Background: Allene oxide synthases (AOS) of the CYP74 family are present in plants, but AOS of fungi have not been characterized.
Results: Expression of dioxygenase-cytochrome P450 fusion proteins of Aspergillus terreus reveals a novel AOS.
Conclusion: AOS of A. terreus forms compound II with catalytic similarities to CYP74 and CYP8A1.
Significance: The fungal AOS protein sequence is unique with little homology to CYP74.
Keywords: Cytochrome P450; Fatty Acid Oxidation; Mutagenesis, Site-specific; Oxygen Radicals; Peroxidase; Class III Cytochrome P450; Linoleate Diol Synthase; Oxylipin
Abstract
Aspergilli oxidize C18 unsaturated fatty acids by dioxygenase-cytochrome P450 fusion proteins to signal molecules involved in reproduction and host-pathogen interactions. Aspergillus terreus expresses linoleate 9R-dioxygenase (9R-DOX) and allene oxide synthase (AOS) activities in membrane fractions. The genome contains five genes (ATEG), which may code for a 9R-DOX-AOS fusion protein. The genes were cloned and expressed, but none of them oxidized 18:2n-6 to 9R-hydroperoxy-10(E),12(Z)-octadecadienoic acid (9R-HPODE). ATEG_02036 transformed 9R-HPODE to an unstable allene oxide, 9(R),10-epoxy-10,12(Z)-octadecadienoic acid. A substitution in the P450 domain (C1073S) abolished AOS activity. The N964V and N964D mutants both showed markedly reduced AOS activity, suggesting that Asn964 may facilitate homolytic cleavage of the dioxygen bond of 9R-HPODE with formation of compound II in analogy with plant AOS (CYP74) and prostacyclin synthase (CYP8A1). ATEG_03992 was identified as 5,8-linoleate diol synthase (5,8-LDS). Replacement of Asn878 in 5,8-LDS with leucine (N878L) mainly shifted ferryl oxygen insertion from C-5 toward C-6, but replacements of Gln881 markedly affected catalysis. The Q881L mutant virtually abolished the diol synthase activity. Replacement of Gln881 with Asn, Glu, Asp, or Lys residues augmented the homolytic cleavage of 8R-HPODE with formation of 10-hydroxy-8(9)-epoxy-12(Z)-octadecenoic acid (erythro/threo, 1–4:1) and/or shifted ferryl oxygen insertion from C-5 toward C-11. We conclude that homolysis and heterolysis of the dioxygen bond with formation of compound II in AOS and compound I in 5,8-LDS are influenced by Asn and Gln residues, respectively, of the I-helices. AOS of A. terreus appears to have evolved independently of CYP74 but with an analogous reaction mechanism.
Introduction
Eicosanoids and oxylipins designate oxidized fatty acids and their metabolites (1–3). In man, eicosanoids are formed from arachidonic acid, which is oxidized by COX,2 LOX, and P450 to a complex series of biological mediators, including prostaglandins, leukotrienes, and epoxyeicosatrienoic acids, which play pivotal roles in reproduction, cancer, allergic inflammation, asthma, and cardiovascular pathophysiology (1, 2, 4). In plants and fungi, C18 fatty acids are oxidized by DOX of the peroxidase gene family, LOX, and P450 to oxylipins involved in host-pathogen interactions, reproduction, and development (3, 5–9). The core reaction mechanisms of these eukaryotic enzymes have either been retained during evolution or may have evolved in parallel. It is therefore not surprising that human, plant, and fungal enzymes can share catalytic similarities but still form different products. Interest in these enzymes is primarily based on their biological importance in human physiology and plant-pathogen interactions but also on their reaction mechanisms, which illustrate fundamental properties of iron oxidation states in biological chemistry (10).
The first step in biosynthesis of oxylipins is hydrogen abstraction by the catalytic base of LOX or by the tyrosyl radical of heme-containing DOX followed by insertion of molecular oxygen (Fig. 1, A and B). In the next step, the formed hydroperoxides and endoperoxides can be transformed by P450, which can be divided into different classes (11, 12). Class II P450 insert an oxygen atom from air into alkyl chains of drugs and endogenous compounds in the presence of electron transport from NAD(P)H via P450 reductase. In contrast, class III P450 do not require molecular oxygen and an electron transport system. This is exemplified by thromboxane synthase (CYP5A1) and prostacyclin synthase (CYP8A1) in humans, AOS (CYP74) in plants, and LDS in fungi (8, 13–17).
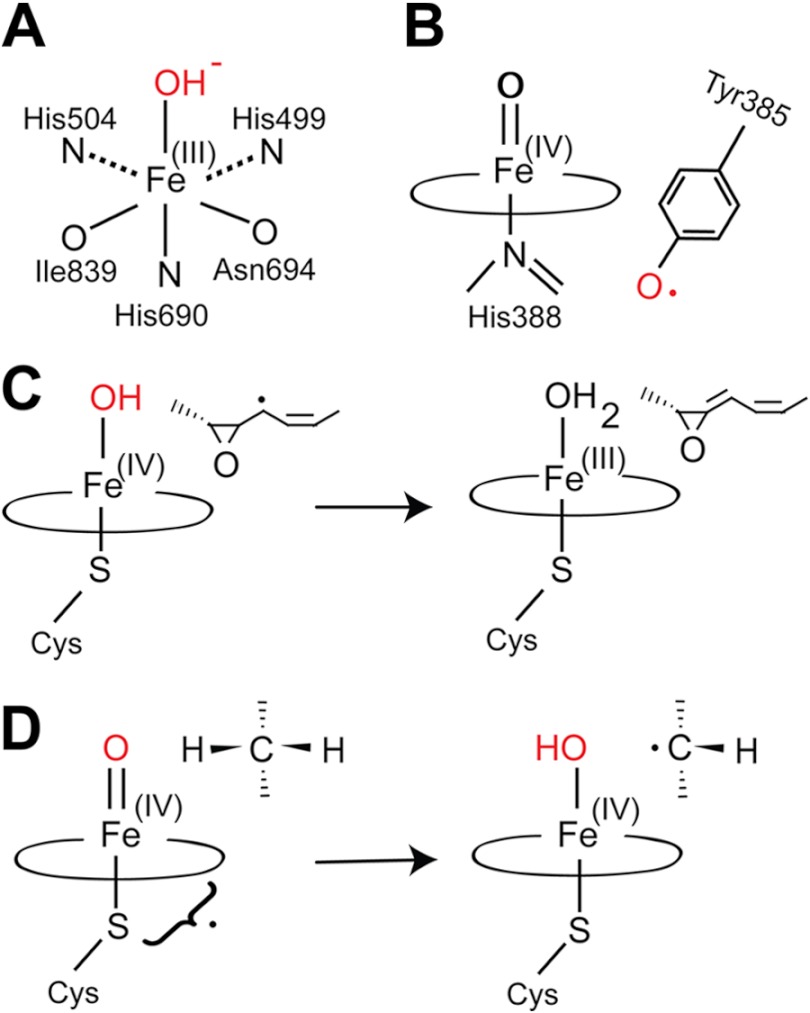
FIGURE 1.
Overview of iron enzymes in biosynthesis of eicosanoids and oxylipins. A, metal center of sLOX-1 in the oxidized form. The three nitrogen and oxygen ligands are indicated. The oxidized form catalyzes proton-coupled electron transfer from bisallylic positions of fatty acids. B, compound I of COX-1 and the catalytic tyrosyl radical. The latter catalyzes proton-coupled electron transfer from bisallylic positions of C20 fatty acids and from allylic positions of C18 fatty acids. LDS, 10R-DOX, and α-DOX use a closely related oxidation mechanism. C, allene oxide synthase. This class III P450 of the CYP74 family catalyzes homolytic cleavage of dioxygen bonds. This forms P450 compound II and a substrate radical, which is dehydrated to an allene oxide with formation of ferric heme. Prostacyclin synthase (CYP8A1) uses a similar reaction mechanism. D, LDS. This class III P450 catalyzes heterolytic cleavage of the dioxygen bond of 8R-HPODE and formation of compound I, which catalyzes hydrogen abstraction from C-7 or C-5 with formation of P450 compound II, a transient substrate radical, and “oxygen rebound” (modified from Ref. 21).
CYP74 catalyzes homolytic cleavage of the dioxygen bond of hydroperoxides (8, 15) with formation of P450 compound II and a carbon-centered radical intermediate. The latter is converted to an allene oxide and P450 compound II is recycled to ferric heme (Fig. 1C). Allene oxides are precursors of JA, which is an important signal molecule in plant defense and development and is present throughout the plant kingdom (18). The P450 domain of LDS transforms 8R-HPODE to diols by intramolecular oxygen insertion following heterolytic cleavage of the hydroperoxide O–O bond with formation of compound I (Por+• Fe(IV)=O) (8, 16, 17, 19). The latter abstracts hydrogen at either C-7 or C-5 and catalyzes oxygen rebound with retention of the configuration (Fig. 1D) (19–21). In this way, LDS transforms 8R-HPODE to 7,8- or 5,8-DiHODE. The latter was identified along with 8R-HODE as a sporulation hormone of Aspergillus nidulans (22).
Oxylipins are formed by plants and plant pathogens in a complex network of interactions (9). Expression of plant LOX is often augmented in defense reactions to fungal infections, e.g. rice 13S-LOX by the rice blast fungus (23). Fungal plant pathogens can secrete LOX, e.g. the root rot fungus of wheat and the stem rot fungus of rice. These enzymes may oxidize plant cellular lipids to reactive oxygen species with detrimental cellular effects (24). JA is a prototype defense molecule of virtually all plants (18), but it can also be formed by fungi. Its biosynthesis by the tropical and devastating plant pathogen Lasiodiplodia theobromae was discovered more than 40 years ago (25, 26). L. theobromae also oxidizes 18:2n-6 sequentially to 9R-HPODE and 9(10)-EODE by its 9R-DOX and AOS activities (27). Research progress in the area has been hampered because of environmental safety restrictions. These regulations do not apply to A. terreus, which oxidizes 18:2n-6 sequentially to 9R-HPODE and 9(10)-EODE in analogy with L. theobromae (28). Plant AOS belong to the CYP74 family and transform hydroperoxy fatty acids formed by 9S- and/or 13S-LOX, but the AOS and 9R-DOX activities of L. theobromae and A. terreus have not been characterized.
