Abstract
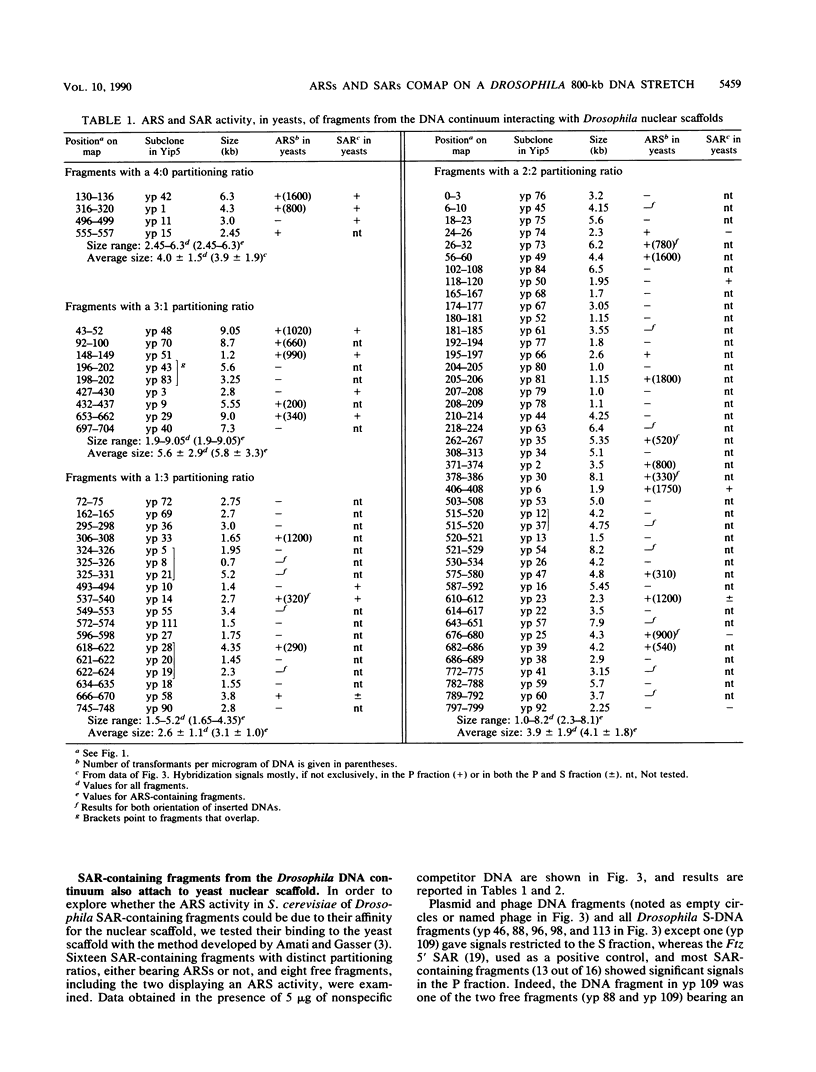
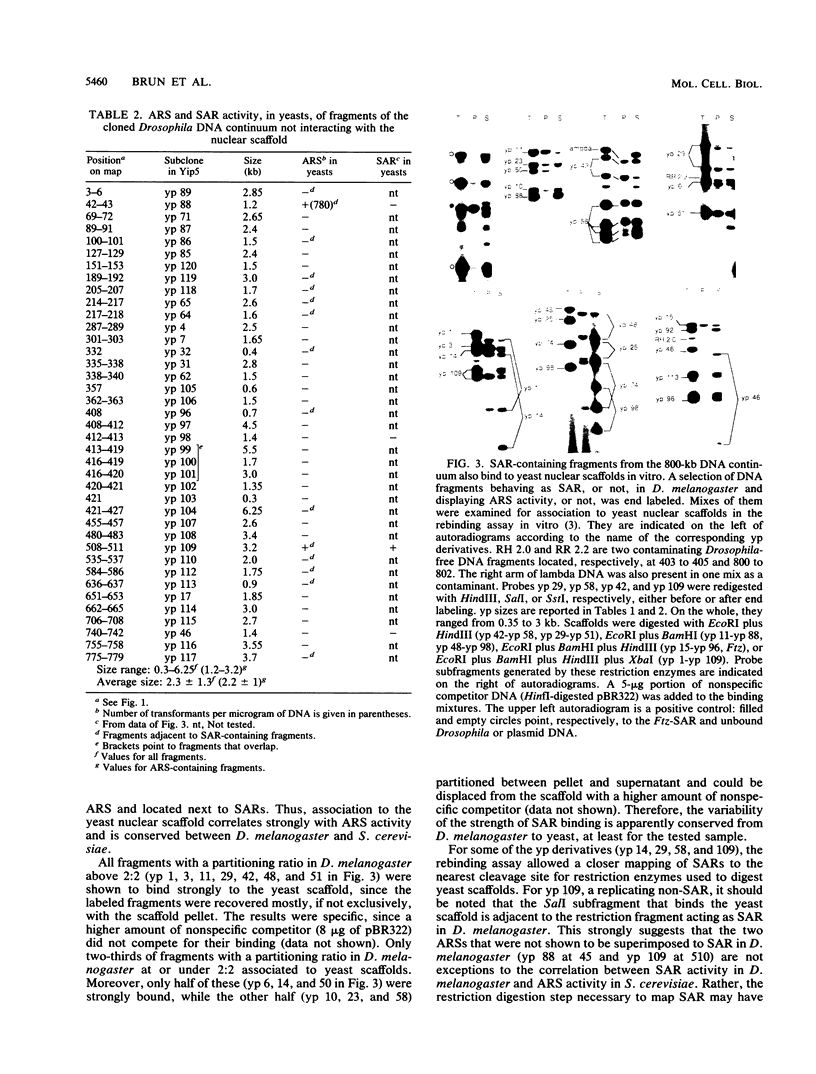
We have previously mapped scaffold-attached regions (SARs) on an 800-kilobase DNA walk from the Drosophila X chromosome. We have also previously shown that the strength of binding, i.e., the ability of SARs to bind to all nuclear scaffolds or only to a fraction of them varied from one SAR to another one. In the present study, 71 of the 85 subfragments that bind scaffolds and 38 fragments that do not bind scaffolds were tested for their ability to promote autonomous replicating sequence (ARS) activity in Saccharomyces cerevisiae. Sixteen SAR-containing fragments from the chromosome walk were also examined for association to yeast nuclear scaffolds in vitro. All identified ARSs (a total of 27) were present on SAR-containing fragments, except two, which were adjacent to SARs. There is thus a correlation between ARS and SAR activities, and this correlation defines a SAR subclass. Moreover, the presence of an ARS on a DNA fragment appeared to be highly correlated with the strength of binding. The binding activity was highly conserved from Drosophila melanogaster to yeast. These data suggest that Drosophila DNA sequences responsible for binding to components of the nuclear scaffold from either D. melanogaster or yeast may be involved in the process of heterologous extrachromosomal replication in yeasts.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adachi Y., Käs E., Laemmli U. K. Preferential, cooperative binding of DNA topoisomerase II to scaffold-associated regions. EMBO J. 1989 Dec 20;8(13):3997–4006. doi: 10.1002/j.1460-2075.1989.tb08582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguinaga M. P., Kiper C. E., Valenzuela M. S. Enriched autonomously replicating sequences in a nuclear matrix-DNA complex isolated from synchronized HeLa cells. Biochem Biophys Res Commun. 1987 Apr 29;144(2):1018–1024. doi: 10.1016/s0006-291x(87)80065-7. [DOI] [PubMed] [Google Scholar]
