Abstract
DNA repair mechanisms are critical for maintaining the integrity of genomic DNA, and their loss is associated with cancer predisposition syndromes. Studies in Saccharomyces cerevisiae have played a central role in elucidating the highly conserved mechanisms that promote eukaryotic genome stability. This review will focus on repair mechanisms that involve excision of a single strand from duplex DNA with the intact, complementary strand serving as a template to fill the resulting gap. These mechanisms are of two general types: those that remove damage from DNA and those that repair errors made during DNA synthesis. The major DNA-damage repair pathways are base excision repair and nucleotide excision repair, which, in the most simple terms, are distinguished by the extent of single-strand DNA removed together with the lesion. Mistakes made by DNA polymerases are corrected by the mismatch repair pathway, which also corrects mismatches generated when single strands of non-identical duplexes are exchanged during homologous recombination. In addition to the true repair pathways, the postreplication repair pathway allows lesions or structural aberrations that block replicative DNA polymerases to be tolerated. There are two bypass mechanisms: an error-free mechanism that involves a switch to an undamaged template for synthesis past the lesion and an error-prone mechanism that utilizes specialized translesion synthesis DNA polymerases to directly synthesize DNA across the lesion. A high level of functional redundancy exists among the pathways that deal with lesions, which minimizes the detrimental effects of endogenous and exogenous DNA damage.
Keywords: base excision repair, nucleotide excision repair, mismatch repair, postreplication repair, translesion synthesis
DNA damage is induced by exposure to environmental agents and is generated spontaneously during normal cellular metabolism (reviewed by Friedberg et al. 2006). Reactive oxygen species (ROS) are an unavoidable by-product of aerobic metabolism and cause both base damage and strand breaks. Additional spontaneous cellular reactions include the hydrolytic loss of bases, especially purines, from the phosphodiester backbone, as well as the deamination and alkylation of bases. In humans, it has been estimated that up to 100,000 spontaneous DNA lesions are generated daily per cell (Hoeijmakers 2009). Environmental DNA-damaging agents include the ultraviolet (UV) component of sunlight, which generates cyclobutane pyrimidine dimers and oxidative base damage; ionizing radiation, which produces clusters of ROS that create double-strand DNA breaks; and base-damaging chemicals such as aflatoxins, benzo(a)pyrene, methyl chloride, and nitrosamines, which alter or destroy base-pairing capacity. Because DNA damage has the potential to inhibit and/or alter fidelity of replication and transcription, there is a need for diverse and highly accurate repair processes. There is also a need for bypass mechanisms that allow unrepaired damage to be tolerated if encountered during replication. An emerging theme in the past 20 years is that there is considerable overlap between the various repair and bypass pathways in terms of the cognate lesions that each can deal with. This functional redundancy is partially a reflection of the very high load of endogenous DNA damage and underscores the importance of these pathways in the maintenance of genome stability.
The first comprehensive review of yeast DNA-repair pathways was published as part of the 1981 Cold Spring Harbor yeast books (Haynes and Kunz 1981). Studies at that time had focused on identifying the genes involved in surviving treatment with UV light and ionizing radiation (RAD genes) and on using epistasis analysis to place the genes into discrete pathways. These early genetic studies identified three discrete pathways, with each being named for the gene whose mutation conferred the most severe phenotype. The RAD3 epistasis group encodes components of the nucleotide excision repair pathway, which is the major pathway for repairing UV-induced lesions; the RAD52 epistasis group encodes components of the homologous recombination pathway and is required for the repair of ionizing radiation-induced damage; and the relatively ill-defined RAD6 postreplication repair pathway encodes components required for the bypass of damages that block replicative DNA polymerases. It should be noted that components of the other major DNA-damage repair pathway—base excision repair—were absent among the early rad mutants and that most were identified biochemically.
The second iteration of the Cold Spring Harbor yeast books was published in 1991, a time when the emphasis was on cloning (usually by functional complementation of the mutant phenotype) and sequencing RAD genes and on purifying the encoded proteins and defining their biochemical properties (Friedberg et al. 1991). The current review will focus on the progress made in the intervening 20 years, which has truly been astounding. The damage-reversal and excision-repair pathways that remove DNA damage will be summarized, with an emphasis on the roles that individual players have within the defined pathway. A major area of new focus will be the mismatch repair system, which is responsible for removing errors made during DNA replication. The only yeast mismatch repair gene known in 1991 was PMS1, and rapid progress has been made in identifying other mismatch repair components and unraveling their molecular mechanisms. In addition, recent studies indicate that ribonucleoside monophosphates are frequently incorporated into genomic DNA, and a pathway for their removal has been described. Finally, the postreplication repair pathway, which is a tolerance/bypass pathway rather than a true repair pathway, will be considered. The most significant advances with relation to this pathway have been the characterization of specialized translesion synthesis DNA polymerases and the discovery that post-translational modification of proliferating cell nuclear antigen (PCNA) regulates alternative mechanisms of lesion bypass. The repair of double-strand breaks, which occurs primarily via homologous recombination in yeast, is covered in another review in this series and will not be considered here. Importantly, all of these pathways exhibit high evolutionary conservation, with discoveries made in the budding yeast Saccharomyces cerevisiae serving as a paradigm for repair processes in higher eukaryotes.
Direct Reversal of DNA Damage
The simplest and most accurate repair mechanism is the direct reversal of damage in a single-step reaction. Direct reversal, however, applies to only a very limited number of DNA lesions. The enzymatic photoreactivation of a cyclobutane pyrimidine dimer (CPD), which is the major product of UVB and UVC radiation, by DNA photolyase is the prototype of this type of reaction. In addition to possessing a CPD-specific photolyase (Phr1), yeast also has a methyltransferase (Mgt1) that removes methyl groups from modified bases.
Phr1, pyrimidine dimer DNA photolyase
The PHR1 gene was identified through the isolation of a mutant unable to photoreactivate CPDs (Resnick 1969) and cloned by restoration of photoreactivation in a phr1 mutant also deficient in nucleotide excision repair (Schild et al. 1984). Transcription of the PHR1 gene is stimulated as a general response to DNA-damaging agents such as UVC radiation and alkylating agents (Sebastian et al. 1990). The Phr1 protein is a monomer that contains stoichiometric amounts of two noncovalently attached chromophores: a catalytic flavin adenine dinucleotide (FADH−) and a methylenetetrahydrofolate (MTHF) “second chromophore” (G. B. Sancar 1985; A. Sancar 2008). Photoreversal occurs by a series of steps initiated when Phr1 binds to CPD-containing DNA in a light-independent reaction. The MTHF of bound Phr1 then absorbs a photon in the near-UV to visible wavelengths (300–500 nm) and transfers its excitation energy to FADH−. Next, the photo-excited FADH− transfers an electron to the CPD to generate an unstable dimer radical anion. The CPD ring splits to restore DNA structure, and a reverse electron transfer restores the functional form of the flavin chromophore.
Mgt1, O6-methylguanine/O4-methylthymine DNA methyltransferase
The mispairing of O6-methylguanine (O6-MeG) with T and O4-methylthymine (O4-MeT) with G gives rise to GC-to-AT and AT-to-GC transitions, respectively. The yeast Mgt1 protein, which reverses both types of damage, was identified biochemically (Sassanfar and Samson 1990), and the corresponding gene was cloned by functional complementation in methyltransferase-deficient Escherichia coli cells (Xiao et al. 1991). Mtg1 repairs O6-MeG by a suicide reaction that irreversibly transfers the methyl group to a cysteine residue in the enzyme active site to generate S-methylcysteine (Sassanfar and Samson 1990; Xiao et al. 1991). Although the protein has much lower affinity for O4-MeT than for O6-MeG in vitro (Sassanfar et al. 1991), O4MeT repair in vivo can be inferred because expression of Mgt1 in E. coli prevents methylation-associated AT-to-GC transitions (Xiao et al. 1991).
Spontaneous mutation rates in Mgt1-deficient strains are enhanced, suggesting the presence of a natural source of SN1-type endogenous or environmental alkylating agents (Xiao and Samson 1992). As expected, disruption of MGT1 enhances sensitivity to the killing and mutagenic effects of SN1-type alkylating agents such as N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) (Xiao and Samson 1992). Transcription of MGT1 is not enhanced by low concentrations of MNNG, suggesting the absence of a bacterial-type adaptive response (Xiao and Samson 1992). The level of Mgt1 is, however, regulated by the Ubr1/Rad6- and Ufd4/Ubc4-mediated protein degradation pathways; loss of both pathways confers hyperresistance to MNNG and hypersensitivity to Mgt1 overexpression (Hwang et al. 2009).
Base Excision Repair
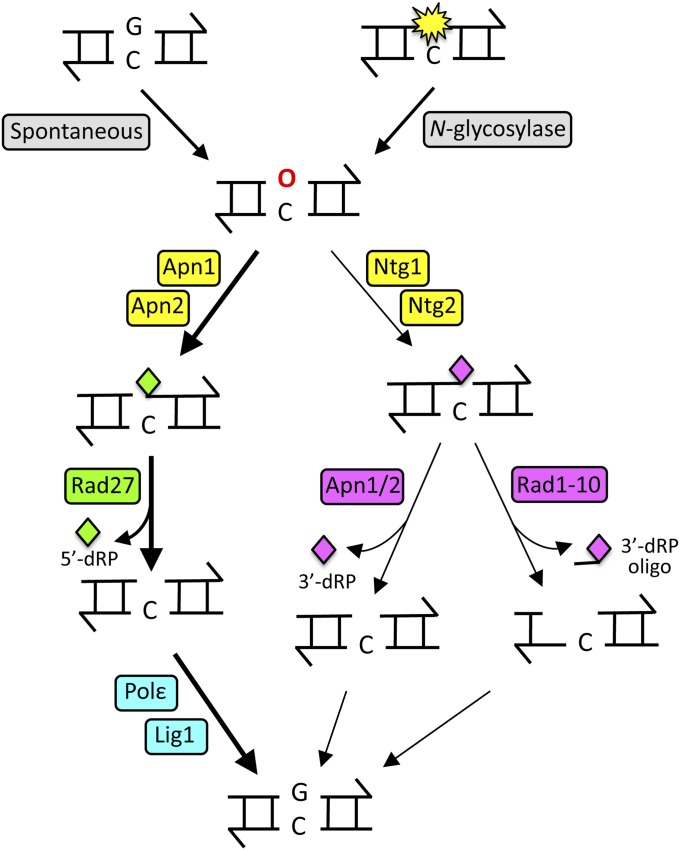
The major endogenous DNA damages result from oxidative stress, hydrolysis, or deamination and are removed by the base excision repair (BER) pathway. BER requires the sequential action of five DNA-modifying activities: (1) a DNA N-glycosylase that releases the base from deoxyribose, (2) an endonuclease/lyase that nicks the DNA backbone at the resulting apyrimidinic/apurinic (AP) site, (3) a 3′- or 5′-phosphodiesterase that removes the remaining deoxyribose phosphate residue, (4) a DNA polymerase that fills the gap thus created, and (5) a DNA ligase to seal the remaining nick (Hoeijmakers 2001). The critical steps in the BER process are illustrated in Figure 1 and properties of the major BER factors are summarized in Table 1.
Figure 1.
The BER pathway. AP sites (red “O”) are generated either by spontaneous base loss or a DNA N-glycosylase. Apn1 and Apn2 nick the backbone on the 5′ side of an AP site to initiate the major pathway for repair; the resulting 5′-dRP is removed by the Rad27 5′-flap endonuclease. AP-site processing can also be initiated by the Ntg1 or Ntg2 lyase, which nicks on the 3′ side of lesion. The resulting 3′-dRP can be removed by the 3′-diesterase activity of Apn1/Apn2 or as part of a Rad1-Rad10 generated oligonucleotide. Finally, the gap is filled by DNA Pol ε, and the backbone is sealed by DNA ligase 1.
Table 1. DNA N-glycosylases, AP endonucleases, and end-processing enzymes.
| Gene name (alternative) | Protein size (kDa) | Properties | Mammalian counterpart |
|---|---|---|---|
| UNG1 | 40.5 | Monofunctional DNA N-glycosylase. Excision of uracil in ssDNA and dsDNA. Nuclear and mitochondrial. | UNG1/2 |
| MAG1 (MMS5) | 34.3 | Monofunctional DNA N-glycosylase. Excision of 3-MeA, 7-MeG, HX, or 1-N6-ethenoA from dsDNA. | AAG |
| NTG1 (SCR1, FUN33, OGG2) | 45.5 | Bifunctional DNA N-glycosylase/AP lyase. Excision of oxidatively damaged pyrimidines and AP sites in dsDNA. Nuclear and mitochondrial. | NTH1 |
| NTG2 (SCR2) | 43.8 | Bifunctional DNA N-glycosylase/AP lyase. Excision of oxidatively damaged pyrimidines and AP sites in dsDNA. Nuclear. | NTH1 |
| OGG1 | 42.8 | Bifunctional DNA N-glycosylase/AP lyase. Excision of 8-oxoG, 8-oxoA, FapyG, and AP sites opposite C in dsDNA. Nuclear and mitochondrial. | OGG1 |
| APN1 | 41.4 | AP endonuclease and 3′-phosphodiesterase. Incision of regular and oxidized AP sites. Excision of 3′-blocked ends. Nuclear and mitochondrial. | APE1 |
| APN2 (ETH1) | 59.4 | AP endonuclease and 3′-phosphodiesterase. Incision of regular and oxidized AP sites. Excision of 3′-blocked ends. Nuclear. | APE2 |
| TPP1 | 27.4 | DNA 3′-phosphatase. | PNPK |
| TDP1 | 62.3 | Hydrolyzes phospho-tyrosyl bond. Repair of trapped topoisomerase I and II. | TDP1 |
| RAD27 (ERC11, RTH1, FEN1) | 43.3 | 5′-Flap endonuclease. Excision of 5′-dRP after cleavage of AP sites by Apn1 or Apn2. | FEN1 |
| HNT3 | 25.8 | Repair of abortive ligation product, 5′-AMP. | APTX |
ssDNA, single-strand DNA; dsDNA, double-strand DNA; 3-MeA, N3-methyladenine; 7-MeG, N7-methylguanine; HX, hypoxanthine; 1-N6-ethenoA, 1,N6-ethenoadenine.
DNA N-glycosylases
BER is initiated by a DNA N-glycosylase that cleaves the N-glycosylic bond between the cognate damaged or unusual base and the sugar moiety to which it is attached. The reaction results in the release of a free base and the formation of an AP site, the central intermediate in the BER pathway (Figure 1). Five DNA N-glycosylases are present in yeast, and these fall into two classes: (1) monofunctional enzymes that only catalyze cleavage of the N-glycosylic bond (Ung1 and Mag1) and (2) bifunctional DNA N-glycosylases/AP lyases that catalyze both cleavage of the N-glycosylic bond and nicking of the phosphodiester backbone at AP sites (Ntg1, Ntg2, and Ogg1).
Ung1, uracil-DNA N-glycosylase 1:
Uracil in DNA arises either by cytosine deamination or through incorporation of dUTP in place of dTTP. Ung1 is highly specific for the removal of uracil in single- or double-strand DNA (Percival et al. 1989); it belongs to the UDG family, which includes the E. coli Ung protein as well as human UNG1/2 (Sousa et al. 2007). The UNG1 gene was originally defined by mutations that allowed successful transformation of uracil-containing DNA into yeast (Burgers and Klein 1986) and was cloned by complementation using an in vitro assay (Percival et al. 1989). UNG1 is cell-cycle regulated at the transcriptional level (Johnston and Johnson 1995), and mutant cells are sensitive to the killing effect of deaminating agents such as sodium bisulfite. Ung1-deficient cells exhibit a moderate spontaneous mutator phenotype (Burgers and Klein 1986), with a strong bias for the GC-to-AT transitions expected to result from cytosine deamination (Impellizzeri et al. 1991; Guillet et al. 2006). Ung1 localizes to the mitochondria as well as to the nucleus, and its loss additionally elevates mutations in mitochondrial DNA (Chatterjee and Singh 2001).
Mag1, methylpurine-DNA N-glycosylase 1:
The MAG1 gene was identified by functional complementation in E. coli, and its disruption confers high sensitivity to the killing effects of alkylating agents such as methyl methanesulfonate (MMS) and MNNG (Chen et al. 1989). MAG1 transcription is induced following exposure to low doses of alkylating agents (Chen et al. 1990), and the purified protein excises N7-methylguanine and N3-methyladenine (Chen et al. 1989, 1990; Berdal et al. 1990), as well as N3-methylguanine, 1,N6-ethenoadenine, and hypoxanthine from DNA (Saparbaev and Laval 1994; Lingaraju et al. 2008). Mag1 also releases normal bases, primarily guanine, at a slow rate from intact DNA (Berdal et al. 1998), and overexpression of the protein confers a strong spontaneous mutator phenotype in cells unable to efficiently repair AP sites (Glassner et al. 1998; Klapacz et al. 2010).
Ntg1 and Ntg2, endonuclease III homologs:
The eNdonuclease Three-like Glycosylase 1 (NTG1) and NTG2 genes were identified based on homology of the encoded proteins to E. coli endonuclease III (Eide et al. 1996; Augeri et al. 1997). Each protein possesses a highly conserved helix-hairpin-helix DNA-binding motif, but only Ntg2 has an endonuclease III-like iron-sulfur (Fe-S) cluster (Eide et al. 1996; Augeri et al. 1997; You et al. 1998; Alseth et al. 1999). The Mms19 protein was recently identified as the protein that delivers Fe-S clusters to Ntg2 and other key proteins involved in DNA metabolism (Stehling et al. 2012). Ntg1 has a positively charged N terminus that serves as a mitochondrial-targeting signal, and the protein localizes primarily to mitochondria (You et al. 1999). By contrast, Ntg2 localizes exclusively to the nucleus (You et al. 1999). An additional difference is that NTG1 is inducible at the transcription level in cells exposed to oxidizing agents, whereas NTG2 is not (Alseth et al. 1999).
Both Ntg1 and Ntg2 excise a variety of oxidized pyrimidines such as 5-hydrouracil, 5-hydroxycytosine, 5-6-dihydrothymine, and thymine glycol, as well as two purine lesions [formamidopyrimidine (Fapy)-Ade and Fapy-Gua] (Senturker et al. 1998). Neither Fapy-Ade nor Fapy-Gua, however, is a substrate for E. coli endonuclease III (Dizdaroglu et al. 1993). Ntg1 also excises 8-oxoguanine (8-oxoG) opposite guanine, but Ntg2 is not active against this lesion (Senturker et al. 1998). Both Ntg1 and Ntg2 are endowed with a robust AP lyase activity that incises DNA on the 3′-side of a regular AP site using a β-elimination reaction, yielding a single-strand break with a 3′-α,β-unsaturated aldehydic (3′-dRP) end (Figure 1) (Meadows et al. 2003).
Cells lacking both Ntg1 and Ntg2 are not unusually sensitive to the killing effects of MMS, γ-radiation, or H2O2, nor do they display a spontaneous mutator phenotype (Gellon et al. 2001). These data suggest alternative activities that can repair oxidized bases and AP sites in yeast. Indeed, inactivation of either the nucleotide excision repair (NER) or homologous recombination pathway in an ntg1Δ ntg2Δ double mutant results in a synergistic increase in sensitivity to the killing and mutagenic effects of H2O2 (Swanson et al. 1999; Gellon et al. 2001). The role of Ntg1 in the maintenance of mitochondrial DNA is controversial; both enhanced and reduced spontaneous mutation frequencies have been reported in an ntg1Δ background (Doudican et al. 2005; Phadnis et al. 2006).
Ogg1, 8-oxoguanine-DNA N-glycosylase 1:
Ogg1 is a bifunctional DNA N-glycosylase/AP lyase that, like Ntg1 and Ntg2, incises AP sites using a β-elimination reaction (Girard et al. 1997). Although it is the functional homolog of the E. coli Fpg (MutM) protein, Ogg1 has no sequence homology to its bacterial counterpart (Boiteux et al. 1987; Van Der Kemp et al. 1996). Instead, Ogg1 is a member of a superfamily of repair proteins that share a common ancestor with endonuclease III of E. coli and, in turn, with Ntg1 and Ntg2.
Ogg1 excises Fapy-Gua and 7,8-dihydro-8-oxoG from γ-irradiated DNA, whereas a wide range of other lesions, including oxidized pyrimidines and adenine lesions, are not substrates (Karahalil et al. 1998). The 8-oxoG N-glycosylase and AP lyase activities of Ogg1 are highly dependent on the identity of the base opposite the lesion, with the enzyme exhibiting a marked preference for cytosine (Girard et al. 1997). Although γ-irradiated DNA contains 8-oxoadenine (8-oxoA) opposite thymine, biochemical data suggest that Ogg1 exclusively excises 8-oxoA opposite cytosine (Girard et al. 1998). With respect to the N-glycosylase and AP lyase reactions, the catalytic mechanisms for Ogg1, Ntg1, and Ntg2 are very similar. The catalytic lysine residue of Ogg1 (K241) attacks the C1′ of the N-glycosylic bond between 8-oxoG and deoxyribose, releasing free 8-oxoG and yielding a covalent imino enzyme-DNA intermediate between Ogg1 and the C1′ of the abasic sugar moiety. A β-elimination reaction then produces a single-strand break with a 3′-dRP end and a restored Ogg1 protein (Boiteux et al. 2002). Although a crystal structure for yeast Ogg1 has not been reported, the strong sequence homology between the yeast and human proteins suggests that structural and mechanistic studies performed with human OGG1 most likely apply to yeast Ogg1 (Boiteux and Radicella 2000; Bruner et al. 2000; Bjoras et al. 2002; Fromme et al. 2004; Dalhus et al. 2009). Human OGG1, like most DNA N-glycosylases, binds the damaged strand and bends the DNA to flip the lesion into the active-site pocket.
Inactivation of OGG1 does not lead to unusual sensitivity to DNA-damaging agents such as H2O2, γ-radiation, MMS, or UV radiation (Thomas et al. 1997). Mutants do, however, exhibit enhanced spontaneous and UVA-induced mutation rates (Thomas et al. 1997; Kozmin et al. 2005). There is a strong bias for GC-to-TA transversions in an ogg1Δ background, consistent with frequent insertion of A opposite a template 8-oxoG (Thomas et al. 1997; Ni et al. 1999; De Padula et al. 2004). Ogg1 additionally plays a role in the maintenance of telomere length homeostasis, reflecting repair of oxidized guanines in telomeric sequences (Lu and Liu 2010). Ogg1 localizes to both the nucleus and the mitochondria and plays an important role in the maintenance of mitochondrial as well as nuclear DNA (Singh et al. 2001; Vongsamphanh et al. 2006).
GO network:
In E. coli, MutT, MutM (Fpg), and MutY limit the mutagenic potential of 8-oxoG, and together are referred to as the “GO” network (Michaels and Miller 1992). Inactivation of either MutM or MutY results in a spontaneous mutator phenotype, which is characterized by specific accumulation of GC-to-TA transversions. MutM (Fpg) excises 8-oxoG opposite cytosine, whereas MutY releases adenine opposite 8-oxoG; together these two glycoslyases form the core of the GO network. The third component of the system is MutT, the loss of which is associated with AT-to-CG transversions. MutT hydrolyzes 8-oxo-dGTP to 8-oxo-dGMP, thereby cleansing the dNTP pool and preventing the incorporation of 8-oxo-dGMP opposite a template adenine. In S. cerevisiae, loss of Ogg1 generates only a moderate mutator phenotype, suggesting the occurrence of a MutY homolog that helps limit 8-oxoG-associated GC-to-TA transversions. Although lacking a MutY-type glycosylase that deals with 8-oxoG:A mispairs, S. cerevisiae nevertheless possesses a biological GO network that prevents the mutagenic action of endogenous 8-oxoG (Figure 2). This network involves the mismatch repair system, which serves a role analogous to that of MutY by removing adenine incorporated opposite a template 8-oxoG (Ni et al. 1999), as well as the Pol η translesion synthesis DNA polymerase, which preferentially inserts C opposite a template 8-oxoG (Haracska et al. 2000). Yeast has no known functional homolog of the bacterial MutT protein, although homologs are present in other eukaryotes.
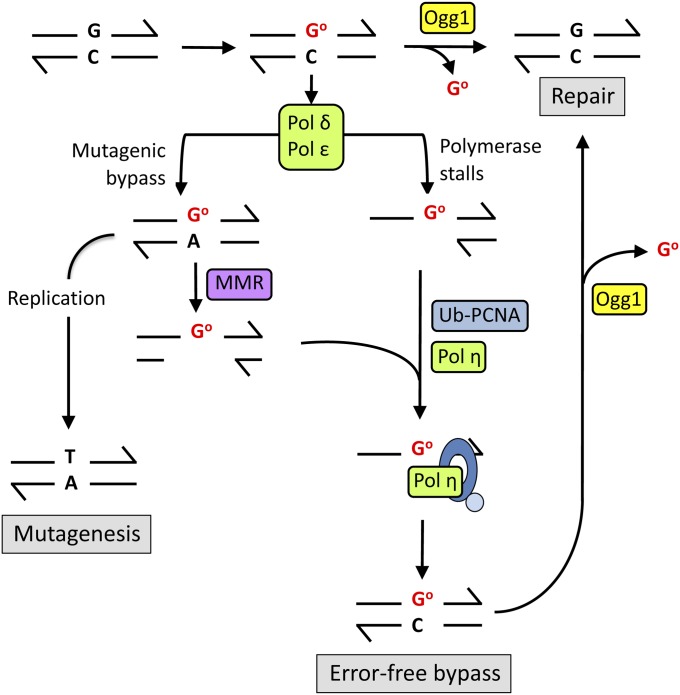
Figure 2.
The GO network. Reactive oxygen species attack guanine base-paired with cytosine to yield 8-oxoG (GO). Ogg1 excises 8-oxoG from the DNA backbone, and the resulting AP site is repaired via BER (“repair”). If encountered during replication, local sequence context will determine whether Pol δ/ε stalls at or bypasses the GO lesion. If Pol δ/ε stalls at 8-oxoG during replication, Pol η is recruited by ubiquitinated PCNA (Ub-PCNA) and preferentially incorporates C opposite the lesion (“error-free bypass”). During bypass by Pol δ/ε, adenine is frequently inserted instead of cytosine to create a GO:A mispair, which is recognized by the MMR machinery. The newly synthesized, A-containing strand is degraded to generate a single-strand gap containing the lesion, and C is incorporated opposite the lesion during a gap-filling reaction, which may involve Pol η. If not repaired, the GO:A mispair will yield a GC-to-TA transversion at the next round of replication (“mutagenesis”).
AP endonucleases
AP endonucleases bind to AP sites in duplex DNA and nick the phosphodiester backbone immediately 5′ of the lesion. In addition, these enzymes process a variety of 3′-blocked termini that would otherwise block DNA polymerization and ligation. Yeast has two AP endonucleases—Apn1 and Apn2—with >95% of in vivo activity attributed to Apn1 (Popoff et al. 1990).
Apn1, AP endonuclease 1:
The APN1 gene was identified through immunological screening of an expression library, and the encoded protein shares extensive homology with the E. coli endonuclease IV (Nfo) protein (Popoff et al. 1990). In addition to its canonical AP-endonuclease activity, which nicks DNA on the 5′-side of a regular or reduced AP site, Apn1 has a 3′-phosphodiesterase activity that excises 3′-blocking groups such as 3′-dRP, 3′-phosphoglycolate (3′-PGA), and 3′-phosphate (3′-P); it possesses a 3′-tyrosyl-DNA phosphodiesterase activity that contributes to the removal of covalently bound topoisomerase 1; and it has 3′-exonuclease activity (reviewed in Boiteux and Guillet 2004). Apn1 also has an endonuclease activity that nicks DNA on the 5′-side of oxidized bases, which initiates an alternative DNA-repair pathway referred to as nucleotide incision repair (Ischenko and Saparbaev 2002). Deletion of APN1 confers moderate sensitivity to the killing effects of oxidizing or alkylating agents (Ramotar et al. 1991), and apn1Δ cells have a spontaneous mutator phenotype, accumulating mostly AT-to-CG events (Kunz et al. 1994). Apn1 is important for repairing alkylation damage in mitochondrial as well as nuclear DNA (Acevedo-Torres et al. 2009), and the protein localizes to both cellular compartments (Ramotar et al. 1993).
Apn2, AP endonuclease 2:
APN2 was identified based on homology to exonuclease III of E. coli (Johnson et al. 1998; Bennett 1999), and its transcription is induced by MMS (Bennett 1999). Loss of APN2 does not sensitize cells to the lethal or mutagenic effects of MMS or H2O2, but apn1Δ apn2Δ double mutants are extremely sensitive to both agents (Johnson et al. 1998; Bennett 1999). In addition to hydrolytic cleavage of DNA on the 5′-side of an AP site, Apn2 is also endowed with X′ to Y′ exonuclease and 3′-phosphodiesterase activities that remove 3′-blocking groups (Unk et al. 2000). Both activities are stimulated by physical interaction with the PCNA sliding clamp, which targets multiple DNA metabolic proteins to nicks/gaps in DNA (Moldovan et al. 2007).
Origin, repair, and biological impact of endogenous AP sites:
AP sites are abundant, endogenous DNA lesions (De Bont and Van Larebeke 2004; Swenberg et al. 2011) that can be lethal and mutagenic (Boiteux and Guillet 2004). Although highly sensitive to alkylating or oxidizing agents, cells lacking both Apn1 and Apn2 are viable and exhibit only a moderate spontaneous mutator phenotype (Johnson et al. 1998; Bennett 1999). This suggests either that AP sites are formed at a much lower rate in vivo than predicted or that backup repair activities manage the effects of persistent AP sites. Indeed, crossing deletions of candidate DNA-repair genes into an apn1Δ apn2Δ background revealed that loss of Rad1-Rad10, a complex best known for its nicking activity during NER, causes cell death (Guillet and Boiteux 2002). In addition, simultaneous inactivation of Apn1, Apn2, Ntg1, and Ntg2 results in a strong spontaneous mutator phenotype dominated by AT-to-CG transversions (Collura et al. 2012). Together, these data confirm that AP sites form under physiological growth conditions and that these sites impair cell viability and genetic stability when DNA repair is compromised.
Although there are likely multiple origins of endogenous AP sites, genetic studies suggest that most are linked to dUTP incorporation, which is normally limited by action of the Dut1 dUTPase (Guillet et al. 2006). If incorporated, however, the uracil is excised by Ung1 to create an AP site (Guillet and Boiteux 2003; Collura et al. 2012). As noted above, inefficient AP-site repair results in mostly AT-to-CG mutations, which presumably reflect bypass of dUMP-derived AP sites by the concerted action of the Rev1 and Pol ζ translesion synthesis DNA polymerases (Figure 3).