A. terreus is infamous for therapy-resistant infections in immunocompromised patients but also renowned for industrial production of lovastatin (29). The secondary metabolism of aspergilli has many industrial and toxicological implications and has attracted considerable attention (30). The genomes of at least eight aspergilli have been sequenced. A. terreus was sequenced in 2005 (for a review, see Ref. 31).
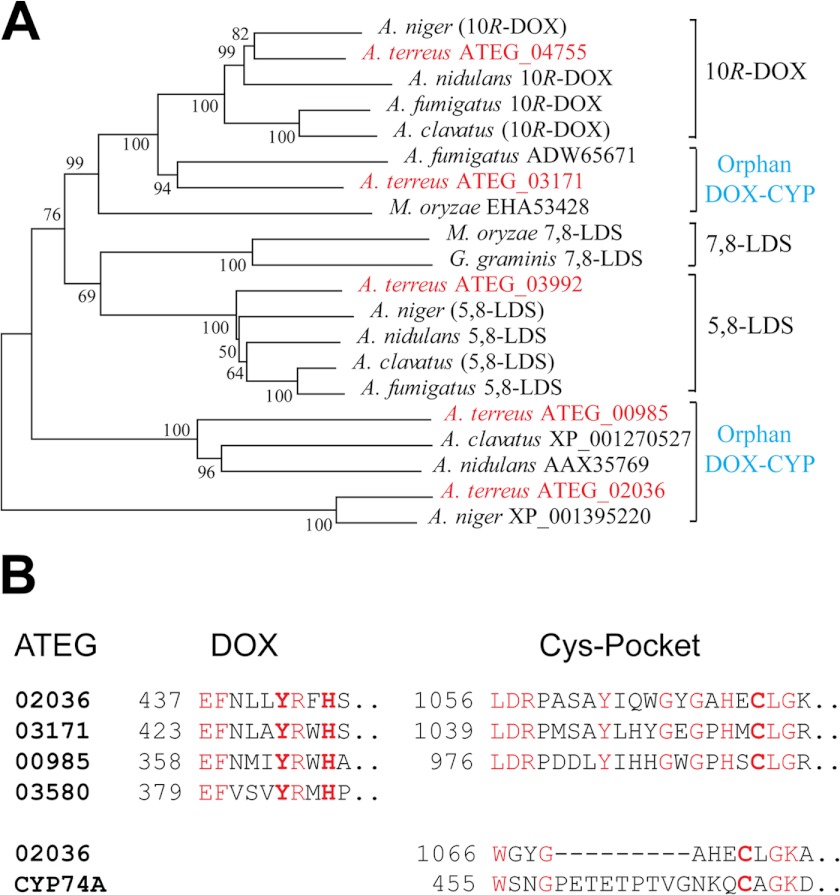
The genome of A. terreus contains five genes with homology to DOX-CYP fusion enzymes of other aspergilli (28). A phylogenetic tree of DOX-CYP fusion proteins is shown in Fig. 2A. Two proteins aligned with the 5,8-LDS and 10R-DOX clusters, indicating their putative catalytic activities. The remaining three proteins appear to be DOX-CYP orphans inasmuch as they do not cluster with characterized enzymes. There is also at least one gene with homology to the DOX domain (ATEG_03580; Fig. 2), but it lacks the P450 domain. In addition, sequenced aspergilli contain about 110–155 P450 genes (32).
FIGURE 2.
Phylogenetic tree and overview of catalytically important amino acid residues of DOX-CYP fusion proteins. A, phylogenetic tree of DOX-CYP fusion proteins (modified from Ref. 54) of Gaeumannomyces graminis, Magnaporthe oryzae, and five aspergilli (Aspergillus clavatus, A. fumigatus, A. nidulans, A. niger, and A. terreus), which express the marked enzyme activities identified by gene expression. Genes within parentheses are deduced by sequence homology and detected enzyme activities. The five DOX-CYP homologues of A. terreus are red. One sequence aligns within the 10R-DOX cluster, and one aligns within the 5,8-LDS cluster, whereas three sequences are orphans. The bootstrap numbers from 1,000 replications are indicated at the branches (MEGA 4). B, alignments of the DOX and P450 domains of DOX-CYP fusion proteins of the three orphans. Left, the proximal heme histidine ligand and the catalytically important Tyr residue of the DOX domains of LDS are bold. Right, the Cys pocket sequences with the heme thiolate ligands of the P450 domains. Conserved residues are red. Bottom, the short sequences around the heme thiolate ligand of AOS of Arabidopsis thaliana (CYP74A) is aligned with the corresponding region of ATEG_02036, and the former contains an insert of nine amino acids.
COX and the N-terminal DOX domain of DOX-CYP fusion proteins contain Tyr and His residues in a characteristic catalytic motif, Tyr-(Arg/His)-Trp-His (33). The conserved His residue in this sequence is the proximal heme ligand, and the Tyr residue likely forms the catalytically important radical (1, 7, 34). This sequence is conserved in two orphans but modified by replacement of the Trp residue by Phe or Met residues in two of the sequences (Fig. 2B). Whether these substitutions affect heme oxidation and formation of the Tyr radical is unknown. The C-terminal domains of the DOX-CYP orphans contain a series of conserved residues in the Cys pocket region, including the heme thiolate ligand, but not the typical insertion loop of CYP74 (Fig. 2B).
We hypothesized that the 9R-DOX and AOS activities could be present in an orphan DOX-CYP fusion protein in analogy with the 8R-DOX and hydroperoxide isomerase activities of LDS. In that case, A. terreus could also provide a model for studies of the tentative evolution of DOX-CYP fusion proteins into 9R-DOX and AOS.
The objective of the present study was to clone and express all five DOX-CYP homologues and one DOX homologue and to determine whether they could catalyze the 9R-DOX or AOS activities of A. terreus. This objective was met as described in the following experiments with identification of the first fungal AOS. This discovery enabled us to compare the active sites of AOS and the P450 domain of 5,8-LDS by site-directed mutagenesis.
EXPERIMENTAL PROCEDURES
Materials
Fatty acids (99%) were from Merck, Sigma, and Larodan and were stored at 50–100 mm in ethanol (−20 °C). [13C18]18:2n-6 (98%) was from Larodan. HPODEs and HPOMEs were prepared by photooxidation and purified by NP-HPLC, and stereoisomers were separated on Reprosil Chiral NR-R (35). Mouse anti-V5 antibody, Superscript III kit, InsectSelect System with pIZ/V5-His, Cellfectin, phleomycin (Zeocin), and Champion pET Directional TOPO kit were from Invitrogen. Restriction enzymes and chemically competent Escherichia coli (NEB5α) were from New England Biolabs, Invitrogen, and Fermentas. Plasmid Midi kit and QIAquick gel extraction kits were from Qiagen. Prestained protein ladder (Page-Ruler) for SDS-PAGE, Pfu DNA polymerase, and CloneJet PCR cloning kit were from Fermentas. Phusion DNA polymerase was from Finnzymes. ECL Advance Western blotting detection kit, dNTPs, and horseradish peroxidase-labeled anti-mouse IgG antibodies were from GE Healthcare. RNase A and ampicillin were from Sigma. SYBR Green Supermix analysis kit was from Bio-Rad. Ex-Cell 420 insect serum-free medium was purchased from SAFC Biosciences (Hampshire, UK). Gentamycin sulfate and Spodoptera frugiperda (Sf21 and Sf9) cells were obtained locally. Primers were obtained from Cybergene (Huddinge, Sweden) and TIB MOLBIOL (Berlin, Germany). Sequencing was performed at Uppsala Genome Center (Rudbeck Laboratories, Uppsala University). A. terreus strain IBT1948 (IBT Culture Collection, Denmark) was provided by Dr. Melin (The Swedish Agricultural University, Uppsala), and strain A1156 (NIH2624) was obtained from the Fungal Genetic Stock Center (Kansas City, MO).
Mycelium of A. terreus
Conidia of A. terreus were added to 2% malt extract, Czapek's Dox, yeast malt extract, Harrold's, or rich medium (36) and incubated at 37 °C in the dark (180 rpm) for 40–48 h. In some experiments, the mycelia were grown for a few additional days at room temperature. Nitrogen powder of mycelia was homogenized in 0.1 m KHPO4 buffer (pH 7.4), 2 mm EDTA, 0.04% Tween 20 and centrifuged (17,000 × g, 5 min). The supernatants and suspended pellets were incubated with 100 μm 18:2n-6 or [13C18]18:2n-6 for 30 min on ice.
Differential Centrifugation
The homogenate of nitrogen powder of mycelia was subjected to differential centrifugation (1,000 × g, 10,000 × g, and 17,000 × g; 5–20 min; +4 °C) and ultracentrifugation (10,000 × g for 20 min followed by 100,000 × g for 60 min, +4 °C). 9R-DOX and AOS activities were assayed with 50–100 μm [13C18]18:2n-6 as substrate in the precipitates and the supernatants.
Assay of Non-competitive D-KIE of 9R-DOX
The supernatant of nitrogen powder of mycelia was prepared as above and incubated in triplicate with 0.1 mm 18:2n-6 or [11,11-2H2]18:2n-6 for 5 or 20 min on ice. 13-Hydroxyoctadecatrienoic acid was added as an internal standard. The D-KIE was assayed by LC-MS/MS analysis of the carboxylate anions after reduction to alcohols (m/z 295–296 → full scan) with monitoring of m/z 171. These experiments were also performed by comparison of [11,11-2H2]18:2n-6 with [13C18]18:2n-6 to correct for the small amounts of endogenous 18:2n-6.
RNA Extraction and cDNA Synthesis
Total RNA was isolated by LiCl extraction as described (37) and treated with RQ1 RNase-free DNase (Promega). First strand cDNA was synthesized using reverse transcriptase (Superscript III) with random hexamer or oligo(dT)20 primers according to the manufacturer's instructions.
