Abstract
Dbf4-dependent kinase (DDK) and cyclin-dependent kinase (CDK) are essential to initiate DNA replication at individual origins. During replication stress, the S-phase checkpoint inhibits the DDK- and CDK-dependent activation of late replication origins. Rad53 kinase is a central effector of the replication checkpoint and both binds to and phosphorylates Dbf4 to prevent late-origin firing. The molecular basis for the Rad53–Dbf4 physical interaction is not clear but occurs through the Dbf4 N terminus. Here we found that both Rad53 FHA1 and FHA2 domains, which specifically recognize phospho-threonine (pT), interacted with Dbf4 through an N-terminal sequence and an adjacent BRCT domain. Purified Rad53 FHA1 domain (but not FHA2) bound to a pT Dbf4 peptide in vitro, suggesting a possible phospho-threonine-dependent interaction between FHA1 and Dbf4. The Dbf4–Rad53 interaction is governed by multiple contacts that are separable from the Cdc5- and Msa1-binding sites in the Dbf4 N terminus. Importantly, abrogation of the Rad53–Dbf4 physical interaction blocked Dbf4 phosphorylation and allowed late-origin firing during replication checkpoint activation. This indicated that Rad53 must stably bind to Dbf4 to regulate its activity.
Keywords: Rad53, replication checkpoint, DNA replication, DDK, Dbf4
THE fidelity of chromosome replication depends on checkpoint mechanisms to stabilize stalled forks, regulate origin activation, and repair DNA damage (Hartwell and Weinert 1989; Bartek et al. 2004; Segurado and Tercero 2009). In response to replication stress, the replication checkpoint maintains replisome stability and prevents late origins from firing, which allows time for DNA repair and the completion of DNA replication prior to chromosome segregation. Incomplete DNA replication or uncoordinated origin firing following DNA damage can result in genomic instability, cancer predisposition, and premature aging (Branzei and Foiani 2010).
In the budding yeast Saccharomyces cerevisiae, activation of the checkpoint sensor kinase Mec1 (vertebrate ATR, Ataxia Telangiectasia and Rad3-related) is triggered at stalled forks or sites of DNA damage (Majka et al. 2006; Labib and De Piccoli 2011). Subsequent signal amplification through the Mrc1 or Rad9 adaptors leads to activation of the checkpoint kinase Rad53 (the ortholog of the human tumor suppressor Chk2) (Branzei and Foiani 2009). Rad53 is an integral transducer of various cellular responses to replication stress or DNA damage. Rad53 induces a series of transcriptional responses through MBF-regulated genes (Bastos de Oliveira et al. 2012; Travesa et al. 2012) and also activates the Dun1 kinase, which promotes the expression of ribonucleotide reductase (RNR) subunits and additional DNA repair genes (Huang et al. 1998). In parallel, Rad53 down-regulates the RNR inhibitor Sml1 to increase deoxyribonucleotide levels and facilitate DNA synthesis (Zhao et al. 2001). In response to replication fork stalling, Rad53 prevents the activation of late replication origins by phosphorylating two proteins required for the initiation of DNA replication: Dbf4 and Sld3 (Lopez-Mosqueda et al. 2010; Zegerman and Diffley 2010; Duch et al. 2011). Dbf4 is the regulatory subunit of Cdc7 kinase, which is required to initiate DNA replication at individual origins by phosphorylating the replicative MCM helicase (Tsuji et al. 2006; Francis et al. 2009; Randell et al. 2010; Sheu and Stillman 2010). Sld3 is also required to activate the MCM helicase by promoting Cdc45–MCM association (Fu and Walter 2010; Boos et al. 2011).
Cdc7 requires the Dbf4 regulatory subunit for kinase activity. Dbf4 is expressed in late G1 phase, peaks during S phase, and is present until early to midmitosis, when it is destroyed by ubiquitin-mediated proteolysis (Cheng et al. 1999; Weinreich and Stillman 1999; Ferreira et al. 2000; Miller et al. 2009). The timing of Dbf4 destruction suggests that Dbf4 has postreplicative functions. Indeed, recent work has shown that Dbf4 prevents premature exit from mitosis and also controls the segregation of homologous chromosomes in meiosis I by a direct interaction with Cdc5, the only Polo-like kinase in budding yeast (Matos et al. 2008; Miller et al. 2009; Chen and Weinreich 2010). Rad53-mediated phosphorylation of Dbf4 postpones late-origin firing during replication stress (Lopez-Mosqueda et al. 2010; Zegerman and Diffley 2010; Duch et al. 2011) but Cdc7-Dbf4 kinase activity is reduced only twofold by Rad53-dependent Dbf4 phosphorylation (Weinreich and Stillman 1999). It is clear that Dbf4 is an in vivo target of Rad53 and interacts with Rad53 (Kihara et al. 2000; Duncker et al. 2002; Matthews et al. 2012), but the molecular details of the Rad53–Dbf4 interaction and how Rad53 phosphorylation of Dbf4 prevents late-origin activation are unclear.
Rad53 is unique in budding yeast in that it contains two fork-head associated (FHA) domains, termed FHA1 and FHA2, which flank a central kinase domain. FHA domains compose a ubiquitous class of protein–protein interaction modules found in >200 different proteins from yeast to mammals (Mahajan et al. 2008). Structural studies show that FHA domains fold into a β-sandwich composed of six-stranded and five-stranded β-sheets (Durocher et al. 2000). Four of the five most conserved residues in the domain are situated in substrate-binding loops and contribute to highly selective binding to phospho-threonine (Liang and Van Doren 2008). Oriented peptide library screening identified consensus phospho-threonine peptides for the FHA1 and FHA2 domains, and the structural basis of their interaction with the Rad53 FHA domains was also determined (Liao et al. 1999; Durocher et al. 2000; Byeon et al. 2001). The FHA1 domain preferentially binds peptides containing the consensus sequence pT-x-x-D, but the FHA2 domain prefers an isoleucine residue at the +3 position, pT-x-x-I. FHA domains also make extensive contacts with additional regions of pT-containing proteins to stabilize binding (reviewed by Mahajan et al. 2008).
Here we have mapped the Dbf4 residues important for a physical interaction with Rad53. We found that a sequence from residues 100–109, which contained a potential FHA1-binding site (T105-x-x-E), and an adjacent BRCA1 carboxyl-terminal (BRCT) domain both interacted with Rad53. Within full-length Rad53, both Rad53 FHA domains were required to bind Dbf4. Biochemical assays showed that the FHA1 domain (but not FHA2) bound to a Dbf4 pT105-X-X-E peptide in a phosphorylation-dependent manner. Finally, abrogation of the Rad53–Dbf4 physical interaction blocked Dbf4 phosphorylation by Rad53 and allowed late-origin firing in the presence of HU. We suggest that the Dbf4 N terminus binds Rad53 using multiple contacts and that Rad53–Dbf4 binding may be phosphorylation dependent. The Rad53 physical interaction with Dbf4 then promotes phosphorylation of Dbf4 at critical downstream sites to inhibit late-origin firing.
Materials and Methods
Construction of yeast strains, plasmids, and baculoviruses
Plasmids and yeast strains used in this study are listed in Supporting Information, Table S1 and Table S2, respectively. PJ69–4a cells (MATa trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2:GAL1-HIS3GAL2-ADE2 met::GAL7-lacZ) were used for two-hybrid experiments. All other strains were derivatives of W303-1A. The natMX4 cassette flanked with DBF4 target sequences was PCR-amplified from p4339 with primers 5′-CTA TCA ACG GCA ATG TTA TTG AAT CAC TTT CTC ATT CAC CCT TGT ACA TGG AGG CCC AGA ATA CC-3′ and 5′-ATG CAA TTG ATA ATA TAT GGA CGA GTA AAT AAG AGT TAA GTC AAT CAG TAT AGC GAC CAG CAT TC-3′ (Goldstein and McCusker 1999), and transformed into M1261 (dbf4-NΔ109). clonNAT-resistant transformants (Werner Bioagents) were confirmed with natMX4 marker and then backcrossed to W303. The epitope-tagged RAD53 strains were made by the method of Longtine et al. (1998). Deletions and point mutations within DBF4 and RAD53 were generated by site-directed mutagenesis using the QuikChange system (Stratagene). PCR-amplified EcoRI-PstI fragments containing the full-length RAD53-coding sequence (1–821), FHA1 domain (1–300), FHA2 domain (483–821), and DBF4-coding sequence (66–227) were cloned into the same sites of pGAD-C1 (Clontech) to give the Gal4 activation domain fusions. Rad53 residues 2–164 were cloned on a BamHI-XhoI into pET24a-GST for expression of the His6-GST-FHA1 domain. Construction of baculovirus plasmids encoding wild-type Dbf4, Dbf4-NΔ109, HA-Cdc7, and 3Myc-Cdc5 was previously described (Gabrielse et al. 2006). An NcoI-PstI fragment containing the full-length RAD53-coding sequence (1–821) was cloned into the baculovirus transfer vector pAcSG2. High-titer baculoviruses were generated by transfection of Sf9 cells using the BaculoGold kit (BD Biosciences) followed by plaque purification and virus amplification.
