Abstract
It is increasingly appreciated that phenotypic stochasticity plays fundamental roles in biological systems at the cellular level and that a variety of mechanisms generates phenotypic interconversion over a broad range of time scales. The ensuing dynamic heterogeneity can be used to understand biological and clinical processes involving diverse phenotypes in different cell populations. The same principles can be applied, not only to populations composed of cells, but also to populations composed of molecules, tissues, and multicellular organisms. Stochastic units generating dynamic heterogeneity can be integrated across various length scales. We propose that a graphical tool we have developed, called a metronomogram, will allow us to identify factors that suitably influence the restoration of homeostatic heterogeneity so as to modulate the consequences of dynamic heterogeneity for desired outcomes.
Keywords: Homeostatic heterogeneity, evolution
1. Introduction
In Liao et al. A, we developed a conceptual tool for understanding and utilizing dynamic heterogeneity in cancer therapy. Collisions between biomolecular components reshuffling in an ongoing way can generate stochasticity at the subcellular level. These stochastic fluctuations in mRNA and protein level can result in the generation of heterogeneity within a cell population, as well as reversible transitions between multiple states. This phenotypic interconversion tends to restore a population to its previous composition after it has been depleted of specific members. We called this tendency homeostatic heterogeneity. We used these insights to develop a tool (metronomogram) to help understand how to optimize therapeutic dosing schedules on a patient-individualized basis when targeting cells undergoing back and forth transitions between phenotypes of relative drug-sensitivity and drug-resistance. For simplicity, we made examples in Liao et al. A specific in three ways. (i) The sources of stochastic phenotypic fluctuations were non-genetic fluctuations in mRNA and protein levels. (ii) We used transitions between drug-sensitive and drug-resistant cells as our primary example of phenotypic interconversion. (iii) Our analysis assumed that stochastic fluctuations occurred within individual cells.
The purpose of this paper is to emphasize that an understanding of dynamic heterogeneity and its biological and clinical consequences is not restricted to these specific examples. In section 2, we describe a collection of unifying categories of mechanisms that can generate phenotypic stochasticity. Non-genetic proteomic fluctuation is only one example in this list. These examples span a broad range of time scales for phenotypic interconversion. In section 3, we demonstrate that the concepts of dynamic heterogeneity and the metronomogram can be used to understand phenotypes besides those directly related to drug-resistance. We discuss phenotypic transitions in biofilms, metastasis and dissemination of tumor cells, and oncogene overexpression. Notably, stochastic fluctuations need not be contained within individual cells. Section 4 provides an example in which the phenotypically interconverting units are individual organisms. Section 5 discusses examples of multicellular systems where the “stochastic units” of interest undergoing phenotypic interconversion may be an integrated collection of cell clusters or tissues or individuals rather than individual cells. Because cells in multicellular communities are connected by a variety of signaling loops, the stochastic fluctuations in one cell can spill over to modulate the phenotypes of other cells in the microenvironment. In other words, stochastic fluctuations can be integrated across various scales of length and population number.
Generalizing our understanding of dynamic heterogeneity in these ways expands the number of mechanisms and molecular targets we can potentially manipulate to control population heterogeneity and population numbers. In section 6, we use the metronomogram to propose a strategy for uncovering factors that modulate the time scales of phenotypic interconversion in a “proliferation-independent” fashion. This particular form of manipulation would allow a system to move between the regions above and below the diagonal of the metronomogram. In this way, the consequences of dynamic heterogeneity for a population can be changed from extinction to long-term survival or vice versa. This strategy suggests an alternative to the “whack-a-mole” approach to cancer treatment. Rather than engineering a therapeutic modality (drug, surgery, radiation, etc.) and schedule to address each potential molecular target that might present itself in the tumor cell population, one could, instead, coax the tumor cell population to schedule the dynamics of its phenotypic fluctuations so that a proposed therapeutic schedule becomes effective. The biologic agents used for such kinetic manipulation need not themselves be traditional cytotoxic or cytostatic chemotherapeutic drugs.
2. Multiple mechanisms can generate phenotypic stochasticity
In Liao et al. A, we offered a perspective on the origins of stochasticity by discussing biochemical reactions taking place in individual cells. These examples are not exclusive. Instead, they are members of a broader collection of physical mechanisms that can dynamically generate heterogeneity in the phenotypes of a population of cells. While specific models vary in molecular detail, many mechanisms for the generation of stochasticity can be understood in terms of the small collection of unifying categories that we now describe using Figure 1.
Figure 1.
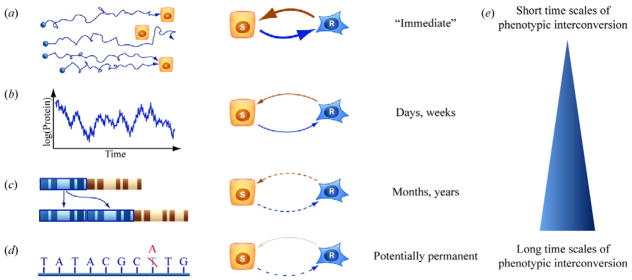
Regeneration of phenotypic heterogeneity occurs according to different mechanisms and different timescales. (a) Stochastic delivery of therapeutic agent. (b) Proteomic fluctuations. (c) Large-scale genetic alterations. (d) Point mutation. (e) From the almost “instantaneous” apparent drug resistance that results from extrinsic exposure to drug to the extended time scale of years observed with gene amplification or other chromosomal changes, the rates of interconversion span a remarkable range of time scales. Some have postulated that this is not by chance but allows a contingency plan for a wide range of challenges.
2.1. Stochastic encounters with (extrinsic) soluble factors
In Liao et al. A, we described variation in phenotype owing to fluctuations in the abundances of molecular species within individual cells. However, variation in perceived phenotype can also be the result of fluctuations in the abundances of molecular species extrinsic to individual cells. In the example of exposure to a drug in Figure 1(a), some cells survive and other cells are killed during a finite duration of exposure to the drug as a consequence of cell-extrinsic stochasticity in drug delivery at the microscopic scale. Tortuous vasculature, heterogeneous blood flow, hypoxia, extracellular acidosis, and high interstitial fluid pressure all challenge homogeneous delivery of drug to tumor cells. As those molecules of drug that do access the tumor undergo Brownian motion, temporary local gaps appear allowing some cells to “slip through” unscathed, at least until the next cloud of drug molecules arrives with a spatial arrangement likely to differ at the microscopic scale. The extracellular fluid can be stirred and washed by mechanisms independent of the cell cycle, so the timescales over which apparently drug-resistant cells can become apparently drug sensitive are potentially “instantaneous.” The mathematical expression of this concept is the famous pharmacologic exp(−kt) law of cell kill [1]. This “law” predicts that cell-death increases arbitrarily with increasing drug concentration given in a fixed time period. In other words, the concept that “more is better” underlying maximum-tolerated dosing strategies referred to in Liao et al. A can be rationalized by assuming that fluctuations between apparent states of drug-sensitivity and drug-resistance are exclusively effected by fluctuations in delivery. As we have described, drug-sensitivity and –resistance are effected by fluctuations other than that of drug exposure. These considerations are not exclusive to drug therapy. A similar view of stochasticity applies to the chance encounters between cells and other soluble signaling factors such as hormones or growth factors.
2.2. Proteomic fluctuations
Figure 1(b) refers to the concept that fluctuations in abundances of molecular species result from stochastic variation in the time intervals between biochemical reactions inside individual cells. This was the primary model in Liao et al. A. As we will explain, time scales for such cell-intrinsic fluctuations can be similar in magnitude to time scales for cell division. Thus, some of these “proteomic” fluctuations, or examples of “non-genetic individuality” as they are sometimes called, can be characterized by time scales measured in days or weeks.
2.2.1. Time scale analysis for proteomic fluctuations
To illustrate these points, we provide three examples of mechanisms that can relate the time scales for the generation of heterogeneity to the time scales for population expansion (Figure 2). We use these examples to develop qualitative estimates of orders of magnitude. In physicists’ language, we are performing “back-of-the-envelope” calculations.
Figure 2.
