Abstract
Applying in silico simulations and in vitro experiments, the amino acid proline was proved to have a profound influence on Streptomyces griseus trypsinogen, and the hydrogen bond between H57 and D102 was found to be crucial for trypsin activity. By introducing an artificial propeptide, IVEF, the titer of trypsin was increased 6.71-fold.
TEXT
Trypsin (EC 3.4.21.4), an archetypal serine protease, has been discovered in many organisms, ranging from bacteria to mammals (1). Bovine trypsinogen, as a representative, has been well studied (2, 3) and functions through the cleavage of an N-terminal hexapeptide (VDDDDK) by autoactivation (4) or hydrolyzation with enterokinase (5). In addition, previous studies have shown that four unusual aspartyl residues (DDDD) in trypsinogen are strictly conserved in mammals, including humans, and may have arisen by selective pressure during the course of evolution. In comparison, although activation of the bacterial Streptomyces griseus trypsinogen also involved removal of the propeptide (APNP), the enzymatic mechanism was considered to be different from that of mammalian trypsinogen (3, 6). Since the 1970s, the amino acid sequence, open reading frame, and crystal structure of S. griseus trypsin have been solved (7–10). However, the activation mechanism of S. griseus trypsinogen remains poorly understood. In the 1990s, Kim et al. deduced that S. griseus trypsin consists of three parts, according to its amino acid sequence, and that activation of S. griseus trypsinogen is nonautocatalytic (8). Recently, we successfully overexpressed S. griseus trypsin in Pichia pastoris (11) by including four residues (YVEF) at its N terminus (Fig. 1A). The four residues were inserted by digestion with restriction enzymes SnaBI and EcoRI and cleavage of the α-factor signal peptide by peptidases KEX2 and STE13 (12–14). However, no activity was detected when substituting the native propeptide with mammalian trypsinogen propeptide VDDDDK or its mutant VDDDDD (11). Recently, many short synthetic amphiphilic peptides were identified and used for improving enzyme properties (15–17). Therefore, artificial modification of the N-terminal propeptide represents a suitable alternative for enzyme engineering.
Fig 1.
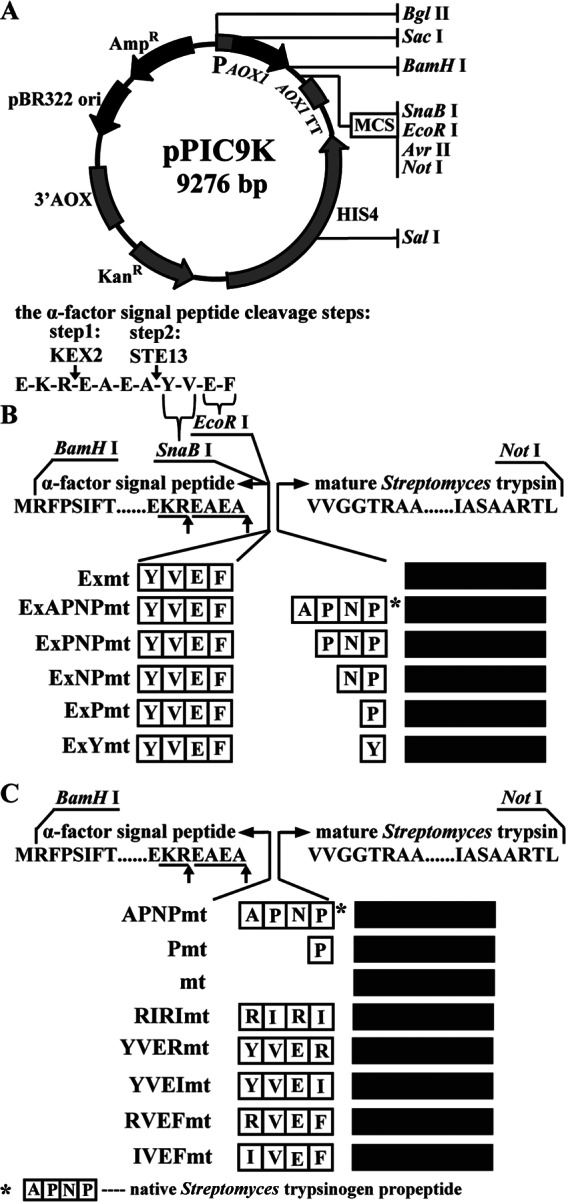
Schematic presentation of the construction of the recombinants with different propeptides. (A) Map of the integrated vector pPIC9k and the cleavage process of the α-factor signal peptide in Pichia pastoris. (B) Structure of the recombinants with different propeptides. YVEF was the residual sequence at the N terminus because of digestion by endonucleases SnaBI and EcoRI and cleavage of the α-factor signal peptide by peptidases KEX2 and STE13. (C) Structure of recombinant trypsin by fusing different designed propeptides.
