Summary
Although they possess a well-characterized ability to porate the bacterial membrane, emerging research suggests that cationic antimicrobial peptides (CAPs) can influence pathogen behavior at levels that are sub-lethal. In this study, we investigated the interaction of polymyxin B and human neutrophil peptide (HNP-1) with the human pathogen Streptococcus pyogenes. At sub-lethal concentrations, these CAPs preferentially targeted the ExPortal, a unique microdomain of the S. pyogenes membrane, specialized for protein secretion and processing. A consequence of this interaction was the disruption of ExPortal organization and a redistribution of ExPortal components into the peripheral membrane. Redistribution was associated with inhibition of secretion of certain toxins, including the SpeB cysteine protease and the Streptolysin O (SLO) cytolysin, but not SIC, a protein that protects S. pyogenes from CAPs. These data suggest a novel function for CAPs in targeting the ExPortal and interfering with secretion of factors required for infection and survival. This mechanism may prove valuable for the design of new types of antimicrobial agents to combat the emergence of antibiotic-resistant pathogens.
Keywords: Streptococcus pyogenes, cationic antimicrobial peptides, polymyxin B, human neutrophil peptide 1, membrane lipid microdomain
Introduction
Cationic antimicrobial peptides (CAPs) are gene-encoded antibacterial peptides produced by nearly all known life-forms. A group of these, the defensins, are expressed by neutrophils and epithelial cells of humans and other mammals, functioning both as broad-spectrum microbicides and as modulators of the immune response (Mestecky, 2005). It is generally accepted that the antibacterial activity of CAPs involves interactions of their charged and hydrophobic residues with the hydrophilic charged head groups and the fatty acyl chains of phospholipids in the bacterial membrane. The consequence of this interaction is the destruction of membrane permeability and bacterial cell death (Boman, 2003, Hancock & Sahl, 2006). However, more recent studies have suggested that many of these peptides may have multiple targets and/or much more complicated mechanisms of action (reviewed by (Hale & Hancock, 2007)). In particular, one aspect of the peptide-pathogen interaction that is not often examined is the manner in which CAPs act on their bacterial targets at levels below their minimum inhibitory concentrations.
Interaction with CAPs plays an important role in host-pathogen interactions for infections caused by Streptococcus pyogenes (group A streptococcus). This Gram-positive pathogen can cause numerous diseases in humans that range from largely superficial infection of the skin and mucous membranes (impetigo, pharyngitis), to highly invasive and life-threatening diseases (necrotizing fasciitis), as well as, serious post-infection sequelae (rheumatic fever, glomerulonephritis, reviewed in (Cunningham, 2000)). Recent studies have shown that multiple human CAPs are lethal to S. pyogenes in vitro (Harder et al., 2001, Braff et al., 2005), and that the CAP cathelicicin LL-37 is highly expressed in severe soft tissue infection in humans at sites coinciding with high tissue burdens of S. pyogenes (Johansson et al., 2008). Several studies have highlighted the important role of CAPs in innate immune defenses against S. pyogenes. These include the observation that soft tissue infection by S. pyogenes in mice deficient in a major CAP of the cathelicidin family (CRAMP) was significantly exacerbated (Nizet et al., 2001) and that overexpression of a cathelicidin in murine skin provides enhanced protection (Lee et al., 2005). It is not surprising then, that much recent research has been directed at identifying streptococcal factors that subvert the lethal effects of CAP activity (Kristian et al., 2005, Gryllos et al., 2008, Frick et al., 2003).
The signature ability of CAPs to bind to the cytoplasmic membrane, offers numerous targets whose functions could be affected at sub-inhibitory concentrations. The cytoplasmic membrane is of particular importance to S. pyogenes, as its ability to cause disease is dependent on the secretion of an extensive network of virulence proteins (Kwinn & Nizet, 2007). Lacking other known secretion systems, these virulence proteins are exported by the general secretory (Sec) pathway, which is highly conserved between Gram-positive and –negative bacteria and eukaryotes (for a review see (de Keyzer et al., 2003)). However, unlike these latter two classes, Gram-positive bacteria lack a specialized cellular compartment for folding proteins following their translocation across the membrane by the Sec system. A solution to this problem used by S. pyogenes and several other species of Gram-positive cocci, is to cluster the Sec translocons at a defined microdomain of the cytoplasmic membrane that has been termed the ExPortal (Rosch & Caparon, 2004, Rosch & Caparon, 2005, Kline et al., 2009, Campo et al., 2004, Hu et al., 2008). The ExPortal is also highly enriched with accessory factors for protein biogenesis, including sortases, and HtrA. The former are involved in the covalent attachment of proteins to the cell wall, while the latter is a multi-function protease and chaperone that is required for the biogenesis of the active form of the SpeB protease. These data suggest that one function of the ExPortal is to spatially couple secretion with protein maturation (Rosch & Caparon, 2004, Rosch & Caparon, 2005, Rosch et al., 2007, Kline et al., 2009, Raz & Fischetti, 2008). This is supported by the observation that mutations causing the mis-localization of HtrA or sortase C outside of the ExPortal microdomain result in a highly reduced efficiency for maturation of secreted proteases and pili, in S. pyogenes and Enterococcus faecalis, respectively (Rosch & Caparon, 2005, Kline et al., 2009).
Due to their cationic nature, CAPs predominantly bind to anionic phospholipids in bacterial membranes (Hancock & Sahl, 2006). Of interest, the ExPortal of S. pyogenes has been shown to have an asymmetric lipid content enriched in anionic phospholipids (Rosch et al., 2007) that may contribute to the preferential retention of certain proteins at the ExPortal vs. the peripheral membrane (Kline et al., 2009). These observations suggest that the ExPortal may be uniquely sensitive to the action of CAPs. In the present study, we examined the interaction between several CAPs and S. pyogenes and report that CAPs preferentially interact with the ExPortal when examined at sub-lethal concentrations and that this results in a re-distribution of ExPortal components into the peripheral membrane. Furthermore, this disruption is associated with an inhibition of secretion of the SpeB cysteine protease and the Streptolysin O (SLO) cytolysin.
Results
Polymyxin B binds to a single, unique site on the S. pyogenes membrane
To test the hypothesis that CAPs may interact with the ExPortal, we examined how the CAP polymyxin B interacts with the S. pyogenes membrane at sub-lethal concentrations. Readily available and well-characterized in its ability to bind anionic lipids (Katz et al., 2003), polymyxin B is a cyclic CAP antibiotic that is highly active against Gram-negative, but not most Gram-positive bacteria (Hancock, 2001). However, S. pyogenes is an exception and is highly susceptible to polymyxin B (see below), which likely reflects that unlike many other Gram-positive species its genome does not contain mprF (Ferretti et al., 2001), which encodes an enzyme that modifies the negative charge of phosphatidyl glycerol via lysinylation to reduce its affinity for binding CAPs (Peschel et al., 2001, Thedieck et al., 2006). The interaction of polymyxin B with S. pyogenes was therefore examined using a biotin-labeled derivative of polymyxin B to treat cultures of the M1 serotype S. pyogenes strain SF370 in a “challenge assay.” In this assay (see “Experimental Procedures”), polymyxin B was added to cultures in the late logarithmic phase of growth (time = 0 h) and samples harvested for analysis after cultures had entered stationary phase (Fig. S3). The concentrations of polymyxin B used had no effect on viability in this assay and did not result in significant poration of membranes (see “Experimental Procedures” and Fig. S1). Bound polymyxin B was detected using a streptavidin-gold conjugate and examined by electron microscopy. This analysis revealed that rather than a uniform pattern of circumferential staining, cells consistently exhibited a single intense focus of gold particles at a discrete location adjacent to the membrane (Fig. 1A). An identical pattern was observed following treatment of the M14 serotype S. pyogenes strain HSC5 (data not shown) and minimal staining with the streptavidin conjugate was observed in the absence of treatment with biotin-labeled polymyxin B (data not shown).
