Gene duplications provide the raw material for evolutionary novelties. Homospermidine synthase (HSS) of pyrrolizidine alkaloid biosynthesis was used as model system to study molecular mechanisms that result in the evolution of novel functions after duplication. HSS is involved in the chemical defense of several plant families and therefore provides an adaptive character under selective pressure.
Abstract
Homospermidine synthase (HSS), the first pathway-specific enzyme of pyrrolizidine alkaloid biosynthesis, is known to have its origin in the duplication of a gene encoding deoxyhypusine synthase. To study the processes that followed this gene duplication event and gave rise to HSS, we identified sequences encoding HSS and deoxyhypusine synthase from various species of the Convolvulaceae. We show that HSS evolved only once in this lineage. This duplication event was followed by several losses of a functional gene copy attributable to gene loss or pseudogenization. Statistical analyses of sequence data suggest that, in those lineages in which the gene copy was successfully recruited as HSS, the gene duplication event was followed by phases of various selection pressures, including purifying selection, relaxed functional constraints, and possibly positive Darwinian selection. Site-specific mutagenesis experiments have confirmed that the substitution of sites predicted to be under positive Darwinian selection is sufficient to convert a deoxyhypusine synthase into a HSS. In addition, analyses of transcript levels have shown that HSS and deoxyhypusine synthase have also diverged with respect to their regulation. The impact of protein–protein interaction on the evolution of HSS is discussed with respect to current models of enzyme evolution.
INTRODUCTION
Plants produce an amazing diversity of secondary metabolites of which several classes of compounds occur only in individual plant lineages. These metabolites are essential for the survival of the plant in its specific environment, as they are involved in processes such as defense against herbivores and pathogens, protection against UV radiation, or attraction of pollinators (Harborne, 1993; Hartmann, 2007). Secondary metabolites thus provide adaptive characters that should be under strong selective pressure, and the biosynthetic pathways of secondary metabolism that generally encompass many, highly specific enzymes are shaped and maintained by selection. Therefore, secondary metabolism and the enzymes involved therein are suitable for the study of molecular evolutionary mechanisms of adaptation and enzyme evolution in particular (Pichersky and Gang, 2000; Grotewold, 2005). In this regard, pyrrolizidine alkaloid (PA) biosynthesis has proved to be a powerful model system (Ober, 2005, 2010; Ober and Kaltenegger, 2009).
PAs are typical compounds of secondary metabolism and encompass more than 350 structures. They are characterized by a nitrogen-containing bicyclic ring system, the necine base, which is esterified with one or more necic acids. Because of the observed feeding deterrence, PAs are regarded as part of the chemical defense of plants (Dreyer et al., 1985; Singer and Stireman, 2003; Hristov and Conner, 2005; Siciliano et al., 2005; Reinhard et al., 2009). PAs occur in several unrelated angiosperm families, one of which is the Convolvulaceae (Hartmann and Witte, 1995; Langel et al., 2011). Sequence identity and examination of the reaction mechanism suggest that the first specific enzyme of the pathway, homospermidine synthase (HSS), evolved from the gene encoding deoxyhypusine synthase (DHS) by gene duplication (Ober and Hartmann, 1999a). DHS, an enzyme of primary metabolism, is involved in the posttranslational activation of the eukaryotic initiation factor 5A (eIF5A). DHS transfers the amino butyl moiety of spermidine to a specific protein-bound Lys residue, forming the rare amino acid deoxyhypusine (Chen and Liu, 1997; Park et al., 1997). DHS, although favoring the eIF5A precursor protein, can also accept putrescine as an amino butyl acceptor, catalyzing the formation of homospermidine. This side activity is regarded as being responsible for the occurrence of small amounts of homospermidine throughout the angiosperms and even in some mosses (Ober et al., 2003a). Kinetic and binding studies have shown that HSS of the PA-producing plant Senecio vernalis (Asteraceae) has lost the ability to bind the eIF5A precursor protein but has maintained the ability to catalyze the formation of homospermidine by binding putrescine as a substrate (Ober et al., 2003b). Homospermidine is not part of the highly dynamic polyamine pool of primary metabolism but is exclusively incorporated into the necine base moiety of PAs (Böttcher et al., 1993, 1994). Phylogenetic analyses of DHS- and HSS-coding cDNA sequences from PA-producing plants of various angiosperm lineages have shown that HSS evolved from several individual duplication events of ancestral dhs genes (Reimann et al., 2004). Two independent gene duplications have been detected early in the lineages of the monocots and the Boraginales, and a further two gene duplications occurred within the Asteraceae. In all these cases of HSS evolution, the recruitment of a dhs gene duplicate was accompanied by the loss of the ability to bind the eIF5A precursor protein to the enzyme’s active site (Reimann et al., 2004).
Gene duplications provide the raw material for evolutionary novelties (Zhang, 2003; Freeling and Thomas, 2006; Wapinski et al., 2007). Analysis of the functional fate of duplicate genes has revealed that duplicated genes rarely diverge with respect to biochemical function but typically diverge with respect to regulatory control (Gu et al., 2002; Wapinski et al., 2007). With regard to the less frequent case in which gene duplications lead to functional innovation by one of the copies, several models are discussed in the literature. The most prominent models are the model of neofunctionalization (Ohno, 1970; Conant and Wolfe, 2008), the subfunctionalization hypothesis including the duplication, degeneration, complementation model (Hughes, 1994, 2002; Force et al., 1999; Lynch and Force, 2000), and the escape from adaptive conflict model (Des Marais and Rausher, 2008). The testing for positive Darwinian selection has been suggested to distinguish between models (Conant and Wolfe, 2008). The most convincing signal of positive selection is an accelerated rate of nonsynonymous nucleotide substitution, a signal that is often obscured by numerous subsequent neutral changes occurring during longer periods of time after the duplication event (Long et al., 2003; Hughes, 2005). Studies of the evolution of HSS by the duplication of the dhs gene might provide insights into the mechanisms that gave rise to the repeated establishment of functional innovation during angiosperm evolution.
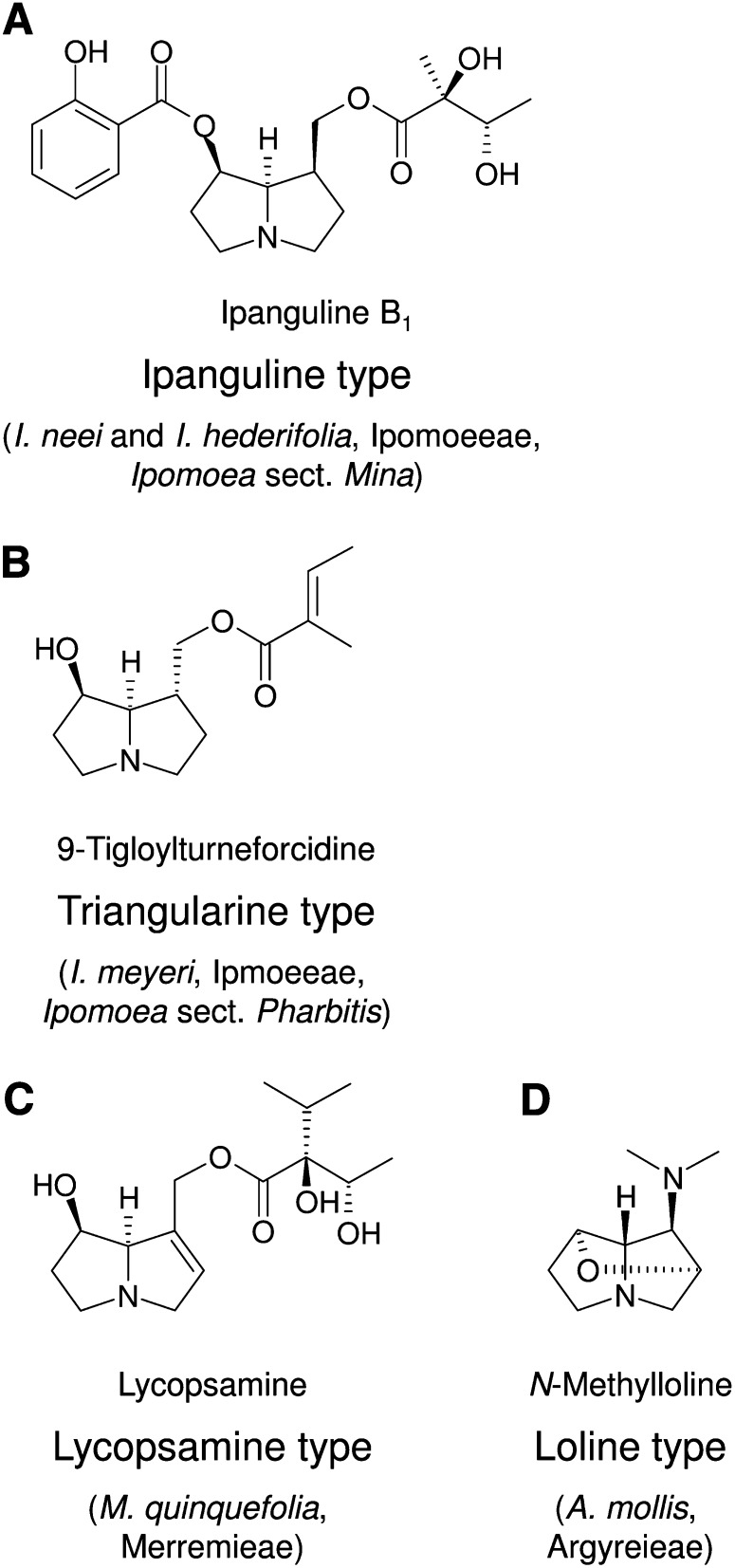
We have chosen to study the morning glory family (Convolvulaceae), which comprises ∼1600 to 1700 species (Mabberley, 1987; Miller et al., 2002; Stefanovic et al., 2003). A screen of ∼150 species from 23 genera of this family, particularly with regard to the occurrence of various classes of alkaloids, has shown that PAs occur only in individual unrelated species of one of the two major clades of this family (Eich, 2008). Of note, the structures of detected PAs are surprisingly diverse, suggesting that, within this family, PA biosynthesis might have evolved several times independently. Such a scenario has been demonstrated for HSS evolution in the Asteraceae, in which independent duplications of the dhs gene and subsequent recruitment of the duplicated gene for PA biosynthesis have been found within the tribes Senecioneae and Eupatorieae. Both tribes are characterized by different types of PAs, the senecionine type and the lycopsamine type, respectively (Reimann et al., 2004; Hartmann and Ober, 2008). The diverse PAs of the Convolvulaceae include (1) the ipangulines and minalobines, which are characterized by 1,2-saturated necine bases forming mono- and diesters with aromatic and aliphatic necic acids (Figure 1A) detected in Ipomoea species of the section Mina (Jenett-Siems et al., 1993, 1998, 2005); (2) PAs of the triangularine type (i.e., necine bases esterified with mainly aliphatic C5-necic acids) (Figure 1B), detected in Ipomoea meyeri, a species of the section Pharbitis series Pharbitis (Tofern, 1999; Eich, 2008); (3) PAs of the lycopsamine type found within Merremia quinquefolia (Figure 1C), the only species within this family in which 1,2-unsaturated PAs have been identified (Mann, 1997; Eich, 2008); and (4) the lolines (Figure 1D), found in Argyreia mollis (Tofern et al., 1999). Lolines are well-studied compounds from certain poaceous species in which they are synthesized by seed-transmitted fungal endophytes (Schardl, 2009). Tracer experiments have revealed that, in contrast with plants, not polyamines but Pro and homoserine are precursors of the lolines (Blankenship et al., 2005). Alkaloid-producing endophytes have also been described to occur in species of the Convolvulaceae (Steiner et al., 2006; Markert et al., 2008). As lolines in Argyreia might be products of as yet undiscovered fungal symbionts (Schardl et al., 2007), we excluded this species from our study and focus on the PA-producing species of the genus Ipomoea (sections Mina and Pharbitis) and Merremia in comparison with other PA-free species. Recent phylogenetic analyses confirm that these three groups of PA-producing species are not closely related within the clade 1 sensu Stefanović (Stefanovic et al., 2002, 2003), one of the two major clades representing most of the species of the Convolvulaceae.
