Abstract
In the plant Arabidopsis thaliana, multiple quantitative trait loci (QTLs), including RFO2, account for the strong resistance of accession Columbia-0 (Col-0) and relative susceptibility of Taynuilt-0 (Ty-0) to the vascular wilt fungus Fusarium oxysporum forma specialis matthioli. We find that RFO2 corresponds to diversity in receptor-like protein (RLP) genes. In Col-0, there is a tandem pair of RLP genes: RFO2/At1g17250 confers resistance while RLP2 does not. In Ty-0, the highly diverged RFO2 locus has one RLP gene conferring weaker resistance. While the endogenous RFO2 makes a modest contribution to resistance, transgenic RFO2 provides strong pathogen-specific resistance. The extracellular leucine-rich repeats (eLRRs) in RFO2 and RLP2 are interchangeable for resistance and remarkably similar to eLRRs in the receptor-like kinase PSY1R, which perceives tyrosine-sulfated peptide PSY1. Reduced infection in psy1r and mutants of related phytosulfokine (PSK) receptor genes PSKR1 and PSKR2 shows that tyrosine-sulfated peptide signaling promotes susceptibility. The related eLRRs in RFO2 and PSY1R are not interchangeable; and expression of the RLP nPcR, in which eLRRs in RFO2 are replaced with eLRRs in PSY1R, results in constitutive resistance. Counterintuitively, PSY1 signaling suppresses nPcR because psy1r nPcR is lethal. The fact that PSK signaling does not similarly affect nPcR argues that PSY1 signaling directly downregulates the expression of nPcR. Our results support a speculative but intriguing model to explain RFO2's role in resistance. We propose that F. oxysporum produces an effector that inhibits the normal negative feedback regulation of PSY1R, which stabilizes PSY1 signaling and induces susceptibility. However, RFO2, acting as a decoy receptor for PSY1R, is also stabilized by the effector and instead induces host immunity. Overall, the quantitative resistance of RFO2 is reminiscent of the better-studied monogenic resistance traits.
Authors Summary
The fungus Fusarium oxysporum causes debilitating vascular infections in plants and is responsible for Fusarium wilt diseases in numerous crop species. To cope with microbial pathogens such as F. oxysporum, plants express variation in resistance genes, which typically facilitate recognition of infection by pathogens and instigate a defense response. Presently, receptor like-proteins (RLPs) are characterized as a minor class of resistance proteins with strong effect. Studying resistance to Fusarium wilt disease in the plant Arabidopsis thaliana, we discover that RFO2, a gene providing modest quantitative resistance, encodes an RLP. Extracellular leucine-rich repeats (eLRRs) of RLPs typically mediate the recognition of infection by pathogens. However, we find that the eLRRs of RFO2 do not specify resistance. The eLRRs of RFO2 and PSY1R, which is the putative receptor for an endogenous tyrosine-sulfated peptide growth regulator PSY1, share remarkable identity. Moreover, we find that PSY1 signaling promotes susceptibility to Fusarium wilt disease. From genetic analysis of a novel RLP gene that we created from both RFO2 and PSY1R, we propose a model that explains the relationship between RFO2 and PSY1R. In our model, RFO2 induces resistance because RFO2 mediates the recognition of F. oxysporum's attempt to manipulate PSY1 signaling.
Introduction
The fungus Fusarium oxysporum largely persists in soil as a saprophyte or in the roots of asymptomatic plants as an endophyte [1], [2]. It is the rarer pathogens of F. oxysporum that are capable of invading and colonizing the vascular system of host plants, and persistence of F. oxysporum in water-conducting xylem vessels is indicative of host susceptibility [1]–[4]. Numerous agricultural crops, notably tomato, cotton and banana, are susceptible to debilitating vascular infection by F. oxysporum and consequently develop wilt disease [2], [3], [5], [6].
Fusarium wilt diseases can be especially destructive to crop monocultures because pathogens are virulent in a narrow range of plant species [3], [7]. In recognition of this host specificity, pathogenic isolates are classified as having special forms, or formae speciales, which typically represent one to several phylogenetic lineages in the F. oxysporum species complex [7]. Pathogens of the same forma specialis infect similar host species, and a commercial host often names the forma specialis. For instance, F. oxysporum forma specialis matthioli (FOM) is isolated from garden stock (Matthiola incana) [8].
Fusarium wilt of Arabidopsis thaliana is an ideal pathosystem for mapping, identifying and characterizing the genes responsible for host resistance to vascular wilt fungi [9]. A. thaliana is the preeminent plant for molecular genetic and genomic studies and is susceptible to infection by FOM and two other crucifer-infecting formae speciales [9], [10]. In the field, F. oxysporum forma specialis conglutinans (FOC) and F. oxysporum forma specialis raphani (FOR) are recovered from diseased cabbage (Brassica species) and radish (Raphanus sativus), respectively [11]. The symptoms and progression of wilt disease in A. thaliana recapitulate the disease syndrome observed in native field hosts [8], [9], [12], [13]. Furthermore, this experimental pathosystem preserves host specificity because A. thaliana remains completely resistant to formae speciales isolated from non-crucifer hosts [9].
Innate resistance to Fusarium wilt as well as other infectious diseases often varies among plants of the same or interbreeding species [9], [14], [15]. Host resistance to the infecting pathogen when available in commercially acceptable varieties and crop rotation when feasible are preferable measures to control soil-borne diseases such as Fusarium wilt because chemical treatment of fields is usually uneconomical or has too negative cost to the environment [3], [14], [16]. However, genetic resistance may be poorly defined or unavailable in acceptable crop varieties.
The response of wild accessions of A. thaliana to infection by FOC, FOM and FOR ranges widely from complete resistance to ready susceptibility [9]. For example, accession Col-0 exhibits complete resistance to a dose of FOM that consistently kills accession Ty-0. On the other hand, Ty-0 exhibits more resistance than Col-0 when accessions are instead infected with FOC race 1. Thus, in large part, variation in resistance is specific to the infecting forma specialis. Most researchers using the Fusarium-Arabidopsis pathosystem infect the common laboratory accession Col-0 with FOC [17]–[19]. Because Col-0 exhibits considerable but partial resistance to FOC, it is possible to observe either enhanced resistance or increased susceptibility in Arabidopsis mutants using the same F. oxysporum pathogen.
To improve the resistance of cultivated varieties, plant breeders exploit the genes controlling natural variation in resistance, so-called resistance genes [20]. In crosses between resistant and susceptible varieties, qualitative resistance may be inherited as multiple quantitative trait loci (QTLs) conferring polygenic resistance or as a simple discontinuous Mendelian trait conferring monogenic resistance [21]. The best-studied resistance genes confer strong monogenic resistance to specific pathogens and typically but not always code for members of the nucleotide binding, leucine-rich repeat (NB-LRR) class of resistance proteins [22], [23]. There are few examples of genes providing polygenic resistance, so it remains unclear whether particular classes of genes with common function are commonly associated with quantitative disease resistance traits [24]–[26].
In A. thaliana, RESISTANCE TO F. OXYSPORUM (RFO) is a polygenic trait [9]. Six RFO QTLs are detected in the recombinant progeny of Col-0 and Ty-0 accessions and account for the strong resistance of Col-0 and susceptibility of Ty-0 to FOM. RFO1, which expresses the strongest resistance among RFO QTLs, is a member of the wall-associated kinase (WAK) family of receptor-like kinase (RLK) genes. The WAK family is one of several RLK gene families whose history, genome organization and expression suggest their involvement in response to pathogens [27]. RFO1 contributes quantitatively to immunity as loss-of-function in rfo1 enhances F. oxysporum infection in the root vascular cylinder [28]. Resistance conferred by RFO2 and two other RFO QTLs appears epistatic to RFO1 and is either enhanced or dependent on the presence of RFO1 [9].
Here we show that the RFO2 QTL corresponds to diversity in receptor-like protein (RLP) genes that have conspicuous sequence similarity to the PSY1 peptide receptor gene PSY1R [29]. We find that, while the native RFO2 in Col-0 expresses modest quantitative resistance, transgenic RFO2 expresses strong, nearly qualitative resistance and confers specific resistance to FOM and no resistance to FOC. In contrast, we find that the RFO2-related PSY1R and phytosulfokine (PSK) receptor genes PSKR1 and PSKR2 promote susceptibility to F. oxysporum infection [30]. From the phenotypes and genetic interactions of chimeric RLP and RLK transgenes, we characterize the resistance function of RFO2 and propose a speculative model that connects the peptide signaling of PSY1R and pathogen-specific resistance of RFO2.
Results
Mapping and identification of RFO2
In previous genetic analysis, resistance to FOM was associated with six RFO loci in recombinant offspring of Col-0 and Ty-0 accessions of A. thaliana [9]. Plants that were heterozygous Col-0/Ty-0 at RFO loci, including RFO2 on chromosome 1, exhibited more resistance than plants that were homozygous Ty-0/Ty-0.
We first confirmed the association of RFO2 with resistance by testing the resistance of progeny of a new cross, in which RFO1 and RFO2 were the only RFO QTLs segregating. RFO1 was included in the cross because previous analysis suggested that the Col-0 allele of RFO1 (RFO1-C) enhances resistance conferred by the Col-0 allele of RFO2 (RFO2-C) [9]. One parent of the cross, plant 4D2, was heterozygous at RFO2 and homozygous Ty-0 at the remaining RFO loci, including RFO1 (RFO1-T/T RFO2-C/T). The other parent was the near isogenic line 1A3, which has a small chromosomal region around RFO1-C introgressed into the Ty-0 genetic background (RFO1-C/C RFO2-T/T). Among the resulting progeny, plants that inherited RFO2-C (RFO1-C/T RFO2-C/T) were more resistant than plants that did not inherit RFO2-C (RFO1-C/T RFO2-T/T), which still exhibited more resistance than Ty-0 presumably because all progeny were RFO1-C/T (Figure 1A). Also, as previously observed, RFO1-C enhanced resistance conferred by RFO2-C. Both the self progeny of RFO1-T/T RFO2-C/T (4D2), which segregated for RFO2-C but lacked RFO1-C, and Ty-0, which lacked both RFO2-C and RFO1-C, exhibited similar severe symptoms (Figure 1A). Thus, RFO1-C was included in crosses to map RFO2.
Figure 1. Resistance of endogenous and transgenic RFO2.

(A) Fractions of n plants, either Ty-0, line 1A3, self progeny of 4D2 (RFO2-C/T), or progeny of cross 1A3×4D2, RFO2-C/T (C/T) or RFO2-T/T (T/T), were susceptible or resistant, or had intermediate resistance, according to HI scores at 18 dpi. The same italicized letter is for genotypes with similar median ranks, according to the Mann-Whitney (M-W) U test (two-tailed p>0.01) of ranks from HI scores at three time points in one week. (B) Fractions of n T1 1A3 harboring Col-0 genomic clone Kpn1.2, Avr2.1, Xba1.1 or Sal1.2 were susceptible or resistant, had intermediate resistance, according to HI scores at 21 dpi. Resistance conferred by subclones Kpn1.2 and Avr2.1, or Xba1.1 and Sal1.2, were different according to M-W U test on rank-ordered T1, two-tailed p = 0.0011, or 0.0008, respectively. (C) Fractions of n FOM-infected (left) or FOC-infected (right) progeny of F1 backcross (1A3+Kpn1.2×Ty-0)×Ty-0 with (+) and without (−) copies of RFO1-C and tRFO2 had the lowest, middle or highest third of ranks, according HI scores from four time points. FOC-infected plants with (+) and without (−) tRFO2 had similar median ranks, according to M-W U test (two-tailed p = 0.72).
Using genetic linkage analysis, we mapped RFO2 to an interval, corresponding to less than 258 kilobasepairs (kb) and fewer than 68 genes, as described in Materials and Methods, which suggested that RFO2 is the effect of a single gene.
To clone the RFO2 gene sequence, we tested whether Col-0 genomic sequence in the final RFO2 interval could enhance the resistance of RFO1-C/C RFO2-T/T (1A3). In total, 19 genomic subclones were stably introduced to line 1A3 using Agrobacterium tumefaciens-mediated transformation, and just two subclones enhanced the resistance of 1A3 (Figure S1). Independent kanamycin-resistant T1 transformants of subclone Kpn1.2 expressed more resistance to FOM than T1 transformants of subclone Avr2.1; and, similarly, T1 transformants of Sal1.2 were more resistant than T1 transformants of Xba1.1 (Figure 1B).
In the two positive genomic subclones, Sal1.2 and Kpn1.2, there were 13.8 kb of overlapping sequence. This overlapping sequence was further subcloned as six restriction fragments (that are mapped in Figure 2 and Figure S1). Only 1A3 transformants harboring constructs Hind3.1, Nsi1.2 and Nsi1.3 that include gene At1g17250 showed enhanced resistance. Meanwhile, 1A3 transformants, harboring subclones without full-length At1g17250, namely Age1, BamH1 and Nsi1.1, were similarly affected by FOM infection as the untransformed line 1A3.
