Abstract
Importin13 (Imp13) is a bidirectional karyopherin that can mediate both import and export of cargoes. Imp13 recognizes several import cargoes, which include the exon junction complex components Mago-Y14 and the E2 SUMO-conjugating enzyme Ubc9, and one known export cargo, the translation initiation factor 1A (eIF1A). To understand how Imp13 can perform double duty, we determined the 3.6-Å crystal structure of Imp13 in complex with RanGTP and with eIF1A. eIF1A binds at the inner surface of the Imp13 C-terminal arch adjacent and concomitantly to RanGTP illustrating how eIF1A can be exported by Imp13. Moreover, the 3.0-Å structure of Imp13 in its unbound state reveals the existence of an open conformation in the cytoplasm that explains export cargo release and completes the export branch of the Imp13 pathway. Finally, we demonstrate that Imp13 is able to bind and export eIF1A in vivo and that its function is essential.
Keywords: eIF1A, export, Importin13, nucleo-cytoplasmic transport
Introduction
In eukaryotic cells, the nuclear envelope selectively separates the nucleus, where transcription and splicing occur, from the cytoplasm, where translation takes place. Proteins and RNAs can cross this barrier through the nuclear pore complexes (NPCs). Small molecules, up to ∼20–40 kDa, can passively diffuse across the NPCs, while other molecules need to be actively transported (Görlich and Kutay, 1999; Mohr et al, 2009). Active nucleo-cytoplasmic transport is mainly mediated by a superfamily of proteins, known as karyopherins. Karyopherins can be divided into two groups based on their directionality: importins translocate cargoes from the cytoplasm to the nucleus, while exportins transport their cargoes from the nucleus into the cytoplasm. The directionality of both import and export is determined by the same driving force: a gradient of the small GTPase Ran. Importins bind their cargoes in the cytoplasm and release them upon binding to the GTP-bound form of Ran (RanGTP), which is confined to the nucleus. Conversely, exportins require RanGTP to stably bind their cargo in the nucleus; the dissociation of the export complex is then triggered in the cytoplasm by the hydrolysis of Ran-bound GTP to GDP (Görlich and Kutay, 1999; Cook et al, 2007). So far, only a few karyopherins including Importin13 (Imp13), Exportin4 and Msn5, have been characterized as bidirectional transport factors being able to both import and export cargoes from the nucleus (Mingot et al, 2001; Yoshida and Blobel, 2001; Gontan et al, 2009).
In Drosophila larvae, Imp13 affects neurotransmitter release at the neuromuscular junctions and homozygous imp13 mutations are lethal (Giagtzoglou et al, 2009). In humans, Imp13 has been involved in the import of the core exon junction complex components Mago-Y14, the E2 SUMO-conjugating enzyme Ubc9 (Mingot et al, 2001), histone fold-containing and paired-type homeodomain transcription factors (Ploski et al, 2004; Kahle et al, 2005; Walker et al, 2009), the glucocorticoid receptor (Tao et al, 2006) and the actin-binding protein myopodin (Liang et al, 2008). The only export cargo of Imp13 known so far is the eukaryotic initiation factor 1A (eIF1A) (Mingot et al, 2001). However, the function and importance of this export pathway have not yet been addressed in vivo.
eIF1A is a protein conserved across eukaryotes with multiple functions in translation initiation (Jackson et al, 2010; Hinnebusch, 2011). Together with another translation initiation factor, eIF1, it directly associates to the small ribosomal subunit and is required for the assembly of the pre-initiation complex (Jackson et al, 2010; Aitken and Lorsch, 2012). The two initiation factors cooperate in promoting an ‘open’, scanning-competent conformation of the 40S subunit (Passmore et al, 2007). eIF1A consists of an oligonucleotide-binding (OB) β-barrel fold followed by an extended helix, and two unstructured tails at the N- and C-termini (Battiste et al, 2000). Due to its small size (17 kDa), eIF1A is thought to passively diffuse through the NPCs; its active export might therefore be required both to deplete eIF1A from the nucleus and to maintain sufficient cytoplasmic levels (Mingot et al, 2001). Despite the importance of eIF1A in translation, limited information is available on its localization.
Several structures of karyopherins have been determined to date, comprising two nuclear import factors (Importin β (Impβ) and Transportin (Tpn)) (Cingolani et al, 1999; Vetter et al, 1999a; Bayliss et al, 2000; Lee et al, 2003, 2005; Cansizoglu and Chook, 2007; Cansizoglu et al, 2007; Cook et al, 2007; Imasaki et al, 2007; Wohlwend et al, 2007; Mitrousis et al, 2008; Bhardwaj and Cingolani, 2010; Forwood et al, 2010; Xu et al, 2010; Zhang and Chook, 2012) and four nuclear export factors (Cse1, Crm1, Expo5 and Xpo-t) (Matsuura and Stewart, 2004; Cook et al, 2005, 2009; Monecke et al, 2009; Okada et al, 2009; Dong et al, 2009a, 2009b; Güttler et al, 2010) but it is still unclear how a transport factor, such as Imp13, can have a mixed transport competence. How RanGTP binding to bidirectional karyopherins can cause opposite effects, dissociation of the cargo to be imported and association of the cargo to be exported, is an open question in the nuclear transport field. Our previous work on the structures of the Imp13-RanGTP intermediate state and the Imp13-Mago-Y14 and Imp13-Ubc9 cargo complexes have provided key insights into import cargo recognition and dissociation by RanGTP (Bono et al, 2010; Grünwald and Bono, 2011). Well-characterized import factors such as Impβ and Tpn normally recognize only a small portion of their cargos as an import signal (Cingolani et al, 1999, 2002; Cansizoglu and Chook, 2007; Imasaki et al, 2007; Wohlwend et al, 2007; Mitrousis et al, 2008; Bhardwaj and Cingolani, 2010; Forwood et al, 2010; Xu et al, 2010; Zhang and Chook, 2012). Remarkably, Imp13 recognizes the folded domains of the import cargoes Ubc9 and Mago-Y14, predominantly via charged and polar residues distributed over the entire proteins. RanGTP binding to Imp13 is similar to RanGTP binding by Impβ and Tpn although Imp13 lacks the acidic loop that is found in the canonical import factors (Cook et al, 2007). Imp13 uses non-overlapping surfaces for the recognition of different import cargoes. As a consequence, the release mechanism of these cargoes is different: Mago-Y14 is released via a steric hindrance mechanism, while Ubc9 and RanGTP directly compete for the same binding surface on Imp13 (Bono et al, 2010; Grünwald and Bono, 2011).
Our structural and biochemical studies of Imp13 in its unbound state and the Imp13-RanGTP-eIF1A trimeric complex shed light on how bidirectional karyopherins can perform double duty and also be regulated in opposite ways by RanGTP. We further address the functional basis by which the nuclear transport factor Imp13 is able to recognize both import and export cargoes and deliver them in the appropriate cellular compartment and the in vivo relevance of Imp13 function.
Results and Discussion
Structure determination and quality
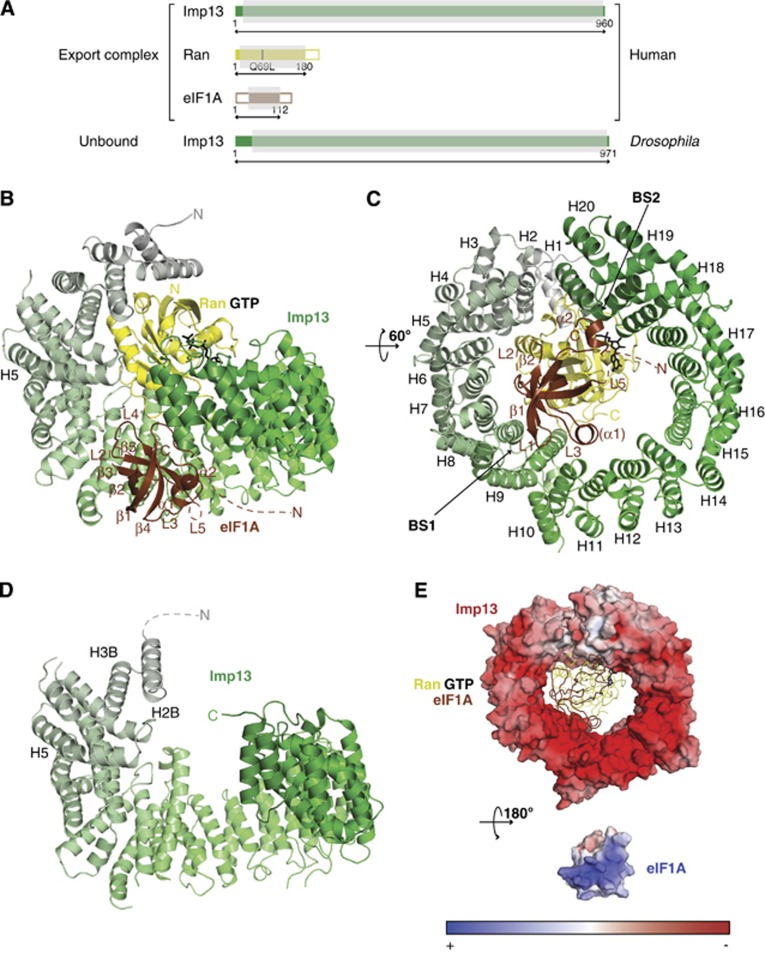
We determined the crystal structures of Imp13 in its nuclear export complex with RanGTP and eIF1A and also in the unbound form that corresponds to a cytosolic state. For the crystallization of the ternary export complex, human (Hs) Imp13, Ran and eIF1A were expressed separately in E. coli. While Imp13 was used as a full-length construct, the small GTPase Ran was truncated to contain the residues 1–180 and a Gln69Leu mutation to inhibit GTP hydrolysis, as previously described (Bischoff et al, 1994; Matsuura and Stewart, 2004; Lee et al, 2005; Cook et al, 2009; Monecke et al, 2009; Bono et al, 2010) (Figure 1A; Supplementary Figure 1A). Attempts to crystallize the trimeric complex in presence of full-length eIF1A were not successful probably due to the two large unstructured regions at the N- and C-termini (Battiste et al, 2000) (NTT and CTT, respectively). Therefore, a truncated version of eIF1A was used that includes the residues from 1 to 112 and lacks the CTT portion of the protein (eIF1AΔC; Figure 1A and Supplementary Figure 1A). The complex was reconstituted in vitro with an excess of RanGTP and eIF1A and purified to homogeneity by size exclusion chromatography (SEC). The export complex crystallized in a centred monoclinic space group (C2). Initial phases for the structure were obtained by molecular replacement (MR) with the Imp13-RanGTP structure as a search model (Bono et al, 2010; pdb id.: 2x19). Although density for eIF1A could be observed, a reliable MR solution could not be obtained when using available structures of eIF1A as search models (Battiste et al, 2000; pdb ids: 2oqk and 2dgy). We manually placed the crystal structure of eIF1A from Cryptosporidium parvum (pdb id.: 2oqk; 75% sequence identity over the region encompassed by our construct) into the electron density using an anomalous Fourier map of a seleno-methionine (SeMet) substituted eIF1A as a guide (Supplementary Figure 1A). This eIF1A was mutated to include a third methionine residue so that the structure could be positioned accurately by using three anomalous difference peaks (Supplementary Figures 1A and 2A). A view of the quality of the electron density of eIF1A is shown in Supplementary Figure 2B. Model building of the complex was also verified by single-wavelength anomalous diffraction (SAD) structure solution of a trimeric complex reconstituted with a SeMet substituted Imp13 (unpublished observation).
