Abstract
Human hemoglobins, the oxygen carriers in the blood, are composed by two α-like and two β-like globin monomers. The β-globin gene cluster located at 11p15.5 comprises one pseudogene and five genes whose expression undergoes two critical switches: the embryonic-to-fetal and fetal-to-adult transition. HBD encodes the δ-globin chain of the minor adult hemoglobin (HbA2), which is assumed to be physiologically irrelevant. Paradoxically, reduced diversity levels have been reported for this gene. In this study, we sought a detailed portrait of the genetic variation within the β-globin cluster in a large human population panel from different geographic backgrounds. We resequenced the coding and noncoding regions of the two adult β-globin genes (HBD and HBB) in European and African populations, and analyzed the data from the β-globin cluster (HBE, HBG2, HBG1, HBBP1, HBD, and HBB) in 1,092 individuals representing 14 populations sequenced as part of the 1000 Genomes Project. Additionally, we assessed the diversity levels in nonhuman primates using chimpanzee sequence data provided by the PanMap Project. Comprehensive analyses, based on classic neutrality tests, empirical and haplotype-based studies, revealed that HBD and its neighbor pseudogene HBBP1 have mainly evolved under purifying selection, suggesting that their roles are essential and nonredundant. Moreover, in the light of recent studies on the chromatin conformation of the β-globin cluster, we present evidence sustaining that the strong functional constraints underlying the decreased contemporary diversity at these two regions were not driven by protein function but instead are likely due to a regulatory role in ontogenic switches of gene expression.
Keywords: β-globin cluster, hemoglobin switch, gene diversity, chromatin interactions
Introduction
Hemoglobin (Hb) is the major protein in the circulating human red blood cells and its main function is to transport oxygen (O2). Human Hb is a tetramer composed of two dimers of α-like and β-like globin chains, which differ according to developmental stage. From shortly after early embryonic development up to adulthood, normal human hemoglobins maintain identical α-globin chains, while β-like chains are replaced, as result of two critical switches in gene expression, the first at the embryonic-to-fetal transition and the second at the fetal-to-adult one (Johnson et al. 2002; Schechter 2008; Sankaran et al. 2010).
The five human β-globin paralogs that code for the different β-like chains are clustered at chromosome 11 together with one pseudogene, being arranged as 5'-ε(HBE)-Gγ(HBG2)-Aγ(HBG1)-ψβ(HBBP1)-δ(HBD)-β(HBB)-3', in a region extending over approximately 80 kb. The stage-specific expression of each of these genes proceeds sequentially from embryonic (HBE), to fetal (HBG2 and HBG1), and finally to adult genes (HBD and HBB) and relies on the interactions with the locus control region (LCR), located from approximately 6 to 18 kb upstream of HBE (fig. 1A) (Bulger and Groudine 1999; Tolhuis et al. 2002; Bank 2006). In adulthood, HBB expression levels are much higher than those of its neighbor, HBD, resulting in two major Hb tetramers: HbA (α2β2), which accounts for approximately 97% of the total Hb, incorporates β-chains produced by HBB, and HbA2 (α2δ2), the minor fraction of adult Hb (generally < 3%), which contains the δ-chains encoded by HBD (Schechter 2008).
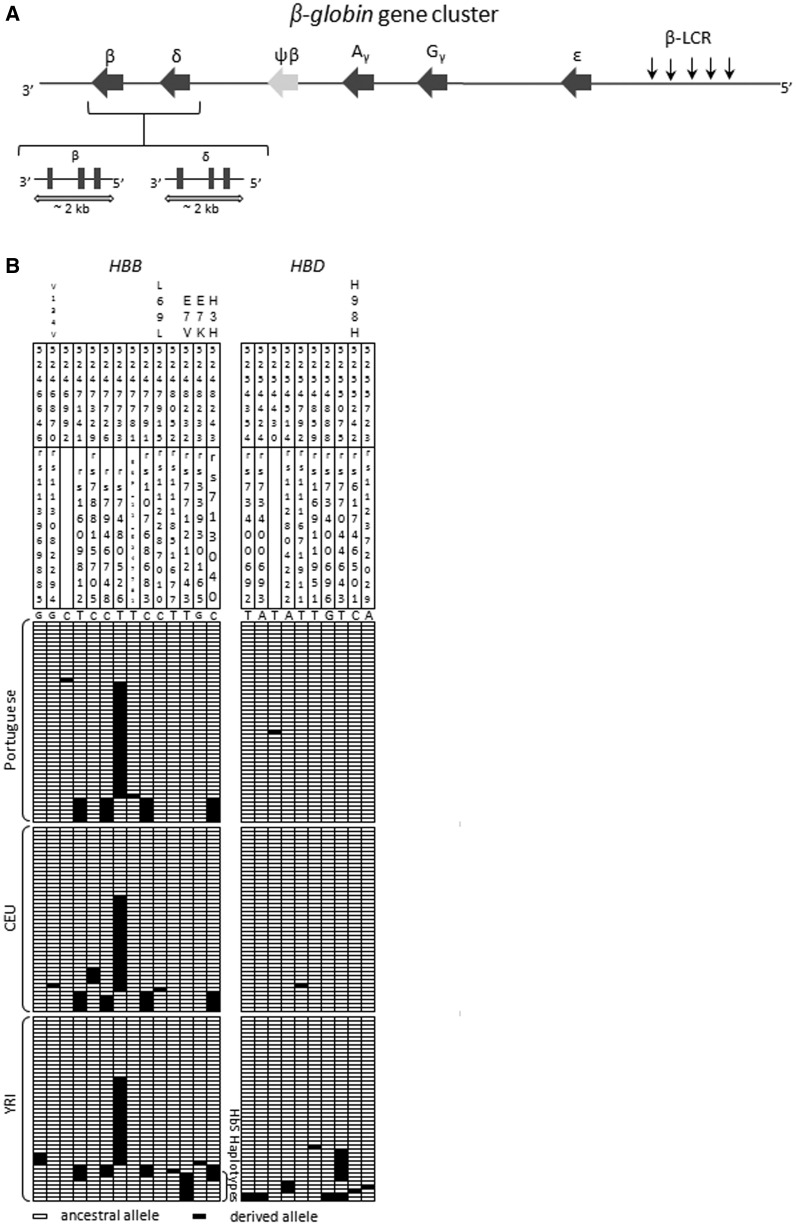
Fig. 1.—
(A) Schematic representation of the β-globin cluster located at chromosome 11p15.5. Top: The relative position of the β-globin genes and the LCR in the cluster. Bottom: δ-globin (HBD) and β-globin (HBB) gene organization (exons are represented by grey squares). The arrows in the lower diagram indicate the extent of the segments surveyed in the resequencing study of the European and African populations. Small black arrows indicate the five DNase I hypersensitive sites (HS) encompassed by the LCR located from approximately 6 to 18 kb upstream of ε- globin (HBE). (B) HBD and HBB haplotypes as inferred by PHASE v.2.02. The ancestral state at each site was inferred from ortholog nonhuman primate sequences. From the 25 sites, only 2 lacked a previously associated reference identification code in public databases (dbSNP and Exome Sequencing Project release ESP5400) and were unique to the Portuguese population. Coding variants are labeled. These include the following: one synonymous amino acid replacement in HBB (H3H) for the PT population; three synonymous replacements in HBB (H3H, L69L, and V134V) for the CEU population; one synonymous replacement in HBD (H98H) and one synonymous in HBB (H3H) and two nonsynonymous replacements (E7K–HbC allele, E7V–HbS allele) in HBB for the YRI population. SNP identifiers as in dbSNP and their chromosomal position based on GRCh37 version are indicated in each column.
