Abstract
Innate effector cells, including innate effector cells of myeloid and lymphoid lineages, are crucial components of various types of immune responses. Bone marrow progenitors differentiate into many subsets of innate effector cells after receiving instructional signals often provided by cytokines. Signal transducer and activator of transcription (STATs) have been shown to be essential in the differentiation of various types of innate effector cells. In this review, we focus specifically on the differentiation of innate effector cells, particularly the role of cytokine signaling in the differentiation of innate effector cells.
Keywords: STATs, innate effector cells, differentiation, cytokine, transcription factor
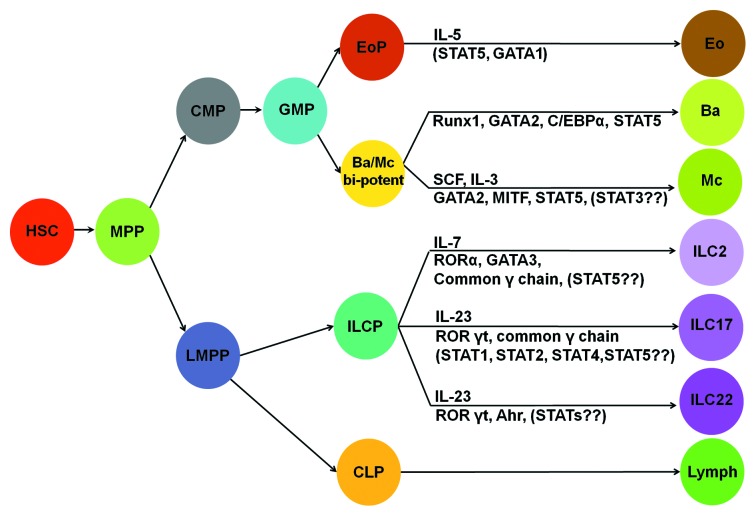
Innate effector cells are important components of T helper cell-mediated immunity, which can be classified into Type-1, Type-2, Type-9, Type-17 and Type-22 immunity (see review in refs. 1 and 2). Innate effector cells include innate lymphoid cells (ILCs) and innate myeloid cells (IMCs). ILCs can be further divided into ILC1, ILC2, ILC17 and ILC22 subsets (Fig. 1).3 We propose to call Th2 cytokine-producing eosinophils, basophils and mast cells as Type-2 innate myeloid cells (IMC2). A cytokine or a specific growth factor has been reported to be critical in the development and function of many types of hematopoietic cells. For example, erythropoietin is needed for erythrocyte differentiation,4,5 IL-5 is imperative for eosinophil differentiation,6 M-CSF is critical for monocyte differentiation7 and thrombopoietin is crucial for megakaryocyte differentiation.8 In this review, we focus on the role of cytokine signaling in the differentiation of innate effector cells.
Figure 1. Lineage relationship of innate myeloid cells and innate lymphoid cells. Cytokines and transcription factors that specify cell fate of a particular subset of innate myeloid cells or innate lymphoid cells are illustrated. Ba/Mc bi-potent, common basophils and mast cell progenitors; eo, eosinophils; ba, basophils; mc, mast cells; lymph, all lymphocytes. The rest of abbreviations are given in the text.
Stem Cells and Progenitors in Hematopoiesis
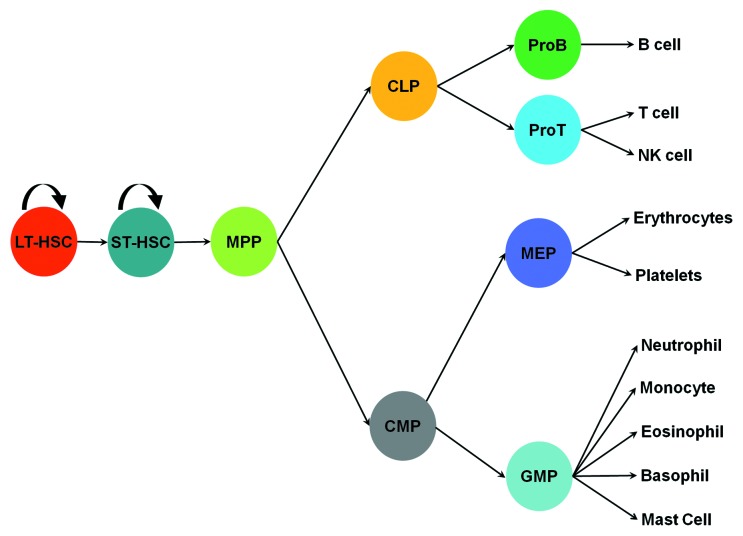
Innate effector cells originate from various hematopoietic progenitors in the bone marrow, where a hematopoietic hierarchy is located. Long-term repopulating hematopoietic stem cells (HSC) are at the top of the hematopoietic hierarchy. These cells possess the capacity for self-renewal and the potential to give rise to all types of blood cells. Long-term HSCs can generate short-term repopulating HSCs, which then give rise to multiple potential progenitors (MPPs). MPPs, in turn, can give rise to common lymphoid progenitors (CLP) and common myeloid progenitors (CMPs). CMPs can differentiate into granulocyte-monocyte progenitors (GMPs).9 GMPs give rise to eosinophil lineage-restricted progenitors (EoPs),10 basophil lineage-restricted progenitors (BaPs),11 neutrophils and macrophages (Fig. 2).
Figure 2. Differentiation of hematopoietic cells. ProB, pro B cells; ProT, pro T cells; MEP, megakaryocyte progenitor. The rest of abbreviations are given in the text.
Differentiation of Type-2 Innate Myeloid Cells
The origin of basophils and mast cells
Basophils and mast cells share many common characteristics, such as the expression of a high affinity immunoglobulin E (IgE) receptor (FcεR), and contain many of the same granules.12,13 These two types of cells also show noticeable differences. For instance, basophils circulate in the blood stream, whereas mast cells reside in tissue. Mature basophils do not proliferate and have a short life of approximately 60 h,14 whereas mature mast cells are proliferative and possess a much longer life span of up to several months.15 Functionally, both basophils and mast cells are the key effectors in Type-2 immunity, which is responsible for mediating allergic disease and provides protection against parasitic infections. Accumulated evidence supports that basophils play non-redundant roles in immune regulation, protective immunity, allergies and autoimmunity.16 The recent success of using anti-IgE antibody to treat various allergic disorders in humans supports the importance of both FcεR-expressing basophils and mast cells in human diseases.17,18 Thus, a more comprehensive understanding of the basophil and mast cell developmental pathway is of high significance.
