Abstract
Helicobacter pylori is a Gram-negative bacterium that colonizes the human stomach and contributes to peptic ulceration and gastric adenocarcinoma. H. pylori secretes a pore-forming exotoxin known as vacuolating toxin (VacA). VacA contains two distinct domains, designated p33 and p55, and assembles into large “snowflake”-shaped oligomers. Thus far, no structural data are available for the p33 domain, which is essential for membrane channel formation. Using single-particle electron microscopy and the random conical tilt approach, we have determined the three-dimensional structures of six VacA oligomeric conformations at ~15-Å resolution. The p55 domain, composed primarily of β-helical structures, localizes to the peripheral arms, while the p33 domain consists of two globular densities that localize within the center of the complexes. By fitting the VacA p55 crystal structure into the electron microscopy densities, we have mapped inter-VacA interactions that support oligomerization. In addition, we have examined VacA variants/mutants that differ from wild-type (WT) VacA in toxin activity and/or oligomeric structural features. Oligomers formed by VacAΔ6–27, a mutant that fails to form membrane channels, lack an organized p33 central core. Mixed oligomers containing both WT and VacAΔ6–27 subunits also lack an organized core. Oligomers formed by a VacA s2m1 chimera (which lacks cell-vacuolating activity) and VacAΔ301–328 (which retains vacuolating activity) each contain p33 central cores similar to those of WT oligomers. By providing the most detailed view of the VacA structure to date, these data offer new insights into the toxin's channel-forming component and the intermolecular interactions that underlie oligomeric assembly.
Keywords: pore-forming toxins, single-particle electron microscopy, oligomerization, pathogenesis
Introduction
Helicobacter pylori is a Gram-negative bacterium that colonizes the human stomach1–3 and can lead to the development of peptic ulcer disease, gastric adenocarcinoma, or gastric lymphoma.4–6 Gastric cancer is the second leading cause of cancer-related deaths worldwide,7 and H. pylori is classified as a type 1 carcinogen by the World Health Organization.
The major secreted exotoxin of H. pylori is known as VacA (vacuolating toxin), named for its ability to induce vacuolation in the cytoplasm of mammalian cells (reviewed in Refs. 8–12). Additionally, VacA causes a slew of cellular responses that include depolarization of membrane potential,13 mitochondrial dysfunction,14–17 autophagy,18 cell death,17,19,20 activation of mitogen-activated protein kinases,21 and inhibition of T cells.22–24 The mechanisms by which these alterations occur are not fully understood, but many require VacA internalization into cells.15,19,25 VacA forms anion-selective channels in planar lipid bilayers and in the plasma membranes of cells,13,26–28 and it is hypothesized that VacA can also form channels in endosomal and mitochondrial membranes (reviewed in Ref. 10).
VacA, secreted as an 88-kDa toxin, consists of two domains, designated p33 and p55. The p33 domain is postulated to form the anion-selective channel, and both the p33 and p55 domains mediate VacA binding to host cells.10,16,29–32 A 2.4-Å three-dimensional (3D) structure of the p55 domain was determined by X-ray crystallography,33 but there is no structural information on the p33 domain.
VacA assembles into a heterogeneous assortment of oligomeric “snowflake” structures, which have been visualized by several approaches.26,31,32,34–38 Mutant VacA proteins that fail to oligomerize lack toxin activity.39–41 The snowflake structures likely represent water-soluble forms of the anion-selective membrane channels formed by VacA. The highest-resolution structures of VacA oligomers thus far (~ 19-Å) were obtained by cryo-negative stain electron microscopy (EM).31 In this previous structural analysis,31 it was not possible to obtain tilt pair images, and therefore the 3D models relied on a number of modeling techniques to generate structures from the heterogeneous images of VacA oligomers. Analysis of the X-ray crystallographic structure of p55 together with EM images of the oligomers has led to a model of VacA oligomer organization in which the p33 domain occupies the inner core of the snowflakes and the p55 domain extends outward from the central core.33 However, the cryo-negative stain EM structure contained no details about p33 organization and did not allow an analysis of how VacA monomers interact to form oligomers.
Sequence variations within VacA influence cytotoxicity and are associated with distinct pathological responses.42,43 Three polymorphic regions have been identified:44 an N-terminal region that is differentially cleaved during toxin export to form either an s1 or an s2 variant,42 an intermediate region (i1 or i2) located at the C-terminus of the p33 domain,43 and a middle region (m1 or m2) in the p55 domain that affects receptor binding.42,45 Type s1m1 and s1m2 VacA variants are considered fully active, although the s1m2 variant binds to a narrower range of cells.45,46 Type s2 VacA variants contain a 12-residue hydrophilic N-terminal extension, do not vacuolate cells, and form membrane channels less efficiently than s1 variants;42,46–48 moreover, s2 forms of VacA are associated with a very low risk of symptomatic gastric disease.42 The mechanism by which these additional residues disrupt VacA activity is not well understood.
Analysis of experimentally constructed VacA mutant proteins has identified specific VacA domains and/or residues required for pore formation and oligomerization. However, the limited structural understanding of how VacA oligomerizes, coupled with the lack of structural information on p33, has made it difficult to elucidate the exact roles of these domains or residues in VacA function. One mutant, VacAΔ6–27, lacks three hydrophobic GXXXG motifs posited to be important for pore formation29 and exhibits a dominant-negative phenotype when mixed with equal molar ratios of acid-activated wild-type (WT) VacA.39 Although this mutant has previously been structurally characterized,31 the low resolution and lack of structural detail in the central p33 region of the 3D maps made it difficult to mechanistically explain how the Δ6–27 mutation disrupts function, and the structural consequences of VacAΔ6–27 interaction with WT VacA were not explored. Another mutant, containing a deletion between the p33/p55 domains of VacA, retains cell-vacuolating activity49,50 and was reported to assemble into predominantly six-sided oligomeric structures instead of seven-sided oligomers.49
VacA does not exhibit homology to other known bacterial toxins; therefore, there is considerable interest in elucidating the structure of this protein and understanding the mechanism of membrane pore formation. Although the toxicity of VacA lies in its ability to form channels, a high-resolution 3D structure of oligomeric VacA has yet to be determined, creating a large gap in our understanding of this toxin and its role in H. pylori pathogenesis. In this study, we report the determination of ~15-Å 3D reconstructions of six oligomeric forms of VacA. These structures provide the first details of p33 organization in the central pore and allow us to generate a structure-based model for how VacA oligomerizes. We have also used EM analysis to characterize VacA variants and mutants that differ from WT VacA in activity or oligomeric structure, providing structural insights into the mechanism by which a dominant-negative mutant inhibits the activity of WT VacA and the mechanisms by which VacA oligomerizes and forms membrane channels.
