Abstract
Stress granules (SG) are cytoplasmic multimeric RNA bodies that form under stress conditions known to inhibit cap-dependent translation. SG contain translation initiation factors, RNA binding proteins, and signaling molecules. SG are known to inhibit apoptotic pathways, thus contributing to chemo- and radioresistance in tumor cells. However, whether stress granule formation involves oncogenic signaling pathways is currently unknown. Here, we report a novel role of the mTORC1-eukaryotic translation initiation factor 4E (eIF4E) pathway, a key regulator of cap-dependent translation initiation of oncogenic factors, in SG formation. mTORC1 specifically drives the eIF4E-mediated formation of SG through the phosphorylation of 4E-BP1, a key factor known to inhibit formation of the mTORC1-dependent eIF4E-eIF4GI interactions. Disrupting formation of SG by inactivation of mTOR with its specific inhibitor pp242 or by depletion of eIF4E or eIF4GI blocks the SG-associated antiapoptotic p21 pathway. Finally, pp242 sensitizes cancer cells to death in vitro and inhibits the growth of chemoresistant tumors in vivo. This work therefore highlights a novel role of the oncogenic mTORC1-eIF4E pathway, namely, the promotion of formation of antiapoptotic SG.
INTRODUCTION
When exposed to environmental stress, cells rapidly activate pathways that induce a coordinated response of mRNA translation and turnover designed to protect cells against stress-induced damage and promote their survival. One of these pathways involves the formation of stress granules (SG). These cytoplasmic RNA bodies are induced by a number of stress inducers such as ionizing radiation (1), hypoxia (2), viral infection (3, 4), and anticancer drugs, including proteasome inhibitors (5–7). Since such stress-inducing SG inhibit translation initiation, it is thought that SG represent sites where translation of specific mRNAs is repressed (8, 9). In addition to mRNAs and associated proteins, these foci also contain small ribosomal subunits (40S) and eukaryotic translation initiation factors (eIFs), including eIF4E and eIF4GI (8, 9), as well as signaling molecules such as RACK1 and TRAF2 which are known to impact on cell death (10, 11).
Induction of SG upon exposure to several stress types can lead to resistance of tumor cells to apoptosis, a phenomenon which appears to involve several mechanisms acting in tandem. The formation of SG under conditions of hypoxia has been shown to inhibit apoptosis mediated by etoposide (10). This effect was attributed to sequestration of the signaling scaffold protein RACK1 into SG, thus leading to the suppression of mitogen-activated protein (MAP) kinase-dependent apoptotic pathways. Furthermore, SG promote tumor cell survival during radiotherapy by preventing the degradation of specific mRNAs encoding key hypoxia-inducible factor 1 (HIF-1)-regulated survival cytokines, thereby leading to their upregulation (1). Recent yeast studies have described the induction of SG formation by the anticancer drug doxorubicin (12), and we have described formation of SG in human cancer cells induced by bortezomib (Velcade) (5). This FDA-approved drug is clinically efficient for the treatment of myelomas and other hematological malignancies (13–15). Solid tumors of various histological origins generally display resistance to this proteasome inhibitor (13–17), and it was observed that such chemoresistance may be associated with the upregulation of the antiapoptotic protein p21 via a mechanism involving the stabilization of p21 mRNA within SG (6). Targeting SG formation might thus contribute to suppress resistance to proteasome inhibitors at least in part by preventing SG-mediated p21 upregulation.
mTOR controls a key signaling pathway that promotes cell survival and tumorigenesis by stimulating cap-dependent translation of oncogenes and growth factors. The mTOR signaling functions are mediated by two complexes, mTORC1 and mTORC2 (18, 19). mTORC1 and mTORC2 share several components and contain unique proteins (20–26). The defining component of mTORC1 is Raptor, which regulates its assembly (27, 28). mTORC1 is a well-characterized rapamycin-sensitive complex that is activated by growth factor stimulation via the PI3K-AKT pathway. Activated mTORC1 kinase upregulates protein synthesis by phosphorylating key regulators of mRNA translation (29), including 4E-BP1, a binding partner of the cap-binding protein eIF4E.
eIF4E is the best-characterized translation initiation factor known to be targeted by mTOR to promote translation of specific oncogenes leading to tumorigenesis (30). eIF4E is responsible for the early recognition of mRNAs by binding to their 5′-m7GpppN cap structure (31), an interaction which is further enhanced by the scaffolding protein eIF4GI and thus stimulates formation of the cap-binding eIF4F complex (32, 33). Binding of 4E-BP1 to eIF4E competes against eIF4E-eIF4GI associations (32, 34) to disrupt eIF4F complexes, thereby counteracting the translation-promoting activity of eIF4E (35–38). mTORC1-mediated phosphorylation of 4E-BP1 releases eIF4E from 4E-BP1, thus activating translation initiation and inducing cell growth and survival (39–42). In cancer cells, free eIF4E levels rise upon increases in mTORC1 signaling or eIF4E expression, which trigger tumorigenesis (30, 43–45). Consequently, targeting mTORC1 signaling inhibits eIF4E activity and associated tumorigenesis (39, 40, 42, 46, 47). This central role of mTOR in controlling key cellular growth and survival pathways has sparked interest in developing mTOR inhibitors that could be used in cancer therapy. Unlike rapamycin, which influences mTOR enzymatic activity indirectly by binding to a domain distinct from the kinase active site, the recently described TOR kinase domain-targeted mTOR inhibitors (or TORKinibs) pp242 and torin 1 are two ATP-competitive inhibitors of mTOR (48, 49) that bind directly to the mTOR kinase domain and suppress its activity. Unlike rapamycin, which partially interferes with the activity of mTORC1, pp242 and torin 1 efficiently inhibit mTORC1-dependent eIF4E activation (48–52). For purposes of cancer therapy, TORKinibs are thus considered to be promising agents that might be used to target the mTORC1-eIF4E oncogenic pathway (48–52).
We show here that inactivating the mTORC1-eIF4E pathway impairs SG formation and sensitizes cancer cells to bortezomib-induced cell death in vitro and inhibits the growth of bortezomib-chemoresistant tumors in vivo. These results identify a novel eIF4E-based mode of SG assembly and suggest a novel role for the mTORC1 pathway in SG formation via eIF4E through phosphorylation of 4E-BP1.
MATERIALS AND METHODS
Cell lines and tissue culture.
HeLa cervical cancer cells and MCF-7 and Hs578T breast cancer cells as well as N2a neuroblastoma cells were obtained from the American Type Culture Collection (ATCC; Manassas, VA). HeLa cells that stably express either short hairpin RNA (shRNA) control or shRNA p21 were generated using HuSH-29 shRNA vectors purchased from OriGene. Cells were cultured at 37°C in Dulbecco's modified Eagle's medium (DMEM; Sigma-Aldrich, St. Louis, MO) supplemented with 10% fetal bovine serum (FBS), penicillin, and streptomycin (all supplements from Sigma-Aldrich).
Antibodies.
Phospho-specific anti-eIF2α, pan-anti-eIF2, anti-4EBP1, anti-phospho-4EBP1, anti-eIF4GI, anti-mTOR, and anti-caspase-3 were purchased from Cell Signaling Technology (Beverly, MA). Anti-FMRP was previously described (6), and anti-FXR1 was obtained from Edward Khandjian (Laval University). Anti-G3BP1 and anti-histone 3 were kindly provided by Imed Gallouzi (McGill University) and Jean-Yves Masson (Laval University), respectively. Anti-eIF4E was obtained from BD Bioscience. Antitubulin and anti-Raptor were purchased from Abcam. Anti-p21 antibodies were purchased from Santa Cruz Biotech (Santa Cruz, CA).
Chemicals and treatment.
PP242 was obtained from Selleck Chemicals, torin 1 was purchased from Tocris Bioscience, 4EGI-1 was purchased from Alexis Biochemicals, and bortezomib was obtained from LC Laboratories. All compounds were dissolved in dimethyl sulfoxide (DMSO) and stored at −20°C as recommended by the suppliers. For treatment, cells (at ∼60% to 70% confluence) were first incubated with DMEM containing 3% to 5% FBS for at least 2 h before addition of drugs.
siRNA experiments.
All specific small interfering RNAs (siRNAs) and nontargeting control siRNAs were purchased from Dharmacon (Lafayette, CO). siRNA transfections were performed essentially as described previously (6, 53), using HiPerFect reagent (Qiagen) and following the manufacturer's protocol. Twenty-four hours before transfection, cells were plated at a density leading to 60% to 70% confluence at the time of transfection. For a 6-well plate, annealed duplexes were used at a final concentration of 10 nM. Forty-eight hours posttransfection, cells were treated with siRNA (5 nM) for an additional 48 h. Cells were then either fixed and processed for immunofluorescence or harvested for protein extraction. The sequences of the siRNAs used are as follows: for siRNA-eIF4E-1, 5′-GGACGAUGGCUAATTACAT-3′ (84, 85); for siRNA-eIF4E-2, 5′-ACACATATAGGGAGGGTAT-3′; for siRNA-mTOR-1, 5′-CATAAGAGGCAGAAGGCAA-3′; for siRNA-mTOR-2, 5′-GCAAAGATCTCATGGGCTT-3′; for siRNA-4EBP1-1, 5′-GACATAGCCCAGAAGATAA-3′; for siRNA-4EBP1-2, 5′-CCGGGAGGTACCAGGATCA-3′; for siRNA-eIF4GI-1, 5′-TGAGAAAGGAGGAGAGGAA-3′; for siRNA-eIF4GI-2, 5′-GGGCTTAGCTGGAAGGAAT-3′; for si-Raptor-1, 5′-TGGCTAGTCTGTTTCGAAA-3′; for si-Raptor-2, 5′-GGGAGAAGCTGGATTATTT-3′; and for si-4E-BP2, 5′-GGGAGGAACTCGAATCATT-3′.
Immunofluorescence.
Following fixation and permeabilization (20 min in 3.7% paraformaldehyde at room temperature followed by a 15-min immersion in MeOH at −20°C), cells were incubated with primary antibodies diluted in 0.1% (vol/vol) Tween 20–phosphate-buffered saline (PBST) for 2 h at room temperature. After being rinsed with PBST, cells were incubated with goat anti-mouse/anti-rabbit IgG (H+L) secondary antibodies conjugated with the Alexa Fluor dye of the appropriate maximum absorption wavelength (−488 or −594) for 1 h, washed, and then mounted.
To detect 4E-BP1 by immunofluoresence, we used a protocol previously described by Svitkin et al. (84). Proteins were visualized using an LSM 700 confocal laser scanning microscope (Zeiss) controlled with ZEN 2009 software for image acquisition and analysis. Images were acquired using the following settings: 63× oil objective (zoom, 1.0), 0.06 μm for pixel size, and 1.00 Airy units as the pinhole.
DNA transfection.
The pcDNA vectors used encoding 4E-BP1 were previously described (85). The pcDNA encoding murine eIF4E was kindly provided by Katherine Borden (Université de Montréal). Vectors encoding p21 were purchased from OriGene. For DNA transfection, HeLa cells were transfected with DNA vectors in a 6-well plate using an Effectene transfection reagent kit (Qiagen). At 48 h later or unless otherwise specified, cells were treated as indicated and then processed for immunofluorescence or collected for Western blot analysis.
Quantitative RT-PCR.
Reverse transcriptase PCRs (RT-PCRs) were performed using a Quantitect reverse transcriptase kit (Qiagen). Each reaction mixture contained 500 ng of RNA (isolated using an RNeasy Plus minikit; Qiagen), 2 μl of 7× genomic DNA wipeout buffer, 4 μl of 5× Quantiscript RT buffer, 1 μl of RT primer mix, and 1 μl of Quantiscript reverse transcriptase.
Real-time PCRs were prepared using Platinum Power SYBR green quantitative PCR (qPCR) Master Mix (Life Technologies, CA) and the following ingredients in a total volume of 25 μl: 12.5 μl of PCR Master Mix, 0.67 μl of forward primer at 3.75 μM, 0.67 μl of reverse primer at 3.75 μM, 9.2 μl of deionized (Milli-Q-grade) water, and 2 μl of RT. Reactions were run, and data then analyzed using a 7500 Fast Real-Time PCR system (Applied Biosystems) with a 4-stage program: stage 1, 20 s at 50°C; stage 2, 10 min at 95°C; stage 3, 40 cycles of a 2-step reaction (95°C for 15 s and 58°C for 60 s); and stage 4, a 3-step reaction (95°C for 15 s, 60°C for 1 min, and 60°C for 15 s).
