Abstract
Previous work has implicated heat shock transcription factor 1 (HSF1) as the primary transcription factor responsible for the transcriptional response to heat stress in mammalian cells. We characterized the heat shock response of mammalian cells by measuring changes in transcript levels and assaying binding of HSF1 to promoter regions for candidate heat shock genes chosen by a combination of genome-wide computational and experimental methods. We found that many heat-inducible genes have HSF1 binding sites (heat shock elements, HSEs) in their promoters that are bound by HSF1. Surprisingly, for 24 heat-inducible genes, we detected no HSEs and no HSF1 binding. Furthermore, of 182 promoters with likely HSE sequences, we detected HSF1 binding at only 94 of these promoters. Also unexpectedly, we found 48 genes with HSEs in their promoters that are bound by HSF1 but that nevertheless did not show induction after heat shock in the cell types we examined. We also studied the transcriptional response to heat shock in fibroblasts from mice lacking the HSF1 gene. We found 36 genes in these cells that are induced by heat as well as they are in wild-type cells. These results provide evidence that HSF1 does not regulate the induction of every transcript that accumulates after heat shock, and our results suggest that an independent posttranscriptional mechanism regulates the accumulation of a significant number of transcripts.
INTRODUCTION
The heat shock response was first described in 1962 as a puffing pattern on Drosophila polytene chromosomes after thermal stress (Ritossa, 1962). Since then, studies of individual genes have shown that the cellular heat shock response is conserved across kingdoms and is characterized by the strong induction of numerous heat shock proteins (HSPs), many of which are chaperone proteins that assist in protein folding.
The heat shock transcription factor (HSF) transcriptionally regulates the induction of many HSPs in Drosophila (Clos et al., 1990) and Saccharomyces (Sorger and Pelham, 1987; Wiederrecht et al., 1988). HSF binds to a DNA sequence motif, the heat shock element (HSE), which is characterized by an array of inverted repeats of the motif nGAAn. Copies of the HSE are found in the promoters of genes encoding several known heat-inducible proteins, including the Drosophila and human hsp70 genes (Sarge et al., 1993; Pirkkala et al., 2001).
Mammalian genomes encode three homologues of HSF: HSF1, HSF2, and HSF4 (reviewed in Pirkkala et al., 2001). Mammalian HSF1 is believed to be the paralog responsible for regulating the heat-induced transcriptional response in mammalian cells (Rabindran et al., 1991; Sarge et al., 1991, 1993). In support of this hypothesis, mouse HSF1 knockout fibroblasts are unable to induce expression of hsp70 in response to heat stress (McMillan et al., 1998), whereas HSF2 knockout fibroblasts induce hsp70 normally (McMillan et al., 2002). The HSF1 knockout mouse has defects in extraembryonic development and postnatal growth, suggesting that the protein is important for other processes in addition to the response to heat stress (Xiao et al., 1999).
The heat shock response is highly conserved in mammals. Although mammals maintain a uniform internal temperature, the response is likely important during fever and hyperthermia (Hasday and Singh, 2000). Expression of heat shock genes is also implicated in developmental transitions, as a mechanism for phenotypic buffering of genetic changes (Queitsch et al., 2002) and in the responses to other types of stress.
In this study, we characterized the role of HSF1 in the regulation of gene expression changes that occur in mammalian cells during heat shock. We generated a list of potential HSF1-regulated genes by using microarray experiments and genomic sequence analysis and characterized the role of HSF1 in regulating their transcripts levels after heat shock. Our results provide evidence that the regulation of the mammalian heat shock response is more complex than previously thought.
MATERIALS AND METHODS
Detailed methods are available at http://microarray-pubs.stanford.edu/HSF1/.
Microarray Data Search for Heat-induced Genes
We selected genes induced by heat shock in cDNA microarray data from three cell lines exposed to diverse stresses (Murray, Whitfield, Trinklein, Myers, Brown, and Botstein, unpublished data); we selected all genes that clustered with known heat shock genes and performed a Wilcoxon rank-sum test (Eisen and Brown, 1999) to find genes whose expression was significantly higher in heat shocked samples than in nonheat shocked samples.
Human Promoter Microarray Combined with Chromatin Immunoprecipitation (ChIP)
We made promoter microarrays with 768 putative human promoters sequences (Trinklein et al., 2003), most of which had HSE sites. We hybridized Cy3-labeled mock immunoprecipitated DNA and Cy5-labeled HSF1 Chromatin IP DNA to these arrays and selected the top 10% for confirmation by quantitative polymerase chain reaction (PCR).
ChIP
We performed chromatin IP as described previously in Trinklein et al. (2004).
Real-Time Reverse Transcription (RT)-PCR Expression Analysis
We compared mRNA from K562 human erythroleukemia cells before treatment and after 2-h recovery from 1 h of 43°C heat shock. We reverse transcribed the mRNA with Superscript reverse transcriptase (Invitrogen, Carlsbad, CA) and measured the abundance of each gene's transcript in the two samples by quantitative PCR with a Bio-Rad (Hercules, CA) Icycler. We used beta-actin and GAPDH as controls.
Real-Time PCR Analysis of ChIP Enrichment
We measured the enrichment of each promoter in the HSF1 ChIP sample relative to mock IP DNA with real-time PCR by using amplicons designed within 400 base pairs of the predicted transcription start site on a Bio-Rad Icycler. We used beta-actin, GAPDH, and histone H2A promoters as negative controls for HSF1 binding.
