Preface
Hsp90 is an essential, abundant, and ubiquitous eukaryotic chaperone that has crucial roles in protein folding and modulates the activities of key regulators. The fungal Hsp90 interactome, which includes numerous client proteins such as receptors, protein kinases and transcription factors, displays a surprisingly high degree of plasticity that depends on environmental conditions. Furthermore, although Hsp90 levels increase following environmental challenges, Hsp90 activity is tightly controlled via post-translational regulation and an autoregulatory loop involving the heat shock transcription factor, Hsf1. In this review, we discuss the roles and regulation of Hsp90. We propose that Hsp90 acts as a biological transistor that modulates the activity of fungal signalling networks in response to environmental cues via this Hsf1-Hsp90 autoregulatory loop.
Introduction
Heat shock protein 90 (Hsp90) was first described amongst a defined set of heat shock proteins (HSPs) that are rapidly induced in fungal, plant and animal cells in response to acute thermal up-shifts1-5. This HSP induction, which underpins the molecular adaptation to thermal insults, represents the heat shock response that is ubiquitous across the bacterial, archaeal and eukaryotic kingdoms1-5. HSPs have been divided into families based on their molecular mass. The Hsp90/htpG, Hsp70/dnaK and Hsp60/groEL families tend to display strong evolutionary conservation from bacteria to humans6,7. Smaller HSPs, including Hsp42 and Hsp26 in the yeast Saccharomyces cerevisiae, display greater evolutionary divergence5,8. Most of these HSPs are protein chaperones, promoting folding and assembly of newly synthesized proteins, and degradation or repair of damaged proteins that have become dissociated or formed aggregates as a result of thermal or chemical stress.
The high evolutionary conservation of the heat shock response across the fungal kingdom is intriguing. This might not seem surprising as fungi occupy highly divergent environmental niches, where they can be exposed to dramatic thermal fluctuations. However, other adaptive responses, to osmotic, oxidative and cell wall stresses for example, have diverged substantially across the fungal kingdom9. Some key stress regulators have been conserved, but many upstream sensors and downstream transcriptional regulators have diverged substantially9. On this basis one might have expected the heat shock response to have diverged in fungi that inhabit thermally buffered niches, such as the clinically important pathogen C. albicans10, which is obligately associated with warm-blooded animals11. Nevertheless, this response is strongly conserved in C. albicans and heat shock adaptation is essential for its virulence10,12. This reflects the fact that the “heat shock” apparatus is essential for cellular adaptation to the subtle or gradual thermal transitions that organisms often experience in the wild, not just to the acute temperature up-shifts that experimentalists tend to examine in the laboratory13. In particular, the strong conservation of Hsp90 attests to the fundamental importance of the cellular functions executed by this essential chaperone in eukaryotic cells. Indeed, Hsp90 is essential for the growth and viability of evolutionarily divergent yeasts such as S. cerevisiae, Schizosaccharomyces pombe and Candida albicans even under normal growth conditions14-16.
Hsp90 is an essential component of the cytoplasmic Hsp90-Hsp70 chaperone network that promotes protein folding and refolding in eukaryotic cells. Hsp70 promotes the initial folding of some nascent polypeptides as they emerge from the ribosome. Some of these proteins are then passed to the Hsp90 machine which facilitates the later stages of their folding and, in some cases, maintains them in a near-native conformational state17. Indeed, Hsp90 has vital roles in the folding and maintenance of a specific subset of proteins in the fungal cell, termed “client proteins”.
As a direct consequence of its role in maintaining the structural integrity of its client proteins, Hsp90 is thought to generate “protein-folding reservoirs” that can buffer the phenotypic impact of mutations in client proteins, thereby facilitating evolutionary change18. Hence Hsp90 has acted as an evolutionary capacitor during eukaryotic evolution19-21. However, this review focuses on the impact of Hsp90 in cellular, rather than evolutionary timescales. We focus on the impact of temperature on the interactions of fungal Hsp90 with its client proteins. We suggest that, while acting as a capacitor in evolutionary timescales, Hsp90 acts as a biological transistor over cellular timescales by modulating the activities of key signalling networks in response to dynamic changes in environmental conditions.
The Hsp90 chaperone cycle
The structure of the Hsp90 chaperone and its conformational dynamics are the subject of recent elegant reviews17,22,23. Briefly, the Hsp90 protein has three domains and operates as a dimer24-26 (Figure 1). The ~180 residue carboxy-terminal domain mediates constitutive dimerization, whilst the amino-terminal domain of around 215 residues contains the ATP binding domain. These domains are separated by a ~260 residue central domain that mediates many Hsp90-client protein interactions. The central and amino-terminal domains are connected by a charged linker, mutations in which affect interactions with some client proteins and co-chaperones.
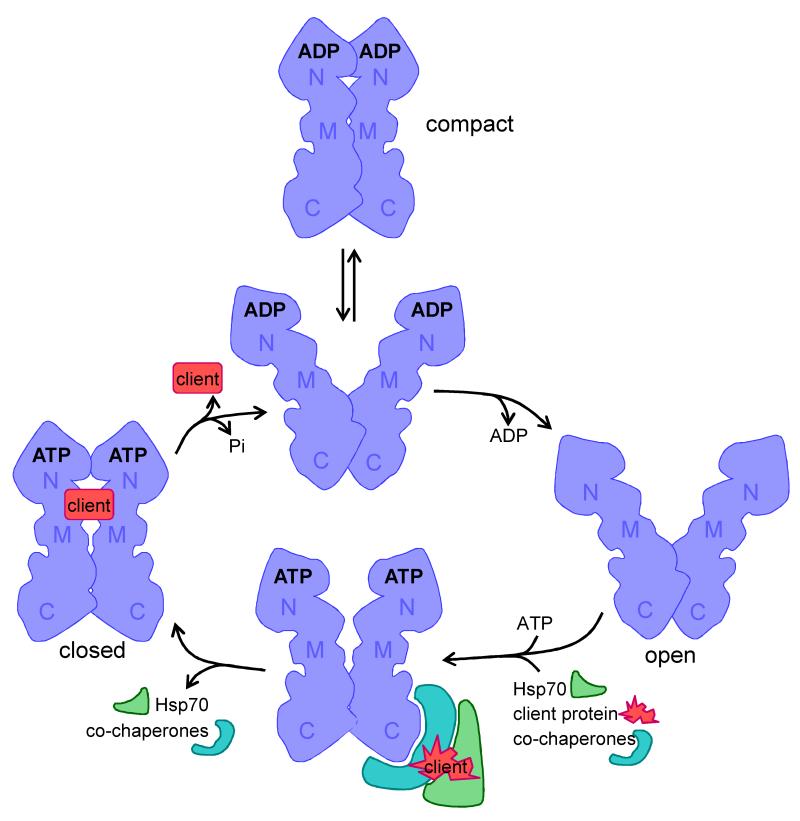
Figure 1. The Hsp90 chaperone cycle.
The shape of the Hsp90 monomer is adapted from Pearl et al. (2008)26, illustrating the amino- (N), middle (M) and carboxy-terminal (C) domains. The Hsp90 chaperone cycle has been reviewed recently17, 23. Hsp90 acts as a dimer that can take up various dynamic conformations in its ADP bound-state in which the amino terminal domains can be apart or closely associated (compact form). The Hsp90 dimer takes up an open state following release of ADP. The rapid association with ATP is associated with interactions with co-chaperones, Hsp70 and client proteins. Different co-chaperones can associate with different domains of the Hsp90 molecule and mediate interactions with distinct client proteins17,23. This is followed by the slow formation of a closed complex and the release of Hsp70 and co-chaperones. Then the folded client protein is released and the ATP is hydrolysed.
The flexible Hsp90 dimer undergoes major conformational shifts during a dynamic chaperone cycle that is driven by ATP hydrolysis17,23-26 (Figure 1). In the absence of ATP binding the dimer takes up an open V-shaped confirmation in which the two amino-terminal domains are separated and the two subunits are held together via their carboxy-terminal domains. ATP binding to the amino-terminal domain stimulates the closing of a lid over the nucleotide binding pocket, and the relatively slow subsequent formation of a closed form in which the two amino-terminal domains in the Hsp90 dimer associate closely together. Hsp90 has weak intrinsic ATPase activity that is modulated by interactions with client proteins and co-chaperones. After ATP hydrolysis, substantial remodelling occurs to regenerate open forms of the protein27. The ATPase cycle is relatively slow, yeast Hsp90 hydrolysing an ATP molecule every 1-2 minutes28,29. This conformational cycle differs between species, but remains crucial for the maturation of client proteins27,30. The classical pharmacological inhibitors of Hsp90, geldanamycin and radicicol, dock at the ATP binding site in the amino-terminal domain17,31, providing useful tools for the dissection of Hsp90 function.
Hsp90 is regulated at multiple levels
Transcriptional control of HSP90
Hsp90 is naturally abundant in fungal cells and is induced to even greater levels by heat shock and other proteotoxic stresses10,15,32-35. Hsp90 protein levels are regulated at transcriptional and post-transcriptional levels (Figure 2).
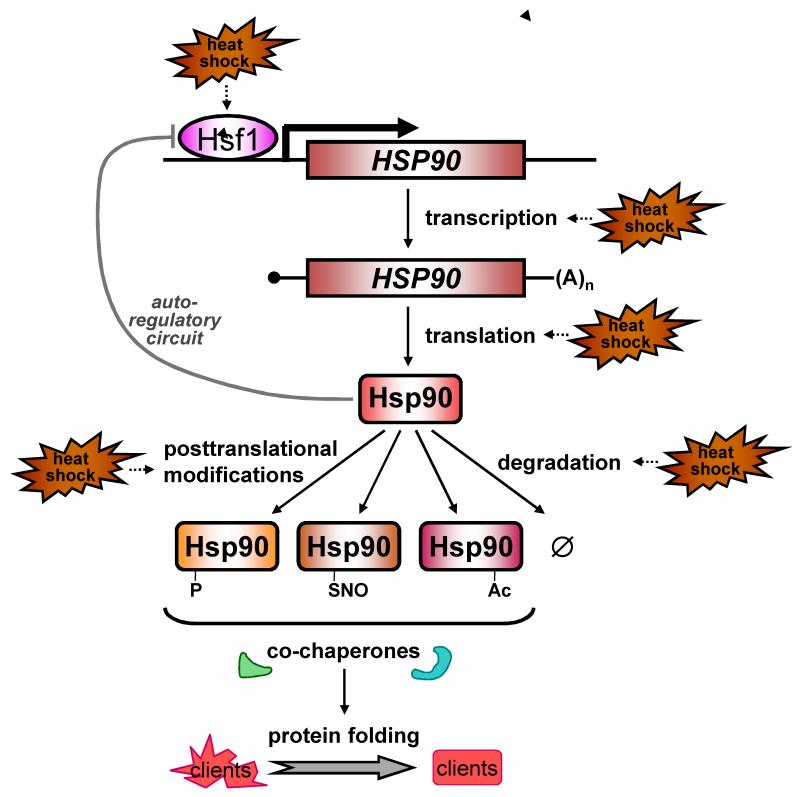
Figure 2. Hsp90 levels and activity are regulated at multiple levels.
(see text). Hsp90 transcription is regulated by the heat shock transcription factor (Hsf1), the activation of which is negatively regulated by Hsp90 via an autoregulatory loop13 that involves a physical interaction between Hsp90 with Hsf1 (Leach et al. unpublished). During heat shock, yeast HSP mRNAs may be preferentially translated and Hsp90 turnover might also be modulated (see text). Hsp90 activity is controlled by posttranslational modifications such as phosphorylation, s-nitrosylation and possibly acetylation. These changes influence Hsp90 interactions with specific co-chaperones, and hence affect the folding of specific subsets of client proteins. Furthermore, after proteotoxic stress changes in Hsp90 availability, mediated by altered HSP90 expression and changes in the amount of Hsp90 associated with unfolded proteins, is predicted to affect interactions with Hsp90 client proteins13.