Real Time PCR Analysis
Gene expression was studied by real time PCR with an iCycler (Bio-Rad) as described (28). cDNA of strain A1156 and primer concentrations of each reaction (25 μl) were 200 and 500 nm, respectively. β-Actin (ATEG_06973) served as the reference gene. Primers are listed in supplemental Table S1 and were designed using the Primer3 (ATEG_04755, 00985, 02036, and 03171) and PrimerQuest software programs (ATEG_03580, 06973, and 03992) allowing distinction of whether amplicons originated from cDNA or contaminating genomic DNA. All samples and negative controls were analyzed in triplicates. Amplification of one single product of the expected size was confirmed by melting point analysis and gel electrophoresis on a 2% agarose gel. Data analysis providing CT values was carried out with the iCycler software (Bio-Rad), and values for primer efficiencies were obtained from the LinRegPCR software (38).
Cloning of ATEGs
ATEG_04755 and 03171 were cloned from A. terreus strain IBT1948, whereas ATEG_03580, 02036, 00985, and 03992 originate from strain A1156 (NIH2624). All genes were cloned by RT-PCR as explained in detail in the supplemental Methods. In short, amplified cDNA segments were ligated in pJet1.2/blunt to build the expression constructs. The open reading frames were ligated into pIZ/V5-His in-frame with the V5 epitope and the His6 tag. All constructs were confirmed by sequencing. All cloning primers are listed in supplemental Table S2.
Expression in Insect Cells
Plasmid-driven expression was performed by transfection of Sf21 or Sf9 cells (33). Cells were harvested after 2 days, suspended in lysis buffer (50 mm KHPO4 buffer (pH 7.4), 1 mm EDTA, 1 mm GSH, 5% glycerol, 0.04% Tween 20) and sonicated (Branson sonifier cell disrupter B15, 20% maximum power, 3 × 5-s pulses or Bioruptor Next Generation, 10 × 30 s, 4 °C). The cell debris was spun down (17,000 × g, 30 min, +4 °C), and the supernatant was used for enzyme assay. Protein expression was detected by Western blot analysis using primary antibodies against the V5 epitopes (33). All constructs were expressed at least in three independent experiments.
Expression in E. coli
Subcloning of open reading frames from pIZ/V5-His to pET101D-TOPO vectors was performed by PCR technology following Invitrogen's instructions. All primers are listed in supplemental Table S3. The amplicons were ligated into pET101/D-TOPO and introduced into BL21 cells by heat shock transformation. Cells were grown to an A600 of 0.6–0.8 in low salt LB medium, and protein expression was induced by 0.1 mm isopropyl 1-thio-β-d-galactopyranoside. Cultures were grown for 5 h at room temperature with moderate shaking (∼100 rpm). Cells were harvested by centrifugation and sonicated (Bioruptor Next Generation, 10 × 30 s, 4 °C) as described (33). At least three independent expressions of each protein were analyzed.
Western Blot Analysis
Recombinant enzymes were separated by SDS-PAGE on 8% polyacrylamide gels prior to blotting to nitrocellulose membranes. For Western blot analysis, a C-terminal anti-V5 antibody served as the primary antibody, and a horseradish peroxidase-conjugated anti-mouse IgG served as the secondary antibody (33). Both antibodies were usually diluted 1:12,500 (in 15% nonfat milk). Chemiluminescence was used for detection (ECL Advance kit, GE Healthcare).
Site-directed Mutagenesis
Site-directed mutagenesis of ATEG_02036 and 5,8-LDS ATEG_03992 was performed according to the QuikChange protocol (Stratagene) in pIZ/V5-His constructs (33). Amplicons were obtained from 10 ng of template by Pfu DNA polymerase (16 cycles) before digestion with DpnI (2 h, 37 °C). Amplification of one distinct PCR product was confirmed by agarose gel electrophoresis before heat shock transformation (NEB5α). Primers containing the designated replacements are listed in supplemental Table S4. All mutations were confirmed by sequencing before subcloning to pET101D-TOPO vectors as described above.
Enzyme Assays
Recombinant proteins expressed in insect cells or in E. coli were incubated with 100 μm fatty acids (18:1n-9, 18:2n-6, 18:3n-3, or 20:4n-6) in triplicate for 30 min on ice. The reaction (0.3–0.5 ml) was terminated with ethanol (2–4 volumes), an internal standard (13-hydroxyoctadecatrienoic acid) was added in some experiments, and proteins were removed by centrifugation. The metabolites were extracted on octadecyl silica (Sep-Pak C18), evaporated to dryness, and diluted in ethanol (40–60 μl), and 5–10 μl were subjected to LC-MS/MS analysis. Conversion of 9R-HPODE (50–100 μm) was assayed in the same way. For trapping of the allene oxide, recombinant ATEG_02036 expressed in E. coli was incubated with 100 μm 9R-HPODE for 1 min on ice and quenched with 60 volumes of methanol (+4 °C, 3 h). The products were extracted and analyzed by LC-MS/MS.
LC-MS/MS Analysis of Oxylipins
Reversed phase HPLC with MS/MS analysis was performed with a Surveyor MS pump (ThermoFisher) and an octadecyl silica column (5 μm, 2.1 × 150 mm, Phenomenex), which was usually eluted with methanol/water/acetic acid (800:200:0.05 or 750:250:0.05) at 0.3 ml/min. The effluent was subjected to electrospray ionization in a linear ion trap mass spectrometer (LTQ, ThermoFisher) with monitoring of carboxylate anions. The heated transfer capillary was set at 315 °C, the ion isolation width was set at 1.5 atomic mass units, the collision energy was set at 35 (arbitrary scale), and the tube lens varied between 90 and 120 V. A trihydroxy fatty acid (prostaglandin F1α; 100 ng/min) was infused for tuning.
NP-HPLC with MS/MS analysis was performed with a silicic acid column (5 μm, Kromasil 100SI, 250 × 2 mm, ChromTech) using hexane/isopropyl alcohol/acetic acid (98:2:0.01) at 0.3 ml/min (P2000, Thermo) for separation of α- and γ-ketols, epoxy alcohols, and hydroxy fatty acids. 5,8- and 8,11-DiHODE were separated by NP-HPLC for steric analysis by hexane/isopropyl alcohol/acetic acid (95:5:0.01) at 0.5 ml/min. The effluent was mixed with isopropyl alcohol/water (60:40) at 0.2–0.3 ml/min from a second pump (Surveyor MS pump). The combined effluents were introduced by electrospray into the ion trap mass spectrometer (LTQ). Preparative separations of HPOME and HPODE were performed on a larger column (5 μm, Reprosil 100Si, 10 × 250 mm), which was eluted at 1–2 ml/min with hexane/isopropyl alcohol/acetic acid (97:3:0.01). Steric analysis of 8- and 10-HODE by chiral phase HPLC-MS/MS was performed with Chiracel-OBH and Reprosil Chiral NR, respectively, and the effluent was mixed with isopropyl alcohol/water (3:2) (37).
Bioinformatics
Proteins were aligned with the ClustalW algorithm (DNA Star software). Phylogenetic trees were constructed with MEGA 4 software with bootstrap tests of the resulting nodes (39). The distance within branches is based on the number of expected substitutions per amino acid position.
Miscellaneous Methods
Fatty acid composition of A. terreus was determined in nitrogen powder of mycelia after alkali treatment (0.5 m KOH in 90% methanol, 70 °C, 1 h) and extractive isolation (CHCl3/methanol) according to Bligh and Dyer (40). The carboxylate anions were analyzed by direct injection.
RESULTS
Bioinformatics
As outlined in Fig. 2, sequence similarities placed ATEG_04755 together with other 10R-DOX. ATEG_03171 can be aligned with 60% identity to ATEG_04755 (ClustalW algorithm), but it is not a member of the 10R-DOX cluster (see Fig. 2). However, it belongs with ATEG_04755 in the CYP6001C subfamily (41). In ATEG_03171, the heme thiolate ligand is retained in contrast to ATEG_04755 and 10R-DOX of Aspergillus fumigatus and A. nidulans (37, 42) (Fig. 2B).
ATEG_03992 aligns with 5,8-LDS and belongs to the CYP6001A subfamily (41). ATEG_00985 can be aligned with 44% amino acid identity to ATEG_03992, but the former is classified as an orphan of the CYP6002 family (41).
The ATEG_02036 sequence is relatively distant from the other DOX-CYP sequences and belongs to the CYP6003 family (41). The conserved DOX motif, Tyr-(Arg/His)-Trp-His, of COX, LDS, and 10R-DOX is modified by replacement of Trp by Phe in ATEG_02036 (Fig. 2B). This Trp residue is replaced by Met in the DOX homologue ATEG_03580.
Studies on Mycelia of A. terreus
A. terreus oxidized 18:2n-6 as reported (28). 9R-HPODE was the main hydroperoxy metabolite followed by 10R-HPODE and 8R-HPODE of nitrogen powder prepared from mycelia in 1.5–2% malt extract (2 days, 37 °C, 180 rpm). Variable amounts of 5,8-DiHODE were also detected. Other growth media appeared to alter the oxylipin profile. 5,8-DiHODE was the main product from mycelia in Czapek's Dox, yeast malt extract, and rich media, whereas 10-DOX activity dominated in mycelia grown in Harrold's medium.
9R-DOX and AOS activities of nitrogen powder preparations were detected in the cytosol (100,000 × g supernatant) and in membrane fractions, respectively (28). Differential centrifugation showed that 9R-DOX-AOS activity was enriched in membrane precipitates (from 1,000 × g to 17,000 × g) in comparison with these supernatants and efficiently linked to AOS as estimated with [13C18]18:2n-6 as a substrate.
9R-DOX catalyzes hydrogen abstraction at C-11 and oxygen insertion at C-9 of 18:2n-6 (28). The D-KIE of 9R-DOX was estimated by comparison of the oxidation of [11,11-2H2]18:2n-6 with 18:2n-6 or [13C18]18:2n-6. The D-KIE averaged 1.3 after 5 min (n = 3) and ranged from 1.6 to 4.1 after 20-min incubations (n = 7) (see supplemental Fig. S1). Hydrogen abstraction at C-11 of linoleic acid is catalyzed by LOX with a large (>25) D-KIE and by COX-1 with an insignificant D-KIE (43–45). We conclude that the D-KIE of 9R-DOX is smaller and similar to the D-KIE of COX-1.
