Abstract
Escherichia coli O157:H7 is a human pathogen that resides asymptomatically in its bovine host. The level of Shiga toxin (Stx) produced is variable in bovine-derived strains in contrast to human isolates that mostly produce high levels of Stx. To understand the genetic basis for varied Stx production, chronological collections of bovine isolates from Wisconsin dairy farms, R and X, were analyzed for multilocus prophage polymorphisms, stx2 subtypes, and the levels of stx2 transcript and toxin. The E. coli O157:H7 that persisted on both farms were phylogenetically distinct and yet produced little to no Stx2 due to gene deletions in Stx2c-encoding prophage (farm R) or insertional inactivation of stx2a by IS1203v (farm X). Loss of key regulatory and lysis genes in Stx2c-encoding prophage abolished stx2c transcription and induction of the prophage and stx2a::IS1203v in Stx2a-encoding prophage generated a truncated stx2a mRNA without affecting phage production. Stx2-producing strains were transiently present (farm R) and became Stx2 negative on farm X (i.e., stx2a::IS1203v). To our knowledge, this is the first study that details the evolution of E. coli O157:H7 and its Stx2-encoding prophage in a chronological collection of natural isolates. The data suggest the bovine and farm environments can be niches where Stx2-negative E. coli O157:H7 emerge and persist, which explains the Stx variability in bovine isolates and may be part of an evolutionary step toward becoming bovine specialists.
INTRODUCTION
Escherichia coli O157:H7 is a leading cause of hemorrhagic colitis and hemolytic-uremic syndrome (HUS) in humans (1–5), with Shiga toxin (Stx)-mediated cytotoxicity being central to pathogenesis. Friedrich et al. showed that the clinical outcome of infection depends on the stx genotype of the infecting strain (6). There are two major antigenic forms (Stx1 and Stx2) that share 56% amino acid identity (7). Purified Stx2 is more toxic than Stx1 in murine infections (8), and the administration of Stx2 to baboons leads to HUS, while comparable amounts of Stx1 do not (9). Clinical strains harbor Stx1 and/or Stx2 or variant forms of the toxins (Stx1c, Stx2, Stx2c, Stx2d, and Stx2e) in various combinations (6, 10), but isolates of E. coli O157:H7 from human cases of HUS are mostly linked to the presence of Stx2 (11).
In most Stx-producing E. coli, stxAB is present in lambdoid prophage and controlled by the phage regulatory system (12, 13). λ remains as a prophage due to cI repression (14), and induction occurs through RecA-mediated cI autocleavage following activation of the bacterial SOS response (15, 16). The early regulators, including the antiterminator Q, control transcription of the late promoter PR′ and the synthesis of polycistronic mRNA encoding for late-phage functions (morphogenesis and lysis). The location of stxAB, downstream of PR′ and upstream of lysis genes, is conserved among most of the sequenced Stx-encoding phages (17). As a result, transcription of stxAB is mainly regulated by the Q-dependent late promoter PR′ (18) and is coupled to the lytic replication of λ (13, 19).
Studies have shown that Stx phage display wide genetic and morphological variation and bacterial hosts of these prophage differ in the production of phage particles and toxin (20, 21). However, the principal integration sites of Stx phage are conserved, i.e., the yehV and wrbA loci for Stx1- and Stx2-encoding phage, respectively (22). Recently, Eppinger et al. (23) further defined that prototypical Stx2-encoding phages (termed BP-933W based on the nomenclature of E. coli O157:H7 strain EDL933) are found at the wrbA locus (or argW), while Stx2c-encoding phages are found next to sbcB (e.g., E. coli O157 strain EC4115).
E. coli O157:H7 has extensive genome heterogeneity among strains. Strain EDL933 contains 18 multigenic prophage regions (24). Ohnishi et al. performed a comparative analysis of O157 strains using whole-genome PCR scanning and determined that structural variation in the prophage regions is a major determinant in genome diversity (25). The presence of multiple lambdoid prophage in a genome with homologous genes facilitates interphage recombination and chromosomal rearrangements (26).
Genome-wide genotyping found that E. coli O157:H7 has diverged into three primary lineages: designated lineage I, lineage I/II, and lineage II (23, 27, 28). These three lineages differ in their association with human disease. Lineage I strains are commonly associated with human infection and have a bovine origin. Lineage I/II strains are predominantly isolated from humans and include the multistate “spinach” outbreak (28, 29). Lineage II strains are primarily bovine isolates from North America (27, 30). Each lineage has a specific combination of stx1 and/or stx2 genes or variant stx genes (23, 31): lineage 1 strains carry both stx1 and stx2 (i.e., EDL933 and Sakai strains), lineage I/II strains carry both stx2 and stx2c genes, and lineage II usually carries stx1 and stx2c genes (30, 31). Zhang et al. demonstrated that lineage I and I/II strains produce significantly more Stx2 than lineage II strains. Furthermore, among lineage I strains, a higher level of Stx2 was produced by isolates from human than from cattle (31), implying that genotypic and phenotypic properties of Shiga toxin production in bovine E. coli O157:H7 appears distinct from clinical isolates.
To further delineate the evolution and genetic variations in Shiga toxin production in bovine isolates, chronological collections of E. coli O157:H7 isolates from two Wisconsin dairy farms (32, 33) were analyzed. Genomic subtyping of isolates using XbaI restriction endonuclease digestion profiles (REDP) found 9 and 10 different O157 REDP groups on farms X and R, respectively. In the present study, 17 strains representing the different REDP groups from both farms were analyzed using a combination of prophage polymorphism-based typing and genetic characterization of Stx2 production. Each farm had a distinct phylogenetic history of E. coli O157:H7 but was commonly predominated by strains impaired in the production of Stx2. Stx2-negative strains resulted from sequential inactivation of Stx2 prophage over time, and these strains persisted in the bovine and farm environments.
MATERIALS AND METHODS
Bacterial strains and culture methods.
Seventeen bovine isolates representing the REDP types identified on farms R and X were selected for analysis (Table 1). Fifteen strains with different REDP were isolated from 1994 to 2001 on farm R (32–34), and 11 were included in this analysis. Likewise, 9 strains with unique REDP were isolated from farm X during 1994 to 1996, and 6 were selected for inclusion in the present study. E. coli O157:H7 EDL933 (ATCC 43895) and E. coli O157:H7 FRIK966 (ATCC BAA-1882) were included as reference strains. In addition, 62 isolates displaying REDP 9 from farm X (33) were analyzed for the presence of stx2::IS.
Table 1.