Figure 2. RFO2 region is highly diverged in Col-0 and Ty-0.
Grey regions between dotted lines depict alignment of most of the longer Col-0 sequence at the RFO2 locus on chromosome 1 with the shorter sequence in Ty-0. At each nucleotide position in Col-0, the percent identity of 37-nucleotides upstream and downstream in the aligned Col-0 and Ty-0 sequences is graphed. Dashed line is at 100 percent identity. Genes and intergenic sequences in the RFO2 region of Col-0 and Ty-0 are depicted above and below, respectively, at scale (see 1 kb at lower right). Arrowheads indicate orientation of genes, wide at exons and narrow at introns. TAIR-annotated genes (filled) are labeled with last three digits of gene number, after ‘At1g17’, and repetitive element (rep) and noncoding degenerate transcription unit (ψ255) are half-filled. Salk identifiers label inverted triangles pointing to T-DNA insertion sites. Endpoints (vertical lines) and extent (horizontal lines) of Col-0 genomic clones are mapped in the RFO2 region of Col-0 (above); and, filled circles indicate that cloned sequence extends outside the RFO2 region. Rfo phenotypes, conferred by genomic clones in transformed 1A3, are listed to the left: either enhanced resistance (R) or no added resistance and susceptible (S).
Many Sal1.2 and Kpn1.2 transgenic lines exhibited unexpectedly strong resistance as compared to the modest RFO1-C-dependent resistance expressed by RFO2 in Col-0 and Ty-0 recombinants. Analysis of a cross, in which both RFO1-C and the putative RFO2-C transgene (tRFO2) were segregating, confirmed that resistance conferred by tRFO2 was in fact independent of RFO1-C and stronger than the resistance of RFO1-C. A Kpn1.2 transgenic line (1A3+tRFO2) and Ty-0 were crossed, and the resulting F1 dihybrid RFO1-C/T tRFO2-(+/−) was then backcrossed to Ty-0 to yield F1BC progeny, (i) without RFO1-C and tRFO2, (ii) with RFO1-C only, (iii) with tRFO2 only, or (iv) with both RFO1-C and tRFO2, in a ratio of 1∶1∶1∶1 that is expected for independent assortment of RFO1 and tRFO2 (Figure 1C). Plants with tRFO2 were more resistant to FOM with or without RFO1-C; and, plants with tRFO2 only were substantially more resistant than plants with RFO1-C only (Figure 1C). When F1BC progeny were infected with FOC instead, plants with tRFO2 were no more resistant than plants without tRFO2 (Figure 1C). Thus, the strong resistance of tRFO2 was specific for FOM.
Because multiple RFO loci contribute to the complete resistance of Col-0, we anticipated that loss-of-function in RFO2 alone might not exhibit loss of resistance in the Col-0 genetic background. Indeed lines homozygous for T-DNA insertions in or adjacent to candidate genes in the RFO2 region were strongly resistant to FOM –see Materials and Methods for details. However, when four insertion lines were crossed to Ty-0, which halved the genetic contribution of Col-0, F1 hybrids could develop obvious wilt symptoms. The F1 hybrids of Salk_051677, in particular, were especially susceptible, and a majority of these F1 hybrids expressed symptoms, while only 20 percent of F1 hybrids of Salk 140524 were similarly affected (Figure 3A). T-DNA insertion in Salk_051677 interrupts At1g17250, the same gene that correlated with enhanced resistance in 1A3 transformants, whereas insertion in Salk_140524 interrupts At1g17200, a gene outside the overlapping sequence in Kpn1.2 and Sal1.2. Salk_051677, which was previously named Atrlp3-1 without a reported phenotype, was renamed rfo2 [31].
Figure 3. T-DNA insertion allele rfo2 abolishes RFO2 QTL.

(A) Fractions of n F1 hybrids of Ty-0 and Salk_014524 (At1g17200) or Salk_051677 (At1g17250) were susceptible or resistant or had intermediate resistance, according to HI scores at 21 dpi. Median ranks of the two F1 hybrids are dissimilar, according to M-W U test (two-tailed p = 0.021). (B) Fractions of n self progeny of rfo1 RFO2/rfo2, either RFO2/RFO2, RFO2/rfo2 or rfo2/rfo2, had the lowest, middle or highest third of ranks at 21 dpi. Same italicized letters above genotypes indicates that median ranks were similar, according to M-W U test (two-tailed p>0.05). (C) Genotypes of 234 F1BC progeny of original cross (Col-0×Ty0)×Ty-0 (top) and 240 F1BC progeny from new cross (rfo2×Ty0)×Ty-0 are either Ty-0/Ty-0 (T) or Col-0/Ty-0 (C), at RFO1-, RFO2- and RFO3-linked markers and marker CHR3.8 that is not linked to a RFO QTL. Fractions of n FOM-infected plants with the lowest, middle or highest third of ranks. Asterisks indicate that alternative genotypes C and T had dissimilar median ranks, according to M-W U test (two-tailed p<0.01).
Although plants with genotype RFO1 rfo2 exhibited strong resistance to FOM, as mentioned above, rfo2 did enhance susceptibility of rfo1 in the double mutant rfo1 rfo2, which was also more susceptible than the rfo1 RFO2/rfo2 heterozygote (Figure 3B). In the Col-0 genetic background, RFO2 expressed resistance in the absence of RFO1 even though resistance conferred by RFO2 showed dependence on RFO1 in the original mapping cross between Col-0 and Ty-0 used to define RFO QTLs (Figure 3C) [9].
In theory, RFO2 might correspond to more than one gene because we discovered RFO2 as a QTL [9]. To address whether At1g17250 alone accounts for the RFO2 QTL, we examined the segregation of resistance in a comparable (rfo2×Ty-0)×Ty-0 mapping population. This new population was similar to our original mapping population with the exception that rfo2 replaced wild type as the Col-0 parent. Specifically, we crossed rfo2 and Ty-0 and then backcrossed the resulting F1 hybrid to Ty-0. As expected, Col-0/Ty-0 heterozygotes and Ty-0/Ty-0 homozygotes appeared in roughly equal proportion with all tested markers (Figure 3C). DNA markers linked to RFO1 and RFO3, which is a third RFO QTL previously detected on chromosome 3 [9], were associated with resistance in both the new and original populations. In contrast, RFO2-C showed significant correlation with resistance only in the original population (Figure 3C). In the (rfo2×Ty-0)×Ty-0 population, resistance at RFO2 instead had a modest correlation with Ty-0 homozygotes (RFO2-T/T).
RFO2 inhibits FOM infection in roots
Up to now, we equated susceptibility with symptom severity in the above ground foliage, where little if any FOM would be present until late in infection [28]. Possibly, quantitative resistance could reflect reduced symptoms in the expressive phase of infection rather than reduced fungal infection in the below ground roots [32]. To distinguish between these possibilities, we compared the effect of RFO1-C and RFO2-C on symptoms in shoots and FOM infection in roots. At 12 dpi, RFO1-C RFO2-C (1A3+tRFO2) exhibited only modest stunting while Ty-0 plants, without the benefit of either RFO1-C or RFO2-C, were severely stunted, and older leaves were yellowing (Figure 4A). Meanwhile, RFO1-C (1A3) developed symptoms that were intermediate to those in Ty-0 and RFO1-C RFO2-C. In situ staining with X-Ara reports F. oxysporum infection as a blue precipitate because F. oxysporum, and not Arabidopsis, expresses detectable arabinofuranosidase (ABF) activity [28]. Blue staining was stronger and more prevalent in roots of RFO1-C than roots of RFO1-C RFO2-C while roots of Ty-0 showed the most extensive staining (Figure 4B); and, uninfected roots of all genotypes remained unstained. The observed differences in X-Ara staining were corroborated by quantifying the accumulation of soluble yellow 4-nitrophenol when roots were incubated with a second substrate of ABF, NP-Ara (Figure 4C).
Figure 4. RFO1-C and RFO2-C restrict infection in roots.

All plants have the genetic background of Ty-0, which lacks both RFO1-C (−) and RFO2-C (−). Line 1A3 and 1A3+tRFO2 also have RFO1-C (+), and only line 1A3+tRFO2 has RFO2-C (+). Plants of each genotype (n = 24) were rank-ordered from most susceptible to most resistant, and plants with median or middle ranks are shown and analyzed. (A) Three representative FOM-infected plants of each genotype are shown at 12 dpi. (B) Roots of FOM-infected plants were stained with X-Ara. (C) Relative Fusarium-derived ABF activity in FOM- (+) and mock- (−) infected roots, in terms of absorbance (OD410 nm) of 4-nitrophenol formed after 16-hr incubation of whole roots, harvested from (n) mock-infected (−) or FOM-infected (+) plants, with NP-Ara at 10 dpi. Values are adjusted to set mean value of mock-infected Ty-0 roots equal to one. Error bars are confidence interval of the mean (α = 0.05). Different italicized letters indicate that means are dissimilar, according to Student's t-test (p<0.05).
RFO2 corresponds to diversity in PSY1R-related RLP genes
In prior surveys of RLP genes in the Col-0 reference genome, At1g17240 and the neighboring gene RFO2/At1g17250 were identified as a tandem pair of highly-related receptor-like protein (RLP) genes and generically named RLP2 and RLP3, respectively [31], [33]. The primary structure of RFO2 is similar to previously characterized RLPs and is comprised of seven domains (Figure S2) [34]: A signal peptide (domains A), four extracellular domains (B through E), a transmembrane domain (F) and a short cytoplasmic tail of nine amino acids (domain G). Most of RFO2 is extracellular and is composed of 23 extracellular leucine-rich repeats (eLRRs, domain C), which are capped at amino-terminal and carboxy-terminal ends by domains B and D, respectively. An acidic domain E joins the extracellular domains to the transmembrane domain. Also, a loop out sequence interrupts the 19th eLRR in domain C.
Genomic sequence in the chromosomal region around RFO2 is highly diverged in Col-0 and Ty-0. In order to characterize the susceptible RFO2-T allele, we obtained an 8,311 bp sequence that spans the RFO2 region in Ty-0 using PCR-sequencing. According to BlastN search of the Col-0 reference genome, the best match for the Ty-0 sequence extended across a 12,878 bp interval that included sequence within and between annotated genes At1g17230 and At1g17260, as depicted in Figure 2. In the shorter Ty-0 sequence, a single RLP gene (RLP2-T) was oriented on the chromosome in the same direction as the head-to-tail pair of RFO2 and RLP2 in Col-0 (Figure 2). Sequence predicted to be intergenic retained remarkably low nucleotide identity in the two accessions, and intergenic sequence between RFO2 and RLP2 could not be aligned to any Ty-0 sequence. Thus, the Col-0 and Ty-0 variants of RFO2 appeared to be ancestral variation in A. thaliana.
The alignment of coding sequences in the single exons of the three RLP genes at the RFO2 locus showed that RLP2 and RLP2-T were more related to each other than RFO2. Specifically, the 1,956-nucleotide sequence starting at the 5′ end of RLP2-T shared more identity with RLP2 (88 percent) than RFO2 (82 percent). However, a shorter 136 bp sequence at the 3′ end of RLP2-T shared more identity with RFO2 (73.5 percent) than RLP2 (55 percent). Interestingly, RLP2 shared most identity (92 percent) with the full-length ortholog AlRLP2 from Arabidopsis lyrata than even a partially aligned RLP2-T.
From BlastP searches of the Arabidopsis genome database, we learned that RFO2-related RLPs share conspicuous similarity with the extracellular regions of the Arabidopsis RLK PSY1R that perceives the small post-translationally modified tyrosine-sulfated peptide hormone PSY1 involved in cell division and expansion [29]. Specifically, alignment of B and C domains of either RFO2 or RLP2 and PSY1R showed that 74 or 80 percent of residues in the eLRRs, respectively, were identical (Figure S3). Remarkably, RFO2 and RLP2 were more similar to PSY1R than they were to each other as just 73 percent of residues were identical in the alignment of B and C domains of RFO2 and RLP2 (Figure S3). However, outside of the eLRRs, there was little or no sequence conservation between RLK and either RLP, and PSY1R poorly aligned to domains D through G of RFO2 or RLP2 (Figure S4).
