PD-L1 decreases anti-viral CD8+ T cell motility and PD-1 blockade restores motility in the presence of high viral loads.
Abstract
Immune responses to persistent viral infections and cancer often fail because of intense regulation of antigen-specific T cells—a process referred to as immune exhaustion. The mechanisms that underlie the induction of exhaustion are not completely understood. To gain novel insights into this process, we simultaneously examined the dynamics of virus-specific CD8+ and CD4+ T cells in the living spleen by two-photon microscopy (TPM) during the establishment of an acute or persistent viral infection. We demonstrate that immune exhaustion during viral persistence maps anatomically to the splenic marginal zone/red pulp and is defined by prolonged motility paralysis of virus-specific CD8+ and CD4+ T cells. Unexpectedly, therapeutic blockade of PD-1–PD-L1 restored CD8+ T cell motility within 30 min, despite the presence of high viral loads. This result was supported by planar bilayer data showing that PD-L1 localizes to the central supramolecular activation cluster, decreases antiviral CD8+ T cell motility, and promotes stable immunological synapse formation. Restoration of T cell motility in vivo was followed by recovery of cell signaling and effector functions, which gave rise to a fatal disease mediated by IFN-γ. We conclude that motility paralysis is a manifestation of immune exhaustion induced by PD-1 that prevents antiviral CD8+ T cells from performing their effector functions and subjects them to prolonged states of negative immune regulation.
Persistent viral infections and tumors often pose a challenge to the immune system by exposing T and B cells to heightened antigenic loads and/or diverse immunoregulatory machinery. Consequently, lymphocytes exposed to these environments are stricken with a state of dysfunction commonly referred to as immune exhaustion (Wherry, 2011). The term exhaustion refers to a state of functional decline that occurs when lymphocytes are chronically exposed to an antigen. During a persistent viral infection, this is operationally defined for T cells as a progressive loss in their ability to lyse target cells and produce important cytokines such as IFN-γ, TNF, and IL-2 (Ahmed and Oldstone, 1988; Zajac et al., 1998; Brooks et al., 2005; Wherry et al., 2003). Exhaustion can in some instances be followed by clonal deletion, resulting in the physical removal of antiviral cells from the immune repertoire (Moskophidis et al., 1993). It is now widely accepted that immune exhaustion contributes to the persistence of many viruses as well as tumors and is maintained by negative immune regulators such as PD-1 (Barber et al., 2006; Velu et al., 2009), IL-10 (Brooks et al., 2006b; Ejrnaes et al., 2006), and CTLA-4 (Kaufmann et al., 2007). Recent studies have shown that therapeutic blockade of negative immune regulators can reverse immune exhaustion and promote clearance of both viruses and tumors (Kim and Ahmed, 2010). Immunoregulatory blockade can also be added to therapeutic vaccination regimens to improve their efficacy (Brooks et al., 2008; Ha et al., 2008). In general, immune regulators are a very promising clinical target, and recent trials have demonstrated that blockade of the PD-1–PD-L1 pathway promotes the clearance of tumors in humans (Brahmer et al., 2012; Topalian et al., 2012).
Although several studies have shed light on the mechanics of lymphocyte exhaustion at a molecular and functional level (Wherry, 2011), little is known about how exhaustion manifests at a dynamic level in living tissues. T cells effectively mount their defense against invading pathogens by moving throughout infected tissues (Hickman et al., 2009; Coombes and Robey, 2010). Studies have revealed that effector T cells can maximize their efficiency by outnumbering infected target cells (Li et al., 2009), engaging multiple targets simultaneously (McGavern et al., 2002) or serially (Bossi et al., 2002; Rothstein et al., 1978), and by participating in short-duration (∼5–15 min) interactions (Stinchcombe et al., 2001; Mempel et al., 2006; Ganusov and De Boer, 2008). These host strategies have evolved to provide a defense against pathogens that replicate exponentially and attempt to outpace the immune system. In fact, whether a pathogen persists or not is often decided within the first week of infection (Althaus et al., 2007), and anything that interferes with the efficiency of immune cell surveillance has the potential to shift the balance in favor of persistence. It is therefore of great importance to understand the factors that influence immune cell dynamics after infection. Recent studies have demonstrated that negative immune regulators such as CTLA-4 (Schneider et al., 2006) and PD-1–PD-L1 (Fife et al., 2009; Yokosuka et al., 2012) can deliver a repulsionary signal to CD4 T cells that overrides TCR-induced stop signals and reduces stable target cell engagement in situations where TCR recognition is attenuated by tolerance mechanisms (Schubert et al., 2012). T cell repulsion is an interesting mechanism to prevent the development of autoimmunity; however, it is unclear whether the same rules apply to antiviral T cells for which TCR recognition is optimal.
To gain novel insights into the anatomy and dynamics of T cell exhaustion, we set out to study antiviral CD8+ and CD4+ T cells in the living spleen (Aoshi et al., 2008) during an acute or persistent viral infection using laser scanning two-photon microscopy (TPM; Zinselmeyer et al., 2009). We exploited two commonly used strains of lymphocytic choriomeningitis virus (LCMV) that differ by only 3 aa (Ahmed and Oldstone, 1988; Bergthaler et al., 2010; Sullivan et al., 2011). After intravenous inoculation, the Armstrong (Arm) strain of LCMV is cleared in 7–10 d, whereas the clone 13 (CL13) strain establishes persistence. The ability of CL13 to persist stems from a mutation in its glycoprotein that enables the virus to bind to its cellular receptor, α-dystroglycan, with higher affinity relative to Arm (Sevilla et al., 2000) and a mutation in the polymerase that gives the virus a replicative advantage (Bergthaler et al., 2010). These mutations afford CL13 the ability to establish systemic persistence, which results in the engagement of immune dampening pathways such as the PD-1 (Barber et al., 2006) and IL-10 (Brooks et al., 2006b; Ejrnaes et al., 2006) that induce T cell exhaustion (Wherry, 2011). It is well described that exhaustion impedes the function of antiviral T cells, and the process of immune exhaustion begins within the first week of a CL13 infection (Ahmed and Oldstone, 1988; Wherry et al., 2003). However, it is not known where functional exhaustion is mediated anatomically or how this state impacts T cell motility. Here, we show using TPM that the initiation of antiviral T cell exhaustion during CL13 persistence is facilitated by PD-1–PD-L1-induced motility paralysis in the splenic red pulp/marginal zone.
RESULTS
Viral persistence causes T cell motility arrest in the splenic red pulp
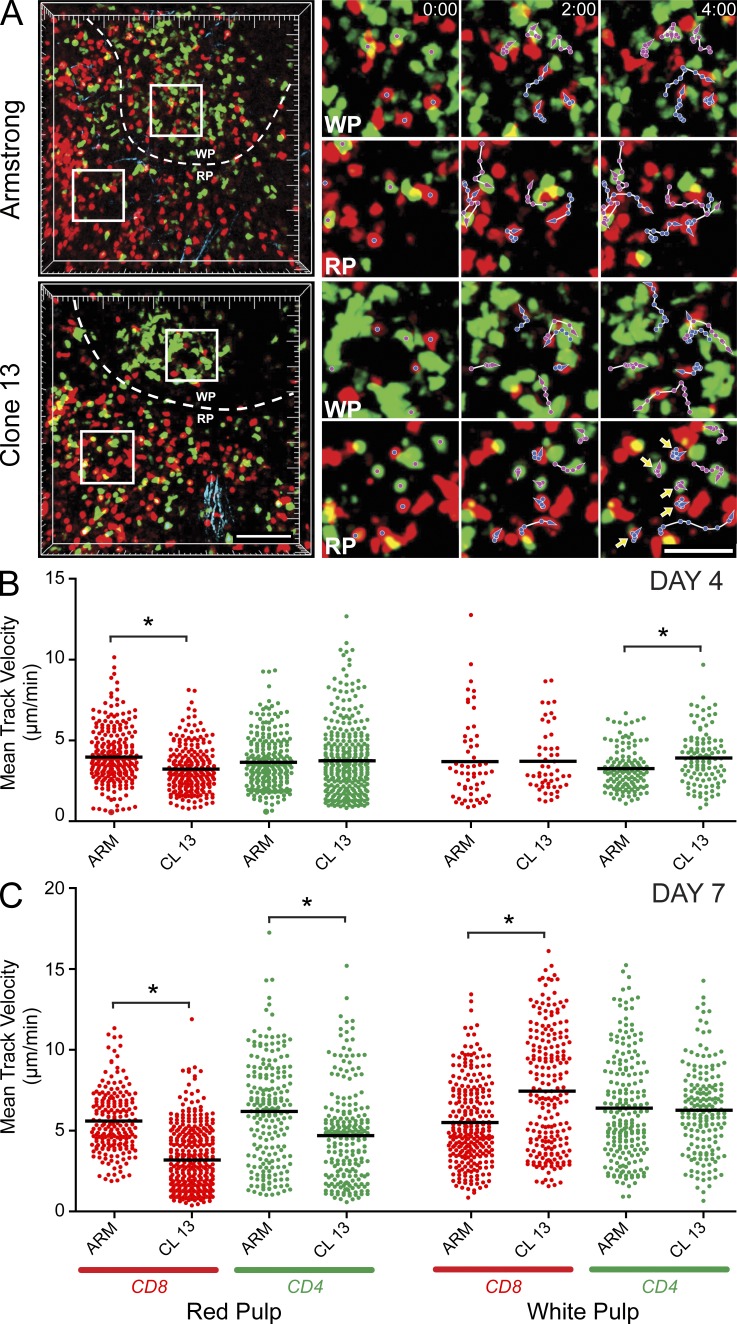
To examine the dynamics of virus-specific CD8+ and CD4+ T cells during acute versus persistent viral infection, we transferred 5,000 cyan fluorescent protein (CFP)–tagged DbGP33-41 TCR transgenic (tg) CD8+ T cells (CFP+ P14 cells) and 5,000 GFP-tagged I-AbGP61-80 TCR-tg CD4+ T cells (GFP+ SMARTA cells) into B6 mice 1 d before i.v. inoculation with LCMV Armstrong (Arm) or clone 13 (CL13). After i.v. inoculation, the spleen is rapidly infected by both viruses; however, only CL13 induces immune exhaustion in T cells and persists (Zajac et al., 1998). We therefore examined P14 (Pircher et al., 1989) and SMARTA (Oxenius et al., 1998) dynamics in the splenic white and red pulp by TPM at two time points after infection (Fig. 1 and Videos 1–3). Day 4 was selected as a time point when Arm and CL13 reach comparable titers in the spleen, and antiviral T cells to both viruses remain functional (Brooks et al., 2006a). In contrast, the fates of Arm and CL13 diverge at day 7 with only CL13 capable of inducing T cell exhaustion and establishing persistence. Day 7 was chosen for analysis because antiviral T cells already show early signs of immune exhaustion (e.g., reductions in cytokine production and cytolytic activity) at this time point (Ahmed and Oldstone, 1988; Wherry et al., 2003). We separately quantified antiviral T cell dynamics in the splenic marginal zone/red pulp and white pulp (Video 1) because we theorized that T cell behaviors would differ in these two different microenvironments after infection (Fig. 1 A and Videos 1–3). For example, at day 4 after infection subtle differences in T cell motility had already emerged between the two models of infection (Fig. 1 B and Video 2). When compared with Arm infection, P14 cells in the red pulp of CL13-infected mice showed reduced mean track velocities (Fig. 1 B) and motility coefficients (a measure of displacement; not depicted), suggesting enhanced CD8 T cell engagement in this splenic region. In contrast, no differences in P14 motility were observed in the splenic white pulp in Arm- versus CL13-infected mice. The only other notable difference at day 4 after infection was the elevated mean track velocity for SMARTA cells in the CL13 white pulp (Fig. 1 B). Collectively, these data support the conclusion that at day 4 after CL13 infection, P14 cells establish more interactions in the red pulp, whereas SMARTA cells engage (or have access to) less targets in the white pulp when compared with Arm infection.
Figure 1.
Antiviral T cell dynamics after acute versus persistent infection. (A) TPM was used to examine the dynamics of CD8+ P14 (red) and CD4+ SMARTA (green) T cells in the spleen after an acute (Arm) or a persistent (CL13) infection (n = 5 mice). Representative 3D reconstructions of two-photon z stacks are shown for infected mice at day 7 after infection (left). The dashed white lines demarcate the border between the splenic red (RP) and white pulp (WP). The white boxes represent regions of interest from the white or red pulp magnified in the panels on the right. The magnified views show antiviral T cell movement over a 5-min time interval. Blue tracks follow the movement of P14 cells, whereas magenta tracks follow SMARTA cells. Note that nearly all highlighted T cells move over the denoted time interval except for P14 and SMARTA cells residing in the red pulp of CL13-infected mice (yellow arrowheads). Bars: (left) 100 µm; (right) 40 µm. See Videos 1–3. (B and C) Mean track velocities (µm/min) of CD8+ P14 (red) and CD4+ SMARTA (green) cells were quantified in the splenic red and white pulp at days 4 (B) and 7 (C) after Arm or CL13 infection (n = 5 mice). Asterisks denote statistically significant differences (P < 0.05). Each dot represents the track of an individual T cell. Horizontal black bars denote the mean of each group. Data are representative of five independent experiments.
