Abstract
Resorcylic acid lactones and dihydroxyphenylacetic acid lactones represent important pharmacophores with heat shock response and immune system modulatory activities. The biosynthesis of these fungal polyketides involves a pair of collaborating iterative polyketide synthases (iPKSs): a highly reducing iPKS with product that is further elaborated by a nonreducing iPKS (nrPKS) to yield a 1,3-benzenediol moiety bridged by a macrolactone. Biosynthesis of unreduced polyketides requires the sequestration and programmed cyclization of highly reactive poly-β-ketoacyl intermediates to channel these uncommitted, pluripotent substrates to defined subsets of the polyketide structural space. Catalyzed by product template (PT) domains of the fungal nrPKSs and discrete aromatase/cyclase enzymes in bacteria, regiospecific first-ring aldol cyclizations result in characteristically different polyketide folding modes. However, a few fungal polyketides, including the dihydroxyphenylacetic acid lactone dehydrocurvularin, derive from a folding event that is analogous to the bacterial folding mode. The structural basis of such a drastic difference in the way a PT domain acts has not been investigated until now. We report here that the fungal vs. bacterial folding mode difference is portable on creating hybrid enzymes, and we structurally characterize the resulting unnatural products. Using structure-guided active site engineering, we unravel structural contributions to regiospecific aldol condensations and show that reshaping the cyclization chamber of a PT domain by only three selected point mutations is sufficient to reprogram the dehydrocurvularin nrPKS to produce polyketides with a fungal fold. Such rational control of first-ring cyclizations will facilitate efforts to the engineered biosynthesis of novel chemical diversity from natural unreduced polyketides.
Fungal polyketides are one of the largest families of structurally diverse natural products with antibiotic, antiproliferative, immunosuppressive, and enzyme inhibitory activities. Importantly, they also provide lead compounds and inspiration for pharmaceutical drug discovery, which is evidenced by the statin cholesterol-lowering agents (1, 2). Fungal polyketides are biosynthesized by multidomain megasynthases [type I iterative polyketide synthases (iPKSs)] that use ketoacyl synthase, acyl transferase, and acyl carrier protein (ACP) domains to catalyze recursive thio-Claisen condensations using malonyl-CoA extender units. Although the architecture of these enzymes is similar to a single module of the bacterial type I modular PKSs (3), fungal iPKSs use a single set of active sites iteratively, analogous to dissociated bacterial type II PKSs (4). Fungal iPKSs may be classified into three subgroups (5). Highly reducing iPKSs (hrPKSs) generate complex linear or nonaromatic cyclic products by reducing the nascent β-ketoacyl intermediates to the β-alcohol, the alkene, or the alkane after each condensation step using their ketoreductase, dehydratase, and enoyl reductase domains to execute a cryptic biosynthetic program (2, 6–8). Partially reducing iPKSs omit enoyl reduction to generate simple cyclic structures (5). Finally, nonreducing iPKSs (nrPKSs) feature no reducing domains and generate a wide variety of aromatic products. nrPKSs select different starter units by a starter unit:ACP transacylase domain (9) and mold the polyketide chains into cyclic products by regiospecific cyclizations. First-ring cyclizations are catalyzed by the product template domains (PTs) (10), whereas the polyketide chains are terminated by Claisen cyclase (11), macrolactone synthase (12) [thioesterase (TE)], or reductive release domains (2).
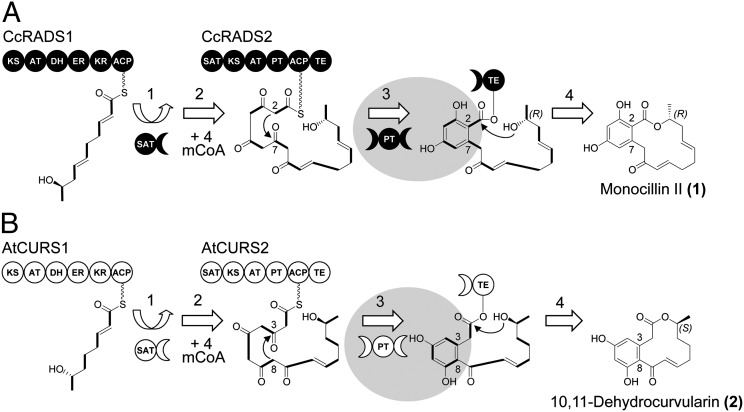
Although the biosynthesis of most fungal polyketides requires a single iPKS enzyme, the assembly of the resorcylic acid lactones (RALs) involves a pair of collaborating hrPKSs and nrPKSs acting in sequence (12–17). Fungal RALs are rich pharmacophores with estrogen agonist (zearalenone), mitogen-activated protein kinase inhibitory (hypothemycin), and heat shock response modulatory activities [radicicol and monocillin II (1)] (Fig. 1) (18, 19). For these RALs, the hrPKS produces a reduced linear polyketide chain that is directly transferred to the nrPKS (9). The nrPKS further extends the polyketide, closes the first six-membered ring by aldol condensation, and releases the RAL product by macrolactone formation (Fig. 1). We have recently shown that the assembly of 10,11-dehydrocurvularin (2), a phytotoxic dihydroxyphenylacetic acid lactone (DAL) from Aspergillus terreus, uses a similar chemical modularity principle (20). Curvularins modulate the mammalian immune system by repressing the inducible nitric oxide synthase (21, 22). In addition, both monocillin II (1) and 10,11-dehydrocurvularin (2) act as promising broad spectrum inhibitors of various cancer cell lines in vitro by overwhelming the heat shock response, an evolutionarily conserved coping mechanism of eukaryotic cells that maintains protein homeostasis (23–26).
