The barley immune receptor MLA mediates disease resistance against powdery mildew fungal pathogens. Upon activation, MLA directly interacts with two antagonistically acting transcription factors, MYB6 and WRKY1. MYB6 positively regulates basal and MLA-mediated immune responses, while MYB6 DNA binding is antagonized by direct association with WRKY1 but enhanced by MLA interaction.
Abstract
The nucleotide binding domain and Leucine-rich repeat (NLR)–containing proteins in plants and animals mediate pathogen sensing inside host cells and mount innate immune responses against microbial pathogens. The barley (Hordeum vulgare) mildew A (MLA) locus encodes coiled-coil (CC)–type NLRs mediating disease resistance against the powdery mildew pathogen Blumeria graminis. Here, we report direct interactions between MLA and two antagonistically acting transcription factors, MYB6 and WRKY1. The N-terminal CC signaling domain of MLA interacts with MYB6 to stimulate its DNA binding activity. MYB6 functions as a positive regulator of basal and MLA-mediated immunity responses to B. graminis. MYB6 DNA binding is antagonized by direct association with WRKY1 repressor, which in turn also interacts with the MLA CC domain. The activated form of full-length MLA10 receptor is needed to release MYB6 activator from WRKY1 repression and to stimulate MYB6-dependent gene expression. This implies that, while sequestered by the WRKY1 repressor in the presence of the resting immune receptor, MYB6 acts as an immediate and positive postactivation signaling component of the active state of MLA during transcriptional reprogramming for innate immune responses.
INTRODUCTION
Plants employ two classes of innate immune receptors for non-self-recognition of microbial pathogens. Extracellular perception of conserved microbe-associated molecular patterns (MAMPs) is mediated by pattern recognition receptors (PRRs) on the cell surface, initiating intracellular signaling pathways that lead to MAMP-triggered immunity (Zipfel, 2009). The detection of strain-specific pathogen effectors that are delivered inside host cells is mediated by intracellular plant nucleotide binding domain and Leucine-rich repeat proteins (NLRs), which results in effector-triggered immunity (ETI) (Chisholm et al., 2006; Jones and Dangl, 2006). The perception of MAMPs by cognate PRRs converges on serial activation of stereotypic cellular responses that are detectable from minutes to days after elicitation (Boller and Felix, 2009). However, NLR-triggered disease resistance in plants is normally accompanied by a localized host cell death (the hypersensitive response) at attempted colonization sites (Shen and Schulze-Lefert, 2007; Maekawa et al., 2011b). Genome-wide transcriptional profiling and the analysis of defense phytohormone signaling mutants in Arabidopsis thaliana suggests the existence of a highly overlapping signaling network in MAMP-triggered immunity and ETI (Tsuda et al., 2009).
Most plant NLRs share a similar modular structure, containing an N-terminal coiled-coil (CC) or TOLL/Interleukin-1 receptor (TIR) domain, a central nucleotide binding (NB) domain, and C-terminal Leucine-rich repeats (LRRs). These NLR receptors are believed to act as molecular switch, driven by NTPase activity mediated by their central NB domain (NB-ARC) to regulate immune responses by intramolecular switching from an inactive (suppressed) form to the active form upon recognition of cognate pathogen effectors (Collier and Moffett, 2009; Lukasik and Takken, 2009). The N-terminal CC or TIR domain of some NLRs functions as a signaling module for triggering host cell death (Swiderski et al., 2009; Krasileva et al., 2010; Bernoux et al., 2011b; Collier et al., 2011; Maekawa et al., 2011a). The crystal structures of the N-terminal CC of barley (Hordeum vulgare) mildew A (MLA) and the TIR domain of flax (Linum usitatissimum) L6 have been solved (Bernoux et al., 2011b; Maekawa et al., 2011a). Both domains form homodimers in solution, and homotypic associations of the corresponding full-length receptors appear to be critical for their disease resistance function.
Plant NLRs localize to various subcellular compartments (i.e., are associated with the plasma membrane or the endomembrane system) (Eitas and Dangl, 2010; Bernoux et al., 2011a) or are dynamically partitioned between the cytoplasm and the nucleus (Deslandes et al., 2002; Burch-Smith et al., 2007; Shen et al., 2007; Wirthmueller et al., 2007; Cheng et al., 2009; Slootweg et al., 2010; Tameling et al., 2010). Subcellular receptor partitioning appears to be linked to compartment-specific NLR signaling activities, as Arabidopsis RPS4 and barley MLA10 were both shown to trigger cell death in the cytoplasm and disease resistance signaling without cell death in the nucleus (Heidrich et al., 2011; Bai et al., 2012). However, how pathogen-activated NLRs connect to downstream immune signaling components in the nucleus and the cytoplasm is largely unknown.
A subset of plant NLRs has been demonstrated to directly interact with transcription factors (TFs) or cofactors to regulate defense gene expression. Barley MLA10 was shown to interact with two WRKY TFs, WRKY1 and WRKY2, through its N-terminal CC domain (Shen et al., 2007). The TIR domain of Arabidopsis SNC1 was shown to mediate the interaction with TPR1, a member of a transcriptional corepressor family (Zhu et al., 2010). Arabidopsis RRS1-R is an unusual NLR receptor consisting of a TIR-NB-LRR structure and a C-terminal WRKY domain that was shown to bind DNA (Deslandes et al., 2002; Noutoshi et al., 2005). These examples point to a likely theme that several NLRs may directly participate in transcriptional reprogramming for defense response initiation upon activation.
The barley MLA locus encodes a large number of mainly allelic CC-type NLRs, each conferring isolate-specific disease resistance against the grass powdery mildew fungus Blumeria graminis f. sp hordei (Bgh) (Seeholzer et al., 2010). MLA1 strain-specific disease resistance to Bgh and cell death signaling of other tested MLA recognition specificities is evolutionarily conserved in different plant lineages (i.e. in transgenic Arabidopsis and upon transient gene expression in Nicotiana benthamiana) (Bai et al., 2012; Maekawa et al., 2012), implying ∼200 million years of functional conservation of the underlying immune mechanism in flowering plants.
Here, we report the identification of a MLA CC domain interactor, barley MYB6, by yeast two-hybrid experiments. MYB6 appears to specifically interact with the homodimer of functional MLA CC domains. Gene silencing and transient gene expression experiments in barley reveal that MYB6 functions as a positive regulator in basal and MLA-triggered disease resistance. Using protein coimmunoprecipitation, yeast three-hybrid, and Arabidopsis protoplast transfection experiments, we show that MYB6 acts as transcriptional activator and interacts directly with barley WRKY1 repressor. The latter TF can suppress the DNA binding activity of MYB6 activator. By introducing an autoactive and a loss-of function mutation in the central ATPase domain of full-length MLA10 receptor, we demonstrate that the activated form of the receptor is needed to release MYB6 from WRKY1 repression to stimulate MYB6-dependent gene expression. We propose that WRKY1 repressor physically sequesters the MYB6 activator in the presence of the resting or inactive forms of MLA. The active state of MLA releases MYB6 from WRKY1 repressor and stimulates its binding to cognate cis-acting elements to initiate disease resistance signaling.
RESULTS
MLA Interacts with MYB6 through the CC Domain
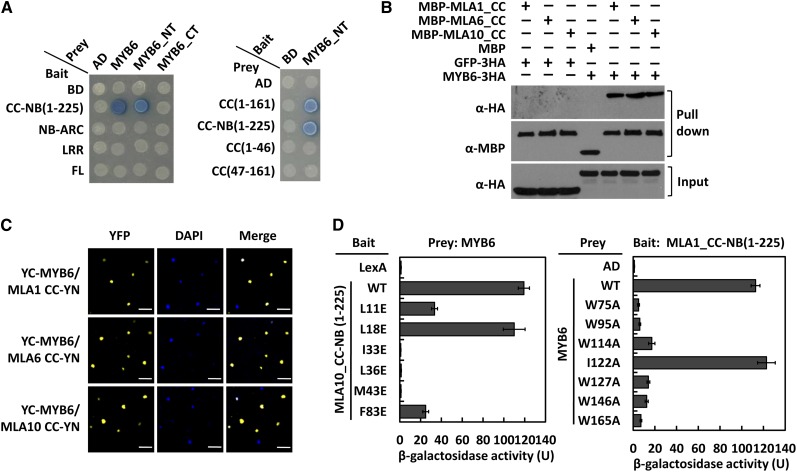
To identify potential postactivation MLA signaling components, we used the CC signaling module containing fragment MLA1_CC-NB(1-225) as bait (Maekawa et al., 2011a) (see Supplemental Table 1 online) to screen a yeast two-hybrid prey library derived from barley leaf epidermis challenged with Bgh. We independently identified three full-length prey clones encoding an R2R3-type MYB TF, designated as MYB6. MLA1 and MYB6 fragments were further tested for direct interactions (Figure 1A; see Supplemental Figure 1A online). Reciprocal tests revealed that MYB6_NT (encoding the N-terminal half of the TF) was necessary and sufficient for interacting with the MLA1 CC-NB(1-225) or CC(1-161) fragment, which encodes the CC module and adjacent linker region (Figure 1A). MYB6 did not interact with MLA1 CC(1-46) or CC(47-161), nor with the central MLA1 NB-ARC or the C-terminal LRR or full-length receptor (Figure1A). Similar protein accumulation of baits and preys in these yeast two-hybrid assays were revealed by protein gel blotting (see Supplemental Figure 1A online).
Figure 1.
MLA and MYB6 Interact through Their N-Terminal Domains.
(A) Analysis of MLA1 and MYB6 interactions in yeast. The LexA DNA binding domain (BD) or the B42 activation domain (AD) was fused at the N terminus of bait or prey, respectively. MYB6_NT and MYB6_CT represent 1 to 190 and 161 to 392 amino acid, respectively.
(B) MLA_CC domain binds MYB6 in pull-down assays. MBP fusions of MLA_CC domain were incubated with GFP-3HA or with MYB6-3HA, respectively. In the MBP pull-down fractions, coprecipitated MYB6-3HA (top panel) and equal loading of MBP fusions (middle panel) were detected; GFP-3HA or MYB6-3HA was detected in the input (bottom panel).
(C) CC domain of MLA1, MLA6, and MLA10 interacts with MYB6 in planta. Indicated pairs were coexpressed in the leaves of N. benthamiana through agroinfiltration for BiFC assays. Confocal images were taken at 30 h after infiltration. 4′,6-Diamidino-2-phenylindole (DAPI) staining revealed the nuclei. Bars = 50 µm.
(D) Interactions between mutant variants of MLA1_CC-NB(1-225) and MYB6 in yeast. MLA10_CC-NB(1-225) variant (left panel) or MYB6 variant (right panel) was used in yeast two-hybrid assays. The β-galactosidase activity was quantified, and data are represented as mean ± se (n = 8). The β-galactosidase experiments were repeated three times with similar results.
To validate the MYB6–MLA1 interaction, we employed the maltose binding protein (MBP) pull-down assay. We constructed N-terminal MBP and C-terminal His-tagged fusions of the CC(1-161) domain of MLA1, MLA6, and MLA10, and these fusions were expressed in Escherichia coli and purified using Ni-affinity chromatography. We also expressed C-terminal 3xHA-tagged fusions of MYB6 or green fluorescent protein (GFP) in N. benthamiana by Agrobacterium tumefaciens–mediated leaf infiltration and obtained crude protein extracts from these plants for pull-down assays. The CC domain of tested MLA1, MLA6, and MLA10 each could pull down MYB6-3HA but not GFP-3HA, indicating that few amino acid polymorphisms in the otherwise conserved CC domain of naturally diversified MLA receptors (Seeholzer et al., 2010) do not affect the association with MYB6 in vitro (Figure 1B). Bimolecular fluorescence complementation (BiFC) assays were conducted to confirm the MYB6 and MLA CC association in N. benthamiana plant cells. We coexpressed fusion pairs of the C-terminal half of yellow fluorescent protein (YFP) fused to MYB6 (YC-MYB6) and the MLA CC domain fused to the YFP N-terminal half (i.e., MLA1 CC-YN, MLA6 CC-YN, or MLA10 CC-YN, respectively). We detected strong YFP fluorescence signals only in the nucleus of N. benthamiana cells coexpressing the aforementioned fusion pairs (Figure 1C). No fluorescence signal was detected with empty vector (EV) controls, although RT-PCR experiments confirmed comparable expression of YN/YC fusion transcripts in all combinations tested (see Supplemental Figures 1B and 1C online).
