Abstract
In Saccharomyces cerevisiae, all H3K4 methylation is performed by a single Set1 Complex (Set1C) that is composed of the catalytic (Set1) and seven other subunits (Swd1, Swd2, Swd3, Bre2, Sdc1, Spp1 and Shg1). It has been known for quite some time that trimethylated H3K4 (H3K4me3) is enriched in the vicinity of meiotic double-strand breaks (DSBs), but the link between H3K4me3 and the meiotic nuclease Spo11 was uncovered only recently. The PHD-containing subunit Spp1, by interacting with H3K4me3 and Mer2, was shown to promote the recruitment of potential meiotic DSB sites to the chromosomal axis allowing their subsequent cleavage by Spo11. Therefore, Spp1 emerged as a key regulator of the H3K4 trimethylation catalyzed by Set1C and of the formation of meiotic DSBs. These findings illustrate the remarkable multifunctionality of Spp1, which not only regulates the catalytic activity of the enzyme (Set1), but also interacts with the deposited mark, and mediates its biological effect (meiotic DSB formation) independently of the complex. As it was previously described for Swd2, and now for Spp1, we anticipate that other Set1C subunits, in addition to regulating H3K4 methylation, may participate in diverse biological functions inside or outside of the complex.
Keywords: Histone lysine 4 methylation, Set1, Set1C, Spp1, meiotic recombination, Swd2, transcription, Mer2
Introduction
In vertebrates, di- and tri-methylation of H3K4 are generally restricted to euchromatin and occur in discrete zones in the proximity of transcriptionally active genes. H3K4 methylation is thought to facilitate transcription through the recruitment of nucleosome remodeling complexes and histone modifying enzymes, or by preventing repressors from binding to chromatin.1,2 Besides transcription, H3K4me3 has been found to influence meiotic recombination,3-5 mammalian V(D)J recombination and pre-mRNA splicing, through the recognition of H3K4me3 by the PHD domain of RAG2 and CHD1, respectively.6,7 This led to realize that the H3K4me3 mark can recruit proteins for a variety of processes, other than transcriptional activation. However, whether these histone marks are the cause or consequence of these processes is not always certain. Here we review the dual role of the PHD-containing Set1C subunit Spp1 in the regulation of H3K4 trimethylation and in the formation of meiotic double-strand breaks (DSBs).
Spp1 as a Key Regulator of H3K4 Trimethylation
The Set1C or COMPASS (for Complex of Proteins Associated with Set1) is assembled around Set1 that acts as a scaffold for seven other subunits (Swd1, Swd2, Swd3, Bre2, Sdc1, Spp1 and Shg1).8-10 Several studies showed that loss of individual Set1C subunits differentially affects Set1 stability, complex integrity, the pattern of global H3K4 methylation, and the distribution of H3K4 methylation marks along active genes.11 Particularly, the absence of the PHD-containing protein Spp1 was shown to strongly decrease H3K4me3 levels.12-14
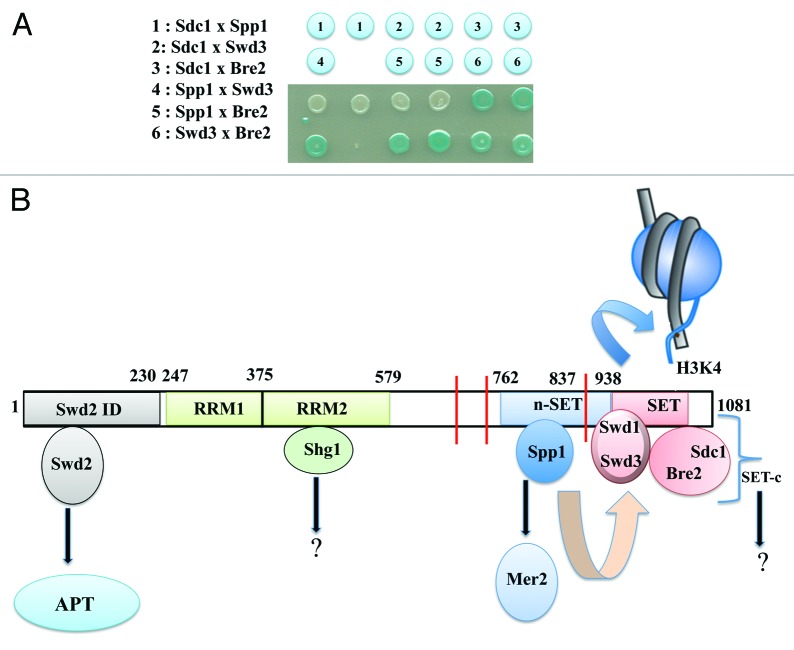
