Abstract
The Lost Hammer (LH) Spring is the coldest and saltiest terrestrial spring discovered to date and is characterized by perennial discharges at subzero temperatures (−5°C), hypersalinity (salinity, 24%), and reducing (≈−165 mV), microoxic, and oligotrophic conditions. It is rich in sulfates (10.0%, wt/wt), dissolved H2S/sulfides (up to 25 ppm), ammonia (≈381 μM), and methane (11.1 g day−1). To determine its total functional and genetic potential and to identify its active microbial components, we performed metagenomic analyses of the LH Spring outlet microbial community and pyrosequencing analyses of the cDNA of its 16S rRNA genes. Reads related to Cyanobacteria (19.7%), Bacteroidetes (13.3%), and Proteobacteria (6.6%) represented the dominant phyla identified among the classified sequences. Reconstruction of the enzyme pathways responsible for bacterial nitrification/denitrification/ammonification and sulfate reduction appeared nearly complete in the metagenomic data set. In the cDNA profile of the LH Spring active community, ammonia oxidizers (Thaumarchaeota), denitrifiers (Pseudomonas spp.), sulfate reducers (Desulfobulbus spp.), and other sulfur oxidizers (Thermoprotei) were present, highlighting their involvement in nitrogen and sulfur cycling. Stress response genes for adapting to cold, osmotic stress, and oxidative stress were also abundant in the metagenome. Comparison of the composition of the functional community of the LH Spring to metagenomes from other saline/subzero environments revealed a close association between the LH Spring and another Canadian high-Arctic permafrost environment, particularly in genes related to sulfur metabolism and dormancy. Overall, this study provides insights into the metabolic potential and the active microbial populations that exist in this hypersaline cryoenvironment and contributes to our understanding of microbial ecology in extreme environments.
INTRODUCTION
Cryoenvironments are defined as permanently subzero or frozen environments, such as permafrost, glaciers, ice sheets, multiyear sea ice, high-elevation Antarctic dry valleys, and some cold saline springs (1–6). Microorganisms inhabiting cryoenvironments must face the challenges of subzero temperatures, low water activity, and, often, high solute concentrations to sustain their viability. The cold saline springs on Axel Heiberg Island (AHI) in the Canadian high Arctic discharge through 500 to 600 m of thick permafrost, maintain a liquid state at subzero temperatures, and offer a unique opportunity to assess microbial adaptations to extremes of both high salinity and subzero temperatures (3, 4, 7–9). These springs occur in an area with an average annual air temperature of −15°C, reaching below −40°C during the winter months, and probably originate from subpermafrost groundwater flow through carboniferous evaporites in areas of diapiric uplift on AHI (10, 11). Other Arctic cold springs, on Ellesmere Island in the Canadian high Arctic and on the Norwegian high-Arctic Svalbard archipelago, have been reported (12–14), although the discharges from these springs are not subzero. Viable microbial communities have been described for all of these Arctic springs (3, 4, 7, 8, 13–15).
The Lost Hammer (LH) Spring, located in the central west region of AHI (79°7′N, 90°21′W) is the coldest and saltiest of all Arctic springs described to date. LH is characterized by a perennial hypersaline (24%) discharge at subzero temperatures (∼−5°C) flowing to the surface through a hollow, 2-m-high cone-shaped salt tufa structure. The discharge waters are microoxic (dissolved oxygen, 0.1 to 1 ppm), highly reducing (≈−165 mV), and neutral (pH ≈7) and contain ammonia (6.87 mg kg−1) and high concentrations of sulfate (10.0%, wt/wt). During the summer months, the spring waters empty from the dome structure, partially exposing the spring sediments to ambient conditions; however, the sediments remain anoxic and highly reducing. Continuous gas emissions from the spring indicate a thermogenic methane source underlying LH (3, 4). On the basis of these properties, this spring is considered a significant astrobiology analogue site (15, 16) for possible habitats currently present on Mars and the cold moons Europa and Enceladus. For example, the widespread distribution of chloride and sulfate minerals on Mars (16, 17), reports of spring-like structures on the Martian surface (18, 19), recent images indicating that liquid brines flowed on Mars during the past decade under mean surface temperatures of −60°C and extensive permafrost (20, 21), and the potential detection of atmospheric methane on Mars (22–24) highlight the importance of cold hypersaline terrestrial environments, such as the LH Spring, as analogue sites for Mars as well as for the icy Saturnian moon Enceladus, where methane, ammonia, and simple organics have been detected in the saline plume features erupting from the surface (25).
In our initial studies of the sediments of the LH Spring outlet (15) and outflow channels (3), microbial activity was detected by using mineralization assays. We also isolated halophilic and cryophilic microbial strains from the sediments of both the spring outlet and outflow channels. Profiling of the microbial community (16S rRNA clone libraries) of LH revealed phylotypes related to halophilic bacteria/archaea, sulfate-reducing archaea/bacteria, methylotrophic/methanotrophic bacteria, and methanogenic archaea (3, 4). The anaerobic methane-oxidizing archaeal group 1 (ANME-1), a clade of anaerobic methane-oxidizing archaea, dominated the archaeal community in the spring outlet sediments (4), while sequences related to Thaumarchaeota dominated the spring channel sediments (3).
Metagenomic analyses of other extremely cold or saline environments have revealed the importance of genes involved in carbon cycling operating in permafrost (26, 27), genes related to stress responses of microorganisms colonizing ice shelves (1), evidence of lateral gene transfer in deep-sea hydrothermal vent biofilms (28–30), and the microbial ecology of an Antarctic meromictic lake (31, 32). Surveys of cDNA of 16S rRNA are currently used to identify potentially active microorganisms in diverse environments (33–36), including a subzero, briny, ice-sealed lake in the Antarctic (36). In the present study, we combined a metagenomic approach with pyrosequencing analyses of cDNA of bacterial and archaeal 16S rRNA genes to assess the functional potential of the LH microbial community and to identify the active community members in the LH Spring and consequently infer their possible ecological functions.
The specific objectives for analyzing the LH metagenomic and cDNA data sets of the spring outlet sediment were (i) to map the microbial metabolic pathways driving biogeochemical cycles, focusing on methane, ammonia, and sulfur cycling, which were expected to play key roles in shaping LH communities based on previous investigations of the LH system (3, 4); (ii) to identify the dominant genes involved in adaptations to cold and high salt concentrations that would allow autochthonous populations to cope with the extreme natural conditions of the site; (iii) to compare the functional potential of the LH metagenome to metagenomes from other cold or saline environments; and (iv) to identify the bacterial and archaeal taxa that may be active in situ.
MATERIALS AND METHODS
Study site and sample collection.
The LH Spring (79°7′N, 90°21′W) is located on Axel Heiberg Island in a valley off the south shore of Strand Fiord. An ∼1.7-m-high dome-like structure composed of precipitated mineral salts surrounds the spring outlet.
Our in situ analyses of geochemical/physical parameters, recorded from 2005 to 2012, indicate that the LH Spring outlet sediment and water environment have remained very consistent (temperature, −5°C; salinity, ∼25%; oxidation-reduction potential [ORP], −160 mV; dissolved oxygen concentration [DOC], 0.1 to 1 ppm) at both the late winter (April/May) and midsummer (July) sampling points. Therefore, we considered that LH samples collected in different years are comparable.
To extract total environmental DNA, about 250 g of sediment was collected (July 2009) approximately 50 mm below the surface by using an ethanol-sterilized Scoopula and was placed in a sterile plastic sampling bottle. LH sediments (15 g) designated for RNA analysis were collected (July 2010) by using an ethanol-sterilized spatula and were stored in sterile 50-ml conical tubes filled with LifeGuard soil preservation solution (Mo Bio Laboratories, Inc., Carlsbad, CA, USA) to a final volume of 50 ml. Both DNA and RNA samples were transported to Montreal, Canada, at temperatures below 5°C, where they were stored at −20°C until further analyses.
Extraction and sequencing of metagenomic DNA.
DNA was extracted from 5 g of LH sediment by using the PowerMax soil DNA isolation kit (Mo Bio Laboratories, Inc., Carlsbad, CA, USA) according to the manufacturer's instructions. To obtain sufficient DNA (a minimum of 500 ng DNA) for metagenomic pyrosequencing, the purified metagenomic DNA was amplified via multiple displacement amplification (MDA) by using a GenomiPhi V2 DNA amplification kit (GE Healthcare, Piscataway, NJ, USA) according to the manufacturer's instructions. One microliter of DNA (concentration, less than 10 ng/μl) was used as the template and was mixed with 9 μl of buffer. The mixed DNA was heated at 95°C for 3 min, cooled to 4°C, and incubated at 30°C for 90 min with 1 μl of enzyme mixture and 9 μl of reaction buffer. To terminate the reaction, the sample was heated at 65°C for 10 min. Control reactions were also performed in parallel by using the positive control provided in the kit and sterile double-distilled H2O (ddH2O) as a negative control. No DNA band was detected in the negative-control sample following MDA. The amplified samples were pooled and were purified using Amicon Ultra 0.5-ml centrifugal filters (Millipore Corporation, Billerica, MA, USA) to a final volume of 21 μl of solution containing 253.4 ng DNA μl−1. The purified sample was sequenced by using a Roche 454 GS FLX Titanium sequencer (454 Life Sciences, Branford, CT, USA), located at the Centre for Applied Genomics, Hospital for Sick Children, Toronto, ON, Canada.
Metagenomic DNA analyses.
