Abstract
Large databases (> 106 sequences) used in metaproteomic and proteogenomic studies present challenges in matching peptide sequences to tandem MS data using database-search programs. Most notably, strict filtering to avoid false positive matches leads to more false negatives, thus constraining the number of peptide matches. To address this challenge, we developed a two-step method wherein matches derived from a primary search against a large database were used to create a smaller subset database. The second search was performed against a target-decoy version of this subset database merged with a host database. High confidence peptide sequence matches (PSMs) were then used to infer protein identities. Applying our two-step method for both metaproteomic and proteogenomic analysis resulted in twice the number of high confidence peptide sequence matches in each case, as compared to the conventional one-step method. The two-step method captured almost all of the same peptides matched by the one-step method, with a majority of the additional matches being false negatives from the one-step method. Furthermore, the two-step method improved results regardless of the database search program used. Our results show that our two-step method maximizes the peptide matching sensitivity for applications requiring large databases, especially valuable for proteogenomics and metaproteomics studies.
Keywords: Two-step workflow, metaproteomics, proteogenomics, sequence database search, mass spectrometry, peptide sequence match
Advances in DNA and RNA sequencing, mass spectrometry and bioinformatics have expanded the scope of proteomics studies. One example is metaproteomics, an emerging field that identifies and characterizes the complement of proteins expressed by a microbial community in an environmental sample [1–2] or a host organism [3]. Another example, proteogenomics, identifies protein sequences that are not yet annotated in the predicted proteome of the organism under study. Proteogenomics provides information for gene annotation and genomic structure -thus enhancing our understanding of genomes [4–5].
Both of these disciplines are based upon large-scale identification of proteins in complex mixtures using tandem mass spectrometry (MS/MS) and sequence database searching, wherein each MS/MS spectrum is potentially matched to a peptide sequence and protein identities are inferred from these matches. The sequence databases used for database searching in these studies are commonly much larger than those used in more traditional proteomics studies. Typically, metagenomes contain hundreds of organisms with a total of more than 5 X 105 sequences. The situation for proteogenomics is more extreme due to use of 6-frame translated genomes or 3-frame translated transcriptomes that generally result in large datasets (more than 2 X 106 sequences). By comparison, the typical human databases used in traditional proteomics studies contain around 7 X 104 sequences or fewer. Determining PSMs by searching against large databases presents challenges because of the increased search space and an increased potential for false positives. This in turn necessitates stringent thresholds to ensure high confidence results [4–6]. Unfortunately, the increased stringency required also increases the number of false negative PSMs, that is true PSMs that fall short of the scoring thresholds employed, resulting in a decreased number of high confidence microbial (for metaproteomic studies) or alternative splice isoform (pASIs) peptides (for proteogenomic studies).
To overcome this challenge, we developed a two-step database searching method, which we have used to characterize the metaproteome of human salivary supernatant [3]. Although our previous work demonstrated the promise of this method for a metaproteomics study, a number of important questions about this method remained unanswered, including: 1) What is the mechanism by which it increases PSMs from large databases? 2) Is this method also effective for other applications, such as proteogenomics? 3) Is the apparent improvement dependent on the database search program? Here we sought to answer these questions, as well as further validate the method, thereby better enabling its use by others.
