Abstract
Nitric oxide (NO) produced by vascular endothelial cells is a potent vasodilator and an antiinflammatory mediator. Regulating production of endothelial-derived NO is a complex undertaking, involving multiple signaling and genetic pathways that are activated by diverse humoral and biomechanical stimuli. To gain a thorough understanding of the rich diversity of responses observed experimentally, it is necessary to account for an ensemble of these pathways acting simultaneously. In this article, we have assembled four quantitative molecular pathways previously proposed for shear-stress-induced NO production. In these pathways, endothelial NO synthase is activated 1), via calcium release, 2), via phosphorylation reactions, and 3), via enhanced protein expression. To these activation pathways, we have added a fourth, a pathway describing actual NO production from endothelial NO synthase and its various protein partners. These pathways were combined and simulated using CytoSolve, a computational environment for combining independent pathway calculations. The integrated model is able to describe the experimentally observed change in NO production with time after the application of fluid shear stress. This model can also be used to predict the specific effects on the system after interventional pharmacological or genetic changes. Importantly, this model reflects the up-to-date understanding of the NO system, providing a platform upon which information can be aggregated in an additive way.
Introduction
One of the most important functions of vascular endothelial cells is to produce nitric oxide (NO). This molecule has a number of different roles in vascular stasis, including acting as a potent vasodilator and a mediator of inflammation (1). Not surprisingly, human vascular endothelial cells have developed multiple pathways by which production of NO is regulated by humoral and biomechnical stimuli via the expression and activation of endothelial nitric oxide synthase (eNOS). Exploring these different pathways one at a time is difficult, because the system is not separable—multiple pathways contribute to the production rate under all physiological circumstances. To understand and model the rich diversity of responses that have been observed experimentally, it is necessary to account for an ensemble of these pathways acting simultaneously over an extensive range of timescales.
The advancements of modern biology and computer science have increasingly enabled researchers to build such multipathway models. In the past two decades, experiments have been conducted that provide quantitative information between molecular species in the cell and their evolution under specific stimuli, facilitating construction of quantitative biochemical pathways that may be used as predictors of cellular response under a wider range of physiological or pathophysiological conditions. This sort of quantitative analysis of molecular pathways provides a valuable tool for assessing biological mechanisms and validating hypothetical mechanisms by comparing simulation results with experimental data.
One of the major hurdles in this process has been the development of in silico models that are sufficiently detailed to describe the complex phenomena observed. The current state of the art is to construct quantitative models based on selected subpaths within a larger molecular pathway. This process is time-consuming, requiring in-depth literature searches, experimentation, and parameter estimation. These isolated subpath models are invaluable and often provide insight into specific biochemical mechanisms. However, these subpathway models are often not independent in vivo or in vitro and have cross-sensitivities due to common species and overlapping reactions. As a result, to address more complex questions, such as the evolution of NO under mechanical shear stress, it is necessary to systematically integrate these subpaths to provide a more comprehensive and accurate purview of cellular mechanisms.
The current process of integrating multiple molecular pathways involves hand curation of individual models into a single monolithic model (see Fig. 1 A). Due to the use of different model coding environments, variable names, pathway separation methodologies, and solution strategies, assembly of monolithic models typically requires substantial rewriting of previously published models. In this process, the links to previously published subpaths become difficult to decipher, and the manual work may be prone to errors, especially for large networks. The process of including new data elements to subpath models that more accurately detail biochemical reaction steps is also nontrivial and often burdensome. Of most importance scientifically, monolithic model integration loses much of the history and progression of pathway determination and development, particularly the detailed experimental condition on which the original models and parameters are based (2).
Figure 1.
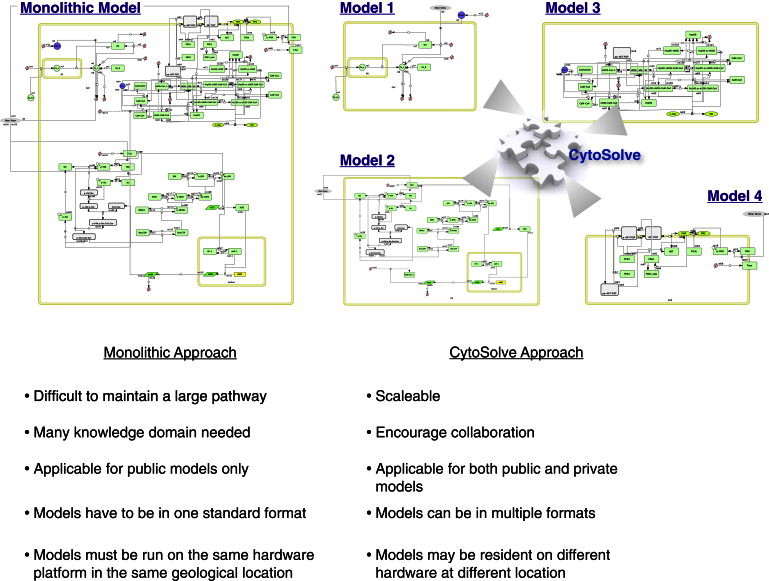
Comparison between the monolithic and CytoSolve approaches in building an integrated model.
In this article, an alternative approach based on the binding-expression concept is adapted (see Fig. 1 B) and the integrated model is viewed as an ontology. Any previously published subpath model is retained in its entirety, allowing it to be updated, extracted, replaced, or removed. Subpathway models are then integrated through bindings that identify common species in and alterations made to each model due to their integration. In our work, the creation of these bindings has been semiautomated through the use of MIRIAM (3) annotations and XML standard formats such as SBML (4) that support computational parsing and reasoning. In this way, common species and reaction pathways can be identified despite variations in nomenclature or number of reactions, thus lowering the complexity bar for the human curator. These tools are made publicly available through CytoSolve (5), a web-accessible interface (http://cytosolve.mit.edu/) capable of model integration and simulation.
In the case of endothelia-derived NO, many pathway models governing its production have been previously established (see Table 1). In this article, we integrate four of these molecular pathway models (see Fig. 2) that modulate the activation of endothelial nitric oxide synthase (eNOS) by shear stress. Specifically, we focus on the calcium-stimulated binding of calmodulin to eNOS, AKT-mediated phosphorylation of eNOS, and upregulation of eNOS transcription through AP-1 and KLF2. These pathways are linked by an additional model describing interaction of eNOS with its protein partners. By integrating these models in CytoSolve, the dynamic regulation and production of NO by eNOS under both shear-stress and static (no-shear-stress) conditions can be investigated and tested. This use of the NO model illustrates the potential of the partitioned model approach and of the CytoSolve tools, which enable simulation of complex problems involving many parallel pathways that cannot be readily isolated experimentally.
Table 1.