To analyze and annotate the metagenomic data, all LH reads were uploaded onto the online metagenomic annotation server MG-RAST (MetaGenome Rapid Annotation with Subsystem Technology) (37). Based on the BLAST-like alignment tool (BLAT algorithm) (38), metagenomic sequences were compared to those of gene and protein-coding databases; the GenBank (http://www.ncbi.nlm.nih.gov/GenBank/) taxonomic database was used for the LH metagenome, while the SEED protein-coding gene database (http://www.theseed.org/wiki/index.php/Home_of_the_SEED) was used for comparison with the putative proteins encoded in the metagenome. Metabolic pathways were mapped using the Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/kegg/) database. For all databases used, only matches of >50 nucleotides and 50% similarity with an E value of ≤10−5 were included for both taxonomy and function analyses. The stringency cutoff was tested with different E values (down to 10−15) at the phylum and subsystem levels for taxonomy and functional classifications, respectively (see Fig. S1 in the supplemental material). The correlations between different E values were tested by the Pearson product-moment correlation coefficient (see Table S1 in the supplemental material).
In addition to automated annotations by MG-RAST, the complete LH metagenome was subjected to additional screenings targeting marker genes of (reverse) methanogenesis (i.e., mcrA, encoding the alpha subunit of the methyl coenzyme M reductase) and methane oxidation (i.e., pmoA and mmoX, encoding the alpha subunits of the particulate and soluble forms of methane monooxygenase). The amino acid sequences of MCRA, PMOA, and MMOX were recovered from the NCBI protein database (on 16 February 2013) and were used as target databases for alignments with the LH metagenome. BLASTX alignments were performed using the BLAST command line application (version 2.2.27+) with default algorithm parameters and an E value cutoff of 10−5. The results were then visualized and proofread in MEGAN (version 4.70.4), and hits with bit scores higher than 50 were considered significant (39). Reads of significant hits were then extracted, subjected to a second set of BLASTX alignments against the complete GenBank nonredundant (nr) database to ascertain their function, and finally reannotated in MEGAN.
Statistical analyses.
We selected 6 other metagenomes publicly available in MG-RAST, generated from the following habitats: the Markham Ice Shelf, the Ward Hunt Ice Shelf, an estuary of the Bay of Fundy, the Lost City hydrothermal system, a hypersaline lagoon in the Galapagos Islands, a microbial mat from the McMurdo Ice Shelf in the Ross Sea sector of Antarctica, an Antarctic saline lake (Ace Lake), and high-Arctic permafrost and active-layer soils from Eureka, Ellesmere Island (MG-RAST identification numbers [ID], 4445126.3, 4445129.3, 4441582.3, 4461585.3, 4441599.3, 4445845.3, 4443684.3, 4443232.3, and 4443231.3, respectively). We also reannotated the assembled Alaskan permafrost metagenome (Integrated Microbial Genomes database [IMG] ID 1618) from Mackelprang and colleagues (27) in MG-RAST to make it comparable in the MG-RAST subsystem. The relative abundance at the “function” level of the SEED hierarchy was used to calculate Bray-Curtis distances between sample pairs using the “vegdist” function of the “vegan” package (http://vegan.r-forge.r-project.org/) in R (version 2.9.0; The R Foundation for Statistical Computing). Principal coordinate analyses (PCoA) were then performed using the “cmdscale” function. Arrows representing the relative abundance at “level 1” of the SEED hierarchy were then superimposed on the ordination as supplementary variables, not involved in the calculation of the ordination (26).
RNA extraction and cDNA analyses.
To obtain total RNA, sediment samples (2 g) from the LH Spring were processed with an RNA PowerSoil total-RNA isolation kit (Mo Bio Laboratories, Inc., Carlsbad, CA, USA) according to the manufacturer's instructions with minor modifications as follows: (i) an additional 1.0 g of 0.1-mm glass beads (Mo Bio Laboratories, Inc., Carlsbad, CA, USA) was added to each reaction tube; (ii) bead-beating time was doubled; and (iii) nucleotide precipitation was performed overnight. The extracted RNA was then treated with amplification-grade DNase I (Invitrogen, Carlsbad, CA, USA) at room temperature for 15 min according to the manufacturer's instructions and was then inactivated by the addition of EDTA at 65°C for 20 min. The treated sample was concentrated and purified using Amicon Ultra 0.5-ml centrifugal filters (Millipore Corporation, Billerica, MA, USA). For the synthesis of cDNA, we used an iScript Select cDNA synthesis kit (Bio-Rad, Hercules, CA, USA) to process the purified RNA samples, using random primers provided in the kit. The cDNA of the 16S rRNA genes was sequenced at the Research and Testing Laboratory (Lubbock, TX, USA) using a Roche 454 GS FLX Titanium sequencer (454 Life Sciences, Branford, CT, USA) system with bacterial (28F, 5′GAGTTTGATCNTGGCTCAG3′; 519R, 5′GTNTTACNGCGGCKGCTG3′) (40) and archaeal (ARCH571F, 5′GCYTAAAGSRNCCGTAGC3′ [41]; ARCH909R [also called 890aR], 5′TTTCAGYCTTGCGRCCGTAC3′ [42]) primers. Tag-encoded pyrosequencing was performed by following established protocols (43).
The cDNA sequences were trimmed, aligned, and dereplicated using the RDP pyrosequencing pipeline (44). The minimum quality score was set to 20, and sequences shorter than 150 bp were excluded from downstream analyses. The trimmed sequences were aligned with the pyrosequencing aligner using the Bacteria/Archaea model. The aligned sequences were then clustered using the complete-linkage clustering method with a maximum distance of 15% and a step size of 1.0. Dereplicated sequences were generated using the representative sequence method with 98% similarity and were then analyzed via the BLASTn algorithm against the online GenBank nr database (45). Neighbor-joining phylogenetic trees of selected sequences were generated with MEGA, version 5 (46), by using a bootstrap method with 1,000 replications and a Jukes-Cantor model.
Nucleotide and metagenome sequence accession numbers.
The sequences of the cDNA of the 16S rRNA genes obtained in this study have been deposited in the GenBank database under accession numbers KC470367 to KC470541. The ID of the LH metagenome deposited in MG-RAST is 4478244.3.
RESULTS AND DISCUSSION
Metagenomic sequencing statistics.
In total, sequencing resulted in 1,032,783 reads containing 341,472,858 bases, with an average length of 330 bp (Table 1). Among all sequences, 751,870 reads (72.8%) passed the quality control (QC) criteria. The numbers of reads representing predicted protein and rRNA features were 403,739 (39.1%) and 50,589 (4.9%), respectively. Of these putative protein- and rRNA-related reads, 187,609 (18.2%) and 220 (2.1%) matched known protein and rRNA sequences, respectively. However, 434,847 (42.1%) reads remained unidentified due to a lack of comparable reference sequences, highlighting the need to further isolate, characterize, and sequence the genomes of new strains from extreme environments such as the LH Spring to complement existing databases, making the annotation of reads from extreme environments more reliable and informative. The average GC content was 45% for the sequences that passed quality control. The total DRISEE (duplicate read inferred sequencing error estimation) error was 0.7% (47), which was deemed acceptable for proceeding to further analyses. The Pearson product-moment correlation coefficients of the cutoff E value (≤10−5) with lower E values (10−10, 10−15, and 10−20) showed that the correlations were highly significant down to 10−15 and 10−20 for taxonomic and functional classifications at the phylum and subsystem levels, respectively (see Table S1 in the supplemental material). Thus, these calculations proved that the E value cutoff we set was appropriate for applying to this metagenome study.
Table 1.
Statistical analyses of the LH metagenome
| Parameter | Value |
|---|---|
| Total no. of sequences | 1,032,783 |
| Total sequence size (bp) | 341,472,858 |
| Shortest sequence length (bp) | 40 |
| Longest sequence length (bp) | 918 |
| Avg sequence length (bp) | 330 |
| No. of sequences that passed QC | 751,870 |
| No. of predicted/identified protein features | 403,739/187,609 |
| No. of predicted/identified rRNA features | 50,589/220 |
| GC content (%) | 45 |
Metagenomic microbial community composition.
Among the total 751,870 sequences that passed the QC criteria, 50.8%, 5.1%, and 0.8% were identified as fragments originating from Bacteria, Eukaryota, and Archaea, respectively. A total of 719,330 sequences (95.7% of the total sequences that passed QC) could be classified and assigned to different phyla by MG-RAST. Sequences related to Cyanobacteria (19.7%), Bacteroidetes (13.3%), and Proteobacteria (6.6%) were the dominant phyla among the sequences classified (Table 2). More than 90% of Cyanobacteria hits belonged to the orders Nostocales (35.5%), Oscillatoriales (23.0%), and Chlorococcales (38.4%). The abundance of sequences related to Cyanobacteria suggests that these microorganisms could potentially carry out photosynthesis, carbon fixation, and nitrogen fixation metabolism at LH. All five classes of Proteobacteria were also detected in the LH metagenome; the major groups present were Gammaproteobacteria (47.6%), Betaproteobacteria (30.7%), and Alphaproteobacteria (12.4%). Methylophilic and methanotrophic genera were detected in both the Gammaproteobacteria and the Betaproteobacteria. Gene fragments related to the ammonia-oxidizing order Nitrosomonadales were identified in the metagenome, which may provide evidence for bacterial ammonia-oxidizing activity at LH, in accordance with the presence of ammonia. In terms of sulfur metabolism, clades related to bacterial sulfate reducers, such as Desulfuromonadales, Desulfovibrionales, and Desulfobacterales, whose metabolic activity would be favorable with the high abundance of sulfate and sulfide as well as the reducing conditions detected at the site, were detected within the Deltaproteobacteria and Betaproteobacteria.
Table 2.