The Set1C has a remarkable mode of assembly since it is initiated during translation in the cytoplasm.15 While the nascent Set1 polypeptide emerges from the ribosome, it is bound by Shg1, Spp1 and Swd1 explaining why Set1 is found associated to its own mRNA and to ribosomal proteins in TAP purifications.15 This study also suggests that Shg1, Spp1 and Swd1 bind directly to the Set1 polypeptide, an assumption further confirmed by recent studies.16,17 The complex is then completed by the subsequent binding of Swd2, Swd3, Bre2 and Sdc1.15 Within the complex, Swd1 and Swd3 as well as Sdc1 and Bre2 interact independently of Set1, and each pair of proteins associates with the SET domain of Set1.8,14 In a recent tour de force study, Roeder and colleagues used purified components of the Set1C to demonstrate that Swd1, Swd3, Bre2 and Sdc1 efficiently co-purify with the isolated SET domain while none of these proteins alone can interact with Set1 suggesting that subunit interactions are required for these four proteins to bind the SET domain.17 Spp1 and Shg1 on their side directly interact with the nSET domain and the second RRM motif of Set1, respectively.14,15,17,18 Strikingly, Roeder and colleagues also showed that the n-SET of Set1 interacts with the SET domain covered by Swd1, Swd3, Bre2 and Sdc1 (here called the SET-c) in a way that depends on the prior binding of Spp1 to the n-SET domain.17 Although Spp1 alone was not shown to interact in vitro with SET-c, a previous TAP-Spp1 purification from the cells expressing Set1 construct carrying a truncation of the SET domain suggested an interaction between Spp1 and the Swd1-Swd3 module.14 Along the same line, bacterial two-hybrid data from our laboratory suggest that Swd3 interacts with Spp1 and Bre2 (shown in Figure 1A). Unless the Swd1-Swd3 module also interacts with the n-SET, one interpretation of these results would be that the communication between the n-SET domain and the SET-domain could be mediated by interactions between Spp1 and Swd3. This model is also consistent with the fact that Spp1, as well as Swd1-Swd3 prevent proteolytic degradation of Set1.14 In summary, Spp1 bound to the n-SET domain may indirectly interact with the SET-c via Swd3 to alter the catalytic pocket of Set1 in a way that allows the trimethylation of H3K4 (Fig. 1B). Monoubiquitylation of H2B would favor the proper positioning of the whole Set1C on the nucleosome or lead to a conformational change in the nucleosome to facilitate Set1 activity.17
Figure 1. Organization of the Set1C and role of its subunits. (A) Protein/protein interactions within the indicated subunits of the Set1C were detected by the E. coli two-hybrid system.14 Interactions are revealed by the presence of β-galactosidase activity visualized on X-gal plates. (B) This scheme depicting the binding of the different Set1C subunits to Set1 is mainly based on8,11,15,17 (see text). The two RRM domains were positioned according to Trésaugues et al.18 Direct binding of Swd2, Shg1 and Spp1 to the indicated regions of Set1 has been established by Kim et al.17 The SET-c comprises Swd1-Swd3 and Bre2-Sdc1 modules that are likely to interact each other17 (see text and Figure 1A). In this model, Spp1 bound to the n-SET domain interacts with the SET-c via Swd3 to alter the enzyme catalytic pocket to facilitate H3K4me3. The SET-c could bind H3 through cooperative interactions between its subunits. Note that the PHD domain of Spp1 is not required for the H3K4 methylase activity of the Set1C. The N-terminal region of Set1 is thought to modulate the Set1C activity in a way that remains to be elucidated. Swd2 and Spp1 are known to interact independently of Set1 with APT and Mer2, respectively. We anticipate that Shg1 and the subunits of the SET-c may in turn establish alternative protein/protein interactions to regulate other processes than H3K4 methylation. Red bars indicate the position of putative PEST sequences. Numbers indicate amino acid residues.
