Abstract
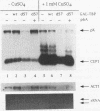
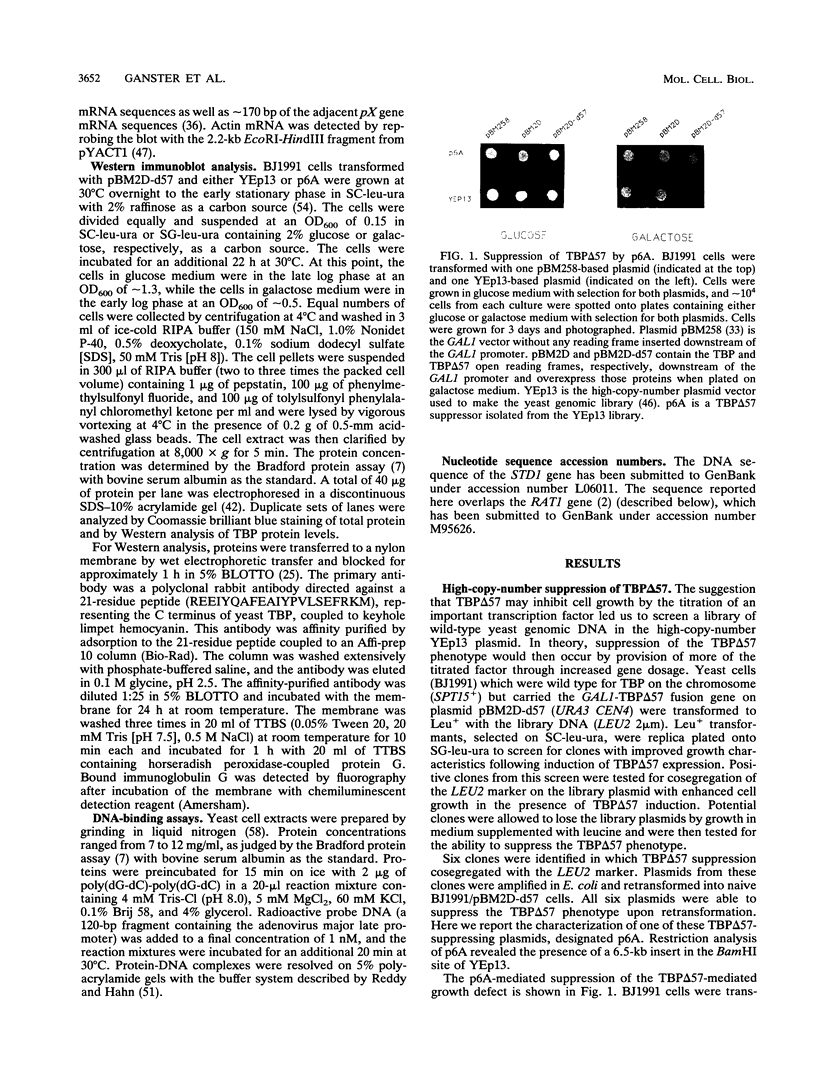
The TATA-binding protein (TBP) is an essential component of the transcriptional machinery of all three nuclear RNA polymerase enzymes. Comparison of the amino acid sequence of TBPs from a number of species reveals a highly conserved 180-residue C-terminal domain. In contrast, the N terminus is variable in both size and amino acid sequence. Overexpression of a TBP protein with a deletion of the nonconserved N terminus (TBP delta 57) in Saccharomyces cerevisiae results in a dominant negative phenotype of extremely slow growth. Associated with the slow-growth phenotype are defects in RNA polymerase II transcription in vivo. We have screened a high-copy-number yeast genomic library for suppression of the slow-growth phenotype and have isolated plasmids which encode suppressors of TBP delta 57 overexpression. Here we report the sequence and initial characterization of one suppressor, designated STD1 for suppressor of TBP deletion. The STD1 gene contains a single continuous open reading frame with the potential to encode a 50.2-kDa protein. Disruption of the STD1 gene indicates that it is not essential for vegetative growth, mating, or sporulation. High-copy-number suppression by the STD1 gene is not the result of a decrease in TBP delta 57 protein accumulation or DNA-binding activity; instead, STD1 suppression is coincident with the elimination of TBP delta 57-induced RNA polymerase II defects in both uninduced and induced transcription in vivo.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amberg D. C., Goldstein A. L., Cole C. N. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 1992 Jul;6(7):1173–1189. doi: 10.1101/gad.6.7.1173. [DOI] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Buratowski S., Zhou H. A suppressor of TBP mutations encodes an RNA polymerase III transcription factor with homology to TFIIB. Cell. 1992 Oct 16;71(2):221–230. doi: 10.1016/0092-8674(92)90351-c. [DOI] [PubMed] [Google Scholar]
- Cavallini B., Faus I., Matthes H., Chipoulet J. M., Winsor B., Egly J. M., Chambon P. Cloning of the gene encoding the yeast protein BTF1Y, which can substitute for the human TATA box-binding factor. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9803–9807. doi: 10.1073/pnas.86.24.9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L., Tanese N., Tjian R. The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL1. Cell. 1992 Mar 6;68(5):965–976. doi: 10.1016/0092-8674(92)90039-f. [DOI] [PubMed] [Google Scholar]
- Cormack B. P., Strubin M., Ponticelli A. S., Struhl K. Functional differences between yeast and human TFIID are localized to the highly conserved region. Cell. 1991 Apr 19;65(2):341–348. doi: 10.1016/0092-8674(91)90167-w. [DOI] [PubMed] [Google Scholar]
- Davison B. L., Egly J. M., Mulvihill E. R., Chambon P. Formation of stable preinitiation complexes between eukaryotic class B transcription factors and promoter sequences. Nature. 1983 Feb 24;301(5902):680–686. doi: 10.1038/301680a0. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue T. F., Cigan A. M. Sequence and structural requirements for efficient translation in yeast. Methods Enzymol. 1990;185:366–372. doi: 10.1016/0076-6879(90)85032-j. [DOI] [PubMed] [Google Scholar]
- Dynlacht B. D., Hoey T., Tjian R. Isolation of coactivators associated with the TATA-binding protein that mediate transcriptional activation. Cell. 1991 Aug 9;66(3):563–576. doi: 10.1016/0092-8674(81)90019-2. [DOI] [PubMed] [Google Scholar]
