Abstract
Phytochromes (Phy) and phytochrome-interacting factor (PIF) transcription factors constitute a major signaling module that controls plant development in response to red and far-red light. A low red:far-red ratio is interpreted as shading by neighbor plants and induces cell elongation—a phenomenon called shade-avoidance syndrome (SAS). PAR1 and its closest homolog PAR2 are negative regulators of SAS; they belong to the HLH transcription factor family that lacks a typical basic domain required for DNA binding, and they are believed to regulate gene expressions through DNA binding transcription factors that are yet to be identified. Here, we show that light signal stabilizes PAR1 protein and PAR1 interacts with PIF4 and inhibits PIF4-mediated gene activation. DNA pull-down and chromatin immunoprecipitation (ChIP) assays showed that PAR1 inhibits PIF4 DNA binding in vitro and in vivo. Transgenic plants overexpressing PAR1 (PAR1OX) are insensitive to gibberellin (GA) or high temperature in hypocotyl elongation, similarly to the pifq mutant. In addition to PIF4, PAR1 also interacts with PRE1, a HLH transcription factor activated by brassinosteroid (BR) and GA. Overexpression of PRE1 largely suppressed the dwarf phenotype of PAR1OX. These results indicate that PAR1–PRE1 and PAR1–PIF4 heterodimers form a complex HLH/bHLH network regulating cell elongation and plant development in response to light and hormones.
Keywords: PAR1, PIF4, PRE1, bHLH, DNA binding
INTRODUCTION
Light is one of the most important environmental factors affecting plant growth and development, not only as an energy source for photosynthesis, but also as a regulatory signal that controls plant development. Plants have an elaborate photoreceptor system that perceives the environmental light conditions; this includes three major photoreceptors: phytochromes, cryptochromes, and phototropins (Quail, 1994; Sakai et al., 2001). Among these, phytochromes perceive red and far-red light and regulate diverse developmental processes including seed germination, hypocotyl elongation, chlorophyll biosynthesis, rhythmic growth, stomata opening, and flowering (Whitelam et al., 1998; Wang et al., 2010). Phytochromes are synthesized in cytoplasm in their inactive red light-absorbing (Pr) form. Upon irradiation with red light, phytochromes are converted from the Pr forms to the active far-red light-absorbing (Pfr) forms, and translocated into the nucleus (Whitelam et al., 1998). In the nucleus, phytochromes directly interact with various transcription factors including PIFs (Quail, 1994; Ni et al., 1998; Huq and Quail, 2002; Huq et al., 2004; Khanna et al., 2004; Oh et al., 2004; Leivar et al., 2008a) and induce global gene expression changes and various light-mediated developments (Huq and Quail, 2002).
The phytochrome-interacting factors (PIFs) are members of the subfamily 15 of the Arabidopsis bHLH transcription factors (Bae and Choi, 2008; Franklin and Quail, 2009). In the dark, four PIFs (PIF1/PIL5, PIF3, PIF4, and PIF5/PIL6) redundantly repress photomorphogenesis (Leivar et al., 2008b), but, under light, activated phytochromes interact with PIFs and induce their phosphorylation and degradation, and thus promote photomorphogenesis. PIFs regulate various light-mediated developmental processes. PIF1 negatively regulates seed germination (Oh et al., 2004); PIF1 and PIF3 inhibit chlorophyll biosynthesis in the etiolated seedlings to prevent photobleaching (Huq et al., 2004; Monte et al., 2004). PIF4 and PIF5 regulate shade-avoidance response and rhythmic hypocotyl growth (Nozue et al., 2007; Lorrain et al., 2008). Recent studies have shown that PIF4 plays a key role in gibberellin (GA) signal transduction and high-temperature-mediated hypocotyl elongation (Ogawa et al., 2003; de Lucas et al., 2008; Koini et al., 2009). Therefore, PIF4 seems to integrate external signals (light and temperature) and internal signal (GA) to regulate growth and development.
Since PIF4 is crucial for optimizing plant growth and development, PIF4 activity is controlled at multiple levels. Light-mediated degradation is a major mechanism regulating PIF4 activity (Nozue et al., 2007). In addition, PIF4 expression is affected by circadian rhythm and temperature (Nozue et al., 2007; Koini et al., 2009). PIF4 activity is also regulated through interaction with negative regulators such as DELLA and HFR1 (de Lucas et al., 2008; Hornitschek et al., 2009; Foreman et al., 2011). DELLA is a major negative regulator in the GA signaling pathway. HFR1, a bHLH transcription factor belonging to the same subfamily 15 as PIF4, is involved in the shade-avoidance, far-red light, and high-temperature responses. Both DELLA and HFR1 directly interact with PIF4 and prevent PIF4 from binding to DNA.
PAR1 and its closest homolog, PAR2, are primary phytochrome signaling target genes that are rapidly induced by shade (Roig-Villanova et al., 2006). Under shade, PAR1 and PAR2 negatively regulate shade-avoidance response to prevent an exaggerated shade response. PAR1 and PAR2 are atypical HLH proteins lacking proper DNA binding domain (Roig-Villanova et al., 2007) and, hence, are not expected to directly bind to DNA. Consistently with this hypothesis, a recent paper reported that only HLH and C-terminal domains are required for the PAR1 function. Furthermore, the transactivation domain fusion form of PAR1 repressed target gene expression, suggesting that PAR1 functions as a transcription cofactor regulating target gene expressions through interaction with canonical transcription factors that directly bind to DNA (Galstyan et al., 2011). However, no such transcription factors have been identified yet.
Here, we show that PAR1 directly interacts with PIF4 and inhibits PIF4 function. DNA pull-down assays indicated that PAR1 inhibits PIF4 binding to DNA. Consistently with in vitro data, PAR1 repressed PIF4-induced gene expression and PIF4-mediated developmental processes such as skotomorphogenesis and hypocotyl elongation responses to GA and high temperature. We also showed that PAR1 interacts with another atypical HLH protein, PRE1, which promotes cell elongation. Furthermore, PAR1 protein stability is increased by light. Our results suggest that PAR1–PIF4 and PAR1–PRE1 heterodimers form a complex HLH network regulating cell elongation and plant development.
RESULTS
PAR1 Is Involved in Photomorphogenesis
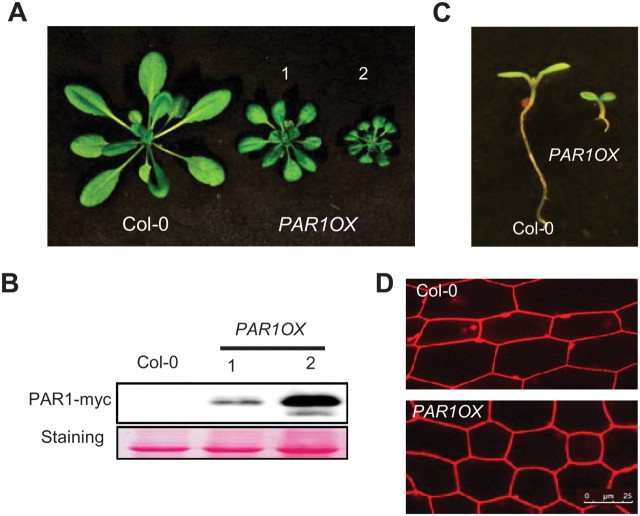
PAR1 and PAR2 act as negative regulators of shade-avoidance response. In addition, both PAR1- and PAR2-overexpressing transgenic plants showed severe dwarfism, indicating that they function as growth repressors under normal conditions (Roig-Villanova et al., 2007). To further investigate the biological functions of PAR1, we generated transgenic plants overexpressing PAR1 fused with the myc tag (PAR1OX). Consistently with previous observations (Roig-Villanova et al., 2007), PAR1OX plants displayed dwarfism with reduced petiole length and small leaves compared with wild-type (Figure 1A), and the severity of the phenotypes correlated with the PAR1-myc levels (Figure 1A and 1B), indicating that PAR1–myc is functional in planta. The PAR1OX seedlings showed small, dark-green cotyledons and short hypocotyl compared with wild-type (Figure 1C). Examination under a confocal microscope showed that the average hypocotyl cell length of PAR1OX was shorter than that of wild-type (Figure 1D), indicating that the short hypocotyl phenotype is at least partially due to reduced cell elongation.