In addition to the role it plays in H3K4 methylation, the WD40 repeat protein Swd2 belongs to the RNA 3′-end processing and termination complex called APT (for associated with Pta1) which co-purifies with the cleavage and polyadenylation factors.8,19-23 The APT complex contributes to transcriptional termination of snoRNAs and other noncoding transcripts that is primarily performed by the Nrd1-Nab3-Sen1 complex.20,21,24-26 Within the Set1C, Swd2 was also recently shown to directly interact with the N-terminal part of Set117 in agreement with previous results showing that Swd2 prevents Set1 degradation and robustly co-purifies with TAP-tagged Set1.8,14,26 Unexpectedly, in vitro omission of Swd2 from the reconstituted Set1C increases the level of H3K4 methylation assayed on recombinant H2Bub chromatin suggesting that Swd2 may counteract a positive action of the N-terminal region of Set1 on the H2Bub-dependent H3K4 methylase activity.17 In an independent study, Swd2 was shown to be ubiquitylated by Rad6/Bre1.27 A Swd2 ubiquitination mutant (K68,69R) exhibited reduced levels of H3K4me3 and a concomitant reduction of Spp1 recruitment to chromatin.27 The mechanism by which ubiquitylation of Swd2 at K68,69 facilitates H3K4me3 remains to be fully elucidated. It opens the possibility of a complex regulation mediated by Set1C subunits to bring in interaction the N- and the C-terminal regions of Set1, as suggested.17,28
Spp1 Links H3k4me3 with Meiotic Double-Strand Break Formation
Meiosis is a specialized cell division, in which a single round of replication is followed by two rounds of chromosome segregation. First, during the reductional division, the homologous chromosomes segregate. Then, the sister chromatids segregate through an equational division leading to the formation of haploid nuclei and gametes. Importantly, the reductional division requires physical links between the homologs mediated by the process of homologous recombination leading to crossovers. Thus, in meiosis, recombination plays a dual role in promoting novel haplotypes and ensuring proper chromosome segregation.29 Meiotic recombination begins with the introduction of DSBs catalyzed by Spo11, a meiosis-specific nuclease highly conserved throughout evolution.30,31 In budding yeast, DSBs cluster in discrete regions called hot spots that are mostly located in nucleosome depleted intergenic regions, near promoters.32 DSB formation also relies upon several Spo11-associated proteins that appear to form sub-complexes but whose interactions are not yet fully elucidated.33 Among them, the sub-complex Mer2/Mei4/Rec114 has been proposed to link the DSB sites located on chromatin loops to the meiosis-specific axial chromosome structures.34
H3K4me3 was shown to be an important mark for the initiation of recombination in yeast and mice.4,5,35 In S. cerevisiae, this assumption was initially based on the fact that Set1 loss severely reduces meiotic DSB frequencies, and that H3K4me3 level is constitutively higher near DSB sites.3,5 In agreement with these observations, deletion of RAD6 as well as the substitution of the ubiquitylation site of histone H2B, both affecting H3K4 methylation, also reduced DSB frequencies at various hot spots.36 However, beyond these correlations, the functional link between H3K4 trimethylation and the activity of Spo11 remained unclear.