The origin of basophils and mast cells has been a long-standing, unsolved and important issue in hematology. By using colony formation assays, two groups have claimed that basophils develop from a common basophil and eosinophil progenitor.19,20 Whether basophils and mast cells are derived from a common progenitor remains a controversial issue in hematology. Galli and colleagues found mast cell lineage-restricted progenitors (MCPs) in the bone marrow and concluded that MCPs are derived from MPPs instead of CMPs or GMPs.21 On the other hand, Akashi and colleagues showed that both basophils and mast cells were derived from CMPs and GMPs;22 they further showed that basophil/mast cell progenitors (BMCPs) found in the spleen, but not in the bone marrow, gave rise to both basophils and mast cells.11 However, whether BMCPs truly are authentic bi-potential basophil/mast cell progenitors has been challenged by a recent study, in which Galli and colleagues demonstrated that BMCPs only gave rise to mast cells.23
STAT5 signaling is essential in basophil differentiation and expansion
It remains unclear which growth factor is required for basophil differentiation. It has been reported that neither IL-3 nor TSLP are required for basophil development.24,25 We also found that IL-4 is not required for basophils development (data not shown). Due to the absence of a specific factor identified for basophil differentiation, a default theory, which states that terminal differentiation of basophils can occur without a specific factor, has been proposed.26 Nevertheless, we found that STAT5 was critical in basophil differentiation. The STAT family consists of seven members—STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B and STAT6. STATs can be activated by JAK kinases through phosphorylation of their tyrosine residues.27,28 Cytokines mainly use the JAK-STAT pathway to exert their biological functions. STAT5 can be activated by many cytokines and growth factors that are important in the development of both lymphoid and myeloid cells.27,29-31 Using radiation chimera mice reconstituted with STAT5-deficient fetal liver cells, we found that STAT5-deficient stem cells failed to differentiate into basophils.32 Recently, we verified this finding by using inducible conditional STAT5 knockout mice (submitted for publication). Because STAT5 is a signaling molecule whose activation is most likely triggered by an external factor, our analysis supports the existence of an external factor that instructs basophil development.
Despite the finding that IL-3 is not required for basophil differentiation, it has been shown that IL-3 is a potent factor in expanding basophils. Evidence supports that IL-3, produced primarily by CD4+ T cells,33-35 is pivotal in expanding basophils. In vitro, IL-3 has been demonstrated to induce basophil differentiation and to enhance acute IL-4 production in mouse basophils.36-38 Galli and colleagues reported that mice lacking IL-3 both failed to show increased numbers of basophils and failed to expel nematode Strongyloides.24 We report that administrating the IL-3 complex (IL-3 plus anti-IL-3 antibody) in vivo greatly facilitates the differentiation of GMPs into BaPs and increases the number of BMCPs in the spleen.32 Binding of cytokine to a respective anti-cytokine antibody that does not interfere with the ability of the cytokine to bind its receptor has been documented to increase the effectiveness of the particular cytokine in vivo, namely by increasing the half-life of the bound cytokine.39 We showed that GMPs, but not CMPs, expressed low levels of IL-3 receptor. IL-3 receptor expression was dramatically upregulated on BaPs, but not on EoPs. We showed that about 38% of BMCPs expressed the IL-3 receptor.32 The IL-3 receptor expression patterns might explain why IL-3 specifically expanded basophils in vivo. We further demonstrated that basophil expansion—specifically induced by the IL-3 complex—depended on STAT5 signaling.32
STAT5 signaling is most likely involved in TSLP-induced basophil expansion. One report showed that daily intraperitoneal injection of recombinant TSLP for 4–7 consecutive days resulted in a 2- to 4-fold increase in basophil numbers in the blood and spleen.40 A second report noted a more modest increase (50 to 100%) in basophil numbers after TSLP injection.23 It was further demonstrated that TSLP stimulated BaP proliferation and mature basophil survival.40 Compared with IL-3, TSLP is less potent in basophil expansion. Because TSLP activated STAT5 via a still unknown kinase, it is reasonable to assume that STAT5 signaling is required for TSLP-induced basophil expansion.
Molecular regulation of basophil differentiation
STAT5,32 Runx1,23 GATA241 and C/EBPα42 have all been implicated as the imperative players in basophil differentiation. The order of expression of GATA2 and C/EBPα has also been suggested as the crucial determinant in basophil fate vs. eosinophil cell fate.42 If GATA2 expression precedes C/EBPα expression at the GMP stage, GATA2 together with C/EBPα will drive basophil differentiation. Conversely, if C/EBPα expression precedes GATA2 expression at the GMP stage, then both C/EBPα and GATA2 will drive eosinophil differentiation.42 However, it remains unknown which of the aforementioned factors is the master determinant for basophil cell fate.