- Amati B. B., Gasser S. M. Chromosomal ARS and CEN elements bind specifically to the yeast nuclear scaffold. Cell. 1988 Sep 23;54(7):967–978. doi: 10.1016/0092-8674(88)90111-0. [DOI] [PubMed] [Google Scholar]
- Amati B., Gasser S. M. Drosophila scaffold-attached regions bind nuclear scaffolds and can function as ARS elements in both budding and fission yeasts. Mol Cell Biol. 1990 Oct;10(10):5442–5454. doi: 10.1128/mcb.10.10.5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benyajati C., Worcel A. Isolation, characterization, and structure of the folded interphase genome of Drosophila melanogaster. Cell. 1976 Nov;9(3):393–407. doi: 10.1016/0092-8674(76)90084-2. [DOI] [PubMed] [Google Scholar]
- Blumenthal A. B., Kriegstein H. J., Hogness D. S. The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harb Symp Quant Biol. 1974;38:205–223. doi: 10.1101/sqb.1974.038.01.024. [DOI] [PubMed] [Google Scholar]
- Bode J., Maass K. Chromatin domain surrounding the human interferon-beta gene as defined by scaffold-attached regions. Biochemistry. 1988 Jun 28;27(13):4706–4711. doi: 10.1021/bi00413a019. [DOI] [PubMed] [Google Scholar]
- Brewer B. J., Fangman W. L. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987 Nov 6;51(3):463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- Brown N. H., King D. L., Wilcox M., Kafatos F. C. Developmentally regulated alternative splicing of Drosophila integrin PS2 alpha transcripts. Cell. 1989 Oct 6;59(1):185–195. doi: 10.1016/0092-8674(89)90880-5. [DOI] [PubMed] [Google Scholar]
- Buongiorno-Nardelli M., Micheli G., Carri M. T., Marilley M. A relationship between replicon size and supercoiled loop domains in the eukaryotic genome. Nature. 1982 Jul 1;298(5869):100–102. doi: 10.1038/298100a0. [DOI] [PubMed] [Google Scholar]
- Citri Y., Colot H. V., Jacquier A. C., Yu Q., Hall J. C., Baltimore D., Rosbash M. A family of unusually spliced biologically active transcripts encoded by a Drosophila clock gene. Nature. 1987 Mar 5;326(6108):42–47. doi: 10.1038/326042a0. [DOI] [PubMed] [Google Scholar]
