Abstract
Most genomic resources available for insects represent the Holometabola, which are insects that undergo complete metamorphosis like beetles and flies. In contrast, the Hemimetabola (direct developing insects), representing the basal branches of the insect tree, have very few genomic resources. We have therefore created a large and publicly available transcriptome for the hemimetabolous insect Gryllus bimaculatus (cricket), a well-developed laboratory model organism whose potential for functional genetic experiments is currently limited by the absence of genomic resources. cDNA was prepared using mRNA obtained from adult ovaries containing all stages of oogenesis, and from embryo samples on each day of embryogenesis. Using 454 Titanium pyrosequencing, we sequenced over four million raw reads, and assembled them into 21,512 isotigs (predicted transcripts) and 120,805 singletons with an average coverage per base pair of 51.3. We annotated the transcriptome manually for over 400 conserved genes involved in embryonic patterning, gametogenesis, and signaling pathways. BLAST comparison of the transcriptome against the NCBI non-redundant protein database (nr) identified significant similarity to nr sequences for 55.5% of transcriptome sequences, and suggested that the transcriptome may contain 19,874 unique transcripts. For predicted transcripts without significant similarity to known sequences, we assessed their similarity to other orthopteran sequences, and determined that these transcripts contain recognizable protein domains, largely of unknown function. We created a searchable, web-based database to allow public access to all raw, assembled and annotated data. This database is to our knowledge the largest de novo assembled and annotated transcriptome resource available for any hemimetabolous insect. We therefore anticipate that these data will contribute significantly to more effective and higher-throughput deployment of molecular analysis tools in Gryllus.
Introduction
The vast majority of existing insect genomic resources are for the Holometabola or “higher insects,” which undergo true metamorphosis. These include disease vectors such as the mosquito Anopheles gambiae [1], agricultural pests such as the flour beetle Tribolium castaneum [2], and the powerful genetic model organism Drosophila melanogaster [3], [4]. However, there are very few complete genome sequences available for the Hemimetabola or “lower insects”, which do not undergo true metamorphosis and branch basally to the Holometabola. Only three of the over 146,000 estimated species of hemimetabolous insects [5] have available genome sequences: the aphid Acyrthosiphon pisum [6], the kissing bug Rhodnius prolixus [7], [8], and the human body louse Pediculus humanus [9]. Moreover, sequence divergence is so great among insects [10] that a specific genome cannot be used as a reference sequence for other insects even within the same order; see for example [11].
Among the Hemimetabola, the basally branching orthopteroid orders of insects are of particular interest to many fields of biology. Orthopterans have served as classical model organisms for neurobiology for several decades [12]. Multiple cricket species have been used for important studies of ecologically relevant polyphenisms (reviewed in [13]), the evolution of endocrine functions and photobiology [14], [15], [16], [17], speciation [18], [19], [20], [21], [22] and the evolution of behavior [23], [24], [25]. Crickets and locusts have also been important for addressing outstanding questions in evolutionary developmental biology, such as the evolution of molecular mechanisms for regeneration, segmentation, and axial patterning [26], [27], [28], [29], [30], [31], [32], [33]. However, de novo genome assembly for organisms with extremely large genome sizes is costly and challenging [34], [35], [36]. Grasshopper genomes can be over twice as large as the human genome [37], and even the genome of the laboratory model cricket Gryllus bimaculatus is estimated at 1.7 Gbp (C. G. Extavour and R. Gregory, unpublished). If orthopteran genome projects are eventually undertaken, their annotation success will be significantly enhanced by the availability of large transcriptomes, but these are also few in number.
To date, only three Sanger-based EST projects and one large de novo assembled transcriptome generated with next-generation sequencing have been reported for orthopterans (Table 1). These projects have focused on specific post-embryonic developmental stages of pest locusts (L. migratoria, S. gregaria) and on the CNS of a cricket (L. kohalensis). Although most functional genetic studies on orthopterans focus on embryonic development (see for example [28], [29], [38], [39]) and neurophysiological studies are increasingly examining the embryonic origins of neural structures and functions (see for example [e.g. 16], [40], [41], [42], [43]), a transcriptome enriched for embryonic developmental transcripts is lacking. Here we present such a transcriptome for the model laboratory cricket, G. bimaculatus.
Table 1. Large-scale Orthopteran transcriptome resources to date.
| Locusta migratoria 1 | Laupala kohalensis 2 | Schistocerca gregaria 3 | Locusta migratoria 4 | Gryllus bimaculatus 5 | |
| Orthopteran Suborder | Caelifera | Ensifera | Caelifera | Caelifera | Ensifera |
| Superfamily | Acridoidea | Grylloidea | Acridoidea | Acridoidea | Grylloidea |
| Family | Acrididae | Gryllidae | Acrididae | Acrididae | Gryllidae |
| Sequencing Platform | Sanger | Sanger | Sanger | Illumina | 454 Titanium |
| Tissue Source(s) | L56 | L5–L8 CNS | L3–L5 & adult CNS | Mainly L4 | Ovaries & embryos |
| Normalized Library | No | Yes | Yes | No | No |
| # Raw Reads | 76,012 | 14,502 | nd | 447,718,464 | 4,248,346 |
| # Reads Used in Assembly | 45,449 | 14,377 | 34,672 | nd | 4,216,721 |
| # bp Used in Assembly | 21,760,812 | 10,121,408 | nd | nd | 1,449,059,795 |
| % Raw Reads Assembled | 59.79% | 99.14% | nd | nd | 99.26% |
| # Contigs or Clusters | 4,550 | 2,575 | 4,785 | 72,977 | 43,321 |
| N50 7 or Mean Contig Length (bp) | 471 | 935 | 750 | 2,275 | 2,133 |
| # Singletons or # Single ESTs 8 | 7,611 | 6,032 | 7,924 | nd | 120,805 |
| % Singletons (of assembled reads) | 16.75% | 41.96% | 22.85% | nd | 2.86% |
| # Total Assembly Products | 12,161 | 8,607 | 12,709 | 72,977 | 142,317 |
| # Unigenes or # Unique BLAST 9 Hits to nr | 12,616 | 8,607 | 12,709 | 11,490 | 19,874 |
Data from [75].
Data from [76].
Data from [72].
Data from this report.
“N50” refers to isotig N50 from the G. bimaculatus de novo transcriptome assembly; mean contig length is shown for all other orthopteran transcriptome resources in this table.
# singletons are shown for the G. bimaculatus de novo transcriptome assembly; # single ESTs (not incorporated into contigs) are shown for all other orthopteran transcriptome resources in this table.
# unique BLAST hits against nr are shown for the G. bimaculatus de novo transcriptome assembly; # unigenes are shown for all other orthopteran transcriptome resources in this table.
G. bimaculatus is a highly tractable orthopteran model for functional genetic studies in the laboratory (Fig. 1). Gene knockdown can be achieved by RNA interference during embryonic, post-embryonic and regenerative development [32], [43], [44]. G. bimaculatus is also the only orthopteran for which stable germ line transgenesis has been established [39]. Moreover, protocols for targeted genome editing using zinc finger nucleases or TALE nucleases have recently been developed [45]. However, all G. bimaculatus genes studied to date have been obtained by degenerate PCR (for example [28], [46]) or from limited Sanger-based EST libraries or RNA-Seq data that are not available in an annotated database (for example [26], [47]).
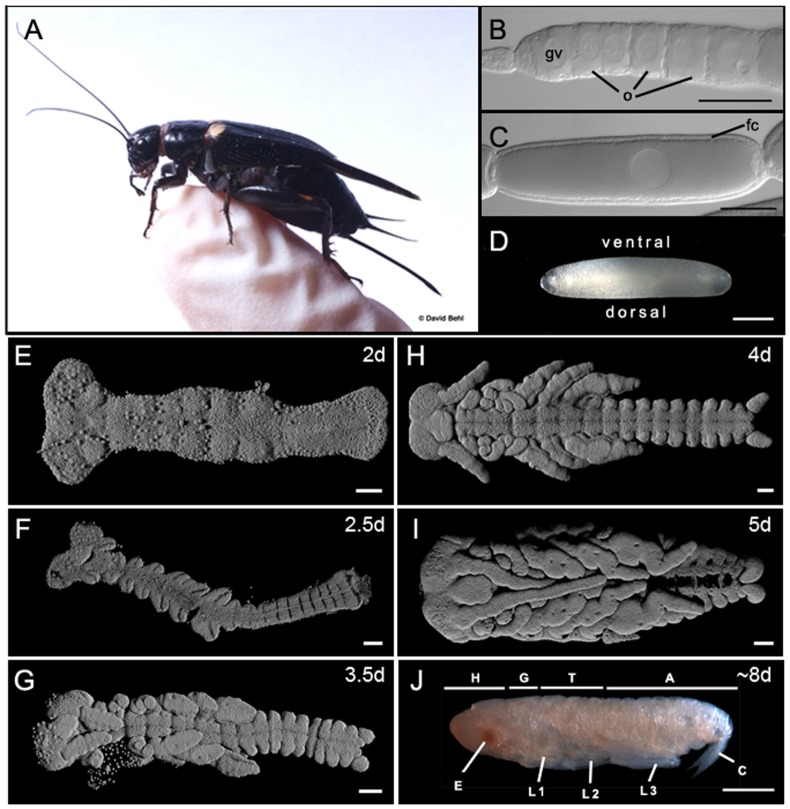
Figure 1. Oogenesis and embryogenesis in the cricket model organism Gryllus bimaculatus.
(A) Adult female cricket perched on a gloved human finger for perspective. (B) Anterior tip of a single ovariole from an adult female ovary, showing oocytes (o) at early previtellogenic stages of oogenesis. A single large germinal vesicle (gv) is distinguishable in each oocyte. Unlike meroistic (containing nurse cells) Drosophila ovaries, G. bimaculatus ovaries are panoistic and lack nurse cells [100]. (C) A single late stage oocyte with a single layer of columnar follicle cells (fc). (D–J) Chronological stages of G. bimaculatus embryogenesis showing the range of embryonic stages represented in the transcriptome presented here. (D) A fertilized egg just after laying. The egg nucleus is distinguishable as a dense patch in the dorsal yolk (arrowhead). Ages are shown as days (d) after egg-laying at 29°C. (E–I) are 3D reconstructions of confocal optical sections of Hoechst 3342-stained embryos dissected free from the egg; (J) is a micrograph of a live embryo dissected free from the chorion. Abbreviations: A = abdomen; C = cerci; E = eye; H = head; G = gnathal segments; L1 = first thoracic leg; L2 = second thoracic leg; L3 = third thoracic leg; T = thorax. Scale bar is 100 µm in (B, C, E–I) and 500 µm in (D, J). Anterior is to the left in all panels. Photo in (A) courtesy of David Behl; photos in (D) and (J) from [101].
In this report we present a de novo assembled and annotated transcriptome for G. bimaculatus oogenesis and embryonic development. We show that this transcriptome contains more putative unique gene transcripts than previous orthopteran transcriptomes, and adds sequence data to known GenBank accessions for G. bimaculatus. We manually annotate over 400 developmental genes, and develop an automated annotation method for the entire transcriptome based on similarity to Drosophila sequences. For predicted transcripts that lack significant similarity to GenBank accessions, we examine specifically those that are more similar to known orthopteran sequences, and find that the most represented predicted protein domains of such “orthopteroid” transcripts are domains of unknown function (DUFs). In contrast, the most represented predicted protein domains of transcripts of the transcriptome overall are zinc finger domains. Finally, we created a publicly accessible repository and database for the transcriptome, which is searchable by BLAST, pre-computed BLAST hits, or putative orthology assignments (gene names) derived from both manual and automated annotation.
Materials and Methods
Animal culture and collection of tissues for cDNA synthesis
G. bimaculatus cultures were maintained as previously described [28], at 28–29°C on a diet of oatmeal, wheat germ, soya protein, corn meal, sugar, yeast, salt, corn oil and Purina Cat Chow. This non-isogenic culture derives from a population of G. bimaculatus obtained from Livefoods Direct (Sheffield, UK), and was maintained as an inbred, self-sustaining culture for four years (or approximately 26 generations) prior to tissue collection. We do not have estimates of genetic polymorphism for this population, so that accurate interpretation of putative SNP data is not possible in the present analysis. Separate egg collections (total mass 781 mg) of 50–100 embryos on each of the first eight days of embryogenesis (approximately 66.7% of development at 28°C) (Figure 1D–J) were washed in distilled water, shock frozen in liquid nitrogen and stored at −80°C. Embryos were collected from cages containing 25–50 females per cage. Ovaries from one adult female (Fig. 1B, C) were dissected from the body cavity, rinsed in 1× PBS, and homogenized in TRIzol (Invitrogen, NY, USA).
cDNA Synthesis
Total RNA was isolated separately from embryos at each day of embryonic development and from ovaries, using TRIzol (Invitrogen, NY, USA) and following manufacturer's instructions. RNA isolation was performed separately from embryonic and ovarian tissues, so that tissue lysis, which can affect the efficiency of subsequent RNA isolation, would be as homogeneous as possible within a sample. A pilot study was first conducted to determine library quality by sequencing ovarian and embryonic cDNA separately. For this pilot sequencing run, cDNA was synthesized using the SMART cDNA synthesis kit (Clontech, CA, USA) and normalized using the Evrogen Trimmer Direct kit (Evrogen, Moscow, Russia) following previously described methods [11]. Results from both libraries were comparable in read length and sequence quality, and all further experiments were carried out with pooled RNA libraries as described below. Raw reads from the pilot studies were incorporated into the final assembly as previously described [11].
To create a pooled cDNA library for large-scale sequencing, 1.5 µg of each of the mixed-stage embryonic RNA pool and ovarian RNA was used as a template for first strand cDNA synthesis. cDNA was synthesized as previously described [11]. Primary amplification proceeded with 10 PCR cycles monitored in real-time via qPCR [22], and secondary amplification began to plateau after 9 cycles. 16 parallel reactions of 0.73 µg each were co-purified into elution buffer using QIAquick PCR purification columns (Qiagen Inc., CA, USA). These 16 parallel reactions were identical, and were performed in individual tubes for the sole reason that a single PCR reaction sufficient to generate the 2 µg of cDNA required for sequencing would have had to be performed in a volume too large to undergo efficient cycling in our PCR machine (Bio-Rad Tetrad 2). We therefore calculated the predicted yield from the largest single PCR reaction that we could perform in our machine, and scaled up the number of reactions in parallel to achieve the required 2 µg total yield.
