Background: Streptococcus pneumoniae BglA-2 is a GH-1 6-phospho-β-glucosidase with specificity toward 1,4-linked 6-phospho-β-glucosides.
Results: BglA-2 and other GH-1 members adopt a similar overall structure and catalytic mechanism.
Conclusion: Tyr126, Tyr303, and Trp338 determine substrate specificity, and Ser424, Lys430, and Tyr432 discriminate phosphorylated from non-phosphorylated substrate. A tryptophan residue discriminates 6-phospho-β-glucosidase from 6-phospho-β-galactosidase activities.
Significance: BglA-2 structures provide new insight into characteristics and substrate specificity of 6-phospho-β-glucosidase.
Keywords: Crystal Structure, Enzyme Catalysis, Enzyme Mechanisms, Pneumonia, Streptococcus, 6-Phospho-β-glucosidase, PEP-PTS, Substrate Specificity, Thiocellobiose 6-Phosphate
Abstract
The 6-phospho-β-glucosidase BglA-2 (EC 3.2.1.86) from glycoside hydrolase family 1 (GH-1) catalyzes the hydrolysis of β-1,4-linked cellobiose 6-phosphate (cellobiose-6′P) to yield glucose and glucose 6-phosphate. Both reaction products are further metabolized by the energy-generating glycolytic pathway. Here, we present the first crystal structures of the apo and complex forms of BglA-2 with thiocellobiose-6′P (a non-metabolizable analog of cellobiose-6′P) at 2.0 and 2.4 Å resolution, respectively. Similar to other GH-1 enzymes, the overall structure of BglA-2 from Streptococcus pneumoniae adopts a typical (β/α)8 TIM-barrel, with the active site located at the center of the convex surface of the β-barrel. Structural analyses, in combination with enzymatic data obtained from site-directed mutant proteins, suggest that three aromatic residues, Tyr126, Tyr303, and Trp338, at subsite +1 of BglA-2 determine substrate specificity with respect to 1,4-linked 6-phospho-β-glucosides. Moreover, three additional residues, Ser424, Lys430, and Tyr432 of BglA-2, were found to play important roles in the hydrolytic selectivity toward phosphorylated rather than non-phosphorylated compounds. Comparative structural analysis suggests that a tryptophan versus a methionine/alanine residue at subsite −1 may contribute to the catalytic and substrate selectivity with respect to structurally similar 6-phospho-β-galactosidases and 6-phospho-β-glucosidases assigned to the GH-1 family.
Introduction
Pathogenic strains of Streptococcus pneumoniae are the primary cause of acute pneumonia, otitis media, and meningitis in humans (1–3). During infection and colonization, abundant carbon sources are needed to support growth of the organism in host cells. For this purpose, 32 carbohydrates have been identified as energy sources for S. pneumoniae, including three-carbon molecules (glycerol), nine hexoses or hexose derivatives, three α-galactosides, two β-galactosides, four α-glucosides, seven β-glucosides, and six polysaccharides (4). In S. pneumoniae, the accumulation of carbohydrates is facilitated by three major types of sugar transporters (5, 6), including phosphoenolpyruvate-dependent phosphotransferase system (PEP-PTS)3 transporters (7–11), cation/proton-coupled transporters (12, 13), and ATP-binding cassette transporters (14–16). Among them, the most ubiquitous PEP-PTS systems in bacteria are responsible for the uptake of various carbohydrates, such as glucose, sucrose, mannitol, fructose, lactose, mannose, and cellobiose (11). During transport via the PEP-PTS, sugars, including the disaccharide cellobiose (Fig. 1), are simultaneously phosphorylated at the C6 hydroxyl moiety during entry into the cell (10). The gene SP_0578 encoding a 6-phospho-β-glucosidase BglA-2 from S. pneumoniae TIGR4 resides in a three-gene operon (Fig. 2A) comprising a transcription anti-terminator LicT (SP_0576) and a downstream gene BglP (SP_0577) encoding a β-glucoside PEP-PTS transporter. This operon is present in a number of species, including Streptococcus mutans, Clostridium longisporum, and Listeria monocytogenes. BglP comprises three domains, EIIA, EIIB, and EIIC, that facilitate the simultaneous translocation and phosphorylation of cellobiose and related β-glycosides (Fig. 2B). The transmembrane permease domain (EIIC) is responsible for recognition and binding of specific substrates, and the incoming sugars are phosphorylated by EIIA and EIIB domains of the PTS (17, 18). BglA-2 hydrolyzes cellobiose 6′-phosphate (cellobiose-6′P) to yield G6P and glucose (Fig. 2B) that are further metabolized by the energy-generating glycolytic pathway (17, 19). Based on the sequence similarity, BglA-2 is assigned to glycoside hydrolase family 1 (GH-1), which includes a variety of glycoside hydrolases, such as 6-phospho-β-glucosidase (EC 3.2.1.86), 6-phospho-β-galactosidase (EC 3.2.1.85), and β-glucosidase (EC 3.2.1.21) (20). Members of the GH-1 family share a common catalytic mechanism and exhibit similar structural folds, including a (β/α)8 TIM-barrel. As described in Koshland's double displacement mechanisms (21, 22), two conserved acidic residues (glutamate or aspartate) are catalytic residues. One of these amino acids functions as a proton donor, and the other functions as a nucleophile. First the proton donor provides a proton to the substrate to protonate the glycosidic oxygen, with the attendant formation of the transient oxocarbenium state. Then the nucleophile residue attacks the protonated glycosidic bond and forms a glycosyl-enzyme intermediate. Finally, a water molecule provides a proton to break the glycosyl-enzyme intermediate, thus restoring the enzyme to its original protonated state.
FIGURE 1.
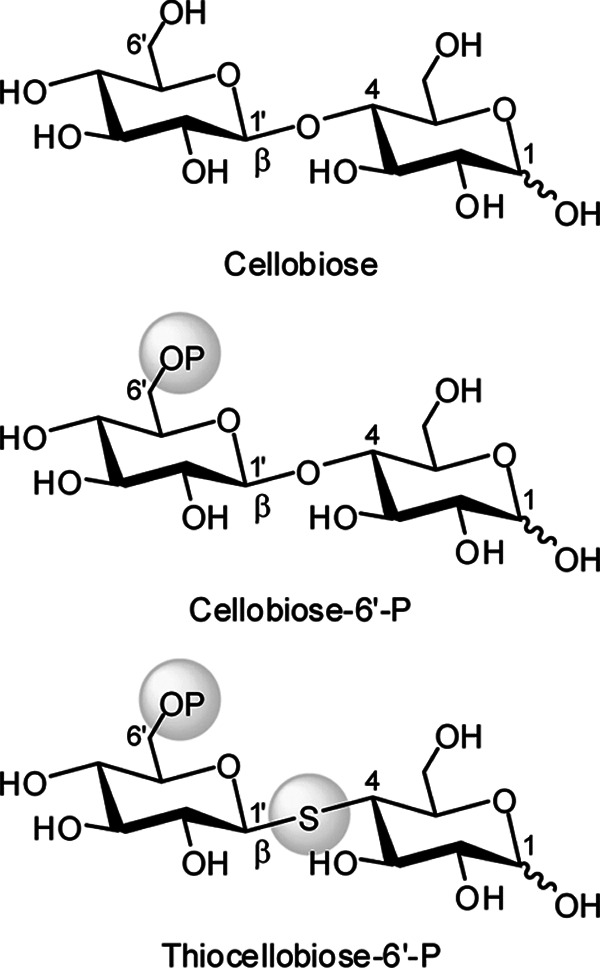
Structural formulas of cellobiose and its derivatives.