Composition of organisms detected in the LH metagenome
| Phylum | No. of hits | % of metagenome |
|---|---|---|
| Unclassified reads | 322,498 | 42.9 |
| Archaea | 6,020 | 0.8 |
| Euryarchaeota | 4,037 | 0.5 |
| Crenarchaeota | 351 | <0.1 |
| Thaumarchaeota | 75 | <0.1 |
| Other archaea | 1,557 | 0.2 |
| Bacteria | 382,205 | 50.8 |
| Cyanobacteria | 148,325 | 19.7 |
| Bacteroidetes | 99,693 | 13.3 |
| Proteobacteria | 49,803 | 6.6 |
| Firmicutes | 4,803 | 0.6 |
| Actinobacteria | 3,469 | 0.5 |
| Verrucomicrobia | 1,615 | 0.2 |
| Other bacteria | 74,470 | 9.9 |
| Eukaryota | 38,233 | 5.1 |
| Others | 2,914 | 0.4 |
| Total | 751,870 |
The small proportion of total archaeal reads within the metagenome included Euryarchaeota (0.5%; 4,037 hits), Crenarchaeota (0.05%; 351 hits), and Thaumarchaeota (0.01%; 75 hits) (Table 2). Among the Crenarchaeota, only Thermoprotei (343 hits) and unclassified Crenarchaeota (8 hits) were detected, while for Thaumarchaeota, sequences related to unclassified Thaumarchaeota (75 hits) were found. These hits for Crenarchaeota and Thaumarchaeota were related primarily to genes involved in DNA duplication, transcription, translation, and electron transport. The hits for Euryarchaeota were more varied; Archaeoglobi (213 hits), Halobacteria (481 hits), Methanomicrobia (1,878 hits), Methanobacteria (420 hits), Thermococci (323 hits), Methanococci (381 hits), Methanopyri (14 hits), Thermoplasmata (85 hits), and unclassified Euryarchaeota (242 hits) were identified. As well, the functional gene categories associated with these hits were more diverse; the most abundant genes were related to potassium channel proteins (potassium metabolism) (145 hits), cold shock DEAD box protein A (RNA metabolism) (84 hits), O-phosphoseryl-tRNA:cysteinyl-tRNA synthase (protein metabolism) (76 hits), and lysyl-tRNA synthetase (protein metabolism) (41 hits).
Diverse methanogenic genera were present within the Euryarchaeota data set, including Methanobrevibacter (29 hits), Methanothermobacter (342 hits), Methanothermus (21 hits), Methanococcus (226 hits), and Methanocorpusculum (39 hits). The detection of these methanogen-related sequences support the idea that at least a small portion of the methane exsolving from the LH Spring may be partly biogenic, although previous carbon and hydrogen isotope analyses indicated that the LH methane is thermogenic in origin (4). The presence of Methanobrevibacter was also reported in the LH Spring outflow channel area by using a 16S rRNA clone library (3). Methanogenic populations have also been found in other cold extreme environments, e.g., Canadian high-Arctic and Alaskan permafrost, melting glaciers, and other AHI saline springs (7, 26, 48), and have been shown to remain active down to −16.5°C in Siberian permafrost (49). Although hypersaline conditions are known to inhibit acetoclastic and hydrogenotrophic methanogenesis above ∼12% NaCl, methanogens relying on “noncompetitive” substrates (i.e., methylated amines, methanol, or dimethyl sulfide) can withstand higher salt concentrations; there have been reports of methanogenesis at salinities of 30% in endoevaporite communities (50, 51). Methanogenesis typically requires a lower redox potential than most other anaerobic bioreactions; considering the anoxic and highly reducing conditions in the LH Spring sediments, methanogens can be expected to be present and active in this ecosystem.
In our previous study of the LH Spring (4), the anaerobic methane-oxidizing archaeal group 1 (ANME-1) was the dominant archaeal clade (46.8%) detected, based on archaeal 16S rRNA clone library results. Although we detected archaeal 16S rRNA sequences identical to the previously identified LH ANME-1 sequences in the MDA-amplified LH DNA prior to metagenome pyrosequencing, no ANME-1 16S rRNA was detected after the metagenomic analysis, due, we suspect, to the low abundance of this population. ANME-1-related DNA fragments were searched by BLAST analysis, using unordered contigs of an ANME-1 genome (GenBank nucleotide database, accession no. FP565147). (52). BLAST analyses in MG-RAST (version 2.0) resulted in 1,000 hits within the LH metagenome with sequence similarities of 80% to 97% to ANME-1-related sequences. Functional annotation by BLASTx against the NCBI nr database classified these ANME-1-related hits as genes encoding the integrase core domain, FAD (flavin adenine dinucleotide)-containing dehydrogenase, Fe-S oxidoreductase related to leucyl-tRNA synthetase, and hypothetical proteins. However, only one gene probably involved in central carbon metabolism in ANME organisms, a putative carbon monoxide dehydrogenase/acetyl coenzyme A (acetyl-CoA) synthase (EC 2.3.1.169) (2 hits; 85% identity), was detected in the LH metagenome. Since the identity of the matches between the LH metagenome sequences and the ANME-1 genome (accession no. FP565147) was generally low, LH ANME-1 organisms most likely belong to a subgroup different from that for the published genome (FP565147). However, we still need more evidence to support this assumption and to better understand the ANME-1 previously detected in the LH Spring sediments.
Functional gene profiles of the LH metagenome.
The functional gene profile revealed that among the 259,557 annotated protein sequences (25.1% of the total reads), the most abundant functional groups were related to housekeeping functions, such as carbohydrate metabolism (10.1%), amino acid biosynthesis (10.0%), and vitamin and pigment metabolism (6.6%). Stress response-related sequences constituted 2.3% of all annotated reads and included a high proportion of oxidative stress (53.1%)- and osmotic stress (11.9%)-related sequences. The abundance of these genes may reflect adaptations to the high salinity and, possibly, the high-salinity-induced oxidative stress at LH, indicating that the LH microorganisms have the potential to survive and remain viable under the prevailing conditions. Descriptions of genes involved in methane, nitrogen, and sulfur metabolism, as well as stress responses, are discussed in the following paragraphs.
Methane metabolism.
Several functional genes directly related to methanogenesis were detected (Table 3). These include genes encoding an F420-dependent methylene-H4 MPT reductase (EC 1.5.99.11) (1 hit), formylmethanofuran dehydrogenases (fmd) (EC 1.2.99.5) (2 hits), CoB-CoM heterodisulfide reductases (EC 1.8.98.1) (2 hits), F420-reducing hydrogenases (EC 1.12.98.1) (30 hits), and methylenetetrahydromethanopterin dehydrogenases (mer) (EC 1.5.99.9) (2 hits). It should be noted, however, that the presence of the gene encoding the enzyme of the last step of methanogenesis, methyl-coenzyme M reductase (MCR), was not confirmed in the LH metagenome. An additional screening of the metagenome against an MCR target database did identify potential (MCR) homologs, but these sequences most closely matched ABC transporters/ATP-binding proteins when compared to the GenBank nr database. Considering the low frequency of other methanogenesis genes recovered, the absence of mcr sequences in the annotated LH data set may result from insufficient sequencing coverage. Similar results have also been found in other metagenomic studies of deep subsurface marine sediments where genes involved in methanogenesis were found but no mcr sequences were recovered (53).
Table 3.
Numbers of different gene variants retrieved in the LH metagenomic data sets for different functions
| Function | No. of hits |
|---|---|
| Methane | |
| F420-dependent methylene-H4 MPT reductase (EC 1.5.99.11) | 1 |
| F420-reducing hydrogenase (EC 1.12.98.1) | 30 |
| CoB-CoM heterodisulfide reductase (EC 1.8.98.1) | 2 |
| Formylmethanofuran dehydrogenase (EC 1.2.99.5) | 2 |
| Methylenetetrahydromethanopterin dehydrogenase (EC 1.5.99.9) | 2 |
| Nitrogen | |
| Nitrogenase (EC 1.18.6.1) | 2 |
| Copper-containing nitrite reductase (EC 1.7.2.1) | 45 |
| Nitric oxide reductase (EC 1.7.99.7) | 14 |
| Nitrite reductase [NAD(P)H] (EC 1.7.1.4) | 48 |
| Nitrous oxide reductase (EC 1.7.99.6) | 16 |
| Respiratory nitrate reductase (EC 1.7.99.4) | 4 |
| Sulfur | |
| Phosphoadenylyl-sulfate reductase (EC 1.8.4.8) | 29 |
| Sulfite reductase (ferredoxin) (EC 1.8.7.1) | 49 |
| Sulfate adenylyltransferase (EC 2.7.7.4) | 222 |
| Adenylyl-sulfate kinase (EC 2.7.1.25) | 172 |
| Sulfur oxidation genes (soxBDHR) | 302 |
Despite the presence of several reads related to known bacterial clades containing methanotrophic members (i.e., Gammaproteobacteria and Betaproteobacteria), no gene hits relating to the key enzyme in aerobic methanotrophy, the particulate or soluble methane monooxygenase (i.e., PMO or MMO), were obtained by either MG-RAST or MEGAN annotation.
Nitrogen metabolism.
