Abstract
The primate lentivirus auxiliary protein Vpx counteracts an unknown restriction factor that renders human dendritic and myeloid cells largely refractory to HIV-1 infection1-6. Here we identify SAMHD1 as this restriction factor. SAMHD1 is a protein involved in Aicardi–Goutières syndrome, a genetic encephalopathy with symptoms mimicking congenital viral infection, that has been proposed to act as a negative regulator of the interferon response7. We show that Vpx induces proteasomal degradation of SAMHD1. Silencing of SAMHD1 in non-permissive cell lines alleviates HIV-1 restriction and is associated with a significant accumulation of viral DNA in infected cells. Concurrently, overexpression of SAMHD1 in sensitive cells inhibits HIV-1 infection. The putative phosphohydrolase activity of SAMHD1 is probably required for HIV-1 restriction. Vpx-mediated relief of restriction is abolished in SAMHD1-negative cells. Finally, silencing of SAMHD1 markedly increases the susceptibility of monocytic-derived dendritic cells to infection. Our results demonstrate that SAMHD1 is an antiretroviral protein expressed in cells of the myeloid lineage that inhibits an early step of the viral life cycle.
Most monocytic cell lines fail to recapitulate the HIV-1 restriction phenotype that is witnessed in primary dendritic cells and to a lesser extent in macrophages. One exception is represented by phorbol 12-myristate 13-acetate (PMA)-differentiated THP-1 monocytic cells1,2,4-6. Transduction of differentiated THP-1 cells by virus-like particles containing Vpx (VLP-Vpx) has been shown to increase their permissiveness to HIV-1 infection1. To identify the restriction factor targeted by Vpx, we generated a stable THP-1 cell line expressing Flag-HA epitope-tagged Vpxmac251 from sooty mangabey (THP-1-Vpx). As expected, THP-1-Vpx cells were 17-fold more permissive to HIV-1 infection than parental cells when exposed to a vesicular stomatitis virus G protein (G)-pseudotyped HIV-1 with the luciferase gene in place of nef (HIV-LUC-G) (Supplementary Fig. 1). After differentiation, extracts were prepared from THP-1 and THP-1-Vpx cells. Flag-HA–Vpxmac251 (F/H-Vpx) was purified using tandem affinity chromatography8. Purified Vpxmac251-associated proteins were resolved by SDS–polyacrylamide gel electrophoresis (PAGE) and silver stained (Fig. 1a). The eluates were further analysed by mass spectrometry to allow for the identification of cellular partners that engage with Vpxmac251 in non-permissive cells. Previously described Vpxmac251 interactants were recovered, including the DDB1–CUL4–DCAF E3 ligase complex9,10, confirming the validity of our approach. The SAM- and HD-domain-containing protein SAMHD1 was identified as a major Vpxmac251-interacting protein (Table 1). SAMHD1 was initially isolated as an interferon (IFN)-γ-induced factor in macrophages and dendritic cells11,12 for which a function in innate immunity has been suggested7,13. Furthermore, mutations in SAMHD1 have been shown to be responsible for 5% of Aicardi–Goutières syndrome cases, a genetically heterogeneous disorder characterized by inappropriate activation of the immune system and aberrant IFN-α secretion, symptoms reminiscent of congenital infection7.
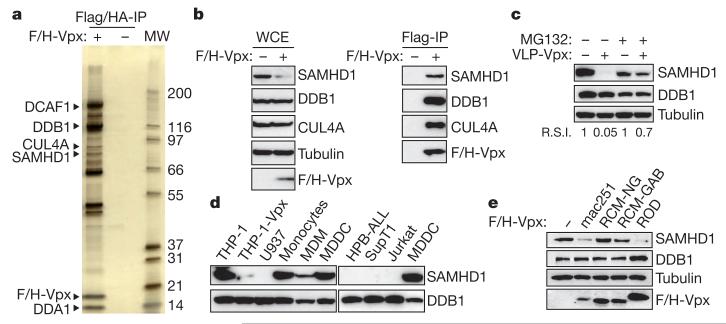
Figure 1. SAMHD1 interacts with Vpx and is degraded by the proteasome.