HSP90 transcription is controlled by the heat shock transcription factor HSF, a key regulator of the heat shock response36,37 that is evolutionarily conserved from S. cerevisiae (Hsf1) to mammals (HSF1/2). Hsf1 is essential for viability in yeasts10,38,39, and is required for the basal expression of HSP genes10,39,40. It acts as a trimer, binding constitutively to heat shock elements (HSEs) in the promoters of HSP genes41-44. Hsf1 is activated by hyperphosphorylation in response to specific environmental cues, which in some fungi include glucose starvation, superoxide, oxygen tension and changes in membrane lipid composition as well as heat shock10,39,45,46. Hsf1 activation drives increased HSP gene transcription10,47,48, primarily via the carboxy-terminal activation domain of Hsf112,49, and this leads to the accumulation of HSPs37.
Notably, fungal Hsf1 is only temporarily activated on thermal upshifts10. Hsf1 is rapidly phosphorylated and then dephosphorylated after a 30-42°C heat shock in C. albicans13, suggesting regulation via a negative feedback loop. Almost three decades ago Lindquist2,50 and Didomenico et al.51 postulated that feedback components down-regulate the heat shock response. Initially, Hsp70 was thought to be the key HSF repressor in mammalian systems52-54, and Hsf1 interacts with Hsp70 family members in S. cerevisiae55. However, yeast Sse1 (an Hsp70) is required for Hsp90-dependent functions56, suggesting that Hsp90 is the Hsf1 repressor.
Pharmacological inhibition of mammalian Hsp90 correlates with HSF1 activation57. Furthermore, human HSF1 interacts physically with Hsp90 complexes57,58, and this is thought to repress HSF1 transcriptional activity. No such physical interaction between Hsf1 and Hsp90 has been demonstrated in the fungal kingdom. However, mutations that interfere with Hsp90 function derepress the expression of Hsf1-dependent reporter genes in S. cerevisiae59, and an Hsf1-Hsp90 autoregulatory loop has now been confirmed in C. albicans (Figure 2). Both pharmacological inhibition and genetic depletion of Hsp90 induces Hsf1 activation in C. albicans (Leach et al, unpublished). Furthermore, a physical interaction between Hsp90 and Hsf1 has been confirmed by co-immunoprecipitation (Leach et al, unpublished). Therefore, Hsp90-mediated repression of Hsf1 is released following heat shock, Hsf1 becomes activated, and this leads to a transcriptionally mediated increase in Hsp90 levels13. This response is then down-regulated when excess Hsp90 binds to Hsf1.
Post-transcriptional control of HSP90
In C. albicans, HSP90 transcript levels increase dramatically on thermal upshifts13, but Hsp90 protein levels do not increase to the same extent. Translational regulation in heat shocked Drosophila cells was reported by Lindquist three decades ago2. Translational regulation is not thought to occur in yeast following heat shock, the patterns of protein synthesis generally reflecting the dynamically changing mRNA populations during heat shock adaptation2. However, yeast heat shock mRNAs generally carry relatively unstructured 5′-leader regions that are less dependent on cap-dependent mechanisms of translation initiation60.
Hsp90 protein levels may also be regulated at the level of protein turnover. S. cerevisiae Hsp90 can be targeted for ubiquitin-mediated degradation by the cell cycle regulated protein kinase, Swe161. Hsp90 stability is probably regulated in C. albicans as Hsp90 levels decline rapidly as cells become adapted to elevated temperatures (Leach et al, unpublished). Clearly, Hsp90 protein levels are tightly regulated at multiple levels.
Post-translational modification of Hsp90
Hsp90 activity is regulated by post-translational modification. It has been known for some time that Hsp90 is phosphorylated in mammalian cells62,63. Similarly, phosphorylation of S. cerevisiae Hsp90 is thought to influence its activity and hence the rate of maturation of specific client proteins64,65. Yeast Hsp90 is phosphorylated by Swe1 and casein kinase 2 (CK2), and is dephosphorylated by Ppt161,64-66. CK2 also phosphorylates Hsp90 in C. albicans67. However, the relationship between Hsp90 phosphorylation and activity is complex. For example, Hsp90 hyperphosphorylation, induced by inactivation of the phosphatase Ppt1, leads to a reduction in the activity of the Hsp90 chaperone system64. Meanwhile, blocking the Swe1-mediated phosphorylation of Hsp90 at Tyr24, which normally occurs in a cell cycle dependent fashion, inhibits Hsp90 interactions with a specific subset of co-chaperones (Aha1) and client proteins (Ste11 and Slt2)61. Also, phosphorylation at Thr22 by casein kinase 2 modulates Hsp90 interactions with the co-chaperones Cdc37 and Aha165. In addition, yeast Hsp90 is S-nitrosylated, which affects its dimerization dynamics and activity68, and by analogy with its mammalian counterpart69, yeast Hsp90 might also be acetylated.
Co-chaperone modulation of Hsp90 function
Hsp90 activity is further regulated by its interactions with various co-factors (or co-chaperones), which influence the binding specificity of Hsp90 for particular client proteins. An association with a specific co-chaperone is thought to fix Hsp90 in a specific open conformation that helps to establish the binding specificity for that Hsp90 molecule and its chaperone complex23,70-73.
A range of Hsp90 co-chaperones have been identified in yeast, most of which are conserved in mammals (Table 1). Hsp70 and Hsp40, the other members of the major cytoplasmic chaperone network, have been described as Hsp90 co-chaperones. Hsp70 associates with Hsp90 via the adapter protein Sti1/Hop174 to form the minimal Hsp90 core complex75. Sti1/Hop1 inhibits the ATPase activity of Hsp9075, as does the co-chaperone Cdc37/p5070. By contrast, Aha1 enhances the ATPase activity of Hsp9072,76, and Sba1/p23 couples this ATPase activity to polypeptide release77. The binding of Sba1/p23, Cdc37/p50 and Aha1 to Hsp90 are thought to be mutually exclusive26. Additional yeast co-chaperones include Tah1 and Pih178, Cns179,80, Sgt181, and Cpr6 and Cpr7 which are homologues of mammalian Cyp4082.
Table 1. Hsp90 Co-chaperones.
| Co-chaperone1 | Essential2 | Function |
|---|---|---|
| Sti1 (Hop1) | Non-essential | Binds to the N- and C-terminal of Hsp90, preventing the N-terminal dimerization reaction required for ATP hydrolysis140 |
| Aha1 (Aha1) | Non-essential | Enhances Hsp90 ATPase activity72,76 |
| Cdc37 (p50) | Essential | Links Hsp90 to client protein kinases70 |
| Pih1 | Non-essential | Interacts with Hsp90 and DNA helicases78 |
| Cns1 (TTC4) | Essential | Interacts with Hsp90 C-terminus141 |
| Tah1 | Non-essential | Interacts with Hsp90 C-terminus and DNA helicases78 |
| Sgt1 (Sgt1) | Essential | Binds non-ATP bound forms of Hsp90, linking Hsp90 to client proteins81 |
| Sba1 (p23) | Non-essential | Stabilises the ATP bound state of Hsp9077 |
| Cpr6/7 (Cyp40) | Non-essential | Enhances Hsp90 ATPase activity82 |
(Name of mammalian orthologue)
Essential for viability in S. cerevisiae
The interaction of Hsp90 with these various co-chaperones is thought to impose distinct architectures on the Hsp90 complex, and this in turn drives interactions with different client proteins23,26. For example, Cdc37/p50 mediates interactions with protein kinases in yeast and mammalian cells83,84. By contrast, Sgt1 contributes to kinetochore assembly85 whilst Tah1 and Pih1 are involved in chromatin remodelling and epigenetic regulation78,86. Zhao and coworkers also demonstrated that inactivation of Tah1 and Pih1 impairs the maturation of client proteins in S. cerevisiae78. This illustrates the breadth of cellular processes that the Hsp90 chaperone complex supervises87. Consequently mammalian and fungal Hsp90s have become major foci for the development of anti-cancer and antifungal drugs88-90.
The Hsp90 interactome
The global Hsp90 interactome (the subset of cellular proteins that interacts with Hsp90) has been defined in fungi and mammalian cells using a range of unbiased, genome-wide approaches. The elaboration of the fungal Hsp90 interactome has combined the proteomic identification of components of tandem affinity purified complexes with molecular two-hybrid-based screens for Hsp90 interacting proteins, genetic screens for mutations that confer hypersensitivity to Hsp90 inhibitors such as geldanamycin and Macbecin II, and screens for mutations that confer synthetic genetic phenotypes in combination with a temperature sensitive hsp90 mutation (hsp82G170D)67,78,91-93. The fungal interactome has been experimentally defined in S. cerevisiae78,92, S. pombe94 and C. albicans67. These data have been merged with experimental Hsp90 interaction datasets from plants, nematodes, flies and mammals to generate a conceptual human Hsp90 chaperone machine database(www.picard.ch/Hsp90Int)95.
The Hsp90 interactome is large, comprising about 10% of the proteome (Figure 3). This is hardly surprising given that Hsp90 is a central component of a large molecular machine and that the role of this machine is to promote the folding of numerous client proteins. Nevertheless, the evidence suggests that, despite the breadth of the Hsp90 interactome, it comprises a specific subset of cellular proteins. For example, the Hsp90 interaction network displays distinct topological features, such as its high density and connectivity that reflect the underlying biological connections of this biological machine95. The re-identification of Hsp90 co-chaperones and cofactors in the various unbiased global screens, that have been performed attests to the validity of their biological outputs67,78,91-93. Furthermore, the validity of numerous Hsp90 client proteins identified in genome wide screens has been confirmed experimentally67,78,92. However, important differences do exist between the Hsp90 clientele of S. cerevisiae and C. albicans67, suggesting considerable evolutionary plasticity in the fungal Hsp90 interactome.
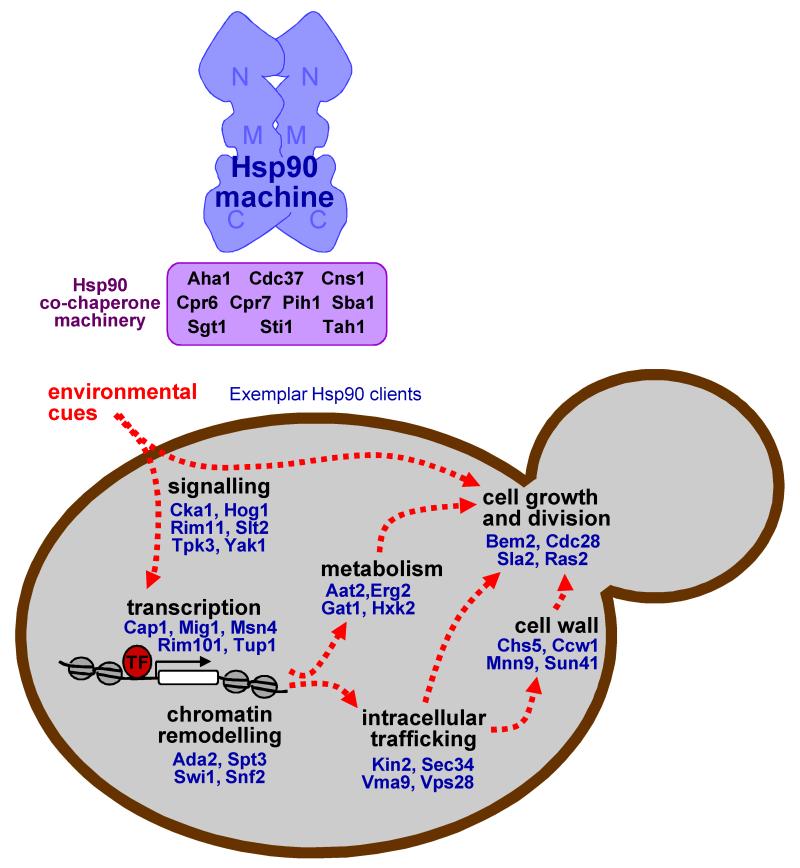
Figure 3. The fungal Hsp90 chaperone machine and exemplar client proteins.