Fig. 1.
Biosynthesis of monocillin II (1) and 10,11-dehydrocurvularin (2). (A) During the biosynthesis of the radicicol intermediate monocillin II (1) in C. chiversii (14), the hrPKS CcRADS1 produces a reduced pentaketide starter unit (13) that is transferred to the nrPKS CcRADS2 by the starter unit:ACP transacylase (SAT) domain (9) (step 1). After another four successive condensation events with malonyl-CoA (mCoA; step 2) catalyzed by the ketoacyl synthase of the nrPKS CcRADS2, the linear ACP-bound polyketide chain undergoes a C2-C7 aldol condensation catalyzed by the PT domain (10) (step 3). This condensation follows an F-type folding mode (28, 29); 1 is released by macrolactone formation catalyzed by the TE domain (2) (step 4). (B) Assembly of 2 in A. terreus AH-02–30-F7 also involves sequentially acting collaborating iPKSs. However, the hrPKS AtCURS1 produces a reduced tetraketide starter, whereas the AtCURS2 PT domain catalyzes aldol condensation in the C8-C3 register using an S-type folding mode (28). C-C bonds in bold indicate intact acetate equivalents (malonate-derived C2 units) incorporated into the polyketide chain by the iPKSs.
A crucial step during the programmed biosynthesis of aromatic polyketide natural products is the cyclization of the first ring, which is catalyzed by the PT domains of the nrPKS (10, 27). This event commits the highly reactive pluripotent poly-β-ketoacyl chains to defined structural classes of the possible polyketide scaffold space. PT-catalyzed cyclizations most often follow an F-type pattern, whereby the benzene ring is assembled from two intact malonate-derived C2 units and two bridging carbons from two additional acetate equivalents (28, 29) (Fig. 1). F-type first-ring cyclizations typically result from aldol condensations in the C2-C7 (as in 1), C4-C9, or C6-C11 register. In contrast, bacterial polyketide cyclase/aromatase enzymes (parts of type II PKS multienzyme complexes) typically direct an S-type folding event, whereby the carbons of the benzene ring are derived from three intact malonate-derived C2 units (1, 28). Nevertheless, a select few fungal polyketides, including DALs like 2, feature a first-ring connectivity that is analogous to the S-type folding mode, resulting from an unorthodox C8-C3 aldol cyclization event (20). The bacterial aromatase/cylcase enzymes show little sequence similarity to fungal PT domains and feature a different protein fold and active site architecture as a prominent example of convergent evolution (10, 30–32). The sequences of fungal PT domains catalyzing F-type cyclization can be classified into seven clades according to their regiospecificity and the length of their product (29, 33). Despite catalyzing an atypical S-type folding, the PT domain of the AtCURS2 curvularin synthase nrPKS is firmly rooted in the C2-C7 clade of PTs, which yield RALs like 1 (20). This result concurs with previous observations that fungal iPKSs evolve orthogonal product specificities primarily by point mutations and not domain shuffling among distinct enzymes (2).
The present work aimed to define how different regiospecific outcomes for first-ring cyclization are programmed into nrPKS enzymes. By exploiting the orthogonal aldol condensation regiospecificities of the related PT domains of the nrPKSs for 1 and 2, we attempted to alter this program to switch F- and S-type cyclization modes. Achieving precise control of regiospecificity during the engineered biosynthesis of fungal polyketides is central to producing biologically active unnatural products, and it may guide efforts to generate novel chemical diversity from natural nonreduced fungal polyketides.
Results and Discussion
PT Domains Are Necessary for Programmed Polyketide Formation.
Throughout this study, we have used an in vivo reconstituted system for polyketide production, whereby recombinant hrPKS + nrPKS pairs are expressed from compatible plasmids in the host Saccharomyces cerevisiae BJ5464-NpgA to produce 1 (9 mg/L, isolated yield), 2 (6 mg/L), and their derivatives (13, 34). Deletion of PT domains has previously been shown in the Tang and Townsend laboratories to yield shunt metabolites with backbones that have undergone spontaneous cyclizations (12, 27, 29). Thus, at the start of this work, we considered it to be possible that PTAtCURS2 is simply an inactive enzyme that does not contribute to the folding of the nascent polyketide chain, and thus, the DAL scaffold of 2 is a serendipitous derailment product retained by evolution. To exclude this possibility, we have deleted the PT domain of AtCURS2 as well as the PT domain of CcRADS2, the radicicol/monocillin II nrPKS of Chaetomium chiversii (14). However, the corresponding yeast expression strains produced no polyketides (Fig. 2). Complementation of the PT-less nrPKSs with their dissected PT domains, expressed as separate ORFs in trans, led to the rescue of the production of the native products 1 (1 mg/L) and 2 (2 mg/L), respectively (Fig. 2). The success of this experiment contrasts previous in trans domain complementation attempts that did not yield observable products in vivo (1, 29) and emphasizes that appropriate protein–protein docking, substrate recognition, and processing still can take place with dissected domains expressed in heterologous hosts. Taken together, these experiments show that the PT-less nrPKSs and the freestanding PT domains are all catalytically active. The absence of RAL/DAL product formation in the absence of PT domains, thus, indicates that these domains fulfill an essential role in both systems and that untemplated poly-β-ketones are not released by these nrPKSs.
Fig. 2.
