Abstract
Alkyl hydroperoxide reductase subunit C (AhpC) is the catalytic subunit responsible for the detoxification of reactive oxygen species that form in bacterial cells or are derived from the host; thus, AhpC facilitates the survival of pathogenic bacteria under environmental stresses or during infection. This study investigates the role of AhpC in the induction and maintenance of a viable but nonculturable (VBNC) state in Vibrio parahaemolyticus. In this investigation, ahpC1 (VPA1683) and ahpC2 (VP0580) were identified in chromosomes II and I of this pathogen, respectively. Mutants with deletions of these two ahpC genes and their complementary strains were constructed from the parent strain KX-V231. The growth of these strains was monitored on tryptic soy agar–3% NaCl in the presence of the extrinsic peroxides H2O2 and tert-butyl hydroperoxide (t-BOOH) at different incubation temperatures. The results revealed that both ahpC genes were protective against t-BOOH, while ahpC1 was protective against H2O2. The protective function of ahpC2 at 4°C was higher than that of ahpC1. The times required to induce the VBNC state (4.7 weeks) at 4°C in a modified Morita mineral salt solution with 0.5% NaCl and then to maintain the VBNC state (4.7 weeks) in an ahpC2 mutant and an ahpC1 ahpC2 double mutant were significantly shorter than those for the parent strain (for induction, 6.2 weeks; for maintenance, 7.8 weeks) and the ahpC1 mutant (for induction, 6.0 weeks; for maintenance, 8.0 weeks) (P < 0.03). Complementation with an ahpC2 gene reversed the effects of the ahpC2 mutation in shortening the times for induction and maintenance of the VBNC state. This investigation identified the different functions of the two ahpC genes and confirmed the particular role of ahpC2 in the VBNC state of V. parahaemolyticus.
INTRODUCTION
Vibrio parahaemolyticus is a halophilic Gram-negative bacterium that causes food-borne gastroenteritis worldwide and is prevalent in some Asian countries (1). This pathogen has acquired global significance since the occurrence of the first pandemic O3:K6 strains in 1996 (2). It inhabits brackish water, where many shellfish are found (3). As with other vibrios in the marine environment, a unique physiological state known as the viable but nonculturable (VBNC) state can be induced in V. parahaemolyticus by incubation at low temperatures in a mineral salt starvation medium (4–6). Cells in this state are viable but unable to form colonies on common agar medium; they can be resuscitated by a temperature upshift treatment (7).
The VBNC state may represent a state of dormancy that improves the survival of nonsporulating bacteria in an adverse environment (8). Drastic morphological and physiological changes have been demonstrated in the VBNC state of V. parahaemolyticus (6, 9). Virulence usually decreases markedly as cells enter the VBNC state (9); nevertheless, virulence genes and pathogenic potential are maintained (8, 10). However, the genetic control of the VBNC state in V. parahaemolyticus and in other bacteria has not been fully clarified.
The involvement of antioxidative factors in the VBNC state has been investigated previously (11–13). The addition of extrinsic catalase or of other antioxidative factors to the culture medium improves the culturability of VBNC cultures in Aeromonas hydrophila (13), Escherichia coli (12), and Vibrio vulnificus (11). In the VBNC suppression mutant of V. vulnificus, glutathione S-transferase has been detected; this is a cytosolic, mitochondrial, and microsomal protein that detoxifies endogenous compounds, such as peroxidized lipids (14). This enzyme is highly expressed by the VBNC-suppressing mutant (about 10-fold changes in 6 to 24 h) under stress due to low temperatures (14). These studies suggest that upon exposure to high nutrient levels, VBNC cells form reactive oxygen species (ROS) but are not able to detoxify these harmful radicals and thus fail to replicate and grow in such high-nutrient media (11, 15).
An oxyR mutant of V. vulnificus that lacks catalase activity has been constructed, and it is nonculturable on solid media (16). That study attributes the cold-induced loss of catalase activity to the induction of the VBNC state. However, OxyR regulates the expression both of catalases and of alkyl hydroperoxide reductase subunit C (AhpC) in several Gram-negative bacteria in response to elevated ROS levels (17, 18). AhpC proteins are the catalytic subunits of alkyl hydroperoxide reductases (19), which are members of a family of peroxidases, collectively called peroxiredoxins, or thiol peroxidases (TPx family), that can be divided into two subgroups depending on the number of conserved cysteines. These ubiquitous enzymes constitute an important component of defense against a wide range of substrates, including H2O2, organic peroxides, and peroxynitrite. They are involved both in the control of endogenous peroxides and in the inducible defense response to exogenous peroxides or general stresses (20). The roles of catalases and AhpC proteins in the VBNC state need to be differentiated.
A search through the genome of V. parahaemolyticus RIMD2210633 (21) revealed the presence of several putative AphC factors (VP0580, VPA1681, VPA1683, VPA1684, VPA1293). By use of BLAST analysis (http://blast.ncbi.nlm.nih.gov/) with homologous genes of Vibrio cholerae, V. vulnificus, and Salmonella enterica (22), the VPA1683 and VP0580 genes of V. parahaemolyticus have been designated ahpC1 and ahpC2, respectively. In our previous study, the induction of the VBNC state in V. parahaemolyticus by prolonged incubation at low temperatures in a nutrient-limited medium was characterized by proteomics, and the level of AhpC2 was increased about 10-fold as determined by 2-dimensional (2-D) gel electrophoresis (23).
On the basis of the functions of antioxidative factors in the VBNC state, mentioned above, we hypothesize that AhpC proteins play important roles in protection against the low-level oxidative stress generated in cells incubated at low temperatures in a starvation medium, consequently slowing down the loss of culturability during the induction of the VBNC state and maintaining the viability of cells that have entered the VBNC state. In this study, the specific roles of ahpC1 and ahpC2 in the induction and maintenance of the VBNC state in V. parahaemolyticus were investigated by means of deletion mutations and gene complementation, while the expression of genes encoding various antioxidative factors and their regulators (RpoS and OxyR) in these mutants and the wild-type strain was monitored.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
V. parahaemolyticus strain KX-V231 (Kanagawa phenomenon positive [KP+; exhibiting β-hemolysis on Wagatsuma blood agar]; serotype O3:K6; authors' lab strain no. 1173), isolated in Thailand from a clinical specimen, was used in this study (Table 1). It was stored frozen at −85°C in beads in Microbank cryovials (Pro-Lab Diagnostics, Austin, TX, USA). KX-V231 was cultured at 37°C on tryptic soy agar (Becton, Dickinson Diagnostic Systems, Sparks, MD, USA) supplemented with 3% sodium chloride (TSA–3% NaCl), or in tryptic soy broth with 3% NaCl (TSB–3% NaCl), in a 50-ml tube, which was shaken at 160 rpm. A 50-μl aliquot of the 16-h broth culture was inoculated into 10 ml of fresh TSB–3% NaCl and was incubated at 37°C with shaking for 2 h, to allow entry into the exponential phase (ca. 108 CFU/ml), and this culture was used as the inoculum in subsequent experiments. Escherichia coli was cultured in Luria broth (LB; Becton, Dickinson) at 37°C with shaking at 160 rpm. Chloramphenicol (final concentration, 6 μg/ml) or chloramphenicol (20 μg/ml)-ampicillin (50 μg/ml) was added to the medium as required for the cultivation of some of the V. parahaemolyticus or E. coli strains, respectively.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristics or sequence | Source or reference |
|---|---|---|
| Strains | ||
| V. parahaemolyticus | ||
| KX-V231 | Wild type; serotype O3:K6; KP+; clinical isolate | This study |
| ΔahpC1 strain | KX-V231 ΔahpC1 | This study |
| ΔahpC2 strain | KX-V231 ΔahpC2 | This study |
| ΔahpC12 strain | KX-V231 ΔahpC1 ΔahpC2 | This study |
| ΔahpC1/C1 strain | ΔahpC1 strain containing pSCB02 | This study |
| ΔahpC2/C2 strain | ΔahpC2 strain containing pSCB03 | This study |
| ΔahpC12/C1 strain | ΔahpC12 strain containing pSCB02 | This study |
| ΔahpC12/C2 strain | ΔahpC12 strain containing pSCB03 | This study |
| ΔahpC12V strain | ΔahpC12 strain containing pSCB01 | This study |
| KX-V231V | KX-V231 containing pSCB01 | This study |
| E. coli | ||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| SM10 λ-pir | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu λ pirR6K; Kmr | 50 |
| Plasmids | ||
| pGEM-T Easy | Cloning vector; Apr | Promega |
| pBR328 | Vector derived from pBR325 and pMB1; Apr Cmr Tcr | Roche |
| pDS132 | R6K ori mobRP4 sacB; Cmr | 51 |
| pSCA01 | pDS132 with ahpC1 deletion | This study |
| pSCA02 | pDS132 with ahpC2 deletion | This study |
| pSCB01 | Derived from pBR328 and pDS132; mobRP4; Apr Cmr Tcr | This study |
| pSCB02 | pSCB01 with Tc::ahpC1 promoter-ahpC1 | This study |
| pSCB03 | pSCB01 with Tc::ahpC2 promoter-ahpC2 | This study |
Construction of deletion mutants.