Determination of the HSE Position-specific Score matrix (PSSM) and Occurrence Scores
We used 280 base pairs of genomic sequence for 46 promoters enriched at least 40-fold by ChIP to create an HSE PSSM with the MEME algorithm (http://meme.sdsc.edu/). We calculated PSSM occurrence scores by multiplying each 14-base pair window sequence by the MEME-derived PSSM and summing this product across all windows on both strands of a sequence.
Luciferase-based Heat Shock Promoter Assays
We cloned 1-kb putative promoter sequences upstream of luciferase and transfected them into the cell lines HeLa, HT1080, 293, and wild-type and HSF1-/- mouse embryonic fibroblasts. We cotransfected a control plasmid (Renilla under the control of the thymidine kinase promoter) (Promega, Madison, WI) and the experimental constructs by using FuGENE6 LipofectAMINE reagent (Roche Applied Science, Indianapolis, IN) in 96-well white tissue culture plates. We measured luciferase and Renilla activity of quadruplicate transfections 24 h after transfection and either 0 or 3 h of heat shock in a 96-well luminometer (Wallac-Perkin Elmer, Boston, MA) with the dual luciferase kit (Promega). The heat-induced activity of each promoter was found by comparing the average luciferase to Renilla ratio in heat shock and control samples.
Mouse Microarray Gene Expression Measurements
We prepared mRNA from heat shocked or untreated HSF1+/+ or HSF1-/- mouse embryonic fibroblasts by using the FastTrack 2.0 kit (Invitrogen) and analyzed using mouse full-genome cDNA microarrays (Stanford Functional Genomics Facility: http://www.microarray.org/sfgf/) as described previously (Troyanskaya et al., 2002), by using a reference consisting of combined mRNA from heat shocked and untreated wild-type fibroblasts. Mouse homologues of human genes were defined through Homologene (http://www.ncbi.nlm.nih.gov/HomoloGene/).
RESULTS
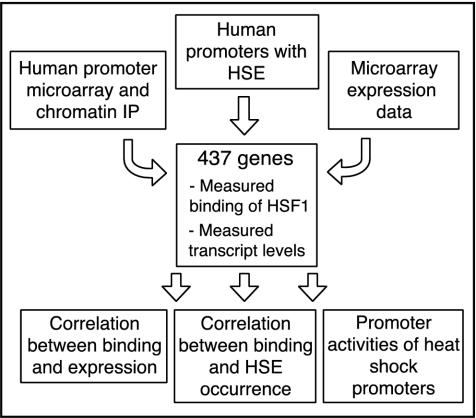
We used three independent approaches to generate a list of 437 genes potentially involved in the heat shock response. First, we searched a database of >20,000 putative human promoters (Trinklein et al., 2003) for occurrences of the HSE. Second, we made human promoter microarrays containing 768 putative promoters, many of which contained sequences similar to the previously defined heat shock element and used these microarrays to detect promoters enriched by ChIP with an HSF1 antibody after heat shock. Finally, we searched a microarray data set describing the changes in gene expression in human cell lines resulting from multiple types of stress for genes induced specifically by heat (Murray, Whitfield, Trinklein, Myers, Brown, and Botstein, unpublished data). For genes selected in the promoter microarray and expression microarray experiments, we did not consider a gene further if we did not confirm significant ChIP enrichment or heat induction, respectively, in our quantitative PCR assay. For the 176 remaining genes for which we did confirm ChIP enrichment or heat induction, we measured transcript levels before and after heat shock by using quantitative RT-PCR, and we measured HSF1 binding by chromatin IP (outlined in Figure 1). The data for all candidate genes is available as Supplemental Table 1.
Figure 1.
Methods for detection of potential HSF1 targets. 437 genes were selected for study based on either enrichment of their promoter in HSF1 chromatin IP as detected on a human promoter microarray containing 768 genes, presence of a sequence with high similarity to the previously described HSE in their promoter, or significant induction in heat shock microarray experiments. HSF1 binding was measured by chromatin IP followed by quantitative PCR and expression by quantitative RT-PCR for each gene, and these data was used to classify genes into HSF1-dependent and -independent as well as heat-inducible and nonheat-inducible subsets. We calculated an optimized HSF1 binding motif based on the promoters that showed HSF1 binding. Finally, we tested promoter sequences of all classes for ability to confer heat-induced expression by using a luciferase reporter system.
Detection of New Heat-inducible Genes by Expression Analysis
Based on a bimodal distribution formed by the RT-PCR gene expression levels (see Supplemental Figure 6), we defined a gene as heat-induced if we measured induction >1.95-fold. Based on this threshold, 93 genes were induced after 1 h of heat shock, with induction levels ranging from 2-fold to 90-fold. Of these genes, 35 encode proteins of unknown function. The heat-induced genes included many genes already known to encode heat shock proteins and other proteins involved in protein folding and degradative pathways (Trinklein et al., 2004), including hsp27, hsp40, hsp60, hsp70, hsp105, hsp110, and members of the Crystallin family. However, the majority of the 58 previously characterized genes were not previously known to be induced by heat shock (see Supplemental Table 1).