Gene Expression Analysis by Real Time PCR
Real time PCR revealed that all six studied genes were transcribed (Table 1). ATEG_06973, coding for β-actin, served as the reference gene. mRNA and cDNA were prepared from nitrogen powder of A. terreus with prominent 9R-DOX and AOS activities.
TABLE 1.
CT values for mRNA of the ATEG genes of A. terreus A1156 and primer efficiencies
Actin (ATEG_06973) mRNA served as reference with a CT value of 24.4 ± 0.6 and primer efficiency of 1.98.
| ATEG mRNA | CT value ±S.D. | Primer efficiency |
|---|---|---|
| ATEG_04755 | 29.0 ± 1.5 | 1.97 |
| ATEG_03992 | 30.1 ± 0.8 | 1.99 |
| ATEG_02036 | 30.0 ± 1.2 | 1.84 |
| ATEG_03171 | 31.9 ± 1.8 | 1.68 |
| ATEG_00985 | 29.0 ± 0.6 | 2.11 |
| ATEG_03580 | 30.4 ± 0.83 | 1.87 |
The values shown in Table 1 represent the median results of three analyses performed on different cDNA preparations and suggest that the genes were transcribed in the following order: ATEG_00985 = 04755 > 02036 ≥ 03992 ≥ 03580 > 03171. The CT value for ATEG_03992 combines the transcripts of both the full length and its splice variant described below. For all primer pairs, the negative controls showed no significant amplification. We conclude that all six genes were transcribed and without large differences in mRNA expression levels (see Table 1).
Summary of Cloning, Expression, and Western Blot Analysis
The five DOX-CYP proteins and the DOX homologue were expressed in insect cells and E. coli. Recombinant proteins of the expected molecular weights were detected by Western blot analysis as illustrated in supplemental Fig. S2. Supplemental Fig. S2 also shows Western blot analysis of a splice variant, ATEG_03992_sv (see below). These data were in agreement with the expected sizes of these proteins with the appended C-terminal His tags and V5 epitopes (ATEG_04755 (129 kDa), ATEG_03171 (126 kDa), ATEG_03992 (124 kDa), ATEG_03992_sv (122 kDa), ATEG_00985 (120 kDa), ATEG_02036 (133 kDa), and ATEG_03580 (74 kDa).
We investigated the recombinant proteins for enzymatic activities. The results are summarized in Table 2. ATEG_02036 transformed 9R-HPODE to an allene oxide, but it did not oxidize 18:2n-6. We could identify 5,8-LDS, a splice variant of 5,8-LDS with 8R-DOX activity, 10R-DOX, and a homologue of 10R-DOX, ATEG_03171. The latter transformed 18:2n-6 to 10R-HPODE only in low yields (supplemental Fig. S3). 5,8-LDS and 10R-DOX are homologues to these enzymes of A. fumigatus and A. nidulans (17, 20). ATEG_00985 formed only traces of 8R-HPODE (supplemental Fig. S3). Oxidation of C18 fatty acids by ATEG_03580 could not be detected. We confirmed that the lack of activity was not due to the absence of heme by adding 1.5 μm hematin back.
TABLE 2.
Overview of DOX-CYP genes, corresponding GenBank accession numbers of the proteins, end products formed with 18:2n-6 or 9R-HPODE as substrates, and assigned P450 families
| ATEG | Protein | Substrate | End product | Enzyme | P450 | Strain |
|---|---|---|---|---|---|---|
| 04755 | AFB71131a | 18:2n-6 | 10R-HPODE | 10R-DOX | 6001C | IBT1948 |
| 03171 | AFB71132b | 18:2n-6 | 10R-HPODE | 10R-DOXc | 6001C | IBT1948 |
| 03992 | AGA95448 | 18:2n-6 | 5S,8R-DiHODE | 5,8-LDS | 6001A | A1156 |
| 03992_sv | AGA95449d | 18:2n-6 | 8R-HPODE | 8R-DOX | A1156 | |
| 02036 | KC525886 | 9R-HPODE | 9 (10)-EODE | AOS | 6003B | A1156 |
| 00985 | KC525887e | 18:2n-6 | (8R-HPODE)f | Orphanf | 6002A | A1156 |
| 03580 | EAU35382g | 18:2n-6 | NDh | Orphan | A1156 |
a This protein sequence differs at six positions from the deduced protein of A1156 (GenBank accession number EAU35202).
b This sequence differs at 18 positions from the deduced protein of A1156 (GenBank accession number EAU36445).
c Only low enzyme activity was detected (see supplemental Fig. S3).
d This sequence differs from the deduced sequence (EAU35794) with an additional intron, leading to a frameshift after the first 963 N-terminal residues.
e This sequence differs in several ways from the deduced protein of A1156 as described in the supplemental Results section 4.
f Weak 8R-DOX activity was noticed (see supplemental Fig. S3).
g The cloned sequence was identical to the deduced sequence of A1156 (GenBank accession number EAU35382).
h Metabolites not detected.
AOS (ATEG_02036)
Cloning of ATEG_02036 yielded a 3432-bp cDNA fragment with 10 introns as predicted by conceptual translation (GenBankTM accession number EAU36998) and without nucleotide substitutions.
Recombinant ATEG_02036 was expressed both in insect cells and E. coli in the catalytically active form. Incubation of the recombinant protein with 9R-HPODE yielded 9-hydroxy-10-oxo-12(Z)-octadecenoic acid (α-ketol) and 13-hydroxy-10-oxo-11(E)-octadecenoic acid (γ-ketol) (Fig. 3A) in analogy with subcellular fractions of A. terreus (28). Small amounts of 11-hydroxy-9(10)-epoxy-12(Z)-octadecenoic acid were also noted. The MS3 spectrum of the α-ketol yielded strong and characteristic fragments (Fig. 3B), whereas the MS/MS spectrum contained weak ions (<10% of base peak, m/z 293 (311 − 18)).
FIGURE 3.
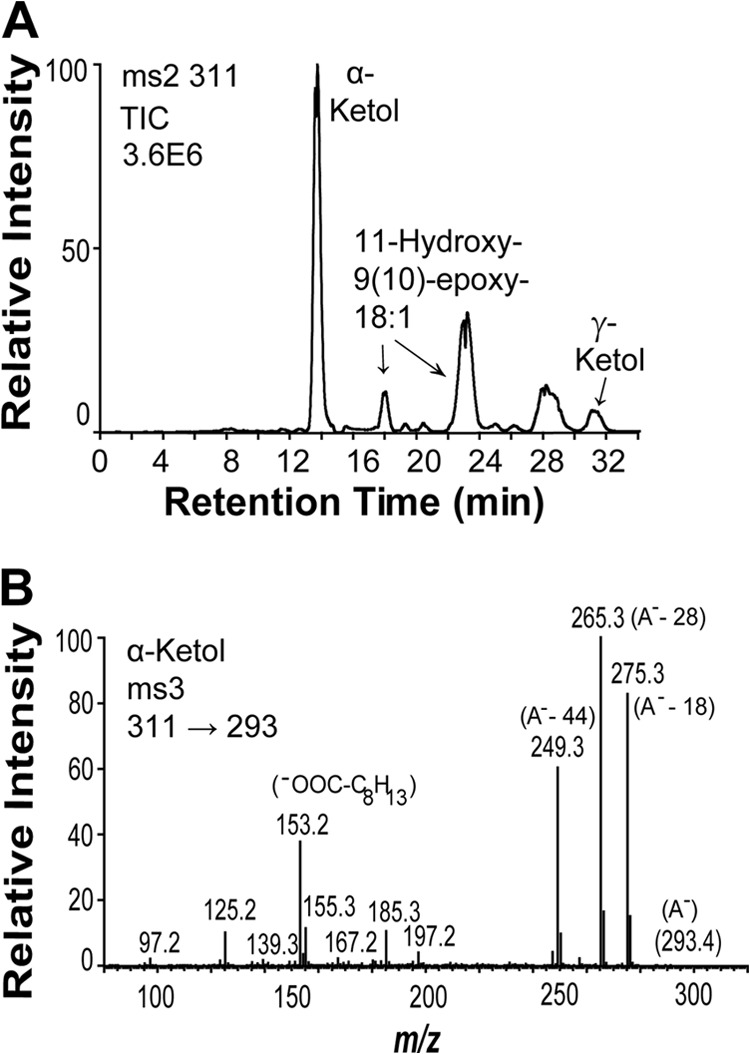
Transformation of 9R-HPODE by recombinant AOS (ATEG_02036). A, NP-HPLC-MS/MS analysis of metabolites. B, MS3 spectrum (m/z 311 → 293 → full scan) of the α-ketol, 9-hydroxy-10-oxo-(12Z)-octadecenoic acid. The intense ions at m/z 265, 185, 155, and 153 corresponded to m/z 282, 195, 165, and 162 in the MS3 spectrum of 9-hydroxy-10-oxo-(12Z)-[13C18]octadecenoic acid. The signal at m/z 155 could be due to the fragment ion −OOC-C8H15 (possibly due to dehydration at C-9 and cleavage between C-9 and C-10), and the signal at m/z 153 (155–2; loss of H2) could be due to −OOC-C8H13 (inset). TIC, total ion current; A−, the carboxylate anion; γ-Ketol, 10-oxy-13-hydroxy-(11E)-octadecenoic acid.
The α- and γ-ketols originate from spontaneous hydrolysis of an unstable allene oxide, 9(10)-EODE (28). This was confirmed by short time incubation and trapping of 9(10)-EODE with methanol. NP-HPLC-MS/MS analysis (m/z 325 → full scan; supplemental Fig. S4) thus showed formation of a product with characteristic signals, inter alia, at m/z 171 and in the upper mass range at m/z 307 (A− −18), 293 (A− −32, loss of methanol), and 275 (293 − 18). This MS/MS spectrum was consistent with 9-hydroxy-10-methoxy-12(Z)-octadecenoic acid.