To prepare templates for p21 mRNA, the oligonucleotide pair used was 5′-GACTTTGTCACCGAGACACC-3′ (forward) and 5′-GACAGGTCCACATGGTCTTC-3′ (reverse). For glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA templates, the oligonucleotide pair used was 5′-ACGACCACTTTGTCAAGCTC-3′ (forward) and 5′-GTTGCTGTAGCCAAATTCGT-3′ (reverse). For 4E-BP2 mRNA templates, we used the following oligonucleotide pair: 5′-CCTTACAGCTTGGTGCAGTT-3′ (forward) and 5′-ATGAGGCATGACACAAAGGT-3′ (reverse).
Preparation of cellular extracts.
Nuclear and cytoplasmic extracts were prepared as described previously (86). The cells were cooled on ice for 5 min, centrifuged at 150 × g for 5 min, and resuspended in ice-cold EBKL buffer containing 25 mM HEPES (pH 7.6), 5 mM MgCl2, 1.5 mM KCl, 2 mM dithiothreitol (DTT), protease inhibitors, and 0.1% NP-40. The cells were then lysed on ice by 20 strokes in a Dounce homogenizer (Sigma) (tight pestle). The nuclei were removed by two 3-min centrifugations at 600 × g. The resulting supernatant, spun at 10,000 × g for 10 min, was labeled as the total cytoplasmic extract.
Polysome preparation.
Polysomes were prepared as follows. Cells were collected in lysis buffer (20 mM Tris-HCl [pH 7.4], 1.25 mM MgCl2, 150 mM NaCl, 1 mM DTT, 1% NP-40, 5 U/ml of RNase inhibitor [Invitrogen]) supplemented with complete Mini EDTA-free protease inhibitor cocktail tablets (Roche). The cell homogenate was then clarified by centrifugation at 12,000 rpm for 10 min at 4°C. The cytoplasmic extract was then loaded onto a 15% to 55% linear sucrose gradient previously generated with an Isco model 160 gradient former (Teledyne Isco, Lincoln, NE) and then separated by sedimentation velocity through centrifugation for 2.5 h at 37,000 rpm using a Sorvall TH-641 ultracentrifuge rotor (Du Pont) at 4°C. The sucrose gradient was processed for fractionation using an Isco type 11 optical unit with 254-nm and 280-nm filters (Teledyne Isco). Equal fractions were collected with continuous monitoring of absorbance at 254 nm using an Isco UA-6 UV-visible light (UV-vis) detector (Teledyne Isco). Fractions were precipitated, resuspended in equal volumes of SDS-PAGE sample buffer, and analyzed by Western blotting.
Cap-binding assays.
Cells were lysed in buffer A (50 mM Tris-HCl [pH 7.4], 100 mM NaCl, 1 mM EDTA, and protease inhibitors [Roche] supplemented with 0.5% NP-40), and cell lysates were incubated for 2 h at 4°C with 30 μl of the mRNA cap analog m7GTP-Sepharose (GE Healthcare) in buffer A. The m7GTP-Sepharose-bound proteins were washed with buffer A, and eIF4E-bound proteins were eluted with SDS loading buffer and resolved by SDS-PAGE and Western blotting.
Annexin V-FITC/PI assay and FACS analysis.
At the end of the experimental period, both adherent and detached cells were harvested. Cells were washed with ice-cold PBS, pelleted again at 1,500 rpm for 10 min at 4°C, and resuspended in ice-cold binding buffer (10 mM HEPES/NaOH [pH 7.4], 140 mM NaCl, 2.5 mM CaCl2). Cells were subsequently stained with annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) for 15 min in the dark. A total of 2 × 104 cells were counted, and dead cells were examined by flow cytometry. For fluorescence-activated cell sorter (FACS) analysis, collected cells were fixed with ethanol, washed with PBS, stained with 4′,6-diamidino-2-phenylindole (DAPI) (1 μg/ml), and analyzed by flow cytometry.
CAM tumor assay.
Day 0 fertilized chicken eggs were purchased from Couvoir Provincial (Victoriaville, QC, Canada). The eggs were incubated for 10 days in a Pro-FI egg incubator fitted with an automatic egg turner before being transferred to a Roll-X static incubator for the rest of the incubation time. The eggs were kept at 37°C in a 60%-relative-humidity atmosphere for the whole incubation period. Using a hobby drill (Dremel, Racine, WI), a hole was drilled on the side of the embryo, and negative pressure was applied to create a new air sac. A window was opened on this new air sac and was covered with transparent adhesive tape to prevent contamination. A freshly prepared cell suspension (40 μl) of HeLa cells (1 × 106 cells/egg) was applied directly onto the freshly exposed chorioallantoic membrane (CAM) tissue through the window. On day 11, the tested drugs were injected intravenously (i.v.) in a small volume (100 μl) into embryos for each experiment. The embryos were incubated until day 17, at which time they were euthanized by transfer at 4°C followed by decapitation. Tumors were collected, and the tumor wet weights were recorded.
RESULTS
Role of eIF4E in SG formation under mild stress conditions.
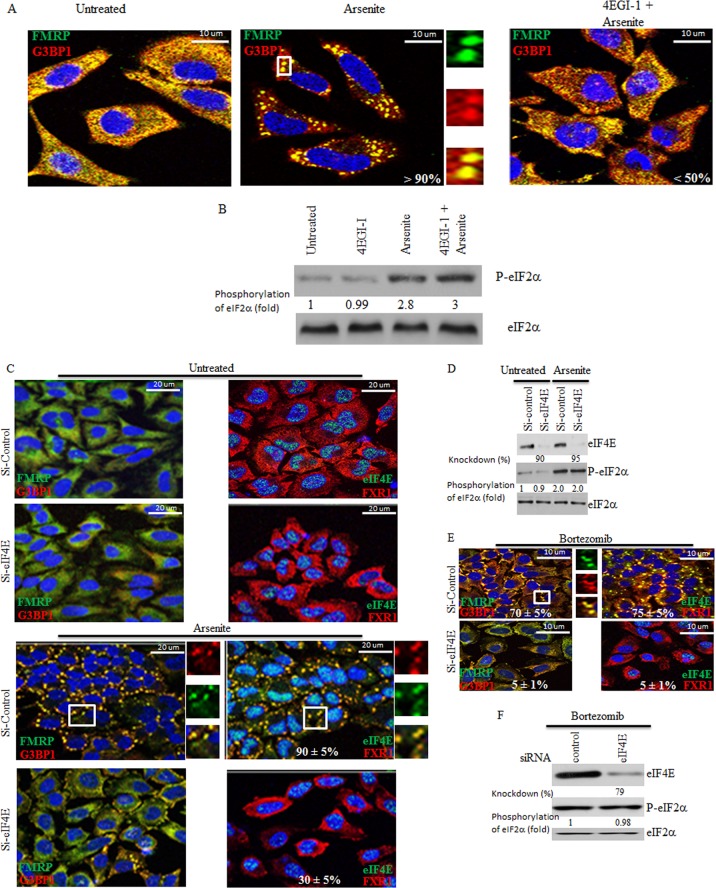
Since formation of SG occurs under conditions that inhibit cap-dependent translation initiation, it was suggested that this inhibition is sufficient to induce SG. Other studies have reported, however, that genetic depletion or chemical inactivation of the cap-binding protein eIF4E does not result in SG formation, thus establishing steps of translation initiation that can be inactivated without inducing SG (53). This result prompted us to investigate if eIF4E plays an active role in SG formation. First, we tested whether 4EGI-1, a compound that inactivates eIF4E by disrupting its interaction with both eIF4GI (54, 55) (see Fig. S1A in the supplemental material) and mRNA (55), alters formation of SG. HeLa cells were treated with 4EGI-1, and SG formation was then assessed upon arsenite (150 μM) treatment, using immunofluorescence with antibodies specific to the two SG markers FMRP and G3BP1 (56, 57). This mild arsenite concentration was chosen as the minimal level inducing SG in >90% of HeLa cells within 1 h. The results show that 4EGI-1 significantly reduced both the number (from 90% in mock-treated cells to ≤50% in 4EGI-1-treated cells) and the size (from ∼2 μm in mock-treated cells to ≤1 μm in 4EGI-1-treated cells) of SG upon arsenite treatment (Fig. 1A). Suppressing SG formation in HeLa by 4EGI-1 is not specific to arsenite, since we obtained similar results upon treatment with bortezomib (2 μM), a mild stressor (data not shown). SG formation that occurs upon either arsenite or bortezomib treatment is known to require phosphorylation of the translation initiation factor eIF2α (5, 6, 58–61), a key event known to inhibit translation initiation under stress situations (62). However, 4EGI-1 did not prevent the phosphorylation of eIF2α induced by either arsenite (Fig. 1B) or bortezomib (data not shown), even though it altered SG formation (Fig. 1A). The latter results exclude the possibility that 4EGI-1 inhibits SG formation under mild stress conditions by altering eIF2α phosphorylation and suggest that eIF4E is required for formation of SG, which is inhibited by 4EGI-1. To further confirm that eIF4E is required for SG formation, we knocked down its expression in HeLa cells with two different eIF4E-targeted siRNAs and assessed SG formation upon arsenite addition (150 μM) using immunofluorescence with antibodies specific to the three SG markers FMRP, G3BP1, and FXR1 as well as to eIF4E. We found that depletion of eIF4E (Fig. 1C and D) significantly reduced both the number (from 90% in mock-depleted cells to ∼30% in eIF4E-depleted cells) and the size (from ∼2 μm to ≤1 μm) of SG upon arsenite treatment (Fig. 1C), thus corroborating the results obtained with 4EGI-1 (Fig. 1A). This negative effect of eIF4E depletion is specific, since formation of SG was rescued in eIF4E-depleted cells by expressing murine eIF4E insensitive to eIF4E siRNAs used to deplete human eIF4E (see Fig. S1B and C in the supplemental material). Depletion of eIF4E or its inactivation by 4EGI-1 in both MCF-7 and Hs578T human breast carcinoma cells also significantly diminished formation of SG (see Fig. S1D to F in the supplemental material; also data not shown), indicating that the role of eIF4E in SG assembly is not limited to HeLa cells. Furthermore, the antagonistic effect of eIF4E depletion on SG formation is not specific to arsenite since we obtained similar results upon treatment with bortezomib. In this case, depletion of eIF4E with siRNAs decreased the proportion of SG-containing cells to ≤5% compared to 75% in mock-depleted HeLa cells (Fig. 1E). Depletion of eIF4E in HeLa cells does not affect the phosphorylation of eIF2α induced by either arsenite or bortezomib (Fig. 1D and F), excluding the possibility that depletion of eIF4E inhibits SG formation by altering eIF2α phosphorylation. Overall, these results unveil a novel requirement of eIF4E in SG formation under our mild stress conditions.
Fig 1.
Inactivating eIF4E or reducing its levels in HeLa cells impairs SG formation without affecting phosphorylation of eIF2α. (A and B) Cells were preincubated with 4EGI-1 (250 μM) for 6 h and then treated with 150 μM arsenite in the presence of 4EGI-1 (250 μM) for one additional hour. (A) Cells were then fixed, permeabilized, and processed for immunofluorescence using antibodies against different SG markers (FMRP in green and G3BP1 in red). DAPI (blue) was used as a nuclear stain. Pictures were taken using a 63× objective with a 1.5 zoom. The percentage of cells harboring SG (>3 granules/cell) from at least 5 different fields and 5 different experiments containing a total of 2 × 103 cells is indicated at the bottom of the merged images. Typical SG are shown in enlarged pictures. (B) Cells were then lysed, and total cell lysates were analyzed by Western blotting for eIF2α phosphorylation using anti-phospho-eIF2α antibodies. Total eIF2α was analyzed using pan-eIF2α antibodies. The amount of phosphorylated eIF2α was determined by densitometry quantitation of the film signal and is expressed as a percentage of total eIF2α. The results are representative of 3 different experiments. (C to F) Cells were treated with nonspecific or eIF4E-selective siRNAs for 96 h. Cells were then incubated with 150 μM arsenite for 1 h (C and D) or with 2 μM bortezomib for 4 h (E and F). (C and E) Cells were fixed, permeabilized, and then processed for immunofluorescence to detect SG using anti-FMRP (green), anti-G3BP1 (red), and anti-FXR1 (red) antibodies. Anti-eIF4E (green) antibodies were used in order to detect SG and to assess eIF4E depletion. DAPI (blue) was used as a nuclear stain. Pictures were taken using a 63× objective with a 1.5 zoom. The percentage of cells harboring SG (>3 granules/cell) from 5 different fields and 5 different experiments for a total of 2 × 103 cells is indicated in the merged images. Typical SG are shown. (D and F) Cells were lysed, and protein extracts were prepared and analyzed by Western blotting to detect eIF4E and phospho-eIF2α. Total eIF2α was analyzed using the pan-eIF2α antibodies and served as a loading control. The percentage of eIF4E knockdown was determined by densitometry quantification of the film signal using Photoshop and expressed as a percentage of total eIF2α. The amount of phosphorylated eIF2α was determined as described for panels A and B. The results are representative of 3 different experiments.
