Abstract
The peopling of the Americas has been the subject of extensive genetic, archaeological and linguistic research; however, central questions remain unresolved1–5. One contentious issue is whether the settlement occurred via a single6–8 or multiple streams of migration from Siberia9–15. The pattern of dispersals within the Americas is also poorly understood. To address these questions at higher resolution than was previously possible, we assembled data from 52 Native American and 17 Siberian groups genotyped at 364,470 single nucleotide polymorphisms. We show that Native Americans descend from at least three streams of Asian gene flow. Most descend entirely from a single ancestral population that we call “First American”. However, speakers of Eskimo-Aleut languages from the Arctic inherit almost half their ancestry from a second stream of Asian gene flow, and the Na-Dene-speaking Chipewyan from Canada inherit roughly one-tenth of their ancestry from a third stream. We show that the initial peopling followed a southward expansion facilitated by the coast, with sequential population splits and little gene flow after divergence, especially in South America. A major exception is in Chibchan-speakers on both sides of the Panama Isthmus, who have ancestry from both North and South America.
The settlement of the Americas occurred at least 15,000 years ago through Beringia, a land bridge between Asia and America that existed during the ice ages1–5. Most analyses of Native American genetic diversity have examined single loci, particularly mitochondrial DNA or the Y-chromosome, and some interpretations of these data model the settlement of America as a single migratory wave from Asia6–8. We assembled Native population samples from Canada to the southern tip of South America, genotyped them on single nucleotide polymorphism (SNP) microarrays, and merged our data with six other datasets. The combined dataset consists of 364,470 SNPs genotyped in 52 Native American populations (493 samples; Figure 1A; Table S1); 17 Siberian populations (245 samples; Figure S1; Table S2); and 57 other populations (1,613 samples) (Note S1).
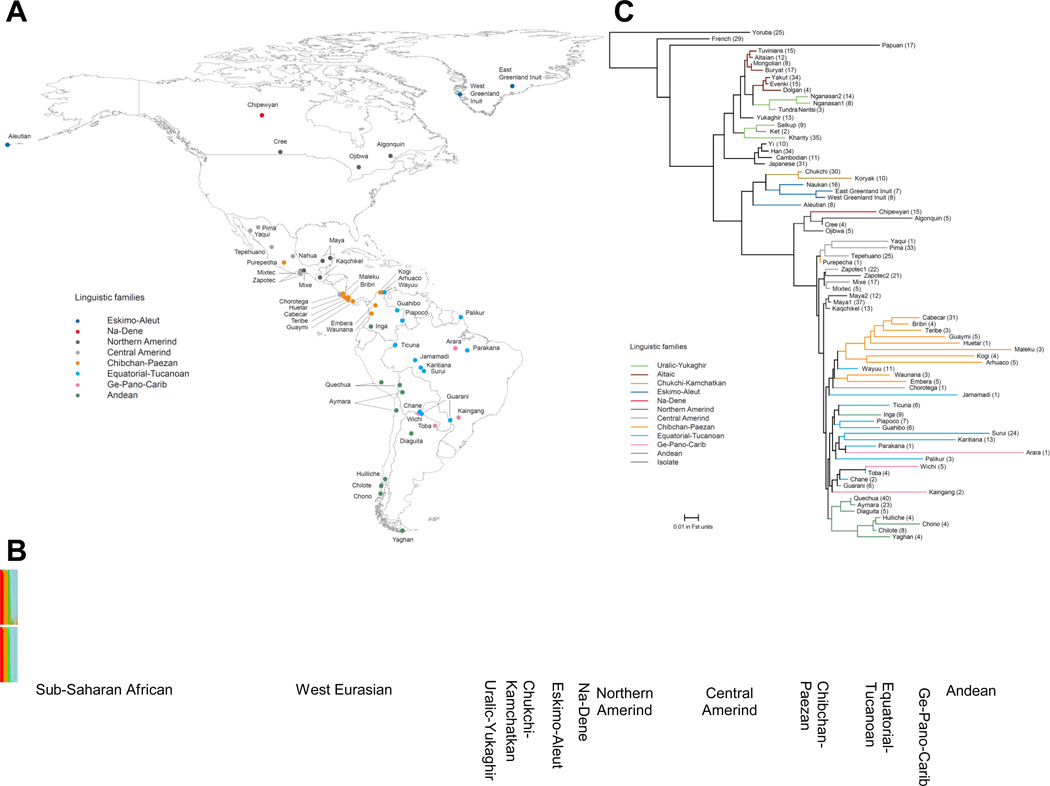
Figure 1. Geographic, linguistic and genetic overview of 52 Native American populations.
(A) Sampling locations of the populations, with colors corresponding to linguistic groups. (B) Cluster-based analysis (k=4) using ADMIXTURE shows evidence of some West Eurasian-related and sub-Saharan-African-related ancestry in many Native Americans prior to masking (top), but little afterward (bottom). Thick vertical lines denote major linguistic groupings, and thin vertical lines separate individual populations. (C) Neighbor-Joining tree based on FST distances relating Native American to selected non-American populations (sample sizes in parentheses). Native American and Siberian data were analyzed after masking but consistent trees were obtained on a subset of completely unadmixed samples (Figure S3). Some populations have evidence for substructure, and we represent these as two different groups (e.g. Maya1 and Maya2).