Figure 1.
PAR1 Inhibits Cell Elongation.
(A) PAR1-overexpressing transgenic plants (PAR1OX) show dwarfism. Plants were grown for 4 weeks in long-day conditions.
(B) Immunoblot analysis of PAR1–myc levels in the PAR1OX plants shown in panel (A).
(C) PAR1OX seedlings exhibit short hypocotyls and dark-green cotyledons compared to wild-type (Col-0). Seedlings were grown for 5 d under white light.
(D) Hypocotyl cells of PAR1OX are shorter than those of wild-type (Col-0). Hypocotyls were stained with propidium iodide and examined by confocal microscopy. Bar = 25 μm.
When grown in the dark, PAR1OX plants showed short hypocotyl length and open apical hook compared to wild-type (Figure 2A and 2B). When grown under different intensities of white light (from 0 to 15 μmol m−2 s−1), PAR1OX showed hypersensitive response to light (Figure 2C). These results suggest that PAR1 promotes photomorphogenesis.
Figure 2.
PAR1 Is Involved in Photomorphogenesis.
(A) PAR1OX showed short hypocotyl and partially opened apical hook in the dark. Seedlings were grown in the dark for 5 d.
(B) Apical hook angle of Col-0, PAR1OX, and the pif1pif3pif4pif5 quadruple mutant (pifq). Seedlings were grown in the dark for 5 d.
(C) PAR1OX is more sensitive to light. Seedlings were grown in the dark or under various intensities of white light for 5 d.
(D) Light increased PAR1 protein stability. PAR1OX were grown in the dark for 5 d and then irradiated with white light for the indicated time. PAR1–myc protein was analyzed by immunoblotting. Ponceau S staining shows equal loading. The numbers below the bands indicate quantification of relative band intensity.
(E) Red, far-red, and blue light increased PAR1 protein stability. Conditions were the same as in (D) but irradiated with different wavelengths of light for 3 h. Ponceau S staining shows equal loading.
Many proteins involved in the light-signaling pathway are regulated post-translationally by light. Therefore, we examined whether PAR1 protein is also affected by light. Transgenic plants overexpressing PAR1–myc from the constitutive 35S promoter were grown in the dark and then treated with white light for various durations. As shown in Figure 2D, PAR1 protein levels were increased when etiolated seedlings were irradiated with white light for 1 h, but prolonged white light (up to 12-h) treatment did not increase PAR1 protein levels further. To test which wavelength of light regulates PAR1 protein stability, we irradiated etiolated seedlings for 3 h with different wavelengths of light and then quantified PAR1 protein levels. Red, far-red, and blue light treatments all increased PAR1 protein levels, indicating that PAR1 protein stability is regulated by multiple photoreceptors (Figure 2E).
PAR1 Interacts with PIF4
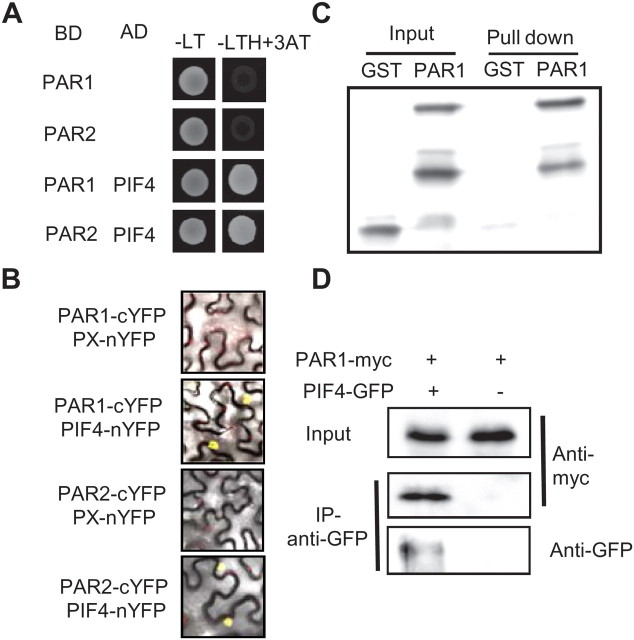
Although PAR1 belongs to the bHLH transcription factor family, the H/K9–E13–R17 DNA binding motif is missing in PAR1. It has been suggested that PAR1 acts as a transcription cofactor that modulates activities of other transcription factors (Galstyan et al., 2011). To identify PAR1-interacting transcription factors, we performed yeast two-hybrid assays of possible PAR1 interaction with various transcription factors involved in the light and shade-avoidance responses in which PAR1 has been shown to play a role. We found that PAR1 interacts with PIF4 (Figure 3A). A PAR1 homolog, PAR2, also interacted with PIF4 in yeast. To confirm this interaction in plant cells, we performed a bimolecular fluorescence complementation (BiFC) assay in tobacco (Nicotiana benthamiana) lower epidermal cells. Consistently with the yeast two-hybrid results, both PAR1 and PAR2 fused with N-terminal half of YFP interacted with PIF4 fused with the C-terminal half of YFP, generating YFP fluorescence in the nucleus (Figure 3B).
Figure 3.
PAR1 and PAR2 Interact with PIF4.
(A) PAR1 and PAR2 interact with PIF4 in yeast. Yeast colonies containing BD (GAL4 binding domain)–PAR1 or PAR2 and AD (GAL4 activation domain)–PIF4 were grown on the –LT or –LTH medium with 1 mM 3AT.
(B) Bimolecular fluorescence complementation (BiFC) assays show interaction between PAR1/PAR2 and PIF4. The indicated plasmid vectors were co-transformed into Nicotiana benthamiana leaves and images show overlay of fluorescence and light view.
(C) In vitro pull-down assays show interaction between PAR1 and PIF4. GST or GST–PAR1 was pulled down by MBP–PIF4 immobilized on maltose-agarose beads. Proteins bound to MBP–PIF4 were detected by immunoblotting using anti-GST antibody.
(D) Co-immunoprecipitation of PAR1–myc and PIF4–GFP using Arabidopsis mesophyll protoplast. 35S::PAR1–myc was transfected alone or co-transfected with 35S::PIF4–GFP into protoplasts and the proteins extracted from the protoplasts were immunoprecipitated by anti-GFP antibody and detected by immunoblotting using anti-myc or anti-GFP antibody.
The interaction between PAR1 and PIF4 was further analyzed by in vitro pull-down assays. As shown in Figure 3C, only GST–PAR1, but not GST itself, was specifically pulled down by MBP–PIF4. In addition, MBP–PIF4 was pulled down by GST–PAR1 much more effectively than MBP alone (Supplemental Figure 1), confirming the interaction between PAR1 and PIF4 in vitro. Next, we conducted co-immunoprecipitation assays by co-transfecting PIF4–GFP and PAR1–myc into Arabidopsis mesophyll protoplasts. The transfected protoplasts were incubated at room temperature for 6 h to express the proteins, and then co-immunoprecipitation was carried out with anti-GFP antibody, and subsequently detected by immunoblot with anti-myc or anti-GFP antibody. Figure 3D shows that PAR1 directly interacts with PIF4 in the mesophyll protoplasts.
