Abstract
Classic ways to determine MHC restriction involve inhibition with locus specific antibodies and antigen presentation assays with panels of cell lines matched or mismatched at the various loci of interest. However, these determinations are often complicated by T-cell epitope degeneracy and promiscuity. We describe selection of 46 HLA DR, DQ and DP specificities that provide worldwide population (phenotypic) coverage of almost 90% at each locus, and account for over 66% of all genes at each locus. This panel afforded coverage of at least four HLA class II alleles in over 95% of the individuals in four study populations of diverse ethnicity from the US and South Africa. Next, a panel of single HLA class II transfected cell lines, corresponding to these 46 allelic variants was assembled, consisting of lines previously developed and 15 novel lines generated for the present study. The novel lines were validated by assessing their HLA class II expression by FACS analysis, the in vitro peptide binding activity of HLA molecules purified from the cell lines, and their antigen presenting capacity to T-cell lines of known restriction. We also show that these HLA class II transfected cell lines can be used to rapidly and unambiguously determine HLA restriction of epitopes recognized by an individual donor in a single experiment. This panel of lines will enable high throughput determination of HLA restriction, enabling better characterization of HLA class II-restricted T-cell responses and facilitating the development of HLA tetrameric staining reagents.
Keywords: HLA Class II, restriction, transfectants, epitopes, population coverage, polymorphism
INTRODUCTION
The accurate identification and characterization of class II restricted epitope-specific responses is important for a variety of applications. These include the study of mechanisms of host-pathogen interactions and the evaluation of vaccine candidates, but also basic studies related to probing different T helper subsets in terms of their lymphokine secretion patterns and expression of phenotypic markers, such as chemokine receptors and memory/activation markers. As of today, this is accomplished primarily by two techniques, either testing epitopes/peptides by ICS and/or staining with tetrameric reagents.
However, practical hurdles have severely limited the widespread applicability of tetramer staining coupled with ICS analysis. Often, only small amounts of blood are available, thereby precluding, in practical terms, the testing of large pools and/or panels of, for example, overlapping peptides spanning the entire sequence of a pathogen or allergen of interest by the ICS assay. In terms of tetramer assays, relatively few HLA allelic variants have been validated at the level of production and functionality. This, as a result, has forced investigators into a rather incomplete level of characterization at the population level, as only select individuals expressing the specific HLA types for which tetramers are available can be studied. But limits are also imposed at the individual level, as responses restricted by just one of the three to eight different HLA molecules expressed in a given individual can typically be characterized. In the context of both ICS and tetramers, the challenge of complete characterization is increased by the fact that usually only a minority of the epitopes restricted by any given HLA are known for a particular antigenic system.
Thus, to enable the widespread use of epitope specific human T cell responses, the challenge that needs to be met is the precise determination of which HLA locus and allele restrict epitope–specific T cells. This is not a trivial issue, since in the context of HLA class II α/β hetero-dimers, most humans express molecules encoded by four different β-chain loci (DRB1, DRB3/4/5, DQB1, DPB1) (Marsh et al. 2000), as well as corresponding α-chain loci (DRA1, DQA1 and DPA1). To further complicate the issue, all loci, with the exception of the DR alpha chain, are extremely polymorphic, and more than 1500 different alleles have been identified to date (Robinson et al. 2011). As a result, most individuals are heterozygotes, and express up to 8 different HLA class II molecules.
Classic ways to determine T cell restriction involve inhibition with locus specific antibodies, followed by antigen presentation assays with panels of cell lines matched or mismatched at the various loci of interest (see, e.g., (Oseroff et al. 2010; Oseroff et al. 2012a; Oseroff et al. 2012b; Wang 2009; Wilson et al. 2001). Since epitope binding to HLA class II molecules is absolutely necessary (but not sufficient) for T cell activation, data from in vitro HLA binding assays has also been useful to narrow down the possible restrictions (Arlehamn et al. 2012b). This is usually accomplished by testing a given epitope for binding to the specific HLA molecules expressed in a specific donor, and eliminating from further consideration HLA molecules to which the epitope does not bind.
Restriction determinations are further complicated by the phenomena of T cell epitope degeneracy and promiscuity (Sinigaglia et al. 1988; Panina-Bordignon et al. 1989a; Panina-Bordignon et al. 1989b; Krieger et al. 1991). In the late 1980s it was shown that multiple HLA class II molecules can bind the same dominant epitope (Roche and Cresswell 1990; Panina-Bordignon et al. 1989b; Busch et al. 1990; O’Sullivan et al. 1991; O’Sullivan et al. 1990), and recent data show that the overlap in the peptide binding repertoires between different alleles (both within and even across different loci) is extensive (Greenbaum et al. 2011; Sidney et al. 2010a, b). Furthermore, it was shown that the same T cell clone could often be activated by a given peptide presented by two different HLA allelic molecules (Krieger et al. 1991; Ho et al. 1990; Panina-Bordignon et al. 1989b; Doherty et al. 1998; Karr et al. 1991). This overlap and cross-reactivity is often referred to as T cell and HLA class II degeneracy, or promiscuity.
Our recent study in the Timothy grass system did indeed show that promiscuous binding and presentation is extensive (Greenbaum et al. 2011; Oseroff et al. 2010). Analysis of the data relating to response magnitude revealed that considering just the top 20 antigenic regions could account for about 80% of the total response against the allergens studied. When the restriction data relating to these epitopic regions was examined, it was revealed that all of these most dominant antigenic regions are promiscuous, in that multiple different HLA class II molecules can bind and present them. This observation provides at least a partial explanation for their dominance, and also suggests that this additional dimension of complexity must be considered in the design of studies utilizing tetrameric reagents. It was further shown that prediction of promiscuous HLA binding capacity could actually be utilized to identify a large fraction of the most dominant epitopes (Oseroff et al. 2010; Oseroff et al. 2012a; Oseroff et al. 2012b).
The phenomenon of epitope promiscuity poses a challenge to restriction determination because in the preponderance of cases, it does not allow clear and decisive assignment of putative restriction on the basis of binding data. Furthermore, in many cases, promiscuity yields inconclusive data when EBV transformed homozygote cell lines are used in antigen presentation assays. For example, the influenza HA 307–319 epitope was known to be recognized equally well by T cells from a specific donor when presented by either DRB1*07:01 or DRB1*04:01. However, mapping restriction using EBV lines would have resulted in it being erroneously assigned to DRB4*01:01 since, because of linkage disequilibrium, this molecule is usually co-expressed with both DRB1*07:01 and DRB1*04:01, and would thus be the only molecule shared between EBV lines also expressing DRB1*07:01 or DRB1*04:01 (Krieger et al. 1991).
Almost 20 years ago, Robert Karr and co-workers (Karr et al. 1991; Klohe et al. 1988; Lair et al. 1988) pioneered the use of L cell lines transfected with a single HLA class II molecule as a way to overcome the problems alluded to above, and that are inherent in the use of EBV cell lines as APCs. In the present study, we have expanded the initial panel of cell lines produced by Karr and co-workers by selecting an additional panel of HLA class II molecules. The genes selected for transfection were of interest to afford high population coverage at the various HLA class II loci. In each case where single HLA class II molecules transfected cells were not available, we generated and validated new cell lines. We further show how this panel of transfected cell lines can be used to quickly and unambiguously determine HLA restriction.
METHODS
HLA typing and human subjects
The HLA typing data utilized for this study was generated in the course of independent studies analyzing T cell responses to common allergens (Oseroff et al. 2012a; Oseroff et al. 2012b) and Mycobacterium tubercolosis ((Arlehamn et al. 2012b) and Scriba et al., unpublished). Genomic DNA isolated from PBMC of the study subjects by standard techniques (QIAmp; Qiagen, Valencia, CA) was used for HLA typing. High resolution Luminex-based typing for HLA Class II was utilized according to the manufacturer’s instructions (Sequence-Specific Oligonucleotides (SSO) typing; One Lambda, Canoga Park, CA). Where indicated, PCR based methods were used to provide high resolution sub-typing. (Sequence-Specific Primer (SSP) typing; One Lambda, Canoga Park, CA).
All studies using human PBMCs were performed following approved protocols from the relevant Research Ethics Committees, and informed consent was obtained from all individual blood donors. These studies, and the Research Ethics Committee which approved the study, included four studies in San Diego, CA (University of CA and LIAI), one study each in Denver, CO (University of Colorado and LIAI), Baltimore, MD (Johns Hopkins University), and Cape Town, South Africa (Boland-Overberg region of the Western Cape Province of South Africa and the Research Ethics Committee of the University of Cape Town). HLA typing was performed on all samples.
Population coverage
Haplotype and phenotype frequencies of individual alleles in the general population represent averages across several major populations, are based on data available at dbMHC and allelefrequencies.net (Middleton et al. 2003; Meyer et al. 2007), and considers prevalence in Europe, North Africa, North-East Asia, the South Pacific (Australia and Oceania), Hispanic North and South America, American Indian, South-East Asia, South-West Asia, and Sub-Saharan Africa. DP, DRB1 and DRB3/4/5 frequencies consider only the beta chain frequency, given that the DRA chain is largely monomorphic, and that differences in DPA are not thought to significantly influence peptide binding. Frequency data are not available for DRB3/4/5 alleles. However, because of linkage with DRB1 alleles, coverage for these specificities may be assumed as follows: DRB3 with DR3, DR11, DR12, DR13 and DR14; DRB4 with DR4, DR7 and DR9; DRB5 with DR15 and DR16. Specific allele frequencies at each B3/B4/B5 locus is based on published associations with the various DRB1 alleles listed above, and assumes only limited allelic variation at the indicated locus (Greenbaum et al. 2011).