Growth conditions, cell-cycle synchronization, and replication intermediate assays
Yeast cells were cultured in YPD or synthetic complete medium (SCM) as described (Gabrielse et al. 2006). To detect replication intermediates (Figure 7), cells were synchronized in G1 phase with 5 μg/ml α-factor for 3 hr and released into 0.2 M HU for the indicated times. The alkaline gel electrophoresis and probes for the replication origins (ARS305, ARS501 and ARS603, Autonomously Replicating Sequence) were previously described (Mantiero et al. 2011). DNA content was analyzed by flow cytometry as previously published (Mantiero et al. 2011).
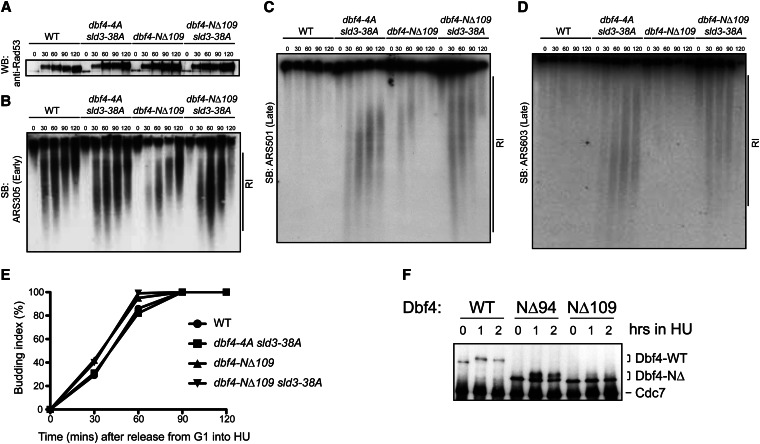
Figure 7.
The dbf4-NΔ109 sld3-38A double mutant allowed late-origin firing in the presence of HU. (A) Wild-type and mutant cells were synchronized in G1 phase and released into medium containing 0.2 mM HU for the indicated times. Total protein extracts were examined by Western blotting for Rad53 to assess Rad53 activation (upper band). (B–D) RIs were separated by alkaline gel electrophoresis and detected by Southern blotting to measure the activity of early (ARS305) and late (ARS501 and ARS603) origins. Flow cytometry assays indicated all strains arrested in early S phase with HU (not shown); the budding indices are shown in E. (F) Wild-type Cdc7-Dbf4 or N-terminal Dbf4 truncation mutants were immunoprecipitated from asynchronous yeast extracts (t = 0) and after 1 or 2 hr exposure to 0.1 M HU, separated on an SDS gel, and then blotted for Cdc7 and Dbf4 proteins.
Two-hybrid analysis
Various DBF4 bait constructs containing the Gal4 DNA-binding domain were transformed with Gal4 activation domain prey plasmids in PJ69–4a and selected on SCM plates lacking tryptophan and leucine. These were spotted at 10-fold serial dilutions on the same plates and also on plates also lacking histidine but containing various concentrations of 3-aminotriazole (3AT) and cultured for 2–3 days at 30°. O-nitrophenyl-β-d-galactoside (Sigma) was used to measure β-galactosidase activity.
Immunoprecipitation from Sf9 cells and Western blotting
Sf9 cells were co-infected with HA-Cdc7, 3Myc-Cdc5, Rad53, and Dbf4 mutants as previously described (Chen and Weinreich 2010). Whole-cell extracts and immunoprecipitates (IPs) were probed with polyclonal antibodies against Cdc7 (1:4000) and Dbf4 (1:1000). Rad53 and 3Myc-Cdc5 were detected with yC-19 (Santa Cruz Biotechnology) and 9E10 antibodies, respectively. Antibodies against Gal4 (729-874) were a gift from K. Melcher (Van Andel Institute).
Protein purification and peptide-binding assays
His6-GST-FHA1 and His6-GST-FHA2 domains were induced in BL21(DE3) cells for 3 hr at 30° using 0.5 mM isopropyl 1-thio-β-d-galactopyranoside. Protein purification and the AlphaScreen luminescence proximity assay (PerkinElmer Life Sciences) were previously described (Chen and Weinreich 2010). All peptides used in this study are listed in Table S3.
Results
Rad53 interacts with a sequence preceding the Dbf4 BRCT domain
Dbf4 is a downstream substrate of the Rad53 kinase in the DNA replication checkpoint (Masai et al. 1999; Weinreich and Stillman 1999; Lopez-Mosqueda et al. 2010; Zegerman and Diffley 2010; Duch et al. 2011). In the presence of HU, Rad53 phosphorylates multiple sites within Dbf4 to inhibit late-origin firing. Our previous study showed that deletion of Dbf4 residues from 66 to 109 prevented Rad53-mediated Dbf4 phosphorylation in HU (Gabrielse et al. 2006), suggesting that these residues, which are N-terminal to a conserved BRCT domain, played a critical role in the Rad53–Dbf4 interaction.
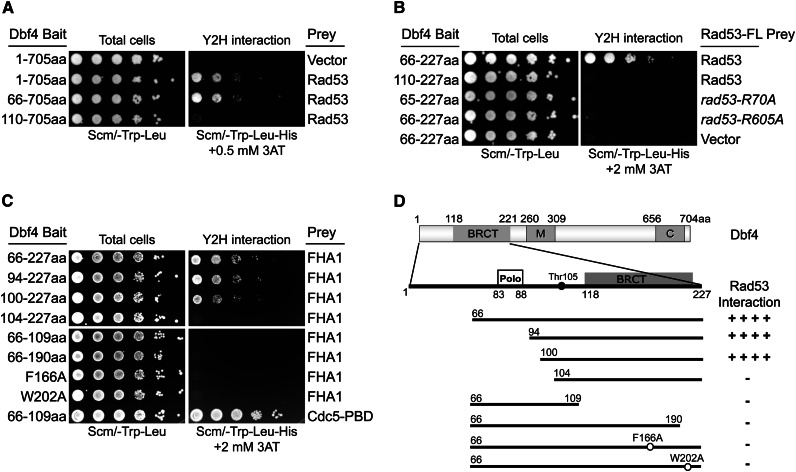
We used a two-hybrid assay to map the Rad53-binding site within Dbf4. Using a series of Dbf4 N-terminal truncations, we found that a deletion through residue 65 retained the Rad53–Dbf4 interaction (Figure 1A). However, a further deletion to residue 109 (just prior to the BRCT domain) resulted in a complete loss of Rad53 binding. Dbf4 N-terminal residues 66–227 were sufficient to interact with Rad53, but, like full-length Dbf4, a deletion to residue 109 abrogated this interaction (Figure 1B). Therefore, the Dbf4 N terminus contains a separate domain (or domains) that interacts with the Rad53 kinase. Also, a sequence preceding the BRCT domain (i.e., within residues 66–109) is required for that interaction.
Figure 1.
Mapping the interaction between Dbf4 and Rad53. (A) Deletion mutants in otherwise full-length Dbf4 were tested for a two-hybrid interaction with full-length Rad53. Tenfold serial dilutions of saturated cultures were spotted onto SCM-Trp-Leu plates to visualize total cells and SCM-Trp-Leu-His + 2 mM 3AT plates to score the two-hybrid interaction. (B) A Dbf4 N-terminal fragment (residues 66–227) was sufficient for the Rad53 interaction, and this interaction required both the FHA domains. (C) Dbf4 residues 100–227 composed the minimal region for Rad53 FHA1-domain binding. (D) Schematic of the features in Dbf4 are shown, including the Polo-like kinase (Cdc5) binding site, a conserved BRCT domain, and motifs M and C, along with a summary of the Dbf4–FHA1 domain interaction.
Both Rad53 FHA domains are required to interact with a Dbf4 N-terminal region spanning residues 100–227
FHA domains are phospho-threonine-specific protein-binding modules, and recognition of the pT residue requires a conserved arginine residue (Durocher et al. 2000; Byeon et al. 2001). Alanine substitutions of the corresponding arginine residues in the FHA1 and FHA2 domains (R70A and R605A, respectively) abolished the Dbf4 interaction (Figure 1B). Mutation of either Rad53 residue did not decrease Rad53-GAD protein stability (see below), as shown previously for endogenous Rad53 (Pike et al. 2003). These results not only indicate that Rad53 binding to the Dbf4 N terminus relies on both FHA domains, but also suggest that the Rad53–Dbf4 interaction involves phosphorylation-dependent FHA contacts.