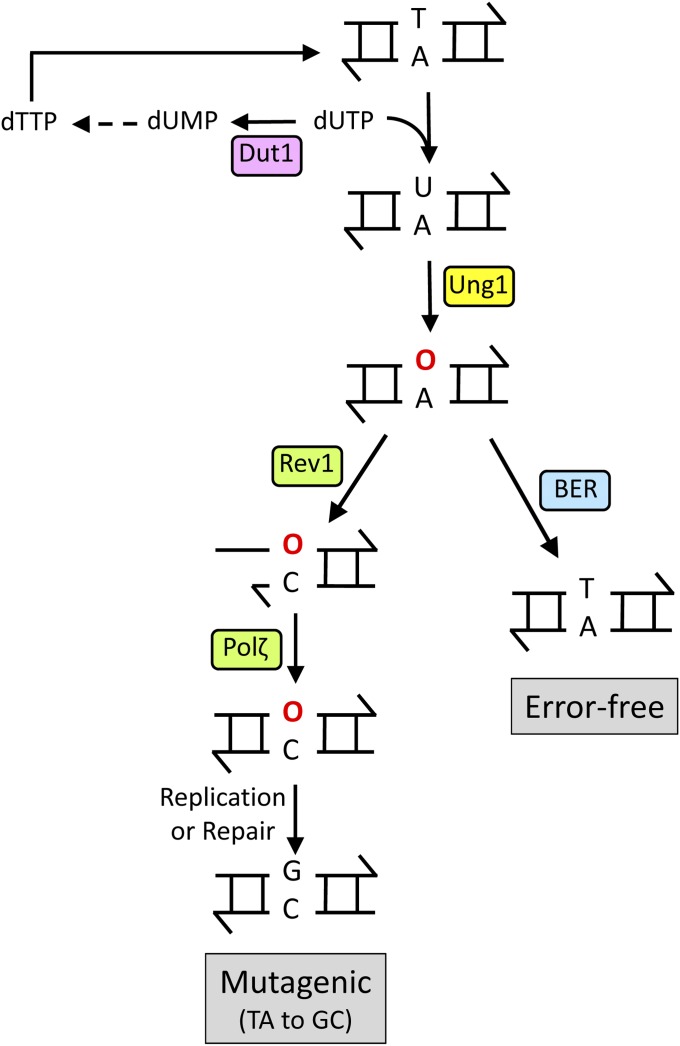
Figure 3.
Bypass of endogenous AP sites. dUTP levels in the nucleotide pool are reduced by Dut1 activity, thereby limiting the incorporation of dUMP into genomic DNA. Most endogenous AP sites (indicated by red “O”) are generated by Ung1 removal of uracil in DNA. The resulting AP site can be repaired by the BER pathway or bypassed by the concerted action of Rev1, which usually inserts cytosine opposite the AP site, and Pol ζ, which extends the O:C terminus.
Single-strand break repair: “dirty end” processing factors
To be a substrate for DNA polymerase and DNA ligase, single-strand breaks must have “clean” ends: a 3′-OH dNMP and a 5′-P dNMP. Genetic and biochemical studies have revealed that there is considerable redundancy with respect to the proteins that can generate such ends during BER (see Figure 1).
Processing 3′-dirty ends:
DNA N-glycosylases/AP lyases incise DNA on the 3′-side of a regular AP site, generating a single-strand break with an abasic 3′-dRP end (Boiteux et al. 2002). Other 3′-blocked ends include the 3′-PGA or 3′-P, formed when DNA is exposed to oxidative stress, as well as the covalent attachment of Top1 via a 3′-phosphotyrosyl linkage (Caldecott 2008). Whereas Tdp1 catalyzes the removal of 3′-Top1 (Pouliot et al. 1999), the major defenses against 3′-dRP, 3′-PGA, and 3′-P are the 3′-DNA phosphatase activity of Tpp1 and the 3′-phosphodiesterase activity of Apn1 and Apn2 (Vance and Wilson 2001a,b). Rad1-Rad10 also has a role in the release of all types of 3′-blocked termini at a single-strand break (Vance and Wilson 2002; Boiteux and Guillet 2004; Guzder et al. 2004). Finally, other structure-dependent endonucleases, such as Mus81-Mms4 and Slx1-Slx4, can participate in the removal of 3′-dirty ends (Guillet and Boiteux 2002; Deng et al. 2005).
Processing 5′-dirty ends:
Apn1 and Apn2 nick 5′ of AP sites, generating nicks with 5′-dRP ends. In mammalian cells, removal of 5′-dRP is accomplished either by the lyase activity of DNA polymerase β or by the 5′-flap endonuclease (FEN) activity of FEN1 (Hoeijmakers 2001). In S. cerevisiae, both DNA Pol4 (Bebenek et al. 2005) and Trf4 (Gellon et al. 2008) possess a 5′-dRP lyase activity, but whether either contributes to BER in vivo has not been clearly established (McInnis et al. 2002; Gellon et al. 2008). The high MMS and H2O2 sensitivity of yeast cells missing Fen1/Rad27 (Reagan et al. 1995; Hansen et al. 2000), and the suppression of the MMS sensitivity of rad27Δ by APN1 deletion (Wu and Wang 1999), suggest that Rad27 is the main activity for removing 5′-dRP (see Figure 1).
The ligation intermediate 5′-AMP, which reflects an abortive attempt to ligate a nick, constitutes another important type of 5′-dirty end. In human cells, 5′-AMP is reversed by Aprataxin, the loss of which causes the severe neurodegenerative disorder ataxia-oculomotor apraxia 1 (Caldecott 2008). Hnt3 is an Aprataxin-like protein in S. cerevisiae, and it can repair 5′-AMP in vitro (Ahel et al. 2006). Although Hnt3-deficient cells are not unusually sensitive to the killing effect of H2O2 or MMS, simultaneous loss of Hnt3 and Rad27 results in a synergistic increase in H2O2 and MMS sensitivity, suggesting redundant roles in the repair of 5′-AMP (Daley et al. 2010).
DNA polymerase and DNA ligase
The removal of 3′- and/or 5′-dirty ends by appropriate BER enzymes generates a small gap in DNA, which is then ready to be filled by a DNA polymerase (Figure 1). DNA polymerase ε (Pol ε) is the major source of BER-associated repair synthesis in nuclear extracts (Wang et al. 1993), and Pol2, the catalytic subunit of Pol ε, binds with high affinity to a BER intermediate in vitro (Sukhanova et al. 2011). Furthermore, the pol2-16 allele, which impairs DNA polymerase activity, confers significant sensitivity to MMS, but not to UV radiation (Kesti et al. 1999). A role of other DNA polymerases such as polymerase δ (Pol δ) in BER has not been excluded, however. DNA repair is completed by the Cdc9 DNA ligase (Lig1), which uses an AMP-lysine intermediate susceptible to generating 5′-AMP damage (Tomkinson et al. 1992).
Nucleotide Excision Repair
Components of the NER pathway belong to the RAD3 epistasis group, and most were identified in genetic screens for mutants with enhanced UV sensitivity (Haynes and Kunz 1981). NER is characterized by an ability to remove a large number of structurally unrelated, helix-distorting lesions that interfere with base pairing and generally impair replication and transcription. This pathway is particularly relevant for preventing the lethal and mutagenic effects of environmental mutagens; cognate lesions include the UV-induced CPD and 6-4 photoproduct [(6-4) PP] as well as chemical carcinogen-induced bulky adducts (Cadet et al. 2005; Friedberg et al. 2006). Loss of NER in humans is associated with the disease xeroderma pigmentosum, which is characterized by an extreme sensitivity to sunlight and cancer predisposition (Friedberg et al. 2006). NER also removes bulky, endogenous oxidative DNA damage that results from intramolecular crosslinking between the C8 position of purines and the 5′ position of deoxyribose (Kuraoka et al. 2000). Finally, NER can provide an alternative mechanism to repair AP sites and oxidized bases (Scott et al. 1999; Swanson et al. 1999; Torres-Ramos et al. 2000; Gellon et al. 2001).
The NER pathway can be divided into two subpathways based on the initial lesion recognition step: global-genome NER (GG-NER) and transcription-coupled NER (TC-NER). In GG-NER, dedicated proteins directly recognize an initiating lesion, and complexes containing Rad7 and Rad16 are unique to this subpathway. By contrast, TC-NER is initiated when a lesion on the transcribed strand blocks RNA polymerase; unique to TC-NER are Rad26 and the Rpb9 subunit of RNA polymerase II (RNA Pol II). Following lesion recognition, the two subpathways converge and dual incisions are made flanking the damage. Although additional proteins are relevant in vivo, biochemical reconstitution of the dual incision reaction on naked, UV-irradiated DNA has identified six essential NER factors: Rad4-Rad23, Rad14, TFIIH, Rad1-Rad10, Rad2, and replication protein A (RPA) (Guzder et al. 1995b). In vivo, there may exist a preassembled “repairosome” that contains all NER factors (Svejstrup et al. 1995; Rodriguez et al. 1998), or, alternatively, NER factors may assemble either one-by-one or as part of discrete subcomplexes (Prakash and Prakash 2000). Recent studies in mammalian cells favor the sequential recruitment of DNA repair factors/subcomplexes during NER (Volker et al. 2001; Mocquet et al. 2008). Following the dual-incision reaction, a lesion-containing single strand of 25–30 nucleotides is released from the helix, the resulting gap is filled by DNA polymerase, and the remaining nick is sealed by ligase. The properties of proteins involved in NER are summarized in Table 2 and are detailed further below in relation to major steps of this repair pathway. Figure 4 presents a complete model of NER that illustrates lesion recognition during GG-NER; Figure 5 presents unique aspects of TC-NER.
Table 2. NER genes.
| Gene name (alternative) | Protein size (kDa) | Properties | Mammalian counterpart |
|---|---|---|---|
| RAD4 | 87.2 | Forms a complex with Rad23 and Rad33 that binds damaged DNA. | XPC |
| RAD23 | 42.4 | Forms a complex with Rad4 that binds damaged DNA. | HRAD23B |
| RAD33 | 20.3 | Forms a complex with Rad4 that binds damaged DNA. | CEN2 |
| RAD7 | 63.8 | Forms a complex with Rad16. | DDB1 |
| RAD16 (PSO5) | 91.4 | Forms a complex with Rad7 that has ATP-dependent binding of damaged DNA, chromatin remodeling activity, and E3 ligase activity. | DDB2 |
| RAD1 (LPB9) | 126.4 | Forms a complex with Rad10 that has structure-dependent endonuclease activity; incises DNA on the 5′-side of lesions. | XPF |
| RAD10 | 24.3 | Forms a complex with Rad1. | ERCC1 |
| RAD2 | 117.8 | Structure-dependent endonuclease; incises DNA on the 3′-side of lesions. | XPG |
| RAD14 | 43.0 | Zinc-finger protein; binds damaged DNA. | XPA |
| RFA1 (BUF2, FUN3, RPA1) | 70.3 | Component of heterotrimeric RPA, the yeast single-strand DNA binding protein. | RPA |
| RFA2 (BUF1, RPA2) | 29.9 | Component of heterotrimeric RPA. | RPA |
| RFA3 (RPA3) | 13.8 | Component of heterotrimeric RPA. | RPA |
| RAD25 (SSL2, LOM3) | 95.3 | TFIIH subunit; DNA-dependent ATPase and X′ to Y′ helicase. | XPB |
| RAD3 (REM1) | 89.8 | TFIIH subunit; DNA dependent ATPase and helicase with Y′ to X′ polarity. | XPD |
| TFB1 | 72.9 | Core TFIIH component. | GTF2H1 |
| SSL1 | 52.3 | Core TFIIH component. | GTF2H2 |
| TFB2 | 58.5 | Transcription initiation factor IIB, a core TFIIH component. | GTF2H4 |
| TFB4 | 37.5 | Transcription factor B subunit 4, a core TFIIH component. | GTF2H3 |
| TFB5 | 8.2 | Core TFIIH component. | TTDA |
| CDC9 (MMS8) | 84.8 | DNA ligase 1. | LIG1 |
| RAD26 | 124.5 | DNA-dependent ATPase required for transcriptional bypass of lesions and for TC-NER. | CSB |
| RAD28 | 58.2 | WD40 repeat protein of unknown function. | CSA |
| RPB9 (SSU73) | 14.3 | Nonessential RNA Pol II subunit required for Rad26-independent TC-NER. | POLR21 |
| POL2 | 255.7 | Catalytic subunit of replicative DNA Pol ε. | p261, POLE |
| POL3 | 124.6 | Catalytic subunit of replicative DNA Pol δ; important for repair synthesis during NER. | p125, POLD1 |
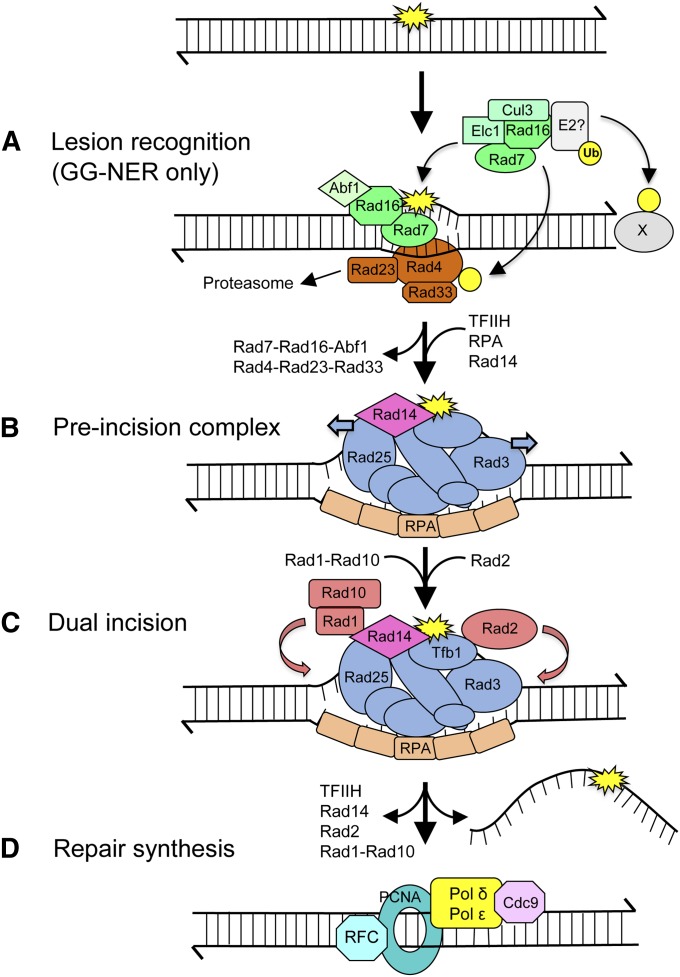
Figure 4.
The NER pathway. (A) During GG-NER, a helix-distorting lesion (yellow star) is recognized by Rad4-Rad23-Rad33 and Rad7-Rad16 complexes. Rad7-Rad16-Abf1 has chromatin-remodeling activity, whereas Rad7-Rad16-Elc1-Cul3-E2 has Ub ligase activity, which modifies Rad4 and additional factors (“X”). These reactions allow efficient recognition of lesions by Rad4, its proper positioning, and opening of the helix ∼10 bp. (B) TFIIH (components are in blue), Rad14, and RPA are recruited to form a pre-incision complex that verifies the lesion and further unwinds DNA. Rad4-Rad23-Rad33 and Rad7-Rad16-Abf1 are released. (C) The structure-specific endonucleases Rad1-Rad10 and Rad2 are positioned to incise 5′ and 3′ of the lesion, respectively. (D) A lesion-containing oligonucleotide (25–30 nt) is released from the duplex, followed by repair synthesis and ligation.
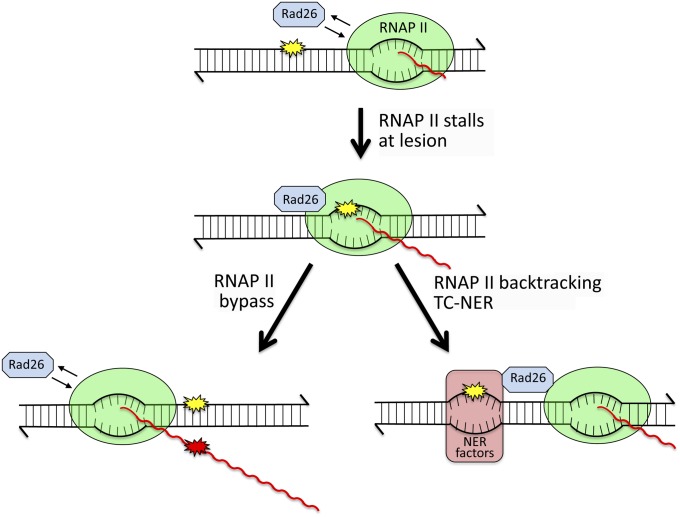
Figure 5.
Responses to stalling of RNA Polymerase II at a template lesion. Stalling of RNA Polymerase II (RNAP II) at a lesion (yellow star) in the transcribed strand of an active gene stabilizes its interaction with Rad26/CSB. Transcriptional bypass of a lesion that is a moderate block to RNAP II is promoted by Rad26/CSB. During such bypass, an incorrect rNMP (red star) can be inserted into the nascent mRNA (red wavy line), which may then specify a mutant protein (“transcriptional mutagenesis”). If the lesion is a strong block to RNAP II, Rad26/CSB and additional factors mediate the backtracking of polymerase, which exposes the lesion and promotes the recruitment of NER factors. Following lesion removal, transcription resumes without loss of the transcript.
Recognition of lesions during GG-NER
The GG-NER pathway repairs lesions without regard to transcription status or associated chromatin structure and is initiated when a trimeric Rad4-Rad23-Rad33 complex senses distortion of the DNA helix (Figure 4A). Recruitment of Rad4-Rad23-Rad33 to sites of DNA damage may be facilitated by interactions between Rad4 and chromatin remodeling complexes such as Ino80 (Sarkar et al. 2010) and SWI/SNF (Gong et al. 2006). The Rad7-Rad16 complex is mandatory for GG-NER in vivo; as described in human cells, this complex, in association with alternative factors, may promote the recruitment of Rad4 through its high affinity for DNA damage and its E3 ubiquitin (Ub) ligase activity (Sugasawa et al. 2005). Similar to the analogous human complex, yeast Rad4-Rad23-Rad33 likely initiates the opening of an ∼10-bp region around the lesion (Tapias et al. 2004).
Rad4-Rad23-Rad33 and Rad34:
Whereas Rad4 is essential for all NER, loss of either Rad23 or Rad33 only reduces repair. Simultaneous deletion of both proteins, however, generates a complete deficiency in NER (Den Dulk et al. 2006). Rad4 and Rad23 form a stable complex that specifically binds to UV-irradiated DNA in vitro (Jansen et al. 1998), and, as noted above, this complex is required for dual incision in the reconstituted system (Guzder et al. 1995b). Although Rad23 is unable to bind damaged DNA by itself, it stimulates Rad4-binding activity and prevents its degradation by the proteasome (Xie et al. 2004). In the crystal structure of a Rad4-Rad23-DNA-CPD complex, Rad4 inserts a β-hairpin through the DNA duplex, causing the two damaged base pairs to flip out of the helix; the damaged nucleotides are exposed to solvent whereas the undamaged ones contact Rad4. These structural data suggest that Rad4 recognizes damage through the sensing of thermodynamically unstable base pairs and thus provide an explanation for its broad substrate specificity (Min and Pavletich 2007). It should be noted that inactivation of Rad4 completely abolishes both GG-NER and TC-NER in yeast. In human cells, however, inactivation of the XPC homolog results in only an intermediate sensitivity to UV radiation, and functional TC-NER is retained (Venema et al. 1991; Verhage et al. 1994).
Rad33 is a an additional NER factor that binds directly to Rad4 (Den Dulk et al. 2006, 2008). Rad4 interacts with Rad23 and Rad33 through independent sites in its C-terminal region, and the roles of these proteins are presumably to modulate Rad4 activity and integrity (Den Dulk et al. 2006). Finally, the Rad34 protein has sequence homology to Rad4 and, like Rad4, interacts with Rad23. Available data suggest a role for Rad34 during NER that occurs in the RNA Pol I-transcribed ribosomal DNA genes (Den Dulk et al. 2005; Tremblay et al. 2008).
Rad7-Rad16-Abf1 and Rad7-Rad16-Cul3-Elc1:
Rad7 and Rad16 are required for CPD removal from unexpressed sequences and for repair of UV-induced damage located on the nontranscribed strand of active genes (Verhage et al. 1994), indicating a specific role in the GG-NER subpathway (Hanawalt and Spivak 2008). Loss of either protein results in an intermediate level of UV sensitivity (Prakash et al. 1993). Rad7 and Rad16 form a stable complex in an ATP-dependent manner, and the complex binds with high affinity to UV-irradiated DNA (Guzder et al. 1997). Rad16 has homology to Snf2, the catalytic subunit of the SWI/SNF chromatin-remodeling complex, but the complex does not have detectable DNA helicase activity. Rad16 facilitates histone H3 acetylation after UV irradiation (Teng et al. 2008), however, and an effect of histone H3 methylation on NER at silenced loci has been reported (Chaudhuri et al. 2009). Although not required for dual cleavage of UV-irradiated DNA in vitro, the Rad7-Rad16 complex markedly stimulates the reaction; combining Rad4-Rad23 and Rad7-Rad16 synergistically enhances binding to UV-damaged DNA (Guzder et al. 1999). Finally, physical interaction between Rad4 and Rad7 allows the formation of a large lesion-recognition complex (Guzder et al. 1999). These data clearly point to a role for Rad7-Rad16 in DNA damage recognition during GG-NER, and other studies suggest an additional role for Rad7-Rad16 in post-incision events (Reed et al. 1998).
Purification of Rad7-Rad16 led to the identification of the essential protein autonomously replicating sequence binding factor 1 (Abf1) as the third component of a trimeric Rad7-Rad16-Abf1 complex. Abf1 plays a direct role in NER; abf1 conditional mutants are defective in the removal of UV damage and exhibit high sensitivity to UV radiation (Reed et al. 1999). Rad7-Rad16-Abf1 can generate superhelical torsion in DNA in vitro (Yu et al. 2004), suggesting a model in which the complex binds to ABF1 sites in DNA in the absence of UV irradiation. Following UV irradiation, the complex uses ATP hydrolysis to translocate on DNA, generating conformational changes and eventually stalling at damaged sites to facilitate the recruitment of Rad4 (Guzder et al. 1998; Yu et al. 2009).
In addition to interacting with Rad4, Rad7-Rad16 interacts with Elc1-Cul3 to form a cullin-based E3 Ub ligase that promotes UV-dependent ubiquitination of Rad4 and other chromatin-associated proteins (Ramsey et al. 2004; Gillette et al. 2006). Rad7-Rad16-based complexes thus have multiple roles in lesion recognition that include binding to DNA damage, interacting with Rad4-Rad23-Rad33, driving conformational changes in DNA, and remodeling chromatin through acetylation and ubiquitination. Although essential for GG-NER in vivo, Rad7-Rad16 is not required for dual incisions in a reconstituted GG-NER system (Guzder et al. 1995b). Most likely, the in vitro levels of other repair factors and DNA damage are high enough to bypass the need for complexes that operate primarily at a chromatin level in vivo.
Formation of an open-structure and pre-incision complex
During GG-NER, the DNA structure generated by Rad4-Rad23-Rad33 allows the recruitment and positioning of the TFIIH transcription factor, which extends opening of the helix using the ATPase/helicase activities of Rad3 and Rad25 (Figure 4B). The Rad4-Rad23-Rad33 recognition complex is released, and Rad14 and RPA are recruited to stabilize the pre-incision complex. In the case of TC-NER, TFIIH is already positioned at the lesion (see below). The presence of the lesion in DNA is then reassessed in a “verification step” mediated by TFIIH, RPA, and Rad14. RPA binds the undamaged strand, whereas Rad14 binds to the DNA lesion (De Laat et al. 1999). In the absence of lesion verification, the NER reaction aborts before dual incision occurs (Sugasawa et al. 2001).
TFIIH:
TFIIH is an essential factor required for the initiation of transcription at RNA Pol II promoters, and it also is required for NER. A role of TFIIH in NER emerged when the RAD3 and RAD25 gene products were identified as components of TFIIH (Feaver et al. 1993). Rad3 is a DNA-dependent ATPase with Y′ to X′ DNA helicase activity (Sung et al. 1987; Harosh et al. 1989); the ATPase activity of Rad3 is essential for NER, but not for transcription (Feaver et al. 1993). Rad25 is also a DNA-dependent ATPase, but has DNA helicase activity with an opposite, X′ to Y′ polarity (Guzder et al. 1994; Sung et al. 1996). In contrast to Rad3, the ATPase/helicase activity of Rad25 is essential during both NER and transcription.
TFIIH is composed of 10 subunits that can be divided into two subcomplexes: a core TFIIH subcomplex with seven subunits (Rad25, Rad3, Tfb1, Tfb2, Ssl1, Tfb4, and Tfb5) and a CAK kinase complex composed of three subunits (Kin28, Ccl1, and Tfb3) (Egly and Coin 2011; Gibbons et al. 2012). The core TFIIH complex alone is highly active in NER (Svejstrup et al. 1995); only Tfb5 is dispensable for viability and for NER (Giglia-Mari et al. 2004; Ranish et al. 2004). Absence of Tfb5, however, results in poor growth, enhanced sensitivity to UV radiation, and greatly reduced NER activity in vitro (Ranish et al. 2004; Zhou et al. 2007). The roles of Tfb1, Ssl1, Tfb2, Tfb4, and Tfb5 in NER are probably to structure the TFIIH core complex via protein–protein interactions. The catalytic role of TFIIH during NER primarily reflects the opposite polarities of the Rad3 and Rad25 ATPase/helicase activities, which unwind DNA flanking the lesion. In addition, TFIIH interacts with Rad4, Rad23, and RPA (Bardwell et al. 1994b; Guzder et al. 1995a). TFIIH is thus a pivotal component of NER because of its intrinsic helicase activities and its interactions with other essential NER components.
Rad14:
Rad14 contains a zinc-finger domain and binds with high affinity to UV-damaged DNA (Guzder et al. 1993). It is essential for incision of UV-damaged DNA in a reconstituted system (Guzder et al. 1995b) and is required for NER in vivo. Rad1-Rad10 forms a complex with Rad14 through a direct interaction between Rad1 and Rad14 (Guzder et al. 1996b); the biological significance of the interaction is suggested by the high UV sensitivity of mutant Rad1 proteins that are unable to interact with Rad14 in vitro (Guzder et al. 2006).
RPA:
RPA is the eukaryotic counterpart of the E. coli single-strand binding protein (SSB) and is composed of three subunits encoded by the RFA1, RFA2, and RFA3 genes. It binds with high affinity to single-strand DNA and is indispensable during NER (Guzder et al. 1995b). RPA also binds to Rad14 and to TFIIH (see below), and these interactions are presumably important during NER (Huang et al. 1998).
Dual incision
Following formation of a pre-incision complex, the Rad2 and Rad1-Rad10 structure-dependent endonucleases are positioned to form a new complex with TFIIH, RPA, and Rad14 (Figure 4C). Because neither Rad2 nor Rad1-Rad10 has specificity for damaged DNA, each must be targeted through interactions with other proteins. Rad2 thus interacts with TFIIH via Tfb1 (Lafrance-Vanasse et al. 2012), and Rad1-Rad10 forms a complex with Rad14 (Guzder et al. 1996b). Rad2 makes an incision 2–8 nt from the lesion on the 3′ side, while Rad1-Rad10 makes an incision 15–24 nt from the lesion on the 5′ side (Evans et al. 1997). The lesion-containing oligonucleotide thus generated is released together with other NER factors. A recent study in human cells suggests that 5′ incision precedes 3′ incision (Staresincic et al. 2009).
Rad1-Rad10 complex:
Rad1 forms a stable complex with Rad10 that degrades circular single-strand DNA (Bailly et al. 1992; Tomkinson et al. 1993) and nicks supercoiled DNA, probably at transient, single-strand regions (Tomkinson et al. 1994). The major activity of Rad1-Rad10, however, is as a structure-dependent endonuclease that recognizes the junction between single- and double-strand DNA. It specifically removes unpaired 3′ tails by nicking within duplex DNA at a position 2–5 nt from the junction (Bardwell et al. 1994a; Davies et al. 1995; Rodriguez et al. 1996). In NER reactions reconstituted from purified proteins, Rad1-Rad10 is essential for the incision of UV-damaged DNA (Guzder et al. 1995b). The properties of purified Rad1-Rad10 are consistent with nicking of the damaged strand on the 5′-side of a lesion after the DNA helix has been locally unwound. It should be noted that the role of Rad1-Rad10 is not limited to NER; the complex is also important for removing 3′ dirty ends generated during BER (see above) and nonhomologous 3′ tails that arise during homologous recombination (Lyndaker and Alani 2009).
Rad2:
The Rad2 protein is endowed with an endonuclease activity that degrades circular, single-strand DNA and a Y′ to X′ exonuclease activity that digests single- or double-strand DNA (Habraken et al. 1993, 1994). Like Rad1-Rad10, Rad2 is a junction-specific endonuclease, but cleaves with opposite polarity. Rad2 thus removes 5′-overhanging tails and processes bubble structures by nicking duplex DNA 1 nt from a single- to double-strand junction (Habraken et al. 1995).
Resynthesis and ligation
In human cells, dual incision and repair synthesis are closely coordinated. Following incision to create a lesion-containing oligonucleotide, RPA and XPG (yeast Rad2) associate with the PCNA clamp and the replication factor C (RFC) clamp loader to form a platform for Pol δ (Mocquet et al. 2008); the recruitment of Pol δ is associated with release of the remaining NER factors (Figure 4D). In human cells, three DNA polymerases (Pol δ, Pol ε, and Pol κ) are involved in repair synthesis (Ogi et al. 2010; Lehmann 2011). Although poorly documented in yeast, data suggest that either Pol δ or Pol ε can carry out repair synthesis (Budd and Campbell 1995). The final ligation reaction is performed by DNA ligase 1, the product of the CDC9 gene (Budd and Campbell 1995).
TC-NER
TC-NER promotes rapid, strand-specific removal of transcription-blocking lesions by targeting the NER apparatus to a stalled RNA Pol II complex (reviewed by Hanawalt and Spivak 2008). TC-NER was discovered in mammalian cells, where repair of CPDs is faster in the expressed DHFR gene than in transcriptionally silent downstream sequences (Bohr et al. 1985). Importantly, the more efficient repair of CPDs in expressed sequences specifically reflects the preferential removal of lesions from the transcribed DNA strand (Mellon et al. 1987). In yeast, CPDs at the transcriptionally active MATα locus are similarly repaired faster than those at the inactive HMLα locus (Terleth et al. 1990), but it was initially not clear whether this reflected a difference in chromatin structure or a specific role of transcription in repair. Preferential repair of CPDs in the transcribed strand (TS) relative to the nontranscribed strand (NTS) was subsequently demonstrated in the RPB2 gene (Sweder and Hanawalt 1992).
As noted previously, all genes that are essential for GG-NER are required for TC-NER, with the exception of RAD7 and RAD16, which are required only for repair of the NTS (Verhage et al. 1994). TC-NER, which is triggered when RNA Pol II stalls, has two subpathways: one dependent on the Rad26 protein and a second dependent on the Rpd9 subunit of RNA Pol II. A connection between TC-NER and mRNP biogenesis/export, which affects transcript elongation, has also emerged (Gaillard et al. 2007), with a screen of the yeast deletion collection for elongation defects revealing additional players within the two major TC-NER subpathways (Gaillard et al. 2009).