Structure-informed amino acid substitutions in the CC domain were found to affect, to different degrees, the self-association of MLA10_CC-NB(1-225) in yeast (Maekawa et al., 2011a). We therefore tested whether these mutations also affect the MLA10_CC-NB(1-225) heteromeric interaction with MYB6 (Figure 1D, left). Notably, MLA10_CC-NB(1-225) variants harboring I33E, L36E, or M43E substitutions that abolish self-association and disease resistance activity against Bgh (Maekawa et al., 2011a) (see Supplemental Table 2 online), completely lost their capacity for heteromeric interaction with MYB6. By contrast, L11E or F83E CC substitution variants that retain residual self-association showed weak interactions with MYB6 in yeast. Moreover, the MLA10_CC-NB(1-225) variant harboring the L18E mutation that retained wild-type-like self-association and disease resistance activity against Bgh retained interactions with MYB6 that are indistinguishable from wild-type MLA10_CC-NB(1-225) (Figure 1D, left panel). We also tested the interaction between MLA10_CC-NB(1-225) and MYB6 variants containing substitutions of conserved Trp residues in the R2R3 repeat of the TF (Figure 1D, right). MLA10_CC-NB(1-225) interacted very weakly with MYB6 variants harboring an Ala substitution at each of the six Trp residue positions in the R2 or R3 repeat (Stracke et al., 2001), compared with the strong interactions seen with wild-type MYB6 or the substitution variant harboring I122A that is located at the junction region between the R2 and R3 repeat, used as a control (Figure 1D, right). All bait or prey fusions accumulated to similar levels as revealed by protein gel blotting (see Supplemental Figure 1D online). Taken together, the MLA CC domain of tested naturally diversified MLA receptors can interact with MYB6 in the nucleus. This interaction requires an intact N-terminal R2R3 repeat and is likely dependent on dimeric self-association of the CC receptor module.
MYB6 Specifically Interacts with Barley WRKY1
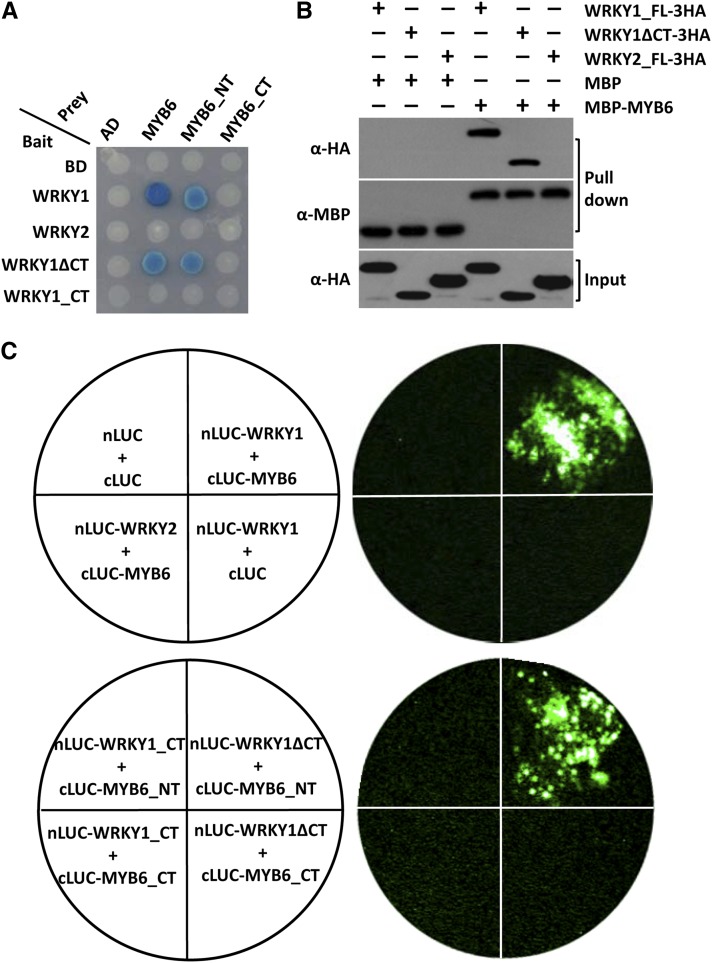
Effector-activated MLA10 receptor interacts in the nucleus with sequence-related WRKY1 and WRKY2 TFs that repress basal defense responses against Bgh and interfere with MLA-dependent immunity in barley (Shen et al., 2007). The conserved C-terminal part of WRKY1 (CT, amino acids 260 to 353) and WRKY2 mediates the interaction with the MLA10 CC domain (Jordan et al., 2011; Maekawa et al., 2011a). We therefore investigated potential interactions between MYB6 and WRKY1 or WRKY2. Unexpectedly, MYB6 strongly interacts with full-length WRKY1 and less strongly with a truncated derivative lacking the CT part (WRKY1ΔCT) in yeast two-hybrid experiments, but not with WRKY2 (Figure 2A). The N-terminal half of MYB6 encoded by the MYB6_NT prey was necessary and sufficient to recapitulate the interactions seen between full-length MYB6 and the tested WRKY1 baits (Figure 2A). The Trp-to-Ala substitutions in the R2 and R3 repeats of MYB6 that compromised interactions with MLA10 CC-NB(1-225) also abrogated interactions with WRKY1 (Figure 1D, right; see Supplemental Figure 2 online), indicating the need of an intact R2R3 repeat in MYB6 for association with both MLA CC and WRKY1. We validated the MYB6–WRKY1 interaction by conducting in vitro MBP binding assays using MBP-MYB6 obtained from E. coli and C-terminal 3xHA-tagged WRKY TFs expressed in N. benthamiana. We observed that MBP-MYB6 could specifically pull down WRKY1_FL-3HA or WRKY1ΔCT-3HA but not WRKY2_FL-3HA (Figure 2B). This confirms the selective association between MYB6 and WRKY1.
Figure 2.
WRKY1, but Not WRKY2, Specifically Interacts with MYB6.
(A) Y2H analysis showing WRKY1 interacts with MYB6 via their N terminus. WRKY1ΔCT and WRKY1_CT represent the N-terminal 1 to 260 and C-terminal 261 to 355 amino acid residues, respectively.
(B) WRKY1_FL or WRKY1ΔCT binds MYB6. Coprecipitated WRKY1_FL-3HA or WRKY1ΔCT-3HA was detected in the pull-down fraction (top panel); equal loading of MBP or MBP-MYB6 fusions is shown (middle panel); HA-tagged WRKY proteins were detected in the input (bottom panel).
(C) LCI assays showing the N terminus of WRKY1 and MYB6 mediates their interaction. The N- or C-terminal fragment of LUC (nLUC or cLUC) was fused with indicated proteins, and indicated pairs were coexpressed in N. benthamiana. The luminescent signal was collected at 60 h after infiltration (Chen et al., 2008 in Supplemental References 1 online).
To verify that these interactions can occur in plant cells, we conducted firefly luciferase complementation imaging (LCI) assays in N. benthamiana, in which the N-terminal part of luciferase (nLUC) was fused to WRKY1 FL or its ΔCT truncation or WRKY2 FL, while the C-terminal part of luciferase (cLUC) was fused to MYB6 FL or its NT fragment. Upon coexpression of all possible pairs by Agrobacterium-mediated leaf infiltration, strong LUC activity signals captured with a charge-coupled device camera were detected when full-length or the N-terminal parts of both WRKY1 and MYB6 were coexpressed, for example, WRKY1/MYB6, WRKY1ΔCT/MYB6_NT (Figure 2C), WRKY1ΔCT/MYB6, and WRKY1/MYB6_NT (see Supplemental Figure 3 online). No signal was detected upon coexpression of WRKY2/MYB6 or any other combination involving WRKY1_CT or MYB6_CT or any negative controls (Figure 2C; see Supplemental Figure 3 online). Taken together, MYB6 specifically interacts with WRKY1 but not WRKY2 in yeast two-hybrid, in vitro pull-down, and in planta LCI assays, and this interaction is likely mediated by the N-terminal parts of MYB6 and WRKY1.
MLA10 CC Abrogates the WRKY1-MYB6 Interaction in a WRKY1_CT-Dependent Manner
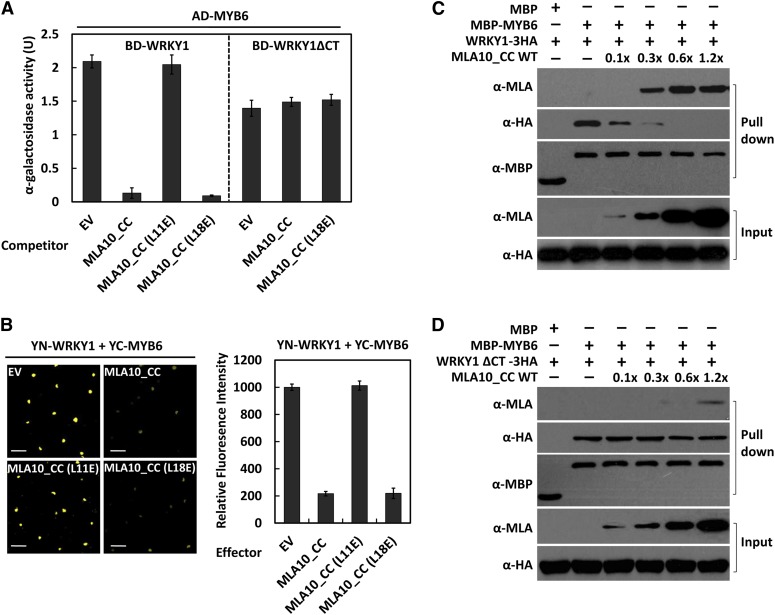
To dissect the identified tripartite interactions among MLA, WRKY1, and MYB6, we used a yeast three-hybrid system (Li et al., 2011). We constructed the DB-WRKY1 bait and the AD-MYB6 prey, and their coexpression with EV in yeast resulted in reporter activity of ∼2.0 units (Figure 3A, left). Further conditional expression of MLA10_CC wild type or its L18E variant, but not of the CC L11E variant, dramatically reduced reporter activity to ∼0.1 unit (Figure 3A, left), possibly resulting from competitive interactions (i.e., CC binding to WRKY1, MYB6, or both). To discriminate between these possibilities, we repeated the assays using the truncated bait BD-WRKY1ΔCT, which interacts with MYB6 (Figure 2) but not the MLA CC module (Jordan et al., 2011; Maekawa et al., 2011a). BD-WRKY1ΔCT expression led to reduced reporter activity (∼1.4 compared with ∼2.0 units for BD-WRKY1; Figure 3A), confirming weaker WRKY1ΔCT interactions with MYB6 than with WRKY1 (Figure 2A). However, further conditional expression of either MLA10 CC or its L18E variant did not reduce reporter activity like before (Figure 3A, right). These data indicate that MLA10_CC or its L18E derivative disrupt the WRKY1-MYB6 association through direct interactions with the WRKY1 C-terminus.
Figure 3.
MLA10_CC Abrogates the WRKY1-MYB6 Association in a WRKY1_CT-Dependent Manner.
(A) MLA10_CC or CC(L18E) variant abrogates WRKY1–MYB6 interaction in yeast. In addition to bait and prey, MLA10_CC wild type or its variant was expressed conditionally as a competitor. Data show average and se from three independent experiments.