Figure 1.
Structural Types of PAs Occurring within the Convolvulaceae.
Ipanguline-type PAs of Ipomoea species (section Mina) characterized by the saturated necine base platynecine forming mono- and diesters with aromatic and aliphatic necic acids (A). Triangularine-type PAs of I. meyeri (section Pharbitis) with saturated necine bases, predominantly turneforcidine, and aliphatic necic acids (B). Lycopsamine-type PAs of M. quinquefolia with the 1,2-unsaturated necine base retronecine and unique C7 necic acids (C). Loline-type PAs of A. mollis characterized by a 2,7 ether bridge and the replacement of C-9 by an amino substituent (D). The studied species in which the PA type was detected (with the exception of the loline-type PAs) are given.
By identifying DHS- and HSS-coding sequences in selected species of the Convolvulaceae and reconstructing the gene evolution by phylogenetic methods, we are able to show that only one gene duplication event in the lineage of the Convolvulaceae gave rise to the evolution of HSS. Furthermore, we present evidence that, in several species, the inability to produce PAs might be attributable to a loss or inactivation of this gene copy, although, in one PA-free species, Ipomoea alba, a functional hss gene has been detected. We have also been able to demonstrate, by statistical methods and by functional analyses of manipulated enzymes, that, after duplication, the evolution of HSS is characterized by time intervals related to various selection pressures, including purifying selection, relaxed functional constraints, and most likely positive Darwinian selection. Our data indicate that various kinds of protein–protein interaction had an impact on the evolution of the new enzymatic activity from an ancestral gene that was at least bifunctional.
RESULTS
Identification of HSS and DHS from PA-Producing Convolvulaceae Species
We used degenerate oligonucleotide-primed PCR to identify sequences with homology to known DHS and HSS sequences within PA-producing species of the Convolvulaceae. Using cDNA preparations of Ipomoea neei and M. quinquefolia of various tissues, we were able to identify two fragments of each species showing a high degree of identity to the previously identified cDNA encoding the DHS of Ipomoea hederifolia (Reimann et al., 2004). After completing these fragments by the rapid amplification of cDNA ends technique, comparisons that included the previously identified DHS of I. hederifolia revealed a high degree of sequence identity between DHS and HSS orthologs, respectively, within the genus Ipomoea. Therefore, we were able to amplify the full-length cDNA sequences coding for DHS of I. meyeri and I. neei and HSS of I. meyeri and I. hederifolia with primers that had been designed for the amplification of the open reading frame (ORF) of the DHS of I. hederifolia and the HSS of I. neei.
As no HSS- or DHS-specific sequence signatures are available, it is impossible to denote a sequence as HSS or DHS merely based on sequence data (Reimann et al., 2004; Nurhayati and Ober, 2005). Therefore, all identified sequences were heterologously expressed in Escherichia coli, affinity-purified, and assayed for HSS and DHS activity. In accordance with Reimann et al. (2004), all proteins with activity in the HSS assay (with only minimal DHS activity) were designated as HSS, whereas proteins with DHS activity (with or without additional HSS activity) were designated as DHS. Table 1 summarizes the specific activities and the resulting annotation of the recombinant proteins as DHS and HSS.
Table 1. Predicted Size of the Gene Products and Specific Activities of DHS- and HSS-Coding cDNA Sequences Identified from Various Convolvulaceae Species.
| Sequence | Subunit Size (kD) | HSS Assay, Aminobutyl Acceptor Putrescine (pkat/mg) | DHS Assay, Aminobutyl Acceptor eIF5A Precursor (pkat/mg) |
|---|---|---|---|
| DHS I. neei | 42.2 | 41 | 45* |
| HSS I. neei | 41.6 | 1040* | 6.3 |
| DHS M. quinquefolia | 42.1 | 100 | 53* |
| HSS M. quinquefolia | 41.3 | 1055* | n.d. |
| DHS I. hederifolia | 42.3 | 318 | 200* |
| HSS I. hederifolia | 41.6 | 2267* | 12 |
| DHS I. meyeri | 42.2 | 146 | 116* |
| HSS I. meyeri | 41.5 | 2295* | 3 |
| DHS I. alba | 42.3 | 318 | 161* |
| HSS I. alba | 41.5 | 788* | 38.5 |
The values for specific activity refer to the affinity-purified recombinant proteins. Values noted with an asterisk are interpreted as main activity. n.d., not detectable.
Identification of dhs and hss Genes from Various Convolvulaceae Species
To investigate the evolutionary fate of dhs and hss genes within the Convolvulaceae, we analyzed PA-producing species (I. neei, I. hederifolia, I. meyeri, and M. quinquefolia) and species that were found in previous studies to be free of PAs (I. alba, Ipomoea purpurea, Convolvulus arvensis, and Convolvulus tricolor; Eich, 2008). The use of genomic DNA enabled us to identify gene fragments or nonactively expressed genes that were homologous to the dhs gene but that might have been silenced after their origin by gene duplication. The length and position of introns was predicted by a comparison of the genomic sequences with the cDNA sequences identified in this project. The determination of the number, position, and phase of introns did not result in the identification of an additional variable property, as these properties are identical for all identified genes from the Convolvulaceae species (see Supplemental Table 1 online) and to the dhs and hss genes of various other angiosperm families (Reimann et al., 2004).
The dhs gene has been recognized as a single-copy gene within many eukaryotic genomes completed so far (Reimann et al., 2004; Nurhayati and Ober, 2005; Nurhayati et al., 2009). Indeed, in the PA-free C. tricolor, we also found only one copy of a putative dhs gene. The same holds true for I. purpurea, although we amplified two homologous sequences. Most likely, they represent two alleles of the dhs gene, as they share a nucleotide sequence identity of 97.8 and 99.3% when the complete genomic sequences (including the introns) and the assembled exons are compared, respectively. However, in C. arvensis, we identified three homologous genes with a shared sequence identity, within the exons, of only 87 to 90%. Comparisons of the sequences with those of the other Convolvulaceae species suggest that one (carv-1) represents the intact dhs gene, whereas carv-2 and carv-3 are pseudogenes (see Supplemental Figure 1 online). In carv-2, the complete exon 5 is deleted so that an intron of 160 bp joins exons 4 and 6. In carv-3, the last 22 bp of exon 5 are lost, probably because of a modified splicing site. This results in a frame shift so that the ORF encodes a truncated protein. Of note, in PA-free I. alba, we have identified two genes with homology to dhs genes that are identical with respect to the number and phase of introns to the genes identified from other Convolvulaceae species and show no signatures of being pseudogenes. Sequence comparison suggests that these genes are orthologs to the dhs and hss genes, as one sequence shares 97 and 87% and the other sequence 87 and 95% sequence identity with the ORFs of the dhs and hss genes of I. neei, respectively.
Characterization of a Functional hss Gene in Non-PA-Producing I. alba
To test whether the two genes identified from I. alba are transcribed, we used RT-PCR with various plant tissues as template. With cDNA of shoots, two full-length cDNA sequences were amplified that showed 100% identity to the exon sequences of the putative dhs and hss genes of I. alba. Both cDNA sequences were heterologously expressed for biochemical characterization. HSS and DHS assays of the recombinant proteins confirmed that the putative dhs gene encoded a functional DHS. The protein encoded by the putative hss gene showed characteristics of HSS. However, the specific activity with putrescine as an aminobutyl acceptor was only slightly increased compared with the DHS, whereas the activity with eIF5A precursor was reduced, but not as much as was the case for the HSS enzymes identified from other Convolvulaceae species (Table 1). As I. alba is a PA-free species, the in vivo function of this enzyme is unknown.