Polymorphic prophage patterns of E. coli O157:H7 isolates examined in this studya
| Phage | Target locus | EDL933 | PPP (strain, REDP) |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Farm R |
Farm X |
||||||||||||||||||
| 1994 |
1995-1996 |
1999 |
2000-2001 |
1994 |
1995-1996 |
||||||||||||||
| 783 25 | 920 1 | 944 3 | 1540 7 | 966 3 | 2417 30 | 2455 31 | 2467 32 | 2065 33 | 2069 34 | 2533 36 | 427 19 | 606 20 | 804 29 | 1275 9 | 1707 12 | 1625 10 | |||
| BP-933V | V1 | + | − | − | − | − | − | − | − | − | − | − | − | − | − | + | + | + | + |
| V2 | + | − | − | − | − | − | − | − | − | − | − | − | + | − | + | + | + | + | |
| CI | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| CII | + | + | + | + | + | + | − | + | + | + | + | + | + | + | + | + | + | + | |
| V3 | + | − | − | − | − | + | − | − | − | − | − | − | − | − | + | + | + | + | |
| stx1 | + | + | + | + | + | + | − | + | + | + | + | + | − | − | + | + | + | + | |
| V4 | + | + | + | + | + | + | − | − | − | − | − | − | + | + | + | + | + | + | |
| BP-933W | W1 | + | − | − | − | − | − | + | − | − | − | − | − | − | − | + | + | + | − |
| W2 | + | + | + | + | + | + | + | − | − | − | − | − | − | − | + | + | + | − | |
| CI | + | + | + | + | + | + | + | − | − | − | − | − | − | − | + | + | + | − | |
| CII | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | |
| stx2 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | |
| W4 | + | − | − | − | − | − | + | − | − | − | − | − | − | − | + | + | + | − | |
| W5 | + | − | − | − | − | − | + | − | − | − | − | − | − | − | + | + | + | − | |
| W6 | + | − | − | − | − | − | + | − | − | − | − | − | − | − | + | + | + | − | |
| CP-933O | O1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − |
| O2 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| O3 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | |
| O4 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | + | |
| O6 | + | − | − | − | − | − | + | − | − | − | − | − | + | + | + | − | − | − | |
| O7 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | |
| O8 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | |
| O9 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | |
| O16 | + | + | + | + | + | + | − | + | + | + | + | + | + | + | + | + | + | + | |
| O17 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | |
| CP-933R | R1 | + | − | − | − | − | − | + | − | − | − | − | − | − | − | + | + | − | + |
| R3 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| R4 | + | − | − | − | − | − | + | − | − | − | − | − | − | − | + | + | − | + | |
| R5 | + | − | − | − | − | − | + | − | − | − | − | − | − | − | + | + | − | + | |
| CP-933C | C1 | + | − | − | − | − | − | + | − | − | − | − | − | − | − | + | + | − | + |
| C2 | + | − | − | − | − | + | + | + | + | + | + | + | − | − | + | + | + | + | |
| C3 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| C4 | + | − | − | − | − | − | + | − | − | − | − | − | − | − | + | + | − | − | |
| C5 | + | − | − | − | − | − | + | − | − | − | − | − | − | − | + | + | − | + | |
| C6 | + | − | − | − | − | − | + | − | − | − | − | − | − | − | + | + | − | + | |
| CP-933N | N1 | + | − | − | − | − | − | + | − | − | − | − | − | − | − | + | + | + | + |
| N2 | + | − | − | − | − | − | + | − | − | − | − | − | − | − | + | + | + | + | |
| N3 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| N4 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| CP-933X | X1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| X2 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| CP-933P | P1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| P2 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| P3 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| CP-933M | M1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| M2 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| CP-933U | U1 | + | + | + | + | + | + | + | − | − | − | − | − | + | + | + | + | + | + |
| U2 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
To determine the polymorphic prophage pattern (PPP), a PCR product of the same size as the reference strain, EDL933, was scored as “+” (present, conserved). Positive results are highlighted for emphasis. –, negative PCR result or different product size. Values are grouped by farm, collection year(s), and “strain, REDP.” Strains are identified by the Food Research Institute-Kaspar (FRIK) culture collection number. The restriction enzyme digestion profile (REDP) was determined by pulsed-field gel electrophoresis (32–34).
Cultures were grown at 37°C in Luria-Bertani (LB) broth, with shaking at 180 rpm, or on LB agar. Unless noted, the strains were initially grown overnight in 2 ml of LB broth, transferred to 5 to 10 ml of LB broth in a 250-ml flask, and incubated to the desired optical density at 600 nm (OD600).
In silico analysis of prophage regions and primer design.
Comparative genomic analysis of EDL933 and other sequenced O157 strains (RefSeq [National Center for Biotechnology Information]) was performed using a synteny map interface in the MaGe genome browser, which schematically aligns local regions of the multiple O157 genomes. Specifically, synteny maps of 10 lambdoid prophage regions, including BP-933V, BP-933W, CP-933O, CP-933R, CP-933C, CP-933N, CP-933X, CP-933P, CP-933M, and CP-933U (24) were generated, and each prophage region was inspected to identify loci or genes that varied between the different O157 genomes. Based on the EDL933 chromosomal sequence, 48 pairs of PCR primers were designed to amplify nonoverlapping segments or genes within the identified variable regions. For prophage that displayed higher variability among genomes (CP-933O, CP-933R, and CP-933C), primer pairs amplifying longer chromosomal segments (3 to 5 kb) were designed to permit the detection of multiple genetic changes. For the remaining prophages, primer pairs targeted a single gene. The 48 pairs of primers are listed in Table S1 in the supplemental material.
PCR analysis of prophage polymorphism.
PCR was carried out using an AccuPrime Pfx SuperMix (Invitrogen, Grand Island, NY) for the amplification of 3- to 5-kb target sequences and BioReady rTaq DNA polymerase (Bulldog Bio, Inc., Portsmouth, NH) for shorter amplicons according to the manufacturer's protocols. PCR conditions were 5 min at 94°C and 30 cycles of 94°C for 30 s, 54°C for 30 s, and 72°C for 1 to 5 min, depending on the amplicon sizes, followed by a 5-min extension at 72°C. Portions (50 ng) of genomic DNA (gDNA) from overnight cultures were used as a template in each PCR. In EDL933, all primer pairs yielded amplicons of the expected sizes when analyzed by gel electrophoresis. For 3- to 5-kb amplicons, additional confirmation of binding of the primer pairs to the correct target sequence was done by analyzing restriction digestion profiles of the PCR products.
The presence or absence of DNA target sequences in strains was determined by comparing the amplified PCR product with the amplicon size obtained with EDL933 DNA. Strains that yielded PCR products of identical size to those obtained with EDL933 were considered conserved. Strains with DNA target sequences that generated no PCR product or one with a different size than EDL933 were considered to have modified targets, such as a deletion or sequence divergence. For 3- to 5-kb amplicons, a restriction enzyme that cut each PCR product once or twice was chosen by sequence analysis and used to confirm PCR products. All PCR-negative reactions were confirmed by repeating the assay.
Sequence analysis of stx2.
The nucleotides stx2-F1 (5′-GGG TCT GGT GCT GAT TAC TTC) and stx2-R1 (5′-GTT ACC CAC ATA CCA CGA ATC AG) were used to amplify a 1,314-bp fragment of the entire stxAB2 sequence in EDL933 (Fig. 1A). stx2-F1 anneals at a 42- to 23-bp upstream region of the ATG start codon of the stxA2 gene. stx2-R1 anneals at a 9- to 31-bp downstream region of TGA stop codon of stxB2. For shorter sequencing templates, PCR with an alternative primer pair, stx2-F1 and stx2-R3 (5′-AAC ATC TTC TTC ATG CTT AAC TCC) was used to amplify a 1,027-bp fragment 5′ proximal to stxAB2. Similarly, a primer pair of stx2-F3 (5′-CAC TCA CTG GTT TCA TCA TAT CTG) and stx2-R1 was used to obtain an 812-bp fragment 3′ proximal to stxAB2. PCR template was prepared from gDNA extract derived from each strain. KOD Hot Start DNA polymerase (Novagen, Darmstadt, Germany) was used according to the manufacturer's protocol. The purified PCR products were directly sequenced in both directions.
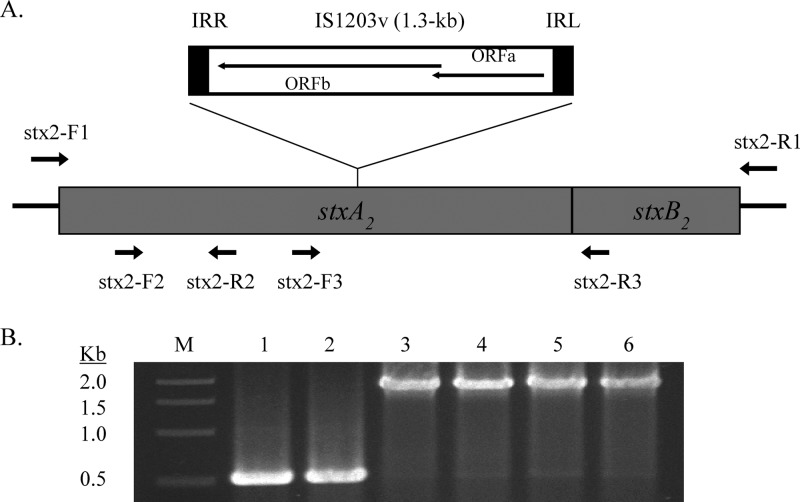
Fig 1.