Figure 1.
Structure of the ternary export complex Imp13-RanGTP-eIF1A and constructs used. (A) Schematic representation of the architecture of the proteins used in this study. Colour-filled areas in the scheme identify structural domains, which include the Ran core (in yellow) and the OB-fold of eIF1A (in brown). The residue numbers corresponding to the constructs used are indicated. The portions of the polypeptides ordered in the three-dimensional structure are shaded in grey. Cartoon view (B) and view rotated 60° along the x axis (C) of the export complex. Imp13 is shown in a colour gradient from grey (N-terminus) to green (C-terminus). Ran is in yellow with GTP as a stick model in black and eIF1A is coloured in brown. Secondary structure elements and HEAT repeats are labelled. (D) Imp13 apo structure in a similar colour code and view as in (A). (E) Surface rendering representation of Imp13 (top) and of eIF1A (bottom) in the export complex. The surface is coloured according to the electrostatic potential. Top, eIF1A and Ran, in the same colour code as above, are shown as loops with GTP as stick model in black. For the electrostatic potential calculation of eIF1A, the backbone was used as a template to include all the side chains with MODELLER (Sali and Blundell, 1993) (r.m.s.d. of 0.277 Å over 1121 atoms). These and all other protein structure figures were generated using PyMOL (http://www.pymol.org).
The asymmetric unit is composed of three distinct complexes. Complexes 1 and 2 present the ternary export complex and are very similar (r.m.s.d. of 0.634 Å over 868 Cα atoms). Complex 3 does not show density for eIF1A and superposes with an r.m.s.d. of 2.0 Å over 855 Cα atoms to complex 1. Complex 1 shows the best quality electron density and therefore, throughout the paper, we will refer to complex 1, unless explicitly stated. The model was refined to 3.6 Å resolution with an Rfree and Rwork of 29.9 and 26.8%, respectively, and good stereochemistry (Table I). Disordered regions are at the very N- and C-termini of Imp13 as well as a long inter-repeat loop at residues 655–672. The NTT of eIF1A is not observed (residues 1–27) together with several loop regions (residues 54–56; 97–100; 109–112) (Figure 1A and B). In complex 2, additional regions of the C-terminal arch of Imp13 and eIF1A are disordered.
Table 1. Crystallographic statistics.
Data collection and refinement statistics of the crystal structure of Imp13-RanGTP-eIF1A complex and of Imp13 apo.
aValues in parentheses correspond to the highest resolution shell.
bMolprobity (Chen et al, 2010).
The Drosophila (Dm) Imp13 apo form crystallized in spacegroup P43212 and the crystals diffracted to 3.0 Å (Figure 1D). The structure was solved by MR using Dm Imp13 as a search model (Bono et al, 2010; pdb id.: 2x1g) fragmented into three portions. The refined model has an Rfree of 31% with good stereochemistry (Table I). For a sample of the electron density map, see Supplementary Figure 2C. In the unbound Imp13 structure, some N-terminal regions (residues 1–53; 78–117; 100–121), intermediate loops as well as the very C-terminus (residues 968–971) are not well defined and could not be modelled (Figures 1D and 6A).
Architecture of Imp13 in the export complex and in the unbound state
As previously shown, Imp13 folds into 20 consecutive HEAT repeats (Bono et al, 2010; Grünwald and Bono, 2011). HEAT repeats are motifs consisting of two helices (A and B) connected by a short loop (intra-repeat). Each HEAT repeat stacks against the following one, via an inter-repeat loop, to generate a superhelical arrangement (Cook et al, 2007). The A helices form the outer surface of the superhelix, while the B helices form the inner concave surface. The C-terminus of Imp13 is stabilized by a HEAT-like repeat motif composed of three helices (A–C). A hinge region around HEAT10 (H10) divides Imp13 into two arches, the N- and C-terminal arches. Two further hinge regions at H4 and H14 increase the protein’s conformational flexibility (Bono et al, 2010; Grünwald and Bono, 2011).
In complex with RanGTP and eIF1A, Imp13 adopts a toroidal conformation with the N- and C-terminal edges offset by 8.2 Å and an inner diameter of ∼58 Å (Figure 1B and C). The C helix of H20 approaches the N-terminal tip of H5 (Figures 1B and 6D). Both arches of Imp13 are engaged in complex formation. As in the Imp13-RanGTP structure, RanGTP binds to the inner concave surface of the N-terminal arch shifted towards the intra-repeat loops (Bono et al, 2010). However, when in complex with the export cargo, the toroidal conformation of Hs Imp13 is less compact than in the Imp13-RanGTP complex, with the C-terminal arch more open to accommodate eIF1A (Figure 1B). In the complex with Imp13 and eIF1A, the structure and conformation of Hs RanGTP is very similar to the structure of RanGTP in complex with RanBD1 (r.m.s.d. of 0.48 Å over 168 Cα atoms) and of RanGTP in the binary complex with Imp13 (r.m.s.d. of 0.34 Å over 147 Cα atoms) and has been extensively described elsewhere (Vetter et al, 1999b; Bono et al, 2010).
In the structure of Dm Imp13 in the unbound state, the karyopherin toroidal structure is in an open conformation (Figure 1D). H1 and the external helices of H2 and 3 are disordered in the structure, suggesting flexibility at the very N-terminus of the protein. Flexibility in this region might be required for the docking of RanGTP in the nucleus to displace the import cargoes (Grünwald and Bono, 2011) (Figures 1D, 6A and B) and to recognize cargoes that bind at the N-terminal arch of Imp13, such as Ubc9.
Export cargo recognition by Imp13 and RanGTP
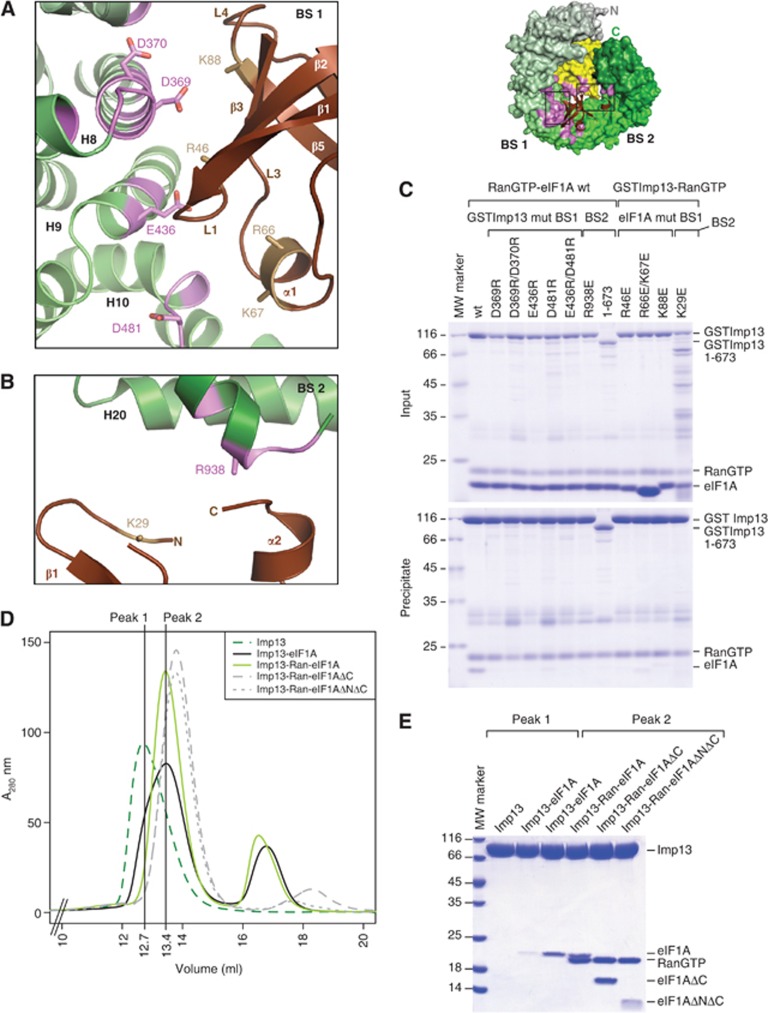
The OB-fold of eIF1A is composed of five β strands (β1–β5 strands of eIF1A: β1E–β5E) forming an elliptic cavity interrupted by a loop between β3E and β4E (L3E) that folds into a small helix (α1E). C-terminal to the β-barrel eIF1A features an additional helix (α2E). Although the NTT of eIF1A was present in the construct that was crystallized, no density for this part of the structure was observed in the trimeric export complex, likely due to conformational flexibility in this region (Battiste et al, 2000). When the full-length solution structure is superposed on the export complex, the NTT points into solvent (Supplementary Figure 6B). To rule out the possibility that the NTT is required for binding and the possibility that the NTT was not observed due to the relatively low resolution map, we performed SEC with truncated versions of eIF1A. These experiments show that the folded part of eIF1A (residues 26–115; ΔNΔC) is sufficient for binding to Imp13 (Figure 2D and E).