ATEG_02036 and CYP6003B2 (41) are designated AOS for simplicity. We observed that C1073S replacement in AOS abolished the catalytic activity and confirmed expression of the mutant by Western blot analysis (supplemental Fig. S2).
9S- and 9R-HPOME and 10S- and 10R-HPOME were transformed by ATEG_02036 to small amounts of epoxy alcohols but without detectable stereospecificity. 13S-, 13R-, and 9S-HPODE were not transformed by AOS.
5,8-LDS (ATEG_03992)
Cloning of ATEG_03992 yielded two different cDNA sequences. One cDNA sequence coded for 1071 amino acids, and it was identical to the sequence predicted by conceptual translation (GenBank accession number EAU35794), and the other coded for a splice variant (ATEG_03992_sv). The last intron was retained in the open reading frame, leading to a frameshift after the first 963 N-terminal amino acids and a stop codon after 1064 amino acid residues. The splice variant lacked the heme thiolate residue of the P450 domain.
The full-length recombinant ATEG_03992 formed 5,8-DiHODE as the main product (Fig. 4A). Small amounts of 8,11-DiHODE (12–14%) and traces of 6,8-DiHODE were also observed. The retention time of 8,11-DiHODE was identical to that of authentic 8R,11S-DiHODE on straight phase HPLC (20). The MS/MS spectrum of 8,11-DiHODE showed only structurally important fragment ions with low intensities (20), whereas the MS3 spectrum contained characteristic and intense ions (Fig. 4B). The MS/MS spectrum of 6,8-DiHODE was as reported (46). 18:1 and 18:3n-3 were also transformed to 5,8-diols.
FIGURE 4.
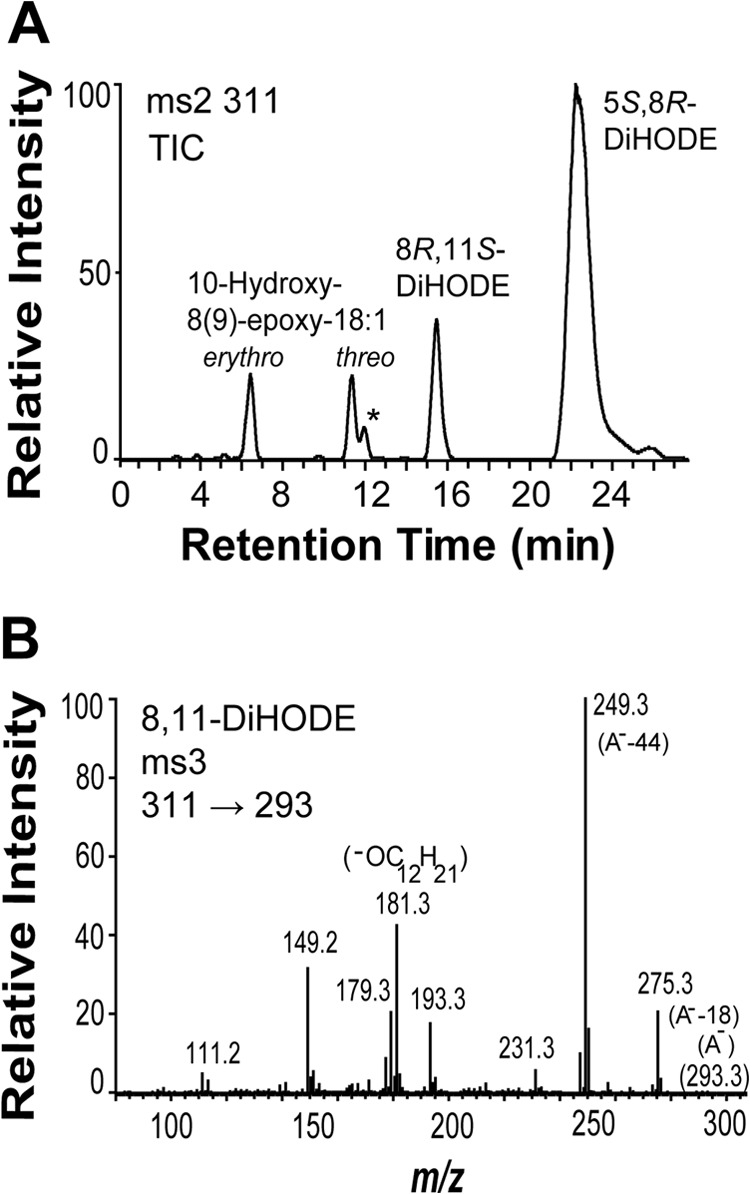
Oxidation of 18:2n-6 by recombinant 5,8-LDS (ATEG_03992). A, NP-HPLC-MS/MS analysis of products formed by 5,8-LDS. The peak marked with an asterisk (*) provided the typical MS/MS spectrum of 6,8-DiHODE with characteristic strong signals at m/z 155 and 129 (46). B, MS3 spectrum (m/z 311 → 293 → full scan) of 8,11-DiHODE. The intense ions at m/z 193, 181, 179, and 149 corresponded to m/z 206, 193, 191, and 159 in the MS3 spectrum of 8,11-[13C18]DiHODE. This suggested a complex fragmentation of the dehydrated 8,11-diol after the first MS selection, e.g. m/z 179 (293 −114; −OC12H19) and m/z 181 (293 −112; possibly −OC12H21; inset). TIC, total ion current; A−, the carboxylate anion.
8R-DOX (ATEG_03992_sv)
ATEG_03992_sv converted 18:1n-9, 18:2n-6, and 18:3n-6 mainly to the 8-hydroxy metabolites, but 5,8-diols were not detected as expected. This confirms that LDS are fusion enzymes with independent DOX and P450 activities (33).
10R-DOX (ATEG_04755)
The deduced amino acid sequence of ATEG_04755 from the strain IBT1948 (GenBank accession number AFB71131) showed six amino acid substitutions to the corresponding sequence of the strain A1156 (GenBank accession number EAU35202): A54S, E160D, F719L, M838L, T946S, and G983E.
ATEG_04755 converted 18:2n-6 and 18:3n-6 mainly to 10-HPODE and 10-hydroperoxyoctadecatrienoic acid, respectively (see supplemental Fig. S3). 10-HPODE consisted mainly of the 10R stereoisomer (data not shown). Oxidation of 18:1n-9 by ATEG_04755 yielded 8-HPOME as the main metabolite in analogy with 10R-DOX of A. fumigatus and A. nidulans (37, 42). 20:4n-6 was transformed to allylic and bisallylic hydroxyeicosatetraenoic acids by ATEG_04755. These results are in concordance with 10R-DOX of A. fumigatus (37). A. terreus, which was grown in malt extract medium, formed only small amounts of C20 fatty acids. The main fatty acid was 18:2, and the relative concentrations of C18 and C16 fatty acids were 18:2 (100%), 18:1 (70%), 16:3 (35%), 18:0 (30%), 18:3 (15%), and C16:1 (10%) (supplemental Fig. S5).
Cloning and Expression of ATEG_00985 (Orphan), 03171 (10R-DOX), and 03580 (Orphan)
The results are summarized in Table 2, and details are provided in the supplemental Methods.
Site-directed Mutagenesis of the I-helices in the P450 Domains
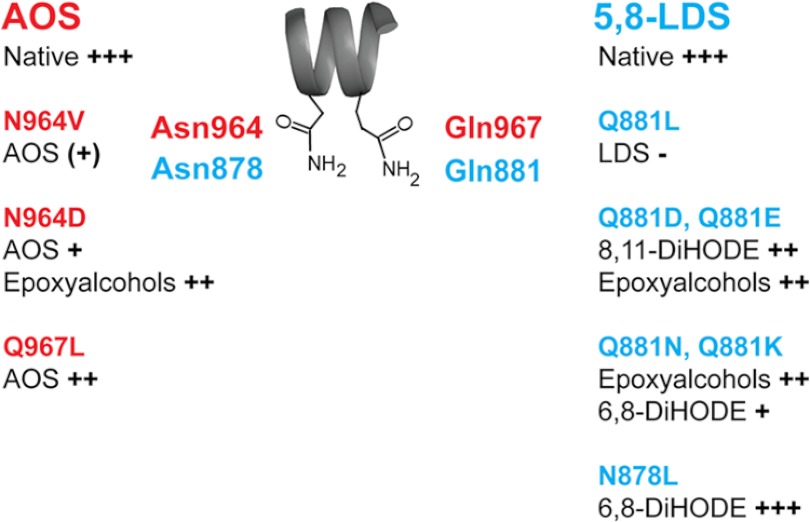
AOS (ATEG_02036) and 5,8-LDS (ATEG_03992) can be aligned with ∼38% amino acid identity (ClustalW) as shown in supplemental Fig. S6. Inspection of this alignment revealed a sequence of 12 amino acids that could be realigned as shown in Fig. 5 with 50% identity, including Asn964 of AOS and Asn878 of 5,8-LDS.
FIGURE 5.
Partial alignment of the I-helices of AOS with 5,8-LDS and CYP74A with CYP8A1, P450cam, and CYP4F8. Residues of AOS and 5,8-LDS of A. terreus marked with asterisks (*) were mutated. Conserved residues are blue, and the Asn residues are red. The alignment of the last four proteins was obtained by ClustalW. The important Asn residues of the two class III P450, CYP74A and CYP8A1, are red, and other conserved residues are blue. The Thr residues of the two class II P450, P450cam and CYP4F8 (prostaglandin H2 19-hydroxylase), align with the two Asn residues. The acid-alcohol pair, Asp-Thr, of P450cam and CYP4F8 in the conserved sequence Gly-Gly-Xaa-Asp-Thr are blue.
CYP74A and CYP8A1 catalyze homolytic cleavage of the O–O bonds. The marked Asn residues of these class III P450 in Fig. 5 are important for this process (8, 47). These two Asn residues align with Thr in P450cam and prostaglandin H 19-hydroxylase (CYP4F8) using ClustalW. The Thr residue with an Asp residue forms the acid-alcohol pair, Asp-Thr, in the conserved sequence (Gly/Ala)-Gly-Xaa-(Asp/Glu)-Thr, which takes part in the heterolysis of O–O bonds by many class II P450 (12).