A complication in studying Native American genetic history is admixture with European and African immigrants since 1492. Cluster analysis16 shows that many of the samples we examined have some non-Native admixture (an average of 8.5%; Figure 1B; Table S1; Table S3). To address this, we validated our inferences using three independent approaches. First, we restricted analyses to 163 Native Americans from 34 populations without evidence of admixture (Note S2). Second, we subtracted the expected contribution of European and African ancestry to the statistics we used to learn about population relationships (Note S3). Third, we inferred the probability of non-Native ancestry at each genomic segment and “masked” segments with more than a negligible probability of this ancestry (Note S4; Figure S2; Figure 1B). Our inferences from these three approaches are concordant (Figure S3, Figure S4).
We built a tree (Figure 1C) using FST distances between pairs of populations, which broadly agrees with geography and linguistic categories17 (trees based on masked and unmasked data are similar; Figure S3). An early split separates Asians from Native Americans and extreme northeastern Siberians (Chukchi, Naukan, Koryak), consistent with studies that have identified pan-American variants shared with northeastern Siberians6,7,10,18. Eskimo-Aleut speakers and northeastern Siberians form a cluster that is separated from other Native American populations by a long internal branch. Within America, the tree shows a series of splits in an approximate north-to-south sequence beginning with the Arctic, followed by northern North America, northern/central and southern Mexico, lower Central America/Colombia, and ending in three South American clusters (the Andes, the Chaco region and eastern South America). This pattern of splits is consistent with a north-to-south population expansion, an inference that is also supported by the negative correlation between heterozygosity and distance from the Bering Strait (r=−0.48, P=0.007). This correlation increases if we use “least cost distances” which consider the coasts as facilitators of migration19–21, and persists if we exclude four Native North American populations with ancestry from later streams of Asian gene flow (Note S5; Figure S5).
Trees provide a simplified model of history that does not accommodate the possibility of gene flow after population separation. Circumstantial evidence that some Native American populations may not fit a simple tree comes from cluster analysis which infers Siberian-related ancestry in some northern North Americans (Figure 1B), and from single locus studies that have identified genetic variants shared between Eurasia and North America that are absent from South America11,22,23. However, these methods cannot distinguish shared ancestry from admixture after population separation. The advent of genome-wide data sets has allowed the development of a 4 Population Test for whether sets of four populations are consistent with a tree. This test is robust to the ascertainment bias affecting SNP arrays24. For each of the 52 Native American populations in turn, we tested the hypothesis that they conform to the tree: ((Test Population, Southern Native American), (Outgroup1, Outgroup2)) for 45 pairs of 10 Asian outgroups. We used a Hotelling T-test to evaluate whether all 4 Population Test “f4” statistics of this form are consistent with the expectation of zero (Note S6). The test is not significant for 47 populations, consistent with their stemming from the same, presumably first wave of American settlement, and we call this ancestry “First American” (Table 1). In contrast, 4 populations from northern North America show highly significant evidence of ancestry from additional streams of gene flow from Asia, subsequent to the initial peopling of America, which we confirm through the Hotelling T-test and a complementary test (Note S6): East Greenland Inuit (P<10−9), West Greenland Inuit (P<10−9), Aleutian Islanders (P=9×10−5) and Chipewyan (P<10−9). The recently sequenced genome of a 4,000 year old Saqqaq Paleo-Eskimo from Greenland25 also has evidence of ancestry that is distinct from more southern Native Americans (P=2×10−9) (Note S6).
Table 1.
Native Americans descend from at least three streams of Asian gene flow
| Population groupings tested | P-value for this many Asian streams being enough to explain the data |
Minimum number of streams of Asian gene flow needed to explain the data |
||
|---|---|---|---|---|
| 1 | 2 | 3 | ||
| E. Greenland Inuit / W. Greenland Inuit / First American | <10−9 | 0.64 | 1 | 2 |
| E. Greenland Inuit / Aleutian / First American | <10−9 | 0.57 | 1 | 2 |
| W. Greenland Inuit / Aleutian / First American | <10−9 | 0.41 | 1 | 2 |
| Chipewyan / E. Greenland Inuit / First American | <10−9 | 0.02 | 1 | 3 |
| Chipewyan / W. Greenland Inuit / First American | <10−9 | 0.006 | 1 | 3 |
| Chipewyan / Aleutian / First American | <10−9 | 0.03 | 1 | 3 |
| Saqqaq / E. Greenland Inuit / First American | <10−9 | 6×10−6 | 1 | 3 |
| Saqqaq / W. Greenland Inuit / First American | <10−9 | 2×10−6 | 1 | 3 |
| Saqqaq / Aleutian / First American | <10−9 | 0.17 | 1 | 2 |
| Saqqaq / Chipewyan / First American | <10−9 | 0.29 | 1 | 2 |
| Saqqaq / Eskimo-Aleut / Chipewyan / First American | <10−9 | 8×10−6 | 0.27 | 3 |
Notes: We use the method described in Note S6 to test formally whether specified groupings of Native American populations are consistent with descending from 1, 2, or 3 streams of gene flow from Asia. We use “First American” to refer to a pool of 43 populations from Meso-America southward, and “Eskimo-Aleut” to refer to a pool of East and West Greenland Inuit and Aleuts. We test either 3 or 4 population groupings (when there are 3 groupings, the maximum number of streams we can reject is 2, and so the P-value for 3 streams is always 1). At least two streams of Asian gene flow are required to explain all rows (P<10−9). The Chipewyan, Eskimo-Aleut and First Americans can only be jointly explained by at least three streams. Analysis of the Saqqaq Paleo-Eskimo (using ~6-fold fewer SNPs than for the other analyses) show that the Asian ancestry in this individual has a component that is different from that in First Americans and Greenland Inuit, but indistinguishable from the Chipewyan.