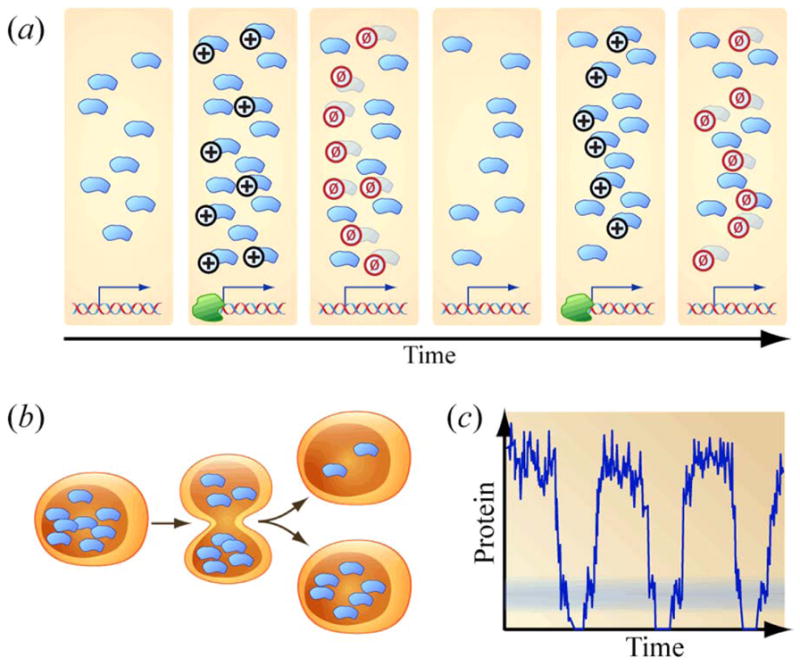
Mechanisms by which timescales for the generation of phenotypic heterogeneity may be similar to timescales for population expansion. (a) Periodic dilution. (b) Partition noise during cell division. (c) Periodic decrease in mRNA and protein levels.
The first example we will describe involves the dilution of proteins owing to cell division. Sigal et al. described the dynamics of the dissipation of fluctuations in single-cell protein levels using an autocorrelation function [2]. In their supplemental theory, the authors calculated the “mixing time” for this auto-correlation function for a simple “birth-death” model of protein translation and degradation. In their example, the time scale for cell-division defined the time scale for protein “memory.” We provide a heuristic for understanding this result in Figure 2(a). At the left, a cell happens to contain 8 copies of a protein inherited from a just-completed mitotic event. In the next snapshot, transcription and translation occur, adding 8 more proteins to the proteomic atmosphere. The cell then enters mitosis, with half of the 16 copies of protein lost to the sister cell not shown. The cell under study again has only 8 copies of protein immediately following this mitotic event. In this cell, biochemistry is stochastic, and whereas the cell has just replenished its proteome with 8 new copies of protein, only 6 new copies of protein will next be generated. For simplicity, we assume in this example that proteins are partitioned precisely during mitosis. This means that 7 of 14 proteins are lost to the sister cell not shown. When the primary mechanism of protein loss is cell division, a proteome that starts out by chance rich, or alternatively, poor, in a particular protein can remain in such an outlier state for roughly a generation before mitosis reduces the copy numbers of cytosolic constituents by a factor of two. Proteins newly synthesized after mitosis are then added alongside these remnant populations of proteins. The molecular composition of the cell now reflects an average of the protein production rates before and after mitosis. This “averaging” process partially dilutes away those fluctuations generated preceding mitosis. The cell-division time provides an order of magnitude for the memory time of the cell. More sophisticated gene regulatory network architectures, i.e. relying on feedback loops, can prolong the protein-level memories of cells, and rapid protein degradation, intervening between cell division events, can hasten the loss of proteomic memory.
We provide a second example involving “partitioning” noise. Huh and Paulsson have remarked that fluctuations in the levels of biological molecules could result from partitioning of molecules between daughter cells according to a binomial (coin-toss) process during mitosis [3]. In Figure 2(b), a cell initially containing 8 copies of a protein divides. The individual copies of protein randomly circulate throughout the increasingly hourglass-shaped cell. At the moment when the daughters separate, there might by chance be 2 copies of protein in the upper daughter and 6 in its sister. Even if the duration of the cell cycle and the rates of transcription and translation were precisely reproducible over generations, partitioning noise would introduce fluctuations into the proteome on a time scale of once a generation. This is beautifully illustrated in the unequal partitioning of double minute chromosomes (DMs) which carry amplified copies of the DHFR gene and confer drug resistance [4]. The examples in Figure 2(a) and (b) are similar. Both ideas are based on (i) an assumption that stochasticity is present and (ii) an assumption that there is a time scale at which proteins (or organelles) from one mother cell are assorted between two daughters. The underlying stochastic events in each differ, but both perspectives lead to the same time scale for proteomic fluctuations. Indeed, Huh and Paulsson explained that their perspective of partitioning noise could quantitatively accommodate experimental measurements previously rationalized in terms of stochastic gene transcription.
In a third example, we discuss the consequences of protein-level fluctuations during the cell cycle. Various protein levels are upregulated and downregulated as the cell moves through the phases of the cell cycle. Any proteins that are highly upregulated during one phase and essentially shut off during another must periodically pass through an intermediate band of small, but finite copy numbers as shaded in Figure 2(c). During these times, the relative magnitude of stochastic fluctuations may be large relative to expected average values. Such small protein copy numbers would be achieved temporarily both while the protein level decreased and while the protein level subsequently increased, providing any downstream molecular networks access to a random number generator, i.e. access to a “roll of dice,” at least twice during each cell cycle. Recent experimental work in synthetic biology suggests that biological circuitry may have evolved in order to introduce cyclic noise in this way. The competence circuit of the bacterium B. subtilis has been modeled as an excitable system with a self-activating protein ComK that inhibits ComS, which itself activates ComK. When the cell enters a “cycle” of excitation, the level of ComS collapses. The average durations and likelihoods of entering competence for this circuit are reproduced in a synthetic circuit topology that, in contrast, causes the regulatory partner of ComK to increase following excitation. Why, then, does the native circuit rely on a protein-level collapse? Çağatay et al. have suggested that the collapse of ComS levels provides a source of molecular noise that leads to variability in competence duration and thus an evolutionary advantage in fluctuating environments [5]. In another example, Spudich et al. have hypothesized that a cyclic decrease in the levels of some proteins could underlie the highly variable duration of the G1-phase of the cell cycle and result in rapid asynchronization of initially synchronized cell populations [6]. Some phenotypes are cell-cycle specific. For example, increased sensitivity to some chemotherapeutic agents requires exiting G1. Randomness in the duration of G1 would manifest as randomness in the time that elapses before a cell acquires any such phenotype. In the supplemental materials, we discuss the consequences of this view for the scheduling of doses of cell-cycle-specific agents using the metronomogram. It may prove fruitful to continue studying oscillatory and excitable motifs in circuits that regulate cell cycle to determine the prevalence of topologies that cyclically reduce the abundance of proteins to copy numbers near unity.
2.2.2. Experimental examples
The preceding heuristic discussions suggest that some fluctuations in protein levels may be associated with time scales similar to the cell-division time. In fact, some time scales for the generation of phenotypic heterogeneity are similar to time scales for proliferation as seen in a variety of experimental examples.
In the study by Sigal et al., the “mixing” times τm for 20 proteins were measured in individual cells from a human lung-cancer cell line [2]. The proteins were involved in diverse functions including apoptosis, transcriptional regulation, chromatin remodeling, and cold response. Thus, it may be unsurprising that the mixing times varied. Interestingly, however, the variation that was reported covered a range from τm = 0.8 to 2.6 generations. The time scale for proteomic fluctuations and the time scale for cell replication shared the same order of magnitude.
In another example, Chang et al. investigated the generation of phenotypic heterogeneity in populations of “EML” progenitors in the hemapoietic system of the mouse. Purified subpopulations with low, intermediate, and high levels of the marker Sca-1 were obtained from an initially broad distribution. The repopulation of the initial distribution from these purified subpopulations was visible within days, with saturation occurring by about 2 weeks [7]. In this example, re-establishment of heterogeneity in Sca-1 levels corresponded to re-establishment of heterogeneity in time rates for realizing different cell fates. In additional examples, rapid return toward homeostatic heterogeneity (well underway within 3 days) has also been observed in studies of cancer “stem” cells in mammary cell lines [8], [9]. Taken together, these reports suggest that various cell populations can achieve time scales for phenotypic fluctuation of the same order of magnitude as the time scales for cell replication, consistent with the heuristic examples from the previous subsection.
We have just considered conceptual and experimental examples suggesting that time scales for cell replication and phenotypic conversion can often be similar. In the supplemental materials, we use this observation, along with equations (1) and (2) from Liao et al. A, to show that high-frequency dosing may in some cases be beneficial. It also should be noted that time scales for exhibiting stochastic fluctuations can sometimes greatly exceed the time scales for cell replication, as is illustrated in Section 2.3.
2.3. Genetic alterations
In the examples above (Figure 1(a)and (b)), we discussed non-genetic sources of cell-cell heterogeneity in drug-response. The mathematical discussion in this paper and Liao et al. A are agnostic to the molecular origins of phenotypic heterogeneity, so the same conceptual lens can be applied to heterogeneity produced through genetic or epigenetic variation.