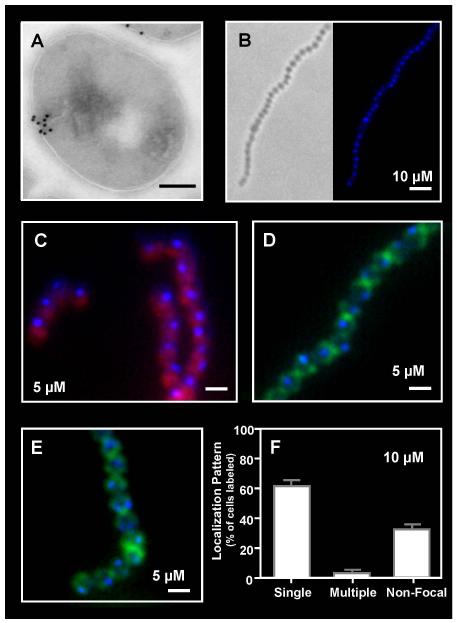
Figure 1. Focal binding of Polymyxin B to the S. pyogenes surface.
The distribution of polymyxin B on the surface of S. pyogenes SF370 following sub-lethal challenge was revealed: (A) by treatment with biotinylated polymyxin B (1:10,000) and immunogold electron microscopy using staining with a streptavidin-gold conjugate (scale bar = 200nm) and (B, C, D, E) by fluorescent microscopy following challenge with dansyl-polymyxin B alone (B) at the concentration indicated in the Figure (scale bar = 1μm) or in cells counterstained with Nile Red (C), fluorescent vancomycin (D) or wheat germ agglutinin Alexa Fluor 488 conjugate (E). Staining patterns following challenge with dansyl-polymyxin B were quantitated as described in the Experimental Procedures (F). Data represents the mean and standard error of the mean (SEM) derived from at least 3 independent experiments and examination of a minimum of 1000 stained cells. The number of cells with a single focus was significantly higher than any other staining pattern (P < 0.0001).
Polymyxin B targets the ExPortal
The pattern of polymyxin B binding was then examined following treatment with a fluorescent derivative of polymyxin B (dansyl-polymyxin B) at sub-inhibitory concentrations. Treatment with ≤60 μM polymyxin B under our challenge assay conditions (see “Experimental Procedures”) did not alter the viability of cultures at the end of the period of incubation as determined by enumeration of colony forming units (Fig. S1A). Examination of cultures treated with a fluorescent probe that is excluded by intact membranes (Live/Dead®) confirmed the viability of polymyxin B treated cultures and demonstrated that most cells had membranes that were not porated (Fig. S1B). Examination by fluorescent microscopy revealed that when treated at concentrations ≤15 μM the fluorescent CAP typically localized to a single discrete site of the membrane (Fig. 1B, C, D, E). Some cells with staining at multiple foci or with a more diffuse distribution around the circumference of the cell were also observed (data not shown). However, when examined quantitatively, the number of cells with single foci exceeded 60% and was significantly higher than that observed for any other staining pattern (P<0.0001, Fig. 1F). To assess whether polymyxin B was targeting the ExPortal, co-staining was conducted to determine if these foci corresponded to the site of secretion of the SpeB cysteine protease, a signature feature of the ExPortal (Rosch & Caparon, 2004, Rosch & Caparon, 2005). This was conducted using an assay that employs a protease substrate that is intramolecularly quenched but becomes active when cleaved to visualize the site of secretion of active SpeB protease in cells that have been embedded in agarose (Rosch & Caparon, 2004, Scott et al., 2001). When a sub-inhibitory concentration of dansyl-polymyxin B (5 μM or 10 μM) was included in this assay about 50% of the cells showed staining with both reagents. However, when co-stained (Fig. 2), the coincidence of the membrane site recognized by dansyl-polymyxin B and the site of SpeB secretion approached 100% (out of at least 200 co-stained cells observed).
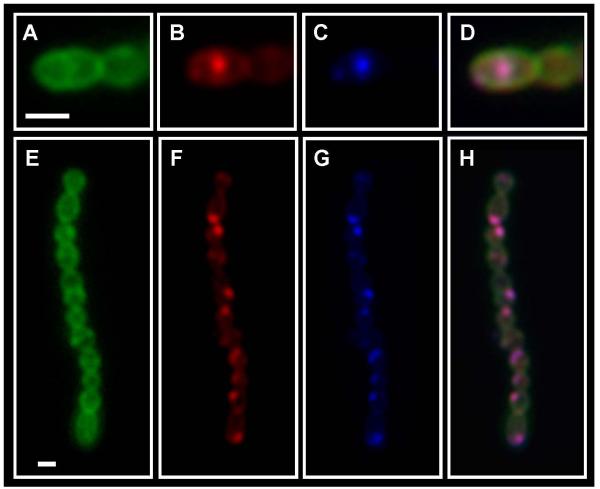
Figure 2. The site of polymyxin B binding is coincident with the site of SpeB secretion.
Cells of S. pyogenes SF370 were challenged with dansyl-polymyxin B (10 μM), fluorescent vancomycin stained to visualize cell wall and subjected to the red protease assay, which monitors cleavage of BODIPY TR-X-casein by the SpeB protease. Stained cells were then examined by fluorescent microscopy. Panels are as follows: (A, E) fluorescent vancomycin, (B, F) BODIPY TR-X-casein, (C, G) Dansyl-polymyxin B and (D, H) Merge of Panels A, B and C. Scale bar = 1μm.
Polymyxin B disrupts ExPortal Lipids
The data above suggest that at sub-lethal concentrations, polymyxin B preferentially targets the ExPortal. The consequence of this interaction was then examined using higher, but still sub-lethal concentrations of the CAP. ExPortal integrity was assessed by staining with 10-nonyl acridine orange (NAO), a fluorescent membrane probe that preferentially binds anionic phospholipids (Rosch et al., 2007, Mileykovskaya & Dowhan, 2000). In the absence of polymyxin B, NAO stained cells at single foci (Fig. 3A), which were previously shown to be co-incident with the ExPortal (Rosch et al., 2007). Following treatment with 30 μM polymyxin B, most cells had either multiple foci or a more diffuse staining pattern (Fig. 3B), while this latter pattern predominated following treatment with 60 μM polymyxin B (Fig. 3C). This result suggested that the CAP was altering the organization of ExPortal-associated anionic phospholipids. Consistent with this, treatment with a higher, but still sub-lethal concentration of biotin-labeled polymyxin B resulted in cells stained at multiple foci or more diffusely around the membrane when examined by electron microscopy, rather than staining at single foci as was observed at lower concentrations (compare Fig. 3E with Fig. 1A,). Similarly, treatment with a higher concentration of dansyl-polymxin B resulted in a majority of cells stained at multiple foci (compare Fig. 3F, G to Fig. 1B, D, E). This shift was examined quantitatively (Fig. 3H), revealing that the number of cells stained at multiple foci was significantly higher than other staining patterns observed. Stressing membranes by subjecting cells to a condition known to induce the heat shock response (42°C, 30 mins.; (Rosch & Caparon, 2005)) did not alter the focal pattern of NAO staining (Fig. 3; compare panels 3D, 3A). Similarly, heat shock did not alter the focal binding pattern of a low sub-lethal concentration of bodipy-labeled polymyxin B (10 μM, Fig. S2C,D), which contrasts with the disruption observed at a higher sub-lethal concentration (45 μM, Fig. S2E,F). Thus, these data suggest that disruption of the ExPortal-associated anionic lipid domain is specific to high sub-lethal concentrations of polymyxin B and does not result from a general membrane stress response.
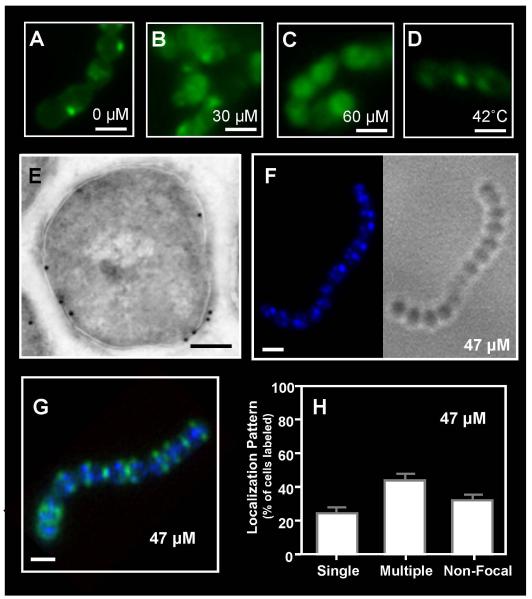
Figure 3. Sub-lethal challenge with polymyxin B alters the distribution of anionic membrane lipids.