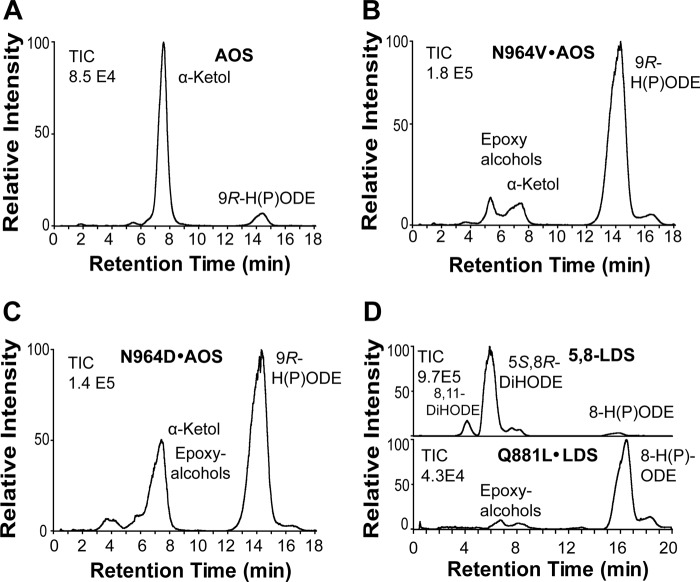
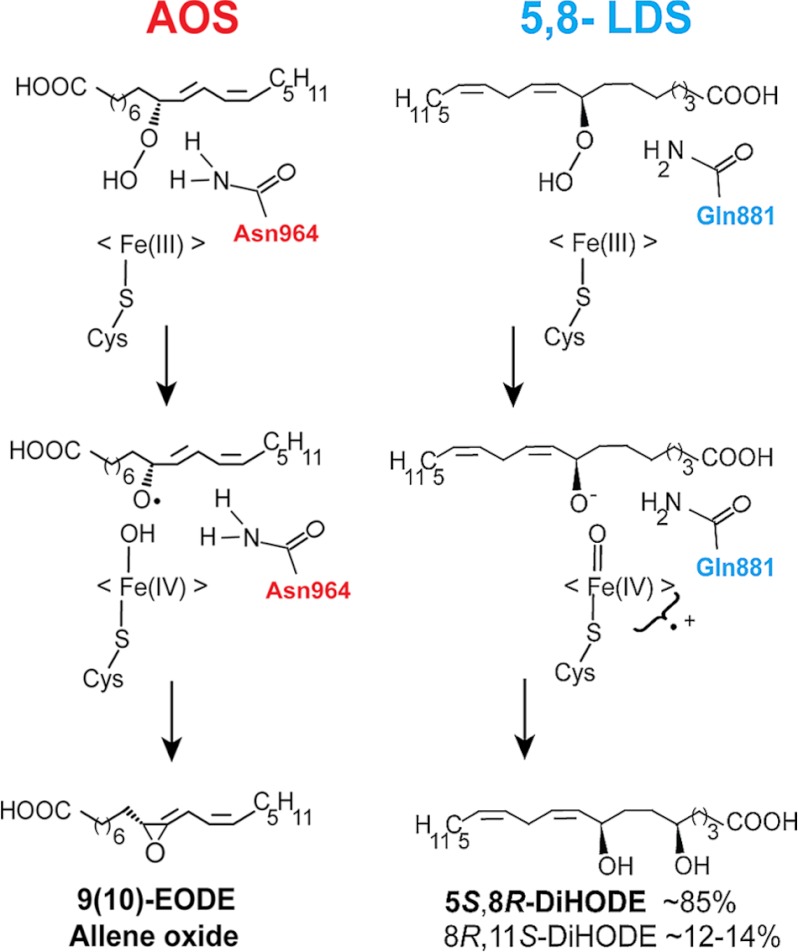
We replaced Asn964 of AOS with Val and Asp, designated N964V·AOS and N964D·AOS, respectively. Both mutants had lost almost all AOS activity in comparison with native AOS (Fig. 6, A–C). Further analysis of N964D·AOS by NP-HPLC revealed that in addition to biosynthesis of the α-ketol there was also formation of epoxy alcohols (Fig. 4 and supplemental Fig. S7). The homolytic cleavage of the hydroperoxide might be partly retained, but the desaturation step in biosynthesis of 9(10)-EODE appeared to be deranged with formation of epoxy alcohols (supplemental Fig. S7).
FIGURE 6.
Effect of site-directed mutagenesis of critical Asn and Gln residues in the I-helices of AOS and 5,8-LDS on the profile of oxylipin formation. A, native recombinant AOS. Transformation of 9R-HPODE by AOS (ATEG_02036) is shown. B, transformation of 9R-HPODE by N964V·AOS. C, transformation of 9R-HPODE by N964D·AOS. D, oxidation of 18:2n-6 by 5,8-LDS and its Q881L mutant. Analyses were carried out by reversed phase HPLC-MS/MS. TIC, total ion current.
Three additional replacements, T957A·AOS, S959A·AOS, and Q967L·AOS, had little influence on the catalytic profile (see supplemental Fig. S7). This was unexpected inasmuch as the homologous Gln881 residue of 5,8-LDS was important for the P450 activity and for the position of ferryl oxygen insertion (see below).
The partial sequence of the I-helix of 5,8-LDS in Fig. 5 contains several polar amino acids of possible catalytic importance, and we analyzed three of them by mutagenesis (Asn878, Gln879, and Gln881). Q879L·5,8-LDS had little influence on the product profile, but replacements of Asn878 and Gln881 resulted in reduced catalytic activities and different product profiles.
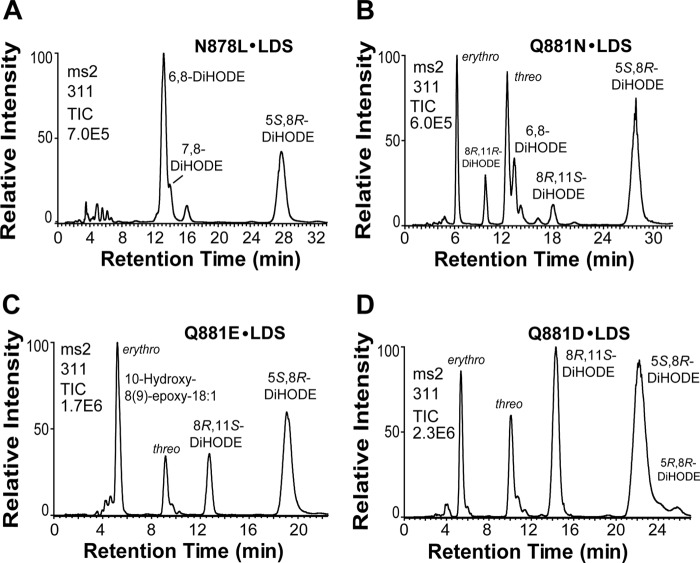
N878L shifted oxygenation from C-5 to C-6 and to C-7 as shown by NP-HPLC (Fig. 7A). The efficient heterolytic cleavage of the O–O bond was not altered as only small amounts of 10-hydroxy-8(9)-epoxy-12(Z)-octadecenoic acid were detected.
FIGURE 7.
NP-HPLC-MS/MS analysis of products formed from 18:2n-6 by four mutants of 5,8-LDS. A, N878L·5,8-LDS transformed 18:2n-6 to 6,8-DiHODE as the main metabolite and to 5,8- and 7,8-DiHODE. B, Q881N·5,8-LDS formed epoxy alcohols and both stereoisomers of 8R,11-DiHODE. C, Q881E·5,8-LDS preferentially formed the erythro isomer of the epoxy alcohol. D, Q881D·5,8-LDS augmented the biosynthesis of 8R,11S-DiHODE. TIC, total ion current of MS/MS analysis.
Replacement of Q881L·5,8-LDS almost abolished the diol synthase activity (Fig. 6D). The dramatic effect of Q881L was investigated by chain shortening of Gln to Asn and by replacement of the amide of Gln with a carboxyl group (Glu), with Asp, and with an amine (Lys) (Fig. 7).
Q881N·5,8-LDS mainly augmented formation of epoxy alcohols and 6,8-DiHODE (Fig. 7B). Q881E·5,8-LDS augmented the biosynthesis of 10-hydroxy-8(9)-epoxy-18:1 (with erythro and threo isomers in a ratio of 4:1) and 8,11-DiHODE (Fig. 7C). The increased biosynthesis of 8,11-DiHODE was even more marked in Q881D·5,8-LDS (Fig. 7D). The products formed by Q881K·5,8-LDS were similar to those of Q881N·5,8-LDS but with an increased formation of 6,8- and 7,8-DiHODE (supplemental Fig. S8).
We conclude that polar substitutions of Gln881 yielded three main effects. First, the efficient heterolytic cleavage of 8R-HPODE can be partly replaced by homolytic cleavage as judged from formation of 10-hydroxy-8(9)-epoxy-12(Z)-octadecenoic acid. Second, the position of oxygen insertion was partly shifted from C-5 to C-11, C-6, and C-7. Third, Q881E formed the erythro and threo isomers of the 8(9)-epoxy alcohol in a ratio of 4:1, indicating enzymatic biosynthesis.
DISCUSSION
We have cloned and expressed the first AOS of fungal origin, which constitutes our major finding. AOS of A. terreus was located in the P450 domain of a DOX-CYP fusion protein (ATEG_02036). One enzyme with 5,8-LDS activity and two with linoleate 10R-DOX activities were also expressed. Site-directed mutagenesis of the I-helices of AOS and 5,8-LDS revealed catalytically important residues for homolytic and heterolytic scission of the O–O bond and for subsequent dehydration or hydroxylation (Figs. 8 and 9).
FIGURE 8.
Schematic view of the homolytic and heterolytic scissions of dioxygen bonds of 9R- and 8R-HPODE in reaction mechanisms of AOS and 5,8-LDS. The Asn964 residue of AOS may facilitate catalysis by the same mechanism as Asn321 of CYP74A (47). Gln881 of LDS may facilitate charge separation and transfer of hydrogen and participate in hydrogen bonding.
FIGURE 9.