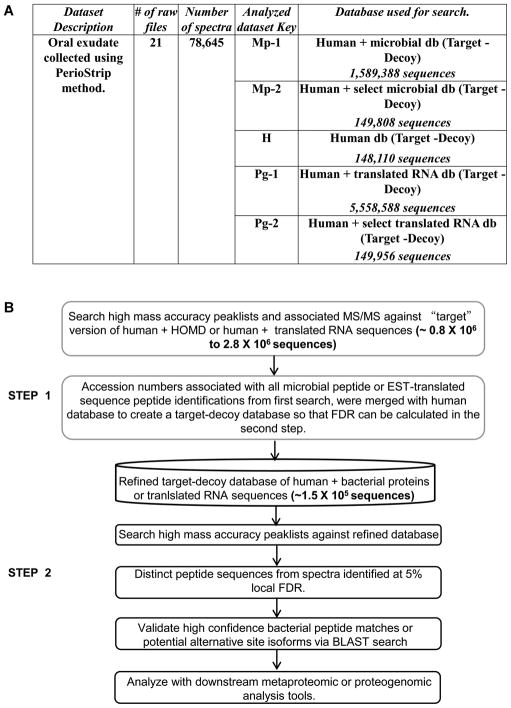
For the described studies (See supporting information S1 for more detailed information), we analyzed proteins from oral mucosa tissue exudates, collected from three individuals using PerioPaper strips and processed for MS-based proteomics using “on-strip” trypsin digestion as previously described [7]. Twenty-one peptide fractions (seven SCX fractions per sample; Figure 1A) were analyzed using online capillary liquid chromatography coupled with an LTQ-Orbitrap XL mass spectrometer (Thermo Scientific, San Jose, CA). Raw MS/MS data was processed through MaxQuant ‘Quant’ module to generate MSM files with highly accurate precursor masses, which increase confidence in PSMs when using large databases [4, 10]. The MSM files, after converting to mgf format, were searched using ProteinPilot ™ v 4.0 (AB SCIEX, Foster City, CA) [8] as described in Jagtap et al 2012 [9].
Figure 1. Overview of dataset and workflow.
A. Dataset, databases and keys used for metaproteomic and proteogenomic analysis.
B. Workflow for the two-step database search workflow.
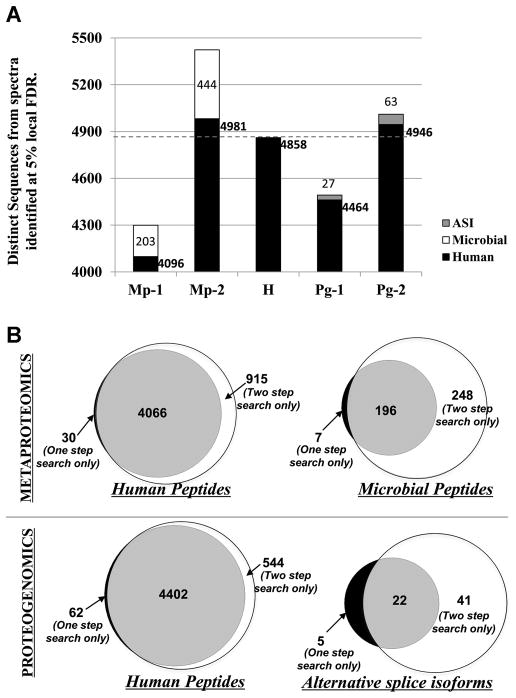
For the metaproteomics analysis, the traditional “one-step” database search method was used on a target-decoy database (Mp-1 in Figure 1A; 1,589,388 target-decoy sequences) of the human oral microbial database (HOMD dated December 2011) [11] concatenated with the human proteome database (UniProt isoform canonical database). We generated a target-decoy version of the database by appending a reversed version of sequences (i.e. the decoy sequences) to the forward sequences (i.e. the target sequences). For the proteogenomics analysis, the traditional one-step method was carried out on a target-decoy database (Pg-1 in Figure 1A; 5,558,588 target-decoy sequences) of the three-frame translated expressed sequence tag (EST) database (dated April 2011 as described in Menon et al 2011 [12]) with the human proteome (UniProt isoform canonical database dated December 2011). Distinct sequences associated with PSMs at 5% local FDR were recorded (Figure2A). Local FDR, a measure available through ProteinPilot, estimates the error rate around a specific PSM and ensures all peptides at or above this threshold are at least this likely to be correct – something not ensured by global FDRs. We have used the local FDR threshold since it reports PSMs with more stringent quality control thresholds [13]. Distinct sequences of human origin, microbial origin (for metaproteomic analysis) and translated EST origin (for proteogenomic analysis) were noted. The traditional one-step search resulted in 4096 human PSMs and 203 microbial PSMs for the metaproteomic analysis (Mp-1 in Figure2A). For the proteogenomics analysis, true pASIs sequences were verified by searching translated EST sequences using BLASTP (v BLASTP 2.2.2.27+) against a database containing 20,468,417 non-redundant GenBank human sequences (See supporting information S1 for details). This resulted in 4464 human peptide sequences and 27 pASIs sequences matched (Pg-1 in Figure2A). We used these results from the one-step method as a baseline to compare against the PSMs from the two-step method.