454 Titanium Pyrosequencing
The samples were nebulized, adaptor-ligated, and pyrosequenced using the 454 GS-FLX platform on pilot embryonic and ovarian cDNA separately, or the 454 GS-FLX Titanium platform for pooled ovarian/embryonic cDNA samples by the Institute for Genome Science and Policy DNA Sequencing Facility (Duke University). All of the raw reads generated in this study have been submitted to the NCBI Short Read Archive (Study Accession Numbers SRX023831, SRX023830, and SRX023832).
Sequence Assembly
Sequences were trimmed and assembled with Newbler v2.5, which was shown to outperform other assemblers for de novo assembly of 454 pyrosequencing reads [48]. Assembly parameters are described in [49], with the exception of the file used for the –vt flag (“Gb Adaptors”), which is available at http://www.extavourlab.com/protocols/index.html. Assembly results are available at http://www.extavourlab.com/resources/index.html and at http://asgard.rc.fas.harvard.edu/download.html).
Sequence Annotation
A nucleotide BLAST database was created using the isotigs and singletons produced by the Newbler assembly. To increase efficiency of BLAST comparison to this database, we first removed redundant isotigs and singletons created due to a combination of putative SNPs, sequencing errors, and low quality reads. Note that these data could in principle yield SNP data, but as we did not use an isogenic G. bimaculatus culture, nor do we have estimates of polymorphism for the culture, an accurate SNP analysis is not performed in the present study. Each assembly product was compared with the BLAST database using the BLASTN algorithm. Individual isotigs and singletons with BLAST hits (>95% identity based on bit score and sequence length) to longer sequences in the assembly, resulting in a high scoring segment pair (HSP) that spans the full length of the sequence, were removed. To identify the number of unique BLAST hits we followed the method described in [49].
To identify members of signaling pathways as described by the KEGG database [50], we manually annotated the G. bimaculatus transcriptome as described in [49]. Briefly, BLAST was used to compare the sequences of D. melanogaster pathway members with the G. bimaculatus transcriptome assembly and the top hit was selected as a putative ortholog with an E-value cutoff of e-10.
To determine whether the de novo assembly contained members of previously known G. bimaculatus GenBank accessions, we used tBLASTn (for 80 protein coding genes) or BLASTn (for 3 ribosomal RNA genes) to query the G. bimaculatus transcriptome assembly.
For automatic annotation of all transcriptome sequences, we designed a custom script called “Gene Predictor” (genePrediction.pl, available at http://www.extavourlab.com/protocols/index.html). This script assigns putative gene orthology based on comparisons with the D. melanogaster proteome, downloaded as described in Table S1. A protein BLAST database was created using the D. melanogaster proteome. A nucleotide BLAST database was created using the non-redundant assembly products (isotigs and singletons) of the G. bimaculatus de novo transcriptome assembly. The top 50 BLAST hits for each sequence of the D. melanogaster proteome compared with the G. bimaculatus transcriptome were obtained using the TBLASTN algorithm and stored in a MySQL database. Reciprocally, the top BLAST hit for each sequence of the G. bimaculatus transcriptome against the D. melanogaster proteome was obtained using the BLASTX algorithm and stored within a separate MySQL database. A custom script then iterates through each of the entries of the D. melanogaster proteome vs. the G. bimaculatus transcriptome MySQL database indices based on query identity and e-value. The same script also checks the G. bimaculatus transcriptome sequence identity against the D. melanogaster proteome MySQL database to confirm if the reciprocal top BLAST hit is the same as the D. melanogaster query. After confirmation of the reciprocal BLAST identity, the script verifies whether any G. bimaculatus transcriptome sequences have already been assigned to the same D. melanogaster protein. If the existing sequence does not overlap with the confirmed sequence for more than 14 amino acids based on their HSP against the D. melanogaster protein, both sequences are recorded as orthologs. Otherwise, the confirmed sequence is further processed to determine whether it is a putative isoform or paralog of the existing sequence. If the confirmed sequence is a singleton or in the same isogroup as the existing sequence based on Newbler prediction, it is designated as an alternate isoform; otherwise, the sequence is annotated as a putative paralog.
A list of all curated D. melanogaster transcription factors was downloaded on March 26th 2011 from http://flytf.org. Each D. melanogaster transcription factor was examined to determine whether it was predicted to have an ortholog in the G. bimaculatus transcriptome using the Gene Predictor script described above. Custom scripts to generate tables based on the ASGARD schema (“ASGARD_NEW_DB.pl”) [51], upload assembled transcriptome sequences into ASGARD tables (“ASGARD_UPLOAD.pl”), upload BLAST results of the D. melanogaster proteome against the assembled transcriptome (“up_DMP.pl”), upload the BLAST results of the assembled transcriptome against the D. melanogaster proteome (“up_vDMP.pl”), and determine the best reciprocal BLAST result for each assembly products (“gene_prediction.pl”) are available at http://www.extavourlab.com/protocols/bio_tools/ASGARD_uploadGene_Predictor.zip).
Determination of sequencing depth and transcript completion
Ortholog hit ratio calculations and subassembly experiments were performed as described in [49]. Briefly, ortholog hit ratios were calculated using a custom script (“OrthologHitRatio.pl” available at http://www.extavourlab.com/protocols/bio_tools/Perl_Transcriptome_Analysis_Scripts.zip) that compares the length of each assembly product with the full length of its putative orthologous mRNA in D. melanogaster, based on the reciprocal best BLAST hit criteria described above. Subassemblies were performed by assembling progressively larger random subsets of all trimmed reads, using the same assembly parameters as those used for the complete assembly.
Protein Domain Analysis
23 proteomes based on completely sequenced genomes and two EST libraries were downloaded as described in Table S1. A protein BLAST database was created from each proteome. All G. bimaculatus assembly products were compared with each database using the BLASTX algorithm with an E-value cutoff of 1e-5. The resulting reports were parsed using the Uniqueblast.pl script as previously described [49] (available at http://www.extavourlab.com/protocols/index.html).
A local installation of EST Scan [52] (ESTSCAN 3.03) was downloaded on April 11th 2011 as a Linux rpm package from http://estscan.sourceforge.net/. All assembly products were screened using ESTSCAN with default parameters, except for the “-l” flag that was used with a value of 20 to restrict the minimum result size to 20 amino acids. The “-t” flag was also used to allow ESTSCAN to produce the predicted protein sequence of each assembly product.
A local installation of InterPro Scan [53], [54] (IPRSCAN 4.7) was downloaded on April 15th 2011 from ftp://ftp.hgc.jp/pub/mirror/ebi/software/iprscan/index.html. The “-cli” flag was used to turn on pipeline mode and suppress html outputs. All assembly products were screened using IPRSCAN against existing protein feature databases [55], [56], [57], [58], [59], [60], [61], [62], [63], [64], [65], [66], and the results were stored in xml format for further analysis.
Welch's t-test (appropriate in this case for use with samples with unequal variance [67]) was used for statistical comparisons of lengths of sequences and predicted protein coding regions in various annotation categories.
Results and Discussion
Collection and preparation of material
We aimed to create a transcriptome containing genes deployed during oogenesis, when maternally deposited factors required for embryogenesis may be synthesized, and during all stages of embryogenesis. We therefore collected ovaries (Figure 1B, C) and embryos from early to late stages of embryogenesis (Figure 1D–J) for mRNA extraction. We pooled these mRNA samples and prepared non-normalized cDNA libraries for 454 Titanium pyrosequencing. We chose to omit normalization in preparing these libraries as our previous studies [11] suggest that at this scale of sequencing, normalization does not significantly aid in gene discovery.
Sequencing and basic transcriptome assembly
We used Newbler v2.5 (Roche) for the de novo assembly of 4,248,348 raw reads (1,483,726,666 bp) obtained by 454 pyrosequencing (Table 1). Using default Newbler assembly parameters, raw reads were screened and trimmed of both 5′ and 3′ adaptors (see Methods), and low quality reads were removed. (Newbler's quality scores are defined as “Phred-like” or “Phred equivalent” [68]. The Phred quality score is a widely used base quality parameter defined by determining qualities of the data used to generate each base call [69], [70]. We used a Newbler quality score cutoff of >20; a Phred score of 20 would indicate a base call accuracy of ≥99%.). 99.26% of all reads passed this quality control process (4,216,721 reads = 1,449,059,795 bp) (Figure S1A, Table 1), and were subsequently used in the sequence alignment process. 88.78% of these reads (3,743,561) were fully assembled, meaning that the entire read sequence was used in a contig. 6.69% (282,259) were partially assembled, meaning that the entire read was not used in a contig (Figure S1B, C). Of the 190,901 good quality reads (4.53%) that were not aligned, 13,416 (0.32%) were too short (<40 bp) to be included in the assembly, 1,989 (0.05%) were predicted to be from a repeat region (meaning that >70% of the read's seeds match at least 70 other reads, or determined to partially overlap a contig; note that portions of reads in this category that overlap unique contigs are still included in the assembly results), 54,691 (1.30%) were considered outliers (e.g. chimeric reads or results of sequencing errors), and 120,805 (2.86%) were preserved as singletons.
Newbler assembly products fall into one of four categories: (1) contigs are groups of assembled reads with significant overlapping regions (we used the Newbler default minimum overlap of 40 bp), which may represent exons; (2) isotigs are continuous paths through a given set of contigs, and represent putative transcripts, including possible splice variants of a given transcription unit; (3) isogroups are groups of isotigs that were assembled from the same contig set, and are the closest to gene predictions as it is possible for a de novo assembly to achieve; and (4) singletons, which are single good quality reads that lack significant overlap with any other read, and therefore are not incorporated into any contig. We use these terms henceforth to refer to the G. bimaculatus assembly products. It is important to note that determination of whether contigs represent true exons, or isotigs true transcripts, would require further validation by sequencing full-length cDNAs and comparison with a fully sequenced genome. For this reason we refer to the G. bimaculatus transcriptome de novo assembly products as “contigs” and “isotigs” or “predicted transcripts” or “putative transcripts” throughout, rather than as “exons” or “transcripts” respectively.
Upon assembly we obtained 43,321 unique contigs using the aligned reads (Table 1). Newbler then further assembled these contigs into 21,512 isotigs that belonged to 16,456 isogroups (Table 2). 13,157 (79.95%) of the isogroups (putative genes) consist of only a single isotig, and on average there are 1.2 isotigs per isogroup (Table 2). 12,701 (62.78%) isotigs consist of a single contig, and on average there are 1.7 contigs per isotig. The isotig N50 is 2,133 bp (Table 1), meaning that the majority of predicted transcripts are over 2 kb in length. FASTA files of all assembly products are available for download from our interactive database (described below).
Table 2. Assembly statistics and BLAST results against nr for the G. bimaculatus de novo transcriptome assembly.
| Parameter1 | Value |
| # bp Raw reads | 1,483,726,666 |
| Maximum raw read length | 803 |
| Minimum raw read length | 13 |
| Median raw read length | 364 |
| Maximum assembled read length | 771 |
| Minimum assembled read length | 20 |
| Median assembled read length | 358 |
| # Isogroups2 (“genes”) | 16,456 |
| Mean # isotigs per isogroup | 1.2 |
| # Isotigs | 21,512 |
| Maximum isotig length | 10,865 |
| Minimum isotig length | 57 |
| Median isotig length | 1,054.5 |
| # Isotigs with BLAST hit against nr 3, E-value cutoff e-10 (% of all isotigs) | 11,135 (51.8%) |
| # Isotigs with BLAST hit against nr, E-value cutoff e-5 (% of all isotigs) | 11,943 (55.5%) |
| Mean # contigs per isotig | 1.7 |
| # Singletons | 120,805 |
| Maximum singleton length | 620 |
| Minimum singleton length | 50 |
| Median singleton length | 250.5 |
| # Singletons with BLAST hit against nr, E-value cutoff e-10 (% of all singletons) | 7,914 (6.6%) |
| # Singletons with BLAST hit against nr, E-value cutoff e-5 (% of all singletons) | 10,815 (9.0%) |
| # Non-redundant assembly products (NRAP) | 142,317 |
| # NRAP with BLAST hit against nr, E-value cutoff e-10 (% of all NRAP) | 19,049 (13.4%) |
| # NRAP with BLAST hit against nr, E-value cutoff e-5 (% of all NRAP) | 22,758 (16.0%) |
| Total # BLAST hits4 (nr) | 22,758 |
| Average coverage/bp | 51.3 |
Values for number of raw reads, number and % of raw reads assembled (passed quality filters described in main text), number of contigs, isotig N50, % of singletons, total number of assembly products, and number of unique BLAST hits against nr, are shown in Table 1.
Because isogroups are collections of isotigs that are hypothesized to originate from the same gene, they do not comprise a single sequence and so cannot be mapped to nr using BLAST.
nr = NCBI non-redundant database.
For BLAST against nr the E-value cutoff was 1e-5. For breakdown of BLAST hits among different classes of assembly sequences, see Table 3.
Assessment of transcript coverage and depth
The average coverage across the assembly is 51.3 reads per base pair; in other words, each base pair of the assembly was sequenced on average over 50 times. This coverage is high compared to other de novo transcriptome assemblies [11], [49], [71], which we attribute largely to the high number of reads used to create the G. bimaculatus transcriptome. We note, however, that the G. bimaculatus transcriptome coverage we obtained is more than twice as high as that of the recently de novo assembled transcriptome for the crustacean Parhyale hawaiensis (25.4 reads/bp), even though the G. bimaculatus transcriptome contained only 1.3 fold more base pairs in raw reads than that of P. hawaiensis, which was also generated from embryonic and ovarian cDNA, and was assembled and annotated identically to the G. bimaculatus transcriptome described in this report [49].
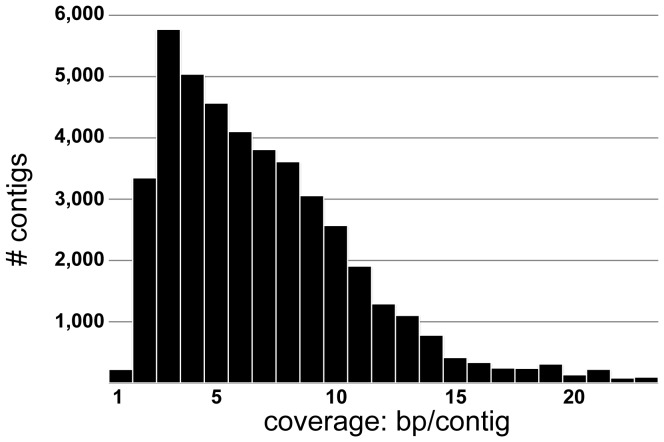
An additional measure of coverage is the average contig read depth (total number of base pairs from all reads aligned to generate a given contig, divided by contig length). This value is 391 bp/contig, with a median value of 16.7 bp/contig. We note that the predicted transcript coverage (number of base pairs of raw reads comprising each contig) is highly variable, suggesting that some genes are represented by many more raw reads than others (Figure 2). 19,093 (43.97%) contigs had a coverage ≤10 bp/contig, and 538 contigs (1.24%) had a coverage ≥10,000 bp/contig.