(A) Map of IS1203v insertion in stxAB2. The stx2-F2, stx2-F3, stx2-R2, and stx2-R3 primers were used in various combinations for PCRs to map the integration site and size of IS1203v. The stx2-F1 and stx2-R1 primers were used for sequencing. IRR and IRL represent inverted repeats flaking IS1203v. (B) PCR screening of 1.3-kb IS1203v insertion in strains. Lanes: M, 1-kb marker (Promega); 1, EDL933; 2, FRIK804; 3, FRIK1275; 4, FRIK1707; 5 and 6, two strains with REDP 9. stx2-F3 and stx2-R3 were used as primers.
PCR analysis for the integration site of Stx2-encoding phages.
A primer pair of wrbA-R (5′-CCA GTA CCG GTG GAA CTA AAG AC-3′) and intstx2a-F (5′-GGT ATT AAC TAC GCG GTA AGC TCC-3′) amplifying the 646-bp region across wrbA and the adjacent integrase gene (int) in EDL933 was used to determine whether the integration site of the Stx2a phage in the farm strains was conserved. A primer pair of sbcB-R (5′-GAC AAT CTC TTC CGC GTA CTG-3′) and intstx2c-R (5′-GTC TGG CAT CCT TCA AAG ACA G-3′) amplifying the 1,805-bp region across sbcB and int of the Stx2c phage in EC4115 was used to determine the integration site of Stx2c phages. Genomic DNA from the farm strains was analyzed by multiplex PCR using both primer pairs (5 μmol each per reaction). The following PCR conditions were used: 5 min at 94°C and then 23 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min, followed by a 5-min extension at 72°C. Amplicons of the wrbA-int and sbcB-int were analyzed by 1% agarose gel electrophoresis.
PCR screening for stx2::IS1203v.
Cultures were grown on LB agar plates overnight at 37°C, and colonies were used as a PCR template. The PCR conditions included 5 min at 94°C and 30 cycles of 94°C for 30 s, 54°C for 30 s, and 72°C for 3 min, followed by a 5-min extension at 72°C. stx2-F3 and stx-R3 (Fig. 1A) were used to screen for the 1.3-kb IS1203v insertion. The size difference between 523 and 1,823 bp (with IS insertion) was analyzed by 1% agarose gel electrophoresis.
RNA extraction.
To analyze the level of stx2 transcript, an overnight culture was diluted 100-fold (OD600 of ∼0.04) and grown to an OD600 of 0.5 to 0.7. At this point, mitomycin C (MMC) was added to the culture at a concentration of 1 μg/ml. Cells from MMC-treated cultures were harvested after 2 h of incubation, while untreated control cells were collected at the same time, pelleted by centrifugation, and stored at −20°C. Total RNA was extracted with TRI Reagent (Sigma-Aldrich, St. Louis, MO) according to the manufacturer's protocol. The final RNA pellet was resuspended in 100 μl of nuclease-free water. The RNA concentration was determined by measuring the OD260. Triple measurements were performed for each RNA sample, and an average value of the concentration was calculated. RNA was digested with RQ1 RNase-free DNase reagent (Promega, Madison, WI) according to the manufacturer's instructions and tested for possible DNA contamination by a control PCR prior to use in reverse transcriptase PCR (RT-PCR). Three independent RNA extracts were analyzed by RT-PCR.
RT-PCR.
Semiquantitative RT-PCR was performed using the Access Quick RT-PCR system (Promega). The RT-PCR (25 μl) contained the following components: 300 μg of DNA-free RNA, 10 pmol of forward and reverse primers, and 0.5 U of RT. cDNA synthesis was conducted at 52°C for 45 min. The following cycle conditions were used for the subsequent PCR: 30 s at 94°C, 30 s at 54°C, and 60 s at 72°C for extension. The annealing temperature and number of cycles varied depending on the primers used and the level of target RNA to ensure that the comparisons were performed in the linear range of the amplification; 23 cycles for stx2 and 18 cycles for 16S rRNA. 16S rRNA was used as an internal control to confirm that equal amounts of RNA were used in reactions. Portions (10 μl) of each RT-PCR product were analyzed by 1.5% agarose gel electrophoresis, and bands were visualized after staining with ethidium bromide. Two different pairs of primers were used to monitor stx2 expression (see Fig. 1A). The primers stx2-F3 (5′-CAC TCA CTG GTT TCA TCA TAT CTG) and stx2-R3 (5′-AAC ATC TTC TTC ATG CTT AAC TCC) were used to amplify the 461- to 985-bp region of stxAB2, which extends to the first 14 bp of the B subunit gene. The primer pair stx2-F2 (5′-ACC GGG CAG TTA TTT TGC TGT GGA) and stx2-R2 (5′-GAA AGT ATT TGT TGC CGT ATT AAC GA) amplified the 201- to 333-bp region of stxA2 in EDL933 and was used for screening truncated stx2 mRNAs due to an IS insertion (see below). The oligonucleotides used to amplify 16S rRNA were 16S-F (5′-ATA CCT TTG CTC ATT GAC GTT ACC) and 16S-R (5′-CCA GTC ATG AAT CAC AAA GTG GTA AG).
Western blot analysis for Stx2.
Protein from cell-free culture supernatants were precipitated by adding a 4× volume of 10% trichloroacetic acid in acetone containing 0.07% β-mercaptoethanol, followed by incubation for 1 to 2 h at −20°C. Protein precipitate was pelleted at 13,000 × g (4°C) and washed with cold acetone. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and subsequently transferred to a polyvinylidene difluoride membrane (0.2-mm pores; Bio-Rad, Hercules, CA) in transfer buffer (25 mM Tris base, 192 mM glycine, 20% [vol/vol] methanol). Membranes were blocked overnight in Tris-buffered saline–Tween 20 (TBST; 150 mM NaCl, 2.7 mM KCl, 25 mM Tris base, 0.05% [vol/vol], Tween 20; pH 7.4) supplemented with 5% (wt/vol) dried skim milk (Difco, Detroit, MI) at 4°C. Membranes were incubated with mouse anti-Stx2 monoclonal antibody (ATCC CRL-1907) in TBST and subsequently incubated with a secondary antibody conjugated to alkaline phosphatase (Promega). The Stx2-antibody complex was visualized with a solution of 0.165 mg of BCIP (5-bromo-4-chloro-3-indolylphosphate) p-toluidine salt (Promega)/ml and 0.33 mg of nitroblue tetrazolium chloride (Promega)/ml in alkaline phosphatase buffer (100 mM Tris-Cl [pH 9.5], 100 mM NaCl, 5 mM MgCl2). DnaK was used as a control to ensure equal loading of protein between samples and was detected by using a monoclonal antibody (Enzo Life Sciences, Farmingdale, NY).
Prophage induction.
Overnight cultures were diluted 1:100 (final OD600 of ∼0.04) and grown to an OD600 of 0.5 to 0.7, and MMC was added to a final concentration of 1 μg/ml. After 18 h of incubation, unlysed cells and debris were removed by centrifugation (6,000 × g, 15 min, 4°C). Cell-free supernatants from uninduced cells were collected as a control. Supernatants were filtered through 0.22-μm-pore-size membrane filter (Millipore, Billerica, MA) and stored at 4°C. The cell-free supernatant was further processed for Stx detection and phage purification as described below.
Phage purification.