Generation of transfected cell lines
The RM3 cell line was utilized for HLA transfections. RM3 is derived from the human B lymphocyte cell line Raji (DRA1*01:01, DRB1*03:01, DRB1*10:01, DRB3*02, DQA1*01:01, DQA1*05:01, DQB1*02:01, DQB1*05:01, DPB1*01:01) following mutagenesis with ethane methylsulfonate and selection for class II negative cells (Calman and Peterlin 1988). Transfection of RM3 cells to produce cell lines expressing a single MHC specificity has been utilized in previous studies (see, e.g., (Dzuris et al. 2001; Giraldo-Vela et al. 2008)).
RM3 and control cell lines were cultured in RPMI 1640 medium supplemented with 2 mM glutamine, 1% (vol/vol) nonessential amino acids, 1% (vol/vol) sodium pyruvate, penicillin (50 U/ml), streptomycin (50 μg/ml) (all from Life Technologies) and 10% heat-inactivated fetal bovine serum (Gemini BioProducts) (R10).
Where available, ORF clones containing the alpha and beta chains used to generate HLA Class II transfectants were obtained from the members of the ORFeome Collaboration (PlasmID, Dana-Farber/Harvard Cancer Center DNA Resource Core, Boston, MA and GeneCopoeia, Rockville, MD). Alternately, synthetic genes based on the coding region sequence information available in the IMGT/HLA Database (http://www.ebi.ac.uk/imgt/hla/) (Robinson et al. 2003; Robinson et al. 2011; Robinson et al. 2000) were constructed (GenScript, Piscataway, NJ). The complete gene sequence was amplified by PCR using sequence specific primers. The PCR product was cloned into the pENTR/D-TOPO vector, and re-inserted into pcDNA-DEST40 vector (beta chains) or pcDNA 6.2/V5 DEST vector (alpha chains) using the Gateway system (Life Technologies). Chemically competent Escherichia coli (OneShot Top10; Life Technologies) were transformed with the plasmid, plated on ampicillin plates, and single colonies were selected and the plasmid was isolated (Qiagen). Sequence integrity was confirmed by sequence determination (Retrogen, San Diego, CA).
RM3 cells (1 × 107 cells) were electroporated with 15 μg of plasmid DNA using Amaxa Cell Line Nucleofector Kit C (Lonza) according to the manufacturer’s instructions. After 24 hours, transfected cells were selected in R10 medium supplemented with a final concentration of 700 μg/ml G418 and 12 μg/ml blasticidin (Life Technologies). After 48 hours, transient expression was evaluated by flow cytometry using anti-DR, -DP, and -DQ antibodies (LB3.1, B7/21, and SPVL3, respectively). Stable transfectants were expanded, and high HLA class II expressing cell populations were selected by cell sorting using the antibodies described previously.
Peptide, MHC purification and HLA binding assays
Peptides utilized in this study were purchased from Mimotopes (Clayton, Victoria, Australia) and/or A and A (San Diego, CA) as crude material on a small (1 mg) scale. Peptides utilized as radiolabeled ligands were synthesized on larger scale, and purified (>95%) by reversed phase HPLC, as previously described (Southwood et al. 1998).
Purification of MHC class II molecules by affinity chromatography has been described in detail elsewhere (Sidney et al. 2001). Briefly, EBV transformed homozygous cell lines, or single MHC allele transfected fibroblast, L, or RM3 (B) cell lines, were used as sources of MHC molecules. HLA molecules were purified from cell pellet lysates by repeated passage over Protein A Sepharose beads conjugated with locus specific monoclonal Abs. Protein purity, concentration, and the effectiveness of depletion steps were monitored by SDS-PAGE and BCA assay.
Assays to quantitatively measure peptide binding to purified class II MHC molecules are based on the inhibition of binding of a high affinity radiolabeled peptide to purified MHC molecules, and were performed essentially as detailed elsewhere (Sidney et al. 2008; Sidney et al. 2010b, a; Greenbaum et al. 2011; Sidney et al. 2001). Briefly, 0.1–1 nM of radiolabeled peptide was co-incubated at room temperature or 37°C with purified MHC in the presence of a cocktail of protease inhibitors. Following a two- to four-day incubation, MHC bound radioactivity was determined by capturing MHC/peptide complexes on monoclonal Ab coated Lumitrac 600 plates (Greiner Bio-one, Frickenhausen, Germany), and measuring bound cpm using the TopCount (Packard Instrument Co., Meriden, CT) microscintillation counter. In the case of competitive assays, the concentration of peptide yielding 50% inhibition of the binding of the radiolabeled peptide was calculated. Under the conditions utilized, where [label] < [MHC] and IC50 ≥ [MHC], the measured IC50 values are reasonable approximations of the true Kd values (Cheng and Prusoff 1973; Gulukota et al. 1997). Each competitor peptide was tested at six different concentrations covering a 100,000-fold dose range in three or more independent experiments. As a positive control, the unlabeled version of the radiolabeled probe was also tested in each experiment.
In vitro expansion of epitope-specific T cells
Epitope specific short-term T cell lines were obtained from thawed PBMCs cultured in RPMI 1640 (V Scientific, Tarzana, CA) supplemented with 5% human serum (Cellgro, Herndon, VA) at a density of 2×106 cells/ml in 24-well plates (BD Biosciences, San Jose, CA) and stimulated with 10 μg/ml of the respective individual epitope. Cells were cultured at 37°C in 5% CO2 and additional IL-2 (10 U/ml; eBioscience, San Diego, CA) was added every 3 days after initial antigenic stimulation. On day 11, cells were harvested and screened for reactivity in antigen presentation assays.
ELISPOT Antigen presentation assays
Short-term T cell lines were incubated with antigen presenting cell lines (APCs) expressing various HLA molecules that had been pulsed with an appropriate peptide epitope, and the production of IL-5 or IFN-γ was determined in ELISPOT assays. Flat-bottom 96-well nitrocellulose plates (Millipore, Bedford, MA) were prepared according to manufacturer’s instructions and coated with 10 μg/ml anti-human IL-5 (Clone TRFK5; Mabtech, Cinncinati, OH) or anti-human IFN-γ (Clone 1-D1K; Mabtech). T cells were then incubated at a density of 1×104/well at a ratio of 1:1 with the various peptide-pulsed APCs. Peptide pulsed autologous PBMCs were utilized as a positive control and the untransfected RM3 cell line was used as negative control. After 24 hours, cells were removed, and plates were incubated with either 2 μg/ml biotinylated anti-human IL-5 Ab (Mabtech) or biotinylated anti-human IFN-γ Ab (Mabtech) at 37°C. After 2 hours, spots corresponding to the biotinylated Ab (IL-5 or IFN-γ) were developed with 3-amino-9-ethylcarvazole solution (Sigma-Aldrich, St. Louis, MO). Spots were counted by computer-assisted image analysis (Zeiss, KS-ELISPOT reader, Munich, Germany). Each assay was performed in triplicate. The level of statistical significance was determined with a Student’s t-test using the mean of triplicate values of the response against relevant peptides versus the response against APC not pulsed with any peptide. Criteria for a positive response to a peptide were ≥ 20 SFC/106 input PBMC, p ≤ 0.05 in the t test, and a stimulation index (SI) ≥ 2.
RESULTS
Selection of a target set of HLA molecules
The basic premise of this study is to generate approaches and reagents to allow definition of epitopes restricted by a broad array of different HLA class II molecules, without any assumption with respect to which particular HLA class II locus (DR, DP or DQ) might be most relevant as a restriction element. Accordingly, we previously defined a panel of 27 different HLA DR, DQ and DP specificities (Greenbaum et al. 2011) that essentially covers 100% of individuals in the general population at the phenotypic level with at least one allele, and encompasses 46 to 77% of all genes, depending on the locus (see Table 1, alleles in bold font). For each of the 27 molecules we have previously developed high-throughput peptide binding assays (Greenbaum et al. 2011; Sidney et al. 2010a, b; Southwood et al. 1998) that were subsequently utilized to derive detailed and quantitative peptide binding motifs and predictive algorithms for each molecule (Greenbaum et al. 2011; Sidney et al. 2010b; Solomon et al. 2010; Wang et al. 2008; Wang et al. 2010). Here, we further supplemented this panel to include 19 additional alleles expressed at relatively lower frequency, but for which single HLA class II transfected cell lines were already available. Together, this panel of 46 HLA DR, DQ and DP specificities provides population coverage of almost 90% at each locus (and over 66% of all genes at each locus) (Table 1). While the 19 additional alleles have only a limited impact on total coverage (Online Resource 1), they do provide a resource to study alleles and sub-types that may be of interest in specific donor cohorts.
Table 1.
