Abstract
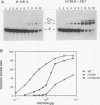
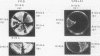
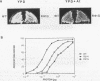
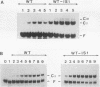
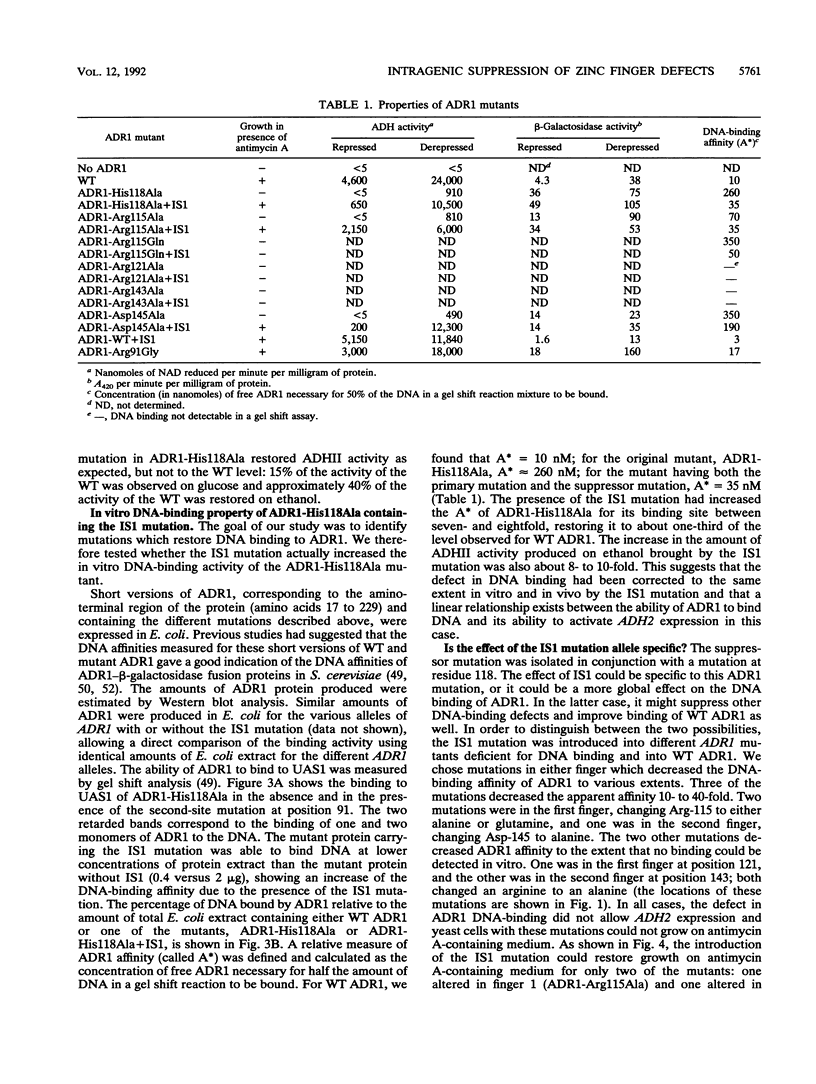
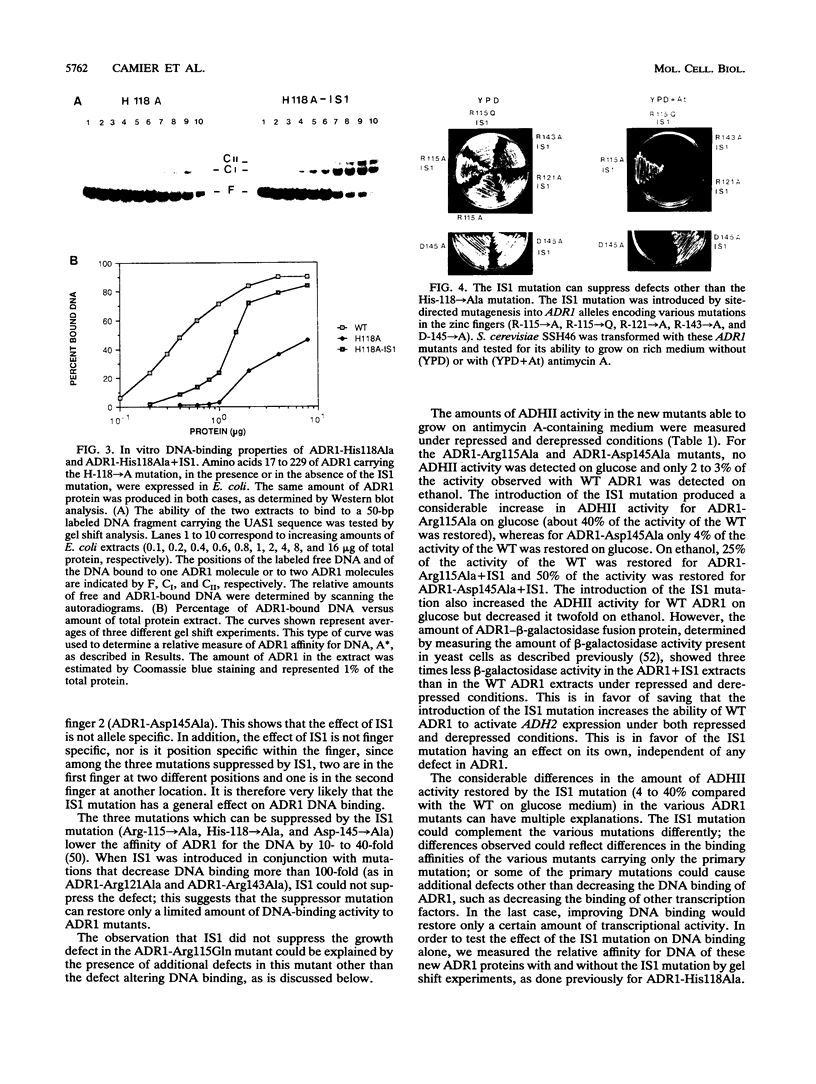
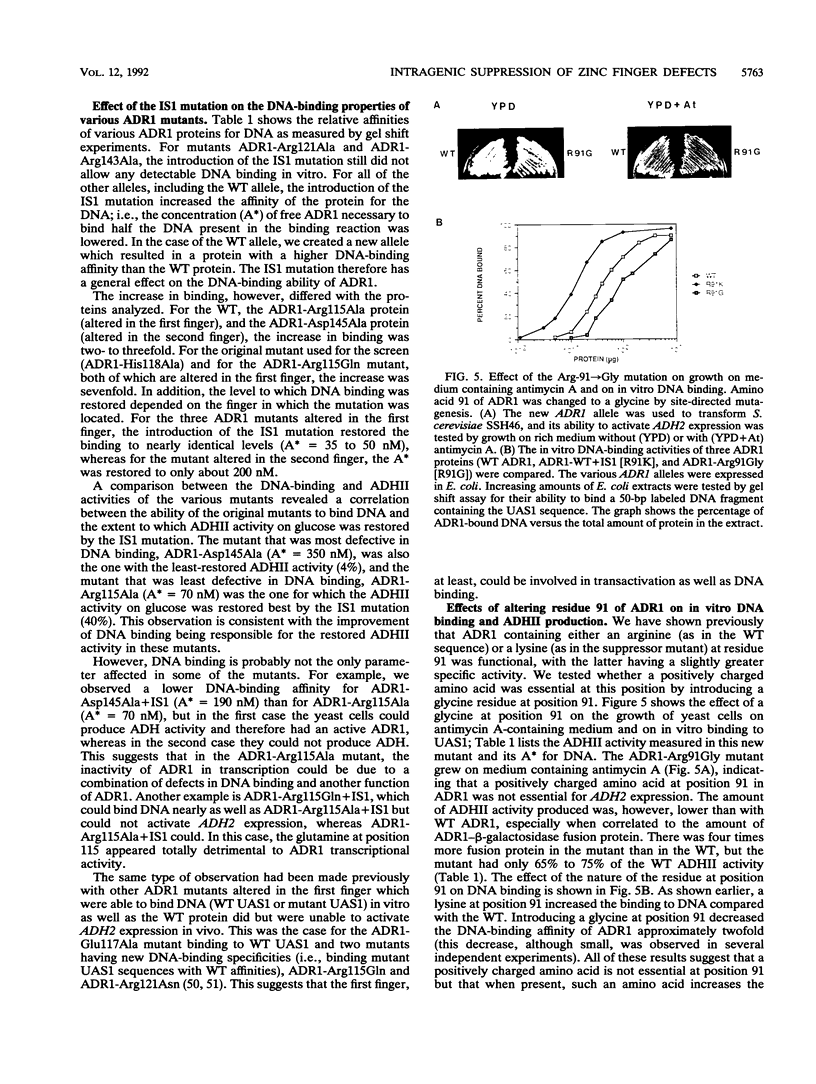
A second-site mutation that restored DNA binding to ADR1 mutants altered at different positions in the two zinc fingers was identified. This mutation (called IS1) was a conservative change of arginine 91 to lysine in a region amino terminal to the two zinc fingers and known from previous experiments to be necessary for DNA binding. IS1 increased binding to the UAS1 sequence two- to sevenfold for various ADR1 mutants and twofold for wild-type ADR1. The change of arginine 91 to glycine decreased binding twofold, suggesting that this arginine is involved in DNA binding in the wild-type protein. The increase in binding by IS1 did not involve protein-protein interactions between the two ADR1 monomers, nor did it require the presence of the sequences flanking UAS1. However, the effect of IS1 was influenced by the sequence of the first finger, suggesting that interactions between the region amino terminal to the fingers and the fingers themselves could exist. A model for the role of the amino-terminal region based on these results and sequence homologies with other DNA-binding motifs is proposed.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bellefroid E. J., Poncelet D. A., Lecocq P. J., Revelant O., Martial J. A. The evolutionarily conserved Krüppel-associated box domain defines a subfamily of eukaryotic multifingered proteins. Proc Natl Acad Sci U S A. 1991 May 1;88(9):3608–3612. doi: 10.1073/pnas.88.9.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg J. M. Zinc fingers and other metal-binding domains. Elements for interactions between macromolecules. J Biol Chem. 1990 Apr 25;265(12):6513–6516. [PubMed] [Google Scholar]
- Blumberg H., Eisen A., Sledziewski A., Bader D., Young E. T. Two zinc fingers of a yeast regulatory protein shown by genetic evidence to be essential for its function. 1987 Jul 30-Aug 5Nature. 328(6129):443–445. doi: 10.1038/328443a0. [DOI] [PubMed] [Google Scholar]