Figure 2.
Detailed view of the interaction between Imp13 and eIF1A. Top right, the export complex is rendered with Imp13 and RanGTP as surface and eIF1A as cartoon in the same colour coding as in Figure 1B. The interaction surface of eIF1A on Imp13-RanGTP is in pink. (A) Close-up view of the eIF1A interaction with Imp13 at BS 1 in a similar view as in Figure 1B. Residues involved in the interaction on eIF1A and Imp13 are shown as sand and pink sticks, respectively. HEAT repeats are labelled in black and secondary structure elements of eIF1A are marked in brown or white. (B) Zoomed in view of the BS 2 interaction site. (C) Protein co-precipitation by GST-tagged Imp13 incubated with RanGTP and eIF1A with either wt or mutant proteins. In this experiment, Ran was His tagged for better separation from eIF1A on gel. For the input control, 1/6 of the samples were kept (upper panel) and the rest was co-precipitated with glutathione sepharose beads (lower panel) and analysed on Coomassie-stained 15% SDS–PAGE. The far left lane was loaded with a molecular weight marker. Some contaminations likely from degradation of GSTImp13 are visible between 25 and 35 kDa. GST controls for this experiment are in Supplementary Figure 6A. (D) Overlay of SEC plots of Imp13 alone or together with different eIF1A constructs with or without RanGTP. (E) Samples of the peaks fractions were analysed on Coomassie-stained SDS–PAGE (Any kD, BIORAD). The far left lane was loaded with a molecular weight marker.
eIF1A can be approximated to a triangle that is recognized at its vertices on three main interaction surfaces, two on Imp13 and one on RanGTP. The two corners at the base of the triangle β2E and L3E, and α2E interact at two opposite surface areas (BS1 and BS2, respectively) that bridge the inner central part of Imp13 and its very C-terminus (Figures 1B, C, 2A, B and 3A). The interaction surface of eIF1A to Imp13 contains several conserved, positively charged residues, which complement the negatively charged inner binding surface of Imp13 (Figure 1E).
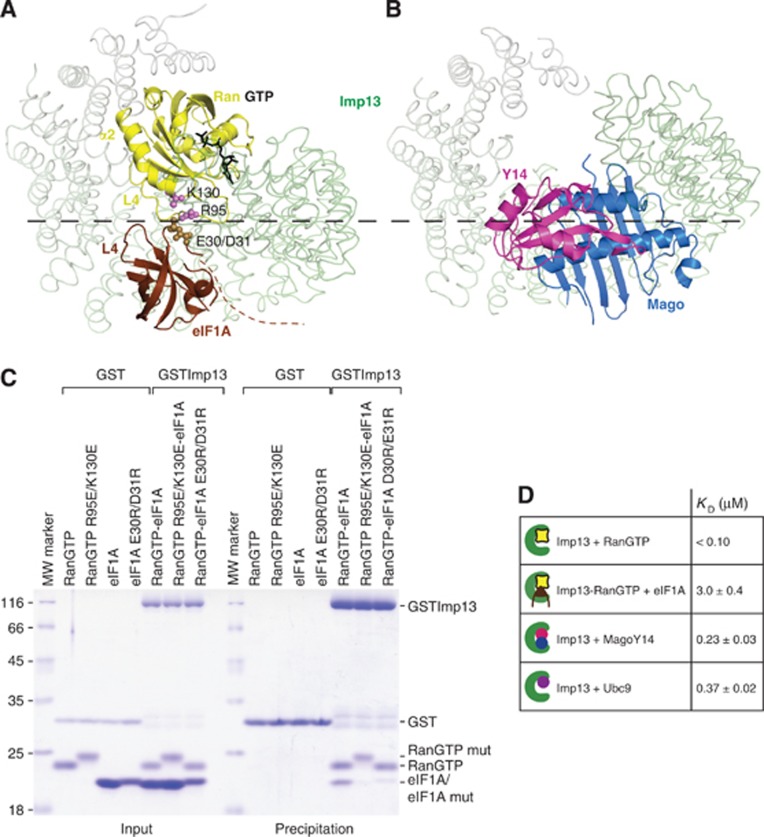
Figure 3.
Structural basis of Imp13 bidirectionality and KD affinities in the Imp13 cycle. (A) RanGTP and eIF1A concomitant interaction in the ternary export complex. Interacting residues on Ran and eIF1A are rendered as spheres in pink and sand, respectively. (B) Imp13-Mago-Y14 structure oriented as in Figure 3A (pdb ids.: 2x1g). Imp13 is represented as loop trace. A dashed line in black marks the boundary between compatible and hindered binding. (C) Protein co-precipitations by GST-tagged Imp13 incubated together with His-RanGTP and eIF1A with either wt or mutant proteins performed as in the previous figure. (D) Table representing KD values determined by DSF from at least three independent experiments.
The larger area of interactions (BS1) is centred around H9 of Imp13 and stretches across the hinge between the N- and C-terminal arches involving H7-11 on the side of the inter-repeat loops. On eIF1A the loop L1E points to the groove between H8 and 9 (Figure 2A); β3E and the N-terminal stretch of L3E contact H8 and H10-11, respectively, while the N-terminal part of β5E and L4E interacts with H7-8. A series of salt bridges are likely to stabilize this interface: Asp369 on H8 of Imp13 (Asp369I) points to Lys88 of eIF1A (Lys88E) on β5E; on H9, Glu436I approaches Arg46E on L1E; Asp481I at the inter-repeat loop between H10-11 is in proximity to Arg66E and Lys67E on L3E.
At the smaller interaction surface, helix α2E packs end-on to the C-terminal helix of Imp13. At this site, Arg938I approaches helix α2E as well as the N-terminal stretch of eIF1A (Figure 2B; Supplementary Figure 1). Consistent with the structural analysis, reverse charge mutations of Asp369I, Asp370I, Glu436I and Asp481I to arginine in BS1 or of Arg938I to glutamate in BS2 resulted in reduction or loss of binding to eIF1A in pull-down assays (Figure 2C; Supplementary Figure 1). Although structurally less well defined (many side chains of eIF1A do not have visible electron density), the contribution of positively charged residues of eIF1A to Imp13 binding could also be confirmed using reverse charge mutations. Residues Lys29E, Arg46E, Arg66/Lys67E and Lys88E are conserved (Supplementary Figure 1A) and mutations to negatively charged residues impeded binding to Imp13, as confirmed by pull-down assays (Figure 2C; Supplementary Figure 6A).
Similarly to the import cargoes Ubc9 and Mago-Y14, eIF1A is primarily recognized by Imp13 as a folded domain. This indicates that both import and export signals recognized by Imp13 are complex ones, therefore different from the linear and semi-linear canonical NLSs and NESs recognized by Impβ, Tpn and Crm1 (Xu et al, 2010; Zhang and Chook, 2012). The structural and mutational analyses show that Imp13 recognizes its cargoes by a combination of shape and charge together with specific contacts at conserved positions on eIF1A.
Imp13 uses shape complementarity of cargoes to perform bidirectional transport
In the export complex, the β-barrel and the very tip of helix α2E of eIF1A bind adjacent to RanGTP at the inner surface of Imp13. The structure strongly suggests that eIF1A is in direct contact with RanGTP in the complex with Imp13. L4 of Ran (L4R, between β5R-6R) is adjacent to L4E and to the N-terminus of eIF1A (Figure 3A). Here, Glu30E and Asp31E point towards the L4R and towards Lys130 of Ran (Lys130R) while L4E approaches Arg95R at helix α2R (Figure 3A; Supplementary Figure 1A). Consistently, mutants of Glu30E and Asp31E to arginine and of Arg95R and Lys130R to glutamate impair binding of Imp13-RanGTP to eIF1A (Figure 3C).
To further understand Imp13 bidirectionality, we compared the mode of binding of Imp13 in the export complex with the structure of Imp13 bound to the import cargo Mago-Y14 (Bono et al, 2010). As we have previously shown, when the latter structures are superposed, RanGTP and Mago-Y14 assume an adjacent but overlapping position that precludes concomitant binding of Mago-Y14 with RanGTP due to steric hindrance (Bono et al, 2010). In the complex of Imp13 with RanGTP and eIF1A, the export cargo is much smaller than Mago-Y14 so that it fits neatly into the hole created by the C-terminal arch. In this case there is not a clash with RanGTP, instead the complementary charge interactions with RanGTP stabilize the binding and increase the binding affinity (Figure 3D). Consistently, SEC experiments of Imp13-eIF1A complexes show only a partial binding of eIF1A in the absence of RanGTP (Figure 2D and E). Imp13 is therefore able to discriminate between cargoes based on their structural features and can therefore promote either cooperative binding (for export) or antagonistic binding (for import).
Comparison of the Imp13-RanGTP-eIF1A complex with the Imp13-Mago-Y14 structure also shows that the binding of Mago-Y14 and eIF1A is mutually exclusive. The release of Mago-Y14 by RanGTP is less efficient in vitro as compared to that of another import cargo, Ubc9, which shows a different mode of binding. We have previously shown that addition of eIF1A together with RanGTP enhances the release of Mago-Y14 from Imp13 (Grünwald and Bono, 2011; Figure 3A and B). In contrast, a comparison of the Imp13 export complex with Imp13-Ubc9 import complex shows no major clashes between the cargoes (Figure 6D and E). However, a simultaneous binding of eIF1A and Ubc9 to Imp13 in vitro is not possible (Grünwald and Bono, 2011). The structural and biochemical data suggest that the shape complementarity of eIF1A and its overlap with the Mago-Y14 binding site is critical for achieving the directionality of Mago-Y14 import. RanGTP likely destabilizes Mago-Y14 binding by preventing optimal association with Imp13. Further association of eIF1A with Imp13 would lock the importin in an export conformation, preventing re-association of Mago-Y14 and imparting directionality to this nuclear import step.