Rad26 and Rad28:
Genes specifically involved in TC-NER were first identified in humans, where their loss is responsible for Cockayne syndrome (CS). CSA- or CSB-deficient cells are very sensitive to UV, a property that allowed cloning of the human CSA and CSB genes (Troelstra et al. 1992; Henning et al. 1995). Based on sequence homology to the encoded proteins, yeast homologs were identified and the corresponding genes were named RAD28 and RAD26, respectively (Van Gool et al. 1994; Bhatia et al. 1996). CSB and Rad26 share strong sequence homology, which includes the seven conserved motifs of DNA/RNA helicases in the SNF2 subfamily. Although both proteins exhibit DNA-dependent ATPase activity, neither has detectable helicase activity (Guzder et al. 1996a). In the case of CSB, the ATPase activity is important for in vivo function (Citterio et al. 1998). CSA/Rad28 is a WD40 repeat protein with no identified catalytic activity and is probably involved in protein interactions (Henning et al. 1995; Bhatia et al. 1996). In contrast to human CS cells, yeast strains lacking either Rad26 or Rad28 are not UV sensitive, which explains why the corresponding genes were not recovered in early mutant screens.
Analysis of strand-specific repair of CPDs demonstrated that repair of the TS is significantly delayed in rad26Δ mutants (Van Gool et al. 1994), but is not affected in rad28Δ cells (Bhatia et al. 1996). Although not evident in a rad26Δ single mutant, an effect of Rad26 on survival after UV irradiation can be observed in the absence of GG-NER, with a rad16Δ rad26Δ double mutant being more UV sensitive than a rad16Δ single mutant (Bhatia et al. 1996; Verhage et al. 1996). A rad16Δ rad26Δ double mutant, however, is still less UV sensitive than a completely NER-deficient strain such as rad14Δ, suggesting residual repair of CPDs on the TS by a Rad26-independent TC-NER subpathway (Verhage et al. 1996).
Recent yeast studies suggest that Rad26 may be associated with RNA Pol II during transcriptional elongation; its “recruitment” to the site of the lesion would thus result from stalling of RNA Pol II in an elongation mode (Malik et al. 2010). Furthermore, Rad26 is subject to Mec1-dependent phosphorylation, which enhances the rate of TC-NER of UV-induced damage in vivo (Taschner et al. 2010). Finally, Rad26 copurifies with Def1, a factor that is involved in the ubiquitination of RNA Pol II and leads to its degradation by the proteasome. Although Def1 does not affect TC-NER of UV-induced DNA damage, genetic data nevertheless suggest a connection between Def1 and NER (Woudstra et al. 2002; Reid and Svejstrup 2004).
Model for Rad26-dependent TC-NER:
The stalling of RNA Pol II at a lesion in the TS is presumed to stabilize the interaction with the Rad26 protein. As illustrated in Figure 5, the stalled complex has two alternative outcomes that depend on whether the lesion stalls the transcription machinery transiently or permanently. Inactivation of Rad26 results in a delay in messenger RNA (mRNA) synthesis, and data suggest that Rad26 can promote elongation through endogenous DNA damages such as 8-oxoG, 3-MeA, and AP sites (Lee et al. 2001, 2002; Yu et al. 2003). In vitro, some lesions can be efficiently bypassed by RNA Pol II with the help of elongation factors such as CSB, elongin, and TFIIS (Charlet-Berguerand et al. 2006). At a moderately blocking lesion, Rad26 is thought to promote bypass and thereby allow transcription to continue. Because such bypass may be associated with the incorporation of incorrect ribonucleotides into the corresponding mRNA, it can lead to the production of aberrant proteins via a process termed “transcriptional mutagenesis” (Bregeon and Doetsch 2011). At strongly blocking lesions, RNA Pol II permanently stalls without the possibility of bypass, and TC-NER is triggered. Rad26 initiates chromatin remodeling to attract additional NER factors, and, in humans, a CSA-containing E3 complex recruits XAB2, HMGN1, and TFIIS (Fousteri and Mullenders 2008; Hanawalt and Spivak 2008). These latter factors allow backtracking of RNA Pol II without dissociation from the template, which exposes the lesion to the NER machinery. Following processing of the lesion by NER, transcription can be rapidly resumed.
It should be noted that the lack of CPD repair on the TS fails to explain the growth and neurological defects associated with CS, as these defects are absent in NER-defective patients (Hanawalt and Spivak 2008). In the absence of Rad26/CSB, permanent stalling of RNA Pol II at endogenous DNA lesions would significantly impair RNA synthesis and might generate a signal for apoptosis and cell death, which could account for the severe growth defects observed in CS patients (Hanawalt and Spivak 2008). In contrast to the situation in human cells, lesion-stalled RNA Pol II complexes might be disassembled by the replication machinery in rapid-cycling yeast cells, leading to only minor, if any, effects on cell survival.
Rpb9 subpathway:
Rpb9, a nonessential subunit of RNA Pol II, mediates a Rad26-independent TC-NER subpathway; rpb9Δ rad26Δ double mutants are completely defective in TC-NER (Li and Smerdon 2002). rad16Δ rad26Δ rpb9Δ triple-mutant cells are extremely sensitive to UV radiation, similar to a rad1Δ strain, and the removal of CPDs in both the TS and the NTS is abolished (Li and Smerdon 2002). These data suggest a direct or indirect recruitment of NER factors by Rpb9, similar to that described for Rad26-dependent TC-NER.
TC-NER at AP sites:
Involvement of NER in the removal of AP sites was inferred by the synergistic increase in the killing and mutagenic effects of MMS observed when both BER and NER are disabled (Swanson et al. 1999; Torres-Ramos et al. 2000). Recent results demonstrate that AP sites likely are not directly recognized by the NER machinery, but rather that their removal is via TC-NER that is triggered when RNA Pol II stalls at an AP site (Kim and Jinks-Robertson 2010).
Additional Remarks Concerning NER Mechanisms
The specific models proposed in Figures 4 and 5 incorporate genetic data from yeast with biochemical data obtained primarily with purified human proteins. While the overall schemes presented for GG-NER and TC-NER are likely correct, details of the reactions will surely be modified with time. As noted, yeast Rad4 is absolutely required for the repair of CPDs in the TS, whereas human xeroderma pigmentosum group C (XPC) protein is not; inactivation of yeast Rad28 does not impair TC-NER, whereas human CSA cells are deficient in TC-NER; inactivation of Rad26 in yeast does not confer UV sensitivity, whereas CSB cells are highly sensitive to UV radiation; and yeast has an alternative Rpb9-dependent TC-NER subpathway that is absent in human cells.
Yeast studies dealing with repair at nucleotide resolution and in the context of chromatin have not been included here; these important aspects of NER have been recently reviewed (Waters et al. 2009; Reed 2011). Also not considered here is the checkpoint-signaling role of single-strand gaps created during NER (Giannattasio et al. 2010) and roles of post-translational protein modifications. Furthermore, it should be noted that the dominant role of the NER pathway in promoting survival following acute exposure to high levels of UV damage becomes secondary to that of the RAD6 lesion-bypass pathway (see below) when cells are exposed to low levels of chronic damage (Hishida et al. 2009). Finally, while the NER pathway promotes genome integrity in dividing cells, it appears to be required for most UV-induced mutagenesis that occurs in nondividing yeast cells (Eckardt and Haynes 1977; James and Kilbey 1977).
Mismatch Repair
The mismatch repair (MMR) pathway removes helical distortions that arise when errors are made during DNA synthesis or when non-identical duplexes exchange strands during recombination (reviewed by Harfe and Jinks-Robertson 2000c; Kunkel and Erie 2005; Hsieh and Yamane 2008). During replication, mismatch correction limits mutagenesis; during recombination, correction of mismatches within heteroduplex DNA intermediates generates gene conversion events. In addition to initiating mismatch removal during recombination, the MMR system monitors identity between interacting molecules, which can limit use of non-identical substrates as repair templates. Finally, loss of MMR sensitizes cells to the antimetabolite 5-fluorouracil (Matuo et al. 2010) and can alter resistance to DNA damage (Bertrand et al. 1998; Durant et al. 1999; Cejka et al. 2005). Importantly, the sensitivity of methyltransferase (Mgt1)-deficient yeast or mammalian cells to MNNG depends on functional MMR (Branch et al. 1993; Cejka et al. 2005). This is thought to reflect repetitive, futile attempts of the MMR machinery to repair the O6-MeG:T mismatches generated during replication of damaged templates. The focus here will be on the replication-related role of the MMR machinery, and relevant genes are summarized in Table 3.
Table 3. MMR genes.
| Gene name (alternative) | Protein size (kDa) | Description | Mammalian counterpart |
|---|---|---|---|
| MSH1 | 109.4 | Mitochondrial MutS homolog. | — |
| MSH2 (PMS5) | 108.9 | Forms MutSα and MutSβ mismatch-recognition heterodimers with Msh6 and Msh3, respectively. | MSH2 |
| MSH3 | 116.5 | Component of MutSβ, which recognizes small and large IDLs. | MSH3 |
| MSH4 | 99.2 | Interacts with Msh5 to form the MutSγ heterodimer, which is meiotic-specific and binds to HJs. | MSH4 |
| MSH5 | 102.2 | Interacts with Msh4 to form the MutSγ heterodimer. | MSH5 |
| MSH6 (PMS3) | 140.1 | Component of MutSα, which recognizes base-base mismatches and small IDLs. | MSH6 |
| PMS1 | 99.4 | Interacts with Mlh1 to form the MutLα heterodimer; has endonuclease activity. | PMS2 |
| MLH1 (PMS2) | 87.1 | Interacts with Pms1 to form the MutLα heterodimer. | MLH1 |
| MLH2 | 78.2 | Interacts with Mlh1 to form the MutLβ heterodimer; important for repair of some frameshift intermediates. | PMS1 |
| MLH3 | 82.0 | Interacts with Mlh1 to form the MutLγ heterodimer; required for meiotic crossover formation and for repair of some frameshift intermediates. | MLH3 |
| POL30 (REV6) | 28.9 | Subunit of PCNA homotrimer; required for MMR and interacts with MutSα, MutSβ, and MutLα. | PCNA |
| EXO1 (DHS1) | 80.2 | Y′ to X′ Double-strand exonuclease. | EXO1 |
Bacterial paradigm
The intellectual framework for eukaryotic MMR derives from the E. coli system, which contains three dedicated mutator or “Mut” proteins (reviewed in Modrich and Lahue 1996). A MutS homodimer binds mismatches, a MutL homodimer coordinates mismatch detection with downstream processing steps, and MutH nicks the nascent strand to initiate removal. Discrimination between nascent and template strands is provided by transient, hemi-methylation of DNA in the wake of replication, with MutH specifically nicking the new, unmethylated strand. The MutH-nicked strand is then degraded by the concerted action of a helicase (UvrC) and one of four single-strand exonucleases, DNA polymerase fills the gap, and ligase seals the remaining nick.
In eukaryotes, the single MutS and MutL homodimers are replaced with heterodimers, each of which has specialized functions. An explanation for the evolution of heterodimeric complexes in eukaryotes emerged when the crystal structure of the bacterial MutS homodimer revealed that it is a structural heterodimer (Lamers et al. 2000; Obmolova et al. 2000). The basic functions of the MutS and MutL homologs are highly conserved within the eukaryotic lineage, with an early link between MMR defects and the genetic instability characteristic of human nonpolyposis colorectal cancer coming from studies of dinucleotide repeat instability in yeast (Strand et al. 1993). A MutH-like protein is absent in eukaryotes, as well as in most bacteria, and methylation does not serve as a strand-discrimination signal during replication. The exact nature of this signal(s) and the mechanism of strand removal has yet to be fully resolved.
MutS homologs
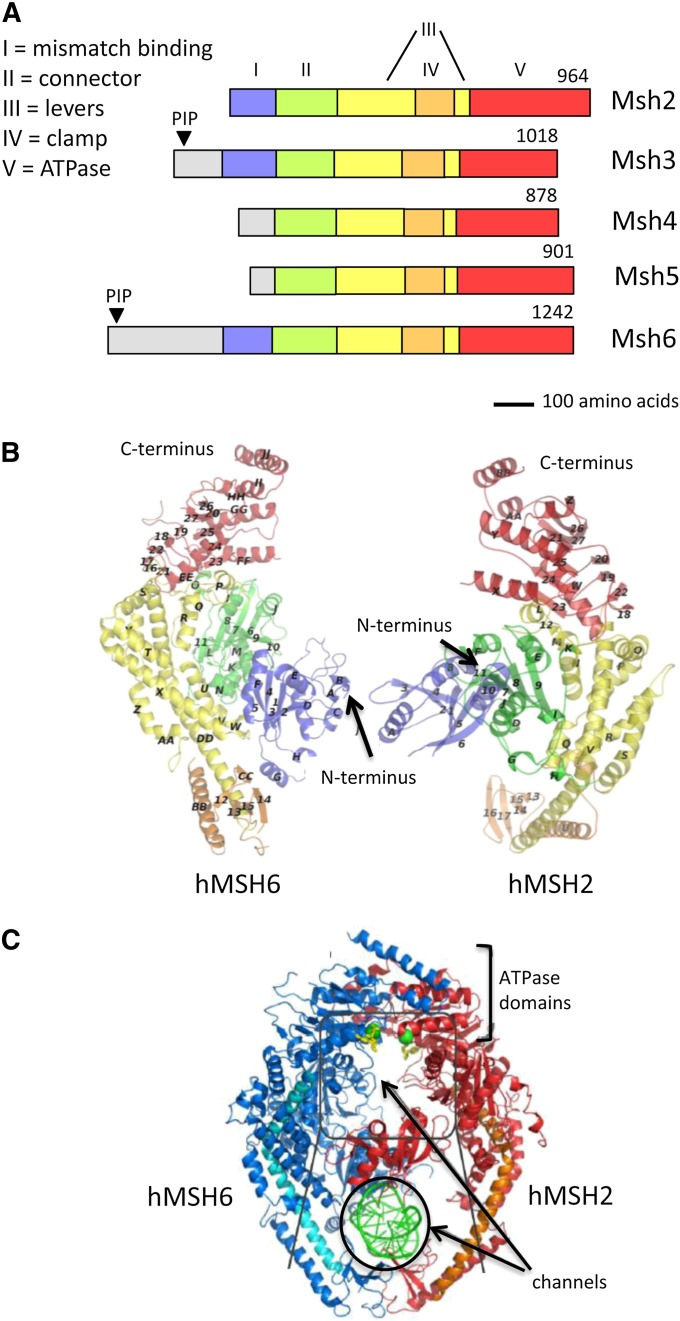
The yeast genome encodes six MutS homologs, four of which (Msh1, Msh2, Msh3, and Msh6) were identified by homology to bacterial MutS proteins (Reenan and Kolodner 1992a,b; New et al. 1993; Marsischky et al. 1996). Msh1 functions exclusively in the mitochondria (Mookerjee et al. 2005; Sia and Kirkpatrick 2005), while Msh2, Msh3, and Msh6 are important for maintaining nuclear genome stability. Deletion of MSH2 completely disables mitotic and meiotic MMR (Reenan and Kolodner 1992b), but msh3Δ or msh6Δ mutants exhibit weaker phenotypes (New et al. 1993; Johnson et al. 1996b; Marsischky et al. 1996). A seminal observation was that an msh3Δ msh6Δ double mutant is phenotypically indistinguishable from an msh2Δ single mutant, leading to the proposal of functionally redundant, MutS-like complexes in which Msh2 partners with either Msh6 or Msh3 (Johnson et al. 1996b; Marsischky et al. 1996). These complexes are referred to as MutSα and MutSβ, respectively. Msh4 and Msh5 form a third, heterodimeric MutS-like complex known as MutSγ, which has meiotic-specific roles (Pochart et al. 1997).
Recognition specificities of MutSα and MutSβ:
Nuclear mismatch-recognition activity is shared between MutSα and MutSβ, with the specificities of the complexes being deduced by comparing the phenotypes of msh6Δ and msh3Δ single mutants, respectively, with those of an msh2Δ single mutant (Detloff et al. 1991; Marsischky et al. 1996; Luhr et al. 1998; Nicholson et al. 2000). The general consensus is that MutSα and MutSβ are specialized to remove base-base mismatches and large insertion-deletion loops (IDLs), respectively, but are largely redundant with respect to small (<4 nt) IDLs (reviewed by Harfe and Jinks-Robertson 2000c). Similar to bacterial MutS, MutSα efficiently initiates repair of all base-base mismatches with the exception of C:C (Detloff et al. 1991). As noted previously, MutSα repairs the mismatch generated when adenine is misincorporated opposite a template 8-oxoG (Ni et al. 1999) and is considered to be the functional homolog of the E. coli MutY DNA glycosylase (Figure 2). Finally, with regard to the “exclusive” role of MutSα in base-base mismatch removal, some transversions increase in an msh3Δ background (Harrington and Kolodner 2007), suggesting that MutSβ functionally replaces MutSα in some contexts.
Forward mutation assays have shown that IDLs are corrected more efficiently than are base–base mismatches (Marsischky et al. 1996; Yang et al. 1999; Lujan et al. 2012). Although there is functional redundancy between MutSα and MutSβ with respect to repairing small IDLs, the relative correction efficiencies of the two complexes can vary dramatically as a function of position of the extrahelical loop, primary sequence, and surrounding sequence context (Strand et al. 1995; Harfe and Jinks-Robertson 1999, 2000a; Marsischky and Kolodner 1999; Gragg et al. 2002). Early studies with tandem repeats suggested a 13- to 16-nt size limit for IDL recognition by MutSβ (Sia et al. 1997), but subsequent analyses suggest that larger loops also may be repaired (Kirkpatrick and Petes 1997; Harfe et al. 2000; Kearney et al. 2001). The removal of larger IDLs, as well as MutSβ-dependent removal of nonhomologous 3′ tails during homologous recombination (Sugawara et al. 1997; reviewed by Lyndaker and Alani 2009), involves the Rad1-Rad10 endonuclease complex, which is essential during NER. A second MMR- and NER-independent pathway for repairing large IDLs has been identified in cell extracts (Sommer et al. 2008), but its physiological relevance is unclear.
Central role of ATP binding/hydrolysis:
MMR is an ATP-dependent process, and a major area of emphasis has been the functional significance of ATP binding/hydrolysis by MutS complexes, which are Walker-type ATPases (reviewed by Kunkel and Erie 2005). In vitro, ATP destabilizes the interaction of MutSα with mismatches that are well repaired in vivo, but fails to destabilize binding to hairpins (Alani 1996), which are refractory to MMR (Nag et al. 1989). This key observation led to the proposal that ATP-dependent dissociation of MutSα from a mismatch licenses the downstream steps required for repair (Alani 1996).
Mutation of the conserved Walker ATP binding/hydrolysis motifs of Msh2 or Msh6 results in a null phenotype (Studamire et al. 1998; Drotschmann et al. 2002), and these alleles exert a dominant-negative effect (Studamire et al. 1998; Das Gupta and Kolodner 2000), which reflects an inability of MutSα to dissociate from mismatches (Hess et al. 2006). Although the role of ATP hydrolysis has been controversial (Blackwell et al. 1998; Gradia et al. 1999; Junop et al. 2001), the current consensus is that ADP-ATP exchange converts a mismatch-bound MutSα complex into a sliding clamp whose movement does not require ATP hydrolysis (Mazur et al. 2006; Hargreaves et al. 2010).
Structural studies of MutS complexes:
Crystal structures of bacterial MutS (Lamers et al. 2000; Obmolova et al. 2000), human MUTSα (hMUTSα; (Warren et al. 2007) and hMUTSβ (Gupta et al. 2012) have revealed five conserved domains (Figure 6, A and B). Each complex has two channels, one of which accommodates duplex DNA; consistent with the structural asymmetry, only one subunit makes mismatch-specific contacts (Figure 6C). In the static crystal structures, DNA is sharply kinked at the position of the mismatch (Lamers et al. 2000; Obmolova et al. 2000). Dynamic DNA conformational changes have been detected by atomic force microscopy, however, when MutS binds to mismatched vs. homoduplex DNA, and these changes may be important for mismatch verification (Wang et al. 2003).
Figure 6.
Alignment of MutS homologs and MutSβ crystal structure. (A) Linear alignment of the yeast nuclear MutS homologs, with domains identified in the MutS crystal structure color-coded and indicated by Roman numerals (Obmolova et al. 2000). (B) Crystal structures of human MSH2 and MSH6, with domains colored as in A. (C) Crystal structure of hMUTSα with a mismatch, with DNA indicated in green. Crystal structures are from Warren et al. (2007).
Bacterial MutS interacts with mismatches through critical phenylalanine and glutamic acid residues, and these residues are conserved only in Msh6. Mutation of the corresponding Phe337 or Glu339 of Msh6 results in a mutator phenotype and abrogates mismatch binding in vitro (Bowers et al. 1999; Drotschmann et al. 2001; Holmes et al. 2007). Msh2 has an aromatic amino acid at a position comparable to that of Phe337 in Msh6, and its mutation is without consequence (Bowers et al. 1999; Drotschmann et al. 2001). In Msh3, the conserved Phe and Glu are replaced with lysine; the corresponding msh3-K187A,K189A allele confers variable effects on IDL repair (Lee et al. 2007).
The crystal structure of hMUTSβ suggests two distinct modes of IDL recognition (Gupta et al. 2012), and two corresponding classes of msh3 and msh2 mutants have been identified: those defective only in the repair of small IDLs and those defective in repairing large IDLs and removing nonhomologous 3′ tails (Studamire et al. 1999; Dowen et al. 2010). Msh3 mismatch specificity can be imparted to yeast MutSα by replacing the Msh6 mismatch-binding domain (MBD) with the presumptive Msh3 MBD; the reverse domain swap, however, does not yield a functional complex (Shell et al. 2007b). A separation-of-function msh2 allele that differentially affects MutSα and MutSβ provides additional evidence that these complexes sense and/or respond to mismatches differently (Lee et al. 2007; Kumar et al. 2011).
MutSγ, the Msh4-Msh5 complex:
Msh4 and Msh5 were identified in screens for meiotic-specific transcripts and recombination defects (Ross-MacDonald and Roeder 1994; Hollingsworth et al. 1995). Although a specific allele of MSH5 was reported to enhance alkylation-damage tolerance (Bawa and Xiao 2003), there has been no confirmation of a mitotic role. Msh4 and Msh5 have overall homology to Msh2, Msh3, and Msh6, but each is missing conserved domain I (Figure 6A), which separates the two channels in the MutS crystal structure (Figure 6, B and C). The Msh4-Msh5 complex thus is predicted to contain a single channel large enough to accommodate two DNA duplexes (Obmolova et al. 2000). hMSH4-MSH5 binds to progenitor and mature Holliday junctions (HJs) that connect recombining duplexes; upon addition of ATP, hMUTSγ forms an ATPase-independent sliding clamp that embraces both duplexes (Snowden et al. 2004). Only the Walker A box of the yeast Msh5 protein has been shown to be required for meiotic crossover formation (Pochart et al. 1997), but both subunits bind ATP in vitro (Snowden et al. 2008). During meiosis, MutSγ is required for detection of the single-end invasion intermediates that mature into Holliday junctions (Börner et al. 2004).
MutL homologs
The yeast genome encodes four MutL homologs: Mlh1, Mlh2, Mlh3, and Pms1. PMS1 was the first eukaryotic MMR gene identified, and its name reflects an inability to repair mismatches in recombination intermediates (Williamson et al. 1985). Cloning revealed strong homology of Pms1 to bacterial MutL proteins (Kramer et al. 1989), a property that was exploited to identify the remaining MutL homologs (Prolla et al. 1994a; Crouse 1998). Mlh1 is the common component of three MutL-like complexes: Mlh1-Pms1, Mlh1-Mlh2, and Mlh1-Mlh3, which are referred to as MutLα, MutLβ, and MutLγ, respectively (Wang et al. 1999). MutLα interacts with MutSα and MutSβ to coordinate most MMR, and MutLγ partners with MutSγ to regulate meiotic crossover formation. In addition to being the common component of three MutL-like complexes, Mlh1 interacts with the Exo1 exonuclease during MMR (see below), the Ntg2 DNA N-glycosylase/lyase, and the Sgs1 helicase via a highly conserved binding site at its C terminus, which is referred to as S2 (Gellon et al. 2002; Dherin et al. 2009).
MutLα, the Mlh1-Pms1 complex:
Initial genetic analyses placed Mlh1 and Pms1 in a common pathway, and the proteins were shown to physically interact (Prolla et al. 1994b; Pang et al. 1997). The N-terminal region of MutL homologs is highly conserved and contains an ATPase domain; although required for dimerization, the C terminus has only weak sequence conservation (reviewed by Kunkel and Erie 2005). Crystal structures of the N-terminal domain of E. coli MutL (Ban and Yang 1998), human PMS2 (Guarne et al. 2001), and yeast Pms1 (Arana et al. 2010) have been solved, as has the crystal structure of the C-terminal domain of E. coli MutL (Guarne et al. 2004). A model for the intact MutL dimer has been proposed in which the N- and C-terminal domains are separated by a proline-rich linker, with the dimer containing a large central cavity (Guarne et al. 2004).
MutL proteins belong to the GHL family of ATPases (Ban and Yang 1998), and there is functional asymmetry between the MutLα subunits. ATP binding/hydrolysis by Mlh1 is essential for MMR, but only a weak mutator phenotype results from loss of ATP binding/hydrolysis by Pms1 (Tran and Liskay 2000). Consistent with the in vivo asymmetry, Mlh1 binds ATP with higher affinity than does Pms1, suggesting that ATP binding is likely sequential (Hall et al. 2002). In two-hybrid studies, the N-terminal fragments of Pms1 and Mlh1 interact only if ATP hydrolysis by both is blocked; additional elimination of ATP binding by either fragment, however, prevents the interaction (Tran and Liskay 2000). These data suggest that conformational changes are associated with the ATP hydrolysis cycle, and such changes in MutLα have been visualized by atomic force microscopy (Sacho et al. 2008). Finally, MutLα binds cooperatively to DNA (Hall et al. 2001), and mutational impairment of DNA binding is associated with a mutator phenotype (Hoffmann et al. 2003).
As noted previously, eukaryotes do not have a MutH-like protein that nicks mismatched DNA, and yet MMR is nick-directed (Constantin et al. 2005). This conundrum was partially resolved by the discovery that the hPMS2, which is the homolog of yeast Pms1, has a latent endonuclease activity (Kadyrov et al. 2006). Site-directed mutation of the endonuclease motif eliminates mitotic function of MutLα (Erdeniz et al. 2007; Kadyrov et al. 2007) without affecting its ATPase activity or ability to form a ternary complex with mismatched DNA and MutSα (Kadyrov et al. 2007). A similar endonuclease motif is present in Mlh3, but is absent in Mlh1 and Mlh2.
Interaction of MutSα and MutSβ with MutLα:
MutSα/MutSβ must interact with MutLα to initiate downstream steps of MMR, and ternary complexes with mismatched DNA have been described (Habraken et al. 1997, 1998). A domain of Msh2 required to mediate interaction with MutLα resides within the connector loop (see Figure 6); site-directed mutagenesis of this region abolishes the MutSα-MutLα interaction in vitro and is associated with a mutator phenotype (Mendillo et al. 2009). Whether this region of Msh2 is similarly important for MutSβ-MutLα interaction has not been examined. Recent studies, however, have identified a region of hMSH3 that facilitates ternary complex formation with hMUTLα and mismatched DNA, and this region overlaps a PCNA interaction peptide (PIP) (see below) motif at the extreme N terminus of hMSH3 (Charbonneau et al. 2009; Iyer et al. 2010). These data may explain why mutation of the Msh3 PIP domain disables MMR to a greater extent that does mutation of the Msh6 PIP box (Clark et al. 2000). In terms of the region of MutLα that mediates interaction with the MutS complexes, the N terminus and C terminus of hMLH1 are important for interaction with hMUTSα (Plotz et al. 2006) and hMSH3 (Charbonneau et al. 2009), respectively.
MutLγ, the Mlh1-Mlh3 complex:
Subtle phenotypes of mlh3Δ (as well as mlh2Δ) mutants have been reported in some frameshift-reversion assays, where the corresponding MutL complex seems to partner with MutSβ (Flores-Rozas and Kolodner 1998; Harfe et al. 2000). The most significant phenotype of mlh3Δ mutants, however, is a deficit in meiotic crossover formation, a phenotype shared with mlh1Δ, msh4Δ, and msh5Δ mutants (reviewed by Hoffmann and Borts 2004). Site-directed mutagenesis of the Mlh3 endonuclease domain results in phenotypes similar to that of a null mutant (Nishant et al. 2008). A recent study demonstrated that MutLγ is the major meiotic HJ resolvase in yeast (Zakharyevich et al. 2012), thereby providing a direct explanation for crossover reduction in mutants. An attractive model is that MutSγ binds to and stabilizes HJs, which are then symmetrically nicked by MutLγ.
Other proteins important for MMR
The E. coli paradigm suggests that repair of replication errors requires (1) a mechanism(s) to discriminate the nascent and template strands and (2) helicases and/or exonucleases to degrade the mismatch-containing segment. The only two proteins implicated unequivocally in these steps are the PCNA sliding clamp and the Exo1 Y′ to X′ exonuclease.
PCNA sliding clamp:
The PCNA sliding clamp is a donut-shaped homotrimer of the Pol30 protein; it encircles duplex DNA and is loaded/unloaded by RFC at primer-template junctions. Originally described as a processivity factor for replicative DNA polymerases, PCNA serves as a landing pad for multiple DNA-metabolic proteins, most of which interact with the intermonomer region of the clamp via a highly conserved PIP domain (reviewed by Moldovan et al. 2007). The involvement of PCNA in MMR was discovered through characterization of pol30 missense alleles that have an associated mutator phenotype (Johnson et al. 1996a; Umar et al. 1996; Lau et al. 2002). PCNA interacts physically with MutSα (Clark et al. 2000; Flores-Rozas et al. 2000), MutSβ (Johnson et al. 1996a), and Mlh1 (Umar et al. 1996; Lee and Alani 2006). Both Msh3 and Msh6 (but not Msh2) have a PIP box near the amino terminus, and its mutation partially disables MMR (Clark et al. 2000; Flores-Rozas et al. 2000). Interaction of MutSα with PCNA stimulates mismatch binding in vitro (Flores-Rozas et al. 2000), and loss of the Msh6 PIP motif reduces colocalization with PCNA in vivo (Hombauer et al. 2011a). A potential PIP box in Mlh1 also has been identified, but whether this mediates interaction with PCNA is unclear (Lee and Alani 2006). In addition to its PIP domain, Msh6 contains an extended N-terminal domain (NTD) that is absent in other MutS proteins (Figure 6A) and forms an unstructured tether to PCNA (Shell et al. 2007a). The NTD of Msh6 additionally has nonspecific DNA-binding activity that is functionally important (Clark et al. 2007).
Interactions with PCNA are assumed to target MMR proteins to sites of new DNA synthesis and to contribute, at least partially, to strand discrimination (Umar et al. 1996). PCNA also may act as a scaffold to coordinate sequential steps during replication-error repair (Lee and Alani 2006). Finally, the tight link that PCNA provides between MMR and replication may circumvent the potential obstacle posed by nucleosomes that assemble behind the replication fork (Gorman et al. 2010).