Results and Discussion
Visualization of VacA oligomers by negative stain EM
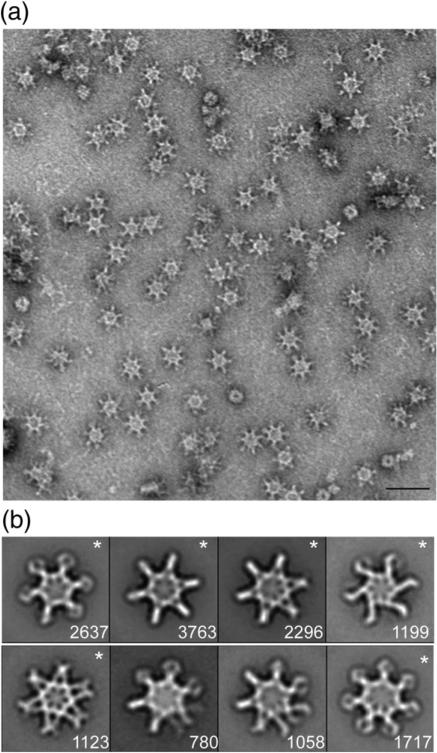
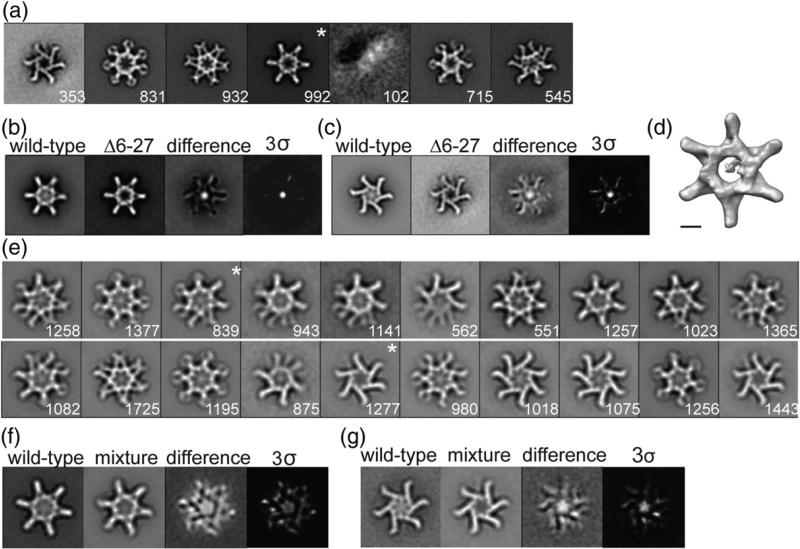
WT VacA was purified from H. pylori culture supernatant and, as expected, acid-activated preparations caused vacuolation of epithelial cells (data not shown). Negative stain EM analysis of purified WT VacA revealed snowflake-like particles (Fig. 1a), corresponding to morphologies observed in other studies.26,31,34–36,38 In contrast to the random orientation of VacA in previously described cryo-negative stain images,31 VacA oligomers adsorbed in a preferred en face orientation on the grids. If the specimen adsorbs on the carbon film in one or a few preferred orientations, the specimen must be tilted in the electron microscope to obtain the additional views required for 3D reconstruction. Using this technique, referred to as the random conical tilt (RCT) approach,51 we calculated the 3D volume by back-projection algorithms. Since the same specimen area is imaged twice in this technique, once at a high tilt angle and then again untilted, the orientation parameters are uniquely defined and the resulting 3D reconstruction is very reliable. This approach is especially useful when examining samples, such as VacA, that exist in more than one structural state since we are able to classify the distinct conformations using the untilted images and then use the tilted images to determine the structure. Image pairs of grids containing negatively stained VacA were recorded at tilt angles of 55° and 0°. A total of 14,573 pairs of particles were selected, and images of the untilted specimens were classified into 20 class averages (Fig. S1a). These class averages showed that while essentially all VacA particles adsorbed to the carbon grid in the same orientation, the oligomer adopts multiple conformations. As seen previously,31 the classes revealed VacA oligomers that contained six chiral arms (Fig. S1a, panels 7 and 8) and oligomers that contained six or seven arms arranged in an achiral fashion. In contrast to previous cryo-negative stain studies,31,38 we did not observe particles with eight or nine arms. Furthermore, all the averages, even the chiral forms, had a visible “spoke”-like density in the center of the oligomers. From the 20 classes, we selected 7 that represent distinct conformations and 1 less well-resolved VacA class (Fig. S1a, marked with an “*”). These were used as references for a cycle of reference-based alignment (Fig. 1b). The results of the reference-based alignment revealed six well-resolved classes (Fig. 1b, marked with an “*”) and two classes that contained particles with disordered arms. Five of the well-resolved classes contained either six or seven achiral arms (Fig. 1b, panels 1, 2, 3, 5, and 8), while one class showed an oligomer with six chiral arms (Fig. 1b, panel 4). As seen in the reference-free alignment, all the reference-based classes had visible central densities that appear structurally organized.
Fig. 1.
Characterization of negatively stained VacA oligomers. (a) Raw image of VacA particles in negative stain. The scale bar represents 50 nm. (b) VacA class averages obtained by reference-based alignment and classification. “*” marks classes that yielded 3D structures (see Fig. 2). The number of particles included in each class is shown in the bottom right corner. Side length of panels, 420 Å.
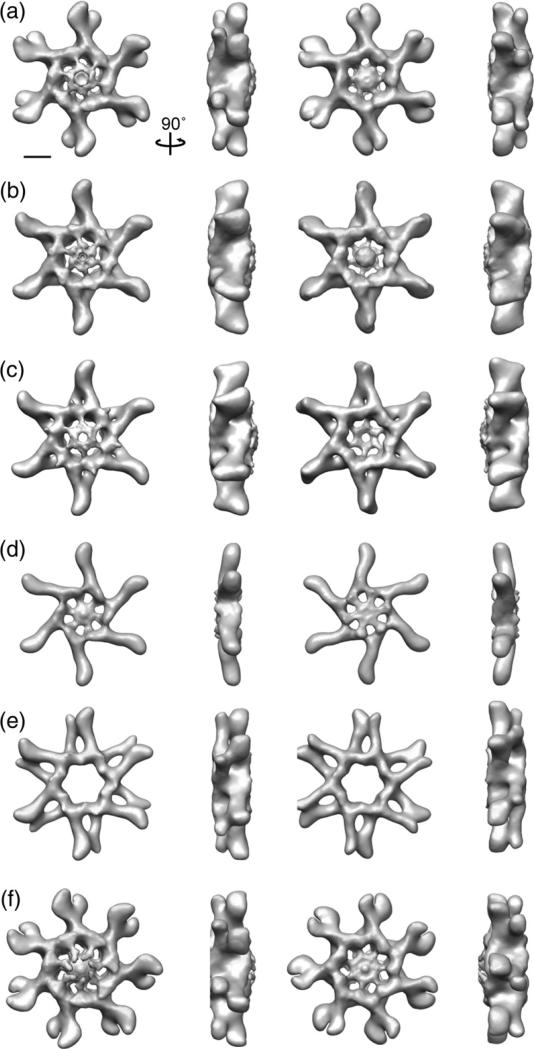
To more carefully analyze the structural organization of the VacA oligomers, we used images from the tilted specimens corresponding to each of the eight classes to calculate 3D reconstructions using RCT52 (Fig. 2). The 3D structures with 6- or 7-fold applied symmetry are shown in Fig. 2, while the same structures without applied symmetry are shown in Fig. S2. Of the eight 3D reconstructions, only tilted particles associated with the six well-resolved class averages (Fig. 1b, marked with an “*”) led to well-defined 3D structures. Structures calculated from particles found in two of the classes looked very similar (Fig. 2b and c) and could easily be aligned. To further improve the reconstruction of a VacA oligomer, we combined the tilted images plus 10% of the highest correlated 0° images from these two classes and used them to calculate a 3D reconstruction either with or without applied 6-fold symmetry (Fig. 3 and Fig. S1c).
