Abstract
Pseudomonas aeruginosa is a metabolically versatile bacterium that is found in a wide range of biotic and abiotic habitats. It is a major human opportunistic pathogen causing numerous acute and chronic infections. The critical traits contributing to the pathogenic potential of P. aeruginosa are the production of a myriad of virulence factors, formation of biofilms and antibiotic resistance. Expression of these traits is under stringent regulation, and it responds to largely unidentified environmental signals. This review is focused on providing a global picture of virulence gene regulation in P. aeruginosa. In addition to key regulatory pathways that control the transition from acute to chronic infection phenotypes, some regulators have been identified that modulate multiple virulence mechanisms. Despite of a propensity for chaotic behaviour, no chaotic motifs were readily observed in the P. aeruginosa virulence regulatory network. Having a ‘birds-eye’ view of the regulatory cascades provides the forum opportunities to pose questions, formulate hypotheses and evaluate theories in elucidating P. aeruginosa pathogenesis. Understanding the mechanisms involved in making P. aeruginosa a successful pathogen is essential in helping devise control strategies.
INTRODUCTION
Pseudomonas aeruginosa is a Gram-negative bacterium that has the ability to thrive in most natural and man-made environments. It is found in diverse habitats, including soil, water, plants and animals, and can infect multiple hosts (1,2). Pseudomonas aeruginosa causes a wide variety of acute (short duration, typically severe) and chronic (persisting for a long time, often refractory to treatment, severity varying with pathogen) human infections, including in patients with severe burn wounds, urinary tract infections, AIDS, lung cancer, chronic obstructive pulmonary disease, bronchiectasis and cystic fibrosis (CF) (3–6).
Metabolic versatility, intrinsic and acquired antibiotic resistance, biofilm formation and production of multiple virulence (disease-causing) factors make P. aeruginosa a formidable pathogen. The virulence machinery of P. aeruginosa comprises both cell-associated determinants (such as lipopolysaccharides, pili, flagella) and numerous secreted factors (such as elastases, proteases, exotoxins, pyocyanin, extracellular polysaccharides). One of the mechanisms by which P. aeruginosa senses external signals is using sensor proteins that, through phosphotransfer or phosphorelay, activate specific transcriptional regulators. These sensor–regulator protein pairs are called two-component systems (TCS). The P. aeruginosa PAO1 genome encodes ∼127 TCS members, compared with 60 in Escherichia coli (7) and 70 in Bacillus subtilis (8), reflecting the adaptability of P. aeruginosa. TCS and their modifications also feed into major regulatory pathways and play a critical role in allowing cells to modulate gene expression in response to environmental conditions (9,10). Many of the secreted virulence factors and phenotypes, such as biofilm formation, are under the control of a cell density recognition mechanism called quorum sensing (QS) that aids in the coordinated expression of genes (11,12). QS is a key to virulence gene expression in many bacteria and serves as an attractive target for antibacterial chemotherapy (13).
In humans, acute P. aeruginosa infections in specific sites, such as the CF lung, eventually lead to chronic inections. This is caused by adaptive modifications in the infecting clonal type, resulting in diverse morphotypes (14). Acute virulence factors include the Type 2 and Type 3 secretion systems, flagella, type IV pili and QS-regulated virulence factors (proteases, elastase, pyocyanin) (15). On establishing a chronic infection, P. aeruginosa overproduces extracellular polysaccharides, forms biofilms and small colony variants and upregulates the Type 6 secretion system (15–18). Antibiotic resistance plays a major role in both types of infection, although the cells display higher levels of resistance in chronic infections (18,19). The transition to a chronic infection phase is the result of numerous changes in cellular physiology in response to external stimuli (20). The changes include downregulation of acute virulence genes with a concomitant upregulation of chronic infection phenotypes and antibiotic resistance, facilitating recalcitrant infections (15,20). Host invasion, establishment of acute infection and the subsequent transition to the chronic phase involves tightly regulated expression of many genes associated with metabolism, virulence and antibiotic resistance. Several key players in these transition processes have been identified and include transcriptional and post-transcriptional regulators (21–23). Many of these will be discussed in subsequent sections of this review.
Gene regulation in P. aeruginosa is a complex process involving numerous transcriptional regulators, regulatory RNAs (rgRNA) and σ factors. The P. aeruginosa genome is >6 Mb (24), approaching that of lower eukaryotes. The genome is plastic and has acquired genes and undergone extensive rearrangements to adapt to specific niches (25). The large genome of P. aeruginosa supports a multitude of regulatory networks, with ∼8% of the total genome dedicated to the regulatory proteins (26). Pseudomonas aeruginosa PAO1 encodes 434 transcriptional regulators, 24 σ factors and 34 small RNAs, many of which remain to be characterized (24,27–29). Moreover, predicted regulatory networks indicate that there is an extensive crosstalk between the different transcriptional regulators (27,30). These networks, however, are based in part on in silico analyses, and their validity needs to be established. This review makes an effort to consolidate the empirically proven major virulence regulatory networks in P. aeruginosa with the hope of providing a framework for future studies to better understand pathogenic processes in P. aeruginosa and in related bacteria.
MAJOR VIRULENCE REGULATORY SYSTEMS IN P. AERUGINOSA
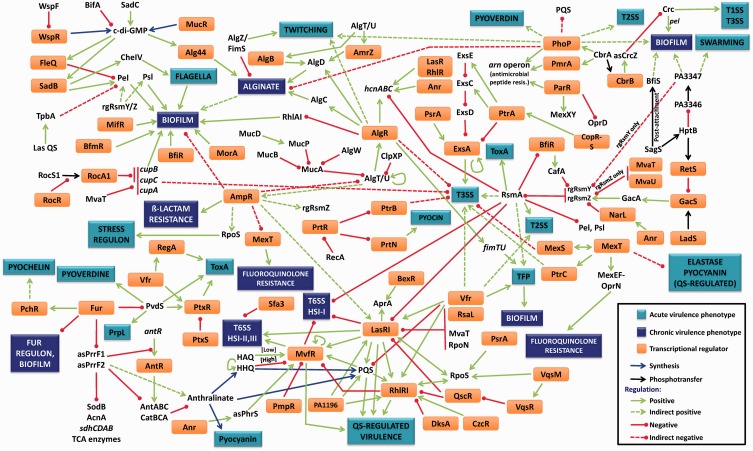
The experimentally established virulence regulatory network in P. aeruginosa is depicted in Figure 1. Our group and others have previously performed in silico analyses of the P. aeruginosa transcriptional regulatory network (27,30,31). Comparing those networks with the network depicted in Figure 1 clearly demonstrates the gap in knowledge between predicted networks and established ones. An important contributing factor to this discrepancy is the fact that the functions of the majority of the genes in the PAO1 genome remain unknown. Deep sequencing, transcriptome metaanalysis (32,33) and complementary studies will aid in assigning functions to the hypothetical genes and undoubtedly narrow this knowledge gap.
Figure 1.
The P. aeruginosa virulence regulatory network. The pathogenic potential of P. aeruginosa is dictated by multiple virulence systems that are regulated transcriptionally, post-transcriptionally and post-translationally. The central mechanism for P. aeruginosa virulence regulation is QS, which controls expression of many virulence factors in a population density-dependent manner. Key activators of this system are LasR, RhlR, MvfR, VqsR, the cAMP receptor protein Vfr and the stationary phase σ factor RpoS. Las system repressors include RsaL, the H-NS protein MvaT, the σ factor RpoN and the sRNA-binding protein RsmA, whereas others like QscR repress both the Las and Rhl systems. Other regulators such as AmpR affect QS genes by an unknown mechanism. QS plays a role in regulating critical pathogenic mechanisms, including biofilm formation, secretion systems, production of numerous virulence factors, efflux pumps, antibiotic resistance and motility. Acute P. aeruginosa infections can lead to chronic infections in response to largely unidentified signals. A key regulatory pathway that controls this lifestyle switch is the RetS–LadS–GacSA–RsmA pathway. RetS and LadS are hybrid sensor proteins that, in response to external signals, either activate or repress the GacSA TCS. The GacA regulator then activates transcription of two rgRNAs, rgRsmZ and rgRsmY that sequester and inhibit activity of the sRNA-binding protein, RsmA. RsmA is a key activator/repressor that post-transcriptionally regulates numerous acute and chronic infection phenotypes, including multiple QS-regulated virulence factors, biofilm formation, Type 2, Type 3 and Type 6 secretion systems and motility. Another major phenotypic change associated with the switch from acute to chronic phases of infection is the formation of biofilm. This is associated with extensive changes in transcription. Three key TCS involved in activating biofilm formation are BfiSR, MifR and BfmSR. Cyclic-di-GMP is another major player influencing this process, whose levels are controlled by diguanylate cyclases and phosphodiesterases. QS and the cup genes enhance biofilms, whereas regulators like AmpR repress it. An important component of P. aeruginosa biofilms are extracellular polysaccharides, such as alginate, Pel and Psl. Alginate production is under the control of the master regulator ECF AlgT/U, whose activity is regulated transcriptionally by AmpR, post-translationally by MucA and MucB and by regulated intermembrane proteolysis involving MucP, AlgW, ClpXP and others. AlgT/U activates the alginate biosynthetic operon through AlgR, AlgB and AmrZ. In addition, biofilm formation is also affected by iron concentration, a process governed by the master repressor of iron uptake, Fur. Fur controls uptake of iron by regulating the σ factor PvdS, thereby modulating sidephore levels. Fur also modulates transcription of two key regulatory RNAs, asPrrF1 and asPrrF2. These two sRNAs are involved not only in regulating iron-uptake-related genes but also enzymes of the trichloroacetic acid cycle and genes involved in anthranilate synthesis. Anthralinate, a precursor for synthesis of PQS, is a key regulatory molecule of the PQS signalling system in P. aeruginosa, which is involved in expression of QS-regulated virulence factors. Details on the individual interactions and the appropriate references can be found in the text. Some of the interactions labelled as indirect are regulated by unknown mechanisms and warrant further investigation. In the figure, some regulators and phenotypes have been mentioned more than once.
Cis regulatory elements (CREs) form a critical part of transcription. CREs are non-coding DNA sequences present in or near a gene, and they often contain binding sites for transcription factors and/or other regulators of transcription (34). The two major CREs are promoters and enhancers (35,36). The promoters contain the binding sites for transcription factors and other regulatory molecules, such as σ factors and regulatory RNAs (37–39). Enhancers, once thought to be part of only eukaryotes, are found widely in prokaryotes also, and they function in conjunction with the σ54-RNA polymerase (40–42). The known P. aeruginosa transcription factor binding sites are listed in Table 1. This section will focus on the transcriptional and post-transcriptional regulation of critical pathways that determine P. aeruginosa pathogenesis.
Table 1.
Cis regulatory elements in P. aeruginosa transcriptional regulation
| Transcription factor | Cis regulatory element | Major virulence phenotype regulated | Reference |
|---|---|---|---|
| AlgR | ACCGTTCGTC | Alginate production, biofilm formation, T3SS | (43) |
| AlgZ | GGCCATTACCAGCC | Alginate production | (44) |
| Anr | TTGATN4ATCAA | Anaerobic regulator of QS | (45) |
| ArgR | TGTCGCN8AA | Carbon and nitrogen catabolism | (46) |
| ExsA | TNAAAANA | T3SS | (47) |
| FleQ | Box 1: CGCCTAAAAATTGACAGTT | Motility, biofilm formation | (48) |
| Box 2: CATTAGATTGACGTTAATC | |||
| Fur | GATAATGATAATCATTATC | Iron uptake | (49) |
| LasR (las box) | NHCTRNSNNDHNDKNNAGNB | QS | (50) |
| MexT | ATCAN5GTCGATN4ACYAT | Antibiotic resistance, T3SS, QS | (51) |
| MvfR | TTCGGACTCCGAA | QS | (52) |
| PsrA | G/CAAACN2-4GTTTG/C | Stress regulon (RpoS), T3SS | (53) |
| RcsBa | TTA-GAAACGTCCTAAA | Fimbriae | (54) |
| RhlR (lux box) | CCTGTGAAT/ATCC/TGGT/CAGTT | QS | (55) |
| Vfr | AATTGACTAATCGTTCACATTTG | QS | (56) |
| VqsR | TCGCCN8GGCGA | QS | (57) |
H = C/T/A; R = A/G; S = C/G; D = G/A/T; K = G/T; B = C/G/T; N = A/C/G/T.
aRcsB is found only in P. aeruginosa PA14, not in P. aeruginosa PAO1.
QS
QS is a signalling mechanism that bacteria use to regulate gene expression in a population density-dependent manner, and it was first demonstrated in Vibrio fischeri (58). In QS, the bacteria produce and secrete small molecules called autoinducers or quoromones. When these molecules reach a concentration threshold, they diffuse back into the cell to elicit a coordinated response promoting group survival (59). Pseudomonas aeruginosa uses QS to regulate production of various virulence determinants, such as extracellular proteases, iron chelators, efflux pump expression, biofilm formation, motility and the response to host immune signals (60). This is achieved using two types of autoinducers, N-acyl-homoserine lactones (AHLs) and 2-alkyl-4 quinolones (AQs) (61).
AHL-mediated QS
Pseudomonas aeruginosa has two canonical AHL QS signalling pathways, the las and rhl systems. Together, these pathways affect expression of ∼10% of the P. aeruginosa transcriptome (62). The lasI (PA1432) and rhlI (PA3476) genes encode the N-3-oxododecanoylhomoserine lactone (3-oxo-C12-AHL) synthetase (63,64) and N-butyrylhomoserine lactone (C4-AHL) synthetase, respectively (65–68). The resulting AHLs then bind and activate their cognate LuxR family regulators, LasR (PA1430) (64) or RhlR (PA3477) (67). LasR and RhlR multimerize in the presence of their cognate AHL (69,70). In in vitro studies, LasR–DNA interaction is cooperative and non-cooperative in the presence or absence of a dyad symmetry in the binding sites, respectively (71). Rhl-regulated promoters have binding sites with a dyad symmetry (72).
AQ-mediated QS
Pseudomonas aeruginosa synthesizes two AQ QS signals, 2-heptyl-3-hydroxy-4-quinolone (PQS) and its precursor, 2-heptyl-4-quinolone (HHQ) (73). Both PQS and HHQ enhance in vitro binding of the LysR-type transcription regulator, MvfR (also known as PqsR, PA1003), to the promoter of the pqsABCDE operon (PA0996–PA1000), suggesting that they function as MvfR effectors (74). Microarray analysis identified 141 genes differentially expressed in an mvfR mutant strain, including lasR, algT/U (PA0762), rsmA (PA0905) and rsaL (PA1431) (75). PQS also acts independently of MvfR to induce expression of the Fur regulon through its ability to bind iron (73,76) and membrane vesicle formation by inducing membrane curvature (77,78). PmpR (PA0964), a YebC member, negatively regulates MvfR (Figure 1) (79).
QS regulation
The las, rhl and PQS/HHQ/MvfR systems exhibit positive feed-forward autoregulation (52,80). In addition, the P. aeruginosa AHL systems function in a hierarchical manner, as the 3-oxo-C12-AHL-LasR complex positively regulates rhlI, rhlR and mvfR expression as well as lasI (81–83). Exceptions to this have been noted. RhlR has been shown to regulate LasR-dependent genes in strains lacking lasR (84), and timing of lasI, lasR, rhlI and rhlR expression can vary drastically depending on growth conditions (85).
Many global regulators have been shown to modulate QS-dependent genes. RpoS (PA3622), the stationary phase σ factor affects ∼40% of the QS regulon (72,86). RpoS binding sites have been identified in several of the QS-dependent promoters. RpoS also affects lasR and rhlR expression, and LasR binding sites have been identified in promoters of other transcriptional regulators in the QS regulon, including PA2588, PA4778, pvdS (PA2426), vqsR (PA2591) and rsaL (87). Chromatin immunoprecipitation studies have shown occupancy by histone-like silencers MvaT (PA4315) and MvaU (PA2667) on lasI, lasR, mvfR, rpoS and rsaL (88). RsaL plays an important role in las signalling homeostasis, by binding to the lasI promoter and preventing LasR-mediated activation (89). In addition to affecting gene expression through las regulation, microarray analyses indicate that RsaL affects expression of 130 genes, including direct regulation of pyocyanin and hydrogen cyanide genes (89). RsaL also seems to be important in regulating the transition from planktonic to a sessile state, as rsaL mutants exhibit increased swarming motility and fail to form biofilms (90). RsaL expression is under the control of LysR-type regulator OxyR (PA5344) (91). The lasI promoter region has also been shown to be bound by CzcR (PA2523), which is required for expression of rhlI and rhlR in addition to lasI (Figure 1) (92). CzcR is part of the CzcRS TCS, which is shown to be involved in carbapenem and heavy metal resistance (93).
VqsR (PA2591), which is induced by H2O2 or human serum (94) and is under LasR regulation (95), regulates QS through inhibition of the LuxR-type regulator, QscR (PA1898, Figure 1) (96). Although QscR binds to 3-oxo-C12-AHL, its specificity is not as stringent as LasR (97). The QscR regulon partially overlaps that ascribed to the las and rhl systems, but also has unique targets (98). In the absence of AHL, QscR can multimerize and form heterodimers with LasR and RhlR (99). QscR also plays a role in LasI homeostasis, as mutations in qscR result in premature lasI expression (100). An AraC family member VqsM (PA2227) regulates VqsR in addition to numerous genes involved in QS, including RsaL, PprB (PA4296), MvfR, RpoS as well as AlgT/U and MexR (PA0424) (101).