- Eisenmann D. M., Arndt K. M., Ricupero S. L., Rooney J. W., Winston F. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev. 1992 Jul;6(7):1319–1331. doi: 10.1101/gad.6.7.1319. [DOI] [PubMed] [Google Scholar]
- Eisenmann D. M., Dollard C., Winston F. SPT15, the gene encoding the yeast TATA binding factor TFIID, is required for normal transcription initiation in vivo. Cell. 1989 Sep 22;58(6):1183–1191. doi: 10.1016/0092-8674(89)90516-3. [DOI] [PubMed] [Google Scholar]
- Elledge S. J., Davis R. W. A family of versatile centromeric vectors designed for use in the sectoring-shuffle mutagenesis assay in Saccharomyces cerevisiae. Gene. 1988 Oct 30;70(2):303–312. doi: 10.1016/0378-1119(88)90202-8. [DOI] [PubMed] [Google Scholar]
- Fikes J. D., Becker D. M., Winston F., Guarente L. Striking conservation of TFIID in Schizosaccharomyces pombe and Saccharomyces cerevisiae. Nature. 1990 Jul 19;346(6281):291–294. doi: 10.1038/346291a0. [DOI] [PubMed] [Google Scholar]
- Fire A., Samuels M., Sharp P. A. Interactions between RNA polymerase II, factors, and template leading to accurate transcription. J Biol Chem. 1984 Feb 25;259(4):2509–2516. [PubMed] [Google Scholar]
- Gasch A., Hoffmann A., Horikoshi M., Roeder R. G., Chua N. H. Arabidopsis thaliana contains two genes for TFIID. Nature. 1990 Jul 26;346(6282):390–394. doi: 10.1038/346390a0. [DOI] [PubMed] [Google Scholar]
- Gill G., Tjian R. A highly conserved domain of TFIID displays species specificity in vivo. Cell. 1991 Apr 19;65(2):333–340. doi: 10.1016/0092-8674(91)90166-v. [DOI] [PubMed] [Google Scholar]
- Haass M. M., Feix G. Two different cDNAs encoding TFIID proteins of maize. FEBS Lett. 1992 Apr 27;301(3):294–298. doi: 10.1016/0014-5793(92)80260-n. [DOI] [PubMed] [Google Scholar]
- Hahn S., Buratowski S., Sharp P. A., Guarente L. Isolation of the gene encoding the yeast TATA binding protein TFIID: a gene identical to the SPT15 suppressor of Ty element insertions. Cell. 1989 Sep 22;58(6):1173–1181. doi: 10.1016/0092-8674(89)90515-1. [DOI] [PubMed] [Google Scholar]
- Hoey T., Dynlacht B. D., Peterson M. G., Pugh B. F., Tjian R. Isolation and characterization of the Drosophila gene encoding the TATA box binding protein, TFIID. Cell. 1990 Jun 29;61(7):1179–1186. doi: 10.1016/0092-8674(90)90682-5. [DOI] [PubMed] [Google Scholar]
- Hoffman A., Sinn E., Yamamoto T., Wang J., Roy A., Horikoshi M., Roeder R. G. Highly conserved core domain and unique N terminus with presumptive regulatory motifs in a human TATA factor (TFIID). Nature. 1990 Jul 26;346(6282):387–390. doi: 10.1038/346387a0. [DOI] [PubMed] [Google Scholar]
- Hoffman C. S., Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57(2-3):267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Hoffmann A., Horikoshi M., Wang C. K., Schroeder S., Weil P. A., Roeder R. G. Cloning of the Schizosaccharomyces pombe TFIID gene reveals a strong conservation of functional domains present in Saccharomyces cerevisiae TFIID. Genes Dev. 1990 Jul;4(7):1141–1148. doi: 10.1101/gad.4.7.1141. [DOI] [PubMed] [Google Scholar]
- Horikoshi M., Wang C. K., Fujii H., Cromlish J. A., Weil P. A., Roeder R. G. Cloning and structure of a yeast gene encoding a general transcription initiation factor TFIID that binds to the TATA box. Nature. 1989 Sep 28;341(6240):299–303. doi: 10.1038/341299a0. [DOI] [PubMed] [Google Scholar]