- Butler G., Thiele D. J. ACE2, an activator of yeast metallothionein expression which is homologous to SWI5. Mol Cell Biol. 1991 Jan;11(1):476–485. doi: 10.1128/mcb.11.1.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill M. E., Suzuki M. 'SPKK' motifs prefer to bind to DNA at A/T-rich sites. EMBO J. 1989 Dec 20;8(13):4189–4195. doi: 10.1002/j.1460-2075.1989.tb08604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriacy M. Isolation and characterization of further cis- and trans-acting regulatory elements involved in the synthesis of glucose-repressible alcohol dehydrogenase (ADHII) in Saccharomyces cerevisiae. Mol Gen Genet. 1979 Nov;176(3):427–431. doi: 10.1007/BF00333107. [DOI] [PubMed] [Google Scholar]
- Clarke N. D., Beamer L. J., Goldberg H. R., Berkower C., Pabo C. O. The DNA binding arm of lambda repressor: critical contacts from a flexible region. Science. 1991 Oct 11;254(5029):267–270. doi: 10.1126/science.254.5029.267. [DOI] [PubMed] [Google Scholar]
- Denis C. L., Ciriacy M., Young E. T. A positive regulatory gene is required for accumulation of the functional messenger RNA for the glucose-repressible alcohol dehydrogenase from Saccharomyces cerevisiae. J Mol Biol. 1981 Jun 5;148(4):355–368. doi: 10.1016/0022-2836(81)90181-9. [DOI] [PubMed] [Google Scholar]
- Descombes P., Chojkier M., Lichtsteiner S., Falvey E., Schibler U. LAP, a novel member of the C/EBP gene family, encodes a liver-enriched transcriptional activator protein. Genes Dev. 1990 Sep;4(9):1541–1551. doi: 10.1101/gad.4.9.1541. [DOI] [PubMed] [Google Scholar]
- Dower W. J., Miller J. F., Ragsdale C. W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988 Jul 11;16(13):6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowzer C. E., Kelly J. M. Analysis of the creA gene, a regulator of carbon catabolite repression in Aspergillus nidulans. Mol Cell Biol. 1991 Nov;11(11):5701–5709. doi: 10.1128/mcb.11.11.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen A., Taylor W. E., Blumberg H., Young E. T. The yeast regulatory protein ADR1 binds in a zinc-dependent manner to the upstream activating sequence of ADH2. Mol Cell Biol. 1988 Oct;8(10):4552–4556. doi: 10.1128/mcb.8.10.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairall L., Harrison S. D., Travers A. A., Rhodes D. Sequence-specific DNA binding by a two zinc-finger peptide from the Drosophila melanogaster Tramtrack protein. J Mol Biol. 1992 Jul 20;226(2):349–366. doi: 10.1016/0022-2836(92)90952-g. [DOI] [PubMed] [Google Scholar]