Both basophils and mast cells are rich sources of cytokines. One of the major characteristics of basophils, as compared with mast cells, is their ability to produce a large quantity of IL-4. STAT6 and GATA3 are critical regulators of Il4 gene expression in CD4+ T cells.43 We profiled mRNA expression of known Th2 transcription factors as well as transcription factors that are pivotal for basophil development, such as GATA3, RBPJ, c-Maf, JunB, C/EBPα, GATA1 and GATA2. We found that C/EBPα was highly expressed in basophils, but not in Th2 cells.44 Our results showed that in response to IgE cross-linking, C/EBPα activated Il4-promoter-driven luciferase-reporter gene transcription, but not other known Th2 or mast cell enhancers, in a basophil-like cell line. We found that the DNA binding domain of C/EBPα and two C/EBPα-binding sites (-44 to -36, and -87 to -79) in the Il4 promoter were required for activating the Il4 promoter. Additionally, our analysis revealed that a mutation in the nuclear factor of activated T cells (NFAT)-binding sites in the Il4 promoter also negated C/EBPα-driven Il4 promoter-luciferase activity. Taken together, these findings within our study demonstrate that C/EBPα directly regulates Il4 gene transcription.44
We examined the role of STAT5 in IL-4 production by mature basophils and found that induced deletion of STAT5 in mature basophils had little to no effect on Il4 mRNA expression or IL-4 protein expression (unpublished data). Thus, it appears that the requirement for STAT5 in basophil differentiation and the requirement for STAT5 in basophil activation differ. Rather, we found that the PI3K pathway and calcineurin were essential in C/EBPα-driven Il4 promoter-luciferase activity.44 Further research is underway to elucidate pathways leading to the activation of transcription of Type-2 cytokine genes in basophils.
Other regulatory regions that confer basophil-specific IL-4 expression have not yet been identified. Using the transgenic approach, Kubo and colleagues tested the regulatory regions known to regulate Il4 gene expression in Th2 cells and demonstrated that a 4 kb long HS4 element, together with a 5′ enhancer [-863 to -5,448 base pair (bp)] and the Il4 promoter (-64 to -827 bp), conferred basophil-specific GFP expression.45 Paradoxically, this study shows that HS4—a recognized silencer of Il4 gene transcription in Th2 cells46—is an enhancer for Il4 gene transcription in basophils. A different set of regulatory regions implies that a different set of transcription factors is used to confer basophil-specific Il4 gene expression.45 A comprehensive examination of histone modifications surrounding the Il4 gene in basophils is needed to further understand how the Il4 gene is regulated in basophils.
Mast cell differentiation
Several cytokines have been shown to regulate mast cell differentiation, survival and function. SCF and IL-3 are critical factors for mast cell development and both cytokines can activate STAT5. A series of experiments have established important roles of STAT5 in mast cell development and survival. In STAT5 deficient mice, mast cells were normal at birth, but were undetectable 12 weeks after birth.47,48 There was a marked decrease in histamine and leukotriene B4 production in STAT5-deficient mast cells. The reduction was the result of altered post-transcriptional control of cytokine mRNA stability in the absence of STAT5.48
IL-10, a suppressive cytokine produced mainly by regulatory T cells, inhibited mast cell functions by downregulating IgE receptor (FcεRIα) expression and signaling through a STAT3-dependent manner.49,50 Like IL-10, IL-4 also inhibited BMMC growth and FcεRIα expression on mouse mast cells.51 But unlike IL-10, IL-4 depended on STAT6 signaling for its function.52 Paradoxically, IL-4 enhances FcεRIα expression on human mast cells.53,54 Explanation for the discrepancy in IL-4 effects on mouse and human mast cells is not yet available. It may be related to differentiation status of mast cells. Finally, IL-4 and IL-10 has also been shown to be an important regulator in mast cell homeostasis. IL-4 and IL-10-induced apoptosis was coupled with decreased expression of bcl-x(L) and bcl-2. While this process occurred independent of the Fas pathway, IL-4 and IL-10 greatly sensitized mast cells to Fas-mediated death.55,56
It is likely that STATs need to cooperate with other transcription factors to promote mast cell functions. In our study, we found a novel population of GMPs that contain highly enriched common basophil and mast cell progenitors. We note that STAT5 signaling is imperative in directing the novel basophil/mast cell progenitors into both basophils and mast cells. We further demonstrate that GATA2 is a downstream molecule of STAT5 and is essential in both basophil and mast cell differentiation (unpublished data). It has been reported that GATA2 and STAT5 can form a complex and regulate target gene transcription in cancer cells.57 However, whether GATA2 and STAT5 cooperate in mast cell is unknown. STAT3 has been demonstrated to enhance MITF activity in melanocyte and mast cell indirectly by taking away PIAS3 from MITF.58,59 MITF is found to associate with PIAS3. When melanocytes and mast cells were activated through gp130 or c-Kit receptor, STAT3 competed for PIAS3 and MITF was free from PIAS3.
Eosinophil differentiation
Eosinophils can cause tissue damage by releasing inflammatory mediators, such as major basic protein, eosinophil cationic protein, eosinophil peroxidase and eosinophil-derived neurotoxin, from their granules.60 Eosinophils are also an excellent source of inflammatory cytokines.61 The importance of IL-4-producing eosinophils in generating protective immunity against parasitic infection has been recently established.62 In the parasitic model, the generation, expansion and maintenance of IL-4-producing eosinophils have been found to be essential for expelling helminth infection.62 We also reported that eosinophils produced the majority of Th2 cytokines in the late-phase response to allergic airway inflammation.63 We have further demonstrated that IL-4 directed bone marrow progenitor cells to differentiate into eosinophil-like cells capable of producing IL-4, IL-5 and IL-13.63,64
IL-5 is a critical factor for eosinophil differentiation, activation, survival and recruitment to the sites of inflammation.65 IL-5 exerts its functions through stimulating the IL-5Rα-chain and a common β-chain, shared by IL-3 and GM-CSF. Stimulation of the IL-5R activates multiple signaling pathways, including the JAK-STAT pathway. IL-5 induces phosphorylation of JAK2, STAT1 and STAT5.66,67 STAT5A and STAT5B exhibit 95% homology in amino acid sequences.68 Deficiency in both the Stat5a and Stat5b genes resulted in a reduction in the number of IL-5-induced eosinophil colonies.69
Our studies demonstrated that IL-4 and IL-5, but not IL-13 or IL-25, directed bone marrow progenitors to differentiate into effector cells that produce Th2 cytokines.64 We further demonstrated that IL-5-driven differentiation depended on STAT5 signaling.64 In addition to STAT5, we found that Erk1 and the transcription factor GATA1 are also critical in regulating Il4 gene expression in eosinophils (unpublished data).