a, SIVmac251 Vpx was tandem-affinity-purified from Flag- and HA-tagged Vpxmac251 (F/H-Vpx)-expressing THP-1 cells (THP-1-Vpx) and peptide-eluted under native conditions. Eluates were separated on SDS–PAGE and silver stained. SAMHD1 and major, previously described, Vpxmac251 interactants identified using tandem mass spectrometry are indicated (MW, protein molecular weight marker in kDa). b, Whole-cell extract (WCE) and Flag-immunoprecipitated F/H-Vpx analysis by western blot against DDB1, CUL4A and SAMHD1. c, THP-1 cells were treated with 50 μM MG132 for 2 h before a 2-h incubation with Vpxmac251 containing virus-like particles (VLP-Vpx). After a further overnight incubation with MG132, whole-cell extracts were prepared and analysed by western blot with the indicated antibodies. R.S.I., relative signal intensity. d, Analysis of the expression profile of SAMHD1 in different cell types by western blot. e, THP-1 cells were transduced with a bicistronic retroviral vector allowing expression of Vpxmac251, VpxROD, VpxRCM-NG and VpxRCM-GAB and a selectable marker. After cell sorting and whole-cell extraction, the ability of Vpx variants to degrade SAMHD1 in THP-1 whole-cell extract was analysed by western blot using the indicated antibodies.
Table 1. Major F/H-Vpx interactants identified by mass spectrometry.
| Protein symbol | Peptide count |
|---|---|
| DDB1 | 620 |
| DCAF1 | 499 |
| SAMHD1 | 290 |
| CUL4A | 55 |
| CUL4B | 49 |
| DDA1 | 18 |
The interaction of SAMHD1 with Vpxmac251 was confirmed by Flag-immunoprecipitation of F/H-Vpx and western blot analysis using a SAMHD1-specific antibody (Fig. 1b). Notably, analysis of whole-cell extracts of THP-1-Vpx as compared to THP-1 reveals that expression of Vpxmac251 correlates with lower expression levels of SAMHD1 (Fig. 1b). Transient delivery of Vpxmac251 into THP-1 cells through VLP-Vpx exposure caused a marked decrease in SAMHD1 levels in THP-1 cells (Fig. 1c). Moreover, treatment of cells with the proteasome inhibitor MG132 restored SAMHD1 protein levels (Fig. 1c), strongly suggesting that Vpxmac251 induces proteasomal degradation of SAMHD1. Additionally, when SAMHD1 was expressed in HeLa cells, Vpx also caused its degradation, demonstrating that Vpx-induced degradation of SAMHD1 is not a cell-type-specific process and excluding a potential transcriptional effect of Vpx on SAMHD1 (Supplementary Fig. 2). These results led us to postulate that SAMHD1 may be the restriction factor that renders dendritic cells refractory to HIV-1 infection. Consistently, SAMHD1 is highly expressed in HIV-1 non-permissive cells such as THP-1, monocytes and monocyte-derived dendritic cells (MDDCs), whereas it is absent from HIV-1-sensitive T-cell lines such as Jurkat, SupT1, human peripheral blood acute lymphoid leukaemia (HPB-ALL) and U937 (Fig. 1d). In correlation with their degree of permissiveness to HIV-1 (refs 1, 4), monocyte-derived macrophages (MDMs) express low levels of SAMHD1 as compared to their highly refractory monocyte precursor (Fig. 1d).
It has been reported that Vpx-mediated enhancement of HIV-1 infection in dendritic cells and myeloid cells is conserved exclusively within the SIVSM (sooty mangabey) and HIV-2 lineage (refs 1, 2 and Supplementary Fig. 2a). Concurrent with this observation, we show that Vpxmac251 and HIV-2ROD Vpx (VpxROD) both caused degradation of SAMHD1, whereas Vpx from SIVRCM of red-capped mangabeys (isolates of Nigerian (VpxRCM-NG) or Gabonese (VpxRCM-GAB) origin) failed to degrade SAMHD1 when expressed in THP-1 (Fig. 1e and Supplementary Fig. 3a, b) or HeLa cells (Supplementary Fig. 3c). Additionally, loss-of-function mutants Vpx(Q76A) and Vpx(F80A) also failed to degrade SAMHD1in differentiated THP-1 cells (Supplementary Fig. 3d).
