Abstract
SUMMARY
The prokaryotic translation elongation factors were identified as essential cofactors for RNA-dependent RNA polymerase activity of the bacteriophage Qβ more than 40 years ago. A growing body of evidence now shows that eukaryotic translation elongation factors (eEFs), predominantly eEF1A, acting in partially characterized complexes sometimes involving additional eEFs, facilitate virus replication. The functions of eEF1A as a protein chaperone and an RNA- and actin-binding protein enable its “moonlighting” roles as a virus replication cofactor. A diverse group of viruses, from human immunodeficiency type 1 and West Nile virus to tomato bushy stunt virus, have adapted to use eEFs as cofactors for viral transcription, translation, assembly, and pathogenesis. Here we review the mechanisms used by viral pathogens to usurp these abundant cellular proteins for their replication.
INTRODUCTION
Human eukaryotic translation elongation factor 1A (eEF1A) is a protein subunit of the eukaryotic translation elongation 1 complex (eEF1), which in metazoans is composed of eEF1A, valyl-tRNA, and the eEF1B complex, comprising eEF1G, eEF1B, and eEF1D. In addition to eEF1A's canonical role in translational elongation by ribosomes, eEF1A has a growing list of functions outside protein synthesis, including protein degradation (1, 2), cellular apoptosis (3) (4), nucleocytoplasmic trafficking (5–7), heat shock (8), and multiple aspects of cytoskeletal regulation (reviewed in reference 9), highlighting its importance in diverse cellular processes (reviewed in reference 10). As an abundant, multifunctional protein, it is not surprising that eEF1A has been subverted by viruses. Studies indicating important roles for eEF1A in virus replication, especially that of RNA viruses, by a variety of different mechanisms, are rapidly accumulating. Here we review current hypotheses of how eEF1A and other eEF1 subunits facilitate virus replication through complex interactions with viral RNA and proteins, evaluating the evidence for the use of eEFs, predominantly eEF1A, by diverse viruses, including retroviruses, flaviviruses, reoviruses, tombusviruses, bacteriophage Qβ, and others, for their replication.
eEF1 SUBUNITS IN TRANSLATION
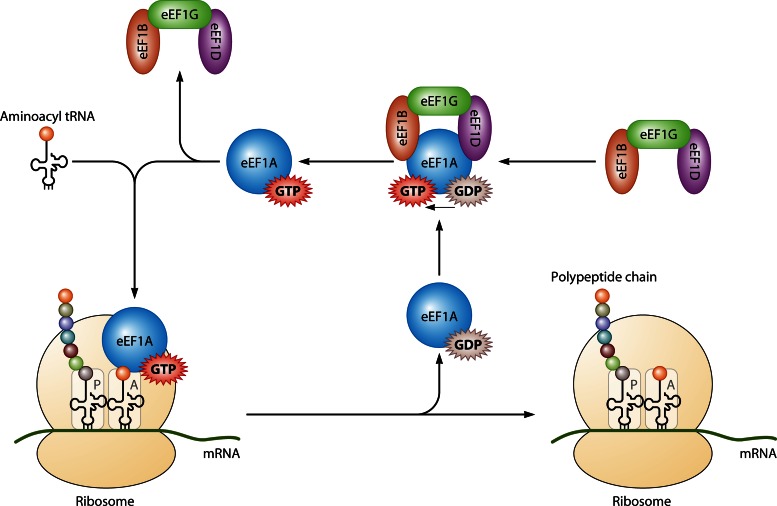
During protein synthesis, eEF1A binds to and delivers aminoacylated tRNAs (aa-tRNAs) to the elongating ribosome (Fig. 1). GTP bound to eEF1A is hydrolyzed upon codon-anticodon match between an aa-tRNA in the A site of the ribosome and mRNA bound to the ribosome. An inactive eEF1A-GDP moiety leaves the ribosome and must be recycled to eEF1A-GTP before binding another aa-tRNA. This GTP exchange process is the function of the nucleotide exchange factor eEF1B complex, which exchanges GDP for GTP to regenerate active eEF1A. The requirement for a guanine nucleotide exchange factor, the eEF1B complex, which in metazoans is composed of the subunits α, δ, and γ (also called eEF1B, eEF1D, and eEF1G, respectively), to reactivate eEF1A is due to the 100-fold higher affinity of eEF1A for GDP than for GTP (reviewed in reference 11). Indeed, in Saccharomyces cerevisiae, although eEF1D is not present and eEF1G is dispensable for viability, deletion of eEF1B is lethal (12). eEF1A has also been described as important for the aminoacylation-dependent tRNA export pathway (reviewed in reference 6).
Fig 1.
Translation elongation (not to scale). Eukaryotic elongation factor 1A (eEF1A; blue circle) in complex with GTP (red star) delivers an aminoacylated tRNA to the A site of the ribosome (yellow). GTP is hydrolyzed when codon-anticodon recognition occurs, and eEF1A-GDP is released from the ribosome. The GTP exchange factor for eEF1A is a complex with three subunits: eEF1B is shown in orange, eEF1G in green, and eEF1D in purple. This complex promotes the exchange of the bound GDP for GTP to regenerate active eEF1A-GTP.
eEF1A ISOFORMS AND STRUCTURE
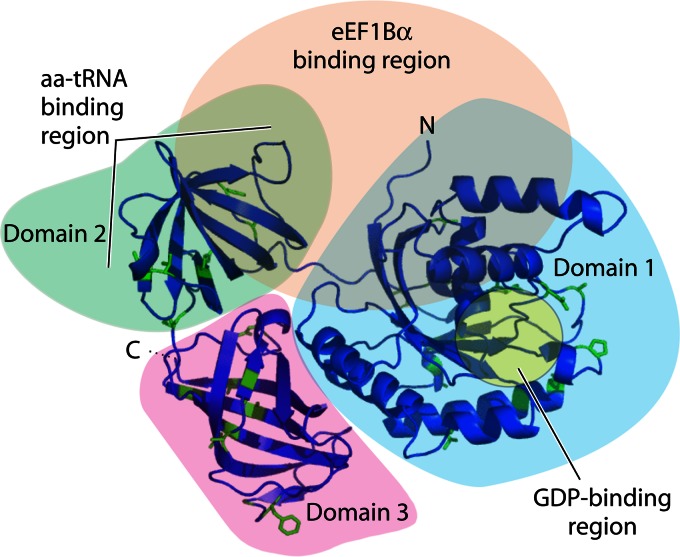
eEF1A is a highly abundant molecule that makes up 3% of the total cellular protein. There are two isoforms of eEF1A, eEF1A1 and eEF1A2, that share 92% amino acid identity (13, 14) (Fig. 2). Structural models of eEF1A1 and eEF1A2 indicate subtle differences in protein surface variant amino acid residues potentially affecting posttranslational modifications, and thus influencing putative functions (15). In contrast to the ubiquitous expression of eEF1A1 in many cell types, eEF1A2 expression is limited to the terminally differentiated cells of the brain, heart, and skeletal muscle (16). The appearance of eEF1A2 in tissues in which the variant is not normally expressed can be coupled to cancer development (17, 18). Deletions of eEF1A in lower organisms (e.g., in yeast) are lethal (19), and while this has not been tested directly, it can be assumed that eEF1A1 is also essential in higher organisms. Deletion of eEF1A2 in mice gives rise to postnatal lethality at the time when eEF1A1 is no longer expressed in muscle and neurons (20). eEF1A proteins have molecular masses of around 50 kDa and have three distinct domains (Fig. 2): domain I (amino acid residues 1 to 240), domain II (amino acid residues 241 to 336), and domain III (amino acid residues 337 to 443) (15). Domain I contains a helix that binds to GTP/GDP, domains I and II bind to the eEF1B complex for nucleotide exchange, domains II and III contain the aa-tRNA binding site, and domain III is an actin bundling domain (21, 22).
Fig 2.
Cartoon representation of a model structure for human eEF1A1 derived from models of the yeast eEF1A homolog. The locations of the C and N termini and domains 1 (blue shading), 2 (green shading), and 3 (pink shading) are indicated. Variant amino acid residues in eEF1A1 compared to eEF1A2 are colored green. The region in domain 1 involved in GDP binding is highlighted in yellow. The aa-tRNA binding region in domain 2 and the eEF1B binding region (peach shading), equivalent to the yeast eEF1Bα homolog, are indicated. (Reprinted from reference 15, which was published under a Creative Commons license.)
GENERAL FEATURES OF RNA VIRUSES: OBLIGATE INTRACELLULAR PARASITES
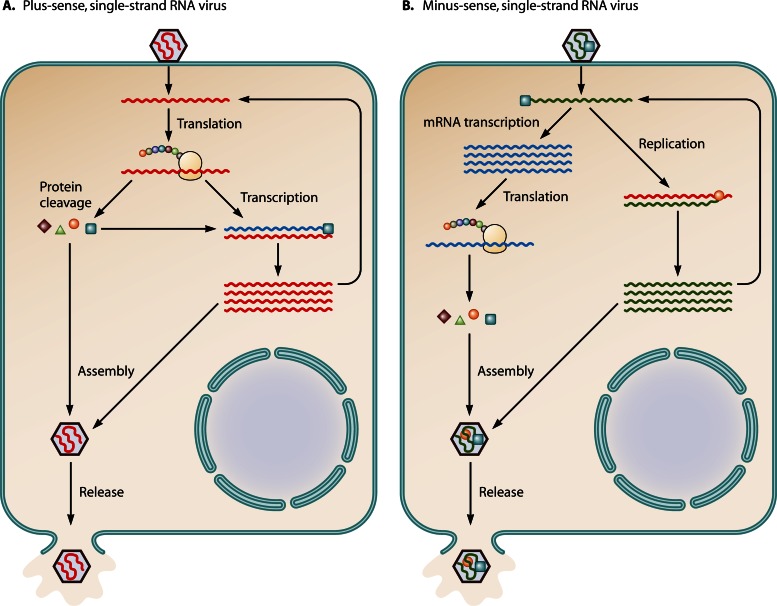
RNA viruses can be highly pathogenic to humans, animals, plants, and even bacteria. The viral RNA genome may be either a double strand or single strand of RNA. Viruses of the latter type are further classified according to the sense or polarity of the RNA into negative (or minus)- or positive (or plus)-sense RNA viruses. Positive-sense viral RNA is similar to mRNA and thus can be translated directly into protein by the cellular translation apparatus (Fig. 3A). Negative-sense viral RNA is complementary to mRNA and must be transcribed into positive-sense RNA by an RNA polymerase before translation (Fig. 3B). Viral RNA genomes are exceptionally small compared to the host genome, and they contain only limited genetic information. In order to complete the virus life cycle, all viruses, including RNA viruses, require host cellular machineries such as those for protein translation, mRNA transcription, membrane biogenesis, and many more functions. As an abundant, multifunctional cellular protein, eEF1A and other subunits of the eEF1 complex have been reported to play important roles in RNA virus replication.
Fig 3.