Additionally, pqsH (PA2587), which encodes the enzyme responsible for oxidation of HHQ to form PQS, is positively regulated by the las system (102,103) and is negatively regulated by the rhl system (52). PQS is derived from anthranilate, which is synthesized by the kynurenine pathway (104). Kynurenine pathway anthranilate is also required for N-decanoyl-homoserine lactone (C10-AHL) dependent signalling, which is independent of las, rhl and qscR (104). The receptor for this signalling is yet to be identified (105). Besides potential heterodimerization with QscR, additional post-transcriptional regulation of QS has been described. In one such mechanism, QteE (PA2593) destabilizes LasR and RhlR, and in the absence of qteE, the quorum threshold-requirement for activation of QS-dependent genes is lost (106). RsmA negatively regulates rhl and las signalling, resulting in reduced AHL levels (107). Moreover, it has been shown the RNA chaperone Hfq (PA4944) positively regulates rhlI translation through rsmY and RsmA (108).
Recently, our laboratory has established a role for the ß-lactamase regulator AmpR (PA4109) in activating QS-regulated genes (23). The production of QS-regulated secreted virulence factors, such as LasA (PA1871) and LasB (PA3724) proteases, and pyocyanin production is significantly impaired in AmpR-deficient strains. Further, loss of ampR reduced virulence in the Caenorhabditis elegans toxicity assay (23,109). In addition, AmpR regulates non–ß-lactam resistance by repressing activity of the MexEF–OprN (PA2493–PA2495) efflux pump, the alginate master regulator AlgT/U (110) and biofilm formation (23), suggesting that it plays a role in maintaining the acute mode of infection.
TCS
TCS are sophisticated signalling mechanisms marked by a highly modular design that have been adapted and integrated into a wide variety of cellular signalling circuits. The archetypical TCS is composed of a membrane integrated sensory histidine kinase (HK) and a cytoplasmic response regulator (RR) (111). The HK contains a periplasmic N-terminal domain that detects specific stimuli (sensing domain) and a C-terminal cytoplasmic transmitter domain that comprises a dimerization domain, a conserved histidine and an adenosine triphosphate catalytic domain (112). HKs can have two or more transmembrane domains with little or no periplasmic domain, whereas others are completely cytoplasmic. The cognate RR contains a conserved receiver domain and a variable output domain (113). On receiving a signal, two HK monomers dimerize and cross-phosphorylate at the conserved histidine residue, and the phosphate is subsequently transferred to an aspartate residue in the receiver domain of the cognate RR (114). The phosphotransfer is catalysed by the receiver domain, and it results in a conformational change that activates the output domain, which often binds DNA and modulates gene expression or enzymatic activity (9,113,115). Variations to this model occur in phosphorelays, where a sensor kinase first transfers the phosphoryl group to an RR that has no output domain. This P∼RR then transfers the phosphoryl group to a histidine-containing phosphotransfer protein, and this in turn serves as a phosphate donor to a terminal RR, which has an output domain mediating a cellular response (10). In other cases, the sensor kinase and the RR lacking an output domain are fused into one protein (hybrid sensor kinase) (116). Other variations include the TCS connectors, a group of proteins that modulate the phosphorylation state and activity of sensor HK and RR and establish regulatory links between otherwise independent signal transduction pathways (117).
Pseudomonas aeruginosa, equipped with 55 HKs, 89 RRs and 14 HK–RR hybrids, possesses one of the largest pool of TCS proteins identified in any microorganism analysed thus far (24). This provides the bacterium with a sophisticated capability to regulate diverse metabolic adaptations, virulence and antibiotic resistance processes that are hallmark of P. aeruginosa infections. One of the critical TCSs is GacSA (GacS-PA0928, GacA-PA2586), which is central to expression of virulence factors, secondary metabolites, biofilm formation and QS (107,118) and is the switch between acute and chronic infections (1,119). GacS is a hybrid sensor HK that contains an HK domain, an RR domain and a histidine phosphotransfer (Hpt) domain (21,120). GacS phosphorylation is under the control of two hybrid sensor kinases, RetS (PA4856) (21) and LadS (PA3974) (22) (Figure 1). RetS can directly interact with GacS and prevent GacS phosphorylation (22,121), whereas LadS phosphorylates GacS (22). Phosphorylated GacA positively regulates the transcription of two small regulatory RNAs, rgRsmZ (PA3621.1) and rgRsmY (PA0527.1), which block the negative regulator RNA-binding protein RsmA (PA0905). RsmA positively regulates genes of the Type 3 secretion system, type IV pili formation and iron homeostasis while repressing QS, Type 6 secretion and potentially other transcription factors (122–124). The GacSA TCS is also involved in antibiotic resistance to three different families of antibiotics, tobramycin, ciprofloxacin and tetracycline (125), apparently through RsmA/rgRsmZ.
In P. aeruginosa, PhoPQ (PA1179–PA1180) together with PmrAB (PA4776–PA4777) are two TCSs that respond to limiting concentrations of cations, and regulate resistance to polymyxin B and cationic antimicrobial peptides through the regulation of the arnBCADTEF-pmrE (PA3552–PA3559) LPS modification operon (126,127). PhoQ is involved in swarming and twitching motility as well as in biofilm formation and is required for virulence without affecting the T3SS or QS systems (Figure 1) (128). The HK PhoQ activates the RR PmrA independently of PmrB, suggesting an interaction between these TCSs (129). In addition, increased resistance to antibiotics, including polymyxin B, aminoglycosides and quinolones in phoQ mutants suggests crosstalk between PhoPQ and other TCSs (130,131).
Formation of biofilms
Biofilms are surface-associated multicellular bacterial communities encapsulated in a self-produced extracellular matrix composed of polysaccharides, proteins and nucleic acids that mediate cell-to-cell and cell-to-surface interactions (132). Pseudomonas aeruginosa biofilms, typically associated with poor patient prognosis, signify the switch from an acute to a chronic infection. Biofilms can be formed on abiotic (environment) or biotic (wounds, surgical implants, CF lung) surfaces (133). Biofilm formation and maintenance is tightly regulated in response to environmental cues, conferring enhanced resistance against antimicrobial agents and immune defence mechanisms on the biofilm bacteria (12). Formation of biofilms is a multi-stage process that is initiated by the surface attachment of planktonic bacteria to form a monolayer, clonal growth/aggregation leading to the formation of microcolonies, maturation to form mushroom-shaped structures and dispersal (134–136). As can be imagined, this complex transition in the bacterial lifestyle is accompanied by drastic changes in gene regulation.
Surface attachment by P. aeruginosa to form microcolonies has been attributed to type IV pili, flagella, free DNA, alginate and Pel and Psl polysaccharides, although pili, alginate and flagella mutants also form biofilms (136,137). Attachment is a reversible process, and the commitment to form biofilms is partly under positive SadB (PA5346) regulation (138). SadB upregulates both Pel polysaccharide production and the chemotaxis-like cluster CheIV (PA0408–PA0417), which is thought to regulate flagellar motion by an unknown mechanism (139).
The Cup fimbriae, encoded by three distinct gene clusters cupA (PA2128–PA2133), cupB (PA4081–PA4086) and cupC (PA0992, PA0993, PA0994) in P. aeruginosa PAO1, have been demonstrated to play a role in different stages of biofilm formation on biotic and abiotic surfaces (140). Regulation of the cup genes is complex, involving a phase variation-dependent repression of cupA expression by an H-NS member MvaT (141,142). MvaT also regulates the cupB and cupC loci to a lesser extent (141). The cupB and cupC clusters are under the primary regulation of the RocS1–RocR–RocA1 (PA3946–PA3948) three-component system (143). This system is similar to the Bordetella pertussis BvgASR system (144) and consists of the hybrid sensor kinase RocS1, the response regulator RocA1 and the RocA1-repressor RocR (143,145). RocR has been hypothesized to bind to c-di-GMP through its EAL (diguanylate phosphodiestrerase) domain and prevents phosphotransfer from RocS1 to RocA1, thus preventing RocA1 activation (143,145). Pseudomonas aeruginosa PA14 has a fourth cup cluster (cupD) on the pathogenicity island PAPI-I, which is controlled positively by the response regulator RcsB (PA4080) and negatively by the EAL-domain containing response regulator PvrR (146). In addition, diguanylate cyclases and phosphodiesterases of the wsp gene cluster (PA3702–PA3708) (147,148) MorA (PA4601) (149) and TpbA–TpbB (PA3885, PA1120) (150) modulate intracellular levels of c-di-GMP to exert a regulatory effect on the cup gene clusters (Figure 1).
The main components of the extracellular polymeric substance matrix of biofilms are Pel and Psl polysaccharides, alginate and free DNA (12,136). Both pel and psl gene loci are post-transcriptionally regulated by the RetS–LadS (PA4856 and PA3974, respectively) system through rsmY and rzmZ (21,22,121) and by c-di-GMP levels, either directly (147) or by binding the transcriptional regulator FleQ (PA1097) (151). The pel operon is also repressed by the las QS system through the tyrosine phosphatase TpbA (PA3885) (150). A membrane-bound sensor, PpyR (PA2663) enhances biofilm formation through the psl operon and virulence through modulating QS (152). Although alginate is a major component of biofilms and affects biofilm structure, it is not essential for biofilm formation (136). Alginate regulation is discussed in a separate section (see later in the text). QS regulates cell lysis in biofilms (153–155), thereby controlling the release of extracellular DNA, a major component of the biofilm matrix (156,157). The QS system also regulates rhamnolipid production (67) that promotes motility and, hence, formation of the cap in the mushroom structure of mature biofilms (158), and maintenance of biofilm channels (159). BfiRS (PA4196–PA4197), BfmRS (PA4101–PA4102) and MifR (PA5511) are TCSs shown to regulate biofilm development and maturation by sequential phosphorylation (160). They activate biofilm formation at different transition stages, reversible to irreversible attachment (BfiRS), irreversible attachment to maturation stage-1 (BfmRS) and maturation stage-1 to mushroom structure formation (MifR) (Figure 1) (160). The BfiRS system may function in conjunction with the GacSA TCS and feed into the Rsm loop of regulation to control biofilm formation (160). SagS (PA2824), the cognate sensor of HptB (PA3345), modulates biofilm development (by controlling BifS phosphorylation) and other virulence phenotypes (by modulating rgsRmZ levels) depending on whether the cells are in the planktonic or biofilm phase (161).
Analyses of clinical isolates reveal a positive correlation between expression of lasR, rhlR and acute virulence factors (162,163), suggesting that QS is required for virulence in vivo. QS is also important when P. aeruginosa grows as biofilms in the CF lung (164). In vivo studies show that lasI and rhlI mutants produce milder chronic lung infections compared with their wild-type counterparts (165) and form more susceptible biofilms (166). However, in some in vitro studies, there was no apparent difference in the biofilms formed by the QS mutants and the wild-type strains (167,168). This discrepancy in the requirement of QS for biofilm formation and establishment of a successful chronic infection is probably not surprising, as QS regulates many different functions. Further, it has been demonstrated that the CF environment selects for strains with lasR mutations, although the rhl system is intact (169). Although lasR is higher up in the QS hierarchy, studies have shown that secondary mutations can re-establish rhl expression in las mutants (170). This suggests that in CF biofilms, the rhl system is more important, and lasR inactivation serves to downregulate the acute virulence factors (171).
A major cause of antibiotic resistance in biofilms has recently been attributed to the phenomenon of persistence. Persister cells are small subpopulations of antibiotic-sensitive cells that have acquired transient antibiotic tolerance (172). When the antibiotic levels drop, the persisters grow into a population of sensitive cells, again with a small sub-population of persisters (173,174). Many genes involved in the formation of persisters have been identified in P. aeruginosa PA14, including two transcriptional regulators AlgR (PA5261) and PilH (PA0409) (175). However, this topic is outside the scope of this review but has been extensively reviewed elsewhere (176,177).
Alginate production
In the lungs, especially in patients with CF, P. aeruginosa can convert from a non-mucoid to an alginate-overproducing mucoid phenotype signalling chronic infection (178). Chronic P. aeruginosa infection seems to be localized to foci within the anaerobic mucus environment in the lung’s respiratory zone (179–182). These foci lead to tissue damage decreasing lung function, and the appearance of the mucoid phenotype correlates with poor patient prognosis (183,184). The exopolysaccharide alginate is a linear polymer of β-d-mannuronic acid and α-l-guluronic acid (185), which stimulates production of IgG and IgA antibodies (186). Although production of alginate is metabolically taxing, it protects the bacteria from phagocytosis and antibodies, thus conferring a survival advantage (187,188). Conversion to mucoidy occurs when biofilms are treated with activated polymorphonuclear leucocytes (189), hydrogen peroxide (189), antibiotics (190) and nutrient starvation (191,192).
A complex regulatory pathway controls alginate biosynthesis. The central player is the σE family extracytoplasmic function σ factor AlgT/U (PA0762) (193,194), whose activity is inhibited post-transcriptionally by the anti-σ factor MucA (PA0763) and by MucB (PA0764) (195–197). Loss of function mutations in mucA or mucB result in a mucoid phenotype (195,198,199) because of release of AlgT/U from MucA by a regulated intramembrane proteolytic pathway [reviewed in (200,201)]. It was recently demonstrated that MucA proteolysis is regulated not only by AlgW (PA4446) but also by MucD (PA0766) by activating the MucP protease (PA3649) (202). In addition, AmpR links alginate production with antibiotic resistance and QS by negatively regulating algT/U expression (Figure 1) (110). AlgT/U regulates alginate production at least in part by autoregulation (193), controlling expression of the transcriptional regulators algR (203,204), algB (198,204), amrZ (205) and the algD (PA3540) alginate biosynthetic operon (204,206,207). AlgB (208), AlgR (43,209,210) and AmrZ (211) directly bind to the algD operon to activate transcription. The alternative σ factor RpoN (PA4462) is also required for high levels of algT/U and algD expression (212).
The algB (PA5483) and algR (PA5261) genes encode NtrC and LytR subfamily, respectively, of TCS RRs (213). Interestingly, aspartic acid phosphorylation in the regulatory domain is not essential for alginate production (214). Transcriptome analysis of a PAOmucA22 mucoid strain (PDO300) (196) identified seven predicted transcriptional regulators, PA1235, PA1261, PA1637 (KdpE), PA2881, PA3420, PA3771 and PA5431, and one sensor kinase, EraS (PA1979), whose expression was downregulated in an algB mutant but not in a strain containing a mutation in its cognate TCS sensor, KinB (PA5484) (208). In addition to regulating the algD operon, AlgR directly activates transcription of algC (PA5322), which encodes a phosphomannomutase/phosphoglucomutase essential for Psl, alginate and rhamnolipid synthesis (Figure 1) (215–218). AlgR also is important for mature biofilm formation, possibly by directly repressing rhl-QS (219), type IV pilus formation by binding to the fimTU–pilVWXY1Y2E promoter (220,221) and hydrogen cyanide production by binding to the hcnA (PA2193) promoter (222). Interestingly, in contrast to alginate production, the phosphorylation site is required for regulating cyanide production and twitching motility (220,223). AlgR has also been shown to indirectly regulate the cyclic AMP/Vfr-dependent pathway (224). The AlgR regulon has been characterized by several transcriptome studies (219,222,225). Two other regulators of alginate production in P. aeruginosa are Alg44 (PA3542) (226) and a diguanylate cyclase, MucR (PA1727) (227). MucR produces a pool of c-di-GMP in the vicinity of the PilZ domain of Alg44 (PA3542), which then positively regulates alginate production (Figure 1) (226–228).
Regulation of iron uptake
Iron is critical for growth of all organisms, and P. aeruginosa is no exception. Transcriptome studies reveal that a large number of genes are regulated in response to iron (229,230). Biologically useful iron (Fe2+) in the environment is scarce and is available mostly in the insoluble Fe3+ form. To help scavenge this free iron, bacteria produce siderophores that bind extracellular iron and transport them back into the cell through TonB-dependent receptors on the cell surface (231). Pseudomonas aeruginosa produces two siderophores, pyoverdine and pyochelin, and can also subvert siderophores produced by other organisms to take up haem (232,233). However, excess free iron in the cell leads to formation of toxic reactive oxygen species, and, therefore, cells tightly regulate the uptake (234). The ferric uptake regulator (Fur, PA4769) is a conserved protein in P. aeruginosa and other Gram-negative bacteria, and it is a major iron acquisition regulator (235). Fur dimerizes rapidly after synthesis, and it takes a minimum of two dimers to bind promoters of genes under Fur regulation in P. aeruginosa (236). Fur controls the iron regulon directly by binding the Fur box (237) and indirectly by modulating expression of other regulators, including the pyochelin uptake regulator PchR (PA4227), ECF σ factors like PvdS, TCS regulators and small regulatory RNAs [asPrrF1 (PA4704.1), asPrrF2 (PA4704.2); Figure 1] (237–239).
Iron concentrations in the cell also influence expression of virulence factors in P. aeruginosa. PvdS, for example, is critical in linking iron and virulence by controlling the production of pyoverdine, an outer membrane pyoverdine receptor [FpvA (PA2398)] and two important extracellular virulence factors [PrpL (PA4175) and exotoxin A (PA1148); Figure 1] (240–242). Also, pvdS mutants showed reduced virulence in a rabbit endocarditis model (243). Human lactoferrin inhibits P. aeruginosa biofilm formation, indicating a role for iron in the process (244). Iron chelation by lactoferrin induces twitching motility in P. aeruginosa negating colonization and ultimately, biofilm formation (244,245). Intracellular iron concentrations are one of the signals for biofilm development in a process involving Fur but not the iron uptake regulatory RNAs asPrrF1 and asPrrF2 (246). Further, high levels of iron suppress the PQS system, release of extracellular DNA and biofilm formation (247).