Phenotype frequencies of transfected HLA class II alleles1
| Allele(s)2 | Genotype frequency | Phenotype frequency | Transfectant line(s)3
|
|
|---|---|---|---|---|
| Name(s) | Host line | |||
| DRB1*0101 | 2.8 | 5.4 | L57.23, L554.3 | DAP.3 |
| DRB1*0301 | 7.1 | 13.7 | TR166, IHW03289 | DAP.3 |
| DRB1*0302 | 1.1 | 2.1 | IHW03290 | DAP.3 |
| DRB1*0401 | 2.3 | 4.6 | IHW03291 | DAP.3 |
| DRB1*0402 | 1.1 | 2.2 | L514.5, IHW03292 | DAP.3 |
| DRB1*0403 | 2.3 | 4.5 | L259.4 | DAP.3 |
| DRB1*0404 | 1.9 | 3.8 | L300.8 | DAP.3 |
| DRB1*0405 | 3.1 | 6.2 | L566M.6 | DAP.3 |
| DRB1*0407 | 2.4 | 4.8 | L331.2 | DAP.3 |
| DRB1*0411 | 1.6 | 3.3 | L527.8 | DAP.3 |
| DRB1*0701 | 7.0 | 13.5 | L605.2, IHW03293 | DAP.3 |
| DRB1*0802 | 2.5 | 4.9 | RR0802.2 | RM3 |
| DRB1*0901 | 3.1 | 6.2 | RR0901.2 | RM3 |
| DRB1*1101 | 6.1 | 11.8 | L581.13, IHW03291 | DAP.3 |
| DRB1*1102 | 1.1 | 2.2 | L625.7, IHW03295 | DAP.3 |
| DRB1*1103 | 0.3 | 0.50 | L624.9, IHW03296 | DAP.3 |
| DRB1*1104 | 1.4 | 2.8 | L537.1, IHW03297 | DAP.3 |
| DRB1*1201 | 2.0 | 3.9 | RR1201.1 | RM3 |
| DRB1*1301 | 3.2 | 6.3 | L597.2, IHW03298 | DAP.3 |
| DRB1*1302 | 3.9 | 7.7 | L650.2, IHW03299 | DAP.3 |
| DRB1*1303 | 1.2 | 2.4 | IHW03300 | DAP.3 |
| DRB1*1304 | 0.1 | 0.2 | IHW03301 | DAP.3 |
| DRB1*1401 | 3.4 | 6.7 | L167.2 | DAP.3 |
| DRB1*1402 | 2.8 | 5.6 | L182.1 | DAP.3 |
| DRB1*1501 | 6.3 | 12.2 | L466.1 | DAP.3 |
| DRB1*1601 | 1.0 | 1.9 | L415.2, L242.5 | DAP.3 |
|
| ||||
| DRB1 Total | 71.1 | 91.7 | ||
|
| ||||
| DRB3*0101 | 14.0 | 26.1 | TR81.19, L575.1 | DAP.3 |
| DRB3*0202 | 18.9 | 34.3 | RRB3.02.2 | RM3 |
| DRB3*0301 | 6.7 | 13.0 | L576.5 | DAP.3 |
| DRB4*0101 | 23.7 | 41.8 | L257.6 | DAP.3 |
| DRB5*0101 | 8.3 | 16.0 | L416.3, IHW03304 | DAP.3 |
| DRB5*0102 | 5.1 | 9.8 | L467.1 | DAP.3 |
|
| ||||
| DRB3/4/5 Total | 76.7 | 94.6 | ||
|
| ||||
| DQA1*0501/DQB1*0201 | 5.8 | 11.3 | RQ0201.x | RM3 |
| DQA1*0201/DQB1*0201 | 5.7 | 11.1 | L21.3 | DAP.3 |
| DQA1*0501/DQB1*0301 | 19.5 | 35.1 | RQ0301.x | RM3 |
| DQA1*0301/DQB1*0302 | 10.0 | 19.0 | RQ0302.3 | RM3 |
| DQA1*0401/DQB1*0402 | 6.6 | 12.8 | RQ0402.3 | RM3 |
| DQA1*0101/DQB1*0501 | 7.6 | 14.6 | RQ0501.3 | RM3 |
| DQA1*0102/DQB1*0502 | 3.5 | 6.9 | RQ0501.3 | RM3 |
| DQA1*0102/DQB1*0602 | 7.6 | 14.6 | RQ0602.3 | RM3 |
|
| ||||
| DQA1/DQB1 Total | 66.3 | 88.7 | ||
|
| ||||
| DPA1*0201/DPB1*0101 | 8.4 | 16.0 | RP0101.2 | RM3 |
| DPA1*0103/DPB1*0201 | 9.2 | 17.5 | L256.12 (1H) | DAP.3 |
| DPA1*0103/DPB1*0401 | 20.1 | 36.2 | RP0401.2 | RM3 |
| DPA1*0103/DPB1*0402 | 23.6 | 41.6 | L25.4 | DAP.3 |
| DPA1*0202/DPB1*0501 | 11.5 | 21.7 | RP0501.2 | RM3 |
| DPA1*0201/DPB1*1401 | 3.8 | 7.4 | RP1401.2 | RM3 |
|
| ||||
| DPB1 Total | 76.5 | 94.5 | ||
Average haplotype and phenotype frequencies for individual alleles are based on data available at dbMHC, and were calculated as described in the Materials and Methods. Alleles comprising the panel previously characterized in detail for binding specificity, as described in (Greenbaum et al. 2011), are highlighted by bold font.
DQ and DP lines were transfected with corresponding alpha and beta chains, as indicated. All DR lines were transfected with the indicated beta chain and the DRA1*0101 alpha chain. Lines generated specifically for the current study are indicated by italicized bold font.
DAP.3 lines were generated as previously described (Karr et al. 1991; Klohe et al. 1988; Lair et al. 1988). RM3 transfected lines were generated for the present study as described in the Materials and Methods.
The select panel allows comprehensive HLA coverage at all loci in diverse populations
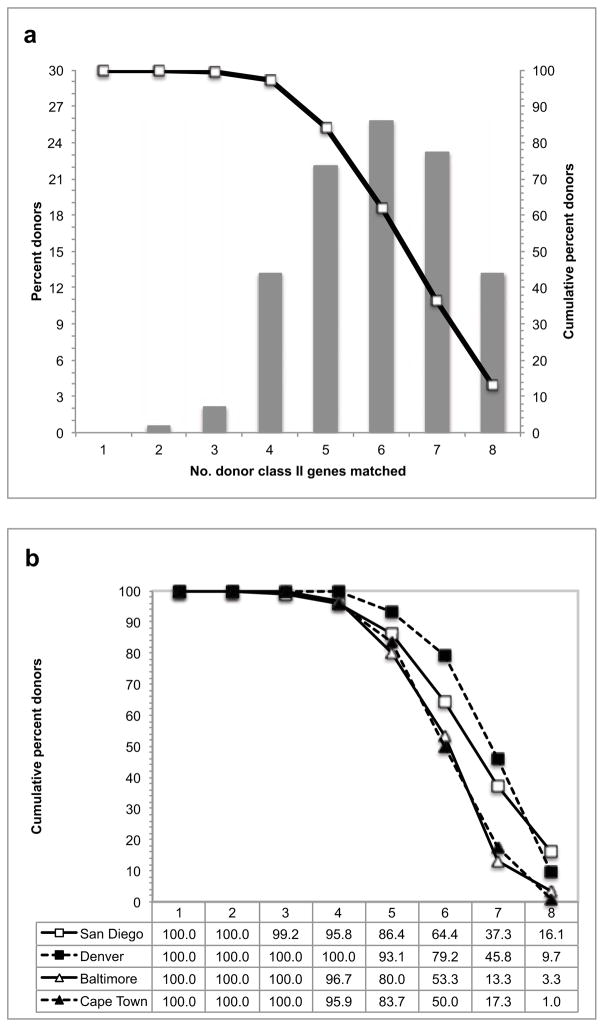
To verify that the selected alleles provide satisfactory coverage in a real-life context, we analyzed data derived from two San Diego-based clinical study cohorts totaling 190 donors, representing a mix of Caucasian, Hispanic, Black and Asian ethnicities (see Online Resource 2). As shown in Figure 1A, the panel of 46 alleles allowed an exact match of 4 or more alleles (out of 8 possible HLA class II expressed per donor) in over 95% of the two cohorts, and for 6 or more out of 8 in over 60%.
Fig. 1. Allelic coverage.
The HLA class II alleles represented in the panel of 46 single transfected cell lines provide coverage of the majority of HLA class II types expressed in a cohort of 190 donors recruited for two different studies in the San Diego area. The fraction of all donors for which the panel provides coverage of 0 to 8 possible class II types expressed is shown in the bar graph (a). The cumulative fraction of donors covered is shown in the line plot (a). This coverage is consistent across different donor cohorts from geographically disparate regions (b).
To verify that the panel would afford coverage in a broader context, to include different geographic locations, we next analyzed data from cohorts of 72 and 30 donors in Denver and Baltimore, respectively. We also analyzed a cohort of 98 donors enrolled in a clinical study in a rural setting outside Cape Town, South Africa. Together, and consistent with our observation with the San Diego based cohort, the panel of alleles selected allowed exact matches for 4 or more donor expressed alleles for about 95% of each cohort, and the overall levels of coverage were generally consistent across the different donor cohorts analyzed (Figure 1B), despite striking differences in ethnic composition between the cohorts. Thus, from these data we conclude that the HLA alleles selected indeed afford broad coverage of different population groups.
Finally, we assessed whether the allele frequencies in the collective donor cohort are largely reflective of frequencies in the general worldwide population. As shown in Online Resource 3, with only a few exceptions, the collective frequencies observed in the cohorts studied are largely reflective of those in the general population, further validating the relevance of the panel of alleles chosen.