Phage particles induced by MMC were precipitated by the addition of 20% polyethylene glycol 8000 (PEG) and 10% NaCl to the cell-free culture supernatants prepared as described above for prophage induction. The mixture was incubated at 4°C overnight and then centrifuged at 10,000 × g for 1 h to pellet the phage particles. The pellets were suspended in SM buffer (0.58% NaCl, 0.25 MgSO4·7H2O, 1 M Tris-Cl [pH 7.5], 0.01% gelatin) (35) and stored at 4°C.
SDS-PAGE and protein band identification.
Phage particles were solubilized in 4× SDS loading buffer, boiled for 5 min, and applied to an SDS–4 to 12% polyacrylamide gel (Invitrogen). The protein bands were visualized by staining with Coomassie blue. To identify protein bands, the band of interest was excised from the gel and digested in gel (http://www.biotech.wisc.edu/facilities/massspec/). The molecular masses of the protein fragments were determined by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS). The protein band was identified by BLAST using the EDL933 genome (http://blast.jcvi.org/cmr-blast/). In cases with multiple homologs in the EDL933 genome, a manual differentiation based on single amino acid polymorphisms was used.
Electron microscopy.
Phage particles collected by PEG-NaCl precipitation were suspended in SM buffer, and a 5-μl drop of the phage suspension was placed on copper grids with carbon-coated Formvar films and negatively stained with 0.8% phosphotungstate (36, 37). Samples were examined at ×30,000 to ×150,000 magnifications using a transmission electron microscope (Hitachi H-600) operated at 75 kV.
RESULTS
Lineage and prevalence of E. coli O157:H7 isolates.
The separation of E. coli O157:H7 strains into different lineages was based upon octamer-based genome scanning (27). Clades of human outbreak strains (lineage I [LI]) and bovine isolates (lineage II [LII]) were identified. In the present study, strains EDL933 and FRIK966 were used as reference LI and LII strains, respectively.
Prophage polymorphisms are effective targets when combined with an open-array PCR platform for subtyping E. coli O157:H7 (38). Using a similar approach, 10 prophage regions that vary in O157 genomes were used to design a set of 48 prophage gene markers to characterize the polymorphic prophage pattern(s) (PPP) of E. coli O157:H7. The PPP of representative strains with different REDP from two Wisconsin farms are shown in Table 1. Farm R strains isolated over an 8-year period had relatively conserved PPP with >91% similarity. These results are consistent with reported similarities in REDP (86 to 100%) among these strains (32–34). Based upon their similarities with the PPP of FRIK966, these strains were assigned to LII. Farm R LII strains have genetic alterations in several prophage regions. In particular, BP-933V and BP-933W loci displayed significant divergence, while maintaining stx1 and stx2 (Table 1). This lineage of strains persisted and remained endemic on farm R for an 8-year period despite the sporadic introduction or appearance of different strains (i.e., FRIK2417). FRIK2417 appears to be distantly related to the LII strains based on its PPP, including negative PCR results for CII, Stx1, and V4 in the phage BP-933V. This strain was transiently present in 1999 but was not isolated in subsequent years (34).
The PPP of E. coli O157:H7 isolates from cattle on farm X were more diverse (Table 1). FRIK427 and FRIK606 represent REDP groups isolated in 1994 (32). These strains exhibited PPP similar to LII from farm R but lacked stx1. Later in the same year, strains exhibiting REDP29 (i.e., FRIK804) dominated (100% of isolates recovered) (32). FRIK804 had a PPP identical to EDL933 (Table 1); thus, it is likely an LI strain. However, E. coli O157:H7 strains exhibiting REDP29 were not isolated in subsequent years, but isolates with different REDP were recovered (i.e., FRIK1275, FRIK1707, and FRIK1625) (33). The REDP of FRIK804, FRIK1275, and FRIK1625 were highly related (33). In addition, the PPP of strains FRIK804, FRIK1275, and FRIK1707 were similar, especially in BP-933W and BP-933V, which suggests that FRIK1275, FRIK1707, and FRIK1625 are likely variants of FRIK804. Furthermore, FRIK1275 and FRIK1707 contained a transposon insertion in stx2 and FRIK1625 lacked BP-933W, including stx2 (described in detail below). Collectively, E. coli O157:H7 from two farms had distinct genotypic histories; LII strains dominated on farm R for 8 years, while farm X featured the introduction or appearance of new strains, as well as genetic turnover from LII to LI.
stx2 subtyping.
Studies have shown that typical LI strains carry canonical stx2 (stx2a for clarity), LII strains have stx2c, and LI/II strains primarily carry both stx2c and stx2a (23, 31). To confirm the lineage assigned by PPP, the stx2 subtype was determined. The entire coding region of stxAB2 in each strain was sequenced. The sequences of stx2 of EDL933 (NP_286976.1) and FRIK966 (ZP_05951333.1) from the NCBI database were used as reference sequences for stx2a and stx2c, respectively. The predicted LI strains—FRIK2417, FRIK804, FRIK1275, and FRIK1707—carried stx2a, while all LII strains from farms R and X carried stx2c (100% nucleotide identity), which confirmed the PPP-based lineage prediction.
stx2 inactivation by IS insertion in LI strains from farm X.
Sequence analyses found that stx2 in FRIK1275 and FRIK1707 from farm X was disrupted by a 1.3-kb IS element (stx2a::IS) (Fig. 1A). The 1.3-kb IS was located 639 bp downstream from the ATG start codon. The nucleotide sequences of the IS showed 100% homology with IS1203v open reading frames a and b (GenBank accession no. GU983682.2).
REDP 9 strains represented by FRIK1275 constituted 97% of the O157:H7 isolates from that study period (33). All 62 REDP 9 isolates analyzed by PCR contained stx2a::IS1203v (Fig. 1B), demonstrating the persistence of this stx2 genotype on farm X.
Gene loss in Stx2c-encoding prophage in LII strains.
PPP indicated that the LII strains significantly vary in their phage gene content and sequence, especially in the Stx prophage in comparison to EDL933 (Table 1). We tested whether the Stx2c prophage present in LII farm strains were related to the prototypical Stx2c phage found in E. coli O157:H7 EC4115 (23). This phage was shown to be distinctive from BP-933W in its gene content and organization, as well as its chromosomal integration site (sbcB). Thus, the presence of the sbcB-int junction characteristic of Stx2c phage integration was initially determined (Fig. 2). The wrbA-int junction indicative of Stx2a phage integration was also analyzed by multiplex PCR (Fig. 2). The data clearly showed that LI strains contain the wrbA-int junction, while LII strains possess the sbcB-int junction. Subsequent in silico analysis revealed that the draft genome of FRIK966 contained about 50 putative orthologues of the 72 Stx2c phage genes found in the genome of EC4115 (D. Park et al., unpublished data). At least half of the orthologues, including integrase and Q genes, displayed 100% nucleotide identity with the EC4115 counterparts. A majority of the missing genes were related to phage lysis, replication, and recombination. Together, these results suggest that the Stx2c phage from LII strains and a prototypical lineage I/II strain EC4115 (23) are related to each other but considerable genetic changes have occurred within Stx2c prophage in LII bovine strains, resulting in functional inactivation of the phages.
Fig 2.
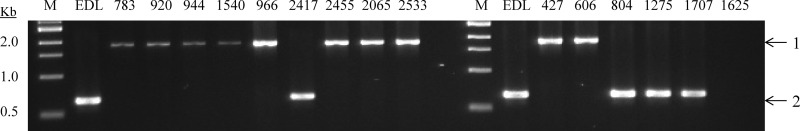
PCR analysis of the integration site of Stx2-encoding phages. Genomic DNA purified from FRIK strains was analyzed by multiplex PCR. A primer pair, intstx2c-R and sbcB-R, was designed to amplify the 1,805-bp region (band 1) across int in a Stx2c-encoding prophage and chromosomal sbcB. Another primer pair, intstx2a-F and wrbA-R, amplified the 646-bp region (band 2) across int in a Stx2a-encoding prophage and chromosomal wrbA. Lanes: M, 1-kb marker (Promega); EDL, 783, 920, etc., FRIK strains.
