Abstract
Single-chain antibody fragments (scFv) expressed in the cytoplasm of mammalian cells, also called intrabodies, have many applications in functional proteomics. These applications are however limited by the aggregation-prone behaviour of many intrabodies. We show here that two scFv with highly homologous sequences and comparable soluble expression levels in E. coli cytoplasm, have different behaviours in mammalian cells. When over-expressed, one of the scFv aggregates in the cytoplasm whereas the second one is soluble and active. When expressed at low levels, using a retroviral vector, as a fusion with the green fluorescent protein (GFP) the former does not form aggregates and is degraded, resulting in weakly fluorescent cells, whereas the latter is expressed as soluble protein, resulting in strongly fluorescent cells. These data suggest that the GFP signal can be used to evaluate the soluble expression of intrabodies in mammalian cells. When applied to a subset of an E. coli optimised intrabody library, we showed that the population of GFP+ cells contains indeed soluble mammalian intrabodies. Altogether, our data demonstrate that the requirements for soluble intrabody expression are different in E. coli and mammalian cells, and that intrabody libraries can be directly optimised in human cells using a simple GFP-based assay.
Keywords: Cell Line, Cytoplasm, metabolism, Escherichia coli, metabolism, Green Fluorescent Proteins, metabolism, Humans, Recombinant Fusion Proteins, metabolism, Single-Chain Antibodies, isolation & purification, Solubility, Tubulin, immunology
Keywords: aggregation, degradation, folding, GFP , intrabody
Introduction
Intrabodies are antibody-derived molecules ectopically expressed inside the cell (Carlson, 1988; Lo et al., 2008). Because intrabodies have shown their ability to modulate the activity of their targets within the cell, they are frequently used as “protein interference” reagents (Visintin et al., 2004a). This approach is similar to RNA interference but at the protein level (Cao and Heng, 2005), allowing a more direct pathway to isolate chemical drugs (Mazuc et al., 2008).
Most of the described intrabodies are single chain Fv (scFv), constituted by the assembly of the variable heavy (VH) and light (VL) chain domains of an immunoglobulin molecule linked together by a flexible peptide. Because of this non-natural assembly and because of the absence of the other domains present in a full immunoglobulin molecule, the scFv can be considered as a heterologous protein when expressed both in E. coli and in mammalian cell.
Interesting targets for intrabodies are usually located in the cell cytoplasm and in the nucleus, which are unnatural environments for immunoglobulin-derived molecules that are normally secreted from the cell. Because of the reducing conditions that pertain in these cell compartments, the conserved disulphide bridge of the antibody domain is not formed, resulting in a decrease in the stability of the scFv and in many cases in protein aggregation (Auf der Maur et al., 2004; Worn et al., 2000). However, even if structural stability is of prime importance for intrabody expression, other factors like folding and aggregation kinetics of folding intermediates, sensitivity to protease degradation, and molecular chaperones have also been shown to affect intrabody expression (Bach et al., 2001; Biocca et al., 1995; Duenas et al., 1994; Kvam et al., 2010; Martineau and Betton, 1999; Philibert and Martineau, 2004), as it is also the case for many over-expressed heterologous proteins (Hartl and Hayer-Hartl, 2009).
Most intrabodies described so far have been identified using a two-step approach. First, antibody fragments are selected against the target protein using either phage-display in E. coli or double hybrid technology in yeast, then tested, as intrabodies, in an appropriate mammalian cell model. Expression of intrabodies in cells can thus be considered as a heterologous protein expression system, since the scFv are not natural mammalian proteins and because they have been first selected for expression in E. coli or yeast.
It is well known that over-expression of mammalian proteins in E. coli frequently results in the accumulation of inclusion bodies (Baneyx and Mujacic, 2004). The reverse has been less studied but is also true in the case of intrabodies, resulting in the accumulation of aggregated materials in mammalian cells even when these intrabodies are able to fold efficiently in E. coli. This constitutes one of the main challenges for the use of intrabodies at a large scale since studies have shown that only between 1% and 10% of the scFv can be expressed at an adequate level in the cytoplasm of mammalian cells (Auf der Maur et al., 2004; Visintin et al., 2004b).
In a previous work, we reported the construction of a scFv library designed and optimised for intracellular expression in E. coli bacterial cells (Philibert et al., 2007). Two highly homologous anti-tubulin scFv were isolated from this library. We show here that, although these scFv display comparable soluble expression levels and do not aggregate in E. coli cytoplasm, they have different behaviours in mammalian cells. When over-expressed in mammalian cells, one of the scFv forms aggregates in the cytoplasm whereas the second one is fully soluble and active, as shown by the strong staining of the microtubule network. When fused to the green fluorescent protein (GFP) and expressed at low level, the former does not form aggregates and is quickly degraded, resulting in weakly fluorescent cells, whereas the latter is expressed as soluble protein, resulting in strongly fluorescent cells. These data suggest that the GFP fusion approach can be used as a protein-solubility assay in mammalian cells. We applied this assay on a subset of our previously described E. coli optimised intrabody library and demonstrated that the population of GFP+ cells contains intrabodies expressed at high soluble levels in the mammalian cytoplasm.
Altogether, we report a straightforward GFP-based assay for the direct selection of soluble antibody fragments in mammalian cytoplasm. We believe that this approach opens the way to new improved intrabody libraries with optimal expression characteristics in mammalian cells.
Materials and methods
Chemicals and enzymes
Chemicals were purchased from Sigma-Aldrich. Restriction enzymes and cloned Taq polymerase were from Fermentas. Phusion® DNA polymerase was purchased from Finnzymes. Plasmid DNA, PCR and agarose separated DNA were purified using Macherey-Nagel kits.
Plasmids
Plasmid pCMV/myc/cyto, obtained from Invitrogen (#V820-20), allows the expression of scFv genes from the strong CMV promoter in the cytoplasm of the cell.
Plasmid pMSCVhygSN is derived from pMSCVhyg plasmid (Clontech) and is used for retroviral expression of the scFv as a N-terminal fusion with a c-myc and a His6 tag. First, a 207 bp PCR fragment was amplified from pAB1 plasmid (Martineau et al., 1998) using primers pelBbamHI2 (CCGCTGGATccTTATTACTC) and M13uni (AGGGTTTTCCCAGTCACGACGTT). After digestion with BamHI and EcoRI enzymes, the 162 bp fragment was inserted in the BglII and EcoRI sites of plasmid pMSCVhyg. Since pMSCVhyg contains two EcoRI sites, partial digestion was performed in order to insert the fragment in the EcoRI site located 18 bp downstream of the BglII site.
