Abstract
Many proteins do not exist in a single rigid conformation. Protein motions, or dynamics, exist and in many cases are important for protein function. The analysis of protein dynamics relies on biophysical techniques that can distinguish simultaneously existing populations of molecules and their rates of interconversion. Hydrogen exchange (HX) detected by mass spectrometry (MS) is contributing to our understanding of protein motions by revealing unfolding and dynamics on a wide timescale, ranging from seconds to hours to days. In this review we discuss HX MS-based analyses of protein dynamics, using our studies of multi-domain kinases as examples. Using HX MS, we have successfully probed protein dynamics and unfolding in the isolated SH3, SH2 and kinase domains of the c-Src and Abl kinase families, as well as the role of inter- and intra-molecular interactions in the global control of kinase function. Coupled with high-resolution structural information, HX MS has proved to be a powerful and versatile tool for the analysis of the conformational dynamics in these kinase systems, and has provided fresh insight regarding the regulatory control of these important signaling proteins. HX MS studies of dynamics are applicable not only to the proteins we illustrate here, but to a very wide range of proteins and protein systems, and should play a role in both classification of and greater understanding of the prevalence of protein motion.
Keywords: Src-family kinase; Hck; Lck; SH3 domain; SH2 domain; Abl; deuterium; HDX; protein dynamics, flexibility
In 1996, we observed something that made us scratch our heads. While working with the hematopoietic cell kinase (Hck) SH3 domain and trying to understand its conformation with hydrogen exchange (HX) mass spectrometry (MS), we realized that parts of this small protein domain were unusually dynamic in solution, but on a rather slow time scale [1]. This observation sent us down a path of trying to understand what this dynamics was all about, how widespread it was, and more importantly, how we could use the then fledgling tool of HX MS to probe all manner of proteins looking for the same phenomenon. We discovered not only that other proteins had such dynamics, but that measuring the partial unfolding with mass spectrometry was an excellent way to investigate inter- and intramolecular protein:protein interaction. In the following pages, we will review what was learned about the proteins in question, the technology used to make the measurements, and the general applicability of the method. While we primarily discuss several specific proteins that we have worked with most closely, it is clear that such experimental methods can be extended to interrogate partial unfolding in any protein of interest
1. Background
The recognized father of hydrogen exchange, Kaj Ulrik Linderstrøm Lang, quite rightly stated many years ago [2] that proteins generally do not exist in a single rigid conformation, but rather are in continuous motion and exist as an ensemble of structures that interconvert [3–5]. These motions, collectively called protein dynamics [5–8], are essential for the function of many proteins (e.g., [9–11]) but can be very difficult to quantify (e.g., [12]). Various methods including hydrogen exchange indicate that there is movement but direct observation of discrete conformations within the ensemble is much more challenging. Realizing such motions exist, visualizing motion has become more desirable, including creation of databases of molecule motions [13,14] and movies thereof [15].
Important examples of these principles are provided by protein-tyrosine kinases of the c-Src family, which are a major focus of this review. Motions within the Src-family kinases, and the structurally related ‘core’ domain of the c-Abl kinase, are critical for their diverse intracellular signaling functions [16–20]. The kinase domains of these proteins are held in a downregulated state by binding of regulatory domains to the “back side”, away from the access point of substrates to the active site. Various components of the proteins help hold the downregulated conformation together, including binding of internal linker sequences, pi-stacking interactions, interaction of phosphorylated residues (both tyrosine and serine) to pockets within the regulatory domains, and insertion of myristic acid into a pocket in the kinase domain. Biophysical characterization of the Src-family kinases and related proteins has included many methods such as NMR, X-ray crystallography, X-ray scattering, and analytical ultracentrifugation. Hydrogen exchange mass spectrometry has also been revealing and has assisted in characterizing a number of mechanistic features of these proteins, as will be described in the following sections.
2. Theory of HX MS
Hydrogen exchange coupled with mass spectrometry is a well-reviewed topic [21–28] which we will not re-review here. In the simplest implementation of HX MS, termed continuous labeling [29], the protein of interest is placed in a buffer solution comprised of >95% D2O and allowed to undergo isotope exchange for defined periods of time. Deuterium in the solution exchanges with labile hydrogen atoms in the protein, most notably the backbone amide hydrogens. At several predetermined times, ranging typically from 10 seconds to 8 hours (and up to days in some circumstances), the labeling is quenched and the extent and location of the deuterium is determined by measuring the mass of the intact protein or measuring the mass of peptides produced from enzymatic digestion of the protein under quench conditions. The labeling reaction favors the forward direction because the amount of D2O is in great excess relative to the amount of H2O in the labeling step. There are multiple technical challenges to making HX MS measurements, as described in the reviews listed above.
The way in which deuterium exchanges into proteins can be summarized with a two process model [6,7]: exchange into folded forms and exchange as a result of localized unfolding. Localized unfolding is the predominate pathway for exchange by proteins under physiological conditions [7] and describes the replacement of hydrogen in proteins with deuterium in solution with three general steps ([30], see also [31] for a description of the history of this equation), as illustrated by Equation 1. In order for replacement to occur, the folded protein containing all
| Eq. 1 |
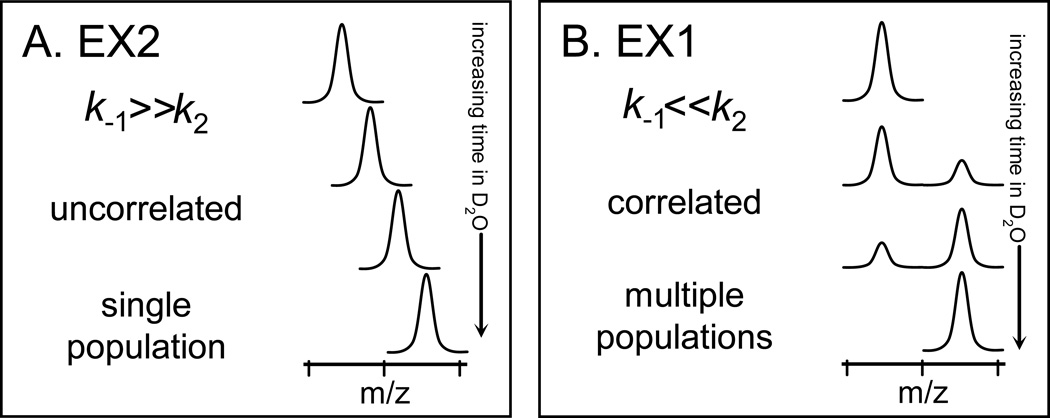
hydrogen (FH) visits an unfolded state (UH) via localized unfolding, hydrogen bonds are broken and positions become exchange competent. In this model, the rate constants k1, k−1, k2 describe unfolding, refolding, and hydrogen exchange, respectively. There are two kinetic limits, termed EX1 and EX2, in this exchange model. EX2 dominates when k−1 ≫ k2; the protein (in part or in whole) experiences many visits to a partially unfolded and exchange competent state before isotopic exchange is complete. When k−1 ≪ k2, EX1 dominates and multiple exchange-competent positions exchange before refolding can occur; that is, isotopic exchange is complete in an unfolded region(s) before the protein refolds. The vast majority of stable proteins in physiological conditions follow EX2 kinetics. By monitoring the rate of deuterium incorporation and the appearance of EX2 or EX1 kinetics, one can learn about the nature of the protein and the biophysical parameters of the protein that must have existed during the labeling process.
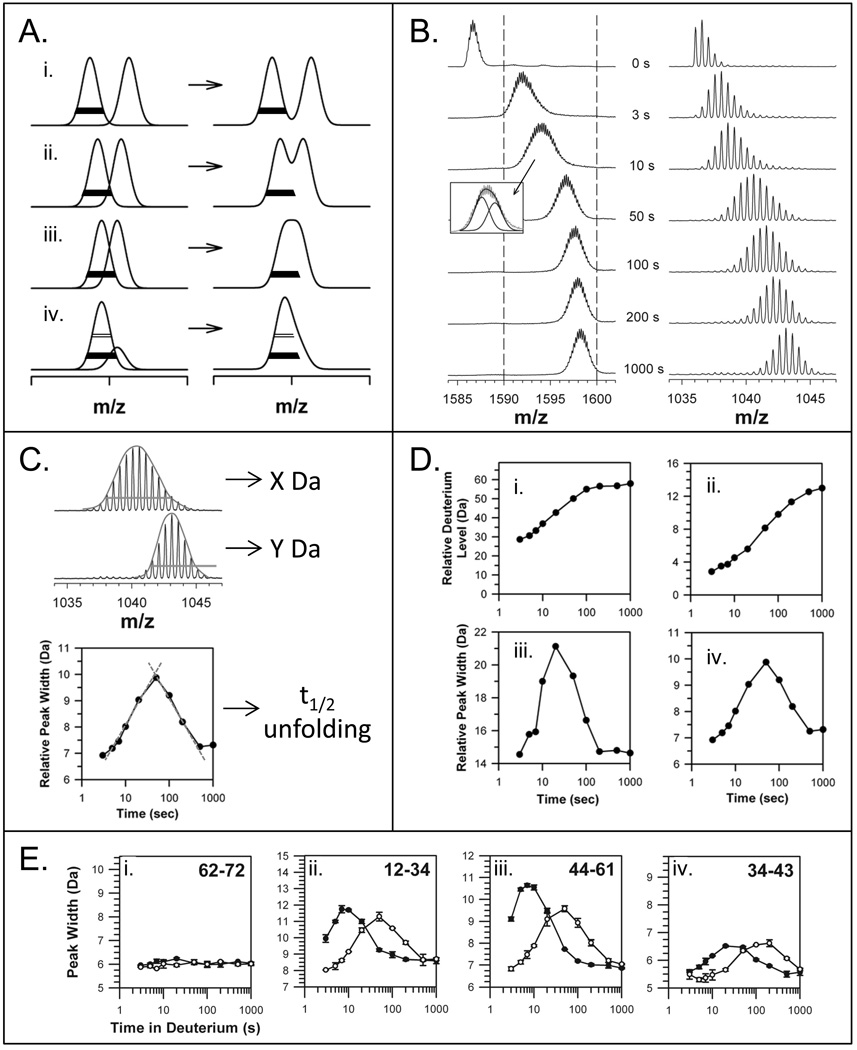
The mass spectra of proteins labeled with deuterium over time (continuous labeling) provides information about the kinetic regime(s) the protein follows, as shown in FIGURE 1 [32]. In contrast to NMR where populations of protein molecules are much more difficult to detect ([33,34], reviewed in [35]), protein populations are easily distinguishable in mass spectra of deuterated proteins. A single binomial isotope distribution (FIGURE 1A), caused by a random distribution of deuterium among the population of sample molecules, indicates HX by EX2 kinetics [7,36]. A bimodal distribution (FIGURE 1B) in the mass spectra indicates EX1 kinetics [23,32,37–39]. In the mass spectra of EX1 kinetics, the lower m/z (molecular weight) isotopic envelope represents proteins in the population that have not yet undergone unfolding and have therefore not been competent to exchange their backbone amide hydrogens for deuterium in the deuterium labeling solution. No deuterium is incorporated into these protected molecules and this population of protein molecules has a lower m/z. On the other hand, the higher m/z isotopic envelope represents the protein molecules in the population that have undergone an unfolding event, were exposed to deuterium, and therefore increased in molecular weight. The mass difference between lower and higher components of the bimodal pattern indicates how many amide hydrogens are involved in the unfolding event. In exclusively EX1 kinetics (as opposed to mixed kinetics where both EX2 and EX1 occur, perhaps in distinct parts of a protein or peptide), the distance on the m/z scale that separates the lower- and higher-mass envelopes remains constant and only the intensity of the peak changes along the time course of deuteration (see FIGURE 1B).
Figure 1.
The distinctive appearance of EX2 and EX1 kinetics in hydrogen exchange mass spectrometry. With increasing amounts of time in D2O, the extremes of the (A) EX2 and (B) EX1 kinetic limits are distinguishable for both proteins and peptides. In EX2, due to random protein fluctuations, the rate of refolding (k−1) (see Equation 1) is much larger than the rate of labeling (k2) and the exchange is classified as uncorrelated. Such exchange produces a single population of molecules that gradually gain mass during the timecourse of the labeling experiment. In EX1, k2 is much larger than k−1 and regions of the protein can all become labeled before refolding (k−1) occurs. This type of exchange is termed correlated as all residues in a region undergoing EX1 appear to have exchanged at the same time. In the mass spectra of EX1, there are two peaks: one for the population that did not yet unfold and become deuterated (at lower m/z) and one for the population that did unfold and become deuterated (at higher m/z). This Figure was taken from [32].
3. The beginning: Partial unfolding in theHck SH3 domain
We originally began to study Src-family kinases with HX MS by measuring HX into the SH3 domain of the Src-family kinase, Hck [1]. Hck, like other Src-family kinases, is a modular, multi-domain protein kinase [20,40–42]. The modular components include an N-terminal anchor domain that localizes the protein to membranes, an ~8 kDa SH3 domain, a ~12.5 kDa SH2 domain, a linker between SH2 and the kinase domain, the ~ 33 kDa kinase domain itself (divided into N-terminal and C-terminal lobes) and a C-terminal tail. In general, SH3 domains are among the best characterized protein-protein interaction modules [40,43–45], and recognize short proline-rich target sequences which form polyproline type II (PPII) helices. In the context of Src family kinases such as Hck, the SH3 domain plays a critical role in kinase downregulation by engaging an internal PPII helix. When HX MS data of Hck SH3 (which has only 67 residues, MW=8,229) were first obtained, it became apparent that part of the protein became deuterated by EX2 kinetics and that part became deuterated by EX1 kinetics. We came to realize that this EX1 pattern indicated partial cooperative unfolding within the Hck SH3 domain, consistent with the model for localized HX (Eq. 1). As shown in FIGURE 2A, intact Hck SH3 left in D2O for 1 minute became approximately 10 daltons heavier. Hydrogen exchange at shorter time points (such as 10 seconds - 1 minute) reports primarily on surface-exposed, non-buried backbone amide hydrogens that are mostly hydrogen bonded to water molecules [46,47] and are not protected from exchange by strong intra-protein hydrogen bonds; this type of exchange occurs generally through an EX2 mechanism. As Hck SH3 remained in deuterium, an EX1 signature appeared beginning around 3–5 minutes of labeling and persisting until approximately 40 minutes of labeling. The original bimodal isotopic pattern is shown in FIGURE 2A, where the two populations are marked with green (low-mass distribution) and red (high-mass distribution) arrows. The partial unfolding of Hck SH3 occurred under physiological conditions (pH 6.9, ~10 mM phosphate buffer, 22 °C) implying that there was some biological relevance to this unfolding.
Figure 2.