In the past decade, computational methods for protein simulation and analysis have been developed and exploited, which in turn has promoted protein research by combining such data with in vitro experimental verification (18–20). In particular, when the crystal structure of the target protein is not available, a computer modeling method can be indispensable for determining the protein conformation. To date, the S. griseus trypsinogen crystal structure has not been resolved. Therefore, computer-aided modeling should offer key information that further directs rational engineering.
By applying the S. griseus ATCC 10137 genome as the template, the fragments encoding trypsinogen or trypsin (Fig. 1) were amplified with designed oligonucleotides (see Table S2 in the supplemental material) and subcloned into pPIC9k (see Table S1 in the supplemental material), which is controlled by the AOX1 promoter and the α-factor signal peptide from Saccharomyces cerevisiae (Fig. 1A). All the recombinants constructed (see Table S1) were further confirmed by PCR amplification (see Fig. S1 in the supplemental material), and trypsin activity was measured as described previously (11). As shown in Fig. 2, the recombinants Exmt, ExYmt, and mt accumulated trypsin to 0.6 U ml−1, 0.32 U ml−1, and 0.21 U ml−1, respectively. However, the recombinants that contained a proline residue showed no activity, despite the observation of expression and secretion (Fig. 3A). The differences between recombinants ExPmt and ExYmt and between Pmt and mt suggested that the proline was vital to the inhibition of trypsin. Moreover, compared with mt, the recombinants Exmt and ExYmt accumulated higher titers of trypsin (Fig. 2).
Fig 2.
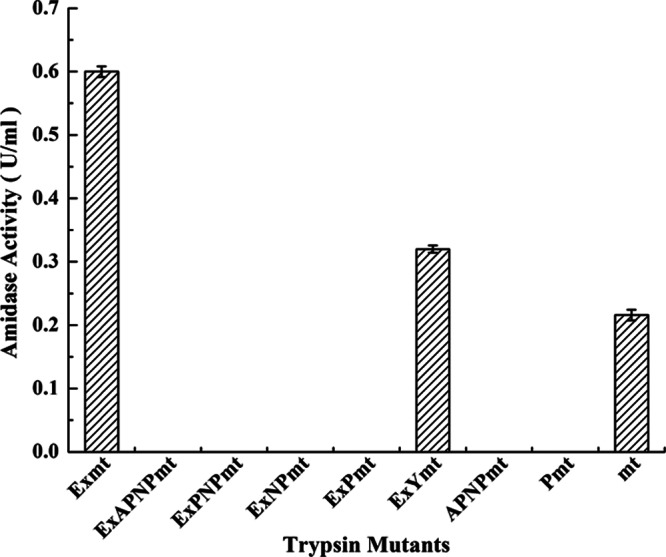
Comparison of the recombinants with different propeptides. Cultivation was performed in baffled shake flasks with 30 ml medium at 30°C for 72 h. Methanol (5 g liter−1) was used as the inducer added to trigger the AOX1 promoter. The results are the averages from three individual experiments.
Fig 3.