For example, gene amplification is reversible and can occur over months and even years (Figure 1(c)) [10], [11]. When present within a homogeneously staining region (HSR) carried on an autosome, the gene copy number fluctuates up and down through mechanisms like homologous recombination and deletion. Furthermore, we also illustrated how increased gene copy numbers carried on fragments of chromatin called “double minute” (DM) chromosomes could also exhibit heterogeneity based on unequal partitioning of these DM chromosomes during mitosis. DM chromosomes lack a centromere and thus lack a mechanism to partition equally at mitosis. The time scales for these two mechanisms is different and these characteristics have been studied as stable drug resistance (when DHFR genes are carried on HSRs) and unstable drug resistance (when the genes are carried on DMs).
Of the mechanisms contributing to drug resistance outlined in Figure 1, genetic point mutations are probably the least likely to reverse and most commonly known (Figure 1(d)). Previous authors have outlined a variety of potential clinical strategies for treating patients in the presence of mutations. As described in Liao et al. A, these strategies include targeting the protein products of mutated genes, exploiting the fitness costs of mutations conferring drug resistance, and identifying cell populations in the microenvironment that retain drug-sensitivity even when the epithelial subpopulation acquires a mutation that confers resistance. We proposed that an understanding of homeostatic heterogeneity would improve our ability to pursue these strategies beneficially.
Additional strategies include prevention of mutation and the use of ecological predation. A frequent goal of computational dose-scheduling studies is to optimize the dosing schedule to minimize the risk of acquiring a mutation that confers resistance [12]. This is often attempted by minimizing the size of the proliferative population for as long as possible. Alternatively, Silva et al. have suggested allowing a drug-sensitive cell population to survive so that it can compete with or control a subpopulation harboring a resistance mutation [13]. For both of these strategies, an understanding of the kinetics of phenotypic interconversion will be necessary, either to most effectively deplete the target population before it acquires a mutation or to avoid completely eradicating a useful cellular subpopulation.
The mechanisms we have described in Figure 1 lead to phenotypic interconversion over a wide variety of time scales, as indicated by the wedge in Figure 1(e). Extrinsic stochasticity in the delivery of soluble factors can occur rapidly compared to the cell-cycle time, or “instantaneously.” Proteomic fluctuations and alterations in gene copy number span intermediate time scales ranging from days, to weeks, to months, and years, while point mutations are potentially permanent. In addition to the variation in time scales seen among mechanisms, there is also variety in the time scales that a given mechanism can generate. To understand why, we use Figure 3 to address a simplification from Liao et al. A. While we previously considered the toy model in which transitions occurred between a pair of phenotypes (Figure 3(a)), many phenotypes can be represented by a continuous spectrum or graded series of discrete states (Figure 3(b)). Suppose that the rate coefficients connecting states 1 and 2 to each other are identical to the rate coefficients connecting states 2 and 3, states 3 and 4, etc. Even in this situation, a variety of time scales can be associated with the system by considering transitions between different pairs of phenotypic states. The time scales for converting from state 1 to 5 and vice versa are slow compared to the time scales for interconversion between states 1 and 2 because no transitions directly connect states 1 and 5. For a cell in state 1 to reach state 5, it must first pass through state 2. Previous experimental studies have explored this relationship and connected these various phenotypic states to clinical consequence [14], [10].
Figure 3.
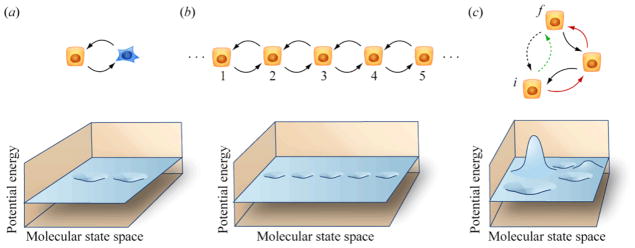
Topologies of transitions that connect interconverting phenotypes. The dynamics of systems in molecular abundance space are often represented by the motions of weights sliding around potential energy surfaces at roughly terminal velocity in an ambient medium, with thermal excitation. (a) Reversible transitions proceed back and forth between a pair of states in a bistable system. This corresponds to two energy valleys. (b) Transitions can occur in stepwise fashion along a continuous spectrum or graded discrete collection of states. (c) Higher-dimensional topologies introduce the possibility of connecting an initial state, i, to a final state, f, through multiple paths (i.e. green path going around energy peak and red path passing through intermediate valley). The typical time required to move from state i to state f along these two paths may differ.
The topology of the connections between phenotypic states is potentially more complicated than illustrated in Figure 3(b). Rather than being represented as a line, the network of phenotypic states may be more accurately characterized as a 2- or even higher-dimensional (Figure 3(c)). This provides a large number of ways to connect pairs of cell states, and thus a variety of time scales over which such conversions can occur.
3. Dynamic heterogeneity can be used to understand diverse biological and clinical processes
We developed the metronomogram in Liao et al. A to study the example of drug kill in a population of interconverting drug-resistant and drug-sensitive cells. However, the mathematical concepts and the broader perspective of phenotypic stochasticity described in Liao et al. A can also be applied to populations of cells undergoing interconversion between other phenotypes. We offer examples with cell adhesion in biofilms, adhesion and proliferative dormancy in metastasis, and tumorigenic phenotypes in oncogene-overexpressing cells.
3.1. Biofilm adhesion and dispersion
We provide an example of phenotypic transitions in biofilms in Figure 4(a). Individual cells in biofilms of the fungus C. neoformans switch between a “smooth” state and a “wrinkled” state [15]. The smooth state more strongly adheres to substrates. If a goal of a biofilm in a natural environment is to colonize distant niches, phenotypic conversion from the adherent to less-adherent state could produce “seeds” to be washed away, perhaps by occasional rainfall. Depending on environmental conditions, dispersing the entire biofilm at once might be unlikely to establish a colony. A more successful strategy may be to maintain a proliferating biofilm at the primary site from which a small non-adherent population can be periodically dispersed. In this case, the survival of the primary biofilm, and thus its ability to shed non-adherent cells long term, depends on exploring the lower half of the metronomogram (the area under the diagonal in Figure 4(b)). The survival of the biofilm occurs when the generation of adhesion heterogeneity is slower than population expansion. Otherwise the biofilm shrinks as it is repeatedly washed away. In analogous financial terms, resisting the temptation to immediately squander a principal can reward the investor with the ability to live off interest long term.
Figure 4.
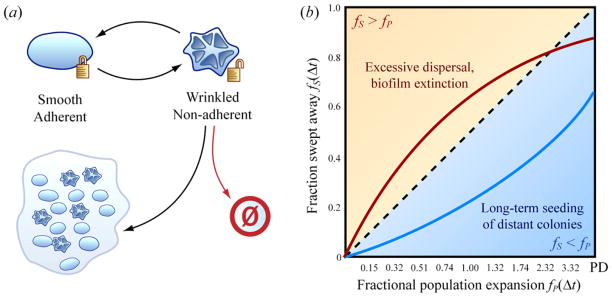
Simplified model for phenotypic switching and survival in biofilm communities. (a) Single cells of the fungus C. neoformans phenotypically switch in biofilms between simplified adherent and non-adherent states. Dispersing non-adherent cells can result in seeding new colonies, but can also result in failure to seed and cell death. (b) Excessively rapid generation of non-adherent cells corresponds to locations above the diagonal (fS > fP) in the metronomogram, corresponding to eventual extinction of the primary biofilm. If the generation of non-adherent cells is slow compared to cell population expansion, the dynamics of the population are described by positions in the metronomogram below the diagonal (fS < fP), corresponding to expansion of the population of the primary biofilm colony.
3.2. Metastasis and dissemination of tumor cells
It has been proposed that primitive multicellular ecologies, i.e. biofilms, offer a model for malignant tissues [16], [17]. Based on these insights, it may prove fruitful to extend the above discussion of biofilms to understand the dissemination of cells from primary tumors. Consistent with this possibility, previous authors have interpreted the inefficiency of metastasis formation in terms of the statistics of rare random events [18]. It has been suggested that the vast majority of disseminated cells may be metastatically unfit. However, by sheer numbers, some of these unfit cells nevertheless beat the odds and establish expanding colonies. Hence, the establishment of secondary colonies may be increased by maintaining a proliferating population at the primary site. It is known that cancer cells can undergo an “epithelial-to-mesenchymal” transition (EMT) toward more mesenchymal states with decreased adhesion to neighboring cells [19]. EMT is conventionally regarded as a deterministic response to external signals. While this can be interpreted as deterministic modulation by the environment of deterministic processes within cells, it is important to consider ways through which stochasticity can also contribute to EMT. Our group has reported evidence that the EMT phenotype is influenced by environmental signals which increase the probability for stochastic events in individual cells [20]. Additionally, the environment can provide, not only deterministic modulation of stochastic events in cells, but also a source of stochasticity itself. Local fluctuations in external signal concentration, as described in section 2.1, are examples of sources of noise that are referred to as gene “extrinsic” [21], [22]. Just as in our discussion of biofilms, primary tumor cell populations may need to “stay below the diagonal of the metronomogram” in order to increase the likelihood of establishing secondary colonies. In principle, this could be achieved by limiting the rate of EMT or by generating transitions in the opposite direction. In fact, the reverse “mesenchymal-epithelial-transition” (MET) occurs and has been considered as a possible prerequisite for disseminated cells to settle down into distant metastatic sites. In light of the current discussion, MET may also provide a cell population with a way to maintain an established primary tumor. A possible direction for continued investigation would be to evaluate the effects of hastening the loss of adhesion on the survival of the primary tumor and the establishment of metastatic lesions.