To assess directly the impact of expression of SAMHD1 on HIV-1 infection, we used short hairpin RNAs (shRNA) to generate SAMHD1-silent THP-1 cells(THP-1-shSAMHD1) or scrambled shRNA (THP-1-scr) (Supplementary Fig. 4). Infection of THP-1-shSAMHD1 cells with HIV-LUC-G results in up to a 12-fold increase in luciferase activity as compared to THP-1-scr cells, demonstrating that depletion of SAMHD1 is sufficient to increase the permissiveness of THP-1 cells to HIV-1 infection (Fig. 2a). As expected, HIV-1 infection of THP-1-shSAMHD1 cells was not further enhanced by treatment with VLP-Vpx (Fig. 2b and Supplementary Fig. 5). Importantly, expression of a shRNA-resistant SAMHD1 mutant (SAMHD1-R) in THP-1-shSAMHD1 cells restored the restriction phenotype in these cells (Fig. 2d and Supplementary Fig. 6). To investigate further whether SAMHD1 possesses an intrinsic restriction activity targeting HIV-1, we stably expressed Flag-HA-tagged SAMHD1 (SAMHD1-F/H) in permissive U937 monocytic cells (U937-SAMHD1). Differentiated U937-SAMHD1 cells were 16-fold less permissive to infection with HIV-LUC-G than parental cells (Fig. 2e and Supplementary Fig. 8). HD domains have an important role in nucleotide metabolism through their nucleotidase and phosphodiesterase activities, where H and D residues are of crucial importance14. Interestingly, U937 cells stably expressing the SAMHD1(HD/AA) (U937-SAMHD1(HD/AA)) mutant did not show restriction activity towards HIV-1 as compared to U937-SAMHD1 cells, indicating that the putative phosphohydrolase activity of SAMHD1 may be required for restriction of HIV-1 (Fig. 2e and Supplementary Figs 7 and 8). Additionally, transient expression of SAMHD1-F/H, but not SAMHD1(HD/AA), in permissive HeLa cells induced restriction of HIV-1 infection (Supplementary Fig. 7).
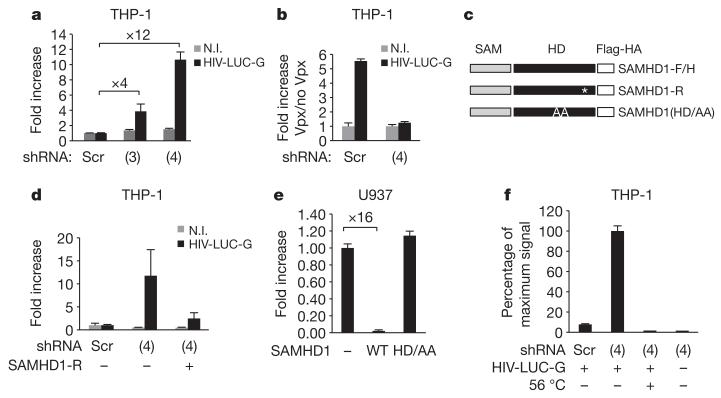
Figure 2. SAMHD1 restricts HIV-1 infection in THP-1 cells.
THP-1 cells were engineered to stably express shRNA 3 (3) or shRNA 4 (4) specifically targeting SAMHD1 (THP-1-shSAMHD1) or scrambled shRNA (THP-1-scr). a, THP-1-shSAMHD1 and THP-1-scr cells were infected with 50 ng of HIV-LUC-G. Luciferase activity was measured 24 h after infection and normalized for protein concentration in analysed samples. Results are expressed as fold increase of luciferase activity in THP-1-shSAMHD1 over THP-1-scr cells. N.I., non-infected. b, THP-1-shSAMHD1 and THP-1-scr cells were treated with VLP-Vpx before infection with 50 ng of HIV-LUC-G. Luciferase activity was measured as in a. Results are expressed as fold increase of luciferase activity in VLP-Vpx-treated over untreated cells. c, Mutants of Flag- and HA-tagged SAMHD1 (SAMHD1-F/H) were generated that are either shSAMHD1-resistant (SAMHD1-R) or mutated in the HD domain (SAMHD1(HD/AA)). These mutants were introduced in an MLV expression vector. Asterisk indicates synonymous mutation. d, THP-1-shSAMHD1 cells were transduced with SAMHD1-R for 48 h or left untreated, differentiated and infected with 100 ng of HIV-LUC-G. Luciferase activity was measured and expressed as in a. e, U937 myeloid cells were transduced with SAMHD1-F/H or SAMHD1(HD/AA) for 24 h. After a further 16-h differentiation step, cells were infected with 10 ng of HIV-LUC-G. Luciferase activity was measured as in a. Results are expressed as fold increase luciferase activity in transduced over parental U937 cells. f, Total viral DNA was quantified by quantitative PCR in THP-1-shSAMHD1 and THP-1-scr cells 24 h after infection with HIV-LUC-G or heat-inactivated virus (56 °C). Results are expressed as per cent maximum signal intensity. All graphs show mean ± standard deviation from a representative experiment (n = 5).