HSS Evolved Only Once within the Convolvulaceae Lineage
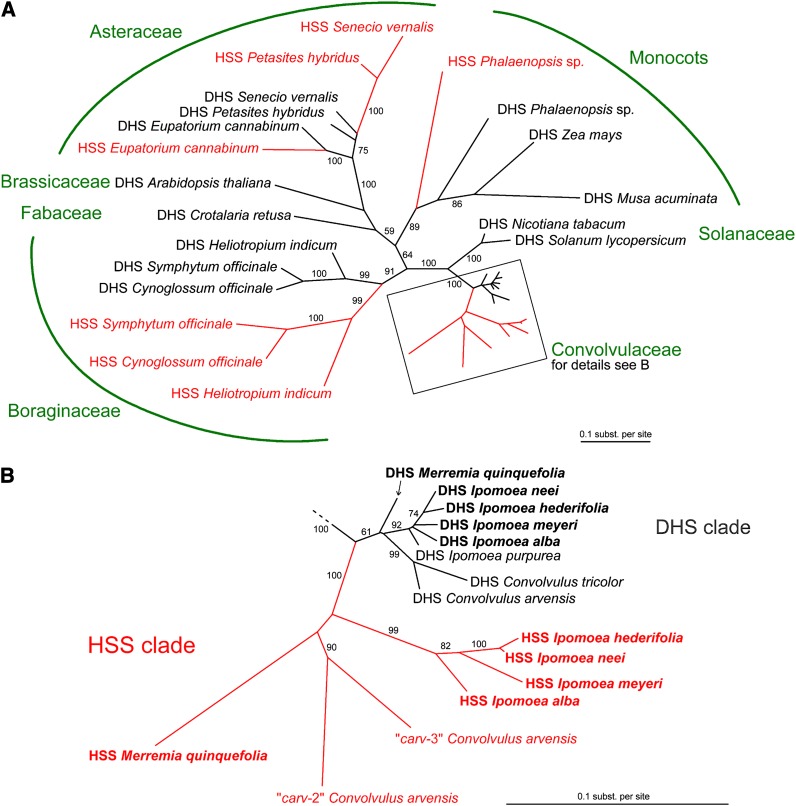
In order to reconstruct the evolution of HSS within the Convolvulaceae, we inferred a gene tree. We used an alignment of cDNA sequences coding for HSS and DHS from various angiosperm species described previously (Reimann et al., 2004; Nurhayati et al., 2009) together with the functionally characterized sequences from this study. In addition, the exon sequences of the genes or gene fragments identified from the PA-free species of the Convolvulaceae were included. Figure 2A shows an unrooted maximum likelihood tree with good support for the monophyletic origin of sequences that belong to the monocots, the Asteraceae, and the Boraginaceae and for the two major families of the Solanales, namely, the Solanaceae and Convolvulaceae. The tree supports a single duplication event of an ancestor of contemporary dhs genes of the Convolvulaceae, resulting in two clearly distinct clusters encompassing DHS- and HSS-coding sequences (DHS clade and HSS clade, Figure 2B). This duplication event is independent of the previously described gene duplications that resulted in HSS-coding sequences within four lineages of the Angiosperms (Reimann et al., 2004). The topology within the DHS and the HSS clade reflects the taxonomic classification of the clade 1 sensu Stefanović of the Convolvulaceae (Miller et al., 2002, 2004; Stefanovic et al., 2002, 2003) with the sequences of Ipomoea species and Convolvulus species forming separate subclades and Merremia being placed on an independent branch. A phylogram obtained using a maximum parsimony algorithm shows a similar topology that supports the single-gene duplication event in the evolution of the species under study (data not shown). The exon sequences of the genomic sequences identified from PA-free species of the Convolvulaceae also fall into the two distinct DHS and HSS clades (Figure 2B). The sequences identified from I. purpurea and C. tricolor and one of the sequences identified from C. arvensis can be grouped with the biochemically characterized DHS-coding sequences, thus supporting their identification as dhs genes. Of note, the two other sequences identified from C. arvensis (i.e., carv-2 and carv-3) clearly fall into the clade of the HSS-coding sequences. Apparently, in this PA-free species, a second duplication event of an ancestral hss-like gene resulted in the two extant paralogous genes that are most likely pseudogenes (see above). Furthermore, the phylogeny in Figure 2B shows that, of the two sequences identified from the PA-free I. alba, the DHS belongs to the DHS cluster, whereas the putative HSS clearly groups with sequences of the HSS cluster, thus confirming the results obtained by quantification of enzymatic activities.
Figure 2.
Unrooted Maximum Likelihood Tree of HSS- and DHS-Encoding cDNA Sequences of Various Angiosperm Species.
Sequences coding for HSS and DHS are shown in red and black, respectively.
(A) The branching pattern supports five independent gene duplication events that give rise to HSS-encoding sequences in the various lineages (i.e., two in the Asteraceae and one in the lineages of the Boraginaceae, the Monocots, and the Convolvulaceae).
(B) Detail of (A) showing the sequence cluster of the Convolvulaceae. Sequences that have been biochemically characterized are given in bold. Bootstrap proportions resulted from 1000 replicates and are given only for values ≥ 50.
Pairwise comparisons of DHS- and HSS-coding cDNA sequences show that amino acid distances (daa) and nucleotide distances (dna) are smaller between orthologous DHS sequences than between paralogous DHS and HSS sequences (see Supplemental Table 2 online). A similar observation has been described previously for other plant lineages that are known to contain PA-producing species (Reimann et al., 2004) and has been interpreted as support for the assumption that the lost necessity of HSS to bind the eIF5A protein results in modified selective forces acting on individual amino acid positions.
Did Relaxed Functional Constraints or Positive Selection Affect the Evolution of PA-Specific HSS?
Increased distance values for paralogs might indicate relaxed functional constraints or even positive selection. To test for this kind of selection pressure, we calculated nonsynonymous substitutions per nonsynonymous site (dN) and synonymous substitutions per synonymous site (dS). Their relative rate (dN/dS) is defined as ω, being a measure of the selective pressure at the protein level. Synonymous mutations are mostly invisible to natural selection, whereas nonsynonymous mutations might be under strong selective pressure. Therefore, ω < 1 indicates purifying selection, ω = 1 neutral evolution, and ω > 1 positive Darwinian selection (Yang et al., 2000).
Based on the DHS- and HSS-coding cDNA sequences identified from the Convolvulaceae and the DHS-coding cDNA sequences of the related solanaceous species tobacco (Nicotiana tabacum) and tomato (Solanum lycopersicum), we created two data sets: (1) data set A excluding the pseudogenes (284 codons, 29 were invariant with identical nucleotides across all species) and (2) data set B including the partial exon sequences of the pseudogenes carv-2 and carv-3 (215 codons, 23 were invariant with identical nucleotides across all species). Using codeml, which is part of the PAML4 software package (Yang, 2007), we calculated, as a first step in pairwise comparisons, averaged ω-values over all sites in both amino acid alignments with similar results. Purifying selection for all sequences was indicated with ω being clearly < 1. However, between HSS orthologs, the ω-values were on average twofold higher (0.11 to 0.39) than between the DHS orthologs (0.06 to 0.17). Pairwise ω-values of HSS and DHS paralogs of the same species ranged from 0.10 to 0.15. These slightly elevated ω-values might result from a relaxation of selective constraints or from positive selection acting on only a subset of sites within the sequence or only during a short postduplication period of adaptive evolution (Yang and Nielsen, 2002; Rodin et al., 2005). To test such scenarios, we compared various models implemented in the codeml software.
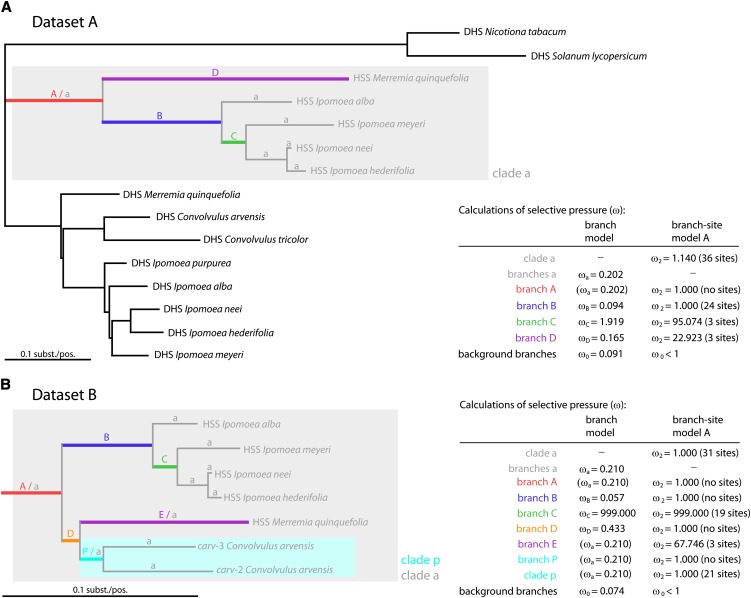
We applied the site model M1a (nearly neutral), which allows sites with ω0 < 1 and ω1 = 1, and the site model M2a (positive selection), which adds an extra category of sites with ω2 > 1. Both models allow the ω-values to vary among sites but average ω over all branches in the phylogeny (Yang et al., 2000). With the log likelihoods of both models being equal, we were unable to detect positive selection (see Supplemental Table 4 online). With the branch model (Yang, 1998; Yang and Nielsen, 1998), which allows the ω ratio to vary among selected branches but averages it over all sites in the protein, we calculated the ω-values for the branches in the HSS clade relative to the background ratio (ω0) for the branches in the DHS clade (Figure 3A, data set A). In detail, separate ω-values in the HSS clade were calculated for the following branches: branch B (ωB), branch C (ωC), and branch D (ωD); ωa was calculated for the rest of the branches in the HSS clade (Figure 3A). This five-ratio model assumes that the ratios for branches A to D are different within the HSS clade and are also different from the background ratio. Under this model, we found slightly elevated ω-values in the HSS clade with ωa = 0.202, ωB = 0.094, and ωD = 0.165, whereas ω0 for the background branches was calculated to be 0.091.Thus, relaxed selective constraints are indicated to have acted in the HSS clade after the duplication event. Moreover, with a ωC-value of 1.919 for branch C (Figure 3A), we have found indications that positive selection has acted in the evolution of HSS in that specific period. In a likelihood ratio test (LRT) according to Yang (1998), we compared the branch model with a model that assumes the same ratio for all branches in the phylogeny (one-ratio or M0 model) and found support for the branch model, indicating that the ω-values are indeed different among lineages (LRT 19.5, df = 4, P < 0.01; see Supplemental Table 4 online, LRT2). Using the more elaborate branch-site model A, we calculated the ω ratios for individual codon sites and along prespecified lineages (the foreground branches) on a phylogenetic tree with the site class ω2 > 1 being defined as positively selected sites (Yang and dos Reis, 2011). With data set A, we separately tested, for positive selection, the four individual branches A to D and the whole clade a, which contains all HSS-coding sequences (Figure 3A). For branches A and B, we found no elevated ω-values. For branch C, the highest ω ratio of 95.074 was calculated to act on 3 out of the 284 codon sites (0.9%; see Supplemental Table 3 online). Additionally, in branch D, we found an elevated ω-value with ω2 = 22.923. To test the statistical significance, we used a LRT (see Supplemental Table 4 online, LRT3) of the branch-site model A and the branch-site model A with ω2 fixed at 1, which has been described by Zhang et al. (2005) as a direct test for positive selection. Of note, when multiple branches on the tree are tested for positive selection using the same data set, a correction for multiple testing is required for the correct interpretation of the data (Yang, 2007). We used two methods: (1) the conservative Bonferroni method (Anisimova and Yang, 2007) and (2) the B-Y method (Narum, 2006). Whereas none of the LRTs was significant according to Bonferroni’s correction (P ≤ 0.01; see Supplemental Table 4 online), the LRT for positive selection in branch C (LRT 5.378, P = 0.0204) was significant according to the B-Y correction (P ≤ 0.0219).
Figure 3.
Branch-Specific Analyses of ω Ratios of DHS- and HSS-Coding Sequences of the Solanales.
Colored lines indicate branches that were defined a priori as foreground branches in separate calculations with the branch-site model. Furthermore, the whole HSS clade, marked with a gray box, was defined as a foreground clade in one calculation using the branch-site model. In the branch model, ω ratios for five categories of branches were calculated: ω0 for branches in the DHS clade, ωa for branch “a” within the HSS clade, and ωB, ωC, and ωD for branches B, C, and D, respectively. The table summarizes the calculations of the branch model and the branch-site model. Note, in the branch-site model, ω ratios (ω2) were calculated for individual codon sites along the prespecified lineages. The number of sites is given in parentheses and is one of the calculated variables of the branch-site model. In the branch model, ω ratios were averaged over all sites in the protein.
(A) Tree topology and summarized results of the analyses with data set A.
(B) Tree topology of the HSS clade and summarized results of the analyses with data set B, which includes the partial exon sequences of two pseudogenes from C. arvensis (carv-2 and carv-3).