Plasmid pMSCVhygSN-EGFP contains unique SfiI and NotI sites for cloning the scFv genes in frame with the gene encoding the enhanced GFP. It is derived from pMSCVhygSN as follows: EGFP gene was amplified from pEGFP-C1 plasmid (Clontech), which contains a red-shifted variant of wild-type GFP, using primers Not_egfp.for (CCGGCGGCCGCCATGGTGAGC) and Eco52i_egfp.back (TGACGGCCGACTTGTACAGCTCGTCCAT). The PCR fragment was digested with Eco52i enzyme and cloned into the unique NotI site of pMSCVhygSN plasmid.
pCMV-SN-EGFP is derived from pcDNA3.1 plasmid (Invitrogen). A 898 bp PCR fragment containing the SfiI and the NotI sites, the EGFP gene and the tags was amplified from pMSCVhygSN-EGFP using primers pMSCV_HindIII. for (TTAGAagCTTATTACTCGCGGCC) and pMSCV_XbaI.rev (ACCCtcTAGAattCTTATTAATGGTGATG). The fragment was digested with HindIII and XbaI enzymes and inserted in the same sites of pcDNA3.1.
When a scFv gene is cloned between the SfiI and the NotI sites of pMSCVhygSN-EGFP or pCMV-SN-EGFP, a protein fusion consisting of the scFv, the EGFP, a c-myc tag recognized by the 9E10 monoclonal antibody (Munro and Pelham, 1986) and a His6 tag is produced.
Expression in E. coli
ScFv cloned in pET23NN plasmid (Philibert and Martineau, 2004) were transfected in BL21(DE3). A single colony was grown in 1 ml of auto-inducible ZYP-5052 medium (Studier, 2005) containing 100 μg/ml of ampicillin at 37°C for 2 h, then incubated for 24 h at 24°C with vigorous shaking. Cells were pelleted by centrifugation, freeze/thawed, resuspended in 250 μl of lysis buffer (Tris 10 mM pH 8.0, EDTA 5 mM, NaCl 30 mM, Hen Lysozyme 0.1 mg/ml), incubated 1 h on ice, then sonicated to complete lysis. Five μl of NaCl 5 M (130 mM final) was added and the soluble fraction was recovered by centrifugation at 16,000 g. The pellet, containing the insoluble material, was washed once with 1 ml of lysis buffer, pelleted by centrifugation, then resuspended and boiled in SDS-PAGE sample buffer (0.01% bromophenol blue, 5% β-mercaptoethanol, 2% SDS, 10% glycerol, 62.5 mM Tris-HCl pH 6.8).
Cell culture
293T, HeLa and MCF7 human cell lines (ATCC) were cultured at 37°C in a 5% CO2 humidified atmosphere in Dulbecco’s Modified Eagle Medium (DMEM) containing Glutamax I™, 10% (v/v) heat-inactivated fetal calf serum (FCS) and an antibiotic/antimycotic solution (Life Technologies). Transient transfection was carried out with the jetPEI transfection reagent (Polyplus Transfection™) according to the manufacturer’s instructions.
Retrovirus production and cell transduction
Stable cell lines expressing the scFv library were obtained by retroviral gene transduction using scFv genes cloned in pMSCVhygSN-EGFP plasmid. Retroviral particules were produced in 293T by transient co-transfection of gag/pol, env-VSV-G, and the indicated viral pMSCV constructs. Briefly, 2.106 cells were seeded on a 10 cm dish the day before transfection. The pMSCV-derived vector (3 μg), the packaging plasmid (1 μg), the amphotropic envelope plasmid (1 μg) and 15 μl of jetPEI transfection reagent (Polyplus Transfection™) were diluted in 500 μl of 150 mM NaCl and left at room temperature for 30 min, then added drop-wise to the cell culture medium. After 5 h, the medium was replaced with 11 ml of fresh medium. The supernatant containing the virus was collected 72 h later. Transduction of 106 MCF7 and Hela cells was performed in 10 cm dishes with 3 ml of virus containing supernatant in the presence of 8 μg/ml polybrene (Sigma-Aldrich) for 12 h. Virus infected MCF7 and Hela cells were selected with 100 and 200 μg/ml of hygromycin, respectively.
Flow cytometry analysis and cell sorting
Single-cell suspension of cell lines expressing scFv-GFP fusions were analysed by collecting at least 10,000 events/sample using an EPICS XL® cytometer (Beckman Coulter).
Hela cells stably transduced with the retroviral scFv-GFP library were sorted for GFP expression with a FACSAria® cell sorter (Beckton Dickinson). One million of cells were resuspended in 1 ml of DMEM with 10% FCS containing Propidium Iodide (5 μg/ml) in order to exclude dying cells. GFP positive cells were sorted and immediately put back in culture.
When required (Figure 3D), proteasome inhibitor MG-132 (Sigma-Aldrich) was added to the cells at a concentration of 10 μM 24 h before flow cytometry analysis.
Fig. 3.