- Cockerill P. N., Garrard W. T. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986 Jan 31;44(2):273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- Cockerill P. N., Garrard W. T. Chromosomal loop anchorage sites appear to be evolutionarily conserved. FEBS Lett. 1986 Aug 11;204(1):5–7. doi: 10.1016/0014-5793(86)81377-1. [DOI] [PubMed] [Google Scholar]
- Cook P. R., Brazell I. A. Supercoils in human DNA. J Cell Sci. 1975 Nov;19(2):261–279. doi: 10.1242/jcs.19.2.261. [DOI] [PubMed] [Google Scholar]
- Dijkwel P. A., Hamlin J. L. Matrix attachment regions are positioned near replication initiation sites, genes, and an interamplicon junction in the amplified dihydrofolate reductase domain of Chinese hamster ovary cells. Mol Cell Biol. 1988 Dec;8(12):5398–5409. doi: 10.1128/mcb.8.12.5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingman C. W. Bidirectional chromosome replication: some topological considerations. J Theor Biol. 1974 Jan;43(1):187–195. doi: 10.1016/s0022-5193(74)80052-4. [DOI] [PubMed] [Google Scholar]
- Freund J. N., Zerges W., Schedl P., Jarry B. P., Vergis W. Molecular organization of the rudimentary gene of Drosophila melanogaster. J Mol Biol. 1986 May 5;189(1):25–36. doi: 10.1016/0022-2836(86)90378-5. [DOI] [PubMed] [Google Scholar]
- Gasser S. M., Laemmli U. K. Cohabitation of scaffold binding regions with upstream/enhancer elements of three developmentally regulated genes of D. melanogaster. Cell. 1986 Aug 15;46(4):521–530. doi: 10.1016/0092-8674(86)90877-9. [DOI] [PubMed] [Google Scholar]
- Gasser S. M., Laemmli U. K. The organisation of chromatin loops: characterization of a scaffold attachment site. EMBO J. 1986 Mar;5(3):511–518. doi: 10.1002/j.1460-2075.1986.tb04240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves D. R., Wilson F. D., Lang G., Kioussis D. Human CD2 3'-flanking sequences confer high-level, T cell-specific, position-independent gene expression in transgenic mice. Cell. 1989 Mar 24;56(6):979–986. doi: 10.1016/0092-8674(89)90631-4. [DOI] [PubMed] [Google Scholar]