To test for relaxed selection in a certain period of HSS evolution, which is indicated by an increase of sites with ω = 1, we used the branch-site model A with ω2 fixed at 1. The site class p2a includes sites that experience purifying selection in the background branches but neutral selection in the foreground branches (see Supplemental Table 3 online). We compared the branch-site model A with ω2 = 1 for different a priori–defined foreground branches with the site model M1a as the null model, which assumes two categories of sites (ω0 ≤ 1; ω1 = 1) averaged over all lineages. When testing single branches in data set A, the LRT4 was not significant for both corrections of multiple hypothesis testing, although the branch-site model A with ω2 = 1 detected sites under relaxed selection for branch B (22 sites, 8%) and C (96 sites, 34%). Of note, in branch A, no sites were calculated to experience relaxed selection, and the log likelihoods of the null model were equal to the model with relaxed selection in branch A (see Supplemental Table 3 online), indicating that, after gene duplication, purifying selection acted to the same degree on both copies and that there was no increase in neutrally evolving sites. However, when the whole HSS clade (clade a) was tested, 34 sites (12%) were calculated, with significance (LRT4 31.2, df = 2, P = 0.0000; see Supplemental Table 4 online), to have experienced relaxation of selective constraints, an observation indicating that, during the evolution of HSS, an increase of neutrally evolving sites occurred, but we are unable to specify a certain period.
When pseudogenes are included in the analyses (data set B, Figure 3B; see Supplemental Tables 5 and 6 online), we obtain similar results. We found statistical support for positive selection acting in branch C when using the B-Y method for corrections of multiple hypothesis testing. The calculated ω2 value for the branch is 999.000, which means infinity, as no synonymous substitutions were calculated to act in branch C. However, we also found strong statistical support for relaxed selection in branch C (LRT 13.1, P = 0.0014) and in the whole HSS clade (LRT 27.9, P = 0.0000) according to both corrections for multiple hypothesis testing (Bonferroni, B-Y method). We also used data set B to estimate the age of the gene inactivation of the pseudogenes carv-2 and carv-3. We analyzed the ω-values with the branch site model A with fixed ω = 1 for branch P, which is the branch before the duplication in the genus Convolvulus and clade p, which comprises branch P and the two branches leading to the duplicated pseudogenes, as foreground branches (Figure 3B). We compared this model again with the nearly neutral site model to test for an increase of neutrally evolving sites (LRT4). The loss of selective constraint after gene inactivation should allow an increase of neutrally evolving sites (Chou et al., 2002), which can be used to estimate the age of the pseudogenization event. In branch P, no increase was detected indicating purifying selection. When the whole clade was tested, we found an increase of neutrally evolving sites (p2a = 9%), but the statistical support was weak (LRT 5.3, P = 0.0707), suggesting recent and independent pseudogenization of the carv-2 and carv-3 genes of Convolvulus.
How to Detect Sites That Are Involved in Optimizing HSS Functionality?
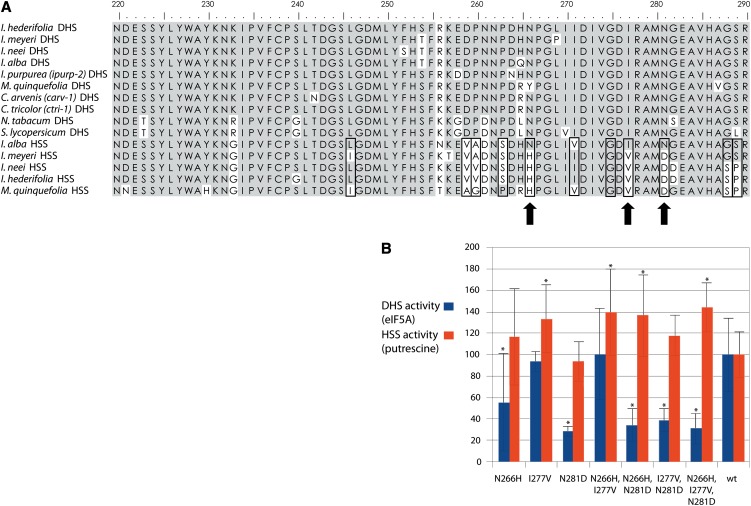
The output of the branch-site model calculation by codeml includes a probability list of sites assumed to be under positive selection (see Supplemental Tables 3 and 5 online). When testing the whole HSS clade (clade a) with data sets A and B, the branch-site model identified several amino acid positions. When the branches C and D, which showed elevated ω ratios, were tested individually, a small subset of these sites was calculated to be under positive selection (i.e., for data set A, Asn-281 and Gly-349 for branch C and Thr-120, Pro-159, and Leu-175 for branch D; numbering of amino acid positions refers to the DHS of I. neei). From the sites predicted by the various tested assumptions, three positions consistently distinguish DHS from HSS sequences (viz, Asn-266, Ile-277, and Asn-281). Exactly these sites were calculated to be under positive selection when branch C was tested for positive selection with data set B. Of note, in these predicted positions, the sequence of HSS from I. alba showed the same amino acids as the DHS enzymes (Figure 4A). As HSS of I. alba shows higher activity with the eIF5A precursor protein than other HSS, these predictions support the hypothesis that positive selection acted in branch C after the HSS of I. alba branched off and might have been responsible for the shift of substrate specificity from the eIF5A precursor protein to putrescine.
Figure 4.
Influence of Sites Predicted to Be under Positive Selection on Substrate Specificity.
(A) Amino acid polymorphisms between DHS and HSS of the Solanales. Shown are amino acids from position 220 to 290. Numbering of the amino acid positions is based on the amino acid sequence of the DHS from I. neei. Position 1 is defined as the start codon in the ORF. Identical amino acids are shaded gray. Sites that are calculated to be under positive selection with the branch-site model are boxed when clade a was tested and marked with an arrow when branch C was tested (Asn-281 with data set A, and Asn-266, Ile-277, and Asn-281 with data set B).
(B) Relative activities of modified DHS proteins from I. neei compared with wild-type DHS (wt). DHS activity is determined by product formation with eIF5A precursor protein as substrate (blue), whereas HSS activity is determined with putrescine as substrate (red). *Significant differences from the activity of the wild type.
[See online article for color version of this figure.]
As shown previously, statistical methods used to predict positive selection often fail to identify sites of functional importance (Nozawa et al., 2009). Therefore, guided by our statistical analyses, we analyzed the impact of amino acid substitutions at the predicted sites on enzyme activity. We postulated that, in the DHS/HSS system, positive selection acted on individual amino acid positions to modify the substrate specificity of DHS by adaptive substitutions. Thus, the eIF5A binding capacity of DHS should be reduced when amino acids in positively selected positions are exchanged against their counterparts. HSS activity should not be decreased by these substitutions, but rather increased. We modified the codons predicted to be the most promising candidates for positively selected sites in the sequence encoding the DHS of I. neei, resulting in the substitutions N266H, I277V, and N281D in single, double, and triple mutants. Comparison of these mutants with the wild-type DHS shows that the mutation N281D is the most effective in reducing DHS activity, while conserving HSS activity of the mutant. The mutation I277V has hardly any effect on DHS activity, but HSS activity is significantly increased. By contrast, in case of mutation N266H, DHS activity is significantly reduced, but HSS activity only slightly increased in comparison with the wild type. The double mutant N266H/I277V suggests that these mutations are responsible for an increase of HSS activity but not for a decrease of DHS activity. This interpretation is supported by the observation that the decrease of DHS activity of the triple mutant is similar to that of the N281D single mutant, whereas HSS activity reaches a value significantly higher than that of the wild-type DHS (Figure 4B). These observations confirm the statistical prediction of positively selected sites by codeml and support the hypothesis that the modification of the three selected amino acids is sufficient for a shift of the enzyme activity from DHS to HSS. However, the HSS activity of the mutant proteins is still lower than wild-type HSS from I. neei, indicating that further substitutions are involved to optimize HSS activity (Table 1).
Tissue-Specific Expression of HSS and DHS in I. neei
To test the hypothesis that the diversification of duplicated genes concerns not only the modification of the structural gene, but also the transcriptional control (Force et al., 1999; Zhang, 2003; Ober and Kaltenegger, 2009, and references therein), we performed RT-PCR experiments to detect the presence or absence of DHS- and HSS-coding cDNAs in various organs of I. neei, I. hederifolia, and I. alba. Supplemental Figure 2 online shows that, in all three species, HSS is transcribed differently in comparison with DHS. Whereas transcripts encoding DHS were detected in all four investigated plant organs, transcripts encoding HSS were detected mainly in young shoots and in roots. Using an affinity-purified antibody against HSS of I. neei, we were able to confirm the tissue-specific expression of HSS in I. neei at the protein level by detecting a signal exclusively in the protein extract of the roots (see Supplemental Figure 2 online).
DISCUSSION
Within the Convolvulaceae, PAs of various structural types occur in only certain isolated species (Eich, 2008). For the Asteraceae, which are also characterized by different PA types, namely, the senecionine type and lycopsamine type, HSS as the first specific enzyme of PA biosynthesis has previously been shown to have been recruited independently within the lineages of the Senecioneae and the Eupatorieae (Reimann et al., 2004). Therefore, speculating that PA biosynthesis also evolved repeatedly within the Convolvulaceae lineage, we identified and characterized HSS- and DHS-encoding cDNA sequences from several PA-producing and non-PA-producing species of this family. We have been able to show (1) that HSS as the first specific enzyme of PA biosynthesis evolved only once in one of the subclades of the Convolvulaceae by duplication of the DHS-coding gene; (2) that, in the genome of some recent PA-free species, no duplicates of the dhs gene can be detected, whereas in others, such as C. arvensis, relics of the hss gene are present that lost their function by pseudogenization; (3) that, in one PA-free species, I. alba, a functional copy of hss has been identified and demonstrated to be expressed; (4) that, according to statistical analyses, directly after the duplication event, purifying selection has acted; (5) that, along individual branches within the HSS sequence cluster, relaxed functional selection and even positive Darwinian evolution might have shaped and optimized the HSS, a statistical prediction supported by molecular experiments; and (6) that HSS and DHS diverged with respect to their tissue-specific expression.
Gain and Loss of HSS-Like Sequences within the Convolvulaceae
A phylogeny based on the cDNA sequences encoding HSS and DHS from various Convolvulaceae species, as identified in this project, together with previously published sequence data from other Angiosperm lineages (Reimann et al., 2004), shows convincingly that HSS evolved from only one gene duplication event in one of the major subclades of this family (Figure 2). In this subclade, which is termed clade 1 sensu Stefanović (Stefanovic et al., 2002, 2003), and which encompasses the tribes Argyreieae, Ipomoeeae, Merremieae, and Convolvuleae, PAs have been detected previously in only individual species (Eich, 2008). Our data suggest that the sporadic occurrence of PAs within this subclade is attributable to several independent losses of a functional gene copy in some tribes and species. Hence, by analyzing genomic DNA from several PA-free species, we have been unable to identify any hss-like sequences in the two species I. purpurea and C. tricolor but have identified two hss-like sequences in C. arvensis, both of which show signatures of pseudogenization. In I. alba, another PA-free species, we have been able to detect a functional HSS-like sequence that is transcribed within the plant. Remarkably, this enzyme, although part of the HSS clade, shows significant DHS activity and a HSS activity that seems to be less optimized than other HSS sequences from this cluster (Table 1). Whether this activity is a relic of the ability to produce PAs in an ancestor of this species or whether it has been recruited for a function other than PA biosynthesis remains open. Of note, I. alba produces indolizidine alkaloids, compounds with a fused six- and five-membered ring containing a heterocyclic nitrogen and showing slight structural resemblance to PAs (Gourley et al., 1969; Ikhiri et al., 1987). In Castanospermum australe (Fabaceae), PAs co-occur with indolizidine alkaloids (Molyneux et al., 1988). Further research should reveal whether the HSS-like enzyme is involved in the biosynthesis of these compounds.