Figure 2. Distribution of average coverage (bp/contig) within contigs produced by de novo assembly of the G. bimaculatus transcriptome.
The coverage within contigs is calculated by dividing the total number of base pairs contained in the reads used to construct a contig by the length of that contig.
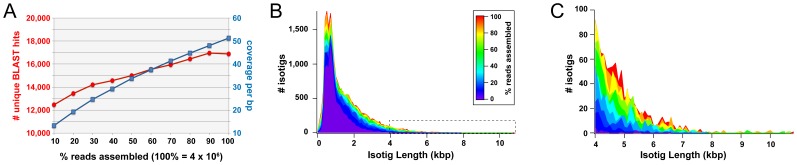
We wished to determine whether similar coverage levels and predicted transcript lengths could have been obtained with fewer reads, and how well our transcriptome had identified all putative transcripts present in our samples. To do this, we created subassemblies using randomly chosen subsets of reads, starting with 10% of reads and adding increments of 10% up to the full complement of trimmed reads. For each subset of reads, we performed an independent assembly with Newbler v2.5. For each of these nine subassemblies, we then assessed both read length distribution and the number of unique BLAST hits against the NCBI non-redundant protein database (nr) with an E-value cutoff of 1e-10. The mean coverage per bp was strongly positively correlated (R2 = 0.96, linear regression) with the number of reads used for the assembly (Figure 3A, blue line). We also found that as the number of reads used in the subassembly increased, the proportion of reads left as singletons decreased from 11.25% for the 10% subassembly, to 2.86% in the full assembly. This is likely because contigs and isotigs increased in length as reads were added (Figure 3B), as we observed an increase in isotig N50 from 1,290 bp with 10% of reads to 2,133 bp with all reads. The distribution of isotig lengths in each subassembly (Figure 3B) indicates the maximum length of assembled isotigs given a certain number of reads. A small proportion of isotigs exceeding 4 kb can be obtained with only 10% of all reads, but by assembling all reads it was possible to obtain predicted transcripts exceeding 10 kb (Figure 3C).
Figure 3. Assessment of gene discovery and read length capacity of the G. bimaculatus de novo assembled transcriptome.
(A) Randomly selected subsets of the trimmed reads were assembled using Newbler v2.5 in 10% increments, up to and including 100% of trimmed reads. For each subassembly, the number of unique BLAST hits against the NCBI non-redundant database (nr) with an E-value cutoff of 1e-10 (red; left axis) and the average coverage per base pair (blue; right axis) was calculated (see text for details). The number of unique BLAST hits did not increase after at least 90% of reads (3,795,085 reads) were assembled, while the coverage per base pair continued to increase as reads were added to the assembly. (B) Isotig length distribution for each subassembly created as described in (A). (C) Isotig length distribution of each subassembly for isotigs ≥4 kb. High numbers (≥50) of isotigs over 4 kb in length are achieved only when ≥40% of reads (1,686,646 reads) are assembled.
The number of unique BLAST hits against nr obtained from all isotigs also increased with the number of reads (Figure 3A, red line), but at a slower rate than that of mean coverage per bp (Figure 3A, blue line). Slightly fewer unique BLAST hits were obtained from isotigs generated with 100% of reads compared to 90%, which may mean that previously unconnected contigs were increasingly incorporated into isotigs as they increased in length and acquired overlapping regions.
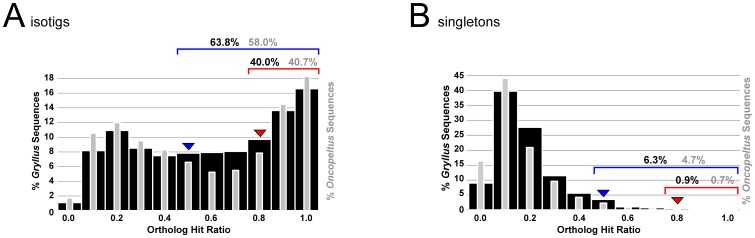
To estimate the degree to which full-length transcripts might be predicted by the transcriptome, we determined the ortholog hit ratio [71] of all assembly products by comparing the BLAST results of the full assembly against the Drosophila melanogaster proteome. The ortholog-hit ratio is calculated as the ratio of the length of a transcriptome assembly product (isotig or singleton) and the full length of the corresponding transcript. Thus, a transcriptome sequence with an ortholog hit ratio of 1 would represent a full-length transcript. In the absence of a sequenced G. bimaculatus genome, for the purposes of this analysis we use the length of the cDNA of the best reciprocal BLAST hit against the D. melanogaster proteome as a proxy for the length of the corresponding transcript. For this reason, we do not claim that an ortholog hit ratio value indicates the true proportion f a full-length transcript, but rather that it is likely to do so. The full range of ortholog hit ratio values for isotigs and singletons is shown in Figure 4. Here we summarize two ortholog hit ratio parameters for both isotigs and singletons: the proportion of sequences with an ortholog hit ratio ≥0.5, and the proportion of sequences with an ortholog hit ratio ≥0.8. We found that 63.8% of G. bimaculatus isotigs likely represented at least 50% of putative full-length transcripts, and 40.0% of isotigs were likely at least 80% full length (Figure 4B). For singletons, 6.3% appeared to represent at least 50% of the predicted full-length transcript, and 0.9% were likely at least 80% full length (Figure 4B). Most ortholog hit ratio values were higher than those obtained for the de novo transcriptome assembly of another hemimetabolous insect, the milkweed bug Oncopeltus fasciatus [11] (Figure 4A, B). We suggest that this may be explained by the fact that the G. bimaculatus de novo transcriptome assembly contains transcript predictions of higher coverage and longer isotigs (N50 = 2,133 compared to 1,735 for O. fasciatus [11]) that are likely closer to predicted full-length transcript sequences, relative to the O. fasciatus de novo transcriptome assembly [11]. However, we cannot exclude the possibility that the higher ortholog hit ratios obtained with the G. bimaculatus transcriptome may be due to its greater sequence similarity with D. melanogaster relative to O. fasciatus. Genome sequences for the two hemimetabolous insects, and rigorous phylogenetic analysis for each predicted gene in both transcriptomes, would be necessary to resolve the origin of the ortholog hit ratio differences that we report here.
Figure 4. Ortholog hit ratio analysis of the G. bimaculatus de novo assembled transcriptome.
The ortholog hit ratio is a comparison of the length of an assembled sequence to the total length of the full length transcript of its putative ortholog [71]. Values close to one suggest that a transcript predicted by the de novo assembly is close to full length. Ortholog hit ratios for the G. bimaculatus transcriptome sequences are compared to those for the previously reported de novo assembled transcriptome of another insect, the milkweed bug Oncopeltus fasciatus [11]. (A) Ortholog hit ratio analysis of assembled isotigs. A majority (63.8%) of all G. bimaculatus isotigs (black bars) have an ortholog hit ratio of ≥0.5 (blue arrowhead), and 40.0% have an ortholog hit ratio of ≥0.8 (red arrowhead). These values are higher than those obtained for the O. fasciatus de novo assembled transcriptome (grey bars) [11]. (B) Ortholog hit ratio analysis of unassembled singletons. As expected, singletons represent much smaller proportions of putative full-length transcripts. 6.3% of G. bimaculatus singletons (black) have an ortholog hit ratio of ≥0.5 (blue arrowhead), while 0.8% have an ortholog hit ratio of ≥0.8 (red arrowhead). As for the isotig analysis, these values are higher than those obtained for the O. fasciatus de novo assembled transcriptome (grey) [11].
Annotation using BLAST against the NCBI non-redundant protein database
All assembly products were compared with the NCBI non-redundant protein database (nr) using BLASTX. We found that 11,943 isotigs (55.52%) and 10,815 singletons (8.95%) were similar to at least one nr sequence with an E-value cutoff of 1e-5 (henceforth called “significant similarity”). The total number of unique BLAST hits against nr for all non-redundant assembly products (isotigs+singletons) was 19,874, which could correspond to the number of unique G. bimaculatus transcripts contained in our sample. The G. bimaculatus transcriptome contains more predicted transcripts than other orthopteran transcriptome projects to date (Table 1). This may be due to the high number of bp incorporated into our de novo assembly, which was generated from approximately two orders of magnitude more reads than previous Sanger-based orthopteran EST projects [72], [73], [74], [75], [76]. However, we note that even a recent Illumina-based locust transcriptome project that assembled over ten times as many base pairs as the G. bimaculatus transcriptome, predicted only 11,490 unique BLAST hits against nr [72]. This may be because the tissues we samples possessed a greater diversity of gene expression than those for the locust project, in which over 75% of the cDNA sequenced was obtained from a single nymphal stage [72]. Although we have used the de novo assembly method that was recommended as outperforming other assemblers in analysis of 454 pyrosequencing data [48], we cannot exclude the possibility that under-assembly of our transcriptome contributes to the high number of predicted transcripts
Since isogroups are groups of isotigs that are assembled from the same group of contigs, the isogroup number of 16,456 may represent the number of G. bimaculatus unique genes represented in the transcriptome. However, because by definition de novo assemblies cannot be compared with a sequenced genome, several issues limit our ability to estimate an accurate transcript or gene number for G. bimaculatus from these ovary and embryo transcriptome data alone.
The number of unique BLAST hits against nr (19,874) or isogroups (16,456) may overestimate the number of unique genes in our samples, because the assembly is likely to contain sequences derived from the same transcript but too far apart to share overlapping sequence; such sequences could not be assembled together into a single isotig and would therefore have been considered “different genes.” If such assembly products were derived from different regions of the same transcript and obtained distinct BLAST hits against nr, then these would be counted as two unique BLAST hits against nr. This limitation is an inevitable result of performing de novo assembly in the absence of a reference genome, and is unavoidable in the case of G. bimaculatus as no orthopteran genomes have yet been sequenced. Conversely, the number of unique BLAST hits against nr could underestimate the number of unique genes, because they cannot include those isotigs (9,569 = 44.5% of all isotigs) and singletons (109,990 = 91.0% of all singletons) that lacked significant BLAST hits against nr. Such sequences could represent non-coding sequences with no matches to the coding-region data contained in nr, or could lack sufficient similarity to known sequences. Finally, because our transcriptome libraries were prepared only from ovarian and embryonic tissue, it is unlikely to contain transcripts of all G. bimaculatus genes, many of which could be expressed exclusively postembryonically and/or in specific nymphal or adult tissue types. Determination of the total gene number for G. bimaculatus must therefore await complete genome sequencing.
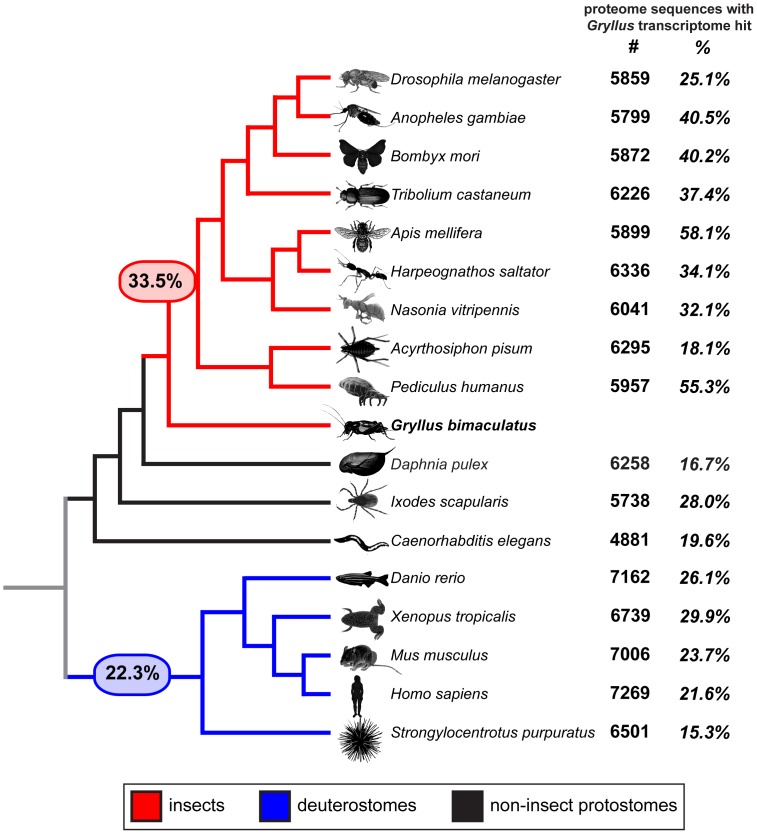
We wished to understand the relative similarities of the G. bimaculatus transcriptome sequences to those from other organisms. Specifically, we asked what proportion of genes found in sequenced animal genomes had putative orthologs in the G. bimaculatus transcriptome. To this end, we used BLAST to compare each non-redundant assembly product (E-value cutoff 1e-5) to the proteomes of several organisms with completely sequenced genomes (Table S1). We found that overall, 33.49% of the sequences contained in insect proteomes had matches in the G. bimaculatus de novo transcriptome assembly, compared to 22.28% of sequences from deuterostome proteomes (Figure 5). Within the insects, the proportion of hits to the D. melanogaster proteome was lower than the proportion of hits to most other insects. This may reflect the relatively greater divergence from a last common insect ancestor, as D. melanogaster belongs to the most derived insect order, the Diptera. However, we noted that the proportion of matches to some insect proteomes appeared unusually low given their phylogenetic relationship to Orthoptera. Specifically, only 18.1% of proteome sequences from the aphid Acyrthosiphon pisum, a hemimetabolous insect, had hits in the G. bimaculatus transcriptome, compared with an average of 36.1% across all holometabolous proteomes surveyed (Figure 5). This is consistent with the description of the A. pisum genome containing many unusual features relative to other insect genomes, including extensive gene family duplications and gene loss [6], [77], [78], [79]. The relatively high proportion of holometabolous proteome sequences with matches in the G. bimaculatus transcriptome suggests that these organisms may share more features derived from a last common insect ancestor than does A. pisum, and highlights the need for further genomic resources in the Hemimetabola. We caution that there are limitations to the biological information that can be derived from these comparisons, as not all animal genomes used for this analysis have comparable levels of coverage or annotation.