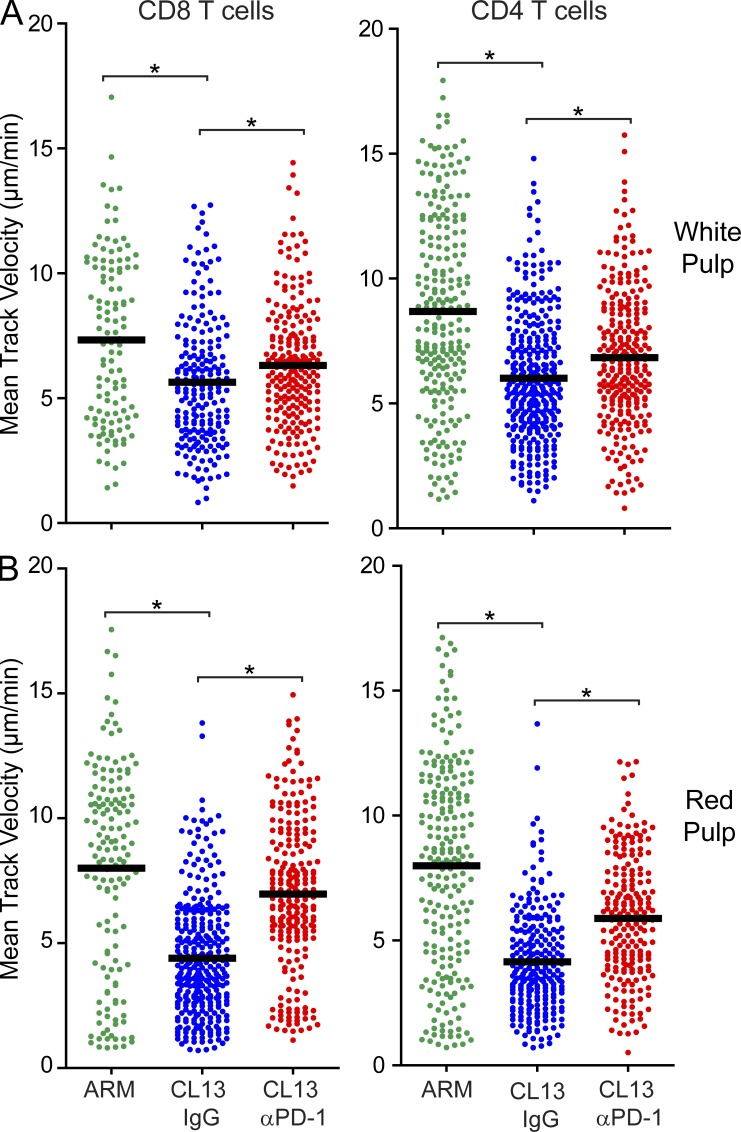
At day 7 after infection, marked decreases in both P14 and SMARTA T cell motility were observed in the red pulp of CL13-infected mice (Fig. 1 C and Video 3). Red pulp P14 velocities (Fig. 1 C), motility coefficients (not depicted), and meandering factors (not depicted) were all significantly decreased in CL13- versus Arm-infected mice. In addition, P14 arrest durations often exceeded 40 min (see also Fig. 3 C), indicating that CL13 infection induced prolonged motility paralysis of antiviral T cells in the red pulp. In contrast, no differences in SMARTA T cell motility were observed in the white pulp, and P14 cells in CL13-infected mice actually showed increased mean track velocities (Fig. 1 C) and motility coefficients (not depicted) in this anatomical region. These data indicate that CL13 infection preferentially impedes antiviral T cell motility in the splenic red pulp.
Figure 3.
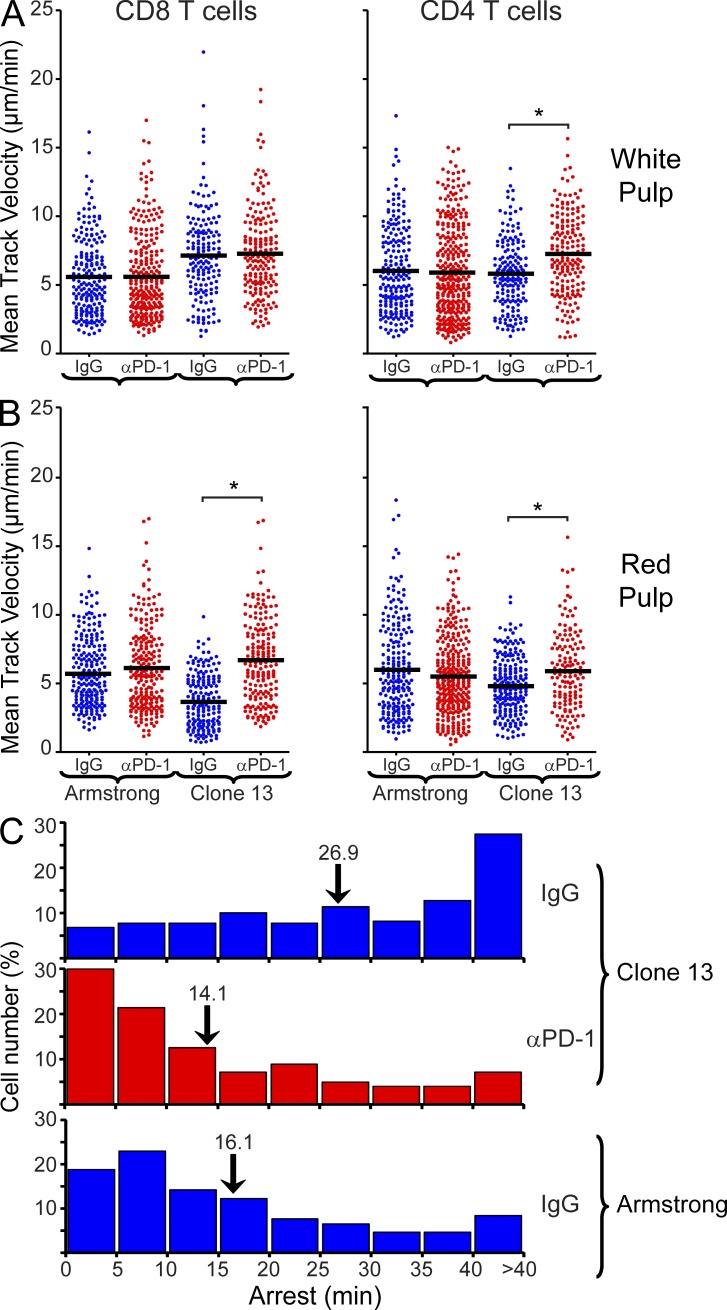
Elevated T cell motility in the red pulp after PD-1 blockade at an early stage of exhaustion. (A and B) Mean track velocities (µm/min) of CD8+ P14 and CD4+ SMARTA cells were quantified in the splenic white (A) and red (B) pulp at day 7 after infection (n = 4 mice). 16 h before imaging, anti–PD-1 or isotype control antibodies (500 µg) were injected intravenously. Asterisks denote statistically significant differences (P < 0.05). Each dot represents the track of an individual T cell. Horizontal black bars denote the mean of each group. (C) Frequency distributions (5 min bins) show the average arrest duration of P14 cells in the splenic red pulp 16 h after injection of anti–PD-1 (224 cells analyzed) or isotype control antibodies (Arm, n = 219 cells analyzed; CL13, n = 261 cells analyzed; n = 4 mice). Numbers above the black arrows denote the mean of each group. Relative to the isotype control group, a statistically significant decrease (P < 0.05) in mean arrest duration was noted after injection of anti–PD-1 into CL13-infected mice. No difference was observed between Arm mice treated with isotype control antibody and CL13 mice treated with anti–PD-1. See corresponding Video 4. Data are representative of four independent experiments.
PD-L1 is highly expressed in splenic red pulp and marginal zone cells during the establishment of viral persistence
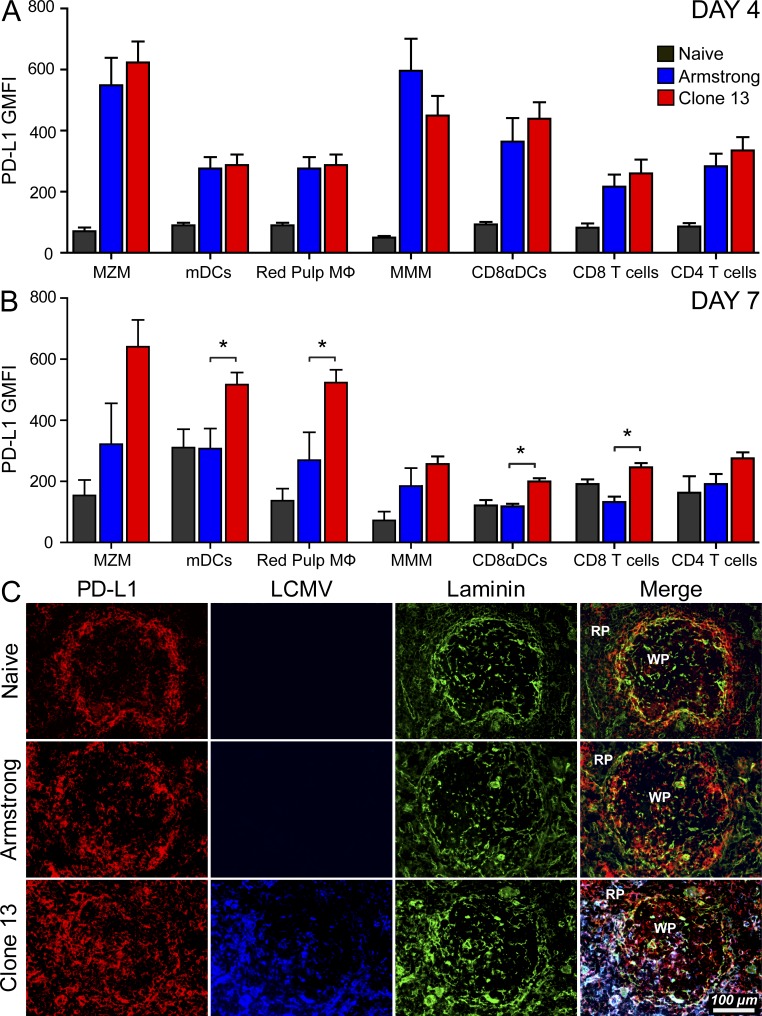
To define the mechanism underlying decreased antiviral T cell motility in the splenic red pulp after CL13 infection, we examined the splenic distribution of LCMV and a known inhibitory ligand (PD-L1; Dong et al., 1999; Freeman et al., 2000). The PD-1–PD-L1 pathway plays an important role in regulating antiviral T cells during persistent viral infection (Sharpe et al., 2007), and blockade of this pathway in the CL13 model restores T cell function and promotes viral clearance (Barber et al., 2006). Both PD-1 and PD-L1 were expressed comparably on splenic P14 and SMARTA cells extracted from Arm versus CL13 mice at day 4 after infection. At day 7 after infection, PD-1 and PD-L1 remained elevated on tg T cells obtained from Arm-infected mice (relative to naive cells); however, levels were significantly higher on cells from CL13 mice (not depicted). The PD-1 receptor is expressed primarily on antiviral T cells after infection, but can also be expressed broadly on many different cell populations (Sharpe et al., 2007; Keir et al., 2008). We examined the splenic distribution of PD-L1 flow cytometrically (Fig. 2, A and B) and immunohistochemically (Fig. 2 C) in naive as well as LCMV-infected mice. As reported (Mueller et al., 2007), PD-L1 was elevated on gp38+ lymphoid reticular cells at day 7 after infection, but was comparably expressed in Arm versus CL13-infected mice (Arm GMFI, 475.0 ± 29.3; CL13, 356.5 ± 31.2; naive, 40.3 ± 28.2). Therefore, we next examined the hematopoietic compartment. At day 4 after infection, PD-L1 was up-regulated comparably in both Arm- and CL13-infected mice on all splenic immune cell populations examined (Fig. 2 A). By day 7, PD-L1 remained elevated in CL13-infected mice but began to decrease in Arm-infected mice (Fig. 2 B). The highest geometric mean fluorescent intensities were observed on CL13 marginal zone macrophages (MZM), myeloid DCs (mDCs), and red pulp macrophages—all populations that reside in the splenic red pulp or marginal zone. We confirmed this immunohistochemically by examining PD-L1 expression on splenic frozen sections (Fig. 2 C). The splenic distribution of PD-L1 in Arm-infected mice at day 7 was similar to that observed in naive mice. LCMV antigen was difficult to detect, and PD-L1 was expressed primarily on cells residing at the laminin rich border between the splenic red and white pulp. In contrast, PD-L1 was heavily expressed in CL13-infected mice, primarily in the marginal zone and red pulp, although some staining was observed in the white pulp as well. Viral antigen was abundant and localized primarily to regions of heightened PD-L1 expression (i.e., the marginal zone/red pulp).
Figure 2.