Partial unfolding in human Hck SH3 as monitored by HX MS. (A) Transformed mass spectra (from Ref. [1]) of Hck SH3 after various times in D2O from 1 minute to 8 hours [including undeuterated (0%) and maximally deuterated (100%) controls]. A bimodal pattern after 22 minutes of labeling is accentuated with two uniform Gaussian distributions: the lower-mass distribution is indicated with a green arrow and represents molecules that have not yet unfolded and become deuterated; the higher-mass distribution is indicated with a red arrow and represents molecules that have undergone partial cooperative unfolding and been labeled with deuterium. These mass spectra were acquired with instrument resolution of ~2,500, which is low relative to more recent data such as in Figure 6 which were obtained with nearly ~10,000 resolution. (B) The area of the lower-mass distribution (indicated with green) of an EX1 pattern is used to monitor the disappearance of the folded/undeuterated species along the time course of the labeling experiment. (C) The rate constant of unfolding and the unfolding half-life for an EX1 event can be determined with the equation shown. The slope of the line fit to the data yields the rate constant. In this example, partial unfolding in Hck SH3 was monitored at the times shown and compared to unfolding in the Hck SH3 domain when bound to the HIV Nef peptide (see [1] and main text for details).
While other SH3 domains were known at the time to be dynamic in solution (e.g. the N-terminal SH3 domain of Drk [48,49]), we did not expect to see such slow unfolding kinetics. The rate of unfolding from EX1 HX MS data was determined by monitoring the change in the relative abundance of the folded species (lower m/z peak, colored green in FIGURE 2B). Fitting the change in abundance to a simple rate expression yields a straight line, the slope of which is the rate constant for unfolding (FIGURE 2C). For the Hck SH3 domain, the unfolding rate constant (k1 from Eq. 1) was 0.04 min−1 (for reference, note that the average k2 at any backbone amide hydrogen is 350 min−1, calculated from [50]). We have subsequently found it easier to express the unfolding in terms of the half-life of unfolding (t1/2), or the point at which half of the population of protein molecules has undergone unfolding and subsequent labeling, and use either t1/2 or the rate of unfolding to describe the event (more below). For Hck SH3, the half-life of unfolding was quite slow (17 minutes) and involved around 15 amino acids (~22% of the SH3 protein). We considered this to be “slow” unfolding as many proteins of this size and complexity, as least as one would imagine, exist in rapidly interconverting states and transition between unfolded and folded states quickly. The half-life of Hck SH3 domain unfolding was later shown to vary depending on the protein concentration, salt composition and ligand binding status (see below). For the remainder of the time spent in D2O, that is after the EX1 kinetic signature was gone (> 40 minutes), the isotopic pattern seen in the Hck SH3 HX MS was binomial (FIGURE 2A)and had a constant width characteristic of EX2 kinetics(see also Section 5.2.1 below and Ref. 32).
4. Measuring protein dynamics aids understanding of Hck
After the initial observation of partial cooperative unfolding in the Hck SH3 domain, we investigated many aspects of the protein dynamics found in Hck SH3. The experiments were divided into several components and were not necessarily performed in the order listed here. First, we made many different constructs that contained Hck SH3 to determine if intramolecular interactions influenced the dynamics (stability) of the Hck SH3 domain. Second, we bound various ligands (peptides, proteins) with different dissociation constants to Hck SH3 and monitored the HX. Third, we extended our studies to SH3 domains from proteins other than Hck to see if partial cooperative unfolding is a general property of SH3domains. Fourth, we tried to determine which residues within SH3 were involved in the partial cooperative unfolding and thereby understand the mechanism of partial unfolding. The outcomes of these four lines of investigation for Hck and other kinases are described in detail in the following sections.
4.1. Intramolecular interactions by HX MS: Divide and conquer
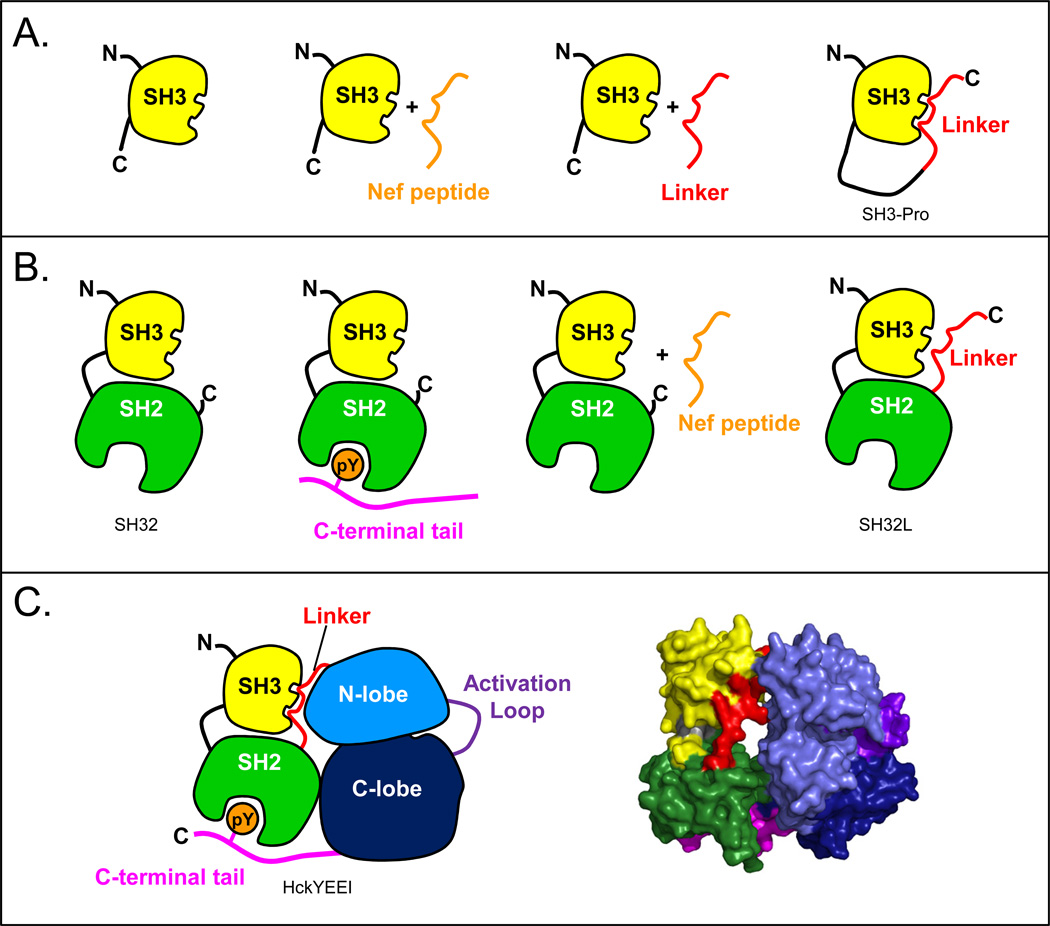
Because Hck, like other Src-family kinases, is a modular, multi-domain protein, it seemed rational to study each domain alone and then in the context of the rest of the protein. In a “divide and conquer” approach, we began to interrogate Hck by studying each domain in isolation, and then slowly built up to the whole protein, measuring HX of various combinations of them along the way. Not only was this easier analytically, as the smaller domains were easier to tackle in the early days of HX MS, but it provided data that could be used to explain inter- and intramolecular interactions. FIGURE 3 shows the cadre of Hck protein constructs that were investigated by HX MS.
Figure 3.
Cartoon models to summarize all of the constructs of human Hck that have been used to study regulation and dynamics by HX MS. (A) SH3 alone with various peptide ligands; (B) the SH32 construct with various ligands; (C) the near full-length protein, HckYEEI, in cartoon form (left) and the crystal structure [60] of the down-regulated form (right).
After SH3 domain unfolding was characterized, we next analyzed a construct including both the SH3 domain and the SH2 (referred to as SH32), joined by their native connector sequence [51], see FIGURE 3B. Both SH3 and SH2, being modular domains [40], retain the same structure as either individual domains or when found within larger proteins; however, their dynamics can be influenced by inter-protein interactions or intra-protein interactions. Measurements of HX in SH3 alone versus in SH32 [51] showed that SH3 domain dynamics were influenced by the presence of the nearby SH2. There were changes in SH3 deuterium incorporation throughout the SH3 domain, and the effects were most concentrated near the SH2 domain interface with SH3. While there were changes in overall SH3 domain dynamics, the rate of partial, cooperative unfolding in the SH3 domain was not influenced by the presence of the SH2 domain leading us to conclude that the unfolding of Hck SH3 was an intrinsic property of the SH3 domain itself that was independent of inter-domain interactions. The SH2 domain by itself [52] was much less dynamic than SH3 and it its dynamics (as read out by deuteration) were not really altered when it was found alongside SH3 in the SH32 construct [51]. One major conclusion from the comparison of individual SH3 and SH2 domains with the larger SH32 construct was that inter-domain interactions can change the intrinsic dynamics of some proteins (e.g., SH3), but not all (e.g., SH2; see also [53]). Further, we demonstrated that changes in dynamics could be detected reliably with HX MS.
4.2. Slowdown factor (reduced dynamics) as an assay for binding
Based in part on the conclusions from the previous section, that dynamics could be influenced by inter-domain interaction, we set out to test associations that were much stronger than just two domains next to each other. We had already measured the effects of binding small peptides to the SH3 domain [1] and shown that an effect could be observed in hydrogen exchange. When the Hck SH3 domain was incubated with a 12-residue peptide from the HIV Nef protein [54], one of the tightest known binders of Hck SH3 (Kd 90 µM), there was a significant slowdown in the kinetics of SH3 domain unfolding. The slowdown was apparent in the HX MS spectra (not shown here, see [1] for data) wherein a bimodal pattern was observed but at much longer deuterium labeling times; reduction in dynamics was also evident in the determination of the rate constant for unfolding (FIGURE 2C). This slowing of protein dynamics (i.e., reduction in protein flexibility and ability to become deuterated) could be quantified by calculating a “slowdown factor”. The slowdown factor is simply the t1/2 of unfolding in the bound state divided by the t1/2 of unfolding in the free state. An example calculation can be derived from FIGURE 2C, where upon binding to the Nef peptide, Hck SH3 unfolding slowed from a t1/2 of ~17 minutes in the free state to a t1/2 of ~63 minutes in the bound state. The slowdown factor for this case of SH3 + Nef peptide was therefore 63/17, or ~3.6 [1]. Slowdown in partial unfolding (EX1 kinetics) is a function of several parameters, including the Kd and therefore the bound concentrations of both protein and ligand. The percent of protein molecules that are bound is important to know when comparing slowdown factors, as illustrated below. Monitoring the slowdown factor provides a sensitive binding assay for any protein or domain that has EX1-related dynamics, both for complexes that are inter-molecular (e.g., an SH3 domain and an exogenous peptide or protein) or in complexes that are intra-molecular (e.g. SH3 domain binding to a peptide sequence within the same protein). The latter case, intra-molecular binding, or internal binding within a protein, is something that is very challenging to measure with methods like isothermal titration calorimetry or surface plasmon resonance; for HX MS, the measurement of intra-molecular binding is as straightforward as it is for inter-molecular binding.
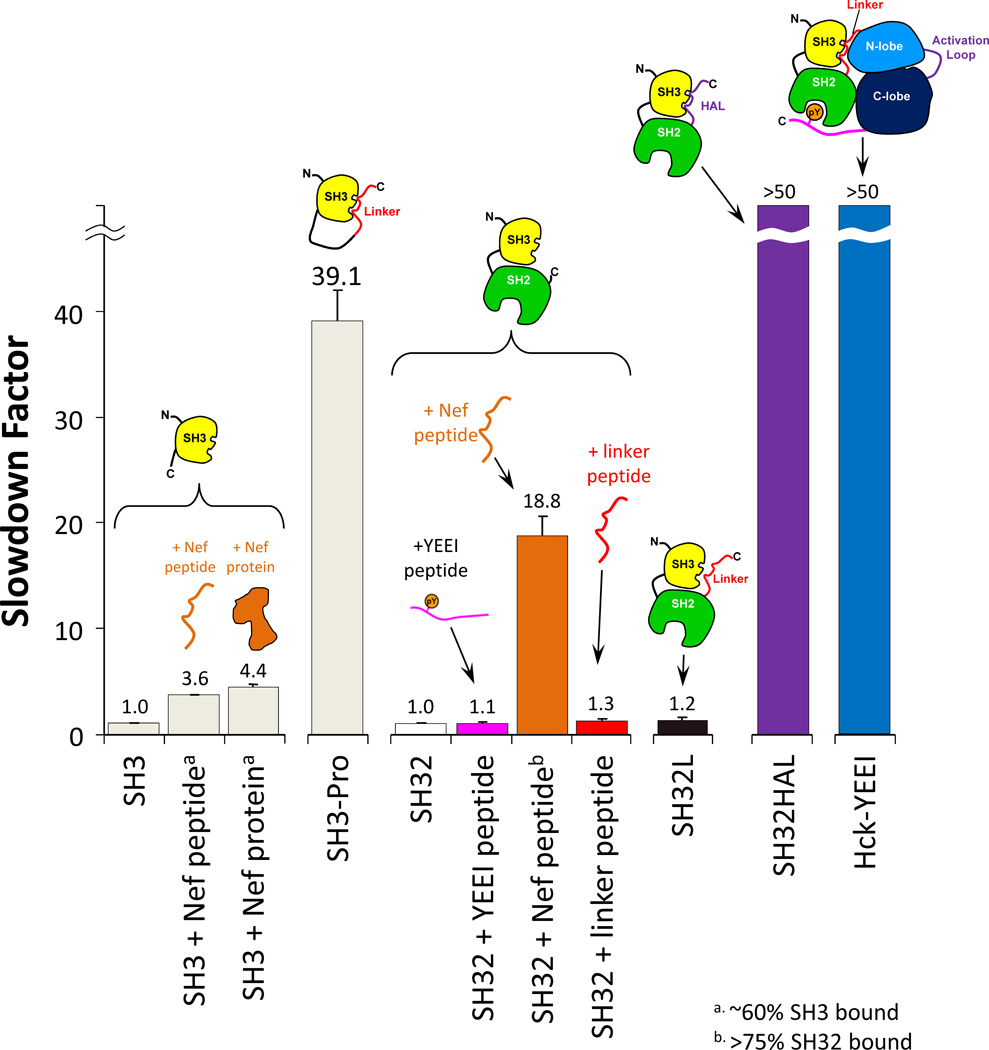
Using measures of the slowdown factor for SH3 dynamics, we tested a number of intra-molecular and inter-molecular binding events for Hck SH3, as summarized in FIGURE 4. The data in this figure have been described in much more detail in Refs. [20,55]. Working from left to right in the figure, one can see that different binding events contribute to different amounts of slowdown, again bearing in mind that more slowdown is indicative of tighter binding/higher occupancy of the binding site such that the protein motions of the SH3 domain are retarded. Both a peptide from the HIV Nef protein and the HIV Nef protein itself led to slowdown factors of around 4 [in separate experiments [56], a number of variant Nef proteins were tested in this same assay]. An important caveat is that in a binding experiments like this, it may not be possible to force SH3 to be bound 100% of the time because to do so may require that the ligand is in vast (>10–100-fold) excess. When this occurs, the excess ligand may (depending on the instrument) saturate the dynamic range of the mass spectrometer, i.e., the strong signals of the highly abundant ligand suppress the weaker signals of the less abundant protein (explained in more detail in [55]). What is actually monitored in a typical HX MS experiment of binding is the mixed population (equilibrium) of bound and free SH3 domains and the slowdown factor is always less than its maximum possible value if the entire population of molecules were bound (see also [57]). An example of the effect is shown for the construct SH3-Pro in which a modest affinity peptide ligand [58] from the Ras-GAP protein was covalently coupled to the SH3 domain by means of a flexible linker [59]. When the peptide ligand was covalently coupled, the occupancy of the binding site could be forced to much higher values, in fact essentially 100% bound, as the peptide ligand could not easily diffuse away from the binding site. The slowdown factor was nearly 40, indicating that there was significant binding and reduction in the dynamics of the SH3 domain when the peptide ligand was forced to be in close proximity to SH3.