Fig. 2.
3D reconstruction of negatively stained VacA oligomers using the RCT approach. (a–f) VacA organizes into a number of oligomeric conformations that include both hexamers, dodecamers, and tetradecamers. These structures have applied symmetry. Structures are rotated along the vertical axis by 90°. The scale bar represents 5 nm.
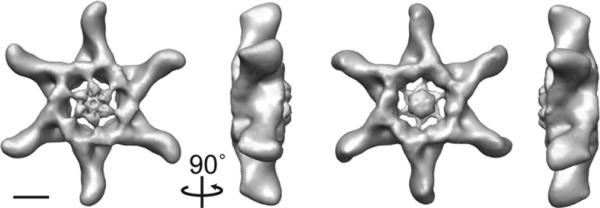
Fig. 3.
Approximately 15-Å 3D reconstruction of VacA dodecamer. The structure contains two prominent features: extended straight “arms” with a slight kink at the distal end and a central spoke-like density composed of two distinct globular domains separated by a thinner connecting density. The structure is rotated around the vertical axis by 90°. The scale bar represents 5 nm.
From the presented structures, it is clear that VacA monomers can oligomerize into achiral double-layer structures containing either six or seven arms (Figs. 2a–c, e, and f and 3) or into a chiral hexameric structure (Fig. 2d). Our statistics show that approximately 8% of the oligomers represent hexamers, 79% represent dodecamers, and 11% represent tetradecamers (Table 1). Interestingly, VacA heptamers, although seen previously under negative stain,32 cryo-negative stain,31 and vitrified ice conditions (data not shown), were not observed in our analysis, even when we expanded the number of classes to 60 (data not shown). One possible explanation stems from our observation that the preferred conformation of VacA oligomers in solution shifts over time towards hexameric, dodecameric, and tetradecameric forms (data not shown). The structures, regardless of oligomeric type, contain two prominent features: extended straight arms with a slight kink at the distal end and central spoke-like densities. The arms clearly represent the VacA p55 domain,33 leaving the central region to represent the p33 domain. The double-layer VacA oligomers contain a well-resolved central region on one side of the structure (Figs. 2a–c and f and 33), although there is central density present on both sides when no symmetry is applied (Fig. S2). Additionally, some of the oligomers appear to contain a “plug” that occludes the central hole (Fig. 2 and Fig. S2). It is possible that the less ordered face is the one that makes contact with the carbon grid and that the presence of a plug represents either unorganized central density or stain accumulation. The exception to this observation is the structure of one VacA dodecamer, which does not contain any well-resolved central density at the molecular weight threshold corresponding to its calculated mass (1056 kDa), regardless of whether or not symmetry is applied (Fig. 2e and Fig. S2e). The six-sided oligomers have a diameter of 290 Å. The extended arms are ~95 Å long, while the central region has a diameter of 100 Å (Figs. 2a–e and 3). The central regions for all the oligomers, with the exception of the dodecamer shown in Fig. 2e, contain two domains separated by a thinner connecting density (Figs. 3 and 4a and c). The innermost, central domain has dimensions of ~15 Å×15 Å×15 Å, while the domain that extends seamlessly from the arm has dimensions of ~30 Å×25 Å×20 Å (Figs. 2 and 3). The tetradecamer is slightly larger than the six-sided oligomers, with an overall diameter of 310 Å and a central region with a diameter of 120 Å (Fig. 2f); however, the size of the arms and central densities are similar to those in the hexameric and dodecameric oligomers.
Table 1.
Comparison of VacA oligomers formed by variants and mutants
| VacA | Hexamers (%) | Dodecamers (%) | Heptamers (%) | Tetradecamers (%) | Disordered (%) | 6-fold (%) | 7-fold (%) | Ratio |
|---|---|---|---|---|---|---|---|---|
| WT | 8 | 79 | — | 11 | 2 | 87 | 11 | 7.9 |
| Δ6–27 | 8 | 59 | — | 18 | 15 | 67 | 18 | 3.7 |
| s2m1 | 17 | 58 | 8 | 11 | 6 | 75 | 17 | 4.4 |
| Δ301–328 | 4 | 84 | — | 5 | 7 | 88 | 5 | 17.6 |
Ratio=%6-fold/%7-fold; “—” not found; “disordered” refers to averages that are difficult to determine whether they represent hexamers, dodecamers, or tetradecamers.
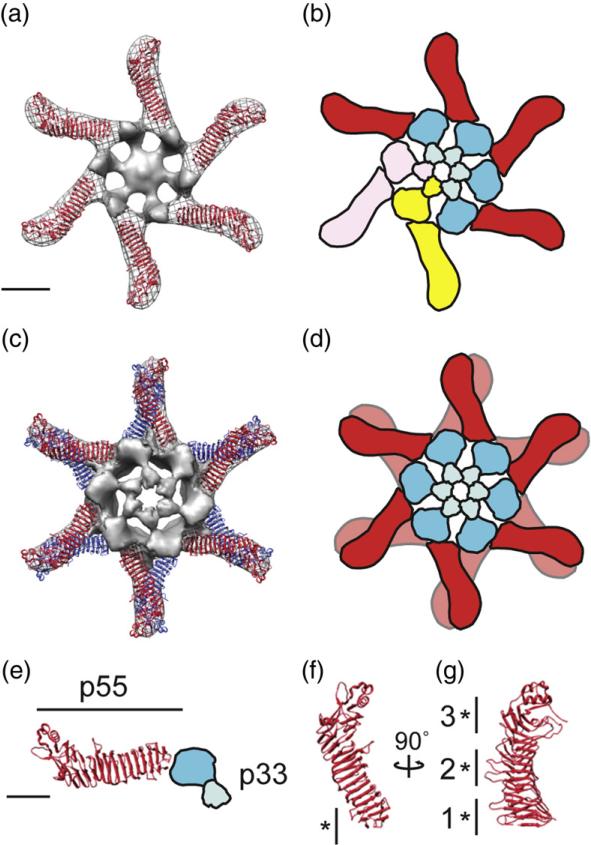
Fig. 4.