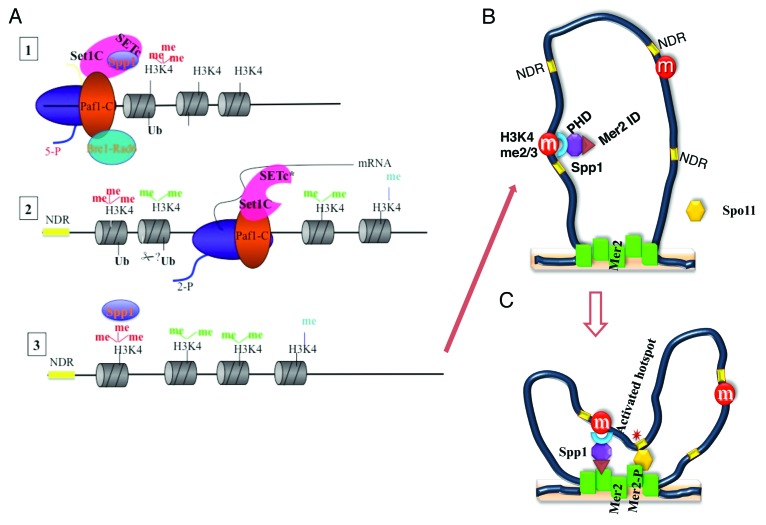
To understand how Set1C might regulate meiotic DSB formation, we decided to tether Set1 and each Set1C subunit to cold regions for meiotic recombination. We showed that the tethering of Spp1 to regions that do not usually experience recombination was sufficient to induce DSB formation even in the absence of H3K4 methylation. Furthermore, we found that Spp1 physically interacts with Mer2 at a time contemporary to DSB formation (Fig. 2). This led us to propose that Spp1, by interacting with both Mer2 and H3K4me3, through its PHD domain, bridges potential meiotic DSB sites to the chromosomal axis where Spo11-associated factors are enriched.37 In a related study, genome-wide localization of Spp1 was shown to be clearly different during meiosis compared with exponentially growing cells.38 While in rich medium, Spp1 is enriched on actively transcribed genes together with RNAP II and Set1,39 during meiosis Spp1 does not co-localize with RNAP II.38 Instead, Spp1 was found along with Mer2 at chromosome axis-associated sites in a way that is, at least partially, independent of Set1.38 Taken together, these results suggested that the interaction between Spp1 and Mer2 could occur independently of Set1 raising the intriguing question of the key regulatory modifications that promote Spp1 association to Mer2 at the expense of the one with the Set1C (Fig. 3). The Spp1 PHD domain is probably not involved in the interaction with Set1 since its inactivation or deletion does not affect H3K4me3.37,38
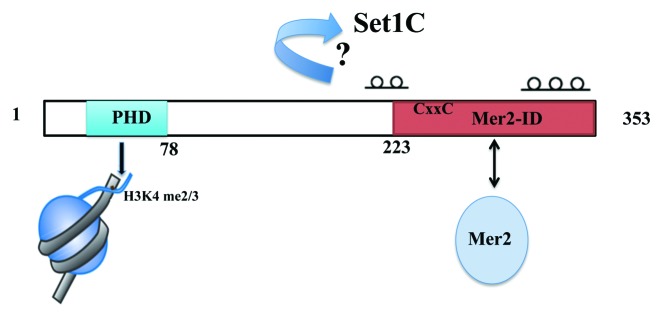
Figure 2. Schematic representation of Spp1. The positions of the PHD and the Mer2 interacting domain of Spp1 are based on.37,55 Deletion of the CxxC (C263GYC266) motif was shown to affect Spp1-Mer2 interaction. Coiled-coil regions are indicated above the scheme.55 The regions of Spp1 responsible for its interaction with Set1 and the Swd1-Swd3 module are unknown.