- Handeli S., Klar A., Meuth M., Cedar H. Mapping replication units in animal cells. Cell. 1989 Jun 16;57(6):909–920. doi: 10.1016/0092-8674(89)90329-2. [DOI] [PubMed] [Google Scholar]
- Hiromi Y., Kuroiwa A., Gehring W. J. Control elements of the Drosophila segmentation gene fushi tarazu. Cell. 1985 Dec;43(3 Pt 2):603–613. doi: 10.1016/0092-8674(85)90232-6. [DOI] [PubMed] [Google Scholar]
- Huberman J. A., Spotila L. D., Nawotka K. A., el-Assouli S. M., Davis L. R. The in vivo replication origin of the yeast 2 microns plasmid. Cell. 1987 Nov 6;51(3):473–481. doi: 10.1016/0092-8674(87)90643-x. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E., Mirkovitch J., Laemmli U. K. Interaction of DNA with nuclear scaffolds in vitro. J Mol Biol. 1988 Mar 5;200(1):111–125. doi: 10.1016/0022-2836(88)90337-3. [DOI] [PubMed] [Google Scholar]
- Jarman A. P., Higgs D. R. Nuclear scaffold attachment sites in the human globin gene complexes. EMBO J. 1988 Nov;7(11):3337–3344. doi: 10.1002/j.1460-2075.1988.tb03205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan P. J., Haase S. B., Calos M. P. Isolation of human sequences that replicate autonomously in human cells. Mol Cell Biol. 1989 Mar;9(3):1026–1033. doi: 10.1128/mcb.9.3.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käs E., Chasin L. A. Anchorage of the Chinese hamster dihydrofolate reductase gene to the nuclear scaffold occurs in an intragenic region. J Mol Biol. 1987 Dec 20;198(4):677–692. doi: 10.1016/0022-2836(87)90209-9. [DOI] [PubMed] [Google Scholar]