Structural model of the VacA oligomerization process. (a) The 2.4-Å crystal structure of p5533 fits into the straight “arms” of the EM map of a VacA hexamer. Subtracting the density of the p55 crystal structure from the EM map highlights p33 (gray, central density and spokes). (b) p88 oligomerizes into hexamers supported by intermolecular interactions between the N-terminal portions of p33 in adjacent protomers, as well as contacts between p33 and an adjacent p55 arm. Blue domains, p33; red domain, p55. Two p88 protomers are colored pink and yellow to show p88 protomer interactions. (c) The 2.4-Å crystal structure of p5533 fits into the straight “arms” of the EM map of a VacA dodecamer. For ease in viewing the model, the p33 density of only the well-organized side is shown. (d) Cartoon of dodecamer formation. Colors are the same as in (b). (a–d) The scale bar represents 5 nm. (e) The C-terminal p55 domain forms a straight arm with a kink at the end, while the N-terminal p33 domain consists of two globular densities connected by a thinner density (blue domains). (f and g) p55 crystal structure (2.4 Å) rotated 90° on the vertical axis. “*” marks regions of p55 involved in (f) hexamer interactions (residues 442–448) and (g) dodecamer and tetradecamer interactions. “1”, residues 395–404 and 421–435; “2”, residues 519–530 and 547–559; and “3”, residues 645–654 and 687–692. (e–g) The scale bar represents 2.5 nm.
VacA structure and oligomerization
To gain insight into how VacA oligomerizes, we placed the 2.4-Å crystal structure of the VacA p55 domain33 into the 3D density maps of the VacA oligomers using the program Chimera53 (Fig. 4a and c and Fig. S3). As is common for negatively stained specimens, VacA oligomers were slightly flattened, causing the tips of the peripheral arms to appear to be touching, a feature that is less pronounced in side views of VacA particles in vitrified ice (Ref. 31 and data not shown). Although the flattening in the z-axis limits our ability to map inter-p55 interactions in double-layered oligomers to specific loops rather than to specific amino acids, the distortion was fairly minor since the p55 structure unambiguously fits into the EM maps even in the double-layered VacA oligomers. Placing the p55 crystal structure into the VacA oligomers (Fig. 4 and Fig. S3) clearly shows that the extended arms represent the p55 domain and the central densities represent the p33 domain along with regions of p55 that have not been crystallized (residues 312–354) (Fig. 4e). As previously predicted,33 the portion of p33 contiguous to the p55 domain likely adopts an extended β-helix fold since there is no clear demarcation in the density map at the p55–p33 intramolecular interface (Fig. 4a, c, and e).
The model of hexameric VacA (Fig. 4a) shows that oligomerization depends on intermolecular interactions between the N-terminal portions of p33 in adjacent protomers, as well as contacts between p33 of one protomer and an adjacent p55 arm of another protomer (Fig. 4b, see p88 pink and yellow protomers). The interactions between the arms can be mapped to a region spanning residues 442–448 of p55 (Fig. 4f) and a region of p33 contiguous with the p55 β-helix in an adjacent arm. The lack of a high-resolution structure of the VacA N-terminus precludes mapping this interaction to specific residues; however, we speculate that this region likely includes residues 346 and 347, which are important in VacA oligomerization.41
VacA forms double layers through interactions mediated by the many loop regions extending from the p55 β-helix (Fig. 4c, d, and g). These loops can be grouped into three major blocks (Fig. 4g) and each oligomeric type uses a different block to support double-layer formation (Fig. 4c and d; Fig. S3). Although this allows for numerous VacA double-layer conformations, the different interactions, with the exception of one conformation (Fig. S3c), do not have any distinct effect on the arrangement of the central p33 domains. This suggests structural flexibility within p88 protomers, likely found at regions around the p33–p55 interface and within the short region connecting the two lobes of the p33 domain (Fig. 4e). The dodecamer lacking a well-defined central density uses regions in the C-terminus of p55 to support double-layer formation (Fig. S3c). Perhaps the angle the arms must adopt in this oligomer exceeds the inherent flexibility found in p88 disrupting ordered N-terminal p33 interactions. From the structural analysis of the numerous VacA oligomer conformations, we propose a model where soluble VacA first oligomerizes into hexamers or heptamers and then these single layers interact to form double-layer structures via structural motifs along the arm domains.
Structural characterization of VacA mutants and variants
A number of VacA variants and mutants have been shown to differ from WT toxin in activity and/or oligomeric structural features. For example, both VacAΔ6–27 and a VacA s2m1 chimera lack cell-vacuolating activity;39,48 VacAΔ301–328 retains vacuolating activity (data not shown), but a similar mutant preferentially formed particles with increased 6-fold symmetry rather than 7-fold symmetry.49 To gain a better mechanistic understanding for why certain VacA mutants and/or variants are more active than others, we examined VacAΔ6–27, VacA s2m1, and VacAΔ301–328 by negative stain EM, and particles from the untilted images were subjected to alignment and classification (Figs. 5 and 6). The classes were compared to WT VacA classes using difference mapping. In cases where significant differences were detected, 3D structures were determined. In addition, we calculated the number of hexameric, heptameric, dodecameric, and tetradecameric forms for these variants and/or mutants (Table 1). In general, WT, Δ6–27, s2m1, and Δ301–328 VacA all formed oligomers with 6-fold symmetry more frequently than 7-fold symmetry and more double-layer oligomers than single-layer oligomers (Table 1). Consistent with a previous report,49 the Δ301–328 mutation leads to an increased proportion of oligomers with 6-fold symmetry, but what was not previously appreciated is that these oligomers also contain 50% fewer single-layer oligomers when compared to WT VacA. Interestingly, s2m1, a VacA variant lacking cell-vacuolating activity, forms single-layer oligomers approximately three times more often than WT VacA (Table 1, 24% versus 8%) and six times more frequently than VacAΔ301–328 (Table 1, 24% versus 4%). These analyses suggest that whether VacA forms single- or double-layer oligomers in solution is not a key determinant of VacA activity.
Fig. 5.