FIGURE 2.
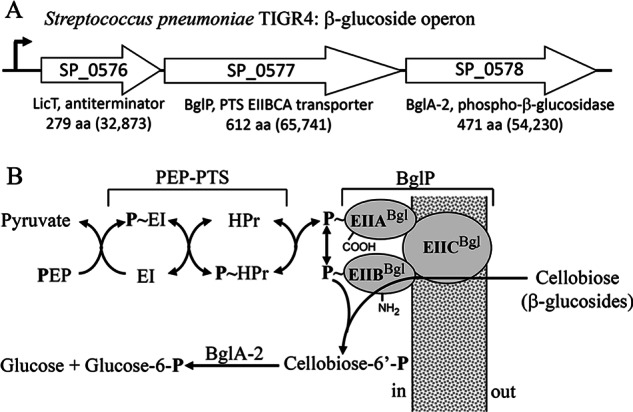
A, genetic organization of the β-glucoside operon in S. pneumoniae TIGR4. SP numbers represent the order of gene loci. Values in parentheses are calculated molecular weights of encoded proteins. B, translocation and phosphorylation of cellobiose by the multicomponent β-glucoside PEP-PTS and BglA-2-catalyzed hydrolysis of cellobiose-6′P.
Several structures of GH-1 members are now available (23–26), but (to our knowledge) there are no reports of the co-crystallization of an intact substrate at the active site of any GH-1 phospho-β-glucosidase. Significantly, the mechanism and determinants of enzyme specificity toward 1,4-linked 6-phospho-β-glucosides remain unclear. In this work, we present the first crystal structures of BglA-2 in both the apo form and in complex with thiocellobiose-6′P at 2.0 and 2.4 Å, respectively. Structural analysis, in combination with the enzymatic data obtained from site-directed mutants, has enabled us to define the structural elements that contribute to enzyme recognition of 1,4-linked 6-phospho-β-glucosides. Importantly, we provide evidence that three key residues, Ser424, Lys430, and Tyr432 of BglA-2, play functional roles in enzyme discrimination between the hydrolysis of phosphorylated and the non-phosphorylated substrates. Finally, results obtained via site-directed mutagenesis show that a tryptophan residue plays an important role in substrate discrimination between 6-phospho-β- galactosidases and 6-phospho-β-glucosidases in the GH-1 family.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of BglA-2 and Its Mutants
The coding sequence of the bglA-2 gene was amplified from the genomic DNA of S. pneumoniae TIGR4. The bglA-2 gene and its mutants were respectively cloned into the pET28a (Novagen) expression vector with an N-terminal His6 tag. Both the wild-type and mutant proteins were overexpressed in Escherichia coli strain BL21(DE3) (Novagen) using 2× YT culture medium (5 g of NaCl, 16 g of Bacto-Tryptone, and 10 g of yeast extract/liter). The transformed cells were grown at 37 °C in 2× YT medium containing 30 μg/ml kanamycin until the A600 nm reached 0.6–0.8. Expression of the recombinant proteins was then induced by the addition of 0.2 mm isopropyl β-d-1-thiogalactopyranoside for 20 h at 16 °C. The cells were collected by centrifugation at 8,000 × g for 10 min and resuspended in 45 ml of lysis buffer (20 mm Tris-Cl, pH 8.0, 100 mm NaCl). After 6 min of sonication and 30 min of centrifugation at 12,000 × g, the supernatant containing the target protein was collected and loaded onto a nickel-nitrilotriacetic acid column (GE Healthcare) equilibrated with the binding buffer (20 mm Tris-Cl, pH 8.0, 100 mm NaCl). The column was washed with binding buffer, and the target protein was then eluted with the same buffer containing 300 mm imidazole. The target protein was then loaded onto a Superdex 200 column (GE Healthcare), and fractions containing the target protein were collected and pooled. The purified protein was concentrated to 10 mg/ml by ultrafiltration (Amicon, Millipore Corp.) for crystallization trials. Samples for enzymatic activity assays were collected at the highest peak fractions without concentration. The purity of protein was assessed by SDS-PAGE, and the protein sample was stored at −80 °C. Site-directed mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) with the plasmid encoding the wild-type BglA-2 serving as template. The mutant proteins were expressed, purified, and stored as described above.
Crystallization, Data Collection, and Processing
Both the apo and complex forms of BglA-2 were concentrated to 10 mg/ml by ultrafiltration for crystallization. All crystals were grown at 16 °C using the sitting drop vapor diffusion technique. Each drop contained 1 μl of protein sample (10 mg/ml protein in buffer containing 20 mm Tris-Cl, pH 8.0, 100 mm NaCl) with an equal volume of the reservoir solution (15% polyethylene glycol 5000MME, 0.1 m sodium citrate tribasic dehydrate, pH 5.6). The crystals were transferred to cryoprotectant (reservoir solution supplemented with 25% glycerol) and flash-cooled with liquid nitrogen. All diffraction data were collected at 100 K in a liquid nitrogen stream at the Shanghai Synchrotron Radiation Facility. The data were integrated with the program Mosflm (27) and scaled with the program Scala in CCP4i (28).
Structure Determination and Refinement
The structure of apo form BglA-2 was solved by molecular replacement with MOLREP using the coordinates of 50% sequence-identical E. coli BglA (Protein Data Bank code 2XHY) as the search model. The complex form of BglA-2 (with thiocellobiose-6′P) was solved by molecular replacement using the apo form BglA-2 as the search model. The initial model was further refined by using the maximum likelihood method implemented in REFMAC5 (29) as part of the CCP4i (28) program suite and rebuilt interactively by using the σA-weighted electron density maps with coefficients 2Fo − Fc and Fo − Fc in the program COOT (30). The final model was evaluated with the programs MOLPROBITY (31) and PROCHECK (32). The data collection and structure refinement statistics of apo form and complex form BglA-2 are listed in Table 1. All of the structure figures were prepared with the program PyMOL (33).
TABLE 1.
Crystal parameters, data collection, and structure refinement
| Apo form | Complex form | |
|---|---|---|
| Data collection | ||
| Space group | C2 | C2 |
| Unit cell | ||
| a, b, c (Å) | 183.28, 65.35, 126.69 | 184.03, 66.65, 133.35 |
| α, β, γ (degrees) | 90.00, 133.38, 90.00 | 90.00, 136.33, 90.00 |
| Resolution range (Å) | 42.9–2.0 | 46.0–2.4 |
| Unique reflections | 72,464 (10,190)a | 42,473 (5,887) |
| Completeness (%) | 98.8 (95.7) | 98.2 (94.6) |
| 〈I/σ(I)〉 | 8.8 (4.5) | 15.3 (6.5) |
| Rmergeb (%) | 8.3 (17.1) | 5.1 (11.2) |
| Average redundancy | 3.4 (3.2) | 3.5 (3.0) |
| Structure refinement | ||
| Resolution range (Å) | 31.7–2.0 | 46.0–2.4 |
| R factorc/R-freed (%) | 17.7/22.2 | 17.5/25.1 |
| No. of protein atoms | 7,504 | 7,522 |
| No. of water atoms | 579 | 202 |
| RMSDe bond lengths (Å) | 0.007 | 0.007 |
| RMSD bond angles (degrees) | 1.159 | 1.143 |
| Mean B factors (Å2) | 20.4 | 44.5 |
| Ramachandran plotf (residues, %) | ||
| Most favored (%) | 99.2 | 98.1 |
| Additional allowed (%) | 0.8 | 1.9 |
| Outliers (%) | 0 | 0 |
| Protein Data Bank entry | 4IPL | 4IPN |
a The values in parentheses refer to statistics in the highest bin.