Exo1 exonuclease:
Exo1 is double-strand-specific exonuclease that degrades single strands in the Y′ to X′ direction (for a review see Tran et al. 2004); its role in mismatch correction was first described in Schizosaccharomyces pombe (Szankasi and Smith 1995). Because Exo1 works on duplex DNA, there is no need for a helicase to unwind the nicked segment; accordingly, no helicases have been implicated in eukaryotic MMR. The budding yeast Exo1 interacts with Msh2 (Tishkoff et al. 1997a) and with Mlh1 (Tran et al. 2001), and it has been suggested to have a structural (Amin et al. 2001; Tran et al. 2001) as well as an enzymatic role during MMR (Sokolsky and Alani 2000). In contrast to the strong mutator phenotype of msh2Δ/mlh1Δ strains, exo1Δ mutants have only a weak mutator phenotype (Tishkoff et al. 1997a). The exo1Δ mutator phenotype reflects a role of Exo1 both during MMR and in the promotion of error-free damage bypass (Tran et al. 2007). Mutating the Mlh1-interacting peptide of Exo1 or the S2 site of Mlh1 results in a modest MMR-dependent mutator phenotype, which is greatly enhanced when combined with hypomorphic mutations of MLH1 or PMS1 (Tran et al. 2007; Dherin et al. 2009). Interestingly, mutating the PIP box of Msh6, which by itself causes only a weak mutator phenotype, completely eliminates MutSα-dependent MMR in an exo1Δ background (Hombauer et al. 2011a).
Are there additional participants in MMR?:
Because of the weak mutator phenotype of exo1Δ mutants and the existence of redundant Y′ to X′ and X′ to Y′ exonucleases in E. coli MMR, an early assumption was that there would be additional nucleases involved in eukaryotic MMR. To date, two candidates have emerged: the Rad27 flap endonuclease and X′ to Y′ exonucleolytic proofreading activities of the replicative DNA polymerases Pol δ and Pol ε. In the case of Rad27, its loss results in dinucleotide repeat instability comparable to that seen in msh2Δ mutants (Johnson et al. 1995), but whether this reflects a direct role in MMR has been questioned (Tishkoff et al. 1997b). An involvement of Pol δ and Pol ε in MMR has been inferred from genetic analyses of homopolymer run instability in exonuclease-defective pol3-01 or pol2-4 strains (Tran et al. 1999). As to whether there might be additional, as yet unidentified, players during MMR, no viable candidates have emerged despite multiple genetic screens (Jeyaprakash et al. 1994; Tran et al. 1999; Amin et al. 2001; Sia et al. 2001; Huang et al. 2003).
Putting it all together: the mechanism(s) of mismatch removal
Interaction of MMR proteins with PCNA is a simple way to focus mismatch correction to regions of new DNA synthesis. How the initiating nicks are targeted specifically to the nascent strand and precisely how the nicked strand is removed remain to be fully elucidated, and differences may exist depending on whether an error arises during leading- vs. lagging-strand synthesis. Indeed, the MMR system more efficiently removes lagging- than leading-strand errors (Pavlov et al. 2003; Kow et al. 2007; Mudrak et al. 2009; Lujan et al. 2012). Most genetic and structure–function studies of eukaryotic MMR have been done in yeast, whereas most attempts to biochemically reconstitute the system have utilized purified human proteins. Below, recent results that provide insight into how MMR likely functions in vivo are discussed, and a model that incorporates current data is presented in Figure 7.
Figure 7.
Proposed mechanisms of MMR. (A) MutSα/β (the red clamp-like structure) is tethered to PCNA (green donut) during replication. Following mismatch recognition/verification by MutSα/β, MutLα (blue heterodimer) interacts to form a ternary complex. (B) . On the lagging strand of replication, the 5′ ends of Okazaki fragments can serve as an entry site for mismatch removal. On either the lagging or leading strand of replication, the asymmetry of PCNA can provide a strand-discrimination signal for MutLα-catalyzed nicking of the nascent strand. On the leading strand, additional nicks may be provided by the activity of RNase H2 at rNMPs incorporated by DNA Pol ε. (C) Removal of the mismatch-containing nicked strand from the 5′ direction can be accomplished by Exo1 (purple pacman). Additional possible removal mechanisms include strand displacement and 5′-flap removal by Rad27 or mismatch excision from the 3′ direction using the proofreading activity of Pol δ/ε (yellow oval). The resulting gap is filled (dashed lines) by DNA polymerase and sealed by DNA ligase. The 3′ ends of single strands are indicated by half-arrowheads.
Temporal and spatial control of MMR:
The general assumption that, as in E. coli, MMR is temporally coupled to replication has been confirmed by limiting MutSα expression to specific phases of the cell cycle (Hombauer et al. 2011b). Although a low level of MMR has been detected in nondividing cells, it lacks strand discrimination, presumably because the normal link with replication is missing (Rodriguez et al. 2012). Such residual MMR, however, may provide a mechanism to introduce a potentially beneficial change into both strands of duplex DNA under stress conditions. A similar, noncanonical MMR mechanism may be relevant to somatic hypermutation of immunoglobulin genes and to methylation-associated mutagenesis in humans (Peña-Diaz et al. 2012).
Visualization of fluorescently tagged MMR proteins has revealed that MutSα foci localize to replication factories and that this requires interaction of Msh6 with PCNA (Hombauer et al. 2011a). The number of these foci is insensitive to the rate of replication errors, however, suggesting that MutSα is a constitutive component of replication factories. Although the formation of MutLα foci is, as expected, dependent on functional MutSα, foci are rarely coincident. In contrast to MutSα foci, the number of MutLα foci increases as a function of replication-error load, suggesting that MutLα foci reflect sites of active MMR. Taken together, these observations suggest that there may be more than one mode of mismatch recognition and that sites of active repair complexes likely contain superstoichiometric numbers of MutLα relative to MutSα complexes.
Origins of nicks:
The 5′ ends of Okazaki fragments are a natural source of nicks on the lagging strand of replication and provide an entry site for Exo1 to initiate Y′ to X′ removal of a mismatch-containing segment. Although this potentially solves the strand-discrimination problem during lagging-strand synthesis, there must be an additional mechanism(s) for generating strand-specific nicks during the relatively continuous synthesis of the leading strand (Figure 7B). The endonuclease activity of MutLα provides at least one source of nicks (Kadyrov et al. 2006, 2007; Erdeniz et al. 2007). In a purified in vitro system, interaction with PCNA is required to activate the endonuclease activity of hMUTLα, and the inherent asymmetry of PCNA with respect to the 3′ end dictates that the “correct” strand is nicked (Pluciennik et al. 2010). It also has been suggested that ribonucleotide monophosphates (rNMPs), which are preferentially incorporated into the leading strand during DNA synthesis (Nick McElhinny et al. 2010b), might provide an additional strand-discrimination signal, through either spontaneous hydrolysis or RNaseH2-dependent cleavage of the DNA backbone (Clark and Kunkel 2010).
Mismatch removal:
Exo1 is the only undisputed protein involved in degrading mismatch-containing DNA in vivo, and yet exo1 null mutants do not have a strong mutator phenotype (Tishkoff et al. 1997a). At least in the purified human system, strand-displacement synthesis provides an alternative way to directionally remove a mismatch-containing segment if a 5′ nick is present (Figure 7C) (Kadyrov et al. 2009). If such a mechanism were to operate in vivo, one would expect Rad27 to be involved in removing the flap thus created. As noted previously, Rad27 was early implicated in yeast MMR (Johnson et al. 1995), but subsequent work revealed that the mutation signature associated with its loss is very different from that associated with MMR defects (Tishkoff et al. 1997b). If Exo1 and Rad27 were to work in redundant pathways for mismatch removal from a 5′ nick, however, an involvement of Rad27 in MMR might have been masked. The inviability of the exoΔ1 rad27Δ double mutant, which is thought to reflect defective Okazaki fragment processing (Tishkoff et al. 1997a; Tran et al. 2002), precludes testing a redundant role of these proteins during MMR. It remains possible that mismatch removal could be effected during DNA synthesis by the 3′–5′ exonuclease activity of replicative DNA polymerases (Figure 7C).
Ribonucleotide Excision Repair
Either RNase H1 or RNase H2 can degrade RNA transcripts that remain stably base paired with the DNA template (R-loops), but only RNase H2 has the ability to incise 5′ of a single rNMP imbedded in duplex DNA (reviewed by Cerritelli and Crouch 2009). The incorporation of rNMPs by DNA polymerases is restricted by an active-site pocket lid, which excludes sugars containing a 2′-OH group (Joyce 1997). Even so, it has been estimated that ∼10,000 rNMPs are incorporated into a haploid yeast genome during each replication cycle, making rNMPs potentially the most abundant noncanonical base/lesion in genomic DNA (Nick McElhinny et al. 2010b). The absence of RNase H2 is associated with replication stress (Nick McElhinny et al. 2010a; Lazzaro et al. 2012) and with a distinctive, topoisomerase 1-dependent mutation signature (Kim et al. 2011a). In yeast cell extracts, the Rad27 flap endonuclease cooperates with RNase H2 to excise an rNMP from duplex DNA, with Rad27 nicking on the 3′ side of the RNase H2-incised rNMP (Rydberg and Game 2002). An entire ribonucleotide excision repair reaction has been reconstituted using purified yeast RNase H2, Rad27 (or Exo1), PCNA, the RFC clamp loader, Pol δ (or Pol ε), and DNA ligase I (Sparks et al. 2012). Defects in human RNase H2 are associated with the neuro-inflammatory disease Aicardi–Goutières syndrome (Crow et al. 2006), and RNase H2 is essential in the mouse (Reijns et al. 2012).
Bypass of DNA Damage
DNA-damage bypass by the RAD6 pathway allows DNA to be synthesized across lesions that otherwise block replicative DNA polymerases. This not only permits the completion of replication, but also allows generation of a complementary, undamaged strand that can be used as a template in a subsequent BER or NER reaction. Although not a true repair pathway, the RAD6 pathway is commonly referred to as the post-replication repair (PRR) pathway, a name originally adopted to describe time-dependent changes in DNA fragment sizes following UV irradiation of NER-deficient cells (Di Caprio and Cox 1981; Prakash 1981).
There are error-free and error-prone subpathways of PRR, with gene assignment based (1) on whether loss results in an increase or decrease in induced mutagenesis, respectively, and (2) on epistatic survival relationships following treatment of single and double mutants with DNA damage (reviewed by Haynes and Kunz 1981). Many genes can be clearly assigned to the error-free or -prone component of PRR, but some reside in both subpathways. Such dual functions largely reflect roles of the encoded proteins in the post-translational modification of PCNA, which directs bypass into the alternative subpathways (reviewed by Moldovan et al. 2007; Ulrich and Walden 2010).
Elimination of the error-free PRR pathway results in a strong mutator phenotype, indicating that it is the major mechanism of lesion bypass. Error-free bypass involves a template switch to the newly synthesized strand of the sister chromatid, thereby allowing an undamaged, complementary strand to direct synthesis over the offending lesion. This process is intimately connected to homologous recombination, which is discussed in detail in another chapter and will be dealt with only peripherally here. The error-prone subpathway employs one or more specialized translesion synthesis (TLS) DNA polymerases, which catalyze DNA synthesis across damage that blocks replicative DNA polymerases. A list of the major genes involved in PRR is given in Table 4.
Table 4. PRR genes.
| Gene name (alternative) | Protein size (kDa) | Description | Mammalian counterpart |
|---|---|---|---|
| RAD6 (UBC2, PSO8) | 19.7 | E2 Ub conjugase that forms a complex with Rad18 (also Bre1 and Ubr1) E3 Ub ligase; Rad18-Rad6 mono-ubiquitinates PCNA on K164. | RAD6A, RAD6B |
| RAD18 | 55.2 | E3 Ub ligase that binds ssDNA; forms a complex with Rad6 to mono-ubiquitinate PCNA at K164. | RAD18 |
| RAD5 (REV2, SNM2) | 134.2 | E3 Ub ligase involved in polyubiquitination of PCNA; ATPase activity. | HTLF, SHPRH |
| MMS2 | 15.5 | Forms a heterodimeric complex with Ubc13; Ub conjugase involved in polyubiquitination of PCNA. | MMS2 |
| UBC13 | 17.5 | Forms a heterodimeric complex with Mms2; Ub conjugase involved in polyubiquitination of PCNA. | UBC13 |
| UBP10 (DOT4) | 88.5 | DUB for PCNA. | USP1 |
| POL30 (REV6) | 28.9 | Subunit of PCNA homotrimer; post-translationally modified to direct PRR subpathways. | PCNA |
| REV1 | 112.2 | dCMP transferase activity; required for Pol ζ-dependent lesion bypass. | REV1L |
| REV3 (PSO1, ANT2, RAD8) | 173.0 | Catalytic subunit of Pol ζ. | REV3L |
| REV7 | 28.8 | Accessory subunit of Pol ζ. | REV7 |
| POL31 (HUS1 SDP5, HYS2) | 55.3 | Subunit of Pol δ and Pol ζ. | B-subunit (p66) |
| POL32 (REV4) | 40.3 | Subunit of Pol δ and Pol ζ. | C-subunit (p50) |
| RAD30 (DBH1) | 71.5 | Pol η TLS polymerase. | POL η |
| UBC9 | 17.9 | E2 SUMO conjugase that SUMOylates PCNA. | UBC9 |
| SIZ1 (ULL1) | 100.8 | E3 SUMO ligase that SUMOylates PCNA. | — |
| SRS2 (HPR5, RADH) | 134.3 | 3′–5′ helicase; translocase that dismantles Rad51 nucleoprotein filaments. | PARI, FBH1 |
Components of error-free bypass
The central players in the error-free subpathway of PRR are the Rad6-Rad18 complex, the Ubc13-Mms2 complex, and Rad5. Rad6 and Rad18 comprise one pair of E2-E3 ubiquitinating enzymes that modify PCNA, while Ubc13-Mms2 and Rad5 constitute a second E2-E3 pair. Rad6-Rad18 adds a single Ub moiety to PCNA, which then can be extended into a regulatory, poly-Ub chain by Rad5 and Ubc13-Mms2. Poly-Ub PCNA coordinates the downstream steps of template switching, the primary candidate mechanisms of which involve either reversal of a blocked replication fork or recombination-mediated invasion of the undamaged sister chromatid (Figure 8, A and B, respectively). Because mono-Ub of PCNA also is required for most TLS, Rad6-Rad18 is required for virtually all lesion bypass.
Figure 8.
Mechanisms of template switching during PRR. Alternative template-switch mechanisms are shown for a lesion blocking leading- vs. lagging-strand replication, but recombination and gap filling are possible on either strand. (A) Lagging-strand synthesis continues when leading-strand synthesis is blocked by a lesion. Helicase-driven reversal of the fork into a “chicken-foot” structure pairs the newly synthesized strands, with the more extensively extended lagging strand providing a template for additional leading-strand synthesis. Resetting of the fork places the lesion back into duplex DNA, where it can be repaired. (B) A lesion-associated gap on the lagging strand is filled when the blocked 3′ end invades the sister chromatid and uses it as template for additional DNA synthesis. Black and red lines are template and nascent DNA, respectively, and 3′ ends are indicated by the half-arrowheads; the red dotted lines indicate DNA synthesized during the bypass reaction. Yellow stars represent DNA lesions.
Rad6-Rad18 complex:
Early genetic studies placed rad6 and rad18 mutants in the same epistasis group, but rad6 mutants had additional growth and sporulation defects (Haynes and Kunz 1981). Purification of Rad6 revealed an E2 Ub-conjugase activity in vitro (Jentsch et al. 1987), and the complex phenotypes of rad6 mutants reflect its interaction with three different E3 Ub ligases: Ubr1, Bre1, and Rad18. Rad6-Ubr1 targets proteins for N-end rule degradation (Dohmen et al. 1991), Rad6-Bre1 modifies chromatin (Wood et al. 2003; Zimmermann et al. 2011), and Rad6-Rad18 adds a single Ub to PCNA monomers (Hoege et al. 2002).
Rad18 has a zinc-coordinated RING finger domain, displays an ATPase activity, and binds to single-strand DNA (Bailly et al. 1994). Rad18 forms a stable complex with Rad6 and self-associates via a separate domain (Ulrich and Jentsch 2000); the significance, if any, of self-association is not known. Because mono-Ub of PCNA is required for TLS, loss of either Rad6 or Rad18 is associated with a reduction in induced mutagenesis. By contrast, rad18Δ strains exhibit a mutator phenotype in spontaneous mutation assays (Liefshitz et al. 1998; Cejka et al. 2001; Minesinger and Jinks-Robertson 2005). These opposing responses highlight a basic difference in how cells respond to acute vs. chronic damage (De Graaf et al. 2009; Hishida et al. 2009). Finally, Rad6-Rad18 has been implicated in the control of an inducible DNA-damage response similar to the SOS response of E. coli (Fu et al. 2008), but this role has been disputed (Davies et al. 2010).
Rad5 protein:
RAD5 was cloned by complementation of the UV sensitivity of mutants, and the encoded protein contains the seven DNA-helicase motifs shared by the SNF2/SWI2 superfamily of ATPases (Johnson et al. 1992). The ATPase activity of Rad5 can drive regression of a model replication-fork structure in vitro (Figure 8A) (Blastyak et al. 2007), but whether it has this activity in vivo is not known. Rad5 self-associates and can interact simultaneously with Rad18 and Ubc13, allowing formation of a larger complex composed of Rad6-Rad18, Rad5, and Ubc13-Mms2 (Ulrich and Jentsch 2000). The domain of Rad18 required for self-association is coincident with that required for Rad5 interaction and is independent of its Rad6-interaction domain (Ulrich and Jentsch 2000). Whether a homodimer of the Rad6-Rad18 complex and/or of Rad5 has physiological function separate from that of the fully assembled complex is not known.
Although they are in the same epistasis group, rad5Δ mutants are less UV sensitive than rad18Δ mutants and generally are not defective in UV-induced mutagenesis (Johnson et al. 1992). This difference reflects the role of mono-Ub PCNA in promoting TLS (see below). While the major function of Rad5 is clearly as part of error-free PRR, a rad5Δ mutant is more sensitive to UV than an mms2Δ or ubc13Δ mutant (Ulrich and Jentsch 2000). Rad5 is necessary for the damage-induced reversion of some alleles (Gangavarapu et al. 2006) and a structural role for Rad5 during mutagenic bypass of defined lesions has been suggested (Pagès et al. 2008a).
Like rad18Δ mutants, rad5Δ strains have a spontaneous mutator phenotype that depends on TLS components. The mutator phenotype is abolished, however, in a rad5Δ rad18Δ double mutant (Liefshitz et al. 1998; Cejka et al. 2001; Minesinger and Jinks-Robertson 2005). The genetic data suggest that, in addition to working together to promote the error-free template switching, Rad18 and Rad5 each independently promote the TLS of a common spontaneous lesion(s). In the absence of either Rad5 or Rad18, template switching cannot occur, and damage is bypassed in a mutagenic manner by the remaining Rad18- or Rad5-dependent TLS pathway, respectively; when both Rad18 and Rad5 are absent, however, neither error-free nor error-prone bypass operates.
Ubc13-Mms2 complex:
MMS2 was identified in a screen for mutations that enhance MMS sensitivity (Prakash and Prakash 1977), and epistasis analysis placed it in the error-free arm of PRR (Broomfield et al. 1998). Mms2 has homology to Ub-conjugating enzymes, but lacks catalytic activity (Broomfield et al. 1998; Xiao et al. 1999; Torres-Ramos et al. 2002). It forms a stable complex with the bona fide E2 conjugase Ubc13, and it is the Ubc13-Mms2 complex that promotes assembly of a regulatory poly-Ub chain on mono-Ub PCNA (Hofmann and Pickart 1999). Deletion of UBC13 or MMS2 produces similar phenotypes in terms of increased damage sensitivity and mutagenesis (Brusky et al. 2000; Ulrich and Jentsch 2000; Xiao et al. 2000).
Error-free PRR and recombination
The existence of discrete PRR and homologous recombination (HR) pathways as defined in early epistasis analyses has become blurred. Even in the initial description of PRR as a mechanism that restores UV-damaged DNA to its full length, a partial requirement for the central recombination protein Rad52 was noted (Prakash 1981). Similarly, error-free bypass of staggered lesions on a plasmid absolutely requires Rad18 and is partially dependent on Rad52 (Zhang and Lawrence 2005). The complex relationship between HR and error-free PRR has been most recently studied using 2D gels, which allow the progression of individual replication forks to be followed in the presence of DNA damage. Fork stalling can be visualized, as well as X-shaped structures that reflect HJs and/or catenated sister chromatids. The formation of the X-structures requires Rad18, Rad5, and Ubc13-Mms2 and, additionally, the Rad51 strand-exchange protein (Branzei et al. 2008; Minca and Kowalski 2010). Genetic studies suggest that there are three distinct genetic pathways of replication-associated recombination: template switching to the sister chromatid promoted by Ubc13-Mms2, fork regression promoted by the Mph1 helicase, and a gap-filling reaction promoted by the Shu complex (Choi et al. 2010). Overall, current data suggest that HR (1) is important for the invasion step of a template switch that occurs in the context of an unbroken replication fork and generates catenated sister chromatids (pseudo-HJs) and (2) is important for the repair of double-strand breaks, which arise from fork collapse or chromosome breakage and generate true HJs.
Components of error-free and error-prone TLS
A mutagenic subpathway of PRR initially was inferred through isolation of “reversionless” (rev) mutants in which the damage-induced reversion of auxotrophic markers was greatly reduced (Lemontt 1971). Of the REV genes thus identified, all have been characterized except that defined by rev5; only REV1, REV3, and REV7 have retained their original names. Sequencing REV3 revealed homology of the encoded protein to the B family of replicative DNA polymerases, leading to the key insight that specialized TLS polymerases uniquely able to replicate over DNA damage likely exist (Morrison et al. 1989). It should be noted that a complete TLS event requires two distinct steps: (1) insertion of a dNMP opposite the lesion and (2) extension of the resulting lesion:base mispair.
Yeast has three TLS polymerases: Pol ζ, which is composed of Rev3 and Rev7, and the Y-family polymerases Pol η, which is encoded by RAD30, and Rev1 (reviewed by Waters et al. 2009). The yeast and most of the ∼15 mammalian TLS polymerases are dispensable; the only known essential enzyme in mammals is Pol ζ (Bemark et al. 2000; Esposito et al. 2000). Whether this reflects a direct requirement for Pol ζ is unclear, however, as the effects of its loss in chicken DT40 cells can be suppressed by additional loss of Pol η (Hirota et al. 2010). Although TLS polymerases are widely referred to as “error-prone,” the fidelity of each relative to replicative polymerases is lesion specific. The current model is that most TLS polymerases are specialized to bypass one or a few naturally occurring lesions in a relatively error-free manner. All, however, are notoriously error-prone on undamaged DNA, lack exonucleolytic proofreading activity, and are nonprocessive.
Assaying TLS:
The in vivo role of a TLS polymerase during lesion bypass can be determined by comparing survival and/or mutagenesis of wild-type and mutant cells following exposure to DNA damage. Because damaging agents indiscriminately generate multiple types of lesions genome-wide, however, it is difficult to link a specific lesion to the genetic readout. By contrast, transformation-based assays can assess the bypass of a single, defined lesion. Lesions have been engineered into gapped plasmids (Nelson et al. 2000; Gibbs et al. 2005), double-strand plasmids (Baynton et al. 1998, 1999; Pagès et al. 2008a,b), and single-strand plasmids (Zhao et al. 2006, 2010). Additionally, a single-strand oligonucleotide containing a defined lesion can be introduced into the genome by transformation, where it serves as a template in the subsequent round of DNA replication (Otsuka et al. 2002a,b, 2005; Kow et al. 2005; Bao and Kow 2009). Finally, in vitro assays with purified proteins afford the opportunity to assess the individual steps of TLS through manipulating primer-template combinations and/or the nucleotide pool. The efficiency of full bypass, the insertion specificity opposite the lesion, and/or the ability to extend a primer from a lesion:base mispair can thus be measured.
Pol ζ, the Rev3-Rev7(-Pol31-Pol32) complex:
REV1, REV3, and REV7 are in the same epistasis group and are required for >90% of induced and at least 50% of spontaneous mutagenesis (reviewed by Lawrence 1994). The polymerase activity of Rev3 is stimulated by Rev7, and these proteins compose the core Pol ζ heterodimer (Nelson et al. 1996a). Although core Pol ζ can accomplish complete bypass (Nelson et al. 1996a; Stone et al. 2011), its primary activity in vitro is as an extender of nucleotides inserted opposite a lesion by another DNA polymerase (Johnson et al. 2000; Guo et al. 2001; Haracska et al. 2001b, 2003; Washington et al. 2004). In addition to DNA-damage bypass, Pol ζ is required for mutagenesis that occurs in strains with a defective replisome, indicating that an unpaired terminus need not involve a lesion (Northam et al. 2010). All four of the yeast B-family DNA polymerases contain an Fe-S cluster (Netz et al. 2012), which is emerging as a common feature of many DNA metabolic proteins.
Pol ζ is important in the bypass of a broad variety of lesions in vivo: methylated bases (Johnson et al. 2007), AP sites (Haracska et al. 2001b; Kim and Jinks-Robertson 2009), oxidative damage (Johnson et al. 2003), interstrand crosslinks (Grossmann et al. 2001; Sarkar et al. 2006), UV-induced lesions (Kozmin et al. 2003; Abdulovic and Jinks-Robertson 2006), and DNA-protein crosslinks (De Graaf et al. 2009; Grogan and Jinks-Robertson 2012). Although most studies have focused on the role of Pol ζ in tolerating nuclear DNA damage, it localizes to and is important in the mitochondria as well (Zhang et al. 2006; Kalifa and Sia 2007).
Most Pol ζ-dependent bypass of induced lesions is Rad18-dependent (see above), but separate Rad18- and Rad5-dependent pathways of spontaneous lesion bypass have been documented using forward and reverse mutation assays (Liefshitz et al. 1998; Cejka et al. 2001; Minesinger and Jinks-Robertson 2005). A multiple-mutation signature produced by Pol ζ in the latter assay indicates that synthesis is limited to tracts of ∼10 nt in vivo (Harfe and Jinks-Robertson 2000b), which is consistent with the distributive behavior of TLS polymerases in vitro. The multiple-mutation signature has been particularly useful for following Pol ζ activity in vivo (Minesinger and Jinks-Robertson 2005; Sabbioneda et al. 2005; Abdulovic and Jinks-Robertson 2006; Minesinger et al. 2006; Lehner and Jinks-Robertson 2009; Grogan and Jinks-Robertson 2012) and has been recapitulated in vitro using undamaged templates (Zhong et al. 2006; Stone et al. 2009).
Cooperation between Pol ζ and the replicative DNA polymerase Pol δ during lesion bypass was early inferred through analysis of non-null catalytic (pol3) mutants (Halas et al. 1997) and by a dependence of Pol ζ-dependent bypass on the Pol32 subunit of Pol δ (Huang et al. 2000; Gibbs et al. 2005; Minesinger and Jinks-Robertson 2005; Hanna et al. 2007; Pagès et al. 2008b; Auerbach and Demple 2010). Recent data, however, have demonstrated that the noncatalytic Pol31 and Pol32 subunits of Pol δ are shared with Pol ζ and that the Pol ζ holoenzyme is actually a four-subunit complex (Baranovskiy et al. 2012; Johnson et al. 2012). Other proteins implicated in Pol ζ-dependent bypass include the Cdc7 kinase (Pessoa-Brandao and Sclafani 2004), the 9-1-1 checkpoint clamp (Sabbioneda et al. 2005), and the Rad24, Rad9, and Mec1 checkpoint proteins (Paulovich et al. 1998; Barbour et al. 2006; Hishida et al. 2009; Pagès et al. 2009).
Rev1, a deoxycytidyl transferase:
Rev1 generally is assumed to be required during Pol ζ-dependent lesion bypass in vivo (reviewed by Lawrence 1994), but there have been reports of differential Rev1 vs. Pol ζ requirements during gapped-plasmid repair (Baynton et al. 1999) for instability in pol3-t background (Mito et al. 2008) and in mitochondrial DNA stability (Zhang et al. 2006). The initial biochemical characterization of purified Rev1 revealed deoxycytidyl (dCMP) transferase activity during bypass of a template AP site (Nelson et al. 1996b), and a strong preference for dCMP insertion in vitro has been confirmed (Pryor and Washington 2011). Rev1 additionally has been defined as a G-template-specific DNA polymerase (Haracska et al. 2002). Indeed, the crystal structure of Rev1 with a template G or AP site reveals that each is evicted from the active-site pocket to allow pairing of the incoming dCTP with an arginine (Nair et al. 2005, 2011). Although it has been argued that Rev1 catalytic activity is not relevant for AP-site bypass in vivo (Haracska et al. 2001b; Pagès et al. 2008b), the consensus is that dCMP is the predominant nucleotide inserted opposite AP sites and that most Pol ζ-dependent bypass of AP sites relies on Rev1 catalytic activity (Otsuka et al. 2002b; Auerbach et al. 2005; Gibbs et al. 2005; Kow et al. 2005; Kim et al. 2011b). According to the two-polymerase model, Rev1 inserts dCMP opposite a lesion, with Pol ζ providing extension activity (see Figure 3). Altogether, the data suggest a specialized, “error-free” role of Rev1 in bypassing guanines either damaged or lost from the DNA backbone.
In addition to AP-site bypass, Rev1 catalytic activity is relevant during bypass of ethenoadenine- (Zhou et al. 2010) and 4-nitroquinilone 1-oxide-induced damage (Wiltrout and Walker 2011a); bypass of other types of lesions, however, depends only on Rev1 presence (Nelson et al. 1996b; Zhou et al. 2010; Wiltrout and Walker 2011a). Rev1 thus has an essential structural role, as well a variable enzymatic role, during Pol ζ-dependent bypass. Rev1 interacts with both Rev7 and Rev3 (Acharya et al. 2005, 2006; D’Souza and Walker 2006), with the Pol32 subunit of Pol δ/ζ (Acharya et al. 2009), with Rad5 (Pagès et al. 2008a), and with mono-Ub PCNA (Wood et al. 2007; D’Souza et al. 2008). Recent work suggests that interaction of Rev1 with Rad5 is required for the noncatalytic function of Rev1 (Kuang et al. 2013). In contrast to mammalian REV1, which interacts with multiple Y-family DNA polymerases via a highly conserved C terminus (Guo et al. 2003), no interaction of yeast Rev1 with Pol η has been detected (Kosarek et al. 2008). Finally, Rev1 is hyper-phosphorylated during mitosis and after DNA damage, suggesting potential post-translational regulation of its activity (Sabbioneda et al. 2007).
Pol η:
The RAD30 gene was identified by homology to the E. coli DinB polymerase (McDonald et al. 1997; Roush et al. 1998). RAD30 is in the RAD6-RAD18 epistasis group, and synergistic sensitivity to UV damage is seen upon additional loss of Rad5 or Rev1/Pol ζ (McDonald et al. 1997). Although loss of Pol η can have opposing, allele-specific effects on UV-induced reversion (McDonald et al. 1997; Roush et al. 1998), it generally is associated with an elevation in Pol ζ-dependent mutagenesis (e.g., Johnson et al. 1999b; Abdulovic and Jinks-Robertson 2006). The TLS field was launched into prominence by the discovery that loss of human POL η is responsible for the variant, NER-proficient form of xeroderma pigmentosum (XP-V). In vitro, Pol η has the unique ability to bypass cis-syn thymine dimers in an error-free manner, thereby providing an explanation for why loss confers XP-V (Johnson et al. 1999a; Masutani et al. 1999).