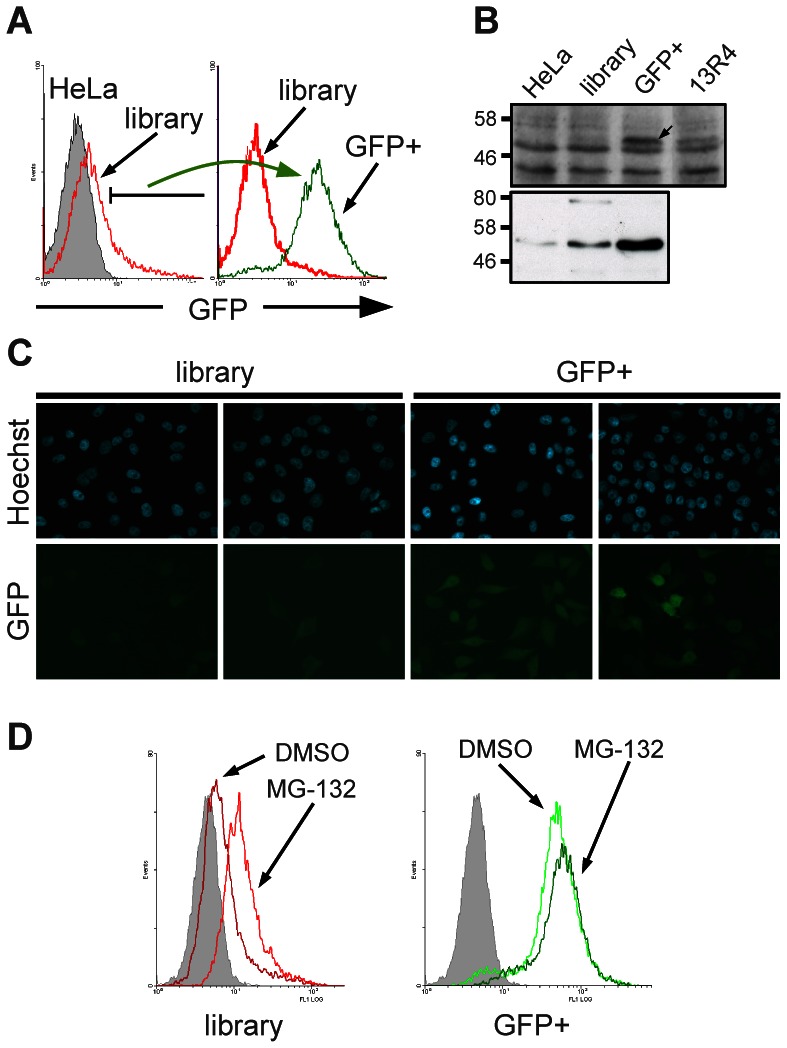
Selection for soluble expression in mammalian cells. A) The GFP fluorescence of the cell populations was analysed by FACS. The library was generated from HeLa cells infected with a subset of an intrabody library described in (Philibert et al., 2007). GFP+ population represents the highly fluorescent population sorted from the library (2% of the library population). B) Expression of the scFv-GFP fusions from the cell populations shown in panel A were detected by Western blotting. Top: total cell extracts revealed using a rabbit polyclonal anti-GFP serum. The arrow shows the position of the scFv-GFP fusion. Unspecific bands are used as a loading control. 13R4 is the evolved scFv used to construct the library and is used as a control of a well expressed/behaved intrabody (Sibler et al., 2003). Bottom: scFv-GFP fusions were captured from the soluble cell extracts by their C-terminal polyHis tag and detected using the anti-c-myc 9E10 monoclonal antibody. C) The expression of the unsorted (library) and the GFP+ cell populations from panel A was analysed by fluorescence microscopy after cell fixation. In the bottom panels (GFP), the analysed signal is the fluorescence of the GFP protein. D) Effect of the proteasome inhibitor MG-132. The cell populations analysed are indicated below each graph (library, GFP+) and are the same than in panel A.
Cell extracts, scFv precipitation and Western Blot analysis
For the recovery of the whole cell protein content, MCF7 and Hela transfected cells were harvested by trypsinization and washed twice in PBS. The pelleted cells were lysed in SDS-PAGE sample buffer.
For scFv precipitation, confluent cell cultures were lysed in a buffer containing 25 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% (w/v) Nonidet-P40, 1 mM Na3VO4, 100 mM NaF, 1 mM PMSF and a protease inhibitor cocktail (Complete EDTA-free, Roche Applied Science). His-tagged proteins were captured from 3 mg of clarified lysates with 20 μl of nickel-loaded beads, either agarose-based (Sigma-Aldrich) or magnetic (Ademtech, France). After incubation for 1 hour at 4°C under constant rotation, beads were washed twice in IMAC buffer (50 mM NaPO4, 500 mM NaCl, pH 7.5) and the protein were eluted in 20 μl of IMAC buffer containing 300 mM imidazol.
Detergent-soluble and -insoluble fractions were prepared as previously described (Kvam et al., 2010). Cells were harvested by trypsinization, counted, then 3.106 cells were lysed in 500 μL of RIPA buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% Nonidet-P40, 0.25% sodium deoxycholate, 0.1% SDS), supplemented with a Complete Protease Inhibitor Cocktail (Roche). Soluble and insoluble materials were separated by centrifugation at 16,000 g for 15 min at 4°C. Pellets were resuspended by sonication in the same volume of RIPA buffer (500 μL).
Cell extracts and nickel-precipitates were analysed by reducing SDS-PAGE (10% w/v polyacrylamide). Proteins were revealed by Western blotting using either a rabbit polyclonal serum against GFP (Santa Cruz, sc-8334) or the 9E10 anti-c-myc monoclonal antibody (Munro and Pelham, 1986), followed by a horseradish peroxidase–conjugated anti-rabbit or anti-mouse secondary antibody (1:15,000; Sigma-Aldrich) in PBS, 0.1% Tween, 5% milk. The signal was revealed using enhanced chemiluminescence and detected with either a Hyperfilm (GE Healthcare) or a camera (G:BOX Chemi, Syngene, Cambridge, UK).
Fluorescence microscopy
Cells were seeded on 12 mm glass coverslips in 6-well plates. Transient transfections were carried out 24 h later. After 2 days, cells were fixed in cold absolute methanol for 10 min at −20°C, then gradually rehydrated with PBS. scFv was revealed using the 9E10 anti-c-myc antibody and an FITC-conjugated anti-mouse IgG antibody diluted 1:12,500 (Sigma-Aldrich). Alternatively, for GFP-tagged scFv, cells were directly visualised after fixation. DNA was visualised using cell-permeant Hoechst 33342 dye at 5 μg/ml. The slides were washed with PBS, mounted in Mowiol and examined with a fluorescence microscope (Leica or Zeiss). To visualise scFv-GFP expression in live cells (Figure 1D), coverslips were washed in PBS, laid on a drop of PBS on a glass slide and rapidly observed to minimize cell death.
Fig. 1.
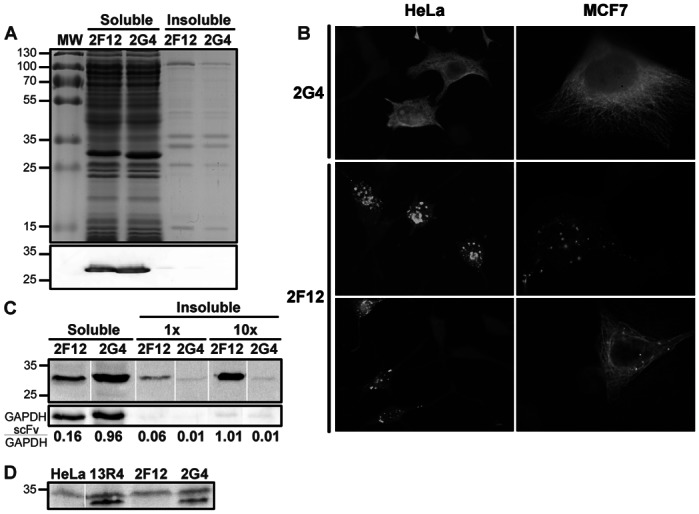
Cytoplasmic expression of c-myc tagged 2F12 and 2G4 intrabodies. A) Expression in E. coli cytoplasm. Top: Coomassie blue stained gel. Bottom: Western blot analysis. 5×107 bacteria/lane. B) Expression of scFv in mammalian cytoplasm. HeLa and MCF7 cells were transfected with c-myc tagged scFv constructs in plasmid pCMV/myc/cyto. After fixation, scFv expression was revealed by immunofluorescence microscopy using 9E10 anti-c-myc monoclonal antibody. C) Soluble and insoluble fractions of transiently-transfected HeLa cells. Transfection efficiency was 80%. Extracts from 150,000 cells were analysed, except for the two lanes labelled 10x that contain the insoluble fractions from 1,500,000 cells. Bands were quantified and the ratio to the GAPDH band is given below the figure. All lanes are from the same gel but were reordered to help the reader. D) Total cell extracts of HeLa cells expressing the scFv cloned in a retroviral vector. In all cases, the scFv was detected by Western blot using the 9E10 anti-c-myc monoclonal antibody.