Cultures of S. pyogenes SF370 were stained with NAO following challenge with 0 μM (A), 30 μM (B) or 60 μM (C) polymyxin B or heat shock at 42°C (D) and examined by fluorescent microscopy (scale bar = 1μm). The distribution of polymyxin B following higher, but still sub-lethal challenge, was revealed by treatment with biotinylated polymyxin B (1:500) and immunogold electron microscopy using staining with a streptavidin-gold conjugate (scale bar = 200nm) (E) and by fluorescent microscopy following challenge with dansyl-polymyxin B alone at the concentration indicated in the Figure (scale bar = 1μm) (F) or costained with fluorescent vancomycin (G). Staining patterns following challenge were quantitated as described previously (H). Data represents the mean and SEM derived from at least 3 independent experiments and examination of a minimum of 1000 stained cells. The number of cells with multiple foci was significantly higher than any other staining pattern (P < 0.0001) at this polymyxin B concentration.
Polymyxin B disrupts ExPortal Organization
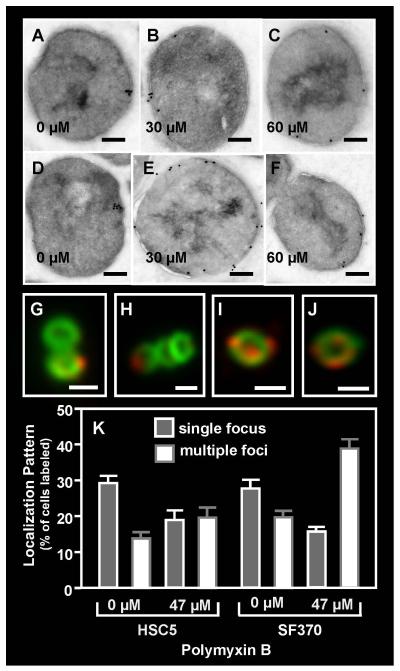
There is evidence to suggest that the asymmetric lipid content of the ExPortal contributes to the retention of some ExPortal-associated membrane proteins (Kline et al., 2009). This suggests that the polymyxin B-mediated disruption of ExPortal lipid structure observed above could result in an overall disruption of ExPortal organization. To test this, the distribution of several proteins known to be enriched at the ExPortal was examined in cells of strain SF370 exposed to a sub-lethal concentration of polymyxin B. The concentrations tested (30-60 μM) were those that produced a high frequency of cells with multiple NAO- or polymyxin B-stained foci (see above). Consistent with prior studies (Rosch & Caparon, 2004, Rosch & Caparon, 2005), in the absence of polymyxin B, the translocon ATPase SecA localized to a single membrane site in a majority of cells when examined by immunogold electron microscopy (Fig. 4A). Similarly, the membrane-associated HtrA serine protease (Rosch & Caparon, 2005) localized to a single membrane site in a majority of cells when examined by immunogold electron microscopy (Fig. 4D) and by immunofluoresent microscopy (Fig. 4G, H). Adding polymyxin B at concentrations <30 μM did not alter this pattern for either SecA or HtrA. However, polymyxin B concentrations of of 30 μM through 60 μM resulted in redistribution of SecA (Fig. 4B,C) and HtrA (Fig. 4E,F, I, J) as demonstrated by a more circumferential staining pattern and a significant increase in the number of cells demonstrating multiple stained foci (P<0.05; Fig. 4K). Similar results were observed with the unrelated strain HSC5 (data not shown). Analysis of culture fractions indicated that the levels of SecA protein were unchanged in peptide-challenged streptococci and that the protein was retained in the same fractions in treated and untreated cells (Fig. S3A), suggesting that only localization of SecA at the membrane is affected. These data demonstrate that sub-lethal polymyxin B treatment can result in a disruption of ExPortal organization.
Figure 4. Redistribution of ExPortal proteins following sub-lethal polymyxin B challenge.
The distribution of SecA (A, B, C) and HtrA (D, E, F) on S. pyogenes SF370 was assessed by immunogold electron microscopy following challenge with polymyxin B at the concentrations indicated in the Figure (scale bar = 200nm). The distribution of HtrA was also assessed by immunofluorescent microscopy in the absence of (G, H) and following challenge with polymyxin B at 47 μM (I, J) and was quantitated as described previously (scale bar = 500nm) (K). Data represents the mean and SEM derived from at least 3 independent experiments and examination of a minimum of 1000 stained cells. In untreated cultures, the number of SF370 or HSC5 cells with a single focus was significantly higher than any other staining pattern (P < 0.05), whereas in polymyxin B-treated cultures, the number of streptococcal cells with multiple foci was significantly higher (P < 0.05).
Polymyxin B inhibits ExPortal-mediated secretion of SpeB
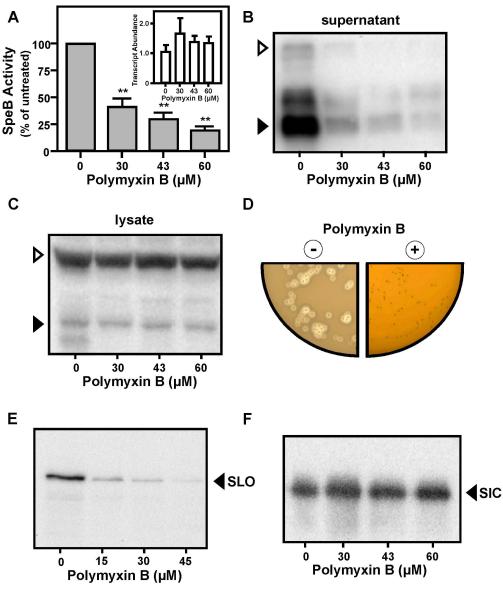
Disruption of ExPortal organization could have a deleterious effect on protein secretion. Particularly vulnerable would be those proteins that require the ExPortal to coordinate the activities of multiple secretion and biogenesis factors. A prominent example of this class is the SpeB cysteine protease, which requires numerous accessory factors, including the ExPortal-localized HtrA, for its secretion and conversion of its 43 kDa zymogen into the 28 kDa mature form (Lyon et al., 1998, Lyon & Caparon, 2004, Rosch et al., 2008, Ma et al., 2009, Rosch & Caparon, 2005). In addition, because the gene encoding SpeB is expressed at the onset of the stationary phase of culture (Loughman & Caparon, 2006b, Kietzman & Caparon, 2010, Kang et al., 2010), its expression would provide a sensitive assessment of the effect of prior challenge with a CAP at the late logarithmic phase of growth. Treatment of strain SF370 with various sub-lethal concentrations of polymyxin B resulted in a dose-dependent reduction in the amount of cysteine protease activity in culture supernatant (Fig. 5A), to a level less than 20% of that observed in the absence of the CAP (Fig. 5A, 60 μM). This reduction correlated with a dose-dependent decrease in the amount of SpeB polypeptide that was detected in treated culture supernatants (Fig. 5B). Secretion of SpeB in strain HSC5 was inhibited at lower concentrations of polymyxin B (Fig. S3B) and challenge with 60 μM polymyxin B resulted in a >95% inhibition of SpeB activity (data not shown). Several lines of evidence indicated that inhibition was not caused by an effect on growth, induction of stress or by suppression of speB transcription. Firstly, at the time of analysis of SpeB in culture supernatants (3 hrs post-challenge, Fig. S3C), the speB transcript was as abundant or present at higher levels as compared to untreated cultures (Fig. 5A, inset). Secondly, when transcription of speB was de-repressed and partially uncoupled from its growth phase-dependent control by mutation of the regulatory gene vfr (Ma et al., 2009, Shelburne et al., 2011), polymyxin B challenge resulted in the same profile of inhibition of SpeB secretion that was observed for the wild type strain (Fig. S3D). Thirdly, sublethal polymyxin B did not induce a general stress response, as treatment had no effect on levels of the HtrA protease (Fig. S3E). Fourthly, while treatment reduced the amount of SpeB present in supernatants, the SpeB polypeptide was still produced and could be detected in cell lysates (Fig. 5C). Finally, incorporation of polymyxin B into protease indicator plates at concentrations that did not reduce numbers of CFU’s, did inhibit expression of protease activity, as indicated by an absence of zones of clearance around colonies on treated media following overnight culture (Fig. 5D), although some activity was evident upon a prolonged period of incubation (not shown). Taken together, these data indicate that polymyxin B challenge inhibits SpeB expression at the level of secretion.