Discovery of New HSF1-binding Sites by Chromatin Immunoprecipitation
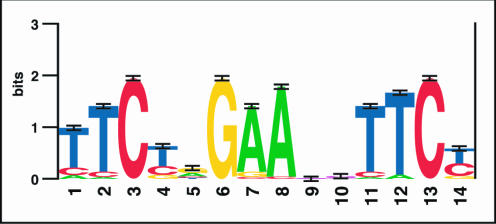
We used chromatin IP to enrich for sequences bound by HSF1 in heat-shocked cells and measured the enrichment of the predicted promoters of our candidate genes with quantitative PCR. We identified 94 genes with an HSF1 ChIP enrichment of fivefold or greater. We categorized these as the genes with HSF1 bound at their promoter based on a bimodal distribution of all ChIP enrichment values (see Supplemental Figure 7). This collection of HSF1-bound sites increases the number of known HSF1-bound sites in the genome by an order of magnitude. We used this large set of new sites to improve our understanding of the HSE. We chose 46 promoters with HSF1-ChIP enrichments of at least 40-fold to define a new HSE consensus sequence. We chose only these highly enriched sequences because the ChIP enrichment is highest for promoters where HSF1 binds closest to the PCR primers used to measure the enrichment (Trinklein et al., 2004). We used MEME, a finite mixture-modeling algorithm (Bailey and Elkan, 1994), to search for overrepresented motifs in 280-base pair segments surrounding the primers we used in detecting enrichment for these 46 promoters. MEME detected an enriched sequence motif similar to the previously defined HSE and refined it in several ways. A graphical representation of the PSSM for this motif is shown in Figure 2. Previously, the HSE consensus has been described as inverted repeats of the motif nGAAn (Sarge et al., 1993; Pirkkala et al., 2001). Our results support the previous finding that the “G” in nGAAn is the most conserved position in the HSE (Xiao and Lis, 1988). Also, in a pair of inverted repeats, a TTC triplet 5′ of a GAA triplet is separated by a pyrimidine-purine dinucleotide, but the two nucleotides separating a GAA triplet 5′ from a TTC triplet seem to be unconstrained.
Figure 2.
Visual representation of the position specific scoring matrix for the newly derived HSE. Overrepresented HSE sequence motifs were identified and aligned by MEME, and the logo-gram was generated at http://www.bio.cam.ac.uk/seqlogo/.
Newly Derived HSE PSSM Improves Prediction of HSF1 Binding
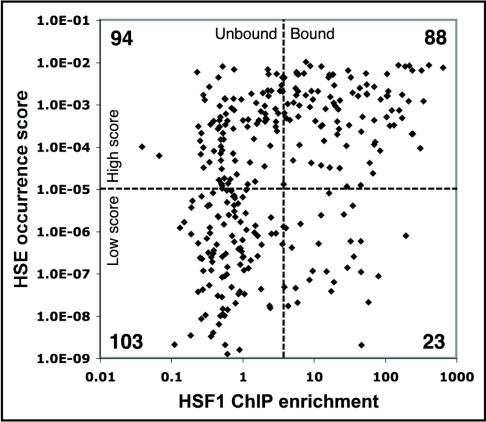
We were interested in determining whether this PSSM could predict HSF1 binding. We used the PSSM to calculate an occurrence score for 2 kb of sequence surrounding the 308 promoters for which we assayed HSF1 binding (see MATERIALS AND METHODS). We classified promoters with scores above 10-5 as high scoring and promoters with scores below 10-5 as low scoring (for the distribution of scores, see Supplemental Figure 8). Of the high-scoring promoters, 48% (88/182) had a ChIP enrichment greater than fivefold and 19% (35/182) had an enrichment >50-fold. Conversely, 18% (23/126) of the low-scoring promoters had a ChIP enrichment of fivefold or greater and only 2% (3/126) had a ChIP enrichment >50-fold; therefore, not all HSF1 bound promoters had a HSE (HSE sequences shown in Supplemental Table 2). Thus, a high HSE PSSM score gives roughly a 50% chance of predicting HSF1 binding, and a low score gives an 82% chance that HSF1 does not bind to that location (Figure 3).
Figure 3.
Relationship between HSE occurrence score and HSF1 binding. The y-axis of the scatter plot is the HSE occurrence score for each promoter as described in the methods. The x-axis is the HSF1 ChIP enrichment determined by quantitative PCR.
We compared the ability of the PSSM to predict HSF1 binding to a simple pattern match with the inverted nGAAn repeats previously used to define the HSE. Two tandem copies of nGAAn were present in 88% of the promoter sequences irrespective of HSF1 binding. When we searched the 2-kb promoter sequences for three copies of the repeat (GAAnnTTCnnGAA), 43% (61/143) had a ChIP enrichment greater than fivefold and 17% (24/143) had an enrichment >50-fold. Conversely, 30% (49/165) of the promoter sequences that did not contain GAAnnTTCnnGAA had a ChIP enrichment of fivefold or greater and 8% (14/165) had a ChIP enrichment >50-fold. Thus, this new scoring matrix predicts HSF1 binding with greater sensitivity and specificity than a pattern match with the canonical motif.
