Abstract
Sulfolobus metallicus is a thermoacidophilic crenarchaeon used in high-temperature bioleaching processes that is able to grow under stressing conditions such as high concentrations of heavy metals. Nevertheless, the genetic and biochemical mechanisms responsible for heavy metal resistance in S. metallicus remain uncharacterized. Proteomic analysis of S. metallicus cells exposed to 100 mM Cu revealed that 18 out of 30 upregulated proteins are related to the production and conversion of energy, amino acids biosynthesis, and stress responses. Ten of these last proteins were also up-regulated in S. metallicus treated in the presence of 1 mM Cd suggesting that at least in part, a common general response to these two heavy metals. The S. metallicus genome contained two complete cop gene clusters, each encoding a metallochaperone (CopM), a Cu-exporting ATPase (CopA), and a transcriptional regulator (CopT). Transcriptional expression analysis revealed that copM and copA from each cop gene cluster were cotranscribed and their transcript levels increased when S. metallicus was grown either in the presence of Cu or using chalcopyrite (CuFeS2) as oxidizable substrate. This study shows for the first time the presence of a duplicated version of the cop gene cluster in Archaea and characterizes some of the Cu and Cd resistance determinants in a thermophilic archaeon employed for industrial biomining.
1. Introduction
Bioleaching is the biological conversion of an insoluble metal compound into a water soluble form [1, 2]. Microbe-based processes have clear economic advantages in the extraction of metals from many low-grade deposits [3], and these metal-extraction processes are usually more environmentally friendly than physical-chemical processes [3–5]. Some ores are refractory to mesophilic leaching and temperatures preferably as high as 75–85°C are required [6, 7]. At high temperatures, biomining consortia are dominated by thermoacidophilic Archaea from the genus Sulfolobus, Acidianus, and Metallosphaera [8].
Metals play an integral role in the life process of microorganisms, but at high levels both essential and nonessential metals can damage cell membranes, alter enzyme specificity, disrupt cellular functions, and damage the structure of DNA [9, 10]. Acid-leaching solutions are characterized by high metal concentrations that are toxic to most life, and as might be expected, microorganisms that grow in mineral-rich environments are, in most cases, remarkably tolerant to a wide range of metal ions [3, 11] and should possess robust metal resistance mechanisms [11–15].
Despite this, only some metal tolerance values have been reported [6] and the genetic and biochemical mechanisms responsible for metal resistance in biomining acidophilic Archaea are just beginning to be characterized [16]. It is therefore important to further understand the mechanisms used by these microorganisms to adapt to and to resist high concentrations of heavy metals.
Related to archaeal copper (Cu) resistance mechanisms, a few metal efflux pumps have been identified from sequenced genomes of some members of this domain [17]. A Cu-resistance (cop) locus has been described in Archaea, which includes genes encoding a new type of archaeal transcriptional regulator (CopT), a putative metal-binding chaperone (CopM), and a putative Cu-transporting P-type ATPase (CopA) [10]. The same Cu-resistance mechanism was described in Sulfolobus solfataricus P2 and Ferroplasma acidarmanus. In both microorganisms, the putative metal chaperones and the ATPase are cotranscribed and their transcriptional levels increase significantly in response to Cu exposure, suggesting that the transport system is operating for Cu efflux [18, 19]. Recently, it was described that the copRTA operon from S. solfataricus strain 98/2 (copTMA in S. solfataricus P2) is cotranscribed at low levels from the copR promoter under all conditions, whereas increased transcription from the copTA promoter took place in the presence of Cu excess. These authors proposed a model for Cu homeostasis in Sulfolobus which relies on Cu efflux and sequestration [20].
In silico studies have further identified a CPx-ATPase which most likely mediates the efflux of heavy metal cations in the biomining archaeon Metallosphaera sedula [21]. This putative protein has significant identity to a P-type ATPase from S. solfataricus (CopA) [19]. Moreover, M. sedula contains ORFs with significant similarity to both CopM (Msed0491) and CopT (Msed0492) from S. solfataricus [21]. Very recently, Maezato et al. [22] have reported a genetic approach to investigate the specific relationship between metal resistance and lithoautotrophy during biotransformation of chalcopyrite by M. sedula. The functional role of its copRTA operon was demonstrated by cross-species complementation of a Cu-sensitive S. solfataricus copR mutant [22].
Cadmium (Cd) is very toxic and probably carcinogenic at low concentrations. However, the biological effects of this metal and the mechanism of its toxicity are not yet clearly understood [23–26]. In some neutrophilic microorganisms, Cd is taken up via the magnesium or manganese uptake systems [23]. Although the mechanisms in acidophiles have not been elucidated, putative Cd resistance operons in some sequenced genomes from acidophilic microorganisms have been identified. The species with the highest homology to the cadA motif were Acidithiobacillus ferrooxidans and Thermoplasma spp. These high similarities suggest that Cd export may be a common resistance mechanism among acidophiles [11]. Recently, a time-dependent transcriptomic analysis using microarrays in the radioresistant archaeon Thermococcus gammatolerans cells exposed to Cd showed the induction of genes related to metal homeostasis, drug detoxification, reoxidation of cofactors, ATP production, and DNA repair [27].
One alternative mechanism proposed for metal tolerance in microorganisms is the sequestration of metal cations by inorganic polyphosphates (polyP) [28], and at the same time the intracellular cations concentration would regulate the hydrolysis of this polymer [29]. S. metallicus can tolerate very high concentrations of copper and accumulates high amounts of polyP granules [14, 30]. Furthermore, the levels of intracellular polyP are greatly decreased when this archaeon is either grown in 200 mM Cu or shifted to 100 mM Cu [14, 30]. An increase in exopolyphosphatase (PPX) activity and Pi efflux due to the presence of Cu suggests a metal tolerance mechanism mediated through polyP [14, 30]. Actual evidence suggests that polyP may provide mechanistic alternatives in tuning microbial fitness for the adaptation under stressful environmental situations and may be of crucial relevance amongst extremophiles. The genes involved in polyP metabolism in Crenarchaeota have been only partially elucidated, as long as a polyP synthase activity is still to be reported and characterized in this kingdom [15].
Thus far, there are no studies on the prospective genetic and biochemical mechanisms that enable S. metallicus to thrive in such high concentrations of Cu and other metals. Understanding these mechanisms could be particularly useful in potential improvement of the bioleaching microorganisms, which could likely increase the efficiency of bioleaching processes in due course. Since the genome of S. metallicus is not currently available, possible genes involved in Cu resistance were searched by using a CODEHOP and “genome walking” approaches and the transcriptional expression of these isolated genes was assessed by real-time RT-PCR. Furthermore, a proteomic approach was used to identify possible proteins involved in resistance to Cu and Cd in S. metallicus. To our knowledge, this is the first paper that shows the occurrence of a duplicated version of the cop gene cluster in Archaea and gives insights into the molecular Cu and Cd resistance determinants in a thermophilic archaeon employed for industrial biomining.
2. Materials and Methods
2.1. Strains and Growth Conditions
S. metallicus DSM 6482 was grown at 65°C in medium 88 (Deutsche Sammlung von Mikroorganismen und Zellkulturen) containing 0.05% (w/v) elemental sulfur and 0.02% (w/v) yeast extract. S. solfataricus DSM 1617 was grown at 70°C in medium 182 (Deutsche Sammlung von Mikroorganismen und Zellkulturen) with 0.1% (w/v) yeast extract and with 0.1% (w/v) Casamino acids. To study Cd tolerance of S. metallicus and S. solfataricus, the microoganisms were grown in their respective media, except that different concentrations of Cd (0.005–5 mM) were present initially, as indicated in the corresponding experiment.
For differential expression assays, S. metallicus cells were grown in the absence of Cu or Cd to the early stationary phase, and after removing the medium from the cells by centrifugation, they were then shifted to a new medium containing 100–200 mM CuSO4 (Cu from now on) or 1 mM CdSO4 (Cd from now on) during 24 h. After this time, cells were treated to obtain protein extracts. Growth was monitored by measuring unstained cells numbers by means of a Petroff-Hausser chamber under a phase contrast microscope.
2.2. Preparation of Protein Extracts from S. metallicus
Cells from 800 mL of a control culture grown to 108 cells/mL (early stationary phase), or cultures shifted to 100–200 mM Cu or 1 mM, were harvested by centrifugation at 7,700 g for 15 min. The pellets were washed with medium 88 to remove the sulfur. Cells were then resuspended in 50 mM Tris-HCl pH 8.15, 10 mM EDTA, 100 μg/mL PMSF buffer (20 μL per mg wet weight), frozen, and sonicated six times for 30 s each time. The lysates were centrifuged at 4,300 g for 5 min to eliminate cellular debris. The protein concentration of supernatants was determined by the method of Bradford (Coomasie Plus protein assay reagent, Pierce). Between 120 and 500 μg of proteins from the protein extracts were mixed with rehydration IEF buffer, as described by Hatzimanikatis et al. [31] and Choe and Lee [32] with some modifications, including urea 8 M, thiourea 2 M, CHAPS 2% (w/v), Bio-Lyte 3–10 0.27% (v/v), Bio-Lyte 5–8 0.13% (v/v), and bromophenol blue 0.001% (w/v), followed by incubation at 25°C for 30 min. DTT (0.03 g) and sterile nanopure water were then added to complete a final volume of 300 μL. The samples were then incubated at room temperature for 1 h.
2.3. Isoelectric Focussing (IEF)
Each sample (300 μL) was loaded onto the pH 3–10 (nonlinear) 17 cm IPG strips (Biorad) in the first dimension chamber and was incubated at room temperature for 1 h. Mineral oil (2.5 mL) was then added to prevent evaporation of the sample and precipitation of urea, and strips were passively rehydrated for 18 h. The isoelectric focusing was performed with the PROTEAN IEF (Biorad) using the following conditions: 250 V for 15 min, 2,000 V for 2 h, 8,000 V for 4 h, 10,000 V for 11 h, and 50 V for 4 h, reaching a total of 120,000 V/h.
2.4. SDS-PAGE
Prior to the second-dimensional electrophoresis, strips were equilibrated as described by Hatzimanikatis et al. [31] with some modifications. Strips were incubated for 15 min with a solution containing 6 M urea, 156 mM DTT, 30% (v/v) glycerol, 2% (w/v) SDS and 24 mM Tris-HCl pH 6.8 and subsequently for 15 more min in a solution containing 6 M urea, 135 mM iodoacetamide, 30% (v/v) glycerol, 2% (p/v) SDS, and 24 mM Tris-HCl pH 6.8. Finally, strips were incubated with electrophoresis buffer containing 192 mM glycine, 1% (w/v) SDS, and 250 mM Tris-HCl pH 8.3 until the second-dimensional run. SDS-PAGE was performed using the PROTEAN II xi cell as described by Laemmli [33], and gels consisted of 11.5 or 15% (w/v) polyacrylamide. Strips were then overlaid onto the second-dimensional gels sealed with 1% (w/v) agarose in electrophoresis buffer containing a trace amount of bromophenol blue. Electrophoresis was carried out at constant 70 V for 15 h. All experiments were performed in triplicate. Gels were stained with silver or Coomasie Blue G-250 as described by Shevchenko et al. [34] and Giavalisco et al. [35], respectively.
