Abstract
Black widow spiders (members of the genus Latrodectus) are widely feared because of their potent neurotoxic venom. α-Latrotoxin is the vertebrate-specific toxin responsible for the dramatic effects of black widow envenomation. The evolution of this toxin is enigmatic because only two α-latrotoxin sequences are known. In this study, ∼4 kb α-latrotoxin sequences and their homologs were characterized from a diversity of Latrodectus species, and representatives of Steatoda and Parasteatoda, establishing the wide distribution of latrotoxins across the mega-diverse spider family Theridiidae. Across black widow species, α-latrotoxin shows ≥94% nucleotide identity and variability consistent with purifying selection. Multiple codon and branch-specific estimates of the nonsynonymous/synonymous substitution rate ratio also suggest a long history of purifying selection has acted on α-latrotoxin across Latrodectus and Steatoda. However, α-latrotoxin is highly divergent in amino acid sequence between these genera, with 68.7% of protein differences involving non-conservative substitutions, evidence for positive selection on its physiochemical properties and particular codons, and an elevated rate of nonsynonymous substitutions along α-latrotoxin’s Latrodectus branch. Such variation likely explains the efficacy of red-back spider, L. hasselti, antivenom in treating bites from other Latrodectus species, and the weaker neurotoxic symptoms associated with Steatoda and Parasteatoda bites. Long-term purifying selection on α-latrotoxin indicates its functional importance in black widow venom, even though vertebrates are a small fraction of their diet. The greater differences between Latrodectus and Steatoda α-latrotoxin, and their relationships to invertebrate-specific latrotoxins, suggest a shift in α-latrotoxin toward increased vertebrate toxicity coincident with the evolution of widow spiders.
Keywords: venom, α-latrotoxin, Latrodectus, toxin evolution, black widow spider
Introduction
Venoms are chemically complex secretions produced by some animals for defense or prey acquisition. Several organismal lineages have independently become venomous and exhibit morphological convergence in venom-injecting organs, as well as similarities at the molecular level in their toxin repertoire due to independent recruitment of related genes for venom production (Fry et al. 2009). Venom toxins have drawn enormous scientific attention because of their applications as pharmaceuticals and as probes for isolating cellular receptors (Adams and Olivera 1994; Lewis and Garcia 2003; Veiseh et al. 2007; Williams et al. 2008). Genes encoding venom toxins are also of significant evolutionary interest because of their direct role in organismal fitness and ecological adaptation. Notably, venom molecular evolution is highly dynamic and appears to be shaped by frequent gene duplications and strong diversifying selection, as well as co-evolution and convergence (Duda and Palumbi 1999; Li et al. 2005; Fry et al. 2006; Aminetzach et al. 2009; Binford et al. 2009; Doley et al. 2009).
The order Araneae (spiders) is the largest clade of venomous organisms, but biochemical characterization of their venoms has been restricted to a small sampling of species (Fletcher et al. 1997; Adams 2004; Ushkaryov et al. 2004; Wullschleger et al. 2004), and evolutionary analyses of spider venoms have been limited (Sollod et al. 2005; Binford et al. 2009). A striking example of this dearth of studies is the molecular composition of black widow spider venom, which has been extensively characterized from only 1 of the 31 Latrodectus species in the family Theridiidae (Latrodectus tredecimguttatus; Knipper et al. 1986; Kiyatkin et al. 1990, Kiyatkin, Dulubova, and Grishin 1993; Dulubova et al. 1996; Volynski, Volkova, et al. 1999). “Black widow” is a common name referring to several Latrodectus species (e.g., L. tredecimguttatus, L. mactans, L. hesperus, and L. variolus), which are widely recognized and feared because of the extreme neurotoxicity of their venom and their abundance in human-inhabited areas (Clark et al. 1992; Muller 1992; Vetter and Isbister 2008). Other Latrodectus species, including the sexually cannibalistic Australian red-back spider (L. hasselti) and highly invasive brown widow (L. geometricus), also inflict painful bites with similar physiological effects to those from black widows but with varying degrees of potency (Muller et al. 1989, 1992; Muller 1992; Isbister and Gray 2003b).
Complementary DNA (cDNAs) for two families of venom protein components—latrodectins and latrotoxins—have been cloned from the venom glands of the Eurasian black widow L. tredecimguttatus. Latrodectins include only two paralogous sequences, with unclear functional roles (Kiyatkin, Dulubova, Chekhovskaya, et al. 1993; Pescatori et al. 1995). By contrast, numerous studies have shown that latrotoxins are the primary neurotoxins in black widow spider venom (Knipper et al. 1986; Kiyatkin et al. 1990, Kiyatkin, Dulubova, and Grishin 1993; Dulubova et al. 1996; Volynski, Volkova, et al. 1999). The four sequenced latrotoxin paralogs encode long polypeptides (1,200–1,400 amino acids) that share 30–60% amino acid sequence identity and an overall similarity in domain organization (Ushkaryov et al. 2004). Although their general role in stimulating neurotransmitter secretion is also similar, experimental evidence suggests the latrotoxins vary in target specificity. For example, α-latroinsectotoxin and δ-latroinsectotoxin appear to selectively affect insect neurons, whereas α-latrocrustotoxin stimulates secretion from crustacean neurons, but not from certain insects (Fritz et al. 1980; Krasnopernov et al. 1991; Magazanik et al. 1992; Kiyatkin et al. 1995; Elrick and Charlton 1999). The fourth paralog, α-latrotoxin, is a vertebrate-specific neurotoxin that causes the extreme pain following black widow bites (Knipper et al. 1986; Kiyatkin et al. 1990; Volynski, Nosyreva, et al. 1999).
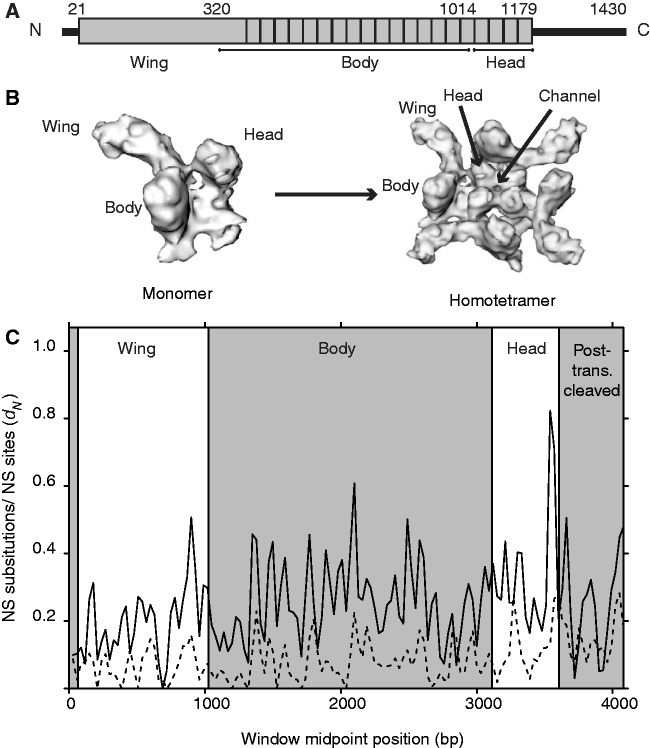
Because of its role in human toxicity and its importance for understanding vertebrate neurosecretion, considerable efforts have focused on the structure–function relationship of α-latrotoxin. The three-dimensional form of α-latrotoxin is a homotetramer resembling a propeller with four “blades” and a central pore, through which calcium ions pass (Orlova et al. 2000). α-Latrotoxin binds to the vertebrate pre-synaptic neuronal receptor neurexin and inserts into the neuronal membrane, becoming a transmembrane channel (Orlova et al. 2000). This leads to an influx of extracellular calcium through α-latrotoxin’s central pore, triggering a massive uncontrolled exocytosis of neurotransmitters. Neurotransmitter release may also be stimulated by α-latrotoxin dimers via intracellular calcium release upon binding to the latrophilin/CIRL receptor (Volynski et al. 2003). The α-latrotoxin monomers have three tandem domains: the wing, body, and head region (320, 694, and 163 amino acids, respectively; Orlova et al. 2000; fig. 1A and B). The wing region is hypothesized to be a receptor-binding domain, whereas the body and head make up the transmembrane channel (Orlova et al. 2000; Ushkaryov et al. 2004).
Fig. 1.
(A) Schematic of the primary structure of α-latrotoxin indicating the functional domains, with numbers indicating amino acid position, boxes showing position of 22 ankyrin repeats, and black lines indicating amino (N)- and carboxy (C)-terminal ends that are posttranslationally cleaved to yield the mature protein. (B) (Left) Three-dimensional structure of α-latrotoxin, and (right) homotetramer indicating position of the protein domains determined from cryo-EM microscopy (modified from Ushkaryov et al. 2004). (C) Sliding window analysis of dN (nonsynonymous substitutions/nonsynonymous site) between α-latrotoxin from Latrodectus tredecimguttatus versus Steatoda grossa (black line) and L. tredecimguttatus versus L. geometricus (dashed line) illustrating variability along gene. Permission for image in fig. 1C received from Elsevier via RightsLink under license number 3078871425016; Publication: Toxicon. Title: The multiple actions of black widow spider toxins and their selective use in neurosecretion studies. Type Of Use: reuse in a journal/magazine.
In contrast to this detailed functional knowledge of α-latrotoxin from L. tredecimguttatus, the evolution of latrotoxins is poorly understood. There is no clear evolutionary link between latrotoxins and any other known protein, and latrotoxins are unknown from spider venoms outside of Latrodectus, suggesting that black widow venom is the product of dramatic molecular evolution. Species in the theridiid genera Steatoda and Parasteatoda can inflict bites with similar, but far less severe, neurotoxic symptoms to those of black widows (Muller et al. 1992; Graudins et al. 2002; Isbister and Gray 2003a). Moreover, median lethal dose values (LD50) in mice vary substantially among venoms from different Latrodectus and Steatoda species (Muller et al. 1989, 1992). The molecular basis for this interspecific diversity may be explained by variation in the phylogenetic distribution, expression, or sequence characteristics of α-latrotoxin. The adaptive significance of a vertebrate-specific toxin in black widow venom is unclear, given that their diet is primarily invertebrate-based, although capture and consumption of small vertebrates by widow spiders is well documented (e.g., geckos, small lizards, snakes, and mice; McCormick and Polis 1982; Hodar and Sanchez-Pinero 2002).
The recent cloning of α-latrotoxin from L. hasselti confirms its presence in other Latrodectus species (Graudins et al. 2012); however, the broader genetic variability and evolution of α-latrotoxin remains largely unknown. Through a combination of genomic polymerase chain reaction (PCR), reverse transcriptase-PCR (RT-PCR) of venom gland cDNA and inverse PCR, we have obtained the coding sequence of the α-latrotoxin gene in its approximate entirety (∼4 kb) from divergent representatives of Latrodectus and Steatoda species. We estimated evolutionary relationships of these sequences to other latrotoxin gene family members and have investigated patterns of variability and selection across α-latrotoxin’s structural domains using multiple methods. Further, we have sequenced a portion of α-latrotoxin spanning parts of the wing and body domain from a denser sampling of Latrodectus species. These data were compared with the mitochondrial gene cytochrome c oxidase I (mt COI) to evaluate the relative rate of α-latrotoxin evolution. Our results indicate a strong functional role for α-latrotoxin in a larger set of species than previously recognized, which has implications for the clinical treatment of widow spider bites, as well as for understanding the evolutionary ecology of black widows.