Characterization of VacAΔ6–27. (a) VacAΔ6–27 class averages obtained by reference-based alignment and classification. “*”, class shown as a 3D volume in (d). (b and c) Difference mapping between WT and similar VacAΔ6–27 class averages. Final panel shown at 3 σ threshold. (a–c) Side length of panels, 573 Å. (d) 3D structure of VacAΔ6–27 corresponding to the “*” average in (a). The structure has no applied symmetry. The scale bar represents 5 nm. (e) Class averages of VacA oligomers generated by mixing WT VacA and VacAΔ6–27. “*”, classes used in difference mapping shown in (f) and (g). (f and g) Difference mapping between WT VacA and mixture of WT:VacAΔ6–27 oligomers. The final panel shown at 3 σ threshold. (e–g) Side length of panels, 420 Å. The number of particles included in each class is shown in the bottom right corner.
Fig. 6.
Reference-based alignment of VacAΔ301–328 and VacA s2m1. (a) VacAΔ301–328 class averages obtained by reference-based alignment and classification. (b) VacA s2m1 class averages obtained by reference-based alignment and classification. Side length of panels, 420 Å. The number of particles included in each class is shown in the bottom right corner.
As seen previously,31 VacAΔ6–27 double-layer oligomers clearly lack density in the p33 region (Fig. 5a–c and d). We now show that the central density is absent in both single- and double-layer oligomers (Fig. 5b and c). To carefully examine the structural consequences of the Δ6–27 mutation, we determined the 3D structure of VacAΔ6–27 using the RCT approach,52 with one representative structure shown in Fig. 5d. For all calculated structures, the central region of each oligomer was no longer structurally organized. We propose a model in which deletion of residues 6–27 leads to the disruption of important interactions between the N-termini of p33 domains in the oligomers, causing the N-terminal globular domains of p33 to no longer adopt an organized central core in any oligomeric form.
VacAΔ6–27 can inhibit WT VacA activity in a dominant-negative manner,39 but the mechanism is not completely understood. It has been proposed that this inhibitory activity arises from interaction of VacAΔ6–27 with WT VacA and formation of mixed oligomers with defective activity.39,48,54 To determine whether VacA oligomers that contain a mixture of WT and VacAΔ6–27 p88 subunits have a structurally organized central density, we mixed WT VacA and VacAΔ6–27 in a 1:1 molar ratio, acidified the preparation to pH 3.5 to promote oligomer disassembly, and then neutralized the pH to promote oligomer formation.39 These oligomers were analyzed by negative stain EM. A total of 22,242 particles were selected, and images were classified into 20 class averages (Fig. 5e). Although these classes show that oligomers containing mixtures of WT and Δ6–27 VacA do seem to have central density, difference maps generated between a common hexamer and dodecamer class (Fig. 5f and g) show that the central density is not as well organized as that found in WT oligomers. This finding provides evidence supporting a model in which the 6–27 deletion disrupts important N-terminal p33 interactions required for organizing the central core and supports a model in which the dominant-negative mutant acts through the formation of mixed oligomers with defects in channel formation.39 To ensure that these changes in the central organization are specific and not simply the result of a general change of structural conformation due to the deletion of residues, we also compared WT VacA averages with averages of VacAΔ301–328 and found no statistical differences (Fig. 6a and data not shown).
To determine whether the lack of an organized central core is a common feature found in variants and mutants of VacA that have defects in pore formation, we compared WT VacA averages with averages of VacA s2m1 and found no statistical differences (Fig. 6b and data not shown). This was especially surprising considering that s2m1 VacA contains 12 extra residues at its N-terminus,46–48 which we expected would disrupt the central core in a manner similar to what was observed with the VacAΔ6–27 mutant. The presence of an intact central core in s2m1 oligomers is notable because many WT H. pylori strains produce s2 forms of VacA.42 These data suggest that, unlike VacAΔ6–27, the lack of vacuolating activity exhibited by VacA s2m1 does not arise from a disorganized p33 region, at least in solution. It is possible that the N-terminal extensions may interfere with important hydrophobic interactions between p33 and lipids that are required for pore formation in the context of a membrane environment.
In summary, we have analyzed how soluble VacA forms single- and double-layer oligomers and have provided the most detailed structural model of VacA to date. Our analysis of different VacA mutants and variants provides further insight into VacA structure–function relationships and the mechanism by which a dominant-negative mutant exhibits inhibitory activity. There have been three published examples of VacA oligomers associated with supported lipid bilayers;26,38,55 however, the low resolution of these studies has made it difficult to conclude which oligomeric type of VacA associates with lipids. It is tempting to speculate that VacA hexamers (Fig. 2d) represent the conformation that associates with lipids and forms pores, while the dodecamers and tetradecamers (Fig. 2a–c, e, and f) represent interactions between hydrophobic regions of p88 that would normally interact with the lipid membrane if present. In future studies, it will be important not only to analyze the structure of VacA oligomers at a higher level of resolution and VacA oligomers associated with lipids but also to investigate structural changes in VacA that are required for the formation of membrane channels.
Methods
Purification of VacA
H. pylori strain 60190 (expressing WT VacA) and strains expressing either VacAΔ6–27 or VacA s2m1 proteins have been described previously.39,48 An H. pylori strain expressing a VacAΔ301–328 mutant was constructed using previously described methodology.56 Strains were grown in broth culture and VacA was isolated in oligomeric form as previously described.35,39 VacA was further purified using Matrex affinity gel and gel-filtration chromatography as previously described.38 Mixed WT VacA: VacAΔ6–27 oligomers were generated by mixing equal molar ratios of WT VacA and VacAΔ6–27 at neutral pH. The pH was acidified to 3.5 to promote oligomer disassembly39 and the tube was lightly vortexed. The pH of the mixture was neutralized so that the mixed oligomers could form.