Examination of the values of the f4 statistics allows us to infer the minimum number of gene flow events from Asia into America consistent with the data. Each stream of gene flow is expected to produce a distinct vector of f4 statistics, constituting a “signature” of how the ancestral migrating population relates to present-day Asian populations. By finding the minimum number of vectors whose linear combinations are necessary to produce the vector observed in each population, we infer that a minimum of three gene flow events from Asia are necessary to explain the data from all Native American populations jointly, including the Saqqaq Paleo-Eskimo (Note S6). These three episodes correspond to First American ancestry (distributed throughout the Americas) and to two additional streams of gene flow detected in a subset of northern North Americans (East Greenland Inuit, West Greenland Inuit, Aleutian Islanders, Chipewyan and Saqqaq). Table 1 shows that f4 statistics in the Inuit and Aleutian islanders are consistent with deriving the non-First American portions of their ancestry from the same later stream of Asian gene flow, providing support for deep shared ancestry between these linguistically linked groups12,26. The Na-Dene speaking Chipewyan have a different pattern of f4 statistics from Eskimo-Aleut speakers, implying that they descend at least in part from a separate stream of Asian gene flow (P<10−9 for comparisons to the Greenland Inuit; Table 1). This is consistent with the hypothesis that Na-Dene languages mark a distinct migration from Asia9,17. Since we only have data from one Na-Dene speaking group, an important direction for future work will be to test if the distinct Asian ancestry we detect in the Chipewyan is a shared signature throughout Na-Dene speakers. Finally, the Saqqaq25 have a vector of f4 statistics consistent with that in the Chipewyan, raising the possibility that the Saqqaq and Chipewyan both carry genetic material from the same later stream of Asian gene flow into the Americas, post-dating the First American migration (Note S6 and Note S7).
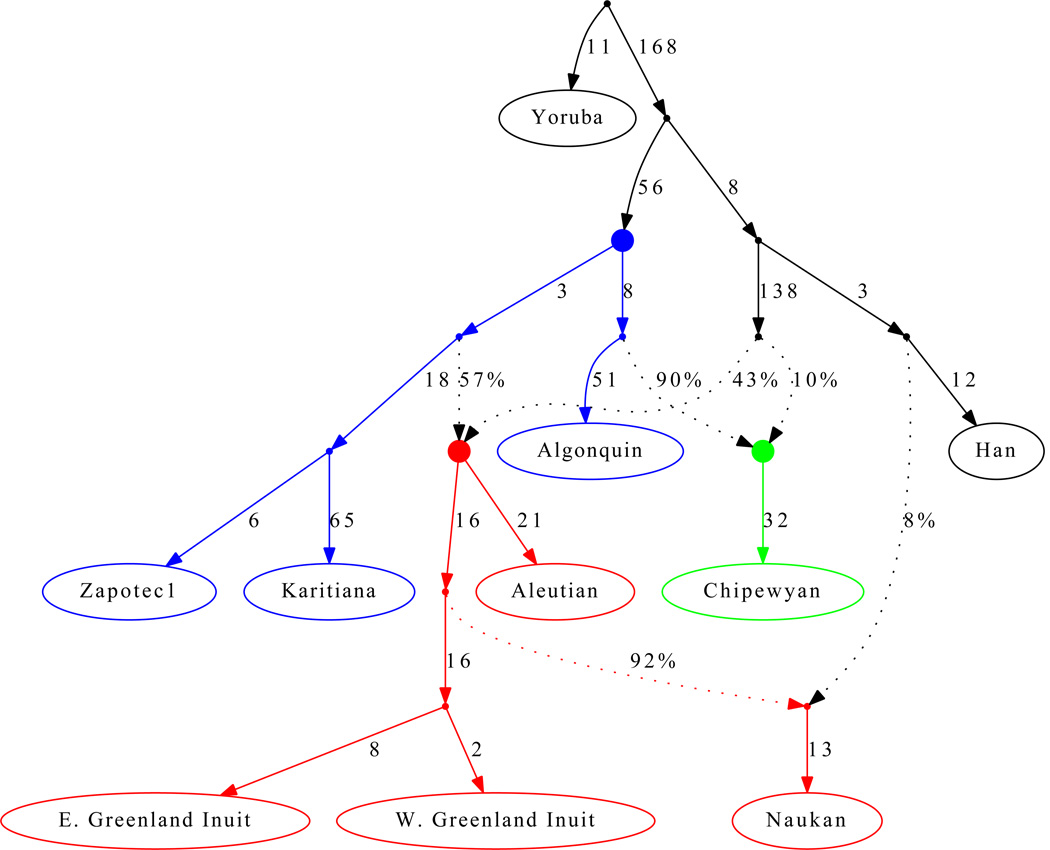
To develop an explicit model for the settlement of the Americas, we used the Admixture Graph (AG) framework24. AGs are generalizations of trees that accommodate the possibility of a limited number of unidirectional gene flow events. They are powerful tools for learning about history because they make predictions about the values of f-statistics (such as f4) that can be used to test the fit of a proposed model24 (Note S7). Figure 2 presents an AG relating selected Native American and Old World populations that is a good fit to the data in the sense that none of the f-statistics predicted by the model are more than 3 standard errors from what is observed. This supports the hypothesis of three deep lineages in Native Americans: the Asian lineage leading to First Americans is the most deeply diverged, while the Asian lineages leading to Eskimo-Aleut speakers and the Na-Dene speaking Chipewyan are more closely related and descend from a putative Siberian ancestral population more closely related to Han (Figure 2). We also arrive at the novel finding that Eskimo-Aleut populations and the Chipewyan derive large proportions of their genomes from First American ancestors: an estimated 57% for Eskimo-Aleut speakers, and 90% in the Chipewyan, likely reflecting major admixture events of the two later streams of Asian migration with the First Americans they encountered after they arrived (Note S7). The high proportion of First American ancestry explains why Eskimo-Aleut and Chipewyan populations cluster with First Americans in trees like Figure 1C despite having some