Results
An E. coli optimised intrabody aggregates in mammalian cells
We recently described several scFv selected for high expression levels in E. coli cytoplasm. Among them, two anti-tubulin scFv, called 2G4 and 2F12, have closely related sequences since their VH domains are identical and their VL domains differ only by the 9 amino acids of the CDR3 loop (Philibert et al., 2007). When expressed in E. coli both scFv accumulated as soluble protein in the cytoplasm and no aggregated material was detected (Figure 1A). To further characterize their solubility in E. coli, we fused the two scFv to the GFP folding reporter protein (Waldo et al., 1999) and measured the fluorescence of induced liquid cultures. At 24°C, both cultures were equally fluorescence, while at 37°C, cells expressing 2F12-GFP fusions were two times more fluorescent than those expressing 2G4-GFP (Supplementary Fig. S1). This demonstrated that 2F12 was at least as soluble as 2G4 when expressed without the GFP tag in E. coli cytoplasm. In addition, we prepared soluble and insoluble cell extracts of the bacteria expressing the two scFv-GFP fusions. At the two temperatures the ratio of soluble to insoluble scFv-GFP protein was comparable for the two scFv. At 24°C, about 80% of the fusions were expressed as soluble protein, showing that the GFP moiety did not strongly decrease the solubility of the two scFv in E. coli.
To evaluate their properties as intrabodies in mammalian cells we expressed both scFv using transient transfection in two frequently used human cell lines, MCF7 and HeLa. When over-expressed, 2G4 was present at high soluble levels whereas 2F12 was mainly found in aggregates in both cell lines (Figure 1B). In addition, 2G4 was active within the cell since it co-localised with its target, as shown by the strong staining of the microtubule network in methanol-fixed cells. The absence of microtubule staining with the 2F12 intrabody is not due to a low affinity of this scFv for this protein network since the scFv, purified from E. coli cytoplasm, is at least as efficient as 2G4 to reveal microtubules by immunofluorescence microscopy of fixed cells (Philibert et al., 2007; Desplancq et al., 2011). It is worth noting that all HeLa cells expressing 2F12 contained aggregated materials whereas some MCF7 cells contained soluble and active 2F12 (Figure 1B, bottom-right), presumably because of a lower expression level. To quantify the amount of soluble and insoluble materials, we prepared detergent-soluble and -insoluble fractions of HeLa cells expressing the two scFv (Figure 1C). No aggregated material was detected by Western blot in the case of 2G4 even when 1.5 millions of cells were analysed. On the opposite, a clear band of aggregated protein was visible in the case of 2F12 and represented between 25% and 40% of the total protein. In addition, the total 2F12 protein (soluble and insoluble fractions) represented only 25% of the total 2G4 protein. This showed that at least 75% of the 2F12 protein was degraded by the cell proteases.
These different behaviours are neither due to the target antigen since it is common to the two scFv, nor to an aggregation-prone nature of the 2F12 since both scFv fold efficiently under the reducing conditions that pertain in E. coli cytoplasm (Figure 1A). These results thus suggest that 2F12 aggregation in eukaryotic cells is due to an improper adaptation of the protein sequence to the mammalian expression and folding machinery.
Using the GFP as a reporter of soluble protein expression in mammalian cells
In an attempt to follow scFv folding in the mammalian cell cytoplasm, we chose to use the GFP from jellyfish as a folding reporter, as previously described in E. coli (Waldo et al., 1999).
The two scFv 2F12 and 2G4 were fused to the EGFP and introduced by transient transfection in HeLa cells. Cells were analysed by FACS (Figure 2A), Western blot (Figure 2A) and fluorescence microscopy (Supplementary Fig. S2). As in the case of the non-fused scFv, 2G4-EGFP was expressed at a much higher level than 2F12-EGFP, showing again that a high proportion (70%) of the 2F12-EGFP fusion was degraded by the cell (Figure 2A). In addition, the proportion of soluble protein was lower than when the scFv were expressed as non fusion proteins, decreasing from 67% to less than 10% in the case of 2F12, and from 100% to 55% in the case of 2G4. Despite this higher proportion of aggregated protein, observation of the cells by fluorescence microscopy only showed small aggregates with 2F12 and no aggregates with 2G4 (Supplementary Fig. S2). This may be due to the size of the aggregates which are smaller than in absence of the GFP moiety (Figure 1B) and thus more difficult to visualise using optical microscopy. To rule out that the non-detection was due to non-fluorescent aggregates, we also observed the same slides by immunofluorescence using the 9E10 antibody as probe. The results were almost identical to those obtained with the GFP signal but with a higher sensitivity. In addition, 2F12-EGFP was detectable in only few cells whereas 2G4-EGFP was present in almost all the cells, in good agreement with the high proportion of degraded 2F12-EGFP protein estimated from the Western blot (Supplementary Fig. S2).
Fig. 2.
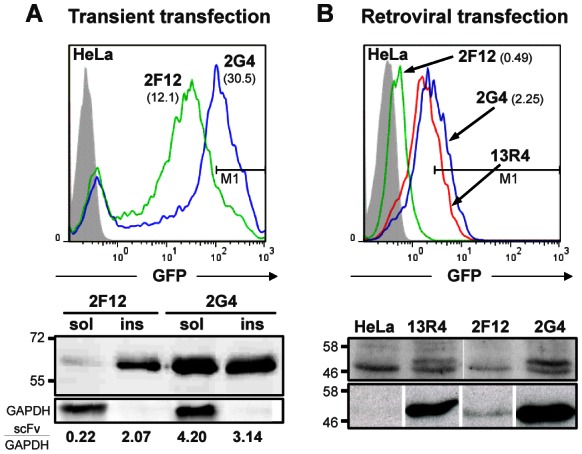
Cytoplasmic expression and whole-cell fluorescence of EGFP tagged 2F12 and 2G4 intrabodies. A) HeLa cells were analysed 24h after transient transfection with plasmid pCMV-SN-EGFP containing the indicated scFv. Top: FACS analysis using the GFP fluorescence signal. The geometric means of the GFP+ cells are 12.1 for 2F12 and 30.5 for 2G4. The region M1 identified by the horizontal line contains 40% of the 2G4 and 10% of the 2F12 population. Bottom: Western blot of detergent-soluble and -insoluble fractions of the same cell populations. The fractions from 150,000 cells were analysed in each lane. The blot was revealed and quantified as in Fig. 1. B) HeLa cells were infected with retrovirus containing 2F12-GFP, 13R4-GFP and 2G4-GFP fusions and the cell populations analysed by: Top: FACS using the GFP fluorescence signal. The geometric means of the GFP+ cell population are, respectively, 0.49 and 2.25 for 2F12 and 2G4. The region M1 contains 40% of the 2G4 and 0.1% of the 2F12 population. Bottom: Western blotting of; Top-Panel: Total cell extracts revealed using a rabbit polyclonal anti-GFP serum; Bottom-Panel: scFv-GFP fusions were captured from soluble cell extracts by their C-terminal polyHis tag and detected using the 9E10 anti-c-myc monoclonal antibody. In all cases, lanes are from the same gel but were reordered to help the reader.
