Abstract
Sophisticated image analysis methods have been developed for the human brain, but such tools still need to be adapted and optimized for quantitative small animal imaging. We propose a framework for quantitative anatomical phenotyping in mouse models of neurological and psychiatric conditions. The framework encompasses an atlas space, image acquisition protocols, and software tools to register images into this space. We show that a suite of segmentation tools (Avants, Epstein et al., 2008) designed for human neuroimaging can be incorporated into a pipeline for segmenting mouse brain images acquired with multispectral magnetic resonance imaging (MR) protocols. We present a flexible approach for segmenting such hyperimages, optimizing registration, and identifying optimal combinations of image channels for particular structures. Brain imaging with T1, T2* and T2 contrasts yielded accuracy in the range of 83% for hippocampus and caudate putamen (Hc and CPu), but only 54% in white matter tracts, and 44% for the ventricles. The addition of diffusion tensor parameter images improved accuracy for large gray matter structures (by >5%), white matter (10%), and ventricles (15%). The use of Markov random field segmentation further improved overall accuracy in the C57BL/6 strain by 6%; so Dice coefficients for Hc and CPu reached 93%, for white matter 79%, for ventricles 68%, and for substantia nigra 80%. We demonstrate the segmentation pipeline for the widely used C57BL/6 strain, and two test strains (BXD29, APP/TTA). This approach appears promising for characterizing temporal changes in mouse models of human neurological and psychiatric conditions, and may provide anatomical constraints for other preclinical imaging, e.g. fMRI and molecular imaging. This is the first demonstration that multiple MR imaging modalities combined with multivariate segmentation methods lead to significant improvements in anatomical segmentation in the mouse brain.
Keywords: Automated segmentation, Magnetic resonance microscopy, Phenotyping, Mouse brain, Alzheimer's mouse model, BXD
Introduction
Multiple imaging modalities have been developed to study brain function and the complex relationships among its structural units, and have spurred interest into building a connectome, both for humans (Sporns, 2011; Van Essen and Ugurbil, 2012) as well as for small animal models (Bohland et al., 2009), such as the mouse. Different hardware may be required to analyze neuronal and vascular circuits at nano (e.g. electron microscopy), or microstructural resolution (e.g. optical tools and protein markers), and the relationship with gene expression patterns, or the transcriptome (French and Pavlidis, 2011; Ng et al., 2009). Such tools focus on discrete brain regions, which later need to be reassembled into a coherent 3D volume. Magnetic resonance imaging (MRI), in turn, offers the advantage of preserving large-scale connectivity and undistorted 3D spatial relationships. Sophisticated methods have been developed for human structural and functional brain image analyses based on MRI, DTI (Smith et al., 2006), and fMRI (Ashburner and Friston, 2000;Chung et al., 2010; Dale et al., 1999; Fischl et al., 1999; Friston et al., 1994; Worsley et al., 2009). These methods can be streamlined into software pipelines, as those built for human brain studies (Rex et al., 2003; Zijdenbos et al., 2002). However, only limited effort has been dedicated to small animal brain segmentation.
Among small animals, the mouse is a valuable resource for understanding the brain. Natural mutations, and interventions using gene targeting (Evans and Kaufman, 1981; Smithies et al., 1985; Thomas and Capecchi, 1987) provide powerful tools to study genetic diseases. Such mouse models help understand the etiology of human neurologic and neuropsychiatric conditions, involving neurodegeneration (Gama Sosa et al., 2012), altered behavior and connectivity (Patterson, 2011), and provide a platform for developing therapies. But the methods for small animal imaging are varied. MRI is well suited for brain imaging because of its excellent soft tissue contrast, three-dimensional, and nondestructive characteristics. The contrast between structures is considerably different than in the clinical domain, and image array sizes are often larger, increasing computational demands. A current rate limiter for high-throughput studies is the availability of automated image analysis tools to deal efficiently with such images of small animal models.
Since manual segmentation of mouse brain compartments is extremely laborious, especially in high-resolution MR images (e.g. 1024 × 512 × 512 image arrays), an automated segmentation method is critical for achieving the throughput required for routine use. To obtain accurate segmentation high contrast-to-noise ratio images are required. However the MR scan duration limits resolution for in vivo studies. At the completion of live animal studies, or as an alternative, one can image fixed specimens — collected at distinct time points during development, aging, or disease progression. The scan time restrictions being removed, MRI can provide a variety of image contrasts at microscopic resolution to emphasize specific aspects of structure in fixed specimens. Modern high field MR technology (Johnson et al., 2007) in conjunction with staining (Johnson et al., 2002; Zhang et al., 2010b), now allow the acquisition of high contrast images of the mouse brain with isotropic resolution of 20–50 µm. Therefore we focus this meta-analysis study on actively stained, fixed mouse brain specimens.
We propose a framework for quantitative assessment of anatomical phenotypes in mouse models of neurological and psychiatric conditions. The framework supports the use of single and multispectral MR protocols, and we identify optimum image channel selections to enhance segmentation accuracy of individual brain regions, including gray matter nuclei, white matter tracts, and the ventricular system.
Most previous studies phenotyping mouse models of neurological disease have relied on a single MR image channel (Badea et al., 2007b;Bock et al., 2006; Borg and Chereul, 2008; Delatour et al., 2006; Lerch et al., 2008; Sawiak et al., 2009; Zhang et al., 2010a). Even if the atlas includes multi-contrast or multi-modal data (Aggarwal et al., 2009b;Badea et al., 2007a;MacKenzie-Graham et al., 2004), computational demands have usually restricted its application to a single image channel (Badea et al., 2009, 2010; Cheng et al., 2011); and the analysis of multimodal biomarkers was done separately, as in Blockx et al. (2011). Recent atlases, however, can provide additional information on both gray and white matter structures (Jiang and Johnson, 2011), based on high-resolution diffusion tensor imaging (DTI). DTI provides more accurate information on the volumes of white matter tracts, as well as their integrity, and has the potential to detect microstructural changes before major tissue loss. Computational tools are therefore required to process large multispectral datasets, to efficiently yield quantitative phenotypes. A recent study used DTI multichannel registration to characterize brain development in mutant mice (Aggarwal et al., 2009a); however the choice of image channels, and impact on mouse brain segmentation performance remain to be addressed. In this study we evaluate the contribution of multiple MR contrasts, including DTI, to segmentation accuracy for select brain structures.
A reference atlas is required to provide priors for brain segmentation. This is usually built from a single animal (Cheng et al., 2011;Maheswaran et al., 2009a,b; Xie et al., 2010; Zhang et al., 2010a), but such priors can be also constructed from multiple animals—imaged live (Aggarwal et al., 2009b; Ma et al., 2008; Maheswaran et al., 2009a,b), or as a fixed specimen (Aggarwal et al., 2009b; Badea et al., 2007a; Kovacevic et al., 2005; Lerch et al., 2008; Ma et al., 2005). In this study we quantify the effect of using multi-atlas priors and a multivariate likelihood model on segmentation accuracy.
The quantitative assessment of automated segmentation accuracy is done with reference to manual labels, and uses various metrics, e.g. volume correlation (Lau et al., 2008), distance-based measures (Zhang et al., 2010a), or voxel overlap i.e. Dice coefficients (Lebenberg et al., 2010; Ma et al., 2005; Maheswaran et al., 2009a,b; Wu et al., 2012). In this paper we present a systematic optimization of the registration process (Avants et al., 2008b, 2011a), based on maximizing Dice coefficients, with the goal to help efficiently select parameters for optimal accuracy. The registration process is followed by an additional step—the use of MRF modeling and Bayesian statistics (Avants et al., 2011b;Held et al., 1997) for classifying voxels. The combined approach uses information from multiple specimens, and multivariate data from different MR protocols, to improve upon the results of single-specimen, single-channel segmentation.
Multiple imaging contrasts and modalities provide complementary information, but also overwhelming data. Today's image data have two or three spatial dimensions, possibly a temporal dimension, and may have originated from multiple sources (such as MRI, SPECT, PET, US, histology, optical, etc.). These sources provide information that needs to be integrated, and efficiently mined. We propose to treat such multispectral information as a hyperimage, which can be used to test a biological hypothesis (e.g. phenotypes of neurological conditions). In this study, a mouse brain hyperimage consists of up to 1024 × 512 × 512 voxels image matrix, with N channels. The channels are T1-, T2- and/or T2*-weighted structural MR images, complemented with images derived from diffusion tensor data, such as fractional anisotropy (FA), radial diffusivity (RD), and trace (Tr). We take advantage of distributed/parallel computation to assess which image channels provide unique information necessary to complete the picture for normal development or aging, a particular brain disease, or toxicological insults. To achieve this we propose and optimize a flexible pipeline for phenotyping mouse neuroanatomy, based on processing the hyperimage with multivariate/multichannel registration and classification. Software scripts and data used to establish accuracy of automated segmentation are shared with the community to aid optimization of other segmentation methods (http://www.civm.duhs.duke.edu/brainpipe2011).
Methods
Animals
Brain images used were from previously published studies (Badea et al., 2007a, 2009, 2010; Jiang and Johnson, 2011; Johnson et al., 2010; Sharief et al., 2008). Animals included C57BL/6 mice, a double transgenic mouse model of Alzheimer's disease (AD) (Jankowsky et al., 2005) and a BXD29 mouse, from the RI family of BXD strains obtained from intercrossing the C57BL/6 and DBA/2 strains. All imaging studies were performed at the Duke Center for In Vivo Microscopy, using animal procedures approved by the Duke University Institutional Animal Care and Use Committee (IACUC). C57BL/6 and BXD mice were obtained from the Jackson Labs (Bar Harbor, ME), and Dr. Joanna Jankowsky (Baylor College of Medicine) donated the AD mice. We selected these strains because of known variability (Badea et al., 2009, 2010). For example, the total volume of BXD29 brain is ~83% of the C57BL/6, while for the AD model is 99% of the C57BL/6 brain volume, but is known to change by about 5–6% relative to age matched controls. Yet in the AD model the volumes of subcortical brain structures change, e.g. the substantia nigra changes with ~17% relative to controls. Absolute hippocampus (Hc) volumes vary from 23.3 µm3 for the AD mouse, to 27.2 µm3 for the BXD29, to 29.9 µm3 for the C57BL/6— a 23% and 10% change relative to the C57BL/6. Relative Hc volumes represent 5.9% of the brain volume for C57BL/6, 5.4% for BXD29, and 5.3% for the AD mouse.
Transcardial perfusion was used to actively stain the specimens with 1:10 ProHance (Bracco Diagnostics Inc., Princeton, NJ), as described in Johnson et al. (2002, 2007). The brain specimens, left in the skull to avoid mechanical distortions, were placed in fomblin-filled tubes for scanning.
Imaging
High-field MR histology of actively stained mouse brains was done in accordance with protocols (Table 1) described in Johnson et al. (2007) for “conventional” imaging, and Jiang and Johnson (2010) for diffusion tensor imaging (DTI). Specimens were scanned on a 9.4 T, 8.9 cm vertical bore Oxford magnet, with shielded coils providing gradients of up to 2000 mT/m (Resonance Research, Inc. Billerica, MA), controlled by a General Electric (GE Healthcare, Milwaukee, WI) EXCITE console (Epic 11.0). We used a solenoid radiofrequency coil built in-house, from a single sheet of microwave substrate (12 mm diameter). Specimens remained in the same position in the magnet during the whole image acquisition protocol. Two distinct image acquisition protocols were used to acquire T1-weighted (T1w) and T2-weighted (T2w), multiple echo frequency-domain image contrast (MEFIC)-enhanced images in C57BL/6 mice (n = 5), as well as for two additional strains: BXD29 (Peirce et al., 2004; Taylor, 1978) (n = 1), and an APP/TTA double transgenic mouse model of Alzheimer's disease (Jankowsky et al., 2005) (n = 1). In addition, fiveC57BL/6micewere scanned using the same T1w sequence, a T2* weighted sequence (T2*w) and a diffusion tensor sequence (DTI) (Jiang and Johnson, 2010).
Table 1.
Summary of protocols for high-resolution magnetic resonance histology for automated anatomical phenotyping.
| Protocol | TR/TE | Matrix | Resolution (mm) | Flip angle a (deg) | Time (h) | Raw size (MB) |
|---|---|---|---|---|---|---|
| T1w | 50/5.2 | 1024×512×512 | 21.5 | 90 | 2 | 512 |
| 512×256×256 | 43 | 90 | 1 | 256 | ||
| T2*w | 50/5.2 | 1024×512×512 | 21.5 | 60 | 2 | 512 |
| 512×256×256 | 43 | 60 | 1 | 256 | ||
| T2w | 400/7.5 | 512×256×256 | 43 | 90 | 4 | 7×256 |
| DTI | 100/11.8; 6 directions b≈1.5×103s/mm2 | 512×256×256 | 43 | 90 | 28 | 1.9×103 |
T1-weighted, T2*-weighted acquisition
The 3D T1w spin echo sequence had the following parameters: echo time (TE) 5.2 ms, repetition time (TR) 50 ms, 62.5 kHz bandwidth, field of view (FOV) of 11 × 11 × 22 mm. A 3D GRASS sequence with TE 4.4 ms, TR 50 ms, and flip angle of 60°, using the same field of view, provided the T2*w images. Fourier space was sampled asymmetrically at 384 × 384 × 768 locations, with zero filling only on one side of the array to 512 × 512 × 1024. The receiver gain was increased at the periphery of Fourier space, to provide an expanded dynamic range and effectively weight (amplify) these higher frequencies during the phase encoding steps. This method allows acquisition of high-resolution images with reduced time (Johnson et al., 2007). Scan times were 2 h 7 min for these protocols, with 21.5 µm isotropic resolution.