Figure 2. Comparison of the one-step, two-step and human database searches.
A. Sequences matched at 5% local FDR in metaproteomic one-step search (Mp-1) and the two-step search (Mp-2), human database search (H), proteogenomic one-step search (Pg-1) and the two-step search (Pg-2). Numbers of matched human peptides, microbial peptides and potential alternative splice isoform site (pASIs) have been denoted next to bar diagrams associated with each search.
B. Venn diagram of human and microbial peptides (for metaproteomic search) or potential alternative splice isoform peptides (for proteogenomic search) matched in the one-step (Mp-1 or Pg-1) and the two-step (Mp-2 or Pg-2) method.
In order to estimate the number of identifiable human proteins in this dataset without effects from the larger metaproteomic or proteogenomic databases, we searched the dataset against a human UniProt database (H in Figure1A; 148,110 target-decoy sequences). This resulted in matching of 4858 distinct peptide sequences at 5% local FDR (H in Figure 2A). Note that in Figure 2A, the number of human sequences in H is higher than Mp-1 or Pg-1 searches, thus demonstrating the overall decrease in matches due to searching against larger databases, ostensibly due to increases in false negative PSMs. It is also noteworthy that the overlap for human peptides between the results from the human-only database search (H) and the Mp-1 (Supporting Information S2) and Pg-1 (Supporting Information S3) results were above 98 %. This demonstrates that although the larger database decreased the overall number of PSMs, the proteins identified between methods were largely the same.
To improve PSM sensitivity we followed a two-step database search method (Figure1B). This method consists of a primary search against only the target version of a large sequence database (Mp-1 for metaproteomics search and Pg-1 for proteogenomics search), followed by the construction of a smaller subset sequence database containing all proteins inferred from PSMs in the primary search. A target-decoy database is then constructed from this refined database and used for a secondary database search in which stringent thresholds are applied to reveal high confidence PSMs. For the metaproteomics analysis, the MS/MS data was first searched against the target version of the HOMD database concatenated with the human proteome database (794,694 sequences); for proteogenomics analysis, the MS/MS data was first searched against the “target” version of three-frame translated EST database concatenated with the human proteome database (2,779,294 sequences). Accession numbers of all peptides matched in the first-step search with microbial (849 microbial proteins for metaproteomics) or EST translated peptides (923 EST-translated proteins for proteogenomics) were used to generate two separate, subset FASTA-formatted sequence databases (See Figure1B and Supporting Information S1 for details). Thus, the two databases were reduced to less than 0.06% and 0.02% relative to the original size for the HOMD and translated EST databases, respectively. The accession numbers corresponding to these selected proteins were merged with the Human UniProt database to construct two separate target-decoy databases for the secondary database search (Mp-2 and Pg-2 in Figure1A).
For both analyses, results from the second database search showed that the PSMs to human peptides (Mp-2 and Pg-2 in Figure 2A and Supporting Information S4 and S5) were restored to numbers comparable to results for the human sequence database alone (H in Figure 2A). For the metaproteomics analysis, the two-step method more than doubled the number of high confidence (95% Conf) microbial PSMs, matching 444 peptides compared to 203 via the onestep method (118% increase in Mp-2 over Mp-1 in Figure 2A). Additionally, Pep2Pro [14] analysis of microbial peptides from the two-step method shows that more non-ambiguous microbial species based on unique peptides (66 species as compared to 18 species) were matched as compared to the one-step method (Supporting Information S6 and S7).
We also evaluated whether the two-step method’s improved performance was dependent on database search program. When using Sequest, OMSSA, X!tandem and Andromeda, the two-step method consistently outperformed the one-step method (Supporting information S8 and data in Tranche). Although the magnitude of increased PSMs was program dependent, on an average the two-step method resulted in more PSMs for metaproteomics analysis for the five programs tested. The reason for variable improvements could be because of variable scoring methods used by each search algorithm. Thus the positive improvements afforded by the two-step method are not limited to a single database search program only.