Generation of a panel of single HLA class II-transfected cell lines
As mentioned in the introduction, and indicated in Table 1, a number of single HLA class II-transfected cell lines had already been generated in previous studies (Karr et al. 1991; Klohe et al. 1988; Lair et al. 1988), and made available to the scientific community. Here, to enable a more comprehensive coverage of the general population, we set out to generate cell lines for several molecules for which single HLA transfectants were unavailable. The experimental scheme utilized for this purpose is shown in Figure 2. Briefly, ORF clones containing HLA Class II alpha or beta chains were obtained from the members of the ORFeome Collaboration (PlasmID, Dana-Farber/Harvard Cancer Center DNA Resource Core, Boston, MA and GeneCopoeia, Rockville, MD). Alternately, synthetic genes based on the coding region sequence information available in the IMGT/HLA Database (http://www.ebi.ac.uk/imgt/hla/) (Robinson et al. 2003; Robinson et al. 2011; Robinson et al. 2000) were constructed. Using the Gateway system, the alpha and beta chain genes were cloned into pcDNA-DEST40 vector (beta chains; G418 selection) or pcDNA 6.2/V5 DEST vector (alpha chains; blasticidin selection). After transfection into RM3 cells (class II expression negative), cells expressing both chains were selected using both G418 and blasticidin antibiotics, and expanded.
Fig. 2. Transfection scheme.
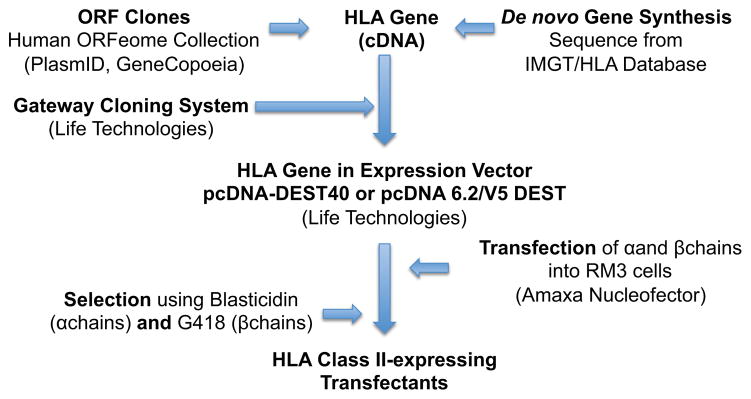
Schematic representation of the process used to generate HLA class II genes and to transfect them into RM3 cell lines.
The level and specificity of expression of the relevant HLA molecule was addressed by fluorescent-activated cell sorting (FACS) using DR, DP and DQ specific antibodies. Representative FACS profiles are shown in Figure 3 for one cell line each representative of the DR (DRB3*02:02), DQ (DQB1*06:02) and DP (DPB1*04:01) loci. As expected, each cell line is positive for expression of the corresponding transfected locus, and negative for the others,
Fig. 3. Specificity of RM3 transfectant HLA expression.
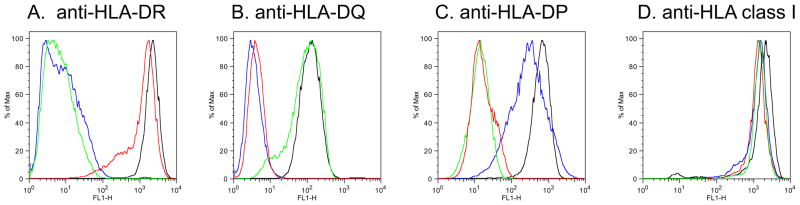
Expression of HLA class II MHC molecules in single transfected RM3 cell lines, as determined by HLA locus specific monoclonal antibodies. Expression was evaluated by flow cytometry using anti-DR (Panel a), -DP (Panel b), and –DQ (Panel c) antibodies (LB3.1, B7/21, and SPVL3, respectively). As a positive control, HLA Class I expression was evaluated using the anti-Class I antibody, W6/32 (Panel d). Black: LG2 EBV cell line (positive control); red: HLA DRB3*02:02 transfected RM3 line RRB3.02.2; green: HLA DQB1*06:02 transfected RM3 line RQ0602.3; blue: HLA DPB1*04:01 transfected RM3 line RP0401.2.
To select for thresholds of expression that would be functionally relevant, we utilized the information derived from the previous panel of transfectants. These cell lines express HLA class II molecules in the 100 to 1000 Mean Fluorescence Intensity (MFI) range, which is functionally relevant since the lines have been successfully utilized in MHC restriction analyses in the context of HLA-DR, -DQ and -DP (see, e.g., (Jaraquemada et al. 1990; Lechler et al. 1988; Jacobson et al. 1989; Sekaly et al. 1988; Austin et al. 1985; Nakatsuji et al. 1987; Larche 2008)). Accordingly, all of the clones selected for the present panel of cell lines expressed the transfected HLA with a MFI of 200 or greater (range 200–1500, average 850). In conclusion, a panel of 46 different single HLA class II transfected cell lines, to include both previous lines and the novel ones reported here, is now available (see Table 1).
Functional validation of the transfected cell lines
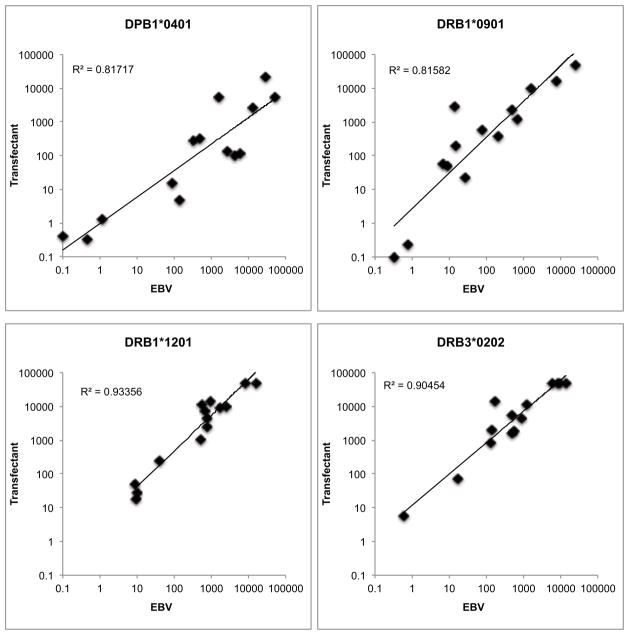
Next, we sought to further validate the newly obtained transfectants by demonstrating their MHC binding specificity and antigen presenting capacity. In a first series of experiments, we purified HLA class II molecules from selected transfectants to compare their peptide binding profile with the binding profile obtained utilizing the same HLA class II allelic variants purified from homozygous EBV transformed B cell lines. For each specificity, panels of 12–14 peptides of varying affinities were tested. Representative results for four alleles (DPB1*04:01, DRB1*09:01, DRB1*12:01 and DRB3*02:02) are shown in Figure 4. In each case, the affinities measured using MHC purified from the single transfectants correlated strongly (R^2 0.81–0.93) with affinities measured using MHC purified from the EBV lines. These data confirm the peptide binding specificity of the MHC molecules produced by the transfected cell lines.
Fig. 4. Functional validation of transfected HLA: evaluation of peptide binding capacity.
HLA class II molecules were purified from single transfected cell lines as described in the Materials and Methods, and then tested for their capacity to bind a panel of peptides representing a range of previously known affinities. The binding affinities of peptides in the panel (Y-axis) were compared to those obtained at the same time using MHC purified from homozygous EBV transformed lines (X-axis) available in the IHWG reference panel. Shown are affinities, expressed in terms of IC50 nM, obtained for four representative preparations. In each case the correlation between affinities measured using MHC from both sources was >0.81, comparable to the correlation observed when using two different preparations of the same molecule purified from the same EBV cell line.
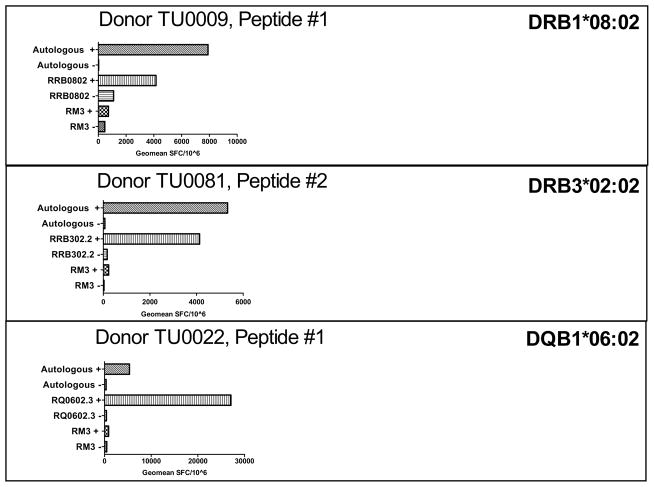
To validate the single HLA class II molecules transfected cell lines at the level of antigen presentation, short-term T cell lines of known epitope specificity and HLA restriction were utilized for antigen presentation assays with specific transfected cell lines. Representative results from this type of experiment are shown in Figure 5 for T cell lines specific for epitopes restricted by DRB1*08:02, DRB3*02:02, and DQB1*06:02. In each case, the response obtained with the antigen presenting cells transfected with the relevant allelic molecule was comparable to that observed with autologous antigen presenting cells, while untransfected antigen presenting cell lines, or cell lines transfected with an irrelevant allele, did not yield significant responses, as defined in the Materials and Methods. Thus, we conclude that the single HLA class II transfected cell lines have been validated at the functional level.