(B) MLA10_CC or CC(L18E) attenuate WRKY1-MYB6 association in planta. YN-WRKY1 and YC-MYB6 pairs were coexpressed with EV or MLA10_CC in N. benthamiana for the BiFC assay. Confocal images show YFP signal in nuclei at 30 h after infiltration (left panel). Bars = 50 µm. The fluorescence intensity was averaged from more than 70 nuclei for each coexpression (right panel), and data are represented as mean ± se. The fluorescence intensity was analyzed four independent times with similar results.
(C) MLA 10_CC competes out WRKY1 for binding MYB6. In the MBP pull-down assays, an increasing amount of MLA10_CC was added into preincubated MBP-MYB6 and WRKY1-3HA mixture. Similar results were obtained from more than five repeats.
(D) MLA 10_CC cannot compete out WRKY1ΔCT for binding MYB6. The same MBP pull-down assays were done as in (C), except that HvWRKY1ΔCT-3HA was used to replace WRKY1-3HA.
[See online article for color version of this figure.]
We conducted BiFC assays in N. benthamiana by expressing MLA10_CC or its L18E variant in addition to YN-WRKY1 and YC-MYB6 (Figure 3B). Confocal imaging showed that nuclear YFP fluorescence signal, resulting from the YN-WRKY1 and YC-MYB6 interaction, could be dramatically reduced by coexpression of MLA10_CC wild type or the L18E variant but not by the L11E variant (Figure 3B, left). These imaging results were quantified by averaging the YFP signal intensity of ∼70 nuclei for each coexpression experiment (Figure 3B, right).
To characterize the antagonistic interactions among WRKY1, MYB6, and MLA10_CC, we expressed WRKY1-3HA and WRKY1ΔCT-3HA in N. benthamiana and purified MBP-MYB6 fusion, MLA10_CC, L11E and L18E variants from E. coli for in vitro binding assays. We added increasing amounts of MLA10_CC to preincubated mixture containing 5 μg of MBP-MYB6 and excess amount of WRKY1-3HA and then subjected the blend to MBP pull-down and immunoblotting to reveal the ratio of each protein in the enriched fraction. When 0.1× MLA10_CC (0.5 μg) was added, no detectable MLA10_CC was retrieved in the MBP pulled-down fraction, although the WRKY1-3HA amount was markedly reduced in the same fraction (Figure 3C, lane 3), suggesting that at this concentration, MLA10_CC could bind to a fraction of WRKY1-3HA and induced its dissociation from MBP-MYB6, but little or no MLA10_CC was left for binding MBP-MYB6. In agreement with this, when more MLA10_CC (0.3×, 1.5 μg) was added, we could detect MLA10_CC in the pull-down fraction and further reduced level of WRKY1-3HA (Figure 3C, lane 4). Addition of more MLA10_CC (0.6× or 1.2×) led to increasing amount of MLA10_CC in the pull-down fraction but undetectable WRKY1-3HA (Figure 3C, lanes 5 and 6). These data suggest a MLA CC dosage-dependent competition with WRKY1-3HA to bind MBP-MYB6. We repeated the experiment using WRKY1ΔCT-3HA instead of WRKY1-3HA (Figure 3D). Remarkably, although the largest amount of MLA10_CC as above (1.2×) was added, we detected only a little MLA10_CC in the MBP-enriched fraction with almost unchanged level of WRKY1ΔCT-3HA (Figure 3D, lanes 2 to 6), indicating again that MLA10_CC disrupted the WRKY1–MYB6 interaction through physical binding to the WRKY1 CT. This interpretation was corroborated by the observation that MLA10_CC L18E could compete out WRKY1 from MYB6-WRKY1 complexes, whereas the L11E variant could not (see Supplemental Figures 4A and 4B online).
Taken together, our results obtained from yeast, in planta, or in vitro protein interaction experiments indicate that the MLA10 CC domain abrogates the WRKY1–MYB6 association through its physical binding to the WRKY1 CT and subsequently forms a complex with MYB6 in the nucleus.
WRKY1 and MLA10 CC Act Antagonistically on MYB6 DNA Binding Activity
MYB TFs bind their cognate cis-elements to regulate gene expression (Yang and Klessig, 1996; Liao et al., 2008). We screened these cis-acting DNA elements by yeast one-hybrid experiments using MYB6 and found it could bind MBS, MBS I, and MRE4 to different levels (see Supplemental Figure 5A online). We employed electrophoretic mobility shift assays (EMSAs) to verify these interactions and detected specific binding of MYB6 only to MBS I (see Supplemental Figure 5B online). We then used MBS I with tandem repeats of the TAACTG consensus sequence to confirm the binding specificity in further EMSA experiments. MYB6 specifically bound MBS I and resulted in a clear signal of DNA-protein complex, which was not seen when the mMBS I probe or MBP protein was used (see Supplemental Figure 5C online).
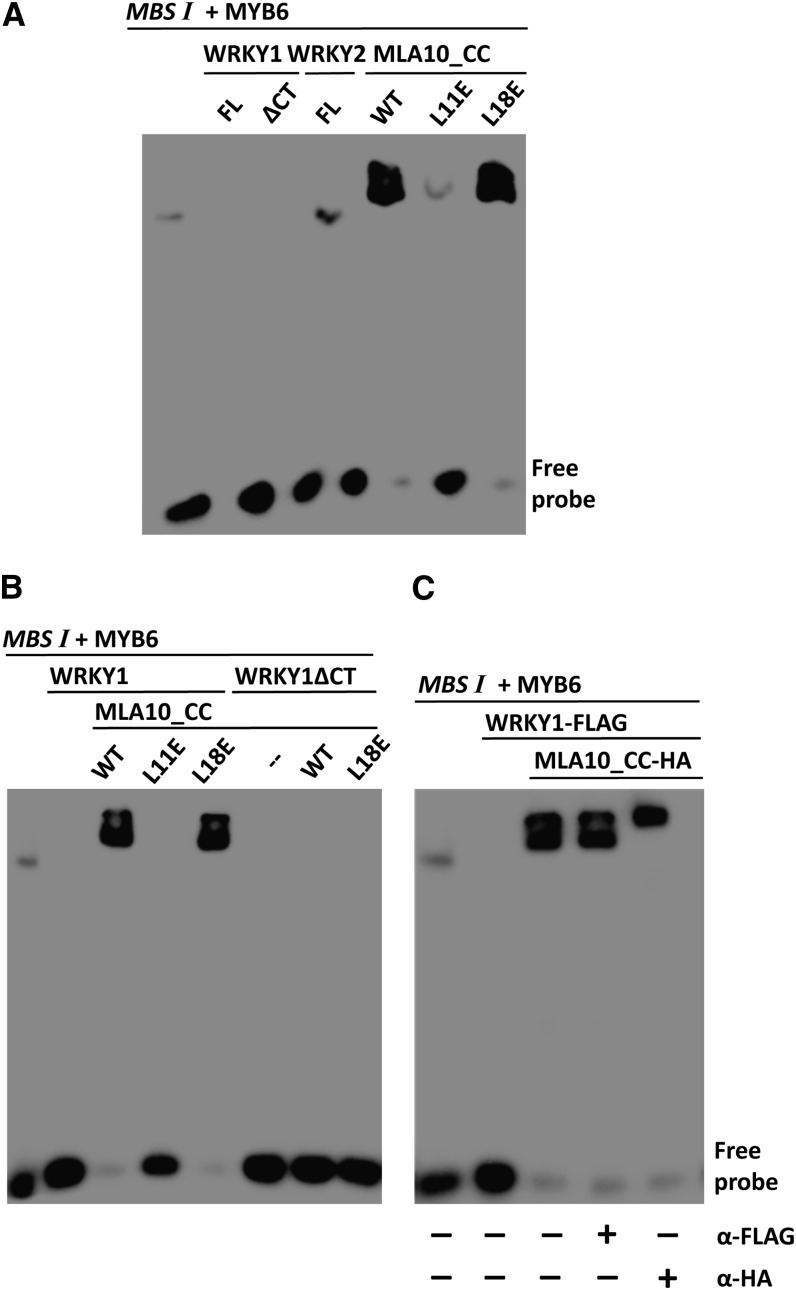
To investigate whether the DNA binding activity of MYB6 could be influenced by WRKYs or MLA10_CC, we prepared recombinant WRKY1_FL, WRKY1ΔCT, WRKY2_FL, MLA10_CC, and L11E or L18E variant. Each of them was tested by EMSA and did not bind the MBS I probe (see Supplemental Figure 5D online). Significantly, addition of WRKY1 or WRKY1ΔCT into the mixture of MBS I and MYB6 resulted in the absence of the DNA-HvMYB6 complex signal, suggesting that WRKY1 FL and WRKY1ΔCT interfere with the DNA binding of MYB6 (Figure 4A, lanes 1 to 3). Addition of WRKY2 did not interfere with the DNA binding of MYB6 (Figure 4A, lane 4). When MLA10_CC or its L18E variant was added to the mixture, we observed a supershifted signal with markedly enhanced intensity (Figure 4A, lanes 5 and 7). Addition of the L11E CC variant also led to a supershifted signal but with little or no changed intensity (Figure 4A, lane 6). Thus, we conclude that WRKY1 abrogates the DNA binding of MYB6; by contrast, MLA10_CC or the L18E variant binds MYB6 and stimulates its DNA binding activity.
Figure 4.
MLA10 CC Releases MYB6 from WRKY1 Suppression and Stimulates the DNA Binding Activity of MYB6.
(A) MYB6 DNA binding was suppressed by WRKY1 but stimulated by MLA10_CC. The MBS I probe was preincubated with MYB6, and then recombinant WRKY1, WRKY2, or MLA10_CC was further added. WT, the wild type. FL, Full length protein.
(B) MLA10_CC or CC(L18E) derepresses MYB6 from WRKY1 suppression. Similar EMSA as in (A), except that, in addition to WRKY1 or HvWRKY1ΔCT, MLA10_CC wild type or variant was further added into the mixture.
(C) MLA10_CC replaces WRKY1 to associate with DNA-MYB6 complex. Similar EMSA as in (B), but WRKY1-FLAG and MLA10_CC-HA fusions were used for incubation. The indicated antibody was added into the mixture before PAGE separation.
All EMSA experiments were repeated three times with similar results.
MLA10 CC Releases MYB6 from WRKY1 Suppression and Enhances Its DNA Binding Activity
MLA10_CC could abrogate the WRKY1–MYB6 interaction in yeast, in vitro, and in planta (Figure 3). To further investigate their antagonistic interactions in vitro, we performed EMSA assays by adding into the mixture of MBS I and MYB6 first WRKY1 and then MLA10_CC, or L11E or L18E variants, respectively (Figure 4B, left). As expected, the addition of WRKY1 alone led to the loss of signal of the DNA-MYB6 complex (Figures 4A and 4B); however, further addition of MLA10_CC wild type or L18E, but not L11E, resulted in reappearance of a supershifted signal with enhanced intensity (Figure 4B, lanes 3 to 5). Again, this supershift was WRKY1 CT dependent since it was not seen when WRKY1ΔCT was used (Figure 4B, lanes 6 to 8). To reveal the components of the supershifted DNA-protein complex, we repeated the EMSA assay using WRKY1-FLAG and MLA10_CC-HA fusions. These fusions were tested not to bind MBS I when mixed sequentially (see Supplemental Figure 5E online). Similar as WRKY1, WRKY1-FLAG suppressed MYB6 DNA binding (Figure 4C, lane 2). Further addition of MLA10_CC-HA resulted again in the reappearance of the supershifted signal with enhanced intensity (Figure 4C, lane 3). However, addition of anti-FLAG antibody did not lead to further band shifting, indicating the absence of WRKY1-FLAG in the DNA-protein complex (Figure 4C, lane 4). By contrast, addition of anti-HA antibody led to further shifting of the complexes, indicating that MLA10_CC-HA resided in the final DNA-protein complex (Figure 4C, lane 5).