To better understand the mechanism of the Imp13 import/export cycle, we measured the KD values by differential scanning fluorimetry (DSF) (Niesen et al, 2007) (Figure 3D; Supplementary Figure 3). The estimated KD for the two import cargoes Mago-Y14 and Ubc9 is in a similar range with a value of 235±30 nM and 370±20 nM, respectively. The KD value for RanGTP binding to Imp13, although not accurately calculated due to the upper detection limit of the method, falls in the low nanomolar or picomolar range (<100 nM) and is consistent with values measured for other karyopherins (Bischoff and Görlich, 1997; Deane et al, 1997; Görlich et al, 1997; Lipowsky et al, 2000). The dissociation constant of the export cargo eIF1A to a preformed Imp13-RanGTP complex is 3±0.38 μM, 10-fold higher than the KD values of the import cargoes (Figure 3D). The rather weak affinity of Imp13-RanGTP for its export cargo eIF1A is comparable to that displayed by Crm1 for most NES, such as HIV Rev (KD of about 0.5 μM) (Askjaer et al, 1999; Paraskeva et al, 1999). One exception is Snurportin that binds to CRM1 with much higher affinity because it is recognized through additional interactions (KD∼10 nM) (Paraskeva et al, 1999). Xpot also binds tRNAs with high affinity (KD∼3 nM) (Kutay et al, 1998; Lipowsky et al, 1999). The binding of CAS/Cse1 to Impα and RanGTP is highly cooperative and cargo binding cannot be disjointed from RanGTP binding (Kutay et al, 1997).
Imp13 exports eIF1A in vivo and its function is required for cell viability
eIF1A is able to passively diffuse through the NPCs and thus reach the nuclear compartment and it has previously been proposed that active export is necessary to deplete eIF1A from the nucleus and contribute in preventing nuclear translation (Mingot et al, 2001). Surprisingly, we observed that endogenous eIF1A is enriched in nucleoli in HeLa cell lines (Supplementary Figure 4A and D). An overexpressed, GFP-tagged version of the protein shows a similar localization in fixed cells (Supplementary Figure 4C); however, the same construct displays a significantly higher cytoplasmic fraction in living cells (Figure 4B), suggesting that fixation affects eIF1A localization to some extent. All together, our results indicate that endogenous eIF1A is localized to both nucleoli and cytoplasm.
Figure 4.
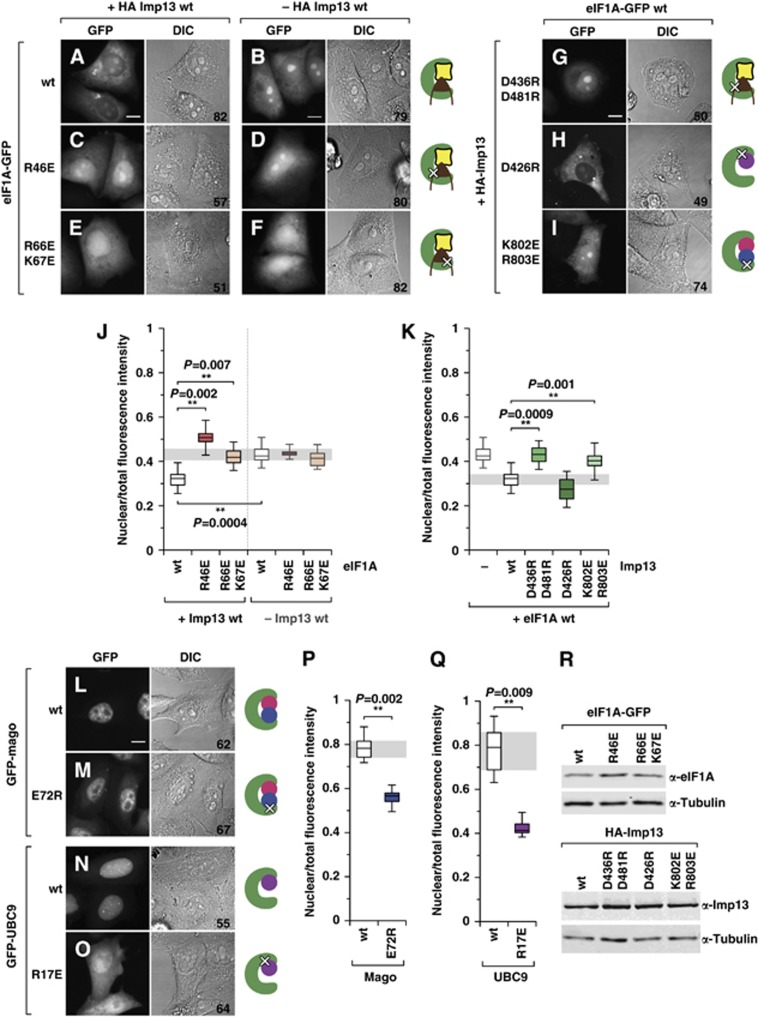
Imp13 binds and exports eIF1A in vivo. (A–F) Fluorescence microscopy images and the corresponding differential interference contrast (DIC) images of living HeLa cells expressing GFP-tagged eIF1A (eIF1A-GFP) wt or mutants. Localization of wt or mutant eIF1A-GFP in the presence (A, C, E) or absence (B, D, F) of HA-Imp13. (G–I) Localization of wt eIF1A-GFP in the presence of the indicated HA-Imp13 mutants. (J, K) Relative quantification of the fluorescence intensity in the nuclear compartment for each construct, in the absence (−) or presence (+) of Imp13 wt or of the indicated Imp13 constructs. (L–O) Fluorescence microscopy images and the corresponding DIC images of living HeLa cells expressing GFP-tagged Mago or Ubc9 wt or mutants. (P, Q) Relative quantification of the fluorescence intensity in the nuclear compartment for each construct. Box plots: the bottom and top of the box depict the first (qu1) and the third (qu3) quartiles, respectively, with the central line indicating the median (qu2); the upper and lower whiskers represent the maximum and minimum values, respectively. At least three independent experiments were performed; the number of analysed cells is indicated for each condition at the bottom right of the corresponding DIC image. Statistical significance was calculated by comparing the means of each experimental replicate with the means of the appropriate control conditions, with one-tail t-tests (α 0.05). Scale bar: 10 μm. (R) Western blot showing the expression levels of HA-Imp13 wt and mutants, or eIF1A-GFP wt and mutants; note that all mutants are expressed at levels similar to the corresponding wt. α-Tubulin served as a loading control.
Overexpression of Imp13 causes an accumulation of wild-type (wt) eIF1A in the cytoplasm (Figure 4A), indicating that Imp13 can bind and export eIF1A in vivo. This redistribution of eIF1A in the cell depends on its ability to bind to Imp13, since Arg46GluE and Arg66Glu/Lys67GluE mutants, which are impaired in their interaction with Imp13 (Figure 2C), do not change their nucleo-cytoplasmic distribution upon Imp13 overexpression (Figure 4C, E and J) despite being expressed at similar levels (Figure 4R). Conversely, only wt Imp13, but not Asp436Arg/Asp481ArgI mutant, which is unable to bind to eIF1A (Figure 2C) but still associates with Mago-Y14 and Ubc9 (Figure 5A), triggers eIF1A depletion from the nucleus (Figure 4G and K). This effect is not due to a lower expression level of the mutants compared to wt Imp13 (Figure 4R). In the absence of Imp13 overexpression, however, eIF1A wt, Arg46GluE and Arg66Glu/Lys67GluE reverse-charge mutants show a similar nucleo-cytoplasmic distribution (Figure 4B, D and F), as shown in the quantification (Figure 4J). These results indicate that other pathways are probably involved in eIF1A localization in vivo.
Figure 5.
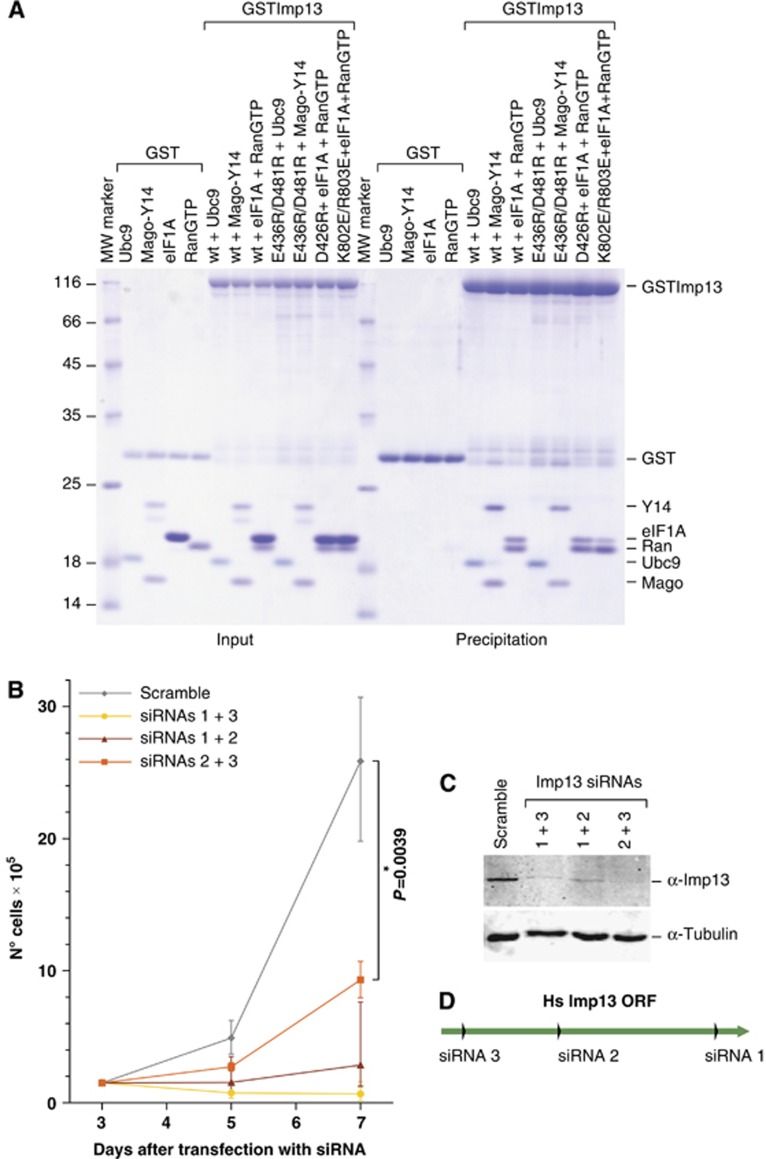
Imp13 function is essential in vivo. (A) Coomassie gel of co-precipitations to demonstrate that the Imp13 mutants impaired in the binding of one of the cargoes analysed are still able to associate to the other two cargoes. (B) HeLa cells were transfected with different siRNA combinations and counted at the indicated time points. Six independent experiments were performed. Statistical significance of the cell counts between knockdown and control condition was assessed with Wilcoxon–Mann–Whitney test (α 0.05). (C) Western blot showing the levels of endogenous Imp13 in the control and the Imp13 knockdown condition; α-tubulin served as a loading control. (D) Schematic representation of Imp13 open reading frame (ORF) showing the target sites of the siRNAs used.