Results
α-Latrotoxin Sequence Variability
We obtained eight approximately 4 kb α-latrotoxin sequences from divergent Latrodectus species and Steatoda grossa. These sequences exhibited minor length variation, with L. geometricus and S. grossa α-latrotoxin having an overlapping 6 bp insertion relative to all other sequences, one additional 3 bp deletion in L. geometricus, and two separate 3 bp deletions in S. grossa. As previously indicated by southern blots (Danilevich and Grishin 2000) and genomic sequence of α-latrotoxin from L. hasselti (Graudins et al. 2012), all α-latrotoxin gene sequences appear to be intronless. Translations of obtained α-latrotoxin sequences did not contain any unexpected stop codons. We also obtained a 618 bp fragment of α-latrotoxin, spanning part of its wing and body domains, from 41 specimens (sampling 18 Latrodectus and two Steatoda species). Translations of the 618 bp fragment of α-latrotoxin from all Latrodectus species also exhibited no length variation or stop codons, but Steatoda capensis had a 9 bp deletion, and two α-latrotoxin paralogs sequenced from Parasteatoda were either 27 or 15 bp shorter than Latrodectus α-latrotoxin. All NCBI Accession numbers for these sequences are listed in supplementary table S1, Supplementary Material online.
Maximal amino acid distance among α-latrotoxin sequences was 35.6% (between S. grossa and L. tredecimguttatus), similar to the distance between the functionally distinct paralogs α-latroinsectotoxin and α-latrocrustotoxin from L. tredecimguttatus (35.5%) but was no more than 16% different within Latrodectus (supplementary table S2, Supplementary Material online). Moreover, 68.7% of all differences between S. grossa and L. tredecimguttatus α-latrotoxin involved nonconservative changes between different physiochemical classes of amino acids (e.g., between hydrophobic and hydrophilic classes), as determined from Livingstone and Barton (1993) categorization of residue properties (supplementary fig. 1, Supplementary Material online). Within Latrodectus, the highest average uncorrected nucleotide distance among α-latrotoxins was 6.1%, which was less than corresponding mt COI distances (13.5%; supplementary table S2, Supplementary Material online). This pattern was reversed when comparing S. grossa with Latrodectus species, with α-latrotoxin average nucleotide distance (27.9%) being more divergent than average mt COI distance (19.3%; supplementary table S2, Supplementary Material online). Likelihood ratio tests of rate homogeneity were rejected for the 4 kb α-latrotoxin sequences (P < 0.01), but could not be rejected for mt COI sequences from the same species (P = 0.08). The greater sequence divergence in S. grossa versus Latrodectus α-latrotoxin relative to mt COI, but the reverse pattern among Latrodectus, could be due to α-latrotoxin having an accelerated evolutionary rate outside of Latrodectus, and/or a deceleration of its substitution rate within the genus. It should be noted that mt COI shows evidence of saturation in third codon positions outside of Latrodectus (Garb et al. 2004), which likely constrains the degree to which it could vary above 19.3% because nearly all changes are restricted to third codon positions. α-Latrotoxin exhibits much greater divergence in first and second codon positions outside of the Latrodectus genus, but it is still half as divergent as mt COI within the genus.
α-Latrotoxin domains exhibited increasingly greater divergence from the N-terminal toward the C-terminal domains (wing < body < head < posttranslationally cleaved domains; supplementary table S2, Supplementary Material online). Pairwise nonsynonymous substitutions per nonsynonymous site (dN) for α-latrotoxin between L. tredecimguttatus versus L. geometricus or versus S. grossa, plotted in a sliding window analysis, similarly shows an overall trend of increasing dN values going from a 5′ to 3′ direction, as well as substantial divergence between (in contrast to within) genera (fig. 1C and supplementary table S2, Supplementary Material online). Some pairwise estimates of the ratio of dN to synonymous substitutions per synonymous site (dS) (ω, dN/dS) for the α-latrotoxin head domain among Latrodectus sequences are as much as 1.69, but never exceed 0.508 in any other domain (supplementary table S2, Supplementary Material online). Moreover, no pairwise estimates of ω greater than 1 were significantly larger than 1, as determined by codon-based Z-tests of selection performed in MEGA5 (Tamura et al. 2011), which failed to reject the hypothesis of dN = dS in favor of the alternative hypothesis of dN > dS for any pairwise comparison. All cysteine residues are 100% conserved across the approximately 4.2 kb α-latrotoxin sequences. In one P. tepidariorum latrotoxin clone, position 393 (a conserved cysteine in all sequences except α-latrocrustotoxin) was an asparagine; mutation of the cysteine in this position in L. tredecimguttatus α-latrotoxin into a serine leads to an interrupted disulfide bond and functional loss of the toxin (Ichtchenko et al. 1998).
BLASTx searches against the UniProtKB database found the greatest similarity between α-latrotoxin and previously reported latrotoxins, followed by ankyrin 1 from Homo sapiens (E score = 2 e−60) or dTRPA1 (transient receptor potential cation channel subfamily A member 1) from Drosophila when limiting the database to arthropods (supplementary table S3, Supplementary Material online).
Phylogenetic Analyses of α-Latrotoxin and mt COI
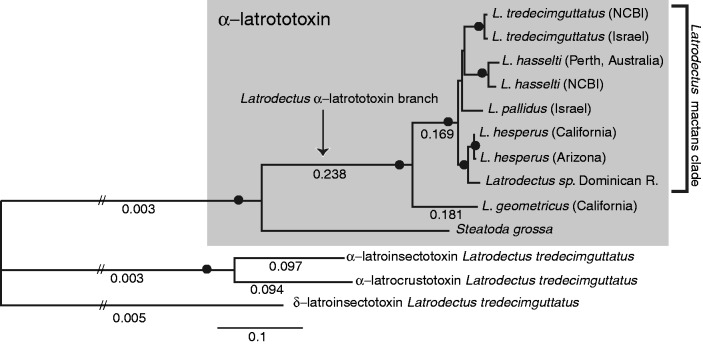
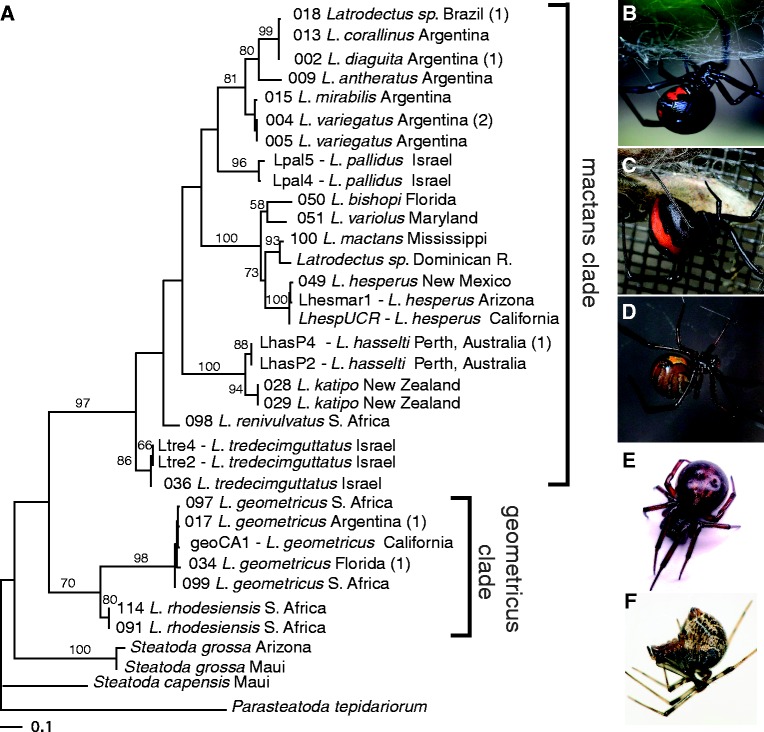
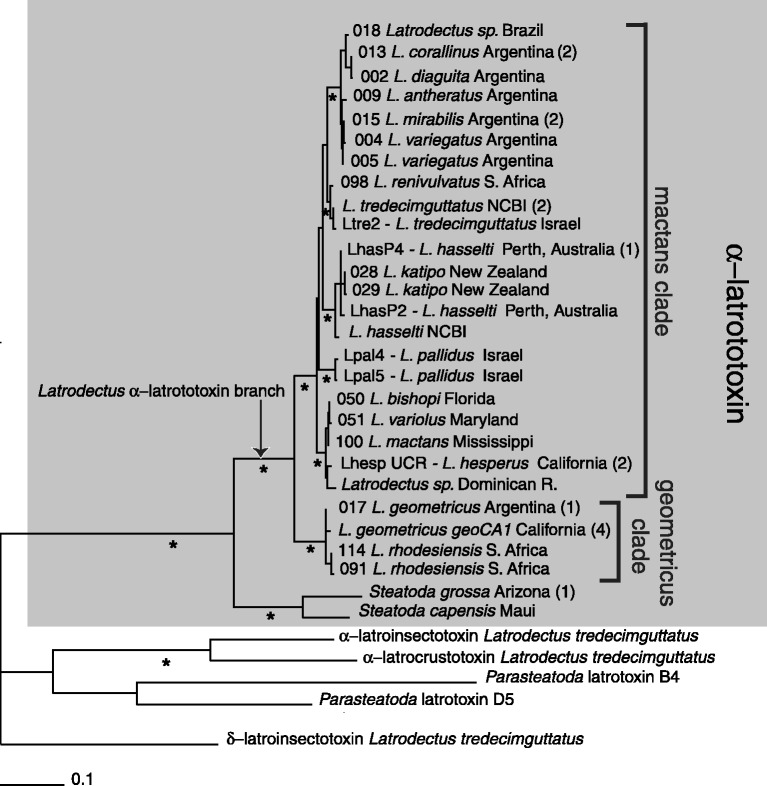
We constructed phylogenetic trees using parsimony, maximum likelihood (ML), and partitioned Bayesian analyses (partitioned by codon position) from nucleotide alignments of the following three data sets: 1) approximately 4 kb of α-latrotoxin and latrotoxin paralogs from a subset of taxa, 2) the conserved 618 bp fragment of α-latrotoxin spanning 28.1% of the wing and 16.4% of the body domains from 41 specimens, and 3) 428–659 bp of mt COI sequences from all specimens, including and excluding third codons positions. The longer latrotoxin sequence alignment (4,233 bp) contained 3,215 variable sites, 2,066 of which were parsimony informative. The Akaike Information Criteria (AIC) in jModeltest selected the GTR + G substitution model for this data set (all model selections for each analysis reported in supplementary table S4, Supplementary Material online). The ML tree (−ln L = 26,441.57532; fig. 2), the partitioned Bayesian consensus tree and the single most parsimonious tree (length = 5,741, consistency index [CI] = 0.835, retention index [RI] = 0.734, rescaled consistency index [RC] = 0.612) were identical in topology with high bootstrap values and posterior probabilities (0.93–1.00 for all nodes), respectively. Relationships of α-latrotoxin largely reflect species relationships previously estimated with all mitochondrial COI codon positions by Garb et al. (2004) as well as with our new data (fig. 3 and supplementary figs. S2 and S3, Supplementary Material online). Within Latrodectus, the mactans clade includes all species other than L. geometricus and L. rhodesiensis and these latter two comprise the geometricus clade. Unlike the ML and parsimony analysis, the partitioned Bayesian consensus of all mt COI codon positions did not recover Latrodectus as monophyletic, instead placing the two Steatoda species as sister to the mactans clade to the exclusion of the geometricus clade with a posterior probability of 0.96 (supplementary fig. S2, Supplementary Material online). Monophyly of Latrodectus was also not supported by parsimony, ML and Bayesian analyses of mt COI excluding third codon positions (supplementary figs. S4 and S5, Supplementary Material online), and these trees were less well resolved than mt COI trees including third codon positions. The limited resolution of these trees is likely due to the decrease in parsimony-informative characters from 220 in all codons positions to 42 in first and second codons positions alone. Analyses of the 618 bp fragment of α-latrotoxin (642 bp in alignment) yielded two ML trees (fig. 4; −ln L = 5,853.97981) that differ only in the arrangement of nearly identical sequences (L. hasselti LhasP2 and LhasP4 relative to the two L. katipo sequences) and both topologies are congruent with the partitioned Bayesian consensus, except for a few nodes with weak support (supplementary fig. S6, Supplementary Material online). Parsimony analyses of this data set resulted in six most parsimonious trees (supplementary fig. S7, Supplementary Material online; length = 5,741, CI = 0.680, RI = 0.723, RC = 0.491). The strict consensus is similar in overall structure to the ML and partitioned Bayesian trees, with sequences falling into well-supported geometricus and mactans clades, but with less resolution among some deeper nodes within the mactans clade. Sequences from P. tepidariorum are highly divergent and not united with the Latrodectus and Steatoda α-latrotoxins, indicating they represent α-latrotoxin paralogs.