- Horikoshi M., Wang C. K., Fujii H., Cromlish J. A., Weil P. A., Roeder R. G. Purification of a yeast TATA box-binding protein that exhibits human transcription factor IID activity. Proc Natl Acad Sci U S A. 1989 Jul;86(13):4843–4847. doi: 10.1073/pnas.86.13.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikoshi M., Yamamoto T., Ohkuma Y., Weil P. A., Roeder R. G. Analysis of structure-function relationships of yeast TATA box binding factor TFIID. Cell. 1990 Jun 29;61(7):1171–1178. doi: 10.1016/0092-8674(90)90681-4. [DOI] [PubMed] [Google Scholar]
- Johnston M., Davis R. W. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Aug;4(8):1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. S., Prakash L. Yeast Saccharomyces cerevisiae selectable markers in pUC18 polylinkers. Yeast. 1990 Sep-Oct;6(5):363–366. doi: 10.1002/yea.320060502. [DOI] [PubMed] [Google Scholar]
- Kao C. C., Lieberman P. M., Schmidt M. C., Zhou Q., Pei R., Berk A. J. Cloning of a transcriptionally active human TATA binding factor. Science. 1990 Jun 29;248(4963):1646–1650. doi: 10.1126/science.2194289. [DOI] [PubMed] [Google Scholar]
- Karin M., Najarian R., Haslinger A., Valenzuela P., Welch J., Fogel S. Primary structure and transcription of an amplified genetic locus: the CUP1 locus of yeast. Proc Natl Acad Sci U S A. 1984 Jan;81(2):337–341. doi: 10.1073/pnas.81.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G. A., Joazeiro C. A., Pisano M., Geiduschek E. P., Colbert T., Hahn S., Blanco J. A. The role of the TATA-binding protein in the assembly and function of the multisubunit yeast RNA polymerase III transcription factor, TFIIIB. Cell. 1992 Dec 11;71(6):1055–1064. doi: 10.1016/0092-8674(92)90399-w. [DOI] [PubMed] [Google Scholar]
- Kelleher R. J., 3rd, Flanagan P. M., Chasman D. I., Ponticelli A. S., Struhl K., Kornberg R. D. Yeast and human TFIIDs are interchangeable for the response to acidic transcriptional activators in vitro. Genes Dev. 1992 Feb;6(2):296–303. doi: 10.1101/gad.6.2.296. [DOI] [PubMed] [Google Scholar]
- Kenna M., Stevens A., McCammon M., Douglas M. G. An essential yeast gene with homology to the exonuclease-encoding XRN1/KEM1 gene also encodes a protein with exoribonuclease activity. Mol Cell Biol. 1993 Jan;13(1):341–350. doi: 10.1128/mcb.13.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koleske A. J., Buratowski S., Nonet M., Young R. A. A novel transcription factor reveals a functional link between the RNA polymerase II CTD and TFIID. Cell. 1992 May 29;69(5):883–894. doi: 10.1016/0092-8674(92)90298-q. [DOI] [PubMed] [Google Scholar]
- Köhrer K., Domdey H. Preparation of high molecular weight RNA. Methods Enzymol. 1991;194:398–405. doi: 10.1016/0076-6879(91)94030-g. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mortimer R. K., Schild D., Contopoulou C. R., Kans J. A. Genetic and physical maps of Saccharomyces cerevisiae. Methods Enzymol. 1991;194:827–863. doi: 10.1016/0076-6879(91)94060-p. [DOI] [PubMed] [Google Scholar]
- Muhich M. L., Iida C. T., Horikoshi M., Roeder R. G., Parker C. S. cDNA clone encoding Drosophila transcription factor TFIID. Proc Natl Acad Sci U S A. 1990 Dec;87(23):9148–9152. doi: 10.1073/pnas.87.23.9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. A., Tatchell K. The structure of transposable yeast mating type loci. Cell. 1980 Mar;19(3):753–764. doi: 10.1016/s0092-8674(80)80051-1. [DOI] [PubMed] [Google Scholar]
- Ng R., Abelson J. Isolation and sequence of the gene for actin in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3912–3916. doi: 10.1073/pnas.77.7.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson M. G., Tanese N., Pugh B. F., Tjian R. Functional domains and upstream activation properties of cloned human TATA binding protein. Science. 1990 Jun 29;248(4963):1625–1630. doi: 10.1126/science.2363050. [DOI] [PubMed] [Google Scholar]