Heat-induced Expression Predicts HSF1 Binding but HSF1 Binding Is Not Sufficient for Induction
We examined the relationship between HSF1-binding to a gene's promoter and the heat-induced expression of that gene. Of the 176 genes for which we measured both binding and expression, 46 were bound and induced, 48 were bound but not induced, 24 were induced but not bound and 58 were not bound or induced (see Supplemental Table 2). Thus, the majority of genes induced by heat shock bound HSF1 at their promoters (46/70 or 65%). The induction of 24 genes for which we did not detect HSF1 binding can be explained either by the presence of binding at sites outside the region we tested, or by the use of an HSF1-independent induction mechanism. We searched for HSE sequences up to 6 kb upstream from all 24 of these genes, and we also assayed for binding further upstream and in the first intron of the DnaJ.B4 and DnaJ.C3 genes. We found no HSEs at any of these genes, and we detected no binding upstream or in the first introns of DnaJ.B4 and DnaJ.C3. Only 48% (46/94) of the genes bound by HSF1 were induced by heat shock. This is consistent with previous finding in Drosophila where it was observed that HSF bound to >100 sites in polytene chromosomes that were not associated with heat induced expression (Westwood et al., 1991). Therefore, our results support the hypothesis that HSF1 binds to many genomic targets without inducing and maybe even repressing transcription.
Microarray Expression Analysis of Heat-shocked HSF1-/- and HSF1+/+ Mouse Embryonic Fibroblasts Identifies HSF1-dependent and HSF1-independent Heat-inducible Genes
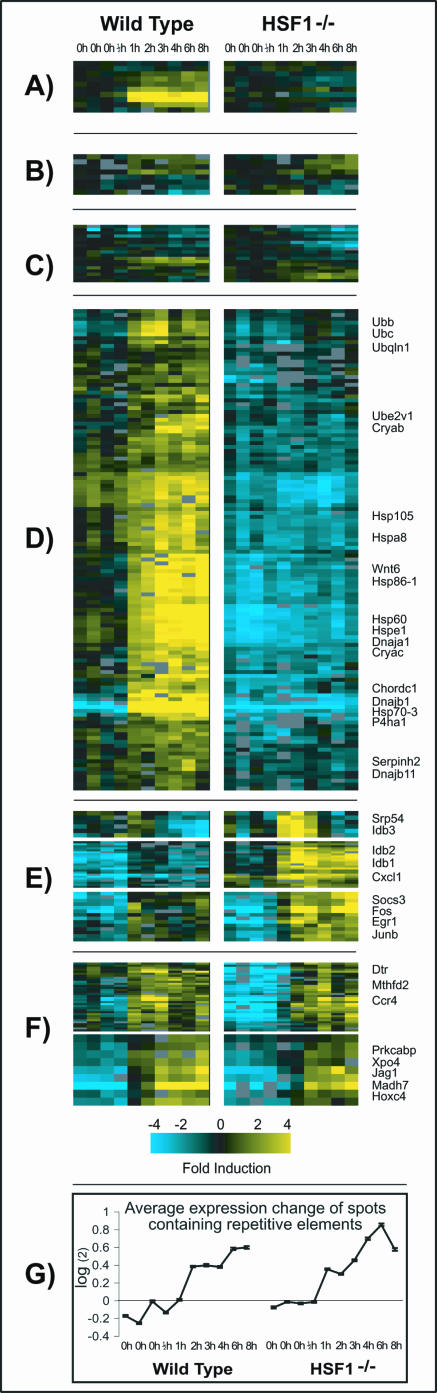
The fact that some genes induced by heat shock apparently do not bind HSF1 suggests an HSF1-independent mechanism of mRNA accumulation after heat shock. To address this possibility, we measured mRNA levels in heat shocked embryonic fibroblasts from wild-type mice and from mice that are missing HSF1 due to a homozygous deletion (HSF1-/-) after 0, 0.5, 1, 2, 3, 4, 6, and 8 h of heat shock with full-genome mouse cDNA microarrays. The data were centered, which allows visualization of differences in gene expression changes between cell lines as well as differences in basal expression during the heat shock time course. The complete clustered microarray data is shown in Supplemental Figure 9, and a summary of the data is shown in Figure 4. Supplemental Figure 10 shows the same genes transformed to show changes relative to untreated samples (eliminating the basal expression).
Figure 4.
Cluster analysis of changes in mRNA levels in heat shocked wild-type and HSF1-/- mouse fibroblasts. The expression changes in both cell types are shown for homologues of a subset of human genes that were HSF1-bound and induced (A), homologues that were bound by HSF1 but not heat induced (B), and homologues that were induced but not bound by HSF1 in the human system (C). The mouse microarray data were also analyzed independent of the human experiments, and these results show genes induced by heat in wild-type but not HSF1-/- fibroblasts (D), genes induced in HSF1-/- fibroblasts (E), and genes induced similarly in wild-type and HSF1-/- fibroblasts (F). (G) Average expression profile for spots containing repetitive elements according to RepeatMasker and the SE of each time point for each cell type.
We extracted the expression data of mouse genes homologous to human genes that we had found to be induced but not bound, bound but not induced, or bound and induced according to Homologene (http://www.ncbi.nlm.nih.gov/HomoloGene/). Homologues of genes that were both bound and induced were induced in the wild-type but not the HSF1-/- fibroblasts (Figure 4A). We observed no induction of transcripts for mouse homologues of human genes for which we detected HSF1 binding but not heat induction (Figure 4B). The results were more variable for homologues of genes that were induced but not bound (Figure 4C). Some of these genes were not induced in the mouse microarray experiments, others were induced in the wild-type cells but not the mutant cells, and some were induced in both wild-type and mutant cells.