b Rmerge = ΣhklΣi|Ii(hkl) − 〈I(hkl)〉|/ΣhklΣiIi(hkl), where Ii(hkl) is the intensity of an observation, and 〈I(hkl)〉 is the mean value for its unique reflection. Summations are over all reflections.
c R factor = Σh|Fo(h) − Fc(h)|/ΣhFo(h), where Fo and Fc are the observed and calculated structure factor amplitudes, respectively.
d R-free was calculated with 5% of the data excluded from the refinement.
e Root mean square deviation from ideal values.
f Categories were defined by Molprobity.
Enzymatic Activity Assays
The kinetic parameters of wild-type BglA-2 and its mutants were determined using chromogenic p-nitrophenyl-β-d-glucopyranoside 6-phosphate (pNPβGlc6P) as substrate (34). All assays were performed at 37 °C in a buffer containing 50 mm Na2HPO4, 50 mm NaH2PO4, pH 7.5, and reactions were initiated by the addition of BglA-2. Changes in absorption at 405 nm (formation of p-nitrophenol) were monitored continuously using a DU800 spectrophotometer (Beckman Coulter, Fullerton, CA). The reaction product p-nitrophenol was calculated from a standard curve of p-nitrophenol, as described by Prag et al. (35). Michaelis-Menten parameters (Vmax and Km) of BglA-2 were extracted from these data by nonlinear fitting to the Michaelis-Menten equation using the program Origin version 7.5.
Preparation of 1-Phenyl-3-methyl-5-pyrazolone (PMP) Derivatives
PMP derivation of saccharides was performed as described previously (36–38) with minor changes. Briefly, 10 μl of reaction mixture was mixed with 10 μl of 0.3 m aqueous NaOH and 10 μl of 0.5 m methanol solution of PMP. The total reaction mixture (30 μl) was maintained at 70 °C for 30 min, cooled to room temperature, and neutralized by the addition of 10 μl of 0.3 m HCl. The solution was further mixed with 100 μl of chloroform. After vigorous shaking and centrifugation, the organic phase was carefully removed to eliminate excess reagents. The extraction procedure was repeated three times. Finally, the aqueous phase containing derivatives was diluted with 40 μl of water prior to HPLC analysis.
HPLC Analysis
The assays with specific substrate were performed at 37 °C in a 10-μl system containing the buffer of 50 mm Na2HPO4, 50 mm NaH2PO4, pH 7.5, and the disaccharide substrate (e.g. cellobiose-6′P) at a range of concentrations. The reactions were initiated by the addition of the purified enzymes and were terminated by the addition of 10 μl of 0.3 m NaOH. After PMP derivation, the reaction product was centrifuged at 12,000 × g for 10 min, and 15 μl of supernatant was analyzed by an HPLC system (Agilent 1200 series). Glucose and G6P standards were quantified by HPLC analysis using various concentrations ranging from 0.1 to 1 mm. The mixing buffer containing 20% acetonitrile and 100 mm Na2HPO4/NaH2PO4, pH 7.0 was processed as described previously (38) for equilibration of the column (Eclipse XDB-C18 column, 4.6 × 150 mm; Agilent), and separation of the components was effected at a flow rate of 1 ml/min. Retention times of monosaccharides were determined by comparison with standard solutions. Kinetic parameters were derived from three independent experiments in order to calculate the means and S.D. for the Km and kcat values.
Preparation of Cellobiose-6′P, Thiocellobiose-6′P, and pNPβGlc6P
Cellobiose was obtained from Pfanstiehl Laboratories, and thiocellobiose was purchased from Toronto Research Chemicals. Phosphorylation of the primary hydroxyl groups of the non-reducing glucose moiety in these O-β-linked disaccharides was as described previously (39). In brief, phosphorylation was effected by incubation of the disaccharides with ATP-dependent β-glucoside kinase (BglK, EC 2.7.1.85) from Klebsiella pneumoniae. Phosphorylated derivatives were first isolated by Ba2+ and ethanol precipitation and further purified by ion exchange and paper chromatography. Structures and product purity were confirmed by thin layer chromatography, mass spectrometry, and NMR spectroscopy. Chromogenic pNPβGlc6P was prepared by phosphorylation of the C6 hydroxyl moiety of pNP-β-d-glucopyranoside with phosphorus oxychloride in trimethyl phosphate containing a small amount of water (34). Lactose-6′-phosphate (lactose-6′P) is not commercially available, and the chromogenic analog o-nitrophenyl-β-d-galactopyranoside 6-phosphate (oNPβGal6P; Sigma-Aldrich) was used as a substitute substrate for kinetics analyses.
RESULTS AND DISCUSSION
Overall Structure
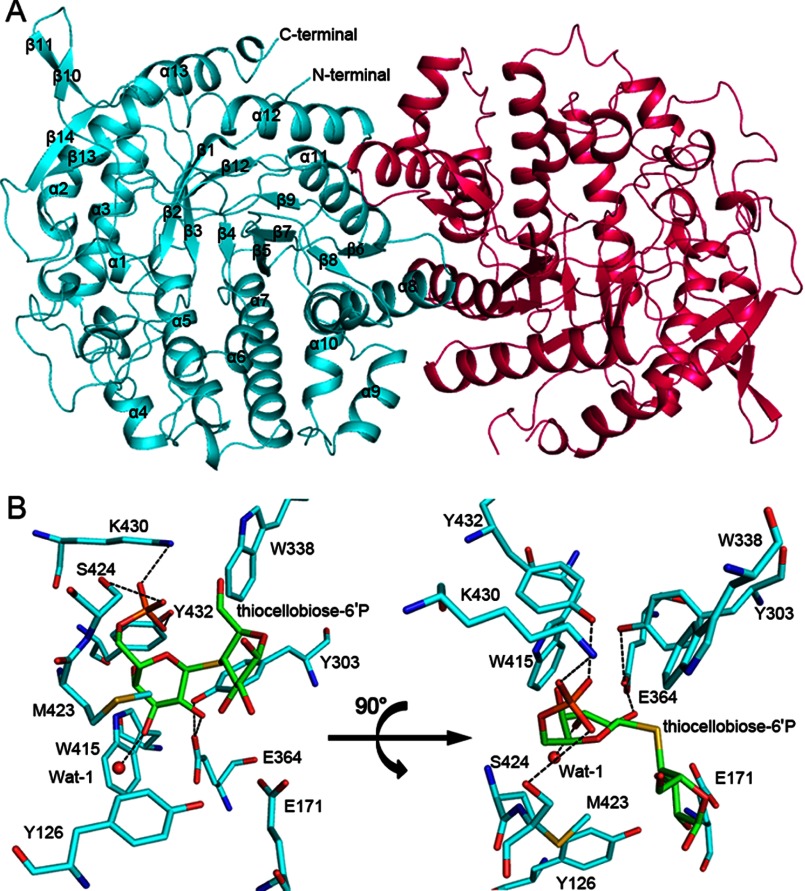
The crystal structure of apo form BglA-2 was determined at 2.0 Å resolution in the space group C2. Each asymmetric unit contains two molecules, which form a stable dimer with a buried interface area of ∼3,300 Å2 (Fig. 3A). The dimerization in the crystal structure is consistent with the results obtained from size exclusion chromatography (supplemental Fig. S1). The dimer interface, composed of four α-helices (α8, α9, α11, and α12) and three loops from one subunit, is stabilized primarily by a number of polar residues via hydrogen bond networks and salt bridge interactions. Similar to other reported GH-1 members, BglA-2 adopts a typical (β/α)8 TIM-barrel: a central eight-stranded (β1–β5, β7, β9, and β12) parallel β-sheet surrounded by eight helices (α2, α3, α5, α7, α8, and α11–13). In addition, the central TIM-barrel is packed by four additional helices (α1, α6, α9, and α10) and six β-strands (β6, β8, β10, β11, β13, and β14). Residues Asn316–Gly324 in both the apo form and complex form are rather flexible and are absent in the final model due to the poor electron density. A molecule of thiocellobiose-6′P is bound at the center of the TIM-barrel in the complex structure. Structural comparison of the apo and complex structures revealed similar conformations, with a root mean square deviation of 0.3 Å over 436 Cα atoms.