Fig. 5. Responses obtained with transfected cell lines are specific and comparable to those observed with autologous antigen presenting cells.
Transfected cell lines were utilized as APCs to probe T cell responses to specific epitopes. T cell assays and cell culture were performed as described in the Materials and Methods. In each assay, T cell responses against autologous APCs, RM3 HLA transfected, and RM3 untransfected cells were determined by measuring IFN (SFC/10^6), both with (+) and without (−) peptide. Shown are representative responses from three individual donors.
The panel of transfected cell lines allows rapid determination of epitope restriction
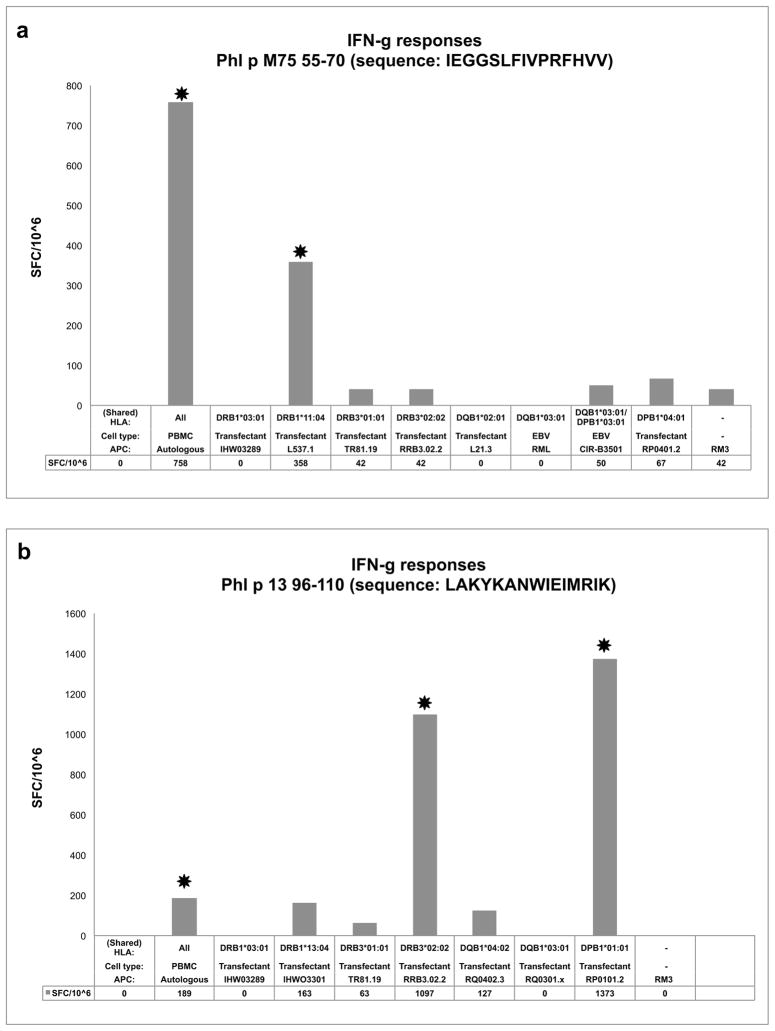
To exemplify the application of the panel of transfected cell lines to determination of epitope specific MHC restriction, we assayed T cell lines derived from two Timothy grass allergic donors (D00089 and U00164) that were specific for two different Timothy grass derived epitopes (Phl p uncharacterized protein [M75] 55–70 and Phl p 13 96–110, respectively).
Since the HLA types of these donors were previously determined as described in the Materials and Methods, the corresponding single HLA transfected cell lines were placed in culture, and used in antigen presentation assays. More specifically, for donor D00089 the DRB1*03:01, DRB1*11:04, DRB3*01:01, DRB3*02:02 DQB1*02:01, DQB1*03:01, DPB1*03:01 and DPB1*04:01 transfected cell lines were put in culture and utilized as APCs in these assays. As shown in Figure 6A, in donor D00089, Phl p M75 55–70 was determined to be monogamously restricted at the DR locus, since IFN- responses were only observed when the peptide was presented by the DRB1*11:04 cell line. Similarly, in the case of donor U00164, the DRB1*03:02, DRB1*13:04, DRB3*01:01, DRB3*02:02, DQB1*03:19, DQB1*04:02, and DPB1*01:01 transfected cell lines, matching the corresponding donor HLA alleles, were placed in culture. In this case, it was found that the Phl p 13 96–110 epitope was promiscuously restricted by DRB3*02:02 and DPB1*01:01 (Figure 6B), with strong IFN- responses observed when the peptide was presented by cell lines expressing these two MHC specificities.
Fig. 6. Efficient determination of HLA restriction using single transfectant cell lines.
Panels of single transfected cell lines matching donor HLA types were utilized as APCs to measure donor responses to specific Timothy grass epitopes. T cell assays were performed, and criteria for positivity utilized, as described in the Materials and Methods. Significant positive responses are indicated by an asterisk. For donor D00089 (a), the Phl p M75 55–70 epitope only induced IFNγ responses (SFC/10^6) when presented in the context of either autologous APCs or the HLA DRB1*11:04 transfectant L537.1 For donor U00164 (b), the Phl p 13 96–110 epitope induced responses when presented by autologous APCs, or the DRB3*02:02 and DPB1*01:01 transfected cell lines (RRB3.02.2 and RP0101.2, respectively).
Taken together, these data demonstrate how the HLA restriction of epitopes recognized by a single donor can be clearly and unambiguously determined in a single experiment. The efficacy of this type of analysis underscores the utility of the cell lines that have been established.
DISCUSSION
The present study details the derivation of a panel of single HLA class II transfected cell lines that, for each of the class II MHC loci, provides coverage for the vast majority of the general population. This panel of cell lines can be used for determining the HLA restriction for most class II restricted T cell epitopes, unequivocally, in a single experiment. By contrast, determination of HLA restriction using a combination of locus specific monoclonal inhibition assays and panels of HLA mismatched EBV lines would entail the performance of multiple assays, and in many cases would not yield decisive information as to exact allelic restriction patterns. Given the broad repertoire overlap that is a hallmark of HLA class II molecules (Greenbaum et al. 2011), and the relative binding promiscuity of HLA class II epitopes (Greenbaum et al. 2011; Oseroff et al. 2010; Sidney et al. 2010a, b), the ability to clearly and decisively determine restriction represents an important advance.
There is ample evidence that each class II MHC locus encodes fully functional molecules, so that this multi-locus and multi-allelic complexity cannot be ignored (Marsh et al. 2000). In a recent survey of HLA class II responses to a common allergen, Timothy grass, utilizing an unbiased approach, the restricting locus for T cells responding to 140 epitopes derived from 10 major Timothy grass allergenic proteins was found to be DR in 61% of the cases, DP in 21%, and DQ in 18% (Oseroff et al. 2010). A similar trend was noted in a recent study characterizing epitopes derived from a panel of common allergens (Oseroff et al. 2012b), where 66, 15 and 19% of the mapped responses were DR, DP and DQ restricted, respectively. And similarly, in a study describing epitopes derived from German cockroach allergens, the rates were 55, 15 and 30%, respectively. These data suggest that while most of the responses are DR restricted, DP and DQ also account for a very appreciable fraction of the total response against recognized epitopes. A similar pattern appears to hold in the case of Mycobacterium tuberculosis (Arlehamn et al. 2012a).
The panel of cell lines described herein provides a resource not just for the commonly probed DRB1 alleles, but also for the most common DRB3/4/5, DQ and DP alleles. Accordingly, the availability of lines for non-DRB1 alleles makes determination of restriction by other loci feasible. Given that expression of DQ and DP alleles is typically lower than DR (Alcaide-Loridan et al. 1999; Peretti et al. 2001; Edwards et al. 1986; Hauber et al. 1995; Guardiola and Maffei 1993; Maurer et al. 1987), determining restriction is more difficult with EBV lines. However, MHC expression in the transfected cell lines is fairly uniform across the different loci, further emphasizing the utility of this panel of transfectants. This is a key feature of our study, because while common DRB3/4/5, DQ and DP alleles are understudied, as the recent data noted above clearly emphasizes, perhaps half of common epitopes are restricted by these loci.
Another important feature of the panel of single MHC transfected cell lines such as we have assembled here is that it allows mapping restriction of promiscuous epitopes. By contrast, without the availability of a panel with the breadth such as described herein, restriction mapping would require initial determination of the restricting locus (or loci) using monoclonal antibody inhibition assays, followed by experiments using panels of HLA mismatched cell lines as APCs. However, given the generally high degree of cross-reactivity between HLA class II molecules (see, e.g., (Greenbaum et al. 2011)), these types of assays, dependent upon decoding often complex panels of mismatched lines, typically lead to ambiguous restriction mapping (see, e.g., (Oseroff et al. 2012a; Oseroff et al. 2012b)).