- Falzon M., Chung S. Y. The expression of rat homeobox-containing genes is developmentally regulated and tissue specific. Development. 1988 Jul;103(3):601–610. doi: 10.1242/dev.103.3.601. [DOI] [PubMed] [Google Scholar]
- Harrison S. C. A structural taxonomy of DNA-binding domains. Nature. 1991 Oct 24;353(6346):715–719. doi: 10.1038/353715a0. [DOI] [PubMed] [Google Scholar]
- Hartshorne T. A., Blumberg H., Young E. T. Sequence homology of the yeast regulatory protein ADR1 with Xenopus transcription factor TFIIIA. Nature. 1986 Mar 20;320(6059):283–287. doi: 10.1038/320283a0. [DOI] [PubMed] [Google Scholar]
- Hecht M. H., Sauer R. T. Phage lambda repressor revertants. Amino acid substitutions that restore activity to mutant proteins. J Mol Biol. 1985 Nov 5;186(1):53–63. doi: 10.1016/0022-2836(85)90256-6. [DOI] [PubMed] [Google Scholar]
- Jordan S. R., Pabo C. O. Structure of the lambda complex at 2.5 A resolution: details of the repressor-operator interactions. Science. 1988 Nov 11;242(4880):893–899. doi: 10.1126/science.3187530. [DOI] [PubMed] [Google Scholar]
- Keller A. D., Maniatis T. Only two of the five zinc fingers of the eukaryotic transcriptional repressor PRDI-BF1 are required for sequence-specific DNA binding. Mol Cell Biol. 1992 May;12(5):1940–1949. doi: 10.1128/mcb.12.5.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knöchel W., Pöting A., Köster M., el Baradi T., Nietfeld W., Bouwmeester T., Pieler T. Evolutionary conserved modules associated with zinc fingers in Xenopus laevis. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6097–6100. doi: 10.1073/pnas.86.16.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The DNA binding domain of the rat liver nuclear protein C/EBP is bipartite. Science. 1989 Mar 31;243(4899):1681–1688. doi: 10.1126/science.2494700. [DOI] [PubMed] [Google Scholar]
- Laughon A. DNA binding specificity of homeodomains. Biochemistry. 1991 Dec 3;30(48):11357–11367. doi: 10.1021/bi00112a001. [DOI] [PubMed] [Google Scholar]
