Abstract
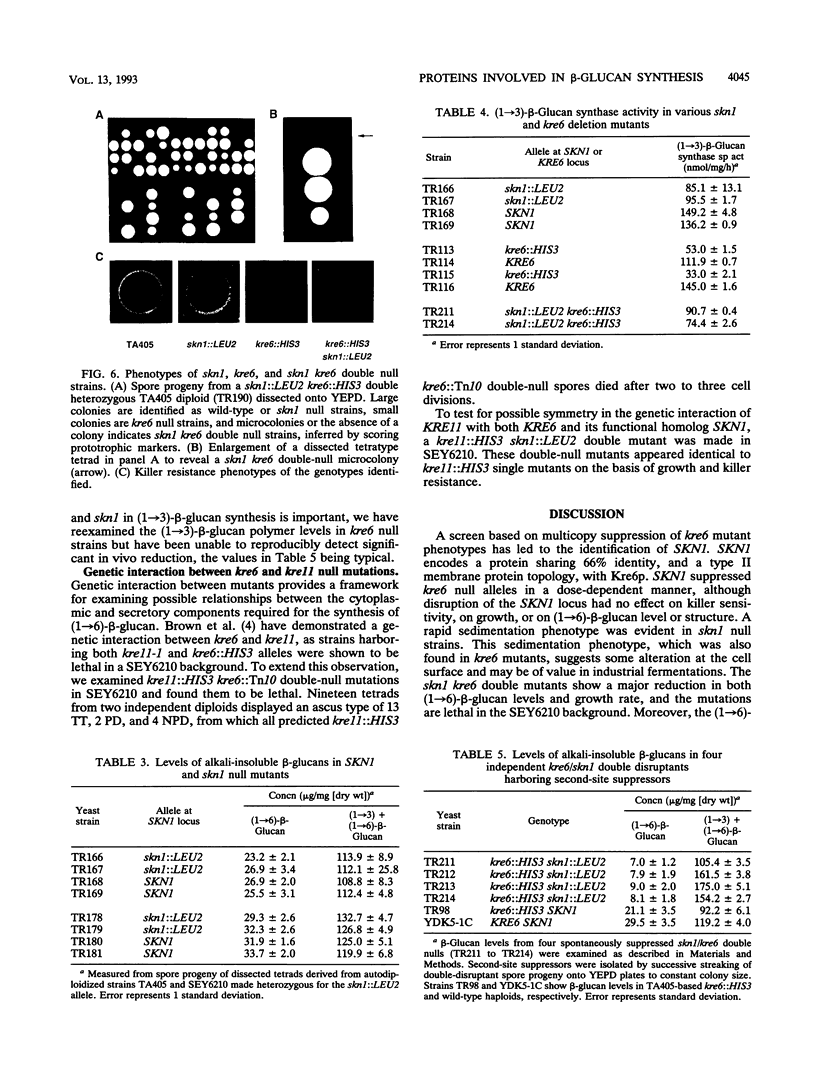
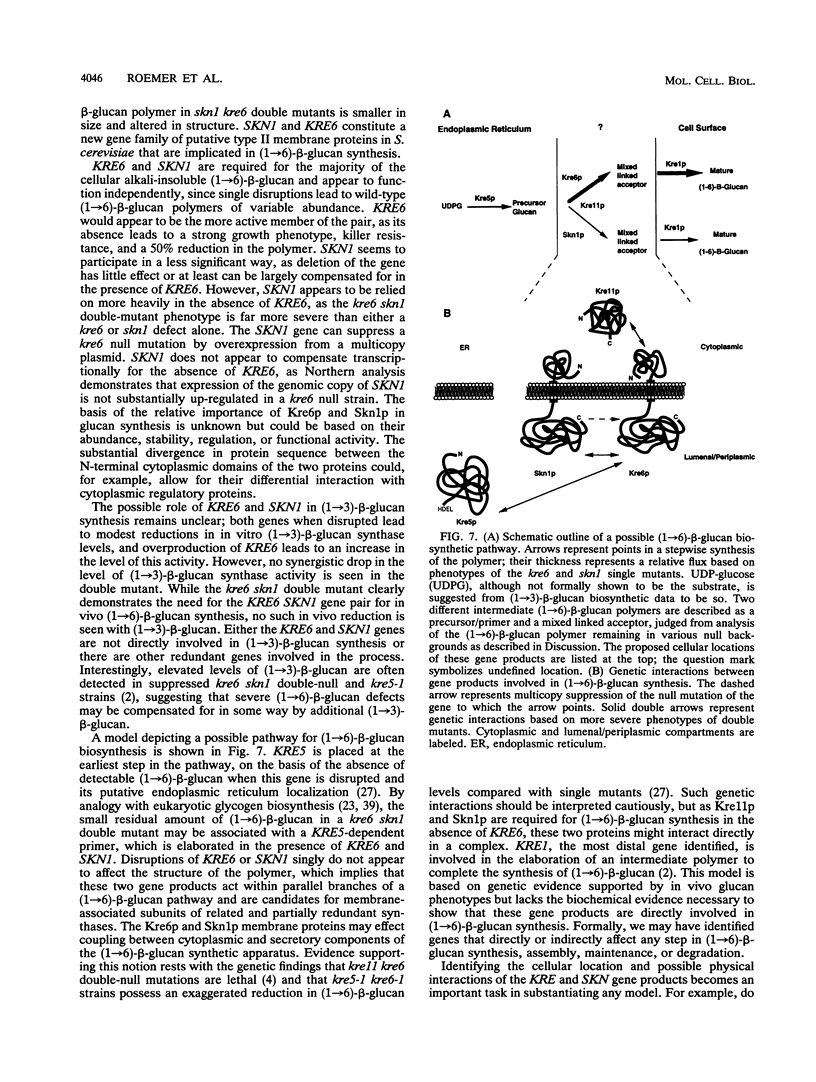
KRE6 encodes a predicted type II membrane protein which, when disrupted, results in a slowly growing, killer toxin-resistant mutant possessing half the normal level of a structurally wild-type cell wall (1-->6)-beta-glucan (T. Roemer and H. Bussey, Proc. Natl. Acad. Sci. USA 88:11295-11299, 1991). The mutant phenotype and structure of the KRE6 gene product, Kre6p, suggest that it may be a beta-glucan synthase component, implying that (1-->6)-beta-glucan synthesis in Saccharomyces cerevisiae is functionally redundant. To examine this possibility, we screened a multicopy genomic library for suppression of both the slow-growth and killer resistance phenotypes of a kre6 mutant and identified SKN1, which encodes a protein sharing 66% overall identity to Kre6p. SKN1 suppresses kre6 null alleles in a dose-dependent manner, though disruption of the SKN1 locus has no effect on killer sensitivity, growth, or (1-->6)-beta-glucan levels. skn1 kre6 double disruptants, however, showed a dramatic reduction in both (1-->6)-beta-glucan levels and growth rate compared with either single disruptant. Moreover, the residual (1-->6)-beta-glucan polymer in skn1 kre6 double mutants is smaller in size and altered in structure. Since single disruptions of these genes lead to structurally wild-type (1-->6)-beta-glucan polymers, Kre6p and Skn1p appear to function independently, possibly in parallel, in (1-->6)-beta-glucan biosynthesis.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BADIN J., JACKSON C., SCHUBERT M. Improved method for determination of plasma polysaccharides with tryptophan. Proc Soc Exp Biol Med. 1953 Nov;84(2):289–291. [PubMed] [Google Scholar]
- Boone C., Sommer S. S., Hensel A., Bussey H. Yeast KRE genes provide evidence for a pathway of cell wall beta-glucan assembly. J Cell Biol. 1990 May;110(5):1833–1843. doi: 10.1083/jcb.110.5.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briza P., Breitenbach M., Ellinger A., Segall J. Isolation of two developmentally regulated genes involved in spore wall maturation in Saccharomyces cerevisiae. Genes Dev. 1990 Oct;4(10):1775–1789. doi: 10.1101/gad.4.10.1775. [DOI] [PubMed] [Google Scholar]