The farm R lineage II strains contained PPP similar to FRIK966, and thus, similar defective Stx prophage. Subsequent isolates had three additional deletions in PPP regions—BP-933V (in the V4 region) and BP-933W (in the W2 and CI regions)—indicating that modification in the Stx prophage continued at a detectable rate.
Stx2 expression.
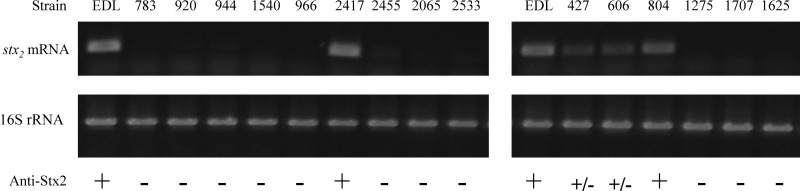
The transcription of stx2 is controlled by the Q-dependent late promoter PR′, and both production and release of the toxin rely upon phage induction and host cell lysis (19, 39). To determine whether the different stx2 genotypes and gene loss in Stx2c prophage affected the level of stx2 transcription, semiquantitative RT-PCR was performed with total RNA recovered after MMC induction. stx2-specific primers were designed for the region conserved in both stx2a and stx2c. stx2-F3/R3 primers (Fig. 1A) were used to detect stxAB2 transcripts. RT-PCR showed that a majority of the farm strains were impaired in stx2 expression; high levels of stx2 mRNA (comparable to that of EDL933) were detected only in FRIK2417 and FRIK804, which encoded for stx2a (Fig. 3). FRIK1275 and FRIK1707, with stx2a::IS1203v, did not produce stxAB2 mRNA, but the presence of a truncated stx2 transcript, upstream of the IS insertion site, was detected by RT-PCR using the stx2-F2 and stx2-R2 primers (Fig. 1A). The levels of truncated stx2 transcript in these strains were comparable to the levels of stx2 mRNA produced by EDL933 and FRIK804 (data not shown). This indicated that the IS insertion did not alter transcription initiation of stx2 but generated truncated transcripts.
Fig 3.
RT-PCR and Western blot analysis of stx2 expression. stx2 mRNAs of EDL933 are shown in the first lanes of each panel. Total RNA was extracted from a log-phase culture incubated for 2 h after mitomycin C addition. For each RT-PCR, 300 ng of total RNA was used to make cDNA, which was subsequently amplified by PCR. The expression of stx2 mRNA was analyzed with stx2-F3 and R3 primer set (see Fig. 1A). 16S rRNA was used as an internal control for total mRNA amount used in each RT-PCR. The presence or absence of Stx2 signal determined by Western blotting are indicated as “+” (“+/−” for basal level production) and “−”, respectively. The experiments were repeated at least three times with similar results.
Downregulation of stx2 expression in FRIK966 was identified previously by microarray analysis (40). Consistently, basal levels of stx2 mRNA were detected in all farm R lineage II strains (Fig. 3). In addition, the two lineage II strains from farm X, FRIK427 and FRIK606, expressed reduced amounts of stx2c. These results demonstrated that stx2c transcription was impaired in lineage II strains and was likely a consequence of gene loss in the Stx2c prophage.
The levels of Stx in cell lysates measured by Western blotting correlated with the transcript levels of stx2 in each strain (Fig. 3). As expected, strains that did not transcribe stx2 did not produce toxin (Fig. 3). Consistent with the level of stx2 transcript, Stx2 was produced by FRIK2417 and FRIK804 at higher levels than FRIK427 and FRIK606. These analyses showed that a majority of the farm strains were impaired in Stx2 production and that IS-mediated inactivation of stx2 (i.e., FRIK1275 and FRIK1707) and gene deletions in Stx2 prophage (i.e., lineage II strains from both farms) were two mechanisms of abolishing or reducing Stx2 production.
Stx2 phage production.
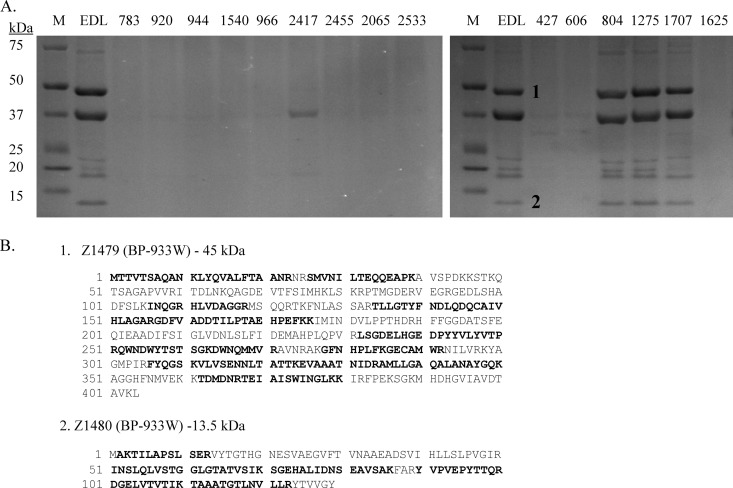
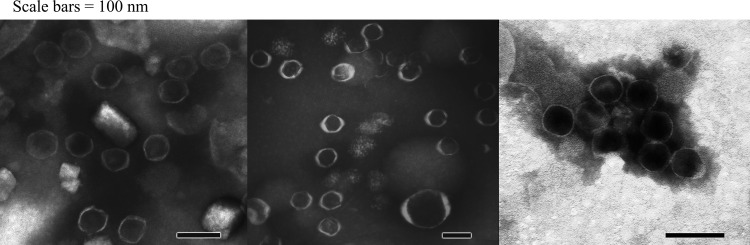
The effects of stx2a::IS1203v and phage gene deletions on phage production were examined. Purified phage particles were analyzed by both SDS-PAGE and transmission electron microscopy (TEM). Phage production was prominent only in EDL933 and strains FRIK804, FRIK1275, and FRIK1707, all of which belonged to LI with identical PPP in BP-933V and BP-933W except for stx2a::IS1203v in FRIK1275 and FRIK1707 (Table 1 and Fig. 1). SDS-PAGE analysis detected ∼10 different phage proteins that were induced in EDL933 and strains FRIK804, FRIK1275, and FRIK1707 but absent in other strains (Fig. 4A). MALDI-TOF analysis identified two prominent bands (1 and 2) present in the four strains examined as 45- and 13.5-kDa proteins encoded by Z1479 and Z1480 of BP-933W, respectively (Fig. 4). TEM showed a single phage produced by EDL933 with a hexagonal head outline approximately 50 to 60 nm in diameter but no tail, a typical morphology of Stx2 encoding phage (35). The phage produced by strains FRIK804, FRIK1275, and FRIK1707 were morphologically identical to the Stx2 phage from EDL933 (Fig. 5). The fact that FRIK1275 and FRIK1707 with stx2::IS1203v produced Stx2 phage indicates that the IS insertion affected stx2 transcription but not downstream phage lysis genes. The farm R LII strains did not produce detectable phage particles, confirming that gene loss within their Stx2 prophage impaired the production of phage. FRIK1625, which lacks Stx2 prophage, did not produce phage particles. A few BP-933W-like phage were detected in PEG samples from strains FRIK2417, FRIK427, and FRIK606 (not shown) but were not investigated. Together, the TEM data indicated that Stx2a prophage with stx2a::IS1203v was inducible, whereas gene deletion in the Stx2c prophage abolished phage production.
Fig 4.