FIGURE 3.
Overall structure and the active site of BglA-2. A, schematic representation of the overall structure of the dimeric BglA-2 (cyan, subunit A; red, subunit B). The secondary structural elements are labeled. B, the active site of BglA-2 from two angles. The active site residues and the thiocellobiose-6′P are shown as cyan and green sticks for the Cα atoms, respectively. A water molecule at the active site is shown as a red sphere. Polar interactions are indicated by dashed lines.
To date, the crystal structures of four 6-phospho-β-glycosidases from the GH-1 family have been solved, including three 6-phospho-β-glucosidases (E. coli BglA, S. mutans putative 6-phospho-β-glucosidase Bgl, and Lactobacillus plantarum Pbg1) and one 6-phospho-β-galactosidase (PGALase) from Lactococcus lactis. These proteins assume a structure similar to that of BglA-2 with an overall root mean square deviation of 2.0, 1.2, 1.3, and 1.4 Å, respectively. The main differences reside in the architecture surrounding the active site pockets. In the complex structure of PGALase with galactose 6-phosphate (galactose-6′P), three antiparallel β-strands and an 11-residue loop (Gln305–Arg334) cover the catalytic pocket, whereas in BglA-2, the corresponding region (Asn316–Gly324) is absent from the structure. In BglA, the loop (Asp38–Pro62) caps the entrance of the substrate tunnel, whereas in BglA-2, the corresponding loop (Leu36–Leu52) is considerably shorter. Despite a similar catalytic mechanism, the variable loops around the active sites may reflect the substrate specificity of each enzyme, which is in agreement with the variety of substrates hydrolyzed by GH-1 members.
The Active Site
As is evident from the BglA-2 complex structure, thiocellobiose-6′P is stabilized at the active site pocket via hydrophilic and hydrophobic interactions (Fig. 3B). In detail, the phosphate group that projects toward the base of the pocket is hydrogen-bonded by the side chains of Ser424, Lys430, and Tyr432. In subsite −1 G6P is anchored by hydrophobic interaction with Trp415 and is also hydrogen-bonded to one of the catalytic residues (Glu364) and a water molecule, Wat-1. The side chain of the catalytic residue Glu171 resides ∼6 Å from the glycosidic bond. Subsite +1 is occupied by glucose and includes three aromatic residues: Tyr126, Tyr303, and Trp338. In addition, Tyr303 makes a hydrogen bond with the carboxyl group of Glu364, presumably to orient and maintain Glu364 in a conformation favorable for catalysis.
When compared with the representative complex structure of PGALase with galactose-6′P in the GH-1 family, many of the active site residues of BglA-2 can be superimposed, especially the two catalytic residues Glu364 and Glu171 (corresponding to Glu375 and Glu160 of PGALase and Glu375 and Glu176 of Bgl) (23). Notably, the distances between the side chains of PGALase Glu160 and Bgl Glu176 and the glycosidic bond are 3.4 and 2.8 Å, respectively, which are favorable for catalysis. Structural comparison of BglA-2 with PGALase and Bgl reveals that the thiocellobiose-6′P adopts a similar position but different conformation. Thus, our structure might be a complex for the interactions of thiocellobiose-6′P with BglA-2 that is not poised for catalysis.
These observations suggest that BglA-2 adopts a catalytic mechanism similar to those of previously documented GH-1 members (21, 22). The fact that mutant proteins E364A, E364Q, E171A, and E171Q lose all catalytic activity toward pNPβGlc6P and cellobiose-6′P confirms the crucial roles of Glu364 and Glu171 in BglA-2 catalysis (Table 2).
TABLE 2.
Kinetic constants of BglA-2 and mutants toward pNPβGlc6P and cellobiose-6′P
| Enzyme | pNPβGlc6P |
Cellobiose-6′P |
||||
|---|---|---|---|---|---|---|
| Km | kcat | kcat/Km ×10−1 | Km | kcat | kcat/Km ×10−1 | |
| μm | s−1 | s−1μm−1 | μm | s−1 | s−1μm−1 | |
| Wild type | 478 ± 4.5 | 195 ± 5.3 | 4.1 ± 0.3 | 1135 ± 9.8 | 170 ± 3.8 | 1.5 ± 0.2 |
| E171A | NDa | ND | ND | ND | ND | ND |
| E171Q | ND | ND | ND | ND | ND | ND |
| E364A | ND | ND | ND | ND | ND | ND |
| E364Q | ND | ND | ND | ND | ND | ND |
| Y126A | ND | ND | ND | ND | ND | ND |
| Y126F | 598 ± 3.8 | 146 ± 6.4 | 2.4 ± 0.2 | 1515 ± 11.8 | 138 ± 7.8 | 0.9 ± 0.1 |
| Y303A | ND | ND | ND | ND | ND | ND |
| Y303F | ND | ND | ND | ND | ND | ND |
| W338A | ND | ND | ND | ND | ND | ND |
| M423A | 512 ± 4.8 | 168 ± 8.3 | 3.3 ± 0.4 | 1320 ± 10.5 | 145 ± 8.9 | 1.1 ± 0.3 |
| S424A | ND | ND | ND | ND | ND | ND |
| K430A | ND | ND | ND | ND | ND | ND |
| Y432F | ND | ND | ND | ND | ND | ND |
a ND, no detectable activity.