Despite this large difference in total and soluble expression levels, the FACS signal was comparable between the two cell lines, 2G4-EGFP expressing cells being only 2.5-fold more fluorescent than those expressing 2F12-EGFP (Figure 2A). In addition, transient transfection resulted in a wide and asymmetric distribution of the GFP signal due to the variable number of plasmids taken up by the cells (Tseng et al., 1997). Furthermore, transient transfection could not be used for the selection of correctly folded proteins from a library, since all the transfected cells would contain several hundred copies of plasmids, each expressing a different scFv. This would result in a weak coupling between genotype and phenotype and in an inefficient selection of mutants. We thus tested the possibility to select for protein folding using a retroviral expression system in which only few copies of the scFv gene are present in each cell.
We first characterized the fluorescence of HeLa and MCF7 cells producing the anti-tubulin 2F12-EGFP and 2G4-EGFP fusions, cloned in a retroviral vector. The original anti-β-galactosidase scFv13R4, used to construct the library, was used as a positive control (13R4-EGFP) (Martineau et al., 1998). None of the scFv aggregated in these conditions, presumably because misfolded proteins were quickly degraded by the cell proteases (Supplementary Fig. S3).
Western blot analysis of total and soluble cell extracts showed that scFv 13R4 and 2G4, fused (Figure 2B) or not (Figure 1D) with the EGFP, were both expressed at high levels in the cell cytoplasm. On the contrary, scFv 2F12, fused (Figure 2B) or not (Figure 1D), was expressed at a very low level since it was not detected in the total cell extracts but still weakly revealed after capture from the soluble extracts (Figure 2B). When analysed by FACS, the GFP signal of the three cell lines correlated with the expression levels of the scFv since 2G4- and 13R4-transfected cells gave a very strong GFP signal whereas the fluorescence of the 2F12-transfected cells was only slightly higher than that of the untransfected HeLa cells. In addition, because of the low fluorescence of the 2F12-EGFP expressing cell line, the fluorescence of the cells expressing 2G4-EGFP was 5-fold higher than those expressing 2F12-EGFP. This is a 2-fold improvement over transient transfection. Furthermore, the FACS signal was more symmetric than in the case of transient transfection, and this allowed to almost perfectly separate 40% of the 2G4-EGFP population from the 2F12-EGFP one. This is shown in Figure 2 by the analysis of the cells present in the region delimited by the horizontal line marked M1. This region contained 40% of the 2G4-EGFP expressing cells in both cases, but 10% and 0.1% of the 2F12-EGFP expressing cell population obtained by transient and retroviral transfection respectively. This demonstrated that the use of the retroviral vector resulted in a 100-fold improvement in the sorting efficiency. Comparable results were obtained in MCF7 cells (data not shown). These data demonstrated that in these three cases, the GFP signal is in good correlation with the soluble cytoplasmic expression levels of the scFv in two mammalian cell lines.
Optimisation of an intrabody library for soluble mammalian cell expression
In order to transpose these preliminary data to a larger number of scFv fragments, we used a subset of our E. coli optimised scFv library (Philibert et al., 2007). This sub-library of about 106 clones was cloned in the retroviral vector in order to express scFv as N-terminal fusions with the EGFP. The new library was transduced in HeLa cells and the mixed population of transfectants was analysed by FACS using the fluorescence of the GFP as marker. As illustrated in Figure 3A, the library showed a slight but clear shift compared to the non infected cell population, showing that most of the cells expressed a soluble scFv, albeit at different and sometimes low levels.
To determine if the fluorescence of the cells correlated with the scFv expression level, we sorted by FACS the cells that presented a high GFP signal. This population was called GFP+ and represented 2% of the library (Figure 3A). The library and the GFP+ populations were analysed in parallel using fluorescence microscopy (Figure 3C). Our data show that more than 50% of the cells belonging to the GFP+ population displayed detectable fluorescence while about 15% of the non-sorted library was detectable in the same experimental conditions. In addition, the average signal was higher for the GFP+ population than for the total library.
Next, the GFP+ population was analysed by Western blot, performed on total and on nickel-captured soluble cell extracts (Figure 3B). Our results show that the GFP activity correlates with the soluble expression levels of the scFv-EGFP fusions. Interestingly, the average expression level of the GFP+ population was even higher than that of the 13R4-EGFP which has been shown to be active and expressed at high levels in the mammalian cytoplasm (Sibler et al., 2003).
It is worth noting that no aggregates were detectable by fluorescence microscopy, presumably as a consequence of the low expression levels of the scFv that allowed the cell to cope with misfolded protein without overloading of the degradation machinery (Betton et al., 1998). Indeed, after treatment with a proteasome inhibitor, the overall fluorescent signal of the whole library population increased, showing that the low signal was mainly due to the degradation of poorly folded scFv by the proteasome machinery (Figure 3D). In the case of the GFP+ population, the fluorescence did not increase after treatment showing that, even in absence of the proteasome inhibitor, most, if not all, of the scFv expressed in this population already escaped degradation because of their correct folding within the mammalian cytoplasm.
To rule out the possibility that the high expression levels of the GFP+ population were due to the insertion site, we measured the mRNA levels of the two populations by qRT-PCR using primers located in the VL framework region, which is identical for all the clones of the library. Our data showed that the whole library and the GFP+ population display similar levels of scFv transcription (data not shown). Finally, the scFv genes from the GFP+ population were amplified by PCR, the VH and VL sequences shuffled together, and the resulting population of shuffled scFv was re-cloned in the same expression system. About 85% of the clones from this new library showed a high GFP signal comparable to that of the initial GFP+ population, demonstrating that we indeed selected for optimised VH and VL sequences and not for the best insertion site or for preferred VH-VL pairs (Figure 4).
Fig. 4.