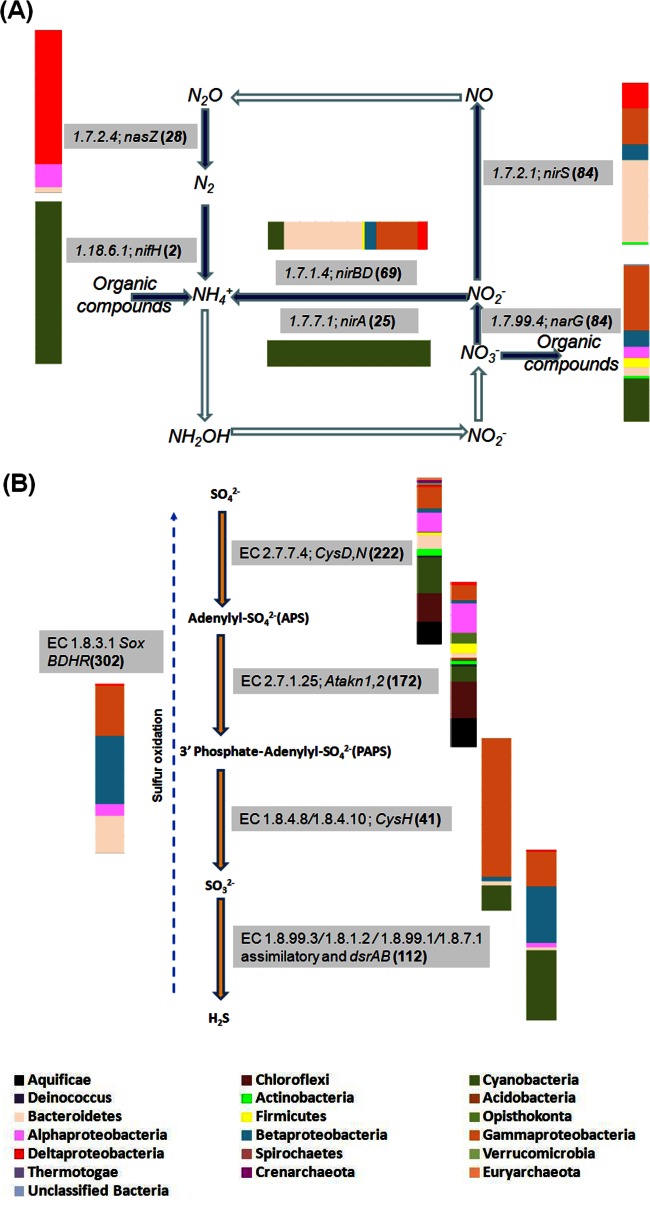
Most genes involved in nitrogen-cycling pathways were detected and were related mainly to Cyanobacteria (Fig. 1A; see also Table S2 in the supplemental material). The nifH gene, which encodes a nitrogenase (EC 1.18.6.1) that converts nitrogen gas to ammonia, was detected and matched cyanobacterial sequences from Cyanothece and Nostoc species. Sequences related to Burkholderia spp., typical denitrifiers in saline environments (54, 55), were detected for two enzymes, NarG (EC 1.7.99.4) and NirS (EC 1.7.2.1). Denitrifiers usually found in saline and freshwater environments, including Kangiella spp. and Flavobacterium spp. (56, 57), respectively, were also detected in the LH metagenome, encoded by genes involved in denitrification pathways. Two enzymes (EC 1.7.1.4 and 1.7.7.1) involved in the reduction of nitrite to ammonia were matched to members of the Cyanobacteria, such as Synechocystis spp., Cyanothece spp., and Nostoc spp. Synechocystis and Cyanothece spp. have been reported to undergo heterotrophic metabolism during the dark phase of their life cycle (58, 59), which may be advantageous for maintaining activity during the long-term darkness of the Arctic winter. Three enzymes that play key roles in nitrogen cycling, nitric oxide reductase (EC 1.7.99.7), ammonia monooxygenase (EC 1.13.12.4), and hydroxylamine oxidase (EC 1.7.3.4), were absent from the LH metagenome. As has been observed in other environments, the function of nitric oxide reductase may be replaced by abiotic processes (60). Although 16S rRNA phylogenetic evidence of Thaumarchaea has been detected previously (3) and in the cDNA of the 16S rRNA genes (see below), the absence of ammonia monooxygenase and hydroxylamine oxidase indicates that the complete ammonia oxidation pathway could not be reconstructed from the LH metagenome, and our analyses cannot yet confirm any metabolic activity of ammonia-oxidizing archaea or bacteria (AOA or AOB) within the LH Spring sediments. We have not been able to detect ammonia oxidation in flask enrichments of LH outlet and channel sediments, either, but we have cloned thaumarchaeal amoA genes from the LH Spring channel sediments (unpublished data). Thus, the occurrence of in situ ammonia oxidation at LH has not yet been experimentally confirmed.
Fig 1.
Phylogenetic profiles for key enzymes in nitrogen cycling (A) and sulfur reduction and oxidation (B). Color-coded bars indicate the percentages of abundance of different genera for each category of enzymes. Each gene or enzyme designation is preceded by the enzyme nomenclature designation and followed by the number of reads annotated from the metagenome (in parentheses). The filled and open arrows indicate the presence and absence of enzymes, respectively. The dashed line shown in Sox pathways indicates that the steps are more complicated than shown.
Sulfur metabolism.
A complete sulfur cycle through reduction and oxidation between sulfur end members and intermediates was identified in the LH metagenome over a high diversity of taxa (Fig. 1B; see also Table S3 in the supplemental material). The enzymes driving sulfate reduction, including sulfate adenylyltransferase (EC 2.7.7.4) (222 hits), adenylyl-sulfate kinase (EC 2.7.1.25) (172 hits), phosphoadenylyl-sulfate reductase (EC 1.8.4.8) (41 hits), and sulfite reductase (EC 1.8.7.1) (112 hits), were all detected; however, pathways were not completely reconstructed for all of the species identified (Fig. 1B). A large number of sulfur oxidation genes (soxB, soxD, soxH, and soxR genes) were recovered (302 hits), with dominant taxa including Thiomicrospira (20% of all sox reads), Thiobacillus, Nitrosococcus, and Roseiflexus. The enzymes for both assimilatory (EC 1.8.99.1) and dissimilatory (EC 1.8.99.3) sulfate reduction were also found, with assimilatory pathways appearing more abundant. The role of anoxygenic photosynthetic green and purple sulfur bacteria was prominent, with an abundance of hits to Chlorobium spp., typical anoxygenic phototrophic sulfide oxidizers, Roseiflexus spp., filamentous low-concentration sulfide oxidizers, and Chloroflexus spp., species containing metabolic features of both purple and green sulfur bacteria, all of which have been found in spring, hypersaline, or sulfur-rich ecosystems (61, 62). Several potential metabolic linkages between the LH nitrogen and sulfur cycles are plausible, with an abundance of hits to species including Thiobacillus denitrificans (54 hits) and Alkalilimnicola (9 hits), and Nitrosococcus (7 hits) species. Thiobacillus denitrificans couples the oxidation of inorganic sulfur compounds to the reduction of oxidized nitrogen compounds (such as nitrate and nitrite) to dinitrogen (63). An Alkalilimnicola sp., an anaerobic, facultatively autotrophic arsenite-oxidizing bacterium, respires nitrate or nitrite or, alternatively, uses sulfide or thiosulfate as the electron donor. Nitrosococcus is a genus of ammonium-oxidizing purple sulfur bacteria (see Table S3 in the supplemental material) that oxidize ammonia to nitrite and reduce sulfate to sulfide (64). The detection of these genes and species involved in both nitrogen and sulfur cycles provides evidence that these two cycles may be synergistically linked by similar species in the LH system.
Stress response.
The presence of stress response-related gene fragments (5,690 hits), all of which were associated with bacterial taxa, likely reflected the potential of LH bacteria to deal with or adapt to stressors in this hypersaline and subzero habitat. Given the stable LH Spring-sediment environment, these genes may be adaptive rather than typical “stress response” genes (i.e., genes responding to cold or heat shock). For example, many cold shock proteins can also be characterized as cold acclimation proteins, i.e., proteins that are present at relatively high levels during growth at constant cold temperatures (65).
The three most abundant groups corresponded to oxidative stress (2,255 hits), heat shock (1,491 hits), and osmotic stress (987 hits) response genes, all possibly linked to natural stressors at the LH Spring. LH outlet sediments are highly reducing and microoxic but are periodically exposed to the air during the summer months. The genes related to oxidative stress in the LH metagenome were associated mainly with Bacteroidetes, Proteobacteria, and Cyanobacteria (see Table S4 in the supplemental material). Anti–oxidative-stress genes may also help in responding to sudden changes in oxygen concentrations under predominantly anoxic conditions (66–69). Many of the genera identified as encoding these enzymes were aerobes, indicating that they may be dormant, or periodically active, in the LH Spring. The presence of genes related to anti–oxidative-stress functions may also be attributed to salinity-induced antioxidant defense responses, since high salinity may also provoke the formation of reactive oxygen species (ROS) as by-products of energy metabolism processes, including photosynthesis (70). For example, the expression of antioxidant enzymes in response to salinity has been observed among Cyanobacteria in Nostoc and Synechocystis species (70, 71), and ROS genes related to these two genera were indeed present in the LH metagenome. Lastly, antioxidant defense against ROS is a requirement for the growth of some species at low temperatures due to a decrease in the requirement for ATP, which results in electron accumulation in the respiratory chain and therefore in an increase in ROS at cold temperatures (72).
Most hits related to osmotic stress involved compatible-solute adaptations, typical in halophilic bacteria. Most hits (767 out of 978 hits) were related to the synthesis of the osmoregulated periplasmic glucan; the gene related to this synthesis may respond to sudden changes in salinity (73), and 131 hits were affiliated with choline and betaine biosynthesis (see Table S5 in the supplemental material). Two enzymes involved in betaine synthesis, choline dehydrogenase (from Cyanobacteria, Actinobacteria, and Gammaproteobacteria) and betaine-aldehyde dehydrogenase (from fungi and Gammaproteobacteria), were detected, suggesting that betaine might be the main osmolyte used by the LH microbial community. Betaine is a well-known osmolyte that plays an important role in balancing the high osmotic pressure exerted on microbial cells in hypersaline environments (74). Other typical adaptive responses to osmotic stress, such as sodium transporters, which are usually used by halophilic archaea to balance the osmotic pressure inside and outside the cells, were not detected.
The heat shock protein genes present in the metagenomic data set probably do not reflect heat shock responses in the permanently cold LH Spring, since these genes were more related to the general chaperone protein DnaK (640 hits) and its interacting protein, DnaJ (31 hits); these proteins are prevalent in microorganisms in cold environments and assist with protein folding (75, 76).
