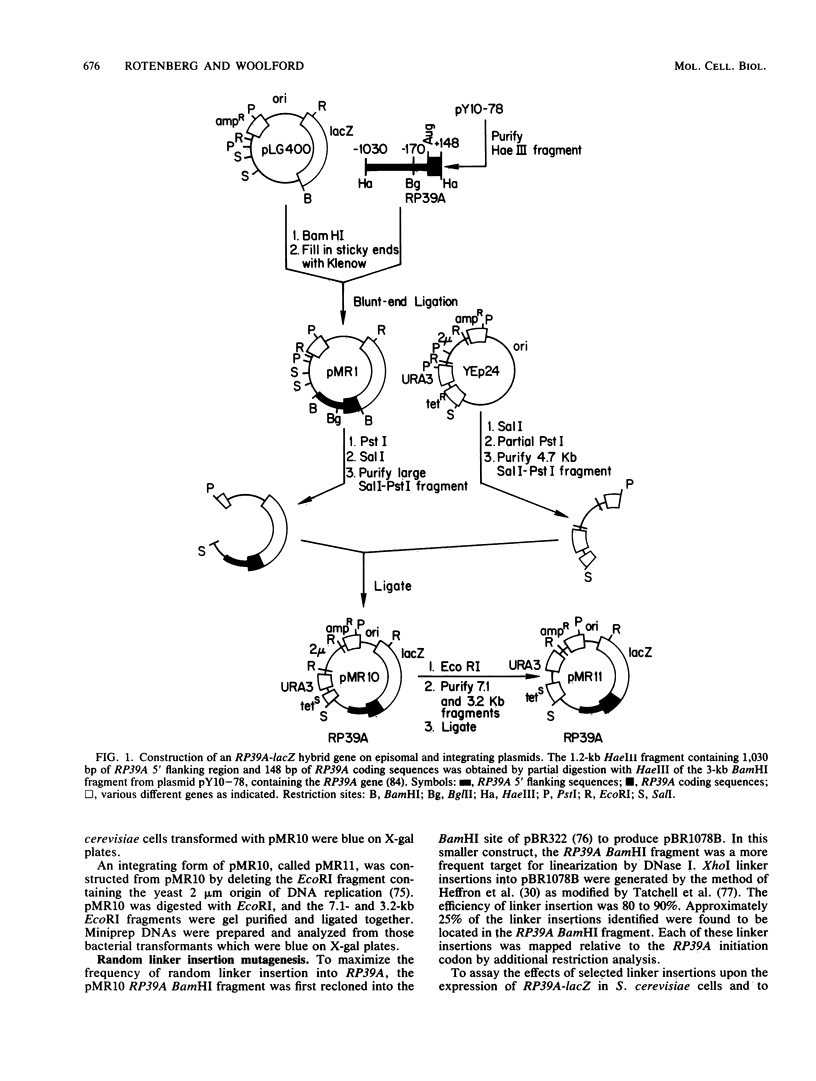
Abstract
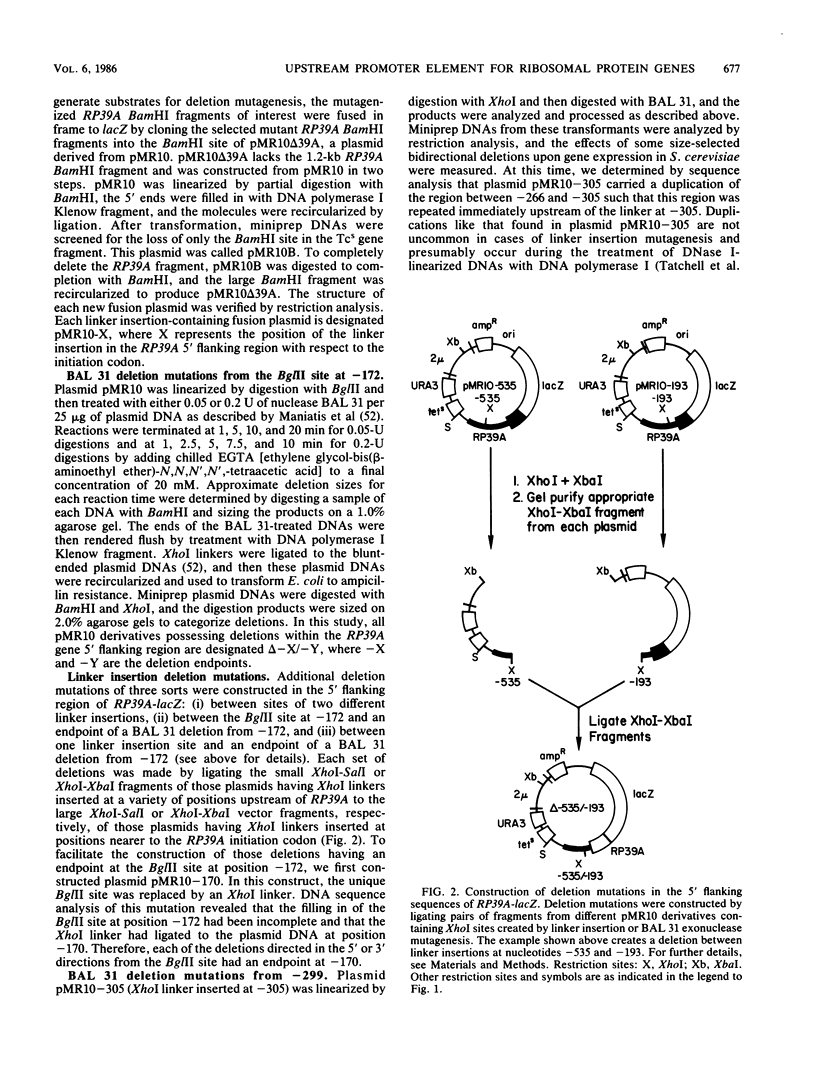
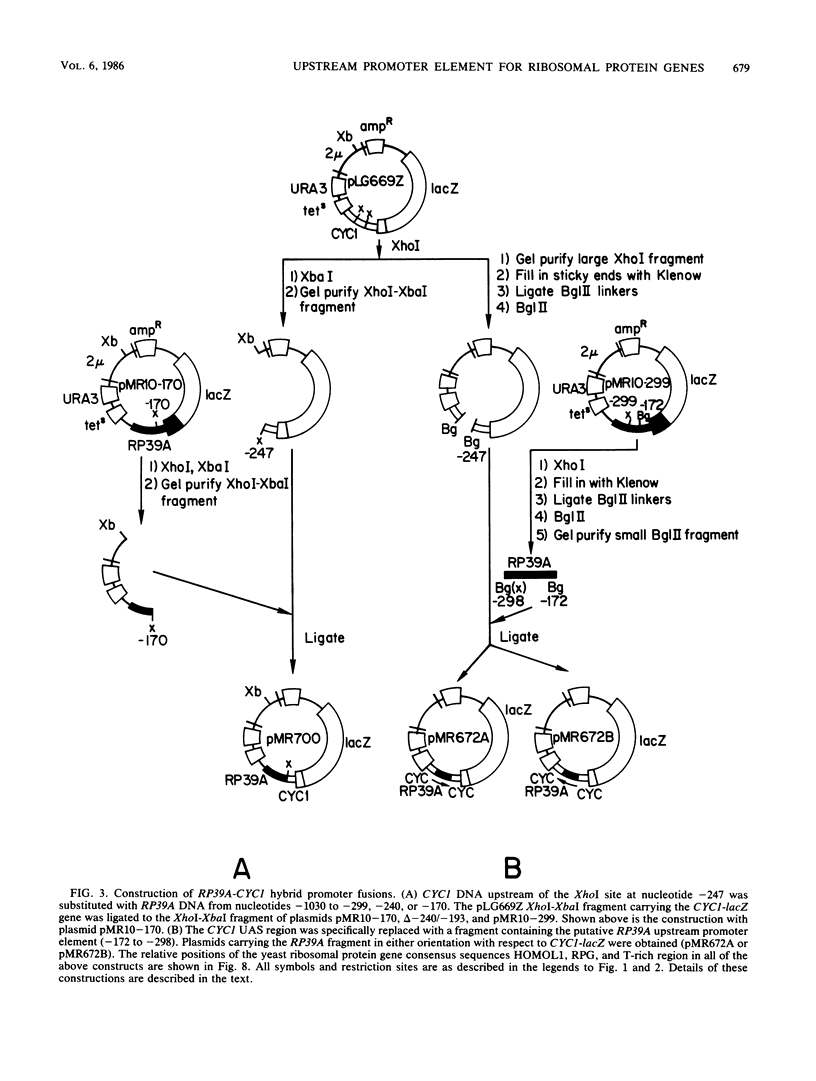
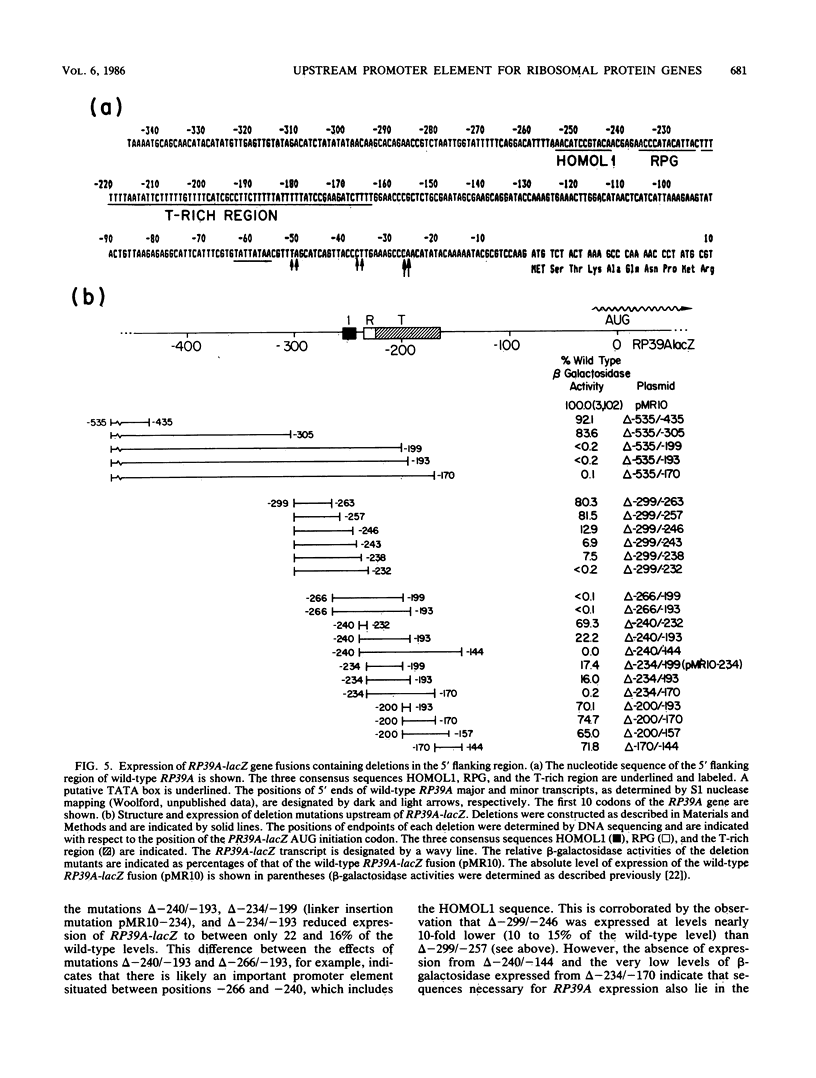
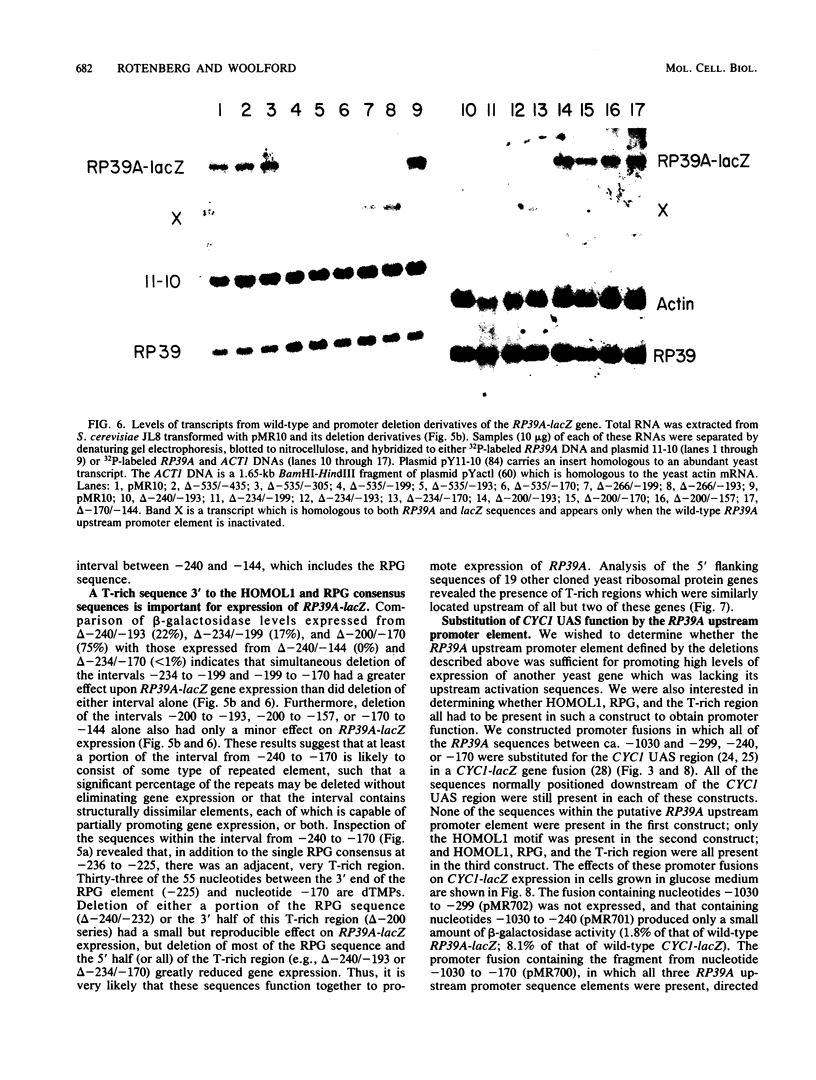
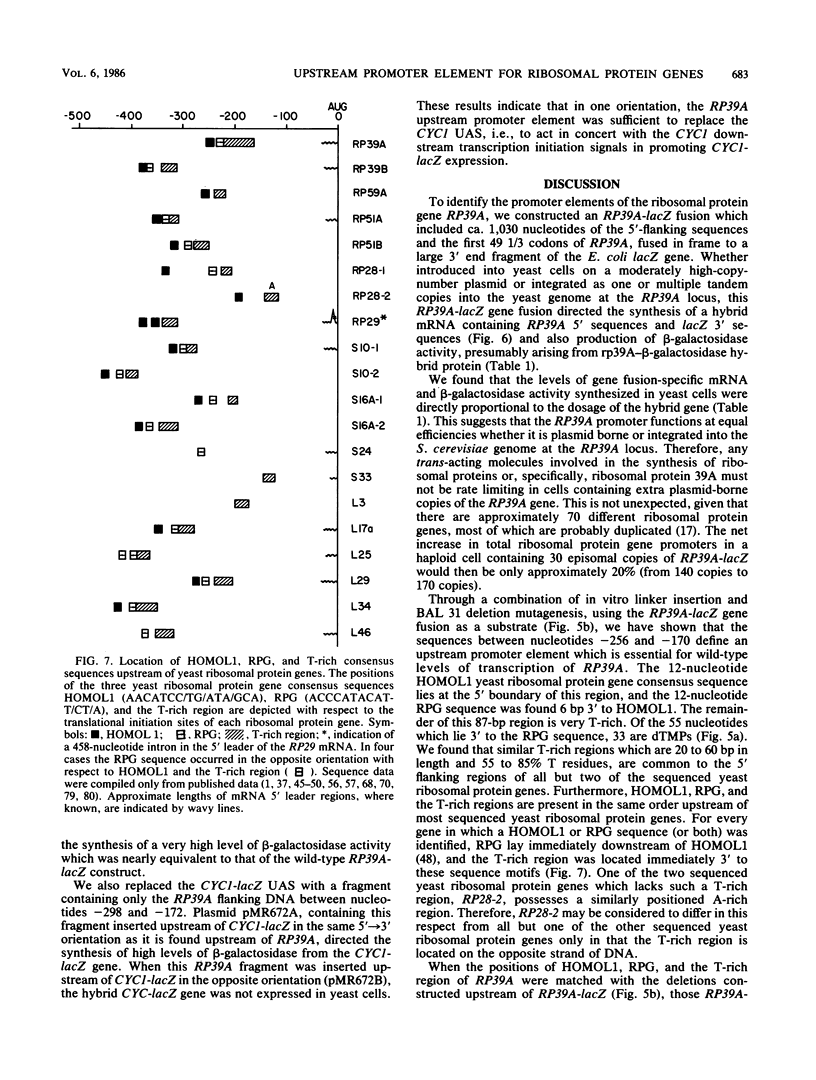
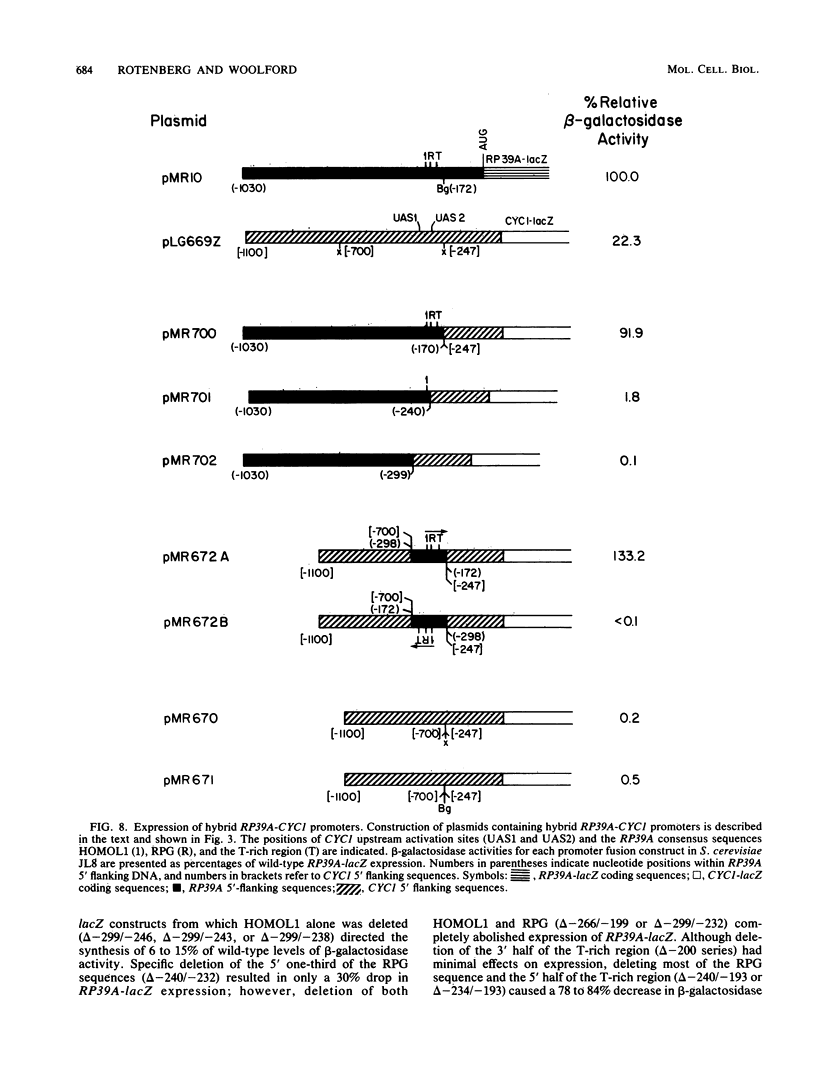
To initiate a genetic analysis of yeast ribosomal protein gene promoters, we have constructed a gene fusion between the yeast ribosomal protein gene RP39A and the Escherichia coli lacZ gene. This gene fusion contains approximately 1,030 nucleotides of the 5' flanking region and the first 49 1/3 codons of RP39A fused in frame to a large 3' end fragment of lacZ. Whether it is introduced into yeast cells on a moderately high-copy-number plasmid, or integrated into the yeast genome at the RP39A locus, this RP39A-lacZ gene directs the synthesis of a hybrid transcript which encodes beta-galactosidase activity. Deletions in the 5' flanking region of RP39A-lacZ were constructed by linker insertion and BAL 31 mutagenesis. The expression of the mutant genes in yeast cells was assayed by measuring RP39A-lacZ mRNA and beta-galactosidase levels. By these means we have shown that the sequences between nucleotides -256 and -170 upstream of RP39A are essential for expression of this gene. Three sequence motifs, HOMOL1, RPG, and a T-rich region, which were found in that order 5'----3' upstream of most yeast ribosomal protein genes, were present within this interval. We found that substitution of the CYC1-lacZ upstream activation site with the fragment from nucleotides -298 to -172 upstream of RP39A, containing the HOMOL1-RPG-T-rich motif in that 5'----3' orientation, fully restored expression of the CYC1-lacZ gene. The essentially of HOMOL1, the RPG sequence, and the T-rich region for wild-type levels of expression of RP39A, the conserved location and order of these sequence motifs in yeast ribosomal protein genes, and the ability of a DNA fragment carrying these three sequence elements to substitute for the upstream activation site regions of CYC1 indicate that these three oligonucleotides may be essential to the transcription of yeast ribosomal protein genes.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abovich N., Rosbash M. Two genes for ribosomal protein 51 of Saccharomyces cerevisiae complement and contribute to the ribosomes. Mol Cell Biol. 1984 Sep;4(9):1871–1879. doi: 10.1128/mcb.4.9.