A copper transport protein affects copper, iron, and phosphate deficiency responses.
Abstract
Copper and iron are essential micronutrients for most living organisms because they participate as cofactors in biological processes, including respiration, photosynthesis, and oxidative stress protection. In many eukaryotic organisms, including yeast (Saccharomyces cerevisiae) and mammals, copper and iron homeostases are highly interconnected; yet, such interdependence is not well established in higher plants. Here, we propose that COPT2, a high-affinity copper transport protein, functions under copper and iron deficiencies in Arabidopsis (Arabidopsis thaliana). COPT2 is a plasma membrane protein that functions in copper acquisition and distribution. Characterization of the COPT2 expression pattern indicates a synergic response to copper and iron limitation in roots. We characterized a knockout of COPT2, copt2-1, that leads to increased resistance to simultaneous copper and iron deficiencies, measured as reduced leaf chlorosis and improved maintenance of the photosynthetic apparatus. We propose that COPT2 could play a dual role under iron deficiency. First, COPT2 participates in the attenuation of copper deficiency responses driven by iron limitation, possibly to minimize further iron consumption. Second, global expression analyses of copt2-1 versus wild-type Arabidopsis plants indicate that low-phosphate responses increase in the mutant. These results open up new biotechnological approaches to fight iron deficiency in crops.
Copper (Cu) and iron (Fe) act as a double-edged sword in living beings, since they are essential redox-active micronutrients but are cytotoxic when in excess. Since both metals participate as catalytic cofactors in multiple metabolic pathways, their homeostasis needs to be strictly controlled. Plants are the basis of trophic chains, and their nutritional deficiencies are often transferred to consumers. The essentiality of Cu and Fe in plants is evidenced by the symptoms that their deficiencies provoke, which affect the yield and nutritional value of crops (Märschner, 2002; Puig et al., 2007). In biological terms, use of Fe preceded Cu due to the high bioavailability of Fe as Fe2+ under anoxic conditions. However, the appearance of oxygen in the atmosphere caused not only reduced Fe bioavailability but also concomitantly increased the bioavailability and use of Cu by biological systems (Crichton and Pierre, 2001). In parallel with the incorporation of Cu into multiple processes requiring higher redox potentials, which temporally coincided with the commencement of pluricellularity, new strategies to solubilize and acquire Fe3+ were developed. In some cases, enzymes that catalyze the same biochemical reaction are coordinately regulated to allow the alternative use of either Cu- or Fe-containing proteins, depending on metal bioavailability. One main example in Arabidopsis (Arabidopsis thaliana) plants is the use of copper/zinc superoxide dismutase (Cu/ZnSOD) versus iron superoxide dismutase (FeSOD), which both play roles in reactive oxygen species detoxification (Abdel-Ghany et al., 2005; Yamasaki et al., 2007; Burkhead et al., 2009; Waters et al., 2012).
In Arabidopsis, deciphering responses to Cu deficiency is now starting (Pilon et al., 2009; Peñarrubia et al., 2010). A family of high-affinity Cu transport proteins, denoted COPTs in plants (CTR in yeast [Saccharomyces cerevisiae] and human), participates in Cu transport toward the cytosol. COPT1 is a plasma membrane-located member of this family that plays a key role in Cu uptake, root growth, and pollen development (Kampfenkel et al., 1995; Sancenón et al., 2004). Recent studies have shown that deregulated Cu transport in COPT1-overexpressing plants affects development under continuous environmental conditions (Andrés-Colás et al., 2010). On the other hand, Cu activates calcium- and potassium-permeable plasma membrane transporters in COPT1-overexpressing plants under 10 μm Cu conditions, which might be caused by the generation of a cytosolic hydroxyl radical (Rodrigo-Moreno et al., 2013). In the COPT family, COPT1 and COPT2 mRNA levels are down-regulated by Cu, and their expression completely rescues the respiratory defect of yeast ctr1Δctr3Δ mutants, which are defective in high-affinity Cu uptake (Sancenón et al., 2003). The Arabidopsis genome encodes a 17-member zinc finger plant-specific transcription factor family named SPL (for SQUAMOSA-promoter binding-like proteins; Birkenbihl et al., 2005). SPL7 has been shown to be essential for the transcriptional activation observed in response to Cu deficiency in vivo through its binding to GTAC motifs within the promoter of Cu-responsive genes, including those encoding COPT1, COPT2, and the FeSOD FSD1 (Yamasaki et al., 2009; Bernal et al., 2012).
Despite its abundance in soils, Fe bioavailability is very limited, which often provokes Fe deficiency symptoms (i.e. chlorosis) and lowers crop yields. The primary response of Arabidopsis plants to Fe deficiency is controlled through coordinated transcriptional activation, including the increased expression of metal reductases and transporters, such as Ferric reduction oxidase2 (FRO2) and Iron-regulated transporter1 (IRT1), respectively, to improve metal bioavailability and acquisition. The Arabidopsis basic helix-loop-helix (bHLH) transcription factor bHLH29/FRU, also known as FIT (for Fe deficiency-induced transcription factor), controls some of the root responses upon Fe limitation at different levels (for review, see Guerinot, 2000; Hindt and Guerinot, 2012; Ivanov et al., 2012).
Fe availability has been shown to play a crucial role in the root architecture changes induced by phosphate (Pi) deficiency in Arabidopsis (Ward et al., 2008). Thus, whereas primary root elongation is greatly inhibited by Pi starvation, root growth is restored under reduced Fe without increasing Pi availability (Ward et al., 2008). Moreover, Pi-starved Arabidopsis plants show elevated Fe accumulation in both shoots and roots (Misson et al., 2005; Ward et al., 2008). Phosphorus is not only an essential macronutrient but also a key component of, among others, membrane phospholipids and is crucial for processes such as signaling cascades (Raghothama, 1999; Chiou and Lin, 2011). It has been suggested that Pi may be sensed indirectly via complex and antagonistic interactions between Pi and Fe availabilities, which still remain to be elucidated (Abel, 2011).
Given their sessile nature, plants are organisms that probably explore the widest variety of responses to environmental nutrient availabilities, and they have developed multiple regulatory mechanisms to respond to metal and nutrient deficiencies. In addition to the aforementioned substitution of specific metalloproteins, improved metal bioavailability and acquisition, including the increased expression of metalloreductases and high-affinity transporters, are among the main strategies to help face metal deficiencies (Jeong and Guerinot, 2009; Palmer and Guerinot, 2009; Puig and Peñarrubia, 2009). Nutrient status information has to be communicated between organs to optimize essential inorganic nutrient allocation, especially in plants growing under suboptimal conditions. Root architecture is differentially modified during nutrient deficiencies. Whereas root elongation is considered an adequate modification under metal deficiencies, possibly to seek metals in underground soil layers, the inhibition of primary root growth and the development of secondary and higher order roots under Pi starvation maximize the interception of the nutrient in top soil layers (Liao et al., 2001). These processes exemplify not only the extensive cross talk between different metals and other nutrient homeostasis networks but also the delicate balance that plant cells have to strike according to the variable nutritional status in the environment.
In this study, we analyzed the function of the high-affinity Cu transport protein COPT2, whose expression in Arabidopsis is up-regulated in roots by both Cu and Fe deficiencies (Sancenón et al., 2003; Colangelo and Guerinot, 2004; Waters et al., 2012), whereas Pi starvation diminishes its expression (Thibaud et al., 2010). The phenotypes and gene expression changes displayed by a copt2-1 line reveal a role for COPT2 in the cross talk among Cu, Fe, and Pi deficiency responses in Arabidopsis.
RESULTS
The High-Affinity Cu Transport Protein COPT2 Localizes to the Plasma Membrane of Arabidopsis Cells
The COPT2 gene (At3g46900) from Arabidopsis encodes a protein with a 78% identity with COPT1 (Kampfenkel et al., 1995; Sancenón et al., 2003; Supplemental Fig. S1A). The hydrophobicity pattern and the topological comparison between COPT2 and other CTR/COPT family members indicate the presence of three transmembrane domains (TMDs) with an external N terminus and a cytosolic C terminus (Supplemental Fig. S1B). Moreover, it possesses the conserved extracellular Met residue (indicated by asterisks in Supplemental Fig. S1) before TMD1, the MxxxM motif in TMD2, and the GxxxG motif in TMD3, which are essential in yeast homologs (Puig et al., 2002; Aller et al., 2004). A CxC motif is also observed in the COPT2 C-terminal domain (Supplemental Fig. S1). Given its ability to fully complement the respiratory defects of yeast ctr1Δctr3Δ mutants and its regulation by environmental Cu in Arabidopsis, it has been previously suggested that both the COPT2 and COPT1 proteins could function in Cu acquisition in the plasma membrane of specific plant cells (Kampfenkel et al., 1995; Sancenón et al., 2003, 2004).
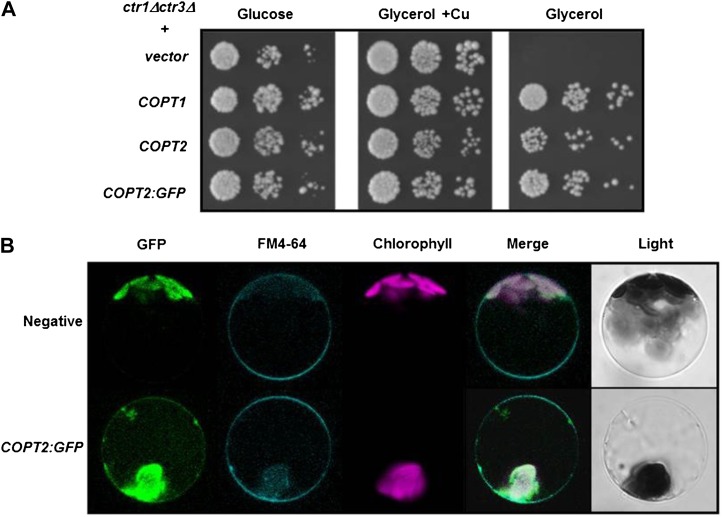
For the purpose of localizing COPT2 at the subcellular level in Arabidopsis cells, its coding sequence was fused to the GFP (COPT2-GFP) under the control of the constitutive CaMV35S promoter. As shown in Figure 1A, the construct COPT:GFP complements the respiratory defect of yeast ctr1Δctr3Δ mutants to a similar extent as COPT2 and COPT1, indicating that addition of the GFP does not interfere with the Cu transport function in yeast. Isolated Arabidopsis protoplasts transiently expressing the PCaMV35S:COPT2:GFP construct were analyzed for the localization of COPT2-GFP, lipophilic styryl dye FM4-64, and chlorophyll fluorescences indicated by green, cyan, and magenta, respectively (Fig. 1B). In addition to the chlorophyll autofluorescence, COPT2-GFP-expressing cells display a signal on the cell surface that is absent in vacuoles, the cytosol, and other discrete subcellular localizations. This signal colocalizes with the FM4-64 marker, which localizes to the cell surface at low temperatures (Fig. 1B). Furthermore, immunofluorescence labeling of stable transgenic Arabidopsis plants expressing the CaMV35S:COPT2-HA construct shows a signal restricted to the peripheral side of the cytoplasm when using anti-hemagglutinin (HA) antibodies (Supplemental Fig. S2). These results strongly suggest that the COPT2 protein localizes to the plasma membrane of Arabidopsis cells.
Figure 1.
COPT2:GFP functionality in yeast, and subcellular localization in Arabidopsis protoplasts. A, Yeast ctr1Δctr3Δ cells were transformed with p426GPD, COPT1, COPT2, and COPT2:GFP and were spotted on synthetic complete-ura medium (Glucose) or yeast extract peptone glycerol (Glycerol) supplemented with 100 μm CuSO4 (Glycerol + Cu). Each spot represents a 1:10 cell culture density dilution, decreasing from left to right. B, Arabidopsis protoplasts were isolated from 30-d-old leaves and were transiently transformed with the PCaMV35S:COPT2:GFP construct, incubated with FM4-64 for 15 min at 4°C, before being analyzed by confocal microscopy at 16 h post transformation. Nontransformed protoplasts were used as a negative control. Green, cyan, and magenta fluorescences are indicative of the localization of the GFP protein, the FM4-64 marker, and chlorophyll, respectively. Representative protoplasts are shown on the same scale, including their merged and light fields.