of their ancestry from later streams of Asian migration, and explains the observation of some genetic mutations that are shared by all Native Americans but are absent elsewhere6,7,10,18. We also infer back-migration of populations related to the Eskimo-Aleut from America into far-northeastern Siberia (we obtain an excellent fit to the data when we model the Naukan and coastal Chukchi as mixtures of groups related to the Greenland Inuit and Asians; Figure 2; Note S7). This explains previous findings of pan-American alleles also in far-northeastern Siberia6,7,10,18.
Figure 2. Distinct streams of gene flow from Asia into America.
We present an Admixture Graph (AG) that gives no evidence of being a poor fit to the data and is consistent with three streams of Asian gene flow into America. Solid points indicate inferred ancestral populations; drift on each lineage is given in units proportional to 1000×FST; and mixture events (dotted lines) are denoted by the percentage of ancestry. The Asian lineage leading to First Americans is the most deeply diverged, while the Asian lineages leading to Eskimo-Aleut speakers and the Na-Dene speaking Chipewyan are more closely related and descend from a common Siberian ancestral population that is a sister group to the Han. The inferred ancestral populations are indicated by filled circles and the lineages descending from them are colored: First American (blue), ancestors of the Na-Dene speaking Chipewyan (green) and Eskimo-Aleut (red). The model also infers a migration of people related to Eskimo-Aleut speakers across the Bering Strait, thus bringing First American genes to Asia (the Naukan are shown, but the Chukchi show a similar pattern; Note S7). Estimated admixture proportions are shown along the dotted lines, and lineage-specific drift estimates are in units proportional to 1000×FST
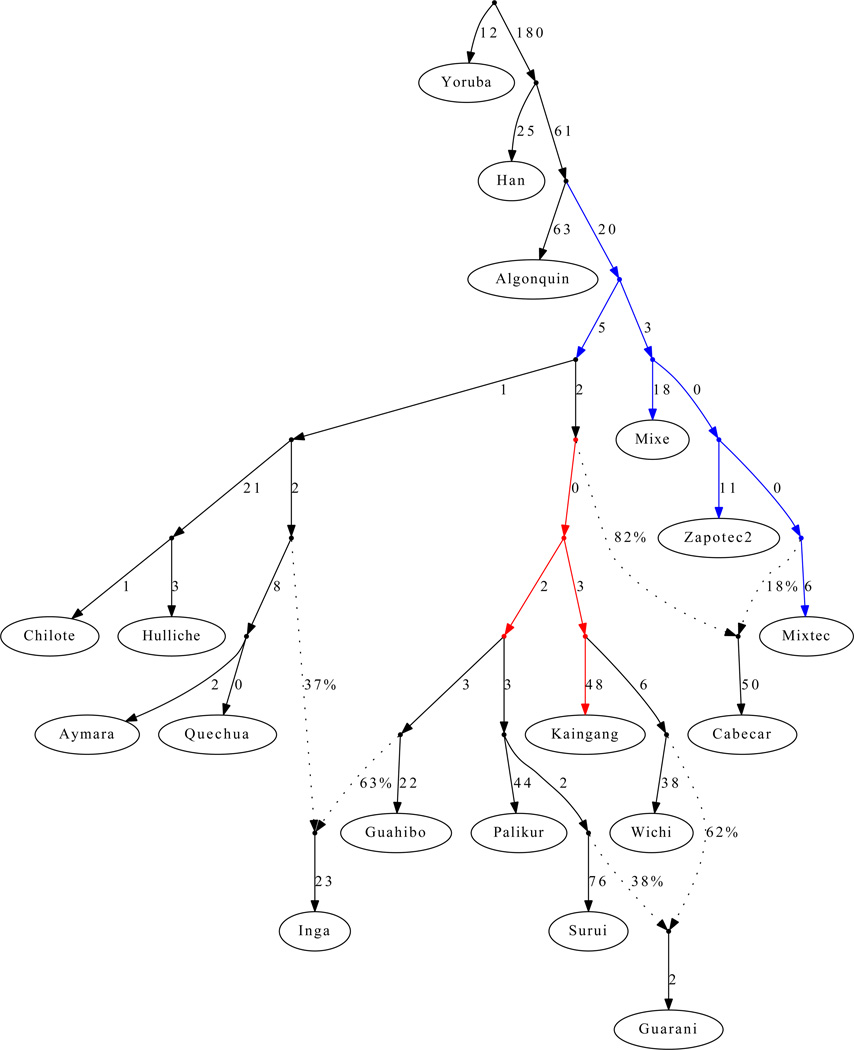
We next used AGs to develop a model for the history of populations who derive all their ancestry from the First American migration, with no ancestry from subsequent streams of Asian gene flow. Figure 3 presents an AG we built for 16 selected Native American populations and 2 outgroups, which is a good fit to the data in that the largest |Z|-score for a difference between the observed and predicted f-statistics greater is 3.2 from among the 11,781 of statistics we tested (Note S7) (The AG of Figure 3 used masked data; however, a consistent set of relationships is inferred for unadmixed samples; Figure S4.) This model provides a greatly improved statistical fit to the data compared with the tree of Figure 1C and leads to several novel inferences. (i) A relatively large fraction of South American populations fit the AG without a need for admixture events, which we hypothesize reflects a history of limited gene flow among these populations since their initial divergence. In contrast, only a small fraction of Meso-American populations fit into the AG, which could reflect either a higher rate of migration among neighboring groups or our denser sampling in Meso-America allowing us to detect more subtle gene flow events. (ii) Some Meso-American populations have experienced very little genetic drift since divergence from the common ancestral population with South Americans (adding up the genetic drifts along the relevant edges of Figure 3 we infer FST=0.014 between the Zapotec and a hypothesized population ancestral to all of Central and South America), suggesting that effective population sizes in Meso-America have been relatively large since settlement of the region. (iii) The model infers three admixture events consistent with geographic locations and linguistic affiliations (Note S7). The Inga have both