2.5. Gels Analysis and Mass Spectrometry
Gel images were digitized by scanning (Epson) and analyzed with the Delta 2D software (Decodon) to identify the spots differentially expressed due to the presence of toxic metals. An estimate of relative quantitative changes was made on the basis of the change in percent volume among silver stained gels. Spots of interest were recovered from Coomasie Blue G-250 stained gels manually and were sent to electron spray tandem ionization mass spectrometric analysis (tandem MS-MS: ESI-QUAD-TOF). The results obtained were analyzed with Mascot algorithm (http://www.matrixscience.com/index.html) by using the MS/MS Ion search, and all genomes available at databases were used as queries. The entire MS analysis was performed at the Cambridge Center for Proteomics, University of Cambridge, UK.
2.6. CODEHOP-PCR
CopA and CopM amino acid sequences from S. acidocaldarius, S. tokodaii and S. solfataricus were obtained from NCBI (http://www.ncbi.nih.gov/). After aligning the sequences by using the CLUSTAL X program to identify areas of homology, consensus-degenerate PCR primers were designed according to the CODEHOP strategy [36, 37], using the WWW access at http://blocks.fhcrc.org/blocks/codehop.html. Two regions of high sequence similarity were identified for both CopA and CopM sequences, respectively (Figure S1 see Supplementry Material
available online at http://dx.doi.org/10.1155/2013/289236) and used to design the consensus-degenerate hybrid oligonucleotide primers (Table 1). Consensus-degenerate hybrid oligonucleotide primers were designed for the N- and C-terminus of CopA, while a pair of degenerate hybrid oligonucleotide primers was designed for CopM (Figure S1).
Table 1.
Oligonucleotides.
| Name | Sequence | Description |
|
| ||
| copA_cdegF | 5′-gatgtagtaatagtaaaaactggagaaataataccngcngaygg | CODEHOP-PCR |
| copA_cdegR | 5′-tcatcagcaaaattagaagaatctccngtngcdat | CODEHOP-PCR |
| copT_cdegF1 | 5′-ctcaaatagaatataaagtattacaaatgttaaaagargaywsnmg | CODEHOP-PCR |
| copT_cdegR1 | 5′-ggattaccatttatttcatttccacartartcrca | CODEHOP-PCR |
| copT_cdegR2 | 5′-cttatcatattcataaatcttccatctattaatttrtarcaytc | CODEHOP-PCR |
| copT_degF1 | 5′-gartgytayaarctnat | DOP-PCR |
| copT_degR1 | 5′-atnagyttrtarcayct | DOP-PCR |
| copM_degF | 5′-gayccngtntgyggnatgga | DOP-PCR |
| copM_degR | 5′-ccnggnttyccntacgg | DOP-PCR |
| AdaptF2 | 5′-cacgcgtcgactagtactagctt | Genome Walking |
| SP1copA1_3′ | 5′-aaggatgagggggaccttatgg | Genome Walking |
| SP2copA1_3′ | 5′-ggagataagaaatggggtaaaagag | Genome Walking |
| SP1copA1_5′ | 5′-tgataccatcatggaacctgtcag | Genome Walking |
| SP2copA1_5′ | 5′-tcctccacaatcccatccgctg | Genome Walking |
| SP1copA1_5′_2 | 5′-gattgtagctaagttaacctcggcctcg | Genome Walking |
| SP2copA1_5′_2 | 5′-cttctcaccctcagtctggttgg | Genome Walking |
| SP1copA2_3′ | 5′-gaaagaggaatatatgcaagggtaaacgg | Genome Walking |
| SP2copA2_3′ | 5′-gtgttaatgggagagctggaggg | Genome Walking |
| SP1copA2_5′ | 5′-cttctctgtggcaacatcataaccagcc | Genome Walking |
| SP2copA2_5′ | 5′-acgcatgtggcgcaatgcattcc | Genome Walking |
| SP1copT1_5′ | 5′-cattcctcgcaccagcttgcacactctc | Genome Walking |
| SP1copT2_5′ | 5′-cctatgaatactagatcttttccctgaac | Genome Walking |
| SP2copT2_5′ | 5′-aacagcttataacactcgtcactttggc | Genome Walking |
| copM2_RT_F | 5′-gatgaaaaaagccaatataagac | RT-PCR |
| copA2_Rv | 5′-gaacactaactaacatcgcc | RT-PCR |
| copM1_RT_Fw | 5′-ctatcgtttttgttccgaagcttg | RT-PCR |
| copA1_Rv | 5′-cagcagcaagaacagagacgcc | RT-PCR |
| *SM16Sf | 5′-acgctctaaaaaggcgtgggaata | RT-qPCR |
| *SM16Sr | 5′-ttgagctcggggtctttaagcagtg | RT-qPCR |
| copA1Sm_qRT_F1 | 5′-gctaaggtaatagagagcgg | RT-qPCR |
| copA1Sm_qRT_R1 | 5′-tgaacaggaatggacagg | RT-qPCR |
| copA2Sm_qRT_F | 5′-tgtgcttgtctccttagcgt | RT-qPCR |
| copA2Sm_qRT_R | 5′-actcttccgtctttcggagt | RT-qPCR |
| copT1Sm_qRT_F | 5′-tgtaggagagtgtgcaagct | RT-qPCRl |
| copT1Sm_qRT_R | 5′-tcgcaagtgagggttatggt | RT-qPCR |
| copT2Sm_qRT_F | 5′-gtgttacggagcttgca | RT-qPCR |
| copT2Sm_qRT_R | 5′-acactcgtcactttggc | RT-qPCR |
All oligonucleotides were synthesized by Invitrogen. *Oligonucleotides designed by Bathe and Norris [39].
Amplification reactions contained 1x thermophilic DNA polymerase buffer with 2 mM MgCl2, 0.2 mM dNTPs, 0.5 μM of each primer, 5 U Taq DNA polymerase, 40 ng of S. metallicus genomic DNA as a template and water to a final volume of 50 μL. The thermal cycling conditions were 3 min at 95°C; following 30 cycles of 95°C for 30 s, 50°C for 30 s, and 72°C for 1 min; 1 final additional cycle at 72°C for 10 min. Products of amplification were applied onto 1.0% (w/v) agarose gels, and main amplification bands were excised, purified, and TA-cloned into pCR2.1-TOPO (Invitrogen) vector and finally sequenced.
2.7. Genome Walking Experiments
Genome walking strategy was performed as described by Acevedo et al. [38]. Thus, a double-stranded oligo-cassette AdaptT adapter was constructed by annealing of the two unphosphorylated primers AdaptF: (5′-CTAGGCCACGCGTCGACTAGTACTA-GCTT-3′) and AdaptR: (5′-AGCTAGTACTAGTCGACGCG-TGGCCTAG-3′). Annealing was performed by heating the primers (10 μM) in a boiling water bath for 5 min, and then slowly cooling to room temperature.
Six different DNA libraries were constructed by means of digesting 1 μg of S. metallicus genomic DNA with the following restriction enzymes: HindIII, BamHI, EcoRI, EcoRV, NcoI, and PstI, respectively. DNA digestion reactions were carried out using 10 U of restriction enzyme, 2 μL of the corresponding enzyme reaction buffer in 20 μL of total reaction volume. The reaction mix was incubated at 16°C during 16 h. To complete the 3′ recessive end of the DNA fragments and to add a 3′ overhanging adenine, 500 ng of the digested and purified DNA were incubated with 5 U of Taq DNA polymerase, 1 μL of 10 mM dNTPs mix, and 5 μL of 10x thermophilic DNA polymerase buffer in 50 μL total volume, at 70°C for 45 min. Seven μL of this mixture was then incubated with 15 pmol of AdaptT oligo-cassette, 1 U T4 DNA ligase (Promega) and 2 μL of 5x ligase buffer, in a total volume of 10 μL. The ligation reaction was incubated at 16°C during 16 h.
Two consecutive amplification reactions were then performed. The first PCR reaction was done with 1x Elongase mix buffer, 1.9 mM MgCl2, 0.2 mM dNTPs, 0.5 μM first specific primer (SP1) (designed from the known sequence of the target gene; a forward primer was used to amplify the 3′ end a reverse primer for 5′ end amplification), 5 μL of the ligated DNA diluted 10-fold and 1 μL of Elongase (Invitrogen) and water to a final volume of 50 μL. The thermal cycling conditions were as follows: 1 cycle at 94°C for 1 min, 20 cycles of 94°C for 32 s and 68°C for 5 min, and one final additional cycle at 70°C for 7 min. The PCR product was diluted 10 fold and 3 μL were used as a DNA template for a second PCR, which was performed using the same conditions as the first PCR with 0.5 μM second specific primer (SP2) and 0.2 μM oligo-cassette-specific primer AdaptF2 (5′-CACGCGTCGACTAGTACTAGCTT-3′) and 1 μL of Elongase. The thermal cycling conditions were 1 cycle at 94°C for 1 min, 35 cycles of 94°C for 32 s, and 68°C for 5 min, and 1 final additional cycle at 70°C for 7 min. PCR products were excised, purified, TA-cloned into pCR2.1-TOPO (Invitrogen) vector, and finally sequenced.
2.8. Isolation of Total RNA
Cell pellets (10 mg wet weight) were collected and diluted in 60 μL TEN buffer (20 mM Tris-HCl pH 8.0, 1 mM EDTA, 100 mM NaCl) followed by addition of 60 μL TENST buffer (20 mM Tris-HCl pH 8.0, 1 mM EDTA, 100 mM NaCl, 1.6% Na-lauroyl sarcosine, and 0.12% Triton X-100). This suspension was incubated at room temperature for 15 min to allow cell lysis. Total RNA was then extracted using the TRIzol reagent (Invitrogen) as recommended by the manufacturer. DNA contamination in RNA preparations was removed by DNase I treatment (Roche), following the manufacturer's instructions. RNA was then purified, precipitated, and finally resuspended in diethylpyrocarbonate (DEPC)-treated water. Total RNA concentrations were estimated by spectrophotometric measurements (OD260) and its quality was evaluated by determining the ratio of absorption at 260 nm and 280 nm, which was within the preferred range of 1.8–2.1.
2.9. Northern Blotting
For differential gene expression by Northern blotting experiments, the media were supplemented with either different concentrations of Cu (5–50 mM), Cd (5 mM), NiSO4 (15 mM), ZnSO4 (50 mM), or Ag2SO4 (0.08 mM), respectively, as indicated. For some experiments, elemental sulfur was replaced by a chalcopyrite (CuFeS2) concentrate, which was used as the only energy source at 1% (w/v).