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. The primary structure of the Saccharomyces cerevisiae gene for alcohol dehydrogenase. J Biol Chem. 1982 Mar 25;257(6):3018–3025. [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollen G. H., Cohen L. H., Mager W. H., Klaassen A. W., Planta R. J. Isolation of cloned ribosomal protein genes from the yeast Saccharomyces carlsbergensis. Gene. 1981 Sep;14(4):279–287. doi: 10.1016/0378-1119(81)90160-8. [DOI] [PubMed] [Google Scholar]
- Bollen G. H., Molenaar C. M., Cohen L. H., van Raamsdonk-Duin M. M., Mager W. H., Planta R. J. Ribosomal protein genes of yeast contain intervening sequences. Gene. 1982 Apr;18(1):29–37. doi: 10.1016/0378-1119(82)90053-1. [DOI] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Broach J. R. Construction of high copy yeast vectors using 2-microns circle sequences. Methods Enzymol. 1983;101:307–325. doi: 10.1016/0076-6879(83)01024-1. [DOI] [PubMed] [Google Scholar]
- Choe J., Kolodrubetz D., Grunstein M. The two yeast histone H2A genes encode similar protein subtypes. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1484–1487. doi: 10.1073/pnas.79.5.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. W., Thomas M., Cameron J., St John T. P., Scherer S., Padgett R. A. Rapid DNA isolations for enzymatic and hybridization analysis. Methods Enzymol. 1980;65(1):404–411. doi: 10.1016/s0076-6879(80)65051-4. [DOI] [PubMed] [Google Scholar]
- Farrelly F. W., Finkelstein D. B. Complete sequence of the heat shock-inducible HSP90 gene of Saccharomyces cerevisiae. J Biol Chem. 1984 May 10;259(9):5745–5751. [PubMed] [Google Scholar]
- Faye G., Leung D. W., Tatchell K., Hall B. D., Smith M. Deletion mapping of sequences essential for in vivo transcription of the iso-1-cytochrome c gene. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2258–2262. doi: 10.1073/pnas.78.4.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried H. M., Nam H. G., Loechel S., Teem J. Characterization of yeast strains with conditionally expressed variants of ribosomal protein genes tcm1 and cyh2. Mol Cell Biol. 1985 Jan;5(1):99–108. doi: 10.1128/mcb.5.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried H. M., Pearson N. J., Kim C. H., Warner J. R. The genes for fifteen ribosomal proteins of Saccharomyces cerevisiae. J Biol Chem. 1981 Oct 10;256(19):10176–10183. [PubMed] [Google Scholar]