PAR1 Inhibits PIF4-Mediated Transcription Activation
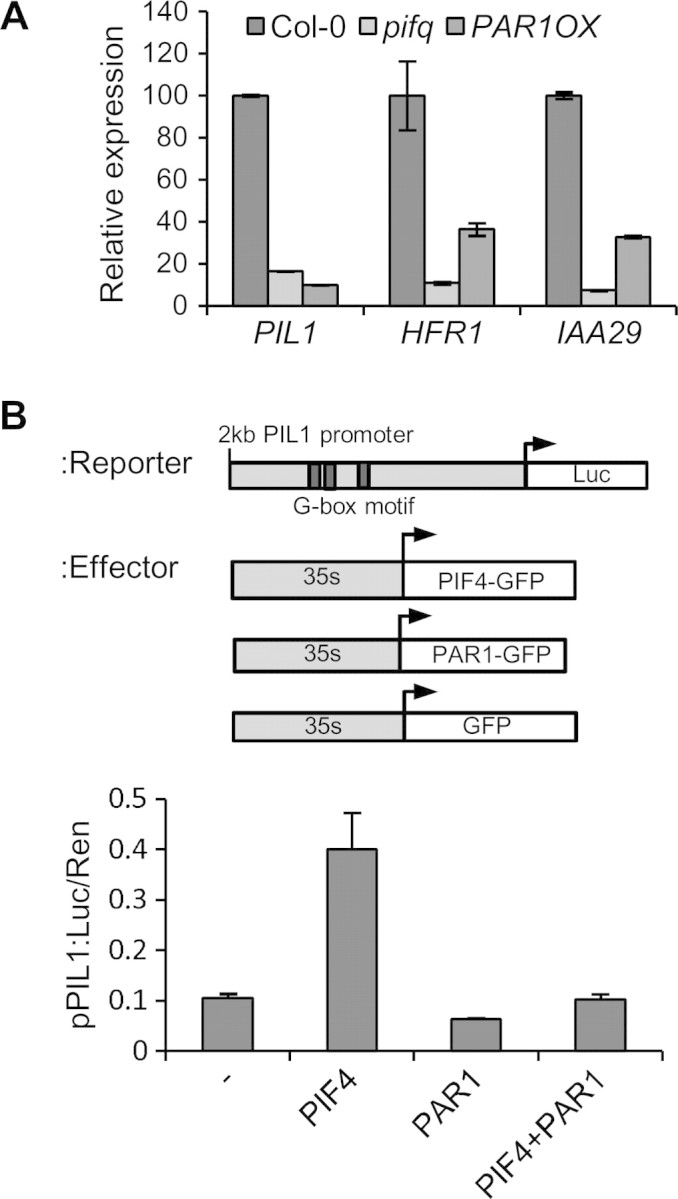
The interaction between PAR1 and PIF4 suggested that PAR1 may regulate PIF4 activity. To test this possibility, we analyzed whether expression levels of previously reported PIF4 direct target genes (PIL1, HFR1, and IAA29) are altered in the PAR1OX plants (Lorrain et al., 2008; Hornitschek et al., 2009; Koini et al., 2009). Consistently with previous studies, expression levels of PIL1, HFR1, and IAA29 were much lower in the pifq mutant than in wild-type (Figure 4A). Although less dramatic, expression levels of these genes were also lower in PAR1OX than Col-0, consistently with PAR1 suppressing PIF4 activity. To see whether PAR1 regulates PIL1 through PIF4, we performed a transient reporter gene assay using a PIL1p::GUS reporter gene containing 2 kb PIL1 promoter, which includes three putative PIF4-binding G-box motifs (Hornitschek et al., 2009) (Figure 4B). PIF4 activated the PIL1 promoter activity about fourfold compared with an empty vector as a negative control (Figure 4B). However, when PIF4 was co-transfected with PAR1, PIL1p::GUS expression was increased less than twofold, indicating that PAR1 repressed PIF4-mediated transcriptional activation of PIL1. Together with our gene expression analysis, this result indicates that PAR1 interacts with PIF4 and inhibits PIF4-mediated transcriptional activation.
Figure 4.
PAR1 Inhibits PIF4-Mediated Transcriptional Activation.
(A) Quantitative RT–PCR analysis of the expression levels of PIL1, HFR1, and IAA29 in the Col-0, pifq, and PAR1OX plants. Seedlings were grown under red light for 5 d. Data were normalized to PP2A gene as internal control. Error bars indicate S.E. of three biological repeats.
(B) PAR1 inhibits PIF4-mediated PIL1 promoter activation. The reporter construct containing a 2-kb PIL1 promoter placed upstream of the luciferase reporter (PIL1p::luc) was co-transformed into protoplasts with the indicated effector vectors. Luc activity was normalized by renilla luciferase activity (internal control). Error bars indicate S.E. of three biological repeats.
PAR1 Blocks PIF4 Binding to DNA
Next, we investigated the molecular mechanism by which PAR1 inhibits PIF4. Since PAR1 has no typical DNA binding domain, it is possible that PAR1 forms a non-DNA binding heterodimer with PIF4, thus preventing PIF4 from binding to DNA. To test this possibility, we performed DNA pull-down assays using biotin-labeled PIL1 promoter containing G-box motifs. Figure 5A shows that PIF4 is pulled down by biotin-labeled PIL1 promoter indicating that PIF4 binds to the PIL1 promoter in vitro as previously reported. PIF4 binding to the PIL1 promoter was reduced when PIF4 was pre-incubated with similar amount of GST–PAR1 and abolished when pre-incubated with fivefold more GST–PAR1, whereas incubation with GST had no effect on PIF4–DNA binding (Figure 5A). These results demonstrate that PAR1 represses PIF4 binding to DNA.
Figure 5.