Since eIF1A is present in the nucleolus, the site of ribosome assembly, we hypothesized that it might be exported together with other complexes. A possible association could be with the small ribosomal subunit, which is exported via a Crm1-dependent export pathway (Thomas and Kutay, 2003) and, at least in yeast, by a recently identified Mex67/Mtr2-dependent pathway (Faza et al, 2012). It has previously been shown that eIF1A cannot directly bind Crm1 (Mingot et al, 2001). We therefore analysed the localization of eIF1A wt and mutants in the presence of leptomycin B (LMB), a specific inhibitor of Crm1 (Wolff et al, 1997). LMB treatment causes a small but consistent and significant nuclear retention of eIF1A, regardless its capacity to bind to Imp13 (Supplementary Figure 5), suggesting that at least a fraction of eIF1A might be exported via a Crm1-dependent pathway reflecting an as yet unidentified association of eIF1A with other nuclear complexes.
We also examined the in vivo localization of two previously characterized Imp13 import cargoes, Mago-Y14 and Ubc9 (Bono et al, 2010; Grünwald and Bono, 2011). As expected, both cargoes are predominantly nuclear in HeLa cells (Kataoka et al, 2000, 2001; Rodriguez et al, 2001; Saitoh et al, 2002) (Figure 4L and N). Mutations previously shown to disrupt their interaction with Imp13 have also a strong impact on the localization of these cargoes (Bono et al, 2010; Grünwald and Bono, 2011) (Figure 4M and O–Q), suggesting that Imp13 is probably the main factor involved in Mago-Y14 and UBC9 import in vivo. Notably, an Imp13 mutant unable to bind to Mago-Y14 (Lys803Glu/Arg803GluI) is also less efficient in exporting eIF1A in vivo (Figure 4I and K), reflecting a reduction in binding to eIF1A in vitro (Figure 5A). Conversely, the Asp426ArgI Imp13 mutant, impaired in Ubc9 binding (Figure 5A), is as efficient as Imp13 wt in exporting eIF1A (Figure 4H and K).
To gain further insights into the role of Imp13-mediated transport in vivo, we depleted endogenous Imp13 in HeLa cells using three different siRNAs targeted to the Imp13 open reading frame (ORF) (Figure 5C and D). With every siRNA and combination of siRNAs, we observed a significant decrease in cell viability (Figure 5B), suggesting that Imp13 might be an essential protein.
Conformational changes of Imp13 in the cytoplasmic and nuclear states
Imp13 shows a marked conformational change when the cytoplasmic and nuclear states are compared (Figure 6). Dm Imp13 in the unbound state adopts an open conformation measuring about 24 Å between H2 and 20 (Figure 6A and F). In the cytoplasmic states of Imp13 bound to Mago-Y14 and Ubc9, the karyopherin also has a more extended conformation (Figure 6C, E and F). In the nucleus where it is associated with RanGTP, Imp13 is more compact with the C-terminal arch approaching the N-terminus (Figure 6B, D and F). Here, maximal compactness is observed in the binary complex with RanGTP (1 Å between H2 and H20). In complex 3, Imp13 shows a conformation similar to the one observed when in complex to RanGTP (Bono et al, 2010) (r.m.s.d. of 0.548 Å over 903 Cα) (Supplementary Figure 7C and D), suggesting that eIF1A is indeed missing in this complex. In the ternary complex with RanGTP and eIF1A, the C-terminus of Imp13 moves slightly away from the N-terminus to fit the export cargo within the C-terminal arch.
Figure 6.
Imp13 samples an ample spectrum of conformations. Cytoplasmic (A, C, E) and nuclear (B, D) states of Imp13 in the same view as in Figure 1B. Imp13 is represented as a tube trace in a colour gradient from grey to green. All cargoes are shown as cartoons with Ubc9 in purple-blue, Mago in blue, Y14 in magenta, Ran in yellow with GTP in black and eIF1A in brown. (F) Graphical representation of the distance from N-terminus to C-terminus measured from the line connecting H2B and H20C as shown in the box (top).
Remarkably, helix A of H5 undergoes partial melting and appears to work as a spacer to sense the RanGTP-bound state. In the RanGTP-bound state, helix H5B contacts the last helix H20C and therefore closes Imp13 into a ring (Figure 6B). In contrast, upon eIF1A binding, the H5A helix of Imp13 folds into a longer helix, breaking the H5B contact and establishing a new contact between the newly formed H5A helix and H20C helix (Figure 6D). Thereby, the toroidal ring of Imp13 opens up slightly at the hinge region at H14 to accommodate eIF1A, which concomitantly contacts the beginning of the H20C helix replacing the H5 interaction. In all Imp13 open conformation states, the import complexes as well as the unbound Imp13, the helix at H5 is also structured.
The spectrum of conformations sampled by Imp13 reflects the cumulative effect of changes distributed over the entire molecule with more prominent changes at the three hinge regions (Grünwald and Bono, 2011).
The structures of several exportins in complex with an export cargo have been solved. A comparison between the different export complexes of Cse1, Crm1, Xpo5 and Xpot with Imp13-RanGTP-eIF1A shows similar topology of Imp13 and Crm1 in the export cargo bound state (Matsuura and Stewart, 2004; Cook et al, 2009; Monecke et al, 2009; Okada et al, 2009; Dong et al, 2009b) (Supplementary Figure 7A and B). Although both karyopherins have a closed ring-like conformation, Imp13 binds the export cargo mainly with the inner concave surface, whereas Crm1 binds its cargoes on its outer surface (Monecke et al, 2009; Dong et al, 2009b). As in most exportins with the exception of Crm1, the positively charged interface of RanGTP directly contacts the cargo. In the apo form, Xpot and Imp13 demonstrate an overall open conformation that closes up upon RanGTP binding (Cook et al, 2009).
Export cargo dissociation and directionality of Imp13 transport
In the nucleus, RanGTP is very abundant and its binding to karyopherins is very tight, thus eIF1A will likely encounter Imp13-RanGTP complex in the closed conformation that will then open at the C-terminal arch to accommodate the cargo. In the cytoplasm, the disassembly of the export cargo-receptor complex is coupled to the activation of the GTPase activity of Ran by RanGAP, a cytoplasmic enhancer of RanGTPase activity (Bischoff et al, 1994). Prior to GTP hydrolysis, RanGTP is likely to be extracted from the complex by the concomitant recognition of the C-terminal region of RanGTP and the Ran binding domain of RanBP1. Similarly to what has been observed with other RanGTP-karyopherin complexes (Hellmuth et al, 1998; Kutay et al, 1998; Vetter et al, 1999b; Koyama and Matsuura, 2010), when the RanGTP-RanBP1 complex is superposed on the export complex, RanBP1 would clash with H14-15 of Imp13 whereas the extended acidic tail at the C-terminus of Ran would clash with H13 (Supplementary Figure 6C). This indicates that the mechanism for removing RanGTP and ending the export cycle is likely similar in Imp13 as in other karyopherins.
Comparison of Imp13 export complex and Imp13 apo structures gives insights into the directionality of the transport. In the apo form, the contact of eIF1A with RanGTP is missing. Moreover, if we optimally superpose the eIF1A binding site at H8-9 of Imp13 in the export complex and in the apo structure (residues 357–438 of Hs Imp13 in complex with RanGTP and eIF1A and 370–452 of Dm Imp13 unbound), the contact at the C-terminus of Imp13 is lost, together with the contact at the inter-loop between H10-11 (Supplementary Figure 6D). The latter includes Asp481I, which is involved in binding to eIF1A (Figure 2A and C) and is displaced in the apo structure. Therefore, in the Imp13 apo form, the complementarity for eIF1A is partially lost, with only the interaction surface at H7-10 being in place. The remaining contacts that compose the intact binding surface in the trimeric complex (at H6 and 11) are also displaced due to a divergence of the N- and C-termini of Imp13 that pushes them away from the eIF1A binding site (Supplementary Figure 6D). Therefore, apo Imp13 has a shape that is incompatible with eIF1A binding. This open form could provide expanded flexibility to allow Imp13 to associate with a variety of different import cargoes that it can recognize in the cytoplasm. It is likely that the tighter binding import cargoes will displace any residual association between eIF1A and Imp13 in the cytoplasm.