Fig. 2.
Phylogram based on ML analysis of 4.2 kb α-latrotoxin alignment, with black dots indicating nodes with 100% bootstrap support in ML and parsimony analyses and with posterior probability values of 1.0 in partitioned Bayesian analyses. Numbers under branches are ω values estimated with codeml using the free ratio model; all other ω values not shown range from 0.000 to 0.556. Hatched lines indicate shortened branches for figure quality. The tree was rooted with the α-latrotoxin latrotoxin paralog δ-latroinsectotoxin.
Fig. 3.
(A) Phylogram based on ML analysis of 659 bp alignment of mt COI sequences (−ln L = 4,085.00413) collected from sampled species including those in figure 4. Numbers above nodes show bootstrap support values from 100 replicates, nodes without numbers received <50% bootstrap support. Numbers in parentheses indicate numbers of additional identical sequences (supplementary table S1, Supplementary Material online: 018 Latrodectus sp. identical to 020 L. corallinus; 002 L. diaguita identical to 003 L. diaguita; 004 L. mirabilis identical to 014 L. mirabilis and 010 Latrodectus sp.; L. hasselti LhasP4 identical to LhasP5; 017 L. geometricus identical to 016 L. geometricus; 034 L. geometricus identical to 033 L. geometricus). Partitioned Bayesian and parsimony consensus trees for these data are shown in supplementary figures 2 and 3, Supplementary Material online, respectively. Numbers before names indicate number assigned to individuals in voucher collection. (B–F) Subset of examined species with images obtained via www.photopin.com (last accessed February 1, 2013) under the Creative Commons license as follows: (B) black widow, L. mactans (http://www.flickr.com/photos/theloushe/5779238278/, last accessed February 1, 2013); (C) red-back spider, L. hasselti, with lizard prey (http://www.flickr.com/photos/ozwildlife/752256755/, last accessed February 1, 2013); (D) brown widow, L. geometricus, (http://www.flickr.com/photos/scorche/3308329124/, last accessed February 1, 2013); (E) cupboard spider, Steatoda grossa (http://www.flickr.com/photos/lofaesofa/2183895562/, last accessed February 1, 2013); (F) common house spider, Parasteatoda tepidariorum (http://www.flickr.com/photos/blueridgekitties/4828847855/, last accessed February 1, 2013). The tree was rooted with the P. tepidariorum mt COI sequence.
Fig. 4.
ML phylogram from conserved 642 bp alignment (618 bp PCR fragment) of α-latrotoxin and latrotoxin homologs. Asterisks indicate nodes supported by ≥88% bootstrap support from 100 replicates (ML) or 1,000 replicates (parsimony), and ≥0.95 clade posterior support values in partitioned Bayesian analysis. Numbers in parentheses indicate numbers of additional identical sequences (supplementary table S1, Supplementary Material online: 013 Latrodectus corallinus identical to 020 L. corallinus and 003 L. diaguita; 015 L. mirabilis identical to 014 L. mirabilis and 010 Latrodectus sp.; L. hasselti LhasP4 identical to LhasP5; L. tredecimguttatus NCBI identical to 036 L. tredecimguttatus and Ltre4; L. hesperus LhespUCR identical to Lhesmar1 and 049 L. hesperus; 017 L. geometricus identical to 016 L. geometricus; L. geometricus geoCA1 identical to 034 L. geometricus, 033 L. geometricus, 097 L. geometricus and 099 L. geometricus; S. grossa Arizona identical to S. grossa Maui). The tree was rooted with the latrotoxin paralog δ-latroinsectotoxin. Partitioned Bayesian and parsimony consensus trees for these data are shown in supplementary figures 6 and 7, Supplementary Material online, respectively.
Tests for Selection on α-Latrotoxin
The 4 kb and 642 bp α-latrotoxin alignments were used to estimate ω (dN/dS) for codons in the alignment and branches of the phylogeny to investigate patterns of selection that have acted on α-latrotoxin using three programs: 1) the codeml program in PAML 4.3 (Yang 2007); 2) multiple modules within the HyPhy package via the www.datamonkey.org (last accessed February 1, 2013) web server (Delport et al. 2010); and 3) the package ADAPTSITE (Suzuki et al. 2001). We also examined codon-specific patterns of radical and conservative amino acid replacements in the α-latrotoxin alignments using ADAPTSITE (Suzuki et al. 2001), as the ratio of these changes can reflect the influence of purifying or positive selection (Suzuki 2007). Collectively, the results of these analyses were consistent in suggesting that α-latrotoxin has largely been subjected to purifying selection across the majority of its codons and along branches of its phylogeny, with some evidence of positive selection acting on a small subset of codons (tables 1 and 2; fig. 2).
Table 1.
Summary of Codon Substitution Models Examined in Codeml for α-Latrotoxin, with Estimates of ω, Model Parameters, Likelihood Values, and Probabilities.a,b
| Model | ln L | Parameter Estimates | χ2 | Df | P |
|---|---|---|---|---|---|
| Branch 4 kb | |||||
| Free ratio | −19,296.84 | ||||
| One ratio | −19,338.7 | ω = 0.143 | 83.26 | 22 | <0.00001 |
| Fixed ratio | −20,232.88 | ω = 1 | 1,788.82 | 1 | <0.00001 |
| Two ratios | −19,334.44 | ω0 = 0.133; ω1 = 0.289 | 8.07 | 1 | <0.005 |
| Branch 642 bp | |||||
| One ratio | −3,259.68 | ω = 0.108 | |||
| Fixed ratio | −3,468.26 | ω = 1 | 417.18 | 1 | <0.00001 |
| Two ratios | −3,259.67 | ω0 = 0.108; ω1 = 0.100 | 0.01 | 1 | 0.92 |
| Sites 4 kb | |||||
| M0 | −19,338.47 | ω = 0.143 | |||
| M3 | −19,147.65 | p(0, 1, 2): 0.192, 0.631, 0.177; ω(0, 1, 2): 0.016, 0.129, 0.501 | 381.63 | 4 | <0.00001 |
| M1a | −19,227.05 | p(0, 1): 0.8568, 0.143; ω(0, 1): 0.11288, 1.00000 | |||
| M2a | −19,227.05 | p(0, 1, 2): 0.857, 0.143, 0.000; ω(0, 1, 2): 0.112, 1.000, 35.731 | 0 | 2 | 1 |
| M7 | −19,153.64 | p = 1.07987; q = 5.23687 | |||
| M8 | −19,149.98 | p0 = 0.966; p = 1.241; q = 6.936 (p1 = 0.035); ω = 1.000 | 7.32 | 2 | 0.02 |
| Sites 642 bp | |||||
| M0 | −3,259.67 | ω = 0.108 | |||
| M3 | −3,224.98 | p(0, 1, 2): 0.242, 0.617, 0.140; ω(0, 1, 2): 0.013, 0.117, 0.301 | 69.38 | 4 | <0.00001 |
| M1a | −3,252.75 | p(0, 1): 0.938, 0.062; ω(0,1): 0.099, 1.000 | |||
| M2a | −3,252.75 | p(0, 1, 2): 0.938, 0.062, 0.000; ω(0, 1, 2): 0.099, 1.000, 21.208 | 0 | 2 | 1 |
| M7 | −3,227.14 | p = 1.25897; q = 9.19405 | |||
| M8 | −3,227.14 | p0 = 0.999; p = 1.259; q = 9.194; (p1 = 0.00001); ω = 1.000 | 0.00 | 2 | 0.999 |
| Branch-sites 4 kb | |||||
| MA (mod.) | −19,216.64 | p(0, 1, 2a + 2b) = 0.760, 0.120, 0.119; ω(2a + 2b, foreground): 2.545 | |||
| MA (fixed) | −19,217.95 | Foreground ω = 1 | 2.61 | 1 | 0.106 |
| Branch-sites 642 bp | |||||
| MA (mod.) | −3,250.71 | p(0, 1, 2a + 2b) = 0.922, 0.062, 0.0167; ω(2a + 2b, foreground): 149.03 | |||
| MA (fixed) | −3,252.06 | Foreground ω = 1 | 2.68 | 1 | 0.100 |
Note.—BEB positively selected residues (posterior probability ω > 1). M8, 4 kb: 468 A 0.644; 524 T 0.596; 711 Q 0.587; 861 N 0.520; 940 S 0.620. Branch-sites, 4.2 kb: 47 A 0.760; 158 L 0.881; 187 E 0.642; 400 E 0.865; 466 T 0.770; 506 S 0.768; 511 F 0.731; 523 V 0.624; 549 Y 0.601; 556 L 0.561; 589 S 0.943; 635 G 0.823; 637 L 0.543; 652 F 0.879; 676 Q 0.554; 697 K 0.580; 712 I 0.568; 739 S 0.548; 751 W 0.873; 857 D 0.547; 892 Q 0.585; 895 R 0.733; 937 S 0.536; 981 I 0.794; 998 H 0.500; 1,035 T 0.642. Branch-sites, 642 bp: 127 T 0.899.
aPAML run with clean data = 1; ω1 is foreground branch, or branch leading to Latrodectus α-latrotoxin, whereas ω0 are for background branches.
bp(0, 1) and p(0, 1, 2) in sites models M3, M1a, and M2a refer to the proportion of sites in each of the two or three estimated ω values, respectively; p0 and p1 in the M8 model refer to the proportion of sites in the two estimated ω values; p and q in sites models M7 and M8 refer to parameters in the beta distribution B(p, q) used to estimate ω in the interval (0, 1).
Table 2.