Our results show that MLA10 CC or its L18E variant can release MYB6 from WRKY1 suppression in a WRKY1 CT-dependent manner and by itself physically associates with MYB6 and stimulates its DNA binding activity.
In Vivo Interplay of MYB6, WRKY1, and MLA10_CC in Arabidopsis Protoplasts
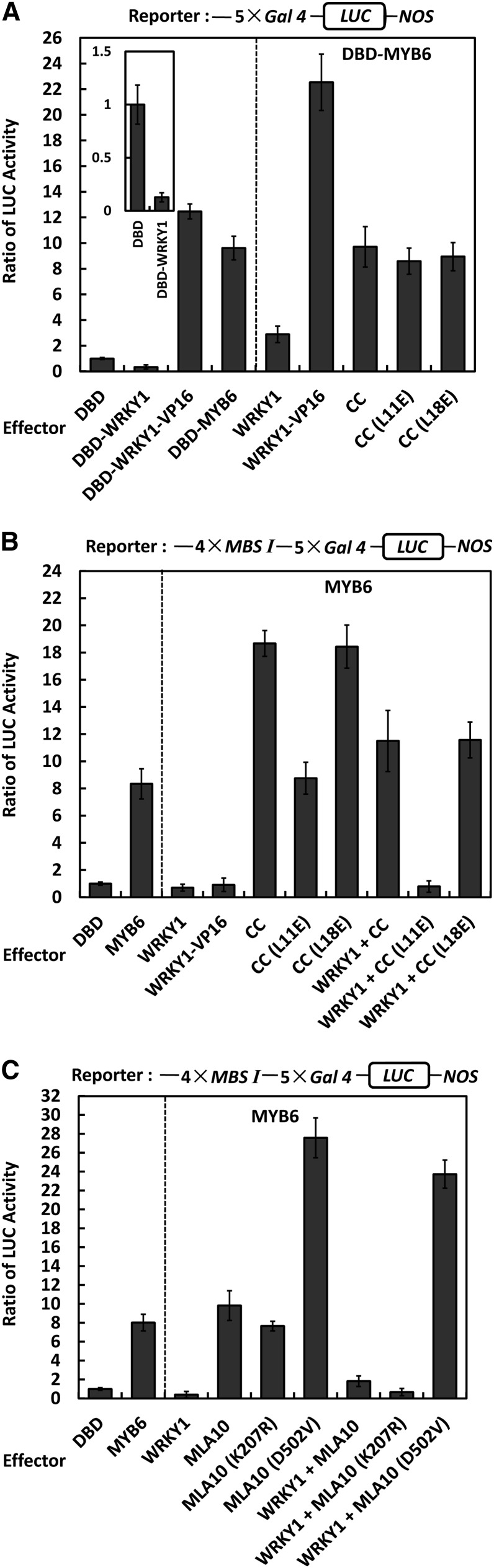
To quantify MYB6 transcriptional activity in plant cells, we used the Arabidopsis leaf protoplast transfection assay (Liao et al., 2008), in which a luciferase (LUC) reporter was cotransfected with an effector construct expressing MYB6 and a control plasmid, to measure reporter LUC activity (LucA) by luminescence (see Supplemental Figure 6A online). The Gal4 DNA binding domain (DBD), which binds the 5xGal4 upstream activating sequences (UAS) in the promoter, was used to determine basal LUC activity, while the DBD-VP16 fusion was used as a transcriptional activator control (Sadowski et al., 1988). Indeed, expression of DBD-VP16 resulted in a very high LucA ratio of ∼14 units (see Supplemental Figure 6A online). Expression of DBD-MYB6 also led to a high LucA ratio of ∼7 to 8 units, suggesting MYB6 acts as a transcriptional activator. The activation domain in MYB6 was delimited to the C-terminal half since expression of DBD-MYB6_CT, but not DBD-MYB6_NT, resulted in a high LucA ratio of ∼11 units (see Supplemental Figure 6A online).
The same assay was used to determine the transcriptional activity of WRKY1. Expression of DBD-WRKY1 consistently resulted in a lower LucA ratio compared with the relative basal LUC activity level, indicating that WRKY1 acts as a transcriptional repressor (Figure 5A, left, lanes 1 and 2 and the inset). An effector was constructed to combine the WRKY1 and VP16 proteins (i.e., DBD-WRKY1-VP16); interestingly, its expression led to a high LucA ratio of ∼12 units, higher than that of DBD-MYB6 (Figure 5A, left, lanes 3 and 4). This indicates that DBD-WRKY1-VP16 acts as a transcriptional activator in this fusion configuration. Furthermore, DBD fusions with MLA10 CC, or L11E and L18E variants did not significantly alter the LucA ratio, suggesting MLA10 CC, L11E, or L18E per se have no transcriptional activity (see Supplemental Figure 6B online).
Figure 5.
Interplay Among MYB6, WRKY1, and MLA10_CC or Full-Length Variants in Arabidopsis Protoplasts.
(A) Reporter activation by DBD-MYB6 was partially repressed by WRKY1 repressor. In the Arabidopsis protoplast transfection assays, the LUC reporter contains 5xGal4 UAS and all effectors were expressed as DBD fusions (left panel); DBD-MYB6 was coexpressed with one more effector (right panel). VP16 is a strong activator. The inset shows the LucA ratio for DBD and DBD-WRKY1 using a different scale.
(B) Reporter activation by MYB6 was fully repressed by WRKY1 and derepressed and stimulated by MLA10_CC. Note that a modified reporter was used here and all effectors were expressed without fused to DBD. MYB6 was expressed alone (left panel) or coexpressed with one or two more proteins (right panel).
(C) Reporter activation by MYB6 was fully repressed by WRKY1 and derepressed and stimulated by MLA10(D502V). Full-length MLA10 wild type or mutant variant was used. Similar settings as in (B).
Error bars represent se of technical triplicates in all figures, and all experiments were repeated at least two times with similar results.
The DBD-MYB6 effector should rely largely on the DBD domain for DNA binding to the reporter in this system (Figure 5A; see Supplemental Figure 6A online). Therefore, it is possible to evaluate the effect of WRKY1 and/or MLA10_CC on the trans-activation activity of MYB6 irrespective of its DNA binding activity. We chose WRKY1, WRKY1-VP16, MLA10_CC wild type, and L11E or L18E variant for coexpression with DBD-MYB6 (Figure 5A, right panel). First, WRKY1 reduced the LucA ratio to ∼3 units (Figure 5A, right, lane 1), suggesting WRKY1 partially suppressed MYB6 trans-activation activity. Surprisingly, WRKY1-VP16 led to a greatly increased LucA ratio of ∼22 units (Figure 5B, right, lane 2), which might reflect cooperative action of the two activators VP16 and MYB6, linked together through WRKY1, on reporter gene expression. Consistent with this, using the firefly LCI assay, we detected in vivo an interaction between WRKY1-VP16 and MYB6 in N. benthamiana (see Supplemental Figure 6C online). Furthermore, MLA10_CC or L11E or L18E variants did not increase the LucA ratio compared with that of DBD-HvMYB6 alone (Figure 5A, right, lanes 3 to 5), suggesting that MLA10_CC per se has no effect on MYB6 trans-activating activity (see also below).
Collectively, these data demonstrate that MYB6 is a transcriptional activator, and WRKY1 is a repressor that can partially suppress the trans-activation activity of MYB6.
MLA10 CC and the MLA10(D502V) Autoactive Receptor Antagonize WRKY1 Suppression on MYB6 and Stimulate Its DNA Binding Activity in Vivo
To extend the above findings and to confirm the effect of WRKY1 and/or MLA10_CC on the DNA binding activity of MYB6 (Figure 4), we modified the reporter by inserting the 4xMBS I element upstream of the 5xGAL4 UAS; importantly, MYB6 was expressed alone without DBD, allowing the TF to bind freely to its cognate cis-element (Figure 5B). Indeed, MYB6 could bind to MBS I and activate the reporter gene in this modified system, leading to a LucA ratio of ∼8 units (Figure 5B, left, lane 2). Next, we expressed WRKY1, WRKY1-VP16, MLA10_CC wild type, and L11E or L18E variant in this system in addition to MYB6 (Figure 5B). Notably, WRKY1 expression could now almost fully suppress reporter gene expression, leading to only basal LucA levels (Figure 5B, right panel, lane 1). Compared with the partial suppression when using DBD-MYB6 (Figure 5A), this implied that WRKY1 may have sequestered MYB6 from MBS I binding. Surprisingly, the expression of WRKY1-VP16 did not enhance the reporter expression like before; on the contrary, it completely suppressed reporter expression, similar as WRKY1 alone (Figure 5B, right panel, lanes 1 and 2). This finding indicated it is very unlikely that WRKY1-VP16 associated with MYB6 bound to the MBS I on the reporter, which would cooperatively activate the reporter like above (Figures 5A and 5B, comparing lane 2 in right panels). Together, this points to a likely scenario in which WRKY1-VP16 or WRKY1 associated with MYB6, thereby preventing MYB6 from binding MBS I to mediate reporter gene expression.
Next, we expressed MLA10_CC, L11E, or L18E variants with MYB6 in this modified reporter expression system. The expression of MLA10_CC or the L18E variant resulted in a markedly increased LucA ratio of ∼18 units (Figure 5B, lanes 3 and 5, right panel). Together with previous findings that MLA10_CC has no transcriptional activity nor has any effect on the trans-activation activity of MYB6 (Figure 5A; see Supplemental Figure 6B online), this strongly suggests that they stimulated MYB6 MBS I binding activity to enhance reporter expression. Again, the MLA10_CC(L11E) variant could not increase the LucA ratio (Figure 5B, right panel, lane 4). Furthermore, upon triple expression including MYB6, plus WRKY1 and respective MLA10_CC variants, we observed a markedly increased LucA ratio to ∼12 units when MLA10_CC or the L18E variant was involved, but only basal LucA ratio when the L11E mutant form was expressed (Figure 5B, right panel, lanes 6 to 8 versus lane 1). In conclusion, WRKY1 sequestered MYB6 from binding MBS I, whereas MLA10_CC or the L18E variant, but not L11E, could abolish WRKY1 suppression and stimulate the MBS I binding activity of MYB6 in vivo.
Since MLA10 full-length protein failed to interact with MYB6 or WRKY1 in yeast (Figure 1A) (Shen et al., 2007), we wondered how MLA10 would behave in the leaf protoplast transfection assay. To address this, we chose MLA10, the MLA10(K207R) P-loop loss-of-function and the MLA10(D502V) autoactive receptor variants for expression assays (Bai et al., 2012). MLA10(D502V) expression frequently resulted in abnormally low luminescence value when measured at 24 h after transfection, indicating MLA10(D502V) might trigger cell death at this time point, consistent with our previous data in N. benthamiana (Bai et al., 2012). We therefore decided to measure the LucA ratio at 16 h rather than 24 h after transfection and could obtain a similar LucA ratio as before for all controls (Figure 5C, bars 1 to 3). The additional expression of MLA10 or MLA10(K207R) only led to a marginally increased or similar LucA ratio compared with MYB6 expression alone (Figure 5C, lanes 4 and 5 versus lane 2). However, expression of the MLA10(D502V) receptor markedly increased the LucA ratio to ∼28 units (Figure 5D, lane 6), suggesting that the autoactive receptor can stimulate the DNA binding activity of MYB6, similar as or even stronger than the CC domain alone. Moreover, in triple expression experiments involving MYB6 and WRKY1 and MLA10, only the expression of MLA10(D502V) regained reporter activation with a LucA value of ∼24 units, while MLA10 wild type or MLA10(K207R) only resulted in low or repressed reporter activity (Figure 5C, bars 7 to 9). Taken together, this suggests that MLA10-mediated stimulation of MYB6 DNA binding activity in vivo and release from WRKY1 repression is a postactivation activity of the immune receptor.