Total RNA (5 μg) was separated by electrophoresis on a 1% formaldehyde agarose gel followed by blotting onto Hybond-N nylon membranes (Amersham Pharmacia Biotech, Bucking-hamshire, UK). Hybridization was conducted as described by Sambrook et al. [40]. DNA probes were labeled by random primer DNA labeling kit (Fermentas) using [α-32 p] dCTP. The hybridization signal was detected and analyzed by using the molecular Imager FX system and Quantity One software. The primer sequences for amplification of these gene probes are listed in Table 1.
2.10. Cotranscriptional Analysis by RT-PCR
To study the expression of adjacent genes copM and copA, cDNA was synthesized by using 0.8 μg of total RNA from a S. metallicus culture grown in the presence of 20 mM Cu and a reverse primer hybridizing to copA sequence. A forward primer annealing on copM sequence was used for the PCR reactions (Table 1). PCR amplifications were performed with 1 μL of a 1/10 dilution of the cDNA and 25 pmol of each primer. Amplification conditions included an initial 3 min of denaturation at 95°C, followed by 35 cycles of 30 s at 95°C, 30 s at 55°C, and 1.5 min at 72°C and finished by 10 min at 72°C. A reverse transcriptase reaction without enzyme was carried out in order to exclude amplification due to genomic DNA contamination.
2.11. Quantitative RT-PCR
Single stranded cDNA was synthesized from 0.8 ng of DNA-free RNA samples using random hexamers (Fermentas) and ImProm-II reverse transcription system (Promega) following manufacturer's instructions. The software tool IDT Scitools (Integrated DNA Technologies) was used to design primers producing amplicons of 150–200 bp (Table 1). qPCR was performed with 2 μL of 1 : 10 diluted cDNA samples, 12.5 μL 2x QuantiFast SYBRGreen PCR Master Mix (Quiagen), 1 μM of each primer, and water to a final volume of 25 μL. The efficiency of each primer pair was calculated from the average slope of a linear regression curve, which resulted from qPCRs using a 10-fold dilution series (10 pg–10 ng) of S. metallicus chromosomal DNA as template. Cq values (quantification cycle) were automatically determined by Real-Time Rotor-gene 6000 PCR software (Corbett Life Sciences) after 40 cycles. Cq values of each transcript of interest was standardized to the Cq value of the 16S rRNA gene [39]. At least 3 biological replicates of each assessed condition and 2 technical replicates per qPCR reaction were performed.
3. Results and Discussion
3.1. Tolerance of S. metallicus and S. solfataricus to Cu and Cd
In a previous work, it was determined that S. metallicus was able to tolerate high Cu concentrations. While the presence of 100 mM Cu did not affect S. metallicus growth kinetics, a decreased in cell biomass of only 30% was observed when exposed to 200 mM Cu [30]. The high tolerance to Cu has been described in other acidophilic Bacteria and Archaea compared mostly with neutrophilic microorganisms [11, 14]. Here, this analysis was further extended to characterize the response of S. metallicus to Cd. Thus, it was determined that S. metallicus growth was not affected in the presence of either 0.5 or 1 mM Cd when compared with the control condition in the absence of the metal (Figure 1(a)). In addition, when S. metallicus was challenged with 2 and 3 mM Cd, it was observed that at the late exponential growth phase, the cell numbers decreased by 30 and 50%, respectively (Figure 1(a)). Moreover, growth of S. metallicus was completely inhibited in the presence of 5 mM Cd (Figure 1(a)). On the other hand, S. solfataricus was not able to grow at Cd concentrations greater than 0.05 mM (Figure 1(b)). At 0.01 mM Cd, S. solfataricus cell numbers decreased around 35% (Figure 1(b)). These results are in agreement with previous reports in which S. solfataricus was shown to be able to grow in up to 0.01 mM Cd [41]. Other acidophilic archaeons involved in bioleaching processes, such as Metallosphaera sedula and Ferroplasma acidarmanus, were found to be able to tolerate up to 0.9 and 9 mM Cd, respectively [18, 42]. The minimal inhibitory concentration (MIC) to Cd has been described to be not higher than 1 mM for several thermophilic and neutrophilic Thermococcus species [43]. Recently, it was reported that the most radioresistant archaeon, Thermococcus gammatolerans, stands Cd concentrations with an MIC of 2 mM [27]. To date, despite the heavy metals tolerance showed by some of these microorganisms, strategies to withstand stress from transition metals have been most widely studied only in haloarchaea [44].
Figure 1.
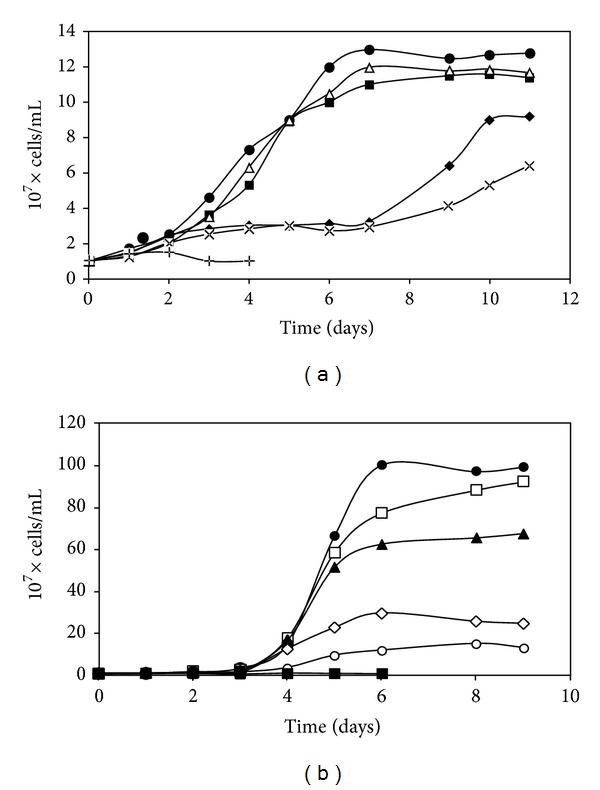
Growth of S. metallicus (a) and S. solfataricus (b) in the presence of Cd. Cells were inoculated in their respective growth media in absence of added Cd (●), or were supplemented with 0.005 mM (□), 0.01 mM (▲), 0.05 mM (◊), 0.1 mM (⚪), 0.5 mM (■), 1 mM (△), 2 mM (♦), 3 mM (×), or 5 mM (+) Cd, and cells were counted daily.
3.2. Effect of Cu and Cd on the Global Proteome of S. metallicus
The proteomic response to Cu and Cd was analyzed to identify proteins which could be involved in heavy metals resistance in S. metallicus. Early stationary phase growing cells were untreated or treated with either 100 or 200 mM Cu or 1 mM Cd during 24 h, and analyzed by comparative two-dimensional gel electrophoresis as indicated in experimental procedures. Twenty-three proteins were found to be down-regulated after 100 mM Cu treatment (Figure 2(a)). A similar pattern was obtained at 200 mM Cu (not shown). Furthermore, by using the Delta 2D software (Decodon), eleven of these proteins were found to be completely absent in the gels and more than 8 proteins decreased their intensity in the range from 1.5-to 5-fold compared with control cells grown in the absence of Cu (Figure 2(a)). Therefore, these results show that a number of proteins became non-detectable or decreased their levels when S. metallicus faced Cu. This kind of response was also seen when the microorganism was exposed to Cd (data not shown). This behavior has also been observed in similar studies where F. acidarmanus was challenged by either As(III) or Cu [16, 18]. In addition, the expression of 30 other proteins was found to be upregulated after Cu treatment (100 or 200 mM) (Figure 2(b)). Most of these proteins could be identified only in the condition with Cu, and spots 5, 13, and 15 were induced 2.6, 3.4, and 5.5-fold, respectively, compared to cells without Cu treatment. Moreover, 3 proteins of low molecular weight (spots 31, 3, and 33) that increased their expression when cells were exposed to 100 mM Cu were also identified (Figure 2, bottom panels). These proteins were only detected when 15% polyacrylamide was used during the second-dimensional separation (SDS-PAGE). On the other hand, a large decrease in overall expression of proteins after 1 mM Cd treatment was observed (not shown). Interestingly, 10 out of the 13 proteins identified as up-regulated when cells were exposed to Cd and also showed increased levels after Cu treatment. Some of them are shown in Figure 3.
Figure 2.
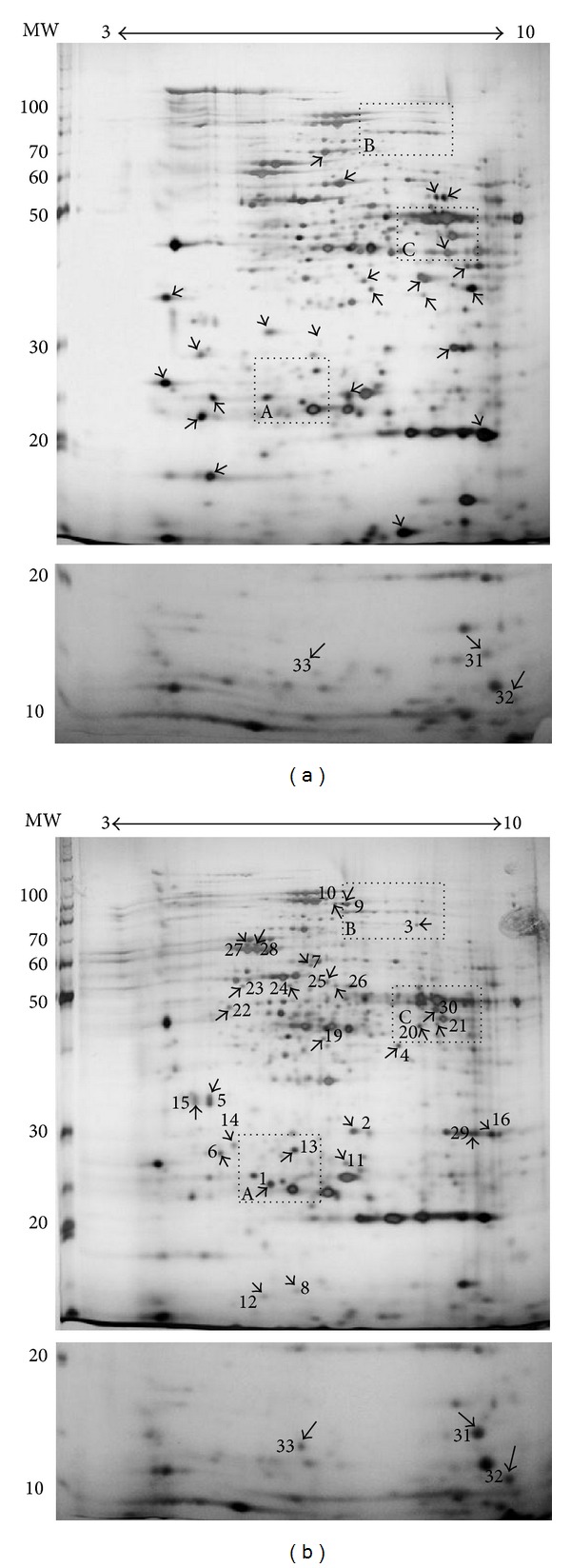
Changes in the proteome of S. metallicus grown in the presence of Cu. Cells were incubated in the absence of any added metal (a) or in the presence of 100 mM Cu (b). Arrows indicate the spots that were downregulated (a) or upregulated (b) in the presence of Cu. Numbers indicate the spots with increased intensity in cells treated with copper. The dashed boxes are shown as enlarged areas that include some proteins upregulated in the presence of Cu and Cd (Figure 3). The bottom panels show low molecular weight proteins upregulated in the presence of Cu separated by using a 15% polyacrylamide gel in the second dimension.