T2-weighted acquisition
The 3D T2w imaging protocol consisted of a Carr-Purcell-Meiboom-Gill (CPMG) sequence with the following parameters: inter-echo spacing 7 ms, 8 echoes, TR 400 ms, 62.5 kHz bandwidth, FOV 11 × 11 × 22 mm, and matrix size 256 × 256 × 512 pixels. The echoes were post-processed using MEFIC implemented in MATLAB (MathWorks, Natick, MA), resulting in a T2w dataset with increased contrast and signal-to-noise-ratio (SNR) (Sharief and Johnson, 2006). The sequence used asymmetric Fourier sampling with expanded dynamic range, filling a 192 × 192 × 512 array, with subsequent zero filling on one side (25%) of each phase encoding axis. The scan time was 4 h 15 min for the T2w protocol, with 43 µm isotropic resolution.
DTI acquisition and processing
The 3D DTI protocol consisted of a diffusion-weighted spin-echo pulse sequence (Jiang and Johnson, 2010) with TE = 11.8 ms, TR = 100 ms, FOV 22 × 11 × 11 mm, matrix size 512 × 256 × 256, and 2 averages (NEX), resulting in 43 µm isotropic voxels. Diffusion encoding was performed using a pair of half-sine gradient pulses (pulse width = 1.3 ms, pulse separation = 6.4 ms, gradient amplitude = 1600 mT/m). One b0 (i.e., b≈0) and 6 diffusion-weighted images (b≈1.5 × 103 s/mm2) sensitized in 6 directions ([1, 1, 0], [1, 0, 1], [0, 1, 1], [−1, 1, 0], [1, 0, −1], [0, −1, 1]) were acquired, the scan time being 28 h.
For the DTI scanned specimens, T1 and T2*w conventional images were also acquired, with the same field of view (FOV) and resolution, with scan time (TR = 50 ms, NEX = 1) of 1 h.
DTI Studio (Jiang et al., 2006) was used for tensor reconstruction, and to calculate fractional (FA) and radial anisotropy (RA), longitudinal (E0) and radial diffusivity (RD), and trace (Tr). These images were aligned into Waxholm Space (WHS) (Johnson et al., 2010), based on the transforms derived from aligning T2*w images of individual specimens to the WHS T2*w channel. We used bilateral filtering (Tomasi and Manduchi, 1998) for edge preserving denoising of DT images.
Reference atlases
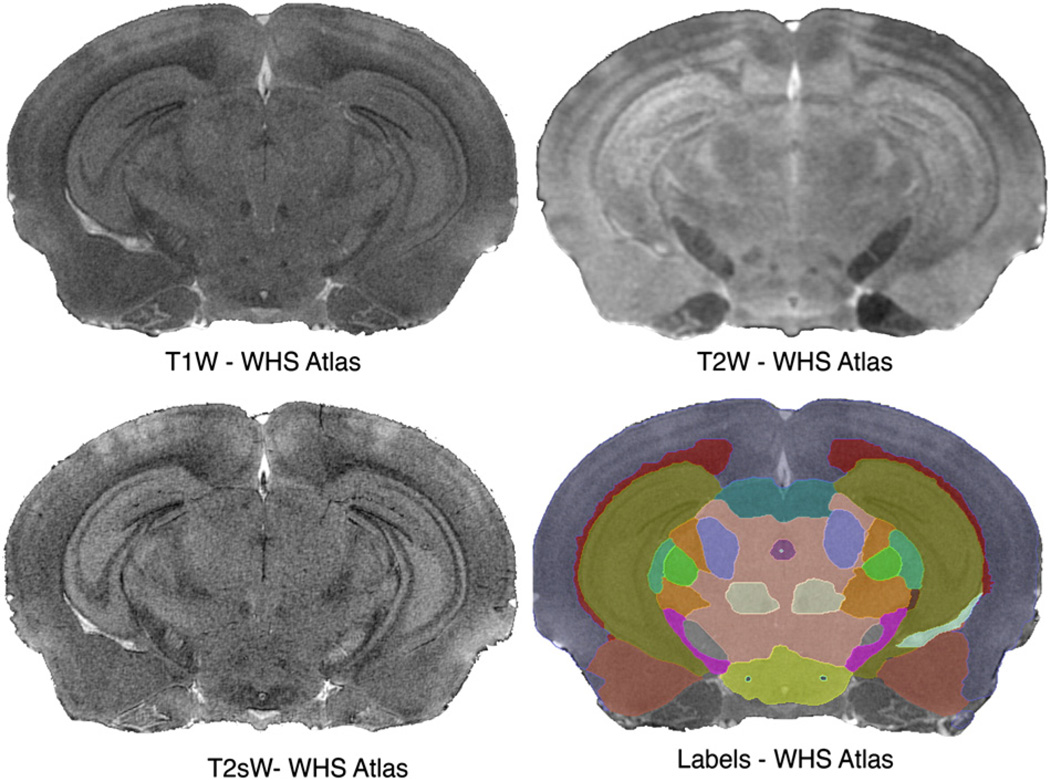
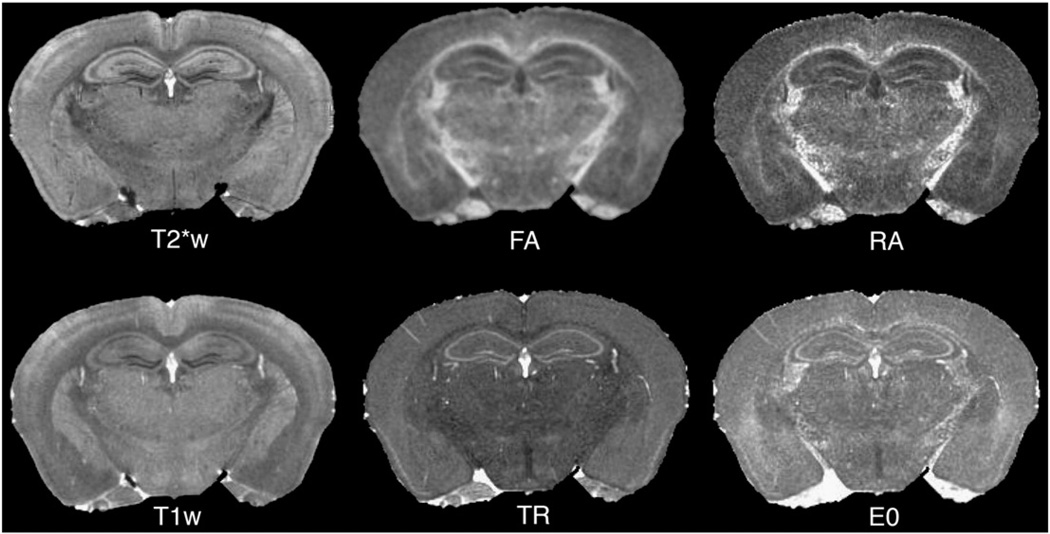
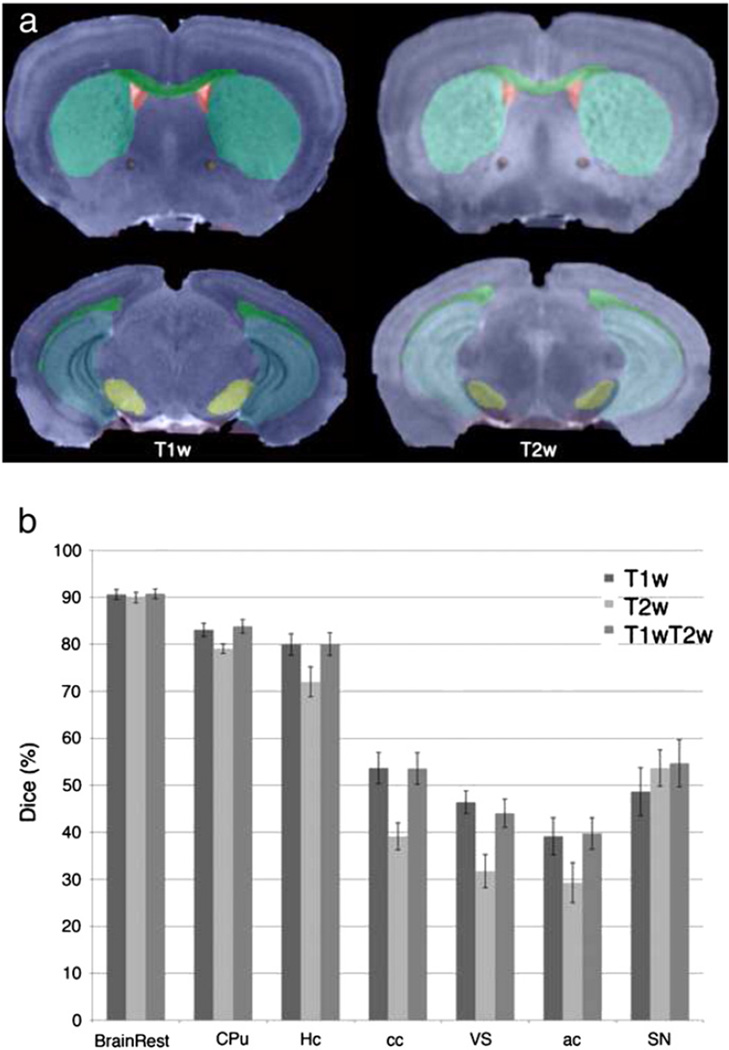
An essential component of segmentation is a mouse brain atlas based on three MR protocols (T1w, T2*w, and T2w), with different contrasts among brain regions (Fig. 1). For brevity, we use the term “conventional” for these data. This atlas defines a common reference, called Waxholm Space (WHS), and includes 37 labeled regions (Johnson, Badea et al., 2010). Other atlases can be used, e.g. the multi-specimen atlas with T1w and T2w contrasts (Badea et al., 2007a), containing 33 labeled regions, or a DTI atlas (Jiang and Johnson, 2011), registered into WHS. Our DTI set is based on this atlas and includes: fractional (FA) and radial anisotropy (RA), trace (Tr), longitudinal (E0) and radial diffusivity (RD) images (Fig. 2).
Fig. 1.
The segmentation pipeline uses an atlas comprising multivariate intensity and label priors. The priors provided by the Waxholm reference are based on an actively stained C57BL/6 mouse brain. Images were acquired using 21.5 µm resolution T1w, T2*w protocols, and 43 µm resolution T2w protocols. The MR images are complemented by a set of 37 labeled regions.
Fig. 2.
Query image sets to be segmented have one or more image channels. It is recommended that at least one of these channels be acquired with the same parameters as the reference. Here a diffusion tensor imaging (DTI) protocol was acquired for a C57BL/6 mouse, in addition to the T1w and T2*w protocols present in the original WHS set. The role of individual DT parameter images can be evaluated with regard to improving segmentation accuracy.
A set of eight regions was manually traced on the T1w, T2*w and DTI set using AMIRA (Visage Imaging, El Camino Real, CA), for validating the automated segmentation. We selected gray matter structures involved in learning and memory, reward and addiction, and Parkinson's disease; major white matter structures; and ventricles, enlarged in brain degeneration. The gray matter structures included the hippocampus (Hc); caudate putamen (CPu) and substantia nigra (SN); white matter structures included corpus callosum (cc) and anterior commissure (cc); and the ventricular space included the lateral and 4th ventricles (LV, 4 V, combined as ventricular system VS). The same regions were manually labeled onto the BXD29 and AD data sets using the T1 and T2w images.