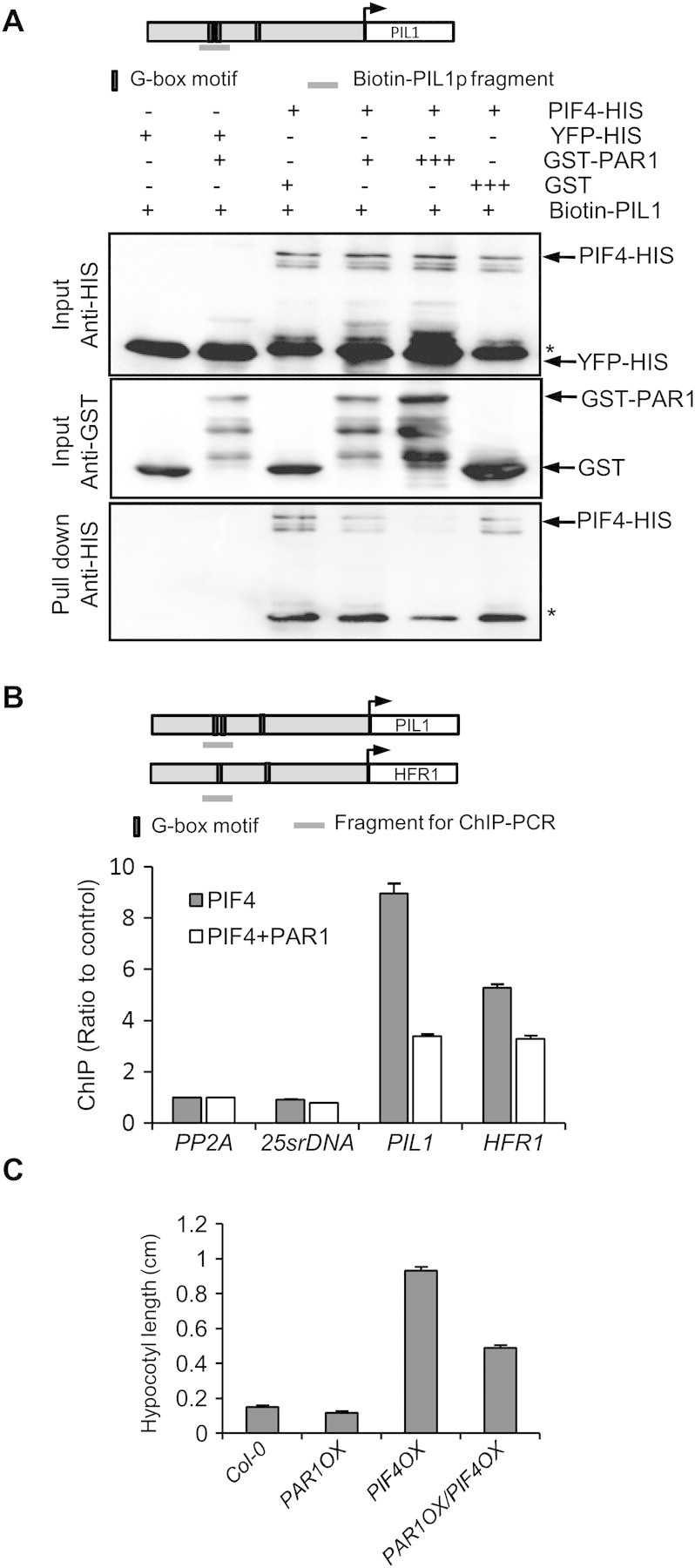
PAR1 Prevents PIF4 from Binding to Its Target DNA and Inhibits PIF4-Promoted Hypocotyl Elongation.
(A) DNA binding assays show that PAR1 inhibits PIF4 DNA binding. The indicated proteins were expressed and purified from E. coli and incubated and pulled down with biotin-labeled PIL1 promoter fragment. The input and pulled-down proteins were analyzed by immunoblotting. Asterisks indicate non-specific band in PIF4–HIS.
(B) ChIP assays show that PAR1 inhibits PIF4 binding to PIL1 and HFR1 promoters. Protoplasts transfected with PIF4–GFP with or without PAR1–myc were analyzed by ChIP using anti-GFP antibody. The PIL1 and HFR1 promoter fragments were analyzed by qPCR and normalized to a fragment of PP2A coding region. Error bars indicate S.D. of two technical repeats. Three independent experiments showed similar patterns.
(C) Seedlings overexpressing PAR1, PIF4, or both were grown under light for 5 d. Bars indicate S.E. of at least 20 seedlings.
To confirm the PAR1 function in inhibition of PIF4 DNA binding in vivo, we performed chromatin immunoprecipitation (ChIP) using Arabidopsis mesophyll protoplasts. Consistently with previous studies showing that PIL1 and HFR1 are direct target genes of PIF4 (Lorrain et al., 2008; Hornitschek et al., 2009), both PIL1 and HFR1 promoter fragments were enriched by immunoprecipitation of PIF4, compared with PP2A (coding region) and 25s rDNA as negative controls (Figure 5B). However, when PAR1 was co-transfected with PIF4, the enrichment of PIL1 and HFR1 promoters was significantly reduced compared with PIF4 alone, supporting that PAR1 inhibits PIF4 binding to target genes in plant cells.
PAR1 Inhibits PIF4-Mediated Hypocotyl Elongation
To test whether PAR1 inhibits PIF4 function in planta, we generated PAR1OX/PIF4OX double transgenic plants and then compared hypocotyl length with each single transgenic plant. While PIF4OX dramatically promoted hypocotyl elongation, PAR1OX partially suppressed the long-hypocotyl phenotype of PIF4OX, consistent with PIF4 being less active in the PAR1OX background (Figure 5C).
PAR1 Is Involved in the High-Temperature and Gibberellins Responses
In addition to hypocotyl inhibition by light, PIF4 also mediates high-temperature-promoted hypocotyl elongation (Koini et al., 2009). Elevated temperature (26–29°C) promotes hypocotyl elongation, partly due to enhanced auxin signaling (Gray et al., 1998). The high-temperature-promoted hypocotyl elongation was not observed in the pif4 mutant, indicating that PIF4 plays a major role in this process (Koini et al., 2009). To see whether PAR1 is also involved in high-temperature-promoted hypocotyl elongation, we measured hypocotyl length of PAR1OX grown at either 22 or 29°C. Hypocotyl lengths of Col-0 were dramatically increased (about four times) by high temperature, but hypocotyl lengths of pifq mutant, which lacks four PIFs including PIF4, were much less increased by high temperature, as previously reported (Koini et al., 2009) (Figure 6A). The hypocotyls of PAR1OX, like those of the pifq mutant, were also less sensitive to high temperature, consistently with PAR1 repressing PIF4-mediated high-temperature-induced hypocotyl elongation.
Figure 6.
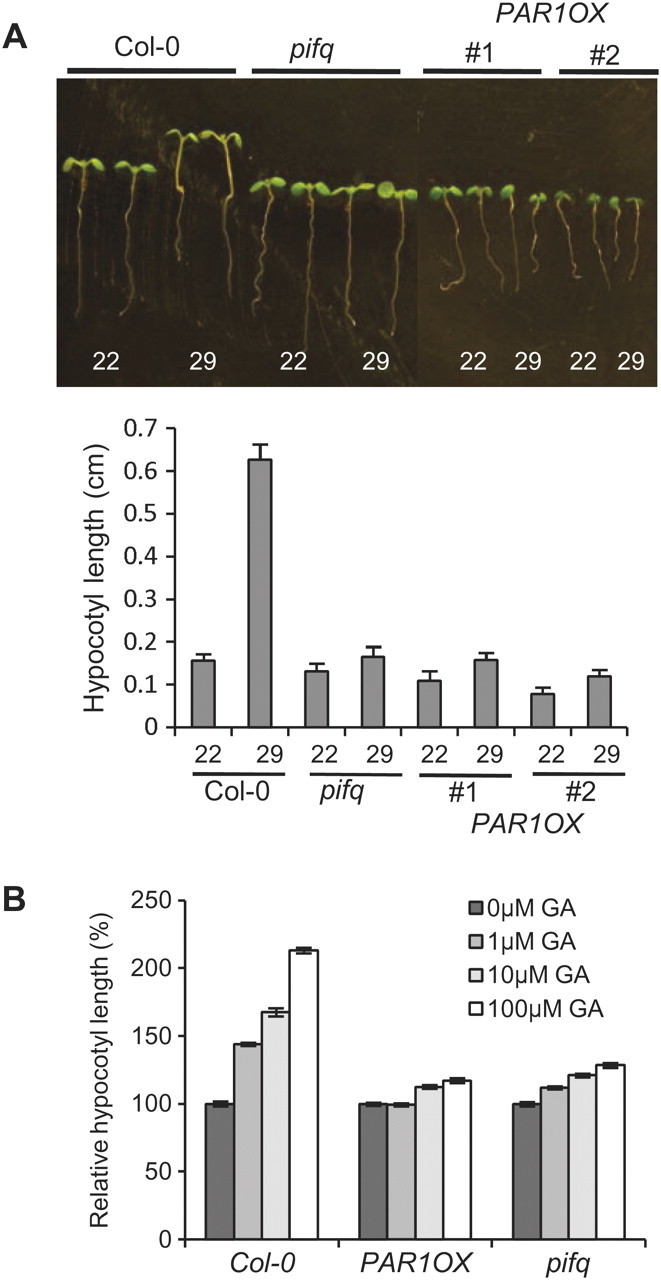
PAR1 Is Involved in the High-Temperature and GA Responses.