Figure 4.
The slowdown factor of unfolding in Hck SH3 for various constructs. Slowdown factor is the ratio of the unfolding rate in bound protein over the unfolding rate in free protein. A small slowdown factor (close to one) means weak or no binding, while a large slowdown factor means tight binding. The constructs involved in each experiment are shown in cartoon form. In cases of interaction with Nef and the Nef-derived peptide, the percentage of SH3-bound molecules is indicated by the superscripts a and b. Further details on the proteins used to produce this data can be found in [20,55].
Further SH3 domain binding studies indicated that in the SH32 protein, binding a ligand to the SH2 domain did nothing to the dynamics of the SH3 domain, again reinforcing the idea that binding to SH3 and SH2 domains is modular and that binding of ligand to one domain does not necessarily influence the other domain allosterically as measured by HX MS. When the amount of Nef peptide binding to the SH3 domain in the SH32 construct was increased relative to that binding to the SH3 domain alone (>75% bound in SH32 versus ~60% bound in SH3 alone), the slowdown factor increased accordingly from 3.6 to 18.6 (FIGURE 4). This change is an effect of the higher relative peptide ligand concentration when more peptide is added, which forces the binding protein (SH32) to be occupied for more of the time (as described above).
4.3. The importance of linker binding in Hck regulation
A remarkable finding for the overall structure of Hck involved the natural linker sequence connecting the SH2 domain to the kinase domain [60,61]. This sequence forms an internal binding site for the SH3 domain in the context of the full-length downregulated kinase. When we added a synthetic peptide with this linker sequence in trans, we could not detect any alterations in SH3 unfolding [55]. This surprising result was significant because it indicated that the linker was not capable of binding to the SH3 domain without the rest of the Hck protein. We feared that the affinity of the linker sequence for SH3 was just too low to detect any changes to the slowdown factor so we covalently attached the linker (as was done for the Ras-GAP peptide in the SH3-Pro construct) to the SH32 construct, producing a protein referred to as SH32L. The kinetics of SH3 domain unfolding in SH32L (FIGURE 4) was identical to that of SH32 and SH3 meaning that the linker was not able to bind to SH3 even with forced proximity that favors high binding-site occupancy. We next made two mutations (replacement of lysine with proline to enhance the polyproline class II conformation of the linker sequence required for SH3 binding) in the linker to turn it into a higher-affinity ligand (HAL, for high-affinity linker) [62]. In this HAL construct of SH32L (called SH32HAL) the unfolding of the Hck SH3 domain was so slow that we could not measure it, which translated to a very large slowdown factor (>50, see FIGURE 4). Additional measurements of the HX of SH32HAL [55] showed that almost all of the changes in exchange were confined to the SH3 domain. These data taken together showed that the native linker sequence of Hck has no affinity for the Hck SH3 domain when the rest of Hck (the kinase domain and C-terminal tail) is missing. However, the SH3 domain is still capable of binding in the absence of the kinase domain since a high-affinity sequence (HAL) essentially bound so tightly that it inhibited SH3 unfolding that we could measure by HX MS. The inability of the native linker sequence to bind the SH3 domain in cis in the absence of the kinase domain was subsequently verified withmuch more complicated and time-consuming measurements than HX MS, including surface plasmon resonance [62] and X-ray crystallography of native SH32L [63].
It was clear from these data that the SH32L construct was missing key regulatory interactions from the kinase domain, as had been hypothesized previously [17]. We went on to measure unfolding of the SH3 domain in near full-length Hck. The construct used for the measurement was HckYEEI, a variant of Hck in which the C-terminal tail has been mutated to a much higher-affinity internal ligand for the SH2 domain [60]. The YEEI mutation makes the protein much more stable and amenable to crystallization, but does not alter the regulatory mechanism [64]. The slowdown factor of SH3 partial unfolding in HckYEEI was, like SH32HAL, too large to be measured. This result meant that when the native linker sequence in the HckYEEI protein – which was identical in every way to the linker sequence tested in SH32L for which there was no change in SH3 domain dynamics – was in the presence of the kinase domain, it adopted a conformation capable of high-affinity interactions with the SH3 domain. Another explanation is that the SH3 domain made additional interactions with the kinase domain that strengthened the association with the linker beyond what was possible with just the linker alone. These HX MS results provided direct biophysical data that supported what had previously been hypothesized [17,65] regarding the necessity of intra-molecular interactions in Src-family kinases for effective downregulation. This example of probing the affinity of binding for a protein domain in the context of other parts of a protein (intra-molecular interactions) illustrates one way that HX MS measurements can access dynamics information that is quite difficult to obtain with other methods.
5. Partial unfolding in other SH3 domains
5.1. Survey of SH3 domains from diverse proteins
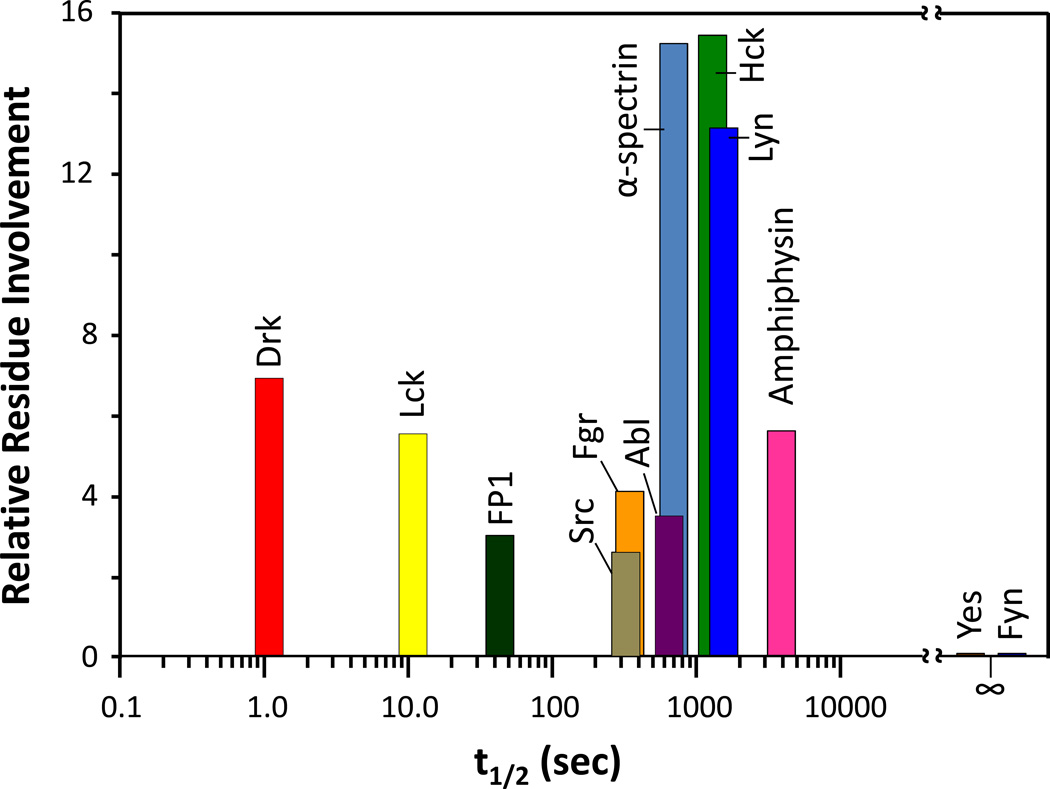
The immediate questions from our work with the Hck SH3 domain were: how commonplace was SH3 domain unfolding? Did all SH3 domains demonstrate these dynamics or was this phenomenon limited to Hck? To answer these questions, we produced recombinant SH3 domains from a variety of proteins and measured their dynamics with HX MS [66]. We discovered a range of dynamic behaviors for the SH3 domains (FIGURE 5) that was as diverse as the source proteins. The SH3 domain from some proteins (e.g., Yes, Fyn) had no unfolding at all, others looked like Hck (e.g., Lyn, α-spectrin), others had very fast dynamics (e.g., Drk, Lck), and some (e.g., Src, Abl) had detectable unfolding that involved far fewer residues than the unfolding in the Hck SH3 domain. As far as we could tell from every angle that we could think of [66], there was no obvious correlation between sequence conservation and unfolding, and no rational explanation for why some domains unfolded quickly and some unfolded slowly. Using pepsin digestion after HX but before MS [24,67], we determined [66] that the regions involved in unfolding were the same in all the proteins tested. Hence, there must be some aspect of this region that held the answers as to why these proteins were dynamic in solution (described in more detail in Section 6 below).
Figure 5.
Partial unfolding of various SH3 domains and measured by HX MS [66]. The measured half-life of unfolding and relative residue involvement were determined as described in Figure 2. Yes and Fyn SH3 domains did not undergo partial unfolding, hence their half-life of unfolding is shown as infinity, with no residue involvement.
5.2. The Lck tyrosine kinase
One of the more interesting proteins that we decided to investigate further was the Src-family kinase Lck. Lck, or lymphocytic cell kinase, is found in T-cell lymphocytes [68] and its domain organization is highly similar to that of Hck. Another interesting aspect of Lck is that it also interacts with viral accessory proteins, just like Hck. Hck binds to the HIV accessory proteins Nef [54] and Vif [69,70] while Lck binds to the Tip protein from Herpesvirus saimiri (HVS) [71]. The partial unfolding of the Lck SH3 domain (FIGURE 5) [72] occurred with a much shorter half-life (~10 seconds) than that of the Hck SH3 domain (~17 mins) [1]. Additionally, far fewer residues in Lck SH3 were involved in unfolding than in Hck SH3, making the detection of EX1 kinetics by HX MS more complex.
5.2.1. Peak width measurements in HX MS characterization of protein dynamics
To address complexities of EX1 measurement for proteins where EX1 was less obvious, we developed an alternative way to describe EX1 unfolding in proteins. We will illustrate the concepts behind the approach with the data from the Lck SH3 domain, and refer to the approach again later for other proteins where it was essential. The complexity in measuring EX1 kinetics for some proteins arises when two peaks describing the two populations (folded and unfolded, see FIGURE 1B) are not sufficiently resolved on the m/z scale to clearly distinguish the two peaks. The problem is illustrated in FIGURE 6A taken from Ref. [32]. When two peaks exhibiting an EX1 kinetic signature are well resolved on the m/z scale (FIGURE 6A,i), processing proceeds by determining the center of mass of each peak and plotting it against deuterium exchange-in time (as in FIGURE 2B,C). When the two peaks are not resolved (FIGURE 6A,ii–iv), the raw mass spectrum that is observed is a sum of the multiple components and the width of this summation is greater than the width of either of the constituents. The solid lines in FIGURE 6A, drawn at 20% peak intensity, are all of the same width, demonstrating that when there are two peaks close to one another in m/z, measuring the width of the total peak feature in the raw spectrum can indicate if EX1 kinetics may be present (i.e., there must be more than one peak of a given width under the raw spectrum).
Figure 6.
Mass spectral peak width as it relates to EX1 kinetics. (A) The various ways in which EX1 can be seen in mass spectra, from well resolved peaks to peak shoulders [32]. The two deconvoluted and resolved peaks are shown on the left and the merged peaks, as observed in raw spectra, are shown on the right. The bars (open at 50% peak intensity and solid at 20% peak intensity) are the same width in all panels. (B) Unresolved EX1 signatures are apparent in the spectra for intact Lck SH3 (left panel) and the Lck SH3 peptide representing residues 44–61 (right panel) [72]. The dotted lines in the left panel are to guide the eye. The inset indicates how two Gaussian distributions fit under the raw data for the intact protein spectra after 10 seconds in deuterium. These mass spectra were acquired with instrument resolution of ~10,000, which higher than older data (Figure 2A) acquired with instrument resolution of ~2,500. (C) The peak width can be used to determine unfolding half-life. The entire width of the isotope distribution is measured, usually at 50% or 20% peak height. In this example, the width at 50% peak intensity is shown by the orange line. The orange line is the same width for both spectra to illustrate how the lower spectrum (Y Da wide) is much more narrow than the upper spectrum (X Da wide). These spectra are taken from part B (top=50 sec, bottom=1000 sec). The peak width (this time at 20% peak intensity) versus time is plotted as shown (bottom graph) and the apex, which represents the t1/2 of unfolding, is determined from the intersection of tangents to the peak on the log scale (red lines) (see [73,105] for details). (D) Deuterium incorporation (i, ii) and peak width-plots (iii, iv) for the unresolved EX1 data on Lck SH3 in part B. These data were taken from Ref. [72]. (E) Examples of using peak width to detect protein:ligand interactions. In this example, the Lck SH3 domain alone (solid symbols) and Lck SH3 bound to a high-affinity peptide from the HVS Tip protein (open symbols) were compared. The residues of each peptide are indicated in the top right of each graph. Lck SH3 domain binding slows unfolding as indicated by the shifts of the peak-width plots to the right upon binding. These data were taken from Ref. [72].
The mass spectra from HX MS experiments for the Lck SH3 domain, both on the whole protein (FIGURE 6B, left) and peptic peptide level (FIGURE 6B, right), indicated that during the time course of deuterium exposure, the width of the isotope distributions increased, and then decreased again back to the width of the earliest time point (3 seconds) [72]. FIGURE 6C shows the mass spectra for a peptide of Lck SH3 (residues 44–61) at two different deuterium exchange times (50 seconds, top; 1000 seconds, bottom). The width of these two isotopic distributions is clearly different. If the width is measured at 20% of maximum peak height and plotted versus exchange time, a peak-width plot is produced (FIGURE 6C,D,E) that indicates a clear deflection from a flat line (note that peak-width plots are made with a log scale on the x-axis). We have described [32] in some detail how much above baseline a peak in a peak-width plot must be in order to be considered significant, and therefore indicative of EX1 kinetics. This description and all the other assumtions that accompany it, was important as there can be some peak widening as a result of deuterium incorporation itself, which is generally minor as we showed with both empirical measurements and kinetic modeling [32]. To conclude that there is EX1 kinetics in a protein based on peak width analysis of intact proteins, there must be an increase in peak width greater than 4 Daltons and for peptides, the increase must be greater than 2 Daltons[32].