Figure 3. Spp1 swaps function. (A) 1- At activated genes, the Paf1 complex mediates the association of Bre1/Rad6 and Set1C to RNAP II39 allowing the transient ubiquitylation of H2BK123 and H3K4 trimethylation of the first nucleosomes of transcribed genes (see 11 for a review). Two and 3- Along the coding regions of the genes, di- and then monomethylation of H3K4 correlate with a gradual reduction of the binding of Set1C.22 We envisage that unknown post-translational modifications facilitate the specific release of Spp1 from the Set1C (B and C) During meiotic differentiation, the interaction of Spp1 with H3K4me3 and the chromatin axis-associated protein Mer2 offers an explanation of the mechanism that select the potential meiotic DSB sites that are brought to the chromosome axis for further cleavage by Spo11.37,38 Our results indicate that H3K4me3 is required for the function of Spp1 probably through its recognition by the Spp1 PHD-domain, a requirement that can be bypassed by tethering Spp1.37 NDR = Nucleosome depleted regions, Mer2 ID = Mer2 interacting domain. Mer2-p = Phosphorylated Mer2.
Other Roles for Set1C Subunits?
The role of the different Set1C subunits in transcription was analyzed by determining genome-wide mRNA expression-profiles of Set1C subunit mutants. In contrast to metazoan, trimethylation loss induced by the inactivation of Spp1 had on its own virtually no effect on steady-state mRNA expression levels. Surprisingly, the combined loss of H3K4me3 and H3K4me2 resulted in a steady-state upregulation of a small group of genes40 that exhibit distinct H3K4 methylation patterns with enrichment of H3K4me3 at their 3′end.40,41 This repression was coupled to Set1-dependent 3′end antisense transcription.40 This observation was in agreement with several other studies indicating that ncRNAs can repress gene expression by influencing the epigenetic state of chromatin.42-46 These studies came up with the concept that H3K4 methylation mark deposited by antisense transcription acts as a repressive mark that signals the recruitment of histone deacetylase thereby attenuating sense transcription.47 Although such complexes may contain PHD finger proteins that bind methylated H3K4, one cannot exclude that Set1C subunits could by themselves play an active role in targeting histone deacetylases.
In this review, we have described the essential role of Spp1 in the regulation of H3K4me3 and in the formation of meiotic DSBs. The Set1C and H3K4 methylation have been involved in addition in multiple other processes such as DNA repair,48-50 telomere position effect and telomere length regulation,48,51 cell wall biogenesis,40,51 chromosome segregation,52 transcription termination,53 and DNA replication.3,54 These pleiotropic effects raise the question of the role of the different Set1C subunits in addition to H3K4 methylation in these various processes. We foresee that subunits other than Spp1 and Swd2 may also have functional roles outside the one they have in regulating H3K4 methylation.
Acknowledgments
VG’s laboratory is supported by “La Ligue contre le Cancer” (équipe labelisée) and by the “Agence Nationale pour la Recherche” (program blanc, UBIGENEX). We thank our collaborators F. Stewart, B. Dichtl, F. Holstege, C. Dargemont, L. Székvölgyi and A. Nicolas for their invaluable interactions and D. Churikov for the critical reading of the manuscript.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/24295
References