Differentiation of Innate Lymphoid Cells
Differentiation of type-2 innate lymphoid cells
Recently, a novel type of innate lymphoid cell capable of producing a large amount of IL-13 and IL-5 in response to IL-25, IL-33 and parasitic infection has been identified.70,71 They can be identified by flow cytometry as Lin− Sca-1+, c-Kit+ (high) or − (low), ST2+, IL-7R+, IL17BR+, CD25+ and CD44+.70-72 Although ILC2 express stem cell/progenitor markers, they represent terminally differentiated effector cells, i.e., they do not possess progenitor activity. The precursor of ILC2 (ILC2P) has been identified.73,74 It has been proposed that ILC2P cells are derived from lymphoid primed multipotent progenitors (LMPPs), which are thought to be upstream progenitors of CLPs (Fig. 1).75 IL-7, common γ chain, retinoic-acid-related orphan receptor (ROR) α and GATA3 have been demonstrated to be critical in the differentiation of ILC2 (Fig. 1).74,76 Another population of IL-25-elicited Lin− Sca-1+ cells in gut-associated lymphoid tissue, named multi-potent progenitor (MPPtype2), has also been reported.77 MPPtype2 contained two types of cells: one was phenotypically defined as Lin− cKit+ IL-4/GFP- (Il4/Gfp gene reporter mice were used) and the other was defined as Lin− c-Kit+ IL-4/GFP+. Lin− c-Kit+ IL-4/GFP+ cells gave rise to mast cells, whereas Lin− c-Kit+ IL-4/GFP− cells gave rise to basophils, mast cells and myeloid cells. Cytokine and the master transcription factor that regulate MPPtype2 differentiation have not been identified.
IL-25, IL-33 and TSLP all induce IL-5 and IL-13 production by ILC2 cells. STAT5 signaling has been implicated in the production of IL-5 and IL-13 because both IL-7 and TSLP are the known activators of STAT5. IL-25 has been shown to activate the NFκB pathway. Which STATs are crucial to ILC2 differentiation and function remains to be documented.
Differentiation of type-17 innate lymphoid cells
The innate counterpart of Th17 cells has been identified in the colon. Type-17 Innate Lymphoid Cells (ILC17) are CD4−CD117−NKp46− Thy1hi and Sca-1+ Lin− cells, which distinguishes them from CD4+CD117+ lymphoid tissue inducer (LTi) cells and NKp46+ ILC Type 22 cells. These cells express IL-23 receptor, ROR-γt and T-bet.78,79 They produce IL-17, IFN-γ and IL-22 in response to IL-23 stimulation. ILC17 are responsible for the pathological changes induced by Helicobacter hepaticus infection.78,79 ILC17 has also been found in humans. They appear to be an overlapping population of cells with LTi cells.3,80 The precursor of ILC17 has not been identified. It has been reported that IL-7, common γ chain and RORγt (encoded by the Rorc gene) are critical for the differentiation of ILC17 (Fig. 1).78 IL-7 is known to activate STAT5. IL-23 has been shown to activate STAT1, STAT2, STAT4 and STAT5.81 Which STATs are crucial for ILC17 differentiation and function also remains to be investigated.
Differentiation of type-22 innate lymphoid cells
An innate counterpart of Th22 has also been identified.82 ILC22 shares some characteristics with LTi cells and NK cells.80,83-86 These cells are distinct from conventional NK cells as they do not possess an ability to kill target cells. Phenotypically, they can be defined as CD56+ NKp46+.3 ILC22 cells are primarily located at mucosal sites in both mice and humans. These cells produce IL-22 and little, if any, IFN-γ. IL-23 can regulate IL-22 production by ILC22 cells in both mice84-86 and humans.83 Cytokines of the common γ chain family (IL-2, IL-7 and IL-15) can also activate proliferation and cytokine production by ILC22 cells.83,87 Combination of IL-12 and IL-18 can also enhance IL-22 production by these cells.88 RORγT-deficient mice do not have ILC22.84-86 The ligand-dependent transcription factor aryl hydrocarbon receptor (AhR) also has a critical role in regulating IL-22 production by mouse T cells89 and human T cells (Fig. 1).90 The exact involvement of STAT signaling requires further study.
Concluding Remarks
STATs play critical roles in the differentiation of Type-2 innate effectors of myeloid lineages. However, it is less clear if STAT is involved in the differentiation of innate lymphoid cells. Further study of molecules downstream to STAT and STAT co-activators will facilitate our understanding of the mechanisms by which STATs direct the differentiation of innate effectors and confer immune functions to innate effectors. Advanced knowledge will lead to more effective interventions and more successful strategies in developing preventions.
Acknowledgments
This work is supported by grants from the National Institutes of Health (RO1AI083986) and the American Recovery and Reinvestment Act administrative supplement (R01AI068083-04S1). We also thank Michael Hubbard for assistance in manuscript preparation.