Inactivation of the mTORC1-eIF4E regulatory pathway impairs SG formation.
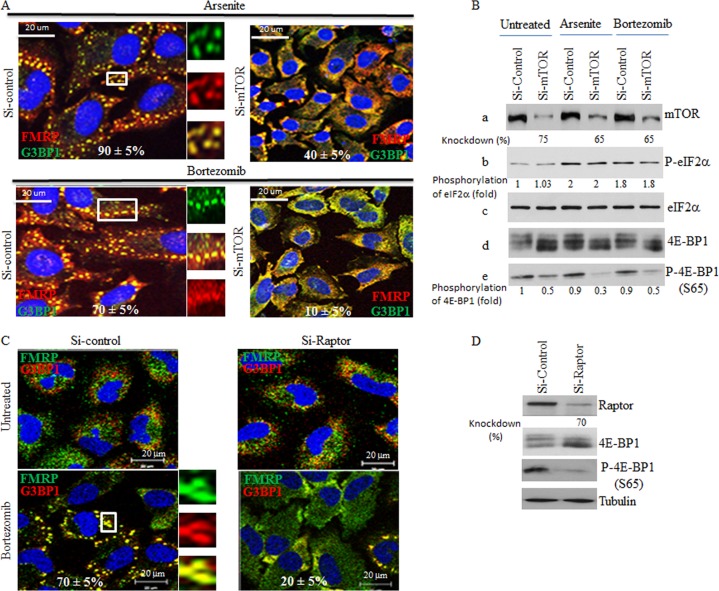
The mTORC1 signaling cascade is the main pathway known to regulate eIF4E-mediated cap-dependent translation in response to mitogens. The results described above clearly suggested a positive role of eIF4E in SG formation. We thus rationalized that mTORC1 might be involved in SG formation through the stimulation of eIF4E activity. To test this hypothesis, we first depleted mTOR using two specific siRNAs and then assessed SG formation in mTOR-depleted cells upon either arsenite (150 μM) or bortezomib (2 μM) treatment. We found that depletion of mTOR in HeLa cells significantly inhibited SG formation (from 90% in mock-depleted cells to ≤40% in mTOR-depleted cells) (Fig. 2A; see also Fig. S2 in the supplemental material), and we obtained similar results using both MCF-7 cells and Hs578T cells (data not shown), thereby unveiling a novel role of mTOR in formation of SG. Depletion of mTOR was assessed by both Western blot and immunofluorescence analysis using anti-mTOR antibodies (Fig. 2B; see also Fig. S2 in the supplemental material). A recent report described a localization of mTOR in SG upon arsenite (500 μM) treatment (63). However, we did not observe significant colocalization of mTOR with FMRP in the majority of SG induced by either arsenite (150 μM) or bortezomib (2 μM), indicating that mTOR was not quantitatively recruited to SG under our stress conditions (see Fig. S2 in the supplemental material). These results indicate that the role of mTOR in SG formation is unlikely to be related to its localization in SG. Control Western blot analysis (Fig. 2B) shows that mTOR depletion (panel a) did not affect either arsenite- or bortezomib-induced eIF2α phosphorylation (panels b and c). As expected, mTOR depletion reduced the phosphorylation of 4E-BP1, thus resulting in the accumulation of hypophosphorylated 4E-BP1 (panels d and e), a binding inhibitor of eIF4E. These results suggest a novel role of mTOR in SG formation via the phosphorylation of its 4E-BP1 target. Phosphorylation of 4E-BP1 is mediated specifically by mTORC1. 4E-BP1 is recruited to mTORC1 through Raptor, a specific component of mTORC1 (27, 28). If mTORC1 is required for formation of SG through phosphorylation of 4E-BP1, then altering mTORC1-mediated phosphorylation of 4E-BP1 through Raptor depletion should diminish formation of SG. We found that depletion of Raptor in HeLa cells using two specific siRNAs significantly inhibited SG formation under bortezomib treatment conditions (from 70% in mock-depleted cells to ≤20% in Raptor-depleted cells) (Fig. 2C). Under these conditions, depletion of rictor, the mTORC2-defining component, had no effect on formation of SG (data not shown). Depletion of Raptor reduced phosphorylation of 4E-BP1 (Fig. 2D) in concordance with previous studies (64). Together, these results suggest that mTORC1 is required for formation of SG through phosphorylation of 4E-BP1.
Fig 2.
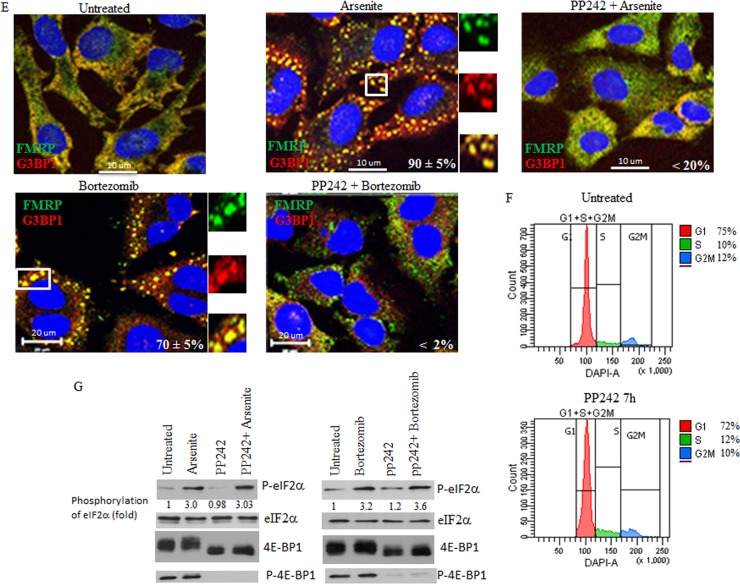
Depletion of either mTOR or Raptor as well as chemical inactivation of mTOR alters SG formation in HeLa cells without affecting eIF2α phosphorylation. (A and B) Cells were treated with nonspecific or mTOR-selective siRNAs for 96 h and then were incubated with either 150 μM arsenite for 1 h or with 2 μM bortezomib for 4 h. (A) Cells were fixed, permeabilized, and then processed for immunofluorescence to detect SG using anti-FMRP and anti-G3BP1. The percentage of cells harboring SG (>3 granules/cell) from 5 different fields and 5 different experiments containing a total of 2 × 103 cells is indicated at the bottom of the merged images. Representative SG are shown in enlarged pictures. (B) Cells were lysed, and protein extracts were prepared and analyzed by Western blotting to detect mTOR, phospho-eIF2α, 4E-BP1, and phospho-4E-BP1 (S65) using the appropriate antibodies. Total eIF2α serves as a loading control. The percentage of mTOR knockdown and the amount of phosphorylated eIF2α and phospho-4E-BP1 were determined as described above. The results are representative of 5 different experiments. (C and D) Depletion of Raptor inhibits formation of SG. Cells were treated with nonspecific or Raptor-selective siRNAs and were then incubated with 2 μM bortezomib for 4 h. (C) Cells were fixed, permeabilized, and then processed for immunofluorescence to detect SG using anti-FMRP and anti-G3BP1, as described above. The percentage of cells harboring SG from 5 different fields and 3 different experiments containing a total of 103 cells is indicated at the bottom of the merged images. (D) Cells were lysed, and protein extracts were prepared and analyzed by Western blotting to detect Raptor, 4E-BP1, phospho-4E-BP1 (S65), and tubulin (loading control) using the appropriate antibodies. The percentage of Raptor knockdown was determined as described above. The results are representative of 3 different experiments. (E) The mTOR inhibitor pp242 is a novel SG suppressor drug. Cells were either pretreated with pp242 (2.5 μM) for 6 h and then incubated with 150 μM arsenite plus 2.5 μM pp242 for 1 h (PP242+Arsenite) or pretreated with pp242 (2.5 μM) for 4 h and then incubated with 2 μM bortezomib for 4 h (PP242+Bortezomib). As controls, cells were treated with pp242 (2.5 μM) for 7 h (PP242), with 150 μM arsenite for 1 h (Arsenite), or with 2 μM bortezomib for 4 h (Bortezomib). Cells were then processed for immunofluorescence to detect SG using anti-FMRP (green) and anti-G3BP1 (red) antibodies. Pictures were taken using a 63× objective with a 1.5 zoom. The percentage of cells harboring SG (>3 granules/cell) from 5 different fields and 5 different experiments containing a total of 2 × 103 cells is indicated at the bottom of merged images. Typical SG are shown in enlarged pictures. (F) Cells were left untreated or were treated with 2.5 μM pp242 for 7 h, collected, washed with PBS, and then fixed with ethanol for 20 min. Cells were washed with PBS, stained with DAPI (1 μg/ml), and analyzed by flow cytometry. (G) Cells were either pretreated with pp242 (2.5 μM) for 6 h and then incubated with 150 μM arsenite plus 2.5 μM pp242 for 1 h (PP242+Arsenite) or were pretreated with pp242 (2.5 μM) for 4 h and then incubated with 2 μM bortezomib for 4 h (PP242+Bortezomib). As controls, cells were treated with pp242 (2.5 μM) for 7 h (PP242), with 150 μM arsenite for 1 h (Arsenite), or with 2 μM bortezomib for 4 h (Bortezomib). Cells were lysed, and protein extracts were prepared and analyzed by Western blotting to detect phospho-eIF2α, 4E-BP1, and phospho-4E-BP1 (S65) using the appropriate antibodies. Total eIF2α was analyzed using the pan-eIF2α antibodies and serves as a loading control. The amount of phosphorylated eIF2α was determined as described above. The results are representative of 5 different experiments.
Several drug inhibitors have been developed that target mTOR. The well-known inhibitor rapamycin reduces mTORC1 activity but is ineffective at preventing SG formation upon arsenite treatment (data not shown), as previously reported (65). A newer mTOR inhibitor, pp242, is more effective in suppressing mTORC1-mediated phosphorylation of 4E-BP1 than rapamycin (48, 51). We thus used pp242 to further assess the role of mTOR in the induction of SG formation. We found that treatment of HeLa cells with pp242 significantly inhibits both the number (from 90% in mock-treated cells to less than 30% in pp242-treated cells) and the size (from ∼2 μm to ≤0.5 μm) of SG upon treatment with arsenite (150 μM). There was a nearly complete abrogation of formation of SG by pp242 (from 70% in mock-treated cells to ∼2% in pp242-treated cells) upon treatment with bortezomib (2 μM) (Fig. 2E). It was previously shown that prolonged (48 h) treatment of mouse embryonic fibroblasts with the mTORC1 inhibitor pp242 induced their accumulation at the G1 phase (66). Our FACS analysis shows, however, that short (7-h) treatment of HeLa cells with pp242 had no effect on the cell cycle (Fig. 2F), although it abrogated formation of SG. These results exclude the possibility that the negative effect of pp242 on formation of SG upon treatment of HeLa with either arsenite or bortezomib is due to a cell cycle defect. The (∼70% to 90%) inhibition of SG formation by pp242 was even more drastic in MCF-7 cells (see Fig. S3A in the supplemental material) as well as in Hs578T and N2a mouse neuroblastoma cells (data not shown). Likewise, torin 1 inhibited the mTORC1-dependent phosphorylation of 4E-BP1 (49, 52). As expected, incubation of MCF-7 with torin 1 significantly (∼70%) reduced SG formation upon treatment with arsenite (250 μM) (data not shown). Torin 1 also reduces both the number and the size of SG induced by bortezomib and arsenite, respectively (see Fig. S3C; also data not shown) in HeLa cells, albeit less efficiently than pp242. These results indicate that pharmacological inhibition mTORC1 activity impairs SG formation in various cancer cell lines, as expected from the results obtained with siRNA-induced mTOR and Raptor depletions (Fig. 2A and C). We obtained similar results using mild heat shock (43°C) as a SG inducer (data not shown), further supporting the idea of a general role of mTORC1 in formation of SG. Moreover, control experiments showed that neither arsenite nor bortezomib had any significant effect on 4E-BP1 phosphorylation (Fig. 2G; see Fig. S3 and S4 in the supplemental material; also data not shown), indicating that under our experimental conditions, these compounds did not inhibit mTORC1-mediated 4E-BP1 phosphorylation. This is consistent with previous results showing that arsenite does not inhibit mTORC1 signaling (67, 68), which might thus promote SG formation, as described above (Fig. 2). As expected, both arsenite and bortezomib induced phosphorylation of eIF2α (Fig. 2B and G; see Fig. S4B), and both treatments induced polysome disassembly in either the absence or presence of pp242 in both HeLa and MCF-7 cells (see Fig. S4A; also data not shown), indicating that translation initiation was disrupted. Thus, although translation initiation was inhibited by either arsenite or bortezomib, mTORC1 remained active under those stress conditions (Fig. 2B and G; see also Fig. S3B and D and S4B) and its inhibition by pp242 (Fig. 2G; see also Fig. S3B) impaired SG formation (Fig. 2E; see also Fig. S2 and S3 in the supplemental material). Neither arsenite- nor bortezomib-induced phosphorylation of eIF2α (Fig. 2G; see also Fig. S4B in the supplemental material) was affected by pp242, ruling out the possibility that mTORC1 inhibition by pp242 suppresses formation of SG by altering the phosphorylation of eIF2α. Moreover, and consistent with previous data (50, 52, 69), mTORC1 inhibition reduced translation initiation, albeit minimally, as evidenced by the slight decrease in polysome peak sizes in pp242-treated HeLa cells (see Fig. S4A) and MCF-7 cells (data not shown). These results exclude the possibility that mTORC1 inhibition by pp242 altered SG formation by completely blocking general translation. Although pp242 (see Fig. S4A) did not dramatically affect general translation initiation, it induced a marked hypophosphorylation of 4E-BP1 in all cell lines tested, independently of the presence of either arsenite or bortezomib (Fig. 2G; see Fig. S3 and S4; also data not shown). These results suggest that mTOR inactivation impairs SG formation through a specific mechanism which likely involves 4E-BP1 hypophosphorylation.