- Fried H. M., Warner J. R. Cloning of yeast gene for trichodermin resistance and ribosomal protein L3. Proc Natl Acad Sci U S A. 1981 Jan;78(1):238–242. doi: 10.1073/pnas.78.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried H. M., Warner J. R. Molecular cloning and analysis of yeast gene for cycloheximide resistance and ribosomal protein L29. Nucleic Acids Res. 1982 May 25;10(10):3133–3148. doi: 10.1093/nar/10.10.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallwitz D., Perrin F., Seidel R. The actin gene in yeast Saccharomyces cerevisiae: 5' and 3' end mapping, flanking and putative regulatory sequences. Nucleic Acids Res. 1981 Dec 11;9(23):6339–6350. doi: 10.1093/nar/9.23.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giniger E., Varnum S. M., Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985 Apr;40(4):767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- Gorenstein C., Warner J. R. Coordinate regulation of the synthesis of eukaryotic ribosomal proteins. Proc Natl Acad Sci U S A. 1976 May;73(5):1547–1551. doi: 10.1073/pnas.73.5.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorenstein C., Warner J. R. Synthesis and turnover of ribosomal proteins in the absence of 60S subunit assembly in Saccharomyces cerevisiae. Mol Gen Genet. 1977 Dec 9;157(3):327–332. doi: 10.1007/BF00268670. [DOI] [PubMed] [Google Scholar]
- Guarente L., Hoar E. Upstream activation sites of the CYC1 gene of Saccharomyces cerevisiae are active when inverted but not when placed downstream of the "TATA box". Proc Natl Acad Sci U S A. 1984 Dec;81(24):7860–7864. doi: 10.1073/pnas.81.24.7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L., Lalonde B., Gifford P., Alani E. Distinctly regulated tandem upstream activation sites mediate catabolite repression of the CYC1 gene of S. cerevisiae. Cell. 1984 Feb;36(2):503–511. doi: 10.1016/0092-8674(84)90243-5. [DOI] [PubMed] [Google Scholar]
- Guarente L., Lauer G., Roberts T. M., Ptashne M. Improved methods for maximizing expression of a cloned gene: a bacterium that synthesizes rabbit beta-globin. Cell. 1980 Jun;20(2):543–553. doi: 10.1016/0092-8674(80)90640-6. [DOI] [PubMed] [Google Scholar]
- Guarente L., Mason T. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell. 1983 Apr;32(4):1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- Guarente L., Ptashne M. Fusion of Escherichia coli lacZ to the cytochrome c gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2199–2203. doi: 10.1073/pnas.78.4.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters: positive and negative elements. Cell. 1984 Apr;36(4):799–800. doi: 10.1016/0092-8674(84)90028-x. [DOI] [PubMed] [Google Scholar]