To identify the FHA-binding sites in Dbf4, we first verified that the FHA1 (Figure 1C) and FHA2 (Figure S1B) domains could bind Dbf4 residues 66–227 independently. We then tested a series of deletion constructs within residues 66–227 for their ability to bind FHA1 and FHA2. Although Dbf4 constructs as short as 100–227 retained FHA binding, deletions beyond residue 100 completely lost FHA1 (and FHA2) binding. This indicates that a Dbf4 sequence following residue 100 is required for the FHA domain interactions. Although critical Dbf4 residues between 66 and 109 were required for the Rad53 interaction (Figure 1A), an ∼40-amino-acid peptide from residues 66 to 109 was not sufficient for the interaction with the FHA1 domain (Figure 1C). This same Dbf4 peptide was sufficient to interact with the Cdc5 Polo-box domain (Figure 1C, bottom), and the Cdc5-binding site has been mapped to Dbf4 residues 83–88 (Miller et al. 2009; Chen and Weinreich 2010). Finally, the Dbf4–FHA domain interaction also required the Dbf4 BRCT domain comprising residues ∼118–224. Any C-terminal deletion that affected the BRCT domain or point mutants in conserved BRCT residues (F166A and W202A) disrupted the Dbf4–FHA domain interaction. To summarize, Dbf4 residues 100–227 compose a minimal region required to bind Rad53 by a two-hybrid assay, and mutation of residues within the BRCT domain or immediately preceding it disrupted that interaction (Figure 1D and below).
Alanine scanning reveals a possible FHA1-binding site in the Dbf4 N terminus
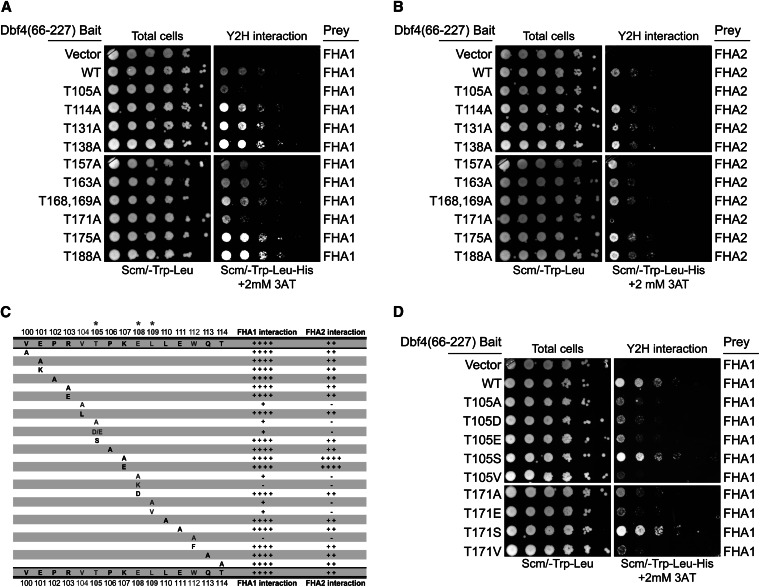
In oriented peptide library screens, the Rad53 FHA1 and FHA2 domains were shown to selectively bind phospho-threonine (Durocher et al. 2000; Byeon et al. 2001). Therefore, we mutated each threonine to alanine within Dbf4 residues 100–227, i.e., the minimal Rad53 binding region that we defined (Figure 2, A and B; Figure S7). We found that the T105A or T171A substitutions significantly impaired the Dbf4–FHA domain interactions. The surrounding sequences of these two threonines (T105-P-K-E and T171-I-V-I) resemble the binding consensus for the FHA1 (pT-x-x-D) and FHA2 domains (pT-x-x-I), respectively (Durocher et al. 2000; Byeon et al. 2001). However, a recent crystal structure of the Dbf4 BRCT domain (Matthews et al. 2012) showed that the T171-I-V-I sequence forms part of the hydrophobic core of the BRCT domain and is not solvent accessible (T171 is only partially buried). So, although the T171-I-V-I motif conforms to a typical FHA2-binding sequence, this motif is buried and is therefore unlikely to interact with the FHA2 domain directly. However, T105 maps just prior to an α-helix adjacent to the BRCT domain and is solvent accessible.
Figure 2.
The Rad53 FHA domains required a T105-x-x-E-L motif in the Dbf4 N terminus for interaction. (A and B) An alanine scan of all Dbf4 threonines within the minimal Rad53-binding region (residues 100–227) using two-hybrid assays against FHA1 and FHA2, respectively. (C) Summary of Dbf4 mutants within residues 100–114 for their effect on the interaction of FHA1 and FHA2 domains. Growth assays are shown in Figure S2. (D) Two-hybrid assays showing the effect of additional T105 and T171 mutations on the Dbf4–FHA1 interaction.
Using a series of point mutants, we determined the Dbf4 residues between 100 and 114 required for binding the FHA1 and FHA2 domains. In addition to T105, we found that alanine substitutions at V104, E108, L109, or W112 disrupted FHA1 and FHA2 domain binding as summarized in Figure 2C (two-hybrid data in Figure S2). The V104A substitution disrupted the interaction, but V104L had little effect, suggesting a structural role or hydrophobic contact for this residue. The E108A mutation strongly impaired FHA binding and E108K abolished FHA binding. However, a conservative E108D mutation retained FHA binding, suggesting that glutamate and aspartate are interchangeable at this position. As expected for an FHA1-binding consensus site, the Dbf4 residues P106 and K107 at the +1 and +2 positions to T105 were not important for binding, consistent with T105-x-x-E being an FHA1-binding site. Our mutagenesis studies also found that several hydrophobic residues near T105 are important. The loss of interaction caused by the W112A mutation was rescued by substituting F, a bulky hydrophobic residue, suggesting that W112 played a structural role for FHA domain binding. Indeed, W112 falls within an α-helix preceding the BRCT domain and makes hydrophobic contacts with the BRCT domain (Matthews et al. 2012). However, L109 may be directly involved in FHA binding, since it is adjacent to E108 and neither the L109A nor L109V mutants interacted with the FHA domains (Figure 2C).
The Rad53 FHA2 domain prefers a pT-x-x-I consensus but can bind to a range of peptides that differ at the +3 position in vitro (Liao et al. 1999; Wang et al. 2000; Byeon et al. 2001). The FHA2 domain interaction with Dbf4 required the same residues as the FHA1 domain although the FHA2–Dbf4 interaction was weaker than the FHA1–Dbf4 interaction, as seen previously (Duncker et al. 2002). Significant exceptions were that the K107A or K107E substitutions (at the +2 position) substantially enhanced FHA2 binding but did not affect FHA1 binding (Figure 2C). Within full-length Rad53, the Dbf4-V104A, -T105A, -E108A mutations also impaired binding to Dbf4 but did not eliminate it (Figure S6C).
FHA domains are highly selective for pT so that even acidic substitutions disrupt FHA binding (Durocher et al. 1999). In agreement with this, we found that T105D or T105E mutations also disrupted the interaction with FHA1 and FHA2 (Figure 2D). In contrast, both T105S and T171S substitutions had little effect on FHA binding (Figure 2D). If the Rad53 FHA domains selectively bind only pT residues in vivo, our mutagenesis data suggest that none of the threonines in the Dbf4 N terminus bind the FHA1/FHA2 domains through a classical pT interaction in yeast.
To further examine the significance of the FHA1–FHA2 interactions, we cloned all 10 remaining FHA domains in the yeast genome and tested their interaction with Dbf4 by the two-hybrid assay. All these proteins were expressed well (not shown); however, none of the 10 domains interacted with Dbf4 (Figure S1C), highlighting the significance of the Rad53 FHA1 and FHA2 interactions. Taken together, our results suggest that both the Rad53 FHA1 and FHA2 domains bind to Dbf4 and that the T105-x-x-E-L sequence is important. However, clearly, other sequences within Rad53 and Dbf4 also contribute to binding.
Dbf4–FHA1 domain interaction is phospho-threonine dependent
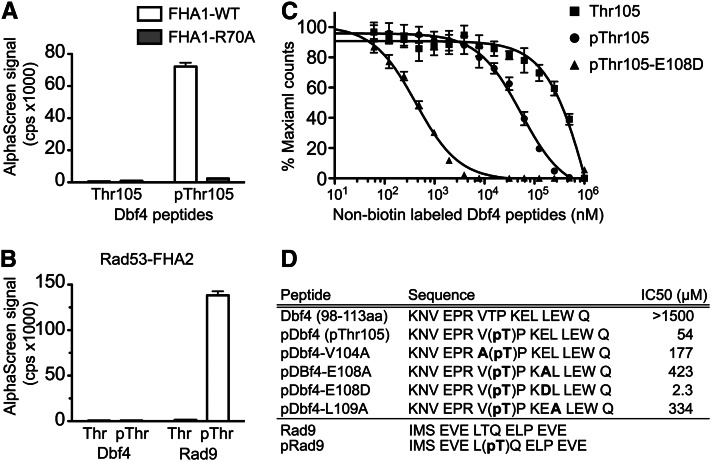
To investigate whether the Dbf4–FHA1 domain interaction required phosphorylation of Dbf4 residue T105, we purified the FHA1 domain and tested its ability to bind synthetic Dbf4 peptides using the AlphaScreen proximity assay (Ullman et al. 1994). The FHA1 domain bound to the biotinylated Dbf4 peptides containing residues 98–113 but only if T105 was phosphorylated (Figure 3A). In addition, mutation of the conserved R70 to A in FHA1 abolished the interaction with the Dbf4 pT105 peptide. These data indicated that the Dbf4–FHA1 domain interaction required T105 phosphorylation. In contrast, although the FHA2 domain bound efficiently to an optimal Rad9 phosphorylated peptide (Byeon et al. 2001), it was unable to bind the same pT105 Dbf4 peptide (Figure 3B). FHA domains bind to pT plus adjacent residues but also make further extensive substrate contacts outside the pT-binding loop (Mahajan et al. 2008). Since neither FHA domain bound to Dbf4 residues 66–109 in the two-hybrid assay unless the BRCT domain was included, FHA1 and FHA2 binding to Dbf4 likely required additional FHA–BRCT contacts.