Engineering first-ring cyclization regiospecificity by domain replacements. (A) Product profiles (HPLC traces recorded at 300 nm) of S. cerevisiae BJ5464-NpgA (13, 34) cotransformed with YEpAtCURS1 and the indicated YEpATCURS2 derivatives (SI Materials and Methods): (i) YEpATCURS2, (ii) YEpATCURS2-ΔPT, (iii) YEpATCURS2-ΔPT+PTAtCURS2, and (iv) YEpATCURS2-PTCcRADS2. The peak in i labeled with the asterisk corresponds to 11-hydroxycurvularin, a spontaneous hydration product of 2. (B) Product profiles (HPLC traces recorded at 300 nm) of S. cerevisiae BJ5464-NpgA (13, 34) cotransformed with YEpCcRADS1 and the indicated YEpCcRADS2 derivatives (SI Materials and Methods): (i) YEpCcRADS2, (ii) YEpCcRADS2-ΔPT, (iii) YEpCcRADS2-ΔPT+PTCcRADS2, and (iv) YEpCcRADS2-PTAtCURS2. Polyketide products were characterized based on their UV, electrospray ionization mass spectrometry (ESI-MS), NMR, and circular dichroism (CD) spectra as well as Mosher’s method (SI Materials and Methods has details on isolation and chemical characterization).
Portability of Aldol Cyclization Programs.
Recent experiments to replace PT domains in nrPKS model systems showed that the register of F-type aldol condensations may be switched (29, 35), with the incoming PTs enforcing a subtle shift of the substrate chains in the catalytic chambers to expose different carbons (C2, C4, or C6) to the catalytic histidine for deprotonation and enolate formation (10). Thus, we were interested to see whether first-ring cyclizations may also be reconfigured between F- and S-type folding modes by exchanging PT domains. This change would require radical rerouting of the polyketide substrate chain to expose a carbon to the catalytic base that is distal (S type) or proximal (F type) to the phosphopantetheine thioester. As shown above and observed previously by others (1, 29, 36), in trans reconstitution of iPKSs from dissected domains incurs a penalty for catalysis in terms of product yield and/or fidelity. Thus, we elected to conduct these experiments with nrPKSs where the heterologous PT domains replace their native equivalents in cis (29). Replacement of the PT domain of AtCURS2 with PTCcRADS2 led to the production of two isocoumarins, both resulting from C2-C7 aldol cyclizations. The priming acyl chain of 3 (0.3 mg/L) corresponds to the expected product of the hrPKS AtCURS1, whereas the major product 4 (3 mg/L) features a carbonyl at C15 (Fig. 2A). The formation of 3 and 4 indicates that PTCcRADS2 is able to process a shorter substrate (an octaketide as opposed to its native nonaketide) while faithfully executing an F-type first-ring closure. Time course analysis of the fermentation with S. cerevisiae BJ5464-NpgA (YEpAtCURS1 and YEpATCURS2-PTCcRADS2) (Fig. 3A) shows that 3 is the primary product 24 h after the induction of polyketide production, but by 48 h, the formerly minor product 4 becomes dominant. Extending the cultivation to 72 h and beyond increased only the production of 4 but did not eliminate 3, and it did not lead to the production of additional polyketide products. Similarly, incubation of purified 3 with the untransformed yeast host strain S. cerevisiae BJ5464-NpgA led to the gradual, albeit not complete, biotransformation of 3 to 4 (Fig. 3B). It was confirmed that the untransformed yeast host strain does not produce 3 or 4, and 3 is not converted to 4 by spontaneous oxidation in the culture medium in the absence of yeast cells (Fig. 3B). Thus, 4 derives from a chance oxidation of the 15-OH of 3 by an endogenous enzyme of the yeast host.
Fig. 3.
Fortuitous oxidation of isocoumarin 3 by S. cerevisiae BJ5464-NpgA. (A) Product profiles (HPLC traces recorded at 300 nm) of S. cerevisiae BJ5464-NpgA (13, 34) cotransformed with YEpAtCURS1 and YEpATCURS2-PTCcRADS2 and cultivated for the indicated amount of time. (B) HPLC analysis of the bioconversion of 3 into 4 by S. cerevisiae BJ5464-NpgA.
The yeast strain coexpressing CcRADS1 and CcRADS2-PTAtCURS2 yielded a unique compound (5) (Fig. 2B), although with a low productivity (0.3 mg/L). Structural characterization of this product revealed that 5 harbors a unique carbon skeleton featuring a C8-C3 dihydroxyphenylacetic acid moiety bridged by an eight-membered lactone (SI Materials and Methods). Thus, PTAtCURS2 is competent to process a longer substrate (a nonaketide as opposed to its native octaketide) while retaining its ability to direct an S-type folding and cyclization event. The 4-oxo-2-oxacyclooctanone ring of 5 may be produced by the facile attack of the C1 carboxyl on C11 of the enone; the involvement of the TE domain in this reaction cannot be excluded at this point.
Collectively, these experiments show that the F- or S-type regiospecificities of first-ring cyclizations are solely programmed into the PT domains of collaborating nrPKSs (29, 35). This programming is portable among nrPKS platforms, without influencing starter unit choice or the number of extensions carried out by the rest of the chassis. The formation of the isocoumarins 3 and 4 and the eight-membered lactone 5 suggests that the nrPKS TE domains may hydrolyze products with switched aldol condensation patterns, but they are unable to form a macrolactone using a carbon chain with an isomeric fold. The low product yield of 5 suggests that CcRADS2 is a more stringent chassis that is less amenable to combinatorial replacement of its domains.
Homology Models of PTAtCURS2 and PTCcRADS2.