COPT2 Expression Is Differentially Regulated by Cu and Fe Deficiencies
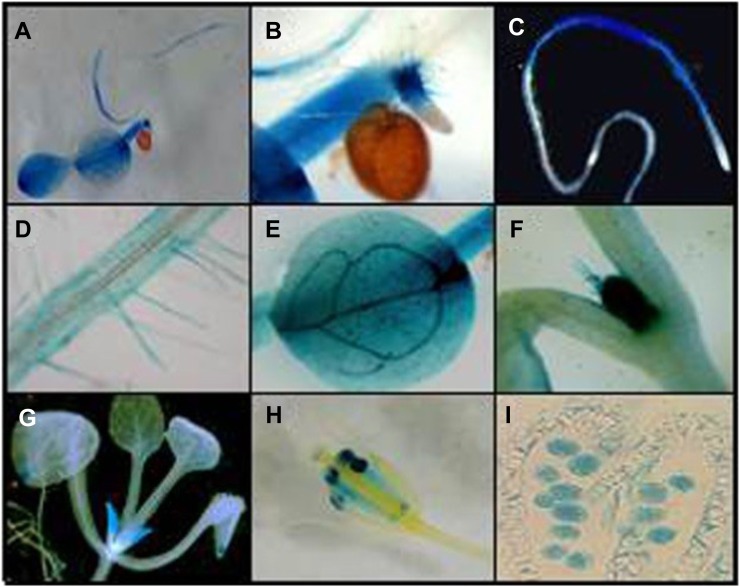
In order to study the COPT2 spatial expression pattern, the promoter region (PCOPT2; covering 1,248 bp upstream from the start codon) was fused to the uidA (GUS) reporter gene. Transgenic Arabidopsis lines harboring the PCOPT2:GUS chimeric gene were obtained. GUS staining in 7-d-old Arabidopsis seedlings grown under Cu-deficient conditions indicated that COPT2 was expressed in most tissues of the seedlings (Fig. 2A). For roots, GUS staining was observed in the differentiation zone but was absent from the elongation and meristematic zones (Fig. 2, B and C). Moreover, COPT2 expression in roots included the lateral roots and root hairs, displaying expression mostly in the epidermis (Fig. 2D). In cotyledons, the highest expression was observed in vascular bundles and hydathodes (Fig. 2E). Histological analyses of seedlings and adult plants showed COPT2 promoter-driven GUS expression in the apical meristem and trichomes (Fig. 2F) and in young leaves (Fig. 2G). During the development of reproductive organs, the GUS staining in anthers (Fig. 2H) indicates that COPT2 is highly expressed in pollen (Fig. 2I). Three other independent PCOPT2:GUS lines were analyzed and showed the same COPT2 tissue pattern expression (data not shown). Schematics of the COPT2, as compared with the COPT1, expression patterns, along with other characteristics of both permeases, are shown in Supplemental Figure S3.
Figure 2.
COPT2 expression pattern in Arabidopsis PCOPT2:GUS transgenic plants under Cu deficiency. Shown are GUS staining in a representative 7-d-old seedling (A), detail of a secondary root tip (B), general view of the main root (C), detail of root hairs (D), cotyledon (E), shoot meristem and trichomes (F), aerial part from a 3-week-old seedling (G), flower (H), and a longitudinal section of an anther with pollen grains (I).
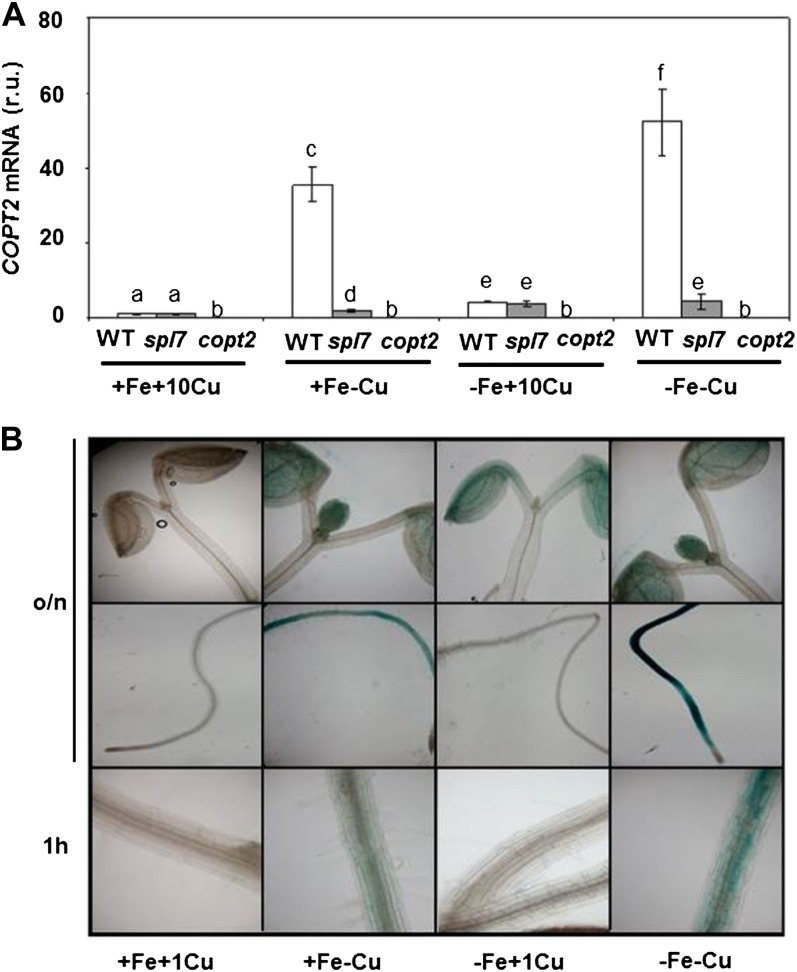
The analysis of the regulatory elements in the PCOPT2 region indicates the presence of four GTAC motifs, which are probably involved in its regulation by Cu (Supplemental Fig. S4; Yamasaki et al., 2009). Interestingly, COPT2 is the COPT family member whose expression was most highly regulated by Cu deficiency (Sancenón et al., 2003; Yamasaki et al., 2009; del Pozo et al., 2010). In addition, a putative E-box consensus, which may be involved in the interaction with bHLH-type transcription factors such as the Fe-responsive FIT protein (Hartmann et al., 2005), was present in its promoter (Supplemental Fig. S4). Accordingly, COPT2 expression has been shown to respond to Fe deficiency in a partially FIT-dependent manner (Colangelo and Guerinot, 2004). To ascertain COPT2 expression under separate Fe and Cu deficiencies, or when both deficiencies are applied simultaneously, wild-type seedlings were grown in the four following media combinations: first, Fe- and Cu-sufficient medium (+Fe+Cu; one-half-strength Murashige and Skoog medium [1/2 MS] supplemented with 10 μm CuSO4); second, Fe-sufficient and Cu-deficient medium (+Fe−Cu; 1/2 MS without CuSO4); third, Fe-deficient and Cu-sufficient medium (−Fe+Cu; 1/2 MS without Fe and supplemented with 10 μm CuSO4 and 300 μm ferrozine, a specific Fe2+ chelator); and fourth, Fe- and Cu-deficient medium (−Fe−Cu; 1/2 MS without Cu and Fe and supplemented with 300 μm ferrozine). The Cu concentration used in this experiment was 10 μm, which was high enough to guarantee a sufficient Cu supply while being far from the deficiency and excess responses (Andrés-Colás et al., 2010). COPT2 expression was induced by Cu deficiency (35.6-fold), by Fe deficiency (4.3-fold), and its expression was further increased under simultaneous Fe and Cu deprivation (52.3-fold; Fig. 3A; Supplemental Table S1). COPT2 expression remained mostly under the control of the SPL7 transcription factor, since the spl7 mutant displayed a significant drop at the mRNA levels (Fig. 3A; Yamasaki et al., 2009; Bernal et al., 2012). However, COPT2 was still expressed at low basal levels under all the conditions tested in the spl7 mutant as compared with a copt2 null mutant (copt2-1 line [see below]; Fig. 3A), indicating that this basal expression level was SPL7 independent. Furthermore, the COPT2 tissue pattern shows that under Cu deficiency (+Fe−Cu), it was highly expressed in roots and that its expression increased under both metal deficiencies (−Fe−Cu; Fig. 3B). In order to further define where COPT2 expression started, root photographs under 1 h at 37°C for GUS staining were obtained, showing cell patches of differentiated root cells (Fig. 3B). Interestingly, high (1 μm) Cu levels (+Fe+Cu) completely abolished COPT2 expression in roots, but a low expression level, which requires overnight GUS staining, remained restricted to cotyledons under Fe deficiency (−Fe+Cu; Fig. 3B). The expression in roots requires Cu deficiency conditions, and it was not observed under Fe deficiency when the Cu levels in the medium were higher than 0.25 μm (Supplemental Fig. S5). However, the low expression in cotyledons was observed at higher Cu levels under overnight GUS-staining conditions (Fig. 3B; Supplemental Fig. S5). Taken together, these results not only demonstrate that COPT2 expression is differentially regulated by low Cu and Fe conditions but suggest a cross talk between both metal deficiencies.
Figure 3.
COPT2 expression under Cu and Fe deficiencies. A, COPT2 expression analysis by qPCR in wild-type (WT; white bars), spl7 (gray bars), and copt2-1 (copt2; dark gray bars) seedlings. Total RNA from 7-d-old seedlings grown under the control (+Fe+Cu; supplemented with 10 μm CuSO4), Cu deficiency (+Fe−Cu), Fe deficiency (−Fe+Cu; supplemented with 10 μm CuSO4), or Fe and Cu deficiency (−Fe−Cu) conditions was isolated and retrotranscribed to complementary DNA. UBQ10 gene expression was used as a loading control. Values are means ± sd of three biological replicates. r.u., Relative units. Different letters above the bars represent significant differences among all the means (P < 0.05). B, GUS staining in 7-d-old seedlings from the PCOPT2:GUS transgenic lines grown under control (+Fe+Cu; supplemented with 1 μm CuSO4), Cu deficiency (+Fe−Cu), Fe deficiency (−Fe+Cu; supplemented with 1 μm CuSO4), or Fe and Cu deficiency (−Fe−Cu) conditions at different incubation times at 37°C (overnight [o/n] and 1 h). [See online article for color version of this figure.]
Characterization of the copt2-1 Line under Cu and Fe Deficiency Conditions
A copt2-1 line (Supplemental Fig. S6) that contains the transfer DNA (T-DNA) insert at −55 bp (in relation to the translation start codon), separating the four putative Cu regulatory GTAC motifs from the COPT2 coding sequence (Supplemental Fig. S6A), shows no COPT2 expression in seedlings grown on different Cu availabilities (100 μm bathocuproinedisulfonic acid disodium and 0, 1, 5, or 10 μm CuSO4; Supplemental Fig. S6C). Moreover under the four conditions previously stated for Cu and Fe availability, the COPT2 expression levels in the copt2-1 line were below those found in the spl7 mutant (Fig. 3A). Therefore, we used the copt2-1 mutant to study the function of this transporter in Arabidopsis.