- 1.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 2.Smith E, Shilatifard A. The chromatin signaling pathway: diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Mol Cell. 2010;40:689–701. doi: 10.1016/j.molcel.2010.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sollier J, Lin W, Soustelle C, Suhre K, Nicolas A, Géli V, et al. Set1 is required for meiotic S-phase onset, double-strand break formation and middle gene expression. EMBO J. 2004;23:1957–67. doi: 10.1038/sj.emboj.7600204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buard J, Barthès P, Grey C, de Massy B. Distinct histone modifications define initiation and repair of meiotic recombination in the mouse. EMBO J. 2009;28:2616–24. doi: 10.1038/emboj.2009.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borde V, Robine N, Lin W, Bonfils S, Géli V, Nicolas A. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J. 2009;28:99–111. doi: 10.1038/emboj.2008.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matthews AG, Kuo AJ, Ramón-Maiques S, Han S, Champagne KS, Ivanov D, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–10. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sims RJ, 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, et al. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28:665–76. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, Aasland R, et al. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 2001;20:7137–48. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, et al. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci U S A. 2001;98:12902–7. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagy PL, Griesenbeck J, Kornberg RD, Cleary ML. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc Natl Acad Sci U S A. 2002;99:90–4. doi: 10.1073/pnas.221596698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dehé PM, Géli V. The multiple faces of Set1. Biochem Cell Biol. 2006;84:536–48. doi: 10.1139/o06-081. [DOI] [PubMed] [Google Scholar]
- 12.Morillon A, Karabetsou N, Nair A, Mellor J. Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol Cell. 2005;18:723–34. doi: 10.1016/j.molcel.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 13.Schneider J, Wood A, Lee JS, Schuster R, Dueker J, Maguire C, et al. Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol Cell. 2005;19:849–56. doi: 10.1016/j.molcel.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 14.Dehé PM, Dichtl B, Schaft D, Roguev A, Pamblanco M, Lebrun R, et al. Protein interactions within the Set1 complex and their roles in the regulation of histone 3 lysine 4 methylation. J Biol Chem. 2006;281:35404–12. doi: 10.1074/jbc.M603099200. [DOI] [PubMed] [Google Scholar]
- 15.Halbach A, Zhang H, Wengi A, Jablonska Z, Gruber IM, Halbeisen RE, et al. Cotranslational assembly of the yeast SET1C histone methyltransferase complex. EMBO J. 2009;28:2959–70. doi: 10.1038/emboj.2009.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mersman DP, Du HN, Fingerman IM, South PF, Briggs SD. Charge-based interaction conserved within histone H3 lysine 4 (H3K4) methyltransferase complexes is needed for protein stability, histone methylation, and gene expression. J Biol Chem. 2012;287:2652–65. doi: 10.1074/jbc.M111.280867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim JA, Kim JA, McGinty RK, Nguyen UT, Muir TW, Allis CD, et al. The n-SET Domain of Set1 Regulates H2B Ubiquitylation-Dependent H3K4 Methylation. Mol Cell. 2013 doi: 10.1016/j.molcel.2013.01.034. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trésaugues L, Dehé PM, Guérois R, Rodriguez-Gil A, Varlet I, Salah P, et al. Structural characterization of Set1 RNA recognition motifs and their role in histone H3 lysine 4 methylation. J Mol Biol. 2006;359:1170–81. doi: 10.1016/j.jmb.2006.04.050. [DOI] [PubMed] [Google Scholar]
- 19.Nedea E, He X, Kim M, Pootoolal J, Zhong G, Canadien V, et al. Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. J Biol Chem. 2003;278:33000–10. doi: 10.1074/jbc.M304454200. [DOI] [PubMed] [Google Scholar]
- 20.Dichtl B, Aasland R, Keller W. Functions for S. cerevisiae Swd2p in 3′ end formation of specific mRNAs and snoRNAs and global histone 3 lysine 4 methylation. RNA. 2004;10:965–77. doi: 10.1261/rna.7090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng H, He X, Moore C. The essential WD repeat protein Swd2 has dual functions in RNA polymerase II transcription termination and lysine 4 methylation of histone H3. Mol Cell Biol. 2004;24:2932–43. doi: 10.1128/MCB.24.7.2932-2943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soares LM, Buratowski S. Yeast Swd2 is essential because of antagonism between Set1 histone methyltransferase complex and APT (associated with Pta1) termination factor. J Biol Chem. 2012;287:15219–31. doi: 10.1074/jbc.M112.341412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vitaliano-Prunier A, Babour A, Hérissant L, Apponi L, Margaritis T, Holstege FC, et al. H2B ubiquitylation controls the formation of export-competent mRNP. Mol Cell. 2012;45:132–9. doi: 10.1016/j.molcel.