The link between iron and QS systems in P. aeruginosa is complex. QS systems are enhanced under limiting iron concentrations (85,248,249), and major QS regulators are also involved in regulating iron responsive genes (75,250,251). MvfR, for example, has been demonstrated to control transcription of iron-related genes, and it has an iron-starvation box in its promoter, a site recognized by PvdS (PA2426) to turn on transcription under low-iron concentrations (229,252). It was recently demonstrated that iron levels affect activity of the MvfR signalling molecule HAQ, adding another layer of complexity to the role of iron in QS (253). Another major QS and virulence regulator, VqsR regulates phenazine production by modulating phnAB expression (94). Moreover, the small regulatory RNAs asPrrF1 and asPrrF2, which are negatively regulated by Fur, positively regulate PQS production (Figure 1) (254). PQS has been shown to accumulate in the outer membrane and in membrane vesicles (77). PQS chelates iron, and this facilitates pyochelin and pyoverdin in scavenging iron (73,76).
Thus, iron uptake regulation in P. aeruginosa is a complex affair and involves multiple regulators that affect expression of numerous genes either by themselves or through other regulators. In addition, the interconnections between iron uptake mechanisms and other virulence systems, such as QS and biofilm formation, demonstrate the versatility of this bacterium in being able to pragmatically read environmental signals to accordingly modulate gene expression.
Toxins and exoproteins
Exoproteins are an important component of bacterial survival not only because they allow the bacteria to interact with their immediate environment and other organisms in the vicinity but also because they play a critical role in virulence. P. aeruginosa has a large complement of secreted proteins and five (type I, II, III, V and VI) of the seven secretion systems characterized in bacteria (255). A majority of the secreted proteins are toxins that aid in P. aeruginosa virulence, most of which, including LasA, LasB, PrpL, ToxA and phospholipases [PlcH (PA0844), PlcN (PA3319), PlcB (PA0026)], is secreted through the Xcp type II secretion system (T2SS) (255). Effector molecules that are crucial for evading the host phagocytic response are secreted through a dedicated T3SS (256), whereas the type I system (T1SS) secretes the alkaline protease AprA (257,258) and the haemophore HasAp (PA3407) (259). Substrates of the recently identified T6SS are just being discovered (260). In addition, c-di-GMP levels, modulated by the diguanylate cyclase WspR (PA3702) is involved in the switch between T3SS and T6SS independent of RetS but is dependent on rgRsmY and rgRsmZ (Figure 1) (261).
HasAp, a T1SS-secreted haem-uptake protein in P. aeruginosa, is under QS control (250). QS is also known to regulate PrpL that targets the human lactoferrin (see previous section) (240). Thus, QS in P. aeruginosa not only regulates enzymes to degrade the human lactoferrin but also produces proteins to retrieve the iron from the degraded lactoferrin. The other known T1SS substrate, AprA, is regulated by a novel LTTR named BexR (PA2432) that controls bistability in P. aeruginosa (Figure 1) (262). Inactivation of QS has been demonstrated to reduce expression of T2SS-secreted proteases, chitinases and lipases (63,263) because of downregulation of the Xcp T2SS (264,265). The TCS PhoBR (PA5360–PA5361) regulates other T2SS-secreted exoproteins, such as PlcH, PlcC, PlcN and the Hxc T2SS secreted alkaline phosphatase LapA (266,267). Microarray analysis revealed that a novel cell-surface signalling system PUMA3 regulates Hxc T2SS genes (268). The three T6SS systems (HSI-I, HSI-II and HSI-III) in P. aeruginosa are differentially regulated by the QS systems (269). Although LasR and MvfR negatively regulate the HSI-I system, they positively regulate expression of the functionally redundant HSI-II and HSI-III (Figure 1) (269). In addition, a putative regulator Sfa3 (SfnR) in P. aeruginosa PA14 (an orthologue of PA2359 in PAO1) potentially regulates the HSI-III cluster (269). HSI-I expression is also regulated by RetS through RsmA (270).
Pseudomonas aeruginosa T3SS is regulated in a complex and multi-tiered process, and it is probably the most well understood (271). Expression of T3SS is regulated transcriptionally and post-transcriptionally in response to host cell contact and environmental Ca2+ levels (272,273). ExsA (PA1713), an AraC member, regulates expression of the 43 genes that form the T3SS in P. aeruginosa by binding as a monomer to an A-rich 8-bp region upstream of the −35 in the promoter of genes under its regulation (272,47). ExsA autoregulates its own expression and is also activated by PsrA (PA3006), a member of the TetR family (274,275). Two anti-activators [ExsD (PA1714) and PtrA (PA2808)] also regulate ExsA-mediated activation (Figure 1). T3SS transcription is coupled to secretion and involves the anti-activator ExsD and the anti–anti-activator ExsC (PA1710) that regulate ExsA function. Under non-inducing conditions (high Ca2+), ExsE (PA1711) binds the anti–anti-activator ExsC, allowing the anti-activator ExsD to bind ExsA and inhibit transcription. Under Ca2+ delimiting conditions, ExsE is secreted, freeing ExsC to bind ExsD. Free ExsA then activates T3SS expression (Figure 1) (276). Although this is the primary mode of control, T3SS can also be triggered by stress because of DNA damage (RecA-mediated activation of PtrB) (277), high salt (278,279), metabolic stress (123,279,280), alginate regulators AlgT/U, AlgR and MucA (281), the MexEF–OprN efflux pump regulator MexT (PA2492) through PtrC (282) and the RetS/LadS/Gac-Rsm TCSs (21,22,283). Under low oxygen conditions, the anaerobic regulator Anr activates the response regulator NarL (PA3879), which in turn represses rgRsmY and rgRsmZ expression, allowing RsmA to activate T3SS (284). Expression of T3SS, however, happens only in a subset of the population even under inducing conditions (279,285). Multiple levels of control allow fine-tuning of T3SS expression, allowing P. aeruginosa to sense various environmental conditions and regulate expression in conjunction with other virulence factors.
Regulatory RNAs in P. aeruginosa virulence
RNAs other than messenger RNAs, transfer RNAs or ribosomal RNAs are termed small RNAs (sRNAs), and they affect all steps in gene expression pathways in both prokaryotes and eukaryotes (286). In general, sRNA-mediated regulation occurs in one of two ways, base pairing with DNA or mRNA, or by affecting the activity of a protein or protein complex (286). Not surprisingly, virulence gene expression in P. aeruginosa also relies on small rgRNA-mediated post-transcriptional regulation. The importance of rgRNAs in regulation of bacterial virulence is well established (39,287,288). Most of the P. aeruginosa rgRNAs that have been characterized play a role in virulence gene regulation (discussed later in the text). A recent study identified ∼150 novel sRNAs using sRNA-Seq in P. aeruginosa PAO1 and PA14, which includes both strain-specific and shared ones (289).
Perhaps the most well characterized system involves the rgRNAs, rgRsmY and rgRsmZ (whose roles in virulence regulation have been discussed in the ‘TCS’ and ‘Toxins and Exoproteins’ sections). These are two functionally redundant rgRNAs in P. aeruginosa that play a critical role in the switch between acute and chronic infections (21,290). GacA of the GacSA TCS positively regulates expression of rgRsmY and rgRsmZ, which then bind to and sequester the sRNA-binding protein RsmA through the GGA motif (291–293), leading to derepression of the genes that RsmA represses (107,124,294,295). The consequences of RsmA sequestration result in dysregulation of the expression of numerous virulence factors, as discussed in the ‘TCS’ section. Given the importance of this regulatory process in P. aeruginosa pathogenesis, regulation of expression of rgRsmY and rgRsmZ is multi-tiered. The histidine phosphotransfer protein HptB is phosphorylated by a phosphorelay involving the three sensor kinases PA2824, PA1611 and PA1976 (296). Phosphorylated HptB then transfers the phosphate to an anti–anti-σ factor PA3374, to negatively regulate expression of rsmY (296,297). In another mode of regulation, the BfiSR TCS activates expression of the ribonuclease CafA (PA4477), which specifically targets rgRsmZ (298). Further regulation is achieved by the global regulators of the H-NS family of proteins MvaT and MvaU, which bind to AT-rich regions upstream of the rsmZ gene repressing their expression (299). In addition to all this, there is a negative autoregulatory feedback mechanism, the details of which have not been elucidated yet (300). On synthesis, rgRsmY is stabilized by Hfq binding, either alone or in conjunction with RsmA (108,301).
Another example of post-transcriptional regulation by sequestering a RNA-binding protein links virulence with metabolism. Pseudomonas aeruginosa Crc (PA5332) is a RNA-binding protein that recognizes CA-motifs around the ribosome binding sites of the mRNA of carbon compound catabolism genes. Crc thus represses genes whose products help utilize less preferred carbon sources (302–304). When less preferred substrates, such as mannitol, have to be utilized, expression of catabolic genes is achieved by sequestration of Crc by the rgRNA, rgCrcZ (305). Expression of rgCrcZ is under the control of the TCS CbrAB (PA4725–PA4726), which in conjunction with Crc plays a role in carbon compound catabolism, biofilm formation, antibiotic resistance, secretion systems and swarming (306–312).
Pseudomonas aeruginosa antisense sRNAs (asRNAs) can also act by base pairing with target mRNAs, thus inhibiting translation (313). One such example is asPhrS (PA3305.1), which plays a role in PQS and pyocyanin expression (314). Transcriptome studies indicate an extensive overlap between the genes that are positively regulated by the transcriptional regulator PqsR (also known as MvfR, PA1003) and asPhrS, suggesting that asPhrS regulates pqsR mRNA (75,314). Interestingly, it was shown that asPhrS specifically targets a region in the RBS of a small ORF (uof), which is present upstream of PqsR (314). As translation of pqsR and uof are coupled, asPhrS regulates pqsR translation by modulating translation of uof (314). Expression of asPhrS is under the control of the oxygen responsive regulator Anr (314). Hfq controls asPhrS expression indirectly by regulating Anr expression, whose mechanism of action is yet to be elucidated (314,315).
Small asRNAs also play a role in regulation of iron uptake and involve base pairing by the sRNAs asPrrF1 and asPrrF2, which are the P. aeruginosa orthologues of E. coli RyhB (237,316). Expression of asPrrF1 and asPrrF2 is repressed by Fur when iron concentrations are high (237). Under iron-starvation conditions, asPrrF1 and asPrrF2 are expressed and base pair with the mRNA of target genes, which include the superoxide dismutase sodB (PA4366), genes involved in the trichloroacetic acid cycle and anthranilate and cathechol degradation (317). Thus, asPrrF1 and asPrrF2 link carbon metabolism, iron uptake and QS-mediated virulence. Another asRNA gene asPrrH is located in the same locus as asPrrF1 and asPrrF2. The asPrrH asRNA (at 325 nt) is longer than asPrrF1 (116 nt) and asPrrF2 (114 nt), and the coding region of asPrrH overlaps with the asPrrF1 terminator, the intergenic region between asPrrF1 and asPrrF2 and the 5′-end of the asPrrF2 ORF (318). The expression of asPrrH is maximal in the stationary phase of growth, similar to asPrrF1 and asPrrF2, and under iron-deplete conditions (318). Haem represses asPrrH expression, and this involves the outer membrane haem receptors PhuR (PA4710) and HasR (PA3408) (318). Interestingly, under conditions of haem starvation, asPrrH expression leads to the repression of achAB and sdhCDAB, which are also targets of the PrrF asRNAs (318). In addition to these targets, asPrrH also represses NirL, a protein involved in biosynthesis of haem, under haem and iron limitation (318).
CONCLUSIONS AND PERSPECTIVES
Pseudomonas aeruginosa is a versatile bacterium that can thrive in a wide range of habitats. This is achieved by an intricately interlinked regulatory system of transcriptional regulators, σ factors, sRNAs and their regulons. The exquisite control of gene expression is exemplified in the virulence regulatory network (Figure 1), which demonstrates that none of the virulence mechanisms are isolated. Expression of individual virulence networks is under transcriptional and post-transcriptional regulation of multiple-regulatory systems, either directly or indirectly. Furthermore, some signalling cascades inversely regulate the acute and chronic virulence phenotypes depending on the signals sensed. The next critical phase of research should focus on the signals that the bacteria recognizes to achieve gene regulation.
The extent of cross-regulation between the transcriptional regulators highlights the global nature of the regulation, where individual subnetworks (such as the QS network, alginate network and so forth) are interlinked to form a hyperconnected network (Figure 1). Given the complexity of the connections, one can expect the response of a cell to be elaborate even when faced with a simple stress condition. A fundamental point in a network setting is that one should evaluate the role of individual players (such as a regulator) not in isolation, but with the knowledge that the entire network will react to what it does. In other words, local changes can have global effects. This, in turn, results in subtle cause–effect relationships. Studying the functions of a regulator by generating deletion or overexpression strains is often performed under the assumption that other regulators will remain static. In reality, however, such modifications can lead to aberrant changes across the network in ways that were initially unintended. Moreover, there is a possibility that such changes can occur because the network connections might not always be obvious. This can be attributed, in part, to as yet uneludicated implicit players in the network that dictate or otherwise influence cellular response. This is a likely explanation for the many ‘global’ regulators in P. aeruginosa and in similar bacteria that have complex regulatory networks. In such cases, many of the phenotypes observed with single regulator mutant strains can be part of a ripple effect that propagates through the network affecting disparate phenotypes.
A simplified model of gene regulatory network treats genes as being on or off, that is, taking binary values. It is, therefore, no surprise that Boolean networks (discrete dynamical network models) have been used to model and study gene regulatory circuits (319–321). Probabilistic Boolean networks, which take into account molecular and genetic noise (322,323), and stochastic Boolean networks, which permit the modelling of gene perturbations (324), provide important insights into the dynamical behaviour of the system. Although they are computationally complex, they are a valuable addition to the numerous other programs that are available to analyse gene regulatory networks (31,325,326).
Dynamical systems theory helps us to analyse the behaviour of complex systems that can frequently be expressed by time-differential equations. When the behaviour of a dynamical system depends sensitively on small changes in initial conditions, then the system is said to be chaotic, that is, capable of exhibiting chaotic behaviour. Researchers have investigated whether regulatory networks can have subsystems that are capable of exhibiting chaotic behaviour (327). It has been shown that competition between two or more subnetworks of comparable importance can lead to chaos (328–330). In fact, chaos has been shown to be possible in biochemical systems with only two feedback loops, and positive feedback is known to be necessary for chaotic behaviour (331). So, one would expect chaotic subsystems in a regulatory network as complex as the one that controls P. aeruginosa virulence (Figure 1). Despite of this predisposition, gene regulatory networks seldom exhibit chaotic behaviour. This could be because the competitions between opposing nodes are not strong enough (332) or that chaotic behaviours are short-lived because of triggering of other pathways, such as cell–cell communication (333). Another possibility is that the natural random variability of biochemical systems masks the chaotic behaviour (332). However, maintaining a low level of chaos in such a complex network is probably a combination of the aforementioned and, potentially, as yet unknown factors.
In gene regulatory networks, a particular dynamical system is characterized by time-evolving variables (chemical concentrations, gene expression and so forth) and by parameters (temperature, ambient chemical concentrations and so forth). A network can exhibit chaotic or non-chaotic behaviour depending on the parameters that influence it (334). Environmental factors, such as the temperature or the nutritional status of the cells, parameterize the relationship between transcription factors and the genes that they regulate. Although it is understood that some choices of parameters can induce chaotic behaviours, the parameter may not even be achievable, such as high temperatures (334). Mutations can also alter relationships in regulatory networks by causing changes in existing links or forming new ones. In a dynamically robust (non-chaotic) system, small finite changes in the parameters lead to only qualitative changes in the dynamical behaviour. However, there are boundaries in the parameter space where the behaviour of the system changes qualitatively and may include the possibility of chaotic dynamics. Predicting whether a network will be stable or chaos-prone under some conditions has been proven to be difficult and remains poorly characterized. Recent work has identified the minimum number, types and interactions among three and four nodes/subnetworks that can lead to chaos in a gene regulatory network (332). Such minimal subnetworks have been termed ‘chaotic motifs’, and networks with these motifs can exhibit chaotic behaviour under the right parameters (332). Analysis of the network in Figure 1 does not readily show such chaotic motifs. This could be because the network is incomplete (lack of data on the interactions among the P. aeruginosa regulators) or because of errors in the inferred interactions. Although P. aeruginosa virulence regulation has been extensively studied, there is yet much to learn. Thus, absence of empirical evidence does not preclude a propensity to chaos and is worth further investigation.
Depending on an elaborate network to achieve gene regulation is likely an adaptive mechanism by P. aeruginosa. Possessing alternate pathways to regulate the same phenotype ensures a rapid response to stimuli even if one of the pathways is affected, thus enhancing survival. Such examples can be seen throughout the network. As discussed in the ‘Toxins and Exoproteins’ section, expression of T3SS genes can be regulated at multiple levels, in response to various different signals and stress conditions, and it is not entirely dependent on any one signal. However, the extent of contributions of the individual regulators and, consequently, the fine balance that exists in some regulatory cascades within the network, are sometimes not easily apparent. Network dependence is also a probable reason for regulator genes being non-essential, in the sense that deleting a transcriptional regulator gene typically does not affect cell viability because of the presence of alternate regulatory mechanisms. Having key regulators modulate different related phenotypes has the added advantage in allowing the cells to adapt to external signals by modulating one or a few regulators instead of individually regulating different virulence systems. A case in point is AmpR that positively regulates acute virulence factors while downregulating chronic infection phenotypes (23). Also of importance is the co-regulation of metabolism and virulence. Studies have identified regulators like CbrB that, with its cognate sensor CbrA, not only regulate carbon metabolism but also virulence phenotypes through the rgRNA, rgCrcZ and the RNA-binding protein Crc (306,307). Moreover, there is crosstalk between CbrA and regulators other than CbrB, highlighting the complexity of the system (306).