PP242-induced hypophosphorylation of 4E-BP1 prevents eIF4E-mediated SG formation.
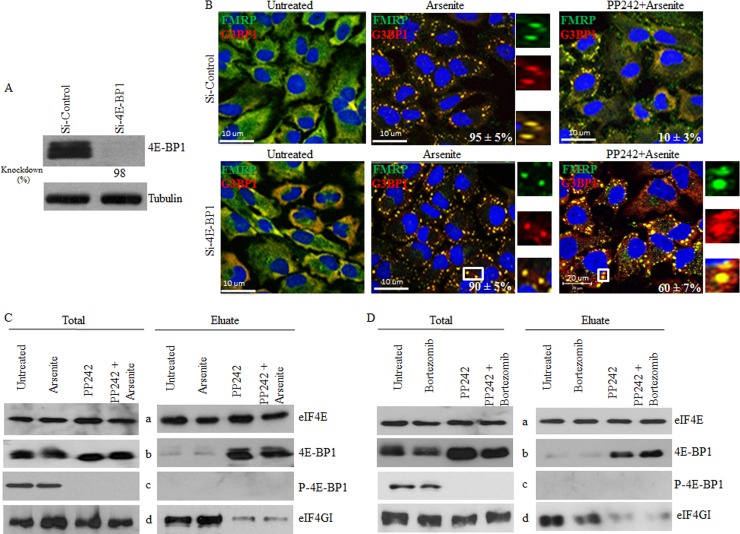
In the absence of mTOR activity, binding of hypophosphorylated 4E-BP1 to eIF4E prevents its association with eIF4GI, thus impairing formation of translation initiation complexes competent for mTORC1-dependent translation initiation. Thus, one possible mechanism by which mTOR inactivation impairs SG formation involves the disruption of eIF4E-eIF4GI associations by 4E-BP1. To test this possibility, we first assessed the role of 4E-BP1 in pp242-mediated SG suppression in HeLa cells. We depleted 4E-BP1 using two different siRNAs and assessed both arsenite- and bortezomib-induced SG formation upon mTOR inactivation with pp242 (Fig. 3; see also Fig. S5A in the supplemental material). Control experiments showed that 4E-BP1 depletion per se (Fig. 3A; see also Fig. S5A) does not prevent SG formation upon either arsenite treatment (Fig. 3B) or bortezomib treatment (see Fig. S5A), excluding a possible positive role of this protein in SG formation under our experimental conditions. This notion is supported by localization studies showing that 4E-BP1 is excluded from SG (see Fig. S5A), confirming a previous report (65). As expected, pp242-mediated mTOR inactivation reduced SG formation in mock-depleted cells (Fig. 3B; see also Fig. S5A). However, this negative effect of mTOR inhibition on arsenite-induced SG formation was significantly rescued by 4E-BP1 depletion (∼60% of pp242-treated cells contained SG) compared to mock-depleted cell results (∼10% of pp242-treated cells formed SG) (Fig. 3B). We obtained similar results upon bortezomib treatment (see Fig. S5A). 4E-BP1 has two homologs, 4E-BP2 and 4E-BP3 (70–72). In addition to 4E-BP1, HeLa cells express 4E-BP2, but they lack detectable 4E-BP3 (72). We found that depletion of 4E-BP2 (see Fig. S5D) does not rescue formation of arsenite-induced SG upon pp242 treatment (see Fig. S5C). Although we do not exclude the possibility of a minimal contribution of 4E-BP2, our results indicate that pp242-induced suppression of SG involves 4E-BP1 hypophosphorylation as a result of mTORC1 inhibition.
Fig 3.
mTOR inactivation-induced hypophosphorylation of 4E-BP1 prevents eIF4E-mediated SG formation by disrupting its interaction with eIF4GI. (A and B) Depletion of 4E-BP1 rescues arsenite-induced SG formation in pp242-treated cells. HeLa cells were treated with nonspecific or 4E-BP1-selective siRNAs for 96 h. (A) Cells were lysed, and protein extracts were prepared and analyzed by Western blotting to detect 4E-BP1 using specific antibodies. Tubulin serves as a loading control. (B) Cells were then left untreated or were treated with pp242 (2. 5 μM) for 7 h or arsenite (150 μM) for 1 h or were incubated with pp242 (2. 5 μM) for 6 h before addition of arsenite (150 μM) together with pp242 (2.5 μΜ) for an additional 1 h. Cells were then processed for immunofluorescence to detect SG using anti-FMRP and anti-G3BP1 antibodies. Pictures were taken using a 63× objective with a 1.5 zoom. The percentage of cells harboring SG (>3 granules/cell) from 5 different fields and 5 different experiments containing a total of 2 × 103 cells is indicated at the bottom of merged images. (C) pp242 disrupts eIF4F assembly, as monitored by cap-binding assays. HeLa cells were left untreated or were treated with pp242 (2. 5 μM) for 7 h or arsenite (150 μM) for 1 h or were incubated with pp242 (2.5 μM) for 6 h before addition of arsenite (150 μM) together with pp242 (2.5 μM) for an additional 1 h. Cells were lysed and then incubated with m7GTP-Sepharose beads. Eluted proteins were analyzed by Western blotting using specific antibodies. Total is 1% of the input used for cap pulldown. (D) HeLa cells were left untreated or were treated with pp242 (2. 5 μM) for 8 h or bortezomib (2 μM) for 4 h or were incubated with pp242 (2.5 μM) for 4 h before addition of bortezomib (2 μM) for an additional 4 h. Cells were lysed and then incubated with m7GTP-Sepharose beads. Eluted proteins were analyzed by Western blotting using specific antibodies. Total is 1% of the input used for cap pulldown.
Using cap-binding assays, we then investigated the possibility that hypophosphorylated 4E-BP1 suppresses SG formation by sequestering eIF4E, thus preventing its interaction with eIF4GI. HeLa cell lysates were incubated with the cap analogue m7GTP bound to Sepharose beads. The cap-binding protein eIF4E along with its bound partners was eluted and analyzed by Western blotting. Control experiments revealed a barely detectable interaction between eIF4E and 4E-BP1 in lysates prepared from either untreated or arsenite-treated cells (Fig. 3C, right, panels a to c). Alone or in combination with arsenite, pp242 induced a strong interaction between hypophosphorylated 4E-BP1 and eIF4E (Fig. 3C, right, panels a and b). As expected, no interaction between phosphorylated 4E-BP1 and eIF4E was detected in these assays under any of the conditions tested (Fig. 3C, right, panels a and c). We obtained similar results in cells treated with pp242 together with bortezomib (Fig. 3D) and in MCF-7 cells (data not shown). Thus, mTORC1 inactivation by pp242 likely contributes to SG suppression by inducing the interaction of eIF4E with its hypophosphorylated 4E-BP1 inhibitory factor. Alone or in combination either with arsenite (Fig. 3C, right, panels a and d) or with bortezomib (Fig. 3D, right, panels a and d), pp242 strongly decreased the amount of eIF4GI, a molecular mimic of 4E-BP1, bound to eIF4E. These results support the assumption that 4E-BP1 antagonizes SG formation by competing against eIF4E-eIF4GI interactions. Moreover, the negative effect of pp242 on eIF4E-4GI interactions was rescued in 4E-BP1-depleted cells (see Fig. S5B in the supplemental material), in keeping with previous data obtained using mouse embryonic fibroblasts (MEFs) lacking 4E-BPs (66). The latter result is consistent with the above-described rescue data (Fig. 3B; see also Fig. S5A), showing the formation of SG in 4E-BP1-depleted cells despite mTOR inhibition with pp242. As a control, depletion of 4E-BP2 does not rescue eIF4E-4GI interactions in pp242-treated cells (see Fig. S5E), in keeping with a recent study showing a dispensable role of 4E-BP2 in disrupting eIF4E-4GI association in torin 1-treated cells (52). This lack of effect of 4E-BP2 depletion on eIF4E-4GI association is also consistent with our results showing a dispensable role of 4E-BP2 in mediating the pp242-negative effects on formation of SG (see Fig. S5C). Overall, these results suggest that targeting mTORC1-induced eIF4E-eIF4GI association by the pp242-mediated hypophosphorylation of 4E-BP1 impairs SG formation.
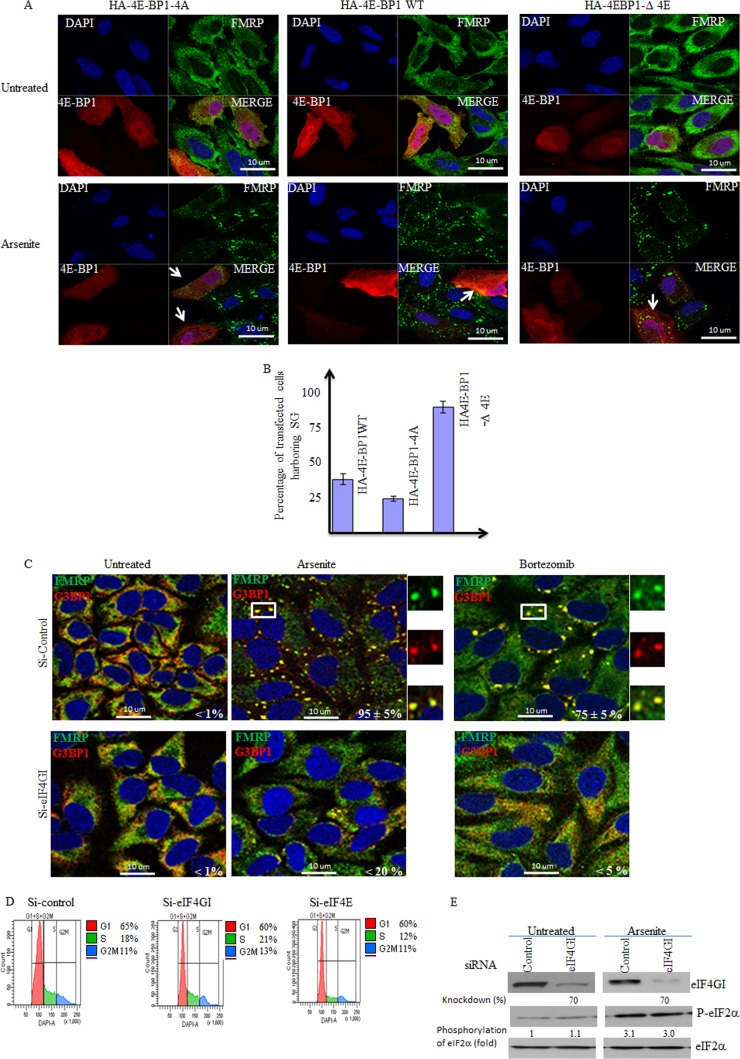
4E-BP1 antagonizes SG formation when it is hypophosphorylated.