For ProteinPilot metaproteomics analysis, the two-step method showed a 97% overlap with those PSMs from the one-step method (Figure 2B). Out of the 7 peptides matched by the one-step method (Mp-1) and not the two-step method (Mp-2), two were not matched in the first step and hence not incorporated into the database for the second step. Although the reason for missing these peptides is not clear, it may stem from the fact that scoring functions for PSMs are in part dependent on sequence database size, resulting in small variations of results when analyzing the same MS/MS data against different databases (See Supporting Information S9 and S13).
For the proteogenomic analysis, the two-step method more than doubled the number of pASIs peptides (confirmed by BLASTP searches described above; Supporting Information S10), matching 63 of these peptides compared to 27 using the one-step method (133% increase in Pg-2 as compared to Pg-1 in Figure 2A). Again, we found a substantial overlap (81%) of pASIs peptides matched by the two methods (Figure 2B – Proteogenomics). Out of the 5 peptides matched only by the one-step method, two were not found in the first step and hence not incorporated into the database for the second step of the two-step method. The reason for this discrepancy between the methods is again most likely due to the dependence on database size for scoring PSMs (See Supporting Information S9 and S13).
To further validate the veracity of the two-step method, for the metaproteomic analysis we took advantage of the fact that we spiked-in E. coli Beta-galactosidase peptides in to the starting human exudate sample in two of the three samples. As expected, no beta galactosidase was identified in the sample containing no spiked in Beta-galactosidase. For the spiked-in samples, we identified Beta-galactosidase using both the one-step method and the two-step, with the two-step method matching 5 more distinct peptides than the one-step (Supporting Information S11). This demonstrated that the improvement in microbial PSMs using the two-step method was due to authentic matches and not a result of random false positives..
As additional validation, spectral annotations were performed for a few representative spectra from both analyses matched exclusively by the two-step method (Table 1). ProteinPilot PSM annotation and manual annotation have been provided in Supporting Information S13. Furthermore, for microbial PSMs or pASIs that were exclusively matched by the two-step method (Mp-2 or Pg-2), we looked at the characteristics of the PSM for these same MS/MS spectra from the one-step method (Mp-1 or Pg-1). Interestingly, spectral assignment characteristics such as number of matching peaks (Sc), delta precursor mass and assigned peptide sequences were identical for ~90% of the spectra regardless of the method used. However, the Conf score which is based on Sc and other features such as modifications, mass accuracy and other factors was not high enough in the one-step method for these to be deemed acceptable at the 5% local FDR threshold. This analysis demonstrates that increased false negatives are one of the main reasons for the lower performance of the one-step method compared to the two-step method.
Table 1.
Spectral assignment characteristics of one-step method false-negative PSMs (for both metaproteomic and proteogenomic studies). Representative PSMs found exclusively in the two-step method were compared with the match from the same MS/MS spectrum from the one-step method in terms of their charge state, theoretical mass to charge ratio (Theor. m/z), difference in mass between the measured and the theoretical precursor ion (Delta mass) and peptide score (Sc; count of the MS/MS peaks that match to a theoretical ions). The PSMs differed only in Peptide Conf - which is a confidence for the peptide match, expressed as a percentage.