Figure 5. High sub-lethal challenge with polymyxin B inhibits secretion of SpeB and SLO, but not SIC.
Expression of the SpeB protease in cultures of S. pyogenes SF370 was determined following challenge with the indicated concentrations of polymyxin B by quantitation of cysteine protease activity in culture supernatant (A), by real-time RT-PCR analysis of speB transcript abundance (A, inset) by Western blot analysis of culture supernatant (B) in whole cell lysates (C) and following overnight culture on protease indicator plates containing (+) or lacking (−) 150 μM polymyxin B (D). Expression of SpeB results in a zone of clearing around colonies. Western blotting was also used to analyze the amount of SLO (E) and SIC (F) present in culture supernatant following challenge with the indicated concentrations of polymyxin B. All samples for SpeB analysis were harvested at 3 hrs post-challenge; samples for SLO and SIC were harvested 2hrs post-challenge. Open and filled triangles indicate the migration of the zymogen and mature form of SpeB, respectively. The migration of the SLO and SIC polypeptides are also indicated.
Polymyxin B inhibits secretion of SLO but not SIC
The Sec pathway delivers presecretory proteins to the translocons via either the post-translational pathway or the co-translational signal peptide recognition (SRP) pathway. While SpeB can be secreted via the post-translational pathway, the cytolysin SLO is targeted via the SRP pathway (Rosch et al., 2008). Given that expression of this cytolysin begins during the early logarithmic phase of growth, the challenge assay was slightly modified (see Experimental Procedures) in order to detect SLO that was secreted post-challenge. Comparison of supernatant harvested from peptide-challenged and untreated cultures of SF370 revealed a dose-dependent decrease in the amount of SLO protein detected in treated cultures (Fig. 5E); an effect that was more pronounced in HSC5 (data not shown). Similar to speB, slo transcript levels in polymyxin B-challenged cultures were as abundant as in untreated cultures at the time of analysis (Fig. S3F). A prominent difference between SF370 and HSC5 is that the former possesses the Streptococcal Inhibitor of Complement (SIC), which has been shown to enhance the resistance of S. pyogenes to CAPs (Frick et al., 2003, Fernie-King et al., 2004). In contrast to SLO, no apparent differences in secreted SIC polypeptide was observed between culture supernatants from untreated and challenged cultures of SF370 (Fig. 5F). Thus, inhibition of toxin secretion is selective for certain toxins and is not a universal feature of sub-lethal polymyxin B treatment.
Human Defensin HNP-1 also disrupts the S. pyogenes ExPortal
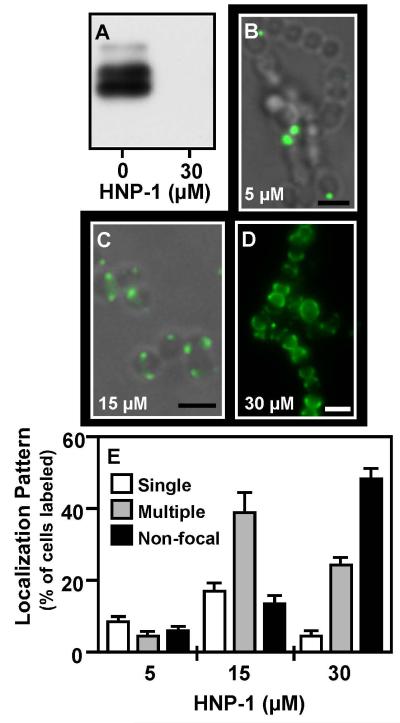
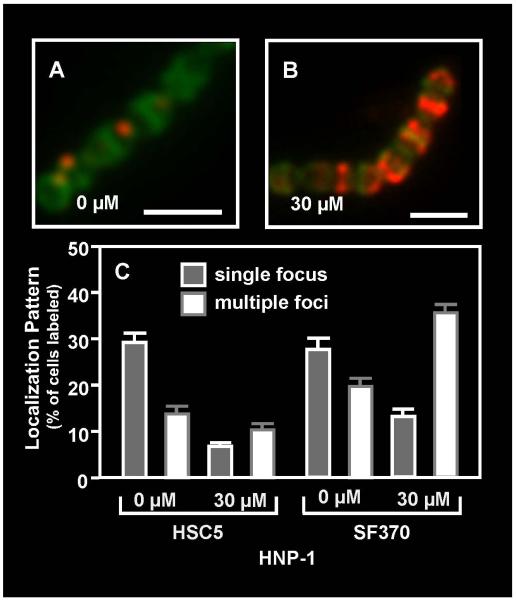
In order to assess if human CAPs could also disrupt ExPortal function, cultures were challenged with sub-lethal concentrations of the α-defensin HNP-1, the ß-defensins hBD-1 and hBD-2, and the cathelicidin LL-37. A Western blot analysis of culture supernatant revealed that of these, only HNP-1 noticeably inhibited secretion of SpeB at the CAP concentrations tested (<50 μM for all CAPs tested), and did so at concentrations lower than those required for similar inhibition by polymyxin B (Fig. 6A). Examination of the staining pattern of a fluorescent derivative of HNP-1 (5-FAM-HNP1) revealed that while staining was observed on a small population of cells at low concentration (5 μM, Fig. 6B), that these predominantly stained at discrete single or multiple foci (Fig. 6E). Increasing concentrations (15 μM) resulted in a higher percentage of cells with multiple foci (Fig. 6C, 6E), while at even higher concentrations (30 μM) the CAP primarily localizes to one hemisphere of the cell (Fig. 6D). Finally, localization of HtrA by immunofluorescence demonstrated that similar to polymyxin B, treatment with HNP-1 at sub-lethal concentrations that inhibited SpeB secretion also significantly decreased the number of cells exhibiting HtrA at discrete foci (P<0.0001, Fig. 7). These results suggest that a CAP to which S. pyogenes may be exposed to inside a human host can disrupt the organization and function of the ExPortal.
Figure 6. Focal localization and inhibition of SpeB expression by HNP-1.
Cultures were challenged with HNP-1 or by 5-FAM-HNP-1 at the concentrations indicated. Expression of SpeB was evaluated by a Western blot analysis (A) and binding to the S. pyogenes surface was assessed using fluorescent microscopy (B, C, D). Presented are overlays of fluorescent and phase images (B, C) or the fluorescent image alone (D) (scale bar = 1μm). Staining patterns were quantitated as described previously (E). Data represents the mean and SEM derived from at least 3 independent experiments and examination of a minimum of 1000 stained cells. At minimal concentration the number of cells with single foci is significantly higher than all other staining patterns, whereas multiple foci and hemisphere staining predominated significantly at higher peptide concentrations (P < 0.05).
Figure 7. Challenge with HNP-1 results in re-distribution of HtrA.
Cultures of S. pyogenes HSC5 were challenged with the indicated concentrations of HNP-1 and subjected to immunofluorescent microscopy to assess the distribution of HtrA (A, B). Cell walls were visualized by staining with fluorescent vancomycin (scale bar = 1μm). Staining patterns were quantitated as described previously (C). Data represents the mean and SEM derived from at least 3 independent experiments and examination of a minimum of 1000 stained cells. Challenge with the peptide significantly decreased the number of SF370 or HSC5 cells exhibiting HtrA at single foci (P<0.0001) and resulted in a significant increase in the number of multiple foci (P<0.0001) in the SF370 strain.
Discussion
The ability of S. pyogenes to cause a wide range of diseases has been linked to its production of virulence factors that neutralize or subvert innate mechanisms of immunity, including the lethal effects of CAPs (for review, see (Cole et al., 2007, von Pawel-Rammingen & Bjorck, 2003, Kwinn & Nizet, 2007)). However, by showing that sub-lethal concentrations of CAPs are able to disrupt the secretory ExPortal, the results of this study suggest that interaction with CAPs may play a more intimate role in S. pyogenes pathogenesis, serving to modulate virulence factor expression at both the transcriptional and post-transcriptional levels. Finally, the observation that disruption of ExPortal organization is associated with a defect in secretion of certain toxins provides additional support for the concept that the ExPortal serves a functional role in facilitating protein secretion.