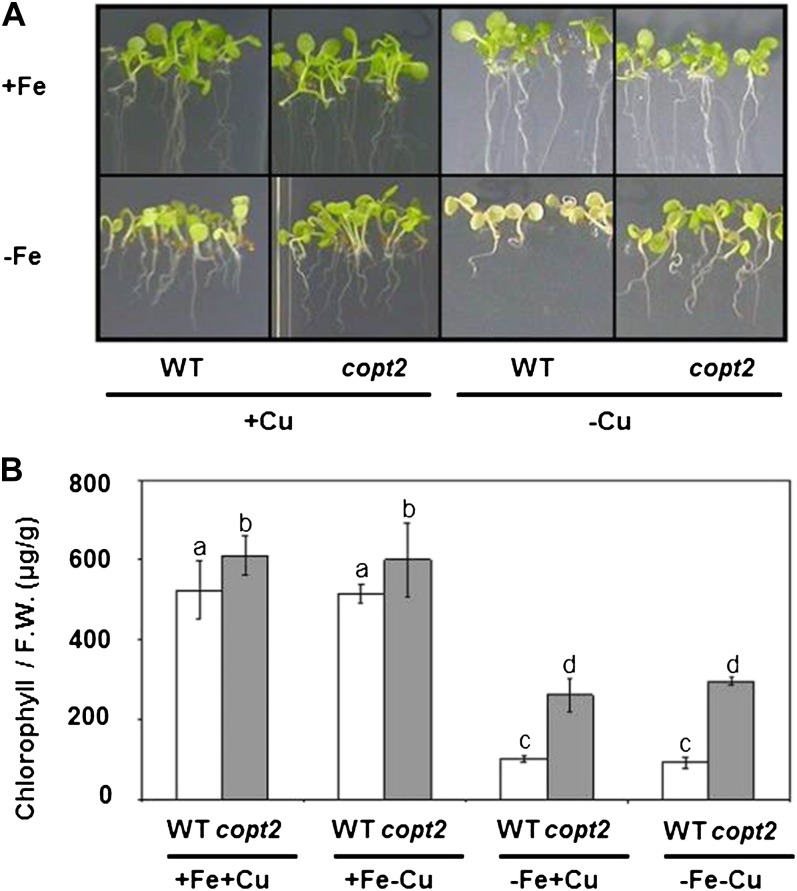
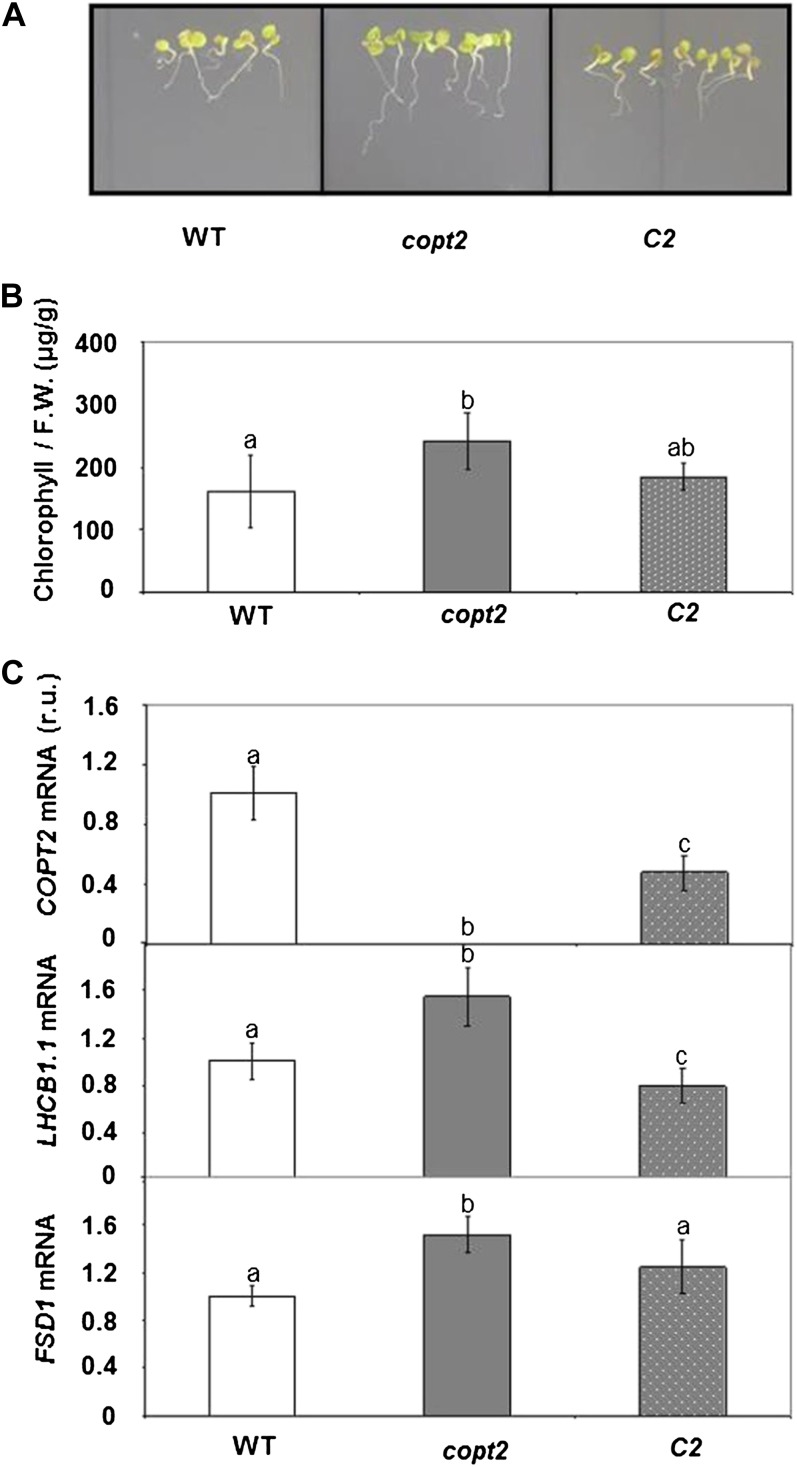
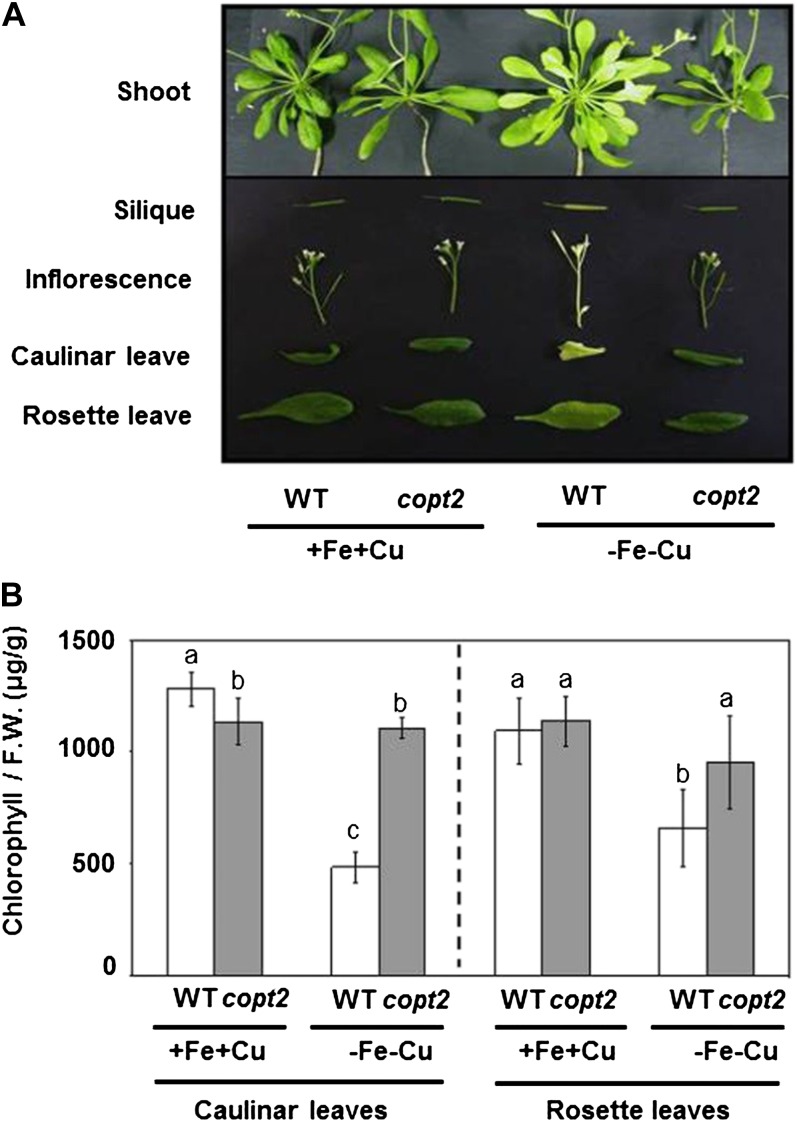
Since COPT2 displays different expression patterns under +Fe−Cu, −Fe+Cu, and −Fe−Cu conditions, we studied the potential copt2-1 phenotypes under these conditions as compared with the +Fe+Cu control medium. Apparently, the mutant did not seem to differ from the wild-type controls when grown on soil or in 1/2 MS with different Cu availabilities. Furthermore, certain parameters, such as root length, seed germination, deetiolation, and hypocotyl length, showed no significant differences between the wild type and the copt2-1 line under the different Cu statuses analyzed (data not shown). However, whereas the wild-type plants grown under simultaneous Cu and Fe deficiency conditions exhibited a light green leaf appearance, a typical symptom of mild chlorosis, the copt2-1 seedlings displayed a slightly increased resistance to Fe deprivation by delaying symptoms, as shown by their greener leaves (Fig. 4A). In order to quantify this copt2-1 phenotype, total chlorophylls were measured in 7-d-old wild-type and mutant seedlings grown under the four Fe/Cu conditions stated above. As Figure 4B illustrates, the copt2-1 line showed increased chlorophylls when compared with the wild-type seedlings, especially under −Fe conditions. Despite no significant changes in fresh weight being detected under Fe deficiency (Supplemental Fig. S7A), a parameter indicative of photosynthetic apparatus integrity such as LHCB1.1 (for light-harvesting complex B1.1) mRNA, showed a slightly higher level in the mutant than in the wild-type plants under Fe deficiency conditions (Supplemental Fig. S7B). In order to ascertain whether these defects were COPT2 specific, we transformed the copt2-1 line with the COPT2 wild-type gene driven by its own promoter (PCOPT2:COPT2:GFP). Importantly, the PCOPT2:COPT2:GFP seedlings grown under both metal deficiencies revealed a partial restoration of COPT2 expression and, accordingly, displayed an increased sensitivity to metal deficiencies, as shown by the chlorophyll content (Fig. 5). Moreover, the LHCB1.1 and FSD1 expression decreased when compared with the copt2-1 line under simultaneous metal deficiency (Fig. 5C), which is in agreement with reduced plant performance in the presence of COPT2 expression. The copt1 mutant also displayed a similar phenotype of resistance to Fe deficiency-induced chlorosis (data not shown), which further corroborates the role of Cu in this process. To investigate whether the better resistance of the mutant to ferric chlorosis under −Fe−Cu conditions was also displayed in adult stages, plants were grown in hydroponic cultures under +Fe+Cu (Hoagland) and −Fe−Cu (Hoagland with no added Cu and Fe) conditions. As indicated for the seedlings grown on agar plates (Figs. 4 and 5), once again the chlorosis under −Fe−Cu conditions in the aerial part was less pronounced or retarded in the copt2-1 line than in the wild-type adult plants (Fig. 6). It is worth noting that the plastocyanin content in the leaves of the adult plants grown under −Fe−Cu conditions was higher in the copt2-1 line than in wild-type plants (Supplemental Fig. S8). Moreover, senescence of the copt2-1 siliques is delayed compared with the wild type (Supplemental Fig. S9A), and subsequently, the mutant produces more seeds (Supplemental Fig. S9B) with a higher germination rate (40%) than wild-type plants (5%) when grown under both metal deficiency conditions. Taken together, these results further support the better or extended maintenance of the photosynthetic apparatus in the copt2-1 line than in the wild-type plants grown under conditions of simultaneous Fe and Cu deficiencies, which leads to improved plant growth and seed production.
Figure 4.
Phenotypes of the copt2-1 seedlings grown under Cu and Fe deficiencies. A, Photographs of 7-d-old seedlings of the wild type (WT) and the copt2-1 line grown under the same conditions described in Figure 3A. B, Chlorophyll contents of 7-d-old seedlings of both the wild type (white bars) and copt2 (gray bars) grown under the same conditions described in Figure 3. Values are means ± sd of at least three biological replicates. F.W., Fresh weight. Different letters above the bars represent significant differences among all the means (P < 0.05).
Figure 5.
Chlorophyll contents and gene expression of complemented copt2-1 seedlings grown under Cu and Fe deficiencies. A, Photographs of 7-d-old seedlings of the wild type (WT), copt2-1 (copt2), and PCOPT2:COPT2:GFP (C2) grown under −Fe−Cu conditions. B, Chlorophyll contents of wild-type (white bars), copt2 (gray bars), and C2 (dotted bars) seedlings. Values are means ± sd of at least four biological replicates. Different letters above the bars represent significant differences among all the means (P < 0.01). F.W., Fresh weight. C, Expression analysis of COPT2, LHCB1.1, and FSD1 genes by qPCR in wild-type (white bars), copt2 (gray bars), and C2 (dotted bars) seedlings, as described in Figure 3. UBQ10 gene expression was used as a loading control. Values are means ± sd of three biological replicates. r.u., Relative units. Different letters above the bars represent significant differences among all the means (P < 0.05). [See online article for color version of this figure.]
Figure 6.
Phenotypes of copt2-1 adult plants grown under Cu and Fe deficiencies. A, Photographs of rosette leaves and aerial part details from the wild type (WT) and the copt2-1 line grown on the +Fe+Cu and −Fe−Cu media were taken 14 d after treatments. B, Chlorophyll contents of cauline and rosette leaves from the wild-type (white bars) and copt2 (gray bars) plants shown in A. Values are means ± sd of four biological replicates. F.W., Fresh weight. Different letters above the bars represent significant differences (P < 0.05) among treatments in each type of leaf.