Codons in the α-Latrotoxin Alignments Identified as Positively Selected Using FEL and/or MEME Methods with Statistical Support, BEB Posterior Probability Values if also Identified in Codeml Models as Positively Selected; Radical/Conservative Substitution Rate Ratio for Those Sites Estimated with ADAPTSITE.
| Codon Positiona | FEL (dN−dS) | FEL (P value) | MEME (ω+)b | MEME (P value) | Codeml M8 BEB Pr (ω > 1) | Codeml MA(mod) BEB Pr (ω > 1) | ADAPTSITE Radical/Conservative |
|---|---|---|---|---|---|---|---|
| 4-kb Alignment | |||||||
| 55 | 0.815 | 0.035 | >100 | 0.039 | — | — | 0.424 |
| 63 | −0.215 | 0.584 | >100 | 0.002 | — | — | 0 |
| 102 | 0.847 | 0.247 | >100 | 0.052 | — | — | 0.244 |
| 158 | 0.133 | 0.253 | >100 | 0.008 | — | 0.881 | 0 |
| 173 | 0.125 | 0.758 | >100 | 0.045 | — | — | 9.312 |
| 181 | 0.240 | 0.507 | 12.337 | 0.089 | — | — | 0 |
| 207 | −0.394 | 0.223 | 26.499 | 0.076 | — | — | 0 |
| 243 | 0.347 | 0.097 | >100 | 0.104 | — | — | 1.602 |
| 276 | −0.711 | (0.051) | >100 | 0.045 | — | — | 0 |
| 295 | −0.288 | 0.765 | 24.546 | 0.023 | — | — | 0.559 |
| 313 | 0.186 | 0.259 | >100 | 0.094 | — | — | 0 |
| 336 | 0.158 | 0.213 | >100 | 0.026 | — | — | 0 |
| 468 | 2.608 | 0.113 | >100 | 0.072 | 0.644 | — | 0.378 |
| 471 | 0.289 | 0.653 | 12.042 | 0.039 | — | — | 0.206 |
| 571 | 0.613 | 0.075 | >100 | 0.109 | — | — | 3.842 |
| 625 | 0.359 | 0.039 | >100 | 0.010 | — | — | 0 |
| 632 | 0.137 | 0.685 | >100 | 0.008 | — | — | 0 |
| 655 | 0.383 | 0.322 | >100 | 0.091 | — | — | 0.358 |
| 657 | 0.790 | 0.096 | >100 | 0.128 | — | — | 0.54 |
| 660 | 1.472 | 0.422 | >100 | 0.016 | — | — | 1.876 |
| 683 | 2.313 | 0.155 | 36.496 | 0.017 | — | — | 0.468 |
| 691 | −0.161 | 0.897 | 7.598 | 0.096 | — | — | 0.35 |
| 725 | 0.574 | 0.093 | >100 | 0.100 | — | — | 0.56 |
| 753 | −0.123 | 0.634 | >100 | 0.065 | — | — | 2.656 |
| 754 | 0.476 | 0.084 | >100 | 0.100 | — | — | 0.591 |
| 756 | 0.444 | 0.095 | >100 | 0.111 | — | — | 1.285 |
| 776 | 0.907 | 0.068 | >100 | 0.090 | — | — | 0.543 |
| 802 | 0.090 | 0.943 | >100 | 0.023 | — | — | 0 |
| 817 | 0.581 | 0.080 | >100 | 0.079 | — | — | 0.748 |
| 881 | 0.397 | 0.254 | >100 | 0.012 | — | — | 0.408 |
| 883 | 0.240 | 0.559 | >100 | 0.023 | — | — | 3.404 |
| 1008 | 0.213 | 0.262 | >100 | 0.092 | — | — | 0.419 |
| 642 bp Alignment | |||||||
| 50 | −0.304 | 0.524 | 21.312 | 0.025 | — | — | 0.618 |
| 127 | 0.036 | 0.886 | 47.663 | 0.097 | — | 0.899 | 1.336 |
Note.—Statistical support for positive selection by either method is indicated by underlined P values, value in parentheses indicates site found to be negatively selected with significance for FEL method. Em dash indicates codons not identified as positively selected by the BEB procedure in Codeml.
aSites found to be under positive selection in more than one method are underlined. For 4-kb alignment, codons 55–295 in wing domain, positions 313–883 in body domain, 1,008 in posttranslationally cleaved region; for 642-bp alignment codon 50 in wing domain, codon 127 in body domain.
bValues indicate inferred ω(β+/α), values where they exceed 100 include sites where α is 0.
The codeml-free ratio branch model of PAML estimated ω along all branches of the 4 kb α-latrotoxin phylogeny as being far less than 1 (fig. 2). Fixed branch model estimates of ω were 0.14 and 0.10 for the 4 kb and 642 bp data sets, respectively, and were a significantly better fit to the data than the branch model fixing ω to one, but a significantly worse fit to the free ratio branch model (table 1). The two-ratio branch model for the 4 kb data set, allowing the branch leading to Latrodectus α-latrotoxin to have a different ω (0.289) than the background ratio (0.133), was a significantly better fit than the one ratio model (P < 0.005). This suggests an overall history of purifying selection on α-latrotoxin, with some temporal variation along branches, including elevated ω along the Latrodectus α-latrotoxin branch. However, the two-ratio branch model was not significantly different from the one-ratio model for the 642 bp data set. For both the 4 kb and 642 bp alignments, comparisons of the M0 and M3 sites models indicated significant variation in ω among codon sites (table 1). The M7 and M8 sites models were significantly different for the 4 kb alignment (P = 0.02), and although five sites undergoing positive selection were identified with the Bayes empirical Bayes (BEB) procedure, there was insufficient statistical support for this result (all five sites with posterior probability ω > 1 being 0.644 or less). Moreover, the M2a model was not a better fit to the data than the M1a model. The MA (modified) branch-sites model for the 4 kb alignment examining evidence for selection at specific sites along the Latrodectus α-latrotoxin branch was also not significantly different than the null model, fixing this branch to an ω of 1. However, the branch-sites model of the 4 kb data set did estimate 11% of sites having a mean ω of 2.54 along the Latrodectus α-latrotoxin branch, and the BEB procedure identified 26 codons as possible targets of positive selection, but only five of the 26 had posterior probabilities between 0.85 and 0.95: 158 L, 400 E, 589 S, 652 F, and 751 W (table 1). The first of these sites is located in the wing domain, while the other four are located in various positions in the body domain. All but the last of these five sites involve changes to amino acids in different physiochemical categories. For the 642 bp alignment, the sites model M8 was not significantly different from M7, nor was the MA (modified) branch-sites model significantly different from the null. However, the BEB procedure following the branch-sites model identified one codon from the body domain (127 T), also identified from the 4 kb branch-sites model (400 E), as having ω > 1 with a posterior probability of 0.899 (table 1).
Estimations of ω (or dN–dS to avoid infinity values) at codon positions and across branches of the 4 kb and 642 bp α-latrotoxin trees were also performed with the single-likelihood ancestor counting (SLAC) method, the fixed effects likelihood (FEL) method, the Fast Unbiased Bayesian AppRoximation (FUBAR) method, and the Mixed Effects Model of Episodic Diversifying Selection (MEME) method of HyPhy using the datamonkey server (table 2; Delport et al. 2010). For the 4 kb α-latrotoxin alignment, the SLAC method for estimating dN–dS at each codon position found no positively selected sites and 130 negatively selected sites (where normalized dN−dS is negative) at a significance level of 0.1. SLAC estimated an overall ω of 0.28 across all codons. FEL found 10 positively selected codons (two in the wing domain and eight in the body domain; where normalized dN−dS is positive), and 370 negatively selected codons at the 0.1 significance level (table 2 and supplementary fig. S8, Supplementary Material online). FUBAR found no sites under diversifying selection and 639 sites with evidence of negative selection with a posterior probability >0.9. MEME, which allows ω to vary across codons as well as across branches of the phylogeny, identified 27 codons with evidence of episodic positive selection at a significance level of 0.1. Eight of the 27 codons detected by MEME undergo only nonsynonymous substitutions on the Latrodectus α-latrotoxin branch (wing domain codons 55 and 158, and body domain codons 313, 468, 660, 725, 754, and 817; table 2). One of these codons (158: from Ala → Leu) is among those identified by the BEB procedure in the MA (modified) branch-sites model of codeml as having ω > 1 on the Latrodectus α-latrotoxin branch with a posterior probability of 0.88. Another one of the 27 codons (468: from Thr → Ala) was identified by the M8 model of codeml as potentially under positive selection, without strong statistical support. However, neither of these amino acid substitutions involves substantial changes in charge or hydrophobicity. Of the 32 codons identified in α-latrotoxin as positively selected with statistical support using any method, 10 are in the wing domain (comprising 3.3% of wing codons) and 21 are in the body domain (1.1% of all body codons). We also ran the GA-Branch program of the HyPhy package on the 4 kb alignment, which partitions branches in a phylogeny into multiple dN/dS rate classes. The GA-Branch analysis on the 4 kb α-latrotoxin tree yielded three dN/dS rate classes: 0.146 over 76% of the tree; 0.232 over 23% of the tree; and 0.475 over 1% of the tree. The Latrodectus α-latrotoxin branch falls into this middle dN/dS rate class of 0.232, which is nearly identical to the free ratio branch model estimate of 0.238 determined for this branch by codeml. The SLAC, FEL, FUBAR, and MEME methods were also applied to the 642 bp α-latrotoxin alignment, and only two codons (codon 50 in the wing and 127 in the body domain) were detected by MEME as having experienced positive selection (table 2). SLAC, FEL, and FUBAR identified 36, 59, and 67 total codons, respectively, as negatively selected. Codon 50 was also identified by the MEME procedure as positively selected in the 4 kb alignment (codon 295), whereas codon 127, though not identified in the 4 kb HyPhy analyses, was identified by the BEB procedure in codeml following the branch-sites model for both the 642 bp and 4 kb alignments (codon 400; table 2).
We calculated the rates of various types of nonsynonymous substitutions with the adaptsite-p and adaptsite-t programs in the ADAPTSITE package to estimate radical substitutions (cr) per radical site (sr), as well as conservative substitutions (cc) per conservative site (sc) at each codon in both alignments (Suzuki et al. 2001). Radical and conservative nonsynonymous substitutions are divided by the difference in charge of the amino acids changed (Suzuki 2007). For the 4 kb alignment, the adaptsite-p analysis showed that the radical over conservative substitution rate ratio [(cr/sr)/(cc/sc)] exceeded 1 at 231 (21.9%) of the 1,051 total codons in the α-latrotoxin alignment (an additional 114 codons with radical substitutions had 0 conservative substitutions). The ratio was less than 1 in 561 (53.3%) of total codons, indicating a greater number of conservative substitutions. Tests of neutrality by adaptsite-t found 92 sites were under negative selection based on the dN/dS ratio, and five codons were under negative selection using the radical/conservative substitution ratio, but no codons were identified as positively selected with statistical support using either criterion. Similarly, analyses of the 642 bp alignment found that (cr/sr)/(cc/sc) exceeded 1 at 28 (22.0%) of the 127 examined codons, an additional 12 codons with radical substitutions had no conservative substitutions, and (cr/sr)/(cc/sc) was less than 1 in 71 (55.9%) of the codons. Twenty-four codons were negatively selected based on the dN/dS ratio, and none were positively selected either by the dN/dS or (cr/sr)/(cc/sc) ratios.