Mutants with deletions of the ahpC genes were constructed by following the published method with modifications (24). PCR-amplified DNA fragments that were used to construct the in-frame deletion mutation of ahpC1 were generated by means of overlap PCR as described previously (25). Two DNA fragments were amplified by PCR with V. parahaemolyticus KX-V231 chromosomal DNA as the template—one with primer pair ahpC1-1 and ahpC1-2 and the other with primer pair ahpC1-3 and ahpC1-4 (see Table S1 in the supplemental material). Phusion high-fidelity DNA polymerase (Finnzymes Oy, Vantaa, Finland) was used in this PCR. These two amplified fragments were then used as the templates for a second PCR with primers ahpC1-1 and ahpC1-4, resulting in the construction of a fragment with a deletion in the ahpC1 gene. This fragment containing the deletion was purified, reacted with Taq at 72°C for 30 min to add 3′ A overhangs to the blunt ends, cloned into the pGEM-T Easy vector, and transformed into E. coli XL1-Blue by following the manufacturer's protocol (Promega Co., Madison, WI, USA). The inserted sequence was verified by sequencing. This fragment was then removed from the pGEM-T Easy vector by digestion using SacI and SphI and was cloned into a suicide vector, pDS132, which contained a chloramphenicol resistance gene and the sacB gene, conferring sensitivity to sucrose. This plasmid (pDS132 with an ahpC1 deletion) was introduced into E. coli SM10 λ-pir, which was then mated with V. parahaemolyticus strain KX-V231. Thiosulfate-citrate-bile-sucrose (TCBS) agar containing chloramphenicol was used to screen the V. parahaemolyticus cells containing the inserted plasmid. The V. parahaemolyticus clones were isolated and cultured in LB (Becton, Dickinson) supplemented with 2% NaCl and chloramphenicol. DNA was extracted from these cultures, and the inserted sequence was detected by PCR using primers ahpC1-1 and ahpC1-4. The culture that contained the pDS132-ahpC1 deletion plasmid was incubated at 37°C for 3 h in LB containing 2% NaCl and was then plated onto an LB agar plate containing 2% NaCl and 10% sucrose. Isolated colonies that were unable to grow on an LB agar plate containing chloramphenicol were selected, and the homologous recombination of the deleted fragment was verified by PCR using primers ahpC1-0 and ahpC1-5. By following the same procedures with different primers, the ahpC2 deletion mutant was also constructed (Table 1). Amplification of the ahpC1 or ahpC2 gene by PCR with primers ahpc1-0 and ahpc1-5 or primers ahpc2-0 and ahpc2-5 yielded amplicons of 1,849 bp or 2,004 bp, respectively, while the in-frame deletion mutants formed amplicons of 1,336 bp or 1,494 bp, respectively. The mutated genes were also verified by nucleotide sequencing of the amplified fragments.
Sequencing service was provided by Genomics BioScience and Technology Co., Inc., Taipei, Taiwan, using Sanger's method with an Applied Biosystems 3730 analyzer.
Construction of complementary strains.
The entire lengths of the ahpC1 and ahpC2 genes were amplified by PCR with V. parahaemolyticus KX-V231 chromosomal DNA as the template using primer pairs (ahpC1-salI and ahpC1-sphI for ahpC1; ahpC2-salI and ahpC2-sphI for ahpC2) with restriction enzyme linkers (SalI, SphI) (see Table S1 in the supplemental material). The amplicons were digested with SalI and SphI and were ligated to the shuttle vector pSCB01, which had been digested with the same enzymes. The shuttle vector pSCB01 (8,123 bp) was constructed by ligating the mobRP4 fragment that was recovered from the HindIII-digested fragments of pDS132 to pBR328 that had been digested by HindIII (Table 1). Plasmids pSCB02 and pSCB03, containing the entire length of the aphC1 or ahpC2 gene, respectively, were propagated in E. coli SM10 λ-pir and were conjugated to the corresponding ahpC mutants to generate complementary strains, which were selected by their chloramphenicol resistance (Table 1). The presence of the entire lengths of the ahpC1 and ahpC2 genes in these strains was verified by PCR.
Effects of peroxides and incubation temperatures on the growth and survival of the ahpC mutants.
Cultures of different V. parahaemolyticus strains in the exponential phase, with an absorbance at 600 nm of 0.5 to 0.6, were diluted 10−1-fold to 10−5-fold (−1 to −5 log10) with TSB–3% NaCl. Ten-microliter aliquots of the diluted suspensions were spotted onto TSA–3% NaCl plates with or without the addition of 250 μM H2O2 or 80 μM tert-butyl hydroperoxide (t-BOOH). The plates were incubated at 25°C for 16 h, and the colony-forming abilities of the inocula were observed (26).
Aliquots (100 μl) of different strains (108 CFU/ml) were also spread onto TSA–3% NaCl plates and were incubated at 22, 30, or 37°C for 24, 15, or 15 h, respectively, and the diameters of colonies were measured. In another experiment, cultures in exponential phase in TSB–3% NaCl were incubated at 4°C, and the survivors were counted by the standard plate count method for 9 days.
Induction of the VBNC state and resuscitation of VBNC cells.
Different strains of V. parahaemolyticus were induced to enter the VBNC state by a previously described method (6, 23). Briefly, bacterial cells in the exponential phase were harvested, washed twice using a modified Morita mineral salt solution (MMS–0.5% NaCl), and resuspended in the same medium at a concentration of 107 cells/ml before being incubated statically at 4°C to induce the VBNC state.
Culturable cells of the cultures were counted at weekly intervals by the standard plate count method. When the cultures entered the VBNC state, they were resuscitated every week by a temperature upshift treatment at 25°C for 48 h prior to plate counting (6, 7). The levels of culturable cells (CFU/ml) were expressed in log10 values.
RT-qPCR.
The expression of antioxidative genes and related regulators was determined by real-time quantitative reverse transcription-PCR (RT-qPCR) (27). Briefly, cells were lysed with TRIzol reagent (Invitrogen, United Kingdom), and RNA samples were extracted by using an RNApure kit (Genesis Biotech Inc., Taipei, Taiwan) according to the manufacturer's instructions. RNA samples were treated with DNase I (TaKaRa Bio Inc., Shiga, Japan) and were then reverse transcribed using a SuperScript III First-Strand Synthesis SuperMix (Invitrogen, United Kingdom) according to the manufacturer's instructions. Primers (see Table S2 in the supplemental material) were designed using Primer Express software (Perkin-Elmer Applied Biosystems, Foster City, CA, USA), and 16S rRNA was used as the internal control. Real-time PCR was performed using the ABI Prism 7300 sequence detection system (Perkin-Elmer Applied Biosystems) with a SYBR green PCR Master Mix and RT-PCR reagents. All the data were normalized to the 16S gene expression levels of the culture at each time point, and the normalized values for each gene were compared. The expression of each gene was compared to its expression in the exponential phase, and the relative values were expressed as log10 fold (log fold) according to the manufacturer's instructions (Perkin-Elmer Applied Biosystems).
The quantity and quality of the RNA samples were assayed by determining the A260/A280 ratio. The integrity of the RNA samples, before and after DNase treatment, was assessed by 1% (wt/vol) agarose gel electrophoresis with ethidium bromide staining. The absence of genomic DNA contamination was verified by PCR amplification using total RNA as the template and primers that were designed for the amplification of a 500-bp DNA fragment (forward primer [F], 5′-CAGGCCTAACACATGCAAGTC; reverse primer, 5′-ATTACCGCGGCTGCTGG) from the 16S rRNA gene and a 72-bp DNA fragment (F, 5′-AGCATCAACCTATTCGTGTT; R, 5′-TAATTCGTAGCGATTGTCTG) from the VP2468 (l,d-carboxypeptidase) gene (see Fig. S1 in the supplemental material). Also, a recombinant plasmid for the VP2468 gene was created to be used as a calibration standard, and a strong correlation (R2, 0.999) between the concentration of this target gene and threshold cycle (CT) values was observed using the present RT-qPCR protocol (see Fig. S2 in the supplemental material) (28, 29).
Statistical analysis.
Triplicate experiments were performed for the induction of the VBNC state and the resuscitation of VBNC cultures. Replicate experiments on gene expression were performed. The statistical significance of data was assessed by Student's t test or analysis of variance (ANOVA) with Duncan's multiple-range test at a significance level (α) of 0.05, using SPSS for Windows, version 11.0 (SPSS Inc., Chicago, IL, USA).