Vpx has been shown to facilitate HIV-1 replication by promoting accumulation of viral DNA1,2,4,5,15. To investigate at which step of the viral replication cycle SAMHD1-dependent restriction operates, we quantified total viral DNA species 24 h after infection of THP-1-shSAMHD1 cells (Fig. 2f). A 13-fold accumulation of total viral DNA was observed in SAMHD1-silenced cells as compared to their THP-1-scr counterpart. This observation locates the restriction operated by SAMHD1 at the reverse transcription step, which has been previously described to be overcome by Vpx in dendritic and myeloid cells2.
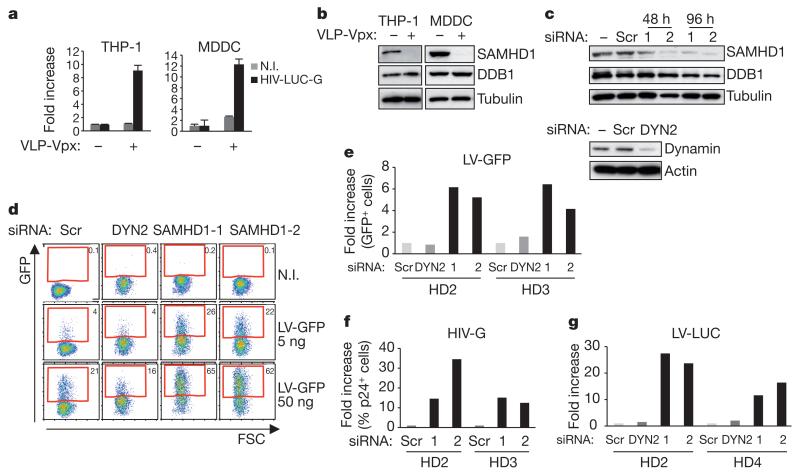
Exposure of differentiated THP-1, MDDCs and MDMs to VLP-Vpx relieves restriction to HIV-1 infection (Fig. 3a and Supplementary Fig. 9) and correlates with a decrease in SAMHD1 levels (Fig. 3b and Supplementary Fig. 9). Lower basal levels of SAMHD1 in primary MDMs (Fig. 1d) may be accountable for the weaker impact of VLP-Vpx treatment of these cells (Supplementary Fig. 9). Next, we asked whether SAMHD1 restricts HIV-1 infection of primary human dendritic cells. Immature MDDCs were prepared from the blood of four healthy donors. To silence SAMHD1 expression, we treated MDDCs with two different SAMHD1-specific siRNAs (si-SAMHD1-1 or si-SAMHD1-2). Scrambled or siRNA targeting dynamin 2 (siDYN2) were used as controls (Fig. 3c and Supplementary Fig. 10). MDDCs were transduced (at 48 h after silencing) with green-fluorescent-protein- or luciferase-encoding lentiviral vectors (LV-GFP or LV-LUC, respectively) or with a VSV-G-pseudotyped HIV (HIV-G). Silencing of SAMHD1 resulted in up to a 6-fold increase of GFP-positive cells with LV-GFP, 25-fold enhanced luciferase activity with LV-LUC, and up to 34-fold increased Gag-positive cells with HIV-G(Fig. 3d-f and Supplementary Fig. 10), demonstrating that SAMHD1 silencing in dendritic cells enhances their susceptibility to HIV-1 infection.