Splenic distribution of PD-L1 expression after infection. (A and B) The geometric mean fluorescent intensity (GMFI) of PD-L1 expression (mean ± SD) was quantified on splenic marginal zone macrophages (CD45+ Thy1.2− F4/80− CD209b+; MZM), myeloid dendritic cells (CD45+ Thy1.2− F4/80− CD169− CD209b− CD11c+ CD11b+; mDCs), red pulp macrophages (CD45+ Thy1.2− CD169− CD209b− CD11c− CD11b+ F4/80+; red pulp Mϕ), metallophilic macrophages (CD45+ Thy1.2− F4/80− CD169+; MMM), CD8α DCs (CD45+ Thy1.2− F4/80− CD169− CD209b− CD11b− CD11c+ CD8α+), CD8 T cells (CD45+ Thy1.2+ CD8α+), and CD4 T cells (CD45+ Thy1.2+ CD4+) at days 4 and 7 after Arm or CL13 infection. Total expression of PD-L1 on the denoted cell populations was defined as PD-L1 GMFI minus the isotype control GMFI. Naive mice were used as a control at both time points (n = 4–5 mice). Asterisks denote statistically significant differences between the Arm and CL13 groups (P < 0.05). (C) Representative 3-color splenic images show the distribution of PD-L1 (red), LCMV (blue), and laminin (green) in naive, day 7 Arm, and day 7 CL13-infected mice (n = 4 mice). All data in this figure are representative of two independent experiments.
Therapeutic blockade of PD-1 and PD-L1 increases antiviral T cell motility and function
Because PD-L1 expression was markedly elevated in the splenic marginal zone/red pulp, we next evaluated the degree to which PD-1–PD-L1 interactions contributed to reduced T cell motility on day 7 (Fig. 3 and Video 4). Anti–PD-1 or isotype control antibodies were injected i.v. and T cell motility was quantified by TPM in the spleen 16 h later. We injected antibodies i.v. because this route of administration is known to deliver molecules to the spleen within seconds of injection (Waite et al., 2011). Importantly, anti–PD-1 administration completely restored the motility of P14 and SMARTA cells in the splenic red pulp at 16 h (Fig. 3 B and Video 4). Mean track velocities, motility coefficients, and meandering factors were comparable to that observed in mice infected with Arm. In addition, the average P14 arrest duration decreased significantly (P < 0.05) from 26.9 to 14.1 min after anti–PD-1 administration, and the majority (>50%) of P14 cells engaged their splenic targets for <15 min, comparable to cells in Arm-infected mice (Fig. 3 C). This contrasted with the long P14 arrest durations observed in the isotype control group (one third exceeded 40 min). These data indicate that PD-1–PD-L1 interactions promote prolonged arrest of antiviral T cells, and blockade of this pathway increases T cell motility despite the presence of persistent viral antigen.
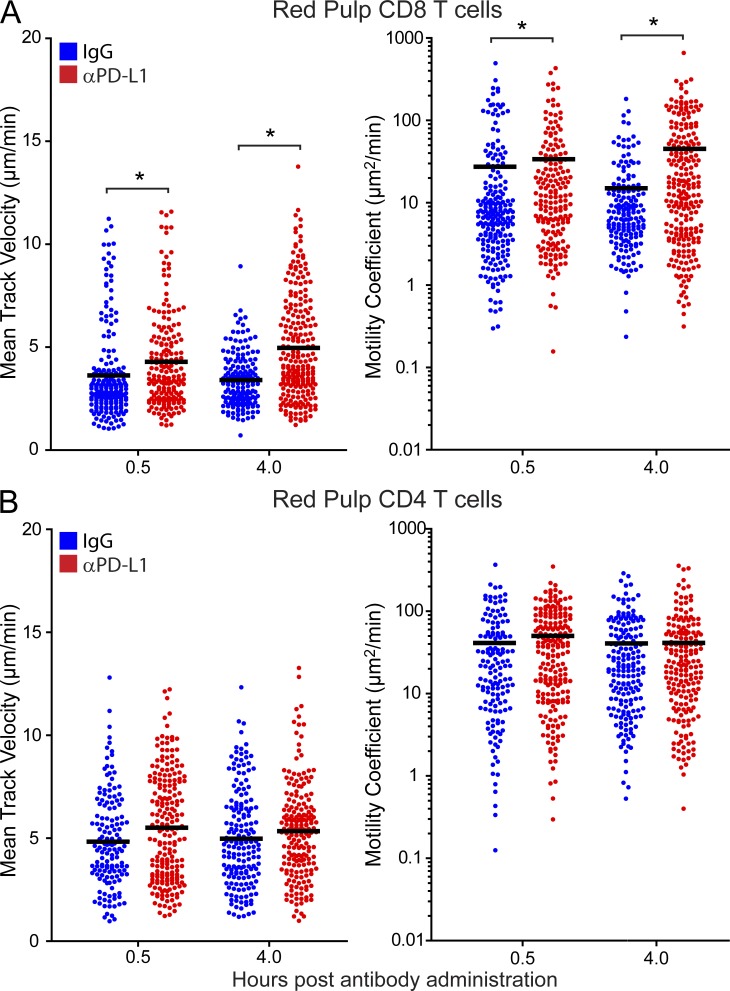
We postulated that prolonged cellular arrest mediated by the PD-1–PD-L1 pathway could be another manifestation of immune exhaustion (Wherry, 2011) that reduced the ability of antiviral T cells to perform their effector functions in vivo. To test this hypothesis, we assessed the kinetics of red pulp T cell motility recovery in relation to cytokine production and viral clearance (Fig. 4). A statistically significant increase in P14 mean track velocity was observed just 30 min after administration of anti–PD-1, and velocity was further increased between 10 and 16 h (Fig. 4 A). In contrast, red pulp SMARTA cells did not show a significant increase in velocity until 16 h (Fig. 4 B). Injecting anti–PD-L1 antibodies and quantifying antiviral T cell motility in the splenic red pulp confirmed these findings. When compared with isotype controls, a statistically significant increase in P14 mean track velocity and motility coefficients was observed at both 30 min and 4 h after injection of anti–PD-L1 (Fig. 5 A). No significant increase in SMARTA motility was observed at either time point (Fig. 5 B). The data demonstrate that the kinetics of motility recovery at day 7 differ between antiviral CD8 and CD4 T cells.
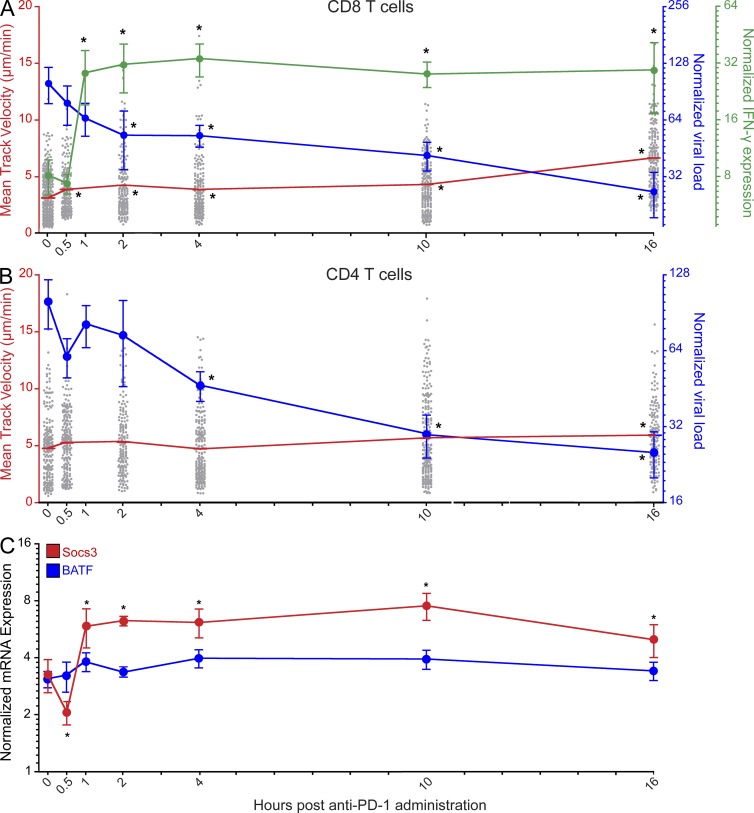
Figure 4.
Enhanced CD8 T cell motility precedes functional restoration and is associated with viral clearance. (A and B) Splenic mean track velocities of CD8+ P14 (A) and CD4+ SMARTA cells (B) in the red pulp (gray dots; red lines; n = 3–4 mice), IFN-γ expression (green line; n = 8 mice), and viral loads as determined by plaque assay (A) or Q-PCR (B; blue lines; n = 8 mice) were quantified at different time points after anti–PD-1 injection on day 7. Asterisks denote statistically significant differences (P < 0.05) from baseline (time point zero). Lines with error bars (green and blue) are plotted as mean ± SEM. The red lines for P14 and SMARTA cell motility represent the mean of each group. Each gray dot represents the track of an individual T cell. Data are representative of 2–4 independent experiments. (C) Socs3 (red) and BATF (blue) mRNA expression were quantified by Q-PCR at different time points after anti–PD-1 injection on day 7 (n = 8 mice). Asterisks denote statistically significant differences (P < 0.05) from baseline. Data are plotted mean ± SEM and are representative of two independent experiments.
Figure 5.
PD-L1 blockade increases virus-specific CD8+ T cell motility. (A and B) Splenic mean track velocities (left) and motility coefficients (right) for CD8+ P14 (A) and CD4+ SMARTA cells (B) in the red pulp were quantified at 0.5 and 4 h after i.v. injection of anti–PD-L1 (red) or isotype control (blue) antibodies on day 7. Asterisks denote statistically significant differences (P < 0.05). Each dot represents the track of an individual T cell. Data are representative of four independent experiments.
At the cytokine level, IFN-γ mRNA markedly increased between 30 min and 1 h after anti–PD-1 administration and remained elevated at all time points thereafter (Fig. 4 A). This was preceded by a significant decline in SOCS3 (a suppressor of STAT3-dependent cytokine signaling; Pellegrini et al., 2011) expression at 30 min; however, SOCS3 rapidly increased by 1 h, and, ultimately, mirrored the pattern of elevated IFN-γ expression (Fig. 4 C). Interestingly, enhanced antiviral T cell motility did not coincide with any changes (up or down) in the expression of BATF (Fig. 4 C), a transcription factor recently linked to PD-1 signaling (Quigley et al., 2010). The sharp increase in IFN-γ expression coincided with a decline in viral loads as measured by plaque assay (Fig. 4 A) and quantitative PCR (Q-PCR; Fig. 4 B). The first statistically significant reduction in virus was observed by plaque assay 2 h after administration of anti–PD-1 and at 4 h by Q-PCR. By 16 h, infectious virus was reduced on average by 74% in the spleen. For example, in one experiment, splenic viral titers declined from 4.75 × 107 (0 min) to 4.64 × 106 (16 h) PFU/g after anti–PD-1 administration– a 90% reduction in infectious virus. These data indicate that PD-1–PD-L1 blockade increases virus-specific CD8 T cell motility shortly after administration (within 30 min), which is followed by elevated IFN-γ transcription, a sharp decline in viral loads, and then an increase in antiviral CD4 T cell motility.
Because day 7 represents an early stage of immune exhaustion, we next examined whether PD-1 blockade similarly increased antiviral T cell motility at a later time point after infection (Fig. 6). We selected day 14 because engagement of immunosuppressive pathways often peaks at this time point after CL13 infection. Interestingly, PD-1 blockade significantly enhanced P14 and SMARTA motility in both the white and red pulp 4 h after anti–PD-1 administration, although the increases were still more substantial in the red pulp. Thus, inhibition of antiviral T cell motility by the PD-1 pathway is not restricted to the early stages of immune exhaustion (day 7). By day 14, the effect is equally prominent and even influences T cells residing in the splenic white pulp.
Figure 6.
PD-1 blockade enhances white and red pulp T cell motility at a later stage of exhaustion. (A and B) Mean track velocities (µm/min) of CD8+ P14 and CD4+ SMARTA cells were quantified in the splenic white (A) and red (B) pulp at day 14 after infection (n = 4 mice). 4 h before imaging, anti–PD-1 or isotype control antibodies (500 µg) were injected intravenously. Asterisks denote statistically significant differences (P < 0.05). Each dot represents the track of an individual T cell. Horizontal black bars denote the mean of each group.