Figure 5. Phylogenetic comparison of proportion of known proteomes represented in the G. bimaculatus de novo assembled transcriptome.
The number (bold) and percentage (bold italics) of proteome sequences with a putative G. bimaculatus ortholog in the de novo transcriptome assembly is shown for selected animals with sequenced genomes (based on top BLAST hit, E-value cutoff 1e-5). Proteomes were predicted from genome sequence sources as shown in Table S1. Numbers in large font in red and blue ovals indicate average proportion of sequences from all tested insect and deuterostome proteomes, respectively, represented in the G. bimaculatus transcriptome.
Manual annotation of conserved developmental genes and members of signaling pathways
G. bimaculatus has been the subject of molecular embryology for over a decade, and as a result over 80 GenBank accessions are available (NCBI accessed 12 August 2012). We asked whether these genes were represented in our transcriptome, and found that 72.3% of them were present (60/83). Moreover, the transcriptome contributed to these accessions by extending their sequences by an average of 737 nucleotides per accession (205.0% on average across all 83 G. bimaculatus GenBank accessions) and in some cases by over 1,700% (Table S2). This shows that the G. bimaculatus transcriptome will be an extremely useful resource for continued research into the function and evolution of most previously cloned genes.
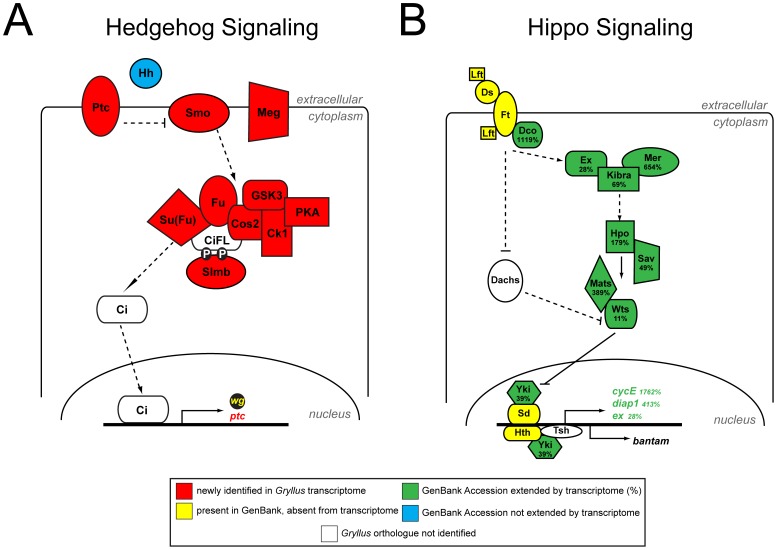
To determine the transcriptome's utility as a source of new gene discovery, we searched for putative orthologs of the 1,168 D. melanogaster transcription factors catalogued in the FlyTF transcription factor database [80]. We found that 542 (46.4%) of them were present, based on the criterion of being the best reciprocal BLAST hit with a D. melanogaster sequence using an E-value cutoff of 1e-5 (Table S3). We also undertook manual annotation of 122 genes from seven conserved metazoan signaling pathways (Table S4), 261 genes involved in male and female gametogenesis in D. melanogaster (Table S5), and 24 additional genes with roles in maternal or zygotic embryonic patterning (Table S6). For the Notch [81], TGF-beta [82], Wnt [83], JAK/STAT [84], MAPK [85] and hedgehog [86] signaling pathways, most G. bimaculatus orthologs of these genes were previously unknown. Our transcriptome newly identified 66 genes participating in these signaling pathways (Table S4, Figure S2), including nearly all members besides the ligand of the hedgehog pathway (Figure 6A). In the case of the Hippo signaling pathway [87], for which most G. bimaculatus core kinase orthologs were already present in GenBank, the G. bimaculatus de novo transcriptome assembly increased the length of known sequences by an average of 323%, and by as much as 1,119% in the case of the discs overgrown (dco) gene (Figure 6B, Table S2).
Figure 6. Sequence extension and gene discovery in the G. bimaculatus Hedgehog and Hippo pathways.
(A) The de novo transcriptome assembly of G. bimaculatus newly identifies most members of the hedgehog pathway (red), from which only the hedgehog ligand (blue) was previously known (GenBank accession AB044709). (B) The transcriptome also adds significant sequence data to the fragments of many genes in the Hippo signaling pathway that had been previously identified (green). Seven genes of the known pathway were not identified in the transcriptome (yellow, white), two of which lack any sequence data in GenBank (white). GenBank accessions for previously identified sequences are as follows: discs overgrown (dco): AB443442; expanded (ex): AB378099; warts (wts): AB300574; cyclin E (cycE): AB378067; hippo (hpo): AB378070; inhibitor of apoptosis protein (diap1): AB378071; mob as tumor suppressor (mats): AB378072; yorkie (yki): AB378076; scaffold protein salvador (sav): AB378074; Merlin (Mer): AB378073; Kibra: DC445461.
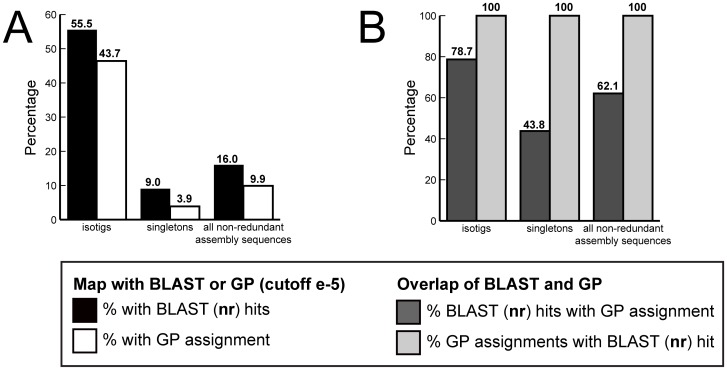
Automated annotation using the custom script “Gene Predictor” identifies 14,130 transcriptome sequences as putatively orthologous to D. melanogaster genes
Although manual annotation proved a highly effective way to identify developmental genes of interest in the G. bimaculatus transcriptome, it is not efficient at large scales. We therefore developed an automated annotation tool that uses the criterion of best reciprocal BLAST hit against the D. melanogaster proteome (E-value cutoff 1e-5) to propose putative orthologs for all assembly products of the transcriptome. This method is not qualitatively different from manual annotation using BLAST with a specific known sequence as a query, but rather simply automates the process of detecting a best reciprocal BLAST hit, which is a method of orthology assignment routinely employed as an annotation method in genomics studies using insect genomes [88], [89], [90]. Using this tool, called Gene Predictor (see Methods), we were able to assign putative orthologs to 43.7% of isotigs, very close to the proportion of isotigs (55.5%) with significant BLAST hits against nr (Figure 7A). Of the 60 known G. bimaculatus GenBank accessions that were identified in the transcriptome by manual annotation (Table S2), 52 have significant BLAST hits to a D. melanogaster gene (the remaining 8 genes have significant similarity only to non D. melanogaster sequences, as determined by BLAST against nr). Gene Predictor correctly identified 36 of these 52 genes (69.2%). Gene Predictor's failure to identify the remaining 16 genes (30.8%) means that while these genes do have significant BLAST hits in the D. melanogaster genome, they are more similar to a non-D. melanogaster gene, and are thus not the reciprocal best BLAST hit of any D. melanogaster gene.
Figure 7. Automated annotation of the G. bimaculatus de novo transcriptome assembly using Gene Predictor.
(A) Comparison of the proportion of non-redundant assembly sequences, isotigs and singletons that obtained a significant BLAST hit against nr (black bars), and those that were assigned a putative orthology by Gene Predictor (GP; white bars), based on the best reciprocal top BLAST hit with the Drosophila melanogaster proteome (see Table S1). (B) Comparison of the proportion of sequences with a significant BLAST hit in nr that also had a putative orthology assignment based on Gene Predictor (dark grey bars). All sequences assigned putative orthologs by Gene Predictor also had significant BLAST hits in nr (light grey bars).
These results suggest that for de novo insect transcriptome assemblies, Gene Predictor could be an efficient annotation tool, as it is nearly as effective as BLAST mapping against the large nr database, but is computationally much less intensive as it relies only on the D. melanogaster proteome of 23,361 predicted proteins. Relative to BLAST mapping against nr, Gene Predictor was more effective at suggesting orthologs for isotigs than for singletons (Figure 7A), likely due to the fact that isotigs are easier to map by any method as they contain more sequence data. Gene Predictor did not, however, assign orthologs to any assembly products that did not already have a significant BLAST hit in nr (Figure 7B), as expected since the D. melanogaster proteome is contained within nr. Conversely, not all assembly sequences with BLAST hits in nr obtained a significant hit with Gene Predictor (Figure 7B), indicating that some of the G. bimaculatus predicted transcripts share greater similarity to sequences other than those in the D. melanogaster proteome, or may represent genes that have been lost in D. melanogaster. The Gene Predictor scripts are freely available at http://www.extavourlab.com/protocols/index.html.
Transcripts lacking significant BLAST hits against nr may encode functional protein domains
The majority (55.5%) of predicted transcripts retrieved a significant BLAST hit against the nr database (Figure 7A). This exceeds the proportion of de novo assembly products typically identifiable by BLAST mapping against nr [71], including the 43.4% and 29.5% of predicted transcripts mapped in this way from two de novo arthropod transcriptome assemblies that we previously constructed using similar methods to those described here [11], [49]. This may be due to the much higher read depth and coverage of the G. bimaculatus transcriptome, which to our knowledge is the largest de novo assembled transcriptome available for the Hemimetabola, and the largest 454-based transcriptome for any organism to date. Even this assembly, however, contains a large proportion (44.5%) of sequences of unknown identity. These sequences could represent contaminants of unknown origin, sequences that are too short to obtain significant hits to nr sequences, non-coding transcripts, non-coding portions of protein-coding transcripts, or clade- or species-specific transcripts that may be unidentifiable due to the paucity of orthopteran genomic data in GenBank. We believe that significant contaminants are unlikely, as less than one percent of all assembly products retrieved BLAST hits to prokaryote, fungal or plant sequences with an E-value cutoff of 1e-10.
We also compared the length (in nucleotides) of sequences with and without significant BLAST hits (Tables 3, 4), and found that unidentified isotigs were significantly shorter than isotigs with BLAST hits (Table 5). The difference was also significant for singletons (Tables 4, 5). This is consistent with the possibility that contig length may play a role in sequence recognizability, also observed with the low proportion of singletons with significant BLAST hits compared to isotigs (9.0% vs 55.5%; Figure 8A, B).
Table 3. Length parameters of isotigs according to BLAST annotation and predicted protein-coding status.
| BLAST hit1/predicted protein coding status | Parameter | Value |
| Significant hit against nr 2 , 3 | Maximum sequence length4 | 10865 |
| Minimum sequence length | 91 | |
| Median sequence length | 1669.50 | |
| Average sequence length | 1927.98 | |
| Significant hit against nr and contains predicted protein-coding region(s) | Maximum sequence length | 10865 |
| Minimum sequence length | 168 | |
| Median sequence length | 1730.5 | |
| Average sequence length | 1997.42 | |
| Maximum predicted peptide length5 | 2076 | |
| Minimum predicted peptide length | 11 | |
| Median predicted peptide length | 317.50 | |
| Average predicted peptide length | 386.82 | |
| No significant hit against nr | Maximum sequence length | 6886 |
| Minimum sequence length | 57 | |
| Median sequence length | 728.50 | |
| Average sequence length | 924.277 | |
| No significant hit against nr and contains predicted protein-coding region(s) | Maximum sequence length | 6686 |
| Minimum sequence length | 60 | |
| Median sequence length | 858.5 | |
| Average sequence length | 1130.16 | |
| Maximum predicted peptide length | 1710 | |
| Minimum predicted peptide length | 7 | |
| Median predicted peptide length | 144.5 | |
| Average predicted peptide length | 197.61 | |
| All NRI 6 containing predicted protein-coding regions | Maximum sequence length | 10865 |
| Minimum sequence length | 60 | |
| Median sequence length | 1544.50 | |
| Average sequence length | 1837.57 | |
| Maximum predicted peptide length | 2076 | |
| Minimum predicted peptide length | 7 | |
| Median predicted peptide length | 282.50 | |
| Average predicted peptide length | 351.95 | |
| All NRI without predicted protein-coding regions | Maximum sequence length | 6677 |
| Minimum sequence length | 57 | |
| Median sequence length | 708.50 | |
| Average sequence length | 878.27 | |
| No significant hit against nr and significant hit against Locusta migratoria sequences 7 | Maximum sequence length | 5287 |
| Minimum sequence length | 124 | |
| Median sequence length | 1093.50 | |
| Average sequence length | 1358.21 | |
| Maximum predicted peptide length | 1710 | |
| Minimum predicted peptide length | 25 | |
| Median predicted peptide length | 244.50 | |
| Average predicted peptide length | 320.84 | |
| No significant hit against nr and significant hit against Laupala kohalensis sequences 8 | Maximum sequence length | 6677 |
| Minimum sequence length | 62 | |
| Median sequence length | 1004.50 | |
| Average sequence length | 1304.64 | |
| Maximum predicted peptide length | 1710 | |
| Minimum predicted peptide length | 16 | |
| Median predicted peptide length | 248.50 | |
| Average predicted peptide length | 315.37 |
BLAST E-value cutoff is e-5 for all hits reported in this table.
nr = NCBI non-redundant database.
Numbers of sequences in each category are shown in Figure 9.
Sequence lengths are reported in base pairs.
Predicted peptide lengths are reported in amino acids.
NRI = all non-redundant isotigs regardless of BLAST results against nr.
Laupala kohalensis sequences used for comparison are from [75].