Replication strategies of plus- and minus-strand RNA viruses. (A) The plus-sense RNA genome is the same sense as mRNA and can be translated using the cellular machinery when it is uncoated in the cell to produce the single polyprotein (represented by West Nile virus, a single-stranded, plus-sense RNA virus). The polyprotein is then processed into each individual protein. The newly made viral polymerase protein, RNA-dependent RNA polymerase, copies the plus-sense genomic RNA into complementary minus-sense RNA. It is unclear how the viral translation switches to transcription using the same genomic RNA as the template. New minus-sense strands serve as the template for new plus-sense strand synthesis. The nascent plus-sense RNA can be the new mRNA for translation, a template for replication, or packaged. (B) The minus-sense RNA genome (represented by VSV) must be transcribed into mRNAs before translation takes place, while the genomic RNA can also be transcribed into a full-length plus-sense RNA that will be the template for nascent minus-sense genomic RNA replication. Transcription is performed by the viral polymerase, which is packaged in the virion. For some minus-strand RNA viruses, for example, influenza virus, primary transcription and replication of viral RNA occur in the nucleus.
eEF1A BINDS TO VIRAL RNA
Since GTP-charged eEF1A can bind aminoacylated tRNAs, among other mammalian RNA species (8), it is not surprising that eEF1A can also bind to various RNA structures found in some untranslated regions (UTRs) of viral genomes. Many elegant studies have revealed how viruses use the RNA binding ability of eEF1A to regulate their life cycle.
eEF1A1 Binds to a tRNA-Like Structure at the 3′UTR in the Genomic RNAs of Some Plant Viruses
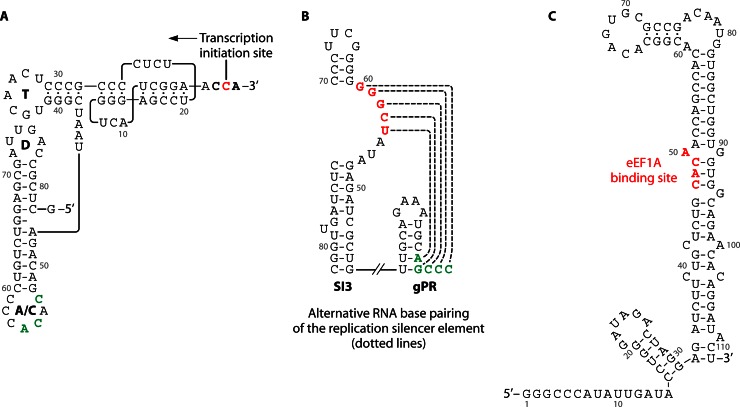
Seminal studies of the single-stranded RNA bacteriophage Qβ demonstrated the unexpected outcome that translation elongation factors associated with the bacteriophage-encoded RNA-dependent RNA polymerase (RdRp), the Qβ replicase (reviewed in reference 23). Many early investigations of bacteriophage RNA noted cloverleaf-shaped tRNA-like structures (TLSs) at the 3′ ends of RNA phages, suggesting that EF-Tu, the prokaryotic homolog of eEF1A, might bind to an RNA structure to facilitate phage replication. Subsequently, eEF1A1 was shown to bind aminoacylatable TLSs in the 3′UTRs of different plant RNA viral genomes (reviewed in reference 24). For example, turnip yellow mosaic virus (TYMV), tobacco mosaic virus (TMV), and brome mosaic virus (BMV) genomes contain a structural RNA element similar to tRNA (Fig. 4A). These TLSs can be aminoacylated by aminoacyl-tRNA synthetases and have important roles in virus replication, such as enhancement of virus protein translation (25), repression of virus minus-strand RNA synthesis in an in vitro system (26), and facilitation of virus RNA encapsidation (27, 28). The interaction of eEF1A with aminoacylated viral RNA so as to enhance viral protein translation while repressing viral minus-strand RNA synthesis has been proposed as a mechanism to regulate virus protein translation and genomic RNA replication (26, 29). Matsuda et al. showed that the TLS of TYMV can be valylated, forming a stable complex with eEF1A-GTP to enable efficient translation. The study showed that mutations of the TLS that decrease the affinity for eEF1A and diminish the RNA aminoacylation efficiency strongly decrease the translation efficiency (25). On the other hand, minus-strand RNA transcription from the valylated positive-strand RNA template can be repressed strongly when eEF1A-GTP is bound (26). Other viral RNA structures may operate independently of the TLS. For example, while eEF1A can bind an aminoacylated TLS of TMV, a conserved upstream pseudoknot domain (UPD) located immediately upstream of the TLS in the 3′UTR of the viral genome also bound eEF1A-GTP independently of a TSL structure, and in the absence of aminoacylation (30, 31). A detailed understanding of the mechanisms controlling the balance between viral protein translation and RNA replication remains an important issue.
Fig 4.
eEF1A can bind to various RNA structures usually located at the 3′UTR of viral genomes. (A) TYMV tRNA-like structure. The secondary structure model emphasizes similarities to canonical tRNA, including a CCA 3′-terminal sequence, a 12-bp acceptor/T arm, interacting D and T loops, and an anticodon loop (A/C) with a valine-specific anticodon (CAC). Unlike canonical tRNAs, however, the acceptor stem consists of an RNA pseudoknot. The 3′ CCA sequence (bold) serves as an initiation box controlling minus-strand synthesis directed by TYMV RdRp, which initiates opposite the 3′-most C residue (red). The nucleotides in the anticodon loop constitute the valine identity elements (green) that direct specific valylation of the 3′ end of the RNA by plant valyl-tRNA synthetase. Nucleotides are numbered from the 3′ end (26). (B) Stem-loop structure at 3′UTR of tombusvirus genomic RNA. The highlighted nucleotides GGGCU, termed the replication silencer element, which can bind to the 3′ end of the genome (AGCCC), are proposed eEF1A binding sites (33). (C) Stem-loop structure of 3′UTR of West Nile virus genomic RNA. The eEF1A binding site on the RNA stem-loop structure is shown (red) (35).
eEF1A Binds to an RSE in the SL Region of the 3′UTR of Tombusvirus Genomic RNA
Tomato bushy stunt virus (TBSV) is a member of the Tombusvirus genus in the Tombusviridae family (32). Like many plus-strand RNA viruses, TBSV is noted for producing many more plus-strand than minus-strand RNA molecules during virus replication. While differential promoter efficiencies in the RNA strand could explain this outcome, TBSV has evolved an alternative mechanism using an RNA element in the viral genome, referred to as a “replication silencer” element (RSE), which can downregulate complementary, minus-strand RNA synthesis relative to plus-strand RNA synthesis (33). The RSE is a 5-nucleotide-long RNA “bulge” sequence located within an internal RNA stem-loop (SL) structure called SL3 (Fig. 4B). The RSE is capable of intrastrand base pairing with the 3′ terminus of the RNA genome, which would otherwise function as a promoter, called gPR, for minus-strand RNA synthesis. Several lines of evidence suggest that eEF1A interacts with the TBSV RSE to stimulate minus-strand RNA synthesis: first, RNA mobility shift assays indicate that recombinant eEF1A interacts with wild-type SL3 but not with a mutated SL3 RNA lacking an RSE sequence; and second, eEF1A is present within a highly purified TBSV replicase complex interacting with p33, a viral replicase component (32). The functional relevance of the interactions is supported by yeast models of TBSV replication, in which mutations that downregulate eEF1A guanine exchange factor requirements significantly reduced the synthesis of minus-strand RNA. This was attributed to decreased stability of the p33 TBSV replicase cofactor in cells (32). Interaction of eEF1A with TBSV genomic RNA was shown to stimulate minus-strand RNA synthesis in an in vitro system (34). Further studies indicate that an additional eEF1 factor, eEF1Bγ (the plant homolog of the metazoan protein eEF1G), can also bind TBSV RNA, possibly by forming protein complexes involving eEF1A and the viral replicase to enable virus minus-strand RNA synthesis. A detailed mechanism describing a molecular interaction between the TBSV replicase complex and eEF1 components or, perhaps, other translation factors remains to be determined.
eEF1A Binds to the 3′SL Region of WNV Genomic RNA
West Nile virus (WNV), a mosquito-borne member of the Flavivirus genus within the family Flaviviridae, has a single-stranded, positive-sense RNA genome that is about 10.7 kb long. The conserved SL structure at the 3′UTR was shown to bind eEF1A specifically (35). The apparent equilibrium dissociation constant for the interaction between eEF1A and WNV 3′ SL RNA was calculated to be 1.1 nM, which is similar to that for the interaction between eEF1A and individual charged tRNA molecules. eEF1A has conserved tyrosine, threonine, and serine residues that are phosphorylated (discussed in reference 15), and dephosphorylation of eEF1A by calf intestinal alkaline phosphatase inhibited binding of eEF1A to WNV 3′SL RNA. Four nucleotides (CACA) at the stem region, which in part form a small bulge in SL, were found to be critical for this binding (Fig. 4C). Mutations of these nucleotides in infectious clones resulted in significantly reduced minus-strand RNA transcription in BHK or C6/36 cells but did not affect the translation efficiency of the viral polyprotein from the genomic RNA. Two lines of evidence suggest that the eEF1A association with WNV RNA is functionally important. First, there is a positive correlation between mutations that decrease eEF1A binding to the 3′ SL RNA in vitro and viral minus-strand RNA synthesis in infected cells. Second, a mutation that increases the efficiency of eEF1A binding to the 3′ SL RNA also increases minus-strand RNA synthesis in transfected cells, resulting in decreased synthesis of genomic RNA. These results strongly suggest that the interaction between eEF1A and the WNV 3′ SL facilitates viral minus-strand synthesis (36). The role played by eEF1A in minus-strand synthesis may be conserved in Flavivirus replication, as eEF1A was shown to bind with similar efficiency to the 3′-terminal SL RNAs of four divergent Flavivirus members, including tick-borne encephalitis virus, dengue virus, and yellow fever virus (36, 37).
eEF1A Binds to RNA Structures Outside the 3′UTR in Some Viral Genomes
Hepatitis delta virus (HDV) is a small human pathogen consisting of a single-stranded RNA molecule. The small, 1,680-nucleotide genome of HDV encodes only two proteins, the large and small hepatitis D antigens (HDAg-L and HDAg-S), and its replication is completely dependent on coinfection with hepatitis B virus for production of the virus structural proteins. Sikora et al. showed that eEF1A could bind to a region of the HDV RNA genome called the right terminal stem-loop domain (38). The right terminal stem-loop domain of the HDV genome is a 199-nucleotide RNA element that contains the proposed initiation site for HDAg mRNA transcription and negative-strand RNA synthesis (39, 40). Three approaches were used to identify eEF1A as one of a few cellular proteins specifically binding to HDV RNA (the other proteins were p54nrb, hnRNPL, glyceraldehyde-3-phosphate dehydrogenase [GAPDH], and ASF/SF2): first, isolation of a UV-cross-linked ribonucleoprotein (RNP) complex and mass spectrometry to identify RNP proteins; second, RNA affinity chromatography using HDV RNA as a ligand; and third, screening of a library of purified RNA binding proteins and confirmation of positive candidate proteins by coimmunoprecipitation (38). However, direct evidence that eEF1A affects HDV transcription and minus-strand RNA synthesis remains to be demonstrated. eEF1A was also shown to interact with the 5′-terminal 110 nucleotides of the polioviral RNA genome, an RNA segment whose secondary structure resembles a cloverleaf and which recruits the poliovirus proteinase 3CDpro. This interaction may be necessary for virus replication, as genetic mutations that disrupt the RNA cloverleaf structure inhibit 3CDpro RNA binding and virus replication (41, 42).