A literature is emerging indicating that individual CAPs have a considerable diversity in how they interact with and kill bacteria using both lytic and non-lytic mechanisms (reviewed in (Jenssen et al., 2006, Brogden, 2005, Wilmes et al., 2011)). However, an initial step common to all pathways analyzed to date, involves the binding of the CAP to anionic lipids in the membrane. While little is currently known about the mechanism(s) by which sub-lethal concentrations of CAPs may affect bacterial physiology, the data presented here indicate that disruption of ExPortal organization and inhibition of SpeB and SLO secretion were also associated with membrane binding by the CAPs. Most models of CAP action postulate that at low sub-lethal concentrations, the peptides bind in a parallel orientation relative to the lipid membrane and begin to aggregate as concentrations increase, until a lethal concentration is reached and the peptides reorient to attack the membrane (reviewed by (Brogden, 2005, Hale & Hancock, 2007)). The observation that CAP binding to the ExPortal leads to redistribution of anionic lipids in the absence of pore formation contrasts with CAP behavior in model membranes, where they typically cluster anionic lipids (reviewed in (Epand & Epand, 2010)). Additionally, this redistribution of anionic lipids does not appear to induce or result from generalized disruption of the cytoplasmic membrane, as our data show that the integrity of the membrane is not compromised. Furthermore, numerous functions that depend on membrane integrity are also not compromised, including energy-dependent processes such as gene transcription and cell-growth, the latter of which also requires the assembly of macromolecular complexes at the membrane. The binding of polymyxin B to the membrane appears to only affect the physical location of anionic lipids and SecA within the membrane relative to each other and to accessory factors like HtrA. This suggests that the initial binding and/or aggregation of the CAPs is sufficient to disrupt the factor(s) that maintains ExPortal organization. If the anionic character of these lipids plays a fundamental role in organization, then it is possible that CAP binding serves to neutralize their charge to promote disruption. Alternatively, the interaction of CAPs with ExPortal lipids may disrupt the insertion or translocation of factors that function to maintain ExPortal lipid organization. The redistribution of ExPortal-associated membrane proteins may then occur subsequent to disruption of lipid structure, as analysis of a bitopic membrane protein (sortase C) has suggested that interaction between anionic lipids and a high density of positive charge in the cytoplasmic tail of its transmembrane helix is responsible for its localization at the ExPortal (Kline et al., 2009).
Many bacterial pathogens segregate charged lipids into microdomains (reviewed in (Matsumoto et al., 2006, Epand & Epand, 2009)) and these support the function of several multi-enzyme complexes required for secretion, membrane protein biogenesis and cell division (van Klompenburg & de Kruijff, 1998, Barak et al., 2008, van Dalen & de Kruijff, 2004). Since some CAPs can kill in the absence of pore formation and can also target charged lipids, it has been proposed that they exert their lethal effect by interfering with the dynamic function of these lipid-associated complexes, referred to as the “sand in a gearbox” mechanism (Pag et al., 2008). This concept is supported by the observation that these CAPs can elicit enhanced expression of some components of these complexes (Pag et al., 2008). By documenting the physical disruption of a multi-component organelle in the absence of poration, the results of the present study provide strong support for this mechanism. Furthermore, they extend this emerging concept to show that disruption can also occur at sub-lethal exposure to CAPs, and as a consequence, significantly influence how the bacterium interacts with its environment.
To date, modulation of signaling via the CovRS (CsrRS) two-component transcription regulator has been the most widely studied consequence of sub-lethal S. pyogenes-CAP interaction. Analysis of this mechanism has shown that from among a large panel of CAPs that included HNP-1, only human LL-37 functioned to induce signaling (Gryllos et al., 2008). Similarly, ExPortal targeting was not a property that was shared by all the CAPs tested in the present study, including LL-37. While the reason for this is not known, it was not unexpected, as the considerable diversity of CAP structure is reflected in differences in selectivity, binding efficiency, mechanism of killing and the physical conditions required for optimum activity (reviewed by (Jenssen et al., 2006)). Furthermore, these behaviors have not been extensively investigated at sub-lethal concentrations for any CAP.
Of the human-derived CAPs tested in the present study, HNP-1 proved more potent that polymyxin B in ExPortal disruption. This α-defensin is expressed constitutively in the azurophilic granules of neutrophils (Mestecky, 2005) and can be detected at concentrations of up to 63 μM (van Belkum et al., 2007) in secretions from nasopharyngeal tissue, a niche that can be colonized by S. pyogenes. Thus, it is likely that S. pyogenes encounters HNP-1 during infection. Also, while HNP-1 targets anionic lipids, it may have additional targets, including the lipid II molecule involved in cell wall synthesis (de Leeuw et al., 2010). Binding to lipid II likely explains why higher concentrations of HNP-1 produced more heterogenous patterns of staining, including a tendency for staining one hemisphere of the cell. In “ovococci” like S. pyogenes, which divide in successive parallel planes perpendicular to their long axis, this pattern is consistent with the pattern of new cell wall synthesis (reviewed by (Zapun et al., 2008)).
The ability of S. pyogenes to secrete numerous toxins likely plays a central role in its ability to cause disease (Mitchell, 2003). Thus, inhibition of SpeB and SLO may represent a physiologically relevant property of sub-lethal CAP concentrations. Since secretion of essential proteins will be required to support viability, it is unlikely that sub-lethal CAP treatment would produce a global blockade of protein secretion. It is known that biogenesis of SpeB requires secretion factors that are not required for other Sec-secreted proteins (Lyon et al., 1998, Lyon & Caparon, 2004, Rosch et al., 2008, Ma et al., 2009). Similarly, as an SRP substrate (Rosch et al., 2008) SLO’s secretion requires coordination between FtsY, the membrane receptor for the SRP, and the Sec translocons (Luirink et al., 2005). In contrast SIC secretion in the SF370 strain was unaffected by CAP treatment, a result that suggests that its secretion may not require additional factors in addition to Sec. If the ExPortal functions to coordinate interactions between various secretion components, then disruption of ExPortal organization could lead to the secretion defect. Alternatively, it has been shown that some CAPs can act on internal targets (reviewed by (Hale & Hancock, 2007)), raising the possibility that sub-lethal CAPs may interfere with secretion factors in the cytoplasmic compartment of the streptococcal cell.
Interestingly, analysis of the recently introduced antibiotic daptomycin has suggested that it principally targets regions of membrane enriched in anionic phospholipids (Straus & Hancock, 2006), suggesting that these domains can be exploited for the development of even more potent antibiotics. Thus, further analysis of CAP-ExPortal interaction will be valuable for uncovering the fundamental organizing principles of the ExPortal, how the ExPortal informs the coordination of protein secretion and maturation and the importance of modulation of secretion in response to host factors during infection.
Experimental Procedures
Strains, plasmids, media and growth conditions
As indicated, experiments utilized S. pyogenes HSC5 (Hanski et al., 1992) or SF370 (Ferretti et al., 2001). Localization of HtrA was analyzed in HSC5 following transformation by pHtrA-HA, which expresses a HA-tagged derivative of HtrA (Rosch & Caparon, 2005). Strain GCP682 contains an in-frame deletion (vfrΔ15-289) in vfr (SPy_0887) (G. Port and M. Caparon in preparation). Routine culture was at 37°C in Todd-Hewitt broth (BBL) supplemented with 0.2% yeast extract (Difco) (THY medium). Functional assays utilized cultures grown in unmodified C medium as described previously (Loughman & Caparon, 2006a). To produce solid media, Bacto Agar (Difco) was added to a final concentration of 1.4%. Expression of the secreted SpeB cysteine protease was evaluated on skimmed milk agar medium (Lyon et al., 1998). Liquid cultures were grown without agitation in closed containers and solid cultures were incubated under anaerobic conditions using a commercial gas generator (GasPak, cat. #260678, BBL) as described (Loughman & Caparon, 2006a). The SpeB protease was inhibited in cultures used to analyze the abundance of secreted streptococcal proteins by including the cysteine protease inhibitor E64 (Lyon et al., 1998). In selected experiments, media were supplemented with CAPs, including polymyxin B (cat. #P0972-50MU, Sigma), Human Neutrophil Peptide-1 (HNP-1, cat. #60743, AnaSpec), Human ß-defensin 1 (hßD-1, cat. #072-53, Phoenix Pharmaceuticals), Human ß-defensin 2 (hßD-2, cat.#072-48, Phoenix Pharmaceuticals), and Human cathelicidin LL-37 (LL-37, cat. #61302, AnaSpec) at the concentrations indicated in the text.