Overview of effects by replacement of Asn and Gln residues in the I-helices of AOS and 5,8-LDS of A. terreus. The two residues are present in the sequence Asn-(Val/Gln)-(Leu/Gly)-Gln (Fig. 5). The native catalytic activities of AOS and 5,8-LDS and the relative changes caused by the mutations are schematically graded from +++ to − for simplicity.
AOS
The amino acid sequence of AOS of A. terreus can be aligned with less than 25% amino acid identities to plant CYP74A and CYP8A1 but with ∼38% identity to 5,8-LDS. It lacks the characteristic insert of nine amino acids in the con-served heme binding sequence of CYP74A (Fig. 2B) (8, 15). Site-directed mutagenesis revealed that the heme thiolate ligand (Cys1073) was absolutely required for AOS catalysis.
The I-helices of AOS, 5,8-LDS, CYP74A, and CYP8A1 can be aligned with a conserved Asn residue (Fig. 5). The effects of site-directed mutagenesis of AOS and 5,8-LDS are summarized in Fig. 9.
Replacement of Asn964 with Val or Asp residues in the Asn964-Val-Leu-Gln sequence markedly reduced the AOS activity. Asn residues of CYP8A1 and CYP74 are known to interact with the dioxygen of prostaglandin H2 and hydroperoxides as judged by three-dimensional structure analysis and site-directed mutagenesis (8, 47). Chain elongation of Asn321 of CYP74A to Gln and replacement with Ala markedly reduced the AOS activity (8). Asn321 of CYP74A has been suggested to form hydrogen bonds with the hydroperoxide and facilitate homolytic cleavage (8). This mechanism could be conserved in the fungal AOS. The activity of N964D·AOS was reduced and formed only traces of 9(10)-EODE (Figs. 6 and 9 and supplemental Fig. S7). Thus, the amide group of Asn964 of the fungal AOS was not absolutely required for catalysis in analogy with Asn321 of CYP74A.
AOS of the CYP74 family can metabolize 9- and/or 13-hydroperoxides of 18:2n-6 and 18:3n-3 (15). The transformation of 13S-hydroperoxyoctadecatrienoic acid to an allene oxide is the first step in the sequential biosynthesis of JA, including transformation by allene-oxide cyclase and β-oxidation (18). In contrast, the fungal biosynthesis of JA remains enigmatic.
The tropical plant pathogen L. theobromae and a few other fungi secrete JAs (25, 26, 48). A key intermediate in plant JA biosynthesis, 12-oxo-10,15(Z)-phytodienoic acid, was recently identified in the culture medium of L. theobromae (49). Plant and fungi also form JA with identical absolute configuration (25, 48, 50). The pathway to JA described in plants seems therefore likely, but key enzymes have not been detected in vitro (27). L. theobromae expresses 5,8-LDS and 9R-DOX linked to AOS in analogy with A. terreus (27). The genome of L. theobromae has not yet been sequenced; therefore, it is unknown whether it contains a homologue of AOS of A. terreus. Homologues with 64–79% amino acid identities to ATEG_02036 are present in Aspergillus niger (see Fig. 2), Aspergillus flavus, Aspergillus oryzae, and Aspergillus kawachii, but the Asn964-Val-Leu-Gln sequence of AOS of A. terreus (Fig. 5) is modified in the position homologous to Asp-Val-Leu-(Gln/Asn) in these proteins. Their catalytic activities remain to be determined.
5,8-LDS
The biological function of oxylipins produced by 5,8-LDS is well established in A. nidulans (7). The first step in biosynthesis of 5,8-DiHODE from 8R-HPODE is heterolytic cleavage of the dioxygen bond. In peroxidases, the distal histidine and an arginine residue are important for heterolysis of peroxides (51). P450cam and over 80% of human P450 contain the conserved sequence (Ala/Gly)-Gly-Xaa-Asp-Thr (12). The two latter residues are known as the “acid-alcohol” pair (12). This pair participates in transfer of hydrogen and in hydrogen bond networks, which facilitate heterolysis of the dioxygen bond (12).
Homolytic cleavage of 8-HPODE with formation of epoxy alcohols can be catalyzed non-enzymatically and to equal amounts of erythro and threo isomers (52). In contrast, P450 transform hydroperoxides after homolytic cleavage to epoxy alcohols with stereospecificity (52). N878L·5,8-LDS altered the substrate position for hydroxylation, and replacements of Gln881 partly shifted the heterolytic scission of the O–O bond to homolytic as summarized in Fig. 9 with formation of erythro and threo isomers of 10-hydroxy-8(9)-epoxy-12(Z)-octadecenoic acid in variable ratios. Gln881 of 5,8-LDS in the Asn-Gln-Gly-Gln881 sequence may have a function similar to that of the acid-alcohol pair Asp-Thr (12, 51).
9R-DOX
9-LOX participates in plant bacterial defense (5), and 9R-DOX of A. terreus may have a similar function in addition to biosynthesis of 9(10)-EODE.
LOX with catalytic iron and DOX of the peroxidase family catalyze antarafacial hydrogen abstraction, and their D-KIEs for hydrogen abstraction at C-11 of 18:2n-6 differ by 1 order of magnitude (43–45). The D-KIE of 9R-DOX was low, consistent with oxidation by a tyrosyl radical and not by a LOX. Furthermore, the genome of A. terreus lacks LOX homologues. Therefore, we expected to find the 9R-DOX activity in one of the expressed DOX, but this was not the case. 9R-DOX catalyzes suprafacial hydrogen abstraction and oxygenation (28) and differs from 5,8-LDS, 10R-DOX, and COX-1 in this respect. 9R-DOX could nevertheless belong to the same DOX family as 8R- and 10R-DOX inasmuch as minor steric changes can alter their product profiles. 8R-DOX of A. fumigatus oxygenates the 9-trans-12-cis isomer of 18:2 by hydrogen abstraction at C-11 and oxygenation at C-9 to 9R-HPODE as the main metabolite (44), and replacement of one amino acid of 10R-DOX shifts oxygenation from C-10 to C-8 (37). In addition, the suprafacial hydrogen abstraction and oxygenation by 9S-MnLOX can be altered by a single amino acid replacement (53).
In conclusion, allene oxides of A. terreus are formed from exogenous 9R-HPODE by the P450 domain of a fusion protein of the DOX-CYP family. The reaction mechanism is closely related to plant AOS with homolytic cleavage of 9-HPODE and a catalytically important Asn residue in the heme environment. In analogy, a Gln residue of 5,8-LDS appears to facilitate heterolytic scission of 8R-HPODE and subsequent hydroxylation.
Supplementary Material
Acknowledgments
We thank Professor Mats Hamberg, the Karolinska Institute, for unfailing cooperation and the generous gift of deuterated linoleic acids. The contributions and assistance of Jordi Jacas i Mateu, Mireia Mensa Vendrell, and Anna Knecht during gene cloning and expression of putative enzymes are gratefully acknowledged.
This work was supported by Vetenskapsrådet Grant 03X-06523, Knut and Alice Wallenberg Foundation Grant KAW 2004.123, and Uppsala University.
- COX
- cyclooxygenase(s)
- AOS
- allene oxide synthase(s)
- CT
- cycle threshold
- CYP
- cytochrome P450
- CYP8A1
- human prostacyclin synthase
- CYP74A
- allene oxide synthase of A. thaliana
- D-KIE
- deuterium kinetic isotope effect
- DiHODE
- dihydroxyoctadecadienoic acid
- DOX
- dioxygenase(s)
- 9(10)-EODE
- 9(10)-epoxy-10,12(Z)-octadecadienoic acid
- HPODE
- hydroperoxyoctadecadienoic acid
- HPOME
- hydroperoxyoctadecenoic acid
- JA
- jasmonic acid
- LDS
- linoleate diol synthase(s)
- LOX
- lipoxygenase(s)
- NP
- normal phase
- P450
- cytochrome(s) P450.