Mutations in the β-chain of human HbA are associated with the most common inherited β-globin gene disorders world-wide (WHO 2011), such as β-thalassemia (Galanello and Origa 2010) and sickle cell disease (Orkin and Higgs 2010). On the contrary, HbA2, the minor adult Hb, is assumed to be physiologically irrelevant and mutations in HBD are per se clinically silent (Steinberg and Adams 1991; Schechter 2008). At the functional level, HbA2 has features that are nearly identical with those of HbA (de Bruin and Janssen 1973) and in the absence of β-chain production, as occurs in patients suffering from β-thalassemia major, HbA2 becomes the predominant oxygen carrier. However, HbA2 never reaches the amount that would be necessary to effectively replace HbA function (Steinberg and Adams 1991; Giambona et al. 2009; Mosca et al. 2009), which implies that its levels are only relevant for the diagnosis of β-globin disorders. Indeed, an elevated HbA2 concentration (≥3.5%) is the most significant parameter in the diagnosis of thalassemia syndromes (Cao and Moi 2000; Thein 2005; Galanello and Origa 2010), justifying the key role that HbA2 measurement plays in β-thalassemia screening programs (Giambona et al. 2009; Mosca et al. 2009).
HBD, encoding the unique δ-globin chain of HbA2, arose via duplication of the HBB gene after the marsupial/eutherian split and is therefore unique to placental mammals (Opazo et al. 2008). HBD has been inactivated or deleted in some lineages (Goodman et al. 1984; Hardies et al. 1984), but maintained an intact open reading frame in a few primate species, namely in humans, apes, and New World monkeys (Martin et al. 1980; Spritz and Giebel 1988). Human HBD and HBB show a high degree of homology (93%), as reflected in the similarity of their encoded proteins that only differ in 10 out of 147 amino acids (Steinberg and Adams 1991). Nonallelic gene conversion is the most commonly accepted explanation for such sequence homogeneity, albeit few gene conversion events have been described in the evolution of HBD and HBB (Papadakis and Patrinos 1999; Borg et al. 2009).
Given the apparent physiological redundancy of the HbA2 protein, the maintenance of HBD in several primate lineages is intriguing. Even though we might expect HBD to be subject to much lower selective pressure than HBB, in a previous study analyzing a small number of African individuals HBD was found to have lower diversity levels than HBB (Webster et al. 2003). In line with this finding, very few HBD mutations causing δ-thalassemia have been described (Steinberg and Adams 1991; Patrinos et al. 2004b). This pattern of sequence conservation, typical of genes under strong evolutionary constraints (Zhang 2003), is puzzling given the low expression levels of HBD and the presumed negligible functional role of HbA2 in oxygen transport (Steinberg and Adams 1991). One possible explanation for this apparent inconsistency could be that genetic variation has been more exhaustively assessed for HBB than for HBD. Even though the β-globin cluster is among the most extensively studied regions in the human genome, current genetic diversity estimates for the HBD and HBB genes across human populations are likely to be biased, because the genetic analysis of HBD and HBB has been performed mainly for diagnostic purposes and often based on a set of pre-ascertained single nucleotide polymorphisms (SNPs) (Morgado et al. 2007; Lacerra et al. 2008; Liu et al. 2009; Phylipsen et al. 2011).
In this study, we sought a better understanding of the evolutionary forces acting on HBD and HBB genes as well as of the physiological relevance of δ-globin conservation in placental mammals. To this end, we performed an unbiased characterization of the genetic diversity at the β-globin cluster based on Sanger sequencing of both HBB and HBD in ethnically diverse samples from Europe and Africa and on the sequence data from the 1000 Genomes Project Consortium (Altshuler et al. 2012). Then, we have evaluated the patterns of sequence variation, by means of allele frequency spectrum, linkage disequilibrium (LD) and haplotype structure analyses. Finally, we also assessed the diversity levels in this cluster in nonhuman primates using chimpanzee sequence data provided by the PanMap Project. Our findings indicate that purifying selection has shaped the evolutionary history of HBD and surprisingly, the same seems to apply to HBBP1. Furthermore, we present evidence that strong functional constraints have contributed to reduce the contemporary diversity of these two regions, probably due to a regulatory role in ontogenic switches of gene expression.
Materials and Methods
Population Samples and Sanger Sequencing
Sequence variation for HBB and HBD, was surveyed in a total of 71 samples: 25 Portuguese samples (PT) were collected in Coimbra’s university hospital (Centro Hospitalar e Universitário de Coimbra), from healthy individuals, under informed consent; 46 samples from the International HapMap Project, 23 CEU (Utah residents with Northern and Western European ancestry from the CEPH collection), and 23 YRI (Yoruba from Ibadan in Nigeria). Two DNA fragments of approximately 2 kb, one spanning the entire HBD gene and the other the HBB gene, were resequenced (fig. 1A). Primers for amplification and sequencing were designed using the latest version of the human genome assembly (GRCh37; http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/index.shtml). DNA fragments were amplified using polymerase chain reaction and sequenced with the BigDye Terminator v.3.1 Cycle Sequencing Kit and run on an Applied Biosystems ABI PRISM 3130xl Genetic Analyzer. All sequences were assembled and analyzed using Geneious version 5.4 created by Biomatters (available from http://www.geneious.com/) and all putative polymorphisms were manually inspected and individually confirmed.
Data Retrieval
Data from the 1000 Genomes Project were retrieved from its website (http://www.1000genomes.org/). Chromosomal locations and genomic segments from HBB (5246599–5248441), HBD (5253972–5255850), HBBP1 (5263085–5265019), HBG1 (5269347–5271272), HBG2 (5274263–5276195), and HBE (5289469–5291330) were obtained using latest version of the human genome assembly GRCh37. Chimpanzee reference sequence was downloaded from the UCSC Genome Browser (UCSC: http://genome.ucsc.edu/) and used as outgroup. Chimpanzee sequence variation was downloaded from PanMap Project website (http://panmap.uchicago.edu/). Additionally, we performed a scan for regulatory elements in HBD–HBBP1 region using data generated by the Encyclopedia of DNA Elements (ENCODE) Consortium (Myers et al. 2011), available in UCSC Genome Browser (http://genome.ucsc.edu/) in the Human GRCh37/hg19 assembly.
Statistical Analysis
The summary statistics of population genetic variation, number of segregating sites (S), nucleotide diversity (π) (Nei and Li 1979), which is based on average number of pairwise differences between sequences; Watterson’s estimator of the population mutation rate parameter (θW) (Watterson 1975), which is based on the number of segregating sites and sample size, and Tajima’s D (Tajima 1989) and Fay and Wu’s H (Fay and Wu 2000), which summarizes information about the spectrum of allele frequencies, were calculated using SLIDER (http://genapps.uchicago.edu/slider/index.html). To assess the statistical significance of Tajima’s D, we ran 100,000 coalescent simulations (Hudson 2002) using the previously estimated S statistic. Simulations were produced with the ‘‘ms’’ program, assuming distinct demographic models including constant population size and African and European best-fit models (Schaffner et al. 2005; Gutenkunst et al. 2009). To assess the statistical significance of Fay and Wu’s H, we computed 10,000 coalescent simulations in DnaSP v.5.10 (Rozas 2009). Haplotypes of HBB and HBD were inferred using the program PHASE v.2.02 (Stephens et al. 2001; Stephens and Donnelly 2003). Haplotype data were then annotated with additional SNP information and ancestral allele. Ancestral allele state was retrieved from dbSNP (http://www.ncbi.nlm.nih.gov/). LD analyses were performed using Haploview v.4.2 (Barrett et al. 2005) and haplotype blocks were identified through the standard algorithm implemented in the software (Gabriel et al. 2002).