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dheur S, Vo TA, Voisinet-Hakil F, Minet M, Schmitter JM, Lacroute F, et al. Pti1p and Ref2p found in association with the mRNA 3′ end formation complex direct snoRNA maturation. EMBO J. 2003;22:2831–40. doi: 10.1093/emboj/cdg253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim M, Vasiljeva L, Rando OJ, Zhelkovsky A, Moore C, Buratowski S. Distinct pathways for snoRNA and mRNA termination. Mol Cell. 2006;24:723–34. doi: 10.1016/j.molcel.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 26.Nedea E, Nalbant D, Xia D, Theoharis NT, Suter B, Richardson CJ, et al. The Glc7 phosphatase subunit of the cleavage and polyadenylation factor is essential for transcription termination on snoRNA genes. Mol Cell. 2008;29:577–87. doi: 10.1016/j.molcel.2007.12.031. [DOI] [PubMed] [Google Scholar]
- 27.Vitaliano-Prunier A, Menant A, Hobeika M, Géli V, Gwizdek C, Dargemont C. Ubiquitylation of the COMPASS component Swd2 links H2B ubiquitylation to H3K4 trimethylation. Nat Cell Biol. 2008;10:1365–71. doi: 10.1038/ncb1796. [DOI] [PubMed] [Google Scholar]
- 28.Schlichter A, Cairns BR. Histone trimethylation by Set1 is coordinated by the RRM, autoinhibitory, and catalytic domains. EMBO J. 2005;24:1222–31. doi: 10.1038/sj.emboj.7600607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zickler D, Kleckner N. Meiotic chromosomes: integrating structure and function. Annu Rev Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
- 30.Keeney S. Spo11 and the Formation of DNA Double-Strand Breaks in Meiosis. Genome Dyn Stab. 2008;2:81–123. doi: 10.1007/7050_2007_026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Székvölgyi L, Nicolas A. From meiosis to postmeiotic events: homologous recombination is obligatory but flexible. FEBS J. 2010;277:571–89. doi: 10.1111/j.1742-4658.2009.07502.x. [DOI] [PubMed] [Google Scholar]
- 32.Pan J, Sasaki M, Kniewel R, Murakami H, Blitzblau HG, Tischfield SE, et al. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell. 2011;144:719–31. doi: 10.1016/j.cell.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maleki S, Neale MJ, Arora C, Henderson KA, Keeney S. Interactions between Mei4, Rec114, and other proteins required for meiotic DNA double-strand break formation in Saccharomyces cerevisiae. Chromosoma. 2007;116:471–86. doi: 10.1007/s00412-007-0111-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panizza S, Mendoza MA, Berlinger M, Huang L, Nicolas A, Shirahige K, et al. Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell. 2011;146:372–83. doi: 10.1016/j.cell.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 35.Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–40. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamashita K, Shinohara M, Shinohara A. Rad6-Bre1-mediated histone H2B ubiquitylation modulates the formation of double-strand breaks during meiosis. Proc Natl Acad Sci U S A. 2004;101:11380–5. doi: 10.1073/pnas.0400078101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Acquaviva L, Székvölgyi L, Dichtl B, Dichtl BS, de La Roche Saint André C, Nicolas A, et al. The COMPASS subunit Spp1 links histone methylation to initiation of meiotic recombination. Science. 2013;339:215–8. doi: 10.1126/science.1225739. [DOI] [PubMed] [Google Scholar]