The results described above suggest that 4E-BP1 plays an antagonistic role in SG formation by disrupting eIF4E-4GI interactions. Therefore, expressing a dominant-negative and constitutively hypophosphorylated 4E-BP1-4A mutant, as well as RNA interference (RNAi)-mediated depletion of eIF4GI, should disrupt eIF4F complexes, thus suppressing SG formation. Using cap-binding assays, we first analyzed eIF4E-eIF4GI interactions in cells overexpressing the constitutively hypophosphorylated 4E-BP1-4A mutant. Control experiments (see lane 1 in Fig. S5F in the supplemental material) showed, as expected, a strong interaction between eIF4E and eIF4GI in mock-transfected cells. While a strong interaction between 4E-BP1-4A and eIF4E was found, eIF4GI was barely recovered in these 4E-BP1-4A-eIF4E complexes (see lane 2 in Fig. S5F), indicating that the eIF4E-eIF4GI interaction is lost in cells expressing the hypophosphorylated 4E-BP1-4A mutant form. We also detected an interaction between wild-type (wt) 4E-BP1 and eIF4E, albeit it was less strong than the 4E-BP1-4A-eIF4E interaction (see lane 3 in Fig. S5F). However, this apparent weaker interaction between wt 4E-BP1 and eIF4E appears to be sufficient to disrupt the eIF4E-eIF4GI interaction, as evidenced by the reduced recovery of eIF4GI in eIF4E complexes (see lane 3 in Fig. S5F). Control experiments showed that expression of an 4E-BP1 mutant (4E-BP1-Δ 4E) which is unable to interact with eIF4E had no effect on eIF4E-eIF4GI association in our assays (see lane 4 in Fig. S5F). Overall, these results show that expression of either wt 4E-BP1 or its hypophosphorylated 4E-BP1-4A mutant disrupts eIF4E-eIF4GI interaction. Expression of either wt 4E-BP1 or its hypophosphorylated 4E-BP1-4A mutant significantly (∼50% to 55% or 75% to 80%, respectively) prevented SG formation upon arsenite treatment (Fig. 4A and B), as well as under conditions of bortezomib treatment (data not shown), compared to mock-transfected cell results. As expected, expression of 4E-BP1-Δ 4E has much less (∼10%) effect on SG formation. These results indicate that 4E-BP1 inhibits SG formation mainly through its hypophosphorylated form, which disrupts formation of mTORC1-dependent translation initiation complexes by binding eIF4E, thereby preventing its association with eIF4GI. Our immunofluorescence (Fig. 4A; see also Fig. S5A in the supplemental material) and Western blot (see Fig. S5G) experiments showed that overexpressed 4E-BP1 localizes in both nuclei and cytoplasm, consistent with previous data (65). Fractionation studies showed, however, that eIF4E is distributed mainly in the cytoplasm where 4E-BP1 is sequestering it (see Fig. S5G). Finally, we assessed whether disruption of eIF4E-eIF4GI through eIF4GI depletion could suppress SG formation. We found that depletion of eIF4GI in both HeLa (Fig. 4C; see also Fig. S6 in the supplemental material) and MCF-7 (data not shown) cells with two specific siRNAs strongly (>70%) inhibits SG formation upon either arsenite or bortezomib treatment, thus identifying eIF4GI as a novel factor required for SG formation. Depletion of eIF4GI did not affect either the cell cycle (Fig. 4D) or phosphorylation of eIF2α upon arsenite (Fig. 4E) or bortezomib (data not shown) treatment, in keeping with the results obtained upon eIF4E depletion (Fig. 1 and 4). These results exclude the possibility that eIF4GI depletion inhibits SG formation by arresting the cell cycle or by altering eIF2α phosphorylation and support the notion of a positive role of the eIF4E-eIF4GI interaction in SG formation. In conclusion, we have uncovered a novel role for mTORC1-induced eIF4E-eIF4GI interactions in SG formation. Furthermore, 4E-BP1 inhibits mTORC1-dependent SG formation by disrupting eIF4E-4GI association.
Fig 4.
eIF4GI is required for formation of SG, which is antagonized by 4E-BP1. (A and B) HeLa cells were transfected with wild-type (WT) HA-4E-BP1 or HA-4E-BP1-4A or with HA-4E-BP1-Δ4E for 48 h and were then treated with arsenite (150 μM) for 1 h. (A) Cells were processed for immunofluorescence to detect 4E-BP1 with anti-4E-BP1 antibodies or SG using anti-FMRP antibodies. (B) The percentage of transfected cells (indicated by arrows) harboring SG (>3 granules/cell) from 5 different fields and 3 different experiments containing a total of 500 cells is indicated. (C to E) eIF4GI is a novel SG-promoting factor. Cells were treated with nonspecific or eIF4GI-specific siRNAs for 96 h. Cells were treated with 150 μM arsenite for 1 h or with 2 μM bortezomib for 4 h. (C) HeLa cells were fixed, permeabilized, and then processed for immunofluorescence to detect SG using anti-FMRP and anti-G3BP1 antibodies. Pictures were taken using a 63× objective with a 1.5 zoom. The percentage of cells harboring SG was calculated as described for Fig. 2. (D) Cells were treated with nonspecific or eIF4E- or eIF4GI-specific siRNAs for 96 h. Cells were collected, washed with PBS, and then fixed with ethanol for 20 min. Cells were washed with PBS, stained with DAPI (1 μg/ml), and analyzed by flow cytometry. (E) Cells were treated with nonspecific or eIF4GI-specific siRNAs for 96 h. Cells were lysed, and protein extracts were prepared and analyzed by Western blotting to detect eIF4GI and phospho-eIF2α. Total eIF2α was analyzed using the pan-eIF2α antibodies and serves as a loading control. The percentage of eIF4GI knockdown and phosphorylated eIF2α was determined as described above by densitometry quantification of the film signal using Photoshop, and results are expressed as a percentage of total eIF2α. The results are representative of 3 different experiments.
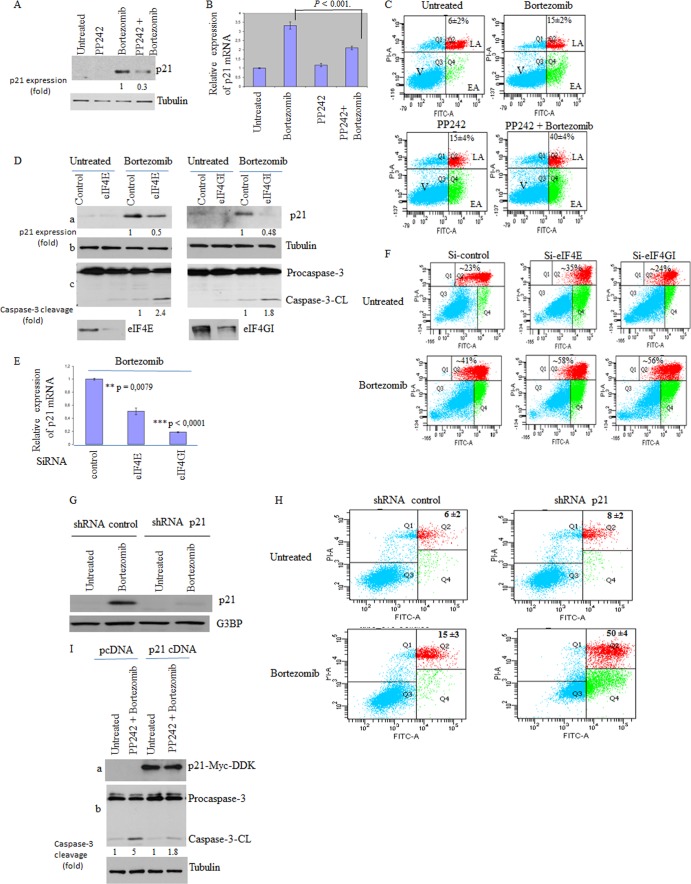
PP242 sensitizes cancer cells to bortezomib-mediated apoptosis in vitro and inhibits tumor growth in vivo.
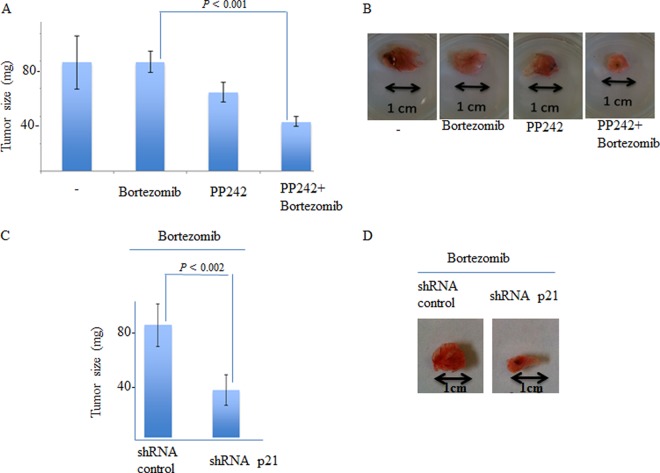
Previous studies have implicated SG in HeLa and MCF-7 cell resistance to bortezomib-mediated apoptosis (5, 6). This SG-mediated resistance to bortezomib involves upregulation of the expression of the anti-apoptotic factor p21WAF1/CIP1, which, in turn, is due to the accumulation of p21 mRNA within SG (6). Moreover, interfering with p21 expression, either by siRNA-mediated p21 mRNA depletion or by inducing p21 mRNA degradation by blocking its recruitment within SG, promoted apoptosis during bortezomib treatment (6). Based on the evidence described above and since SG inhibit apoptosis (73), we hypothesized that impairing SG formation either through mTORC1 inactivation with pp242 or by depleting eIF4E and eIF4GI sensitizes cancer cells to bortezomib-induced apoptosis. First, we assessed whether the inhibition of SG formation by pp242 or through depletion of eIF4E and eIF4GI might prevent SG-associated p21 upregulation following bortezomib treatment. p21 mRNA and its protein product have a short half-life and are thus barely detectable under normal growth conditions, as confirmed in control experiments (Fig. 5A, B, and D). Neither the level of p21 mRNA nor its expression in HeLa cells (Fig. 5A and B) or in MCF-7 cells (see Fig. S7A and B in the supplemental material) was affected by pp242, indicating that p21 mRNA is unlikely to be a direct target of mTOR signaling. In agreement with previous data (6), bortezomib induced an upregulation of p21 expression in both HeLa and MCF-7 cells at both the protein level (Fig. 5A and D; see also Fig. S7A) and the mRNA level (Fig. 5B and E; see also Fig. S7B). Since p21 mRNA accumulation induced by bortezomib is related to its stabilization within SG (6), we reasoned that disrupting SG formation with pp242 or by depleting eIF4E or eIF4GI or mTOR should prevent p21 mRNA accumulation, thereby reducing its expression. We found that pp242 significantly (by 3-fold) abrogated bortezomib-induced p21 expression (Fig. 5A; see also Fig. S7A). Depleting eIF4E, eIF4GI, or mTOR also reduced p21 expression under bortezomib conditions (Fig. 5D, panels a, and data not shown), corroborating the pp242 results. This negative effect on bortezomib-mediated p21 upregulation was due, at least in part, to a reduced steady-state p21 mRNA level (Fig. 5B and E; see Fig. S7B; also data not shown), which reflects the inhibition of SG formation under these conditions (Fig. 1 and 2). These results indicate that pp242 impairs SG formation and blocks its downstream p21 upregulation pathway, which together could contribute in sensitizing cancer cells to bortezomib-mediated apoptosis. To test this assumption, HeLa and MCF-7 cells were treated with pp242 and exposed to bortezomib (2 μM), and apoptosis was then measured with an annexin V assay. Treatment with pp242 did not significantly induce apoptosis in either HeLa cells (Fig. 5C) or MCF-7 cells (see Fig. S7C in the supplemental material), indicating that mTOR inactivation is at best a weak inducer of apoptosis, in keeping with previous data (50). Following incubation with bortezomib, however, pp242 promoted apoptosis in a high percentage (∼40%) of both HeLa (Fig. 5C) and MCF-7 (see Fig. S7C) cells compared to incubation with bortezomib alone (∼15% to 20%). Downregulating eIF4E or eIF4GI also promoted apoptosis during bortezomib treatment of HeLa cells, as assessed by caspase-3 cleavage and annexin V experiments (panels c of Fig. 5D and F), and we obtained similar results upon mTOR depletion (data not shown). These results suggest that suppression of SG either with pp242 or though depletion of eIF4E and eIF4GI sensitizes cancer cells to bortezomib-mediated apoptosis, which could involve p21 downregulation, although disruption of additional antiapoptotic pathways is likely to be involved. In keeping with our assumption, however, we found that depletion of p21 using specific shRNAs (Fig. 5G) significantly (∼40%) promoted apoptosis under bortezomib treatment conditions, corroborating the pp242 results (Fig. 5H). Moreover, overexpressing p21 (we used a plasmid-encoding p21 that lacks regulatory 5′ and 3′ [UTR] to ensure its constitutive expression) (Fig. 5I, panel a) partially reduced bortezomib-mediated apoptosis of HeLa cells, despite TORKinibs treatment (Fig. 5I, panel b). Overall, these results indicate that pp242 sensitizes cancer cells to bortezomib-mediated cell death in vitro, at least in part by downregulating the SG-associated p21 antiapoptotic pathway. The results described above may also indicate a negative effect of this mTOR/SG inhibitor on the growth of chemoresistant tumors. We assessed this hypothesis in chick embryos using the chick chorioallantoic membrane (CAM) in vivo assay (74–77). This in vivo model for tumor growth benefits from the fact that the CAM, a highly vascularized tissue that surrounds the chicken embryo, is in an external and easily accessed position. Moreover, CAM assays allow rapid assessment of the effects of chemotherapeutic drugs on tumor formation by the use of human cancer cells grafted onto the CAM. Grafting of HeLa cells onto the CAM resulted of tumor formation in nearly all of the 100 embryos used for the experiments. Control experiments show that bortezomib alone does not affect HeLa tumor formation in CAM assays (Fig. 6A and B). This result is consistent with previous data showing that bortezomib is inefficient for treating solid tumors (16). pp242 treatment has a slight effect on tumor growth, corroborating recent results obtained using mouse models (69). There was, however, an ∼2-fold reduction in the growth of HeLa tumors in the CAM assay when the treatment also combined bortezomib and pp242 (Fig. 6A and B), indicating that mTOR inactivation may synergistically inhibit solid-tumor growth with bortezomib. We then tested if depleting p21, one among the other possible downstream effectors of mTORC1-SG pathway, could also inhibit the growth of solid tumors the CAM assay. For this, we used our HeLa cells expressing either control shRNA or p21 shRNA (Fig. 5G), which we grafted onto the CAM. Grafting of HeLa cells expressing either control shRNA or p21 shRNA onto the CAM resulted in the formation of tumors of similar sizes in nearly all of the 100 embryos used for the experiments (data not shown). Under bortezomib treatment conditions, however, there was an ∼2-fold reduction in the growth of HeLa tumors expressing p21 shRNAs in the CAM assay when they were treated with bortezomib compared to the growth of HeLa tumors expressing control shRNAs (Fig. 6C and D). We obtained similar results using HeLa cells stably expressing different p21 shRNAs (data not shown). These results indicate that p21 downregulation synergistically inhibits solid-tumor growth with bortezomib treatment, further suggesting that it may contribute to the effect of pp242 in inhibiting the growth of solid tumors when such treatment is combined with bortezomib treatment. Overall, our studies show that mTOR inactivation by pp242 inhibits bortezomib-induced SG formation and resistance to apoptosis in vitro and reduces tumor formation in vivo when such treatment is combined with bortezomib treatment.