Microorganisms that sustain metabolic processes at cold temperatures produce cold acclimation and cold shock proteins involved in DNA replication (GyrA, RecA, and DnaA) (77–79), transcription (NusA), RNA unwinding (cold shock DEAD box protein A [CsdA] and cold shock protein A [CspA]) (80–84), protein folding (prolyl isomerase) (79, 85–87), pyruvate metabolism (AceE and AceF) (88, 89), and unsaturated fatty acid metabolism (fatty acid desaturases; DnaJ) (1, 90–92). Genes encoding such proteins were identified as containing cold adaptation features within the LH metagenome and originated mostly from Proteobacteria, Bacteroidetes, and Cyanobacteria (see Table S6 in the supplemental material). These genes were also detected in these three phyla in both Antarctic and Arctic metagenomic samples from the McMurdo Ice Shelf, the Ward Hunt Ice Shelf, and the Markham Ice Shelf (1). The ubiquity of these genes among similar taxa in other polar habitat metagenomes strongly suggests that such adaptive genetic systems are a global feature in cryoenvironments.
Comparison with other metagenomes.
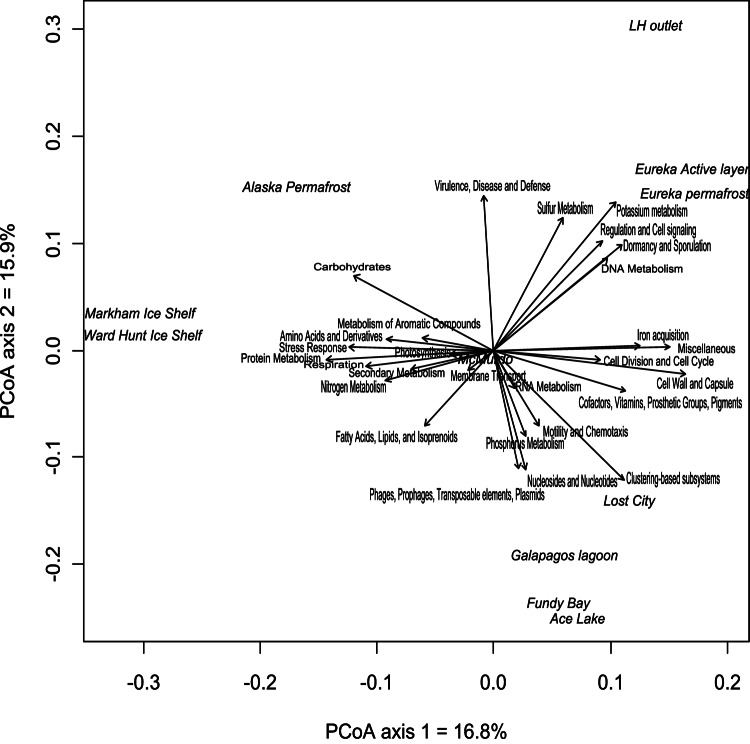
Based on the relative abundance of functional genes in different MG-RAST subsystems, we created an ordination of the LH metagenome together with other metagenomes of cold and salty environments. The ordination produced by PCoA (Fig. 2) shows the similarity between samples (the closer the samples, the more functionally similar they are). The arrows point toward the samples that have the highest relative abundance of a particular subsystem. The PCoA revealed that the LH metagenome clustered with the metagenomes from permafrost and active-layer samples from the Canadian high Arctic and was clearly distinct from the other samples. These patterns can perhaps be explained by higher relative abundances of genes related to dormancy and sporulation, reflecting possible adaptations to the extreme conditions of the LH Spring and Arctic permafrost environments at Eureka (93, 94), which are from the same geological region, about 70 to 80 km away from each other. A total of 82,711 genetic hits (Cyanobacteria [66.3%], Bacteroidetes [30.0%], Proteobacteria [3.5%], Firmicutes [0.2%]) related to dormancy and sporulation were present in the LH metagenome. In addition, the LH Spring water flows through permafrost before reaching the surface, which would result in the transfer of permafrost microbial communities to the LH Spring outlet sediments. Although the Markham Ice Shelf and the Ward Hunt Ice Shelf are also located in the same geographical region, the biomat metagenomes from these two ice shelves were not closely related to the LH metagenome, probably due to the difference in habitat conditions between the ice shelf mat habitats (freshwater, oxic) and the LH Spring system. The metagenome originating from the biofilm of the deep-sea hydrothermal field Lost City was also associated relatively closely with the LH metagenome, perhaps reflecting similarities with respect to methane and oxygen concentrations, as well as sulfur metabolism (4, 30, 95).
Fig 2.
Functional community composition of the LH Spring sediment and other extremely cold or saline environments, based on principal coordinate analysis of the relative abundances of all MG-RAST subsystems. Comparative sites include the Markham and Ward Hunt Ice Shelves (Arctic) and the McMurdo Ice Shelf (Antarctica), Alaskan and Eurekan (Canadian Arctic) permafrost, the Lost City hydrothermal system, a hypersaline lagoon in the Galapagos Islands, the Bay of Fundy, and Ace Lake (Antarctica).
Active profiling of the LH microbial community based on pyrosequencing of the cDNA of the 16S rRNA genes.
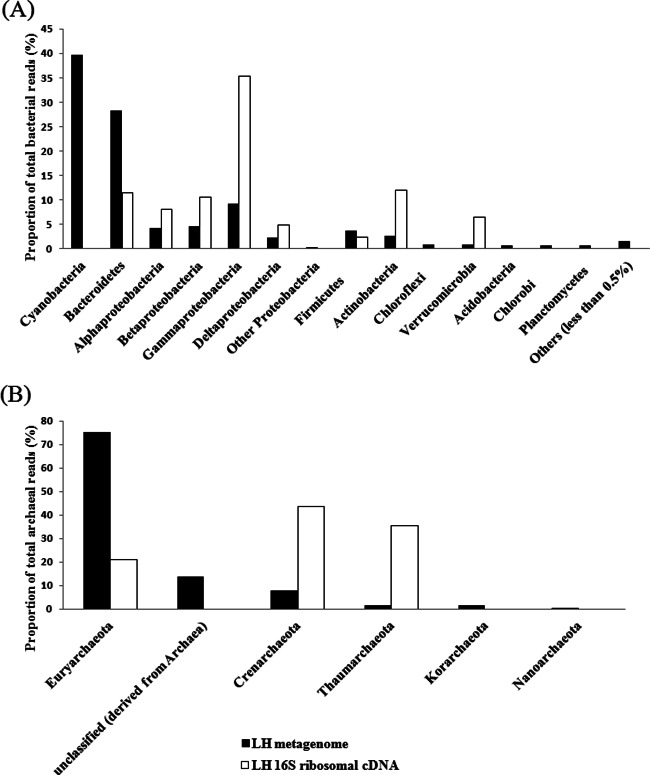
In an attempt to reveal which microorganisms are active at the LH Spring, we pyrosequenced the reverse-transcribed bacterial and archaeal 16S rRNA genes from extracted sediment RNA and compared the cDNA profile obtained with the LH metagenomic data set. Comparison of the two data sets revealed similar trends for bacterial and archaeal abundances at the phylum and class levels (Fig. 3; see also Fig. S4 and S5 in the supplemental material). With regard to bacterial abundance, Bacteroidetes, Proteobacteria, Firmicutes, Actinobacteria, and Verrucomicrobia were the most abundant phylotypes detected in the cDNA data set (Fig. 3A; see also Fig. S4). Whereas the Gammaproteobacteria, Actinobacteria, and Verrucomicrobia were overrepresented by a >50% increase in relative abundance in the cDNA data set compared to the metagenome, the Bacteroidetes and Firmicutes showed a relative decrease (Fig. 3A). The Gammaproteobacteria consisted of four genera (Marinobacter, Stenotrophomonas, Pseudomonas, and Enterobacter) in the cDNA library (see Table S7 in the supplemental material). The reads belonging to the same orders as these four genera (i.e., the orders Alteromonadales, Pseudomonadales, Xanthomonadales, and Enterobacteriales) made up 60.0% of the total gammaproteobacterial reads in the LH metagenome. The high proportion of gammaproteobacterial reads present in the cDNA library, and the taxonomic similarities between those reads and those present in the metagenome, suggest that the LH Gammaproteobacteria may be adapted to the hypersaline and cold conditions of the site and, as such, active in situ. It should be pointed out that Marinobacter and Pseudomonas species have also been detected and isolated at other perennial cold saline springs on AHI (4, 7, 8).
Fig 3.
Proportions of different clades of bacterial (A) and archaeal (B) reads from the LH metagenome and cDNA libraries. The classification was based on phyla, except for the Proteobacteria, which are shown at the class level. The total reads of bacteria were 662,411 and 1,092 in the LH metagenome and cDNA library, respectively. The total reads of archaea were 943 and 8,604 in the LH metagenome and cDNA library, respectively.
A striking difference between the two data sets, however, was that no Cyanobacteria reads were detected by cDNA pyrosequencing, while this was the most abundant phylum in the LH metagenome. This difference might have been caused by amplification biases, since the primer pair used for cDNA pyrosequencing matched only 61% of the Cyanobacteria sequences present in the RDP database used for annotation. Yet since this primer pair can amplify Synechococcus, Cyanothece, and Trichodesmium reads (which accounted for 41.3% of the total cyanobacterial reads in the LH metagenome), the results obtained in the cDNA library suggest that Cyanobacteria were probably dormant at the LH Spring, a possibility supported by the finding that up to 66.3% of the genes related to dormancy found in the LH metagenome were of cyanobacterial origin.
The data set for the cDNA of the bacterial 16S rRNA genes also contained microorganisms involved in nitrogen cycling. For example, the detection of Pseudomonas- and Stenotrophomonas-related sequences in the cDNA data set suggests that denitrification may be driven by these microorganisms (see Fig. S2 in the supplemental material). The presence of sequences associated with Desulfobulbus suggests that active sulfate reduction occurs within the LH outlet sediments (see Fig. S2 in the supplemental material).