MYB6 Positively Regulates Basal and MLA-Triggered Disease Resistance against B. graminis in Barley
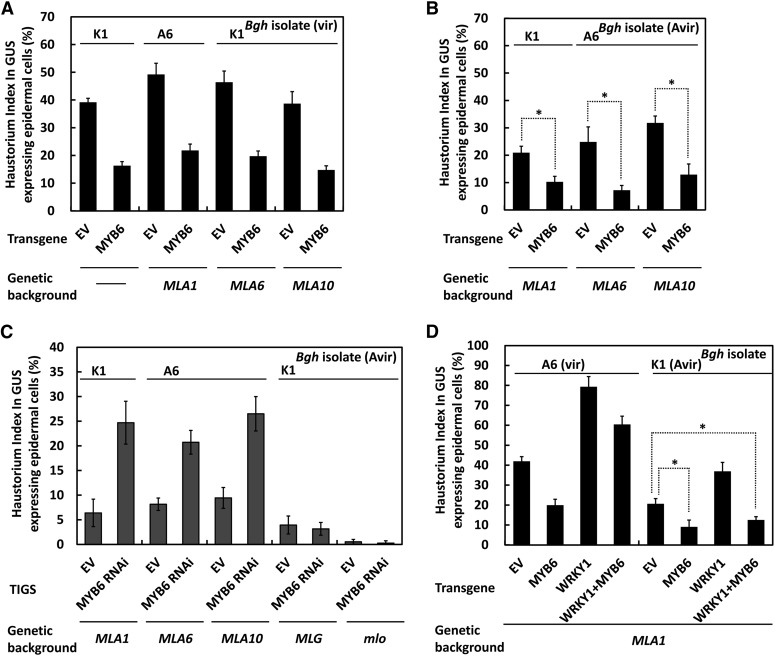
To elucidate the function of MYB6 in disease resistance against the Bgh fungus, we conducted single-cell transient gene expression assays using detached leaves. In this assay, MYB6 was coexpressed with a β-glucuronidase (GUS) reporter in barley leaf epidermal cells by particle delivery, and upon Bgh conidiospore inoculation, the frequency of fungal haustorium formation in the transformed cells was scored as haustorium index (HI%) (Shen et al., 2003). Overexpression of MYB6 in several MLA-containing near-isogenic lines resulted in marked HI% reduction to ∼15 to 20%, compared with ∼40 to 50% for EV controls, upon inoculation of respective virulent Bgh isolates (Figure 6A). We observed further HI% reduction to ∼10% compared with ∼20 to 32% for EV in these MLA-containing lines upon inoculation of the avirulent isolates (Figure 6B). Together, these results suggest MYB6 overexpression can enhance barley basal as well as MLA-triggered immune responses against the Bgh fungus.
Figure 6.
MYB6 Acts as a Positive Regulator in Basal and MLA-Mediated Disease Resistance against Bgh in Barley.
(A) MYB6 overexpression enhances basal disease resistance. HI% was scored in barley leaf epidermal cells expressing EV or MYB6 in MLA-containing near-isogenic lines as indicated. The virulent Bgh isolate was used for inoculation respectively.
(B) MYB6 overexpression enhances MLA-mediated disease resistance against Bgh. Indicated avirulent Bgh isolate was used for inoculation.
(C) TIGS of MYB6 compromises MLA-mediated disease resistance. TIGS RNA interference vector harboring MYB6 sense and antisense fragment (MYB6 RNAi) was used to trigger cellular RNA interference in indicated barley lines. Spores of the avirulent Bgh isolate were inoculated at 60 h after particle delivery.
(D) Activated MLA1 derepresses MYB6 from WRKY1 and potentiates resistance responses against Bgh in barley. Indicated virulent or avirulent Bgh isolate was used for inoculation, noting that vir or Avir in brackets represents virulent or avirulent isolate, respectively.
At least 50 cells were analyzed in one experiment, and the data shown are all average of three independent experiments ± se. Asterisk indicates a significant difference (P < 0.01).
The requirement of MYB6 in MLA-triggered immunity was verified by silencing experiments. First, we employed a transient-induced gene silencing system (TIGS) (Douchkov et al., 2005) to silence MYB6 in barley leaf epidermal cells. Significantly, silencing of MYB6 in MLA1-, MLA6-, or MLA10-containing backgrounds compromised MLA-triggered resistance to Bgh, resulting in increased HI% of ∼20 to 25% compared with ∼6 to 9% for EV controls (Figure 6C, bars 1 to 6). Notably, the MLG-triggered race-specific and the mlo-mediated broad spectrum resistance to Bgh were not affected by MYB6 silencing (Figure 6C, bars 7 to 10) (Büschges et al., 1997; Görg et al., 2005). Second, we performed virus-induced gene silencing using a barley stripe mosaic virus (BSMV) vector (Yuan et al., 2011) to silence MYB6 or MLA12 in a MLA12-containing barley line. While in leaves infected with BSMV-EV the obtained Bgh microcolony index (MI%) was typically around ∼8%, the MI% scored in BSMV-MLA12as or BSMV-MYB6as infected seedlings was markedly increased to ∼83 or ∼62%, respectively (see Supplemental Figure 7A online). Quantitative RT-PCR analysis confirmed effective knockdown of MYB6 transcripts in BSMV-MYB6as infected seedlings (see Supplemental Figure 7B online), and visual inspections also revealed more Bgh microcolony formation on the leaf surface of MLA12- or MYB6-silenced seedlings than that of EV control (see Supplemental Figure 7C online). The data from different gene silencing assays strongly suggest that MYB6 is essential for disease resistance responses to Bgh upon activation of different MLA recognition specificities.
Since WRKY1 suppresses MYB6-dependent gene expression and the derepression likely requires the activated MLA receptor in Arabidopsis protoplasts (Figure 5C), we wondered whether this reflects the activity of WRKY1, MYB6, and MLA in barley in the context of disease resistance against Bgh. To this end, we expressed MYB6, WRKY1, or both in an MLA1-containing barley line and inoculated with the virulent or avirulent Bgh isolate (Figure 6D). The overexpression of WRKY1 rendered barley supersusceptible to the virulent Bgh isolate A6, resulting in a HI% of ∼80% compared with that in the EV control (∼41%) or the MYB6 overexpression (∼20%), respectively (Figure 6D, lanes 1 to 3, right panel). Interestingly, coexpression of WRKY1 and MYB6 resulted in HI% ∼60%. Although this HI% is lower than that of WRKY1 expression alone, it is significantly higher than that of MYB6 expression alone (∼20%) (Figure 6D, left panel, lane 4). These findings suggest that WRKY1 acts as a suppressor of PAMP-mediated basal immunity and this activity can only be partially negated by MYB6 overexpression during compatible interactions (Figure 6D, left panel). The overexpression of WRKY1 in incompatible interactions also resulted in significantly higher HI% compared with that of EV or MYB6 expression alone (Figure 6D, right panel, lanes 1 to 3), indicating MLA1-mediated Bgh resistance was suppressed by WRKY1; however, coexpression of MYB6 and WRKY1 led to markedly reduced HI% to ∼12%, similar to that of MYB6 expression alone (Figure 6D, right panel, lanes 2 and 4). Taken together, these data suggest that upon immune receptor activation endogenous MLA1 can antagonize WRKY1 suppression and potentiate MYB6-dependent immune responses against the Bgh fungus in barley plants.
DISCUSSION
Plant cells undergo extensive transcriptional reprogramming upon pathogen recognition by PRRs or NLRs (Tao et al., 2003; Boller and Felix, 2009; Zipfel, 2009). However, signaling components acting immediately downstream of activated immune receptors are not well characterized. This is particularly true for NLR-mediated ETI pathways (Dodds and Rathjen, 2010; Maekawa et al., 2011b). Our previous study proposed that activated MLA interferes with two nuclear WRKY repressors of PRR-triggered basal defense, thereby potentiating immune responses (Shen et al., 2007). However, it remained unclear how MLA abrogated WRKY repressor activity. Here, we report the identification of MYB6 as a direct target of activated MLA and describe functional antagonistic interactions among MLA, WRKY1/2, and MYB6.
The MLA CC domain is necessary and sufficient for interacting with MYB6, whereas full-length MLA is unable to interact with MYB6 in yeast (Figure 1). Similarly, we showed previously that MLA CC but not full-length MLA interacts with WRKY1/2 in yeast, and, importantly, Fluorescence lifetime imaging-Förster resonance energy transfer (FLIM-FRET) analyses detected an AVRA effector-dependent association of full-length MLA10 with WRKY2 in the nucleus of barley cells (Shen et al., 2007). Along the same line, the NB-ARC MHD motif variant MLA10(D502V) that mimics the activated form of MLA10 (Bai et al., 2012) can antagonize WRKY1 and activate MYB6-dependent reporter gene expression in Arabidopsis protoplasts (Figure 5). Together, these findings are consistent with the hypothesis that NLRs are activated by intramolecular conformational switching upon pathogen effector detection (Takken and Goverse, 2012). In addition, our data imply that NLR activation-dependent receptor conformation is critical for efficient docking to immediate downstream signaling components (Figure 5C). Thus, it is possible that surface residues of the MLA CC homodimer, needed for MYB6 or WRKY1/2 contacts, are buried in the resting state of MLA by intramolecular interdomain interactions as recently proposed for flax TIR-type NLRs (Maekawa et al., 2011a; Ravensdale et al., 2012). In this context, it is interesting to understand the possible configurations of the MLA CC domain or the activated MLA full-length protein when associating with the MYB6 or WRKY1 TF. According to our mutagenesis analyses of the MLA10 N terminus in this study (Figure 1D) and previously (Maekawa et al., 2011a), it is likely that activated MLA adopts a distinctive conformation as homodimer for subsequent association with the TFs. MYB TFs can form homo- or heterodimers, and this can stimulate their DNA binding activity and modulate DNA binding specificity (Dubos et al., 2010). Presumably, the MLA homodimer assists and/or stabilizes MYB6 homodimer formation that allows these TFs to recognize DNA with higher affinity and specificity (Dubos et al., 2010). This may explain why the MLA CC domain could dramatically enhance the DNA binding activity of MYB6 (Figure 4).
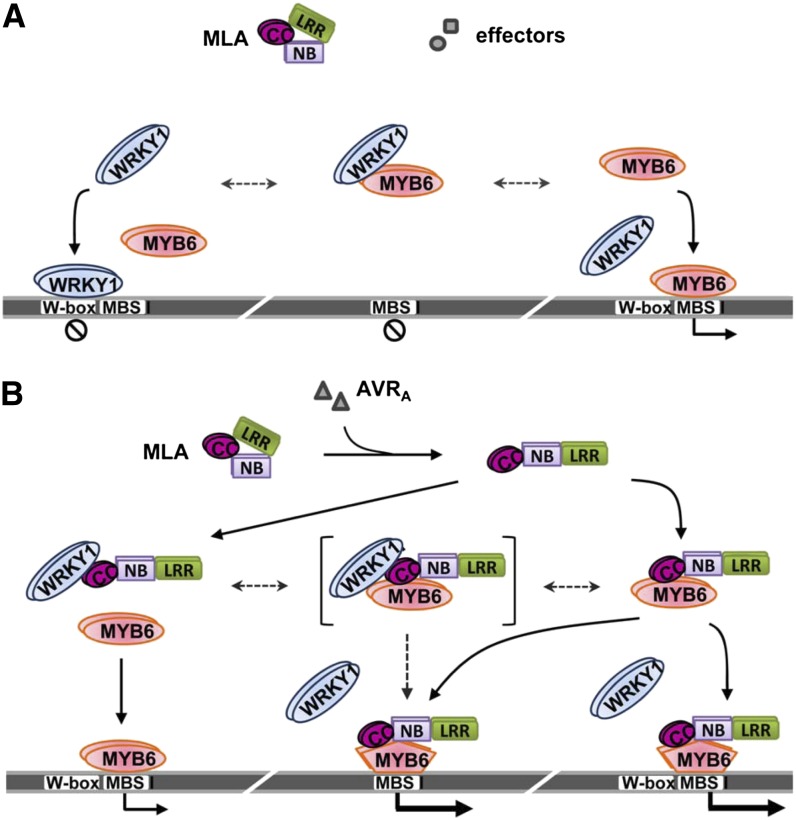
Regarding the tripartite interactions among MLA and the TFs, our current data do not exclusively favor independent or simultaneous binding of MYB6 and WRKY1. As shown in Figure 4C, MLA-HA releases MYB6 from WRKY1-FLAG suppression and then engages in DNA-MYB6 complex formation; importantly, the resulting DNA-MYB6-MLA complexes do not contain WRKY1-FLAG, suggesting MLA associates with MYB6 independently from WRKY1 binding at least in these complexes. Furthermore, in the Arabidopsis protoplast transfection assays (Figures 5B and 5C) MLA10 CC or MLA10(D502V) seemingly can enhance MYB6 DNA binding and reporter gene expression at maximal levels, whereas in the presence of WRKY1, only partially enhanced reporter gene expression was observed. This may reflect the existence of transient tripartite WRKY1-MLA-MYB6 complexes whose DNA binding activity or transactivation activity is impaired compared with the binary MLA-MYB6 complex. A model for possible protein–protein and protein–DNA interactions and their consequences in MLA-mediated immune responses is presented in Figure 7.