- Brown J. L., Kossaczka Z., Jiang B., Bussey H. A mutational analysis of killer toxin resistance in Saccharomyces cerevisiae identifies new genes involved in cell wall (1-->6)-beta-glucan synthesis. Genetics. 1993 Apr;133(4):837–849. doi: 10.1093/genetics/133.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulawa C. E. CSD2, CSD3, and CSD4, genes required for chitin synthesis in Saccharomyces cerevisiae: the CSD2 gene product is related to chitin synthases and to developmentally regulated proteins in Rhizobium species and Xenopus laevis. Mol Cell Biol. 1992 Apr;12(4):1764–1776. doi: 10.1128/mcb.12.4.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulawa C. E., Slater M., Cabib E., Au-Young J., Sburlati A., Adair W. L., Jr, Robbins P. W. The S. cerevisiae structural gene for chitin synthase is not required for chitin synthesis in vivo. Cell. 1986 Jul 18;46(2):213–225. doi: 10.1016/0092-8674(86)90738-5. [DOI] [PubMed] [Google Scholar]
- Bussey H. K1 killer toxin, a pore-forming protein from yeast. Mol Microbiol. 1991 Oct;5(10):2339–2343. doi: 10.1111/j.1365-2958.1991.tb02079.x. [DOI] [PubMed] [Google Scholar]
- Bussey H., Sacks W., Galley D., Saville D. Yeast killer plasmid mutations affecting toxin secretion and activity and toxin immunity function. Mol Cell Biol. 1982 Apr;2(4):346–354. doi: 10.1128/mcb.2.4.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabib E., Kang M. S. Fungal 1,3-beta-glucan synthase. Methods Enzymol. 1987;138:637–642. doi: 10.1016/0076-6879(87)38057-7. [DOI] [PubMed] [Google Scholar]
- Cabib E., Sburlati A., Bowers B., Silverman S. J. Chitin synthase 1, an auxiliary enzyme for chitin synthesis in Saccharomyces cerevisiae. J Cell Biol. 1989 May;108(5):1665–1672. doi: 10.1083/jcb.108.5.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas I., Hardy T. A., DePaoli-Roach A. A., Roach P. J. Isolation of the GSY1 gene encoding yeast glycogen synthase and evidence for the existence of a second gene. J Biol Chem. 1990 Dec 5;265(34):20879–20886. [PubMed] [Google Scholar]
- Farkas I., Hardy T. A., Goebl M. G., Roach P. J. Two glycogen synthase isoforms in Saccharomyces cerevisiae are coded by distinct genes that are differentially controlled. J Biol Chem. 1991 Aug 25;266(24):15602–15607. [PubMed] [Google Scholar]
- HALVORSON H. Studies on protein and nucleic acid turnover in growing cultures of yeast. Biochim Biophys Acta. 1958 Feb;27(2):267–276. doi: 10.1016/0006-3002(58)90333-0. [DOI] [PubMed] [Google Scholar]
- Hartmann E., Rapoport T. A., Lodish H. F. Predicting the orientation of eukaryotic membrane-spanning proteins. Proc Natl Acad Sci U S A. 1989 Aug;86(15):5786–5790. doi: 10.1073/pnas.86.15.5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman C. S., Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57(2-3):267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Hutchins K., Bussey H. Cell wall receptor for yeast killer toxin: involvement of (1 leads to 6)-beta-D-glucan. J Bacteriol. 