Challenge with CAPs
Unless otherwise indicated, bacteria were cultured overnight in liquid THY medium, diluted 1:100 in fresh C medium and then cultured to the late-logarithmic phase of growth (Fig. S3C, 0h). The CAPs were then added to the various final concentrations indicated in the text and incubation continued until cultures reached stationary phase (3 hrs post-treatment, Fig. S3C) as determined by monitoring optical density (OD600). Control cultures were grown in parallel, but were not treated with any CAPs; control and treated cultures entered stationary phase at approximately the same time (Fig. S3C). For this challenge assay, the minimum concentration of polymyxin B and HNP-1 that resulted in a greater than one-log reduction in the number of SF370 viable cells (determined by enumeration of colony forming units as described below) were 145 μM and 67 μM, respectively. This assay differs from a conventional assay for determining the minimal inhibitory concentration, which employs an inoculum of stationary phase cells from an overnight culture at a low cell density. As expected, SF370 was more sensitive to polymyxin B in the conventional assay (22 μM). In contrast, SF370 was more resistant to polymyxin B on protease indicator medium (see Fig. 5), which is likely due to its increased lipid content. For analysis of proteins whose expression begins in early logarithmic phase, the challenge assay was modified in order to remove proteins secreted into the supernatant prior to challenge as follows: cultures were grown to mid-logarithmic phase (Fig. S3C), harvested by centrifugation, immediately resuspended in an equivalent volume of warm fresh medium and then challenged with CAPs as indicated in the text. Samples were collected and processed for assessment of viability, for microscopy and for analysis of protein secretion, as described below. Data presented are representative of those obtained over the range of sub-lethal concentrations of CAPs.
Analysis of Viability
At selected time points following challenge with CAPs, aliquots were removed from cultures and viability assessed by determination of CFUs following brief sonication to disrupt streptococcal chains, serial dilution in PBS and plating on C medium agar as described previously (Brenot et al., 2004). Viability was also assessed by staining with a fluorescent vital dye (LIVE/DEAD®, BacLight™) as recommended by the manufacturer (Invitrogen). Viability was quantitated by examination using a fluorescent microscope (Leica model DM IRE 2) with enumeration of the percentage of live bacteria in randomly chosen microscopic fields totaling >1000 cells for each condition examined. Data reported represent the mean and standard error of the mean derived from a minimum of 3 independent experiments. Differences between calculated means were evaluated for significance using a one-way Analysis of Variance (ANOVA).
Cellular staining and fluorescent microscopy
Streptococcal cultures were challenged with various CAPs as described above, stained with various fluorescent reagents and then analyzed by fluorescent microscopy as follows: Analysis of the location and integrity of a membrane microdomain enriched in anionic phospholipids was assessed by staining with 10-nonyl acridine orange (NAO, cat. #A7847 Sigma) as described (Rosch et al., 2007). Localization of the discrete membrane site of secretion of the SpeB cysteine protease was conducted using the red protease assay (Rosch & Caparon, 2004). Analysis of CAP binding involved the substitution of native CAPs in the challenge assay with sub-lethal concentrations of fluorescent derivatives, including dansyl-polymyxin B (5, 10, 15 and 47μM, cat. #P13238, Invitrogen), polymyxin B BODIPY®FL conjugate (10 and 45μM, cat. #P13235, Invitrogen) or 5-FAM-HNP-1 (5, 15 and 30μM, custom synthesis by AnaSpec). Samples were examined using a Leica model DM IRE 2 fluorescent microscope and images captured using a QImaging Retiga 1350 EX charged-coupled device camera and Openlab software (Improvision). Where indicated, simultaneous treatment with 2 reagents was conducted in order to assess co-localization of staining. In these experiments streptococcal cells walls were visualized by staining with fluorescent vancomycin (1 μg ml−1, cat. #V34850, Invitrogen), or wheat germ agglutinin Alexa Fluor 488 conjugate (5 μg ml−1, cat. #W11261, Invitrogen) and neutral membrane lipids were visualized by staining with Nile Red (2.5 ug ml−1, cat. #N1142, Invitrogen). Focal localization in images of cells was quantitated as described in detail elsewhere (Kline et al., 2009) and scored as staining at a unique focus, multiple foci, or non-specifically (staining was of homogeneous intensity around the cellular circumference). Co-localization in the red protease assay was quantitated as the percentage of vancomycin-stained cells that exhibited focal staining with each individual reagent where the two foci were superimposable. Data presented for each condition represents the mean and standard error of the mean derived from at least 3 independent experiments and examination of a minimum of 1000 stained cells. Images were processed for publication using Adobe Photoshop CS3.
Immunofluorescent microscopy
The localization of native and HA-tagged HtrA protein (in SF370 and HSC5, respectively) was detected by immunofluorescent microscopy, as described (Rosch & Caparon, 2005). Aliquots from untreated or CAP-treated (47 μM polymyxin B or 29 μM HNP-1) challenge assay cultures (see above) were treated with PlyC lysin (prepared as described, (Nelson et al., 2006)) and fixed according to the method of Raz and Fischetti (Raz & Fischetti, 2008). Antisera used included a polyclonal rabbit anti-HtrA antiserum (a gift from Mark Walker, University of Wollongong, Australia) used at a dilution of 1:100 that was detected using an AlexaFluor 594-lableled goat anti-rabbit IgG (Invitrogen) at 1:500 and an AlexaFluor 594-conjugated rabbit IgG anti-HA epitope antiserum (Invitrogen) used at 1:500. Wheat germ agglutinin (WGA)-AlexaFluor 488 (Invitrogen) at a final concentration of 5 μg ml−1 was used to visualize the streptococcal cell walls. Slides were mounted in an anti-fade reagent (Prolong Gold, Invitrogen) and images captured and staining patterns quantitated as described above. Differences between means were analyzed for significance using a two-tailed Student’s t-test.
Immunogold electron microscopy
Localization of polymyxin B binding was examined by electron microscopy, as follows: Cultures in the assay described above were challenged with biotin-conjugated polymyxin B (HyCult Biotechnologies) at the various dilutions indicated in the text. At 1 hr post-challenge, aliquots were removed and prepared for immunoelectron microscopy as described (Rosch & Caparon, 2004, Rosch & Caparon, 2005). Sections were stained using a streptavidin-gold conjugate (20nm, BBI International) and examined by electron microscopy as detailed (Rosch & Caparon, 2004, Rosch & Caparon, 2005). Localization of SecA and HtrA following treatment in the challenge assay with the concentrations of polymyxin B indicated in the text was determined at 2 hr post-challenge by immunogold electron microscopy (Rosch & Caparon, 2004, Rosch & Caparon, 2005). Focal staining was defined and quantitiated as described in detail elsewhere (Kline et al., 2009).
Analysis of protein expression and secretion
Expression of the transcript for the SpeB cysteine protease was determined at various time points following challenge with polymyxin B by real time RT-PCR as described (Brenot et al., 2005, Loughman & Caparon, 2006a). Data represents the mean and standard error of the mean derived from 3 independent experiments conducted on different days, with each sample analyzed in quadruplicate. Supernantant fractions from cultures challenged with polymyxin B were analyzed for SpeB proteolytic activity as previously described (Rosch & Caparon, 2005) via the relative increase in fluorescence generated by the cleavage of fluorescein isothiocyanate-casein (cat. #C3777, Sigma). Supernatant, cell wall and protoplast fractions from cultures challenged with CAPs were prepared and analyzed for the presence of SpeB and HtrA by Western blotting as described (Rosch et al., 2008). Blots were developed using a Chemidoc XRS imager (BioRad) and relative protein concentrations determined using Quantity One software (BioRad, version 4.6.7). Data are expressed relative to untreated cultures and were derived from a minimum of 3 independent experiments. Differences between means were evaluated for significance using one-way ANOVA.
Supplementary Material
Acknowledgements
We thank Andy Kau and Jason Rosch for their input and ideas that served as the inspiration for this work. We also thank Kim Kine for many valuable discussions and are indebted to Wandy Beatty for her skill with EM imaging. This work was supported by Public Health Service Grant AI46433 from the NIH (to M. C.) and 1F31AI081504-01A1 from NIAID (to L.V.).