The shape of the Hsp90 machine is adapted from Pearl et al. (2008)26, illustrating the amino- (N), middle (M) and carboxy-terminal (C) domains. The specificity of an Hsp90 complex depends on which co-chaperone it binds. Co-chaperones can bind different faces of Hsp90. Examples of Hsp90 client proteins that are representative of the major cellular processes that Hsp90 influences are illustrated in blue.
Members of this Hsp90 interactome are divisible into two main classes: Hsp90 client proteins, and the Hsp90 co-chaperones and cofactors that promote the conformational integrity of these client proteins. There is considerable overlap between fungal and mammalian cells with regard to the categories of Hsp90 client proteins that have been defined to date. The two main categories, protein kinases/phosphatases and transcription factors, are clearly ubiquitous in eukaryotic systems95 (Supplementary Figure). The third category is less well defined, but is also conserved. This category clusters structurally unrelated mammalian Hsp90 client proteins that include viral-replication proteins and receptors involved in innate immunity26. In fungal systems, this category probably includes Hsp90 clients involved in secretion, vesicular transport and mitochondrial membrane components78,92. It is significant that fungal Hsp90 client proteins are highly enriched for regulators that have crucial roles in the control of growth, cell division, environmental adaptation and development (Figure 3). A more comprehensive listing of Hsp90 client proteins is provided in the Supplementary Figure, and examples of these three categories of Hsp90 client proteins are discussed below. Given the focus of our review on the modulation of cell signalling by Hsp90, our emphasis here is on protein kinases and phosphatases.
Kinases/phosphatases as clients
The protein phosphatase calcineurin (Cna1) is an Hsp90 client protein in 5 yeasts96,97. Calcineurin has a major role in calcium signalling in eukaryotic systems. In yeasts, calcineurin signalling contributes to antifungal drug tolerance, with inhibitors such as cyclosporin and FK506 dramatically enhancing the sensitivity of yeasts to antifungal drugs such as fluconazole and caspofungin98,99. More recently, genetic or pharmacological attenuation of Hsp90 function in S. cerevisiae97,100 and C. albicans96,101 was shown to block the activation of calcineurin. Furthermore, a physical interaction between Hsp90 and calcineurin has been demonstrated in both yeasts96,97, thereby confirming that this important regulatory molecule is an Hsp90 client protein. Calcineurin is stabilized by Hsp90 and becomes activated following its dissociation from the chaperone97. This is important from a therapeutic point of view because Hsp90 inhibitors provide a potent mechanism for increasing the sensitivity of pathogenic fungi, including C. albicans, Aspergillus fumigatus and Aspergillus terreus, to clinically important antifungal drugs20,102.
The S. cerevisiae mitogen activated protein (MAP) kinase Slt2, which is an essential component of the cell integrity signalling pathway, is an Hsp90 client protein91. Hsp90 interacts with the phosphorylated, activated form of Slt2. This interaction is essential for Slt2-mediated activation of the transcription factor Rlm191, which regulates key cell wall biosynthetic enzymes required for the maintenance of cell wall integrity when cells are exposed to cell wall stresses such as antifungal drugs103,104. This chaperone-client relationship has been evolutionarily conserved in C. albicans. The C. albicans orthologue of Slt2, Mkc1, also is regulated by Hsp90, with depletion of this chaperone leading to destabilization of this crucial MAP kinase and inactivation of the cell integrity signalling pathway101. Therefore, the increased sensitivity of yeasts to antifungal drugs imparted by Hsp90 inhibitors is mediated by down-regulation of Mkc1 and cell integrity signalling, as well as through inhibition of calcineurin signalling.
The highly conserved stress activated protein kinase, Hog1/Sty1, has crucial roles in stress adaptation in S. cerevisiae, S. pombe and C. albicans32,105-109, and in C. albicans, this key stress regulator contributes to virulence110. Therefore, the fact that Hog1 has been identified as an Hsp90 client protein67,111 is important in terms of fungal stress adaptation and pathogenicity. It infers that Hsp90 might modulate the activity of this key stress signalling pathway. However, whilst Sty1 is activated by mild heat shocks in S. pombe112, Hog1 signalling decreases following equivalent up-shifts in C. albicans108. Therefore Hsp90 might differentially modulate Hog1/Sty1 activity in these evolutionarily divergent yeasts.
Hsp90 also contributes to cell cycle regulation. In S. cerevisiae, the Hsp90 co-chaperone Cdc37, as well as Hsp90 itself modulates the function of the crucial cell cycle regulator, Cdc2861,113. Furthermore, in C. albicans, this cyclin-dependent kinase interacts physically with, and is stabilized by Hsp90114. Importantly, cell cycle and morphogenesis are tightly regulated, and Hsp90 has been implicated for the thermal regulation of morphogenesis115, a key virulence factor in this pathogen.
Transcription factors as clients
The Hsp90 interactome includes numerous transcription factors and chromatin remodelling components67,78,91-93,116 (Supplementary Figure). Tah1 and Pih1 were identified through genome wide screens as novel Hsp90 co-chaperones in yeast, and their interactions with Hsp90 have been confirmed experimentally78. Tah1 and Pih1 promote Hsp90-mediated maturation of the glucocorticoid receptor in yeast (a model Hsp90 substrate), of Rvb1 and Rvb2, which are essential components of the yeast Ino80 chromatin remodelling complex (which modulates the expression of about 5% of yeast genes) and the folding of subunits of the SWR-C chromatin remodelling complex (which controls gene expression close to heterochromatic regions)78. The list of Hsp90 clients also includes numerous S. cerevisiae and C. albicans transcription factors that are involved in transcriptional reprogramming in response to environmental cues (Supplementary Figure)67, 78, 91-93, 116, reinforcing the view that Hsp90 underpins fungal adaptation and pathogenicity.
Other types of client
The genome wide screens for Hsp90 interacting proteins revealed other types of Hsp90 client protein involved in processes such as secretion and intracellular trafficking, the cell wall and metabolism67,78,91-93,116 (Figure 3, Supplementary Figure). Indeed McClellan and co-workers have reported that of the 25 S. cerevisiae deletion mutants that displayed the greatest sensitivity to the Hsp90 inhibitor macbecin II at 30°C, fourteen carried mutations in transport-related genes that included numerous vacuolar protein sorting genes and the four lobe B components of the bilobular conserved oligomeric Golgi complex92. Consistent with this, Hsp90 inhibition was shown to reduce secretion without affecting glycosylation, suggesting that Hsp90 influences the functions of proteins involved in the secretory pathway92. These findings indicate that Hsp90 also has an impact on the fungal cell surface and thereby is likely to affect interactions between fungal pathogens and their hosts.
Plasticity of the Hsp90 interactome
Clearly, Hsp90 interacts specifically with a range of key cellular regulators and modulates their activities under normal conditions, even in the absence of an environmental stress. However, the various genomic characterizations of the fungal Hsp90 interactome have highlighted a second important point – the fungal Hsp90 interactome displays input-dependent plasticity, i.e. the Hsp90 interactome responds to environmental conditions.
The various proteomic, genetic and chemical genomic screens have provided the notion of a robust fungal Hsp90 interactome67,78,91-93. This view is reinforced by the reproducible identification of key co-chaperones in these screens, the common functional categories that are substantially enriched in their outputs, and the fact that the resultant Hsp90 interaction networks display topological features that distinguish them clearly from randomly selected networks95. However, the overlap between these experimentally determined Hsp90 interaction networks is limited when the growth conditions are changed. For example, substantial differences are observed between the Hsp90 interactomes of C. albicans cells grown at 30°C or 37°C67, with more protein kinases appearing in the interactome when cells are grown at elevated temperatures. Similarly, in S. cerevisiae more proteins involved in signal transduction, cell cycle, cytokinesis and budding are observed in the 37°C Hsp90 interactome compared with the 30°C interactome92. Analogous differences are observed under other stress conditions. For example, cell wall stresses strengthen the interactions between the Slt2 MAP kinase and Hsp90 in S. cerevisiae91, and encourage many new proteins to become Hsp90 clients in C. albicans67.
Clearly the Hsp90 interactome displays substantial environmental contingency67. Although considerable energy has been devoted to the characterization of the Hsp90 interactome, we suggest that much remains to be discovered about its environmental plasticity and in particular, the temporal dynamics of this plasticity. For example, the underrepresentation of functions associated with “stress response” and “protein folding” in the global Hsp90 interactome95 probably reflects the fact that most of the genome wide screens have been performed essentially under steady-state conditions. The true plasticity of the Hsp90 interactome is likely to be best observed under transient conditions, when the Hsp90 chaperone machine is contributing to dynamic cellular adaptation to environmental insults.
Hsp90 circuitry – dynamic behaviours
Our understanding of the organization of cellular networks and their dynamic behaviours can be substantially improved by quantitative analyses and mathematical modelling of these processes and their responses to external perturbations. Systems biology models have increased our understanding of, for example, the dynamics of the S. cerevisiae cell cycle117, the highly complex response of osmotically stressed cells that involves cell signalling, gene regulation and metabolic adaptation118, as well as the impact of osmotic stress on cell cycle progression119.
Given its essentiality and ubiquity, the heat shock response is a good candidate for mathematical modelling. At the network level, Mihalik and Csermely analysed the effects of heat shock on the organization of the yeast interactome revealing partial disintegration of the interactome in response to this stress120. In the absence of global protein-protein interaction datasets generated under heat shock conditions, these authors exploited transcriptomic datasets to show that during heat shock there is a substantial decrease in the degree of overlap and the frequency of connections between the modules of the yeast interactome. For example, two central ribosomal modules, which reflect the major role of protein synthesis in unstressed growing yeast cells, display decreased community centrality in the interaction network after heat shock. By contrast the metabolism module retains its central position in the network. Whilst the general connectivity of network modules decreases during heat shock, heat shock proteins contribute to the integration of this partially decoupled interaction network, reflecting their major influence on protein function under these conditions120.
Ebong and colleagues analysed the kinetics of Hsp90 complex formation with its chaperones121. Mass spectrometric analyses of complexes formed in vitro by recombinant human Hsp90, Hsp70, Hop, and FKBP52 allowed the authors to construct an interaction network for these factors and to predict dominant complexes that are formed during the Hsp90 cycle. The relative similarity of the binding constants for the species studied supported the view that the Hsp90 chaperone machinery is complex and dynamic121.
Also, the role of Hsp70 and Hsp90 in protein homeostasis has been modelled dynamically122. This study examined the relationships between Hsp90, Hsp70, JNK and p38 in the context of neurodegeneration. The simulations suggested that following the imposition of a proteotoxic stress, protein homeostasis can be maintained for short periods, but that in the longer term the chaperone system becomes overwhelmed leading to an increased probability of protein aggregation and cell death122.