SDS-PAGE and MALDI-TOF analysis of phage proteins. Phage particles were obtained by PEG precipitation of cell lysate from mitomycin C-treated culture. (A) SDS-PAGE of Coomassie blue-stained phage proteins. Protein bands that were identified by MALDI-TOF are labeled as “1” and “2.” (B) Genes encoding the sequenced protein. Z1479 and Z1480 are encoded in BP-933W. Matched peptides are shown in boldface. The 37-kDa protein band was identified as OmpC, a major outer membrane protein (derived from outer membrane copurified with phage particles).
Fig 5.
TEM of Stx2a phage particles. The phage from strains FRIK804 and EDL933 were similar, as predicted from the genotype data. Also, the phage from strain FRIK1275 harboring stx2a::IS1203v was similar morphologically to the wild-type Stx2a phage (strains EDL933 and FRIK804). Phage particles were obtained by PEG precipitation of cell lysate from mitomycin C-treated cultures.
In summary, two distinct genetic defects were identified in prophage within persistent bovine isolates of E. coli O157 examined in the present study, stx2a::IS1203v and loss of Stx2c prophage gene(s), both of which impaired Stx production. These findings demonstrate that Stx production of isolates cannot be predicted solely on the presence of stx but must be determined by phenotypic analyses. In addition, Stx2 phage lysis is not always associated with toxin release as shown in the E. coli O157:H7 isolates with stx2a::IS1203v.
DISCUSSION
Human infection by E. coli O157:H7 varies in clinical manifestations and severity depending upon host factors and the genetic makeup of the bacterium. The phylogeny of E. coli O157:H7 is comprised of three lineages (I, II, and I/II) with lineage-specific virulence signatures, such as stx subtypes (23, 27, 28, 30). Strains of bovine and human origin differ in their genomic makeup (23, 27, 40), expression of virulence genes (41), and pathology in animal models (42). A complete understanding of the genetic stability and expression of virulence factors (e.g., Stx expression) in E. coli O157:H7 in different hosts and environments is needed to predict the virulence potential and emergence of new variants of pathogenic E. coli.
This study defined the changes in prophage and Stx production within a chronological collection of E. coli O157:H7 isolates from two Wisconsin farms, which is summarized in Fig. 6 and Table S2 in the supplemental material. Each farm had a distinct phylogenetic history, but both were populated by strains impaired in the production of Stx2. The data indicated that Stx-negative strains resulted from sequential deletion or inactivation of stx or prophage genes and that these strains persisted despite the presence of strains with fully intact and functional stx2.
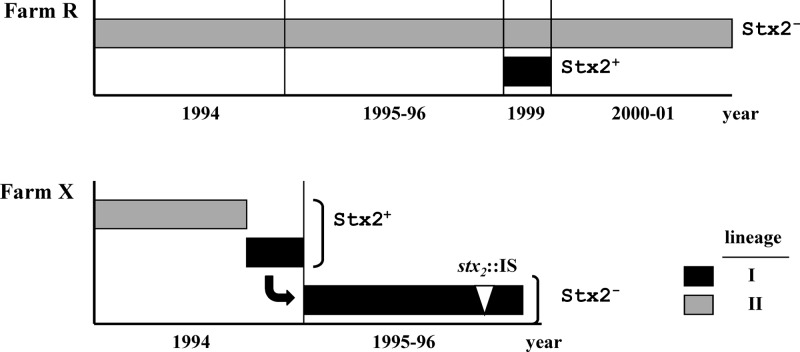
Fig 6.
Genetic dynamics of E. coli O157:H7 on farms R and X. Both farms had dominant lineage II strains until new Stx2-positive lineage I strains emerged in 1999 on farm R and 1994 on farm X, respectively. The Stx2-positive strain on farm R disappeared and on farm X was modified (stx2::IS genotype) during the following years. This model highlights how Stx2-negative E. coli O157:H7 became prevalent on the farms over time at the strain and genetic levels.
Strain EDL933 is a prototypical Shiga toxigenic E. coli O157 strain that produces Stx2 phage particles and Stx2. As shown in Table S2 in the supplemental material, FRIK804 was the only strain isolated with genotypic and phenotypic traits of Stx identical to EDL933 of the 17 REDP O157:H7 groups isolated from these farms. FRIK804 was transiently present on farm X but replaced by Stx2-negative strains from other REDP groups. Overall, there was a trend among sequential isolates for the loss of stx expression and this genetic bias may represent the evolutionary path of E. coli O157:H7 in bovine and farm environments.
Lineage II (LII) strains are over-represented by bovine isolates (23, 27). Isolates collected over an 8-year period from farm R belonged to LII (10 of the 11 O157:H7 REDP groups) with the exception of strain FRIK2417 (Fig. 6 and see Table S2 in the supplemental material). Stx genotypes of the LII strains were distinct from those of the LI and carried stx2c and defective Stx2c prophage. Gene deletions in Stx2c prophage abolished phage induction and reduced transcription and production of Stx2. LII strains persisted on farm R for at least 8 years (33, 34). In contrast, strain FRIK2417 produced Stx and was present on farm R for less than 1 year. These results indicated that the persistence of E. coli O157:H7 in cattle was independent of Stx production. In addition, the loss of two additional Stx2 prophage regions (CI and W2) was observed in farm R LII strains isolated during the later years of the strain collection (Table 1). Given the genome-wide diversity in LII strains (23, 40), the long-term persistence of LII strains in cattle may indicate evolution toward “bovine specialists” and the loss of Stx prophage may be a part of this process.
Similar to the findings with farm R strains, strains defective in Stx2 production predominated on farm X; however, the underlying genetic basis in the Stx2-negative strains and their emergence differed from farm R strains (Fig. 6 and see Table S2 in the supplemental material). Strains 427 and 606 producing Stx2 at low levels (presumably due to defects in Stx2c prophage) were present initially on the farm and were replaced by an Stx2-producing strain (i.e., FRIK804). Strain 804 had a PPP identical to the reference LI strain EDL933 and produced high levels of Stx2 and the Stx2 phage; however, this strain was transiently present (<6 months) and was replaced by Stx2-negative strains during the following years. FRIK1275 (REDP 9), which contained an IS insertion in stx2, comprised 97% of the O157:H7 isolates during this period (33). FRIK804 and FRIK1275 have common features, including an inducible BP-933W (Fig. 4 and 5) and similar REDP (33). Thus, it seems likely that REDP 9 strains evolved from FRIK804. The emergence and dissemination of the strain with stx2::IS suggested there was selection for strains impaired in Stx2 production. Based on the similarities in REDP (33) and genome-wide IS insertion profiles (E. Stanton et al., unpublished data), FRIK1625 is likely another variant of FRIK804 that is stx2 negative, but in this case, because of the nearly complete deletion of BP-933W genes (Table 1). Collectively, these data indicated that the Stx2 prophage were deleted or that stx2 was inactivated during their presence in cattle and farm environments. Such genetic changes may contribute to long-term fitness of E. coli O157:H7 within its bovine host.
The first viable Stx2c-encoding prophage found in E. coli O157:NM CB2851, a human fecal isolate (43) was phage 2851. It differs from the prototypical Stx2a-encoding phages BP-933W in its chromosomal integration site, phage morphology, and superinfection immunity (43, 44). The Stx2c prophage found in lineage I/II strains (i.e., EC4115) have gene content and architecture similar to phage 2851 (23). In silico sequence comparison of the Stx2c prophage from EC4115 and the prophage in FRIK966 revealed 100% nucleotide identities in key genes encoding for Stx2c, Q antiterminator, and integrase despite other missing genes in the Stx2c prophage from FRIK966. Using PCR screening methods, we confirmed that the Stx2c prophages in the lineage II strains from both farms are located in the sbcB locus, the preferential integration site of Stx2c phages (23, 43). These findings indicated that the defective Stx2c prophage present in lineage II strains may have resulted from the acquisition of a 2851-like Stx2 phage, followed by the loss of phage genes.