| METAPROTEOMICS | ||||||||
|---|---|---|---|---|---|---|---|---|
| Spectrum information | Identification | Peptide Conf | ||||||
| Sample | Fraction | Charge | Theor m/z | Δ Mass | Sequence | Sc | One Step Method | Two Step Method |
| 1 | 2 | 2 | 437.255 | −0.0003 | EVAKEIGK | 9 | 12.3 | 98.6 |
| 6 | 6 | 2 | 468.761 | −0.0072 | MLFGELVK | 9 | 64.9 | 98.4 |
| 6 | 6 | 2 | 441.251 | 0.0007 | QSLGAHLR | 9 | 80.1 | 98.3 |
| 1 | 3 | 2 | 407.226 | −0.0001 | LNVENPK | 9 | 87.4 | 98.1 |
| 1 | 3 | 2 | 634.853 | 0.0003 | WLSLPGETRPL | 11 | 88.1 | 96.6 |
| 6 | 6 | 2 | 511.751 | 0.0002 | EHFAIYKD | 8 | 66.1 | 95.5 |
| PROTEOGENOMICS | ||||||||
|---|---|---|---|---|---|---|---|---|
| Spectrum information | Identification | Peptide Conf | ||||||
| Sample | Fraction | Charge | Theor m/z | Δ Mass | Sequence | Sc | One Step Method | Two Step Method |
| 6 | 3 | 2 | 510.232 | 0.0061 | MDNAIGDQR | 9 | 66.1 | 96.7 |
| 4 | 3 | 2 | 498.774 | −0.0005 | MSPHLQRK | 9 | 92.1 | 96.2 |
| 6 | 5 | 2 | 574.2341 | 0.0019 | GEEEGEGGGGGGR | 9 | 84.8 | 97.5 |
| 6 | 5 | 2 | 679.8591 | −0.0093 | RTSVCPRTWPR | 10 | 73.85 | 98.1 |
Large databases searches have presented a challenges for metaproteomic and proteogenomic studies in terms of search times and number of PSMs [4–6]. These studies have made use of synthetic translated metagenomes [3, 15] or matched metagenomes [10] for metaproteomic studies and 6-frame genome translations or 3-frame transcriptome translations for proteogenomic studies [4]. Various strategies have been suggested to address sensitivity and accuracy of PSMs in large database searches. This includes use of various gene annotation and gene assembly strategies for matched metaproteomes [10]; database reduction methods such as the iterative searching method on synthetic genomes [15]; use of specific and smaller databases from human proteome [16] and bacterial proteome [17]; and use of HiRIEF fractionation methods and in silico proteogenomic databases [18].
Our two-step method provides a straightforward alternative to these other methods that effectively addresses the challenges of MS proteomics-based studies employing large sequence databases. We demonstrate that the two-step method increases the number of high confidence PSMs significantly, both for metaproteomic and proteogenomic studies. We also demonstrate that the mechanism for this improvement stems largely from a decrease in false negatives using the two-step method. Importantly, we observed improvements using the two-step method regardless of the sequence database-searching program used. We believe our method should improve the sensitivity of microbial peptide (for metaproteomic analyses) and novel pASIs PSMs (for proteogenomic analyses). Although in its current state the two-step method is manual, we are working on automating the method for more general use across a wide variety of applications.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health (U.S.) Grant 1R01 DE17734 and NSF grant 1147079 to T.J.G. The authors recognize the assistance from LeeAnn Higgins from the Center for Mass Spectrometry and Proteomics (CMSP) at the University of Minnesota (supporting agencies listed here: http://www.cbs.umn.edu/msp/about.shtml) and John Chilton from Minnesota Supercomputing Institute.
ABBREVIATIONS
- pASIs
potential alternative splice isoform sites
- LTQ
linear trap quadrupole
- MGF
Mascot generic format
- HOMD
Human Oral Microbiome Database
- EST
Expressed Sequence Tags
- FDR
False discovery rate
- PSM
Peptide Sequence Match
- HiRIEF
High Resolution Peptide Isoelectric Focusing
Footnotes
CONFLICT OF INTEREST STATEMENT
All authors declare that there is no financial / commercial conflict of interest.
Additional tables, text and figures are described in the text are included in the supporting information. The data associated with this manuscript can be downloaded from ProteomeCommons.org Tranche by using associated hash code and the passphrase mentioned below.