More recently Leach and colleagues13 used an ODE (ordinary differential equation) model to investigate the dynamics of the Hsf1-Hsp90 autoregulatory loop (Figure 2). The model focuses on the Hsp90-Hsf1 interaction because the available data suggest that Hsp90 is the main chaperone that represses Hsf113,59. In this model, Hsp90 interacts with and represses Hsf1. After heat shock, Hsp90 becomes sequestered in complexes with unfolded and damaged proteins, and therefore is less available temporarily for complex formation with Hsf1. As a result, the released Hsf1 becomes activated and induces HSP90 gene transcription, thereby leading to the production of more Hsp90. Ultimately excess Hsp90 rebinds Hsf1, blocking further transcriptional activation13. Therefore, this model predicted that alterations in ambient temperature lead to changes in the concentration of free Hsp90 and influenced the interactions of Hsp90 with one of its client proteins, Hsf1. Aspects of this model have been confirmed in vivo12 and Hsf1 has now been shown to be an Hsp90 client protein (Leach et al, unpublished). Given the observed environmental plasticity of the Hsp90 interactome67,92, thermal fluctuations are also likely to influence the interactions of Hsp90 with other client proteins.
In principle, how might changes in ambient temperature affect the Hsp90 interactome? As described above, the binding specificities and affinities of the Hsp90 chaperone machine for its client proteins are driven by its co-chaperones. However, for the purpose of this discussion, let us simplify things by considering merely that Hsp90 displays differing affinities for different client proteins. As described above, Hsp90 abundance increases following heat shock (and in response to other environmental insults). However, the abundance of free Hsp90 that is available for interactions with client proteins declines transiently as Hsp90 becomes sequestered in complexes with unfolded proteins. Modelling of a simple conceptual Hsp90 interactome (Figure 4), in which two client proteins (A and B) are present at differing levels, is sufficient to demonstrate that changes in Hsp90 availability will substantially affect its interactions with these client proteins (Figure 4C).
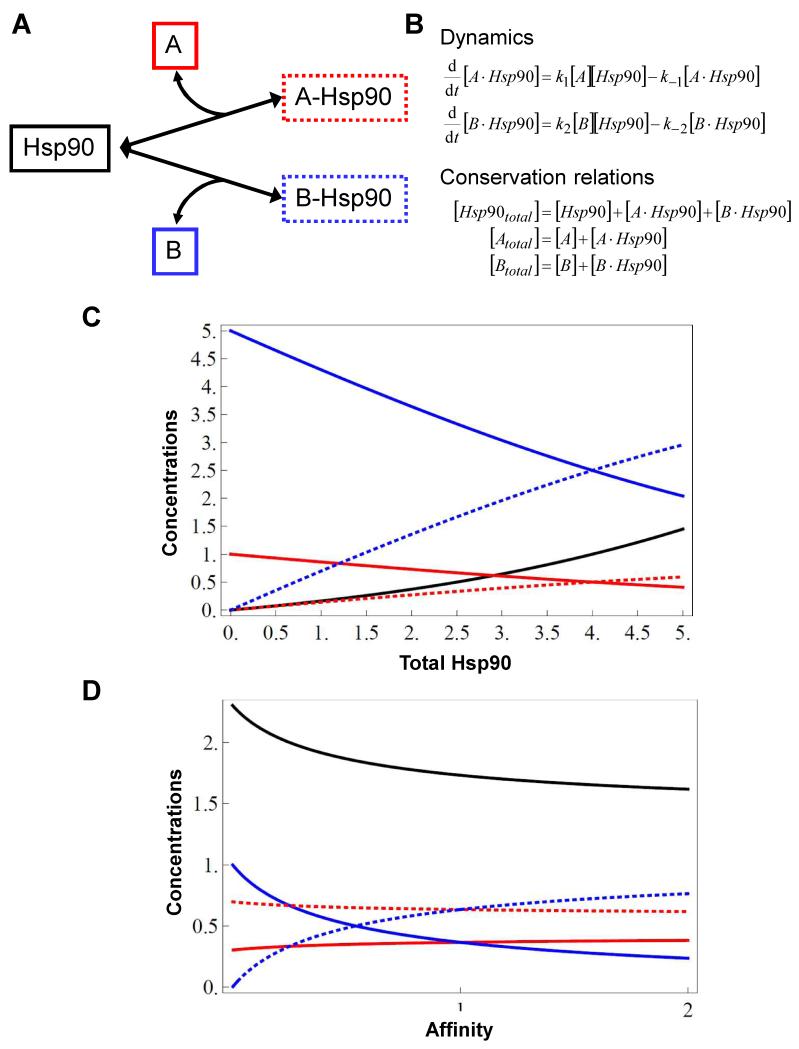
Figure 4. Alterations in Hsp90 concentrations are predicted to differentially affect interactions with specific Hsp90 client proteins.
(A) A simple conceptual network illustrating the interactions between Hsp90 (black) and two distinct client proteins, A (red) and B (blue). The solid boxes surround the free forms of these proteins, and the dotted boxes surround the A-Hsp90 and B-Hsp90 client-chaperone complexes. (B) Equations describing the dynamic relationships between these proteins and complexes in this conceptual network, and their conservation in the network. In steady state, set the two dynamic equations equal to zero. (C) Graph showing the impact of changing total Hsp90 levels on the concentrations of the free and Hsp90-bound forms of the client proteins A and B. In this case the affinities of Hsp90 for A and B are the same, but the total concentration of A is five-fold lower than B. Clearly Hsp90 availability influences the proportions of A and B that are incorporated into Hsp90 complexes. (D) Graph depicting the effects of altering the affinity of Hsp90 for B (from 0 to 2), whilst the affinity of Hsp90 for A remains constant (=1) (p1 = k1/k−1 = 1; p2 = k2/k−2 = 0…2; [Atotal] = 1; [Btotal] = 1). As the affinity of Hsp90 for B increases, the concentration of the B-Hsp90 complex increases (dotted blue line) and the levels of free B (solid blue line) and free Hsp90 (solid black line) decrease. By contrast, the concentration of the A-Hsp90 complex remains relatively unaffected (dotted red line). Hence the proportions of A and B that are chaperoned by Hsp90 change as a result of this altered affinity for one of the client proteins. Solid and dotted red, blue and black lines correspond to those in (A).
Let us then, for the sake of discussion, make the likely presumptions that Hsp90 displays differing affinities for different client proteins, and that changes in ambient temperature affect the affinity of Hsp90 for some client proteins more than others. In this case, our model of this simple Hsp90 interactome shows that an increase in the affinity of Hsp90 for client protein B will result in a substantial increase in the proportion of B that complexes with Hsp90 relative to client protein A (Figure 4D). If Hsp90 binding activates these client proteins, this increased affinity for B will result in a dramatic shift from a situation where pathway A is more active than pathway B, to a new situation where B is more active than A.
Therefore, modelling of this simple Hsp90 interactome indicates that the kinetics of binding between Hsp90 and its client proteins is likely to be substantially affected by changes in Hsp90 abundance, binding affinities, relative concentrations and on/off rates. Clearly thermal fluctuations will exert dramatic effects on the Hsp90 interactome, having an important role in the dynamic competition between binding partners. Three major aspects are relevant here: the relative affinity of client and unfolded proteins for Hsp90; the relative abundance of these different ligands; and whether specific client proteins are activated or inhibited through their interactions with Hsp90. We suggest that Hsp90 essentially acts as a biological transistor, tuning the activities of key signalling pathways in response to thermal inputs and other proteotoxic environmental cues.
Cellular consequences
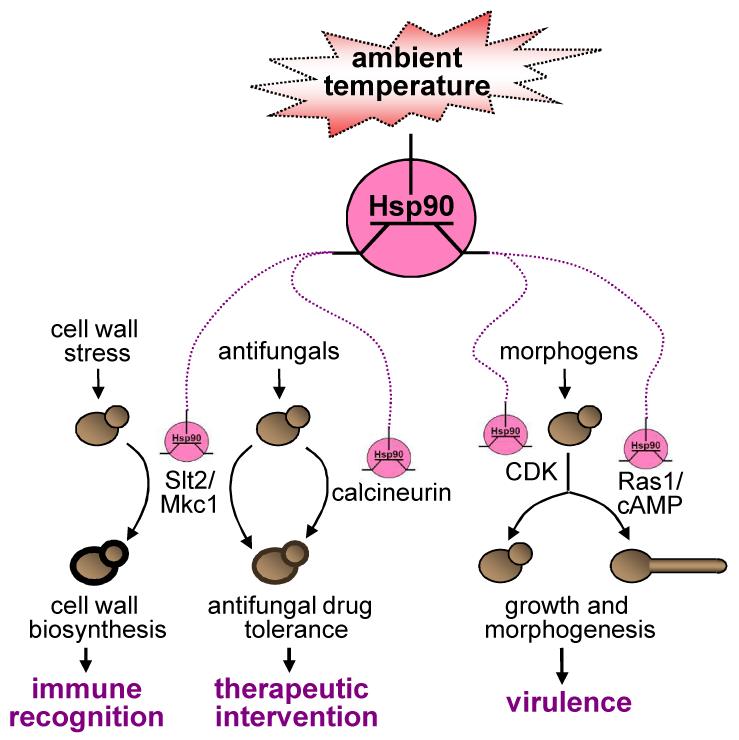
What are the likely cellular consequences of Hsp90 modulating the activity of global signalling networks in response to changes in ambient temperature through differential client interactions? Thermal modulation of Hsp90 availability promotes the activation of some signalling pathways in fungal cells. Hsp90 regulates morphogenesis in C. albicans by repressing Ras1-PKA signalling at relatively low temperatures115. At elevated temperatures (circa 37°C) this Hsp90 repression is relieved, leading to filamentation. Hence, through the Hsp90 transistor, changes in ambient temperature influence cellular morphology, a key virulence determinant in this pathogen (Figure 5).
Figure 5. Hypothesis: the Hsp90 transistor tunes multiple cellular outputs in response to thermal input.
Thermal fluctuations influence Hsp90 availability and probably the affinity of the Hsp90 chaperone machine for certain client proteins (see text). Hsp90 modulates the activity of many client proteins, which include regulators in key signalling pathways. Hence the Hsp90 chaperone machine is proposed to act like a transistor that modulates the activity of these signalling pathways in response to thermal (and other proteotoxic) inputs. As a result, temperature modulates cell division, adaptation, growth and morphogenesis through the Hsp90 transistor.
Hsp90 also has evolutionarily conserved roles in cell cycle progression. Several key cell cycle regulators have been identified as Hsp90 interactors in S. cerevisiae, including Swe1, Cdc28, Cdc50 and Cdc6061,92,123. Furthermore, Hsp90 has recently been implicated in cell cycle progression in C. albicans. Cell cycle arrest is believed to promote filamentation in response to compromised Hsp90 function; Cdc28 has been identified as an Hsp90 client, and key effectors for morphogenesis induced by compromised Hsp90 or elevated temperature include Pho85 and Pcl1 providing possible links between Hsp90, cell cycle regulation and morphogenesis114,124.
Hsp90 also has a profound impact on responses to drug-induced cellular stress, in this case through activation of other signalling pathways. For example, cell integrity and Ca++-calmodulin signalling are down-regulated following Hsp90 depletion via destabilization of Slt2/Mkc191,101 and calcineurin96, respectively. These pathways play major parts in antifungal drug tolerance, and the impact of Hsp90 on this phenotype has been well characterized in both C. albicans and S. cerevisiae. For example, Hsp90 depletion phenocopies the azole sensitivity of calcineurin and protein kinase C mutants96,101. Importantly, elevated temperatures also recapitulate the antifungal drug sensitivity caused by Hsp90 inhibition20. Therefore, thermal upshifts down-regulate antifungal drug resistance via the effects of the Hsp90 transistor on cell integrity and Ca++-calmodulin signalling (Figure 5).