RESULTS
It is hypothesized that the change in culturability of V. parahaemolyticus cells incubated under VBNC induction conditions depends on the presence of toxic ROS and the function of the protecting antioxidative AhpC proteins. In this study, we demonstrated only the antioxidative properties of these AhpC proteins and their roles in determining the induction time and length of maintenance of the VBNC state.
Antioxidative activity of the ahpC mutants.
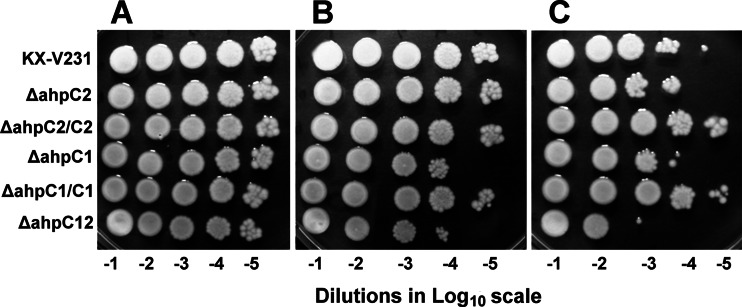
The ahpC1 and ahpC2 deletion mutants and their corresponding complementary strains were examined for their colony-forming abilities on an agar medium under the influence of extrinsic peroxides. The extents of growth of the wild-type, mutant, and complementary strains, when spotted at different dilutions on TSA–3% NaCl with no extrinsic peroxide added, were similar (Fig. 1A). In the presence of inorganic H2O2, the colony-forming abilities of the ahpC1 mutant and the ahpC1 ahpC2 double mutant (the ΔahpC12 strain) were reduced, and this inhibition was reversed by complementation of the ahpC1 gene (in the ΔahpC1/C1 strain). The colony-forming abilities of the ahpC2 mutant and its complementary strain were similar to that of the wild-type strain (Fig. 1B). The presence of 80 μM t-BOOH reduced the colony-forming abilities of the wild-type strain, and this inhibition was further enhanced in the ahpC1, ahpC2, and double mutants. The presence of the complementary ahpC genes in the single ahpC mutants substantially reversed this inhibition (Fig. 1C). These results demonstrated the antioxidative functions of these two ahpC genes.
Fig 1.
Colony-forming abilities of wild-type and ahpC mutant strains of Vibrio parahaemolyticus. The wild-type strain (KX-V231), the ahpC1 mutant and its complementary (ΔahpC1/C1) strain, the ahpC2 mutant and its complementary (ΔahpC2/C2) strain, and the ahpC1 ahpC2 (ΔahpC12) double mutant were diluted 10−1- to 10−5-fold (−1 to −5 on the log scale), spotted onto TSA–3% NaCl containing either no extrinsic peroxides (A), 250 μM H2O2 (B), or 80 μM t-BOOH (C), and incubated at 25°C for 16 h.
Growth and survival of ahpC mutants at different incubation temperatures.
When the wild-type strain KX-V231, the ahpC mutants, and their complementary strains were spread on rich agar medium (TSA–3% NaCl) and incubated at 22, 30, or 37°C, the ahpC1 ahpC2 double mutant (the ΔahpC12 strain) formed the smallest colonies at these temperatures, while the ahpC1 mutant formed colonies significantly smaller than those of the ahpC2 mutant or the wild-type strain (P < 0.05). The influence of ahpC1 on the diameters of the colonies was enhanced at incubation temperatures of 22 and 30°C but not at 37°C. Chloramphenicol was added to the agar medium to maintain the complementary plasmids or cloning vector, and it affected the sizes of the colonies. Colonies of the double mutant containing the cloning vector (the ΔahpC12V strain), which was grown on agar medium containing chloramphenicol, were markedly smaller than those of the ΔahpC12 strain, while addition of the complementary ahpC1 or ahpC2 gene to the corresponding mutant relieved the inhibition of growth, and these complementary strains formed larger colonies on the same agar medium (Table 2).
Table 2.
Growth of V. parahaemolyticus strains on agar medium upon incubation at different temperatures
| Straina | Diam of colonies (mm) at the following temp (°C): |
||
|---|---|---|---|
| 22 | 30 | 37 | |
| KX-V231 | 5.8 ± 0.84 a | 15.2 ± 1.30 a | 2.8 ± 0.45 a |
| ΔahpC1 strain | 3.8 ± 0.45 b | 6.2 ± 1.48 d | 2.6 ± 0.55 ab |
| ΔahpC2 strain | 5.2 ± 0.45 a | 13.4 ± 1.52 b | 2.8 ± 0.45 a |
| ΔahpC12 strain | 1.6 ± 0.55 c | 4.8 ± 0.84 d | 2.0 ± 0 bc |
| ΔahpC12/C1 strain | 1.2 ± 0.45 cd | 9.4 ± 0.89 c | 2.8 ± 0.50 ab |
| ΔahpC12/C2 strain | 1.2 ± 0.45 cd | 9.2 ± 0.84 c | 2.6 ± 0.55 ab |
| ΔahpC12V strain | 0.67 ± 0.06 d | 3.0 ± 1.58 e | 1.6 ± 0.55 c |
Different strains of V. parahaemolyticus were spread on TSA–3% NaCl and were incubated at 22, 30, or 37°C for 24, 15, or 15 h, respectively. The medium for the culture of the ΔahpC12/C1, ΔahpC12/C2, and ΔahpC12V strains contained chloramphenicol. The sizes of colonies were measured. The sizes at various temperatures were analyzed separately by ANOVA with Duncan's multiple-range test, and those followed by different letters were significantly different from each other (P < 0.05).
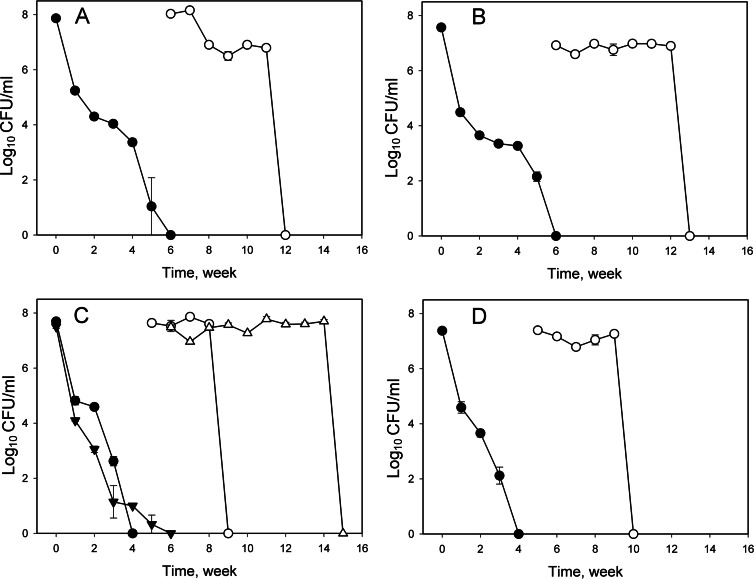
When the V. parahaemolyticus cultures in TSB–3% NaCl were incubated at 4°C, the culturability of KX-V231 and the single ahpC mutants gradually declined at similar rates (about 22 to 28% survival in 9 days), while the culturability of the double mutant (the ΔahpC12 strain) declined rapidly to less than 1% in 9 days (Fig. 2A). When the double mutant containing the cloning vector (the ΔahpC12V strain) was incubated in broth containing chloramphenicol at 4°C, its culturability declined rapidly; the low rate of decline (about 24% survival in 9 days) was restored in the presence of a complementary ahpC2 gene (the ΔahpC12/C2 strain). Complementation with ahpC1 (the ΔahpC12/C1 strain) did not alter the culturability of this double mutant (Fig. 2B). These results demonstrated that ahpC2 was important for the survival of V. parahaemolyticus at refrigeration temperatures.
Fig 2.