References
- Barak I, Muchova K, Wilkinson AJ, O’Toole PJ, Pavlendova N. Lipid spirals in Bacillus subtilis and their role in cell division. Mol Microbiol. 2008;68:1315–1327. doi: 10.1111/j.1365-2958.2008.06236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boman HG. Antibacterial peptides: basic facts and emerging concepts. J Intern Med. 2003;254:197–215. doi: 10.1046/j.1365-2796.2003.01228.x. [DOI] [PubMed] [Google Scholar]
- Braff MH, Zaiou M, Fierer J, Nizet V, Gallo RL. Keratinocyte production of cathelicidin provides direct activity against bacterial skin pathogens. Infect Immun. 2005;73:6771–6781. doi: 10.1128/IAI.73.10.6771-6781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenot A, King KY, Caparon MG. The PerR regulon in peroxide resistance and virulence of Streptococcus pyogenes. Mol Microbiol. 2005;55:221–234. doi: 10.1111/j.1365-2958.2004.04370.x. [DOI] [PubMed] [Google Scholar]
- Brenot A, King KY, Janowiak B, Griffith O, Caparon MG. Contribution of glutathione peroxidase to the virulence of Streptococcus pyogenes. Infect Immun. 2004;72:408–413. doi: 10.1128/IAI.72.1.408-413.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- Campo N, Tjalsma H, Buist G, Stepniak D, Meijer M, Veenhuis M, Westermann M, Muller JP, Bron S, Kok J, Kuipers OP, Jongbloed JD. Subcellular sites for bacterial protein export. Mol Microbiol. 2004;53:1583–1599. doi: 10.1111/j.1365-2958.2004.04278.x. [DOI] [PubMed] [Google Scholar]
- Cole JN, Aquilina JA, Hains PG, Henningham A, Sriprakash KS, Caparon MG, Nizet V, Kotb M, Cordwell SJ, Djordjevic SP, Walker MJ. Role of group A Streptococcus HtrA in the maturation of SpeB protease. Proteomics. 2007;7:4488–4498. doi: 10.1002/pmic.200700626. [DOI] [PubMed] [Google Scholar]
- Cunningham MW. Pathogenesis of group A streptococcal infections. Clin Microbiol Rev. 2000;13:470–511. doi: 10.1128/cmr.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Keyzer J, van der Does C, Driessen AJ. The bacterial translocase: a dynamic protein channel complex. Cell Mol Life Sci. 2003;60:2034–2052. doi: 10.1007/s00018-003-3006-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw E, Li C, Zeng P, Diepeveen-de Buin M, Lu WY, Breukink E, Lu W. Functional interaction of human neutrophil peptide-1 with the cell wall precursor lipid II. FEBS Lett. 2010;584:1543–1548. doi: 10.1016/j.febslet.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epand RM, Epand RF. Lipid domains in bacterial membranes and the action of antimicrobial agents. Biochim Biophys Acta. 2009;1788:289–294. doi: 10.1016/j.bbamem.2008.08.023. [DOI] [PubMed] [Google Scholar]
- Epand RM, Epand RF. Bacterial membrane lipids in the action of antimicrobial agents. J Pept Sci. 2010 doi: 10.1002/psc.1319. [DOI] [PubMed] [Google Scholar]
- Fernie-King BA, Seilly DJ, Lachmann PJ. The interaction of streptococcal inhibitor of complement (SIC) and its proteolytic fragments with the human beta defensins. Immunology. 2004;111:444–452. doi: 10.1111/j.0019-2805.2004.01837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti JJ, McShan WM, Ajdic D, Savic DJ, Savic G, Lyon K, Primeaux C, Sezate S, Suvorov AN, Kenton S, Lai HS, Lin SP, Qian Y, Jia HG, Najar FZ, Ren Q, Zhu H, Song L, White J, Yuan X, Clifton SW, Roe BA, McLaughlin R. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc Natl Acad Sci U S A. 2001;98:4658–4663. doi: 10.1073/pnas.071559398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick IM, Akesson P, Rasmussen M, Schmidtchen A, Bjorck L. SIC, a secreted protein of Streptococcus pyogenes that inactivates antibacterial peptides. J Biol Chem. 2003;278:16561–16566. doi: 10.1074/jbc.M301995200. [DOI] [PubMed] [Google Scholar]
- Gryllos I, Tran-Winkler HJ, Cheng MF, Chung H, Bolcome R, 3rd, Lu W, Lehrer RI, Wessels MR. Induction of group A Streptococcus virulence by a human antimicrobial peptide. Proc Natl Acad Sci U S A. 2008;105:16755–16760. doi: 10.1073/pnas.0803815105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale JD, Hancock RE. Alternative mechanisms of action of cationic antimicrobial peptides on bacteria. Expert Rev Anti Infect Ther. 2007;5:951–959. doi: 10.1586/14787210.5.6.951. [DOI] [PubMed] [Google Scholar]
- Hancock RE. Cationic peptides: effectors in innate immunity and novel antimicrobials. Lancet Infect Dis. 2001;1:156–164. doi: 10.1016/S1473-3099(01)00092-5. [DOI] [PubMed] [Google Scholar]
- Hancock RE, Sahl HG. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol. 2006;24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- Hanski E, Horwitz PA, Caparon MG. Expression of protein F, the fibronectin-binding protein of Streptococcus pyogenes JRS4, in heterologous streptococcal and enterococcal strains promotes their adherence to respiratory epithelial cells. Infect Immun. 1992;60:5119–5125. doi: 10.1128/iai.60.12.5119-5125.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder J, Bartels J, Christophers E, Schroder JM. Isolation and characterization of human beta -defensin-3, a novel human inducible peptide antibiotic. J Biol Chem. 2001;276:5707–5713. doi: 10.1074/jbc.M008557200. [DOI] [PubMed] [Google Scholar]
- Hu P, Bian Z, Fan M, Huang M, Zhang P. Sec translocase and sortase A are colocalised in a locus in the cytoplasmic membrane of Streptococcus mutans. Arch Oral Biol. 2008;53:150–154. doi: 10.1016/j.archoralbio.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Jenssen H, Hamill P, Hancock RE. Peptide antimicrobial agents. Clin Microbiol Rev. 2006;19:491–511. doi: 10.1128/CMR.00056-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson L, Thulin P, Sendi P, Hertzen E, Linder A, Akesson P, Low DE, Agerberth B, Norrby-Teglund A. Cathelicidin LL-37 in severe Streptococcus pyogenes soft tissue infections in humans. Infect Immun. 2008;76:3399–3404. doi: 10.1128/IAI.01392-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SO, Caparon MG, Cho KH. Virulence gene regulation by CvfA, a putative RNase: the CvfA-enolase complex in Streptococcus pyogenes links nutritional stress, growth-phase control, and virulence gene expression. Infect Immun. 2010;78:2754–2767. doi: 10.1128/IAI.01370-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz M, Tsubery H, Kolusheva S, Shames A, Fridkin M, Jelinek R. Lipid binding and membrane penetration of polymyxin B derivatives studied in a biomimetic vesicle system. Biochem J. 2003;375:405–413. doi: 10.1042/BJ20030784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kietzman CC, Caparon MG. CcpA and LacD.1 affect temporal regulation of Streptococcus pyogenes virulence genes. Infect Immun. 2010;78:241–252. doi: 10.1128/IAI.00746-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline KA, Kau AL, Chen SL, Lim A, Pinkner JS, Rosch J, Nallapareddy SR, Murray BE, Henriques-Normark B, Beatty W, Caparon MG, Hultgren SJ. Mechanism for sortase localization and the role of sortase localization in efficient pilus assembly in Enterococcus faecalis. J Bacteriol. 2009;191:3237–3247. doi: 10.1128/JB.01837-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristian SA, Datta V, Weidenmaier C, Kansal R, Fedtke I, Peschel A, Gallo RL, Nizet V. D-alanylation of teichoic acids promotes group a streptococcus antimicrobial peptide resistance, neutrophil survival, and epithelial cell invasion. J Bacteriol. 2005;187:6719–6725. doi: 10.1128/JB.187.19.6719-6725.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwinn LA, Nizet V. How group A Streptococcus circumvents host phagocyte defenses. Future Microbiol. 2007;2:75–84. doi: 10.2217/17460913.2.1.75. [DOI] [PubMed] [Google Scholar]