Whether the cytoplasmic import cargo-bound form of Imp13, after RanGTP hydrolysis and export cargo release, occurs through an unbound or eIF1A-bound intermediate is unclear. Our binding studies show that eIF1A binds poorly to Imp13 in the absence of RanGTP (Figure 2D and E). In addition, an Imp13 mutant with a C-terminal deletion (residues 1–673) is impaired in the binding to eIF1A indicating that the C-terminal contact of Imp13 to eIF1A is important for the binding (Figure 2C). However, our SEC data indicate that eIF1A remains partially associated with Imp13 in the absence of RanGTP, and so association with import cargoes might be necessary to clear residual eIF1A from Imp13 in the cytoplasm. This would imply that Imp13 could take two pathways after entry to the cytoplasm, one where binding of an import cargo is directly coupled to eIF1A ejection and one where Imp13 is released in an apo state before binding to other cargoes (Figure 7).
Figure 7.
The Imp13 pathway. Schematic display of the dynamic interactions that take place between Imp13 and its protein partners in the cell. In the cytoplasm, Imp13 in the unbound state adopts an open conformation that probably facilitates association of the cargoes to be imported. In the binary import complexes with Mago-Y14 or Ubc9, Imp13 maintains a rather open conformation and translocates through the NPCs into the nucleus. Here, high concentration and the high affinity binding of RanGTP to Imp13 promotes release of the import cargoes. Mago-Y14 is then free to be re-incorporated into an EJC on a newly transcribed and processed mRNP, while Ubc9 promotes SUMOylation of a large number of nuclear targets. In the binary complex with RanGTP, Imp13 adopts a compact conformation that favours the docking of eIF1A. A slight opening of Imp13 conformation at the C-terminal arch accommodates eIF1A side-by-side with RanGTP. The trimeric export complex crosses the NPCs to deliver the export cargo in the cytoplasm upon RanGTP extraction and hydrolysis. Some residual- or re-binding of eIF1A to Imp13 in the absence of RanGTP might occur in the cytoplasm. In this case, import cargo association to Imp13 will competitively displace eIF1A. Due to its small size that allows its diffusion through the NPCs (or possibly via a yet unknown active import mechanism) eIF1A is found in the nucleus at steady state. eIF1A export from the nucleus is not unique to Imp13 and might involve other export factors, such as Crm1, and might occur in complex with other proteins.
Conclusions
Imp13 is an essential import factor that also has a critical role as an export factor. The structure of the export complex of Imp13 with RanGTP and eIF1A shows how Imp13 can perform this dual duty. Imp13 in this complex adopts a compact conformation where eIF1A recognizes the inner surface of the C-terminal arch of Imp13 and is engaged in a stabilizing contact with RanGTP. eIF1A association with Imp13 is compatible with the presence of RanGTP, since the export cargo overall shape/structure does not clash with Ran and the binding sites do not overlap. Contacts on Imp13 and on RanGTP stabilize cargo binding for export. The binding mode of eIF1A to Imp13 is similar and spatially overlapping to the binding mode of Mago-Y14 to Imp13. In the latter case, however, the association with RanGTP is mutually exclusive. Due to the higher affinity of binding to Imp13 and high nuclear concentration of the small GTPase, the import cargo is released in the nucleus. At each stage of the import/export cycle Imp13 adopts a shape that allows complementary cargoes to compete for binding in the same compartment. The combination of shape, charge and size of the bound cargo will drive the concomitant or exclusive interactions with RanGTP and ultimately determine the directionality of the transport and the delivery of the cargoes in the appropriate compartment. The mechanism by which Imp13 can act as a bidirectional transport factor could be exploited by more karyopherins, suggesting that the range of bifunctional receptors might be larger than believed so far.
The apo structure explains the directionality of eIF1A export. In this state of Imp13, the binding site for eIF1A is partially distorted and the shape complementarity for eIF1A is lost. These data suggest two pathways for importin association in the cytoplasm: either Imp13 is released as a free apo molecule after RanGTP has been extracted, or import cargoes directly displace the export cargo concomitantly with RanGTP release (Figure 7).
We demonstrate that eIF1A is exported by Imp13 in vivo. However, the intracellular localization of eIF1A mutants designed based on the structure show that Imp13 export is functionally redundant and that a Crm1-dependent pathway might be important for eIF1A localization. We also show that Imp13 function is required for viability in human cells, though this may relate to its role in other transport pathways. Further investigations will clarify eIF1A function in the nucleus and eIF1A alternative export pathways.
Materials and methods
DNA constructs
For mammalian expression, Hs Imp13, Mago, Y14 and Ubc9 were cloned in a pEGFP-C1 vector (Clontech); to obtain HA-tagged fusion proteins, the EGFP sequence was deleted and substituted with one encoding for the HA epitope (YPYDVPDYA). Hs eIF1A was cloned in a pEGFP-N3 vector (Clontech). Hs fibrillarin (FBL) was amplified from HeLa cDNA and cloned into a modified pEGFP-C1 vector, in which the sequence encoding EGFP was replaced by the ORF of mCherry (pmCherry-C1). GFP-NES was obtained by inserting an oligonucleotide encoding for the nuclear export signal (NES) of the protein kinase A inhibitor (PKI) (Güttler et al, 2010) into the XhoI/HindIII sites of the pEGFP-C1 vector.
Protein expression and purification
Recombinant Hs and Dm Imp13 were expressed as described before (Grünwald and Bono, 2011). Hs Ran Q69L (1–180) and Hs eIF1A full-length and truncated constructs were cloned in a pETMCN vector (Diebold et al, 2011) with an N-terminal hexahistidin tag. The recombinant Hs Ran was prepared similarly to the yeast orthologue (Bono et al, 2010). The eIF1A constructs were expressed in the E. coli strain BL21 (DE3) Gold in autoinducing medium (Studier, 2005) at 20°C overnight. The cleared cell lysates were affinity purified by Ni2+ NTA in buffer A (20 mM Tris pH 7.5, 300 mM NaCl, 5 mM imidazole and 1 mM β-mercaptoethanol) with a gradient of 5–250 mM imidazole, followed by dialysis in the presence of TEV protease in buffer B (20 mM Tris pH 7.5, 300 mM NaCl, 1 mM DTT). The protein was further purified by a second step of Ni2+ NTA chromatography and the collected flow-through applied to a cation-exchange column in buffer B with a gradient of 300–1000, mM NaCl.
Site-directed mutagenesis was used to introduce a third methionine at residue 65 in pETMCN His eIF1A (1–112). To produce eIF1A Leu65Met 1–112 SeMet-labelled protein, the construct was expressed in the E. coli strain DL41 auxotroph for methionine in M9 medium and starved before addition of SeMet. The protein was purified as described above for the unmodified protein.
For complex formation, proteins were mixed in a 1:1.5:2 ratio of Imp13-RanGTP-eIF1A in complex buffer A (20 mM Tris pH 7.5, 50 mM NaCl, 4 mM MgCl2 and 1 mM DTT) and incubated for 1 h at 4°C. The complexes were further purified by SEC and concentrated to 15 mg/ml prior to setting up crystallization trials.
Analytical SEC
For analytical SEC, 1 mg of Imp13 was injected directly or after incubation for 1 h at 4°C with 1.5 × excess of RanGTP and/or 2 × excess of different eIF1A constructs. The runs were carried out with a Superdex 200 10/300GL gel filtration column (GE Healthcare) using the UV absorbance at 280 nm. A sample of each peak fraction was analysed by 15% SDS–PAGE. Proteins were visualized by Coomassie staining.
Crystallization, data collection and analysis
Optimized crystals of the ternary export complex were obtained in 6% PEG 8000, 0.1 M Na-citrate, pH 5.8 grown at 18°C by vapour diffusion. For data collection, crystals were cryo-protected with mother liquor supplemented with 30% glycerol and flash frozen in liquid nitrogen. The crystals diffracted to 3.6 Å resolution and belong to the spacegroup C2 with cell dimensions of a=186.50 Å, b=100.4 Å, c=274.72 Å, α=γ=90° β=90.909°. The asymmetric unit contains three independent complexes. Data were processed and scaled using XDS (Kabsch, 1993). The structure was solved by MR using PHASER (McCoy et al, 2007) and the Imp13-RanGTP structure (pdb id.: 2x19) as a search model. For placing eIF1A into the electron density, SAD data of the SeMet substituted eIF1A Leu65Met in complex with Imp13 and RanGTP were collected and an anomalous Fourier map was calculated. Refinement was carried out using iterative cycles of model building in COOT (Emsley and Cowtan, 2004) and restrained refinement including NCS in BUSTER (Smart et al, 2012) and B-group refinement in PHENIX (Adams et al, 2010).
Crystals of unbound Dm Imp13 grew in 0.1 M NaoAc, pH 4.9 with addition of 0.8 M ammonium tartrate at 18°C by vapour diffusion method (8 mg/ml). The crystals diffracted to 3 Å and belonged to spacegroup P43212 with cell dimensions of a=b=167.72, c=95.630 and α=β=γ=90°. The structure was solved by MR using PHASER and Dm Imp13 as a search model (pdb id.: 2x1g) fragmented into three regions encompassing H2-8, H9-12; H15-20. Iterative cycles of model building and restrained refinement were carried out in COOT and PHENIX (Emsley and Cowtan, 2004; Adams et al, 2010).
All diffraction data were collected at the PXII beamline of the Swiss Light Source (Villigen, Switzerland).
In vitro binding assays
For binding assays, GST-tagged Imp13 was incubated with purified binding partners (7 μg of eIF1A or 4 μg of each of the other proteins) in binding buffer (20 mM HEPES pH 7.5, 50 mM NaCl, 4 mM MgCl2, 10% glycerol, 0.01% (v/v) Nonidet P40 and 1 mM DTT) in a final volume of 60 μl for 1 h at 4°C. The GST-tagged protein was immobilized on 15 μl of glutathione agarose beads (Macherey-Nagel) and incubated for 1 h at 4°C. The beads were washed three times with 500 μl of binding buffer and eluted with 8 μl of SDS loading buffer. The eluted proteins were analysed by 15% SDS–PAGE and visualized by Coomassie staining.