For the Lck SH3 domain, the peak width changes were clearly large enough to be considered significant. FIGURE 6D shows several deuterium incorporation and peak width plots for Lck SH3. For Lck SH3 and its peptides, there was an obvious and significant deflection in the peak-width plot for both the intact protein (FIGURE 6D,iii) and for peptides derived from it (FIGURE 6D,iv). The apex of the peak in a peak-width plot provides the approximate t1/2 of unfolding, which can be found by a triangulation method [73], as illustrated with red lines in FIGURE 6C, bottom. The relative height of the peak provides information about the number of residues that participate in the EX1 unfolding event (taller peaks mean more separation between the two components of the EX1 distribution, a wider combined peak of the folded and unfolded populations, and therefore more residues involved). Inspection of the plots can reveal changes to protein unfolding rates and protein dynamics based on how the peak shifts: left or right, up or down, as illustrated in FIGURE 6E for several peptides of the Lck SH3 domain [72]. In the experiments shown in FIGURE 6E, Lck SH3 was exposed to deuterium in the presence or absence of a peptide ligand derived from the Tip protein [74]. This peptide binds to Lck SH3 with a dissociation constant of ~8.7 µM and we calculated that 88% of the Lck SH3 molecules were bound to the peptide under the conditions used [72]. In the bound state, the unfolding of the Lck SH3 domain was slowed by a factor of five from a half-life of ~10 seconds to a half-life of ~50 seconds. This slowing can be clearly seen in the shift in the peak-width plots to the right upon binding (FIGURE 6E). So to summarize, peak width plots produced from HX MS data provide information about binding when there are EX1 unfolding events in proteins. The magnitude of the slowing of unfolding as a result of peptide binding is observed as changes in the time at which the maximum of the peak in a peak-width plot appears and the amount of residues that are involved in the unfolding is provided by the height of the peak.
5.2.2. Comparison of Hck and Lck: Conserved intramolecular interactions?
It was striking that the Hck SH3 domain has slow unfolding and the Lck SH3 domain had fast unfolding. We asked ourselves if this could have some functional implications for the two proteins, or perhaps it is a feature of the sequence, hydrophobic and electrostatic nature of the proteins. To try to answer these questions, we used the divide-and-conquer HX MS approach for Lck that we had used for Hck to investigate larger and larger constructs of Lck until we were analyzing nearly the entire kinase. The goal of these studies was to understand how intra-protein regulation worked in Lck (similar to what we had done with Hck), if its regulation was different from that of Hck and then how the viral protein Tip interfered with regulation.
The HVS Tip protein is known to cause very potent kinase activation of Lck (see [72] and references contained therein) and this is presumed to occur via structural rearrangement that removes the down-regulatory influences of the SH3 and SH2 domains on the back side of the kinase domain. We incubated a near-full length version of Lck (LckYEEI [75,76], similar to HckYEEI [60] from above) with various Tip peptides known to bind Lck SH3 [72] and recombinant, full-length HVS Tip protein which was shown to be intrinsically disordered yet functional [75]. It is important to note here that Lck, like all Src-family kinases, becomes activated when a key tyrosine in the activation loop is phosphorylated (reviewed in [20] and data not shown).
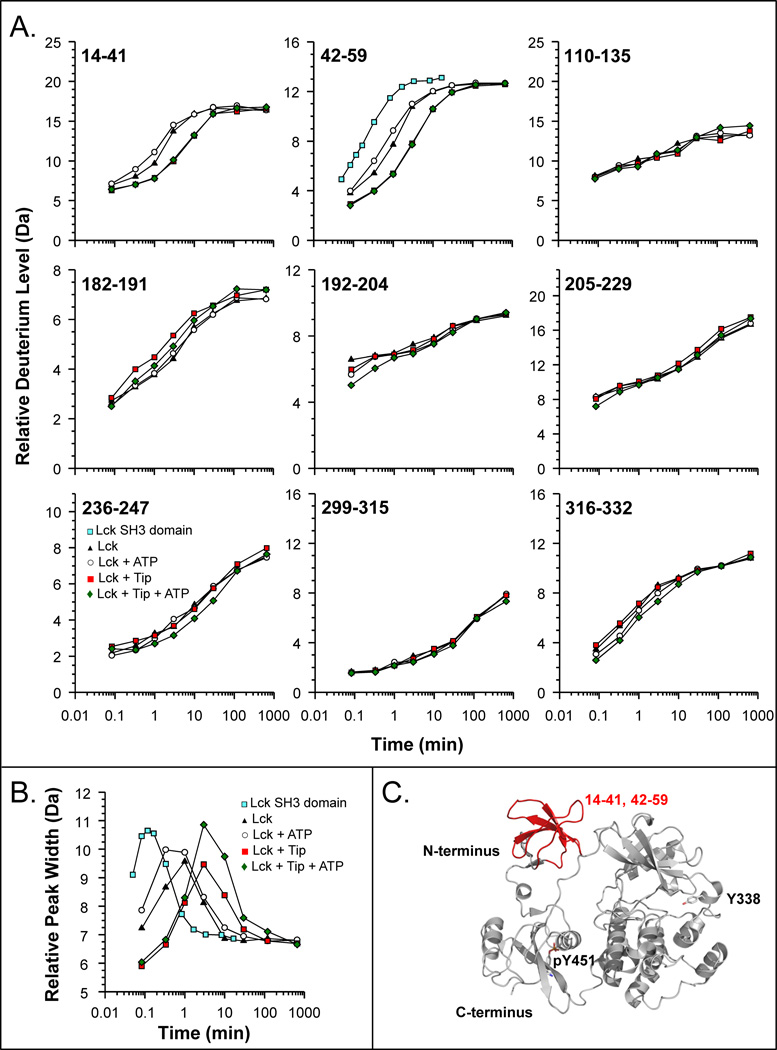
We measured the effects of phosphorylation, and therefore kinase activation, on the HX of LckYEEI and the results are shown in FIGURE 7. From deuterium incorporation graphs for various parts of Lck, it is clear that the majority of changes to Lck conformation and dynamics occur in the SH3 domain. There were little to no changes in other parts of the protein, including in the activation loop (peptide 316–332). These results suggest that in Lck, the SH3 domain plays a major role in regulation, as one would expect based on related kinases [20]. The unfolding of the Lck SH3 domain in the context of the whole LckYEEI protein was measured, it was found to be about five times slower than the Lck SH3 domain alone (moving from a t1/2 of about 10 seconds for Lck SH3 alone to a t1/2 of near 50 seconds for SH3 in LckYEEI). These results were similar to what was observed for slowdown as a result of binding the isolated Lck SH3 to a high-affinity ligand in trans (Ref. [72] and FIGURE 6 above). Upon incubation with ATP and kinase activation, there were only slight changes in the deuterium incorporation, specifically in the unfolding rate of the SH3 domain. Rather than the unfolding returning to the rate when SH3 was unbound, it stayed essentially the same, suggesting that there can be kinase activation without SH3 domain displacement. Such a phenomenon was described before for the Hck SH3 domain [62]. Further, when LckYEEI was bound to the full-length Tip protein, the SH3 domain unfolding rate became the slowest we have ever observed for Lck. Unfolding slowed from a half-life of near 50 seconds to a half-life of near 300 seconds. These results indicated that the Tip:SH3 domain interaction was very strong indeed and that Tip binding led to significant reduction in the dynamics of the SH3 domain. Surprisingly, at least according to HX MS, the rest of the kinase remained essentially unchanged in HX (note that some conformational motions may not be reported by HX, including things like whole domain motion or changes to regions in which exchange was already completed by the shortest time point of interrogation, in this case, 5 seconds). We concluded that the role of Tip binding was primarily to bind to and displace the SH3 domain and not to interact with other regions of the kinase. Given the much greater affinity of the Tip protein for SH3 over that of the internal linker sequence (as read out by the changes in SH3 dynamics, FIGURE 7B), it is unlikely that Tip would release the SH3 domain and allow Lck to return to a down-regulated conformation in which the SH3 domain again bound to internal linker sequence. Lck bound to Tip, therefore would be incapable of being down-regulated; Lck kinase activity would be continuous, and Lck would phosphorylate virtually any substrate in its path. These latter points are known to be the case for Tip-bound Lck from biological studies [77–82]; therefore, these HX MS data provide direct biophysical evidence in support of the constitutive activation model for Tip-mediated Lck activation.
Figure 7.
HX MS of the Lck protein. (A) Deuterium uptake curves for selected peptic peptides of Lck (solid triangles), Lck + ATP (open circles), Lck + Tip (solid red squares), and Lck + Tip + ATP (green diamonds). Also shown for peptide 42–59 is the deuterium uptake curve for the free SH3 domain of Lck (blue squares) [72]. The residues of each peptide are indicated in the upper left corner of each plot. The maximum of the vertical axis in each graph is the maximum number of exchangeable amide hydrogens. In all these experiments, LckYEEI [56,75] was used. (B) Peak width plot for Lck peptide 42–59, measured at 20% of peak maximum. (C) Regions of the Lck SH3 domain that experienced alterations in deuterium incorporation in response to Tip and/or ATP binding. The structure shown is a homology model for Lck based on the structure of down-regulated Hck (PDB 1QCF, [60]).
5.3. The Abl tyrosine kinase
Another protein for which we made extensive use of the HX MS measurements of protein unfolding is the Abl kinase [73,83,84]. The Abelson (c-abl) proto-oncogene encodes a non-receptor protein-tyrosine kinase (c-Abl) that is tightly downregulated in cells [19]. In contrast, the oncoprotein Bcr-Abl, which results from a chromosomal translocation that fuses Bcr sequences to the N-terminal region of c-Abl, is constitutively active [85,86]. The enhanced tyrosine kinase activity of Bcr-Abl fusion proteins is linked to chronic myelogenous leukemia (CML) and other forms of leukemia [86]. Most but not all of the c-Abl protein sequence is retained in the context of Bcr-Abl.
The domain organization of the ‘core’ region of c-Abl is similar to Src-family kinases (including Hck and Lck described above) in that it has SH3, SH2 and kinase domain; there are however, a few important differences. On the N-terminal side of the SH3 domain, Abl contains a sequence known as the N-terminal cap (NCap) region [19,87–91]. Unlike the Src-family kinases, there is no C-terminal tail that interacts with the SH2 domain. Multiple intramolecular interactions involving the regulatory SH3 and SH2 domains have been observed in the crystal structures of the downregulated c-Abl core [89,90]. As in Hck and Lck, the Abl SH3 domain binds the SH2-kinase linker, an interaction necessary to suppress kinase activity [92]. The NCap region is also essential for c-Abl downregulation [88–91]. The glycine residue at position 2 in the NCap is myristoylated and binds to a deep pocket in the C-lobe of the kinase domain, thereby latching SH2 and SH3 in their downregulatory positions at the back of the kinase domain and stabilizing the intramolecular interactions between SH3/SH2 and the kinase domain [19,89].
We used the HX MS methods described above to investigate several aspects of Abl dynamics: 1) what was the unfolding rate of the Abl SH3 domain and was it similar to the other SH3 domains that had been investigated; 2) was the linker sequence between the SH2 domain and the kinase domain also unable to bind to the SH3 domain without the presence of the kinase domain, as was found for Hck (FIGURE 4); 3) what was the role of the NCap in SH3 domain dynamics; and 4) how did phosphorylation of Abl alter the unfolding dynamics and therefore the regulatory ability of the Abl SH3 domain.
Initial experiments [66], described in Section 5.1 above (see also FIGURE 5), showed that the Abl SH3 domain did have some partial unfolding and that the half-life for this unfolding was approximately 10 minutes. The number of residues involved in the unfolding was small (~ 4 amide hydrogens) and this made detection of the unfolding challenging. Fortunately, peak width plots clearly revealed the unfolding event for both the intact protein [73] and for several peptides, which we later used to monitor unfolding [83,84]. Abl dynamics were also very sensitive to binding: unfolding could be slowed in the presence of a peptide called BP1 that was known to have a high affinity (Kd = 2 µM, [93]) for the Abl SH3 domain. Given that we were able to assess the dynamics of Abl SH3 based on its EX1 kinetic signature in HX MS, and that the rate of these dynamics were highly sensitive to binding, HX MS could be used – as it was for Hck and for Lck – as a sensitive probe of both intra- and intermolecular interactions for the Abl SH3 and all constructs of Abl containing an SH3 domain.
5.3.1. Linker interactions and additional regulatory elements (NCap)
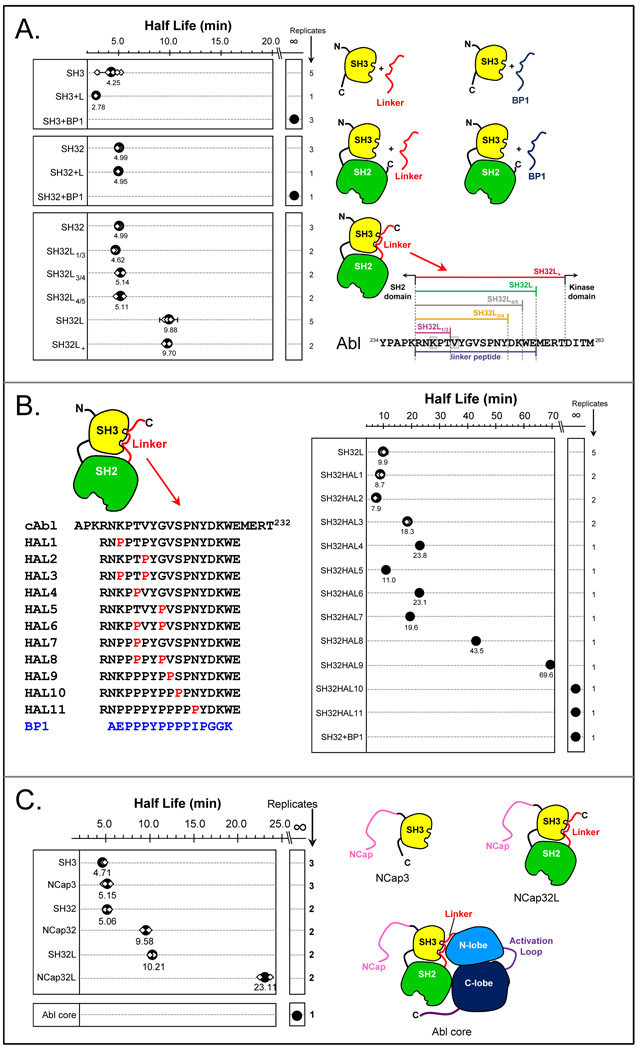
HX MS had clearly shown that the Hck kinase linker was unable to bind to the Hck SH3 domain without assistance from the kinase domain (summarized in FIGURE 4) and we wondered whether the same was true for Abl. In Abl, since there is no C-terminal tail to engage the SH2 domain, the hypothesis was that the SH3-kinase linker interaction must be stronger than the comparable interaction in Hck in order to compensate for the lack of the C-terminal tail. Using HX MS, we characterized the Abl linker in some detail [73]. FIGURE 8A summarizes the most important findings using another mode of display that we have also adopted to explain HX MS results: half-life graphs. Half-life graphs simply plot the relative unfolding half-life (see FIGURE 2) found from HX MS data that displays EX1 kinetics. The half-life is plotted for various constructs on the same scale, all relative to one another, such that comparisons of the half-life of unfolding between the protein constructs in the graph can be made. For all determinations of half-life in Abl proteins, we made use of the peak-width plots (as described above for Lck, see FIGURE 6) and found the apex of the peak in each peak-width graph. Consider the top panel of FIGURE 8A. In this series of HX MS experiments, the half-life of unfolding in Abl SH3 was compared when Abl SH3 was alone, incubated with a peptide with the same sequence as the Abl linker, and incubated with the high-affinity peptide BP1. The half-life of unfolding in the Abl SH3 domain was determined multiple times (small white symbols in FIGURE 8) and the average plotted (large black dots in FIGURE 8). What was readily apparent (FIGURE 8A) was that the Abl SH3 domain has no affinity for the linker peptide (the half-life of unfolding was unchanged versus SH3 alone) but bound tightly to the BP1 peptide (the half-life of unfolding was so long it could not be measured – we call this ‘infinity’ for the purposes of comparison). The same results were obtained for a larger construct consisting of the SH3 and SH2 domains (Abl SH32; FIGURE 8A, middle panel). When the linker peptide sequence was covalently attached to Abl SH32, there was significant and reproducible reduction to Abl SH3 domain unfolding but only when the linker was beyond a certain length (see FIGURE 8A, lower right). These experiments allowed us to decipher exactly how the Abl linker is different from that found in Hck [73].