In addition to looking at the mouse orthologues of the new human heat shock genes we identified, we looked for novel expression patterns by analyzing the full mouse microarray data set with hierarchical clustering (Eisen et al., 1998). As expected, the expression of a cluster of genes, including many known heat-inducible chaperones, was induced strongly by heat in wild-type cells (Figure 4D). Most of these genes showed no significant changes after heat shock in the knockout; however, a few had subtly increased mRNA levels after heat shock. For hsp70, it has previously been shown that increased mRNA stability also plays a role in heat-induced transcript accumulation (DiDomenico et al., 1982; Theodorakis and Morimoto, 1987; Kaarniranta et al., 1998). Also, the basal expression levels of several of these HSF1-dependent heat-inducible genes were higher in the HSF1 wild-type cells compared with the HSF1 knockout cells. The most likely explanation is that a small fraction of cells in a given culture system stochastically exhibit the heat shock response, and this contributes to the basal signal seen in our zero time points. However, the possibility exists that HSF1 plays a role in the basal expression of certain heat shock genes.
However, we also found multiple clusters of genes induced at least as strongly in the HSF-/- cells as they were in the wild-type cells (Figure 4, E-G). Some genes were induced more strongly in HSF1-/- cells than in the wild-type cells, including many immediate-early genes such as JUNB, JUND, EGR1, IER5, GADD45g, and three members of the Inhibitor of DNA Binding family of transcriptional repressors. There were two clusters with a total of 36 genes that were induced similarly in both cell lines, which is consistent with our discovery of heat-inducible genes not bound by HSF1. One cluster included metabolic genes (such as methylene tetrahydrofolate dehydrogenase and carbon catabolite repressor) and signaling molecules (such as the diphtheria toxin receptor), and the second cluster had many signal transduction proteins (Jagged 1, Hoxc4, Protein kinase C alpha binding protein, and MAD homologue 7). Also, many spots containing repetitive sequences were induced with nearly identical patterns in the wild-type and HSF1-/- cells (Figure 4G). A substantial number of genes were repressed irrespective of HSF1 status, including many genes associated with growth and cell division in fibroblasts such as actin, alpha and beta tubulin, other extracellular matrix components, ribosomal proteins, and cell cycle genes. This suggests that heat-induced down-regulation of the cellular growth rate occurs independent of HSF1.
Only Promoters of Heat-inducible and HSF1-bound Genes Have Intrinsic Heat-induced Activity
To further understand why some genes were not induced by heat shock despite binding of HSF1 to their promoters, and why other genes were induced by heat shock but lacked HSF1 binding at their promoters, we created reporter vectors expressing luciferase under the control of 33 predicted promoters of genes from each class. We transfected each reporter construct into three human cell lines (HeLa, HT1080, and 293) and into wild-type and HSF1-/- mouse embryonic fibroblasts. For each cell line, we measured the heat inducibility of luciferase driven by each experimental promoter relative to that of a cotransfected control vector (containing Renilla driven by the thymidine kinase promoter). We also measured the heat inducibility of 48 random promoters (Trinklein et al., 2003) in HT1080 cells.
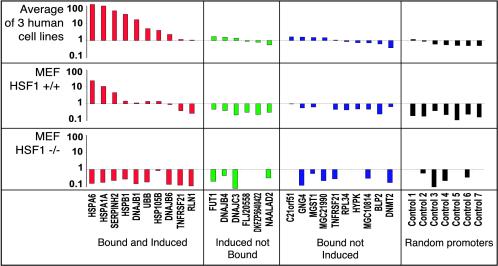
Promoters of genes whose endogenous transcripts are heat-induced and bound by HSF1 showed induced luciferase activity by as much as 60-fold after heat shock (Figure 5). The induction of luciferase activity by these transfected promoters was comparable with the previously observed induction of the endogenous genes corresponding to these promoters, and did not occur in HSF1-/- fibroblasts, confirming that the induction is HSF1 dependent. In contrast, 48 random human promoters showed an average 5.4-fold repression of luciferase activity after heat shock (see Supplemental Figure 11), which suggests that there is a general down-regulation of transcription or translation during heat shock. This occurred in the HSF1-/- fibroblasts and thus is not dependent on HSF1.
Figure 5.
Promoter activities before and after heat shock in three different human cell lines and HSF1+/+ mouse fibroblasts and HSF1-/- mouse fibroblasts. The average heat-induced promoter activity are shown for the three human cell lines, wild-type mouse fibroblasts, and mouse HSF1-/- fibroblasts for 10 promoters defined as HSF1 bound and heat induced (red), seven promoters from genes that were induced but not bound by HSF1 (green), seven promoters defined as HSF1-bound but not induced (blue), and seven random promoters (black).
In contrast, promoters bound by HSF1 where the endogenous gene was not heat induced showed no heat-inducible luciferase activity, and promoters not bound by HSF1 where the expression of the endogenous gene was heat induced were also not able to induce luciferase activity (Figure 5).
DISCUSSION
Implications of HSF1 Binding without Transcript Induction
Although much of the heat-induced gene expression can be explained by HSF1 binding, we did not find perfect correlation between binding and induction. There are several possible explanations for why one-half of the HSF1-bound promoters are not induced by heat. Information intrinsic to the 1-kb putative promoter sequence might somehow distinguish among sites that should cause inductions and sites that should not. For instance, additional positive or negative factors may modulate the transcriptional induction of HSF1-bound genes, However, the identity of such factors is unknown. Alternatively, features of the binding site may directly modulate the activity of bound HSF1. This hypothesis implies a two-step process where HSF1 binds to an HSE and then recognizes some other structural or sequence motif before it is able to activate transcription.