Glossary
Abbreviations:
- IMCs
innate myeloid cells
- ILCs
innate lymphoid cells
- STATs
signal transducer and activator of transcription
- HSC
hematopoietic stem cells
- MPPs
multiple potential progenitors
- CMPs
common myeloid progenitors
- GMPs
granulocyte-monocyte progenitors
- EoPs
eosinophil lineage-restricted progenitors
- BaPs
basophil lineage-restricted progenitors
- FcεRIα
immunoglobulin E receptor α chain
- MCPs
mast cell lineage-restricted progenitors
- BMCPs
basophil/mast cell progenitors
- NFAT
nuclear factor of activated T cells
- PIAS3
protein inhibitor of activated STAT3
- MITF
microphthalmin-associated transcription factor
- ROR
retinoic-acid related orphan receptor
- ILC17
type-17 innate lymphoid cells
- and LTi
lymphoid tissue inducer
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/jak-stat/article/23531
References
- 1.Paul WE, Zhu J. How are T(H)2-type immune responses initiated and amplified? Nat Rev Immunol. 2010;10:225–35. doi: 10.1038/nri2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*) Annu Rev Immunol. 2010;28:445–89. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spits H, Di Santo JP. The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling. Nat Immunol. 2011;12:21–7. doi: 10.1038/ni.1962. [DOI] [PubMed] [Google Scholar]
- 4.Wu H, Liu X, Jaenisch R, Lodish HF. Generation of committed erythroid BFU-E and CFU-E progenitors does not require erythropoietin or the erythropoietin receptor. Cell. 1995;83:59–67. doi: 10.1016/0092-8674(95)90234-1. [DOI] [PubMed] [Google Scholar]
- 5.Kaushansky K. Lineage-specific hematopoietic growth factors. N Engl J Med. 2006;354:2034–45. doi: 10.1056/NEJMra052706. [DOI] [PubMed] [Google Scholar]
- 6.Rothenberg ME, Hogan SP. The eosinophil. Annu Rev Immunol. 2006;24:147–74. doi: 10.1146/annurev.immunol.24.021605.090720. [DOI] [PubMed] [Google Scholar]
- 7.Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327:656–61. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaushansky K, Broudy VC, Lin N, Jorgensen MJ, McCarty J, Fox N, et al. Thrombopoietin, the Mp1 ligand, is essential for full megakaryocyte development. Proc Natl Acad Sci U S A. 1995;92:3234–8. doi: 10.1073/pnas.92.8.3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kondo M, Wagers AJ, Manz MG, Prohaska SS, Scherer DC, Beilhack GF, et al. Biology of hematopoietic stem cells and progenitors: implications for clinical application. Annu Rev Immunol. 2003;21:759–806. doi: 10.1146/annurev.immunol.21.120601.141007. [DOI] [PubMed] [Google Scholar]
- 10.Iwasaki H, Mizuno S, Mayfield R, Shigematsu H, Arinobu Y, Seed B, et al. Identification of eosinophil lineage-committed progenitors in the murine bone marrow. J Exp Med. 2005;201:1891–7. doi: 10.1084/jem.20050548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arinobu Y, Iwasaki H, Gurish MF, Mizuno S, Shigematsu H, Ozawa H, et al. Developmental checkpoints of the basophil/mast cell lineages in adult murine hematopoiesis. Proc Natl Acad Sci U S A. 2005;102:18105–10. doi: 10.1073/pnas.0509148102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galli SJ, Franco CB. Basophils are back! Immunity. 2008;28:495–7. doi: 10.1016/j.immuni.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 13.Marone G, Galli SJ, Kitamura Y. Probing the roles of mast cells and basophils in natural and acquired immunity, physiology and disease. Trends Immunol. 2002;23:425–7. doi: 10.1016/S1471-4906(02)02274-3. [DOI] [PubMed] [Google Scholar]
- 14.Ohnmacht C, Voehringer D. Basophil effector function and homeostasis during helminth infection. Blood. 2009;113:2816–25. doi: 10.1182/blood-2008-05-154773. [DOI] [PubMed] [Google Scholar]
- 15.Galli SJ, Tsai M, Piliponsky AM. The development of allergic inflammation. Nature. 2008;454:445–54. doi: 10.1038/nature07204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karasuyama H, Mukai K, Obata K, Tsujimura Y, Wada T. Nonredundant roles of basophils in immunity. Annu Rev Immunol. 2011;29:45–69. doi: 10.1146/annurev-immunol-031210-101257. [DOI] [PubMed] [Google Scholar]
- 17.Busse WW, Morgan WJ, Gergen PJ, Mitchell HE, Gern JE, Liu AH, et al. Randomized trial of omalizumab (anti-IgE) for asthma in inner-city children. N Engl J Med. 2011;364:1005–15. doi: 10.1056/NEJMoa1009705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holgate S, Casale T, Wenzel S, Bousquet J, Deniz Y, Reisner C. The anti-inflammatory effects of omalizumab confirm the central role of IgE in allergic inflammation. J Allergy Clin Immunol. 2005;115:459–65. doi: 10.1016/j.jaci.2004.11.053. [DOI] [PubMed] [Google Scholar]
- 19.Leary AG, Ogawa M. Identification of pure and mixed basophil colonies in culture of human peripheral blood and marrow cells. Blood. 1984;64:78–83. [PubMed] [Google Scholar]
- 20.Denburg JA, Telizyn S, Messner H, Lim B, Jamal N, Ackerman SJ, et al. Heterogeneity of human peripheral blood eosinophil-type colonies: evidence for a common basophil-eosinophil progenitor. Blood. 1985;66:312–8. [PubMed] [Google Scholar]
- 21.Chen CC, Grimbaldeston MA, Tsai M, Weissman IL, Galli SJ. Identification of mast cell progenitors in adult mice. Proc Natl Acad Sci U S A. 2005;102:11408–13. doi: 10.1073/pnas.0504197102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arinobu Y, Iwasaki H, Akashi K. Origin of basophils and mast cells. Allergol Int. 2009;58:21–8. doi: 10.2332/allergolint.08-RAI-0067. [DOI] [PubMed] [Google Scholar]