Figure 3.
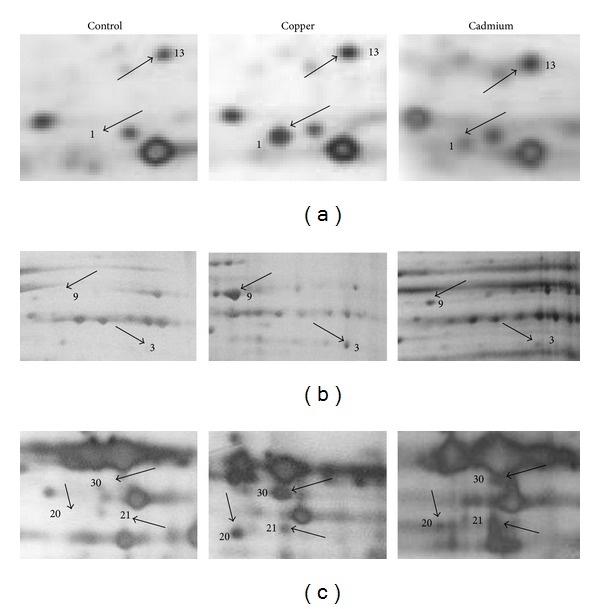
Comparison of selected proteins of S. metallicus cells treated with Cu or Cd. Protein extracts were obtained from cells treated without metals and with 100 mM Cu or 1 mM Cd for 24 h and the proteins were separated by 2D-PAGE. The segments (a), (b), and (c) are the enlarged dashed boxes in Figure 2 under the three conditions indicated. Numbers show some of the spots with increased intensity in cells treated with Cu or Cd.
3.3. Identification of Proteins Upregulated in Cells Treated with Cu and Cd in S. metallicus
A total of 18 S. metallicus proteins whose levels were found to be up-regulated in response to Cu were analyzed by mass spectrometry. The identified proteins included functions related to the production and transport of energy, biosynthesis of amino acids, stress responses, and transcription regulation (Table 2). Amongst the proteins related to production and transport of energy, a putative ATP synthase subunit B (spot 23) that has been described as playing a fundamental role in ATP synthesis was identified (Table 2). When cells are subjected to some stressing conditions such as the presence of heavy metals, a greater cellular demand for energy has been reported to occur [18]. Most likely, this phenomenon might be due to ATP-driven Cu transport via ATPases that have a substantial interplay during metal detoxification [45]. Furthermore, the ATP synthase subunit B was also identified as up-regulated when S. metallicus was treated with Cd (Table 2).
Table 2.
Proteins upregulated in S. metallicus cells exposed to 100 mM Cu or 1 mM Cd.
| Spot | Molecular weight (kDa) | Cu induction levels | Putative function | Microorganism related | Accesion number |
|---|---|---|---|---|---|
| 1† | 26.6 | ∞ | Proteasome subunit | S. solfataricus | AAK41034 |
| 3† | 70.1 | ∞ | Ferredoxin oxidoreductase | S. solfataricus | AAK42926 |
| 4† | 42.9 | ∞ | Alcohol dehydrogenase | S. solfataricus | AAK43154 |
| 5 | 35 | 2.6 | Hypothetical protein | S. acidocaldarius | YP_254976 |
| 6 | 27 | ∞ | Hypothetical protein | P. aerophilum | NP_559889 |
| 9† | 57 | ∞ | Phosphoglycerate dehydrogenase | M. thermautotrophicus | AAB85466 |
| 13† | 27 | 3.4 | Unknown | — | — |
| 14 | 28 | ∞ | Unknown | — | — |
| 15 | 35 | 5.5 | Unknown | — | — |
| 20† | 46.1 | ∞ | Glutamate dehydrogenase | S. solfataricus | AAK42230 |
| 21† | 45 | ∞ | Hypothetical protein | M. acetivorans | NP_618380.1 |
| 22 | 49 | ∞ | Hypothetical protein | P. torridus | YP_022807 |
| 23† | 51 | ∞ | ATP synthase subunit B | S. solfataricus | AAK40880 |
| 24 | 58.5 | ∞ | HSP60 subunit | S. shibatae | AAG37273.1 |
| 30† | 48 | ∞ | Unknown | — | — |
| 31 | 16 | ND | Unknown | — | — |
| 32 | 14.4 | ND | Putative transcription regulator | S. solfataricus | AAK40413 |
| 33 | 13.8 | ND | Transcription factor nusA | S. solfataricus | AAK40563 |
Spots refer to those numbered in Figure 2. Proteins in Figure 2 which are not included in this table were not subjected to sequencing due to their low intensities in the gels.
†Spot up-regulated in cells exposed to Cu and Cd.
ND: not determined.
Infinity symbol indicates that proteins were expressed only in presence of copper or cadmium.
M. thermoautotrophicus: Methanobacterium thermoautotrophicus; M. acetivorans: Methanosarcina acetivorans; S. shibatae: Sulfolobus shibatae; P. torridus: Picrophilus torridus; P. aerophilum: Pyrobaculum aerophilum; S. acidocaldarius: Sulfolobus acidocaldarius.
Two other proteins corresponded to putative oxidoreductases such as ferredoxin oxidoreductase (spot 3) and alcohol dehydrogenase (spot 4), that have been generally involved in electron transporter chains and use NAD+ as an electron acceptor. The levels of ferredoxin oxidoreductase were also up-regulated in response to Cd (Table 2). Several previous reports have suggested that oxidoreductases contribute to an oxidative protection in both Bacteria and Archaea in response to heavy metals [46–48]. In the neutrophilic microorganisms Escherichia coli and Staphylococcus aureus, it has been described that oxidases and dehydrogenases contribute to oxidative protection, Cu homeostasis, and stress responses [46, 47]. Furthermore, some proteins involved in oxidative damage repair, such as NADH-dependent oxidases and thioredoxin reductases, were expressed in cells of F. acidarmanus exposed to As(III) [16]. The expression of this group of proteins has also been observed when the same microorganism was exposed to Cu [18]. Therefore, oxidoreductases play an important role against oxidative stress and may eliminate reactive oxygen species, which constitute the major component of the stress caused by transition metals [44, 49, 50].
The enzymes related to biosynthesis of amino acids were found as commonly up-regulated either in response to Cu or Cd. They corresponded to a phosphoglycerate dehydrogenase (spot 9) and a glutamate dehydrogenase (spot 20). The former one catalyzes the NAD+-dependent oxidation of 3-phosphoglycerate into 3-phosphohydroxypyruvate, a branch point from the glycolytic pathway and the initial reaction in L-serine biosynthesis [51]. Glutamate dehydrogenase catalyzes the oxidative deamination of glutamate to produce 2-oxoglutarate and ammonia with reduction of NAD+ [52]. Proteins involved in amino acids synthesis were expressed in cells of F. acidarmanus exposed to As(III) [16]. Moreover, increased levels of proteins involved in the biosynthesis of sulfur-containing amino acids have been observed in Saccharomyces cerevisiae cells exposed to Cd [24], suggesting that these proteins might also have a role in the cellular response to adverse conditions.
Two proteins showed identity to components of the stress response mechanism such as one subunit of the HSP60 chaperonin (spot 24) and one subunit of the proteasome (spot 1). In Archaea, the stress protein HSP60 is directly involved in protein folding processes [53]. Proteomic analysis of F. acidarmanus cells exposed to As(III) and Cd revealed the expression of proteins involved in protein folding and DNA repair, including HSP60 chaperonin and DnaK heat-shock protein (HSP70); thereby the authors suggested that this microorganism uses multiple mechanisms to resist high levels of Cu [16, 18]. On the other hand, proteasomes are described as nonspecific proteolytic nanomachines associated with protein catabolism. Proteasomes are known to be closely involved in maintaining protein quality control by degrading miss folded and denatured proteins in response to cell stress in all three domains of life [54]. A proteomic analysis in the thermoacidophilic archaeon Thermoplasma acidophilum showed that proteasomes and chaperone-related proteins were highly induced against stress conditions, indicating a high turnover rate of proteins [55]. In P. furiosus, the expression of the β1 subunit of the proteasome was induced in cells subjected to heat shock stress [56]. Additionally, the proteasome subunit was also found to be up-regulated when S. metallicus was subjected to Cd stress (Table 2).
The transcriptional regulation-related proteins corresponded to a putative transcription regulator (spot 32) and the transcription factor NusA (spot 33) involved in the intrinsic termination of transcription [57]. NusA has been also reported to display important functions in the cellular response to adverse factors [58]. The expression of two transcriptional regulators related to amino acids biosynthesis and metal-dependent genetic repression were found to be induced in P. furiosus cells subjected to oxidative stress [48]. Moreover, the expression of several genes encoding predicted transcriptional regulators were induced following Cd exposure in T. gammatolerans cells [27].
Spots 5, 6, 13, 14, 15, 21, 22, 30, and 31 (Table 2) did not show significant scores with any protein sequences currently available in the data bases. Interestingly, spots 2, 4, 9, 13, 21, and 30 were found to be commonly upregulated when S. metallicus cells were treated with either Cu or Cd (Figure 3, Table 2).
The results presented here are consistent with previous work describing global protein expression profiles in response to heavy metals as reported for F. acidarmanus [16, 18], microbial-biofilm communities in an acid mine drainage site [59], and other prokaryotic and eukaryotic microorganisms [24, 60]. Therefore, a coordinated expression of different groups of genes suggests the existence of regulatory networks such as stress response mechanisms and respiratory chain adjustments to cope with the presence of heavy metal ions [61].
3.4. Cu-Resistance (Cop) Genes Cluster Is Duplicated in S. metallicus Genome
A Cu-resistance (cop) locus has been described to be highly conserved in archaeal genomes, which consists of genes encoding a new type of archaeal transcriptional regulator (CopT), a metal-binding chaperone (CopM), and a Cu-transporting P-type ATPase (CopA) [19]. Since the genome sequence of S. metallicus is not yet available, we searched for the presence of cop genes by using consensus degenerate hybrid oligonucleotide primers-based PCR (CODEHOP-PCR). CopA and CopM amino acid sequence alignments from S. acidocaldarius, S. tokodaii, and S. solfataricus were used to identify conserved sequence blocks and therefore to design either CODEHOP or DOP (degenerate hybrid oligonucleotide) primers (Figure S1,Table 1). Thereby PCR assays using S. metallicus genomic DNA as template yielded amplicons of ca. 150 bp and ca. 950 bp for copM and copA CODEHOP pair primers, respectively (Figures 4(a), 4(b)). As estimated from primer pair's position on amino acid sequence alignments, the obtained PCR products corresponded to the expected sizes, suggesting the presence of copM- and copA-like genes in S. metallicus genome. In order to determine the identity of the amplified DNA fragments, they were TA-cloned and sequenced. Blastx sequence analysis showed that the 150 bp DNA fragment coded for an incomplete ORF sharing 63% homology with S. solfataricus CopM (SSO10823), whereas the analysis of the 950 pb DNA fragment yielded a partial ORF sharing 54% homology with S. solfataricus CopA protein (SSO2651).