Figure 3.
The Rad53 FHA1 domain directly bound to a T105 phosphorylated Dbf4 peptide. (A) Biotinylated Dbf4 peptides (residues 98–113) were tested for interaction with the purified 6His-FHA1 domain using the AlphaScreen Assay. Data represent the average of three independent experiments ± SEM. (B) Purified 6His-FHA2 domain does not interact with the pThr105 Dbf4 peptide, but does selectively bind a Rad9-phosphorylated peptide. (C) The Dbf4–FHA1 domain interaction was competed by nonbiotinylated, T105-phosphorylated Dbf4 peptide (pThr105), a peptide containing the optimal FHA1-binding sequence (pT105-E108D), but not by the T105 (nonphosphorylated) Dbf4 peptide. (D) Summary of peptide sequences and the IC50 values determined by competition assays.
To test whether the additional residues discovered in the two-hybrid screen (V104, E108, and L109) were important for the FHA1–Dbf4 peptide interaction, we used nonbiotinylated peptides to compete FHA1::biotin-pT105 peptide binding. The FHA1::biotin-pT105 interaction was competed by an identical pT105 peptide but not by a nonphosphorylated T105 peptide or by an unrelated serine phosphorylated peptide (Figure 3C and Figure S3A), indicating that the interaction was specific for phospho-T105. The FHA1-pT105 peptide competed with an IC50 of 50–60 µM, indicating a moderate FHA1-binding affinity to this peptide. In the yeast two-hybrid assays, we found that E108 and the hydrophobic residues immediately adjacent to the pT105-x-x-E motif were critical for the FHA1 interaction. In agreement with these data, a pT105-x-x A peptide was significantly impaired in its ability to compete the FHA1::biotin-pT105 peptide interaction (Figure S3A). Similarly, alanine substitutions of V104 or L109 within otherwise identical pT105 peptides reduced the ability to compete the FHA1::biotin-pT05 peptide interaction (Figure S3B). Finally, the E108D mutation, which did not affect the Rad53–Dbf4 interaction in the two-hybrid assay and matched the optimal binding sequence for the FHA1 domain, competed the interaction but with a much higher binding affinity (1–5 µM) as shown in Figure 3C. Based on the two-hybrid and biochemical assays, the Rad53 FHA1 domain selectively bound a pT-x-x-E sequence, which closely conforms to an FHA1-binding consensus sequence.
Rad53 and Cdc5 interact with Dbf4-dependent kinase through the Dbf4 N terminus and form a ternary protein complex
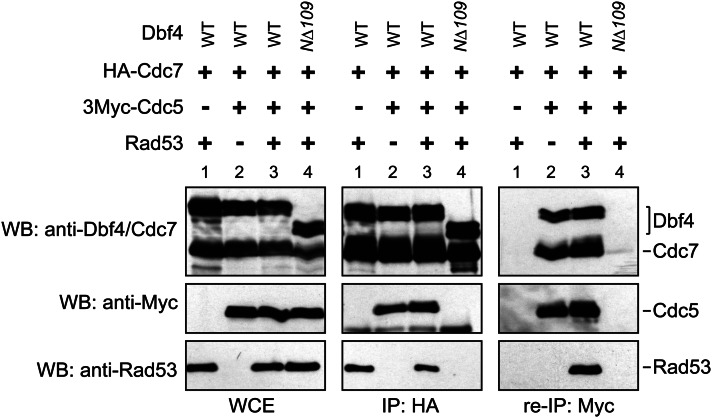
Although Dbf4 is well known for its essential role in binding and activating Cdc7 to initiate DNA replication, we recently proposed that Dbf4 also functions as a molecular scaffold to bind and regulate Cdc5 kinase. Dbf4 residues 83–88 directly interact with the Polo-box domain of Cdc5 kinase and are required for Dbf4-dependent kinase (DDK) to bind Cdc5 (Miller et al. 2009; Chen and Weinreich 2010). DDK inhibits Cdc5 in the mitotic exit network and also is critical for the spindle position checkpoint (Miller et al. 2009; Chen and Weinreich 2010). Once we had defined a distinct binding site for the Rad53 kinase in the Dbf4 N terminus in close proximity to the Cdc5-binding site, we wondered whether Dbf4 could form a ternary complex with the Rad53, Cdc5, and Cdc7 kinases.
To examine the DDK interaction with Cdc5 and Rad53, we employed a baculovirus system to express Rad53, Cdc5, Cdc7, and various Dbf4 derivatives in Sf9 cells. Consistent with previous reports (Miller et al. 2009; Chen and Weinreich 2010), Cdc5 was co-immunoprecipitated with wild-type Cdc7-Dbf4 but not with the Cdc7-Dbf4-NΔ109 truncation derivative (Figure 4, middle). Similarly, Rad53 bound to wild-type Cdc7-Dbf4, but not to Cdc7-Dbf4-NΔ109. Both Rad53 and Cdc5 also bound the Cdc7-Dbf4-NΔ65 derivative (Miller et al. 2009; Chen and Weinreich 2010; data not shown). These results indicate that the association of Rad53 and Cdc5 with Cdc7-Dbf4 depended on Dbf4 residues from 66 to 109, which contain the Cdc5-binding site (residues 83–88) and a Rad53-binding site (residues 104–109).
Figure 4.
Rad53 and Cdc5 interact with DDK through the Dbf4 N terminus and form a ternary protein complex. HA-Cdc7-Dbf4 complexes were immunoprecipitated from baculovirus-infected Sf9 cells using 12CA5 antibodies and examined for co-immunoprecipitation of Rad53 and 3Myc-Cdc5 by Western blotting (middle). Following 12CA5 immunoprecipitation, proteins were eluted from the beads using HA peptide and subjected to another round of immunoprecipitation by 9E10 antibodies. Rad53 was co-immunoprecipitated with 3Myc-Cdc5 and wild-type DDK but not if Dbf4-NΔ109 was expressed (right).
The co-immunoprecipitation results suggest two different possibilities. Either DDK exists in two distinct protein complexes (DDK-Rad53 and DDK-Cdc5), or, alternatively, DDK can bind to Cdc5 and Rad53 simultaneously. To clarify this, we asked whether Rad53 bound the DDK–Cdc5 complex by performing a sequential co-immunoprecipitation. We expressed all four proteins in Sf9 cells and immunoprecipitated DDK using the HA tag on the Cdc7 subunit. This procedure immunoprecipitates protein bound to DDK, which includes DDK-(Myc-Cdc5), DDK-Rad53, and presumptive DDK-(Myc-Cdc5)-Rad53 complexes. We then eluted the bound proteins using 1 mM HA peptide and performed a second round of immunoprecipitation using 9E10 monoclonal antibodies to immunoprecipitate only the DDK-(Myc-Cdc5) complexes. Rad53 was present in the second IP (Figure 4, right), indicating that Rad53 forms a ternary complex with Cdc5 and DDK. Together, these results demonstrate that the Dbf4 N terminus acts as a docking site for both Rad53 and Cdc5 and that both kinases can simultaneously associate with DDK.
Rad53 checkpoint defect together with loss of specific Dbf4 N-terminal residues results in synthetic lethality
DDK and rad53 mutants show a series of complex genetic interactions (Desany et al. 1998; Dohrmann et al. 1999; Dohrmann and Sclafani 2006; Gabrielse et al. 2006). We previously reported that the dbf4-NΔ109 mutant was synthetically lethal with the rad53-1 hypomorphic mutant (Gabrielse et al. 2006). This is interesting since the dbf4-NΔ109 mutant exhibits an apparently normal S phase, is not defective for activating early or late replication origins, and is not sensitive to genotoxic agents (Gabrielse et al. 2006). Although the Dbf4-NΔ109 protein both binds to and activates Cdc7 normally (Gabrielse et al. 2006; Harkins et al. 2009), it is defective for binding Cdc5 (Miller et al. 2009; Chen and Weinreich 2010) and Rad53 (this study). Therefore, we tested whether the synthetic lethality between dbf4-NΔ109 and rad53-1 was due to the loss of the Dbf4–Cdc5 or Dbf4–Rad53 interactions.