List of main mechanisms of shear-stress-regulated eNOS activation
| Main mechanisms of shear-stress-regulated eNOS activation | ||||
|---|---|---|---|---|
| Transcriptional regulation | ||||
| Key proteins | Known pathway | Note | References | Model inclusion |
| AP-1 | Shc → Grb2-Sos → Ras → JNK → AP-1 | Transient | (17,48) | eNOS expression |
| NFκB | Akt → IκK → NFκB | Transient (complex) | (16,49,50) | Not included |
| KLF2 | ? → MEK5 → ERK5 → MEF2 → KLF2 | Long term | (23) | eNOS expression |
| Posttranslational regulation | ||||
| Phosphorylation | ||||
| Key kinases | Known pathway | Phosphorylation site | References | Model inclusion |
| Akt | PI3K → AKT | Ser-1177 | (10) | eNOS phosphorylation |
| PKA | ? → [cAMP] → PKA | Ser-635 (bovine) | (12) | Not included |
| AMPK | ? → [AMP] → AMPK | Ser-1177 | (22) | Not included |
| Protein partners | ||||
| Key partners | Effect on eNOS | References | Model inclusion | |
| Caveolin-1 | Inactivation | (51) | NO production | |
| CaM | Activation, facilitate recruitment of Hsp90 | (28,52) | Calcium influx, NO production | |
| Hsp90 | Activation, facilitate recruitment of Akt | (27,53) | NO production | |
Figure 2.
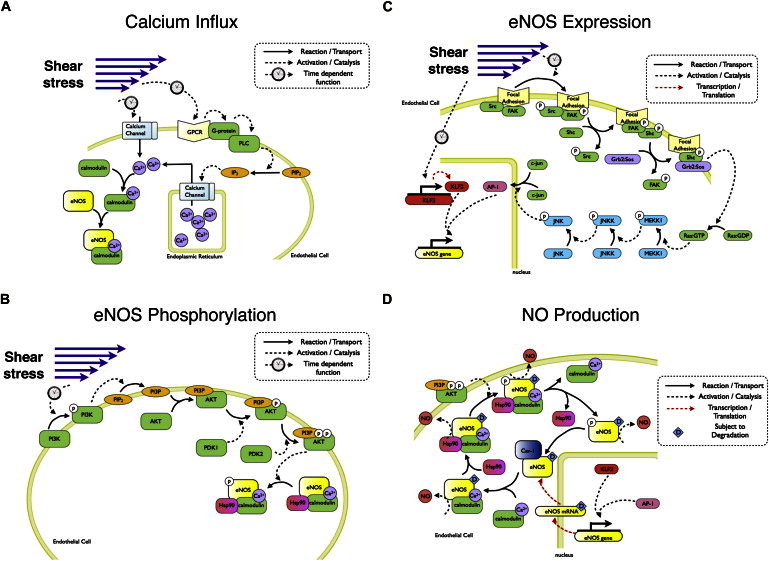
The four models of the shear-stress-induced NO production system. (A) The calcium influx model. (B) The eNOS phosphorylation model. (C) The eNOS expression model. (D) The NO production model.
Methods
In this section, we discuss the individual well-characterized pathway models that regulate eNOS and, as a result, NO production. These models are linked to a new model describing the interactions of eNOS and its binding partners. The section concludes with a description of the tools used to bind the individual models together, creating a partitioned NO pathway model capable of describing the multiple phenomena that regulate NO production.
Mechanisms of shear-stress-induced NO production
Several key signaling pathways have been identified that modulate the activity of eNOS—the primary source of NO production in vascular endothelial cells. In this section, we introduce three pathway models that alter eNOS activation or regulate eNOS protein expression. To link these models, an additional model was constructed that describes the binding of calcium, calmodulin (CaM), heat-shock protein 90 (Hsp90), eNOS, and phosphorylated eNOS, as well as the resulting enzymatic production of NO. Parameters of individual models were optimized to fit the experimental observations (details of the model schemes, inputs, species, reactions, parameters, and parameter optimization steps are described in the Supporting Material).
Shear-stress-induced calcium influx and eNOS activation
In response to increased fluid shear stress, endothelial cells exhibit a transient increase in cytosolic free calcium (see Fig. 2 A). The influx of calcium is due to mechanisms such as activation of stress-sensitive calcium channels and activation of G-protein pathways (6). A calcium channel is directly activated by fluid shear stress, and this leads to intracellular calcium influx. G-protein-coupled receptors can also be activated by shear stress (7). Activated G-protein induces activity of phospholipase C and production of inositol 1,4,5-trisphosphate (IP3). IP3 binds to its receptor on the surface of the endoplasmic reticulum and promotes calcium release from this intracellular storage. The increased intracellular Ca2+ then rapidly binds to CaM, a calcium-binding protein that significantly upregulates the activity of eNOS. The elevated intracellular calcium level leads to increased calcium export via the sodium-calcium exchanger and reuptake in intracellular stores, making increase in intracellular Ca2+ a transient (∼5-min) event (8).
To describe the calcium dynamics in response to shear stress, a mathematical model published by Wiesner et al. was used (8,9). This model assumes a step change in calcium influx mediated by the stress-sensitive calcium channel at the onset of shear stress (10 dynes/cm2). The resulting concentration profile of calcium transient is shown in Fig. 3 A.
Figure 3.
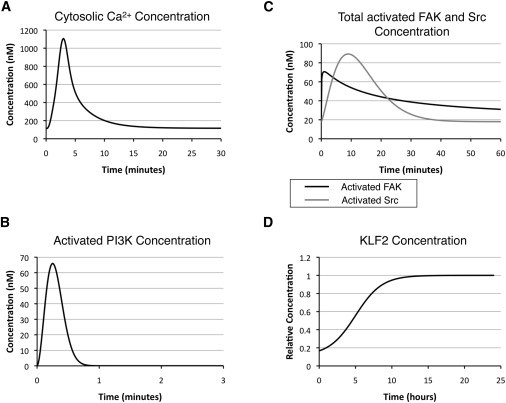
Simulation profiles of model inputs upon initiation of shear stress. The differential equations behind these simulation profiles were calculated based on time-dependent functions fit from experimental data (see Supporting Material for a detailed description of how these equations were generated). (A) Cytosolic Ca2+. (B) Activated PI3K. (C) Total activated FAK and activated Src. (D) KLF2.
Shear-stress-induced AKT and eNOS phosphorylation
In addition to its regulation by calcium-dependent CaM binding, eNOS activity can also be regulated by posttranslational modifications, the most important of these being phosphorylation reactions. Some key phosphorylation sites include the activity-inducing serines 1177, 635, and 617, and activity inhibitors serine 116 and threonine 495. It is important to note that phosphorylation of serine 1177 is thought to be a key indicator of eNOS activity under shear-stress conditions (10,11). In this model, we focused solely on the phosphorylation of this particular site. The reaction of serine 1177 phosphorylation is catalyzed by several protein kinases, including AKT, PKA, and AMPK (1). The mechanism of how shear stress activates AMPK is still unclear, but shear-stress-induced AKT and PKA activation has been shown to be phosphoinositide 3-kinase (PI3K)-dependent (10,12). Phosphorylation of serine 1177 is significantly decreased when endothelial cells are treated with PI3K inhibitor, Ly294002, or transfected with dominant-negative AKT (10,13). These data suggest that the PI3K-AKT pathway plays a critical role in shear-stress-induced eNOS phosphorylation. Based on this experimental observation, it is assumed in our model that PI3K-dependent AKT activation is the main pathway responsible for eNOS phosphorylation. PI3K activation leads to production of phosphatidylinositol (3–5)-trisphosphate (PIP3). PIP3 then recruits cytosolic free AKT to the membrane, where it is phosphorylated by both PDK1 and PDK2 (see Fig. 2 B). Finally, phosphorylated AKT phosphorylates eNOS on serine 1177. It is important to note also that due to the lack of detailed kinetic parameters for the phosphorylation reactions for PKA and AMPK on serine 1177 of eNOS, the model focuses only on the AKT-dependent pathway.