The program TreeSAAP v.3.2 (Woolley et al. 2003; McClellan and Ellison 2010) was also used to detect positive selection on the 4 kb α-latrotoxin alignment based on physiochemical changes in protein sequences. The sliding window analysis in TreeSAAP identified 15 physiochemical categories in which α-latrotoxin had a significant Z score (≥3.09) for a magnitude 8 (the most extreme) change, indicating evidence of positive selection. Only the following three categories were significant for larger windows (≥20 bp windows): isoelectric point, equilibrium constant, and mean root mean square (r.m.s.) fluctuation displacement. Significance scores for these three categories, along with surrounding hydrophobicity, were plotted along the length of α-latrotoxin (supplementary fig. S9, Supplementary Material online), which indicate significant changes in hydrophobicity concentrated in the α-latrotoxin wing domain, whereas significant changes in isoelectric point and mean r.m.s. fluctuation displacement are concentrated in the body and head domains. Sites affected by positive selection, when mapped on the Latrodectus branch in the α-latrotoxin phylogeny, affected 4, 10, or 1 residues in the wing, body, and head domains, respectively (supplementary table S5, Supplementary Material online). Two of these residues (554 and 760) were identified by the codeml M8 model as positively selected, but without statistical support (BEB posterior probability ω > 1 between 0.587 and 0.596).
Discussion
α-Latrotoxin Distribution, Diversity, and Biomedical Significance
α-Latrotoxin is a fundamental tool in the study of vertebrate neurosecretion and is the molecule responsible for the suite of symptoms (latrodectism) resulting from black widow spider envenomation in humans. Despite its biomedical importance, research on this toxin has overwhelmingly focused on the functional mechanisms of α-latrotoxin from L. tredecimguttatus. Recently, Graudins et al. (2012) published a second α-latrotoxin sequence from L. hasselti (red-back spider), and provided evidence of its presence (along with other latrotoxins) from short peptide sequences obtained through mass spectrometry from two other Latrodectus species (L. mactans and L. hesperus). The limited sequence data have precluded molecular evolutionary analyses of α-latrotoxin, or a determination of its wider phylogenetic distribution. In this study, we provide evidence of α-latrotoxin from a larger sampling of Latrodectus species, including members of the divergent geometricus clade (containing brown widows), as well as from species in the genus Steatoda. We have also isolated fragments of latrotoxin paralogs from the theridiid species Parasteatoda tepidariorum (common house spider). Morphological- and molecular-based phylogenies of the family Theridiidae show Steatoda as a possible sister genus of Latrodectus, whereas Parasteatoda is in a distantly related subfamily within Theridiidae (Agnarsson 2004; Arnedo et al. 2004). This distribution indicates the widespread occurrence of latrotoxins across this extremely large spider family (2,351 spp; Platnick 2013), and places the origin of α-latrotoxin at the common ancestor of Latrodectus and Steatoda or earlier.
Although the cDNA data presented here show the expression of α-latrotoxin in Steatoda venom, the latrotoxin paralogs identified from Parasteatoda were isolated from genomic DNA, and it is unknown whether they encode venom components, or proteins performing different functions in other tissues. Some clinical and neurophysiological work suggests the presence of latrotoxins in Parasteatoda venom (Gillingwater et al. 1999; Isbister and Gray 2003a). Most theridiid species are much smaller than Latrodectus species (Ubick et al. 2005), and are rarely the cause of human envenomation. Further investigation into the venoms of these numerous, more cryptic theridiid species will provide more detailed insights into the origin and functional diversification of latrotoxins.
The lack of stop codons, limited variability, conserved cysteine residues, similar lengths, and posttranslational processing signals of Latrodectus α-latrotoxin sequences, suggest that all species in the genus express a form of this vertebrate-specific toxin with strong functional similarity to the ortholog from L. tredecimguttatus. This finding has important implications for treating widow spider bites globally (Graudins et al. 2002, 2012; Daly et al. 2007). Black widow spiders are one of the two most clinically significant types of spiders worldwide (Vetter and Isbister 2008), with at least 5,000 envenomations reported annually in Australia for the red-back spider alone (Isbister and White 2004). The limited sequence variability of α-latrotoxin across the mactans clade of Latrodectus (maximum pairwise nucleotide distance = 5.8%), explains why red-back spider antivenom (RBSAV) is broadly effective in treating bites from other Latrodectus species (Daly et al. 2007; Graudins et al. 2012).
In contrast to the high conservation of α-latrotoxin across black widow species, the level of protein distance between S. grossa and all other Latrodectus α-latrotoxins (as much as 35.6%) equals the distance between functionally distinct latrotoxin paralogs characterized from L. tredecimguttatus (34.7% between α-latroinsectotoxin and α-latrocrustotoxin), and 68.7% of these differences involve nonconservative amino acid substitutions. This greater sequence divergence, as well the substantial divergence in α-latrotoxin between the mactans clade and the geometricus clade, most likely explains why bites from both Steatoda species and L. geometricus are less severe than bites from species from the mactans clade (Muller 1992; Graudins et al. 2002). Specifically, L. geometricus bites elicit neurologically similar symptoms (pain and nausea) to bites from Latrodectus species in the mactans clade, but L. geometricus bites are usually not as debilitating (Muller 1992). Bites from some Steatoda species can also produce local pain and nausea, but are far less severe than bites from Latrodectus species, including L. geometricus (Graudins et al. 2002). However, RBSAV has also been effective in neutralizing the effect of S. grossa venom in humans, and experimentally in vertebrate animal preparations (Graudins et al. 2002, 2012). Because α-latrotoxin is the only component from black widow spider venom associated with neurotoxic effects in vertebrates, this suggests RBSAV is binding to some epitopes in S. grossa α-latrotoxin despite its substantial divergence from Latrodectus α-latrotoxin.
α-Latrotoxin Phylogeny and Evolutionary Origins
The phylogeny of α-latrotoxin provides a framework for examining changing patterns of selection in this venom toxin among lineages over time, and also contributes to our understanding of relationships among Latrodectus species. Several species in the Latrodectus mactans clade are difficult to diagnose using morphological characters, and for many years, multiple geographically widespread species were synonymized as L. mactans (Levi 1959). Recent analyses of mitochondrial DNA identified substantial genetic divergence among these previously synonymized species and determined various levels of species relationships (Garb et al. 2004; Griffiths et al. 2005; Vink et al. 2008). More molecular markers are needed to fully resolve the Latrodectus phylogeny and rapidly evolving markers will be especially important for tracking the spread of the invasive species L. geometricus and L. hasselti (Brown et al. 2008; Vincent et al. 2008; Vink et al. 2008). α-Latrotoxin’s lack of introns and its limited variability within species (e.g., identical sequences between North American and Argentine L. geometricus) suggests it may not be especially helpful for determining population structure without a more thorough sampling of allelic diversity. However, it does hold promise as a marker for diagnosing species and determining their relationships. The α-latrotoxin gene tree provides additional support for some nodes previously identified with mitochondrial COI (Garb et al. 2004), such as the split between the geometricus and mactans clades, as well as much stronger support for monophyly of Latrodectus than does mt COI. However, relationships among major lineages within the mactans clade exhibit discordance between the mt COI and α-latrotoxin trees (figs. 2–4). For example, mt COI shows L. tredecimguttatus as an early branching lineage, and L. pallidus as more closely related to the South American Latrodectus, whereas α-latrotoxin shows North American Latrodectus species as an early branching lineage, but L. tredecimguttatus being more closely related to the South American species (figs. 3 and 4). In any cases of discordance, those branches did not receive significant support, and the discordance is likely due to insufficient phylogenetic signal in one or both of these genes. Moreover, caution should be exercised when using members of gene families encoding venom toxins for generating species trees, as it may be difficult to identify orthologs and detect gene loss (Casewell et al. 2011). However, as of yet, there is no evidence for highly similar copies of α-latrotoxin within Latrodectus genomes.
α-Latrotoxin is most likely descended from a gene duplicate of an ancestral latrotoxin with insecticidal function, but determining the closest relative of α-latrotoxin requires additional homologs to confidently root the gene family tree. BLAST searches of L. tredecimguttatus α-latrotoxin against NCBI and UniProtKB databases did not uncover sequences similar enough to latrotoxins to be incorporated in the phylogenetic analyses. Holz and Habener (1998) identified two short (10–12 amino acid) regions of α-latrotoxin similar to extendin-4, a toxin isolated from the venom of the Gila monster Heloderma suspectum. Extendin-4 is in the family of secretagogic hormones that includes glucagon-like peptide 1 (GLP-1). The extracellular domain of GLP-1’s receptor (GLP1-R), which extendin-4 is an agonist of, also shares sequence similarity to a short region of latrophilin (CIRL), one of the receptors of α-latrotoxin. However, the similarity of α-latrotoxin to extendin-4 is considered the result of molecular mimicry (convergence) of the authentic ligand of latrophilin and GLP1-R, respectively (Holz and Habener 1998).
BLAST searches conducted for this study found greatest similarity of α-latrotoxin to different ankyrin motif (ANK) rich proteins, including ankyrin 1 and ankyrin 2,3/unc 44 as well as to Drosophila transient receptor potential cation channel subfamily A member 1 (dTRPA1) (supplementary table S3, Supplementary Material online). Both ankyrin 1 and ankyrin 2,3/unc 44 are members of a protein family containing 14–20 ANK repeats that link integral cell membrane proteins to the spectrin-based membrane skeleton (Bennet and Baines 2001), but are substantially longer than latrotoxins. When limiting BLAST searches to arthropod sequences, α-latrotoxin shows greatest similarity to dTRPA1, a sequence of similar length to α-latrotoxin with 16–17 ANK repeats. dTRPA1, also known as the Wasabi receptor, functions as a calcium permeable transmembrane channel primarily expressed in sensory neurons and underlies sensitivity to temperature and chemical irritants (Cordero-Morales et al. 2011). Given α-latrotoxin’s activity in forming a calcium permeable ion channel, this result suggests another potential origin of latrotoxins as paralogs of endogenous spider TRPA1, which is yet to be characterized.
Ecological Implications of α-Latrotoxin Molecular Evolution
Venom molecules are widely known for their rapid evolution (Duda and Palumbi 1999; Gibbs and Rossiter 2008), and the unexpected conservation of α-latrotoxin across black widow species contrasts with its divergence from the brown widow ortholog, and much greater divergence from the Steatoda ortholog. This divergence in α-latrotoxin’s primary sequence likely explains the functional variation observed across these species’ venoms. In comparison with Latrodectus species from the mactans clade, bites from L. geometricus and S. grossa are less severe and their venoms have greater LD50 (median lethal dose) values in mice, indicating a reduced vertebrate toxicity relative to black widow venom (Muller et al. 1989, 1992). Moreover, venoms from L. geometricus and Steatoda are unable to elicit as much neurotransmitter release from vertebrate neurons as do venoms from species in the mactans clade, which contains black widows and red-back spiders (Muller et al. 1989, 1992; Graudins et al. 2002, 2012). Yet, bites from L. geometricus still produce more severe symptoms than do bites from Steatoda species, and L. geometricus venom elicits more neurotransmitter release from vertebrate neurons than does venom from Steatoda species. The phylogenetic relationships among these species suggest a shift toward increased vertebrate toxicity in the venom of Latrodectus, and particularly in the mactans clade. However, establishing the reduced vertebrate toxicity, and possible greater insecticidal activity of S. grossa α-latrotoxin will require functional assays of recombinant toxin.
Across Latrodectus and Steatoda, the dominant mode of selection operating on α-latrotoxin appears to be purifying selection, as estimates of ω using multiple methods found the majority of codons and branches of the phylogeny with ω < 1. However, several methods (FEL, MEME, codeml M8 sites, and MA [modified] branch-sites models) identified a small fraction of codons as having been subjected to positive selection, and TreeSAAP analyses also detected evidence of positive selection on α-latrotoxin for several physiochemical/biochemical characteristics. In addition, both the codeml MA (modified) branch-sites model and the GA-Branch method of HyPhy suggested a significant increase in ω along the Latrodectus α-latrotoxin branch and some pairwise estimates of ω for α-latrotoxin’s head domain exceeded 1, suggesting past episodes of more rapid evolution, as well as variation among domains and codons in their functional constraints. These patterns suggest an earlier, more rapid evolution of α-latrotoxin, followed by stronger purifying selection, coincident with the origin of the Latrodectus genus and a probable shift in α-latrotoxin toward increased vertebrate toxicity.