Given the impact of the cell integrity and Ca++-calmodulin pathways on the C. albicans cell wall, a clear prediction is that ambient temperature will also affect the architecture of the cell surface and hence immune recognition by the host (Figure 5). Indeed, C. albicans cells grown at 37°C versus 25°C contain longer mannans125. Mannans from C. albicans cell wall have the ability to stimulate cytokine production126,127, and induce dendritic cell maturation128.
These effects of Hsp90 on cellular processes will not be restricted to acute heat shocks. Mathematical modelling has strongly suggested that cells are constantly tuning Hsp90 levels to subtle changes in ambient temperature. Hence it is highly likely that the Hsp90 transistor is constantly tuning the activity of the cellular signalling network to subtle thermal fluctuations. Furthermore, the Hsp90 transistor probably modulates fungal signalling networks in response to other proteotoxic stresses that influence Hsp90 availability.
Conclusions
In conclusion, Hsp90 has a central role in tuning cellular outputs to thermal inputs, discussed here primarily in the context of yeast species. However, as Hsp90 is one of the most ancient and highly conserved regulators of cellular signalling, this is likely to have broad relevance across the eukaryotic kingdom. There is some plasticity in the Hsp90 chaperone machine, though key co-chaperones are conserved across all eukaryotes87. Also, many features of the Hsp90 interactome are highly conserved from yeast to humans despite substantial network rewiring over evolutionary time. These conserved features include a preponderance of protein kinases and transcription factors as client proteins67,78,91-93. As a consequence of its extensive connectivity in interaction networks and its profound impact on signal transducers, the Hsp90 transistor tunes physiological responses to environmental conditions.
Hsp90 also modulates the phenotypic effects of genetic variation thereby influencing evolution. By governing the activation of diverse regulators of cellular signalling, Hsp90 can influence the phenotypic effects of genetic variation in an environmentally responsive manner in at least two distinct ways. First, Hsp90 can buffer the effects of genetic or epigenetic variation, keeping it silent until released when the chaperone reservoir is compromised in response to stress. This role as a capacitor for variation has been observed in diverse eukaryotes including flies, plants, and fungi18,19,129-134. Second, Hsp90 can enable the phenotypic effects of new mutations either by stabilizing mutant regulators or by stabilizing non-mutant regulators that mediate signalling required for adaptation. This role as a potentiator for genetic variation has been observed in fungi as well as mammalian cancer cells20,88,135,136. A recent study exploiting the genetic tractability of S. cerevisiae, revealed that Hsp90 influences approximately 20% of natural genetic variation serving both to maintain phenotypic robustness and promote diversification18. Furthermore, the link between Hsp90, Hsf1 and environmental stress has been reinforced by the recent observation that stimulation of a stress response either by heat shock or increased expression of HSF-1 reduces the phenotypic effects of mutations in the nematode Caenorhabditis elegans137.
Thus, Hsp90 has a multitude of impacts on cellular circuitry: as a capacitor that buffers genetic variation until released by stress; as a potentiator that enables the phenotypic effects of new mutations; and as a transistor that tunes cellular outputs to environmental inputs. This illustrates the stunning complexity of ways in which stress response circuitry orchestrates adaptation on both physiological and evolutionary timescales.
An elegant combination of molecular, genetic and genomic approaches has dramatically increased our understanding of Hsp90 as an evolutionary capacitor. However these approaches have only provided glimpses of the role of Hsp90 as a cellular transistor. Understanding this role represents a major challenge for the future. How does the Hsp90 chaperone machine modulate the functions of the key cellular regulators that drive physiological adaptation during the minutes that follow an environmental challenge? To address this question we need high throughput biochemical, biophysical and mathematical tools to define and understand the short term dynamism of the Hsp90 interactome as well as its long term plasticity.
Supplementary Material
Text Box 1. Modelling the structure and dynamics of regulatory networks.
Systems biologists use different modelling approaches, depending on the specific scientific question they wish to address, the particular system of interest, the nature and amount of available experimental data, and simply taste (for an introduction138). Network oriented approaches enumerate compounds such as proteins or metabolites and analyse the type and frequency of their interactions. This allows the identification of patterns or statistical correlations that are not obvious from analyses of the individual components. By contrast, dynamic models are usually based on the characterization of the network and its interactions, describing their changes over time. These changes might arise, for example, through inherently dynamic processes such as cell cycle or circadian rhythms, or as a result of external perturbation of the system, such as environmental stresses or nutrient changes. An important class of these models describes the dynamics of their components using systems of ordinary differential equations (ODE), i.e.
where xi with i = 1,..,n are the concentrations of compounds and complexes, fi are the (usually non-linear) functions describing their changes over time, which can be represented as the sum over stoichiometric coefficients nij multiplied by the rates of the individual reactions, vj, with j = 1,..,r. The rates depend on current compound concentrations and parameter values, pl, such as kinetic constants, binding constants, maximal rates, or Hill coefficients.
Although confidence about the wiring of networks increases through, for example, high-throughput or dedicated protein-protein interaction studies or through metabolic reconstructions139, the precise form of rate expressions and the values of kinetic parameters are often elusive, and their choice depends on the available experimental data and laborious parameter estimation exercises. Achieving the goal of models that are firmly based on quantitative data and that have predictive value, is hindered by limitations in the ability to describe the whole cell and all its multifarious levels of regulation. Thus, defining the boundaries of the system being modelled, necessitates assumptions about some processes, and simplification of the system whilst considering the major players relevant to the scientific question. Descriptions are frequently restricted to compounds that are amenable to experimental quantification and, hence, reactions and regulatory steps between them are often compressed into single equations.
Acknowledgements
We are grateful to numerous colleagues, and Joe Heitman in particular, for insightful discussions. MDL is a Sir Henry Wellcome Postdoctoral Fellow (Wellcome Trust 096072). EK is supported by grants from the German Research Council (GRK 1772, SFB618, SFB740), German Ministry of Education and Research (0315786A, 0315584B) and European Commission (FINSysB, PITN-GA-2008-214004, Unicellsys HEALTH-2007-201142). LEC is supported by a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund, a Canada Research Chair in Microbial Genomics and Infectious Disease, a Ministry of Research and Innovation Early Researcher Award, a Natural Sciences & Engineering Research Council Discovery Grant # 355965, and a Canadian Institutes of Health Research Grants MOP-86452 and MOP-119520. AJPB is supported by grants from the UK Biotechnology and Biological Research Council (BB/D009308/1; BB/F00513X/1), Wellcome Trust (080088; 097377) and European Commission (FINSysB, PITN-GA-2008-214004; STRIFE, ERC-2009-AdG-249793).
Glossary
- Proteotoxic stress
Cellular stress conditions that prompt the accumulation of unfolded or damaged proteins, or the formation of protein aggregates.
- Heat shock elements (HSEs)
The consensus sequence present in the promoter regions of heat shock genes that is bound by the transcription factor Hsf1 thereby activating their expression.
- Hyperphosphorylation
The phosphorylation of a target protein at multiple residues.
- S-nitrosylation
Post-translational modification of a protein by covalent attachment of a nitrosyl group on the thiol moiety of cysteine residues, usually as a result of nitrosative stress.
- Synthetic genetic phenotypes
Phenotypes that are not apparent as a result of a single perturbation, but that are revealed by combining two mutations or genetic and pharmacological perturbations.
- Chemical genomic screens
Screens that combine small molecule inhibitors or activators with genome-wide mutant collections to identify mutations that confer sensitivity or resistance to these molecules.
- Filamentation
A collective term for elongated cellular phenotypes such as yeast cells that have not undergone cell separation, pseudohyphae and hyphae.
Author Biographies
MICHELLE D. LEACH
Michelle Leach is a Sir Henry Wellcome Postdoctoral Fellow. Her four-year fellowship is centred at the University of Aberdeen. Presently, she is working with Leah Cowen at the University of Toronto. She will then study with Joe Heitman at Duke University before returning to Al Brown’s laboratory at the University of Aberdeen. Before starting her postdoctoral fellowship, Michelle completed her B.Sc. in Genetics and Immunology at the University of Aberdeen. She was then awarded a Caledonian Scholarship from the Carnegie Trust to undertake her Ph.D in Molecular Biology with Al Brown at the Aberdeen Fungal Group. Her research is aimed at determining the mechanisms by which pathogenic fungi such as Candida albicans and Cryptococcus neoformans sense and adapt to temperature, enabling them to be successful as pathogens.
EDDA KLIPP
Edda Klipp got her diploma in Biophysics after studies in Berlin and Moscow, and her doctoral degree in Theoretical Biophysics at Humboldt-Universität Berlin, Germany. She was Group Leader for Kinetic Modeling and for Computational Systems Biology at the Max-Planck-Institute for Molecular Genetics, Berlin. Since 2008, she is Full Professor for Theoretical Biophysics at Humboldt-Universität Berlin. In 2009, she was awarded a Doctor honoris causa at Göteborg University. She studies regulatory processes such as signaling pathways, cell cycle or gene expression in different cell types and develops mathematical models and computational approaches. She is co-author of “Systems Biology. A Textbook” and “Systems Biology in Practice”. She is a member of several European and national consortia for systems biology and different applications. She coordinates the DFG-funded Research Training Group for Computational Systems Biology.
LEAH E. COWEN
Leah Cowen is an Assistant Professor in the Department of Molecular Genetics at the University of Toronto and Canada Research Chair in Microbial Genomics and Infectious Disease. She completed her B.Sc. in Microbiology and Immunology at the University of British Columbia and pursued her Ph.D. with Jim Anderson and Linda Kohn at the University of Toronto, focused on the genomic architecture of adaptation to antifungal drugs. As a postdoctoral fellow with Susan Lindquist at the Whitehead Institute, she investigated how the molecular chaperone Hsp90 impacts fungal evolution and phenotypic diversity. She assumed her faculty position in 2007 and is recognized with a Burroughs Wellcome Fund Career Award in the Biomedical Sciences and a Merck Irving S. Sigal Memorial Award. Her research focuses on molecular mechanisms by which cellular signaling and stress responses govern fungal evolution, development, drug resistance, and pathogenesis and further explores how these mechanisms can be harnessed for treating fungal infectious disease.
ALISTAIR J. P. BROWN
Al Brown gained undergraduate and postgraduate degrees at Aberdeen University, and then studied at the Brewing Industry Research Foundation, Surrey, UK, and the Massachusetts Institute of Technology, Cambridge, USA. He gained his first faculty position at Glasgow University before moving to Aberdeen University, Scotland where he now holds a personal chair in molecular and cell biology. Together with Neil Gow, he founded the Aberdeen Fungal Group. He coordinates the European FINSysB Network and leads the CRISP systems biology consortium (Combinatorial Responses In Stress Pathways). He holds an Advanced Grant from the European Research Council and fellowships of the Royal Society of Edinburgh, the American Academy of Microbiology and the Institute of Biology. He studies the molecular regulation of environmental adaptation in Candida albicans, focusing on responses that are relevant to the infection process, such as yeast–hypha morphogenesis, stress responses and metabolic adaptation.
Online Summary
Hsp90 is an essential, abundant, and ubiquitous eukaryotic chaperone that has crucial roles in the folding of its client proteins. Hsp90 has been shown to stabilize client proteins, buffering the phenotypic impact of mutations in these proteins, thereby acting as an evolutionary capacitor during fungal evolution.