The plethora of transcriptome studies using microarrays or deep sequencing will add to the database of genes that are differentially expressed in response to regulator mutations or specific growth conditions. Differentiating the direct effect of a change from a ripple effect can, at least partly, be achieved by meta-analysis studies that look at multiple transcriptomes, identifying effects unique to each condition and differentiating them from the so-called ripple (32,33). Network analyses can help us understand the relationship between different regulators, group them based on function and, more importantly, help identify critical nodes and prominent players. This can serve as a means of target identification in attempting to deal with P. aeruginosa infections. In Figure 1, we see that some parts of the network are more densely connected than others, with central cores containing most of the links. A case in point is LasR of the QS subnetwork. It is well known that QS is central to virulence regulation in P. aeruginosa and targeting key regulators will have a better chance of therapeutic success. Recently, inhibitors of a key QS regulator were shown to reduce pathogenicity in Vibrio cholera (335).
With the extensive use of high-throughput transcriptomics, gene regulation studies are now focusing on the role of non-coding RNAs in bacteria. rgRNAs have been shown to be extensively involved in gene regulation in P. aeruginosa and other bacteria (124,237,299,305,314,336). Techniques such as RNA-seq allow for the entire transcriptome to be sequenced, giving us an unprecedented insight into non-coding RNAs, asRNAs and sRNAs involved in regulation. Preliminary studies using prediction software and complementary experiments have already advanced our understanding (29,108,124,299,314,337). Given the many different ways in which small RNAs can modulate gene expression (313) and potentially undiscovered ones, we can look forward to exciting new discoveries in bacterial gene regulation in the coming years.
FUNDING
National Institutes of Health-Minority Medical Research Support SCORE [S06 GM08205 and 5SC1AI081376 to K.M.]; FIU Research Assistantship (Herbert Werthiem College of Medicine to D.B.). Funding for open access charge: National Institutes of Health-Minority Medical Research Support SCORE [5SC1AI081376 to K.M.].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors are extremely grateful to Giri Narasimhan (Florida International University) and Edward Celarier (NASA Goddard) for extensive discussions and critical comments on chaos theory.
REFERENCES
- 1.Rahme LG, Ausubel FM, Cao H, Drenkard E, Goumnerov BC, Lau GW, Mahajan-Miklos S, Plotnikova J, Tan MW, Tsongalis J, et al. Plants and animals share functionally common bacterial virulence factors. Proc. Natl Acad. Sci. USA. 2000;97:8815–8821. doi: 10.1073/pnas.97.16.8815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mahajan-Miklos S, Rahme LG, Ausubel FM. Elucidating the molecular mechanisms of bacterial virulence using non-mammalian hosts. Mol. Microbiol. 2000;37:981–988. doi: 10.1046/j.1365-2958.2000.02056.x. [DOI] [PubMed] [Google Scholar]
- 3.Kerr KG, Snelling AM. Pseudomonas aeruginosa: a formidable and ever-present adversary. J. Hosp. Infect. 2009;73:338–344. doi: 10.1016/j.jhin.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 4.Valderrey AD, Pozuelo MJ, Jimenez PA, Macia MD, Oliver A, Rotger R. Chronic colonization by Pseudomonas aeruginosa of patients with obstructive lung diseases: cystic fibrosis, bronchiectasis, and chronic obstructive pulmonary disease. Diagn. Microbiol. Infect. Dis. 2010;68:20–27. doi: 10.1016/j.diagmicrobio.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 5.Bouza E, Burillo A, Munoz P. Catheter-related infections: diagnosis and intravascular treatment. Clin. Microbiol. Infect. 2002;8:265–274. doi: 10.1046/j.1469-0691.2002.00385.x. [DOI] [PubMed] [Google Scholar]
- 6.Manfredi R, Nanetti A, Ferri M, Chiodo F. Pseudomonas spp. complications in patients with HIV disease: an eight-year clinical and microbiological survey. Eur. J. Epidemiol. 2000;16:111–118. doi: 10.1023/a:1007626410724. [DOI] [PubMed] [Google Scholar]
- 7.Mizuno T. Compilation of all genes encoding two-component phosphotransfer signal transducers in the genome of Escherichia coli. DNA Res. 1997;4:161–168. doi: 10.1093/dnares/4.2.161. [DOI] [PubMed] [Google Scholar]
- 8.Fabret C, Feher VA, Hoch JA. Two-component signal transduction in Bacillus subtilis: how one organism sees its world. J. Bacteriol. 1999;181:1975–1983. doi: 10.1128/jb.181.7.1975-1983.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gooderham WJ, Hancock RE. Regulation of virulence and antibiotic resistance by two-component regulatory systems in Pseudomonas aeruginosa. FEMS Microbiol. Rev. 2009;33:279–294. doi: 10.1111/j.1574-6976.2008.00135.x. [DOI] [PubMed] [Google Scholar]
- 10.Buelow DR, Raivio TL. Three (and more) component regulatory systems - auxiliary regulators of bacterial histidine kinases. Mol. Microbiol. 2010;75:547–566. doi: 10.1111/j.1365-2958.2009.06982.x. [DOI] [PubMed] [Google Scholar]
- 11.Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harmsen M, Yang L, Pamp SJ, Tolker-Nielsen T. An update on Pseudomonas aeruginosa biofilm formation, tolerance, and dispersal. FEMS Immunol. Med. Microbiol. 2010;59:253–268. doi: 10.1111/j.1574-695X.2010.00690.x. [DOI] [PubMed] [Google Scholar]
- 13.Njoroge J, Sperandio V. Jamming bacterial communication: new approaches for the treatment of infectious diseases. EMBO Mol. Med. 2009;1:201–210. doi: 10.1002/emmm.200900032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burns JL, Gibson RL, McNamara S, Yim D, Emerson J, Rosenfeld M, Hiatt P, McCoy K, Castile R, Smith AL, et al. Longitudinal assessment of Pseudomonas aeruginosa in young children with cystic fibrosis. J. Infect. Dis. 2001;183:444–452. doi: 10.1086/318075. [DOI] [PubMed] [Google Scholar]
- 15.Hogardt M, Heesemann J. Microevolution of Pseudomonas aeruginosa to a chronic pathogen of the cystic fibrosis lung. Curr. Top. Microbiol. Immunol. 2012 doi: 10.1007/82_2011_199. Feb 8 (doi: 10.1007/82_2011_199; epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 16.Coggan KA, Wolfgang MC. Global regulatory pathways and cross-talk control Pseudomonas aeruginosa environmental lifestyle and virulence phenotype. Curr. Issues Mol. Biol. 2012;14:47–70. [PMC free article] [PubMed] [Google Scholar]
- 17.Hoboth C, Hoffmann R, Eichner A, Henke C, Schmoldt S, Imhof A, Heesemann J, Hogardt M. Dynamics of adaptive microevolution of hypermutable Pseudomonas aeruginosa during chronic pulmonary infection in patients with cystic fibrosis. J. Infect. Dis. 2009;200:118–130. doi: 10.1086/599360. [DOI] [PubMed] [Google Scholar]
- 18.Hogardt M, Hoboth C, Schmoldt S, Henke C, Bader L, Heesemann J. Stage-specific adaptation of hypermutable Pseudomonas aeruginosa isolates during chronic pulmonary infection in patients with cystic fibrosis. J. Infect. Dis. 2007;195:70–80. doi: 10.1086/509821. [DOI] [PubMed] [Google Scholar]
- 19.Oliver A, Mena A. Bacterial hypermutation in cystic fibrosis, not only for antibiotic resistance. Clin. Microbiol. Infect. 2010;16:798–808. doi: 10.1111/j.1469-0691.2010.03250.x. [DOI] [PubMed] [Google Scholar]
- 20.Hassett DJ, Sutton MD, Schurr MJ, Herr AB, Caldwell CC, Matu JO. Pseudomonas aeruginosa hypoxic or anaerobic biofilm infections within cystic fibrosis airways. Trends Microbiol. 2009;17:130–138. doi: 10.1016/j.tim.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 21.Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell. 2004;7:745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 22.Ventre I, Goodman AL, Vallet-Gely I, Vasseur P, Soscia C, Molin S, Bleves S, Lazdunski A, Lory S, Filloux A. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc. Natl Acad. Sci. USA. 2006;103:171–176. doi: 10.1073/pnas.0507407103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balasubramanian D, Schneper L, Merighi M, Smith R, Narasimhan G, Lory S, Mathee K. The regulatory repertoire of Pseudomonas aeruginosa AmpC ß-lactamase regulator AmpR includes virulence genes. PLoS One. 2012;7:e34067. doi: 10.1371/journal.pone.0034067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, et al. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 25.Mathee K, Narasimhan G, Valdes C, Qiu X, Matewish JM, Koehrsen M, Rokas A, Yandava CN, Engels R, Zeng E, et al. Dynamics of Pseudomonas aeruginosa genome evolution. Proc. Natl Acad. Sci. USA. 2008;105:3100–3105. doi: 10.1073/pnas.0711982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winsor GL, Lam DK, Fleming L, Lo R, Whiteside MD, Yu NY, Hancock RE, Brinkman FS. Pseudomonas genome database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 2011;39:D596–D600. doi: 10.1093/nar/gkq869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balasubramanian D, Murugapiran SK, Silva-Herzog E, Schneper L, Yang X, Tatke G, Narasimhan G, Mathee K. In: Bacterial Gene Regulation and Transcriptional Networks. Babu MM, editor. United Kingdom: Caiser Academic Press; 2013. [Google Scholar]
- 28.Potvin E, Sanschagrin F, Levesque RC. Sigma factors in Pseudomonas aeruginosa. FEMS Microbiol. Rev. 2008;32:38–55. doi: 10.1111/j.1574-6976.2007.00092.x. [DOI] [PubMed] [Google Scholar]
- 29.Livny J, Brencic A, Lory S, Waldor MK. Identification of 17 Pseudomonas aeruginosa sRNAs and prediction of sRNA-encoding genes in 10 diverse pathogens using the bioinformatic tool sRNAPredict2. Nucleic Acids Res. 2006;34:3484–3493. doi: 10.1093/nar/gkl453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galan-Vasquez E, Luna B, Martinez-Antonio A. The regulatory network of Pseudomonas aeruginosa. Microb. Inform. Exp. 2011;1:3. doi: 10.1186/2042-5783-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Babu MM, Teichmann SA, Aravind L. Evolutionary dynamics of prokaryotic transcriptional regulatory networks. J. Mol. Biol. 2006;358:614–633. doi: 10.1016/j.jmb.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 32.Balasubramanian D, Mathee K. Comparative transcriptome analyses of Pseudomonas aeruginosa. Hum. Genomics. 2009;3:349–361. doi: 10.1186/1479-7364-3-4-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goodman AL, Lory S. Analysis of regulatory networks in Pseudomonas aeruginosa by genomewide transcriptional profiling. Curr. Opin. Microbiol. 2004;7:39–44. doi: 10.1016/j.mib.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 34.Ong CT, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat. Rev. Genet. 2011;12:283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levine M. Transcriptional enhancers in animal development and evolution. Curr. Biol. 2010;20:R754–R763. doi: 10.1016/j.cub.2010.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144:327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ishihama A. Prokaryotic genome regulation: multifactor promoters, multitarget regulators and hierarchic networks. FEMS Microbiol. Rev. 2010;34:628–645. doi: 10.1111/j.1574-6976.2010.00227.x. [DOI] [PubMed] [Google Scholar]
- 38.Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell. 2011;43:880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reitzer LJ, Magasanik B. Transcription of glnA in E. coli is stimulated by activator bound to sites far from the promoter. Cell. 1986;45:785–792. doi: 10.1016/0092-8674(86)90553-2. [DOI] [PubMed] [Google Scholar]
- 41.Popham DL, Szeto D, Keener J, Kustu S. Function of a bacterial activator protein that binds to transcriptional enhancers. Science. 1989;243:629–635. doi: 10.1126/science.2563595. [DOI] [PubMed] [Google Scholar]
- 42.Buck M, Gallegos MT, Studholme DJ, Guo Y, Gralla JD. The bacterial enhancer-dependent sigma(54) (sigma(N)) transcription factor. J. Bacteriol. 2000;182:4129–4136. doi: 10.1128/jb.182.15.4129-4136.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mohr CD, Leveau JH, Krieg DP, Hibler NS, Deretic V. AlgR-binding sites within the algD promoter make up a set of inverted repeats separated by a large intervening segment of DNA. J. Bacteriol. 1992;174:6624–6633. doi: 10.1128/jb.174.20.6624-6633.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramsey DM, Baynham PJ, Wozniak DJ. Binding of Pseudomonas aeruginosa AlgZ to sites upstream of the algZ promoter leads to repression of transcription. J. Bacteriol. 2005;187:4430–4443. doi: 10.1128/JB.187.13.4430-4443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winteler HV, Haas D. The homologous regulators ANR of Pseudomonas aeruginosa and FNR of Escherichia coli have overlapping but distinct specificities for anaerobically inducible promoters. Microbiology. 1996;142:685–693. doi: 10.1099/13500872-142-3-685. [DOI] [PubMed] [Google Scholar]
- 46.Lu CD, Winteler H, Abdelal A, Haas D. The ArgR regulatory protein, a helper to the anaerobic regulator ANR during transcriptional activation of the arcD promoter in Pseudomonas aeruginosa. J. Bacteriol. 1999;181:2459–2464. doi: 10.1128/jb.181.8.2459-2464.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hovey AK, Frank DW. Analyses of the DNA-binding and transcriptional activation properties of ExsA, the transcriptional activator of the Pseudomonas aeruginosa exoenzyme S regulon. J. Bacteriol. 1995;177:4427–4436. doi: 10.1128/jb.177.15.4427-4436.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baraquet C, Murakami K, Parsek MR, Harwood CS. The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c-di-GMP. Nucleic Acids Res. 2012;40:7207–7218. doi: 10.1093/nar/gks384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ochsner UA, Vasil AI, Vasil ML. Role of the ferric uptake regulator of Pseudomonas aeruginosa in the regulation of siderophores and exotoxin A expression: purification and activity on iron-regulated promoters. J. Bacteriol. 1995;177:7194–7201. doi: 10.1128/jb.177.24.7194-7201.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 2003;185:2080–2095. doi: 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tian ZX, Fargier E, Mac Aogain M, Adams C, Wang YP, O'Gara F. Transcriptome profiling defines a novel regulon modulated by the LysR-type transcriptional regulator MexT in Pseudomonas aeruginosa. Nucleic Acids Res. 2009;37:7546–7559. doi: 10.1093/nar/gkp828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiao G, He J, Rahme LG. Mutation analysis of the Pseudomonas aeruginosa mvfR and pqsABCDE gene promoters demonstrates complex quorum-sensing circuitry. Microbiology. 2006;152:1679–1686. doi: 10.1099/mic.0.28605-0. [DOI] [PubMed] [Google Scholar]
- 53.Kojic M, Aguilar C, Venturi V. TetR family member psrA directly binds the Pseudomonas rpoS and psrA promoters. J. Bacteriol. 2002;184:2324–2330. doi: 10.1128/JB.184.8.2324-2330.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nicastro GG, Boechat AL, Abe CM, Kaihami GH, Baldini RL. Pseudomonas aeruginosa PA14 cupD transcription is activated by the RcsB response regulator, but repressed by its putative cognate sensor RcsC. FEMS Microbiol. Lett. 2009;301:115–123. doi: 10.1111/j.1574-6968.2009.01803.x. [DOI] [PubMed] [Google Scholar]
- 55.Pearson JP, Pesci EC, Iglewski BH. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 1997;179:5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dasgupta N, Ferrell EP, Kanack KJ, West SE, Ramphal R. fleQ, the gene encoding the major flagellar regulator of Pseudomonas aeruginosa, is sigma70 dependent and is downregulated by Vfr, a homolog of Escherichia coli cyclic AMP receptor protein. J. Bacteriol. 2002;184:5240–5250. doi: 10.1128/JB.184.19.5240-5250.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liang H, Deng X, Ji Q, Sun F, Shen T, He C. The Pseudomonas aeruginosa global regulator VqsR directly inhibits QscR to control quorum-sensing and virulence gene expression. J. Bacteriol. 2012;194:3098–3108. doi: 10.1128/JB.06679-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nealson KH, Platt T, Hastings JW. Cellular control of the synthesis and activity of the bacterial luminescent system. J. Bacteriol. 1970;104:313–322. doi: 10.1128/jb.104.1.313-322.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stevens AM, Schuster M, Rumbaugh KP. Working together for the common good: cell-cell communication in bacteria. J. Bacteriol. 2012;194:2131–2141. doi: 10.1128/JB.00143-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Williams P, Camara M. Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: a tale of regulatory networks and multifunctional signal molecules. Curr. Opin. Microbiol. 2009;12:182–191. doi: 10.1016/j.mib.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 61.Jimenez PN, Koch G, Thompson JA, Xavier KB, Cool RH, Quax WJ. The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 2012;76:46–65. doi: 10.1128/MMBR.05007-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schuster M, Greenberg EP. A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int. J. Med. Microbiol. 2006;296:73–81. doi: 10.1016/j.ijmm.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 63.Passador L, Cook JM, Gambello MJ, Rust L, Iglewski BH. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science. 1993;260:1127–1130. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- 64.Pearson JP, Gray KM, Passador L, Tucker KD, Eberhard A, Iglewski BH, Greenberg EP. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc. Natl Acad. Sci. USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brint JM, Ohman DE. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J. Bacteriol. 1995;177:7155–7163. doi: 10.1128/jb.177.24.7155-7163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Latifi A, Winson MK, Foglino M, Bycroft BW, Stewart GS, Lazdunski A, Williams P. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol. Microbiol. 1995;17:333–343. doi: 10.1111/j.1365-2958.1995.mmi_17020333.x. [DOI] [PubMed] [Google Scholar]