Other explanations are possible but seem unlikely based on our results. One is that epigenetic modifications or structural features of the genomic context of a gene repress or activate HSF1 activity. Because promoters bound by HSF1 whose endogenous transcripts showed no heat induction were not heat inducible in the luciferase assay, which is independent of genomic context, it seems unlikely that the lack of induction we saw for some HSF1-bound genes depends on the large-scale genomic context.
A small fraction of the genes we called HSF1 bound but not induced (∼10%) seemed to be subtly induced, but less than our twofold threshold. The subtle induction of some of these genes was confirmed in the mouse microarray experiment and in the human microarray data set. However, the data are clear that the majority of genes that we called bound but not induced were not induced. In fact, for some of these genes, it seems that HSF1 may act as a transcriptional repressor. The promoters for DNA methyltransferase-2, peroxiredoxin 3, and interferon, alpha 16 all contain an HSE, are all bound by HSF1, and their promoter activities are repressed after heat shock. The endogenous transcripts from these genes are repressed almost twofold after heat shock, as measured by RT-PCR, suggesting that these genes may be targets of HSF1-mediated transcriptional repression. This is consistent with previous studies that show that HSF1 can act as a transcriptional repressor of some genes (Cahill et al., 1996; Xie et al., 2002). If this is the case, HSF1 will be a powerful model system to use for understanding how transcription factors can act positively or negatively depending on the specific regulatory sequence to which they are bound.
Evidence for an HSF1-independent Mechanism of Heat-induced Gene Expression
We discovered 24 human genes that were induced by heat but had no detectable HSF1 binding. That result was confirmed by the mouse microarray experiment, which demonstrated that transcripts for 36 genes are induced by heat shock, even in an HSF1-/- cell line. The promoters of heat-inducible genes that lack HSF1 binding were indistinguishable from control promoters in the luciferase heat inducibility assay. It is unlikely that we missed the true promoter for all of these genes because the majority showed significant basal expression above background in our luciferase assays. Because our promoter activity results indicate that the induction of these genes in not due to an increased rate of transcription, it seems likely that a posttranscriptional mechanism such as changes in mRNA processing or mRNA stability may contribute to the induced transcript levels of these genes after heat shock. Alternatively, distant enhancer elements could be responsible for the induction of some of these genes.
Our analysis of the mouse microarray data as a whole revealed many genes that were induced by heat shock in the HSF-/- cell line. Most of the genes induced in the knockout cells were induced less than twofold or not at all in the wild-type cells. These included several genes involved in proliferation control, including p21-activated kinase 2, kinetochore component SUGT1, tumor necrosis factor receptor superfamily member 21, and the apoptosis inducer DEDD2. These genes could be directly induced by heat stress or they could be part of a secondary response that occurs if the cell fails to adequately respond to the heat stress. A smaller subset of genes that is induced by heat shock in both cell types and with similar kinetics possibly includes genes that are involved in the primary heat shock response and that are regulated in an HSF1-independent manner. All the microarray features that contained repetitive elements, showed a striking upregulation in both the wild-type and HSF1-/- cells. It was previously known that heat shock induces expression of these elements (Liu et al., 1997; Li et al., 1999), and our results indicate that this expression is independent of HSF1. These results provide evidence for a mechanism of heat induced gene expression independent of HSF1 binding, but it is difficult to know for certain for each gene whether it is involved in the primary heat shock response or a secondary stress response.
Explanations for HSF1 Binding to Only a Subset of HSE Sites
It is interesting that HSF1 binds to some HSEs but not to other HSEs that are nearly identical in sequence and position relative to the start of transcription. HSF1 binding may be blocked by the chromatin conformation in that specific region or by other structural features of the genome. Alternatively, the binding of HSF1 to some high-scoring HSEs but not to others could mean that our description of the HSE is incomplete or that other nearby sequence elements bind factors that are necessary for stable HSF1 binding. Recent results have shown that the transcription factors Msn2p and Msn4p contribute to the regulation of heat-induced transcription in yeast in addition to HSF (Boy-Marcotte et al.,1999). Also, in Caenorhabditis elegans, a second motif, the heat shock-associated site (5′-GGGTGTC-3′) seems to work cooperatively with the HSE in heat shock-mediated gene induction (GuhaThakurta et al., 2002). Our search for overrepresented sequence motifs in the human HSF1-bound sequences with MEME did not find any sequences similar to this site and its occurrence in our human promoters did not improve our ability to predict HSF1-binding (or heat-induced expression). This suggests that there are other cis-elements or other interacting factors that stabilize HSF1 binding at some heat shock elements.
Evidence for Alternative and Bidirectional Promoters in HSF1-mediated Regulation
Fifty-eight of the 93 genes induced by heat shock code for proteins with annotated functions but were not previously known to be induced by heat shock (see Supplemental Table 2). For example, we discovered that the basement membrane collagen COL4A6 and two collagen biosynthetic genes, SERPINH2 and FLJ13063, were heat induced. This is surprising, especially considering that the experiments were done in an erythroleukemia cell line, which we did not expect to express basement membrane collagen. It is possible that the heat induction of collagen is selected for in some other cell type and that the induction occurs spuriously in blood cells because it has a relatively low energetic cost, or this could represent a novel biological function of this collagen protein. Interestingly, COL4A6 is expressed as two isoforms, each of which uses a distinct promoter (Sugimoto et al., 1994). We detected HSF1 binding at only one of these two promoters, which suggests that this gene uses a heat-inducible alternative promoter. Therefore, the possibility exists that one promoter confers tissue specificity for COL4A6's role in the basement membrane and the other promoter regulates that heat inducibility of the gene for its potentially novel role in the heat shock response.