The mathematical model used to describe the AKT activation process is taken from Koh et al. (14), a model originally established to study the cross talk between AKT and MAPK pathways upon binding of receptors to growth factors. This model provides a detailed illustration of the PI3K-AKT pathway, which we assumed to be conserved across different human cell types. In our model, PI3K activation was assigned to be the input signal based on a time-dependent function fit from experimental data by Go (15). Because the mechanism by which shear stress leads to activation of molecular pathways is still poorly understood, time-dependent functions were used as model inputs throughout the NO system as proxies for the mechanotransduction process (see Supporting Material for more details of the generation of time-dependent functions). In this study, laminar shear stress with a magnitude of 5 dynes/cm2 was used in experiments. In other works used to generate model inputs for subsequent models, laminar shear stress or oscillatory shear stress with a mean magnitude of 12 dynes/cm2 was applied. Here, we made a general assumption that eNOS activation in endothelial cells respond similarly given a shear-stress stimulus in the range 5–12 dynes/cm2.
Shear-stress-induced eNOS expression
A third mechanism leading to an overall increase in NO production is upregulation of eNOS expression. Key transcription factors governing shear-stress-induced eNOS promoter activity include AP-1, NFκB, and KLF2 (11). The role of NFκB on eNOS expression remains controversial, as recent study indicates that expression of NFκB and eNOS is negatively correlated under shear stress (16). Therefore, in our model, we focused on simulating the effects of AP-1 and KLF2 on eNOS transcription (Fig. 2 C). In this model, it is assumed that there is no interaction between these two transcription factors and that they have no synergistic effect on eNOS transcription.
AP-1, a Jun-Jun homodimer or a Jun-Fos heterodimer, is involved in shear-stress-induced eNOS expression. A qualitative pathway model describing how shear stress leads to AP-1 nucleus translocation has been established previously (17). In the proposed model, shear stress activates the focal adhesion site and leads to phsophorylation of focal adhesion kinase (FAK), Src kinase, and the adaptor protein Shc. Activation of these kinases leads to formation of the first complex, FAK-Shc, then a second complex between FAK-Shc and Grb2-Sos. The second complex activates Ras protein, and initiates the MAP kinase cascade through MEKK1, JNKK, and JNK. JNK phosphorylates Jun and eventually leads to Jun dimer association to form AP-1, which translocates to nucleus and facilitates eNOS expression.
To quantitatively model the contribution of AP-1 in regulating eNOS expression, two existing mathematical models were used as the bases. The first model, excerpted from Hatakeyama et al. (18), describes the activation pathway from Src, FAK to Ras. However, since the upstream mechanical activation of Src and FAK molecules is not well understood, the kinetics of these two molecules (Fig. 3 C) were based on time-dependent experimental measures observed by Li et al. (19) and Jalali et al. (20). The shear-stress experiments in those two articles used a laminar shear stress of 12 dynes/cm2. The second model, modified from the Kholodenko study (21), illustrates the kinetics of how Ras initiates the MAP kinase cascade. These two models were combined and integrated with reactions including JNK-mediated Jun phosphorylation/dimerization, AP-1 nuclear translocation, AP-1-mediated eNOS transcription, eNOS translation, and eNOS mRNA degradation. The rates of these reactions were estimated based on experimental observation and previously established models. Many parameters of this model were optimized to fit the experimental observations. Further details can be found in the Supporting Material.
KLF2, the third transcription factor responsible for eNOS expression, is characterized as leading to long-term upregulated eNOS transcription. Compared to the fast and transient nuclear translocation of AP-1 in response to shear stress, the increase in KLF2 concentration inside the nucleus is relatively slow but sustained (22). The upstream mechanosensors for KLF2 expression are still poorly understood, but its expression is known to be dependent on MEK5, ERK5, and MEF2 (23). Due to the limited amount of experimental data available to construct a complete model, KLF2 dynamics is simulated based on data from a time-course shear-stress experiment (1 Hz oscillatory shear stress of 12 ± 4 dynes/cm2) by Young et al. (22) (Fig. 3 D).
Shear-stress-induced NO production
The previously described models establish the concentration profile of cytosolic free calcium, phosphorylated eNOS, and total eNOS expression in response to shear stress. However, to integrate these pathways and understand NO production, one additional model was necessary to characterize the interactions of eNOS and its binding partners (Fig. 2 D).
The biphasic binding of Ca2+ to CaM is well-documented in the literature by Bayley et al. (24) and others (25). This has been shown to occur due to the very rapid dissociation of Ca2+ from the N-ter EF-hand pair compared to its dissociation from the C-ter EF-hand pair, although some evidence suggests cooperative binding of Ca2+ to CaM (25). Black et al. have shown that a number of sequential kinetic models can predict binding response (26). Based on their results, Ca2+ binding to CaM was modeled using a four-step process, with two fast and two slow steps. In our model, we assumed that the fast steps are much faster than the slow steps and that therefore CaM(Ca2+)2 and CaM(Ca2+)4 are the only stable CaM- Ca2+ forms. Both species were assumed to bind to eNOS.
Besides CaM, another key regulator for eNOS activation under shear stress is Hsp90. Hsp90 does not bind to eNOS under static conditions, but significant binding was detected just 15 min after initiation of shear stress (27). CaM-bound eNOS has been shown to significantly increase the efficiency of Hsp90 recruitment (28). Studies have also shown that formation of the eNOS-CaM-Hsp90 complex is required for Akt-mediated eNOS phosphorylation on serine 1177 (29). Once phosphorylated, eNOS is stable in the active state, with enhanced NO production efficiency, until the phosphate group is removed. A quantitative model is created based on this scheme. All rate constants were either derived from existing models or optimized based on experimental data (see Supporting Material).
Model integration
All individual models were built using CellDesigner 4.1 (http://www.CellDesigner.org), a visual design tool for cell models and molecular pathways. Each model was coded in SBML, an XML-based format that is widely used to encode biomolecular pathways. SBML (4) was selected due to its open standard, the available programming interface, the LibSBML library (31), and its wide usage in the molecular modeling community and model repositories (such as http://www.Biomodels.net (30)). All models were encoded using the MIRIAM (minimum information requested in annotation of biochemical models) guidelines (3), which provide a rigorous set of information that mathematical models should include so that they can be reused.