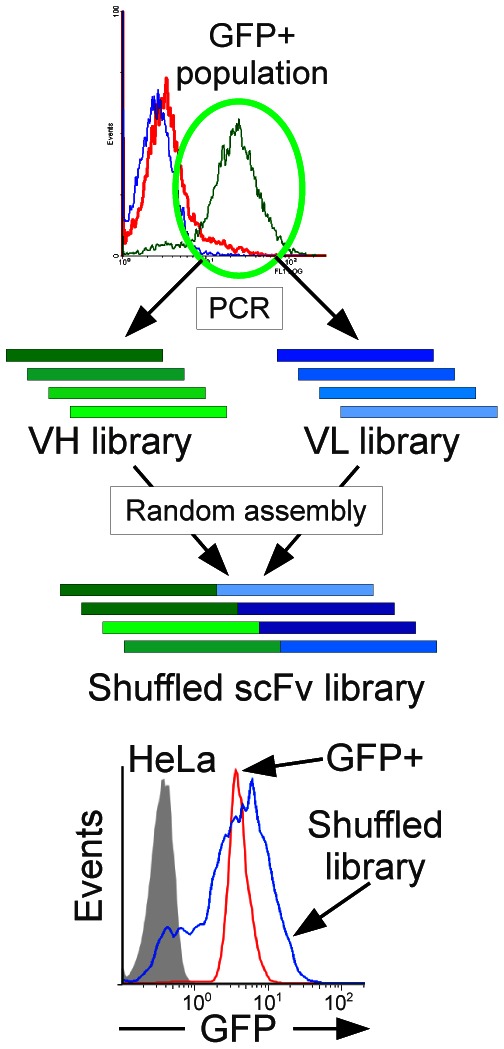
Chain shuffling of the GFP+ population. The VH and VL genes from the GFP+ population of Fig. 3A were amplified by PCR, then assembled at random to generate a new shuffled library. This “Shuffled library” was analysed by FACS and compared to the untransfected HeLa cells and the GFP+ population.
Altogether, our data demonstrate that, in our system, soluble expression levels in mammalian cells correlate with GFP activity and that this approach can therefore be used to select for proteins with improved folding properties and lower degradation sensitivity.
Discussion
Our results show that a scFv particularly well expressed in E. coli cytoplasm is mainly found as insoluble material when transiently expressed at high levels in the mammalian cytoplasm (Figure 1). This mammalian aggregation behaviour is due to few residues of its sequence since another scFv with 96.6% identity (258/267 amino acids) is fully soluble and active in the cytoplasm of mammalian cells and does not form any aggregated material. These different behaviours are not due to the nature of the target since both scFv are directed against the same antigen. These data demonstrate that subtle differences in proteins may influence the output of the in vivo folding process and that this depends strongly on the cellular environment.
To measure the level of soluble scFv expression in mammalian cytoplasm, we developed a detection system that uses the GFP as a folding reporter, as already described in E. coli (Waldo et al., 1999). In E. coli, the whole-cell fluorescence of the bacteria expressing the GFP fusion is proportional to the amount of non-aggregated passenger protein expressed without the GFP. Since we only tested two proteins, it is not possible to validate or invalidate such a correlation in mammalian cells. However, the large difference of solubility of the two non-fused proteins (Figure 1C) only resulted in a small 2.5-fold difference in the FACS signal that did not allow a clear separation between the two populations (Figure 2A). In addition, using transient transfection is not a suitable approach in mammalian cells. Indeed, when transfected with a plasmid library, mammalian cells contain several thousand different copies of the mutated gene, making the selection difficult, if not impossible, since several different scFv are expressed by a single cell.
We thus used a retroviral vector to express only few different scFv-GFP fusion sequences in each cell (1-3 copies/cell, data not shown). The low copy number of the scFv gene resulted in a low expression level of the scFv protein compared to the plasmid-based transfection system. In the case of the scFv 2F12, which aggregated when over-expressed, this resulted in a complete degradation of the protein instead of an aggregation, both for the non- and the EGFP-fused proteins. Since in the case of the 2F12-EGFP almost no protein was present in the cell, the resulting fluorescence signal was very low, resulting in a higher difference in the FACS signal between 2F12- and 2G4-EGFP expressing cells (Figure 2). This difference was high enough to sort the most fluorescent cells, allowing to isolate clones expressing soluble scFv-EGFP fusions from an E. coli optimised intrabody library (Figure 3). An alternative strategy to remove the background could have been to use the new and improved GFP reporters that allow tunable sensitivity and extended dynamic range (Cabantous et al., 2008).
Our results fit well with a folding process as depicted in Figure 5. It is indeed well known that, in many cases, inclusion bodies are formed from aggregation-prone folding intermediates (Hartl and Hayer-Hartl, 2009; Roodveldt et al., 2005), as we have shown for the scFv 13R4 expressed in the cytoplasm of E. coli (Martineau and Betton, 1999). Since proteases have also a strong preference for unfolded or partially unfolded proteins it has been proposed that in vivo aggregation and degradation may proceed from identical or closely related folding intermediates (Betton et al., 1998; Philibert and Martineau, 2004). This is also underlined by the strong link between the molecular chaperones and the degradation machinery (Hartl and Hayer-Hartl, 2009). Thus, selecting for low degradation at low levels of expression, as we did, resulted in the selection of proteins that follow a folding trajectory without aggregation-prone intermediates and that did not aggregate within the cell.
Fig. 5.
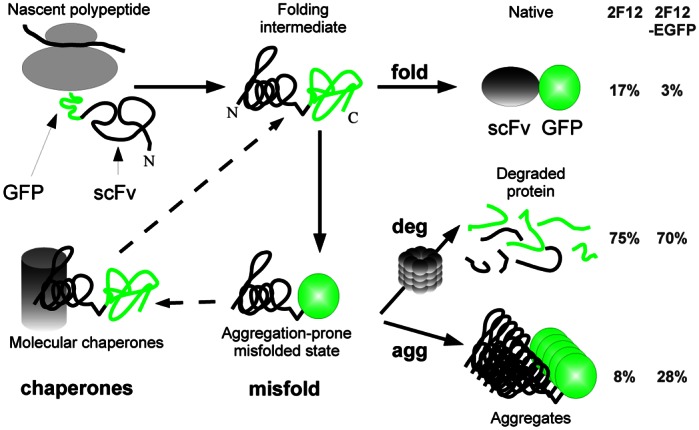
Schematic view of the in vivo folding fate of scFv-GFP fusions. In this scheme, the fusion protein may follow folding (fold), aggregation (agg) and degradation (deg) pathways. The kinetic competition between the pathways are supposed to be affected by the cellular environment, represented here by molecular chaperones (chaperones). Degradation and aggregation are though to proceed from the same or a closely-related misfolded intermediate (misfold). Both native protein and aggregates are fluorescent. The relative magnitudes of the fluxes, estimated from the Western blots in Fig. 1C & 2A, are given at the right of the figure, both for the untagged and the EGFP-tagged aggregation-prone scFv2F12 expressed at high levels using transient transfection.