Another interesting class of heat-inducible genes consists of six pairs of genes organized in a head-to-head orientation with <1 kb separating their transcription initiation sites (see Supplemental Table 3). We detected clear induction for both members of three pairs of genes. All but four of 12 total bidirectional genes are hypothetical proteins with no known function. This kind of head-to-head gene organization was reported recently for hsp60 and hsp10 (Hansen et al., 2003), and our data suggest that regulation of pairs of genes from bidirectional inducible promoters is a common feature in HSF1-induced gene expression.
General Implications for Transcriptional Regulation
By beginning to comprehensively identify genes and promoters whose transcription is affected by heat in mammalian cells, this work has significantly increased our understanding of the complexity of heat shock regulation, especially the role of HSF1. One striking result was that HSF1-binding by itself does not confer heat-inducibility of a gene. The implication is that HSF1 may bind to these genes, but these genes may not be targets in the sense that they are not transcriptionally induced by HSF1. This has broad implications in how we should define targets of other transcription factors, because other studies have looked for binding sites for c-Myc (Fernandez et al., 2003) and E2F (Ren et al., 2002; Weinmann et al., 2002). Our results support the idea that transcription initiation might often require the assembly of many different factors at the promoter. Therefore, the localization of a single factor is unlikely to predict with certainty the expression of that gene considering the enormous potential of combinatorial complexity in the transcriptional apparatus. All of the data underlying this work is freely available for download at http://microarraypubs.stanford.edu/HSF1/.
Supplementary Material
Acknowledgments
We thank the members of the Myers and Botstein laboratories for helpful discussion and support; Bob Kingston's and Ivor Benjamin's laboratories for sharing the HSF1-/- mouse cell line; Stuart Kim and John Wang for use of the microarray printer and for help printing human promoter arrays; and Mike Fero and the Stanford Functional Genomics Facility for mouse arrays. N.D.T. and S.J.H. are supported by the Stanford Genome Training Program (grant 5 T32 HG00044) and J.I.M. is a Howard Hughes Medical Institute Predoctoral Fellow.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-10-0738. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-10-0738.
Online version of this article contains supplementary material for some figures. Online version available at www.molbiolcell.org.
References
- Bailey, T.L., and Elkan, C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Second International Conference on Intelligent Systems for Molecular Biology, 28-36. [PubMed]
- (1999). The heat shock response in yeast: differential regulations and contributions of the Msn2p/Msn4p and Hsf1p regulons. Mol. Microbiol. 33, 274-283. [DOI] [PubMed] [Google Scholar]
- Cahill, C.M., Waterman, W.R., Xie, Y., Auron, P.E., and Calderwood, S.K. (1996). Transcriptional repression of the prointerleukin 1beta gene by heat shock factor 1. J. Biol. Chem. 271, 24874-24879. [PubMed] [Google Scholar]
- Clos, J., Westwood, J.T., Becker, P.B., Wilson, S., Lambert, K., and Wu, C. (1990). Molecular cloning and expression of a hexameric Drosophila heat shock factor subject to negative regulation. Cell 63, 1085-1097. [DOI] [PubMed] [Google Scholar]
- DiDomenico, B.J., Bugaisky, G.E., and Lindquist, S. (1982). The heat shock response is self-regulated at both the transcriptional and posttranscriptional levels. Cell 31, 593-603. [DOI] [PubMed] [Google Scholar]
- Eisen, M.B., and Brown, P.O. (1999). DNA arrays for analysis of gene expression. Methods Enzymol. 303, 179-205. [DOI] [PubMed] [Google Scholar]
- Eisen, M.B., Spellman, P.T., Brown, P.O., and Botstein, D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95, 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez, P.C., Frank, S.R., Wang, L., Schroeder, M., Liu, S., Greene, J., Cocito, A., and Amati, B. (2003). Genomic targets of the human c-Myc protein. Genes Dev. 17, 1115-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GuhaThakurta, D., Palomar, L., Stormo, G.D., Tedesco, P., Johnson, T.E., Walker, D.W., Lithgow, G., Kim, S., and Link, C.D. (2002). Identification of a novel cis-regulatory element involved in the heat shock response in Caenorhabditis elegans using microarray gene expression and computational methods. [erratum appears in Genome Res 2002 Aug;12(8):1301]. Genome Res. 12, 701-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, J.J., Bross, P., Westergaard, M., Nielsen, M.N., Eiberg, H., Borglum, A.D., Mogensen, J., Kristiansen, K., Bolund, L., and Gregersen, N. (2003). Genomic structure of the human mitochondrial chaperonin genes: HSP60 and HSP10 are localised head to head on chromosome 2 separated by a bidirectional promoter. Human Genet. 112, 71-77. [DOI] [PubMed] [Google Scholar]
- Hasday, J.D., and Singh, I.S. (2000). Fever and the heat shock response: distinct, partially overlapping processes. Cell Stress Chaperones 5, 471-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaarniranta, K., Elo, M., Sironen, R., Lammi, M.J., Goldring, M.B., Eriksson, J.E., Sistonen, L., and Helminen, H.J. (1998). Hsp70 accumulation in chondrocytic cells exposed to high continuous hydrostatic pressure coincides with mRNA stabilization rather than transcriptional activation. Proc. Natl. Acad. Sci. USA 95, 2319-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, T., Spearow, J., Rubin, C.M., and Schmid, C.W. (1999). Physiological stresses increase mouse short interspersed element (SINE) RNA expression in vivo. Gene 239, 367-372. [DOI] [PubMed] [Google Scholar]