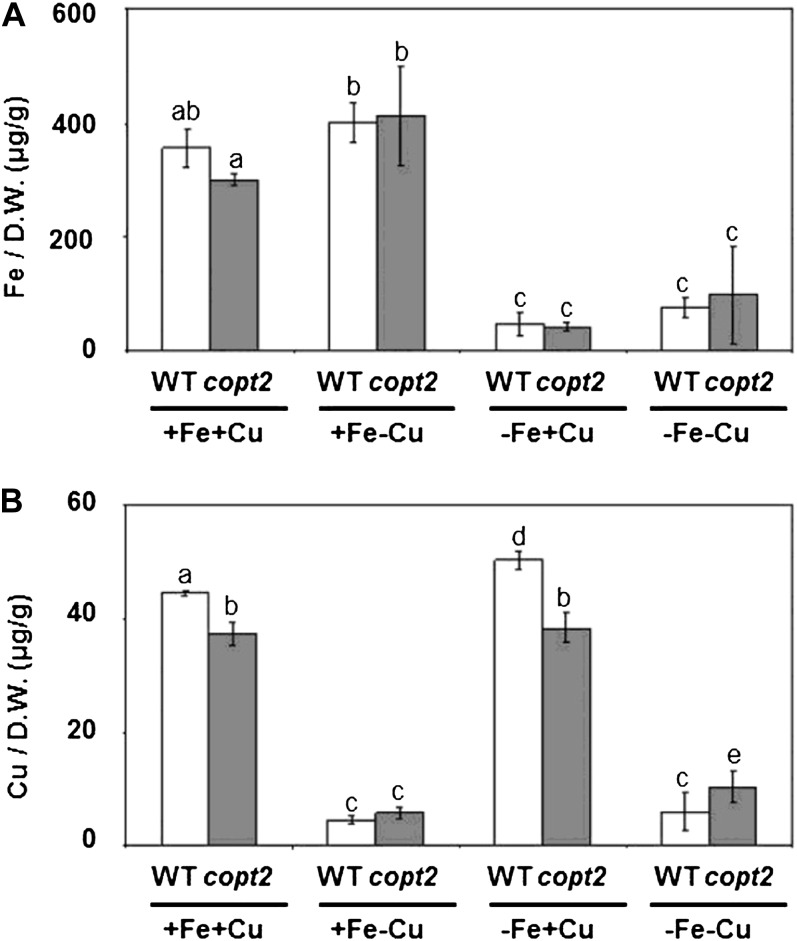
In order to determine whether the total endogenous metal content in these plants was responsible for the phenotypes observed in the copt2-1 line, the Fe and Cu contents from the 7-d-old seedlings grown under the four Fe/Cu conditions described above were determined by inductively coupled plasma mass spectroscopy (ICP-MS). No significant differences in Fe content were observed between wild-type and copt2-1 plants (Fig. 7A). Although a slight decrease in Cu content was detected in the mutant under high Cu (Fig. 7B), COPT2 was expressed at low levels under these conditions (Fig. 3A), questioning a putative COPT2 role in Cu uptake from the medium under high Cu. Moreover, no significant differences in Cu content between the wild-type and copt2-1 plants grown under either hydroponic or Cu deficiency conditions were found in different organs, such as roots, rosette leaves, stems, or inflorescences (data not shown). These results suggest that the total endogenous metal content in seedlings is not responsible for the phenotype of the simultaneous resistance to Fe and Cu deficiencies displayed by the copt2-1 line.
Figure 7.
Endogenous Fe and Cu contents in wild-type and copt2-1 seedlings. Fe (A) and Cu (B) contents were measured by ICP-MS from whole 7-d-old wild-type (WT; white bars) and copt2-1 (gray bars) plants. Seedlings were grown under the same conditions described in Figure 3. Values are means ± sd of at least three biological replicates. D.W., Dry weight. Different letters above the bars represent significant differences among all the means (P < 0.05).
The Negative Effects of Fe Starvation on Cu Deficiency Responses Are Altered in the copt2-1 Line
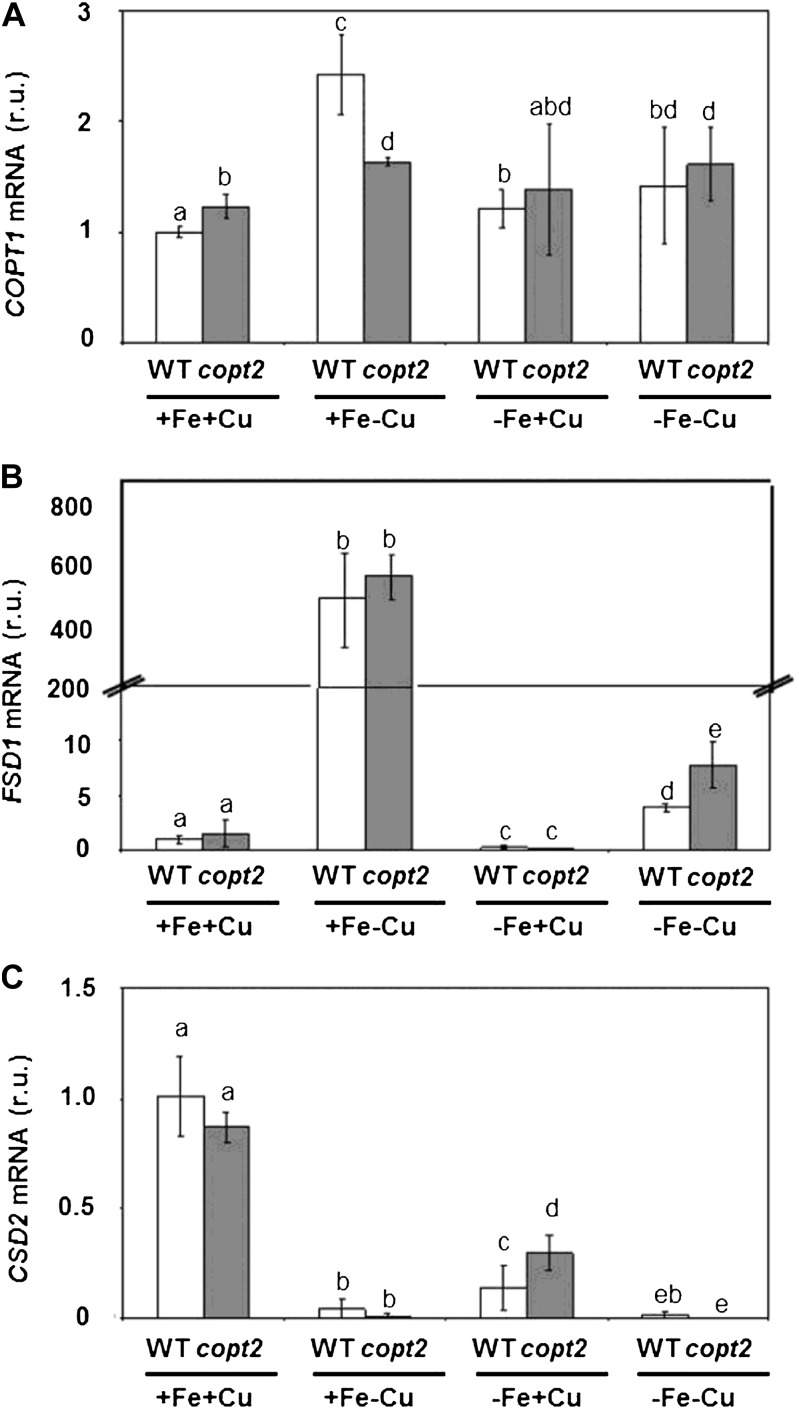
In order to understand the molecular reasons underlying the copt2-1 phenotype, the mRNA levels of selected genes regulated by Cu deficiency (COPT1, FSD1, and CSD2) were determined by quantitative PCR (qPCR) in 7-d-old wild-type and mutant seedlings grown under the four Fe/Cu conditions described above (Fig. 8). We observed that COPT1 expression was up-regulated by Cu deficiency in both wild-type and copt2-1 lines (Fig. 8A, +Fe+Cu versus +Fe−Cu; Sancenón et al., 2003). It is interesting that when Fe was limited (−Fe conditions), no COPT1 up-regulation was observed in response to Cu deficiency (Fig. 8A, −Fe+Cu versus −Fe−Cu). These results suggest that Fe deficiency negatively affects COPT1 induction by low Cu in both the wild-type and copt2-1 lines. To further address this observation, we determined the mRNA levels of Cu-regulated genes FSD1 and CSD2, which encode for FeSOD and Cu/ZnSOD, respectively. As Figure 8, B and C, depicts, the CSD2/FSD1 substitution was normally observed with Cu deficiency under Fe-sufficient conditions (+Fe+Cu versus +Fe−Cu). However, when Fe deficiency was imposed, the increased FSD1 mRNA levels were greatly compromised (Fig. 8B, −Fe+Cu versus −Fe−Cu), and the copt2-1 line showed a slightly increased FSD1 expression under Cu and Fe deficiencies (Fig. 8B). To check whether this slight increase in superoxide dismutase (SOD) expression implies a general enhancement in oxidative stress protection in the copt2-1 line, SOD activities were measured in gel and no significant differences were found between the wild type and the mutant (Supplemental Fig. S10A). Moreover, when subjected to oxidative treatments, such as hydrogen peroxide (500 μm) or paraquat (0.1 μm), no phenotypical differences were observed (Supplemental Fig. S10B), indicating that the copt2-1 line does not show more resistance to general oxidative stress conditions.
Figure 8.
Gene expression pattern of Cu deficiency marker genes in copt2-1 seedlings. Expression analysis is shown for the Cu transporter COPT1 (A) and the FeSOD and Cu/ZnSOD genes FSD1 (B) and CSD2 (C), respectively, by qPCR in wild-type (WT; white bars) and copt2-1 (gray bars) seedlings, grown under the same conditions described in Figure 3. UBQ10 gene expression was used as a loading control. Values are means ± sd of three biological replicates. r.u., Relative units. Different letters above the bars represent significant differences among all the means (P < 0.05).
Since Fe starvation attenuated the Cu deficiency-induced up-regulation of COPT1 and FSD1 (Fig. 8, A and B), we wondered how Cu deficiency could affect plant responses to Fe scarcity. For this purpose, the expression patterns of two well-known Fe deficiency genes, the metalloreductase FRO2 and the Fe transporter IRT1, were analyzed under the four previously assayed Fe/Cu conditions. Although both genes were activated in response to Fe deficiency (Supplemental Fig. S11, A and B, +Fe versus −Fe), as reported previously (Colangelo and Guerinot, 2004), no major differences were observed in the expression of FRO2 and IRT1 in copt2-1 plants (Supplemental Fig. S11).
Global Analysis of Gene Expression Changes under Fe and Cu Deficiencies Indicate That copt2-1 Plants Display Pi Starvation Responses
With the aim of characterizing at the molecular level the causes of enhanced resistance to Fe deficiency and induced chlorosis in the copt2-1 mutant, a global profiling analysis of gene expression under Fe and Cu deficiency was performed. To that end, wild-type and copt2-1 seedlings were grown for 7 d under Fe- and Cu-sufficient conditions (+Fe+Cu) and compared with seedlings grown on medium without Fe and Cu (−Fe−Cu). Moreover, medium without Fe (−Fe+Cu) was used as a control for Cu-complemented expression changes in the copt2-1 line. In order to validate these growth conditions, we previously confirmed by qPCR analysis the expression pattern of two genes previously known to be up-regulated by Cu deficiency, COPT2 (Sancenón et al., 2003; Fig. 3A; Supplemental Fig. S12A) and ZIP2, a ZRT and IRT-like protein2 transporter (Wintz et al., 2003; Supplemental Fig. S12B).