Cell culture and transfection
HeLa cells were maintained in Dulbecco’s Modified Eagle’s medium (DMEM) supplemented with fetal bovine serum (10%), L-glutamine (2 mM), penicillin (100 units/ml) and streptomycin (100 μg/ml; all from Invitrogen). Cells were grown in a humidified incubator with 5% CO2 at 37°C. Transfections were performed using Lipofectamine 2000 (Invitrogen), following manufacturer’s recommendations.
Knockdown efficiency was assessed by western blot using anti-Imp13 antibody (produced in-house) and anti-tubulin as a loading control (1:10 000; Sigma).
For immunofluorescence, the cells were grown directly on glass coverslips, in 24-well plates. The transfection mixtures contained 600 ng of one or more plasmids encoding GFP-, mCherry- or HA-protein fusions. The cells were fixed 48 h after transfection.
For live-cell imaging, the cells were grown in 24-well plates. The transfection mixtures contained 600 ng of one or more plasmids encoding GFP-, mCherry- or HA-protein fusions. The day after transfection, the cells were transferred in 35 mm glass-bottom dishes (Greiner Bio-One). The cells were imaged 48 h after transfection. Where indicated, the cells were treated with 20 ng/ml Leptomycin B (LMB; Sigma-Aldrich) for 5 h before imaging.
Fluorescence microscopy, image processing and data analysis
For immunofluorescence, HeLa cells were fixed with 4% paraformaldehyde in PBS for 10 min, and permeabilized for 10 min with PBS containing 0.5% Triton X-100. Anti-eIF1AY (1:50, #Y055246, abm) and anti-Imp13 were used for detection. Incubation with the primary antibodies was performed in 0.1 M Tris–HCl pH 8 with 5% donkey serum, for 60 min at room temperature. Cells were then washed 2 × in 0.1 M Tris–HCl pH 8 and 3 × in 0.1 M Tris–HCl pH 8 with 1 M NaCl. Appropriate Alexa Fluor 488- or Alexa Fluor 555-coupled secondary antibodies (Molecular Probes) were diluted 1:1.000 in 0.1 M Tris–HCl pH 8 with 1 M NaCl and incubated 45 min at room temperature; where stated, Alexa 633-phalloidin (Molecular Probes) was also included. DNA was stained with Hoechst 33342 (Molecular Probes). Samples were mounted using Fluoromount-G (Southern Biotechnology Associates, Inc.). For live imaging, HeLa cells grown in 35 mm glass-bottom dishes were maintained in CO2-independent medium (Gibco) supplemented with fetal bovine serum (1%).
Images were acquired on an Olympus FluoView FV1000 confocal microscope, equipped with a UPLSAPO 60X NA:1.20 water objective. Z-stacks were taken to scan the cell volume, with slices every 0.49 μm. Where indicated, DIC images were also acquired to define the boundaries of cytoplasm and nucleus. Image processing and quantification was performed with ImageJ. Briefly, all the slices of a Z-stack were summed and, after background subtraction, total fluorescence intensity was measured for each cellular compartment. The values obtained for the nuclear compartment were then expressed as a fraction of the corresponding total fluorescence. Statistical representation and analysis were performed with gnumeric (http://projects.gnome.org/gnumeric/).
Differential scanning fluorimetry
Previous attempts at measuring binding affinities with ITC and Biacore proved unsuccessful due to aggregation and precipitation of Imp13. For this reason, we used DSF to determine the dissociation constants of Imp13 with various cargoes (Niesen et al, 2007). The cargo was titrated in 18–20 steps to a fixed concentration of Imp13 in buffer containing 20 mM HEPES pH 7.5, 100 mM NaCl, 4 mM MgCl2 and 15% glycerol in a 96-well plate (BIOZYME). For Mago-Y14 and RanGTP Q69L (residues 1–180), a fixed concentration of 0.5 μM Imp13 was used, while Ubc9 was titrated to 1 μM Imp13. eIF1A was titrated to 1 μM of preformed and purified Imp13-RanGTP complex. To each titration step, 5 × SYPRO Orange (SIGMA) was added, the wells were sealed with Flat Caps Strips (BIOZYME) and incubated for 30 min at RT. Melting curves were measured with BIO-RAD DNA Engine Chromo4 with a temperature gradient from 25 to 85 °C with an increment of 1°C/s and an adjustment time of 5 s. Two channels, SYBR and CY3, were used for excitation and absorption. The melting temperature (Tm) was calculated with the program GraphPad PRISM5 by fitting the Boltzman equation to the measured melting curves. For the calculation of the KD, the Tm was plotted against the concentration of the cargo and fitted with an exponential equation.
Accession numbers
The coordinates and structure factors have been deposited in the Macromolecular Structure Database of European Bioinformatic Institute (EBI) with ID code 3zjy and 3zkv for Imp13 ternary export complex and unbound form, respectively.
Supplementary Material
Acknowledgments
We wish to thank J Basquin, K Valer-Saldana and S Pleyer at the MPI-Martinsried crystallization facility and C Basquin and Y Grömping for advice on biophysical measurements. We also thank E Izaurralde (MPI-Tübingen) for plasmids, W Antonin (MPI-Tübingen) and F Baumann (Hertie Institut, Tuebingen) for cell lines, R Neher for advice on statistical analysis and the staff at the Swiss Light Source synchrotron for assistance during data collection. We are grateful to C Liebig and G Jékely for help in fluorescence microscopy data acquisition and analysis and to S Wachter for help with protein purification. We thank A Cook and E Conti for discussion and critical reading of the manuscript. This study was supported by the Max Planck Gesellshaft and by the DFG grant BO3588/1-1.
Author contributions: Biochemical, biophysical and structural work was done by MG; cell-based assays by DL; FB supervised the project. All authors wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken CE, Lorsch JR (2012) A mechanistic overview of translation initiation in eukaryotes. Nat Struct Mol Biol 19: 568–576 [DOI] [PubMed] [Google Scholar]
- Askjaer P, Bachi A, Wilm M, Bischoff FR, Weeks DL, Ogniewski V, Ohno M, Niehrs C, Kjems J, Mattaj IW, Fornerod M (1999) RanGTP-regulated interactions of CRM1 with nucleoporins and a shuttling DEAD-box helicase. Mol Cell Biol 19: 6276–6285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battiste JL, Pestova TV, Hellen CU, Wagner G (2000) The eIF1A solution structure reveals a large RNA-binding surface important for scanning function. Mol Cell 5: 109–119 [DOI] [PubMed] [Google Scholar]
- Bayliss R, Littlewood T, Stewart M (2000) Structural basis for the interaction between FxFG nucleoporin repeats and importin-beta in nuclear trafficking. Cell 102: 99–108 [DOI] [PubMed] [Google Scholar]
- Bhardwaj A, Cingolani G (2010) Conformational selection in the recognition of the snurportin importin beta binding domain by importin beta. Biochemistry 49: 5042–5047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff FR, Görlich D (1997) RanBP1 is crucial for the release of RanGTP from importin beta-related nuclear transport factors. FEBS Lett 419: 249–254 [DOI] [PubMed] [Google Scholar]
- Bischoff FR, Klebe C, Kretschmer J, Wittinghofer A, Ponstingl H (1994) RanGAP1 induces GTPase activity of nuclear Ras-related Ran. Proc Natl Acad Sci USA 91: 2587–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono F, Cook AG, Grünwald M, Ebert J, Conti E (2010) Nuclear import mechanism of the EJC component Mago-Y14 revealed by structural studies of importin 13. Mol Cell 37: 211–222 [DOI] [PubMed] [Google Scholar]
- Cansizoglu AE, Chook YM (2007) Conformational heterogeneity of karyopherin beta2 is segmental. Structure (London, England: 1993) 15: 1431–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cansizoglu AE, Lee BJ, Zhang ZC, Fontoura BMA, Chook YM (2007) Structure-based design of a pathway-specific nuclear import inhibitor. Nat Struct Mol Biol 14: 452–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66: 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani G, Bednenko J, Gillespie MT, Gerace L (2002) Molecular basis for the recognition of a nonclassical nuclear localization signal by importin beta. Mol Cell 10: 1345–1353 [DOI] [PubMed] [Google Scholar]
- Cingolani G, Petosa C, Weis K, Müller CW (1999) Structure of importin-beta bound to the IBB domain of importin-alpha. Nature 399: 221–229 [DOI] [PubMed] [Google Scholar]
- Cook A, Bono F, Jinek M, Conti E (2007) Structural biology of nucleocytoplasmic transport. Annu Rev Biochem 76: 647–671 [DOI] [PubMed] [Google Scholar]
- Cook A, Fernandez E, Lindner D, Ebert J, Schlenstedt G, Conti E (2005) The structure of the nuclear export receptor Cse1 in its cytosolic state reveals a closed conformation incompatible with cargo binding. Mol Cell 18: 355–367 [DOI] [PubMed] [Google Scholar]
- Cook AG, Fukuhara N, Jinek M, Conti E (2009) Structures of the tRNA export factor in the nuclear and cytosolic states. Nature 461: 60–65 [DOI] [PubMed] [Google Scholar]
- Deane R, Schäfer W, Zimmermann HP, Mueller L, Görlich D, Prehn S, Ponstingl H, Bischoff FR (1997) Ran-binding protein 5 (RanBP5) is related to the nuclear transport factor importin-beta but interacts differently with RanBP1. Mol Cellular Biol 17: 5087–5096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebold M-L, Fribourg S, Koch M, Metzger T, Romier C (2011) Deciphering correct strategies for multiprotein complex assembly by co-expression: application to complexes as large as the histone octamer. J Struct Biol 175: 178–188 [DOI] [PubMed] [Google Scholar]