- Guarente L., Yocum R. R., Gifford P. A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7410–7414. doi: 10.1073/pnas.79.23.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron F., So M., McCarthy B. J. In vitro mutagenesis of a circular DNA molecule by using synthetic restriction sites. Proc Natl Acad Sci U S A. 1978 Dec;75(12):6012–6016. doi: 10.1073/pnas.75.12.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hereford L. M., Rosbash M. Number and distribution of polyadenylated RNA sequences in yeast. Cell. 1977 Mar;10(3):453–462. doi: 10.1016/0092-8674(77)90032-0. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G., Fink G. R. Repeated DNA sequences upstream from HIS1 also occur at several other co-regulated genes in Saccharomyces cerevisiae. J Biol Chem. 1983 Apr 25;258(8):5238–5247. [PubMed] [Google Scholar]
- Hinnebusch A. G., Lucchini G., Fink G. R. A synthetic HIS4 regulatory element confers general amino acid control on the cytochrome c gene (CYC1) of yeast. Proc Natl Acad Sci U S A. 1985 Jan;82(2):498–502. doi: 10.1073/pnas.82.2.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M., Davis R. W. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Aug;4(8):1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones E. W., Lam K. B. Mutations affecting levels of tetrahydrofolate interconversion enzymes in Saccharomyces cerevisiae. II. Map positions on chromosome VII of ade3-41 and ADE15. Mol Gen Genet. 1973 Jul 2;123(3):209–218. doi: 10.1007/BF00271239. [DOI] [PubMed] [Google Scholar]
- Khoury G., Gruss P. Enhancer elements. Cell. 1983 Jun;33(2):313–314. doi: 10.1016/0092-8674(83)90410-5. [DOI] [PubMed] [Google Scholar]
- Kief D. R., Warner J. R. Coordinate control of syntheses of ribosomal ribonucleic acid and ribosomal proteins during nutritional shift-up in Saccharomyces cerevisiae. Mol Cell Biol. 1981 Nov;1(11):1007–1015. doi: 10.1128/mcb.1.11.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C. H., Warner J. R. Messenger RNA for ribosomal proteins in yeast. J Mol Biol. 1983 Mar 25;165(1):79–89. doi: 10.1016/s0022-2836(83)80243-5. [DOI] [PubMed] [Google Scholar]