Figure 8.
Measurements of unfolding half-life in the Abl SH3 domain for various constructs and mutants. In all these plots, large solid black symbols indicate the average unfolding half-life from the individual determinations (small white diamonds). The number of replicates of each measurement is shown on the right of each graph. (A) Unfolding was monitored for the constructs shown, bound to various ligands as indicated in the cartons on the left-hand side. For various lengths of linker, the sequence and linker nomenclature is indicated. These results were adapted from [73]. (B) Unfolding in various mutants of the Abl kinase linker. The wild-type linker sequence is shown on the top, indicated with cAbl. Each of the HAL linker sequences is shown. BP1 is a high-affinity Abl SH3 binding peptide (Kd = 2 µM, [93]). (C) Unfolding in various versions of Abl constructs containing the NCap (see [89] for crystal structure). These data were adapted from Ref. [83].
We had found and used [62] a high-affinity linker mutant for Hck (see FIGURE 4) to help us understand the binding of the linker sequence to the Hck SH3 domain. For Hck SH3, the poly-proline sequence of the natural Hck linker is not ideal for SH3 binding and requires other forces (from the kinase domain) to change the linker conformation to one compatible with SH3 binding. In the Hck linker, two mutations (from lysine to proline) created a high-affinity linker (HAL) mutant with the necessary linker conformation for high-affinity binding even in the absence of the kinase domain [62]. To test if analogous mutations would convert the Abl linker to a high affinity sequence, we mutated the corresponding residues of the Abl linker to proline and tested the ability of the mutant linker to bind to the Abl SH3 domain (in the context of an Abl SH32-linker protein). The HAL3 mutant (see FIGURE 8B) of Abl SH3 bears the analogous mutations to those that created the Hck HAL mutant. Interestingly, we did not observe the same dramatic ability of HAL3 to bind to the Abl SH3 domain as we had seen in the comparable HAL mutant in Hck. The half-life of Abl SH3 unfolding in HAL3 was only 2-fold slower than for wild-type Abl SH3 whereas the Hck HAL mutant slowed partial unfolding in the Hck SH3 domain by well over 50-fold (FIGURE 4). We went on to discover that many more residues in the natural Abl linker had to be mutated to create a high-affinity linker sequence. We slowly converted the native Abl linker sequence, by successive rounds of single proline substitutions, to the sequence of BP1 (FIGURE 8B) and measured the half-life of unfolding in Abl SH3 when bound to each of the HAL1 through HAL11 linker sequences. Multiple proline substitutions were required (HAL8-11) in order to slow the unfolding of the Abl SH3 to that observed when Abl SH3 was bound to the high affinity peptide BP1.
The role of the NCap sequence in Abl was also studied with HX MS [83], as summarized in FIGURE 8C. Constructs starting with just the SH3 domain all the way up to the whole Abl core protein were studied by HX MS. The NCap sequence did have some stabilizing effects on the Abl SH3 dynamics, effectively doubling the unfolding half-life in both SH32 vs. NCapSH32 (shifted the unfolding half-life of ~5 minutes to a half-life of ~10 minutes) and in SH32L vs. NCapSH32L (shifting the unfolding half-life from ~10 minutes to ~20 minutes). We further showed that unfolding of Abl SH3 was essentially turned off in the Abl core protein (a construct containing the NCap, SH3, SH2, linker, and kinase domain), presumably due to interactions of the NCap with the kinase domain (as shown in the crystal structure [89]).
To understand the role of intra-domain interactions within the Abl protein and how these interactions were tied to function, subsequent experiments in vitro and in vivo were performed on Abl core constructs containing each of the HAL sequences (data not shown, manuscript under review). The results showed that full-length Abl core proteins with HAL sequences 1, 4, 6, 8 and 9 (as in FIGURE 8B) were functionally downregulated as expected whereas Abl core proteins with HAL substitutions 2, 3, 5, and 7 showed much higher phosphorylation of autoregulatory tyrosine sites and a concomitant increase in cellular phosphotyrosine content. The HAL10 core protein was also upregulated, albeit to a lesser extent. While the HX MS studies showed that the HAL7 and HAL10 substitutions in a SH32L construct both strongly enhanced SH3 domain interaction, in the whole Abl core where the kinase domain was also present they must also have produced additional changes to the linker structure that interfered with its ability to effectively downregulate kinase activity. We next conducted an extensive characterization of the HAL9 Abl core protein, because this HAL substitution strengthened SH3 interaction to the greatest extent without affecting the downregulated state. Both cellular assays and HX MS (data not shown, manuscript under review), allowed us to conclude that strengthening the Abl SH3 domain interaction with its native linker in the whole Abl protein was effective in downregulating the kinase, and compensated for the activating effects of mutations in other regions of the protein. Taken together, we learned that the natural Abl linker sequence was a poor ligand for the Abl SH3 domain and concluded that other forces outside of the Abl SH3-linker interaction must be required to hold the Abl kinase in a downregulated conformation, especially in the context of the entire Abl core protein. From the point of view of the protein, this may make some sense because if the affinity were too strong, it would be hard to disengage the SH3 domain from its regulatory binding to the linker and alter protein activity. Requiring other forces in the protein besides just the SH3:linker interaction also open the possibility of allosteric control of activity from other parts of the molecule, something we have shown with other HX MS studies not described here [94–96]. Were it not for the HX MS analyses of the Abl SH3 interaction with its native linker, it would not have been possible to understand the mechanistic details of the linker:SH3 domain interactions and its subsequent role in the regulation of kinase activity.
5.3.2 Phosphorylation disrupts internal binding: Assay by HX MS
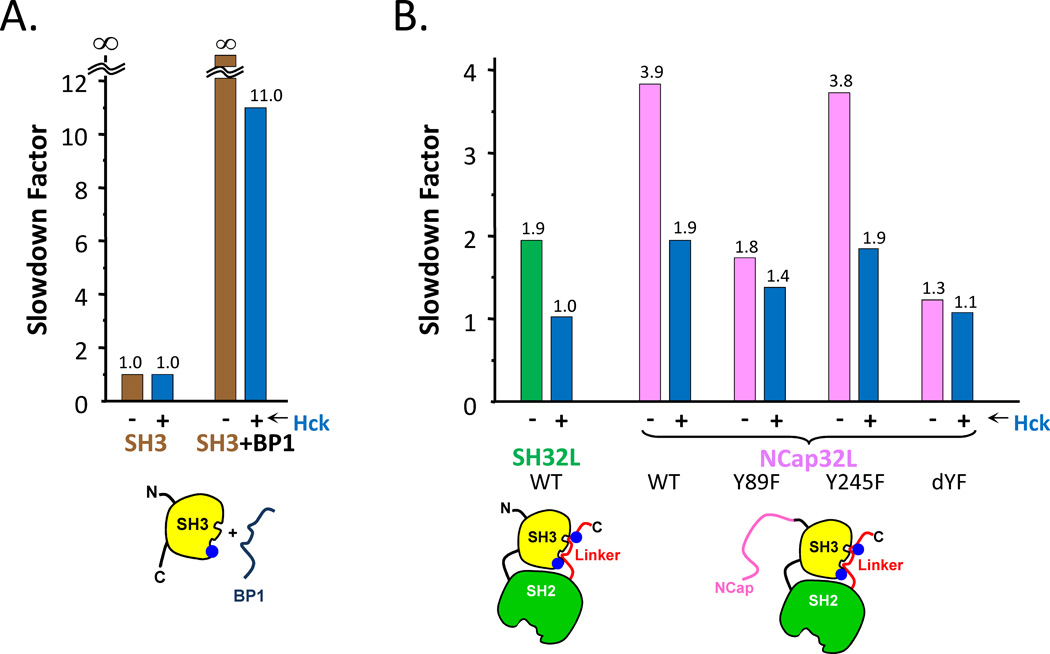
The effects of phosphorylation on Abl SH3 unfolding were also studied [84]. It was shown previously by our laboratories that the Src-family tyrosine kinases Hck, Lyn, and Fyn phosphorylate Bcr-Abl in the Abl SH3 and SH2 domains [97]. Tyr89 (Abl 1b numbering) in the Abl SH3 domain was found to be the most prominent phosphorylation site, both in vitro and in CML cells. Phosphorylation of Tyr89 was shown to be necessary for the full biological activity of Bcr-Abl, as substitution of this tyrosine residue with phenylalanine reduced the transforming potential of Bcr-Abl in a cytokine-dependent myeloid cell line [98]. The crystal structure of the c-Abl core [19] localizes Tyr89 to the binding surface between the SH3 domain and the SH2-kinase linker, a region important for maintaining the inactive, down-regulated state. The prediction was that phosphorylation of this residue by Src-family kinases would prevent the Abl linker from interacting with the Abl SH3 domain and thereby prevent full downregulation of the kinase activity. In terms of an HX MS readout, linker association would slow the unfolding of the Abl SH3 domain and if phosphorylation prevented linker association, the slowdown in unfolding as a result of binding, i.e., the slowdown factor, would be smaller. To test this idea, HX MS was measured for a number of Abl constructs, including those shown in FIGURE 9 [84]. In the Abl SH3 domain (FIGURE 9A), phosphorylation of Tyr89 prevented high affinity BP1 binding and reduced the slowdown factor from infinity (i.e., not measurable) to 11. In a control experiment without BP1, there was no effect on Abl SH3 domain unfolding solely as a result of phosphorylation. Similar to the results described above for FIGURE 4, the interpretation is that unfolding in the SH3 domain was slowed by binding of the ligand (BP1 in this first experiment, the natural linker in those that followed); preventing interaction of the ligand via phosphorylation reduces the slowdown factor. In the larger SH32L and NCapSH32L constructs of Abl, phosphorylation of Abl by the Hck kinase reduced the slowdown factor from 2 to 1 for SH32L and from 4 to 2 for NCapSH32L (FIGURE 9B). In the larger constructs with the linker, there was also the possibility of phosphorylation at position Tyr245, which is in the linker itself. To determine whether phosphorylation of Tyr89 or Tyr245 (or both) influenced linker binding to the Abl SH3 domain, we created single and double mutants of these sites. Mutation of Tyr245 had no effect on reducing the slowdown factor (preventing binding) whereas the single Tyr89 mutant or the double mutants both showed reduced linker association. These HX MS results provided strong evidence that phosphorylation at Tyr89 physically prevents the Abl linker from binding to the Abl SH3 domain. Upon phosphorylation and subsequent interference with linker binding, downregulation of the entire protein is not possible. Src-family kinases that phosphorylate the Bcr-Abl SH3 domain are likely to alter the Abl core region in a similar way to that measured for the Abl core protein, thus contributing to the constitutive kinase activity of Bcr-Abl.
Figure 9.
Summary of the effects of phosphorylation on Abl SH3 unfolding for various constructs. (A). Comparison of slowdown factor of free SH3 and SH3 incubated with BP1, both with and without phosphorylation by Hck. When Hck is present (as indicated with the -/+ symbols) Abl is phosphorylated at the positions indicated (see also [84]) by the blue dots in the cartoons at the bottom – Tyr89 (in SH3) and Tyr245 (in the SH2-kinase linker). (B). Comparison of slowdown factor as a function of phosphorylation in the constructs indicated. Mutants are indicated, with dYF being the double mutant of Y89F,Y245F. This figure is taken from Ref. [84].
6. Which parts of SH3 participate in local unfolding, and why
While HX MS was used in many experiments to understand kinases and dissect the intricacies of their inner workings, one important question still remained: precisely which residues in the SH3 domains were participating in the unfolding, and why? With HX MS experiments described thus far, it was not possible to answer these questions. Hydrogen exchange can be detected by mass spectrometry on the whole protein level or deuterated proteins can be digested, under exchange quench conditions, into peptide fragments and the mass of the fragments measured [67,99,100]. Digestion increases the spatial resolution of the exchange information but the peptides that are produced cannot be predicted from sequence and the investigator has no control over where in the protein the enzyme (usually pepsin, an aspartic protease) will cut [24]. Generally, the peptides are between 5–12 residues long and this dictates the resolution of the deuterium incorporation measurements.
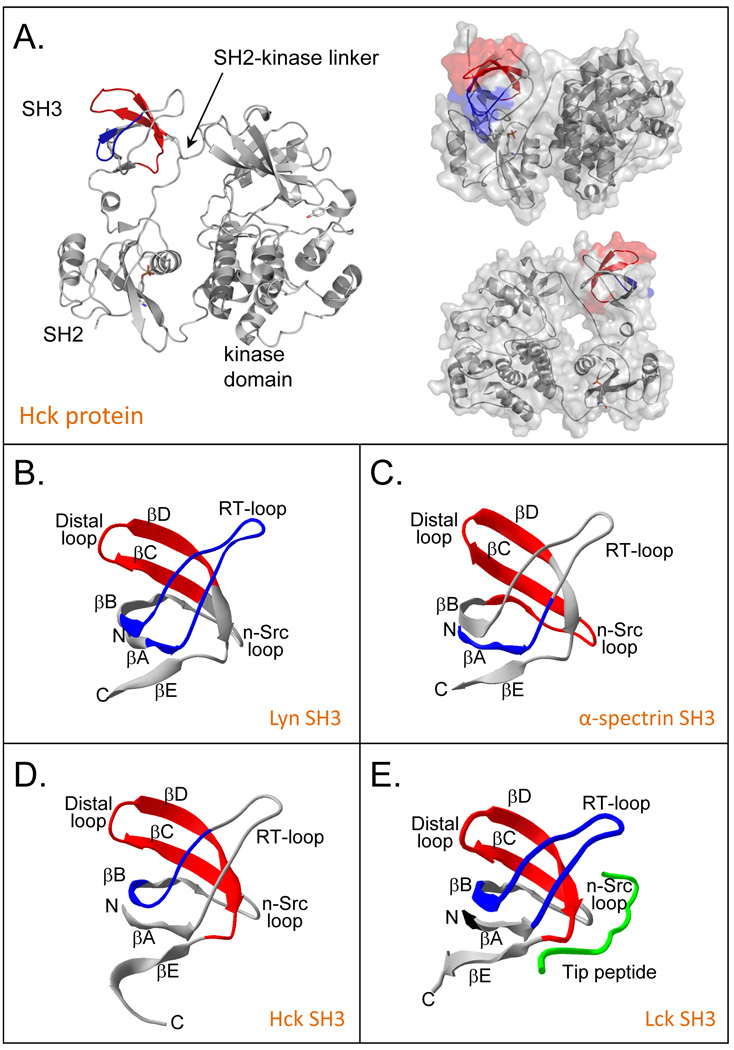
The initial experiments using pepsin digestion localized the unfolding in Hck SH3 to two regions [1]. These two regions encompassed residues 98–105 (SFQKGDQM) and 115–132 (WKARSLATRKEGYIPSNY) (numbering according to the Hck crystal structure as in Ref. [60]). It was evident that these were the regions involved because the mass spectra for peptides in these two regions (see [1,51] for the actual mass spectra) also displayed EX1 kinetics at the same time as that for the whole protein (as in FIGURE 2A). High resolution structures of Hck SH3 were not available when we first reported the partial unfolding in Hck SH3 [1], but several structures were later published including solution NMR structures of isolated Hck SH3 [101,102], the x-ray crystal structure of isolated Hck SH3 [103], the structure of a combined Hck SH3-SH2 construct [63] and near full-length intact Hck [60,61]. With new structural details of Hck SH3, it became possible to better define the region of SH3 unfolding. The two parts of SH3 with EX1 signatures were close in tertiary structure but distant in primary structure: residues 115–132 form two strands of the small beta sheet within Hck SH3 while residues 98–105 form a short hairpin turn that is located directly across from the turn separating the two strands of residues 115–132 (see FIGURE 10). The regions involved were also described in terms of hydrogen bonding and solvent accessibility [104] and the region of unfolding was narrowed to include only residues 98–105 and 115–124 based on this analysis and some, by today’s standards, crude MS/MS experiments (see below). The location of the majority of the residues in these two peptides is actually away from the interaction surface for the natural Hck linker (FIGURE 10A).