Specimen preparation and EM
Uranyl formate (0.7% w/v) was used for conventional negative staining as previously described.57 For further details, see Supplemental Data. Images of WT VacA, VacA Δ301–328, VacA s2m1, and WT:VacΔ6–27 were taken using an F20 electron microscope (FEI) equipped with a field emission gun at an acceleration voltage of 200 kV under low-dose conditions at a magnification of 62,000× using a defocus value of –1.5 μm and recorded on a 4k×4k Gatan CCD camera. Images were converted to mrc (mixed raster content) format and binned by a factor of 2, resulting in final images with 3.5 Å/pixel. Images of WT VacA (used for difference mapping with VacAΔ6–27 particles) and VacAΔ6–27 were recorded using a Tecnai T12 electron microscope (FEI) equipped with a LaB6 filament and operated at an acceleration voltage of 120 kV. Images were taken under low-dose conditions at a magnification of 67,000× using a defocus value of –1.5 μm. Images were recorded on DITABIS digital imaging plates (Pforzheim, Germany). The plates were scanned on a DITABIS micron scanner (Pforzheim, Germany), converted to mrc format, and binned by a factor of 2, yielding final images with 4.48 Å/pixel. All binned MRC files were converted to SPIDER format using EM2EM. Image analysis for all VacA samples was carried out with SPIDER and the associated display program WEB.58
Classification and RCT of negatively stained WT, variant, and mutant VacA particles
Micrograph tilt pairs of WT and Δ6–27 VacA were recorded at 55° and 0°. Images for VacA s2m1, and VacAΔ301–328 and the mixture of VacA:VacAΔ6–27 particles were collected at 0°. Particle pairs (14,573 for WT VacA and 4470 for VacAΔ6–27) and particles for s2m1, VacAΔ301–328, and VacA:VacAΔ6–27 (7032 for VacA s2m1, 6342 for VacAΔ301–328, and 22,242 for VacA:VacAΔ6–27) were selected interactively using WEB. VacA particles were windowed into 120×120 pixel images (3.5 Å/pixel for WT, VacA s2m1, VacAΔ301–328, and the VacA:VacAΔ6–27 mixture; 4.48 Å/pixel for VacAΔ6–27). The untilted images were rotationally and translationally aligned and subjected to 10 cycles of multi-reference alignment and K-means classification. Particles of WT VacA were grouped into 20 classes (Fig. S1a). The references used for the first multi-reference alignment were randomly chosen from the raw images. From the class averages, eight representative projections were chosen and used as references for another cycle of reference-based alignment (Fig. S1a marked with an “*”). Particles of VacAΔ6–27 were grouped into 10 classes (Fig. S4a), and from these class averages, seven representative projections were chosen and used as references for another cycle of reference-based alignment (Fig. S4a marked with an “*”). Particles of VacA s2m1 and VacAΔ301–328 were grouped into 60 classes (Fig. S4b and c), and from these class averages, 10 representative projections were chosen and used as references for another cycle of reference-based alignment (Fig. S4b and c marked with an “*”). Particles of VacA:VacAΔ6–27 were grouped into 20 classes (Fig. 5e). Difference images were calculated using both Diffmap.exe and SPIDER.58 Results were similar using either program.
The tilted images corresponding to each of the WT and VacAΔ6–27 classes generated from reference-based alignment were used to calculate initial 3D reconstructions by back projection using the in-plane rotation angles determined by rotational alignment and the preselected tilt angle of 55° implemented in the processing package SPIDER.57 The density maps were improved by back projection and angular refinement in SPIDER. Ten percent of the particles selected from the images of the untilted specimens in each class were included in the data set and angular refinement was repeated. During the angular refinement process, either 6- or 7-fold symmetry was applied to the final reconstructions as determined by the number of “arms” in each oligomeric form. Structures with no applied symmetry are shown in Fig. S2. For further details about structural methods used in this analysis, refer to Cheng et al.59
For WT VacA, two of the WT structures were very similar (Fig. 2b and c), and therefore, the 6059 particles associated with these structures were combined and a new structure was calculated using angular refinement (Fig. 3). The Fourier shell correlation curve corresponds to normalized cross-correlation coefficients of Fourier shells from even and odd particles within the data set. Using a 0.5 Fourier shell correlation criterion, the resolution is ~10 Å (Fig. S1b); however, the lack of any secondary structural details in our map suggests that the resolution more likely falls closer to 15 Å. This resolution corresponds to the appearance of the crystal structure of VacA p5533 when filtered to 15 Å resolution (data not shown). For display purposes, structures were filtered in chimera using the “Hide Dust” command to diminish “salt and pepper” noise from the maps by removing single voxels that were unconnected to the main volume of the 3D density.53 Contouring thresholds were chosen to match the molecular weight for each VacA oligomer. Surface rendering of the structure was performed with the program Chimera.53
Supplementary Material
Acknowledgements
This work was supported by T32 GM08320 (to S.E.C.); T32 AI007281 (to C.G.-R.); NIH AI39657, CA116087, and the Department of Veterans Affairs (to T.L.C.); a Burroughs Wellcome Investigator in the Pathogenesis of Infectious Disease award (to D.B.L.); and Vanderbilt Development funds (to M.D.O.).
Abbreviations used
- 3D
three-dimensional
- EM
electron microscopy
- RCT
random conical tilt
- VacA
vacuolating toxin
- WT
wild-type
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jmb.2012.11.020
References
- 1.Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;1:1311–1315. doi: 10.1016/s0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- 2.Cover TL, Blaser MJ. Helicobacter pylori in health and disease. Gastroenterology. 2009;136:1863–1873. doi: 10.1053/j.gastro.2009.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atherton JC, Blaser MJ. Coadaptation of Helicobacter pylori and humans: ancient history, modern implications. J. Clin. Invest. 2009;119:2475–2487. doi: 10.1172/JCI38605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suerbaum S, Michetti P. Helicobacter pylori infection. N. Engl. J. Med. 2002;347:1175–1186. doi: 10.1056/NEJMra020542. [DOI] [PubMed] [Google Scholar]
- 5.Atherton JC. The pathogenesis of Helicobacter pylori-induced gastro-duodenal diseases. Annu. Rev. Pathol. 2006;1:63–96. doi: 10.1146/annurev.pathol.1.110304.100125. [DOI] [PubMed] [Google Scholar]
- 6.Amieva MR, El-Omar EM. Host–bacterial interactions in Helicobacter pylori infection. Gastroenterology. 2008;134:306–323. doi: 10.1053/j.gastro.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 7.Fuchs CS, Mayer RJ. Gastric carcinoma. N. Engl. J. Med. 1995;333:32–41. doi: 10.1056/NEJM199507063330107. [DOI] [PubMed] [Google Scholar]