Figure 7.
Model for MLA Interference with Two Antagonistically Acting TFs and Initiation of Defense Signaling.
(A) During compatible interactions with B. graminis, barley MLA is in the resting state. The WRKY1 repressor binds to cognate W-box DNA binding sites to suppress defense gene expression and physically sequesters MYB6 activator from binding to MBS I cis-elements. This negative regulation is thought to limit excessive activation of defense responses in MAMP-triggered immunity.
(B) Upon perception of cognate AVRA effectors, MLA adopts an active conformation with its N-terminal signaling domain exposed, which is capable of interacting with WRKY1 to release MYB6 from WRKY1 suppression. Released MYB6 may directly bind MBS I to initiate gene expression. Activated MLA can also directly interact with free MYB6 to enhance its MBS I binding activity and thus potentiate defense gene expression and immune responses. MLA, WRKY1, and MYB6 are depicted in homodimer configurations according to the data of this study or previous reports (Shen et al., 2007; Dubos et al., 2010; Maekawa et al., 2011a). A tripartite MLA-WRKY1-MYB6 complex may exist transiently and is depicted by brackets. MYB6 shown as pentagon represents the conformation whose DNA binding activity is enhanced by the binding of activated MLA. Dashed arrows indicate hypothetic equilibrium transitions between protein complexes. Black circles containing a diagonal line below the DNA (thick gray line) denote inactive defense genes.
WRKY1 and WRKY2 belong to the same WRKY subgroup and thus contain the same domain structure and motifs and share high sequence similarity (Shen et al., 2007). Nevertheless, it was unclear whether they act together or separately to regulate target gene expression in response to Bgh infection in barley. Their Arabidopsis homologs, At-WRKY18, At-WRKY40, and At-WRKY60, have both redundant and distinct functions in negatively regulating resistance responses to bacterial and powdery mildew fungal pathogens (Xu et al., 2006; Shen et al., 2007; Pandey et al., 2010). We show here that WRKY1, but not WRKY2, interacts with MYB6 to interfere with its DNA binding activity, probably by restricting MYB6 binding to the promoter of its target genes (Figures 2, 4, and 5). Thus, WRKY1 and WRKY2 appear to exert separable repressor functions by targeting different genes, although it is possible that these TFs may also coregulate another subset of target genes. Although it seems likely that a subset of genes in the barley genome share binding sites for WRKY1 and MYB6 in their regulatory sequences, the detected direct association between these two unrelated TFs suggests the existence of an additional layer of TF regulation. Accordingly, part of WRKY1 repressor activity could be explained by sequestration of MYB6 away from its DNA binding site. Future experimentation will define all in planta target genes whose expression is regulated by WRKY1/MYB6 and/or WRKY2.
Similar to the functionally diversified family of WRKY TFs (Rushton et al., 2010), the MYB TF family expanded in plants with particularly large numbers of the R2R3-type members being evolved in diverse lineages (Stracke et al., 2001; Dubos et al., 2010). Some plant R2R3-type MYB TFs have been demonstrated to play pivotal roles in transcriptional regulation of plant-specific biological processes (Dubos et al., 2010). Arabidopsis At-MYB30 was shown to regulate hypersensitive cell death signaling in response to bacterial pathogens (Raffaele et al., 2008); interestingly, its transcriptional activity and resistance function is negatively regulated not only by a host protein but also by a Xanthomonas Type III effector through physical association in the nucleus (Froidure et al., 2010; Canonne et al., 2011). Since the rice (Oryza sativa) genome encodes 95 R2R3-type MYB genes (Feller et al., 2011), the barley genome likely contains a similar range of these MYB genes that may have evolved diversified functions. Our phylogeny analysis revealed that the most closely related R2R3-type MYB in barley shares with MYB6 only 42 or 18% sequence identity at the nucleotide or amino acid level, respectively (see Supplemental Figures 8A and 8B online). This suggests the absence of MYB6 subgroup members in barley and, hence, a functional redundancy through closely related MYB6 members appears unlikely. Furthermore, we could identify in wheat (Triticum aestivum), Brachypodium, or in the rice genome each a more closely related R2R3-type MYB of Hv-MYB6 (see Supplemental Figure 8C online). Sequence alignment revealed that Hv-MYB6 shares 95% sequence identity to a likely wheat homolog, Ta-MYB22, and less sequence relatedness to a Brachypodium or rice MYB (51 or 32% identity, respectively) (see Supplemental Figure 8C online). On the other hand, we could not identify any close homologs of barley Hv-MYB6 in Arabidopsis (see Supplemental Figure 8D and Supplemental Data Set 1 online). These data together indicate that Hv-MYB6–like homologs might be a grass or Triticeae-specific innovation.
The overexpression of MYB6 may have the pitfall of unspecific promoter binding and transcription initiation resulting from TF overloading in the transformed barley cells. However, the silencing of MYB6 via two different gene silencing approaches strongly support a critical role of MYB6 in disease resistance activated by the tested MLA recognition specificities against Bgh isolates in leaf tissue (Figure 6). Although we cannot fully exclude the possibility of cosilencing a highly sequence related MYB family member in our silencing experiments, our further BLASTN search of the barley genome database using the C-terminal Hv-MYB6 fragment did not retrieve any sequence with high sequence similarity other than that of Hv-MYB6 (see Supplemental Table 3 online), suggesting off-targeting silencing was unlikely to occur in our MYB6 silencing experiments. Moreover, retained MLG-specified race-specific and mlo-mediated broad-spectrum resistance to Bgh following MYB6 silencing supports its specific engagement in MLA-triggered resistance signaling (Figure 6).
Our EMSA experiments showed that MLA CC binding to MYB6 strongly stimulates its binding to the cognate DNA binding site (Figure 4), and a MLA CC amount of only one-third of that of MYB6 was sufficient to detect MLA CC-MYB6 complexes (Figure 3C, top panel), while MLA CC of two-thirds of that of MYB6 was enough to release all WRKY1 from WRKY1-MYB6 complexes in pull-down experiments (Figure 3C, second panel). Consistent with these data, upon challenge with the avirulent Bgh isolate, endogenous levels of MLA1 are sufficient to enhance disease resistance against Bgh infection when both MYB6 and WRKY1 are overexpressed (Figure 6D, right panel). The activity is likely mediated through a fraction of MYB6 that was not associated with WRKY1 or was released from WRKY1. Because the expression of MLA, WRKY1/2, and MYB6 is rapidly induced upon pathogen challenge (see Supplemental Figure 9 online) (Halterman et al., 2003; Shen et al., 2007), but with differential kinetics, the stoichiometry of the four proteins in plant cells will probably change over time after pathogen attack. Consequently, the ratio of antagonistically acting MLA-WRKY1 and MLA-MYB6 complexes in plant cells might be subject to spatio-temporal dynamics in cells in physical contact with the pathogen and in neighboring host cells, which could also provide a mechanism to turn off MLA signaling.
Together with recent findings for TIR-type NLR receptors (García et al., 2010; Bhattacharjee et al., 2011; Heidrich et al., 2011), we envisage direct links between the nuclear pool of plant NLRs to the transcriptional network and the transcriptional machinery as an important step in the initiation of plant immune responses against the invasion of diverse pathogens.
METHODS
Plant and Fungal Materials and Yeast Strains
The barley (Hordeum vulgare) lines used in this study include ‘Pallas’ (recurrent parent for the following near isogenic lines), ‘P01’ (near-isogenic line containing Mla1), ‘P03’ (near-isogenic line containing Mla6), ‘P09’ (near-isogenic line containing Mla10), ‘P21’ (near-isogenic line containing MLG), ‘P22’ (near-isogenic line containing mlo), and ‘I10’ (near-isogenic line containing Mla12 in ‘Ingrid’ background). All barley seedlings were grown in a growth chamber under a 16-h/8-h, 20°C/18°C day/night cycle with 70% relative humidity. Nicotiana benthamiana plants were grown in greenhouse at 23 ± 1°C with a 16-h/8-h light/dark cycle.
The Blumeria graminis f. sp hordei (BGH) isolates K1 (AvrMla1; virMla6, virMla10, virMla12) and A6 (AvrMla6, AvrMla10, AvrMla12; virMla1) used for this assay were maintained on the plants of barley cultivar I10 and P01, respectively, and kept at 70% relative humidity and a 20°C day/18°C night cycle.
The yeast strains EGY48 [MATa, his3, trp1, ura3, LexAop(x6)-LEU2 plus p8op-lacZ] and YM4271 [MATa, ura3-52, his3-200, lys2-801, ade2-101, ade5, trp1-901, leu2-3, 112, tyr1-501, gal4-D512, gal80-D538, ade5::hisG] were used for yeast two-hybrid assays and maintained on SD/-Ura and YPAD plates, respectively. The yeast strain AH109 (MATa, trp1-901, leu2-3, 112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2::GAL1UAS-GAL1TATA-HIS3, GAL2UAS-GAL2TATA-ADE2, URA3::MEL1UAS-MEL1TATA-lacZ) was used for yeast three-hybrid assays and kept on YPAD plate.
Yeast Experiments
For yeast one-hybrid assay, the pHIS2 derivatives (harboring the MBS, MBS I, MBS II, MYBR, MRE, MRE1, MRE2, MRE3, and MRE4 cis-elements) were cotransformed with the construct pAD-MYB6 into the yeast strain Y187. EV pHIS2 cotransformed with pAD-MYB6 was used as a negative control. The transformants were analyzed on the medium SD-Trp-Leu-His supplemented with 30 or 60 mM 3-Amino-1,2,4-Triazole. For yeast two-hybrid screening, a total of 2 × 107 transformants were screened for growth on the SD/Gal/Raf/-Ura/-His/-Trp/-Leu medium. The potential positive clones were further verified using the same selective media supplemented with X-Gal, according to the yeast protocol handbook (Clontech). For yeast three-hybrid assay, bait and prey were cotransformed with a competitor conditionally expressed under the control of MET25 promoter and grown on the SD/-Leu/-Trp/-Met +125 ng/mL AbA (Takara; D9000) medium (Li et al., 2011). The interaction strength was quantified using a α-galactosidase assay (see below). For yeast protein accumulation analysis, yeast cultures were grown in SD selection media for overnight to OD600 ∼1.5 and harvested by centrifugation at 3000 rpm for 5 min, and the pellet was boiled for 5 min. The protein sample was then mixed with the equal volume of 2× Laemmli buffer and boiled for 5 min before SDS-PAGE running. The expression of the bait or prey was detected with anti-LexA antiserum (Santa Cruz Biotechnology sc-7544) or anti-HA antibody (Roche 11867423001).