The magnitudes of the flux in Figure 5 can be estimated from the Western blot quantifications of the detergent-soluble and insoluble fractions of the transiently transfected cells. If we use the total 2G4 expression level as the 100%, we can estimate the degraded, soluble and insoluble fractions of the 2F12 scFv to 75%, 17% and 8%. The results are comparable in the case of 2F12-EGFP fusion, but with an inverse proportion of soluble and insoluble protein (70%, 3%, and 28%, respectively). This seems to be different from E. coli where the fusion with the GFP more strongly decreases the passenger protein solubility (Pédelacq et al., 2006). When expressed at a low level using the retroviral vector, close to 100% of the 2F12 protein, fused or not to the EGFP, was degraded by the cell, contrary to the 2G4 which was expressed and soluble at a level comparable to the original optimised 13R4 scFv. The degradation of fusion proteins, when one of the two partners is badly folded, has been already described and exploited to construct a system to reversibly tune protein expression in living mammalian cells (Banaszynski et al., 2006).
The low stability of reduced scFv has been the most frequent explanation for intrabody aggregation (Auf der Maur et al., 2004; Worn et al., 2000). Intrinsic stability of reduced scFv is of premium importance for intrabody soluble expression but the different folding behaviours of the anti-tubulin intrabody 2F12 in E. coli and in mammalian cells argues in favour, at least in the present case, of a predominant role of the cell folding machinery. Several differences between E. coli and mammalian cells may explain these differences in the in vivo folding properties of the scFv 2F12 and other proteins. It is indeed well known that chaperone systems have a common organization but differences in their mechanisms between E. coli and mammalian cells (Esser et al., 2004; Hartl and Hayer-Hartl, 2009; Herman and D’Ari, 1998; Levy et al., 2007). It has been proposed that the folding machineries have evolved differently because of the five-times faster translation rate in E. coli than in mammalian cells and because of the larger size and the multi-domain nature of most mammalian proteins (Hartl and Hayer-Hartl, 2009; Netzer and Hartl, 1997). An important but still debated (Tsai et al., 1999; Nicola et al., 1999) consequence is that the folding process is mainly post-translational in E. coli and co-translational in mammalian cells. This has been shown to have strong consequences on the folding efficiency of GFP fusions in bacteria and in eukaryotic cells (Chang et al., 2005). Indeed, by comparing the folding efficiency in E. coli and in yeast of several fusions, the authors showed that the efficiency of the co-translational folding of multi-domain proteins in eukaryotes avoids the interference during folding between the two domains. If the passenger protein aggregates, the GFP moiety may however fold into its active conformation, resulting in fluorescent aggregates. This not only true in our system but also when the GFP is fused to the highly aggregation-prone polyQ tract from the human Huntingtin protein (Suhr et al., 2001). In addition, independent folding of the two partners may allow the GFP to interact with cellular chaperones and escape aggregation as depicted on the left of Figure 5. This is particularly relevant for GFP since it has been proposed that interactions with chaperones may explain the different folding trajectories in E. coli and in mammalian cells, resulting in a better expression of E. coli optimised GFP variants in the bacterium and of the wild-type GFP in mammalian cells (Sacchetti et al., 2001). Further experiments will be needed to estimate the relative importance of these possible mechanisms in the in vivo folding process of the scFv-GFP fusions. Furthermore, the importance of other factors, different in the two cellular environments and that may influence the protein output of the transcription/translation machinery, such as the translation rate, the differences in pH (Schuhmann et al., 1997; Zilberstein et al., 1984) or the nature of the molecular crowding of the cytoplasm (Minton, 1992), will also have to be evaluated.
Our results advocate for a direct optimisation of proteins in the cellular system in which they are supposed to be expressed. Intrabodies are often expressed in mammalian cells with the purpose of inducing a phenotypic knock-out at the protein level (Visintin et al., 2004a). Even if approaches based on double-hybrid technology in yeast may have a greater chance to produce mammalian-efficient intrabodies (Visintin et al., 2004b), optimisation in mammalian cells may be desirable to obtain the best possible sequences. The mammalian selection system developed here could be adapted to make large libraries tailored for intracellular expression in the mammalian cytoplasm. As we did in E. coli (Philibert et al., 2007), we could first eliminate in mammalian cells the badly expressed VH and VL before assembly of the final library. Indeed, after selection in mammalian cells, the shuffling of the selected VH and VL resulted in highly fluorescent clones (Figure 4), demonstrating that we did not select for specific pairs of VH and VL but for robust folding chains. This is of premium importance for the construction of large and diverse libraries since this shuffling step re-introduces diversity and partially compensates for the loss of variability associated with the solubility selection. Further experiments will be required to show that it is indeed possible to make a functional library using this tool. However, small scale sequencing of the GFP+ pool showed that some diversity was maintained since 72% of the scFv sequences were unique. Among the 25 sequenced clones, 12 VH and 17 VL were unique, allowing to potentially produce 204 (12×17) different scFv using the shuffling approach presented in Figure 4. Extrapolated to a whole library, this shows that the final recombination of the VH and VL pools should indeed almost compensate for the diversity loss due to the solubility selection. More generally, the GFP-based system presented here could be used with any protein and might help to study and characterise protein folding in mammalian cells.
Supplementary Material
Acknowledgments
Authors are grateful to the Montpellier Rio Imaging (MRI) cytometry facilities, and particularly to the technical assistance of Christophe Duperray.
Funding
This work was supported by grants from the Institut National de la Santé et de la Recherche Médicale and the University of Montpellier 1.