An attractive feature of the SBML standard, combined with MIRIAM, is the ability to include the resource description framework (RDF) statements. These enable the unique identification of biomolecular components across multiple models irrespective of an individual model’s notation. This is achieved by labeling elements (i.e., species, reactions, etc.) in external resources (ontologies or databases) that provide a mechanism for identifying common species and reactions across models. For a generic SBML- and MIRIAM-compliant model, it is possible to associate RDF statements to species, to reactions, and to the model itself, providing a means for performing more advanced processing and model merging. For example, a digital object identifier number or PubMed article identification provides a unique link to a published article with the model details, and the universal resource identifier links to elements in the Systems Biology Ontology (32) or Chemical Entities of Biological Interest ontology (33) provide extensive information on individual biochemical species used within the model.
Writing each SBML model to be MIRIAM-compliant requires additional effort, but the RDF annotations enable models to be parsed and merged by a suitable logical reasoner. For this work, we used the ontology reasoning engine for molecular pathways (OREMP) computational code (34), which can automatically identify duplicate species across models. Moreover, OREMP can detect potential redundant reactions or reaction series that are shared across models. Identification of overlapping reactions is critical, because a hidden synergistic action of two or more separate mathematical statements of the same reaction leads to erroneous simulation results. The use of both species and reaction annotations becomes very useful in this process, as they enable the automatic match of cross-model components, minimizing user input. Using an ontological approach, the properties of each model discovered by the OREMP software can be appended to the description of the submodels, thus archiving these steps for future use.
The duplicate species and reactions between submodels provide the relevant information for model integration. They act as bindings that provide the map between individual model species and reactions within a single model and their interactions in the entire merged model. As newer models become available, they can also be integrated, either augmenting the current model or replacing redundant paths. This process differs significantly from the monolithic process, where each individual model is incorporated into a single model, requiring significantly more user effort.
Solving the model pathway
The partitioned models and their bindings provide the necessary information to simulate the global behavior of all interacting models. Models are aligned using CytoSolve and OREMP in combination, as outlined above, providing the detected duplicates as an editable list to the user. Once the bindings between models have been constructed, the user may then set up the necessary initial conditions, measured experimentally or estimated from computer optimization, for simulating the molecular pathway. Simulation is handled using libSBML to parse the original SBML models and SOSLib with SUNDIALS (35) to compute the evolution of submodels through time. CytoSolve solves the joint model not as a monolithic model but as a separated system of models. The merging of concentrations of individual model species is handled via a mass-balance controller, which ensures both that aligned bound species maintain the same concentration throughout the simulation and that the time steps taken are small enough to guarantee convergence of the separable solution to the true (monolithic) solution (see Ayyadurai and Dewey (5) and Nordsletten et al. (36) for further details).
Results
A simulation of the integrated shear-stress-induced NO production model
When endothelial cells are exposed to shear stress, one of the first events is influx of calcium from extracellular space and intracellular storage. Fig. 3 A illustrates the concentration profile of intracellular calcium governed by the calcium influx model. The calcium level increases within the first 3 min after onset of shear stress; this transient response lasts for 10 min and quickly goes back to the resting-state level.
Another early event observed after onset of shear stress is activation of PI3K. The concentration profile (Fig. 3 B) of PI3K is simulated based on a time-dependent function generated from experimental data. Activation of PI3K is short and transient, but accumulation of PIP3 results in downstream Akt phosphorylation (Fig. 4 A). This result is consistent with experimental observations, where fully active AKT reaches the peak level within 30 min after onset of shear stress and gradually decays back to the initial state in hours (12,37).
Figure 4.
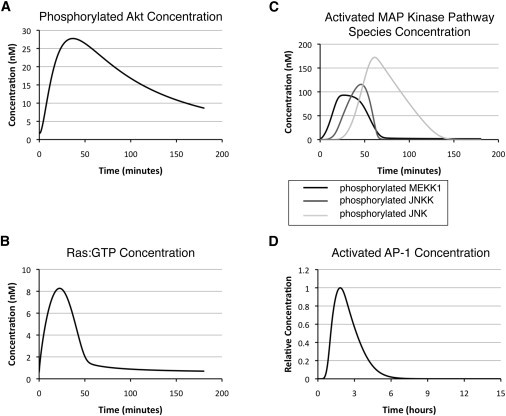
Simulation profiles of intermediate species upon initiation of shear stress. (A) Phosphorylated Akt (pp-Akt). (B) Ras/GTP. (C) Activated MAP kinase pathway species (p-MEKK1, pp-JNKK, and pp-JNK). (D) Activated AP-1.
A third early event after initiation of shear stress is activation of the focal adhesion complex, including phosphorylation of both focal adhesion kinase (FAK) and Src kinase (Fig. 3 C). Phosphorylation of the two proteins leads to downstream activation of Ras (Fig. 4 B) and the MAP kinase pathway proteins (Fig. 4 C) and subsequent AP-1 formation and nuclear translocation (Fig. 4 D). This process is transient, with a time span of a few hours, and is responsible for the fast-responding upregulation of eNOS mRNA and proteins after the cells experience a change in hemodynamic environment. Besides AP-1, the concentration profile for KLF2, another important transcription factor for eNOS, is shown in Fig. 3 D. KLF2 is responsible for long-term upregulation of eNOS mRNA and protein.
The above data describe the simulation results of individual pathways. These pathways interact with each other to control the dynamics of various eNOS species. Under the static (no-shear-stress) condition, eNOS primarily binds to Cav-1. After the onset of shear stress, calcium is transported to the cell and bound to CaM. Four calcium ions bind to each CaM to make the active form of CaM, which associates with eNOS to enhance its catalytic activity to produce NO (Fig. 5 A). In the meantime, the CaM-eNOS complex recruits Hsp90, which stabilizes the complex and facilitates Akt-mediated eNOS phosphorylation. The simulated concentration profile of eNOS phosphorylated on Ser-1177 (Fig. 5 B) is consistent with existing experimental observations (12) and depicts a biphasic pattern. In the first 10 min, when the phosphorylated Akt (enzyme) concentration is low but the CaM-eNOS-Hsp90 (substrate) concentration is high, there is rapid eNOS phosphorylation due to high substrate concentration. From 10 to 40 min, even though the substrate availability becomes low due to lower calcium concentration, the phosphorylated eNOS level stays high as a result of increasing phosphorylated Akt.
Figure 5.
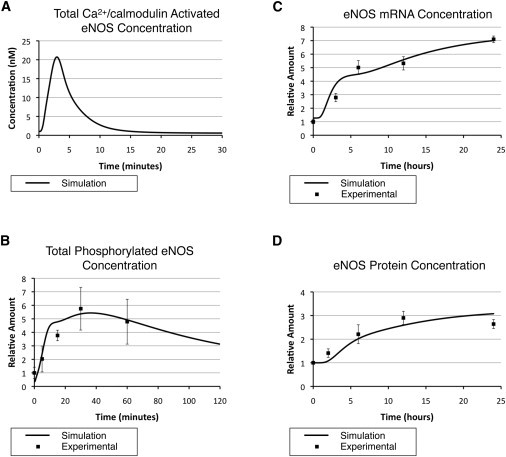
Simulation profiles of eNOS species. (A) Total Ca2+/CaM-activated eNOS. (B) Total phosphorylated eNOS (Ser-1177). The simulated data are compared with experimental observations from Boo et al. (12). (C) eNOS mRNA. The simulated data are compared with our experimental observations. (D) Total eNOS protein. The simulated data are compared with experimental observations from Li et al. (38).