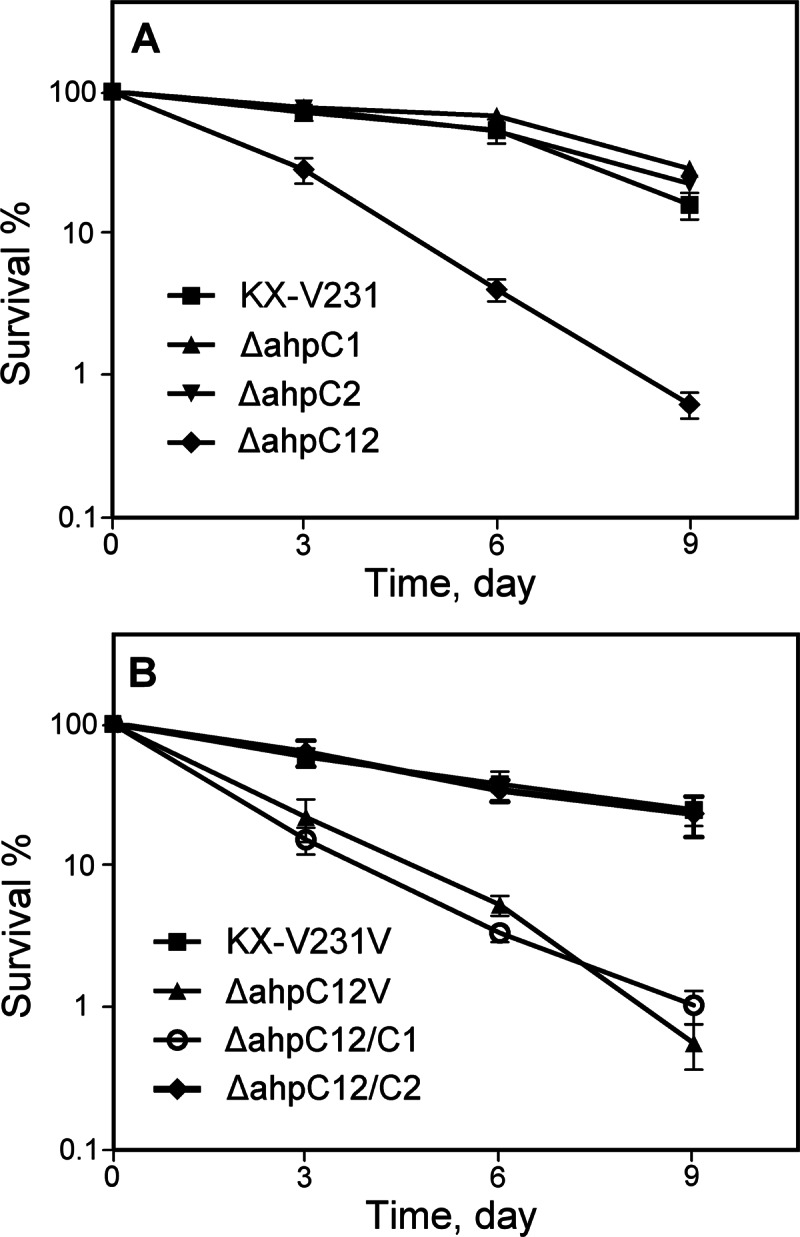
Survival of different V. parahaemolyticus strains incubated in TSB–3% NaCl at 4°C for different periods. (A) The wild-type strain (KX-V231), ahpC1 mutant, ahpC2 mutant, and ahpC1 ahpC2 double mutant (the ΔahpC12 strain). (B) Strains containing the cloning vector pSCB01 (KX-V231V, the ΔahpC12V strain), a complementary ahpC1 gene (ΔahpC12/C1), or a complementary ahpC2 gene (the ΔahpC12/C2 strain).
Induction of the VBNC state and resuscitation in the ahpC mutants.
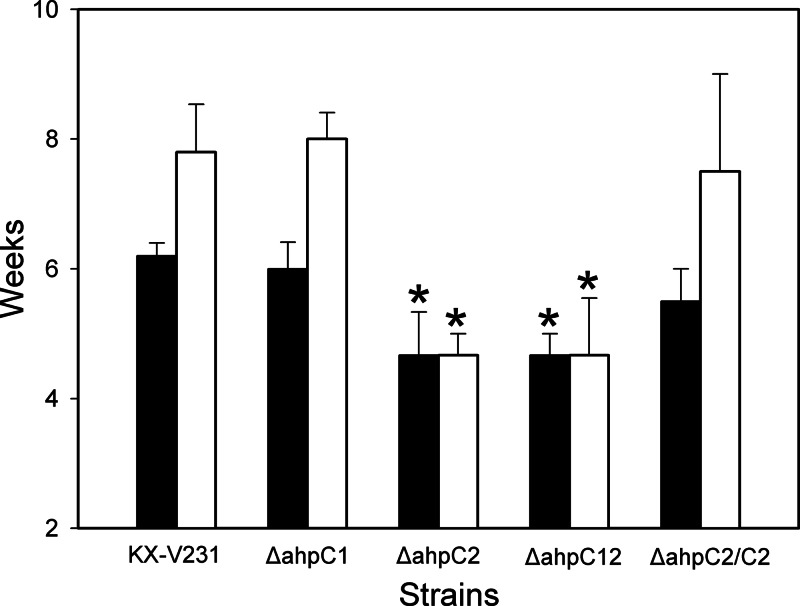
This study demonstrated, by gene deletion and complementation, that ahpC2 has a greater role than ahpC1 in the VBNC state of V. parahaemolyticus and in the maintenance of viability in that state (Fig. 3 and 4). The VBNC state was successfully induced in the wild-type strain in approximately 6 weeks, while the induction times for the ahpC2 mutant and the ahpC1 ahpC2 double mutant (the ΔahpC12 strain) were significantly shorter, about 4.6 weeks (P ≤ 0.05) (Fig. 3 and 4). The VBNC induction time of the ahpC2 mutant in the presence of its complementary gene (the ΔahpC2/C2 strain) was significantly longer (about 5.5 weeks) than that of the uncomplemented ahpC2 mutant and was approximately equal to that for the wild-type strain (P < 0.05). The induction time of the VBNC state in the ahpC1 mutant was not significantly different from that of the wild-type strain (P > 0.05).
Fig 3.
Induction of the VBNC state in different strains of Vibrio parahaemolyticus and their resuscitation. V. parahaemolyticus strains in exponential phase were incubated at 4°C in MMS–0.5% NaCl medium, and culturable cells were enumerated at different intervals. (A) The wild-type strain KX-V231; (B) the ahpC1 mutant; (C) the ahpC2 mutant (circles) and its complementary strain (inverted and upright triangles); (D)the ahpC1 ahpC2 double mutant. Filled symbols, levels of culturable cells during induction of the VBNC state; open symbols, levels of culturable cells of VBNC cultures after temperature upshift treatment.
Fig 4.
Times required by different strains of Vibrio parahaemolyticus to enter the VBNC state and the maximum lengths of time at which the VBNC cultures could be resuscitated. Shown are results for the wild type (strain KX-V231), the ahpC1 mutant, the ahpC2 mutant, the ahpC1 ahpC2 double mutant (the ΔahpC12 strain), and the ahpC2 mutant with a complementary ahpC2 gene (the ΔahpC2/C2 strain). Filled bars, times required to enter the VBNC state; open bars, maximum lengths of time at which the cells in the VBNC state could be resuscitated. Asterisks indicate significant differences (P < 0.05) from corresponding values for the wild-type strain.
Within 5 to 7 weeks in the VBNC state, the levels of culturable cells in the resuscitated cultures (about 7 log CFU/ml) were similar to those at the beginning of induction of the VBNC state (Fig. 3). The wild-type strain remained viable and, when maintained in the VBNC state for an average of 7.8 weeks, could be resuscitated. This time was significantly shortened, to 4.7 weeks, for the ahpC2 mutant and the ahpC1 ahpC2 double mutant (P ≤ 0.05). The times for maintenance of the cultures in the VBNC state were not significantly different for the ahpC1 mutant (8.0 weeks), the complemented ahpC2 mutant (the ΔahpC2/C2 strain) (7.5 weeks), and the wild-type strain (7.8 weeks) (Fig. 4) (P > 0.05). These results revealed that ahpC2 was markedly associated with the induction and maintenance of the VBNC state in V. parahaemolyticus.
Gene expression.