Structural studies have revealed that the active-site pocket of Pol η is large relative to that of replicative DNA polymerases (Trincao et al. 2001). This facilitates bypass of bulky lesions, but also makes Pol η one of the most inaccurate TLS polymerases on undamaged DNA templates (Matsuda et al. 2000; Washington et al. 2001). In contrast to the rigid steric fit used by replicative DNA polymerases for dNTP discrimination, Pol η relies more on Watson–Crick hydrogen bonding. This latter property allows the error-free bypass of lesions such as thymine dimers, which retain base-pairing capacity (Washington et al. 2003; Alt et al. 2007).
Numerous in vitro studies have reported bypass of defined lesions by purified Pol η, but only three for which there is accompanying genetic data will be mentioned here: UV-induced lesions, AP sites, and 8-oxoG. Bypass of UV-induced cis-syn thymine dimers by Pol η is error-free (see above), as is that of the more distorting (6-4) CC or (6-4) TC (Yu et al. 2001). By contrast, Pol η preferentially inserts a G opposite the 3′-T of (6-4) TT in vitro (Johnson et al. 2001). This specificity can explain why UV-induced reversion of the arg4-17 allele, which occurs primarily via T-to-C transitions, is reduced in a rad30Δ background (Zhang and Siede 2002). With regard to an AP site, bypass by Pol η is poor in vitro, presumably because the enzyme requires a template base opposite the incoming dNTP (Haracska et al. 2001c). No significant involvement of Pol η has been observed in a gap-filling assay (Gibbs et al. 2005) or during bypass of genomic AP sites (Otsuka et al. 2002b; Auerbach and Demple 2010; Kim et al. 2011b). With respect to 8-oxoG, Pol η has a strong preference for inserting C rather than A in vitro (Haracska et al. 2000; McCulloch et al. 2009), and there is a strong increase in 8-oxoG-associated mutagenesis upon deletion of the RAD30 gene (Haracska et al. 2000; De Padula et al. 2004; Mudrak et al. 2009; Van Der Kemp et al. 2009). As noted previously, Pol η is part of the yeast GO network (Figure 2).
Pol η interacts with PCNA via a C-terminal PIP domain, and its activity is stimulated by PCNA in vitro (Haracska et al. 2001a). The PIP domain is essential for survival following UV irradiation (Haracska et al. 2001a) and for error-free 8-oxoG bypass (Mudrak et al. 2009). Pol η additionally has a Ub-binding (UBZ) domain that further facilitates interaction with mono-Ub PCNA and is required for in vivo function (De Padula et al. 2004; Parker et al. 2007; Pabla et al. 2008; Van Der Kemp et al. 2009). In vitro, mono-Ub PCNA regulates a polymerase exchange between Pol δ and Pol η during lesion bypass (Zhuang et al. 2008). Finally, Pol η itself is mono-Ub, and its UBZ domain is required for this as well (Parker et al. 2007). Mono-Ub of Pol η decreases in S phase or following UV irradiation, suggesting a regulatory role for this modification (Pabla et al. 2008).
Post-translational modification of PCNA
Regulation of error-free vs. error-prone PRR through post-translational modification of PCNA was reported in a landmark study published in 2002 (Hoege et al. 2002). Following a sublethal dose of DNA damage, Rad6 and Rad18 are required to attach a single Ub moiety to lysine 164 (K164) of Pol30, which can be extended into a regulatory, K63-linked poly-Ub chain in the presence of Mms2, Ubc13, and Rad5. As noted previously, mono-Ub of PCNA promotes TLS (Stelter and Ulrich 2003) while poly-Ub is required for the error-free template-switch pathway (Hoege et al. 2002). During S phase or following a very high dose of damage, however, a single small ubiquitin-like modifier (SUMO) moiety is attached to K164, and SUMO additionally is attached to K127 (Hoege et al. 2002). SUMOylation of PCNA requires the E2 SUMO conjugase Ubc9 and the Siz1 E3 SUMO ligase and helps promote error-free PRR (see below). PCNA also can be poly-SUMOylated (Parker et al. 2008; Windecker and Ulrich 2008), but the significance of this is not known. Ubiquitination of PCNA at K164 and its key role in promoting PRR is highly conserved among eukaryotes, but SUMOylation is not (Ulrich and Walden 2010). Although not relevant to PRR, ligase 1 defects trigger PCNA ubiquitination at K107; this requires Rad5 and Msm2-Ubc4 and is important in checkpoint activation (Das-Bradoo et al. 2010). The post-translational modifications to PCNA are summarized in Figure 9.
Figure 9.
Post-translational modifications to PCNA and PRR regulation. (A) Crystal structure of the human PCNA homotrimer with the subunits indicated in different colors (http://en.wikipedia.org/wiki/Proliferating_cell_nuclear_antigen). The approximate positions of the lysines (K) modified by Ub or SUMO (yellow and red circles, respectively) are indicated. (B) The proteins/complexes involved in modifying PCNA to direct the appropriate response are indicated and are described in the text.
The pol30-K164R single-, pol30-K127R single-, and pol30-K127R,K164R double-mutant alleles have been widely used to assess how Ub and/or SUMO affects PRR (e.g., Hoege et al. 2002; Pfander et al. 2005). One intriguing observation made in early studies was that a pol30-K127R,K164R double mutant is less sensitive to DNA damage than a pol30-K164R single mutant (Hoege et al. 2002). Survival of a pol30-K164R mutant also is increased by loss of the Srs2 X′–Y′ helicase (Hoege et al. 2002), consistent with the long-known suppression of rad6 and rad18 alleles by srs2 alleles (Aboussekhra et al. 1989; Schiestl et al. 1990; Rong et al. 1991). The requirement of homologous recombination for srs2 suppression of rad6 (Schiestl et al. 1990), together with the hyper-recombination phenotype srs2 mutants (Rong et al. 1991), suggested that one role of Srs2 is to limit recombination. Indeed, Srs2 has “strippase” activity that dismantles the Rad51 nucleoprotein filaments required for the strand-invasion step that initiates recombination (Krejci et al. 2003; Veaute et al. 2003; reviewed by Marini and Krejci 2010). The demonstration that SUMOylated PCNA physically interacts with the C terminus of Srs2 brought the genetic and biochemical observations into a coherent model in which SUMOylated PCNA recruits Srs2 to dismantle Rad51 nucleoprotein filaments, thereby preventing unrestrained/inappropriate recombination (Papouli et al. 2005; Pfander et al. 2005). In the absence of Srs2, PRR defects are thus partially rescued because damage is bypassed by the alternative, homologous recombination pathway.
In principle, the Pol30 monomers that compose the PCNA homotrimer can be differentially modified and/or interact with different proteins. With regard to the former, template switching appears to occur most efficiently when PCNA is simultaneously SUMOylated and polyubiquitinated (Branzei et al. 2008). Both modifications can exist on the same Pol30 monomer (Windecker and Ulrich 2008) and recent work suggests that Rad18 is actually a SUMO-targeted ubiquitin ligase (Parker and Ulrich 2012). With regard to the latter, replicative and TLS polymerases can potentially interact simultaneously with the clamp, providing a mechanism to effect bypass by switching between polymerases (the “tool belt” model). The crystal structure of a Ub-Pol30 fusion protein places the PIP-binding domain and Ub on the back face of the homotrimer (i.e., the side away from the primer end), with Pol δ interacting on the front face (Freudenthal et al. 2010). The solution structure of Ub-PCNA indicates that the association of Ub with PCNA is dynamic, however, with Ub transitioning between different docking sites on the clamp (Tsutakawa et al. 2011). This suggests a model in which Ub holds a TLS polymerase in reserve on the back face of PCNA and then transitions it to a side position for lesion bypass.
In vitro data suggest that the switch from Pol η back to Pol δ during lesion bypass requires either a de-ubiquitinating enzyme (a “DUB”) or unloading of Ub-PCNA by a clamp loader (Zhuang et al. 2008). Ubp10 has been biochemically identified as the DUB for PCNA, removing both mono- and di-Ub (Gallego-Sanchez et al. 2012). Elg1, which is part of an alternative clamp loader, promotes genome stability and interacts with PCNA, preferring the SUMOylated form of the clamp. Srs2 accumulates on DNA in the absence of Elg1, and loss of Srs2 and Elg1 confers a synthetic fitness defect that is suppressed by blocking SUMOylation of PCNA (Parnas et al. 2010). These observations suggest that persistence of SUMO-PCNA on DNA is toxic and that Elg1 may be important either for removing SUMO from PCNA or for unloading SUMO-PCNA from DNA.
When and where does PRR occur?
In Xenopus extracts, PCNA ubiquitination is triggered when the replicative DNA polymerase and helicase are uncoupled at a replication fork (Chang et al. 2006). This is expected to generate tracts of single-strand DNA bound by RPA; indeed, yeast RPA interacts with Rad18, and this interaction is required for damage-associated PCNA ubiquitination (Davies et al. 2008). PRR presumably evolved as a mechanism to circumvent problems that arise during replication, but whether bypass occurs directly at a stalled fork or as a gap-filling process behind the fork has not been resolved. During “discontinuous” lagging-strand synthesis, the constant repriming of Okazaki fragments provides a ready mechanism to generate gaps that could be subsequently filled by TLS or a template switch (Figure 8B). By contrast, the “continuous” nature of leading-strand synthesis led to an early assumption that bypass would have to occur directly at the fork to prevent replication arrest (Figure 8A). Following high levels of damage, however, gaps have been detected by electron microscopy on both arms of yeast replication forks (Lopes et al. 2006). In addition, cell-cycle-specific induction of key PRR proteins has demonstrated that PRR can be temporally separated from replication and hence need not occur directly at a stalled fork (Daigaku et al. 2010; Karras and Jentsch 2010). Even though PRR can occur completely outside the context of replication, this does not necessarily mean that this is how it normally occurs. It remains possible that there are inherent differences in how lesions are bypassed on the leading vs. lagging strand of replication, reflecting the relatively continuous and discontinuous modes, respectively, of new DNA synthesis (Gangavarapu et al. 2007; Minca and Kowalski 2010).
With regard to the specific timing of Pol ζ-dependent bypass, there is 50-fold more Rev1 in G2/M than in G1/S (Waters and Walker 2006), and this reflects primarily regulation via proteosomal degradation (Wiltrout and Walker 2011b). Rev3 and Rev7 levels, however, are constant throughout the cell cycle (D’Souza and Walker 2006). Maximal expression of Rev1 outside of S phase has led to the suggestion that most Pol ζ-dependent lesion bypass occurs via gap-filling reactions that occur well behind the replication fork. Consistent with this, spontaneous, Pol ζ-dependent lesion bypass is refractory to correction by the MMR machinery (Lehner and Jinks-Robertson 2009). The higher mutation rate of late-replicating DNA also is reduced in rev1Δ background (Lang and Murray 2011), consistent with the cell-cycle regulation of Rev1 and suggesting that there may be temporal separation of error-free and error-prone lesion bypass.
Summary and Future Directions
There has been remarkable progress in unraveling the diverse mechanisms that deal with damage to the DNA double helix and with errors introduced during DNA synthesis. Genetic studies have elucidated the high degree of redundancy built into these systems, with the net effect being the maintenance of an extraordinarily stable genome in the face of constant internal and external assaults. Importantly, the lessons learned in yeast have been useful for understanding basic principles that operate in all eukaryotes and for understanding the origins of sporadic and inherited cancers. The focus here has largely been on the individual players involved in specific pathways and on underlying biochemical mechanisms. Understanding how the cellular environment affects repair/bypass mechanisms remains a future challenge: the composition and cellular locations of machines/factories, the roles of post-translational modifications, the effect of the cell cycle, and the roles of chromatin modifications. There are also expected to be novel connections between proteins and pathways that emerge from recently developed systems approaches. At least in yeast, however, genetic screens will continue to be a useful tool. There is much to keep us busy in the next 20 years, when perhaps even new and unanticipated pathways and mechanisms for promoting genome stability will be discovered.
Acknowledgments
S.B. thanks C. Dhérin-Boiteux and B. Castaing for critical reading of the manuscript. S.J.-R. thanks past and present lab members for their intellectual and experimental contributions. S.J.-R. also thanks the National Institutes of Health for financial support. S.B. thanks the Centre National pour la Recherche Scientifique and the Comité de Radioprotection of Electricité de France for financial support.
Footnotes
Communicating editor: R. Rothstein
Literature Cited
- Abdulovic A. L., Jinks-Robertson S., 2006. The in vivo characterization of translesion synthesis across UV-induced lesions in Saccharomyces cerevisiae: insights into Polζ- and Polη-dependent frameshift mutagenesis. Genetics 172: 1487–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aboussekhra A., Chanet R., Zgaga Z., Cassier-Chauvat C., Heude M., et al. , 1989. RADH, a gene of Saccharomyces cerevisiae encoding a putative DNA helicase involved in DNA repair: characteristics of radH mutants and sequence of the gene. Nucleic Acids Res. 17: 7211–7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acevedo-Torres K., Fonseca-Williams S., Ayala-Torres S., Torres-Ramos C. A., 2009. Requirement of the Saccharomyces cerevisiae APN1 gene for the repair of mitochondrial DNA alkylation damage. Environ. Mol. Mutagen. 50: 317–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya N., Haracska L., Johnson R. E., Unk I., Prakash S., et al. , 2005. Complex formation of yeast Rev1 and Rev7 proteins: a novel role for the polymerase-associated domain. Mol. Cell. Biol. 25: 9734–9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya N., Johnson R. E., Prakash S., Prakash L., 2006. Complex formation with Rev1 enhances the proficiency of Saccharomyces cerevisiae DNA polymerase ζ for mismatch extension and for extension opposite from DNA lesions. Mol. Cell. Biol. 26: 9555–9563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya N., Johnson R. E., Pages V., Prakash L., Prakash S., 2009. Yeast Rev1 protein promotes complex formation of DNA polymerase ζ with Pol32 subunit of DNA polymerase δ. Proc. Natl. Acad. Sci. USA 106: 9631–9636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahel I., Rass U., El-Khamisy S. F., Katyal S., Clements P. M., et al. , 2006. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature 443: 713–716. [DOI] [PubMed] [Google Scholar]
- Alani E., 1996. The Saccharomyces cerevisiae Msh2 and Msh6 proteins form a complex that specifically binds to duplex oligonucleotides containing mismatched DNA base pairs. Mol. Cell. Biol. 17: 2436–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alseth I., Eide L., Pirovano M., Rognes T., Seeberg E., et al. , 1999. The Saccharomyces cerevisiae homologues of endonuclease III from Escherichia coli, Ntg1 and Ntg2, are both required for efficient repair of spontaneous and induced oxidative DNA damage in yeast. Mol. Cell. Biol. 19: 3779–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt A., Lammens K., Chiocchini C., Lammens A., Pieck J. C., et al. , 2007. Bypass of DNA lesions generated during anticancer treatment with cisplatin by DNA polymerase η. Science 318: 967–970. [DOI] [PubMed] [Google Scholar]
- Amin N. S., Nguyen M. N., Oh S., Kolodner R. D., 2001. exo1-Dependent mutator mutations: model system for studying functional interactions in mismatch repair. Mol. Cell. Biol. 21: 5142–5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arana M. E., Holmes S. F., Fortune J. M., Moon A. F., Pedersen L. C., et al. , 2010. Functional residues on the surface of the N-terminal domain of yeast Pms1. DNA Repair (Amst.) 9: 448–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach P. A., Demple B., 2010. Roles of Rev1, Pol ζ, Pol32 and Pol η in the bypass of chromosomal abasic sites in Saccharomyces cerevisiae. Mutagenesis 25: 63–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach P., Bennett R. A., Bailey E. A., Krokan H. E., Demple B., 2005. Mutagenic specificity of endogenously generated abasic sites in Saccharomyces cerevisiae chromosomal DNA. Proc. Natl. Acad. Sci. USA 102: 17711–17716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augeri L., Lee Y. M., Barton A. B., Doetsch P. W., 1997. Purification, characterization, gene cloning, and expression of Saccharomyces cerevisiae redoxyendonuclease, a homolog of Escherichia coli endonuclease III. Biochemistry 36: 721–729. [DOI] [PubMed] [Google Scholar]
- Bailly V., Sommers C. H., Sung P., Prakash L., Prakash S., 1992. Specific complex formation between proteins encoded by the yeast DNA repair and recombination genes RAD1 and RAD10. Proc. Natl. Acad. Sci. USA 89: 8273–8277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly V., Lamb J., Sung P., Prakash S., Prakash L., 1994. Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev. 8: 811–820. [DOI] [PubMed] [Google Scholar]
- Ban C., Yang W., 1998. Crystal structure and ATPase activity of MutL: implications for DNA repair and mutagenesis. Cell 95: 541–552. [DOI] [PubMed] [Google Scholar]
- Bao G., Kow Y. W., 2009. Effect of sequence context and direction of replication on AP site bypass in Saccharomyces cerevisiae. Mutat. Res. 669: 147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranovskiy A. G., Lada A. G., Siebler H. M., Zhang Y., Pavlov Y. I., et al. , 2012. DNA polymerase δ and ζ switch by sharing accessory subunits of DNA polymerase δ. J. Biol. Chem. 287: 17281–17287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour L., Ball L. G., Zhang K., Xiao W., 2006. DNA damage checkpoints are involved in postreplication repair. Genetics 174: 1789–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell A. J., Bardwell L., Tomkinson A. E., Friedberg E. C., 1994a Specific cleavage of model recombination and repair intermediates by the yeast Rad1-Rad10 DNA endonuclease. Science 265: 2082–2085. [DOI] [PubMed] [Google Scholar]
- Bardwell L., Bardwell A. J., Feaver W. J., Svejstrup J. Q., Kornberg R. D., et al. , 1994b Yeast RAD3 protein binds directly to both SSL2 and SSL1 proteins: implications for the structure and function of transcription/repair factor b. Proc. Natl. Acad. Sci. USA 91: 3926–3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bawa S., Xiao W., 2003. A single amino acid substitution in MSH5 results in DNA alkylation tolerance. Gene 315: 177–182. [DOI] [PubMed] [Google Scholar]
- Baynton K., Bresson-Roy A., Fuchs R. P. P., 1998. Analysis of damage tolerance pathways in Saccharomyces cerevisiae: a requirement for Rev3 DNA polymerase in translesion synthesis. Mol. Cell. Biol. 18: 960–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baynton K., Bresswon-Roy A., Fuchs R. P. P., 1999. Distinct roles for Rev1p and Rev7p during translesion synthesis in Saccharomyces cerevisiae. Mol. Microbiol. 34: 124–133. [DOI] [PubMed] [Google Scholar]
- Bebenek K., Garcia-Diaz M., Patishall S. R., Kunkel T. A., 2005. Biochemical properties of Saccharomyces cerevisiae DNA polymerase IV. J. Biol. Chem. 280: 20051–20058. [DOI] [PubMed] [Google Scholar]
- Bemark M., Khamlichi A. A., Davies S. L., Neuberger M. S., 2000. Disruption of mouse polymerase ζ (Rev3) leads to embryonic lethality and impairs blastocyst development in vitro. Curr. Biol. 10: 1213–1216. [DOI] [PubMed] [Google Scholar]
- Bennett R. A., 1999. The Saccharomyces cerevisiae ETH1 gene, an inducible homolog of exonuclease III that provides resistance to DNA-damaging agents and limits spontaneous mutagenesis. Mol. Cell. Biol. 19: 1800–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdal K. G., Bjoras M., Bjelland S., Seeberg E., 1990. Cloning and expression in Escherichia coli of a gene for an alkylbase DNA glycosylase from Saccharomyces cerevisiae: a homologue to the bacterial alkA gene. EMBO J. 9: 4563–4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdal K. G., Johansen R. F., Seeberg E., 1998. Release of normal bases from intact DNA by a native DNA repair enzyme. EMBO J. 17: 363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand P., Tishkoff D. X., Filosi N., Dasgupta R., Kolodner R. D., 1998. Physical interaction between components of DNA mismatch repair and nucleotide excision repair. Proc. Natl. Acad. Sci. USA 95: 14278–14283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia P. K., Verhage R. A., Brouwer J., Friedberg E. C., 1996. Molecular cloning and characterization of Saccharomyces cerevisiae RAD28, the yeast homolog of the human Cockayne syndrome A (CSA) gene. J. Bacteriol. 178: 5977–5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjoras M., Seeberg E., Luna L., Pearl L. H., Barrett T. E., 2002. Reciprocal “flipping” underlies substrate recognition and catalytic activation by the human 8-oxo-guanine DNA glycosylase. J. Mol. Biol. 317: 171–177. [DOI] [PubMed] [Google Scholar]
- Blackwell L. J., Martik D., Bjornson K. P., Bjornson E. S., Modrich P., 1998. Nucleotide-promoted release of hMutSα from heteroduplex DNA is consistent with an ATP-dependent translocation mechanism. J. Biol. Chem. 48: 32055–32062. [DOI] [PubMed] [Google Scholar]
- Blastyak A., Pinter L., Unk I., Prakash L., Prakash S., et al. , 2007. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol. Cell 28: 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohr V. A., Smith C. A., Okumoto D. S., Hanawalt P. C., 1985. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell 40: 359–369. [DOI] [PubMed] [Google Scholar]
- Boiteux S., Guillet M., 2004. Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair (Amst.) 3: 1–12. [DOI] [PubMed] [Google Scholar]
- Boiteux S., Radicella J. P., 2000. The human OGG1 gene: structure, functions, and its implication in the process of carcinogenesis. Arch. Biochem. Biophys. 377: 1–8. [DOI] [PubMed] [Google Scholar]
- Boiteux S., O’Connor T. R., Laval J., 1987. Formamidopyrimidine-DNA glycosylase of Escherichia coli: cloning and sequencing of the fpg structural gene and overproduction of the protein. EMBO J. 6: 3177–3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiteux S., Gellon L., Guibourt N., 2002. Repair of 8-oxoguanine in Saccharomyces cerevisiae: interplay of DNA repair and replication mechanisms. Free Radic. Biol. Med. 32: 1244–1253. [DOI] [PubMed] [Google Scholar]
- Börner G. V., Kleckner N., Hunter N., 2004. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117: 29–45. [DOI] [PubMed] [Google Scholar]
- Bowers J., Sokolsky T., Quach T., Alani E., 1999. A mutation in the MSH6 subunit of the Sacchaormyces cerevisiae MSH2–MSH6 complex disrupts mismatch recognition. J. Biol. Chem. 274: 16115–16125. [DOI] [PubMed] [Google Scholar]
- Branch P., Aquilina G., Bignami M., Karran P., 1993. Defective mismatch binding and a mutator phenotype in cells tolerant to DNA damage. Nature 362: 652–654. [DOI] [PubMed] [Google Scholar]
- Branzei D., Vanoli F., Foiani M., 2008. SUMOylation regulates Rad18-mediated template switch. Nature 456: 915–920. [DOI] [PubMed] [Google Scholar]
- Bregeon D., Doetsch P. W., 2011. Transcriptional mutagenesis: causes and involvement in tumour development. Nat. Rev. Cancer 11: 218–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broomfield S., Chow B. L., Xiao W., 1998. MMS2, encoding a ubiquitin-conjugating-enzyme-like protein, is a member of the yeast error-free postreplication repair pathway. Proc. Natl. Acad. Sci. USA 95: 5678–5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruner S. D., Norman D. P., Verdine G. L., 2000. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature 403: 859–866. [DOI] [PubMed] [Google Scholar]
- Brusky J., Zhu Y., Xiao W., 2000. UBC13, a DNA-damage-inducible gene, is a member of the error-free postreplication repair pathway in Saccharomyces cerevisiae. Curr. Genet. 37: 168–174. [DOI] [PubMed] [Google Scholar]
- Budd M. E., Campbell J. L., 1995. DNA polymerases required for repair of UV-induced damage in Saccharomyces cerevisiae. Mol. Cell. Biol. 15: 2173–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers P. M., Klein M. B., 1986. Selection by genetic transformation of a Saccharomyces cerevisiae mutant defective for the nuclear uracil-DNA-glycosylase. J. Bacteriol. 166: 905–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadet J., Sage E., Douki T., 2005. Ultraviolet radiation-mediated damage to cellular DNA. Mutat. Res. 571: 3–17. [DOI] [PubMed] [Google Scholar]
- Caldecott K. W., 2008. Single-strand break repair and genetic disease. Nat. Rev. Genet. 9: 619–631. [DOI] [PubMed] [Google Scholar]
- Cejka P., Vondrejs V., Storchova Z., 2001. Dissection of the functions of the Saccharomyces cerevisiae RAD6 postreplicative repair group in mutagenesis and UV sensitivity. Genetics 159: 953–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P., Mojas N., Gillet L., Schar P., Jiricny J., 2005. Homologous recombination rescues mismatch-repair-dependent cytotoxicity of SN1-type methylating agents in S. cerevisiae. Curr. Biol. 15: 1395–1400. [DOI] [PubMed] [Google Scholar]
- Cerritelli S. M., Crouch R. J., 2009. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 276: 1494–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D. J., Lupardus P. J., Cimprich K. A., 2006. Monoubiquitination of proliferating cell nuclear antigen induced by stalled replication requires uncoupling of DNA polymerase and mini-chromosome maintenance helicase activities. J. Biol. Chem. 281: 32081–32088. [DOI] [PubMed] [Google Scholar]
- Charbonneau N., Amunugama R., Schmutte C., Yoder K., Fishel R., 2009. Evidence that hMLH3 functions primarily in meiosis and in hMSH2-hMSH3 mismatch repair. Cancer Biol. Ther. 8: 1411–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlet-Berguerand N., Feuerhahn S., Kong S. E., Ziserman H., Conaway J. W., et al. , 2006. RNA polymerase II bypass of oxidative DNA damage is regulated by transcription elongation factors. EMBO J. 25: 5481–5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A., Singh K. K., 2001. Uracil-DNA glycosylase-deficient yeast exhibit a mitochondrial mutator phenotype. Nucleic Acids Res. 29: 4935–4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri S., Wyrick J. J., Smerdon M. J., 2009. Histone H3 Lys79 methylation is required for efficient nucleotide excision repair in a silenced locus of Saccharomyces cerevisiae. Nucleic Acids Res. 37: 1690–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Derfler B., Maskati A., Samson L., 1989. Cloning a eukaryotic DNA glycosylase repair gene by the suppression of a DNA repair defect in Escherichia coli. Proc. Natl. Acad. Sci. USA 86: 7961–7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Derfler B., Samson L., 1990. Saccharomyces cerevisiae 3-methyladenine DNA glycosylase has homology to the AlkA glycosylase of E. coli and is induced in response to DNA alkylation damage. EMBO J. 9: 4569–4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K., Szakal B., Chen Y. H., Branzei D., Zhao X., 2010. The Smc5/6 complex and Esc2 influence multiple replication-associated recombination processes in Saccharomyces cerevisiae. Mol. Biol. Cell 21: 2306–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citterio E., Rademakers S., Van Der Horst G. T., Van Gool A. J., Hoeijmakers J. H., et al. , 1998. Biochemical and biological characterization of wild-type and ATPase-deficient Cockayne syndrome B repair protein. J. Biol. Chem. 273: 11844–11851. [DOI] [PubMed] [Google Scholar]
- Clark A. B., Kunkel T. A., 2010. The importance of being DNA. Cell Cycle 9: 4422–4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A. B., Valle F., Drotschmann K., Gary R. K., Kunkel T. A., 2000. Functional interaction of proliferating cell nuclear antigen with MSH2–MSH6 and MSH2–MSH3 complexes. J. Biol. Chem. 275: 36498–36501. [DOI] [PubMed] [Google Scholar]
- Clark A. B., Deterding L., Tomer K. B., Kunkel T. A., 2007. Multiple functions for the N-terminal region of Msh6. Nucleic Acids Res. 35: 4114–4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collura A., Kemp P. A., Boiteux S., 2012. Abasic sites linked to dUTP incorporation in DNA are a major cause of spontaneous mutations in absence of base excision repair and Rad17-Mec3-Ddc1 (9–1-1) DNA damage checkpoint clamp in Saccharomyces cerevisiae. DNA Repair (Amst.) 11: 294–303. [DOI] [PubMed] [Google Scholar]
- Constantin N., Dzantiev L., Kadyrov F. A., Modrich P., 2005. Human mismatch repair: reconstitution of a nick-directed bidirectional reaction. J. Biol. Chem. 280: 39752–39761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouse, G. F., 1998 Mismatch repair systems in Saccharomyces cerevisiae, pp. 411–448 in DNA Damage and Repair: DNA Repair in Prokaryotes and Lower Eukaryotes, Vol. 1, edited by J. A. Nickoloff and M. F. Hoekstra. Humana Press, Totowa, NJ. [Google Scholar]
- Crow Y. J., Leitch A., Hayward B. E., Garner A., Parmar R., et al. , 2006. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutières syndrome and mimic congenital viral brain infection. Nat. Genet. 38: 910–916. [DOI] [PubMed] [Google Scholar]
- Daigaku Y., Davies A. A., Ulrich H. D., 2010. Ubiquitin-dependent DNA damage bypass is separable from genome replication. Nature 465: 951–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley J. M., Wilson T. E., Ramotar D., 2010. Genetic interactions between HNT3/Aprataxin and RAD27/FEN1 suggest parallel pathways for 5′ end processing during base excision repair. DNA Repair (Amst.) 9: 690–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalhus B., Laerdahl J. K., Backe P. H., Bjoras M., 2009. DNA base repair–recognition and initiation of catalysis. FEMS Microbiol. Rev. 33: 1044–1078. [DOI] [PubMed] [Google Scholar]
- Das-Bradoo S., Nguyen H. D., Wood J. L., Ricke R. M., Haworth J. C., et al. , 2010. Defects in DNA ligase I trigger PCNA ubiquitylation at Lys 107. Nat. Cell Biol. 12: 74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Gupta R., Kolodner R. D., 2000. Novel dominant mutations in Saccharomyces cerevisiae MSH6. Nat. Genet. 24: 53–57. [DOI] [PubMed] [Google Scholar]
- Davies A. A., Friedberg E. C., Tomkinson A. E., Wood R. D., West S. C., 1995. Role of the Rad1 and Rad10 proteins in nucleotide excision repair and recombination. J. Biol. Chem. 270: 24638–24641. [DOI] [PubMed] [Google Scholar]
- Davies A. A., Huttner D., Daigaku Y., Chen S., Ulrich H. D., 2008. Activation of ubiquitin-dependent DNA damage bypass is mediated by Replication Protein A. Mol. Cell 29: 625–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies A. A., Neiss A., Ulrich H. D., 2010. Ubiquitylation of the 9–1-1 checkpoint clamp is independent of Rad6-Rad18 and DNA damage. Cell 141: 1080–1087. [DOI] [PubMed] [Google Scholar]