- Lee M. S., Gippert G. P., Soman K. V., Case D. A., Wright P. E. Three-dimensional solution structure of a single zinc finger DNA-binding domain. Science. 1989 Aug 11;245(4918):635–637. doi: 10.1126/science.2503871. [DOI] [PubMed] [Google Scholar]
- Miller J., McLachlan A. D., Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai K., Nakaseko Y., Nasmyth K., Rhodes D. Zinc-finger motifs expressed in E. coli and folded in vitro direct specific binding to DNA. Nature. 1988 Mar 17;332(6161):284–286. doi: 10.1038/332284a0. [DOI] [PubMed] [Google Scholar]
- Nardelli J., Gibson T. J., Vesque C., Charnay P. Base sequence discrimination by zinc-finger DNA-binding domains. Nature. 1991 Jan 10;349(6305):175–178. doi: 10.1038/349175a0. [DOI] [PubMed] [Google Scholar]
- Nehlin J. O., Ronne H. Yeast MIG1 repressor is related to the mammalian early growth response and Wilms' tumour finger proteins. EMBO J. 1990 Sep;9(9):2891–2898. doi: 10.1002/j.1460-2075.1990.tb07479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson H. C., Sauer R. T. Lambda repressor mutations that increase the affinity and specificity of operator binding. Cell. 1985 Sep;42(2):549–558. doi: 10.1016/0092-8674(85)90112-6. [DOI] [PubMed] [Google Scholar]
- O'Neil K. T., Shuman J. D., Ampe C., DeGrado W. F. DNA-induced increase in the alpha-helical content of C/EBP and GCN4. Biochemistry. 1991 Sep 17;30(37):9030–9034. doi: 10.1021/bi00101a017. [DOI] [PubMed] [Google Scholar]
- O'Shea E. K., Klemm J. D., Kim P. S., Alber T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science. 1991 Oct 25;254(5031):539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Lewis M. The operator-binding domain of lambda repressor: structure and DNA recognition. Nature. 1982 Jul 29;298(5873):443–447. doi: 10.1038/298443a0. [DOI] [PubMed] [Google Scholar]
- Pavletich N. P., Pabo C. O. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991 May 10;252(5007):809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Párraga G., Horvath S. J., Eisen A., Taylor W. E., Hood L., Young E. T., Klevit R. E. Zinc-dependent structure of a single-finger domain of yeast ADR1. Science. 1988 Sep 16;241(4872):1489–1492. doi: 10.1126/science.3047872. [DOI] [PubMed] [Google Scholar]