PD-L1 localizes to the cSMAC and stabilizes immunological synapse formation
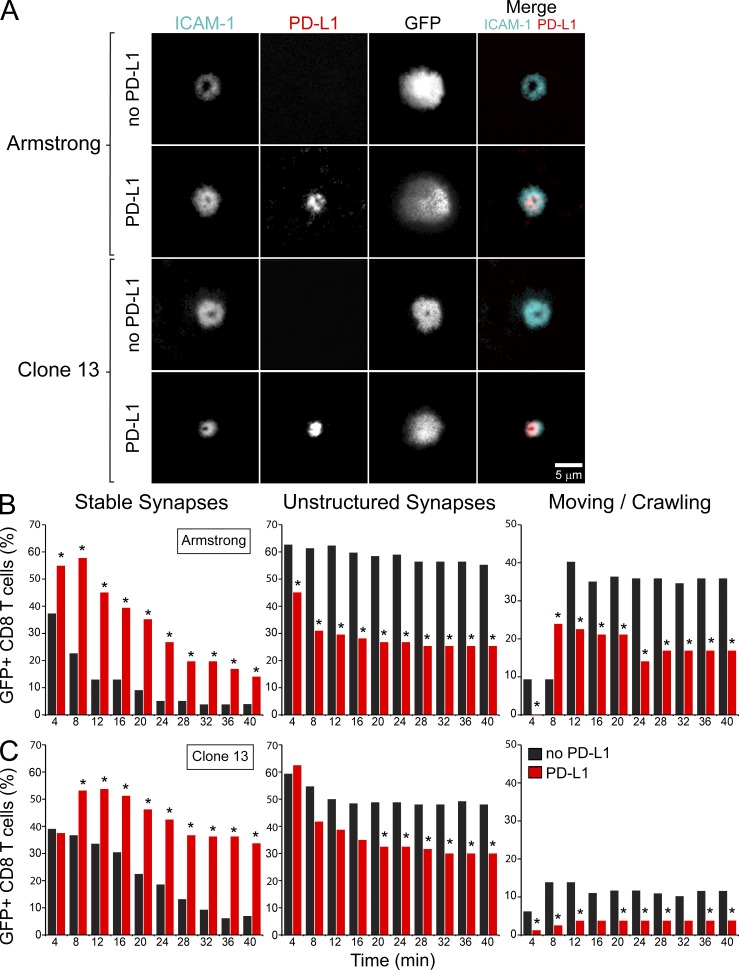
Recent studies using in vitro–activated T cells have demonstrated that PD-1 localizes to inhibitory microclusters upon engagement of PD-L1 and may destabilize CD4 T cell interactions (Yokosuka et al., 2012). During persistent viral infection, our TPM data suggested that PD-1–PD-L1 actually promoted antiviral T cell arrest in the splenic red pulp. To provide additional support for this theory, we developed a reductionist system to study the direct impact of PD-L1 on in vivo generated LCMV-specific CD8+ T cell interactions. To this end, we examined the motility and immunological synapses of day 7 Arm and CL13 splenic P14 cells on planar lipid bilayers (Grakoui et al., 1999) loaded with ICAM-1 and cognate peptide-MHC I (H-2DbGP33-41) in the presence or absence of PD-L1 (Fig. 7; and Videos 5 and 6). We chose to focus our efforts primarily on LCMV-specific CD8+ (rather than CD4+) T cells because of the profound impact that PD-1–PD-L1 had on stabilizing P14 cells in the splenic red pulp (Fig. 3–6). During immunological synapse formation, PD-L1 was found in the central supramolecular activation complex (cSMAC; Monks et al., 1998) for both Arm and CL13 P14 cells (Fig. 7 A and Videos 5 and 6). ICAM-1 localized primarily to the peripheral supramolecular activation complex (pSMAC; Monks et al., 1998), but was more closely concentrated around the cSMAC in the presence of PD-L1, particularly for CL13 P14 cells (Fig. 7 A). This conclusion was supported by radial intensity measurements demonstrated that ICAM-1 signal was actually elevated in the synapses of P14 cells that engaged PD-L1–loaded bilayers (Fig. 8 C).
Figure 7.
PD-L1 promotes attachment and synapse formation of virus-specific CD8+ T cells on planar bilayers. Real-time TIRFM imaging was used to assess the ability of GFP+ CD8+ P14 cells to form stable synapses on planar bilayers and to examine the distribution of ICAM-1/PD-L1 after synapse formation. (A) Purified GFP+ CD8+ P14 cells from day 7 spleens of Arm or CL13-infected mice were loaded onto planar lipid bilayers containing ICAM-1AF405 (cyan, 200 molecules/µm2) and H-2DbGP33-41 (unlabeled, 185 molecules/µm2), in the presence or absence PD-L1AF568 (red, 275 molecules/µm2). Representative images show P14 cell synapses ∼15 min after they were placed on the bilayers. Images represent data from at least three independent experiments per condition. See corresponding Video 5. (B and C) Histograms show the frequency of day 7 Arm (B) or CL13 (C) GFP+ P14 cells that formed synapses, unstructured synapses, or were moving after placement on bilayers with or without PD-L1. P14 cells were incubated on the bilayers for 10 min before imaging, and thus the denoted time points reflect the number of min after this initial incubation period. Histograms reflect the number of GFP+ P14 cells per condition: Arm no PD-L1 (n = 78 cells), Arm PD-L1 (n = 71 cells), CL13 no PD-L1 (n = 130 cells), CL13 PD-L1 (n = 80 cells). Asterisks denote statistically significant differences between the two conditions (no PD-L1 vs. PD-L1) at each time point (P < 0.05). Data are representative of two independent experiments.
Figure 8.
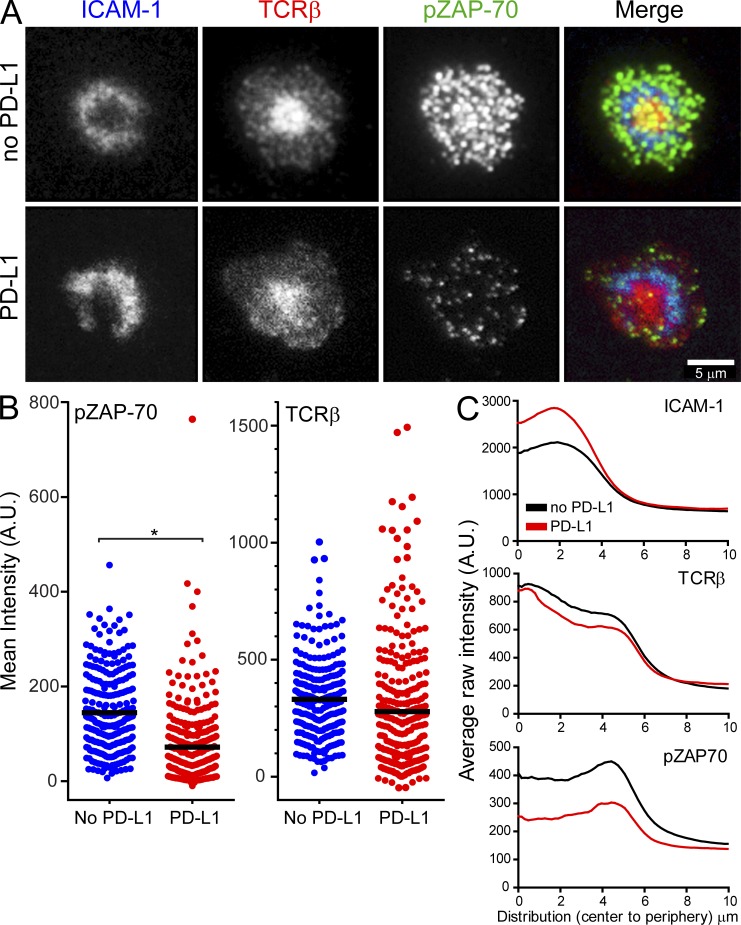
PD-L1 diminishes pZAP-70 clusters in synapse-forming virus-specific CD8+ T cells on planar bilayers. Fixed sample TIRFM imaging was used to assess proximal TCR signaling by measuring fluorescence intensity and distribution of phosphorylated ZAP-70 and TCR microclusters on synapse-forming GFP+ CD8+ P14 cells. (A) Purified GFP+ CD8+ P14 cells from day 7 spleens of CL13-infected mice were incubated with TCRβ (H57) FabAF568 (red) and loaded onto planar lipid bilayers containing ICAM-1AF405 (blue, 200 molecules/µm2) and H-2DbGP33-41 (unlabeled, 185 molecules/µm2), in the absence or presence of PD-L1 (unlabeled, 275 molecules/µm2). P14 cells were fixed 5 min after placement on the bilayers and stained with anti-pZAP-70AF647 (green). (B) Dot plots show quantification of mean intensity fluorescence (background corrected raw mean intensities) for p-ZAP-70 and TCRβ. The asterisk denotes a statistically significant difference (P < 0.05). Each dot represents an individual P14 T cell for the two conditions (no PD-L1, n = 255 cells; PD-L1, n = 305 cells). Horizontal black bars denote the mean of each group. Data are representative of two independent experiments. (C) Radial distributions of after raw intensities (fluorescence) for ICAM-1, TCRβ, and p-ZAP-70 were calculated and plotted for day 7 CL13 GFP+ CD8+ P14 cells interacting with bilayers. Intensity distributions are shown from cell center to periphery (µm) in all graphs. For illustration purposes, P14 cells with a spreading area >93 µm2 are shown. Note that the presence of PD-L1 in the bilayers decreases p-ZAP-70 and increases ICAM-1, but has no impact on TCRβ expression. Graphs are representative of two independent experiments.
Using the bilayer system, we next addressed whether PD-L1 promoted or impeded P14 synapse formation. Quantitative analyses revealed that PD-L1 enhanced stable mature synapse formation for P14 cells obtained from both Arm- (Fig. 7 B) and CL13-infected (Fig. 7 C) mice. In the absence of PD-L1, P14 cells formed more asymmetric, motile junctions referred to as kinapses (Dustin, 2008), as well as more unstructured synapses lacking defined SMACs. In general, CL13 P14 cells (compared with Arm cells) showed decreased motility when placed on the bilayers, and this motility was reduced even further in the presence of PD-L1 (Fig. 7, B and C). Dephosphorylation of proximal TCR signaling molecules like p-ZAP-70 is thought to be the mechanism by which PD-L1 blocks TCR-induced stop signals and destabilizes synapse formation for antigen-specific CD4+ T cells (Yokosuka et al., 2012). Because PD-L1 promoted rather than destabilized the synapse formation of antiviral P14 cells, we quantified synaptic p-ZAP-70 expression in the presence or absence of PD-L1. Interestingly, PD-L1 had little impact on TCR-β concentration in the cSMAC, but markedly reduced p-ZAP-70 in microclusters (Fig. 8, A–C). These data indicate that PD-L1 can promote P14 synapse formation despite impairing proximal TCR signaling.
PD-1 blockade restores antiviral T cell signaling and induces fatal disease
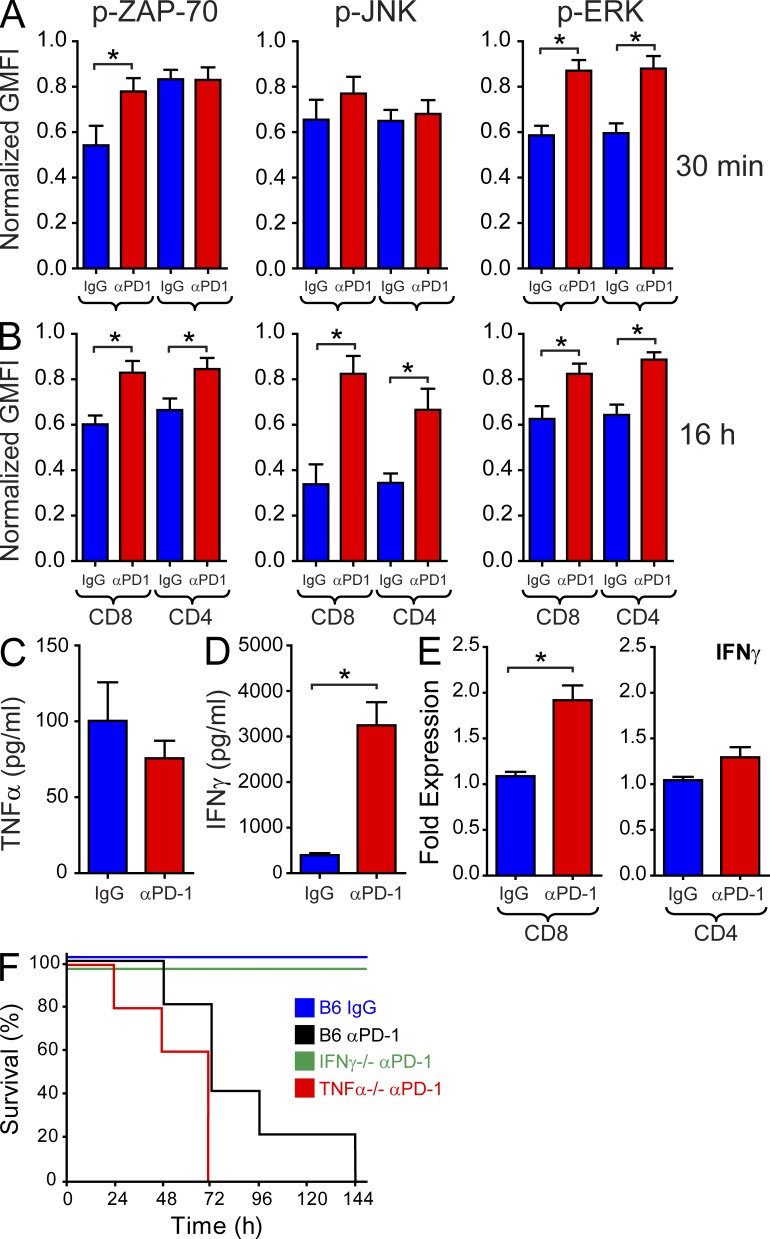
During viral persistence, stabilization of antiviral T cells in a state of arrest would provide target cells with ample time to deactivate key signaling pathways and promote functional exhaustion. This would, in turn, protect the host from developing severe immunopathology. To determine the direct impact of the PD-1 pathway on antiviral T cells during viral persistence, we quantified proximal and distal downstream T cell signaling at 30 min and 16 h following anti–PD-1 administration at day 7 after CL13 infection. These time points were selected to reflect the early versus late events associated with recovery of antiviral CD8 and CD4 T cells after administration of anti–PD-1. Interestingly, increased P14 motility at 30 min (Fig. 4 A) coincided with elevated TCR signaling, as indicated by ZAP-70 phosphorylation and downstream p-ERK signaling (Fig. 9 A). SMARTA cells had enhanced p-ERK levels at 30 min but did not show evidence of TCR signaling (p-ZAP-70), fitting with their failure to regain motility at this early time point. At 16 h after PD-1 blockade, a large increase in both P14 and SMARTA cell motility was observed (Figs. 3 and 4); this was associated with a marked recovery of TCR (p-ZAP-70) and downstream T cell signaling (p-JNK and p-ERK; Fig. 9 B). However, this period of functional recovery did not proceed without consequence to the persistently infected host. IFN-γ (but not TNF) protein levels increased 8.2-fold in the blood 24 h after anti–PD-1 administration (Fig. 9, C and D), and 100% of the treated mice succumbed to fatal disease over the ensuing 6 d (Fig. 9 F). A statistically significant increase in IFN-γ mRNA expression was observed in flow cytometrically sorted P14, but not SMARTA cells following PD-1 blockade (Fig. 9 E), suggesting that antiviral CD8+ T cells are in part responsible for the elevated IFN-γ production. Importantly, genetic deletion of IFN-γ (but not TNF) completely prevented all fatalities (Fig. 9 F), indicating that anti–PD-1 can induce a fatal disease mediated by IFN-γ.