- De Bont R., Van Larebeke N., 2004. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis 19: 169–185. [DOI] [PubMed] [Google Scholar]
- De Graaf B., Clore A., McCullough A. K., 2009. Cellular pathways for DNA repair and damage tolerance of formaldehyde-induced DNA-protein crosslinks. DNA Repair (Amst.) 8: 1207–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Laat W. L., Jaspers N. G., Hoeijmakers J. H., 1999. Molecular mechanism of nucleotide excision repair. Genes Dev. 13: 768–785. [DOI] [PubMed] [Google Scholar]
- Den Dulk B., Brandsma J. A., Brouwer J., 2005. The Rad4 homologue YDR314C is essential for strand-specific repair of RNA polymerase I-transcribed rDNA in Saccharomyces cerevisiae. Mol. Microbiol. 56: 1518–1526. [DOI] [PubMed] [Google Scholar]
- Den Dulk B., Sun S. M., De Ruijter M., Brandsma J. A., Brouwer J., 2006. Rad33, a new factor involved in nucleotide excision repair in Saccharomyces cerevisiae. DNA Repair (Amst.) 5: 683–692. [DOI] [PubMed] [Google Scholar]
- Den Dulk B., Van Eijk P., De Ruijter M., Brandsma J. A., Brouwer J., 2008. The NER protein Rad33 shows functional homology to human Centrin2 and is involved in modification of Rad4. DNA Repair (Amst.) 7: 858–868. [DOI] [PubMed] [Google Scholar]
- Deng C., Brown J. A., You D., Brown J. M., 2005. Multiple endonucleases function to repair covalent topoisomerase I complexes in Saccharomyces cerevisiae. Genetics 170: 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Padula M., Slezak G., Auffret Van Der Kemp P., Boiteux S., 2004. The post-replication repair RAD18 and RAD6 genes are involved in the prevention of spontaneous mutations caused by 7,8-dihydro-8-oxoguanine in Saccharomyces cerevisiae. Nucleic Acids Res. 32: 5003–5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detloff P., Sieber J., Petes T. D., 1991. Repair of specific base pair mismatches formed during meiotic recombination in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 11: 737–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dherin C., Gueneau E., Francin M., Nunez M., Miron S., et al. , 2009. Characterization of a highly conserved binding site of Mlh1 required for exonuclease I-dependent mismatch repair. Mol. Cell. Biol. 29: 907–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Caprio L., Cox B. S., 1981. DNA synthesis in UV-irradiated yeast. Mutat. Res. 82: 69–85. [DOI] [PubMed] [Google Scholar]
- Dizdaroglu M., Laval J., Boiteux S., 1993. Substrate specificity of the Escherichia coli endonuclease III: excision of thymine- and cytosine-derived lesions in DNA produced by radiation-generated free radicals. Biochemistry 32: 12105–12111. [DOI] [PubMed] [Google Scholar]
- Dohmen R. J., Madura K., Bartel B., Varshavsky A., 1991. The N-end rule is mediated by the UBC2(RAD6) ubiquitin-conjugating enzyme. Proc. Natl. Acad. Sci. USA 88: 7351–7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudican N. A., Song B., Shadel G. S., Doetsch P. W., 2005. Oxidative DNA damage causes mitochondrial genomic instability in Saccharomyces cerevisiae. Mol. Cell. Biol. 25: 5196–5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen J. M., Putnam C. D., Kolodner R. D., 2010. Functional studies and homology modeling of Msh2-Msh3 predict that mispair recognition involves DNA bending and strand separation. Mol. Cell. Biol. 30: 3321–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drotschmann K., Yang W., Brownewell F. E., Kool E. T., Kunkel T. A., 2001. Asymmetric recognition of DNA local distortion. Structure-based functional studies of eukaryotic Msh2-Msh6. J. Biol. Chem. 276: 46225–46229. [DOI] [PubMed] [Google Scholar]
- Drotschmann K., Yang W., Kunkel T. A., 2002. Evidence for sequential action of two ATPase active sites in yeast Msh2-Msh6. DNA Repair (Amst.) 1: 743–753. [DOI] [PubMed] [Google Scholar]
- D’Souza S., Walker G. C., 2006. Novel role for the C terminus of Saccharomyces cerevisiae Rev1 in mediating protein-protein interactions. Mol. Cell. Biol. 26: 8173–8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza S., Waters L. S., Walker G. C., 2008. Novel conserved motifs in Rev1 C-terminus are required for mutagenic DNA damage tolerance. DNA Repair (Amst.) 7: 1455–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durant S. T., Morris M. M., Illand M., McKay H. J., McCormick C., et al. , 1999. Dependence on RAD52 and RAD1 for anticancer drug resistance mediated by inactivation of mismatch repair genes. Curr. Biol. 9: 51–54. [DOI] [PubMed] [Google Scholar]
- Eckardt F., Haynes R. H., 1977. Induction of pure and sectored mutant clones in excision-proficient and deficient strains of yeast. Mutat. Res. 43: 327–338. [DOI] [PubMed] [Google Scholar]
- Egly J. M., Coin F., 2011. A history of TFIIH: two decades of molecular biology on a pivotal transcription/repair factor. DNA Repair (Amst.) 10: 714–721. [DOI] [PubMed] [Google Scholar]
- Eide L., Bjoras M., Pirovano M., Alseth I., Berdal K. G., et al. , 1996. Base excision of oxidative purine and pyrimidine DNA damage in Saccharomyces cerevisiae by a DNA glycosylase with sequence similarity to endonuclease III from Escherichia coli. Proc. Natl. Acad. Sci. USA 93: 10735–10740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdeniz N., Nguyen M., Deschenes S. M., Liskay R. M., 2007. Mutations affecting a putative MutLα endonuclease motif impact multiple mismatch repair functions. DNA Repair (Amst.) 6: 1463–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito G., Godin I., Klein U., Yaspo M.-L., Cumano A., et al. , 2000. Disruption of the REV3I-encoded catalytic subunit of polymerase ζ in mice results in early embryonic lethality. Curr. Biol. 10: 1221–1224. [DOI] [PubMed] [Google Scholar]
- Evans E., Moggs J. G., Hwang J. R., Egly J. M., Wood R. D., 1997. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J. 16: 6559–6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feaver W. J., Svejstrup J. Q., Bardwell L., Bardwell A. J., Buratowski S., et al. , 1993. Dual roles of a multiprotein complex from S. cerevisiae in transcription and DNA repair. Cell 75: 1379–1387. [DOI] [PubMed] [Google Scholar]
- Flores-Rozas H., Kolodner R. D., 1998. The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proc. Natl. Acad. Sci. USA 95: 12404–12409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Rozas H., Clark D., Kolodner R. D., 2000. Proliferating cell nuclear antigen and Msh2p-Msh6p interact to form an active mispair recognition complex. Nat. Genet. 26: 375–378. [DOI] [PubMed] [Google Scholar]
- Fousteri M., Mullenders L. H., 2008. Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Res. 18: 73–84. [DOI] [PubMed] [Google Scholar]
- Freudenthal B. D., Gakhar L., Ramaswamy S., Washington M. T., 2010. Structure of monoubiquitinated PCNA and implications for translesion synthesis and DNA polymerase exchange. Nat. Struct. Mol. Biol. 17: 479–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg E. C., Siede W., Cooper A. J., 1991. Cellular responses to DNA damage in yeast, pp. 147–192 in The Molecular and Cellular Biology of the Yeast Saccharomyces cerevisiae: Genome Dynamics, Protein Synthesis, and Energetics, edited by Broach J. R., Pringle J. R., Jones E. W. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Friedberg E. C., Walker G. C., Siede W., Wood R. D., Schultz R. A., et al. , 2006. DNA Repair and Mutagenesis. ASM Press, Washington, DC. [Google Scholar]
- Fromme J. C., Banerjee A., Verdine G. L., 2004. DNA glycosylase recognition and catalysis. Curr. Opin. Struct. Biol. 14: 43–49. [DOI] [PubMed] [Google Scholar]
- Fu Y., Zhu Y., Zhang K., Yeung M., Durocher D., et al. , 2008. Rad6-Rad18 mediates a eukaryotic SOS response by ubiquitinating the 9–1-1 checkpoint clamp. Cell 133: 601–611. [DOI] [PubMed] [Google Scholar]
- Gaillard H., Wellinger R. E., Aguilera A., 2007. A new connection of mRNP biogenesis and export with transcription-coupled repair. Nucleic Acids Res. 35: 3893–3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard H., Tous C., Botet J., Gonzalez-Aguilera C., Quintero M. J., et al. , 2009. Genome-wide analysis of factors affecting transcription elongation and DNA repair: a new role for PAF and Ccr4-not in transcription-coupled repair. PLoS Genet. 5: e1000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Sanchez A., Andres S., Conde F., San-Segundo P. A., Bueno A., 2012. Reversal of PCNA ubiquitylation by Ubp10 in Saccharomyces cerevisiae. PLoS Genet. 8: e1002826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangavarapu V., Haracska L., Unk I., Johnson R. E., Prakash S., et al. , 2006. Mms2-Ubc13-dependent and -independent roles of Rad5 ligase in postreplication repair and translesion DNA synthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 26: 7783–7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangavarapu V., Prakash S., Prakash L., 2007. Requirement of RAD52 group genes for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 27: 7758–7764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellon L., Barbey R., Auffret Van Der Kemp P., Thomas D., Boiteux S., 2001. Synergism between base excision repair, mediated by the DNA glycosylases Ntg1 and Ntg2, and nucleotide excision repair in the removal of oxidatively damaged DNA bases in Saccharomyces cerevisiae. Mol. Genet. Genomics 265: 1087–1096. [DOI] [PubMed] [Google Scholar]
- Gellon L., Werner M., Boiteux S., 2002. Ntg2p, a Saccharomyces cerevisiae DNA N-glycosylase/apurinic or apyrimidinic lyase involved in base excision repair of oxidative DNA damage, interacts with the DNA mismatch repair protein Mlh1p. Identification of a Mlh1p binding motif. J. Biol. Chem. 277: 29963–29972. [DOI] [PubMed] [Google Scholar]
- Gellon L., Carson D. R., Carson J. P., Demple B., 2008. Intrinsic 5′-deoxyribose-5-phosphate lyase activity in Saccharomyces cerevisiae Trf4 protein with a possible role in base excision DNA repair. DNA Repair (Amst.) 7: 187–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannattasio M., Follonier C., Tourriere H., Puddu F., Lazzaro F., et al. , 2010. Exo1 competes with repair synthesis, converts NER intermediates to long ssDNA gaps, and promotes checkpoint activation. Mol. Cell 40: 50–62. [DOI] [PubMed] [Google Scholar]
- Gibbons B. J., Brignole E. J., Azubel M., Murakami K., Voss N. R., et al. , 2012. Subunit architecture of general transcription factor TFIIH. Proc. Natl. Acad. Sci. USA 109: 1949–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs P. E., McDonald J., Woodgate R., Lawrence C. W., 2005. The relative roles in vivo of Saccharomyces cerevisiae Polη, Polζ, Rev1 protein and Pol32 in the bypass and mutation induction of an abasic site, T-T (6–4) photoadduct and T-T cis-syn cyclobutane dimer. Genetics 169: 575–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglia-Mari G., Coin F., Ranish J. A., Hoogstraten D., Theil A., et al. , 2004. A new, tenth subunit of TFIIH is responsible for the DNA repair syndrome trichothiodystrophy group A. Nat. Genet. 36: 714–719. [DOI] [PubMed] [Google Scholar]
- Gillette T. G., Yu S., Zhou Z., Waters R., Johnston S. A., et al. , 2006. Distinct functions of the ubiquitin-proteasome pathway influence nucleotide excision repair. EMBO J. 25: 2529–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard P. M., Guibourt N., Boiteux S., 1997. The Ogg1 protein of Saccharomyces cerevisiae: a 7,8-dihydro-8-oxoguanine DNA glycosylase/AP lyase whose lysine 241 is a critical residue for catalytic activity. Nucleic Acids Res. 25: 3204–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard P. M., d’Ham C., Cadet J., Boiteux S., 1998. Opposite base-dependent excision of 7,8-dihydro-8-oxoadenine by the Ogg1 protein of Saccharomyces cerevisiae. Carcinogenesis 19: 1299–1305. [DOI] [PubMed] [Google Scholar]
- Glassner B. J., Rasmussen L. J., Najarian M. T., Posnick L. M., Samson L. D., 1998. Generation of a strong mutator phenotype in yeast by imbalanced base excision repair. Proc. Natl. Acad. Sci. USA 95: 9997–10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong F., Fahy D., Smerdon M. J., 2006. Rad4-Rad23 interaction with SWI/SNF links ATP-dependent chromatin remodeling with nucleotide excision repair. Nat. Struct. Mol. Biol. 13: 902–907. [DOI] [PubMed] [Google Scholar]
- Gorman J., Plys A. J., Visnapuu M. L., Alani E., Greene E. C., 2010. Visualizing one-dimensional diffusion of eukaryotic DNA repair factors along a chromatin lattice. Nat. Struct. Mol. Biol. 17: 932–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradia S., Subramanian D., Wilson T., Acharya S., Makhov A., et al. , 1999. hMSH2-hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Mol. Cell 3: 255–261. [DOI] [PubMed] [Google Scholar]
- Gragg H., Harfe B. D., Jinks-Robertson S., 2002. Base composition of mononucleotide runs affects DNA polymerase slippage and removal of frameshift intermediates by mismatch repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 22: 8756–8762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grogan D., Jinks-Robertson S., 2012. Formaldehyde-induced mutagenesis in Saccharomyces cerevisiae: molecular properties and the roles of repair and bypass systems. Mutat. Res. 731: 92–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann K. F., Ward A. M., Matkovic M. E., Folias A. E., Moses R. E., 2001. S. cerevisiae has three pathways for DNA interstrand crosslink repair. Mutat. Res. 487: 73–83. [DOI] [PubMed] [Google Scholar]
- Guarne A., Junop M. S., Yang W., 2001. Structure and function of the N-terminal 40 kDa fragment of human PMS2: a monomeric GHL ATPase. EMBO J. 20: 5521–5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarne A., Ramon-Maiques S., Wolff E. M., Ghirlando R., Hu X., et al. , 2004. Structure of the MutL C-terminal domain: a model of intact MutL and its roles in mismatch repair. EMBO J. 23: 4134–4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillet M., Boiteux S., 2002. Endogenous DNA abasic sites cause cell death in the absence of Apn1, Apn2 and Rad1/Rad10 in Saccharomyces cerevisiae. EMBO J. 21: 2833–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillet M., Boiteux S., 2003. Origin of endogenous DNA abasic sites in Saccharomyces cerevisiae. Mol. Cell. Biol. 23: 8386–8394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillet M., Van Der Kemp P. A., Boiteux S., 2006. dUTPase activity is critical to maintain genetic stability in Saccharomyces cerevisiae. Nucleic Acids Res. 34: 2056–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C., Fischhaber P. L., Luk-Paszyc M. J., Masuda Y., Zhou J., et al. , 2003. Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. EMBO J. 22: 6621–6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D., Wu X., Rajpal D. K., Taylor J. S., Wang Z., 2001. Translesion synthesis by yeast DNA polymerase ζ from templates containing lesions of ultraviolet radiation and acetylaminofluorene. Nucleic Acids Res. 29: 2875–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Gellert M., Yang W., 2012. Mechanism of mismatch recognition revealed by human MutSβ bound to unpaired DNA loops. Nat. Struct. Mol. Biol. 19: 72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzder S. N., Sung P., Prakash L., Prakash S., 1993. Yeast DNA-repair gene RAD14 encodes a zinc metalloprotein with affinity for ultraviolet-damaged DNA. Proc. Natl. Acad. Sci. USA 90: 5433–5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzder S. N., Sung P., Bailly V., Prakash L., Prakash S., 1994. RAD25 is a DNA helicase required for DNA repair and RNA polymerase II transcription. Nature 369: 578–581. [DOI] [PubMed] [Google Scholar]
- Guzder S. N., Bailly V., Sung P., Prakash L., Prakash S., 1995a Yeast DNA repair protein RAD23 promotes complex formation between transcription factor TFIIH and DNA damage recognition factor RAD14. J. Biol. Chem. 270: 8385–8388. [DOI] [PubMed] [Google Scholar]
- Guzder S. N., Habraken Y., Sung P., Prakash L., Prakash S., 1995b Reconstitution of yeast nucleotide excision repair with purified Rad proteins, replication protein A, and transcription factor TFIIH. J. Biol. Chem. 270: 12973–12976. [DOI] [PubMed] [Google Scholar]
- Guzder S. N., Habraken Y., Sung P., Prakash L., Prakash S., 1996a RAD26, the yeast homolog of human Cockayne’s syndrome group B gene, encodes a DNA-dependent ATPase. J. Biol. Chem. 271: 18314–18317. [DOI] [PubMed] [Google Scholar]
- Guzder S. N., Sung P., Prakash L., Prakash S., 1996b Nucleotide excision repair in yeast is mediated by sequential assembly of repair factors and not by a pre-assembled repairosome. J. Biol. Chem. 271: 8903–8910. [DOI] [PubMed] [Google Scholar]
- Guzder S. N., Sung P., Prakash L., Prakash S., 1997. Yeast Rad7-Rad16 complex, specific for the nucleotide excision repair of the nontranscribed DNA strand, is an ATP-dependent DNA damage sensor. J. Biol. Chem. 272: 21665–21668. [DOI] [PubMed] [Google Scholar]
- Guzder S. N., Sung P., Prakash L., Prakash S., 1998. The DNA-dependent ATPase activity of yeast nucleotide excision repair factor 4 and its role in DNA damage recognition. J. Biol. Chem. 273: 6292–6296. [DOI] [PubMed] [Google Scholar]
- Guzder S. N., Sung P., Prakash L., Prakash S., 1999. Synergistic interaction between yeast nucleotide excision repair factors NEF2 and NEF4 in the binding of ultraviolet-damaged DNA. J. Biol. Chem. 274: 24257–24262. [DOI] [PubMed] [Google Scholar]
- Guzder S. N., Torres-Ramos C., Johnson R. E., Haracska L., Prakash L., et al. , 2004. Requirement of yeast Rad1-Rad10 nuclease for the removal of 3′-blocked termini from DNA strand breaks induced by reactive oxygen species. Genes Dev. 18: 2283–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzder S. N., Sommers C. H., Prakash L., Prakash S., 2006. Complex formation with damage recognition protein Rad14 is essential for Saccharomyces cerevisiae Rad1-Rad10 nuclease to perform its function in nucleotide excision repair in vivo. Mol. Cell. Biol. 26: 1135–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habraken Y., Sung P., Prakash L., Prakash S., 1993. Yeast excision repair gene RAD2 encodes a single-stranded DNA endonuclease. Nature 366: 365–368. [DOI] [PubMed] [Google Scholar]
- Habraken Y., Sung P., Prakash L., Prakash S., 1994. A conserved 5′ to 3′ exonuclease activity in the yeast and human nucleotide excision repair proteins RAD2 and XPG. J. Biol. Chem. 269: 31342–31345. [PubMed] [Google Scholar]
- Habraken Y., Sung P., Prakash L., Prakash S., 1995. Structure-specific nuclease activity in yeast nucleotide excision repair protein Rad2. J. Biol. Chem. 270: 30194–30198. [DOI] [PubMed] [Google Scholar]
- Habraken Y., Sung P., Prakash L., Prakash S., 1997. Enhancement of MSH2–MSH3-mediated mismatch recognition by the yeast MLH1–PMS1 complex. Curr. Biol. 7: 790–793. [DOI] [PubMed] [Google Scholar]
- Habraken Y., Sung P., Prakash L., Prrakash S., 1998. ATP-dependent assembly of a ternary complex consisting of a DNA mismatch and the yeast MSH2–MSH6 and MLH1–PMS1 protein complexes. J. Biol. Chem. 273: 9837–9841. [DOI] [PubMed] [Google Scholar]
- Halas A., Baranowska H., Policinska Z., Jachymczyk W. J., 1997. Involvement of the REV3 gene in the methylated base-excision repair system. Co-operation of two DNA polymerases, δ and Rev3p, in the repair of MMS-induced lesions in the DNA of Saccharomyces cerevisiae. Curr. Genet. 31: 292–301. [DOI] [PubMed] [Google Scholar]
- Hall M. C., Wang H., Erie D. A., Kunkel T. A., 2001. High affinity cooperative DNA binding by the yeast Mlh1-Pms1 heterodimer. J. Mol. Biol. 312: 637–647. [DOI] [PubMed] [Google Scholar]
- Hall M. C., Shcherbakova P. V., Kunkel T. A., 2002. Differential ATP binding and intrinsic ATP hydrolysis by amino-terminal domains of the yeast Mlh1 and Pms1 proteins. J. Biol. Chem. 277: 3673–3679. [DOI] [PubMed] [Google Scholar]
- Hanawalt P. C., Spivak G., 2008. Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 9: 958–970. [DOI] [PubMed] [Google Scholar]
- Hanna M., Ball L. G., Tong A. H., Boone C., Xiao W., 2007. Pol32 is required for Pol ζ-dependent translesion synthesis and prevents double-strand breaks at the replication fork. Mutat. Res. 625: 164–176. [DOI] [PubMed] [Google Scholar]
- Hansen R. J., Friedberg E. C., Reagan M. S., 2000. Sensitivity of a S. cerevisiae RAD27 deletion mutant to DNA-damaging agents and in vivo complementation by the human FEN-1 gene. Mutat. Res. 461: 243–248. [DOI] [PubMed] [Google Scholar]
- Haracska L., Yu S. L., Johnson R. E., Prakash L., Prakash S., 2000. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase η. Nat. Genet. 25: 458–461. [DOI] [PubMed] [Google Scholar]
- Haracska L., Kondratick C. M., Unk I., Prakash S., Prakash L., 2001a Interaction with PCNA is essential for yeast DNA polymerase η function. Mol. Cell 8: 407–415. [DOI] [PubMed] [Google Scholar]
- Haracska L., Unk I., Johnson R. E., Johansson E., Burgers P. M., et al. , 2001b Roles of yeast DNA polymerases δ and ζ and of Rev1 in the bypass of abasic sites. Genes Dev. 15: 945–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L., Washington M. T., Prakash S., Prakash L., 2001c Inefficient bypass of an abasic site by DNA polymerase η. J. Biol. Chem. 276: 6861–6866. [DOI] [PubMed] [Google Scholar]
- Haracska L., Prakash S., Prakash L., 2002. Yeast Rev1 protein is a G template-specific DNA polymerase. J. Biol. Chem. 277: 15546–15551. [DOI] [PubMed] [Google Scholar]
- Haracska L., Prakash S., Prakash L., 2003. Yeast DNA polymerase ζ is an efficient extender of primer ends opposite from 7,8-dihydro-8-oxoguanine and O6-methylguanine. Mol. Cell. Biol. 23: 1453–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe B. D., Jinks-Robertson S., 1999. Removal of frameshift intermediates by mismatch repair proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 4766–4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe B. D., Jinks-Robertson S., 2000a Sequence composition and context effects on the generation and repair of frameshift intermediates in mononucleotide runs in Saccharomyces cerevisiae. Genetics 156: 571–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe B. D., Jinks-Robertson S., 2000b DNA polymerase ζ introduces multiple mutations when bypassing spontaneous DNA damage in Saccharomyces cerevisiae. Mol. Cell 6: 1491–1499. [DOI] [PubMed] [Google Scholar]
- Harfe B. D., Jinks-Robertson S., 2000c DNA mismatch repair and genetic instability. Annu. Rev. Genet. 34: 359–399. [DOI] [PubMed] [Google Scholar]
- Harfe B. D., Minesinger B. K., Jinks-Robertson S., 2000. Discrete in vivo roles for the MutL homologs Mlh2p and Mlh3p in the removal of frameshift intermediates in budding yeast. Curr. Biol. 10: 145–148. [DOI] [PubMed] [Google Scholar]
- Hargreaves V. V., Shell S. S., Mazur D. J., Hess M. T., Kolodner R. D., 2010. Interaction between the Msh2 and Msh6 nucleotide-binding sites in the Saccharomyces cerevisiae Msh2-Msh6 complex. J. Biol. Chem. 285: 9301–9310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harosh I., Naumovski L., Friedberg E. C., 1989. Purification and characterization of Rad3 ATPase/DNA helicase from Saccharomyces cerevisiae. J. Biol. Chem. 264: 20532–20539. [PubMed] [Google Scholar]
- Harrington J. M., Kolodner R. D., 2007. Saccharomyces cerevisiae Msh2-Msh3 acts in repair of base-base mispairs. Mol. Cell. Biol. 27: 6546–6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes R. H., Kunz B. A., 1981. DNA repair and mutagenesis in yeast, pp. 371–414 in The Molecular Biology of theYeast Saccharomyces: Life Cycle and Inheritance, edited by Strathern J. N., Jones E. W., Broach J. R. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Henning K. A., Li L., Iyer N., McDaniel L. D., Reagan M. S., et al. , 1995. The Cockayne syndrome group A gene encodes a WD repeat protein that interacts with CSB protein and a subunit of RNA polymerase II TFIIH. Cell 82: 555–564. [DOI] [PubMed] [Google Scholar]
- Hess M. T., Mendillo M. L., Mazur D. J., Kolodner R. D., 2006. Biochemical basis for dominant mutations in the Saccharomyces cerevisiae MSH6 gene. Proc. Natl. Acad. Sci. USA 103: 558–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K., Sonoda E., Kawamoto T., Motegi A., Masutani C., et al. , 2010. Simultaneous disruption of two DNA polymerases, Polη and Polζ, in avian DT40 cells unmasks the role of Polη in cellular response to various DNA lesions. PLoS Genet. 6: e1001151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hishida T., Kubota Y., Carr A. M., Iwasaki H., 2009. RAD6–RAD18–RAD5-pathway-dependent tolerance to chronic low-dose ultraviolet light. Nature 457: 612–615. [DOI] [PubMed] [Google Scholar]
- Hoege C., Pfander B., Moldovan G. L., Pyrowolakis G., Jentsch S., 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers J. H., 2001. Genome maintenance mechanisms for preventing cancer. Nature 411: 366–374. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers J. H., 2009. DNA damage, aging, and cancer. N. Engl. J. Med. 361: 1475–1485. [DOI] [PubMed] [Google Scholar]
- Hoffmann E. R., Borts R. H., 2004. Meiotic recombination intermediates and mismatch repair proteins. Cytogenet. Genome Res. 107: 232–248. [DOI] [PubMed] [Google Scholar]
- Hoffmann E. R., Shcherbakova P. V., Kunkel T. A., Borts R. H., 2003. MLH1 mutations differentially affect meiotic functions in Saccharomyces cerevisiae. Genetics 163: 515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann R. M., Pickart C. M., 1999. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96: 645–653. [DOI] [PubMed] [Google Scholar]
- Hollingsworth N. M., Ponte L., Halsey C., 1995. MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev. 9: 1728–1739. [DOI] [PubMed] [Google Scholar]
- Holmes S. F., Scarpinato K. D., McCulloch S. D., Schaaper R. M., Kunkel T. A., 2007. Specialized mismatch repair function of Glu339 in the Phe-X-Glu motif of yeast Msh6. DNA Repair (Amst.) 6: 293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hombauer H., Campbell C. S., Smith C. E., Desai A., Kolodner R. D., 2011a Visualization of eukaryotic DNA mismatch repair reveals distinct recognition and repair intermediates. Cell 147: 1040–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hombauer H., Srivatsan A., Putnam C. D., Kolodner R. D., 2011b Mismatch repair, but not heteroduplex rejection, is temporally coupled to DNA replication. Science 334: 1713–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh P., Yamane K., 2008. DNA mismatch repair: molecular mechanism, cancer, and ageing. Mech. Ageing Dev. 129: 391–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M.-E., De Calignon A., Nicolas A., Galibert F., 2000. POL32, a subunit of the Saccharomyces cerevisiae DNA polymerase δ, defines a link between DNA replication and the mutagenic bypass repair pathway. Curr. Genet. 38: 178–187. [DOI] [PubMed] [Google Scholar]
- Huang M. E., Rio A. G., Nicolas A., Kolodner R. D., 2003. A genomewide screen in Saccharomyces cerevisiae for genes that suppress the accumulation of mutations. Proc. Natl. Acad. Sci. USA 100: 11529–11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Feaver W. J., Tomkinson A. E., Friedberg E. C., 1998. The N-degron protein degradation strategy for investigating the function of essential genes: requirement for replication protein A and proliferating cell nuclear antigen proteins for nucleotide excision repair in yeast extracts. Mutat. Res. 408: 183–194. [DOI] [PubMed] [Google Scholar]
- Hwang C. S., Shemorry A., Varshavsky A., 2009. Two proteolytic pathways regulate DNA repair by cotargeting the Mgt1 alkylguanine transferase. Proc. Natl. Acad. Sci. USA 106: 2142–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impellizzeri K. J., Anderson B., Burgers P. M., 1991. The spectrum of spontaneous mutations in a Saccharomyces cerevisiae uracil-DNA-glycosylase mutant limits the function of this enzyme to cytosine deamination repair. J. Bacteriol. 173: 6807–6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischenko A. A., Saparbaev M. K., 2002. Alternative nucleotide incision repair pathway for oxidative DNA damage. Nature 415: 183–187. [DOI] [PubMed] [Google Scholar]
- Iyer R. R., Pluciennik A., Genschel J., Tsai M. S., Beese L. S., et al. , 2010. MutLα and proliferating cell nuclear antigen share binding sites on MutSβ. J. Biol. Chem. 285: 11730–11739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James A. P., Kilbey B. J., 1977. The timing of UV mutagenesis in yeast: a pedigree analysis of induced recessive mutation. Genetics 87: 237–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen L. E., Verhage R. A., Brouwer J., 1998. Preferential binding of yeast Rad4•Rad23 complex to damaged DNA. J. Biol. Chem. 273: 33111–33114. [DOI] [PubMed] [Google Scholar]
- Jentsch S., McGrath J. P., Varshavsky A., 1987. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature 329: 131–134. [DOI] [PubMed] [Google Scholar]
- Jeyaprakash A., Welch J. W., Fogel S., 1994. Mutagenesis of yeast MW104–1B strain has identified the uncharacterized PMS6 DNA mismatch repair gene locus and additional alleles of existing PMS1, PMS2 and MSH2 genes. Mutat. Res. 325: 21–29. [DOI] [PubMed] [Google Scholar]
- Johnson R. E., Henderson S. T., Petes T. D., Prakash S., Bankmann M., et al. , 1992. Saccharomyces cerevisiae RAD5-encoded DNA repair protein contains DNA helicase and zinc-binding sequence motifs and affects the stability of simple repetitive sequences in the genome. Mol. Cell. Biol. 12: 3807–3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. E., Kovvali G. K., Prakash L., Prakash S., 1995. Requirement of the yeast RTH1 5′ to 3′ exonuclease for the stability of simple repetitive DNA. Science 269: 238–240. [DOI] [PubMed] [Google Scholar]
- Johnson R. E., Kovvali G. K., Guzder S. N., Amin N. S., Holm C., et al. , 1996a Evidence for involvement of yeast proliferating cell nuclear antigen in DNA mismatch repair. J. Biol. Chem. 271: 27987–27990. [DOI] [PubMed] [Google Scholar]
- Johnson R. E., Kovvali G. K., Prakash L., Prakash S., 1996b Requirement of the yeast MSH3 and MSH6 genes for MSH2-dependent genomic stability. J. Biol. Chem. 271: 7285–7288. [DOI] [PubMed] [Google Scholar]
- Johnson R. E., Torres-Ramos C. A., Izumi T., Mitra S., Prakash S., et al. , 1998. Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites. Genes Dev. 12: 3137–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. E., Kondratick C. M., Prakash S., Prakash L., 1999a hRAD30 mutations in the variant form of xeroderma pigmentosum. Nature 285: 263–265. [DOI] [PubMed] [Google Scholar]
- Johnson R. E., Prakash S., Prakash L., 1999b Requirement of DNA polymerase activity of yeast Rad30 protein for its biological function. J. Biol. Chem. 274: 15975–15977. [DOI] [PubMed] [Google Scholar]
- Johnson R. E., Washington M. T., Haracska L., Prakash S., Prakash L., 2000. Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature 406: 1015–1019. [DOI] [PubMed] [Google Scholar]