Fig 5.
PP242 and depletion of either eIF4E or eIF4GI downregulate the SG-associated p21 pathway and sensitize cancer cells to bortezomib. (A) HeLa cells were incubated with pp242 (2.5 μM) or with bortezomib (2 μM) or with both compounds for 16 h. Cells were then lysed and proteins resolved on SDS-PAGE and analyzed by Western blotting for p21 expression. Tubulin serves as a loading control. (B) Quantitative RT-PCR (qRT-PCR) of p21 mRNA. Following treatment with bortezomib (2 μM) or with pp242 (2.5 μM) or with both compounds for 4 h, cells were collected and total RNA content was then isolated. The amount of p21 was quantified by qRT-PCR relative to GAPDH mRNA using the threshold cycle (ΔΔCT) method. Results are expressed as the means ± SEM (error bars) of triplicate measurements. (C) HeLa cells were treated as described for panel A, collected, and then stained with annexin V-FITC and PI and analyzed by flow cytometry. The percentage of total dead or dying cells (indicated at the top of each panel) was defined as the sum of early (lower right box) and late (upper right box) apoptosis and corresponds to the means ± SEM of the results of 3 independent experiments. V, viable cells; EA, early apoptosis; LA, late apoptosis. (D to F) HeLa cells were treated with nonspecific or eIF4E- or eIF4GI-specific siRNAs for 96 h. Cells were treated with 2 μM bortezomib for 16 h. (D) Cells were then lysed, and extracted proteins were analyzed by Western blotting for the expression of p21, eIF4E, and eIF4GI as well as for caspase-3 cleavage. Tubulin serves as a loading control. (E) qRT-PCR of p21 mRNA as described above. (F) Cells were stained with annexin V-FITC and PI as described above and analyzed by flow cytometry. (G and H) Depletion of p21 promotes bortezomib-mediated apoptosis. (G) HeLa cells stably expressing either shRNA control or shRNA p21 were treated with bortezomib and then collected. Proteins extracts were then prepared and analyzed for p21 expression using anti-p21 antibodies. G3BP1 serves as a loading control. (H) HeLa cells stably expressing either shRNA control (left panel) or shRNA p21 (right panel) were treated with bortezomib for 16 h and then analyzed by staining with annexin V-FITC and PI and flow cytometry. The percentage of apoptotic cells is indicated at the top of each panel, and the values represent the means ± standard errors of the means (SEM) of the results of 3 independent experiments. (I) Overexpression of p21 reduces the effects of pp242 in promoting bortezomib-mediated apoptosis. HeLa cells transfected with either p21-myc-DDK or pcDNA for 20 h were treated with bortezomib (2 μM) or with pp242 (2.5 μM) and bortezomib (2 μM) for 16 h. Cells were then lysed, and extracted proteins were analyzed by Western blotting for p21 expression and caspase-3 cleavage. Tubulin serves as a loading control.
Fig 6.
Both depletion of p21 and depletion of pp242 inhibit the formation of bortezomib-resistant tumors in the chick CAM assay. (A and B) HeLa cells (1 × 106 cells/egg) were inoculated directly onto the CAM tissue of 10-day-old embryos. Bortezomib (60 ng/egg) or pp242 (80 ng/egg) or both were then injected i.v. on day 11 into a total of 100 embryos as described in Materials and Methods. At day 17, embryos were euthanized and decapitated and the tumor wet weight was recorded. (B) Representative tumors are shown. (C and D) p21 depletion inhibits the formation of bortezomib-resistant tumors in the chick CAM assay. HeLa cells that were stably expressing either shRNA control (left panel) or shRNA p21 (right panel) (1 × 106 cells/egg) were inoculated directly onto the CAM tissue of 10-day-old embryos. Bortezomib (60 ng/egg) was then injected i.v. on day 11 in a total of 100 embryos as described in Materials and Methods. (C) At day 17, embryos were euthanized and decapitated and the tumor wet weight was recorded. Representative tumors are shown in panel D.
DISCUSSION
The role of the mTOR-eIF4E pathway in regulating SG formation has never been documented to date. Here we established a novel mechanism of SG formation which relies on mTORC1 activity. First, we identified the cap-binding protein eIF4E and its interactor eIF4GI as two novel factors involved in SG formation. mTORC1 specifically promotes this eIF4E-4GI-mediated formation of SG via 4E-BP1 phosphorylation. In support of this result, overexpression of hypophosphorylated 4E-BP1 suppressed SG formation by preventing eIF4E interactions with eIF4GI, and depletion of 4E-BP1 rescued both SG formation and the eIF4E-4GI interaction in pp242-treated cells. Finally, disrupting formation of SG by depletion of eIF4E or eIF4GI blocks the SG-associated antiapoptotic p21 pathway and sensitizes cancer cells to bortezomib-mediated cell death in vitro. We recapitulated these results by inactivating mTOR with pp242, which prevented tumor formation in vivo.
The eIF4E-eIF4GI interaction promotes SG formation under mild stress conditions: role of mTORC1.
The composition of SG comprises most translation initiation factors, including the three eIF4F components 4G, 4E, and 4A. However, the requirement of translation initiation factors in SG formation is still largely unknown. Our present study has identified eIF4E and eIF4GI as two novel factors required for formation of SG under mild stress conditions. eIF4E is responsible for the early recognition of capped mRNAs during translation initiation, and this interaction is stabilized by eIF4GI, resulting in the activation of translation initiation. The interaction between eIF4E and eIF4GI is mainly regulated by mTORC1-mediated phosphorylation of 4E-BP1. Our results established SG formation as representing a novel role of the mTORC1-mediated eIF4E-eIF4GI interaction in stress response. First, we found that depletion of either the mTORC1-defining Raptor component or mTOR itself significantly alters SG formation under mild stress conditions in different cancer cell lines (Fig. 2; see also Fig. S1 in the supplemental material). A recent report described the localization of mTORC1 components in mammalian SG induced by severe stress conditions such as the presence of 500 μM arsenite (63). Under the mild stress conditions used here, however, mTOR is barely detected in SG (see Fig. S2 in the supplemental material), making unlikely the possibility that mTORC1 could serve by itself as a scaffolding complex for formation of SG. Second, pp242-mediated mTORC1 inactivation abrogates both eIF4E-eIF4GI interactions and SG formation (Fig. 2 and 3). Both SG formation and eIF4E-4GI interactions are, however, rescued in 4E-BP1-depleted cells (Fig. 3A and B; see also Fig. S5A and B in the supplemental material), demonstrating that this effect of pp242 on SG formation is likely due to eIF4E sequestration by 4E-BP1, which competitively disrupts eIF4E-eIF4GI interactions. Our results thus suggest a novel role of hypophosphorylated 4E-BP1 in inhibiting formation of SG via eIF4E sequestration, a notion which is further supported by overexpression experiments. As expected, expression of 4E-BP1 inhibited formation of SG upon arsenite treatment (Fig. 4A and B). Expression of constitutively hypophosphorylated 4E-BP1 had a more drastic effect in preventing SG formation. This indicates that the antagonistic effect of 4E-BP1 on SG formation is mediated by its hypophosphorylated form, which sequesters eIF4E, thereby preventing its interaction with eIF4GI (see Fig. S5 in the supplemental material). Finally, expression of a 4E-BP1 mutant that cannot bind eIF4E fails to inhibit eIF4E-eIF4GI interactions (see Fig. S5F in the supplemental material), thereby preserving formation of SG upon exposure to stress (Fig. 4A). We thus speculate that pp242-mediated hypophosphorylation of 4E-BP1 suppresses SG formation through disruption of the eIF4E-eIF4GI interaction. In a recent study, it was reported that treatment of cells with selenite induces the sequestration of eIF4E by 4E-BP1 and promotes formation of noncanonical SG. These SG were referred by those authors as type 2 SG, as opposed to the canonical SG induced by arsenite (78). Selenite-induced type 2 SG lack several canonical SG components and have a reduced size compared to type 1 canonical SG. Formation of selenite-induced type 2 SG was shown to be reinforced by phosphorylation of eIF2α but reduced upon 4E-BP1 depletion, suggesting a possible role of phosphorylation of both eIF2α and 4E-BP1 in promoting formation of selenite-induced type 2 SG. However, this depletion of 4E-BP1 does not prevent formation of SG induced by arsenite (Fig. 3), corroborating previous data (78). Depletion of 4E-BP1 also had no effect on bortezomib-induced SG (see Fig. S5A in the supplemental material), ruling out an essential role for this protein in promoting formation of SG under both arsenite and bortezomib treatment conditions. Finally, 4E-BP1 is excluded from SG induced by either arsenite (Fig. 4) or bortezomib (see Fig. S5 in the supplemental material), which is consistent with a dispensable role of this protein in the formation of SG. Overall, our study identified 4E-BP1 as a novel SG-antagonizing factor under conditions of mild treatment with arsenite and bortezomib, whose activation by mTORC1 inhibitor pp242 prevents formation of SG.