Segmentation pipeline
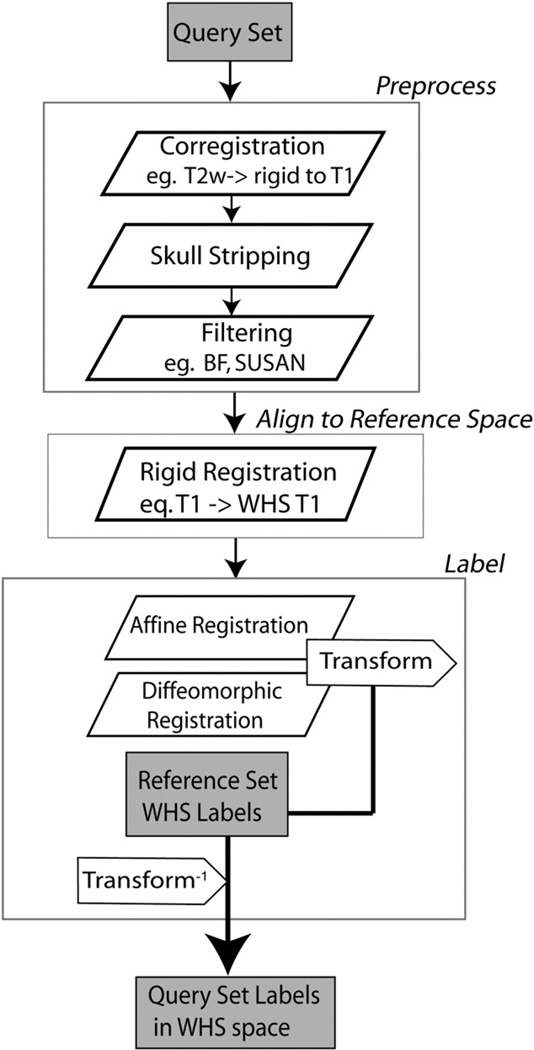
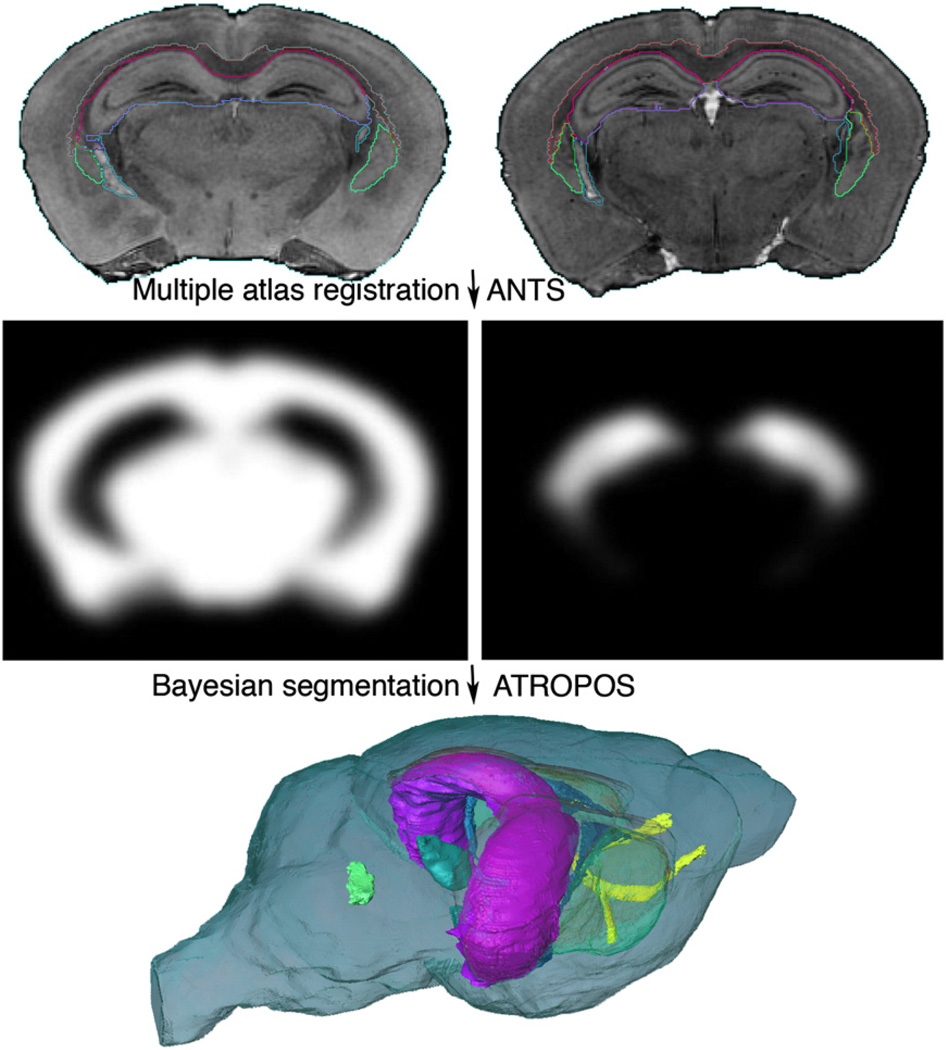
The processing pipeline resides on a dedicated workstation (8 cores, 2 Intel Xeon processors, 2.66 GHz, Mac Pro4, equipped with 64 GB memory), and is implemented in object oriented Perl, with a modular structure. The modules coordinated by the pipeline automatically perform: a) raw image retrieval from an archive database and conversion to NIFTI format; b) stripping extra cranial tissue using mathematical morphology tools in MATLAB; c) multivariate registration between the atlas and new brain specimens. The last step is performed using the ANTs registration software (Avants et al., 2008b, 2011a), as shown in Fig. 3. ANTs has been used sequentially for: 1) rigid coregistration among images acquired for one specimen; 2) rigid registration to WHS; 3) affine registration between the WHS atlas and the query set, i.e. all images acquired for one specimen, to remove global differences; 4) diffeomorphic registration between the WHS atlas and the query set, to compensate for local differences. Bias field correction was performed for new strains using N4ITK (Tustison et al., 2010).
Fig. 3.
Segmentation flow chart for labeling a mouse brain specimen using multispectral MR images. Optional preprocessing steps include bias field correction and noise reduction filters. At first, all images for one specimen are coregistered, e.g. to the T2*w image; then skull stripped. The resulting image set is rigidly registered into the coordinate system defined by the Waxholm space (WHS). Finally the query set is mapped to the WHS reference atlas with a combined affine and diffeomorphic transform. The inverted transforms are applied to the reference atlas labels (WHS). Resulting label volumes are used for anatomical phenotyping, and statistical characterization of treatment effects.
In essence, we used ANTs to derive the mapping between the reference atlas (e.g. WHS) and the query set. The mapping is composed of an affine and a diffeomorphic transform, symmetric and invertible, and is computed in a multiresolution scheme to improve convergence speed. At each iteration the diffeomorphic transform (f), is updated toward a point midway toward images I and J, as described in Avants et al. (2008b):
where δ is a gradient step parameter (referred to as SyN parameter from now on), K is a regularization term (chosen as Gaussian), and Π is a similarity metric. We used mutual information as the similarity metric for the affine transform, and cross correlation for the diffeomorphic transform. The mutual information (MI) between two images I and J is evaluated based on the marginal probability density functions p1(i) and p2(j), and the joint probability density function p(i,j), derived from the marginal histogram of the two images and the joint histogram, with a limited number of bins (200) and sampling points (32,000):
The cross-correlation (CC) between two images I and J is evaluated using a kernel centered in x, of radius r and with i elements, with mean values over the kernel µI, µJ, and is integrated over the image by moving the center of the kernel across the image:
The composite affine and diffeomorphic transforms (that bring the WHS and query image set into register) are applied to the WHS label set to produce new labels for each query set. In addition to registration based segmentation, classification using multivariate intensity and label priors, a Bayesian framework, and Markov random field (MRF) modeling was performed using ATROPOS (Avants et al., 2011b). To classify voxels, the posterior Gaussian probability was modeled by the Euclidian distance in the univariate case, and the Mahalanobis distance in the multivariate case.
All software modules are coordinated by the Perl pipeline, producing a customizable application for segmenting mouse brain images, ideally acquired using one or more of the atlas acquisition protocols.
Evaluation and statistics
We tested the segmentation pipeline for brains of the C57BL/6 strain, a double transgenic AD mouse model (Jankowsky et al., 2005), and a BXD29 mouse. To evaluate the segmentation performance, we used single or multiple-specimen manually labeled atlases. These included the WHS (Johnson et al., 2010) (Fig. 1) with T1w, T2*w, and T2w image channels, the multi-specimen atlas (Badea et al., 2007a), and DTI atlases for the C57BL/6. The atlas labels and images provided priors for segmenting new datasets, and the results of automated segmentation were evaluated using the Dice coefficient (Dice, 1945), illustrating the coincidence of a label in the manual (M) and automated (A) segmentation, in the same subject space.
Results are expressed in percentages, as mean±standard error of the mean. We used t test and one-way analysis of variance (ANOVA) with Dice coefficient values as the dependent variable and segmentation strategy (registration/MRF; or channel combinations as independent variables), and Tukey multiple comparison corrections. A p value<0.05 was considered significant.
Results
We have developed streamlined protocols for quantitative neuroanatomical phenotyping. The results of automated segmentation can be used to produce population atlases, phenotype mouse models, or determine genetic influences on brain structure (Badea et al., 2009). Starting from an atlas based on the C57BL/6 strain we segmented mouse brains of the same strain, as well as new strains — such as the BXD29, and the AD model (Jankowsky et al., 2005).
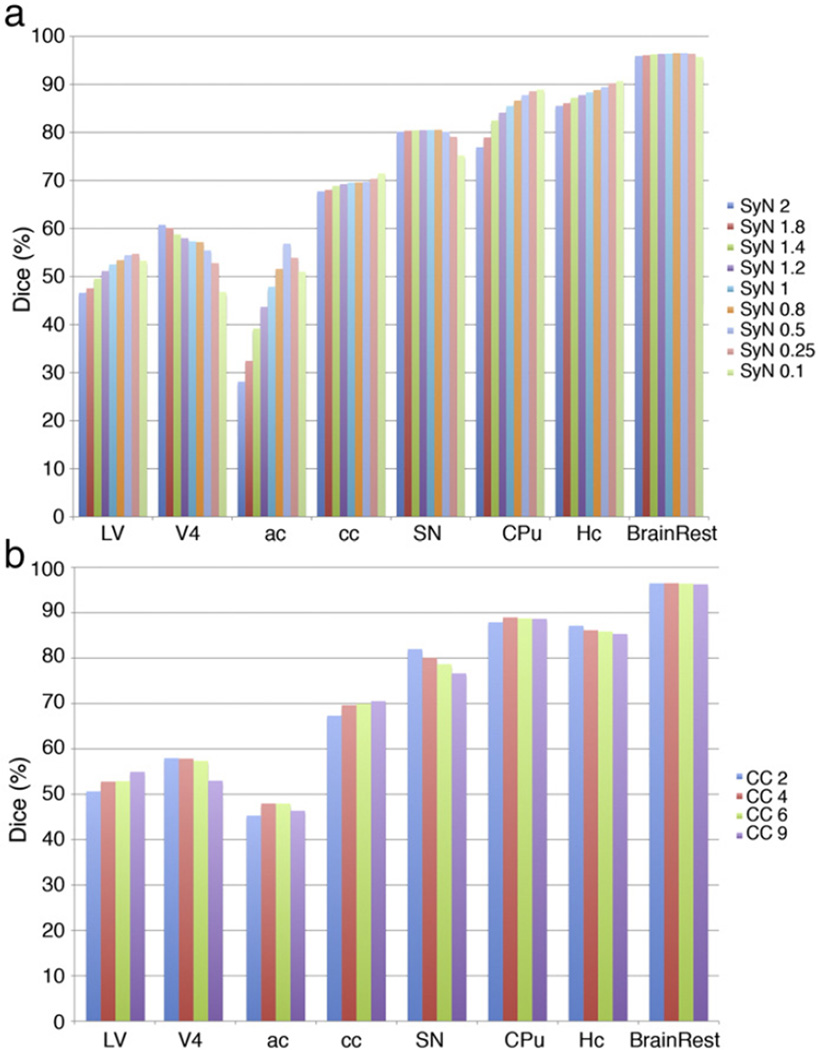
In an initial optimization study we used images from C57BL/6 mice (Fig. 4) to select parameters for the registration and classification processes, including: imaging protocol, metric, deformation, and regularization. At first, single channel, T2*w images were used to examine relative segmentation performance. The pipeline was initialized with the mutual information metric (MI) for affine registration, because of significant gains in computation speed relative to the cross correlation (CC) metric. The CC metric was chosen for deformable registration, for its efficiency, and the effect of kernel size on the accuracy of segmentation was evaluated. The CC kernel size was varied between 2 and 9 voxels (of 43 µm size), as shown in Fig. 4b. A 4 voxel kernel radius (i.e. 172 µm) was chosen for the C57BL/6 set, based on maximum performance for white matter tracts, hippocampus and 4th ventricles. The gradient step parameter for the symmetric normalization was varied between 0.1 and 2 voxels (Fig. 4a), and finally chosen as 0.5 voxels based on the good performance for the anterior commissure and corpus callosum, as well as lateral ventricles, and the reasonable computation cost (increasing as the step gets smaller). A grid search was run for the parameters of the Gaussian regularization factor, varying the total regularization kernel size from 0, 1 and 3 voxels, and the deformation regularization from0, 0.2, and 0.5 voxels. Optimal results were obtained for Gauss [1,0.5].
Fig. 4.
Optimization of deformable registration parameters for C57BL/6 brains for: (a) SyN parameter; (b) cross correlation kernel. Our data suggest using a kernel size of 4, and SyN of 0.5 voxels is optimal, but registration parameters may need to be balanced against computational cost, and may vary in other mouse strains/models.