Table 4. Length parameters of singletons according to BLAST annotation and predicted protein-coding status.
| BLAST hit1/predicted protein coding status | Parameter | Value |
| Significant hit against nr 2,3 | Maximum sequence length4 | 582 |
| Minimum sequence length | 66 | |
| Median sequence length | 340.00 | |
| Average sequence length | 334.25 | |
| Significant hit against nr and contains predicted protein-coding region(s) | Maximum sequence length | 574 |
| Minimum sequence length | 68 | |
| Median sequence length | 343.5 | |
| Average sequence length | 337.54 | |
| Maximum predicted peptide length5 | 192 | |
| Minimum predicted peptide length | 8 | |
| Median predicted peptide length | 103.50 | |
| Average predicted peptide length | 103.28 | |
| No significant hit against nr | Maximum sequence length | 620 |
| Minimum sequence length | 50 | |
| Median sequence length | 243.50 | |
| Average sequence length | 251.67 | |
| No significant hit against nr and contains predicted protein-coding region(s) | Maximum sequence length | 586 |
| Minimum sequence length | 50 | |
| Median sequence length | 231.5 | |
| Average sequence length | 243.16 | |
| Maximum predicted peptide length | 189 | |
| Minimum predicted peptide length | 5 | |
| Median predicted peptide length | 60.50 | |
| Average predicted peptide length | 65.02 | |
| All NRS 6 containing predicted protein-coding region(s) | Maximum sequence length | 586 |
| Minimum sequence length | 50 | |
| Median sequence length | 255.5 | |
| Average sequence length | 268.89 | |
| Maximum predicted peptide length | 192 | |
| Minimum predicted peptide length | 5 | |
| Median predicted peptide length | 71.5 | |
| Average predicted peptide length | 75.45 | |
| All NRS without predicted protein-coding regions | Maximum sequence length | 620 |
| Minimum sequence length | 50 | |
| Median sequence length | 249.50 | |
| Average sequence length | 255.51 | |
| No significant hit against nr and significant hit against Locusta migratoria sequences 7 | Maximum sequence length | 552 |
| Minimum sequence length | 52 | |
| Median sequence length | 299 | |
| Average sequence length | 283.97 | |
| Maximum predicted peptide length | 176 | |
| Minimum predicted peptide length | 17 | |
| Median predicted peptide length | 74.50 | |
| Average predicted peptide length | 75.08 | |
| No significant hit against nr and significant hit against Laupala kohalensis sequences 8 | Maximum sequence length | 597 |
| Minimum sequence length | 52 | |
| Median sequence length | 286.50 | |
| Average sequence length | 280.55 | |
| Maximum predicted peptide length | 188 | |
| Minimum predicted peptide length | 11 | |
| Median predicted peptide length | 77.5 | |
| Average predicted peptide length | 77.40 |
BLAST E-value cutoff is e-5 for all hits reported in this table.
nr = NCBI non-redundant database.
Numbers of sequences in each category are shown in Figure 9.
Sequence lengths are reported in base pairs.
Predicted peptide lengths are reported in amino acids.
NRS = all non-redundant singletons regardless of BLAST results against nr.
Laupala kohalensis sequences used for comparison are from [75].
Table 5. Statistical comparison of isotig and singleton nucleotide sequence lengths according to BLAST annotation and predicted protein-coding status.
| BLAST hit1/predicted protein coding status2 | Significant hit against nr2 | Significant hit against nr and contains predicted coding regions | No significant hit against nr | No significant hit against nr and contains predicted coding regions | All NRAS3 containing predicted protein-coding regions | All NRAS without predicted protein-coding regions | No significant hit against nr and significant hit against Locusta migratoria sequences | No significant hit against nr and significant hit against Laupala kohalensis sequences |
| ISOTIGS 4 | ||||||||
| Significant hit against nr | 0.9998 | *** | *** | *** | *** | *** | *** | |
| Significant hit against nr and contains predicted coding regions | 1 | *** | 1 | 1 | 1 | 1 | ||
| No significant hit against nr | 1 | 1 | *** | 1 | 1 | |||
| No significant hit against nr and contains predicted coding regions | *** | 1 | *** | *** | ||||
| All NRAS containing predicted protein-coding regions | *** | *** | *** | |||||
| All NRAS without predicted protein-coding regions | 1 | 1 | ||||||
| No significant hit against nr and significant hit against Locusta migratoria sequences | 0.2268 | |||||||
| No significant hit against nr and significant hit against Laupala kohalensis sequences | ||||||||
| SINGLETONS | ||||||||
| Significant hit against nr | 0.9798 | *** | *** | *** | *** | *** | *** | |
| Significant hit against nr and contains predicted coding regions | 1 | *** | 1 | 1 | 1 | 1 | ||
| No significant hit against nr | *** | 1 | *** | 0.4208 | 1 | |||
| No significant hit against nr and contains predicted coding regions | *** | *** | * | *** | ||||
| All NRAS containing predicted protein-coding regions | *** | *** | 0.0969 | |||||
| All NRAS without predicted protein-coding regions | 0.1358 | 0.9985 | ||||||
| No significant hit against nr and significant hit against Locusta migratoria sequences | 0.9967 | |||||||
| Significant hit against Laupala kohalensis sequences | ||||||||
Values shown are p≥0.05 value results of a Welch's t-test.
= p<0.0001;
p<0.05.
BLAST E-value cutoff is e-5 for all hits reported in this table.
nr = NCBI non-redundant database.
NRAS = all non-redundant assembly products (isotigs or singletons) regardless of BLAST results against nr.
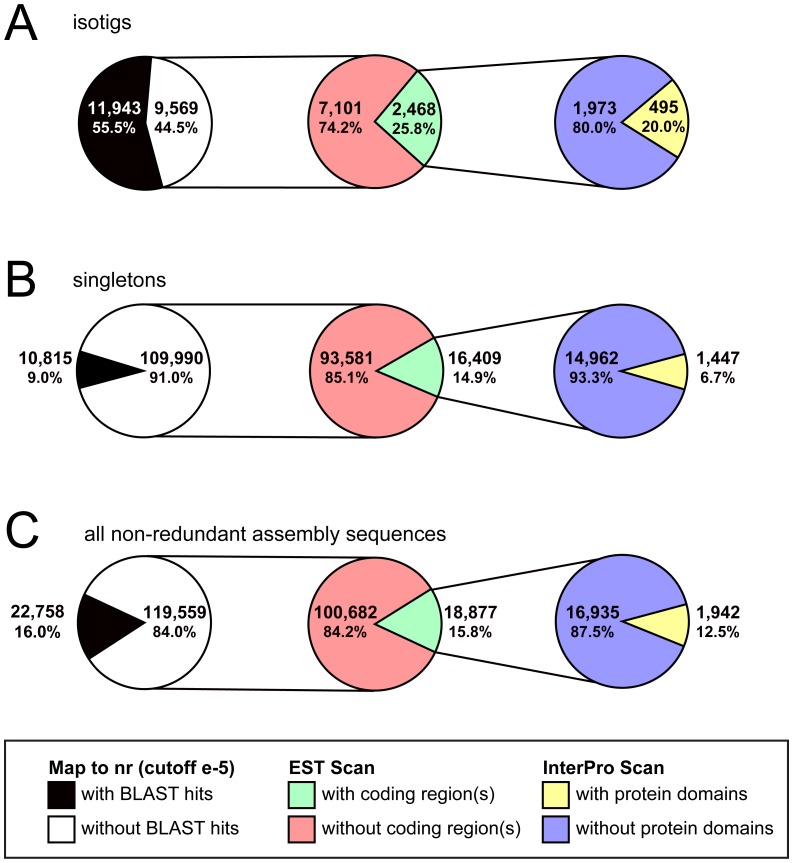
Figure 8. Coding region analysis of G. bimaculatus de novo transcriptome assembly sequences without significant BLAST hits in nr.
Assembly products that failed to obtain significant BLAST hits in nr (white) were examined for the presence of coding regions (green) using EST Scan [52]. Assembly sequences thus predicted to contain coding regions were examined for the presence of known coding domains (yellow) using InterPro Scan [53], [54]. Results are shown separately for isotigs (A), singletons (B) and all non-redundant assembly products (C). See also Table 3.
To obtain additional biological information about sequences that failed to obtain significant BLAST hits against nr, we therefore applied EST Scan analysis to determine whether these sequences potentially encoded unknown proteins. EST Scan uses known differences in hexanucleotide usage between coding and non-coding regions to detect potential coding regions in DNA sequences, without requiring open reading frames [52]. We found that 2,468 (25.8%) unidentified isotigs and 16,409 (14.9%) unidentified singletons were predicted to contain protein-coding regions (Figure 8). Isotigs without predicted coding regions were significantly shorter than sequences with predicted coding regions (Tables 3, 5); the difference was also significant for singletons (Tables 4, 5). Previously unidentified isotigs that were protein-coding were significantly shorter that isotigs with significant BLAST hits, and encoded significantly fewer amino acids (Tables 3, 5, 6). This may mean that significant BLAST hits were not obtained for some of these sequences either because of insufficient contig lengths, or because they contained relatively less protein-coding content, or both. These observations demonstrate that although these 18,877 sequences are not significantly similar to known proteins in the NCBI nr database, they may nevertheless represent portions of coding rather than non-coding transcripts.
Table 6. Statistical comparison of isotig and singleton predicted coding sequence lengths according to BLAST annotation status.
| BLAST hit1/predicted protein coding status2 | Significant hit against nr2 | No significant hit against nr | All NRAS3 | No significant hit against nr and significant hit against Locusta migratoria sequences | No significant hit against nr and significant hit against Laupala kohalensis sequences |
| ISOTIGS 4 | |||||
| Significant hit against nr | *** | 1 | ** | *** | |
| No significant hit against nr | *** | 1 | 1 | ||
| All NRAS | * | 0.0059 | |||
| No significant hit against nr and significant hit against Locusta migratoria sequences | 0.4052 | ||||
| No significant hit against nr and significant hit against Laupala kohalensis sequences | |||||
| SINGLETONS | |||||
| Significant hit against nr | *** | 1 | *** | *** | |
| No significant hit against nr | *** | 1 | 1 | ||
| All NRAS | 0.4091 | 0.9235 | |||
| No significant hit against nr and significant hit against Locusta migratoria sequences | 0.8685 | ||||
| No significant hit against nr and significant hit against Laupala kohalensis sequences | |||||
Values shown are p≥0.05 value results of a Welch's t-test.
= p<0.0001;
p<0.001;
p<0.05.
BLAST E-value cutoff is e-5 for all hits reported in this table.
nr = NCBI non-redundant database.
NRAS = all non-redundant assembly products regardless of BLAST results against nr.
We then used InterPro Scan [53], [54] to query predicted coding regions for predicted functional protein domains. InterPro Scan queries the InterPro consortium databases (ProDom [55], PRINTS [91], SMART [57], TIGRFAMs [58], Pfam [59], PROSITE [60], PIRSF [61], SUPERFAMILY [62], CATH [63], PANTHER [64], SignalPHMM [65], and Transmembrane [66]) for signatures of protein domains of known function. It also identifies evolutionarily conserved protein domains that are predicted to be functional based on their conservation but have no described molecular function to date, called Domains of Unknown Function (DUFs) [92]. This analysis revealed that of those protein-coding sequences of unknown identity, 495 (20.0%) isotigs and 1,447 (6.7%) singletons were predicted to contain functional protein domains. These results show that 1,942 sequences from the de novo transcriptome assembly that could not be identified based on BLAST against nr alone may nonetheless encode functional proteins present during G. bimaculatus oogenesis and embryogenesis.
Taxonomic bias of the nr database can limit gene identification in de novo assembled transcriptomes
Because orthopteran sequence data are poorly represented in nr, we asked whether at least some of the G. bimaculatus transcriptome sequences that appeared to lack significant similarity to known genes might show similarity to sequences from other orthopterans available in the form of EST collections. To determine this, we compared the 9,569 isotigs (44.5% of all isotigs) and 109,990 singletons (91.0% of all singletons) from the G. bimaculatus transcriptome that lacked significant nr hits, with the EST collections for the orthopterans L. migratoria and L. kohalensis. L. migratoria of the suborder Caelifera (grasshoppers and locusts) is a migratory locust that is widespread throughout Asia, Africa, and Australasia [93], and is heavily studied due to its impact as an agricultural pest (see for example [94], [95]). The available sequence collections for this locust sampled transcripts from larval stages L4 and L5 [72], [73], [74], which is when transition between the solitary and gregarious (swarming) behavior of these locusts becomes irreversible [74], [96]. L. kohalensis belongs to the suborder Ensifera (katydids and crickets), and is a Hawaiian species that has been used extensively for studies of the physiology and evolution of speciation and acoustic preference (see for example [23], [97], [98]). The EST library available for this cricket contains sequences derived from transcripts of the larval central nervous system [75]. Because these data are derived from EST collections, they are available through GenBank but are not included in nr.
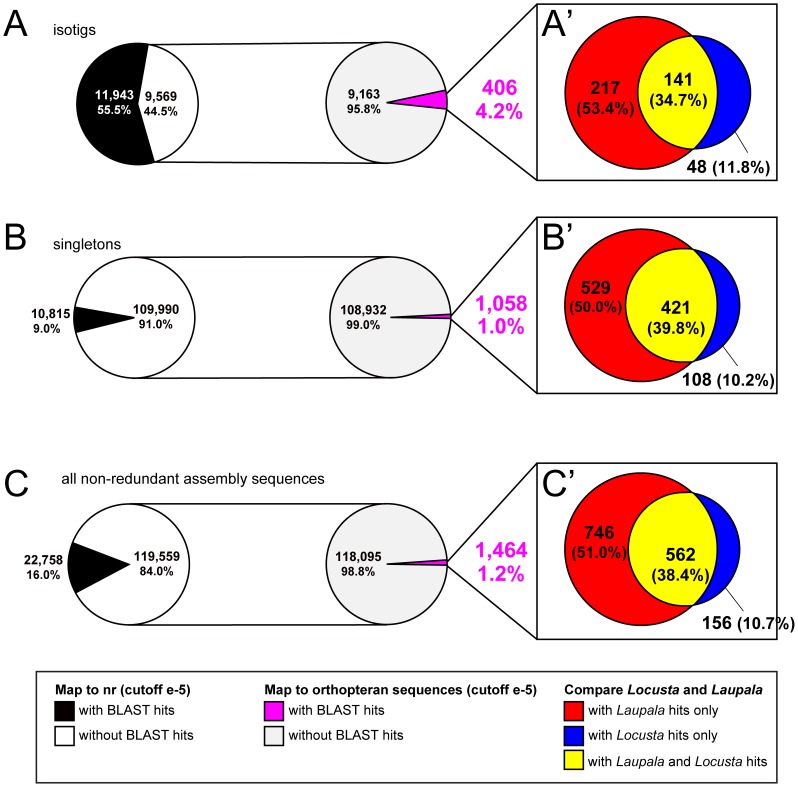
Using BLAST with an E-value cutoff of e-5, we found that the majority of previously unidentified G. bimaculatus transcriptome sequences also lacked significant similarity to L. migratoria or L. kohalensis sequences. This may be due to the difference in starting material for the libraries compared, as the G. bimaculatus transcriptome contains transcripts from ovaries and embryos, while the other two libraries represent exclusively post-embryonic transcripts, and the L. kohalensis library is further restricted only to transcripts from the nervous system. However, 406 isotigs (4.24%) and 1,058 singletons (0.96%) did display significant similarity (Figure 9A, B), suggesting that these transcripts could represent “orthopteroid” genes. However, we noted that sequences of both isotigs and singletons in this category contained significantly fewer nucleotides (Tables 3–5) and encoded significantly fewer amino acids on average (Tables 3, 4, 6) than transcriptome sequences with BLAST hits to nr (Tables 3–6). An alternative explanation for these apparent “orthopteroid” sequences is thus that these sequences, as well as their matches from L kohalensis and L. migratoria, might prove significantly similar to other sequences from nr, if their transcript sequences were longer.