Stx-mediated systemic vascular damage in receptor-bearing, endothelial cells is linked to HUS in humans. The preferred receptor for Stx is globotriaosylceramide (Gb3) present on the kidney and brain cells of humans. The tolerance of calves and adult cattle to E. coli O157:H7 infection is partially due to the lack of Gb3 (45). Pathology in infected neonatal calves requires the locus enterocyte effacement (LEE) and is independent of stx2 carriage (46). Furthermore, colonization of the terminal rectal mucosa requires intimin, Tir, and pO157, but not Stx2 (47). The lack of selective pressure for Stx in cattle may result in stx loss during long-term persistence in cattle and farm environments as demonstrated by LII strains from farm R. Alternatively, the absence of Stx may be favorable for dissemination in cattle as observed on farm X, where stx2 was inactivated by IS insertion. Stx causes immunosuppression (48) and enhances LEE activity in bovines (49), which may not be optimal to this asymptomatic host-microbe interaction.
To our knowledge, this is the first study that details the evolution of E. coli O157:H7 and Stx encoding prophage in a natural host and an environmental setting (i.e., a dairy herd). Although Stx2 is involved in human pathogenesis, Stx2 appears dispensable for the long-term persistence of E. coli O157:H7 in the bovine and farm environments. Genetic events (e.g., deletion and IS insertion) responsible for inactivation of Stx2 phage were elucidated that explain the variability in Stx production by bovine E. coli O157:H7. The emergence of Stx2-negative E. coli O157:H7 may be part of an evolutionary step toward commensalism in bovines. Additional investigations of sequential isolates of E. coli O157:H7 from cattle and other sources will determine whether the evolutionary pattern of prophage and stx inactivation is a general or bovine-specific process.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Heather A. Owen for providing technical expertise with the electron microscopy and to Nydia Morales-Soto for sharing her knowledge of bacteriophage. We also thank Helen Piontkivska for supportive distant tree analysis.
This study was supported by the National Science Foundation grant NSF-EF-091342.
Footnotes
Published ahead of print 28 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03158-12.
REFERENCES
- 1. Caprioli A, Luzzi I, Rosmini F, Pasquini P, Cirrincione R, Gianviti A, Matteucci MC, Rizzoni G. 1992. Hemolytic-uremic syndrome and Vero cytotoxin-producing Escherichia coli infection in Italy. J. Infect. Dis. 166:154–158 [DOI] [PubMed] [Google Scholar]
- 2. Griffin PM, Tauxe RV. 1991. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic-uremic syndrome. Epidemiol. Rev. 13:60–98 [DOI] [PubMed] [Google Scholar]
- 3. Karmali MA. 2004. Infection by Shiga toxin-producing Escherichia coli: an overview. Mol. Biotechnol. 26:117–122 [DOI] [PubMed] [Google Scholar]
- 4. Riley LW, Remis RS, Helgerson SD, McGee HB, Wells JG, Davis BR, Hebert RJ, Olcott ES, Johnson LM, Hargrett NT, Blake PA, Cohen ML. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308:681–685 [DOI] [PubMed] [Google Scholar]
- 5. Rowe PC, Orrbine E, Lior H, Wells GA, McLaine PN. 1993. A prospective study of exposure to verotoxin-producing Escherichia coli among Canadian children with haemolytic-uraemic syndrome. Epidemiol. Infect. 110:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Friedrich AW, Bielaszewska M, Zhang WL, Pulz M, Kuczius T, Ammon A, Karch H. 2002. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185:74–84 [DOI] [PubMed] [Google Scholar]
- 7. Strockbine NA, Marques LR, Newland JW, Smith HW, Holmes RK, O'Brien AD. 1986. Two toxin-converting phages from Escherichia coli O157:H7 strain 933 encode antigenically distinct toxins with similar biologic activities. Infect. Immun. 53:135–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tesh VL, Burris JA, Owens JW, Gordon VM, Wadolkowski EA, O'Brien AD, Samuel JE. 1993. Comparison of the relative toxicities of Shiga-like toxins type I and type II for mice. Infect. Immun. 61:3392–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Siegler RL, Obrig TG, Pysher TJ, Tesh VL, Denkers ND, Taylor FB. 2003. Response to Shiga toxin 1 and 2 in a baboon model of hemolytic-uremic syndrome. Pediatr. Nephrol. 18:92–96 [DOI] [PubMed] [Google Scholar]
- 10. Zhang W, Bielaszewska M, Kuczius T, Karch H. 2002. Identification, characterization, and distribution of a Shiga toxin 1 gene variant (stx1c) in Escherichia coli strains isolated from humans. J. Clin. Microbiol. 40:1441–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boerlin P, McEwen SA, Boerlin-Petzold F, Wilson JB, Johnson RP, Gyles CL. 1999. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J. Clin. Microbiol. 37:497–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Neely MN, Friedman DI. 1998. Arrangement and functional identification of genes in the regulatory region of lambdoid phage H-19B, a carrier of a Shiga-like toxin. Gene 223:105–113 [DOI] [PubMed] [Google Scholar]
- 13. Plunkett G, 3rd, Rose DJ, Durfee TJ, Blattner FR. 1999. Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product. J. Bacteriol. 181:1767–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Oppenheim AB, Kobiler O, Stavans J, Court DL, Adhya S. 2005. Switches in bacteriophage lambda development. Annu. Rev. Genet. 39:409–429 [DOI] [PubMed] [Google Scholar]
- 15. Craig NL, Roberts JW. 1980. Escherichia coli RecA protein-directed cleavage of phage lambda repressor requires polynucleotide. Nature 283:26–30 [DOI] [PubMed] [Google Scholar]
- 16. Phizicky EM, Roberts JW. 1981. Induction of SOS functions: regulation of proteolytic activity of Escherichia coli RecA protein by interaction with DNA and nucleoside triphosphate. Cell 25:259–267 [DOI] [PubMed] [Google Scholar]
- 17. Karch H, Schmidt H, Janetzki-Mittmann C, Scheef J, Kroger M. 1999. Shiga toxins even when different are encoded at identical positions in the genomes of related temperate bacteriophages. Mol. Gen. Genet. 262:600–607 [DOI] [PubMed] [Google Scholar]
- 18. Wagner PL, Neely MN, Zhang X, Acheson DW, Waldor MK, Friedman DI. 2001. Role for a phage promoter in Shiga toxin 2 expression from a pathogenic Escherichia coli strain. J. Bacteriol. 183:2081–2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Neely MN, Friedman DI. 1998. Functional and genetic analysis of regulatory regions of coliphage H-19B: location of Shiga-like toxin and lysis genes suggest a role for phage functions in toxin release. Mol. Microbiol. 28:1255–1267 [DOI] [PubMed] [Google Scholar]
- 20. Muniesa M, Blanco JE, De Simon M, Serra-Moreno R, Blanch AR, Jofre J. 2004. Diversity of stx2 converting bacteriophages induced from Shiga-toxin-producing Escherichia coli strains isolated from cattle. Microbiology 150:2959–2971 [DOI] [PubMed] [Google Scholar]
- 21. Osawa R, Iyoda S, Nakayama SI, Wada A, Yamai S, Watanabe H. 2000. Genotypic variations of Shiga toxin-converting phages from enterohaemorrhagic Escherichia coli O157: H7 isolates. J. Med. Microbiol. 49:565–574 [DOI] [PubMed] [Google Scholar]