ANTs was used with multivariate inputs to test for the optimal combination of images, restricting the search to two channels, in the face of memory limitation, and to maintain reasonable computation time (Fig. 5). The similarity metric was CC with a 4 voxels kernel (172 µm), and the transform was symmetric normalization with SyN step of 0.8 voxels (34.4 µm). The combination of conventional + DTI channels improved segmentation significantly. Specifically, the combination of either of the conventional channels and the radial anisotropy yielded best results. As expected, univariate ANOVA indicated that accuracy was different for different image channels, and higher for white matter, such as the anterior commissure [ac: F(12,39) = 8.0, p = 2.98 × 10−7] and corpus callosum [cc: F(12,39) = 6.6, p = 0.004)].
Fig. 5.
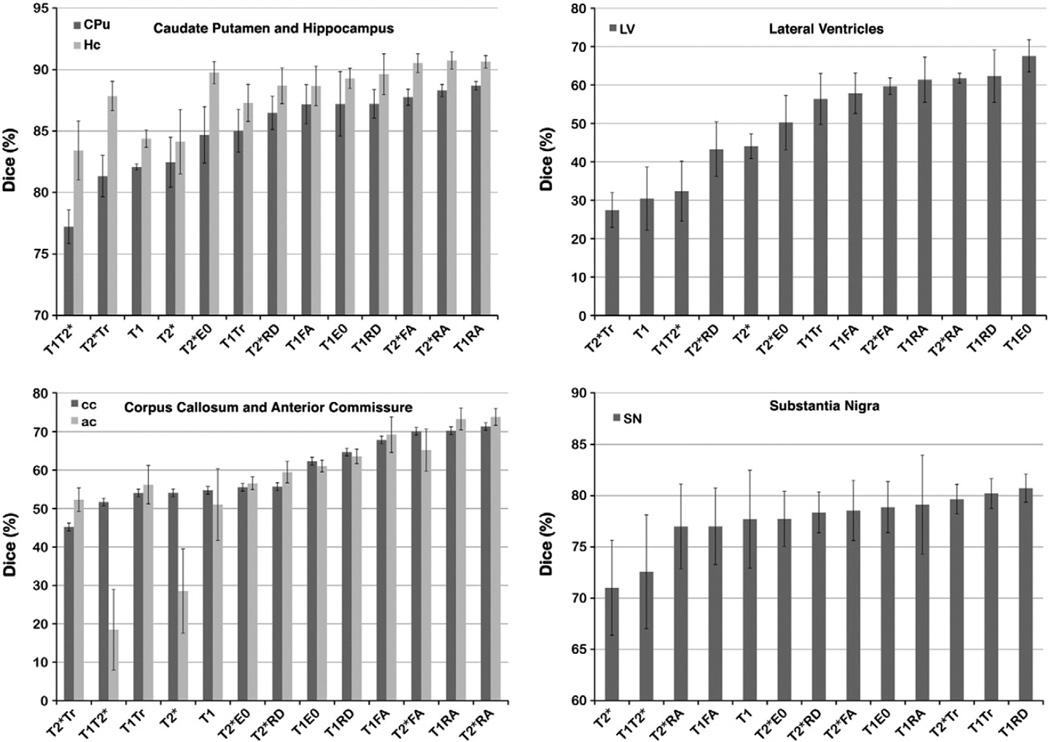
Dice coefficients indicate that combining information from T1 or T2*w images with information from DTI parameter images improves the accuracy of registration-based segmentation. The improvement between single channel (T1w or T2*w), and T1RA or T2*RA combined channel segmentation was >6% for gray matter structures like CPu and Hc. As expected, accuracy improved for white matter structures, >26% for ac and cc in T1RA, T2*RA relative to T1T2*, and 16% relative to T1 or T2*. Dice coefficients also increased for small nuclei such as SN (~10% higher in T1RD and T1Tr than in T2*, 3% higher in T1RD than in T1), as well as for ventricles (>16% higher in T2RD than in T2*, 30% higher in T2Tr relative to T1). While for SN the combination T1RD and T1Tr produced the most accurate segmentation (80 ± 1%), and T1E0 for LV (68 ± 4%), the combination of T2*RA produced the most accurate segmentation for most structures (88 ± 2% for CPu, and 90 ± 1% for Hc).
Details on the size of differences, and the statistics are presented in Tables 2A–2E. In summary, Dice coefficients for corpus callosum (cc) increased from 54 ± 4% (mean ± SEM) for T2*w based segmentation, to 71 ± 1% for T2*RA and 70 ± 1% for T2*FA (Table 2A). Dice values for combined conventional and DTI channels, like RA and FA, were greater relative to single channel based segmentation. A posthoc analysis (Table 2B) indicated that Dice values for combined conventional and DTI channels were larger relative to the T2*Tr, and differences were significant. The combination of T1 and T2* with RA provided better accuracy than the T1T2* combination; and T2*FA provided better accuracy compared to T1, T2* and T1T2* (>16%, ns after multiple comparison correction).
Table 2.
| A | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Summary of segmentation evaluation for select structures: brain rest, hippocampus (Hc), caudate putamen (CPu), substantia nigra (SN), anterior commissure (ac), corpus callosum (cc), lateral ventricles (LV). Data represent mean Dice coefficients±SEM. Minimum and maximum values are shown in bold characters. | |||||||||||||||
| Channel selection impacted the accuracy of segmentation. | |||||||||||||||
| C57BL/6 | T1 (%) | T2* (%) | T1T2* (%) | T1Tr (%) | T1FA (%) | T1RA (%) | T2*FA (%) | T2*RA (%) | T2*Tr (%) | T1RD (%) | T2*RD (%) | T1E0 (%) | T2*E0(%) | F(12,39) | p |
| Brain rest | 95.47±0.58 | 94.94±0.64 | 95.84±0.21 | 96.57±0.21 | 96.41±0.67 | 97.24±0.03 | 97.01±0.17 | 97.18±0.04 | 96.16±0.17 | 96.76±0.23 | 96.34±0.25 | 96.76±0.12 | 96.51±0.21 | 3.79 | 7.62E−04 |
| Hc | 84.37±0.71 | 84.14±2.62 | 83.43±2.40 | 87.29±1.51 | 88.67±1.62 | 90.64±0.50 | 90.53±0.77 | 90.75±0.71 | 87.85±1.19 | 89.62±1.67 | 88.69±1.46 | 89.29±0.83 | 89.75±0.91 | 3.13 | 0.0034 |
| CPu | 82.08±0.26 | 82.47±2.03 | 77.21±1.37 | 85.02±1.75 | 87.18±1.59 | 88.68±0.35 | 87.75±0.66 | 88.30±0.50 | 81.34±1.68 | 87.22±1.15 | 84.11±1.35 | 87.21±1.56 | 84.69±1.66 | 4.88 | 7.58E−05 |
| SN | 77.69±4.78 | 71.01±4.64 | 72.58±5.55 | 80.21±1.44 | 77.01±3.73 | 79.12±4.84 | 78.55±2.93 | 77.00±4.14 | 79.66±1.45 | 80.74±1.37 | 78.36±2.00 | 78.88±2.50 | 77.73±2.69 | 0.64 | 0.7945 |
| ac | 51.02±9.29 | 28.52±10.95 | 18.43±10.48 | 56.20±5.01 | 69.24±4.63 | 73.31±2.81 | 65.21±5.47 | 73.83±2.21 | 52.30±3.04 | 63.60±1.89 | 59.48±2.81 | 61.05±1.56 | 56.56±1.66 | 7.99 | 2.98E−07 |
| cc | 54.74±4.43 | 54.10±3.64 | 51.67±2.11 | 54.07±5.13 | 67.87±6.68 | 70.28±2.40 | 70.10±1.19 | 71.33±1.28 | 45.21±4.16 | 64.66±7.34 | 55.72±7.94 | 62.34±3.76 | 55.50±6.10 | 3.04 | 0.0042 |
| LV | 30.49±8.21 | 44.09±3.24 | 32.38±7.84 | 56.41±6.60 | 57.89±5.29 | 61.42±5.90 | 59.72±2.18 | 61.80±1.29 | 27.46±4.55 | 62.33±6.83 | 43.32±7.12 | 67.60±4.19 | 50.26±7.07 | 5.46 | 2.39E−05 |
| B | |||||
|---|---|---|---|---|---|
| Multichannel segmentation of C57BL/6mice incorporating DTI outperformed single channel segmentation using conventional contrast. | |||||
| Factor 1 | Factor 2 | Diff (%) | CI1 | CI2 | p |
| Hc | |||||
| T2*RA | T1T2* | 7.32 | 0.11 | 14.53 | 0.04* |
| T1RA | T1T2* | 7.21 | 0.00 | 14.42 | 0.05 |
| T2*FA | T1T2* | 7.10 | −0.11 | 14.31 | 0.06 |
| T2*RA | T2* | 6.61 | −0.60 | 13.82 | 0.10 |
| T2*RA | T1 | 6.37 | −0.84 | 13.58 | 0.13 |
| CPu | |||||
| T1RA | T1T2* | 11.48 | 3.82 | 19.13 | 0.0003* |
| T2*RA | T1T2* | 11.09 | 3.44 | 18.74 | 0.0006* |
| T2*FA | T1T2* | 10.55 | 2.89 | 18.20 | 0.0012* |
| T1RD | T1T2* | 10.01 | 2.36 | 17.66 | 0.003* |
| T1E0 | T1T2* | 10.00 | 2.35 | 17.65 | 0.003* |
| T1FA | T1T2* | 9.97 | 2.32 | 17.63 | 0.003* |
| T2*RD | T1T2* | 9.26 | 1.61 | 16.91 | 0.01* |
| T1Tr | T1T2* | 7.81 | 0.16 | 15.46 | 0.04* |
| T2*E0 | T1T2* | 7.48 | −0.17 | 15.13 | 0.06 |
| T1RA | T2*Tr | 7.35 | −0.30 | 15.00 | 0.07 |
| T2*RA | T2*Tr | 6.96 | −0.69 | 14.61 | 0.11 |
| LV | |||||
| T1E0 | T2*Tr | 40.14 | 11.22 | 69.06 | 0.001* |
| T1E0 | T1 | 37.11 | 8.19 | 66.03 | 0.003* |
| T1E0 | T1T2* | 35.22 | 6.30 | 64.14 | 0.006* |
| T1RD | T2*Tr | 34.87 | 5.95 | 63.79 | 0.007* |
| T2*RA | T2*Tr | 34.34 | 5.42 | 63.26 | 0.01* |
| T1RA | T2*Tr | 33.96 | 5.04 | 62.88 | 0.01* |
| T2*FA | T2*Tr | 32.26 | 3.34 | 61.18 | 0.02* |
| T1RD | T1 | 31.84 | 2.92 | 60.76 | 0.02* |
| T2*RA | T1 | 31.31 | 2.39 | 60.23 | 0.02* |
| T1RA | T1 | 30.93 | 2.01 | 59.85 | 0.03* |
| T1FA | T2*Tr | 30.43 | 1.51 | 59.35 | 0.03* |
| T1RD | T1T2* | 29.96 | 1.03 | 58.88 | 0.04* |
| T2*RA | T1T2* | 29.42 | 0.50 | 58.34 | 0.04* |
| T2*FA | T1 | 29.23 | 0.31 | 58.15 | 0.046* |
| T1RA | T1T2* | 29.04 | 0.12 | 57.96 | 0.048* |
| T1Tr | T2*Tr | 28.95 | 0.03 | 57.87 | 0.05 |
| T1FA | T1 | 27.40 | −1.52 | 56.32 | 0.08 |
| T2*FA | T1T2* | 27.34 | −1.58 | 56.26 | 0.08 |
| ac | |||||
| T2*RA | T1T2* | 55.40 | 26.70 | 84.10 | <.0001* |
| T1RA | T1T2* | 54.88 | 26.18 | 83.59 | <.0001* |
| T1FA | T1T2* | 50.81 | 22.11 | 79.51 | <.0001* |
| T2*FA | T1T2* | 46.78 | 18.08 | 75.49 | <.0001* |
| T2*RA | T2* | 45.31 | 16.61 | 74.01 | 0.0001* |
| T1RD | T1T2* | 45.17 | 16.47 | 73.87 | 0.0001* |
| T1RA | T2* | 44.79 | 16.09 | 73.49 | 0.0002* |
| T1E0 | T1T2* | 42.62 | 13.92 | 71.32 | 0.0004* |
| T2*RD | T1T2* | 41.05 | 12.35 | 69.75 | 0.001* |
| T1FA | T2* | 40.72 | 12.02 | 69.42 | 0.001* |
| T2*E0 | T1T2* | 38.13 | 9.43 | 66.83 | 0.002* |
| T1Tr | T1T2* | 37.77 | 9.07 | 66.47 | 0.002* |
| T2*FA | T2* | 36.69 | 7.99 | 65.40 | 0.004* |
| T1RD | T2* | 35.08 | 6.37 | 63.78 | 0.006* |
| T2*Tr | T1T2* | 33.87 | 5.17 | 62.58 | 0.009* |
| T1 | T1T2* | 32.59 | 3.88 | 61.29 | 0.01* |
| T1E0 | T2* | 32.53 | 3.82 | 61.23 | 0.01* |
| T2*RD | T2* | 30.96 | 2.25 | 59.66 | 0.02* |
| T2*E0 | T2* | 28.04 | −0.66 | 56.74 | 0.06 |
| T1Tr | T2* | 27.68 | −1.03 | 56.38 | 0.07 |
| cc | |||||
| T2*RA | T2*Tr | 26.13 | 2.08 | 50.17 | 0.02* |
| T1RA | T2*Tr | 25.08 | 1.03 | 49.12 | 0.03* |
| T2*FA | T2*Tr | 24.90 | 0.85 | 48.94 | 0.04* |
| T1FA | T2*Tr | 22.66 | −1.39 | 46.71 | 0.08 |
| Brain rest | |||||
| T1RA | T2* | 2.30 | 0.60 | 4.01 | 0.002* |
| T2*RA | T2* | 2.24 | 0.53 | 3.95 | 0.003* |
| T2*FA | T2* | 2.07 | 0.36 | 3.78 | 0.01* |
| T1E0 | T2* | 1.82 | 0.11 | 3.53 | 0.03* |
| T1RD | T2* | 1.82 | 0.11 | 3.53 | 0.03* |
| T1RA | T1 | 1.77 | 0.06 | 3.48 | 0.04* |