The carboxy-end of RFO2 specifies resistance to FOM
By using the same constitutive promoter to express RFO2, RLP2 and RLP2-T, we tested whether differences in transcription could explain why RFO2 conferred resistance while its homologs RLP2 and RLP2-T did not. Coding sequences of the three RLP genes were fused downstream of the constitutive promoter ENTCUP2, and these promoter-gene fusions were introduced to line 1A3 by stable genetic transformation [35], [36]. Phosphinothricin (Ppt)-resistant T1 transformants of 1A3 harboring the three promoter-gene fusions exhibited the same wild-type appearance as the untransformed line 1A3. Independent T2 lines harboring constitutively-expressed RFO2 (cRFO2) exhibited strong resistance to FOM while, in the same infection assays, independent T2 lines harboring constitutively-expressed RLP2 (cRLP2) appeared similar to the untransformed parental line 1A3 (Figure 5A and 5B). Meanwhile, a T2 line with constitutive expression of RLP2-T (cRLP2-T) exhibited marginally more resistance than the parental line 1A3, and this resistance was modest in comparison to the resistance conferred by cRFO2 in the same assay (Figure 5C).
Figure 5. Constitutively-expressed and chimeric RLP genes.
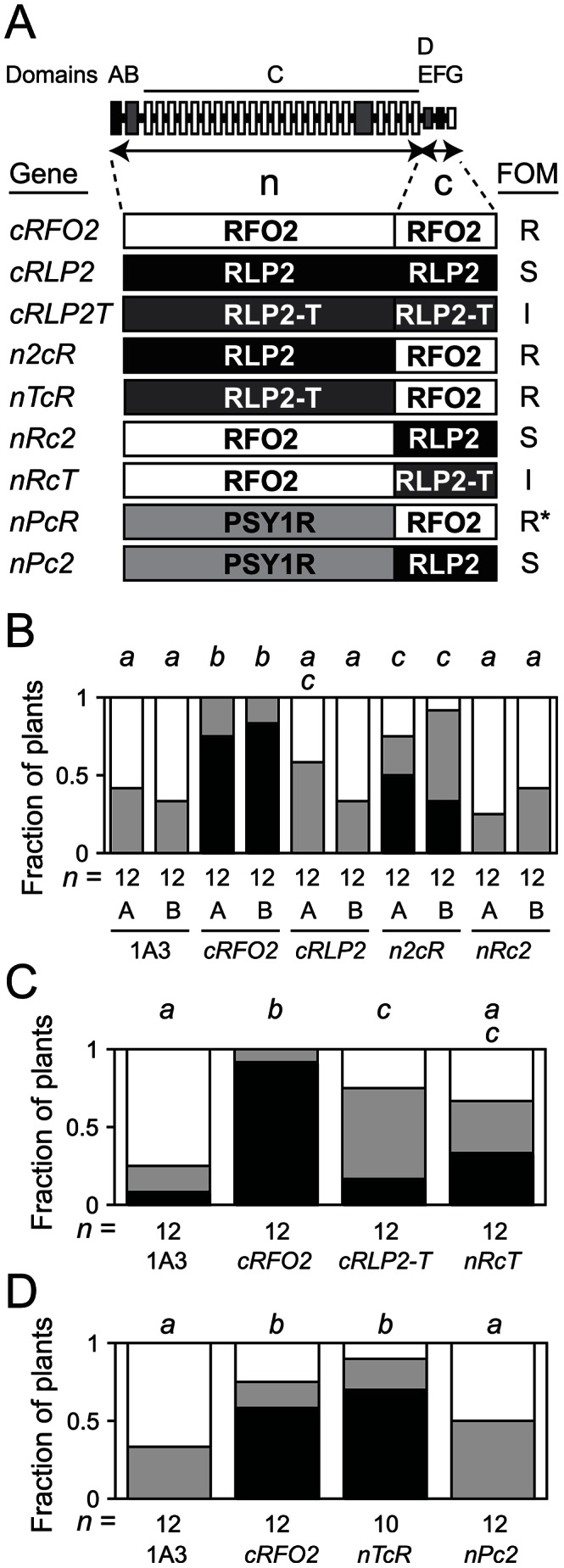
(A) Domain structure of RFO2-related RLPs is shown as a series of boxes. From left to right, signal/anchor peptide (A, filled), N-cap (B, half-filled), eLRR subdomains (open) and loop out subdomain (half-filled) of domain C, C-cap/acidic domain (D/E, half-filled), transmembrane domain (F, filled) and cytoplasmic tail (G, open). Amino-terminal domains A through C (n) and carboxy-terminal domains D through G (c) of RFO2-related RLPs RFO2 (R), RLP2 (2) and RLP2-T (T) and the n-domain of PSY1R (P) are combined in the constitutive expression vector ORE-E3 to make the three original and six chimeric RLP transgenes as listed with names to the left. On the right, Rfo phenotypes of listed RLP transgenes: strong resistance (R), intermediate resistance (I) or no effect on susceptibility of line 1A3 (S) to FOM infection. nPcR confers strong resistance to FOC, FOM and FOR (*). (B) Experiments A and B were performed with independent T2 lines, and plants in separate experiments were rank-ordered at 24 dpi. Fractions of n FOM-infected untransformed 1A3 or Ppt-resistant 1A3 T2, transformed with the constitutively expressed RLP transgenes (above), with the lowest, middle, highest third of ranks were susceptible (open), had intermediate resistance (half-filled) or were resistant (filled). Same italicized letters above columns indicates that median ranks were similar according to M-W U test, p>0.05. (C and D) Fractions of n FOM-infected untransformed 1A3 or Ppt-resistant 1A3 T2, transformed with constitutively-expressed RLP transgenes, with the lowest, middle or highest third of ranks at 24 dpi were susceptible (open), had intermediate resistance (half-filled) or were resistant (filled).
Because the resistance of RFO2 was not a consequence of differences in promoter expression of the three RLP genes, we next examined whether resistance could be localized to the PSY1R-related amino-terminal (n-) domains (A through C) or the more diverged carboxy-terminal (c-) domains (D through G) of the three RLPs. Coding sequences for the n- and c-domains were recombined to make chimeric RLP genes. A unique SpeI restriction site was introduced as a silent mutation in coding sequence of cRFO2, cRLP2 and cRLP2-T that joins domains C and D (Figure S4). Sequences on either side of the SpeI site, coding for n- or c-domains, were swapped (as depicted in Figure 5A), and the resulting chimeric RLP genes were stably introduced to line 1A3 by genetic transformation. Ppt-resistant T2 plants harboring in vitro constructed chimeric genes with the n-domains of either RLP2 or RLP2-T and the c-domains of RFO2 in Figure 5B or 5D, respectively, expressed strong resistance to FOM; henceforth, these chimeric genes are referred to as n2cR and nTcR, respectively. Meanwhile, Ppt-resistant T2 plants harboring transgenes with the reciprocal exchanges, coding sequence of the n-domains of RFO2 fused to sequence of the c-domains of RLP2 (nRc2) or RLP2-T (nRcT), expressed resistance to FOM similar to the untransformed parental line 1A3 and cRLP2 (in Figure 5B) or the original cRLP2-T (in Figure 5C), respectively. Thus, the shorter, less conserved sequence of the c-domains of RFO2 specified resistance to FOM, and the function of the conserved eLRRs of the three RLP homologs was roughly equivalent for resistance.
PSY1R, PSKR1, and PSKR2 promote susceptibility
The sequence conservation of eLRRs in RFO2 and PSY1R prompted us to examine wilt disease progression in loss-of-function mutant psy1r [29]. The function of PSY1R overlaps with the function of two closely related RLK genes PSKR1 and PSKR2 that perceive the tyrosine-sulfated peptide PSK [29], [30]. PSK accumulates in cell culture medium and is a key factor permitting the dedifferentiation and redifferentiation of plant cells in culture [37]. Signaling by peptides PSY1 and PSK negatively regulates stress response and senescence, and addition of PSK and PSY1 to agar medium promotes elongation of roots of Arabidopsis seedlings [29], [30], [37]. The functional overlap of PSY1 and PSK signaling prompted us to test the infection of pskr1 and pskr2 as well as psy1r.
When plants that are insensitive to PSY1 (psy1r), insensitive to PSK (double mutant pskr1 pskr2) or insensitive to both PSY1 and PSK (psy1r pskr1 double mutant and psy1r pskr1 pskr2 triple mutant) were infected with FOM, all mutants were completely resistant. However, the receptor mutants have the Col-0 genetic background, and Col-0 is already completely resistant to FOM. When plants were instead infected with FOC (Figure 6A and 6B) or FOR (Figure 6C), two formae speciales to which Col-0 normally expresses incomplete resistance, mutants were noticeably more resistant than wild type. The triple mutant that is insensitive to both PSK and PSY1 peptides showed the strongest suppression of disease symptoms while mutants that are insensitive to either PSK or PSY1 showed a more modest suppression of disease. X-Ara staining of FOC-infected roots of the triple mutant suggested that initial infection of root tips was normal but indicated that subsequent infection of xylem by F. oxysporum was suppressed. We quantified the diminished fungal infection in roots using NP-Ara (Figure 6D), and two-fold less F. oxysporum-derived ABF activity was measured in roots of the triple mutant than wild type.
Figure 6. PSY1 and PSK promotes susceptibility to Fusarium wilt.

(A) Representative FOC-infected (+) or mock-infected (−) wild type (Col-0) and mutants having median HI scores (n = 6 for each genotype) are shown at 24 dpi. (B) Fraction of FOC-infected or (C) FOR-infected plants (n = 10 for each genotype) with the lowest, middle, highest third of ranks at 18 dpi. Different italicized letters indicates that median ranks of genotypes were dissimilar (M-W U test, p<0.05). (D) Relative Fusarium-derived ABF activity in FOC- (+) and mock- (−) infected roots, in terms of absorbance (OD410 nm) of 4-nitrophenol formed after 20-hr incubation with NP-Ara, was different at 10 dpi, according to Student's t-test (n = 4; two-tailed p = 0.0001). Error bars are confidence interval of the mean (α = 0.05). (E) Representative FOC-infected (+) or mock-infected (−) wild type (Col-0) and tpst having median HI scores (n = 10 for each genotype) are shown at 18 dpi.
We next examined whether perception of endogenous PSY1 and PSK peptides was critical for the susceptibility that peptide hormone receptor genes expressed in wild type. Arabidopsis has a single tyrosyl-protein sulfotransferase gene TPST, and tpst produces only unsulfated and inactive PSY1 and PSK peptides [38], [39]. FOC-infected tpst expressed strong resistance that was comparable to the enhanced resistance of psy1r pskr1 pskr2 (Figure 6E), which suggested that endogenous peptide signaling suppressed resistance to F. oxysporum.
Resistance of a PSY1R-RFO2 chimeric RLP
Because the eLRRs of RFO2 and PSY1R have remarkable similarity, we examined whether their eLRRs also share a common, interchangeable function by reciprocally swapping their homologous n-domains (Figure S3) and testing the resulting chimeric RLP and RLK genes for function in place of RFO2 and PSY1R, respectively. On the one hand, the chimeric RLP gene nPcR was the fusion of sequence coding for the extracellular n-domains of PSY1R (including domains A through C) to sequence coding for the membrane proximal c-domains of RFO2 (including domains D through G, as depicted in Figure 5A). On the other hand, the chimeric RLK gene nRcP was the fusion of sequences coding for n-domains of RFO2 and c-domains of PSY1R.
Even though amino acid similarity in n-domains of RFO2 and PSY1R is comparable to the similarity in functionally equivalent n-domains of RFO2, RLP2 and RLP2-T (Figure S3), n-domains in RFO2 and PSY1R proved to have dissimilar function. For the sake of comparison, we also fused full-length coding sequence of PSY1R downstream of the constitutive promoter ENTCUP2 to make cPSY1R. Endogenous PSY1 promotes the growth of roots, and the PSY1-insensitive roots of psy1r are shorter than wild-type roots on agar plates [29]. Stable transformation of psy1r with cPSY1R appreciably enhanced root growth while roots of independent transformants of psy1r harboring the chimeric RLK gene nRcP were not significantly longer than untransformed psy1r roots (Figure 7A).
Figure 7. Phenotypes of cRFO2 and cPSY1R chimeric RLP and RLK genes.

(A) Mean root length of psy1r, nRcP psy1r and cPSY1R psy1r after two weeks of growth on vertical PN agar plates. Error bars are the confidence interval of the mean (α = 0.05). Same italicized letter indicates that means were similar, according to Student's t test (p>0.05; for all genotypes, n = 15). Similar results were reproduced with independent T2 lines. Typical phenotypes of representative nPcR transgenic line 1E9: (B) smaller rosette leaves of four-week-old nPcR homozygote (right) as compared to leaves of its wild-type Col-0 parent (left); (C) senescence of leaves (before senescence of leaves of comparable wild type); (D) malformed, misshapened rosette leaf; and, (E) macroscopic necrotic lesion at leaf margin. (F) Severity of pleiotropy cosegregated with herbicide resistance marker linked to nPcR in self progeny of 1E9 hemizygote: Progeny of plants with normal appearance (wild type, Wt) were all Ppt-sensitive (−/−), progeny of the most affected were all Ppt-resistant (+/+), and Ppt resistance and sensitivity segregated (+/−) in progeny of plants with intermediate phenotypes.