- Qian Y. Q., Billeter M., Otting G., Müller M., Gehring W. J., Wüthrich K. The structure of the Antennapedia homeodomain determined by NMR spectroscopy in solution: comparison with prokaryotic repressors. Cell. 1989 Nov 3;59(3):573–580. doi: 10.1016/0092-8674(89)90040-8. [DOI] [PubMed] [Google Scholar]
- Record M. T., Jr, Lohman M. L., De Haseth P. Ion effects on ligand-nucleic acid interactions. J Mol Biol. 1976 Oct 25;107(2):145–158. doi: 10.1016/s0022-2836(76)80023-x. [DOI] [PubMed] [Google Scholar]
- Ron D., Brasier A. R., Habener J. F. Angiotensinogen gene-inducible enhancer-binding protein 1, a member of a new family of large nuclear proteins that recognize nuclear factor kappa B-binding sites through a zinc finger motif. Mol Cell Biol. 1991 May;11(5):2887–2895. doi: 10.1128/mcb.11.5.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer R. T., Jordan S. R., Pabo C. O. Lambda repressor: a model system for understanding protein-DNA interactions and protein stability. Adv Protein Chem. 1990;40:1–61. doi: 10.1016/s0065-3233(08)60286-7. [DOI] [PubMed] [Google Scholar]
- Simon M., Adam G., Rapatz W., Spevak W., Ruis H. The Saccharomyces cerevisiae ADR1 gene is a positive regulator of transcription of genes encoding peroxisomal proteins. Mol Cell Biol. 1991 Feb;11(2):699–704. doi: 10.1128/mcb.11.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz T. A. Structural studies of protein-nucleic acid interaction: the sources of sequence-specific binding. Q Rev Biophys. 1990 Aug;23(3):205–280. doi: 10.1017/s0033583500005552. [DOI] [PubMed] [Google Scholar]
- Stillman D. J., Bankier A. T., Seddon A., Groenhout E. G., Nasmyth K. A. Characterization of a transcription factor involved in mother cell specific transcription of the yeast HO gene. EMBO J. 1988 Feb;7(2):485–494. doi: 10.1002/j.1460-2075.1988.tb02836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M. SPXX, a frequent sequence motif in gene regulatory proteins. J Mol Biol. 1989 May 5;207(1):61–84. doi: 10.1016/0022-2836(89)90441-5. [DOI] [PubMed] [Google Scholar]
- Tanaka I., Appelt K., Dijk J., White S. W., Wilson K. S. 3-A resolution structure of a protein with histone-like properties in prokaryotes. Nature. 1984 Aug 2;310(5976):376–381. doi: 10.1038/310376a0. [DOI] [PubMed] [Google Scholar]