Figure 9.
Anti–PD-1 treatment restores antiviral T cell signaling and cytokine production, resulting in death. (A and B) Upstream (p-ZAP-70) and downstream (p-JNK; p-ERK) T cell signaling was quantified immediately ex vivo 30 min (A) or 16 h (B) after administration of anti–PD-1 or isotype control antibodies (n = 10 mice). Signaling was quantified simultaneously in P14 and SMARTA cells. Asterisks denote a statistically significant (P < 0.05) increase in signaling when compared with the corresponding isotype control group. (C and D) Serum concentrations of TNF (C) and IFN-γ (D) were measured by ELISA 24 h after administration of anti–PD-1 or isotype control antibodies (n = 5 mice). The asterisk denotes a statistically significant (P < 0.05) difference from the control group. (E) Q-PCR was used to quantify IFN-γ expression in flow cytometrically sorted CD8+ P14 and CD4+ SMARTA cells 16 h after injection of anti–PD-1 or isotype control antibodies. The asterisk denotes a statistically significant (P < 0.05) difference from the control group. (F) Survival was monitored in B6 (black), IFN-γ−/− (green), and TNF−/− (red) mice after injection of anti–PD-1 antibody on day 7 (n = 5 mice). B6 receiving isotype control antibody (blue) served as a control for this experiment. All mice receiving anti–PD-1 expired except for IFN-γ−/− mice. The data presented in this figure are representative of 2–4 independent experiments.
DISCUSSION
The establishment of a persistent viral infection often coincides temporally with a decline or loss of T cell function (Wherry, 2011). In our study, we sought mechanistic insights into the anatomy and dynamics of antiviral T cells as they mounted a defense against an acute or persistent viral infection. Our real-time splenic imaging studies uncovered several novel findings that advance our understanding of T cell exhaustion and the factors that give rise to viral persistence. First, we observed that the early stages (day 7) of T cell exhaustion are induced in the splenic marginal zone/red pulp of CL13-infected mice and are associated with a state of prolonged motility arrest. In fact, the most marked reductions in antiviral CD8+ and CD4+ T cell motility were observed in this splenic region at both time points analyzed. Notably, CD8+ T cell arrest durations often exceeded 40 min, which we postulate has a negative impact on killing efficiency. That motility paralysis was first observed in the splenic marginal zone/red pulp rather than white pulp is best explained by the gradient of viral replication observed in the CL13-infected spleen. Viral antigen at day 7 was most abundant in the marginal zone/red pulp, likely facilitating frequent T cell encounters with cognate peptide-MHC. The splenic marginal zone/red pulp also showed the highest expression of PD-L1, which supports our conclusion that negative immune regulation would be most pronounced in this anatomical region. Indeed, therapeutic blockade of the PD-1–PD-L1 pathway through intravenous antibody administration resulted in an unexpected elevation in antiviral T cell motility. At day 7 after CL13 infection, red pulp CD8+ T cells increased in speed within just 30 min, whereas CD4+ T cells required 16 h. These data suggest that the PD-1–PD-L1 interactions stabilize rather than repel antiviral T cells. This theory is supported by our lipid bilayer data showing that PD-L1 localizes to the cSMAC and promotes stable, mature immunological synapse formation in LCMV-specific CD8+ T cells despite reduced phosphorylation of proximal TCR signaling molecules like ZAP-70. We propose that motility paralysis during persistent viral infection decreases T cell scanning efficiency and facilitates engagement of regulatory pathways that negatively impact antiviral T cell function. Importantly, blockade of PD-1 resulted in a rapid recovery of T cell signaling (particularly for CD8+ T cells), an increase in antiviral cytokine production (IFN-γ), and the induction of a fatal disease. Collectively, these data demonstrate that T cell motility paralysis is a manifestation of immune exhaustion.
From an adaptive immune perspective, the key to a successful antiviral defense (particularly against an aggressive pathogen) is optimization of target cell scanning and effector molecule utilization. For example, it was recently demonstrated that pathogen-specific CD8 T cells use a chemokine-driven persistent migration interspersed with local searching to more rapidly find target cells (Harris et al., 2012). This general strategy, referred to as a Levy walk and commonly used by predators and foraging animals (Sims et al., 2008; Viswanathan et al., 1999), has been adopted by T cells to facilitate pathogen clearance. T cells also perform best when engaging their target cells for no more than 10–15 min (Stinchcombe et al., 2001; Mempel et al., 2006; Ganusov and De Boer, 2008). This allows T cells to quickly move from one target to the next, thus maximizing pathogen elimination. Through massive clonal expansion (Murali-Krishna et al., 1998) and rapid target cell acquisition (Ganusov and De Boer, 2008), T cells endow their host with a defense that favors pathogen clearance; however, this efficiency sometimes gives rise to intolerable immunopathology, especially when a pathogen transitions into a state of persistence. For this reason, the immune system is equipped with dampening mechanisms that induce T cell exhaustion (Wherry, 2011). The molecular signature of T cell exhaustion during viral persistence is characterized by a plethora of phenotypic and functional changes that mitigate cellular activity (Zajac et al., 1998; Wherry et al., 2007). These changes include alterations in the pathways responsible for cytolytic capacity, cytokine production, chemotaxis, adhesion, and metabolism, for example (Wherry et al., 2007). At the dynamic level, we observed that the establishment of viral persistence induces T cell motility paralysis. We propose that prolonged states of cellular arrest (as observed in the CL13 model) reduce T cell killing efficiency by preventing the timely engagement of multiple targets (Rothstein et al., 1978; Stinchcombe et al., 2001), and should, therefore, be considered another manifestation of immune exhaustion. In support of this idea, cytotoxic lymphocytes have been observed engaging tumor cells for up to 6 h (Breart et al., 2008). Tumors expose T cells to chronic antigenic loads similar to persistent viral infections and routinely induce states of functional exhaustion (Kim and Ahmed, 2010).
Anatomically, T cell motility paralysis mapped initially to the splenic marginal zone/red pulp. Mean track velocities and motility coefficients were significantly reduced for virus-specific CD8+ and CD4+ T cells in this anatomical region 7 d after CL13 infection, whereas no alterations in motility parameters were observed in the splenic white pulp. For CD8+ T cells, a reduction in motility was observed in the marginal zone/red pulp as early as day 4 after infection, which is when the immune response to Arm versus CL13 begins to diverge (Brooks et al., 2006a; Althaus et al., 2007). Based on these observations, we propose that splenic T cell exhaustion is initially induced in the marginal zone/red pulp of CL13-infected mice. It is known that high viral loads contribute to exhaustion in the LCMV model (Mueller and Ahmed, 2009). Supporting this theory, we observed that viral antigen, PD-L1 expression, and T cell motility arrest were highest in the splenic marginal zone/red pulp at day 7. Moreover, marginal zone macrophages, red pulp macrophages, and myeloid DCs (all residents of this anatomical region) expressed the highest levels of PD-L1. Macrophages residing in the splenic marginal zone are responsible for trapping blood-borne pathogens (Aichele et al., 2003) and share similarities with subcapsular sinus macrophages in draining lymph nodes (Junt et al., 2007). After intravenous injection, LCMV localizes to the marginal zone (Borrow et al., 1995) and likely spreads from there. Interestingly, we found that PD-L1 is expressed highly in the marginal zone even before infection, suggesting that a heightened level of regulation is required in this anatomical region under steady-state conditions. In infected mice, LCMV CL13 disseminates rapidly and overtakes most of the splenic red pulp within a week of infection, whereas Arm is controlled during this time period. CL13 persistence induces the expression of several immune regulators, such as IL-10 (Brooks et al., 2006b; Ejrnaes et al., 2006) and PD-L1 (Barber et al., 2006), both by hematopoietic and nonhematopoietic cells (Mueller et al., 2010). Within the spleen it was recently shown that these regulators are often produced simultaneously by the same antigen-presenting cell after CL13 infection (Wilson et al., 2012). Thus, we propose that antiviral T cells are subjected to multiple inhibitory pathways while locked in prolonged states of arrest in the marginal zone/red pulp. Prolonged arrest likely facilitates exhaustion by enhancing the probability that antiviral T cells will be exposed to multiple regulatory pathways.
T cell exhaustion during CL13 persistence is known to be cognate peptide-MHC–dependent (Blattman et al., 2009; Mueller and Ahmed, 2009). Because peptide-MHC recognition induces T cell arrest (Dustin et al., 1997), it is reasonable to assume that the overabundance of LCMV antigen in the splenic marginal zone/red pulp contributes to reductions in antiviral T cell motility. However, this region also had elevated PD-L1 expression, which was recently shown to destabilize immunological synapse formation (Yokosuka et al., 2012) and serve as a repulsionary signal that inhibits the arrest of self-reactive CD4+ T cells (Fife et al., 2009). This mechanism was inconsistent with the long duration antiviral T cell interactions we observed in CL13-infected mice. We, therefore, administered PD-1 and PD-L1 antibodies intravenously to determine the impact that the PD-1–PD-L1 pathway had on antiviral T cell dynamics during the establishment of a persistent viral infection. Unexpectedly, both treatments resulted in increased T cell motility in the splenic marginal zone/red pulp. This occurred within 30 min for CD8+ T cells and within 16 h for CD4+ T cells. This suggests that PD-1–PD-L1 signaling differentially impacts CD8 versus CD4 T cell motility in vivo. Because increased CD8+ T cell motility preceded IFN-γ transcription and a significant reduction in viral load, we hypothesized that PD-1–PD-L1 directly participated in their motility arrest. To test this theory, we conducted ex vivo analyses of antiviral CD8+ T cells engaging lipid bilayers loaded with peptide-MHC I, ICAM-1, and PD-L1. Using this reductionist system, we observed that PD-L1 localizes to the cSMAC and promotes stable immunological synapse formation. More unstructured synapses and motile cells were observed in the absence of PD-L1. This finding is in contrast to recent studies with AND-TCR-tg CD4+ T cells, which revealed that PD-1–PD-L1 interactions destabilize synapse formation by recruiting SHP-2 phosphatases to TCR microclusters that dephosphorylate proximal signaling molecules like ZAP-70 (Yokosuka et al., 2012). However, with virus-specific CD8+ T cells exposed to planar bilayers, we observed that PD-L1 can actually promote mature synapse formation despite dephosphorylation of ZAP-70. Au-Yeung et al. (2010) recently demonstrated that ZAP-70 kinase activity is not required for activation of LFA-1 in regulatory T cells. The discrepancy between our findings and those published by Yokosuka et al. (2012) could be caused by the differential impact of PD-L1 on antigen-specific CD8+ versus CD4+ T cells. At day 7 after infection, we observed that PD-1 blockade rapidly increased the motility of antiviral CD8+ but not CD4+ T cells. Thus, it is conceivable that the PD-1 pathway only directly promotes arrest of antiviral CD8+ T cells and that increased motility of CD4+ T cells is secondary to other factors, such as viral clearance. Nevertheless, our bilayer studies clearly demonstrate that PD-L1 can promote CD8+ T cell synapse formation and motility arrest while impeding proximal TCR signaling. Thus, degradation of TCR signaling does not always override the stop signal. It will be important in future studies to identify all of the factors that promote T cell motility versus arrest using different in vivo models.