- 8.Montecucco C, de Bernard M. Molecular and cellular mechanisms of action of the vacuolating cytotoxin (VacA) and neutrophil-activating protein (HP-NAP) virulence factors of Helicobacter pylori. Microbes Infect. 2003;5:715–721. doi: 10.1016/s1286-4579(03)00124-2. [DOI] [PubMed] [Google Scholar]
- 9.Fischer W, Gebert B, Haas R. Novel activities of the Helicobacter pylori vacuolating cytotoxin: from epithelial cells towards the immune system. Int. J. Med. Microbiol. 2004;293:539–547. doi: 10.1078/1438-4221-00300. [DOI] [PubMed] [Google Scholar]
- 10.Cover TL, Blanke SR. Helicobacter pylori VacA, a paradigm for toxin multifunctionality. Nat. Rev. Microbiol. 2005;3:320–332. doi: 10.1038/nrmicro1095. [DOI] [PubMed] [Google Scholar]
- 11.Rassow J, Meinecke M. Helicobacter pylori VacA: a new perspective on an invasive chloride channel. Microbes Infect. 2012 doi: 10.1016/j.micinf.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Boquet P, Ricci V. Intoxication strategy of Helicobacter pylori VacA toxin. Trends Microbiol. 2012;20:165–174. doi: 10.1016/j.tim.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 13.Szabo I, Brutsche S, Tombola F, Moschioni M, Satin B, Telford JL, et al. Formation of anion-selective channels in the cell plasma membrane by the toxin VacA of Helicobacter pylori is required for its biological activity. EMBO J. 1999;18:5517–5527. doi: 10.1093/emboj/18.20.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galmiche A, Rassow J, Doye A, Cagnol S, Chambard JC, Contamin S, et al. The N-terminal 34 kDa fragment of Helicobacter pylori vacuolating cytotoxin targets mitochondria and induces cytochrome c release. EMBO J. 2000;19:6361–6370. doi: 10.1093/emboj/19.23.6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willhite DC, Blanke SR. Helicobacter pylori vacuolating cytotoxin enters cells, localizes to the mitochondria, and induces mitochondrial membrane permeability changes correlated to toxin channel activity. Cell. Microbiol. 2004;6:143–154. doi: 10.1046/j.1462-5822.2003.00347.x. [DOI] [PubMed] [Google Scholar]
- 16.Domanska G, Motz C, Meinecke M, Harsman A, Papatheodorou P, Reljic B, et al. Helicobacter pylori VacA toxin/subunit p34: targeting of an anion channel to the inner mitochondrial membrane. PLoS Pathog. 2010;6:e1000878. doi: 10.1371/journal.ppat.1000878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jain P, Luo ZQ, Blanke SR. Helicobacter pylori vacuolating cytotoxin A (VacA) engages the mitochondrial fission machinery to induce host cell death. Proc. Natl Acad. Sci. USA. 2011;108:16032–16037. doi: 10.1073/pnas.1105175108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Terebiznik MR, Raju D, Vazquez CL, Torbricki K, Kulkarni R, Blanke SR, et al. Effect of Helicobacter pylori's vacuolating cytotoxin on the autophagy pathway in gastric epithelial cells. Autophagy. 2009;5:370–379. doi: 10.4161/auto.5.3.7663. [DOI] [PubMed] [Google Scholar]
- 19.Calore F, Genisset C, Casellato A, Rossato M, Codolo G, Esposti MD, et al. Endosome–mitochondria juxtaposition during apoptosis induced by H. pylori VacA. Cell Death Differ. 2010;17:1707–1716. doi: 10.1038/cdd.2010.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radin JN, Gonzalez-Rivera C, Ivie SE, McClain MS, Cover TL. Helicobacter pylori VacA induces programmed necrosis in gastric epithelial cells. Infect. Immun. 2011;79:2535–2543. doi: 10.1128/IAI.01370-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakayama M, Kimura M, Wada A, Yahiro K, Ogushi K, Niidome T, et al. Helicobacter pylori VacA activates the p38/activating transcription factor 2-mediated signal pathway in AZ-521 cells. J. Biol. Chem. 2004;279:7024–7028. doi: 10.1074/jbc.M308898200. [DOI] [PubMed] [Google Scholar]
- 22.Gebert B, Fischer W, Weiss E, Hoffmann R, Haas R. Helicobacter pylori vacuolating cytotoxin inhibits T lymphocyte activation. Science. 2003;301:1099–1102. doi: 10.1126/science.1086871. [DOI] [PubMed] [Google Scholar]
- 23.Sundrud MS, Torres VJ, Unutmaz D, Cover TL. Inhibition of primary human T cell proliferation by Helicobacter pylori vacuolating toxin (VacA) is independent of VacA effects on IL-2 secretion. Proc. Natl. Acad. Sci. USA. 2004;101:7727–7732. doi: 10.1073/pnas.0401528101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sewald X, Fischer W, Haas R. Sticky socks: Helicobacter pylori VacA takes shape. Trends Microbiol. 2008;16:89–92. doi: 10.1016/j.tim.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 25.Gauthier NC, Monzo P, Gonzalez T, Doye A, Oldani A, Gounon P, et al. Early endosomes associated with dynamic F-actin structures are required for late trafficking of H. pylori VacA toxin. J. Cell Biol. 2007;177:343–354. doi: 10.1083/jcb.200609061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Czajkowsky DM, Iwamoto H, Cover TL, Shao Z. The vacuolating toxin from Helicobacter pylori forms hexameric pores in lipid bilayers at low pH. Proc. Natl Acad. Sci. USA. 1999;96:2001–2006. doi: 10.1073/pnas.96.5.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iwamoto H, Czajkowsky DM, Cover TL, Szabo G, Shao Z. VacA from Helicobacter pylori: a hexameric chloride channel. FEBS Lett. 1999;450:101–104. doi: 10.1016/s0014-5793(99)00474-3. [DOI] [PubMed] [Google Scholar]
- 28.Tombola F, Carlesso C, Szabo I, de Bernard M, Reyrat JM, Telford JL, et al. Helicobacter pylori vacuolating toxin forms anion-selective channels in planar lipid bilayers: possible implications for the mechanism of cellular vacuolation. Biophys. J. 1999;76:1401–1409. doi: 10.1016/S0006-3495(99)77301-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McClain MS, Iwamoto H, Cao P, Vinion-Dubiel AD, Li Y, Szabo G, et al. Essential role of a GXXXG motif for membrane channel formation by Helicobacter pylori vacuolating toxin. J. Biol. Chem. 2003;278:12101–12108. doi: 10.1074/jbc.M212595200. [DOI] [PubMed] [Google Scholar]
- 30.Torres VJ, Ivie SE, McClain MS, Cover TL. Functional properties of the p33 and p55 domains of the Helicobacter pylori vacuolating cytotoxin. J. Biol. Chem. 2005;280:21107–21114. doi: 10.1074/jbc.M501042200. [DOI] [PubMed] [Google Scholar]