The α-galactosidase assay was performed as described with some modifications (Li et al., 2011). Briefly, the yeast culture was grown in the SD/-Leu/-Trp/-His/-Met medium to OD600 0.2 to 0.5 and pelleted by centrifugation at 3500 rpm. Medium aliquot of 300 μL was mixed with assay buffer of 900 μL (0.33 M sodium acetate, pH 4.3, and 33 mM p-nitrophenyl-α-d-galactopyranoside). The reaction was stopped by the addition of 300 μL of 2 M Na2CO3. The α-galactosidase activity was calculated as the absorbance measured at 410 nm. Quantification of β-galactosidase activities was performed as described in the Yeast Protocol Handbook (Clontech). Briefly, overnight yeast culture was grown in the SD/Gal/Raf/-Ura/-His/-Trp medium to OD600 ∼1.5 and pelleted by centrifugation at 3500 rpm. The yeast cells were then broken open with several rounds of freeze/thaw cycles and mixed with o-Nitrophenyl β-D-Galactopyranoside (ONPG) in Z-buffer. The reaction was stopped with Na2CO3. The β-galactosidase activities were calculated by the supernatant absorbance at 420 and 600 nm.
BiFC and LCI Assays in N. benthamiana
For BiFC assay, the CTAP-YN/YC derivative constructs carrying fragments of the genes of interest were coexpressed in N. benthamiana leaves by agroinfiltration. Briefly, overnight agrobacteria cultures containing the indicated constructs were harvested and resuspended to OD600 = 0.7 in the infiltration buffer (0.5% Murashige and Skoog, 2% Suc, 100 µM acetosyringone, and 10 mM MES), and equal volume of resuspensions of agrobacteria carrying CTAP-YN or CTAP-YC derivatives were mixed and infiltrated into the N. benthamiana leaves. For the expression of the third protein (i.e., MLA10_CC wild type or variants), the same volume of agrobacteria suspension carrying the respective CTAP derivatives was coinfiltrated with equal volume of agrobacteria containing CTAP-YN or CTAP-YC. The infiltrated area was examined for YFP fluorescence 28 to 60 h after infiltration with confocal microscopy (Carl Zeiss 710). The leaves were infiltrated with 4,6-diamidino-2-phenylindole solution (2 μg/mL) for nuclei staining 2 h before microscopy. For LCI assay, N. benthamiana leaves were coinfiltrated with the agrobacteria culture transformed with the nLUC and cLUC derivative constructs. The infiltrated leaves were analyzed for the LUC activity 60 h after infiltration using a cooled charge-coupled device imaging apparatus (Andor iXon).
Protein Expression and Purification from Escherichia coli
The pMAL and pET32a constructs were transformed into Escherichia coli strain BL21DE3. The expression and purification of the recombinant proteins in the E. coli were performed according to the manual provided by Novagen. Briefly, E. coli was first grown in liquid Luria-Bertani medium to reach OD600 0.2 to 0.8, and this culture was treated with 20 mM Isopropyl β-D-1-Thiogalactopyranoside (IPTG) (Takara D9030B) and further grown to an OD600 of ∼2.0. The E. coli culture was then harvested and the recombinant protein was purified using Nickel-Nitrilotriacetic acid (NI-NTA) agarose according to the manufacturer’s instructions (Qiagen 30230).
In Vitro Protein–Protein Interaction Assay
The in vitro protein–protein interaction assay was performed as described (Qi et al., 2011) with modifications. Briefly, ∼5 μg purified MBP and MBP fusion protein was incubated with 150 μL amylose resin (New England Biolabs) for 3 h at 4°C. The amylose resin beads was washed with RB buffer (100 mM NaCl, 50 mM Tris-Cl, pH 7.8, 25 mM imidazole, 0.1% Tween 20, 10% glycerol, EDTA-free complete miniprotease inhibitor cocktail, and 20 mM β-mercaptoethanol) and incubated for 2 h with 150 μL protein isolated from 3.0 g N. benthamiana leaves expressing protein of interest by agroinfiltration. For in vitro protein competition binding assay, after washing with RB buffer, an increasing amount of a third protein (e.g., MLA10_CC wild type or variants) was added and incubated further for 5 min. After washing with RB buffer, the precipitates was washed with RB buffer and resuspended in 2× Laemmli buffer and boiled for 5 min before SDS-PAGE separation. The coprecipitated MLA or HA-tagged protein was analyzed by immunoblotting using mouse anti-MLA27 antibody (Bai et al., 2012) and rat anti-HA antibody (Roche 11867423001), respectively. The same amount of protein mixtures was blotted with anti-MBP antibody to show that equal amount of MBP and MBP fusion protein was present in the precipitates.
EMSAs
The EMSA assay was conducted as described (Lai et al., 2011). Briefly, the double-stranded probe was generated by annealing the two single-stranded nucleotide chains and labeled with [α-32P]dATP using T4 polynucleotide kinase (New England Biolabs M0201L). The binding reactions (20 μL) contained 2.0 ng of labeled oligo DNA, 5.0 mg of poly(dIdC), and 1.0 μg of MBP-MYB6 recombinant protein. A total of 1.0 μg of the effectors (i.e., WRKY1) was added 20 min after the preincubation of the MBP-MYB6-DNA mixture, and 1.0 μg of the second effectors [i.e., MLA10_CC, MLA10_CC(L18E), etc.] was added and incubated for further 5 min. The resulting DNA-protein complexes were resolved on a 4% polyacrylamide gel and exposed with a phosphor screen for 12 h.
Arabidopsis thaliana Protoplast Transactivation Assay
The isolation and transformation of Arabidopsis protoplast and the protoplast transactivation assay were described previously (Liao et al., 2008). Briefly, Arabidopsis protoplasts were transfected with a mixture of effector, a LUC reporter, and an internal control in a ratio of 6:6:1. The LUC activity was then examined using a Promega dual-LUC reporter assay system (E1910). Each transfection, including three replicates, was repeated two or three times using independently prepared protoplasts.
Single-Cell Transient Gene Expression Assay
The single-cell transient gene expression was conducted essentially as described (Shen et al., 2007). Briefly, a GUS reporter plasmid and plasmids expressing candidate of interests were mixed and delivered into leaf epidermal cells by particle bombardment. Transformed cells were stained using GUS staining solution and scored for fungal haustorium index upon inoculation with fungal spores (see Supplemental Methods 1, 2 and 3 online).
TIGS and BSMV Virus-Induced Gene Silencing Assay
For TIGS, a similar procedure was performed as above in transient gene expression assay, but a plasmid containing sense and antisense fragments (e.g., TIGS-MYB6) was used to trigger cellular RNA silencing of the candidate gene (Douchkov et al., 2005), and fungal spores inoculation was done later than above, allowing efficient gene silencing. BSMV-induced gene silencing assay was described (Yuan et al., 2011) (see Supplemental Methods 1, 2 and 3 online).
RT-PCR and Real-Time PCR Analysis
For RT-PCR analysis in the BiFC or LUC assay, total RNA of the N. benthamiana leaves coinfiltrated with the agrobacteria containing respective constructs were extracted using TRizol solution (Invitrogen 15596-026) and treated with RNase-free DNase I (Takara 2270B) to remove the potential DNA contamination. The first-strand cDNA was synthesized using 1 μg of the total RNA. The resulted cDNAs were used in the quantitative RT-PCR as a template to detect the expression of the constructs in the N. benthamiana. The real-time PCR assay was performed using the ABI step-one real-time PCR system with the GoTaq qPCR Master Mix (Promega A6001). The expression of ACTIN was set as the internal control, and the expression of MYB6 was analyzed using the primer 5′-AGGGAGCGGTCTGTCAAT-3′/5′-ACTCTTCACCG TGGCAAT-3′. The real-time PCR assay was repeated in three independent biological replicates.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: MYB6 (KC311227), WRKY1 (AJ536667), WRKY2 (AJ853838), MLA1 (AY009939), MLA6 (AJ302292), and MLA10 (AY266445).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Protein Accumulation of Baits and Preys and BiFC Controls for MLA_CC and MYB6 Interaction Analysis.
Supplemental Figure 2. Y2H Analysis between WRKY1 and the Mutant Variants of MYB6.
Supplemental Figure 3. Firefly LCI Assays for WRKY1–MYB6 Interaction.
Supplemental Figure 4. MBP Pull-Down Assays Testing MLA10_CC Variants Competing with WRKY1 to Bind MYB6.
Supplemental Figure 5. MYB6 Specifically Binds the MBS I cis-Element, Whereas WRKY1, WRKY2, or MLA10_CC Do Not.
Supplemental Figure 6. MYB6 Is a Transcription Activator, MLA10_CC Has No Transcriptional Activity, and WRKY1-VP16 Fusion Interacts with MYB6 in LCI Assays.
Supplemental Figure 7. BSMV-VIGS Silencing of MYB6 Compromises MLA12-Mediated Disease Resistance.
Supplemental Figure 8. Sequence Comparison among Hv-MYB6 and Its Most Closely Related R2R3-Type MYB TFs from Barley, Wheat, Brachypodium, and Rice and Phylogeny Analysis with the Arabidopsis R2R3-Type MYB Proteins.
Supplemental Figure 9. MYB6 Expression Is Induced by Bgh Infection with the Maximum Increases by the Avirulent Isolate.
Supplemental Table 1. Plasmids Constructed in This Study.
Supplemental Table 2. Plasmids from Other References Used in This Study.
Supplemental Table 3. Results of BLASTN Search of the Barley Genome Database Using the Fragment for HvMYB6 Silencing.
Supplemental Data Set 1. At-R2R3–Type MYB Alignments with Hv-MYB6.
Supplemental Methods 1. Plasmid Construction.
Supplemental Methods 2. Single-Cell Transient Gene Expression, Transient-Induced Gene Silencing Assay, and BSMV-Mediated Virus-Induced Gene Silencing.
Supplemental Methods 3. Phylogenetics Tree Construction.
Supplemental References 1. References cited mainly in Supplemental Data.
Supplementary Material
Acknowledgments
We thank Imre Somssich (Max Planck Institute for Plant Breeding Research) for reading the article, Shouyi Chen (Chinese Academy of Sciences) for vectors used in yeast one-hybrid and Arabidopsis transfection assays, Yule Liu (Tsinghua University, China) and Jian-Min Zhou (IGDB, Chinese Academy of Sciences) for the vectors used in BiFC and LCI assays, Wei Qian (Chinese Academy of Sciences) for technical advice on EMSA assays, and Pengya Xue for phylogeny tree constructions. This work was funded by the National Basic Research Program of China (2011CB100700), the National Natural Science Foundation of China (31030007 and 30971876), and the Chinese Academy of Sciences (KSCX2-EW-N-06 and KXCX-YW-N-075). P.S.-L. is supported by the German Research Foundation in the collaborative research centre SFB670.
AUTHOR CONTRIBUTIONS
C.C. and Q.-H.S. designed the research. P.S.-L. and Q.-H.S. conceived the project. C.C., D.Y., J.J., and S.J. performed the experiments with most contributions from C.C. C.C. and Q.-H.S. analyzed the data. C.C., P.S.-L., and Q.-H.S. wrote the article.