To provide a temporal dimension to the phylogenetic relationships among haplotypes and to estimate the coalescent times and ages of relevant mutations, we used GENETREE v.9.0 (Griffiths and Tavare 1994). Since GENETREE assumes no recombination two incompatible haplotypes were removed from the analysis of HBB. Time, scaled in 2Ne generations, was derived from θ = 4Neµ. The mutation rate (µ) per gene, per generation, was deduced from the average number of nucleotide substitutions per site between human and chimpanzee reference sequences (Dxy), calculated with DnaSP v.5.10 (Rozas 2009). Time estimates in generations were converted into years using a 25-year generation time. Human/chimpanzee divergence was assumed to have occurred 5.4 Ma (Patterson et al. 2006).
To infer cladistic (network) relationships among the haplotypes, we used Network v.4.6, applying the Median-Joining method (Bandelt et al. 1999).
The evolutionary rates per site, per year, and per generation, were deduced from Jukes and Cantor distance calculated with DnaSP v.5.10 (Rozas 2009) and assuming the same human/chimpanzee divergence time.
Results
HBD and HBB Sequence Variation
We characterized the patterns of variation of the HBD and HBB genes by surveying two DNA fragments, each one spanning approximately 2 kb covering coding, noncoding, and the flanking 5′ and 3′ regions (fig. 1). Both segments were resequenced in a total of 71 samples belonging to populations from different geographic origins: Europeans (PT and CEU) and Africans (YRI). Overall, we identified 25 polymorphic sites, including a 2-bp deletion in HBD intron 2, which was excluded from further analyses (fig. 1B). In HBD, we observed 10 SNPs: 1 synonymous and 9 noncoding; and in HBB, we found 14 SNPs including 2 nonsynonymous, 2 synonymous, and 10 noncoding. The two nonsynonymous replacements, identified only in the YRI population, were the HbS (HBB:c.20A > T) and HbC (HBB:c.19G > A) alleles, which are known to confer resistance to Plasmodium falciparum and to occur at the highest frequencies in Africa, in endemic areas of malaria (Kwiatkowski 2005). Contrary to HBD in which the low frequency variants (singletons and doubletons) represented 80% of polymorphic sites, the same category of variants in HBB included only 36% of the sites. The frequencies for HbS and HbC alleles were 15% and 2%, respectively. The HbS alleles were linked to divergent haplotypes, which are likely to correspond to 3 out of the 5 “classical” βs haplotype backgrounds that are named according to their putative geographical origins (Benin, Bantu, Cameron, Senegal, and Arab) (Pagnier et al. 1984).
HBD and HBB Polymorphism Levels and Neutrality Tests
Standard population statistics based on the polymorphism levels as summarized by nucleotide diversity (π), and by the estimator of the population mutation rate parameter θw (Watterson 1975) are shown in table 1. Tajima’s D (Tajima 1989) tends to be slightly negative in populations of African descent while it is frequently associated to more positive values in populations of European descent (Wall and Przeworski 2000; Frisse et al. 2001; Akey et al. 2004; Stajich and Hahn 2005; Voight et al. 2005). In our data set, Tajima’s D statistics at HBD differs from the common trend showing significantly negative values in all populations analyzed PT, CEU, and YRI (table 1). This result is mainly due to a skew toward low frequency variants in YRI and to the lack of variation in CEU and PT. On the other hand, the Tajima’s D values estimated for HBB are similar in the three populations, moderately negative but nonsignificant (table 1).
Table 1.
Summary Statistics of Population Variation for the Two Adult β-globin Genes
| Population | Na |
HBD |
HBB |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Lb | Sc | πd | θwe | TDf | L | S | π | θw | TD | ||
| PT | 50 | 1,879 | 1 | 0.21 | 1.18 | −1.10* | 1,843 | 7 | 7.82 | 8.49 | −0.21 |
| CEU | 46 | 1,879 | 1 | 0.23 | 1.21 | −1.11* | 1,843 | 8 | 8.23 | 9.89 | −0.46 |
| YRI | 46 | 1,879 | 8 | 4.55 | 9.66 | −1.53** | 1,843 | 9 | 8.11 | 11.12 | −0.77 |
aNumber of chromosomes.
bTotal number of sites surveyed.
cPolymorphic sites.
dNucleotide diversity (×104) (Nei and Li 1979).
eWatterson θ per site (×104) (Watterson 1975).
fTajima’s D statistic (Tajima 1989).
*P ≤ 0.001 according to the constant size model, to the best fit model from Schaffner et al. (2005) and to the best fit model from Gutenkunst et al. (2009).
**P ≤ 0.05 according to the constant size model and to the best fit model from Gutenkunst et al. (2009).
To confirm the significant departure of HBD from the expectations under the neutral equilibrium we generated theoretical null distributions for calibrated models of human demography by coalescent simulations (Schaffner et al. 2005; Gutenkunst et al. 2009). The significant results obtained with such tests (table 1) provide arguments for a nondemographic interpretation of the low variation levels at HBD. In addition, the diversity patterns observed at the adjacent gene (HBB) seem to favor a selective hypothesis for the evolution of HBD rather than a population expansion that would have affected both genes equally. Two alternative selective hypotheses could explain the significant departure from neutrality of HBD: first, strong purifying selection that would purge any new deleterious mutations and second, a complete selective sweep in which an advantageous variant reached fixation. Considering the functional redundancy of HbA2, it is difficult to accept a protein modification as the likely target for positive selection. The single fixed difference in HBD between humans and great apes replaces two functionally equivalent amino acids, a valine by methionine (V127M) and it is already present at the Denisova sequence (Reich et al. 2011; Meyer et al. 2012). This gives a minimum time frame of 600,000–800,000 years for the origin of M127, which would be incompatible as well with a recent complete selective sweep. The possibility of a noncoding variant under positive selection cannot be excluded; however in that scenario, we would expect other typical features of selection like long and homogeneous haplotypes, high levels of population differentiation and unusual patterns of diversity given the observed divergence, which were not detected (Hudson et al. 1987; Fay and Wu 2000; Sabeti et al. 2002; Zeng et al. 2006; Mathias et al. 2012), as further described below.
Gene Genealogy and Age Estimates of HBD and HBB
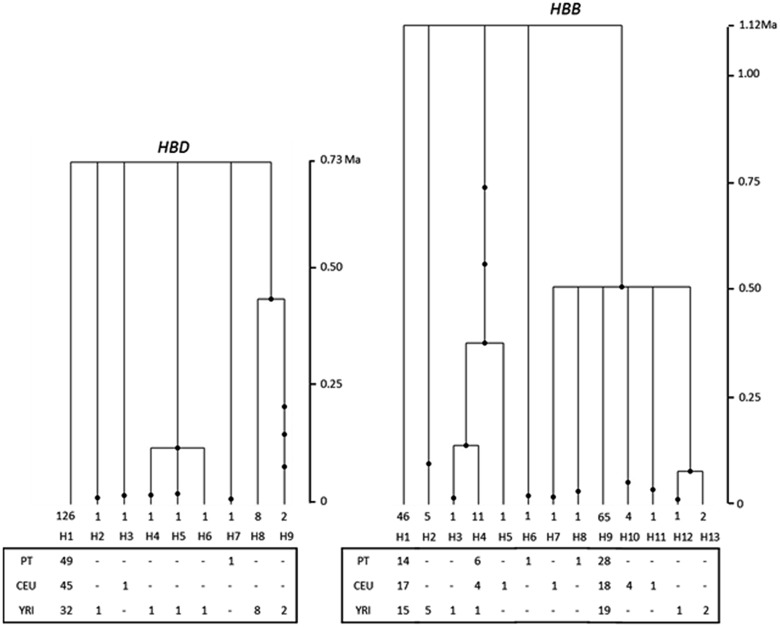
We reconstructed the gene genealogy of HBD and HBB, and estimated the time to the most recent common ancestor (TMRCA) using a maximum likelihood coalescent analysis (Griffiths and Tavare 1994). The results represented in figure 2, reveal that the tree of HBD differs sharply in its topology from that of HBB. Theoretically, HBD tree shape could be attributed either to population expansion, to positive or background selection (Harpending et al. 1998; Bamshad and Wooding 2003; Tishkoff and Verrelli 2003). The estimated TMRCA of HBD, 0.73 ± 0.33 Myr, is much younger than the estimated for HBB (1.12 ± 0.41 Myr), the latter being in full agreement with both observed and expected TMRCA for human autosomal genes (Excoffier 2002; Tishkoff and Verrelli 2003; Garrigan and Hammer 2006; Kim et al. 2010). Given the previous findings and the rejection of a demographic hypothesis, these results provide further support for a nonneutral interpretation of HBD evolution. The contrasting tree structure and recent TMRCA of HBD relative to HBB, can instead be more parsimoniously attributed to strong purifying selection, which is expected to show more recent coalescence times than under neutrality (Bamshad and Wooding 2003).