Key Residues Contributing to Specificity at Subsite +1
GH-1 enzymes with known structures are reported to specifically catalyze the hydrolysis of 1,4-linked non-phosphorylated β-glycosides or 1,4-linked 6-phospho-β-glycosides. β-Glucosidase A from Bacillus polymyxa reportedly hydrolyzes β-1,4-linked oligosaccharides composed of more than 2 units of glucose (25), and human cytosolic β-glucosidase hydrolyzes certain flavonoid glucosides (26). The enzyme PGALase from L. lactis was reported to hydrolyze lactose-6′P, formed during transport and phosphorylation via the lactose PEP-PTS (23). The model of lactose-6′P bound to PGALase predicted that Trp347 at the channel entrance and Tyr299 and Trp421 at the substrate cavity were involved in guiding and binding the substrate by hydrophobic interactions (24). Compared with BglA-2, Trp421 of PGALase corresponds to subsite −1 Trp415, whereas Tyr299 and Trp347 of PGALase are aligned with subsite +1 Tyr303 and Trp338, respectively. Although the structures of β-glucosidase A, cytosolic β-glucosidase, and PGALase are known, a lack of enzymatic assays and 6-phospho-β- glucoside complexed structure leaves the substrate specificity of 6-phospho-β-glucosidases still ambiguous. To address the structural basis of the substrate specificity toward 1,4-linked 6-phospho-β-glucosides, concerted efforts were made to obtain crystals of BglA-2 in complex with cellobiose-6′P, pNPβGlc6P, glucose, G6P, and thiocellobiose-6′P. Fortuitously, phospho-β-glucosidases in family GH-1 are unable to hydrolyze this phosphorylated cellobiose analog. (By contrast, phospho-β-glucosidases assigned to family GH-4 readily cleave thiocellobiose-6′P by a catalytically unique series of oxidation-elimination-addition and reduction reactions (40, 41).) After numerous attempts, the structure of BglA-2 in complex with thiocellobiose-6′P was successfully solved. Inspection of the complex shows that the hexose ring at the subsite +1 moiety is orientated almost perpendicular to that of the G6P in subsite −1. The glucose moiety at subsite +1 is sandwiched by residues Tyr303 and Trp338 on one side and on the opposite side by Tyr126 (Fig. 3B). Among the three subsite +1 residues, Tyr126 is newly identified in our structure and corresponds to Phe137 in PGALase. Multiple-sequence alignment among 6-phospho-β-glycosidases in the GH-1 family shows that Tyr126, Tyr303, and Trp338 are generally conserved in bacteria, besides the only protozoa, Leishmania infantum (Fig. 4). To clarify the roles of Tyr126, Tyr303, and Trp338 in BglA-2 catalysis, we first determined the equilibrium dissociation constants (Kd) of both wild-type and mutant proteins of BglA-2 (Y126A, Y303A, Y303F, and W338A) toward cellobiose-6′P by fluorescence spectrometry. The Kd values of mutants increased ∼6–7-fold compared with the wild type (supplemental Table S1), suggesting that the mutants had much lower cellobiose-6′P binding affinities compared with the wild type. Subsequently, we compared the enzymatic activities of these mutants with that of the wild type. The mutant Y126A was devoid of all activity, whereas Y126F retained ∼59% of that of the wild type, indicative of the important role of Tyr126 in this hydrophobic pocket. Neither the mutant Y303A nor Y303F showed any enzymatic activity (Table 2). These results show that Tyr303 is essential for activity, not only by contributing to the hydrophobicity of the pocket but also by stabilizing the orientation of the catalytically functional carboxyl group of Glu364. This result is in accordance with the complex structure, in which Tyr303 makes a hydrogen bond with the carboxyl group of Glu364 (Fig. 3B). The mutant W338A was also completely inactive.
FIGURE 4.
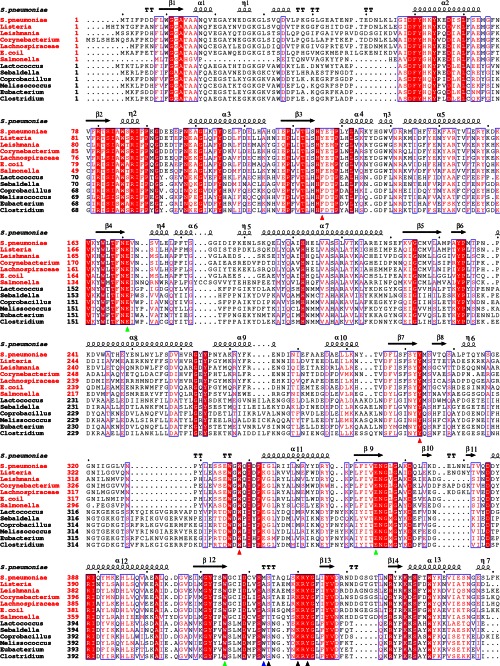
Multiple-sequence alignment of BglA-2 and homologs. The 6-phospho-β-glucosidases and 6-phospho-β-galactosidases are labeled with red and black titles, respectively. Two conserved catalytic glutamate residues and the subsite −1 residue Trp415 are depicted by green triangles. The subsite +1 residues and the phosphate-binding residues are marked with red and black triangles, respectively. The tryptophan residue discriminating 6-phospho-β-galactosidase from 6-phospho-β-glucosidase in GH-1 family is indicated by a blue triangle.
To further study the subsite +1 specificity, we investigated the activity of the wild type and mutants of BglA-2 toward two stereoisomers of cellobiose-6′P, namely gentiobiose-6′P (β-1,6-linked glucose 6-phosphate and glucose) and maltose-6′P (α-1,4-linked glucose 6-phosphate and glucose). Neither wild-type nor mutant proteins hydrolyzed gentiobiose-6′P or maltose-6′P (supplemental Fig. S2). These results established the specificity of BglA-2 toward cellobiose-6′P. Furthermore, it is evident that the active site residues Tyr126, Tyr303, and Trp338 play important roles not only in substrate binding but also in recognition of the unique spatial orientation of glucose (in cellobiose-6′P) that permits this moiety to occupy subsite +1 of BglA-2.
Key Residues That Dictate Specificity toward the Phosphate Group of Cellobiose-6′P
As described previously, the phosphate-binding residues Ser428, Lys435, and Tyr437 in PGALase discriminate the phosphorylated from the non-phosphorylated sugar (24). In BglA-2, three residues (Ser424, Lys430, and Tyr432) make hydrogen bonds with the phosphate group of thiocellobiose-6′P (Fig. 3B). Comparison of the complex structures of BglA-2 and PGALase shows that residues Lys430 and Tyr432 of BglA-2 adopt a position similar to Lys435 and Tyr437 of PGALase, respectively. However, Ser424 of BglA-2 corresponds to Ser430 rather than Ser428 of PGALase. Sequence analysis suggested that Lys430 and Tyr432 of BglA-2 are conserved in GH-1 6-phospho-β-glycosidases (Fig. 4). However, they are substituted by non-polar residues in other GH-1 glycosidases hydrolyzing non-phosphorylated substrates, such as β-glucosidase A from B. polymyxa (25). Indeed, BglA-2 shows no hydrolytic activity toward non-phosphorylated cellobiose. To verify the roles of the three residues, we measured the enzymatic activities toward cellobiose-6′P through use of several mutants. The results showed that mutations S424A, K430A, and Y432F abolished all activity, thereby confirming the key roles of Ser424, Lys430, and Tyr432 of BglA-2 in determining specificity toward phosphorylated substrate (Table 2).
Sequence alignment of 6-phospho-β- glycosidases in the GH-1 family indicates that two catalytic residues (Glu364 and Glu171); the subsite +1 residues Tyr126, Tyr303, and Trp338; and the phosphate-binding residues Lys430 and Tyr432 are exclusively conserved. The observations suggest that these homologs might adopt a similar overall structure and hydrolyze phosphorylated substrates (Fig. 4). Our structural analyses provide new insight into the substrate binding patterns and determinants of specificity of GH-1 family phospho-β-glycosidases.