We and others (Chemnitz et al., 2004; Yokosuka et al., 2012) have observed that PD-1 promotes dephosphorylation of proximal TCR signals that ultimately results in decreased downstream signaling. When analyzed temporally, we demonstrated that PD-1 blockade promotes ZAP-70 and ERK phosphorylation within 30 min for CD8+ T cells. This was accompanied by a reduction in SOCS3 transcription, which was shown recently to be involved in suppressing cytokine signaling in the CL13 model (Pellegrini et al., 2011). Increased CD8+ T cell signaling and motility was followed by a rapid restoration in IFN-γ transcription (within 1 h) and a reduction in viral loads (within 2–4 h). In fact, a 74% decrease in splenic viral titers was observed 16 h after anti–PD-1 administration. It was at this time point that antiviral CD4+ T cells became more motile, suggesting a link between decreased viral loads and increased motility for CD4+ T cells. All of the changes over the 16-h time window occurred without alterations in the expression of BATF, a transcription factor recently linked to PD-1 signaling in exhausted T cells (Quigley et al., 2010). Unfortunately, the restoration in antiviral T cell motility, signaling, and effector function after PD-1 blockade did not proceed without a consequence. A fatal IFN-γ–mediated disease was observed in all CL13-infected mice treated with anti–PD-1, and this was likely caused by increased synthesis of IFN-γ by virus-specific CD8+ T cells. A recent study also demonstrated that PD-1 knockout mice infected with CL13 succumb to CD8+ T cell–mediated collapse of the circulatory system (Frebel et al., 2012). Thus, it is important that anti–PD-1 be administered with caution in a clinical setting because immune exhaustion is a state imposed by an infected host to prevent severe immunopathology. Releasing T cells from this exhausted state holds great promise for the treatment of viruses and tumors, but can come with undesirable side effects.
In conclusion, our data demonstrate that during the establishment of a persistent viral infection, PD-1–PD-L1 mediate their inhibitory effects by locking antiviral T cells into a state of prolonged motility arrest by localizing to the cSMAC and promoting stable, mature immunological synapse formation. This occurs most notably for antiviral CD8+ T cells and maps anatomically to the splenic marginal zone/red pulp at early stages of infection in CL13-infected mice. Prolonged motility arrest is an excellent host strategy to decrease T cell efficiency and likely facilitates exposure to multiple regulatory pathways. PD-1–PD-L1 blockade is known to restore function to virus-specific (Barber et al., 2006; Day et al., 2006; Velu et al., 2009) and tumor-specific (Iwai et al., 2002; Curran et al., 2010) T cells, and has shown some promise in recent clinical trials (Brahmer et al., 2012; Topalian et al., 2012). Our in vivo studies offer a novel view of how the PD-1 pathway impedes antiviral T cell function, but it remains to be determined whether this pathway similarly promotes the lengthy T cell arrest times observed in tumors (Breart et al., 2008). In general, there is still a great deal to be learned about the factors that influence T cell dynamics during states of immunosuppression, and therapeutic enhancement of T cell motility provides a simple means to improve antigen-specific T cell efficiency in vivo.
MATERIALS AND METHODS
Mice.
C57BL/6 (B6), B6.129S7-Ifngtm1Ts/J (IFNγ−/−), and B6;129S-Tnftm1Gkl/J (TNF−/−) mice were purchased from The Jackson Laboratory. B6 actin-CFP+ DbGP33-41 TCR-tg (CFP+ P14), B6 actin-GFP+ DbGP33-41 TCR-tg (GFP+ P14), and B6 actin-GFP+ I-AbGP61-80 TCR-tg (GFP+ SMARTA) mice were bred and maintained in a closed breeding facility at The National Institutes of Health (NIH). All mice were housed under specific pathogen–free conditions and handled in accordance with the National Institute of Neurological Disorders and Stroke Animal Care and Use Committee.
Virus.
Adult mice at 6–8 wk of age were infected i.v. with 2 × 106 PFU of LCMV Armstrong (Arm) clone 53b or clone 13 (CL13). Stocks were prepared by a single passage on BHK-21 cells, and viral titers were determined by plaque formation on Vero cells.
Mononuclear cell isolations.
To obtain cell suspensions for flow cytometric analysis and cell transfers, spleens were harvested from donor mice, and, afterward, single-cell suspensions were prepared by mechanical disruption through a 100-µm filter. Spleen cells were treated with red blood cell lysis buffer (0.14 M NH4Cl and 0.017 M Tris-HCl, pH 7.2), washed twice, and resuspended in PBS containing 1% FBS.
P14 and SMARTA cell transfers.
CD8+ T cells were purified from naive CFP+ P14 mouse splenocytes by negative selection (Stem Cell Technologies). CD4+ T cells were also purified by negative selection from naive GFP+ SMARTA mice. The purity after enrichment was determined to be >98%. For imaging and mRNA isolation studies, 5,000 purified CFP+ P14 CD8+ T cells and 5,000 GFP+ SMARTA CD4+ T cells were injected i.v. into naive mice. 1 d later, the mice were challenged i.v. with LCMV Arm or CL13. For planar bilayer experiments, CD8+ T cells were purified by MACs+ selection (Miltenyi Biotec) before flow cytometric sorting of GFP+ P14 cells was performed. For mRNA isolation experiments, splenic Thy1.2+ T cells were purified by positive selection before simultaneous flow cytometric sorting of GFP+ P14 and CFP+ SMARTA cells.
In vivo blocking antibodies.
The following antibodies (BioXCell) diluted in PBS were injected i.v. for in vivo blocking experiments: rat anti-CD279 (500 µg; clone RMP1-14) and rat anti-CD274 (500 µg; clone 10F.9G2). The following were used as isotype control antibodies: rat IgG2a (clone 2A3) and IgG2b (clone LTF-2).
Flow cytometry.
Splenocytes were harvested and blocked with 3.3 µg/ml anti–mouse CD16/CD32 (Fc Block; BD) in PBS containing 1% FBS and 0.1% sodium azide for 15 min on ice. After the Fc block, cells were stained with cocktails of the following conjugated antibodies: CD279 biotin (RMP1-30), CD274 biotin (10F.962), F4/80 PerCP (BMB), CD45.2 APC (104), CD11c APC Cy7 (N418), CD45.1 Alexa Fluor 700 (A20), CD45.2 Alexa Fluor 700 (104; BioLegend), CD4 FITC (RM4-5), Thy1.1 PerCP (OX-7), CD8 PE (53–6.7), Thy1.2 PE (53–2.1; BD), CD11b PE Cy7 (M1/70), CD209b Alexa Fluor 647 (22D1; eBioscience), CD169 FITC (3D6.112; Serotec), CD8 Pacific Blue (5H10; Invitrogen), and gp38 PE (8.1.1; BioLegend). Biotinylated antibodies were detected with streptavidin-Qdot 605nm (Invitrogen). Rat IgG2b-biotin (RTK4530; BioLegend) was used as an isotype control antibody for the CD279 and CD274 stains. For phospho-flow experiments (Krutzik and Nolan, 2003), 106 splenocytes were fixed with 1.5% formaldehyde for 10 min at room temperature. Cells were washed, incubated in ice cold methanol for an additional 10 min at 4°C, washed, and then stained with surface antibodies combined with the following cell signaling antibodies: pZAP-70 Alexa Fluor 647 (pY319; clone 17a/P-ZAP70; BD), pERK1/2 Alexa Fluor 647 (pThr202/Tyr404; clone D13.14.4E; Cell Signaling Technology), and pJNK Alexa Fluor 647 (pThr183/Tyr185; clone G9; Cell Signaling Technology). Mouse IgG1 Alexa Fluor 647 (clone MOPC-21; BD) and rabbit IgG Alexa Fluor 647 (Cell Signaling Technology) were used as isotype controls for the phospho-flow experiments. All cells were acquired using a digital flow cytometer (Digital LSR II; BD), and flow cytometric data were analyzed with FlowJo software (Tree Star).
Immunohistochemistry.
To examine the splenic distribution of PD-L1, fresh spleen tissue was frozen on dry ice in optimal cutting temperature (OCT; Tissue-Tek). 6-µm frozen sections were cut, fixed with 4% paraformaldehyde for 15 min, blocked with an avidin/biotin-blocking kit (Vector Laboratories), and stained at 4°C overnight with biotinylated rat anti-CD274 (1:250; clone 10F.962; BioLegend), polyclonal guinea pig anti-LCMV (1:1500), and rabbit anti-laminin (1:500; Sigma-Aldrich). Primary antibodies were detected by a 1-h, room temperature incubation with streptavidin rhodamine red-X (1:250), donkey anti–guinea pig DyLight 649 (1:250), and donkey anti–rabbit DyLight 488 (1:250; Jackson ImmunoResearch Laboratories), respectively. Sections were additionally stained with 1 µg/ml DAPI (Sigma-Aldrich) for 3 min at room temperature to visualize cell nuclei. All working stocks of primary and secondary reagents were diluted in PBS containing 2% FBS. Images were captured using a Zeiss Axio Observer Z1 microscope fitted with a 20× objective and an AxioCam HRc Rev. 3 digital camera.
Real-time PCR.
Spleens were homogenized and subsequently treated with TRIzol before performing a column-based RNA purification using the PureLink RNA mini kit (Invitrogen). RNA from flow cytometrically sorted GFP+ P14 and CFP+ SMARTA cells was isolated in a similar manner. Total splenic RNA was further treated with amplification grade DNase I (Invitrogen) before reverse transcription. cDNAs were synthesized from 1.0 µg of DNase I–treated RNA using iScript (Bio-Rad Laboratories) reverse-transcription reagent kit without any gene specific primers. For determination of absolute LCMV genome copy number, 10 ng of cDNA was used for quantitative real-time polymerase chain reactions (Q-PCR) in which serial dilutions of known quantities of linearized LCMV S-segment plasmid DNA was used as a reference (McCausland and Crotty, 2008). The following primers specific to the LCMV glycoprotein were used as described previously (McCausland and Crotty, 2008): GP-Fwd 5′-CATTCACCTGGACTTTGTCAGACTC-3′, GP-Rev 5′-GCAACTGCTGTGTTCCCGAAAC-3′. For gene-specific Q-PCRs, the housekeeping gene β-actin was used as reference. All Q-PCR reactions were performed using SsoFast EvaGreen SYBR green supermix (Bio-Rad Laboratories) in a 96-well optic tray on a CFX96 Real-Time PCR detection system (Bio-Rad Laboratories). The reactions were performed in triplicate, and RNA samples without reverse transcription were used as a negative control for each gene candidate. Q-PCR reactions were run with an initial denaturation temperature of 95°C for 3 min, which was followed by 40 cycles of a three-step amplification reaction. Annealing temperatures for gene specific primers were optimized individually on positive controls before performing experimental runs. The following primers were used: Actb (NM_007393), Fwd, 5′-AGCTCATTGTAGAAGGTGTGG-3′ and Rev, 5′-GTGGGAATGGGTCAGAAG-3′; Ifn-γ (NM_008337), Fwd, 5′-TCAAGTGGCATAGATGTGGAA) and Rev, 5′-TGGCTCTGCAGGATTTTCATG-3′; Batf (NM_016767), Fwd, 5′-GCAGTGACTCCAGCTTCAG-3′ and Rev, 5′-TGTCGGCTTTCTGTGTCTG-3′; Socs3 (NM_007707), Fwd, 5′-CCTATGAGAAAGTGACCCAGC-3′ and Rev, 5′-TTTGTGCTTGTGCCATGTG-3′.
Planar lipid bilayers.
Glass-supported planar lipid bilayers were prepared in sterile “Ibidi” chambers (Sticky-Slide I, 0.2 mm Luer; Ibidi) using a variation on previously described methods (Grakoui et al., 1999). In brief, liposome bilayers consisting of 12.5% DOGS-NTA Nickel in DOPC (Avanti) were applied to positively charged glass coverslips that had been treated with 70% H2SO4/30% H2O2 (acid Piranha). This treatment allows lipid bilayers to form on the glass with optimal lateral mobility. After blocking with 5% casein, ICAM-1-12-His labeled with Alexa Fluor 405 (AF405; Invitrogen; 200 molecules/µm2) was reacted with lipid bilayers in combination with H-2DbGP33-41-6-His (NIH tetramer core facility, Emory University; 185 molecules/um2). The GP33-41 epitope (KAVYNFATM) from LCMV is recognized by tg CD8+ P14 T cells. For real-time imaging experiments to assess the impact of PD-L1 on immunological synapse formation, hPD-L1-6-His purified from CHO cells was labeled with Alexa Fluor 568 (AF568; Invitrogen) and combined with ICAM-1AF405 and H-2DbGP33-41-6-His whenever noted (275 molecules/µm2). Ibidi chambers were prewarmed to 37°C, and, afterward, 1–2 × 106 flow cytometrically sorted day 7 GFP+CD8+ P14 T cells in 100 µl of prewarmed Hepes-buffered saline containing 1% human serum albumin (HBS/HSA) were injected into chambers. The GFP+ P14 cells were imaged immediately or 10 min after injection. Images were collected for 40 min at 37°C on a Nikon Eclipse automated inverted fluorescence total internal reflection microscope (Morrell Instruments), with a custom laser illumination system (Solamere Technologies). Protein densities on the planar bilayers were quantified by loading liposome-containing 5-µm silica beads (Bangs Laboratories Inc.) with the denoted proteins at the same concentrations present on the planar bilayers. After labeling the specific proteins with FITC-conjugated antibodies, densities were calculated by flow cytometry (FACSCalibur) using Quantum FITC-5 MESF calibration beads (Bangs Laboratories). For fixed sample imaging experiments conducted to assess the extent of pZAP-70 and TCR signaling in the presence of PD-L1, lipid bilayers containing ICAM-1-12-HisAF405, H-2DbGP33-41-6-His, and unlabeled hPDL1-6-His were prepared exactly as described above. Ibidi chambers were prewarmed to 37°C, and, afterward, 1–2 × 106 flow cytometrically sorted day 7 GFP+CD8+ P14 T cells were incubated for 2 min with 4 µg of Alexa Fluor 568 TCRβ (H57) Fab (BioLegend) in 100 µl of prewarmed HBS/HSA and subsequently injected into chambers. Chambers were incubated at 37°C for a time-course of 5 min and cells were immediately fixed in 4% paraformaldehyde for 10 min at room temperature. After washing in HBS/HSA, cells were permeabilized in 0.4% Triton X-100 (5 min, room temperature), rinsed, and blocked in 5% casein (20 min, room temperature). Cells were stained overnight at 4°C with Alexa Fluor 647 pZAP-70 (Y319 BD; 50 µl per chamber), rinsed, and images collected on the Nikon TIRF microscope described above.