- Loc P. V., Strätling W. H. The matrix attachment regions of the chicken lysozyme gene co-map with the boundaries of the chromatin domain. EMBO J. 1988 Mar;7(3):655–664. doi: 10.1002/j.1460-2075.1988.tb02860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughney K., Kreber R., Ganetzky B. Molecular analysis of the para locus, a sodium channel gene in Drosophila. Cell. 1989 Sep 22;58(6):1143–1154. doi: 10.1016/0092-8674(89)90512-6. [DOI] [PubMed] [Google Scholar]
- Marilley M., Gassend-Bonnet G. Supercoiled loop organization of genomic DNA: a close relationship between loop domains, expression units, and replicon organization in rDNA from Xenopus laevis. Exp Cell Res. 1989 Feb;180(2):475–489. doi: 10.1016/0014-4827(89)90074-8. [DOI] [PubMed] [Google Scholar]
- McNabb S. L., Beckendorf S. K. Cis-acting sequences which regulate expression of the Sgs-4 glue protein gene of Drosophila. EMBO J. 1986 Sep;5(9):2331–2340. doi: 10.1002/j.1460-2075.1986.tb04501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkovitch J., Gasser S. M., Laemmli U. K. Scaffold attachment of DNA loops in metaphase chromosomes. J Mol Biol. 1988 Mar 5;200(1):101–109. doi: 10.1016/0022-2836(88)90336-1. [DOI] [PubMed] [Google Scholar]
- Mirkovitch J., Mirault M. E., Laemmli U. K. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell. 1984 Nov;39(1):223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- Pardoll D. M., Vogelstein B., Coffey D. S. A fixed site of DNA replication in eucaryotic cells. Cell. 1980 Feb;19(2):527–536. doi: 10.1016/0092-8674(80)90527-9. [DOI] [PubMed] [Google Scholar]
- Paulson J. R., Laemmli U. K. The structure of histone-depleted metaphase chromosomes. Cell. 1977 Nov;12(3):817–828. doi: 10.1016/0092-8674(77)90280-x. [DOI] [PubMed] [Google Scholar]
- Razin S. V. DNA interactions with the nuclear matrix and spatial organization of replication and transcription. Bioessays. 1987 Jan;6(1):19–23. doi: 10.1002/bies.950060106. [DOI] [PubMed] [Google Scholar]