- Kim C. H., Warner J. R. Mild temperature shock alters the transcription of a discrete class of Saccharomyces cerevisiae genes. Mol Cell Biol. 1983 Mar;3(3):457–465. doi: 10.1128/mcb.3.3.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraig E., Haber J. E., Rosbash M. Sporulation and rna2 lower ribosomal protein mRNA levels by different mechanisms in Saccharomyces cerevisiae. Mol Cell Biol. 1982 Oct;2(10):1199–1204. doi: 10.1128/mcb.2.10.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käufer N. F., Fried H. M., Schwindinger W. F., Jasin M., Warner J. R. Cycloheximide resistance in yeast: the gene and its protein. Nucleic Acids Res. 1983 May 25;11(10):3123–3135. doi: 10.1093/nar/11.10.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin J. C., Woolford J. L., Jr Molecular cloning and analysis of the CRY1 gene: a yeast ribosomal protein gene. Nucleic Acids Res. 1983 Jan 25;11(2):403–420. doi: 10.1093/nar/11.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Last R. L., Stavenhagen J. B., Woolford J. L., Jr Isolation and characterization of the RNA2, RNA3, and RNA11 genes of Saccharomyces cerevisiae. Mol Cell Biol. 1984 Nov;4(11):2396–2405. doi: 10.1128/mcb.4.11.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leer R. J., Van Raamsdonk-Duin M. M., Mager W. H., Planta R. J. Conserved sequences upstream of yeast ribosomal protein genes. Curr Genet. 1985;9(4):273–277. doi: 10.1007/BF00419955. [DOI] [PubMed] [Google Scholar]
- Leer R. J., van Raamsdonk-Duin M. M., Hagendoorn M. J., Mager W. H., Planta R. J. Structural comparison of yeast ribosomal protein genes. Nucleic Acids Res. 1984 Sep 11;12(17):6685–6700. doi: 10.1093/nar/12.17.6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leer R. J., van Raamsdonk-Duin M. M., Kraakman P., Mager W. H., Planta R. J. The genes for yeast ribosomal proteins S24 and L46 are adjacent and divergently transcribed. Nucleic Acids Res. 1985 Feb 11;13(3):701–709. doi: 10.1093/nar/13.3.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leer R. J., van Raamsdonk-Duin M. M., Mager W. H., Planta R. J. The primary structure of the gene encoding yeast ribosomal protein L16. FEBS Lett. 1984 Oct 1;175(2):371–376. doi: 10.1016/0014-5793(84)80771-1. [DOI] [PubMed] [Google Scholar]