Total internal reflection microscopy (TIRFM).
All TIRF imaging was performed on a Nikon Eclipse Ti inverted fluorescence microscope using an Apo TIRF 100x Oil 1.49 NA objective. Coherent diode lasers at 405, 488, 561, and 641 nm were used for TIRF illumination and a Xenon arc lamp (Newport) filtered at 490 nm was used for reflection imaging. The microscope and EMCCD camera (Andor DU-897 X-4654) were fully automated using NIS-Elements software. For real-time experiments, images were acquired every 4 min for overall time-courses of 40 min (10 time points). Between 20 and 28 fields were acquired for each time point using the automatic stage moving feature. For fixed sample experiments, between 50 and 75 fields were acquired using the automatic stage moving feature. In all experiments, focus was stabilized using the Nikon perfect focus system and was adjusted to compensate for chromatic aberration (400 nm offset) between 405 nm (violet) and 561 nm (red) channels during acquisition. Between 20 and 28 fields were acquired for each time point using the automatic stage moving feature. Microscope stage temperature was maintained at 37°C with a Nikon Livecell forced air heater. NIS-Elements Viewer 3.20/4.0 (Nikon) and ImageJ 1.45q/1.47d35 (National Institutes of Health) software were used for image analysis. The following parameters were scored for the real-time experiments: (a) “mature synapses,” frequency of P14 cells that formed stable, classical immunological synapses, as defined by the symmetric distribution of ICAM-1 in the pSMAC around a central cSMAC; (b) “unstructured synapses,” frequency of cells that accumulated ICAM-1 and were nonmobile, but lacked a pSMAC and cSMAC; (c) “moving/crawling” (or kinapses), frequency of cells moving or crawling on the bilayer across an xy field at any given time-point and not achieving attachment (Dustin, 2008). The following parameters were scored for fixed sample experiments: mean fluorescent intensities of designated ROIs for TCR and pZAP-70 (raw mean intensity values with local background subtracted) as well as raw mean intensity radial profiles for TCR, pZAP-70, and ICAM-1.
TPM.
For splenic imaging experiments, spleens were glued to a plastic coverslip and cut longitudinally with a Leica vibratome. The coverslip was then attached with plumbers grease into a flow chamber and maintained at 37°C by perfusion with warm, high-glucose DMEM bubbled with a mixture of 95% O2 and 5% CO2 (Aoshi et al., 2008). 4D datasets were captured using a Leica SP5 two photon imaging system (Leica Microsystems) equipped with an 8000 Hz resonant scanner, a 20×/1.0 NA dipping objective, a Mai Tai HP DeepSee Laser (Spectraphysics) tuned to 905 nm, and an external NDD4 detector. 3D stacks consisting of between 18 and 28 planes (2.5 µm step size) were captured every 30 s. To enhance signal contrast, we averaged 10 video frames for each plane in resonance scanning mode. Fluorescence emission was passed through customized filters (Semrock) in the NDD4 detector. The primary dichroic mirror (495 nm-LP) was followed by two secondary dichroics (458 nm-LP and 552 nm-LP). The collagen structure of the spleen was visualized by collecting the second harmonic signal at emission wavelengths below 458 nm. Light from 458 to 495 nm was collected for CFP emission, and 495 to 552 nm for GFP emission. After acquisition, images were spectrally unmixed using the Leica Application Suite AF software before final processing and analysis of the 4D data with Imaris 7.0 software (Bitplane). Mean track velocities (µm/min) and motility coefficients (µm2/min) were calculated for individual tracks spanning 11 time points (or 5 min). The motility coefficient was defined as the slope of the fitted line of displacement squared versus time. The meandering factor was calculated by dividing the displacement of a cell by its total path length. All track calculations were performed using T cell Analyser version 1.7.0 software written by J. Dempster (University of Strathclyde, Glasgow, Scotland; Zinselmeyer et al., 2009).
Statistical analysis.
Statistical significance (P < 0.05) was determined using a Student’s t test (two groups) or a one way ANOVA (more than two groups). An ANOVA on ranks was used for datasets with a non-Gaussian distribution and more than two groups. Statistical significance for planar bilayer data was determined using a two population proportion test of equality. All statistical analyses were performed using Sigma Plot 11.0 or SYSTAT13 software.
Online supplemental material.
Video 1 shows the methodology used to quantify antiviral T cell dynamics in the spleen. Video 2 shows an analysis of early splenic antiviral T cell dynamics after acute versus persistent viral infection. Video 3 shows splenic antiviral T cell dynamics during acute versus persistent viral infection. Video 4 shows anti–PD-1 enhances the motility of antiviral T cells during persistent viral infection. Video 5 shows PD-L1 promotes CD8+ T cell synapse formation. Video 6 shows PD-L1 centralizes to the cSMAC during synapse formation. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20121416/DC1.
Supplementary Material
Acknowledgments
We would like to thank Dr. John Dempster at the University of Strathclyde for generously providing customized T cell motility analysis software, the NIH tetramer core facility (Emory University) for providing H-2DbGP33-41, Judy Hua Zhong (NYU Division of Biostatistics) for advice on statistical analysis, as well as Patrick Wu and Theo Roth (NIH) for their assistance in generating some of the T cell motility data.
This work was supported by National Institutes of Health (NIH) intramural program (D.B. McGavern) and grant P01AI080192 (M.L. Dustin, C. Sacristán, and M. Cammer), as well as by the Wellcome Trust (S.J. Davis). D. Nayak is presently supported by a NIH Intramural Competitive Fellowship.
The authors have no further conflicts of interest.
Footnotes
Abbreviations used:
- LCMV
- lymphocytic choriomeningitis virus
- TPM
- two-photon microscopy
References
- Ahmed R., Oldstone M.B. 1988. Organ-specific selection of viral variants during chronic infection. J. Exp. Med. 167:1719–1724 10.1084/jem.167.5.1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aichele P., Zinke J., Grode L., Schwendener R.A., Kaufmann S.H., Seiler P. 2003. Macrophages of the splenic marginal zone are essential for trapping of blood-borne particulate antigen but dispensable for induction of specific T cell responses. J. Immunol. 171:1148–1155 [DOI] [PubMed] [Google Scholar]
- Althaus C.L., Ganusov V.V., De Boer R.J. 2007. Dynamics of CD8+ T cell responses during acute and chronic lymphocytic choriomeningitis virus infection. J. Immunol. 179:2944–2951 [DOI] [PubMed] [Google Scholar]
- Aoshi T., Zinselmeyer B.H., Konjufca V., Lynch J.N., Zhang X., Koide Y., Miller M.J. 2008. Bacterial entry to the splenic white pulp initiates antigen presentation to CD8+ T cells. Immunity. 29:476–486 10.1016/j.immuni.2008.06.013 [DOI] [PubMed] [Google Scholar]
- Au-Yeung B.B., Levin S.E., Zhang C., Hsu L.Y., Cheng D.A., Killeen N., Shokat K.M., Weiss A. 2010. A genetically selective inhibitor demonstrates a function for the kinase Zap70 in regulatory T cells independent of its catalytic activity. Nat. Immunol. 11:1085–1092 10.1038/ni.1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber D.L., Wherry E.J., Masopust D., Zhu B., Allison J.P., Sharpe A.H., Freeman G.J., Ahmed R. 2006. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 439:682–687 10.1038/nature04444 [DOI] [PubMed] [Google Scholar]
- Bergthaler A., Flatz L., Hegazy A.N., Johnson S., Horvath E., Löhning M., Pinschewer D.D. 2010. Viral replicative capacity is the primary determinant of lymphocytic choriomeningitis virus persistence and immunosuppression. Proc. Natl. Acad. Sci. USA. 107:21641–21646 10.1073/pnas.1011998107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattman J.N., Wherry E.J., Ha S.J., van der Most R.G., Ahmed R. 2009. Impact of epitope escape on PD-1 expression and CD8 T-cell exhaustion during chronic infection. J. Virol. 83:4386–4394 10.1128/JVI.02524-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrow P., Evans C.F., Oldstone M.B. 1995. Virus-induced immunosuppression: immune system-mediated destruction of virus-infected dendritic cells results in generalized immune suppression. J. Virol. 69:1059–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi G., Trambas C., Booth S., Clark R., Stinchcombe J., Griffiths G.M. 2002. The secretory synapse: the secrets of a serial killer. Immunol. Rev. 189:152–160 10.1034/j.1600-065X.2002.18913.x [DOI] [PubMed] [Google Scholar]
- Brahmer J.R., Tykodi S.S., Chow L.Q., Hwu W.J., Topalian S.L., Hwu P., Drake C.G., Camacho L.H., Kauh J., Odunsi K., et al. 2012. Safety and Activity of Anti-PD-L1 Antibody in Patients with Advanced Cancer. N. Engl. J. Med. 366:2455–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breart B., Lemaître F., Celli S., Bousso P. 2008. Two-photon imaging of intratumoral CD8+ T cell cytotoxic activity during adoptive T cell therapy in mice. J. Clin. Invest. 118:1390–1397 10.1172/JCI34388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks D.G., Teyton L., Oldstone M.B., McGavern D.B. 2005. Intrinsic functional dysregulation of CD4 T cells occurs rapidly following persistent viral infection. J. Virol. 79:10514–10527 10.1128/JVI.79.16.10514-10527.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks D.G., McGavern D.B., Oldstone M.B. 2006a. Reprogramming of antiviral T cells prevents inactivation and restores T cell activity during persistent viral infection. J. Clin. Invest. 116:1675–1685 10.1172/JCI26856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks D.G., Trifilo M.J., Edelmann K.H., Teyton L., McGavern D.B., Oldstone M.B. 2006b. Interleukin-10 determines viral clearance or persistence in vivo. Nat. Med. 12:1301–1309 10.1038/nm1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks D.G., Lee A.M., Elsaesser H., McGavern D.B., Oldstone M.B. 2008. IL-10 blockade facilitates DNA vaccine-induced T cell responses and enhances clearance of persistent virus infection. J. Exp. Med. 205:533–541 10.1084/jem.20071948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemnitz J.M., Parry R.V., Nichols K.E., June C.H., Riley J.L. 2004. SHP-1 and SHP-2 associate with immunoreceptor tyrosine-based switch motif of programmed death 1 upon primary human T cell stimulation, but only receptor ligation prevents T cell activation. J. Immunol. 173:945–954 [DOI] [PubMed] [Google Scholar]
- Coombes J.L., Robey E.A. 2010. Dynamic imaging of host-pathogen interactions in vivo. Nat. Rev. Immunol. 10:353–364 10.1038/nri2746 [DOI] [PubMed] [Google Scholar]
- Curran M.A., Montalvo W., Yagita H., Allison J.P. 2010. PD-1 and CTLA-4 combination blockade expands infiltrating T cells and reduces regulatory T and myeloid cells within B16 melanoma tumors. Proc. Natl. Acad. Sci. USA. 107:4275–4280 10.1073/pnas.0915174107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day C.L., Kaufmann D.E., Kiepiela P., Brown J.A., Moodley E.S., Reddy S., Mackey E.W., Miller J.D., Leslie A.J., DePierres C., et al. 2006. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 443:350–354 10.1038/nature05115 [DOI] [PubMed] [Google Scholar]
- Dong H., Zhu G., Tamada K., Chen L. 1999. B7-H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat. Med. 5:1365–1369 10.1038/70932 [DOI] [PubMed] [Google Scholar]
- Dustin M.L. 2008. Hunter to gatherer and back: immunological synapses and kinapses as variations on the theme of amoeboid locomotion. Annu. Rev. Cell Dev. Biol. 24:577–596 10.1146/annurev.cellbio.24.110707.175226 [DOI] [PubMed] [Google Scholar]