Models for the Evolution of an Optimized HSS after Duplication of the dhs Gene
Despite the several cases of gene loss and pseudogenization detected in this study, the duplicated gene has escaped this extremely common fate of gene duplications in a number of species and has been successfully recruited as HSS for PA biosynthesis. Several models are discussed in the literature to explain the means by which gene duplication gives rise to new enzymatic functions (Long et al., 2003; Conant and Wolfe, 2008; Ober, 2010). The classical neofunctionalization model proposed by Ohno (1970) suggests that, after gene duplication, one copy is redundant and freed from functional constraints. This copy collects randomly selectively neutral substitutions and might develop, just by chance, a new function. Other models suggest that the new function of one copy is an old property of an ancestor gene that is at least bifunctional or promiscuous and that has been co-opted to a primary role after the duplication (Depristo, 2007; Conant and Wolfe, 2008). In contrast with nonadaptive models of gene evolution, which do not explain novel functions, the “escape of adaptive conflict model” postulates that a single-copy gene is selected to perform a novel function while maintaining its ancestral function. The resulting bifunctionality should result in a reduction of the copy’s ability to perform the original function. Gene duplication resolves this adaptive conflict, as each copy is free to specialize by subfunctionalization and by positive selection on one of these functions (Piatigorsky and Wistow, 1991; Hittinger and Carroll, 2007; Des Marais and Rausher, 2008; Deng et al., 2010).
Our data suggest that the evolution of HSS within the Convolvulaceae has signatures of various models. DHS occurs ubiquitously within eukaryotes, mostly as a single-copy gene, and has been shown to be essential for the viability of the cell (Park et al., 1998). Therefore, the ancestor gene that was duplicated in the Convolvulaceae lineage most likely had the characteristics of a DHS, suggesting that one of the gene copies remained almost unchanged to guarantee DHS functionality, whereas the other copy diversified. This is a characteristic postulate of the neofunctionalization model of Ohno (1970). On the other hand, functional analyses of the DHS proteins from various angiosperm species (Reimann et al., 2004) and of this study demonstrate that this enzyme catalyzes the formation of homospermidine (Table 1). Therefore, DHS catalyzes, as an inherent activity, the reaction that was selected in one of the copies after gene duplication. Such a specialization of a gene copy to one of the functions of the ancestral protein is a central feature of the subfunctionalization theory (Conant and Wolfe, 2008), which can be regarded as an “escape” of an adaptive conflict.
For the evolution of HSS, protein–protein interactions also have to be considered. Human DHS is a homotetramer organized as a dimer of two dimers, as two subunits at a time interact closely and form two active sites at the interphase of the subunits (Liao et al., 1998; Umland et al., 2004). Biochemical comparisons confirm that native HSS is also a tetramer (Ober and Hartmann, 1999a). After duplication of the dhs gene, protein–protein interactions might affect the fate of the duplicate in two ways. First, the protein encoded by the gene copy might still interact with the eIF5A substrate protein and compete with the second copy that ensures DHS functionality. Therefore, mutations in the gene copy that compromise the function but not the binding would interfere with the essential activity of DHS. Second, the protein subunits encoded by the diverging gene copies might form heterotetramers that are probably less efficient or even inactive (Wagner, 2003; Bridgham et al., 2008). Indeed, our branch-specific analyses of selection pressures show that purifying selection directly after the duplication of the dhs gene delayed adaptive evolution. This selection-driven preservation of the two gene copies should have resulted in a longer half-life of the duplicated gene pair and in an increased likelihood for the evolution of a new function (Bridgham et al., 2008). The situation changes later in the HSS clade. Along branch B, after the HSS of Merremia branched off, the branch model also suggests strong purifying selection (ω = 0.094), but the branch-site model detects 24 sites with ω = 1, giving hints of relaxed functional constraints (Figure 3). In branch C (i.e., in the evolution of the HSS of I. meyeri, I. neei, and I. hederifolia), the branch model and the branch-site model indicate that positive Darwinian selection has acted on the genes, most probably to reduce DHS activity while improving HSS activity. However, the branch-site model additionally predicts relaxed selection in branch C, when data set B, which includes the pseudogenes, is analyzed.
Several examples are described in the literature giving indications for the involvement of positive selection in the evolution of plant metabolic diversity, such as flower pigmentation, flower scent, and defense compounds (Bishop et al., 2000; Barkman, 2003; Lu and Rausher, 2003; Rodríguez-Trelles et al., 2003; Benderoth et al., 2006; Smith et al., 2013). To establish the causality of this statistical prediction in the case of HSS evolution, we substituted three of the predicted sites in the amino acid sequence of the DHS of I. neei and have been able to show that these substitutions are indeed sufficient in the triple mutant to reduce DHS activity to almost one-third of that of the native DHS, while improving HSS activity (Figure 4B). A recent analysis of the impact of substitutions on protein functionality suggests that the most prominent effects are observed when several sites that are part of a collectively evolving network can flip simultaneously (McLaughlin et al., 2012). Such a scenario might also be the reason for the strong effect of the triple mutant of HSS, with single and double mutants being less effective.
Relaxation of functional constraints should not be possible as long as the above-mentioned adaptive conflict is effective, because of protein–protein interactions, even though two copies exist. As an escape from this conflict, the separation of gene expression should allow each copy to be freed from their limited ability to evolve and to specialize with respect to its regulation. Transcriptional divergence is described in the literature as the most frequent fate of gene copies after the duplication event and is considered as a mechanism that allows the evolution of tissue and developmental specialization (Gu et al., 2002; Li et al., 2005; Duarte et al., 2006; Wapinski et al., 2007). In the case of HSS evolution, this optimization affects essentially one of the two copies (i.e., the gene encoding the developing HSS). Our analyses of the transcript localization in various tissues of I. neei, I. hederifolia, and I. alba show that, in these three species, the hss gene transcript is detectable mainly in young shoots and the roots. An immunoblot analysis of HSS of I. neei defines the roots as being the predominant site of protein expression (see Supplemental Figure 2 online). The dhs gene transcript is detectable in all analyzed tissues, a pattern that is also described in other angiosperm lineages (Moll et al., 2002; Anke et al., 2004; Nurhayati and Ober, 2005), suggesting that only the regulation of one copy was modified. Indeed, HSS expression is highly variable in various PA-producing lineages, making this enzyme one of the most diverse expressed proteins described in the literature (Niemüller et al., 2012). Thus, for DHS and HSS, the spatial separation of expression might have been selected leading to the release of the two copies from the molecular constraints. As a consequence, the separation of expression should also allow the gene copies to diverge with respect to enzymatic function.
METHODS
Plant Material, Nucleic Acid Isolation, and cDNA Synthesis
Seeds were collected from natural habitats or obtained from the Botanical Garden, Kiel (see Supplemental Table 9 online). Plants were grown under axenic conditions in vitro on a Murashige and Skoog medium modified according to von Borstel and Hartmann (1986) under a light-dark regime (16 h/8 h) at 25°C or in the greenhouse of the Botanical Garden (19 to 22°C without artificial light). Total RNA was isolated from fresh or stored (−80°C) plant tissues using either the RNeasy Plant Mini Kit (Qiagen) or TRIzol reagent (Invitrogen) according to the manufacturer’s protocols. First-strand cDNA synthesis of 1 µg total RNA was oligo(dT) primed (P1; all primers are given in Supplemental Table 7 online) using reverse transcriptases, such as Superscript III reverse transcriptase (Invitrogen). Genomic DNA was isolated using either the DNeasy Plant Mini Kit (Qiagen) or the NucleoSpin Plant II kit (Macherey-Nagel) according to the manufacturer’s instructions.
Identification and Heterologous Expression of cDNAs Coding for HSS and DHS
To identify cDNA sequences that code for HSS and DHS in Ipomoea neei and Merremia quinquefolia, a PCR approach with a pair of degenerate primers (P2/P3) was used as described previously (Ober and Hartmann, 1999a; Nurhayati and Ober, 2005). The rapid amplification of cDNA ends technique was used to identify the 3′- and 5′-ends of the cDNA sequences coding for HSS of I. neei and HSS and DHS of M. quinquefolia according to Ober and Hartmann (1999b). Based on the resulting sequence data, we designed the primer pairs for amplification of the complete ORFs. The amplification strategies are summarized in Supplemental Table 8A online. A high sequence identity between orthologs of DHS and HSS within the genus Ipomoea allowed us to amplify the ORFs of both orthologs from Ipomoea meyeri and Ipomoea alba and the DHS from I. neei with the primer pairs designed for the amplification of the DHS encoding the ORF of Ipomoea hederifolia (P16/P17) and the HSS of I. neei (P18/P19). Sequences containing the complete ORF were cloned into the expression vector pET22b (Novagen), which encoded an artificial C-terminal hexahistidine (6xHis) tag extension. The resulting constructs were sequenced and used for expression in Escherichia coli BL21(DE3) according to Ober and Hartmann (1999a). Recombinant proteins were purified using nickel-nitrilotriacetic acid metal affinity chromatography (Qiagen) according to the manufacturer’s protocol. For biochemical characterization, the purified proteins were concentrated and rebuffered to standard buffer (100 mM Gly-NaOH buffer, pH 9, 1 mM DTT, and 0.1 mM EDTA). Successful expression and purity were monitored by SDS-PAGE analysis. Protein quantities were estimated according to Bradford (1976).
Biochemical Characterization of Recombinant Proteins
Enzyme assays with radioactively labeled substrates were used to analyze the substrate specificity of the recombinant proteins. Two aminobutyl acceptors were tested: putrescine to test for HSS activity (HSS assay) and recombinant eIF5A precursor protein of Senecio vernalis to test for DHS activity (DHS assay). The recombinant eIF5A precursor protein from S. vernalis has been shown to be accepted by DHS proteins from various angiosperm species and to be a suitable substrate to estimate DHS activity (Reimann et al., 2004). Assays contained 2.5 to 20 µg of purified recombinant protein and were incubated at 30°C. Aliquots were taken after 2, 4, 8, and 16 min to ensure linearity of product formation. Enzyme reactions and product quantification followed the basic protocol described by Ober and Hartmann (1999b).