A Tryptophan Residue Discriminates 6-Phospho-β-galactosidase from 6-Phospho-β-glucosidase in GH-1 Family
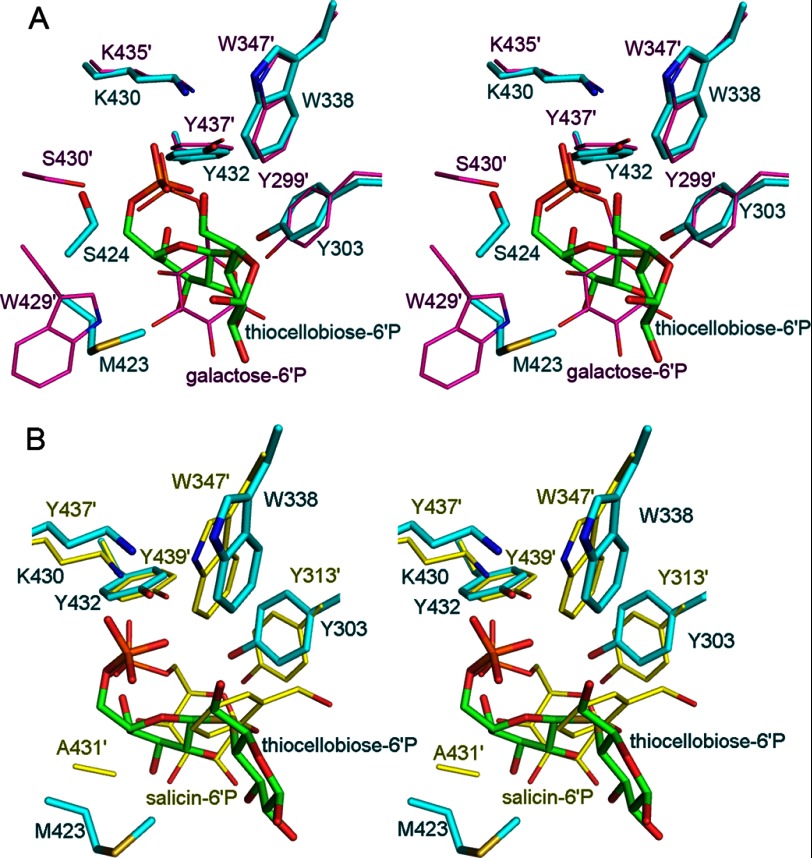
In GH-1 family, two types of enzymes can hydrolyze phosphorylated substrates. 6-Phospho-β-glucosidase and 6-phospho-β-galactosidase are distinguished by their subsite −1 sugars, with G6P for 6-phospho-β-glucosidase and galactose-6′P for 6-phospho-β-galactosidase. Whereas BglA-2 hydrolyzes cellobiose-6′P, PGALase hydrolyzes lactose-6′P. The major difference between cellobiose-6′P and lactose-6′P resides in the orientation of the C4 hydroxyl group of the hexose-6P compounds at subsite −1. In the case of cellobiose-6′P, the equatorial C4 hydroxyl lies on the opposite side of the C1 hydroxyl, whereas in lactose-6′P, the axial C4 hydroxyl moiety lies on the same side as the C1 hydroxyl group. In BglA-2, the subsite −1 sugar (G6P) shifts 2.4 Å apart and rotates about 67° from that of Gal6P in PGALase. This difference between the substrates is in accordance with the active site conformations. In PGALase, the C4 hydroxyl moiety is hydrogen-bonded by Trp429 (24), which is substituted by Met423 in BglA-2 or Ala431 in Bgl (Fig. 5, A and B). However, Met423 in BglA-2 and Ala431 in Bgl have no interaction with the subsite −1 sugar. Indeed, no residue was found to stabilize the C4 hydroxyl group of BglA-2 and Bgl. The M423A and M423W mutants of BglA-2 retain about 80 and 67% of the activity of the wild-type enzyme respectively, suggesting a non-essential role for Met423 in catalysis (Table 2 and supplemental Table S2). In further studies, the activities of the wild-type BglA-2 and M423W mutant were determined using oNPβGal6P as a chromogenic substitute for lactose-6′P. As expected, the wild-type BglA-2 has no detectable activity toward oNPβGal6P. Remarkably, the single mutation M423W elicited hydrolytic activity toward oNPβGal6P with a kcat/Km value of 8.6 ± 2.1 × 10−5 s−1 μm−1 (supplemental Table S2). These results show that a tryptophan residue of PGALase determines that subsite −1 of this 6-phospho-β-galactosidase is occupied by galactose-6′P.
FIGURE 5.
Stereo representation of active site comparison of BglA-2 (cyan) with L. lactis PGALase (pink; Protein Data Bank code 4PBG) (A) and S. mutans Bgl (yellow; Protein Data Bank code 4F79) (B).
Sequence analysis revealed an ∼50% sequence homology between these two types of enzymes, suggesting a similar structural fold of a (β/α)8 TIM-barrel and a common catalytic mechanism (Fig. 4). However, Trp429 of PGALase is exclusively conserved in 6-phospho-β-galactosidases but is usually alanine in 6-phospho-β-glucosidases (Fig. 4). We hypothesize that both enzyme species evolved from a common ancestor, but at some point 6-phospho-β-galactosidases evolved independently, such that a tryptophan residue was acquired in order to accommodate galactose-6′P (rather than G6P) at the active site of subsite −1.
6-Phospho-β-glycosidases Provide Alternative (but Non-essential) Pathways for the Utilization of β-Linked Disaccharides
Multiple-sequence alignment among 6-phospho-β-glycosidases in GH-1 family shows that they are generally conserved in bacteria, with only the protozoa L. infantum as the exception (Fig. 4). Abundant bacterial growth is dependent upon the uptake of various carbohydrates from the environment. As noted previously, there are several pathways for the uptake of carbohydrates in bacterial species, including PEP-PTS transporters (7–11), cation/proton-coupled transporters (12, 13), and ATP-binding cassette transporters (14–16). Among these, the group translocation PEP-PTS system is perhaps the most ubiquitous. Many species possess operons that encode genes for a glycosidase and the appropriate PEP-PTS (4). The PEP-PTS system enables organisms such as S. pneumoniae to obtain their metabolic energy via the simultaneous transport and phosphorylation of disaccharides. Intracellular 6-phospho-β-glycosidases hydrolyze these phosphorylated compounds to metabolizable monosaccharides that furnish the requisite energy for growth of microorganisms. With the exception of L. infantum, 6-phospho-β-glycosidases exist only in bacteria, and it is likely that BglA-2 and other 6-phospho-β-glycosidases in the GH-1 family have evolved in bacteria simultaneously with the PEP-PTS to permit the dissimilation of environmental disaccharides. The exceptional case of L. infantum may be the result of gene transfer from bacteria, as reported previously (42).