Unexpectedly, most Ppt-resistant T1 transformants harboring the chimeric RLP gene nPcR exhibited obvious pleiotropy. Phenotypes of nPcR were reminiscent of the constitutive resistance that is displayed by activated resistance genes in mutants or the autoimmunity of hybrid necrosis, resulting from crosses between particular Arabidopsis accessions [40]–[43]; and, nPcR transformants of Col-0 or line 1A3 were similarly affected. Indeed, nPcR conferred complete resistance to FOC, FOM and FOR as nPcR plants in F. oxysporum-infected and mock-infected soil were indistinguishable; and, X-Ara staining detected no vascular infection in nPcR roots at 12 dpi. nPcR transformants had smaller (Figure 7B), often misshapen rosette leaves (Figure 7D) that had macroscopic lesions (Figure 7E) and were prone to senesce before wild-type leaves (Figure 7C). nPcR inflorescences were stunted and occasionally arrested by necrosis at their apices. nPcR pleiotropy was dose dependent as phenotypes were consistently less and more severe among nPcR hemizygotes and nPcR homozygotes in the same transgenic line, respectively (Figure 7F); and, phenotypes were more and less severe at high (30°C) and low (22°C) temperatures, respectively. We never observed similar phenotypes in transformants harboring other RLP constructs, including cRFO2.
To test whether the c-domains of RFO2 were critical for expresfsion of nPcR-related phenotypes, coding sequences of n-domains of PSY1R and c-domains of RLP2 were fused in the chimeric RLP gene nPc2 (Figure 5A). Ppt-resistant T1 plants harboring nPc2 had wild-type appearance, and T2 plants showed no enhanced resistance to FOM (Figure 5D). Thus, both strong resistance to FOM and visible pleiotropy required the c-domains of RFO2.
Considering that RLPs and RLKs may self-associate as dimers or in oligomeric complexes, we tested whether RFO2 and PSY1R contributed to the pleiotropy of nPcR by examining the effect of rfo2 and psy1r on nPcR [44], [45]. In a representative nPcR transgenic line 1E9, Ppt-resistance and pleiotropy cosegregated as a single locus. Pure-breeding 1E9 (nPcR) was crossed to both rfo2 and the double mutant pskr2 psy1r, and self-crosses of the resulting F1 plants generated F2 progeny. Among the Ppt-resistant F2 of cross nPcR×rfo2, the three possible genotypes of RFO2 segregated with the expected ratio of 1∶2∶1 (Table 1). When F2 were rank-ordered by size, the median ranks of nPcR/– rfo2 and nPcR/– RFO2 were comparable (two-tailed p = 0.52, using Mann-Whitney U test), and thus rfo2 had no effect on the small stature of nPcR. Among the Ppt-resistant F2 of cross nPcR×pskr2 psy1r, PSKR2 segregated with the expected ratio of 1∶2∶1 (Table 1). However, among the 48 Ppt-resistant (nPcR/–) F2, there were no psy1r homozygotes, and the observed segregation of PSY1R significantly deviated from the expected ratio of 1∶2∶1 (p = 0.0003, Table 1); in fact, numbers of wild-type homozygotes (PSY1R/PSYR1) and PSY1R/psy1r heterozygotes approximated the ratio (1∶2∶0) expected for a recessive lethal condition (p = 0.54). To confirm that PSY1R and nPcR were unlinked and psy1r homozygotes were viable in the absence of nPcR, we genotyped 32 Ppt-sensitive F2 and obtained the expected ratio of 1∶2∶1 for genotypes at PSY1R (Table 1). Thus, viability of nPcR-expressing plants required PSY1R.
Table 1. F2 segregation of mutants in crosses with transgene nPcR.
| Parental cross | nPcR a | Geneb | N c | Genotype of F2 | p d | |||||
| Observede | Expectedf | |||||||||
| W | H | M | W | H | M | |||||
| nPcR×rfo2 | + | rfo2 | 51 | 14 | 22 | 15 | 13 | 25 | 13 | 0.69 |
| nPcR×pskr2 psyr1 | + | pskr2 | 45 | 12 | 23 | 10 | 11 | 23 | 11 | 0.91 |
| + | psy1r | 48 | 18 | 30 | 0 | 12 | 24 | 12 | 0.0003 | |
| − | psy1r | 32 | 6 | 20 | 6 | 8 | 16 | 8 | 0.37 | |
| nPcR×tspt | + | tpst | 19 | 7 | 12 | 0 | 5 | 9 | 5 | 0.033 |
| − | tpst | 24 | 6 | 13 | 5 | 6 | 12 | 6 | 0.88 | |
F2 progeny were selected for Ppt resistance (+), which cosegregates with nPcR, or sensitivity (−).
See Materials and Methods for codominant DNA markers used to genotype particular genes.
Number of F2 plants genotyped.
Probability from chi-squared test that observed genotypes fit expected 1∶2∶1 ratio, derived from random segregation of wild-type and mutant alleles. Degrees freedom = 2. p values less than 0.05 are in bold type.
Observed genotypes were wild-type homozygotes (W), mutant homozygotes (M) and heterozygotes (H).
Expected monohybrid segregation ratio is 1 wild type : 2 heterozygotes : 1 mutant.
Because PSY1R is the putative receptor of PSY1, we tested whether the viability of nPcR-expressing plants also required the presence of active PSY1. As PSY1 is unsulfated and inactive in tpst, we crossed nPcR and tpst. In the self F2 progeny of cross nPcR×tpst, only wild-type homozygotes and TPST/tpst heterozygotes were identified among 19 herbicide-resistant (nPcR/–) F2, and their numbers approximated the 1∶2∶0 ratio (p = 0.62) and not the 1∶2∶1 ratio (p = 0.033), whereas TPST genotypes among 24 herbicide-sensitive F2 approximated the 1∶2∶1 ratio (p = 0.88, Table 1). Thus, nPcR-expressing plants depended on the presence of active, sulfated peptides, including PSY1, and the presence of PSY1R.
Discussion
Phylogenic analysis implicates most of the 90 and 57 RLP gene sequences in the reference genomes of rice and Arabidopsis, respectively, in host response to biotic stress [34]. Most RLP genes are members of species-specific clades and (76 percent in rice and 58 percent in Arabidopsis) are clustered at loci with two or more related genes [33]. Species-specific genes and gene clustering are also features of the NB-LRR family of resistance genes and imply that lineages of RLP genes are expanding, contracting and diversifying to meet the evolving challenge of infectious disease [46], [47]. Indeed, reverse genetic approaches show that loss-of-function mutations in three Arabidopsis RLP genes quantitatively compromise innate immunity to virulent and nonhost pathogens [31], [48], [49].
However, diversity in RLP genes has not been associated with a disease resistance trait in Arabidopsis or rice until now. In cultivated species such as tomato, RLP resistance traits are typically the result of interspecific breeding or the inadvertent propagation of loss-of-function polymorphisms [50], [51]. The highly diverged RFO2 alleles suggest that diversity in RLP genes contributes to quantitative variation in resistance in wild species.
Although monogenic resistance traits are usually associated with NB-LRR genes, several RLP genes confer strong monogenic resistance to specific pathogens as well [52]. The strong pathogen-specific resistance of transgenic RFO2 is reminiscent of such gene-for-gene resistance. The best-studied RLP genes are in the Cf clade and mediate resistance to specific races of the foliar fungal pathogen Cladosporium fulvum that express corresponding avirulence genes [50]. Meanwhile, apple Vfa1 and Vfa2 confer resistance to five races of the obligate fungal pathogen Venturia inaequalis, tomato LeEix2 confers recognition of an ethylene-inducing xylanase from biocontrol fungus Trichoderma viride, oilseed rape LepR3 confers resistance to Leptosphaeria maculans expressing AvrLm1, and tomato Ve1 confers strong resistance to races of Verticillium species expressing avirulence gene Ave1 [53]–[56]. Interestingly, Ve1 also confers modest quantitative resistance to virulent F. oxysporum forma specialis lycopersici [53]. Likewise, we presume that RFO2 perceived an extracellular Fusarium-derived signal that was present in FOM infection and absent in FOC infection [57]. However, we cannot discount that FOC infection suppressed RFO2's perception of a signal that was present in all F. oxysporum infections. In either case, once induced, RFO2 was effective against all three crucifer-infecting formae speciales as the constitutive resistance of nPcR lacked specificity.
PSY1 and PSK signaling compromised resistance to vascular infection by F. oxysporum. Recently, Igarashi et al. reported that pskr1 (but not pskr2) is more resistant to leaf infection by virulent P. syringae [58]. PSY1R, PSKR1 and PSKR2 were identified and characterized for perception of PSY1 and PSK and for the effects that this perception has on root growth, cell proliferation and senescence [29], [30]. Igarashi et al. suggest that PSK signaling directs allocation of resources between energy-intensive processes, toward growth and away from immunity [58]. However, PSK and PSY1 signaling more fully influences the longevity and growth potential of mature differentiated cells [29], and absence of peptide signaling in the triple mutant arguably had a more modest effect on plant mass than wilt resistance (in Figure 6A). Natural resistance traits, such as RFO1, RFO2 and tomato Immunity genes, promote resistance to Fusarium wilt by inhibiting infection in the vascular cylinder [2], [28]. Because PSK depresses stress responses in general and immunity in particular, we suspect that peptide signaling depressed the considerable but incomplete resistance of Col-0 to FOC and FOR [9], [37], [58], [59]. The strong resistance of tpst suggests that endogenous PSY1 and PSK depressed immunity, though the expression of other proteins with tyrosine sulfation, including root meristem growth factors, is also affected by tpst [60]. The strong wilt resistance of the receptor triple mutant does not tell us whether FOC or FOR normally exploits PSY1 and/or PSK signaling to induce susceptibility; however, it does demonstrate that manipulation of even basal signaling would be a fruitful target for pathogen effectors.
Amino acid identity in the eLRRs of RFO2 and PSY1R is conspicuous because RLPs and RLKs usually lack meaningful sequence conservation beyond the structural constraints of the eLRR motif [33]. Premature termination of translation in an RLK gene, such as Xa21D, may give rise to a residual RLP-like gene [61]. However, RFO2 is not simply a truncation of PSY1R as the RFO2-related RLPs and PSY1R have little if any sequence conservation outside of the eLRRs (see Figure S4). The regular presence of PSY1R-related RLK genes and sporadic distribution of RFO2-like RLP genes in plant genomes in the Phytozome v8.0 database presumably reflects the distinct roles of these genes in peptide signaling and defense response, respectively (A.D., unpublished data) [62].
In spite of the relatedness of eLRRs of RFO2 and PSY1R, we failed to connect RFO2 to a role in PSY1 signaling in normal root growth. PSY1 supplementation enhances root growth, and roots of psy1r are shorter than wild-type (Figure 7A) [29]. However, neither transgenic expression (tRFO2 and cRFO2) nor deficiency (rfo2) of RFO2 affected root length; and, we found that root lengths of psy1r and psy1r rfo2 were comparable (Y.S., unpublished data).
RFO2's similarity to PSY1R and lack of function in PSY1 signaling are consistent with the decoy model for perception of pathogen effectors [63]. In theory, effectors that target PSY1R might select for a decoy receptor, such as RFO2, that mimics the interaction between effectors and PSY1R but lacks the function that effectors are targeting. Because PSY1 signaling suppressed immunity to F. oxysporum infection, the relevant effector would be an agonists or positive regulator of PSYR1. Although how PSY1R perceives PSY1 is unknown, PSK directly bind to the PSK receptor, and a photo-activated analog of PSK preferentially labels the loop out sequence within eLRRs [64].
Considering this decoy model, we were surprised that the c-domains of RFO2 were responsible for resistance to FOM. We anticipated that sequence coding for the n-domains, including eLRRs, would distinguish RFO2 from homologs (RLP2 and RLP2-T) that failed to confer resistance. Instead, n-domains of RFO2, RLP2 and RLP2-T were functionally equivalent. Contrast this with similar domain-swapping experiments that invariably map recognition of specific C. fulvum effectors to sequence in eLRRs of Cf proteins [50]. Interestingly, the critical role for membrane-proximal c-domains of RFO2 is consistent with the observation that cRLP2-T, unlike cRLP2, expressed some resistance (albeit weaker than cRFO2). While RLP2-T is generally more related to RLP2, the c-domains of RLP2-T and RFO2 are more similar to each other and dissimilar to RLP2 (Figure S4). That RLP2 is a nonfunctional pseudogene could explain the lack of resistance from RLP2, just as a loss-of-function polymorphism accounts for the susceptible allele of Ve1 [51]. However, Wang et al. report that RLP2 is functional [65]. When the CLV2 promoter is used to ectopically-expresses RLP2, wild-type carpel number and pedicel length is restored to clv2. Suppression of clv2 by RLP2, though an abnormal gain of function, clearly shows that RLP2 can compensate for loss of another RLP gene. Possibly, resistance occurs with RFO2 and not RLP2 because defense signaling is engaged by the c-domains of RFO2 and not by the c-domains of RLP2.