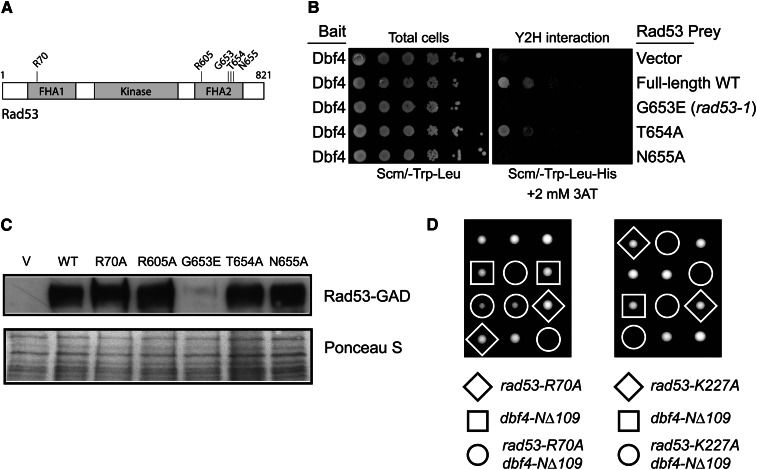
We first sequenced the rad53-1 gene (Weinert et al. 1994) and found a single G653E point mutation, which is identical to that reported for the rad53-11 allele (Dohrmann and Sclafani 2006). G653 falls within a loop between the β6 and β7 strands of the FHA2 domain (Figure 5A) and is adjacent to the conserved N655 residue, which plays an important role in substrate recognition (Byeon et al. 2001). The rad53-1 (G653E) or N655A full-length Rad53 mutants were unable to bind the Dbf4 N terminus in the two-hybrid assay like the Rad53-R605A mutant (Figure 1B), but mutation of an adjacent nonconserved residue (T654A) had no effect (Figure 5B). The FHA2 mutants were expressed similarly to wild type with the exception of G653E (Figure 5C), confirming the importance of the FHA2 domain for the Dbf4 interaction. Similar to rad53-1, we found that the rad53-R70A mutant, which also did not interact with Dbf4 (Figure 1B), was synthetically sick or lethal with dbf4-NΔ109 but obviously not with DBF4 (Figure 5D). We also observed synthetic lethality between dbf4-NΔ109 and the rad53-K227A (kinase dead) allele (Figure 5D). Since the rad53-R70A and rad53-G653E (rad53-1) mutants are defective for interacting with DDK to begin with, their synthetic lethality with dbf4-NΔ109 cannot be due to the further loss of only the Rad53-binding site on Dbf4. The synthetic lethality is likely caused by compromised Rad53 function coupled with loss of a Rad53-independent function of Dbf4 present in the N-terminal 109 residues. We know that this function is not the ability to bind Cdc5, since a dbf4-Δ82-88 mutant, which is completely defective for binding Cdc5 (Miller et al. 2009; Chen and Weinreich 2010), was not synthetically lethal with rad53-1 or a rad53Δ (Figure S4).
Figure 5.
dbf4-NΔ109 was synthetically lethal with rad53-R70A, rad53-K227A, and rad53-G653E. (A) Schematic diagram of Rad53 showing the mutations studied within the FHA1 and FHA2 domains. (B) The rad53-G653E and rad53-N655A mutants did not interact with Dbf4 in yeast two-hybrid assays. (C) Western blot of the full-length Rad53-GAD fusion used for two-hybrid analyses and the corresponding FHA domain mutants. The Ponceau S-stained blot is shown below as a loading control. (D) Representative tetrads from diploid strains of genotype DBF4/dbf4-NΔ109 RAD53/rad53-R70A and DBF4/dbf4-NΔ109 RAD53/rad53-K227A were sporulated and dissected onto YPD plates. Recombinant genotypes are indicated.
Dbf4–Rad53 interaction is separable from a Dbf4 BRCT domain interaction with Msa1
Using the N terminus of Dbf4 as bait (pCG60), we have also identified Msa1 as a Dbf4-interacting protein (Figure 6A). MSA1 encodes a transcription factor that regulates the timing of G1-specific gene expression (Ashe et al. 2008). MSA1 was simultaneously reported as a high copy suppressor of temperature-sensitive mutations in sld2 and dbp11 (Li et al. 2008). Both Sld2 and Dpb11 are required for the initiation of DNA replication and act together with DDK and Sld3 to promote Cdc45 and GINS binding to the MCM helicase, a critical step in MCM helicase activation (Labib 2010).
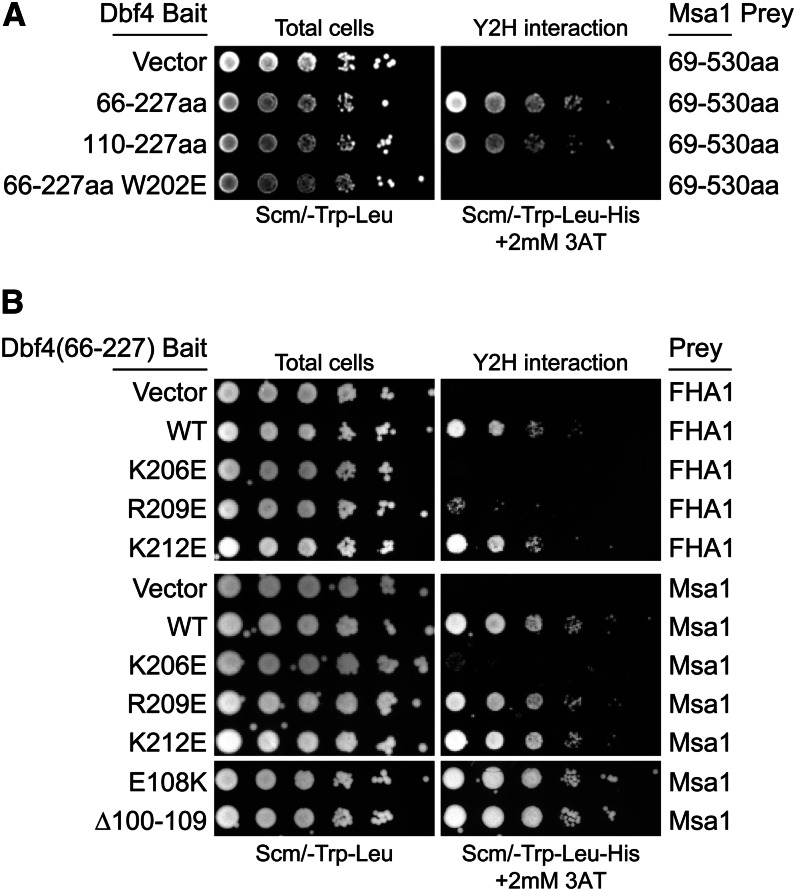
Figure 6.
Both Msa1 and Rad53 interacted with the Dbf4 BRCT domain, but the Rad53 interaction was specifically disrupted by an R209E mutation. (A) Two-hybrid assays with indicated Dbf4 bait and Msa1 prey vectors, spotted as in Figure 1. Msa1 interacts with the BRCT domain alone (Dbf4-110-227). (B) Charge reversal mutations of lysine and arginine residues in the BRCT α3 helix (Matthews et al. 2012) identify R209E as specifically affecting the Rad53 interaction. Dbf4 residues 100–109 are also uniquely required for the Rad53 interaction (Fig. S6C) but not for the Msa1 interaction.
We found that Dbf4 interacted with Msa1 through its BRCT domain, since both Dbf4 (66–227) and Dbf4 (110–227) proteins interacted with Msa1 similarly, but a W202E (or W202A, not shown) mutation within the BRCT domain blocked binding to Msa1 (Figure 6A). This is in contrast to Rad53, which does not interact with the BRCT domain alone [i.e., Dbf4 (110–227) in Figure 1, B and C]. We used the Msa1–Dbf4 BRCT interaction as a control to identify specific BRCT residues that contact Rad53. α-Helix 3 (α3) of the Dbf4 BRCT domain is spatially close to α0, which contains the E108, L109, and W212 residues important for binding Rad53 (Figure 2), and directly precedes the BRCT domain (Matthews et al. 2012). Therefore, we mutated three positively charged residues within α3 to glutamate and tested the interaction with both Msa1 and Rad53. A K206E mutation disrupted both the Msa1 and Rad53 interactions, and a K212E mutation disrupted neither interaction (Figure 6B). However, the R209E mutation selectively blocked the Dbf4 interaction with Rad53 and thus defined a unique BRCT residue that was important for interaction with Rad53 but not with Msa1 (Figure 6B). Similarly, the Dbf4-E108K and Dbf4-Δ100-109 mutations disrupted the Rad53 interaction (Figure S6C) but have no effect on the Msa1 interaction. Thus, although both Msa1 and Rad53 bind the Dbf4 N terminus, Dbf4 residues from 100 to 109 and R209 are uniquely required for interaction with Rad53.