1983 Apr;154(1):161–169. doi: 10.1128/jb.154.1.161-169.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang P. K., Tugendreich S., Fletterick R. J. Molecular analysis of GPH1, the gene encoding glycogen phosphorylase in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Apr;9(4):1659–1666. doi: 10.1128/mcb.9.4.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin D. E., Bartlett-Heubusch E. Mutants in the S. cerevisiae PKC1 gene display a cell cycle-specific osmotic stability defect. J Cell Biol. 1992 Mar;116(5):1221–1229. doi: 10.1083/jcb.116.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomako J., Lomako W. M., Whelan W. J. The nature of the primer for glycogen synthesis in muscle. FEBS Lett. 1990 Jul 30;268(1):8–12. doi: 10.1016/0014-5793(90)80959-m. [DOI] [PubMed] [Google Scholar]
- Manners D. J., Masson A. J., Patterson J. C., Björndal H., Lindberg B. The structure of a beta-(1--6)-D-glucan from yeast cell walls. Biochem J. 1973 Sep;135(1):31–36. doi: 10.1042/bj1350031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinac B., Zhu H., Kubalski A., Zhou X. L., Culbertson M., Bussey H., Kung C. Yeast K1 killer toxin forms ion channels in sensitive yeast spheroplasts and in artificial liposomes. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6228–6232. doi: 10.1073/pnas.87.16.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaden P., Hill K., Wagner J., Slipetz D., Sommer S. S., Bussey H. The yeast KRE5 gene encodes a probable endoplasmic reticulum protein required for (1----6)-beta-D-glucan synthesis and normal cell growth. Mol Cell Biol. 1990 Jun;10(6):3013–3019. doi: 10.1128/mcb.10.6.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paravicini G., Cooper M., Friedli L., Smith D. J., Carpentier J. L., Klig L. S., Payton M. A. The osmotic integrity of the yeast cell requires a functional PKC1 gene product. Mol Cell Biol. 1992 Nov;12(11):4896–4905. doi: 10.1128/mcb.12.11.4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks G. D., Lamb R. A. Topology of eukaryotic type II membrane proteins: importance of N-terminal positively charged residues flanking the hydrophobic domain. Cell. 1991 Feb 22;64(4):777–787. doi: 10.1016/0092-8674(91)90507-u. [DOI] [PubMed] [Google Scholar]