(A) PAR1OX is less sensitive to high temperature, like pifq mutant. Seedlings were grown at 22°C for 7 d (22) or at 22°C for 4 d and then at 29°C for 3 d. Error bars indicate S.E. of 20 seedlings.
(B) PAR1OX is less sensitive to GA. Seedlings were grown on the medium containing various concentrations of GA. Hypocotyl lengths were normalized by hypocotyl length of seedlings grown on medium without GA. Error bars indicate S.E. of 20 seedlings.
According to recent studies, PIF4 is also involved in the GA signaling pathway by directly interacting with DELLA proteins, which are negative regulators of GA signaling (Feng et al., 2008). Thus, we tested whether PAR1 also regulates GA responses. As shown in Figure 6B, GA treatment significantly promoted hypocotyl elongation in Col-0, whereas the hypocotyls of pifq were much less sensitive to GA, consistently with PIFs playing a key role in GA responses. Similarly, PAR1OX showed reduced responsiveness to GA with regard to hypocotyl elongation (Figure 6B). Taken together, PAR1OX showed defects in both high-temperature and GA responses, which is consistent with our observation that PAR1 represses PIF4 activity by preventing PIF4 from binding to DNA, and with previous reports that PIF4 plays key roles in these responses.
PAR1 Interacts with PRE1
In addition to PIF4, we also found that PAR1 interacts with PRE1 in yeast two-hybrid assays (Figure 7A). Like PAR1, PRE1 is a small HLH transcription factor and does not have a conserved H/K9–E13–R17 motif for DNA binding. Thus, it has been thought that PRE1 regulates gene expression through other transcription factors capable of binding to DNA. To confirm the interaction between PAR1 and PRE1, we performed the BiFC assays. When PAR1–cYFP was co-transformed with PRE1–nYFP into tobacco leaves, a strong YFP signal was observed in the nucleus, indicating that PAR1 interacts with PRE1 in planta (Figure 7B). Previously, it was reported that PRE1 and its close homolog, ATBS1, interact with both IBH1 and AIF1 and compromise their function in inhibition of cell elongation. Because PAR1 also inhibits cell elongation (Figure 1D), it is highly possible that PRE1 inhibits PAR1 activity in a manner similar to the mechanism by which PRE1 (or ATBS1) inhibits IBH1 (or AIF1) activity. To test this possibility, we crossed the PAR1OX showing dwarf phenotype with PRE1OX and examined the phenotype of PAR1OX/PRE1OX double transgenic plants. PAR1OX/PRE1OX plants showed long petioles and expanded leave blades, similarly to PRE1OX plants (Figure 7C). The PAR1OX/PRE1OX double transgenic plants accumulated a slightly reduced level of PRE1 and increased level of PAR1 protein compared to each parental line (Figure 7D). The reason for such changes of protein level is unclear, but the changes are opposite to that expected from the phenotype, and thus indicate that the long-petiole phenotype of PAR1OX/PRE1OX plants was caused by altered activities rather than accumulation levels of PAR1 or PRE1 proteins. These results support our hypothesis that PRE1 directly suppresses PAR1, leading to de-repression of PIF4 and activation of cell elongation.
Figure 7.
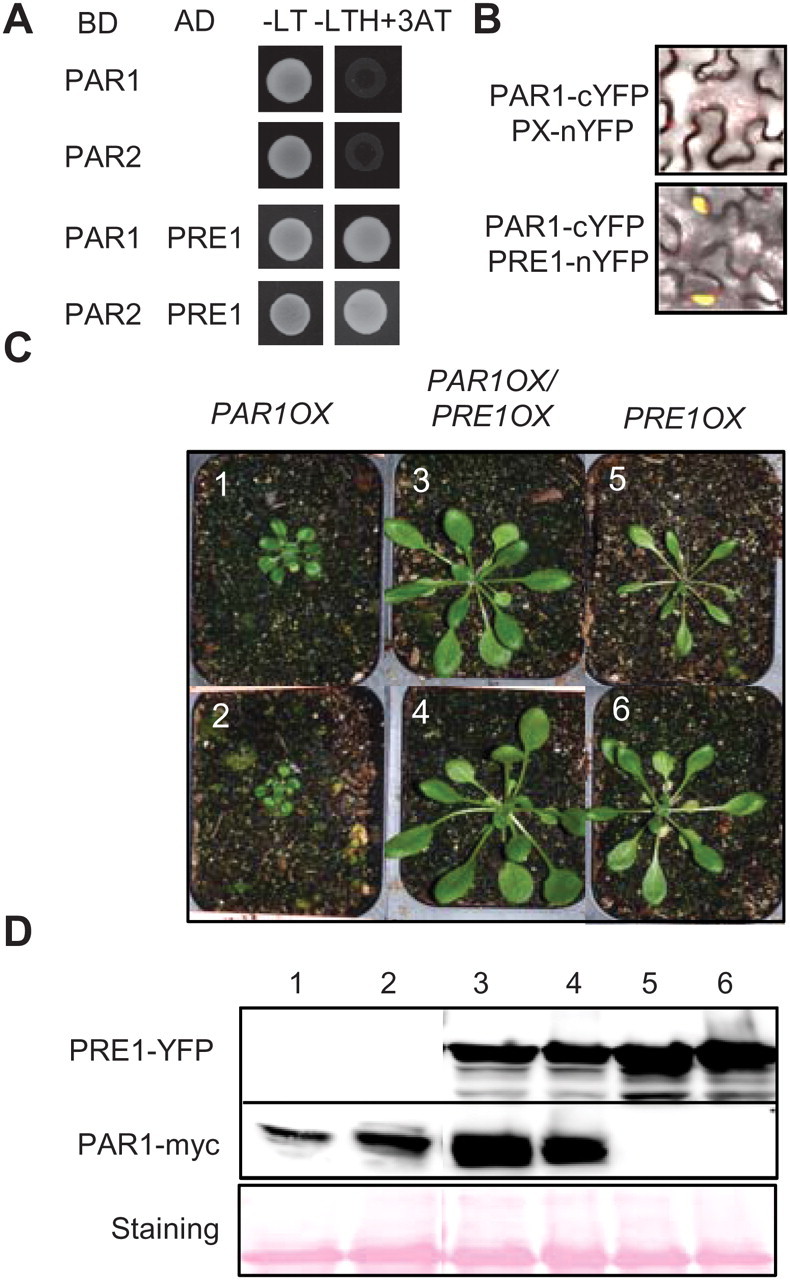
PRE1 Interacts with PAR1 and Suppresses Dwarfism of PAR1OX.
(A) PAR1 interacts with PRE1 in yeast two-hybrid assays.
(B) Bimolecular fluorescence complementation (BiFC) assays in Nicotiana benthamiana leaves show interaction between PAR1 and PRE1.
(C) PRE1OX suppresses dwarfism of PAR1OX. Plants were grown for 4 weeks in long-day conditions.
(D) PRE1–YFP and PAR1–myc levels of plants shown in panel (C), analyzed by immunoblotting. Ponceau S staining shows equal loading.