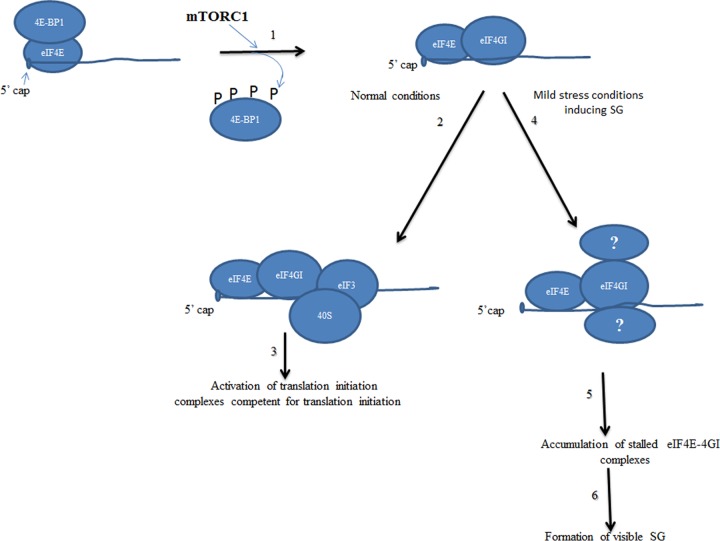
Neither pp242 nor torin 1 induces phosphorylation of eIF2α, and both have a minimal effect on general translation initiation (see Fig. S4 in the supplemental material), in keeping with previous data showing that mTORC1 inhibition by either drug selectively impairs the translation of a subset of mRNAs (52, 64, 79, 80). The best-characterized mTORC1-mRNA targets are those encoding factors of cell proliferation and protein synthesis (52, 64). Translation of mTORC1-mRNA targets is highly dependent on the eIF4E-eIF4GI interaction, and loss of this interaction with pp242 reduces the capacity of eIF4E to bind mTORC1-mRNA targets much more than that seen with other mRNAs, explaining why mTORC1 inactivation selectively suppresses their translation. mRNAs being core components of SG, one likely mechanism by which inactivation of mTORC1 prevents formation of SG could involve the release of mTORC1-mRNA targets from eIF4E, which otherwise would promote recruitment of specific SG-promoting factors. eIF4E-4GI complexes might also be required for formation of SG through recruitment of key SG-promoting factors by direct interactions. This is consistent with recent studies reporting interactions between specific SG-promoting factors, many of which are translation repressors, with either eIF4E or yeast eIF4G (81–83). We thus propose as a model (Fig. 7) such that under stress conditions, eIF4E-eIF4GI complexes could serve as a scaffold for recruitment of specific translation repressors, not identified yet, in an mTORC1-dependent manner. This binding would then stall the translation initiation complexes whose accumulation leads to SG formation. Future experiments are needed to establish how mTORC1-mediated eIF4E-eIF4GI interactions contribute to the formation of SG.
Fig 7.
Our working model of mTORC1-eIF4E-4GI-dependent mode of SG assembly. Under both normal and SG-inducing stress conditions, mTORC1 drives formation of eIF4E-eIF4GI translation initiation complexes through phosphorylation of its 4E-BP1 target (1). Under normal growth conditions, eIF4E-4GI complexes are joined by 40S ribosomes at an early step of translation initiation (2 and 3). Under mild stress conditions inducing SG, eIF4E-4GI complexes may serve as scaffolding for the recruitment of unidentified factors in an mTORC1-dependent manner (4). This binding then stalls eIF4E-4GI complexes in an inactive status (5) and results in accumulation leading to formation of SG (6).
mTOR inhibition as a possible strategy to circumvent chemoresistance.
Our results established that depletion of either eIF4E or eIF4GI, as well as inhibition of the oncogenic mTOR pathway, impairs bortezomib-induced SG. This suggests that mTORC1-dependent eIF4E-eIF4GI complex formation plays an essential role in SG assembly under conditions of treatment with a therapeutically relevant proteasome inhibitor. SG formation has recently been shown to suppress cancer cell death in response to chemotherapeutic and genotoxic drugs (5, 6, 10), which could explain the previously described role of SG in promoting tumor cell radioresistance (1). We have now shown that disrupting the mTORC1-eIF4E-4GI pathway either using pp242 or through depletion of eIF4E and eIF4GI or mTOR itself prevents bortezomib-induced SG (Fig. 2 and 4), thereby downregulating its associated p21 antiapoptotic pathway in cancer cells. In keeping with these results, both the presence of pp242 and depletion of eIF4E, eIF4GI, or mTOR sensitize cancer cells to apoptosis in vitro (Fig. 5 and data not shown). We attempted to rescue apoptosis resistance to bortezomib in eIF4E- or eIF4GI-depleted cells by overexpressing p21. However, and despite our efforts, we could not achieve conditions that allowed reproducible efficient depletion of either eIF4E or eIF4GI upon overexpression of p21, precluding statistical analysis of rescuing apoptosis resistance to bortezomib. Nevertheless, and in agreement with our results showing that p21 downregulation sensitizes cancer cells to bortezomib in vitro (Fig. 5), overexpression of p21 partially reduced bortezomib-mediated apoptosis despite treatment with pp242. Thus, downregulation of p21 contributes to the effects of pp242 in sensitizing cancer cells to bortezomib, although additional pathways could be involved. Finally, and in keeping with the results described above, pp242 suppresses formation of bortezomib-resistant tumors in chick embryos (Fig. 6). The present work has thus identified a novel role for the oncogenic mTOR-eIF4E-eIF4GI pathway in promoting resistance to bortezomib. mTORC1-mediated formation of SG might constitute an additional pathway by which mTOR induces such chemoresistance. It will be interesting to determine whether targeting the mTORC1-eIF4E-eIF4GI pathway might sensitize cancer cells to other therapeutic conditions resulting in SG formation, such as radiation therapy. In addition to bortezomib, other chemotherapeutic drugs are likely to induce SG, whose formation could contribute to apoptosis resistance. Further investigations will help to determine if mTORC1 has a general role in promoting the induction of SG formation by relevant chemotherapeutic drugs and associated chemoresistance. These findings should encourage a broader evaluation of the efficacy of TORKinibs in sensitizing chemoresistant tumors to various drugs.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Imed Gallouzi (McGill University) for providing the anti-G3BP1 and to Edward Khandjian and Jean-Yves Masson (Laval University) for their gift of anti-FXR1 and anti-histone 3 antibodies, respectively. We are thankful to Anne Cammas and Sergio Di Marco (McGill University) for their helpful advice with polysomes. We also thank Yves Labelle and Richard Poulin (CRCHU) for editing the manuscript.
This work was supported by a Canadian Institute Health Research (CIHR) grant (MOP-79334) to R.C.-G. and by both CIHR grant MOP-IC093226-CBT and Canadian Foundation for Innovation grant MOP-GF091050 to R.M. R.M. is a recipient of a CIHR New Investigator Scholarship award.
We declare that no competing interests exist.
Footnotes
Published ahead of print 1 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01517-12.
REFERENCES
- 1. Moeller BJ, Cao Y, Li CY, Dewhirst MW. 2004. Radiation activates HIF-1 to regulate vascular radiosensitivity in tumors: role of reoxygenation, free radicals, and stress granules. Cancer Cell 5:429–441 [DOI] [PubMed] [Google Scholar]
- 2. Gardner LB. 2008. Hypoxic inhibition of nonsense-mediated RNA decay regulates gene expression and the integrated stress response. Mol. Cell. Biol. 28:3729–3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mazroui R, Sukarieh R, Bordeleau ME, Kaufman RJ, Northcote P, Tanaka J, Gallouzi I, Pelletier J. 2006. Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2alpha phosphorylation. Mol. Biol. Cell 17:4212–4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McInerney GM, Kedersha NL, Kaufman RJ, Anderson P, Liljestrom P. 2005. Importance of eIF2alpha phosphorylation and stress granule assembly in alphavirus translation regulation. Mol. Biol. Cell 16:3753–3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fournier MJ, Gareau C, Mazroui R. 2010. The chemotherapeutic agent bortezomib induces the formation of stress granules. Cancer Cell Int. 10:12 doi:10.1186/1475-2867-10-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gareau C, Fournier MJ, Filion C, Coudert L, Martel D, Labelle Y, Mazroui R. 2011. p21(WAF1/CIP1) upregulation through the stress granule-associated protein CUGBP1 confers resistance to bortezomib-mediated apoptosis. PLoS One 6:e20254 doi:10.1371/journal.pone.0020254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mazroui R, Di Marco S, Kaufman RJ, Gallouzi IE. 2007. Inhibition of the ubiquitin-proteasome system induces stress granule formation. Mol. Biol. Cell 18:2603–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Anderson P, Kedersha N. 2006. RNA granules. J. Cell Biol. 172:803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Anderson P, Kedersha N. 2008. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 33:141–150 [DOI] [PubMed] [Google Scholar]
- 10. Arimoto K, Fukuda H, Imajoh-Ohmi S, Saito H, Takekawa M. 2008. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat. Cell Biol. 10:1324–1332 [DOI] [PubMed] [Google Scholar]
- 11. Kim WJ, Back SH, Kim V, Ryu I, Jang SK. 2005. Sequestration of TRAF2 into stress granules interrupts tumor necrosis factor signaling under stress conditions. Mol. Cell. Biol. 25:2450–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Morita T, Satoh R, Umeda N, Kita A, Sugiura R. 2012. The stress granule protein Vgl1 and poly(A)-binding protein Pab1 are required for doxorubicin resistance in the fission yeast Schizosaccharomyces pombe. Biochem. Biophys. Res. Commun. 417:399–403 [DOI] [PubMed] [Google Scholar]
- 13. McConkey DJ, Zhu K. 2008. Mechanisms of proteasome inhibitor action and resistance in cancer. Drug Resist. Updat. 11:164–179 [DOI] [PubMed] [Google Scholar]
- 14. Richardson PG, Mitsiades C, Schlossman R, Ghobrial I, Hideshima T, Munshi N, Anderson KC. 2008. Bortezomib in the front-line treatment of multiple myeloma. Expert Rev. Anticancer Ther. 8:1053–1072 [DOI] [PubMed] [Google Scholar]
- 15. Sterz J, von Metzler I, Hahne JC, Lamottke B, Rademacher J, Heider U, Terpos E, Sezer O. 2008. The potential of proteasome inhibitors in cancer therapy. Expert Opin. Invest. Drugs 17:879–895 [DOI] [PubMed] [Google Scholar]
- 16. Caravita T, de Fabritiis P, Palumbo A, Amadori S, Boccadoro M. 2006. Bortezomib: efficacy comparisons in solid tumors and hematologic malignancies. Nat. Clin. Pract. Oncol. 3:374–387 [DOI] [PubMed] [Google Scholar]
- 17. Codony-Servat J, Tapia MA, Bosch M, Oliva C, Domingo-Domenech J, Mellado B, Rolfe M, Ross JS, Gascon P, Rovira A, Albanell J. 2006. Differential cellular and molecular effects of bortezomib, a proteasome inhibitor, in human breast cancer cells. Mol. Cancer Ther. 5:665–675 [DOI] [PubMed] [Google Scholar]
- 18. Guertin DA, Sabatini DM. 2009. The pharmacology of mTOR inhibition. Sci. Signal. 2:pe24 doi:10.1126/scisignal.267pe24 [DOI] [PubMed] [Google Scholar]
- 19. Laplante M, Sabatini DM. 2012. mTOR signaling in growth control and disease. Cell 149:274–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, Hall MN. 2004. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 6:1122–1128 [DOI] [PubMed] [Google Scholar]
- 21. Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, Gray NS, Sabatini DM. 2009. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell 137:873–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. 2004. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 14:1296–1302 [DOI] [PubMed] [Google Scholar]
- 23. Takahara T, Hara K, Yonezawa K, Sorimachi H, Maeda T. 2006. Nutrient-dependent multimerization of the mammalian target of rapamycin through the N-terminal HEAT repeat region. J. Biol. Chem. 281:28605–28614 [DOI] [PubMed] [Google Scholar]
- 24. Wang L, Rhodes CJ, Lawrence JC., Jr 2006. Activation of mammalian target of rapamycin (mTOR) by insulin is associated with stimulation of 4EBP1 binding to dimeric mTOR complex 1. J. Biol. Chem. 281:24293–24303 [DOI] [PubMed] [Google Scholar]
- 25. Yip CK, Murata K, Walz T, Sabatini DM, Kang SA. 2010. Structure of the human mTOR complex I and its implications for rapamycin inhibition. Mol. Cell 38:768–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang HH, Lipovsky AI, Dibble CC, Sahin M, Manning BD. 2006. S6K1 regulates GSK3 under conditions of mTOR-dependent feedback inhibition of Akt. Mol. Cell 24:185–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K. 2002. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110:177–189 [DOI] [PubMed] [Google Scholar]
- 28. Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. 2002. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110:163–175 [DOI] [PubMed] [Google Scholar]
- 29. Zhou H, Huang S. 2010. The complexes of mammalian target of rapamycin. Curr. Protein Pept. Sci. 11:409–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Richter JD, Sonenberg N. 2005. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433:477–480 [DOI] [PubMed] [Google Scholar]
- 31. Sonenberg N. 2008. eIF4E, the mRNA cap-binding protein: from basic discovery to translational research. Biochem. Cell Biol. 86:178–183 [DOI] [PubMed] [Google Scholar]