The panel now available provides coverage of the majority of alleles expressed in most donors. In total, the selection of alleles covers about two-thirds of all genes at each of the four class II loci. With only one exception (DPB1*03:01), the panel includes all alleles present with gene frequencies >5%. Future studies will enable inclusion of additional alleles. However, it should be noted that while additional reagents will be valuable in specific cases, they would provide diminishing return in terms of overall population coverage. For example, in the case of the DQ locus, the 8 haplotypes included in the panel cumulatively cover about 66% of DQ genes. Inclusion of the 8 next most common haplotypes would only provide an additional 15% coverage. Similarly, doubling the panel of DP specificities, which presently covers about 76% of the genes, would provide only an additional 16%. Indeed, expanding the panel from the 27 most common alleles to 46, as described here, had only a limited impact on the depth of coverage. Specifically, while the full panel allowed an exact match at 4 or more alleles (out of 8 possible HLA class II expressed per donor) in over 95% of donors in the two San Diego cohorts studied, it is notable that the sub-panel of 27 already allowed a similar match for 88% (Online Resource 1).
Another feature of the panel is that it provides uniform coverage across populations of very different ethnic composition. Specifically, coverage of about 65% of the genes at each locus, and 95% of individuals in each population examined, was achieved. This high level of coverage was obtained in a cohort from Cape Town, comprised largely of Cape Mixed Ancestry, noted as perhaps having the highest level of mixed ancestry in the world (Tishkoff et al. 2009), a cohort of largely North American Blacks from Baltimore, a largely Caucasian cohort from Denver, and a relatively mixed (Caucasian, Asian and Hispanic) cohort from San Diego. Based on publically available population frequency data (Meyer et al. 2007; Middleton et al. 2003), we expect that this high degree of coverage would be maintained in most, if not all, major populations worldwide. Thus, the present panel represents a broadly applicable tool for characterizing epitope responses, and one that would be of particular value for cohorts of diverse ethnic background, such as, for example, the general US population. Further, because of the breadth of coverage afforded, the panel would also be applicable for characterizing donor samples from studies and clinical trials pertaining to disease indications such as HIV, tuberculosis, and malaria, to name three main infectious diseases disproportionally affecting non-Caucasians.
A large amount of data and assay validation is already available for the panel of molecules selected and described herein. HLA binding assays utilizing purified HLA molecules in vitro have been validated for 29 of the 46 alleles, and predictive algorithms are available on the IEDB website (www.IEDB.org (Vita et al. 2010)) for all of them. Finally, HLA tetramers are available or under development for 32 of the 46 alleles.
Over the last 15 years, an impressive wealth of information has been generated by taking advantage of tetrameric staining reagents (Constantin et al. 2002; Nepom et al. 2002; Kwok 2003; Nepom 2012). Importantly, tetramers provide a means for accurate enumeration of T cells specific for a given epitope, irrespective of their functional state or phenotype, and also allow sophisticated phenotyping of the responding cells, often without in vitro manipulations. Recent advances in the technology have involved the development of tetramer enrichment protocols, which allow the detection of cells present at very low frequencies, and even studying the repertoire of naïve T cells available for a given epitope specificity (Moon et al. 2007; Moon et al. 2009; Kotturi et al. 2008; James et al. 2007). In the past few years, several techniques have been developed to allow for multiplex tetrameric staining, making it possible to map antigenic epitopes for multiple class II alleles simultaneously (Newell et al. 2012; Alivisatos et al. 2005; Yang et al. 2006). Furthermore, the generation of HLA class II tetramers, albeit considerably more challenging than generation of class I tetramers, has become more approachable (Nepom 2012; Kwok 2003)
Our laboratory has been designing a systematic approach to address determination of HLA restriction and empower ICS and tetramer staining studies. Our vision is to assemble, or enable assembling, repositories of epitopes for each antigenic system, with well-defined HLA restriction. In practice, considering by way of example a typical epitope identification study, following donor recruitment and collection and processing of blood donations, donor PBMC could be screened using high-throughput ELISPOT assays for responses against candidate epitopes (for recent examples from our laboratory, see (Oseroff et al. 2010; Oseroff et al. 2012a; Oseroff et al. 2012b; Arlehamn et al. 2012b). After identification of specific epitopes inducing donor T cell responses, the actual HLA class II allele restricting the response could then be determined in a single follow-up assay utilizing single transfected cell lines as APCs. The end result of this activity would be the availability of repositories of epitopes for each antigenic system, with defined HLA restriction for each of the alleles of interest. For any study probing vaccine performance or host pathogen interactions, we then envision that donors would be HLA typed, and the epitopes restricted by the HLA types expressed in the particular donor pooled and used in ICS assays, allowing use of minimal amounts of blood. In the case of tetrameric reagents, all corresponding tetramers could be either pooled, or used in multiplex configurations. Either way, the ability to simultaneously characterize epitope specific reactivity restricted by multiple HLA and multiple epitopes in a given donor is expected to increase, by additive effects, the frequency of detectable antigen specific cells, thus reducing the amount of blood sample required in the studies.
In conclusion, the work summarized here has led to the establishment of a panel of HLA class II single allele transfected cell lines. The panel provides coverage of the most common class II alleles, and as such provides a valuable tool for characterizing the immune response to disease specific epitopes in the general population.
Supplementary Material
Online Resource 1. Comparison of population coverage afforded by the full panel and the sub-panel of the 27 most common alleles
The collective frequencies provided by the HLA class II alleles represented in the panel of 46 single transfected cell lines, and the sub-panel of the 27 most common alleles, are compared. Coverage was examined in a cohort of 190 donors recruited for two different studies in the San Diego area. The cumulative percent of all donors for which the panels provide coverage of 0 to 8 possible class II types expressed is shown.
Online Resource 2. Ethnic breakdown of four donor cohorts from different geographical areas
Representation of different ethnic groups, in terms of percent of individuals, in the San Diego, Denver, Baltimore and Cape Town cohorts. Ethnicity is as described by donors during clinical enrollment.
Online Resource 3. Correspondence between HLA frequencies in the donor cohort and the general population
The collective frequencies of alleles present in the San Diego, Denver, Baltimore and Cape Town cohorts examined largely match those found in the general worldwide population, based on HLA frequency data obtained from dbMHC (Middleton et al. 2003; Meyer et al. 2007).
Acknowledgments
We thank Victoria Tripple, Duy Le and Ryan Henderson for technical assistance. We thank Dr. Robert Karr for providing us with a large panel of single transfected HLA class II cell lines, and Dr. Karr and Dr. Howard Grey for their helpful comments. This work was supported by National Institutes of Health contract Nr. N01-AI-900044C, AI-900048C, AI-100275 (to A.S.) and Bill and Melinda Gates Foundation grant OPP1021972-3 (to W.H.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute Of Allergy And Infectious Diseases or the National Institutes of Health.