To maintain long-term NO production, a third mechanism employed by cells is increasing eNOS protein expression. Fig. 5, C and D, demonstrates the increase in eNOS mRNA and protein as catalyzed by the two transcription factors AP-1 and KLF2. The simulated expression of eNOS mRNA and protein under shear stress is shown with the experimental data from our lab and that of Li et al. (38), respectively. The concentration profile of eNOS mRNA also reveals a biphasic pattern as a result of early transcription by AP-1 and later transcription by KLF2. This biphasic effect is smoothed out after the eNOS translation process (Fig. 5 D).
Finally, the total NO production from eNOS is simulated under both static and shear-stress conditions. Fig. 6 shows the accumulated NO production over time from the integrated model. We used relative units for NO, since its experimentally observed concentration varies depending on the cell confluency and media volume of individual experimental setup. The simulated NO production profile resembles the experimental data measured by Florian et al. (39). Under static conditions, there is low NO production from background level of CaM-activated eNOS and phosphorylated eNOS. Under the shear-stress condition, calcium influx in the first few minutes leads to a quick burst of NO production from CaM-activated eNOS. As calcium goes back to the basal level, phosphorylated eNOS kicks in to support NO production in the first few hours. The effect of eNOS expression does not come in until a few hours later (described in more detail in the next section).
Figure 6.
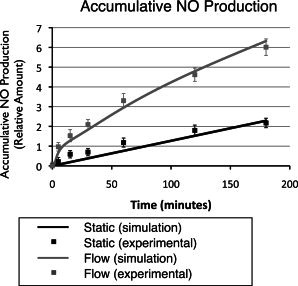
Comparison of the simulation results regarding cumulative NO production with experimental data (39) under static (no-shear-stress) and shear-stress conditions.
The model integration approach provides insight into the system that could not be easily gathered experimentally
Having established a system model that allows us to simulate shear-stress-induced NO production comparable to that observed experimentally, we next explored several aspects of the system that can be simulated easily but are difficult to test experimentally. First, we analyzed the contribution of individual pathways to the overall NO production. The integrated modeling approach allows investigation of the relative importance of individual pathways instantaneously. Fig. 7 A demonstrates the cumulative NO production contributed by different eNOS species. The data show that almost all of the NO produced in the first 10 min comes from Ca2+/CaM-activated eNOS, with later production of NO mostly contributed by phosphorylated eNOS. In contrast, the NO produced by the intermediate species, Ca2+/CaM-activated phosphorylated eNOS, is not significant.
Figure 7.
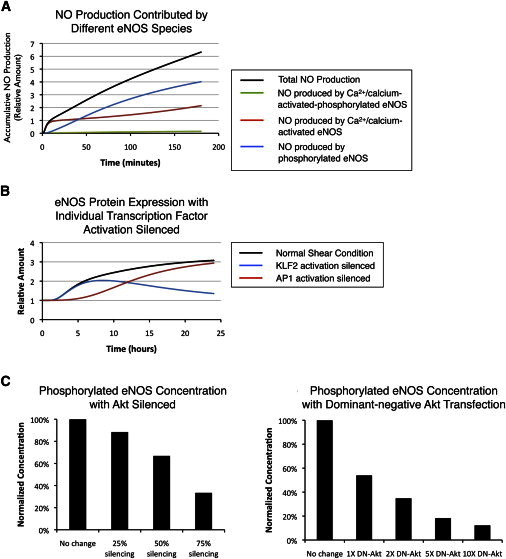
The integrated model allows us to easily assess the contribution of individual eNOS species or simulate the condition where one pathway is modified. (A) Contribution of NO production by different eNOS species. (B) eNOS protein expression with individual transcription-factor activation silenced. Concentrations of the specific transcription factor are fixed at the static level. (C) Normalized concentration of total phosphorylated eNOS with addition of Akt siRNA (left) or dominant-negative Akt (right) 1 h after onset of shear stress. In the Akt siRNA simulation, total Akt concentration was reduced based on the specific silencing efficiency. In the dominant-negative Akt (DN-Akt) simulation, DN-Akt follows the exact kinetics of wild-type Akt except that it loses its catalytic ability to phosphorylate eNOS. The amount of wild-type Akt remains constant, whereas the amount of DN-Akt is 1×, 2×, 5×, and 10× the amount of wild-type Akt.
Second, we simulate the small-interfering RNA (siRNA) gene-silencing approaches by selectively silencing shear-stress-induced activation of individual pathways. This process can be easily illustrated by removing or modifying species in the system, giving reasonable predictions while saving tremendous resources. In the NO system, we assess the effect of modifying individual pathways on overall NO production. To research how an individual transcription factor affects overall eNOS protein expression, AP-1 and KLF2 activation were blocked (Fig. 7 B). Blocking AP-1 activation yields a delayed response in eNOS expression under shear stress, whereas blocking KLF2 activation leads to no shear-stress-induced eNOS expression after 24 h.
Finally, we attempt to predict the effect of transfecting endothelial cells with Akt siRNA or dominant-negative Akt on eNOS phosphorylation (Fig. 7 C) under 1 h of shear stress. Our simulation data suggest that the relationship between silencing efficiency and the resulting decrease in eNOS phosphorylation is not linear. An Akt knockdown efficiency of 25% has little effect on eNOS phosphoryation, a 50% efficiency still retains >60% of phosphorylated eNOS; it is not until a 75% silencing efficiency is achieved that we observe <40% eNOS phosphorylation. A similar effect of decreasing eNOS phosphorylation can be achieved with an alternative approach. Fisslthaler et al. have demonstrated that transfecting the cells with dominant-negative Akt (DN-Akt) decreases shear-stress-induced eNOS phosphorylation (13). Here, we simulate the condition in which there are various amounts of DN-Akt (1×, 2×, 5×, and 10× relative to wild-type) in the system in addition to wild-type eNOS. DN-Akt competes with the wild-type Akt for the binding site on the plasma membrane and significantly reduces shear-stress-activated eNOS phosphorylation.
It is important to note that when a known pathway is knocked down, which, for a given pathway, would yield no shear-stress-induced NO production, the integrated model shows robustness to the knockdown of that specific pathway (e.g., Akt) but fails to take into account alternative ones (e.g., PKA, AMPK). This result emphasizes the importance of systems biology for achieving a quantitative understanding of macroscopic cellular response processes, as well as the power of such analyses for more comprehensive pathway assessment. However, this result also highlights a major limitation of the approach, which is the absolute dependence of previously reported and characterized data on defined signaling pathways that can be incorporated into a model (e.g., Akt versus PKA versus AMPK).