Amazonian and Andean ancestry, consistent with them speaking a Quechuan language but living in the eastern Andean slopes of Colombia and thus interacting with groups in the neighboring Amazonian lowlands. The Guarani stem from two distinct strands of ancestry within eastern South America. The most striking admixture event is in the Costa Rican Cabecar (Figure 3) and other Chibchan-speaking populations (Note S7) from the Isthmo-Colombian area. One of the lineages that we detect in these populations occurs definitively within the radiation of South American populations, and so the presence of these populations in lower Central America suggests that there was reverse gene flow across the Panama isthmus after the initial settlement of South America. There has been controversy about whether Chibchan-speakers of lower Central America represent direct descendants of the first settlers in the region or more recent migration across the isthmus, and our results support the view that more recent migration has contributed most of these populations’ ancestry27.
Figure 3. A model fitting populations of entirely First American ancestry.
We show an Admixture Graph (AG) depicting the relationships among 16 selected Native American populations with entirely First American ancestry along with 2 outgroups (Yoruba and Han). The Colombian Inga are modeled as a mixture of Andean and Amazonian ancestry. The Paraguayan Guarani are fit as a mixture of separate strands of ancestry from eastern South America. The Central American Cabecar are modeled as a mixture of strands of ancestry related to South Americans and to North Americans, supporting back-migration from South into Central America. The coloring of edges indicates alternative insertion points for the admixing lineages leading to the Cabecar that produce a similar fit to the data in the sense that the χ2 statistic is within 3.84 of the AG shown. The red coloring shows that the South American lineage contributing to the Cabecar split off after the divergence of the Andean populations, and the blue coloring shows that the other lineage present in the Cabecar diverged before the separation of Andeans.
This is the most comprehensive survey of genetic diversity in Native Americans to date, and the first to account for recent non-Native admixture. Our analyses show that the great majority of Native American populations—from Canada to the southern tip of Chile—derive their ancestry from a homogeneous “First American” ancestral population, presumably the one that crossed the Bering Strait more than 15,000 years ago6–8. We also document at least two additional streams of Asian gene flow into America, allowing us to reject the view that all present-day Native Americans stem from a single migration wave6–8, consistent with more complex scenarios proposed by other studies9–15. In particular, the three distinct Asian lineages we detect: "First American", "Eskimo-Aleut," and a separate one in the Na-Dene speaking Chipewyan, are consistent with a three wave model proposed by Greenberg, Turner and Zegura based mostly on dental morphology and a controversial interpretation of the linguistic data9. However, our analyses also document extensive admixture between First Americans and the subsequent streams of Asian migrants, which was not predicted by the model of Greenberg and colleagues, such that Eskimo-Aleut speakers and the Chipewyan derive more than half their ancestry from First Americans. Further insights into Native American history will benefit from the application of analyses similar to those performed here to whole genome sequences and to data from the many admixed populations in the Americas that do not self-identify as Native28–30.