DISCUSSION
PAR1 has been identified as a negative regulator of shade-avoidance response (Roig-Villanova et al., 2007); however, the molecular mechanism by which PAR1 inhibits cell elongation has remained unclear. In the present work, we show that PAR1 directly interacts with a positive regulator of shade-avoidance response, PIF4, and inhibits PIF4-mediated transcriptional activation by preventing PIF4 from binding to its target genes, including PIL1 and HFR1. In addition to shade-avoidance responses, PIF4 is also involved in the light regulation of seedling development, and the responses to high temperature and GA. We show that all of these processes are oppositely regulated by PIF4 and PAR1, which is consistent with PAR1 inactivation of PIF4 at the molecular level. Therefore, our data suggest that PAR1 regulates diverse developmental responses by inhibiting PIF4 activity (Figure 8).
Figure 8.
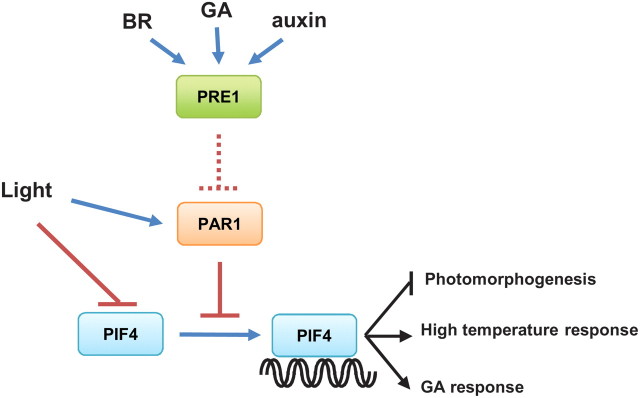
A Model of PAR1 and PIF4 Functions in Light and Hormone Responses.
In the absence of PAR1, PIF4 binds to DNA to inhibit photomorphogenesis and promote high-temperature and GA responses. Light induces degradation of PIF4 and stabilizes PAR1. Accumulated PAR1 inhibits PIF4 activity by preventing PIF4 from binding to DNA. PRE1, which is induced by BR, GA, and auxin, interacts with PAR1, and possibly prevents its interaction with PIF4.
Previous studies using overexpression and RNAi suppression have shown that PAR1 and its close homolog PAR2 negatively regulate cell elongation, particularly the elongation induced by shade (Roig-Villanova et al., 2007). Suppression of both PAR1 and PAR2 by RNAi increased cell elongation, and the par2-1 knockout mutant also showed slight but significant increase in cell elongation. It is believed that PAR1 and PAR2 play redundant or overlapping roles. We showed that PAR2 also interacts with PIF4. However, the par2-1 (Salk_109270) single mutant did not show significantly altered responsiveness to light, high temperature, or GA (Supplemental Figure 2), suggesting that lack of PAR2 activity is compensated by other redundant factors like PAR1 under our experimental conditions. Future analysis of par1/par2 double mutant will be required to determine whether PAR1 and PAR2 are functionally redundant and to evaluate their roles in light-regulated responses.
Our results strongly suggest that PAR1 promotes photomorphogenesis by inhibiting PIF4, which is a key negative regulator of photomorphogenesis. First, PAR1 directly interacts with PIF4 (Figure 3) and blocks PIF4 binding to DNA. Second, PAR1OX reduces PIF4 binding to target promoters in vivo (Figure 5) and has similar effects to pifq on the expression of PIF4 target genes (PIL1, HFR1, and IAA29). Third, PIF4 transcriptional activation of PIL1 is compromised by PAR1 in transient assays (Figure 4). Finally, PAR1OX reduces the hypocotyl length of PIF4OX (Figure 5C). Our results indicate that PAR1 forms a non-DNA-binding heterodimer with PIF4 to inhibit PIF4 function.
The RNA levels of PAR1 and PAR2 are rapidly regulated by changing light conditions, and thus PAR1 and PAR2 were previously considered as downstream of light signaling. Our finding that PAR1 protein stability is regulated by light supports that PAR1 mediates light signaling. Light induces PIF4 degradation, but stabilizes PAR1 protein (Figure 2D). Therefore, light inhibits PIF4 activity through two distinct mechanisms to promote photomorphogenesis: light induces phytochrome-mediated PIF4 protein degradation by 26s proteasome and light also increased accumulation of PAR1, which inhibits PIF4 DNA binding. Red, far-red, and blue light all increased PAR1 protein level. Therefore, PAR1 and PIF4 appear to be involved in the general light signaling pathways, rather than a specific photoreceptor pathway. Consistently with this notion, previous studies showed that PIF4 is involved in the red, far-red, and blue light signaling (Huq and Quail, 2002; Kang et al., 2005; Kunihiro et al., 2010).
Light stabilizes many proteins through inhibiting the ring finger-type E3 ubiquitin ligase, COP1. In the dark, COP1 is located in the nucleus and interacts with various positive regulators of photomorphogenesis such as HY5 and HFR1 (Osterlund et al., 2000; Jang et al., 2005), inducing their degradation by the 26s proteasome. In contrast, light suppresses COP1 activity partially by promoting its translocation into cytoplasm (Vonarnim and Deng, 1994), resulting in accumulation of HY5 and HFR1 in the nucleus. Because light treatments also stabilize PAR1, it is possible that PAR1 protein stability is also regulated by COP1. This will need to be tested by future study.
In addition to PIF4, PAR1 also interacted with the small HLH protein PRE1, and PRE1OX completely suppressed the growth defect of PAR1OX (Figure 7). PRE1 was first identified as a GA signaling component because PRE1OX rescued the dwarfism of a GA-deficient mutant and reduced plants’ sensitivity to paclobutrazol, a GA biosynthesis inhibitor (Lee et al., 2006). Recent studies showed that PRE1 and its homolog, ATBS1, mediate BR-induced cell elongation by inhibiting bHLH proteins IBH1 and AIF1, respectively (Wang et al., 2009; Zhang et al., 2009). PRE1 is a small HLH protein and does not have a DNA binding domain, like PAR1. It was previously suggested that PRE1 inactivates IBH1, which inhibits cell elongation. Our results indicate that PRE1 also inhibits PAR1 activity.
Given that PAR1 binds to PIF4 to prevent PIF4 binding to DNA, PRE1 interaction with PAR1 may prevent PAR1 from interacting with PIF4. In addition, PRE1 was reported to interact with HFR1, which, like PAR1, also interacts with PIF4 and inhibits its DNA binding (Hornitschek et al., 2009). Thus, PRE1 may sequester both PAR1 and HFR1, and hence allow PIF4 to activate target genes. PRE1 expression is increased by GA and BR. The stabilities of PAR1, HFR1, and PIF4 proteins are regulated by light. Thus, hormones and light may control the balance between two heterodimers, PRE1–PAR1 (or HFR1) and PAR1–PIF4, to optimize growth and development. The hormones increase PRE1 to inhibit PAR1 and thus de-repress PIF4, whereas light reduces PIF4 accumulation and also stabilizes PAR1, which further inactivates PIF4. Therefore, PAR1 interaction with PIF4 and PRE1 provides another important link for integrating the light and hormone pathways (Figure 8). This mechanism should contribute to the control of overlapping transcriptomes by BR and light (Song et al., 2009; Luo et al., 2010). Future study of these molecular interactions will advance our understanding of how growth and development are co-regulated by environmental and endogenous signals.
METHODS
Plant Materials and Growth Conditions
Seeds were surface-sterilized for 15 min with 75% ethanol and plated on ½ Murashige and Skoog basal salt medium (PhytoTechnology Laboratories) supplemented with 0.7% phytoagar. After 3 d of incubation at 4°C to promote germination, seedlings were grown under either white light, red, blue, or far-red light for further analysis.