- Dong X, Biswas A, Chook YM (2009a) Structural basis for assembly and disassembly of the CRM1 nuclear export complex. Nat Struct Mol Biol 16: 558–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Biswas A, Süel KE, Jackson LK, Martinez R, Gu H, Chook YM (2009b) Structural basis for leucine-rich nuclear export signal recognition by CRM1. Nature 458: 1136–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Faza MB, Chang Y, Occhipinti L, Kemmler S, Panse VG (2012) Role of Mex67-Mtr2 in the Nuclear Export of 40S Pre-Ribosomes. PLoS Genet 8: e1002915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forwood JK, Lange A, Zachariae U, Marfori M, Preast C, Grubmüller H, Stewart M, Corbett AH, Kobe B (2010) Quantitative structural analysis of importin-β flexibility: paradigm for solenoid protein structures. Structure (London, England: 1993) 18: 1171–1183 [DOI] [PubMed] [Google Scholar]
- Giagtzoglou N, Lin YQ, Haueter C, Bellen HJ (2009) Importin 13 regulates neurotransmitter release at the Drosophila neuromuscular junction. J Neurosci 29: 5628–5639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontan C, Guttler T, Engelen E, Demmers J, Fornerod M, Grosveld FG, Tibboel D, Gorlich D, Poot RA, Rottier RJ (2009) Exportin 4 mediates a novel nuclear import pathway for Sox family transcription factors. J Cell Biol 185: 27–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünwald M, Bono F (2011) Structure of Importin13-Ubc9 complex: nuclear import and release of a key regulator of sumoylation. EMBO J 30: 427–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Dabrowski M, Bischoff FR, Kutay U, Bork P, Hartmann E, Prehn S, Izaurralde E (1997) A novel class of RanGTP binding proteins. J Cell Biol 138: 65–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Kutay U (1999) Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol 15: 607–660 [DOI] [PubMed] [Google Scholar]
- Güttler T, Madl T, Neumann P, Deichsel D, Corsini L, Monecke T, Ficner R, Sattler M, Görlich D (2010) NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1. Nat Struct Mol Biol 17: 1367–1376 [DOI] [PubMed] [Google Scholar]
- Hellmuth K, Lau DM, Bischoff FR, Künzler M, Hurt E, Simos G (1998) Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol Cell Biol 18: 6374–6386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG (2011) Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol Mol Biol Rev 75: 434–467 first page of table of contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imasaki T, Shimizu T, Hashimoto H, Hidaka Y, Kose S, Imamoto N, Yamada M, Sato M (2007) Structural basis for substrate recognition and dissociation by human transportin 1. Mol Cell 28: 57–67 [DOI] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CUT, Pestova TV (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11: 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Cryst 26: 795–800 [Google Scholar]
- Kahle J, Baake M, Doenecke D, Albig W (2005) Subunits of the heterotrimeric transcription factor NF-Y are imported into the nucleus by distinct pathways involving importin beta and importin 13. Mol Cell Biol 25: 5339–5354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka N, Diem MD, Kim VN, Yong J, Dreyfuss G (2001) Magoh, a human homolog of Drosophila mago nashi protein, is a component of the splicing-dependent exon-exon junction complex. EMBO J 20: 6424–6433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka N, Yong J, Kim VN, Velazquez F, Perkinson RA, Wang F, Dreyfuss G (2000) Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol Cell 6: 673–682 [DOI] [PubMed] [Google Scholar]
- Koyama M, Matsuura Y (2010) An allosteric mechanism to displace nuclear export cargo from CRM1 and RanGTP by RanBP1. EMBO J 29: 2002–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U, Bischoff FR, Kostka S, Kraft R, Görlich D (1997) Export of importin alpha from the nucleus is mediated by a specific nuclear transport factor. Cell 90: 1061–1071 [DOI] [PubMed] [Google Scholar]
- Kutay U, Lipowsky G, Izaurralde E, Bischoff FR, Schwarzmaier P, Hartmann E, Görlich D (1998) Identification of a tRNA-specific nuclear export receptor. Mol Cell 1: 359–369 [DOI] [PubMed] [Google Scholar]
- Lee SJ, Matsuura Y, Liu SM, Stewart M (2005) Structural basis for nuclear import complex dissociation by RanGTP. Nature 435: 693–696 [DOI] [PubMed] [Google Scholar]
- Lee SJ, Sekimoto T, Yamashita E, Nagoshi E, Nakagawa A, Imamoto N, Yoshimura M, Sakai H, Chong KT, Tsukihara T, Yoneda Y (2003) The structure of importin-beta bound to SREBP-2: nuclear import of a transcription factor. Science (New York, NY) 302: 1571–1575 [DOI] [PubMed] [Google Scholar]
- Liang J, Ke G, You W, Peng Z, Lan J, Kalesse M, Tartakoff AM, Kaplan F, Tao T (2008) Interaction between importin 13 and myopodin suggests a nuclear import pathway for myopodin. Mol Cell Biochem 307: 93–100 [DOI] [PubMed] [Google Scholar]
- Lipowsky G, Bischoff FR, Izaurralde E, Kutay U, Schäfer S, Gross HJ, Beier H, Görlich D (1999) Coordination of tRNA nuclear export with processing of tRNA. RNA (New York, NY) 5: 539–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipowsky G, Bischoff FR, Schwarzmaier P, Kraft R, Kostka S, Hartmann E, Kutay U, Görlich D (2000) Exportin 4: a mediator of a novel nuclear export pathway in higher eukaryotes. EMBO J 19: 4362–4371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura Y, Stewart M (2004) Structural basis for the assembly of a nuclear export complex. Nature 432: 872–877 [DOI] [PubMed] [Google Scholar]
- McCoy A, Grosse-Kunstleve R, Adams P, Winn M, Storoni L, Read R (2007) Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingot JM, Kostka S, Kraft R, Hartmann E, Görlich D (2001) Importin 13: a novel mediator of nuclear import and export. EMBO J 20: 3685–3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrousis G, Olia AS, Walker-Kopp N, Cingolani G (2008) Molecular basis for the recognition of snurportin 1 by importin beta. J Biol Chem 283: 7877–7884 [DOI] [PubMed] [Google Scholar]
- Mohr D, Frey S, Fischer T, Güttler T, Görlich D (2009) Characterisation of the passive permeability barrier of nuclear pore complexes. EMBO J 28: 2541–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monecke T, Güttler T, Neumann P, Dickmanns A, Görlich D, Ficner R (2009) Crystal structure of the nuclear export receptor CRM1 in complex with Snurportin1 and RanGTP. Science (New York, NY) 324: 1087–1091 [DOI] [PubMed] [Google Scholar]
- Niesen FH, Berglund H, Vedadi M (2007) The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc 2: 2212–2221 [DOI] [PubMed] [Google Scholar]
- Okada C, Yamashita E, Lee SJ, Shibata S, Katahira J, Nakagawa A, Yoneda Y, Tsukihara T (2009) A High-Resolution Structure of the Pre-microRNA Nuclear Export Machinery. Science (New York, NY) 326: 1275–1279 [DOI] [PubMed] [Google Scholar]
- Paraskeva E, Izaurralde E, Bischoff FR, Huber J, Kutay U, Hartmann E, Lührmann R, Görlich D (1999) CRM1-mediated recycling of snurportin 1 to the cytoplasm. J Cell Biol 145: 255–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V (2007) The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell 26: 41–50 [DOI] [PubMed] [Google Scholar]
- Ploski JE, Shamsher MK, Radu A (2004) Paired-type homeodomain transcription factors are imported into the nucleus by karyopherin 13. Mol Cell Biol 24: 4824–4834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez MS, Dargemont C, Hay RT (2001) SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem 276: 12654–12659 [DOI] [PubMed] [Google Scholar]
- Saitoh H, Pizzi MD, Wang J (2002) Perturbation of SUMOlation enzyme Ubc9 by distinct domain within nucleoporin RanBP2/Nup358. J Biol Chem 277: 4755–4763 [DOI] [PubMed] [Google Scholar]
- Sali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234: 779–815 [DOI] [PubMed] [Google Scholar]
- Smart OS, Womack TO, Flensburg C, Keller P, Paciorek W, Sharff A, Vonrhein C, Bricogne G (2012) Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr D Biol Crystallogr 68: 368–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW (2005) Protein production by auto-induction in high density shaking cultures. Protein Expr Purif 41: 207–234 [DOI] [PubMed] [Google Scholar]
- Tao T, Lan J, Lukacs GL, Haché RJG, Kaplan F (2006) Importin 13 regulates nuclear import of the glucocorticoid receptor in airway epithelial cells. Am J Respir Cell Mol Biol 35: 668–680 [DOI] [PubMed] [Google Scholar]
- Thomas F, Kutay U (2003) Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J Cell Sci 116: 2409–2419 [DOI] [PubMed] [Google Scholar]
- Vetter IR, Arndt A, Kutay U, Görlich D, Wittinghofer A (1999a) Structural view of the Ran-Importin beta interaction at 2.3 A resolution. Cell 97: 635–646 [DOI] [PubMed] [Google Scholar]
- Vetter IR, Nowak C, Nishimoto T, Kuhlmann J, Wittinghofer A (1999b) Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: implications for nuclear transport. Nature 398: 39–46 [DOI] [PubMed] [Google Scholar]
- Walker P, Doenecke D, Kahle J (2009) Importin 13 mediates nuclear import of histone fold containing chrac heterodimers. J Biol Chem 284: 11652–11662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlwend D, Strasser A, Dickmanns A, Ficner R (2007) Structural basis for RanGTP independent entry of spliceosomal U snRNPs into the nucleus. J Mol Biol 374: 1129–1138 [DOI] [PubMed] [Google Scholar]
- Wolff B, Sanglier JJ, Wang Y (1997) Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem Biol 4: 139–147 [DOI] [PubMed] [Google Scholar]
- Xu D, Farmer A, Chook YM (2010) Recognition of nuclear targeting signals by Karyopherin-β proteins. Curr Opin Struct Biol 20: 782–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Blobel G (2001) The karyopherin Kap142p/Msn5p mediates nuclear import and nuclear export of different cargo proteins. J Cell Biol 152: 729–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZC, Chook YM (2012) Structural and energetic basis of ALS-causing mutations in the atypical proline-tyrosine nuclear localization signal of the Fused in Sarcoma protein (FUS). Proc Natl Acad Sci USA 109: 12017–12021 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.