Fig. 2.—
HBD and HBB gene genealogies as estimated by Genetree. Time is scaled in millions of years (Myr). Solid circles represent nucleotide substitutions. The number below each branch of the trees represents the chromosomes observed for each haplotype, and in the lower diagram this information is split by population.
β-Globin Gene Cluster Diversity Patterns
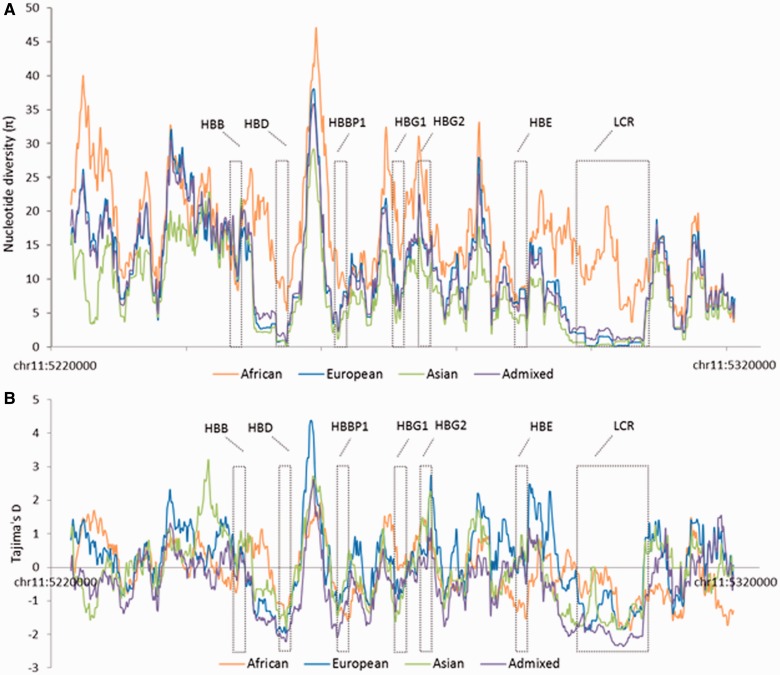
To obtain a more comprehensive assessment of the patterns of diversity on the entire β-globin gene cluster, we analyzed the data available from the 1000 Genomes Project phase 1 release v.3 (Altshuler et al. 2012), generated from the genome sequencing of 1,092 individuals belonging to 14 populations and five major populations groups: African, European, American, and East and South Asian (supplementary table S1, Supplementary Material online). We first analyzed the patterns of LD in the 80 kb region encompassing the entire β-globin cluster, in the populations resequenced in our study (CEU and YRI) (supplementary fig. S1, Supplementary Material online). We observed in the full β-globin cluster two distinct regions with strong LD, one that contains HBB (LD region 1) and the other extending from HBD to the LCR (LD region 2). These two regions are separated by a segment that encompasses one of the first recombination hotspots identified in humans (Chakravarti et al. 1984; Smith et al. 1998; Wall et al. 2003). Then, we evaluated the polymorphism levels for the five β-globin cluster genes comprised in the LD region 2: HBD, HBE, HBG2, HBG1, and HBBP1, and for HBB, included in LD region 1. As shown in table 2, summary statistics of CEU and YRI populations are in agreement with the results from our resequencing study. In the complete 1000 Genomes data set, higher nucleotide diversity levels were observed in African populations (YRI, LWK, and ASW) (table 2 and supplementary file S1, Supplementary Material online). Notwithstanding, HBD consistently displayed reduced levels of nucleotide diversity and strongly negative Tajima’s D values when compared with the remaining β-globin cluster genes, independently of the population studied (table 2 and supplementary file S1, Supplementary Material online). Interestingly, HBBP1 sequence, 7 kb upstream of HBD, is also one of the less diverse regions in the cluster, only surpassed by HBD and HBE, the latter encoding the extremely conserved embryonic globin. Thus, diversity patterns observed for HBD and HBBP1 suggest strong evolutionary constraints not related to protein function, as both are either marginally or not transcribed at all. Finally, we used the 1000 Genomes data to perform a sliding-window analysis of variation over the genomic region covering the entire β-globin cluster. As shown in figure 3, the genomic regions corresponding to HBD and HBBP1 presented the lowest values of both nucleotide diversity and Tajima’s D, only comparable with those obtained for the LCR region. Noteworthy, the intergenic region flanked by HBBP1 and HBD shows the opposite trend, with high levels of nucleotide diversity and positive Tajima’s D values, suggesting a complex evolutionary history possibly shaped by a noncanonical gene function.
Table 2.
Summary Statistics of Population Variation for the β-globin Cluster Genes Using the 1,000 Genomes Project Data
| La | CEU |
YRI |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Nb | Sc | πd | θwe | TDf | Na | Sc | πd | θwe | TDf | ||
| HBB | 1,843 | 170 | 7 | 8.28 | 6.65 | 0.53 | 176 | 11 | 8.49 | 10.39 | −0.44 |
| HBD | 1,879 | 4 | 0.31 | 3.73 | −1.67 | 10 | 6.53 | 9.26 | −0.70 | ||
| HBBP1 | 1,935 | 11 | 8.75 | 9.96 | −0.29 | 14 | 6.15 | 12.59 | −1.30 | ||
| HBG1 | 1,926 | 10 | 11.79 | 9.09 | 0.71 | 18 | 21.78 | 16.27 | 0.90 | ||
| HBG2 | 1,933 | 11 | 17.47 | 9.97 | 1.83 | 16 | 22.09 | 14.41 | 1.39 | ||
| HBE | 1,862 | 4 | 5.39 | 3.76 | 0.79 | 13 | 7.39 | 12.15 | −0.98 | ||
aTotal number of sites surveyed.
bNumber of chromosomes.
cPolymorphic sites.
dNucleotide diversity (×104) (Nei and Li 1979).
eWatterson θ per site (×104) (Watterson 1975).
fTajima’s D statistic (Tajima 1989).
Fig. 3.—
Sliding window analysis of the genomic region encompassing the β-globin gene cluster. Data were obtained from 1000 Genomes Project, representing 1,092 individuals from 14 populations: three African (ASW, LWK, and YRI), five European (CEU, FIN, GBR, IBS, and TSI), three Asian (CHB, CHS, and JPT) and three American-admixed populations (CLM, MXL, and PUR). Both π (A) and Tajima’s D (B) were calculated in 2 kb windows with increments of 150 bp.