- Razin S. V., Kekelidze M. G., Lukanidin E. M., Scherrer K., Georgiev G. P. Replication origins are attached to the nuclear skeleton. Nucleic Acids Res. 1986 Oct 24;14(20):8189–8207. doi: 10.1093/nar/14.20.8189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stief A., Winter D. M., Strätling W. H., Sippel A. E. A nuclear DNA attachment element mediates elevated and position-independent gene activity. Nature. 1989 Sep 28;341(6240):343–345. doi: 10.1038/341343a0. [DOI] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surdej P., Got C., Miassod R. Developmental expression pattern of a 800 kb DNA continuum cloned from the Drosophila X chromosome 14B-15B region. Biol Cell. 1990;68(2):105–118. doi: 10.1016/0248-4900(90)90295-e. [DOI] [PubMed] [Google Scholar]
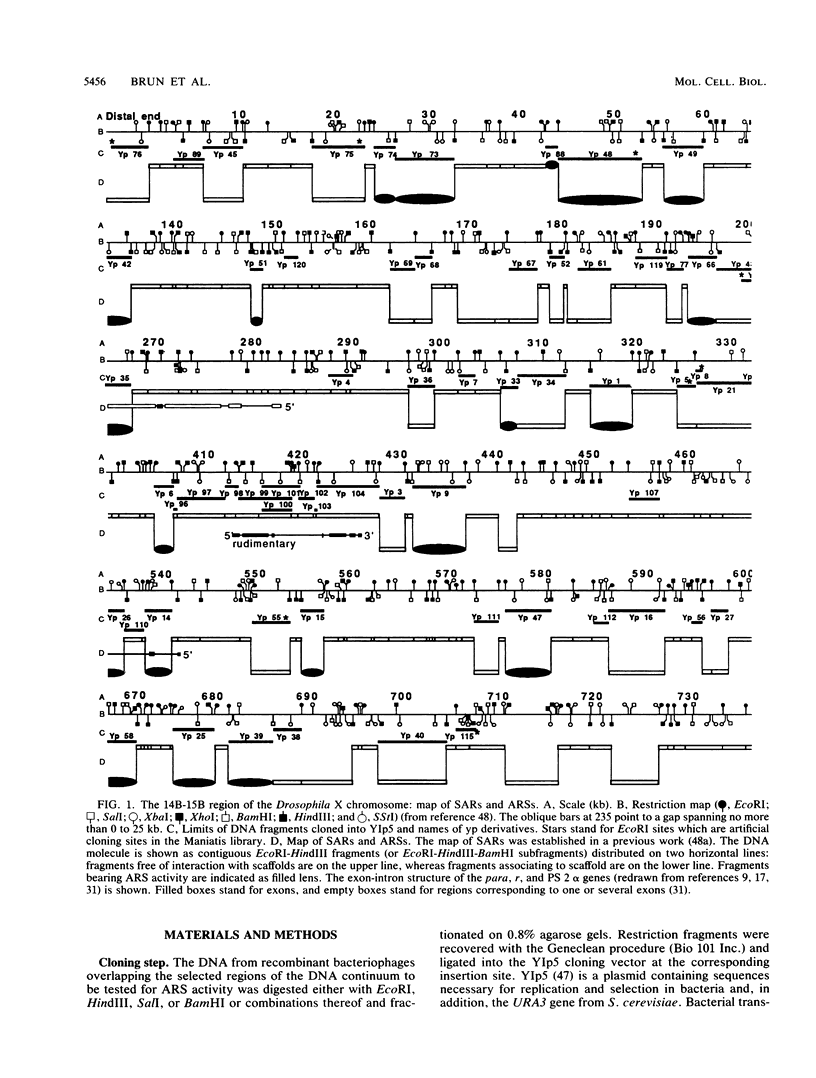
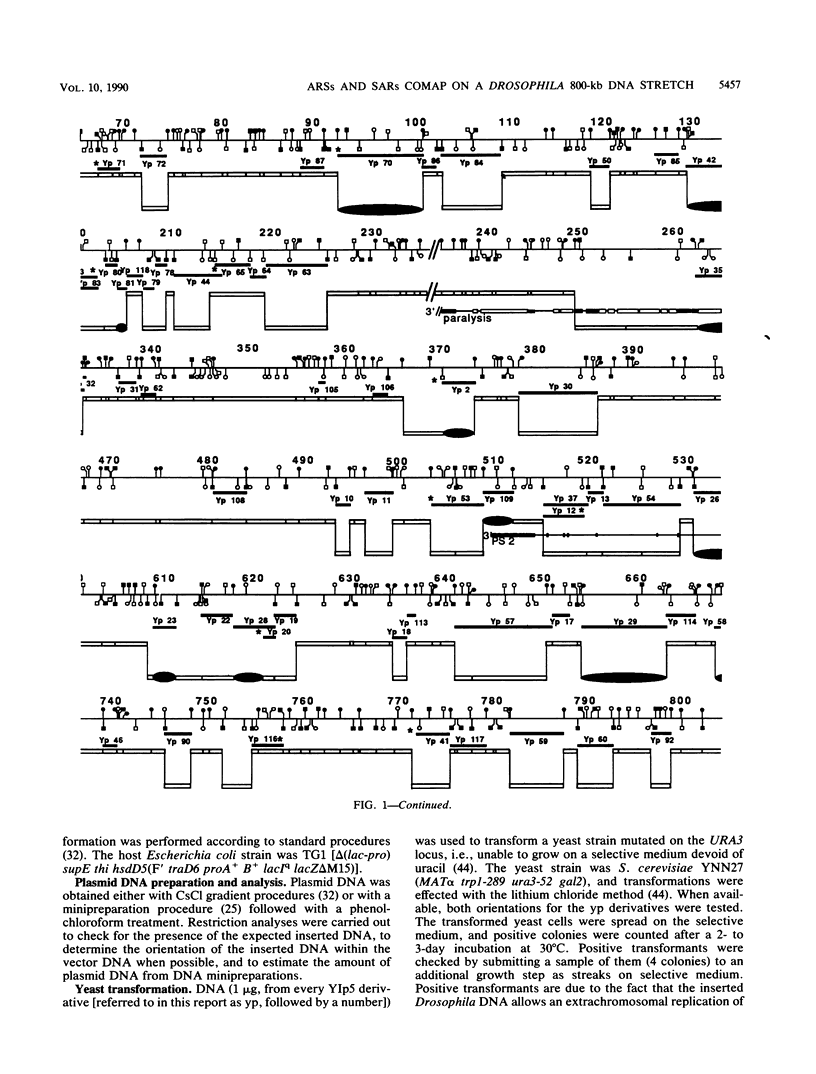
- Surdej P., Got C., Rosset R., Miassod R. Supragenic loop organization: mapping in Drosophila embryos, of scaffold-associated regions on a 800 kilobase DNA continuum cloned from the 14B-15B first chromosome region. Nucleic Acids Res. 1990 Jul 11;18(13):3713–3722. doi: 10.1093/nar/18.13.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes R. C., Lin D., Hwang S. J., Framson P. E., Chinault A. C. Yeast ARS function and nuclear matrix association coincide in a short sequence from the human HPRT locus. Mol Gen Genet. 1988 May;212(2):301–309. doi: 10.1007/BF00334700. [DOI] [PubMed] [Google Scholar]
- Umek R. M., Linskens M. H., Kowalski D., Huberman J. A. New beginnings in studies of eukaryotic DNA replication origins. Biochim Biophys Acta. 1989 Jan 23;1007(1):1–14. doi: 10.1016/0167-4781(89)90123-1. [DOI] [PubMed] [Google Scholar]
- Vogelstein B., Pardoll D. M., Coffey D. S. Supercoiled loops and eucaryotic DNA replicaton. Cell. 1980 Nov;22(1 Pt 1):79–85. doi: 10.1016/0092-8674(80)90156-7. [DOI] [PubMed] [Google Scholar]
- van der Velden H. M., van Willigen G., Wetzels R. H., Wanka F. Attachment of origins of replication to the nuclear matrix and the chromosomal scaffold. FEBS Lett. 1984 Jun 4;171(1):13–16. doi: 10.1016/0014-5793(84)80451-2. [DOI] [PubMed] [Google Scholar]