An experimental study of “tumor self-seeding” by Kim et al. suggests another situation in which staying “below the diagonal” could confer a survival benefit to a tumor cell population [23]. In this study, temporary dissemination and then re-infiltration by tumor cells can confer increased primary tumor growth and recruitment of supportive stroma. Retaining a portion of the primary tumor in situ provides homing signals for the circulating subpopulation.
Metastasis and tumor cell dissemination provide additional potential opportunities to apply the concept of homeostatic heterogeneity. Deakin et al. reported on phenotypic switching between an amoeboid mode of cell movement allowing movement through existing gaps in extracellular matrix and a mesenchymal mode involving proteolysis of extracellular proteins [24]. The authors suggest that phenotypic interconversion between these states, rather than permanent residence in either one state alone, increases invasiveness and establishment of metastases. In another example, Heyn et al. have suggested that some solitary disseminated cells may remain in a nonproliferative state before developing into late metastatic colonies [25]. Even when the dissemination of cells from a primary tumor occurs in a short-term burst, the timings of attempts to establish metastases may effectively be spaced out by stochastic variation in the durations that single cells remain in such “dormant” states. This may allow a disseminated population to sample a variety of time periods when it is not possible to determine which intervals of time would be conducive to establishing a metastatic colony. The term “dynamic heterogeneity” has been historically associated with the spontaneous interconversion of cells between highly-metastatic and non-metastatic phenotypes at rates much higher than “generally associated with point mutations and deletions” [26].
3.3. Oncogene overexpression: HER2 expression in cancer cells
In a third example, we consider the possibility of fluctuations in the single-cell levels of in HER2 (erbB2) surface receptor. HER2 is well-known as the target of the monoclonal antibody Trastuzumab [27]. However, its biologic role was known before it was selected as a drug target. Overexpression of HER2 with breast cancer cells is associated with aggressive cellular phenotypes including increased cell proliferation and survival, decreased dependence on estrogen for proliferation, poorer differentiation, increased invasiveness and motility, and increased angiogenesis [28], [29]. HER2 overexpression also correlates with decreased survival at the patient level [30].
Overexpression of the receptor is found in approximately 20–30% of human breast carcinomas, with 90–95% of these cases corresponding to amplification of the wild-type gene [29], [31] and a minority corresponding to transcriptional and translational mechanisms in the absence of gene amplification [27]. As we have discussed, both gene amplification and transcriptional and translational mechanisms can underlie phenotypic stochasticity and reversible transitions between phenotypes. Savelyeva et al. have noted the possibility that amplification “varies among members of the tumor cell populations” [31]. Immunohistochemistry also shows cell-cell variation in staining for the receptor [32].
Because increases in HER2 are associated with more aggressive phenotypes, it may be natural to recognize phenotypic conversion to HER2-overexpressing states as beneficial for the survival of tumor cells. However, phenotypic transitions that lower HER2 expression may also provide a survival benefit. HER2 overexpressing cells respond to heregulin ligand with biphasic growth [33]. Modest concentrations of heregulin increase proliferation and colony formation in vitro, but high concentrations inhibit growth. This inhibition is only seen in cells with HER2 overexpression. By contributing to a homeostatic heterogeneity that contains a low-HER2 subpopulation, phenotypic interconversion between relatively low- and high-HER2 level states may reserve a subpopulation of low-HER2 cells prepared to continue expanding even during pulses of heregulin that inhibit expansion of high-HER2 cells. Indeed, immunohistochemistry of heregulin shows spatial heterogeneity of ligand staining, consistent with the idea that the levels of heregulin in the microenvironments of single cells can vary [32]. Patients with lymphoid infiltration have a better prognosis if they are HER2 positive instead of HER2 negative, suggesting that HER2 may be targeted by the immune system [30]. Conversion of high-HER2 cells to transient states of low HER2 expression may allow some cells to escape immune clearance. As discussed in the examples for biofilms and metastasis, excessive motility may also counterintuitively impose a survival disadvantage for a multicellular colony. The ability to convert some cells with high-HER2 to a state with lower HER2 expression may rein in motile cells to prevent the tumor from “evaporating” from a primary site with established stromal support. The example of HER2 overexpression provides a reminder that it may be overly simplistic to assume that only one direction of phenotypic conversion provides a survival advantage for a cell population. Survival advantage may be provided by virtue of a combination of directions of phenotypic interconversion.
4. The stochastic units generating dynamic heterogeneity can be found at many population scales
4.1. Social modulation of task switching rates in harvester ants
In the three examples we provided above, we considered the consequences of dynamic heterogeneity for populations of cells. As we noted in Liao et al. A, these concepts apply to populations composed of molecules, cells, individuals, etc. In a fourth example, we discuss dynamic heterogeneity within populations composed of multicellular organisms. In ecological studies, various species of harvester ants can convert between phenotypes at different levels. “Task switching” can occur between states including patrolling, nest maintenance, foraging, and midden work (cleaning up debris) [34], [35]. The rates of transitions depend on the initial and final single-ant phenotype under consideration. In the species P. barbatus, the conversion of midden ants to foraging ants and the conversion of ants performing nest maintenance to patrolling ants are unidirectional. However, phenotypic conversion occurs both from patrolling to foraging and in the reverse direction. Phenotypic conversion also proceeds at a finer scale. Not all “foragers” in P. barbatus forage every day. Ants in the “inactive” state remain in the nest [36].
How might phenotypic switching provide a survival benefit to a population of ants? The dispersal of foragers can provide the benefit of acquiring food. However, the act of foraging outside of the nest also risks loss of ants to wind and predation. To remain “under the diagonal” in the metronomogram, the rate of converting to the actively foraging state must not be excessive. Because environmental conditions change, a conversion rate that barely avoids extinction in one situation could, in other cases, be unnecessarily cautious, forfeiting the efficient acquisition of food. This suggests that a species that adjusts the rates of phenotypic switching according to dynamic environmental conditions enjoys a survival advantage over a species with more primitive, non-social phenotypic switching. In fact, the rates at which individual members of P. barbatus convert from the inactive foraging state to the active foraging state is modulated non-linearly by the time-frequency of encounters with patrollers [36]. Low frequencies of encounters (up to once every 45 seconds) could indicate that patrollers have been swept away, suggesting that foragers should remain inactive. High frequencies of encounters (i.e. once every second) may indicate that all of the patrollers have hurried back to the nest to evade a threat and that under these circumstances the foragers should also remain inactive. Intermediate frequencies (i.e. once every 10 seconds) may indicate relative safety and signal that the foragers should activate foraging. These are the frequencies for which the rate of converting to the actively foraging state peaks.
In this discussion, social modulation of phenotypic conversion rates can confer a survival benefit, but populations of ants displaying phenotypic conversion in the absence of social modulation might also survive, though with less efficient collection of food. One may ask whether switching between active and inactive foraging states evolved at the same time as social mechanisms for modulating interconversion rates, or whether, instead, primitive switching evolved first, to be subsequently refined by the addition of social control. This is an example of how the concepts described in Liao et al. A can point to directions for research, not only in the clinical treatment of human disease, but also in ecology and evolutionary biology.
5. The stochastic units generating dynamic heterogeneity can be integrated
In the Markov model illustrated using roulettes in figure 2(d) of Liao et al. A, stochastic fluctuations are depicted as though they occur independently in different cells. One cell’s spin of a wheel of fortune is not affected by the spins of other wheels at the same time, and vice versa. In other words, stochasticity is cell intrinsic (Figure 5(a)). However, this perspective is likely to be often an oversimplification. Just because a cell is depicted in a cartoon as a well-defined container does not mean that it is “statistically” isolated.
Figure 5.