HBBP1, HBD, and HBB Haplotype Analysis
We next focused on the comparison of the haplotype structure of the two adult genes, HBD and HBB. HBBP1 was also included in the analysis because of the unusual low levels of diversity presented by this pseudogene. We reconstructed haplotype genealogies for HBB, HBD, and HBBP1 using the full data set of the 1000 Genomes project. For HBD, a single common haplotype with an 88% frequency was identified. The star-shaped structure observed in the HBD network (supplementary figs. S2 and S3, Supplementary Material online) is in agreement with the atypical genetree of HBD built with our own resequencing data (CEU, PT, and YRI). Overall, these results give further support to the hypothesis of strong purifying selection operating on HBD, irrespectively of the diverse population demographic histories. The haplotype network for HBBP1 might be consistent with purifying selection as well, even though its footprints are somewhat weaker than those at HBD.
Interspecies Comparisons
To gain further insight into the possible functional constraints that have been shaping the evolutionary history of this genomic region, we used human and chimpanzee sequences to calculate and compare the rate of divergence, in exons and introns, across HBD, HBB, and HBBP1 (table 3). In HBB, introns display an evolutionary rate ∼7 times higher than exons. Although higher divergence rates are expected for introns, in the case of HBB this likely reflects an increased intronic mutation rate as reported for β-globin genes (1.89%), when compared with the intronic average (1.03%) (Chen and Li 2001). Remarkably, HBD and HBBP1 Homo–Pan nucleotide differences are more homogeneously distributed between exons and introns, and the overall substitution rate is nearly half of that observed for HBB introns (supplementary table S2, Supplementary Material online), indicating again that these genes are under higher evolutionary constraints.
Table 3.
Evolutionary Rates Based on Jukes–Cantor Distance
| Exons |
Introns |
|||||
|---|---|---|---|---|---|---|
| Diva | Mutation Rateb |
Diva | Mutation Rateb |
|||
| Year | Generation | Year | Generation | |||
| HBD | 0.80 | 0.74 × 10−9 | 1.84 × 10−8 | 0.99 | 0.91 × 10−9 | 2.28 × 10−8 |
| HBB | 0.26 | 0.24 × 10−9 | 0.60 × 10−8 | 1.75 | 1.62 × 10−9 | 4.06 × 10−8 |
| HBBP1 | 1.45 | 1.34 × 10−9 | 3.36 × 10−8 | 1.04 | 0.96 × 10−9 | 2.41 × 10−8 |
aAverage nucleotide divergence between human and chimpanzee.
bMutation rate per site.
We next evaluated the haplotype diversity for these 3 genes in chimpanzees, by using sequence variation data from 10 Western chimpanzees (Pan troglodytes verus) generated by the PanMap Project (supplementary table S2, Supplementary Material online), which has revealed that HBD and HBBP1 presented lower nucleotide diversity relative to HBB. These results resemble those obtained for the different human populations, and therefore it seems likely that HBD and HBBP1 are under purifying selection also in chimpanzees.
Scan for Regulatory Elements in HBD–HBBP1 Region
The low levels of diversity found in HBD and HBBP1 suggest that both sequences are evolving under purifying selection which cannot be attributed to constraints on a functional protein. An alternative explanation is that HBD and HBBP1 lie within crucial regions for the regulation of gene transcription, in which selective pressures would be acting to maintain the nucleotide sequence. To explore this possibility, we analyzed the data generated by the Encyclopedia of DNA Elements (ENCODE) Consortium (Myers et al. 2011), available through the UCSC Genome Browser (http://genome.ucsc.edu/). A large number of transcription factor binding sites have been experimentally detected by ChIP-seq throughout the β-globin cluster, some of which with an established role in chromatin remodeling, namely the zinc finger protein (CTCF–CCCTC-binding factor), the C2H2 type zinc-finger protein (BCL11A) and the globin transcription factor 1 (Gata-1) (Vakoc et al. 2005; Splinter et al. 2006; Hou et al. 2010; Xu et al. 2010). Previous studies have also analyzed the long-range chromatin interactions in the β-globin cluster by Chromosome Conformation Capture Carbon Copy (5C) (Dostie et al. 2006; Sanyal et al. 2012) (supplementary fig. S4A, Supplementary Material online). Noteworthy, significant interactions were detected between a segment comprising both HBD and HBBP1, and different regions upstream HBE, which overlap the LCR. Moreover, these interactions were detected specifically in the erythropoietic cell line K562, in which β-globin genes are actively transcribed. Interestingly, in this cell line both the LCR and HBD lie in regions of open chromatin, as determined by DNAse I hypersensitivity and FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) assays, two different methods to identify nucleosome-depleted regions of the genome (supplementary fig. S4B, Supplementary Material online). The specific conformation observed in K562 cells suggests that this structure has a role in maintaining an active transcriptional state of the β-globin cluster in this cell line. In fact, both HBD and HBBP1 may act as anchor regions in LCR-driven chromatin looping, a crucial mechanism for temporal coordination of gene expression in the human β-globin cluster (Bulger and Groudine 1999; Patrinos et al. 2004a).
Discussion
Unusual low levels of diversity have been described for human HBD, which are difficult to reconcile with the negligible function of HbA2 in oxygen transport (Webster et al. 2003). In primates HBD exhibits a surprisingly high level of sequence conservation relative to functional paralogs (Steinberg and Adams 1991), suggesting an important role of HBD in those lineages. Here, we gain insight into the evolutionary history of HBD and its potential function in gene regulation by performing a detailed analysis of the sequence diversity and divergence of the β-globin cluster in humans and chimpanzees. Our results demonstrate that the HBD lack of diversity is a common trend across human populations that cannot be due neither to a systematic bias of clinical studies nor to an effect of population history. Even though recent evidence point to a human explosive growth over the past 10,000 years leading to an increment in very low frequency variants (Keinan and Clark 2012), the significance of the negative Tajima’s D values obtained under the best-fit models discourage a demographic interpretation of HBD diversity. Furthermore, the findings of strong negative Tajima’s D values in the absence of other features of positive selection, pinpoint HBD as a target of purifying selection like previously reported for innate-immunity genes under strong functional constraints (Barreiro et al. 2009; Mukherjee et al. 2009; Wlasiuk and Nachman 2010).
An atypical tree topology was identified for HBD consistent with the loss of mild deleterious variants by negative selective pressures and a low TMRCA of 0.73 ± 0.33 Myr (or 0.58 ± 0.26 Myr for a generation time of 20 years), amongst the most recent estimates obtained for autosomal genes (0.20–0.31 Myr for a generation time of 20 years), was confirmed (Fullerton et al. 2000; Martinez-Arias et al. 2001; Excoffier 2002; Tishkoff and Verrelli 2003; Webster et al. 2003; Garrigan and Hammer 2006; Kim et al. 2010). Conversely, the polymorphism levels and TMRCA estimate obtained for HBB are in agreement with the expectations under a neutral model and do not differ from previous studies (Harding et al. 1997).
Taken together, the results obtained for the β-globin cluster cannot be reconciled with other explanatory hypotheses rather than purifying selection: the low values of nucleotide diversity are confined to HBD and HBBP1 and do not extend into the flanking regions which display contrastingly high levels of variation; the haplotype structure of HBD and HBBP1 are similar across worldwide populations; furthermore, chimpanzee HBD and HBBP1 also exhibit lower haplotype diversity than HBB and their mutation rates, as inferred from human–chimpanzee divergence, are considerably reduced when compared with HBB and to other β-globin noncoding regions (Chen and Li 2001). Evidence here presented suggests a long-term effect of purifying selection probably predating modern human origins (200,000 years), with the same strong functional constrains acting across different primate species for at least 5 Myr.