Inspection of the 2.16-megabase genome of S. pneumoniae TIGR4 reveals three PEP-PTS systems (SP_0303-10, SP_0577, and SP_2021-4), and a sugar efflux transporter that may participate in the utilization and metabolism of cellobiose by this organism (4, 5, 43). For the PEP-PTS systems, cellobiose should be phosphorylated to cellobiose-6′P prior to hydrolysis by 6-phospho-β-glucosidases, such as BglA-2 of the SP_0577 PTS operon and SP_0303 of the SP_0303–10 PTS operon. Alternatively, cellobiose may be transported into the cell via a sugar efflux transporter and hydrolyzed intracellularly by putative cellobiase(s) encoded by SP_2021 and SP_0265. As reported previously (4), growth of S. pneumoniae on cellobiose is essentially abolished upon deletion of the transporter gene SP_0310 of the SP_0305–10 PTS, suggesting that this PEP-PTS may be essential for the uptake of cellobiose. By contrast, the ΔSP_0577 strain shows a doubled generation time (195 min) and a prolonged lag period compared with the wild-type strain on methyl-β-glucoside, suggesting that SP_0577 PTS is responsible for the uptake of this and related β-glucosides (4, 18).
Each of the three PEP-PTS systems has a cellobiose/cellobiose-6′P hydrolase that is encoded by the SP_0303, BglA-2, and SP_2021 gene, respectively. Structure-based sequence alignment confirms that SP_0303 and BglA-2 are 6-phospho-β-glucosidases, whereas SP_2021 is most likely a cellobiase. To test whether SP_0303 and/or BglA-2 are essential for the utilization of cellobiose, we constructed ΔBglA-2 and ΔBglA-2&ΔSP_0303 strains and recorded the growth profiles. The results showed no significant difference of growth rate on cellobiose between the wild-type and deletion strains (supplemental Fig. S3). Although a functional 6-phospho-β-glucosidase may not be essential for the utilization of cellobiose, 6-phospho-β-glycosidases encoded by PEP-PTS systems provide S. pneumoniae with alternate pathways for dissimilation of this carbohydrate and other β-linked disaccharides.
In summary, we have solved the crystal structure of BglA-2 in complex with the non-metabolizable analog, thiocellobiose-6′P. Based on structural analysis and enzymatic assays, we have clarified issues pertaining to the architectural environments of subsites −1 and +1 and have identified residues that interact with the phosphate moiety of G6P at subsite −1 of 6-phospho-β-glucosidase. Importantly, we present evidence that a tryptophan residue plays a functional role in the differentiation of the catalytic properties of 6-phospho-β-galactosidases and those of 6-phospho-β-glucosidases assigned to GH-1 of the glycoside hydrolase family.
Supplementary Material
Acknowledgments
We thank the staff at the Shanghai Synchrotron Radiation Facility for expert technical assistance. We also appreciate the efforts of Kang Zhou with data collection and thank the developers of the CCP4 Suite, ESPript, TreeView, and PyMOL programs. We thank Drs. Josef Deutscher and Stefan Immel for assistance in compiling Figs. 1 and 2.
This work was supported, in whole or in part, by the National Institutes of Health, NIDCR, Intramural Research Program. This work was also supported by Ministry of Science and Technology of China Grant 2009CB918800, National Natural Science Foundation of China Grant 31270781, and Research Fund for the Doctoral Program of Higher Education of China Grant 20123402110004.
The atomic coordinates and structure factors (codes 4IPL and 4IPN) have been deposited in the Protein Data Bank (http://wwpdb.org/).

This article contains supplemental Tables S1 and S2 and Figs. S1–S3.
- PEP-PTS
- phosphoenolpyruvate-dependent phosphotransferase system
- PTS
- phosphotransferase system
- cellobiose-6′P
- β-1,4-linked cellobiose 6-phosphate
- G6P
- glucose 6-phosphate
- GH-1
- glycoside hydrolase family 1
- pNPβGlc6P
- p-nitrophenyl-β-d-glucopyranoside 6-phosphate
- PMP
- 1-phenyl-3-methyl-5-pyrazolone
- thiocellobiose-6′P
- thiocellobiose 6-phosphate
- lactose-6′P
- lactose 6-phosphate
- galactose-6′P
- galactose 6-phosphate
- oNPβGal6P
- o-nitrophenyl-β-d-galactopyranoside 6-phosphate
- PGALase
- 6-phospho-β-galactosidase
- gentiobiose 6′P
- β-1,6-linked glucose 6-phosphate and glucose
- maltose-6′P
- α-1,4-linked glucose 6-phosphate and glucose.
REFERENCES
- 1. Kadioglu A., Weiser J. N., Paton J. C., Andrew P. W. (2008) The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat. Rev. Microbiol. 6, 288–301 [DOI] [PubMed] [Google Scholar]
- 2. Cartwright K. (2002) Pneumococcal disease in western Europe. Burden of disease, antibiotic resistance and management. Eur. J. Pediatr. 161, 188–195 [DOI] [PubMed] [Google Scholar]
- 3. Steinfort C., Wilson R., Mitchell T. (1989) Effects of Streptococcus pneumoniae on human respiratory epithelium in vitro. Infect. Immun. 57, 2006–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bidossi A., Mulas L., Decorosi F., Colomba L., Ricci S., Pozzi G., Deutscher J., Viti C., Oggioni M. R. (2012) A functional genomics approach to establish the complement of carbohydrate transporters in Streptococcus pneumoniae. PloS One 7, e33320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Saier M. H., Jr. (2000) Families of transmembrane sugar transport proteins. Mol. Microbiol. 35, 699–710 [DOI] [PubMed] [Google Scholar]
- 6. Ajdić D., Pham V. T. (2007) Global Transcriptional analysis of Streptococcus mutans sugar transporters using microarrays. J. Bacteriol. 189, 5049–5059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kundig W., Ghosh S., Roseman S. (1964) Phosphate bound to histidine in a protein as an intermediate in a novel phosphotransferase system. Proc. Natl. Acad. Sci. U.S.A. 52, 1067–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Reizer J., Saier M. H., Jr., Deutscher J., Grenier F., Thompson J., Hengstenberg W. (1988) The phosphoenolpyruvate:sugar phosphotransferase system in gram-positive bacteria. Properties, mechanism, and regulation. Crit. Rev. Microbiol. 15, 297–338 [DOI] [PubMed] [Google Scholar]
- 9. Roseman S. (1989) Sialic acid, serendipity, and sugar transport. Discovery of the bacterial phosphotransferase system. FEMS Microbiol. Rev. 5, 3–11 [DOI] [PubMed] [Google Scholar]
- 10. Meadow N. D., Fox D. K., Roseman S. (1990) The bacterial phosphoenol-pyruvate. Glycose phosphotransferase system. Annu. Rev. Biochem. 59, 497–542 [DOI] [PubMed] [Google Scholar]
- 11. Postma P. W., Lengeler J. W., Jacobson G. R. (1993) Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57, 543–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Henderson P. J., Baldwin S. A., Cairns M. T., Charalambous B. M., Dent H. C., Gunn F., Liang W. J., Lucas V. A., Martin G. E., McDonald T. P. (1992) Sugar-cation symport systems in bacteria. Int. Rev. Cytol. 137, 149–208 [PubMed] [Google Scholar]