REFEENCES
- Alcaide-Loridan C, Lennon AM, Bono MR, Barbouche R, Dellagi K, Fellous M. Differential expression of MHC class II isotype chains. Microbes and infection / Institut Pasteur. 1999;1 (11):929–934. doi: 10.1016/s1286-4579(99)00224-5. [DOI] [PubMed] [Google Scholar]
- Alivisatos AP, Gu W, Larabell C. Quantum dots as cellular probes. Annu Rev Biomed Eng. 2005;7:55–76. doi: 10.1146/annurev.bioeng.7.060804.100432. [DOI] [PubMed] [Google Scholar]
- Arlehamn CS, Gerasimova A, Mele F, Henderson R, Swann J, Greenbaum JA, Kim Y, Sidney J, James EA, Taplitz R, McKinney DM, Kwok WW, Grey H, Sallusto F, Peters B, Sette A. Memory T cells in Latent Mycobacterium tuberculosis Infection are Directed Against Three Antigenic Islands and Largely Contained in a CXCR3+CCR6+ Th1 Subset. PLoS Pathog. 2012a doi: 10.1371/journal.ppat.1003130. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlehamn CS, Sidney J, Henderson R, Greenbaum JA, James EA, Moutaftsi M, Coler R, McKinney DM, Park D, Taplitz R, Kwok WW, Grey H, Peters B, Sette A. Dissecting Mechanisms of Immunodominance to the Common Tuberculosis Antigens ESAT-6, CFP10, Rv2031c (hspX), Rv2654c (TB7.7), and Rv1038c (EsxJ) J Immunol. 2012b;188(10):5020–5031. doi: 10.4049/jimmunol.1103556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin P, Trowsdale J, Rudd C, Bodmer W, Feldmann M, Lamb J. Functional expression of HLA-DP genes transfected into mouse fibroblasts. Nature. 1985;313 (5997):61–64. doi: 10.1038/313061a0. [DOI] [PubMed] [Google Scholar]
- Busch R, Strang G, Howland K, Rothbard JB. Degenerate binding of immunogenic peptides to HLA-DR proteins on B cell surfaces. Int Immunol. 1990;2 (5):443–451. doi: 10.1093/intimm/2.5.443. [DOI] [PubMed] [Google Scholar]
- Calman AF, Peterlin BM. Evidence for a trans-acting factor that regulates the transcription of class II major histocompatibility complex genes: genetic and functional analysis. Proc Natl Acad Sci U S A. 1988;85 (23):8830–8834. doi: 10.1073/pnas.85.23.8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22 (23):3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- Constantin CM, Bonney EE, Altman JD, Strickland OL. Major histocompatibility complex (MHC) tetramer technology: an evaluation. Biol Res Nurs. 2002;4 (2):115–127. doi: 10.1177/1099800402238332. [DOI] [PubMed] [Google Scholar]
- Doherty DG, Penzotti JE, Koelle DM, Kwok WW, Lybrand TP, Masewicz S, Nepom GT. Structural basis of specificity and degeneracy of T cell recognition: pluriallelic restriction of T cell responses to a peptide antigen involves both specific and promiscuous interactions between the T cell receptor, peptide, and HLA-DR. J Immunol. 1998;161 (7):3527–3535. [PubMed] [Google Scholar]
- Dzuris JL, Sidney J, Horton H, Correa R, Carter D, Chesnut RW, Watkins DI, Sette A. Molecular determinants of peptide binding to two common rhesus macaque major histocompatibility complex class II molecules. J Virol. 2001;75 (22):10958–10968. doi: 10.1128/JVI.75.22.10958-10968.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JA, Durant BM, Jones DB, Evans PR, Smith JL. Differential expression of HLA class II antigens in fetal human spleen: relationship of HLA-DP, DQ, and DR to immunoglobulin expression. J Immunol. 1986;137 (2):490–497. [PubMed] [Google Scholar]
- Giraldo-Vela JP, Rudersdorf R, Chung C, Qi Y, Wallace LT, Bimber B, Borchardt GJ, Fisk DL, Glidden CE, Loffredo JT, Piaskowski SM, Furlott JR, Morales-Martinez JP, Wilson NA, Rehrauer WM, Lifson JD, Carrington M, Watkins DI. The major histocompatibility complex class II alleles Mamu-DRB1*1003 and -DRB1*0306 are enriched in a cohort of simian immunodeficiency virus-infected rhesus macaque elite controllers. J Virol. 2008;82(2):859–870. doi: 10.1128/JVI.01816-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum J, Sidney J, Chung J, Brander C, Peters B, Sette A. Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics. 2011;63(6):325–335. doi: 10.1007/s00251-011-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guardiola J, Maffei A. Control of MHC class II gene expression in autoimmune, infectious, and neoplastic diseases. Crit Rev Immunol. 1993;13 (3–4):247–268. [PubMed] [Google Scholar]
- Gulukota K, Sidney J, Sette A, DeLisi C. Two complementary methods for predicting peptides binding major histocompatibility complex molecules. J Mol Biol. 1997;267 (5):1258–1267. doi: 10.1006/jmbi.1997.0937. [DOI] [PubMed] [Google Scholar]
- Hauber I, Gulle H, Wolf HM, Maris M, Eggenbauer H, Eibl MM. Molecular characterization of major histocompatibility complex class II gene expression and demonstration of antigen-specific T cell response indicate a new phenotype in class II-deficient patients. J Exp Med. 1995;181 (4):1411–1423. doi: 10.1084/jem.181.4.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho PC, Mutch DA, Winkel KD, Saul AJ, Jones GL, Doran TJ, Rzepczyk CM. Identification of two promiscuous T cell epitopes from tetanus toxin. Eur J Immunol. 1990;20(3):477–483. doi: 10.1002/eji.1830200304. [DOI] [PubMed] [Google Scholar]
- Jacobson S, Sekaly RP, Jacobson CL, McFarland HF, Long EO. HLA class II-restricted presentation of cytoplasmic measles virus antigens to cytotoxic T cells. J Virol. 1989;63 (4):1756–1762. doi: 10.1128/jvi.63.4.1756-1762.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James EA, Bui J, Berger D, Huston L, Roti M, Kwok WW. Tetramer-guided epitope mapping reveals broad, individualized repertoires of tetanus toxin-specific CD4+ T cells and suggests HLA-based differences in epitope recognition. Int Immunol. 2007;19(11):1291–1301. doi: 10.1093/intimm/dxm099. [DOI] [PubMed] [Google Scholar]
- Jaraquemada D, Martin R, Rosen-Bronson S, Flerlage M, McFarland HF, Long EO. HLA-DR2a is the dominant restriction molecule for the cytotoxic T cell response to myelin basic protein in DR2Dw2 individuals. J Immunol. 1990;145 (9):2880–2885. [PubMed] [Google Scholar]
- Karr RW, Panina-Bordignon P, Yu WY, Lanzavecchia A. Antigen-specific T cells with monogamous or promiscuous restriction patterns are sensitive to different HLA-DR beta chain substitutions. J Immunol. 1991;146 (12):4242–4247. [PubMed] [Google Scholar]
- Klohe EP, Watts R, Bahl M, Alber C, Yu WY, Anderson R, Silver J, Gregersen PK, Karr RW. Analysis of the molecular specificities of anti-class II monoclonal antibodies by using L cell transfectants expressing HLA class II molecules. J Immunol. 1988;141 (6):2158–2164. [PubMed] [Google Scholar]
- Kotturi MF, Scott I, Wolfe T, Peters B, Sidney J, Cheroutre H, von Herrath MG, Buchmeier MJ, Grey H, Sette A. Naive precursor frequencies and MHC binding rather than the degree of epitope diversity shape CD8+ T cell immunodominance. J Immunol. 2008;181(3):2124–2133. doi: 10.4049/jimmunol.181.3.2124. 181/3/2124 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger JI, Karr RW, Grey HM, Yu WY, O’Sullivan D, Batovsky L, Zheng ZL, Colon SM, Gaeta FC, Sidney J, et al. Single amino acid changes in DR and antigen define residues critical for peptide-MHC binding and T cell recognition. J Immunol. 1991;146 (7):2331–2340. [PubMed] [Google Scholar]
- Kwok WW. Challenges in staining T cells using HLA class II tetramers. Clin Immunol. 2003;106 (1):23–28. doi: 10.1016/s1521-6616(02)00018-9. [DOI] [PubMed] [Google Scholar]
- Lair B, Alber C, Yu WY, Watts R, Bahl M, Karr RW. A newly characterized HLA-DP beta-chain allele. Evidence for DP beta heterogeneity within the DPw4 specificity. J Immunol. 1988;141 (4):1353–1357. [PubMed] [Google Scholar]
- Larche M. Determining MHC restriction of T-cell responses. Methods Mol Med. 2008;138:57–72. doi: 10.1007/978-1-59745-366-0_6. [DOI] [PubMed] [Google Scholar]
- Lechler RI, Bal V, Rothbard JB, Germain RN, Sekaly R, Long EO, Lamb J. Structural and functional studies of HLA-DR restricted antigen recognition by human helper T lymphocyte clones by using transfected murine cell lines. J Immunol. 1988;141 (9):3003–3009. [PubMed] [Google Scholar]
- Marsh SGE, Parham P, Barber LD. The HLA FactsBook. Academic Press; London: 2000. [Google Scholar]
- Maurer DH, Hanke JH, Mickelson E, Rich RR, Pollack MS. Differential presentation of HLA-DR, DQ, and DP restriction elements by interferon-gamma-treated dermal fibroblasts. J Immunol. 1987;139 (3):715–723. [PubMed] [Google Scholar]
- Meyer D, Singe R, Mack S, Lancaster A, Nelson M, Erlich H, Frenandez-Vina M, Thomson G. Single Locus Polymorphism of Classical HLA Genes. Immunobiology of the Human MHC: Proceedings of the 13th International Histocompatibility Workshop and Conference; Seattle, WA. 2007. pp. 653–704. [Google Scholar]
- Middleton D, Menchaca L, Rood H, Komerofsky R. New allele frequency database. Tissue Antigens. 2003;61(5):403–407. doi: 10.1034/j.1399-0039.2003.00062.x. http://www.allelefrequencies.net. 062 [pii] [DOI] [PubMed] [Google Scholar]
- Moon JJ, Chu HH, Hataye J, Pagan AJ, Pepper M, McLachlan JB, Zell T, Jenkins MK. Tracking epitope-specific T cells. Nat Protoc. 2009;4(4):565–581. doi: 10.1038/nprot.2009.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon JJ, Chu HH, Pepper M, McSorley SJ, Jameson SC, Kedl RM, Jenkins MK. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity. 2007;27(2):203–213. doi: 10.1016/j.immuni.2007.07.007. S1074-7613(07)00366-4 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuji T, Inoko H, Ando A, Sato T, Koide Y, Tadakuma T, Yoshida TO, Tsuji K. The role of transfected HLA-DQ genes in the mixed lymphocyte reaction-like condition. Immunogenetics. 1987;25 (1):1–6. doi: 10.1007/BF00768826. [DOI] [PubMed] [Google Scholar]