Several decades ago, a hypothesis was formulated holding an important regulatory role of HBD and HBBP1 in the Hb fetal-to-adult switch that matches quite well the assumption of strong negative selective forces acting on these sequences (Ottolenghi et al. 1979; Bank et al. 1980; Chang and Slightom 1984; Goodman et al. 1984). Over the past years, the β-globin cluster has been regarded as a complex genetic system and a paradigm of gene expression regulation. More recently, a boost of studies on the β-globin cluster have contributed to a better understanding of the mechanisms underlying the regulation of each gene in the cluster (Harju et al. 2002; Chakalova et al. 2005; Noordermeer and de Laat 2008; Sankaran et al. 2010). Remarkably, chromosome conformation (3C and 5C) analyses for the β-globin locus disclosed strong interactions between the LCR and the region encompassing both HBD and HBBP1 (Dostie et al. 2006; Sanyal et al. 2012). Furthermore, distinct spatial interactions of the LCR in fetal and adult stages were uncovered by another study based only in 3C assay in which HBD sequence was proposed to be enrolled in the maintenance of a transcriptionally competent structure at the adult stage (Beauchemin and Trudel 2009). These recent findings suggest that HBD and HBBP1 might be involved in chromatin looping in the human β-globin cluster, a crucial mechanism for temporal coordination of gene expression (Holwerda and De Laat 2012). Importantly, one SNP (rs10128556) in HBBP1 has been also identified as a modulator of HbF levels reinforcing the idea that this genomic region is indeed involved in the Hb fetal-to-adult switch (Galarneau et al. 2010). Considering that the mechanism of Hb switch is common to all simian primates (Johnson et al. 2002), we might expect to find similar patterns of conservation and diversity in orthologous sequences of HBD and HBBP1 and to detect signatures of purifying selection at the β-globin cluster over 40 Myr of primate evolution.
In contrast to the low diversity levels of HBD and HBBP1 are the high levels found in the HBD-HBBP1 intergenic region. This pattern has been previously observed in a study involving 23 individuals from the Luo population (Kenya, Africa) (Webster et al. 2003). There, the authors found a significant positive Tajima’s D and two divergent haplotypes “R” and “T” regarded either as relics of a subdivided ancestral hominid population or as a signature of balancing selection (Maeda et al. 1983; Webster et al. 2002, 2003). It is known that the HBD–HBBP1 intergenic region contains a small segment of 1 kb flanked by two opposite orientated Alu elements, which is expected to increase genetic instability and local mutation rate (Wang and Vasquez 2006), therefore providing an explanation for the observed high diversity levels. Also of note, is the fact that a 3.5 kb segment upstream of HBD was proven to be necessary for HBG1 and HBG2 gene silencing since its deletion causes an increment in HbF production throughout adulthood (Sankaran et al. 2011). This segment overlaps a binding region of BCL11A, which is a biochemically validated and fundamental switching factor (Sankaran et al. 2008, 2009), and contains other functional elements, such as a polypyrimidine tract that can serve as a binding site for multiprotein chromatin remodeling complexes (Bank et al. 2005). Furthermore, it has been demonstrated that the binding of protein complexes in this region induces conformational changes in the β-globin cluster, which in turn mediate long-range physical interactions between distal regulatory elements located in the LCR (Xu et al. 2010). Therefore, we propose that the two alternative haplotype configurations “R” and “T” may represent nucleotide combinations that preserve specific motifs for binding of protein complexes involved in transcriptional regulation and chromatin remodeling.
Collectively, these findings illustrate the complexity of the regulatory mechanisms within the cluster and provide a rationale for the heterogeneity in diversity levels observed within the HBD–HBBP1 region. In the yeast genome, higher conservation was observed in nucleosome free regions, compared with regions with high nucleosome occupancy, irrespective of their overlap with coding sequences, which underscores the correlation between chromatin structure and evolutionary constraints (Nikolaou et al. 2010). Accordingly, a recent study integrating ENCODE and 1000 Genomes data uncovered a widespread signature of purifying selection on regulatory regions in humans, including promoters, enhancers, insulators and other regions with different chromatin conformational states (Ward and Kellis 2012). The authors also noticed that many of these regions are lineage specific and a fraction of these arose in the common ancestor of primates. We hypothesize that the clear signature of purifying selection at HBD and HBBP1 may reflect constraints on local chromatin conformation and the maintenance of a nucleosome free region available for frequent interactions with the LCR.
In summary, the results here presented indicate that purifying selection is driving not only HBD evolution but also its neighbor pseudogene, HBBP1. In the light of recent advances in the characterization of the β-globin cluster, we propose that the complex patterns of diversity observed in this genomic region arose from distinct functional constraints related with the intricate process of chromatin and protein interactions coordinating the differential expression of genes at the β-globin cluster during development.
Supplementary Material
Supplementary tables S1 and S2, file S1, and figures S1–S4 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Dr Letícia Ribeiro for her collaboration in providing the Portuguese samples. This work was supported by the Portuguese Foundation for Science and Technology (FCT) fellowship (SFRH/BD/73508/2010 and SFRH/BPD/73366/2010) to A.M. and A.M.L., respectively, and by the POPH-QREN – Promotion of scientific employment, the European Social Fund, and national funds of the Ministry of Education and Science grants to S.S. IPATIMUP is an Associate Laboratory of the Portuguese Ministry of Education and Science and is partially supported by FCT.