- 13. Poolman B., Knol J., van der Does C., Henderson P. J., Liang W. J., Leblanc G., Pourcher T., Mus-Veteau I. (1996) Cation and sugar selectivity determinants in a novel family of transport proteins. Mol. Microbiol. 19, 911–922 [DOI] [PubMed] [Google Scholar]
- 14. Davidson A. L. (2002) Mechanism of coupling of transport to hydrolysis in bacterial ATP-binding cassette transporters. J. Bacteriol. 184, 1225–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Davidson A. L., Chen J. (2004) ATP-binding cassette transporters in bacteria. Annu. Rev. Biochem. 73, 241–268 [DOI] [PubMed] [Google Scholar]
- 16. Patzlaff J. S., van der Heide T., Poolman B. (2003) The ATP/substrate stoichiometry of the ATP-binding cassette (ABC) transporter OpuA. J. Biol. Chem. 278, 29546–29551 [DOI] [PubMed] [Google Scholar]
- 17. Thompson J., Pikis A., Ruvinov S. B., Henrissat B., Yamamoto H., Sekiguchi J. (1998) The gene glvA of Bacillus subtilis 168 encodes a metal-requiring, NAD(H)-dependent 6-phospho-α-glucosidase. Assignment to family 4 of the glycosylhydrolase superfamily. J. Biol. Chem. 273, 27347–27356 [DOI] [PubMed] [Google Scholar]
- 18. Cote C. K., Cvitkovitch D., Bleiweis A. S., Honeyman A. L. (2000) A novel β-glucoside-specific PTS locus from Streptococcus mutans that is not inhibited by glucose. Microbiology 146, 1555–1563 [DOI] [PubMed] [Google Scholar]
- 19. Tettelin H., Nelson K. E., Paulsen I. T., Eisen J. A., Read T. D., Peterson S., Heidelberg J., DeBoy R. T., Haft D. H., Dodson R. J., Durkin A. S., Gwinn M., Kolonay J. F., Nelson W. C., Peterson J. D., Umayam L. A., White O., Salzberg S. L., Lewis M. R., Radune D., Holtzapple E., Khouri H., Wolf A. M., Utterback T. R., Hansen C. L., McDonald L. A., Feldblyum T. V., Angiuoli S., Dickinson T., Hickey E. K., Holt I. E., Loftus B. J., Yang F., Smith H. O., Venter J. C., Dougherty B. A., Morrison D. A., Hollingshead S. K., Fraser C. M. (2001) Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293, 498–506 [DOI] [PubMed] [Google Scholar]
- 20. Henrissat B. (1991) A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280, 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Koshland D. E. (1953) Stereochemistry and the mechanism of enzymic reactions. Biol. Rev. Camb. Philos. Soc. 28, 416–436 [Google Scholar]
- 22. Henrissat B., Callebaut I., Fabrega S., Lehn P., Mornon J. P., Davies G. (1995) Conserved catalytic machinery and the prediction of a common fold for several families of glycosyl hydrolases. Proc. Natl. Acad. Sci. U.S.A. 92, 7090–7094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wiesmann C., Beste G., Hengstenberg W., Schulz G. E. (1995) The three-dimensional structure of 6-phospho-β-galactosidase from Lactococcus lactis. Structure 3, 961–968 [DOI] [PubMed] [Google Scholar]
- 24. Wiesmann C., Hengstenberg W., Schulz G. E. (1997) Crystal structures and mechanism of 6-phospho-β-galactosidase from Lactococcus lactis. J. Mol. Biol. 269, 851–860 [DOI] [PubMed] [Google Scholar]
- 25. Sanz-Aparicio J., Hermoso J. A., Martínez-Ripoli M., Lequerica J. L., Polaina J. (1998) Crystal structure of β-glucosidase A from Bacillus polymyxa. Insights into the catalytic activity in family 1 glycosyl hydrolases. J. Mol. Biol. 275, 491–502 [DOI] [PubMed] [Google Scholar]
- 26. Tribolo S., Berrin J. G., Kroon P. A., Czjzek M., Juge N. (2007) The crystal structure of human cytosolic β-glucosidase unravels the substrate aglycone specificity of a family 1 glycoside hydrolase. J. Mol. Biol. 370, 964–975 [DOI] [PubMed] [Google Scholar]
- 27. Leslie A. G. (1999) Integration of macromolecular diffraction data. Acta Crystallogr. D Biol. Crystallogr. 55, 1696–1702 [DOI] [PubMed] [Google Scholar]
- 28. Collaborative Computational Project, Number 4 (1994) The CCP4 suite. Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 29. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 30. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 31. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity. All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Laskowski R. A., Macarthur M. W., Moss D. S., Thornton J. M. (1993) Procheck. A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 33. DeLano W. (2010) The PyMol Molecular Graphics System, version 1.3r1, Schrodinger LLC, New York [Google Scholar]
- 34. Thompson J., Robrish S. A., Bouma C. L., Freedberg D. I., Folk J. E. (1997) Phospho-β-glucosidase from Fusobacterium mortiferum. Purification, cloning, and inactivation by 6-phosphoglucono-δ-lactone. J. Bacteriol. 179, 1636–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Prag G., Papanikolau Y., Tavlas G., Vorgias C. E., Petratos K., Oppenheim A. B. (2000) Structures of chitobiase mutants complexed with the substrate Di-N-acetyl-d-glucosamine. The catalytic role of the conserved acidic pair, aspartate 539 and glutamate 540. J. Mol. Biol. 300, 611–617 [DOI] [PubMed] [Google Scholar]
- 36. Yang X., Zhao Y., Wang Q., Wang H., Mei Q. (2005) Analysis of the monosaccharide components in Angelica polysaccharides by high performance liquid chromatography. Anal. Sci. 21, 1177–1180 [DOI] [PubMed] [Google Scholar]
- 37. Honda S., Akao E., Suzuki S., Okuda M., Kakehi K., Nakamura J. (1989) High-performance liquid-chromatography of reducing carbohydrates as strongly ultraviolet-absorbing and electrochemically sensitive 1-phenyl-3-methyl-5-pyrazolone derivatives. Anal. Biochem. 180, 351–357 [DOI] [PubMed] [Google Scholar]
- 38. Jiang Y. L., Yu W. L., Zhang J. W., Frolet C., Di Guilmi A. M., Zhou C. Z., Vernet T., Chen Y. (2011) Structural basis for the substrate specificity of a novel β-N-acetylhexosaminidase StrH protein from Streptococcus pneumoniae R6. J. Biol. Chem. 286, 43004–43012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Thompson J., Lichtenthaler F. W., Peters S., Pikis A. (2002) β-Glucoside kinase (BglK) from Klebsiella pneumoniae. Purification, properties, and preparative synthesis of 6-phospho-β-d-glucosides. J. Biol. Chem. 277, 34310–34321 [DOI] [PubMed] [Google Scholar]
- 40. Yip V. L., Varrot A., Davies G. J., Rajan S. S., Yang X., Thompson J., Anderson W. F., Withers S. G. (2004) An unusual mechanism of glycoside hydrolysis involving redox and elimination steps by a Family 4 β-glycosidase from Thermotoga maritima. J. Am. Chem. Soc. 126, 8354–8355 [DOI] [PubMed] [Google Scholar]
- 41. Yip V. L. Y., Withers S. G. (2006) Family 4 glycosidases carry out efficient hydrolysis of thioglycosides by an α,β-elimination mechanism. Angew. Chem. Int. Ed. 45, 1–5 [DOI] [PubMed] [Google Scholar]
- 42. Puri V., Goyal A., Sankaranarayanan R., Enright A. J., Vaidya T. (2011) Evolutionary and functional insights into Leishmania META1. Evidence for lateral gene transfer and a role for META1 in secretion. BMC Evol. Biol. 11, 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McKessar S. J, Hakenbeck R. (2007) The two-component regulatory system TCS08 is involved in cellobiose metabolism of Streptococcus pneumoniae R6. J. Bacteriol. 189, 1342–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.