Figure 3. SAMHD1 restricts HIV-1 infection in primary MDDCs.
a, THP-1 and monocyte-derived dendritic cells (MDDCs) from healthy donors (HD) were treated with VLP-Vpx for 2 h and infected with 100 ng of HIV-LUC-G. Luciferase activity was measured at 48 h after infection and normalized for protein concentration. Results are expressed as fold increase luciferase activity in VLP-Vpx treated over untreated cells. Graphs show mean ± standard deviation from a representative experiment (n = 4). b, Cells from a were subjected to whole-cell extraction before analysis by western blot using indicated antibodies. c, MDDCs from HD1 were mock transfected or transfected with siRNA targeting SAMHD1 (siRNA 1 and 2) or dynamin 2 (DYN2) for 48 and 96 h before whole-cell extraction and analysis by western blot using indicated antibodies. d, Cells from HD2 were transduced at 48 h with GFP encoding lentiviral vector (LV-GFP, 50 ng) and analysed by flow cytometry after 4 days. e, HD2 and HD3 were transduced as in d, except that a concentration of 5 ng of LV-GFP was used. The percentage of GFP-positive cells from HD2 and HD3 was quantified and expressed as fold increase GFP-positive cells relative to scrambled siRNA-treated cells. f, Transfected MDDCs from HD2 and HD3 were infected with 10 ng of HIV-G. Results are expressed as fold increase of p24-positive cells relative to scrambled siRNA-treated cells. g, MDDCs from HD2 and HD4 were transfected as in c before infection with 10 ng of luciferase-expressing lentiviral vector (LV-LUC). Results are expressed as in Fig. 2a.
So far, three restriction factors (TRIM5α, APOBEC-3G and tetherin) have been identified that could have constituted a major hindrance to HIV-1 replication, had the virus not developed ways to counteract or to escape their action16-18. These restriction factors are part of the intrinsic immunity that is circumvented by HIV-1 mainly through the action of its auxiliary proteins. However, the cell-type-specific restriction factor SAMHD1 is not counteracted by HIV-1, resulting in poorly efficient replication in dendritic cells. Indeed, in the normal course of HIV-1 infection and because dendritic cells are non-permissive, these cells rather facilitate viral dissemination through trans-enhancement of infection, eventually favouring CD4+ T-cell depletion19,20. Poor HIV-1 replication in dendritic cells may also allow for avoidance of a recently described cryptic viral sensor that would otherwise elicit antiviral interferon-induced immune responses20,21. Similarly, in productively infected CD4+ T cells, through the action of the cellular DNase TREX1, HIV-1 avoids the induction of type 1 IFN production that could result from accumulation of viral DNA20,22. Of note, both TREX1 and SAMHD1 deficiencies lead to Aicardi–Goutières syndrome, indicating that they have an impact on the same pathway of cell-intrinsic antiviral response7. SAMHD1, through the putative nucleotidase activity, may degrade or prevent accumulation of HIV DNA. It will be worth determining further the relative roles of TREX1 and SAMHD1 in the control of virus infection and in the triggering of innate antiviral and inflammatory responses.
Our findings position SAMHD1 as pivotal to the fate of infection by HIV-1 in cells of the myeloid lineage. Thus, modulating SAMHD1 function could render human hosts more prone to develop appropriate innate and adaptive immune responses19,22-24. Our findings should be integrated in the development of dendritic-cell-targeted vaccines against HIV/AIDS.
METHODS
Cell lines
Adherent and suspension cells were cultured in DMEM or RPMI supplemented with 10% fetal calf serum (FCS), ultraglutamine and antibiotics. All cell culture reagents were purchased from Lonza. Cell lines expressing Flag- and HA-tagged proteins were constructed using the previously described MMLV-based retroviral constructs8,26 that contain a bicistronic transcriptional unit allowing for expression of a selectable marker (IL-2 receptor-α chain (pOZ-IL2Rα) or puromycin resistance gene (pOZ-puro)). IL2Rα-selected cell lines were selected using magnetic beads. Puromycin-selected cell lines were cultured in appropriate media supplemented with 1 μg ml−1 puromycin. shRNA-silenced cell lines were generated according to the manufacturer’s instructions (Openbiosystem) and selected for resistance to puromycin. U937 and THP-1 cells were differentiated overnight with 30 ng ml−1 PMA (Sigma).