- Johnson R. E., Haracska L., Prakash S., Prakash L., 2001. Role of DNA polymerase η in the bypass of a (6–4) TT photoproduct. Mol. Cell. Biol. 21: 3558–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. E., Yu S. L., Prakash S., Prakash L., 2003. Yeast DNA polymerase ζ is essential for error-free replication past thymine glycol. Genes Dev. 17: 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. E., Yu S. L., Prakash S., Prakash L., 2007. A role for yeast and human translesion synthesis DNA polymerases in promoting replication through 3-methyl adenine. Mol. Cell. Biol. 27: 7198–7205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. E., Prakash L., Prakash S., 2012. Pol31 and Pol32 subunits of yeast DNA polymerase δ are also essential subunits of DNA polymerase ζ. Proc. Natl. Acad. Sci. USA 109: 12455–12460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston L. H., Johnson A. L., 1995. The DNA repair genes RAD54 and UNG1 are cell cycle regulated in budding yeast but MCB promoter elements have no essential role in the DNA damage response. Nucleic Acids Res. 23: 2147–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce C. M., 1997. Choosing the right sugar: how polymerases select a nucleotide substrate. Proc. Natl. Acad. Sci. USA 94: 1619–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junop M. S., Obmolova G., Rausch K., Hsieh P., Yang W., 2001. Composite active site of an ABC ATPase: MutS uses ATP to verify mismatch recognition and authorize DNA repair. Mol. Cell 7: 1–12. [DOI] [PubMed] [Google Scholar]
- Kadyrov F. A., Dzantiev L., Constantin N., Modrich P., 2006. Endonucleolytic function of MutLα in human mismatch repair. Cell 126: 297–308. [DOI] [PubMed] [Google Scholar]
- Kadyrov F. A., Holmes S. F., Arana M. E., Lukianova O. A., O’Donnell M., et al. , 2007. Saccharomyces cerevisiae MutLα is a mismatch repair endonuclease. J. Biol. Chem. 282: 37181–37190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyrov F. A., Genschel J., Fang Y., Penland E., Edelmann W., et al. , 2009. A possible mechanism for exonuclease 1-independent eukaryotic mismatch repair. Proc. Natl. Acad. Sci. USA 106: 8495–8500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalifa L., Sia E. A., 2007. Analysis of Rev1p and Polζ in mitochondrial mutagenesis suggests an alternative pathway of damage tolerance. DNA Repair (Amst.) 6: 1732–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karahalil B., Girard P. M., Boiteux S., Dizdaroglu M., 1998. Substrate specificity of the Ogg1 protein of Saccharomyces cerevisiae: excision of guanine lesions produced in DNA by ionizing radiation- or hydrogen peroxide/metal ion-generated free radicals. Nucleic Acids Res. 26: 1228–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karras G. I., Jentsch S., 2010. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell 141: 255–267. [DOI] [PubMed] [Google Scholar]
- Kearney H. M., Kirkpatrick D. T., Gerton J. L., Petes T. D., 2001. Meiotic recombination involving heterozygous large insertions in Saccharomyces cerevisiae: formation and repair of large, unpaired DNA loops. Genetics 158: 1457–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesti T., Flick K., Keranen S., Syvaoja J. E., Wittenberg C., 1999. DNA polymerase ε catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol. Cell 3: 679–685. [DOI] [PubMed] [Google Scholar]
- Kim N., Jinks-Robertson S., 2009. dUTP incorporation into genomic DNA is linked to transcription in yeast. Nature 459: 1150–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N., Jinks-Robertson S., 2010. Abasic sites in the transcribed strand of yeast DNA are removed by transcription-coupled nucleotide excision repair. Mol. Cell. Biol. 30: 3206–3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N., Huang S. Y., Williams J. S., Li Y. C., Clark A. B., et al. , 2011a Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science 332: 1561–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N., Mudrak S. V., Jinks-Robertson S., 2011b The dCMP transferase activity of yeast Rev1 is biologically relevant during the bypass of endogenously generated AP sites. DNA Repair (Amst.) 10: 1262–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick D. T., Petes T. D., 1997. Repair of DNA loops involves DNA-mismatch and nucleotide-excision repair proteins. Nature 387: 929–931. [DOI] [PubMed] [Google Scholar]
- Klapacz J., Lingaraju G. M., Guo H. H., Shah D., Moar-Shoshani A., et al. , 2010. Frameshift mutagenesis and microsatellite instability induced by human alkyladenine DNA glycosylase. Mol. Cell 37: 843–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosarek J. N., Woodruff R. V., Rivera-Begeman A., Guo C., D’Souza S., et al. , 2008. Comparative analysis of in vivo interactions between Rev1 protein and other Y-family DNA polymerases in animals and yeasts. DNA Repair (Amst.) 7: 439–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kow Y. W., Bao G., Minesinger B., Jinks-Robertson S., Siede W., et al. , 2005. Mutagenic effects of abasic and oxidized abasic lesions in Saccharomyces cerevisiae. Nucleic Acids Res. 33: 6196–6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kow Y. W., Bao G., Reeves J. W., Jinks-Robertson S., Crouse G. F., 2007. Oligonucleotide transformation of yeast reveals mismatch repair complexes to be differentially active on DNA replication strands. Proc. Natl. Acad. Sci. USA 104: 11352–11357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozmin S. G., Pavlov Y. I., Kunkel T. A., Sage E., 2003. Roles of Saccharomyces cerevisiae DNA polymerases Polη and Polζ in response to irradiation by simulated sunlight. Nucleic Acids Res. 31: 4541–4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozmin S., Slezak G., Reynaud-Angelin A., Elie C., De Rycke Y., et al. , 2005. UVA radiation is highly mutagenic in cells that are unable to repair 7,8-dihydro-8-oxoguanine in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 102: 13538–13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer W., Kramer B., Williamson M. S., Fogel S., 1989. Cloning and nucleotide sequence of DNA mismatch repair gene PMS1 from Saccharomyces cerevisiae: homology of PMS1 to procaryotic MutL and HexB. J. Bacteriol. 171: 5339–5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci L., Van Komen S., Li Y., Villemain J., Reddy M. S., et al. , 2003. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423: 305–309. [DOI] [PubMed] [Google Scholar]
- Kuang L., Kou H., Xie Z., Zhou Y., Feng X., et al. , 2013. A noncatalytic function of Rev1 in translesion DNA synthesis and mutagenesis is mediated by its stable interaction with Rad5. DNA Repair 12: 27–37. [DOI] [PubMed] [Google Scholar]
- Kumar C., Piacente S. C., Sibert J., Bukata A. R., O’Connor J., et al. , 2011. Multiple factors insulate Msh2-Msh6 mismatch repair activity from defects in Msh2 domain I. J. Mol. Biol. 411: 765–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Erie D. A., 2005. DNA mismatch repair. Annu. Rev. Biochem. 74: 681–710. [DOI] [PubMed] [Google Scholar]
- Kunz B. A., Henson E. S., Roche H., Ramotar D., Nunoshiba T., et al. , 1994. Specificity of the mutator caused by deletion of the yeast structural gene (APN1) for the major apurinic endonuclease. Proc. Natl. Acad. Sci. USA 91: 8165–8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraoka I., Bender C., Romieu A., Cadet J., Wood R. D., et al. , 2000. Removal of oxygen free-radical-induced 5′,8-purine cyclodeoxynucleosides from DNA by the nucleotide excision-repair pathway in human cells. Proc. Natl. Acad. Sci. USA 97: 3832–3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafrance-Vanasse J., Arseneault G., Cappadocia L., Chen H. T., Legault P., et al. , 2012. Structural and functional characterization of interactions involving the Tfb1 subunit of TFIIH and the NER factor Rad2. Nucleic Acids Res. 40: 5739–5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers M. H., Perrakis A., Enzlin J. H., Winterwerp H. H. K., De Wind N., et al. , 2000. The crystal structure of DNA mismatch repair protein MutS binding to a G:T mismatch. Nature 407: 711–717. [DOI] [PubMed] [Google Scholar]
- Lang G. I., Murray A. W., 2011. Mutation rates across budding yeast chromosome VI are correlated with replication timing. Genome Biol. Evol. 3: 799–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau P. J., Flores-Rozas H., Kolodner R. D., 2002. Isolation and characterization of new proliferating cell nuclear antigen (POL30) mutator mutants that are defective in DNA mismatch repair. Mol. Cell. Biol. 22: 6669–6680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C., 1994. The RAD6 DNA repair pathway in Saccharomyces cerevisiae: What does it do, and how does it do it? Bioessays 16: 253–258. [DOI] [PubMed] [Google Scholar]
- Lazzaro F., Novarina D., Amara F., Watt D. L., Stone J. E., et al. , 2012. RNase H and postreplication repair protect cells from ribonucleotides incorporated in DNA. Mol. Cell 45: 99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. D., Alani E., 2006. Analysis of interactions between mismatch repair initiation factors and the replication processivity factor PCNA. J. Mol. Biol. 355: 175–184. [DOI] [PubMed] [Google Scholar]
- Lee S. D., Surtees J. A., Alani E., 2007. Saccharomyces cerevisiae MSH2–MSH3 and MSH2–MSH6 complexes display distinct requirements for DNA binding domain I in mismatch recognition. J. Mol. Biol. 366: 53–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. K., Yu S. L., Prakash L., Prakash S., 2001. Requirement for yeast RAD26, a homolog of the human CSB gene, in elongation by RNA polymerase II. Mol. Cell. Biol. 21: 8651–8656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. K., Yu S. L., Prakash L., Prakash S., 2002. Yeast RAD26, a homolog of the human CSB gene, functions independently of nucleotide excision repair and base excision repair in promoting transcription through damaged bases. Mol. Cell. Biol. 22: 4383–4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann A. R., 2011. DNA polymerases and repair synthesis in NER in human cells. DNA Repair (Amst.) 10: 730–733. [DOI] [PubMed] [Google Scholar]
- Lehner K., Jinks-Robertson S., 2009. The mismatch repair system promotes Polζ-dependent translesion synthesis in yeast. Proc. Natl. Acad. Sci. USA 106: 5749–5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemontt J. F., 1971. Mutants of yeast defective in mutation induced by ultraviolet light. Genetics 68: 21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Smerdon M. J., 2002. Rpb4 and Rpb9 mediate subpathways of transcription-coupled DNA repair in Saccharomyces cerevisiae. EMBO J. 21: 5921–5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liefshitz B., Steinlauf R., Friedl A., Eckardt-Schupp F., Kupiec M., 1998. Genetic interactions between mutants of the ‘error-prone’ repair group of Saccharomyces cerevisiae and their effect on recombination and mutagenesis. Mutat. Res. 407: 135–145. [DOI] [PubMed] [Google Scholar]
- Lingaraju G. M., Kartalou M., Meira L. B., Samson L. D., 2008. Substrate specificity and sequence-dependent activity of the Saccharomyces cerevisiae 3-methyladenine DNA glycosylase (Mag). DNA Repair (Amst.) 7: 970–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M., Foiani M., Sogo J. M., 2006. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell 21: 15–27. [DOI] [PubMed] [Google Scholar]
- Lu J., Liu Y., 2010. Deletion of Ogg1 DNA glycosylase results in telomere base damage and length alteration in yeast. EMBO J. 29: 398–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhr B., Scheller J., Meyer P., Kramer W., 1998. Analysis of in vivo correction of defined mismatches in the DNA mismatch repair mutants msh2, msh3 and msh6 of Saccharomyces cerevisiae. Mol. Gen. Genet. 257: 362–367. [DOI] [PubMed] [Google Scholar]
- Lujan S. A., Williams J. S., Pursell Z. F., Abdulovic-Cui A. A., Clark A. B., et al. , 2012. Mismatch repair balances leading and lagging strand DNA replication fidelity. PLoS Genet. 8: e1003016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyndaker A. M., Alani E., 2009. A tale of tails: insights into the corrdination of 3′ end processing during homologous recombination. Bioessays 31: 315–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S., Chaurasia P., Lahudkar S., Durairaj G., Shukla A., et al. , 2010. Rad26p, a transcription-coupled repair factor, is recruited to the site of DNA lesion in an elongating RNA polymerase II-dependent manner in vivo. Nucleic Acids Res. 38: 1461–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini V., Krejci L., 2010. Srs2: the “Odd-Job Man” in DNA repair. DNA Repair (Amst.) 9: 268–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsischky G. T., Kolodner R. D., 1999. Biochemical characterization of the interaction between the Saccharomyces cerevisiae MSH2–MSH6 complex and mispaired bases in DNA. J. Biol. Chem. 274: 26668–26682. [DOI] [PubMed] [Google Scholar]
- Marsischky G. T., Filosi N., Kane M. F., Kolodner R., 1996. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 10: 407–420. [DOI] [PubMed] [Google Scholar]
- Masutani C., Kusumoto R., Yamada A., Dohmae N., Yokoi M., et al. , 1999. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature 399: 700–704. [DOI] [PubMed] [Google Scholar]
- Matsuda T., Bebenek K., Masutani C., Hanaoka F., Kunkel T. A., 2000. Low fidelity DNA synthesis by human DNA polymerase η. Nature 404: 1011–1013. [DOI] [PubMed] [Google Scholar]
- Matuo R., Sousa F. G., Escargueil A. E., Soares D. G., Grivicich I., et al. , 2010. DNA repair pathways involved in repair of lesions induced by 5-fluorouracil and its active metabolite FdUMP. Biochem. Pharmacol. 79: 147–153. [DOI] [PubMed] [Google Scholar]
- Mazur D. J., Mendillo M. L., Kolodner R. D., 2006. Inhibition of Msh6 ATPase activity by mispaired DNA induces a Msh2(ATP)-Msh6(ATP) state capable of hydrolysis-independent movement along DNA. Mol. Cell 22: 39–49. [DOI] [PubMed] [Google Scholar]
- McCulloch S. D., Kokoska R. J., Garg P., Burgers P. M., Kunkel T. A., 2009. The efficiency and fidelity of 8-oxo-guanine bypass by DNA polymerases δ and η. Nucleic Acids Res. 37: 2830–2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald J. P., Levine A. S., Woodgate R., 1997. The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics 147: 1557–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnis M., O’Neill G., Fossum K., Reagan M. S., 2002. Epistatic analysis of the roles of the RAD27 and POL4 gene products in DNA base excision repair in S. cerevisiae. DNA Repair (Amst.) 1: 311–315. [DOI] [PubMed] [Google Scholar]
- Meadows K. L., Song B., Doetsch P. W., 2003. Characterization of AP lyase activities of Saccharomyces cerevisiae Ntg1p and Ntg2p: implications for biological function. Nucleic Acids Res. 31: 5560–5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellon I., Spivak G., Hanawalt P. C., 1987. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell 51: 241–249. [DOI] [PubMed] [Google Scholar]
- Mendillo M. L., Hargreaves V. V., Jamison J. W., Mo A. O., Li S., et al. , 2009. A conserved MutS homolog connector domain interface interacts with MutL homologs. Proc. Natl. Acad. Sci. USA 106: 22223–22228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels M. L., Miller J. H., 1992. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine). J. Bacteriol. 174: 6321–6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J. H., Pavletich N. P., 2007. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature 449: 570–575. [DOI] [PubMed] [Google Scholar]
- Minca E. C., Kowalski D., 2010. Multiple Rad5 activities mediate sister chromatid recombination to bypass DNA damage at stalled replication forks. Mol. Cell 38: 649–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minesinger B. K., Jinks-Robertson S., 2005. Roles of RAD6 epistasis group members in spontaneous Polζ-dependent translesion synthesis in Saccharomyces cerevisiae. Genetics 169: 1939–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minesinger B. K., Abdulovic A. L., Ou T. M., Jinks-Robertson S., 2006. The effect of oxidative metabolism on spontaneous Pol ζ-dependent translesion synthesis in Saccharomyces cerevisiae. DNA Repair (Amst.) 5: 226–234. [DOI] [PubMed] [Google Scholar]
- Mito E., Mokhnatkin J. V., Steele M. C., Buettner V. L., Sommer S. S., et al. , 2008. Mutagenic and recombinagenic responses to defective DNA polymerase δ are facilitated by the Rev1 protein in pol3-t mutants of Saccharomyces cerevisiae. Genetics 179: 1795–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocquet V., Laine J. P., Riedl T., Yajin Z., Lee M. Y., et al. , 2008. Sequential recruitment of the repair factors during NER: the role of XPG in initiating the resynthesis step. EMBO J. 27: 155–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrich P., Lahue R., 1996. Mismatch repair in replication fidelity, genetic recombination and cancer biology. Annu. Rev. Biochem. 65: 101–133. [DOI] [PubMed] [Google Scholar]
- Moldovan G. L., Pfander B., Jentsch S., 2007. PCNA, the maestro of the replication fork. Cell 129: 665–679. [DOI] [PubMed] [Google Scholar]
- Mookerjee S. A., Lyon H. D., Sia E. A., 2005. Analysis of the functional domains of the mismatch repair homologue Msh1p and its role in mitochondrial genome maintenance. Curr. Genet. 47: 84–99. [DOI] [PubMed] [Google Scholar]
- Morrison A., Christensen R. B., Alley J., Beck A. K., Bernstine E. G., et al. , 1989. REV3, a Saccharomyces cerevisiae gene whose function is required for induced mutagenesis, is predicted to encode a nonessential DNA polymerase. J. Bacteriol. 171: 5659–5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudrak S. V., Welz-Voegele C., Jinks-Robertson S., 2009. The polymerase η translesion synthesis DNA polymerase acts independently of the mismatch repair system to limit mutagenesis caused by 7,8-dihydro-8-oxoguanine in yeast. Mol. Cell. Biol. 29: 5316–5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag D. K., White M. A., Petes T. D., 1989. Palindromic sequences in heteroduplex DNA inhibit mismatch repair in yeast. Nature 340: 318–320. [DOI] [PubMed] [Google Scholar]
- Nair D. T., Johnson R. E., Prakash L., Prakash S., Aggarwal A. K., 2005. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science 309: 2219–2222. [DOI] [PubMed] [Google Scholar]
- Nair D. T., Johnson R. E., Prakash L., Prakash S., Aggarwal A. K., 2011. DNA synthesis across an abasic lesion by yeast REV1 DNA polymerase. J. Mol. Biol. 406: 18–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J. R., Lawrence C. W., Hinkle D. C., 1996a Thymine-thymine dimer bypass by yeast DNA polymerase ζ. Science 272: 1646–1649. [DOI] [PubMed] [Google Scholar]
- Nelson J. R., Lawrence C. W., Hinkle D. C., 1996b Deoxycytidyl transferase activity of yeast REV1 protein. Nature 382: 729–731. [DOI] [PubMed] [Google Scholar]
- Nelson J. R., Gibbs P. E. M., Nowicka A. M., Hinkle D. C., Lawrence C. W., 2000. Evidence for a second function for Saccharomyces cerevisiae Rev1p. Mol. Microbiol. 37: 549–554. [DOI] [PubMed] [Google Scholar]
- Netz D. J., Stith C. M., Stumpfig M., Kopf G., Vogel D., et al. , 2012. Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat. Chem. Biol. 8: 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- New L., Liu K., Crouse G. F., 1993. The yeast gene MSH3 defines a new class of eukaryotic MutS homologues. Mol. Gen. Genet. 239: 97–108. [DOI] [PubMed] [Google Scholar]
- Ni T. T., Marsischky G. T., Kolodner R. D., 1999. MSH2 and MSH6 are required for removal of adenine misincorporated opposite 8-oxo-guanine in S. cerevisiae. Mol. Cell 4: 439–444. [DOI] [PubMed] [Google Scholar]
- Nicholson A., Hendrix M., Jinks-Robertson S., Crouse G. F., 2000. Regulation of mitotic homeologous recombination in yeast: functions of mismatch repair and nucleotide excision repair genes. Genetics 154: 133–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny S. A., Kumar D., Clark A. B., Watt D. L., Watts B. E., et al. , 2010a Genome instability due to ribonucleotide incorporation into DNA. Nat. Chem. Biol. 6: 774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny S. A., Watts B. E., Kumar D., Watt D. L., Lundstrom E. B., et al. , 2010b Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl. Acad. Sci. USA 107: 4949–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishant K. T., Plys A. J., Alani E., 2008. A mutation in the putative MLH3 endonuclease domain confers a defect in both mismatch repair and meiosis in Saccharomyces cerevisiae. Genetics 179: 747–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northam M. R., Robinson H. A., Kochenova O. V., Shcherbakova P. V., 2010. Participation of DNA polymerase ζ in replication of undamaged DNA in Saccharomyces cerevisiae. Genetics 184: 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obmolova G., Ban C., Hsieh P., Yang W., 2000. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature 407: 703–710. [DOI] [PubMed] [Google Scholar]
- Ogi T., Limsirichaikul S., Overmeer R. M., Volker M., Takenaka K., et al. , 2010. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol. Cell 37: 714–727. [DOI] [PubMed] [Google Scholar]
- Otsuka C., Kobayashi K., Kawaguchi N., Kunitomi N., Moriyama K., et al. , 2002a Use of yeast transformation by oligonucleotides to study DNA lesion bypass in vivo. Mutat. Res. 502: 53–60. [DOI] [PubMed] [Google Scholar]
- Otsuka C., Sanadai S., Hata Y., Okuto H., Noskov V. N., et al. , 2002b Difference between deoxyribose- and tetrahydrofuran-type abasic sites in the in vivo mutagenic responses in yeast. Nucleic Acids Res. 30: 5129–5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka C., Kunitomi N., Iwai S., Loakes D., Negishi K., 2005. Roles of the polymerase and BRCT domains of Rev1 protein in translesion DNA synthesis in yeast in vivo. Mutat. Res. 578: 79–87. [DOI] [PubMed] [Google Scholar]
- Pabla R., Rozario D., Siede W., 2008. Regulation of Saccharomyces cerevisiae DNA polymerase η transcript and protein. Radiat. Environ. Biophys. 47: 157–168. [DOI] [PubMed] [Google Scholar]
- Pagès V., Bresson A., Acharya N., Prakash S., Fuchs R. P., et al. , 2008a Requirement of Rad5 for DNA polymerase ζ-dependent translesion synthesis in Saccharomyces cerevisiae. Genetics 180: 73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagès V., Johnson R. E., Prakash L., Prakash S., 2008b Mutational specificity and genetic control of replicative bypass of an abasic site in yeast. Proc. Natl. Acad. Sci. USA 105: 1170–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagès V., Santa Maria S. R., Prakash L., Prakash S., 2009. Role of DNA damage-induced replication checkpoint in promoting lesion bypass by translesion synthesis in yeast. Genes Dev. 23: 1438–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Q., Prolla T. A., Liskay R. M., 1997. Functional domains of the Saccharomyces cerevisiae Mlh1p and Pms1p DNA mismatch repair proteins and their relevance to human hereditary nonpolyposis colorectal cancer-associated mutations. Mol. Cell. Biol. 17: 4465–4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papouli E., Chen S., Davies A. A., Huttner D., Krejci L., et al. , 2005. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell 19: 123–133. [DOI] [PubMed] [Google Scholar]
- Parker J. L., Ulrich H. D., 2012. A SUMO-interacting motif activates budding yeast ubiquitin ligase RAD18 towards SUMO-modified PCNA. Nucleic Acids Res. 40: 11380–11388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J. L., Bielen A. B., Dikic I., Ulrich H. D., 2007. Contributions of ubiquitin- and PCNA-binding domains to the activity of Polymerase η in Saccharomyces cerevisiae. Nucleic Acids Res. 35: 881–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J. L., Bucceri A., Davies A. A., Heidrich K., Windecker H., et al. , 2008. SUMO modification of PCNA is controlled by DNA. EMBO J. 27: 2422–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnas O., Zipin-Roitman A., Pfander B., Liefshitz B., Mazor Y., et al. , 2010. Elg1, an alternative subunit of the RFC clamp loader, preferentially interacts with SUMOylated PCNA. EMBO J. 29: 2611–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulovich A. G., Armour C. D., Hartwell L. H., 1998. The Saccharomyces cerevisiae RAD9, RAD17, Rad24 and MEC3 genes are required for tolerating irreparable, ultraviolet-induced DNA damage. Genetics 150: 75–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov Y. I., Mian I. M., Kunkel T. A., 2003. Evidence for preferential mismatch repair of lagging strand DNA replication errors in yeast. Curr. Biol. 13: 744–748. [DOI] [PubMed] [Google Scholar]
- Peña-Diaz J., Bregenhorm S., Ghodgaonkar M., Follonier C., Artola-Borán M., et al. , 2012. Noncanonical mismatch repair as a source of genomic instability in human cells. Mol. Cell 47: 669–680. [DOI] [PubMed] [Google Scholar]
- Percival K. J., Klein M. B., Burgers P. M., 1989. Molecular cloning and primary structure of the uracil-DNA-glycosylase gene from Saccharomyces cerevisiae. J. Biol. Chem. 264: 2593–2598. [PubMed] [Google Scholar]
- Pessoa-Brandao L., Sclafani R. A., 2004. CDC7/DBF4 functions in the translesion synthesis branch of the RAD6 epistasis group in Saccharomyces cerevisiae. Genetics 167: 1597–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfander B., Moldovan G. L., Sacher M., Hoege C., Jentsch S., 2005. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436: 428–433. [DOI] [PubMed] [Google Scholar]
- Phadnis N., Mehta R., Meednu N., Sia E. A., 2006. Ntg1p, the base excision repair protein, generates mutagenic intermediates in yeast mitochondrial DNA. DNA Repair (Amst.) 5: 829–839. [DOI] [PubMed] [Google Scholar]
- Plotz G., Welsch C., Giron-Monzon L., Friedhoff P., Albrecht M., et al. , 2006. Mutations in the MutSα interaction interface of MLH1 can abolish DNA mismatch repair. Nucleic Acids Res. 34: 6574–6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluciennik A., Dzantiev L., Iyer R. R., Constantin N., Kadyrov F. A., et al. , 2010. PCNA function in the activation and strand direction of MutLα endonuclease in mismatch repair. Proc. Natl. Acad. Sci. USA 107: 16066–16071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pochart P., Woltering D., Hollingsworth N. M., 1997. Conserved properties between functionally distinct MutS homologs in yeast. J. Biol. Chem. 272: 30345–30349. [DOI] [PubMed] [Google Scholar]
- Popoff S. C., Spira A. I., Johnson A. W., Demple B., 1990. Yeast structural gene (APN1) for the major apurinic endonuclease: homology to Escherichia coli endonuclease IV. Proc. Natl. Acad. Sci. USA 87: 4193–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouliot J. J., Yao K. C., Robertson C. A., Nash H. A., 1999. Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science 286: 552–555. [DOI] [PubMed] [Google Scholar]
- Prakash L., 1981. Characterization of postreplication repair in Saccharomyces cerevisiae and effects of rad6, rad18, rev3 and rad52 mutations. Mol. Gen. Genet. 184: 471–478. [DOI] [PubMed] [Google Scholar]
- Prakash L., Prakash S., 1977. Isolation and characterization of MMS-sensitive mutants of Saccharomyces cerevisiae. Genetics 86: 33–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S., Prakash L., 2000. Nucleotide excision repair in yeast. Mutat. Res. 451: 13–24. [DOI] [PubMed] [Google Scholar]
- Prakash S., Sung P., Prakash L., 1993. DNA repair genes and proteins of Saccharomyces cerevisiae. Annu. Rev. Genet. 27: 33–70. [DOI] [PubMed] [Google Scholar]
- Prolla T. A., Christie D.-M., Liskay R. M., 1994a Dual requirement in yeast DNA mismatch repair for MLH1 and PMS1, two homologs of the bacterial mutL gene. Mol. Cell. Biol. 14: 407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prolla T. A., Pang Q., Alani E., Kolodner R. D., Liskay R. M., 1994b MLH1, PMS1, and MSH2 interactions during the initiation of DNA mismatch repair in yeast. Science 265: 1091–1093. [DOI] [PubMed] [Google Scholar]
- Pryor J. M., Washington M. T., 2011. Pre-steady state kinetic studies show that an abasic site is a cognate lesion for the yeast Rev1 protein. DNA Repair (Amst.) 10: 1138–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramotar D., Popoff S. C., Gralla E. B., Demple B., 1991. Cellular role of yeast Apn1 apurinic endonuclease/3′-diesterase: repair of oxidative and alkylation DNA damage and control of spontaneous mutation. Mol. Cell. Biol. 11: 4537–4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramotar D., Kim C., Lillis R., Demple B., 1993. Intracellular localization of the Apn1 DNA repair enzyme of Saccharomyces cerevisiae. Nuclear transport signals and biological role. J. Biol. Chem. 268: 20533–20539. [PubMed] [Google Scholar]
- Ramsey K. L., Smith J. J., Dasgupta A., Maqani N., Grant P., et al. , 2004. The NEF4 complex regulates Rad4 levels and utilizes Snf2/Swi2-related ATPase activity for nucleotide excision repair. Mol. Cell. Biol. 24: 6362–6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranish J. A., Hahn S., Lu Y., Yi E. C., Li X. J., et al. , 2004. Identification of TFB5, a new component of general transcription and DNA repair factor IIH. Nat. Genet. 36: 707–713. [DOI] [PubMed] [Google Scholar]
- Reagan M. S., Pittenger C., Siede W., Friedberg E. C., 1995. Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J. Bacteriol. 177: 364–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed S. H., 2011. Nucleotide excision repair in chromatin: damage removal at the drop of a HAT. DNA Repair (Amst.) 10: 734–742. [DOI] [PubMed] [Google Scholar]
- Reed S. H., You Z., Friedberg E. C., 1998. The yeast RAD7 and RAD16 genes are required for postincision events during nucleotide excision repair. In vitro and in vivo studies with rad7 and rad16 mutants and purification of a Rad7/Rad16-containing protein complex. J. Biol. Chem. 273: 29481–29488. [DOI] [PubMed] [Google Scholar]
- Reed S. H., Akiyama M., Stillman B., Friedberg E. C., 1999. Yeast autonomously replicating sequence binding factor is involved in nucleotide excision repair. Genes Dev. 13: 3052–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reenan R. G., Kolodner R. D., 1992a Isolation and characterization of two Saccharomyces cerevisiae genes encoding homologs of the bacterial HexA and MutS mismatch repair proteins. Genetics 132: 963–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reenan R. G., Kolodner R. D., 1992b Characterization of insertion mutations in the Saccharomyces cerevisiae MSH1 and MSH2 genes: evidence for separate mitochondrial and nuclear functions. Genetics 132: 975–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid J., Svejstrup J. Q., 2004. DNA damage-induced Def1-RNA polymerase II interaction and Def1 requirement for polymerase ubiquitylation in vitro. J. Biol. Chem. 279: 29875–29878. [DOI] [PubMed] [Google Scholar]
- Reijns M. A., Rabe B., Rigby R. E., Mill P., Astell K. R., et al. , 2012. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell 149: 1008–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick M. A., 1969. A photoreactivationless mutant of Saccharomyces cerevisiae. Photochem. Photobiol. 9: 307–312. [DOI] [PubMed] [Google Scholar]