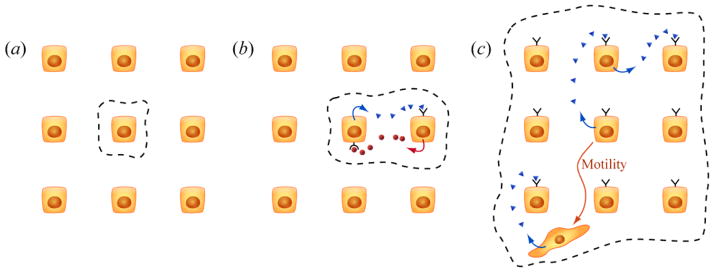
Stochastic fluctuations can be integrated at various scales. (a) Some fluctuations in the abundances of some molecules may be localized to individual cells. These fluctuations are cell-intrinsic. (b) Local signaling may propagate the effects of fluctuations in molecular levels in small clusters of cells. (c) A phenotypic fluctuation originating in a single cell may cause fluctuations in the phenotypes of other cells in the tissue at large. Molecular signals may be relayed by non-motile cells, as well as be carried to distant sites by motile cells.
Molecular fluctuations may propagate through clusters of cells connected by paracrine signaling (Figure 5(b)). Kim et al. have described a paracrine signaling loop in Wnt1-induced mouse mammary tumors where luminal cells provide Wnt1 signaling for basal cells presenting the Lrp5 receptor [37]. In principle, transient fluctuations in Wnt1 signaling secreted by the luminal subpopulation could manifest cell-extrinsic effects, including transient losses in tumor-initiating capability. In a study of human embryonic stem cells (hESCs), Bendall et al. proposed a model in which hESCs differentiated into a fibroblast-like population (hdFs). The hdFs in turn secrete signaling factors, such as IGF-II, that sustain the hESCs in a self-renewing, pluripotent state [38]. In these two examples, a transient loss of signaling from one cell type could result in a loss of stem-like phenotypes in another cell type, causing the system to “differentiate out.”
Fordyce et al. have shown that DNA damage stress in primary human mammary epithelial cells increases the secretion of Activin A, which can increase the levels of Activin A in surrounding cells [39]. Human mammary epithelial cells (HMEC) respond by secreting molecules (prostaglandins) that increase the motility of surrounding epithelial cells. Thus fluctuations in Activin A may ripple outward in a bed of stationary cells, as well as be carried along by newly mobilized vehicles (Figure 5(c)). In the presence of cell-cell signaling and cell motility, the fundamental “stochastically fluctuating units” most relevant to consider for therapy may be cell communities in a tissue, rather than individual cells in a population.
For the particular case of metronomic therapy, this perspective offers a direction for increasing our understanding of the role of the microenvironment. As discussed in Liao et al. A, rationales that have historically been associated with high-frequency therapeutic dosing schedules have included targeting “non-epithelial” populations and processes such as angiogenesis, carcinoma-associated fibroblasts, and immune modulation. Our current discussion suggests going beyond simply regarding stromal cells as secondary targets for metronomic therapy. We propose that combinations of the constituents of the microenvironment and the frank carcinoma may need to be regarded together as the basic, cohesive units, in which stochastic fluctuations appear, propagate, and integrate.
6. Manipulating populations undergoing dynamic heterogeneity
To conclude, in this section, we outline a strategy whereby we can understand the effects of manipulating the components of a system displaying dynamic heterogeneity. Our purpose is to modulate the consequences of dynamic heterogeneity for a designed outcome.
6.1. Proliferation-independent vs. proliferation-dependent modulation of the time scales of restoring homeostatic heterogeneity
For our discussion, we will discuss the particular example of periodic drug kill, though the insights developed can be applied to mechanisms that cell populations could have evolved to control their own duration of survival.
Previous authors have suggested that the generation of phenotypic heterogeneity is itself a target for therapy [40]. Because phenotypic interconversion can maintain a drug-resistant subpopulation, Sharma et al. have suggested blocking interconversion [41]. In a study of TRAIL-induced apoptosis, Spencer et al. suggested “reducing the impact of cell-to-cell variability … through co-drugging” [42]. However, because phenotypic interconversion can also generate drug-sensitivity, we would ask whether, as an alternative, accelerating the generation of phenotypic heterogeneity could be beneficial. Because the finite time scale for the generation of heterogeneity in drug-sensitivity can impose a bottleneck on the speed with which we deplete the target cell population (Liao et al. A), we should find strategies for accelerating the acquisition of drug-sensitivity. In this subsection, we describe the potential use of the metronomogram to evaluate the ability of a second biologic agent to hasten the acquisition of sensitivity to a cytotoxic drug. The approach we will describe is an example of an emerging strategy: the use of agents that might themselves be neither cytotoxic nor cytostatic, but which may, nevertheless, improve the efficacy of more traditional cytostatic and cytotoxic drugs along with which they are delivered.
To accomplish the goal stated above, we must seek a particular kind of acceleration of the generation of sensitivity to cytotoxic drug. Specifically, we seek biologic agents that increase the drug-sensitive fraction generated in a given time interval without a concomitant increase in population expansion. To clarify our goal, we illustrate hypothetical measurements in Figure 6. We use Figure 6(a) to provide an example of the kind of dynamical behavior our analysis is designed to reject. Figure 6(b) provides an example of the kind of dynamical behavior we seek.
Figure 6.
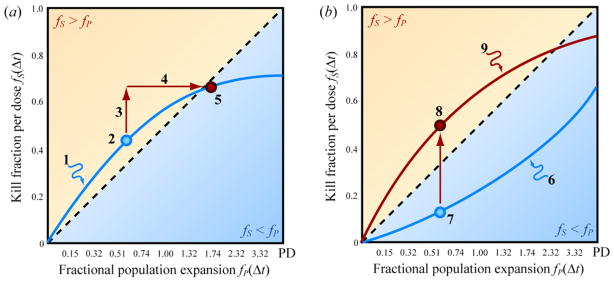
Using a metronomogram to identify therapies that accelerate the generation of drug-sensitivity in a proliferation-independent fashion. (a) “Proliferation-dependent” acceleration. (b) “Proliferation-independent” acceleration.
Figure 6(a) illustrates an example of a biologic agent that produces “proliferation-dependent” acceleration of the acquisition of drug-sensitivity. Curve 1 represents the effectiveness of various administration frequencies for a cytotoxic drug applied at a particular dose. Circle 2 represents one dosing frequency. Upon the addition of a second biologic agent, the same dosing frequency now corresponds to circle 5. The biologic agent under investigation indeed produces an increase in the drug-sensitive fraction fS generated in the given interdose period Δt. The displacement from circle 2 to 5 has a vertical component (arrow 3). However, the displacement also has a horizontal component (arrow 4), which represents an increase in the raw population number generated in the time interval Δt. In this example, the acceleration of the generation of drug-sensitivity is not worth the concomitant acceleration in population expansion. The horizontal movement is sufficient to push circle 5 into the region below the diagonal, where the target cell population is expected to expand. Circle 5 has merely explored another position of the curve to which circle 2 already belonged. Circle 5 does not move to a different curve. This can result when the rate coefficients in (1) and (2) from Liao et al. A are uniformly multiplied by the same scale factor. This changes the values of laboratory clock time to which different points on curve 1 correspond, but the shape of the parametric plot on the metronomogram is unchanged. This mathematical picture is consistent with the hypothesis that the biologic agent accelerates the acquisition of sensitivity to drug by virtue of increasing the number of proliferation events. This increases the number of opportunities for stochastic transitions between states of drug-sensitivity and drug-resistance. However, the efficiency with which a given number of proliferation events generates transitions to the drug-sensitive state is unchanged. This is why we call this category of acceleration of the acquisition of drug-sensitivity “proliferation-dependent.”
Figure 6(b) is a second example in which a different biologic agent provides, instead, “proliferation-independent” acceleration of the acquisition of drug-sensitivity. Curve 6 represents the efficacy of a variety of dosing frequencies for a cytotoxic drug applied at a particular dose. Circle 7 represents one dosing frequency. Upon administration of the biologic agent, the dosing frequency that previously corresponded to circle 7 now corresponds to circle 8. The biologic agent maps curve 6 below to curve 9 above. The absence of horizontal movement indicates that the biologic agent has not caused an increase in population expansion. We seek vertical movement because it increases the range of drug-dosing frequencies that lie above the fS = fP diagonal. A larger proportion of curve 9, as compared to curve 6, resides in the region fS > fP. When evaluating candidate biologic agents to be combined with cytotoxic drugs, we seek the vertical motion in Figure 6(b). This is in contrast to the proliferation-dependent motion in Figure 6(a), where an increase in proliferation negates the acceleration of the acquisition of drug-sensitivity.