In cellular timescales, fungal Hsp90 has been shown to interact with and modulate the activities of client proteins. These include key regulators such as protein kinases and transcription factors which control fungal growth, environmental adaptation and pathogenicity.
Fungal Hsp90 activity is tightly regulated and induced in response to heat shock and other proteotoxic stresses: Hsp90 synthesis is controlled by an autoregulatory circuit involving the heat shock transcription factor (Hsf1); and Hsp90 binding specificity is modulated by post-transcriptional modification.
Straightforward mathematical modelling predicts that the degree to which Hsp90 binds specific client proteins depends on Hsp90 availability and the relative affinities of the Hsp90 chaperone for these client proteins. This prediction is consistent with the experimental observation that the fungal Hsp90 interactome displays considerable environmental plasticity.
This plasticity infers that environmental challenges promote transient changes in the profile of regulators bound by Hsp90, and hence modulate the activities of the corresponding signalling pathways. We propose that Hsp90 acts as a biological transistor that tunes the activity of fungal signalling networks to environmental conditions.
Footnotes
Subject Categories
Fungi, Genomics, Cellular Microbiology, Pathogens, signal transduction, protein folding
References
- 1.Ritossa F. A new puffing pattern induced by temperature shock and DNP in Drosophila. Cell. Mol. Life Sci. 1962;18:571–573. [Google Scholar]
- 2.Lindquist S. Regulation of protein synthesis during heat shock. Nature. 1981;293:311–314. doi: 10.1038/293311a0. [DOI] [PubMed] [Google Scholar]
- 3.Lindquist S. The heat-shock response. Annu. Rev. Biochem. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- 4.Key JL, Lin CY, Chen YM. Heat shock proteins of higher plants. Proceedings of the National Academy of Sciences. 1981;78:3526–3530. doi: 10.1073/pnas.78.6.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindquist S, Craig EA. The heat-shock proteins. Annu. Rev. Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- 6.Ang D, Liberek K, Skowyra D, Zylicz M, Georgopoulos C. Biological role and regulation of the universally conserved heat shock proteins. J. Biol. Chem. 1991;266:24233–6. [PubMed] [Google Scholar]
- 7.Young JC, Agashe VR, Siegers K, Hartl FU. Pathways of chaperone-mediated protein folding in the cytosol. Nat Rev Mol Cell Biol. 2004;5:781–791. doi: 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]
- 8.Haslbeck M, et al. Hsp42 is the general small heat shock protein in the cytosol of Saccharomyces cerevisiae. EMBO J. 2004;23:638–49. doi: 10.1038/sj.emboj.7600080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nikolaou E, et al. Phylogenetic diversity of stress signalling pathways in fungi. BMC Evol Biol. 2009;9:44. doi: 10.1186/1471-2148-9-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicholls S, Leach MD, Priest CL, Brown AJ. Role of the heat shock transcription factor, Hsf1, in a major fungal pathogen that is obligately associated with warm-blooded animals. Mol. Microbiol. 2009;74:844–61. doi: 10.1111/j.1365-2958.2009.06883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Odds FC, editor. Candida and Candidosis. Bailliere Tindall; London, United Kingdom: 1988. [Google Scholar]
- 12.Nicholls S, et al. Activation of the heat shock transcription factor Hsf1 is essential for the full virulence of the fungal pathogen Candida albicans. Fungal Genet. Biol. 2011;48:297–305. doi: 10.1016/j.fgb.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leach MD, Tyc KM, Brown AJP, Klipp E. Modelling the regulation of thermal adaptation in Candida albicans, a major fungal pathogen of humans. PLoS ONE. 2012;7:e32467. doi: 10.1371/journal.pone.0032467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borkovich KA, Farrelly FW, Finkelstein DB, Taulien J, Lindquist S. Hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol. Cell. Biol. 1989;9:3919–30. doi: 10.1128/mcb.9.9.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swoboda RK, et al. Structure and regulation of the HSP90 gene from the pathogenic fungus Candida albicans. Infect. Immun. 1995;63:4506–14. doi: 10.1128/iai.63.11.4506-4514.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim D-U, et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotech. 2010;28:617–623. doi: 10.1038/nbt.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nature Reviews. Molecular Cell Biology. 2010;11:515–528. doi: 10.1038/nrm2918. Excellent review of the structural dynamics of Hsp90 and its regulation
- 18.Jarosz DF, Lindquist S. Hsp90 and environmental stress transform the adaptive value of natural genetic variation. Science. 2010;330:1820–1824. doi: 10.1126/science.1195487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. The description of Hsp90 as a capcitor of evolutionary change in eukaryotes.
- 20.Cowen LE, Lindquist S. Hsp90 potentiates the rapid evolution of new traits: drug resistance in diverse fungi. Science. 2005;309:2185–2189. doi: 10.1126/science.1118370. Hsp90 and the evolution of antifungal drug resistance.
- 21.Chen G, Bradford WD, Seidel CW, Li R. Hsp90 stress potentiates rapid cellular adaptation through induction of aneuploidy. Nature. 2012;482:246–50. doi: 10.1038/nature10795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wandinger SK, Richter K, Buchner J. The Hsp90 chaperone machinery. J. Biol. Chem. 2008 doi: 10.1074/jbc.R800007200. [DOI] [PubMed] [Google Scholar]
- 23.Li J, Soroka J, Buchner J. The Hsp90 chaperone machinery: Conformational dynamics and regulation by co-chaperones. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2012;1823:624–635. doi: 10.1016/j.bbamcr.2011.09.003. Excellent review on the Hsp90 chaperone cycle and its co-chaperones.
- 24.Pearl LH, Prodromou C. Structure and in vivo function of Hsp90. Curr. Opin. Struct. Biol. 2000;10:46–51. doi: 10.1016/s0959-440x(99)00047-0. [DOI] [PubMed] [Google Scholar]
- 25.Ali MM, et al. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 2006;440:1013–1017. doi: 10.1038/nature04716. The structural characterization of an Hsp90-co-chaperone complex.
- 26.Pearl LH, Prodromou C, Workman P. The Hsp90 molecular chaperone: an open and shut case for treatment. Biochem. J. 2008;410:439–53. doi: 10.1042/BJ20071640. [DOI] [PubMed] [Google Scholar]
- 27.Southworth DR, Agard DA. Species-dependent ensembles of conserved conformational states define the Hsp90 chaperone ATPase cycle. Mol. Cell. 2008;32:631–640. doi: 10.1016/j.molcel.2008.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panaretou B, et al. ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. EMBO J. 1998;17:4829–4836. doi: 10.1093/emboj/17.16.4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scheibel T, Weikl T, Buchner J. Two chaperone sites in Hsp90 differing in substrate specificity and ATP dependence. PNAS. 1998;95:1495–1499. doi: 10.1073/pnas.95.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siligardi G, et al. Co-chaperone regulation of conformational switching in the Hsp90 ATPase cycle. J. Biol. Chem. 2004;279:51989–51998. doi: 10.1074/jbc.M410562200. [DOI] [PubMed] [Google Scholar]
- 31.Stebbins CE, et al. Crystal structure of an Hsp90-geldanamycin complex: targeting of a protein chaperone by an antitumor agent. Cell. 1997;89:239–250. doi: 10.1016/s0092-8674(00)80203-2. [DOI] [PubMed] [Google Scholar]
- 32.Chen D, et al. Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell. 2003;14:214–229. doi: 10.1091/mbc.E02-08-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steen BR, et al. Cryptococcus neoformans gene expression during experimental cryptococcal meningitis. Eukaryotic Cell. 2003;2:1336–1349. doi: 10.1128/EC.2.6.1336-1349.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Albrecht D, Guthke R, Brakhage AA, Kniemeyer O. Integrative analysis of the heat shock response in Aspergillus fumigatus. BMC Genomics. 2010;11:32. doi: 10.1186/1471-2164-11-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minchiotti G, Gargano S, Maresca B. The intron-containing hsp82 gene of the dimorphic pathogenic fungus Histoplasma capsulatum is properly spliced in severe heat shock conditions. Mol. Cell. Biol. 1991;11:5624–30. doi: 10.1128/mcb.11.11.5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sorger PK. Heat shock factor and the heat shock response. Cell. 1991;65:363–366. doi: 10.1016/0092-8674(91)90452-5. [DOI] [PubMed] [Google Scholar]
- 37.Wu C. Heat shock transcription factors: structure and regulation. Annu Rev Cell and Dev Biol. 1995;11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- 38.Wiederrecht G, Seto D, Parker CS. Isolation of the gene encoding the S. cerevisiae heat shock transcription factor. Cell. 1988;54:841–53. doi: 10.1016/s0092-8674(88)91197-x. [DOI] [PubMed] [Google Scholar]
- 39.Sorger PK, Pelham HRB. Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell. 1988;54:855–864. doi: 10.1016/s0092-8674(88)91219-6. [DOI] [PubMed] [Google Scholar]
- 40.Sakurai H, Ota A. Regulation of chaperone gene expression by heat shock transcription factor in Saccharomyces cerevisiae: Importance in normal cell growth, stress resistance, and longevity. FEBS Lett. 2011;585:2744–2748. doi: 10.1016/j.febslet.2011.07.041. [DOI] [PubMed] [Google Scholar]
- 41.Jakobsen BK, Pelham HR. Constitutive binding of yeast heat shock factor to DNA in vivo. Mol. Cell. Biol. 1988;8:5040–2. doi: 10.1128/mcb.8.11.5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McDaniel D, et al. Basal-level expression of the yeast HSP82 gene requires a heat shock regulatory element. Mol. Cell. Biol. 1989;9:4789–4798. doi: 10.1128/mcb.9.11.4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sorger PK, Nelson HCM. Trimerization of a yeast transcriptional activator via a coiled-coil motif. Cell. 1989;59:807–813. doi: 10.1016/0092-8674(89)90604-1. [DOI] [PubMed] [Google Scholar]
- 44.Gross DS, English KE, Collins KW, Lee SW. Genomic footprinting of the yeast HSP82 promoter reveals marked distortion of the DNA helix and constitutive occupancy of heat shock and TATA elements. J. Mol. Biol. 1990;216:611–31. doi: 10.1016/0022-2836(90)90387-2. [DOI] [PubMed] [Google Scholar]
- 45.Sewell AK, et al. Mutated yeast heat shock transcription factor exhibits elevated basal transcriptional activation and confers metal resistance. J. Biol. Chem. 1995;270:25079–25086. doi: 10.1074/jbc.270.42.25079. [DOI] [PubMed] [Google Scholar]
- 46.Liu XD, Thiele DJ. Oxidative stress induced heat shock factor phosphorylation and HSF-dependent activation of yeast metallothionein gene transcription. Genes Dev. 1996;10:592–603. doi: 10.1101/gad.10.5.592. [DOI] [PubMed] [Google Scholar]