- 31.El-Bez C, Adrian M, Dubochet J, Cover TL. High resolution structural analysis of Helicobacter pylori VacA toxin oligomers by cryo-negative staining electron microscopy. J. Struct. Biol. 2005;151:215–228. doi: 10.1016/j.jsb.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 32.Gonzalez-Rivera C, Gangwer KA, McClain MS, Eli IM, Chambers MG, Ohi MD, et al. Reconstitution of Helicobacter pylori VacA toxin from purified components. Biochemistry. 2010;49:5743–5752. doi: 10.1021/bi100618g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gangwer KA, Mushrush DJ, Stauff DL, Spiller B, McClain MS, Cover TL, Lacy DB. Crystal structure of the Helicobacter pylori vacuolating toxin p55 domain. Proc. Natl Acad. Sci. USA. 2007;104:16293–16298. doi: 10.1073/pnas.0707447104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lupetti P, Heuser JE, Manetti R, Massari P, Lanzavecchia S, Bellon PL, et al. Oligomeric and subunit structure of the Helicobacter pylori vacuolating cytotoxin. J. Cell Biol. 1996;133:801–807. doi: 10.1083/jcb.133.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cover TL, Hanson PI, Heuser JE. Acid-induced dissociation of VacA, the Helicobacter pylori vacuolating cytotoxin, reveals its pattern of assembly. J. Cell Biol. 1997;138:759–769. doi: 10.1083/jcb.138.4.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lanzavecchia S, Bellon PL, Lupetti P, Dallai R, Rappuoli R, Telford JL. Three-dimensional reconstruction of metal replicas of the Helicobacter pylori vacuolating cytotoxin. J. Struct. Biol. 1998;121:9–18. doi: 10.1006/jsbi.1997.3941. [DOI] [PubMed] [Google Scholar]
- 37.Reyrat JM, Lanzavecchia S, Lupetti P, de Bernard M, Pagliaccia C, Pelicic V, et al. 3D imaging of the 58 kDa cell binding subunit of the Helicobacter pylori cytotoxin. J. Mol. Biol. 1999;290:459–470. doi: 10.1006/jmbi.1999.2877. [DOI] [PubMed] [Google Scholar]
- 38.Adrian M, Cover TL, Dubochet J, Heuser JE. Multiple oligomeric states of the Helicobacter pylori vacuolating toxin demonstrated by cryo-electron microscopy. J. Mol. Biol. 2002;318:121–133. doi: 10.1016/S0022-2836(02)00047-5. [DOI] [PubMed] [Google Scholar]
- 39.Vinion-Dubiel AD, McClain MS, Czajkowsky DM, Iwamoto H, Ye D, Cao P, et al. A dominant negative mutant of Helicobacter pylori vacuolating toxin (VacA) inhibits VacA-induced cell vacuolation. J. Biol. Chem. 1999;274:37736–37742. doi: 10.1074/jbc.274.53.37736. [DOI] [PubMed] [Google Scholar]
- 40.Genisset C, Galeotti CL, Lupetti P, Mercati D, Skibinski DA, Barone S, et al. A Helicobacter pylori vacuolating toxin mutant that fails to oligomerize has a dominant negative phenotype. Infect. Immun. 2006;74:1786–1794. doi: 10.1128/IAI.74.3.1786-1794.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ivie SE, McClain MS, Torres VJ, Algood HM, Lacy DB, Yang R, et al. Helicobacter pylori VacA subdomain required for intracellular toxin activity and assembly of functional oligomeric complexes. Infect. Immun. 2008;76:2843–2851. doi: 10.1128/IAI.01664-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Atherton JC, Cao P, Peek RM, Jr, Tummuru MK, Blaser MJ, Cover TL. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori association of specific vacA types with cytotoxin production and peptic ulceration. J. Biol. Chem. 1995;270:17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 43.Rhead JL, Letley DP, Mohammadi M, Hussein N, Mohagheghi MA, Eshagh Hosseini M, Atherton JC. A new Helicobacter pylori vacuolating cytotoxin determinant, the intermediate region, is associated with gastric cancer. Gastroenterology. 2007;133:926–936. doi: 10.1053/j.gastro.2007.06.056. [DOI] [PubMed] [Google Scholar]
- 44.Gangwer KA, Shaffer CL, Suerbaum S, Lacy DB, Cover TL, Bordenstein SR. Molecular evolution of the Helicobacter pylori vacuolating toxin gene vacA. J. Bacteriol. 2010;192:6126–6135. doi: 10.1128/JB.01081-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pagliaccia C, de Bernard M, Lupetti P, Ji X, Burroni D, Cover TL, et al. The m2 form of the Helicobacter pylori cytotoxin has cell type-specific vacuolating activity. Proc. Natl Acad. Sci. USA. 1998;95:10212–10217. doi: 10.1073/pnas.95.17.10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Letley DP, Rhead JL, Twells RJ, Dove B, Atherton JC. Determinants of non-toxicity in the gastric pathogen Helicobacter pylori. J. Biol. Chem. 2003;278:26734–26741. doi: 10.1074/jbc.M304071200. [DOI] [PubMed] [Google Scholar]
- 47.Letley DP, Atherton JC. Natural diversity in the N terminus of the mature vacuolating cytotoxin of Helicobacter pylori determines cytotoxin activity. J. Bacteriol. 2000;182:3278–3280. doi: 10.1128/jb.182.11.3278-3280.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McClain MS, Cao P, Iwamoto H, Vinion-Dubiel AD, Szabo G, Shao Z, Cover TL. A 12-amino-acid segment, present in type s2 but not type s1 Helicobacter pylori VacA proteins, abolishes cytotoxin activity and alters membrane channel formation. J. Bacteriol. 2001;183:6499–6508. doi: 10.1128/JB.183.22.6499-6508.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burroni D, Lupetti P, Pagliaccia C, Reyrat JM, Dallai R, Rappuoli R, Telford JL. Deletion of the major proteolytic site of the Helicobacter pylori cytotoxin does not influence toxin activity but favors assembly of the toxin into hexameric structures. Infect. Immun. 1998;66:5547–5550. doi: 10.1128/iai.66.11.5547-5550.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tombola F, Pagliaccia C, Campello S, Telford JL, Montecucco C, Papini E, Zoratti M. How the loop and middle regions influence the properties of Helicobacter pylori VacA channels. Biophys. J. 2001;81:3204–3215. doi: 10.1016/S0006-3495(01)75956-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Radermacher M, Wagenknecht T, Verschoor A, Frank J. Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. J. Microsc. 1987;146:113–136. doi: 10.1111/j.1365-2818.1987.tb01333.x. [DOI] [PubMed] [Google Scholar]
- 52.Radermacher M. Three-dimensional reconstruction from random projections: orientational alignment via Radon transforms. Ultramicroscopy. 1994;53:121–136. doi: 10.1016/0304-3991(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 53.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 54.Torres VJ, McClain MS, Cover TL. Mapping of a domain required for protein-protein interactions and inhibitory activity of a Helicobacter pylori dominant-negative VacA mutant protein. Infect. Immun. 2006;74:2093–2101. doi: 10.1128/IAI.74.4.2093-2101.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Geisse NA, Cover TL, Henderson RM, Edwardson JM. Targeting of Helicobacter pylori vacuolating toxin to lipid raft membrane domains analysed by atomic force microscopy. Biochem. J. 2004;381:911–917. doi: 10.1042/BJ20031719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gonzalez-Rivera C, Algood HM, Radin JN, McClain MS, Cover TL. The intermediate region of Helicobacter pylori VacA is a determinant of toxin potency in a Jurkat T cell assay. Infect. Immun. 2012;80:2578–2588. doi: 10.1128/IAI.00052-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ohi M, Li Y, Cheng Y, Walz T. Negative staining and image classification—powerful tools in modern electron microscopy. Biol. Proced. Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 59.Cheng Y, Wolf E, Larvie M, Zak O, Aisen P, Grigorieff N, et al. Single particle reconstructions of the transferrin–transferrin receptor complex obtained with different specimen preparation techniques. J. Mol. Biol. 2006;355:1048–1065. doi: 10.1016/j.jmb.2005.11.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.