6.2. Different factors modulate the kinetics of the generation of heterogeneity in proliferation-dependent and proliferation-independent ways
The idea of “proliferation-independent” and “proliferation-dependent” acceleration of phenotypic conversion has been applied in regenerative biology. In a study of the generation of induced pluripotent stem cells (iPSCs) from differentiated cells in a murine Nanog-GFP system, Hanna et al. found that some candidate molecular manipulations (p21KD, p53KD, Lin28OE) accelerated the generation of iPSCs relative to laboratory clock time by virtue of accelerating proliferation [43]. In contrast, overexpression of Nanog accelerated iPSC generation significantly through an increase in the probability of acquiring pluripotency during each cell division. This is proliferation-independent acceleration.
Candidate methods to identify effective agents to accelerate phenotypic conversion could use screening strategies [44], as well as analysis of the effect of stress signaling factors which indicate a more unpredictable environment. For example, the cytokine IL-6 increases the rate of conversion from non-stem to “stem-like” cancer cells, as described by Iliopoulos et al. [8]. Combined use of agents that target degradation and synthesis of mRNA and protein provide an additional approach. This strategy can be described using the particular example of translational bursts [45]. By decreasing the average copy number of mRNA while increasing the average rate of translation, the distribution of single-cell protein levels can be widened while maintaining the same mean value. The same idea could be applied in other molecular cascades. Identification of agents through these strategies will allow modulation of dynamic heterogeneity that can then be deconstructed through a mathematical analysis.
7. Summary
In sections 2–5, we explored a variety of mechanisms that can generate phenotypic stochasticity, a variety of phenotypes for which phenotypic interconversion can be biologically and clinically consequential, and ranges of population scales and integration over which stochastic fluctuations can arise and propagate. Finally, we outlined strategies for optimizing modulation of dynamic heterogeneity. Detailed study of examples like these will undoubtedly elucidate molecules and pathways that participate in the return of a population of units to homeostatic heterogeneity and provide clinical utility.
Supplementary Material
Acknowledgments
The research described here was supported by award U54CA143803 from the US National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the US National Cancer Institute or the US National Institutes of Health.
References
- 1.Kamen BA, Rubin E, Aisner J, Glatstein E. High-time chemotherapy or high time for low dose. J Clin Oncol. 2000;18:2935–2937. doi: 10.1200/JCO.2000.18.16.2935. [DOI] [PubMed] [Google Scholar]
- 2.Sigal A, Milo R, Cohen A, Geva-Zatorsky N, Klein Y, Liron Y, Rosenfeld N, Danon T, Perzov N, Alon U. Variability and memory of protein levels in human cells. Nature. 2006;444:643–646. doi: 10.1038/nature05316. [DOI] [PubMed] [Google Scholar]
- 3.Huh D, Paulsson J. Non-genetic heterogeneity from stochastic partitioning at cell division. Nat Genet. 2011;43:95–100. doi: 10.1038/ng.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaufman RJ, Brown PC, Schimke RT. Loss and stabilization of amplified dihydrofolate reductase genes in mouse sarcoma S-180 cell lines. Mol Cell Biol. 1981;1:1084–1093. doi: 10.1128/mcb.1.12.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Çaatay T, Turcotte M, Elowitz MB, Garcia-Ojalvo J, Süel GM. Architecture-dependent noise discriminates functionally analogous differentiation circuits. Cell. 2009;139:512–522. doi: 10.1016/j.cell.2009.07.046. [DOI] [PubMed] [Google Scholar]
- 6.Spudich JL, Koshland DE., Jr Non-genetic individuality: chance in the single cell. Nature. 1976;262:467–471. doi: 10.1038/262467a0. [DOI] [PubMed] [Google Scholar]
- 7.Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453:544–547. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iliopoulos D, Hirsch HA, Wang G, Struhl K. Inducible formation of breast cancer stem cells and their dynamic equilibrium with non-stem cancer cells via IL6 secretion. Proc Natl Acad Sci USA. 2011;108:1397–1402. doi: 10.1073/pnas.1018898108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta PB, Fillmore CM, Jiang G, Shapira SD, Tao K, Kuperwasser C, Lander ES. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146:633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 10.Johnston RN, Beverley SM, Schimke RT. Rapid spontaneous dihydrofolate reductase gene amplification shown by fluorescence-activated cell sorting. Proc Natl Acad Sci USA. 1983;80:3711–3715. doi: 10.1073/pnas.80.12.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimmel M, Axelrod DE. Mathematical models of gene amplification with applications to cellular drug resistance and tumorigenicity. Genetics. 1990;125:633–644. doi: 10.1093/genetics/125.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foo J, Michor F. Evolution of resistance to targeted anti-cancer therapies during continuous and pulsed administration strategies. PLoS Comput Biol. 2009;5:e1000557. doi: 10.1371/journal.pcbi.1000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silva AS, Gatenby RA. A theoretical quantitative model for evolution of cancer chemotherapy resistance. Biol Direct. 2010;5(25):1–17. doi: 10.1186/1745-6150-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rath H, Tlsty T, Schimke RT. Rapid emergence of methotrexate resistance in cultured mouse cells. Cancer Res. 1984;44:3303–3306. [PubMed] [Google Scholar]
- 15.Martinez LR, Ibom DC, Casadevall A, Fries BC. Characterization of phenotypic switching in Cryptococcus neoformans biofilms. Mycopathologia. 2008;166:175–180. doi: 10.1007/s11046-008-9133-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lambert G, Estévez-Salmeron L, Oh S, Liao D, Emerson BM, Tlsty TD, Austin RH. An analogy between the evolution of drug resistance in bacterial communities and malignant tissues. Nat Rev Cancer. 2011;11:375–382. doi: 10.1038/nrc3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies PCW, Lineweaver CH. Cancer tumors as Metazoa 1.0: tapping genes of ancient ancestors. Phys Biol. 2011;8:015001. doi: 10.1088/1478-3975/8/1/015001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weiss L. Random and nonrandom processes in metastasis, and metastatic inefficiency. Invasion Metastasis. 1983;3:193–207. [PubMed] [Google Scholar]
- 19.Neito MA, Cano A. The epithelial-mesenchymal transition under control: Global programs to regulate epithelial plasticity. Semin Cancer Biol. 2012 doi: 10.1016/j.semcancer.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 20.Dumont N, Wilson MB, Crawford YG, Reynolds PA, Sigaroudinia M, Tlsty TD. Sustained induction of epithelial to mesenchymal transition activates DNA methylation of genes silenced in basal-like breast cancers. Proc Natl Acad Sci USA. 2008;105:14867–14872. doi: 10.1073/pnas.0807146105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 22.Maheshri N, O’Shea EK. Living with noisy genes: How cells function reliably with inherent variability in gene expression. Annu Rev Biophys Biomol Struct. 2007;36:413–434. doi: 10.1146/annurev.biophys.36.040306.132705. [DOI] [PubMed] [Google Scholar]
- 23.Kim MY, Oskarsson T, Acharyya S, Nguyen DX, Zhang XHF, Norton L, Massague J. Tumor self-seeding by circulating cancer cells. Cell. 2009;139:1315–1326. doi: 10.1016/j.cell.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deakin NO, Turner CE. Distinct roles for paxillin and Hic-5 in regulating breast cancer cell morphology, invasion, and metastasis. Mol Biol Cell. 2011;22:327–341. doi: 10.1091/mbc.e10-09-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heyn C, et al. In vivo MRI of cancer cell fate at the single-cell level in a mouse model of breast cancer metastasis to the brain. Magn Reson Med. 2006;56:1001–1010. doi: 10.1002/mrm.21029. [DOI] [PubMed] [Google Scholar]
- 26.Ling V, Chambers AF, Harris JF, Hill RP. Dynamic heterogeneity and metastasis. J Cell Physio Suppl. 1984;3:99–103. doi: 10.1002/jcp.1041210412. [DOI] [PubMed] [Google Scholar]