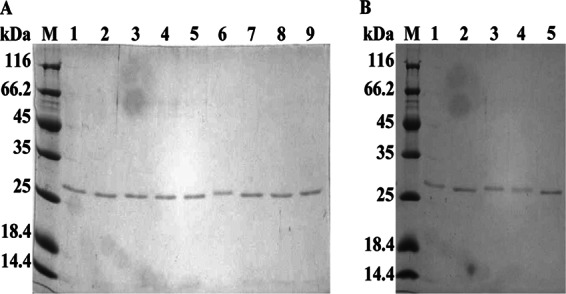
SDS-PAGE analysis of the purified recombinants with different propeptides. (A) Recombinants with the native propeptide and mutants constructed by stepwise deletion. Lanes 1 to 9 contain the purified recombinants Exmt, ExAPNPmt, ExPNPmt, ExNPmt, ExPmt, ExYmt, APNPmt, Pmt, and mt, respectively. (B) Mutants with different designed propeptides. Lanes 1 to 5 contain the recombinants RIRImt, YVERmt, YVERmt, RVEFmt, and IVEFmt, respectively. M, molecular markers (Thermo, USA). All the purified enzymes were concentrated by ultrafiltration.
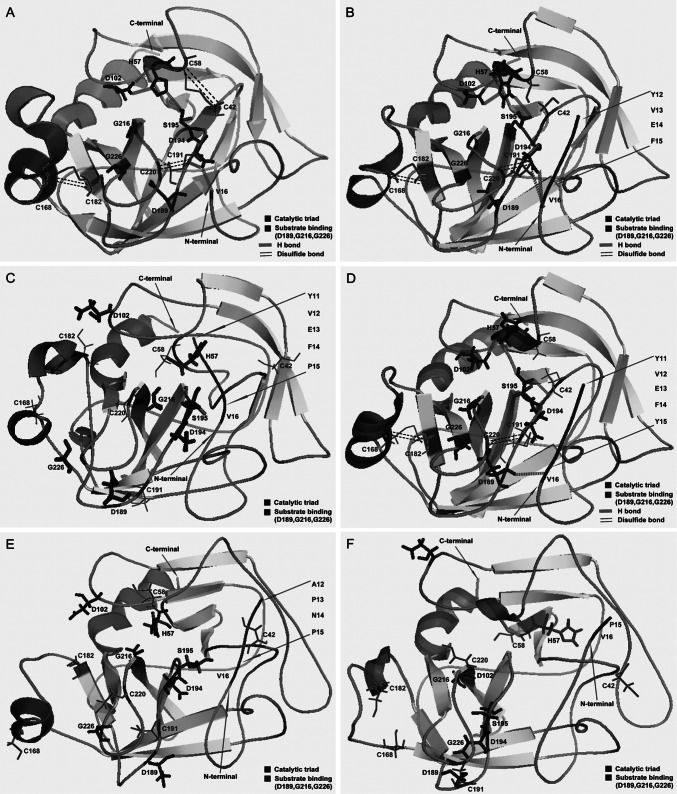
To study the inhibition mechanism of the native propeptide for the S. griseus trypsinogen, especially the role of the residue proline, we carried out molecular dynamics. Molecular dynamics (18) of the recombinants were simulated by the NAMD software with the CHARMM force field (http://www.ks.uiuc.edu/Research/namd) and comparatively analyzed with the active S. griseus trypsin (PDB ID, 1SGT). Total electrostatic energy in a particle mesh Ewald periodic box was calculated by the Ewald summation method, and the whole system was minimized using the descent method plus the conjugate gradient method. As shown in Fig. 4A, the major features of native trypsin were characterized. First, three disulfide bonds between residues C168 and C182, residues C191 and C220, and residues C42 and C58 held the substrate binding pocket rigid, and the correct fold was observed. Second, three hydrogen (H) bonds among the catalytic triad (H57, D102, and S195) maintained the accurate conformation of the catalytic center (4). Third, one H bond had formed between V16 and D194, and this interaction stabilized the structure of trypsin. In contrast, although ExPmt, APNPmt, and Pmt were found to be expressed (Fig. 3A), their structures did not resemble the crystal structure (Fig. 4C, E, and F). Consequently, it could be concluded that the propeptide, especially proline, affected the formation of disulfide bonds and H bonds, which eventually resulted in a native inactive trypsinogen. In contrast, both Exmt (YVEF) and ExYmt (YVEFY) were actively overexpressed. Furthermore, compared with the native trypsin, the recombinants Exmt and ExYmt appeared to be much more flexible, presumably because of the missing disulfide bond (Fig. 4A, B, and D), suggesting