Figure 4.
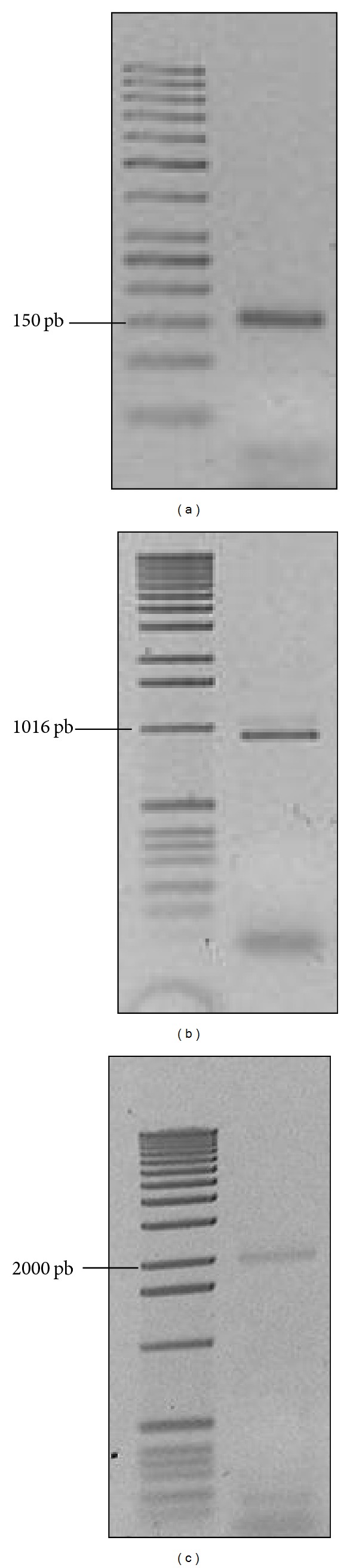
CODEHOP-based PCR for the amplification of the putative genes copA and copM from S. metallicus. Amplification products, (a) using degenerate PCR primers (DOP) copM_degF and copM_degR designed from amino acid sequence conserved blocks of Sulfolobales CopM proteins, (b) using primers copA_cdegF and copA_cdegR designed by CODEHOP strategy for amplification of a copA-like gene. (c) Primers copM_degF and copA_cdegR were assesed for the amplification of a copMA-like DNA region. Those primers were tested with genomic S. metallicus DNA samples. Amplicons expected sizes were as follows: ca. 150 bp for copM, ca. 950 bp for copA, and ca. 2,000 bp for copMA. PCR products were excised and cloned for later sequencing.
The genomic organization of the putative cop genes in S. metallicus was analyzed to find out its similarity with those previously described in S. solfataricus P2 and F. acidarmanus, where the cop cluster consists of tandem-orientated genes as copTMA [18, 19]. Thus, using copM_degF (forward) and copA_cdegR (reverse) primers, we attempted to amplify a putative copMA DNA region. This PCR yielded a product of ca. 2,000 pb, corresponding to the expected size, and resulted in 2 ORFs showing high sequence homology with CopM and CopA from S. solfataricus (Figure 4(c)). Interestingly, when aligning this copMA sequence with the copA and copM sequences obtained previously, they were not identical, which strongly indicated the presence of duplicated putative cop genes encoding for both Cu-ATPases (copA1 and copA2) and metallochaperones (copM1 and copM2) in the S. metallicus genome.
To isolate the entire cop gene sequences, the genome walking method described by Acevedo et al. [38] was used. Six different DNA libraries were constructed from S. metallicus genomic DNA digested with several restriction enzymes, including 5′ overhang and blunt end restriction enzymes. The DNA libraries were used as templates in 2 successive PCR reactions. From the previously known sequences, two specific sense primers for the 3′ end amplification and two specific antisense primers for the 5′ end amplification were designed (abbreviated SP1 and SP2). Therefore, the 5′ and 3′ ends of the putative genes copA1 and copA2 that had been partially identified through the genome walking technique were amplified.
By overlapping the sequence isolated by degenerate PCR and the lateral sequences obtained by genome walking (Figure S2), it was possible to complete the whole nucleotide sequence of the putative gene copA1. Additionally, it was possible to confirm the presence of the upstream copM1 gene and determine the occurrence of a partial copT1 gene tandem-orientated upstream of copM1 (Figure 5). Through additional genome walking experiments, the partial copT1 sequence could be further completed. The same was true when isolating the whole nucleotide sequence of the putative gene copA2 as the presence of copM2 and copT2 genes were confirmed upstream of copA2. However, it was not possible to complete the whole 5′ end of the putative gene copT2 as its determined length corresponded to only 90% of the full length when compared with S. solfataricus copT gene (Figure 5).
Figure 5.
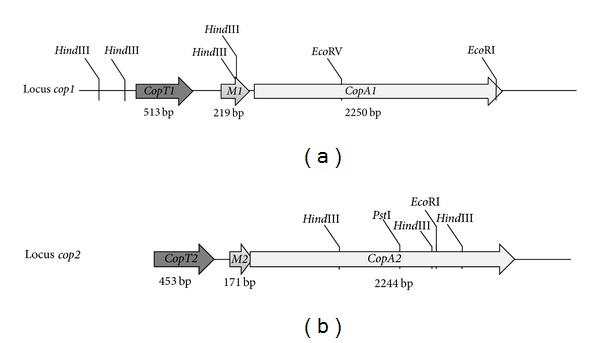
Schematic representation of the two cop gene clusters isolated from S. metallicus genome. Each cluster codes for an Archaea-specific transcriptional regulator (copT), a metallochaperone (copM), and a P-type Cu-exporting ATPase (copA). Lengths of each gene are indicated. Locus cop1 corresponds to 4,346 sequenced base pairs and shows an intergenic region copT1-M1 of 232 base pairs. Locus cop2 corresponds to 3,435 base pairs and shows 89 base pairs copT2-M2 intergenic region. Genes copM2 and copA2 overlapped in 11 base pairs. copT2 was not fully isolated.
In conclusion, degenerate PCR together with genome walking experiments allowed to describe the occurrence in S. metallicus genome of 2 cop genes loci (named as locus cop1 and locus cop2) coding for paralogous genes whose products may be involved in Cu-resistance (Figure 5). Although some archaeal genomes exhibit two copies of putative Cu-P-type ATPases as described for S. solfataricus [62], the genomic arrangement displayed as a cop gene cluster (copTMA) has been reported to be represented in only one copy for many archaeal genomes [10, 19, 21]. Moreover, we further updated this analysis by searching for duplications of the cop genes cluster in all available archaeal genome sequences up to date (October, 2012), retrieving only one cop locus in each case. Thereby we propose that the discovery of this cop gene cluster duplication in the genome of S. metallicus constitutes so far an unprecedented feature for a representative of the Archaea domain and might contribute to its high metal resistance.
In this context, it has been widely reported that increased gene copy number can increase gene expression allowing prokaryotic to thrive under growth limiting conditions [13, 63–68]. However, alongside the augmented gene copy, one cannot exclude that the cop operon duplication in S. metallicus may offer among others: a wider repertoire of Cu-ATPases, which might differ in terms of metal affinities and/or specificity, efflux rates, and/or differences in their abundance regulation.
Throughout amino acid sequence analysis, it was further determined that each polypeptide encoded by S. metallicus, with the exception of CopT1, showed to be highly homologous to their orthologous counterparts encoded by others Sulfolobales (Table 3). Moreover, the 3 ORFs encoded by the locus cop2 shared about 90% identity with the corresponding S. solfataricus orthologous peptides (Table 3). S. metallicus cop genes products showed characteristic domains, referred to as critical for their respective proposed biological activities. S. metallicus paralogous gene products CopA1 (749 aa′) and CopA2 (747 aa′) shared 51.3% of identity. These two putative Cu-ATPases (CopA1 and CopA2) contain the amino acid sequence motif CPCALGLA which has been proposed to confer Cu-transporting specificity in CPx-ATPases [48] and has been also found in other CopA-like proteins from biomining Bacteria and Archaea [14].
Table 3.
Sequence identity comparison of S. metallicus Cop proteins to those in other Sulfolobales.
| S. metallicus protein | % Identity to homologue in other Sulfolobales | |||
|---|---|---|---|---|
| S. solfataricus P2 | S. acidocaldarius | S. tokodaii | M. sedula | |
| CopA1 | 51 | 43 | 44 | 45 |
| CopA2 | 88 | 46 | 49 | 50 |
| CopM1 | 63 | 69 | 71 | 71 |
| CopM2 | 89 | 72 | 70 | 77 |
| CopT1 | 36 | 27 | 30 | 32 |
| CopT2* | 90 | 52 | 52 | 53 |
Boldface type indicates to which organism the S. metallicus homologue shows the highest sequence identity.
*Refers to the analysis obtained using the uncompleted amino acid sequence of CopT2, which represents ~90% in length when compared with S. solfataricus CopT sequence.
On the other hand, CopM1 (71 aa) and CopM2 (56 aa), sharing 66% of identity, present the proposed metal binding domain TRASH (trafficking, resistance, and sensing of heavy metals) [10]. Additionally, when analyzing the putative transcriptional regulators, the presence of a C-terminal TRASH domain in both CopT1 and CopT2 was also found. CopT1 sequence also contains an entire N-terminal helix-turn-helix (HTH) motif (not shown) that resembles the DNA-binding motifs of prokaryotic transcriptional regulators, such as Lrp-like proteins [67]. The HTH motif was only partially identified in CopT2 since the 5′ region of copT1 gene has not been yet fully isolated.
3.5. Transcriptional Analysis of S. metallicus Cop Genes
To get insight into the role of the two cop loci, the expression of the corresponding genes was analyzed under various conditions. S. metallicus was first grown in presence of different heavy metals (Cu, Zn, Cd, Ni o Ag) at a given concentration that did not affect growth kinetics [30, 42]. Subsequently, total RNA was isolated from late exponentially grown cultures and Northern blot experiments were carried out in order to determine the expression of both copA1 and copA2 genes. As depicted in Figure 6, transcription of both copA1 and copA2 messengers were specifically induced in the presence of Cu and Cd ions. This gene expression pattern is in good agreement with what has been described for S. solfataricus, where the bicistronic copMA transcript levels were also found to be up-regulated in response to both Cu and Cd [19].
Figure 6.