- 67.Ochsner UA, Reiser J. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA. 1995;92:6424–6428. doi: 10.1073/pnas.92.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pearson JP, Passador L, Iglewski BH, Greenberg EP. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA. 1995;92:1490–1494. doi: 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kiratisin P, Tucker KD, Passador L. LasR, a transcriptional activator of Pseudomonas aeruginosa virulence genes, functions as a multimer. J. Bacteriol. 2002;184:4912–4919. doi: 10.1128/JB.184.17.4912-4919.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lamb JR, Patel H, Montminy T, Wagner VE, Iglewski BH. Functional domains of the RhlR transcriptional regulator of Pseudomonas aeruginosa. J. Bacteriol. 2003;185:7129–7139. doi: 10.1128/JB.185.24.7129-7139.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schuster M, Urbanowski ML, Greenberg EP. Promoter specificity in Pseudomonas aeruginosa quorum sensing revealed by DNA binding of purified LasR. Proc. Natl Acad. Sci. USA. 2004;101:15833–15839. doi: 10.1073/pnas.0407229101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schuster M, Greenberg EP. Early activation of quorum sensing in Pseudomonas aeruginosa reveals the architecture of a complex regulon. BMC Genomics. 2007;8:287. doi: 10.1186/1471-2164-8-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Diggle SP, Matthijs S, Wright VJ, Fletcher MP, Chhabra SR, Lamont IL, Kong X, Hider RC, Cornelis P, Camara M, et al. The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem. Biol. 2007;14:87–96. doi: 10.1016/j.chembiol.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 74.Xiao G, Deziel E, He J, Lepine F, Lesic B, Castonguay MH, Milot S, Tampakaki AP, Stachel SE, Rahme LG. MvfR, a key Pseudomonas aeruginosa pathogenicity LTTR-class regulatory protein, has dual ligands. Mol. Microbiol. 2006;62:1689–1699. doi: 10.1111/j.1365-2958.2006.05462.x. [DOI] [PubMed] [Google Scholar]
- 75.Deziel E, Gopalan S, Tampakaki AP, Lepine F, Padfield KE, Saucier M, Xiao G, Rahme LG. The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation: multiple quorum sensing-regulated genes are modulated without affecting lasRI, rhlRI or the production of N-acyl-L-homoserine lactones. Mol. Microbiol. 2005;55:998–1014. doi: 10.1111/j.1365-2958.2004.04448.x. [DOI] [PubMed] [Google Scholar]
- 76.Bredenbruch F, Geffers R, Nimtz M, Buer J, Haussler S. The Pseudomonas aeruginosa quinolone signal (PQS) has an iron-chelating activity. Environ. Microbiol. 2006;8:1318–1329. doi: 10.1111/j.1462-2920.2006.01025.x. [DOI] [PubMed] [Google Scholar]
- 77.Mashburn LM, Whiteley M. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature. 2005;437:422–425. doi: 10.1038/nature03925. [DOI] [PubMed] [Google Scholar]
- 78.Schertzer JW, Whiteley M. A bilayer-couple model of bacterial outer membrane vesicle biogenesis. MBio. 2012;3:e00297–11. doi: 10.1128/mBio.00297-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liang H, Li L, Dong Z, Surette MG, Duan K. The YebC family protein PA0964 negatively regulates the Pseudomonas aeruginosa quinolone signal system and pyocyanin production. J. Bacteriol. 2008;190:6217–6227. doi: 10.1128/JB.00428-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wagner VE, Li LL, Isabella VM, Iglewski BH. Analysis of the hierarchy of quorum-sensing regulation in Pseudomonas aeruginosa. Anal. Bioanal. Chem. 2007;387:469–479. doi: 10.1007/s00216-006-0964-6. [DOI] [PubMed] [Google Scholar]
- 81.Latifi A, Foglino M, Tanaka K, Williams P, Lazdunski A. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhIR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol. Microbiol. 1996;21:1137–1146. doi: 10.1046/j.1365-2958.1996.00063.x. [DOI] [PubMed] [Google Scholar]
- 82.McGrath S, Wade DS, Pesci EC. Dueling quorum sensing systems in Pseudomonas aeruginosa control the production of the Pseudomonas quinolone signal (PQS) FEMS Microbiol. Lett. 2004;230:27–34. doi: 10.1016/S0378-1097(03)00849-8. [DOI] [PubMed] [Google Scholar]
- 83.Pesci EC, Pearson JP, Seed PC, Iglewski BH. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 1997;179:3127–3132. doi: 10.1128/jb.179.10.3127-3132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dekimpe V, Deziel E. Revisiting the quorum-sensing hierarchy in Pseudomonas aeruginosa: the transcriptional regulator RhlR regulates LasR-specific factors. Microbiology. 2009;155:712–723. doi: 10.1099/mic.0.022764-0. [DOI] [PubMed] [Google Scholar]
- 85.Duan K, Surette MG. Environmental regulation of Pseudomonas aeruginosa PAO1 Las and Rhl quorum-sensing systems. J. Bacteriol. 2007;189:4827–4836. doi: 10.1128/JB.00043-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schuster M, Hawkins AC, Harwood CS, Greenberg EP. The Pseudomonas aeruginosa RpoS regulon and its relationship to quorum sensing. Mol. Microbiol. 2004;51:973–985. doi: 10.1046/j.1365-2958.2003.03886.x. [DOI] [PubMed] [Google Scholar]
- 87.Gilbert KB, Kim TH, Gupta R, Greenberg EP, Schuster M. Global position analysis of the Pseudomonas aeruginosa quorum-sensing transcription factor LasR. Mol. Microbiol. 2009;73:1072–1085. doi: 10.1111/j.1365-2958.2009.06832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Castang S, McManus HR, Turner KH, Dove SL. H-NS family members function coordinately in an opportunistic pathogen. Proc. Natl Acad. Sci. USA. 2008;105:18947–18952. doi: 10.1073/pnas.0808215105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rampioni G, Schuster M, Greenberg EP, Bertani I, Grasso M, Venturi V, Zennaro E, Leoni L. RsaL provides quorum sensing homeostasis and functions as a global regulator of gene expression in Pseudomonas aeruginosa. Mol. Microbiol. 2007;66:1557–1565. doi: 10.1111/j.1365-2958.2007.06029.x. [DOI] [PubMed] [Google Scholar]
- 90.Rampioni G, Schuster M, Greenberg EP, Zennaro E, Leoni L. Contribution of the RsaL global regulator to Pseudomonas aeruginosa virulence and biofilm formation. FEMS Microbiol. Lett. 2009;301:210–217. doi: 10.1111/j.1574-6968.2009.01817.x. [DOI] [PubMed] [Google Scholar]
- 91.Wei Q, Le Minh PN, Dotsch A, Hildebrand F, Panmanee W, Elfarash A, Schulz S, Plaisance S, Charlier D, Hassett D, et al. Global regulation of gene expression by OxyR in an important human opportunistic pathogen. Nucleic Acids Res. 2012;40:4320–4333. doi: 10.1093/nar/gks017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dieppois G, Ducret V, Caille O, Perron K. The transcriptional regulator CzcR modulates antibiotic resistance and quorum sensing in Pseudomonas aeruginosa. PLoS One. 2012;7:e38148. doi: 10.1371/journal.pone.0038148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Perron K, Caille O, Rossier C, Van Delden C, Dumas JL, Kohler T. CzcR-CzcS, a two-component system involved in heavy metal and carbapenem resistance in Pseudomonas aeruginosa. J. Biol. Chem. 2004;279:8761–8768. doi: 10.1074/jbc.M312080200. [DOI] [PubMed] [Google Scholar]
- 94.Juhas M, Wiehlmann L, Huber B, Jordan D, Lauber J, Salunkhe P, Limpert AS, von Gotz F, Steinmetz I, Eberl L, et al. Global regulation of quorum sensing and virulence by VqsR in Pseudomonas aeruginosa. Microbiology. 2004;150:831–841. doi: 10.1099/mic.0.26906-0. [DOI] [PubMed] [Google Scholar]
- 95.Li LL, Malone JE, Iglewski BH. Regulation of the Pseudomonas aeruginosa quorum-sensing regulator VqsR. J. Bacteriol. 2007;189:4367–4374. doi: 10.1128/JB.00007-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liang H, Deng X, Ji Q, Sun F, Shen T, He C. The Pseudomonas aeruginosa global regulator VqsR directly inhibits QscR to control quorum-sensing and virulence gene expression. J. Bacteriol. 2012;194:3098–3108. doi: 10.1128/JB.06679-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lee JH, Lequette Y, Greenberg EP. Activity of purified QscR, a Pseudomonas aeruginosa orphan quorum-sensing transcription factor. Mol. Microbiol. 2006;59:602–609. doi: 10.1111/j.1365-2958.2005.04960.x. [DOI] [PubMed] [Google Scholar]
- 98.Lequette Y, Lee JH, Ledgham F, Lazdunski A, Greenberg EP. A distinct QscR regulon in the Pseudomonas aeruginosa quorum-sensing circuit. J. Bacteriol. 2006;188:3365–3370. doi: 10.1128/JB.188.9.3365-3370.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ledgham F, Ventre I, Soscia C, Foglino M, Sturgis JN, Lazdunski A. Interactions of the quorum sensing regulator QscR: interaction with itself and the other regulators of Pseudomonas aeruginosa LasR and RhlR. Mol. Microbiol. 2003;48:199–210. doi: 10.1046/j.1365-2958.2003.03423.x. [DOI] [PubMed] [Google Scholar]
- 100.Chugani SA, Whiteley M, Lee KM, D'Argenio D, Manoil C, Greenberg EP. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA. 2001;98:2752–2757. doi: 10.1073/pnas.051624298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dong YH, Zhang XF, Xu JL, Tan AT, Zhang LH. VqsM, a novel AraC-type global regulator of quorum-sensing signalling and virulence in Pseudomonas aeruginosa. Mol. Microbiol. 2005;58:552–564. doi: 10.1111/j.1365-2958.2005.04851.x. [DOI] [PubMed] [Google Scholar]
- 102.Deziel E, Lepine F, Milot S, He J, Mindrinos MN, Tompkins RG, Rahme LG. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc. Natl Acad. Sci. USA. 2004;101:1339–1344. doi: 10.1073/pnas.0307694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gallagher LA, McKnight SL, Kuznetsova MS, Pesci EC, Manoil C. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J. Bacteriol. 2002;184:6472–6480. doi: 10.1128/JB.184.23.6472-6480.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Farrow JM, 3rd, Pesci EC. Two distinct pathways supply anthranilate as a precursor of the Pseudomonas quinolone signal. J. Bacteriol. 2007;189:3425–3433. doi: 10.1128/JB.00209-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chugani S, Greenberg EP. LuxR homolog-independent gene regulation by acyl-homoserine lactones in Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA. 2010;107:10673–10678. doi: 10.1073/pnas.1005909107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Siehnel R, Traxler B, An DD, Parsek MR, Schaefer AL, Singh PK. A unique regulator controls the activation threshold of quorum-regulated genes in Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA. 2010;107:7916–7921. doi: 10.1073/pnas.0908511107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pessi G, Williams F, Hindle Z, Heurlier K, Holden MT, Camara M, Haas D, Williams P. The global posttranscriptional regulator RsmA modulates production of virulence determinants and N-acylhomoserine lactones in Pseudomonas aeruginosa. J. Bacteriol. 2001;183:6676–6683. doi: 10.1128/JB.183.22.6676-6683.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sonnleitner E, Schuster M, Sorger-Domenigg T, Greenberg EP, Blasi U. Hfq-dependent alterations of the transcriptome profile and effects on quorum sensing in Pseudomonas aeruginosa. Mol. Microbiol. 2006;59:1542–1558. doi: 10.1111/j.1365-2958.2006.05032.x. [DOI] [PubMed] [Google Scholar]
- 109.Kong KF, Jayawardena SR, Indulkar SD, Del Puerto A, Koh CL, Hoiby N, Mathee K. Pseudomonas aeruginosa AmpR is a global transcriptional factor that regulates expression of AmpC and PoxB beta-lactamases, proteases, quorum sensing, and other virulence factors. Antimicrob. Agents Chemother. 2005;49:4567–4575. doi: 10.1128/AAC.49.11.4567-4575.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Balasubramanian D, Kong KF, Jayawardena SR, Leal SM, Sautter RT, Mathee K. Co-regulation of ß-lactam resistance, alginate production and quorum sensing in Pseudomonas aeruginosa. J. Med. Microbiol. 2011;60:147–156. doi: 10.1099/jmm.0.021600-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rodrigue A, Quentin Y, Lazdunski A, Mejean V, Foglino M. Two-component systems in Pseudomonas aeruginosa: why so many? Trends Microbiol. 2000;8:498–504. doi: 10.1016/s0966-842x(00)01833-3. [DOI] [PubMed] [Google Scholar]
- 112.Raghavan V, Groisman EA. Orphan and hybrid two-component system proteins in health and disease. Curr. Opin. Microbiol. 2010;13:226–231. doi: 10.1016/j.mib.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu. Rev. Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 114.Laub MT, Goulian M. Specificity in two-component signal transduction pathways. Annu. Rev. Genet. 2007;41:121–145. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- 115.Mascher T, Helmann JD, Unden G. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol. Mol. Biol. Rev. 2006;70:910–938. doi: 10.1128/MMBR.00020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gao R, Stock AM. Molecular strategies for phosphorylation-mediated regulation of response regulator activity. Curr. Opin. Microbiol. 2010;13:160–167. doi: 10.1016/j.mib.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mitrophanov AY, Groisman EA. Signal integration in bacterial two-component regulatory systems. Genes Dev. 2008;22:2601–2611. doi: 10.1101/gad.1700308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kitten T, Kinscherf TG, McEvoy JL, Willis DK. A newly identified regulator is required for virulence and toxin production in Pseudomonas syringae. Mol. Microbiol. 1998;28:917–929. doi: 10.1046/j.1365-2958.1998.00842.x. [DOI] [PubMed] [Google Scholar]
- 119.Coleman FT, Mueschenborn S, Meluleni G, Ray C, Carey VJ, Vargas SO, Cannon CL, Ausubel FM, Pier GB. Hypersusceptibility of cystic fibrosis mice to chronic Pseudomonas aeruginosa oropharyngeal colonization and lung infection. Proc. Natl Acad. Sci. USA. 2003;100:1949–1954. doi: 10.1073/pnas.0437901100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hrabak EM, Willis DK. The lemA gene required for pathogenicity of Pseudomonas syringae pv. syringae on bean is a member of a family of two-component regulators. J. Bacteriol. 1992;174:3011–3020. doi: 10.1128/jb.174.9.3011-3020.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Goodman AL, Merighi M, Hyodo M, Ventre I, Filloux A, Lory S. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev. 2009;23:249–259. doi: 10.1101/gad.1739009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Heeb S, Haas D. Regulatory roles of the GacS/GacA two-component system in plant-associated and other Gram-negative bacteria. Mol. Plant Microbe Interact. 2001;14:1351–1363. doi: 10.1094/MPMI.2001.14.12.1351. [DOI] [PubMed] [Google Scholar]
- 123.Wolfgang MC, Lee VT, Gilmore ME, Lory S. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev. Cell. 2003;4:253–263. doi: 10.1016/s1534-5807(03)00019-4. [DOI] [PubMed] [Google Scholar]
- 124.Brencic A, Lory S. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol. Microbiol. 2009;72:612–632. doi: 10.1111/j.1365-2958.2009.06670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Linares JF, Gustafsson I, Baquero F, Martinez JL. Antibiotics as intermicrobial signaling agents instead of weapons. Proc. Natl Acad. Sci. USA. 2006;103:19484–19489. doi: 10.1073/pnas.0608949103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.McPhee JB, Bains M, Winsor G, Lewenza S, Kwasnicka A, Brazas MD, Brinkman FS, Hancock RE. Contribution of the PhoP-PhoQ and PmrA-PmrB two-component regulatory systems to Mg2+-induced gene regulation in Pseudomonas aeruginosa. J. Bacteriol. 2006;188:3995–4006. doi: 10.1128/JB.00053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Macfarlane EL, Kwasnicka A, Hancock RE. Role of Pseudomonas aeruginosa PhoP-PhoQ in resistance to antimicrobial cationic peptides and aminoglycosides. Microbiology. 2000;146:2543–2554. doi: 10.1099/00221287-146-10-2543. [DOI] [PubMed] [Google Scholar]
- 128.Gooderham WJ, Gellatly SL, Sanschagrin F, McPhee JB, Bains M, Cosseau C, Levesque RC, Hancock RE. The sensor kinase PhoQ mediates virulence in Pseudomonas aeruginosa. Microbiology. 2009;155:699–711. doi: 10.1099/mic.0.024554-0. [DOI] [PubMed] [Google Scholar]
- 129.McPhee JB, Lewenza S, Hancock RE. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol. Microbiol. 2003;50:205–217. doi: 10.1046/j.1365-2958.2003.03673.x. [DOI] [PubMed] [Google Scholar]