Methods Summary
The DNA samples we analyzed were collected over several decades. For each sample, we verified that informed consent was obtained consistent with studies of population history and that institutional approval had been obtained in the country of collection. Ethical oversight and approval for this project was provided by the NHS National Research Ethics Service, Central London committee (Ref # 05/Q0505/31). The dataset is based on merging Illumina SNP array data newly generated for this study (including 273 Native American samples) with data from six other studies. We applied stringent data curation and validation procedures to the merged data set. We used local ancestry inference software to identify genome segments in each Native American and Siberian sample without evidence of recent European or African admixture, and created a dataset that masked segments of potentially non-Native origin. Most of analyses are performed on the masked data set; however, we confirmed major inferences on a subset of 163 Native American samples that had no evidence of European or African admixture. We used model-based clustering and neighbor-joining trees to obtain an overview of population relationships, and then tested whether proposed sets of four populations were consistent with having a simple tree relationship using the 4 Population Test, which we generalized via a Hotelling T-test. We analyzed the correlation in allele frequency differences across populations to infer the minimum number of gene flow events that occurred between Asia and America. We fit the patterns of correlation in allele frequency differences to proposed models of history—Admixture Graphs—that can incorporate population splits and mixtures.
Methods
DNA Samples
The samples analyzed here were collected for previous studies over several decades. We reviewed the documentation available for each population to confirm that all samples were collected with informed consent encompassing genetic studies of population history. Institutional approval for use of each set of samples in such research was obtained prior to this study in the country of collection. Approval for this study was also provided by the NHS National Research Ethics Service, Central London REC 4 (Ref # 05/Q0505/31).
Genotyping
All samples were genotyped using Illumina arrays, and the data set analyzed here is the result of merging data from seven different sources (Note S1). The genotyping that was carried out specifically for this study was performed at the Broad Institute of Harvard and MIT, with the exception of 10 Chipewyan samples that were genotyped at McGill University (no systematic differences were observed between these and the 5 Chipewyan samples genotyped at the Broad Institute). Table S3 specifies details for each of the 493 Native American samples. A total of 419 samples were genotyped from genomic DNA, and 74 from whole genome amplified (WGA) material prepared used the Qiagen REPLI-g midi kit.
Data curation
We required >95% genotyping completeness for each SNP and sample. We merged the data specifically obtained for this study with six other datasets. We further removed samples that were outliers in PCA relative to others from their group, showed an excess rate of heterozygotes compared to the expected rate from the frequency in the population, or had evidence of being a second degree relative or closer to another sample in the study (Note S1). Genetic analyses summarized in Note S1 found substructure in some populations (Maya, Zapotec and Nganasan); we use labels like “Maya1” and “Maya2” to indicate the subgroups.
Masking of genomic segments containing non-Native American ancestry
For each Native American individual, we used HAPMIX31 to model their haplotypes with two ancestral panels: (i) “Old World” populations (a pool of 408 Europeans and 130 West Africans) and (ii) “Native” populations, a pool of all Native American and Siberian populations. Haplotype phase in the ancestral panel, which is necessary for HAPMIX, was determined by phasing both pools of samples together using Beagle32. We masked genome segments that had an expected number of >0.01 non-Native American chromosomes according to HAPMIX, thus retaining segments with an extremely high nominal probability of being homozygous for Native ancestry. Multiple analyses reported in the supplementary materials indicate that our masking procedure produces inferences about history that are consistent with those based on unadmixed samples.
Population structure analysis, FST and Neighbor Joining tree
We used EIGENSOFT to carry out PCA and compute pair-wise population FST33. Clustering was performed using ADMIXTURE16. A Neighbor Joining34 tree based on FST was built using POWERMARKER35.
Linguistic categories
We used Greenberg’s classification17,36. We considered using alternative classifications; however, others (such as Campbell’s37) do not hypothesize links among languages at a deep enough level to compare to genetic relationships on a continent-wide scale.
Correlating geography with population diversity
Euclidean distances from the Bering Strait (64.8N 177.8E) and the location of each population (Table S1) were calculated using great arc distances based on a Lambert azimuthal equal area projection. Least-cost distances between the same points were computed using PATHMATRIX19, which allows us to build a spatial cost map incorporating the coastal outline of the Americas. We compared the following coastal/inland relative costs: 1:2, 1:5, 1:10, 1:20, 1:30, 1:40, 1:50, 1:100, 1:200, 1:300, 1:400, and 1:500. We computed a Pearson correlation coefficient between heterozygosity for each population and their least cost distance from the Bering Strait (Note S5).
Documentation of at least three streams of gene flow from Asia to America
We used the 4 Population Test to assess whether proposed sets of four populations were consistent with a tree. For each of 52 Test Populations, we assessed their consistency with deriving from the same Asian source population as southern Native Americans by studying statistics of the form f4(Southern Native American, Test Population; Outgroup1, Outgroup2), where the two outgroups are the 45(=10×9/2) possible pairs of 10 Asian outgroups (Han Chinese and 9 Siberian populations with at least ten samples each and not including the Naukan and Chukchi who we showed have some First American ancestry due to back-migration across the Bering Strait, making them inappropriate as outgroups (Note S6 and Note S7)). We applied a Hotelling T-test to assess whether the ensemble of all possible f4 statistics was consistent with zero after taking into account their correlation structure, resulting in a single hypothesis test for whether the Test Population is consistent with having the same relationship to the panel of Asian populations as the set of Southern Native American samples used as a reference group. We also generalized this test by studying the matrix of all f4 statistics simultaneously and computing statistics that measure whether the f4 statistics seen in proposed sets of Native American populations are consistent with deriving from a specified number of Asian migrations. In Note S6 we show that if there have been N distinct streams of gene flow from Asia into the Americas, then the matrix of all possible f4 statistics can have rank no more than N-1 (ignoring sampling noise). The case N=1 reduces to calculating a Hotelling T2 statistic. We also developed a likelihood ratio test, generalizing the Hotelling T-test, to evaluate the statistical evidence for larger values of N, allowing us to estimate the minimum number of exchanges between Asia and America that are needed to explain the genetic data.