that the disulfide bond between residues C42 and C58 could be ignored, yet trypsin activity is retained. In the native trypsin and the recombinant Exmt, there are three H bonds in the catalytic triad. Nevertheless, only one H bond formed between H57 and D102 in ExYmt, suggesting that the H bond between H57 and D102 may be essential for active trypsin. Furthermore, in Exmt and the native trypsin, one H bond formed between V16 and D194, whereas in ExYmt, the H bond formed between V16 and D189. As a result, it could be deduced that the H bond between V16 and the loop (D189-D194) is important for active trypsin. In comparison with the native trypsin, the recombinants contained fewer H bonds and ion pairs, which indicated the profound influence of the N-terminal propeptide (see Table S3 in the supplemental material).
Fig 4.
Simulated structure of the S. griseus trypsinogen recombinants by NAMD molecular calculation. The crystal structure of S. griseus trypsin was used as the template. (A) Crystal structure of S. griseus trypsin; (B to F) simulated structures of the recombinants Exmt, ExPmt, ExYmt, APNPmt, and Pmt, respectively.
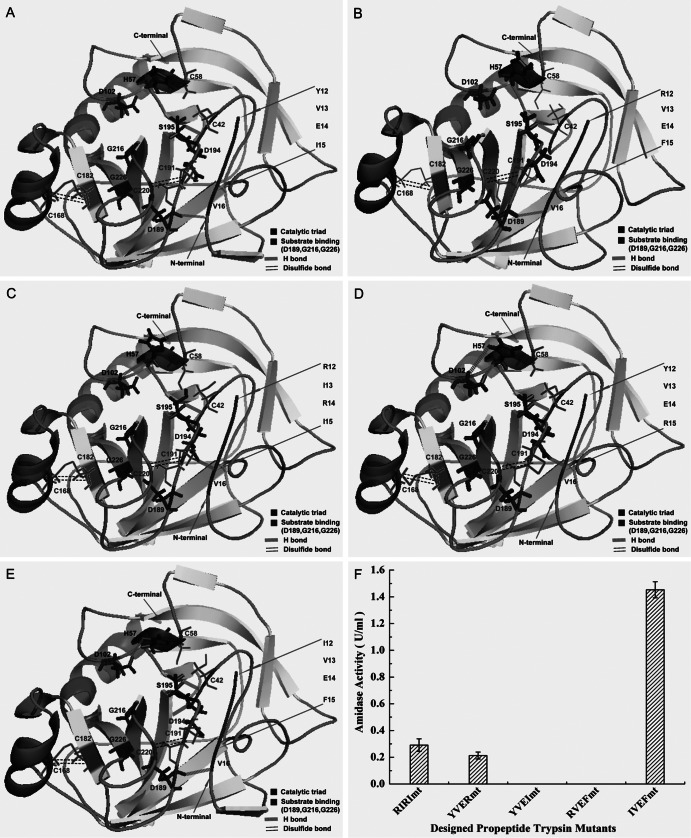
After exploring the inhibitory mechanism of the propeptide, we evaluated the significance of the three factors identified above. First, we designed and constructed four recombinants, YVEImt, RVEFmt, RIRImt, and YVERmt (Fig. 1C), and simulated their structures. Interestingly, introduction of the peptide YVEI or RVEF resulted in successful formation of disulfide bonds (C168-C182 and C191-C220) and a hydrogen bond (V16-D189) (Fig. 5A and B). However, no activity of YVEImt and RVEFmt was detected (Fig. 5F), although secretory expression was observed (Fig. 3B). In contrast, the recombinants RIRImt and YVERmt exhibited clear activity (0.29 U ml−1 and 0.22 U ml−1, respectively) (Fig. 5F). Consequently, by comparing the results of in silico simulations and in vitro experiments, it could be concluded the formation of the H bond between residues H57 and D102 is essential for trypsin activity. Subsequently, we further designed a novel peptide IVEF and fused it at the N terminus of the native trypsin (Fig. 1C). By in silico simulation, the recombinant IVEFmt formed two disulfide bonds (C168-C182 and C191-C220) and two H bonds (H57-D102 and V16-D189) (Fig. 5E), which indicated