The nPcR pleiotropy is reminiscent of constitutive activation of resistance in a number of laboratory mutants and transgenic plants [41], [43], and nPcR showed complete resistance to infection by FOC, FOM and FOR. However, it should be noted that nPcR roots, like nPcR shoots, had stunted and irregular growth. Nevertheless, roots of tir3 are stunted and have irregular growth too, and FOC infection of tir3 and wild type is similar [28].
The critical role of the c-domains of RFO2 in both the FOM-specific resistance of cRFO2 and constitutive resistance of nPcR suggests that the pleiotropy of nPcR is the aberrant, constitutive activation of resistance that FOM normally induces via RFO2. When the c-domains in RLP2, which expressed no resistance to FOM, replaced c-domains of RFO2 in cRFO2 and nPcR, the resulting RLPs nRc2 and nPc2 expressed neither resistance to FOM nor visible pleiotropy. If pleiotropy were simply the consequence of expressing a truncated PSY1R without a kinase domain, nPc2 should also express visible pleiotropy.
In an attempt to explain the constitutive resistance of nPcR, we recalled that PSY1R perceived endogenous PSY1 while RFO2 appeared insensitive. When the n-domains of RFO2 and PSY1R were swapped, the n-domains of RFO2 appeared insensitive to PSY1 in the chimeric RLK nRcP, which failed to suppress the reduced root growth of psy1r (Figure 7A). We reasoned that RFO2 was only activated by an FOM-derived signal, while nPcR was continuously activated by endogenous PSY1. If this hypothesis were correct, simply removing endogenous PSY1 should abolish the pleiotropy of nPcR.
Contrary to expectation, absence of active sulfated peptides, including PSY1, (in tpst) as well as loss of the PSY1 receptor (in psy1r) exacerbated the phenotype of nPcR. We imagine that plants with the tpst nPcR and psy1r nPcR genotypes were not recovered because constitutive resistance had attained a lethal level of expression. This would be consistent with the obvious effect that a two-fold difference in nPcR copy number in hemizygotes and transgene homozygotes had on nPcR phenotypes (in Figure 7F) [66], [67]. Genetic analysis discounted trivial explanations for why tpst nPcR and psy1r nPcR were not recovered, such as linkage between the nPcR transgene and mutation. Thus, paradoxically, we found that PSY1 was not required to activate resistance via nPcR, rather PSY1 signaling was negatively regulating the constitutive resistance of nPcR.
We considered the possibility that PSY1 and PSK signaling indirectly suppressed the constitutive resistance of nPcR in wild type. Because psy1r pskr1 pskr2 and tpst strongly enhanced resistance to F. oxysporum infection, it was possible that an enhanced defense response in tpst made the constitutive resistance of nPcR lethal. On the other hand, psy1r had a much more modest effect on resistance to F. oxysporum than the triple receptor mutant or tpst, so it seemed remarkable that psy1r would have as profound an effect on viability as tpst. Nevertheless, if basal signaling of PSY1 and PSK were suppressing the constitutive resistance of nPcR, we reasoned that inducing PSK signaling by supplementation with PSK should counteract constitutive resistance and improve growth of nPcR seedlings, including roots. According to Igarashi et al., exogenous PSK can suppress elicitor-induced root growth inhibition [58]. However, we found that added PSK failed to have an appreciable effect on the abbreviated root growth of nPcR seedlings or improve the appearance of nPcR seedlings, even as PSK was able to stimulate the root growth of wild-type and psy1r seedlings (Figure S6). Furthermore, pskr2 nPcR, which has a partial deficiency in PSK signaling, was recovered from crosses and was indistinguishable from nPcR siblings, though both pskr2 and psy1r contributed to resistance to F. oxysporum (in Figure 6). Because manipulation of PSK signaling failed to alleviate or exacerbate the pleiotropy of nPcR, PSY1 signaling appeared to be intimately associated with negative regulation of nPcR.
To account for the PSY1-dependent negative regulation of nPcR, we hypothesize that an activity-dependent negative feedback mechanism that normally controls the expression of PSY1R also controls the expression of nPcR (Figure 8A). In this scenario, engagement of PSY1 and PSY1R has two consequences, (i) transduction of PSY1 signaling and (ii) downregulation of PSY1-activated PSY1R. Engagement of PSY1 and nPcR, on the other hand, targets nPcR for downregulation but does not transduce the PSY1 signal. Interestingly, endocytosis is proposed to have a prominent role in attenuating PAMP signaling [68]. Downregulation of the flagellin receptor FLS2 by its synthetic peptide ligand flg22 is a clear precedent for negative feedback in RLK signaling in plants [69]. Engagement of FLS2 and flg22 recruits the coreceptor BAK1, which concomitently promotes FLS2 signaling as well as proteosome-mediated degradation of FLS2 [70].
Figure 8. Model for RFO2-mediate resistance.

In our model, the extracellular LRRs of PSY1R are activated by PSY1, and the intracellular kinase domain of PSY1-activated PSY1R then (i) tranduces PSY1 signaling, depressing resistance to F. oxysporum and (ii) promotes downregulation of PSY1-activated PSYR1. Downregulation has an extracellular component (X) because the eLRRs of PSY1R are presumed to be the target of downregulation in nPcR. RFO2 is a decoy receptor, mimicking the PSY1-activated eLRRs of PSY1R that are competent for downregulation by X. Extracellular (out) and intracellular (in) compartments are on either side of the cell membrane. (A) In nPcR, nPcR is incompletely downregulated by PSY1 negative feedback regulation. Consequently, stable expression of the c-domains (R) of RFO2 in nPcR induces resistance constitutively. (B) In tpst nPcR or pyr1r nPcR, expression of nPcR is further stabilized by loss of all downregulation, by loss of PSY1 activation (in tpst) or by loss of PSY1-activated PSY1R (in psy1r). Expression of resistance exceeds a lethal threshold without downregulation. Nevertheless, X downregulates RFO2 independent of PSY1 signaling. (C) In infected rfo2, FOM inhibits X and downregulation of PSY1-activated PSY1R. Stabilized PSY1-activated PSY1R upregulates PSY1 signaling and exaggerates normal suppression of resistance. s(D) In infected RFO2, FOM's inhibition of X also stabilizes RFO2 expression and thereby induces a robust resistance response.
Importantly, the apparent negative regulation of nPcR by PSY1 signaling suggests a connection between PSY1 signaling and RFO2-mediated resistance. The mechanism that normally downregulates PSY1-activated nPcR presumably targets a common structural feature in PSY1R and nPcR, which is the extracellular n-domains of PSY1R. Conservation of the corresponding n-domains of RFO2 and PSY1R implies that RFO2 is also a target of the same negative regulation.
Because constitutive resistance of nPcR was not dependent on PSY1 (in tpst nPcR), we needed an alternative explanation for the different phenotypes of cRFO2 and nPcR. Both cRFO2 and nPcR encode the same c-domains of RFO2, so it must be the n-domains of nPcR that constitutively provide resistance and the n-domains of RFO2 that constitutively provide no resistance (in the absence of FOM infection). Considering this, we hypothesize that the eLRRs of RFO2, acting as a decoy, mimic a state of the eLRRs of PSY1R that is already PSY1-activated and competent for downregulation. Intrinsically-activated eLRRs in RFO2 would account for their insensitivity to PSY1. Being competent for downregulation, RFO2 would be constitutively downregulated, even if no PSY1 signaling were present, and thus would normally express no resistance. On the other hand, nPcR, which needs activation by PSY1 in order to be downregulated, would constitutively express resistance in those tissues where, and at times when, there is insufficient PSY1 to fully downregulate nPcR (Figure 8B).
We present a model to explain how an effector could induce PSY1 signaling without directly engaging PSY1R, and how RFO2 could directly or indirectly perceive such an effector. By expressing an effector that inhibits the PSY1-dependent negative feedback mechanism, FOM could stabilize PSY1-activated PSY1R (Figure 8C). Just as psy1r and tpst were able to upregulated the expression of resistance by nPcR by abolishing PSY1-dependent downregulation, an effector could upregulate PSY1 signaling by inhibiting the downregulation of PSY1-activated PSY1R and thereby suppress host immunity. However, in plants expressing RFO2, the effector would likewise stabilize RFO2, acting as a decoy for the downregulation-competent PSY1R. Stabilized and upregulated RFO2 would induce robust defense response (Figure 8D).
There are two appealing aspects to this model. For one, existence of negative feedback in PSY1 signaling explains why a pathogen targets this mechanism rather than secretes a PSY1-like ligand. Chronic PSY1 signaling would be achieved more effectively by stabilizing endogenous PSY1-activated PSY1R if perception of excess PSY1-like ligand were suppressed by downregulation of the receptor. For two, RFO2 behaves as a guard protein and does not need to directly engage the effector that it recognizes. If an effector were to inhibit any component of the negative feedback mechanism, RFO2 would be activated. Thus, RFO2 functions as a guard protein for the negative feedback mechanism.
Simply changing the transcriptional context or copy number of RFO2 in transgenic plants converted a modest quantitative resistance trait into a strong resistance gene. No aberrant visible phenotype accompanied the stronger resistance of transgenic RFO2, and resistance remained specific to FOM. We wonder whether gene expression rather than protein structure limits the strength of other resistance QTLs as well. Effectiveness of some qualitative gene-for-gene resistance traits is restricted, for instance, to a developmental stage, which suggests a partial deficiency in gene expression [71], [72]. Some opinion holds that resistance QTLs are in fact weak gene-for-gene resistance traits [21], [26]. If our experience of limited gene expression were commonplace, the potential utility of genes underlying resistance QTLs might be underappreciated.
Finally, our initial analysis of RFO2 has produced a testable model to account for RFO2-mediated resistance. Future work should establish the biochemical nature of PSY1-dependent negative regulation of nPcR. In addition, identification of the relevant F. oxysporum PAMP(s) and/or effector(s) should prove especially useful for molecular characterization of this resistance mechanism.
Materials and Methods
Arabidopsis and plant growth conditions
Salk insertion lines and BAC DNA clones, F6I1, F20D23 and F28G4 were obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus, OH). Seeds of psy1r, psy1r pskr1, pskr1 pskr2, psy1r pskr1 pskr2 and tpst were provided by Dr. Y. Matsubayashi (Nagoya University, Nagoya, Japan). Ty-0, rfo1 and lines 1A3 and 4D2 were derived from F1BC plants in [9]. Plants were grown on Jiffy7 peat pellets (Growers Solution, Cookeville, TN) under cool white fluorescent lighting with moderate intensity with 12-hr daylength and 28°C daytime and 26°C nighttime temperatures. Seedlings were grown from bleach-sterilized seeds on Petri plates with plant nutrient (PN) minimal medium and 0.8% agar alone or, for antibiotic selection, with 0.5% sucrose [73]. Transgenic seeds were selected with kanamycin (50 mg/L) or phosphinothricin (Ppt, 20 mg/L). Phytosulfokine-α was purchased from PolyPeptide Laboratories, Inc., Torrance, CA. The Arabidopsis Information Resource (TAIR, www.arabidopsis.org) provided reference genome sequence and annotation.
Infections with F. oxysporum
FOC, FOR and FOM originate from P.H. Williams through H.C. Kistler [11], [74]. F. oxysporum cultures were grown on Czapek-Dox minimal medium (Oxoid Ltd., Hampshire, UK), and conidia were harvested from 3- to 5-d shaken cultures, washed 3 times and resuspended in sterile water. For infection, conidial density was adjusted to between 106 to 108 conidia/mL, using a hemacytometer, and 2- to 3-wk old plants were irrigated with conidial suspension or water (for mock infection). Disease symptoms were scored between 12 and 24 days post infection (dpi) using a health index (HI), previously called a disease index in [9], with ordinal ratings of one (dead) to five (unaffected) in steps of 0.5. Plants that were deemed susceptible typically had HI<3, or resistant had HI≥4 or were scored as having intermediate resistance if 3≤HI<4. For statistical analysis, plants were rank-ordered, from most susceptible to most resistance, and Mann-Whitney U test was used to evaluate the ranks of different genotypes. Sometimes rank-order was derived from multiple HI scores, recorded at two or more time points, in which case later HI scores had priority over earlier scores. From rank-order, plants with the lowest, highest or middle third of ranks were arbitrarily deemed susceptible, resistant or had intermediate resistance, respectively.