Discussion
The power of automatic model integration
Quantitative modeling of molecular pathways provides a powerful tool for simulating and predicting function. With more and more comprehensive experimental data, it is rapidly becoming possible to construct more complete models of molecular pathways. A major bottleneck in this process, however, is the current model paradigm where models are manually integrated into a single complex model, obscuring the link to previously published pathways. In this article, we introduced an alternative model-binding approach in which individual models are written and retained as is standard in MIRIAM-compliant SBML format. The alterations, duplicate species, and duplicate reactions are then detailed within the model bindings, providing clarity on how past models are incorporated into the current model. The tools for this merger process, as well as the simulation of merged models, have been made available, as part of our continued work, by CytoSolve.
In this study, we considered four primary pathways that govern the activation and transcription/translation of eNOS, the NO catalyst. All these paths have been shown to work in tandem to govern the transient NO response of cells to shear stress. Indeed, we show that the NO response consists of three primary phases that act on varying timescales. The transient response is thus governed by the ensemble of molecular pathways and cannot be accurately modeled by considering any component individually. In addition to illustrating the need for model integration, the introduced pathways demonstrate the power of the model-binding approach. As the system is composed of individual models, additional pathway models based on new experimental data can be quickly incorporated into the system without rewriting the existing model. It also allows us to easily investigate the relative contribution of each pathway and conduct in silico experiments as demonstrated in Fig. 7.
Utilizing MIRIAM standards and references to web-accessible ontologies, this study also introduced an approach for automating model integration. This strategy is the opposite of the monolithic approach in which many models are manually assembled to compose a single new model that is more difficult to further edit, generally resulting in valued work with limited reusability. In contrast to this approach, the partitioned approach introduced in this work provides an additive model-creation paradigm, where previous knowledge, data, and effort can easily be managed, curated, and used by the wider scientific community. This is accomplished by the model-binding approach, where individual models retain their original identity. The bindings between individual models provide the necessary interface for both defining how an original model is used and altered, preserving the original model and its lineage and making the modeling process more straightforward. In addition, the integration of models via the partitioned approach makes much more straightforward the process of updating the model while at the same time preserving the extant bindings and models.
Limitations in shear-stress-induced NO pathway modeling
In this article, we collect the relevant pathways, species, and reactions to reflect the state-of-the-art understanding of NO production. Although the NO system provides a quantitative link between shear stress and NO production through activation and transcription pathways that match well with published experimental data, further work is required to improve and enhance the model. Due to limitations in available data, some model components were based on experiments using different endothelial cell types, as well as different experimental conditions (variations in culture conditions and mechanisms for applying shear). Although these issues are not unique to this model, but are common to a number of cellular models, they warrant further experimental investigation and validation. These weaknesses can be annotated in the model and made transparent for future improvement. Despite these limitations, the model does demonstrate the general dynamics of multiple pathways acting in concert to upregulate NO production.
To further improve the integrated NO model, incorporation of other mechanotransduction pathways is necessary. For example, the endothelial glycocalyx has been shown to be an important mechanosensor for shear-stress-induced NO production. Treating the endothelial surface with heparanase to remove heparan sulfate, one major glycosaminoglycan of the glycocalyx layer, significantly reduces NO production resulting from shear stress (39,40). The specific signaling pathways that trigger the models used here are still not known. Moreover, we have used a simple heuristic model based on experimental data to represent the upregulation of KLF2 by shear stress. KLF2, a key regulator for eNOS expression, is also a shear-responding transcription factor leading to antiinflammatory and antithrombotic phenotypes (41). KLF2 expression is known to follow the MEK5-ERK5-MEF2 pathway, but the mechanosensors that lead to the activation of this pathway are still unclear (23). These additions are currently limited by incomplete knowledge of pathway mechanisms and lack of critical kinetic data. However, the current integrated model creates a platform to identify deficiencies in our current understanding, provide more suitable parameters, and embed additional pathways or information in an additive way. This process acts as a communal way of documenting what is understood about cellular mechanisms of NO production in endothelial cells.
Future model integration tools and development
This article outlines the use of CytoSolve in facilitating model integration. Although this tool automates the model integration process with minimal input from the user, further development could dramatically improve the effectiveness of the integration process, store relevant changes at various stages in development, and provide tools for incorporating input from the wider scientific community.
One of the major aims of this work was to demonstrate that the combination of separate biological models to generate a new, larger predictive model is difficult but becomes tractable with the right tools. Because a model that exists completely independently of other models is of limited use to the research community, it behooves the community to define processes by which existing and new models can be augmented, combined, and increased in complexity so that existing work is properly assimilated.
It is also important to recognize that different models have different aims and often operate at different timescales, spatial scales, and initial conditions; most of the models have species exchanges on the order of μM/min or nM/min, but this is not universally suitable for every objective. Because SBML makes it possible to define arbitrary units by composing IS units, it is among CytoSolve's goals to resolve and normalize differing units in a transparent manner. A new algorithm will extend the current mass-balance, introducing a unit-conversion routine that will be called at run time, keeping the merged solution independent of any individual model's representation.
Another future addition will be the extension of the actual cross-model information-sharing process to show users other reactions that may be of interest to them. This will provide an autocompletion for models to include reactions seen in other models. The goal of this functionality is to accelerate the information-sharing and model-composition process even more: building models on top of others, reducing unnecessary duplicates, and informing researchers of known existing pathways that may be relevant to their study.
Conclusion
In this study, we simulated the process of shear-stress-induced NO production in endothelial cells by combining a number of existing published pathway models to describe and predict the complex interactions that occur between them at multiple timescales. The program we used, CytoSolve, is specifically designed to facilitate the federation of individual biological pathways in a manner that allows them to run as a combined monolithic model without losing their individuality and the metadata attached to them. The integrated model reflects the state-of-the-art understanding of the NO system, and the simulation data are able to describe experimental observations resulting from complex interactions between multiple pathways. The system-level simulation approach can also provide researchers with useful insights into the system that have traditionally only been achieved with challenging and time-consuming experiments. It is important to note that this approach to biological pathway integration is not only helpful for our understanding of biological system, but it also provides a platform to aggregate information in an additive way, which eventually could allow us to predict biology.
Acknowledgments
We thank Akila Surendran, Manuel Legrand, Jacqueline Wentz, and John Yazbek for help creating the preliminary models.
This study was funded by a grant from the National Institutes of Health (R01HL090856). We also acknowledge support from the Singapore-MIT Computational and Systems Biology Program.