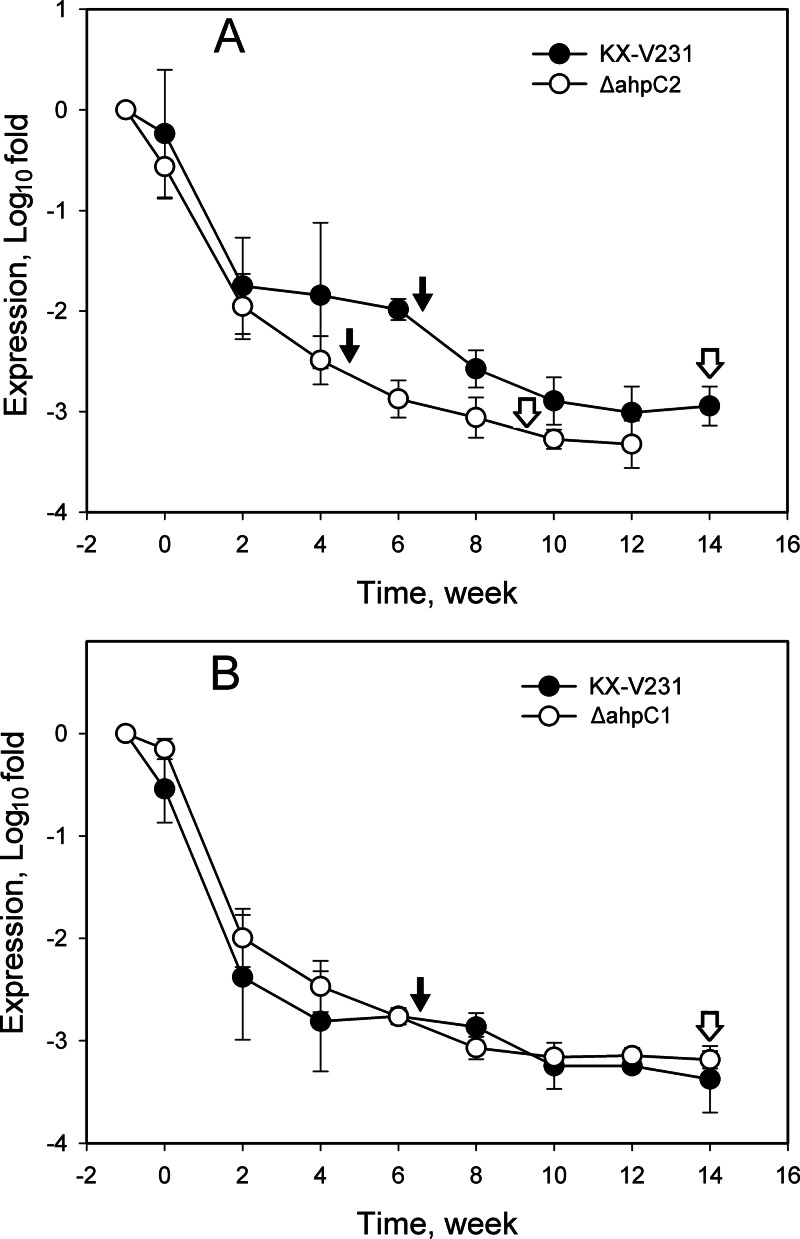
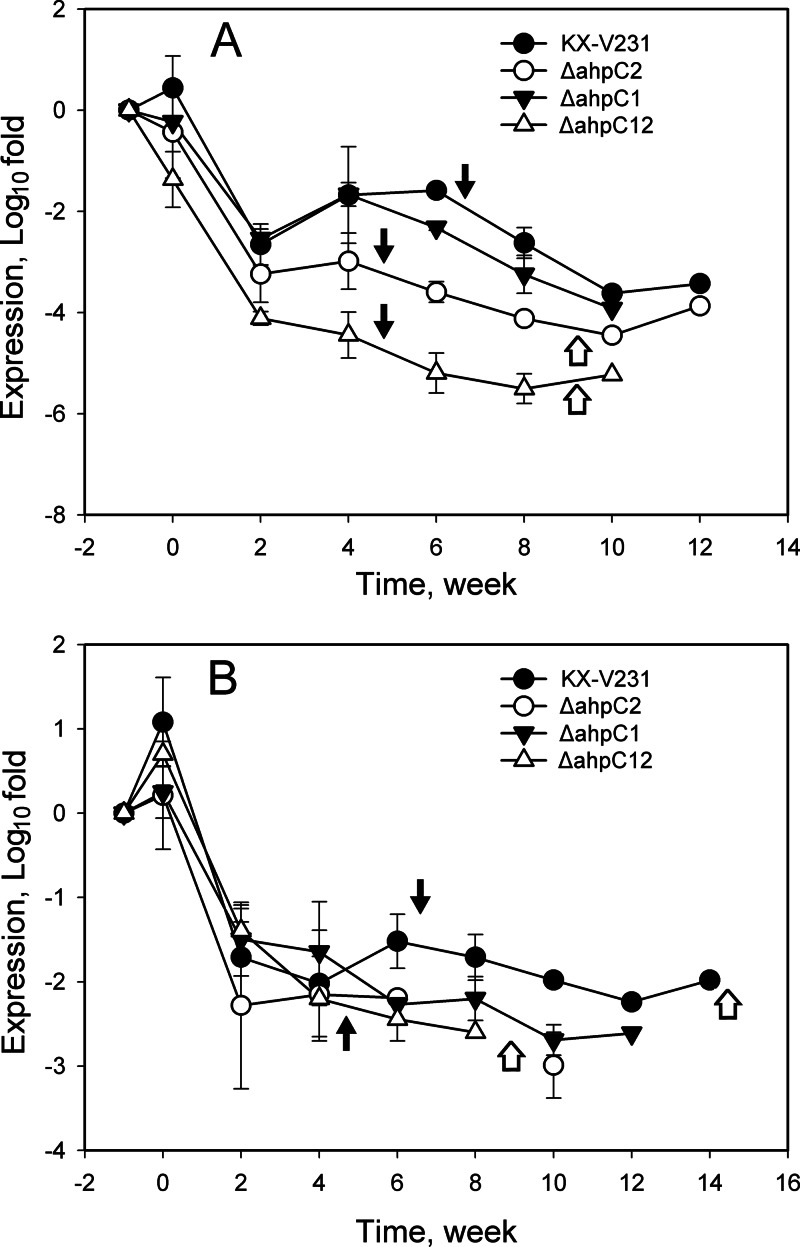
The expression of antioxidative genes (encoding 4 catalases and 2 AhpC proteins) and related regulators (RpoS and OxyR) in the induction of the VBNC state and the resuscitation of VBNC cultures was monitored by RT-qPCR. The relative quantities of ahpC1 and ahpC2 transcripts in the wild-type strain declined rapidly during the first 2 weeks (from 0 to −2.38 log fold changes for ahpC1; from 0 to −1.75 log fold changes for ahpC2) of induction of the VBNC state, and thereafter, they declined gradually (−3.01 log fold changes at 12 weeks for ahpC1; −3.25 log fold changes for ahpC2) (Fig. 5). The relative quantity of the ahpC1 transcript in the ahpC2 mutant (−2.88 to −3.06 log fold changes) was significantly lower than that in the wild-type strain (−1.99 to −2.58 log fold changes) upon incubation under VBNC induction conditions for 6 to 8 weeks (P < 0.05) (Fig. 5A). The relative quantities of the ahpC2 transcript in the wild type and the ahpC1 mutant were not significantly different throughout the monitoring period (P > 0.05) (Fig. 5B). These results suggested that expression of the ahpC genes was markedly affected in the ahpC2 mutant.
Fig 5.
Expression of ahpC genes in different strains of Vibrio parahaemolyticus incubated under VBNC-inducing conditions. V. parahaemolyticus cultures in exponential phase were incubated at 4°C in MMS–0.5% NaCl medium. Relative quantities of ahpC1 (A) and ahpC2 (B) transcripts were determined by RT-qPCR, and the calculated quantities were compared to those in exponential phase, specified as −1 week in the graph and presented on a log scale. Filled arrows indicate the times of entry into the VBNC state, while open arrows indicate the times after which the VBNC cells failed to be resuscitated.
The expression of four catalase genes (VPA0305, VPA0453, VPA0768, and VPA1418) was monitored, and the results revealed two patterns of change in the relative quantities of their transcripts. The patterns for VPA0305 and VPA1418 were similar, while the patterns for VPA0453 and VPA0768 were similar. The patterns of VPA0305 and VPA0453 are presented in Fig. 6. Relative quantities of the transcripts of all these catalase genes declined rapidly in the first 2 weeks and declined gradually for the rest of the time. The relative quantities of catalase VPA0305 in the wild type (−1.68 to −2.63 log fold changes) and the ahpC1 mutant (−1.67 to −3.25 log fold changes) were not significantly different (P > 0.05) and were significantly higher than those in the ahpC2 mutant (−2.99 to −4.12 log fold changes) and the ahpC1 ahpC2 double mutant (−4.45 to −5.51 log fold changes) from 4 to 8 weeks (P ≤ 0.05) (Fig. 6A). The expression patterns of VPA0453 in different strains were not significantly different, whereas the relative quantities of the transcripts of VPA0453 declined to about −2 log fold changes in 2 weeks and gradually declined for the rest of the time (P > 0.05) (Fig. 6B). These results suggested that the expression of some catalase genes was also markedly affected in the ahpC2 mutant and the ahpC1 ahpC2 mutant.
Fig 6.
Expression of catalase genes VPA0305 (A) and VPA0453 (B) in different strains of Vibrio parahaemolyticus under VBNC-inducing conditions. V. parahaemolyticus strains in exponential phase were incubated at 4°C in MMS–0.5% NaCl medium. Relative quantities of transcripts of the VPA0305 and VPA0453 catalase genes were determined by RT-qPCR, and the calculated quantities were compared to those in exponential phase, specified as −1 week in the graph and presented on a log scale. Filled arrows indicate times to entry into the VBNC state, while open arrows indicate the times after which the VBNC cells failed to be resuscitated.