- 23.Mukai K, BenBarak MJ, Tachibana M, Nishida K, Karasuyama H, Taniuchi I, et al. Critical role of P1-Runx1 in mouse basophil development. Blood. 2012;120:76–85. doi: 10.1182/blood-2011-12-399113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lantz CS, Boesiger J, Song CH, Mach N, Kobayashi T, Mulligan RC, et al. Role for interleukin-3 in mast-cell and basophil development and in immunity to parasites. Nature. 1998;392:90–3. doi: 10.1038/32190. [DOI] [PubMed] [Google Scholar]
- 25.Siracusa MC, Saenz SA, Hill DA, Kim BS, Headley MB, Doering TA, et al. TSLP promotes interleukin-3-independent basophil haematopoiesis and type 2 inflammation. Nature. 2011;477:229–33. doi: 10.1038/nature10329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arock M, Schneider E, Boissan M, Tricottet V, Dy M. Differentiation of human basophils: an overview of recent advances and pending questions. J Leukoc Biol. 2002;71:557–64. [PubMed] [Google Scholar]
- 27.Leonard WJ, O’Shea JJ. Jaks and STATs: biological implications. Annu Rev Immunol. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- 28.Schindler C, Darnell JE., Jr. Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu Rev Biochem. 1995;64:621–51. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 29.Bradley HL, Hawley TS, Bunting KD. Cell intrinsic defects in cytokine responsiveness of STAT5-deficient hematopoietic stem cells. Blood. 2002;100:3983–9. doi: 10.1182/blood-2002-05-1602. [DOI] [PubMed] [Google Scholar]
- 30.Bunting KD, Bradley HL, Hawley TS, Moriggl R, Sorrentino BP, Ihle JN. Reduced lymphomyeloid repopulating activity from adult bone marrow and fetal liver of mice lacking expression of STAT5. Blood. 2002;99:479–87. doi: 10.1182/blood.V99.2.479. [DOI] [PubMed] [Google Scholar]
- 31.Snow JW, Abraham N, Ma MC, Abbey NW, Herndier B, Goldsmith MA. STAT5 promotes multilineage hematolymphoid development in vivo through effects on early hematopoietic progenitor cells. Blood. 2002;99:95–101. doi: 10.1182/blood.V99.1.95. [DOI] [PubMed] [Google Scholar]
- 32.Ohmori K, Luo Y, Jia Y, Nishida J, Wang Z, Bunting KD, et al. IL-3 induces basophil expansion in vivo by directing granulocyte-monocyte progenitors to differentiate into basophil lineage-restricted progenitors in the bone marrow and by increasing the number of basophil/mast cell progenitors in the spleen. J Immunol. 2009;182:2835–41. doi: 10.4049/jimmunol.0802870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ihle JN. Interleukin-3 and hematopoiesis. Chem Immunol. 1992;51:65–106. doi: 10.1159/000319080. [DOI] [PubMed] [Google Scholar]
- 34.Min B, Prout M, Hu-Li J, Zhu J, Jankovic D, Morgan ES, et al. Basophils produce IL-4 and accumulate in tissues after infection with a Th2-inducing parasite. J Exp Med. 2004;200:507–17. doi: 10.1084/jem.20040590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen T, Kim S, Do JS, Wang L, Lantz C, Urban JF, et al. T cell-derived IL-3 plays key role in parasite infection-induced basophil production but is dispensable for in vivo basophil survival. Int Immunol. 2008;20:1201–9. doi: 10.1093/intimm/dxn077. [DOI] [PubMed] [Google Scholar]
- 36.Valent P, Schmidt G, Besemer J, Mayer P, Zenke G, Liehl E, et al. Interleukin-3 is a differentiation factor for human basophils. Blood. 1989;73:1763–9. [PubMed] [Google Scholar]
- 37.Ben-Sasson SZ, Le Gros G, Conrad DH, Finkelman FD, Paul WE. Cross-linking Fc receptors stimulate splenic non-B, non-T cells to secrete interleukin 4 and other lymphokines. Proc Natl Acad Sci U S A. 1990;87:1421–5. doi: 10.1073/pnas.87.4.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le Gros G, Ben-Sasson SZ, Conrad DH, Clark-Lewis I, Finkelman FD, Plaut M, et al. IL-3 promotes production of IL-4 by splenic non-B, non-T cells in response to Fc receptor cross-linkage. J Immunol. 1990;145:2500–6. [PubMed] [Google Scholar]
- 39.Finkelman FD, Madden KB, Morris SC, Holmes JM, Boiani N, Katona IM, et al. Anti-cytokine antibodies as carrier proteins. Prolongation of in vivo effects of exogenous cytokines by injection of cytokine-anti-cytokine antibody complexes. J Immunol. 1993;151:1235–44. [PubMed] [Google Scholar]
- 40.Siracusa MC, Saenz SA, Hill DA, Kim BS, Headley MB, Doering TA, et al. TSLP promotes interleukin-3-independent basophil haematopoiesis and type 2 inflammation. Nature. 2011;477:229–33. doi: 10.1038/nature10329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsai FY, Orkin SH. Transcription factor GATA-2 is required for proliferation/survival of early hematopoietic cells and mast cell formation, but not for erythroid and myeloid terminal differentiation. Blood. 1997;89:3636–43. [PubMed] [Google Scholar]
- 42.Iwasaki H, Mizuno S, Arinobu Y, Ozawa H, Mori Y, Shigematsu H, et al. The order of expression of transcription factors directs hierarchical specification of hematopoietic lineages. Genes Dev. 2006;20:3010–21. doi: 10.1101/gad.1493506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol. 2006;24:607–56. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- 44.Qi X, Nishida J, Chaves L, Ohmori K, Huang H. CCAAT/enhancer-binding protein alpha (C/EBPalpha) is critical for interleukin-4 expression in response to FcepsilonRI receptor cross-linking. J Biol Chem. 2011;286:16063–73. doi: 10.1074/jbc.M110.213389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yagi R, Tanaka S, Motomura Y, Kubo M. Regulation of the Il4 gene is independently controlled by proximal and distal 3′ enhancers in mast cells and basophils. Mol Cell Biol. 2007;27:8087–97. doi: 10.1128/MCB.00631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ansel KM, Greenwald RJ, Agarwal S, Bassing CH, Monticelli S, Interlandi J, et al. Deletion of a conserved Il4 silencer impairs T helper type 1-mediated immunity. Nat Immunol. 2004;5:1251–9. doi: 10.1038/ni1135. [DOI] [PubMed] [Google Scholar]