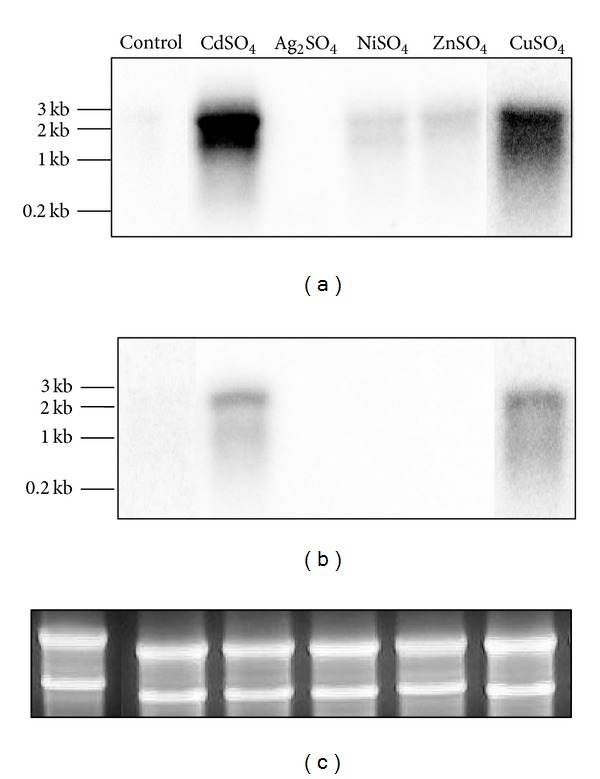
Northern blot analysis to determine the expression of S. metallicus copA1 (a) and copA2 (b) putative genes in response to various metals. S. metallicus total RNA was extracted from cultures grown either in the absence (control) or the presence of CuSO4 (50 mM), Ag2SO4 (0.08 mM), NiSO4 (15 mM), ZnSO4 (50 mM), and CdSO4 (2 mM). P32-radioactivelly labelled DNA fragments annealing to copA1 and copA2 sequences, respectively, were used as probes. (c) shows rRNAs as total RNA loading control.
Northern blot analysis showed that the sizes seen for both copA transcripts (~2.5 Kb) did not correspond exactly with the individual genes length (each of about 2.25 Kb) (Figure 6). This strongly suggested that the transcripts could also include the respective copM gene located upstream of copA in each cop loci. To confirm this assumption, a co-transcription experiment was done (Figure 7) in which the cDNAs were obtained by using RNA extracted from a culture grown in the presence of 20 mM Cu and using a reverse primer hybridizing with copA1 (Figure 7(a)) or copA2 (Figure 7(b)) gene sequence, respectively. PCR amplifications were carried out by using the corresponding cDNAs as templates and each pair of primers lying in adjacent genes (copM). The presence of an amplicon of the expected size in each case indicated the adjacent genes were part of polycistronic messengers (Figure 7). These results clearly show that in S. metallicus each couple of copMA genes was expressed in the form of transcriptional units, as reported in F. acidarmanus and S. solfataricus [18, 19]. The coexpression of gene pair's copMA may suggest a coordinated and dependent function for the respective encoded proteins. In the bacterium E. hirae, the metallochaperone CopZ (CopM in S. metallicus) fulfills a pivotal role in the mechanism of Cu homeostasis [45]. Thus, it was shown that this protein interacts directly with the Cu-ATPase CopB (CopA in S. metallicus), handing the Cu for subsequent removal. In this context, one might expect that proteins CopM1 and CopM2 from S. metallicus have a similar role to that described for E. hirae. Furthermore, quantitative RT-PCR experiments were carried out in order to determine differential expression of the cop genes in response to different Cu concentrations. The expression of copA1, copA2, copT1 and copT2 was tested relative to the transcript levels of rRNA 16S gene since its levels were not significantly affected in all assessed conditions (data not shown). As shown in Figure 8(a), copA2 and copA1 transcript levels were found to be concomitantly increased with the increasing Cu concentrations present in the medium. Higher transcripts levels of copA1 were seen in all tested conditions when compared with copA2 levels. copA1 transcript levels were found to be 32.5-fold up-regulated when comparing 50 mM Cu condition versuscontrol (absence of metal), whereas copA2 transcript levels showed an increment of only 17.5-fold (Figure 8(a)). The finding that Cu-ATPases mRNA levels were significantly increased in response to Cu ions exposure suggests that the transport system may operate for Cu efflux in S. metallicus.
Figure 7.
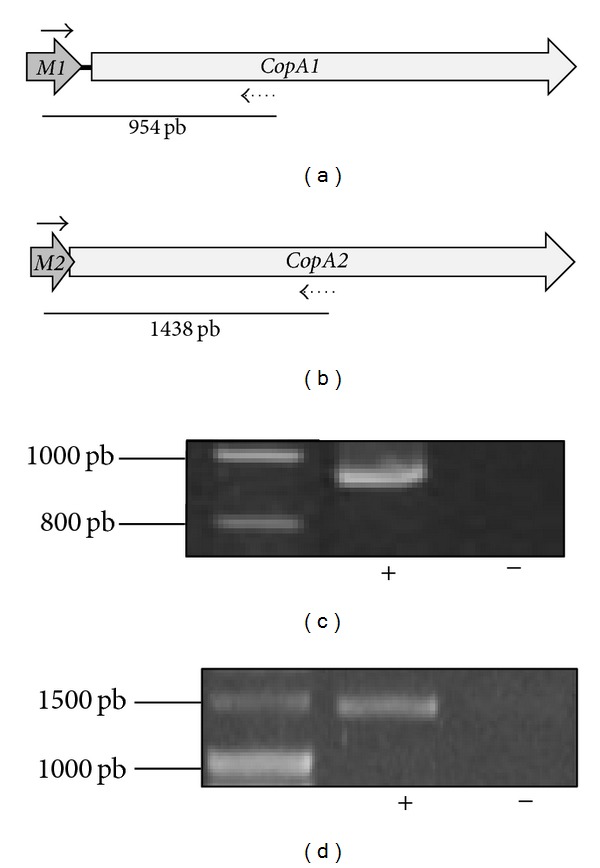
Cotranscription analysis of copMA1 and copMA2 genes. cDNA was synthesized with a reverse primer (dotted arrows) hybridizing toward the 3′ end of either copA1 (a) or copA2 (b). S. metallicus total RNA was extracted at the late exponential phase from a culture growing in presence of 20 mM Cu. PCR amplifications were carried out with these cDNAs and each corresponding primer pair (black arrows) as listed in Table 1. (c) and (d) show RT-PCR products obtained for the copMA1 and copMA2 intergenic regions, respectively. Reverse transcriptase reactions with (+) and without (−) the Improm II reverse transcriptase enzyme were carried out in order to exclude amplification due to genomic DNA contamination. Expected sizes (in base pairs) for the corresponding PCR products are given in (a) and (b).
Figure 8.
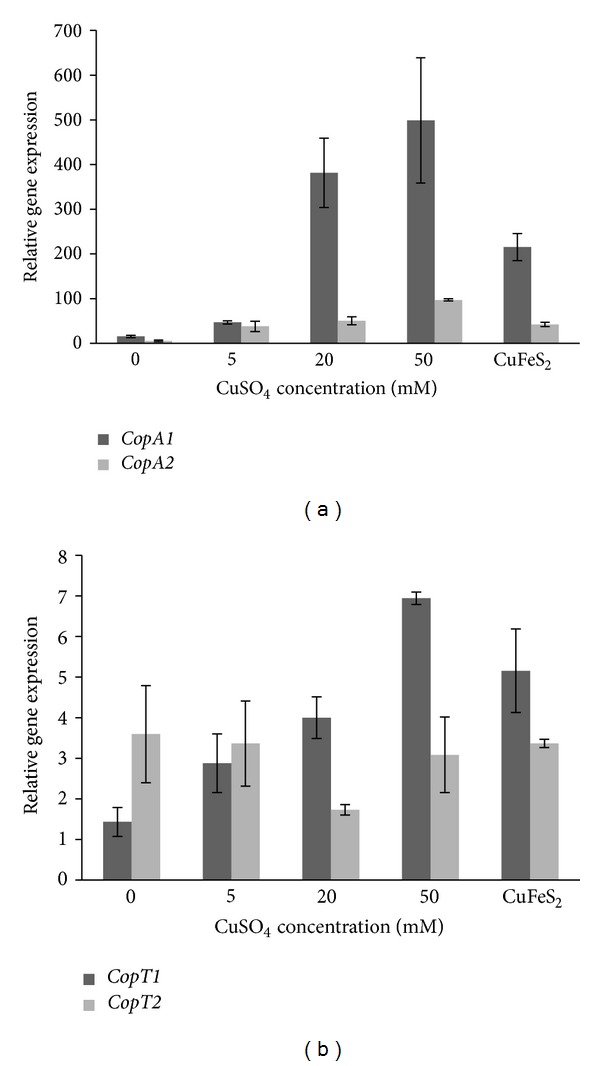
Relative expression levels of S. metallicus genes copA1 and copA2 (a) copT1 and copT2 (b). Expression of S. metallicus cop genes wasassessed by qRT-PCR and normalized against 16S rRNA gene expression in individual cultures. Mean values and standard deviations are from analyses of three independent cultures grown in the presence of 0, 5, 20, 50 mM Cu or in 1% of a chalcopyrite ore (CuFeS2) concentrate.
Moreover, copA2 and copA1 gene expression was quantified when S. metallicus was grown using chalcopyrite (CuFeS2) ore as an oxidizable substrate (Figure 8(a)). Mineral oxidation mediated by the microorganism generates a progressive increase in Cu ions concentration in the medium. To find out whether the amount of solubilized Cu induced the expression of the Cu-ATPases genes, total RNA was extracted from a S. metallicus culture grown to late exponential phase and in the presence of 1% CuFeS2. It was clear that an up-regulation of the Cu-ATPases encoding genes also took place when S. metallicus was grown in the presence of CuFeS2 (Figure 8(a)). copA1 transcript levels increased 14-fold compared with the control condition in the absence of Cu, while copA2 gene expression showed an increase of 7.6 fold. Along with this, by means of atomic absorption spectrometry (AAS) analysis, an overall amount of solubilized Cu ions (Cu2+/Cu1+) of 14.4 ± 2.1 mM was determined to be present in the medium, indicating that copA2 and copA1 gene expression was most likely in response to the Cu present in the culture due to CuFeS2 microbial-solubilization.
The finding that copA1 was highly expressed compared with copA2 in all tested conditions may suggest a possible physiological hierarchy between the two ATPases when overcoming either Cu or Cd stress. In this regard, by means of a genetic approach it was demonstrated in S. solfataricus that while CopA was an effective Cu efflux transporter at low and high Cu concentrations, the other Cu-ATPase (CopB) only appeared to be a low-affinity Cu export ATPase [68]. Moreover, by using a M. sedula copA deletion mutant it was demonstrated that this strain compromised metal resistance and consequently abolished chalcopyrite oxidation [22], highlighting the role of Cu detoxification mechanisms during a given bioleaching process. Our attempts to show the functionality of CopA1 and CopA2 from S. metallicus by using E. coli as a heterologous host were not successful. Apparently, the overexpression of these ORFs had a toxic effect on E. coli that compromised its viability. Although gene disruption tools have not been yet developed for S. metallicus the functional role of both copTMA locimight be further studied by cross-species complementation of a copper sensitive S. solfataricus copR mutant as it was described by Maezato et al. [22]. It will be of great interest to establish in future studies the possible functionality of the isolated putative transporters from S. metallicus.