Mapping RFO2
Linkage analysis of 80 FOM-infected progeny from the cross of 1A3 and 4D2 mapped RFO2 between flanking SSLP markers CIW12 and F21M12 [9]. To confirm RFO2 genotypes of plants with informative recombination breakpoints, we tested the co-segregation of Rfo2 phenotype (resistance to FOM) and genotype of an RFO2-linked marker in 25 to 50 progeny. If Rfo2 phenotype and marker genotype cosegregated, the genotype was RFO2-C/T; and, if Rfo2 phenotype and marker genotype assorted independently, the genotype was RFO2-T/T or RFO2-C/C. A fine-map position for RFO2 was obtained by screening for recombination breakpoints in the CIW12-F21M12 interval among 200 uninfected progeny from cross (1A3×4D2) and the 248 original F1BC progeny (see Table S1 for description of SSLP markers for fine-mapping) [9]. In particular, Rfo2 phenotype co-segregated with RFO2-linked markers in progeny of 4E3 and 1B9 that have breakpoints on either side of RFO2 (see Figure S5A). The interval between breakpoints in 4E3 and 1B9 was less than the 258 kb between SSLPs F11A6 and F17F16 (see Figure S5B).
Cloning RFO2
In total, 25 Col-0 genomic restriction fragments of 3 BAC clones F6I1, F28G4 and F20D23, representing 50 of 68 genes in the RFO2 interval (see Figure S1), were subcloned into binary vector pPZP212 [75] for Agrobacterium tumefaciens-mediated transformation of line 1A3. Kanamycin resistance selected for stable integration of Col-0 genomic subclones. Relative HI scores of multiple FOM-infected T1 and/or T2 transformants as well as untransformed 1A3 were used to assign Rfo2 phenotypes to subclones. A summary of all Col-0 genomic subclones and their Rfo2 phenotypes is in Figure S1.
Sequencing RFO2 in Ty-0
RFO2-T sequence (Genbank accession HQ141412) was a contig assembled from PCR-sequencing. Both strands of four overlapping PCR products amplified from Ty-0 DNA were sequenced. Primer sequences, sizes of PCR products and lengths of sequence overlap are in Table S2. Best-matched sequence to RFO2-T in TAIR10 reference genome database was between nucleotides 5,893,811 to 5,906,689 on chromosome 1, according to BlastN 2.2.8 search function at TAIR. DNA similarity search tool YASS (http://bioinfo.lifl.fr/yass/) assisted the hand-edited alignment of intergenic regions in Col-0 and Ty-0 sequences [76]. The percent nucleotide identity was calculated in a 75-nucleotide window centered at a nucleotide position, and mismatched nucleotides and gaps of any length in the alignment of Col-0 and Ty-0 sequences were discounted equally. Sequence of the Arabidopsis lyrata ortholog AlRLP2 (gene 920636) was from the Phytozome v8.0 plant genome database.
Genotyping
DNA markers were PCR-amplified from crude leaf preparations and analyzed as in [77]. PCR primers for genotyping tpst, rfo2, pskr2 and psy1r are in Table S3.
Genotypic and phenotypic analysis of rfo2
At least five plants for each of 30 homozygous Salk T-DNA lines (listed in Table S4) were infected with 5×107 FOM conidia/mL. Four lines (Salk_014524, Salk_051677, Salk_049366 and Salk_097514) were crossed to Ty-0, and resulting F1 were infected with 107 FOM conidia/mL. F1BC progeny of (rfo2×Ty-0)×Ty-0 as well as the original (Col-0×Ty-0)×Ty-0 population [9] were genotyped with RFO1-, RFO2-, RFO3-linked and RFO-unlinked Col-0-specific dominant markers (Table S5). Dominant marker primers were used with the Qiagen Multiplex PCR kit (Qiagen Inc., Valencia, CA). FOM-infected F1BC populations were rank-ordered using HI scores recorded at 12, 15 and 18 dpi. Lowest, middle and highest third of ranks were designated susceptible, intermediate resistance and resistant, respectively.
Chimeric RLP and RLK transgenes
BamHI and SpeI, or SpeI and NotI, sites were introduced to 5′ and 3′ ends of PCR-amplified sequence coding for n- or c-domains, respectively, using PCR primers with restriction sites at 5′ ends (Table S6). Sequences coding for n- and c-domains of RFO2, RLP2 and PSY1R or RLP2-T were PCR-amplified from Col-0 or Ty-0 DNA. DNA sequencing was used to verify the sequence of PCR-amplified subclones. Restriction fragments coding for n- and c-domains were ligated to BamHI- and NotI-digested binary vector pORE-E3 to make cRFO2, cRLP2, cRLP2-T and cPSY1R expression constructs [36]. To make chimeric constructs, BamHI- and SpeI-digested DNA for n-domains in cRFO2, cRLP2, cRLP2-T and cPSY1R expression constructs were exchanged using DNA ligation. In pORE binary vectors, gene constructs were located in T-DNA and were ready for transfer to plants using A. tumefaciens GV3101 [36], [78]. Resistance to Ppt selected for seedlings with stably integrated constructs, and the presence of chimeric gene sequences in transformed Ppt-resistant plants was verified by PCR.
Visualizing and quantifying glycosidase activity in roots
Cleaning and staining of roots with 5-bromo-4-chloro-3-indoxyl-α-L-arabinofuranoside (X-Ara), 4-nitrophenyl-α-L-arabinofuranoside (NP-Ara), purchased from Gold Biotechnologies Inc. (St. Louis, MO) is described in [28]. To quantify Fusarium-derived arabinofuranosidase activity, freshly harvested roots were incubated with 0.04% NP-ARA for 16 h at 28°C in 30-fold excess staining solution.
Phylogenic analysis
Coding sequences and translated sequences of RFO2, RLP2, RLP2-T, PSY1R (Atg1g72300) and PSKR1 (At2g02220) in the TAIR10 genome and proteome databases were aligned using the Clustal method and default settings in MEGA5 [79].
Supporting Information
Resistance phenotype of Col-0 subclones in the RFO2 interval. The RFO2 interval (258 kbp between nucleotides 5,766,000 and 6,023,500 in TAIR10 reference sequence for chromosome 1) was defined by a recombinant breakpoint in lines 4E3 between SSLPs F11A6 (at nucleotide 5,766,000) and F20D23 (at nucleotide 5,820,000), on the low end, and a recombination breakpoint in line 1B9 between SSLPs F28G4 (at nucleotide 5,943,000) and F17F16 (at nucleotide 6,023,500), on the high end. Resistance phenotypes (Rfo) of Col-0 genomic clones were tested in T1 and/or T2 transformants of line 1A3. Horizontal bars are proportional to the sequence length of subcloned Col-0 DNA and extend across their respective positions in the genomic interval below. Bars are labeled with the subclone names and, in paratheses, the sizes and gene content of subcloned sequence (to the right). Fifty of the 68 genes in the RFO2 interval were included in at least one construct: Xba1.5 includes nucleotides 5777587 to 5789031 and genes AT1G16900, AT1G16905, AT1G16910, AT1G16916 and AT1G16920; Xba1.4 includes nucleotides 5791422 to 5801901 and genes AT1G16940, AT1G16950 and AT1G16960; Pac1.4 includes nucleotides 5810084 to 5824597 and genes AT1G17000, AT1G17010, AT1G17020 and AT1G17030; Pac1.3 includes nucleotides 5824895 to 5833233 and genes AT1G17040 and AT1G17050; Kpn1.4 includes nucleotides 5833071 to 5841463 and genes AT1G17060, AT1G17070 and AT1G17080; Xba1.3 includes nucleotides 5837955 to 5843587 and genes AT1G17070, AT1G17080 and AT1G17090; Kpn1.3 includes nucleotides 5846018 to 5854394 and genes AT1G17110 and AT1G17120; Xba1.2 includes nucleotides 5843587 to 5855420 and genes AT1G17100, AT1G17110 and AT1G17120; Pac1.2includes nucleotides 5850646 to 5866025 and genes AT1G17120, AT1G17130, AT1G17140, AT1G17145 and AT1G17147; Pac1.1 includes nucleotides 5866025 to 5878981 and genes AT1G17150, AT1G17160, AT1G17170, AT1G17180 and AT1G17190; Sal1.4 includes nucleotides 5867417 to 5882030 and genes AT1G17160, AT1G17170, AT1G17180, AT1G17190 and AT1G17200; Sal1.3 includes nucleotides 5882030 to 5891121 and genes AT1G17210 and AT1G17220; Xba1.1 includes nucleotides 5878230 to 5891727 and genes AT1G17200, AT1G17210 and AT1G17220; Kpn1.2 includes nucleotides 5886702 to 5904885 and genes AT1G17220, AT1G17232, AT1G17230, AT1G17235, AT1G17240 and AT1G17250; Sal1.2 includes nucleotides 5891121 to 5914913 and genes AT1G17232, AT1G17230, AT1G17235, AT1G17240, AT1G17250, AT1G17260, AT1G17270 and AT1G17275; Kpn1.1 includes nucleotides 5905771 to 5921349 and genes AT1G17260, AT1G17270, AT1G17275, AT1G17277, AT1G17280 and AT1G17285; Sal1.1 includes nucleotides 5933219 to 5946989 and genes AT1G17340, AT1G17345 and AT1G17350; Xma1.1 includes nucleotides 5970942 to 5986501 and genes AT1G17420, AT1G17430 and AT1G17440; Avr2.1 includes nucleotides 6007806 to 6004198 and genes no full-length gene; Nsi1.3 includes nucleotides 5898644 to 5889883 and genes AT1G17232, AT1G17230, AT1G17235 and AT1G17240; BamH1.1 includes nucleotides 5900529 to 5891012 and genes AT1G17232, AT1G17230, AT1G17235 and AT1G17240; Age1.1 includes nucleotides 5902722 to 5891905 and genes AT1G17232, AT1G17230, AT1G17235, AT1G17240 and AT1G17250; Hind3.1 includes nucleotides 5904051 to 5895229 and genes AT1G17235, AT1G17240 and AT1G17250; Nsi1.1 includes nucleotides 5907976 to 5898644 and genes AT1G17250 and AT1G17260; and, Nsi1.2 includes nucleotides 5904885 to 5898644 and gene AT1G17250.
(PDF)
Domain structure of RFO2. The amino acid sequence of RFO2, in single-letter code, is divided into seven alphabetically named domains. Sequence in the C domain is further subdivided into 23 leucine-rich repeats (LRRs) and a loop out sequence. The number of amino acid residues (Len) in each domain and a brief comment about each sequence are to the right of sequences. Residues corresponding to the eLRR consensus (LxxLxxLxxLxLxxNxLxGxIPxx, where x represents a nonconserved residue between conserved residues) are highlight by underlined bold type [34]. Potential N-glycosylation sites are highlighted by half-filled bold type. Conserved cysteine residues in the N- and C-cap (domains B and D, respectively) are highlighted in bold [34]. In domain E, acidic residues are highlighted in bold. In domain F, a predicted transmembrane sequence is underlined, and a conserved GxxxG motif is highlighted in bold [33].
(PDF)
Alignment of eLRRs (domain C) of PSY1R-like proteins and PSKR1. Alignment of the translated amino acid sequences of PSY1R, RFO2, RLP2 (RLP2c), RLP2-T (RLP2t) and PSKR1, encoding domain C, are shown in single-letter code. All residues that are identical to PSY1R are highlighted by white type on black background. The amino acid position from the start codon is given for the leftmost residue in each line.
(PDF)
Alignment of PSY1R and RFO2-like RLP sequences at the carboxy-terminal ends of RLPs. Translated sequences of RFO2, RLP2 (RLP2c) and RLP2-T (RLP2t), encoding carboxy-terminal ends of RLPs, are aligned to the translated sequence of PSY1R in single-letter code. Amino acid residues that are identical in >50 percent of sequences are highlighted by white type on black background. The amino acid position from the start codon is given for the leftmost residue, and asterisks are stop codons. A SpeI restriction site (5′-ACT-AGT-3′), which codes for the threonine (T) and serine (S) residues at the arrow, was introduced as a silent mutation into coding sequences for the creation of chimeric fusions among RLP and RLK genes.
(PDF)
Recombination breakpoints defining RFO2 map position. (A) Fractions of n F2 from cross 1A3×4D2 that were susceptible (HI scores <2, open column), had intermediate resistance (2≤HI scores <4, half-filled) or were resistant (HI scores ≥4, filled) at 18 dpi. Only F2 of F1 plants 4E3, 1B9 and 5E1 from cross 1A3×4D2 that were either homozygous Ty-0 (T/T) or Col-0/Ty-0 heterzygotes (C/T) at RFO2-linked markers as well as C/T at RFO1-linked marker F19K16 are shown. (B) Genotypes, either Ty-0 (T) or Col-0 (C), at RFO2-linked markers (above) on the single recombinant chromosomes in F1 plants 4E3, 5E1 and 1B9 from cross 1A3×4D2. Marker intervals with a crossover are marked with ‘X’. Number of TAIR10 annotated open reading frames (ORFs) in marker intervals is given below. Rfo2 phenotype of F1 plants (on the right) was evaluated in F2 progeny (in A).