- Rodriguez G. P., Romanova N. V., Bao G., Rouf N. C., Kow Y. W., et al. , 2012. Mismatch repair-dependent mutagenesis in nondividing cells. Proc. Natl. Acad. Sci. USA 109: 6153–6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez K., Wang Z., Friedberg E. C., Tomkinson A. E., 1996. Identification of functional domains within the RAD1•RAD10 repair and recombination endonuclease of Saccharomyces cerevisiae. J. Biol. Chem. 271: 20551–20558. [DOI] [PubMed] [Google Scholar]
- Rodriguez K., Talamantez J., Huang W., Reed S. H., Wang Z., et al. , 1998. Affinity purification and partial characterization of a yeast multiprotein complex for nucleotide excision repair using histidine-tagged Rad14 protein. J. Biol. Chem. 273: 34180–34199. [DOI] [PubMed] [Google Scholar]
- Rong L., Pallandino F., Aguilera A., Klein H. L., 1991. The hyper-gene conversion hpr5–1 mutation in Saccharomyces cerevisiae is an allele of the SRS2/RADH gene. Genetics 127: 75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-MacDonald P., Roeder G. S., 1994. Mutation of a meiosis-specific MutS homolog decreases crossing over but not mismatch correction. Cell 79: 1069–1080. [DOI] [PubMed] [Google Scholar]
- Roush A. A., Suarez M., Friedberg E. C., Radman M., Siede W., 1998. Deletion of the Saccharomyces cerevisiae gene RAD30 encoding an Escherichia coli DinB homolog confers UV radiation sensitivity and altered mutability. Mol. Gen. Genet. 257: 686–692. [DOI] [PubMed] [Google Scholar]
- Rydberg B., Game J., 2002. Excision of misincorporated ribonucleotides in DNA by RNase H (type 2) and FEN-1 in cell-free extracts. Proc. Natl. Acad. Sci. USA 99: 16654–16659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbioneda S., Minesinger B. K., Giannattasio M., Plevani P., Muzi-Falconi M., et al. , 2005. The 9–1-1 checkpoint clamp physically interacts with Polζ and is partially required for spontaneous Polζ-dependent mutagenesis in Saccharomyces cerevisiae. J. Biol. Chem. 280: 38657–38665. [DOI] [PubMed] [Google Scholar]
- Sabbioneda S., Bortolomai I., Giannattasio M., Plevani P., Muzi-Falconi M., 2007. Yeast Rev1 is cell cycle regulated, phosphorylated in response to DNA damage and its binding to chromosomes is dependent upon MEC1. DNA Repair (Amst.) 6: 121–127. [DOI] [PubMed] [Google Scholar]
- Sacho E. J., Kadyrov F. A., Modrich P., Kunkel T. A., Erie D. A., 2008. Direct visualization of asymmetric adenine-nucleotide-induced conformational changes in MutLα. Mol. Cell 29: 112–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., 2008. Structure and function of photolyase and in vivo enzymology: 50th anniversary. J. Biol. Chem. 283: 32153–32157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar G. B., 1985. Sequence of the Saccharomyces cerevisiae PHR1 gene and homology of the PHR1 photolyase to E. coli photolyase. Nucleic Acids Res. 13: 8231–8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saparbaev M., Laval J., 1994. Excision of hypoxanthine from DNA containing dIMP residues by the Escherichia coli, yeast, rat, and human alkylpurine DNA glycosylases. Proc. Natl. Acad. Sci. USA 91: 5873–5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S., Davies A. A., Ulrich H. D., McHugh P. J., 2006. DNA interstrand crosslink repair during G1 involves nucleotide excision repair and DNA polymerase ζ. EMBO J. 25: 128512–128594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S., Kiely R., McHugh P. J., 2010. The Ino80 chromatin-remodeling complex restores chromatin structure during UV DNA damage repair. J. Cell Biol. 191: 1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassanfar M., Samson L., 1990. Identification and preliminary characterization of an O6-methylguanine DNA repair methyltransferase in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 265: 20–25. [PubMed] [Google Scholar]
- Sassanfar M., Dosanjh M. K., Essigmann J. M., Samson L., 1991. Relative efficiencies of the bacterial, yeast, and human DNA methyltransferases for the repair of O6-methylguanine and O4-methylthymine. Suggestive evidence for O4-methylthymine repair by eukaryotic methyltransferases. J. Biol. Chem. 266: 2767–2771. [PubMed] [Google Scholar]
- Schiestl R. H., Prakash S., Prakash L., 1990. The SRS2 suppressor of rad6 mutations of Saccharomyces cerevisiae acts by channeling DNA lesions into the RAD52 DNA repair pathway. Genetics 124: 817–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild D., Johnston J., Chang C., Mortimer R. K., 1984. Cloning and mapping of Saccharomyces cerevisiae photoreactivation gene PHR1. Mol. Cell. Biol. 4: 1864–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A. D., Neishabury M., Jones D. H., Reed S. H., Boiteux S., et al. , 1999. Spontaneous mutation, oxidative DNA damage, and the roles of base and nucleotide excision repair in the yeast Saccharomyces cerevisiae. Yeast 15: 205–218. [DOI] [PubMed] [Google Scholar]
- Sebastian J., Kraus B., Sancar G. B., 1990. Expression of the yeast PHR1 gene is induced by DNA-damaging agents. Mol. Cell. Biol. 10: 4630–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senturker S., Auffret Van Der Kemp P., You H. J., Doetsch P. W., Dizdaroglu M., et al. , 1998. Substrate specificities of the Ntg1 and Ntg2 proteins of Saccharomyces cerevisiae for oxidized DNA bases are not identical. Nucleic Acids Res. 26: 5270–5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shell S. S., Putnam C. D., Kolodner R. D., 2007a The N terminus of Saccharomyces cerevisiae Msh6 is an unstructured tether to PCNA. Mol. Cell 26: 565–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shell S. S., Putnam C. D., Kolodner R. D., 2007b Chimeric Saccharomyces cerevisiae Msh6 protein with an Msh3 mispair-binding domain combines properties of both proteins. Proc. Natl. Acad. Sci. USA 104: 10956–10961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia E. A., Kirkpatrick D. T., 2005. The yeast MSH1 gene is not involved in DNA repair or recombination during meiosis. DNA Repair (Amst.) 4: 253–261. [DOI] [PubMed] [Google Scholar]
- Sia E. A., Kokoska R. J., Dominska M., Greenwell P., Petes T. D., 1997. Microsatellite instability in yeast: dependence on repeat unit size and DNA mismatch repair genes. Mol. Cell. Biol. 17: 2851–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia E. A., Dominska M., Stefanovic L., Petes T. D., 2001. Isolation and characterization of point mutations in mismatch repair genes that destabilize microsatellites in yeast. Mol. Cell. Biol. 21: 8157–8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K. K., Sigala B., Sikder H. A., Schwimmer C., 2001. Inactivation of Saccharomyces cerevisiae OGG1 DNA repair gene leads to an increased frequency of mitochondrial mutants. Nucleic Acids Res. 29: 1381–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden T., Acharya S., Butz C., Berardini M., Fishel R., 2004. hMSH4-hMSH5 recognizes Holliday Junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol. Cell 15: 437–451. [DOI] [PubMed] [Google Scholar]
- Snowden T., Shim K. S., Schmutte C., Acharya S., Fishel R., 2008. hMSH4-hMSH5 adenosine nucleotide processing and interactions with homologous recombination machinery. J. Biol. Chem. 283: 145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolsky T., Alani E., 2000. EXO1 and MSH6 are high-copy suppressors of conditional mutations in the MSH2 mismatch repair gene of Saccharomyces cerevisiae. Genetics 155: 589–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer D., Stith C. M., Burgers P. M., Lahue R. S., 2008. Partial reconstitution of DNA large loop repair with purified proteins from Saccharomyces cerevisiae. Nucleic Acids Res. 36: 4699–4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa M. M., Krokan H. E., Slupphaug G., 2007. DNA-uracil and human pathology. Mol. Aspects Med. 28: 276–306. [DOI] [PubMed] [Google Scholar]
- Sparks J. L., Chon H., Cerritelli S. M., Kunkel T. A., Johansson E., et al. , 2012. RNase H2-initiated ribonucleotide excision repair. Mol. Cell 47: 980–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staresincic L., Fagbemi A. F., Enzlin J. H., Gourdin A. M., Wijgers N., et al. , 2009. Coordination of dual incision and repair synthesis in human nucleotide excision repair. EMBO J. 28: 1111–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehling O., Vashisht A. A., Mascarenhas J., Jonsson Z. O., Sharma T., et al. , 2012. MMS19 assembles iron-sulfur proteins required for DNA metabolism and genomic integrity. Science 337: 195–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelter P., Ulrich H. D., 2003. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425: 188–191. [DOI] [PubMed] [Google Scholar]
- Stone J. E., Kissling G. E., Lujan S. A., Rogozin I. B., Stith C. M., et al. , 2009. Low-fidelity DNA synthesis by the L979F mutator derivative of Saccharomyces cerevisiae DNA polymerase ζ. Nucleic Acids Res. 37: 3774–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone J. E., Kumar D., Binz S. K., Inase A., Iwai S., et al. , 2011. Lesion bypass by S. cerevisiae Polζ alone. DNA Repair (Amst.) 10: 826–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand M., Prolla T. A., Liskay R. M., Petes T. D., 1993. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature 365: 274–276. [DOI] [PubMed] [Google Scholar]
- Strand M., Earley M. C., Crouse G. F., Petes T. D., 1995. Mutations in the MSH3 gene preferentially lead to deletions within tracts of simple repetitive DNA in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 92: 10418–10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studamire B., Quach T., Alani E., 1998. Saccharomyces cerevisiae Msh2p and Msh6p ATPase activities are both required during mismatch repair. Mol. Cell. Biol. 18: 7590–7601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studamire B., Price G., Sugawara N., Haber J. E., Alani E., 1999. Separation-of-function mutations in Saccharomyces cerevisiae MSH2 that confer mismatch repair defects but do not affect nonhomologous-tail removal during recombination. Mol. Cell. Biol. 19: 7558–7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa K., Okamoto T., Shimizu Y., Masutani C., Iwai S., et al. , 2001. A multistep damage recognition mechanism for global genomic nucleotide excision repair. Genes Dev. 15: 507–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa K., Okuda Y., Saijo M., Nishi R., Matsuda N., et al. , 2005. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell 121: 387–400. [DOI] [PubMed] [Google Scholar]
- Sugawara N., Paques F., Colaiacovo M., Haber J. E., 1997. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proc. Natl. Acad. Sci. USA 94: 9214–9219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukhanova M. V., d’Herin C., Van Der Kemp P. A., Koval V. V., Boiteux S., et al. , 2011. Ddc1 checkpoint protein and DNA polymerase varepsilon interact with nick-containing DNA repair intermediate in cell free extracts of Saccharomyces cerevisiae. DNA Repair (Amst.) 10: 815–825. [DOI] [PubMed] [Google Scholar]
- Sung P., Prakash L., Matson S. W., Prakash S., 1987. RAD3 protein of Saccharomyces cerevisiae is a DNA helicase. Proc. Natl. Acad. Sci. USA 84: 8951–8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P., Guzder S. N., Prakash L., Prakash S., 1996. Reconstitution of TFIIH and requirement of its DNA helicase subunits, Rad3 and Rad25, in the incision step of nucleotide excision repair. J. Biol. Chem. 271: 10821–10826. [DOI] [PubMed] [Google Scholar]
- Svejstrup J. Q., Wang Z., Feaver W. J., Wu X., Bushnell D. A., et al. , 1995. Different forms of TFIIH for transcription and DNA repair: holo-TFIIH and a nucleotide excision repairosome. Cell 80: 21–28. [DOI] [PubMed] [Google Scholar]
- Swanson R. L., Morey N. J., Doetsch P. W., Jinks-Robertson S., 1999. Overlapping specificities of base excision repair, nucleotide excision repair, recombination, and translesion synthesis pathways for DNA base damage in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 2929–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweder K. S., Hanawalt P. C., 1992. Preferential repair of cyclobutane pyrimidine dimers in the transcribed strand of a gene in yeast chromosomes and plasmids is dependent on transcription. Proc. Natl. Acad. Sci. USA 89: 10696–10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenberg J. A., Lu K., Moeller B. C., Gao L., Upton P. B., et al. , 2011. Endogenous vs. exogenous DNA adducts: their role in carcinogenesis, epidemiology, and risk assessment. Toxicol. Sci. 120(Suppl. 1): S130–S145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szankasi P., Smith G. R., 1995. A role for exonuclease I from S. pombe in mutation avoidance and mismatch correction. Science 267: 1166–1169. [DOI] [PubMed] [Google Scholar]
- Tapias A., Auriol J., Forget D., Enzlin J. H., Scharer O. D., et al. , 2004. Ordered conformational changes in damaged DNA induced by nucleotide excision repair factors. J. Biol. Chem. 279: 19074–19083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner M., Harreman M., Teng Y., Gill H., Anindya R., et al. , 2010. A role for checkpoint kinase-dependent Rad26 phosphorylation in transcription-coupled DNA repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 30: 436–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng Y., Liu H., Gill H. W., Yu Y., Waters R., et al. , 2008. Saccharomyces cerevisiae Rad16 mediates ultraviolet-dependent histone H3 acetylation required for efficient global genome nucleotide-excision repair. EMBO Rep. 9: 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terleth C., Schenk P., Poot R., Brouwer J., Van De Putte P., 1990. Differential repair of UV damage in rad mutants of Saccharomyces cerevisiae: a possible function of G2 arrest upon UV irradiation. Mol. Cell. Biol. 10: 4678–4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D., Scot A. D., Barbey R., Padula M., Boiteux S., 1997. Inactivation of OGG1 increases the incidence of G•C→T•A transversions in Saccharomyces cerevisiae: evidence for endogenous oxidative damage to DNA in eukaryotic cells. Mol. Gen. Genet. 254: 171–178. [DOI] [PubMed] [Google Scholar]
- Tishkoff D. X., Boerger A. L., Bertrand P., Filosi N., Gaida G. M., et al. , 1997a Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc. Natl. Acad. Sci. USA 94: 7487–7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff D. X., Filosi N., Gaida G. M., Kolodner R. D., 1997b A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell 88: 253–263. [DOI] [PubMed] [Google Scholar]
- Tomkinson A. E., Tappe N. J., Friedberg E. C., 1992. DNA ligase I from Saccharomyces cerevisiae: physical and biochemical characterization of the CDC9 gene product. Biochemistry 31: 11762–11771. [DOI] [PubMed] [Google Scholar]
- Tomkinson A. E., Bardwell A. J., Bardwell L., Tappe N. J., Friedberg E. C., 1993. Yeast DNA repair and recombination proteins Rad1 and Rad10 constitute a single-stranded-DNA endonuclease. Nature 362: 860–862. [DOI] [PubMed] [Google Scholar]
- Tomkinson A. E., Bardwell A. J., Tappe N., Ramos W., Friedberg E. C., 1994. Purification of Rad1 protein from Saccharomyces cerevisiae and further characterization of the Rad1/Rad10 endonuclease complex. Biochemistry 33: 5305–5311. [DOI] [PubMed] [Google Scholar]
- Torres-Ramos C. A., Johnson R. E., Prakash L., Prakash S., 2000. Evidence for the involvement of nucleotide excision repair in the removal of abasic sites in yeast. Mol. Cell. Biol. 20: 3522–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Ramos C. A., Prakash S., Prakash L., 2002. Requirement of RAD5 and MMS2 for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 22: 2419–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H. T., Gordenin D. A., Resnick M. A., 1999. The 3′-5′ exonucleases of DNA polymerases δ and ε and the 5′-3′ exonuclease Exo1 have major roles in postreplication mutation avoidance in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 2000–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran P. T., Liskay M., 2000. Functional studies on the candidate ATPase domains of Saccharomyces cerevisiae MutLα. Mol. Cell. Biol. 20: 6390–6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran P. T., Simon J. A., Liskay R. M., 2001. Interactions of Exo1p with components of MutLα in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 98: 9760–9765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran P. T., Erdeniz N., Dudley S., Liskay R. M., 2002. Characterization of nuclease-dependent functions of Exo1p in Saccharomyces cerevisiae. DNA Repair (Amst.) 1: 895–912. [DOI] [PubMed] [Google Scholar]
- Tran P. T., Erdeniz N., Symington L. S., Liskay R. M., 2004. EXO1 - A multi-tasking eukaryotic nuclease. DNA Repair (Amst.) 3: 1549–1559. [DOI] [PubMed] [Google Scholar]
- Tran P. T., Fey J. P., Erdeniz N., Gellon L., Boiteux S., et al. , 2007. A mutation in EXO1 defines separable roles in DNA mismatch repair and post-replication repair. DNA Repair (Amst.) 6: 1572–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay M., Teng Y., Paquette M., Waters R., Conconi A., 2008. Complementary roles of yeast Rad4p and Rad34p in nucleotide excision repair of active and inactive rRNA gene chromatin. Mol. Cell. Biol. 28: 7504–7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trincao J., Johnson R. E., Escalante C. R., Prakash S., Prakash L., et al. , 2001. Structure of the catalytic core of S. cerevisiae DNA polymerase η: implications for translesion DNA synthesis. Mol. Cell 8: 417–426. [DOI] [PubMed] [Google Scholar]
- Troelstra C., Van Gool A., De Wit J., Vermeulen W., Bootsma D., et al. , 1992. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne’s syndrome and preferential repair of active genes. Cell 71: 939–953. [DOI] [PubMed] [Google Scholar]
- Tsutakawa S. E., Van Wynsberghe A. W., Freudenthal B. D., Weinacht C. P., Gakhar L., et al. , 2011. Solution X-ray scattering combined with computational modeling reveals multiple conformations of covalently bound ubiquitin on PCNA. Proc. Natl. Acad. Sci. USA 108: 17672–17677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich H. D., Jentsch S., 2000. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 19: 3388–3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich H. D., Walden H., 2010. Ubiquitin signalling in DNA replication and repair. Nat. Rev. Mol. Cell Biol. 11: 479–489. [DOI] [PubMed] [Google Scholar]
- Umar A., Buermeyer A. B., Simon J. A., Thomas D. C., Clark A. B., et al. , 1996. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell 87: 65–73. [DOI] [PubMed] [Google Scholar]
- Unk I., Haracska L., Johnson R. E., Prakash S., Prakash L., 2000. Apurinic endonuclease activity of yeast Apn2 protein. J. Biol. Chem. 275: 22427–22434. [DOI] [PubMed] [Google Scholar]
- Vance J. R., Wilson T. E., 2001a Repair of DNA strand breaks by the overlapping functions of lesion-specific and non-lesion-specific DNA 3′ phosphatases. Mol. Cell. Biol. 21: 7191–7198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance J. R., Wilson T. E., 2001b Uncoupling of 3′-phosphatase and 5′-kinase functions in budding yeast. Characterization of Saccharomyces cerevisiae DNA 3′-phosphatase (TPP1). J. Biol. Chem. 276: 15073–15081. [DOI] [PubMed] [Google Scholar]
- Vance J. R., Wilson T. E., 2002. Yeast Tdp1 and Rad1-Rad10 function as redundant pathways for repairing Top1 replicative damage. Proc. Natl. Acad. Sci. USA 99: 13669–13674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Kemp P. A., Thomas D., Barbey R., De Oliveira R., Boiteux S., 1996. Cloning and expression in Escherichia coli of the OGG1 gene of Saccharomyces cerevisiae, which codes for a DNA glycosylase that excises 7,8-dihydro-8-oxoguanine and 2,6-diamino-4-hydroxy-5-N-methylformamidopyrimidine. Proc. Natl. Acad. Sci. USA 93: 5197–5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Kemp P. A., De Padula M., Burguiere-Slezak G., Ulrich H. D., Boiteux S., 2009. PCNA monoubiquitylation and DNA polymerase η ubiquitin-binding domain are required to prevent 8-oxoguanine-induced mutagenesis in Saccharomyces cerevisiae. Nucleic Acids Res. 37: 2549–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gool A. J., Verhage R., Swagemakers S. M., Van De Putte P., Brouwer J., et al. , 1994. RAD26, the functional S. cerevisiae homolog of the Cockayne syndrome B gene ERCC6. EMBO J. 13: 5361–5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veaute X., Jeusset J., Soustelle C., Kowalczykowski S. C., Le Cam E., et al. , 2003. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423: 309–312. [DOI] [PubMed] [Google Scholar]
- Venema J., Van Hoffen A., Karcagi V., Natarajan A. T., Van Zeeland A. A., et al. , 1991. Xeroderma pigmentosum complementation group C cells remove pyrimidine dimers selectively from the transcribed strand of active genes. Mol. Cell. Biol. 11: 4128–4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhage R., Zeeman A. M., De Groot N., Gleig F., Bang D. D., et al. , 1994. The RAD7 and RAD16 genes, which are essential for pyrimidine dimer removal from the silent mating type loci, are also required for repair of the nontranscribed strand of an active gene in Saccharomyces cerevisiae. Mol. Cell. Biol. 14: 6135–6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhage R. A., Van Gool A. J., De Groot N., Hoeijmakers J. H., Van De Putte P., et al. , 1996. Double mutants of Saccharomyces cerevisiae with alterations in global genome and transcription-coupled repair. Mol. Cell. Biol. 16: 496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volker M., Mone M. J., Karmakar P., Van Hoffen A., Schul W., et al. , 2001. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell 8: 213–224. [DOI] [PubMed] [Google Scholar]
- Vongsamphanh R., Wagner J. R., Ramotar D., 2006. Saccharomyces cerevisiae Ogg1 prevents poly(GT) tract instability in the mitochondrial genome. DNA Repair (Amst.) 5: 235–242. [DOI] [PubMed] [Google Scholar]
- Wang H., Yang Y., Schofield M. J., Du C., Fridman Y., et al. , 2003. DNA bending and unbending by MutS govern mismatch recognition and specificity. Proc. Natl. Acad. Sci. USA 100: 14822–14827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T.-F., Kleckner N., Hunter N., 1999. Functional specificity of MutL homologs in yeast: evidence for three Mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proc. Natl. Acad. Sci. USA 96: 13914–13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wu X., Friedberg E. C., 1993. DNA repair synthesis during base excision repair in vitro is catalyzed by DNA polymerase ε and is influenced by DNA polymerases α and δ in Saccharomyces cerevisiae. Mol. Cell. Biol. 13: 1051–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren J. J., Pohlhaus T. J., Changela A., Iyer R. R., Modrich P. L., et al. , 2007. Structure of the human MutSα DNA lesion recognition complex. Mol. Cell 26: 579–592. [DOI] [PubMed] [Google Scholar]
- Washington M. T., Johnson R. E., Prakash S., Prakash L., 2001. Mismatch extension ability of yeast and human DNA polymerase η. J. Biol. Chem. 276: 2263–2266. [DOI] [PubMed] [Google Scholar]
- Washington M. T., Helquist S. A., Kool E. T., Prakash L., Prakash S., 2003. Requirement of Watson-Crick hydrogen bonding for DNA synthesis by yeast DNA polymerase η. Mol. Cell. Biol. 23: 5107–5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington M. T., Minko I. G., Johnson R. E., Haracska L., Harris T. M., et al. , 2004. Efficient and error-free replication past a minor-groove N2-guanine adduct by the sequential action of yeast Rev1 and DNA polymerase ζ. Mol. Cell. Biol. 24: 6900–6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters L. S., Walker G. C., 2006. The critical mutagenic translesion DNA polymerase Rev1 is highly expressed during G(2)/M phase rather than S phase. Proc. Natl. Acad. Sci. USA 103: 8971–8976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters L. S., Minesinger B. K., Wiltrout M. E., D’Souza S., Woodruff R. V., et al. , 2009. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol. Mol. Biol. Rev. 73: 134–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters R., Teng Y., Yu Y., Yu S., Reed S. H., 2009. Tilting at windmills? The nucleotide excision repair of chromosomal DNA. DNA Repair (Amst.) 8: 146–152. [DOI] [PubMed] [Google Scholar]
- Williamson M. S., Game J. C., Fogel S., 1985. Meiotic gene conversion mutants in Saccharomyces cerevisiae. I. Isolation and characterization of pms1–1 and pms1–2. Genetics 110: 609–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltrout M. E., Walker G. C., 2011a The DNA polymerase activity of Saccharomyces cerevisiae Rev1 is biologically significant. Genetics 187: 21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltrout M. E., Walker G. C., 2011b Proteasomal regulation of the mutagenic translesion DNA polymerase, Saccharomyces cerevisiae Rev1. DNA Repair (Amst.) 10: 169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windecker H., Ulrich H. D., 2008. Architecture and assembly of poly-SUMO chains on PCNA in Saccharomyces cerevisiae. J. Mol. Biol. 376: 221–231. [DOI] [PubMed] [Google Scholar]
- Wood A., Krogan N. J., Dover J., Schneider J., Heidt J., et al. , 2003. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell 11: 267–274. [DOI] [PubMed] [Google Scholar]
- Wood A., Garg P., Burgers P. M., 2007. A ubiquitin-binding motif in the translesion DNA polymerase Rev1 mediates its essential functional interaction with ubiquitinated proliferating cell nuclear antigen in response to DNA damage. J. Biol. Chem. 282: 20256–20263. [DOI] [PubMed] [Google Scholar]
- Woudstra E. C., Gilbert C., Fellows J., Jansen L., Brouwer J., et al. , 2002. A Rad26-Def1 complex coordinates repair and RNA pol II proteolysis in response to DNA damage. Nature 415: 929–933. [DOI] [PubMed] [Google Scholar]
- Wu X., Wang Z., 1999. Relationships between yeast Rad27 and Apn1 in response to apurinic/apyrimidinic (AP) sites in DNA. Nucleic Acids Res. 27: 956–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W., Samson L., 1992. The Saccharomyces cerevisiae MGT1 DNA repair methyltransferase gene: its promoter and entire coding sequence, regulation and in vivo biological functions. Nucleic Acids Res. 20: 3599–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W., Derfler B., Chen J., Samson L., 1991. Primary sequence and biological functions of a Saccharomyces cerevisiae O6-methylguanine/O4-methylthymine DNA repair methyltransferase gene. EMBO J. 10: 2179–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W., Chow B. L., Fontanie T., Ma L., Bacchetti S., et al. , 1999. Genetic interactions between error-prone and error-free postreplication repair pathways in Saccharomyces cerevisiae. Mutat. Res. 435: 1–11. [DOI] [PubMed] [Google Scholar]
- Xiao W., Chow B. L., Broomfield S., Hanna M., 2000. The Saccharomyces cerevisiae RAD6 group is composed of an error-prone and two error-free postreplicational repair pathways. Genetics 155: 1633–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Liu S., Zhang Y., Wang Z., 2004. Roles of Rad23 protein in yeast nucleotide excision repair. Nucleic Acids Res. 32: 5981–5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Karthikeyan R., Mack S. E., Vonarx E. J., Kunz B. A., 1999. Analysis of yeast pms1, msh2, and mlh1 mutators points to differences in mismatch correction efficiencies between prokaryotic and eukaryotic cells. Mol. Gen. Genet. 261: 777–787. [DOI] [PubMed] [Google Scholar]
- You H. J., Swanson R. L., Doetsch P. W., 1998. Saccharomyces cerevisiae possesses two functional homologues of Escherichia coli endonuclease III. Biochemistry 37: 6033–6040. [DOI] [PubMed] [Google Scholar]
- You H. J., Swanson R. L., Harrington C., Corbett A. H., Jinks-Robertson S., et al. , 1999. Saccharomyces cerevisiae Ntg1p and Ntg2p: broad specificity N-glycosylases for the repair of oxidative DNA damage in the nucleus and mitochondria. Biochemistry 38: 11298–11306. [DOI] [PubMed] [Google Scholar]
- Yu S.-L., Johnson R. E., Prakash S., Prakash L., 2001. Requirement of DNA polymerase η for error-free bypass of UV-induced CC and TC photoproducts. Mol. Cell. Biol. 21: 185–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S. L., Lee S. K., Johnson R. E., Prakash L., Prakash S., 2003. The stalling of transcription at abasic sites is highly mutagenic. Mol. Cell. Biol. 23: 382–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Owen-Hughes T., Friedberg E. C., Waters R., Reed S. H., 2004. The yeast Rad7/Rad16/Abf1 complex generates superhelical torsion in DNA that is required for nucleotide excision repair. DNA Repair (Amst.) 3: 277–287. [DOI] [PubMed] [Google Scholar]
- Yu S., Smirnova J. B., Friedberg E. C., Stillman B., Akiyama M., et al. , 2009. ABF1-binding sites promote efficient global genome nucleotide excision repair. J. Biol. Chem. 284: 966–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharyevich K., Tang S., Ma Y., Hunter N., 2012. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell 149: 334–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Lawrence C. W., 2005. The error-free component of the RAD6/RAD18 DNA damage tolerance pathway of budding yeast employs sister-strand recombination. Proc. Natl. Acad. Sci. USA 102: 15954–15959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Siede W., 2002. UV-induced T→C transition at a TT photoproduct site is dependent on Saccharomyces cerevisiae polymerase η in vivo. Nucleic Acids Res. 30: 1262–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Chatterjee A., Singh K. K., 2006. Saccharomyces cerevisiae polymerase ζ functions in mitochondria. Genetics 172: 2683–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Wang J., Geacintov N. E., Wang Z., 2006. Polη, Polζ and Rev1 together are required for G to T transversion mutations induced by the (+)- and (−)-trans-anti-BPDE-N2-dG DNA adducts in yeast cells. Nucleic Acids Res. 34: 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X., Garg P., Stith C. M., Nick McElhinny S. A., Kissling G. E., et al. , 2006. The fidelity of DNA synthesis by yeast DNA polymerase ζ alone and with accessory proteins. Nucleic Acids Res. 34: 4731–4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Kou H., Wang Z., 2007. Tfb5 interacts with Tfb2 and facilitates nucleotide excision repair in yeast. Nucleic Acids Res. 35: 861–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Wang J., Zhang Y., Wang Z., 2010. The catalytic function of the Rev1 dCMP transferase is required in a lesion-specific manner for translesion synthesis and base damage-induced mutagenesis. Nucleic Acids Res. 38: 5036–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Z., Johnson R. E., Haracska L., Prakash L., Prakash S., et al. , 2008. Regulation of polymerase exchange between Polη and Polδ by monoubiquitination of PCNA and the movement of DNA polymerase holoenzyme. Proc. Natl. Acad. Sci. USA 105: 5361–5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann C., Chymkowitch P., Eldholm V., Putnam C. D., Lindvall J. M., et al. , 2011. A chemical-genetic screen to unravel the genetic network of CDC28/CDK1 links ubiquitin and Rad6-Bre1 to cell cycle progression. Proc. Natl. Acad. Sci. USA 108: 18748–18753. [DOI] [PMC free article] [PubMed] [Google Scholar]