Dbf4–Rad53 physical interaction is required to inhibit late-origin firing during replication checkpoint activation
In response to replication fork arrest, Rad53 phosphorylates Dbf4 and Sld3 to inhibit late-origin firing, but phosphorylation of either protein is sufficient for this inhibition (Lopez-Mosqueda et al. 2010; Zegerman and Diffley 2010). Both studies mapped Rad53 phosphorylation sites on Dbf4 in vitro. A dbf4-4A mutant that changes 4 serine and threonine Rad53 phosphorylation sites to alanine is sufficient to allow late-origin activation when combined with an sld3-38A mutant containing alanine mutations in 38 Rad53 phosphorylation sites (Zegerman and Diffley 2010). We hypothesized that Rad53 regulation of Dbf4 in the replication checkpoint depended on its physical interaction with the Dbf4 N terminus. To test this, we examined whether the combination of a dbf4-NΔ109 mutant (defective for Rad53 binding, Figure 4) and the sld3-38A mutant, which cannot be phosphorylated by Rad53, would allow late-origin firing in the presence of HU. Yeast cells were synchronized in G1 phase using mating pheromone and then released into S phase in the presence of 0.2 M HU to stall replication forks from early origins. At different time points following release from the G1 arrest, replication intermediates (RI) near ARSs were separated on alkaline gels and detected by Southern blotting with ARS-specific probes to measure replication origin activity. As a control, Rad53 was activated (evidenced by the phosphorylation-dependent mobility shift) in both wild-type and mutant cells following HU treatment (Figure 7A), indicating that neither the dbf4 nor the sld3 mutations affect Rad53 checkpoint activation.
The early origin, ARS305, was active in the wild type, dbf4-NΔ109, dbf4-4A sld3-38A, and dbf4-NΔ109 sld3-38A mutant strains, indicating that induction of the replication checkpoint does not interfere with early origin firing in these cells (Figure 7B). Although Rad53 activation inhibited the firing of late origins ARS501 and ARS603 in the wild-type and the single mutants as expected (Lopez-Mosqueda et al. 2010; Zegerman and Diffley 2010; Duch et al. 2011), replication intermediates were detected at late origins in both the sld3-38A dbf4-4A and sld3-38A dbf4-NΔ109 double mutants to a similar extent (Figure 7, C and D). Thus, the dbf4-NΔ109 mutant defective for Dbf4-Rad53 binding was similarly defective in preventing late-origin firing in HU as the dbf4-4A phosphorylation site allele.
We also tested the effect of Dbf4 N-terminal truncations on the Rad53-dependent phosphorylation of Dbf4 in yeast (seen as a mobility shift in SDS-PAGE gels) after exposure to HU (Weinreich and Stillman 1999; Gabrielse et al. 2006; Lopez-Mosqueda et al. 2010; Zegerman and Diffley 2010). Log-phase yeast cells (t = 0) were treated with HU for 1 and 2 hr to arrest replication forks and induce the replication checkpoint. DDK was immunoprecipitated at each time point and probed for Cdc7 and Dbf4 proteins by Western blotting. Both the wild-type Dbf4 and Dbf4-NΔ94 proteins that retained binding to Rad53 were shifted upon exposure to HU; however, the Dbf4-NΔ109 protein that did not bind Rad53 was not shifted (Figure 7F). These data indicate that loss of Rad53 binding to Dbf4 mediated by critical Dbf4 residues between 95 and 109 caused a significant defect in Dbf4 phosphorylation upon replication fork arrest. Together, these data show that Rad53 must stably interact with Dbf4 through its N-terminal binding site to phosphorylate Dbf4 and inhibit late-origin firing in response to HU.
Discussion
Rad53 FHA domains interact with the Dbf4 N terminus
Multiple groups have reported genetic and physical interactions between S. cerevisiae Dbf4 and Rad53 (Dohrmann et al. 1999; Weinreich and Stillman 1999; Kihara et al. 2000; Duncker et al. 2002; Gabrielse et al. 2006; Matthews et al. 2012) and Schizosaccharomyces pombe Dfp1 and Cds1 (Takeda et al. 2001; Fung et al. 2002). Furthermore, in response to DNA damage human and Xenopus DDK are downstream targets of ATR signaling (Costanzo et al. 2003; Lee et al. 2012). The Dbf4 regulation by checkpoint kinases is broadly conserved since it likely promotes genome stability. Here we have mapped a Rad53-binding site in the Dbf4 N terminus and have shown that a Rad53–Dbf4 physical interaction is critical for regulating late replication origin firing. A minimal Rad53-binding region corresponds to Dbf4 residues 100–227, which compose the Dbf4 BRCT domain (∼118–224) and residues immediately N-terminal to this domain. Mutations in either conserved BRCT residues or residues within 100–109 caused defects in Rad53 binding in the two-hybrid assay. Therefore, both the BRCT domain and the region preceding it contributed to Rad53 binding. Despite their different consensus peptide-binding sites, both Rad53 FHA domains interacted with this 100–109 region independently and apparently using the same Dbf4 residues (see below). Mutations that impair phospho-threonine binding in either Rad53 FHA domain blocked interaction with Dbf4, supporting the contention that the Rad53–Dbf4 interaction is mediated by phosphorylation and is multivalent.
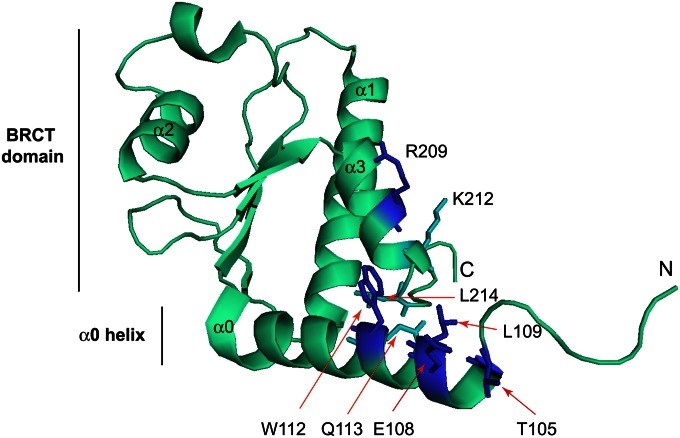
Very recently, a crystal structure, the Dbf4 BRCT domain (that included residues 98–221), was described (Matthews et al. 2012). These authors also showed that Rad53 interacted with the Dbf4 BRCT domain plus the preceding α-helix using two-hybrid assays; however, none of threonines contained within the structure was shown to directly interact with Rad53. Our study found that residues V104, T105, E108, L109, W112, and R209 were important for the Dbf4–Rad53 interaction and that FHA1 specifically interacted with a T105 phosphorylated Dbf4 peptide in vitro. The structure of the Dbf4 N terminus allows us to rationalize this data (Figure 8). T105 is solvent-exposed and occurs within a sequence (T-x-x-E) that closely matches a FHA1-binding site. The E108 residue has the same spatial orientation as T105, and L109 is directly adjacent to E108. The W112 residue packs against L214 present at the C terminus of the BRCT α3 helix. This hydrophobic interaction presumably helps stabilize the α0–α3 orientation and would explain why a W112A mutation disrupts the Dbf4–Rad53 interaction but W112F does not. Finally, R209 on the α3 helix is solvent-exposed and is suitably oriented to interact with an FHA domain bound to α0 or, alternatively, to mediate BRCT–BRCT domain interactions. In a tandem BRCT-BRCT dimer, the α2 helix from one monomer packs against the α1 and α3 helices from the second monomer (Glover et al. 2004). Mutation of R209 (but not K212, which is oriented orthogonally to R209 and away from α0) abolished the Dbf4–FHA1 two-hybrid interaction (Figure 6). Since the purified Rad53 FHA1 domain bound only to T105 phosphorylated Dbf4 peptides, these data raise the possibility that the Dbf4 pT105-x-x-E-L motif binds to Rad53 but the Rad53 interaction is further stabilized by additional BRCT domain contacts. T105 phosphorylation is not essential for the Rad53–Dbf4 physical interaction since a Dbf4 quadruple mutant protein (S84A S92A T95A T105A) still underwent a Rad53-dependent shift in HU (Gabrielse et al. 2006). Also, a T105A mutation diminished the Dbf4 two-hybrid interaction with full-length Rad53 but did not eliminate it (Figure S6C).
Figure 8.
Structural representation of the budding yeast BRCT domain and a preceding α-helix (α0) (both in cyan) using PDB coordinates 3QBZ (Matthews et al. 2012). The helices within the BRCT domain are numbered α1, α2, and α3. Five residues that are important for interaction with Rad53 (T105, E108, L109, W112, R209) are colored blue. L214 and Q113, which form a hydrophobic surface with W112, are colored in cyan. K212 is also colored in cyan but is not important for the Rad53 interaction.
Although the Rad53 FHA2 domain bound to the same sequence as FHA1 in the two-hybrid assay (Figure 2), it did not bind to the 16mer pT-x-x-E peptide in vitro, and this sequence does not match the optimal FHA2-binding site consensus. The FHA2 domain might bind to Dbf4 indirectly in the two-hybrid assay, but the FHA2 interaction still occurs in a strain deleted for Rad53 (data not shown), and so it is not mediated by endogenous Rad53. Importantly, none of the other 10 FHA domains encoded in the yeast genome can bind to this Dbf4 sequence (Figure S1C), suggesting again that the FHA2 interaction is biologically relevant. Since a previous study also demonstrated an interaction between FHA2 and Dbf4 using two-hybrid and GST-pull-down assays in yeast (Duncker et al. 2002), we suggest that the FHA2 domain interaction with Dbf4 is stabilized in vivo by additional contacts within the BRCT domain.