| T2*RA | T1 | 1.70 | −0.01 | 3.41 | 0.05 |
| T1Tr | T2* | 1.63 | −0.08 | 3.34 | 0.08 |
| T2*E0 | T2* | 1.57 | −0.14 | 3.28 | 0.10 |
| C | |||||
|---|---|---|---|---|---|
| Multichannel segmentation in test strains showed that T1 and T1T2w combinations have similar performance, and outperformed T2w in segmenting the cc (T1 vs T2w: CI [4.7 81.9]; T2w vs T1T2w: CI [−81.6 −4.3]) and VS (T1 vs T2w: CI [4.7 85.8]; T2w vs T1T2w: CI [−83.8 −2.7]). | |||||
| T1 (%) | T2(%) | T1T2w (%) | F | P | |
| Brain rest | 90.60±1.07 | 89.92±1.14 | 90.76±1.03 | 0.02 | 0.98 |
| Hc | 79.95±2.27 | 72.02±3.18 | 80.04±2.42 | 0.07 | 0.93 |
| CPu | 83.08±1.41 | 79.07±1.03 | 83.82±1.45 | 0.18 | 0.84 |
| SN | 48.63±5.11 | 53.69±3.88 | 54.70±5.00 | 0.04 | 0.96 |
| ac | 39.14±3.97 | 29.28±4.25 | 39.74±3.35 | 0.64 | 0.54 |
| cc | 53.67±3.32 | 39.14±2.88 | 53.57±3.36 | 5.29 | 0.01* |
| VS | 46.42±2.41 | 31.75±3.54 | 44.07±3.01 | 5.05 | 0.02* |
| D | |||||||
|---|---|---|---|---|---|---|---|
| For C57BL/6 miceMRF classification following deformable registration (Diff), using T2* and RA images outperformed Diff only (using T2* and RA images), and differences were significant in 5 out of 8 structures labeled. | |||||||
| C57BL/6 | Diff(%) | MRF(%) | Difference (%) | t | CI1 | CI2 | p |
| Brain rest | 95.97±0.70 | 95.13±0.63 | −0.84 | −0.89 | −2.94 | 1.26 | 0.39 |
| Hc | 91.39±0.39 | 93.61±0.28 | 2.22 | 4.65 | 1.16 | 3.29 | 0.001* |
| CPu | 88.82±0.60 | 92.02±0.54 | 3.2 | 3.99 | 1.41 | 4.99 | 0.003* |
| SN | 72.56±2.35 | 80.86±1.12 | 8.3 | 3.18 | 2.49 | 14.11 | 0.01* |
| ac | 74.90±3.18 | 77.59±5.17 | 2.69 | 0.44 | −10.83 | 16.21 | 0.67 |
| cc | 73.47±1.86 | 79.96±1.75 | 6.49 | 2.54 | 0.8 | 12.18 | 0.03* |
| LV | 62.65±3.29 | 73.92±3.13 | 11.26 | 2.48 | 1.14 | 21.38 | 0.03* |
| V4 | 56.92±5.55 | 63.30±5.55 | 6.39 | 0.81 | −11.11 | 23.88 | 0.44 |
| E | |||||||
|---|---|---|---|---|---|---|---|
| For test strains there was a trend for MRF classification following deformable registration (Diff) to outperform Diff, with the exception of the ac, which was not reliably segmented. | |||||||
| Test strains | Diff (%) | MRF (%) | Difference (%) | t | CI1 | CI2 | p |
| Brain rest | 90.76±0.42 | 90.12±0.30 | −0.64 | 1.24 | −1.57 | 2.85 | 0.34 |
| Hc | 80.04±1.88 | 87.37±1.91 | 7.32 | −2.74 | −18.84 | 4.19 | 0.11 |
| CPu | 83.82±1.15 | 86.23±4.61 | 2.41 | −0.51 | −22.87 | 18.05 | 0.66 |
| SN | 54.70±4.74 | 66.62±3.64 | 11.92 | −1.99 | −37.64 | 13.80 | 0.18 |
| ac | 39.74±1.98 | 16.83±1.69 | −22.90 | 8.79 | 11.69 | 34.11 | 0.01 |
| cc | 53.57±6.51 | 59.81±6.77 | 6.24 | −0.66 | −46.64 | 34.16 | 0.57 |
| VS | 44.07±7.15 | 53.08±16.12 | 9.01 | −0.51 | −84.87 | 66.85 | 0.66 |
indicates significance at a p<0.05 level
A similar trend was observed for the anterior commissure (ac), where Dice coefficients increased from 29 ± 11% for T2*w, and 51 ± 10% for T1w, to 74 ± 2% for T2*RA, and 73 ± 3% for T1RA. A posthoc analysis indicated that Dice coefficients for T1FA, T1RA, T2*RA, T1RD, T2*RD and T1E0 were larger, and differences were statistically significant (p<0.05), relative to T2*w, or T1T2*w.
Unexpectedly, not only white matter tract segmentation accuracy benefited from the use of DTI channels in a multivariate context, but also gray matter nuclei. SN accuracy increased from 71 ± 5% for T2*w to 78 ± 5% for T1w, and 80 ± 1% for T1Tr, and 81 ± 1% for T1RD, but differences were not significant (ns). However, for Hc accuracy increased from 84 ± 1% and 84 ± 3% for T1w and T2*w respectively, to 91 ± 1% for T1RA and T2*RA. Differences were statistically significant (p = 0.003); and a posthoc analysis revealed that T2*RA accuracy was larger relative to T1T2*, and approaching significance relative to T2*w (p = 0.1). Similarly, for CPu accuracy increased from 82 ± 0.3% for T1w, and 83 ± 2% for T2*w, to 89 ± 0.4% for T1RA and 88 ± 0.5% for T2*RD. Differences among groups were statistically significant (p = 7.6 × 10−5), and post hoc analysis revealed significant differences between the following groups: T1T2* vs T1RA, T2*RA, T1RD, T2*RD, T1FA and T2*FA; T1RA, T2*RA, T1E0, T1Tr; while T2*Tr vs T1RA and T2*RA approached significance (p = 0.1). Of the DTI channels Tr combined with T2* was the least favorable. However, channel combinations involving T1w, or T2*w and a DTI channel were in general superior to T1T2* driven segmentation.
Even though the ventricles were challenging to segment, because of their variability, and susceptibility effects at high-field MR, the maximum accuracy was obtained for T1E0 (68 ± 4%), and T2*RA combination (62 ± 1%), and differences between groups were significant (p = 2.4 × 10−5). The post hoc analysis revealed that differences were statistically significant between the following groups: T1 vs T1E0, T1RA, T2*RA, T2*FA, T1RD; T1T2* vs T1E0, T1RD, T2*RA, T1RA, T2*FA. T2*Tr gave inferior results compared to T1E0, T1RD, T2*RA, T1FA, T1RA, T2*RA, T2*FA (p<0.05).
The rest of the brain was segmented accurately (>95%) with any of the image channel combinations. Groups gave however different accuracies (p = 7.6 × 10−4). In essence single channel T2*w based segmentation underperformed relative to combinations involving T1 and RA, RD, or E0; T2*RA; or T2*FA. T1RA was better than T1 based segmentation (p<0.05).
The accuracy for segmenting other strains (Fig. 6, and Table 2C) was lower compared to C57BL/6,when using the same settings for ANTs, but only T1w and T2w channels (no DTI). Correcting for bias field nonuniformity (data not shown) improved results with 2.8% on average, most importantly for cc (4%), and ac (9%). With the exception of cc and ventricular system (VS), differences were not significant. T1w and the combined T1T2w (70% contribution from T1w, and 30% from T2w) gave superior results to T2w based segmentation, with the exception of SN, where T2w yielded better accuracy. The larger nuclei were segmented with accuracy of 85 ± 2% for CPu, and 78 ± 5% for Hc in the BXD29 mouse, and 83 ± 0.6% and 82 ± 0.5% respectively, in the AD mouse. The penalties were greater for smaller nuclei like SN (50 ± 8% in BXD29, 59 ± 6% in AD), white matter tracts like cc (47 ± 5% in BXD29, 60 ± 1% in AD), and ac (38 ± 4 in BXD29, 42 ± 6% in AD), and ventricles (37 ± 2% in BXD29, and 51 ± 3% in AD).
Fig. 6.
(a) Qualitative evaluation of automated segmentation in test strains (BXD29M shown) in T1w (21.5 µm voxels) and T2w images (43 µm voxels); (b) Quantitative evaluation of segmentation accuracy, based on 8 labels, for both test strains using T1w, T2w, and a combination of the two channels indicates better performance for T1w and combined T1wT2w channel segmentation in general, with the exception of SN, where T2w provides higher accuracy than T1w (ns).
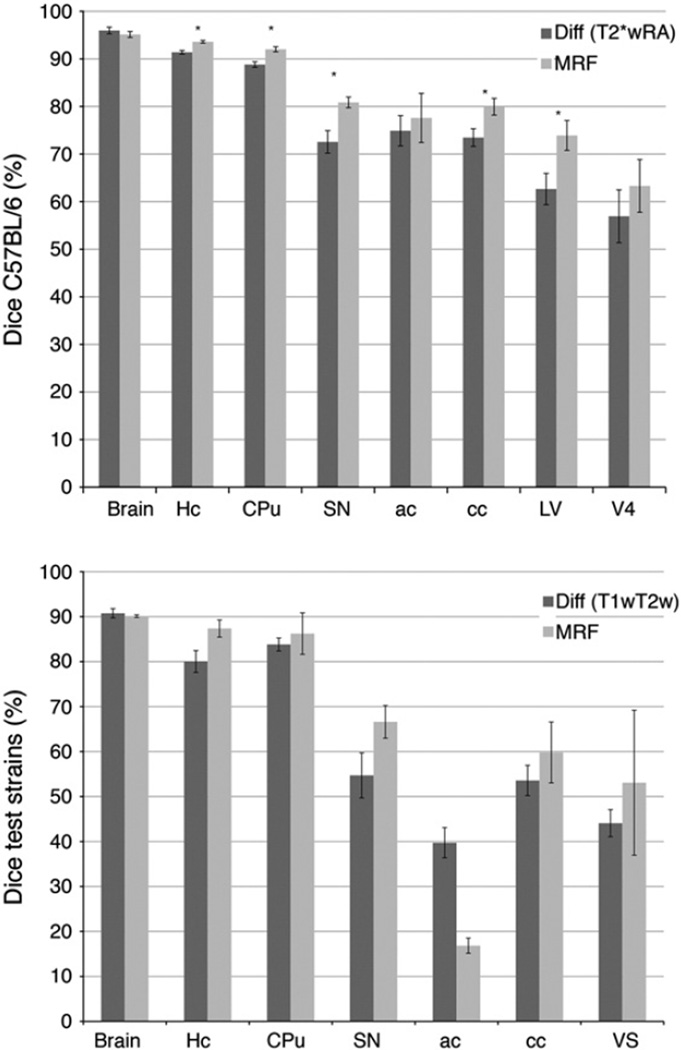
The addition of MRF modeling and Bayesian classification based on priors constructed from multiple atlases (Fig. 7), and the T2*w and RA image channels, improved the results of segmentation relative to the single atlas reference, using optimal registration parameters (SyN 0.5 voxels, cross correlation kernel 4 voxels, regularization [1,0.5]) (Fig. 8). The improvement was significant in C57BL/6 mice for 5 out of the 8 segmented structures, i.e. the Hc, CPu, SN, cc and LV. Among gray matter structures Hc accuracy increased from 91 ± 1% to 94 ± 1% for MRF (p = 0.001), CPu increased from 89 ± 2% to 92 ± 1% (p = 0.003); SN from 73 ± 2% to 81 ± 1% (p = 0.01). Both white matter structures were segmented more accurately; ac increased from 75 ± 3% to 78 ± 5% (ns), but only cc increased significantly, from 74 ± 2% to 80 ± 2% (p = 0.03). The lateral ventricle increased from63 ± 3% to 74 ± 3% (p = 0.03). In the BXD29 and AD strains, MRF improved (ns) segmentation in all tested structures but the whole brain and the anterior commissure. The CPu and cc were segmented with higher accuracy (by 2, and 6% respectively), but the improvements were largest for Hc (by 7%), ventricles (by 9%), and substantia nigra (by 11%) (Fig. 8).