- 47.Nieto-Sotelo J, Wiederrecht G, Okuda A, Parker CS. The yeast heat shock transcription factor contains a transcriptional activation domain whose activity is repressed under nonshock conditions. Cell. 1990;62:807–817. doi: 10.1016/0092-8674(90)90124-w. [DOI] [PubMed] [Google Scholar]
- 48.Sorger PK, Lewis MJ, Pelham HR. Heat shock factor is regulated differently in yeast and HeLa cells. Nature. 1987;329:81–4. doi: 10.1038/329081a0. [DOI] [PubMed] [Google Scholar]
- 49.Morano KA, Santoro N, Koch KA, Thiele DJ. A trans-activation domain in yeast heat shock transcription factor is essential for cell cycle progression during stress. Mol. Cell. Biol. 1999;19:402–411. doi: 10.1128/mcb.19.1.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lindquist S. Varying patterns of protein synthesis in Drosophila during heat shock: implications for regulation. Dev. Biol. 1980;77:463–79. doi: 10.1016/0012-1606(80)90488-1. [DOI] [PubMed] [Google Scholar]
- 51.DiDomenico BJ, Bugaisky GE, Lindquist S. The heat shock response is self-regulated at both the transcriptional and posttranscriptional levels. Cell. 1982;31:593–603. doi: 10.1016/0092-8674(82)90315-4. [DOI] [PubMed] [Google Scholar]
- 52.Abravaya K, Myers MP, Murphy SP, Morimoto RI. The human heat shock protein Hsp70 interacts with Hsf1, the transcription factor that regulates heat shock gene expression. Genes Dev. 1992;6:1153–64. doi: 10.1101/gad.6.7.1153. [DOI] [PubMed] [Google Scholar]
- 53.Baler R, Welch WJ, Voellmy R. Heat shock gene regulation by nascent polypeptides and denatured proteins: Hsp70 as a potential autoregulatory factor. J. Cell Biol. 1992;117:1151–9. doi: 10.1083/jcb.117.6.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mosser DD, Duchaine J, Massie B. The DNA-binding activity of the human heat shock transcription factor is regulated in vivo by Hsp70. Mol. Cell. Biol. 1993;13:5427–38. doi: 10.1128/mcb.13.9.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bonner JJ, et al. Complex regulation of the yeast heat shock transcription factor. Mol. Biol. Cell. 2000;11:1739–1751. doi: 10.1091/mbc.11.5.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu X-D, Morano KA, Thiele DJ. The yeast Hsp110 family member, Sse1, is an Hsp90 cochaperone. J. Biol. Chem. 1999;274:26654–26660. doi: 10.1074/jbc.274.38.26654. [DOI] [PubMed] [Google Scholar]
- 57.Zou J, Guo Y, Guettouche T, Smith DF, Voellmy R. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell. 1998;94:471–480. doi: 10.1016/s0092-8674(00)81588-3. [DOI] [PubMed] [Google Scholar]
- 58.Guo Y, et al. Evidence for a mechanism of repression of heat shock factor 1 transcriptional activity by a multichaperone complex. J. Biol. Chem. 2001;276:45791–45799. doi: 10.1074/jbc.M105931200. [DOI] [PubMed] [Google Scholar]
- 59.Duina AA, Kalton HM, Gaber RF. Requirement for Hsp90 and a CyP-40-type cyclophilin in negative regulation of the heat shock response. J. Biol. Chem. 1998;273:18974–18978. doi: 10.1074/jbc.273.30.18974. [DOI] [PubMed] [Google Scholar]
- 60.Gerstel B, Tuite MF, McCarthy JEG. The effects of 5′-capping, 3′-polyadenylation and leader composition upon the translation and stability of mRNA in a cell-free extract derived from the yeast Saccharomyces cerevisiae. Mol. Microbiol. 1992;6:2339–2348. doi: 10.1111/j.1365-2958.1992.tb01409.x. [DOI] [PubMed] [Google Scholar]
- 61.Mollapour M, et al. Swe1Wee1-dependent tyrosine phosphorylation of Hsp90 regulates distinct facets of chaperone function. Mol. Cell. 2010;37:333–343. doi: 10.1016/j.molcel.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dougherty JJ, Puri RK, Toft DO. Phosphorylation in vivo of chicken oviduct progesterone receptor. J. Biol. Chem. 1982;257:14226–30. [PubMed] [Google Scholar]
- 63.Dougherty JJ, Rabideau DA, Iannotti AM, Sullivan WP, Toft DO. Identification of the 90 kDa substrate of rat liver type II casein kinase with the heat shock protein which binds steroid receptors. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 1987;927:74–80. doi: 10.1016/0167-4889(87)90067-x. [DOI] [PubMed] [Google Scholar]
- 64.Wandinger SK, Suhre MH, Wegele H, Buchner J. The phosphatase Ppt1 is a dedicated regulator of the molecular chaperone Hsp90. EMBO J. 2006;25:367–376. doi: 10.1038/sj.emboj.7600930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mollapour M, et al. Threonine 22 phosphorylation attenuates Hsp90 interaction with cochaperones and affects Its chaperone activity. Mol. Cell. 2011;41:672–681. doi: 10.1016/j.molcel.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Scroggins BT, Neckers L. Post-translational modification of heat-shock protein 90: impact on chaperone function. Expert Opinion on Drug Discovery. 2007;2:1403–1414. doi: 10.1517/17460441.2.10.1403. [DOI] [PubMed] [Google Scholar]
- 67.Diezmann S, Michaut M, Shapiro RS, Bader GD, Cowen LE. Mapping the Hsp90 genetic interaction network in Candida albicans reveals environmental contingency and rewired circuitry. PLoS Genetics. 2012;8:e1002562. doi: 10.1371/journal.pgen.1002562. First description of the Hsp90 interactome and its plasticity in a fungal pathogen.
- 68.Retzlaff M, et al. Hsp90 is regulated by a switch point in the C-terminal domain. EMBO Reports. 2009;10:1147–1153. doi: 10.1038/embor.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Scroggins BT, et al. An acetylation site in the middle domain of Hsp90 regulates chaperone function. Mol. Cell. 2007;25:151–159. doi: 10.1016/j.molcel.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Siligardi G, et al. Regulation of Hsp90 ATPase activity by the co-chaperone Cdc37p/p50cdc37. J. Biol. Chem. 2002;277:20151–20159. doi: 10.1074/jbc.M201287200. [DOI] [PubMed] [Google Scholar]
- 71.Roe SM, et al. The mechanism of Hsp90 regulation by the protein kinase-specific cochaperone p50cdc37. Cell. 2004;116:87–98. doi: 10.1016/s0092-8674(03)01027-4. [DOI] [PubMed] [Google Scholar]
- 72.Panaretou B, et al. Activation of the ATPase activity of Hsp90 by the stress-regulated cochaperone Aha1. Mol. Cell. 2002;10:1307–1318. doi: 10.1016/s1097-2765(02)00785-2. [DOI] [PubMed] [Google Scholar]
- 73.Zhang W, et al. Biochemical and structural studies of the interaction of Cdc37 with Hsp90. J. Mol. Biol. 2004;340:891–907. doi: 10.1016/j.jmb.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 74.Hernandez MP, Sullivan WP, Toft DO. The assembly and intermolecular properties of the hsp70-Hop-hsp90 molecular chaperone complex. J. Biol. Chem. 2002;277:38294–304. doi: 10.1074/jbc.M206566200. [DOI] [PubMed] [Google Scholar]
- 75.Prodromou C, et al. Regulation of Hsp90 ATPase activity by tetratricopeptide repeat (TPR)-domain co-chaperones. EMBO J. 1999;18:754–762. doi: 10.1093/emboj/18.3.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lotz GP, Lin H, Harst A, Obermann WMJ. Aha1 binds to the middle domain of Hsp90, contributes to client protein activation, and stimulates the ATPase activity of the molecular chaperone. J. Biol. Chem. 2003;278:17228–17235. doi: 10.1074/jbc.M212761200. [DOI] [PubMed] [Google Scholar]
- 77.Young JC, Hartl FU. Polypeptide release by Hsp90 involves ATP hydrolysis and is enhanced by the co-chaperone p23. EMBO J. 2000;19:5930–5940. doi: 10.1093/emboj/19.21.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhao R, et al. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the Hsp90 chaperone. Cell. 2005;120:715–727. doi: 10.1016/j.cell.2004.12.024. This work maps the physical and genetic interactions of Hsp90 in S. cerevisiae.
- 79.Nathan DF, Vos MH, Lindquist S. Identification of SSF1, CNS1, and HCH1 as multicopy suppressors of a Saccharomyces cerevisiae Hsp90 loss-of-function mutation. Proceedings of the National Academy of Sciences. 1999;96:1409–1414. doi: 10.1073/pnas.96.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tesic M, Marsh JA, Cullinan SB, Gaber RF. Functional interactions between Hsp90 and the co-chaperones Cns1 and Cpr7 in Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:32692–32701. doi: 10.1074/jbc.M304315200. [DOI] [PubMed] [Google Scholar]
- 81.Catlett MG, Kaplan KB. Sgt1p is a unique co-chaperone that acts as a client adaptor to link Hsp90 to Skp1p. J. Biol. Chem. 2006;281:33739–33748. doi: 10.1074/jbc.M603847200. [DOI] [PubMed] [Google Scholar]
- 82.Mayr C, Richter K, Lilie H, Buchner J. Cpr6 and Cpr7, two closely related Hsp90-associated immunophilins from Saccharomyces cerevisiae, differ in their functional properties. J. Biol. Chem. 2000;275:34140–34146. doi: 10.1074/jbc.M005251200. [DOI] [PubMed] [Google Scholar]
- 83.Kimura Y, et al. Cdc37 is a molecular chaperone with specific functions in signal transduction. Genes Dev. 1997;11:1775–85. doi: 10.1101/gad.11.14.1775. [DOI] [PubMed] [Google Scholar]
- 84.Ni J, Gao Y, Liu H, Chen J. Candida albicans Cdc37 interacts with the Crk1 kinase and is required for Crk1 production. FEBS Lett. 2004;561:223–230. doi: 10.1016/S0014-5793(04)00172-3. [DOI] [PubMed] [Google Scholar]
- 85.Bansal PK, Abdulle R, Kitagawa K. Sgt1 associates with Hsp90: an initial step of assembly of the core kinetochore complex. Mol. Cell. Biol. 2004;24:8069–8079. doi: 10.1128/MCB.24.18.8069-8079.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Eckert K, et al. The Pih1-Tah1 cochaperone complex inhibits Hsp90 molecular chaperone ATPase activity. J. Biol. Chem. 2010;285:31304–31312. doi: 10.1074/jbc.M110.138263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Johnson J, Brown C. Plasticity of the Hsp90 chaperone machine in divergent eukaryotic organisms. Cell Stress Chaperones. 2009;14:83–94. doi: 10.1007/s12192-008-0058-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nature Reviews. Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 89.Mahalingam D, et al. Targeting HSP90 for cancer therapy. Br. J. Cancer. 2009;100:1523–1529. doi: 10.1038/sj.bjc.6605066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dolgin E, Motluk A. Heat shock and awe. Nat. Med. 2011;17:646–649. doi: 10.1038/nm0611-646. [DOI] [PubMed] [Google Scholar]
- 91.Millson SH, et al. A two-hybrid screen of the yeast proteome for Hsp90 interactors uncovers a novel Hsp90 chaperone requirement in the activity of a stress-activated mitogen-activated protein kinase, Slt2p (Mpk1p) Eukaryotic Cell. 2005;4:849–860. doi: 10.1128/EC.4.5.849-860.2005. The interaction of Hsp90 with a fungal MAPK client is influenced by environmental conditions.
- 92.McClellan AJ, et al. Diverse cellular functions of the Hsp90 molecular chaperone uncovered using systems approaches. Cell. 2007;131:121–135. doi: 10.1016/j.cell.2007.07.036. This work reveals the plasticity of the S. cerevisiae Hsp90 interactome in response to stress.