Identification of hss and dhs Genes
For identification of genomic sequences with homology to DHS- and HSS-coding genes in PA-producing and PA-free species of the Convolvulaceae, we performed PCR amplification with genomic DNA as template and AccuTaq LA DNA polymerase (Sigma-Aldrich) according to the manufacturer’s instructions. Because of a high degree of sequence identity, we were able to amplify dhs- and hss-like genes with the gene-specific primers P16/P17 and P18/P19, which had been designed for amplification of the complete ORF of the DHS of I. hederifolia and the HSS of I. neei from various closely related species. For the amplification of more divergent sequences, degenerate primers specific for the Convolvulaceae (P24 to P27) were designed based on an alignment of the HSS- and DHS-coding cDNA sequences of I. neei, I. hederifolia, I. meyeri, and M. quinquefolia and the DHS-coding sequences of tobacco (Nicotiana tabacum) and tomato (Solanum lycopersicum). For identification of the hss gene of I. neei, three overlapping fragments were generated (i.e., the 5′- and 3′-ends) using the primer pairs P18/P30 and P5/P19, respectively, and the internal fragment by an inverse PCR approach according to Ochman et al. (1988) using primer pair P28/P29. As a template for inverse PCR, genomic DNA from I. neei was digested with KpnI. All fragments were cloned into pGEM T Easy vector (Promega) or pCR-XL-TOPO vector (Invitrogen). The amplification strategies and the length of the identified fragments are summarized in Supplemental Table 8B online.
Sequence and Phylogenetic Analyses
DNA sequences were analyzed by the Wisconsin Software Package of the Genetics Computer Group (version 11.1; Accelrys). Using ClustalX 1.4 (Thompson et al., 1997), the amino acid sequences of DHS and HSS were aligned and subsequently reconverted to a nucleotide alignment with BioEdit 7.0.5.3 (Hall, 1999). The resulting alignments were employed to estimate phylogenies with the following software of the PHYLIP program package (Felsenstein, 2005): DNAML as a maximum likelihood method with randomized input order. SEQBOOT and CONSENSE were used to estimate bootstrap values of 1000 replicates. To calculate distances of individual sequence pairs, DNADIST was used for nucleotide sequences with the two-parameter model of Kimura and a transition/transversion ratio of 2 (Kimura, 1980), and PROTDIST was used for amino acid sequences with the Jones-Taylor-Thornton model (Jones et al., 1992).
Analysis of Codon Substitution Patterns to Test for Positive Selection
The CODEML program from the PAML 4.1 package (Yang, 2007) was used to analyze codon substitution patterns with a maximum likelihood approach to test for potential positive selection. The data set included all identified HSS- and DHS-coding sequences of the Convolvulaceae and the DHS-coding cDNA sequences from tobacco and tomato. Two alignments, namely, data set A excluding the pseudogenes (852 bp length after deleting gaps) and data set B including the coding regions of pseudogenes carv-2 and carv-3 (645 bp length after deleting gaps), were used to calculate a maximum likelihood tree using PAUP version 4.0b10 (Swofford, 1998). A heuristic search was conducted with 100 random sequence additions and tree bisection-reconnection branch swapping. The topology of the tree and the alignment were used for the CODEML calculations. Model comparisons were performed as recommended recently (Yang et al., 2005; Yang and dos Reis, 2011). Briefly, the M0 model, which allows one dN/dS ratio (ω) for all branches, was calculated as the simplest model and was compared with the more sophisticated models for consistency (Yang, 1997, 2007). Furthermore, the site models M1a (nearly neutral) and M2a (positive selection) were calculated, which allowed the ω ratio to vary among sites. Positive selection on different branches was tested by the branch model, which allowed the ω ratio to vary among branches in the phylogeny, and by the more elaborate branch-site model A (Yang and Nielsen, 2002; Zhang et al., 2005), which is aimed at detecting episodic positive selection that acts on a subset of sites and on a specific branch. The last-mentioned model divides the phylogeny a priori into foreground and background lineages. Only foreground lineages might have experienced positive selection (Zhang et al., 2005). It assumes four site classes: p0 as sites under purifying selection in background and foreground branches, p1 as sites under neutral selection (ω1 = 1) in background and foreground branches, p2a as sites under purifying selection, and p2b under neutral selection in the background branches but ω2 ≥ 1 in the foreground branches. Thus, p2a includes sites that have experienced positive selection within a certain time period compared with the background. The LRT was used to test for the statistical significance of the signal of selection (Anisimova et al., 2001) in the various models. The following LRTs were performed: LRT1, the branch model against the M0 model; LRT2, the site model M2a (positive selection) against the M1a (nearly neutral) (Yang, 1997; Yang and Nielsen, 2002); LRT3, the branch-site model A against the branch-site model A with ω2 fixed at 1 in the background branches; and LRT4, to test for neutral selection (Zhang et al., 2005). In LRT3, the null model allows sites evolving under negative selection on the background lineages to be released from constraints and to evolve neutrally on the foreground lineages. Thus, the test is a direct test for positive selection on the foreground lineages (Zhang et al., 2005). In LRT4, the branch-site model A with ω2 fixed at 1 was compared with the M1a (nearly neutral) site model as the null model. The M1a model calculates sites evolving under neutral selection (p1, ω1 = 1) averaged over all branches, whereas the branch-site model A with ω2 fixed at 1 detects an increase of neutrally evolving sites in the foreground branches (p2a) compared with the background branches, considered as a signature of relaxed selection. For multiple hypothesis testing, we used (1) the Bonferroni method to control the family-wise error rate according to (Anisimova and Yang, 2007) and (2) the procedure described by Benjamini and Yekutieli (2001), which controls the false discovery rate, referred to as the B-Y method. Whereas the Bonferroni method is known to be most conservative (Anisimova and Yang, 2007), the B-Y method is considered to deliver more biologically meaningful results (Narum, 2006).
Site-Specific Mutagenesis
For the modification of DHS of I. neei, the pET22b-based expression construct containing the cDNA coding for DHS was used as the template. The mutations were introduced by a PCR-based strategy according to the instructions of the Quick-Change site-directed mutagenesis kit (Stratagene). For the single mutations of N266H, I277V, and N281D, primer pairs P31/P32, P35/P36, and P37/P38 were used. To introduce simultaneously the mutations I277V and N281D, the primer pair P33/P34 was used. After control sequencing, the mutated genes were expressed in E. coli BL21(DE3), purified by nickel-nitrilotriacetic acid agarose metal chelate chromatography, concentrated, and rebuffered to standard buffer for biochemical characterization in comparison with the wild-type DHS of I. neei. DHS and HSS activities result from at least six independent replicates. We calculated q-q plots to assign a normal distribution of data points, Levene tests to determine homogeneity of variance, and t tests to test for significant differences in activities.
RT-PCR for Detection of Gene Expression
Plant organs (young shoot, shoot, leaves, and root) were sampled from plants grown in the greenhouse, and total RNA was isolated and reverse transcribed as described above. For I. neei and I. hederifolia, primer pairs P16/P17 and P18/P19 were used to amplify the full-length cDNA of HSS and DHS, respectively. For the RT-PCR of I. alba, gene-specific primers P39/P40 and P41/P42 were designed to amplify an internal fragment of HSS and DHS, respectively. Aliquots of the reactions were analyzed after 35 cycles by agarose gel electrophoresis.
Polyclonal Antibody Preparation and Protein Gel Blot Analysis
To generate polyclonal antibodies, recombinant HSS from I. neei was expressed in 500-mL E. coli BL21(DE3) cultures and purified as described above. The protein was rebuffered to 5 mM potassium phosphate, pH 8.0, using Millipore (Amicon Ultra-4) concentrators. Freeze-dried aliquots (1× 500 μg; 2× 250 μg) were used to raise polyclonal antibodies in rabbits by repeated subcutaneous injections (Sequence Laboratories). The antibodies were affinity purified from the serum as described previously (Moll et al., 2002). Plant organs were sampled from I. neei plants grown in the greenhouse, and soluble proteins were extracted, separated on 12% SDS-PAGE gels, and electroblotted onto a polyvinylidene difluoride membrane (RotiPVDF; Roth) as described previously (Anke et al., 2004). To prevent protein degradation, proteinase inhibitors (10 μM PMSF, 0.5 mM EDTA, 1 μM benzamidine, 1 μg/mL pepstatin A, 1 μg/mL leupeptin, and 5 μg/mL aprotinin) were added to the extraction buffer. After being blotted and blocked, the membrane was incubated with the affinity-purified polyclonal antibody (OD280 = 2.4 × 10−4) for 1 h. Antibody-antigen reactions were detected using a goat-anti-rabbit secondary antibody conjugated to horseradish peroxidase (Dianova).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers given in Supplemental Table 8 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignment of the Genomic Sequences with Similarity to dhs Genes Identified from Convolvulus arvensis.
Supplemental Figure 2. Tissue-Specific Expression of DHS and HSS in Various Plant Organs.
Supplemental Table 1. Genomic Organization of dhs and hss Genes of the Convolvulaceae Species I. neei, I. alba, I. purpurea, C. tricolor, and C. arvensis.
Supplemental Table 2. Distances from Pairwise Comparisons of Selected Pairs of DHS- and HSS-Coding cDNA Sequences from the Convolvulaceae.
Supplemental Table 3. Analyses of Codon Substitution Patterns for Dataset A.
Supplemental Table 4. Likelihood Ratio Test Statistics and P Values for Various Tests of Positive Selection for Dataset A.
Supplemental Table 5. Analyses of Codon Substitution Patterns for Dataset B.
Supplemental Table 6. Likelihood Ratio Test Statistics and P Values for Various Tests of Positive Selection for Dataset B.
Supplemental Table 7. Nucleotide Sequence of Primers.
Supplemental Table 8. Amplification Strategy of DHS- and HSS-Coding cDNAs and dhs and hss Genes and Accession Numbers of Sequences Identified in This Project and Taken from the Databases.
Supplemental Table 9. Species Studied in This Project with Year and Country of Collection.
Supplemental Data Set 1. Text File of Sequence Alignment Corresponding to Phylogenetic Analysis in Figure 2.
Supplementary Material
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (grant to D.O.) and by the Austrian Academy of Sciences (scholarship to E.K.). We thank Brigitte Schemmerling for her excellent technical assistance and the staff of the Botanic Gardens, Kiel, for their support in culturing the Convolvulaceae species. We thank the Institute of Clinical Molecular Biology in Kiel for providing Sanger sequencing as support in part by the Deutsche Forschungsgemeinschaft Cluster of Excellence “Inflammation at Interfaces” and “Future Ocean.” We thank the technicians Sandra Greve and Stefanie Arndt for technical support.
AUTHOR CONTRIBUTIONS
E.K. performed and designed the research, analyzed and interpreted the data, and wrote the article. D.O. designed the research, analyzed and interpreted the data, and wrote the article. E.E. contributed seeds and data of the species analyzed in this study and interpreted the data.