Fig. 7.
Multiple specimen atlases were used to generate probabilistic priors. The location, intensity, and label priors can be used in a Bayesian segmentation framework to improve upon the accuracy of single atlas based segmentation. Five specimens were used to generate priors using ANTs; subsequently used for MRF segmentation using ATROPOS. The accuracy of multi-atlas segmentation was evaluated for 6 labeled regions including the anterior commissure (yellow), corpus callosum (orange), hippocampus (magenta), striatum (transparent green), substantia nigra (cyan), and ventricles (LV: blue; V4: solid green).
Fig. 8.
MRF segmentation of C57BL/6 mice using ATROPOS provided more accurate results than registration based segmentation, differences that are statistically significant are indicated by asterisk (*). T2*w and RA parameter images provided multivariate priors for MRF segmentation. The accuracy was reduced in test strains other than the C57BL/6, which provided the priors, and in the absence of DTI parameter images. MRF improved however overall accuracy relative to registration based segmentation.
We used the mouse brain segmentation pipeline (MOBS) on an Intel Mac Pro with 2 Quad Core Processor, and 64 GB RAM. Segmentation time was approximately 26 h for a dual channel (512 × 512 × 256 × 2) structural and DTI parameter images, filtered with the bilateral filter, using full diffeomorphism with time, and a four levels multiresolution scheme; and 36 h for unfiltered T1w and T2w combinations with four levels resolution scheme. A 1024 × 512 × 512 single channel image (at 21.5 µm resolution), using greedy SyN, and a 4 levels resolution scheme needed ~20 h to complete. A single contrast T2*w data set at 512 × 256 × 256 (at 43 µm resolution) required ~16 h.
Discussion
Automated or semi-automated pipelines are essential for phenotyping mouse models of neurological and neuropsychiatric human diseases, and assessing efficacy of treatments. We describe automated pipelines for single and multivariate segmentation of the mouse brain in a widely used strain, C57BL/6, as well as two other strains, BXD29 and APP/TTA double transgenic mouse of Alzheimer's disease (AD); and evaluate the contributions of different imaging and registration protocols, as well as the use of multiple atlases to the success of segmentation.
The novelty of our approach consists in using multiple MR protocols. We performed multivariate registration and segmentation using recently developed tools, available to the community (Avants et al., 2008b, 2011a,b). While additional evaluation may be necessary to confirm the suitability of the proposed pipelines for quantitative phenotyping, we present a framework in which such studies can be performed efficiently, in a streamlined fashion, and we assess performance of such pipelines executed on a multitude of imaging protocols. Our initial studies have been performed on an exhaustive data set so that we can determine the best performance one might derive in automated segmentation. These results are now informing us on image acquisition and optimal registration parameters. For example, improvements in RF coils and a streamlined acquisition protocol have reduced the acquisition time for a T2*w sequence to ~14 min. A new gradient insert and Agilent console on one of our 7 T magnets will allow to simultaneously acquire DTI images on 8 specimens using multiple RF coils/channels. The pipeline and data here will provide the baseline against which we can gauge these new strategies.
We propose using existing atlases as references for automated segmentation, such as (Badea et al., 2007a; Jiang and Johnson, 2011;Johnson et al., 2010; Kovacevic et al., 2005; Ma et al., 2005) for fixed specimens, and (Ma et al., 2008) for in vivo scans. These atlases may be adapted to new studies (Lee et al., 2009), or new atlases may be required if image acquisition protocols differ dramatically, or if there are large differences in the brain anatomy. We have used commercial software programs, such as AMIRA (Visage Imaging Inc., San Diego, CA) or Avizo (VSG, Burlington, MA), for manual labeling. These incorporate segmentation tools and have the capability to work with large data sets; however, they require a license. Excellent free and open source tools, such as ITK-SNAP (Yushkevich et al., 2006), are promising alternative for producing future atlases. It is recommended that reference atlases be rigidly aligned into the Waxholm space, using intensity based methods or anatomical markers such as the anterior commissure, posterior commissure, or tip of 4th ventricles.
While our protocols can be used with lower resolution MR images from actively stained brain specimens, they are not optimized for lower resolution in vivo MR images. Our study was aimed toward higher resolution, contrast-enhanced MRM images of the mouse brain, where ~40 structures can be identified, but some researchers prefer the benefits of in vivo images, which avoid shrinkage associated with fixation, and offer the possibility of doing longitudinal studies. While the segmentation algorithms are amenable to process images obtained from such protocols, the lower resolution and contrast require new atlases and anatomical priors, for a reduced set of structures. Directly using the tools with the provided atlases may result in reduced segmentation accuracy. Existing in vivo MR atlases can potentially replace WHS as a reference in the pipeline, producing labels for 12 (Scheenstra et al., 2009), or 20 structures (Ma et al., 2008).
There are other neuroimaging workflows and pipelines that could be adapted for mouse brain segmentation. The Soma Workflow (Laguitton et al., 2011) is a Python application dedicated to the execution of low level workflows on parallel resources. It has been used with pipelines such as BrainVisa (Cointepas et al., 2010) to optimize segmentation of white matter, gray matter and CSF on a database of 80 human brain images, and dramatically reduced computation costs on multiple core clusters (1 h) relative to single core (23 h). In comparison, our pipeline runs single jobs; however, several instances can be run in parallel.
Another open source effort, Nipype (Gorgolewski et al., 2011) provides an environment that allows exploration of algorithms from different packages (e.g., SPM, FSL, FreeSurfer, Camino, AFNI, Slicer), and is mostly dedicated to functional brain image analysis, but could potentially be used for mouse brains. Similarly, the LONI pipeline (Dinov et al., 2009; Rex et al., 2003) bundles resources that can be pipelined using a graphical user interface, and has been used for mouse brain phenotyping. Registration (e.g. AIR, Woods et al., 1998) and segmentation tools - (Tu et al., 2008) are incorporated, but these need to be retrained when translating to the mouse. These tools have been used for analyzing ex vivo (MacKenzie-Graham et al., 2006), and in vivo images (for 3–4 regions) of EAE mouse models (Mackenzie-Graham et al., 2012). A close alternative, PSOM has been designed to process large numbers of brain data sets, ultimately for fMRI analysis, using Octave and MATLAB (Bellec et al., 2012), using parallel and distributed computing. Such pipelines will process workflows for structural analysis, as the one we present, but need to be configured appropriately. Furthermore, unlike the ANTs segmentation tool, these pipelines do not explicitly support multivariate segmentation.
Our mouse brain segmentation pipeline (MOBS) integrates software from different packages for image preprocessing, registration, segmentation and quantization, and can run on multiple platforms (e.g. OSX, LINUX). Similarly with Nipype, it lacks a graphical user interface present in other pipelines and segmentation programs (Beg et al., 2005; Lee et al., 2009; Rex et al., 2003), but offers in return ease of scripting (lacking in GUI based pipelines, such as the LONI pipeline) — to automate processing of multiple studies, and runs locally rather than on a remote cluster, limited by bandwidth of the connection, or resource availability. The segmentation pipeline is implemented using a Perl wrapper for in house built scripts, and a suite of published registration methods (Avants et al., 2008a,b;Avants et al., 2011a). These were tuned for high-resolution multivariate MR images of the mouse brain. The pipeline can be executed as a sequence of steps, or restarted for just individual steps allowing for optimization at each step. Similar parameters as in human neuroimaging can be used for mouse brain studies (Avants et al., 2011a), while edge preserving filtering techniques, such as the bilateral filter (Tomasi and Manduchi, 1998), may be used to reduce noise and improve convergence speed, particularly important for DTI. In our experience the cross-correlation (CC) metric provided faster convergence, and more accurate results compared to mutual information (MI) (see Supplementary material at http://www.civm.duhs.duke.edu/brainpipe2011). However CC can be used for similar images, such as those in the standard operating protocols defined for the WHS, while different image acquisition protocols may benefit from using the mutual information metric. The diffeomorphic registration with a SyN parameter of 0.5 provided optimum results for C57BL/6 and the test strains under consideration. A radius of 4 provided optimum results overall for a subset of 8 structures manually segmented in DTI, in combination with structural imaging for C57BL/6, and similarly in test strains, although a larger radius may provide optimal results for select structures, such as SN in the BXD29 and AD strains.
While it is difficult to compare accuracy across studies, since metrics differ (landmarks distance, volume correlation, surface distance, Jaccard or Dice overlap), and usually only select structures are manually segmented, the hippocampus is present in most evaluation studies because of its importance for normal brain function, as well as diseases involving learning and memory deficits. The accuracy of registration-based segmentation for hippocampus in test strains, using conventional imaging (BXD and AD mouse model), compared well (80% before, and 84% after N4 corrections) with other studies on BXD (~78%; Sharief et al., 2008), and in AD mouse models (~75%; Maheswaran et al., 2009a,b). Values were higher when multiple atlases were used to provide statistical priors (87%), and increased for the same strain registration using DTI (91% for C57BL/6), and 94% after MRF. In this case Dice coefficients for Hc exceeded 90% for registration and MRF segmentation, comparable to other studies (Ma et al., 2005). These values were comparable to those reported for human brains (~80%; Fischl et al., 2002; 83% Avants et al., 2011a). Another structure segmented consistently well, with similar accuracy is the caudate putamen. Accuracy ranged from 84% after registration, to 86% after MRF, in test strains, based on conventional imaging. Accuracy increased to 89% in C57BL/6 when using DTI, and 92% after MRF (Table 2D). In humans accuracy has been reported to be 80% after registration (Avants et al., 2011a), and 90% after MRF (Avants et al., 2011b; Fischl et al., 2002).
Among the segmented mouse brain regions, particularly challenging were the anterior commissure (Ali et al., 2005; Ma et al., 2005) and ventricles, because of their complex thin shapes, and in the latter case, high variability and possible perfusion caused changes. In comparison, in vivo human studies reported higher accuracy, from 66% (Avants et al., 2011b), up to 90% (Fischl et al., 2002).
While the use of bilateral filtration increased convergence speed due to noise reduction in DTI parametric images, the segmentation process may still require ~30 h on an Intel Mac Pro with 2 Quad Core Processor, using 64 GB RAM. Multiple instances can however be run in parallel (6 processes using a 16 hyper-threaded cores, with 64 GB memory system), to expedite the results for a particular study. Parallel processing can speed up population studies, or may offer the advantage of using multiple atlases to segment a single target brain. In comparison, manual segmentation for the 43 µm resolution data requires on average ~4 h/structure, at least 3 workdays for a set of 8 structures, and substantially more for a larger set of structures (i.e. delineating 37 structures in the WHS atlas required ~2 months). Automated segmentation time, in turn, is not affected by the number of labels associated with the images, but increases with image size.
While high resolution may appear as the ultimate pursuit in magnetic resonance histology, it is not the sole factor determining the success of brain segmentation, be it manual or automated. Contrast is also of primary importance. We have shown qualitatively (Johnson, Badea et al., 2010) that specific brain structures are better emphasized in different acquisitions, i.e. thalamic or vestibular nuclei in T2w acquisitions, and cortical and hippocampal subfields in T1w or T2*w acquisitions. We hypothesized that delineation of white matter tracts can be disambiguated based on DTI protocols, benefiting from directional information in color-coded FA images. This is particularly important in areas where multiple white matter tracts are in close proximity, but DTI acquisitions are substantially longer. It is therefore a continuous challenge to select protocols that offer the best trade-off between imaging time, resolution and contrast to resolve structures of interest.
Multispectral image channel combinations were evaluated with respect to producing optimal segmentations. Because of limited computation power and memory, we used two image channels for both registration and classification. For the test strains these were T1w and T2w (MEFIC processed) images, and the higher resolution T1w channel provided overall better accuracy than the lower resolution T2w image channel. This was particularly true for white matter structures, and larger nuclei, at least partly white matter bound; however T2w was more suitable for segmenting nuclei such as SN. Because of similar contrast of thalamic nuclei (notice e.g. the medial geniculate in Fig. 6), we believe the T2w contribution to be essential in these cases as well. When used together, the two channels provided similar or more accurate results than either of the two. With the exception of the ac, where priors registration was particularly challenging, accuracy was further increased after the MRF-based classification in the test strains (Table 2E).