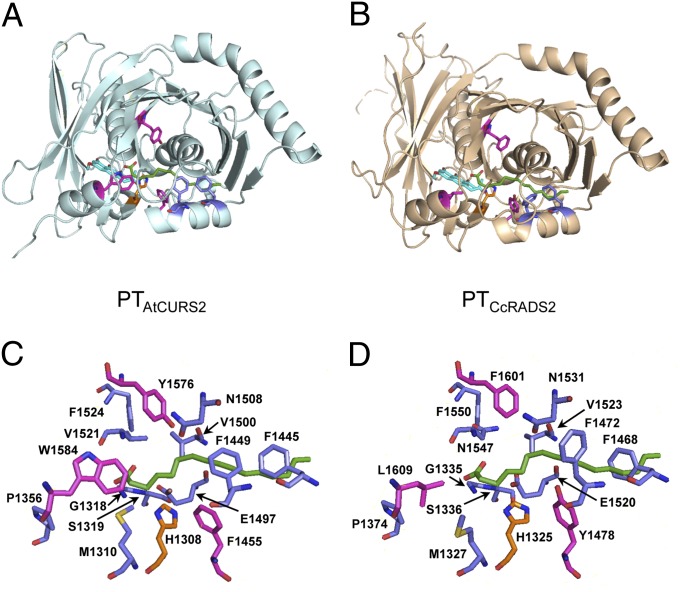
To identify the structural basis of the programming of regiospecificity in PT domains, we have created homology models of the PTAtCURS2 and the PTCcRADS2 domains based on the experimentally determined structure of the PT domain of NSAS from Aspergillus parasiticus (PTNSAS; Protein Data Bank ID codes 3HRR and 3HRQ) (10). The nrPKS NSAS catalyzes the formation of norsorolinic acid, a C20 polyketide primed with hexanoic acid, with the PTNSAS directing an F-type folding mode first-ring cyclization event (28) in the C4-C9 register followed by a second-ring closure at C2-C11.
Despite relatively low sequence similarities with PTNSAS (PTAtCURS2: 22% identity and 41% similarity; PTCcRADS2: 20% identity and 40% similarity), combined structural evaluation and fold recognition scores of 3.6 (PTCcRADS2) and 3.47 (PTAtCURS2) obtained from the reliability assessment engine PCONS5 (37) indicated that the fold recognition is reliable for both PTAtCURS2 and PTCcRADS2 (a PCONS score > 2.17 is considered reliable, and a score > 1.5 is considered significant). PTNSAS features a long, straight cyclization chamber and a hydrophobic hexyl-binging region that accommodates the starter unit, with the substrate bound in an extended conformation (10). In contrast, homology modeling had suggested that PKS4, the zearalenone nrPKS from Gibberella fujikuroi (10, 16, 38), contains a PT domain with a wider, curved catalytic chamber, where the substrate adopts a bent conformation (10). Similar to the model proposed for PTPKS4, the hydrophobic hexyl-binding region present in PTNSAS was found to be closed off in both PTAtCURS2 and PTCcRADS2 by the bulky side chains of two phenylalanine residues (Fig. 4). First, a replacement of M1495 of NSAS by phenylalanine (AtCURS2: F1449; CcRADS2: F1472) narrows the hexyl-binding region. Next, the side chain of another phenylalanine residue in place of G1491 (AtCURS2: F1445; CcRADS2: F1468) directly clashes with the tail of the palmitic acid that occupies this pocket in structure 3HRR. This latter phenylalanine is also conserved in not only all other RAL PT domains (14–16, 38) but also clades II, III, and V of characterized PT domains (29). Thus, the hrPKS-derived reduced acyl chains of the nascent intermediates for 1 and 2 may not be sequestered in a deep, buried pocket (10) in these enzymes.
Fig. 4.
Homology models of PTCcRADS2 and PTAtCURS2. Cartoon views of the homology models and stick representation of the amino acids lining the cyclization chambers of (A and C) PTAtCURS2 and (B and D) PTCcRADS2. The side chains of residues discussed in the text are shown as golden (catalytic histidine), magenta (residues implicated in differentiating substrate orientation), and blue (other significant residues) sticks. Green sticks show the substrate analog palmitic acid resident in the PTNSAS structure 3HRQ (10).
The RAL/DAL PT models retain the large substrate binding chamber where cyclization occurs (10). This cyclization chamber seems constricted at the residues corresponding to V1347 and A1397 of NSAS (AtCURS2: M1310 and P1356; CcRADS2: M1327 and P1374), but this narrowing is compensated by substitutions with less bulky side chains corresponding to P1355 and W1571 (AtCURS2: G1318 and F1524; CcRADS2: G1335 and F1550) (Fig. 4). The cyclization chamber is proposed to feature an active site dyad (AtCURS2: H1308, E1497; CcRADS2: H1325, E1520), where the aspartic acid (D1543) that polarizes the catalytic base (H1345) in NSAS is replaced by glutamic acid. This functionally conserved replacement is present in all known RAL and DAL PT domains (14–16, 38) but not clades II–V of functionally characterized PT domains (29). The same D to E replacement is, nonetheless, common in dehydratases with a fold similar to the fold of PT domains, and it was also found in some type II PKS aromatase/cyclase enzymes (32). After deprotonation of C8 (AtCURS2) or C2 (CcRADS2) by the catalytic base, the enolate intermediate is thought to be stabilized by the backbone amine of V1521 (AtCURS2) or N1547 (CcRADS2) (10). After the collapse of the enolate and aldol addition to the carbonyl, the oxyanion may be stabilized by a network of water molecules that are coordinated in NSAS by S1356, D1543, T1546, and N1568. Only some of these residues are conserved in AtCURS2 (S1319, E1497, V1500, and V1521) and CcRADS2 (S1336, E1520, V1523, and N1547), and similar replacements are also present in all RAL PT domains (14–16, 38). The second one-half of the oxyanion hole is provided by the backbone amine of a glycine in dehydratase domains and hydratases with which PT domains share a double hot dog fold and a proposed evolutionary origin (10). For NSAS, this glycine was seen to be replaced by P1355, but this residue is restored to glycine in all known RAL PT domains (14–16, 38) (AtCURS2: G1318; CcRADS2: G1335). The corresponding position is occupied by serine in clades II, III, and V of PT domains (29). The electrophilic carbonyl that takes part in the aldol cyclization is polarized by hydrogen bonding through the same water network, whereas hydrogen bonding with an asparagine that is conserved in all functionally characterized PT domains (29) may help to orient the substrate in the chamber (NSAS: N1554; AtCURS2: N1508; CcRADS2: N1531).