A shift toward increased vertebrate toxicity in black widow spider venom may be explained by changes in the ecology of these species. Female theridiids range in total body size from 0.8 to 13.0 mm, with Latrodectus females being the largest in size for the family (6–13 mm; Kaston 1970; Ubick et al. 2005). Latrodectus females also build large, robust webs that can intercept and retain small vertebrates. Although widow spiders are generalist predators of arthropods, there are many published accounts of these spiders consuming small vertebrate prey such as geckos, small lizards, snakes, and mice (McCormick and Polis 1982; Hodar and Sanchez-Pinero 2002). Immobilization of vertebrate prey by widow spiders is undoubtedly facilitated by α-latrotoxin’s presence in their venom. The possibility that α-latrotoxin could serve a defensive function for black widows is suggested by the apparent aposematic coloration of the females, which in the mactans clade have varying levels of bright red pigmentation on top of a contrasting black background on their abdomens (Kaston 1970). Brown widow females and species in closely related genera have much less contrast in their color patterning or are a relatively uniform color (fig. 3).
Long-term purifying selection acting on Latrodectus α-latrotoxin implies that it is a functional venom component, and that the maintenance of this vertebrate neurotoxin must have important fitness consequences. It could be that α-latrotoxin is utilized by widow spiders in vertebrate predation or in defense from vertebrates more frequently than previously recognized. Another possibility is that in addition to vertebrate toxicity, Latrodectus α-latrotoxin also has an insecticidal function. α-latrotoxin’s lack of insecticidal activity was determined from a small number of species (locust, blowfly, or cabbage looper moth larvae; Knipper et al. 1986; Magazanik et al. 1992; Kiyatkin et al. 1995). It is possible that neurophysiological application of α-latrotoxin to other insects might indicate a broader phyletic spectrum of toxicity. Finally, α-latrotoxin may interact with other venom components in such a way that increases the insecticidal activity of the venom cocktail. Although this study reveals a history of persistent selection on α-latrotoxin in black widow spider venom, further investigation of its function in nature is needed to understand its role in the ecology of widow species.
The hypothesis that α-latrotoxin was driven by positive selection during the early evolution of Latrodectus, but that advantageous alleles useful in prey acquisition or defense were subsequently maintained by purifying selection, is one of several scenarios that could explain the observed patterns of variability in this gene. For example, relaxation of purifying selection, as opposed to positive selection, due to a population bottleneck during the origin of Latrodectus, is an alternative possibility to explain the increased rate of nonsynonymous substitutions on the α-latrotoxin Latrodectus branch. Background selection, where purifying selection is acting on a gene linked to α-latrotoxin, could also account for the limited variation in α-latrotoxin across black widow species. Given the lack of similar studies of Latrodectus nuclear genes, it is difficult to comprehensively evaluate these hypotheses. It will therefore be important to consider patterns of selection on other black widow spider genomic regions, to further determine whether molecular evolution of the vertebrate neurotoxin α-latrotoxin is atypical relative to other venom and nonvenom genes. More population-level molecular and ecological studies of Latrodectus species are also needed to illuminate their demographic histories, such as historical changes in population size.
This article provides a critical step in identifying the phylogenetic range of species harboring the dangerous human neurotoxin α-latrotoxin. Moreover, we provide a substantially greater understanding of the variability and evolution of α-latrotoxin, which is essential for predicting the spectrum of spider bites that can be effectively treated with current antivenoms and broadens the available set of probes for studies of vertebrate neurosecretion.
Materials and Methods
Taxon Sampling
A total of 38 individuals representing 16 Latrodectus species were sampled to cover broad phylogenetic and geographic diversity across the genus. We also sampled two species from the putative sister genus Steatoda, as well as a more distantly related theridiid, Parasteatoda tepidariorum (common house spider) (Agnarsson 2004; Arnedo et al. 2004). A complete list of specimens, species identification and collecting localities are in supplementary table S1, Supplementary Material online.
Genomic PCR
Two sets of α-latrotoxin sequences were amplified from genomic DNA from the sampled specimens. Genomic DNA was extracted from each specimen using the QIAGEN DNeasy Tissue kit (QIAGEN, Inc.). For the first set, nearly the entire α-latrotoxin coding sequence (∼4 kb) was amplified from eight specimens representing divergent Latrodectus species. Sequences were amplified in successively overlapping fragments, using a total of 2–23 primer pair combinations per species. All primer sequences, primer combinations used, and PCR conditions used in this study are listed in supplementary tables S6 and S7, Supplementary Material online. Reactions were primarily conducted using nested PCR reactions, with an initial touchdown PCR reaction, followed by amplification of the product using internal nested primers. A second set of sequences spanning 618 bp of α-latrotoxin’s wing and body domains were collected from 41 specimens (from 18 Latrodectus and two Steatoda species). Because the directly sequenced PCR product from Parasteatoda tepidariorum yielded a poor-quality chromatogram with messy peaks and many ambiguous bases, it was cloned into pCR2.1-TOPO plasmids (Invitrogen). Plasmid inserts were sequenced and two α-latrotoxin paralogs were recovered. A 428–659 bp fragment of the mt COI was amplified and sequenced from all specimens using primers and conditions reported in Garb et al. (2004). All latrotoxin and COI PCR products were directly sequenced in both directions.
RT-PCR and Inverse PCR
Genomic PCR amplification of α-latrotoxin using the designed primers was largely unsuccessful outside of Latrodectus. To complete the α-latrotoxin coding sequence from Steatoda grossa, a portion of the conserved region that could be amplified with genomic PCR was used to design primers for RT-PCR of the downstream region using methods modified from Binford et al. (2009). First, venom was collected from 12 S. grossa individuals using electro-stimulation to promote gene transcription in venom glands. Venom glands were then dissected from these individuals and flash frozen at −80°C two days after venom collection. Total RNA was extracted from the glands by homogenization with TRIzol (Invitrogen). Single-stranded (ss) cDNA was synthesized from venom gland RNA, using the SuperScript III reverse transcriptase protocol (Invitrogen), with an adaptor oligo-dT primer targeting the 3′-end of mRNAs (3′ RACE Adapter 5′-ggccacgcgtcgactagtactttttttttttttttttv-3′). This ss cDNA (1:10 dilution) was used as a PCR template with gene-specific forward primer 1,781-f (5′-cttcaaacaccdytgcacttggc-3′), a primer matching cDNA 3′ ends (3′UniPrimer 5′-gccacgcgtcgactagtac-3′), and AccuPrime Taq DNA Polymerase (Invitrogen). Resulting bands were gel-purified and cloned using the pCR4-TOPO vector (Invitrogen). Positive clones were PCR screened, purified and four were sequenced in their entirety.
To obtain the 5′ end of α-latrotoxin from S. grossa, inverse (i) PCR was performed with high molecular weight DNA from one S. grossa using the protocol of Sambrook and Russell (2000). Aliquots of the DNA were separately digested with the restriction enzymes, Xbal, PstI, and HindIII. Each completed digest was incubated with T4 DNA ligase to promote circularization. The self-ligated DNAs were templates in nested PCR reactions with primer pairs directed away from each other to amplify the unknown α-latrotoxin 5′ region. The first primer pair was 5′ROUT (5′-gcatctgctttcgtgtcttttctac-3′) + 3′F OUT (5′-gcattgctatatggtactcagactt-3′). The product of this PCR was the template in the nested reaction with primer pair 5′R-In (5′-gctgccagcttggtaatttcct-3′) and 3′-F-In (5′-gactttaaatcgtcacttgtgg-3′). Resulting bands were gel-purified and directly sequenced. The continuity of the S. grossa fragments was verified through PCR of the entire coding region. Double peaks found in chromatograms of forward and reserve directly sequenced PCR products were coded as heterozygosities using IUPAC ambiguity codes.
Phylogenetic Reconstruction
Protein translations of the following sequence data sets were aligned with ClustalX: 1) ∼4 kb of α-latrotoxin from a subset of taxa, 2) the conserved 618 bp fragment of α-latrotoxin from all specimens, and 3) 428–659 bp of mt COI sequences from all specimens. The paralog δ-latroinsectotoxin was included in the α-latrotoxin alignments for rooting the analyses. The resulting alignments were used to guide alignments of the encoding nucleotides. AIC in jModelTest 0.1 (Posada 2008) was used to determine models of nucleotide substitutions for each data set. ML and parsimony phylogenetic trees were generated with PAUP 4.0 (Swofford 2006), using 1,000 random taxon addition replicates and gaps treated as missing data. Bootstrap values were generated from 1,000 replicates. Bayesian trees were constructed using partitioned analyses in Mr.Bayes 3.2.1 (Ronquist et al. 2011) with separate models for each of the three codon positions selected by the AIC in MrModeltest 2.3 (Nylander 2004) and model parameters being unlinked across partitions. Bayesian tree searches were run for five million generations, sampling trees every 1,000 generations and the first 25% of samples were discarded as burn-in. Rate homogeneity of the 4 kb α-latrotoxin sequences and mt COI sequences was tested using the likelihood ratio test, comparing ML topologies with and without an enforced molecular clock (Huelsenbeck and Rannala 1997).
Molecular Evolutionary Analyses
For a visual representation of functional divergence along the length of α-latrotoxin, pairwise dN between aligned nucleotides was computed using a sliding window approach, with a window size of 60 bp taken at 20 bp intervals. Within each window, the nonsynonymous substitution rate (dN) was computed using the Nei and Gojobori (1986) method with the R package SeqinR (Charif and Lobry 2007). Pairwise dN, dS, and ω values were also computed for all data sets using the Nei and Gojobori (1986) method in PAML.
In the codeml program of PAML, the M0, M1a, M2a, M3, M7, and M8 sites models were used to determine ML estimates of ω for both the 4 kb and 642 bp α-latrotoxin sequence data sets under the assumption of no site variation (M0) in comparison with variable ω among sites (M3), and to compare models to test for positive selection (M1a vs. M2a and M7 vs. M8; Yang et al. 2000). The α-latrotoxin data sets were used to examine variable ω across branches of the phylogeny, comparing the free-ratio with the one-ratio model, as well as the free-ratio model in comparison with when ω was fixed to 1, and comparing the estimated one-ratio model with a two-ratio model, allowing the α-latrotoxin branch connecting to crown Latrodectus to have a variable foreground ratio (Yang 1998). The MA (modified) branch-sites Model A was also compared with the MA (fixed) model to detect positive selection that may have affected specific sites along the α-latrotoxin branch connecting to crown Latrodectus (Yang and Nielsen 2002). Statistical differences among models were estimated using a likelihood ratio test, comparing twice the difference between log-likelihood scores under each model.
In the HyPhy package via datamonkey.org, we ran the SLAC, FEL and FUBAR modules that use differing methods to estimate (dN−dS) at every codon in both the 4 kb and 642 bp α-latrotoxin alignments and report which codons show evidence of positive or negative selection, using default significance levels. We also ran the MEME module to detect diversifying episodic selection acting on codons, and the GA-Branch module to fit dN/dS rate classes to branches of the tree (Delport et al. 2010). For all modules of HyPhy, analyses were run using the nucleotide evolution model selected by the AIC in the datamonkey model selection tool, and providing the ML tree topology derived from the corresponding alignment.