For the C57BL/6 set, the T1w and T2*w structural images were complemented by DTI parameter images such as fractional (FA) and radial anisotropy (RA), longitudinal (E0) and radial diffusivity (RD), and trace (Tr). Overall the optimal combination was given by T2* and RA channels, followed by T1 and RA, for most of the manually segmented regions, with the exception of the lateral ventricles and substantia nigra, where the combination of T1E0 for ventricles, and T1RD and T1Tr for substantia nigra, provided the highest accuracy. This suggests that image channels may contribute differently to segmentation accuracy of particular brain regions.
Limitations of the segmentation approach using C57BL/6 priors were encountered when labeling other strains, resulting in lower Dice coefficients for registration-based segmentation. However, registration followed with Bayesian classification was found to improve overall segmentation accuracy, relative to registration alone. Accuracy increased by more than 10% in lateral ventricles, and 8% in the substantia nigra. The improvement was less significant in different strains, probably due to reduced accuracy of the priors. The nonparametric non-uniformity correction filter (Sled et al., 1998; Tustison et al., 2010) also increased Dice coefficients, although differences were not significant, by as much as 9% in the anterior commissure. It is reasonable to expect that further optimizing the filter can lead to better results in rodent imaging, as demonstrated in human studies (Zheng et al., 2009).
The use of multiple atlases and MRF segmentation improved results mostly in homogeneous structures like the substantia nigra and ventricles, but also in other structures like the hippocampus. Further optimization may be required for mouse models presenting anatomical phenotypes that deviate from those of the C57BL/6, and other methods may help improve accuracy. For example one may resort to masking regions and/or locally varying the weight of the priors. Individual image channels may give better results for distinct structures; so drawing heavily weighted priors from one channel versus another may improve results. In more extreme cases, largely different from the C57BL/6 characteristic anatomy, population specific atlases may be required to provide optimal results.
While using multiple atlases increases the accuracy of segmentation, a lot of effort goes into generating multiple priors. The time cost can average more than 1 month per brain, and at least 5–10 atlases are usually needed. If large mouse brain data banks become available, one can use strategies developed for human brain segmentation to select and rank atlases based on meta-information (e.g. age and strain) and image similarity. These strategies have been shown to perform better relative to non-selectively using the available atlases (Aljabar et al., 2009). But, only a small number of manually labeled atlases are available for the mouse brain. However, recent studies suggest that it is possible to use automatically generated templates, based on the query sets themselves, to effectively reduce the number of needed atlases (Mallar Chakravarty et al., 2012). This comes at the expense of computational cost due to increasing the number of registration/voting instances. On the other hand, efficient acquisition protocols, and coil arrays dedicated to concurrent imaging of multiple mice (Bock et al., 2003; Dazai et al., 2011) are being developed to speed up the effective scan time. As suggested by Bai et al. (2012) the results of using complex segmentation models such as MRF and multi-atlas priors are still influenced by registration accuracy. If different MR acquisitions better emphasize different structures, and improve registration, these should provide value also in the context of probabilistic atlases, and help understand the biological bases of MR image contrast.
Our work confirmed that, as suggested by other studies in human (Avants et al., 2008a) and mouse (Aggarwal et al., 2009a, 2012) brains, the conventional and DTI derived information are complementary and may help improve segmentation results. Moreover, DTI parameter changes may signal early effects in diseases such as AD (Gold et al., 2010), before significant GM loss can be detected, or account for age related differences independently of GM changes (Moy et al., 2011). For these reasons DTI imaging justifies investing the time for preclinical and translational imaging, to detect early changes indicative of disease, and subtle changes with treatment.
Neuroanatomical phenotypes have been identified in mouse models of Alzheimer's disease (Lau et al., 2008), Huntington disease (Carroll et al., 2011), or multiple sclerosis (Mackenzie-Graham et al., 2012) based on high-resolution ex-vivo and in-vivo MR imaging using similar software tools. These studies mostly address imaging of the adult mouse brain (Lee et al., 2009), but may as well be extended for segmenting embryos (Zamyadi et al., 2010). Dynamic changes may be relevant in studying plastic changes associated with learning (Lerch et al., 2011), or neurodevelopment (Aggarwal et al., 2009b). More recently multimodal studies (Aggarwal et al., 2009b) have gained popularity, providing means for comprehensive phenotyping. The accurate implementation of segmentation pipelines for such multivariate images will facilitate the characterization of temporal changes in mouse models of human neurological and psychiatric conditions, may reduce the burden of functional image analysis by providing anatomical constraints, and will help to objectively and quantitatively assess the effects of putative therapies in preclinical stages.
Conclusion
We present streamlined protocols for quantitative analysis of mouse brain hyperimages using multivariate registration and classification. The framework for automated segmentation encompasses a high-resolution, MR-based atlas space, and uses a suite of previously published software tools (including ITK (Ibanez et al., 2003), ANTs (Avants et al., 2011a), and ATROPOS (Avants et al., 2008b)) to spatially normalize new datasets into this reference space, assign voxels to predefined classes, and derive quantitative neuroanatomical phenotypes. We show that a suite of segmentation tools designed for human neuroimaging studies can be adapted for the segmentation of mouse brain images acquired with multiple MR imaging protocols. We use a simple command line invocation of the software pipeline, and a modular, flexible approach that allows restarting and quality control at every step. We identify optimal combinations of image channels and registration parameters.
One of the major benefits of magnetic resonance histology is the ability to provide routine measures of multiple subregions in the tissue specimen (in this case the brain). As acquisition protocols have become more efficient, post processing will become the rate-limiting step. Processing pipelines will be essential. The generalized approach and results demonstrate the utility of multispectral protocols, define the accuracy of the several multispectral combinations, and point to new directions for improving the method.
Acknowledgments
We thank Dr. Yi Jiang for supplying the DTI mouse brain images, Yi Qi for help with specimen preparation, Gary Cofer for his MR expertise, Darin Clark for the initial implementation of the bilateral filter, and Sally Zimney for editorial assistance. The study was supported by grants to GAJ for NIH/NCRR/NIBIB National Biomedical Technology Research Center (P41 EB015897) and Small Animal Imaging Resource Program (U24 CA092656).
Contributor Information
Alexandra Badea, Email: alexandra.badea@duke.edu.
Sally Gewalt, Email: sally.gewalt@duke.edu.
Brian B. Avants, Email: avants@grasp.cis.upenn.edu.
James J. Cook, Email: james.j.cook@dm.duke.edu.
G. Allan Johnson, Email: gjohnson@duke.edu.
References
- Aggarwal M, Ye X, et al. Quantitative phenotype characterization of developing mouse brains by diffusion tensor imaging: application for the Frizzled-4−/−mutant mice. Proceedings 17th Scientific Meeting, International Society for Magnetic Resonance in Medicine; Honolulu, HI. 2009a. p. 2009. [Google Scholar]
- Aggarwal M, Zhang J, et al. Magnetic resonance imaging and micro-computed tomography combined atlas of developing and adult mouse brains for stereotaxic surgery. Neuroscience. 2009b;162(4):1339–1350. doi: 10.1016/j.neuroscience.2009.05.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal M, Jones MV, et al. Probing mouse brain microstructure using oscillating gradient diffusion MRI. Magn. Reson. Med. 2012;67(1):98–109. doi: 10.1002/mrm.22981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali AA, Dale AM, et al. Automated segmentation of neuroanatomical structures in multispectral MR microscopy of the mouse brain. NeuroImage. 2005;27(2):425–435. doi: 10.1016/j.neuroimage.2005.04.017. [DOI] [PubMed] [Google Scholar]
- Aljabar P, Heckemann RA, et al. Multi-atlas based segmentation of brain images: atlas selection and its effect on accuracy. NeuroImage. 2009;46(3):726–738. doi: 10.1016/j.neuroimage.2009.02.018. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Voxel-based morphometry—the methods. NeuroImage. 2000;11(6 Pt 1):805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- Avants B, Duda JT, et al. Multivariate analysis of structural and diffusion imaging in traumatic brain injury. Acad. Radiol. 2008a;15(11):1360–1375. doi: 10.1016/j.acra.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Epstein CL, et al. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal. 2008b;12(1):26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Tustison NJ, et al. A reproducible evaluation of ANTs similarity metric performance in brain image registration. NeuroImage. 2011a;54(3):2033–2044. doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Tustison NJ, et al. An open source multivariate framework for n-tissue segmentation with evaluation on public data. Neuroinformatics. 2011b;9(4):381–400. doi: 10.1007/s12021-011-9109-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea A, Ali-Sharief AA, et al. Morphometric analysis of the C57BL/6J mouse brain. NeuroImage. 2007a;37(3):683–693. doi: 10.1016/j.neuroimage.2007.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea A, Nicholls PJ, et al. Neuroanatomical phenotypes in the reeler mouse. NeuroImage. 2007b;34(4):1363–1374. doi: 10.1016/j.neuroimage.2006.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea A, Johnson GA, et al. Genetic dissection of the mouse brain using high-field magnetic resonance microscopy. NeuroImage. 2009;45(4):1067–1079. doi: 10.1016/j.neuroimage.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea A, Johnson GA, et al. Remote sites of structural atrophy predict later amyloid formation in a mouse model of Alzheimer's disease. NeuroImage. 2010;50(2):416–427. doi: 10.1016/j.neuroimage.2009.12.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J, Trinh TL, et al. Atlas-based automatic mouse brain image segmentation revisited: model complexity vs. image registration. Magn. Reson. Imaging. 2012;30(6):789–798. doi: 10.1016/j.mri.2012.02.010. [DOI] [PubMed] [Google Scholar]
- Beg MF, Miller MI, et al. Computing Large Deformation Metric Mappings via Geodesic Flows of Diffeomorphisms. Int. J. Comput. Vision. 2005;61(2):139–157. [Google Scholar]
- Bellec P, Lavoie-Courchesne S, et al. The pipeline system for Octave and Matlab (PSOM): a lightweight scripting framework and execution engine for scientific workflows. Front Neuroinformatics. 2012;6:7. doi: 10.3389/fninf.2012.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blockx I, Van Camp N, et al. Genotype specific age related changes in a transgenic rat model of Huntington's disease. NeuroImage. 2011;58(4):1006–1016. doi: 10.1016/j.neuroimage.2011.07.007. [DOI] [PubMed] [Google Scholar]
- Bock NA, Konyer NB, et al. Multiple-mouse MRI. Magn. Reson. Med. 2003;49(1):158–167. doi: 10.1002/mrm.10326. [DOI] [PubMed] [Google Scholar]
- Bock NA, Kovacevic N, et al. In vivo magnetic resonance imaging and semiautomated image analysis extend the brain phenotype for cdf/cdf mice. J. Neurosci. 2006;26(17):4455–4459. doi: 10.1523/JNEUROSCI.5438-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohland JW, Wu C, et al. A proposal for a coordinated effort for the determination of brainwide neuroanatomical connectivity in model organisms at a mesoscopic scale. PLoS Comput. Biol. 2009;5(3):e1000334. doi: 10.1371/journal.pcbi.1000334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg J, Chereul E. Differential MRI patterns of brain atrophy in double or single transgenic mice for APP and/or SOD. J. Neurosci. Res. 2008;86(15):3275–3284. doi: 10.1002/jnr.21778. [DOI] [PubMed] [Google Scholar]
- Carroll JB, Lerch JP, et al. Natural history of disease in the YAC128 mouse reveals a discrete signature of pathology in Huntington disease. Neurobiol. Dis. 2011;43(1):257–265. doi: 10.1016/j.nbd.2011.03.018. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Peng Q, et al. Structural MRI detects progressive regional brain atrophy and neuroprotective effects in N171-82Q Huntington's disease mouse model. NeuroImage. 2011;56(3):1027–1034. doi: 10.1016/j.neuroimage.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung MK, Worsley KJ, et al. General multivariate linear modeling of surface shapes using SurfStat. NeuroImage. 2010;53(2):491–505. doi: 10.1016/j.neuroimage.2010.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cointepas YT, Geffroy D, et al. The BrainVISA project: a shared software development infrastructure for biomedical imaging research. Proceedings 16th HBM.2010. [Google Scholar]