(PDF)
Root growth of nPcR is unaffected by PSK peptide. Two-week old seedlings of Col-0 (wild type), psy1r, psy1r, pskr1pskr2 and nPcR (line 1E9) were grown from seeds sown on vertically-oriented PN agar plates with (+) or without (−) added PSK (0.1 µM). (A) Lengths of PSK-treated wild-type and psy1r roots (n = 20) were longer than untreated roots (n = 20), according to Student's t test (two-tailed p = 0.044 and 0.015, respectively) while length of PSK-treated and untreated psy1r pskr1 pskr2 roots (n = 20) had similar lengths (p = 0.60). (B) Length of PSK-treated wild-type roots (n = 20) were longer than untreated roots (n = 20), according to Student's t test (two-tailed p = 0.005) while length of PSK-treated and untreated nPcR roots (n = 20) had similar lengths (p = 0.16).
(PDF)
PCR primers for mapping and genotyping RFO2 and RFO1.
(PDF)
PCR primers and products for sequencing RFO2 in Ty-0.
(PDF)
PCR primers for genotyping Salk insertions.
(PDF)
Homozygous T-DNA insertion lines in RFO2 genetic interval.
(PDF)
PCR primers for dominant multiplex PCR markers.
(PDF)
PCR primers for constructing RLP and RLK genes.
(PDF)
Acknowledgments
We thank K. Hirschi and F. Ausubel for critical reading of manuscript drafts, UCLA undergraduate student Kristen Guiang for assistance, and Y. Matsubayashi for providing seeds.
Funding Statement
This work was paid by start up funds by the University of California Los Angeles. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Beckman CH (1987) The nature of wilt diseases of plants. St. Paul: American Phytopathological Society.
- 2. Beckman CH, Roberts EM (1995) On the nature and genetic basis for resistance and tolerance to fungal wilt diseases of plants. Adv Bot Res 21: 35–77. [Google Scholar]
- 3.Mace ME, Bell AA, Beckman CH, editors (1981) Fungal wilt diseases of plants. New York: Academic Press.
- 4. Talboys PW (1972) Resistance to vascular wilt fungi. Proc R Soc Lond Ser B Biol Sci 181: 319–332. [Google Scholar]
- 5. Liu J, Bell AA, Wheeler MH, Stipanovic RD, Puckhaber LS (2011) Phylogeny and pathogenicity of Fusarium oxysporum isolates from cottonseed imported from Australia into California for dairy cattle feed. Can J Microbiol 57: 874–886. [DOI] [PubMed] [Google Scholar]
- 6. Fourie G, Steenkamp ET, Gordon TR, Viljoen A (2009) Evolutionary relationships among the Fusarium oxysporum f. sp. cubense vegetative compatibility groups. Appl Environ Microbiol 75: 4770–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kistler HC (1997) Genetic diversity in the plant-pathogenic fungus Fusarium oxysporum. Phytopathology 87: 474–479. [DOI] [PubMed] [Google Scholar]
- 8. Baker KF (1948) Fusarium wilt of garden stock (Mathiola incana). Phytopathology 38: 399–403. [Google Scholar]
- 9. Diener AC, Ausubel FM (2005) RESISTANCE TO FUSARIUM OXYSPORUM 1, a dominant Arabidopsis disease-resistance gene, is not race specific. Genetics 171: 305–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Koornneef M, Meinke D (2010) The development of Arabidopsis as a model plant. Plant J 61: 909–921. [DOI] [PubMed] [Google Scholar]
- 11. Bosland PW, Williams PH (1987) An evaluation of Fusarium oxysporum from crucifers based on pathogenicity isozyme polymorphism, vegetative compatibility and geographic origin. Can J Bot 65: 2067–2073. [Google Scholar]
- 12. Kendrick JB, Snyder WC (1942) Fusarium wilt of radish. Phytopathology 32: 1031–1033. [Google Scholar]
- 13. Joseph C, Gilman JC (1916) Cabbage Yellows and the Relation of Temperature to Its Occurrence. Ann Mo Bot Gard 3: 25–84. [Google Scholar]
- 14. Sherbakoff CD (1949) Breeding for resistance to Fusarium and Verticillium wilts. Bot Rev 15: 377–422. [Google Scholar]
- 15. Laine A-L, Burdon JJ, Dodds PN, Thrall PH (2011) Spatial variation in disease resistance: from molecules to metapopulations. J Ecol 99: 96–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goicoechea N (2009) To what extent are soil amendments useful to control Verticillium wilt? Pest Manag Sci 65: 831–839. [DOI] [PubMed] [Google Scholar]
- 17. Michielse CB, Rep M (2009) Pathogen profile update: Fusarium oxysporum. Mol Plant Pathol 10: 311–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Anderson JP, Badruzsaufari E, Schenk PM, Manners JM, Desmond OJ, et al. (2004) Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16: 3460–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Trusov Y, Rookes JE, Chakravorty D, Armour D, Schenk PM, et al. (2006) Heterotrimeric G proteins facilitate Arabidopsis resistance to necrotrophic pathogens and are involved in jasmonate signaling. Plant Physiol 140: 210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Boyd LA, Ridout C, O'Sullivan DM, Leach JE, Leung H (2012) Plant-pathogen interactions: disease resistance in modern agriculture. Trends Genet doi:10.1016/j.tig.2012.10.011 [DOI] [PubMed] [Google Scholar]
- 21. St Clair DA (2010) Quantitative disease resistance and quantitative resistance Loci in breeding. Annu Rev Phytopathol 48: 247–268. [DOI] [PubMed] [Google Scholar]
- 22. Rafiqi M, Bernoux M, Ellis JG, Dodds PN (2009) In the trenches of plant pathogen recognition: Role of NB-LRR proteins. Semin Cell Dev Biol 20: 1017–1024. [DOI] [PubMed] [Google Scholar]
- 23. Bent AF, Mackey D (2007) Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu Rev Phytopathol 45: 399–436. [DOI] [PubMed] [Google Scholar]
- 24. Kou Y, Wang S (2012) Toward an understanding of the molecular basis of quantitative disease resistance in rice. J Biotechnol 159: 283–290. [DOI] [PubMed] [Google Scholar]
- 25. Zhang Y, Lubberstedt T, Xu M (2013) The genetic and molecular basis of plant resistance to pathogens. J Genet Genomics 40: 23–35. [DOI] [PubMed] [Google Scholar]
- 26. Poland JS, Balint-Kurti PJ, Wisser RJ, Pratt RC, Nelson RJ (2009) Shades of gray: the world of quantitative disease resistance. Trends Plant Sci 14: 21–29. [DOI] [PubMed] [Google Scholar]
- 27. Lehti-Shiu MD, Zou C, Hanada K, Shiu SH (2009) Evolutionary history and stress regulation of plant receptor-like kinase/pelle genes. Plant Physiol 150: 12–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Diener A (2012) Visualizing and quantifying Fusarium oxysporum in the plant host. Mol Plant Microbe Interact 25: 1531–1541. [DOI] [PubMed] [Google Scholar]
- 29. Amano Y, Tsubouchi H, Shinohara H, Ogawa M, Matsubayashi Y (2007) Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proc Natl Acad Sci USA 104: 18333–18338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Matsubayashi Y, Ogawa M, Kihara H, Niwa M, Sakagami Y (2006) Disruption and overexpression of Arabidopsis phytosulfokine receptor gene affects cellular longevity and potential for growth. Plant Physiol 142: 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang G, Ellendorff U, Kemp B, Mansfield JW, Forsyth A, et al. (2008) A genome-wide functional investigation into the roles of receptor-like proteins in Arabidopsis. Plant Physiol 147: 503–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Thatcher LF, Manners JM, Kazan K (2009) Fusarium oxysporum hijacks COI1-mediated jasmonate signaling to promote disease development in Arabidopsis. Plant J 58: 927–939. [DOI] [PubMed] [Google Scholar]
- 33. Fritz-Laylin LK, Krishnamurthy N, Tör M, Sjölander KV, Jones JD (2005) Phylogenomic analysis of the receptor-like proteins of rice and Arabidopsis. Plant Physiol 138: 611–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jones DA, Jones JD (1997) The role of leucine-rich repeat proteins in plant defences. Adv Bot Res 24: 89–167. [Google Scholar]
- 35. Malik K, Wu K, Li XQ, Martin-Heller T, Hu M, et al. (2002) A constitutive gene expression system derived from the tCUP cryptic promoter elements. Theor Appl Genet 105: 505–514. [DOI] [PubMed] [Google Scholar]
- 36. Coutu C, Brandle J, Brown D, Brown K, Miki B, et al. (2007) pORE: a modular binary vector series suited for both monocot and dicot plant transformation. Transgenic Res 16: 771–781. [DOI] [PubMed] [Google Scholar]
- 37. Motose H, Iwamoto K, Endo S, Demura T, Sakagami Y, et al. (2009) Involvement of phytosulfokine in the attenuation of stress response during the transdifferentiation of zinnia mesophyll cells into tracheary elements. Plant Physiol 150: 437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Komori R, Amano Y, Ogawa-Ohnishi M, Matsubayashi Y (2009) Identification of tyrosylprotein sulfotransferase in Arabidopsis. Proc Natl Acad Sci USA 106: 15067–15072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhou W, Wei L, Xu J, Zhai Q, Jiang H, et al. (2010) Arabidopsis Tyrosylprotein sulfotransferase acts in the auxin/PLETHORA pathway in regulating postembryonic maintenance of the root stem cell niche. Plant Cell 22: 3692–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bomblies K, Lempe J, Epple P, Warthmann N, Lanz C, et al. (2007) Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol 5: e236 doi:10.1371/journal.pbio.0050236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yang S, Yang H, Grisafi P, Sanchatjate S, Fink GR, et al. (2006) The BON/CPN gene family represses cell death and promotes cell growth in Arabidopsis. Plant J 45: 166–179. [DOI] [PubMed] [Google Scholar]
- 42. Yang H, Shi Y, Liu J, Guo L, Zhang X, et al. (2010) A mutant CHS3 protein with TIR-NB-LRR-LIM domains modulates growth, cell death and freezing tolerance in a temperature-dependent manner in Arabidopsis. Plant J 63: 283–296. [DOI] [PubMed] [Google Scholar]
- 43. Zhang Y, Yang Y, Fang B, Gannon P, Ding P, et al. (2010) Arabidopsis snc2-1D activates receptor-like protein-mediated immunity transduced through WRKY70. Plant Cell 22: 3153–3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bleckmann A, Weidtkamp-Peters S, Seidel CA, Simon R (2010) Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiol 152: 166–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Russinova E, Borst JW, Kwaaitaal M, Caño-Delgado A, Yin Y, et al. (2004) Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 16: 3216–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Leister D (2004) Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance gene. Trends Genet 20: 116–122. [DOI] [PubMed] [Google Scholar]
- 47. Pan Q, Liu YS, Budai-Hadrian O, Sela M, Carmel-Goren L, et al. (2000) Comparative genetics of nucleotide binding site-leucine rich repeat resistance gene homologues in the genomes of two dicotyledons: tomato and Arabidopsis. Genetics 155: 309–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ramonell K, Berrocal-Lobo M, Koh S, Wan J, Edwards H, et al. (2005) Loss-of-function mutations in chitin responsive genes show increased susceptibility to the powdery mildew pathogen Erysiphe cichoracearum. Plant Physiol 138: 1027–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang Y, Yang Y, Fang B, Gannon P, Ding P, et al. (2010) Arabidopsis snc2-1D activates receptor-like protein-mediated immunity transduced through WRKY70. Plant Cell 22: 3153–3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wulff BB, Chakrabarti A, Jones DA (2009) Recognitional specificity and evolution in the tomato-Cladosporium fulvum pathosystem. Mol Plant Microbe Interact 22: 1191–1202. [DOI] [PubMed] [Google Scholar]
- 51. Fradin EF, Zhang Z, Juarez Ayala JC, Castroverde CD, et al. (2009) Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol 150: 320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kruijt M, de Kock MJ, de Wit PJ (2005) Receptor-like proteins involved in plant disease resistance. Mol Plant Pathol 6: 85–97. [DOI] [PubMed] [Google Scholar]
- 53. de Jonge R, van Esse HP, Maruthachalam K, Bolton MD, Santhanam P, et al. (2012) Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc Natl Acad Sci USA 109: 5110–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Malnoy M, Xu M, Borejsza-Wysocka E, Korban SS, Aldwinckle HS (2008) Two receptor-like genes, Vfa1 and Vfa2, confer resistance to the fungal pathogen Venturia inaequalis inciting apple scab disease. Mol Plant Microbe Interact 21: 448–458. [DOI] [PubMed] [Google Scholar]