- 47.Shelburne CP, McCoy ME, Piekorz R, Sexl V, Roh KH, Jacobs-Helber SM, et al. Stat5 expression is critical for mast cell development and survival. Blood. 2003;102:1290–7. doi: 10.1182/blood-2002-11-3490. [DOI] [PubMed] [Google Scholar]
- 48.Barnstein BO, Li G, Wang Z, Kennedy S, Chalfant C, Nakajima H, et al. Stat5 expression is required for IgE-mediated mast cell function. J Immunol. 2006;177:3421–6. doi: 10.4049/jimmunol.177.5.3421. [DOI] [PubMed] [Google Scholar]
- 49.Bailey DP, Kashyap M, Bouton LA, Murray PJ, Ryan JJ. Interleukin-10 induces apoptosis in developing mast cells and macrophages. J Leukoc Biol. 2006;80:581–9. doi: 10.1189/jlb.0405201. [DOI] [PubMed] [Google Scholar]
- 50.Kennedy Norton S, Barnstein B, Brenzovich J, Bailey DP, Kashyap M, Speiran K, et al. IL-10 suppresses mast cell IgE receptor expression and signaling in vitro and in vivo. J Immunol. 2008;180:2848–54. doi: 10.4049/jimmunol.180.5.2848. [DOI] [PubMed] [Google Scholar]
- 51.Speiran K, Bailey DP, Fernando J, Macey M, Barnstein B, Kolawole M, et al. Endogenous suppression of mast cell development and survival by IL-4 and IL-10. J Leukoc Biol. 2009;85:826–36. doi: 10.1189/jlb.0708448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ryan JJ, DeSimone S, Klisch G, Shelburne C, McReynolds LJ, Han K, et al. IL-4 inhibits mouse mast cell Fc epsilonRI expression through a STAT6-dependent mechanism. J Immunol. 1998;161:6915–23. [PubMed] [Google Scholar]
- 53.Macey MR, Sturgill JL, Morales JK, Falanga YT, Morales J, Norton SK, et al. IL-4 and TGF-beta 1 counterbalance one another while regulating mast cell homeostasis. J Immunol. 2010;184:4688–95. doi: 10.4049/jimmunol.0903477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Toru H, Ra C, Nonoyama S, Suzuki K, Yata J, Nakahata T. Induction of the high-affinity IgE receptor (Fc epsilon RI) on human mast cells by IL-4. Int Immunol. 1996;8:1367–73. doi: 10.1093/intimm/8.9.1367. [DOI] [PubMed] [Google Scholar]
- 55.Yeatman CF, 2nd, Jacobs-Helber SM, Mirmonsef P, Gillespie SR, Bouton LA, Collins HA, et al. Combined stimulation with the T helper cell type 2 cytokines interleukin (IL)-4 and IL-10 induces mouse mast cell apoptosis. J Exp Med. 2000;192:1093–103. doi: 10.1084/jem.192.8.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bouton LA, Ramirez CD, Bailey DP, Yeatman CF, Yue J, Wright HV, et al. Costimulation with interleukin-4 and interleukin-10 induces mast cell apoptosis and cell-cycle arrest: the role of p53 and the mitochondrion. Exp Hematol. 2004;32:1137–45. doi: 10.1016/j.exphem.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 57.Kumar MS, Hancock DC, Molina-Arcas M, Steckel M, East P, Diefenbacher M, et al. The GATA2 transcriptional network is requisite for RAS oncogene-driven non-small cell lung cancer. Cell. 2012;149:642–55. doi: 10.1016/j.cell.2012.02.059. [DOI] [PubMed] [Google Scholar]
- 58.Levy C, Nechushtan H, Razin E. A new role for the STAT3 inhibitor, PIAS3: a repressor of microphthalmia transcription factor. J Biol Chem. 2002;277:1962–6. doi: 10.1074/jbc.M109236200. [DOI] [PubMed] [Google Scholar]
- 59.Sonnenblick A, Levy C, Razin E. Interplay between MITF, PIAS3, and STAT3 in mast cells and melanocytes. Mol Cell Biol. 2004;24:10584–92. doi: 10.1128/MCB.24.24.10584-10592.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rothenberg ME. Eosinophilia. N Engl J Med. 1998;338:1592–600. doi: 10.1056/NEJM199805283382206. [DOI] [PubMed] [Google Scholar]
- 61.Wardlaw AJ, Moqbel R, Kay AB. Eosinophils: biology and role in disease. Adv Immunol. 1995;60:151–266. doi: 10.1016/S0065-2776(08)60586-6. [DOI] [PubMed] [Google Scholar]
- 62.Shinkai K, Mohrs M, Locksley RM. Helper T cells regulate type-2 innate immunity in vivo. Nature. 2002;420:825–9. doi: 10.1038/nature01202. [DOI] [PubMed] [Google Scholar]
- 63.Chen L, Grabowski KA, Xin JP, Coleman J, Huang Z, Espiritu B, et al. IL-4 induces differentiation and expansion of Th2 cytokine-producing eosinophils. J Immunol. 2004;172:2059–66. doi: 10.4049/jimmunol.172.4.2059. [DOI] [PubMed] [Google Scholar]
- 64.Zhu Y, Chen L, Huang Z, Alkan S, Bunting KD, Wen R, et al. Cutting edge: IL-5 primes Th2 cytokine-producing capacity in eosinophils through a STAT5-dependent mechanism. J Immunol. 2004;173:2918–22. doi: 10.4049/jimmunol.173.5.2918. [DOI] [PubMed] [Google Scholar]
- 65.Adachi T, Alam R. The mechanism of IL-5 signal transduction. Am J Physiol. 1998;275:C623–33. doi: 10.1152/ajpcell.1998.275.3.C623. [DOI] [PubMed] [Google Scholar]
- 66.Sato S, Katagiri T, Takaki S, Kikuchi Y, Hitoshi Y, Yonehara S, et al. IL-5 receptor-mediated tyrosine phosphorylation of SH2/SH3-containing proteins and activation of Bruton’s tyrosine and Janus 2 kinases. J Exp Med. 1994;180:2101–11. doi: 10.1084/jem.180.6.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ogata N, Kikuchi Y, Kouro T, Tomonaga M, Takatsu K. The activation of the JAK2/STAT5 pathway is commonly involved in signaling through the human IL-5 receptor. Int Arch Allergy Immunol. 1997;114(Suppl 1):24–7. doi: 10.1159/000237712. [DOI] [PubMed] [Google Scholar]
- 68.Liu X, Robinson GW, Gouilleux F, Groner B, Hennighausen L. Cloning and expression of Stat5 and an additional homologue (Stat5b) involved in prolactin signal transduction in mouse mammary tissue. Proc Natl Acad Sci U S A. 1995;92:8831–5. doi: 10.1073/pnas.92.19.8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Teglund S, McKay C, Schuetz E, van Deursen JM, Stravopodis D, Wang D, et al. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell. 1998;93:841–50. doi: 10.1016/S0092-8674(00)81444-0. [DOI] [PubMed] [Google Scholar]