Models for Rad53 binding to Dbf4
We propose several models to explain how Rad53 interacts with Dbf4. Rad53 could use each FHA domain to bind two separate sites within Dbf4, or both Rad53 FHA domains could bind to the same T-x-x-E-L sequence but on different Dbf4 subunits within a Dbf4 dimer. Alternatively, the T-x-x-E-L sequence might help mediate dimerization or other structural changes in the BRCT domain necessary to promote Rad53 binding.
Although both Rad53 FHA domains required the same T-x-x-E-L sequence for binding, the BRCT domain is also critical for binding Rad53. It is possible that the Rad53 FHA1 domain binds to pT105-x-x-E –L and that the FHA2 domain binds to another phosphorylated residue in the BRCT domain. This seems unlikely, however, since mutation of every other threonine (Figure 2) or tyrosine residue (Figure S1) (Liao et al. 1999; Wang et al. 2000; Byeon et al. 2001) in the BRCT domain had no effect on FHA1 or FHA2 binding, with the exception of T171 discussed above. Since both the T105S and T171S Dbf4 mutants bound the FHA domains normally, these residues may not interact with Rad53 through “canonical” FHA-pT interactions in vivo.
A second model is that the Dbf4 N termini form a dimer using the BRCT domains, and then this Dbf4 dimer provides two T105-x-x-E108 sites for the binding of the FHA1 and FHA2 domains separately. DDK or Dbf4 oligomerization has been suggested previously (Shellman et al. 1998; Matthews et al. 2012). Furthermore, tandem BRCT domains form dimers that bind phospho-S/T motifs (Caldecott 2003; Rodriguez and Songyang 2008), and intermolecular dimerization between BRCT domains has also been described for the DNA repair proteins XRCC1 and Ligase III (Cuneo et al. 2011). In support of this model, we saw a significant but weak interaction between Dbf4 N termini using the yeast two-hybrid assay (Figure S5). Substitutions of conserved residues in the Dbf4 BRCT domain as well as deletion of residues 100–109 disrupted the Dbf4–Dbf4 two-hybrid interaction (Figure S5), supporting the idea of BRCT domain dimerization. Arguing against this model is the lack of biochemical data supporting an interaction between the Rad53 FHA2 domain and the pT105-x-x-E peptide; however, as stated above, FHA2 is likely to make additional contacts with the BRCT domain.
Finally, the involvement of residues 100–109 and the T-x-x-E-L motif in particular for Rad53 binding could reflect a requirement for these residues to mediate BRCT structural changes needed for Rad53 binding. Clearly, further biochemical or structural data of FHA1-Dbf4 or Rad53-Dbf4 complexes will be needed to determine exactly how Rad53 interacts with Dbf4.
Dbf4-Rad53 binding is critical for regulation of late-origin activation
Upon sensing DNA damage or replication fork stalling, Rad53 directly phosphorylates Dbf4 and Sld3 to inhibit late-origin firing (Lopez-Mosqueda et al. 2010; Zegerman and Diffley 2010). We demonstrated that the dbf4-NΔ109 mutant defective in the Rad53–Dbf4 interaction coupled with the sld3-38A mutant allows late-origin firing in the presence of HU (Figure 7). Since Dbf4-NΔ94 was phosphorylated in HU but the Dbf4-NΔ109 mutant was not (Gabrielse et al. 2006) (Figure 7), both pieces of data reveal the importance of residues 95–109 for interaction with Rad53. Together, these data strongly suggest that the Rad53-mediated Dbf4 phosphorylation during the replication checkpoint depends on the physical interaction between Dbf4 and Rad53. Interestingly, mutation of the Rad53 FHA1 domain also impairs late-origin regulation in HU (Pike et al. 2004), raising the possibility that Rad53 FHA1 interactions are critical for binding both Dbf4 and Sld3.
Regulation of DDK in the replication checkpoint may involve two phosphorylation events. First, phosphorylation of Dbf4 residue T105 by an unknown kinase could promote the Rad53–DDK interaction. In support of this, T105 was identified as a Rad53 site in vitro, and so Rad53 may phosphorylate this site to promote its own binding to Dbf4 (Lopez-Mosqueda et al. 2010). Determining whether T105 is phosphorylated in vivo and, if so, determining the kinase that phosphorylates T105 is an important future goal. Second, Rad53 then phosphorylates Dbf4 at critical sites downstream of its binding site. Since Rad53 cannot bind to or significantly phosphorylate Dbf4-NΔ109, our data raise the possibility that stable binding of Rad53 to other targets may be needed for efficient phosphorylation. This is similar, for example, to DDK itself, which is targeted to Mcm4 through an N-terminal sequence (Sheu and Stillman 2010).
Supplementary Material
Acknowledgments
We thank FuJung Chang (Van Andel Research Institute) and Feng-Ling Tsai (University of Pittsburgh) for technical help or advice. We also thank John Diffley and Jörg Heierhorst for yeast strains.
Footnotes
Communicating editor: J. A. Nickoloff
Literature Cited
- Ashe M., de Bruin R. A., Kalashnikova T., McDonald W. H., Yates J. R., III, et al. , 2008. The SBF- and MBF-associated protein Msa1 is required for proper timing of G1-specific transcription in Saccharomyces cerevisiae. J. Biol. Chem. 283: 6040–6049. [DOI] [PubMed] [Google Scholar]
- Bartek J., Lukas C., Lukas J., 2004. Checking on DNA damage in S phase. Nat. Rev. Mol. Cell Biol. 5: 792–804. [DOI] [PubMed] [Google Scholar]
- Bastos de Oliveira F. M., Harris M. R., Brazauskas P., de Bruin R. A., Smolka M. B., 2012. Linking DNA replication checkpoint to MBF cell-cycle transcription reveals a distinct class of G1/S genes. EMBO J. 31: 1798–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boos D., Sanchez-Pulido L., Rappas M., Pearl L. H., Oliver A. W., et al. , 2011. Regulation of DNA replication through Sld3-Dpb11 interaction is conserved from yeast to humans. Curr. Biol. 21: 1152–1157. [DOI] [PubMed] [Google Scholar]
- Branzei D., Foiani M., 2009. The checkpoint response to replication stress. DNA Repair (Amst.) 8: 1038–1046. [DOI] [PubMed] [Google Scholar]
- Branzei D., Foiani M., 2010. Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol. 11: 208–219. [DOI] [PubMed] [Google Scholar]
- Byeon I. J., Yongkiettrakul S., Tsai M. D., 2001. Solution structure of the yeast Rad53 FHA2 complexed with a phosphothreonine peptide pTXXL: comparison with the structures of FHA2-pYXL and FHA1-pTXXD complexes. J. Mol. Biol. 314: 577–588. [DOI] [PubMed] [Google Scholar]
- Caldecott K. W., 2003. Cell signaling. The BRCT domain: Signaling with friends? Science 302: 579–580. [DOI] [PubMed] [Google Scholar]
- Chen Y. C., Weinreich M., 2010. Dbf4 regulates the Cdc5 Polo-like kinase through a distinct non-canonical binding interaction. J. Biol. Chem. 285: 41244–41254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L., Collyer T., Hardy C. F., 1999. Cell cycle regulation of DNA replication initiator factor Dbf4p. Mol. Cell. Biol. 19: 4270–4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo V., Shechter D., Lupardus P. J., Cimprich K. A., Gottesman M., et al. , 2003. An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol. Cell 11: 203–213. [DOI] [PubMed] [Google Scholar]
- Cuneo M. J., Gabel S. A., Krahn J. M., Ricker M. A., London R. E., 2011. The structural basis for partitioning of the XRCC1/DNA ligase III-alpha BRCT-mediated dimer complexes. Nucleic Acids Res. 39: 7816–7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desany B. A., Alcasabas A. A., Bachant J. B., Elledge S. J., 1998. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 12: 2956–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohrmann P. R., Sclafani R. A., 2006. Novel role for checkpoint Rad53 protein kinase in the initiation of chromosomal DNA replication in Saccharomyces cerevisiae. Genetics 174: 87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohrmann P. R., Oshiro G., Tecklenburg M., Sclafani R. A., 1999. RAD53 regulates DBF4 independently of checkpoint function in Saccharomyces cerevisiae. Genetics 151: 965–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duch A., Palou G., Jonsson Z. O., Palou R., Calvo E., et al. , 2011. A Dbf4 mutant contributes to bypassing the Rad53-mediated block of origins of replication in response to genotoxic stress. J. Biol. Chem. 286: 2486–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncker B. P., Shimada K., Tsai-Pflugfelder M., Pasero P., Gasser S. M., 2002. An N-terminal domain of Dbf4p mediates interaction with both origin recognition complex (ORC) and Rad53p and can deregulate late origin firing. Proc. Natl. Acad. Sci. USA 99: 16087–16092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durocher D., Henckel J., Fersht A. R., Jackson S. P., 1999. The FHA domain is a modular phosphopeptide recognition motif. Mol. Cell 4: 387–394. [DOI] [PubMed] [Google Scholar]
- Durocher D., Taylor I. A., Sarbassova D., Haire L. F., Westcott S. L., et al. , 2000. The molecular basis of FHA domain:phosphopeptide binding specificity and implications for phospho-dependent signaling mechanisms. Mol. Cell 6: 1169–1182. [DOI] [PubMed] [Google Scholar]