Likewise, quantitative RT-PCR experiments showed that copT1 transcript levels increased concomitantly with increasing Cu concentrations, whereas copT2 showed relatively similar transcripts levels, most likely indicating a constitutive expression profile (Figure 8(b)). Furthermore, whereas copT1 transcript levels were found to be increased 3.6 fold in CuFeS2 grown cultures, copT2 levels remained unchanged in comparison with the control in the absence of Cu (Figure 8(b)). The results obtained for S. metallicus copT2 gene expression are consistent with those reported for S. solfataricus. CopT transcriptional regulator has been proposed to function as a repressor in S. solfataricus, showing a constitutive expression that in the presence of Cu loses its affinity for the promoter region of copMA allowing the expression of this polycistron [19]. In contrast, the increased transcripts levels of both copA1 and copT1 concomitant with higher Cu concentrations in the environment suggest that the putative transcriptional regulator CopT1 may act by activating both copMA1 and probably itself (Figure 8). Regarding this, it was recently reported that CopR (CopT in S. metallicus) from S. solfataricus strain 98/2 acts as an activator of copT (copM in S. metallicus) and copA expression [69]. Nevertheless, additional experiments would be required to test this possibility in S. metallicus.
4. Concluding Remarks
We previously reported that S. metallicus resists extremely high Cu concentrations, which was mediated to some extent by the use of a possible metal resistance system based on inorganic polyphosphate hydrolysis and consequently Cu-PO4 2−efflux [15, 29, 30]. Here we have addressed the question whether this microorganism coded for other determinants that might help to explain its high heavy metal tolerance. As the genomic sequence of this microorganism is not yet available, we jointly used CODEHOP-PCR and genome walking approaches and were able to establish the occurrence of two nonidentical homologous cop loci into the genome of S. metallicus (cop1 and cop2). Each cop locus codes for an archaeal transcriptional regulator (CopT), a metal-binding chaperone (CopM) and a Cu-transporting P-type ATPase (CopA). High levels of the polycistronic mRNAscopMA of each cop locus were observed after treatment with either Cu or Cd, suggesting that the encoded ATPases efflux heavy metals out in order to detoxify the intracellular environment. Altogether, previous reports and the results obtained in this study allow us to suggest that some key elements that may explain the high resistance to Cu in S. metallicus is the duplication of the Cu resistance cop genes, a defensive response to stress and a polyP-based accumulation mechanism. In the case of Cd, although some similar stress responses were observed, whether comparable Cu-responsive elements also participate in Cd responses remains to be seen.
Supplementary Material
CODEHOP-based PCR details for the amplification of the putative genes copA and copM from S. metallicus are provided in the supplementary material.
Conflict of Interests
The authors declare that the research was done in the absence of any commercial or financial relationships that could be construed as a potential conflict of interests.
Acknowledgments
This work was supported by Grant no. FONDECYT 1110214 and in part by ICM P-05-001-F project.
References
- 1.Schippers A. Microorganisms involved in bioleaching and nucleic acid-based molecular methods for their identification and quantification. In: Donati RE, Sand W, editors. Microbial Processing of Metal Sulfides. Berlin, Germany: Elsevier; 2007. pp. 3–33. [Google Scholar]
- 2.Watling HR. The bioleaching of sulphide minerals with emphasis on copper sulphides: a review. Hydrometallurgy. 2006;84(1-2):81–108. [Google Scholar]
- 3.Rawlings DE, Dew D, Du Plessis C. Biomineralization of metal-containing ores and concentrates. Trends in Biotechnology. 2003;21(1):38–44. doi: 10.1016/s0167-7799(02)00004-5. [DOI] [PubMed] [Google Scholar]
- 4.Valenzuela L, Chi A, Beard S, et al. Genomics, metagenomics and proteomics in biomining microorganisms. Biotechnology Advances. 2006;24(2):197–211. doi: 10.1016/j.biotechadv.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 5.Jerez CA. Bioleaching and biomining for the industrial recovery of metals. In: Moo-Young M, editor. Comprehensive Biotechnology. 2nd edition. Vol. 3. Amstaerdam, The Netherlands: Elsevier; 2011. pp. 717–729. [Google Scholar]
- 6.Norris PR, Owen JP. Mineral sulphide oxidation by enrichment cultures of novel thermoacidophilic bacteria. FEMS Microbiology Reviews. 1993;11(1–3):51–56. [Google Scholar]
- 7.Rawlings DE, Johnson DB. The microbiology of biomining: development and optimization of mineral-oxidizing microbial consortia. Microbiology. 2007;153(2):315–324. doi: 10.1099/mic.0.2006/001206-0. [DOI] [PubMed] [Google Scholar]
- 8.Norris PR, Burton NP, Foulis NAM. Acidophiles in bioreactor mineral processing. Extremophiles. 2000;4(2):71–76. doi: 10.1007/s007920050139. [DOI] [PubMed] [Google Scholar]
- 9.Bruins MR, Kapil S, Oehme FW. Microbial resistance to metals in the environment. Ecotoxicology and Environmental Safety. 2000;45(3):198–207. doi: 10.1006/eesa.1999.1860. [DOI] [PubMed] [Google Scholar]
- 10.Ettema TJG, Huynen MA, De Vos WM, Van Der Oost J. TRASH: a novel metal-binding domain predicted to be involved in heavy-metal sensing, trafficking and resistance. Trends in Biochemical Sciences. 2003;28(4):170–173. doi: 10.1016/S0968-0004(03)00037-9. [DOI] [PubMed] [Google Scholar]
- 11.Dopson M, Baker-Austin C, Koppineedi PR, Bond PL. Growth in sulfidic mineral environments: metal resistance mechanisms in acidophilic micro-organisms. Microbiology. 2003;149(8):1959–1970. doi: 10.1099/mic.0.26296-0. [DOI] [PubMed] [Google Scholar]
- 12.Franke S, Rensing C. Acidophiles. Mechanisms to tolerate metal and acid toxicity. In: Gerday C, Glansdorff N, editors. Physiology and Biochemistry of Extremophiles. Washington, DC, USA: ASM Press; 2007. pp. 271–278. [Google Scholar]
- 13.Navarro CA, Orellana LH, Mauriaca C, Jerez CA. Transcriptional and functional studies of Acidithiobacillus ferrooxidans genes related to survival in the presence of copper. Applied and Environmental Microbiology. 2009;75(19):6102–6109. doi: 10.1128/AEM.00308-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orell A, Navarro CA, Arancibia R, Mobarec JC, Jerez CA. Life in blue: copper resistance mechanisms of bacteria and Archaea used in industrial biomining of minerals. Biotechnology Advances. 2010;28(6):839–848. doi: 10.1016/j.biotechadv.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 15.Orell A, Navarro CA, Rivero M, Aguilar JS, Jerez CA. Inorganic polyphosphates in extremophiles and their posible functions. Extremophiles. 2012;16(4):573–583. doi: 10.1007/s00792-012-0457-9. [DOI] [PubMed] [Google Scholar]
- 16.Baker-Austin C, Dopson M, Wexler M, et al. Extreme arsenic resistance by the acidophilic archaeon “Ferroplasma acidarmanus” Fer1. Extremophiles. 2007;11(3):425–434. doi: 10.1007/s00792-006-0052-z. [DOI] [PubMed] [Google Scholar]
- 17.Pedone E, Bartolucci S, Fiorentino G. Sensing and adapting to environmental stress: the archaeal tactic. Frontiers in Bioscience. 2004;9:2909–2926. doi: 10.2741/1447. [DOI] [PubMed] [Google Scholar]
- 18.Baker-Austin C, Dopson M, Wexler M, Sawers RG, Bond PL. Molecular insight into extreme copper resistance in the extremophilic archaeon “Ferroplasma acidarmanus” Fer1. Microbiology. 2005;151(8):2637–2646. doi: 10.1099/mic.0.28076-0. [DOI] [PubMed] [Google Scholar]
- 19.Ettema TJG, Brinkman AB, Lamers PP, Kornet NG, de Vos WM, van der Oost J. Molecular characterization of a conserved archaeal copper resistance (cop) gene cluster and its copper-responsive regulator in Sulfolobus solfataricus P2. Microbiology. 2006;152(7):1969–1979. doi: 10.1099/mic.0.28724-0. [DOI] [PubMed] [Google Scholar]
- 20.Villafane AA, Voskoboynik Y, Cuebas M, Ruhl I, Bini E. Response to excess copper in the hyperthermophile Sulfolobus solfataricus strain 98/2. Biochemical and Biophysical Research Communications. 2009;385(1):67–71. doi: 10.1016/j.bbrc.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Auernik KS, Maezato Y, Blum PH, Kelly RM. The genome sequence of the metal-mobilizing, extremely thermoacidophilic archaeon Metallosphaera sedula provides insights into bioleaching-associated metabolism. Applied and Environmental Microbiology. 2008;74(3):682–692. doi: 10.1128/AEM.02019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maezato Y, Johnson T, McCarthy S, Dana K, Blum P. Metal resistance and lithoautotrophy in the extreme thermoacidophile Metalosphaera sedula . Journal of Bacteriology. 2012;194(24):6856–6863. doi: 10.1128/JB.01413-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nies DH. Microbial heavy-metal resistance. Applied Microbiology and Biotechnology. 1999;51(6):730–750. doi: 10.1007/s002530051457. [DOI] [PubMed] [Google Scholar]
- 24.Vido K, Spector D, Lagniel G, Lopez S, Toledano MB, Labarre J. A proteome analysis of the cadmium response in Saccharomyces cerevisiae . The Journal of Biological Chemistry. 2001;276(11):8469–8474. doi: 10.1074/jbc.M008708200. [DOI] [PubMed] [Google Scholar]
- 25.Bertin G, Averbeck D. Cadmium: cellular effects, modifications of biomolecules, modulation of DNA repair and genotoxic consequences (a review) Biochimie. 2006;88(11):1549–1559. doi: 10.1016/j.biochi.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 26.Joe MH, Jung SW, Im SH, et al. Genome-wide response of Deinococcus radiodurans on cadmium toxicity. Journal of Microbiology and Biotechnology. 2011;21(4):438–447. [PubMed] [Google Scholar]
- 27.Lagorce A, Fourçans A, Dutertre M, Bouyssiere B, Zivanovic Y, Confalonieri F. Genome-wide transcriptional response of the archeon Thermococcus gammatolerans to cadmium. PLOS ONE. 2012;7(7) doi: 10.1371/journal.pone.0041935.e41935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kornberg A, Rao NN, Ault-Riché D. Inorganic polyphosphate: a molecule of many functions. Annual Review of Biochemistry. 1999;68:89–125. doi: 10.1146/annurev.biochem.68.1.89. [DOI] [PubMed] [Google Scholar]