These studies demonstrate that for a variety of different RNA viruses, eEF1A can play a pivotal role in viral RNA replication through direct binding to RNA elements. Often this is through binding to the 3′ UTR of the viral genome. It is likely that eEF1A can recruit or somehow stabilize the viral replicase complex, most likely in conjunction with other cellular factors. Further analysis of viral replicase complex proteins and their interactions with host cell factors will be required to confirm this possibility. In addition, the interaction of eEF1A with virus genomic RNAs and the difference between this interaction and that of eEF1A with tRNA would be worth further investigation. Such structure-based studies would provide insight into the critical interactions of eEF1A with RNA and into subsequent changes in protein conformation (43).
THE SUBUNITS OF eEF1 INTERACT WITH VIRAL POLYMERASES AND OTHER PROTEINS
The subunits of eEF1 have been reported to affect the replication of a variety of viruses from several families through interactions with viral proteins. One predominant finding is the ability of eEF1A to interact with viral polymerase proteins.
Subunits of eEF1, such as eEF1A, Interact with Viral Polymerase Proteins and Support the Formation of Virus Replication Complexes (RCs)
A requirement for translation elongation factors in virus polymerase activity and polymerase complex formation was first identified for the Qβ bacteriophage (44). EF-Tu and EF-Ts (the Escherichia coli counterparts of eukaryotic eEF1A and the eEF1B complex) were copurified and determined to be integral cofactors in Qβ replicase activity. Qβ replicase contains a virally encoded RdRp, the β-subunit, whose activity was lost or regained when EF-Tu and EF-Ts were removed or restored, respectively. Transcription of RNA by RNA polymerases is generally biphasic, involving an initiation reaction by an RdRp followed by elongation of the nascent RNA strand. It was demonstrated that the Qβ-replicase complex, which also includes Hfq and the ribosomal protein S1 (45, 46), was capable of efficient RNA elongation upon removal of EF-Tu and EF-Ts, leading to the conclusion that EF-Tu and EF-Ts are important for initiation of RNA transcription by Qβ replicase (23, 47). Recently, structural studies have shown that hydrophobic interactions between the β-subunit and EF-Tu and -Ts are required for the maintenance of the β-subunit catalytic core crevasse of Qβ replicase and for the expression and assembly of the complex in infected cells (48, 49) (see eEF1A Works in a Viral Protein Complex). A similar study by another group further demonstrated that the hydrophobic interactions of the β-subunit and EF-Tu and -Ts take place between the finger and thumb domains of the β-subunit and domain 2 of EF-Tu and the coiled-coil motif of EF-Ts, respectively (50), thereby potentially greatly increasing the stability and efficiency of the replicase complex.
The interaction of eEF1A with viral RdRps has been studied extensively in plant RNA viruses such as TMV. TMV is a single-stranded, positive-sense RNA virus belonging to the genus Tobamovirus in the family Virgaviridae. Studies have shown that the plant eEF1A homolog interacts with both subunits of the active TMV RdRp, the 126-kDa and 183-kDa proteins, called 126K and 183K, respectively (51). The 183K subunit is made by readthrough of the amber termination codon of 126K. The 126K and 183K subunits have methyltransferase (M), helicase (H), and internal (I) domains, while 183K has a polymerase (P) domain as well. Although eEF1A can bind to the 3′UTR of TMV genomic RNA (31), its interaction with the RdRp is independent of viral RNA (51). The binding of TMV RdRp and eEF1A requires the RdRp M domain, and downregulating the levels of eEF1A in plants by gene silencing resulted in dramatically reduced levels of TMV RNA in infected plants and decreased the spread of infection, indicating that the interaction is biologically relevant (52). It is worth noting that TMV RdRp complexes are associated with membrane structures (53–55). Similarly, eEF1A was also reported to interact with the RdRp of turnip mosaic virus (TuMV), a member of the family Potyviridae (56). Following TuMV infection, eEF1A [as well as the translation factors eIF4E and poly(A)-binding protein] was significantly enriched in membrane fractions of plant lysates and could be copurified with TuMV RdRp by a tandem affinity isolation method. It is possible that association with cellular membranes and binding of eEF1A by viral RdRp are features common to RNA viruses, such as Flavivirus members, that replicate in specialized virus-induced cellular convoluted membranes/paracrystalline arrays and vesicle packets (57, 58).
Elegant studies by Li et al. used a powerful yeast model system to identify eEF1A mutations that could negatively affect TBSV replication. In these two studies, they described an interaction between eEF1A and the TBSV p33 replicase component that results in its increased stability, and they found that eEF1A also binds to the TBSV p92pol replication protein (32, 34). They showed that overexpression of an eEF1A mutant (T22S) could result in a reduced half-life of p33 and significantly lowered TBSV RNA replication. Importantly, this was not attributable to defects in protein translation by the eEF1A mutant, because cellular levels of the viral RdRp p92pol were not affected. Exactly how eEF1A supports p33 stability in cells and if eEF1A protects p33 from protein degradation are unclear. Li et al. also identified several eEF1A mutations that substantially increased viral minus-strand RNA synthesis, which they showed was due to increased replicase activity and, again, was not due to a general effect on translation. Using methods that reconstituted replicase activity, they demonstrated that chemical inhibitors of eEF1A, didemnin and gamendazole, which act on eEF1A by unique mechanisms, both inhibited replicase activity in vitro (34). By comparing the levels of abortive viral transcripts, i.e., from very short RNA transcripts to elongated RNA products, they demonstrated that eEF1A affected only the initiation of minus-strand transcription. They concluded that eEF1A acted primarily to recruit and stabilize the viral RdRp complex, indicating that an eEF1A chaperone-like function was important to support virus replication (2, 59).
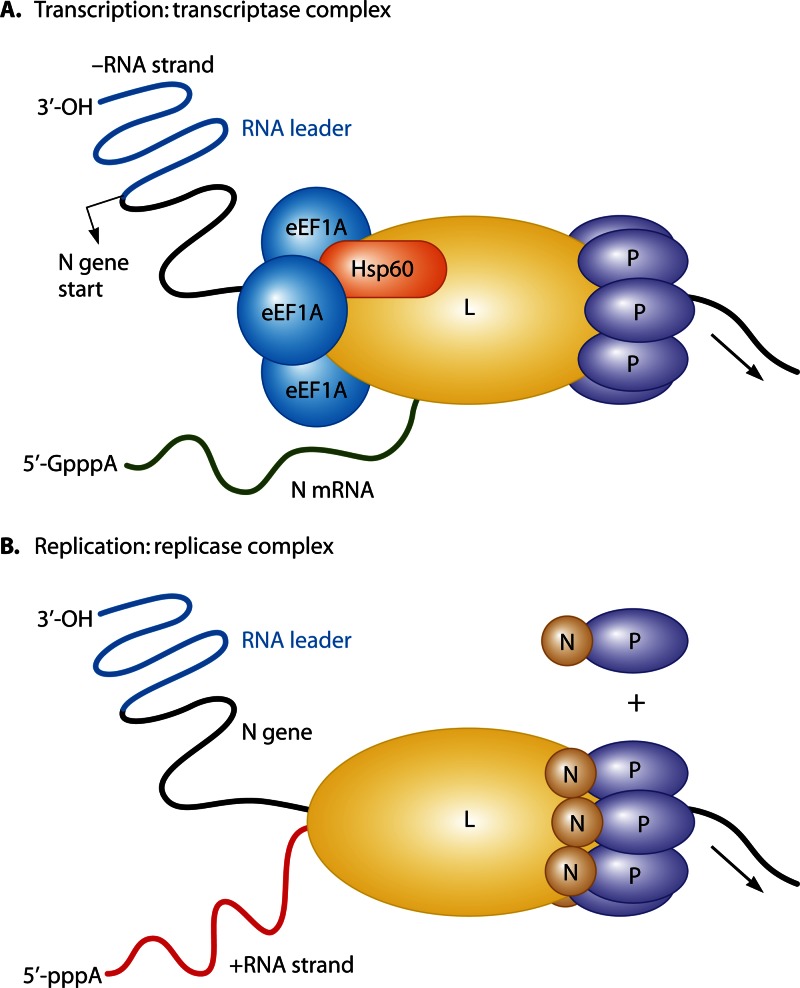
eEF1A and other subunits of eEF1 have been reported to interact with viral polymerases from a variety of animal viruses. An early study by Banerjee's group found that eEF1A was tightly associated with the RNA polymerase large protein (L protein) of vesicular stomatitis virus (VSV), a prototypic nonsegmented negative-strand RNA virus of the Vesiculovirus genus in the family Rhabdoviridae, and that all the subunits of the eEF1A complex, i.e., α, β, and γ (corresponding to eEF1A1, eEF1B, and eEF1D, respectively), were necessary for L protein activity (60). A later study from the same group isolated two different highly purified RNA polymerase complexes by immunoaffinity chromatography (61). The group proposed that VSV forms two different viral RNA polymerase complexes in infected cells: the transcriptase and the replicase (Fig. 5). The role of the transcriptase complex is to synthesize capped mRNAs by initiating transcription at the first gene (N) start site, whereas the replicase complex initiates genomic minus-strand RNA synthesis at the precise 3′ end of the plus-strand RNA. The transcriptase was described as a protein complex containing the virus-encoded RdRp (called L) and phosphoprotein (called P), the cellular proteins eEF1A and heat shock protein 60, and a submolar amount of cellular mRNA cap guanylyltransferase. The replicase complex was described as containing the viral proteins L, P, and nucleocapsid (N) but not eEF1A, heat shock protein 60, or the guanylyltransferase. Hence, it was proposed that eEF1A is important for transcription of viral mRNAs but not for genomic RNA replication in VSV.
Fig 5.
eEF1A is a component of the VSV transcriptase complex. The schematic is based on proposed models of VSV replicase and transcriptase complexes (61, 74). (A) Transcription of the VSV minus-strand RNA by the transcriptase complex. The minus-strand RNA includes an RNA leader region (blue) followed by the N gene (black) transcription start site. The viral transcriptase, composed of the VSV L and P proteins and host cellular factors eEF1A and Hsp60, synthesizes viral mRNA (green). (B) The first step of virus RNA replication is shown. The viral replicase complex is composed of the L protein and N-P complex proteins that initiate transcription from a promoter located in the RNA leader.
With respect to members of the Flaviviridae family, investigation of WNV indicates that eEF1A plays an important role in minus-strand RNA synthesis through interaction with the viral replication complex. As previously discussed (see “eEF1A Binds to the 3′ SL Region of WNV Genomic RNA”), eEF1A has been reported to bind WNV RNA SL structures, an observation supported by mutational analysis of RNA binding, which downregulates eEF1A binding to RNA and sharply inhibits replication of WNV. Importantly, eEF1A was found to colocalize with WNV and dengue virus NS3 (a viral protease/helicase) and NS5b (the viral RdRp) proteins in infected cells, and both NS3 and NS5 could be coimmunoprecipitated using an anti-eEF1A antibody (36). The combined evidence suggests that eEF1A is important for minus-strand RNA synthesis through interactions with viral RNA and the replication complex proteins, including NS3 and NS5. This is similar to the model proposed by Li et al. for minus-strand RNA synthesis by TBSV (34).