- 27.Park JW, Neve RM, Szollosi J, Benz CC. Unraveling the biologic and clinical complexities of HER2. Clin Breast Cancer. 2008;8:392–401. doi: 10.3816/CBC.2008.n.047. [DOI] [PubMed] [Google Scholar]
- 28.Dean-Colomb W, Esteva FJ. Her2-positive breast cancer: Herceptin and beyond. Eur J Cancer. 2008;44:2806–2812. doi: 10.1016/j.ejca.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 29.Zaczek A, Brandt B, Bielawski KP. The diverse signaling network of EGFR, HER2, HER3 and HER4 tyrosine kinase receptors and the consequences for therapeutic approaches. Histol Histopathol. 2005;20:1005–1015. doi: 10.14670/HH-20.1005. [DOI] [PubMed] [Google Scholar]
- 30.Ménard S, Fortis S, Castiglioni F, Agresti R, Balsari A. HER2 as a prognostic factor in breast cancer. Oncology. 2001;61(suppl 2):67–72. doi: 10.1159/000055404. [DOI] [PubMed] [Google Scholar]
- 31.Savelyeva L, Schwab M. Amplification of oncogenes revisited: from expression profiling to clinical application. Cancer Lett. 2001;167:115–123. doi: 10.1016/s0304-3835(01)00472-4. [DOI] [PubMed] [Google Scholar]
- 32.Esteva FJ, Hortobagyi GN, Sahin AA, Smith TL, Chin DM, Liang SY, Pusztai L, Buzdar AU, Bacus SS. Expressio nof erB/HER receptors, heregulin and p38 in primary breast cancer using quantitative immunohistochemistry. Pathol Oncol Res. 2001;7:171–177. doi: 10.1007/BF03032345. [DOI] [PubMed] [Google Scholar]
- 33.Lupu R, Cardillo M, Cho C, Harris L, Hijazi M, Perez C, Rosenberg K, Yang D, Tang C. The significance of heregulin in breast cancer tumor progression and drug resistance. Breast Cancer Res Treat. 1996;38:57–66. doi: 10.1007/BF01803784. [DOI] [PubMed] [Google Scholar]
- 34.Gordon DM. The persistence of role in exterior workers of the harvester ant, Pogonomyrmex badius. Psyche. 1984;91:251–265. [Google Scholar]
- 35.Gordon DM. Dynamics of task switching in harvester ants. Anim Behav. 1989;38:194–204. [Google Scholar]
- 36.Greene MJ, Gordon DM. Interaction rate informs harvester ant task decisions. Behav Ecol. 2007;18:451–455. [Google Scholar]
- 37.Kim S, Goel S, Alexander CM. Differentiaton generates paracrine cell pairs that maintain basaloid mouse mammary tumors: Proof of concept. PLoS One. 2011;6:e19310. doi: 10.1371/journal.pone.0019310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bendall SC, et al. IGF and GFG cooperatively establish the regulatory stem cell niche of pluripotent human cells in vitro. Nature. 2007;448:1015–1021. doi: 10.1038/nature06027. [DOI] [PubMed] [Google Scholar]
- 39.Fordyce C, Fessenden T, Pickering C, Jung J, Singla V, Berman H, Tlsty T. DNA damage drives an Activin A-dependent induction of Cyclooxygenase-2 in premalignant cells and lesions. Cancer Prev Res. 2010;3:190–201. doi: 10.1158/1940-6207.CAPR-09-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niepel M, Spencer SL, Sorger PK. Non-genetic cell-to-cell variability and the consequences for pharmacology. Curr Opin Chem Biol. 2009;13:556–561. doi: 10.1016/j.cbpa.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spencer SL, Gaudet S, Albeck JG, Burke JM, Sorger PK. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature. 2009;459:428–432. doi: 10.1038/nature08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, van Oudenaarden A, Jaenisch R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dykxhoorn DM, Lieberman J. The silent revolution: RNA interference as basic biology, research tool, and therapeutic. Annu Rev Med. 2005;56:401–423. doi: 10.1146/annurev.med.56.082103.104606. [DOI] [PubMed] [Google Scholar]
- 45.Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Regulation of noise in the expression of a single gene. Nat Genet. 2002;31:69–73. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- 46.Charlebois DA, Abdennur N, Kaern M. Gene expression noise facilitates adaptation and drug resistance independently of mutation. Phys Rev Lett. 2011;107:218101. doi: 10.1103/PhysRevLett.107.218101. [DOI] [PubMed] [Google Scholar]
- 47.Eisen M, Schiller J. Stability analysis of normal and neoplastic growth. Bull Math Biol. 1977;39:597–605. doi: 10.1007/BF02461205. [DOI] [PubMed] [Google Scholar]
- 48.Quastler H, Sherman FG. Cell population kinetics in the intestinal epithelium of the mouse. Exp Cell Res. 1959;17:420–438. doi: 10.1016/0014-4827(59)90063-1. [DOI] [PubMed] [Google Scholar]
- 49.Webb GF. A cell population model of periodic chemotherapy treatment. In: Eisenfeld J, Levine DS, Witten M, editors. Biomedical Modeling and Simulation. Amsterdam: Elsevier Science Publishers B.V; 1992. pp. 83–92. [Google Scholar]
- 50.Panetta JC. A mathematical model of breast and ovarian cancer treated with paclitaxel. Math Biosci. 1997;146:89–113. doi: 10.1016/s0025-5564(97)00077-1. [DOI] [PubMed] [Google Scholar]
- 51.Camitta BM, Kamen BA. Role of methotrexate in the treatment of acute lymphoblastic leukemia. In: Pui CH, editor. Current Clinical Oncology: Treatment of Acute Leukemias: New Directions for Clinical Research. Totowa, New Jersey: Humana Press Ltd; 2003. pp. 357–364. [Google Scholar]
- 52.Silva J, Smith A. Capturing pluripotency. Cell. 2008;132:532–536. doi: 10.1016/j.cell.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Borst P, Cross GAM. Molecular basis for trypanosome antigenic variation. Cell. 1982;29:291–303. doi: 10.1016/0092-8674(82)90146-5. [DOI] [PubMed] [Google Scholar]
- 54.Raj A, vanOudenaarden A. Nature, nurture, or chance: Stochastic gene expression and its consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raser JM, O’Shea EK. Noise in gene expression: Origins, consequences, and control. Science. 2005;309:2010–2013. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.MacArthur BD, Ma’ayan A, Lemischka IR. Systems biology of stem cell fate and cellular reprogramming. Nature. 2009;10:672–681. doi: 10.1038/nrm2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 58.Levin BR. Noninherited resistance to antibiotics. Science. 2004;305:1578–1579. doi: 10.1126/science.1103077. [DOI] [PubMed] [Google Scholar]
- 59.Huang S. Non-genetic heterogeneity of cells in development: more than just noise. Development. 2009;136:3853–3862. doi: 10.1242/dev.035139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y. MET amplification leads to Gefitinib resistance in lung cancer by activating ErbB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 61.Schrödinger E. What is Life? Based on lectures at the Dublin Institute for Advances Studies at Trinity College 1943; 1944; Dublin. [Google Scholar]
- 62.Balázsi G, van Oudenaarden A, Collins JJ. Cellular decision making and biological noise: From microbes to mammals. Cell. 2011;144:910–925. doi: 10.1016/j.cell.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Strogatz SH. Nonlinear Dynamics and Chaos: With Applications to Physics, Biology, Chemistry, and Engineering. Boulder, CO: Westview Press; 1994. [Google Scholar]
- 64.Boyce WE, DiPrima RC. Elementary Differential Equations and Boundary Value Problems. 7. New York: John Wiley and Sons; 2001. [Google Scholar]
- 65.Bernoulli J. Ars Conjectandi. Basel: Thurneysen Brothers; 1713. [Google Scholar]
- 66.Skipper HE, Schabel FMJ, Mellett LB, Montgomery JA, Wilkoff LJ, Lloyd HH, Brockman RW. Implications of biochemical, cytokinetic, pharmacologic, and toxicologic relationships in the design of optimal therapeutic schedules. Cancer Chemother Rep. 1970;54:431–450. [PubMed] [Google Scholar]
- 67.Hernández-Bronchud M, Molife R. Pharmacology and principles of high dose chemotherapy. In: Lorigan P, Vandenberghe E, editors. High Dose Chemotherapy: Principles and Practice. London: Martin Dunitz; 2002. pp. 15–28. [Google Scholar]
- 68.Pui CH, Evans WE. Treatment of acute lymphoblastic leukemia. N Engl J Med. 2006;354:166–178. doi: 10.1056/NEJMra052603. [DOI] [PubMed] [Google Scholar]
- 69.Kamen BA, Glod J, Cole PD. Metronomic therapy from a pharmacologist’s view. J Pediatr Hematol Oncol. 2006;28:325–327. doi: 10.1097/00043426-200606000-00001. [DOI] [PubMed] [Google Scholar]