- 38.Sommermeyer V, Béneut C, Chaplais E, Serrentino ME, Borde V. Spp1, a member of the Set1 Complex, promotes meiotic DSB formation in promoters by tethering histone H3K4 methylation sites to chromosome axes. Mol Cell. 2013;49:43–54. doi: 10.1016/j.molcel.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 39.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–19. doi: 10.1016/S1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 40.Margaritis T, Oreal V, Brabers N, Maestroni L, Vitaliano-Prunier A, Benschop JJ, et al. Two distinct repressive mechanisms for histone 3 lysine 4 methylation through promoting 3′-end antisense transcription. PLoS Genet. 2012;8:e1002952. doi: 10.1371/journal.pgen.1002952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kirmizis A, Santos-Rosa H, Penkett CJ, Singer MA, Vermeulen M, Mann M, et al. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature. 2007;449:928–32. doi: 10.1038/nature06160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Camblong J, Beyrouthy N, Guffanti E, Schlaepfer G, Steinmetz LM, Stutz F. Trans-acting antisense RNAs mediate transcriptional gene cosuppression in S. cerevisiae. Genes Dev. 2009;23:1534–45. doi: 10.1101/gad.522509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Houseley J, Rubbi L, Grunstein M, Tollervey D, Vogelauer MA. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol Cell. 2008;32:685–95. doi: 10.1016/j.molcel.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pinskaya M, Gourvennec S, Morillon A. H3 lysine 4 di- and tri-methylation deposited by cryptic transcription attenuates promoter activation. EMBO J. 2009;28:1697–707. doi: 10.1038/emboj.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim T, Buratowski S. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell. 2009;137:259–72. doi: 10.1016/j.cell.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim T, Xu Z, Clauder-Münster S, Steinmetz LM, Buratowski S. Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics. Cell. 2012;150:1158–69. doi: 10.1016/j.cell.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pinskaya M, Morillon A. Histone H3 lysine 4 di-methylation: a novel mark for transcriptional fidelity? Epigenetics. 2009;4:302–6. doi: 10.4161/epi.4.5.9369. [DOI] [PubMed] [Google Scholar]
- 48.Corda Y, Schramke V, Longhese MP, Smokvina T, Paciotti V, Brevet V, et al. Interaction between Set1p and checkpoint protein Mec3p in DNA repair and telomere functions. Nat Genet. 1999;21:204–8. doi: 10.1038/5991. [DOI] [PubMed] [Google Scholar]
- 49.Schramke V, Neecke H, Brevet V, Corda Y, Lucchini G, Longhese MP, et al. The set1Delta mutation unveils a novel signaling pathway relayed by the Rad53-dependent hyperphosphorylation of replication protein A that leads to transcriptional activation of repair genes. Genes Dev. 2001;15:1845–58. doi: 10.1101/gad.193901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Faucher D, Wellinger RJ. Methylated H3K4, a transcription-associated histone modification, is involved in the DNA damage response pathway. PLoS Genet. 2010;6:e1001082. doi: 10.1371/journal.pgen.1001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nislow C, Ray E, Pillus L. SET1, a yeast member of the trithorax family, functions in transcriptional silencing and diverse cellular processes. Mol Biol Cell. 1997;8:2421–36. doi: 10.1091/mbc.8.12.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang K, Lin W, Latham JA, Riefler GM, Schumacher JM, Chan C, et al. The Set1 methyltransferase opposes Ipl1 aurora kinase functions in chromosome segregation. Cell. 2005;122:723–34. doi: 10.1016/j.cell.2005.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Terzi N, Churchman LS, Vasiljeva L, Weissman J, Buratowski S. H3K4 trimethylation by Set1 promotes efficient termination by the Nrd1-Nab3-Sen1 pathway. Mol Cell Biol. 2011;31:3569–83. doi: 10.1128/MCB.05590-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rizzardi LF, Dorn ES, Strahl BD, Cook JG. DNA replication origin function is promoted by H3K4 di-methylation in Saccharomyces cerevisiae. Genetics. 2012;192:371–84. doi: 10.1534/genetics.112.142349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Murton BL, Chin WL, Ponting CP, Itzhaki LS. Characterising the binding specificities of the subunits associated with the KMT2/Set1 histone lysine methyltransferase. J Mol Biol. 2010;398:481–8. doi: 10.1016/j.jmb.2010.03.036. [DOI] [PubMed] [Google Scholar]