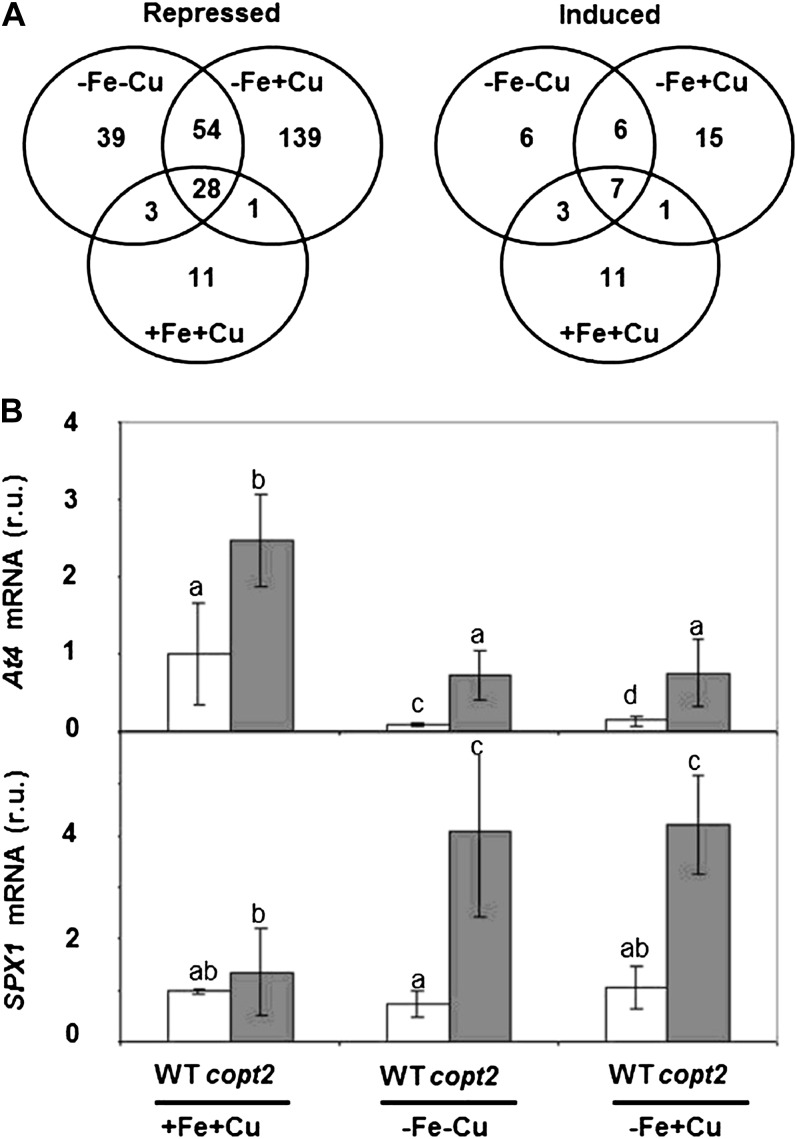
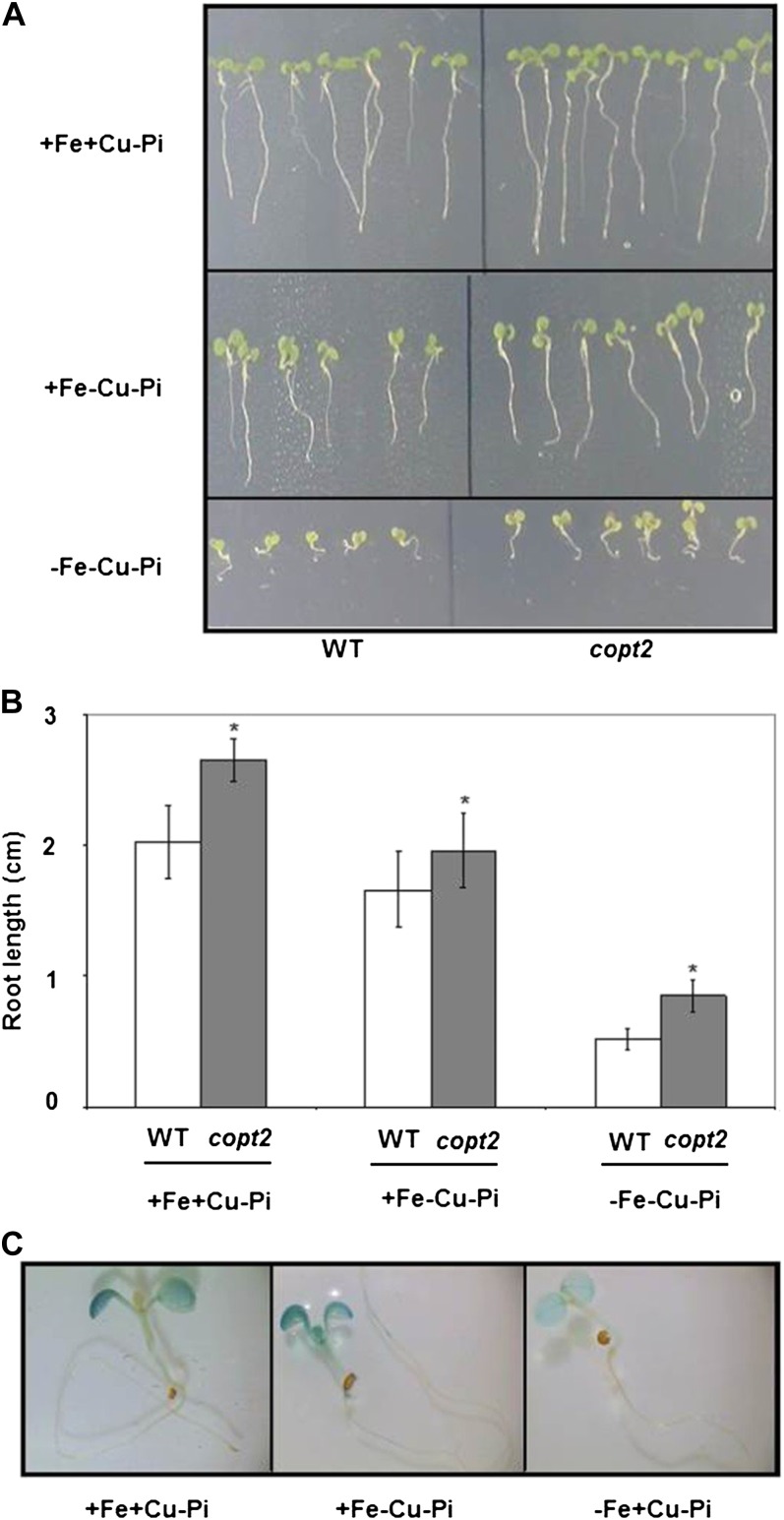
Global profiling analysis of copt2-1 and wild-type lines was carried out from four biological replicates grown in the three different conditions (+Fe+Cu, −Fe−Cu, and −Fe+Cu). A median log2 ratio of 1 (2-fold difference in expression) was used as a cutoff criterion to compare the mutant with the wild type under each condition. We identified 324 differentially expressed genes (Supplemental Tables S1 and S2) distributed in 49 induced (ratio > 1) and 275 repressed (ratio < 1) genes in the copt2-1 line in the three growth conditions (Fig. 9A). Repressed genes are more abundant than induced genes in all three conditions. The condition −Fe+Cu shows the largest number of genes in which expression is affected. Gene Ontology analysis indicated that repressed genes include those of the abiotic stress and detoxifying processes, maybe reflecting the consequences of dismantling the photosynthetic apparatus taking place in wild-type plants (Supplemental Table S3), which may be delayed in the mutant. Interestingly, a significant percentage (35%) of the induced genes are related to Pi starvation responses based on the literature (Table I). Overrepresentation of this category in all three nutritional conditions indicated a general effect of COPT2 function in Pi starvation (Supplemental Table S4). The riboregulators At4, IPS1, and several SPX domain proteins are among the best-known genes induced in Pi-starved plants, and they have been shown to participate in Pi homeostasis and signaling (Franco-Zorrilla et al., 2007; Duan et al., 2008; Thibaud et al., 2010; Chiou and Lin, 2011). Therefore, At4 and SPX1 expression was analyzed in the copt2-1 line, confirming the microarray results (Fig. 9B). Moreover, these data underscore the intricate interactions between Fe and Pi deficiencies, since whereas At4 is down-regulated by Fe deficiency in the wild type, SPX1 expression remains mostly unaffected (Fig. 9B). To check how Cu is implicated in Pi starvation, we grew wild-type and copt2-1 seedlings in Pi-deficient medium and under different Cu and Fe regimes. As shown in Figure 10, A and B, copt2-1 displays slightly larger roots than wild-type plants in all growth conditions, indicating a general insensitivity to Pi deficiency. Moreover, the COPT2 expression pattern remains unaffected under Pi starvation (Fig. 10C). Taken together, our results are compatible with a model where COPT2 is involved in the antagonistic responses of metals and Pi deficiencies (Fig. 11).
Figure 9.
Venn diagrams of gene expression changes in wild-type versus copt2-1 seedlings under different metal statuses. A, Values indicate the number of induced/repressed genes grown under the same conditions described in Figure 3B in copt2-1 seedlings: control (+Fe+Cu), Fe deficiency (−Fe+Cu), or Fe and Cu deficiency (−Fe−Cu). B, Expression analysis by qPCR of At4 and SPX1 in wild-type (WT; white bars) and copt2-1 (gray bars) seedlings, as described in A. UBQ10 gene expression was used as a loading control. Values are means ± sd of at least three biological replicates. r.u., Relative units. Different letters above the bars represent significant differences among all the means (P < 0.05).
Table I. Pi-starvation biological process overrepresented in the copt2-1 line.
Common Pi-starvation- and copt2-1-induced genes compared with the wild type are indicated (Thibaud et al., 2010; asterisks indicate other sources). Munich Information Center for Protein Sequences (MIPS) codes, gene description, gene ontology cellular function, and the microarray values under +Fe+Cu, –Fe–Cu, and –Fe+Cu are indicated. The genes used in qPCR quantitation are in boldface; the genes that are statistically significant with values of less than 1 are italic; and genes that are not statistically significant are indicated as n.s.
| MIPS Code | Gene Description | Gene Ontology Cellular Function | +Fe+Cu | –Fe–Cu | –Fe+Cu |
| At2g32960* | PFA-DSP2 | Catalytic activity, phosphatase activity, protein Tyr phosphatase activity | 2.524 | 2.873 | 2.652 |
| At2g04460 | Unknown | Unknown | 2.205 | 2.495 | 2.300 |
| At5g03545* | At4 | Unknown | 1.383 | 1.986 | 1.855 |
| At5g20150 | SPX1 | Unknown | 1.145 | 1.309 | 1.477 |
| At1g73010 | PPsPase1 | Phosphoric monoester hydrolase activity | 1.073 | 1.222 | 1.165 |
| At2g11810 | MGD3 | 1,2-Diacylglycerol 3-β-galactosyltransferase activity | 1.306 | 1.052 | 0.738 |
| At5g20790 | Unknown | Unknown | 0.865 | 1.811 | 1.515 |
| At3g09922* | IPS1 | Unknown | 0.614 | 1.354 | 1.469 |
| At1g73220 | OCT1 | Carbohydrate and carnitine transporter activity | 2.067 | 0.782 | 0.733 |
| At3g03530 | NPC4 | Hydrolase and phospholipase C activity | 1.112 | 0.489 | 0.607 |
| At1g17710 | Unknown | Phosphoric monoester hydrolase activity | 1.448 | 1.289 | n.s. |
| At1g08310 | Unknown | Galactolipid biosynthetic process, negative regulation of transcription, DNA dependent | 1.000 | 1.441 | n.s. |
| At4g26530* | Unknown | Fru-bisphosphate aldolase activity | 1.123 | 0.219 | n.s. |
| At2g45130 | SPX3 | Unknown | 2.069 | n.s. | n.s. |
| At2g30540 | Unknown | Thiol-disulfide exchange intermediate activity | 1.039 | n.s. | n.s. |
| At4g11800* | Unknown | Protein Ser/Thr phosphatase activity | n.s. | 1.129 | n.s. |
| At1g21980* | PIP5K1 | 1-Phosphatidylinositol-4-P 5-kinase activity | n.s. | n.s. | 1.111 |
Figure 10.
Phenotypes of copt2-1 seedlings grown under Pi, Cu, and Fe deficiencies. A, Representative photographs of 7-d-old seedlings from the wild type (WT) and the copt2-1 line grown under Pi starvation (−Pi) and the conditions described in Figure 3A with regard to Cu deficiency (−Cu) and Cu and Fe deficiencies (−Cu−Fe). B, Root lengths from the wild type (white bars) and copt2 (gray bars) measured in three biological replicates ± sd in the samples described in A. Asterisks above the bars represent significant differences among all the means (P < 0.05) compared with the wild type. C, Overnight GUS staining in 7-d-old seedlings from the PCOPT2:GUS transgenic line grown under Pi deficiency (+Fe+Cu−Pi), Cu and Pi deficiency (+Fe−Cu−Pi), and Fe and Pi deficiency (−Fe+Cu−Pi). [See online article for color version of this figure.]
Figure 11.
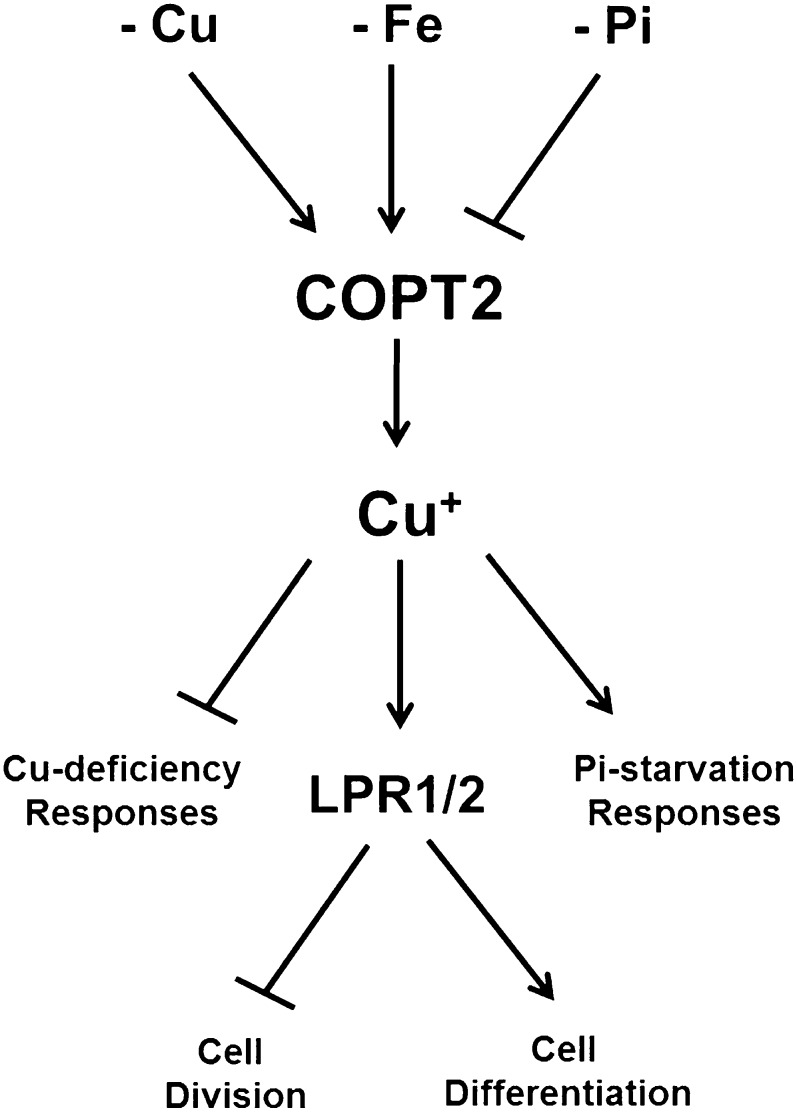
Model of the COPT2-mediated interactions among Cu and Fe deficiencies and Pi starvation responses. Cu, Fe, and Pi deficiencies display antagonistic effects on COPT2 expression. Cu+ uptake mediated by COPT2-attenuated Cu deficiency responses could participate in Cu delivery to low-Pi local sensing and systemic signaling. Local sensing mediated by the multicopper oxidases LPR1 and LPR2 potentiates cell differentiation versus cell division.