- Leer R. J., van Raamsdonk-Duin M. M., Molenaar C. M., Cohen L. H., Mager W. H., Planta R. J. The structure of the gene coding for the phosphorylated ribosomal protein S10 in yeast. Nucleic Acids Res. 1982 Oct 11;10(19):5869–5878. doi: 10.1093/nar/10.19.5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leer R. J., van Raamsdonk-Duin M. M., Schoppink P. J., Cornelissen M. T., Cohen L. H., Mager W. H., Planta R. J. Yeast ribosomal protein S33 is encoded by an unsplit gene. Nucleic Acids Res. 1983 Nov 25;11(22):7759–7768. doi: 10.1093/nar/11.22.7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusty C. J., Widgren E. E., Broglie K. E., Nyunoya H. Yeast carbamyl phosphate synthetase. Structure of the yeast gene and homology to Escherichia coli carbamyl phosphate synthetase. J Biol Chem. 1983 Dec 10;258(23):14466–14477. [PubMed] [Google Scholar]
- Martinez-Arias A., Yost H. J., Casadaban M. J. Role of an upstream regulatory element in leucine repression of the Saccharomyces cerevisiae leu2 gene. Nature. 1984 Feb 23;307(5953):740–742. doi: 10.1038/307740b0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A. M., MacKay V. L., Nasmyth K. A. Identification and comparison of two sequence elements that confer cell-type specific transcription in yeast. Nature. 1985 Apr 18;314(6012):598–603. doi: 10.1038/314598a0. [DOI] [PubMed] [Google Scholar]
- Mitra G., Warner J. R. A yeast ribosomal protein gene whose intron is in the 5' leader. J Biol Chem. 1984 Jul 25;259(14):9218–9224. [PubMed] [Google Scholar]
- Molenaar C. M., Woudt L. P., Jansen A. E., Mager W. H., Planta R. J., Donovan D. M., Pearson N. J. Structure and organization of two linked ribosomal protein genes in yeast. Nucleic Acids Res. 1984 Oct 11;12(19):7345–7358. doi: 10.1093/nar/12.19.7345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moye W. S., Amuro N., Rao J. K., Zalkin H. Nucleotide sequence of yeast GDH1 encoding nicotinamide adenine dinucleotide phosphate-dependent glutamate dehydrogenase. J Biol Chem. 1985 Jul 15;260(14):8502–8508. [PubMed] [Google Scholar]
- Moye W. S., Zalkin H. Deletion mapping the yeast TRP5 control region. J Biol Chem. 1985 Apr 25;260(8):4718–4723. [PubMed] [Google Scholar]
- Nagata S., Nagashima K., Tsunetsugu-Yokota Y., Fujimura K., Miyazaki M., Kaziro Y. Polypeptide chain elongation factor 1 alpha (EF-1 alpha) from yeast: nucleotide sequence of one of the two genes for EF-1 alpha from Saccharomyces cerevisiae. EMBO J. 1984 Aug;3(8):1825–1830. doi: 10.1002/j.1460-2075.1984.tb02053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng R., Abelson J. Isolation and sequence of the gene for actin in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3912–3916. doi: 10.1073/pnas.77.7.