Plasmids
SIV3+ was a gift from N. Manel. HIV-LUC, VSV-G, MMLV packaging and A-MLV envelope have been previously described27. shRNA constructs were purchased from Openbiosystem. VpxROD, VpxRCM-NG and VpxRCM-GAB were synthesized by MWG biotech. The SAMHD1 molecular clone was purchased from Invitrogen. All Vpxmac251, VpxROD, VpxRCM-NG and VpxRCM-GAB were subcloned in pOZ-IL2Rα expression vector with tags at the N terminus, and SAMHD1 and SAMHD1 mutants were subcloned in pOZ-puro expression vector with tags at the C terminus according to standard ligation procedures. SAMHD1(HD/AA) and SAMHD1-R mutants were generated using the Quickchange lightning kit (Agilent technologies) according the manufacturer’s recommendations.
Virus production
Viral particles were produced from 293Tcells using the standard phosphate calcium transfection protocol. Briefly, for HIV-LUC-G production, 293T cells were transfected with 8 μg HIV-LUC and 2 μg VSV-G encoding plasmid; for shRNA production, 293T cells were transfected with 4 μg shRNA construct, 4 μg packaging plasmid, 2 μg VSV-G encoding plasmid; for MLV transduction particles, 293T cells were transfected with 5 μg pOZ construct, 2.5 μg packaging plasmid and 2.5 μg A-MLV envelope encoding plasmid; and for VLP-Vpx production, 8 μg SIV3+ was co-transfected or not with 2 μg VSV-G encoding plasmid. Media was replaced 16 h after transfection and viruses were harvested 24 h later, filtered at 0.45 μm. When required, p24 concentration was measured by ELISA (Innogenetics).
Infection
Infection of THP-1, U937 and HeLa cells was performed by addition of 100, 50, 10, 5 or 1 ng p24 of HIV-LUC-G depending on the experiment. Viruses were added to cells and luciferase activity was measured 24 h after infection. VLP-Vpx treatment was performed for 2 h before infection by addition of RPMI-diluted VLP-Vpx to cells.
Cell extract preparation and western blot analysis
Whole-cell extracts were prepared with buffer containing 0.5% Triton X-100, 150 mM NaCl, 10 mM KCL, 1.5 mM MgCl2, 0.5 mM EDTA 10 mM β-mercaptoethanol, 0.5 mM PMSF. Mouse anti-SAMHD1, anti-CUL4A and anti-DDB1 were purchased from Abcam. HA (11 clone 16B12) and tubulin antibodies were from Covance/Eurogentec and Sigma, respectively.
Immunopurification
Vpxmac251 was purified from 5 × 109 differentiated THP-1 cells stably expressing Flag- and HA-tagged Vpxmac251 (F/H-Vpx) by two-step affinity chromatography according to the standard method8. Extracts were first incubated with anti-Flag antibody conjugated agarose beads (Sigma), and the bound polypeptides were eluted with Flag peptide (Sigma) under native conditions. The Flag affinity-purified material was further immunopurified by affinity chromatography using anti-HA antibody conjugated agarose beads (Santa Cruz) and eluted under native conditions using HA peptide (Roche). Five per cent of immunoaffinity-purified F/H-Vpx or mock were resolved on SDS-PAGE and stained with the Silverquest kit (Invitrogen). The remainder of the eluate was stained with colloidal blue. Individual Coomassie-stained bands, or for closely migrating bands, regions of the gel were excised and subsequently analysed by tandem mass spectrometry at the Harvard Medical School Taplin Biological Mass Spectrometry facility.
Quantification of HIV-1 total DNA
Before infection, viral stocks were treated for 1 h at 37 °C with 100 U ml−1 of DNaseI (Roche). 3 × 105 THP-1-scr or THP-1-shSAMHD1 cells were infected with 100 ng of HIV-LUC-Gor with heat-inactivated HIV-LUC-G. Cells were washed twice in PBS 2 h after infection, harvested 22 h later, washed twice in PBS, and DNA was extracted using the QIAamp Blood DNA Minikit (Qiagen). Quantification of viral DNA was performed by quantitative PCR (qPCR) using previously described probes27 allowing for amplification of the luciferase gene on a LightCycler 480 system (Roche). Viral DNA was normalized using 7SK-specific probes.