- Poon D., Schroeder S., Wang C. K., Yamamoto T., Horikoshi M., Roeder R. G., Weil P. A. The conserved carboxy-terminal domain of Saccharomyces cerevisiae TFIID is sufficient to support normal cell growth. Mol Cell Biol. 1991 Oct;11(10):4809–4821. doi: 10.1128/mcb.11.10.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh B. F., Tjian R. Mechanism of transcriptional activation by Sp1: evidence for coactivators. Cell. 1990 Jun 29;61(7):1187–1197. doi: 10.1016/0092-8674(90)90683-6. [DOI] [PubMed] [Google Scholar]
- Reddy P., Hahn S. Dominant negative mutations in yeast TFIID define a bipartite DNA-binding region. Cell. 1991 Apr 19;65(2):349–357. doi: 10.1016/0092-8674(91)90168-x. [DOI] [PubMed] [Google Scholar]
- Reinberg D., Horikoshi M., Roeder R. G. Factors involved in specific transcription in mammalian RNA polymerase II. Functional analysis of initiation factors IIA and IID and identification of a new factor operating at sequences downstream of the initiation site. J Biol Chem. 1987 Mar 5;262(7):3322–3330. [PubMed] [Google Scholar]
- Schmidt M. C., Kao C. C., Pei R., Berk A. J. Yeast TATA-box transcription factor gene. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7785–7789. doi: 10.1073/pnas.86.20.7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz M. C., Choe S. Y., Reeder R. H. Specific initiation by RNA polymerase I in a whole-cell extract from yeast. Proc Natl Acad Sci U S A. 1991 Feb 1;88(3):1004–1008. doi: 10.1073/pnas.88.3.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz M. C., Reeder R. H., Hahn S. Variants of the TATA-binding protein can distinguish subsets of RNA polymerase I, II, and III promoters. Cell. 1992 May 15;69(4):697–702. doi: 10.1016/0092-8674(92)90233-3. [DOI] [PubMed] [Google Scholar]
- Tamura T., Sumita K., Fujino I., Aoyama A., Horikoshi M., Hoffmann A., Roeder R. G., Muramatsu M., Mikoshiba K. Striking homology of the 'variable' N-terminal as well as the 'conserved core' domains of the mouse and human TATA-factors (TFIID). Nucleic Acids Res. 1991 Jul 25;19(14):3861–3865. doi: 10.1093/nar/19.14.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele D. J. ACE1 regulates expression of the Saccharomyces cerevisiae metallothionein gene. Mol Cell Biol. 1988 Jul;8(7):2745–2752. doi: 10.1128/mcb.8.7.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada K., Wada Y., Ishibashi F., Gojobori T., Ikemura T. Codon usage tabulated from the GenBank genetic sequence data. Nucleic Acids Res. 1992 May 11;20 (Suppl):2111–2118. doi: 10.1093/nar/20.suppl.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R. J., Jackson S. P., Rigby P. W. A role for the TATA-box-binding protein component of the transcription factor IID complex as a general RNA polymerase III transcription factor. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1949–1953. doi: 10.1073/pnas.89.5.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J. M., Liu F., Bateman E. Cloning and expression of the Acanthamoeba castellanii gene encoding transcription factor TFIID. Gene. 1992 Aug 1;117(1):91–97. doi: 10.1016/0378-1119(92)90494-a. [DOI] [PubMed] [Google Scholar]
- Zawel L., Reinberg D. Advances in RNA polymerase II transcription. Curr Opin Cell Biol. 1992 Jun;4(3):488–495. doi: 10.1016/0955-0674(92)90016-6. [DOI] [PubMed] [Google Scholar]
- Zhou Q. A., Schmidt M. C., Berk A. J. Requirement for acidic amino acid residues immediately N-terminal to the conserved domain of Saccharomyces cerevisiae TFIID. EMBO J. 1991 Jul;10(7):1843–1852. doi: 10.1002/j.1460-2075.1991.tb07710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]