Figure 10.
Locations of peptides involved in SH3 domain unfolding. (A) Peptides 98–105 (blue) and 115–132 (red) of Hck SH3 in the context of the near full length down-regulated Hck kinase (PDB 1QCF, [60]). (B) Lyn SH3 (PDB 1W1F, [127]) residues 69–86 (blue), 100–111 (red). (C) α-spectrin SH3 (PDB 1U06, [128]) residues 964–977 (blue) and 995–1014 (red). (D) Hck SH3, derived from panel A (PDB 1QCF, [60]) residues 98–105 (blue) and 115–132 (red) and (E) Lck SH3 (PDB 1LCK, [129]) residues 16–33 (blue) and 44–61 (red). Parts of this figure were derived from Refs. [66] and [72].
6.1. Comparisons of SH3 domains
With analysis of unfolding in other proteins (e.g., [66]), it became clear that the location of unfolding in the Hck SH3 domain was similar to the location of unfolding found in diverse SH3 domains. As shown in FIGURE 10B–E, simple pepsin digestion HX MS experiments and monitoring for EX1 kinetics localized the unfolding in the SH3 domains from α-spectrin and Lyn [66], Lck [72] and Abl [83,84,105]. In all cases, unfolding involved the same two β strands before and after the distal loop, as well as part of the RT-loop. While these parts of SH3 were influenced by binding, as revealed in the EX1 kinetics described above, these parts of SH3 were not directly in the SH3 domain binding site.
Based on the EX1 signatures, not all positions in the peptides identified become deuterated during partial unfolding. From intact protein analyses, the number of residues involved was variable (see FIGURE 5) with Hck, α-spectrin and Lyn having the most residues involved. While there were initial attempts at refining the location with tandem MS experiments [104], early experiments failed to definitively identify which residues were participating for a number of reasons. Tandem MS experiments, in which the deuterium labeled peptides are further fragmented into pieces within the mass spectrometer, have the potential to increase the resolution even further, even to the single amino-acid level. The concept is that a protein is labeled with deuterium, the labeling quenched, the protein digested into small fragments, the fragments are introduced to the mass spectrometer via chromatography, and then each of the peptide ions (precursor ions) are fragmented into smaller peptides (product ions) within the mass spectrometer. By comparing deuterium levels in all the product ions, one can elucidate which amino acids contained deuterium at the backbone amide hydrogen positions.
6.2. MS/MS measurements using ETD
Early MS/MS experiments in HX MS used collision induced dissociation (CID) of deuterium containing peptides [106–110]. Unfortunatley, CID causes deuterium “scrambling” via deuteron migration during the fragmentation process [111]. If ions are given too much internal energy during fragmentation, deuterium might migrate making the tandem MS/MS data of deuterium position meaningless. Recent descriptions of electron transfer dissociation (ETD) under controlled analysis conditions that minimize scrambling [112] indicate that single-residue resolution in HX MS can be achieved in multiple types of mass analyzers [113–115]. While ETD HX MS experiments are far from routine, we have very recently revisited the question of where in Hck SH3 unfolding occurs using ETD fragmentation methods. Identifying which residues participate is important to understanding the physical mechanism by which the partial unfolding occurs in SH3 domains.
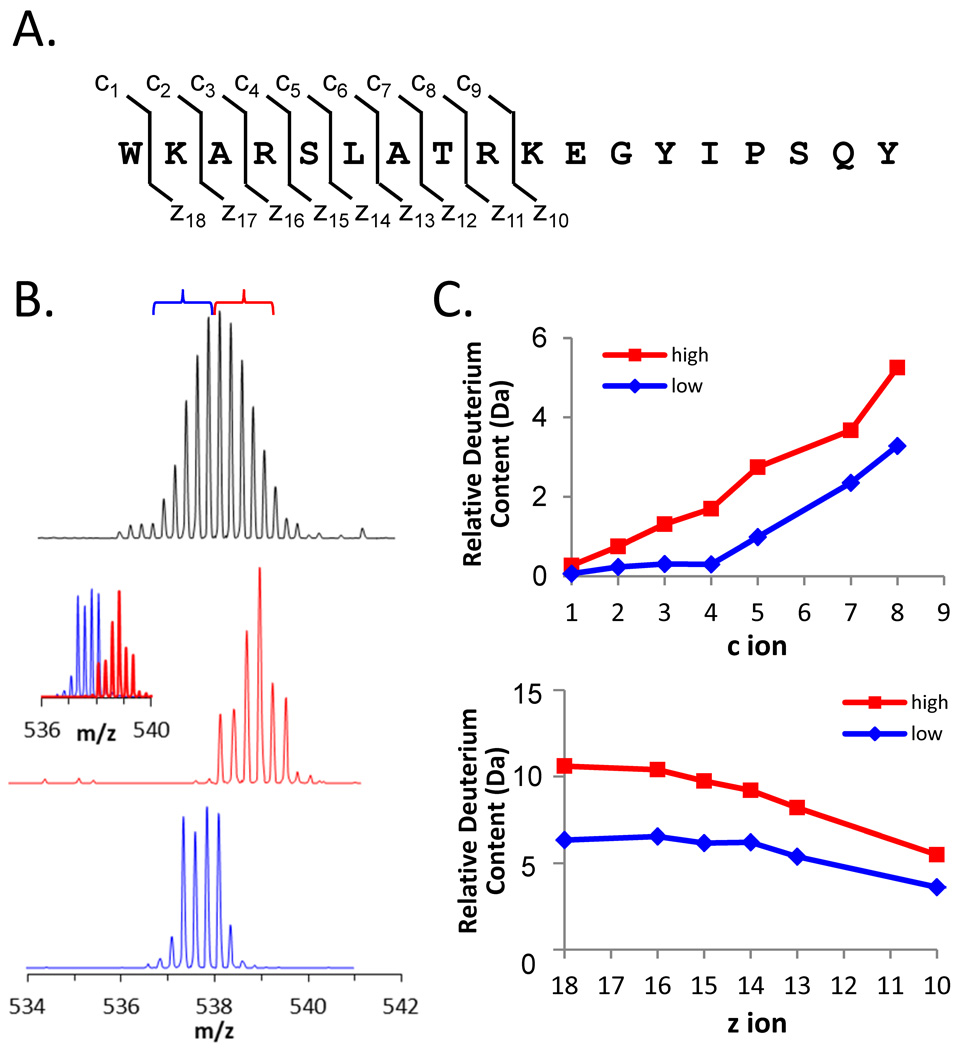
ETD was first performed on an undeuterated sample of Hck SH3 that had been digested into peptic peptides. This experiment was designed to determine the ETD dissociation patterns and ETD spectra that would result from fragmentation of each peptide involved in unfolding. ETD was most efficient on the +4 charge state of the 115–132 peptide although it gave reliable c and z fragments for only the first 9 residues of the peptide (FIGURE 11A). The only charge state observed for the 98–105 peptide was the +2 charge state and ETD fragmentation of this peptide ion gave a mixture of c and z ions that, when combined, covered most of the peptide (data not shown). Because ETD analysis was combined with chromatographic separation of the peptides resulting from pepsin digestion, rather than infusion of a peptide, typically only 3–4 scans were taken over each chromatographic peak. Despite the limited number of mass spectra taken for each peptide, good ETD intensity of the fragments was obtained.
Figure 11.
ETD analysis of deuteration in the peptide 115–132 of Hck SH3. (A) The sequence of residues 115–132 with the c and z ion cleavage positions indicated. These c and z product ions were the only ones for which reliable data could be obtained; fragmentation of the C-terminal part of the peptide was not efficient. (B) Mass spectra of the +4 charge state of peptide 115–132 after 15 minutes of deuterium exchange (top). The low-mass region (blue bracket) and the high-mass region (red bracket) of the entire peak were independently selected for ETD using a quadrupole LM resolution setting of 15. The isolated ions are shown in the middle (red) and lower (blue) spectra; the combination of the red and blue spectra is shown in the inset. All spectra were obtained with a Waters Synapt G2 equipped with ETD using mild conditions that minimize deuterium scrambling [113]. (C) Deuterium content of the high- and low-mass selections according to the product ions of the c-type (top) and z-type (bottom).
Hck SH3 was deuterated for 15 minutes and the ETD experiments repeated. The mass spectra of the unfragmented deuterated species of peptides 98–105 and 115–132 were as expected (spectra for 115–132 peptide are shown in FIGURE 11B, top) and indicated the characteristic EX1 pattern that was always observed for the unfolding event in these peptides of Hck SH3. The high-mass and low-mass components of the isotopic distribution for each peptide were mass selected using the quadrupole mass filter of the Waters SynaptG2 mass spectrometer, see FIGURE 11B for peptide 115–132. The quadrupole selection process does not excite ions and allows for the use of a narrow selection window (approx. 2.0 m/z, as demonstrated in FIGURE 11B); other ion selection methods such as with traps require a much wider precursor selection window (approx. 10 m/z) to avoid deuterium scrambling from side-band excitation [115–117] and would therefore not be able to make the fine selection of the lower- and upper-mass envelopes as we describe here. The low-mass region of the isotope distribution for peptide 115–132 was found to incorporate 6.5 deuterium while the high-mass region incorporated 10.6 deuterium. Incorporation into the low-mass part of the EX1 distribution was a result of deuterium incorporation in the peptide that did not involve local unfolding, or EX1 (part of the peptide undergoes exchange via EX2 and part undergoes exchange via EX1). The ETD spectra were analyzed and the results (FIGURE 11C) indicated that in residues 115–132, about half of the deuterium incorporated during localized unfolding was in the first half (WKARSLAT…) of the peptide (WKARSLATRKEGYIPSQY). The first two amino acids do not retain deuterium during analysis, but after that the deuterium content was uniformly distributed. Likewise, difference plots for the 98–105 peptide (data not shown) also indicated that the deuterium was uniformly distributed through the last five amino acids (…KGDQM) of the peptide (SFQKGDQM).
Contrary to what was expected, it was not as easy to define the location of the residues involved in EX1 kinetics as expected. Back-exchange during LC MS analysis, which can be minimized but never eliminated [24], meant that some of the label we were trying to detect was not present by the time each peptide fragment made it to the detector. Cleavage by pepsin did not produce the ideal ions for ETD and this problem could be mitigated by the use of other aspartic proteases often employed in HX MS [118]. Finally, it is not trivial to perform ETD fragmentation on peptide ions eluting in very narrow peaks from ultra-high performance chromatography particularly when the intensity of the signals is weak. At the maximum, there may be the opportunity to acquire 2–3 good scans of the fragmentation pattern under these conditions, as opposed to the many scans that can be acquired of proteins that are infused into the instrument rather than studied by chromatography.
6.3. Additional information from mutagenesis
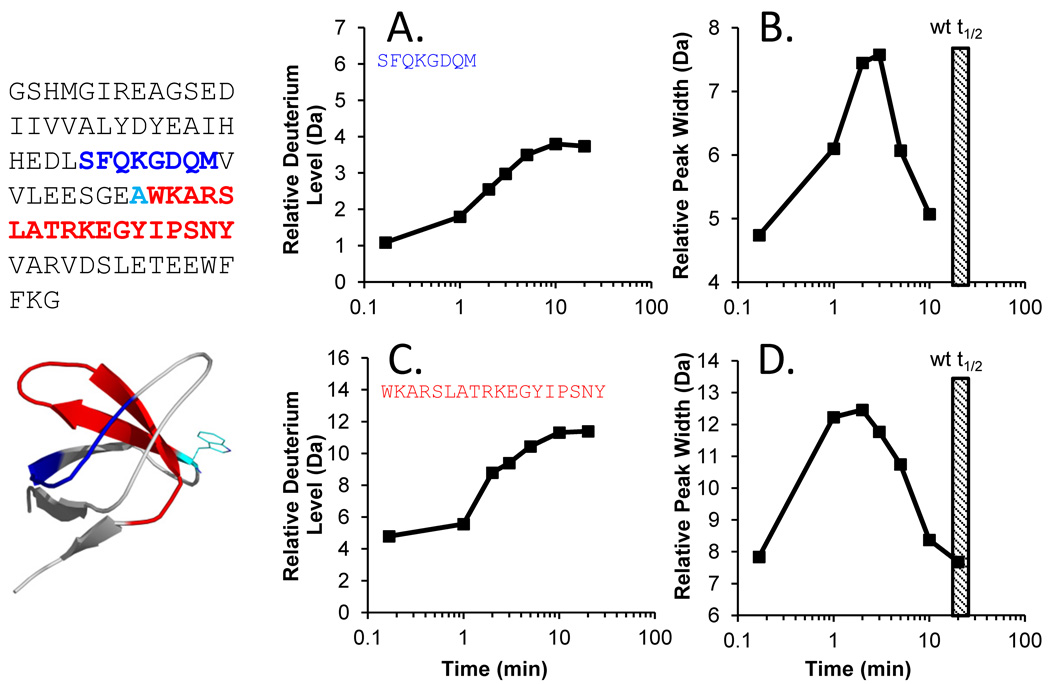
Although the location was slightly better refined with MS/MS, it still remains unclear why this part of SH3 domains has partial unfolding. Comparative analyses of various SH3 domains with diverse sequences [66] attempted to answer this question but did not offer definitive answers as to why this part of SH3 underwent partial unfolding. For other reasons, we had characterized the unfolding of a mutant of Hck SH3 wherein position 93 was changed from a tryptophan to an alanine. The so-called W93A mutant of Hck is not capable of interaction with and activation of substrates, including Vif [69,119], Nef [120] and STAT3 [121]. Surprisingly, partial unfolding in the Hck SH3 W93A mutant was much faster than that of wild-type Hck SH3 (FIGURE 12). While wild-type Hck SH3 had a half-life of unfolding around 18–20 minutes, the W93A mutant of Hck SH3 underwent partial unfolding with a half-life of 2–3 minutes, 10 times faster. This result was surprising given that the position of W93 is at the beginning of the 115–132 peptide that had garnered so much attention. Apparently changing the hydrophobic character and packing of this single amino acid side chain residue is enough to destabilize the local dynamics of the Hck SH3 domain such that its unfolding speed is much faster. It will be interesting to make additional mutants in this and surrounding positions to further understand how packing forces influence the partial unfolding reaction. Readout from HX MS will be instrumental in these studies.