- Dustin M.L., Bromley S.K., Kan Z., Peterson D.A., Unanue E.R. 1997. Antigen receptor engagement delivers a stop signal to migrating T lymphocytes. Proc. Natl. Acad. Sci. USA. 94:3909–3913 10.1073/pnas.94.8.3909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejrnaes M., Filippi C.M., Martinic M.M., Ling E.M., Togher L.M., Crotty S., von Herrath M.G. 2006. Resolution of a chronic viral infection after interleukin-10 receptor blockade. J. Exp. Med. 203:2461–2472 10.1084/jem.20061462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fife B.T., Pauken K.E., Eagar T.N., Obu T., Wu J., Tang Q., Azuma M., Krummel M.F., Bluestone J.A. 2009. Interactions between PD-1 and PD-L1 promote tolerance by blocking the TCR-induced stop signal. Nat. Immunol. 10:1185–1192 10.1038/ni.1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frebel H., Nindl V., Schuepbach R.A., Braunschweiler T., Richter K., Vogel J., Wagner C.A., Loffing-Cueni D., Kurrer M., Ludewig B., Oxenius A. 2012. Programmed death 1 protects from fatal circulatory failure during systemic virus infection of mice. J. Exp. Med. 209:2485–2499 10.1084/jem.20121015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman G.J., Long A.J., Iwai Y., Bourque K., Chernova T., Nishimura H., Fitz L.J., Malenkovich N., Okazaki T., Byrne M.C., et al. 2000. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J. Exp. Med. 192:1027–1034 10.1084/jem.192.7.1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganusov V.V., De Boer R.J. 2008. Estimating in vivo death rates of targets due to CD8 T-cell-mediated killing. J. Virol. 82:11749–11757 10.1128/JVI.01128-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grakoui A., Bromley S.K., Sumen C., Davis M.M., Shaw A.S., Allen P.M., Dustin M.L. 1999. The immunological synapse: a molecular machine controlling T cell activation. Science. 285:221–227 10.1126/science.285.5425.221 [DOI] [PubMed] [Google Scholar]
- Ha S.J., Mueller S.N., Wherry E.J., Barber D.L., Aubert R.D., Sharpe A.H., Freeman G.J., Ahmed R. 2008. Enhancing therapeutic vaccination by blocking PD-1-mediated inhibitory signals during chronic infection. J. Exp. Med. 205:543–555 10.1084/jem.20071949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T.H., Banigan E.J., Christian D.A., Konradt C., Tait Wojno E.D., Norose K., Wilson E.H., John B., Weninger W., Luster A.D., et al. 2012. Generalized Lévy walks and the role of chemokines in migration of effector CD8+ T cells. Nature. 486:545–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman H.D., Bennink J.R., Yewdell J.W. 2009. Caught in the act: intravital multiphoton microscopy of host-pathogen interactions. Cell Host Microbe. 5:13–21 10.1016/j.chom.2008.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai Y., Ishida M., Tanaka Y., Okazaki T., Honjo T., Minato N. 2002. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc. Natl. Acad. Sci. USA. 99:12293–12297 10.1073/pnas.192461099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junt T., Moseman E.A., Iannacone M., Massberg S., Lang P.A., Boes M., Fink K., Henrickson S.E., Shayakhmetov D.M., Di Paolo N.C., et al. 2007. Subcapsular sinus macrophages in lymph nodes clear lymph-borne viruses and present them to antiviral B cells. Nature. 450:110–114 10.1038/nature06287 [DOI] [PubMed] [Google Scholar]
- Kaufmann D.E., Kavanagh D.G., Pereyra F., Zaunders J.J., Mackey E.W., Miura T., Palmer S., Brockman M., Rathod A., Piechocka-Trocha A., et al. 2007. Upregulation of CTLA-4 by HIV-specific CD4+ T cells correlates with disease progression and defines a reversible immune dysfunction. Nat. Immunol. 8:1246–1254 10.1038/ni1515 [DOI] [PubMed] [Google Scholar]
- Keir M.E., Butte M.J., Freeman G.J., Sharpe A.H. 2008. PD-1 and its ligands in tolerance and immunity. Annu. Rev. Immunol. 26:677–704 10.1146/annurev.immunol.26.021607.090331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim P.S., Ahmed R. 2010. Features of responding T cells in cancer and chronic infection. Curr. Opin. Immunol. 22:223–230 10.1016/j.coi.2010.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzik P.O., Nolan G.P. 2003. Intracellular phospho-protein staining techniques for flow cytometry: monitoring single cell signaling events. Cytometry A. 55:61–70 10.1002/cyto.a.10072 [DOI] [PubMed] [Google Scholar]
- Li Q., Skinner P.J., Ha S.J., Duan L., Mattila T.L., Hage A., White C., Barber D.L., O’Mara L., Southern P.J., et al. 2009. Visualizing antigen-specific and infected cells in situ predicts outcomes in early viral infection. Science. 323:1726–1729 10.1126/science.1168676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCausland M.M., Crotty S. 2008. Quantitative PCR technique for detecting lymphocytic choriomeningitis virus in vivo. J. Virol. Methods. 147:167–176 10.1016/j.jviromet.2007.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGavern D.B., Christen U., Oldstone M.B. 2002. Molecular anatomy of antigen-specific CD8(+) T cell engagement and synapse formation in vivo. Nat. Immunol. 3:918–925 10.1038/ni843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mempel T.R., Pittet M.J., Khazaie K., Weninger W., Weissleder R., von Boehmer H., von Andrian U.H. 2006. Regulatory T cells reversibly suppress cytotoxic T cell function independent of effector differentiation. Immunity. 25:129–141 10.1016/j.immuni.2006.04.015 [DOI] [PubMed] [Google Scholar]
- Monks C.R., Freiberg B.A., Kupfer H., Sciaky N., Kupfer A. 1998. Three-dimensional segregation of supramolecular activation clusters in T cells. Nature. 395:82–86 10.1038/25764 [DOI] [PubMed] [Google Scholar]
- Moskophidis D., Lechner F., Pircher H., Zinkernagel R.M. 1993. Virus persistence in acutely infected immunocompetent mice by exhaustion of antiviral cytotoxic effector T cells. Nature. 362:758–761 10.1038/362758a0 [DOI] [PubMed] [Google Scholar]
- Mueller S.N., Ahmed R. 2009. High antigen levels are the cause of T cell exhaustion during chronic viral infection. Proc. Natl. Acad. Sci. USA. 106:8623–8628 10.1073/pnas.0809818106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S.N., Matloubian M., Clemens D.M., Sharpe A.H., Freeman G.J., Gangappa S., Larsen C.P., Ahmed R. 2007. Viral targeting of fibroblastic reticular cells contributes to immunosuppression and persistence during chronic infection. Proc. Natl. Acad. Sci. USA. 104:15430–15435 10.1073/pnas.0702579104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S.N., Vanguri V.K., Ha S.J., West E.E., Keir M.E., Glickman J.N., Sharpe A.H., Ahmed R. 2010. PD-L1 has distinct functions in hematopoietic and nonhematopoietic cells in regulating T cell responses during chronic infection in mice. J. Clin. Invest. 120:2508–2515 10.1172/JCI40040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murali-Krishna K., Altman J.D., Suresh M., Sourdive D.J., Zajac A.J., Miller J.D., Slansky J., Ahmed R. 1998. Counting antigen-specific CD8 T cells: a reevaluation of bystander activation during viral infection. Immunity. 8:177–187 10.1016/S1074-7613(00)80470-7 [DOI] [PubMed] [Google Scholar]
- Oxenius A., Bachmann M.F., Zinkernagel R.M., Hengartner H. 1998. Virus-specific MHC-class II-restricted TCR-transgenic mice: effects on humoral and cellular immune responses after viral infection. Eur. J. Immunol. 28:390–400 [DOI] [PubMed] [Google Scholar]
- Pellegrini M., Calzascia T., Toe J.G., Preston S.P., Lin A.E., Elford A.R., Shahinian A., Lang P.A., Lang K.S., Morre M., et al. 2011. IL-7 engages multiple mechanisms to overcome chronic viral infection and limit organ pathology. Cell. 144:601–613 10.1016/j.cell.2011.01.011 [DOI] [PubMed] [Google Scholar]
- Pircher H., Bürki K., Lang R., Hengartner H., Zinkernagel R.M. 1989. Tolerance induction in double specific T-cell receptor transgenic mice varies with antigen. Nature. 342:559–561 10.1038/342559a0 [DOI] [PubMed] [Google Scholar]
- Quigley M., Pereyra F., Nilsson B., Porichis F., Fonseca C., Eichbaum Q., Julg B., Jesneck J.L., Brosnahan K., Imam S., et al. 2010. Transcriptional analysis of HIV-specific CD8+ T cells shows that PD-1 inhibits T cell function by upregulating BATF. Nat. Med. 16:1147–1151 10.1038/nm.2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein T.L., Mage M., Jones G., McHugh L.L. 1978. Cytotoxic T lymphocyte sequential killing of immobilized allogeneic tumor target cells measured by time-lapse microcinematography. J. Immunol. 121:1652–1656 [PubMed] [Google Scholar]
- Schneider H., Downey J., Smith A., Zinselmeyer B.H., Rush C., Brewer J.M., Wei B., Hogg N., Garside P., Rudd C.E. 2006. Reversal of the TCR stop signal by CTLA-4. Science. 313:1972–1975 10.1126/science.1131078 [DOI] [PubMed] [Google Scholar]
- Schubert D.A., Gordo S., Sabatino J.J., Jr, Vardhana S., Gagnon E., Sethi D.K., Seth N.P., Choudhuri K., Reijonen H., Nepom G.T., et al. 2012. Self-reactive human CD4 T cell clones form unusual immunological synapses. J. Exp. Med. 209:335–352 10.1084/jem.20111485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevilla N., Kunz S., Holz A., Lewicki H., Homann D., Yamada H., Campbell K.P., de La Torre J.C., Oldstone M.B. 2000. Immunosuppression and resultant viral persistence by specific viral targeting of dendritic cells. J. Exp. Med. 192:1249–1260 10.1084/jem.192.9.1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe A.H., Wherry E.J., Ahmed R., Freeman G.J. 2007. The function of programmed cell death 1 and its ligands in regulating autoimmunity and infection. Nat. Immunol. 8:239–245 10.1038/ni1443 [DOI] [PubMed] [Google Scholar]
- Sims D.W., Southall E.J., Humphries N.E., Hays G.C., Bradshaw C.J., Pitchford J.W., James A., Ahmed M.Z., Brierley A.S., Hindell M.A., et al. 2008. Scaling laws of marine predator search behaviour. Nature. 451:1098–1102 10.1038/nature06518 [DOI] [PubMed] [Google Scholar]
- Stinchcombe J.C., Bossi G., Booth S., Griffiths G.M. 2001. The immunological synapse of CTL contains a secretory domain and membrane bridges. Immunity. 15:751–761 10.1016/S1074-7613(01)00234-5 [DOI] [PubMed] [Google Scholar]
- Sullivan B.M., Emonet S.F., Welch M.J., Lee A.M., Campbell K.P., de la Torre J.C., Oldstone M.B. 2011. Point mutation in the glycoprotein of lymphocytic choriomeningitis virus is necessary for receptor binding, dendritic cell infection, and long-term persistence. Proc. Natl. Acad. Sci. USA. 108:2969–2974 10.1073/pnas.1019304108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalian S.L., Hodi F.S., Brahmer J.R., Gettinger S.N., Smith D.C., McDermott D.F., Powderly J.D., Carvajal R.D., Sosman J.A., Atkins M.B., et al. 2012. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 366:2443–2454 10.1056/NEJMoa1200690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velu V., Titanji K., Zhu B., Husain S., Pladevega A., Lai L., Vanderford T.H., Chennareddi L., Silvestri G., Freeman G.J., et al. 2009. Enhancing SIV-specific immunity in vivo by PD-1 blockade. Nature. 458:206–210 10.1038/nature07662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan G.M., Buldyrev S.V., Havlin S., da Luz M.G., Raposo E.P., Stanley H.E. 1999. Optimizing the success of random searches. Nature. 401:911–914 10.1038/44831 [DOI] [PubMed] [Google Scholar]
- Waite J.C., Leiner I., Lauer P., Rae C.S., Barbet G., Zheng H., Portnoy D.A., Pamer E.G., Dustin M.L. 2011. Dynamic imaging of the effector immune response to listeria infection in vivo. PLoS Pathog. 7:e1001326 10.1371/journal.ppat.1001326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry E.J. 2011. T cell exhaustion. Nat. Immunol. 12:492–499 10.1038/ni.2035 [DOI] [PubMed] [Google Scholar]
- Wherry E.J., Blattman J.N., Murali-Krishna K., van der Most R., Ahmed R. 2003. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J. Virol. 77:4911–4927 10.1128/JVI.77.8.4911-4927.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry E.J., Ha S.J., Kaech S.M., Haining W.N., Sarkar S., Kalia V., Subramaniam S., Blattman J.N., Barber D.L., Ahmed R. 2007. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 27:670–684 10.1016/j.immuni.2007.09.006 [DOI] [PubMed] [Google Scholar]
- Wilson E.B., Kidani Y., Elsaesser H., Barnard J., Raff L., Karp C.L., Bensinger S., Brooks D.G. 2012. Emergence of distinct multiarmed immunoregulatory antigen-presenting cells during persistent viral infection. Cell Host Microbe. 11:481–491 10.1016/j.chom.2012.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokosuka T., Takamatsu M., Kobayashi-Imanishi W., Hashimoto-Tane A., Azuma M., Saito T. 2012. Programmed cell death 1 forms negative costimulatory microclusters that directly inhibit T cell receptor signaling by recruiting phosphatase SHP2. J. Exp. Med. 209:1201–1217 10.1084/jem.20112741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajac A.J., Blattman J.N., Murali-Krishna K., Sourdive D.J., Suresh M., Altman J.D., Ahmed R. 1998. Viral immune evasion due to persistence of activated T cells without effector function. J. Exp. Med. 188:2205–2213 10.1084/jem.188.12.2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinselmeyer B.H., Dempster J., Wokosin D.L., Cannon J.J., Pless R., Parker I., Miller M.J. 2009. Chapter 16. Two-photon microscopy and multidimensional analysis of cell dynamics. Methods Enzymol. 461:349–378 10.1016/S0076-6879(09)05416-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.