During HIV-1 DNA synthesis by reverse transcription, in vitro studies demonstrated that eEF1A supports significantly improved late HIV-1 DNA synthesis but does not affect the initiation of the reaction (62). While the precise mechanism remains to be elucidated, studies from our group, using both in vitro and cell-based approaches, demonstrated that HIV-1 reverse transcriptase (RT) associated with eEF1A. eEF1A and eEF1G were copurified with RT from reverse transcription complexes (RTCs) by gradient centrifugation and colocalized with RT following infection of cells (62). Importantly, HIV-1 reverse transcription was inhibited when target cells were pretreated with a small interfering RNA (siRNA) that downregulated steady-state levels of eEF1A or eEF1G, an outcome that was attributed to the instability of the viral RTCs in siRNA-treated cells.
Interactions Involving eEF1A and Other Viral Structural and Nonstructural Proteins
Table 1 shows eEF1A viral interaction proteins other than RNA polymerases. Using a yeast two-hybrid screen, the bovine viral diarrhea virus (BVDV) NS5A protein was identified as interacting with eEF1A (63). BVDV is a positive-sense single-stranded RNA virus of the genus Pestivirus in the Flaviviridae family. The eEF1A-NS5A interaction, which was confirmed using in vitro methods, was observed for NS5As from all BVDV strains that were tested. However, NS5A of hepatitis C virus (HCV), of the genus Hepacivirus in the Flaviviridae family, was unable to interact with bovine eEF1A (63). One possibility is that an interaction between eEF1A and NS5A is species specific. While the exact role of NS5A in the BVDV replicase remains unclear, it is obviously important for virus replication (71). The BVDV NS5A protein is a large, hydrophilic phosphoprotein of approximately 56 to 58 kDa. Like HCV NS5A, the protein is associated with cellular membranes via an amino-terminal helix that inserts itself into the luminal leaflet of endoplasmic reticulum (ER)-like membranes (72), and the HCV NS5A and BVDV NS5A proteins have similar structures (71, 73). Overall, a biological role for eEF1A in BVDV replication requires further confirmation.
TABLE 1.
eEF1A interacts with viral proteins other than RNA polymerases
| Virus | Virus family | Viral protein interacting with eEF1A | Reference |
|---|---|---|---|
| Bovine viral diarrhea virus | Flaviviridae | NS5A | 63 |
| Hepatitis C virus | Flaviviridae | NS4 | 64 |
| Poliovirus | Picornaviridae | 3CDpro | 65 |
| HIV-1 | Retroviridae | MA, CA | 66 |
| Integrase | 62, 67 | ||
| Acetylated integrase | 68 | ||
| HPV38 | Papillomaviridae | Oncoprotein E7 | 69 |
| HBV | Hepadnaviridae | X protein | 70 |
| TBSV | Tombusviridae | p33 | 34 |
Although HCV NS5A failed to interact with bovine eEF1A, the HCV nonstructural protein 4A (NS4A) was reported to interact with eEF1A in in vitro glutathione S-transferase (GST) pulldown assays (64). HCV NS4A is a small, 54-amino-acid protein that acts as a cofactor of the NS3 serine protease and can inhibit protein synthesis in cells. In this case, the NS4A N-terminal region, amino acid residues 1 to 34, was required to bind eEF1A, while amino acid residues 21 to 34 imparted an inhibitory effect on both cap-dependent and HCV internal ribosome entry site (IRES)-mediated translation. The translation inhibitory effect of NS4A could be relieved by the addition of purified recombinant eEF1A in an in vitro translation system, indicating the role of eEF1A in controlling IRES-mediated HCV translation (64).
An N-terminally derived truncated form of eEF1A was also reported to interact with the 3CDpro protein, the precursor of the mature 3C protease and 3D polymerase of poliovirus, in the Enterovirus genus of the Picornaviridae family (65). Both intact eEF1A and a C-terminally truncated eEF1A, p36, were identified in chromatographically purified proteins assayed by gel shift assay and identified by protein microsequencing. The 5′ region of poliovirus RNA includes a 110-nucleotide region of RNA whose secondary structure resembles a cloverleaf, which can bind to a p36-3CDpro complex but not to 3CDpro alone (65). The 5′ cloverleaf structure is important for several aspects of poliovirus replication, such as preventing viral RNA degradation and minus-strand RNA synthesis (75). While a clear role for p36 in the replication of poliovirus RNA requires confirmation, cellular factors such as p36 are likely to be important for RNA replication (76).
Several HIV-1 structural and nonstructural proteins interact with eEF1A, including the HIV-1 Gag protein. Gag is an HIV-1 precursor protein that is processed by HIV-1 protease into the matrix (MA), capsid (CA), nucleocapsid (NC), and two smaller spacer peptides. eEF1A was identified as an MA binding protein by use of a yeast two-hybrid screen of a library made from human HeLa cells (66), an outcome that was confirmed using complementary in vitro pulldown assays. The interaction required the MA basic region. HIV-1 NC was also identified as an eEF1A binding protein, and the basic amino acid residues, but not the NC zinc finger domains, were required for interaction (66). The interaction was mediated by RNA, and addition of purified MA inhibited in vitro cap-dependent and -independent translation reactions. The significance of these outcomes is unclear, as Gag has not been reported to have similar effects when overexpressed in cells. One possibility is that eEF1A is a very abundant cellular protein, making inhibition difficult in a cellular context. However, HIV-1 Gag is synthesized on cytosolic polysomes, and it was speculated that eEF1A may play a role in releasing viral RNA from polysomes, thereby permitting genomic RNA to be packaged into virions (66). Analysis of additional mutant Gag proteins that do not interact with eEF1A could help to test this hypothesis.
Another HIV-1 protein interacting with eEF1A is integrase (IN). IN is the viral enzyme that catalyzes integration of viral DNA into chromosomes. One study used IN affinity chromatography to identify HSP60, PCK1, CCT4, TEF1α (the yeast homolog of eEF1A), and TEF2 as IN binding proteins (67). Subsequent studies found that HIV-1 IN is acetylated by a cellular histone acetyltransferase, p300, at its C terminus and that this acetylation is necessary for IN function (77) and modulates the interaction with eEF1A (68).
Recently, eEF1A was shown to interact with the HIV-1 Nef protein in in vitro and cell-based assays (78). Amino acid residues 1 to 74 of eEF1A (domain I) were required for interaction with the Nef C-terminal region (amino acid residues 55 to 204). Analysis of nuclear and cytoplasmic cell lysate fractions indicated that Nef regulated the nucleocytoplasmic localization of eEF1A, which is a cofactor for exportin-t-mediated export of tRNA from the nucleus (6). Importantly, this Nef-mediated mislocalization of eEF1A resulted in decreased stress-induced apoptosis in primary human monocyte-derived macrophages (MDMs). This may indicate another important role of eEF1A in HIV-1 replication, and further detailed study is needed. However, the study did not include analysis of MDMs infected with HIV-1 or Nef-deleted HIV-1, which could confirm the biological relevance of this observation.
eEF1A was also reported to interact with the viral oncoproteins E6 and E7 of cutaneous human papillomavirus (HPV) type 38 (69) and with the X protein of hepatitis B virus (HBV) (70). Both HPV and HBV are double-stranded DNA viruses and are from the Papillomaviridae and Hepadnaviridae families, respectively. The interaction of eEF1A with the E6, E7, and HBV X proteins plays a role in the pathogenesis of the respective viruses (see the section on pathogenesis below).
eEF1A AND eEF1G FACILITATE VIRUS RC ASSEMBLY
eEF1A has been shown to be important for virus replication in cells infected with dengue virus (35), West Nile virus (79), bacteriophage Qβ (80), tomato mosaic virus (81), and tomato bushy stunt virus (32), facilitating virus replication complex formation.
In WNV-infected cells, eEF1A was found to colocalize with viral replication complexes (RCs), and antibodies to eEF1A coimmunoprecipitated viral RC proteins (36), suggesting a role for eEF1A in facilitating virus RC formation.
By purifying the reverse transcription complex from HEK293T cells infected with HIV, Warren et al. found reduced levels of HIV reverse transcription products and reverse transcription complexes when eEF1A or eEF1G was downregulated using siRNA (62), showing that eEF1A and eEF1G are crucial for HIV reverse transcription complex formation, stability, and function. However, whether eEF1A and eEF1G act in concert as part of a novel complex or independently during reverse transcription remains to be determined.
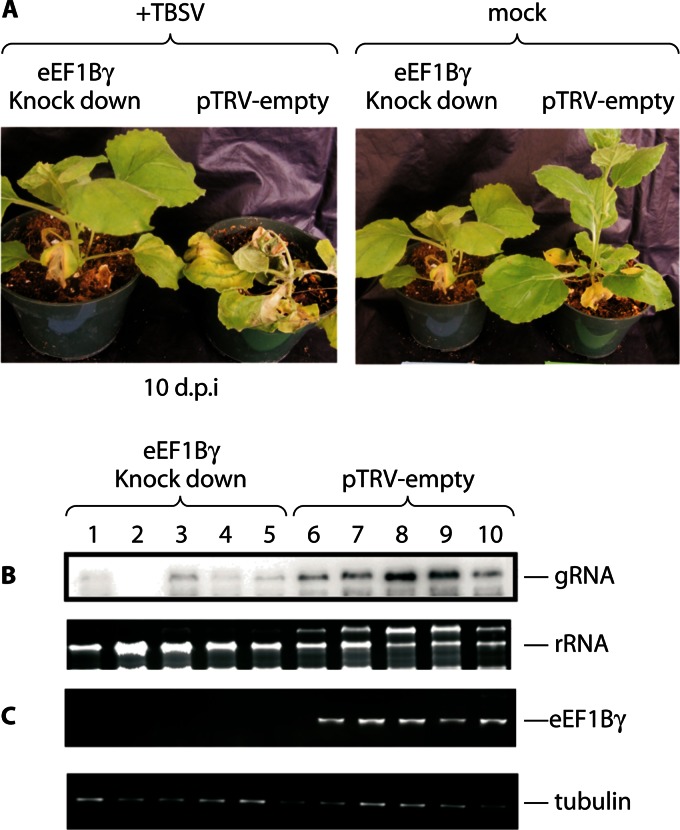
eEF1A has been shown to interact with the components of the TBSV replicase, including the virus genome replication proteins p33 and p92pol (32). These interactions were shown to be important for virus replicase complex assembly and minus-strand RNA synthesis in an in vitro system. Moreover, in the presence of the eEF1A inhibitors didemnin B and gamendazole, the assembly of the viral replicase activity was sharply inhibited. However, the drugs did not inhibit preformed replicase complexes, suggesting that eEF1A is required for replicase assembly rather than RNA synthesis (34). More recently, Sasvari et al. provided strong evidence that eEF1Bγ (the plant homologue of metazoan eEF1G) is a cellular cofactor for TBSV replicase activity (82). For example, downregulation of eEF1Bγ expression in plants inhibited TBSV replication and pathogenesis (Fig. 6A to C), as eEF1Bγ is important for RNA synthesis by the TBSV replicase. The outcomes suggest that eEF1A and eEF1Bγ, in conjunction with other cellular proteins (described in references 83 and 84), act concertedly to affect TBSV RNA synthesis.