- Thukral S. K., Eisen A., Young E. T. Two monomers of yeast transcription factor ADR1 bind a palindromic sequence symmetrically to activate ADH2 expression. Mol Cell Biol. 1991 Mar;11(3):1566–1577. doi: 10.1128/mcb.11.3.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thukral S. K., Morrison M. L., Young E. T. Alanine scanning site-directed mutagenesis of the zinc fingers of transcription factor ADR1: residues that contact DNA and that transactivate. Proc Natl Acad Sci U S A. 1991 Oct 15;88(20):9188–9192. doi: 10.1073/pnas.88.20.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thukral S. K., Morrison M. L., Young E. T. Mutations in the zinc fingers of ADR1 that change the specificity of DNA binding and transactivation. Mol Cell Biol. 1992 Jun;12(6):2784–2792. doi: 10.1128/mcb.12.6.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thukral S. K., Tavianini M. A., Blumberg H., Young E. T. Localization of a minimal binding domain and activation regions in yeast regulatory protein ADR1. Mol Cell Biol. 1989 Jun;9(6):2360–2369. doi: 10.1128/mcb.9.6.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman J., Gönczy P., Vashishtha M., Harris E., Desplan C. A single amino acid can determine the DNA binding specificity of homeodomain proteins. Cell. 1989 Nov 3;59(3):553–562. doi: 10.1016/0092-8674(89)90038-x. [DOI] [PubMed] [Google Scholar]
- Turner R., Tjian R. Leucine repeats and an adjacent DNA binding domain mediate the formation of functional cFos-cJun heterodimers. Science. 1989 Mar 31;243(4899):1689–1694. doi: 10.1126/science.2494701. [DOI] [PubMed] [Google Scholar]
- Ward A. C. Single-step purification of shuttle vectors from yeast for high frequency back-transformation into E. coli. Nucleic Acids Res. 1990 Sep 11;18(17):5319–5319. doi: 10.1093/nar/18.17.5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolberger C., Vershon A. K., Liu B., Johnson A. D., Pabo C. O. Crystal structure of a MAT alpha 2 homeodomain-operator complex suggests a general model for homeodomain-DNA interactions. Cell. 1991 Nov 1;67(3):517–528. doi: 10.1016/0092-8674(91)90526-5. [DOI] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis of DNA fragments cloned into M13 vectors. Methods Enzymol. 1983;100:468–500. doi: 10.1016/0076-6879(83)00074-9. [DOI] [PubMed] [Google Scholar]