Figure 9. Comparison of sequences lacking significant BLAST hits to nr, with Laupala kohalensis and Locusta migratoria databases.
(A–C) Assembly products that failed to obtain significant BLAST hits to nr (white) were examined for significant similarity (magenta) to transcripts from at least one of L. migratoria or L. kohalensis [72], [73], [74], [75]. (A′–C′) Assembly sequences thus identified were parsed into sequences with significant hits among only L. kohalensis sequences (red), only L. migratoria sequences (blue), or both (yellow). Results are shown separately for isotigs (A, A′), singletons (B, A′) and all non-redundant assembly products (C, A′).
Because Ensifera and Caelifera are believed to have diverged 300 Mya [5], we predicted that we would find greater similarity between sequences from the two crickets, than between G. bimaculatus and the locust. Accordingly, of the putative “orthopteroid sequences,” 746 (51.0%) G. bimaculatus sequences yielded hits exclusively to L. kohalensis sequences, compared to 156 (10.7%) sequences with exclusive hits among L. migratoria sequences (Figure 9C′). This likely reflects the closer phylogenetic relationship between the two crickets, which are both within the same family of Gryllidae.
Putative orthopteroid-specific sequences contain a high proportion of predicted protein coding domains of unknown function (DUFs)
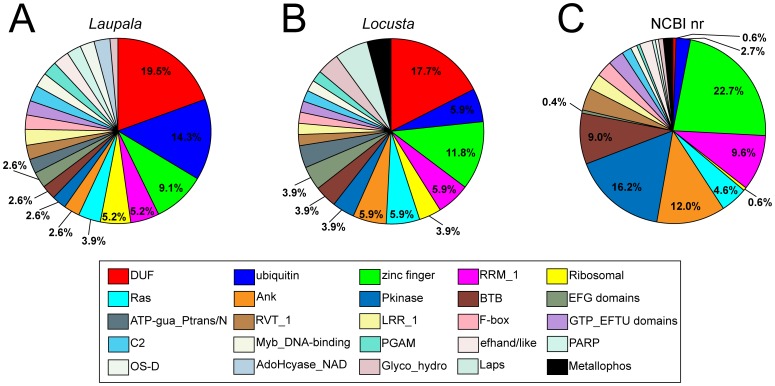
Finally, we asked whether these “orthopteroid sequences” shared any characteristics that might aid in understanding their putative clade-specific functions. We used InterPro Scan [53] to determine the distribution of recognizable protein domains among transcriptome sequences with significant L. kohalensis or L. migratoria hits, and compared them with those of all transcriptome sequences with significant BLAST hits to nr. We found that the number of distinct domains was similar for L. kohalensis-like sequences (77 different protein domains) and all other transcriptome sequences with significant BLAST hits (83 different protein domains), but considerably lower for L. migratoria-like sequences (55 different protein domains). Given the small number of sequences examined here (Figure 9C), this is unlikely to represent true differences in protein type between the three datasets.
However, the datasets differed strikingly in the relative proportions of different protein domains encoded. Considering the top 25 most frequently represented protein domains within each dataset, the most abundant domains in both orthopteran-like groups were domains of unknown function (DUFs, 18.8% of both orthopteran matches combined), followed by ubiquitin family domains (Pfam PF00240, 10.9%), zinc finger domains (multiple Pfam categories combined, 10.2%), and RNA recognition motifs (Pfam PF00076, 5.5%) (Figure 10A, B). In contrast, transcriptome sequences with significant BLAST hits to nr encoded proteins principally containing zinc finger domains (multiple Pfam categories combined, 22.7%), protein kinase domains (Pfam 00069, 16.2%), and ankyrin repeat domains (Pfam PF00023, 12.0%), followed by RNA recognition motifs (Pfam PF00076, 9.6%) and BTB/POZ domains (Pfam PF00651, 9.0%) (Figure 10C). These differing proportions of predicted protein domains between orthopteran-matched and nr-matched G. bimaculatus sequences were observed even when all predicted protein domains were considered (Figure S3). We speculate that the “orthopteroid-like” proteins predicted to be present in the G. bimaculatus transcriptome might share greater functional similarity with orthopteran proteins than with proteins from other organisms represented in nr. Moreover, the high proportion of DUFs predicted in these “orthopteroid-like” proteins may mean that some of these DUFs serve clade-specific functions. The specific roles of these genes in G. bimaculatus and other orthopterans are currently unknown, and will require functional genetic testing to be elucidated. However, the present analysis demonstrates that even for de novo assembled transcriptome sequences that are not easily identifiable based on GenBank comparisons, it may be possible to extract potentially meaningful biological and evolutionary information, and with further refinement, perhaps even to define new or clade-specific DUFs as candidates for future functional testing.
Figure 10. Principal protein domain composition of G. bimaculatus transcriptome sequences with highest similarity to Laupala kohalensis or Locusta migratoria sequences.
Relative proportions of the top 25 protein domains coded by G. bimaculatus transcriptome sequences with significant similarity to sequences from L. kohalensis (A), L. migratoria (B), or sequences from nr (C). Protein domain nomenclature from Pfam [102] as follows: AdoHcyase_NAD: PF00670; Ank: PF00023; ATP-gua_Ptrans/N: PF02807; BTB/POZ: PF00651; C2: PF00168; DUF (combined): n/a; EFG domains (combined): n/a; efhand/like: PF09279; F-box: PF00646; Glyco_hydro (combined): n/a; GTP_EFTU domains: PF00009; Laps: PF10169; LRR_1: PF00560; Metallophos: PF00149; Myb_DNA-binding (combined): n/a; OS-D: PF03392; PARP: PF00644; PGAMP: PF07644; Pkinase: PF00069; Ras: PF00071; Ribosomal (combined): n/a; RRM_1: PF00076; RVT_1: PF00078; ubiquitin: PF00240; zinc finger (combined): n/a. “Combined” indicates that multiple Pfam accessions are combined.
Creation of a searchable database to house arthropod de novo assembled transcriptomes
The volume of high-throughput transcriptome data available for all organisms is rapidly increasing, but many of these datasets are not publicly available in an easily searchable format. The NCBI Short Read Archive [99] provides a repository for raw read data from transcriptome projects, but a searchable interface for de novo assembled transcriptomes that do not have an associated genome sequence or previously developed community web interface is lacking. Like EST collections, transcriptome assemblies can be made public through the NCBI Transcriptome Shotgun Assembly Sequence Database (TSA: http://www.ncbi.nlm.nih.gov/genbank/tsa), but annotation of these data is not required, and they are not included in nr. To maximize the public utility of our data, we therefore created a searchable database that facilitates access to the annotated G. bimaculatus de novo assembled transcriptome reported here. The Assembled Searchable Giant Arthropod Read Database (ASGARD) includes all nr BLAST, manual annotation, and Gene Predictor annotation results for the G. bimaculatus transcriptome. Details of the design and database schema of ASGARD have been previously described [51]. This database also contains two additional de novo assembled transcriptomes that we constructed previously, for the milkweed bug Oncopeltus fasciatus [11] and the amphipod crustacean Parhyale hawaiensis [49]. The O. fasciatus transcriptome, which was originally assembled with Newbler v2.3 [11], was re-assembled with Newbler 2.5, which was used to assemble the P. hawaiensis and G. bimaculatus transcriptomes. Complete updated assembly files in FASTA format for all three transcriptomes can be downloaded via ASGARD. We also processed the O. fasciatus and P. hawaiensis transcriptomes with the EST Scan, InterPro Scan, and the Gene Predictor script, so that they could be searched in the same way as the G. bimaculatus transcriptome. ASGARD allows users to search these de novo assembled transcriptomes in four ways: (1) for putative orthologs to known D. melanogaster genes (based on Gene Predictor results); (2) by searching the text of the top 50 significant BLAST hits for the name of any gene of interest (based on nr BLAST mapping results); (3) by searching for transcripts with a given GO term assignment; and (4) by read name if the unique identifier of a given assembly product is known (this information is provided in the results of the previous three searches). All search result output pages allow users to view and download the nucleotide sequences of matching assembly products, the pre-computed results of a BLAST search of that sequence against nr (E-value cutoff 1e-5), their predicted translation products if applicable (determined using EST Scan), and any predicted functional protein domains (determined using InterPro Scan). Finally, ASGARD also contains a BLAST interface that allows users to search any or all transcriptomes using the BLASTN, TBLASTN or TBLASTX algorithms. ASGARD is available at http://asgard.rc.fas.harvard.edu.
Supporting Information
Comparison of read lengths from de novo assembly of the G. bimaculatus transcriptome. (A) Distribution of read lengths before (black) and after (blue) trimming to remove low quality reads (see text for details). (B) Distribution of trimmed read lengths before (blue) and after (red) assembly with Newbler v2.5. The assembly yielded assembled reads of over 10,000 bp. (C) Distribution of read lengths of the shortest assembled (red) and raw (blue) reads.
(TIF)
Schematics of conserved metazoan signal transduction pathways showing components identified in the G. bimaculatus transcriptome. BLAST was used to search for signaling pathway genes in the G. bimaculatus transcriptome (see Table S4); genes with newly identified putative orthologs are indicated in red. Genes outlined in grey with grey typeface indicate genes without D. melanogaster homologs. Pathway schematics are modified from KEGG pathway model images (http://www.genome.jp/kegg/kegg1.html). (A) Notch pathway. (B) TGF-βeta pathway. (C) Wnt pathway. (D) Janus Kinase (JAK)-signal transducer and activator of transcription (STAT) pathway. (E) Mitogen-activated protein Kinase (MAPK) pathway.
(TIF)
Complete protein domain composition of G. bimaculatus transcriptome sequences with highest similarity to Laupala kohalensis or Locusta migratoria sequences. Relative proportions of all protein domains coded by G. bimaculatus transcriptome sequences with significant similarity to sequences from L. kohalensis (A), L. migratoria (B), or sequences from nr (C). Protein domain nomenclature from Pfam [102] and SMART [103] databases as follows: 5_nucleotid_C: PF2872; Abhydrolase_1: PF00561; adh_short: PF00106; ADK: OF00406; AdoHcyase_NAD: PF00670; Amidohydro_1: PF01979; Ank: PF00023; AP_endonuc_2_N: PF07582; Asparaginase_2: PF01112; ATP-gua_Ptrans/N: PF02807; BAH: PF01426; BTB/POZ: PF00651; Btz: SM 01044; bZIP_2: PF07716; C2: PF00168; CBM_14: PF01607; COesterase: PF00135; Cyclin_N: PF00134; Cys_Met_Meta_PP: PF01053; DEAD: PF00270; DUF (combined): n/a; EFG domains (combined): n/a; efhand/like: PF09279; eIF-5_eIF-2B: PF01873; ELM2: PF01448; ELO: PF01151; EMP70: PF02990; ETF_alpha: PF00766; Exo_endo_phos: PF03372; F-box: PF00646; fn3: PF00041; G-patch: PF01858; GATA: PF00320; GCV_H: PF01597; GHMP_kinases_N: PF00288; Glyco_hydro (combined): n/a; GTP_EFTU domains: PF00009; HECT: PF00632; Hemocyanin_N: PF03722; HSP90: PF00183; IF-2B: PF01008; IPP-2: PF04979; JHBP: PF06585; Laps: PF10169; Ldl_recept_a: PF00057; Lectin_C: PF00059; LRR_1: PF00560; MA3: PF00560; MADF_DNA_bdg: PF10545; MAP65_ASE1: PF03999; Metallophos: PF00149; MIF4G: PF02854; Myb_DNA-binding (combined): n/a; NAC: PF01849; NAP: PF00956; NDUF_B8: PF05821; NIPSNAP: PF07978; Nucleoplasmin: PF03066; OS-D: PF03392; p450: PF00067; PABP: PF00658; PARP: PF00644; Peptidase_M17: PF00883; PGAMP: PF07644; PH: PF00169; PI-PLC-X/Y: PF00378/8; Pkinase: PF00069; PTPS: PF01242; Ras: PF00071; Ribophorin_I: PF04597; Ribosomal (combined): n/a; RNA_pol_A_bac: PF01000; RnaseH: PF00075; RRM_1: PF00076; RVT_1: PF00078; SAM_1: PF00536; Sedlin_N: PF04628; Serpin: PF00079; SH2: PF00017; SH3_1: PF00018; SNase: PF00565; Stathmin: PF008310; Synaptobrevin: PF00957; Thioredoxin: PF00085; Thymosin: PF01290; TRAP-gamma: PF07074; TRM: PF02005; TUDOR: PF00567; ubiquitin: PF00240; W2: PF02020; WD40: PF00400; zinc finger (combined): n/a. “Combined” indicates that multiple Pfam accessions are combined.
(TIF)
Sources of proteome sequences from animals with sequenced genomes used for comparison with the G. bimaculatus de novo transcriptome assembly. Sequences were used for ortholog hit ratio analyses (Figure 3) and phylogenetic comparisons of proportion of proteome sequences for which putative G. bimaculatus orthologs were found (Figure 4).
(PDF)
Contribution of the G. bimaculatus transcriptome to GenBank accessions. Sequences of G. bimaculatus developmental genes from GenBank were used as a query to BLAST the de novo transcriptome assembly. Matches in the transcriptome were found among both assembled reads and singletons.
(PDF)
FlyTF transcription factor orthologs identified in the G. bimaculatus transcriptome. BLAST (E-cutoff 1e-5) was used to search the G. bimaculatus transcriptome for orthologs to the transcription factors belonging to the FlyTF database [80].
(PDF)
Selected signaling pathway genes identified in the G. bimaculatus transcriptome. Hit ID indicates if gene hits were found assembled reads (A) or singletons (S). Length (range) indicates the shortest and longest A or S hit sequences for each gene. Query organism was D. melanogaster for all cases.
(PDF)
Selected gametogenesis genes identified in the G. bimaculatus transcriptome. Hit ID indicates if gene hits found were assembled reads (A) or singletons (S). Length (range) indicates the shortest and longest A or S hit sequences for each gene. Groups of hits of a given color indicate transcriptome sequences that mapped to the same overlapping region of the BLAST target (putative SNPs or isoforms); hits of different colors indicate transcriptome sequences that map to different, non-overlapping regions of the BLAST target. Query organism was D. melanogaster for all cases.