- Dale AM, Fischl B, et al. Cortical surface-based analysis: I. segmentation and surface reconstruction. NeuroImage. 1999;9(2):179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- Dazai J, Spring S, et al. Multiple-mouse neuroanatomical magnetic resonance imaging. J. Vis. Exp. 2011:48. doi: 10.3791/2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delatour B, Guegan M, et al. In vivo MRI and histological evaluation of brain atrophy in APP/PS1 transgenic mice. Neurobiol. Aging. 2006;27(6):835–847. doi: 10.1016/j.neurobiolaging.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Dice LR. Measures of the amount of ecologic association between species. Ecology. 1945;26(3):297–302. [Google Scholar]
- Dinov I, Van Horn JD, et al. Efficient, distributed and interactive neuroimaging data analysis using the LONI pipeline. Front Neuroinformatics. 2009;3(22):1. doi: 10.3389/neuro.11.022.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292(5819):154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, et al. Cortical surface-based analysis: II: inflation, flattening, and a surface-based coordinate system. NeuroImage. 1999;9(2):195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- Fischl B, Salat DH, et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33(3):341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- French L, Pavlidis P. Relationships between gene expression and brain wiring in the adult rodent brain. PLoS Comput. Biol. 2011;7(1):e1001049. doi: 10.1371/journal.pcbi.1001049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJ, Holmes AP, et al. Statistical parametric maps in functional imaging: a general linear approach. Hum. Brain Mapp. 1994;2(4):189–210. [Google Scholar]
- Gama Sosa MA, De Gasperi R, et al. Modeling human neurodegenerative diseases in transgenic systems. Hum. Genet. 2012;131(4):535–563. doi: 10.1007/s00439-011-1119-1. [DOI] [PubMed] [Google Scholar]
- Gold BT, Powell DK, et al. Alterations in multiple measures of white matter integrity in normal women at high risk for Alzheimer's disease. NeuroImage. 2010;52(4):1487–1494. doi: 10.1016/j.neuroimage.2010.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgolewski K, Burns CD, et al. Nipype: a flexible, lightweight and extensible neuroimaging data processing framework in Python. Front Neuroimformatics. 2011;5:13. doi: 10.3389/fninf.2011.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held K, Rota Kops E, et al. Markov random field segmentation of brain MR images. IEEE Trans. Med. Imaging. 1997;16(6):878–886. doi: 10.1109/42.650883. [DOI] [PubMed] [Google Scholar]
- Ibanez L, Schroeder W, et al. The ITK Software Guide. Kitare, Inc.; 2003. [Google Scholar]
- Jankowsky JL, Slunt HH, et al. Persistent amyloidosis following suppression of Abeta production in a transgenic model of Alzheimer disease. PLoS Med. 2005;2(12):e355. doi: 10.1371/journal.pmed.0020355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Johnson GA. Microscopic diffusion tensor imaging of the mouse brain. NeuroImage. 2010;50(2):465–471. doi: 10.1016/j.neuroimage.2009.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Johnson GA. Microscopic diffusion tensor atlas of the mouse brain. NeuroImage. 2011;56(3):1235–1243. doi: 10.1016/j.neuroimage.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, van Zijl PC, et al. DTIStudio: resource program for diffusion tensor computation and fiber bundle tracking. Comput. Methods Programs Biomed. 2006;81(2):106–116. doi: 10.1016/j.cmpb.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Johnson GA, Cofer GP, et al. Morphologic phenotyping with MR microscopy: the visible mouse. Radiology. 2002;222(3):789–793. doi: 10.1148/radiol.2223010531. [DOI] [PubMed] [Google Scholar]
- Johnson GA, Ali-Sharief A, et al. High-throughput morphologic phenotyping of the mouse brain with magnetic resonance histology. NeuroImage. 2007;37(1):82–89. doi: 10.1016/j.neuroimage.2007.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GA, Badea A, et al. Waxholm space: an image-based reference for coordinating mouse brain research. NeuroImage. 2010;53(2):365–372. doi: 10.1016/j.neuroimage.2010.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacevic N, Henderson JT, et al. A three-dimensional MRI atlas of the mouse brain with estimates of the average and variability. Cereb. Cortex. 2005;15(5):639–645. doi: 10.1093/cercor/bhh165. [DOI] [PubMed] [Google Scholar]
- Laguitton S, Rivière D, et al. Soma-workflow: a unified and simple interface to parallel computing resources. MICCAI Workshop on High Performance and Distributed Computing for Medical Imaging. MICCAI Workshop on High Performance and Distributed Computing for Medical Imaging. Toronto, CA: 2011. [Google Scholar]
- Lau JC, Lerch JP, et al. Longitudinal neuroanatomical changes determined by deformation-based morphometry in a mouse model of Alzheimer's disease. NeuroImage. 2008;42(1):19–27. doi: 10.1016/j.neuroimage.2008.04.252. [DOI] [PubMed] [Google Scholar]
- Lebenberg J, Herard AS, et al. Validation of MRI-based 3D digital atlas registration with histological and autoradiographic volumes: an anatomofunctional transgenic mouse brain imaging study. NeuroImage. 2010;51(3):1037–1046. doi: 10.1016/j.neuroimage.2010.03.014. [DOI] [PubMed] [Google Scholar]
- Lee J, Jomier J, et al. Evaluation of atlas based mouse brain segmentation. Proc. SPIE. 2009;7259:725943–725949. doi: 10.1117/12.812762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch JP, Carroll JB, et al. Automated deformation analysis in the YAC128 Huntington disease mouse model. NeuroImage. 2008;39(1):32–39. doi: 10.1016/j.neuroimage.2007.08.033. [DOI] [PubMed] [Google Scholar]
- Lerch JP, Yiu AP, et al. Maze training in mice induces MRI-detectable brain shape changes specific to the type of learning. NeuroImage. 2011;54(3):2086–2095. doi: 10.1016/j.neuroimage.2010.09.086. [DOI] [PubMed] [Google Scholar]
- Ma Y, Hof PR, et al. A three-dimensional digital atlas database of the adult C57BL/6J mouse brain by magnetic resonance microscopy. Neuroscience. 2005;135(4):1203–1215. doi: 10.1016/j.neuroscience.2005.07.014. [DOI] [PubMed] [Google Scholar]
- Ma Y, Smith D, et al. In vivo 3D digital atlas database of the adult C57BL/6J mouse brain by magnetic resonance microscopy. Front. Neuroanat. 2008;2:1. doi: 10.3389/neuro.05.001.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie-Graham A, Lee EF, et al. A multimodal, multidimensional atlas of the C57BL/6J mouse brain. J. Anat. 2004;204(2):93–102. doi: 10.1111/j.1469-7580.2004.00264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie-Graham A, Tinsley MR, et al. Cerebellar cortical atrophy in experimental autoimmune encephalomyelitis. NeuroImage. 2006;32(3):1016–1023. doi: 10.1016/j.neuroimage.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Mackenzie-Graham A, Rinek GA, et al. Cortical atrophy in experimental autoimmune encephalomyelitis: in vivo imaging. NeuroImage. 2012;60(1):95–104. doi: 10.1016/j.neuroimage.2011.11.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheswaran S, Barjat H, et al. Analysis of serial magnetic resonance images of mouse brains using image registration. NeuroImage. 2009a;44(3):692–700. doi: 10.1016/j.neuroimage.2008.10.016. [DOI] [PubMed] [Google Scholar]
- Maheswaran S, Barjat H, et al. Longitudinal regional brain volume changes quantified in normal aging and Alzheimer's APP×PS1 mice using MRI. Brain Res. 2009b;1270:19–32. doi: 10.1016/j.brainres.2009.02.045. [DOI] [PubMed] [Google Scholar]
- Mallar Chakravarty M, Steadman P, et al. Performing label-fusion-based segmentation using multiple automatically generated templates. Hum. Brain Mapp. 2012 doi: 10.1002/hbm.22092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy G, Millet P, et al. Magnetic resonance imaging determinants of intraindividual variability in the elderly: combined analysis of grey and white matter. Neuroscience. 2011;186:88–93. doi: 10.1016/j.neuroscience.2011.04.028. [DOI] [PubMed] [Google Scholar]
- Ng L, Bernard A, et al. An anatomic gene expression atlas of the adult mouse brain. Nat. Neurosci. 2009;12(3):356–362. doi: 10.1038/nn.2281. [DOI] [PubMed] [Google Scholar]
- Patterson PH. Modeling autistic features in animals. Pediatr. Res. 2011;69(5 Pt 2):34R–40R. doi: 10.1203/PDR.0b013e318212b80f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirce JL, Lu L, et al. A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genet. 2004;5:7. doi: 10.1186/1471-2156-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rex DE, Ma JQ, et al. The LONI pipeline processing environment. NeuroImage. 2003;19(3):1033–1048. doi: 10.1016/s1053-8119(03)00185-x. [DOI] [PubMed] [Google Scholar]
- Sawiak SJ, Wood NI, et al. Voxel-based morphometry in the R6/2 transgenic mouse reveals differences between genotypes not seen with manual 2D morphometry. Neurobiol. Dis. 2009;33(1):20–27. doi: 10.1016/j.nbd.2008.09.016. [DOI] [PubMed] [Google Scholar]
- Scheenstra AE, van de Ven RC, et al. Automated segmentation of in vivo and ex vivo mouse brain magnetic resonance images. Mol. Imaging. 2009;8(1):35–44. [PubMed] [Google Scholar]
- Sharief AA, Johnson GA. Enhanced T2 contrast for MR histology of the mouse brain. Magn. Reson. Med. 2006;56(4):717–725. doi: 10.1002/mrm.21026. [DOI] [PubMed] [Google Scholar]
- Sharief AA, Badea A, et al. Automated segmentation of the actively stained mouse brain using multi-spectral MR microscopy. NeuroImage. 2008;39(1):136–145. doi: 10.1016/j.neuroimage.2007.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, et al. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans. Med. Imaging. 1998;17(1):87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, et al. Tract-based spatial statistics: voxelwise analysis of multi-subject diffusion data. NeuroImage. 2006;31(4):1487–1505. doi: 10.1016/j.neuroimage.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Smithies O, Gregg RG, et al. Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature. 1985;317(6034):230–234. doi: 10.1038/317230a0. [DOI] [PubMed] [Google Scholar]
- Sporns O. The human connectome: a complex network. Ann. N. Y. Acad. Sci. 2011;1224:109–125. doi: 10.1111/j.1749-6632.2010.05888.x. [DOI] [PubMed] [Google Scholar]
- Taylor B. Origins of Inbred Mice. New York: Academic Press; 1978. Recombinant inbred strains: use in gene mapping. [Google Scholar]
- Thomas KR, Capecchi MR. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51(3):503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- Tomasi C, Manduchi R. Bilateral filtering for gray and color images. 6th Int. Conf. Computer Vision; New Delhi, India. 1998. [Google Scholar]
- Tu Z, Narr K, et al. Brain anatomical structure segmentation by hybrid discriminative/ generative models. IEEE Trans. Med. Imaging. 2008;27(4):495–508. doi: 10.1109/TMI.2007.908121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tustison NJ, Avants BB, et al. N4ITK: improved N3 bias correction. IEEE Trans. Med. Imaging. 2010;29(6):1310–1320. doi: 10.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Ugurbil K. The future of the human connectome. Neuroimage. 2012;62(2):1299–1310. doi: 10.1016/j.neuroimage.2012.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods RP, Grafton ST, et al. Automated image registration: I. General methods and intrasubject, intramodality validation. J. Comput. Assist. Tomogr. 1998;22(1):139–152. doi: 10.1097/00004728-199801000-00027. [DOI] [PubMed] [Google Scholar]
- Worsley KJ, Taylor JE, et al. SurfStat: a Matlab toolbox for the statistical analysis of univariate and multivariate surface and volumetric data using linear mixed effects models and random field theory. NeuroImage. 2009;47(Supplement 1)(0):S102. [Google Scholar]
- Wu T, Bae MH, et al. A prior feature SVM-MRF based method for mouse brain segmentation. NeuroImage. 2012;59(3):2298–2306. doi: 10.1016/j.neuroimage.2011.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Yang D, et al. Characterizing the regional structural difference of the brain between tau transgenic (rTg4510) and wild-type mice using MRI. Med. Image Comput. Comput. Assist. Interv. 2010;13(Pt 1):308–315. doi: 10.1007/978-3-642-15705-9_38. [DOI] [PubMed] [Google Scholar]
- Yushkevich PA, Piven J, et al. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. NeuroImage. 2006;31(3):1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Zamyadi M, Baghdadi L, et al. Mouse embryonic phenotyping by morphometric analysis of MR images. Physiol. Genomics. 2010;(2):42A. 89–95. doi: 10.1152/physiolgenomics.00091.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Peng Q, et al. Longitudinal characterization of brain atrophy of a Huntington's disease mouse model by automated morphological analyses of magnetic resonance images. NeuroImage. 2010a;49(3):2340–2351. doi: 10.1016/j.neuroimage.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Bearer EL, et al. Increased anatomical detail by in vitro MR microscopy with a modified Golgi impregnation method. Magn. Reson. Med. 2010b;63(5):1391–1397. doi: 10.1002/mrm.22322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Chee MW, et al. Improvement of brain segmentation accuracy by optimizing non-uniformity correction using N3. NeuroImage. 2009;48(1):73–83. doi: 10.1016/j.neuroimage.2009.06.039. [DOI] [PubMed] [Google Scholar]
- Zijdenbos AP, Forghani R, et al. Automatic “pipeline” analysis of 3-D MRI data for clinical trials: application to multiple sclerosis. IEEE Trans. Med. Imaging. 2002;21(10):1280–1291. doi: 10.1109/TMI.2002.806283. [DOI] [PubMed] [Google Scholar]