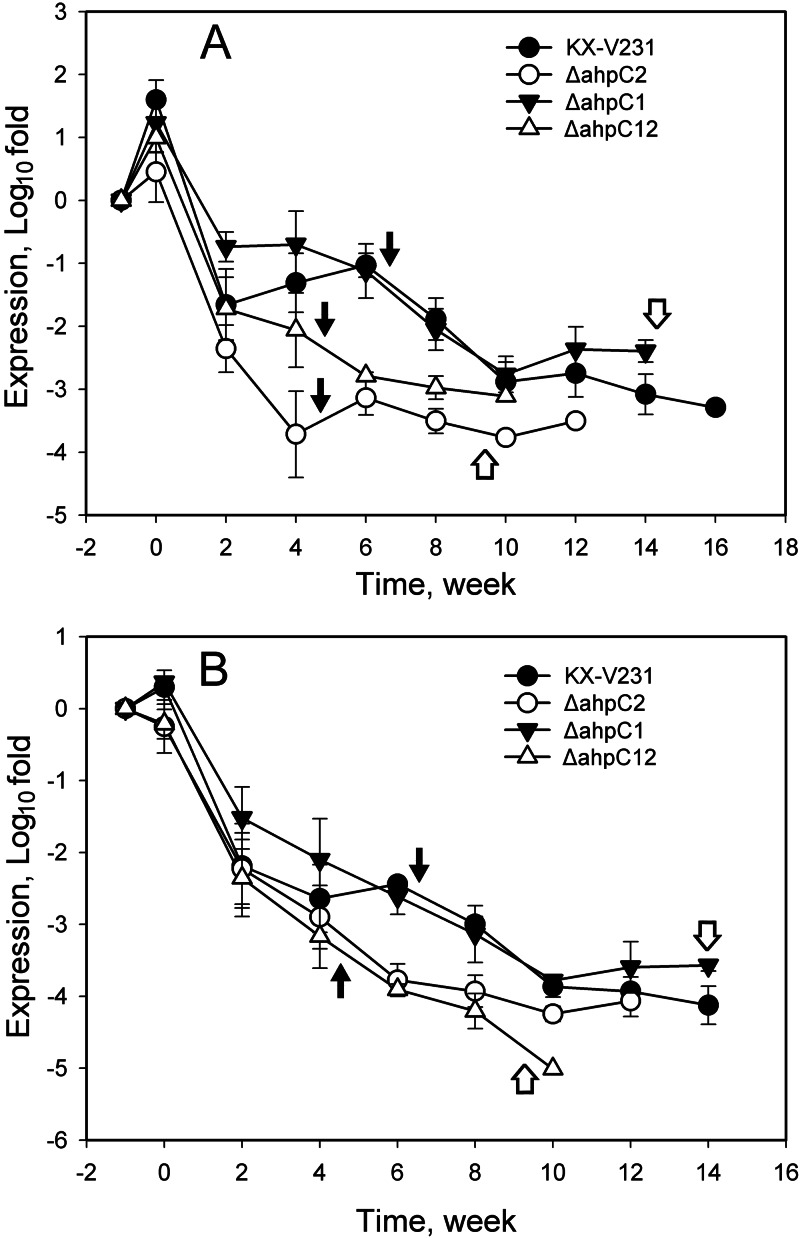
The relative quantities of rpoS and oxyR regulators declined at different rates in different mutant strains incubated under VBNC induction conditions. At 6 to 8 weeks, the expression of both regulators in the wild-type strain (−1.03 to −1.89 log fold changes for rpoS; −2.44 to −2.99 log fold changes for oxyR) and the ahpC1 mutant (−1.12 to −2.05 log fold changes for rpoS; −2.62 to −3.14 log fold changes for oxyR) was significantly higher than that in the ahpC2 mutant (−3.14 to −3.51 log fold changes for rpoS; −3.78 to −3.93 log fold changes for oxyR) and the ahpC1 ahpC2 double mutant (−2.79 to −2.98 log fold changes for rpoS; −3.91 to −4.21 log fold changes for oxyR) (P < 0.05) (Fig. 7). These results suggested that the expression of these two regulator genes was also markedly affected in the ahpC2 mutant and the ahpC1 ahpC2 mutant.
Fig 7.
Expression of the regulators rpoS (A) and oxyR (B) in different strains of Vibrio parahaemolyticus under VBNC-inducing conditions. V. parahaemolyticus strains in the exponential phase were incubated at 4°C in MMS–0.5% NaCl medium. Relative quantities of rpoS and oxyR transcripts in different strains were determined by RT-qPCR, and the calculated quantities were compared to those in exponential phase, specified as −1 week in the graph and presented on a log scale. Filled arrows indicate times to entry into the VBNC state, while open arrows indicate the times after which the VBNC cells failed to be resuscitated.
The expression of these genes was also monitored during the resuscitation of VBNC cultures. After a temperature upshift treatment, the transcripts for all of these genes in the mutant strains were increased to levels similar to those in the wild-type strain (0 to 2 log fold increases over levels in the exponential phase). Expression remained at low levels (−3 to −4 log fold) when the VBNC cultures failed to be resuscitated (data not shown).
Under the VBNC induction conditions for several weeks, the relative quantities of transcripts for these antioxidants and regulator genes remained low (−3 to −5 log fold changes), and in most cases, these transcripts remained detectable when the VBNC cultures could no longer be resuscitated (Fig. 4 to 6).
DISCUSSION
The VBNC state is typically induced in vibrios and some other Gram-negative bacteria in a nutrient-limited medium at low temperatures (9, 30, 31). As a survival strategy, bacterial cells in the VBNC state undergo substantial changes in their morphologies and metabolic activities (27, 32), while the pathogenic potential is maintained in this state (8, 10). Thus, the VBNC state of pathogenic vibrios remains a consistent threat to public health. A microarray was utilized to analyze a total of 3,707 genes of V. cholerae; 100 genes were found to be expressed in the VBNC state, and the distribution of genes among the various functional groups in the VBNC state was similar to that obtained for stationary-phase cells, suggesting that VBNC bacteria may retain the functions that are essential for viability (33). The metabolic activities of VBNC bacteria generate endogenous ROS, which damage the cell (34) and may cause a loss of culturability and viability, especially in cells with weakened antioxidative activity, whereas the sodC gene (VC1583) was the only antioxidative gene expressed among the superoxide dismutase (SOD), catalase, and AhpC genes (33).
Our previous proteomic study suggested the specific role of ahpC2 in the VBNC state of V. parahaemolyticus (23), and in the present study, the involvement of ahpC2 was further supported by gene deletion and complementation (Fig. 3 and 4). The role of ahpC2 in the VBNC state was also indicated by the change of gene expression in the mutant strains. The expression of ahpC genes, catalase genes, and related regulators declined rapidly by 3 to 5 orders of magnitude as culturability declined and remained low in V. parahaemolyticus under the VBNC state induction conditions, while the expression of these genes was even lower in the ahpC2 mutant and the ahpC1 ahpC2 double mutant than in the ahpC1 mutant (Fig. 5 to 7).
Different species of peroxiredoxins have been identified in bacteria, including the fusion protein of thiol peroxidase and glutaredoxin in V. cholerae (35). Functions of different AhpC species have been reported in a few studies. Legionella pneumophila and Coxiella burnetii each contain two ahpC genes, ahpC1 and ahpC2. The ahpC2 gene is adjacent to an alkyl hydroperoxide reductase partner (ahpD), which is also found in Mycobacterium species. ahpC1 is expressed following the exponential phase, and ahpC2 is expressed during the early-exponential phase. Levels of ahpC1 mRNA are approximately 7 to 10 times those of ahpC2D mRNA. The expression of ahpC2D is significantly increased in the ahpC1 mutant, whereas ahpC1 expression is unchanged in the ahpC2D mutant, indicating the compensatory activity of the ahpC2D system in response to oxidative stress (36). In V. vulnificus, AhpC1 is a typical NADH-dependent peroxiredoxin and forms a peroxide reductase system with AhpF (26), while AhpC2 aids in the growth of V. vulnificus under high salinity by scavenging ROS in cells (37). This study shows that both ahpC1 and ahpC2 of V. parahaemolyticus were active against t-BOOH, which is commonly used to mimic lipid hydroperoxides, and that ahpC1 was more effective than ahpC2 against H2O2 (Fig. 1).
The unique role of ahpC2 in inducing and maintaining the VBNC state of V. parahaemolyticus may be associated with its enhanced expression/activity at low incubation temperatures in a starvation medium and/or with the inhibition of ahpC1 under such conditions. In an Anabaena sp., ahpC is associated with protection against high temperatures (47°C), high salinity, toxic pesticides (carbofuran), heavy metals, and UV irradiation (38). However, in Xanthomonas campestris, ahpC performs no protective function against heat treatment (39). The expression of AhpC proteins in Acidithiobacillus ferrooxidans (40) and Shewanella putrefaciens (41) increases when they are incubated at low temperatures of 2 to 15°C. In another study, the promoter of the putative ahpC gene in the Shewanella species is highly responsive to incubation at a low temperature of 4°C (42). These investigations reveal that different species of AhpC in these bacteria may respond differently to high and low incubation temperatures.