(PDF)
Selected developmental process genes identified in the G. bimaculatus de novo transcriptome assembly. Hit ID indicates if gene hits found were assembled reads (A) or singletons (S). Length (range) indicates the shortest and longest A or S hit sequences for each gene. Groups of hits of a given color indicate transcriptome sequences that mapped to the same overlapping region of the BLAST target (putative SNPs or isoforms); hits of different colors indicate transcriptome sequences that map to different, non-overlapping regions of the BLAST target. Query organism was D. melanogaster for all cases.
(PDF)
Acknowledgments
Thanks to Sumihare Noji for encouragement, Evelyn E. Schwager and Franz Kainz for assistance with embryo collection, Lisa Bukovnik for administration of the sequencing, the Harvard Faculty of Arts and Sciences Research Computing group for database discussions, and the Extavour lab for discussions of the results.
Funding Statement
This work was partially supported by Harvard Stem Cell Institute Seed Grant SG-0057-10-00, Ellison Medical Foundation New Scholar Award AG-NS-07010-10, and National Science Foundation (NSF) Grant IOS-0817678 to CE, an NSF Predoctoral Fellowship to BEC, a Fletcher Family award from Bowdoin College to HH, DFG Collaborative Research Centre 680 funds to SR, and JSPS KAKENHI 22124003/22370080/23687033 to TM. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, et al. (2002) The genome sequence of the malaria mosquito Anopheles gambiae . Science 298: 129–149. [DOI] [PubMed] [Google Scholar]
- 2. Brown SJ, Denell R, Gibbs R, Klingler M, Lorenzen M, et al. (2008) The genome of the model beetle and pest Tribolium castaneum . Nature 452: 949–955. [DOI] [PubMed] [Google Scholar]
- 3. Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, et al. (2000) The genome sequence of Drosophila melanogaster . Science (New York, NY) 287: 2185–2195. [DOI] [PubMed] [Google Scholar]
- 4. Myers EW, Sutton GG, Delcher AL, Dew IM, Fasulo DP, et al. (2000) A whole-genome assembly of Drosophila . Science (New York, NY) 287: 2196–2204. [DOI] [PubMed] [Google Scholar]
- 5.Grimaldi D, Engel MS (2005) Evolution of the Insects. Cambridge: Cambridge University Press. 772 p.
- 6. Consortium IAG (2010) Genome sequence of the pea aphid Acyrthosiphon pisum . PLoS Biology 8: e1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huebner E (2007) The Rhodnius Genome Project: The promises and challenges it affords in our understanding of reduviid biology and their role in Chagas' transmission. Comparative Biochemistry and Physiology, Part A 148: S130. [Google Scholar]
- 8. Megy K, Emrich SJ, Lawson D, Campbell D, Dialynas E, et al. (2012) VectorBase: improvements to a bioinformatics resource for invertebrate vector genomics. Nucleic Acids Res 40: D729–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kirkness EF, Haas BJ, Sun W, Braig HR, Perotti MA, et al. (2010) Genome sequences of the human body louse and its primary endosymbiont provide insights into the permanent parasitic lifestyle. Proceedings of the National Academy of Sciences 107: 12168–12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zdobnov EM, Bork P (2007) Quantification of insect genome divergence. Trends in Genetics 23: 16–20. [DOI] [PubMed] [Google Scholar]
- 11. Ewen-Campen B, Shaner N, Panfilio KA, Suzuki Y, Roth S, et al. (2011) The maternal and embryonic transcriptome of the milkweed bug Oncopeltus fasciatus . BMC Genomics 12: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huber F, Moore TE, Loher W, editors (1989) Cricket Behavior and Neurobiology. Ithaca, NY: Cornell University Press. 571 p.
- 13.Hartfelder K, Emlen DJ (2011) Endocrine control of insect polyphenism. In: Gilbert LI, editor. Insect Endocrinology: Elsevier. pp. 464–522.
- 14. Zera AJ (2006) Evolutionary genetics of juvenile hormone and ecdysteroid regulation in Gryllus: a case study in the microevolution of endocrine regulation. Comp Biochem Physiol A Mol Integr Physiol 144: 365–379. [DOI] [PubMed] [Google Scholar]
- 15. Tomioka K, Matsumoto A (2010) A comparative view of insect circadian clock systems. Cellular and Molecular Life Sciences 67: 1397–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Danbara Y, Sakamoto T, Uryu O, Tomioka K (2010) RNA interference of timeless gene does not disrupt circadian locomotor rhythms in the cricket Gryllus bimaculatus . Journal of Insect Physiology 56: 1738–1745. [DOI] [PubMed] [Google Scholar]
- 17. Tomioka K, Uryu O, Kamae Y, Umezaki Y, Yoshii T (2012) Peripheral circadian rhythms and their regulatory mechanism in insects and some other arthropods: a review. Journal of Comparative Physiology B, Biochemical, Systemic, and Environmental Physiology 182: 729–740. [DOI] [PubMed] [Google Scholar]
- 18. Shaw KL, Parsons YM, Lesnick SC (2007) QTL analysis of a rapidly evolving speciation phenotype in the Hawaiian cricket Laupala. Mol Ecol 16: 2879–2892. [DOI] [PubMed] [Google Scholar]
- 19. Howard DJ, Marshall JL, Hampton DD, Britch SC, Draney ML, et al. (2002) The genetics of reproductive isolation: a retrospective and prospective look with comments on ground crickets. American Naturalist 159 Suppl 3: S8–S21. [DOI] [PubMed] [Google Scholar]
- 20. Ellison CK, Wiley C, Shaw KL (2011) The genetics of speciation: genes of small effect underlie sexual isolation in the Hawaiian cricket Laupala. J Evol Biol 24: 1110–1119. [DOI] [PubMed] [Google Scholar]
- 21. Maroja LS, Clark ME, Harrison RG (2008) Wolbachia plays no role in the one-way reproductive incompatibility between the hybridizing field crickets Gryllus firmus and G. pennsylvanicus . Heredity 101: 435–444. [DOI] [PubMed] [Google Scholar]
- 22. Andres JA, Maroja LS, Harrison RG (2008) Searching for candidate speciation genes using a proteomic approach: seminal proteins in field crickets. Proceedings of the Royal Society of London Series B: Biological Sciences 275: 1975–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shaw KL, Lesnick SC (2009) Genomic linkage of male song and female acoustic preference QTL underlying a rapid species radiation. Proceedings of the National Academy of Sciences of the United States of America 106: 9737–9742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fedorka KM, Mousseau TA (2004) Female mating bias results in conflicting sex-specific offspring fitness. Nature 429: 65–67. [DOI] [PubMed] [Google Scholar]
- 25. Bussiere LF, Hunt J, Jennions MD, Brooks R (2006) Sexual conflict and cryptic female choice in the black field cricket, Teleogryllus commodus . Evolution 60: 792–800. [PubMed] [Google Scholar]
- 26. Bando T, Hamada Y, Kurita K, Nakamura T, Mito T, et al. (2011) Lowfat, a mammalian Lix1 homologue, regulates leg size and growth under the Dachsous/Fat signaling pathway during tissue regeneration. Developmental Dynamics 240: 1440–1453. [DOI] [PubMed] [Google Scholar]
- 27. Lynch JA, Peel AD, Drechsler A, Averof M, Roth S (2010) EGF signaling and the origin of axial polarity among the insects. Current Biology 20: 1042–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kainz F, Ewen-Campen B, Akam M, Extavour CG (2011) Delta/Notch signalling is not required for segment generation in the basally branching insect Gryllus bimaculatus . Development 138: 5015–5026. [DOI] [PubMed] [Google Scholar]
- 29. Mito T, Shinmyo Y, Kurita K, Nakamura T, Ohuchi H, et al. (2011) Ancestral functions of Delta/Notch signaling in the formation of body and leg segments in the cricket Gryllus bimaculatus . Development 138: 3823–3833. [DOI] [PubMed] [Google Scholar]
- 30. Mito T, Kobayashi C, Sarashina I, Zhang H, Shinahara W, et al. (2007) even-skipped has gap-like, pair-rule-like, and segmental functions in the cricket Gryllus bimaculatus, a basal, intermediate germ insect (Orthoptera). Developmental Biology 303: 202–213. [DOI] [PubMed] [Google Scholar]
- 31. Mito T, Inoue Y, Kimura S, Miyawaki K, Niwa N, et al. (2002) Involvement of hedgehog, wingless, and dpp in the initiation of proximodistal axis formation during the regeneration of insect legs, a verification of the modified boundary model. Mechanisms of Development 114: 27–35. [DOI] [PubMed] [Google Scholar]
- 32. Nakamura T, Mito T, Bando T, Ohuchi H, Noji S (2007) Dissecting insect leg regeneration through RNA interference. Cellular and Molecular Life Sciences 65: 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nakamura T, Mito T, Miyawaki K, Ohuchi H, Noji S (2008) EGFR signaling is required for re-establishing the proximodistal axis during distal leg regeneration in the cricket Gryllus bimaculatus nymph. Developmental Biology 319: 46–55. [DOI] [PubMed] [Google Scholar]
- 34. Feuillet C, Leach JE, Rogers J, Schnable PS, Eversole K (2011) Crop genome sequencing: lessons and rationales. Trends in Plant Science 16: 77–88. [DOI] [PubMed] [Google Scholar]
- 35. Gregory TR (2005) Synergy between sequence and size in large-scale genomics. Nature Reviews Genetics 6: 699–708. [DOI] [PubMed] [Google Scholar]
- 36. Schatz MC, Delcher AL, Salzberg SL (2010) Assembly of large genomes using second-generation sequencing. Genome Research 20: 1165–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hanrahan SJ, Johnston JS (2011) New genome size estimates of 134 species of arthropods. Chromosome Res 19: 809–823. [DOI] [PubMed] [Google Scholar]
- 38. Takagi A, Kurita K, Terasawa T, Nakamura T, Bando T, et al. (2012) Functional analysis of the role of eyes absent and sine oculis in the developing eye of the cricket Gryllus bimaculatus . Development, Growth and Differentiation 54: 227–240. [DOI] [PubMed] [Google Scholar]
- 39. Nakamura T, Yoshizaki M, Ogawa S, Okamoto H, Shinmyo Y, et al. (2010) Imaging of transgenic cricket embryos reveals cell movements consistent with a syncytial patterning mechanism. Current Biology 20: 1641–1647. [DOI] [PubMed] [Google Scholar]
- 40. Meier T, Chabaud F, Reichert H (1991) Homologous patterns in the embryonic development of the peripheral nervous system in the grasshopper Schistocerca gregaria and the fly Drosophila melanogaster . Development 112: 241–253. [DOI] [PubMed] [Google Scholar]
- 41. Meier T, Reichert H (1991) Serially homologous development of the peripheral nervous system in the mouthparts of the grasshopper. Journal of Comparative Neurology 305: 201–214. [DOI] [PubMed] [Google Scholar]
- 42. Meier T, Reichert H (1989) Embryonic Development and Evolutionary Origin of the Orthopteran Auditory Organs. Journal of Neurobiology 21: 592–610. [DOI] [PubMed] [Google Scholar]
- 43. Takahashi T, Hamada A, Miyawaki K, Matsumoto Y, Mito T, et al. (2009) Systemic RNA interference for the study of learning and memory in an insect. Journal of Neuroscience Methods 179: 9–15. [DOI] [PubMed] [Google Scholar]
- 44. Miyawaki K, Mito T, Sarashina I, Zhang H, Shinmyo Y, et al. (2004) Involvement of Wingless/Armadillo signaling in the posterior sequential segmentation in the cricket, Gryllus bimaculatus (Orthoptera), as revealed by RNAi analysis. Mechanisms of Development 121: 119–130. [DOI] [PubMed] [Google Scholar]
- 45. Watanabe T, Ochiai H, Sakuma T, HOrch HW, Hamaguchi N, et al. (2012) Non-transgenic genome modifications in a hemimetabolous insect using zinc-finger and TAL effector nucleases. Nature Communications in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Inoue Y, Niwa N, Mito T, Ohuchi H, Yoshioka H, et al. (2002) Expression patterns of hedgehog, wingless, and decapentaplegic during gut formation of Gryllus bimaculatus (cricket). Mechanisms of Development 110: 245–248. [DOI] [PubMed] [Google Scholar]
- 47. Bando T, Ishimaru Y, Kida T, Hamada Y, Matsuoka Y, et al. (2013) Analysis of RNA-Seq data reveals involvement of JAK/STAT signalling during leg regeneration in the cricket Gryllus bimaculatus. Development 140: 959–964. [DOI] [PubMed] [Google Scholar]
- 48. Kumar S, Blaxter ML (2010) Comparing de novo assemblers for 454 transcriptome data. BMC Genomics 11: 571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zeng V, Villanueva KE, Ewen-Campen B, Alwes F, Browne WE, et al. (2011) De novo assembly and characterization of a maternal and developmental transcriptome for the emerging model crustacean Parhyale hawaiensis . BMC Genomics 12: 581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M (2010) KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Research 38: D355–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zeng V, Extavour CG (2012) ASGARD: an open-access database of annotated transcriptomes for emerging model arthropod species. Database 2012: bas048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Iseli C, Jongeneel CV, Bucher P (1999) ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proceedings of the International Conference on Intelligent Systems for Molecular Biology 138–148. [PubMed] [Google Scholar]
- 53. Zdobnov EM, Apweiler R (2001) InterProScan–an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17: 847–848. [DOI] [PubMed] [Google Scholar]
- 54. Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, et al. (2005) InterProScan: protein domains identifier. Nucleic Acids Research 33: W116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bru C, Courcelle E, Carrere S, Beausse Y, Dalmar S, et al. (2005) The ProDom database of protein domain families: more emphasis on 3D. Nucleic Acids Res 33: D212–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Attwood TK, Bradley P, Flower DR, Gaulton A, Maudling N, et al. (2003) PRINTS and its automatic supplement, prePRINTS. Nucleic Acids Res 31: 400–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Letunic I, Goodstadt L, Dickens NJ, Doerks T, Schultz J, et al. (2002) Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res 30: 242–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Haft DH, Selengut JD, White O (2003) The TIGRFAMs database of protein families. Nucleic Acids Res 31: 371–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bateman A, Coin L, Durbin R, Finn RD, Hollich V, et al. (2004) The Pfam protein families database. Nucleic Acids Res 32: D138–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hulo N, Sigrist CJ, Le Saux V, Langendijk-Genevaux PS, Bordoli L, et al. (2004) Recent improvements to the PROSITE database. Nucleic Acids Res 32: D134–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wu CH, Nikolskaya A, Huang H, Yeh LS, Natale DA, et al. (2004) PIRSF: family classification system at the Protein Information Resource. Nucleic Acids Res 32: D112–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gough J, Karplus K, Hughey R, Chothia C (2001) Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J Mol Biol 313: 903–919. [DOI] [PubMed] [Google Scholar]
- 63. Pearl FM, Lee D, Bray JE, Sillitoe I, Todd AE, et al. (2000) Assigning genomic sequences to CATH. Nucleic Acids Research 28: 277–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mi H, Lazareva-Ulitsky B, Loo R, Kejariwal A, Vandergriff J, et al. (2005) The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Research 33: D284–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. Journal of Molecular Biology 340: 783–795. [DOI] [PubMed] [Google Scholar]
- 66. Sonnhammer EL, von Heijne G, Krogh A (1998) A hidden Markov model for predicting transmembrane helices in protein sequences. Proceedings of the International Conference on Intelligent Systems for Molecular Biology 6: 175–182. [PubMed] [Google Scholar]
- 67. Welch BL (1947) The generalisation of “student's” problems when several different population variances are involved. Biometrika 34: 28–35. [DOI] [PubMed] [Google Scholar]
- 68.Roche (2011) 454 Sequencing System Software Manual version 2.6. Part C: GS De Novo Assembler, GS Reference Mapper, SFF Tools: Roche.