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson N. J., Fried H. M., Warner J. R. Yeast use translational control to compensate for extra copies of a ribosomal protein gene. Cell. 1982 Jun;29(2):347–355. doi: 10.1016/0092-8674(82)90151-9. [DOI] [PubMed] [Google Scholar]
- Pearson N. J., Haber J. E. Changes in regulation of ribosome synthesis during different stages of the life cycle of Saccharomyces cerevisiae. Mol Gen Genet. 1977 Dec 14;158(1):81–91. doi: 10.1007/BF00455122. [DOI] [PubMed] [Google Scholar]
- Roeder G. S., Beard C., Smith M., Keranen S. Isolation and characterization of the SPT2 gene, a negative regulator of Ty-controlled yeast gene expression. Mol Cell Biol. 1985 Jul;5(7):1543–1553. doi: 10.1128/mcb.5.7.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosbash M., Harris P. K., Woolford J. L., Jr, Teem J. L. The effect of temperature-sensitive RNA mutants on the transcription products from cloned ribosomal protein genes of yeast. Cell. 1981 Jun;24(3):679–686. doi: 10.1016/0092-8674(81)90094-5. [DOI] [PubMed] [Google Scholar]
- Rose M., Casadaban M. J., Botstein D. Yeast genes fused to beta-galactosidase in Escherichia coli can be expressed normally in yeast. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2460–2464. doi: 10.1073/pnas.78.4.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz L. D., Friesen J. D. Nucleotide sequence of the tcml gene (ribosomal protein L3) of Saccharomyces cerevisiae. J Bacteriol. 1983 Jul;155(1):8–14. doi: 10.1128/jb.155.1.8-14.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman R. W., Warner J. R. Ribosomal RNA transcription in a mutant of Saccharomyces cerevisiae defective in ribosomal protein synthesis. Mol Gen Genet. 1978 May 3;161(2):221–223. doi: 10.1007/BF00274191. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Struhl K. Genetic properties and chromatin structure of the yeast gal regulatory element: an enhancer-like sequence. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7865–7869. doi: 10.1073/pnas.81.24.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. The yeast his3 promoter contains at least two distinct elements. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7385–7389. doi: 10.1073/pnas.79.23.7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. pBR322 restriction map derived from the DNA sequence: accurate DNA size markers up to 4361 nucleotide pairs long. Nucleic Acids Res. 1978 Aug;5(8):2721–2728. doi: 10.1093/nar/5.8.