Admixture Graphs
We used the Admixture Graph (AG) framework24 to fit models of population separation followed by mixture to the data. An AG makes predictions about the correlations in allele frequency differentiation statistics (f-statistics) that will be observed among all pairs, triples, and quadruples of populations24, and these can be compared to the observed values (along with a standard error from a Block Jackknife) to test hypotheses about population relationships (Note S7). We do not have a formal goodness-of-fit test for whether a given AG fits the data correcting for the number of hypotheses tested and number of degrees of freedom, but use two approximations. First, we examine individual f-statistics, searching for ones that are >3 standard errors from expectation indicative of a poor fit. Second, we compute a χ2 statistic for the match between the observed and predicted f-statistics, taking into account the empirical covariance matrix among the f-statistics computed based on a Block Jackknife. This results in a nominal P-value, but it is unclear to us at present whether the empirical covariance matrix that we obtain can be equated with the theoretical covariance matrix that is needed to compute a formal P-value. For a fixed graph complexity (number of drift edges and admixture weights), however, we can compare the χ2 value for different admixture graphs to obtain a formal test for whether some topologies are significantly better fits; this results in the coloring of edges in Figure 3 showing which shows alternative insertion points for admixture edges are equally good fits.
Supplementary Material
Acknowledgments
We are grateful to the volunteers who provided the samples that made this study possible. We thank E.D. Ruiz for assistance in the collection involving the Mixtec, Zapotec and Mixe; A. Carnevale, M. Crawford, M. Metspalu, F.C. Nielsen, X. Soberon, R. Villems and E. Willerslev for facilitating sharing of data from Mexican, Siberian and Arctic populations; and C. Stevens and A. Crenshaw for assistance with genotyping. We thank P. Bellwood, D. Bolnick, K. Bryc, J. Diamond, T. Dillehay, R. Gonzalez-José, M. Hammer, J. Hill, B. Kemp, S. LeBlanc, D. Meltzer, P. Moorjani, A. Moreno-Estrada, B. Pakendorf, J. Pickrell, M. Ruhlen, D.G. Smith, M. Stoneking, N. Tuross and A. Williams for thoughtful critiques and valuable discussions. Support was provided by NIH grants NS043538 (A.R.-L.), NS037484 and MH075007 (N.B.F.), GM079558 (A.D.), GM079558-S1 (A.D.), GM057672 (K.K.K. & J.R.K.), HG006399 (D.R., N.P. & A.L.P); by an NSF HOMINID grant 1032255 (D.R. & N.P.); by a Canadian Institutes of Health Research grant (D.L.); by a Universidad de Antioquia CODI grant (G.B.); by a FIS grant PS09/02368 (A.C.); by a MICINN grant SAF2011-26983 (A.S.); by a Wenner-Gren Foundation Grant ICRG-65 (A.D. & R.S.); by Russian Foundation for Basic Research Grants 06-04-048182 (R.S.) and 02-06-80524a (L.O.); by a Siberian Branch Russian Academy of Sciences Field Grant (L.O.); by a PIR CNRS Amazonie grant (J.-M.D.); and by startup funds from Harvard Medical School (D.R.) and the Harvard School of Public Health (A.L.P.).
Footnotes
Author contributions. D.R., N.B.F., A.L.P. and A.R-L. conceived the project. D.R., N.P., D.C., A.T., S.M., N.R. and A.R-L. performed analyses. D.R. and A. R.-L. wrote the paper with input from all the co-authors. A.R.-L. assembled the sample collection, directed experimental work, and coordinated the study. All other authors contributed to collection of samples and data.
Data access. The data analyzed here are available for non-profit research on population history under an inter-institutional data access agreement with the Universidad de Antioquia, Colombia. Queries regarding data access should be sent jointly to G.B. (gbedoya@quimbaya.udea.edu.co) and A.R.-L. (a.ruizlin@ucl.ac.uk).