- 70.Moro K, Yamada T, Tanabe M, Takeuchi T, Ikawa T, Kawamoto H, et al. Innate production of T(H)2 cytokines by adipose tissue-associated c-Kit(+)Sca-1(+) lymphoid cells. Nature. 2010;463:540–4. doi: 10.1038/nature08636. [DOI] [PubMed] [Google Scholar]
- 71.Neill DR, Wong SH, Bellosi A, Flynn RJ, Daly M, Langford TKA, et al. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature. 2010;464:1367–70. doi: 10.1038/nature08900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brickshawana A, Shapiro VS, Kita H, Pease LR. Lineage(-)Sca1+c-Kit(-)CD25+ cells are IL-33-responsive type 2 innate cells in the mouse bone marrow. J Immunol. 2011;187:5795–804. doi: 10.4049/jimmunol.1102242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Halim TY, MacLaren A, Romanish MT, Gold MJ, McNagny KM, Takei F. Retinoic-acid-receptor-related orphan nuclear receptor alpha is required for natural helper cell development and allergic inflammation. Immunity. 2012;37:463–74. doi: 10.1016/j.immuni.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 74.Hoyler T, Klose CS, Souabni A, Turqueti-Neves A, Pfeifer D, Rawlins EL, et al. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity. 2012;37:634–48. doi: 10.1016/j.immuni.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Adolfsson J, Borge OJ, Bryder D, Theilgaard-Mönch K, Astrand-Grundström I, Sitnicka E, et al. Upregulation of Flt3 expression within the bone marrow Lin(-)Sca1(+)c-kit(+) stem cell compartment is accompanied by loss of self-renewal capacity. Immunity. 2001;15:659–69. doi: 10.1016/S1074-7613(01)00220-5. [DOI] [PubMed] [Google Scholar]
- 76.Mjösberg J, Bernink J, Golebski K, Karrich JJ, Peters CP, Blom B, et al. The transcription factor GATA3 is essential for the function of human type 2 innate lymphoid cells. Immunity. 2012;37:649–59. doi: 10.1016/j.immuni.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 77.Saenz SA, Siracusa MC, Perrigoue JG, Spencer SP, Urban JF, Jr., Tocker JE, et al. IL25 elicits a multipotent progenitor cell population that promotes T(H)2 cytokine responses. Nature. 2010;464:1362–6. doi: 10.1038/nature08901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Buonocore S, Ahern PP, Uhlig HH, Ivanov II, Littman DR, Maloy KJ, et al. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature. 2010;464:1371–5. doi: 10.1038/nature08949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Geremia A, Arancibia-Cárcamo CV, Fleming MP, Rust N, Singh B, Mortensen NJ, et al. IL-23-responsive innate lymphoid cells are increased in inflammatory bowel disease. J Exp Med. 2011;208:1127–33. doi: 10.1084/jem.20101712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cupedo T, Crellin NK, Papazian N, Rombouts EJ, Weijer K, Grogan JL, et al. Human fetal lymphoid tissue-inducer cells are interleukin 17-producing precursors to RORC+ CD127+ natural killer-like cells. Nat Immunol. 2009;10:66–74. doi: 10.1038/ni.1668. [DOI] [PubMed] [Google Scholar]
- 81.Watford WT, Hissong BD, Bream JH, Kanno Y, Muul L, O’Shea JJ. Signaling by IL-12 and IL-23 and the immunoregulatory roles of STAT4. Immunol Rev. 2004;202:139–56. doi: 10.1111/j.0105-2896.2004.00211.x. [DOI] [PubMed] [Google Scholar]
- 82.Sonnenberg GF, Monticelli LA, Alenghat T, Fung TC, Hutnick NA, Kunisawa J, et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science. 2012;336:1321–5. doi: 10.1126/science.1222551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cella M, Fuchs A, Vermi W, Facchetti F, Otero K, Lennerz JK, et al. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 2009;457:722–5. doi: 10.1038/nature07537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Luci C, Reynders A, Ivanov II, Cognet C, Chiche L, Chasson L, et al. Influence of the transcription factor RORgammat on the development of NKp46+ cell populations in gut and skin. Nat Immunol. 2009;10:75–82. doi: 10.1038/ni.1681. [DOI] [PubMed] [Google Scholar]
- 85.Sanos SL, Bui VL, Mortha A, Oberle K, Heners C, Johner C, et al. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat Immunol. 2009;10:83–91. doi: 10.1038/ni.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Satoh-Takayama N, Vosshenrich CA, Lesjean-Pottier S, Sawa S, Lochner M, Rattis F, et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008;29:958–70. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 87.Crellin NK, Trifari S, Kaplan CD, Satoh-Takayama N, Di Santo JP, Spits H. Regulation of cytokine secretion in human CD127(+) LTi-like innate lymphoid cells by Toll-like receptor 2. Immunity. 2010;33:752–64. doi: 10.1016/j.immuni.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 88.Satoh-Takayama N, Dumoutier L, Lesjean-Pottier S, Ribeiro VS, Mandelboim O, Renauld JC, et al. The natural cytotoxicity receptor NKp46 is dispensable for IL-22-mediated innate intestinal immune defense against Citrobacter rodentium. J Immunol. 2009;183:6579–87. doi: 10.4049/jimmunol.0901935. [DOI] [PubMed] [Google Scholar]
- 89.Veldhoen M, Hirota K, Westendorf AM, Buer J, Dumoutier L, Renauld JC, et al. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–9. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- 90.Trifari S, Kaplan CD, Tran EH, Crellin NK, Spits H. Identification of a human helper T cell population that has abundant production of interleukin 22 and is distinct from T(H)-17, T(H)1 and T(H)2 cells. Nat Immunol. 2009;10:864–71. doi: 10.1038/ni.1770. [DOI] [PubMed] [Google Scholar]