- 32. Haghighat A, Sonenberg N. 1997. eIF4G dramatically enhances the binding of eIF4E to the mRNA 5′-cap structure. J. Biol. Chem. 272:21677–21680 [DOI] [PubMed] [Google Scholar]
- 33. Haghighat A, Svitkin Y, Novoa I, Kuechler E, Skern T, Sonenberg N. 1996. The eIF4G-eIF4E complex is the target for direct cleavage by the rhinovirus 2A proteinase. J. Virol. 70:8444–8450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Haghighat A, Mader S, Pause A, Sonenberg N. 1995. Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 14:5701–5709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R, Sonenberg N. 1999. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 13:1422–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mamane Y, Petroulakis E, Rong L, Yoshida K, Ler LW, Sonenberg N. 2004. eIF4E—from translation to transformation. Oncogene 23:3172–3179 [DOI] [PubMed] [Google Scholar]
- 37. Rong L, Livingstone M, Sukarieh R, Petroulakis E, Gingras AC, Crosby K, Smith B, Polakiewicz RD, Pelletier J, Ferraiuolo MA, Sonenberg N. 2008. Control of eIF4E cellular localization by eIF4E-binding proteins, 4E-BPs. RNA 14:1318–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rousseau D, Gingras AC, Pause A, Sonenberg N. 1996. The eIF4E-binding proteins 1 and 2 are negative regulators of cell growth. Oncogene 13:2415–2420 [PubMed] [Google Scholar]
- 39. Averous J, Proud CG. 2006. When translation meets transformation: the mTOR story. Oncogene 25:6423–6435 [DOI] [PubMed] [Google Scholar]
- 40. Huang S, Houghton PJ. 2003. Targeting mTOR signaling for cancer therapy. Curr. Opin. Pharmacol. 3:371–377 [DOI] [PubMed] [Google Scholar]
- 41. Janus A, Robak T, Smolewski P. 2005. The mammalian target of the rapamycin (mTOR) kinase pathway: its role in tumourigenesis and targeted antitumour therapy. Cell. Mol. Biol. Lett. 10:479–498 [PubMed] [Google Scholar]
- 42. Petroulakis E, Mamane Y, Le Bacquer O, Shahbazian D, Sonenberg N. 2006. mTOR signaling: implications for cancer and anticancer therapy. Br. J. Cancer 94:195–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Graff JR, Konicek BW, Carter JH, Marcusson EG. 2008. Targeting the eukaryotic translation initiation factor 4E for cancer therapy. Cancer Res. 68:631–634 [DOI] [PubMed] [Google Scholar]
- 44. Graff JR, Konicek BW, Vincent TM, Lynch RL, Monteith D, Weir SN, Schwier P, Capen A, Goode RL, Dowless MS, Chen Y, Zhang H, Sissons S, Cox K, McNulty AM, Parsons SH, Wang T, Sams L, Geeganage S, Douglass LE, Neubauer BL, Dean NM, Blanchard K, Shou J, Stancato LF, Carter JH, Marcusson EG. 2007. Therapeutic suppression of translation initiation factor eIF4E expression reduces tumor growth without toxicity. J. Clin. Invest. 117:2638–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Konicek BW, Dumstorf CA, Graff JR. 2008. Targeting the eIF4F translation initiation complex for cancer therapy. Cell Cycle 7:2466–2471 [DOI] [PubMed] [Google Scholar]
- 46. Mamane Y, Petroulakis E, LeBacquer O, Sonenberg N. 2006. mTOR, translation initiation and cancer. Oncogene 25:6416–6422 [DOI] [PubMed] [Google Scholar]
- 47. Nathan CO, Amirghahari N, Rong X, Giordano T, Sibley D, Nordberg M, Glass J, Agarwal A, Caldito G. 2007. Mammalian target of rapamycin inhibitors as possible adjuvant therapy for microscopic residual disease in head and neck squamous cell cancer. Cancer Res. 67:2160–2168 [DOI] [PubMed] [Google Scholar]
- 48. Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, Ruggero D, Shokat KM. 2009. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 7:e38 doi:10.1371/journal.pbio.1000038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. 2009. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 284:8023–8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Grosso S, Pesce E, Brina D, Beugnet A, Loreni F, Biffo S. 2011. Sensitivity of global translation to mTOR inhibition in REN cells depends on the equilibrium between eIF4E and 4E-BP1. PLoS One 6:e29136 doi:10.1371/journal.pone.0029136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hsieh AC, Costa M, Zollo O, Davis C, Feldman ME, Testa JR, Meyuhas O, Shokat KM, Ruggero D. 2010. Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP-eIF4E. Cancer Cell 17:249–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. 2012. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485:109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mokas S, Mills JR, Garreau C, Fournier MJ, Robert F, Arya P, Kaufman RJ, Pelletier J, Mazroui R. 2009. Uncoupling stress granule assembly and translation initiation inhibition. Mol. Biol. Cell 20:2673–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen L, Aktas BH, Wang Y, He X, Sahoo R, Zhang N, Denoyelle S, Kabha E, Yang H, Freedman RY, Supko JG, Chorev M, Wagner G, Halperin JA. 2012. Tumor suppression by small molecule inhibitors of translation initiation. Oncotarget 3:869–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Moerke NJ, Aktas H, Chen H, Cantel S, Reibarkh MY, Fahmy A, Gross JD, Degterev A, Yuan J, Chorev M, Halperin JA, Wagner G. 2007. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell 128:257–267 [DOI] [PubMed] [Google Scholar]
- 56. Mazroui R, Huot ME, Tremblay S, Filion C, Labelle Y, Khandjian EW. 2002. Trapping of messenger RNA by Fragile X Mental Retardation protein into cytoplasmic granules induces translation repression. Hum. Mol. Genet. 11:3007–3017 [DOI] [PubMed] [Google Scholar]
- 57. Tourrière H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, Tazi J. 2003. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160:823–831 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58. Kedersha N, Chen S, Gilks N, Li W, Miller IJ, Stahl J, Anderson P. 2002. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell 13:195–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P. 2000. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 151:1257–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kedersha NL, Gupta M, Li W, Miller I, Anderson P. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 147:1431–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. McEwen E, Kedersha N, Song B, Scheuner D, Gilks N, Han A, Chen JJ, Anderson P, Kaufman RJ. 2005. Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J. Biol. Chem. 280:16925–16933 [DOI] [PubMed] [Google Scholar]
- 62. Holcik M, Sonenberg N. 2005. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 6:318–327 [DOI] [PubMed] [Google Scholar]
- 63. Wippich F, Bodenmiller B, Trajkovska MG, Wanka S, Aebersold R, Pelkmans L. 2013. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell 152:791–805 [DOI] [PubMed] [Google Scholar]
- 64. Dowling RJ, Topisirovic I, Fonseca BD, Sonenberg N. 2010. Dissecting the role of mTOR: lessons from mTOR inhibitors. Biochim. Biophys. Acta 1804:433–439 [DOI] [PubMed] [Google Scholar]
- 65. Sukarieh R, Sonenberg N, Pelletier J. 2009. The eIF4E-binding proteins are modifiers of cytoplasmic eIF4E relocalization during the heat shock response. Am. J. Physiol. Cell Physiol. 296:C1207–C1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dowling RJ, Topisirovic I, Alain T, Bidinosti M, Fonseca BD, Petroulakis E, Wang X, Larsson O, Selvaraj A, Liu Y, Kozma SC, Thomas G, Sonenberg N. 2010. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science 328:1172–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Patel J, McLeod LE, Vries RG, Flynn A, Wang X, Proud CG. 2002. Cellular stresses profoundly inhibit protein synthesis and modulate the states of phosphorylation of multiple translation factors. Eur. J. Biochem. 269:3076–3085 [DOI] [PubMed] [Google Scholar]
- 68. Wu XN, Wang XK, Wu SQ, Lu J, Zheng M, Wang YH, Zhou H, Zhang H, Han J. 2011. Phosphorylation of Raptor by p38beta participates in arsenite-induced mammalian target of rapamycin complex 1 (mTORC1) activation. J. Biol. Chem. 286:31501–31511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Alain T, Morita M, Fonseca BD, Yanagiya A, Siddiqui N, Bhat M, Zammit D, Marcus V, Metrakos P, Voyer LA, Gandin V, Liu Y, Topisirovic I, Sonenberg N. 2012. eIF4E/4E-BP ratio predicts the efficacy of mTOR targeted therapies. Cancer Res. 72:6468–6476 [DOI] [PubMed] [Google Scholar]
- 70. Lin TA, Kong X, Haystead TA, Pause A, Belsham G, Sonenberg N, Lawrence JC., Jr 1994. PHAS-I as a link between mitogen-activated protein kinase and translation initiation. Science 266:653–656 [DOI] [PubMed] [Google Scholar]
- 71. Pause A, Belsham GJ, Gingras AC, Donze O, Lin TA, Lawrence JC, Jr, Sonenberg N. 1994. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature 371:762–767 [DOI] [PubMed] [Google Scholar]
- 72. Poulin F, Gingras AC, Olsen H, Chevalier S, Sonenberg N. 1998. 4E-BP3, a new member of the eukaryotic initiation factor 4E-binding protein family. J. Biol. Chem. 273:14002–14007 [DOI] [PubMed] [Google Scholar]
- 73. Buchan JR, Parker R. 2009. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36:932–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Bragado P, Estrada Y, Sosa MS, Avivar-Valderas A, Cannan D, Genden E, Teng M, Ranganathan AC, Wen HC, Kapoor A, Bernstein E, Aguirre-Ghiso JA. 2012. Analysis of marker-defined HNSCC subpopulations reveals a dynamic regulation of tumor initiating properties. PLoS One 7:e29974 doi:10.1371/journal.pone.0029974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gloesenkamp CR, Nitzsche B, Ocker M, Di Fazio P, Quint K, Hoffmann B, Scherubl H, Hopfner M. 2012. AKT inhibition by triciribine alone or as combination therapy for growth control of gastroenteropancreatic neuroendocrine tumors. Int. J. Oncol. 40:876–888 [DOI] [PubMed] [Google Scholar]
- 76. Petitclerc E, Deschesnes RG, Cote MF, Marquis C, Janvier R, Lacroix J, Miot-Noirault E, Legault J, Mounetou E, Madelmont JC, C-Gaudreault R. 2004. Antiangiogenic and antitumoral activity of phenyl-3-(2-chloroethyl)ureas: a class of soft alkylating agents disrupting microtubules that are unaffected by cell adhesion-mediated drug resistance. Cancer Res. 64:4654–4663 [DOI] [PubMed] [Google Scholar]
- 77. Ribatti D, Nico B, Cimpean AM, Raica M, Crivellato E, Ruggieri S, Vacca A. 19 April 2012. B16-F10 melanoma cells contribute to the new formation of blood vessels in the chick embryo chorioallantoic membrane through vasculogenic mimicry. Clin. Exp. Med. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 78. Fujimura K, Sasaki AT, Anderson P. 2012. Selenite targets eIF4E-binding protein-1 to inhibit translation initiation and induce the assembly of non-canonical stress granules. Nucleic Acids Res. 40:8099–8110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ, Wang S, Ren P, Martin M, Jessen K, Feldman ME, Weissman JS, Shokat KM, Rommel C, Ruggero D. 2012. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 485:55–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Larsson O, Morita M, Topisirovic I, Alain T, Blouin MJ, Pollak M, Sonenberg N. 2012. Distinct perturbation of the translatome by the antidiabetic drug metformin. Proc. Natl. Acad. Sci. U. S. A. 109:8977–8982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hilliker A, Gao Z, Jankowsky E, Parker R. 2011. The DEAD-box protein Ded1 modulates translation by the formation and resolution of an eIF4F-mRNA complex. Mol. Cell 43:962–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Rajyaguru P, She M, Parker R. 2012. Scd6 targets eIF4G to repress translation: RGG motif proteins as a class of eIF4G-binding proteins. Mol. Cell 45:244–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Shih JW, Wang WT, Tsai TY, Kuo CY, Li HK, Wu Lee YH. 2012. Critical roles of RNA helicase DDX3 and its interactions with eIF4E/PABP1 in stress granule assembly and stress response. Biochem. J. 441:119–129 [DOI] [PubMed] [Google Scholar]
- 84. Svitkin YV, Herdy B, Costa-Mattioli M, Gingras AC, Raught B, Sonenberg N. 2005. Eukaryotic translation initiation factor 4E availability controls the switch between cap-dependent and internal ribosomal entry site-mediated translation. Mol. Cell. Biol. 25:10556–10565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Yanagiya A, Suyama E, Adachi H, Svitkin YV, Aza-Blanc P, Imataka H, Mikami S, Martineau Y, Ronai ZA, Sonenberg N. 2012. Translational homeostasis via the mRNA cap-binding protein, eIF4E. Mol. Cell 46:847–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Di Marco S, Hel Z, Lachance C, Furneaux H, Radzioch D. 2001. Polymorphism in the 3′-untranslated region of TNFalpha mRNA impairs binding of the posttranscriptional regulatory protein HuR to TNFalpha mRNA. Nucleic Acids Res. 29:863–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.