Conversion of S- and F-Type Folding Modes by Structure-Based Site-Directed Mutagenesis.
A superimposition of the models for PTAtCURS2 and PTCcRADS2 (47% sequence identity and 65% similarity) showed that the active sites and the cyclization chambers of these two enzymes are highly conserved. Nevertheless, PTAtCURS2 forms a first ring in the C8-C3 register with a folding mode analogous to the S type, whereas PTCcRADS2 catalyzes an F-type folding mode (28) first-ring closure in the C2-C7 register. We have identified three key differences that we hypothesized would result in a change in substrate orientation in the binding pocket and lead to the orthogonal cyclization regiospecificities observed in 1 vs. 2 (Fig. 5A). First, L1609 of PTCcRADS2 is replaced by W1584 near the substrate entrance in PTAtCURS2. The bulky side chain of this tryptophan narrows the entrance of the cyclization chamber of PTAtCURS2 and may serve to direct C2 of the penetrating acyl chain away from the catalytic histidine. Leucine is strictly conserved at this position in all characterized RAL PTs (14–16, 38) and predominates (with methionine as an alternative) in clades II–V PTs catalyzing various F-type cyclizations (29). Next, both PTAtCURS2 and PTCcRADS2 (as well as all other characterized RAL PTs) (29) display a tyrosine-phenylalanine residue pair on opposing faces at the rear of the binding pocket. Remarkably, these residues are inverted in PTAtCURS2 vs. all known RAL PT domains (PTAtCURS2: F1455 and Y1576; PTCcRADS2: Y1478 and F1601). By participating in hydrogen bond networks (Y) or contributing to a hydrophobic surface of the pocket (F), these residues may help to position the chain such that either C8 (PTAtCURS2) or C2 (PTCcRADS2) would be presented to the catalytic base (PTAtCURS2: H1308; PTCcRADS2: H1325), leading to S- (PTAtCURS2) or F-type (PTCcRADS2) cyclization outcomes. Notably, all three distinguishing residues (W1584, F1455, and Y1576) are conserved between PTAtCURS2 and the PT domain of an orphan hrPKS–nrPKS system in the genome of Pyrenophora tritici-repentis PT-1C-BFP (GenBank accession nos. EDU47225 and EDU47223): this putative DAL synthase is the closest ortholog of the dehydrocurvularin synthase AtCURS1-AtCURS2 (20). Although the highlighted residues are positioned such that substrate binding is expected to be affected, verification of the specific contacts must await the determination of the crystal structures of these domains with bound substrates/products. In the meantime, these three residues may serve as distinguishing sequence signatures to predict RAL vs. DAL formation by orphan biosynthetic systems found in sequenced fungal genomes.
Fig. 5.
Reprogramming first-ring cyclization regiospecificity by site-directed mutagenesis. (A) Proposed mechanism of regiospecific cyclizations catalyzed by the PTAtCURS2 and PTCcRADS2 domains. Enolate formation by deprotonation of C8 (AtCURS2) or C2 (CcRADS2) is promoted by the histidine catalytic base that is polarized by a conserved glutamic acid (AtCURS2: H1308 and E1497; CcRADS2: H1325 and E1520). The substrates are proposed to thread through alternate routes as directed by different gating residues at the entrance of the cyclization chamber (AtCURS2: W1584; CcRADS2: L1609) and further oriented by an inverted hydrophobic/H-bond donor residue pair lining the rear end of the cyclization chamber (AtCURS2: F1455 and Y1576; CcRADS2: Y1478 and F1601). (B) Product profiles (HPLC traces recorded at 300 nm) of S. cerevisiae BJ5464-NpgA (13, 34) cotransformed with YEpAtCURS1 and the YEpATCURS2 derivatives encoding the indicated PTAtCURS2 domain mutations in AtCURS2.
To test our structural analysis, we systematically replaced one, two, or all three of the identified residues (F1455, Y1576, and W1584) in AtCURS2 with their counterparts of CcRADS2 and coexpressed these enzymes with AtCURS1 in yeast. Single mutations did not alter the regiospecificity of first-ring closure in the product (Fig. 5B). Double mutations either eliminated product formation (F1455Y + W1584L) or yielded a mixture of 2 and 4, indicating a relaxed aldol condensation regiospecificity for some of these mutant enzymes. Finally, the enzyme with all three mutations produced only 3 and 4 (0.2 and 2 mg/L, respectively), with no detectable 2. Thus, these three selected point mutations were sufficient to completely transform the native C8-C3 (S-type) regiospecificity of PTAtCURS2 to C2-C7 (F type) (Fig. 5B). The converse experiment (replacing Y1478, F1601, and L1609 in CcRADS2 with the corresponding residues of AtCURS2 in all combinations) reduced the yield or completely eliminated the production of 1, but it did not provide 5 or any other detectable C8-C3 DAL (SI Materials and Methods). The absence of the expected product may not be surprising if we consider the low yield of 5, even with CcRADS2-PTAtCURS2, where the incoming PTAtCURS2 domain has presumably been optimized by evolution for the effective synthesis of C8-C3 products. A similar recalcitrance to alteration of stereocontrol has been noted for ketoreductase and enoyl reductase domains during site-directed mutagenesis (but not during complete domain exchanges) in the context of modular PKS systems. Presumably, any proofreading activity from downstream domains (e.g., the TE in our system) may override the effects of subtle alterations of the active site architecture (3, 39, 40).