The data presented here reveal that the functions of ahpC1 and ahpC2 in V. parahaemolyticus may be influenced by the incubation temperature and the richness of the culture medium. AhpC1 is probably the chief functional peroxidase in V. parahaemolyticus that is cultured in a rich medium at appropriate incubation temperatures (Table 2). When the cultures were incubated at 4°C in a rich medium, the ahpC1 and ahpC2 mutants and the parental wild-type strain exhibited similar survival rates (Fig. 2A), suggesting the compensatory activities of these two ahpC genes in a rich medium. However, when these mutants were cultured in a starvation medium at 4°C (under VBNC state-inducing conditions), ahpC1 failed to compensate for the normal function of ahpC2 in the ahpC2 mutant (Fig. 4 and 5). We postulate that ahpC2 may be activated at 4°C regardless of the culture medium, whereas ahpC1 may be activated only in the rich medium. Different regulators may be critical for sensing the peroxides and regulating the expression of these ahpC genes.
Two regulators (rpoS, oxyR) associated with the function of peroxidases in V. parahaemolyticus were identified. The catalase genes and ahpC genes have been demonstrated to exhibit a compensatory expression pattern in bacteria under the influence of the OxyR regulon (17), whereas rpoS is a general stress response regulator (43, 44). However, in this study, the expression of catalase genes was reduced in the ahpC2 mutant, rather than being stimulated through a typical compensatory expression pattern (Fig. 7). The compensatory function of catalases may fail at low temperatures, under starvation conditions, or under a combination of both of these stresses, which are commonly applied to induce the VBNC state.
In addition to rpoS and oxyR, other regulators may also be involved in the temperature-regulated activity. The native ahpC1 gene compensated for the normal function of ahpC2 in the ahpC2 mutant when V. parahaemolyticus was incubated at 4°C in a rich medium (Fig. 2A), while the extrachromosomal complementary ahpC1 gene did not (Fig. 2B). The function of the native ahpC1 gene at low temperatures may depend on a regulator that is associated with it. A search of the genome of V. parahaemolyticus (21) shows that VPA1682, which is adjacent to ahpC1 (VPA1683), may be an ohrR-like regulator that is associated with temperature-regulated activities (36). VPA1682 is a putative MarR family protein and is homologous to the organic peroxide sensor and transcriptional regulator of both Bacillus subtilis (45) and some Gram-negative bacteria (46, 47). OxyR and OhrR are commonly known as the positive (48) and negative (49) regulators, respectively, of the functions of catalases and AhpC proteins. Coordinated activity of these regulators may be responsible for the normal functioning of the peroxiredoxins in the VBNC state of V. parahaemolyticus.
Since the transcripts of both ahpC1 and ahpC2 similarly declined to low levels by approximately 3 orders of magnitude in the induction of the VBNC state (Fig. 4), while an enhanced quantity of AhpC2 protein was detected (23), enhanced translation efficiency and protein stability may also result in the increased quantity of this antioxidative protein that was produced. Certain ahpC genes may be better expressed than other antioxidative genes and may function better at low temperatures (40, 42). The lowered expression and weaker function of catalases under VBNC-inducing conditions (16) may also strengthen the involvement of AhpC proteins, since both detoxify peroxides.
In conclusion, this investigation demonstrated the antioxidative activities of ahpC1 and ahpC2 against H2O2 and organic peroxide in V. parahaemolyticus, whereas ahpC2 alone played a significant role in the induction and maintenance of the VBNC state in this pathogen. This study facilitates the understanding of the roles of AhpC proteins in the growth and survival of V. parahaemolyticus under environmental stresses. Direct evidence, such as quantification of ROS and antioxidative activities during the induction of the VBNC state, may be needed in further studies to define the role of ROS in this state.
Supplementary Material
ACKNOWLEDGMENTS
We thank the National Science Council of the Republic of China for financially supporting this research under contracts NSC 95-2313-B-031-002 and NSC 97-2313-B-031-001-MY3.
Ted Knoy is appreciated for editorial assistance.
Footnotes
Published ahead of print 5 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00560-13.
REFERENCES
- 1. Anonymous 2011. Risk assessment of Vibrio parahaemolyticus in seafood: interpretative summary and technical report. Microbiological Risk Assessment Series no. 16. World Health Organization and Food and Agriculture Organization of the United Nations, Rome, Italy [Google Scholar]
- 2. Nair GB, Ramamurthy T, Bhattacharya SK, Dutta B, Takeda Y, Sack DA. 2007. Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin. Microbiol. Rev. 20:39–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wong HC, Ting SH, Shieh WR. 1992. Incidence of toxigenic vibrios in foods available in Taiwan. J. Appl. Bacteriol. 73:197–202 [DOI] [PubMed] [Google Scholar]
- 4. Bates TC, Oliver JD. 2004. The viable but nonculturable state of Kanagawa positive and negative strains of Vibrio parahaemolyticus. J. Microbiol. 42:74–79 [PubMed] [Google Scholar]
- 5. Jiang XP, Chai TJ. 1996. Survival of Vibrio parahaemolyticus at low temperatures under starvation conditions and subsequent resuscitation of viable, nonculturable cells. Appl. Environ. Microbiol. 62:1300–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wong HC, Wang P. 2004. Induction of viable but non-culturable state in Vibrio parahaemolyticus and its susceptibility to environmental stresses. J. Appl. Microbiol. 96:359–366 [DOI] [PubMed] [Google Scholar]
- 7. Wong HC, Wang P, Chen SY, Chiu SW. 2004. Resuscitation of viable but non-culturable Vibrio parahaemolyticus in a minimum salt medium. FEMS Microbiol. Lett. 233:269–275 [DOI] [PubMed] [Google Scholar]
- 8. Oliver JD. 2010. Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol. Rev. 34:415–425 [DOI] [PubMed] [Google Scholar]
- 9. Wong HC, Shen CT, Chang CN, Lee YS, Oliver JD. 2004. Biochemical and virulence characterization of viable but nonculturable cells of Vibrio parahaemolyticus. J. Food Prot. 67:2430–2435 [DOI] [PubMed] [Google Scholar]
- 10. Rahman I, Shahamat M, Chowdhury MA, Colwell RR. 1996. Potential virulence of viable but nonculturable Shigella dysenteriae type 1. Appl. Environ. Microbiol. 62:115–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bogosian G, Aardema ND, Bourneuf EV, Morris PJ, O'Neil JP. 2000. Recovery of hydrogen peroxide-sensitive culturable cells of Vibrio vulnificus gives the appearance of resuscitation from a viable but nonculturable state. J. Bacteriol. 182:5070–5075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mizunoe Y, Wai SN, Takade A, Yoshida S. 1999. Restoration of culturability of starvation-stressed and low-temperature-stressed Escherichia coli O157 cells by using H2O2-degrading compounds. Arch. Microbiol. 172:63–67 [DOI] [PubMed] [Google Scholar]
- 13. Wai SN, Mizunoe Y, Takade A, Yoshida S. 2000. A comparison of solid and liquid media for resuscitation of starvation- and low-temperature-induced nonculturable cells of Aeromonas hydrophila. Arch. Microbiol. 173:307–310 [DOI] [PubMed] [Google Scholar]
- 14. Abe A, Ohashi E, Ren H, Hayashi T, Endo H. 2007. Isolation and characterization of a cold-induced nonculturable suppression mutant of Vibrio vulnificus. Microbiol. Res. 162:130–138 [DOI] [PubMed] [Google Scholar]
- 15. Bloomfield SF, Stewart GS, Dodd CE, Booth IR, Power EG. 1998. The viable but non-culturable phenomenon explained? Microbiology 144(Part 1):1–3 [DOI] [PubMed] [Google Scholar]
- 16. Kong IS, Bates TC, Hulsmann A, Hassan H, Smith BE, Oliver JD. 2004. Role of catalase and oxyR in the viable but nonculturable state of Vibrio vulnificus. FEMS Microbiol. Ecol. 50:133–142 [DOI] [PubMed] [Google Scholar]
- 17. Charoenlap N, Eiamphungporn W, Chauvatcharin N, Utamapongchai S, Vattanaviboon P, Mongkolsuk S. 2005. OxyR mediated compensatory expression between ahpC and katA and the significance of ahpC in protection from hydrogen peroxide in Xanthomonas campestris. FEMS Microbiol. Lett. 249:73–78 [DOI] [PubMed] [Google Scholar]