- 70.Seidman AD, et al. Randomized Phase III trial of weekly compared with every-3-weeks paclitaxel for metastatic breast cancer, with trastuzumab for all HER-2 overexpressors and random assignment to trastuzumab or not in HER-2 nonoverexpressors. J Clin Oncol. 2008;26:1642–1649. doi: 10.1200/JCO.2007.11.6699. [DOI] [PubMed] [Google Scholar]
- 71.Kerbel RS, Kamen BA. The anti-antiogenic basis of metronomic chemotherapy. Nat Rev Can. 2004;4:423–436. doi: 10.1038/nrc1369. [DOI] [PubMed] [Google Scholar]
- 72.Klingebiel T, et al. Treatment of children with metastatic soft tissue sarcoma with oral maintenance compared to high dose chemotherapy: Report of the HD CWS-96 trial. Pediatr Blood Cancer. 2008;50:739–745. doi: 10.1002/pbc.21494. [DOI] [PubMed] [Google Scholar]
- 73.Baltali E, Altundag K, Ozisik Y, Guler N, Tekuzman G. Weekly paclitaxel in pretreated metastatic breast cancer: Retrospective analysis of 52 patients. Tohoku J Exp Med. 2004;203:205–210. doi: 10.1620/tjem.203.205. [DOI] [PubMed] [Google Scholar]
- 74.Alvarez A, Mickiewicz E, Brosio C, Giglio R, Cinat G, Suarez A. Weekly taxol (T) in patients who had relapsed or remained stable with T in a 21 day schedule. Proc Am Soc Clin Oncol. 1998;17:188a. [Google Scholar]
- 75.Toyoda Y, Manabe A, Tsuchida M, Hanada R, Ikuta K, Okimoto Y, Ohara A, Ohkawa Y, Mori T. Six months of maintenance chemotherapy after intensified treatment for acute lymphoblastic leukemia of childhood. J Clin Oncol. 2000;18:1508–1516. doi: 10.1200/JCO.2000.18.7.1508. [DOI] [PubMed] [Google Scholar]
- 76.Cassidy . Drug development and study design. In: Shellens JHM, McLeod HL, Newell DR, editors. Cancer Clinical Pharmacology. Oxford: University Press; 2005. pp. 41–50. [Google Scholar]
- 77.Möricke A, et al. Long-term results of five consecutive trials in childhood acute lymphoblastic leukemia performed by the ALL-BFM study group from 1981 to 2000. Leukemia. 2010;24:265–284. doi: 10.1038/leu.2009.257. [DOI] [PubMed] [Google Scholar]
- 78.de Bont JM, van der Holt B, Dekker AW, van der Does-van den Berg A, Sonneveld P, Pieters R. Significant difference in outcome for adolescents with acute lymphoblastic leukemia treated on pediatric vs adult protocols in the Netherlands. Leukemia. 2004;18:2032–2035. doi: 10.1038/sj.leu.2403538. [DOI] [PubMed] [Google Scholar]
- 79.Klement GL, Kamen BA. Nontoxic, fiscally responsible, future of oncology: Could it be beginning in the third world? J Pediatr Hematol Oncol. 2011;33:1–3. doi: 10.1097/MPH.0b013e3182024918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kalmar T, Lim C, Hayward P, Muñoz-Descalzo Nichols J, Garcia-Ojalvo J, Arias MAA. Regulated fluctuations in Nanog expression mediate cell fate decisions in embryonic stem cells. PLoS Biol. 2009;7:e1000149. doi: 10.1371/journal.pbio.1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cross GAM. Cellular and genetic aspects of antigenic variation in trypanosomes. Annu Rev Immunol. 1990;8:83–110. doi: 10.1146/annurev.iy.08.040190.000503. [DOI] [PubMed] [Google Scholar]
- 82.Dhar N, McKinney JD. Microbial phenotypic heterogeneity and antibiotic tolerance. Curr Opin Microbiol. 2007;10:30–38. doi: 10.1016/j.mib.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 83.Brock A, Chang H, Huang S. Non-genetic heterogeneity -- a mutation-independent driving force for the somatic evolution of tumours. Nat Rev Genet. 2009;10:336–342. doi: 10.1038/nrg2556. [DOI] [PubMed] [Google Scholar]
- 84.Kepler TB, Elston TC. Stochasticity in transcriptional regulation: Origins, consequences, and mathematical representations. Biophys J. 2001;81:3116–3136. doi: 10.1016/S0006-3495(01)75949-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Waks Z, Silver PA. Nuclear origins of cell-to-cell variability. Cold Spring Harb Symp Quant Biol. 2010;75:1–8. doi: 10.1101/sqb.2010.75.027. [DOI] [PubMed] [Google Scholar]
- 86.Gillespie DT. Exact stochastic simulation of coupled chemical reactions. J Phys Chem. 1977;81:2340–2361. [Google Scholar]
- 87.Bartholomay AF. Stochastic models for chemical reactions: I. Theory of the unimolecular reaction process. Bull Math Biophys. 1958;20:175–190. [Google Scholar]
- 88.Thattai M, van Oudenaarden A. Intrinsic noise in gene regulatory networks. Proc Natl Acad Sci USA. 2001;98:8614–8619. doi: 10.1073/pnas.151588598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McAdams HH, Arkin A. It’s a noisy business! Genetic regulation at the nanomolar scale. Trends Genet. 1999;15:65–69. doi: 10.1016/s0168-9525(98)01659-x. [DOI] [PubMed] [Google Scholar]
- 90.Bricmont J. Science of chaos or chaos in science? Physicalia Mag. 1995;17:159–208. [Google Scholar]
- 91.Bueche F. Principles of Physics. 4. New York: McGraw-Hill; 1982. [Google Scholar]
- 92.Aguirre-Ghiso JA. Models, mechanisms and clinical evidence for cancer dormancy. Nat Rev Can. 2007;7:834–846. doi: 10.1038/nrc2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Klein CA. Parallel progression of primary tumours and metastases. Nat Rev Can. 2009;9:302–312. doi: 10.1038/nrc2627. [DOI] [PubMed] [Google Scholar]
- 94.Michor F, Hughes TP, Iwasa Y, Branford S, Shah NP, Sawyers CL, Nowak MA. Dynamics of chronic myeloid leukemia. Nature. 2005;435:1267–1270. doi: 10.1038/nature03669. [DOI] [PubMed] [Google Scholar]
- 95.Meng S, et al. Circulating tumor cells in patients with breast cancer dormancy. Clin Cancer Res. 2004;10:8152–8162. doi: 10.1158/1078-0432.CCR-04-1110. [DOI] [PubMed] [Google Scholar]
- 96.Nagrath S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cerussi AE, Tanamai VW, Mehta RS, Hsiang D, Butler J, Tromberg BJ. Frequent optical imaging during breast cancer neoadjuvant chemotherapy reveals dynamic tumor physiology in an individual patient. Acad Radiol. 2010;17:1031–1039. doi: 10.1016/j.acra.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lardinois D, Weder W, Hany TF, Kamel EM, Korom S, Seifert B, von Schulthess GK, Steinert HC. Staging of non-small-cell lung cancer with integrated positron-emission tomography and computed tomography. N Engl J Med. 2003;348:2500–2507. doi: 10.1056/NEJMoa022136. [DOI] [PubMed] [Google Scholar]
- 99.Marom EM, Sarvis S, Herndon JE, II, Patz EF. T1 lung cancers: Sensitivity of diagnosis with fluorodeoxyglucose PET. Radiology. 2002;223:453–459. doi: 10.1148/radiol.2232011131. [DOI] [PubMed] [Google Scholar]
- 100.Satti J. The emerging low-dose therapy for advanced cancers. Dose-Response. 2009;7:208–220. doi: 10.2203/dose-response.08-010.Satti. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hughes TP, et al. Long-term prognostic significance of early molecular response to imatinib in newly diagnosed chronic meyloid leukemia: an analysis from the International Randomized Study of Interferon and STI571 (IRIS) Blood. 2010;116:3758–3765. doi: 10.1182/blood-2010-03-273979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Samaga R, Saez-Rodriguez J, Alexopoulos LG, Sorger PK, Klamt S. The logic of EGFR/ErbB signaling: Theoretical properties and analysis of high-throughput data. PLoS Comput Biol. 2009;5:e1000438. doi: 10.1371/journal.pcbi.1000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Browder T, Butterfield CE, Kräling BM, Shi B, Marshall B, O’Reilly MS, Folkman J. Antiangiogenic scheduling of chemotherapy improves efficacy against expeirmental drug-resistant cancer. Cancer Res. 2000;60:1878–1886. [PubMed] [Google Scholar]
- 104.Li KL, Henry RG, Wilmes LJ, Gibbs J, Zhu X, Lu Y, Hylton NM. Kinetic assessment of breast tumors using high spatial resolution Signal Enhancement Ratio (SER) imaging. Magn Reson Med. 2007;58:572–581. doi: 10.1002/mrm.21361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zen K, Zhang CY. Circulating microRNAs: A novel class of biomarkers to diagnose and monitor human cancers. Med Res Rev. 2012;32:326–348. doi: 10.1002/med.20215. [DOI] [PubMed] [Google Scholar]
- 106.Duffy MJ, Evoy D, McDermott EW. CA 15–3: Uses and limitation as a biomarker for breast cancer. Clin Chim Acta. 2010;411:1869–1874. doi: 10.1016/j.cca.2010.08.039. [DOI] [PubMed] [Google Scholar]
- 107.Gomella LG, Liu XS, Trabulsi EJ, Kelly WK, Myers R, Showalter T, Dicker A, Wender R. Screening for prostate cancer: the current evidence and guidelines controversy. Can J Urol. 2011;18:5875–5883. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