- Liu, X.D., Liu, P.C., Santoro, N., and Thiele, D.J. (1997). Conservation of a stress response: human heat shock transcription factors functionally substitute for yeast HSF. EMBO J. 16, 6466-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan, D.R., Christians, E., Forster, M., Xiao, X., Connell, P., Plumier, J.C., Zuo, X., Richardson, J., Morgan, S., and Benjamin, I.J. (2002). Heat shock transcription factor 2 is not essential for embryonic development, fertility, or adult cognitive and psychomotor function in mice. Mol. Cell. Biol. 22, 8005-8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan, D.R., Xiao, X., Shao, L., Graves, K., and Benjamin, I.J. (1998). Targeted disruption of heat shock transcription factor 1 abolishes thermotolerance and protection against heat-inducible apoptosis. J. Biol. Chem. 273, 7523-7528. [DOI] [PubMed] [Google Scholar]
- Pirkkala, L., Nykanen, P., and Sistonen, L. (2001). Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 15, 1118-1131. [DOI] [PubMed] [Google Scholar]
- Queitsch, C., Sangster, T.A., and Lindquist, S. (2002). Hsp90 as a capacitor of phenotypic variation. Nature 417, 618-624. [DOI] [PubMed] [Google Scholar]
- Rabindran, S.K., Giorgi, G., Clos, J., and Wu, C. (1991). Molecular cloning and expression of a human heat shock factor, HSF1. Proc. Natl. Acad. Sci. USA 88, 6906-6910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, B., Cam, H., Takahashi, Y., Volkert, T., Terragni, J., Young, R.A., and Dynlacht, B.D. (2002). E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 16, 245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritossa, F. (1962). A new puffing pattern induced by temperature shock and DNP in Drosophila. Experientia 18, 571-573. [Google Scholar]
- Sarge, K.D., Murphy, S.P., and Morimoto, R.I. (1993). Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Mol. Cell. Biol. 13, 1392-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarge, K.D., Zimarino, V., Holm, K., Wu, C., and Morimoto, R.I. (1991). Cloning and characterization of two mouse heat shock factors with distinct inducible and constitutive DNA-binding ability. Genes Dev. 5, 1902-1911. [DOI] [PubMed] [Google Scholar]
- Sorger, P.K., and Pelham, H.R. (1987). Purification and characterization of a heat-shock element binding protein from yeast. EMBO J. 6, 3035-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto, M., Oohashi, T., and Ninomiya, Y. (1994). The genes COL4A5 and COL4A6, coding for basement membrane collagen chains alpha 5(IV) and alpha 6(IV), are located head-to-head in close proximity on human chromosome Xq22 and COL4A6 is transcribed from two alternative promoters. Proc. Natl. Acad. Sci. USA 91, 11679-11683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodorakis, N.G., and Morimoto, R.I. (1987). Posttranscriptional regulation of hsp70 expression in human cells: effects of heat shock, inhibition of protein synthesis, and adenovirus infection on translation and mRNA stability. Mol. Cell. Biol. 7, 4357-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinklein, N.D., Aldred, S.J., Saldanha, A.J., and Myers, R.M. (2003). Identification and functional analysis of human transcriptional promoters. Genome Res. 13, 308-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinklein, N.D., Chen, W.C., Kingston, R.E., and Myers, R.M. (2004) Transcriptional regulation and binding of HSF1 and HSF2 to 32 human heat shock genes during thermal stress and differentiation. Cell Stress Chaperones (in press). [DOI] [PMC free article] [PubMed]
- Troyanskaya, O.G., Garber, M.E., Brown, P.O., Botstein, D., and Altman, R.B. (2002). Nonparametric methods for identifying differentially expressed genes in microarray data. Bioinformatics 18, 1454-1461. [DOI] [PubMed] [Google Scholar]
- Weinmann, A.S., Yan, P.S., Oberley, M.J., Huang, T.H., and Farnham, P.J. (2002). Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes Dev. 16, 235-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westwood, J.T., J. Clos, and Wu, C. (1991). Stress-induced oligomerization and chromosomal relocalization of heat-shock factor. Nature 353, 822-827. [DOI] [PubMed] [Google Scholar]
- Wiederrecht, G., Seto, D., and Parker, C.S. (1988). Isolation of the gene encoding the S. cerevisiae heat shock transcription factor. Cell 54, 841-853. [DOI] [PubMed] [Google Scholar]
- Xiao, H., and Lis, J.T. (1988). Germline transformation used to define key features of heat-shock response elements. Science 239, 1139-1142. [DOI] [PubMed] [Google Scholar]
- Xiao, X., Zuo, X., Davis, A.A., McMillan, D.R., Curry, B.B., Richardson, J.A., and Benjamin, I.J. (1999). HSF1 is required for extra-embryonic development, postnatal growth and protection during inflammatory responses in mice. EMBO J. 18, 5943-5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Y., Chen, C., Stevenson, M.A., Auron, P.E., and Calderwood, S.K. (2002). Heat shock factor 1 represses transcription of the IL-1beta gene through physical interaction with the nuclear factor of interleukin 6. J. Biol. Chem. 277, 11802-11810. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.