Figure 12.
Deuterium uptake plots and peak width analysis for peptides from the Hck SH3 W93A mutant. The sequence of the protein is shown at the left, with the 98–105 peptide colored in blue and the 115–132 peptide colored in red. The location of the mutation is shown in cyan – note the numbering of the mutation (W93A) is not in the same numbering scheme as the rest of the article, see Ref. [123]. (A,C) Deuterium uptake plots and (B,D) peak width plots for the two peptides, as indicated. The hatched box in panels B and D represents the time of the peak width maximum for wild type Hck SH3.
7. Partial unfolding in other proteins
While partial cooperative unfolding in SH3 domains has been studied in great depth, as described above and in the many publications that accompany this summary, what about other proteins? Just how widespread is EX1-related protein dynamics? We have only begun to scratch the surface of this field as the use of HX MS is not as widespread as needed to broadly characterize the prevalence of partial unfolding in proteins. Several additional examples of EX1 kinetic signatures in other proteins, along with ways in which these mass spectral signatures are useful, will be described in the next sections.
One important aspect of observing EX1 kinetics in HX MS experiments is being sure that the mass spectral pattern is actually derived from the protein and not from experimental artifact. We previously described [122] how several factors can produce false EX1 kinetic signatures, including peptide carryover, and protein aggregation. These possibilities must be eliminated before any conclusions about protein-related EX1 kinetics are drawn. Careful control of analytical conditions (as described [122]) allow one to remove parameters that lead to false EX1.
The dissociation constant for interactions can be derived from HX MS measurements involving EX1. One simply needs to titrate the mixture of protein and ligand and monitor the reduction in EX1-related unfolding as the concentration of ligand changes. One recent example from our laboratories of applying this methodology involved the Elongin BC complex and the HIV Vif protein [123]. When the HIV Vif protein, which is largely unstructured [119], binds to the complex of Elongin B and Elongin C, protection occurs in the Elongin C protein. This protection changes as the concentration of the Vif partner increases. Using various concentrations and mutants of Vif, it was possible to extract a dissociation constant for the interaction. While other biophysical methods are better in many circumstances for measuring equilibrium dissociation constants, there are some advantages to using HX MS for this measurement including the relatively small amount of material, no requirement for immobilization of one partner, and determination of the value for a single member of a multi-protein complex. The big disadvantage is that HX MS can best be utilized for proteins that experience some kind of EX1 event, but the majority of proteins do not undergo exchange via this kinetic regime.
In a study of the processivity clamp β from E. coli, we observed a significant degree of EX1 kinetics [124]. Processivity clamps form a planar ring that encircles DNA during translation and DNA repair, and reduce the possibility of the DNA replication machinery falling off the DNA (for more details and crystal structures see [125,126]. Two β clamp monomers join together to form the clamp ring structure in E. coli. EX1 kinetics was found in the first domain of the monomer and parts of the third domain. The regions involved form the interface which opens when the clamp is loaded onto the DNA. The half-life of unfolding in this protein was found to be near 4 hours. Other processivity clamps have been analyzed (data not shown) and indicate there is wide variability in the partial unfolding kinetics in these proteins.
8. Conclusions
HX MS is a unique technique which permits detection of simultaneously existing populations of molecules in various folded states. Partial cooperative unfolding in Hck SH3 was initially investigated using HX MS and similar phenomena were examined in other SH3 domains from other proteins. Via systematic investigation of the parts of a larger system (e.g., buildup from SH3 to SH32, to SH32L and interaction with linkers, binders, etc.), a more complete picture of protein interactions and stability can be obtained, as we illustrated for Hck, Lck and Abl kinases. In these proteins, SH3 was receptive to modulation by the presence of nearby domains like SH2 and the kinase domain, but the relationship was not reciprocal. To understand the detailed regulatory mechanism of these kinases, HX MS analysis of SH3 domain dynamics became a powerful tool to probe both inter-molecular and intra-molecular interactions. Not only was local unfolding an interesting biophysical phenomenon of SH3 domains, but also turned out to be an invaluable probe for SH3-ligand interaction strength. In the end, perhaps SH3 domain dynamics plays a role in fine-tuning of kinase regulation: SH3 needs to be dynamic to transiently uncouple from the kinase and associate with activating ligands, an essential part of the physiological role of these signaling proteins.
Our ultimate conclusion is that the actual prevalence of partial cooperative unfolding in proteins remains unknown. To determine just how widespread, and thereby perhaps physiologically relevant, such protein flexibility really is, it will be important to study many more proteins with HX MS.
Acknowledgements
This work has been graciously supported over the years by the NIH [GM070590, GM086507 and GM101135 to JRE; AI057083 and CA81398 to TES], a research collaboration with the Waters Corporation (JRE), funding from Biogen-Idec, Johnson & Johnson, and the Dana-Farber Cancer Institute.
References
- 1.Engen JR, Smithgall TE, Gmeiner WH, Smith DL. Biochemistry. 1997;36(47):14384. doi: 10.1021/bi971635m. [DOI] [PubMed] [Google Scholar]
- 2.Linderstrom-Lang KU, Schellman JA. In: The Enzymes. Boyer PD, Lardy H, Myrbäck K, editors. Vol. 1. New York: Academic; 1959. [Google Scholar]
- 3.Berger A, Linderstrom-Lang K. Arch Biochem Biophys. 1957;69:106. doi: 10.1016/0003-9861(57)90478-2. [DOI] [PubMed] [Google Scholar]
- 4.Frauenfelder H, Parak F, Young RD. Annu Rev Biophys Biophys Chem. 1988;17:451. doi: 10.1146/annurev.bb.17.060188.002315. [DOI] [PubMed] [Google Scholar]
- 5.Frauenfelder H, Sligar SG, Wolynes PG. Science. 1991;254(5038):1598. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 6.Woodward C, Simon I, Tuchsen E. Mol Cell Biochem. 1982;48(3):135. doi: 10.1007/BF00421225. [DOI] [PubMed] [Google Scholar]
- 7.Englander SW, Kallenbach NR. Q Rev Biophys. 1983;16(4):521. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]
- 8.Daniel RM, Dunn RV, Finney JL, Smith JC. Annu Rev Biophys Biomol Struct. 2003;32:69. doi: 10.1146/annurev.biophys.32.110601.142445. [DOI] [PubMed] [Google Scholar]
- 9.Berendsen HJ, Hayward S. Curr Opin Struct Biol. 2000;10(2):165. doi: 10.1016/s0959-440x(00)00061-0. [DOI] [PubMed] [Google Scholar]
- 10.Kern D, Zuiderweg ER. Curr Opin Struct Biol. 2003;13(6):748. doi: 10.1016/j.sbi.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 11.Eisenmesser EZ, Millet O, Labeikovsky W, Korzhnev DM, Wolf-Watz M, Bosco DA, Skalicky JJ, Kay LE, Kern D. Nature. 2005;438(7064):117. doi: 10.1038/nature04105. [DOI] [PubMed] [Google Scholar]
- 12.Artymiuk P. Nature. 1988;332(6165):582. doi: 10.1038/332582a0. [DOI] [PubMed] [Google Scholar]
- 13.Gerstein M, Krebs W. Nucleic Acids Res. 1998;26(18):4280. doi: 10.1093/nar/26.18.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Echols N, Milburn D, Gerstein M. Nucleic Acids Res. 2003;31(1):478. doi: 10.1093/nar/gkg104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maiti R, Van Domselaar GH, Wishart DS. Nucleic Acids Res. 2005;33(Web Server issue):W358. doi: 10.1093/nar/gki485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Courtneidge SA, Fumagalli S, Koegl M, Superti-Furga G, Twamley-Stein GM. Dev Suppl. 1993:57. [PubMed] [Google Scholar]
- 17.Superti-Furga G. FEBS Lett. 1995;369(1):62. doi: 10.1016/0014-5793(95)00636-n. [DOI] [PubMed] [Google Scholar]
- 18.Superti-Furga G, Gonfloni S. BioEssays. 1997;19(6):447. doi: 10.1002/bies.950190602. [DOI] [PubMed] [Google Scholar]
- 19.Hantschel O, Superti-Furga G. Nat Rev Mol Cell Biol. 2004;5(1):33. doi: 10.1038/nrm1280. [DOI] [PubMed] [Google Scholar]
- 20.Engen JR, Wales TE, Hochrein JM, Meyn MA, 3rd, Banu Ozkan S, Bahar I, Smithgall TE. Cell Mol Life Sci. 2008;65(19):3058. doi: 10.1007/s00018-008-8122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith DL, Deng Y, Zhang Z. J Mass Spectrom. 1997;32(2):135. doi: 10.1002/(SICI)1096-9888(199702)32:2<135::AID-JMS486>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 22.Engen JR, Smith DL. Anal Chem. 2001;73(9):256A. doi: 10.1021/ac012452f. [DOI] [PubMed] [Google Scholar]
- 23.Hoofnagle AN, Resing KA, Ahn NG. Annu Rev Biophys Biomol Struct. 2003;32:1. doi: 10.1146/annurev.biophys.32.110601.142417. [DOI] [PubMed] [Google Scholar]
- 24.Wales TE, Engen JR. Mass Spectrom Rev. 2006;25(1):158. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 25.Yan X, Maier CS. Methods Mol Biol. 2009;492:255. doi: 10.1007/978-1-59745-493-3_15. [DOI] [PubMed] [Google Scholar]
- 26.Engen JR. Anal Chem. 2009;81(19):7870. doi: 10.1021/ac901154s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marcsisin SR, Engen JR. Anal Bioanal Chem. 2010;397(3):967. doi: 10.1007/s00216-010-3556-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Konermann L, Pan J, Liu YH. Chem Soc Rev. 2011;40(3):1224. doi: 10.1039/c0cs00113a. [DOI] [PubMed] [Google Scholar]
- 29.Deng Y, Zhang Z, Smith DL. J Am Soc Mass Spectrom. 1999;10(8):675. doi: 10.1016/S1044-0305(99)00038-0. [DOI] [PubMed] [Google Scholar]
- 30.Hvidt A. C R Trav Lab Carlsberg. 1964;34:299. [PubMed] [Google Scholar]
- 31.Baldwin RL. Proteins. 2011;79(7):2021. doi: 10.1002/prot.23039. [DOI] [PubMed] [Google Scholar]
- 32.Weis DD, Wales TE, Engen JR, Hotchko M, Ten Eyck LF. J Am Soc Mass Spectrom. 2006;17(11):1498. doi: 10.1016/j.jasms.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 33.Miranker A, Robinson CV, Radford SE, Aplin RT, Dobson CM. Science. 1993;262(5135):896. doi: 10.1126/science.8235611. [DOI] [PubMed] [Google Scholar]
- 34.Arrington CB, Teesch LM, Robertson AD. J Mol Biol. 1999;285(3):1265. doi: 10.1006/jmbi.1998.2338. [DOI] [PubMed] [Google Scholar]
- 35.Engen JR, Wales TE, Shi X. In: Encyclopedia of Analytical Chemistry. Meyers RA, editor. Wiley; 2011. [Google Scholar]
- 36.Englander SW. Annu Rev Biophys Biomol Struct. 2000;29:213. doi: 10.1146/annurev.biophys.29.1.213. [DOI] [PubMed] [Google Scholar]
- 37.Engen JR, Smith DL. Methods Mol Biol. 2000;146:95. doi: 10.1385/1-59259-045-4:95. [DOI] [PubMed] [Google Scholar]
- 38.Sivaraman T, Robertson AD. Methods Mol Biol. 2001;168:193. doi: 10.1385/1-59259-193-0:193. [DOI] [PubMed] [Google Scholar]
- 39.Kaltashov IA, Eyles SJ. Mass Spectrom Rev. 2002;21(1):37. doi: 10.1002/mas.10017. [DOI] [PubMed] [Google Scholar]
- 40.Kuriyan J, Cowburn D. Curr. Opin. Struct. Biol. 1993;3(6):828. [Google Scholar]
- 41.Lim WA. Curr Opin Struct Biol. 2002;12(1):61. doi: 10.1016/s0959-440x(02)00290-7. [DOI] [PubMed] [Google Scholar]
- 42.Boggon TJ, Eck MJ. Oncogene. 2004;23(48):7918. doi: 10.1038/sj.onc.1208081. [DOI] [PubMed] [Google Scholar]
- 43.Musacchio A, Gibson T, Lehto VP, Saraste M. FEBS Lett. 1992;307(1):55. doi: 10.1016/0014-5793(92)80901-r. [DOI] [PubMed] [Google Scholar]
- 44.Smithgall TE, Pharmacol J. Toxicol. Methods. 1995;34(3):125. doi: 10.1016/1056-8719(95)00082-7. [DOI] [PubMed] [Google Scholar]
- 45.Kuriyan J, Cowburn D. Annu. Rev. Biophys. Biomol. Struct. 1997;26:259. doi: 10.1146/annurev.biophys.26.1.259. [DOI] [PubMed] [Google Scholar]
- 46.Dharmasiri K, Smith DL. Anal Chem. 1996;68(14):2340. doi: 10.1021/ac9601526. [DOI] [PubMed] [Google Scholar]
- 47.Mandell JG, Falick AM, Komives EA. Proc. Natl. Acad. Sci. USA. 1998;95(25):14705. doi: 10.1073/pnas.95.25.14705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang O, Forman-Kay JD. Biochemistry. 1995;34(20):6784. doi: 10.1021/bi00020a025. [DOI] [PubMed] [Google Scholar]
- 49.Farrow NA, Zhang O, Forman-Kay JD, Kay LE. Biochemistry. 1995;34(3):868. doi: 10.1021/bi00003a021. [DOI] [PubMed] [Google Scholar]