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatchell K., Nasmyth K. A., Hall B. D., Astell C., Smith M. In vitro mutation analysis of the mating-type locus in yeast. Cell. 1981 Nov;27(1 Pt 2):25–35. doi: 10.1016/0092-8674(81)90357-3. [DOI] [PubMed] [Google Scholar]
- Taussig R., Carlson M. Nucleotide sequence of the yeast SUC2 gene for invertase. Nucleic Acids Res. 1983 Mar 25;11(6):1943–1954. doi: 10.1093/nar/11.6.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teem J. L., Abovich N., Kaufer N. F., Schwindinger W. F., Warner J. R., Levy A., Woolford J., Leer R. J., van Raamsdonk-Duin M. M., Mager W. H. A comparison of yeast ribosomal protein gene DNA sequences. Nucleic Acids Res. 1984 Nov 26;12(22):8295–8312. doi: 10.1093/nar/12.22.8295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teem J. L., Rosbash M. Expression of a beta-galactosidase gene containing the ribosomal protein 51 intron is sensitive to the rna2 mutation of yeast. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4403–4407. doi: 10.1073/pnas.80.14.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udem S. A., Warner J. R. Ribosomal RNA synthesis in Saccharomyces cerevisiae. J Mol Biol. 1972 Mar 28;65(2):227–242. doi: 10.1016/0022-2836(72)90279-3. [DOI] [PubMed] [Google Scholar]
- West R. W., Jr, Yocum R. R., Ptashne M. Saccharomyces cerevisiae GAL1-GAL10 divergent promoter region: location and function of the upstream activating sequence UASG. Mol Cell Biol. 1984 Nov;4(11):2467–2478. doi: 10.1128/mcb.4.11.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford J. L., Jr, Hereford L. M., Rosbash M. Isolation of cloned DNA sequences containing ribosomal protein genes from Saccharomyces cerevisiae. Cell. 1979 Dec;18(4):1247–1259. doi: 10.1016/0092-8674(79)90236-8. [DOI] [PubMed] [Google Scholar]
- Woolford J. L., Jr, Rosbash M. Ribosomal protein genes rp 39(10 - 78), rp 39(11 - 40), rp 51, and rp 52 are not contiguous to other ribosomal protein genes in the Saccharomyces cerevisiae genome. Nucleic Acids Res. 1981 Oct 10;9(19):5021–5036. doi: 10.1093/nar/9.19.5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalkin H., Yanofsky C. Yeast gene TRP5: structure, function, regulation. J Biol Chem. 1982 Feb 10;257(3):1491–1500. [PubMed] [Google Scholar]