- Ferreira M. F., Santocanale C., Drury L. S., Diffley J. F., 2000. Dbf4p, an essential S phase-promoting factor, is targeted for degradation by the anaphase-promoting complex. Mol. Cell. Biol. 20: 242–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis L. I., Randell J. C., Takara T. J., Uchima L., Bell S. P., 2009. Incorporation into the prereplicative complex activates the Mcm2–7 helicase for Cdc7-Dbf4 phosphorylation. Genes Dev. 23: 643–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. V., Walter J. C., 2010. DNA replication: metazoan Sld3 steps forward. Curr. Biol. 20: R515–R517. [DOI] [PubMed] [Google Scholar]
- Fung A. D., Ou J., Bueler S., Brown G. W., 2002. A conserved domain of Schizosaccharomyces pombe dfp1(+) is uniquely required for chromosome stability following alkylation damage during S phase. Mol. Cell. Biol. 22: 4477–4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrielse C., Miller C. T., McConnell K. H., DeWard A., Fox C. A., et al. , 2006. A Dbf4p BRCA1 C-terminal-like domain required for the response to replication fork arrest in budding yeast. Genetics 173: 541–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover J. N., Williams R. S., Lee M. S., 2004. Interactions between BRCT repeats and phosphoproteins: tangled up in two. Trends Biochem. Sci. 29: 579–585. [DOI] [PubMed] [Google Scholar]
- Goldstein A. L., McCusker J. H., 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15: 1541–1553. [DOI] [PubMed] [Google Scholar]
- Harkins V., Gabrielse C., Haste L., Weinreich M., 2009. Budding yeast Dbf4 sequences required for Cdc7 kinase activation and identification of a functional relationship between the Dbf4 and Rev1 BRCT domains. Genetics 183: 1269–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H., Weinert T. A., 1989. Checkpoints: controls that ensure the order of cell cycle events. Science 246: 629–634. [DOI] [PubMed] [Google Scholar]
- Huang M., Zhou Z., Elledge S. J., 1998. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94: 595–605. [DOI] [PubMed] [Google Scholar]
- Kihara M., Nakai W., Asano S., Suzuki A., Kitada K., et al. , 2000. Characterization of the yeast Cdc7p/Dbf4p complex purified from insect cells. Its protein kinase activity is regulated by Rad53p. J. Biol. Chem. 275: 35051–35062. [DOI] [PubMed] [Google Scholar]
- Labib K., 2010. How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev. 24: 1208–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K., De Piccoli G., 2011. Surviving chromosome replication: the many roles of the S-phase checkpoint pathway. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366: 3554–3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A. Y., Chiba T., Truong L. N., Cheng A. N., Do J., et al. , 2012. Dbf4 is direct downstream target of ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and Rad3-related (ATR) protein to regulate intra-S-phase checkpoint. J. Biol. Chem. 287: 2531–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. M., Tetzlaff M. T., Elledge S. J., 2008. Identification of MSA1, a cell cycle-regulated, dosage suppressor of drc1/sld2 and dpb11 mutants. Cell Cycle 7: 3388–3398. [DOI] [PubMed] [Google Scholar]
- Liang X., Van Doren S. R., 2008. Mechanistic insights into phosphoprotein-binding FHA domains. Acc. Chem. Res. 41: 991–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H., Byeon I. J., Tsai M. D., 1999. Structure and function of a new phosphopeptide-binding domain containing the FHA2 of Rad53. J. Mol. Biol. 294: 1041–1049. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., III, Demarini D. J., Shah N. G., Wach A., et al. , 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Lopez-Mosqueda J., Maas N. L., Jonsson Z. O., Defazio-Eli L. G., Wohlschlegel J., et al. , 2010. Damage-induced phosphorylation of Sld3 is important to block late origin firing. Nature 467: 479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan A., Yuan C., Lee H., Chen E. S., Wu P. Y., et al. , 2008. Structure and function of the phosphothreonine-specific FHA domain. Sci. Signal. 1: re12. [DOI] [PubMed] [Google Scholar]
- Majka J., Niedziela-Majka A., Burgers P. M., 2006. The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Mol. Cell 24: 891–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantiero D., Mackenzie A., Donaldson A., Zegerman P., 2011. Limiting replication initiation factors execute the temporal programme of origin firing in budding yeast. EMBO J. 30: 4805–4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H., Sato N., Takeda T., Arai K., 1999. CDC7 kinase complex as a molecular switch for DNA replication. Front. Biosci. 4: D834–D840. [DOI] [PubMed] [Google Scholar]
- Matos J., Lipp J. J., Bogdanova A., Guillot S., Okaz E., et al. , 2008. Dbf4-dependent CDC7 kinase links DNA replication to the segregation of homologous chromosomes in meiosis I. Cell 135: 662–678. [DOI] [PubMed] [Google Scholar]
- Matthews L. A., Jones D. R., Prasad A. A., Duncker B. P., Guarne A., 2012. Saccharomyces cerevisiae Dbf4 has unique fold necessary for interaction with Rad53 kinase. J. Biol. Chem. 287: 2378–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. T., Gabrielse C., Chen Y. C., Weinreich M., 2009. Cdc7p-Dbf4p regulates mitotic exit by inhibiting Polo kinase. PLoS Genet. 5: e1000498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike B. L., Yongkiettrakul S., Tsai M. D., Heierhorst J., 2003. Diverse but overlapping functions of the two forkhead-associated (FHA) domains in Rad53 checkpoint kinase activation. J. Biol. Chem. 278: 30421–30424. [DOI] [PubMed] [Google Scholar]
- Pike B. L., Tenis N., Heierhorst J., 2004. Rad53 kinase activation-independent replication checkpoint function of the N-terminal forkhead-associated (FHA1) domain. J. Biol. Chem. 279: 39636–39644. [DOI] [PubMed] [Google Scholar]
- Randell J. C., Fan A., Chan C., Francis L. I., Heller R. C., et al. , 2010. Mec1 is one of multiple kinases that prime the Mcm2–7 helicase for phosphorylation by Cdc7. Mol. Cell 40: 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M. C., Songyang Z., 2008. BRCT domains: phosphopeptide binding and signaling modules. Front. Biosci. 13: 5905–5915. [DOI] [PubMed] [Google Scholar]
- Segurado M., Tercero J. A., 2009. The S-phase checkpoint: targeting the replication fork. Biol. Cell 101: 617–627. [DOI] [PubMed] [Google Scholar]
- Shellman Y. G., Schauer I. E., Oshiro G., Dohrmann P., Sclafani R. A., 1998. Oligomers of the Cdc7/Dbf4 protein kinase exist in the yeast cell. Mol. Gen. Genet. 259: 429–436. [DOI] [PubMed] [Google Scholar]
- Sheu Y. J., Stillman B., 2010. The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature 463: 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T., Ogino K., Tatebayashi K., Ikeda H., Arai K., et al. , 2001. Regulation of initiation of S phase, replication checkpoint signaling, and maintenance of mitotic chromosome structures during S phase by Hsk1 kinase in the fission yeast. Mol. Biol. Cell 12: 1257–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travesa A., Kuo D., de Bruin R. A., Kalashnikova T. I., Guaderrama M., et al. , 2012. DNA replication stress differentially regulates G1/S genes via Rad53-dependent inactivation of Nrm1. EMBO J. 31: 1811–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji T., Ficarro S. B., Jiang W., 2006. Essential role of phosphorylation of MCM2 by Cdc7/Dbf4 in the initiation of DNA replication in mammalian cells. Mol. Biol. Cell 17: 4459–4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullman E. F., Kirakossian H., Singh S., Wu Z. P., Irvin B. R., et al. , 1994. Luminescent oxygen channeling immunoassay: measurement of particle binding kinetics by chemiluminescence. Proc. Natl. Acad. Sci. USA 91: 5426–5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Byeon I. J., Liao H., Beebe K. D., Yongkiettrakul S., et al. , 2000. II. Structure and specificity of the interaction between the FHA2 domain of Rad53 and phosphotyrosyl peptides. J. Mol. Biol. 302: 927–940. [DOI] [PubMed] [Google Scholar]
- Weinert T. A., Kiser G. L., Hartwell L. H., 1994. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 8: 652–665. [DOI] [PubMed] [Google Scholar]
- Weinreich M., Stillman B., 1999. Cdc7p-Dbf4p kinase binds to chromatin during S phase and is regulated by both the APC and the RAD53 checkpoint pathway. EMBO J. 18: 5334–5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P., Diffley J. F., 2010. Checkpoint-dependent inhibition of DNA replication initiation by Sld3 and Dbf4 phosphorylation. Nature 467: 474–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Chabes A., Domkin V., Thelander L., Rothstein R., 2001. The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J. 20: 3544–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.