The current work affirms that first-ring cyclization regiospecificity in fungal collaborating iPKSs is programmed in the composition and geometry of the cyclization chambers of the PT domains. Exploiting structural information on PT domains, we have gained insight into the origins of the programming of this folding specificity. Replacement of just three select residues of the product cyclization chambers in a keyhole surgery-like approach converted a PT domain from an atypical, C8-C3–specific, S-type folding mode cyclase into a typical, C2-C7–selective, F-type folding mode enzyme as predicted. The identified signature residues may be used to predict polyketide folding modes in orphan RAL/DAL biosynthetic systems. More importantly, rational reprogramming of polyketide folding modes in fungal iPKSs opens new possibilities for the engineered biosynthesis of novel unnatural polyketides with isomeric folds, including cancer cell proliferation inhibitors and immune system modulators.
Materials and Methods
Strains and Culture Conditions.
Escherichia coli DH10B and plasmid pJET1.2 (Fermentas) were used for routine cloning and sequencing. S. cerevisiae BJ5464-NpgA (MATα ura3-52 his3-Δ200 leu2-Δ1 trp1 pep4::HIS3 prb1 Δ1.6R can1 GAL) (34, 41) was maintained on yeast extract peptone dextrose agar (Difco) and transformed using the small-scale lithium chloride protocol (42). The yeast–E. coli shuttle vectors YEpADH2p-FLAG-URA and YEpADH2p-FLAG-TRP (20) are based on the YEpADH2p vectors with the URA3 or the TRP1 selectable markers (13). Primers used in this study and details on the construction of gene variants and expression constructs are described in SI Materials and Methods.
For each recombinant yeast strain, three to five independent transformants were analyzed for the production of polyketides by small-scale fermentation, and fermentations with representative isolates were repeated at least three times to confirm results.
Small-Scale Fermentation and Analysis of Products.
Yeast strains were cultured in 50 mL 0.67% (wt/vol) yeast nitrogen base, 2% (wt/vol) glucose, and 0.72 g/L Trp/Ura DropOut supplement at 30 °C with shaking at 250 rpm. When the OD600 reached 0.6, an equal volume of an equal volume of a solution of 1% (wt/vol) yeast extract and 2% (wt/vol) peptone was added to the cultures, and the fermentation was continued at 30 °C with shaking at 250 rpm for an additional 2 d. The cultures were adjusted to pH 5.0 and extracted with equal volumes of ethyl acetate three times. The collected organic extracts were evaporated to dryness and analyzed by reversed-phase HPLC (Kromasil C18 column, 5 µm, 4.6 × 250 mm; eluted with 5% aqueous acetonitrile for 5 min followed by a linear gradient of 5–95% CH3CN over 10 min and 95% CH3CN for 10 min at a flow rate of 0.8 mL/min; detection at 300 nm). Analysis of the time course of the production of 3 and 4, biotransformation of 3 to 4 by S. cerevisiae BJ5464-NpgA, and scale-up of fermentations and isolation of polyketide products for structure elucidation are described in SI Materials and Methods.
Chemical Characterization of Polyketide Products.
Optical rotations were recorded on a Rudolph Autopol IV polarimeter with a 10-cm cell. CD spectra were acquired with a JASCO J-810 instrument using a path length of 1 cm. 1H, 13C, and 2D NMR [homonuclear correlation spectroscopy (COSY), heteronuclear single-quantum correlation spectroscopy (HSQC), heteronuclear multiple-bond correlation spectroscopy (HMBC), and rotating frame nuclear Overhauser effect spectroscopy (ROESY)] spectra were recorded in DMSO-d6, CD3OD, or C5D5N on a JEOL ECX-300 Spectrometer. ESI-MS data were collected on an Agilent 6130 Single Quad LC-MS. SI Materials and Methods has details.
Homology Modeling.
Sequences of the PTAtCURS2 and PTCcRADS2 domains were submitted to the BioInfoBank Meta Server (3DJury) (43). Models based on the Research Collaboratory for Structural Bioinformatics Protein Data Bank PKsA structures 3HRQ and 3HRR (10) were generated by several homology modeling servers. Carbon-α models generated by the SAM-HMM server (44) were ranked by 3DJury as the most representative and chosen for use. All atom models were created using MODELER (45). There were no significant differences between the models based on 3HRQ and the models based on 3HRR. PCONS5 (37) was used to evaluate fold recognition.
Supplementary Material
Acknowledgments
We thank professors N. A. Da Silva (University of California, Irvine, CA) and Y. Tang (University of California, Los Angeles, CA) for providing the yeast host strain and expression vectors. This work was supported by National Institutes of Health Grants HL062969 (to W.R.M.) and AI065357 (to J. Zhan), National Research and Development Project of Transgenic Crops of China 2011ZX08009-003-002 (M.L.), National Basic Research 973 Program of China 2013CB733903 (M.L.), the Utah State University Seed Program to Advance Research Collaborations (J. Zhan), and National Science Foundation Grant MCB-0948751 (to I.M.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1301201110/-/DCSupplemental.