Literature Cited
- Akey JM, et al. Population history and natural selection shape patterns of genetic variation in 132 genes. PLoS Biol. 2004;2:7. doi: 10.1371/journal.pbio.0020286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler DM, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad M, Wooding SP. Signatures of natural selection in the human genome. Nat Rev Genet. 2003;4:99–111. doi: 10.1038/nrg999. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Bank A. Regulation of human fetal hemoglobin: new players, new complexities. Blood. 2006;107:435–443. doi: 10.1182/blood-2005-05-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bank A, Mears J, Ramirez F. Disorders of human hemoglobin. Science. 1980;207:486–493. doi: 10.1126/science.7352255. [DOI] [PubMed] [Google Scholar]
- Bank A, et al. Role of intergenic human γ-δ-globin sequences in human hemoglobin switching and reactivation of fetal hemoglobin in adult erythroid cells. Ann N Y Acad Sci. 2005;1054:48–54. doi: 10.1196/annals.1345.057. [DOI] [PubMed] [Google Scholar]
- Barreiro LB, et al. Evolutionary dynamics of human toll-like receptors and their different contributions to host defense. PLoS Genet. 2009;5:e1000562. doi: 10.1371/journal.pgen.1000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Beauchemin H, Trudel M. Evidence for a bigenic chromatin subdomain in regulation of the fetal-to-adult hemoglobin switch. Mol Cell Biol. 2009;29:1635–1648. doi: 10.1128/MCB.01735-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg J, Georgitsi M, Aleporou-Marinou V, Kollia P, Patrinos GP. Genetic recombination as a major cause of mutagenesis in the human globin gene clusters. Clin Biochem. 2009;42:1839–1850. doi: 10.1016/j.clinbiochem.2009.07.014. [DOI] [PubMed] [Google Scholar]
- Bulger M, Groudine M. Looping versus linking: toward a model for long-distance gene activation. Genes Dev. 1999;13:2465–2477. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]
- Cao A, Moi P. Genetic modifying factors in β-thalassemia. Clin Chem Lab Med. 2000;38:123–132. doi: 10.1515/CCLM.2000.019. [DOI] [PubMed] [Google Scholar]
- Chakalova L, et al. Developmental regulation of the β-globin gene locus. In: Jeanteur P, editor. Epigenetics and chromatin. Berlin (Germany): Springer; 2005. pp. 183–206. [DOI] [PubMed] [Google Scholar]
- Chakravarti A, et al. Nonuniform recombination within the human beta-globin gene cluster. Am J Hum Genet. 1984;36:1239–1258. [PMC free article] [PubMed] [Google Scholar]
- Chang LYE, Slightom JL. Isolation and nucleotide sequence analysis of the β-type globin pseudogene from human, gorilla and chimpanzee. J Mol Biol. 1984;180:767–783. doi: 10.1016/0022-2836(84)90256-0. [DOI] [PubMed] [Google Scholar]
- Chen FC, Li WH. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am J Hum Genet. 2001;68:444–456. doi: 10.1086/318206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin SH, Janssen LHM. Comparison of the oxygen and proton binding behavior of human hemoglobin A and A2. Biochim Biophys Acta. 1973;295:490–494. doi: 10.1016/0005-2795(73)90044-5. [DOI] [PubMed] [Google Scholar]
- Dostie J, et al. Chromosome conformation capture carbon copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L. Human demographic history: refining the recent African origin model. Curr Opin Genet Dev. 2002;12:675–682. doi: 10.1016/s0959-437x(02)00350-7. [DOI] [PubMed] [Google Scholar]
- Fay JC, Wu CI. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisse L, et al. Gene conversion and different population histories may explain the contrast between polymorphism and linkage disequilibrium levels. Am J Hum Genet. 2001;69:831–843. doi: 10.1086/323612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullerton SM, et al. Apolipoprotein E variation at the sequence haplotype level: implications for the origin and maintenance of a major human polymorphism. Am J Hum Genet. 2000;67:881–900. doi: 10.1086/303070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel SB, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Galanello R, Origa R. Beta-thalassemia. Orphanet J Rare Dis. 2010;5:11. doi: 10.1186/1750-1172-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galarneau G, et al. Fine-mapping at three loci known to affect fetal hemoglobin levels explains additional genetic variation. Nat Genet. 2010;42:1049–1051. doi: 10.1038/ng.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigan D, Hammer MF. Reconstructing human origins in the genomic era. Nat Rev Genet. 2006;7:669–680. doi: 10.1038/nrg1941. [DOI] [PubMed] [Google Scholar]
- Giambona A, Passarello C, Renda D, Maggio A. The significance of the hemoglobin A2 value in screening for hemoglobinopathies. Clin Biochem. 2009;42:1786–1796. doi: 10.1016/j.clinbiochem.2009.06.026. [DOI] [PubMed] [Google Scholar]
- Goodman M, Koop BF, Czelusniak J, Weiss ML, Slightom JL. The eta-globin gene: its long evolutionary history in the beta-globin gene family of mammals. J Mol Biol. 1984;180:803–823. doi: 10.1016/0022-2836(84)90258-4. [DOI] [PubMed] [Google Scholar]
- Griffiths RC, Tavare S. Sampling theory for neutral alleles in a varying environment. Philos Trans R Soc Lond B Biol Sci. 1994;344:403–410. doi: 10.1098/rstb.1994.0079. [DOI] [PubMed] [Google Scholar]
- Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 2009;5:e1000695. doi: 10.1371/journal.pgen.1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardies SC, Edgell MH, Hutchison CA. Evolution of the mammalian beta-globin gene cluster. J Biol Chem. 1984;259:3748–3756. [PubMed] [Google Scholar]
- Harding RM, et al. Archaic African and Asian lineages in the genetic ancestry of modern humans. Am J Hum Genet. 1997;60:772–789. [PMC free article] [PubMed] [Google Scholar]
- Harju S, McQueen KJ, Peterson KR. Chromatin structure and control of β-like globin gene switching. Exp Biol Med. 2002;227:683–700. doi: 10.1177/153537020222700902. [DOI] [PubMed] [Google Scholar]
- Harpending HC, et al. Genetic traces of ancient demography. Proc Natl Acad Sci U S A. 1998;95:1961–1967. doi: 10.1073/pnas.95.4.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holwerda S, De Laat W. Chromatin loops, gene positioning and gene expression. Front Genet. 2012;3:217. doi: 10.3389/fgene.2012.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou C, Dale R, Dean A. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc Natl Acad Sci U S A. 2010;107:3651–3656. doi: 10.1073/pnas.0912087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- Hudson RR, Kreitman M, Aguadé M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RM, Gumucio D, Goodman M. Globin gene switching in primates. Comp Biochem Physiol A Mol Integr Physiol. 2002;133:877–883. doi: 10.1016/s1095-6433(02)00205-2. [DOI] [PubMed] [Google Scholar]
- Keinan A, Clark AG. Recent explosive human population growth has resulted in an excess of rare genetic variants. Science. 2012;336:740–743. doi: 10.1126/science.1217283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HL, Igawa T, Kawashima A, Satta Y, Takahata N. Divergence, demography and gene loss along the human lineage. Philos Trans R Soc B: Biol Sci. 2010;365:2451–2457. doi: 10.1098/rstb.2010.0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski DP. How malaria has affected the human genome and what human genetics can teach us about malaria. Am J Hum Genet. 2005;77:171–192. doi: 10.1086/432519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacerra G, et al. Molecular evidences of single mutational events followed by recurrent crossing-overs in the common delta-globin alleles in the Mediterranean area. Gene. 2008;410:129–138. doi: 10.1016/j.gene.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Liu L, et al. High-density SNP genotyping to define beta-globin locus haplotypes. Blood Cells Mol Dis. 2009;42:16–24. doi: 10.1016/j.bcmd.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda N, Bliska JB, Smithies O. Recombination and balanced chromosome polymorphism suggested by DNA sequences 5' to the human delta-globin gene. Proc Natl Acad Sci U S A. 1983;80:5012–5016. doi: 10.1073/pnas.80.16.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SL, Zimmer EA, Kan YW, Wilson AC. Silent delta-globin gene in Old World monkeys. Proc Natl Acad Sci U S A. 1980;77:3563–3566. doi: 10.1073/pnas.77.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Arias R, et al. Sequence variability of a human pseudogene. Genome Res. 2001;11:1071–1085. doi: 10.1101/gr.167701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias RA, et al. Adaptive evolution of the FADS gene cluster within Africa. PLoS One. 2012;7:e44926. doi: 10.1371/journal.pone.0044926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, et al. A high-coverage genome sequence from an Archaic Denisovan individual. Science. 2012;338:222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgado A, et al. Mutational spectrum of delta-globin gene in the Portuguese population. Eur J Haematol. 2007;79:422–428. doi: 10.1111/j.1600-0609.2007.00949.x. [DOI] [PubMed] [Google Scholar]