REFERENCES
- 1. Smith W. L., Urade Y., Jakobsson P. J. (2011) Enzymes of the cyclooxygenase pathways of prostanoid biosynthesis. Chem. Rev. 111, 5821–5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Haeggström J. Z., Funk C. D. (2011) Lipoxygenase and leukotriene pathways: biochemistry, biology, and roles in disease. Chem. Rev. 111, 5866–5898 [DOI] [PubMed] [Google Scholar]
- 3. Andreou A., Feussner I. (2009) Lipoxygenases—structure and reaction mechanism. Phytochemistry 70, 1504–1510 [DOI] [PubMed] [Google Scholar]
- 4. Imig J. D. (2012) Epoxides and soluble epoxide hydrolase in cardiovascular physiology. Physiol. Rev. 92, 101–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vicente J., Cascón T., Vicedo B., García-Agustín P., Hamberg M., Castresana C. (2012) Role of 9-lipoxygenase and α-dioxygenase oxylipin pathways as modulators of local and systemic defense. Mol. Plant 5, 914–928 [DOI] [PubMed] [Google Scholar]
- 6. Hamberg M., Ponce de Leon I., Rodriguez M. J., Castresana C. (2005) α-Dioxygenases. Biochem. Biophys. Res. Commun. 338, 169–174 [DOI] [PubMed] [Google Scholar]
- 7. Brodhun F., Feussner I. (2011) Oxylipins in fungi. FEBS J. 278, 1047–1063 [DOI] [PubMed] [Google Scholar]
- 8. Lee D. S., Nioche P., Hamberg M., Raman C. S. (2008) Structural insights into the evolutionary paths of oxylipin biosynthetic enzymes. Nature 455, 363–368 [DOI] [PubMed] [Google Scholar]
- 9. Christensen S. A., Kolomiets M. V. (2011) The lipid language of plant-fungal interactions. Fungal Genet. Biol. 48, 4–14 [DOI] [PubMed] [Google Scholar]
- 10. Yin G. C. (2010) Active transition metal oxo and hydroxo moieties in nature's redox, enzymes and their synthetic models: structure and reactivity relationships. Coord. Chem. Rev. 254, 1826–1842 [Google Scholar]
- 11. Werck-Reichhart D., Feyereisen R. (2000) Cytochromes P450: a success story. Genome Biol. 1, REVIEWS3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Denisov I. G., Makris T. M., Sligar S. G., Schlichting I. (2005) Structure and chemistry of cytochrome P450. Chem. Rev. 105, 2253–2277 [DOI] [PubMed] [Google Scholar]
- 13. Ullrich V., Brugger R. (1994) Prostacyclin and thromboxane synthase—new aspects of hemethiolate catalysis. Angew. Chem. Int. Ed. Engl. 33, 1911–1919 [Google Scholar]
- 14. Song W. C., Funk C. D., Brash A. R. (1993) Molecular cloning of an allene oxide synthase: a cytochrome P450 specialized for the metabolism of fatty acid hydroperoxides. Proc. Natl. Acad. Sci. U.S.A. 90, 8519–8523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brash A. R. (2009) Mechanistic aspects of CYP74 allene oxide synthases and related cytochrome P450 enzymes. Phytochemistry 70, 1522–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brodowsky I. D., Hamberg M., Oliw E. H. (1992) A linoleic acid (8R)-dioxygenase and hydroperoxide isomerase of the fungus Gaeumannomyces graminis. Biosynthesis of (8R)-hydroxylinoleic acid and (7S,8S)-dihydroxylinoleic acid from (8R)-hydroperoxylinoleic acid. J. Biol. Chem. 267, 14738–14745 [PubMed] [Google Scholar]
- 17. Brodhun F., Göbel C., Hornung E., Feussner I. (2009) Identification of PpoA from Aspergillus nidulans as a fusion protein of a fatty acid heme dioxygenase/peroxidase and a cytochrome P450. J. Biol. Chem. 284, 11792–11805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wasternack C. (2007) Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 100, 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hamberg M., Zhang L. Y., Brodowsky I. D., Oliw E. H. (1994) Sequential oxygenation of linoleic acid in the fungus Gaeumannomyces graminis: stereochemistry of dioxygenase and hydroperoxide isomerase reactions. Arch. Biochem. Biophys. 309, 77–80 [DOI] [PubMed] [Google Scholar]
- 20. Garscha U., Jernerén F., Chung D., Keller N. P., Hamberg M., Oliw E. H. (2007) Identification of dioxygenases required for Aspergillus development. Studies of products, stereochemistry, and the reaction mechanism. J. Biol. Chem. 282, 34707–34718 [DOI] [PubMed] [Google Scholar]
- 21. Rittle J., Green M. T. (2010) Cytochrome P450 compound I: capture, characterization, and C-H bond activation kinetics. Science 330, 933–937 [DOI] [PubMed] [Google Scholar]
- 22. Champe S. P., el-Zayat A. A. (1989) Isolation of a sexual sporulation hormone from Aspergillus nidulans. J. Bacteriol. 171, 3982–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Peng Y. L., Shirano Y., Ohta H., Hibino T., Tanaka K., Shibata D. (1994) A novel lipoxygenase from rice. Primary structure and specific expression upon incompatible infection with rice blast fungus. J. Biol. Chem. 269, 3755–3761 [PubMed] [Google Scholar]
- 24. Wennman A., Oliw E. H. (2013) Secretion of two novel enzymes, manganese 9S-lipoxygenase and epoxy alcohol synthase, by the rice pathogen Magnaporthe salvinii. J. Lipid Res. 54, 762–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aldridge D., Galt S., Giles D., Turner W. (1971) Metabolites of Lasiodiplodia theobromae. J. Chem. Soc. C 1623–1627 [Google Scholar]
- 26. Cross B. E., Webster G. R. B. (1970) New metabolites of Gibberella fujikuroi. J. Chem. Soc. C 1838–1842 [DOI] [PubMed] [Google Scholar]
- 27. Jernerén F., Eng F., Hamberg M., Oliw E. H. (2012) Linolenate 9R-dioxygenase and allene oxide synthase activities of Lasiodiplodia theobromae. Lipids 47, 65–73 [DOI] [PubMed] [Google Scholar]
- 28. Jernerén F., Hoffmann I., Oliw E. H. (2010) Linoleate 9R-dioxygenase and allene oxide synthase activities of Aspergillus terreus. Arch. Biochem. Biophys. 495, 67–73; Correction (2010) Arch. Biochem. Biophys.500, 210 [DOI] [PubMed] [Google Scholar]
- 29. Endo A. (1992) The discovery and development of HMG-CoA reductase inhibitors. J. Lipid Res. 33, 1569–1582 [PubMed] [Google Scholar]
- 30. Sanchez J. F., Somoza A. D., Keller N. P., Wang C. C. (2012) Advances in Aspergillus secondary metabolite research in the post-genomic era. Nat. Prod. Rep. 29, 351–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Andersen M. R., Nielsen J. (2009) Current status of systems biology in aspergilli. Fungal Genet. Biol. 46, S180–S190 [DOI] [PubMed] [Google Scholar]
- 32. Nazmul Hussain Nazir K. H., Ichinose H., Wariishi H. (2010) Molecular characterization and isolation of cytochrome P450 genes from the filamentous fungus Aspergillus oryzae. Arch. Microbiol. 192, 395–408 [DOI] [PubMed] [Google Scholar]
- 33. Hoffmann I., Jernerén F., Garscha U., Oliw E. H. (2011) Expression of 5,8-LDS of Aspergillus fumigatus and its dioxygenase domain. A comparison with 7,8-LDS, 10-dioxygenase, and cyclooxygenase. Arch. Biochem. Biophys. 506, 216–222 [DOI] [PubMed] [Google Scholar]
- 34. Su C., Sahlin M., Oliw E. H. (1998) A protein radical and ferryl intermediates are generated by linoleate diol synthase, a ferric hemeprotein with dioxygenase and hydroperoxide isomerase activities. J. Biol. Chem. 273, 20744–20751 [DOI] [PubMed] [Google Scholar]
- 35. Oliw E. H., Wennman A., Hoffmann I., Garscha U., Hamberg M., Jernerén F. (2011) Stereoselective oxidation of regioisomeric octadecenoic acids by fatty acid dioxygenases. J. Lipid Res. 52, 1995–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zain M. E., Razak A. A., El-Sheikh H. H., Soliman H. G., Khalil A. M. (2009) Influence of growth medium on diagnostic characters of Aspergillus and Penicillium species. Afr. J. Microbiol. Res. 3, 280–286 [Google Scholar]
- 37. Garscha U., Oliw E. H. (2009) Leucine/valine residues direct oxygenation of linoleic acid by (10R)- and (8R)-dioxygenases: expression and site-directed mutagenesis of (10R)-dioxygenase with epoxyalcohol synthase activity. J. Biol. Chem. 284, 13755–13765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ramakers C., Ruijter J. M., Deprez R. H., Moorman A. F. (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339, 62–66 [DOI] [PubMed] [Google Scholar]
- 39. Tamura K., Dudley J., Nei M., Kumar S. (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 [DOI] [PubMed] [Google Scholar]
- 40. Bligh E. G., Dyer W. J. (1959) A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 41. Nelson D. R. (2009) The cytochrome p450 homepage. Hum. Genomics 4, 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brodhun F., Schneider S., Göbel C., Hornung E., Feussner I. (2010) PpoC from Aspergillus nidulans is a fusion protein with one active heme. Biochem. J. 425, 553–565 [DOI] [PubMed] [Google Scholar]
- 43. Klinman J. P. (2006) Linking protein structure and dynamics to catalysis: the role of hydrogen tunnelling. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361, 1323–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hoffmann I., Hamberg M., Lindh R., Oliw E. H. (2012) Novel insights into cyclooxygenases, linoleate diol synthases, and lipoxygenases from deuterium kinetic isotope effects and oxidation of substrate analogs. Biochim. Biophys. Acta 1821, 1508–1517 [DOI] [PubMed] [Google Scholar]
- 45. Wu G., Lü J. M., van der Donk W. A., Kulmacz R. J., Tsai A. L. (2011) Cyclooxygenase reaction mechanism of prostaglandin H synthase from deuterium kinetic isotope effects. J. Inorg. Biochem. 105, 382–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jernerén F., Sesma A., Franceschetti M., Hamberg M., Oliw E. H. (2010) Gene deletion of 7,8-linoleate diol synthase of the rice blast fungus: studies on pathogenicity, stereochemistry, and oxygenation mechanisms. J. Biol. Chem. 285, 5308–5316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chiang C. W., Yeh H. C., Wang L. H., Chan N. L. (2006) Crystal structure of the human prostacyclin synthase. J. Mol. Biol. 364, 266–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Miersch O., Bohlmann H., Wasternack C. (1999) Jasmonates and related compounds from Fusarium oxysporum. Phytochemistry 50, 517–523 [Google Scholar]
- 49. Tsukada K., Takahashi K., Nabeta K. (2010) Biosynthesis of jasmonic acid in a plant pathogenic fungus, Lasiodiplodia theobromae. Phytochemistry 71, 2019–2023 [DOI] [PubMed] [Google Scholar]
- 50. Miersch O., Preiss A., Sembdner G., Schreiber K. (1987) (+)-7-Iso-jasmonic acid and related compounds from Botryodiplodia theobromae. Phytochemistry 26, 1037–1039 [Google Scholar]
- 51. Poulos T. L. (2010) Thirty years of heme peroxidase structural biology. Arch. Biochem. Biophys. 500, 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chang M. S., Boeglin W. E., Guengerich F. P., Brash A. R. (1996) Cytochrome P450-dependent transformations of 15R- and 15S-hydroperoxyeicosatetraenoic acids: stereoselective formation of epoxy alcohol products. Biochemistry 35, 464–471 [DOI] [PubMed] [Google Scholar]
- 53. Wennman A., Jernerén F., Hamberg M., Oliw E. H. (2012) Catalytic convergence of Mn- and Fe-lipoxygenases by replacement of a single amino acid. J. Biol. Chem. 287, 31757–31765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jernerén F., Garscha U., Hoffmann I., Hamberg M., Oliw E. H. (2010) Reaction mechanism of 5,8-linoleate diol synthase, 10R-dioxygenase and 8,11-hydroperoxide isomerase of Aspergillus clavatus. Biochim. Biophys. Acta 1801, 503–507 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.