- Nepom GT. MHC class II tetramers. J Immunol. 2012;188(6):2477–2482. doi: 10.4049/jimmunol.1102398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepom GT, Buckner JH, Novak EJ, Reichstetter S, Reijonen H, Gebe J, Wang R, Swanson E, Kwok WW. HLA class II tetramers: tools for direct analysis of antigen-specific CD4+ T cells. Arthritis Rheum. 2002;46(1):5–12. doi: 10.1002/1529-0131(200201)46:1<5::AID-ART10063>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Newell EW, Sigal N, Bendall SC, Nolan GP, Davis MM. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity. 2012;36(1):142–152. doi: 10.1016/j.immuni.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan D, Arrhenius T, Sidney J, Del Guercio MF, Albertson M, Wall M, Oseroff C, Southwood S, Colon SM, Gaeta FC, et al. On the interaction of promiscuous antigenic peptides with different DR alleles. Identification of common structural motifs. J Immunol. 1991;147 (8):2663–2669. [PubMed] [Google Scholar]
- O’Sullivan D, Sidney J, Appella E, Walker L, Phillips L, Colon SM, Miles C, Chesnut RW, Sette A. Characterization of the specificity of peptide binding to four DR haplotypes. J Immunol. 1990;145 (6):1799–1808. [PubMed] [Google Scholar]
- Oseroff C, Sidney J, Kotturi MF, Kolla R, Alam R, Broide DH, Wasserman SI, Weiskopf D, McKinney DM, Chung JL, Petersen A, Grey H, Peters B, Sette A. Molecular determinants of T cell epitope recognition to the common Timothy grass allergen. J Immunol. 2010;185(2):943–955. doi: 10.4049/jimmunol.1000405. jimmunol.1000405 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oseroff C, Sidney J, Tripple V, Grey H, Wood R, Broide DH, Greenbaum J, Kolla R, Peters B, Pomes A, Sette A. Analysis of T Cell Responses to the Major Allergens from German Cockroach: Epitope Specificity and Relationship to IgE Production. J Immunol. 2012a;189(2):679–688. doi: 10.4049/jimmunol.1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oseroff C, Sidney J, Vita R, Tripple V, McKinney DM, Southwood S, Brodie TM, Sallusto F, Grey H, Alam R, Broide D, Greenbaum JA, Kolla R, Peters B, Sette A. T Cell Responses to Known Allergen Proteins Are Differently Polarized and Account for a Variable Fraction of Total Response to Allergen Extracts. J Immunol. 2012b doi: 10.4049/jimmunol.1200850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panina-Bordignon P, Demotz S, Corradin G, Lanzavecchia A. Study on the immunogenicity of human class-II-restricted T-cell epitopes: processing constraints, degenerate binding, and promiscuous recognition. Cold Spring Harb Symp Quant Biol. 1989a;54(Pt 1):445–451. doi: 10.1101/sqb.1989.054.01.053. [DOI] [PubMed] [Google Scholar]
- Panina-Bordignon P, Tan A, Termijtelen A, Demotz S, Corradin G, Lanzavecchia A. Universally immunogenic T cell epitopes: promiscuous binding to human MHC class II and promiscuous recognition by T cells. Eur J Immunol. 1989b;19 (12):2237–2242. doi: 10.1002/eji.1830191209. [DOI] [PubMed] [Google Scholar]
- Peretti M, Villard J, Barras E, Zufferey M, Reith W. Expression of the three human major histocompatibility complex class II isotypes exhibits a differential dependence on the transcription factor RFXAP. Mol Cell Biol. 2001;21 (17):5699–5709. doi: 10.1128/MCB.21.17.5699-5709.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J, Malik A, Parham P, Bodmer JG, Marsh SG. IMGT/HLA database--a sequence database for the human major histocompatibility complex. Tissue Antigens. 2000;55 (3):280–287. doi: 10.1034/j.1399-0039.2000.550314.x. [DOI] [PubMed] [Google Scholar]
- Robinson J, Mistry K, McWilliam H, Lopez R, Parham P, Marsh SG. The IMGT/HLA database. Nucleic Acids Res. 2011;39(Database issue):D1171–1176. doi: 10.1093/nar/gkq998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J, Waller MJ, Parham P, de Groot N, Bontrop R, Kennedy LJ, Stoehr P, Marsh SG. IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res. 2003;31 (1):311–314. doi: 10.1093/nar/gkg070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche PA, Cresswell P. High-affinity binding of an influenza hemagglutinin-derived peptide to purified HLA-DR. J Immunol. 1990;144 (5):1849–1856. [PubMed] [Google Scholar]
- Sekaly RP, Jacobson S, Richert JR, Tonnelle C, McFarland HF, Long EO. Antigen presentation to HLA class II-restricted measles virus-specific T-cell clones can occur in the absence of the invariant chain. Proc Natl Acad Sci U S A. 1988;85 (4):1209–1212. doi: 10.1073/pnas.85.4.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidney J, Assarsson E, Moore C, Ngo S, Pinilla C, Sette A, Peters B. Quantitative peptide binding motifs for 19 human and mouse MHC class I molecules derived using positional scanning combinatorial peptide libraries. Immunome Res. 2008;4:2. doi: 10.1186/1745-7580-4-2. 1745–7580-4-2 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidney J, Southwood S, Oseroff C, del Guercio MF, Sette A, Grey HM. Measurement of MHC/peptide interactions by gel filtration. Curr Protoc Immunol. 2001;Chapter 18(Unit 18):13. doi: 10.1002/0471142735.im1803s31. [DOI] [PubMed] [Google Scholar]
- Sidney J, Steen A, Moore C, Ngo S, Chung J, Peters B, Sette A. Divergent motifs but overlapping binding repertoires of six HLA-DQ molecules frequently expressed in the worldwide human population. J Immunol. 2010a;185(7):4189–4198. doi: 10.4049/jimmunol.1001006. jimmunol.1001006 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidney J, Steen A, Moore C, Ngo S, Chung J, Peters B, Sette A. Five HLA-DP molecules frequently expressed in the worldwide human population share a common HLA supertypic binding specificity. J Immunol. 2010b;184(5):2492–2503. doi: 10.4049/jimmunol.0903655. jimmunol.0903655 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinigaglia F, Guttinger M, Kilgus J, Doran DM, Matile H, Etlinger H, Trzeciak A, Gillessen D, Pink JR. A malaria T-cell epitope recognized in association with most mouse and human MHC class II molecules. Nature. 1988;336 (6201):778–780. doi: 10.1038/336778a0. [DOI] [PubMed] [Google Scholar]
- Solomon C, Southwood S, Hoof I, Rudersdorf R, Peters B, Sidney J, Pinilla C, Marcondes MC, Ling B, Marx P, Sette A, Mothe BR. The most common Chinese rhesus macaque MHC class I molecule shares peptide binding repertoire with the HLA-B7 supertype. Immunogenetics. 2010;62(7):451–464. doi: 10.1007/s00251-010-0450-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwood S, Sidney J, Kondo A, del Guercio MF, Appella E, Hoffman S, Kubo RT, Chesnut RW, Grey HM, Sette A. Several common HLA-DR types share largely overlapping peptide binding repertoires. J Immunol. 1998;160 (7):3363–3373. [PubMed] [Google Scholar]
- Tishkoff SA, Reed FA, Friedlaender FR, Ehret C, Ranciaro A, Froment A, Hirbo JB, Awomoyi AA, Bodo JM, Doumbo O, Ibrahim M, Juma AT, Kotze MJ, Lema G, Moore JH, Mortensen H, Nyambo TB, Omar SA, Powell K, Pretorius GS, Smith MW, Thera MA, Wambebe C, Weber JL, Williams SM. The genetic structure and history of Africans and African Americans. Science. 2009;324(5930):1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vita R, Zarebski L, Greenbaum JA, Emami H, Hoof I, Salimi N, Damle R, Sette A, Peters B. The immune epitope database 2.0. Nucleic Acids Res. 2010;38(Database issue):D854–862. doi: 10.1093/nar/gkp1004. gkp1004 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Sidney J, Dow C, Mothe B, Sette A, Peters B. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput Biol. 2008;4(4):e1000048. doi: 10.1371/journal.pcbi.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Sidney J, Kim Y, Sette A, Lund O, Nielsen M, Peters B. Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinformatics. 2010;11:568. doi: 10.1186/1471-2105-11-568. 1471–2105-11-568 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RF. Molecular cloning and characterization of MHC class I- and II-restricted tumor antigens recognized by T cells. In: Coligan John E, et al., editors. Current protocols in immunology. Unit 20. Chapter 20. 2009. p. 10. [DOI] [PubMed] [Google Scholar]
- Wilson CC, Palmer B, Southwood S, Sidney J, Higashimoto Y, Appella E, Chesnut R, Sette A, Livingston BD. Identification and Antigenicity of Broadly Cross-Reactive and Conserved Human Immunodeficiency Virus Type 1-Derived Helper T-Lymphocyte Epitopes. J Virol. 2001;75(9):4195–4207. doi: 10.1128/jvi.75.9.4195-4207.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, James EA, Huston L, Danke NA, Liu AW, Kwok WW. Multiplex mapping of CD4 T cell epitopes using class II tetramers. Clin Immunol. 2006;120(1):21–32. doi: 10.1016/j.clim.2006.03.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Online Resource 1. Comparison of population coverage afforded by the full panel and the sub-panel of the 27 most common alleles
The collective frequencies provided by the HLA class II alleles represented in the panel of 46 single transfected cell lines, and the sub-panel of the 27 most common alleles, are compared. Coverage was examined in a cohort of 190 donors recruited for two different studies in the San Diego area. The cumulative percent of all donors for which the panels provide coverage of 0 to 8 possible class II types expressed is shown.
Online Resource 2. Ethnic breakdown of four donor cohorts from different geographical areas
Representation of different ethnic groups, in terms of percent of individuals, in the San Diego, Denver, Baltimore and Cape Town cohorts. Ethnicity is as described by donors during clinical enrollment.
Online Resource 3. Correspondence between HLA frequencies in the donor cohort and the general population
The collective frequencies of alleles present in the San Diego, Denver, Baltimore and Cape Town cohorts examined largely match those found in the general worldwide population, based on HLA frequency data obtained from dbMHC (Middleton et al. 2003; Meyer et al. 2007).