- 18. Hishinuma S, Yuki M, Fujimura M, Fukumori F. 2006. OxyR regulated the expression of two major catalases, KatA and KatB, along with peroxiredoxin, AhpC in Pseudomonas putida. Environ. Microbiol. 8:2115–2124 [DOI] [PubMed] [Google Scholar]
- 19. Mongkolsuk S, Whangsuk W, Vattanaviboon P, Loprasert S, Fuangthong M. 2000. A Xanthomonas alkyl hydroperoxide reductase subunit C (ahpC) mutant showed an altered peroxide stress response and complex regulation of the compensatory response of peroxide detoxification enzymes. J. Bacteriol. 182:6845–6849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hofmann B, Hecht, HJ, Flohe. L. 2002. Peroxiredoxins. Biol. Chem. 383:347–364 [DOI] [PubMed] [Google Scholar]
- 21. Makino K, Oshima K, Kurokawa K, Yokoyama K, Uda T, Tagomori K, Iijima Y, Najima M, Nakano M, Yamashita A, Kubota Y, Kimura S, Yasunaga T, Honda T, Shinagawa H, Hattori M, Iida T. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet 361:743–749 [DOI] [PubMed] [Google Scholar]
- 22. Kang JH, Lee JH, Park JH, Huh SH, Kong IS. 1998. Cloning and identification of a phospholipase gene from Vibrio mimicus. Biochim. Biophys. Acta 1394:85–89 [DOI] [PubMed] [Google Scholar]
- 23. Lai CJ, Chen SY, Lin IH, Chang CH, Wong HC. 2009. Change of protein profiles in the induction of the viable but nonculturable state of Vibrio parahaemolyticus. Int. J. Food Microbiol. 135:118–124 [DOI] [PubMed] [Google Scholar]
- 24. Okada N, Iida T, Park KS, Goto N, Yasunaga T, Hiyoshi H, Matsuda S, Kodama T, Honda T. 2009. Identification and characterization of a novel type III secretion system in trh-positive Vibrio parahaemolyticus strain TH3996 reveal genetic lineage and diversity of pathogenic machinery beyond the species level. Infect. Immun. 77:904–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Park KS, Ono T, Rokuda M, Jang MH, Okada K, Iida T, Honda T. 2004. Functional characterization of two type III secretion systems of Vibrio parahaemolyticus. Infect. Immun. 72:6659–6665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Baek WK, Lee HS, Oh MH, Koh MJ, Kim KS, Choi SH. 2009. Identification of the Vibrio vulnificus ahpCl gene and its influence on survival under oxidative stress and virulence. J. Microbiol. 47:624–632 [DOI] [PubMed] [Google Scholar]
- 27. Chen SY, Jane WN, Chen YS, Wong HC. 2009. Morphological changes of Vibrio parahaemolyticus under cold and starvation stresses. Int. J. Food Microbiol. 129:157–165 [DOI] [PubMed] [Google Scholar]
- 28. Lessard MH, Belanger G, St-Gelais D, Labrie S. 2012. The composition of Camembert cheese-ripening cultures modulates both mycelial growth and appearance. Appl. Environ. Microbiol. 78:1813–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Marum L, Miguel A, Ricardo CP, Miguel C. 2012. Reference gene selection for quantitative real-time PCR normalization in Quercus suber. PLoS One 7:e35113 doi: 10.1371/journal.pone.0035113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Oliver JD, Hite F, McDougald D, Andon NL, Simpson LM. 1995. Entry into, and resuscitation from, the viable but nonculturable state by Vibrio vulnificus in an estuarine environment. Appl. Environ. Microbiol. 61:2624–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yaron S, Matthews KR. 2002. A reverse transcriptase-polymerase chain reaction assay for detection of viable Escherichia coli O157:H7: investigation of specific target genes. J. Appl. Microbiol. 92:633–640 [DOI] [PubMed] [Google Scholar]
- 32. Chaiyanan S, Chaiyanan S, Grim C, Maugel T, Huq A, Colwell RR. 2007. Ultrastructure of coccoid viable but non-culturable Vibrio cholerae. Environ. Microbiol. 9:393–402 [DOI] [PubMed] [Google Scholar]
- 33. Asakura H, Ishiwa A, Arakawa E, Makino S, Okada Y, Yamamoto S, Igimi S. 2007. Gene expression profile of Vibrio cholerae in the cold stress-induced viable but non-culturable state. Environ. Microbiol. 9:869–879 [DOI] [PubMed] [Google Scholar]
- 34. Dubbs JM, Mongkolsuk S. 2012. Peroxide-sensing transcriptional regulators in bacteria. J. Bacteriol. 194:5495–5503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cha MK, Hong SK, Lee DS, Kim IH. 2004. Vibrio cholerae thiol peroxidase-glutaredoxin fusion is a 2-Cys TSA/AhpC subfamily acting as a lipid hydroperoxide reductase. J. Biol. Chem. 279:11035–11041 [DOI] [PubMed] [Google Scholar]
- 36. Klomsiri C, Panmanee W, Dharmsthiti S, Vattanaviboon P, Mongkolsuk S. 2005. Novel roles of ohrR-ohr in Xanthomonas sensing, metabolism, and physiological adaptive response to lipid hydroperoxide. J. Bacteriol. 187:3277–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Koh MJ, Lee HS, Rhee JE, Choi SH. 2010. Evidence that Vibrio vulnificus ahpC2 is essential for survival under high salinity by modulating intracellular level of ROS. J. Microbiol. 48:129–133 [DOI] [PubMed] [Google Scholar]
- 38. Mishra Y, Chaurasia N, Rai LC. 2009. AhpC (alkyl hydroperoxide reductase) from Anabaena sp. PCC 7120 protects Escherichia coli from multiple abiotic stresses. Biochem. Biophys. Res. Commun. 381:606–611 [DOI] [PubMed] [Google Scholar]
- 39. Buranajitpakorn S, Piwkam A, Charoenlap N, Vattanaviboon P, Mongkolsuk S. 2011. Genes for hydrogen peroxide detoxification and adaptation contribute to protection against heat shock in Xanthomonas campestris pv. campestris. FEMS Microbiol. Lett. 317:60–66 [DOI] [PubMed] [Google Scholar]
- 40. Mykytczuk NC, Trevors JT, Foote SJ, Leduc LG, Ferroni GD, Twine SM. 2011. Proteomic insights into cold adaptation of psychrotrophic and mesophilic Acidithiobacillus ferrooxidans strains. Antonie Van Leeuwenhoek 100:259–277 [DOI] [PubMed] [Google Scholar]
- 41. Leblanc L, Leboeuf C, Leroi F, Hartke A, Auffray Y. 2003. Comparison between NaCl tolerance response and acclimation to cold temperature in Shewanella putrefaciens. Curr. Microbiol. 46:157–162 [DOI] [PubMed] [Google Scholar]
- 42. Miyake R, Kawamoto J, Wei YL, Kitagawa M, Kato I, Kurihara T, Esaki N. 2007. Construction of a low-temperature protein expression system using a cold-adapted bacterium, Shewanella sp. strain Ac10, as the host. Appl. Environ. Microbiol. 73:4849–4856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hulsmann A, Rosche TM, Kong IS, Hassan HM, Beam DM, Oliver JD. 2003. RpoS-dependent stress response and exoenzyme production in Vibrio vulnificus. Appl. Environ. Microbiol. 69:6114–6120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tan HJ, Liu SH, Oliver JD, Wong HC. 2010. Role of RpoS in the susceptibility of low salinity-adapted Vibrio vulnificus to environmental stresses. Int. J. Food Microbiol. 137:137–142 [DOI] [PubMed] [Google Scholar]
- 45. Fuangthong M, Atichartpongkul S, Mongkolsuk S, Helmann JD. 2001. OhrR is a repressor of ohrA, a key organic hydroperoxide resistance determinant in Bacillus subtilis. J. Bacteriol. 183:4134–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen H, Hu J, Chen PR, Lan L, Li Z, Hicks LM, Dinner AR, He C. 2008. The Pseudomonas aeruginosa multidrug efflux regulator MexR uses an oxidation-sensing mechanism. Proc. Natl. Acad. Sci. U. S. A. 105:13586–13591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Newberry KJ, Fuangthong M, Panmanee W, Mongkolsuk S, Brennan RG. 2007. Structural mechanism of organic hydroperoxide induction of the transcription regulator OhrR. Mol. Cell 28:652–664 [DOI] [PubMed] [Google Scholar]
- 48. Italiani VC, da Silva Neto JF, Braz VS, Marques MV. 2011. Regulation of catalase-peroxidase KatG is OxyR dependent and Fur independent in Caulobacter crescentus. J. Bacteriol. 193:1734–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sukchawalit R, Loprasert S, Atichartpongkul S, Mongkolsuk S. 2001. Complex regulation of the organic hydroperoxide resistance gene (ohr) from Xanthomonas involves OhrR, a novel organic peroxide-inducible negative regulator, and posttranscriptional modifications. J. Bacteriol. 183:4405–4412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1:784–791 [Google Scholar]
- 51. Philippe N, Alcaraz JP, Coursange E, Geiselmann J, Schneider D. 2004. Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid 51:246–255 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.