Fig 6.
Downregulation of eEF1Bγ expression inhibits RNA virus replication in plants. (A to C) Nicotiana benthamiana infection with TBSV (single-stranded positive-sense RNA virus). (A) Silencing of eEF1Bγ (the plant homolog of metazoan eEF1G) decreased virus replication compared to that in untreated plants. (B) Northern blot analysis of viral genomic RNA (gRNA) levels compared to levels of rRNA. (C) RT-PCR analysis of steady-state RNA levels for eEF1Bγ compared to tubulin. (Adapted from reference 82, which was published under a Creative Commons license.)
The accumulating evidence points to roles for eEF1A and eEF1G in the replication of a variety of viruses, by interacting with viral polymerases, structural and nonstructural proteins, and viral genomic RNA to stimulate or otherwise enable viral RNA or DNA synthesis. In some instances, viral proteins may recruit eEF1A to assist or hinder translation, but further investigation is required to discover whether eEF1A and any other subunits of eEF1 drive replicase or transcriptase complex formation or are passively involved in the process. Clearly, eEF1A and eEF1G are abundant cellular resources that play important roles in viral genomic replication, transcription, or DNA synthesis and may also be required by other viral pathogens.
eEF1A WORKS IN A VIRAL PROTEIN COMPLEX
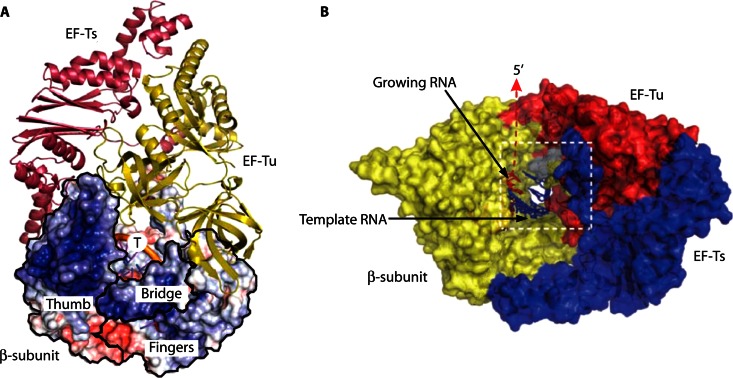
Studies of Qβ bacteriophage replicase showed that the anti-EF-Tu and -EF-Ts association reagents kirromycin and GDP could effectively inhibit Qβ replicase activity (85). Also, EF-Tu cross-linked to EF-Ts is far more effective at renaturation of Qβ replicase activity than untreated EF-Tu and EF-Ts in vitro (86), indicating that the elongation factors act as a complex with Qβ replicase (23). Most recently, a crystal structure of the active Qβ replicase, comprising the β-subunit (RdRp), EF-Tu, and EF-Ts, revealed that the tight binary complex maintains the structure of the catalytic core crevasse of the β-subunit (Fig. 7), thus having a chaperone-like function in the assembly and maintenance of the Qβ replicase structure (48–50).
Fig 7.
Structure of Qβ replicase core complex. (A) Cartoon representation of EF-Tu (yellow) and EF-Ts (magenta) overlaid with a surface representation of the β-subunit (βS) displaying the electrostatic potential of βS. Structural domains of βS, including the thumb, fingers, and bridge, are indicated by black outlines. The position of the RNA template strand (T) is indicated. (B) Surface model of Qβ replicase showing the β-subunit (yellow), EF-Tu (red), and EF-Ts (blue). The viral RNA template (blue) and the growing, nascent RNA (red) are indicated. (Adapted from references 48 and 49 by permission from Macmillan Publishers Ltd.)
The canonical eukaryotic translation elongation factor complex, eEF1, consists in part of two eEF1A molecules bound to the eEF1B complex, which itself comprises up to three proteins: eEF1G, eEF1B, and eEF1D (reviewed in reference 11). Investigation of cellular factors required to stimulate late steps of HIV-1 reverse transcription indicated that the eEF1 complex is important (62, 87, 88). Two components of the eEF1 complex, eEF1A and eEF1G, copurified with the viral RTC and colocalized with HIV-1 RT after infection. However, it is not known whether the HIV-1 RTC associates with an intact eEF1 complex or a novel protein complex involving eEF1A, eEF1G, and other cellular factors.
Other studies also demonstrated that plant and animal viruses require components of the eEF1 complex for replication. For example, the VSV L protein, an RdRp subunit, was shown to require eEF1A, eEF1B, and eEF1G for activity in vitro (60). As previously discussed, the TBSV replicase complex requires both eEF1A and eEF1Bγ (the plant homolog of metazoan eEF1G) for replicase activity in vitro and for virus replication in plants and yeast models of TBSV replicase function (32, 34, 82, 84).
eEF1A FACILITATES VIRUS PARTICLE ASSEMBLY
eEF1A is found in highly purified particles of different viruses from various families, including vesicular stomatitis virus (Rhabdoviridae) (89), vaccinia virus (Poxviridae) (90, 91), cytomegalovirus (Herpesviridae) (92, 93), severe acute respiratory syndrome coronavirus (SARS-CoV) (Coronaviridae) (94), and HIV-1 (Retroviridae) (66, 95). It is critical to determine whether encapsidation into virus particles is simply a consequence of the relative abundance of eEF1A or if encapsidation is biologically important for virus replication.
eEF1A has been detected specifically in purified HIV-1 virions but not in Moloney murine leukemia virus virions (66, 95, 96). eEF1A can be cleaved by HIV-1 protease. Cimarelli and Luban found both full-length eEF1A and a truncated form, with an apparent size of 34 to 36 kDa, in virions (66). Although eEF1A can bind to actin, its incorporation into virions was not inhibited by cytochalasin D, a potent inhibitor of actin polymerization (66, 97), and was dependent on an interaction with HIV Gag. Virion production and tRNA incorporation were sharply reduced using a Gag mutant that was unable to bind eEF1A, while steady-state Gag levels in cells were unchanged (66, 98).
The interactions of eEF1A with viral structural proteins, the virus replication complex, and the cytoskeleton suggest that eEF1A may have an active role in virion assembly and budding (96). However, direct evidence for an active role for eEF1A in assembly or budding and for the incorporation of eEF1A into different viruses by specific mechanisms remains to be provided.
ROLES OF eEF1A IN VIRUS PATHOGENESIS OF BOTH RNA AND DNA VIRUSES
As discussed above, eEF1A assists in the replication of many different viruses and is a necessary factor in viral pathogenesis. However, a “moonlighting” function of eEF1A in actin bundling may be specifically targeted by viruses, leading to cellular abnormalities and transformation. Examples have been described for a few viruses.
HPVs are small double-stranded DNA viruses from the Papillomaviridae family that are capable of causing lesions or tumors by infecting basal keratinocytes. Ghittoni et al. found that approximately a dozen HPV types, including HPV16 and HPV18, are capable of cellular immortalization and transformation to produce tumor cells through expression of two early genes, called E6 and E7 (99). Dong et al. found that E6 and E7 perform the same function in HPV36 (100). Recently, the oncoprotein E7 from HPV36 was reported to induce cellular immortalization and transformation of primary keratinocytes through an interaction with eEF1A (69). eEF1A presents two actin binding sites, one in domain 1 and the other in domain 3 (101). The interaction of HPV38 E7 with domain 3 of eEF1A contributes to decreased binding of eEF1A to actin, causing disruption of actin stress fiber formation and impaired actin cytoskeleton remodeling (69), a key event associated with tumorigenesis.
The HBV-encoded X protein is a multifunctional regulatory protein that modulates not only HBV genes but also a number of host genes involved in cell proliferation, cell cycle progression, genetic stability, and apoptosis (102, 103). eEF1A was identified as an X protein binding partner in HBV-infected hepatocytes. In Huh-7 hepatoma cells, the X protein inhibits eEF1A1 dimer formation, an event shown to be important for the role of eEF1A in filamentous actin bundling (70). The role of dimerization of eEF1A in terms of protein synthesis is unknown. However, considering the important role of the actin cytoskeleton in cell shape, cell division, adhesion, motility, signal transduction, and protein sorting (104), the interference of X protein with eEF1A and F-actin may be a crucial factor in HBV pathogenesis.
Coronaviruses (CoVs) are among the largest of the single-stranded positive-sense RNA viruses, with about 30,000 nucleotides. CoVs, including the human pathogen SARS-CoV, are etiological agents of serious respiratory and enteric diseases in humans and animals. A screen of a yeast two-hybrid human protein library with the SARS-CoV N protein as bait identified eEF1A as a binding partner, a result confirmed using several in vitro and cell-based assays (105). The N protein is a highly phosphorylated, basic RNA binding protein (106) which is important for viral RNA packaging, viral RNA transcription, translation, and budding. Using a coimmunoprecipitation method with epitope-tagged eEF1A and green fluorescent protein (GFP) or a GFP-N fusion protein, a sharp increase in high-molecular-weight eEF1A aggregates in cell lysates was observed. Expression of the N protein in cells had significant negative effects on translation, cytokinesis, and cell proliferation, which arguably may be connected to N protein effects on the role of eEF1A in actin bundle formation and on its canonical role in protein synthesis (105).
A recent paper showed that eEF1A can be found in a stoichiometric complex with activation-induced cytidine deaminase (AID) in chicken and mammalian cells (107, 108). The central role of AID is to target immunoglobulin loci for rearrangement during development, but as it is a potent mutator, it is essential that it be kept in the cytoplasm when it is not playing a role in antibody diversity. This cytoplasmic retention is brought about at least partially through the action of eEF1A. In some circumstances, AID is an antiviral agent (109) that can be induced by CD40 ligand incorporated into the HIV envelope (110). We note that another member of the AID family of proteins, APOBEC3, has been shown to have broad antiretroviral activities (111). The significance of the interaction of eEF1A and AID warrants further investigation. For instance, is it possible that an eEF1A-AID interaction is enhanced or inhibited by viral infection?
SURPRISES AND SPECULATIONS
More than 40 years ago, seminal reports demonstrated a role for prokaryotic translation elongation factors in Qβ RdRp activity (112–114). Blumenthal and Carmichael commented that the discovery was “originally perceived as bizarre” (23). Today, the discovery of host and virus protein interactions involves very sensitive detection methods, and too often cellular proteins such as eEF1A, representing ∼3% of total cellular protein, are labeled as possible cocontaminants. However, viruses must use host factors for their replication, and many use eEF1A. It is very likely that the list of viruses using the eEF1 complex or its subunits will continue to grow. Many of eEF1A's “moonlighting” roles fit well into virus replication strategies. eEF1A acts as a protein chaperone, an RNA binding protein, and an actin binding protein, matching the requirements of virus replication for many viruses. Perhaps the ability of eEF1A to interact with diverse cellular proteins in different pathways underlies its utility in virus replication.