- Ribas J. C., Diaz M., Duran A., Perez P. Isolation and characterization of Schizosaccharomyces pombe mutants defective in cell wall (1-3)beta-D-glucan. J Bacteriol. 1991 Jun;173(11):3456–3462. doi: 10.1128/jb.173.11.3456-3462.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roemer T., Bussey H. Yeast beta-glucan synthesis: KRE6 encodes a predicted type II membrane protein required for glucan synthesis in vivo and for glucan synthase activity in vitro. Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):11295–11299. doi: 10.1073/pnas.88.24.11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Schiestl R. H., Gietz R. D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989 Dec;16(5-6):339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Shaw J. A., Mol P. C., Bowers B., Silverman S. J., Valdivieso M. H., Durán A., Cabib E. The function of chitin synthases 2 and 3 in the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1991 Jul;114(1):111–123. doi: 10.1083/jcb.114.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shematek E. M., Braatz J. A., Cabib E. Biosynthesis of the yeast cell wall. I. Preparation and properties of beta-(1 leads to 3)glucan synthetase. J Biol Chem. 1980 Feb 10;255(3):888–894. [PubMed] [Google Scholar]
- Silverman S. J. Similar and different domains of chitin synthases 1 and 2 of S. cerevisiae: two isozymes with distinct functions. Yeast. 1989 Nov-Dec;5(6):459–467. doi: 10.1002/yea.320050605. [DOI] [PubMed] [Google Scholar]
- Smythe C., Villar-Palasi C., Cohen P. Structural and functional studies on rabbit liver glycogenin. Eur J Biochem. 1989 Jul 15;183(1):205–209. doi: 10.1111/j.1432-1033.1989.tb14914.x. [DOI] [PubMed] [Google Scholar]
- Szaniszlo P. J., Kang M. S., Cabib E. Stimulation of beta(1----3)glucan synthetase of various fungi by nucleoside triphosphates: generalized regulatory mechanism for cell wall biosynthesis. J Bacteriol. 1985 Mar;161(3):1188–1194. doi: 10.1128/jb.161.3.1188-1194.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivieso M. H., Mol P. C., Shaw J. A., Cabib E., Durán A. CAL1, a gene required for activity of chitin synthase 3 in Saccharomyces cerevisiae. J Cell Biol. 1991 Jul;114(1):101–109. doi: 10.1083/jcb.114.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernet T., Dignard D., Thomas D. Y. A family of yeast expression vectors containing the phage f1 intergenic region. Gene. 1987;52(2-3):225–233. doi: 10.1016/0378-1119(87)90049-7. [DOI] [PubMed] [Google Scholar]
- Zhang G. F., Staehelin L. A. Functional compartmentation of the Golgi apparatus of plant cells : immunocytochemical analysis of high-pressure frozen- and freeze-substituted sycamore maple suspension culture cells. Plant Physiol. 1992 Jul;99(3):1070–1083. doi: 10.1104/pp.99.3.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Bussey H. Mutational analysis of the functional domains of yeast K1 killer toxin. Mol Cell Biol. 1991 Jan;11(1):175–181. doi: 10.1128/mcb.11.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]