Glossary
- PA
pyrrolizidine alkaloid
- HSS
homospermidine synthase
- DHS
deoxyhypusine synthase
- ORF
open reading frame
- LRT
likelihood ratio test
References
- Anisimova M., Bielawski J.P., Yang Z. (2001). Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Mol. Biol. Evol. 18: 1585–1592 [DOI] [PubMed] [Google Scholar]
- Anisimova M., Yang Z. (2007). Multiple hypothesis testing to detect lineages under positive selection that affects only a few sites. Mol. Biol. Evol. 24: 1219–1228 [DOI] [PubMed] [Google Scholar]
- Anke S., Niemüller D., Moll S., Hänsch R., Ober D. (2004). Polyphyletic origin of pyrrolizidine alkaloids within the Asteraceae. Evidence from differential tissue expression of homospermidine synthase. Plant Physiol. 136: 4037–4047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkman T.J. (2003). Evidence for positive selection on the floral scent gene isoeugenol-O-methyltransferase. Mol. Biol. Evol. 20: 168–172 [DOI] [PubMed] [Google Scholar]
- Benderoth M., Textor S., Windsor A.J., Mitchell-Olds T., Gershenzon J., Kroymann J. (2006). Positive selection driving diversification in plant secondary metabolism. Proc. Natl. Acad. Sci. USA 103: 9118–9123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Yekutieli D. (2001). The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29: 1165–1188 [Google Scholar]
- Bishop J.G., Dean A.M., Mitchell-Olds T. (2000). Rapid evolution in plant chitinases: Molecular targets of selection in plant-pathogen coevolution. Proc. Natl. Acad. Sci. USA 97: 5322–5327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenship J.D., Houseknecht J.B., Pal S., Bush L.P., Grossman R.B., Schardl C.L. (2005). Biosynthetic precursors of fungal pyrrolizidines, the loline alkaloids. ChemBioChem 6: 1016–1022 [DOI] [PubMed] [Google Scholar]
- Böttcher F., Adolph R.D., Hartmann T. (1993). Homospermidine synthase, the first pathway-specific enzyme in pyrrolizidine alkaloid biosynthesis. Phytochemistry 32: 679–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher F., Ober D., Hartmann T. (1994). Biosynthesis of pyrrolizidine alkaloids: putrescine and spermidine are essential substrates of enzymatic homospermidine formation. Can. J. Chem. 72: 80–85 [Google Scholar]
- Bradford M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Bridgham J.T., Brown J.E., Rodríguez-Marí A., Catchen J.M., Thornton J.W. (2008). Evolution of a new function by degenerative mutation in cephalochordate steroid receptors. PLoS Genet. 4: e1000191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K.Y., Liu A.Y. (1997). Biochemistry and function of hypusine formation on eukaryotic initiation factor 5A. Biol. Signals 6: 105–109 [DOI] [PubMed] [Google Scholar]
- Chou H.-H., Hayakawa T., Diaz S., Krings M., Indriati E., Leakey M., Paabo S., Satta Y., Takahata N., Varki A. (2002). Inactivation of CMP-N-acetylneuraminic acid hydroxylase occurred prior to brain expansion during human evolution. Proc. Natl. Acad. Sci. USA 99: 11736–11741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant G.C., Wolfe K.H. (2008). Turning a hobby into a job: How duplicated genes find new functions. Nat. Rev. Genet. 9: 938–950 [DOI] [PubMed] [Google Scholar]
- Deng C., Cheng C.-H.C., Ye H., He X., Chen L. (2010). Evolution of an antifreeze protein by neofunctionalization under escape from adaptive conflict. Proc. Natl. Acad. Sci. USA 107: 21593–21598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depristo M.A. (2007). The subtle benefits of being promiscuous: Adaptive evolution potentiated by enzyme promiscuity. HFSP J. 1: 94–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Des Marais D.L., Rausher M.D. (2008). Escape from adaptive conflict after duplication in an anthocyanin pathway gene. Nature 454: 762–765 [DOI] [PubMed] [Google Scholar]
- Dreyer D.L., Jones K.C., Molyneux R.J. (1985). Feeding deterrency of some pyrrolizidine, indolizidine, and quinolizidine alkaloids towards pea aphid (Acyrthosiphon pisum) and evidence for phloem transport of indolizidine alkaloid swainsonine. J. Chem. Ecol. 11: 1045–1051 [DOI] [PubMed] [Google Scholar]
- Duarte J.M., Cui L., Wall P.K., Zhang Q., Zhang X., Leebens-Mack J., Ma H., Altman N., dePamphilis C.W. (2006). Expression pattern shifts following duplication indicative of subfunctionalization and neofunctionalization in regulatory genes of Arabidopsis. Mol. Biol. Evol. 23: 469–478 [DOI] [PubMed] [Google Scholar]
- Eich, E. (2008). Solanaceae and Convolvulaceae: Secondary Metabolites. Biosynthesis, Chemotaxonomy, Biological and Economic Significance. A Handbook. (Berlin, Heidelberg: Springer). [Google Scholar]
- Felsenstein, J. (2005). PHYLIP, Phylogeny Inference Package, Version 3.6. (Seattle, WA: University of Washington). [Google Scholar]
- Force A., Lynch M., Pickett F.B., Amores A., Yan Y.L., Postlethwait J. (1999). Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M., Thomas B.C. (2006). Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res. 16: 805–814 [DOI] [PubMed] [Google Scholar]
- Gourley J.M., Heacock R.A., McInnes A.G., Nikolin B., Smith D.G. (1969). The structure of ipalbine, a new hexahydroindolizine alkaloid, isolated from Ipomoea alba L. J. Chem. Soc. Chem. Commun. 709–710 [Google Scholar]
- Grotewold E. (2005). Plant metabolic diversity: A regulatory perspective. Trends Plant Sci. 10: 57–62 [DOI] [PubMed] [Google Scholar]
- Gu Z., Nicolae D., Lu H.H.S., Li W.-H. (2002). Rapid divergence in expression between duplicate genes inferred from microarray data. Trends Genet. 18: 609–613 [DOI] [PubMed] [Google Scholar]
- Hall T.A. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41: 95–98 [Google Scholar]
- Harborne, J.B. (1993). Introduction to Ecological Biochemistry. (San Diego, CA: Academic Press). [Google Scholar]
- Hartmann T. (2007). From waste products to ecochemicals: fifty years research of plant secondary metabolism. Phytochemistry 68: 2831–2846 [DOI] [PubMed] [Google Scholar]
- Hartmann, T., and Ober, D. (2008). Defense by pyrrolizidine alkaloids: Developed by plants and recruited by insects. In Induced Plant Resistance to Herbivory, A. Schaller, ed (Berlin: Springer Science and Business Media), pp. 213–231. [Google Scholar]
- Hartmann, T., and Witte, L. (1995). Chemistry, biology and chemoecology of the pyrrolizidine alkaloids. In Alkaloids: Chemical and Biological Perspectives, S.W. Pelletier, ed (Oxford, UK: Pergamon Press), pp. 155–233. [Google Scholar]
- Hittinger C.T., Carroll S.B. (2007). Gene duplication and the adaptive evolution of a classic genetic switch. Nature 449: 677–681 [DOI] [PubMed] [Google Scholar]
- Hristov N., Conner W.E. (2005). Effectiveness of tiger moth (Lepidoptera, Arctiidae) chemical defenses against an insectivorous bat (Eptesicus fuscus). Chemoecology 15: 105–113 [Google Scholar]
- Hughes A.L. (1994). The evolution of functionally novel proteins after gene duplication. Proc. Biol. Sci. 256: 119–124 [DOI] [PubMed] [Google Scholar]
- Hughes A.L. (2002). Adaptive evolution after gene duplication. Trends Genet. 18: 433–434 [DOI] [PubMed] [Google Scholar]
- Hughes A.L. (2005). Gene duplication and the origin of novel proteins. Proc. Natl. Acad. Sci. USA 102: 8791–8792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikhiri K., Koulodo D.D.D., Garba M., Mamane S., Ahond A., Poupat C., Potier P. (1987). Noveaux alcaloides indiliziniques isolés de Ipomoea alba. J. Nat. Prod. 50: 152–156 [Google Scholar]
- Jenett-Siems K., Kaloga M., Eich E. (1993). Ipangulines, the first pyrrolizidine alkaloids from the Convolvulaceae. Phytochemistry 34: 437–440 [Google Scholar]
- Jenett-Siems K., Ott S.C., Schimming T., Siems K., Müller F., Hilker M., Witte L., Hartmann T., Austin D.F., Eich E. (2005). Ipangulines and minalobines, chemotaxonomic markers of the infrageneric Ipomoea taxon subgenus Quamoclit, section Mina. Phytochemistry 66: 223–231 [DOI] [PubMed] [Google Scholar]
- Jenett-Siems K., Schimming T., Kaloga M., Eich E., Siems K., Gupta M.P., Witte L., Hartmann T. (1998). Pyrrolizidine alkaloids of Ipomoea hederifolia and related species. Phytochemistry 47: 1551–1560 [Google Scholar]
- Jones D.T., Taylor W.R., Thornton J.M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8: 275–282 [DOI] [PubMed] [Google Scholar]
- Kimura M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16: 111–120 [DOI] [PubMed] [Google Scholar]
- Langel D., Ober D., Pelser P. (2011). The evolution of pyrrolizidine alkaloid biosynthesis and diversity in the Senecioneae. Phytochem. Rev. 10: 3–74 [Google Scholar]
- Li W.-H., Yang J., Gu X. (2005). Expression divergence between duplicate genes. Trends Genet. 21: 602–607 [DOI] [PubMed] [Google Scholar]
- Liao D.I., Wolff E.C., Park M.H., Davies D.R. (1998). Crystal structure of the NAD complex of human deoxyhypusine synthase: an enzyme with a ball-and-chain mechanism for blocking the active site. Structure 6: 23–32 [DOI] [PubMed] [Google Scholar]
- Long M., Betrán E., Thornton K., Wang W. (2003). The origin of new genes: Glimpses from the young and old. Nat. Rev. Genet. 4: 865–875 [DOI] [PubMed] [Google Scholar]
- Lu Y., Rausher M.D. (2003). Evolutionary rate variation in anthocyanin pathway genes. Mol. Biol. Evol. 20: 1844–1853 [DOI] [PubMed] [Google Scholar]
- Lynch M., Force A. (2000). The probability of duplicate gene preservation by subfunctionalization. Genetics 154: 459–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabberley, D.J. (1987). The Plant Book. (Cambridge, UK: Cambridge University Press). [Google Scholar]
- Mann, P. (1997). Zur Phytochemie und Chemotaxonomie Tropischer und Mediterraner Convolvulaceen unter Besonderer Berücksichtigung des Alkaloid-Vorkommens. PhD dissertation (Berlin: Fachbereich Pharmazie, Freie Universität). [Google Scholar]
- Markert A., Steffan N., Ploss K., Hellwig S., Steiner U., Drewke C., Li S.-M., Boland W., Leistner E. (2008). Biosynthesis and accumulation of ergoline alkaloids in a mutualistic association between Ipomoea asarifolia (Convolvulaceae) and a clavicipitalean fungus. Plant Physiol. 147: 296–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin R.N., Jr, Poelwijk F.J., Raman A., Gosal W.S., Ranganathan R. (2012). The spatial architecture of protein function and adaptation. Nature 491: 138–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R.E., Buckley T.R., Manos P.S. (2002). An examination of the monophyly of morning glory taxa using Bayesian phylogenetic inference. Syst. Biol. 51: 740–753 [DOI] [PubMed] [Google Scholar]