Evidence is accumulating that eEF1A1 is not the only cellular factor subverted for viral RNA transcription and replication. For example, both nucleophosmin (a nuclear/nucleolar trafficking protein; also called B23) (115) and HAX1 (a protein with antiapoptotic properties) (116) colocalize with viral RNA and are important for viral RNA transcription and/or replication in the nucleus. In this case, eEF1A has not been implicated as an influenza virus RNA replication cofactor, although it is interesting that eEF1A, while predominantly cytoplasmic, is also found in the nucleus. It is quite possible that viral strategies to increase the efficiency of viral RNA transcription and replication in eEF1A1-independent mechanisms occur.
Another important question is whether the identification of other translation factors, such as the eEF1B subunits, as proteins subverted by the viral machinery sheds any light on the role played by eEF1A. Factors such as eEF1G have fewer, if any, noncanonical roles ascribed to them, and there is no evidence that eEF1A is associated with its cofactors when participating in noncanonical functions. Indeed, the first clue that eEF1A might have additional roles was the observation that it is present in excess over other translation factors. If a viral function uses more than one elongation factor, as is the case for HIV RT and the TBSV replicase, is this because the two factors are in a complex anyway and are both abundant? Disentangling the canonical and noncanonical roles of eEF1A and the extent to which each contributes to viral function is essential if eEF1A is to be targeted therapeutically. There are already a number of agents that target eEF1A, but clearly drugs that can disturb the viral subversion of eEF1A without affecting its crucial cellular role in protein synthesis would be essential. Given that the eEF1 complex is essential for cellular metabolism, targeting it as an antiviral strategy will be challenging, but precise targeting of specific domains may be a promising avenue for further research.
ACKNOWLEDGMENTS
This work was supported by a grant from the National Health and Medical Research Council to D.H.
We thank Kylie Warren for graphical support and Haran Sivakumaran for helpful advice.
Biographies

Dongsheng Li is a research virologist with the Queensland Institute of Medical Research (QIMR) in Brisbane, Australia. He received his Ph.D. from the School of Molecular and Microbial Science, University of Queensland, Australia, in 2004. He has worked on various human viruses, including hepatitis A, B, C, D, E, and G viruses, human respiratory viruses, dengue virus, and HIV, for more than 20 years, at the Henan Provincial Centre of Disease Control, China, and Monash University, Queensland University of Technology, Australia. His research areas include molecular virology, public health, epidemiology, and viral disease control. He started his work at QIMR in 2011, and his research focus is to characterize cellular factors in HIV replication.

Ting Wei is currently a senior research officer at the Queensland Institute of Medical Research, Australia. She obtained her bachelor's and master's degrees in veterinary medicine in China and a Ph.D. in biological science at the University of Auckland, New Zealand. During the past 20 years, she has worked in different areas of host and pathogen interaction, disease diagnosis and control, and technology development, in different countries, including the China Animal CDC, Hong Kong University, the National University of Singapore, the University of Auckland, Investigation & Diagnostic Centre, and Biosecurity of New Zealand, Ministry of Agriculture and Forestry, New Zealand. She has published 35 papers and 2 books. Her research interests include virology, molecular biology, cell biology, and disease control. Her current research is focused on investigating molecular mechanisms and pathways of HIV replication and development of inhibitors thereof.

Cathy Abbott is Professor of Mammalian Molecular Genetics at the University of Edinburgh. She did her Ph.D. work at the MRC Mammalian Genetics Unit at Harwell, jointly with biochemistry studies at the University of Reading. After a postdoc and lectureship at University College London, she moved to the MRC Human Genetics Unit in 1993 and then to the University of Edinburgh in 1996, to the newly opened Molecular Medicine Centre. There she carries out research on the molecular genetics of translation elongation factors and their roles in motor neuron disease and cancer. She is the Director of Postgraduate Studies for the School of Molecular Genetics and Population Health Sciences and Director of the IGMM Graduate School and is on the board of Medical Research Scotland.

David Harrich received a Ph.D. in experimental pathology from the University of California, Los Angeles, in 1994, focusing on human immunodeficiency virus type 1. After postdoctoral training at the University of Texas Southwestern Medical Center, he was appointed Head of the HIV Unit at the Sir Albert Sakzewski Virus Research Centre in Brisbane, Australia, in 1997. In 2002, he was appointed Lab Head of the HIV Molecular Virology Laboratory at the Queensland Institute of Medical Research (QIMR), and he was appointed Group Leader of Molecular Virology at QIMR in 2011. He also holds an appointment as an Australian Research Council Future Fellow. The impact and death toll of the HIV/AIDS pandemic inspired his interest in HIV-1 replication, and his work focuses on host cell and viral protein interactions, an interest he has pursued for over 20 years.
REFERENCES
- 1. Gonen H, Smith CE, Siegel NR, Kahana C, Merrick WC, Chakraburtty K, Schwartz AL, Ciechanover A. 1994. Protein synthesis elongation factor EF-1 alpha is essential for ubiquitin-dependent degradation of certain N alpha-acetylated proteins and may be substituted for by the bacterial elongation factor EF-Tu. Proc. Natl. Acad. Sci. U. S. A. 91:7648–7652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hotokezaka Y, Tobben U, Hotokezaka H, Van Leyen K, Beatrix B, Smith DH, Nakamura T, Wiedmann M. 2002. Interaction of the eukaryotic elongation factor 1A with newly synthesized polypeptides. J. Biol. Chem. 277:18545–18551 [DOI] [PubMed] [Google Scholar]
- 3. Ruest LB, Marcotte R, Wang E. 2002. Peptide elongation factor eEF1A-2/S1 expression in cultured differentiated myotubes and its protective effect against caspase-3-mediated apoptosis. J. Biol. Chem. 277:5418–5425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chang R, Wang E. 2007. Mouse translation elongation factor eEF1A-2 interacts with Prdx-I to protect cells against apoptotic death induced by oxidative stress. J. Cell. Biochem. 100:267–278 [DOI] [PubMed] [Google Scholar]
- 5. Khacho M, Mekhail K, Pilon-Larose K, Pause A, Cote J, Lee S. 2008. eEF1A is a novel component of the mammalian nuclear protein export machinery. Mol. Biol. Cell 19:5296–5308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kohler A, Hurt E. 2007. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 8:761–773 [DOI] [PubMed] [Google Scholar]
- 7. Murthi A, Shaheen HH, Huang HY, Preston MA, Lai TP, Phizicky EM, Hopper AK. 2010. Regulation of tRNA bidirectional nuclear-cytoplasmic trafficking in Saccharomyces cerevisiae. Mol. Biol. Cell 21:639–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shamovsky I, Ivannikov M, Kandel ES, Gershon D, Nudler E. 2006. RNA-mediated response to heat shock in mammalian cells. Nature 440:556–560 [DOI] [PubMed] [Google Scholar]
- 9. Kim S, Coulombe PA. 2010. Emerging role for the cytoskeleton as an organizer and regulator of translation. Nat. Rev. Mol. Cell Biol. 11:75–81 [DOI] [PubMed] [Google Scholar]
- 10. Mateyak MK, Kinzy TG. 2010. eEF1A: thinking outside the ribosome. J. Biol. Chem. 285:21209–21213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Le Sourd F, Boulben S, Le Bouffant R, Cormier P, Morales J, Belle R, Mulner-Lorillon O. 2006. eEF1B: at the dawn of the 21st century. Biochim. Biophys. Acta 1759:13–31 [DOI] [PubMed] [Google Scholar]
- 12. Hiraga K, Suzuki K, Tsuchiya E, Miyakawa T. 1993. Cloning and characterization of the elongation factor EF-1 beta homologue of Saccharomyces cerevisiae. EF-1 beta is essential for growth. FEBS Lett. 316:165–169 [DOI] [PubMed] [Google Scholar]
- 13. Ann DK, Moutsatsos IK, Nakamura T, Lin HH, Mao PL, Lee MJ, Chin S, Liem RK, Wang E. 1991. Isolation and characterization of the rat chromosomal gene for a polypeptide (pS1) antigenically related to statin. J. Biol. Chem. 266:10429–10437 [PubMed] [Google Scholar]
- 14. Knudsen SM, Frydenberg J, Clark BF, Leffers H. 1993. Tissue-dependent variation in the expression of elongation factor-1 alpha isoforms: isolation and characterisation of a cDNA encoding a novel variant of human elongation-factor 1 alpha. Eur. J. Biochem. 215:549–554 [DOI] [PubMed] [Google Scholar]
- 15. Soares DC, Barlow PN, Newbery HJ, Porteous DJ, Abbott CM. 2009. Structural models of human eEF1A1 and eEF1A2 reveal two distinct surface clusters of sequence variation and potential differences in phosphorylation. PLoS One 4:e6315 doi: 10.1371/journal.pone.0006315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee S, Francoeur AM, Liu S, Wang E. 1992. Tissue-specific expression in mammalian brain, heart, and muscle of S1, a member of the elongation factor-1 alpha gene family. J. Biol. Chem. 267:24064–24068 [PubMed] [Google Scholar]
- 17. Anand N, Murthy S, Amann G, Wernick M, Porter LA, Cukier IH, Collins C, Gray JW, Diebold J, Demetrick DJ, Lee JM. 2002. Protein elongation factor EEF1A2 is a putative oncogene in ovarian cancer. Nat. Genet. 31:301–305 [DOI] [PubMed] [Google Scholar]
- 18. Tomlinson VA, Newbery HJ, Wray NR, Jackson J, Larionov A, Miller WR, Dixon JM, Abbott CM. 2005. Translation elongation factor eEF1A2 is a potential oncoprotein that is overexpressed in two-thirds of breast tumours. BMC Cancer 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sandbaken MG, Culbertson MR. 1988. Mutations in elongation factor EF-1 alpha affect the frequency of frameshifting and amino acid misincorporation in Saccharomyces cerevisiae. Genetics 120:923–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chambers DM, Peters J, Abbott CM. 1998. The lethal mutation of the mouse wasted (wst) is a deletion that abolishes expression of a tissue-specific isoform of translation elongation factor 1alpha, encoded by the eEF1A2 gene. Proc. Natl. Acad. Sci. U. S. A. 95:4463–4468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Andersen GR, Valente L, Pedersen L, Kinzy TG, Nyborg J. 2001. Crystal structures of nucleotide exchange intermediates in the eEF1A-eEF1Bα complex. Nat. Struct. Biol. 8:531–534 [DOI] [PubMed] [Google Scholar]
- 22. Gaucher EA, Das UK, Miyamoto MM, Benner SA. 2002. The crystal structure of eEF1A refines the functional predictions of an evolutionary analysis of rate changes among elongation factors. Mol. Biol. Evol. 19:569–573 [DOI] [PubMed] [Google Scholar]