Preparation of MDDCs and MDMs
Human buffy coats were obtained from Etablissement Français du Sang (EFS). Monocytes were purified from total peripheral blood mononuclear cells after Ficoll gradient separation with CD14 MicroBeads (Miltenyi Biotec) or using the standard adhesion protocol. Human monocyte-derived dendritic cells (MDDCs) were generated by incubation of CD14 purified monocytes in IMDM medium supplemented with 10% FCS, 2 mM l-glutamine, 100 IU ml−1 penicillin, 100 mg ml−1 streptomycin, 10 mM HEPES, 1% non-essential amino acids, 1 mM sodium pyruvate, 10 ng ml−1 GM-CSF and 50 ng ml−1 IL-4 (Miltenyi Biotec). On day 4, two-thirds of the culture medium was replaced by fresh medium containing GM-CSF and IL-4. Immature MDDCs (iDCs) were harvested and further used at day 6.
MDMs were generated by 7 days of stimulation with 50 ng ml−1 of recombinant human granulocyte-macrophage colony-stimulating factor (rhGM-CSF) (Immunotools).
Efficient differentiation of MDM and MDDCs was verified by flow cytometry using antibodies against CD1a, CD14, CD80, CD83 (BD Bioscience) and CD86 (B72 clone, Invitrogen). MDDCs were more than 98% DC-SIGN positive.
siRNA transfection of MDDCs
scrambled siRNA, siSAMHD1-1 (5′-GAUUCAUUGUGGCCAUAUA-3′) and siSAMHD1-3 (5′-CAACCAGAGCUGCAGAUAA-3′) were synthesized by MWGoperon. siDYN2 was from Qiagen. HiPerFect Reagent (Qiagen) was used for transfection in accordance with the manufacturer’s recommendations. In brief, 5 × 105 iDCs were transfected with 100 nM siRNA in 500 ml of IMDM/1% FCS medium in 12-well plates. A second round of transfection was performed 24 h later. Specific gene knockdowns were assessed by immunoblot.
Transduction of MDDCs
One day after the second round of silencing, MDDCs were transduced with a lentiviral vector expressing the GFP or luciferase gene (LV-GFP and LV-LUC, respectively) or VSV-G-pseudotyped HIV (HIV-G). Briefly, 2.5 × 105 MDDCs were seeded in 24-well plates and exposed to the indicated doses of lentiviral vectors or HIV-G (from 1 to 100 ng p24 per ml). After overnight incubation, the medium was replaced with fresh medium containing GM-CSF and IL-4. Four days after transduction, GFP or p24 expression was analysed by flow cytometry using a FacsCalibur (Becton Dickinson) with FlowJo software or luciferase activity was measured using a Mithras luminometer (Berthold technologies).
Supplementary Material
Acknowledgements
We wish to thank members of the Molecular Virology laboratory for critical reading of the manuscript, N. Manel for SIV3+ molecular clone and J. Luban for SIV delta Vpx and Vpx mutants. This work was supported by grants from the ERC (250333), Sidaction, ANRS and FRM ’équipe labéllisée’ to M.B. N.L was supported by ANRS and SIDACTION fellowships; B.S. by ANRS fellowship; M.R. by CNRS/région Languedoc Roussillon fellowship. O.S. and N.C. are supported by grants from ANRS, Sidaction, ANR, European FP7 contract 201412 and Institut Pasteur.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
The authors declare no competing financial interests.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
References
- 1.Goujon C, et al. Characterization of simian immunodeficiency virus SIVSM/human immunodeficiency virus type 2 Vpx function in human myeloid cells. J. Virol. 2008;82:12335–12345. doi: 10.1128/JVI.01181-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goujon C, et al. SIVSM/HIV-2 Vpx proteins promote retroviral escape from a proteasome-dependent restriction pathway present in human dendritic cells. Retrovirology. 2007;4:2. doi: 10.1186/1742-4690-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirsch VM, et al. Vpx is required for dissemination and pathogenesis of SIVSM PBj: evidence of macrophage-dependent viral amplification. Nature Med. 1998;4:1401–1408. doi: 10.1038/3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaushik R, Zhu X, Stranska R, Wu Y, Stevenson M. A cellular restriction dictates the permissivity of nondividing monocytes/macrophages to lentivirus and gammaretrovirus infection. Cell Host Microbe. 2009;6:68–80. doi: 10.1016/j.chom.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharova N, et al. Primate lentiviral Vpx commandeers DDB1 to counteract a macrophage restriction. PLoS Pathog. 2008;4:e1000057. doi: 10.1371/journal.ppat.1000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ayinde D, Maudet C, Transy C, Margottin-Goguet F. Limelight on two HIV/SIV accessory proteins in macrophage infection: is Vpx overshadowing Vpr? Retrovirology. 2010;7:35. doi: 10.1186/1742-4690-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rice GI, et al. Mutations involved in Aicardi-Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nature Genet. 2009;41:829–832. doi: 10.1038/ng.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakatani Y, Ogryzko V. Immunoaffinity purification of mammalian protein complexes. Methods Enzymol. 2003;370:430–444. doi: 10.1016/S0076-6879(03)70037-8. [DOI] [PubMed] [Google Scholar]