- Miller R.E., McDonald J.A., Manos P.S. (2004). Systematics of Ipomoea subgenus Quamoclit (Convolvulaceae) based on ITS sequence data and a Bayesian phylogenetic analysis. Am. J. Bot. 91: 1208–1218 [DOI] [PubMed] [Google Scholar]
- Moll S., Anke S., Kahmann U., Hänsch R., Hartmann T., Ober D. (2002). Cell-specific expression of homospermidine synthase, the entry enzyme of the pyrrolizidine alkaloid pathway in Senecio vernalis, in comparison with its ancestor, deoxyhypusine synthase. Plant Physiol. 130: 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneux R.J., Benson M., Wong R.Y., Tropea J.E., Elbein A.D. (1988). Australine a novel pyrrolizidine alkaloid glucosidase inhibitor from Castanospermum australe. J. Nat. Prod. 51: 1198–1206 [DOI] [PubMed] [Google Scholar]
- Narum S. (2006). Beyond Bonferroni: Less conservative analyses for conservation genetics. Conserv. Genet. 7: 783–787 [Google Scholar]
- Niemüller D., Reimann A., Ober D. (2012). Distinct cell-specific expression of homospermidine synthase involved in pyrrolizidine alkaloid biosynthesis in three species of the Boraginales. Plant Physiol. 159: 920–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa M., Suzuki Y., Nei M. (2009). Reliabilities of identifying positive selection by the branch-site and the site-prediction methods. Proc. Natl. Acad. Sci. USA 106: 6700–6705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurhayati N., Gondé D., Ober D. (2009). Evolution of pyrrolizidine alkaloids in Phalaenopsis orchids and other monocotyledons: Identification of deoxyhypusine synthase, homospermidine synthase and related pseudogenes. Phytochemistry 70: 508–516 [DOI] [PubMed] [Google Scholar]
- Nurhayati N., Ober D. (2005). Recruitment of alkaloid-specific homospermidine synthase (HSS) from ubiquitous deoxyhypusine synthase: Does Crotalaria possess a functional HSS that still has DHS activity? Phytochemistry 66: 1346–1357 [DOI] [PubMed] [Google Scholar]
- Ober D. (2005). Seeing double: Gene duplication and diversification in plant secondary metabolism. Trends Plant Sci. 10: 444–449 [DOI] [PubMed] [Google Scholar]
- Ober D. (2010). Gene duplications and the time thereafter - Examples from plant secondary metabolism. Plant Biol (Stuttg) 12: 570–577 [DOI] [PubMed] [Google Scholar]
- Ober D., Gibas L., Witte L., Hartmann T. (2003a). Evidence for general occurrence of homospermidine in plants and its supposed origin as by-product of deoxyhypusine synthase. Phytochemistry 62: 339–344 [DOI] [PubMed] [Google Scholar]
- Ober D., Harms R., Witte L., Hartmann T. (2003b). Molecular evolution by change of function. Alkaloid-specific homospermidine synthase retained all properties of deoxyhypusine synthase except binding the eIF5A precursor protein. J. Biol. Chem. 278: 12805–12812 [DOI] [PubMed] [Google Scholar]
- Ober D., Hartmann T. (1999a). Homospermidine synthase, the first pathway-specific enzyme of pyrrolizidine alkaloid biosynthesis, evolved from deoxyhypusine synthase. Proc. Natl. Acad. Sci. USA 96: 14777–14782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober D., Hartmann T. (1999b). Deoxyhypusine synthase from tobacco. cDNA isolation, characterization, and bacterial expression of an enzyme with extended substrate specificity. J. Biol. Chem. 274: 32040–32047 [DOI] [PubMed] [Google Scholar]
- Ober D., Kaltenegger E. (2009). Pyrrolizidine alkaloid biosynthesis, evolution of a pathway in plant secondary metabolism. Phytochemistry 70: 1687–1695 [DOI] [PubMed] [Google Scholar]
- Ochman H., Gerber A.S., Hartl D.L. (1988). Genetic applications of an inverse polymerase chain reaction. Genetics 120: 621–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno, S. (1970). Evolution by Gene Duplication. (Berlin: Springer). [Google Scholar]
- Park M.H., Joe Y.A., Kang K.R. (1998). Deoxyhypusine synthase activity is essential for cell viability in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 273: 1677–1683 [DOI] [PubMed] [Google Scholar]
- Park M.H., Lee Y.B., Joe Y.A. (1997). Hypusine is essential for eukaryotic cell proliferation. Biol. Signals 6: 115–123 [DOI] [PubMed] [Google Scholar]
- Piatigorsky J., Wistow G. (1991). The recruitment of crystallins: New functions precede gene duplication. Science 252: 1078–1079 [DOI] [PubMed] [Google Scholar]
- Pichersky E., Gang D.R. (2000). Genetics and biochemistry of secondary metabolites in plants: An evolutionary perspective. Trends Plant Sci. 5: 439–445 [DOI] [PubMed] [Google Scholar]
- Reimann A., Nurhayati N., Backenköhler A., Ober D. (2004). Repeated evolution of the pyrrolizidine alkaloid-mediated defense system in separate angiosperm lineages. Plant Cell 16: 2772–2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard A., Janke M., von der Ohe W., Kempf M., Theuring C., Hartmann T., Schreier P., Beuerle T. (2009). Feeding deterrence and detrimental effects of pyrrolizidine alkaloids fed to honey bees (Apis mellifera). J. Chem. Ecol. 35: 1086–1095 [DOI] [PubMed] [Google Scholar]
- Rodin S.N., Parkhomchuk D.V., Rodin A.S., Holmquist G.P., Riggs A.D. (2005). Repositioning-dependent fate of duplicate genes. DNA Cell Biol. 24: 529–542 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Trelles F., Tarrío R., Ayala F.J. (2003). Convergent neofunctionalization by positive Darwinian selection after ancient recurrent duplications of the xanthine dehydrogenase gene. Proc. Natl. Acad. Sci. USA 100: 13413–13417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schardl, C.L. (2009). Fungal endophytes in Lolium and Festuca species. In Molecular Breeding of Forage and Turf, T. Yamada and G. Spangenberg, eds (New York: Springer), pp. 285–298. [Google Scholar]
- Schardl C.L., Grossman R.B., Nagabhyru P., Faulkner J.R., Mallik U.P. (2007). Loline alkaloids: Currencies of mutualism. Phytochemistry 68: 980–996 [DOI] [PubMed] [Google Scholar]
- Siciliano T., Leo M.D., Bader A., Tommasi N.D., Vrieling K., Braca A., Morelli I. (2005). Pyrrolizidine alkaloids from Anchusa strigosa and their antifeedant activity. Phytochemistry 66: 1593–1600 [DOI] [PubMed] [Google Scholar]
- Singer M.S., Stireman J.O., III (2003). Does anti-parasitoid defense explain host-plant selection by a polyphagous caterpillar? Oikos 100: 554–562 [Google Scholar]
- Smith S.D., Wang S., Rausher M.D. (2013). Functional evolution of an anthocyanin pathway enzyme during a flower color transition. Mol. Biol. Evol. 30: 602–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic S., Austin D.F., Olmstead Richard G. (2003). Classification of Convolvulaceae: A phylogenetic approach. Syst. Bot. 28: 791–806 [Google Scholar]
- Stefanovic S., Krueger L., Olmstead R.G. (2002). Monophyly of the Convolvulaceae and circumscription of their major lineages based on DNA sequences of multiple chloroplast loci. Am. J. Bot. 89: 1510–1522 [DOI] [PubMed] [Google Scholar]
- Steiner U., et al. (2006). Molecular characterization of a seed transmitted clavicipitaceous fungus occurring on dicotyledoneous plants (Convolvulaceae). Planta 224: 533–544 [DOI] [PubMed] [Google Scholar]
- Swofford, D.L. (1998). Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4.0b8. (Sunderland, MA: Sinauer). [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tofern, B. (1999). Neue und seltene Sekundärstoffe des Phenylpropan-, Terpen- und Alkaloid-Stoffwechsels aus tropischen Convolvulaceen. PhD dissertation (Berlin: Fachbereich Pharmazie, Freie Universität). [Google Scholar]
- Tofern B., Kaloga M., Witte L., Hartmann T., Eich E. (1999). Occurrence of loline alkaloids in Argyreia mollis (Convolvulaceae). Phytochemistry 51: 1177–1180 [Google Scholar]
- Umland T.C., Wolff E.C., Park M.H., Davies D.R. (2004). A new crystal structure of deoxyhypusine synthase reveals the configuration of the active enzyme and of an enzyme-NAD-inhibitor ternary complex. J. Biol. Chem. 279: 28697–28705 [DOI] [PubMed] [Google Scholar]
- von Borstel K., Hartmann T. (1986). Selective uptake of pyrrolizidine N-oxides by cell suspension cultures from pyrrolizidine alkaloid producing plants. Plant Cell Rep. 5: 39–42 [DOI] [PubMed] [Google Scholar]
- Wagner A. (2003). How the global structure of protein interaction networks evolves. Proc. Biol. Sci. 270: 457–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski I., Pfeffer A., Friedman N., Regev A. (2007). Natural history and evolutionary principles of gene duplication in fungi. Nature 449: 54–61 [DOI] [PubMed] [Google Scholar]
- Yang Z. (1997). PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13: 555–556 [DOI] [PubMed] [Google Scholar]
- Yang Z. (1998). Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol. Biol. Evol. 15: 568–573 [DOI] [PubMed] [Google Scholar]
- Yang Z. (2007). PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24: 1586–1591 [DOI] [PubMed] [Google Scholar]
- Yang Z., dos Reis M. (2011). Statistical properties of the branch-site test of positive selection. Mol. Biol. Evol. 28: 1217–1228 [DOI] [PubMed] [Google Scholar]
- Yang Z., Nielsen R. (1998). Synonymous and nonsynonymous rate variation in nuclear genes of mammals. J. Mol. Evol. 46: 409–418 [DOI] [PubMed] [Google Scholar]
- Yang Z., Nielsen R. (2002). Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 19: 908–917 [DOI] [PubMed] [Google Scholar]
- Yang Z., Nielsen R., Goldman N., Pedersen A.M.K. (2000). Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155: 431–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Wong W.S.W., Nielsen R. (2005). Bayes empirical bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22: 1107–1118 [DOI] [PubMed] [Google Scholar]
- Zhang J. (2003). Evolution by gene duplication: An update. Trends Ecol. Evol. 18: 292–298 [Google Scholar]
- Zhang J., Nielsen R., Yang Z. (2005). Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 22: 2472–2479 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.