For hypocotyl measurement, seedlings were grown for 5 d on medium, imaged with a scanner, and the hypocotyl lengths were measured using the ImageJ software (http://rsb.info.nih.gov/ij).
Plasmid Construction and Plant Transformation
To construct the PAR1 overexpression construct, full-length PAR1 coding sequence was amplified by PCR and cloned into the gateway-compatible pGWB17 vector (Nakagawa et al., 2007). For the constructs expressing PAR1–GFP and PIF4–GFP fusion proteins for protoplast transient expression assay, the coding sequence of PAR1 or PIF4 was cloned into the pBI222–GFP vector. Transgenic plants were generated using the Agrobacterium tumefaciens vacuum infiltration method.
Western Blot Analysis
Plant tissues were harvested and ground in liquid nitrogen. Proteins were extracted with 2X SDS buffer (100 mM Tris-HCl, pH 6.8, 25% glycerol, 2% SDS, 0.01% Bromphenol blue, add β-Mercaptoethanol to 10% before use). Western blot analysis was performed to check PAR1–myc or PRE1–GFP protein levels using anti-myc or anti-GFP antibodies, respectively.
Yeast Two-Hybrid Assay
The full-length coding sequences were cloned into gateway-compatible pBD–GAL4 and pAD–GAL4 vectors (Stratagene) and then transformed into yeast strain AH109, as previously reported (Fields and Song, 1989).
Protoplast Extraction and Analysis of Gene Expression
Protoplast isolation and PEG transformation were performed according to the methods described by Yoo et al. (2007). Plasmid DNAs used in dual-luciferase assay were prepared using the Plasmid Maxi Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. A dual-luciferase reporter assay system was performed as previously described (Hellens et al., 2005; Liu et al., 2008). Three biological repeats were measured for each sample. The reporter vector, pGreen–0800II–luc, contains a 35s promoter upstream of the renilla gene, which is served as internal normalization standard, and a reporter promoter that drives firefly luciferase gene.
Co-Immunoprecipitation Assay
For Co-IP assay, 1 × 106 mesophyll protoplasts were transfected with 20 μg of DNA and incubated overnight. The protoplast cells were harvested by centrifugation and lysed in 200 μl of IP lysis buffer (100 mM NaCl, 500 mM Tris-HCl, 5 mM EDTA, 1% Triton X-100, 1 mM DTT, 5% glycerol). The lysates were incubated with anti-GFP antibody and protein agarose beads for 2 h, thoroughly washed twice with IP lysis buffer, and the eluted samples were analyzed by SDS–PAGE.
RNA Analysis and Quantitative PCR
Total RNA was extracted from 5-day-old Arabidopsis seedlings using a Plant Total RNA extraction kit (Sigma, USA). M-MLV reverse transcriptase (Fermatas, USA) was used to synthesize first-strand cDNA from RNA. Real-time PCR was performed using the Bioline SYBR master mix (Bioline, USA). The conditions for PCR amplification were as follows: 98°C for 10 min; 45 cycles of 98°C for 30 s; 65°C for 45 s and 72°C for 30 s; one cycle 72°C for 10 min. PP2A was used to as an internal reference gene. Gene-specific primers are listed in Supplemental Table 1.
Bimolecular Fluorescence Complementation Assays
Tobacco transformation followed the protocol by Sparkes et al. (2006). Full-length coding sequences of PAR1/2, PIF4, and PRE1 were cloned into the pX–nYFP or pX–cYFP gateway-compatible vectors (Gampala et al., 2007). For expression in the Nicotiana benthamiana, the bacterial suspension was injected into leaves from lower epidermis. Tobacco plants were kept in the greenhouse for at least 36 hours at 22°C to allow the expression of the transfected DNA. All fluorescence observations were made with a Leica microscope.
In Vitro Protein Pull-Down Assay
For in vitro pull-down assays, recombinant GST–PAR1 and MBP–PIF4 were expressed in Escherichia coli and purified using glutathione beads or maltose-agarose beads, respectively. Reaction components were incubated in 1× TEN buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM EDTA) at 4°C for 1 h. The beads were then washed three times with NETN buffer (200 mM NaCl, 1 mM EDTA, 0.5% NonidetP-40, 20 mM Tris-HCl, pH 8.0) and eluted samples were analyzed by SDS–PAGE.
In Vitro DNA:Protein Pull-Down Assay
To produce PIF4-his protein, full-length PIF4 coding sequence was amplified and cloned into pET28a vector. The recombinant PIF4-his protein was purified using NI-NTA resin according to the manufacturer’s instructions (Qiagen, USA). Biotinylated PIL1 promoter DNA was produced by PCR using biotinylated oligos, bound to the streptavidin-coated agarose beads (Sigma, USA) for at least 1 h in advance at 4°C. The DNA-beads were incubated with purified protein for 1 h and then collected by centrifugation and washed three times with DNA binding buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 5 mM EDTA, 0.05% Nonidet P-40). Eluted samples were analyzed by SDS–PAGE.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assay was performed as described previously (Oh et al., 2009) and modified for Arabidopsis protoplasts. After co-transfection and incubation at room temperature overnight, protoplasts were fixed by formaldehyde (final concentration: 1%) for 20 min. The chromatin complex was fragmented by sonication and incubated with anti-GFP antibody-Protein A beads overnight. After washing, beads were treated at 65°C for 6 h to reverse cross-linking and the co-precipitated DNA was purified using a DNA purification kit (Fermantas, USA). The DNA was analyzed by quantitative real-time PCR using the Bioline SYBR master mix (Bioline, USA). Primers for the ChIP–PCR are listed in Supplemental Table 1.
SUPPLEMENTARY DATA
Supplementary Data are available at Molecular Plant Online.
FUNDING
This study was supported by grants from the NIH (R01GM066258) to Z.Y.W. Y.H. was supported by the China Scholarship Council. No conflict of interest declared.