- 50.Bai Y, Milne JS, Mayne L, Englander SW. Proteins. 1993;17(1):75. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Engen JR, Smithgall TE, Gmeiner WH, Smith DL. J Mol Biol. 1999;287(3):645. doi: 10.1006/jmbi.1999.2619. [DOI] [PubMed] [Google Scholar]
- 52.Engen JR, Gmeiner WH, Smithgall TE, Smith DL. Biochemistry. 1999;38(28):8926. doi: 10.1021/bi982611y. [DOI] [PubMed] [Google Scholar]
- 53.Lecomte JT, Falzone CJ. Nat Struct Biol. 1999;6(7):605. doi: 10.1038/10643. [DOI] [PubMed] [Google Scholar]
- 54.Lee CH, Leung B, Lemmon MA, Zheng J, Cowburn D, Kuriyan J, Saksela K. Embo J. 1995;14(20):5006. doi: 10.1002/j.1460-2075.1995.tb00183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hochrein JM, Lerner EC, Schiavone AP, Smithgall TE, Engen JR. Protein Sci. 2006;15(1):65. doi: 10.1110/ps.051782206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trible RP, Emert-Sedlak L, Wales TE, Ayyavoo V, Engen JR, Smithgall TE. J Mol Biol. 2007;374(1):121. doi: 10.1016/j.jmb.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Henkels CH, Oas TG. J Am Chem Soc. 2006;128(24):7772. doi: 10.1021/ja057279+. [DOI] [PubMed] [Google Scholar]
- 58.Briggs SD, Bryant SS, Jove R, Sanderson SD, Smithgall TE. J Biol Chem. 1995;270(24):14718. doi: 10.1074/jbc.270.24.14718. [DOI] [PubMed] [Google Scholar]
- 59.Gmeiner WH, Xu I, Horita DA, Smithgall TE, Engen JR, Smith DL, Byrd RA. Cell Biochem Biophys. 2001;35(2):115. doi: 10.1385/cbb:35:2:115. [DOI] [PubMed] [Google Scholar]
- 60.Sicheri F, Moarefi I, Kuriyan J. Nature. 1997;385(6617):602. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- 61.Schindler T, Sicheri F, Pico A, Gazit A, Levitzki A, Kuriyan J. Mol Cell. 1999;3(5):639. doi: 10.1016/s1097-2765(00)80357-3. [DOI] [PubMed] [Google Scholar]
- 62.Lerner EC, Trible RP, Schiavone AP, Hochrein JM, Engen JR, Smithgall TE. J Biol Chem. 2005;280(49):40832. doi: 10.1074/jbc.M508782200. [DOI] [PubMed] [Google Scholar]
- 63.Alvarado JJ, Betts L, Moroco JA, Smithgall TE, Yeh JI. J Biol Chem. 2010;285(46):35455. doi: 10.1074/jbc.M110.145102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moarefi I, LaFevre-Bernt M, Sicheri F, Huse M, Lee C-H, Kuriyan J, Miller WT. Nature. 1997 Feb 13;385:650. doi: 10.1038/385650a0. 1997. [DOI] [PubMed] [Google Scholar]
- 65.Superti-Furga G, Courtneidge SA. BioEssays. 1995;17(4):321. doi: 10.1002/bies.950170408. [DOI] [PubMed] [Google Scholar]
- 66.Wales TE, Engen JR. J Mol Biol. 2006;357(5):1592. doi: 10.1016/j.jmb.2006.01.075. [DOI] [PubMed] [Google Scholar]
- 67.Zhang Z, Smith DL. Protein Sci. 1993;2(4):522. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Palacios EH, Weiss A. Oncogene. 2004;23(48):7990. doi: 10.1038/sj.onc.1208074. [DOI] [PubMed] [Google Scholar]
- 69.Hassaine G, Courcoul M, Bessou G, Barthalay Y, Picard C, Olive D, Collette Y, Vigne R, Decroly E. J Biol Chem. 2001;276(20):16885. doi: 10.1074/jbc.M009076200. [DOI] [PubMed] [Google Scholar]
- 70.Douaisi M, Dussart S, Courcoul M, Bessou G, Lerner EC, Decroly E, Vigne R. Biochem Biophys Res Commun. 2005;329(3):917. doi: 10.1016/j.bbrc.2005.02.057. [DOI] [PubMed] [Google Scholar]
- 71.Fickenscher H, Fleckenstein B. Philos Trans R Soc Lond B Biol Sci. 2001;356(1408):545. doi: 10.1098/rstb.2000.0780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weis DD, Kjellen P, Sefton BM, Engen JR. Protein Sci. 2006;15(10):2402. doi: 10.1110/ps.052016406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen S, Brier S, Smithgall TE, Engen JR. Protein Sci. 2007;16(4):572. doi: 10.1110/ps.062631007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schweimer K, Hoffmann S, Bauer F, Friedrich U, Kardinal C, Feller SM, Biesinger B, Sticht H. Biochemistry. 2002;41(16):5120. doi: 10.1021/bi015986j. [DOI] [PubMed] [Google Scholar]
- 75.Mitchell JL, Trible RP, Emert-Sedlak LA, Weis DD, Lerner EC, Applen JJ, Sefton BM, Smithgall TE, Engen JR. J Mol Biol. 2007;366(4):1282. doi: 10.1016/j.jmb.2006.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Trible RP, Emert-Sedlak L, Smithgall TE. J Biol Chem. 2006;281(37):27029. doi: 10.1074/jbc.M601128200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Biesinger B, Tsygankov AY, Fickenscher H, Emmrich F, Fleckenstein B, Bolen JB, Broker BM. J Biol Chem. 1995;270(9):4729. doi: 10.1074/jbc.270.9.4729. [DOI] [PubMed] [Google Scholar]
- 78.Jung JU, Lang SM, Friedrich U, Jun T, Roberts TM, Desrosiers RC, Biesinger B. J. Biol. Chem. 1995;270(35):20660. doi: 10.1074/jbc.270.35.20660. [DOI] [PubMed] [Google Scholar]
- 79.Wiese N, Tsygankov AY, Klauenberg U, Bolen JB, Fleischer B, Broker BM. J Biol Chem. 1996;271(2):847. doi: 10.1074/jbc.271.2.847. [DOI] [PubMed] [Google Scholar]
- 80.Hartley DA, Amdjadi K, Hurley TR, Lund TC, Medveczky PG, Sefton BM. Virology. 2000;276(2):339. doi: 10.1006/viro.2000.0570. [DOI] [PubMed] [Google Scholar]
- 81.Hartley DA, Hurley TR, Hardwick JS, Lund TC, Medveczky PG, Sefton BM. J Biol Chem. 1999;274(29):20056. doi: 10.1074/jbc.274.29.20056. [DOI] [PubMed] [Google Scholar]
- 82.Kjellen P, Amdjadi K, Lund TC, Medveczky PG, Sefton BM. Virology. 2002;297(2):281. doi: 10.1006/viro.2002.1419. [DOI] [PubMed] [Google Scholar]
- 83.Chen S, Dumitrescu TP, Smithgall TE, Engen JR. Biochemistry. 2008;47(21):5795. doi: 10.1021/bi800446b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen S, O'Reilly LP, Smithgall TE, Engen JR. J Mol Biol. 2008;383(2):414. doi: 10.1016/j.jmb.2008.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.de Klein A, van Kessel AG, Grosveld G, Bartram CR, Hagemeijer A, Bootsma D, Spurr NK, Heisterkamp N, Groffen J, Stephenson JR. Nature. 1982;300(5894):765. doi: 10.1038/300765a0. [DOI] [PubMed] [Google Scholar]
- 86.Heisterkamp N, Stephenson JR, Groffen J, Hansen PF, de Klein A, Bartram CR, Grosveld G. Nature. 1983;306(5940):239. doi: 10.1038/306239a0. [DOI] [PubMed] [Google Scholar]
- 87.Dorey K, Engen JR, Kretzschmar J, Wilm M, Neubauer G, Schindler T, Superti-Furga G. Oncogene. 2001;20(56):8075. doi: 10.1038/sj.onc.1205017. [DOI] [PubMed] [Google Scholar]
- 88.Hantschel O, Nagar B, Guettler S, Kretzschmar J, Dorey K, Kuriyan J, Superti-Furga G. Cell. 2003;112(6):845. doi: 10.1016/s0092-8674(03)00191-0. [DOI] [PubMed] [Google Scholar]
- 89.Nagar B, Hantschel O, Seeliger M, Davies JM, Weis WI, Superti-Furga G, Kuriyan J. Mol Cell. 2006;21(6):787. doi: 10.1016/j.molcel.2006.01.035. [DOI] [PubMed] [Google Scholar]
- 90.Nagar B, Hantschel O, Young MA, Scheffzek K, Veach D, Bornmann W, Clarkson B, Superti-Furga G, Kuriyan J. Cell. 2003;112(6):859. doi: 10.1016/s0092-8674(03)00194-6. [DOI] [PubMed] [Google Scholar]
- 91.Pluk H, Dorey K, Superti-Furga G. Cell. 2002;108(2):247. doi: 10.1016/s0092-8674(02)00623-2. [DOI] [PubMed] [Google Scholar]
- 92.Brasher BB, Van Etten RA. J Biol Chem. 2000;275(45):35631. doi: 10.1074/jbc.M005401200. [DOI] [PubMed] [Google Scholar]
- 93.Rickles RJ, Botfield MC, Weng Z, Taylor JA, Green OM, Brugge JS, Zoller MJ. Embo J. 1994;13(23):5598. doi: 10.1002/j.1460-2075.1994.tb06897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Iacob RE, Pene-Dumitrescu T, Zhang J, Gray NS, Smithgall TE, Engen JR. Proc Natl Acad Sci U S A. 2009;106(5):1386. doi: 10.1073/pnas.0811912106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang J, Adrian FJ, Jahnke W, Cowan-Jacob SW, Li AG, Iacob RE, Sim T, Powers J, Dierks C, Sun F, Guo GR, Ding Q, et al. Nature. 2010;463(7280):501. doi: 10.1038/nature08675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Iacob RE, Zhang J, Gray NS, Engen JR. PLoS ONE. 2011;6(1):e15929. doi: 10.1371/journal.pone.0015929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Meyn MA, 3rd, Wilson MB, Abdi FA, Fahey N, Schiavone AP, Wu J, Hochrein JM, Engen JR, Smithgall TE. J Biol Chem. 2006;281(41):30907. doi: 10.1074/jbc.M605902200. [DOI] [PubMed] [Google Scholar]
- 98.Warmuth M, Bergmann M, Priess A, Hauslmann K, Emmerich B, Hallek M. J Biol Chem. 1997;272(52):33260. doi: 10.1074/jbc.272.52.33260. [DOI] [PubMed] [Google Scholar]
- 99.Rosa JJ, Richards FM. J Mol Biol. 1979;133:399. doi: 10.1016/0022-2836(79)90400-5. [DOI] [PubMed] [Google Scholar]
- 100.Englander JJ, Rogero JR, Englander SW. Anal Biochem. 1985;147:234. doi: 10.1016/0003-2697(85)90033-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Horita DA, Baldisseri DM, Zhang W, Altieri AS, Smithgall TE, Gmeiner WH, Byrd RA. J. Mol. Biol. 1998;278:253. doi: 10.1006/jmbi.1998.1690. [DOI] [PubMed] [Google Scholar]
- 102.Schmidt H, Hoffmann S, Tran T, Stoldt M, Stangler T, Wiesehan K, Willbold D. J Mol Biol. 2007;365(5):1517. doi: 10.1016/j.jmb.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 103.Arold S, O'Brien R, Franken P, Strub MP, Hoh F, Dumas C, Ladbury JE. Biochemistry. 1998;37(42):14683. doi: 10.1021/bi980989q. [DOI] [PubMed] [Google Scholar]
- 104.Engen JR. Ph.D. thesis. University of Nebraska-Lincoln; 1999. [Google Scholar]
- 105.Chen S. Ph.D. thesis. Northeastern University; 2008. [Google Scholar]
- 106.Deng Y, Pan H, Smith DL. J Am Chem Soc. 1999;121:1966. [Google Scholar]
- 107.Kim MY, Maier CS, Reed DJ, Deinzer ML. J. Am. Chem. Soc. 2001;123(40):9860. doi: 10.1021/ja010901n. [DOI] [PubMed] [Google Scholar]
- 108.Demmers JA, Haverkamp J, Heck AJ, Koeppe RE, 2nd, Killian JA. Proc. Natl. Acad. Sci. U S A. 2000;97(7):3189. doi: 10.1073/pnas.050444797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Demmers JA, Rijkers DT, Haverkamp J, Killian JA, Heck AJ. J. Am. Chem. Soc. 2002;124(37):11191. doi: 10.1021/ja0125927. [DOI] [PubMed] [Google Scholar]
- 110.Hoerner JK, Xiao H, Dobo A, Kaltashov IA. J. Am. Chem. Soc. 2004;126(24):7709. doi: 10.1021/ja049513m. [DOI] [PubMed] [Google Scholar]
- 111.Jorgensen TJ, Gardsvoll H, Ploug M, Roepstorff P. J Am Chem Soc. 2005;127(8):2785. doi: 10.1021/ja043789c. [DOI] [PubMed] [Google Scholar]
- 112.Rand KD, Zehl M, Jensen ON, Jorgensen TJ. Anal Chem. 2009;81(14):5577. doi: 10.1021/ac9008447. [DOI] [PubMed] [Google Scholar]
- 113.Rand KD, Pringle SD, Morris M, Engen JR, Brown JM. J Am Soc Mass Spectrom. 2011;22(10):1784. doi: 10.1007/s13361-011-0196-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Huang RY, Garai K, Frieden C, Gross ML. Biochemistry. 2011;50(43):9273. doi: 10.1021/bi2010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Landgraf RR, Chalmers MJ, Griffin PR. J Am Soc Mass Spectrom. 2012;23(2):301. doi: 10.1007/s13361-011-0298-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rand KD, Adams CM, Zubarev RA, Jorgensen TJ. J Am Chem Soc. 2008;130(4):1341. doi: 10.1021/ja076448i. [DOI] [PubMed] [Google Scholar]
- 117.Zehl M, Rand KD, Jensen ON, Jorgensen TJ. J Am Chem Soc. 2008;130(51):17453. doi: 10.1021/ja805573h. [DOI] [PubMed] [Google Scholar]
- 118.Cravello L, Lascoux D, Forest E. Rapid Commun. Mass Spectrom. 2003;17(21):2387. doi: 10.1002/rcm.1207. [DOI] [PubMed] [Google Scholar]
- 119.Marcsisin SR, Narute PS, Emert-Sedlak LA, Kloczewiak M, Smithgall TE, Engen JR. J Mol Biol. 2011;410(5):1008. doi: 10.1016/j.jmb.2011.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Poe JA, Smithgall TE. J Mol Biol. 2009;394(2):329. doi: 10.1016/j.jmb.2009.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Schreiner SJ, Schiavone AP, Smithgall TE. J Biol Chem. 2002;277(47):45680. doi: 10.1074/jbc.M204255200. [DOI] [PubMed] [Google Scholar]
- 122.Fang J, Rand KD, Beuning PJ, Engen JR. Int J Mass Spectrom. 2011;302(1–3):19. doi: 10.1016/j.ijms.2010.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Marcsisin SR, Engen JR. J Mol Biol. 2010;402(5):892. doi: 10.1016/j.jmb.2010.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fang J, Engen JR, Beuning PJ. Biochemistry. 2011;50(26):5958. doi: 10.1021/bi200580b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kong XP, Onrust R, O'Donnell M, Kuriyan J. Cell. 1992;69(3):425. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- 126.Oakley AJ, Prosselkov P, Wijffels G, Beck JL, Wilce MC, Dixon NE. Acta Crystallogr D Biol Crystallogr. 2003;59(Pt 7):1192. doi: 10.1107/s0907444903009958. [DOI] [PubMed] [Google Scholar]
- 127.Bauer F, Schweimer K, Meiselbach H, Hoffmann S, Rosch P, Sticht H. Protein Sci. 2005;14(10):2487. doi: 10.1110/ps.051563605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chevelkov V, Faelber K, Diehl A, Heinemann U, Oschkinat H, Reif B. J Biomol NMR. 2005;31(4):295. doi: 10.1007/s10858-005-1718-z. [DOI] [PubMed] [Google Scholar]
- 129.Eck MJ, Atwell SK, Shoelson SE, Harrison SC. Nature. 1994;368(6473):764. doi: 10.1038/368764a0. [DOI] [PubMed] [Google Scholar]