References
- 1.Crawford JM, Townsend CA. New insights into the formation of fungal aromatic polyketides. Nat Rev Microbiol. 2010;8(12):879–889. doi: 10.1038/nrmicro2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chooi YH, Tang Y. Navigating the fungal polyketide chemical space: From genes to molecules. J Org Chem. 2012;77(22):9933–9953. doi: 10.1021/jo301592k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keatinge-Clay AT. The structures of type I polyketide synthases. Nat Prod Rep. 2012;29(10):1050–1073. doi: 10.1039/c2np20019h. [DOI] [PubMed] [Google Scholar]
- 4.Zhan J. Biosynthesis of bacterial aromatic polyketides. Curr Top Med Chem. 2009;9(17):1598–1610. doi: 10.2174/156802609789941906. [DOI] [PubMed] [Google Scholar]
- 5.Cox RJ. Polyketides, proteins and genes in fungi: Programmed nano-machines begin to reveal their secrets. Org Biomol Chem. 2007;5(13):2010–2026. doi: 10.1039/b704420h. [DOI] [PubMed] [Google Scholar]
- 6.Zhou H, et al. A fungal ketoreductase domain that displays substrate-dependent stereospecificity. Nat Chem Biol. 2012;8(4):331–333. doi: 10.1038/nchembio.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisch KM, et al. Rational domain swaps decipher programming in fungal highly reducing polyketide synthases and resurrect an extinct metabolite. J Am Chem Soc. 2011;133(41):16635–16641. doi: 10.1021/ja206914q. [DOI] [PubMed] [Google Scholar]
- 8.Yakasai AA, et al. Nongenetic reprogramming of a fungal highly reducing polyketide synthase. J Am Chem Soc. 2011;133(28):10990–10998. doi: 10.1021/ja204200x. [DOI] [PubMed] [Google Scholar]
- 9.Foulke-Abel J, Townsend CA. Demonstration of starter unit interprotein transfer from a fatty acid synthase to a multidomain, nonreducing polyketide synthase. ChemBioChem. 2012;13(13):1880–1884. doi: 10.1002/cbic.201200267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crawford JM, et al. Structural basis for biosynthetic programming of fungal aromatic polyketide cyclization. Nature. 2009;461(7267):1139–1143. doi: 10.1038/nature08475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korman TP, et al. Structure and function of an iterative polyketide synthase thioesterase domain catalyzing Claisen cyclization in aflatoxin biosynthesis. Proc Natl Acad Sci USA. 2010;107(14):6246–6251. doi: 10.1073/pnas.0913531107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou H, Zhan J, Watanabe K, Xie X, Tang Y. A polyketide macrolactone synthase from the filamentous fungus Gibberella zeae. Proc Natl Acad Sci USA. 2008;105(17):6249–6254. doi: 10.1073/pnas.0800657105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou H, Qiao K, Gao Z, Vederas JC, Tang Y. Insights into radicicol biosynthesis via heterologous synthesis of intermediates and analogs. J Biol Chem. 2010;285(53):41412–41421. doi: 10.1074/jbc.M110.183574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang S, et al. Functional characterization of the biosynthesis of radicicol, an Hsp90 inhibitor resorcylic acid lactone from Chaetomium chiversii. Chem Biol. 2008;15(12):1328–1338. doi: 10.1016/j.chembiol.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 15.Reeves CD, Hu Z, Reid R, Kealey JT. Genes for the biosynthesis of the fungal polyketides hypothemycin from Hypomyces subiculosus and radicicol from Pochonia chlamydosporia. Appl Environ Microbiol. 2008;74(16):5121–5129. doi: 10.1128/AEM.00478-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim YT, et al. Two different polyketide synthase genes are required for synthesis of zearalenone in Gibberella zeae. Mol Microbiol. 2005;58(4):1102–1113. doi: 10.1111/j.1365-2958.2005.04884.x. [DOI] [PubMed] [Google Scholar]
- 17.Gaffoor I, et al. Functional analysis of the polyketide synthase genes in the filamentous fungus Gibberella zeae (anamorph Fusarium graminearum) Eukaryot Cell. 2005;4(11):1926–1933. doi: 10.1128/EC.4.11.1926-1933.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winssinger N, Fontaine JG, Barluenga S. Hsp90 inhibition with resorcyclic acid lactones (RALs) Curr Top Med Chem. 2009;9(15):1419–1435. doi: 10.2174/156802609789895665. [DOI] [PubMed] [Google Scholar]
- 19.Winssinger N, Barluenga S. Chemistry and biology of resorcylic acid lactones. Chem Commun (Camb) 2007;2007(1):22–36. doi: 10.1039/b610344h. [DOI] [PubMed] [Google Scholar]
- 20.Xu Y, et al. Characterization of the biosynthetic genes for 10,11-dehydrocurvularin, a heat-shock response modulator anticancer fungal polyketide from Aspergillus terreus. Appl Environ Microbiol. 2013;79(6):2038–2047. doi: 10.1128/AEM.03334-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt N, et al. Transcriptional and post-transcriptional regulation of iNOS expression in human chondrocytes. Biochem Pharmacol. 2010;79(5):722–732. doi: 10.1016/j.bcp.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elzner S, et al. Inhibitors of inducible NO synthase expression: Total synthesis of (S)-curvularin and its ring homologues. ChemMedChem. 2008;3(6):924–939. doi: 10.1002/cmdc.200800022. [DOI] [PubMed] [Google Scholar]