- Mosca A, Paleari R, Ivaldi G, Galanello R, Giordano PC. The role of haemoglobin A2 testing in the diagnosis of thalassaemias and related haemoglobinopathies. J Clin Pathol. 2009;62:13–17. doi: 10.1136/jcp.2008.056945. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Sarkar-Roy N, Wagener DK, Majumder PP. Signatures of natural selection are not uniform across genes of innate immune system, but purifying selection is the dominant signature. Proc Natl Acad Sci U S A. 2009;106:7073–7078. doi: 10.1073/pnas.0811357106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers RM, et al. A user's guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:19. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaou C, Althammer S, Beato M, Guigo R. Structural constraints revealed in consistent nucleosome positions in the genome of S. cerevisiae. Epigenetics Chromatin. 2010;3:1756–8935. doi: 10.1186/1756-8935-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordermeer D, de Laat W. Joining the loops: beta-globin gene regulation. IUBMB Life. 2008;60:824–833. doi: 10.1002/iub.129. [DOI] [PubMed] [Google Scholar]
- Opazo JC, Hoffmann FG, Storz JF. Genomic evidence for independent origins of β-like globin genes in monotremes and therian mammals. Proc Natl Acad Sci U S A. 2008;105:1590–1595. doi: 10.1073/pnas.0710531105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Higgs DR. Sickle cell disease at 100 years. Science. 2010;329:291–292. doi: 10.1126/science.1194035. [DOI] [PubMed] [Google Scholar]
- Ottolenghi S, et al. Globin gene deletion in HPFH, δ° β° thalassaemia and Hb Lepore disease. Nature. 1979;278:654–657. doi: 10.1038/278654a0. [DOI] [PubMed] [Google Scholar]
- Pagnier J, et al. Evidence for the multicentric origin of the sickle cell hemoglobin gene in Africa. Proc Natl Acad Sci U S A. 1984;81:1771–1773. doi: 10.1073/pnas.81.6.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadakis MN, Patrinos GP. Contribution of gene conversion in the evolution of the human β-like globin gene family. Hum Genet. 1999;104:117–125. doi: 10.1007/s004390050923. [DOI] [PubMed] [Google Scholar]
- Patrinos GP, et al. Multiple interactions between regulatory regions are required to stabilize an active chromatin hub. Genes Dev. 2004a;18:1495–1509. doi: 10.1101/gad.289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrinos GP, et al. Improvements in the HbVar database of human hemoglobin variants and thalassemia mutations for population and sequence variation studies. Nucleic Acids Res. 2004b;32:D537–D541. doi: 10.1093/nar/gkh006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Richter DJ, Gnerre S, Lander ES, Reich D. Genetic evidence for complex speciation of humans and chimpanzees. Nature. 2006;441:1103–1108. doi: 10.1038/nature04789. [DOI] [PubMed] [Google Scholar]
- Phylipsen M, Gallivan MVE, Arkesteijn SGJ, Harteveld CL, Giordano PC. Occurrence of common and rare δ-globin gene defects in two multiethnic populations: thirteen new mutations and the significance of δ-globin gene defects in β-thalassemia diagnostics. Int J Lab Hematol. 2011;33:85–91. doi: 10.1111/j.1751-553X.2010.01255.x. [DOI] [PubMed] [Google Scholar]
- Reich D, et al. Denisova admixture and the first modern human dispersals into Southeast Asia and Oceania. Am J Hum Genet. 2011;89:516–528. doi: 10.1016/j.ajhg.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J. DNA sequence polymorphism analysis using DnaSP. Methods Mol Biol. 2009;537:337–350. doi: 10.1007/978-1-59745-251-9_17. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- Sankaran VG, et al. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science. 2008;322:1839–1842. doi: 10.1126/science.1165409. [DOI] [PubMed] [Google Scholar]
- Sankaran VG, et al. A functional element necessary for fetal hemoglobin silencing. N Engl J Med. 2011;365:807–814. doi: 10.1056/NEJMoa1103070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaran VG, Xu J, Orkin SH. Advances in the understanding of haemoglobin switching. Br J Haematol. 2010;149:181–194. doi: 10.1111/j.1365-2141.2010.08105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaran VG, et al. Developmental and species-divergent globin switching are driven by BCL11A. Nature. 2009;460:1093–1097. doi: 10.1038/nature08243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner SF, et al. Calibrating a coalescent simulation of human genome sequence variation. Genome Res. 2005;15:1576–1583. doi: 10.1101/gr.3709305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechter AN. Hemoglobin research and the origins of molecular medicine. Blood. 2008;112:3927–3938. doi: 10.1182/blood-2008-04-078188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RA, Ho PJ, Clegg JB, Kidd JR, Thein SL. Recombination breakpoints in the human beta-globin gene cluster. Blood. 1998;92:4415–4421. [PubMed] [Google Scholar]
- Splinter E, et al. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spritz R, Giebel L. The structure and evolution of the spider monkey delta-globin gene. Mol Biol Evol. 1988;5:21–29. doi: 10.1093/oxfordjournals.molbev.a040474. [DOI] [PubMed] [Google Scholar]
- Stajich JE, Hahn MW. Disentangling the effects of demography and selection in human history. Mol Biol Evol. 2005;22:63–73. doi: 10.1093/molbev/msh252. [DOI] [PubMed] [Google Scholar]
- Steinberg M, Adams JG., 3rd Hemoglobin A2: origin, evolution, and aftermath. Blood. 1991;78:2165–2177. [PubMed] [Google Scholar]
- Stephens M, Donnelly P. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thein S. Genetic modifiers of beta-thalassemia. Haematologica. 2005;90:649–660. [PubMed] [Google Scholar]
- Tishkoff SA, Verrelli BC. Patterns of human genetic diversity: implications for human evolutionary history and disease. Annu Rev Genomics Hum Genet. 2003;4:293–340. doi: 10.1146/annurev.genom.4.070802.110226. [DOI] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- Vakoc CR, et al. Proximity among distant regulatory elements at the β-globin locus requires GATA-1 and FOG-1. Mol Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Voight BF, et al. Interrogating multiple aspects of variation in a full resequencing data set to infer human population size changes. Proc Natl Acad Sci U S A. 2005;102:18508–18513. doi: 10.1073/pnas.0507325102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JD, Frisse LA, Hudson RR, Di Rienzo A. Comparative linkage-disequilibrium analysis of the beta-globin hotspot in primates. Am J Hum Genet. 2003;73:1330–1340. doi: 10.1086/380311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JD, Przeworski M. When did the human population size start increasing? Genetics. 2000;155:1865–1874. doi: 10.1093/genetics/155.4.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Vasquez KM. Non-B DNA structure-induced genetic instability. Mutat Res. 2006;598:103–119. doi: 10.1016/j.mrfmmm.2006.01.019. [DOI] [PubMed] [Google Scholar]
- Ward LD, Kellis M. Evidence of abundant purifying selection in humans for recently acquired regulatory functions. Science. 2012;337:1675–1678. doi: 10.1126/science.1225057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson GA. On the number of segregating sites in genetical models without recombination. Theor Popul Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Webster MT, Clegg JB, Harding RM. Common 5' ß-globin RFLP haplotypes harbour a surprising level of ancestral sequence mosaicism. Hum Genet. 2003;113:123–139. doi: 10.1007/s00439-003-0954-0. [DOI] [PubMed] [Google Scholar]
- Webster MT, Wells RS, Clegg JB. Analysis of variation in the human β-globin gene cluster using a novel DHPLC technique. Mutat Res. 2002;501:99–103. doi: 10.1016/s0027-5107(02)00017-9. [DOI] [PubMed] [Google Scholar]
- Wlasiuk G, Nachman MW. Adaptation and constraint at Toll-like receptors in primates. Mol Biol Evol. 2010;27:2172–2186. doi: 10.1093/molbev/msq104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. World malaria report. Geneva (Switzerland): World Health Organization; 2011. [Google Scholar]
- Xu J, et al. Transcriptional silencing of {gamma}-globin by BCL11A involves long-range interactions and cooperation with SOX6. Genes Dev. 2010;24:783–798. doi: 10.1101/gad.1897310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng K, Fu Y, Shi S, Wu C. Statistical tests for detecting positive selection by utilizing high-frequency variants. Genetics. 2006;174:1431–1439. doi: 10.1534/genetics.106.061432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Evolution by gene duplication: an update. Trends Ecol Evol. 2003;18:292–298. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.