Footnotes
This is an Open Access article distributed under the terms of the Creative Commons-Attribution Noncommercial License (http://creativecommons.org/licenses/by-nc/2.0/), which permits unrestricted noncommercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Supporting Material
References
- 1.Sessa W.C. eNOS at a glance. J. Cell Sci. 2004;117:2427–2429. doi: 10.1242/jcs.01165. [DOI] [PubMed] [Google Scholar]
- 2.Niederer S.A., Fink M., Smith N.P. A meta-analysis of cardiac electrophysiology computational models. Exp. Physiol. 2009;94:486–495. doi: 10.1113/expphysiol.2008.044610. [DOI] [PubMed] [Google Scholar]
- 3.Le Novère N., Finney A., Wanner B.L. Minimum information requested in the annotation of biochemical models (MIRIAM) Nat. Biotechnol. 2005;23:1509–1515. doi: 10.1038/nbt1156. [DOI] [PubMed] [Google Scholar]
- 4.Hucka M., Finney A., Wang J., SBML Forum The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 5.Ayyadurai V.A., Dewey C.F. CytoSolve: a scalable computational method for dynamic integration of multiple molecular pathway models. Cell Mol. Bioeng. 2011;4:28–45. doi: 10.1007/s12195-010-0143-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies P.F. Flow-mediated endothelial mechanotransduction. Physiol. Rev. 1995;75:519–560. doi: 10.1152/physrev.1995.75.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chachisvilis M., Zhang Y.L., Frangos J.A. G protein-coupled receptors sense fluid shear stress in endothelial cells. Proc. Natl. Acad. Sci. USA. 2006;103:15463–15468. doi: 10.1073/pnas.0607224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiesner T.F., Berk B.C., Nerem R.M. A mathematical model of the cytosolic-free calcium response in endothelial cells to fluid shear stress. Proc. Natl. Acad. Sci. USA. 1997;94:3726–3731. doi: 10.1073/pnas.94.8.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wiesner T.F., Berk B.C., Nerem R.M. A mathematical model of cytosolic calcium dynamics in human umbilical vein endothelial cells. Am. J. Physiol. 1996;270:C1556–C1569. doi: 10.1152/ajpcell.1996.270.5.C1556. [DOI] [PubMed] [Google Scholar]
- 10.Dimmeler S., Fleming I., Zeiher A.M. Activation of nitric oxide synthase in endothelial cells by Akt-dependent phosphorylation. Nature. 1999;399:601–605. doi: 10.1038/21224. [DOI] [PubMed] [Google Scholar]
- 11.Balligand J.-L., Feron O., Dessy C. eNOS activation by physical forces: from short-term regulation of contraction to chronic remodeling of cardiovascular tissues. Physiol. Rev. 2009;89:481–534. doi: 10.1152/physrev.00042.2007. [DOI] [PubMed] [Google Scholar]
- 12.Boo Y.C., Sorescu G., Jo H. Shear stress stimulates phosphorylation of endothelial nitric-oxide synthase at Ser1179 by Akt-independent mechanisms: role of protein kinase A. J. Biol. Chem. 2002;277:3388–3396. doi: 10.1074/jbc.M108789200. [DOI] [PubMed] [Google Scholar]
- 13.Fisslthaler B., Dimmeler S., Fleming I. Phosphorylation and activation of the endothelial nitric oxide synthase by fluid shear stress. Acta Physiol. Scand. 2000;168:81–88. doi: 10.1046/j.1365-201x.2000.00627.x. [DOI] [PubMed] [Google Scholar]
- 14.Koh G., Teong H.F.C., Thiagarajan P.S. A decompositional approach to parameter estimation in pathway modeling: a case study of the Akt and MAPK pathways and their crosstalk. Bioinformatics. 2006;22:e271–e280. doi: 10.1093/bioinformatics/btl264. [DOI] [PubMed] [Google Scholar]
- 15.Go Y.M., Park H., Jo H. Phosphatidylinositol 3-kinase γ mediates shear stress-dependent activation of JNK in endothelial cells. Am. J. Physiol. 1998;275:H1898–H1904. doi: 10.1152/ajpheart.1998.275.5.H1898. [DOI] [PubMed] [Google Scholar]
- 16.Won D., Zhu S.N., Cybulsky M.I. Relative reduction of endothelial nitric-oxide synthase expression and transcription in atherosclerosis-prone regions of the mouse aorta and in an in vitro model of disturbed flow. Am. J. Pathol. 2007;171:1691–1704. doi: 10.2353/ajpath.2007.060860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen K.D., Li Y.S., Shyy J.Y. Mechanotransduction in response to shear stress. Roles of receptor tyrosine kinases, integrins, and Shc. J. Biol. Chem. 1999;274:18393–18400. doi: 10.1074/jbc.274.26.18393. [DOI] [PubMed] [Google Scholar]
- 18.Hatakeyama M., Kimura S., Konagaya A. A computational model on the modulation of mitogen-activated protein kinase (MAPK) and Akt pathways in heregulin-induced ErbB signalling. Biochem. J. 2003;373:451–463. doi: 10.1042/BJ20021824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li S., Kim M., Shyy J.Y. Fluid shear stress activation of focal adhesion kinase. Linking to mitogen-activated protein kinases. J. Biol. Chem. 1997;272:30455–30462. doi: 10.1074/jbc.272.48.30455. [DOI] [PubMed] [Google Scholar]
- 20.Jalali S., Li Y.S., Shyy J.Y. Shear stress activates p60src-Ras-MAPK signaling pathways in vascular endothelial cells. Arterioscler. Thromb. Vasc. Biol. 1998;18:227–234. doi: 10.1161/01.atv.18.2.227. [DOI] [PubMed] [Google Scholar]
- 21.Kholodenko B.N. Negative feedback and ultrasensitivity can bring about oscillations in the mitogen-activated protein kinase cascades. Eur. J. Biochem. 2000;267:1583–1588. doi: 10.1046/j.1432-1327.2000.01197.x. [DOI] [PubMed] [Google Scholar]
- 22.Young A., Wu W., García-Cardeña G. Flow activation of AMP-activated protein kinase in vascular endothelium leads to Krüppel-like factor 2 expression. Arterioscler. Thromb. Vasc. Biol. 2009;29:1902–1908. doi: 10.1161/ATVBAHA.109.193540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parmar K.M., Larman H.B., García-Cardeña G. Integration of flow-dependent endothelial phenotypes by Kruppel-like factor 2. J. Clin. Invest. 2006;116:49–58. doi: 10.1172/JCI24787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bayley P., Ahlström P., Forsen S. The kinetics of calcium binding to calmodulin: Quin 2 and ANS stopped-flow fluorescence studies. Biochem. Biophys. Res. Commun. 1984;120:185–191. doi: 10.1016/0006-291x(84)91431-1. [DOI] [PubMed] [Google Scholar]
- 25.Linse S., Helmersson A., Forsén S. Calcium binding to calmodulin and its globular domains. J. Biol. Chem. 1991;266:8050–8054. [PubMed] [Google Scholar]
- 26.Black D.J., Selfridge J.E., Persechini A. The kinetics of Ca2+-dependent switching in a calmodulin-IQ domain complex. Biochemistry. 2007;46:13415–13424. doi: 10.1021/bi700774s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.García-Cardeña G., Fan R., Sessa W.C. Dynamic activation of endothelial nitric oxide synthase by Hsp90. Nature. 1998;392:821–824. doi: 10.1038/33934. [DOI] [PubMed] [Google Scholar]