- 23.Santagata S, et al. Using the heat-shock response to discover anticancer compounds that target protein homeostasis. ACS Chem Biol. 2012;7(2):340–349. doi: 10.1021/cb200353m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McLellan CA, et al. A rhizosphere fungus enhances Arabidopsis thermotolerance through production of an HSP90 inhibitor. Plant Physiol. 2007;145(1):174–182. doi: 10.1104/pp.107.101808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Workman P, Burrows F, Neckers L, Rosen N. Drugging the cancer chaperone HSP90: Combinatorial therapeutic exploitation of oncogene addiction and tumor stress. Ann N Y Acad Sci. 2007;1113:202–216. doi: 10.1196/annals.1391.012. [DOI] [PubMed] [Google Scholar]
- 26.McDonald E, Workman P, Jones K. Inhibitors of the HSP90 molecular chaperone: Attacking the master regulator in cancer. Curr Top Med Chem. 2006;6(11):1091–1107. doi: 10.2174/156802606777812004. [DOI] [PubMed] [Google Scholar]
- 27.Crawford JM, et al. Deconstruction of iterative multidomain polyketide synthase function. Science. 2008;320(5873):243–246. doi: 10.1126/science.1154711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas R. A biosynthetic classification of fungal and streptomycete fused-ring aromatic polyketides. ChemBioChem. 2001;2(9):612–627. doi: 10.1002/1439-7633(20010903)2:9<612::AID-CBIC612>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 29.Li Y, et al. Classification, prediction, and verification of the regioselectivity of fungal polyketide synthase product template domains. J Biol Chem. 2010;285(30):22764–22773. doi: 10.1074/jbc.M110.128504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee MY, Ames BD, Tsai SC. Insight into the molecular basis of aromatic polyketide cyclization: Crystal structure and in vitro characterization of WhiE-ORFVI. Biochemistry. 2012;51(14):3079–3091. doi: 10.1021/bi201705q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ames BD, et al. Crystal structure and functional analysis of tetracenomycin ARO/CYC: Implications for cyclization specificity of aromatic polyketides. Proc Natl Acad Sci USA. 2008;105(14):5349–5354. doi: 10.1073/pnas.0709223105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ames BD, et al. Structural and biochemical characterization of ZhuI aromatase/cyclase from the R1128 polyketide pathway. Biochemistry. 2011;50(39):8392–8406. doi: 10.1021/bi200593m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahuja M, et al. Illuminating the diversity of aromatic polyketide synthases in Aspergillus nidulans. J Am Chem Soc. 2012;134(19):8212–8221. doi: 10.1021/ja3016395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma SM, et al. Complete reconstitution of a highly reducing iterative polyketide synthase. Science. 2009;326(5952):589–592. doi: 10.1126/science.1175602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vagstad AL, et al. Combinatorial domain swaps provide insights into the rules of fungal polyketide synthase programming and the rational synthesis of non-native aromatic products. Angew Chem Int Ed Engl. 2013;52(6):1718–1721. doi: 10.1002/anie.201208550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vagstad AL, Bumpus SB, Belecki K, Kelleher NL, Townsend CA. Interrogation of global active site occupancy of a fungal iterative polyketide synthase reveals strategies for maintaining biosynthetic fidelity. J Am Chem Soc. 2012;134(15):6865–6877. doi: 10.1021/ja3016389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wallner B, Elofsson A. Pcons5: Combining consensus, structural evaluation and fold recognition scores. Bioinformatics. 2005;21(23):4248–4254. doi: 10.1093/bioinformatics/bti702. [DOI] [PubMed] [Google Scholar]
- 38.Gaffoor I, Trail F. Characterization of two polyketide synthase genes involved in zearalenone biosynthesis in Gibberella zeae. Appl Environ Microbiol. 2006;72(3):1793–1799. doi: 10.1128/AEM.72.3.1793-1799.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwan DH, Leadlay PF. Mutagenesis of a modular polyketide synthase enoylreductase domain reveals insights into catalysis and stereospecificity. ACS Chem Biol. 2010;5(9):829–838. doi: 10.1021/cb100175a. [DOI] [PubMed] [Google Scholar]
- 40.Kwan DH, Tosin M, Schläger N, Schulz F, Leadlay PF. Insights into the stereospecificity of ketoreduction in a modular polyketide synthase. Org Biomol Chem. 2011;9(7):2053–2056. doi: 10.1039/c1ob00022e. [DOI] [PubMed] [Google Scholar]
- 41.Lee KK, Da Silva NA, Kealey JT. Determination of the extent of phosphopantetheinylation of polyketide synthases expressed in Escherichia coli and Saccharomyces cerevisiae. Anal Biochem. 2009;394(1):75–80. doi: 10.1016/j.ab.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 42.Gietz RD, Schiestl RH. Quick and easy yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc. 2007;2(1):35–37. doi: 10.1038/nprot.2007.14. [DOI] [PubMed] [Google Scholar]
- 43.Ginalski K, Elofsson A, Fischer D, Rychlewski L. 3D-Jury: A simple approach to improve protein structure predictions. Bioinformatics. 2003;19(8):1015–1018. doi: 10.1093/bioinformatics/btg124. [DOI] [PubMed] [Google Scholar]
- 44.Karplus K. SAM-T08, HMM-based protein structure prediction. Nucleic Acids Res. 2009;37(Web Server issue):W492–W497. doi: 10.1093/nar/gkp403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eswar N, et al. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics. 2006;15:5.6.1–5.6.30. doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.