OnPkqAkLKfcf1WGDk1tjAcCX9tQ916NHNl3IlHhq8rBFCRH5J+xrtpyDN6Rc2ahmxhN93QhnamBP7JbG69J1L/F/HfUAAAAAAAADAQ==
Passphrase to access data: twostep
References
- 1.Siggins A, Gunnigle E, Abram F. Exploring mixed microbial community functioning: recent advances in metaproteomics. FEMS Microbiol Ecol. 2012;80(2):265–280. doi: 10.1111/j.1574-6941.2011.01284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hettich RL, Sharma R, Chourey K, Giannone RJ. Microbial metaproteomics: identifying the repertoire of proteins that microorganisms use to compete and cooperate in complex environmental communities. Curr Opin Microbiol. 2012 doi: 10.1016/j.mib.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 3.Jagtap P, McGowan T, Bandhakavi S, Tu ZJ, et al. Deep metaproteomic analysis of human salivary supernatant. Proteomics. 2012;12:992–1001. doi: 10.1002/pmic.201100503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Renuse S, Chaerkady R, Pandey A. Proteogenomics. Proteomics. 2011;11(4):620–630. doi: 10.1002/pmic.201000615. [DOI] [PubMed] [Google Scholar]
- 5.Castellana N, Bafna V. Proteogenomics to discover the full coding content of genomes: a computational perspective. J Proteomics. 2010;73(11):2124–2135. doi: 10.1016/j.jprot.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cargile BJ, Bundy JL, Stephenson JL., Jr Potential for false positive identifications from large databases through tandem mass spectrometry. J Proteome Res. 2004;3(5):1082–1085. doi: 10.1021/pr049946o. [DOI] [PubMed] [Google Scholar]
- 7.Kooren JA, Rhodus NL, Tang C, Jagtap PD, et al. Evaluating the potential of a novel oral lesion exudate collection method coupled with mass spectrometry-based proteomics for oral cancer biomarker discovery. Clin Proteomics. 2011;8:13. doi: 10.1186/1559-0275-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shilov IV, Seymour SL, Patel AA, Loboda A, et al. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol Cell Proteomics. 2007:1638–1655. doi: 10.1074/mcp.T600050-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Jagtap P, Bandhakavi S, Higgins L, McGowan T, et al. Workflow for analysis of high mass accuracy salivary data set using MaxQuant and ProteinPilot search algorithm. Proteomics. 2012;12(11):1726–1730. doi: 10.1002/pmic.201100097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cantarel BL, Erickson AR, VerBerkmoes NC, Erickson BK, et al. Strategies for metagenomic-guided whole-community proteomics of complex microbial environments. PLoS One. 2011;6(11):e27173. doi: 10.1371/journal.pone.0027173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen T, Yu WH, Izard J, Baranova OV, et al. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford) 2010:baq013. doi: 10.1093/database/baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Menon R, Omenn GS. Identification of alternatively spliced transcripts using a proteomic informatics approach. Methods Mol Biol. 2011;696:319–326. doi: 10.1007/978-1-60761-987-1_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang WH, Shilov IV, Seymour SL. Nonlinear fitting method for determining local false discovery rates from decoy database searches. J Proteome Res. 2008;7(9):3661–3667. doi: 10.1021/pr070492f. [DOI] [PubMed] [Google Scholar]
- 14.Askenazi M, Marto JA, Linial M. The complete peptide dictionary—a meta-proteomics resource. Proteomics. 2010;10(23):4306–4310. doi: 10.1002/pmic.201000270. [DOI] [PubMed] [Google Scholar]
- 15.Rooijers K, Kolmeder C, Juste C, Doré J, et al. An iterative workflow for mining the human intestinal metaproteome. BMC Genomics. 2011;12:6. doi: 10.1186/1471-2164-12-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prakash A, Ahmad S, Sarracino D, Krastins B, et al. Small, Standardized Protein Database Provides Rapid and Statistically Significant Peptide Identifications for Targeted Searches Using Percolator. 2012; Proceedings of the 60th ASMS Conference on Mass Spectrometry and Allied Topics; Vancouver (Canada). [Google Scholar]
- 17.Beck DAC, Hendrickson EL, Wang T, Hackett M. Protein database construction for microbial community proteomics and impacts on peptide and protein assignments, confidence, quantitation and differential analysis. 2012; Proceedings of the 60th ASMS Conference on Mass Spectrometry and Allied Topics; Vancouver (Canada). [Google Scholar]
- 18.Granholm V, Branca RM, Orre LM, Johansson HJ, et al. Identification of novel gene models: Matching mass spectrometry data against a 6-frame translation of the human genome. 2012; Proceedings of the 60th ASMS Conference on Mass Spectrometry and Allied Topics; Vancouver (Canada). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.