References
- Auf der Maur A, Tissot K, Barberis A. Methods. 2004;34:215–224. doi: 10.1016/j.ymeth.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Bach H, Mazor Y, Shaky S, Shoham-Lev A, Berdichevsky Y, Gutnick DL, Benhar I. J Mol Biol. 2001;312:79–93. doi: 10.1006/jmbi.2001.4914. [DOI] [PubMed] [Google Scholar]
- Banaszynski LA, Chen L, Maynard-Smith LA, Ooi AGL, Wandless TJ. Cell. 2006;126:995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baneyx F, Mujacic M. Nat Biotechnol. 2004;22:1399–1408. doi: 10.1038/nbt1029. [DOI] [PubMed] [Google Scholar]
- Betton JM, Sassoon N, Hofnung M, Laurent M. J Biol Chem. 1998;273:8897–8902. doi: 10.1074/jbc.273.15.8897. [DOI] [PubMed] [Google Scholar]
- Biocca S, Ruberti F, Tafani M, Pierandrei-Amaldi P, Cattaneo A. Biotechnology (NY) 1995;13:1110–1115. doi: 10.1038/nbt1095-1110. [DOI] [PubMed] [Google Scholar]
- Cabantous S, Rogers Y, Terwilliger TC, Waldo GS. PLoS ONE. 2008;3:e2387. doi: 10.1371/journal.pone.0002387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao T, Heng BC. Ann Clin Lab Sci. 2005;35:227–229. [PubMed] [Google Scholar]
- Carlson JR. Mol Cell Biol. 1988;8:2638–2646. doi: 10.1128/mcb.8.6.2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H, Kaiser CM, Hartl FU, Barral JM. J Mol Biol. 2005;353:397–409. doi: 10.1016/j.jmb.2005.08.052. [DOI] [PubMed] [Google Scholar]
- Desplancq D, Rinaldi A, Stoessel A, Sibler A, Busso D, Oulad-Abdelghani M, Van Regenmortel MH, Weiss E. J Immunol Methods. 2011;369:42–50. doi: 10.1016/j.jim.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Duenas M, Vazquez J, Ayala M, Soderlind E, Ohlin M, Perez L, Borrebaeck CA, Gavilondo JV. BioTechniques. 1994;16:476–7. 480–3. [PubMed] [Google Scholar]
- Esser C, Alberti S, Höhfeld J. Biochim Biophys Acta. 2004;1695:171–188. doi: 10.1016/j.bbamcr.2004.09.020. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. Nat Struct Mol Biol. 2009;16:574–581. doi: 10.1038/nsmb.1591. [DOI] [PubMed] [Google Scholar]
- Herman C, D’Ari R. Curr Opin Microbiol. 1998;1:204–209. doi: 10.1016/s1369-5274(98)80012-x. [DOI] [PubMed] [Google Scholar]
- Kvam E, Sierks MR, Shoemaker CB, Messer A. Protein Eng Des Sel. 2010;23:489–498. doi: 10.1093/protein/gzq022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy R, Molineux IJ, Iverson BL, Georgiou G. J Immunol Methods. 2007;321:164–173. doi: 10.1016/j.jim.2007.01.017. [DOI] [PubMed] [Google Scholar]
- Lo AS, Zhu Q, Marasco WA. Handb Exp Pharmacol. 2008;181:343–373. doi: 10.1007/978-3-540-73259-4_15. [DOI] [PubMed] [Google Scholar]
- Martineau P, Betton JM. J Mol Biol. 1999;292:921–929. doi: 10.1006/jmbi.1999.3105. [DOI] [PubMed] [Google Scholar]
- Martineau P, Jones P, Winter G. J Mol Biol. 1998;280:117–127. doi: 10.1006/jmbi.1998.1840. [DOI] [PubMed] [Google Scholar]
- Mazuc E, Villoutreix BO, Malbec O, Roumier T, Fleury S, Leonetti J, Dombrowicz D, Daëron M, Martineau P, Dariavach P. J Allergy Clin Immunol. 2008;122:188–94. doi: 10.1016/j.jaci.2008.04.026. [DOI] [PubMed] [Google Scholar]
- Minton AP. Biophys J. 1992;63:1090–1100. doi: 10.1016/S0006-3495(92)81663-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S, Pelham HR. Cell. 1986;46:291–300. doi: 10.1016/0092-8674(86)90746-4. [DOI] [PubMed] [Google Scholar]
- Netzer WJ, Hartl FU. Nature. 1997;388:343–349. doi: 10.1038/41024. [DOI] [PubMed] [Google Scholar]
- Nicola AV, Chen W, Helenius A. Nat Cell Biol. 1999;1:341–345. doi: 10.1038/14032. [DOI] [PubMed] [Google Scholar]
- Pédelacq J, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Nat Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- Philibert P, Martineau P. Microb Cell Fact. 2004;3:16. doi: 10.1186/1475-2859-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philibert P, Stoessel A, Wang W, Sibler A, Bec N, Larroque C, Saven JG, Courtete J, Weiss E, Martineau P. BMC Biotechnol. 2007;7:81. doi: 10.1186/1472-6750-7-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roodveldt C, Aharoni A, Tawfik DS. Curr Opin Struct Biol. 2005;15:50–56. doi: 10.1016/j.sbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Sacchetti A, Cappetti V, Marra P, Dell’ Arciprete R, El Sewedy T, Crescenzi C, Alberti S. J Cell Biochem Suppl. 2001;36:117–128. doi: 10.1002/jcb.1091. [DOI] [PubMed] [Google Scholar]
- Schuhmann K, Voelker C, Höfer GF, Pflügelmeier H, Klugbauer N, Hofmann F, Romanin C, Groschner K. FEBS Lett. 1997;408:75–80. doi: 10.1016/s0014-5793(97)00391-8. [DOI] [PubMed] [Google Scholar]
- Sibler A, Nordhammer A, Masson M, Martineau P, Trave G, Weiss E. Exp Cell Res. 2003;286:276–287. doi: 10.1016/s0014-4827(03)00093-4. [DOI] [PubMed] [Google Scholar]
- Studier FW. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Suhr ST, Senut MC, Whitelegge JP, Faull KF, Cuizon DB, Gage FH. J Cell Biol. 2001;153:283–294. doi: 10.1083/jcb.153.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CJ, Maizel JVJ, Nussinov R. Protein Sci. 1999;8:1591–1604. doi: 10.1110/ps.8.8.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng WC, Haselton FR, Giorgio TD. J Biol Chem. 1997;272:25641–25647. doi: 10.1074/jbc.272.41.25641. [DOI] [PubMed] [Google Scholar]
- Visintin M, Meli GA, Cannistraci I, Cattaneo A. J Immunol Methods. 2004a;290:135–153. doi: 10.1016/j.jim.2004.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin M, Quondam M, Cattaneo A. Methods. 2004b;34:200–214. doi: 10.1016/j.ymeth.2004.04.008. [DOI] [PubMed] [Google Scholar]
- Waldo GS, Standish BM, Berendzen J, Terwilliger TC. Nat Biotechnol. 1999;17:691–695. doi: 10.1038/10904. [DOI] [PubMed] [Google Scholar]
- Worn A, Auf der Maur A, Escher D, Honegger A, Barberis A, Pluckthun A. J Biol Chem. 2000;275:2795–2803. doi: 10.1074/jbc.275.4.2795. [DOI] [PubMed] [Google Scholar]
- Zilberstein D, Agmon V, Schuldiner S, Padan E. J Bacteriol. 1984;158:246–252. doi: 10.1128/jb.158.1.246-252.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