DISCUSSION
The conserved family of CTR/COPT proteins mediates high-affinity Cu transport in eukaryotic organisms (for review, see Puig et al., 2002; Kim et al., 2008; Nevitt et al., 2012). Distinct COPT family members have been characterized in Arabidopsis. Whereas COPT1 is a plasma membrane protein that functions in root Cu uptake and pollen development, COPT5 is intracellularly localized and mediates Cu mobilization under severe deficiency conditions (Sancenón et al., 2004; Garcia-Molina et al., 2011; Klaumann et al., 2011). Previous studies have shown that COPT2 is the most similar COPT family protein to COPT1, that both genes fully complement the Cu transport defect of yeast ctr1Δctr3Δ mutants, and that they are up-regulated in response to Cu deficiency in an SPL7-dependent manner, probably by SPL7 binding to GTAC motifs in the COPT1 and COPT2 promoter regions (Sancenón et al., 2003; Yamasaki et al., 2009; Bernal et al., 2012). Here, we show that, similar to COPT1, the COPT2 protein localizes to the plasma membrane of Arabidopsis cells. Furthermore, both genes display a similar aerial expression pattern under slight Cu-deficient conditions (1/2 MS medium), including expression in cotyledons from young seedlings, trichomes, anthers, and mature pollen (Sancenón et al., 2004). These observations suggest that COPT1 and COPT2 might exhibit a partially redundant function in Cu homeostasis in the aerial part. A notable difference between COPT1 and COPT2 concerns their root expression patterns. COPT1 is exclusively expressed in primary and secondary root tips (Sancenón et al., 2004), where overexpression activates plasma membrane hidroxyl radical-sensitive calcium and potassium channels and subsequent root apex signaling (Rodrigo-Moreno et al., 2013). However, COPT2 is expressed in subapical root regions (Fig. 2), where the activation of these channels is prevented (Rodrigo-Moreno et al., 2013), suggesting local and specific functions and subsequent signaling events of these transporters in Arabidopsis roots. Moreover, another key difference is the regulation of both genes under Fe deficiency. In low-Cu medium, whereas COPT1 is down-regulated (Fig. 8A), COPT2 expression is increased by Fe deficiency (Fig. 3A).
In this sense, it is noteworthy that previous genome-wide expression studies in Fe-deficient roots were mostly performed under low Cu levels (Colangelo and Guerinot, 2004; Buckhout et al., 2009; Yang et al., 2010; Stein and Waters, 2012). These studies have shown that COPT2 expression is up-regulated in response to Fe deficiency, which has been further corroborated herein (Fig. 3; Supplemental Fig. S5). However, the Fe deficiency-induced COPT2 expression in roots is abolished by high Cu in medium, although it is still present in shoots (Fig. 3B; Supplemental Fig. S5), suggesting a role for Cu in FIT-mediated responses in Arabidopsis roots. Recently, Waters et al. (2012) reported increased COPT2 expression in roots but a decrease in rosettes for Fe deficiency under low-Cu conditions. The remaining COPT2 expression in cotyledons under −Fe+Cu (Fig. 3B; Supplemental Fig. S5) may be attributed to another putative Fe deficiency network that differs from FIT and is not affected by high Cu levels. It is believed that the increase in endogenous Cu levels when Fe is low (Fig. 7B) may occur to favor the cofactor supply to Cu enzymes (e.g. Cu/ZnSOD), which substitute their Fe counterparts (e.g. FeSOD) to optimize the utilization of low Fe available in more important or irreplaceable Fe-dependent functions (Waters et al., 2012).
Surprisingly, parameters indicative of the photosynthetic apparatus status, such as chlorophyll content, are higher in copt2-1 than in wild-type plants (Figs. 4 and 5), indicating that the absence of COPT2 expression results in better plant performance, which is more relevant under both metal deficiencies (Supplemental Fig. S9). A putative explanation for this interesting phenotype is based on the observation that copt2-1 plants display higher FSD1 mRNA levels than wild-type plants for combined Fe and Cu deficiencies (Fig. 8B). However, a concomitant disadvantageous effect of COPT2 expression resulting from diminished oxidative protection is not observed in the copt2-1 line (Supplemental Fig. S10). Instead, the analysis of global expression changes in the copt2-1 mutant suggests a complex scenario where different cuproproteins and maybe specific COPT-mediated signaling processes could be the basis of the observed phenotype. One of the altered categories in the copt2-1 line is the response to low Pi, which is mostly independent of the metal status (Fig. 9B; Supplemental Tables S1 and S2). Our results reveal a role of COPT2-mediated Cu transport in Pi starvation signaling, which is in agreement with the down-regulated COPT2 expression observed under Pi starvation conditions (Thibaud et al., 2010). Potential connections between Pi starvation responses and the homeostasis of other ions have been described (Abel, 2011; Chiou and Lin, 2011). Root responses to Pi starvation have been suggested to be an outcome of the complex interactions between Pi and other nutrients, essentially Fe (Svistoonoff et al., 2007; Ward et al., 2008). With simultaneous Fe and Pi deficiencies, a recovery in primary root elongation has been reported (Ward et al., 2008). Our results add Cu homeostasis to those interactions in which Fe deficiency and Pi starvation have antagonistic effects by up-regulating and down-regulating COPT2 expression, respectively (Figs. 3A and 11; Thibaud et al., 2010).
COPT2 could participate in Pi sensing by Cu delivery to cuproproteins, such as the multicopper oxidases LPR1 and LPR2 (for low phosphate root), which have been involved in root growth responses to low Pi (Svistoonoff et al., 2007). Our results are compatible with a role of COPT2-mediated Cu transport in supplying Cu to these enzymes. In this sense, a slight increase in root length is observed in the copt2-1 line as compared with the wild type under low-Pi conditions (Fig. 10), which is consistent with the phenotype observed in lpr1 and lpr2 mutants (Svistoonoff et al., 2007). In addition, these data reveal a putative complex COPT2-mediated role in Pi starvation systemic signaling, such as At4 and SPX1 (named after the yeast Syg1 and Pho81 and human XPR1 proteins) expression (Fig. 9B), which still remains to be elucidated.
A putative explanation for the better maintenance of chlorophylls observed in the mutant can be postulated as an indirect effect through Pi starvation responses. Indeed, under Pi-limiting conditions, plants substitute phosphoglycerolipids by activating the genes for galactolipid biosynthesis (Kobayashi et al., 2009). Monogalactosyldiacylglycerol is an abundant lipid in chloroplast membranes (Shimojima and Ohta, 2011). Mutants affected in monogalactosyldiacylglycerol synthase show reduced chlorophyll content (Jarvis et al., 2000). Changes in lipid composition have already been shown to be involved in Cu deficiency responses in Chlamydomonas reinhardtii (Castruita et al., 2011). The genes induced in copt2-1 plants suggest that phospholipid substitution for galactolipids could take place in the mutant (Table I; Supplemental Tables S2 and S4). Alternatively, a putative COPT2-mediated signaling event could be involved in chlorophyll degradation, and consequently, this process would be retarded in the copt2-1 line. In agreement with this suggestion, other degradation processes, such as seed protein mobilization, are also inhibited in the mutant (Supplemental Tables S1 and S3). In addition, both the influence of Cu on ethylene perception (Hirayama et al., 1999) and the recently postulated role of a multicopper oxidase in Fe homeostasis (Bernal et al., 2012) also indicate different Cu functions, which could affect plant responses under Fe and Pi starvation (Romera and Alcantara, 1994; Hirsch et al., 2006).
Although more studies will be needed to further ascertain the relevance of COPT2 in these processes, this study opens up novel possibilities to help develop strategies to improve the growth, yield, and nutritional quality of crops under environmental metal deficiencies. Along these lines, recent studies in rice (Oryza sativa) implicate certain COPT family members, which are up-regulated by both Cu and Fe deficiency (Yuan et al., 2011), as potential targets for biotechnological improvement.
MATERIALS AND METHODS
Plant Growth Conditions and Treatments
Seeds of Arabidopsis (Arabidopsis thaliana), ecotype Columbia-0, were surface sterilized and stratified for 2 d at 4°C and then germinated on 1/2 MS plates either including 1% Suc (Murashige and Skoog, 1962) or supplemented with the indicated concentrations of metal ions. For severe Cu-deficient conditions, the 1/2 MS was supplemented with 100 μm bathocuproinedisulfonic acid disodium. In order to know the effects of both Cu and Fe deficiencies in plants, the components of the 1/2 MS were prepared separately according to the following conditions: macronutrients (10 mm NH4NO3, 9.4 mm KNO3, 0.37 mm MgSO4 7H20, 0.62 mm KH2PO4, and 1.13 mm CaCl2), micronutrients (50 μm H3BO3, 36.6 μm MnSO4 H20, 15 μm ZnSO4 7H20, 0.57 μm NaMoO4 2H2O, and 0.05 μm CoCl2 6H20), 50 μm Fe-EDTA, 0.25 mm potassium iodide, 1 μm CuSO4 5H20, 0.05% MES, 1% Suc, and 0.8% phytoagar, pH 5.7 to 5.8. Seven-day-old seedlings were grown in Fe- and Cu-sufficient medium (+Fe+Cu), Fe-sufficient and Cu-deficient medium (+Fe−Cu), Fe-deficient and Cu-sufficient medium (−Fe+Cu), and Fe- and Cu-deficient medium (−Fe−Cu). Cu-excess medium was supplemented with 10 μm CuSO4 5H20. Fe-deficient medium was supplemented with 300 μm ferrozine. With the Pi-deficient medium, KH2PO4 was not included in the macronutrient solution. To study sensitivity to others stresses, seedlings were grown in paraquat and hydrogen peroxide solution (0.1 and 500 μm, respectively). Seedlings were grown for 7 d with a 12-h photoperiod (65 μmol m−2 cool-white fluorescent light) at a 23°C/16°C temperature cycle. Hydroponic cultures were performed in the same photoperiod conditions from three- to four-true-leaf seedlings grown in commercial soil, which were transferred to black boxes containing standard Hoagland solution (0.1×), pH 5.8, as described by Hermans et al. (2005). After a 14-d adaptation, the −Fe−Cu treatment (corresponding to Hoagland medium without Cu and Fe sources) commenced. Media were changed weekly for 4 to 5 weeks. The chlorophyll content of the Arabidopsis seedlings and leaves from adult plants was determined by the trichlorometric method (Parsons and Strickland, 1962). Root length was measured using ImageJ 1.42q software (http://rsb.info.nih.gov./ij).
Functional Complementation Assays in Yeast
The yeast (Saccharomyces cerevisiae) ctr1Δctr3Δ mutant strain was transformed with p426GPD or with a vector containing COPT1, COPT2, or COPT2:GFP and was assayed for growth on Glc (1% yeast extract, 2% bactopeptone, and 2% Glc), glycerol (1% yeast extract, 2% bactopeptone, 2% ethanol, and 3% glycerol), and glycerol + Cu (1% yeast extract, 2% bactopeptone, 2% ethanol, 3% glycerol, and 100 µm CuSO4) media solidified with 1.5% agar (Puig et al., 2002).
Metal Accumulation Measurements
Fresh Arabidopsis material was dried at 65°C for 2 d and digested with 65% (v/v) HNO3 at 80°C to 90°C. Digested samples were then diluted with Millipore water (Purelab Ultra), and Cu and Fe contents were determined by ICP-MS at the Servei Central d'Instrumentació Científica (Universitat Jaume I).
SOD Activity Assay
Plant material was homogenized in an equal volume of ice-cold grinding buffer (50 mm potassium phosphate, pH 7.4, 0.1% bovine serum albumin [BSA], 0.1% ascorbate, 0.05% β-mercaptoethanol, and 0.2% Triton X-100) and was clarified by centrifugation at 14,000 rpm for 15 min. Total protein was measured by the protein-dye binding assay (Bradford, 1976) with the Bio-Rad Protein Assay reagent. Samples (20 µg) were separated on a nondenaturing 12.5% polyacrylamide gel at 100 V for 3 to 4 h. SOD activity was detected on these gels using the in situ staining technique of Beauchamp and Fridovich (1971). Briefly, the gel was kept in 1 mg mL−1 nitroblue tetrazolium for 10 min in the dark and then in developing buffer (33 mm potassium phosphate, pH 7.8, 28 µm riboflavin, and 28 mm N,N,N′,N′-tetramethylethylenediamine) for another 20-min period before exposure to light.