- 9.Bergamaschi A, et al. The human immunodeficiency virus type 2 Vpx protein usurps the CUL4A-DDB1 DCAF1 ubiquitin ligase to overcome a postentry block in macrophage infection. J. Virol. 2009;83:4854–4860. doi: 10.1128/JVI.00187-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Srivastava S, et al. Lentiviral Vpx accessory factor targets VprBP/DCAF1 substrate adaptor for cullin 4 E3 ubiquitin ligase to enable macrophage infection. PLoS Pathog. 2008;4:e1000059. doi: 10.1371/journal.ppat.1000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li N, Zhang W, Cao X. Identification of human homologue of mouse IFN-γ induced protein from human dendritic cells. Immunol. Lett. 2000;74:221–224. doi: 10.1016/s0165-2478(00)00276-5. [DOI] [PubMed] [Google Scholar]
- 12.Liao W, Bao Z, Cheng C, Mok YK, Wong WS. Dendritic cell-derived interferon-γ-induced protein mediates tumor necrosis factor-α stimulation of human lung fibroblasts. Proteomics. 2008;8:2640–2650. doi: 10.1002/pmic.200700954. [DOI] [PubMed] [Google Scholar]
- 13.Zhao D, Peng D, Li L, Zhang Q, Zhang C. Inhibition of G1P3 expression found in the differential display study on respiratory syncytial virus infection. Virol. J. 2008;5:114. doi: 10.1186/1743-422X-5-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zimmerman MD, Proudfoot M, Yakunin A, Minor W. Structural insight into the mechanism of substrate specificity and catalytic activity of an HD-domain phosphohydrolase: the 5′-deoxyribonucleotidase YfbR from Escherichia coli. J. Mol. Biol. 2008;378:215–226. doi: 10.1016/j.jmb.2008.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gramberg T, Sunseri N, Landau NR. Evidence for an activation domain at the amino terminus of simian immunodeficiency virus Vpx. J. Virol. 2010;84:1387–1396. doi: 10.1128/JVI.01437-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stremlau M, et al. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 17.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 18.Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 19.Altfeld M, Fadda L, Frleta D, Bhardwaj N. DCs and NK cells: critical effectors in the immune response to HIV-1. Nature Rev. Immunol. 2011;11:176–186. doi: 10.1038/nri2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan N, Lieberman J. Gaining a foothold: how HIV avoids innate immune recognition. Curr. Opin. Immunol. 2011;23:21–28. doi: 10.1016/j.coi.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manel N, et al. A cryptic sensor for HIV-1 activates antiviral innate immunity in dendritic cells. Nature. 2010;467:214–217. doi: 10.1038/nature09337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borrow P, Shattock RJ, Vyakarnam A. Innate immunity against HIV: a priority target for HIV prevention research. Retrovirology. 2010;7:84. doi: 10.1186/1742-4690-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cobb A, et al. Development of a HIV-1 lipopeptide antigen pulsed therapeutic dendritic cell vaccine. J. Immunol. Methods. 2011;365:27–37. doi: 10.1016/j.jim.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 24.Lepelley A, et al. Innate sensing of HIV-infected cells. PLoS Pathog. 2011;7:e1001284. doi: 10.1371/journal.ppat.1001284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blanchet FP, et al. Human immunodeficiency virus-1 inhibition of immunoamphisomes in dendritic cells impairs early innate and adaptive immune responses. Immunity. 2010;32:654–669. doi: 10.1016/j.immuni.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar D, Shadrach JL, Wagers AJ, Lassar AB. Id3 is a direct transcriptional target of Pax7 in quiescent satellite cells. Mol. Biol. Cell. 2009;20:3170–3177. doi: 10.1091/mbc.E08-12-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sobhian B, et al. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol. Cell. 2010;38:439–451. doi: 10.1016/j.molcel.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.