- 22. Shaikh N, Tarr PI. 2003. Escherichia coli O157:H7 Shiga toxin-encoding bacteriophages: integrations, excisions, truncations, and evolutionary implications. J. Bacteriol. 185:3596–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eppinger M, Mammel MK, Leclerc JE, Ravel J, Cebula TA. 2011. Genomic anatomy of Escherichia coli O157:H7 outbreaks. Proc. Natl. Acad. Sci. U. S. A. 108:20142–20147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Perna NT, Plunkett G, III, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Posfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta ET, Potamousis KD, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA, Blattner FR. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529–533 [DOI] [PubMed] [Google Scholar]
- 25. Ohnishi M, Terajima J, Kurokawa K, Nakayama K, Murata T, Tamura K, Ogura Y, Watanabe H, Hayashi T. 2002. Genomic diversity of enterohemorrhagic Escherichia coli O157 revealed by whole genome PCR scanning. Proc. Natl. Acad. Sci. U. S. A. 99:17043–17048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Iguchi A, Iyoda S, Terajima J, Watanabe H, Osawa R. 2006. Spontaneous recombination between homologous prophage regions causes large-scale inversions within the Escherichia coli O157:H7 chromosome. Gene 372:199–207 [DOI] [PubMed] [Google Scholar]
- 27. Kim J, Nietfeldt J, Benson AK. 1999. Octamer-based genome scanning distinguishes a unique subpopulation of Escherichia coli O157:H7 strains in cattle. Proc. Natl. Acad. Sci. U. S. A. 96:13288–13293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Laing CR, Buchanan C, Taboada EN, Zhang Y, Karmali MA, Thomas JE, Gannon VP. 2009. In silico genomic analyses reveal three distinct lineages of Escherichia coli O157:H7, one of which is associated with hyper-virulence. BMC Genomics 10:287 doi:10.1186/1471-2164-10-287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kulasekara BR, Jacobs M, Zhou Y, Wu Z, Sims E, Saenphimmachak C, Rohmer L, Ritchie JM, Radey M, McKevitt M, Freeman TL, Hayden H, Haugen E, Gillett W, Fong C, Chang J, Beskhlebnaya V, Waldor MK, Samadpour M, Whittam TS, Kaul R, Brittnacher M, Miller SI. 2009. Analysis of the genome of the Escherichia coli O157:H7 2006 spinach-associated outbreak isolate indicates candidate genes that may enhance virulence. Infect. Immun. 77:3713–3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ziebell K, Steele M, Zhang Y, Benson A, Taboada EN, Laing C, McEwen S, Ciebin B, Johnson R, Gannon V. 2008. Genotypic characterization and prevalence of virulence factors among Canadian Escherichia coli O157:H7 strains. Appl. Environ. Microbiol. 74:4314–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang Y, Laing C, Zhang Z, Hallewell J, You C, Ziebell K, Johnson RP, Kropinski AM, Thomas JE, Karmali M, Gannon VP. 2010. Lineage and host source are both correlated with levels of Shiga toxin 2 production by Escherichia coli O157:H7 strains. Appl. Environ. Microbiol. 76:474–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Faith NG, Shere JA, Brosch R, Arnold KW, Ansay SE, Lee MS, Luchansky JB, Kaspar CW. 1996. Prevalence and clonal nature of Escherichia coli O157:H7 on dairy farms in Wisconsin. Appl. Environ. Microbiol. 62:1519–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shere JA, Bartlett KJ, Kaspar CW. 1998. Longitudinal study of Escherichia coli O157:H7 dissemination on four dairy farms in Wisconsin. Appl. Environ. Microbiol. 64:1390–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jeong KC, Hiki O, Kang MY, Park D, Kaspar CW. 2012. Prevalent and persistent Escherichia coli O157:H7 strains on farms are selected by bovine passage. Vet. Microbiol. doi:10.1016/j.vetmic.2012.11.034 [DOI] [PubMed] [Google Scholar]
- 35. Asadulghani M, Ogura Y, Ooka T, Itoh T, Sawaguchi A, Iguchi A, Nakayama K, Hayashi T. 2009. The defective prophage pool of Escherichia coli O157: prophage-prophage interactions potentiate horizontal transfer of virulence determinants. PLoS Pathog. 5:e1000408 doi:10.1371/journal.ppat.1000408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brenner S, Horne RW. 1959. A negative staining method for high resolution electron microscopy of viruses. Biochim. Biophys. Acta 34:103–110 [DOI] [PubMed] [Google Scholar]
- 37. De Carlo S, Harris JR. 2011. Negative staining and cryo-negative staining of macromolecules and viruses for TEM. Micron 42:117–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gonzales TK, Kulow M, Park DJ, Kaspar CW, Anklam KS, Pertzborn KM, Kerrish KD, Ivanek R, Dopfer D. 2011. A high-throughput open-array qPCR gene panel to identify, virulotype, and subtype O157 and non-O157 enterohemorrhagic Escherichia coli. Mol. Cell. Probes 25:222–230 [DOI] [PubMed] [Google Scholar]
- 39. Tyler JS, Mills MJ, Friedman DI. 2004. The operator and early promoter region of the Shiga toxin type 2-encoding bacteriophage 933W and control of toxin expression. J. Bacteriol. 186:7670–7679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dowd SE, Crippen TL, Sun Y, Gontcharova V, Youn E, Muthaiyan A, Wolcott RD, Callaway TR, Ricke SC. 2010. Microarray analysis and draft genomes of two Escherichia coli O157:H7 lineage II cattle isolates FRIK966 and FRIK2000 investigating lack of Shiga toxin expression. Foodborne Pathog. Dis. 7:763–773 [DOI] [PubMed] [Google Scholar]
- 41. Vanaja SK, Springman AC, Besser TE, Whittam TS, Manning SD. 2010. Differential expression of virulence and stress fitness genes between Escherichia coli O157:H7 strains with clinical or bovine-biased genotypes. Appl. Environ. Microbiol. 76:60–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Baker DR, Moxley RA, Francis DH. 1997. Variation in virulence in the gnotobiotic pig model of O157:H7 Escherichia coli strains of bovine and human origin. Adv. Exp. Med. Biol. 412:53–58 [DOI] [PubMed] [Google Scholar]
- 43. Strauch E, Hammerl JA, Konietzny A, Schneiker-Bekel S, Arnold W, Goesmann A, Puhler A, Beutin L. 2008. Bacteriophage 2851 is a prototype phage for dissemination of the Shiga toxin variant gene 2c in Escherichia coli O157:H7. Infect. Immun. 76:5466–5477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Strauch E, Schaudinn C, Beutin L. 2004. First-time isolation and characterization of a bacteriophage encoding the Shiga toxin 2c variant, which is globally spread in strains of Escherichia coli O157. Infect. Immun. 72:7030–7039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pruimboom-Brees IM, Morgan TW, Ackermann MR, Nystrom ED, Samuel JE, Cornick NA, Moon HW. 2000. Cattle lack vascular receptors for Escherichia coli O157:H7 Shiga toxins. Proc. Natl. Acad. Sci. U. S. A. 97:10325–10329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dean-Nystrom EA, Bosworth BT, Moon HW, O'Brien AD. 1998. Escherichia coli O157:H7 requires intimin for enteropathogenicity in calves. Infect. Immun. 66:4560–4563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sheng H, Lim JY, Knecht HJ, Li J, Hovde CJ. 2006. Role of Escherichia coli O157:H7 virulence factors in colonization at the bovine terminal rectal mucosa. Infect. Immun. 74:4685–4693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hoffman MA, Menge C, Casey TA, Laegreid W, Bosworth BT, Dean-Nystrom EA. 2006. Bovine immune response to Shiga-toxigenic Escherichia coli O157:H7. Clin. Vaccine Immunol. 13:1322–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Robinson CM, Sinclair JF, Smith MJ, O'Brien AD. 2006. Shiga toxin of enterohemorrhagic Escherichia coli type O157:H7 promotes intestinal colonization. Proc. Natl. Acad. Sci. U. S. A. 103:9667–9672 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.