We ran the adaptsite-p and adaptsite-t programs in the ADAPTSITE package to estimate the dN/dS rate ratio and the radical/conservative [(cr/sr)/(cc/sc)] amino acid substitution rate ratio at each codon in both the 4 kb and 642 bp α-latrotoxin alignments and to determine the probability of those values under selective neutrality (Suzuki et al. 2001). Following the program requirements, all codons in the alignment with gaps and ambiguities were removed (equivalent to cleandata = 1 matrix used in codeml analyses) and a neighbor-joining tree, constructed by the program njtree was supplied. Substitutions were estimated based on the transition/transversion rate ratio of 2.2 estimated from the α-latrotoxin alignment in codeml of PAML.
The 4 kb α-latrotoxin alignment was also analyzed using the program TreeSAAP v.3.2 (Woolley et al. 2003; McClellan and Ellison 2010), but excluding α-latrotoxin paralogs. A sliding windows analysis of eight magnitude categories for changes in physicochemical properties was conducted using a window size of 20 sites wide taken over each site. All magnitude category 6–8 changes with P values ≤ 0.001, indicating a significant physicochemical change and positive selection, were mapped over the length of the sequence and each significant site was mapped onto the Latrodectus α-latrotoxin branch.
To identify possible latrotoxin homologs in other organisms, α-latrotoxin sequences were also subjected to various BLAST searches (BLASTx and BLASTp with psiBLAST) against the NCBI protein database and Swissprot, or with tBLASTx against the NCBI EST database (dbEST). Searches were performed with entire databases and also limiting the database to Arthropoda.
Supplementary Material
Supplementary tables S1–S7 and figures S1–S9 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank the following individuals for providing the specimens used in this study: M. Andrade, S. Crews, R. Gillespie, A. Gonzalez, M. Harvey, M. Hedin, P. Krushelnycky, C. Kristensen, A. Leroy, L. Lotz, Y. Lubin, M. Segoli, R. Vetter, and C. Vink. The authors also thank N. Ayoub, G. Binford, and K. Zinsmaier for providing advice and assistance in data collection. A. Lancaster provided assistance with the sliding window analysis and ADAPTSITE. They also thank N. Ayoub, M. McGowen, and J. Starrett for critical review of this manuscript. This work was supported by the National Institutes of Health grants (NIGMS 1F32GM083661-01 and 1R15GM097714-01) to J.E.G.
References
- Adams ME. Agatoxins: ion channel specific toxins from the American funnel web spider, Agelenopsis aperta. Toxicon. 2004;43:509–525. doi: 10.1016/j.toxicon.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Adams ME, Olivera BM. Neurotoxins: overview of an emerging research technology. Trends Neurosci. 1994;17:151–155. doi: 10.1016/0166-2236(94)90092-2. [DOI] [PubMed] [Google Scholar]
- Agnarsson I. Morphological phylogeny of cobweb spiders and their relatives (Araneae, Araneoidea, Theridiidae) Zool J Linn Soc. 2004;141:447–626. [Google Scholar]
- Aminetzach YT, Srouji JR, Kong CY, Hoekstra HE. Convergent evolution of novel protein function in shrew and lizard venom. Curr Biol. 2009;22:1925–1931. doi: 10.1016/j.cub.2009.09.022. [DOI] [PubMed] [Google Scholar]
- Arnedo MA, Coddington J, Agnarsson I, Gillespie RG. From a comb to a tree: phylogenetic relationships of the combfooted spiders (Araneae, Theridiidae) inferred from nuclear and mitochondrial genes. Mol Phylogenet Evol. 2004;31:225–245. doi: 10.1016/S1055-7903(03)00261-6. [DOI] [PubMed] [Google Scholar]
- Bennet V, Baines AJ. Spectrin and ankyrin-based pathways: metazoan inventions for integrating cells into tissues. Phys Rev. 2001;81:1353–1392. doi: 10.1152/physrev.2001.81.3.1353. [DOI] [PubMed] [Google Scholar]
- Binford GJ, Bodner MR, Cordes MH, Baldwin KL, Rynerson MR, Burns SN, Zobel-Thropp PA. Molecular evolution, functional variation, and proposed nomenclature of the gene family that includes sphingomyelinase D in sicariid spider venoms. Mol Biol Evol. 2009;26:547–566. doi: 10.1093/molbev/msn274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KS, Necaise JS, Goddard J. Additions to the known U.S. distribution of Latrodectus geometricus (Araneae: Theridiidae) J Med Entomol. 2008;45:959–962. doi: 10.1603/0022-2585(2008)45[959:ATTKUD]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Casewell NR, Wagstaff SC, Harrison RA, Wuster W. Gene tree parsimony of multilocus snake venom protein families reveals species tree conflict as a result of multiple parallel gene loss. Mol Biol Evol. 2011;28:1157–1172. doi: 10.1093/molbev/msq302. [DOI] [PubMed] [Google Scholar]
- Charif D, Lobry JR. SeqinR 1.0-2: a contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis. In: Bastolla U, Porto M, Roman HE, Vendruscolo M, editors. Structural approaches to sequence evolution: molecules, networks, populations. Berlin (Germany): Springer; 2007. pp. 207–232. [Google Scholar]
- Clark RF, Wethern-Kestner S, Vance MV, Gerkin R. Clinical presentation and treatment of black widow spider envenomation: a review of 163 cases. Ann Emerg Med. 1992;21:782–787. doi: 10.1016/s0196-0644(05)81021-2. [DOI] [PubMed] [Google Scholar]
- Cordero-Morales JF, Gracheva EO, Julius D. Cytoplasmic ankyrin repeats of transient receptor potential A1 (TRPA1) dictate sensitivity to thermal and chemical stimuli. Proc Natl Acad Sci U S A. 2011;108:E1184–E1191. doi: 10.1073/pnas.1114124108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly FFS, Daly F, Hill RE, Bogdan GM, Dart RC. Neutralization of Latrodectus mactans and L. hesperus venom by redback spider (L. hasselti) antivenom. Clin Toxicol. 2007;39:119–123. doi: 10.1081/clt-100103826. [DOI] [PubMed] [Google Scholar]
- Danilevich VN, Grishin EV. The chromosomal genes for black widow spider neurotoxins do not contain introns. Bioorg Khim. 2000;26:933–939. doi: 10.1023/a:1026666606311. [DOI] [PubMed] [Google Scholar]
- Delport W, Poon AF, Frost SD, Kosakovsky Pond SL. Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics. 2010;26:2455–2457. doi: 10.1093/bioinformatics/btq429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doley R, Mackessy S, Manjunatha RM. Role of accelerated segment switch in exons to alter targeting (ASSET) in the molecular evolution of snake venom proteins. BMC Evol Biol. 2009;9:146. doi: 10.1186/1471-2148-9-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda TF, Jr, Palumbi SR. Molecular genetics of ecological diversification: duplication and rapid evolution of toxin genes of the venomous gastropod Conus. Proc Natl Acad Sci U S A. 1999;96:6820–6823. doi: 10.1073/pnas.96.12.6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova IE, Krasnoperov VG, Khvotchev MV, Pluzhnikov KA, Volkova TM, Grishin EV, Vais H, Bell DR, Usherwood PN. Cloning and structure of δ-latroinsectotoxin, a novel insect-specific member of the latrotoxin family: functional expression requires C-terminal truncation. J Biol Chem. 1996;271:7535–7543. doi: 10.1074/jbc.271.13.7535. [DOI] [PubMed] [Google Scholar]
- Elrick DB, Charlton MP. α-Latrocrustatoxin increases neurotransmitter release by activating a calcium influx pathway at crayfish neuromuscular junction. J Neurophysiol. 1999;82:3550–3562. doi: 10.1152/jn.1999.82.6.3550. [DOI] [PubMed] [Google Scholar]
- Fletcher JI, Smith R, O’Donoghue SI, Nilges M, Connor M, Howden MEH, Christie MJ, King GF. The structure of a novel insecticidal neurotoxin, ω-atracotoxin-HV1, from the venom of an Australian funnel web spider. Nat Struct Biol. 1997;4:559–566. doi: 10.1038/nsb0797-559. [DOI] [PubMed] [Google Scholar]
- Fritz LC, Tzeng M-C, Mauro A. Different components of black widow spider venom mediate transmitter release at vertebrate and lobster neuromuscular junctions. Nature. 1980;283:486–487. doi: 10.1038/283486a0. [DOI] [PubMed] [Google Scholar]
- Fry BG, Roelants K, Champagne DE, et al. (9 co-authors) The toxicogenomic multiverse: convergent recruitment of proteins into animal venoms. Annu Rev Genomics Hum Genet. 2009;10:483–511. doi: 10.1146/annurev.genom.9.081307.164356. [DOI] [PubMed] [Google Scholar]
- Fry BG, Vidal N, Norman JA, et al. (11 co-authors) Early evolution of the venom system in lizards and snakes. Nature. 2006;439:584–588. doi: 10.1038/nature04328. [DOI] [PubMed] [Google Scholar]
- Garb JE, Gonzalez A, Gillespie RG. The black widow spider genus Latrodectus: phylogeny, biogeography and invasion history. Mol Phylogenet Evol. 2004;31:1127–1142. doi: 10.1016/j.ympev.2003.10.012. [DOI] [PubMed] [Google Scholar]
- Gibbs HL, Rossiter W. Rapid evolution by positive selection and gene gain and loss: PLA(2) venom genes in closely related Sistrurus rattlesnakes with divergent diets. J Mol Evol. 2008;66:151–166. doi: 10.1007/s00239-008-9067-7. [DOI] [PubMed] [Google Scholar]
- Gillingwater TH, Kalikulov D, Ushkaryov Y, Ribchester RR. Comparison of effects of α-latrotoxin with a partially purified toxin from another theridiid spider, Steatoda paykulliana, on exocytosis at mouse neuromuscular junctions. J Physiol. 1999;520P:40. [Google Scholar]
- Graudins A, Gunja N, Broady KW, Nicholson GM. Clinical and in vitro evidence for the efficacy of Australian red-back spider (Latrodectus hasselti) antivenom in the treatment of envenomation by a Cupboard spider (Steatoda grossa) Toxicon. 2002;40:767–775. doi: 10.1016/s0041-0101(01)00280-x. [DOI] [PubMed] [Google Scholar]
- Graudins A, Little MJ, Pineda SS, Hains PG, King GF, Broady KW, Nicholson GM. Cloning and activity of a novel α-latrotoxin from red-back spider venom. Biochem Pharmacol. 2012;83:170–183. doi: 10.1016/j.bcp.2011.09.024. [DOI] [PubMed] [Google Scholar]
- Griffiths JW, Paterson AM, Vink CJ. Molecular insights into the biogeography and species status of New Zealand’s endemic Latrodectus spider species; L. katipo and L. atritus. J Arachnol. 2005;33:776–784. [Google Scholar]
- Hodar JA, Sanchez-Pinero F. Feeding habits of the black widow spider Latrodectus lilianae (Araneae: Theridiidae) in an arid zone of south-east Spain. J Zool. 2002;257:101–109. [Google Scholar]