- Lee PH, Ohtake T, Zaiou M, Murakami M, Rudisill JA, Lin KH, Gallo RL. Expression of an additional cathelicidin antimicrobial peptide protects against bacterial skin infection. Proc Natl Acad Sci U S A. 2005;102:3750–3755. doi: 10.1073/pnas.0500268102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughman JA, Caparon M. Regulation of SpeB in Streptococcus pyogenes by pH and NaCl: a model for in vivo gene expression. J Bacteriol. 2006a;188:399–408. doi: 10.1128/JB.188.2.399-408.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughman JA, Caparon MG. A novel adaptation of aldolase regulates virulence in Streptococcus pyogenes. Embo J. 2006b;25:5414–5422. doi: 10.1038/sj.emboj.7601393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luirink J, von Heijne G, Houben E, de Gier JW. Biogenesis of inner membrane proteins in Escherichia coli. Annu Rev Microbiol. 2005;59:329–355. doi: 10.1146/annurev.micro.59.030804.121246. [DOI] [PubMed] [Google Scholar]
- Lyon WR, Caparon MG. Role for serine protease HtrA (DegP) of Streptococcus pyogenes in the biogenesis of virulence factors SpeB and the hemolysin streptolysin S. Infect Immun. 2004;72:1618–1625. doi: 10.1128/IAI.72.3.1618-1625.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon WR, Gibson CM, Caparon MG. A role for trigger factor and an rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. Embo J. 1998;17:6263–6275. doi: 10.1093/emboj/17.21.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Bryant AE, Salmi DB, McIndoo E, Stevens DL. vfr, a novel locus affecting cysteine protease production in Streptococcus pyogenes. J Bacteriol. 2009;191:3189–3194. doi: 10.1128/JB.01771-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K, Kusaka J, Nishibori A, Hara H. Lipid domains in bacterial membranes. Mol Microbiol. 2006;61:1110–1117. doi: 10.1111/j.1365-2958.2006.05317.x. [DOI] [PubMed] [Google Scholar]
- Mestecky J. Mucosal immunology. Elsevier Academic Press; Amsterdam ; Boston: 2005. p. 2. v. (lvi, 1868, [1850] p., [1820] p. of plates) [Google Scholar]
- Mileykovskaya E, Dowhan W. Visualization of phospholipid domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-N-nonyl acridine orange. J Bacteriol. 2000;182:1172–1175. doi: 10.1128/jb.182.4.1172-1175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell TJ. The pathogenesis of streptococcal infections: from tooth decay to meningitis. Nat Rev Microbiol. 2003;1:219–230. doi: 10.1038/nrmicro771. [DOI] [PubMed] [Google Scholar]
- Nelson D, Schuch R, Chahales P, Zhu S, Fischetti VA. PlyC: a multimeric bacteriophage lysin. Proc Natl Acad Sci U S A. 2006;103:10765–10770. doi: 10.1073/pnas.0604521103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizet V, Ohtake T, Lauth X, Trowbridge J, Rudisill J, Dorschner RA, Pestonjamasp V, Piraino J, Huttner K, Gallo RL. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature. 2001;414:454–457. doi: 10.1038/35106587. [DOI] [PubMed] [Google Scholar]
- Pag U, Oedenkoven M, Sass V, Shai Y, Shamova O, Antcheva N, Tossi A, Sahl HG. Analysis of in vitro activities and modes of action of synthetic antimicrobial peptides derived from an alpha-helical ‘sequence template’. J Antimicrob Chemother. 2008;61:341–352. doi: 10.1093/jac/dkm479. [DOI] [PubMed] [Google Scholar]
- Peschel A, Jack RW, Otto M, Collins LV, Staubitz P, Nicholson G, Kalbacher H, Nieuwenhuizen WF, Jung G, Tarkowski A, van Kessel KP, van Strijp JA. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J Exp Med. 2001;193:1067–1076. doi: 10.1084/jem.193.9.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz A, Fischetti VA. Sortase A localizes to distinct foci on the Streptococcus pyogenes membrane. Proc Natl Acad Sci U S A. 2008;105:18549–18554. doi: 10.1073/pnas.0808301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosch J, Caparon M. A microdomain for protein secretion in Gram-positive bacteria. Science. 2004;304:1513–1515. doi: 10.1126/science.1097404. [DOI] [PubMed] [Google Scholar]
- Rosch JW, Caparon MG. The ExPortal: an organelle dedicated to the biogenesis of secreted proteins in Streptococcus pyogenes. Mol Microbiol. 2005;58:959–968. doi: 10.1111/j.1365-2958.2005.04887.x. [DOI] [PubMed] [Google Scholar]
- Rosch JW, Hsu FF, Caparon MG. Anionic lipids enriched at the ExPortal of Streptococcus pyogenes. J Bacteriol. 2007;189:801–806. doi: 10.1128/JB.01549-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosch JW, Vega LA, Beyer JM, Lin A, Caparon MG. The signal recognition particle pathway is required for virulence in Streptococcus pyogenes. Infect Immun. 2008;76:2612–2619. doi: 10.1128/IAI.00239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott ME, Dossani ZY, Sandkvist M. Directed polar secretion of protease from single cells of Vibrio cholerae via the type II secretion pathway. Proc Natl Acad Sci U S A. 2001;98:13978–13983. doi: 10.1073/pnas.241411198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne SA, 3rd, Olsen RJ, Makthal N, Brown NG, Sahasrabhojane P, Watkins EM, Palzkill T, Musser JM, Kumaraswami M. An amino-terminal signal peptide of Vfr protein negatively influences RopB-dependent SpeB expression and attenuates virulence in Streptococcus pyogenes. Mol Microbiol. 2011;82:1481–1495. doi: 10.1111/j.1365-2958.2011.07902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straus SK, Hancock RE. Mode of action of the new antibiotic for Gram-positive pathogens daptomycin: comparison with cationic antimicrobial peptides and lipopeptides. Biochim Biophys Acta. 2006;1758:1215–1223. doi: 10.1016/j.bbamem.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Thedieck K, Hain T, Mohamed W, Tindall BJ, Nimtz M, Chakraborty T, Wehland J, Jansch L. The MprF protein is required for lysinylation of phospholipids in listerial membranes and confers resistance to cationic antimicrobial peptides (CAMPs) on Listeria monocytogenes. Mol Microbiol. 2006;62:1325–1339. doi: 10.1111/j.1365-2958.2006.05452.x. [DOI] [PubMed] [Google Scholar]
- van Belkum A, Emonts M, Wertheim H, de Jongh C, Nouwen J, Bartels H, Cole A, Cole A, Hermans P, Boelens H, Toom NL, Snijders S, Verbrugh H, van Leeuwen W. The role of human innate immune factors in nasal colonization by Staphylococcus aureus. Microbes Infect. 2007;9:1471–1477. doi: 10.1016/j.micinf.2007.08.003. [DOI] [PubMed] [Google Scholar]
- van Dalen A, de Kruijff B. The role of lipids in membrane insertion and translocation of bacterial proteins. Biochim Biophys Acta. 2004;1694:97–109. doi: 10.1016/j.bbamcr.2004.03.007. [DOI] [PubMed] [Google Scholar]
- van Klompenburg W, de Kruijff B. The role of anionic lipids in protein insertion and translocation in bacterial membranes. J Membr Biol. 1998;162:1–7. doi: 10.1007/s002329900336. [DOI] [PubMed] [Google Scholar]
- von Pawel-Rammingen U, Bjorck L. IdeS and SpeB: immunoglobulin-degrading cysteine proteinases of Streptococcus pyogenes. Curr Opin Microbiol. 2003;6:50–55. doi: 10.1016/s1369-5274(03)00003-1. [DOI] [PubMed] [Google Scholar]
- Wilmes M, Cammue BP, Sahl HG, Thevissen K. Antibiotic activities of host defense peptides: more to it than lipid bilayer perturbation. Nat Prod Rep. 2011;28:1350–1358. doi: 10.1039/c1np00022e. [DOI] [PubMed] [Google Scholar]
- Zapun A, Vernet T, Pinho MG. The different shapes of cocci. FEMS Microbiol Rev. 2008;32:345–360. doi: 10.1111/j.1574-6976.2007.00098.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.