- 29.Keasling JD. Regulation of intracellular toxic metals and other cations by hydrolysis of polyphosphate. Annals of the New York Academy of Sciences. 1997;829:242–249. doi: 10.1111/j.1749-6632.1997.tb48579.x. [DOI] [PubMed] [Google Scholar]
- 30.Remonsellez F, Orell A, Jerez CA. Copper tolerance of the thermoacidophilic archaeon Sulfolobus metallicus: possible role of polyphosphate metabolism. Microbiology. 2006;152(1):59–66. doi: 10.1099/mic.0.28241-0. [DOI] [PubMed] [Google Scholar]
- 31.Hatzimanikatis V, Choe LH, Lee KH. Proteomics: theoretical and experimental considerations. Biotechnology Progress. 1999;15(3):312–318. doi: 10.1021/bp990004b. [DOI] [PubMed] [Google Scholar]
- 32.Choe LH, Lee KH. A comparison of three commercially available isoelectric focusing units for proteome analysis: the Multiphor, the IPGphor and the Protean IEF cell. Electrophoresis. 2000;21(5):993–1000. doi: 10.1002/(SICI)1522-2683(20000301)21:5<993::AID-ELPS993>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 33.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 34.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Analytical Chemistry. 1996;68(5):850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 35.Giavalisco P, Nordhoff E, Kreitler T, et al. Proteome analysis of Arabidopsis thaliana by two-dimensional gel electrophoresis and matrix-assisted laser desorption/ionisation-time of flight mass spectrometry. Proteomics. 2005;5(7):1902–1913. doi: 10.1002/pmic.200401062. [DOI] [PubMed] [Google Scholar]
- 36.Rose TM, Schultz ER, Henikoff JG, Pietrokovski S, McCallum CM, Henikoff S. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Research. 1998;26(7):1628–1635. doi: 10.1093/nar/26.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rose TM, Henikoff JG, Henikoff S. CODEHOP (Consensus-Degenerate Hybrid Oligonucleotide Primer) PCR primer design. Nucleic Acids Research. 2003;31(13):3763–3766. doi: 10.1093/nar/gkg524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Acevedo JP, Reyes F, Parra LP, Salazar O, Andrews BA, Asenjo JA. Cloning of complete genes for novel hydrolytic enzymes from Antarctic sea water bacteria by use of an improved genome walking technique. Journal of Biotechnology. 2008;133(3):277–286. doi: 10.1016/j.jbiotec.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 39.Bathe S, Norris PR. Ferrous iron- and sulfur-induced genes in Sulfolobus metallicus . Applied and Environmental Microbiology. 2007;73(8):2491–2497. doi: 10.1128/AEM.02589-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. New York, NY, USA: Cold Spring Harbor Press; 1989. [Google Scholar]
- 41.Miller KW, Sass Risanico S, Risatti JB. Differential tolerance of Sulfolobus strains to transition metals. FEMS Microbiology Letters. 1992;93(1):69–73. [Google Scholar]
- 42.Huber G, Spinnler C, Gambacorta A, Stetter KO. Metallosphaera sedula gen. and sp. nov. represents a new genus of aerobic, metal-mobilizing, thermoacidophilic archaebacteria. Systematic and Applied Microbiology. 1989;12:38–47. [Google Scholar]
- 43.Llanos J, Capasso C, Parisi E, Prieur D, Jeanthon C. Susceptibility to heavy metals and cadmium accumulation in aerobic and anaerobic thermophilic microorganisms isolated from deep-sea hydrothermal vents. Current Microbiology. 2000;41(3):201–205. doi: 10.1007/s00284431056. [DOI] [PubMed] [Google Scholar]
- 44.Kaur A, Pan M, Meislin M, Facciotti MT, El-Gewely R, Baliga NS. A systems view of haloarchaeal strategies to withstand stress from transition metals. Genome Research. 2006;16(7):841–854. doi: 10.1101/gr.5189606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Solioz M, Stoyanov JV. Copper homeostasis in Enterococcus hirae . FEMS Microbiology Reviews. 2003;27(2-3):183–195. doi: 10.1016/S0168-6445(03)00053-6. [DOI] [PubMed] [Google Scholar]
- 46.Sitthisak S, Howieson K, Amezola C, Jayaswal RK. Characterization of a multicopper oxidase gene from Staphylococcus aureus . Applied and Environmental Microbiology. 2005;71(9):5650–5653. doi: 10.1128/AEM.71.9.5650-5653.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodríguez-Montelongo L, Volentini SI, Farías RN, Massa EM, Rapisarda VA. The Cu(II)-reductase NADH dehydrogenase-2 of Escherichia coli improves the bacterial growth in extreme copper concentrations and increases the resistance to the damage caused by copper and hydroperoxide. Archives of Biochemistry and Biophysics. 2006;451(1):1–7. doi: 10.1016/j.abb.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 48.Williams E, Lowe TM, Savas J, DiRuggiero J. Microarray analysis of the hyperthermophilic archaeon Pyrococcus furiosus exposed to gamma irradiation. Extremophiles. 2007;11(1):19–29. doi: 10.1007/s00792-006-0002-9. [DOI] [PubMed] [Google Scholar]
- 49.Gaetke LM, Chow CK. Copper toxicity, oxidative stress, and antioxidant nutrients. Toxicology. 2003;189(1-2):147–163. doi: 10.1016/s0300-483x(03)00159-8. [DOI] [PubMed] [Google Scholar]
- 50.Davies MJ. The oxidative environment and protein damage. Biochimica et Biophysica Acta. 2005;1703(2):93–109. doi: 10.1016/j.bbapap.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 51.Thompson JR, Bell JK, Bratt J, Grant GA, Banaszak LJ. Vmax regulation through domain and subunit changes. The active form of phosphoglycerate dehydrogenase. Biochemistry. 2005;44(15):5763–5773. doi: 10.1021/bi047944b. [DOI] [PubMed] [Google Scholar]
- 52.Consalvi V, Chiaraluce R, Politi L, Gambacorta A, De Rosa M, Scandurra R. Glutamate dehydrogenase from the thermoacidophilic archaebacterium Sulfolobus solfataricus . European Journal of Biochemistry. 1991;196(2):459–467. doi: 10.1111/j.1432-1033.1991.tb15837.x. [DOI] [PubMed] [Google Scholar]
- 53.Macario AJL, Lange M, Ahring BK, Conway De Macario E. Stress genes and proteins in the archaea. Microbiology and Molecular Biology Reviews. 1999;63(4):923–967. doi: 10.1128/mmbr.63.4.923-967.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maupin-Furlow JA, Gil MA, Humbard MA, et al. Archaeal proteasomes and other regulatory proteases. Current Opinion in Microbiology. 2005;8(6):720–728. doi: 10.1016/j.mib.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 55.Sun N, Beck F, Wilhelm Knispel R, et al. Proteomics analysis of Thermoplasma acidophilum with a focus on protein complexes. Molecular and Cellular Proteomics. 2007;6(3):492–502. doi: 10.1074/mcp.M600322-MCP200. [DOI] [PubMed] [Google Scholar]
- 56.Madding LS, Michel JK, Shockley KR, et al. Role of the β1 subunit in the function and stability of the 20S proteasome in the hyperthermophilic Archaeon Pyrococcus furiosus . Journal of Bacteriology. 2007;189(2):583–590. doi: 10.1128/JB.01382-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shibata R, Bessho Y, Shinkai A, et al. Crystal structure and RNA-binding analysis of the archaeal transcription factor NusA. Biochemical and Biophysical Research Communications. 2007;355(1):122–128. doi: 10.1016/j.bbrc.2007.01.119. [DOI] [PubMed] [Google Scholar]
- 58.Bae W, Xia B, Inouye M, Severinov K. Escherichia coli CspA-family RNA chaperones are transcription antiterminators. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(14):7784–7789. doi: 10.1073/pnas.97.14.7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ram RJ, VerBerkmoes NC, Thelen MP, et al. Microbiology: community proteomics of a natural microbial biofilm. Science. 2005;308(5730):1915–1920. [PubMed] [Google Scholar]
- 60.Noël-Georis I, Vallaeys T, Chauvaux R, et al. Global analysis of the Ralstonia metallidurans proteome: prelude for the large-scale study of heavy metal response. Proteomics. 2004;4(1):151–179. doi: 10.1002/pmic.200300551. [DOI] [PubMed] [Google Scholar]
- 61.Novo MTM, Da Silva AC, Moreto R, et al. Thiobacillus ferrooxidans response to copper and other heavy metals: growth, protein synthesis and protein phosphorylation. Antonie van Leeuwenhoek. 2000;77(2):187–195. doi: 10.1023/a:1002462701671. [DOI] [PubMed] [Google Scholar]
- 62.She Q, Singh RK, Confalonieri F, et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(14):7835–7840. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anderson RP, Roth JR. Tandem genetic duplications in phage and bacteria. Annual Review of Microbiology. 1977;31:473–505. doi: 10.1146/annurev.mi.31.100177.002353. [DOI] [PubMed] [Google Scholar]
- 64.Gevers D, Vandepoele K, Simillion C, Van De Peer Y. Gene duplication and biased functional retention of paralogs in bacterial genomes. Trends in Microbiology. 2004;12(4):148–154. doi: 10.1016/j.tim.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 65.Reams AB, Neidle EL. Selection for gene clustering by tandem duplication. Annual Review of Microbiology. 2004;58:119–142. doi: 10.1146/annurev.micro.58.030603.123806. [DOI] [PubMed] [Google Scholar]
- 66.Orellana LH, Jerez CA. A genomic island provides Acidithiobacillus ferrooxidans ATCC 53993 additional copper resistance: a possible competitive advantage. Applied Microbiology and Biotechnology. 2011;92(4):761–767. doi: 10.1007/s00253-011-3494-x. [DOI] [PubMed] [Google Scholar]
- 67.Brinkman AB, Ettema TJG, de Vos WM, van der Oost J. The Lrp family of transcriptional regulators. Molecular Microbiology. 2003;48(2):287–294. doi: 10.1046/j.1365-2958.2003.03442.x. [DOI] [PubMed] [Google Scholar]
- 68.Vollmecke C, Drees SL, Reimann J, Albers SV, Lubben M. The ATPases CopA and CopB both contribute to copper resistance of the thermoacidophilic archaeon Sulfolobus solfataricus . Microbiology. 2012;158(6):1622–1633. doi: 10.1099/mic.0.055905-0. [DOI] [PubMed] [Google Scholar]
- 69.Villafane A, Voskoboynik Y, Ruhl I, et al. CopR of Sulfolobus solfataricus represents a novel class of archaeal-specific copper-responsive activators of transcription. Microbiology. 2011;157(10):2808–2817. doi: 10.1099/mic.0.051862-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CODEHOP-based PCR details for the amplification of the putative genes copA and copM from S. metallicus are provided in the supplementary material.