- 69. Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Research 8: 175–185. [DOI] [PubMed] [Google Scholar]
- 70. Ewing B, Green P (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Research 8: 186–194. [PubMed] [Google Scholar]
- 71. O'Neil ST, Dzurisin JDK, Carmichael RD, Lobo NF, Emrich SJ, et al. (2010) Population-level transcriptome sequencing of nonmodel organisms Erynnis propertius and Papilio zelicaon . BMC Genomics 11: 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chen S, Yang P, Jiang F, Wei Y, Ma Z, et al. (2010) De novo analysis of transcriptome dynamics in the migratory locust during the development of phase traits. PLoS ONE 5: e15633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ma Z, Yu J, Kang L (2006) LocustDB: a relational database for the transcriptome and biology of the migratory locust (Locusta migratoria). BMC Genomics 7: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kang L, Chen X, Zhou Y, Liu B, Zheng W, et al. (2004) The analysis of large-scale gene expression correlated to the phase changes of the migratory locust. Proc Natl Acad Sci U S A 101: 17611–17615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Danley PD, Mullen SP, Liu F, Nene V, Quackenbush J, et al. (2007) A cricket Gene Index: a genomic resource for studying neurobiology, speciation, and molecular evolution. BMC Genomics 8: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Badisco L, Huybrechts J, Simonet G, Verlinden H, Marchal E, et al. (2011) Transcriptome analysis of the desert locust central nervous system: production and annotation of a Schistocerca gregaria EST database. PLoS ONE 6: e17274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lu HL, Tanguy S, Rispe C, Gauthier JP, Walsh T, et al. (2011) Expansion of genes encoding piRNA-associated Argonaute proteins in the pea aphid: diversification of expression profiles in different plastic morphs. PLoS ONE 6: e28051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lin G-w, Chang C-c (2009) Cloning and developmental characterization of four vasa genes, Apvasa1–4, in the parthenogenetic and viviparous pea aphid Acyrthosiphon pisum . Mechanisms of Development 126: S252 215–P017. [Google Scholar]
- 79. Shigenobu S, Bickel RD, Brisson JA, Butts T, Chang C-C, et al. (2010) Comprehensive survey of developmental genes in the pea aphid, Acyrthosiphon pisum: frequent lineage-specific duplications and losses of developmental genes. Insect Molecular Biology 19 Suppl 2: 47–62. [DOI] [PubMed] [Google Scholar]
- 80. Pfreundt U, James DP, Tweedie S, Wilson D, Teichmann SA, et al. (2010) FlyTF: improved annotation and enhanced functionality of the Drosophila transcription factor database. Nucleic Acids Research 38: D443–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kopan R, Ilagan MX (2009) The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137: 216–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kitisin K, Saha T, Blake T, Golestaneh N, Deng M, et al. (2007) Tgf-Beta signaling in development. Science STKE 2007: cm1. [DOI] [PubMed] [Google Scholar]
- 83. Nusse R, Varmus H (2012) Three decades of Wnts: a personal perspective on how a scientific field developed. EMBO Journal 31: 2670–2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Schindler C, Levy DE, Decker T (2007) JAK-STAT signaling: from interferons to cytokines. J Biol Chem 282: 20059–20063. [DOI] [PubMed] [Google Scholar]
- 85. Shaul YD, Seger R (2007) The MEK/ERK cascade: from signaling specificity to diverse functions. Biochimica et Biophysica Acta 1773: 1213–1226. [DOI] [PubMed] [Google Scholar]
- 86. Ingham PW, Nakano Y, Seger C (2011) Mechanisms and functions of Hedgehog signalling across the metazoa. Nature Reviews Genetetics 12: 393–406. [DOI] [PubMed] [Google Scholar]
- 87. Tordjmann T (2011) Hippo signalling: Liver size regulation and beyond. Clinics and Research in Hepatology and Gastroenterology [DOI] [PubMed] [Google Scholar]
- 88. Liu D, Finley RL Jr (2010) Cyclin Y is a novel conserved cyclin essential for development in Drosophila . Genetics 184: 1025–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Yandell M, Mungall CJ, Smith C, Prochnik S, Kaminker J, et al. (2006) Large-scale trends in the evolution of gene structures within 11 animal genomes. PLoS Comput Biol 2: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Chen F, Mackey AJ, Vermunt JK, Roos DS (2007) Assessing performance of orthology detection strategies applied to eukaryotic genomes. PLoS One 2: e383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Attwood TK, Coletta A, Muirhead G, Pavlopoulou A, Philippou PB, et al. (2012) The PRINTS database: a fine-grained protein sequence annotation and analysis resource–its status in 2012. Database 2012: bas019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Jaroszewski L, Li Z, Krishna SS, Bakolitsa C, Wooley J, et al. (2009) Exploration of uncharted regions of the protein universe. PLoS Biology 7: e1000205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ma C, Yang P, Jiang F, Chapuis MP, Shali Y, et al. (2012) Mitochondrial genomes reveal the global phylogeography and dispersal routes of the migratory locust. Molecular Ecology [DOI] [PubMed] [Google Scholar]
- 94. Ceccato P, Cressman K, Giannini A, Trzaska S (2007) The Desert Locust Upsurge in West Africa (2003–2005): Information on The Desert Locust Early Warning System, and The Prospects for Seasonal Climate Forecasting. International Journal of Pest Management 53: 7–13. [Google Scholar]
- 95. Lomer CJ, Bateman RP, Johnson DL, Langewald J, Thomas M (2001) Biological control of locusts and grasshoppers. Annual Review of Entomology 46: 667–702. [DOI] [PubMed] [Google Scholar]
- 96. Nolte DJ (1963) A pheremone for melanization of locusts. Nature 200: 660–661. [Google Scholar]
- 97. Shaw KL, Parsons YM, Lesnick SC (2007) QTL analysis of a rapidly evolving speciation phenotype in the Hawaiian cricket Laupala . Molecular Ecology 16: 2879–2892. [DOI] [PubMed] [Google Scholar]
- 98. Ellison CK, Wiley C, Shaw KL (2011) The genetics of speciation: genes of small effect underlie sexual isolation in the Hawaiian cricket Laupala . Journal of Evolutionary Biology 24: 1110–1119. [DOI] [PubMed] [Google Scholar]
- 99. Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, et al. (2007) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 35: D5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Büning J (1994) The Insect Ovary: ultrastructure, previtellogenic growth and evolution. London: Chapman and Hall. 400 p.
- 101.Kainz F (2009) Cell communication during patterning: Notch and FGF signalling in Gryllus bimaculatus and their role in segmentation [PhD]. Cambridge: University of Cambridge. 161 p.
- 102. Bateman A, Coin L, Durbin R, Finn RD, Hollich V, et al. (2004) The Pfam protein families database. Nucleic Acids Research 32: D138–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Letunic I, Goodstadt L, Dickens NJ, Doerks T, Schultz J, et al. (2002) Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Research 30: 242–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of read lengths from de novo assembly of the G. bimaculatus transcriptome. (A) Distribution of read lengths before (black) and after (blue) trimming to remove low quality reads (see text for details). (B) Distribution of trimmed read lengths before (blue) and after (red) assembly with Newbler v2.5. The assembly yielded assembled reads of over 10,000 bp. (C) Distribution of read lengths of the shortest assembled (red) and raw (blue) reads.
(TIF)
Schematics of conserved metazoan signal transduction pathways showing components identified in the G. bimaculatus transcriptome. BLAST was used to search for signaling pathway genes in the G. bimaculatus transcriptome (see Table S4); genes with newly identified putative orthologs are indicated in red. Genes outlined in grey with grey typeface indicate genes without D. melanogaster homologs. Pathway schematics are modified from KEGG pathway model images (http://www.genome.jp/kegg/kegg1.html). (A) Notch pathway. (B) TGF-βeta pathway. (C) Wnt pathway. (D) Janus Kinase (JAK)-signal transducer and activator of transcription (STAT) pathway. (E) Mitogen-activated protein Kinase (MAPK) pathway.
(TIF)
Complete protein domain composition of G. bimaculatus transcriptome sequences with highest similarity to Laupala kohalensis or Locusta migratoria sequences. Relative proportions of all protein domains coded by G. bimaculatus transcriptome sequences with significant similarity to sequences from L. kohalensis (A), L. migratoria (B), or sequences from nr (C). Protein domain nomenclature from Pfam [102] and SMART [103] databases as follows: 5_nucleotid_C: PF2872; Abhydrolase_1: PF00561; adh_short: PF00106; ADK: OF00406; AdoHcyase_NAD: PF00670; Amidohydro_1: PF01979; Ank: PF00023; AP_endonuc_2_N: PF07582; Asparaginase_2: PF01112; ATP-gua_Ptrans/N: PF02807; BAH: PF01426; BTB/POZ: PF00651; Btz: SM 01044; bZIP_2: PF07716; C2: PF00168; CBM_14: PF01607; COesterase: PF00135; Cyclin_N: PF00134; Cys_Met_Meta_PP: PF01053; DEAD: PF00270; DUF (combined): n/a; EFG domains (combined): n/a; efhand/like: PF09279; eIF-5_eIF-2B: PF01873; ELM2: PF01448; ELO: PF01151; EMP70: PF02990; ETF_alpha: PF00766; Exo_endo_phos: PF03372; F-box: PF00646; fn3: PF00041; G-patch: PF01858; GATA: PF00320; GCV_H: PF01597; GHMP_kinases_N: PF00288; Glyco_hydro (combined): n/a; GTP_EFTU domains: PF00009; HECT: PF00632; Hemocyanin_N: PF03722; HSP90: PF00183; IF-2B: PF01008; IPP-2: PF04979; JHBP: PF06585; Laps: PF10169; Ldl_recept_a: PF00057; Lectin_C: PF00059; LRR_1: PF00560; MA3: PF00560; MADF_DNA_bdg: PF10545; MAP65_ASE1: PF03999; Metallophos: PF00149; MIF4G: PF02854; Myb_DNA-binding (combined): n/a; NAC: PF01849; NAP: PF00956; NDUF_B8: PF05821; NIPSNAP: PF07978; Nucleoplasmin: PF03066; OS-D: PF03392; p450: PF00067; PABP: PF00658; PARP: PF00644; Peptidase_M17: PF00883; PGAMP: PF07644; PH: PF00169; PI-PLC-X/Y: PF00378/8; Pkinase: PF00069; PTPS: PF01242; Ras: PF00071; Ribophorin_I: PF04597; Ribosomal (combined): n/a; RNA_pol_A_bac: PF01000; RnaseH: PF00075; RRM_1: PF00076; RVT_1: PF00078; SAM_1: PF00536; Sedlin_N: PF04628; Serpin: PF00079; SH2: PF00017; SH3_1: PF00018; SNase: PF00565; Stathmin: PF008310; Synaptobrevin: PF00957; Thioredoxin: PF00085; Thymosin: PF01290; TRAP-gamma: PF07074; TRM: PF02005; TUDOR: PF00567; ubiquitin: PF00240; W2: PF02020; WD40: PF00400; zinc finger (combined): n/a. “Combined” indicates that multiple Pfam accessions are combined.
(TIF)
Sources of proteome sequences from animals with sequenced genomes used for comparison with the G. bimaculatus de novo transcriptome assembly. Sequences were used for ortholog hit ratio analyses (Figure 3) and phylogenetic comparisons of proportion of proteome sequences for which putative G. bimaculatus orthologs were found (Figure 4).
(PDF)
Contribution of the G. bimaculatus transcriptome to GenBank accessions. Sequences of G. bimaculatus developmental genes from GenBank were used as a query to BLAST the de novo transcriptome assembly. Matches in the transcriptome were found among both assembled reads and singletons.
(PDF)
FlyTF transcription factor orthologs identified in the G. bimaculatus transcriptome. BLAST (E-cutoff 1e-5) was used to search the G. bimaculatus transcriptome for orthologs to the transcription factors belonging to the FlyTF database [80].
(PDF)
Selected signaling pathway genes identified in the G. bimaculatus transcriptome. Hit ID indicates if gene hits were found assembled reads (A) or singletons (S). Length (range) indicates the shortest and longest A or S hit sequences for each gene. Query organism was D. melanogaster for all cases.
(PDF)
Selected gametogenesis genes identified in the G. bimaculatus transcriptome. Hit ID indicates if gene hits found were assembled reads (A) or singletons (S). Length (range) indicates the shortest and longest A or S hit sequences for each gene. Groups of hits of a given color indicate transcriptome sequences that mapped to the same overlapping region of the BLAST target (putative SNPs or isoforms); hits of different colors indicate transcriptome sequences that map to different, non-overlapping regions of the BLAST target. Query organism was D. melanogaster for all cases.
(PDF)
Selected developmental process genes identified in the G. bimaculatus de novo transcriptome assembly. Hit ID indicates if gene hits found were assembled reads (A) or singletons (S). Length (range) indicates the shortest and longest A or S hit sequences for each gene. Groups of hits of a given color indicate transcriptome sequences that mapped to the same overlapping region of the BLAST target (putative SNPs or isoforms); hits of different colors indicate transcriptome sequences that map to different, non-overlapping regions of the BLAST target. Query organism was D. melanogaster for all cases.
(PDF)