- 23. Blumenthal T, Carmichael GG. 1979. RNA replication: function and structure of Qbeta-replicase. Annu. Rev. Biochem. 48:525–548 [DOI] [PubMed] [Google Scholar]
- 24. Dreher TW. 2009. Role of tRNA-like structures in controlling plant virus replication. Virus Res. 139:217–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Matsuda D, Dreher TW. 2004. The tRNA-like structure of Turnip yellow mosaic virus RNA is a 3′-translational enhancer. Virology 321:36–46 [DOI] [PubMed] [Google Scholar]
- 26. Matsuda D, Yoshinari S, Dreher TW. 2004. eEF1A binding to aminoacylated viral RNA represses minus strand synthesis by TYMV RNA-dependent RNA polymerase. Virology 321:47–56 [DOI] [PubMed] [Google Scholar]
- 27. Annamalai P, Rao AL. 2007. In vivo packaging of brome mosaic virus RNA3, but not RNAs 1 and 2, is dependent on a cis-acting 3′ tRNA-like structure. J. Virol. 81:173–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Choi YG, Dreher TW, Rao AL. 2002. tRNA elements mediate the assembly of an icosahedral RNA virus. Proc. Natl. Acad. Sci. U. S. A. 99:655–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dreher TW, Uhlenbeck OC, Browning KS. 1999. Quantitative assessment of EF-1α.GTP binding to aminoacyl-tRNAs, aminoacyl-viral RNA, and tRNA shows close correspondence to the RNA binding properties of EF-Tu. J. Biol. Chem. 274:666–672 [DOI] [PubMed] [Google Scholar]
- 30. Takamatsu N, Watanabe Y, Meshi T, Okada Y. 1990. Mutational analysis of the pseudoknot region in the 3′ noncoding region of tobacco mosaic virus RNA. J. Virol. 64:3686–3693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zeenko VV, Ryabova LA, Spirin AS, Rothnie HM, Hess D, Browning KS, Hohn T. 2002. Eukaryotic elongation factor 1A interacts with the upstream pseudoknot domain in the 3′ untranslated region of tobacco mosaic virus RNA. J. Virol. 76:5678–5691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li Z, Pogany J, Panavas T, Xu K, Esposito AM, Kinzy TG, Nagy PD. 2009. Translation elongation factor 1A is a component of the tombusvirus replicase complex and affects the stability of the p33 replication co-factor. Virology 385:245–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pogany J, Fabian MR, White KA, Nagy PD. 2003. A replication silencer element in a plus-strand RNA virus. EMBO J. 22:5602–5611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li Z, Pogany J, Tupman S, Esposito AM, Kinzy TG, Nagy PD. 2010. Translation elongation factor 1A facilitates the assembly of the tombusvirus replicase and stimulates minus-strand synthesis. PLoS Pathog. 6:e1001175 doi: 10.1371/journal.ppat.1001175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Blackwell JL, Brinton MA. 1997. Translation elongation factor-1 alpha interacts with the 3′ stem-loop region of West Nile virus genomic RNA. J. Virol. 71:6433–6444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Davis WG, Blackwell JL, Shi PY, Brinton MA. 2007. Interaction between the cellular protein eEF1A and the 3′-terminal stem-loop of West Nile virus genomic RNA facilitates viral minus-strand RNA synthesis. J. Virol. 81:10172–10187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. De Nova-Ocampo M, Villegas-Sepulveda N, del Angel RM. 2002. Translation elongation factor-1alpha, La, and PTB interact with the 3′ untranslated region of dengue 4 virus RNA. Virology 295:337–347 [DOI] [PubMed] [Google Scholar]
- 38. Sikora D, Greco-Stewart VS, Miron P, Pelchat M. 2009. The hepatitis delta virus RNA genome interacts with eEF1A1, p54(nrb), hnRNP-L, GAPDH and ASF/SF2. Virology 390:71–78 [DOI] [PubMed] [Google Scholar]
- 39. Abrahem A, Pelchat M. 2008. Formation of an RNA polymerase II preinitiation complex on an RNA promoter derived from the hepatitis delta virus RNA genome. Nucleic Acids Res. 36:5201–5211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gudima S, Wu SY, Chiang CM, Moraleda G, Taylor J. 2000. Origin of hepatitis delta virus mRNA. J. Virol. 74:7204–7210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Andino R, Rieckhof GE, Achacoso PL, Baltimore D. 1993. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 12:3587–3598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Andino R, Rieckhof GE, Baltimore D. 1990. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell 63:369–380 [DOI] [PubMed] [Google Scholar]
- 43. Kanibolotsky DS, Novosyl'na OV, Abbott CM, Negrutskii BS, El'skaya AV. 2008. Multiple molecular dynamics simulation of the isoforms of human translation elongation factor 1A reveals reversible fluctuations between “open” and “closed” conformations and suggests specific for eEF1A1 affinity for Ca2+-calmodulin. BMC Struct. Biol. 8:4 doi: 10.1186/1472-6807-8-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Blumenthal T, Landers TA, Weber K. 1972. Bacteriophage Q replicase contains the protein biosynthesis elongation factors EF Tu and EF Ts. Proc. Natl. Acad. Sci. U. S. A. 69:1313–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ahlquist P, Noueiry AO, Lee WM, Kushner DB, Dye BT. 2003. Host factors in positive-strand RNA virus genome replication. J. Virol. 77:8181–8186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Su Q, Schuppli D, Tsui Hc T, Winkler ME, Weber H. 1997. Strongly reduced phage Qbeta replication, but normal phage MS2 replication in an Escherichia coli K12 mutant with inactivated Qbeta host factor (hfq) gene. Virology 227:211–214 [DOI] [PubMed] [Google Scholar]
- 47. Landers TA, Blumenthal T, Weber K. 1974. Function and structure in ribonucleic acid phage Q beta ribonucleic acid replicase. The roles of the different subunits in transcription of synthetic templates. J. Biol. Chem. 249:5801–5808 [PubMed] [Google Scholar]
- 48. Kidmose RT, Vasiliev NN, Chetverin AB, Andersen GR, Knudsen CR. 2010. Structure of the Qβ replicase, an RNA-dependent RNA polymerase consisting of viral and host proteins. Proc. Natl. Acad. Sci. U. S. A. 107:10884–10889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Takeshita D, Tomita K. 2012. Molecular basis for RNA polymerization by Qbeta replicase. Nat. Struct. Mol. Biol. 19:229–237 [DOI] [PubMed] [Google Scholar]
- 50. Takeshita D, Tomita K. 2010. Assembly of Qβ viral RNA polymerase with host translational elongation factors EF-Tu and -Ts. Proc. Natl. Acad. Sci. U. S. A. 107:15733–15738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yamaji Y, Kobayashi T, Hamada K, Sakurai K, Yoshii A, Suzuki M, Namba S, Hibi T. 2006. In vivo interaction between Tobacco mosaic virus RNA-dependent RNA polymerase and host translation elongation factor 1A. Virology 347:100–108 [DOI] [PubMed] [Google Scholar]
- 52. Yamaji Y, Sakurai K, Hamada K, Komatsu K, Ozeki J, Yoshida A, Yoshii A, Shimizu T, Namba S, Hibi T. 2010. Significance of eukaryotic translation elongation factor 1A in tobacco mosaic virus infection. Arch. Virol. 155:263–268 [DOI] [PubMed] [Google Scholar]
- 53. Hagiwara Y, Komoda K, Yamanaka T, Tamai A, Meshi T, Funada R, Tsuchiya T, Naito S, Ishikawa M. 2003. Subcellular localization of host and viral proteins associated with tobamovirus RNA replication. EMBO J. 22:344–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mas P, Beachy RN. 1999. Replication of tobacco mosaic virus on endoplasmic reticulum and role of the cytoskeleton and virus movement protein in intracellular distribution of viral RNA. J. Cell Biol. 147:945–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Osman TA, Buck KW. 1996. Complete replication in vitro of tobacco mosaic virus RNA by a template-dependent, membrane-bound RNA polymerase. J. Virol. 70:6227–6234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Thivierge K, Cotton S, Dufresne PJ, Mathieu I, Beauchemin C, Ide C, Fortin MG, Laliberte JF. 2008. Eukaryotic elongation factor 1A interacts with Turnip mosaic virus RNA-dependent RNA polymerase and VPg-Pro in virus-induced vesicles. Virology 377:216–225 [DOI] [PubMed] [Google Scholar]
- 57. Uchil PD, Satchidanandam V. 2003. Architecture of the flaviviral replication complex. Protease, nuclease, and detergents reveal encasement within double-layered membrane compartments. J. Biol. Chem. 278:24388–24398 [DOI] [PubMed] [Google Scholar]
- 58. Westaway EG, Mackenzie JM, Kenney MT, Jones MK, Khromykh AA. 1997. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J. Virol. 71:6650–6661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Caldas T, Laalami S, Richarme G. 2000. Chaperone properties of bacterial elongation factor EF-G and initiation factor IF2. J. Biol. Chem. 275:855–860 [DOI] [PubMed] [Google Scholar]
- 60. Das T, Mathur M, Gupta AK, Janssen GM, Banerjee AK. 1998. RNA polymerase of vesicular stomatitis virus specifically associates with translation elongation factor-1 alphabetagamma for its activity. Proc. Natl. Acad. Sci. U. S. A. 95:1449–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Qanungo KR, Shaji D, Mathur M, Banerjee AK. 2004. Two RNA polymerase complexes from vesicular stomatitis virus-infected cells that carry out transcription and replication of genome RNA. Proc. Natl. Acad. Sci. U. S. A. 101:5952–5957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Warren K, Wei T, Li D, Qin F, Warrilow D, Lin MH, Sivakumaran H, Apolloni A, Abbott CM, Jones A, Anderson JL, Harrich D. 2012. Eukaryotic elongation factor 1 complex subunits are critical HIV-1 reverse transcription cofactors. Proc. Natl. Acad. Sci. U. S. A. 109:9587–9592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Johnson CM, Perez DR, French R, Merrick WC, Donis RO. 2001. The NS5A protein of bovine viral diarrhoea virus interacts with the alpha subunit of translation elongation factor-1. J. Gen. Virol. 82:2935–2943 [DOI] [PubMed] [Google Scholar]
- 64. Kou YH, Chou SM, Wang YM, Chang YT, Huang SY, Jung MY, Huang YH, Chen MR, Chang MF, Chang SC. 2006. Hepatitis C virus NS4A inhibits cap-dependent and the viral IRES-mediated translation through interacting with eukaryotic elongation factor 1A. J. Biomed. Sci. 13:861–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Harris KS, Xiang W, Alexander L, Lane WS, Paul AV, Wimmer E. 1994. Interaction of poliovirus polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J. Biol. Chem. 269:27004–27014 [PubMed] [Google Scholar]
- 66. Cimarelli A, Luban J. 1999. Translation elongation factor 1-alpha interacts specifically with the human immunodeficiency virus type 1 Gag polyprotein. J. Virol. 73:5388–5401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Parissi V, Calmels C, De Soultrait VR, Caumont A, Fournier M, Chaignepain S, Litvak S. 2001. Functional interactions of human immunodeficiency virus type 1 integrase with human and yeast HSP60. J. Virol. 75:11344–11353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Allouch A, Cereseto A. 2011. Identification of cellular factors binding to acetylated HIV-1 integrase. Amino Acids 41:1137–1145 [DOI] [PubMed] [Google Scholar]