Verrucomicrobia sequences were also detected in both data sets (Fig. 3; see also Fig. S2 in the supplemental material) and occupied an important/large proportion (16.8%) of the bacterial cDNA library (see Fig. S4 in the supplemental material). However, their potential function could not determined; no sequence of the verrucomicrobial pmoA gene was present in the LH metagenome, nor did the verrucomicrobial sequences from the cDNA library cluster with any known culture representatives (see Fig. S6 in the supplemental material). Therefore, the ecological status of the LH Verrucomicrobia is unknown.
With regard to archaeal abundance, the metagenome data set included all five known archaeal phyla, including Korarchaeota and Nanoarchaeota. On the other hand, the data set for the cDNA of the 16S rRNA genes comprised only sequences related to Euryarchaeota (20.1%), Crenarchaeota (42.4%), and Thaumarchaeota (37.5%) (Fig. 3B; see also Fig. S5 and Table S8 in the supplemental material). Although Euryarchaeota sequences were the most abundant ones in the archaeal metagenome data set, Crenarchaeota- and Thaumarchaeota-related sequences dominated the archaeal cDNA library, supporting some metagenomic interpretations of active S, N, and C biogeochemical cycles in situ. The detection of methanogenic archaea in both the metagenomic and cDNA archaeal data sets suggests that at least some of the methane exsolving from the LH outlet is biogenic (see Table S8 in the supplemental material).
The high proportion of Crenarchaoeta in the cDNA library was unexpected, considering that they usually represent hyperthermophilic archaea. These sequences had greater sequence identities to the thermophilic crenarchaea than to the “mesophilic crenarchaea,” which were reclassified as Thaumarchaeota after 2008 (96). Thus, the detection of these thermophilic crenarchaea at the LH Spring provides evidence of cold-adapted crenarchaea at LH. The crenarchaeal phylotypes recovered from the cDNA of the 16S rRNA genes were most closely related to Thermoprotei species (a Vulcanisaeta sp. [97% identity], a Thermoproteus sp. [98% identity], and a Pyrobaculum sp. [99% identity]) (see Table S8 and Fig. S5 in the supplemental material). All of these genera are anaerobic sulfur/sulfide-oxidizing microorganisms and may participate in sulfur cycling at LH (97–99). Among these, the Thermoproteus sp., a sulfide oxidizer, may also be involved in inorganic carbon fixation; the Pyrobaculum sp., a sulfur oxidizer and denitrifier, may utilize sulfur as an electron acceptor anaerobically (98, 99). In addition, it was surprising that no halophilic archaea were detected in the active-community profile, considering the hypersalinity in situ and the fact that they were detected previously (3, 4). This result perhaps reflects extraction and/or amplification biases. On the other hand, halophilic archaea are mostly aerobic microorganisms and might not be suitable for inhabiting the microoxic sediments of the LH outlet, indicating that the halophilic archaea were inactive in the LH sediments.
Conclusion.
Here we report the first metagenomic and cDNA pyrosequencing study of the unique hypersaline subzero Lost Hammer Spring. Metagenomic data combined with analyses of the cDNA of the 16S rRNA genes provides insight into the complex metabolic and functional genes present in this extreme hypersaline cryoenvironment, where active C, N, and S cycling appears to be functioning in situ and probably synergistically. The metagenomic data set also contains a large diversity of genes belonging to taxa not known to inhabit cryoenvironments and may reflect a large pool of dormant microorganisms or novel cold-adapted representatives of these taxa, such as the Crenarchaeota, capable of surviving in halophilic cryoenvironments. An abundance of active aerobic taxa also provides evidence that seasonal or microoxic environments may support the activity of allochthonous bacteria and archaea that are deposited in the anoxic LH Spring sediments. Genes involved in oxidative and osmotic stress, present in greater abundance than other stress-related genes, probably represent adaptations to the multiple stressors intrinsic to the subzero temperatures and high salinity in this habitat. Overall, investigations of the hypersaline subzero LH Spring system expand our knowledge of microbial life in extreme cryoenvironments on earth (3, 4) and provide evidence of how microbial life could inhabit the subzero briny environments thought to exist on Mars (21) and Enceladus (25).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Canadian Astrobiology Training Program (CATP), the National Sciences and Engineering Research Council of Canada (NSERC), the Canadian Space Agency (CSA), the Fonds de Recherche du Québec—Nature et Technologies (FQRNT), the Canada Foundation of Innovation (CFI), the Polar Continent Shelf Program (PCSP), and the Northern Scientific Training Program (NSTP).
The Alaskan permafrost sequence data were produced by the U.S. Department of Energy Joint Genome Institute (http://www.jgi.doe.gov/) in collaboration with the user community.
Footnotes
Published ahead of print 5 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00153-13.
REFERENCES
- 1. Varin T, Lovejoy C, Jungblut AD, Vincent WF, Corbeil J. 2012. Metagenomic analysis of stress genes in microbial mat communities from Antarctica and the high Arctic. Appl. Environ. Microbiol. 78:549–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Steven B, Pollard WH, Greer CW, Whyte LG. 2008. Microbial diversity and activity through a permafrost/ground ice core profile from the Canadian high Arctic. Environ. Microbiol. 10:3388–3403 [DOI] [PubMed] [Google Scholar]
- 3. Lay CY, Mykytczuk NC, Niederberger TD, Martineau C, Greer CW, Whyte LG. 2012. Microbial diversity and activity in hypersaline high Arctic spring channels. Extremophiles 16:177–191 [DOI] [PubMed] [Google Scholar]
- 4. Niederberger TD, Perreault NN, Tille S, Lollar BS, Lacrampe-Couloume G, Andersen D, Greer CW, Pollard W, Whyte LG. 2010. Microbial characterization of a subzero, hypersaline methane seep in the Canadian high Arctic. ISME J. 4:1326–1339 [DOI] [PubMed] [Google Scholar]
- 5. Cary SC, McDonald IR, Barrett JE, Cowan DA. 2010. On the rocks: the microbiology of Antarctic Dry Valley soils. Nat. Rev. Microbiol. 8:129–138 [DOI] [PubMed] [Google Scholar]
- 6. Bowman JS, Rasmussen S, Blom N, Deming JW, Rysgaard S, Sicheritz-Ponten T. 2012. Microbial community structure of Arctic multiyear sea ice and surface seawater by 454 sequencing of the 16S RNA gene. ISME J. 6:11–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Perreault NN, Andersen DT, Pollard WH, Greer CW, Whyte LG. 2007. Characterization of the prokaryotic diversity in cold saline perennial springs of the Canadian high Arctic. Appl. Environ. Microbiol. 73:1532–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Perreault NN, Greer CW, Andersen DT, Tille S, Lacrampe-Couloume G, Lollar BS, Whyte LG. 2008. Heterotrophic and autotrophic microbial populations in cold perennial springs of the high Arctic. Appl. Environ. Microbiol. 74:6898–6907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pollard W, Haltigin T, Whyte L, Niederberger T, Andersen D, Omelon C, Nadeau J, Ecclestone M, Lebeuf M. 2009. Overview of analogue science activities at the McGill Arctic Research Station, Axel Heiberg Island, Canadian High Arctic. Planet Space Sci. 57:646–659 [Google Scholar]
- 10. Andersen DT, Pollard WH, McKay CP, Heldmann J. 2002. Cold springs in permafrost on Earth and Mars. J. Geophys Res. Planets 107(E3):4-1-4-7 [Google Scholar]
- 11. Pollard W, Omelon C, Andersen D, McKay C. 1999. Perennial spring occurrence in the Expedition Fiord area of western Axel Heiberg Island, Canadian High Arctic. Can. J. Earth Sci. 36:105–120 [Google Scholar]
- 12. Grasby SE, Allen CC, Longazo TG, Lisle JT, Griffin DW, Beauchamp B. 2003. Supraglacial sulfur springs and associated biological activity in the Canadian high Arctic—signs of life beneath the ice. Astrobiology 3:583–596 [DOI] [PubMed] [Google Scholar]
- 13. Reigstad LJ, Jorgensen SL, Lauritzen SE, Schleper C, Urich T. 2011. Sulfur-oxidizing chemolithotrophic Proteobacteria dominate the microbiota in high Arctic thermal springs on Svalbard. Astrobiology 11:665–678 [DOI] [PubMed] [Google Scholar]
- 14. Gleeson DF, Williamson C, Grasby SE, Pappalardo RT, Spear JR, Templeton AS. 2011. Low temperature S-0 biomineralization at a supraglacial spring system in the Canadian High Arctic. Geobiology 9:360–375 [DOI] [PubMed] [Google Scholar]
- 15. Steven B, Niederberger TD, Bottos EM, Dyen MR, Whyte LG. 2007. Development of a sensitive radiorespiration method for detecting microbial activity at subzero temperatures. J. Microbiol. Methods 71:275–280 [DOI] [PubMed] [Google Scholar]
- 16. Gendrin A, Mangold N, Bibring JP, Langevin Y, Gondet B, Poulet F, Bonello G, Quantin C, Mustard J, Arvidson R, LeMouelic S. 2005. Sulfates in Martian layered terrains: the OMEGA/Mars Express view. Science 307:1587–1591 [DOI] [PubMed] [Google Scholar]