that the expressed protein would be active. As predicted, the recombinant IVEFmt was overexpressed (Fig. 3B), and a remarkable activity of 1.45 U ml−1 was obtained (Fig. 5F). Compared with parameters between the native trypsin and the recombinant IVEFmt, we found that although H bonds and ion pairs decreased in the recombinant IVEFmt, π interactions and the proportion of the random coil increased, which may give the recombinant IVEFmt more flexibility (see Table S3 in the supplemental material).
Fig 5.
Comparison of the recombinants with different designed propeptides. (A to E) Simulated structures of the recombinants YVEImt, RVEFmt, RIRImt, YVERmt, and IVEFmt, respectively. (F) Secretory trypsin accumulation in cultured supernatants. Cultivation was performed in baffled shake flasks with 30 ml medium at 30°C for 72 h. Methanol (5 g liter−1) was used as the inducer to trigger the AOX1 promoter. The results are the averages from three individual experiments.
In conclusion, by applying the in silico simulations and in vitro verification, we explored the inhibitory mechanism of the propeptide to the S. griseus trypsinogen and interpreted the crucial role of the proline. The disulfide and H bonds were found to be the major factors involved in trypsin regulation. In particular, the H bond between residues H57 and D102 was crucial for trypsin activity. More importantly, by rationally designing an artificial propeptide IVEF, the titer of trypsin was increased 6.71-fold. Furthermore, our work demonstrated that the combination of in silico simulations and in vitro experiments would be favorable to protein engineering.
Supplementary Material
ACKNOWLEDGMENTS
We thank Byong Lee at Jiangnan University for his discussion and revision.
This work was financially supported by the National High Technology Research and Development Program of China (863 Program, 2011AA100905), Program for Changjiang Scholars and Innovative Research Team in University (no. IRT1135), the National Natural Science Foundation of China (31200020, 31000031), the Independent Innovation Program of Jiangnan University (JUSRP111A23), the Doctor Candidate Foundation of Jiangnan University (JUDCF10014), the 111 Project, and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Footnotes
Published ahead of print 5 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00376-13.
