B., Wang D., Lippard S. J., Sharp P. A. (2002) Transcription-coupled and DNA damage-dependent ubiquitination of RNA polymerase II in vitro. Proc. Natl. Acad. Sci. U.S.A. 99, 4239–4244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Inukai N., Yamaguchi Y., Kuraoka I., Yamada T., Kamijo S., Kato J., Tanaka K., Handa H. (2004) A novel hydrogen peroxide-induced phosphorylation and ubiquitination pathway leading to RNA polymerase II proteolysis. J. Biol. Chem. 279, 8190–8195 [DOI] [PubMed] [Google Scholar]
- 53. Arima Y., Nitta M., Kuninaka S., Zhang D., Fujiwara T., Taya Y., Nakao M., Saya H. (2005) Transcriptional blockade induces p53-dependent apoptosis associated with translocation of p53 to mitochondria. J. Biol. Chem. 280, 19166–19176 [DOI] [PubMed] [Google Scholar]
- 54. Ljungman M., Zhang F., Chen F., Rainbow A. J., McKay B. C. (1999) Inhibition of RNA polymerase II as a trigger for the p53 response. Oncogene 18, 583–592 [DOI] [PubMed] [Google Scholar]
- 55. Champoux J. J. (2000) Structure-based analysis of the effects of camptothecin on the activities of human topoisomerase I. Ann. N.Y. Acad. Sci. 922, 56–64 [DOI] [PubMed] [Google Scholar]
- 56. Fiorani P., Reid R. J., Schepis A., Jacquiau H. R., Guo H., Thimmaiah P., Benedetti P., Bjornsti M. A. (2004) The deubiquitinating enzyme Doa4p protects cells from DNA topoisomerase I poisons. J. Biol. Chem. 279, 21271–21281 [DOI] [PubMed] [Google Scholar]
- 57. Zhang H. F., Tomida A., Koshimizu R., Ogiso Y., Lei S., Tsuruo T. (2004) Cullin 3 promotes proteasomal degradation of the topoisomerase I-DNA covalent complex. Cancer Res. 64, 1114–1121 [DOI] [PubMed] [Google Scholar]
- 58. Hamilton N. K., Maizels N. (2010) MRE11 function in response to topoisomerase poisons is independent of its function in double strand break repair in Saccharomyces cerevisiae. PLoS One 5, e15387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kerzendorfer C., Whibley A., Carpenter G., Outwin E., Chiang S. C., Turner G., Schwartz C., El-Khamisy S., Raymond F. L., O'Driscoll M. (2010) Mutations in Cullin 4B result in a human syndrome associated with increased camptothecin-induced topoisomerase I-dependent DNA breaks. Hum. Mol. Genet. 19, 1324–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. O'Connell B. C., Adamson B., Lydeard J. R., Sowa M. E., Ciccia A., Bredemeyer A. L., Schlabach M., Gygi S. P., Elledge S. J., Harper J. W. (2010) A genome-wide camptothecin sensitivity screen identifies a mammalian MMS22L-NFKBIL2 complex required for genomic stability. Mol. cell 40, 645–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wu J., Liu L. F. (1997) Processing of topoisomerase I cleavable complexes into DNA damage by transcription. Nucleic Acids Res. 25, 4181–4186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhang H., Wang J. C., Liu L. F. (1988) Involvement of DNA topoisomerase I in transcription of human ribosomal RNA genes. Proc. Natl. Acad. Sci. U.S.A. 85, 1060–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Durand-Dubief M., Persson J., Norman U., Hartsuiker E., Ekwall K. (2010) Topoisomerase I regulates open chromatin and controls gene expression in vivo. EMBO J. 29, 2126–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lyu Y. L., Wang J. C. (2003) Aberrant lamination in the cerebral cortex of mouse embryos lacking DNA topoisomerase IIβ. Proc. Natl. Acad. Sci. U.S.A. 100, 7123–7128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tsutsui K., Tsutsui K., Hosoya O., Sano K., Tokunaga A. (2001) Immunohistochemical analyses of DNA topoisomerase II isoforms in developing rat cerebellum. J. Comp. Neurol. 431, 228–239 [DOI] [PubMed] [Google Scholar]
- 66. Li L., Bhatia R. (2011) Stem cell quiescence. Clin. Cancer Res. 17, 4936–4941 [DOI] [PMC free article] [PubMed] [Google Scholar]