- 55. Ron M, Avni A (2004) The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 16: 1604–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rouxel T, Balesdent M-H (2013) From model to crop plant–pathogen interactions: cloning of the first resistance gene to Leptosphaeria maculans in Brassica napus. New Phytol 197: 356–358. [DOI] [PubMed] [Google Scholar]
- 57. Thomma BP, Nürnberger T, Joosten MH (2011) Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell 23: 4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Igarashi D, Tsuda K, Katagiri F (2012) The peptide growth factor, phytosulfokine, attenuates pattern-triggered immunity. Plant J 71: 194–204. [DOI] [PubMed] [Google Scholar]
- 59. Yamakawa S, Matsubayashi Y, Sakagami Y, Kamada H, Satoh S (1999) Promotive effects of the peptidyl plant growth factor, phytosulfokine-alpha, on the growth and chlorophyll content of Arabidopsis seedlings under high night-time temperature conditions. Biosci Biotechnol Biochem 63: 2240–2243. [DOI] [PubMed] [Google Scholar]
- 60. Matsuzaki Y, Ogawa-Ohnishi M, Mori A, Matsubayashi Y (2010) Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science 329: 1065–1067. [DOI] [PubMed] [Google Scholar]
- 61. Wang GL, Ruan DL, Song WY, Sideris S, Chen L, et al. (1998) Xa21D encodes a receptor-like molecule with a leucine-rich repeat domain that determines race-specific recognition and is subject to adaptive evolution. Plant Cell 10: 765–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, et al. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. van der Hoorn RA, Kamoun S (2008) From Guard to Decoy: a new model for perception of plant pathogen effectors. Plant Cell 20: 2009–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Shinohara H, Ogawa M, Sakagami Y, Matsubayashi Y (2007) Identification of ligand binding site of phytosulfokine receptor by on-column photoaffinity labeling. J Biol Chem 282: 124–131. [DOI] [PubMed] [Google Scholar]
- 65. Wang G, Long Y, Thomma BP, de Wit PJ, Angenent GC, et al. (2010) Functional analyses of the CLAVATA2-like proteins and their domains that contribute to CLAVATA2 specificity. Plant Physiol 152: 320–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li Y, Pennington BO, Hua J (2009) Multiple R-like genes are negatively regulated by BON1 and BON3 in arabidopsis. Mol Plant Microbe Interact 22: 840–848. [DOI] [PubMed] [Google Scholar]
- 67. Yang S, Yang H, Grisafi P, Sanchatjate S, Fink GR, et al. (2006) The BON/CPN gene family represses cell death and promotes cell growth in Arabidopsis. Plant J 45: 166–179. [DOI] [PubMed] [Google Scholar]
- 68. Altenbach D, Robatzek S (2007) Pattern recognition receptors: from the cell surface to intracellular dynamics. Mol Plant Microbe Interact 20: 1031–1039. [DOI] [PubMed] [Google Scholar]
- 69. Robatzek S, Chinchilla D, Boller T (2006) Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev 20: 537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lu D, Lin W, Gao X, Wu S, Cheng C, et al. (2011) Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332: 1439–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Horvath H, Rostoks N, Brueggeman R, Steffenson B, von Wettstein D, Kleinhofs A (2003) Genetically engineered stem rust resistance in barley using the Rpg1 gene. Proc Natl Acad Sci U S A 100: 364–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Whalen MC (2005) Host defence in a developmental context. Mol Plant Pathol 6: 347–360. [DOI] [PubMed] [Google Scholar]
- 73. Haughn GW, Somerville C (1986) Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol Gen Genet 204: 430–434. [Google Scholar]
- 74. Kistler HC, Momol EA, Benny U (1991) Repetitive genomic sequences for determining relatedness among strains of Fusarium oxysporum. Phytopathology 81: 331–336. [Google Scholar]
- 75. Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25: 989–994. [DOI] [PubMed] [Google Scholar]
- 76. Noé L, Kucherov G (2005) YASS: enhancing the sensitivity of DNA similarity search. Nucleic Acids Res 33: W540–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Celenza JL Jr, Grisafi PL, Fink GR (1995) A pathway for lateral root formation in Arabidopsis thaliana. Genes Dev 9: 2131–2142. [DOI] [PubMed] [Google Scholar]
- 78. Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743. [DOI] [PubMed] [Google Scholar]
- 79. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Resistance phenotype of Col-0 subclones in the RFO2 interval. The RFO2 interval (258 kbp between nucleotides 5,766,000 and 6,023,500 in TAIR10 reference sequence for chromosome 1) was defined by a recombinant breakpoint in lines 4E3 between SSLPs F11A6 (at nucleotide 5,766,000) and F20D23 (at nucleotide 5,820,000), on the low end, and a recombination breakpoint in line 1B9 between SSLPs F28G4 (at nucleotide 5,943,000) and F17F16 (at nucleotide 6,023,500), on the high end. Resistance phenotypes (Rfo) of Col-0 genomic clones were tested in T1 and/or T2 transformants of line 1A3. Horizontal bars are proportional to the sequence length of subcloned Col-0 DNA and extend across their respective positions in the genomic interval below. Bars are labeled with the subclone names and, in paratheses, the sizes and gene content of subcloned sequence (to the right). Fifty of the 68 genes in the RFO2 interval were included in at least one construct: Xba1.5 includes nucleotides 5777587 to 5789031 and genes AT1G16900, AT1G16905, AT1G16910, AT1G16916 and AT1G16920; Xba1.4 includes nucleotides 5791422 to 5801901 and genes AT1G16940, AT1G16950 and AT1G16960; Pac1.4 includes nucleotides 5810084 to 5824597 and genes AT1G17000, AT1G17010, AT1G17020 and AT1G17030; Pac1.3 includes nucleotides 5824895 to 5833233 and genes AT1G17040 and AT1G17050; Kpn1.4 includes nucleotides 5833071 to 5841463 and genes AT1G17060, AT1G17070 and AT1G17080; Xba1.3 includes nucleotides 5837955 to 5843587 and genes AT1G17070, AT1G17080 and AT1G17090; Kpn1.3 includes nucleotides 5846018 to 5854394 and genes AT1G17110 and AT1G17120; Xba1.2 includes nucleotides 5843587 to 5855420 and genes AT1G17100, AT1G17110 and AT1G17120; Pac1.2includes nucleotides 5850646 to 5866025 and genes AT1G17120, AT1G17130, AT1G17140, AT1G17145 and AT1G17147; Pac1.1 includes nucleotides 5866025 to 5878981 and genes AT1G17150, AT1G17160, AT1G17170, AT1G17180 and AT1G17190; Sal1.4 includes nucleotides 5867417 to 5882030 and genes AT1G17160, AT1G17170, AT1G17180, AT1G17190 and AT1G17200; Sal1.3 includes nucleotides 5882030 to 5891121 and genes AT1G17210 and AT1G17220; Xba1.1 includes nucleotides 5878230 to 5891727 and genes AT1G17200, AT1G17210 and AT1G17220; Kpn1.2 includes nucleotides 5886702 to 5904885 and genes AT1G17220, AT1G17232, AT1G17230, AT1G17235, AT1G17240 and AT1G17250; Sal1.2 includes nucleotides 5891121 to 5914913 and genes AT1G17232, AT1G17230, AT1G17235, AT1G17240, AT1G17250, AT1G17260, AT1G17270 and AT1G17275; Kpn1.1 includes nucleotides 5905771 to 5921349 and genes AT1G17260, AT1G17270, AT1G17275, AT1G17277, AT1G17280 and AT1G17285; Sal1.1 includes nucleotides 5933219 to 5946989 and genes AT1G17340, AT1G17345 and AT1G17350; Xma1.1 includes nucleotides 5970942 to 5986501 and genes AT1G17420, AT1G17430 and AT1G17440; Avr2.1 includes nucleotides 6007806 to 6004198 and genes no full-length gene; Nsi1.3 includes nucleotides 5898644 to 5889883 and genes AT1G17232, AT1G17230, AT1G17235 and AT1G17240; BamH1.1 includes nucleotides 5900529 to 5891012 and genes AT1G17232, AT1G17230, AT1G17235 and AT1G17240; Age1.1 includes nucleotides 5902722 to 5891905 and genes AT1G17232, AT1G17230, AT1G17235, AT1G17240 and AT1G17250; Hind3.1 includes nucleotides 5904051 to 5895229 and genes AT1G17235, AT1G17240 and AT1G17250; Nsi1.1 includes nucleotides 5907976 to 5898644 and genes AT1G17250 and AT1G17260; and, Nsi1.2 includes nucleotides 5904885 to 5898644 and gene AT1G17250.
(PDF)
Domain structure of RFO2. The amino acid sequence of RFO2, in single-letter code, is divided into seven alphabetically named domains. Sequence in the C domain is further subdivided into 23 leucine-rich repeats (LRRs) and a loop out sequence. The number of amino acid residues (Len) in each domain and a brief comment about each sequence are to the right of sequences. Residues corresponding to the eLRR consensus (LxxLxxLxxLxLxxNxLxGxIPxx, where x represents a nonconserved residue between conserved residues) are highlight by underlined bold type [34]. Potential N-glycosylation sites are highlighted by half-filled bold type. Conserved cysteine residues in the N- and C-cap (domains B and D, respectively) are highlighted in bold [34]. In domain E, acidic residues are highlighted in bold. In domain F, a predicted transmembrane sequence is underlined, and a conserved GxxxG motif is highlighted in bold [33].
(PDF)
Alignment of eLRRs (domain C) of PSY1R-like proteins and PSKR1. Alignment of the translated amino acid sequences of PSY1R, RFO2, RLP2 (RLP2c), RLP2-T (RLP2t) and PSKR1, encoding domain C, are shown in single-letter code. All residues that are identical to PSY1R are highlighted by white type on black background. The amino acid position from the start codon is given for the leftmost residue in each line.
(PDF)
Alignment of PSY1R and RFO2-like RLP sequences at the carboxy-terminal ends of RLPs. Translated sequences of RFO2, RLP2 (RLP2c) and RLP2-T (RLP2t), encoding carboxy-terminal ends of RLPs, are aligned to the translated sequence of PSY1R in single-letter code. Amino acid residues that are identical in >50 percent of sequences are highlighted by white type on black background. The amino acid position from the start codon is given for the leftmost residue, and asterisks are stop codons. A SpeI restriction site (5′-ACT-AGT-3′), which codes for the threonine (T) and serine (S) residues at the arrow, was introduced as a silent mutation into coding sequences for the creation of chimeric fusions among RLP and RLK genes.
(PDF)
Recombination breakpoints defining RFO2 map position. (A) Fractions of n F2 from cross 1A3×4D2 that were susceptible (HI scores <2, open column), had intermediate resistance (2≤HI scores <4, half-filled) or were resistant (HI scores ≥4, filled) at 18 dpi. Only F2 of F1 plants 4E3, 1B9 and 5E1 from cross 1A3×4D2 that were either homozygous Ty-0 (T/T) or Col-0/Ty-0 heterzygotes (C/T) at RFO2-linked markers as well as C/T at RFO1-linked marker F19K16 are shown. (B) Genotypes, either Ty-0 (T) or Col-0 (C), at RFO2-linked markers (above) on the single recombinant chromosomes in F1 plants 4E3, 5E1 and 1B9 from cross 1A3×4D2. Marker intervals with a crossover are marked with ‘X’. Number of TAIR10 annotated open reading frames (ORFs) in marker intervals is given below. Rfo2 phenotype of F1 plants (on the right) was evaluated in F2 progeny (in A).
(PDF)
Root growth of nPcR is unaffected by PSK peptide. Two-week old seedlings of Col-0 (wild type), psy1r, psy1r, pskr1pskr2 and nPcR (line 1E9) were grown from seeds sown on vertically-oriented PN agar plates with (+) or without (−) added PSK (0.1 µM). (A) Lengths of PSK-treated wild-type and psy1r roots (n = 20) were longer than untreated roots (n = 20), according to Student's t test (two-tailed p = 0.044 and 0.015, respectively) while length of PSK-treated and untreated psy1r pskr1 pskr2 roots (n = 20) had similar lengths (p = 0.60). (B) Length of PSK-treated wild-type roots (n = 20) were longer than untreated roots (n = 20), according to Student's t test (two-tailed p = 0.005) while length of PSK-treated and untreated nPcR roots (n = 20) had similar lengths (p = 0.16).
(PDF)
PCR primers for mapping and genotyping RFO2 and RFO1.
(PDF)
PCR primers and products for sequencing RFO2 in Ty-0.
(PDF)
PCR primers for genotyping Salk insertions.
(PDF)
Homozygous T-DNA insertion lines in RFO2 genetic interval.
(PDF)
PCR primers for dominant multiplex PCR markers.
(PDF)
PCR primers for constructing RLP and RLK genes.
(PDF)