- 130.Kwon DH, Lu CD. Polyamines induce resistance to cationic peptide, aminoglycoside, and quinolone antibiotics in Pseudomonas aeruginosa PAO1. Antimicrob. Agents Chemother. 2006;50:1615–1622. doi: 10.1128/AAC.50.5.1615-1622.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kwon DH, Lu CD. Polyamine effects on antibiotic susceptibility in bacteria. Antimicrob. Agents Chemother. 2007;51:2070–2077. doi: 10.1128/AAC.01472-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Flemming HC, Wingender J. The biofilm matrix. Nat. Rev. Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 133.Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 135.Lopez D, Vlamakis H, Kolter R. Biofilms. Cold Spring Harb Perspect. Biol. 2010;2:a000398. doi: 10.1101/cshperspect.a000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Stapper AP, Narasimhan G, Ohman DE, Barakat J, Hentzer M, Molin S, Kharazmi A, Hoiby N, Mathee K. Alginate production affects Pseudomonas aeruginosa biofilm development and architecture, but is not essential for biofilm formation. J. Med. Microbiol. 2004;53:679–690. doi: 10.1099/jmm.0.45539-0. [DOI] [PubMed] [Google Scholar]
- 137.Klausen M, Heydorn A, Ragas P, Lambertsen L, Aaes-Jorgensen A, Molin S, Tolker-Nielsen T. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 2003;48:1511–1524. doi: 10.1046/j.1365-2958.2003.03525.x. [DOI] [PubMed] [Google Scholar]
- 138.Caiazza NC, O'Toole GA. SadB is required for the transition from reversible to irreversible attachment during biofilm formation by Pseudomonas aeruginosa PA14. J. Bacteriol. 2004;186:4476–4485. doi: 10.1128/JB.186.14.4476-4485.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Merritt JH, Brothers KM, Kuchma SL, O'Toole GA. SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J. Bacteriol. 2007;189:8154–8164. doi: 10.1128/JB.00585-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Vallet I, Olson JW, Lory S, Lazdunski A, Filloux A. The chaperone/usher pathways of Pseudomonas aeruginosa: identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc. Natl Acad. Sci. USA. 2001;98:6911–6916. doi: 10.1073/pnas.111551898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Vallet I, Diggle SP, Stacey RE, Camara M, Ventre I, Lory S, Lazdunski A, Williams P, Filloux A. Biofilm formation in Pseudomonas aeruginosa: fimbrial cup gene clusters are controlled by the transcriptional regulator MvaT. J. Bacteriol. 2004;186:2880–2890. doi: 10.1128/JB.186.9.2880-2890.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Vallet-Gely I, Donovan KE, Fang R, Joung JK, Dove SL. Repression of phase-variable cup gene expression by H-NS-like proteins in Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA. 2005;102:11082–11087. doi: 10.1073/pnas.0502663102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kulasekara HD, Ventre I, Kulasekara BR, Lazdunski A, Filloux A, Lory S. A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol. Microbiol. 2005;55:368–380. doi: 10.1111/j.1365-2958.2004.04402.x. [DOI] [PubMed] [Google Scholar]
- 144.Merkel TJ, Barros C, Stibitz S. Characterization of the bvgR locus of Bordetella pertussis. J. Bacteriol. 1998;180:1682–1690. doi: 10.1128/jb.180.7.1682-1690.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kuchma SL, Connolly JP, O'Toole GA. A three-component regulatory system regulates biofilm maturation and type III secretion in Pseudomonas aeruginosa. J. Bacteriol. 2005;187:1441–1454. doi: 10.1128/JB.187.4.1441-1454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mikkelsen H, Ball G, Giraud C, Filloux A. Expression of Pseudomonas aeruginosa CupD fimbrial genes is antagonistically controlled by RcsB and the EAL-containing PvrR response regulators. PLoS One. 2009;4:e6018. doi: 10.1371/journal.pone.0006018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hickman JW, Tifrea DF, Harwood CS. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl Acad. Sci. USA. 2005;102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Guvener ZT, Harwood CS. Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol. Microbiol. 2007;66:1459–1473. doi: 10.1111/j.1365-2958.2007.06008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Choy WK, Zhou L, Syn CK, Zhang LH, Swarup S. MorA defines a new class of regulators affecting flagellar development and biofilm formation in diverse Pseudomonas species. J. Bacteriol. 2004;186:7221–7228. doi: 10.1128/JB.186.21.7221-7228.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ueda A, Wood TK. Connecting quorum sensing, c-di-GMP, pel polysaccharide, and biofilm formation in Pseudomonas aeruginosa through tyrosine phosphatase TpbA (PA3885) PLoS Pathog. 2009;5:e1000483. doi: 10.1371/journal.ppat.1000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hickman JW, Harwood CS. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol. Microbiol. 2008;69:376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Attila C, Ueda A, Wood TK. PA2663 (PpyR) increases biofilm formation in Pseudomonas aeruginosa PAO1 through the psl operon and stimulates virulence and quorum-sensing phenotypes. Appl. Microbiol. Biotechnol. 2008;78:293–307. doi: 10.1007/s00253-007-1308-y. [DOI] [PubMed] [Google Scholar]
- 153.D'Argenio DA, Calfee MW, Rainey PB, Pesci EC. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J. Bacteriol. 2002;184:6481–6489. doi: 10.1128/JB.184.23.6481-6489.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Allesen-Holm M, Barken KB, Yang L, Klausen M, Webb JS, Kjelleberg S, Molin S, Givskov M, Tolker-Nielsen T. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 2006;59:1114–1128. doi: 10.1111/j.1365-2958.2005.05008.x. [DOI] [PubMed] [Google Scholar]
- 155.Heurlier K, Denervaud V, Haenni M, Guy L, Krishnapillai V, Haas D. Quorum-sensing-negative (lasR) mutants of Pseudomonas aeruginosa avoid cell lysis and death. J. Bacteriol. 2005;187:4875–4883. doi: 10.1128/JB.187.14.4875-4883.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 157.Dominiak DM, Nielsen JL, Nielsen PH. Extracellular DNA is abundant and important for microcolony strength in mixed microbial biofilms. Environ. Microbiol. 2010;13:710–721. doi: 10.1111/j.1462-2920.2010.02375.x. [DOI] [PubMed] [Google Scholar]
- 158.Pamp SJ, Tolker-Nielsen T. Multiple roles of biosurfactants in structural biofilm development by Pseudomonas aeruginosa. J. Bacteriol. 2007;189:2531–2539. doi: 10.1128/JB.01515-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Davey ME, Caiazza NC, O'Toole GA. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2003;185:1027–1036. doi: 10.1128/JB.185.3.1027-1036.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Petrova OE, Sauer K. A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS Pathog. 2009;5:e1000668. doi: 10.1371/journal.ppat.1000668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Petrova OE, Sauer K. SagS contributes to the motile-sessile switch and acts in concert with BfiSR to enable Pseudomonas aeruginosa biofilm formation. J. Bacteriol. 2011;193:6614–6628. doi: 10.1128/JB.00305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Janjua HA, Segata N, Bernabo P, Tamburini S, Ellen A, Jousson O. Clinical populations of Pseudomonas aeruginosa isolated from acute infections show a wide virulence range partially correlated with population structure and virulence gene expression. Microbiology. 2012;158:2089–2098. doi: 10.1099/mic.0.056689-0. [DOI] [PubMed] [Google Scholar]
- 163.Storey DG, Ujack EE, Rabin HR, Mitchell I. Pseudomonas aeruginosa lasR transcription correlates with the transcription of lasA, lasB, and toxA in chronic lung infections associated with cystic fibrosis. Infect. Immun. 1998;66:2521–2528. doi: 10.1128/iai.66.6.2521-2528.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 2000;407:762–764. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- 165.Wu H, Song Z, Givskov M, Doring G, Worlitzsch D, Mathee K, Rygaard J, Hoiby N. Pseudomonas aeruginosa mutations in lasI and rhlI quorum sensing systems result in milder chronic lung infection. Microbiology. 2001;147:1105–1113. doi: 10.1099/00221287-147-5-1105. [DOI] [PubMed] [Google Scholar]
- 166.Bjarnsholt T, Jensen PO, Burmolle M, Hentzer M, Haagensen JA, Hougen HP, Calum H, Madsen KG, Moser C, Molin S, et al. Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology. 2005;151:373–383. doi: 10.1099/mic.0.27463-0. [DOI] [PubMed] [Google Scholar]
- 167.Purevdorj B, Costerton JW, Stoodley P. Influence of hydrodynamics and cell signaling on the structure and behavior of Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 2002;68:4457–4464. doi: 10.1128/AEM.68.9.4457-4464.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Heydorn A, Ersboll B, Kato J, Hentzer M, Parsek MR, Tolker-Nielsen T, Givskov M, Molin S. Statistical analysis of Pseudomonas aeruginosa biofilm development: impact of mutations in genes involved in twitching motility, cell-to-cell signaling, and stationary-phase sigma factor expression. Appl. Environ. Microbiol. 2002;68:2008–2017. doi: 10.1128/AEM.68.4.2008-2017.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D'Argenio DA, Miller SI, Ramsey BW, Speert DP, Moskowitz SM, et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl Acad. Sci. USA. 2006;103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Van Delden C, Pesci EC, Pearson JP, Iglewski BH. Starvation selection restores elastase and rhamnolipid production in a Pseudomonas aeruginosa quorum-sensing mutant. Infect. Immun. 1998;66:4499–4502. doi: 10.1128/iai.66.9.4499-4502.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kirisits MJ, Parsek MR. Does Pseudomonas aeruginosa use intercellular signalling to build biofilm communities? Cell. Microbiol. 2006;8:1841–1849. doi: 10.1111/j.1462-5822.2006.00817.x. [DOI] [PubMed] [Google Scholar]
- 172.Lewis K. Multidrug tolerance of biofilms and persister cells. Curr. Top. Microbiol. Immunol. 2008;322:107–131. doi: 10.1007/978-3-540-75418-3_6. [DOI] [PubMed] [Google Scholar]
- 173.Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 2004;230:13–18. doi: 10.1016/S0378-1097(03)00856-5. [DOI] [PubMed] [Google Scholar]
- 174.Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 2004;186:8172–8180. doi: 10.1128/JB.186.24.8172-8180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.De Groote VN, Verstraeten N, Fauvart M, Kint CI, Verbeeck AM, Beullens S, Cornelis P, Michiels J. Novel persistence genes in Pseudomonas aeruginosa identified by high-throughput screening. FEMS Microbiol. Lett. 2009;297:73–79. doi: 10.1111/j.1574-6968.2009.01657.x. [DOI] [PubMed] [Google Scholar]
- 176.Fauvart M, De Groote VN, Michiels J. Role of persister cells in chronic infections: clinical relevance and perspectives on anti-persister therapies. J. Med. Microbiol. 2011;60:699–709. doi: 10.1099/jmm.0.030932-0. [DOI] [PubMed] [Google Scholar]
- 177.Lewis K. Persister cells. Annu. Rev. Microbiol. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 178.Pressler T, Bohmova C, Conway S, Dumcius S, Hjelte L, Hoiby N, Kollberg H, Tummler B, Vavrova V. Chronic Pseudomonas aeruginosa infection definition: EuroCareCF Working Group report. J. Cyst. Fibros. 2011;10:S75–S78. doi: 10.1016/S1569-1993(11)60011-8. [DOI] [PubMed] [Google Scholar]
- 179.de Jong PA, Nakano Y, Lequin MH, Mayo JR, Woods R, Pare PD, Tiddens HA. Progressive damage on high resolution computed tomography despite stable lung function in cystic fibrosis. Eur. Respir. J. 2004;23:93–97. doi: 10.1183/09031936.03.00006603. [DOI] [PubMed] [Google Scholar]
- 180.Meyer KC, Sharma A. Regional variability of lung inflammation in cystic fibrosis. Am. J. Respir. Crit. Care Med. 1997;156:1536–1540. doi: 10.1164/ajrccm.156.5.9701098. [DOI] [PubMed] [Google Scholar]
- 181.Tiddens HA. Detecting early structural lung damage in cystic fibrosis. Pediatr. Pulmonol. 2002;34:228–231. doi: 10.1002/ppul.10134. [DOI] [PubMed] [Google Scholar]
- 182.Worlitzsch D, Tarran R, Ulrich M, Schwab U, Cekici A, Meyer KC, Birrer P, Bellon G, Berger J, Weiss T, et al. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Invest. 2002;109:317–325. doi: 10.1172/JCI13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Doggett RG, Harrison GM, Carter RE. Mucoid Pseudomonas aeruginosa in patients with chronic illnesses. Lancet. 1971;1:236–237. doi: 10.1016/s0140-6736(71)90973-1. [DOI] [PubMed] [Google Scholar]
- 184.Hoiby N. Pseudomonas aeruginosa infection in cystic fibrosis. Relationship between mucoid strains of Pseudomonas aeruginosa and the humoral immune response. Acta. Pathol. Microbiol. Scand. [B] Microbiol. Immunol. 1974;82:551–558. [PubMed] [Google Scholar]
- 185.Evans LR, Linker A. Production and characterization of the slime polysaccharide of Pseudomonas aeruginosa. J. Bacteriol. 1973;116:915–924. doi: 10.1128/jb.116.2.915-924.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Pedersen SS, Espersen F, Hoiby N, Jensen T. Immunoglobulin A and immunoglobulin G antibody responses to alginates from Pseudomonas aeruginosa in patients with cystic fibrosis. J. Clin. Microbiol. 1990;28:747–755. doi: 10.1128/jcm.28.4.747-755.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Pier GB, Coleman F, Grout M, Franklin M, Ohman DE. Role of alginate O acetylation in resistance of mucoid Pseudomonas aeruginosa to opsonic phagocytosis. Infect. Immun. 2001;69:1895–1901. doi: 10.1128/IAI.69.3.1895-1901.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Simpson JA, Smith SE, Dean RT. Alginate inhibition of the uptake of Pseudomonas aeruginosa by macrophages. J. Gen. Microbiol. 1988;134:29–36. doi: 10.1099/00221287-134-1-29. [DOI] [PubMed] [Google Scholar]
- 189.Mathee K, Ciofu O, Sternberg C, Lindum PW, Campbell JI, Jensen P, Johnsen AH, Givskov M, Ohman DE, Molin S, et al. Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology. 1999;145:1349–1357. doi: 10.1099/13500872-145-6-1349. [DOI] [PubMed] [Google Scholar]
- 190.Govan JR, Fyfe JA. Mucoid Pseudomonas aeruginosa and cystic fibrosis: resistance of the mucoid form to carbenicillin, flucloxacillin and tobramycin and the isolation of mucoid variants in vitro. J. Antimicrob. Chemother. 1978;4:233–240. doi: 10.1093/jac/4.3.233. [DOI] [PubMed] [Google Scholar]
- 191.Speert DP, Farmer SW, Campbell ME, Musser JM, Selander RK, Kuo S. Conversion of Pseudomonas aeruginosa to the phenotype characteristic of strains from patients with cystic fibrosis. J. Clin. Microbiol. 1990;28:188–194. doi: 10.1128/jcm.28.2.188-194.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Terry JM, Pina SE, Mattingly SJ. Environmental conditions which influence mucoid conversion Pseudomonas aeruginosa PAO1. Infect. Immun. 1991;59:471–477. doi: 10.1128/iai.59.2.471-477.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.DeVries CA, Ohman DE. Mucoid-to-nonmucoid conversion in alginate-producing Pseudomonas aeruginosa often results from spontaneous mutations in algT, encoding a putative alternate sigma factor, and shows evidence for autoregulation. J. Bacteriol. 1994;176:6677–6687. doi: 10.1128/jb.176.21.6677-6687.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Martin DW, Holloway BW, Deretic V. Characterization of a locus determining the mucoid status of Pseudomonas aeruginosa: AlgU shows sequence similarities with a Bacillus sigma factor. J. Bacteriol. 1993;175:1153–1164. doi: 10.1128/jb.175.4.1153-1164.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Martin DW, Schurr MJ, Mudd MH, Deretic V. Differentiation of Pseudomonas aeruginosa into the alginate-producing form: inactivation of mucB causes conversion to mucoidy. Mol. Microbiol. 1993;9:497–506. doi: 10.1111/j.1365-2958.1993.tb01711.x. [DOI] [PubMed] [Google Scholar]
- 196.Mathee K, McPherson CJ, Ohman DE. Posttranslational control of the algT (algU)-encoded sigma22 for expression of the alginate regulon in Pseudomonas aeruginosa and localization of its antagonist proteins MucA and MucB (AlgN) J. Bacteriol. 1997;179:3711–3720. doi: 10.1128/jb.179.11.3711-3720.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Mathee K, Kharazami AA, Hoiby N. In: Molecular Ecology of Biofilms. McLean RJC, editor. Norfolk: Horizon; 2002. pp. 23–55. [Google Scholar]
- 198.Goldberg JB, Gorman WL, Flynn JL, Ohman DE. A mutation in algN permits trans activation of alginate production by algT in Pseudomonas species. J. Bacteriol. 1993;175:1303–1308. doi: 10.1128/jb.175.5.1303-1308.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Martin DW, Schurr MJ, Mudd MH, Govan JR, Holloway BW, Deretic V. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc. Natl Acad. Sci. USA. 1993;90:8377–8381. doi: 10.1073/pnas.90.18.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Ohman D. In: Alginates: Biology and Applications. Rehm BHA, editor. Vol. 13. Berlin/Heidelberg: Springer; 2009. pp. 117–133. [Google Scholar]