- 69. Yue J, Shukla R, Accardi R, Zanella-Cleon I, Siouda M, Cros MP, Krutovskikh V, Hussain I, Niu Y, Hu S, Becchi M, Jurdic P, Tommasino M, Sylla BS. 2011. Cutaneous human papillomavirus type 38 E7 regulates actin cytoskeleton structure for increasing cell proliferation through CK2 and the eukaryotic elongation factor 1A. J. Virol. 85:8477–8494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lin WS, Jiao BY, Wu YL, Chen WN, Lin X. 2012. Hepatitis B virus X protein blocks filamentous actin bundles by interaction with eukaryotic translation elongation factor 1 alpha 1. J. Med. Virol. 84:871–877 [DOI] [PubMed] [Google Scholar]
- 71. Tellinghuisen TL, Paulson MS, Rice CM. 2006. The NS5A protein of bovine viral diarrhea virus contains an essential zinc-binding site similar to that of the hepatitis C virus NS5A protein. J. Virol. 80:7450–7458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sapay N, Montserret R, Chipot C, Brass V, Moradpour D, Deleage G, Penin F. 2006. NMR structure and molecular dynamics of the in-plane membrane anchor of nonstructural protein 5A from bovine viral diarrhea virus. Biochemistry 45:2221–2233 [DOI] [PubMed] [Google Scholar]
- 73. Macdonald A, Harris M. 2004. Hepatitis C virus NS5A: tales of a promiscuous protein. J. Gen. Virol. 85:2485–2502 [DOI] [PubMed] [Google Scholar]
- 74. Chuang JL, Perrault J. 1997. Initiation of vesicular stomatitis virus mutant polR1 transcription internally at the N gene in vitro. J. Virol. 71:1466–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Barton DJ, O'Donnell BJ, Flanegan JB. 2001. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 20:1439–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lin JY, Chen TC, Weng KF, Chang SC, Chen LL, Shih SR. 2009. Viral and host proteins involved in picornavirus life cycle. J. Biomed. Sci. 16:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Cereseto A, Manganaro L, Gutierrez MI, Terreni M, Fittipaldi A, Lusic M, Marcello A, Giacca M. 2005. Acetylation of HIV-1 integrase by p300 regulates viral integration. EMBO J. 24:3070–3081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Abbas W, Khan KA, Tripathy MK, Dichamp I, Keita M, Rohr O, Herbein G. 2012. Inhibition of ER stress-mediated apoptosis in macrophages by nuclear-cytoplasmic relocalization of eEF1A by the HIV-1 Nef protein. Cell Death Dis. 3:e292 doi: 10.1038/cddis.2012.32 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 79. Brinton MA. 2001. Host factors involved in West Nile virus replication. Ann. N. Y. Acad. Sci. 951:207–219 [DOI] [PubMed] [Google Scholar]
- 80. Blumenthal T, Young RA, Brown S. 1976. Function and structure in phage Qbeta RNA replicase. Association of EF-Tu-Ts with the other enzyme subunits. J. Biol. Chem. 251:2740–2743 [PubMed] [Google Scholar]
- 81. Nishikiori M, Dohi K, Mori M, Meshi T, Naito S, Ishikawa M. 2006. Membrane-bound tomato mosaic virus replication proteins participate in RNA synthesis and are associated with host proteins in a pattern distinct from those that are not membrane bound. J. Virol. 80:8459–8468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Sasvari Z, Izotova L, Kinzy TG, Nagy PD. 2011. Synergistic roles of eukaryotic translation elongation factors 1Bγ and 1A in stimulation of tombusvirus minus-strand synthesis. PLoS Pathog. 7:e1002438 doi: 10.1371/journal.ppat.1002438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Nagy PD, Pogany J. 2010. Global genomics and proteomics approaches to identify host factors as targets to induce resistance against Tomato bushy stunt virus. Adv. Virus Res. 76:123–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Pogany J, Nagy PD. 2012. p33-independent activation of a truncated p92 RNA-dependent RNA polymerase of Tomato bushy stunt virus in yeast cell-free extract. J. Virol. 86:12025–12038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Brown S, Blumenthal T. 1976. Function and structure in ribonucleic acid phage Qbeta ribonucleic acid replicase. Effect of inhibitors of EF-Tu on ribonucleic acid synthesis and renaturation of active enzyme. J. Biol. Chem. 251:2749–2753 [PubMed] [Google Scholar]
- 86. Brown S, Blumenthal T. 1976. Reconstitution of Qbeta RNA replicase from a covalently bonded elongation factor Tu-Ts complex. Proc. Natl. Acad. Sci. U. S. A. 73:1131–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Warrilow D, Meredith L, Davis A, Burrell C, Li P, Harrich D. 2008. Cell factors stimulate human immunodeficiency virus type 1 reverse transcription in vitro. J. Virol. 82:1425–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Warrilow D, Warren K, Harrich D. 2010. Strand transfer and elongation of HIV-1 reverse transcription is facilitated by cell factors in vitro. PLoS One 5:e13229 doi: 10.1371/journal.pone.0013229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Moerdyk-Schauwecker M, Hwang SI, Grdzelishvili VZ. 2009. Analysis of virion associated host proteins in vesicular stomatitis virus using a proteomics approach. Virol. J. 6:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Chung CS, Chen CH, Ho MY, Huang CY, Liao CL, Chang W. 2006. Vaccinia virus proteome: identification of proteins in vaccinia virus intracellular mature virion particles. J. Virol. 80:2127–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Resch W, Hixson KK, Moore RJ, Lipton MS, Moss B. 2007. Protein composition of the vaccinia virus mature virion. Virology 358:233–247 [DOI] [PubMed] [Google Scholar]
- 92. Kattenhorn LM, Mills R, Wagner M, Lomsadze A, Makeev V, Borodovsky M, Ploegh HL, Kessler BM. 2004. Identification of proteins associated with murine cytomegalovirus virions. J. Virol. 78:11187–11197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Varnum SM, Streblow DN, Monroe ME, Smith P, Auberry KJ, Pasa-Tolic L, Wang D, Camp DG, 2nd, Rodland K, Wiley S, Britt W, Shenk T, Smith RD, Nelson JA. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 78:10960–10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Neuman BW, Joseph JS, Saikatendu KS, Serrano P, Chatterjee A, Johnson MA, Liao L, Klaus JP, Yates JR, 3rd, Wuthrich K, Stevens RC, Buchmeier MJ, Kuhn P. 2008. Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J. Virol. 82:5279–5294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Chertova E, Chertov O, Coren LV, Roser JD, Trubey CM, Bess JW, Jr, Sowder RC, 2nd, Barsov E, Hood BL, Fisher RJ, Nagashima K, Conrads TP, Veenstra TD, Lifson JD, Ott DE. 2006. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J. Virol. 80:9039–9052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Ott DE, Coren LV, Johnson DG, Kane BP, Sowder RC, 2nd, Kim YD, Fisher RJ, Zhou XZ, Lu KP, Henderson LE. 2000. Actin-binding cellular proteins inside human immunodeficiency virus type 1. Virology 266:42–51 [DOI] [PubMed] [Google Scholar]
- 97. Sasaki H, Nakamura M, Ohno T, Matsuda Y, Yuda Y, Nonomura Y. 1995. Myosin-actin interaction plays an important role in human immunodeficiency virus type 1 release from host cells. Proc. Natl. Acad. Sci. U. S. A. 92:2026–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Poon DT, Wu J, Aldovini A. 1996. Charged amino acid residues of human immunodeficiency virus type 1 nucleocapsid p7 protein involved in RNA packaging and infectivity. J. Virol. 70:6607–6616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Ghittoni R, Accardi R, Hasan U, Gheit T, Sylla B, Tommasino M. 2010. The biological properties of E6 and E7 oncoproteins from human papillomaviruses. Virus Genes 40:1–13 [DOI] [PubMed] [Google Scholar]
- 100. Dong W, Kloz U, Accardi R, Caldeira S, Tong WM, Wang ZQ, Jansen L, Durst M, Sylla BS, Gissmann L, Tommasino M. 2005. Skin hyperproliferation and susceptibility to chemical carcinogenesis in transgenic mice expressing E6 and E7 of human papillomavirus type 38. J. Virol. 79:14899–14908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Liu G, Grant WM, Persky D, Latham VM, Jr, Singer RH, Condeelis J. 2002. Interactions of elongation factor 1alpha with F-actin and beta-actin mRNA: implications for anchoring mRNA in cell protrusions. Mol. Biol. Cell 13:579–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Lee YH, Yun Y. 1998. HBx protein of hepatitis B virus activates Jak1-STAT signaling. J. Biol. Chem. 273:25510–25515 [DOI] [PubMed] [Google Scholar]
- 103. Tang H, Oishi N, Kaneko S, Murakami S. 2006. Molecular functions and biological roles of hepatitis B virus X protein. Cancer Sci. 97:977–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Dominguez R, Holmes KC. 2011. Actin structure and function. Annu. Rev. Biophys. 40:169–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Zhou B, Liu J, Wang Q, Liu X, Li X, Li P, Ma Q, Cao C. 2008. The nucleocapsid protein of severe acute respiratory syndrome coronavirus inhibits cell cytokinesis and proliferation by interacting with translation elongation factor 1alpha. J. Virol. 82:6962–6971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Macneughton MR, Davies HA. 1978. Ribonucleoprotein-like structures from coronavirus particles. J. Gen. Virol. 39:545–549 [DOI] [PubMed] [Google Scholar]
- 107. Hasler J, Rada C, Neuberger MS. 2011. Cytoplasmic activation-induced cytidine deaminase (AID) exists in stoichiometric complex with translation elongation factor 1alpha (eEF1A). Proc. Natl. Acad. Sci. U. S. A. 108:18366–18371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Hasler J, Rada C, Neuberger MS. 2012. The cytoplasmic AID complex. Semin. Immunol. 24:273–280 [DOI] [PubMed] [Google Scholar]
- 109. Gourzi P, Leonova T, Papavasiliou FN. 2006. A role for activation-induced cytidine deaminase in the host response against a transforming retrovirus. Immunity 24:779–786 [DOI] [PubMed] [Google Scholar]
- 110. Epeldegui M, Thapa DR, de la Cruz J, Kitchen S, Zack JA, Martinez-Maza O. 2010. CD40 ligand (CD154) incorporated into HIV virions induces activation-induced cytidine deaminase (AID) expression in human B lymphocytes. PLoS One 5:e11448 doi: 10.1371/journal.pone.0011448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99–103 [DOI] [PubMed] [Google Scholar]
- 112. Kamen R. 1970. Characterization of the subunits of Q-beta replicase. Nature 228:527–533 [DOI] [PubMed] [Google Scholar]
- 113. Kamen R, Kondo M, Romer W, Weissmann C. 1972. Reconstitution of Q replicase lacking subunit with protein-synthesis-interference factor I. Eur. J. Biochem. 31:44–51 [DOI] [PubMed] [Google Scholar]
- 114. Kondo M, Gallerani R, Weissmann C. 1970. Subunit structure of Q-beta replicase. Nature 228:525–527 [DOI] [PubMed] [Google Scholar]
- 115. Mayer D, Molawi K, Martinez-Sobrido L, Ghanem A, Thomas S, Baginsky S, Grossmann J, Garcia-Sastre A, Schwemmle M. 2007. Identification of cellular interaction partners of the influenza virus ribonucleoprotein complex and polymerase complex using proteomic-based approaches. J. Proteome Res. 6:672–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Hsu WB, Shih JL, Shih JR, Du JL, Teng SC, Huang LM, Wang WB. 2013. Cellular protein HAX1 interacts with the influenza A virus PA polymerase subunit and impedes its nuclear translocation. J. Virol. 87:110–123 [DOI] [PMC free article] [PubMed] [Google Scholar]