Plasmid Constructs
The entire COPT2 open reading frame was obtained by PCR using the following specific primers: COPT2-SalI-F (5′-CATGTCGACATCATGGATCATGATCACATGCAT-3′) and COPT2-NcoI-R (5′-TCTCCATGGTACAAACGCACCCTGAAGACGGCGGAA-3′). The COPT2 C terminus was fused in frame to the GFP with the CaMV35S promoter through its insertion into transient expression vector P35SΩsGFP(S65T) (Miras et al., 2002). COPT2-GFP, obtained from the previous construct by PCR using the specific primers COPT2-HindII-F (5′-CATAAGCTTATGGATCATGATCACATGCAT-3′) and GFP-R-SalI (5′-CATGTCGACTTACTTGTACAGCTCGTCCAT-3′) was cloned into the p426GDP plasmid for the yeast functionality assay. Plasmids p426GDP-COPT1 and p426GDP-COPT2 were described by Sancenón et al. (2003). The uidA coding sequence and 1,248 bp from the COPT2 transcription start site (PCOPT2; arrows in Supplemental Fig. S4) were amplified and cloned into the pFP101 plasmid (Bensmihen et al., 2005) without its CaMV35S promoter to obtain the PCOPT2:GUS construct, used to determine the COPT2 spatial expression pattern by a GUS assay. A PCR product containing 1,248 bp of the PCOPT2 and the entire COPT2 coding sequence was cloned into the pFP101 vector and introduced into the copt2-1 line by floral dipping (Clough and Bent 1998). Homozygous transgenic plants (PCOPT2:COPT2) were selected based on seed fluorescence. The entire COPT2 coding sequence was tagged with the human influenza virus HA epitope with the specific primers COPT2-XbaIF (5′-CATTCTAGAATGGATCATGATCACATGCAT-3′) and COPT2HA-SalI-R (5′-ATGTCGACTCAAGCATAATCTGGAACATCGTATGGATAACAAACGCAGCCTGAAGACGGCGGAA-3′) and cloned into the pFP101 vector. Transgenic lines were selected based on the fluorescence of the seeds.
Computer-Assisted Sequence Analyses
The hydrophobic profile of the COPT2 protein was obtained by the TMHHM application (www.cbs.dtu.dk/services). The theoretical analysis of the promoter sequences was performed with the PLACE Web Signal Scan (www.dna.affrc.go.jp/PLACE/signalscan.html) and Patmatch from The Arabidopsis Information Resource (www.arabidopsis.org).
Subcellular Localization of the GFP Fusion Proteins
The Arabidopsis protoplasts from fresh leaf tissue of 30-d-old plants grown in soil were transiently transformed with COPT2-GFP as described previously (Abdel-Ghany et al., 2005). After 16 h under continuous light at 23°C in the wash solution, protoplasts were incubated with FM4-64 dye (Invitrogen) at a concentration of 50 μm for 15 min at 4°C before analyzing. Confocal images were obtained using fluorescence confocal microscope TCS SP, vertical (DM-R; Leica), equipped with argon ion (458 and 488 nm), helium-neon I (543 nm), and helium-neon II (633 nm) excitation laser systems and a 40× to 60× objective lens. Fluorescence signals were detected at 500 to 530 nm for GFP, 650 to 750 nm for chlorophyll, and 560 to 650 nm for FM4-64 after exciting at 488, 633, and 543 nm, respectively.
Immunohistochemical Techniques and Microscopy
Samples of Arabidopsis leaves were fixed in Karnousky reagent. They were dehydrated in a successive ethanol series (20%, 40%, 60%, 80%, 95%, and 100%) and embedded in polyethylene glycol for fluorescence microscopy. Semithin sections (2 µm) were placed on slides and rehydrated with xylene and a successive ethanol series (100%, 95%, 70%, and 0%), washed with PBSII (PBSI [137 mm NaCl, 2.7 mm KCl, 9 mm Na2HPO4, and 1.5 mm KH2PO4, pH 7.2] + 0.1% BSA and 0.05% Na2N3), and dried. Slides were blocked in PBSII with 2% BSA and 1% powder milk for 20 min and dried. Then, they were labeled overnight at 4°C and in humidity with rat monoclonal antibody to the HA epitope (clone 3F10; Roche) diluted 1:50 in PBSII. Subsequently, they were washed with PBSI, blocked in PBSII with 2% BSA and 1% powder milk for 20 min, and dried. Then, an anti-rat IgG antibody conjugated with AlexaFluor546 (Molecular Probes) diluted 1:500 with 0.1 m Gly was used for 1 to 4 h at 4°C in humidity and darkness. Slides were then washed with PBSI and stained with 1 µg mL−1 4′,6-diamino-phenylindole in PBSI for 5 min. Finally, the slides were washed with PBSI and mounted in Citifluor. The fluorescence of immunolabeled COPT2-HA and of 4′,6-diamino-phenylindole-stained nuclei was visualized with a fluorescence microscope (Axioskop 2; Zeiss) using the appropriate filter combinations. Micrographs were taken by a SPOT camera (Diagnostic Instruments) and were processed through the Photoshop program (Adobe Systems).
Protein Analysis by Western Blot
Crude extracts from wild-type and copt2-1 Arabidopsis leaves grown on plates were obtained, and the amount of protein was determined by bicinchoninic acid with the BCA Protein Assay Kit (Thermo). Next, 40 μg of protein was analyzed by SDS-PAGE, transferred to a nitrocellulose membrane, and blotted with an antibody against plastocyanin (Agrisera). Coomassie blue staining was used as a loading control.
GUS Expression Analyses
Assays were performed as described (Jefferson et al., 1987). Briefly, the seedlings and organs from the adult PCOPT2:GUS plants were embedded with the substrate solution [100 mm NaPO4, pH 7.2, 0.5 mm K3Fe(CN)6, 0.5 mm K4Fe(CN)6, 0.1% (v/v) Triton X-100, 0.5 mm 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (AppliChem), and 10 mm EDTA, pH 7.2]. Reactions took place at 37°C and were stopped with ethanol (70%).
copt2-1 Mutant Line Analyses and Complementation
The Arabidopsis T-DNA insertion line AL770147 was obtained from the GABI-Kat Project. Plants were self-pollinated, and a homozygous mutant was obtained (denoted the copt2-1 line). The COPT2-LB (F) and COPT2 RB (R) primers were used to amplify the wild-type alleles, whereas the COPT2-LB (F) and GKAT-PCR (S) primers amplified the inserted alleles in the copt2-1 line (Supplemental Table S6). The location of the T-DNA insert was obtained by PCR amplification and sequencing. Several independent lines obtained from different Arabidopsis databases were checked, but none carried a T-DNA insertion in the coding region, and the copt2-1 line was the closest insertion to the COPT2 coding sequence found among the lines analyzed. A PCR fragment containing PCOPT2::COPT2 in frame with the GFP reporter gene was cloned into the pFP101 plasmid and introduced in the copt2-1 line by floral dipping to generate a complemented mutant line.
Microarrays and Bioinformatics
Four biological replicates (7-d-old seedlings of wild-type and copt2-1 plants grown in a 12-h-light/12-h-dark photoperiod) were obtained for each treatment (+Fe+Cu, −Fe+Cu, and −Fe−Cu). Fe-deficient medium was supplemented with 300 μm ferrozine. Total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen), and antisense RNA was amplified using the MessageAmp II aRNA Amplification kit (Ambion). Long oligonucleotide microarrays were provided by Dr. David Galbraith (University of Arizona; http://www.ag.arizona.edu/microarray/). The hybridization and analysis were performed as described elsewhere (Bueso et al., 2007). The expression values (log2) were obtained using the GenePix Pro 6.0 microarray analysis software (Molecular Devices) and normalized with GenePix Pro 6.0 and Acuity 4.0 software (Molecular Devices). Differential genes were identified with significance analysis of microarray (Tusher et al., 2001) with a false discovery rate of less than 6% and 2-fold change (log2 ≤ |1|). Biological processes were identified with the Gene Ontology annotation (Ashburner et al., 2000), performed with GeneCodis2.0 (http://genecodis.dacya.ucm.es/; Carmona-Saez et al., 2007; Nogales-Cadenas et al., 2009) programs (Table 1). The total microarray differentially regulated genes are shown in Supplemental Tables S1 and S2. The microarray raw data were deposited in the National Center for Biotechnology Information Gene Expression Omnibus (Edgar et al., 2002) and are accessible through accession number GSE42642.
Gene Expression by Semiquantitative and Real-Time qPCR
Total Arabidopsis RNA was extracted with Trizol Reagent (Ambion), and reverse transcription-PCR was performed with SuperScript II (Invitrogen), as described previously (Andrés-Colás et al., 2006). RNA was quantified by UV spectrophotometry; its integrity was visually assessed on ethidium bromide-stained agarose gels and was treated with DNase I Amp Grade (Invitrogen). Semiquantitative PCR was carried out with specific oligonucleotides for ACT1 and COPT2 (Supplemental Table S6). Real-time qPCR was carried out with SYBR-Green qPCR Super-Mix-UDG with ROX (Invitrogen), and the specific primers detailed in Supplemental Table S6 were used in the CFX96 Touch Real Time PCR Detection System (Bio-Rad) with one cycle of 95°C for 2 min and 40 cycles consisting in 95°C for 30 s and 60°C for 30 s. Values were normalized to the UBQ10 mRNA levels, and in control conditions the wild type was used as a reference.
Statistical Analysis
Statistical analysis of relative expression was performed by comparing the relative expression of the genes (reverse transcription PCR) based on the pairwise fixed reallocation randomization test (P < 0.05; Pfaffl et al., 2002); for the remaining parameters, it was carried out using two-way ANOVA with the means compared by the Duncan test (P ≤ 0.05) using the InfoStat software, version 2008 (http://www.infostat.com.ar).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number At3g46900.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. COPT2 protein sequence and topology.
Supplemental Figure S2.. COPT2 subcellular localization in Arabidopsis COPT2:HA plants.
Supplemental Figure S3. Comparative COPT1 and COPT2 characteristics.
Supplemental Figure S4. Theoretical cis-elements in the COPT2 promoter region.
Supplemental Figure S5. The COPT2 expression pattern in Arabidopsis PCOPT2:GUS transgenic plants under different Cu content.
Supplemental Figure S6. The COPT2 gene expression dramatically diminished in the Arabidopsis copt2-1 line.
Supplemental Figure S7. Fresh weight and the LHCB1.1 gene expression in the copt2-1 seedlings.
Supplemental Figure S8. Plastocyanin content of the copt2-1 adult plants grown under Cu and Fe deficiencies.
Supplemental Figure S9. Phenotype of the siliques from the copt2-1 mutant plants.
Supplemental Figure S10. SODs activity and phenotype of copt2-1 seedlings grown under several oxidative stresses.
Supplemental Figure S11. The gene expression pattern of the Fe-deficiency marker genes in copt2-1 mutant seedlings.
Supplemental Figure S12. The gene expression pattern in copt2-1 seedlings.
Supplemental Table S1. Differential genes repressed in copt2-1 seedlings.
Supplemental Table S2. Differential genes induced in copt2-1 seedlings.
Supplemental Table S3. Biological processes overrepresented in the differential genes repressed in copt2-1 seedlings.
Supplemental Table S4. Biological processes overrepresented in the differential genes induced in copt2-1 seedlings.
Supplemental Table S5. Oligonucleotides used in the qPCR reactions.
Supplemental Table S6. Oligonucleotides used in the semiquantitative PCR reactions.
Acknowledgments
We thank the Servei Central de Suport a la Investigació Experimental (Universitat de València) for sequencing and confocal microscopy services. We are grateful to Dr. Marinus Pilon for the GFP plasmid and to Dr. Toshiharu Shikanai for the spl7 mutant. Finally, we thank Drs. Amparo Sanz and Javier Paz-Ares for critically reading the manuscript.