- 201.Damron FH, Goldberg JB. Proteolytic regulation of alginate overproduction in Pseudomonas aeruginosa. Mol. Microbiol. 2012;84:595–607. doi: 10.1111/j.1365-2958.2012.08049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Damron FH, Yu HD. Pseudomonas aeruginosa MucD regulates the alginate pathway through activation of MucA degradation via MucP proteolytic activity. J. Bacteriol. 2011;193:286–291. doi: 10.1128/JB.01132-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Martin DW, Schurr MJ, Yu H, Deretic V. Analysis of promoters controlled by the putative sigma factor AlgU regulating conversion to mucoidy in Pseudomonas aeruginosa: relationship to sigma E and stress response. J. Bacteriol. 1994;176:6688–6696. doi: 10.1128/jb.176.21.6688-6696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Wozniak DJ, Ohman DE. Transcriptional analysis of the Pseudomonas aeruginosa genes algR, algB, and algD reveals a hierarchy of alginate gene expression which is modulated by algT. J. Bacteriol. 1994;176:6007–6014. doi: 10.1128/jb.176.19.6007-6014.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Wozniak DJ, Sprinkle AB, Baynham PJ. Control of Pseudomonas aeruginosa algZ expression by the alternative sigma factor AlgT. J. Bacteriol. 2003;185:7297–7300. doi: 10.1128/JB.185.24.7297-7300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Chitnis CE, Ohman DE. Genetic analysis of the alginate biosynthetic gene cluster of Pseudomonas aeruginosa shows evidence of an operonic structure. Mol. Microbiol. 1993;8:583–593. doi: 10.1111/j.1365-2958.1993.tb01602.x. [DOI] [PubMed] [Google Scholar]
- 207.Deretic V, Gill JF, Chakrabarty AM. Gene algD coding for GDPmannose dehydrogenase is transcriptionally activated in mucoid Pseudomonas aeruginosa. J. Bacteriol. 1987;169:351–358. doi: 10.1128/jb.169.1.351-358.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Leech AJ, Sprinkle A, Wood L, Wozniak DJ, Ohman DE. The NtrC family regulator AlgB, which controls alginate biosynthesis in mucoid Pseudomonas aeruginosa, binds directly to the algD promoter. J. Bacteriol. 2008;190:581–589. doi: 10.1128/JB.01307-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Kato J, Chakrabarty AM. Purification of the regulatory protein AlgR1 and its binding in the far upstream region of the algD promoter in Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA. 1991;88:1760–1764. doi: 10.1073/pnas.88.5.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Mohr CD, Martin DW, Konyecsni WM, Govan JR, Lory S, Deretic V. Role of the far-upstream sites of the algD promoter and the algR and rpoN genes in environmental modulation of mucoidy in Pseudomonas aeruginosa. J. Bacteriol. 1990;172:6576–6580. doi: 10.1128/jb.172.11.6576-6580.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Baynham PJ, Brown AL, Hall LL, Wozniak DJ. Pseudomonas aeruginosa AlgZ, a ribbon-helix-helix DNA-binding protein, is essential for alginate synthesis and algD transcriptional activation. Mol. Microbiol. 1999;33:1069–1080. doi: 10.1046/j.1365-2958.1999.01550.x. [DOI] [PubMed] [Google Scholar]
- 212.Damron FH, Qiu D, Yu HD. The Pseudomonas aeruginosa sensor kinase KinB negatively controls alginate production through AlgW-dependent MucA proteolysis. J. Bacteriol. 2009;191:2285–2295. doi: 10.1128/JB.01490-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Wood LF, Leech AJ, Ohman DE. Cell wall-inhibitory antibiotics activate the alginate biosynthesis operon in Pseudomonas aeruginosa: Roles of sigma (AlgT) and the AlgW and Prc proteases. Mol. Microbiol. 2006;62:412–426. doi: 10.1111/j.1365-2958.2006.05390.x. [DOI] [PubMed] [Google Scholar]
- 214.Ma S, Selvaraj U, Ohman DE, Quarless R, Hassett DJ, Wozniak DJ. Phosphorylation-independent activity of the response regulators AlgB and AlgR in promoting alginate biosynthesis in mucoid Pseudomonas aeruginosa. J. Bacteriol. 1998;180:956–968. doi: 10.1128/jb.180.4.956-968.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Zegans ME, Wozniak D, Griffin E, Toutain-Kidd CM, Hammond JH, Garfoot A, Lam JS. Pseudomonas aeruginosa exopolysaccharide Psl promotes resistance to the biofilm inhibitor polysorbate 80. Antimicrob. Agents Chemother. 2012;56:4112–4122. doi: 10.1128/AAC.00373-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Fujiwara S, Zielinski NA, Chakrabarty AM. Enhancer-like activity of A1gR1-binding site in alginate gene activation: positional, orientational, and sequence specificity. J. Bacteriol. 1993;175:5452–5459. doi: 10.1128/jb.175.17.5452-5459.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Ye RW, Zielinski NA, Chakrabarty AM. Purification and characterization of phosphomannomutase/phosphoglucomutase from Pseudomonas aeruginosa involved in biosynthesis of both alginate and lipopolysaccharide. J. Bacteriol. 1994;176:4851–4857. doi: 10.1128/jb.176.16.4851-4857.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Zielinski NA, Maharaj R, Roychoudhury S, Danganan CE, Hendrickson W, Chakrabarty AM. Alginate synthesis in Pseudomonas aeruginosa: environmental regulation of the algC promoter. J. Bacteriol. 1992;174:7680–7688. doi: 10.1128/jb.174.23.7680-7688.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Morici LA, Carterson AJ, Wagner VE, Frisk A, Schurr JR, Honer zu Bentrup K, Hassett DJ, Iglewski BH, Sauer K, Schurr MJ. Pseudomonas aeruginosa AlgR represses the Rhl quorum-sensing system in a biofilm-specific manner. J. Bacteriol. 2007;189:7752–7764. doi: 10.1128/JB.01797-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Belete B, Lu H, Wozniak DJ. Pseudomonas aeruginosa AlgR regulates type IV pilus biosynthesis by activating transcription of the fimU-pilVWXY1Y2E operon. J. Bacteriol. 2008;190:2023–2030. doi: 10.1128/JB.01623-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Lizewski SE, Lundberg DS, Schurr MJ. The transcriptional regulator AlgR is essential for Pseudomonas aeruginosa pathogenesis. Infect. Immun. 2002;70:6083–6093. doi: 10.1128/IAI.70.11.6083-6093.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Carterson AJ, Morici LA, Jackson DW, Frisk A, Lizewski SE, Jupiter R, Simpson K, Kunz DA, Davis SH, Schurr JR, et al. The transcriptional regulator AlgR controls cyanide production in Pseudomonas aeruginosa. J. Bacteriol. 2004;186:6837–6844. doi: 10.1128/JB.186.20.6837-6844.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Cody WL, Pritchett CL, Jones AK, Carterson AJ, Jackson D, Frisk A, Wolfgang MC, Schurr MJ. Pseudomonas aeruginosa AlgR controls cyanide production in an AlgZ-dependent manner. J. Bacteriol. 2009;191:2993–3002. doi: 10.1128/JB.01156-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Jones AK, Fulcher NB, Balzer GJ, Urbanowski ML, Pritchett CL, Schurr MJ, Yahr TL, Wolfgang MC. Activation of the Pseudomonas aeruginosa AlgU regulon through mucA mutation inhibits cyclic AMP/Vfr signaling. J. Bacteriol. 2010;192:5709–5717. doi: 10.1128/JB.00526-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Lizewski SE, Schurr JR, Jackson DW, Frisk A, Carterson AJ, Schurr MJ. Identification of AlgR-regulated genes in Pseudomonas aeruginosa by use of microarray analysis. J. Bacteriol. 2004;186:5672–5684. doi: 10.1128/JB.186.17.5672-5684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Remminghorst U, Rehm BH. Alg44, a unique protein required for alginate biosynthesis in Pseudomonas aeruginosa. FEBS Lett. 2006;580:3883–3888. doi: 10.1016/j.febslet.2006.05.077. [DOI] [PubMed] [Google Scholar]
- 227.Hay ID, Remminghorst U, Rehm BH. MucR, a novel membrane-associated regulator of alginate biosynthesis in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2009;75:1110–1120. doi: 10.1128/AEM.02416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Merighi M, Lee VT, Hyodo M, Hayakawa Y, Lory S. The second messenger bis-(3'-5')-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol. Microbiol. 2007;65:876–895. doi: 10.1111/j.1365-2958.2007.05817.x. [DOI] [PubMed] [Google Scholar]
- 229.Ochsner UA, Wilderman PJ, Vasil AI, Vasil ML. GeneChip expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol. Microbiol. 2002;45:1277–1287. doi: 10.1046/j.1365-2958.2002.03084.x. [DOI] [PubMed] [Google Scholar]
- 230.Palma M, Worgall S, Quadri LE. Transcriptome analysis of the Pseudomonas aeruginosa response to iron. Arch. Microbiol. 2003;180:374–379. doi: 10.1007/s00203-003-0602-z. [DOI] [PubMed] [Google Scholar]
- 231.Visca P, Imperi F, Lamont IL. Pyoverdine siderophores: from biogenesis to biosignificance. Trends Microbiol. 2007;15:22–30. doi: 10.1016/j.tim.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 232.Poole K, McKay GA. Iron acquisition and its control in Pseudomonas aeruginosa: many roads lead to Rome. Front. Biosci. 2003;8:d661–d686. doi: 10.2741/1051. [DOI] [PubMed] [Google Scholar]
- 233.Ochsner UA, Johnson Z, Vasil ML. Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiology. 2000;146:185–198. doi: 10.1099/00221287-146-1-185. [DOI] [PubMed] [Google Scholar]
- 234.Touati D. Iron and oxidative stress in bacteria. Arch. Biochem. Biophys. 2000;373:1–6. doi: 10.1006/abbi.1999.1518. [DOI] [PubMed] [Google Scholar]
- 235.Vasil ML, Ochsner UA. The response of Pseudomonas aeruginosa to iron: genetics, biochemistry and virulence. Mol. Microbiol. 1999;34:399–413. doi: 10.1046/j.1365-2958.1999.01586.x. [DOI] [PubMed] [Google Scholar]
- 236.Pohl E, Haller JC, Mijovilovich A, Meyer-Klaucke W, Garman E, Vasil ML. Architecture of a protein central to iron homeostasis: crystal structure and spectroscopic analysis of the ferric uptake regulator. Mol. Microbiol. 2003;47:903–915. doi: 10.1046/j.1365-2958.2003.03337.x. [DOI] [PubMed] [Google Scholar]
- 237.Wilderman PJ, Sowa NA, FitzGerald DJ, FitzGerald PC, Gottesman S, Ochsner UA, Vasil ML. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc. Natl Acad. Sci. USA. 2004;101:9792–9797. doi: 10.1073/pnas.0403423101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Cornelis P, Matthijs S, Van Oeffelen L. Iron uptake regulation in Pseudomonas aeruginosa. Biometals. 2009;22:15–22. doi: 10.1007/s10534-008-9193-0. [DOI] [PubMed] [Google Scholar]
- 239.Ochsner UA, Vasil ML. Gene repression by the ferric uptake regulator in Pseudomonas aeruginosa: cycle selection of iron-regulated genes. Proc. Natl Acad. Sci. USA. 1996;93:4409–4414. doi: 10.1073/pnas.93.9.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Wilderman PJ, Vasil AI, Johnson Z, Wilson MJ, Cunliffe HE, Lamont IL, Vasil ML. Characterization of an endoprotease (PrpL) encoded by a PvdS-regulated gene in Pseudomonas aeruginosa. Infect. Immun. 2001;69:5385–5394. doi: 10.1128/IAI.69.9.5385-5394.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Ochsner UA, Johnson Z, Lamont IL, Cunliffe HE, Vasil ML. Exotoxin A production in Pseudomonas aeruginosa requires the iron-regulated pvdS gene encoding an alternative sigma factor. Mol. Microbiol. 1996;21:1019–1028. doi: 10.1046/j.1365-2958.1996.481425.x. [DOI] [PubMed] [Google Scholar]
- 242.Gaines JM, Carty NL, Tiburzi F, Davinic M, Visca P, Colmer-Hamood JA, Hamood AN. Regulation of the Pseudomonas aeruginosa toxA, regA and ptxR genes by the iron-starvation sigma factor PvdS under reduced levels of oxygen. Microbiology. 2007;153:4219–4233. doi: 10.1099/mic.0.2007/011338-0. [DOI] [PubMed] [Google Scholar]
- 243.Xiong YQ, Vasil ML, Johnson Z, Ochsner UA, Bayer AS. The oxygen- and iron-dependent sigma factor pvdS of Pseudomonas aeruginosa is an important virulence factor in experimental infective endocarditis. J. Infect. Dis. 2000;181:1020–1026. doi: 10.1086/315338. [DOI] [PubMed] [Google Scholar]
- 244.Singh PK, Parsek MR, Greenberg EP, Welsh MJ. A component of innate immunity prevents bacterial biofilm development. Nature. 2002;417:552–555. doi: 10.1038/417552a. [DOI] [PubMed] [Google Scholar]
- 245.Singh PK. Iron sequestration by human lactoferrin stimulates P. aeruginosa surface motility and blocks biofilm formation. Biometals. 2004;17:267–270. doi: 10.1023/b:biom.0000027703.77456.27. [DOI] [PubMed] [Google Scholar]
- 246.Banin E, Vasil ML, Greenberg EP. Iron and Pseudomonas aeruginosa biofilm formation. Proc. Natl Acad. Sci. USA. 2005;102:11076–11081. doi: 10.1073/pnas.0504266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Yang L, Barken KB, Skindersoe ME, Christensen AB, Givskov M, Tolker-Nielsen T. Effects of iron on DNA release and biofilm development by Pseudomonas aeruginosa. Microbiology. 2007;153:1318–1328. doi: 10.1099/mic.0.2006/004911-0. [DOI] [PubMed] [Google Scholar]
- 248.Bollinger N, Hassett DJ, Iglewski BH, Costerton JW, McDermott TR. Gene expression in Pseudomonas aeruginosa: evidence of iron override effects on quorum sensing and biofilm-specific gene regulation. J. Bacteriol. 2001;183:1990–1996. doi: 10.1128/JB.183.6.1990-1996.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Kim EJ, Wang W, Deckwer WD, Zeng AP. Expression of the quorum-sensing regulatory protein LasR is strongly affected by iron and oxygen concentrations in cultures of Pseudomonas aeruginosa irrespective of cell density. Microbiology. 2005;151:1127–1138. doi: 10.1099/mic.0.27566-0. [DOI] [PubMed] [Google Scholar]
- 250.Schuster M, Lostroh CP, Ogi T, Greenberg EP. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 2003;185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Juhas M, Wiehlmann L, Salunkhe P, Lauber J, Buer J, Tummler B. GeneChip expression analysis of the VqsR regulon of Pseudomonas aeruginosa TB. FEMS Microbiol. Lett. 2005;242:287–295. doi: 10.1016/j.femsle.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 252.Visca P, Leoni L, Wilson MJ, Lamont IL. Iron transport and regulation, cell signalling and genomics: lessons from Escherichia coli and Pseudomonas. Mol. Microbiol. 2002;45:1177–1190. doi: 10.1046/j.1365-2958.2002.03088.x. [DOI] [PubMed] [Google Scholar]
- 253.Hazan R, He J, Xiao G, Dekimpe V, Apidianakis Y, Lesic B, Astrakas C, Deziel E, Lepine F, Rahme LG. Homeostatic interplay between bacterial cell-cell signaling and iron in virulence. PLoS Pathog. 2010;6:e1000810. doi: 10.1371/journal.ppat.1000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Oglesby AG, Farrow JM, 3rd, Lee JH, Tomaras AP, Greenberg EP, Pesci EC, Vasil ML. The influence of iron on Pseudomonas aeruginosa physiology: a regulatory link between iron and quorum sensing. J. Biol. Chem. 2008;283:15558–15567. doi: 10.1074/jbc.M707840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Bleves S, Viarre V, Salacha R, Michel GP, Filloux A, Voulhoux R. Protein secretion systems in Pseudomonas aeruginosa: a wealth of pathogenic weapons. Int. J. Med. Microbiol. 2010;300:534–543. doi: 10.1016/j.ijmm.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 256.Hauser AR. The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat. Rev. Microbiol. 2009;7:654–665. doi: 10.1038/nrmicro2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Guzzo J, Pages JM, Duong F, Lazdunski A, Murgier M. Pseudomonas aeruginosa alkaline protease: evidence for secretion genes and study of secretion mechanism. J. Bacteriol. 1991;173:5290–5297. doi: 10.1128/jb.173.17.5290-5297.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Matsumoto K. Role of bacterial proteases in pseudomonal and serratial keratitis. Biol. Chem. 2004;385:1007–1016. doi: 10.1515/BC.2004.131. [DOI] [PubMed] [Google Scholar]
- 259.Wandersman C, Delepelaire P. Bacterial iron sources: from siderophores to hemophores. Annu. Rev. Microbiol. 2004;58:611–647. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- 260.Hood RD, Singh P, Hsu F, Guvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe. 2010;7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Moscoso JA, Mikkelsen H, Heeb S, Williams P, Filloux A. The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environ. Microbiol. 2011;13:3128–3138. doi: 10.1111/j.1462-2920.2011.02595.x. [DOI] [PubMed] [Google Scholar]
- 262.Turner KH, Vallet-Gely I, Dove SL. Epigenetic control of virulence gene expression in Pseudomonas aeruginosa by a LysR-type transcription regulator. PLoS Genet. 2009;5:e1000779. doi: 10.1371/journal.pgen.1000779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Pearson JP, Pesci EC, Iglewski BH. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 1997;179:5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Chapon-Herve V, Akrim M, Latifi A, Williams P, Lazdunski A, Bally M. Regulation of the xcp secretion pathway by multiple quorum-sensing modulons in Pseudomonas aeruginosa. Mol. Microbiol. 1997;24:1169–1178. doi: 10.1046/j.1365-2958.1997.4271794.x. [DOI] [PubMed] [Google Scholar]
- 265.Lazdunski AM, Ventre I, Sturgis JN. Regulatory circuits and communication in Gram-negative bacteria. Nat. Rev. Microbiol. 2004;2:581–592. doi: 10.1038/nrmicro924. [DOI] [PubMed] [Google Scholar]
- 266.Filloux A, Bally M, Soscia C, Murgier M, Lazdunski A. Phosphate regulation in Pseudomonas aeruginosa: cloning of the alkaline phosphatase gene and identification of phoB- and phoR-like genes. Mol. Gen. Genet. 1988;212:510–513. doi: 10.1007/BF00330857. [DOI] [PubMed] [Google Scholar]