References
- 1.Cavalli-Sforza LL, Menozzi P, Piazza A. The History and Geography of Human Genes. Princeton, UP: 1994. [Google Scholar]
- 2.Meltzer DJ. First peoples in a new world : colonizing ice age America. University of California Press; 2009. [Google Scholar]
- 3.Goebel T, Waters MR, O'Rourke DH. The late Pleistocene dispersal of modern humans in the Americas. Science. 2008;319:1497–1502. doi: 10.1126/science.1153569. [DOI] [PubMed] [Google Scholar]
- 4.Dillehay TD. Probing deeper into first American studies. Proc. Natl. Acad. Sci. U. S. A. 2009;106:971–978. doi: 10.1073/pnas.0808424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Rourke DH, Raff JA. The human genetic history of the Americas: the final frontier. Curr. Biol. 2010;20:R202–R207. doi: 10.1016/j.cub.2009.11.051. [DOI] [PubMed] [Google Scholar]
- 6.Tamm E, et al. Beringian standstill and spread of Native American founders. PLos ONE. 2007:1–6. doi: 10.1371/journal.pone.0000829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kitchen A, Miyamoto MM, Mulligan CJ. A three-stage colonization model for the peopling of the Americas. PLoS ONE. 2008;3:e1596. doi: 10.1371/journal.pone.0001596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fagundes NJ, et al. Mitochondrial population genomics supports a single pre-Clovis origin with a coastal route for the peopling of the Americas. Am. J. Hum. Genet. 2008;82:583–592. doi: 10.1016/j.ajhg.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greenberg JH, Turner CG, Zegura SL. The Settlement of the Americas: A Comparison of the Linguistic, Dental, and Genetic Evidence. Curr. Anthrop. 1986;27:477–497. [Google Scholar]
- 10.Lell JT, et al. The dual origin and Siberian affinities of Native American Y chromosomes. Am J.Hum.Genet. 2002;70:192–206. doi: 10.1086/338457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bortolini MC, et al. Y-chromosome evidence for differing ancient demographic histories in the Americas. Am. J. Hum. Genet. 2003;73:524–539. doi: 10.1086/377588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Volodko NV, et al. Mitochondrial genome diversity in arctic Siberians, with particular reference to the evolutionary history of Beringia and Pleistocenic peopling of the Americas. Am J Hum Genet. 2008;82:1084–1100. doi: 10.1016/j.ajhg.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ray N, et al. A statistical evaluation of models for the initial settlement of the american continent emphasizes the importance of gene flow with Asia. Mol. Biol. Evol. 2010;27:337–345. doi: 10.1093/molbev/msp238. [DOI] [PubMed] [Google Scholar]
- 14.de Azevedo S, et al. Evaluating microevolutionary models for the early settlement of the New World: the importance of recurrent gene flow with Asia. Am. J. Phys. Anthropol. 2011;146:539–552. doi: 10.1002/ajpa.21564. [DOI] [PubMed] [Google Scholar]
- 15.Perego UA, et al. Distinctive Paleo-Indian migration routes from Beringia marked by two rare mtDNA haplogroups. Curr. Biol. 2009;19:1–8. doi: 10.1016/j.cub.2008.11.058. [DOI] [PubMed] [Google Scholar]
- 16.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruhlen M. A Guide to the World's Languages. Stanford University Press; 1991. [Google Scholar]
- 18.Schroeder KB, et al. A private allele ubiquitous in the Americas. Biol. Lett. 2007;3:218–223. doi: 10.1098/rsbl.2006.0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ray N. PATHMATRIX: a geographical information system tool to compute effective distances among samples. Mol. Ecol. Notes. 2005;5:177–180. [Google Scholar]
- 20.Wang S, et al. Genetic variation and population structure in native Americans. PLoS Genet. 2007;3:e185. doi: 10.1371/journal.pgen.0030185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang NN, et al. Contrasting patterns of nuclear and mtDNA diversity in Native American populations. Ann Hum Genet. 2010;74:525–538. doi: 10.1111/j.1469-1809.2010.00608.x. [DOI] [PubMed] [Google Scholar]
- 22.Brown MD, et al. mtDNA haplogroup X: An ancient link between Europe/Western Asia and North America? Am.J.Hum.Genet. 1998;63:1852–1861. doi: 10.1086/302155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karafet TM, et al. Ancestral Asian source(s) of new world Y-chromosome founder haplotypes. Am.J.Hum.Genet. 1999;64:817–831. doi: 10.1086/302282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461:489–494. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rasmussen M, et al. Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature. 2010;463:757–762. doi: 10.1038/nature08835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balter M. Archaeology. The peopling of the Aleutians. Science. 2012;335:158–161. doi: 10.1126/science.335.6065.158. [DOI] [PubMed] [Google Scholar]
- 27.Cooke R. Prehistory of native Americans on the Central American land bridge: Colonization, dispersal, and divergence. J Archaeol. Res. 2005;13:129–187. [Google Scholar]
- 28.Wang S, et al. Geographic patterns of genome admixture in Latin American Mestizos. PLoS Genet. 2008;4:e1000037. doi: 10.1371/journal.pgen.1000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bryc K, et al. Colloquium paper: genome-wide patterns of population structure and admixture among Hispanic/Latino populations. Proc. Natl. Acad. Sci. U. S. A. 2010;107(Suppl 2):8954–8961. doi: 10.1073/pnas.0914618107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wall JD, et al. Genetic variation in Native Americans, inferred from Latino SNP and resequencing data. Mol. Biol. Evol. 2011;28:2231–2237. doi: 10.1093/molbev/msr049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Price AL, et al. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet. 2009;5:e1000519. doi: 10.1371/journal.pgen.1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:2074–2093. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol.Biol.Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 35.Liu K, Muse SV. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21:2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- 36.Greenberg JH. Language in the Americas. Stanford University Press; 1987. [Google Scholar]
- 37.Campbell L. American Indian languages: the historical linguistics of Native America. Oxford University Press; 1997. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.