Supplementary Material
References
- Bae G, Choi G. Decoding of light signals by plant phytochromes and their interacting proteins. Annu. Rev. Plant Biol. 2008;59:281–311. doi: 10.1146/annurev.arplant.59.032607.092859. [DOI] [PubMed] [Google Scholar]
- de Lucas M, Davière J-M, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008;451:475–479. doi: 10.1038/nature06448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S, Song OK. A novel genetic system to detect protein–protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Foreman J, Johansson H, Hornitschek P, Josse EM, Fankhauser C, Halliday KJ. Light receptor action is critical for maintaining plant biomass at warm ambient temperatures. Plant J. 2011;65:441–452. doi: 10.1111/j.1365-313X.2010.04434.x. [DOI] [PubMed] [Google Scholar]
- Franklin KA, Quail PH. Phytochrome functions in Arabidopsis development. J. Exp. Bot. 2009;61:11–24. doi: 10.1093/jxb/erp304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galstyan A, Cifuentes-Esquivel N, Bou-Torrent J, Martinez-Garcia JF. The shade avoidance syndrome in Arabidopsis: a fundamental role for atypical basic helix-loop-helix proteins as transcriptional cofactors. Plant J. 2011;66:258–267. doi: 10.1111/j.1365-313X.2011.04485.x. [DOI] [PubMed] [Google Scholar]
- Gampala SS, et al. An essential role for 14–3–3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell. 2007;13:177–189. doi: 10.1016/j.devcel.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Ostin A, Sandberg G, Romano CP, Estelle M. High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc. Natl Acad. Sci. U S A. 1998;95:7197–7202. doi: 10.1073/pnas.95.12.7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Meth. 2005;1:13. doi: 10.1186/1746-4811-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P, Lorrain S, Zoete V, Michielin O, Fankhauser C. Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 2009;28:3893–3902. doi: 10.1038/emboj.2009.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E, Al-Sady B, Hudson M, Kim CH, Apel M, Quail PH. PHYTOCHROME-INTERACTING FACTOR 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science. 2004;305:1937–1941. doi: 10.1126/science.1099728. [DOI] [PubMed] [Google Scholar]
- Huq E, Quail PH. PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 2002;21:2441–2450. doi: 10.1093/emboj/21.10.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang IC, Yang Y, Seo HS, Chua N-H. HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Gene Dev. 2005;19:593–602. doi: 10.1101/gad.1247205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X, Chong J, Ni M. HYPERSENSITIVE TO RED AND BLUE 1, a ZZ-type zinc finger protein, regulates phytochrome B-mediated red and cryptochrome-mediated blue light responses. Plant Cell. 2005;17:822–835. doi: 10.1105/tpc.104.029165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R, Huq E, Kikis EA, Al-Sady B, Lanzatella C, Quail PH. A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell. 2004;16:3033–3044. doi: 10.1105/tpc.104.025643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koini MA, Alvey L, Allen T, Tilley CA, Harberd NP, Whitelam GC, Franklin KA. High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr. Biol. 2009;19:408–413. doi: 10.1016/j.cub.2009.01.046. [DOI] [PubMed] [Google Scholar]
- Kunihiro A, Yamashino T, Mizuno T. PHYTOCHROME-INTERACTING FACTORS PIF4 and PIF5 are implicated in the regulation of hypocotyl elongation in response to blue light in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2010;74:2538–2541. doi: 10.1271/bbb.100586. [DOI] [PubMed] [Google Scholar]
- Lee S, Lee S, Yang K-Y, Kim Y-M, Park S-Y, Kim SY, Soh M-S. Overexpression of PRE1 and its homologous genes activates gibberellin-dependent responses in Arabidopsis thaliana. Plant Cell Physiol. 2006;47:591–600. doi: 10.1093/pcp/pcj026. [DOI] [PubMed] [Google Scholar]
- Leivar P, Monte E, Al-Sady B, Carle C, Storer A, Alonso JM, Ecker JR, Quail PH. The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell. 2008a;20:337–352. doi: 10.1105/tpc.107.052142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH. Multiple phytochrome-interacting bHLH transcription factors repress premature photomorphogenesis during early seedling development in darkness. Curr. Biol. 2008b;18:1815–1823. doi: 10.1016/j.cub.2008.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, Lin C. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science. 2008;322:1535–1539. doi: 10.1126/science.1163927. [DOI] [PubMed] [Google Scholar]
- Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 2008;53:312–323. doi: 10.1111/j.1365-313X.2007.03341.x. [DOI] [PubMed] [Google Scholar]
- doi: 10.1016/j.devcel.2010.10.023. Luo, X-M., Lin, W-H., Zhu, S-W., Zhu, J-Y., Sun, Y., Fan, X-Y., Cheng, M., Hao, Y., Oh, E., Tian, M., Liu, L., Zhang, M., Xie, Q., Chong, K., and Wang, Z-Y. (2010). Integration of light- and brassinosteroid-signaling pathways by a GATA transcription factor in Arabidopsis. Dev. Cell. 19, 872–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte E, Tepperman JM, Al-Sady B, Kaczorowski KA, Alonso JM, Ecker JR, Li X, Zhang YL, Quail PH. The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc. Natl Acad. Sci. U S A. 2004;101:16091–16098. doi: 10.1073/pnas.0407107101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 2007;104:34–41. doi: 10.1263/jbb.104.34. [DOI] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH. PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell. 1998;95:657–667. doi: 10.1016/s0092-8674(00)81636-0. [DOI] [PubMed] [Google Scholar]
- Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, Maloof JN. Rhythmic growth explained by coincidence between internal and external cues. Nature. 2007;448:358–361. doi: 10.1038/nature05946. [DOI] [PubMed] [Google Scholar]
- Ogawa M, Hanada A, Yamauchi Y, Kuwahara A, Kamiya Y, Yamaguchi S. Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell. 2003;15:1591–1604. doi: 10.1105/tpc.011650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Kang H, Yamaguchi S, Park J, Lee D, Kamiya Y, Choi G. Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell. 2009;21:403–419. doi: 10.1105/tpc.108.064691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Kim J, Park E, Kim JI, Kang C, Choi G. PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell. 2004;16:3045–3058. doi: 10.1105/tpc.104.025163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000;405:462–466. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- Quail PH. Photosensory perception and signal transduction in plants. Curr. Opin. Genet. Dev. 1994;4:652–661. doi: 10.1016/0959-437x(94)90131-l. [DOI] [PubMed] [Google Scholar]
- Roig-Villanova I, Bou J, Sorin C, Devlin P, Martínez-García J. Identification of primary target genes of phytochrome signaling: early transcriptional control during shade avoidance responses in Arabidopsis. Plant Physiol. 2006;141:85–96. doi: 10.1104/pp.105.076331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig-Villanova I, Bou-Torrent J, Galstyan A, Carretero-Paulet L, Portoles S, Rodriguez-Conception M, Martinez-Garcia JF. Interaction of shade avoidance and auxin responses: a role for two novel atypical bHLH proteins. EMBO J. 2007;26:4756–4767. doi: 10.1038/sj.emboj.7601890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Kagawa T, Kasahara M, Swartz TE, Christie JM, Briggs WR, Wada M, Okada K. Arabidopsis nph1 and npl1: blue light receptors that mediate both phototropism and chloroplast relocation. Proc. Natl Acad. Sci. U S A. 2001;98:6969–6974. doi: 10.1073/pnas.101137598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- doi: 10.1093/mp/ssp039. Song, L., Zhou, X.Y., Li, L., Xue, L.J., Yang, X., and Xue, H.W. (2009). Genomewide analysis revealed the complex regulatory network of brassinosteroid effects in photomorphogenesis. Mol. Plant. 2, 755–772. [DOI] [PubMed] [Google Scholar]
- Sparkes IA, Runions J, Kearns A, Hawes C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protocol. 2006;1:2019–2025. doi: 10.1038/nprot.2006.286. [DOI] [PubMed] [Google Scholar]
- Vonarnim AG, Deng XW. Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell. 1994;79:1035–1045. doi: 10.1016/0092-8674(94)90034-5. [DOI] [PubMed] [Google Scholar]
- doi: 10.1093/mp/ssp097. Wang, F.F., Lian, H.L., Kang, C.Y., and Yang, H.Q. (2010). Phytochrome B is involved in mediating red light-induced stomatal opening in Arabidopsis thaliana. Mol. Plant. 3, 246–259. [DOI] [PubMed] [Google Scholar]
- Wang H, Zhu Y, Fujioka S, Asami T, Li J. Regulation of Arabidopsis brassinosteroid signaling by atypical basic helix-loop-helix proteins. Plant Cell. 2009;21:3781–3791. doi: 10.1105/tpc.109.072504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam GC, Patel S, Devlin PF. Phytochromes and photomorphogenesis in Arabidopsis. Phil. Trans. Roy. Soc. Lond. B. 1998;353:1445–1453. doi: 10.1098/rstb.1998.0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S-D, Cho Y-H, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protocol. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- Zhang LY, et al. Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell. 2009;21:3767–3780. doi: 10.1105/tpc.109.070441. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.