- 267.Ball G, Durand E, Lazdunski A, Filloux A. A novel type II secretion system in Pseudomonas aeruginosa. Mol. Microbiol. 2002;43:475–485. doi: 10.1046/j.1365-2958.2002.02759.x. [DOI] [PubMed] [Google Scholar]
- 268.Llamas MA, van der Sar A, Chu BC, Sparrius M, Vogel HJ, Bitter W. A novel extracytoplasmic function (ECF) sigma factor regulates virulence in Pseudomonas aeruginosa. PLoS Pathog. 2009;5:e1000572. doi: 10.1371/journal.ppat.1000572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Lesic B, Starkey M, He J, Hazan R, Rahme LG. Quorum sensing differentially regulates Pseudomonas aeruginosa type VI secretion locus I and homologous loci II and III, which are required for pathogenesis. Microbiology. 2009;155:2845–2855. doi: 10.1099/mic.0.029082-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordonez CL, Lory S, et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Yahr TL, Wolfgang MC. Transcriptional regulation of the Pseudomonas aeruginosa type III secretion system. Mol. Microbiol. 2006;62:631–640. doi: 10.1111/j.1365-2958.2006.05412.x. [DOI] [PubMed] [Google Scholar]
- 272.Frank DW. The exoenzyme S regulon of Pseudomonas aeruginosa. Mol. Microbiol. 1997;26:621–629. doi: 10.1046/j.1365-2958.1997.6251991.x. [DOI] [PubMed] [Google Scholar]
- 273.Vallis AJ, Yahr TL, Barbieri JT, Frank DW. Regulation of ExoS production and secretion by Pseudomonas aeruginosa in response to tissue culture conditions. Infect. Immun. 1999;67:914–920. doi: 10.1128/iai.67.2.914-920.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Shen DK, Filopon D, Kuhn L, Polack B, Toussaint B. PsrA is a positive transcriptional regulator of the type III secretion system in Pseudomonas aeruginosa. Infect. Immun. 2006;74:1121–1129. doi: 10.1128/IAI.74.2.1121-1129.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Hogardt M, Roeder M, Schreff AM, Eberl L, Heesemann J. Expression of Pseudomonas aeruginosa exoS is controlled by quorum sensing and RpoS. Microbiology. 2004;150:843–851. doi: 10.1099/mic.0.26703-0. [DOI] [PubMed] [Google Scholar]
- 276.Dasgupta N, Lykken GL, Wolfgang MC, Yahr TL. A novel anti-anti-activator mechanism regulates expression of the Pseudomonas aeruginosa type III secretion system. Mol. Microbiol. 2004;53:297–308. doi: 10.1111/j.1365-2958.2004.04128.x. [DOI] [PubMed] [Google Scholar]
- 277.Wu W, Jin S. PtrB of Pseudomonas aeruginosa suppresses the type III secretion system under the stress of DNA damage. J. Bacteriol. 2005;187:6058–6068. doi: 10.1128/JB.187.17.6058-6068.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Hornef MW, Roggenkamp A, Geiger AM, Hogardt M, Jacobi CA, Heesemann J. Triggering the ExoS regulon of Pseudomonas aeruginosa: a GFP-reporter analysis of exoenzyme (Exo) S, ExoT and ExoU synthesis. Microb. Pathog. 2000;29:329–343. doi: 10.1006/mpat.2000.0398. [DOI] [PubMed] [Google Scholar]
- 279.Rietsch A, Mekalanos JJ. Metabolic regulation of type III secretion gene expression in Pseudomonas aeruginosa. Mol. Microbiol. 2006;59:807–820. doi: 10.1111/j.1365-2958.2005.04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Dacheux D, Epaulard O, de Groot A, Guery B, Leberre R, Attree I, Polack B, Toussaint B. Activation of the Pseudomonas aeruginosa type III secretion system requires an intact pyruvate dehydrogenase aceAB operon. Infect. Immun. 2002;70:3973–3977. doi: 10.1128/IAI.70.7.3973-3977.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Wu W, Badrane H, Arora S, Baker HV, Jin S. MucA-mediated coordination of type III secretion and alginate synthesis in Pseudomonas aeruginosa. J. Bacteriol. 2004;186:7575–7585. doi: 10.1128/JB.186.22.7575-7585.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Jin Y, Yang H, Qiao M, Jun S. MexT regulates the type III secretion system through MexS and PtrC in Pseudomonas aeruginosa. J. Bacteriol. 2011;193:399–410. doi: 10.1128/JB.01079-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Mulcahy H, O'Callaghan J, O'Grady EP, Adams C, O'Gara F. The posttranscriptional regulator RsmA plays a role in the interaction between Pseudomonas aeruginosa and human airway epithelial cells by positively regulating the type III secretion system. Infect. Immun. 2006;74:3012–3015. doi: 10.1128/IAI.74.5.3012-3015.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.O'Callaghan J, Reen FJ, Adams C, O'Gara F. Low oxygen induces the type III secretion system in Pseudomonas aeruginosa via modulation of the small RNAs rsmZ and rsmY. Microbiology. 2011;157:3417–3428. doi: 10.1099/mic.0.052050-0. [DOI] [PubMed] [Google Scholar]
- 285.Dubnau D, Losick R. Bistability in bacteria. Mol. Microbiol. 2006;61:564–572. doi: 10.1111/j.1365-2958.2006.05249.x. [DOI] [PubMed] [Google Scholar]
- 286.Storz G, Altuvia S, Wassarman KM. An abundance of RNA regulators. Annu. Rev. Biochem. 2005;74:199–217. doi: 10.1146/annurev.biochem.74.082803.133136. [DOI] [PubMed] [Google Scholar]
- 287.Papenfort K, Vogel J. Multiple target regulation by small noncoding RNAs rewires gene expression at the post-transcriptional level. Res. Microbiol. 2009;160:278–287. doi: 10.1016/j.resmic.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 288.Gottesman S. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu. Rev. Microbiol. 2004;58:303–328. doi: 10.1146/annurev.micro.58.030603.123841. [DOI] [PubMed] [Google Scholar]
- 289.Ferrara S, Brugnoli M, De Bonis A, Righetti F, Delvillani F, Deho G, Horner D, Briani F, Bertoni G. Comparative profiling of Pseudomonas aeruginosa strains reveals differential expression of novel unique and conserved small RNAs. PLoS One. 2012;7:e36553. doi: 10.1371/journal.pone.0036553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Heurlier K, Williams F, Heeb S, Dormond C, Pessi G, Singer D, Camara M, Williams P, Haas D. Positive control of swarming, rhamnolipid synthesis, and lipase production by the posttranscriptional RsmA/RsmZ system in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2004;186:2936–2945. doi: 10.1128/JB.186.10.2936-2945.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Heeb S, Blumer C, Haas D. Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. J. Bacteriol. 2002;184:1046–1056. doi: 10.1128/jb.184.4.1046-1056.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Valverde C, Heeb S, Keel C, Haas D. RsmY, a small regulatory RNA, is required in concert with RsmZ for GacA-dependent expression of biocontrol traits in Pseudomonas fluorescens CHA0. Mol. Microbiol. 2003;50:1361–1379. doi: 10.1046/j.1365-2958.2003.03774.x. [DOI] [PubMed] [Google Scholar]
- 293.Kay E, Dubuis C, Haas D. Three small RNAs jointly ensure secondary metabolism and biocontrol in Pseudomonas fluorescens CHA0. Proc. Natl Acad. Sci. USA. 2005;102:17136–17141. doi: 10.1073/pnas.0505673102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Heeb S, Kuehne SA, Bycroft M, Crivii S, Allen MD, Haas D, Camara M, Williams P. Functional analysis of the post-transcriptional regulator RsmA reveals a novel RNA-binding site. J. Mol. Biol. 2006;355:1026–1036. doi: 10.1016/j.jmb.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 295.Kay E, Humair B, Denervaud V, Riedel K, Spahr S, Eberl L, Valverde C, Haas D. Two GacA-dependent small RNAs modulate the quorum-sensing response in Pseudomonas aeruginosa. J. Bacteriol. 2006;188:6026–6033. doi: 10.1128/JB.00409-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Bordi C, Lamy MC, Ventre I, Termine E, Hachani A, Fillet S, Roche B, Bleves S, Mejean V, Lazdunski A, et al. Regulatory RNAs and the HptB/RetS signalling pathways fine-tune Pseudomonas aeruginosa pathogenesis. Mol. Microbiol. 2010;76:1427–1443. doi: 10.1111/j.1365-2958.2010.07146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Hsu JL, Chen HC, Peng HL, Chang HY. Characterization of the histidine-containing phosphotransfer protein B-mediated multistep phosphorelay system in Pseudomonas aeruginosa PAO1. J. Biol. Chem. 2008;283:9933–9944. doi: 10.1074/jbc.M708836200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Petrova OE, Sauer K. The novel two-component regulatory system BfiSR regulates biofilm development by controlling the small RNA rsmZ through CafA. J. Bacteriol. 2010;192:5275–5288. doi: 10.1128/JB.00387-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Brencic A, McFarland KA, McManus HR, Castang S, Mogno I, Dove SL, Lory S. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol. Microbiol. 2009;73:434–445. doi: 10.1111/j.1365-2958.2009.06782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Lapouge K, Schubert M, Allain FH, Haas D. Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol. Microbiol. 2008;67:241–253. doi: 10.1111/j.1365-2958.2007.06042.x. [DOI] [PubMed] [Google Scholar]
- 301.Sorger-Domenigg T, Sonnleitner E, Kaberdin VR, Blasi U. Distinct and overlapping binding sites of Pseudomonas aeruginosa Hfq and RsmA proteins on the non-coding RNA RsmY. Biochem. Biophys. Res. Commun. 2007;352:769–773. doi: 10.1016/j.bbrc.2006.11.084. [DOI] [PubMed] [Google Scholar]
- 302.Moreno R, Marzi S, Romby P, Rojo F. The Crc global regulator binds to an unpaired A-rich motif at the Pseudomonas putida alkS mRNA coding sequence and inhibits translation initiation. Nucleic Acids Res. 2009;37:7678–7690. doi: 10.1093/nar/gkp825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Moreno R, Rojo F. The target for the Pseudomonas putida Crc global regulator in the benzoate degradation pathway is the BenR transcriptional regulator. J. Bacteriol. 2008;190:1539–1545. doi: 10.1128/JB.01604-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Moreno R, Ruiz-Manzano A, Yuste L, Rojo F. The Pseudomonas putida Crc global regulator is an RNA binding protein that inhibits translation of the AlkS transcriptional regulator. Mol. Microbiol. 2007;64:665–675. doi: 10.1111/j.1365-2958.2007.05685.x. [DOI] [PubMed] [Google Scholar]
- 305.Sonnleitner E, Abdou L, Haas D. Small RNA as global regulator of carbon catabolite repression in Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA. 2009;106:21866–21871. doi: 10.1073/pnas.pnas.0910308106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Yeung AT, Bains M, Hancock RE. The sensor kinase CbrA is a global regulator that modulates metabolism, virulence, and antibiotic resistance in Pseudomonas aeruginosa. J. Bacteriol. 2011;193:918–931. doi: 10.1128/JB.00911-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Abdou L, Chou HT, Haas D, Lu CD. Promoter recognition and activation by the global response regulator CbrB in Pseudomonas aeruginosa. J. Bacteriol. 2011;193:2784–2792. doi: 10.1128/JB.00164-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Li W, Lu CD. Regulation of carbon and nitrogen utilization by CbrAB and NtrBC two-component systems in Pseudomonas aeruginosa. J. Bacteriol. 2007;189:5413–5420. doi: 10.1128/JB.00432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Nishijyo T, Haas D, Itoh Y. The CbrA-CbrB two-component regulatory system controls the utilization of multiple carbon and nitrogen sources in Pseudomonas aeruginosa. Mol. Microbiol. 2001;40:917–931. doi: 10.1046/j.1365-2958.2001.02435.x. [DOI] [PubMed] [Google Scholar]
- 310.Rietsch A, Wolfgang MC, Mekalanos JJ. Effect of metabolic imbalance on expression of type III secretion genes in Pseudomonas aeruginosa. Infect. Immun. 2004;72:1383–1390. doi: 10.1128/IAI.72.3.1383-1390.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Linares JF, Moreno R, Fajardo A, Martinez-Solano L, Escalante R, Rojo F, Martinez JL. The global regulator Crc modulates metabolism, susceptibility to antibiotics and virulence in Pseudomonas aeruginosa. Environ. Microbiol. 2010;12:3196–3212. doi: 10.1111/j.1462-2920.2010.02292.x. [DOI] [PubMed] [Google Scholar]
- 312.O'Toole GA, Gibbs KA, Hager PW, Phibbs PV, Jr, Kolter R. The global carbon metabolism regulator Crc is a component of a signal transduction pathway required for biofilm development by Pseudomonas aeruginosa. J. Bacteriol. 2000;182:425–431. doi: 10.1128/jb.182.2.425-431.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Sonnleitner E, Romeo A, Blasi U. Small regulatory RNAs in Pseudomonas aeruginosa. RNA Biol. 2012;9:364–371. doi: 10.4161/rna.19231. [DOI] [PubMed] [Google Scholar]
- 314.Sonnleitner E, Gonzalez N, Sorger-Domenigg T, Heeb S, Richter AS, Backofen R, Williams P, Huttenhofer A, Haas D, Blasi U. The small RNA PhrS stimulates synthesis of the Pseudomonas aeruginosa quinolone signal. Mol. Microbiol. 2011;80:868–885. doi: 10.1111/j.1365-2958.2011.07620.x. [DOI] [PubMed] [Google Scholar]
- 315.Moll I, Afonyushkin T, Vytvytska O, Kaberdin VR, Blasi U. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA. 2003;9:1308–1314. doi: 10.1261/rna.5850703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Masse E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl Acad. Sci. USA. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Vasil ML. How we learnt about iron acquisition in Pseudomonas aeruginosa: a series of very fortunate events. Biometals. 2007;20:587–601. doi: 10.1007/s10534-006-9067-2. [DOI] [PubMed] [Google Scholar]
- 318.Oglesby-Sherrouse AG, Vasil ML. Characterization of a heme-regulated non-coding RNA encoded by the prrF locus of Pseudomonas aeruginosa. PLoS One. 2010;5:e9930. doi: 10.1371/journal.pone.0009930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Bornholdt S. Boolean network models of cellular regulation: prospects and limitations. J. R. Soc. Interface. 2008;5:S85–S94. doi: 10.1098/rsif.2008.0132.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Kauffman SA. Metabolic stability and epigenesis in randomly constructed genetic nets. J. Theor. Biol. 1969;22:437–467. doi: 10.1016/0022-5193(69)90015-0. [DOI] [PubMed] [Google Scholar]
- 321.Thomas R. Boolean formalization of genetic control circuits. J. Theor. Biol. 1973;42:563–585. doi: 10.1016/0022-5193(73)90247-6. [DOI] [PubMed] [Google Scholar]
- 322.Shmulevich I, Dougherty ER, Zhang W. Gene perturbation and intervention in probabilistic Boolean networks. Bioinformatics. 2002;18:1319–1331. doi: 10.1093/bioinformatics/18.10.1319. [DOI] [PubMed] [Google Scholar]
- 323.Shmulevich I, Dougherty ER, Kim S, Zhang W. Probabilistic Boolean Networks: a rule-based uncertainty model for gene regulatory networks. Bioinformatics. 2002;18:261–274. doi: 10.1093/bioinformatics/18.2.261. [DOI] [PubMed] [Google Scholar]
- 324.Liang J, Han J. Stochastic Boolean networks: an efficient approach to modeling gene regulatory networks. BMC Syst. Biol. 2012;6:113. doi: 10.1186/1752-0509-6-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Babu MM. Early Career Research Award Lecture. Structure, evolution and dynamics of transcriptional regulatory networks. Biochem. Soc. Trans. 2010;38:1155–1178. doi: 10.1042/BST0381155. [DOI] [PubMed] [Google Scholar]
- 326.Babu MM, Lang B, Aravind L. Methods to reconstruct and compare transcriptional regulatory networks. Methods Mol. Biol. 2009;541:163–180. doi: 10.1007/978-1-59745-243-4_8. [DOI] [PubMed] [Google Scholar]
- 327.Oestreicher C. A history of chaos theory. Dialogues Clin. Neurosci. 2007;9:279–289. doi: 10.31887/DCNS.2007.9.3/coestreicher. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Goldbeter A. Biochemical Oscillations and Cellular Rythms. Cambridge, UK: Cambridge University Press; 1996. [Google Scholar]
- 329.Novak B, Tyson JJ. Design principles of biochemical oscillators. Nat. Rev. Mol. Cell. Biol. 2008;9:981–991. doi: 10.1038/nrm2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Leloup JC, Goldbeter A. Chaos and birhythmicity in a model for circadian oscillations of the PER and TIM proteins in drosophila. J. Theor. Biol. 1999;198:445–459. doi: 10.1006/jtbi.1999.0924. [DOI] [PubMed] [Google Scholar]
- 331.Suguna C, Chowdhury KK, Sinha S. Minimal model for complex dynamics in cellular processes. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Topics. 1999;60:5943–5949. doi: 10.1103/physreve.60.5943. [DOI] [PubMed] [Google Scholar]
- 332.Zhang Z, Ye W, Qian Y, Zheng Z, Huang X, Hu G. Chaotic motifs in gene regulatory networks. PLoS One. 2012;7:e39355. doi: 10.1371/journal.pone.0039355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Trosko JE, Ruch RJ. Cell-cell communication in carcinogenesis. Front Biosci. 1998;3:d208–d236. doi: 10.2741/a275. [DOI] [PubMed] [Google Scholar]
- 334.Thomas R, D'Ari R. Biological Feedback. Boca Raton, FL: CRC Press; 1990. [Google Scholar]
- 335.Ng WL, Perez L, Cong J, Semmelhack MF, Bassler BL. Broad spectrum pro-quorum-sensing molecules as inhibitors of virulence in vibrios. PLoS Pathog. 2012;8:e1002767. doi: 10.1371/journal.ppat.1002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 337.Sonnleitner E, Sorger-Domenigg T, Madej MJ, Findeiss S, Hackermuller J, Huttenhofer A, Stadler PF, Blasi U, Moll I. Detection of small RNAs in Pseudomonas aeruginosa by RNomics and structure-based bioinformatic tools. Microbiology. 2008;154:3175–3187. doi: 10.1099/mic.0.2008/019703-0. [DOI] [PubMed] [Google Scholar]