- Holz GG, Habener JF. Black widow spider α-latrotoxin: a presynaptic neurotoxin that shares structural homology with the glucagon-like peptide-1 family of insulin secretagogic hormones. Comp Biochem Physiol B: Biochem Mol Biol. 1998;121:177–184. doi: 10.1016/s0305-0491(98)10088-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Rannala B. Phylogenetic methods come of age: testing hypotheses in an evolutionary context. Science. 1997;276:227–232. doi: 10.1126/science.276.5310.227. [DOI] [PubMed] [Google Scholar]
- Ichtchenko K, Khvotchev M, Kiyatkin N, Simpson L, Sugita S, Südhof TC. α-Latrotoxin action probed with recombinant toxin: receptors recruit α-latrotoxin but do not transduce an exocytotic signal. EMBO J. 1998;17:6188–6199. doi: 10.1093/emboj/17.21.6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isbister G, White J. Clinical consequences of spider bites: recent advances in our understanding. Toxicon. 2004;43:477–492. doi: 10.1016/j.toxicon.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Isbister GK, Gray MR. Effects of envenoming by comb-footed spiders of the genera Steatoda and Achaeranea (family Theridiidae: Araneae) in Australia. J Toxicol Clin Toxicol. 2003a;41:809–819. doi: 10.1081/clt-120025346. [DOI] [PubMed] [Google Scholar]
- Isbister GK, Gray MR. Latrodectism: a prospective cohort study of bites by formally identified redback spiders. Med J Aust. 2003b;179:88–91. doi: 10.5694/j.1326-5377.2003.tb05442.x. [DOI] [PubMed] [Google Scholar]
- Kaston BJ. Comparative biology of American black widow spiders. San Diego Soc Nat Hist. 1970;16:33–82. [Google Scholar]
- Kiyatkin NI, Dulubova IE, Chekhovskaya IA, Grishin EV. Cloning and structure of cDNA encoding α-latrotoxin from black widow spider venom. FEBS Lett. 1990;270:127–131. doi: 10.1016/0014-5793(90)81250-r. [DOI] [PubMed] [Google Scholar]
- Kiyatkin N, Dulubova I, Chekhovskaya I, Lipkin A, Grishin E. Structure of the low molecular weight protein copurified with α-latrotoxin. Toxicon. 1993;30:771–774. doi: 10.1016/0041-0101(92)90012-t. [DOI] [PubMed] [Google Scholar]
- Kiyatkin N, Dulubova I, Grishin E. Cloning and structural analysis of α-latroinsectotoxin cDNA abundance of ankyrin-like repeats. Eur J Biochem. 1993;213:121–127. doi: 10.1111/j.1432-1033.1993.tb17741.x. [DOI] [PubMed] [Google Scholar]
- Kiyatkin NI, Kulikovskaya IM, Grishin EV, Beadle DJ, King LA. Functional characterization of black widow spider neurotoxin synthesized in insect cells. Eur J Biochem. 1995;230:854–859. doi: 10.1111/j.1432-1033.1995.tb20628.x. [DOI] [PubMed] [Google Scholar]
- Knipper M, Madeddu L, Breer H, Meldolesi J. Black widow spider venom-induced release of neurotransmitters: mammalian synaptosomes are stimulated by a unique venom component (α-latrotoxin), insect synaptosomes by multiple components. Neuroscience. 1986;19:55–62. doi: 10.1016/0306-4522(86)90005-9. [DOI] [PubMed] [Google Scholar]
- Krasnoperov VG, Shamotienko OG, Grishin EV. Interaction of alpha-[125-I]-latrocrustotoxin with nerve cell membranes from the river crab Astacus astacus. Bioorg Khim. 1991;17:716–718. [PubMed] [Google Scholar]
- Levi HW. The spider genus Latrodectus (Araneae: Theridiidae) Trans Am Microsc Soc. 1959;77:7–43. [Google Scholar]
- Lewis RJ, Garcia ML. Therapeutic potential of venom peptides. Nat Rev Drug Discov. 2003;2:790–802. doi: 10.1038/nrd1197. [DOI] [PubMed] [Google Scholar]
- Li M, Fry BG, Manjunatha Kini R. Putting the brakes on snake venom evolution: the unique molecular evolutionary patterns of Aipysurus eydouxii (marbled sea snake) Phospholipase A2 toxins. Mol Biol Evol. 2005;22:934–941. doi: 10.1093/molbev/msi077. [DOI] [PubMed] [Google Scholar]
- Livingstone CD, Barton GJ. Protein sequence alignments: a strategy for the hierarchical analysis of residue conservation. Comput Appl Biosci. 1993;9:745–756. doi: 10.1093/bioinformatics/9.6.745. [DOI] [PubMed] [Google Scholar]
- Magazanik LG, Fedorova IM, Kovalevskaya GI, Pashkov VN, Bulgakov OV, Grishin EV. Selective presynaptic insectotoxin (α-latroinsectotoxin) isolated from black widow spider venom. Neuroscience. 1992;46:181–188. doi: 10.1016/0306-4522(92)90017-v. [DOI] [PubMed] [Google Scholar]
- McClellan DA, Ellison DD. Assessing and improving the accuracy of detecting protein adaptation with the TreeSAAP analytical software. Int J Bioinform Res Appl. 2010;6:120–133. doi: 10.1504/IJBRA.2010.032116. [DOI] [PubMed] [Google Scholar]
- McCormick S, Polis GA. Arthropods that prey on vertebrates. Biol Rev. 1982;57:29–58. [Google Scholar]
- Muller GJ. Black and brown widow spider bites in South Africa. S Afr Med J. 1992;83:399–405. [PubMed] [Google Scholar]
- Muller GJ, Koch HM, Kriegler AB, van der Walt BJ, van Jaarsveld PP. The relative toxicity and polypeptide composition of two Southern African widow spider species: Latrodectus indistinctus and Latrodectus geometricus. S Afr J Sci. 1989;85:44–46. [Google Scholar]
- Muller GJ, Kriegler AB, van Zyl JM, van der Walt BJ, Dippenaar AS, van Jaarsveld PP. Comparison of the toxicity, neurotransmitter releasing potency and polypeptide composition of the venoms from Stetoda foravae, Latrodectus indistinctus, and L. geometricus (Araneae: Theridiidae) S Afr J Sci. 1992;88:113–116. [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Nylander JAA. Uppsala (Sweden): Evolutionary Biology Centre, Uppsala University; 2004. MrModeltest v2. Program distributed by the author. [Google Scholar]
- Orlova EV, Rahman MA, Gowen B, Volynski KE, Ashton AC, Manser C, van Heel M, Ushkaryov YA. Structure of α-latrotoxin oligomers reveals that divalent cation-dependent tetramers form membrane pores. Nat Struct Biol. 2000;7:48–53. doi: 10.1038/71247. [DOI] [PubMed] [Google Scholar]
- Pescatori M, Bradbury A, Bouet F, Gargano N, Mastrogiacomo A, Grasso A. The cloning of a cDNA encoding a protein (latrodectin) which co-purifies with the α-latrotoxin from the black widow spider Latrodectus tredecimguttatus (Theridiidae) Eur J Biochem. 1995;230:322–328. doi: 10.1111/j.1432-1033.1995.tb20566.x. [DOI] [PubMed] [Google Scholar]
- Platnick NI, [Internet] The world spider catalog, version 13.5. American Museum of Natural History. 2013 Available from: http://research.amnh.org/iz/spiders/catalog, last accessed February 1, 2013. [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2011;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell D. Molecular cloning: a laboratory manual. 3rd ed. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- Sollod BL, Wilson D, Zhaxybayeva O, Gogarten JP, Drinkwater R, King GF. Were arachnids the first to use combinatorial peptide libraries? Peptides. 2005;26:131–139. doi: 10.1016/j.peptides.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Suzuki Y. Inferring natural selection operating on conservative and radical substitution at single amino acid sites. Genes Genet Syst. 2007;82:341–360. doi: 10.1266/ggs.82.341. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Gojobori T, Nei M. ADAPTSITE: detecting natural selection at single amino acid sites. Bioinformatics. 2001;17:660–661. doi: 10.1093/bioinformatics/17.7.660. [DOI] [PubMed] [Google Scholar]
- Swofford D. PAUP*: phylogenetic analysis using parsimony (*and other methods). Version 4. Sunderland (MA): Sinauer Associates; 2006. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubick D, Paquin P, Cushing PE, Roth V. Spiders of North America: an identification manual. American Arachnological Society; 2005. [Google Scholar]
- Ushkaryov YA, Volynski KE, Ashton AC. The multiple actions of black widow spider toxins and their selective use in neurosecretion studies. Toxicon. 2004;43:527–542. doi: 10.1016/j.toxicon.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Veiseh M, Gabikian P, Bahrami SB, et al. (17 co-authors) Tumor paint: a chlorotoxin: Cy5.5 bioconjugate for intraoperative visualization of cancer foci. Cancer Res. 2007;67:6882–6888. doi: 10.1158/0008-5472.CAN-06-3948. [DOI] [PubMed] [Google Scholar]
- Vetter RS, Isbister GK. Medical aspects of spider bites. Ann Rev Entomol. 2008;53:409–429. doi: 10.1146/annurev.ento.53.103106.093503. [DOI] [PubMed] [Google Scholar]
- Vincent LS, Vetter RS, Wrenn WJ, Kempf JK, Berrian JE. The brown widow spider Latrodectus geometricus C.L. Koch, 1841, in southern California. Pan-Pacific Entomol. 2008;84:344–349. [Google Scholar]
- Vink CJ, Sirvid PJ, Malumbres-Olarte J, Griffiths JW, Paquin P, Paterson AM. Species status and conservation issues of New Zealand's endemic Latrodectus spider species (Araneae: Theridiidae) Invertebr Syst. 2008;22:589–604. [Google Scholar]
- Volynski KE, Capogna M, Ashton AC, Thomson D, Orlova EV, Manser CF, Ribchester RR, Ushkaryov YA. Mutant α-latrotoxin (LTXN4C) does not form pores and causes secretion by receptor stimulation. This action does not require neurexins. J Biol Chem. 2003;278:31058–31066. doi: 10.1074/jbc.M210395200. [DOI] [PubMed] [Google Scholar]
- Volynski KE, Nosyreva ED, Ushkaryov YA, Grishin EV. Functional expression of α-latrotoxin in baculovirus system. FEBS Lett. 1999;442:25–28. doi: 10.1016/s0014-5793(98)01624-x. [DOI] [PubMed] [Google Scholar]
- Volynski KE, Volkova TM, Galkina TG, Krasnoperov VG, Pluzhnikov KA, Khvoshchev MV, Grishin EV. Molecular cloning and primary structure of cDNA fragment for α-latrocrustatoxin from black widow spider venom. Bioorg Khim. 1999;25:25–30. [PubMed] [Google Scholar]
- Williams JA, Day M, Heavner JE. Ziconotide: an update and review. Expert Opin Pharmacother. 2008;9:1575–1583. doi: 10.1517/14656566.9.9.1575. [DOI] [PubMed] [Google Scholar]
- Woolley S, Johnson J, Smith MJ, Crandall KA, McClellan DA. TreeSAAP: selection on amino acid properties using phylogenetic trees. Bioinformatics. 2003;19:671–672. doi: 10.1093/bioinformatics/btg043. [DOI] [PubMed] [Google Scholar]
- Wullschleger B, Kuhn-Netwig L, Tromp J, Kampfer U, Schaller J, Schurch S, Nentwig W. CSTX-13, a highly synergistically acting two-chain neurotoxic enhancer in the venom of the spider Cupiennius salei (Ctenidae) Proc Natl Acad Sci U S A. 2004;101:11251–11256. doi: 10.1073/pnas.0402226101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R, Goldman N, Krabbe Pedersen A-M. Codon substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol. 2002;19:908–917. doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.