REFERENCES
- 1. Walsh KA. 1970. Trypsinogens and trypsins of various species. Methods Enzymol. 19:41–63 [Google Scholar]
- 2. Bode W, Huber R. 1978. Crystal structure analysis and refinement of two variants of trigonal trypsinogen: trigonal trypsin and PEG (polyethylene glycol) trypsinogen and their comparison with orthorhombic trypsin and trigonal trypsinogen. FEBS Lett. 90:265–269 [DOI] [PubMed] [Google Scholar]
- 3. Chen J-M, Kukor Z, Le Maréchal C, Toth M, Tsakiris L, Raguénès O, Férec C, Sahin-Tóth M. 2003. Evolution of trypsinogen activation peptides. Mol. Biol. Evol. 20:1767–1777 [DOI] [PubMed] [Google Scholar]
- 4. Davie EW, Neurath H. 1955. Identification of a peptide released during autocatalytic activation of zymogen. J. Biol. Chem. 212:515–530 [PubMed] [Google Scholar]
- 5. Yamashina I. 1956. The action of enterokinase on trypsinogen. Acta Chem. Scand. 10:739–743 [DOI] [PubMed] [Google Scholar]
- 6. Roach JC, Wang K, Gan L, Hood L. 1997. The molecular evolution of the vertebrate trypsinogens. J. Mol. Evol. 45:640–652 [DOI] [PubMed] [Google Scholar]
- 7. Olafson RW, Jurášek L, Carpenter MR, Smillie LB. 1975. Amino acid sequence of Streptomyces griseus trypsin. Cyanogen bromide fragments and complete sequence. Biochemistry 14:1168–1177 [DOI] [PubMed] [Google Scholar]
- 8. Kim JC, Cha SH, Jeong ST, Oh SK, Byun SM. 1991. Molecular cloning and nucleotide sequence of Streptomyces griseus trypsin gene. Biochem. Biophys. Res. Commun. 181:707–713 [DOI] [PubMed] [Google Scholar]
- 9. Ohnishi Y, Ishikawa J, Hara H, Suzuki H, Ikenoya M, Ikeda H, Yamashita A, Hattori M, Horinouchi S. 2008. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J. Bacteriol. 190:4050–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Read R, James JMNG. 1988. Refined crystal structure of Streptomyces griseus trypsin at 1.7 Å resolution. J. Mol. Biol. 200:523–551 [DOI] [PubMed] [Google Scholar]
- 11. Ling Z, Ma T, Li J, Du G, Kang Z, Chen J. 2012. Functional expression of trypsin from Streptomyces griseus by Pichia pastoris. J. Ind. Microbiol. Biotechnol. 39:1651–1662 [DOI] [PubMed] [Google Scholar]
- 12. Brake AJ, Merryweather JP, Coit DG, Heberlein UA, Masiarz FR, Mullenbach GT, Urdea MS, Valenzuela P, Barr PJ. 1984. α-Factor-directed synthesis and secretion of mature foreign proteins in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 81:4642–4646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oka C, Tanaka M, Muraki M, Harata K, Suzuki K, Jigami Y. 1999. Human lysozyme secretion increased by alpha-factor pro-sequence in Pichia pastoris. Biosci. Biotechnol. Biochem. 63:1977–1983 [DOI] [PubMed] [Google Scholar]
- 14. Werten MWT, de Wolf FA. 2005. Reduced proteolysis of secreted gelatin and Yps1-mediated alpha-factor leader processing in a Pichia pastoris kex2 disruptant. Appl. Environ. Microbiol. 71:2310–2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ge B, Yang F, Yu D, Liu S, Xu H. 2010. Designer amphiphilic short peptides enhance thermal stability of isolated photosystem-I. PLoS One 5:e10233 doi: 10.1371/journal.pone.0010233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhao X, Nagai Y, Reeves PJ, Kiley P, Khorana HG, Zhang S. 2006. Designer short peptide surfactants stabilize G protein-coupled receptor bovine rhodopsin. Proc. Natl. Acad. Sci. U. S. A. 103:17707–17712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dai Z, Fu H, Zhang Y, Zeng J, Tang B, Tang X-F. 2012. Insights into the maturation of hyperthermophilic pyrolysin and the roles of its N-terminal propeptide and long C-terminal extension. Appl. Environ. Microbiol. 78:4233–4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. 2005. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26:1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Eswar N, Eramian D, Webb B, Shen Sali M-YA. 2008. Protein structure modeling with MODELLER. Methods Mol. Biol. 426:145–159 [DOI] [PubMed] [Google Scholar]
- 20. Zhang Z, Norris J, Schwartz C, Alexov E. 2011. In silico and in vitro investigations of the mutability of disease-causing missense mutation sites in spermine synthase. PLoS One 6:e20373 doi: 10.1371/journal.pone.0020373 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.