- 28.Gratton J.P., Fontana J., Sessa W.C. Reconstitution of an endothelial nitric-oxide synthase (eNOS), hsp90, and caveolin-1 complex in vitro. Evidence that hsp90 facilitates calmodulin stimulated displacement of eNOS from caveolin-1. J. Biol. Chem. 2000;275:22268–22272. doi: 10.1074/jbc.M001644200. [DOI] [PubMed] [Google Scholar]
- 29.Dudzinski D.M., Michel T. Life history of eNOS: partners and pathways. Cardiovasc. Res. 2007;75:247–260. doi: 10.1016/j.cardiores.2007.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le Novère N., Bornstein B., Hucka M. BioModels Database: a free, centralized database of curated, published, quantitative kinetic models of biochemical and cellular systems. Nucleic Acids Res. 2006;34(Database issue):D689–D691. doi: 10.1093/nar/gkj092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bornstein B.J., Keating S.M., Hucka M. LibSBML: an API library for SBML. Bioinformatics. 2008;24:880–881. doi: 10.1093/bioinformatics/btn051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Courtot M., Juty N., Le Novère N. Controlled vocabularies and semantics in systems biology. Mol. Syst. Biol. 2011;7:543. doi: 10.1038/msb.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Degtyarenko K., de Matos P., Ashburner M. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res. 2008;36(Database issue):D344–D350. doi: 10.1093/nar/gkm791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Umeton R., Nicosia G., Dewey C.F., Jr. OREMPdb: a semantic dictionary of computational pathway models. BMC Bioinformatics. 2012;13(Suppl 4):S6. doi: 10.1186/1471-2105-13-S4-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Machné R., Finney A., Flamm C. The SBML ODE Solver Library: a native API for symbolic and fast numerical analysis of reaction networks. Bioinformatics. 2006;22:1406–1407. doi: 10.1093/bioinformatics/btl086. [DOI] [PubMed] [Google Scholar]
- 36.Nordsletten D.A., Yankama B., Dewey C.F., Jr. Multiscale mathematical modeling to support drug development. IEEE Trans. Biomed. Eng. 2011;58:3508–3512. doi: 10.1109/TBME.2011.2173245. [DOI] [PubMed] [Google Scholar]
- 37.Dimmeler S., Assmus B., Zeiher A.M. Fluid shear stress stimulates phosphorylation of Akt in human endothelial cells: involvement in suppression of apoptosis. Circ. Res. 1998;83:334–341. doi: 10.1161/01.res.83.3.334. [DOI] [PubMed] [Google Scholar]
- 38.Li Y., Zheng J., Magness R.R. Effects of pulsatile shear stress on signaling mechanisms controlling nitric oxide production, endothelial nitric oxide synthase phosphorylation, and expression in ovine fetoplacental artery endothelial cells. Endothelium. 2005;12:21–39. doi: 10.1080/10623320590933743. [DOI] [PubMed] [Google Scholar]
- 39.Florian J.A., Kosky J.R., Tarbell J.M. Heparan sulfate proteoglycan is a mechanosensor on endothelial cells. Circ. Res. 2003;93:e136–e142. doi: 10.1161/01.RES.0000101744.47866.D5. [DOI] [PubMed] [Google Scholar]
- 40.Pahakis M.Y., Kosky J.R., Tarbell J.M. The role of endothelial glycocalyx components in mechanotransduction of fluid shear stress. Biochem. Biophys. Res. Commun. 2007;355:228–233. doi: 10.1016/j.bbrc.2007.01.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dekker R.J., van Soest S., Horrevoets A.J. Prolonged fluid shear stress induces a distinct set of endothelial cell genes, most specifically lung Krüppel-like factor (KLF2) Blood. 2002;100:1689–1698. doi: 10.1182/blood-2002-01-0046. [DOI] [PubMed] [Google Scholar]
- 42.Yamada S., Shiono S., Yoshimura A. Control mechanism of JAK/STAT signal transduction pathway. FEBS Lett. 2003;534:190–196. doi: 10.1016/s0014-5793(02)03842-5. [DOI] [PubMed] [Google Scholar]
- 43.Yee K.L., Weaver V.M., Hammer D.A. Integrin-mediated signalling through the MAP-kinase pathway. IET Syst. Biol. 2008;2:8–15. doi: 10.1049/iet-syb:20060058. [DOI] [PubMed] [Google Scholar]
- 44.Weber M., Hagedorn C.H., Searles C.D. Laminar shear stress and 3′ polyadenylation of eNOS mRNA. Circ. Res. 2005;96:1161–1168. doi: 10.1161/01.RES.0000170651.72198.fa. [DOI] [PubMed] [Google Scholar]
- 45.Kuchan M.J., Frangos J.A. Role of calcium and calmodulin in flow-induced nitric oxide production in endothelial cells. Am. J. Physiol. 1994;266:C628–C636. doi: 10.1152/ajpcell.1994.266.3.C628. [DOI] [PubMed] [Google Scholar]
- 46.Liu J., García-Cardeña G., Sessa W.C. Biosynthesis and palmitoylation of endothelial nitric oxide synthase: mutagenesis of palmitoylation sites, cysteines-15 and/or -26, argues against depalmitoylation-induced translocation of the enzyme. Biochemistry. 1995;34:12333–12340. doi: 10.1021/bi00038a029. [DOI] [PubMed] [Google Scholar]
- 47.Schwarz G., Droogmans G., Nilius B. Shear stress induced membrane currents and calcium transients in human vascular endothelial cells. Pflugers Arch. 1992;421:394–396. doi: 10.1007/BF00374230. [DOI] [PubMed] [Google Scholar]
- 48.Wedgwood S., Mitchell C.J., Black S.M. Developmental differences in the shear stress-induced expression of endothelial NO synthase: changing role of AP-1. Am. J. Physiol. Lung Cell. Mol. Physiol. 2003;284:L650–L662. doi: 10.1152/ajplung.00252.2002. [DOI] [PubMed] [Google Scholar]
- 49.Wang Y., Chang J., Chien S. Shear stress and VEGF activate IKK via the Flk-1/Cbl/Akt signaling pathway. Am. J. Physiol. Heart Circ. Physiol. 2004;286:H685–H692. doi: 10.1152/ajpheart.00237.2003. [DOI] [PubMed] [Google Scholar]
- 50.Davis M.E., Grumbach I.M., Harrison D.G. Shear stress regulates endothelial nitric-oxide synthase promoter activity through nuclear factor κB binding. J. Biol. Chem. 2004;279:163–168. doi: 10.1074/jbc.M307528200. [DOI] [PubMed] [Google Scholar]
- 51.García-Cardeña G., Fan R., Sessa W.C. Endothelial nitric oxide synthase is regulated by tyrosine phosphorylation and interacts with caveolin-1. J. Biol. Chem. 1996;271:27237–27240. doi: 10.1074/jbc.271.44.27237. [DOI] [PubMed] [Google Scholar]
- 52.Förstermann U., Pollock J.S., Murad F. Calmodulin-dependent endothelium-derived relaxing factor/nitric oxide synthase activity is present in the particulate and cytosolic fractions of bovine aortic endothelial cells. Proc. Natl. Acad. Sci. USA. 1991;88:1788–1792. doi: 10.1073/pnas.88.5.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fontana J., Fulton D., Sessa W.C. Domain mapping studies reveal that the M domain of hsp90 serves as a molecular scaffold to regulate Akt-dependent phosphorylation of endothelial nitric oxide synthase and NO release. Circ. Res. 2002;90:866–873. doi: 10.1161/01.res.0000016837.26733.be. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