Glossary
- Cu
copper
- Fe
iron
- Cu/ZnSOD
copper/zinc superoxide dismutase
- FeSOD
iron superoxide dismutase
- Pi
phosphate
- HA
hemagglutinin
- 1/2 MS
one-half-strength Murashige and Skoog medium
- ICP-MS
inductively coupled plasma mass spectroscopy
- qPCR
quantitative PCR
- BSA
bovine serum albumin
- SOD
superoxide dismutase
- T-DNA
transfer DNA
References
- Abdel-Ghany SE, Müller-Moulé P, Niyogi KK, Pilon M, Shikanai T. (2005) Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell 17: 1233–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel S. (2011) Phosphate sensing in root development. Curr Opin Plant Biol 14: 303–309 [DOI] [PubMed] [Google Scholar]
- Aller SG, Eng ET, De Feo CJ, Unger VM. (2004) Eukaryotic CTR copper uptake transporters require two faces of the third transmembrane domain for helix packing, oligomerization, and function. J Biol Chem 279: 53435–53441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés-Colás N, Perea-García A, Puig S, Peñarrubia L. (2010) Deregulated copper transport affects Arabidopsis development especially in the absence of environmental cycles. Plant Physiol 153: 170–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés-Colás N, Sancenón V, Rodríguez-Navarro S, Mayo S, Thiele DJ, Ecker JR, Puig S, Peñarrubia L. (2006) The Arabidopsis heavy metal P-type ATPase HMA5 interacts with metallochaperones and functions in copper detoxification of roots. Plant J 45: 225–236 [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. (2000) Gene Ontology: tool for the unification of biology. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchamp C, Fridovich I. (1971) Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal Biochem 44: 276–287 [DOI] [PubMed] [Google Scholar]
- Bensmihen S, Giraudat J, Parcy F. (2005) Characterization of three homologous basic leucine zipper transcription factors (bZIP) of the ABI5 family during Arabidopsis thaliana embryo maturation. J Exp Bot 56: 597–603 [DOI] [PubMed] [Google Scholar]
- Bernal M, Casero D, Singh V, Wilson GT, Grande A, Yang H, Dodani SC, Pellegrini M, Huijser P, Connolly EL, et al. (2012) Transcriptome sequencing identifies SPL7-regulated copper acquisition genes FRO4/FRO5 and the copper dependence of iron homeostasis in Arabidopsis. Plant Cell 24: 738–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbihl RP, Jach G, Saedler H, Huijser P. (2005) Functional dissection of the plant-specific SBP-domain: overlap of the DNA-binding and nuclear localization domains. J Mol Biol 352: 585–596 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Buckhout TJ, Yang TJ, Schmidt W. (2009) Early iron-deficiency-induced transcriptional changes in Arabidopsis roots as revealed by microarray analyses. BMC Genomics 10: 147–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueso E, Alejandro S, Carbonell P, Perez-Amador MA, Fayos J, Bellés JM, Rodriguez PL, Serrano R. (2007) The lithium tolerance of the Arabidopsis cat2 mutant reveals a cross-talk between oxidative stress and ethylene. Plant J 52: 1052–1065 [DOI] [PubMed] [Google Scholar]
- Burkhead JL, Reynolds KA, Abdel-Ghany SE, Cohu CM, Pilon M. (2009) Copper homeostasis. New Phytol 182: 799–816 [DOI] [PubMed] [Google Scholar]
- Carmona-Saez P, Chagoyen M, Tirado F, Carazo JM, Pascual-Montano A. (2007) GENECODIS: a Web-based tool for finding significant concurrent annotations in gene lists. Genome Biol 8: R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castruita M, Casero D, Karpowicz SJ, Kropat J, Vieler A, Hsieh SI, Yan W, Cokus S, Loo JA, Benning C, et al. (2011) Systems biology approach in Chlamydomonas reveals connections between copper nutrition and multiple metabolic steps. Plant Cell 23: 1273–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou TJ, Lin SI. (2011) Signaling network in sensing phosphate availability in plants. Annu Rev Plant Biol 62: 185–206 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colangelo EP, Guerinot ML. (2004) The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16: 3400–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crichton RR, Pierre JL. (2001) Old iron, young copper: from Mars to Venus. Biometals 14: 99–112 [DOI] [PubMed] [Google Scholar]
- del Pozo T, Cambiazo V, González M. (2010) Gene expression profiling analysis of copper homeostasis in Arabidopsis thaliana. Biochem Biophys Res Commun 393: 248–252 [DOI] [PubMed] [Google Scholar]
- Duan K, Yi K, Dang L, Huang H, Wu W, Wu P. (2008) Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J 54: 965–975 [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J. (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39: 1033–1037 [DOI] [PubMed] [Google Scholar]
- Garcia-Molina A, Andrés-Colás N, Perea-García A, Del Valle-Tascón S, Peñarrubia L, Puig S. (2011) The intracellular Arabidopsis COPT5 transport protein is required for photosynthetic electron transport under severe copper deficiency. Plant J 65: 848–860 [DOI] [PubMed] [Google Scholar]
- Guerinot ML. (2000) The ZIP family of metal transporters. Biochim Biophys Acta 1465: 190–198 [DOI] [PubMed] [Google Scholar]
- Hartmann U, Sagasser M, Mehrtens F, Stracke R, Weisshaar B. (2005) Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol Biol 57: 155–171 [DOI] [PubMed] [Google Scholar]
- Hermans C, Bourgis F, Faucher M, Strasser RJ, Delrot S, Verbruggen N. (2005) Magnesium deficiency in sugar beets alters sugar partitioning and phloem loading in young mature leaves. Planta 220: 541–549 [DOI] [PubMed] [Google Scholar]
- Hindt MN, Guerinot ML. (2012) Getting a sense for signals: regulation of the plant iron deficiency response. Biochim Biophys Acta 1823: 1521–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T, Kieber JJ, Hirayama N, Kogan M, Guzman P, Nourizadeh S, Alonso JM, Dailey WP, Dancis A, Ecker JR. (1999) RESPONSIVE-TO-ANTAGONIST1, a Menkes/Wilson disease-related copper transporter, is required for ethylene signaling in Arabidopsis. Cell 97: 383–393 [DOI] [PubMed] [Google Scholar]
- Hirsch J, Marin E, Floriani M, Chiarenza S, Richaud P, Nussaume L, Thibaud MC. (2006) Phosphate deficiency promotes modification of iron distribution in Arabidopsis plants. Biochimie 88: 1767–1771 [DOI] [PubMed] [Google Scholar]
- Ivanov R, Brumbarova T, Bauer P. (2012) Fitting into the harsh reality: regulation of iron-deficiency responses in dicotyledonous plants. Mol Plant 5: 27–42 [DOI] [PubMed] [Google Scholar]
- Jarvis P, Dörmann P, Peto CA, Lutes J, Benning C, Chory J. (2000) Galactolipid deficiency and abnormal chloroplast development in the Arabidopsis MGD synthase 1 mutant. Proc Natl Acad Sci USA 97: 8175–8179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J, Guerinot ML. (2009) Homing in on iron homeostasis in plants. Trends Plant Sci 14: 280–285 [DOI] [PubMed] [Google Scholar]
- Kampfenkel K, Kushnir S, Babiychuk E, Inzé D, Van Montagu M. (1995) Molecular characterization of a putative Arabidopsis thaliana copper transporter and its yeast homologue. J Biol Chem 270: 28479–28486 [DOI] [PubMed] [Google Scholar]
- Kim BE, Nevitt T, Thiele DJ. (2008) Mechanisms for copper acquisition, distribution and regulation. Nat Chem Biol 4: 176–185 [DOI] [PubMed] [Google Scholar]
- Klaumann S, Nickolaus SD, Fürst SH, Starck S, Schneider S, Ekkehard Neuhaus H, Trentmann O. (2011) The tonoplast copper transporter COPT5 acts as an exporter and is required for interorgan allocation of copper in Arabidopsis thaliana. New Phytol 192: 393–404 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Awai K, Nakamura M, Nagatani A, Masuda T, Ohta H. (2009) Type-B monogalactosyldiacylglycerol synthases are involved in phosphate starvation-induced lipid remodeling, and are crucial for low-phosphate adaptation. Plant J 57: 322–331 [DOI] [PubMed] [Google Scholar]
- Liao H, Rubio G, Yan X, Cao A, Brown KM, Lynch JP. (2001) Effect of phosphorus availability on basal root shallowness in common bean. Plant Soil 232: 69–79 [PubMed] [Google Scholar]
- Märschner H (2002) Mineral Nutrition in Higher Plants. Academic Press, London
- Miras S, Salvi D, Ferro M, Grunwald D, Garin J, Joyard J, Rolland N. (2002) Non-canonical transit peptide for import into the chloroplast. J Biol Chem 277: 47770–47778 [DOI] [PubMed] [Google Scholar]
- Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, et al. (2005) A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA 102: 11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15: 473–497 [Google Scholar]
- Nevitt T, Ohrvik H, Thiele DJ. (2012) Charting the travels of copper in eukaryotes from yeast to mammals. Biochim Biophys Acta 1823: 1580–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales-Cadenas R, Carmona-Saez P, Vazquez M, Vicente C, Yang X, Tirado F, Carazo JM, Pascual-Montano A. (2009) GeneCodis: interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res 37: W317–W322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer CM, Guerinot ML. (2009) Facing the challenges of Cu, Fe and Zn homeostasis in plants. Nat Chem Biol 5: 333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons TR, Strickland JD. (1962) Oceanic detritus. Science 136: 313–314 [DOI] [PubMed] [Google Scholar]
- Peñarrubia L, Andrés-Colás N, Moreno J, Puig S. (2010) Regulation of copper transport in Arabidopsis thaliana: a biochemical oscillator? J Biol Inorg Chem 15: 29–36 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon M, Cohu CM, Ravet K, Abdel-Ghany SE, Gaymard F. (2009) Essential transition metal homeostasis in plants. Curr Opin Plant Biol 12: 347–357 [DOI] [PubMed] [Google Scholar]
- Puig S, Andrés-Colás N, García-Molina A, Peñarrubia L. (2007) Copper and iron homeostasis in Arabidopsis: responses to metal deficiencies, interactions and biotechnological applications. Plant Cell Environ 30: 271–290 [DOI] [PubMed] [Google Scholar]
- Puig S, Lee J, Lau M, Thiele DJ. (2002) Biochemical and genetic analyses of yeast and human high affinity copper transporters suggest a conserved mechanism for copper uptake. J Biol Chem 277: 26021–26030 [DOI] [PubMed] [Google Scholar]
- Puig S, Peñarrubia L. (2009) Placing metal micronutrients in context: transport and distribution in plants. Curr Opin Plant Biol 12: 299–306 [DOI] [PubMed] [Google Scholar]
- Raghothama KG. (1999) Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol 50: 665–693 [DOI] [PubMed] [Google Scholar]
- Rodrigo-Moreno A, Andrés-Colás N, Poschenrieder C, Gunsé B, Peñarrubia L, Shabala S. (2013) Calcium- and potassium-permeable plasma membrane transporters are activated by copper in Arabidopsis root tips: linking copper transport with cytosolic hydroxyl radical production. Plant Cell Environ 36: 844–855 [DOI] [PubMed] [Google Scholar]
- Romera FJ, Alcantara E. (1994) Iron-deficiency stress responses in cucumber (Cucumis sativus L.) roots (a possible role for ethylene?). Plant Physiol 105: 1133–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancenón V, Puig S, Mateu-Andrés I, Dorcey E, Thiele DJ, Peñarrubia L. (2004) The Arabidopsis copper transporter COPT1 functions in root elongation and pollen development. J Biol Chem 279: 15348–15355 [DOI] [PubMed] [Google Scholar]
- Sancenón V, Puig S, Mira H, Thiele DJ, Peñarrubia L. (2003) Identification of a copper transporter family in Arabidopsis thaliana. Plant Mol Biol 51: 577–587 [DOI] [PubMed] [Google Scholar]
- Shimojima M, Ohta H. (2011) Critical regulation of galactolipid synthesis controls membrane differentiation and remodeling in distinct plant organs and following environmental changes. Prog Lipid Res 50: 258–266 [DOI] [PubMed] [Google Scholar]
- Stein RJ, Waters BM. (2012) Use of natural variation reveals core genes in the transcriptome of iron-deficient Arabidopsis thaliana roots. J Exp Bot 63: 1039–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svistoonoff S, Creff A, Reymond M, Sigoillot-Claude C, Ricaud L, Blanchet A, Nussaume L, Desnos T. (2007) Root tip contact with low-phosphate media reprograms plant root architecture. Nat Genet 39: 792–796 [DOI] [PubMed] [Google Scholar]
- Thibaud MC, Arrighi JF, Bayle V, Chiarenza S, Creff A, Bustos R, Paz-Ares J, Poirier Y, Nussaume L. (2010) Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. Plant J 64: 775–789 [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JT, Lahner B, Yakubova E, Salt DE, Raghothama KG. (2008) The effect of iron on the primary root elongation of Arabidopsis during phosphate deficiency. Plant Physiol 147: 1181–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters BM, McInturf SA, Stein RJ. (2012) Rosette iron deficiency transcript and microRNA profiling reveals links between copper and iron homeostasis in Arabidopsis thaliana. J Exp Bot 63: 5903–5918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintz H, Fox T, Wu YY, Feng V, Chen W, Chang HS, Zhu T, Vulpe C. (2003) Expression profiles of Arabidopsis thaliana in mineral deficiencies reveal novel transporters involved in metal homeostasis. J Biol Chem 278: 47644–47653 [DOI] [PubMed] [Google Scholar]
- Yamasaki H, Abdel-Ghany SE, Cohu CM, Kobayashi Y, Shikanai T, Pilon M. (2007) Regulation of copper homeostasis by micro-RNA in Arabidopsis. J Biol Chem 282: 16369–16378 [DOI] [PubMed] [Google Scholar]
- Yamasaki H, Hayashi M, Fukazawa M, Kobayashi Y, Shikanai T. (2009) SQUAMOSA Promoter Binding Protein-Like7 is a central regulator for copper homeostasis in Arabidopsis. Plant Cell 21: 347–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TJ, Lin WD, Schmidt W. (2010) Transcriptional profiling of the Arabidopsis iron deficiency response reveals conserved transition metal homeostasis networks. Plant Physiol 152: 2130–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Li X, Xiao J, Wang S. (2011) Molecular and functional analyses of COPT/Ctr-type copper transporter-like gene family in rice. BMC Plant Biol 11: 69–81 [DOI] [PMC free article] [PubMed] [Google Scholar]