- 93.Wu Z, Moghaddas Gholami A, Kuster B. Systematic identification of the HSP90 regulated proteome. Molecular and Cellular Proteomics. 2012;11 doi: 10.1074/mcp.M111.016675. M111.016675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stark C, et al. The BioGRID Interaction Database: 2011 update. Nucleic Acids Res. 2011;39:D698–D704. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Echeverría PC, Bernthaler A, Dupuis P, Mayer B, Picard D. An interaction network predicted from public data as a discovery tool: application to the Hsp90 molecular chaperone machine. PLoS ONE. 2011;6:e26044. doi: 10.1371/journal.pone.0026044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Singh SD, et al. Hsp90 governs echinocandin resistance in the pathogenic yeast Candida albicans via calcineurin. PLoS Pathog. 2009;5:e1000532. doi: 10.1371/journal.ppat.1000532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Imai J, Yahara I. Role of HSP90 in salt stress tolerance via stabilization and regulation of calcineurin. Mol. Cell. Biol. 2000;20:9262–9270. doi: 10.1128/mcb.20.24.9262-9270.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Edlind T, Smith L, Henry K, Katiyar S, Nickels J. Antifungal activity in Saccharomyces cerevisiae is modulated by calcium signalling. Mol. Microbiol. 2002;46:257–268. doi: 10.1046/j.1365-2958.2002.03165.x. [DOI] [PubMed] [Google Scholar]
- 99.Sanglard D, Ischer F, Marchetti O, Entenza J, Bille J. Calcineurin A of Candida albicans: involvement in antifungal tolerance, cell morphogenesis and virulence. Mol. Microbiol. 2003;48:959–976. doi: 10.1046/j.1365-2958.2003.03495.x. [DOI] [PubMed] [Google Scholar]
- 100.Cowen LE, Carpenter AE, Matangkasombut O, Fink GR, Lindquist S. Genetic architecture of Hsp90-dependent drug resistance. Eukaryotic Cell. 2006;5:2184–2188. doi: 10.1128/EC.00274-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.LaFayette SL, et al. PKC signaling regulates drug resistance of the fungal pathogen Candida albicans via circuitry comprised of Mkc1, calcineurin, and Hsp90. PLoS Pathog. 2010;6:e1001069. doi: 10.1371/journal.ppat.1001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cowen LE, et al. Harnessing Hsp90 function as a powerful, broadly effective therapeutic strategy for fungal infectious disease. Proc. Natl. Acad. Sci. U. S. A. 2009;106:2818–23. doi: 10.1073/pnas.0813394106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Watanabe Y, Takaesu G, Hagiwara M, Irie K, Matsumoto K. Characterization of a serum response factor-like protein in Saccharomyces cerevisiae, Rlm1, which has transcriptional activity regulated by the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol. Cell. Biol. 1997;17:2615–23. doi: 10.1128/mcb.17.5.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Heinisch JJ, Lorberg A, Schmitz HP, Jacoby JJ. The protein kinase C-mediated MAP kinase pathway involved in the maintenance of cellular integrity in Saccharomyces cerevisiae. Mol. Microbiol. 1999;32:671–80. doi: 10.1046/j.1365-2958.1999.01375.x. [DOI] [PubMed] [Google Scholar]
- 105.San Jose C, Monge RA, Perez-Diaz R, Pla J, Nombela C. The mitogen-activated protein kinase homolog HOG1 gene controls glycerol accumulation in the pathogenic fungus Candida albicans. J. Bacteriol. 1996;178:5850–2. doi: 10.1128/jb.178.19.5850-5852.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Toone WM, Jones N. Stress-activated signalling pathways in yeast. Genes Cells. 1998;3:485–498. doi: 10.1046/j.1365-2443.1998.00211.x. [DOI] [PubMed] [Google Scholar]
- 107.Hohmann S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 2002;66:300–72. doi: 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Smith DA, Nicholls S, Morgan BA, Brown AJ, Quinn J. A conserved stress-activated protein kinase regulates a core stress response in the human pathogen Candida albicans. Mol. Biol. Cell. 2004;15:4179–90. doi: 10.1091/mbc.E04-03-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Smith DA, Morgan BA, Quinn J. Stress signalling to fungal stress-activated protein kinase pathways. FEMS Microbiol. Lett. 2010;306:1–8. doi: 10.1111/j.1574-6968.2010.01937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Alonso-Monge R, et al. Role of the mitogen-activated protein kinase Hog1p in morphogenesis and virulence of Candida albicans. J. Bacteriol. 1999;181:3058–68. doi: 10.1128/jb.181.10.3058-3068.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hawle P, et al. Cdc37p is required for stress-induced high-osmolarity glycerol and protein kinase C mitogen-activated protein kinase pathway functionality by interaction with Hog1p and Slt2p (Mpk1p) Eukaryotic Cell. 2007;6:521–532. doi: 10.1128/EC.00343-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nguyen AN, Shiozaki K. Heat shock-induced activation of stress MAP kinase is regulated by threonine- and tyrosine-specific phosphatases. Genes Dev. 1999;13:1653–1663. doi: 10.1101/gad.13.13.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gerber MR, Farrell A, Deshaies RJ, Herskowitz I, Morgan DO. Cdc37 is required for association of the protein kinase Cdc28 with G1 and mitotic cyclins. Proceedings of the National Academy of Sciences. 1995;92:4651–4655. doi: 10.1073/pnas.92.10.4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Senn H, Shapiro RS, Cowen LE. Cdc28 provides a molecular link between Hsp90, morphogenesis, and cell cycle progression in Candida albicans. Mol. Biol. Cell. 2012;23:268–283. doi: 10.1091/mbc.E11-08-0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shapiro RS, et al. Hsp90 orchestrates temperature-dependent Candida albicans morphogenesis via Ras1-PKA signaling. Curr. Biol. 2009;19:621–9. doi: 10.1016/j.cub.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Stark C, et al. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chen KC, et al. Integrative analysis of cell cycle control in budding yeast. Mol. Biol. Cell. 2004;15:3841–3862. doi: 10.1091/mbc.E03-11-0794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Klipp E, Nordlander B, Kruger R, Gennemark P, Hohmann S. Integrative model of the response of yeast to osmotic shock. Nat. Biotechnol. 2005;23:975–982. doi: 10.1038/nbt1114. [DOI] [PubMed] [Google Scholar]
- 119.Adrover MA, et al. Time-dependent quantitative multicomponent control of the G1-S network by the stress-activated protein kinase Hog1 upon osmostress. Science Signaling. 2011;4:ra63. doi: 10.1126/scisignal.2002204. [DOI] [PubMed] [Google Scholar]
- 120.Mihalik Á , Csermely P. Heat shock partially dissociates the overlapping modules of the yeast protein-protein interaction network: a systems level model of adaptation. PLoS Computational Biology. 2011;7:e1002187. doi: 10.1371/journal.pcbi.1002187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ebong I.-o., et al. Heterogeneity and dynamics in the assembly of the Heat Shock Protein 90 chaperone complexes. Proceedings of the National Academy of Sciences. 2011 doi: 10.1073/pnas.1106261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Proctor CJ, Lorimer IAJ. Modelling the role of the Hsp70/Hsp90 system in the maintenance of protein homeostasis. PLoS ONE. 2011;6:e22038. doi: 10.1371/journal.pone.0022038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Aligue R, Akhavan-Niak H, Russell P. A role for Hsp90 in cell cycle control: Wee1 tyrosine kinase activity requires interaction with Hsp90. EMBO J. 1994;13:6099–106. doi: 10.1002/j.1460-2075.1994.tb06956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shapiro RS, et al. Pho85, Pcl1, and Hms1 signaling governs Candida albicans morphogenesis induced by high temperature or Hsp90 compromise. Curr. Biol. 2012;22:461–470. doi: 10.1016/j.cub.2012.01.062. [DOI] [PubMed] [Google Scholar]
- 125.Jabra-Rizk MA, et al. Candida dubliniensis and Candida albicans display surface variations consistent with observed intergeneric coaggregation. Rev Iberoam Micol. 1999;16:187–93. [PubMed] [Google Scholar]
- 126.Vecchiarelli A, Puliti M, Torosantucci A, Cassone A, Bistoni F. In Vitro production of tumor necrosis factor by murine splenic macrophages stimulated with mannoprotein constituents of Candida albicans cell wall. Cell. Immunol. 1991;134:65–76. doi: 10.1016/0008-8749(91)90331-5. [DOI] [PubMed] [Google Scholar]
- 127.Garner R, Rubanowice K, Sawyer R, Hudson J. Secretion of TNF-a by alveolar macrophages in response to Candida albicans mannan. J. Leukoc. Biol. 1994;55:161–168. doi: 10.1002/jlb.55.2.161. [DOI] [PubMed] [Google Scholar]
- 128.Pietrella D, Bistoni G, Corbucci C, Perito S, Vecchiarelli A. Candida albicans mannoprotein influences the biological function of dendritic cells. Cellular Microbiology. 2006;8:602–612. doi: 10.1111/j.1462-5822.2005.00651.x. [DOI] [PubMed] [Google Scholar]
- 129.Jarosz DF, Taipale M, Lindquist S. Protein homeostasis and the phenotypic manifestation of genetic diversity: principles and mechanisms. Annu. Rev. Genet. 2010;44:189–216. doi: 10.1146/annurev.genet.40.110405.090412. [DOI] [PubMed] [Google Scholar]
- 130.Queitsch C, Sangster TA, Lindquist S. Hsp90 as a capacitor of phenotypic variation. Nature. 2002;417:618–624. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- 131.Sangster TA, Lindquist S, Queitsch C. Under cover: causes, effects and implications of Hsp90-mediated genetic capacitance. Bioessays. 2004;26:348–362. doi: 10.1002/bies.20020. [DOI] [PubMed] [Google Scholar]
- 132.Sangster TA, et al. HSP90-buffered genetic variation is common in Arabidopsis thaliana. Proceedings of the National Academy of Sciences. 2008;105:2969–2974. doi: 10.1073/pnas.0712210105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sangster TA, et al. HSP90 affects the expression of genetic variation and developmental stability in quantitative traits. Proceedings of the National Academy of Sciences. 2008;105:2963–2968. doi: 10.1073/pnas.0712200105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sollars V, et al. Evidence for an epigenetic mechanism by which Hsp90 acts as a capacitor for morphological evolution. Nat. Genet. 2003;33:70–4. doi: 10.1038/ng1067. [DOI] [PubMed] [Google Scholar]
- 135.Cowen LE. The evolution of fungal drug resistance: modulating the trajectory from genotype to phenotype. Nature reviews.Microbiology. 2008;6:187–198. doi: 10.1038/nrmicro1835. [DOI] [PubMed] [Google Scholar]
- 136.Xu Y, Lindquist S. Heat-shock protein hsp90 governs the activity of pp60v-src kinase. Proc. Natl. Acad. Sci. U. S. A. 1993;90:7074–8. doi: 10.1073/pnas.90.15.7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Casanueva MO, Burga A, Lehner B. Fitness trade-offs and environmentally induced mutation buffering in isogenic C. elegans. Science. 2012;335:82–85. doi: 10.1126/science.1213491. [DOI] [PubMed] [Google Scholar]
- 138.Klipp E, et al., editors. Systems biology - a textbook. Wiley-VCH; Weinheim: 2009. [Google Scholar]
- 139.Herrgard MJ, et al. A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology. Nat Biotech. 2008;26:1155–1160. doi: 10.1038/nbt1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Richter K, Muschler P, Hainzl O, Reinstein J, Buchner J. Sti1 is a non-competitive inhibitor of the Hsp90 ATPase. J. Biol. Chem. 2003;278:10328–10333. doi: 10.1074/jbc.M213094200. [DOI] [PubMed] [Google Scholar]
- 141.Marsh JA, Kalton HM, Gaber RF. Cns1 is an essential protein associated with the Hsp90 chaperone complex in Saccharomyces cerevisiae that can restore cyclophilin 40-dependent functions in cpr7D cells. Mol. Cell. Biol. 1998;18:7353–9. doi: 10.1128/mcb.18.12.7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.