- 17. Hecht MH, Kounaves SP, Quinn RC, West SJ, Young SMM, Ming DW, Catling DC, Clark BC, Boynton WV, Hoffman J, DeFlores LP, Gospodinova K, Kapit J, Smith PH. 2009. Detection of perchlorate and the soluble chemistry of Martian soil at the Phoenix lander site. Science 325:64–67 [DOI] [PubMed] [Google Scholar]
- 18. Allen CC, Oehler DZ. 2008. A case for ancient springs in Arabia Terra, Mars. Astrobiology 8:1093–1112 [DOI] [PubMed] [Google Scholar]
- 19. Rossi AP, Neukum G, Pondrelli M, van Gasselt S, Zegers T, Hauber E, Chicarro A, Foing B. 22 August 2008. Large-scale spring deposits on Mars? J. Geophys. Res. Planets 113:E08016 doi: 10.1029/2007JE003062 [DOI] [Google Scholar]
- 20. Malin MC, Edgett KS, Posiolova LV, McColley SM, Dobrea EZ. 2006. Present-day impact cratering rate and contemporary gully activity on Mars. Science 314:1573–1577 [DOI] [PubMed] [Google Scholar]
- 21. McEwen AS, Ojha L, Dundas CM, Mattson SS, Byrne S, Wray JJ, Cull SC, Murchie SL, Thomas N, Gulick VC. 2011. Seasonal flows on warm Martian slopes. Science 333:740–743 [DOI] [PubMed] [Google Scholar]
- 22. Keppler F, Vigano I, McLeod A, Ott U, Fruchtl M, Rockmann T. 2012. Ultraviolet-radiation-induced methane emissions from meteorites and the Martian atmosphere. Nature 486:93–96 [DOI] [PubMed] [Google Scholar]
- 23. Mumma MJ, Villanueva GL, Novak RE, Hewagama T, Bonev BP, Disanti MA, Mandell AM, Smith MD. 2009. Strong release of methane on Mars in northern summer 2003. Science 323:1041–1045 [DOI] [PubMed] [Google Scholar]
- 24. Zahnle K, Freedman RS, Catling DC. 2011. Is there methane on Mars? Icarus 212:493–503 [Google Scholar]
- 25. Postberg F, Schmidt J, Hillier J, Kempf S, Srama R. 2011. A salt-water reservoir as the source of a compositionally stratified plume on Enceladus. Nature 474:620–622 [DOI] [PubMed] [Google Scholar]
- 26. Yergeau E, Hogues H, Whyte LG, Greer CW. 2010. The functional potential of high Arctic permafrost revealed by metagenomic sequencing, qPCR and microarray analyses. ISME J. 4:1206–1214 [DOI] [PubMed] [Google Scholar]
- 27. Mackelprang R, Waldrop MP, Deangelis KM, David MM, Chavarria KL, Blazewicz SJ, Rubin EM, Jansson JK. 2011. Metagenomic analysis of a permafrost microbial community reveals a rapid response to thaw. Nature 480:368–371 [DOI] [PubMed] [Google Scholar]
- 28. Brazelton WJ, Baross JA. 2010. Metagenomic comparison of two Thiomicrospira lineages inhabiting contrasting deep-sea hydrothermal environments. PLoS One 5:e13530 doi: 10.1371/journal.pone.0013530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xie W, Wang F, Guo L, Chen Z, Sievert SM, Meng J, Huang G, Li Y, Yan Q, Wu S, Wang X, Chen S, He G, Xiao X, Xu A. 2011. Comparative metagenomics of microbial communities inhabiting deep-sea hydrothermal vent chimneys with contrasting chemistries. ISME J. 5:414–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brazelton WJ, Baross JA. 2009. Abundant transposases encoded by the metagenome of a hydrothermal chimney biofilm. ISME J. 3:1420–1424 [DOI] [PubMed] [Google Scholar]
- 31. Lauro FM, DeMaere MZ, Yau S, Brown MV, Ng C, Wilkins D, Raftery MJ, Gibson JAE, Andrews-Pfannkoch C, Lewis M, Hoffman JM, Thomas T, Cavicchioli R. 2011. An integrative study of a meromictic lake ecosystem in Antarctica. ISME J. 5:879–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ng C, DeMaere MZ, Williams TJ, Lauro FM, Raftery MJ, Gibson JAE, Andrews-Pfannkoch C, Lewis M, Hoffman JM, Thomas T, Cavicchioli R. 2010. Metaproteogenomic analysis of a dominant green sulfur bacterium from Ace Lake, Antarctica. ISME J. 4:1002–1019 [DOI] [PubMed] [Google Scholar]
- 33. Rodriguez-Blanco A, Ghiglione JF, Catala P, Casamayor EO, Lebaron P. 2009. Spatial comparison of total vs. active bacterial populations by coupling genetic fingerprinting and clone library analyses in the NW Mediterranean Sea. FEMS Microbiol. Ecol. 67:30–42 [DOI] [PubMed] [Google Scholar]
- 34. Burow LC, Woebken D, Bebout BM, McMurdie PJ, Singer SW, Pett-Ridge J, Prufert-Bebout L, Spormann AM, Weber PK, Hoehler TM. 2012. Hydrogen production in photosynthetic microbial mats in the Elkhorn Slough estuary, Monterey Bay. ISME J. 6:863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jones SE, Lennon JT. 2010. Dormancy contributes to the maintenance of microbial diversity. Proc. Natl. Acad. Sci. U. S. A. 107:5881–5886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Murray AE, Kenig F, Fritsen CH, McKay CP, Cawley KM, Edwards R, Kuhn E, McKnight DM, Ostrom NE, Peng V, Ponce A, Priscu JC, Samarkin V, Townsend AT, Wagh P, Young SA, Yung PT, Doran PT. 2012. Microbial life at −13°C in the brine of an ice-sealed Antarctic lake. Proc. Natl. Acad. Sci. U. S. A. 109:20626–20631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, Wilkening J, Edwards RA. 2008. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9:386 doi: 10.1186/1471-2105-9-386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kent WJ. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12:656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC. 2011. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 21:1552–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Handl S, Dowd SE, Garcia-Mazcorro JF, Steiner JM, Suchodolski JS. 2011. Massive parallel 16S rRNA gene pyrosequencing reveals highly diverse fecal bacterial and fungal communities in healthy dogs and cats. FEMS Microbiol. Ecol. 76:301–310 [DOI] [PubMed] [Google Scholar]
- 41. Baker GC, Smith JJ, Cowan DA. 2003. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 55:541–555 [DOI] [PubMed] [Google Scholar]
- 42. Burggraf S, Huber H, Stetter KO. 1997. Reclassification of the crenarchaeal orders and families in accordance with 16S rRNA sequence data. Int. J. Syst. Bacteriol. 47:657–660 [DOI] [PubMed] [Google Scholar]
- 43. Bailey MT, Dowd SE, Parry NMA, Galley JD, Schauer DB, Lyte M. 2010. Stressor exposure disrupts commensal microbial populations in the intestines and leads to increased colonization by Citrobacter rodentium. Infect. Immun. 78:1509–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37(Database issue):D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Keegan KP, Trimble WL, Wilkening J, Wilke A, Harrison T, D'Souza M, Meyer F. 2012. A platform-independent method for detecting errors in metagenomic sequencing data: DRISEE. PLoS Comput. Biol. 8:e1002541 doi: 10.1371/journal.pcbi.1002541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Walter-Anthony KM, Anthony P, Grosse G, Chanton J. 2012. Geologic methane seeps along boundaries of Arctic permafrost thaw and melting glaciers. Nat. Geosci. 5:419–426 [Google Scholar]
- 49. Rivkina E, Laurinavichius K, McGrath J, Tiedje J, Shcherbakova V, Gilichinsky D. 2004. Microbial life in permafrost. Adv. Space Res. 33:1215–1221 [DOI] [PubMed] [Google Scholar]
- 50. Oren A. 2011. Thermodynamic limits to microbial life at high salt concentrations. Environ. Microbiol. 13:1908–1923 [DOI] [PubMed] [Google Scholar]
- 51. Tazaz AM, Bebout BM, Kelley CA, Poole J, Chanton JP. 15 June 2012. Redefining the isotopic boundaries of biogenic methane: methane from endoevaporites. Icarus. doi: 10.1016/j.icarus.2012.06.008 [DOI] [Google Scholar]
- 52. Meyerdierks A, Kube M, Kostadinov I, Teeling H, Glockner FO, Reinhardt R, Amann R. 2010. Metagenome and mRNA expression analyses of anaerobic methanotrophic archaea of the ANME-1 group. Environ. Microbiol. 12:422–439 [DOI] [PubMed] [Google Scholar]
- 53. Teske A, Biddle JF. 2008. Analysis of deep subsurface microbial communities by functional genes and genomics, p 159–176 In Dilek Y, Furnes H, Muehlenbachs K. (ed), Modern approaches in solid earth sciences, vol 4 Links between geological processes, microbial activities & evolution of life. Springer, Berlin, Germany [Google Scholar]
- 54. Steward GF, Zehr JP, Jellison R, Montoya JP, Hollibaugh JT. 2004. Vertical distribution of nitrogen-fixing phylotypes in a meromictic, hypersaline lake. Microb. Ecol. 47:30–40 [DOI] [PubMed] [Google Scholar]
- 55. Ferrer M, Guazzaroni ME, Richter M, Garcia-Salamanca A, Yarza P, Suarez-Suarez A, Solano J, Alcaide M, van Dillewijn P, Molina-Henares MA, Lopez-Cortes N, Al-Ramahi Y, Guerrero C, Acosta A, de Eugenio LI, Martinez V, Marques S, Rojo F, Santero E, Genilloud O, Perez-Perez J, Rossello-Mora R, Ramos JL. 2011. Taxonomic and functional metagenomic profiling of the microbial community in the anoxic sediment of a sub-saline shallow lake (Laguna de Carrizo, Central Spain). Microb. Ecol. 62:824–837 [DOI] [PubMed] [Google Scholar]
- 56. Auclair J, Parent S, Villemur R. 2012. Functional diversity in the denitrifying biofilm of the methanol-fed marine denitrification system at the Montreal Biodome. Microb. Ecol. 63:726–735 [DOI] [PubMed] [Google Scholar]
- 57. Qu JH, Yuan HL, Li HF, Deng CP. 2009. Flavobacterium cauense sp. nov., isolated from sediment of a eutrophic lake. Int. J. Syst. Evol. Microbiol. 59:2666–2669 [DOI] [PubMed] [Google Scholar]