Glossary
- NLR
nucleotide binding domain and Leucine-rich repeat protein
- MAMP
microbe-associated molecular pattern
- PRR
pattern recognition receptor
- ETI
effector-triggered immunity
- CC
coiled-coil
- TIR
TOLL/Interleukin-1 receptor
- NB
nucleotide binding
- LRR
Leucine-rich repeat
- TF
transcription factor
- Bgh
Blumeria graminis f. sp hordei
- BiFC
bimolecular fluorescence complementation
- YFP
yellow fluorescent protein
- LCI
luciferase complementation imaging
- GUS
β-glucuronidase
- TIGS
transient-induced gene silencing system
- EMSA
electrophoretic mobility shift assay
References
- Bai S., Liu J., Chang C., Zhang L., Maekawa T., Wang Q., Xiao W., Liu Y., Chai J., Takken F.L.W., Schulze-Lefert P., Shen Q.H. (2012). Structure-function analysis of barley NLR immune receptor MLA10 reveals its cell compartment specific activity in cell death and disease resistance. PLoS Pathog. 8: e1002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernoux M., Ellis J.G., Dodds P.N. (2011a). New insights in plant immunity signaling activation. Curr. Opin. Plant Biol. 14: 512–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernoux M., Ve T., Williams S., Warren C., Hatters D., Valkov E., Zhang X., Ellis J.G., Kobe B., Dodds P.N. (2011b). Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host Microbe 9: 200–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee S., Halane M.K., Kim S.H., Gassmann W. (2011). Pathogen effectors target Arabidopsis EDS1 and alter its interactions with immune regulators. Science 334: 1405–1408 [DOI] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Burch-Smith T.M., Schiff M., Caplan J.L., Tsao J., Czymmek K., Dinesh-Kumar S.P. (2007). A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol. 5: e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büschges R., et al. (1997). The barley Mlo gene: A novel control element of plant pathogen resistance. Cell 88: 695–705 [DOI] [PubMed] [Google Scholar]
- Canonne J., Marino D., Jauneau A., Pouzet C., Brière C., Roby D., Rivas S. (2011). The Xanthomonas type III effector XopD targets the Arabidopsis transcription factor MYB30 to suppress plant defense. Plant Cell 23: 3498–3511 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Cheng Y.T., Germain H., Wiermer M., Bi D., Xu F., García A.V., Wirthmueller L., Després C., Parker J.E., Zhang Y., Li X. (2009). Nuclear pore complex component MOS7/Nup88 is required for innate immunity and nuclear accumulation of defense regulators in Arabidopsis. Plant Cell 21: 2503–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm S.T., Coaker G., Day B., Staskawicz B.J. (2006). Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 124: 803–814 [DOI] [PubMed] [Google Scholar]
- Collier S.M., Hamel L.-P., Moffett P. (2011). Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein. Mol. Plant Microbe Interact. 24: 918–931 [DOI] [PubMed] [Google Scholar]
- Collier S.M., Moffett P. (2009). NB-LRRs work a “bait and switch” on pathogens. Trends Plant Sci. 14: 521–529 [DOI] [PubMed] [Google Scholar]
- Deslandes L., Olivier J., Theulières F., Hirsch J., Feng D.X., Bittner-Eddy P., Beynon J., Marco Y. (2002). Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc. Natl. Acad. Sci. USA 99: 2404–2409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds P.N., Rathjen J.P. (2010). Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11: 539–548 [DOI] [PubMed] [Google Scholar]
- Douchkov D., Nowara D., Zierold U., Schweizer P. (2005). A high-throughput gene-silencing system for the functional assessment of defense-related genes in barley epidermal cells. Mol. Plant Microbe Interact. 18: 755–761 [DOI] [PubMed] [Google Scholar]
- Dubos C., Stracke R., Grotewold E., Weisshaar B., Martin C., Lepiniec L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15: 573–581 [DOI] [PubMed] [Google Scholar]
- Eitas T.K., Dangl J.L. (2010). NB-LRR proteins: Pairs, pieces, perception, partners, and pathways. Curr. Opin. Plant Biol. 13: 472–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller A., Machemer K., Braun E.L., Grotewold E. (2011). Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 66: 94–116 [DOI] [PubMed] [Google Scholar]
- Froidure S., Canonne J., Daniel X., Jauneau A., Brière C., Roby D., Rivas S. (2010). AtsPLA2-α nuclear relocalization by the Arabidopsis transcription factor AtMYB30 leads to repression of the plant defense response. Proc. Natl. Acad. Sci. USA 107: 15281–15286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görg R., Hollricher K., Schulze-Lefert P. (2005). Functional analysis and RFLP-mediated mapping of the Mlg resistance locus in barley. Plant J. 3: 857–866 [Google Scholar]
- García A.V., Blanvillain-Baufumé S., Huibers R.P., Wiermer M., Li G., Gobbato E., Rietz S., Parker J.E. (2010). Balanced nuclear and cytoplasmic activities of EDS1 are required for a complete plant innate immune response. PLoS Pathog. 6: e1000970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halterman D.A., Wei F., Wise R.P. (2003). Powdery mildew-induced Mla mRNAs are alternatively spliced and contain multiple upstream open reading frames. Plant Physiol. 131: 558–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidrich K., Wirthmueller L., Tasset C., Pouzet C., Deslandes L., Parker J.E. (2011). Arabidopsis EDS1 connects pathogen effector recognition to cell compartment-specific immune responses. Science 334: 1401–1404 [DOI] [PubMed] [Google Scholar]
- Jones J.D.G., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Jordan T., Seeholzer S., Schwizer S., Töller A., Somssich I.E., Keller B. (2011). The wheat Mla homologue TmMla1 exhibits an evolutionarily conserved function against powdery mildew in both wheat and barley. Plant J. 65: 610–621 [DOI] [PubMed] [Google Scholar]
- Krasileva K.V., Dahlbeck D., Staskawicz B.J. (2010). Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. Plant Cell 22: 2444–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z., Li Y., Wang F., Cheng Y., Fan B., Yu J.Q., Chen Z. (2011). Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. Plant Cell 23: 3824–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Distelfeld A., Comis A., Dubcovsky J. (2011). Wheat flowering repressor VRN2 and promoter CO2 compete for interactions with NUCLEAR FACTOR-Y complexes. Plant J. 67: 763–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Zou H.F., Wang H.W., Zhang W.K., Ma B., Zhang J.S., Chen S.Y. (2008). Soybean GmMYB76, GmMYB92, and GmMYB177 genes confer stress tolerance in transgenic Arabidopsis plants. Cell Res. 18: 1047–1060 [DOI] [PubMed] [Google Scholar]
- Lukasik E., Takken F.L.W. (2009). STANDing strong, resistance proteins instigators of plant defence. Curr. Opin. Plant Biol. 12: 427–436 [DOI] [PubMed] [Google Scholar]
- Maekawa T., et al. (2011a). Coiled-coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Microbe 9: 187–199 [DOI] [PubMed] [Google Scholar]
- Maekawa T., Kracher B., Vernaldi S., Ver Loren van Themaat E., Schulze-Lefert P. (2012). Conservation of NLR-triggered immunity across plant lineages. Proc. Natl. Acad. Sci. USA 109: 20119–20123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T., Kufer T.A., Schulze-Lefert P. (2011b). NLR functions in plant and animal immune systems: So far and yet so close. Nat. Immunol. 12: 817–826 [DOI] [PubMed] [Google Scholar]
- Noutoshi Y., Ito T., Seki M., Nakashita H., Yoshida S., Marco Y., Shirasu K., Shinozaki K. (2005). A single amino acid insertion in the WRKY domain of the Arabidopsis TIR-NBS-LRR-WRKY-type disease resistance protein SLH1 (sensitive to low humidity 1) causes activation of defense responses and hypersensitive cell death. Plant J. 43: 873–888 [DOI] [PubMed] [Google Scholar]
- Pandey S.P., Roccaro M., Schön M., Logemann E., Somssich I.E. (2010). Transcriptional reprogramming regulated by WRKY18 and WRKY40 facilitates powdery mildew infection of Arabidopsis. Plant J. 64: 912–923 [DOI] [PubMed] [Google Scholar]
- Qi T., Song S., Ren Q., Wu D., Huang H., Chen Y., Fan M., Peng W., Ren C., Xie D. (2011). The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23: 1795–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele S., Vailleau F., Léger A., Joubès J., Miersch O., Huard C., Blée E., Mongrand S., Domergue F., Roby D. (2008). A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 20: 752–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravensdale M., Bernoux M., Ve T., Kobe B., Thrall P.H., Ellis J.G., Dodds P.N. (2012). Intramolecular interaction influences binding of the Flax L5 and L6 resistance proteins to their AvrL567 ligands. PLoS Pathog. 8: e1003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton P.J., Somssich I.E., Ringler P., Shen Q.J. (2010). WRKY transcription factors. Trends Plant Sci. 15: 247–258 [DOI] [PubMed] [Google Scholar]
- Sadowski I., Ma J., Triezenberg S., Ptashne M. (1988). GAL4-VP16 is an unusually potent transcriptional activator. Nature 335: 563–564 [DOI] [PubMed] [Google Scholar]
- Seeholzer S., Tsuchimatsu T., Jordan T., Bieri S., Pajonk S., Yang W., Jahoor A., Shimizu K.K., Keller B., Schulze-Lefert P. (2010). Diversity at the Mla powdery mildew resistance locus from cultivated barley reveals sites of positive selection. Mol. Plant Microbe Interact. 23: 497–509 [DOI] [PubMed] [Google Scholar]
- Shen Q.H., Saijo Y., Mauch S., Biskup C., Bieri S., Keller B., Seki H., Ulker B., Somssich I.E., Schulze-Lefert P. (2007). Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315: 1098–1103 [DOI] [PubMed] [Google Scholar]
- Shen Q.H., Schulze-Lefert P. (2007). Rumble in the nuclear jungle: Compartmentalization, trafficking, and nuclear action of plant immune receptors. EMBO J. 26: 4293–4301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q.H., Zhou F.S., Bieri S., Haizel T., Shirasu K., Schulze-Lefert P. (2003). Recognition specificity and RAR1/SGT1 dependence in barley Mla disease resistance genes to the powdery mildew fungus. Plant Cell 15: 732–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slootweg E., et al. (2010). Nucleocytoplasmic distribution is required for activation of resistance by the potato NB-LRR receptor Rx1 and is balanced by its functional domains. Plant Cell 22: 4195–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R., Werber M., Weisshaar B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4: 447–456 [DOI] [PubMed] [Google Scholar]
- Swiderski M.R., Birker D., Jones J.D.G. (2009). The TIR domain of TIR-NB-LRR resistance proteins is a signaling domain involved in cell death induction. Mol. Plant Microbe Interact. 22: 157–165 [DOI] [PubMed] [Google Scholar]
- Takken F.L.W., Goverse A. (2012). How to build a pathogen detector: Structural basis of NB-LRR function. Curr. Opin. Plant Biol. 15: 375–384 [DOI] [PubMed] [Google Scholar]
- Tameling W.I.L., Nooijen C., Ludwig N., Boter M., Slootweg E., Goverse A., Shirasu K., Joosten M.H.A.J. (2010). RanGAP2 mediates nucleocytoplasmic partitioning of the NB-LRR immune receptor Rx in the Solanaceae, thereby dictating Rx function. Plant Cell 22: 4176–4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., Xie Z., Chen W., Glazebrook J., Chang H.S., Han B., Zhu T., Zou G., Katagiri F. (2003). Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 15: 317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K., Sato M., Stoddard T., Glazebrook J., Katagiri F. (2009). Network properties of robust immunity in plants. PLoS Genet. 5: e1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirthmueller L., Zhang Y., Jones J.D.G., Parker J.E. (2007). Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr. Biol. 17: 2023–2029 [DOI] [PubMed] [Google Scholar]
- Xu X., Chen C., Fan B., Chen Z. (2006). Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18: 1310–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Klessig D.F. (1996). Isolation and characterization of a tobacco mosaic virus-inducible myb oncogene homolog from tobacco. Proc. Natl. Acad. Sci. USA 93: 14972–14977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan C., Li C., Yan L., Jackson A.O., Liu Z., Han C., Yu J., Li D. (2011). A high throughput barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PLoS ONE 6: e26468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Xu F., Zhang Y., Cheng Y.T., Wiermer M., Li X., Zhang Y. (2010). Arabidopsis resistance protein SNC1 activates immune responses through association with a transcriptional corepressor. Proc. Natl. Acad. Sci. USA 107: 13960–13965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C. (2009). Early molecular events in PAMP-triggered immunity. Curr. Opin. Plant Biol. 12: 414–420 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.