- 58. Vernotte C, Picaud M, Kirilovsky D, Olive J, Ajlani G, Astier C. 1992. Changes in the photosynthetic apparatus in the cyanobacterium Synechocystis sp. PCC 6714 following light-to-dark and dark-to-light transitions. Photosynth. Res. 32:42–57 [DOI] [PubMed] [Google Scholar]
- 59. Reddy KJ, Haskell JB, Sherman DM, Sherman LA. 1993. Unicellular, aerobic nitrogen-fixing Cyanobacteria of the genus Cyanothece. J. Bacteriol. 175:1284–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Venterea RT. 2007. Nitrite-driven nitrous oxide production under aerobic soil conditions: kinetics and biochemical controls. Global Change Biol. 13:1798–1809 [Google Scholar]
- 61. Nubel U, Bateson MM, Madigan MT, Kuhl M, Ward DM. 2001. Diversity and distribution in hypersaline microbial mats of bacteria related to Chloroflexus spp. Appl. Environ. Microbiol. 67:4365–4371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kompantseva EI, Sorokin DY, Gorlenko VM, Namsaraev BB. 2005. The phototrophic community found in Lake Khilganta (an alkaline saline lake located in the southeastern Transbaikal region). Microbiology 74:352–361 [PubMed] [Google Scholar]
- 63. Kelly DP, Shergill JK, Lu WP, Wood AP. 1997. Oxidative metabolism of inorganic sulfur compounds by bacteria. Antonie Van Leeuwenhoek 71:95–107 [DOI] [PubMed] [Google Scholar]
- 64. Klotz MG, Arp DJ, Chain PSG, El-Sheikh AF, Hauser LJ, Hommes NG, Larimer FW, Malfatti SA, Norton JM, Poret-Peterson AT, Vergez LM, Ward BB. 2006. Complete genome sequence of the marine, chemolithoautotrophic, ammonia-oxidizing bacterium Nitrosococcus oceani ATCC 19707. Appl. Environ. Microbiol. 72:6299–6315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Feller G, Gerday C. 2003. Psychrophilic enzymes: hot topics in cold adaptation. Nat. Rev. Microbiol. 1:200–208 [DOI] [PubMed] [Google Scholar]
- 66. Kawakami R, Sakuraba H, Kamohara S, Goda S, Kawarabayasi Y, Ohshima T. 2004. Oxidative stress response in an anaerobic hyperthermophilic archaeon: presence of a functional peroxiredoxin in Pyrococcus horikoshii. J. Biochem. 136:541–547 [DOI] [PubMed] [Google Scholar]
- 67. Jean D, Briolat V, Reysset G. 2004. Oxidative stress response in Clostridium perfringens. Microbiology 150:1649–1659 [DOI] [PubMed] [Google Scholar]
- 68. Briolat V, Reysset G. 2002. Identification of the Clostridium perfringens genes involved in the adaptive response to oxidative stress. J. Bacteriol. 184:2333–2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rocha ER, Selby T, Coleman JP, Smith CJ. 1996. Oxidative stress response in an anaerobe, Bacteroides fragilis: a role for catalase in protection against hydrogen peroxide. J. Bacteriol. 178:6895–6903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Srivastava AK. 2010. Assessment of salinity-induced antioxidative defense system of diazotrophic cyanobacterium Nostoc muscorum. J. Microbiol. Biotechnol. 20:1506–1512 [DOI] [PubMed] [Google Scholar]
- 71. Latifi A, Ruiz M, Zhang CC. 2009. Oxidative stress in Cyanobacteria. FEMS Microbiol. Rev. 33:258–278 [DOI] [PubMed] [Google Scholar]
- 72. Chattopadhyay MK. 2002. Low temperature and oxidative stress. Curr. Sci. 83:109 (Letter.) [Google Scholar]
- 73. Bohin JP. 2000. Osmoregulated periplasmic glucans in Proteobacteria. FEMS Microbiol. Lett. 186:11–19 [DOI] [PubMed] [Google Scholar]
- 74. Sleator RD, Hill C. 2002. Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol. Rev. 26:49–71 [DOI] [PubMed] [Google Scholar]
- 75. Ting L, Williams TJ, Cowley MJ, Lauro FM, Guilhaus M, Raftery MJ, Cavicchioli R. 2010. Cold adaptation in the marine bacterium, Sphingopyxis alaskensis, assessed using quantitative proteomics. Environ. Microbiol. 12:2658–2676 [DOI] [PubMed] [Google Scholar]
- 76. Panoff J-M, Thammavongs B, Laplace J-M, Hartke A, Boutibonnes P, Auffray Y. 1995. Cryotolerance and cold adaptation in Lactococcus lactis subsp. lactis IL1403. Cryobiology 32:516–520 [Google Scholar]
- 77. Atlung T, Hansen FG. 1999. Low-temperature-induced DnaA protein synthesis does not change initiation mass in Escherichia coli K-12. J. Bacteriol. 181:5557–5562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Merrin J, Kumar P, Libchaber A. 2011. Effects of pressure and temperature on the binding of RecA protein to single-stranded DNA. Proc. Natl. Acad. Sci. U. S. A. 108:19913–19918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Yamanaka K. 1999. Cold shock response in Escherichia coli. J. Mol. Microbiol. Biotechnol. 1:193–202 [PubMed] [Google Scholar]
- 80. Bakermans C, Bergholz PW, Ayala-del-Río H, Tiedje J. 2009. Genomic insights into cold adaptation of permafrost bacteria. Soil Biol. 16:159–168 [Google Scholar]
- 81. Hunger K, Beckering CL, Wiegeshoff F, Graumann PL, Marahiel MA. 2006. Cold-induced putative DEAD box RNA helicases CshA and CshB are essential for cold adaptation and interact with cold shock protein B in Bacillus subtilis. J. Bacteriol. 188:240–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Py B, Higgins CF, Krisch HM, Carpousis AJ. 1996. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature 381:169–172 [DOI] [PubMed] [Google Scholar]
- 83. Ray S, Lee C, Hou TY, Bhakat KK, Brasier AR. 2010. Regulation of signal transducer and activator of transcription 3 enhanceosome formation by apurinic/apyrimidinic endonuclease 1 in hepatic acute phase response. Mol. Endocrinol. 24:391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Jones PG, Mitta M, Kim Y, Jiang WN, Inouye M. 1996. Cold shock induces a major ribosomal-associated protein that unwinds double-stranded RNA in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 93:76–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Cavicchioli R, Thomas T, Curmi PM. 2000. Cold stress response in Archaea. Extremophiles 4:321–331 [DOI] [PubMed] [Google Scholar]
- 86. de la Cruz J, Kressler D, Linder P. 1999. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem. Sci. 24:192–198 [DOI] [PubMed] [Google Scholar]
- 87. Suzuki Y, Haruki M, Takano K, Morikawa M, Kanaya S. 2004. Possible involvement of an FKBP family member protein from a psychrotrophic bacterium Shewanella sp. SIB1 in cold-adaptation. Eur. J. Biochem. 271:1372–1381 [DOI] [PubMed] [Google Scholar]
- 88. Scherer S, Neuhaus K. 2006. Life at low temperatures, p 210–262 In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E. (ed), The Prokaryotes, 3rd ed, vol 2 Ecophysiology and biochemistry. Springer Science, New York, NY [Google Scholar]
- 89. Wouters JA, Rombouts FM, Kuipers OP, de Vos WM, Abee T. 2001. The role of cold-shock proteins in low-temperature adaptation, p 43–56 In Storey KB, Storey JM. (ed), Protein adaptations and signal transduction. Elsevier Science, Amsterdam, Netherlands [Google Scholar]
- 90. Rodrigues DF, Tiedje JM. 2008. Coping with our cold planet. Appl. Environ. Microbiol. 74:1677–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Kenny JG, Ward D, Josefsson E, Jonsson IM, Hinds J, Rees HH, Lindsay JA, Tarkowski A, Horsburgh MJ. 2009. The Staphylococcus aureus response to unsaturated long chain free fatty acids: survival mechanisms and virulence implications. PLoS One 4:e4344 doi: 10.1371/journal.pone.0004344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Thieringer HA, Jones PG, Inouye M. 1998. Cold shock and adaptation. Bioessays 20:49–57 [DOI] [PubMed] [Google Scholar]
- 93. Nicholson WL, Munakata N, Horneck G, Melosh HJ, Setlow P. 2000. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 64:548–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Sachidanandham R, Yew-Hoong Gin K. 2009. A dormancy state in nonspore-forming bacteria. Appl. Microbiol. Biotechnol. 81:927–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Brazelton WJ, Schrenk MO, Kelley DS, Baross JA. 2006. Methane- and sulfur-metabolizing microbial communities dominate the Lost City hydrothermal field ecosystem. Appl. Environ. Microbiol. 72:6257–6270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. 2008. Mesophilic crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat. Rev. Microbiol. 6:245–252 [DOI] [PubMed] [Google Scholar]
- 97. Itoh T, Suzuki K, Nakase T. 2002. Vulcanisaeta distributa gen. nov., sp. nov., and Vulcanisaeta souniana sp. nov., novel hyperthermophilic, rod-shaped crenarchaeotes isolated from hot springs in Japan. Int. J. Syst. Evol. Microbiol. 52:1097–1104 [DOI] [PubMed] [Google Scholar]
- 98. Strauss G, Eisenreich W, Bacher A, Fuchs G. 1992. 13C-NMR study of autotrophic CO2 fixation pathways in the sulfur-reducing Archaebacterium Thermoproteus neutrophilus and in the phototrophic Eubacterium Chloroflexus aurantiacus. Eur. J. Biochem. 205:853–866 [DOI] [PubMed] [Google Scholar]
- 99. Selig M, Schönheit P. 1994. Oxidation of organic compounds to CO2 with sulfur or thiosulfate as electron acceptor in the anaerobic hyperthermophilic archaea Thermoproteus tenaxT and Pyrobaculum islandicum proceeds via the citric acid cycle. Arch. Microbiol. 162:286–294 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.